Metabolic rates
Last updated: 2020-12-09
Checks: 7 0
Knit directory: exp_evol_respiration/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190703) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 2642c27. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Untracked files:
Untracked: data/3.metabolite_data.csv
Untracked: output/brms_metabolite_PCA_SEM.rds
Untracked: output/brms_metabolite_SEM.rds
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/metabolites.Rmd) and HTML (docs/metabolites.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 2642c27 | lukeholman | 2020-12-09 | More work |
| html | 4f5ee28 | lukeholman | 2020-12-04 | Build site. |
| Rmd | d441b69 | lukeholman | 2020-12-04 | Luke metabolites analysis |
| Rmd | c8feb2d | lukeholman | 2020-11-30 | Same page with Martin |
| html | 3fdbcb2 | lukeholman | 2020-11-30 | Tweaks Nov 2020 |
Load packages
library(tidyverse)── Attaching packages ────────────────────────────────────────────────────────────────────────────── tidyverse 1.3.0 ──✓ ggplot2 3.3.2 ✓ purrr 0.3.4
✓ tibble 3.0.1 ✓ dplyr 1.0.0
✓ tidyr 1.1.0 ✓ stringr 1.4.0
✓ readr 1.3.1 ✓ forcats 0.5.0── Conflicts ───────────────────────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
x dplyr::filter() masks stats::filter()
x dplyr::lag() masks stats::lag()library(GGally)Registered S3 method overwritten by 'GGally':
method from
+.gg ggplot2library(gridExtra)
Attaching package: 'gridExtra'The following object is masked from 'package:dplyr':
combinelibrary(ggridges)
library(brms)Loading required package: RcppLoading 'brms' package (version 2.14.4). Useful instructions
can be found by typing help('brms'). A more detailed introduction
to the package is available through vignette('brms_overview').
Attaching package: 'brms'The following object is masked from 'package:stats':
arlibrary(tidybayes)
Attaching package: 'tidybayes'The following objects are masked from 'package:ggridges':
scale_point_color_continuous, scale_point_color_discrete, scale_point_colour_continuous, scale_point_colour_discrete,
scale_point_fill_continuous, scale_point_fill_discrete, scale_point_size_continuouslibrary(DT)
library(kableExtra)
Attaching package: 'kableExtra'The following object is masked from 'package:dplyr':
group_rowslibrary(knitrhooks) # install with devtools::install_github("nathaneastwood/knitrhooks")Loading required package: knitroutput_max_height() # a knitrhook option
options(stringsAsFactors = FALSE)Load metabolite composition data
This analysis set out to test whether sexual selection treatment had an effect on metabolite composition of flies. We measured fresh and dry fly weight in milligrams, plus the weights of five metabolites which together equal the dry weight. These are:
Lipid_conc(i.e. the weight of the hexane fraction, divided by the full dry weight),Carbohydrate_conc(i.e. the weight of the aqueous fraction, divided by the full dry weight),Protein_conc(i.e. \(\mu\)g of protein per milligram as measured by the bicinchoninic acid protein assay),Glycogen_conc(i.e. \(\mu\)g of glycogen per milligram as measured by the hexokinase assay), andChitin_conc(estimated as the difference between the initial and final dry weights)
We expect body weight to vary between the sexes and potentially between treatments. In turn, we expect body weight to affect our five response variables of interest. Larger flies will have more lipids, carbs, etc., and this may vary by sex and treatment both directly and indirectly.
metabolites <- read_csv('data/3.metabolite_data.csv') %>%
mutate(sex = ifelse(sex == "m", "Male", "Female"),
line = paste(treatment, line, sep = ""),
treatment = ifelse(treatment == "M", "Monogamy", "Polyandry")) %>%
# log transform glycogen since it shows a long tail (others are reasonably normal-looking)
mutate(Glycogen_ug_mg = log(Glycogen_ug_mg)) %>%
# There was a technical error with flies collected on day 1,
# so they are excluded from the whole paper. All the measurements analysed are of 3d-old flies
filter(time == '2') %>%
select(-time)Parsed with column specification:
cols(
sex = col_character(),
treatment = col_character(),
line = col_double(),
time = col_double(),
fwt_mg = col_double(),
dwt_mg = col_double(),
Hex_frac = col_double(),
p_hex_frac = col_double(),
Aq_frac = col_double(),
p_Aq_frac = col_double(),
Protein = col_double(),
Protein_ug_mg = col_double(),
Glycogen = col_double(),
Glycogen_ug_mg = col_double(),
Chitin = col_double(),
Chitin_mg_mg = col_double()
)scaled_metabolites <- metabolites %>%
# Find proportional metabolites as a proportion of total dry weight
mutate(
Dry_weight = dwt_mg,
Lipid_conc = Hex_frac / Dry_weight,
Carbohydrate_conc = Aq_frac / Dry_weight,
Protein_conc = Protein_ug_mg,
Glycogen_conc = Glycogen_ug_mg,
Chitin_conc = Chitin_mg_mg) %>%
select(sex, treatment, line, Dry_weight, ends_with("conc")) %>%
mutate_at(vars(ends_with("conc")), ~ as.numeric(scale(.x))) %>%
mutate(Dry_weight = as.numeric(scale(Dry_weight))) %>%
mutate(sextreat = paste(sex, treatment),
sextreat = replace(sextreat, sextreat == "Male Monogamy", "M males"),
sextreat = replace(sextreat, sextreat == "Male Polyandry", "P males"),
sextreat = replace(sextreat, sextreat == "Female Monogamy", "M females"),
sextreat = replace(sextreat, sextreat == "Female Polyandry", "P females"),
sextreat = factor(sextreat, c("M males", "P males", "M females", "P females")))Inspect the raw data
Raw numbers
All variables are shown in standard units (i.e. mean = 0, SD = 1).
my_data_table <- function(df){
datatable(
df, rownames=FALSE,
autoHideNavigation = TRUE,
extensions = c("Scroller", "Buttons"),
options = list(
dom = 'Bfrtip',
deferRender=TRUE,
scrollX=TRUE, scrollY=400,
scrollCollapse=TRUE,
buttons =
list('csv', list(
extend = 'pdf',
pageSize = 'A4',
orientation = 'landscape',
filename = 'Apis_methylation')),
pageLength = 50
)
)
}
scaled_metabolites %>%
select(-sextreat) %>%
mutate_if(is.numeric, ~ format(round(.x, 3), nsmall = 3)) %>%
my_data_table()Simple plots
The following plot shows how each metabolite varies between sexes and treatments, and how the consecration of each metabolite co-varies with dry weight across individuals.
levels <- c("Carbohydrate", "Chitin", "Glycogen", "Lipid", "Protein", "Dryweight")
cols <- c("M females" = "pink",
"P females" = "red",
"M males" = "skyblue",
"P males" = "blue")
grid.arrange(
scaled_metabolites %>%
rename_all(~ str_remove_all(.x, "_conc")) %>%
mutate(sex = factor(sex, c("Male", "Female"))) %>%
reshape2::melt(id.vars = c('sex', 'treatment', 'sextreat', 'line', 'Dry_weight')) %>%
mutate(variable = factor(variable, levels)) %>%
ggplot(aes(x = sex, y = value, fill = sextreat)) +
geom_hline(yintercept = 0, linetype = 2) +
geom_boxplot() +
facet_grid( ~ variable) +
theme_bw() +
xlab("Sex") + ylab("Concentration") +
theme(legend.position = 'top') +
scale_fill_manual(values = cols, name = ""),
scaled_metabolites %>%
rename_all(~ str_remove_all(.x, "_conc")) %>%
reshape2::melt(id.vars = c('sex', 'treatment', 'sextreat', 'line', 'Dry_weight')) %>%
mutate(variable = factor(variable, levels)) %>%
ggplot(aes(x = Dry_weight, y = value, colour = sextreat, fill = sextreat)) +
geom_smooth(method = 'lm', se = TRUE, aes(colour = NULL, fill = NULL), colour = "grey20", size = .4) +
geom_point(pch = 21, colour = "grey20") +
facet_grid( ~ variable) +
theme_bw() +
xlab("Dry weight (mg)") + ylab("Concentration") +
theme(legend.position = 'none') +
scale_colour_manual(values = cols, name = "") +
scale_fill_manual(values = cols, name = ""),
heights = c(0.55, 0.45)
)`geom_smooth()` using formula 'y ~ x'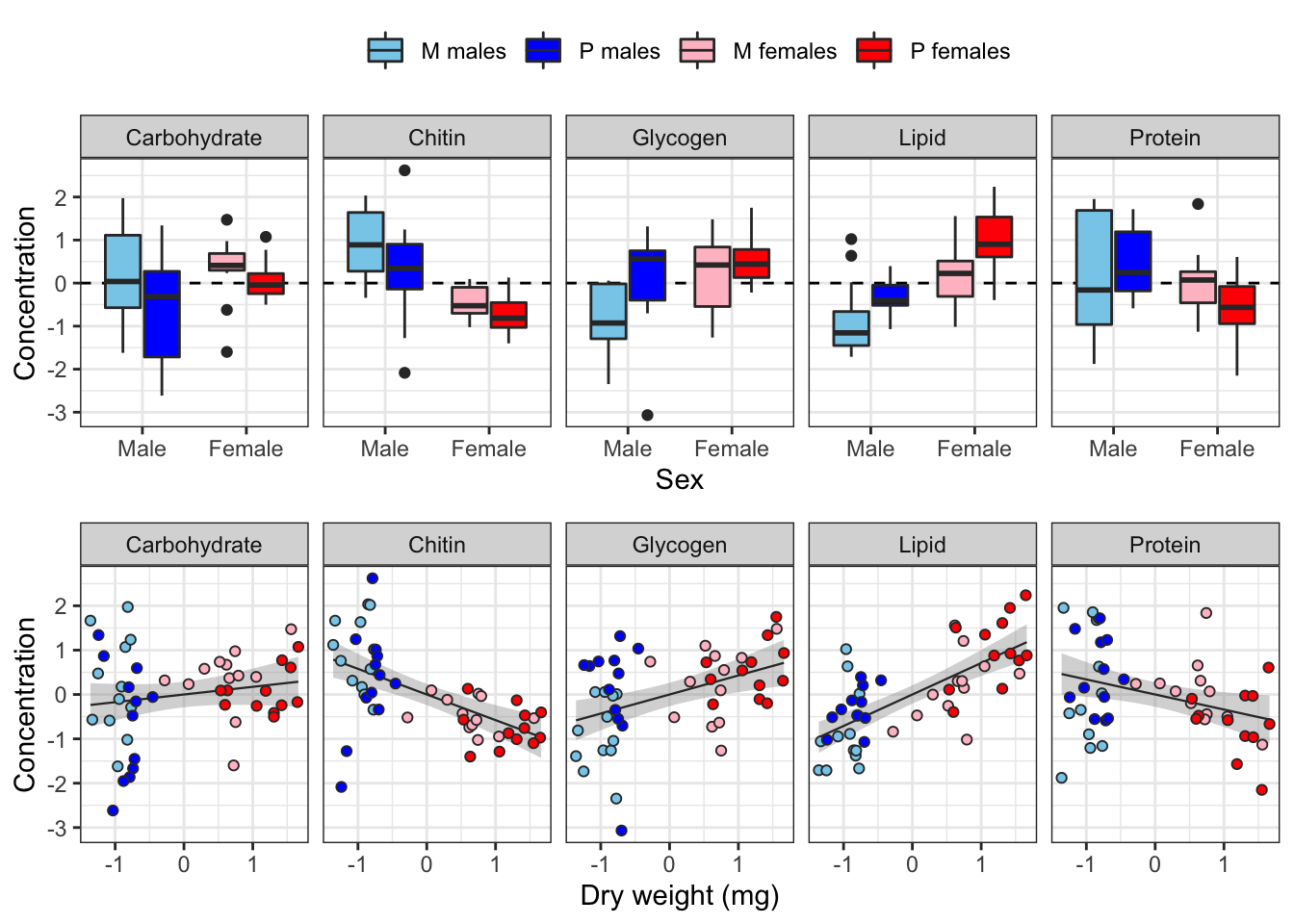
| Version | Author | Date |
|---|---|---|
| 4f5ee28 | lukeholman | 2020-12-04 |
Plot of correlations between variables
Some of the metabolites, especially lipid concentration, are correlated with dry weight. There is also a large difference in dry weight between sexes (and treatments, to a less extent), and sex and treatment effects are evident for some of the metabolites in the raw data. Some of the metabolites are weakly correlated with other metabolites, e.g. lipid and glycogen concentration.
