Clustering of TDP-HA experiment and the D96 samples
Katharina Hembach
28/01/2021
Last updated: 2021-03-11
Checks: 6 1
Knit directory: neural_scRNAseq/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it's best to always run the code in an empty environment.
The command set.seed(20200522) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
- clustering
- dimension-reduction
- dr-individual-clusters
- dr-plots
- DR-QC
- merge-so
- read-data
- session-info-chunk-inserted-by-workflowr
- unnamed-chunk-1
- unnamed-chunk-2
- unnamed-chunk-3
- unnamed-chunk-4
- variable-features-scaling
To ensure reproducibility of the results, delete the cache directory TDP-08-00-clustering-HA-D96_cache and re-run the analysis. To have workflowr automatically delete the cache directory prior to building the file, set delete_cache = TRUE when running wflow_build() or wflow_publish().
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 864cc2d. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: ._.DS_Store
Ignored: ._Rplots.pdf
Ignored: ._Unfiltered.pdf
Ignored: .__workflowr.yml
Ignored: ._coverage.pdf
Ignored: ._coverage_sashimi.pdf
Ignored: ._coverage_sashimi.png
Ignored: ._neural_scRNAseq.Rproj
Ignored: ._pbDS_cell_level.pdf
Ignored: ._pbDS_top_expr_umap.pdf
Ignored: ._pbDS_upset.pdf
Ignored: ._sashimi.pdf
Ignored: ._stmn2.pdf
Ignored: ._tdp.pdf
Ignored: analysis/.DS_Store
Ignored: analysis/.Rhistory
Ignored: analysis/._.DS_Store
Ignored: analysis/._01-preprocessing.Rmd
Ignored: analysis/._01-preprocessing.html
Ignored: analysis/._02.1-SampleQC.Rmd
Ignored: analysis/._03-filtering.Rmd
Ignored: analysis/._04-clustering.Rmd
Ignored: analysis/._04-clustering.knit.md
Ignored: analysis/._04.1-cell_cycle.Rmd
Ignored: analysis/._05-annotation.Rmd
Ignored: analysis/._Lam-0-NSC_no_integration.Rmd
Ignored: analysis/._Lam-01-NSC_integration.Rmd
Ignored: analysis/._Lam-02-NSC_annotation.Rmd
Ignored: analysis/._NSC-1-clustering.Rmd
Ignored: analysis/._NSC-2-annotation.Rmd
Ignored: analysis/.__site.yml
Ignored: analysis/._additional_filtering.Rmd
Ignored: analysis/._additional_filtering_clustering.Rmd
Ignored: analysis/._index.Rmd
Ignored: analysis/._organoid-01-1-qualtiy-control.Rmd
Ignored: analysis/._organoid-01-clustering.Rmd
Ignored: analysis/._organoid-02-integration.Rmd
Ignored: analysis/._organoid-03-cluster_analysis.Rmd
Ignored: analysis/._organoid-04-group_integration.Rmd
Ignored: analysis/._organoid-04-stage_integration.Rmd
Ignored: analysis/._organoid-05-group_integration_cluster_analysis.Rmd
Ignored: analysis/._organoid-05-stage_integration_cluster_analysis.Rmd
Ignored: analysis/._organoid-06-1-prepare-sce.Rmd
Ignored: analysis/._organoid-06-conos-analysis-Seurat.Rmd
Ignored: analysis/._organoid-06-conos-analysis-function.Rmd
Ignored: analysis/._organoid-06-conos-analysis.Rmd
Ignored: analysis/._organoid-06-group-integration-conos-analysis.Rmd
Ignored: analysis/._organoid-07-conos-visualization.Rmd
Ignored: analysis/._organoid-07-group-integration-conos-visualization.Rmd
Ignored: analysis/._organoid-08-conos-comparison.Rmd
Ignored: analysis/._organoid-0x-sample_integration.Rmd
Ignored: analysis/01-preprocessing_cache/
Ignored: analysis/02-1-SampleQC_cache/
Ignored: analysis/02-quality_control_cache/
Ignored: analysis/02.1-SampleQC_cache/
Ignored: analysis/03-filtering_cache/
Ignored: analysis/04-clustering_cache/
Ignored: analysis/04.1-cell_cycle_cache/
Ignored: analysis/05-annotation_cache/
Ignored: analysis/06-clustering-all-timepoints_cache/
Ignored: analysis/07-cluster-analysis-all-timepoints_cache/
Ignored: analysis/Lam-01-NSC_integration_cache/
Ignored: analysis/Lam-02-NSC_annotation_cache/
Ignored: analysis/NSC-1-clustering_cache/
Ignored: analysis/NSC-2-annotation_cache/
Ignored: analysis/TDP-01-preprocessing_cache/
Ignored: analysis/TDP-02-quality_control_cache/
Ignored: analysis/TDP-03-filtering_cache/
Ignored: analysis/TDP-04-clustering_cache/
Ignored: analysis/TDP-05-00-filtering-plasmid-QC_cache/
Ignored: analysis/TDP-05-plasmid_expression_cache/
Ignored: analysis/TDP-06-cluster_analysis_cache/
