Update Meeting: 25-08-2021
Jenny Sjaarda
2021-08-24
Last updated: 2021-09-15
Checks: 7 0
Knit directory: proxyMR/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it's best to always run the code in an empty environment.
The command set.seed(20210602) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version f7fc22e. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: FP_data.RData
Ignored: _targets/
Ignored: analysis/bgenie_GWAS/
Ignored: analysis/data_setup/
Ignored: analysis/process_Neale.out
Ignored: analysis/traitMR/
Ignored: data/Neale_SGG_directory_12_07_2021.csv
Ignored: data/Neale_SGG_directory_15_07_2021.csv
Ignored: data/processed/
Ignored: data/raw/
Ignored: household_MR_exhaustive_summary.rds
Ignored: output/figures/
Ignored: output/tables/traitMR/
Ignored: proxyMR_comparison.RData
Ignored: proxyMR_figure_data.RData
Ignored: proxymr_100_clustermq.out
Ignored: proxymr_101_clustermq.out
Ignored: proxymr_102_clustermq.out
Ignored: proxymr_103_clustermq.out
Ignored: proxymr_104_clustermq.out
Ignored: proxymr_105_clustermq.out
Ignored: proxymr_106_clustermq.out
Ignored: proxymr_107_clustermq.out
Ignored: proxymr_108_clustermq.out
Ignored: proxymr_109_clustermq.out
Ignored: proxymr_10_clustermq.out
Ignored: proxymr_110_clustermq.out
Ignored: proxymr_111_clustermq.out
Ignored: proxymr_112_clustermq.out
Ignored: proxymr_113_clustermq.out
Ignored: proxymr_114_clustermq.out
Ignored: proxymr_115_clustermq.out
Ignored: proxymr_116_clustermq.out
Ignored: proxymr_117_clustermq.out
Ignored: proxymr_118_clustermq.out
Ignored: proxymr_119_clustermq.out
Ignored: proxymr_11_clustermq.out
Ignored: proxymr_120_clustermq.out
Ignored: proxymr_121_clustermq.out
Ignored: proxymr_122_clustermq.out
Ignored: proxymr_123_clustermq.out
Ignored: proxymr_124_clustermq.out
Ignored: proxymr_125_clustermq.out
Ignored: proxymr_126_clustermq.out
Ignored: proxymr_127_clustermq.out
Ignored: proxymr_128_clustermq.out
Ignored: proxymr_129_clustermq.out
Ignored: proxymr_12_clustermq.out
Ignored: proxymr_130_clustermq.out
Ignored: proxymr_131_clustermq.out
Ignored: proxymr_132_clustermq.out
Ignored: proxymr_133_clustermq.out
Ignored: proxymr_134_clustermq.out
Ignored: proxymr_135_clustermq.out
Ignored: proxymr_136_clustermq.out
Ignored: proxymr_137_clustermq.out
Ignored: proxymr_138_clustermq.out
Ignored: proxymr_139_clustermq.out
Ignored: proxymr_13_clustermq.out
Ignored: proxymr_140_clustermq.out
Ignored: proxymr_14_clustermq.out
Ignored: proxymr_15_clustermq.out
Ignored: proxymr_16_clustermq.out
Ignored: proxymr_17_clustermq.out
Ignored: proxymr_18_clustermq.out
Ignored: proxymr_19_clustermq.out
Ignored: proxymr_1_clustermq.out
Ignored: proxymr_20_clustermq.out
Ignored: proxymr_21_clustermq.out
Ignored: proxymr_22_clustermq.out
Ignored: proxymr_23_clustermq.out
Ignored: proxymr_24_clustermq.out
Ignored: proxymr_25_clustermq.out
Ignored: proxymr_26_clustermq.out
Ignored: proxymr_27_clustermq.out
Ignored: proxymr_28_clustermq.out
Ignored: proxymr_29_clustermq.out
Ignored: proxymr_2_clustermq.out
Ignored: proxymr_30_clustermq.out
Ignored: proxymr_31_clustermq.out