modified_densityDiag <- function(data, mapping, ...) {
ggally_densityDiag(data, mapping, colour = "grey10", ...) +
scale_fill_manual(values = cols) +
scale_x_continuous(guide = guide_axis(check.overlap = TRUE))
}
modified_points <- function(data, mapping, ...) {
ggally_points(data, mapping, pch = 21, colour = "grey10", ...) +
scale_fill_manual(values = cols) +
scale_x_continuous(guide = guide_axis(check.overlap = TRUE))
}
modified_facetdensity <- function(data, mapping, ...) {
ggally_facetdensity(data, mapping, ...) +
scale_colour_manual(values = cols)
}
modified_box_no_facet <- function(data, mapping, ...) {
ggally_box_no_facet(data, mapping, colour = "grey10", ...) +
scale_fill_manual(values = cols)
}
pairs_plot <- scaled_metabolites %>%
arrange(sex, treatment) %>%
select(-line, -sex, -treatment) %>%
rename(`Sex and treatment` = sextreat) %>%
rename_all(~ str_replace_all(.x, "_", " ")) %>%
ggpairs(aes(colour = `Sex and treatment`, fill = `Sex and treatment`),
diag = list(continuous = wrap(modified_densityDiag, alpha = 0.7),
discrete = wrap("blank")),
lower = list(continuous = wrap(modified_points, alpha = 0.7, size = 1.1),
discrete = wrap("blank"),
combo = wrap(modified_box_no_facet, alpha = 0.7)),
upper = list(continuous = wrap(modified_points, alpha = 0.7, size = 1.1),
discrete = wrap("blank"),
combo = wrap(modified_box_no_facet, alpha = 0.7, size = 0.5)))
pairs_plot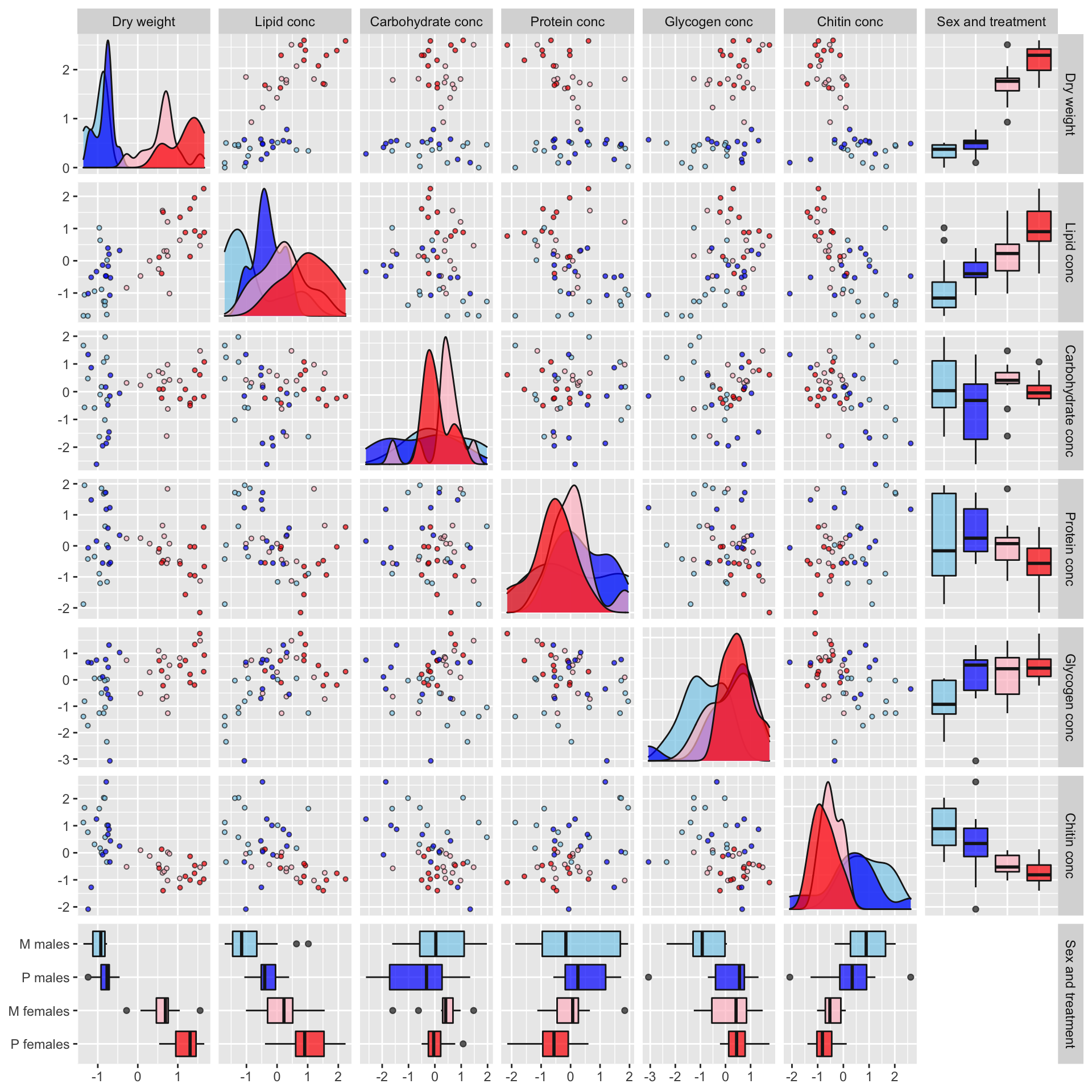
Directed acyclic graph (DAG)
This directed acyclic graph (DAG) illustrates the causal pathways that we observed between the experimental or measured variables (square boxes) and latent variables (ovals). We hypothesise that sex and mating system potentially influence dry weight as well as the metabolite composition (which we assessed by estimating the concentrations of carbohydrates, chitin, glycogen, lipids and protein). Additionally, dry weight is likely correlated with the metabolome, and so dry weight acts as a ‘mediator variable’ between the metabolome, and sex and treatment. The structural equation model below is built with this DAG in mind.
DiagrammeR::grViz('digraph {
graph [layout = dot, rankdir = LR]
# define the global styles of the nodes. We can override these in box if we wish
node [shape = rectangle, style = filled, fillcolor = Linen]
"Metabolite\ncomposition" [shape = oval, fillcolor = Beige]
# edge definitions with the node IDs
"Mating system\ntreatment (M vs P)" -> {"Dry weight"}
"Mating system\ntreatment (M vs P)" -> {"Metabolite\ncomposition"}
"Sex\n(Female vs Male)" -> {"Dry weight"} -> {"Metabolite\ncomposition"}
"Sex\n(Female vs Male)" -> {"Metabolite\ncomposition"}
{"Metabolite\ncomposition"} -> "Carbohydrates"
{"Metabolite\ncomposition"} -> "Chitin"
{"Metabolite\ncomposition"} -> "Glycogen"
{"Metabolite\ncomposition"} -> "Lipids"
{"Metabolite\ncomposition"} -> "Protein"
}')Fit brms structural equation model
Here we fit a model of the five metabolites, which includes dry body weight as a mediator variable. That is, our model estimates the effect of treatment, sex and line (and all the 2- and 3-way interactions) on dry weight, and then estimates the effect of those some predictors (plus dry weight) on the five metabolites. The model assumes that although the different sexes, treatment groups, and lines may differ in their dry weight, the relationship between dry weight and the metabolites does not vary by sex/treatment/line. This assumption was made to constrain the number of parameters in the model, and to reflect out prior beliefs about allometric scaling of metabolites.
