getting_started
karltayeb
2022-05-16
Last updated: 2022-05-16
Checks: 7 0
Knit directory: logistic-susie-gsea/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20220105) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version a8038f9. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: cache/
Ignored: library/
Ignored: renv/library/
Ignored: renv/staging/
Ignored: staging/
Untracked files:
Untracked: .ipynb_checkpoints/
Untracked: _targets.R
Untracked: _targets.html
Untracked: _targets.md
Untracked: _targets/
Untracked: additive.l5.gonr.aggregate.scores
Untracked: analysis/alpha_ash_v_point_normal.Rmd
Untracked: analysis/de_droplet_noshrink.Rmd
Untracked: analysis/de_droplet_noshrink_logistic_susie.Rmd
Untracked: analysis/fetal_reference_cellid_gsea.Rmd
Untracked: analysis/fixed_intercept.Rmd
Untracked: analysis/gsea_made_simple.Rmd
Untracked: analysis/iDEA_examples.Rmd
Untracked: analysis/latent_gene_list.Rmd
Untracked: analysis/linear_method_failure_modes.Rmd
Untracked: analysis/linear_regression_failure_regime.Rmd
Untracked: analysis/linear_v_logistic_pbmc.Rmd
Untracked: analysis/logistic_susie_veb_boost_vs_vb.Rmd
Untracked: analysis/logistic_susie_vis.Rmd
Untracked: analysis/logistic_variational_bound.Rmd
Untracked: analysis/logsitic_susie_template.Rmd
Untracked: analysis/pcb_scratch.Rmd
Untracked: analysis/redundancy_is_a_problem.Rmd
Untracked: analysis/references.bib
Untracked: analysis/roadmap.Rmd
Untracked: analysis/simulations.Rmd
Untracked: analysis/simulations_l1.Rmd
Untracked: analysis/tccm_vs_logistic_susie.Rmd
Untracked: analysis/template.Rmd
Untracked: analysis/test.Rmd
Untracked: build_site.R
Untracked: code/html_tables.R
Untracked: code/latent_logistic_susie.R
Untracked: code/logistic_susie_data_driver.R
Untracked: code/marginal_sumstat_gsea_collapsed.R
Untracked: code/point_normal.R
Untracked: code/sumstat_gsea.py
Untracked: code/susie_gsea_queries.R
Untracked: data/adipose_2yr_topsnp.txt
Untracked: data/de-droplet/
Untracked: data/deng/
Untracked: data/fetal_reference_cellid_gene_sets.RData
Untracked: data/human_chimp_eb/
Untracked: data/pbmc-purified/
Untracked: data/wenhe_baboon_diet/
Untracked: docs.zip
Untracked: export/
Untracked: index.md
Untracked: l1.sim.aggregate.scores
Untracked: linear_v_logistic.png
Untracked: presentations/
Untracked: references.bib
Untracked: simulation_targets/
Unstaged changes:
Modified: .gitignore
Deleted: _simulation_targets.R
Modified: _targets.Rmd
Modified: _targets.yaml
Modified: analysis/alpha_for_single_cell.Rmd
Modified: analysis/baboon_diet.Rmd
Modified: analysis/example_pbmc.Rmd
Modified: analysis/gseabenchmark_tcga.Rmd
Modified: analysis/human_chimp_eb_de_example.Rmd
Modified: analysis/linear_v_logistic_start.Rmd
Modified: analysis/single_cell_pbmc.Rmd
Modified: analysis/single_cell_pbmc_l1.Rmd
Deleted: analysis/summary_stat_gsea_univariate_simulations.Rmd
Modified: analysis/the_big_geneset.Rmd
Modified: code/fit_baselines.R
Modified: code/fit_logistic_susie.R
Modified: code/fit_mr_ash.R
Modified: code/fit_susie.R
Deleted: code/logistic_susie_vb.R
Deleted: code/logistic_susie_veb_boost.R
Modified: code/marginal_sumstat_gsea.R
Deleted: code/simulate_gene_lists.R
Modified: code/tccm_ebnm.R
Modified: code/utils.R
Deleted: target_components/factories.R
Deleted: target_components/methods.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/getting_started.Rmd) and HTML (docs/getting_started.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | a8038f9 | karltayeb | 2022-05-16 | wflow_publish(“analysis/getting_started.Rmd”) |
Introduction
Here we demonstrate the basics of how to use the gseasusie package.