Ignored: proxymr_32_clustermq.out
Ignored: proxymr_33_clustermq.out
Ignored: proxymr_34_clustermq.out
Ignored: proxymr_35_clustermq.out
Ignored: proxymr_36_clustermq.out
Ignored: proxymr_37_clustermq.out
Ignored: proxymr_38_clustermq.out
Ignored: proxymr_39_clustermq.out
Ignored: proxymr_3_clustermq.out
Ignored: proxymr_40_clustermq.out
Ignored: proxymr_41_clustermq.out
Ignored: proxymr_42_clustermq.out
Ignored: proxymr_43_clustermq.out
Ignored: proxymr_44_clustermq.out
Ignored: proxymr_45_clustermq.out
Ignored: proxymr_46_clustermq.out
Ignored: proxymr_47_clustermq.out
Ignored: proxymr_48_clustermq.out
Ignored: proxymr_49_clustermq.out
Ignored: proxymr_4_clustermq.out
Ignored: proxymr_50_clustermq.out
Ignored: proxymr_51_clustermq.out
Ignored: proxymr_52_clustermq.out
Ignored: proxymr_53_clustermq.out
Ignored: proxymr_54_clustermq.out
Ignored: proxymr_55_clustermq.out
Ignored: proxymr_56_clustermq.out
Ignored: proxymr_57_clustermq.out
Ignored: proxymr_58_clustermq.out
Ignored: proxymr_59_clustermq.out
Ignored: proxymr_5_clustermq.out
Ignored: proxymr_60_clustermq.out
Ignored: proxymr_61_clustermq.out
Ignored: proxymr_62_clustermq.out
Ignored: proxymr_63_clustermq.out
Ignored: proxymr_64_clustermq.out
Ignored: proxymr_65_clustermq.out
Ignored: proxymr_66_clustermq.out
Ignored: proxymr_67_clustermq.out
Ignored: proxymr_68_clustermq.out
Ignored: proxymr_69_clustermq.out
Ignored: proxymr_6_clustermq.out
Ignored: proxymr_70_clustermq.out
Ignored: proxymr_71_clustermq.out
Ignored: proxymr_72_clustermq.out
Ignored: proxymr_73_clustermq.out
Ignored: proxymr_74_clustermq.out
Ignored: proxymr_75_clustermq.out
Ignored: proxymr_76_clustermq.out
Ignored: proxymr_77_clustermq.out
Ignored: proxymr_78_clustermq.out
Ignored: proxymr_79_clustermq.out
Ignored: proxymr_7_clustermq.out
Ignored: proxymr_80_clustermq.out
Ignored: proxymr_81_clustermq.out
Ignored: proxymr_82_clustermq.out
Ignored: proxymr_83_clustermq.out
Ignored: proxymr_84_clustermq.out
Ignored: proxymr_85_clustermq.out
Ignored: proxymr_86_clustermq.out
Ignored: proxymr_87_clustermq.out
Ignored: proxymr_88_clustermq.out
Ignored: proxymr_89_clustermq.out
Ignored: proxymr_8_clustermq.out
Ignored: proxymr_90_clustermq.out
Ignored: proxymr_91_clustermq.out
Ignored: proxymr_92_clustermq.out
Ignored: proxymr_93_clustermq.out
Ignored: proxymr_94_clustermq.out
Ignored: proxymr_95_clustermq.out
Ignored: proxymr_96_clustermq.out
Ignored: proxymr_97_clustermq.out
Ignored: proxymr_98_clustermq.out
Ignored: proxymr_99_clustermq.out
Ignored: proxymr_9_clustermq.out
Ignored: renv/library/
Ignored: renv/staging/
Ignored: traits_corr2_update.rds
Untracked files:
Untracked: .nfs0000000200133c860035c099
Untracked: analysis/figures_scratch.Rmd
Untracked: analysis/group_meeting_pres_figures_080921.Rmd
Unstaged changes:
Modified: analysis/AM_MR_summary.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/update_meeting_25_08_2021.Rmd) and HTML (docs/update_meeting_25_08_2021.html) files. If you've configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | f7fc22e | Jenny Sjaarda | 2021-09-15 | update all update meetings Rmds |