Define Priors
We use set Normal priors on all fixed effect parameters, mean = 0, sd = 1, which ‘regularises’ the estimates towards zero – this is conservative (because it makes large posterior effect sizes more improbable), and it also helps the model to converge. Similarly, we set a somewhat conservative half-cauchy prior (mean 0, scale 0.01) on the random effects for line (i.e. we consider large differences between lines – in terms of means and treatment effects – to be possible but improbable). We leave all other priors at the defaults used by brms.
prior1 <- c(set_prior("normal(0, 1)", class = "b", resp = 'Lipid'),
set_prior("normal(0, 1)", class = "b", resp = 'Carbohydrate'),
set_prior("normal(0, 1)", class = "b", resp = 'Protein'),
set_prior("normal(0, 1)", class = "b", resp = 'Glycogen'),
set_prior("normal(0, 1)", class = "b", resp = 'Chitin'),
set_prior("normal(0, 1)", class = "b", resp = 'Dryweight'),
set_prior("cauchy(0,0.01)", class = "sd", resp = 'Lipid', group = "line"),
set_prior("cauchy(0,0.01)", class = "sd", resp = 'Carbohydrate', group = "line"),
set_prior("cauchy(0,0.01)", class = "sd", resp = 'Protein', group = "line"),
set_prior("cauchy(0,0.01)", class = "sd", resp = 'Glycogen', group = "line"),
set_prior("cauchy(0,0.01)", class = "sd", resp = 'Chitin', group = "line"),
set_prior("cauchy(0,0.01)", class = "sd", resp = 'Dryweight', group = "line"))
prior1prior class coef group resp dpar nlpar bound source normal(0, 1) b Lipid user normal(0, 1) b Carbohydrate user normal(0, 1) b Protein user normal(0, 1) b Glycogen user normal(0, 1) b Chitin user normal(0, 1) b Dryweight user cauchy(0,0.01) sd line Lipid user cauchy(0,0.01) sd line Carbohydrate user cauchy(0,0.01) sd line Protein user cauchy(0,0.01) sd line Glycogen user cauchy(0,0.01) sd line Chitin user cauchy(0,0.01) sd line Dryweight user
Define the six sub-models
The fixed effects formula is sex * treatment + Dryweight (or sex * treatment in the case of the model of dry weight). The random effects part of the formula indicates that the 8 independent selection lines may differ in their means, and that the treatment effect may vary in sign/magnitude between lines. The notation | p | means that the model estimates the correlations in line effects (both slopes and intercepts) between the 6 response variables. Finally, the notation set_rescor(TRUE) means that the model should estimate the residual correlations between the response variables.
brms_formula <-
# Sub-models of the 5 metabolites
bf(mvbind(Lipid, Carbohydrate, Protein, Glycogen, Chitin) ~
sex*treatment + Dryweight + (treatment | p | line)) +
# dry weight sub-model
bf(Dryweight ~ sex*treatment + (treatment | p | line)) +
# Allow for (and estimate) covariance between the residuals of the difference response variables
set_rescor(TRUE)
brms_formulaLipid ~ sex * treatment + Dryweight + (treatment | p | line) Carbohydrate ~ sex * treatment + Dryweight + (treatment | p | line) Protein ~ sex * treatment + Dryweight + (treatment | p | line) Glycogen ~ sex * treatment + Dryweight + (treatment | p | line) Chitin ~ sex * treatment + Dryweight + (treatment | p | line) Dryweight ~ sex * treatment + (treatment | p | line)
Running the model
The model is run over 4 chains with 5000 iterations each (with the first 2500 discarded as burn-in), for a total of 2500*4 = 10,000 posterior samples.
if(!file.exists("output/brms_metabolite_SEM.rds")){
brms_metabolite_SEM <- brm(
brms_formula,
data = scaled_metabolites %>% # brms does not like underscores in variable names
rename(Dryweight = Dry_weight) %>%
rename_all(~ gsub("_conc", "", .x)),
iter = 5000, chains = 4, cores = 1,
prior = prior1,
control = list(max_treedepth = 20,
adapt_delta = 0.99)
)
saveRDS(brms_metabolite_SEM, "output/brms_metabolite_SEM.rds")
} else {
brms_metabolite_SEM <- readRDS('output/brms_metabolite_SEM.rds')
}Posterior predictive check of model fit
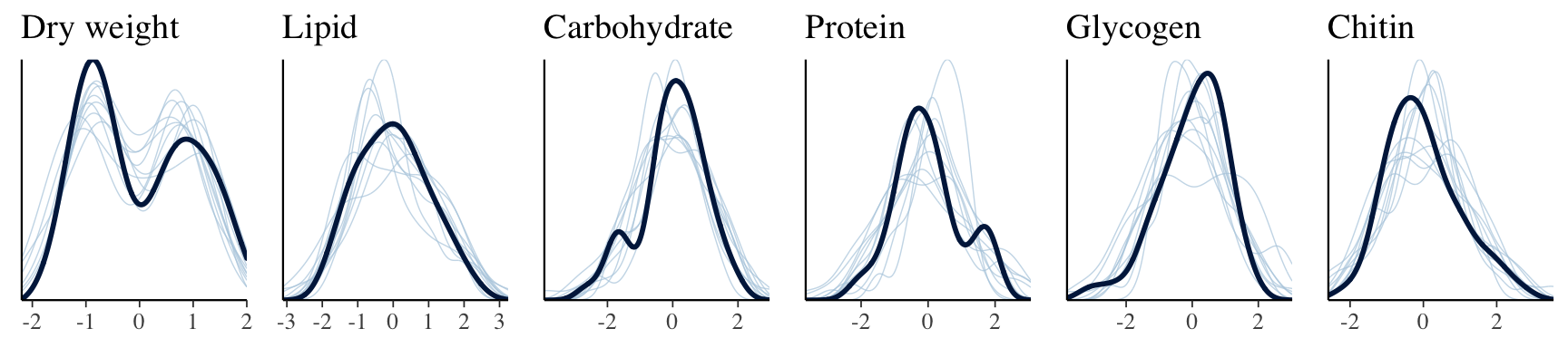
The plot below shows that the fitted model is able to produce posterior predictions that have a similar distribution to the original data, for each of the response variables, which is a necessary condition for the model to be used for statistical inference.
grid.arrange(
pp_check(brms_metabolite_SEM, resp = "Dryweight") +
ggtitle("Dry weight") + theme(legend.position = "none"),
pp_check(brms_metabolite_SEM, resp = "Lipid") +
ggtitle("Lipid") + theme(legend.position = "none"),
pp_check(brms_metabolite_SEM, resp = "Carbohydrate") +
ggtitle("Carbohydrate") + theme(legend.position = "none"),
pp_check(brms_metabolite_SEM, resp = "Protein") +
ggtitle("Protein") + theme(legend.position = "none"),
pp_check(brms_metabolite_SEM, resp = "Glycogen") +
ggtitle("Glycogen") + theme(legend.position = "none"),
pp_check(brms_metabolite_SEM, resp = "Chitin") +
ggtitle("Chitin") + theme(legend.position = "none"),
nrow = 1
)
| Version | Author | Date |
|---|---|---|
| 4f5ee28 | lukeholman | 2020-12-04 |
Table of model parameter estimates
Formatted table
This tables shows the fixed effects estimates for the treatment, sex, their interaction, as well as the slope associated with dry weight (where relevant), for each of the six response variables. The p column shows 1 - minus the “probability of direction”, i.e. the posterior probability that the reported sign of the estimate is correct given the data and the prior; subtracting this value from one gives a Bayesian equivalent of a one-sided p-value. For brevity, we have omitted all the parameter estimates involving the predictor variable line, as well as the estimates of residual (co)variance. Click the next tab to see a complete summary of the model and its output.
vars <- c("Lipid", "Carbohydrate", "Glycogen", "Protein", "Chitin")
tests <- c('_Dryweight', '_sexMale',
'_sexMale:treatmentPolyandry',
'_treatmentPolyandry')
hypSEM <- data.frame(expand_grid(vars, tests) %>%
mutate(est = NA,
err = NA,
lwr = NA,
upr = NA) %>%
# bind body weight on the end
rbind(data.frame(
vars = rep('Dryweight', 3),
tests = c('_sexMale',
'_treatmentPolyandry',
'_sexMale:treatmentPolyandry'),
est = NA,
err = NA,
lwr = NA,
upr = NA)))
for(i in 1:nrow(hypSEM)) {
result = hypothesis(brms_metabolite_SEM,
paste0(hypSEM[i, 1], hypSEM[i, 2], ' = 0'))$hypothesis
hypSEM[i, 3] = round(result$Estimate, 3)
hypSEM[i, 4] = round(result$Est.Error, 3)
hypSEM[i, 5] = round(result$CI.Lower, 3)
hypSEM[i, 6] = round(result$CI.Upper, 3)
}
pvals <- bayestestR::p_direction(brms_metabolite_SEM) %>%
as.data.frame() %>%
mutate(vars = map_chr(str_split(Parameter, "_"), ~ .x[2]),
tests = map_chr(str_split(Parameter, "_"), ~ .x[3]),
tests = str_c("_", str_remove_all(tests, "[.]")),
tests = replace(tests, tests == "_sexMaletreatmentPolyandry", "_sexMale:treatmentPolyandry")) %>%
filter(!str_detect(tests, "line")) %>%
mutate(p_val = 1 - pd, star = ifelse(p_val < 0.05, "*", "")) %>%
select(vars, tests, p_val, star)
hypSEM <- hypSEM %>% left_join(pvals, by = c("vars", "tests"))
hypSEM %>%
mutate(Parameter = c(rep(c('Dry weight', 'Sex (M)',
'Sex (M) x Treatment (P)',
'Treatment (P)'), 5),
'Sex (M)', 'Treatment (P)', 'Sex (M) x Treatment (P)')) %>%
mutate(Parameter = factor(Parameter, c("Dry weight", "Sex (M)", "Treatment (P)", "Sex (M) x Treatment (P)")),
vars = factor(vars, c("Carbohydrate", "Chitin", "Glycogen", "Lipid", "Protein", "Dryweight"))) %>%
arrange(vars, Parameter) %>%
select(Parameter, Estimate = est, `Est. error` = err,
`CI lower` = lwr, `CI upper` = upr, `p` = p_val, star) %>%
rename(` ` = star) %>%
kable() %>%
kable_styling(full_width = FALSE) %>%
group_rows("Carbohydrates", 1, 4) %>%
group_rows("Chitin", 5, 8) %>%
group_rows("Glycogen", 9, 12) %>%
group_rows("Lipids", 13, 16) %>%
group_rows("Protein", 17, 20) %>%
group_rows("Dry weight", 21, 23)| Parameter | Estimate | Est. error | CI lower | CI upper | p | |
|---|---|---|---|---|---|---|
| Carbohydrates | ||||||
| Dry weight | 0.159 | 0.485 | -0.781 | 1.115 | 0.3740 | |
| Sex (M) | 0.128 | 0.789 | -1.416 | 1.671 | 0.4367 | |
| Treatment (P) | -0.314 | 0.449 | -1.183 | 0.555 | 0.2371 | |
| Sex (M) x Treatment (P) | -0.472 | 0.509 | -1.461 | 0.536 | 0.1705 | |
| Chitin | ||||||
| Dry weight | -0.471 | 0.465 | -1.387 | 0.423 | 0.1566 | |
| Sex (M) | 0.552 | 0.763 | -0.966 | 2.026 | 0.2335 | |
| Treatment (P) | -0.064 | 0.399 | -0.822 | 0.745 | 0.4348 | |
| Sex (M) x Treatment (P) | -0.448 | 0.451 | -1.347 | 0.432 | 0.1577 | |
| Glycogen | ||||||
| Dry weight | 0.335 | 0.468 | -0.571 | 1.243 | 0.2383 | |
| Sex (M) | -0.430 | 0.770 | -1.906 | 1.104 | 0.2852 | |
| Treatment (P) | 0.175 | 0.412 | -0.625 | 0.976 | 0.3353 | |
| Sex (M) x Treatment (P) | 0.604 | 0.484 | -0.367 | 1.552 | 0.1054 | |
| Lipids | ||||||
| Dry weight | 0.610 | 0.446 | -0.256 | 1.488 | 0.0868 | |
| Sex (M) | -0.025 | 0.728 | -1.455 | 1.375 | 0.4873 | |
| Treatment (P) | 0.437 | 0.382 | -0.314 | 1.189 | 0.1215 | |
| Sex (M) x Treatment (P) | -0.034 | 0.419 | -0.875 | 0.777 | 0.4677 | |
| Protein | ||||||
| Dry weight | -0.222 | 0.470 | -1.145 | 0.707 | 0.3151 | |
| Sex (M) | -0.128 | 0.776 | -1.617 | 1.417 | 0.4367 | |
| Treatment (P) | -0.378 | 0.441 | -1.232 | 0.484 | 0.1913 | |
| Sex (M) x Treatment (P) | 0.586 | 0.517 | -0.434 | 1.584 | 0.1258 | |
| Dry weight | ||||||
| Sex (M) | -1.591 | 0.141 | -1.864 | -1.312 | 0.0000 |
|
| Treatment (P) | 0.546 | 0.157 | 0.233 | 0.848 | 0.0012 |
|
| Sex (M) x Treatment (P) | -0.395 | 0.195 | -0.779 | -0.012 | 0.0223 |
|
Complete output from summary.brmsfit()
- ‘Group-Level Effects’ (also called random effects): This shows the (co)variances associated with the line-specific intercepts (which have names like
sd(Lipid_Intercept)) and slopes (e.g.sd(Dryweight_treatmentPolyandry)), as well as the correlations between these effects (e.g.cor(Lipid_Intercept,Protein_Intercept)is the correlation in line effects on lipids and proteins) - ‘Population-Level Effects:’ (also called fixed effects): These give the estimates of the intercept (i.e. for female M flies) and the effects of treatment, sex, dry weight, and the treatment \(\times\) sex interaction, for each response variable.