Ignored: analysis/TDP-07-01-STMN2_expression_cache/
Ignored: analysis/TDP-07-cluster_12_cache/
Ignored: analysis/TDP-08-00-clustering-HA-D96_cache/
Ignored: analysis/TDP-08-01-HA-D96-expression-changes_cache/
Ignored: analysis/TDP-08-clustering-timeline-HA_cache/
Ignored: analysis/additional_filtering_cache/
Ignored: analysis/additional_filtering_clustering_cache/
Ignored: analysis/figure/
Ignored: analysis/organoid-01-1-qualtiy-control_cache/
Ignored: analysis/organoid-01-clustering_cache/
Ignored: analysis/organoid-02-integration_cache/
Ignored: analysis/organoid-03-cluster_analysis_cache/
Ignored: analysis/organoid-04-group_integration_cache/
Ignored: analysis/organoid-04-stage_integration_cache/
Ignored: analysis/organoid-05-group_integration_cluster_analysis_cache/
Ignored: analysis/organoid-05-stage_integration_cluster_analysis_cache/
Ignored: analysis/organoid-06-conos-analysis_cache/
Ignored: analysis/organoid-06-conos-analysis_test_cache/
Ignored: analysis/organoid-06-group-integration-conos-analysis_cache/
Ignored: analysis/organoid-07-conos-visualization_cache/
Ignored: analysis/organoid-07-group-integration-conos-visualization_cache/
Ignored: analysis/organoid-08-conos-comparison_cache/
Ignored: analysis/organoid-0x-sample_integration_cache/
Ignored: analysis/sample5_QC_cache/
Ignored: analysis/timepoints-01-organoid-integration_cache/
Ignored: data/.DS_Store
Ignored: data/._.DS_Store
Ignored: data/._.smbdeleteAAA17ed8b4b
Ignored: data/._Lam_figure2_markers.R
Ignored: data/._Reactive_astrocytes_markers.xlsx
Ignored: data/._known_NSC_markers.R
Ignored: data/._known_cell_type_markers.R
Ignored: data/._metadata.csv
Ignored: data/._virus_cell_tropism_markers.R
Ignored: data/._~$Reactive_astrocytes_markers.xlsx
Ignored: data/data_sushi/
Ignored: data/filtered_feature_matrices/
Ignored: output/.DS_Store
Ignored: output/._.DS_Store
Ignored: output/._NSC_cluster2_marker_genes.txt
Ignored: output/._TDP-06-no_integration_cluster12_marker_genes.txt
Ignored: output/._TDP-06-no_integration_cluster13_marker_genes.txt
Ignored: output/._organoid_integration_cluster1_marker_genes.txt
Ignored: output/Lam-01-clustering.rds
Ignored: output/NSC_1_clustering.rds
Ignored: output/NSC_cluster1_marker_genes.txt
Ignored: output/NSC_cluster2_marker_genes.txt
Ignored: output/NSC_cluster3_marker_genes.txt
Ignored: output/NSC_cluster4_marker_genes.txt
Ignored: output/NSC_cluster5_marker_genes.txt
Ignored: output/NSC_cluster6_marker_genes.txt
Ignored: output/NSC_cluster7_marker_genes.txt
Ignored: output/TDP-06-no_integration_cluster0_marker_genes.txt
Ignored: output/TDP-06-no_integration_cluster10_marker_genes.txt
Ignored: output/TDP-06-no_integration_cluster11_marker_genes.txt
Ignored: output/TDP-06-no_integration_cluster12_marker_genes.txt
Ignored: output/TDP-06-no_integration_cluster13_marker_genes.txt
Ignored: output/TDP-06-no_integration_cluster14_marker_genes.txt
Ignored: output/TDP-06-no_integration_cluster15_marker_genes.txt
Ignored: output/TDP-06-no_integration_cluster16_marker_genes.txt
Ignored: output/TDP-06-no_integration_cluster17_marker_genes.txt
Ignored: output/TDP-06-no_integration_cluster1_marker_genes.txt
Ignored: output/TDP-06-no_integration_cluster2_marker_genes.txt
Ignored: output/TDP-06-no_integration_cluster3_marker_genes.txt
Ignored: output/TDP-06-no_integration_cluster4_marker_genes.txt
Ignored: output/TDP-06-no_integration_cluster5_marker_genes.txt
Ignored: output/TDP-06-no_integration_cluster6_marker_genes.txt
Ignored: output/TDP-06-no_integration_cluster7_marker_genes.txt
Ignored: output/TDP-06-no_integration_cluster8_marker_genes.txt
Ignored: output/TDP-06-no_integration_cluster9_marker_genes.txt
Ignored: output/TDP-06_scran_markers.rds
Ignored: output/additional_filtering.rds
Ignored: output/conos/
Ignored: output/conos_organoid-06-conos-analysis.rds
Ignored: output/conos_organoid-06-group-integration-conos-analysis.rds
Ignored: output/figures/
Ignored: output/organoid_integration_cluster10_marker_genes.txt
