Analysis of Experiment 3
Last updated: 2020-04-21
Checks: 7 0
Knit directory: social_immunity/
This reproducible R Markdown analysis was created with workflowr (version 1.6.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20191017) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 1ce9e19 | lukeholman | 2020-04-21 | First commit 2020 |
| html | 1ce9e19 | lukeholman | 2020-04-21 | First commit 2020 |
| Rmd | aae65cf | lukeholman | 2019-10-17 | First commit |
| html | aae65cf | lukeholman | 2019-10-17 | First commit |
Load data and R packages
# All but 1 of these packages can be easily installed from CRAN.
# However it was harder to install the showtext package. On Mac, I did this:
# installed 'homebrew' using Terminal: ruby -e "$(curl -fsSL https://raw.githubusercontent.com/Homebrew/install/master/install)"
# installed 'libpng' using Terminal: brew install libpng
# installed 'showtext' in R using: devtools::install_github("yixuan/showtext")
library(showtext)
library(brms)
library(bayesplot)
library(tidyverse)
library(gridExtra)
library(kableExtra)
library(bayestestR)
library(cowplot)
library(tidybayes)
source("code/helper_functions.R")
# set up nice font for figure
nice_font <- "PT Serif"
font_add_google(name = nice_font, family = nice_font, regular.wt = 400, bold.wt = 700)
showtext_auto()
files <- c("data/data_collection_sheets/hiveA_touching.csv",
"data/data_collection_sheets/hiveB_touching.csv",
"data/data_collection_sheets/hiveC_touching.csv",
"data/data_collection_sheets/hiveD_touching.csv")
experiment3 <- map(
files,
~ read_csv(.x) %>%
gather(minute, touching, -tube) %>%
mutate(treatment = ifelse(substr(tube, 1, 1) %in% c("A", "B"), "AB", "CD"),
minute = as.numeric(minute),
touching = as.integer(touching),
hive = gsub("hive", "", str_extract(.x, "hive[ABCD]")),
tube = paste(hive, tube, sep = "_"))
) %>% bind_rows() %>%
mutate(treatment = replace(treatment,
treatment == "AB" & hive %in% c("A", "B"),
"Ringers"),
treatment = replace(treatment,
treatment == "AB" & hive %in% c("C", "D"),
"LPS"),
treatment = replace(treatment,
treatment == "CD" & hive %in% c("A", "B"),
"LPS"),
treatment = replace(treatment,
treatment == "CD" & hive %in% c("C", "D"),
"Ringers")) %>%
mutate(hive = replace(hive, hive == "A", "Garden"),
hive = replace(hive, hive == "B", "Skylab"),
hive = replace(hive, hive == "C", "Arts"),
hive = replace(hive, hive == "D", "Zoology"))
expt3_counts <- experiment3 %>%
group_by(treatment, tube, hive) %>%
summarise(n_touching = sum(touching),
n_not_touching = sum(touching== 0),
percent = n_touching / (n_touching + n_not_touching)) %>%
ungroup() %>%
filter(!is.na(n_touching)) %>%
mutate(hive = C(factor(hive), sum)) # sum coding for hiveInspect the raw data
Sample sizes by treatment
expt3_counts %>%
group_by(treatment) %>%
summarise(n = n()) %>%
kable() %>% kable_styling(full_width = FALSE)| treatment | n |
|---|---|
| LPS | 219 |
| Ringers | 220 |
Sample sizes by treatment and hive
expt3_counts %>%
group_by(hive, treatment) %>%
summarise(n = n()) %>%
kable() %>% kable_styling(full_width = FALSE)| hive | treatment | n |
|---|---|---|
| Arts | LPS | 50 |
| Arts | Ringers | 50 |
| Garden | LPS | 70 |
| Garden | Ringers | 70 |
| Skylab | LPS | 49 |
| Skylab | Ringers | 50 |
| Zoology | LPS | 50 |
| Zoology | Ringers | 50 |
Inspect the results
expt3_counts %>%
group_by(hive, treatment) %>%
summarise(pc = mean(100 * percent),
SE = sd(percent) /sqrt(n()),
n = n()) %>%
rename(`% observations in which bees were in close contact` = pc,
Hive = hive, Treatment = treatment) %>%
kable(digits = 3) %>% kable_styling(full_width = FALSE) %>%
column_spec(3, width = "2in")| Hive | Treatment | % observations in which bees were in close contact | SE | n |
|---|---|---|---|---|
| Arts | LPS | 72.547 | 0.033 | 50 |
| Arts | Ringers | 69.509 | 0.036 | 50 |
| Garden | LPS | 68.895 | 0.037 | 70 |
| Garden | Ringers | 76.752 | 0.033 | 70 |
| Skylab | LPS | 65.653 | 0.034 | 49 |
| Skylab | Ringers | 70.509 | 0.040 | 50 |
| Zoology | LPS | 73.019 | 0.037 | 50 |
| Zoology | Ringers | 84.075 | 0.021 | 50 |
Plotted as a density histogram
Note that bees more often spent close to 100% of the observation period in contact in the control group, relative to the group treated with LPS.
