Last updated: 2021-12-09
Checks: 7 0
Knit directory: cacoaAnalysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it's best to always run the code in an empty environment.
The command set.seed(20211123) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version b3ebfb3. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.Rhistory
Ignored: analysis/de_figure_ep.nb.html
Ignored: analysis/prepare_cacoa_results.nb.html
Ignored: analysis/preprocess.nb.html
Ignored: analysis/report_asd.nb.html
Ignored: analysis/report_az.nb.html
Ignored: analysis/report_ep.nb.html
Ignored: analysis/report_ms.nb.html
Ignored: analysis/report_pf.nb.html
Ignored: analysis/report_scc.nb.html
Ignored: analysis/report_template.nb.html
Ignored: analysis/simulation_distances.nb.html
Ignored: analysis/simulation_note.nb.html
Ignored: analysis/simulation_variance.nb.html
Ignored: cache/
Ignored: data/ASD/
Ignored: data/AZ/
Ignored: data/EP/
Ignored: data/MS/
Ignored: data/PF/
Ignored: data/SCC/
Ignored: man/
Untracked files:
Untracked: analysis/cluster_free_de_pf.Rmd
Untracked: analysis/de_figure_ep.Rmd
Untracked: analysis/fig_cluster_free_de.Rmd
Untracked: analysis/prepare_cacoa_results.Rmd
Untracked: analysis/report_pf.Rmd
Untracked: analysis/report_scc.Rmd
Untracked: analysis/report_template.Rmd
Untracked: analysis/simulation_note.Rmd
Unstaged changes:
Modified: analysis/report_asd.Rmd
Modified: analysis/report_ep.Rmd
Modified: analysis/report_ms.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/preprocess.Rmd) and HTML (docs/preprocess.html) files. If you've configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | b3ebfb3 | viktor_petukhov | 2021-12-09 | Filtration of samples in SCC preprocessing |
| html | 58bdad8 | viktor_petukhov | 2021-12-01 | Updated the preprocessing notebook |
| Rmd | 3f1371c | viktor_petukhov | 2021-11-30 | Cleaner preprocessing |
| Rmd | 1d802ad | viktor_petukhov | 2021-11-30 | Force rebuild in preprocess.Rmd |
| Rmd | 2212721 | viktor_petukhov | 2021-11-30 | Small fix in SCC preprocessing |
| html | 3085bf5 | viktor_petukhov | 2021-11-30 | Build preprocessing.html |
| Rmd | 70d2759 | viktor_petukhov | 2021-11-30 | Preprocessing notebook |
Libraries
library(tidyverse)
library(magrittr)
library(sccore)
library(pagoda2)
library(conos)
library(dataorganizer)
library(Matrix)
library(reticulate)
devtools::load_all()
N_CORES <- 45
FORCE <- FALSEvirtualenv_create("r-scrublet")virtualenv: r-scrubletvirtualenv_install("r-scrublet", c("scrublet", "matplotlib"))Using virtual environment 'r-scrublet' ...use_virtualenv("r-scrublet")
mpl <- import("matplotlib")
mpl$use('Agg') # Otherwise it shows Qt error in RStudio
scrublet <- import("scrublet")
estimateDoubletInfo <- function(mats, progress=FALSE) {
dub.info <- sccore::plapply(mats, function(m) {
suppressMessages(scrublet$Scrublet(t(m), random_state=as.integer(42))$scrub_doublets()) %>%
lapply(setNames, colnames(m))
}, n.cores=1, progress=progress)
lapply(c(scores=1, mask=2), function(i) {
lapply(dub.info, `[[`, i) %>% unname() %>% unlist()
})
}Alzheimer
Cells are already filtered by mit. fraction, and doublets are removed. We filter only by minimum 500 UMIs per cell and scrublet scores.