Ignored: output/organoid_integration_cluster11_marker_genes.txt
Ignored: output/organoid_integration_cluster12_marker_genes.txt
Ignored: output/organoid_integration_cluster13_marker_genes.txt
Ignored: output/organoid_integration_cluster14_marker_genes.txt
Ignored: output/organoid_integration_cluster15_marker_genes.txt
Ignored: output/organoid_integration_cluster16_marker_genes.txt
Ignored: output/organoid_integration_cluster17_marker_genes.txt
Ignored: output/organoid_integration_cluster1_marker_genes.txt
Ignored: output/organoid_integration_cluster2_marker_genes.txt
Ignored: output/organoid_integration_cluster3_marker_genes.txt
Ignored: output/organoid_integration_cluster4_marker_genes.txt
Ignored: output/organoid_integration_cluster5_marker_genes.txt
Ignored: output/organoid_integration_cluster6_marker_genes.txt
Ignored: output/organoid_integration_cluster7_marker_genes.txt
Ignored: output/organoid_integration_cluster8_marker_genes.txt
Ignored: output/organoid_integration_cluster9_marker_genes.txt
Ignored: output/sce_01_preprocessing.rds
Ignored: output/sce_02_quality_control.rds
Ignored: output/sce_03_filtering.rds
Ignored: output/sce_03_filtering_all_genes.rds
Ignored: output/sce_06-1-prepare-sce.rds
Ignored: output/sce_TDP_01_preprocessing.rds
Ignored: output/sce_TDP_02_quality_control.rds
Ignored: output/sce_TDP_03_filtering.rds
Ignored: output/sce_TDP_03_filtering_all_genes.rds
Ignored: output/sce_organoid-01-clustering.rds
Ignored: output/sce_preprocessing.rds
Ignored: output/so_04-group_integration.rds
Ignored: output/so_04-stage_integration.rds
Ignored: output/so_04_1_cell_cycle.rds
Ignored: output/so_04_clustering.rds
Ignored: output/so_06-clustering_all_timepoints.rds
Ignored: output/so_08-00_clustering_HA_D96.rds
Ignored: output/so_08-clustering_timeline_HA.rds
Ignored: output/so_0x-sample_integration.rds
Ignored: output/so_TDP-06-cluster-analysis.rds
Ignored: output/so_TDP_04_clustering.rds
Ignored: output/so_TDP_05_plasmid_expression.rds
Ignored: output/so_additional_filtering_clustering.rds
Ignored: output/so_integrated_organoid-02-integration.rds
Ignored: output/so_merged_organoid-02-integration.rds
Ignored: output/so_organoid-01-clustering.rds
Ignored: output/so_sample_organoid-01-clustering.rds
Ignored: scripts/.DS_Store
Ignored: scripts/._.DS_Store
Ignored: scripts/._bu_Rcode.R
Ignored: scripts/._plasmid_expression.sh
Untracked files:
Untracked: Filtered.pdf
Untracked: Rplots.pdf
Untracked: Unfiltered
Untracked: Unfiltered.pdf
Untracked: analysis/Lam-0-NSC_no_integration.Rmd
Untracked: analysis/TDP-07-01-STMN2_expression copy.Rmd
Untracked: analysis/additional_filtering.Rmd
Untracked: analysis/additional_filtering_clustering.Rmd
Untracked: analysis/organoid-01-1-qualtiy-control.Rmd
Untracked: analysis/organoid-06-conos-analysis-Seurat.Rmd
Untracked: analysis/organoid-06-conos-analysis-function.Rmd
Untracked: analysis/organoid-07-conos-visualization.Rmd
Untracked: analysis/organoid-07-group-integration-conos-visualization.Rmd
Untracked: analysis/organoid-08-conos-comparison.Rmd
Untracked: analysis/organoid-0x-sample_integration.Rmd
Untracked: analysis/sample5_QC.Rmd
Untracked: coverage.pdf
Untracked: coverage_sashimi.pdf
Untracked: coverage_sashimi.png
Untracked: data/Homo_sapiens.GRCh38.98.sorted.gtf
Untracked: data/Kanton_et_al/
Untracked: data/Lam_et_al/
Untracked: data/Sep2020/
Untracked: data/reference/
Untracked: data/virus_cell_tropism_markers.R
Untracked: data/~$Reactive_astrocytes_markers.xlsx
Untracked: pbDS_cell_level.pdf
Untracked: pbDS_heatmap.pdf
Untracked: pbDS_top_expr_umap.pdf
Untracked: pbDS_upset.pdf
Untracked: sashimi.pdf
Untracked: scripts/bu_Rcode.R
Untracked: scripts/salmon-latest_linux_x86_64/
Untracked: stmn2.pdf
Untracked: tdp.pdf
Unstaged changes:
Modified: analysis/05-annotation.Rmd
Modified: analysis/Lam-02-NSC_annotation.Rmd
Modified: analysis/TDP-04-clustering.Rmd
Modified: analysis/TDP-06-cluster_analysis.Rmd
Modified: analysis/TDP-08-01-HA-D96-expression-changes.Rmd
Modified: analysis/_site.yml
Modified: analysis/organoid-02-integration.Rmd