raw_histogram <- expt3_counts %>%
filter(treatment == "Ringers") %>%
ggplot(aes(100 * percent,
fill = treatment)) +
geom_histogram(data = expt3_counts %>%
filter(treatment == "LPS"),
mapping = aes(y=..density..),
alpha = 0.5, bins = 11, colour = "black", linetype = 2) +
geom_histogram(mapping = aes(y=..density..),
alpha = 0.5, bins = 11,
colour = "black") +
scale_fill_brewer(palette = "Set1",
direction = 1, name = "Treatment") +
xlab("% Time in close contact") + ylab("Density") +
theme_bw() +
theme(legend.position = c(0.27, 0.8),
text = element_text(family = nice_font))
raw_histogram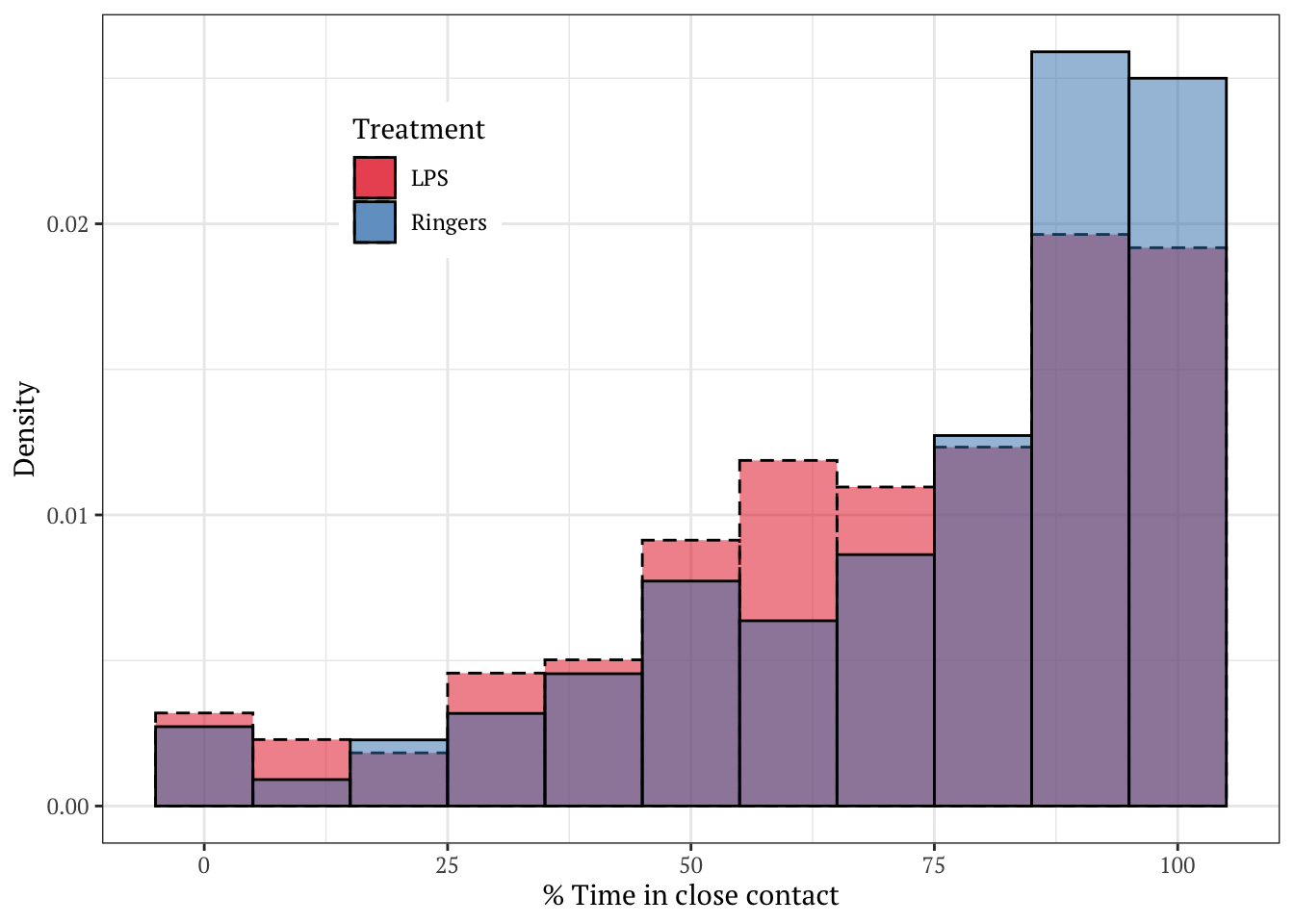
| Version | Author | Date |
|---|---|---|
| 1ce9e19 | lukeholman | 2020-04-21 |
Binomial model of time spent in contact
Run the models
Fit a binomial model, where the response is either a 0 (if bees were not in contact) or 1 (if they were). To assess the effects of our predictor variables, we compare 5 models with different fixed factors, ranking them by posterior model probability.
# new verison....
if(!file.exists("output/exp3_model.rds")){
exp3_model_v1 <- brm(
n_touching | trials(n) ~ treatment * hive + (1 | tube),
data = expt3_counts %>% mutate(n = n_touching + n_not_touching),
prior = c(set_prior("normal(0, 3)", class = "b")),
family = "binomial", save_all_pars = TRUE, sample_prior = TRUE,
chains = 4, cores = 1, iter = 10000, seed = 1)
exp3_model_v2 <- brm(
n_touching | trials(n) ~ treatment + hive + (1 | tube),
data = expt3_counts %>% mutate(n = n_touching + n_not_touching),
prior = c(set_prior("normal(0, 3)", class = "b")),
family = "binomial", save_all_pars = TRUE, sample_prior = TRUE,
chains = 4, cores = 1, iter = 10000, seed = 1)
exp3_model_v3 <- brm(