con <- readOrCreate(DataPath('ASD/con.rds'), function() {
mat <- DataPath("AZ/cell_counts.csv") %>% data.table::fread(sep=",") %>%
{set_rownames(mltools::sparsify(.[, 2:ncol(.)]), .$V1)} %>%
.[rowSums(. > 0) >= 10,]
cell.metadata <- DataPath("AZ/cell_metadata.csv") %>% read_delim(delim='\t') %>%
rename(cell=sampleID, sample=patient) %>%
select(cell, batch, sample, sex, cellType, subclustID) %>%
filter(!grepl("un", sample), !(cellType %in% c('doublet', 'unID')))
sample.metadata <- group_by(cell.metadata, batch, sample, sex) %>%
summarise(n=n()) %>% select(-n) %>% lapply(setNames, .$sample)
cell.metadata %<>% lapply(setNames, .$cell)
mat.per.samp <- splitMatrixByFactor(mat, cell.metadata$sample)
dub.info <- estimateDoubletInfo(mat.per.samp)
p2s <- plapply(mat.per.samp, createPagoda, min.transcripts.per.cell=500, dub.scores=dub.info$scores,
dub.threshold=0.3, mc.preschedule=TRUE, n.cores=N_CORES, progress=FALSE)
createConos(p2s, sample.meta=sample.metadata, cell.meta=cell.metadata, n.cores=N_CORES)
}, force=FORCE) %>% Conos$new()
con$plotGraph(groups=con$misc$cell_metadata$cellType, size=0.2, alpha=0.2)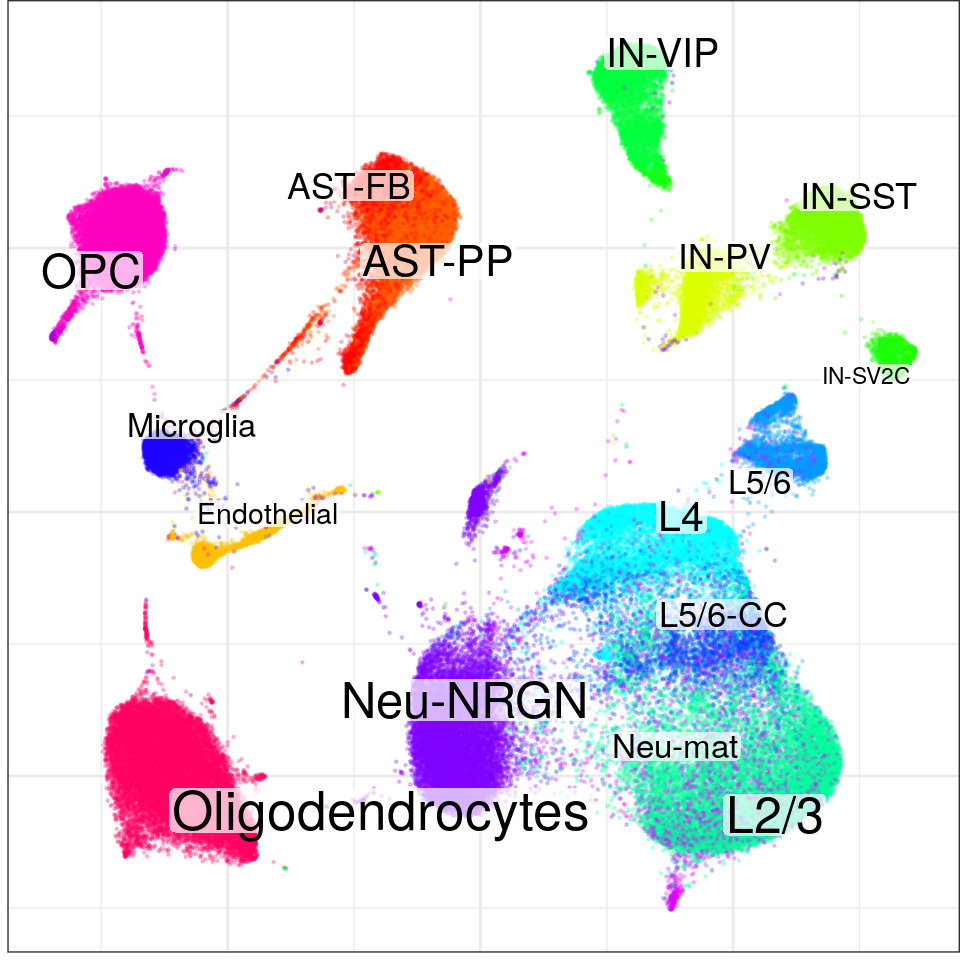
rm(con); gc(); used (Mb) gc trigger (Mb) max used (Mb)
Ncells 4392598 234.6 8418368 449.6 8418368 449.6
Vcells 8814994 67.3 744389120 5679.3 906841008 6918.7Autism
Cells are already filtered by mitochondrial fraction of 0.05 and UMI threshold ~500, no additional filtration is needed.