| Rmd | 8cd643a | jennysjaarda | 2021-08-24 | intial update meeeting 25-08-21 |
| html | 8cd643a | jennysjaarda | 2021-08-24 | intial update meeeting 25-08-21 |
1 Last meeting summary.
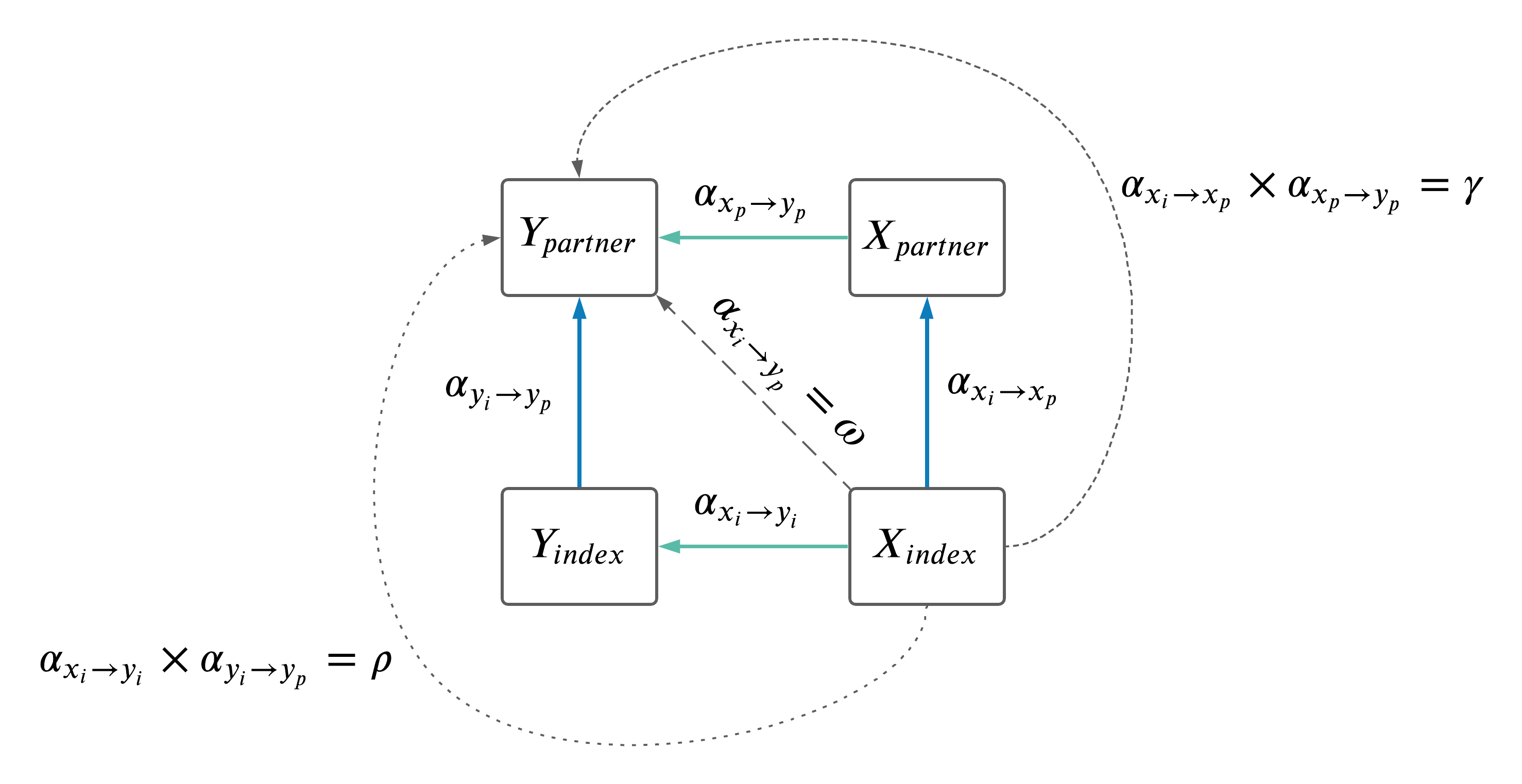
- Discussed summary of pathways from \(X\) in index (\(X_i\)) to \(Y\) in the partner (\(Y_p\)).
- Need for running the same individual MR (i.e. simply the standard MR) so we can compare the two pathways (see DAG below).
- Multiply perpendicular paths together and to calculate the variance between them use the formula here.
