Last updated: 2021-12-08
Checks: 7 0
Knit directory: cacoaAnalysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it's best to always run the code in an empty environment.
The command set.seed(20211123) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 91efca9. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.Rhistory
Ignored: analysis/de_figure_ep.nb.html
Ignored: analysis/figure/
Ignored: analysis/preprocess.nb.html
Ignored: analysis/report_ms.nb.html
Ignored: analysis/report_template.nb.html
Ignored: analysis/simulation_distances.nb.html
Ignored: analysis/simulation_note.nb.html
Ignored: analysis/simulation_variance.nb.html
Ignored: cache/
Ignored: data/ASD/
Ignored: data/AZ/
Ignored: data/EP/
Ignored: data/MS/
Ignored: data/PF/
Ignored: data/SCC/
Ignored: man/
Untracked files:
Untracked: analysis/cluster_free_de_pf.Rmd
Untracked: analysis/de_figure_ep.Rmd
Untracked: analysis/fig_cluster_free_de.Rmd
Untracked: analysis/prepare_cacoa_results.Rmd
Untracked: analysis/report_template.Rmd
Untracked: analysis/simulation_note.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/report_ms.Rmd) and HTML (docs/report_ms.html) files. If you've configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 91efca9 | viktor_petukhov | 2021-12-08 | MS report |
library(cacoa)
library(dataorganizer)
library(ggplot2)
library(dplyr)
library(cowplot)
library(Matrix)
library(dplyr)
library(magrittr)
library(readr)
library(org.Hs.eg.db)
theme_set(theme_bw())cao <- read_rds(DataPath("MS/cao.rds")) %>% Cacoa$new()
cao$plot.params <- list(size=0.1, alpha=0.1, font.size=c(2, 3))Dependency on metadata
lapply(c("coda", "expression.shifts"), function(sp) {
cao$plotSampleDistances(space=sp, legend.position=c(1, 1))
}) %>%
plot_grid(plotlist=., ncol=2, labels=c("CoDA", "Expression"), hjust=0, label_x=0.02, label_y=0.99)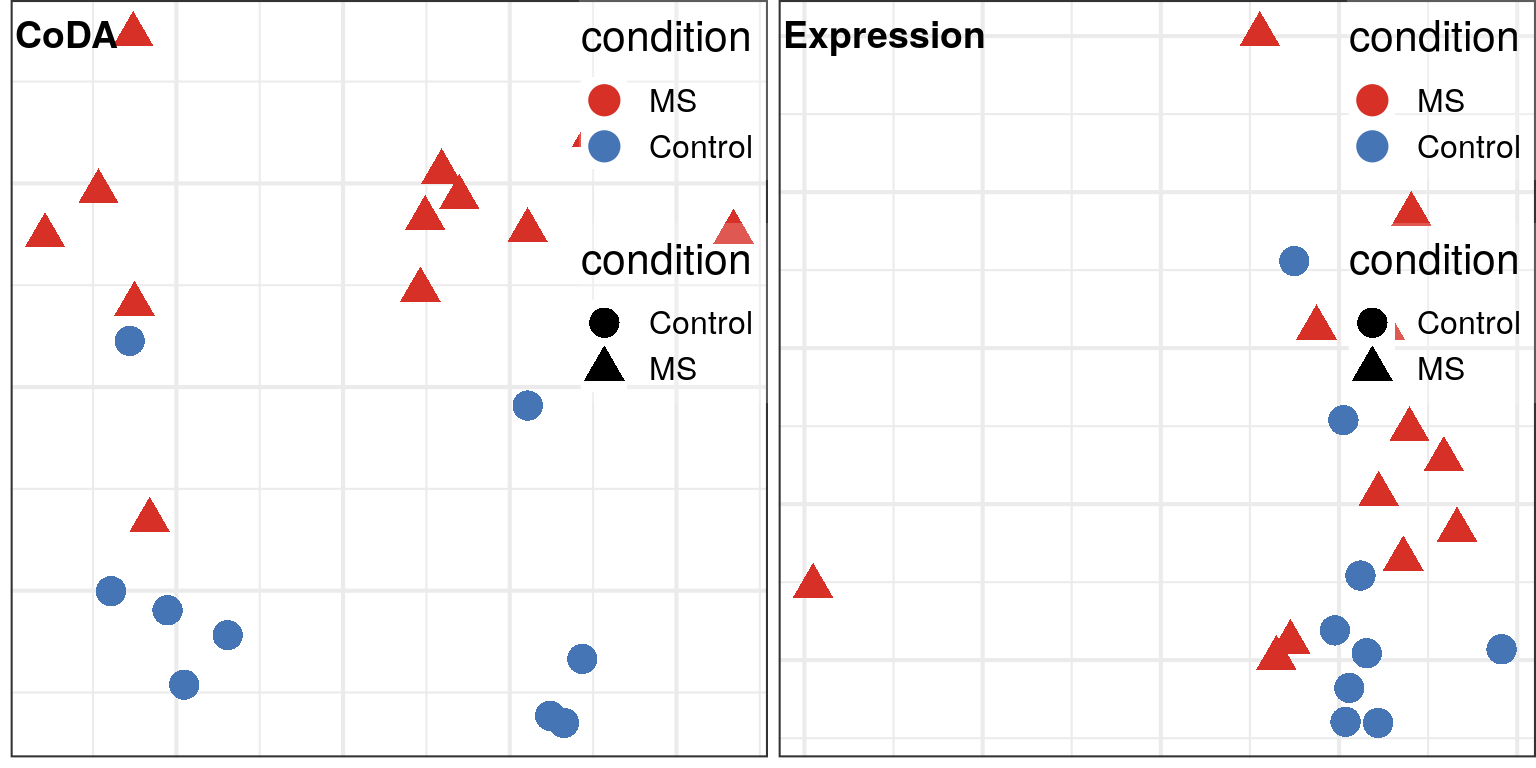
sample_meta <- cao$data.object$misc$sample_metadata
lapply(c("coda", "expression.shifts"), function(sp) {
smd <- as.data.frame(sample_meta) %>% dplyr::select(-sample, -diagnosis)
sep.res <- cao$estimateMetadataSeparation(smd, space=sp, dist="l1", name=paste0("md.", sp),
show.warning=FALSE)
(-log10(sep.res$padjust)) %>% {tibble(Type=names(.), value=.)} %>%
cacoa:::plotMeanMedValuesPerCellType(type="bar", yline=-log10(0.05),
ylab="-log10(separation p-value)") +
scale_y_continuous(expand=c(0, 0, 0.05, 0)) +
scale_fill_manual(values=rep("#2b8cbe", length(sample_meta))) +
theme(axis.title.y=element_text(size=13))
}) %>%
plot_grid(plotlist=., ncol=2, labels=c("CoDA", "Expression"), hjust=0, label_x=0.2, label_y=0.98)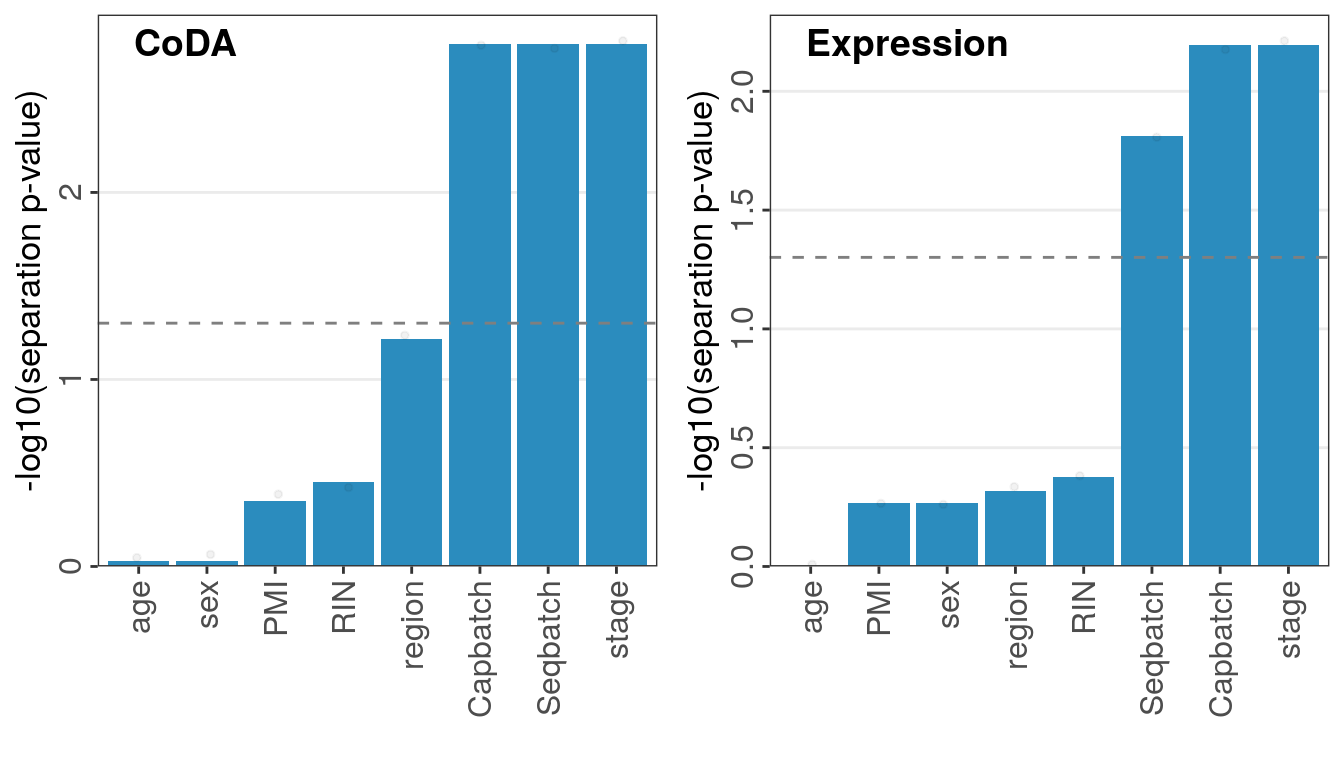
There is significant separation by Capbatch, Seqbatch and stage. Capbatch and Seqbatch are very similar, so we show only the former.