con <- readOrCreate(DataPath('ASD/con.rds'), function() {
mat <- Seurat::Read10X(DataPath("ASD")) %>% .[rowSums(. > 0) >= 10,]
meta <- read_delim(DataPath("ASD/meta.txt"), delim='\t') %>%
rename(cellType=cluster, PMI=`post-mortem interval (hours)`) %>%
mutate(cellType=gsub("-I(I)?", "", cellType))
sample.metadata <- meta %>%
group_by(sample, individual, region, age, sex, diagnosis, Capbatch, Seqbatch) %>%
summarise(PMI=median(PMI)) %>% lapply(setNames, .$sample)
sample.metadata$region_hr <- sample.metadata$sample %>% strsplit('_') %>% sapply(`[[`, 2)
cell.metadata <- meta %>% lapply(setNames, .$cell)
mat.per.samp <- splitMatrixByFactor(mat, cell.metadata$sample)
mat.per.cap <- splitMatrixByFactor(mat, cell.metadata$Capbatch)
dub.info <- estimateDoubletInfo(mat.per.cap)
p2s <- plapply(mat.per.samp, createPagoda, dub.scores=dub.info$scores, dub.threshold=0.17,
mc.preschedule=TRUE, n.cores=N_CORES, progress=FALSE)
createConos(p2s, sample.meta=sample.metadata, cell.meta=cell.metadata, n.cores=N_CORES)
}, force=FORCE) %>% Conos$new()
con$plotGraph(groups=con$misc$cell_metadata$cellType, size=0.1, alpha=0.1)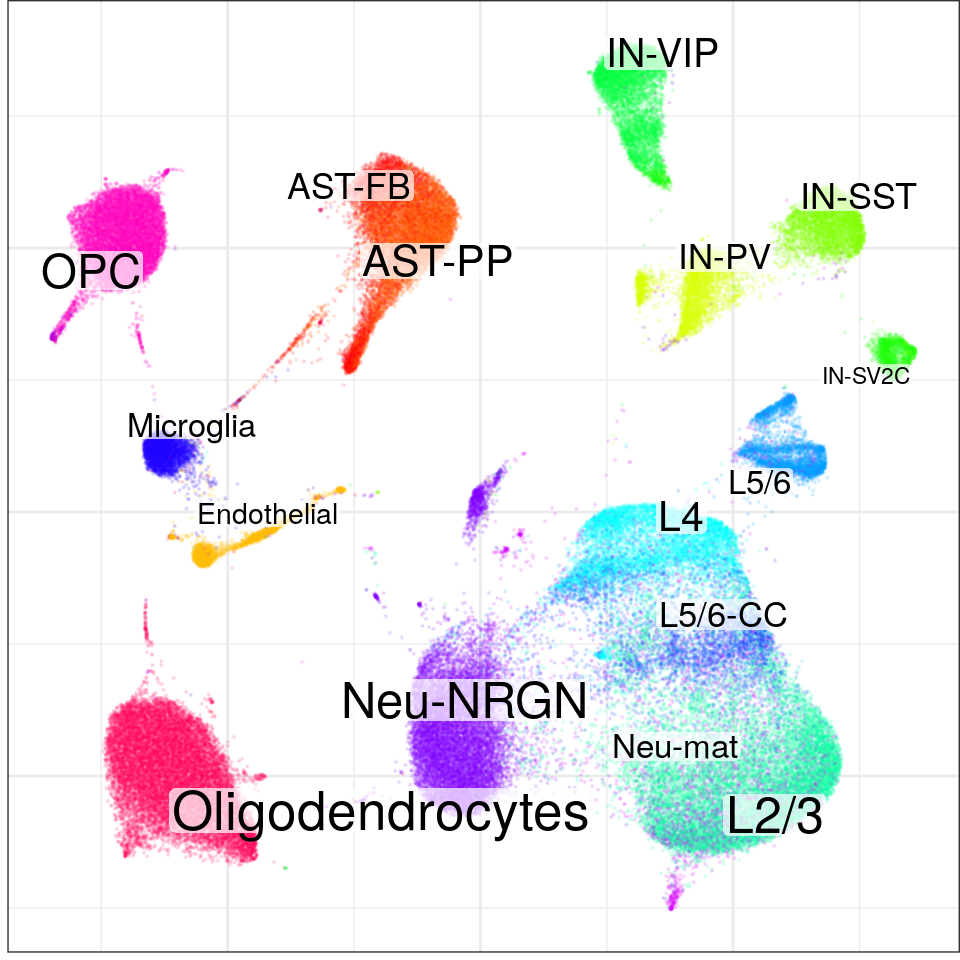
