Analysis of Experiment 1
Last updated: 2019-10-17
Checks: 7 0
Knit directory: LPS_honeybees/
This reproducible R Markdown analysis was created with workflowr (version 1.4.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190701) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/.DS_Store
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 8c02129 | lukeholman | 2019-10-17 | Commit for thesis |
| Rmd | 367c1bd | lukeholman | 2019-09-30 | First big commit |
Load data and R packages
library(brms)
library(bayesplot)
library(tidyverse)
library(gridExtra)
library(kableExtra)
library(bayestestR)
library(tidybayes)
source("code/helper_functions.R")
exp1_treatments <- c("Intact control", "Ringers", "Heat-treated LPS", "LPS")
durations <- read_csv("data/data_collection_sheets/experiment_durations.csv") %>%
filter(experiment == 1) %>% select(-experiment)
outcome_tally <- read_csv(file = "data/clean_data/experiment_1_outcome_tally.csv") %>%
mutate(outcome = replace(outcome, outcome == "Left of own volition", "Left voluntarily")) %>%
mutate(outcome = factor(outcome, levels = c("Stayed inside the hive", "Left voluntarily", "Forced out")),
treatment = factor(treatment, levels = exp1_treatments))
# Re-formatted version of the same data, where each row is an individual bee. We need this format to run the brms model.
data_for_categorical_model <- outcome_tally %>%
mutate(id = 1:n()) %>%
split(.$id) %>%
map(function(x){
if(x$n[1] == 0) return(NULL)
data.frame(
treatment = x$treatment[1],
hive = x$hive[1],
colour = x$colour[1],
outcome = rep(x$outcome[1], x$n))
}) %>% do.call("rbind", .) %>% as_tibble() %>%
arrange(hive, treatment) %>%
mutate(outcome_numeric = as.numeric(outcome),
hive = as.character(hive),
treatment = factor(treatment, levels = exp1_treatments)) %>%
left_join(durations, by = "hive")Inspect the raw data
Table format
outcome_tally %>%
kable(digits = 3) %>% kable_styling() %>%
scroll_box(height = "380px")| hive | treatment | colour | outcome | n |
|---|---|---|---|---|
| Garden | Heat-treated LPS | Green | Stayed inside the hive | 34 |
| Garden | Heat-treated LPS | Green | Left voluntarily | 0 |
| Garden | Heat-treated LPS | Green | Forced out | 0 |
| Garden | Intact control | Blue | Stayed inside the hive | 75 |
| Garden | Intact control | Blue | Left voluntarily | 0 |
| Garden | Intact control | Blue | Forced out | 2 |
| Garden | LPS | Red | Stayed inside the hive | 37 |
| Garden | LPS | Red | Left voluntarily | 0 |
| Garden | LPS | Red | Forced out | 0 |