devtools::install_github('karltayeb/gseasusie', force=TRUE)Downloading GitHub repo karltayeb/gseasusie@HEADtibble (3.1.6 -> 3.1.7 ) [CRAN]
RcppArmad... (0.11.0.0.0 -> 0.11.1.1.0) [CRAN]
openssl (2.0.0 -> 2.0.1 ) [CRAN]
reticulate (1.24 -> 1.25 ) [CRAN]
bigmemory (4.6.0 -> 4.6.1 ) [CRAN]
ggplot2 (3.3.5 -> 3.3.6 ) [CRAN]
httr (1.4.2 -> 1.4.3 ) [CRAN]
furrr (0.2.3 -> 0.3.0 ) [CRAN]Installing 8 packages: tibble, RcppArmadillo, openssl, reticulate, bigmemory, ggplot2, httr, furrrInstalling packages into '/Users/karltayeb/Research/logistic-susie-gsea/renv/library/R-4.1/x86_64-apple-darwin17.0'
(as 'lib' is unspecified)
There is a binary version available but the source version is later:
binary source needs_compilation
RcppArmadillo 0.11.0.0.0 0.11.1.1.0 TRUE
The downloaded binary packages are in
/var/folders/fz/nt8ny7791qb6hztvtd2q8dkh0000gn/T//RtmpYJj6aY/downloaded_packagesinstalling the source package 'RcppArmadillo'* checking for file ‘/private/var/folders/fz/nt8ny7791qb6hztvtd2q8dkh0000gn/T/RtmpYJj6aY/remotes1636e15d95645/karltayeb-gseasusie-6413c58/DESCRIPTION’ ... OK
* preparing ‘gseasusie’:
* checking DESCRIPTION meta-information ... OK
Warning: /private/var/folders/fz/nt8ny7791qb6hztvtd2q8dkh0000gn/T/RtmphRLmii/Rbuild1664839ae4faf/gseasusie/man/report_susie_credible_sets.Rd:15: unexpected section header '\alias'
* checking for LF line-endings in source and make files and shell scripts
* checking for empty or unneeded directories
* building ‘gseasusie_0.0.0.9000.tar.gz’Installing package into '/Users/karltayeb/Research/logistic-susie-gsea/renv/library/R-4.1/x86_64-apple-darwin17.0'
(as 'lib' is unspecified)library(gseasusie)
library(tidyverse)── Attaching packages ─────────────────────────────────────── tidyverse 1.3.1 ──✔ ggplot2 3.3.6 ✔ purrr 0.3.4
✔ tibble 3.1.6 ✔ dplyr 1.0.9
✔ tidyr 1.2.0 ✔ stringr 1.4.0
✔ readr 2.1.2 ✔ forcats 0.5.1devtools::load_all('~/R/gseasusie/')ℹ Loading gseasusiegseasusie ships with lots of gene set databases! There are other packages that curate geneset databases WebGestaltR, msigdbr, and stephenslab/pathways All I did here was write a few functions to load them into a standard format.
Each gene set list contains three elements: * X: a gene x gene set indicator matrx * geneSet: a two column data.frame (geneSet and gene) mapping genes to gene sets * geneSetDes: a written description of what the gene set does
gseasusie::load_gene_sets lets you load multiple gene sets. Note: the first time you try to load gene sets it might take a while because we need to build the indicator matrices (and the implimentation is far from optimized). After your fit it onces it will cache to a folder ./cache/resources/ relative to your working directory/project directory
eventually, the package will ship with some data
If you have multiple experiments that you want to run enrichment for, I’ve found it useful to format as follows. data is a list of data frames, one for each experiment. The dataframes can have whatever information you’d like, but they must have the following columns: * ENTREZID: the gene sets above use ENTREZID, so map your gene names to this! * threshold.on: this might be a p-value, adjusted pvalue, effect size, etc * and beta:: right now this is only used for sign information, so if you don’t care specifically about up or down regulated genes just make a dummy column with 1s.