lapply(c('coda', 'expression'), function(sp) {
lapply(c("Seqbatch", "stage"), function(n) {
cao$plotSampleDistances(space=sp, legend.position=c(0, 1), sample.colors=sample_meta[[n]])
}) %>% plot_grid(plotlist=., nrow=1)
}) %>% plot_grid(plotlist=., ncol=1)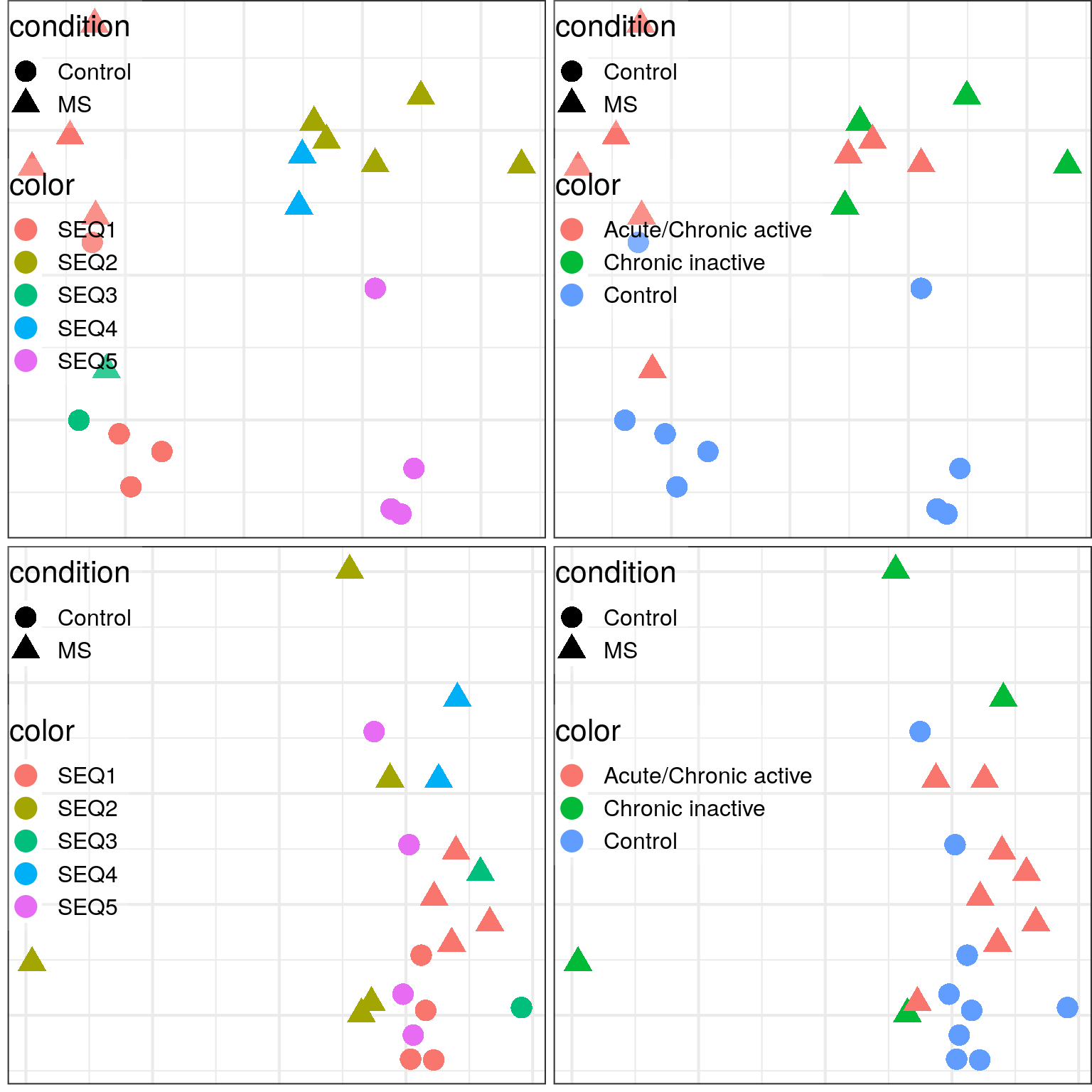
CoDA space (top row) has even stronger separation by batch than expression space.
Compositional differences
Cluster-based