| Garden | Ringers | White | Stayed inside the hive | 41 |
| Garden | Ringers | White | Left voluntarily | 0 |
| Garden | Ringers | White | Forced out | 0 |
| SkyLab | Heat-treated LPS | Blue | Stayed inside the hive | 47 |
| SkyLab | Heat-treated LPS | Blue | Left voluntarily | 6 |
| SkyLab | Heat-treated LPS | Blue | Forced out | 5 |
| SkyLab | Intact control | Red | Stayed inside the hive | 102 |
| SkyLab | Intact control | Red | Left voluntarily | 3 |
| SkyLab | Intact control | Red | Forced out | 0 |
| SkyLab | LPS | Green | Stayed inside the hive | 35 |
| SkyLab | LPS | Green | Left voluntarily | 6 |
| SkyLab | LPS | Green | Forced out | 2 |
| Zoology | Heat-treated LPS | White | Stayed inside the hive | 59 |
| Zoology | Heat-treated LPS | White | Left voluntarily | 1 |
| Zoology | Heat-treated LPS | White | Forced out | 7 |
| Zoology | Intact control | Green | Stayed inside the hive | 105 |
| Zoology | Intact control | Green | Left voluntarily | 0 |
| Zoology | Intact control | Green | Forced out | 1 |
| Zoology | LPS | Blue | Stayed inside the hive | 60 |
| Zoology | LPS | Blue | Left voluntarily | 2 |
| Zoology | LPS | Blue | Forced out | 5 |
| Zoology | Ringers | Red | Stayed inside the hive | 74 |
| Zoology | Ringers | Red | Left voluntarily | 1 |
| Zoology | Ringers | Red | Forced out | 5 |
Plotted as a bar chart
all_hives <- outcome_tally %>%
group_by(treatment, outcome) %>%
summarise(n = sum(n)) %>%
ungroup() %>% mutate(hive = "All hives")
outcome_tally %>%
group_by(treatment, hive, outcome) %>%
summarise(n = sum(n)) %>%
ungroup() %>%
mutate(hive = paste(hive, "hive")) %>%
full_join(all_hives, by = c("treatment", "hive", "outcome", "n")) %>%
mutate(prop = n / sum(n)) %>%
ggplot(aes(outcome, n , fill = treatment)) +
geom_bar(stat = "identity", position = "dodge", colour = "grey15") +
facet_wrap( ~ hive, nrow = 1, scales = "free_y") +
scale_fill_brewer(palette = "Set3", name = "Treatment") +
xlab("Outcome") + ylab("Number of bees") +
theme(axis.text.x = element_text(angle = 45, hjust=1),
legend.position = "top")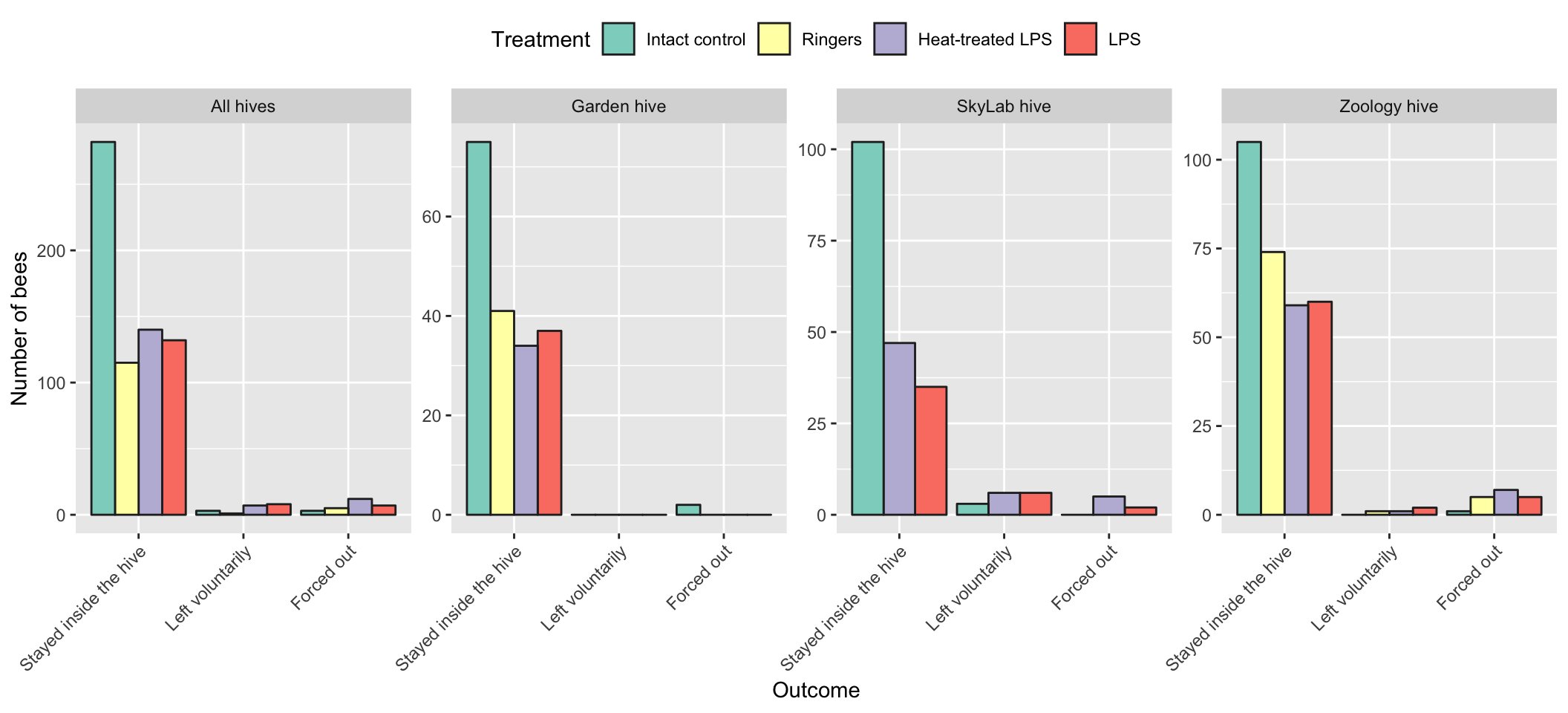
Multinomial model of outcome
Run the models
Fit a multinomial logisitic model, with 3 possible outcomes describing what happened to each bee introduced to the hive: stayed inside, left of its own volition, or forced out by the other workers. To assess the effects of our predictor variables, we compare 5 models with different fixed factors, ranking them by posterior model probability.