Modified: analysis/organoid-04-group_integration.Rmd
Modified: analysis/organoid-06-conos-analysis.Rmd
Modified: analysis/timepoints-01-organoid-integration.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/TDP-08-00-clustering-HA-D96.Rmd) and HTML (docs/TDP-08-00-clustering-HA-D96.html) files. If you've configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 864cc2d | khembach | 2021-03-11 | DR with cells colored by individual clustering |
| html | d20fa77 | khembach | 2021-01-28 | Build site. |
| Rmd | e6766a6 | khembach | 2021-01-28 | cluster TDP experiment together with D96 samples |
Load packages
library(BiocParallel)
library(ggplot2)
library(dplyr)
library(cowplot)
library(ggplot2)
library(Seurat)
library(SingleCellExperiment)
library(future)
library(ComplexHeatmap)
library(RColorBrewer)
library(viridis)Load data
## Seurat objects with normalized data
so_tdp <- readRDS(file.path("output", "so_TDP_05_plasmid_expression.rds"))
so_tdp$group_id <- "TDP"
# so_timeline <- readRDS(file.path("output", "so_06-clustering_all_timepoints.rds"))
so_d96 <- readRDS(file.path("output", "so_04_clustering.rds"))
## select only the D96 cells
so_d96 <- subset(so_d96, subset = group_id == "D96")We merge the samples from the two data sets into a Seurat object.
## merge the two Seurat objects
so <- merge(so_tdp, y = so_d96, add.cell.ids = c("tdp_ha", "D96"),
project = "neural_cultures", merge.data = TRUE)
so$group_id <- factor(so$group_id, levels = c("D96", "TDP"))Variable features
so <- FindVariableFeatures(so, nfeatures = 2000,
selection.method = "vst", verbose = FALSE)
so <- ScaleData(so, verbose = FALSE, vars.to.regress = c("sum",
"subsets_Mt_percent"))Dimension reduction
We perform dimension reduction with t-SNE and UMAP based on PCA results.
so <- RunPCA(so, npcs = 30, verbose = FALSE)
so <- RunTSNE(so, reduction = "pca", dims = seq_len(20),
seed.use = 1, do.fast = TRUE, verbose = FALSE)
so <- RunUMAP(so, reduction = "pca", dims = seq_len(20),
seed.use = 1, verbose = FALSE)Plot PCA results
# top genes that are associated with the first two PCs
VizDimLoadings(so, dims = 1:2, reduction = "pca")
## PCA plot
DimPlot(so, reduction = "pca", group.by = "sample_id")
# elbow plot with the ranking of PCs based on the % of variance explained
ElbowPlot(so, ndims = 30)
Clustering
We cluster the cells using the reduced PCA dimensions.
so <- FindNeighbors(so, reduction = "pca", dims = seq_len(20), verbose = FALSE)
for (res in c(0.2, 0.4, 0.8, 1))
so <- FindClusters(so, resolution = res, random.seed = 1, verbose = FALSE)Dimension reduction plots
We plot the dimension reduction (DR) and color by sample, group and cluster ID
thm <- theme(aspect.ratio = 1, legend.position = "none")
ps <- lapply(c("sample_id", "group_id", "ident"), function(u) {
p1 <- DimPlot(so, reduction = "tsne", group.by = u) + thm
p2 <- DimPlot(so, reduction = "umap", group.by = u)
lgd <- get_legend(p2)
p2 <- p2 + thm
list(p1, p2, lgd)
plot_grid(p1, p2, lgd, nrow = 1,
rel_widths = c(1, 1, 0.5))
})
plot_grid(plotlist = ps, ncol = 1)
QC on DR plots
cs <- sample(colnames(so), 1e4) ## subsample cells
.plot_features <- function(so, dr, id) {
FeaturePlot(so, cells = cs, features = id, reduction = dr, pt.size = 0.4,
cols = c("grey", "blue")) +
guides(col = guide_colourbar()) +
theme_void() + theme(aspect.ratio = 1)
}
ids <- c("sum", "detected", "subsets_Mt_percent", "ENSG00000120948.TARDBP",
"ENSG00000120948.TARDBP-alevin", "TDP43-HA")
for (id in ids) {
cat("### ", id, "\n")
p1 <- .plot_features(so, "tsne", id)
lgd <- get_legend(p1)
p1 <- p1 + theme(legend.position = "none") + ggtitle("tSNE")
p2 <- .plot_features(so, "umap", id) + theme(legend.position = "none") +
ggtitle("UMAP")
ps <- plot_grid(plotlist = list(p1, p2), nrow = 1)
p <- plot_grid(ps, lgd, nrow = 1, rel_widths = c(1, 0.2))
print(p)
cat("\n\n")
}sum

detected

subsets_Mt_percent

ENSG00000120948.TARDBP

ENSG00000120948.TARDBP-alevin

TDP43-HA

DR with cells colored by individial clustering
We show the cluster membership of the individual clustering in the combined UMAP.