Eventually, beta and se columns will be important for the summary stat enrichment analaysis.
source('code/load_data.R')
data <- load_sc_pbmc_deseq2()Loading required package: DESeq2Loading required package: S4VectorsWarning: package 'S4Vectors' was built under R version 4.1.3Loading required package: methodsLoading required package: stats
Attaching package: 'stats'The following objects are masked from 'package:dplyr':
filter, lagLoading required package: stats4Loading required package: BiocGenericsLoading required package: graphics
Attaching package: 'graphics'The following object is masked from 'package:stats4':
plot
Attaching package: 'BiocGenerics'The following objects are masked from 'package:stats':
IQR, mad, sd, var, xtabsThe following objects are masked from 'package:dplyr':
combine, intersect, setdiff, unionThe following objects are masked from 'package:base':
anyDuplicated, append, as.data.frame, basename, cbind, colnames,
dirname, do.call, duplicated, eval, evalq, Filter, Find, get, grep,
grepl, intersect, is.unsorted, lapply, Map, mapply, match, mget,
order, paste, pmax, pmax.int, pmin, pmin.int, Position, rank,
rbind, Reduce, rownames, sapply, setdiff, sort, table, tapply,
union, unique, unsplit, which.max, which.min
Attaching package: 'S4Vectors'The following objects are masked from 'package:dplyr':
first, renameThe following object is masked from 'package:tidyr':
expandThe following objects are masked from 'package:base':
expand.grid, I, unnameLoading required package: IRanges
Attaching package: 'IRanges'The following objects are masked from 'package:dplyr':
collapse, desc, sliceThe following object is masked from 'package:purrr':
reduceLoading required package: GenomicRangesLoading required package: GenomeInfoDbLoading required package: SummarizedExperimentLoading required package: MatrixGenericsLoading required package: matrixStats
Attaching package: 'matrixStats'The following object is masked from 'package:dplyr':
count
Attaching package: 'MatrixGenerics'The following objects are masked from 'package:matrixStats':
colAlls, colAnyNAs, colAnys, colAvgsPerRowSet, colCollapse,
colCounts, colCummaxs, colCummins, colCumprods, colCumsums,
colDiffs, colIQRDiffs, colIQRs, colLogSumExps, colMadDiffs,
colMads, colMaxs, colMeans2, colMedians, colMins, colOrderStats,
colProds, colQuantiles, colRanges, colRanks, colSdDiffs, colSds,
colSums2, colTabulates, colVarDiffs, colVars, colWeightedMads,
colWeightedMeans, colWeightedMedians, colWeightedSds,
colWeightedVars, rowAlls, rowAnyNAs, rowAnys, rowAvgsPerColSet,
rowCollapse, rowCounts, rowCummaxs, rowCummins, rowCumprods,
rowCumsums, rowDiffs, rowIQRDiffs, rowIQRs, rowLogSumExps,
rowMadDiffs, rowMads, rowMaxs, rowMeans2, rowMedians, rowMins,
rowOrderStats, rowProds, rowQuantiles, rowRanges, rowRanks,
rowSdDiffs, rowSds, rowSums2, rowTabulates, rowVarDiffs, rowVars,
rowWeightedMads, rowWeightedMeans, rowWeightedMedians,
rowWeightedSds, rowWeightedVarsLoading required package: BiobaseWelcome to Bioconductor
Vignettes contain introductory material; view with
'browseVignettes()'. To cite Bioconductor, see
'citation("Biobase")', and for packages 'citation("pkgname")'.
