Log2022
Last updated: 2022-01-19
Checks: 7 0
Knit directory: GradLog/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20201014) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 9976af6. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Ignored: analysis/test_log.Rmd
Ignored: code/.DS_Store
Ignored: data/.DS_Store
Ignored: output/.DS_Store
Untracked files:
Untracked: analysis/references.bib
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/Log2022.Rmd) and HTML (docs/Log2022.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 9976af6 | llw | 2022-01-19 | new week |
| html | bd69a21 | llw | 2022-01-13 | Build site. |
| Rmd | 28d0a2b | llw | 2022-01-13 | new week |
| html | ec0a2af | llw | 2022-01-04 | Build site. |
| Rmd | 9e5c2e0 | llw | 2022-01-04 | new week |
| html | 97b4ca1 | llw | 2022-01-03 | Build site. |
| html | 4c90bd8 | llw | 2022-01-03 | Build site. |
| Rmd | 6dc3f9c | llw | 2022-01-03 | add a new year |
Jan 19
1. Comprehensive analysis of coloc regions of trait MS
Add MS’s result to previous coloc analysis (dark green coloc regions)
See earlier section.
dark blue coloc regions
There are 4 dark blue coloc regions, i.e. coloc regions with module-QTL lead pvalue < 1e-5.
| Region | nSNPs | Pval | PP4 |
|---|---|---|---|
| module121:19:10423815 | 140 | 1.5e-06 | 0.98 |