## combined clustering so$RNA_snn_res.0.4
## individual clustering so$integrated_snn_res.0.4 (D96)
## so_tdp$RNA_snn_res.0.4 (TDP-HA)
# we lost the individual clustering of the TDP-HA samples,
# and will add them to the so object
so$TDPHA_snn_res.0.4 <- NA
## iterate throuth each sample and add the corresponding cluster ids
for( s in unique(so_tdp$sample_id)){
ind <- so$sample_id == s
ind_tdp <- so_tdp$sample_id == s
so$TDPHA_snn_res.0.4[ind] <- as.character(so_tdp$RNA_snn_res.0.4)[match(so$barcode[ind],
so_tdp$barcode[ind_tdp])]
}
so$TDPHA_snn_res.0.4 <- factor(so$TDPHA_snn_res.0.4,
levels = as.character(0:16))
so$integrated_snn_res.0.4 <- factor(so$integrated_snn_res.0.4,
levels = as.character(0:16))
cs <- sample(colnames(so), 1e4)
.plot_dr <- function(so, dr, id)
DimPlot(so, cells = cs, group.by = id, reduction = dr, pt.size = 0.4) +
guides(col = guide_legend(nrow = 11,
override.aes = list(size = 3, alpha = 1))) +
theme_void() + theme(aspect.ratio = 1)
ids <- c("group_id", "sample_id", "ident", "TDPHA_snn_res.0.4", "integrated_snn_res.0.4")
for (id in ids) {
cat("## ", id, "\n")
p1 <- .plot_dr(so, "tsne", id)
lgd <- get_legend(p1)
p1 <- p1 + theme(legend.position = "none")
p2 <- .plot_dr(so, "umap", id) + theme(legend.position = "none")
ps <- plot_grid(plotlist = list(p1, p2), nrow = 1)
p <- plot_grid(ps, lgd, nrow = 1, rel_widths = c(1, 0.2))
print(p)
cat("\n\n")
}group_id

sample_id

ident

TDPHA_snn_res.0.4

integrated_snn_res.0.4

How are the individal clusters distributed in the combined clustering?
so$RNA_snn_res.0.4 %>% table.
0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15
6178 6176 4782 3671 3470 3203 2232 2229 1971 1295 1248 1142 675 538 142 51 so$integrated_snn_res.0.4 %>% table.
0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15