Attaching package: 'Biobase'The following object is masked from 'package:MatrixGenerics':
rowMediansThe following objects are masked from 'package:matrixStats':
anyMissing, rowMedians'select()' returned 1:many mapping between keys and columnsdata$`CD19+ B` %>% head() ENSEMBL ENTREZID baseMean log2FoldChange lfcSE pvalue
1 ENSG00000237683 <NA> 2.327879e-03 0.29159244 0.3257749 8.206061e-06
2 ENSG00000228463 728481 9.995371e-05 1.05065835 1.0336767 4.722355e-02
3 ENSG00000228327 <NA> 2.304559e-03 -0.47955339 0.4029420 5.056276e-02
4 ENSG00000237491 105378580 9.536208e-04 0.03026135 0.3774938 8.526449e-01
5 ENSG00000225880 79854 9.567947e-03 0.68820333 0.1604846 5.072878e-11
6 ENSG00000230368 284593 5.068608e-04 1.78293901 0.5604303 2.269509e-04
padj beta se threshold.on
1 1.781359e-05 0.29159244 0.3257749 8.206061e-06
2 6.266937e-02 1.05065835 1.0336767 4.722355e-02
3 6.659266e-02 -0.47955339 0.4029420 5.056276e-02
4 8.561610e-01 0.03026135 0.3774938 8.526449e-01
5 1.568993e-10 0.68820333 0.1604846 5.072878e-11
6 4.258453e-04 1.78293901 0.5604303 2.269509e-04Now we will binarize the data by thresholding on threshold.on We also need to format our gene set matrix and our gene list: (subset to intersection genes, check genes are consistently ordered, etc). gseasusie::prep_binary_data helps us with this!
db <- 'c2' # name of gene set database to use in `genesets`
experiment = 'CD19+ B' # name of experiment to use in `data`
thresh = 1e-4 # threshold for binarizing the data
bin.data <- gseasusie::prep_binary_data(genesets[[db]], data[[experiment]], thresh)
X <- bin.data$X
y <- bin.data$yNow we can fit our enrichment models. NOTE: The marginal regressions are implimented in python (basilisk will spin up a conda environment with necessary dependencies, which will take some time the first time you run it, but will run quickly after!)