3 Results.
3.1 Heterogeneity amonst different groups.
3.1.1 Sex heterogeneity
For each proxyMR (\(Y_{p} \sim X_i\)), analyses were run in each sex separately. The results below display the results that differ among sexes, after BF correction (i.e. 0.05 / (131 * 131))
However in this case, it may be more sensible to only consider same traits (i.e. testing for more of an "assortative mating" effect), reducing the number of tests to 131. However, none pass BF-significance (p < 0.05/131 = 0.0003817). The most significant results is lifetime number of sexual partners.
3.1.2 Binned heterogeneity
For each proxyMR (\(Y_{p} \sim X_i\)), MR analyses were run in the full sample as well as in 5 roughly equal sized bins according to time_together and age. Specifically, the outcome data set was split into 5 bins, and the SNP-outcome effect was estimated in each bin separately. These outcome-SNP effects were then used to generate bin-specific MR estimates, using the same SNP-exposure effects from Neale.
To estimate the difference among bins, used two approaches:
- The Cochran's Q test to test for heterogeneity in meta-analyses.
- Tested the significance of the slope of linear model of median bin (either
ageortime together) versus the bin-specific MR estimate.
The two results are shown below, restricted to only same_traits. Note that each statistic was computed in each sex separately and meta-analyzed.
3.1.2.1 Q-statistic results