cao$plotCellLoadings()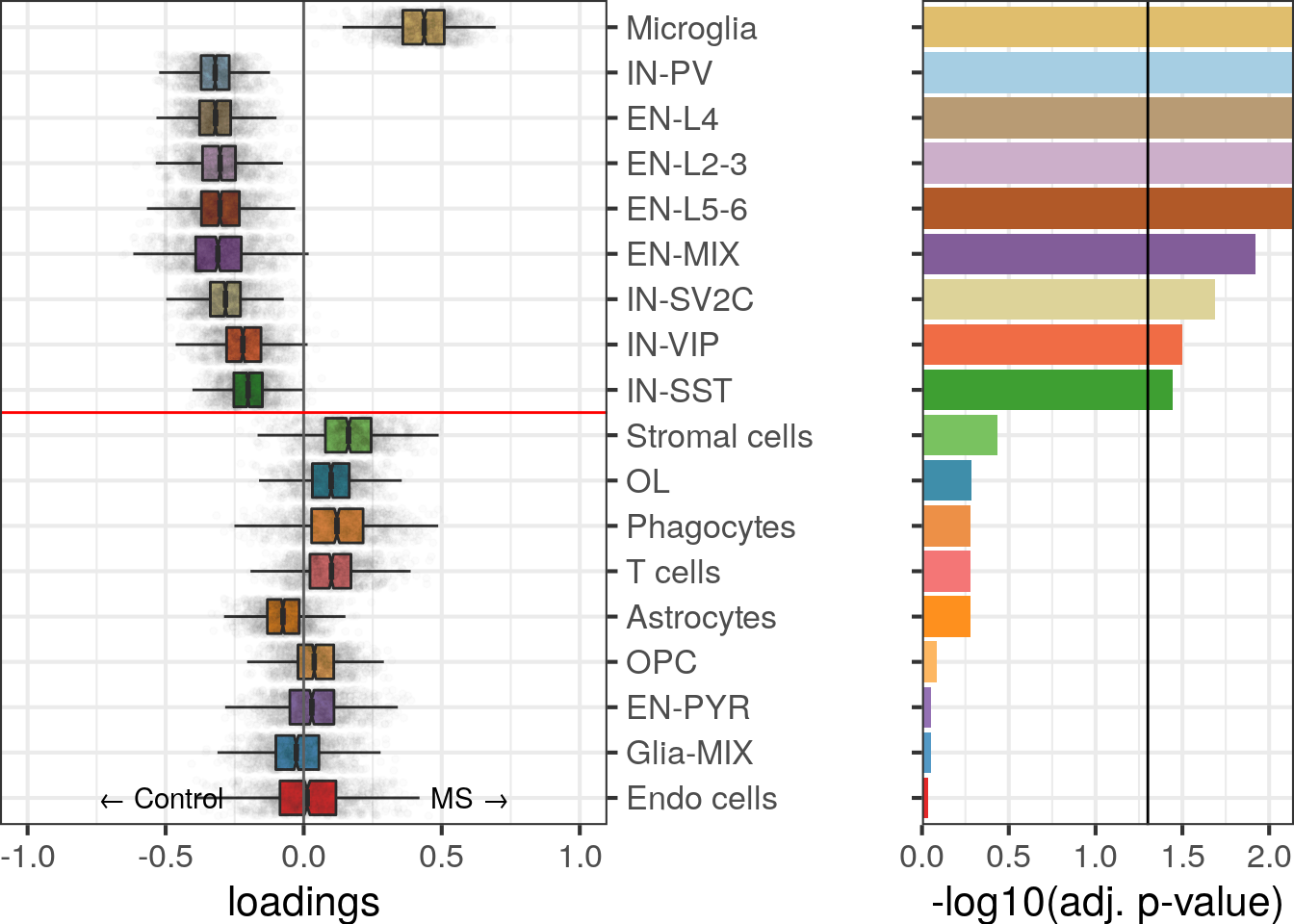
cao$plotContrastTree()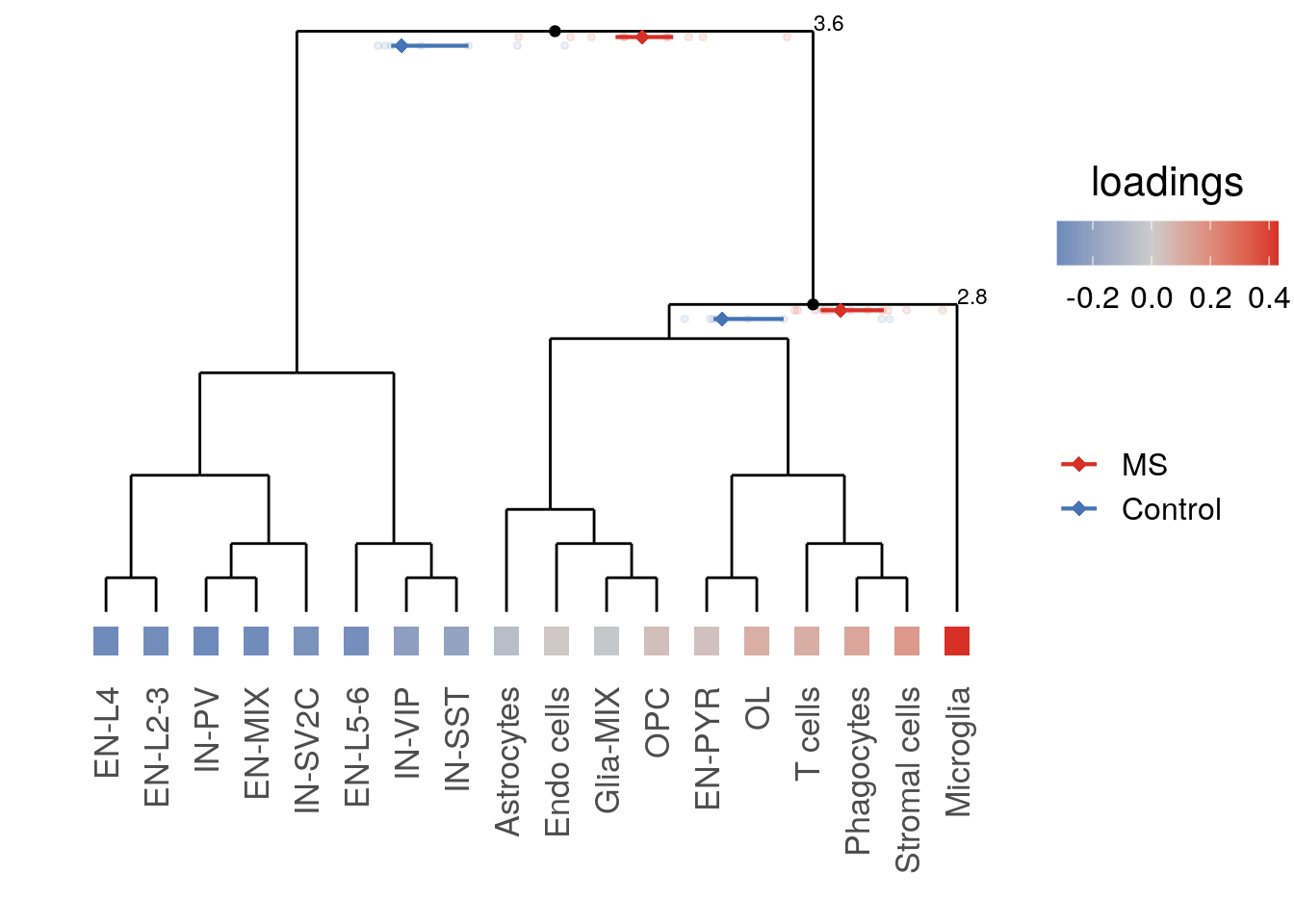
We can see that most neuronal subtypes go to the left branch of the tree, while all glial and immune types go to the right.
Cluster-free
g0 <- cao$plotEmbedding(color.by='cell.groups')
g0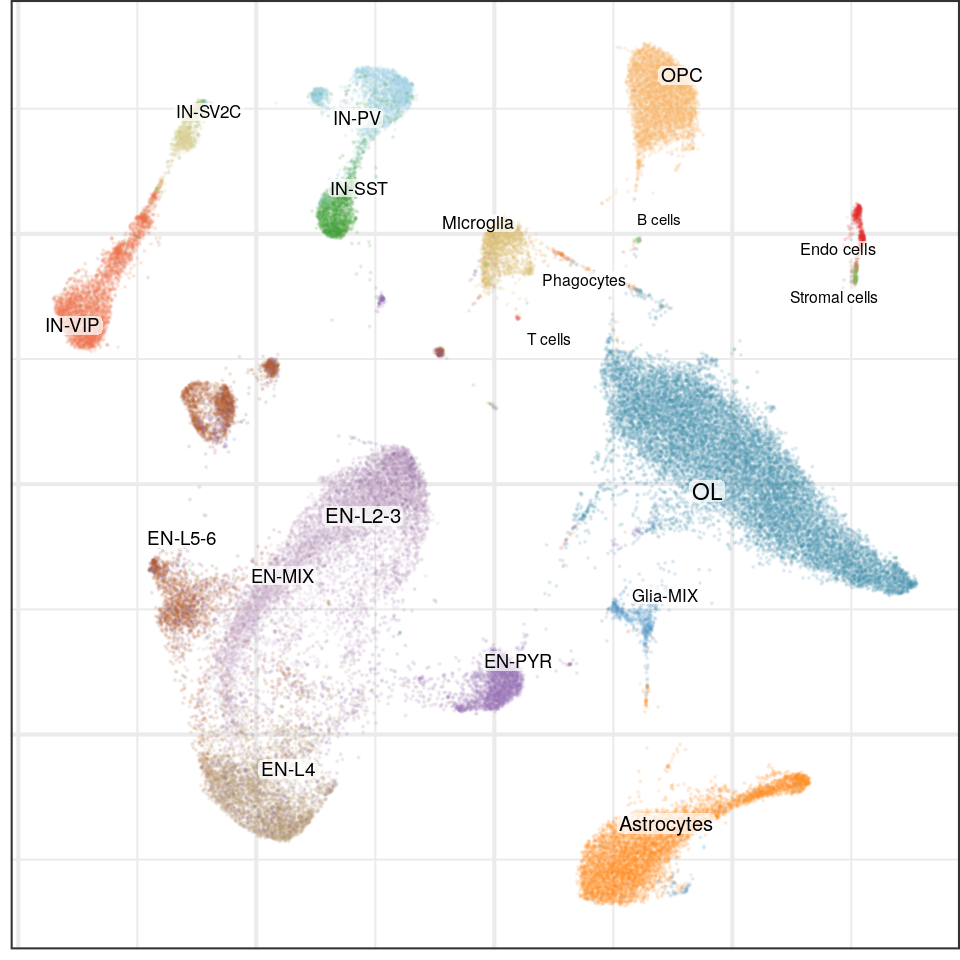
KDE-based:
cao$plotCellDensity(name='cell.density.kde') %>% plot_grid(plotlist=., ncol=2)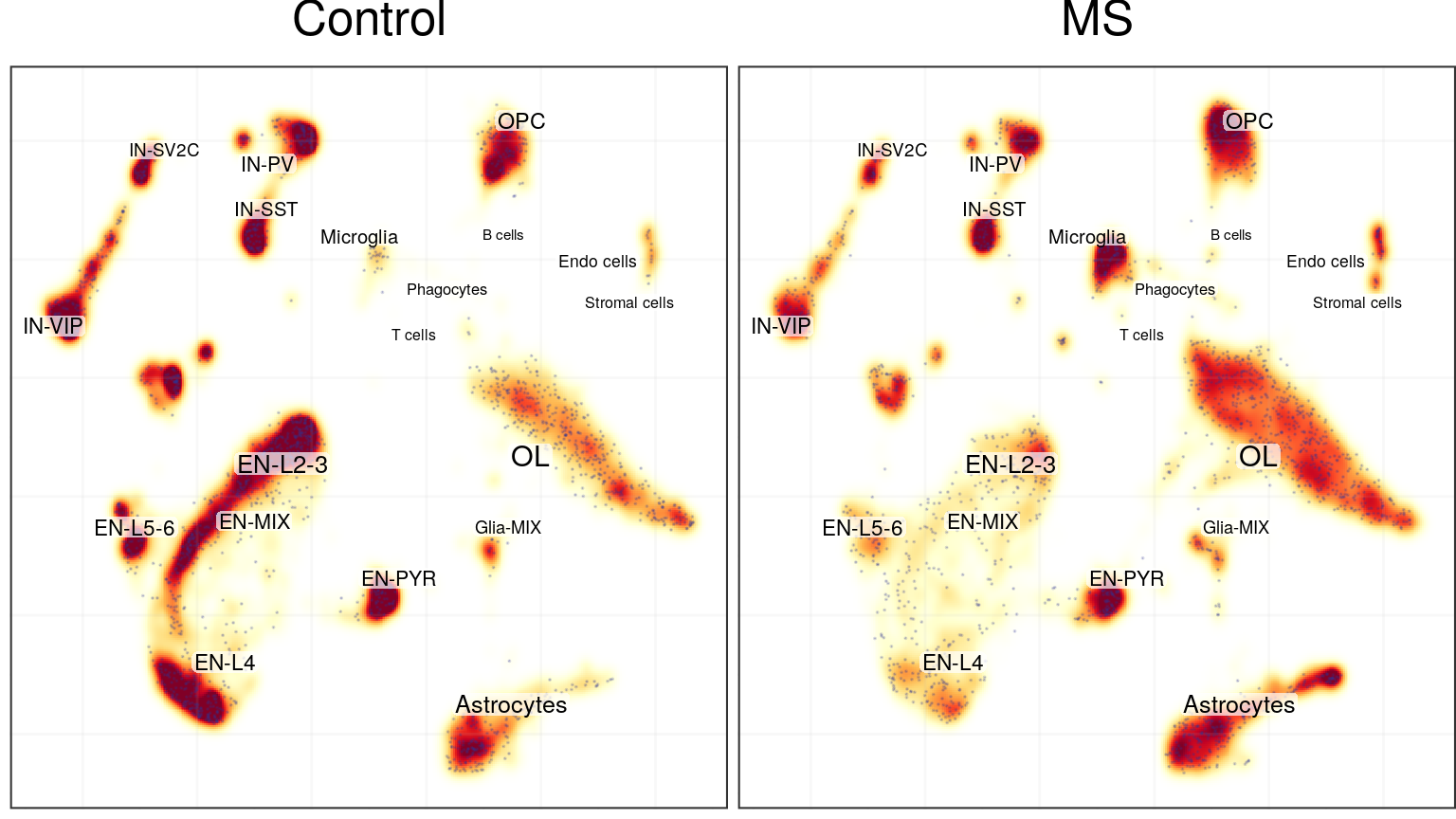