268 1023 29 703 1270 639 670 639 521 464 215 384 501 303 328 123
16
53 so$TDPHA_snn_res.0.4 %>% table.
0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15
4974 4858 3809 3014 2586 2835 2370 1570 1859 1046 858 269 374 200 154 56
16
38 ## D96
## check if cells from the same cluster are still in the same cluster
(n_clusters <- table(so$RNA_snn_res.0.4, so$integrated_snn_res.0.4))
0 1 2 3 4 5 6 7 8 9 10 11 12 13 14
0 186 7 0 46 1270 2 4 2 22 9 107 16 5 1 1
1 3 960 0 0 0 6 27 3 0 24 0 27 3 21 0
2 0 8 0 0 0 0 489 296 0 0 0 38 0 0 0
3 7 33 0 0 0 6 18 5 0 49 1 7 485 19 14
4 0 5 0 0 0 194 0 9 0 239 1 47 0 0 1
5 0 1 0 0 0 392 119 10 0 0 0 3 0 0 1
6 2 0 0 0 0 16 3 312 0 1 0 32 0 0 0
7 6 0 1 1 0 0 0 0 490 0 16 0 0 0 0
8 2 9 0 0 0 0 0 0 0 4 0 1 5 260 0
9 5 0 0 0 0 5 0 0 0 126 0 2 2 0 7
10 55 0 20 656 0 1 1 0 9 7 65 8 0 1 1
11 1 0 0 0 0 4 0 0 0 0 0 6 0 0 143
12 1 0 0 0 0 0 0 0 0 0 0 1 0 0 159
13 0 0 0 0 0 13 9 2 0 4 25 196 1 1 1
14 0 0 8 0 0 0 0 0 0 0 0 0 0 0 0
15 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0
15 16
0 120 0
1 0 0
2 0 0
3 0 0
4 0 0
5 0 0
6 0 0
7 0 0
8 0 0
9 0 0
10 3 53
11 0 0
12 0 0
13 0 0
14 0 0
15 0 0fqs <- prop.table(n_clusters, margin = 2)
mat <- as.matrix(unclass(fqs))
Heatmap(mat,
col = rev(brewer.pal(11, "RdGy")[-6]),
name = "Frequency",
cluster_rows = FALSE,
cluster_columns = FALSE,
row_names_side = "left",
row_title = "combined clusters",
column_title = "individual clusters",
column_title_side = "bottom",
rect_gp = gpar(col = "white"),
cell_fun = function(i, j, x, y, width, height, fill)
grid.text(round(mat[j, i] * 100, 2), x = x, y = y,
gp = gpar(col = "white", fontsize = 8)))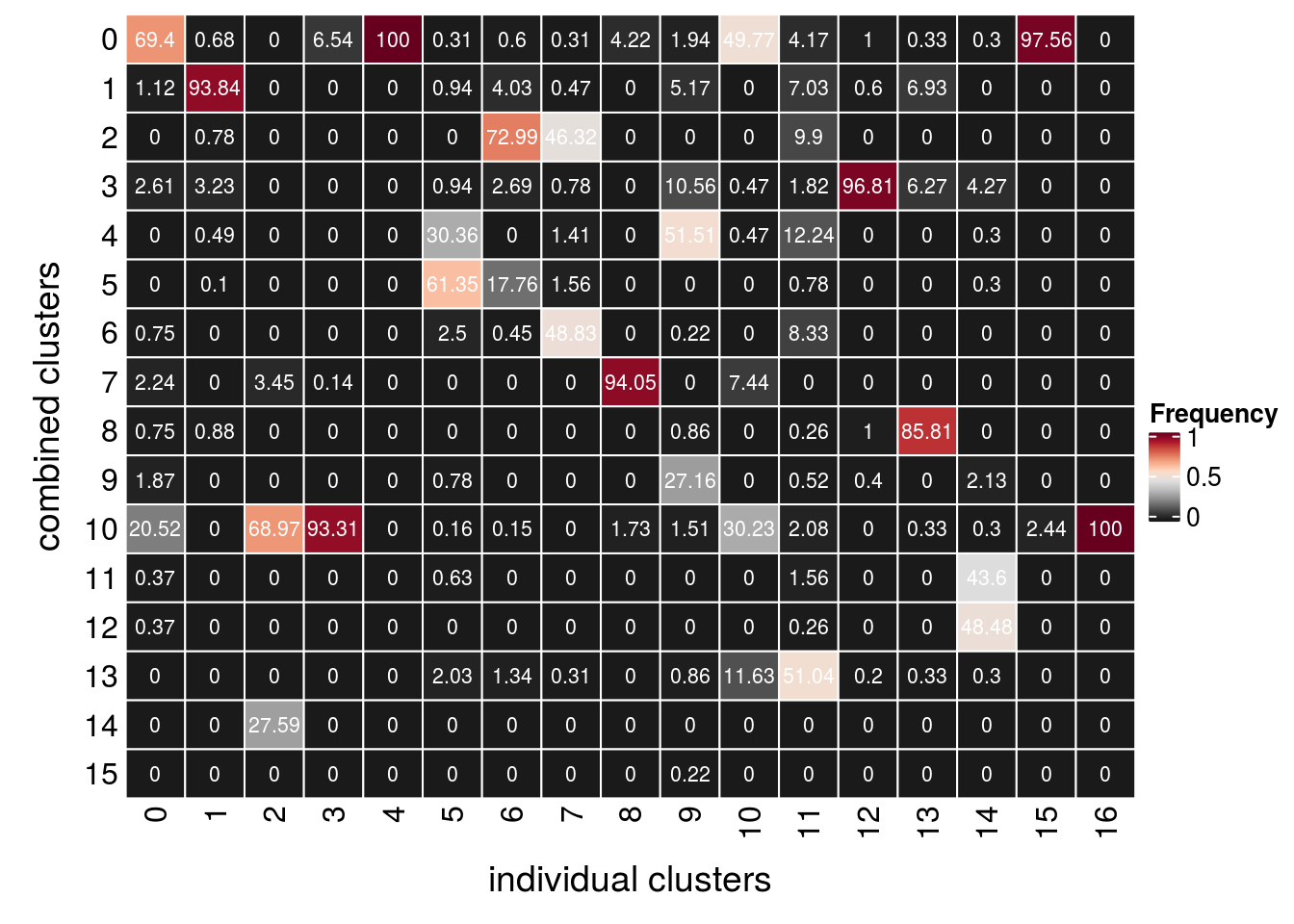
## TDP-HA experiment
(n_clusters <- table(so$RNA_snn_res.0.4, so$TDPHA_snn_res.0.4))
0 1 2 3 4 5 6 7 8 9 10 11 12 13 14
0 531 1572 406 288 288 311 278 162 196 107 99 32 38 20 39