- ‘Family Specific Parameters’: This is the parameter sigma for the residual variance for each response variable
- ‘Residual Correlations:’ This give the correlations between the residuals for each pairs of response variables.
Note that the model has converged (Rhat = 1) and the posterior is adequately samples (high ESS values).
brms_metabolite_SEM Family: MV(gaussian, gaussian, gaussian, gaussian, gaussian, gaussian)
Links: mu = identity; sigma = identity
mu = identity; sigma = identity
mu = identity; sigma = identity
mu = identity; sigma = identity
mu = identity; sigma = identity
mu = identity; sigma = identity
Formula: Lipid ~ sex * treatment + Dryweight + (treatment | p | line)
Carbohydrate ~ sex * treatment + Dryweight + (treatment | p | line)
Protein ~ sex * treatment + Dryweight + (treatment | p | line)
Glycogen ~ sex * treatment + Dryweight + (treatment | p | line)
Chitin ~ sex * treatment + Dryweight + (treatment | p | line)
Dryweight ~ sex * treatment + (treatment | p | line)
Data: scaled_metabolites %>% rename(Dryweight = Dry_weig (Number of observations: 48)
Samples: 4 chains, each with iter = 5000; warmup = 2500; thin = 1;
total post-warmup samples = 10000
Group-Level Effects:
~line (Number of levels: 8)
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sd(Lipid_Intercept) 0.09 0.15 0.00 0.51 1.00 2095 2341
sd(Lipid_treatmentPolyandry) 0.03 0.07 0.00 0.22 1.00 13332 6125
sd(Carbohydrate_Intercept) 0.07 0.14 0.00 0.52 1.00 3623 2636
sd(Carbohydrate_treatmentPolyandry) 0.05 0.14 0.00 0.47 1.00 8612 4337
sd(Protein_Intercept) 0.06 0.13 0.00 0.47 1.00 5854 3298
sd(Protein_treatmentPolyandry) 0.03 0.08 0.00 0.23 1.00 16484 6105
sd(Glycogen_Intercept) 0.03 0.07 0.00 0.24 1.00 12198 5266
sd(Glycogen_treatmentPolyandry) 0.03 0.06 0.00 0.15 1.00 18288 6178
sd(Chitin_Intercept) 0.07 0.13 0.00 0.47 1.00 3937 3874
sd(Chitin_treatmentPolyandry) 0.04 0.11 0.00 0.35 1.00 10361 4796
sd(Dryweight_Intercept) 0.05 0.07 0.00 0.25 1.00 3428 5432
sd(Dryweight_treatmentPolyandry) 0.03 0.06 0.00 0.21 1.00 8815 5238
cor(Lipid_Intercept,Lipid_treatmentPolyandry) 0.00 0.28 -0.54 0.55 1.00 26792 7112
cor(Lipid_Intercept,Carbohydrate_Intercept) 0.01 0.28 -0.53 0.54 1.00 20852 7354
cor(Lipid_treatmentPolyandry,Carbohydrate_Intercept) -0.00 0.28 -0.53 0.53 1.00 16765 6360
cor(Lipid_Intercept,Carbohydrate_treatmentPolyandry) 0.00 0.28 -0.53 0.54 1.00 25816 6812
cor(Lipid_treatmentPolyandry,Carbohydrate_treatmentPolyandry) 0.01 0.28 -0.53 0.54 1.00 17444 7043
cor(Carbohydrate_Intercept,Carbohydrate_treatmentPolyandry) -0.00 0.28 -0.52 0.54 1.00 12013 6962
cor(Lipid_Intercept,Protein_Intercept) 0.00 0.28 -0.53 0.53 1.00 27063 7109
cor(Lipid_treatmentPolyandry,Protein_Intercept) 0.00 0.28 -0.52 0.52 1.00 15083 7506
cor(Carbohydrate_Intercept,Protein_Intercept) 0.01 0.28 -0.53 0.54 1.00 13647 7267
cor(Carbohydrate_treatmentPolyandry,Protein_Intercept) -0.00 0.28 -0.54 0.53 1.00 11023 7498
cor(Lipid_Intercept,Protein_treatmentPolyandry) 0.00 0.28 -0.54 0.53 1.00 29517 6597
cor(Lipid_treatmentPolyandry,Protein_treatmentPolyandry) 0.00 0.28 -0.54 0.54 1.00 17380 6929
cor(Carbohydrate_Intercept,Protein_treatmentPolyandry) -0.00 0.28 -0.53 0.52 1.00 10970 6920
cor(Carbohydrate_treatmentPolyandry,Protein_treatmentPolyandry) -0.00 0.28 -0.54 0.54 1.00 10098 6571
cor(Protein_Intercept,Protein_treatmentPolyandry) 0.00 0.28 -0.53 0.55 1.00 9332 7782
cor(Lipid_Intercept,Glycogen_Intercept) 0.01 0.28 -0.52 0.54 1.00 27042 6990
cor(Lipid_treatmentPolyandry,Glycogen_Intercept) 0.00 0.28 -0.53 0.54 1.00 17579 7248
cor(Carbohydrate_Intercept,Glycogen_Intercept) -0.00 0.28 -0.54 0.54 1.00 14215 6487
cor(Carbohydrate_treatmentPolyandry,Glycogen_Intercept) -0.00 0.28 -0.53 0.52 1.00 10812 7605
cor(Protein_Intercept,Glycogen_Intercept) 0.00 0.28 -0.53 0.53 1.00 8863 7173
cor(Protein_treatmentPolyandry,Glycogen_Intercept) -0.00 0.27 -0.53 0.53 1.00 7004 7339
cor(Lipid_Intercept,Glycogen_treatmentPolyandry) -0.00 0.27 -0.52 0.53 1.00 25535 7050
cor(Lipid_treatmentPolyandry,Glycogen_treatmentPolyandry) -0.00 0.28 -0.53 0.52 1.00 18911 6298
cor(Carbohydrate_Intercept,Glycogen_treatmentPolyandry) -0.00 0.27 -0.52 0.52 1.00 13791 6655
cor(Carbohydrate_treatmentPolyandry,Glycogen_treatmentPolyandry) -0.00 0.28 -0.54 0.52 1.00 10659 7370
cor(Protein_Intercept,Glycogen_treatmentPolyandry) -0.01 0.28 -0.53 0.53 1.00 8603 7328
cor(Protein_treatmentPolyandry,Glycogen_treatmentPolyandry) 0.00 0.28 -0.54 0.53 1.00 7730 7307
cor(Glycogen_Intercept,Glycogen_treatmentPolyandry) 0.00 0.28 -0.53 0.54 1.00 6596 7278
cor(Lipid_Intercept,Chitin_Intercept) 0.00 0.28 -0.54 0.53 1.00 20916 6278
cor(Lipid_treatmentPolyandry,Chitin_Intercept) -0.00 0.28 -0.53 0.53 1.00 15610 7353
cor(Carbohydrate_Intercept,Chitin_Intercept) -0.00 0.28 -0.53 0.53 1.00 12856 7884
cor(Carbohydrate_treatmentPolyandry,Chitin_Intercept) -0.01 0.28 -0.55 0.53 1.00 10129 6817
cor(Protein_Intercept,Chitin_Intercept) 0.00 0.27 -0.51 0.52 1.00 8901 8412
cor(Protein_treatmentPolyandry,Chitin_Intercept) -0.00 0.28 -0.54 0.53 1.00 7775 6917
cor(Glycogen_Intercept,Chitin_Intercept) 0.00 0.28 -0.54 0.54 1.00 6654 7446
cor(Glycogen_treatmentPolyandry,Chitin_Intercept) 0.00 0.28 -0.53 0.54 1.00 6415 7770
cor(Lipid_Intercept,Chitin_treatmentPolyandry) -0.01 0.28 -0.53 0.52 1.00 23692 7056
cor(Lipid_treatmentPolyandry,Chitin_treatmentPolyandry) -0.00 0.28 -0.54 0.53 1.00 18295 6580
cor(Carbohydrate_Intercept,Chitin_treatmentPolyandry) -0.00 0.28 -0.54 0.53 1.00 14640 7469
cor(Carbohydrate_treatmentPolyandry,Chitin_treatmentPolyandry) -0.00 0.28 -0.53 0.55 1.00 10164 6719
cor(Protein_Intercept,Chitin_treatmentPolyandry) 0.00 0.28 -0.54 0.54 1.00 8566 7279
cor(Protein_treatmentPolyandry,Chitin_treatmentPolyandry) 0.00 0.28 -0.52 0.53 1.00 7797 7033
cor(Glycogen_Intercept,Chitin_treatmentPolyandry) -0.01 0.28 -0.53 0.53 1.00 7348 7423
cor(Glycogen_treatmentPolyandry,Chitin_treatmentPolyandry) -0.00 0.28 -0.54 0.53 1.00 5695 7100
cor(Chitin_Intercept,Chitin_treatmentPolyandry) -0.01 0.28 -0.54 0.53 1.00 6015 7267