rm(con); gc(); used (Mb) gc trigger (Mb) max used (Mb)
Ncells 4392731 234.6 8418368 449.6 8418368 449.6
Vcells 8815553 67.3 716309282 5465.1 906841008 6918.7Epilepsy
con <- readOrCreate(DataPath('EP/con.rds'), function() {
con.pap <- read_rds(DataPath("EP/con_filt_samples.rds")) %>% conos::Conos$new()
cell.metadata <- DataPath("EP/annotation.csv") %>% read_csv() %>%
rename(cellType=l4) %>% lapply(setNames, .$cell)
sample.metadata <- DataPath("EP/sample_info.csv") %>% read_csv() %>% lapply(setNames, .$Alias)
con.pap$samples %>% lapply(pagoda2::Pagoda2$new) %>%
createConos(sample.meta=sample.metadata, cell.meta=cell.metadata, k=40, n.cores=N_CORES)
}, force=FORCE) %>% Conos$new()
con$plotGraph(groups=con$misc$cell_metadata$cellType, size=0.1, alpha=0.1, font.size=c(2, 3))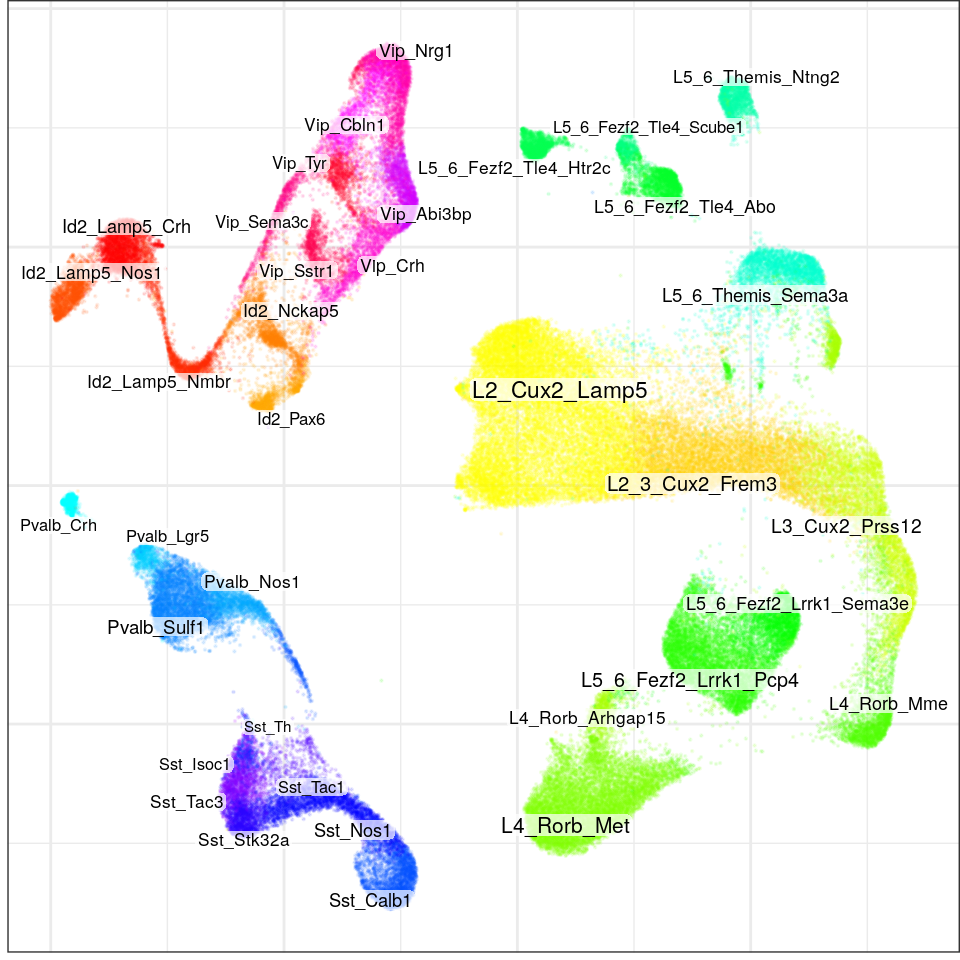
rm(con); gc(); used (Mb) gc trigger (Mb) max used (Mb)
Ncells 4393292 234.7 8418368 449.6 8418368 449.6
Vcells 8920047 68.1 696311263 5312.5 906841008 6918.7MS
The threshold on transcripts here is set only because there was 1 almost empty cell reported, all other cells already had enough transcripts.