Differential cell density:
plot_grid(g0, cao$plotDiffCellDensity(name='cell.density.kde', legend.position=c(0, 1)), ncol=2)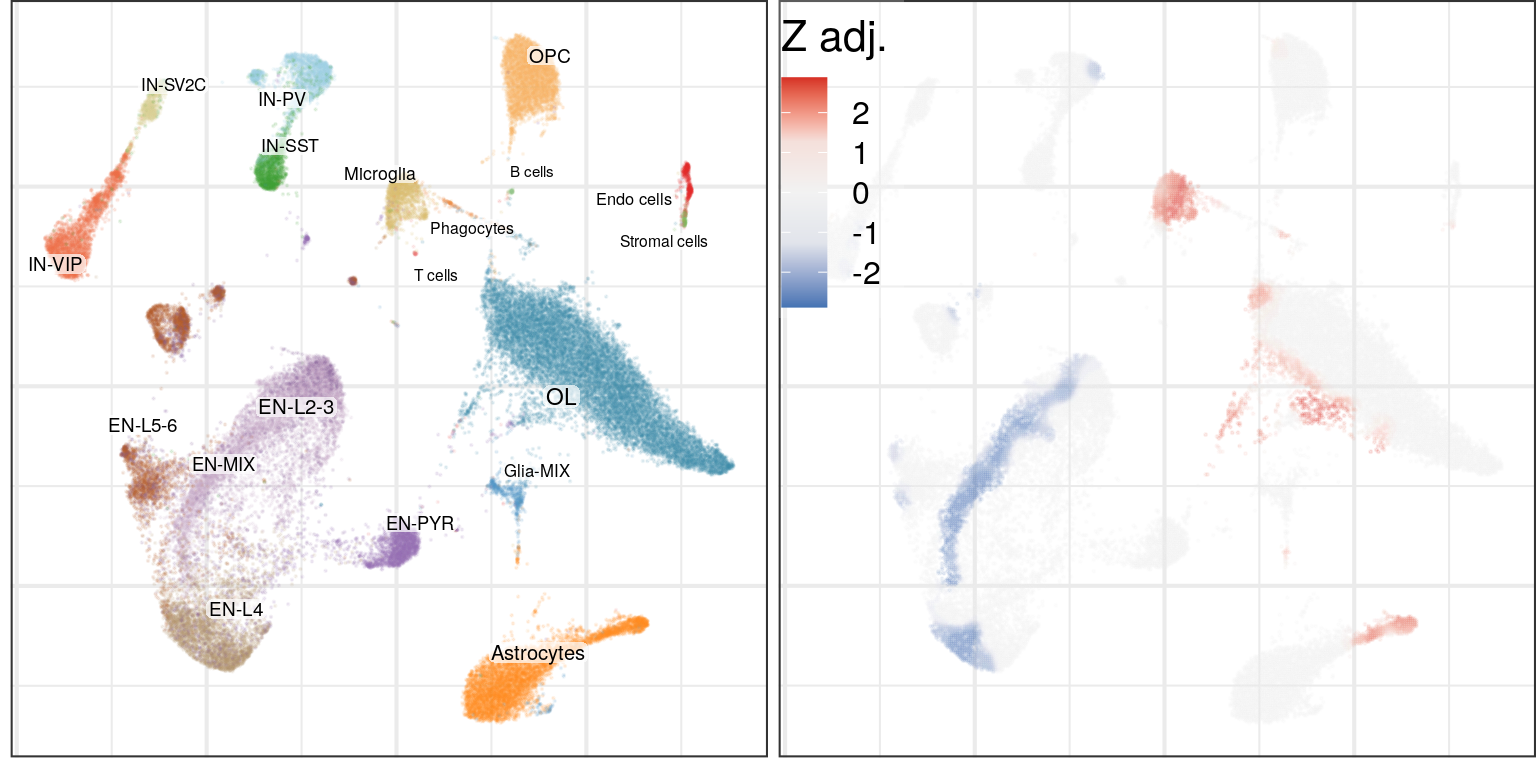
Graph-based:
cao$plotCellDensity(name='cell.density.graph') %>% plot_grid(plotlist=., ncol=2)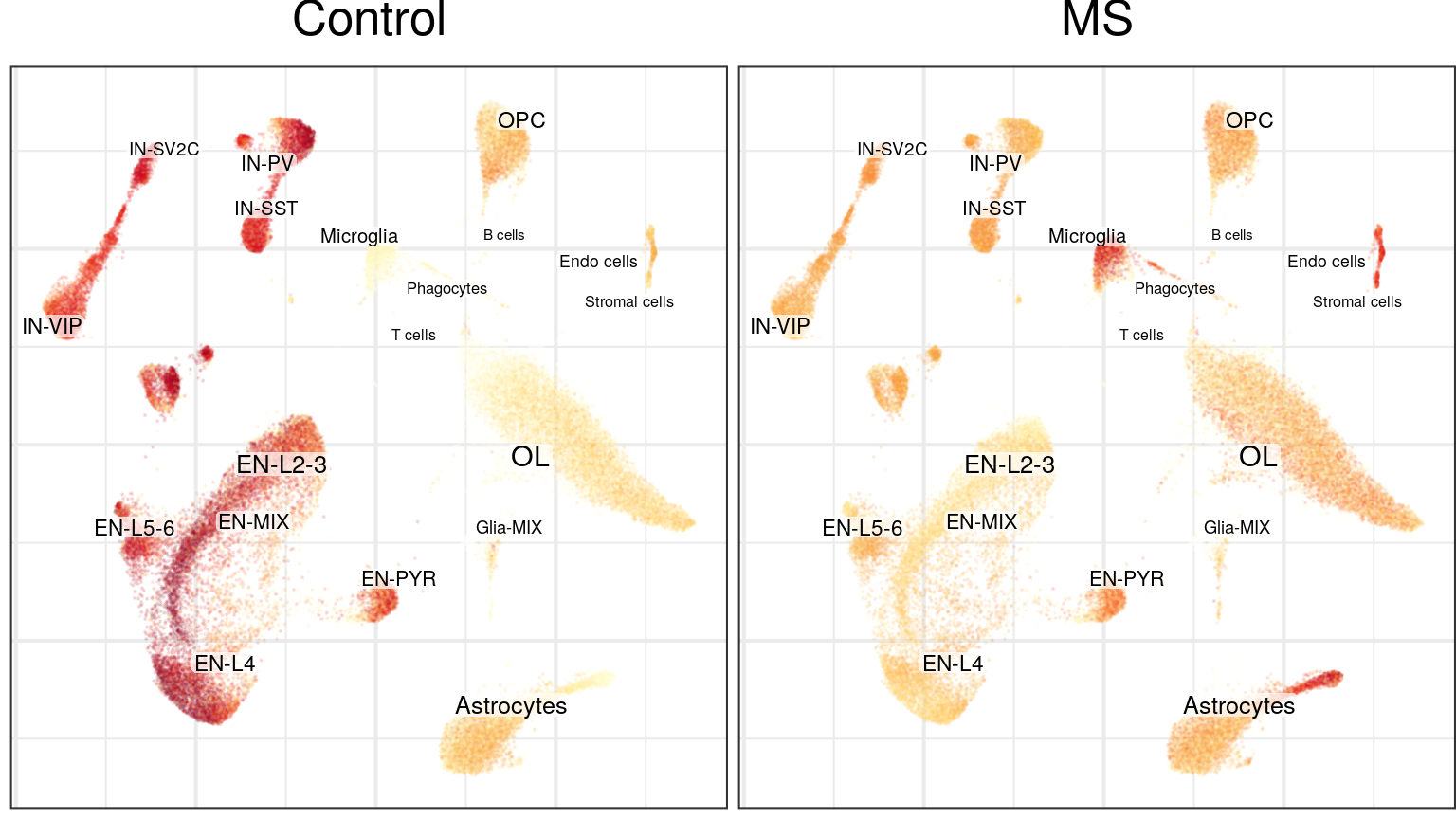
Differential cell density:
plot_grid(g0, cao$plotDiffCellDensity(name='cell.density.graph', legend.position=c(0, 1)), ncol=2)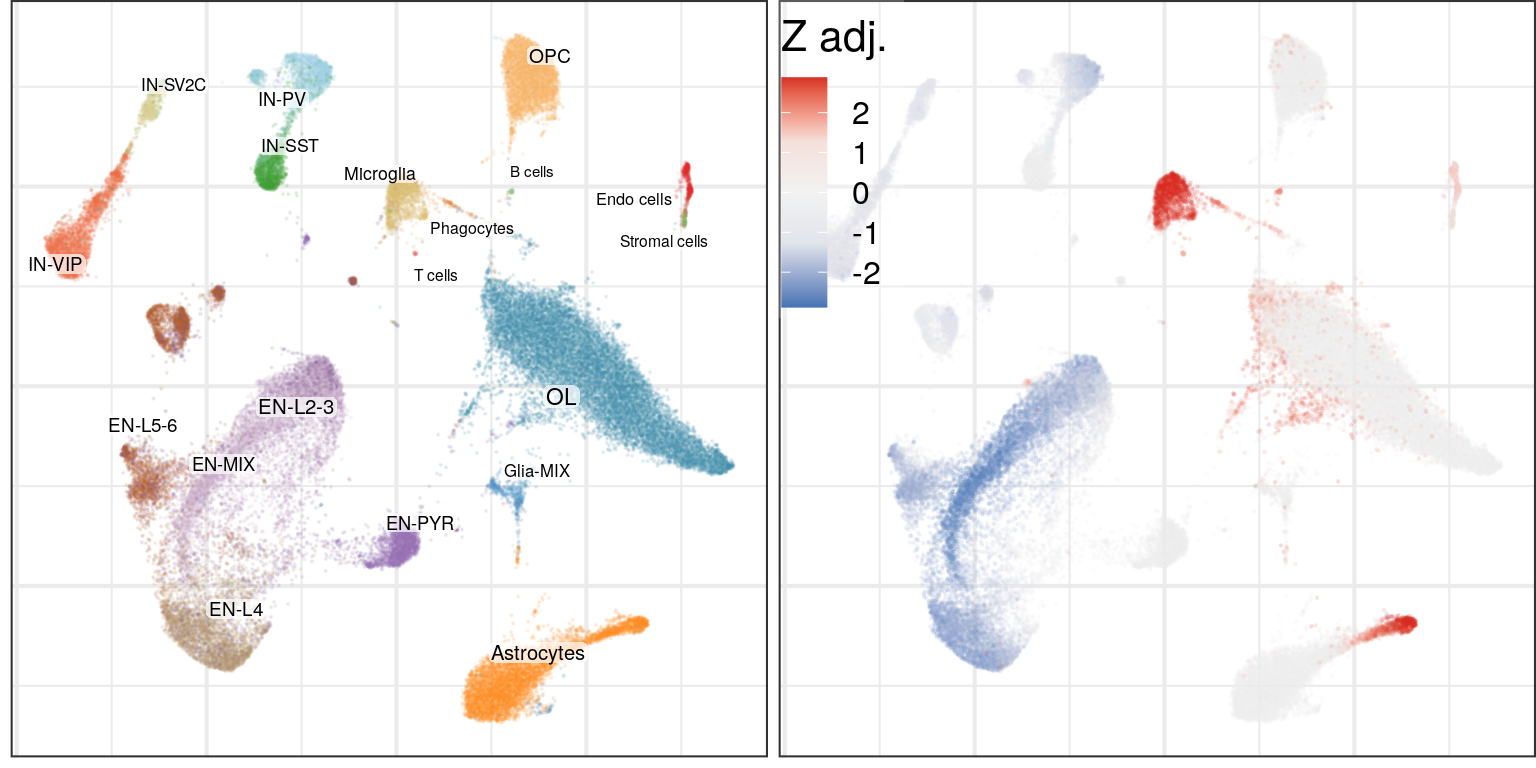
Here, graph-based method shows higher sensitivity, and its results mostly match to the cluster-based version. It also detects that in Astrocytes only one subtype is affected.
Expression differences
Cluster-based
All genes:
cao$plotExpressionShiftMagnitudes()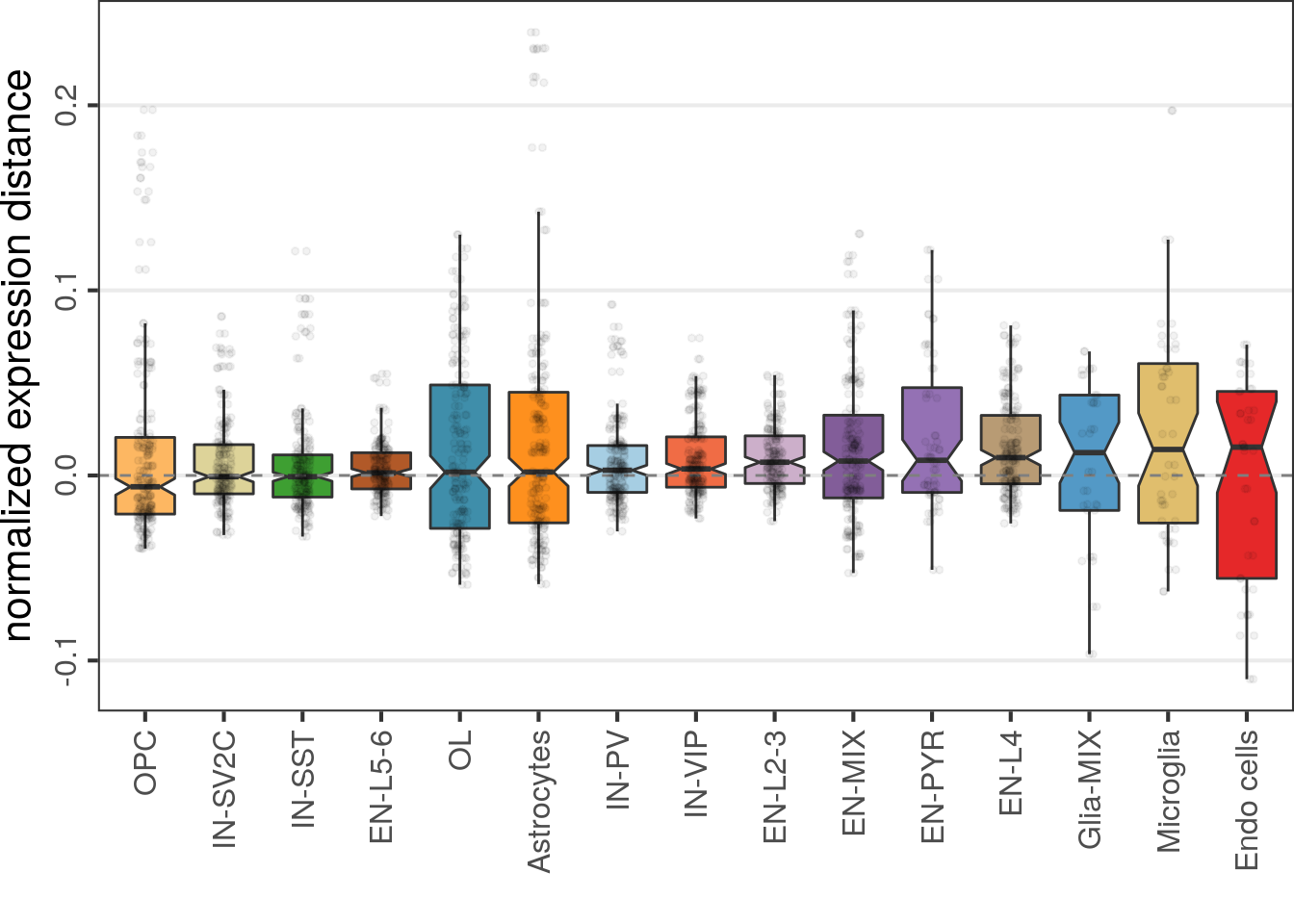
No significant changes detected.