n_touching | trials(n) ~ hive + (1 | tube),
data = expt3_counts %>% mutate(n = n_touching + n_not_touching),
prior = c(set_prior("normal(0, 3)", class = "b")),
family = "binomial", save_all_pars = TRUE, sample_prior = TRUE,
chains = 4, cores = 1, iter = 10000, seed = 1)
posterior_model_probabilities <- tibble(
Model = c("treatment * hive + observation_time_minutes",
"treatment + hive + observation_time_minutes",
"hive + observation_time_minutes"),
post_prob = as.numeric(post_prob(exp3_model_v1,
exp3_model_v2,
exp3_model_v3))) %>%
arrange(-post_prob)
saveRDS(exp3_model_v2, "output/exp3_model.rds") # save the top model, treatment + hive
saveRDS(posterior_model_probabilities, "output/exp3_post_prob.rds")
}
exp3_model <- readRDS("output/exp3_model.rds")
model_probabilities <- readRDS("output/exp3_post_prob.rds")Posterior model probabilites
model_probabilities %>%
kable(digits = 3) %>% kable_styling(full_width = FALSE)| Model | post_prob |
|---|---|
| hive + observation_time_minutes | 0.794 |
| treatment + hive + observation_time_minutes | 0.205 |
| treatment * hive + observation_time_minutes | 0.001 |
Fixed effects from the top non-null model
get_fixed_effects_with_p_values(exp3_model) %>%
kable(digits = 3) %>% kable_styling()| Parameter | Estimate | Est.Error | lower_95_CI | upper_95_CI | Rhat | Bulk_ESS | Tail_ESS | p | |
|---|---|---|---|---|---|---|---|---|---|
| Intercept | 1.304 | 0.139 | 1.030 | 1.579 | 1.009 | 644 | 1105 | 0.000 | *** |
| treatmentRingers | 0.371 | 0.194 | -0.023 | 0.750 | 1.009 | 644 | 1245 | 0.031 | * |
| hive1 | -0.194 | 0.177 | -0.537 | 0.156 | 1.005 | 457 | 706 | 0.131 | |
| hive2 | 0.146 | 0.164 | -0.176 | 0.477 | 1.016 | 563 | 1057 | 0.188 | |