con <- readOrCreate(DataPath('MS/con.rds'), function() {
mat <- Seurat::Read10X(DataPath("MS")) %>% .[rowSums(. > 0) >= 10,]
meta <- read_delim(DataPath("MS/meta.txt"), delim='\t') %>%
mutate(cell_type=gsub("-(Cntl|MS(-1|-2)?)", "", x=cell_type)) %>%
rename(cellType=cell_type)
sample.metadata <- meta[,5:14] %>% split(.$sample) %>% lapply(`[`, 1,) %>%
do.call(rbind, .) %>% lapply(setNames, .$sample)
cell.metadata <- meta %>% lapply(setNames, .$cell)
mat.per.samp <- splitMatrixByFactor(mat, cell.metadata$sample)
mat.per.cap <- splitMatrixByFactor(mat, cell.metadata$Capbatch)
dub.info <- estimateDoubletInfo(mat.per.cap)
p2s <- plapply(mat.per.samp, createPagoda, min.transcripts.per.cell=800, dub.scores=dub.info$scores,
dub.threshold=0.2, mc.preschedule=TRUE, n.cores=N_CORES, progress=FALSE)
createConos(p2s, sample.meta=sample.metadata, cell.meta=cell.metadata, n.cores=N_CORES)
}, force=FORCE) %>% Conos$new()
con$plotGraph(groups=con$misc$cell_metadata$cellType, size=0.1, alpha=0.1)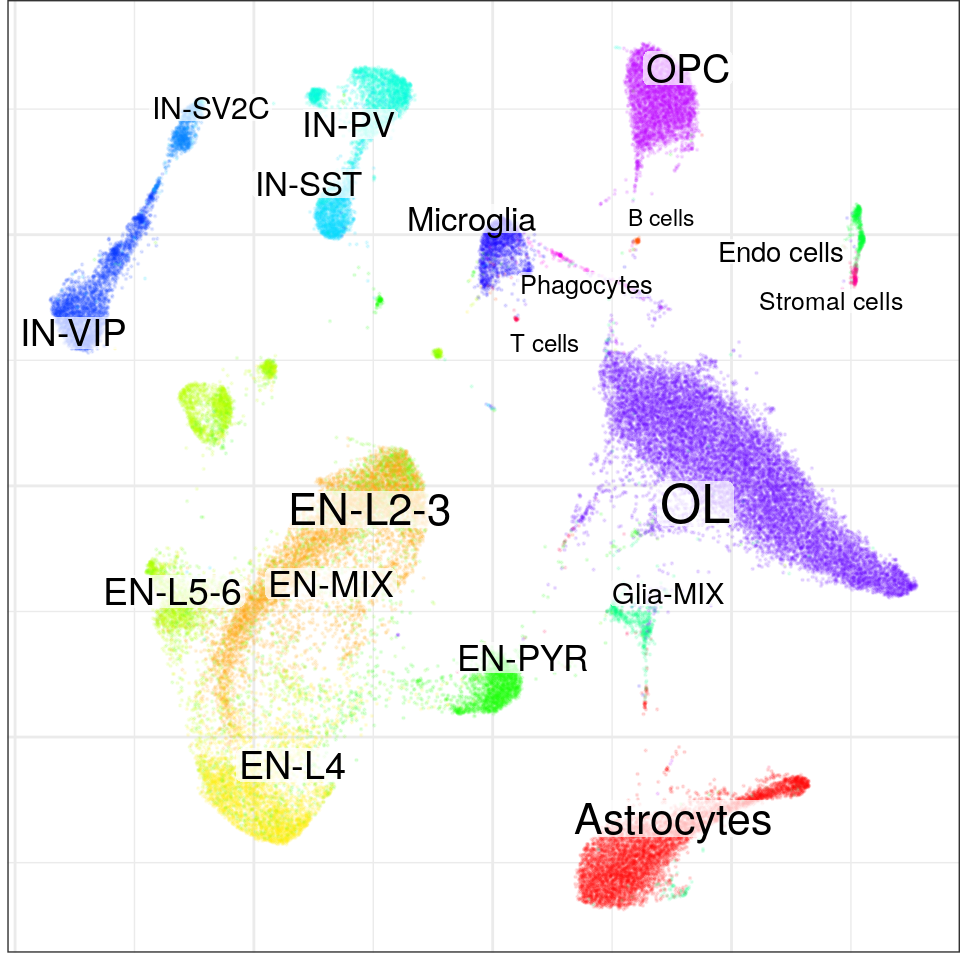
rm(con); gc(); used (Mb) gc trigger (Mb) max used (Mb)
Ncells 4341247 231.9 8418368 449.6 8418368 449.6
Vcells 8113629 62.0 557049011 4250.0 906841008 6918.7PF
The paper already performed the filtration
con <- readOrCreate(DataPath('PF/con.rds'), function() {
cell.metadata <- read_csv(DataPath("PF/cell_metadata.csv")) %>%
dplyr::rename(cell=X1) %>% dplyr::filter(Diagnosis %in% c("Control", "IPF")) %>%
dplyr::rename(sample=Sample_Name, cellType=celltype)