# fit logistic susie
logistic.fit <- gseasusie::fit_logistic_susie_veb_boost(X, y, L=20)fitting logistic susie (via VEB.BoostELBO: -9062.304
9.506 sec elapsed# fit linear susie
# (all of the functions that work with logistic susie fits should work with regular susie fits)
linear.fit <- susieR::susie(X, y)
# compute odds ratios, and pvalues under hypergeometric (one-sided) and fishers exact (two-sided) tests
ora <- gseasusie::fit_ora(X, y)computin ORA statistics...6.197 sec elapsed# like ORA but performed implimented as a univariate logistic regression
marginal_regression <- gseasusie::fit_marginal_regression_jax(X, y)fitting marginal logistic regressions...38.765 sec elapsed# redo the logistic regression ORA, conditional on enrichments found by logistic susie
# in practice, we introduce logistic susie predictions as an offset in the new model
residual_regression <- gseasusie::fit_residual_regression_jax(X, y, logistic.fit)fitting residual logistic regressions...30.208 sec elapsedres = list(
fit=logistic.fit,
ora=ora,
marginal_reg = marginal_regression,
residual_reg = residual_regression
)We can visualize our results with a volcano plot (more plots coming)
The large circles highlight gene sets in a 95% credible set. The color indicates which SuSiE component)
gseasusie::enrichment_volcano(logistic.fit, ora)Joining, by = "geneSet"
Joining, by = c("geneSet", "component")
Joining, by = c("geneSet", "component")
Joining, by = "geneSet"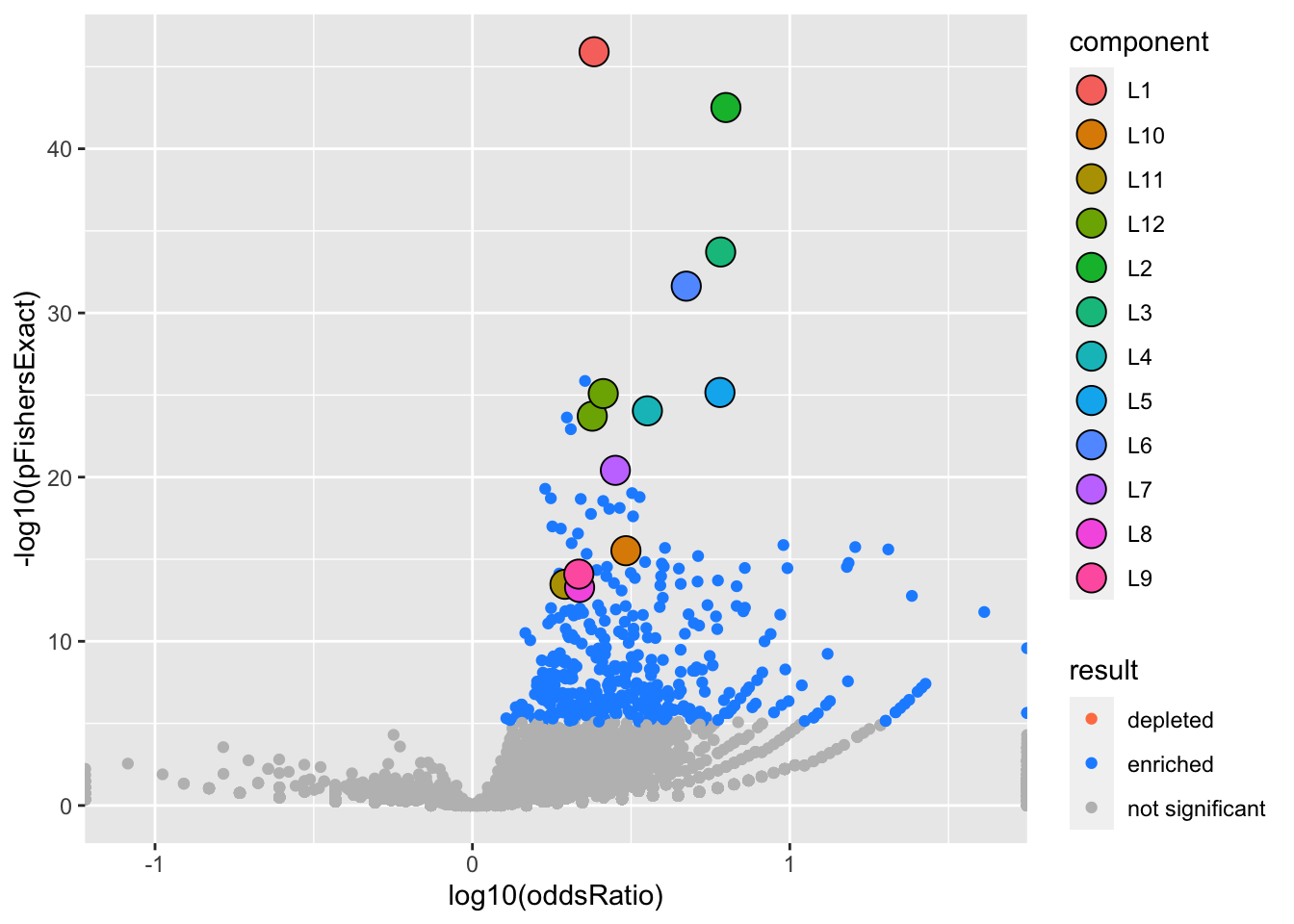
If we account for the predictions made by (logitic) SuSiE, we can see that a lot (but not all of) the enrichment signal has been accounted for.
gseasusie::residual_enrichment_histogram(marginal_regression, residual_regression)`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.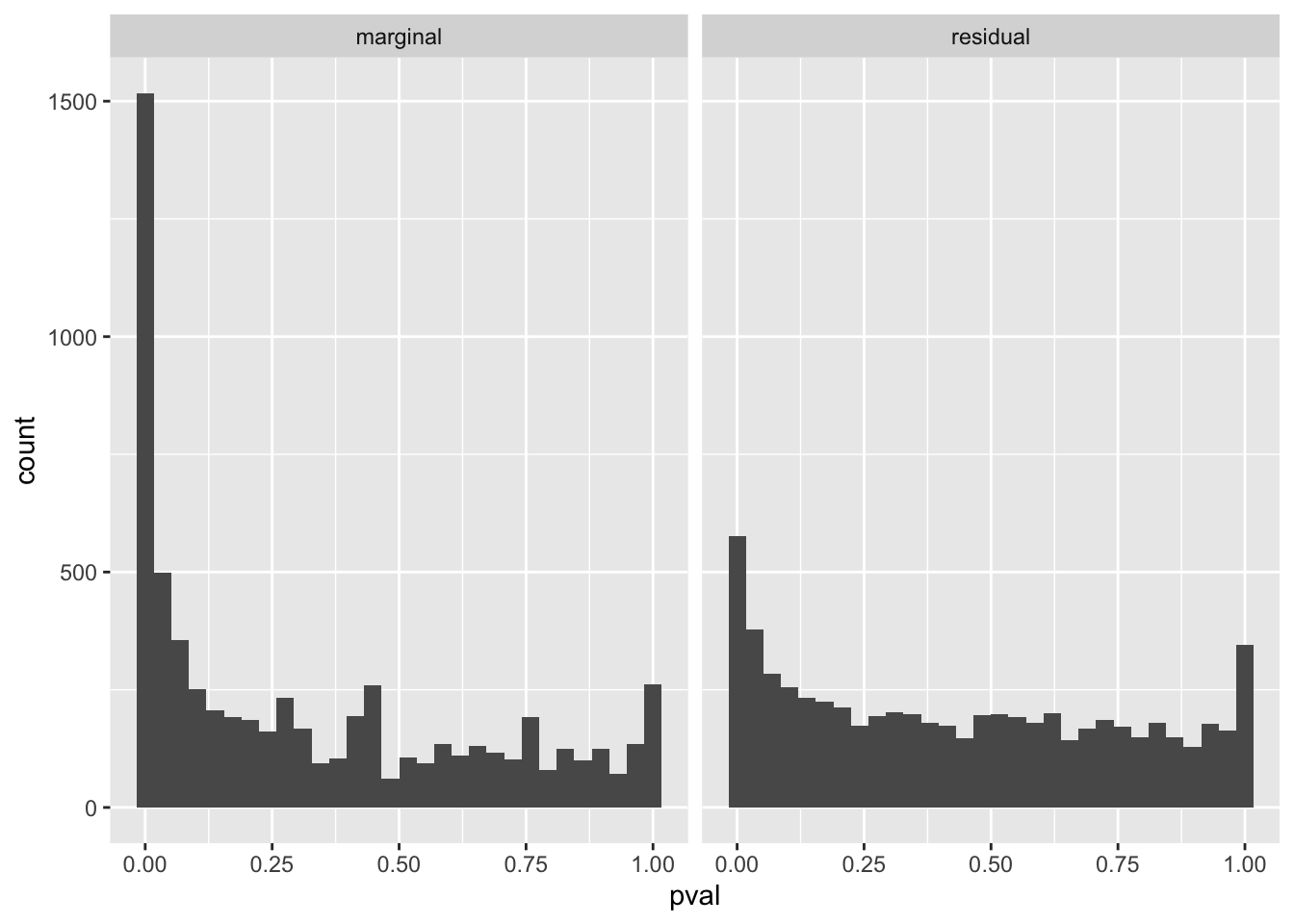