cor(Lipid_Intercept,Dryweight_Intercept) -0.00 0.27 -0.52 0.52 1.00 20265 8172
cor(Lipid_treatmentPolyandry,Dryweight_Intercept) 0.00 0.28 -0.53 0.53 1.00 13293 7369
cor(Carbohydrate_Intercept,Dryweight_Intercept) 0.01 0.28 -0.53 0.53 1.00 13575 7373
cor(Carbohydrate_treatmentPolyandry,Dryweight_Intercept) 0.01 0.28 -0.53 0.55 1.00 10101 7729
cor(Protein_Intercept,Dryweight_Intercept) 0.00 0.27 -0.52 0.52 1.00 9004 8106
cor(Protein_treatmentPolyandry,Dryweight_Intercept) -0.00 0.28 -0.53 0.53 1.00 7796 7585
cor(Glycogen_Intercept,Dryweight_Intercept) -0.00 0.28 -0.53 0.54 1.00 6844 8068
cor(Glycogen_treatmentPolyandry,Dryweight_Intercept) -0.00 0.28 -0.53 0.52 1.00 6176 7400
cor(Chitin_Intercept,Dryweight_Intercept) -0.01 0.28 -0.55 0.53 1.00 6064 8432
cor(Chitin_treatmentPolyandry,Dryweight_Intercept) -0.00 0.28 -0.53 0.53 1.00 5531 7765
cor(Lipid_Intercept,Dryweight_treatmentPolyandry) 0.00 0.28 -0.53 0.54 1.00 24837 7010
cor(Lipid_treatmentPolyandry,Dryweight_treatmentPolyandry) 0.00 0.28 -0.53 0.54 1.00 17859 6856
cor(Carbohydrate_Intercept,Dryweight_treatmentPolyandry) 0.01 0.28 -0.53 0.54 1.00 12990 6799
cor(Carbohydrate_treatmentPolyandry,Dryweight_treatmentPolyandry) 0.01 0.28 -0.55 0.55 1.00 12074 6870
cor(Protein_Intercept,Dryweight_treatmentPolyandry) -0.00 0.28 -0.54 0.53 1.00 9697 7393
cor(Protein_treatmentPolyandry,Dryweight_treatmentPolyandry) 0.00 0.28 -0.53 0.53 1.00 6890 6938
cor(Glycogen_Intercept,Dryweight_treatmentPolyandry) -0.00 0.28 -0.53 0.53 1.00 6833 7580
cor(Glycogen_treatmentPolyandry,Dryweight_treatmentPolyandry) -0.00 0.28 -0.52 0.53 1.00 6143 7092
cor(Chitin_Intercept,Dryweight_treatmentPolyandry) -0.00 0.28 -0.52 0.53 1.00 5544 7050
cor(Chitin_treatmentPolyandry,Dryweight_treatmentPolyandry) -0.01 0.27 -0.54 0.52 1.00 5326 7371
cor(Dryweight_Intercept,Dryweight_treatmentPolyandry) 0.01 0.27 -0.52 0.53 1.00 4403 5906
Population-Level Effects:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
Lipid_Intercept -0.20 0.35 -0.86 0.48 1.00 7669 7609
Carbohydrate_Intercept 0.21 0.39 -0.55 0.99 1.00 8009 8804
Protein_Intercept 0.11 0.39 -0.68 0.87 1.00 10507 8790
Glycogen_Intercept -0.02 0.37 -0.78 0.69 1.00 9087 8075
Chitin_Intercept -0.13 0.36 -0.83 0.60 1.00 7618 8104
Dryweight_Intercept 0.62 0.11 0.40 0.84 1.00 10491 8371
Lipid_sexMale -0.03 0.73 -1.46 1.38 1.00 6329 7775
Lipid_treatmentPolyandry 0.44 0.38 -0.31 1.19 1.00 7294 7481
Lipid_Dryweight 0.61 0.45 -0.26 1.49 1.00 5591 7401
Lipid_sexMale:treatmentPolyandry -0.03 0.42 -0.88 0.78 1.00 9360 7929
Carbohydrate_sexMale 0.13 0.79 -1.42 1.67 1.00 6341 8036
Carbohydrate_treatmentPolyandry -0.31 0.45 -1.18 0.56 1.00 8096 7855
Carbohydrate_Dryweight 0.16 0.48 -0.78 1.12 1.00 5358 7022
Carbohydrate_sexMale:treatmentPolyandry -0.47 0.51 -1.46 0.54 1.00 10099 8210
Protein_sexMale -0.13 0.78 -1.62 1.42 1.00 7981 8072
Protein_treatmentPolyandry -0.38 0.44 -1.23 0.48 1.00 8943 8389
Protein_Dryweight -0.22 0.47 -1.14 0.71 1.00 6639 7473
Protein_sexMale:treatmentPolyandry 0.59 0.52 -0.43 1.58 1.00 10436 8306
Glycogen_sexMale -0.43 0.77 -1.91 1.10 1.00 7808 7720
Glycogen_treatmentPolyandry 0.17 0.41 -0.62 0.98 1.00 9241 8627
Glycogen_Dryweight 0.33 0.47 -0.57 1.24 1.00 6423 7220
Glycogen_sexMale:treatmentPolyandry 0.60 0.48 -0.37 1.55 1.00 11663 8133
Chitin_sexMale 0.55 0.76 -0.97 2.03 1.00 6451 7695
Chitin_treatmentPolyandry -0.06 0.40 -0.82 0.74 1.00 7461 8412
Chitin_Dryweight -0.47 0.47 -1.39 0.42 1.00 5563 7405
Chitin_sexMale:treatmentPolyandry -0.45 0.45 -1.35 0.43 1.00 9817 8535
Dryweight_sexMale -1.59 0.14 -1.86 -1.31 1.00 11621 7897
Dryweight_treatmentPolyandry 0.55 0.16 0.23 0.85 1.00 9733 8020
Dryweight_sexMale:treatmentPolyandry -0.40 0.19 -0.78 -0.01 1.00 9984 7861
Family Specific Parameters:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
sigma_Lipid 0.75 0.09 0.59 0.95 1.00 8716 7203
sigma_Carbohydrate 1.01 0.12 0.81 1.27 1.00 9670 8036
sigma_Protein 1.03 0.12 0.82 1.30 1.00 12465 6467
sigma_Glycogen 0.95 0.11 0.77 1.19 1.00 14771 7813
sigma_Chitin 0.85 0.11 0.67 1.10 1.00 8525 8148
sigma_Dryweight 0.36 0.04 0.29 0.46 1.00 11112 8214
Residual Correlations:
Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
rescor(Lipid,Carbohydrate) -0.32 0.15 -0.59 -0.02 1.00 7428 8208
rescor(Lipid,Protein) -0.05 0.15 -0.35 0.25 1.00 15061 8297
rescor(Carbohydrate,Protein) 0.04 0.15 -0.25 0.32 1.00 12117 8047
rescor(Lipid,Glycogen) 0.03 0.15 -0.28 0.33 1.00 6829 7772
rescor(Carbohydrate,Glycogen) 0.00 0.15 -0.29 0.28 1.00 13511 7694
rescor(Protein,Glycogen) -0.14 0.15 -0.42 0.15 1.00 13684 8006
rescor(Lipid,Chitin) -0.07 0.16 -0.37 0.24 1.00 10168 8151
rescor(Carbohydrate,Chitin) -0.42 0.13 -0.65 -0.14 1.00 10054 8030
rescor(Protein,Chitin) 0.08 0.15 -0.23 0.37 1.00 11453 7923
rescor(Glycogen,Chitin) -0.02 0.15 -0.31 0.27 1.00 9727 7695
rescor(Lipid,Dryweight) 0.12 0.23 -0.35 0.55 1.00 5503 7273
rescor(Carbohydrate,Dryweight) -0.02 0.22 -0.45 0.40 1.00 5119 7464
rescor(Protein,Dryweight) -0.03 0.21 -0.42 0.37 1.00 6002 7734
rescor(Glycogen,Dryweight) 0.01 0.21 -0.41 0.42 1.00 5646 6996
rescor(Chitin,Dryweight) 0.24 0.22 -0.21 0.63 1.00 4934 6998
Samples were drawn using sampling(NUTS). For each parameter, Bulk_ESS
and Tail_ESS are effective sample size measures, and Rhat is the potential
scale reduction factor on split chains (at convergence, Rhat = 1).
Posterior effect size of treatment on metabolite abundance, for each sex
Here, we use the model to predict the mean concentration of each metabolite (in standard units) in each treatment and sex (averaged across the eight replicate selection lines). We then calculate the effect size of treatment by subtracting the (sex-specific) mean for the M treatment from the mean for the P treatment; thus a value of 1 would mean that the P treatment has a mean that is larger by 1 standard deviation. Thus, the y-axis in the following graphs essentially shows the posterior estimate of standardised effect size (Cohen’s d), from the model shown above.