sample.metadata <- cell.metadata %>%
group_by(sample, Sample_Source, Diagnosis, Status, orig.ident) %>%
summarise(n=n()) %>% dplyr::select(-n) %>%
lapply(setNames, .$sample)
mat <- DataPath("PF") %>% Seurat::Read10X(gene.column=1) %>%
.[,cell.metadata$cell] %>% .[rowSums(. > 0) >= 10,]
cell.metadata %<>% lapply(setNames, .$cell)
mat.per.samp <- splitMatrixByFactor(mat, cell.metadata$sample)
p2s <- plapply(mat.per.samp, createPagoda, mc.preschedule=TRUE, n.cores=N_CORES, progress=FALSE)
createConos(p2s, sample.meta=sample.metadata, cell.meta=cell.metadata, n.cores=N_CORES)
}, force=FORCE) %>% Conos$new()
con$plotGraph(groups=con$misc$cell_metadata$cellType, size=0.1, alpha=0.1)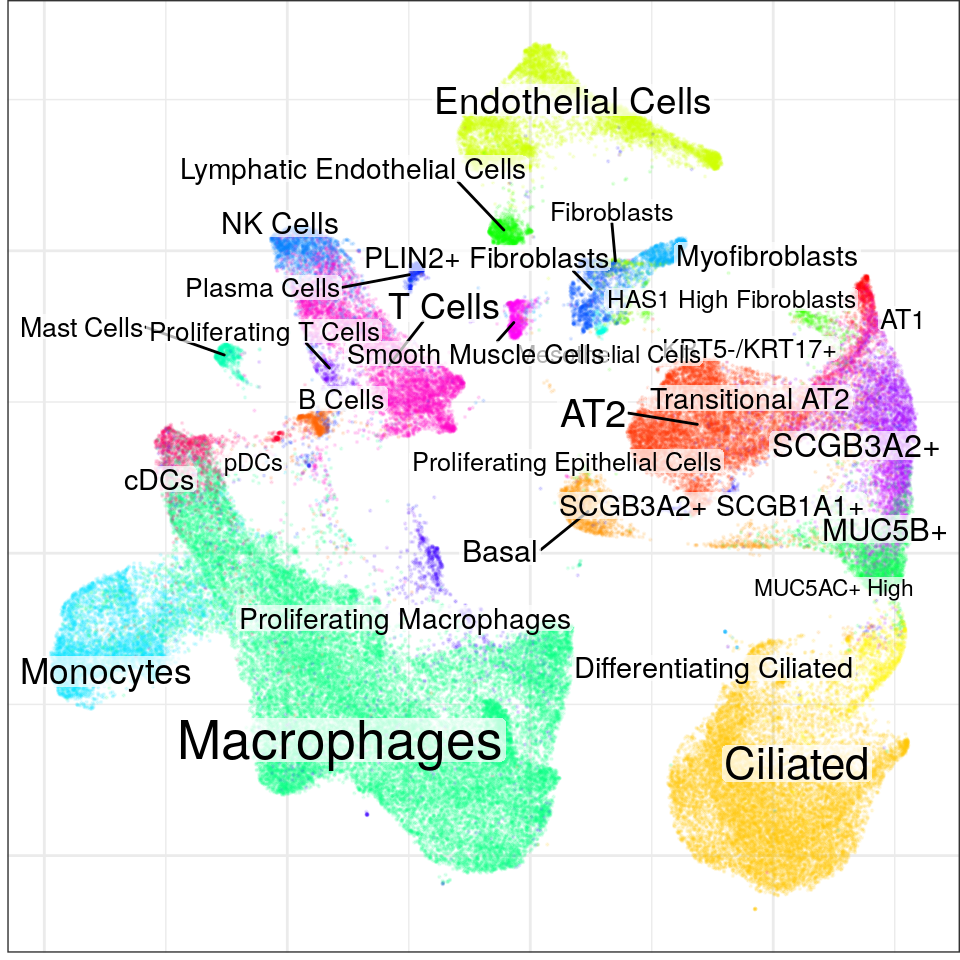
rm(con); gc(); used (Mb) gc trigger (Mb) max used (Mb)
Ncells 4387056 234.3 8418368 449.6 8418368 449.6
Vcells 8786562 67.1 648038600 4944.2 906841008 6918.7SCC
con <- readOrCreate(DataPath('SCC/con.rds'), function() {
mat <- data.table::fread(DataPath("SCC/counts.txt"), sep="\t") %>%
{set_rownames(mltools::sparsify(.[3:nrow(.), 2:ncol(.)]), .$V1[3:nrow(.)])}
cell.metadata <- read_delim(DataPath('SCC/cell_metadata.txt'), delim='\t') %>%
filter(!(level3_celltype %in% c('Multiplet', 'Keratinocyte'))) %>%
rename(cell=nCount_RNA, cellType=level3_celltype)
cell.metadata$sample <- cell.metadata$cell %>% strsplit("_") %>%
sapply(function(x) paste(x[1:2], collapse='_'))
cell.metadata$cellType %<>% gsub("(Normal|Tumor)_", "", .)