| module108:7:50308811 | 122 | 1.6e-06 | 0.98 |
| module50:7:149331042 | 265 | 2.0e-06 | 0.94 |
| module44:7:50308527 | 69 | 0.0e+00 | 0.89 |
In earlier section, the region “module108:7:50308811” coloc with MS is included in these dark blue regions.
2. More traits, from UKBB
Extracted traits
- How?
I extracted specific traits from ukbb traits manifest file, based on:
Traits from eQTLGen traits
Traits from Mu et al paper
Powerful (and interesting) ukbb GWASs
Sorted by case number of EUR
n_cases_EUR.
- What traits?
26 ukbb GWASs in total, including height, BMI, Cholesterol, Hypertension, Asthma, Type 2 diabetes etc.
Exclude height, as not GWASed for EUR.
Use height sum stat from another meta analysis instead. It also has another BMI sum stat.
Meta analyze inds from GIANT and ukbb.
Meta-analysis of genome-wide association studies for height and body mass index in ~700,000 individuals of European ancestry.
Thus, as a result, 27 GWASs. 22 unique traits.
- Visualize the sample sizes of selected traits
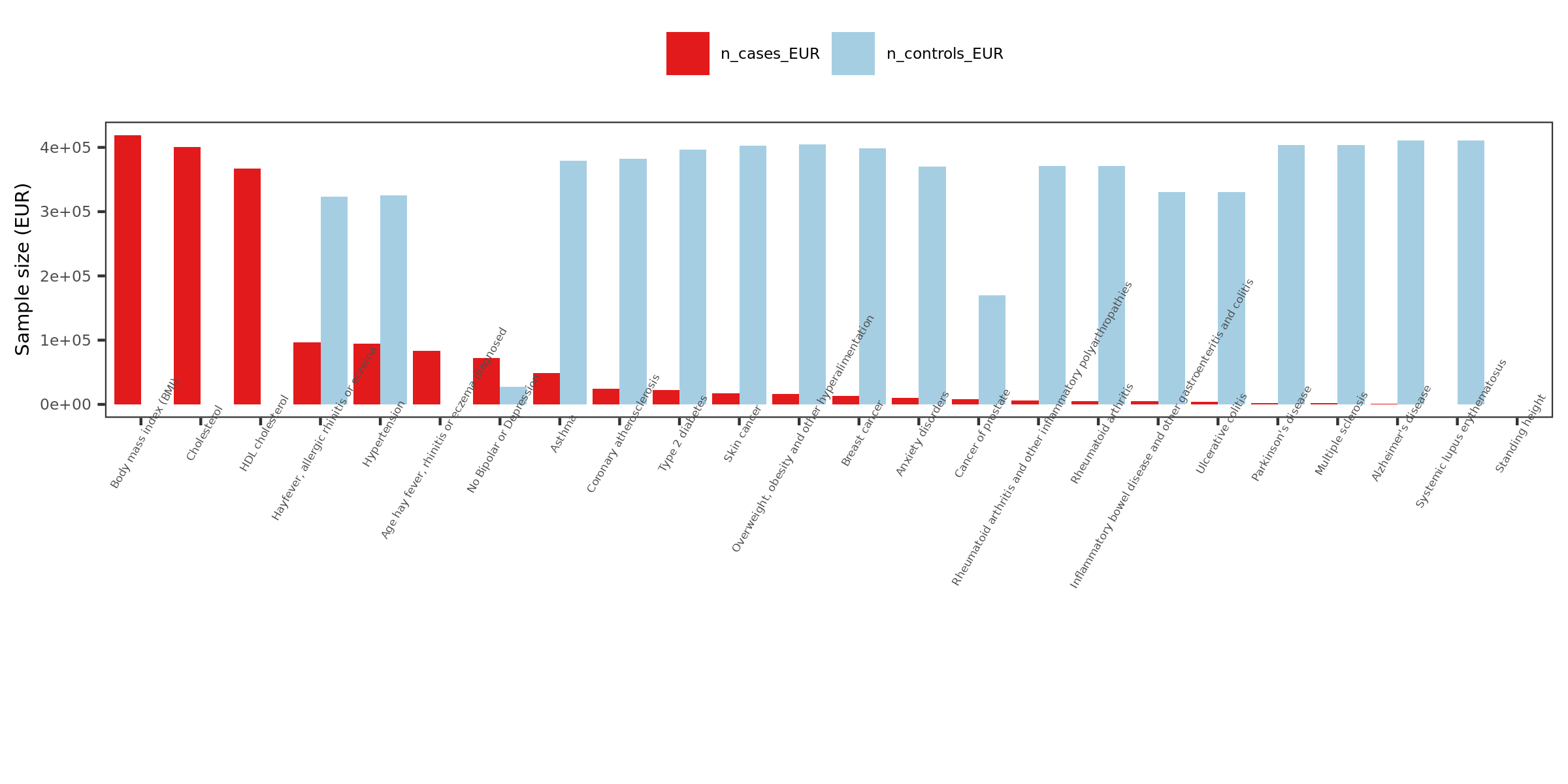
| Version | Author | Date |
|---|---|---|
| 1b40fde | llw | 2022-01-19 |
coloc results
- Visualize
See figure.
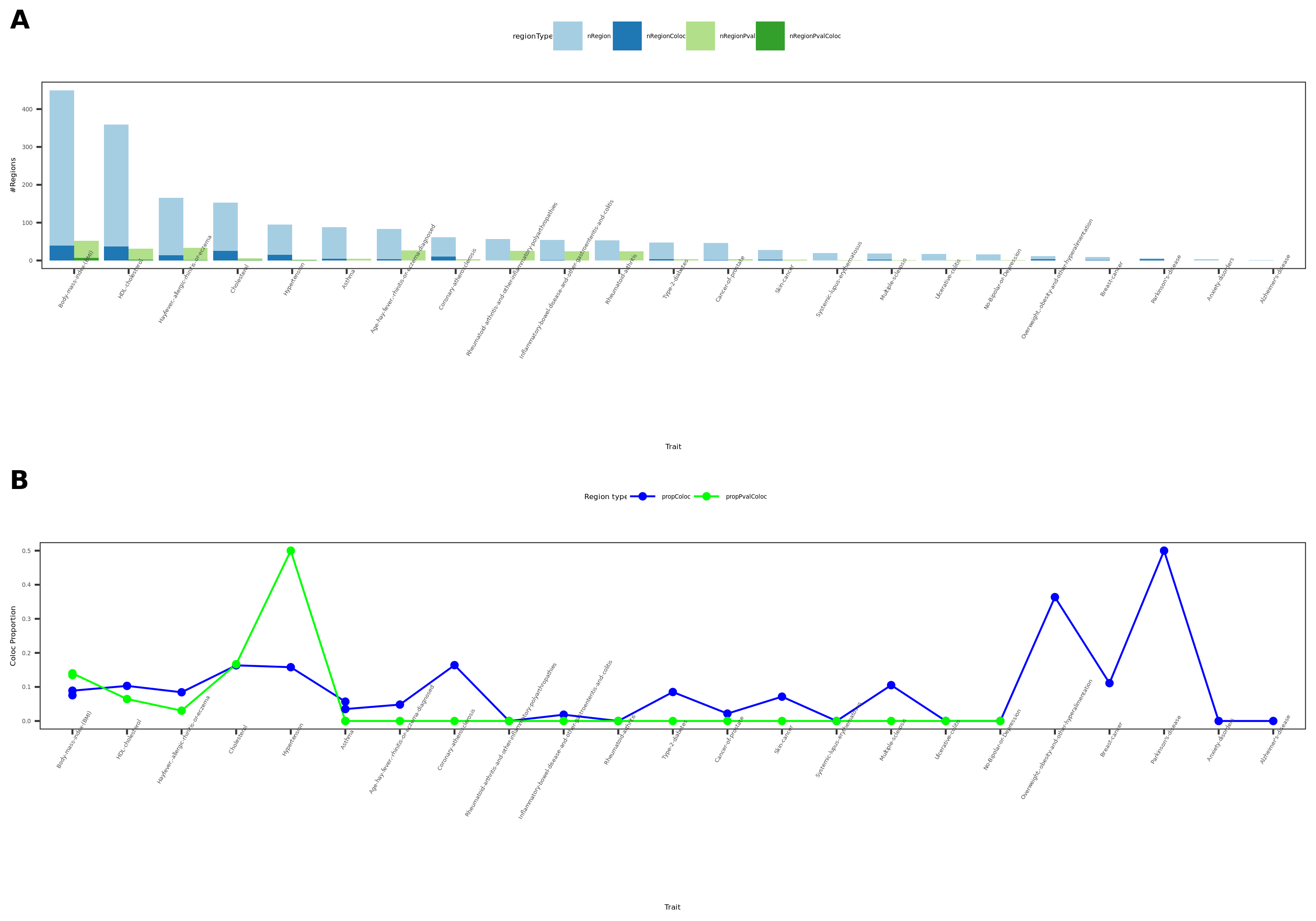
| Version | Author | Date |
|---|---|---|
| 1b40fde | llw | 2022-01-19 |
For detailed numbers, see file.