if(!file.exists("output/model_probabilities_expt1.rds")){
my_prior <- c(set_prior("normal(0, 2)", class = "b"))
run_brms_categorical <- function(formula){
formula <- gsub(" ", "", paste("outcome_numeric ~", formula))
brm(formula,
data = data_for_categorical_model,
prior = my_prior,
family = "categorical",
save_all_pars = TRUE,
control = list(adapt_delta = 0.999, max_treedepth = 15),
chains = 4, cores = 4, iter = 40000, seed = 1)
}
formula_list <- c(
"treatment:hive + treatment + hive + observation_time_minutes",
"treatment + hive + observation_time_minutes",
"treatment + observation_time_minutes",
"hive + observation_time_minutes",
"observation_time_minutes")
# run all 5 models, sequentially
model_list_expt1 <- lapply(formula_list, run_brms_categorical)
# Estimate the log marginal likelihood by bridge sampling, and use to determine the posterior model probabilities
# See: vignette("bridgesampling_example_stan")
model_probabilities <- post_prob(model_list_expt1[[1]],
model_list_expt1[[2]],
model_list_expt1[[3]],
model_list_expt1[[4]],
model_list_expt1[[5]])
top_index <- which.max(unname(model_probabilities))
model_probabilities <- tibble(
`Fixed effects` = formula_list,
`Posterior model probability` = unname(model_probabilities)) %>%
arrange(-`Posterior model probability`)
model_probabilities <- model_probabilities %>%
mutate(`% of top model probability` = round(100 * `Posterior model probability` / `Posterior model probability`[1], 1),
`% of top model probability` = c("-", `% of top model probability`[2:length(`% of top model probability`)]))
saveRDS(model_list_expt1[[1]], "output/full_model_expt1.rds")
saveRDS(model_list_expt1[[3]], "output/trt_model_expt1.rds")
saveRDS(model_list_expt1[[top_index]], "output/top_model_expt1.rds")
saveRDS(model_probabilities, "output/model_probabilities_expt1.rds")
}
top_model <- readRDS("output/top_model_expt1.rds")
full_model <- readRDS("output/full_model_expt1.rds")
model_probabilities <- readRDS("output/model_probabilities_expt1.rds")Posterior model probabilites
This table shows the estimated probability that each of the 5 models we compared is the top model within this set of models. Note that the predictor variable treatment is not present in either of the top 2 models, but that the top models are not much more likely than the remaining models.
model_probabilities %>%
kable(digits =3 ) %>% kable_styling()| Fixed effects | Posterior model probability |
|---|---|
| observation_time_minutes | 0.463 |
| hive + observation_time_minutes | 0.450 |
| treatment + observation_time_minutes | 0.032 |
| treatment + hive + observation_time_minutes | 0.031 |
| treatment:hive + treatment + hive + observation_time_minutes | 0.025 |
Fixed effects from the full model
The model output is not especially informative on its own, but is included here for completeness. See Hypothesis testing and effect sizes below for more useful statistical results.
