OutOfAfrica_2T12
Joseph Marcus
2020-05-13
Last updated: 2020-05-13
Checks: 7 0
Knit directory: drift-workflow/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190211) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 11de659. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .snakemake/
Ignored: data/datasets/
Ignored: data/raw/
Ignored: data/simulations/
Ignored: nb-log-1272639.err
Ignored: nb-log-1272639.out
Ignored: notebooks/.ipynb_checkpoints/
Ignored: output/
Ignored: sandbox/.ipynb_checkpoints/
Unstaged changes:
Modified: analysis/OutOfAfrica_3G09.Rmd
Modified: analysis/index.Rmd
Modified: snakefiles/coalsim/Snakefile
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/OutOfAfrica_2T12.Rmd) and HTML (docs/OutOfAfrica_2T12.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 11de659 | Joseph Marcus | 2020-05-13 | wflow_publish(“OutOfAfrica_2T12.Rmd”) |
Here I visualize population structure with simulated data from the OutOfAfrica_2T12 scenario. See Fu et al. 2013 and Tennessen et al. 2012 for details.
Imports
Import the required libraries and scripts:
suppressMessages({
library(lfa)
library(flashier)
library(drift.alpha)
library(ggplot2)
library(reshape2)
library(tidyverse)
library(alstructure)
})Data
data_path <- "../output/simulations/OutOfAfrica_2T12/rep1.txt"
Y <- t(as.matrix(read.table(data_path, sep=" ")))