- In-depth visualization for selected traits (dark blue regions)
Jan 12
1. Make a story out of the trans- signals
More concrete examples
- Trait, gene, trans- signal
SLE signals in Fig. 5.
reflecting the involvement of interferon signaling in SLE pathophysiology
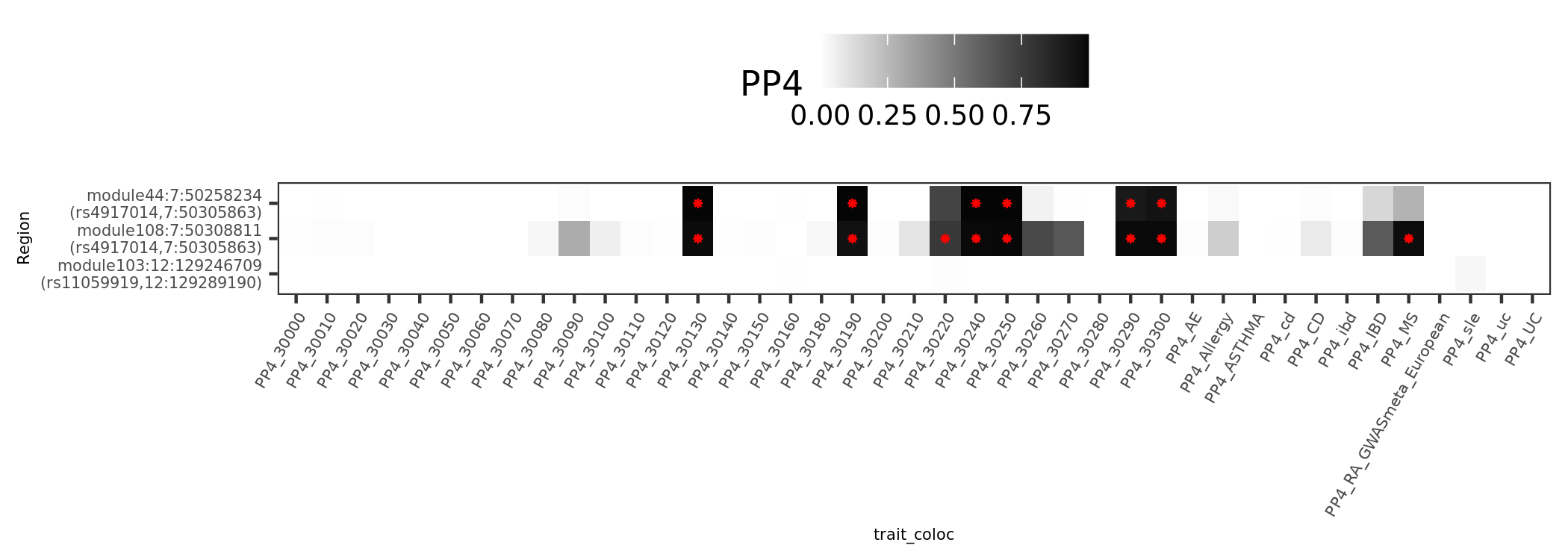
| Version | Author | Date |
|---|---|---|
| 664d0d7 | llw | 2022-01-13 |
- additional examples of trans-eQTL variants associated with traits
ZNF131 locus, age of menarche
Supplementary Figure 11A, rs1532331
FADS1/FADS2 locus, lipid levels
Supplementary Figure 11B, rs174574
IFIH1 locus, inflammatory bowel disease and SLE
Supplementary Figure 11C, rs1990760
GSDMB locus, asthma
Supplementary Figure 11D, rs7216389
CLOCK locus, height
Supplementary Figure 11E, rs1311351834
eQTSs genes GO enrichment
eQTSs genes associated with HDL
Jan 05
1. trans-eQTL and complex disease in eQTLGen, Võsa, Franke et al
Question: are there any trans-eQTLs that are GWAS hits?