| hive3 | -0.223 | 0.177 | -0.567 | 0.131 | 1.012 | 432 | 981 | 0.106 |
movement_files <- c("data/data_collection_sheets/hiveA_movement.csv",
"data/data_collection_sheets/hiveB_movement.csv",
"data/data_collection_sheets/hiveC_movement.csv",
"data/data_collection_sheets/hiveD_movement.csv")
movement <- map(1:4, ~ read_csv(movement_files[.x]) %>%
mutate(hive = c("A", "B", "C", "D")[.x]) %>%
gather(video, movement, -hive, -tube)) %>%
bind_rows() %>%
mutate(treatment = ifelse(substr(tube, 1, 1) %in% c("A", "B"), "AB", "CD")) %>%
mutate(treatment = replace(treatment, treatment == "AB" & hive %in% c("A", "B"), "Ringers"),
treatment = replace(treatment, treatment == "AB" & hive %in% c("C", "D"), "LPS"),
treatment = replace(treatment, treatment == "CD" & hive %in% c("A", "B"), "LPS"),
treatment = replace(treatment, treatment == "CD" & hive %in% c("C", "D"), "Ringers")) %>%
split(.$hive) %>%
map(~ .x %>% arrange(video) %>%
mutate(video = as.numeric(factor(video, unique(video))))) %>%
bind_rows() %>%
mutate(hive = replace(hive, hive == "A", "Garden"),
hive = replace(hive, hive == "B", "Skylab"),
hive = replace(hive, hive == "C", "Arts"),
hive = replace(hive, hive == "D", "Zoology"))
movement %>%
mutate(movement = factor(movement)) %>%
group_by(movement, treatment) %>%
summarise(n=n()) %>%
ggplot(aes(movement, n, fill = treatment)) +
geom_bar(stat = "identity", colour = "grey20", position = "dodge") +
scale_fill_brewer(palette="Pastel2", direction = 1)
library(brms)
mod <- brm(movement | cat(3) ~ treatment + (1 | tube),
data = movement %>% mutate(movement = movement + 1), family = cumulative())
marginal_effects(mod, categorical = T)
new <- expand.grid(hive = c("Arts", "Garden", "Skylab", "Zoology"),
treatment = c("Ringers", "LPS"))