n <- nrow(Y)
maf <- colSums(Y) / (2 * n)
# filter out too rare and too common SNPs
Y <- Y[,((maf>=.05) & (maf <=.95))]
p <- ncol(Y)
Z <- scale(Y)
print(n)[1] 80print(p)[1] 35925# sub-population labels from stdpop
labs <- rep(c("AFR", "EUR"), each=40)we end up with 80 individuals and ~35925 SNPs.
PCA
Lets run PCA on the centered and scaled genotype matrix:
svd_res <- lfa:::trunc.svd(Z, 5)
L_hat <- svd_res$u
plot_loadings(L_hat, labs)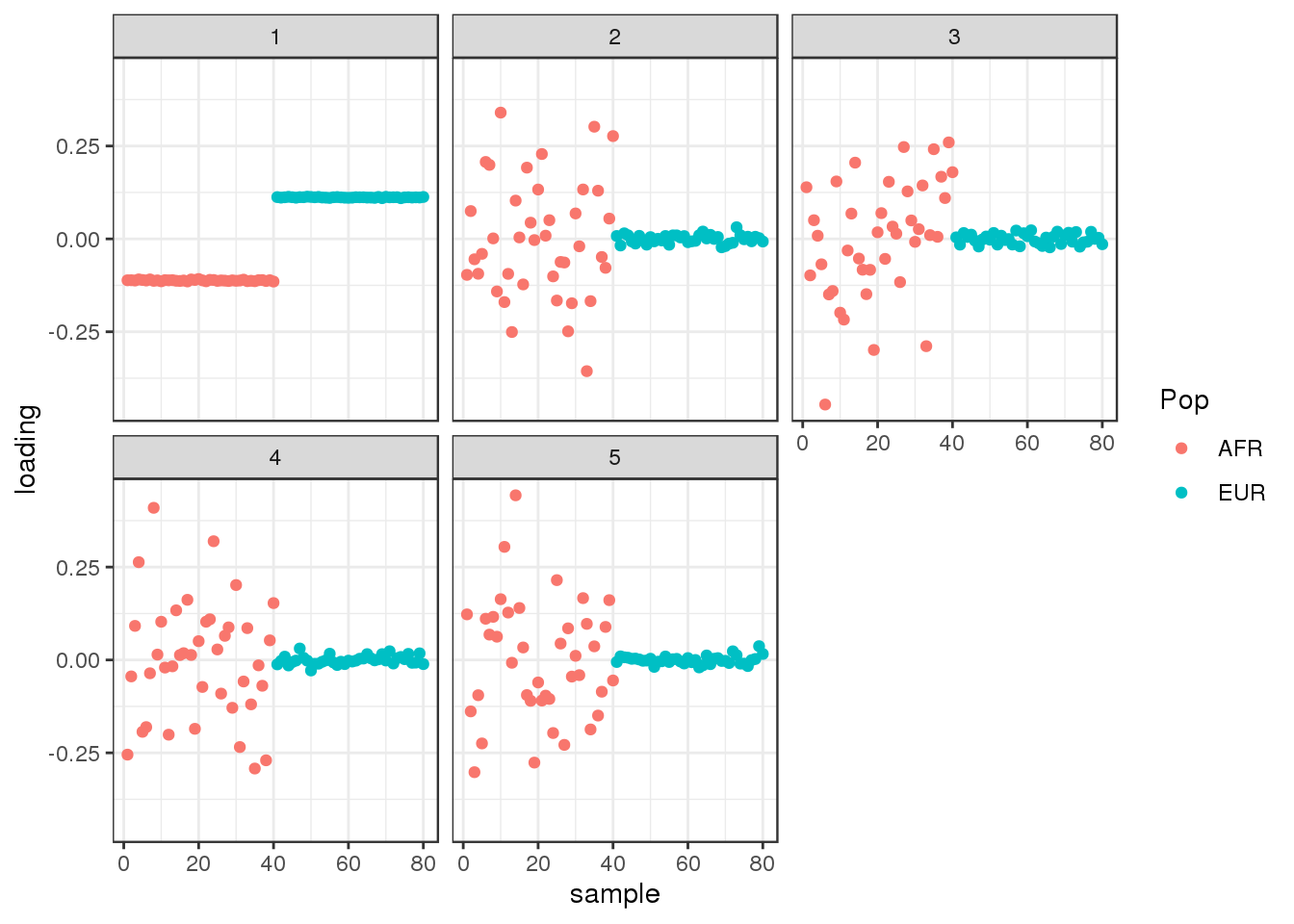
plot the first two factors against each other:
qplot(L_hat[,1], L_hat[,2], color=labs) +
xlab("PC1") +
ylab("PC2") +
theme_bw()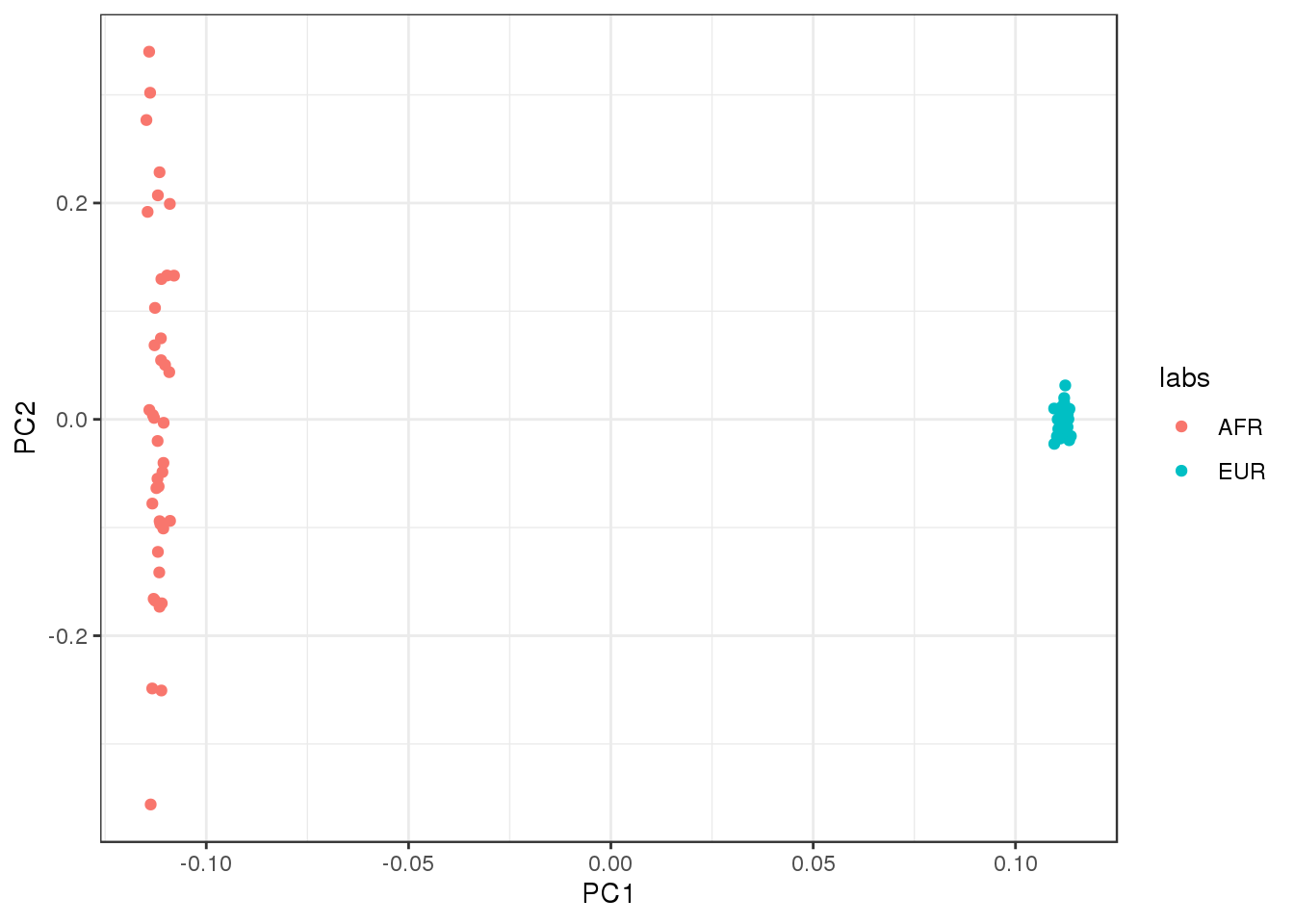