We can produce an interactive table to explore the enrichment results.s
gseasusie::interactive_table(logistic.fit, ora)Joining, by = "geneSet"
Joining, by = c("geneSet", "component")
Joining, by = c("geneSet", "component")
Joining, by = "geneSet"
Joining, by = "geneSet"
sessionInfo()R version 4.1.2 (2021-11-01)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Big Sur 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.1/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.1/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] graphics stats4 stats methods utils base
other attached packages:
[1] DESeq2_1.34.0 SummarizedExperiment_1.24.0
[3] Biobase_2.54.0 MatrixGenerics_1.6.0
[5] matrixStats_0.62.0 GenomicRanges_1.46.1
[7] GenomeInfoDb_1.30.1 IRanges_2.28.0
[9] S4Vectors_0.32.4 BiocGenerics_0.40.0
[11] gseasusie_0.0.0.9000 testthat_3.1.4
[13] forcats_0.5.1 stringr_1.4.0
[15] dplyr_1.0.9 purrr_0.3.4
[17] readr_2.1.2 tidyr_1.2.0
[19] tibble_3.1.6 ggplot2_3.3.6
[21] tidyverse_1.3.1
loaded via a namespace (and not attached):
[1] utf8_1.2.2 reticulate_1.25 tidyselect_1.1.2
[4] htmlwidgets_1.5.4 RSQLite_2.2.13 AnnotationDbi_1.56.2
[7] grid_4.1.2 BiocParallel_1.28.3 devtools_2.4.3
[10] munsell_0.5.0 codetools_0.2-18 future_1.25.0
[13] withr_2.5.0 colorspace_2.0-3 WebGestaltR_0.4.4
[16] filelock_1.0.2 highr_0.9 knitr_1.39
[19] uuid_1.1-0 rstudioapi_0.13 listenv_0.8.0
[22] labeling_0.4.2 git2r_0.30.1 GenomeInfoDbData_1.2.7
[25] mixsqp_0.3-43 farver_2.1.0 bit64_4.0.5
[28] rprojroot_2.0.3 basilisk_1.6.0 parallelly_1.31.1
[31] vctrs_0.4.1 generics_0.1.2 xfun_0.30
[34] R6_2.5.1 doParallel_1.0.17 locfit_1.5-9.5
[37] RcppZiggurat_0.1.6 bitops_1.0-7 spatstat.utils_2.3-0
[40] cachem_1.0.6 reshape_0.8.9 DelayedArray_0.20.0
[43] assertthat_0.2.1 promises_1.2.0.1 scales_1.2.0