Observations from the paper:
Meta-analysis using up to 31,684 blood samples from 37 eQTLGen Consortium cohorts.
For trans, they focused on 10,317 trait-associated SNPs.
The paper linked trans-eQTLs with traits in two ways to find the potential driver genes for traits.
- trans-eQTL analysis
One-third of trait-associated variants have distal effects.
Identified 59,786 trans-eQTL, representing 3,853 SNPs (37% of tested GWAS SNPs) and 6,298 genes (32% of tested genes).
The largest previous trans-eQTL meta-analysis in blood (N = 5,311) identified trans-eQTL for 8% of tested SNPs.
Identify genes that are coordinately affected by multiple independent trait-associated SNPs.
Identified 47 GWAS traits for which at least four independent variants affected the same gene in trans (Supplementary Tables 10). Examples genes affected by at least three SLE-associated genetic variants.
But, Individual trans-eQTL effects too weak to detect. Another way to look for the potential driver genes for traits and the “core” genes:
- eQTSs (associations between PGSs and gene expression, Fig. 6a, 6b)
Individual trait-associated SNPs are combined into a PGS that is associated with gene expression.
when the PGS for a trait correlates with the expression of a gene, trans-eQTL effects of individual risk variants converge on that gene, and it can be prioritized as a putative driver of the disease.
1,263 traits in total. 18,210 eQTSs representing 689 unique traits (55% of tested traits) and 2,568 genes (13% of tested genes).
Of these genes, 719 (28%) were not identified in the trans-eQTL analysis.
Therefore, all 3,853 trans-eQTLs eQTLGen identified are GWAS hits.
Question: if so, can we replicate them?
Among 10,317 trait-associated SNPs, 9,056 (~89%) are included in DGN SNPs. Among 3,853 eQTLGen trans- eQTLs, 27 (~0.7%) are replicated in DGN signals (1,863, \(p<1e-8\)).
In the paper, another trans- study was mentioned,
The largest previous trans-eQTL meta-analysis in blood (N = 5,311) identified trans-eQTL for 8% of tested SNPs.
2. gene SENP7
the cross mappability of SENP7 and the genes in the trans modules
As noted in earlier section, we observed,
- SENP7 is the neasrest gene of the lead SNPs of coloc regions.
- Gene ZNF90P1 is located within SENP7.
- Modules 25, 51, 153, 156 have coloc regions with immune traits and consist of many zinc finger genes.
Therefore, we wanted to know,
- Does gene ZNF90P1 have expression data?
I looked at the expression matrix of 13634 genes. ZNF90P1 doesn’t have expression data in this matrix.
- What is the cross mappability between SENP7 and the zinc finger genes in the modules?
See files for the cross mappability for module 25, module 51, module 153, module 156.
The columns include: “score” for cross mappability score, “score_map1” for mappability score of gene1, “score_map2” for mappability score of gene2.
- For the genes in the trans modules,
See a previous file here.
highlighting SENP7 as a trans-meQTL
https://www-nature-com.proxy.uchicago.edu/articles/s41588-021-00969-x Another paper highlighting SENP7 as a trans-meQTL.
3. pQTL resource
R version 4.1.0 (2021-05-18)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Big Sur 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.1/Resources/lib/libRblas.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.1/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rcpp_1.0.7 whisker_0.4 knitr_1.33 magrittr_2.0.1
[5] R6_2.5.1 rlang_0.4.12 fastmap_1.1.0 fansi_0.5.0
[9] highr_0.9 stringr_1.4.0 tools_4.1.0 xfun_0.23
[13] utf8_1.2.2 git2r_0.28.0 jquerylib_0.1.4 htmltools_0.5.2
[17] ellipsis_0.3.2 rprojroot_2.0.2 yaml_2.2.1 digest_0.6.29
[21] tibble_3.1.6 lifecycle_1.0.1 crayon_1.4.2 later_1.3.0
[25] sass_0.4.0 vctrs_0.3.8 promises_1.2.0.1 fs_1.5.2
[29] glue_1.6.0 evaluate_0.14 rmarkdown_2.10 stringi_1.7.6
[33] bslib_0.3.1 compiler_4.1.0 pillar_1.6.4 jsonlite_1.7.2
[37] httpuv_1.6.4 pkgconfig_2.0.3