Top DE genes:
cao$estimateExpressionShiftMagnitudes(n.permutations=5000, top.n.genes=500, n.pcs=8,
min.samp.per.type=4, name='es.top.de', verbose=FALSE)cao$plotExpressionShiftMagnitudes(name='es.top.de')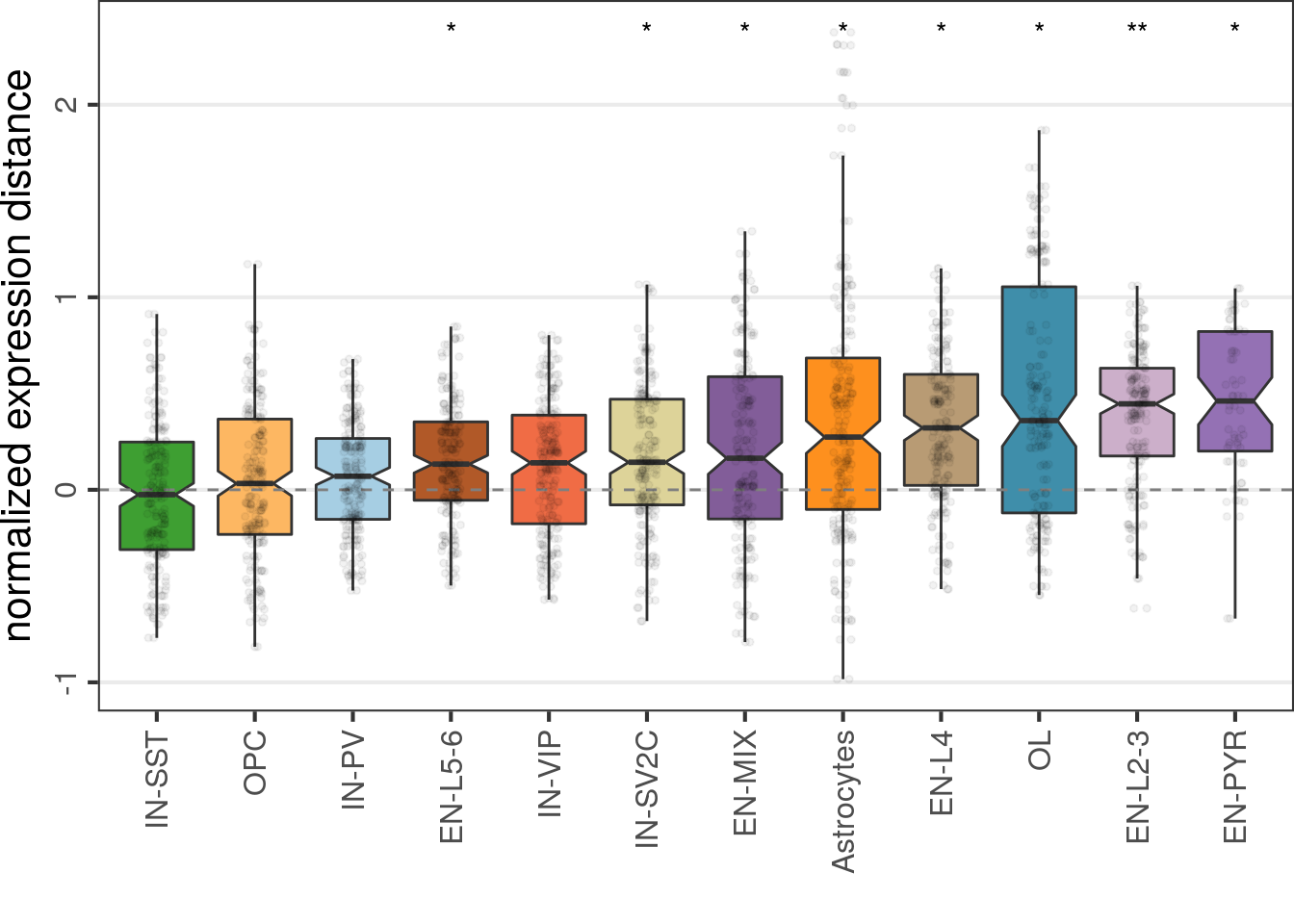
Number of DE genes:
cao$estimateDEPerCellType(
independent.filtering=TRUE, test='DESeq2.Wald', verbose=FALSE, resampling.method='fix.samples',
fix.n.samples=6, n.cells.subsample=30, name='de.fix.samples', n.resamplings=50
)cao$plotNumberOfDEGenes(
name="de.fix.samples", type="box", show.resampling.results=TRUE, jitter.alpha=0.5,
show.jitter=TRUE, y.offset=1
) + scale_y_log10(labels=c(0, 10, 100), breaks=c(1, 11, 101), expand=c(0, 0), limits=c(1, 300))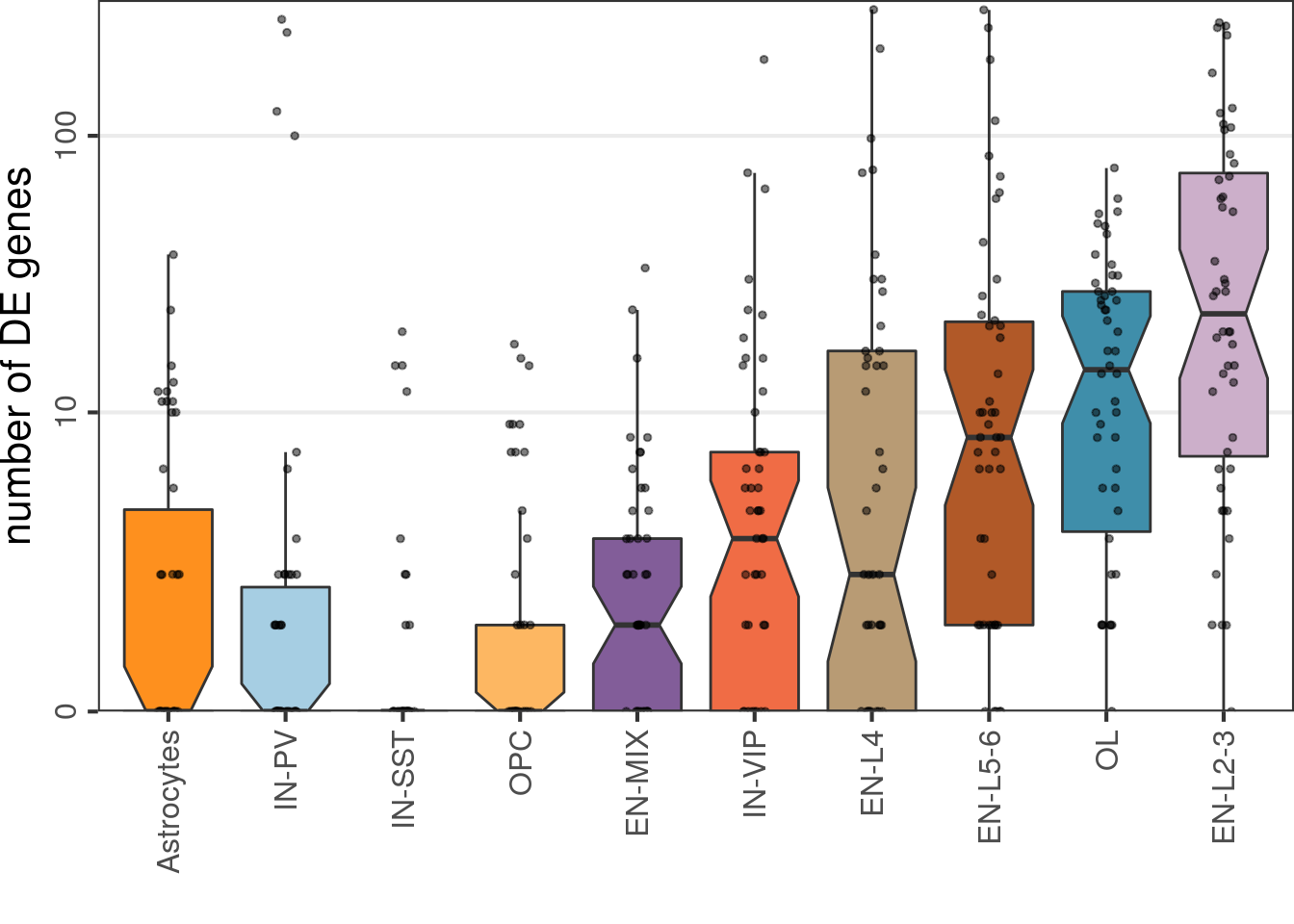
Cluster-free
cao$plotClusterFreeExpressionShifts(legend.position=c(0, 1), font.size=c(1, 2))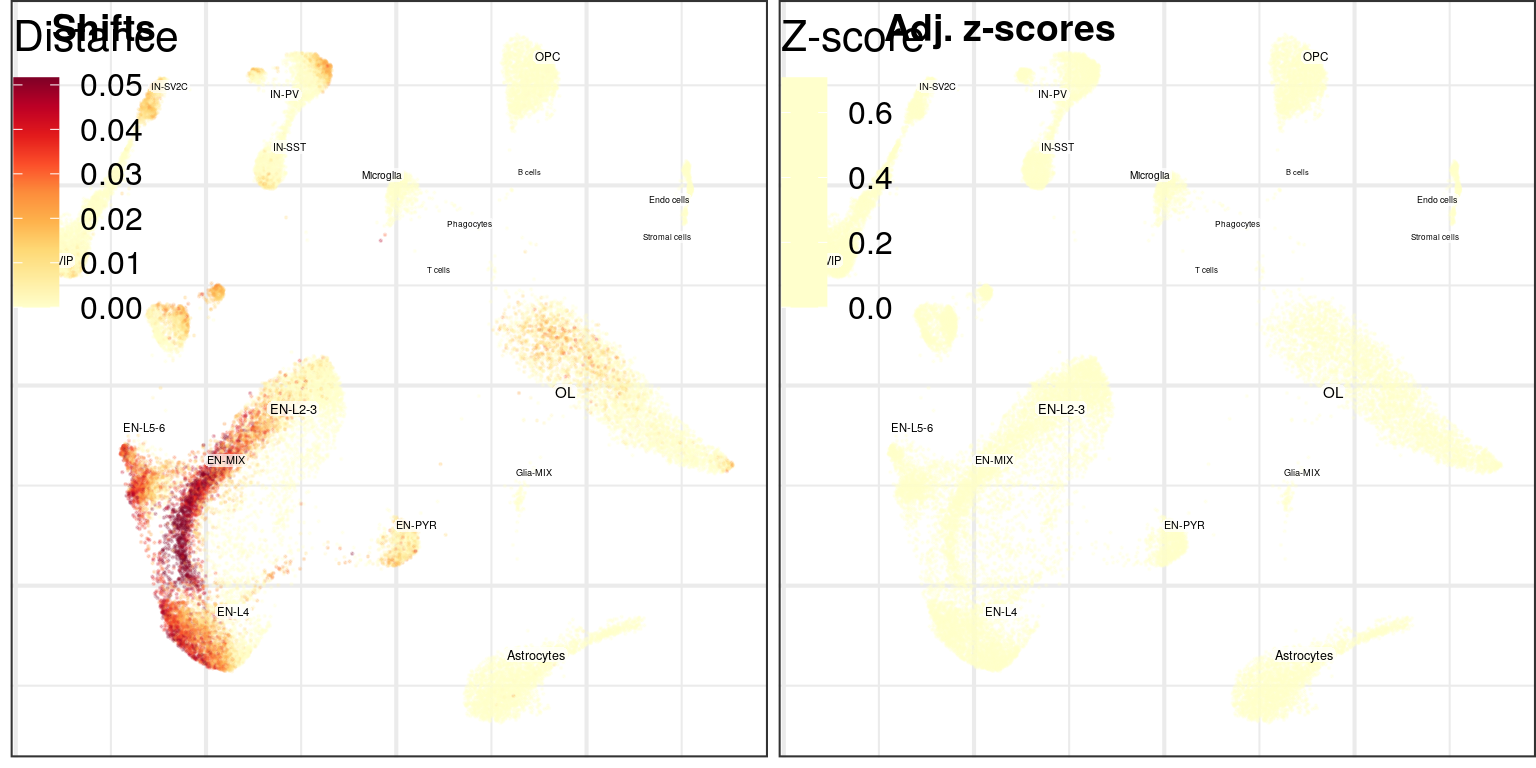
Functional interpretation
cao$plotVolcano(xlim=c(-3, 3), ylim=c(0, 3.5), lf.cutoff=1)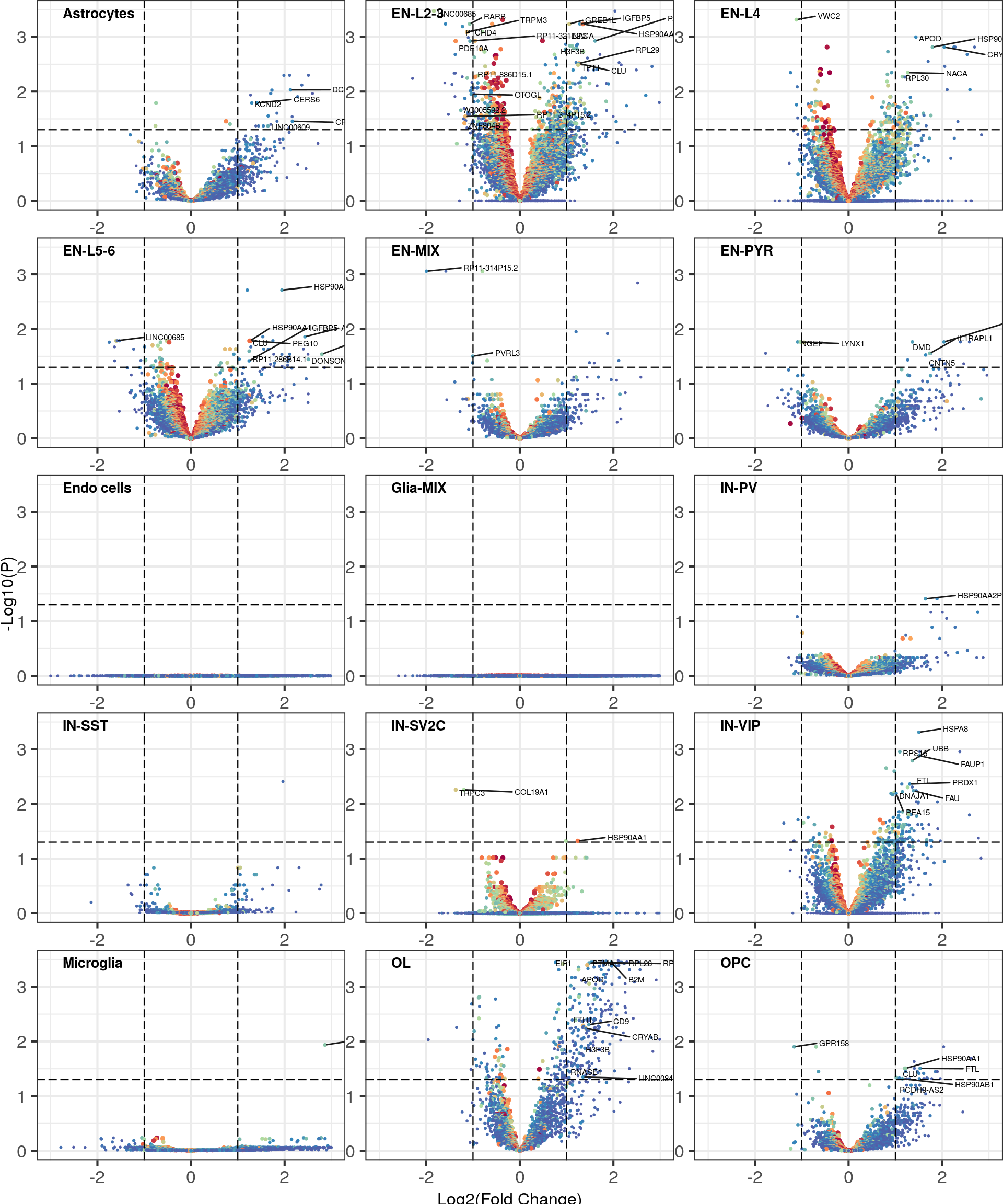
cao$plotOntologyHeatmapCollapsed(name="GSEA", genes="all", n=30, clust.method="ward.D")Loading required package: DOSEDOSE v3.16.0 For help: https://guangchuangyu.github.io/software/DOSE
If you use DOSE in published research, please cite:
Guangchuang Yu, Li-Gen Wang, Guang-Rong Yan, Qing-Yu He. DOSE: an R/Bioconductor package for Disease Ontology Semantic and Enrichment analysis. Bioinformatics 2015, 31(4):608-609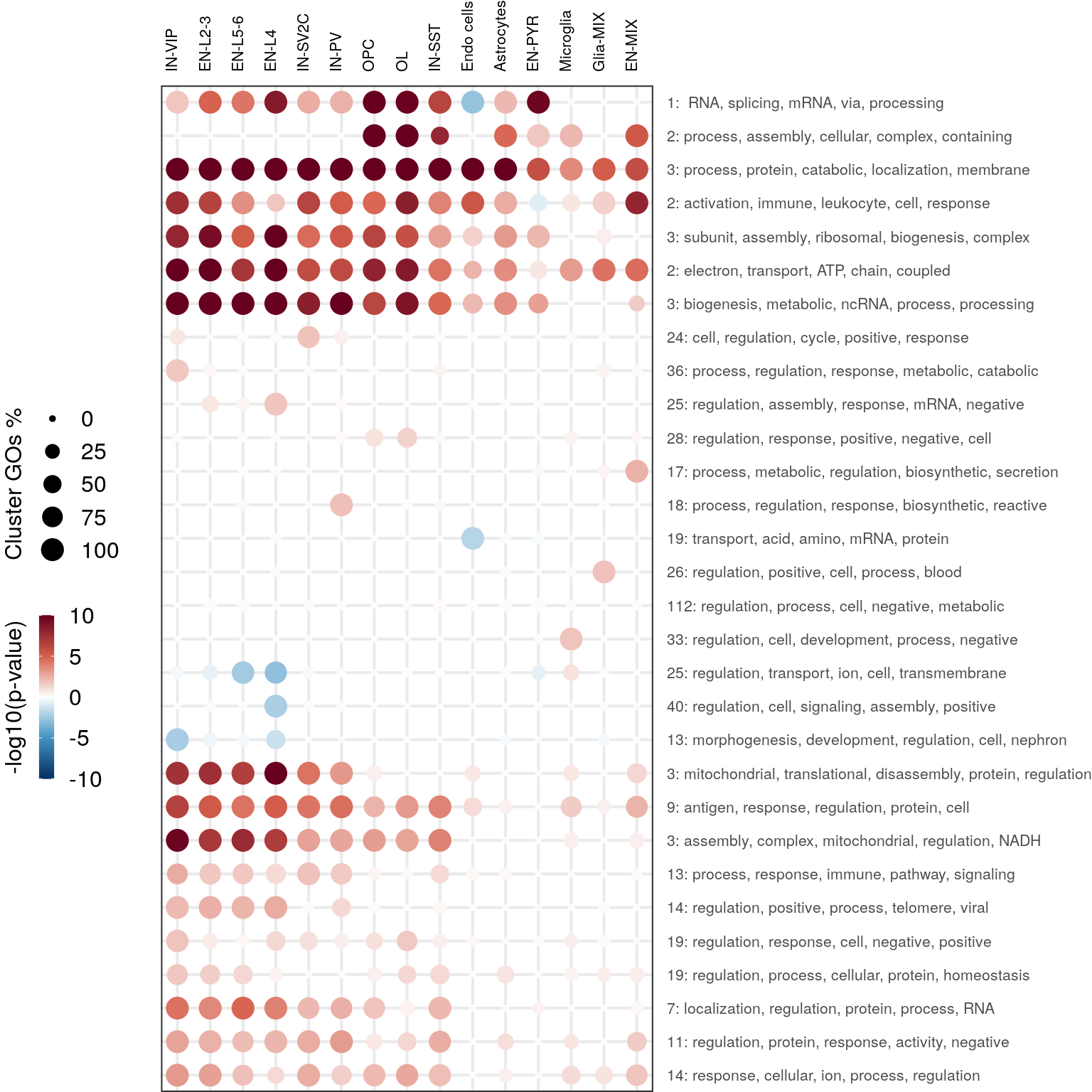