[46] gtable_0.3.0 mr.ash.alpha_0.1-42 globals_0.15.0
[49] processx_3.5.3 workflowr_1.7.0 tictoc_1.0.1
[52] rlang_1.0.2 fastglm_0.0.2 genefilter_1.76.0
[55] systemfonts_1.0.4 splines_4.1.2 broom_0.8.0
[58] BiocManager_1.30.17 yaml_2.3.5 modelr_0.1.8
[61] crosstalk_1.2.0 backports_1.4.1 Rfast_2.0.6
[64] httpuv_1.6.5 tools_4.1.2 usethis_2.1.5
[67] ellipsis_0.3.2 jquerylib_0.1.4 RColorBrewer_1.1-3
[70] sessioninfo_1.2.2 Rcpp_1.0.8.3 plyr_1.8.7
[73] zlibbioc_1.40.0 RCurl_1.98-1.6 ps_1.7.0
[76] basilisk.utils_1.6.0 prettyunits_1.1.1 reactR_0.4.4
[79] haven_2.5.0 here_1.0.1 fs_1.5.2
[82] furrr_0.3.0 apcluster_1.4.9 magrittr_2.0.3
[85] reprex_2.0.1 mvtnorm_1.1-3 whisker_0.4
[88] pkgload_1.2.4 reactable_0.2.3 hms_1.1.1
[91] evaluate_0.15 xtable_1.8-4 XML_3.99-0.9
[94] readxl_1.4.0 compiler_4.1.2 crayon_1.5.1
[97] htmltools_0.5.2 later_1.3.0 tzdb_0.3.0
[100] geneplotter_1.72.0 lubridate_1.8.0 DBI_1.1.2
[103] dbplyr_2.1.1 data.tree_1.0.0 Matrix_1.4-1
[106] brio_1.1.3 cli_3.3.0 datasets_4.1.2
[109] grDevices_4.1.2 parallel_4.1.2 igraph_1.3.1
[112] pkgconfig_2.0.3 bigmemory.sri_0.1.3 dir.expiry_1.2.0
[115] xml2_1.3.3 foreach_1.5.2 svglite_2.1.0
[118] annotate_1.72.0 bslib_0.3.1 rngtools_1.5.2
[121] XVector_0.34.0 rvest_1.0.2 doRNG_1.8.2
[124] callr_3.7.0 digest_0.6.29 Biostrings_2.62.0
[127] rmarkdown_2.14 cellranger_1.1.0 curl_4.3.2
[130] lifecycle_1.0.1 jsonlite_1.8.0 modules_0.10.0
[133] bigmemory_4.6.1 desc_1.4.1 fansi_1.0.3
[136] pillar_1.7.0 susieR_0.11.92 lattice_0.20-45
[139] VEB.Boost_0.0.0.9037 KEGGREST_1.34.0 fastmap_1.1.0
[142] httr_1.4.3 pkgbuild_1.3.1 survival_3.3-1
[145] glue_1.6.2 remotes_2.4.2 png_0.1-7
[148] iterators_1.0.14 emulator_1.2-21 bit_4.0.4
[151] stringi_1.7.6 sass_0.4.1 blob_1.2.3
[154] org.Hs.eg.db_3.14.0 memoise_2.0.1 renv_0.15.4
[157] irlba_2.3.5