3.1.2.2 Slope results
3.2 Standard MR
To complete all MR anlayses in the DAG above, needed to generate same-person MR estimates (i.e. standard MR). These were also run sex-specific, using Neale summary statistics. As expected, there are many BF-significant summary MR results. Significance was determined based on the meta-analyzed result across sexes. Thus the number of tests was equal to: 131^2 - 131 = 17030 (number of traits =131`). The BF-significant results can be seen below.
Number of BF-signifcant MR pairs: 8542.
3.3 Paths from X-index to Y-partner
There are three distinct paths from \(X_{index}\) to \(Y_{partner}\) as illustrated in the DAG above:
- Assortative mating through \(X\): \(\gamma\)
- Assortative mating through \(Y\): \(\rho\)
- Indirectly (i.e. through a latent variable which either increases or decreases the observed effect of \(X_{index}\) to \(Y_{partner}\)): \(\omega\)
The estimates of \(\gamma\), \(\rho\) and \(\omega\) can be seen below, meta-analyzed across sex.
3.3.1 Heterogeneity among paths
Next, we wanted to compare the three paths from \(X_index\) to \(Y_partner\). This was done using a standard heterogeneity test.
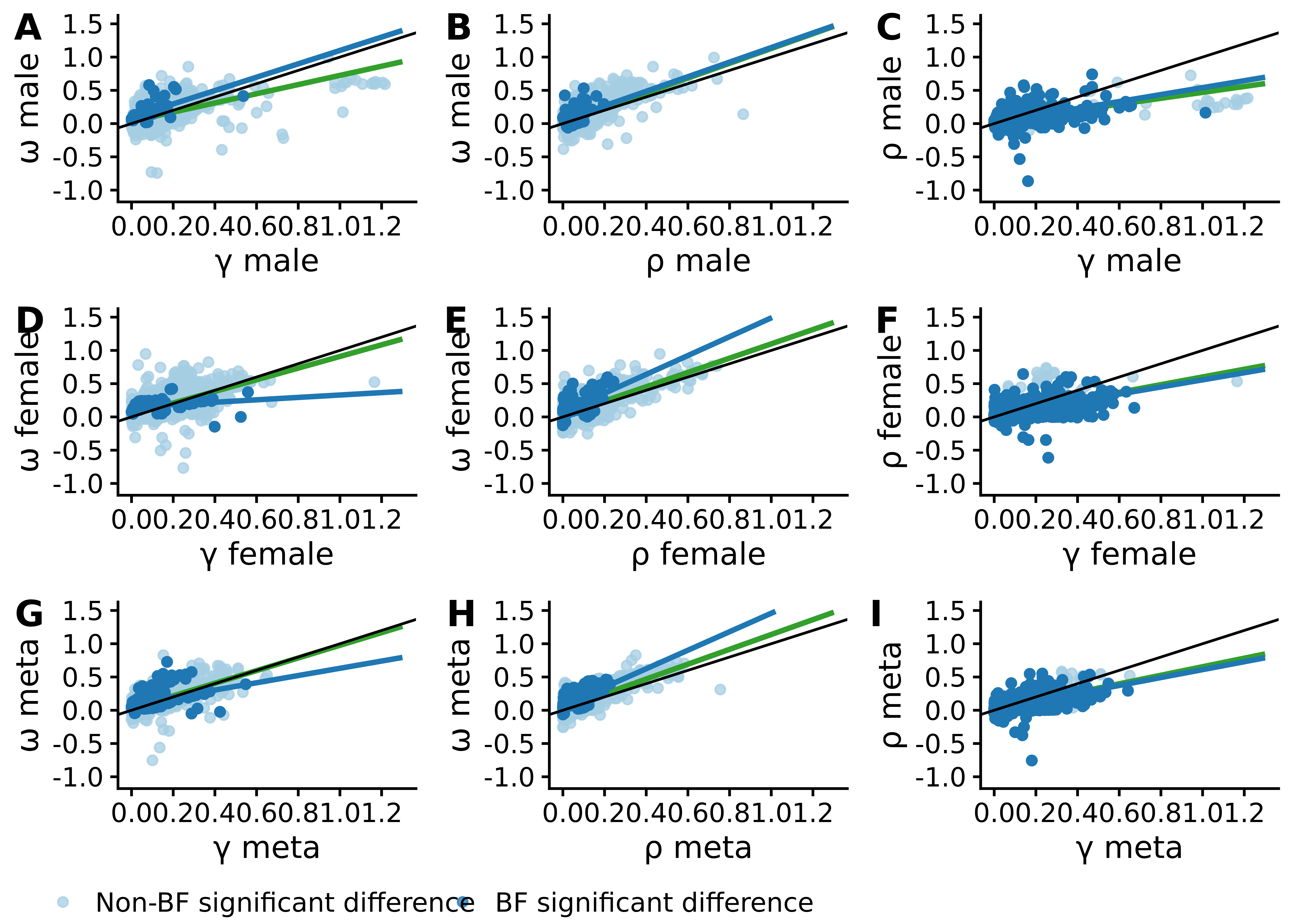
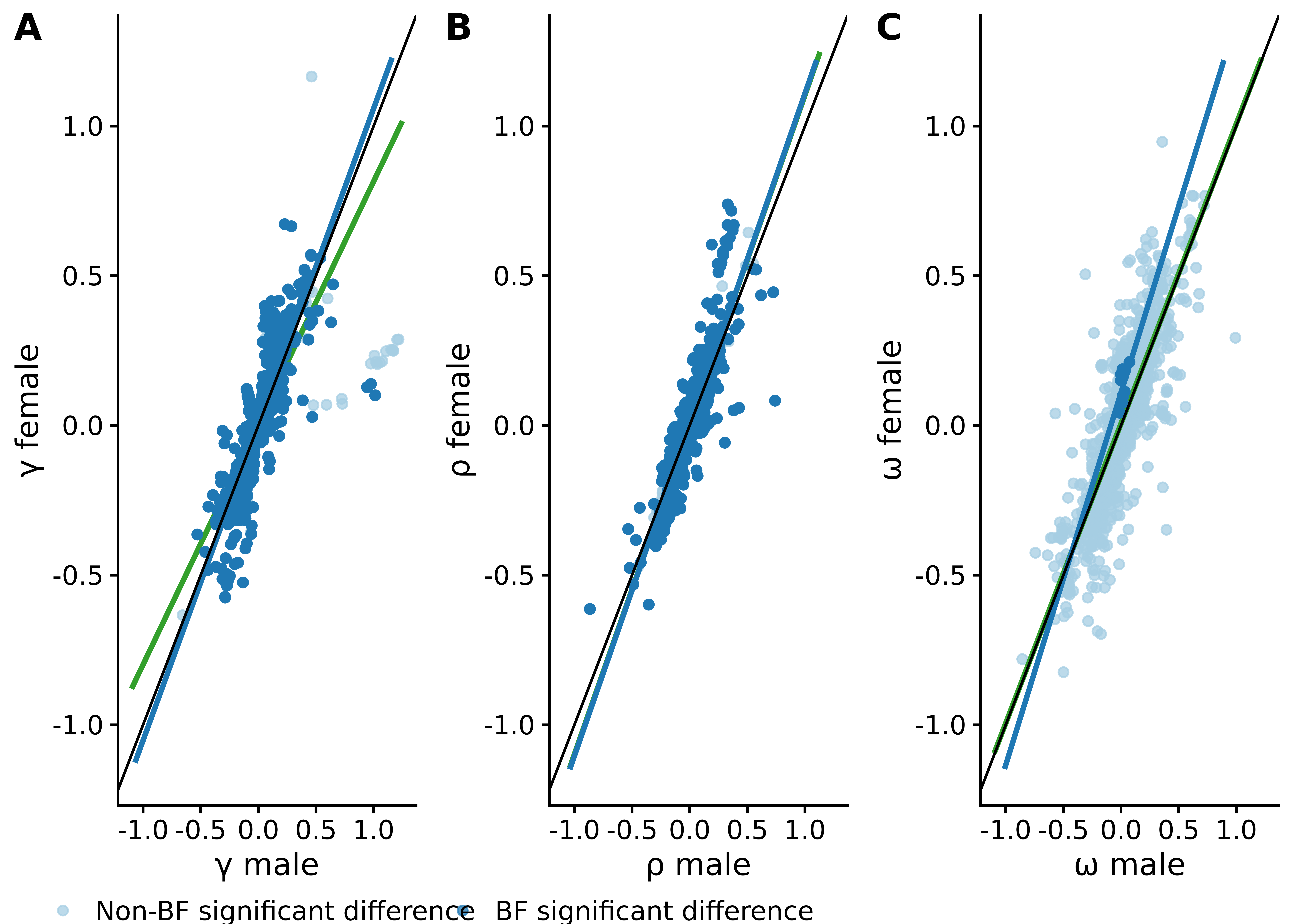
3.3.2 Sex heterogeneity amongst paths
3.3.3 Hierarchical clustering
Working on a figure like the one here and here.
sessionInfo()R version 4.1.0 (2021-05-18)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS: /data/sgg2/jenny/bin/R-4.1.0/lib64/R/lib/libRblas.so