Because the model contains dry weight as a mediator variable, we created these predictions two different ways, and display the answer for both using tabs in the following figures/tables. Firstly, we predicted the means controlling for differences in dry weight between sexes and treatments; this was done by deriving the predictions dry weight set to its global mean, for both sexes and treatments. Secondly, we derived predictions without controlling for dry weight. This was done by deriving the predictions with dry weight set to its average value for the appropriate treatment-sex combination.
Figure
Controlling for differences in dry weight between sexes and treatments
new <- expand_grid(sex = c("Male", "Female"),
treatment = c("Monogamy", "Polyandry"),
Dryweight = NA, line = NA) %>%
mutate(type = 1:n())
levels <- c("Carbohydrate", "Chitin", "Glycogen", "Lipid", "Protein", "Dryweight")
# Estimate mean dry weight for each of the 4 sex/treatment combinations
evolved_mean_dryweights <- data.frame(
new[,1:2],
fitted(brms_metabolite_SEM, re_formula = NA,
newdata = new %>% select(-Dryweight),
summary = TRUE, resp = "Dryweight")) %>%
as_tibble()
# Find the mean dry weight across all sexes/treatments
grand_mean_dryweight <- mean(evolved_mean_dryweights$Estimate)
new_metabolites <- bind_rows(
expand_grid(sex = c("Male", "Female"),
treatment = c("Monogamy", "Polyandry"),
Dryweight = grand_mean_dryweight, line = NA) %>%
mutate(type = 1:4),
evolved_mean_dryweights %>% select(sex, treatment, Dryweight = Estimate) %>%
mutate(line = NA, type = 5:8)
)
# Predict data from the SEM of metabolites...
# Because we use sum contrasts for "line" and line=NA in the new data,
# this function predicts at the global means across the 4 lines (see ?posterior_epred)
fitted_values <- posterior_epred(
brms_metabolite_SEM, newdata = new_metabolites, re_formula = NA,
summary = FALSE, resp = c("Carbohydrate", "Chitin", "Glycogen", "Lipid", "Protein")) %>%
reshape2::melt() %>% rename(draw = Var1, type = Var2, variable = Var3) %>%
as_tibble() %>%
left_join(new_metabolites, by = "type") %>%
select(draw, variable, value, sex, treatment, Dryweight) %>%
mutate(variable = factor(variable, levels))
treat_diff_standard_dryweight <- fitted_values %>%
filter(Dryweight == grand_mean_dryweight) %>%
spread(treatment, value) %>%
mutate(`Difference in means (Poly - Mono)` = Polyandry - Monogamy)
treat_diff_actual_dryweight <- fitted_values %>%
filter(Dryweight != grand_mean_dryweight) %>%
select(-Dryweight) %>%
spread(treatment, value) %>%
mutate(`Difference in means (Poly - Mono)` = Polyandry - Monogamy)
summary_dat1 <- treat_diff_standard_dryweight %>%
filter(variable != 'Dryweight') %>%
rename(x = `Difference in means (Poly - Mono)`) %>%
group_by(variable, sex) %>%
summarise(`Difference in means (Poly - Mono)` = median(x),
`Lower 95% CI` = quantile(x, probs = 0.025),
`Upper 95% CI` = quantile(x, probs = 0.975),
p = 1 - as.numeric(bayestestR::p_direction(x)),
` ` = ifelse(p < 0.05, "*", ""),
.groups = "drop")
summary_dat2 <- treat_diff_actual_dryweight %>%
filter(variable != 'Dryweight') %>%
rename(x = `Difference in means (Poly - Mono)`) %>%
group_by(variable, sex) %>%
summarise(`Difference in means (Poly - Mono)` = median(x),
`Lower 95% CI` = quantile(x, probs = 0.025),
`Upper 95% CI` = quantile(x, probs = 0.975),
p = 1 - as.numeric(bayestestR::p_direction(x)),
` ` = ifelse(p < 0.05, "*", ""),
.groups = "drop")
sampled_draws <- sample(unique(fitted_values$draw), 100)
treat_diff_standard_dryweight %>%
filter(variable != 'Dryweight') %>%
ggplot(aes(x = sex, y = `Difference in means (Poly - Mono)`,fill = sex)) +
geom_hline(yintercept = 0, linetype = 2) +
stat_halfeye() +
geom_line(data = treat_diff_standard_dryweight %>%
filter(draw %in% sampled_draws) %>%
filter(variable != 'Dryweight'),
alpha = 0.8, size = 0.12, colour = "black", aes(group = draw)) +
geom_point(data = summary_dat1, pch = 21, colour = "black", size = 3.1) +
scale_fill_brewer(palette = 'Pastel1', direction = 1, name = "") +
scale_colour_brewer(palette = 'Pastel1', direction = 1, name = "") +
facet_wrap( ~ variable, nrow = 1) +
theme_bw() +
theme(legend.position = 'none',
strip.background = element_blank(),
panel.grid.major.x = element_blank()) +
coord_cartesian(ylim = c(-2.2, 2.2)) +
ylab("Difference in means between\nselection treatments (P - M)") + xlab("Sex")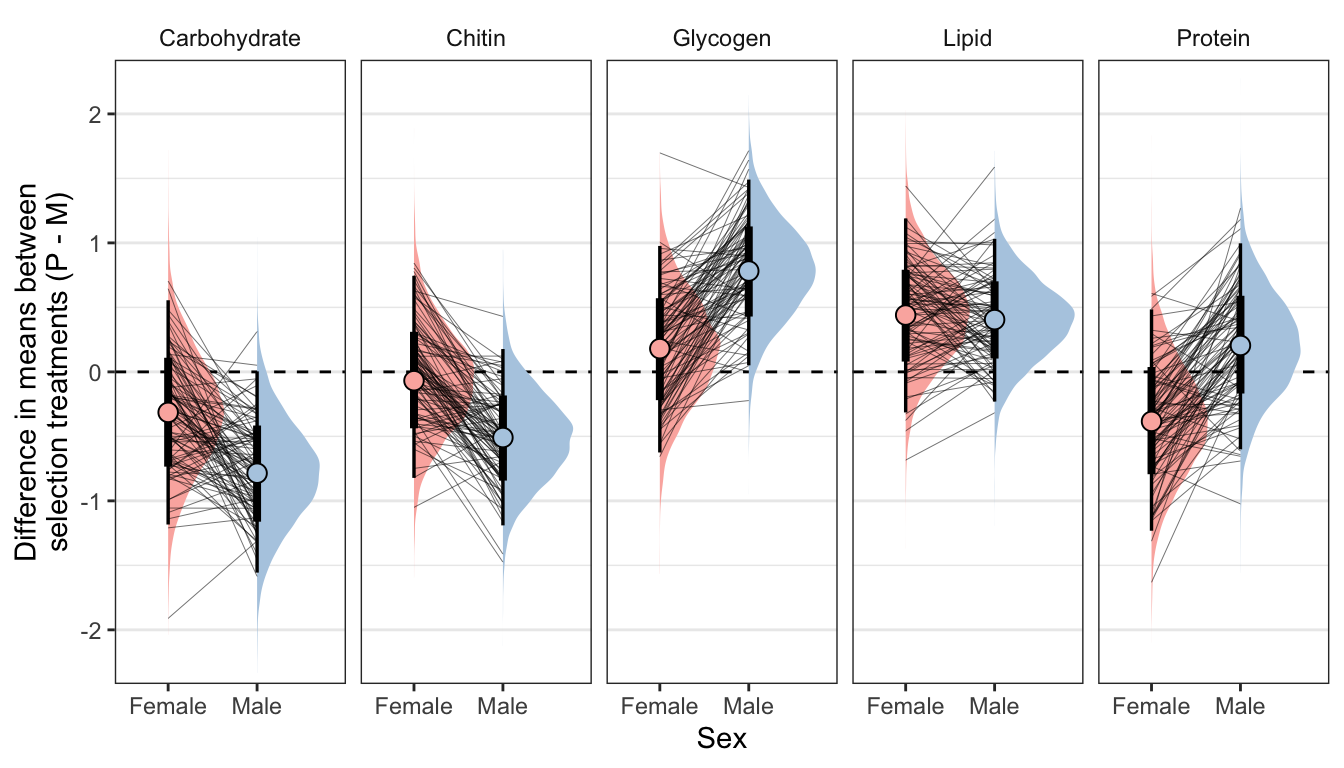
Not controlling for differences in dry weight between sexes and treatments
treat_diff_actual_dryweight %>%
filter(variable != 'Dryweight') %>%
ggplot(aes(x = sex, y = `Difference in means (Poly - Mono)`,fill = sex)) +