PC1 represents “Out of Africa” and PC2 represents within Africa variation.
ALStructure
Run ALStructure with \(K=2\):
admix_res <- alstructure::alstructure(t(Y), d_hat=2)
Qhat <- t(admix_res$Q_hat)
plot_loadings(Qhat, labs)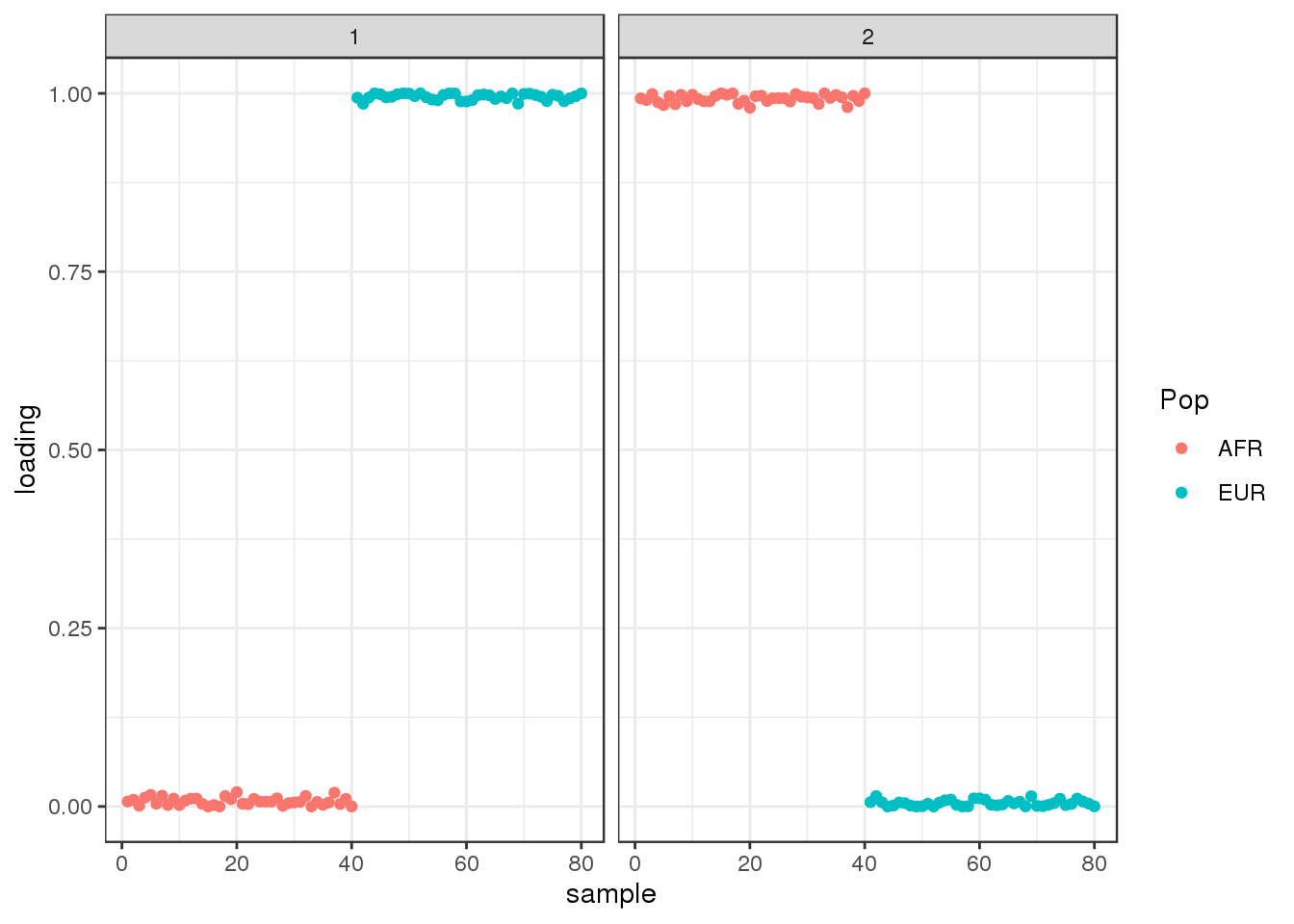
the PSD model assigns two distinct clusters.
flash [greedy]
Run the greedy algorithm:
fl <- flash(Y,
greedy.Kmax=5,
prior.family=c(prior.bimodal(), prior.normal()))Adding factor 1 to flash object...
Adding factor 2 to flash object...
Adding factor 3 to flash object...
Adding factor 4 to flash object...
Factor doesn't significantly increase objective and won't be added.
Wrapping up...
Done.
Nullchecking 3 factors...