get_fixed_effects_with_p_values(full_model) %>%
kable(digits = 3) %>% kable_styling()| Parameter | Estimate | Est.Error | lower_95_CI | upper_95_CI | Rhat | Bulk_ESS | Tail_ESS | p | |
|---|---|---|---|---|---|---|---|---|---|
| mu2_Intercept | -199.536 | 144.749 | -543.710 | -15.110 | 1 | 33464 | 35459 | 0.000 | * |
| mu3_Intercept | -5.946 | 2.792 | -11.619 | -0.712 | 1 | 55189 | 55851 | 0.012 | * |
| mu2_treatmentRingers | 0.453 | 1.604 | -2.692 | 3.593 | 1 | 76261 | 60870 | 0.388 | |
| mu2_treatmentHeatMtreatedLPS | 0.551 | 1.612 | -2.608 | 3.695 | 1 | 79773 | 63419 | 0.365 | |
| mu2_treatmentLPS | 1.026 | 1.570 | -2.044 | 4.098 | 1 | 74871 | 62042 | 0.258 | |
| mu2_hiveZoology | 0.038 | 2.005 | -3.875 | 3.955 | 1 | 116443 | 58337 | 0.492 | |
| mu2_observation_time_minutes | 1.613 | 1.207 | 0.076 | 4.484 | 1 | 33357 | 35600 | 0.005 | * |
| mu2_treatmentRingers:hiveZoology | 0.475 | 1.604 | -2.679 | 3.624 | 1 | 74550 | 61971 | 0.383 | |
| mu2_treatmentHeatMtreatedLPS:hiveZoology | 0.567 | 1.617 | -2.609 | 3.731 | 1 | 81257 | 63922 | 0.362 | |
| mu2_treatmentLPS:hiveZoology | 1.037 | 1.573 | -2.027 | 4.113 | 1 | 74825 | 63492 | 0.256 | |
| mu3_treatmentRingers | -0.242 | 1.197 | -2.686 | 2.017 | 1 | 63191 | 57193 | 0.430 | |
| mu3_treatmentHeatMtreatedLPS | 0.023 | 1.188 | -2.396 | 2.266 | 1 | 63644 | 57899 | 0.482 | |
| mu3_treatmentLPS | -0.137 | 1.205 | -2.593 | 2.149 | 1 | 61756 | 55827 | 0.465 | |
| mu3_hiveZoology | 0.011 | 2.008 | -3.925 | 3.952 | 1 | 62322 | 58839 | 0.498 | |
| mu3_observation_time_minutes | 0.016 | 0.038 | -0.058 | 0.090 | 1 | 56063 | 57181 | 0.342 | |
| mu3_treatmentRingers:hiveZoology | 1.421 | 1.261 | -0.995 | 3.960 | 1 | 61103 | 56084 | 0.129 | |
| mu3_treatmentHeatMtreatedLPS:hiveZoology | 1.759 | 1.248 | -0.618 | 4.267 | 1 | 63425 | 58508 | 0.076 | |
| mu3_treatmentLPS:hiveZoology | 1.519 | 1.268 | -0.910 | 4.086 | 1 | 59890 | 54745 | 0.113 |
Plotting estimates from the model
Derive prediction from the posterior
new <- expand.grid(treatment = levels(data_for_categorical_model$treatment),
hive = "Zoology",
observation_time_minutes = 120)
preds <- fitted(full_model, newdata = new, summary = FALSE)
dimnames(preds) <- list(NULL, new[,1], NULL)
plotting_data <- rbind(