1 1783 587 508 396 340 349 313 204 239 133 101 32 57 29 19
2 510 469 1293 288 270 250 213 150 207 104 76 32 39 28 15
3 401 404 284 232 792 237 178 119 159 77 63 17 30 11 14
4 374 384 287 913 154 206 174 115 133 80 80 18 22 16 11
5 326 305 248 207 156 443 144 119 541 68 52 16 27 7 11
6 215 214 184 150 134 535 125 78 90 53 43 13 18 4 8
7 161 193 121 117 102 111 659 77 69 32 30 8 14 8 8
8 223 224 150 132 114 118 97 433 88 37 33 12 12 10 3
9 140 147 111 97 79 91 67 35 47 288 22 10 4 6 3
10 50 148 30 21 15 32 18 9 13 9 3 2 3 2 0
11 134 112 94 94 77 70 48 37 39 23 230 7 9 9 3
12 80 52 53 50 39 44 36 19 25 19 18 68 2 2 6
13 28 31 22 19 13 21 10 8 10 7 6 1 95 0 13
14 15 9 11 7 7 14 8 3 2 3 2 1 1 48 1
15 3 7 7 3 6 3 2 2 1 6 0 0 3 0 0
15 16
0 9 4
1 8 4
2 2 5
3 5 4
4 4 3
5 2 5
6 1 1
7 2 3
8 3 1
9 1 0
10 13 0
11 1 1
12 1 0
13 2 0
14 2 0
15 0 7fqs <- prop.table(n_clusters, margin = 2)
mat <- as.matrix(unclass(fqs))
Heatmap(mat,
col = rev(brewer.pal(11, "RdGy")[-6]),
name = "Frequency",
cluster_rows = FALSE,
cluster_columns = FALSE,
row_names_side = "left",
row_title = "combined clusters",
column_title = "individual clusters",
column_title_side = "bottom",
rect_gp = gpar(col = "white"),
cell_fun = function(i, j, x, y, width, height, fill)
grid.text(round(mat[j, i] * 100, 2), x = x, y = y,
gp = gpar(col = "white", fontsize = 8)))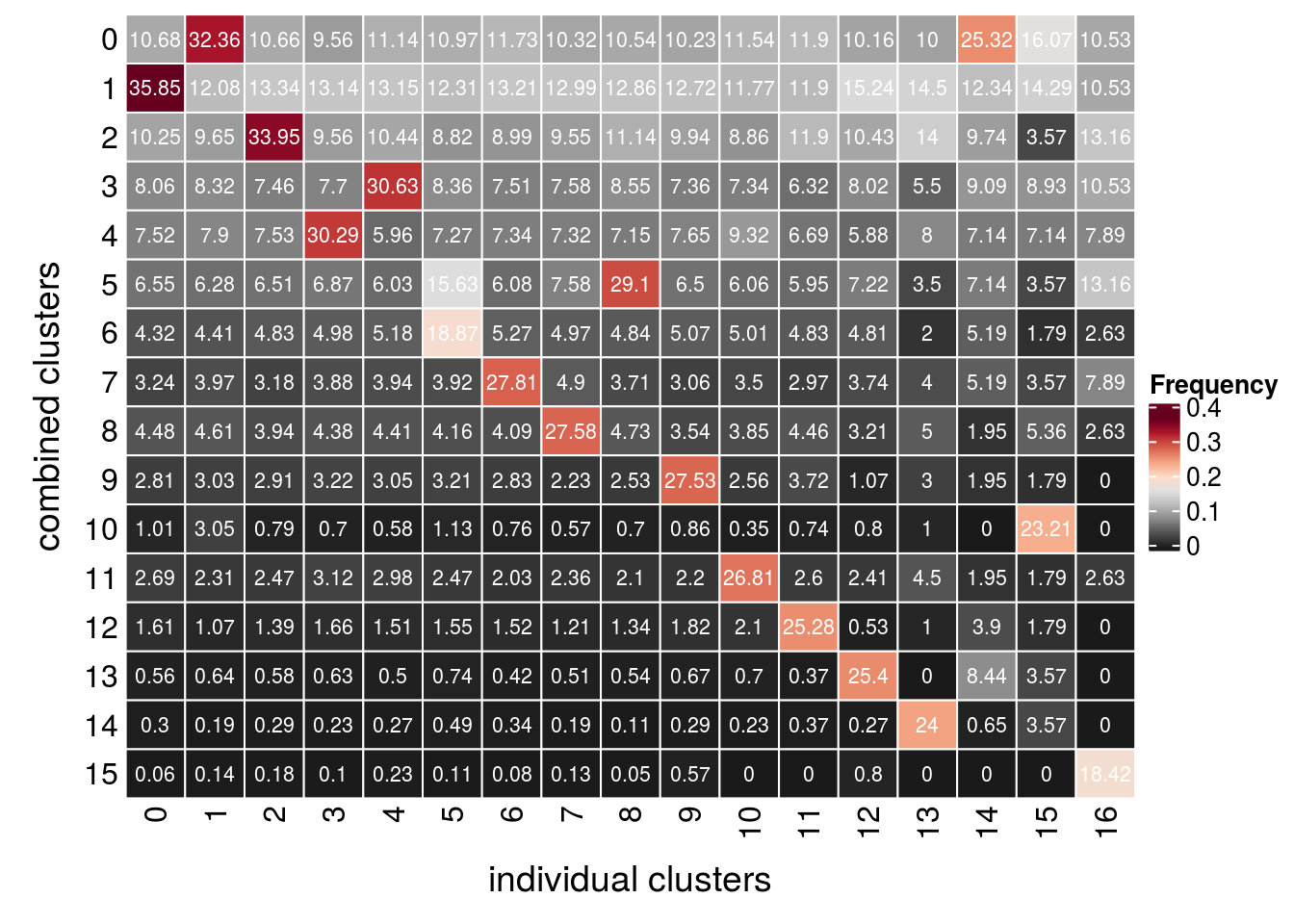
Save Seurat object to RDS
saveRDS(so, file.path("output", "so_08-00_clustering_HA_D96.rds"))
sessionInfo()R version 4.0.0 (2020-04-24)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 16.04.6 LTS
Matrix products: default
BLAS: /usr/local/R/R-4.0.0/lib/libRblas.so
LAPACK: /usr/local/R/R-4.0.0/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] future_1.17.0 SingleCellExperiment_1.10.1