Done.plot_loadings(fl$flash.fit$EF[[1]], labs)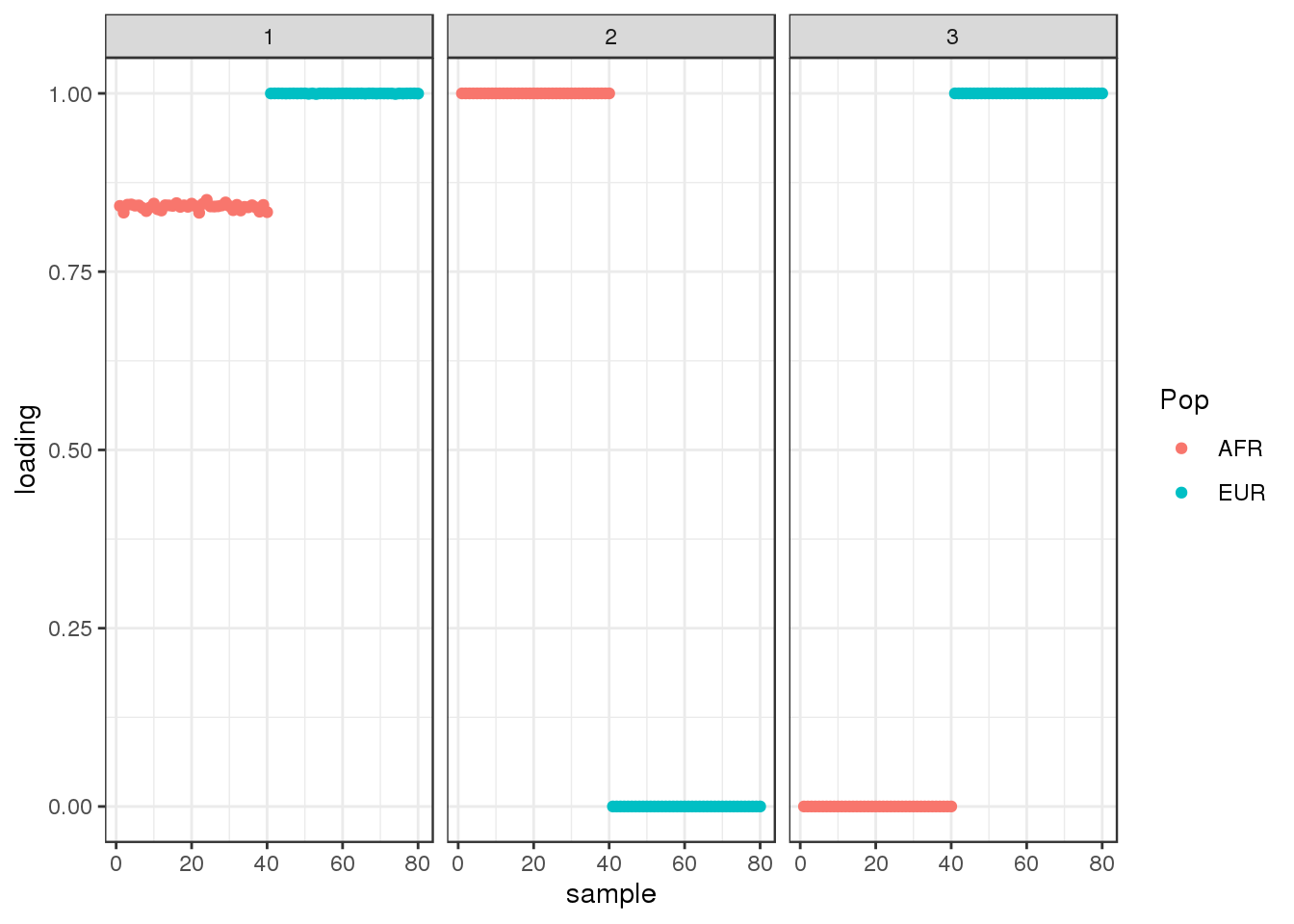
the greedy algorithm learns a shared factor and then finds two population specific factors.
flash [backfit]
Run flash [backfit] initializing from the greedy solution:
flbf <- fl %>%
flash.backfit() %>%
flash.nullcheck(remove=TRUE)Backfitting 3 factors (tolerance: 4.28e-02)...
Difference between iterations is within 1.0e+02...
Difference between iterations is within 1.0e+01...
Difference between iterations is within 1.0e+00...
Difference between iterations is within 1.0e-01...
Wrapping up...
Done.
Nullchecking 3 factors...
Wrapping up...