data.frame(new, rbind(fitted(mod, newdata = new, re_formula = NA)[,,1],
fitted(mod, newdata = new, re_formula = NA)[,,2],
fitted(mod, newdata = new, re_formula = NA)[,,3])) %>%
mutate(movement = factor(rep(0:2, each = nrow(new)))) %>%
ggplot(aes(movement, Estimate, ymin = Q2.5, ymax=Q97.5, colour = treatment)) +
geom_errorbar(position = position_dodge(0.2), width = 0) +
geom_point(position = position_dodge(0.2)) +
facet_wrap(~ hive)Plotting estimates from the model
# new test...
new <- expt3_counts %>%
select(treatment) %>% distinct() %>%
mutate(n = 100, key = paste("V", 1:n(), sep = ""),
hive = NA)
plotting_data <- as.data.frame(fitted(exp3_model,
newdata=new, re_formula = NA, summary = FALSE))
names(plotting_data) <- c("LPS", "Ringers")
plotting_data <- plotting_data %>% gather(treatment, percent_time_in_contact)
cols <- c("#34558b", "#4ec5a5", "#ffaf12")
dot_plot <- plotting_data %>%
mutate(treatment = factor(treatment, c("Ringers", "LPS"))) %>%
ggplot(aes(percent_time_in_contact, treatment)) +
stat_dotsh(quantiles = 100, fill = "grey40", colour = "grey40") +
stat_pointintervalh(
mapping = aes(colour = treatment, fill = treatment),
.width = c(0.5, 0.95),
position = position_nudge(y = -0.07),
point_colour = "grey26", pch = 21, stroke = 0.4) +
xlab("Mean % time in close contact") + ylab("Treatment") +
scale_colour_brewer(palette = "Pastel1",
direction = -1, name = "Treatment") +
scale_fill_brewer(palette = "Pastel1",
direction = -1, name = "Treatment") +
theme_bw() +
coord_cartesian(ylim=c(1.4, 2.4)) +
theme(
text = element_text(family = nice_font),
strip.background = element_rect(fill = "#eff0f1"),
panel.grid.major.y = element_blank(),
legend.position = "none"
)
# positive effect = odds of this outcome are higher for trt2 than trt1 (put control as trt1)
get_log_odds <- function(trt1, trt2){
log((trt2 / (1 - trt2) / (trt1 / (1 - trt1))))
}
LOR <- plotting_data %>%
mutate(posterior_sample = rep(1:(n()/2), 2)) %>%
spread(treatment, percent_time_in_contact) %>%
mutate(LOR = get_log_odds(Ringers/100, LPS/100)) %>%
select(LOR)
LOR_plot <- LOR %>%
ggplot(aes(LOR, y =1)) +
geom_vline(xintercept = 0, linetype = 2) +
stat_dotsh(quantiles = 100, fill = "grey40", colour = "grey40") +
stat_pointintervalh(
colour = "orange", fill = "orange",
.width = c(0.5, 0.95),
position = position_nudge(y = -0.1),
point_colour = "grey26", pch = 21, stroke = 0.4) +
coord_cartesian(ylim=c(0.86, 2)) +
xlab("Effect of LPS on % time\nin close contact (LOR)") +