as.data.frame(preds[,, 1]) %>% mutate(outcome = "Stayed inside the hive", posterior_sample = 1:n()),
as.data.frame(preds[,, 2]) %>% mutate(outcome = "Left voluntarily", posterior_sample = 1:n()),
as.data.frame(preds[,, 3]) %>% mutate(outcome = "Forced out", posterior_sample = 1:n())) %>%
gather(treatment, prop, `Intact control`, Ringers, `Heat-treated LPS`, LPS) %>%
mutate(outcome = factor(outcome, c("Stayed inside the hive", "Left voluntarily", "Forced out"))) %>%
as_tibble() %>% arrange(treatment, outcome)
stats_data <- plotting_data
plotting_data$treatment[plotting_data$treatment == "Intact control"] <- "Intact\ncontrol"
plotting_data$treatment[plotting_data$treatment == "Heat-treated LPS"] <- "Heat-treated\nLPS"
plotting_data$treatment <- factor(plotting_data$treatment, c("Intact\ncontrol", "Ringers", "Heat-treated\nLPS", "LPS"))Make Figure 1
make_plot <- function(dat, lim){
dat %>%
ggplot(aes(treatment, 100 * prop)) +
geom_eye(aes(fill = treatment), size_range = c(0.5, 1.1), .width = c(0.5, 0.95)) +
scale_fill_brewer(palette = "Pastel2", direction = -1) +
facet_wrap( ~ outcome, scales = "free_y") +
coord_cartesian(ylim = lim) +
ylab(NULL) + xlab(NULL) +
theme_bw() +
theme(legend.position = "none")
}
grid.arrange(
plotting_data %>% filter(outcome == "Stayed inside the hive") %>% make_plot(c(75, 100)),
plotting_data %>% filter(outcome == "Left voluntarily") %>% make_plot(c(0, 10)),
plotting_data %>% filter(outcome == "Forced out") %>% make_plot(c(0, 25)),
left = "% bees leaving the hive this way", bottom = "Treatment", nrow = 1)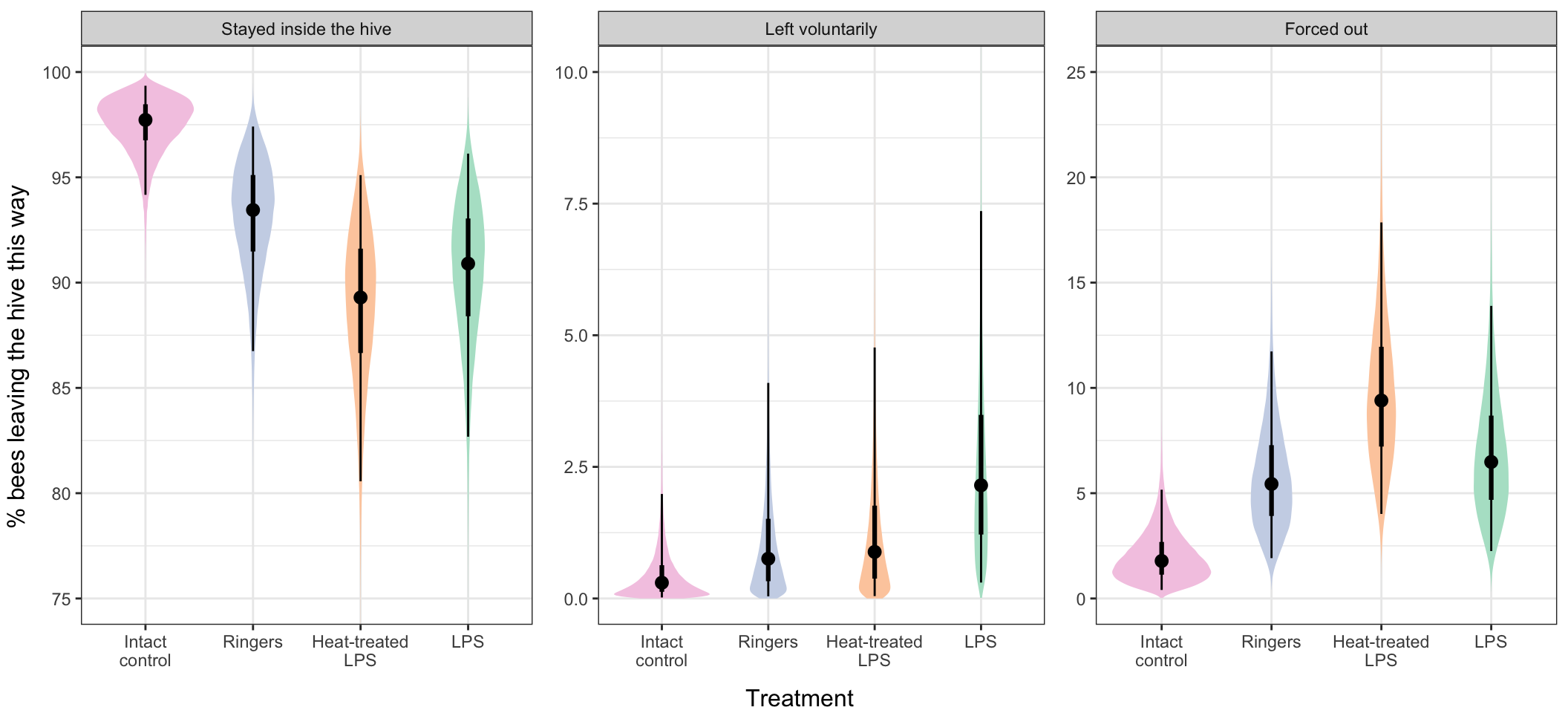
Figure 1: This figure shows the estimated % bees from each treatment group that either stayed inside the hive (left), left the hive voluntarily (middle), or were forced out by their nestmates (right). The points show the median of the posterior distribution of estimates from the model, while the thick and thin error bars show the 50% and 95% credible intervals of this posterior distribution, respectively. The coloured area shows the whole posterior. Bees in the ‘intact control’ group were more likely to stay inside the hive, and were less likely to be forced out, than bees in either heat-treated LPS or LPS treatments (see Table XX).