Done.plot_loadings(flbf$flash.fit$EF[[1]], labs)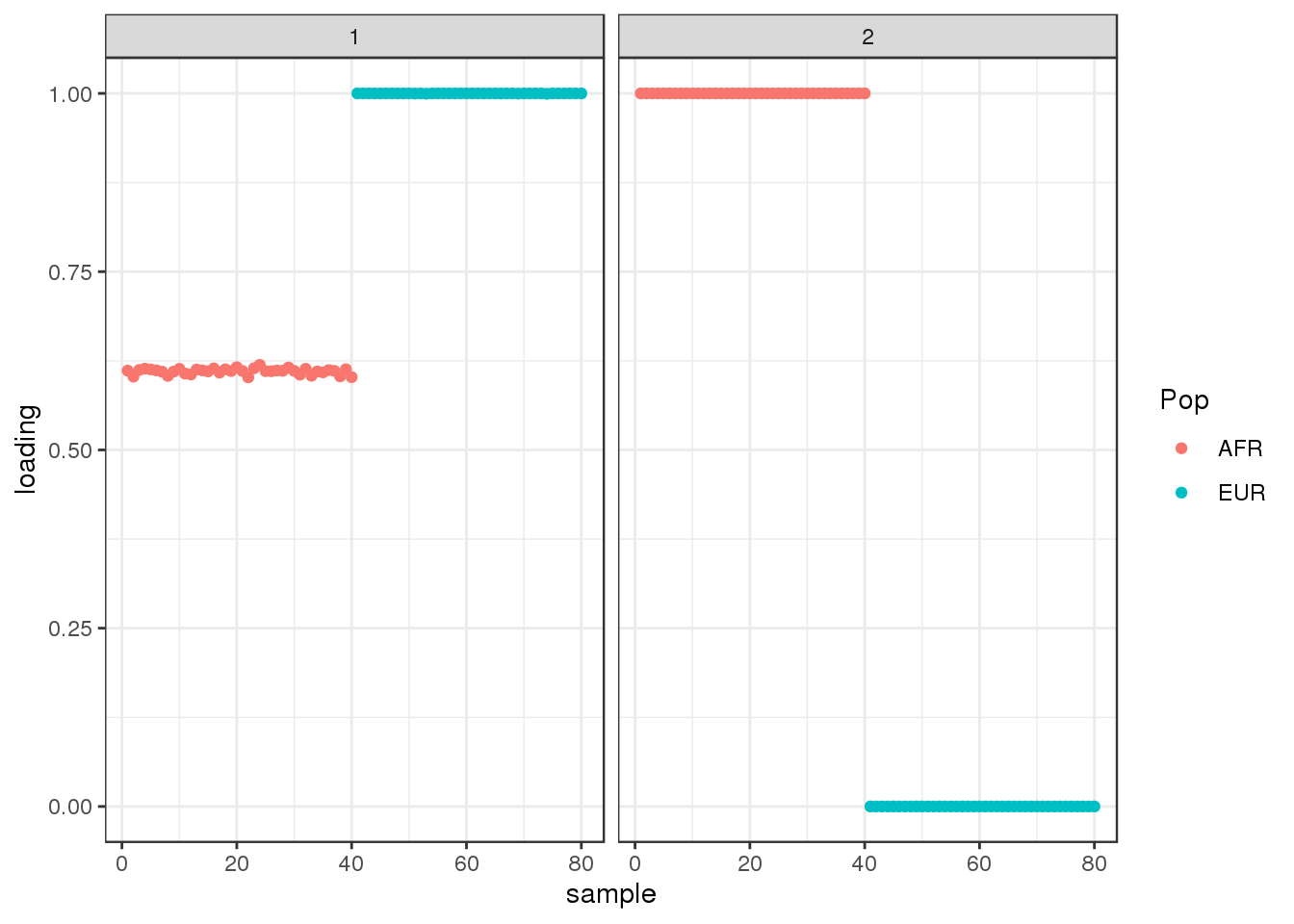
interestingly, flash finds the sparse representation of the the two population tree that we have seen before.