geom_hline(yintercept = 0, linetype = 2) +
stat_halfeye() +
geom_line(data = treat_diff_actual_dryweight %>%
filter(draw %in% sampled_draws) %>%
filter(variable != 'Dryweight'),
alpha = 0.8, size = 0.12, colour = "black", aes(group = draw)) +
geom_point(data = summary_dat2, pch = 21, colour = "black", size = 3.1) +
scale_fill_brewer(palette = 'Pastel1', direction = 1, name = "") +
scale_colour_brewer(palette = 'Pastel1', direction = 1, name = "") +
facet_wrap( ~ variable, nrow = 1) +
theme_bw() +
theme(legend.position = 'none',
strip.background = element_blank(),
panel.grid.major.x = element_blank()) +
coord_cartesian(ylim = c(-1.8, 1.8)) +
ylab("Difference in means between\nselection treatments (P - M)") + xlab("Sex")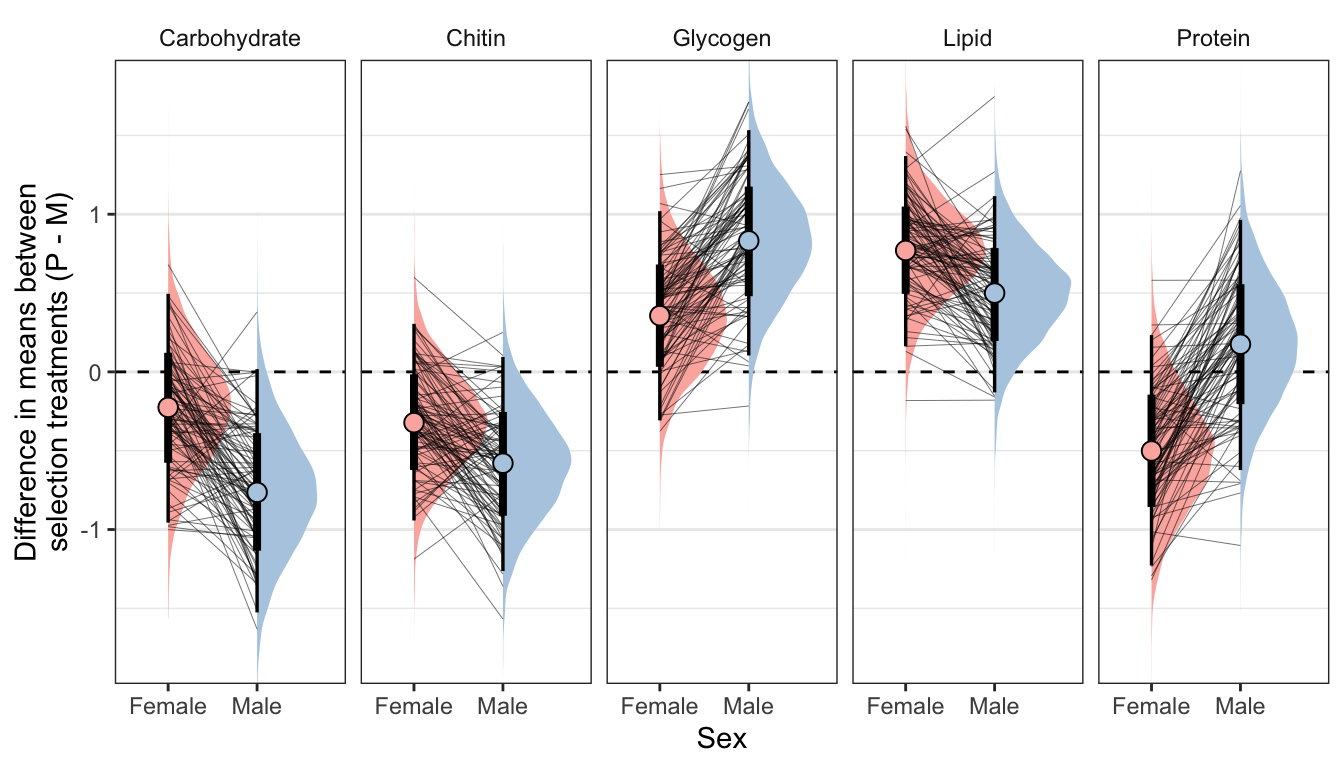
Table
Controlling for differences in dry weight between sexes and treatments
summary_dat1 %>%
kable(digits=3) %>%
kable_styling(full_width = FALSE)| variable | sex | Difference in means (Poly - Mono) | Lower 95% CI | Upper 95% CI | p | |
|---|---|---|---|---|---|---|
| Carbohydrate | Female | -0.314 | -1.183 | 0.555 | 0.237 | |
| Carbohydrate | Male | -0.785 | -1.556 | 0.004 | 0.025 |
|
| Chitin | Female | -0.068 | -0.822 | 0.745 | 0.435 | |
| Chitin | Male | -0.509 | -1.190 | 0.177 | 0.069 | |
| Glycogen | Female | 0.180 | -0.625 | 0.976 | 0.335 | |
| Glycogen | Male | 0.782 | 0.051 | 1.491 | 0.018 |
|
| Lipid | Female | 0.440 | -0.314 | 1.189 | 0.122 | |
| Lipid | Male | 0.405 | -0.230 | 1.031 | 0.103 | |
| Protein | Female | -0.384 | -1.232 | 0.484 | 0.191 | |
| Protein | Male | 0.205 | -0.600 | 0.997 | 0.304 |
Not controlling for differences in dry weight between sexes and treatments
summary_dat2 %>%
kable(digits=3) %>%
kable_styling(full_width = FALSE)| variable | sex | Difference in means (Poly - Mono) | Lower 95% CI | Upper 95% CI | p | |
|---|---|---|---|---|---|---|
| Carbohydrate | Female | -0.225 | -0.956 | 0.495 | 0.267 | |
| Carbohydrate | Male | -0.764 | -1.526 | 0.018 | 0.027 |
|
| Chitin | Female | -0.322 | -0.943 | 0.305 | 0.158 | |
| Chitin | Male | -0.580 | -1.264 | 0.095 | 0.044 |
|
| Glycogen | Female | 0.356 | -0.307 | 1.019 | 0.146 | |
| Glycogen | Male | 0.831 | 0.104 | 1.533 | 0.013 |
|
| Lipid | Female | 0.770 | 0.162 | 1.369 | 0.009 |
|
| Lipid | Male | 0.500 | -0.130 | 1.115 | 0.058 | |
| Protein | Female | -0.502 | -1.229 | 0.234 | 0.092 | |
| Protein | Male | 0.175 | -0.623 | 0.964 | 0.332 |
Posterior difference in treatment effect size between sexes
This section essentially examines the treatment \(\times\) sex interaction term, by calculating the difference in the effect size of the P/M treatment between sexes, for each of the five metabolites. We find no strong evidence for a treatment \(\times\) sex interaction, i.e. the treatment effects did not differ detectably between sexes. However, the effect of the Polyandry treatment on glycogen concentration appears to be marginally more positive in males than females (probability of direction = 92.5%, similar to a one-sided p-value of 0.075).
Figure
Controlling for differences in dry weight between sexes and treatments
treatsex_interaction_data1 <- treat_diff_standard_dryweight %>%
select(draw, variable, sex, d = `Difference in means (Poly - Mono)`) %>%
arrange(draw, variable, sex) %>%
group_by(draw, variable) %>%
summarise(`Difference in effect size between sexes` = d[2] - d[1],
.groups = "drop") # males - females
treatsex_interaction_data1 %>%
filter(variable != 'Dryweight') %>%
ggplot(aes(x = `Difference in effect size between sexes`, y = 1, fill = stat(x < 0))) +
geom_vline(xintercept = 0, linetype = 2) +
stat_halfeyeh() +
facet_wrap( ~ variable) +
scale_fill_brewer(palette = 'Pastel2', direction = 1, name = "") +
theme_bw() +
theme(legend.position = 'none',
strip.background = element_blank()) +
ylab("Posterior density")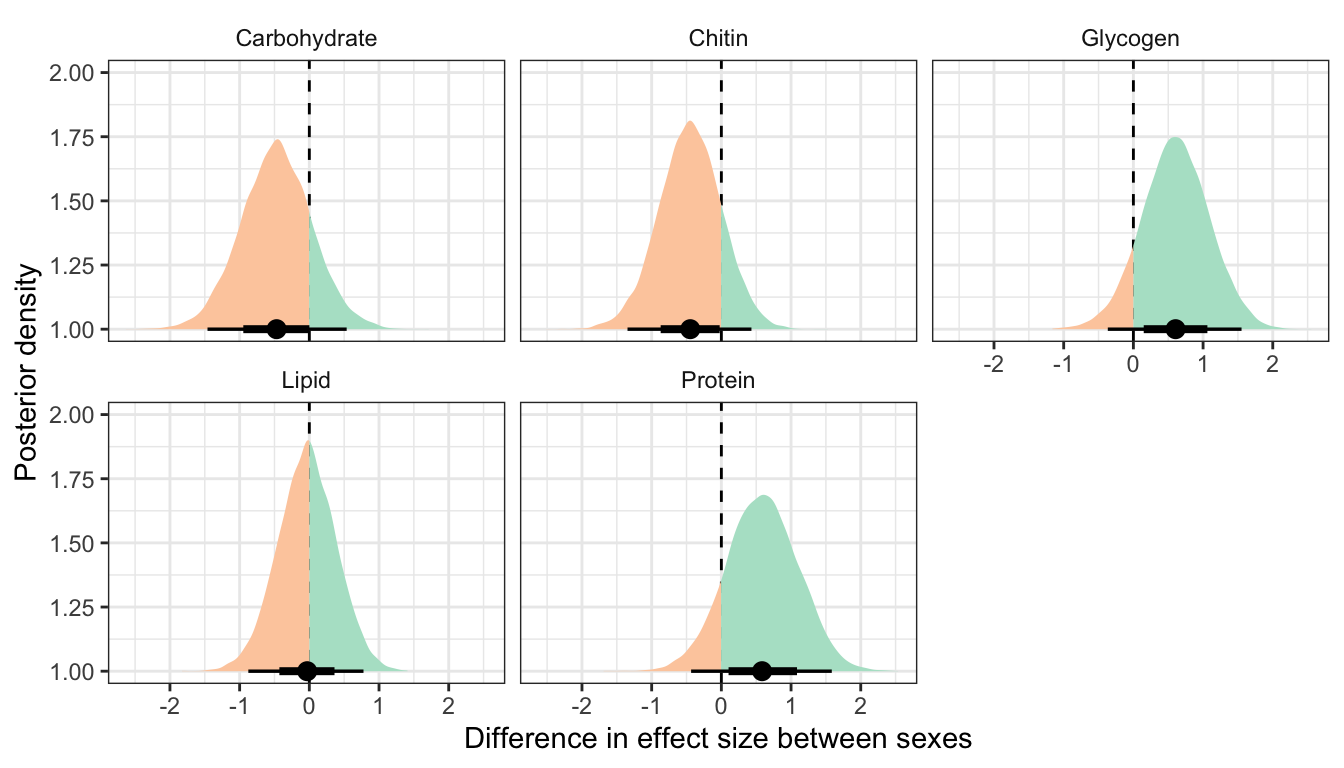
Not controlling for differences in dry weight between sexes and treatments
treatsex_interaction_data2 <- treat_diff_actual_dryweight %>%
select(draw, variable, sex, d = `Difference in means (Poly - Mono)`) %>%
arrange(draw, variable, sex) %>%
group_by(draw, variable) %>%
summarise(`Difference in effect size between sexes` = d[2] - d[1],
.groups = "drop") # males - females
treatsex_interaction_data2 %>%
filter(variable != 'Dryweight') %>%
ggplot(aes(x = `Difference in effect size between sexes`, y = 1, fill = stat(x < 0))) +
geom_vline(xintercept = 0, linetype = 2) +
stat_halfeyeh() +
facet_wrap( ~ variable) +
scale_fill_brewer(palette = 'Pastel2', direction = 1, name = "") +
theme_bw() +
theme(legend.position = 'none',