Hypothesis testing and effect sizes
This section calculates the posterior difference in treatment group means, in order to perform some null hypothesis testing, calculate effect size, and calculate the 95% credible intervals on the effect size.
We regard both the “Intact control” treatment, and the “Ringers injection” treatment as control groups. The heat-treated LPS group was intended as a control, but post-hoc inspection of the results suggests that the heat-treated LPS had a similar effect on behaviour as did regular LPS (this might explain why heat-treated LPS is very seldom used as a control in insect studies). Therefore, we computed the effect sizes and “Bayesian p-values” (i.e. the fraction of the posterior difference in means that lies on the opposite side of zero from the median) for comparisons between each control and each LPS treatment. We do this three times, once for each of the 3 possible outcomes.
% bees exiting the hive
my_summary <- function(df, columns) {
lapply(columns, function(x){
p <- df %>% pull(!! x) %>% bayestestR::p_direction() %>% as.numeric()
p <- (100 - p) / 100
df %>% pull(!! x) %>% posterior_summary() %>% as_tibble() %>%
mutate(p=p) %>% mutate(Metric = x) %>% select(Metric, everything())
}) %>% do.call("rbind", .)
}
make_stats_table <- function(dat, groupA, groupB,
comparison,
metric = "Absolute difference in % bees exiting the hive"){
output <- dat %>%
spread(treatment, prop) %>%
mutate(
metric_here = 100 * (!! enquo(groupA) - !! enquo(groupB)),
`Log10 odds ratio` = log10(!! enquo(groupA) / !! enquo(groupB))) %>%
my_summary(c("metric_here", "Log10 odds ratio")) %>%
mutate(p = c(" ", format(round(p[2], 4), nsmall = 4)),
Comparison = comparison) %>%
select(Comparison, everything()) %>%
mutate(Metric = replace(Metric, Metric == "metric_here", metric))
names(output)[names(output) == "metric_here"] <- metric
output
}
stats_table <- rbind(
stats_data %>%
filter(outcome == "Stayed inside the hive") %>%
make_stats_table(`Intact control`, `Heat-treated LPS`, "Intact control vs Heat-treated LPS"),
stats_data %>%
filter(outcome == "Stayed inside the hive") %>%
make_stats_table(`Ringers`, `Heat-treated LPS`, "Ringers control vs Heat-treated LPS"),
stats_data %>%
filter(outcome == "Stayed inside the hive") %>%
make_stats_table(`Intact control`, `LPS`, "Intact control vs LPS"),
stats_data %>%
filter(outcome == "Stayed inside the hive") %>%
make_stats_table(`Ringers`, `LPS`, "Ringers control vs LPS")
) %>% as_tibble()
stats_table[c(2,4,6,8), 1] <- " "
stats_table %>%
mutate(` ` = ifelse(p < 0.05, "\\*", ""),
` ` = replace(` `, p < 0.01, "**"),
` ` = replace(` `, p < 0.001, "***"),
` ` = replace(` `, p == " ", "")) %>%
kable(digits = 3) %>% kable_styling() %>%
row_spec(c(0,2,4,6,8), extra_css = "border-bottom: solid;")| Comparison | Metric | Estimate | Est.Error | Q2.5 | Q97.5 | p | |
|---|---|---|---|---|---|---|---|
| Intact control vs Heat-treated LPS | Absolute difference in % bees exiting the hive | 8.556 | 3.908 | 1.814 | 17.065 | ||
| Log10 odds ratio | 0.040 | 0.019 | 0.008 | 0.083 | 0.0050 | ** | |
| Ringers control vs Heat-treated LPS | Absolute difference in % bees exiting the hive | 4.181 | 4.617 | -4.592 | 13.679 | ||
| Log10 odds ratio | 0.020 | 0.023 | -0.022 | 0.067 | 0.1784 | ||