ylab("Posterior density") +
theme_bw() +
theme(
text = element_text(family = nice_font),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
panel.grid.major.y = element_blank(),
panel.grid.minor.y = element_blank(),
legend.position = "none"
)
p <- cowplot::plot_grid(raw_histogram,
dot_plot, LOR_plot, labels = c("A", "B", "C"),
nrow = 1, align = 'h', axis = 'l')
ggsave(plot = p, filename = "figures/fig3.pdf", height = 3.2, width = 8.6)
p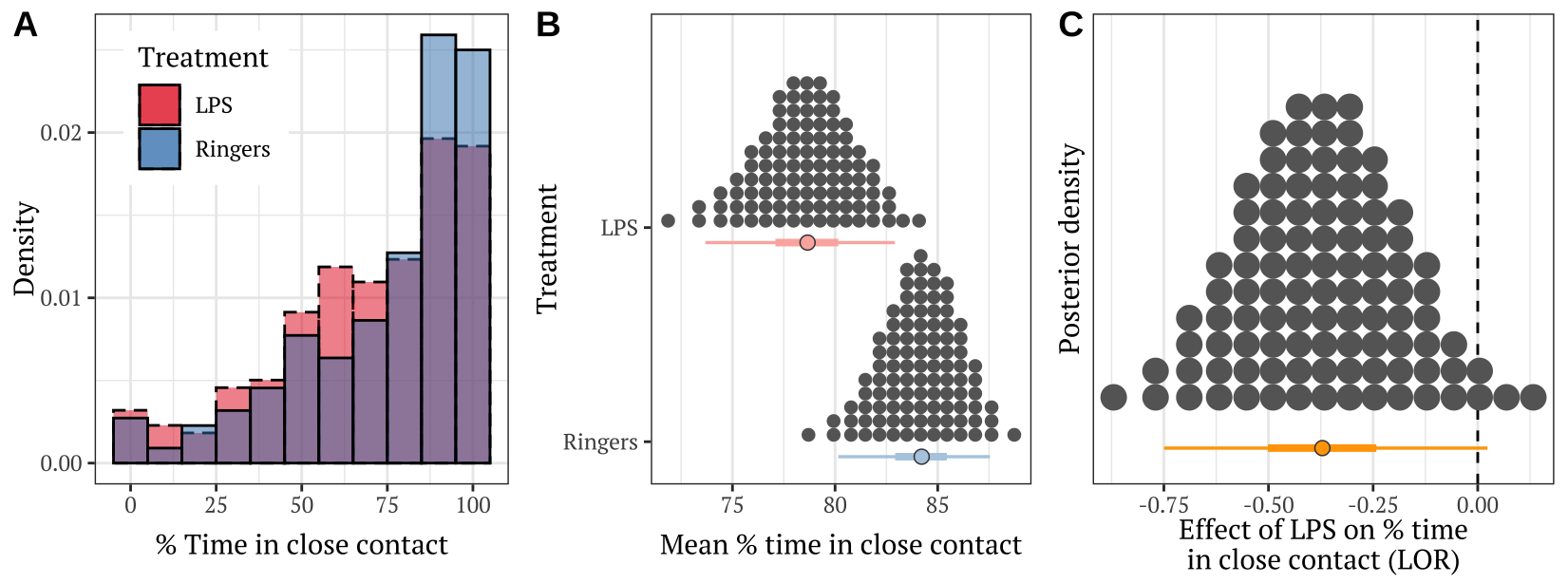
| Version | Author | Date |
|---|---|---|
| 1ce9e19 | lukeholman | 2020-04-21 |
Figure 3: Panel A shows the frequency distribution of the % time in close contact, for pairs of bees from the LPS treatment and the Ringers control. Panel B shows the posterior estimates of the mean % time spent in close contact; the details of the quantile dot plot and error bars are the same as described for Figure 1. Panel C shows the effect size (LOR; log odds ratio) associated with the difference in means in Panel B.
Hypothesis testing and effect sizes
get_log_odds <- function(trt1, trt2){
log((trt2 / (1 - trt2) / (trt1 / (1 - trt1))))
}
my_summary <- function(df) {
diff <- (df %>% pull(Ringers)) - (df %>% pull(LPS))
LOR <- get_log_odds((df %>% pull(Ringers))/100,
(df %>% pull(LPS))/100)
p <- 1 - (diff %>% bayestestR::p_direction() %>% as.numeric())
diff <- diff %>% posterior_summary() %>% as_tibble()
LOR <- LOR %>% posterior_summary() %>% as_tibble()
output <- rbind(diff, LOR) %>%
mutate(p=p,
Metric = c("Absolute difference in % time in close contact",
"Log odds ratio")) %>%
select(Metric, everything()) %>%
mutate(p = format(round(p, 4), nsmall = 4))
output$p[1] <- " "
output
}
plotting_data %>%
as_tibble() %>%
mutate(sample = rep(1:(n() / 2), 2)) %>%
spread(treatment, percent_time_in_contact) %>%
mutate(difference = LPS - Ringers) %>%
my_summary() %>%
mutate(` ` = ifelse(p < 0.05, "\\*", ""),
` ` = replace(` `, p < 0.01, "**"),
` ` = replace(` `, p < 0.001, "***"),
` ` = replace(` `, p == " ", "")) %>%
kable(digits = 3) %>% kable_styling() | Metric | Estimate | Est.Error | Q2.5 | Q97.5 | p | |
|---|---|---|---|---|---|---|
| Absolute difference in % time in close contact | 5.567 | 2.937 | -0.336 | 11.403 | ||
| Log odds ratio | -0.371 | 0.194 | -0.750 | 0.023 | 0.0306 | * |
Table XX: Pairs in which one bee had received LPS were observed in close contact less frequently than pairs in which one bee had received Ringers solution.