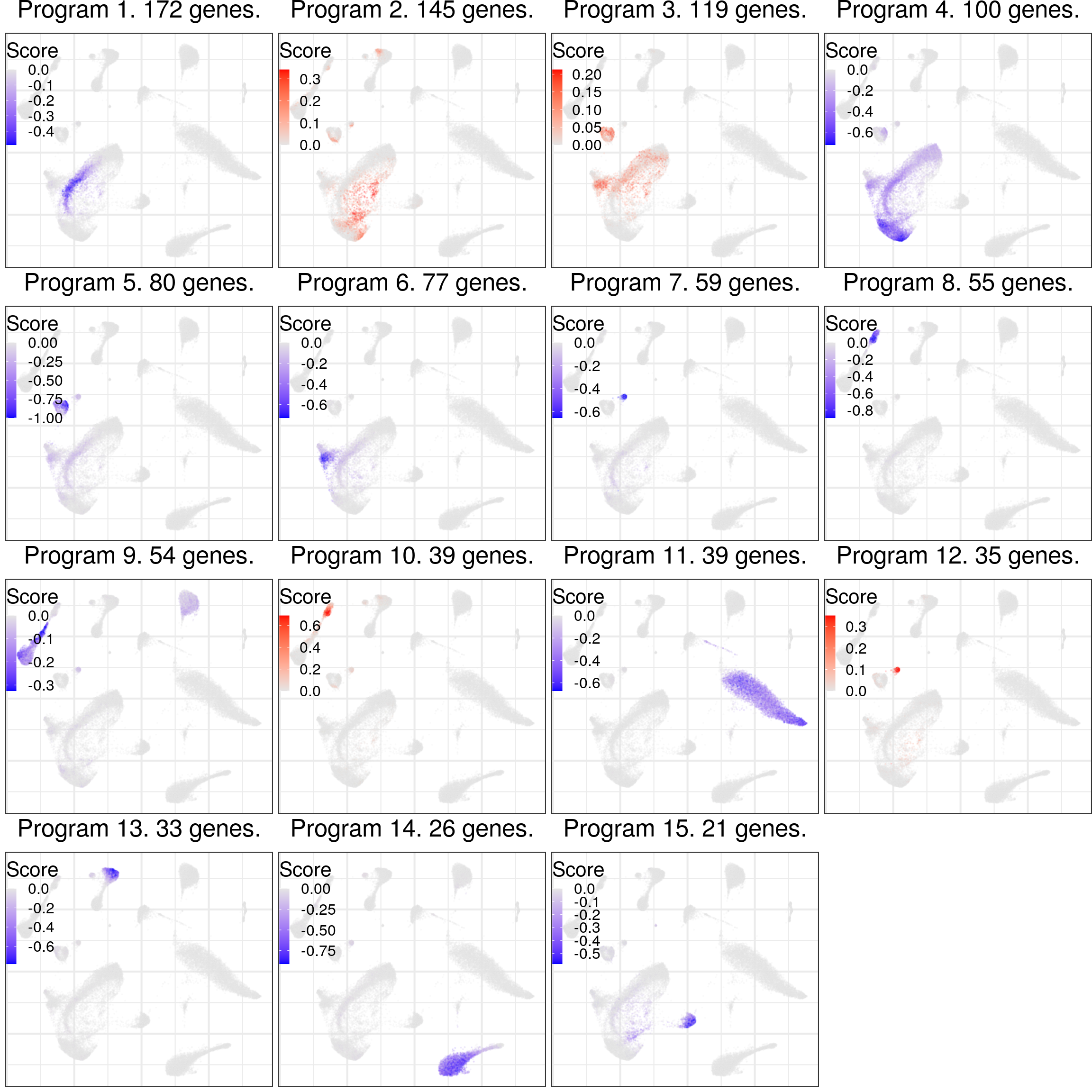
cao$estimateGenePrograms(method="leiden", z.adj=TRUE, smooth=FALSE)cao$plotGeneProgramScores(
legend.position=c(0, 1), plot.na=FALSE,
adj.list=theme(legend.key.width=unit(8, "pt"), legend.key.height=unit(12, "pt"))
)
sessionInfo()R version 4.0.3 (2020-10-10)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 18.04.6 LTS
Matrix products: default
BLAS: /usr/local/R/R-4.0.3/lib/R/lib/libRblas.so
LAPACK: /usr/local/R/R-4.0.3/lib/R/lib/libRlapack.so
locale:
[1] C
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] DOSE_3.16.0 org.Hs.eg.db_3.12.0 AnnotationDbi_1.52.0
[4] IRanges_2.24.1 S4Vectors_0.28.1 Biobase_2.50.0
[7] BiocGenerics_0.36.1 readr_1.4.0 magrittr_2.0.1
[10] cowplot_1.1.1 dplyr_1.0.7 ggplot2_3.3.5
[13] dataorganizer_0.1.0 cacoa_0.2.0 Matrix_1.2-18
[16] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] N2R_0.1.1 circlize_0.4.13 fastmatch_1.1-0
[4] plyr_1.8.6 igraph_1.2.6 lazyeval_0.2.2
[7] splines_4.0.3 BiocParallel_1.24.1 urltools_1.7.3
[10] digest_0.6.28 foreach_1.5.1 htmltools_0.5.2
[13] GOSemSim_2.16.1 GO.db_3.12.1 fansi_0.5.0
[16] RMTstat_0.3 memoise_2.0.0 cluster_2.1.0
[19] doParallel_1.0.16 ComplexHeatmap_2.9.4 extrafont_0.17
[22] matrixStats_0.61.0 R.utils_2.10.1 extrafontdb_1.0
[25] sccore_1.0.0 colorspace_2.0-2 blob_1.2.2
[28] ggrepel_0.9.1 pagoda2_1.0.7 xfun_0.26
[31] crayon_1.4.1 brew_1.0-6 iterators_1.0.13