| Intact control vs LPS | Absolute difference in % bees exiting the hive | 6.966 | 3.673 | 0.681 | 15.010 | ||
| Log10 odds ratio | 0.032 | 0.018 | 0.003 | 0.072 | 0.0146 | * | |
| Ringers control vs LPS | Absolute difference in % bees exiting the hive | 2.592 | 4.450 | -5.946 | 11.731 | ||
| Log10 odds ratio | 0.012 | 0.021 | -0.028 | 0.057 | 0.2767 |
% bees that were forced out
stats_table2 <- rbind(
stats_data %>%
filter(outcome == "Forced out") %>%
make_stats_table(`Intact control`, `Heat-treated LPS`,
"Intact control vs Heat-treated LPS",
metric = "Absolute difference in % bees that were forced out"),
stats_data %>%
filter(outcome == "Forced out") %>%
make_stats_table(`Ringers`, `Heat-treated LPS`,
"Ringers control vs Heat-treated LPS",
metric = "Absolute difference in % bees that were forced out"),
stats_data %>%
filter(outcome == "Forced out") %>%
make_stats_table(`Intact control`, `LPS`,
"Intact control vs LPS",
metric = "Absolute difference in % bees that were forced out"),
stats_data %>%
filter(outcome == "Forced out") %>%
make_stats_table(`Ringers`, `LPS`,
"Ringers control vs LPS",
metric = "Absolute difference in % bees that were forced out")
) %>% as_tibble()
stats_table2[c(2,4,6,8), 1] <- " "
stats_table2 %>%
mutate(` ` = ifelse(p < 0.05, "\\*", ""),
` ` = replace(` `, p < 0.01, "**"),
` ` = replace(` `, p < 0.001, "***"),
` ` = replace(` `, p == " ", "")) %>%
kable(digits = 3) %>% kable_styling() %>%
row_spec(c(0,2,4,6,8), extra_css = "border-bottom: solid;")| Comparison | Metric | Estimate | Est.Error | Q2.5 | Q97.5 | p | |
|---|---|---|---|---|---|---|---|
| Intact control vs Heat-treated LPS | Absolute difference in % bees that were forced out | -7.750 | 3.723 | -15.991 | -1.399 | ||
| Log10 odds ratio | -0.733 | 0.321 | -1.407 | -0.145 | 0.0067 | ** | |
| Ringers control vs Heat-treated LPS | Absolute difference in % bees that were forced out | -3.998 | 4.365 | -13.076 | 4.245 | ||
| Log10 odds ratio | -0.242 | 0.260 | -0.766 | 0.257 | 0.1734 | ||
| Intact control vs LPS | Absolute difference in % bees that were forced out | -4.860 | 3.223 | -12.038 | 0.536 | ||
| Log10 odds ratio | -0.568 | 0.339 | -1.267 | 0.071 | 0.0414 | * | |
| Ringers control vs LPS | Absolute difference in % bees that were forced out | -1.108 | 3.971 | -9.330 | 6.589 | ||
| Log10 odds ratio | -0.076 | 0.286 | -0.640 | 0.491 | 0.3925 |
% bees that left voluntarily
stats_table3 <- rbind(
stats_data %>%
filter(outcome == "Left voluntarily") %>%
make_stats_table(`Intact control`, `Heat-treated LPS`,
"Intact control vs Heat-treated LPS",
metric = "Absolute difference in % bees that left voluntarily"),
stats_data %>%
filter(outcome == "Left voluntarily") %>%
make_stats_table(`Ringers`, `Heat-treated LPS`,
"Ringers control vs Heat-treated LPS",
metric = "Absolute difference in % bees that left voluntarily"),
stats_data %>%
filter(outcome == "Left voluntarily") %>%
make_stats_table(`Intact control`, `LPS`,
"Intact control vs LPS",
metric = "Absolute difference in % bees that left voluntarily"),
stats_data %>%
filter(outcome == "Left voluntarily") %>%
make_stats_table(`Ringers`, `LPS`,
"Ringers control vs LPS",
metric = "Absolute difference in % bees that left voluntarily")
) %>% as_tibble()
stats_table3[c(2,4,6,8), 1] <- " "
stats_table3 %>%