cell.metadata %<>% lapply(setNames, .$cell)
mat <- mat[rowSums(mat > 0) >= 10, cell.metadata$cell]
mat.per.samp <- splitMatrixByFactor(mat, cell.metadata$sample)
mat.per.samp %<>% .[sapply(., ncol) > 500]
p2s <- plapply(mat.per.samp, createPagoda, min.transcripts.per.cell=800,
mc.preschedule=TRUE, n.cores=N_CORES, progress=FALSE)
createConos(p2s, sample.meta=NULL, cell.meta=cell.metadata, n.cores=N_CORES)
}, force=FORCE) %>% Conos$new()
con$plotGraph(groups=con$misc$cell_metadata$cellType, size=0.1, alpha=0.1)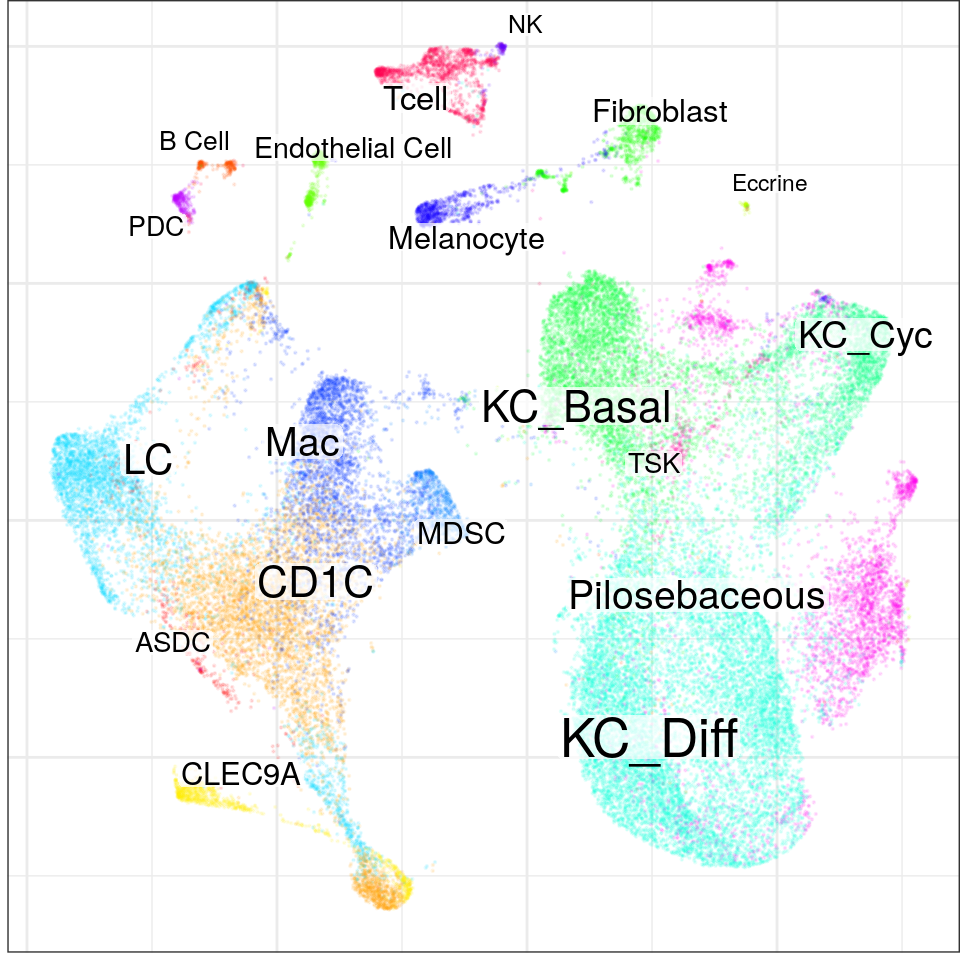
rm(con); gc(); used (Mb) gc trigger (Mb) max used (Mb)
Ncells 4336815 231.7 8418368 449.6 8418368 449.6
Vcells 8007877 61.1 518430880 3955.4 906841008 6918.7
sessionInfo()R version 4.0.3 (2020-10-10)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 18.04.6 LTS
Matrix products: default
BLAS: /usr/local/R/R-4.0.3/lib/R/lib/libRblas.so
LAPACK: /usr/local/R/R-4.0.3/lib/R/lib/libRlapack.so
locale:
[1] C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] cacoaAnalysis_0.1.0 reticulate_1.22 dataorganizer_0.1.0
[4] conos_1.4.4 pagoda2_1.0.7 igraph_1.2.6
[7] Matrix_1.2-18 sccore_1.0.0 magrittr_2.0.1
[10] forcats_0.5.1 stringr_1.4.0 dplyr_1.0.7
[13] purrr_0.3.4 readr_1.4.0 tidyr_1.1.4
[16] tibble_3.1.5 ggplot2_3.3.5 tidyverse_1.3.0
[19] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] readxl_1.3.1 N2R_0.1.1 backports_1.2.1
[4] circlize_0.4.13 splines_4.0.3 usethis_1.6.3
[7] urltools_1.7.3 digest_0.6.28 foreach_1.5.1
[10] htmltools_0.5.2 fansi_0.5.0 RMTstat_0.3
[13] memoise_2.0.0 cluster_2.1.0 doParallel_1.0.16
[16] remotes_2.2.0 ComplexHeatmap_2.9.4 modelr_0.1.8