[34] ape_5.5 glue_1.4.2 gtable_0.3.0
[37] GetoptLong_1.0.5 proj4_1.0-10.1 leidenAlg_0.1.0
[40] Rook_1.1-1 Rttf2pt1_1.3.8 shape_1.4.6
[43] maps_3.3.0 abind_1.4-5 scales_1.1.1
[46] DBI_1.1.1 Rcpp_1.0.7 tmvnsim_1.0-2
[49] clue_0.3-59 tidytree_0.3.4 bit_4.0.4
[52] fgsea_1.16.0 RColorBrewer_1.1-2 ellipsis_0.3.2
[55] pkgconfig_2.0.3 R.methodsS3_1.8.1 farver_2.1.0
[58] utf8_1.2.2 tidyselect_1.1.1 labeling_0.4.2
[61] rlang_0.4.11 reshape2_1.4.4 later_1.1.0.1
[64] munsell_0.5.0 tools_4.0.3 cachem_1.0.6
[67] generics_0.1.0 RSQLite_2.2.8 evaluate_0.14
[70] stringr_1.4.0 fastmap_1.1.0 ggdendro_0.1.22
[73] yaml_2.2.1 knitr_1.36 bit64_4.0.5
[76] fs_1.5.0 purrr_0.3.4 nlme_3.1-149
[79] whisker_0.4 ash_1.0-15 ggrastr_1.0.0
[82] R.oo_1.24.0 grr_0.9.5 DO.db_2.9
[85] compiler_4.0.3 beeswarm_0.4.0 png_0.1-7
[88] tibble_3.1.5 stringi_1.7.5 highr_0.9
[91] drat_0.1.8 ggalt_0.4.0 lattice_0.20-41
[94] psych_2.1.6 vctrs_0.3.8 pillar_1.6.3
[97] lifecycle_1.0.1 triebeard_0.3.0 jquerylib_0.1.4
[100] GlobalOptions_0.1.2 data.table_1.14.2 irlba_2.3.3
[103] Matrix.utils_0.9.8 qvalue_2.22.0 httpuv_1.5.4
[106] conos_1.4.4 R6_2.5.1 promises_1.1.1
[109] KernSmooth_2.23-17 gridExtra_2.3 vipor_0.4.5
[112] codetools_0.2-16 MASS_7.3-53 assertthat_0.2.1
[115] rprojroot_2.0.2 rjson_0.2.20 withr_2.4.2
[118] mnormt_2.0.2 EnhancedVolcano_1.8.0 mgcv_1.8-33
[121] hms_1.1.1 grid_4.0.3 tidyr_1.1.4
[124] rmarkdown_2.11 dendsort_0.3.3 Rtsne_0.15
[127] git2r_0.27.1 ggbeeswarm_0.6.0