mutate(` ` = ifelse(p < 0.05, "\\*", ""),
` ` = replace(` `, p < 0.01, "**"),
` ` = replace(` `, p < 0.001, "***"),
` ` = replace(` `, p == " ", "")) %>%
kable(digits = 3) %>% kable_styling() %>%
row_spec(c(0,2,4,6,8), extra_css = "border-bottom: solid;")| Comparison | Metric | Estimate | Est.Error | Q2.5 | Q97.5 | p | |
|---|---|---|---|---|---|---|---|
| Intact control vs Heat-treated LPS | Absolute difference in % bees that left voluntarily | -0.806 | 1.345 | -4.278 | 1.122 | ||
| Log10 odds ratio | -0.445 | 0.664 | -1.742 | 0.888 | 0.2412 | ||
| Ringers control vs Heat-treated LPS | Absolute difference in % bees that left voluntarily | -0.184 | 1.687 | -3.958 | 3.213 | ||
| Log10 odds ratio | -0.062 | 0.720 | -1.498 | 1.380 | 0.4609 | ||
| Intact control vs LPS | Absolute difference in % bees that left voluntarily | -2.106 | 1.906 | -6.878 | 0.483 | ||
| Log10 odds ratio | -0.864 | 0.592 | -2.082 | 0.254 | 0.0638 | ||
| Ringers control vs LPS | Absolute difference in % bees that left voluntarily | -1.484 | 2.162 | -6.520 | 2.378 | ||
| Log10 odds ratio | -0.480 | 0.619 | -1.801 | 0.662 | 0.2132 |
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS High Sierra 10.13.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_AU.UTF-8/en_AU.UTF-8/en_AU.UTF-8/C/en_AU.UTF-8/en_AU.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] tidybayes_1.1.0 bayestestR_0.2.2 kableExtra_0.9.0 gridExtra_2.3
[5] forcats_0.4.0 stringr_1.4.0 dplyr_0.8.3 purrr_0.3.2
[9] readr_1.1.1 tidyr_0.8.2 tibble_2.1.3 ggplot2_3.1.0
[13] tidyverse_1.2.1 bayesplot_1.6.0 brms_2.10.0 Rcpp_1.0.2
loaded via a namespace (and not attached):
[1] colorspace_1.3-2 ggridges_0.5.0
[3] rsconnect_0.8.8 rprojroot_1.3-2
[5] ggstance_0.3.1 markdown_1.0
[7] base64enc_0.1-3 fs_1.3.1
[9] rstudioapi_0.10 rstan_2.19.2
[11] svUnit_0.7-12 DT_0.4
[13] mvtnorm_1.0-11 lubridate_1.7.4
[15] xml2_1.2.0 bridgesampling_0.4-0
[17] knitr_1.23 shinythemes_1.1.1
[19] jsonlite_1.6 workflowr_1.4.0
[21] broom_0.5.0 shiny_1.3.2
[23] compiler_3.5.1 httr_1.4.0
[25] backports_1.1.2 assertthat_0.2.1
[27] Matrix_1.2-14 lazyeval_0.2.2
[29] cli_1.1.0 later_0.8.0
[31] htmltools_0.3.6 prettyunits_1.0.2
[33] tools_3.5.1 igraph_1.2.1
[35] coda_0.19-2 gtable_0.2.0
[37] glue_1.3.1.9000 reshape2_1.4.3
[39] cellranger_1.1.0 nlme_3.1-137
[41] crosstalk_1.0.0 insight_0.3.0
[43] xfun_0.8 ps_1.3.0
[45] rvest_0.3.2 mime_0.7
[47] miniUI_0.1.1.1 gtools_3.8.1
[49] zoo_1.8-3 scales_1.0.0
[51] colourpicker_1.0 hms_0.4.2
[53] promises_1.0.1 Brobdingnag_1.2-5
[55] parallel_3.5.1 inline_0.3.15
[57] shinystan_2.5.0 RColorBrewer_1.1-2
[59] yaml_2.2.0 loo_2.1.0
[61] StanHeaders_2.19.0 stringi_1.4.3
[63] highr_0.8 dygraphs_1.1.1.6
[65] pkgbuild_1.0.2 rlang_0.4.0
[67] pkgconfig_2.0.2 matrixStats_0.54.0
[69] evaluate_0.14 lattice_0.20-35
[71] rstantools_2.0.0 htmlwidgets_1.3
[73] labeling_0.3 processx_3.2.1
[75] tidyselect_0.2.5 plyr_1.8.4
[77] magrittr_1.5 R6_2.4.0
[79] pillar_1.3.1.9000 haven_1.1.2
[81] whisker_0.3-2 withr_2.1.2
[83] xts_0.11-0 abind_1.4-5
[85] modelr_0.1.2 crayon_1.3.4
[87] arrayhelpers_1.0-20160527 rmarkdown_1.13
[89] grid_3.5.1 readxl_1.1.0
[91] callr_2.0.4 git2r_0.23.0
[93] threejs_0.3.1 digest_0.6.20
[95] xtable_1.8-4 httpuv_1.5.1
[97] stats4_3.5.1 munsell_0.5.0
[99] viridisLite_0.3.0 shinyjs_1.0