[19] matrixStats_0.61.0 R.utils_2.10.1 prettyunits_1.1.1
[22] colorspace_2.0-2 rappdirs_0.3.3 rvest_0.3.6
[25] ggrepel_0.9.1 haven_2.4.1 xfun_0.26
[28] callr_3.5.1 crayon_1.4.1 jsonlite_1.7.2
[31] brew_1.0-6 iterators_1.0.13 glue_1.4.2
[34] gtable_0.3.0 GetoptLong_1.0.5 leidenAlg_0.1.0
[37] pkgbuild_1.1.0 Rook_1.1-1 shape_1.4.6
[40] BiocGenerics_0.36.1 scales_1.1.1 DBI_1.1.1
[43] Rcpp_1.0.7 clue_0.3-59 stats4_4.0.3
[46] httr_1.4.2 RColorBrewer_1.1-2 ellipsis_0.3.2
[49] farver_2.1.0 pkgconfig_2.0.3 R.methodsS3_1.8.1
[52] dbplyr_2.0.0 here_1.0.1 utf8_1.2.2
[55] labeling_0.4.2 tidyselect_1.1.1 rlang_0.4.11
[58] later_1.1.0.1 munsell_0.5.0 cellranger_1.1.0
[61] tools_4.0.3 cachem_1.0.6 cli_3.0.1
[64] generics_0.1.0 devtools_2.3.2 broom_0.7.9
[67] evaluate_0.14 fastmap_1.1.0 yaml_2.2.1
[70] processx_3.4.5 knitr_1.36 fs_1.5.0
[73] nlme_3.1-149 whisker_0.4 ggrastr_1.0.0
[76] R.oo_1.24.0 grr_0.9.5 xml2_1.3.2
[79] compiler_4.0.3 rstudioapi_0.13 beeswarm_0.4.0
[82] png_0.1-7 testthat_3.0.0 reprex_0.3.0
[85] stringi_1.7.5 highr_0.9 ps_1.4.0
[88] drat_0.1.8 desc_1.3.0 lattice_0.20-41
[91] vctrs_0.3.8 pillar_1.6.3 lifecycle_1.0.1
[94] triebeard_0.3.0 jquerylib_0.1.4 GlobalOptions_0.1.2
[97] irlba_2.3.3 Matrix.utils_0.9.8 httpuv_1.5.4
[100] R6_2.5.1 promises_1.1.1 gridExtra_2.3
[103] vipor_0.4.5 IRanges_2.24.1 sessioninfo_1.1.1
[106] codetools_0.2-16 MASS_7.3-53 assertthat_0.2.1
[109] pkgload_1.2.1 rprojroot_2.0.2 rjson_0.2.20
[112] withr_2.4.2 S4Vectors_0.28.1 mgcv_1.8-33
[115] parallel_4.0.3 hms_1.1.1 grid_4.0.3
[118] rmarkdown_2.11 dendsort_0.3.3 Rtsne_0.15
[121] git2r_0.27.1 lubridate_1.7.9.2 ggbeeswarm_0.6.0