sessionInfo()R version 3.6.3 (2020-02-29)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Catalina 10.15.4
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
locale:
[1] en_AU.UTF-8/en_AU.UTF-8/en_AU.UTF-8/C/en_AU.UTF-8/en_AU.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] tidybayes_2.0.1 cowplot_1.0.0 bayestestR_0.5.1 kableExtra_1.1.0
[5] gridExtra_2.3 forcats_0.5.0 stringr_1.4.0 dplyr_0.8.5
[9] purrr_0.3.3 readr_1.3.1 tidyr_1.0.2 tibble_3.0.0
[13] ggplot2_3.3.0 tidyverse_1.3.0 bayesplot_1.7.1 brms_2.12.0
[17] Rcpp_1.0.3 showtext_0.7-1 showtextdb_2.0 sysfonts_0.8
[21] workflowr_1.6.0
loaded via a namespace (and not attached):
[1] colorspace_1.4-1 ellipsis_0.3.0 ggridges_0.5.2
[4] rsconnect_0.8.16 rprojroot_1.3-2 markdown_1.1
[7] base64enc_0.1-3 fs_1.3.1 rstudioapi_0.11
[10] farver_2.0.3 rstan_2.19.3 svUnit_0.7-12
[13] DT_0.13 fansi_0.4.1 mvtnorm_1.1-0
[16] lubridate_1.7.8 xml2_1.3.1 bridgesampling_1.0-0
[19] knitr_1.28 shinythemes_1.1.2 jsonlite_1.6.1
[22] broom_0.5.4 dbplyr_1.4.2 shiny_1.4.0
[25] compiler_3.6.3 httr_1.4.1 backports_1.1.6
[28] assertthat_0.2.1 Matrix_1.2-18 fastmap_1.0.1
[31] cli_2.0.2 later_1.0.0 htmltools_0.4.0
[34] prettyunits_1.1.1 tools_3.6.3 igraph_1.2.5
[37] coda_0.19-3 gtable_0.3.0 glue_1.4.0
[40] reshape2_1.4.4 cellranger_1.1.0 vctrs_0.2.4
[43] nlme_3.1-147 crosstalk_1.1.0.1 insight_0.8.1
[46] xfun_0.13 ps_1.3.0 rvest_0.3.5
[49] mime_0.9 miniUI_0.1.1.1 lifecycle_0.2.0
[52] gtools_3.8.2 zoo_1.8-7 scales_1.1.0
[55] colourpicker_1.0 hms_0.5.3 promises_1.1.0
[58] Brobdingnag_1.2-6 parallel_3.6.3 inline_0.3.15
[61] RColorBrewer_1.1-2 shinystan_2.5.0 curl_4.3
[64] yaml_2.2.1 loo_2.2.0 StanHeaders_2.19.2
[67] stringi_1.4.6 highr_0.8 dygraphs_1.1.1.6
[70] pkgbuild_1.0.6 rlang_0.4.5 pkgconfig_2.0.3
[73] matrixStats_0.56.0 evaluate_0.14 lattice_0.20-41
[76] labeling_0.3 rstantools_2.0.0 htmlwidgets_1.5.1
[79] processx_3.4.2 tidyselect_1.0.0 plyr_1.8.6
[82] magrittr_1.5 R6_2.4.1 generics_0.0.2
[85] DBI_1.1.0 pillar_1.4.3 haven_2.2.0
[88] whisker_0.4 withr_2.1.2 xts_0.12-0
[91] abind_1.4-5 modelr_0.1.5 crayon_1.3.4
[94] arrayhelpers_1.1-0 rmarkdown_2.1 grid_3.6.3
[97] readxl_1.3.1 callr_3.4.3 git2r_0.26.1
[100] threejs_0.3.3 reprex_0.3.0 digest_0.6.25
[103] webshot_0.5.2 xtable_1.8-4 httpuv_1.5.2
[106] stats4_3.6.3 munsell_0.5.0 viridisLite_0.3.0
[109] shinyjs_1.1