strip.background = element_blank()) +
ylab("Posterior density")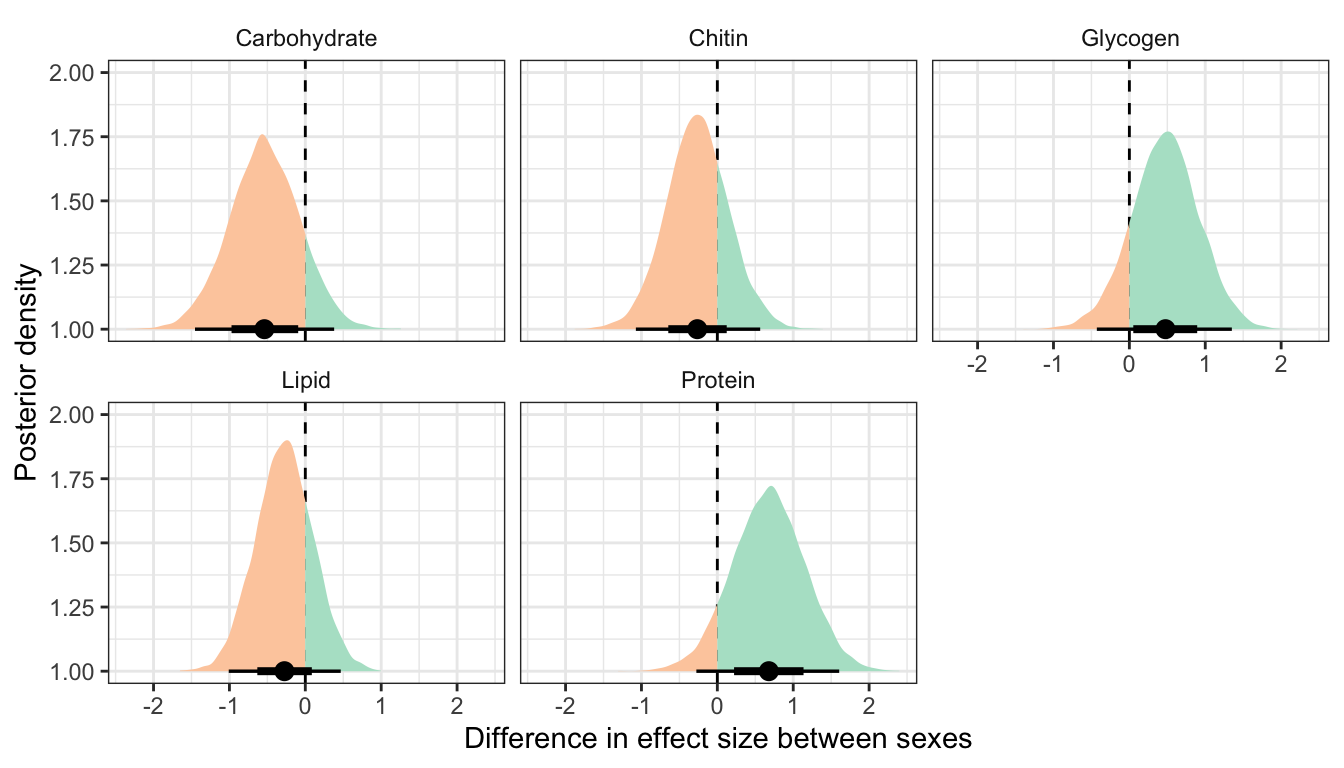
Table
Controlling for differences in dry weight between sexes and treatments
treatsex_interaction_data1 %>%
filter(variable != 'Dryweight') %>%
rename(x = `Difference in effect size between sexes`) %>%
group_by(variable) %>%
summarise(`Difference in effect size between sexes` = median(x),
`Lower 95% CI` = quantile(x, probs = 0.025),
`Upper 95% CI` = quantile(x, probs = 0.975),
p = 1 - as.numeric(bayestestR::p_direction(x)),
` ` = ifelse(p < 0.05, "*", ""),
.groups = "drop") %>%
kable(digits=3) %>%
kable_styling(full_width = FALSE)| variable | Difference in effect size between sexes | Lower 95% CI | Upper 95% CI | p | |
|---|---|---|---|---|---|
| Carbohydrate | -0.470 | -1.461 | 0.536 | 0.170 | |
| Chitin | -0.446 | -1.347 | 0.432 | 0.158 | |
| Glycogen | 0.606 | -0.367 | 1.552 | 0.105 | |
| Lipid | -0.031 | -0.875 | 0.777 | 0.468 | |
| Protein | 0.584 | -0.434 | 1.584 | 0.126 |
Not controlling for differences in dry weight between sexes and treatments
treatsex_interaction_data2 %>%
filter(variable != 'Dryweight') %>%
rename(x = `Difference in effect size between sexes`) %>%
group_by(variable) %>%
summarise(`Difference in effect size between sexes` = median(x),
`Lower 95% CI` = quantile(x, probs = 0.025),
`Upper 95% CI` = quantile(x, probs = 0.975),
p = 1 - as.numeric(bayestestR::p_direction(x)),
` ` = ifelse(p < 0.05, "*", ""),
.groups = "drop") %>%
kable(digits=3) %>%
kable_styling(full_width = FALSE)| variable | Difference in effect size between sexes | Lower 95% CI | Upper 95% CI | p | |
|---|---|---|---|---|---|
| Carbohydrate | -0.539 | -1.454 | 0.380 | 0.124 | |
| Chitin | -0.265 | -1.074 | 0.565 | 0.257 | |
| Glycogen | 0.476 | -0.429 | 1.350 | 0.142 | |
| Lipid | -0.275 | -1.009 | 0.467 | 0.232 | |
| Protein | 0.679 | -0.276 | 1.606 | 0.081 |
sessionInfo()R version 4.0.3 (2020-10-10) Platform: x86_64-apple-darwin17.0 (64-bit) Running under: macOS Catalina 10.15.4 Matrix products: default BLAS: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRblas.dylib LAPACK: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRlapack.dylib locale: [1] en_AU.UTF-8/en_AU.UTF-8/en_AU.UTF-8/C/en_AU.UTF-8/en_AU.UTF-8 attached base packages: [1] stats graphics grDevices utils datasets methods base other attached packages: [1] knitrhooks_0.0.4 knitr_1.30 kableExtra_1.1.0 DT_0.13 tidybayes_2.0.3 brms_2.14.4 Rcpp_1.0.4.6 ggridges_0.5.2 gridExtra_2.3 [10] GGally_1.5.0 forcats_0.5.0 stringr_1.4.0 dplyr_1.0.0 purrr_0.3.4 readr_1.3.1 tidyr_1.1.0 tibble_3.0.1 ggplot2_3.3.2 [19] tidyverse_1.3.0 workflowr_1.6.2 loaded via a namespace (and not attached): [1] readxl_1.3.1 backports_1.1.7 plyr_1.8.6 igraph_1.2.5 svUnit_1.0.3 splines_4.0.3 crosstalk_1.1.0.1 [8] TH.data_1.0-10 rstantools_2.1.1 inline_0.3.15 digest_0.6.25 htmltools_0.5.0 rsconnect_0.8.16 fansi_0.4.1 [15] magrittr_2.0.1 modelr_0.1.8 RcppParallel_5.0.1 matrixStats_0.56.0 xts_0.12-0 sandwich_2.5-1 prettyunits_1.1.1 [22] colorspace_1.4-1 blob_1.2.1 rvest_0.3.5 haven_2.3.1 xfun_0.19 callr_3.4.3 crayon_1.3.4 [29] jsonlite_1.7.0 lme4_1.1-23 survival_3.2-7 zoo_1.8-8 glue_1.4.2 gtable_0.3.0 emmeans_1.4.7 [36] webshot_0.5.2 V8_3.4.0 pkgbuild_1.0.8 rstan_2.21.2 abind_1.4-5 scales_1.1.1 mvtnorm_1.1-0 [43] DBI_1.1.0 miniUI_0.1.1.1 viridisLite_0.3.0 xtable_1.8-4 stats4_4.0.3 StanHeaders_2.21.0-3 htmlwidgets_1.5.1 [50] httr_1.4.1 DiagrammeR_1.0.6.1 threejs_0.3.3 arrayhelpers_1.1-0 RColorBrewer_1.1-2 ellipsis_0.3.1 farver_2.0.3 [57] pkgconfig_2.0.3 reshape_0.8.8 loo_2.3.1 dbplyr_1.4.4 labeling_0.3 tidyselect_1.1.0 rlang_0.4.6 [64] reshape2_1.4.4 later_1.0.0 visNetwork_2.0.9 munsell_0.5.0 cellranger_1.1.0 tools_4.0.3 cli_2.0.2 [71] generics_0.0.2 broom_0.5.6 evaluate_0.14 fastmap_1.0.1 yaml_2.2.1 processx_3.4.2 fs_1.4.1 [78] nlme_3.1-149 whisker_0.4 mime_0.9 projpred_2.0.2 xml2_1.3.2 compiler_4.0.3 bayesplot_1.7.2 [85] shinythemes_1.1.2 rstudioapi_0.11 gamm4_0.2-6 curl_4.3 reprex_0.3.0 statmod_1.4.34 stringi_1.5.3 [92] highr_0.8 ps_1.3.3 Brobdingnag_1.2-6 lattice_0.20-41 Matrix_1.2-18 nloptr_1.2.2.1 markdown_1.1 [99] shinyjs_1.1 vctrs_0.3.0 pillar_1.4.4 lifecycle_0.2.0 bridgesampling_1.0-0 estimability_1.3 insight_0.8.4 [106] httpuv_1.5.3.1 R6_2.4.1 promises_1.1.0 codetools_0.2-16 boot_1.3-25 colourpicker_1.0 MASS_7.3-53 [113] gtools_3.8.2 assertthat_0.2.1 rprojroot_1.3-2 withr_2.2.0 shinystan_2.5.0 multcomp_1.4-13 bayestestR_0.6.0 [120] mgcv_1.8-33 parallel_4.0.3 hms_0.5.3 grid_4.0.3 coda_0.19-3 minqa_1.2.4 rmarkdown_2.5 [127] git2r_0.27.1 shiny_1.4.0.2 lubridate_1.7.8 base64enc_0.1-3 dygraphs_1.1.1.6