LAPACK: /data/sgg2/jenny/bin/R-4.1.0/lib64/R/lib/libRlapack.so
locale:
[1] en_CA.UTF-8
attached base packages:
[1] stats graphics grDevices datasets utils methods base
other attached packages:
[1] knitr_1.33 DT_0.18.1 forcats_0.5.1 stringr_1.4.0
[5] dplyr_1.0.7 purrr_0.3.4 readr_1.4.0 tidyr_1.1.3
[9] tibble_3.1.2 ggplot2_3.3.4 tidyverse_1.3.1 targets_0.5.0.9001
[13] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rcpp_1.0.6 lubridate_1.7.10 ps_1.6.0 assertthat_0.2.1
[5] rprojroot_2.0.2 digest_0.6.27 utf8_1.2.1 R6_2.5.0
[9] cellranger_1.1.0 backports_1.2.1 reprex_2.0.0 evaluate_0.14
[13] highr_0.9 httr_1.4.2 pillar_1.6.1 rlang_0.4.11
[17] readxl_1.3.1 rstudioapi_0.13 data.table_1.14.0 whisker_0.4
[21] callr_3.7.0 rmarkdown_2.9 htmlwidgets_1.5.3 igraph_1.2.6
[25] munsell_0.5.0 broom_0.7.7 compiler_4.1.0 httpuv_1.6.1
[29] modelr_0.1.8 xfun_0.24 pkgconfig_2.0.3 htmltools_0.5.1.1
[33] tidyselect_1.1.1 codetools_0.2-18 fansi_0.5.0 crayon_1.4.1
[37] dbplyr_2.1.1 withr_2.4.2 later_1.2.0 grid_4.1.0
[41] jsonlite_1.7.2 gtable_0.3.0 lifecycle_1.0.0 DBI_1.1.1
[45] git2r_0.28.0 magrittr_2.0.1 scales_1.1.1 cli_2.5.0
[49] stringi_1.6.2 renv_0.13.2-62 fs_1.5.0 promises_1.2.0.1
[53] xml2_1.3.2 ellipsis_0.3.2 generics_0.1.0 vctrs_0.3.8
[57] cowplot_1.1.1 tools_4.1.0 glue_1.4.2 crosstalk_1.1.1
[61] hms_1.1.0 processx_3.5.2 yaml_2.2.1 colorspace_2.0-1
[65] rvest_1.0.0 haven_2.4.1