[3] SummarizedExperiment_1.18.1 DelayedArray_0.14.0
[5] matrixStats_0.56.0 Biobase_2.48.0
[7] GenomicRanges_1.40.0 GenomeInfoDb_1.24.2
[9] IRanges_2.22.2 S4Vectors_0.26.1
[11] BiocGenerics_0.34.0 Seurat_3.1.5
[13] cowplot_1.0.0 dplyr_1.0.2
[15] ggplot2_3.3.2 BiocParallel_1.22.0
[17] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] nlme_3.1-148 tsne_0.1-3 bitops_1.0-6
[4] fs_1.4.2 RcppAnnoy_0.0.16 RColorBrewer_1.1-2
[7] httr_1.4.1 rprojroot_1.3-2 sctransform_0.2.1
[10] tools_4.0.0 backports_1.1.9 R6_2.4.1
[13] irlba_2.3.3 KernSmooth_2.23-17 uwot_0.1.8
[16] lazyeval_0.2.2 colorspace_1.4-1 withr_2.2.0
[19] tidyselect_1.1.0 gridExtra_2.3 compiler_4.0.0
[22] git2r_0.27.1 plotly_4.9.2.1 labeling_0.3
[25] scales_1.1.1 lmtest_0.9-37 ggridges_0.5.2
[28] pbapply_1.4-2 rappdirs_0.3.1 stringr_1.4.0
[31] digest_0.6.25 rmarkdown_2.3 XVector_0.28.0
[34] pkgconfig_2.0.3 htmltools_0.5.0 htmlwidgets_1.5.1
[37] rlang_0.4.7 farver_2.0.3 generics_0.0.2
[40] zoo_1.8-8 jsonlite_1.7.0 ica_1.0-2
[43] RCurl_1.98-1.2 magrittr_1.5 GenomeInfoDbData_1.2.3
[46] patchwork_1.0.1 Matrix_1.2-18 Rcpp_1.0.5
[49] munsell_0.5.0 ape_5.4 reticulate_1.16
[52] lifecycle_0.2.0 stringi_1.4.6 whisker_0.4
[55] yaml_2.2.1 zlibbioc_1.34.0 MASS_7.3-51.6
[58] Rtsne_0.15 plyr_1.8.6 grid_4.0.0
[61] listenv_0.8.0 promises_1.1.1 ggrepel_0.8.2
[64] crayon_1.3.4 lattice_0.20-41 splines_4.0.0
[67] knitr_1.29 pillar_1.4.6 igraph_1.2.5
[70] future.apply_1.6.0 reshape2_1.4.4 codetools_0.2-16
[73] leiden_0.3.3 glue_1.4.2 evaluate_0.14
[76] data.table_1.12.8 vctrs_0.3.4 png_0.1-7
[79] httpuv_1.5.4 gtable_0.3.0 RANN_2.6.1
[82] purrr_0.3.4 tidyr_1.1.0 xfun_0.15
[85] rsvd_1.0.3 RSpectra_0.16-0 later_1.1.0.1
[88] survival_3.2-3 viridisLite_0.3.0 tibble_3.0.3
[91] cluster_2.1.0 globals_0.12.5 fitdistrplus_1.1-1
[94] ellipsis_0.3.1 ROCR_1.0-11