drift
Run drift initializing from the greedy solution:
init <- init_from_flash(fl)
dr <- drift(init, miniter=2, maxiter=500, tol=0.01, verbose=TRUE) 1 : -2312309.293
2 : -2310400.853
3 : -2310030.016
4 : -2309946.385
5 : -2309916.972
6 : -2309899.156
7 : -2309885.123
8 : -2309873.347
9 : -2309863.424
10 : -2309855.130
11 : -2309848.251
12 : -2309842.565
13 : -2309837.852
14 : -2309833.914
15 : -2309830.579
16 : -2309827.663
17 : -2309824.947
18 : -2309822.390
19 : -2309819.965
20 : -2309817.645
21 : -2309815.482
22 : -2309813.664
23 : -2309812.136
24 : -2309810.853
25 : -2309809.779
26 : -2309808.880
27 : -2309808.129
28 : -2309807.502
29 : -2309806.979
30 : -2309806.544
31 : -2309806.182
32 : -2309805.882
33 : -2309805.632
34 : -2309805.425
35 : -2309805.253
36 : -2309805.110
37 : -2309804.992
38 : -2309804.895
39 : -2309804.814
40 : -2309804.747
41 : -2309804.692
42 : -2309804.646
43 : -2309804.608
44 : -2309804.577
45 : -2309804.551
46 : -2309804.530
47 : -2309804.512
48 : -2309804.498
49 : -2309804.486
50 : -2309804.476 plot_loadings(dr$EL, labs)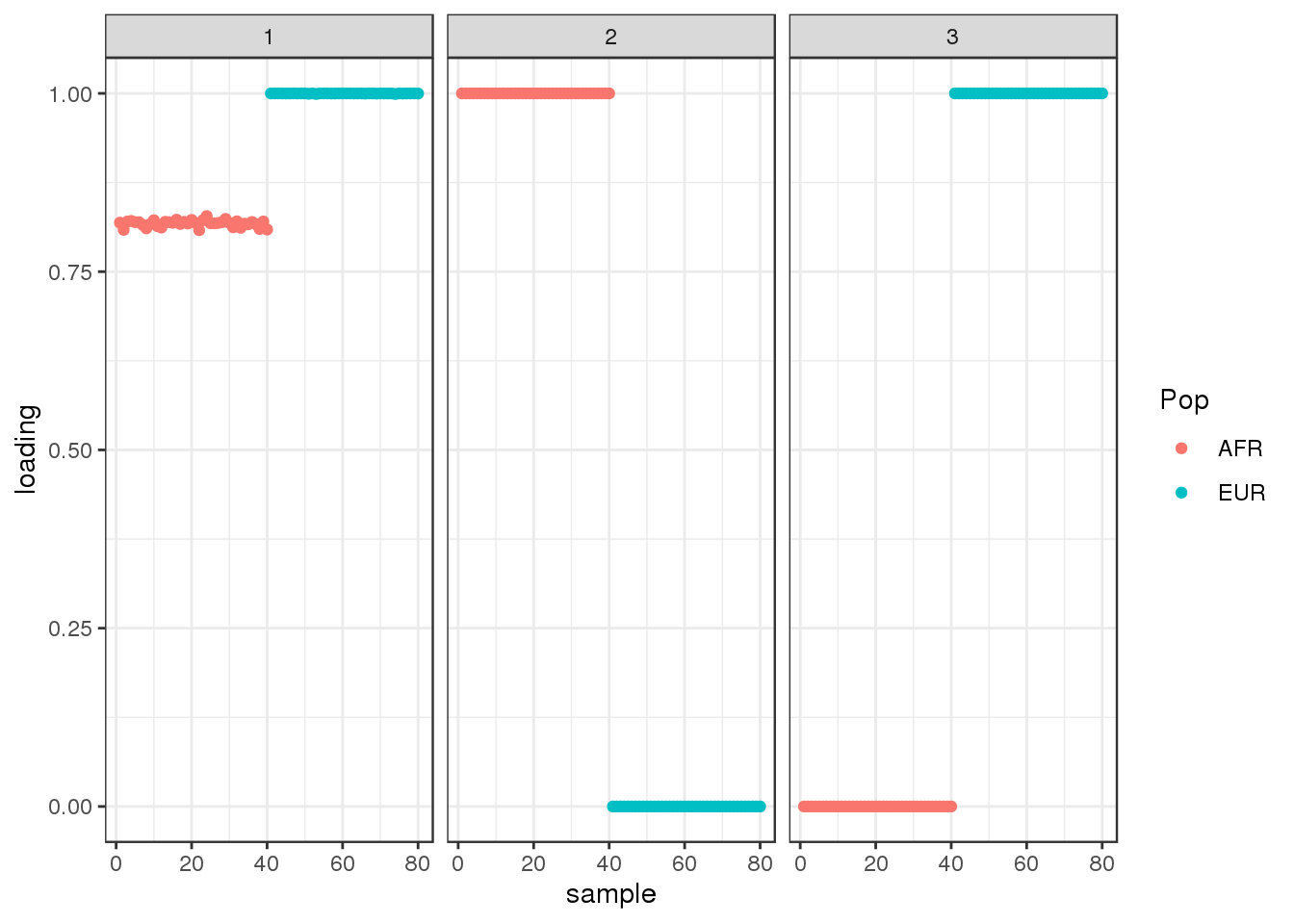
drift looks similar to the greedy algorithm.
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] alstructure_0.1.0 forcats_0.5.0 stringr_1.4.0
[4] dplyr_0.8.5 purrr_0.3.4 readr_1.3.1
[7] tidyr_1.0.2 tibble_3.0.1 tidyverse_1.3.0
[10] reshape2_1.4.3 ggplot2_3.3.0 drift.alpha_0.0.9
[13] flashier_0.2.4 lfa_1.9.0
loaded via a namespace (and not attached):
[1] httr_1.4.1 jsonlite_1.6 modelr_0.1.6 assertthat_0.2.1
[5] mixsqp_0.3-17 cellranger_1.1.0 yaml_2.2.0 ebnm_0.1-24
[9] pillar_1.4.3 backports_1.1.6 lattice_0.20-38 glue_1.4.0
[13] digest_0.6.25 promises_1.0.1 rvest_0.3.5 colorspace_1.4-1
[17] htmltools_0.3.6 httpuv_1.4.5 Matrix_1.2-15 plyr_1.8.4
[21] pkgconfig_2.0.3 invgamma_1.1 broom_0.5.6 haven_2.2.0
[25] corpcor_1.6.9 scales_1.1.0 whisker_0.3-2 later_0.7.5
[29] git2r_0.26.1 farver_2.0.3 generics_0.0.2 ellipsis_0.3.0
[33] withr_2.2.0 ashr_2.2-50 cli_2.0.2 magrittr_1.5
[37] crayon_1.3.4 readxl_1.3.1 evaluate_0.14 fs_1.3.1
[41] fansi_0.4.1 nlme_3.1-137 xml2_1.3.2 truncnorm_1.0-8
[45] tools_3.5.1 hms_0.5.3 lifecycle_0.2.0 munsell_0.5.0
[49] reprex_0.3.0 irlba_2.3.3 compiler_3.5.1 rlang_0.4.5
[53] grid_3.5.1 rstudioapi_0.11 labeling_0.3 rmarkdown_1.10
[57] gtable_0.3.0 DBI_1.0.0 R6_2.4.1 lubridate_1.7.4
[61] knitr_1.20 workflowr_1.6.1 rprojroot_1.3-2 stringi_1.4.6
[65] parallel_3.5.1 SQUAREM_2020.2 Rcpp_1.0.4.6 vctrs_0.2.4
[69] dbplyr_1.4.3 tidyselect_1.0.0