Cormotifcluster_analysis
ERM
2023-04-18
Last updated: 2023-04-18
Checks: 7 0
Knit directory: Cardiotoxicity/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230109) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 6f6aaf4. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/Clamp_Summary.csv
Ignored: data/Cormotif_24_k1-5_raw.RDS
Ignored: data/DAgostres24.RDS
Ignored: data/DAtable1.csv
Ignored: data/DDE_reQTL.txt
Ignored: data/DDEresp_list.csv
Ignored: data/DEG_cormotif.RDS
Ignored: data/DF_Plate_Peak.csv
Ignored: data/Da24counts.txt
Ignored: data/Dx24counts.txt
Ignored: data/Dx_reQTL_specific.txt
Ignored: data/Ep24counts.txt
Ignored: data/GOplots.R
Ignored: data/K_cluster
Ignored: data/K_cluster_kisthree.csv
Ignored: data/K_cluster_kistwo.csv
Ignored: data/Mt24counts.txt
Ignored: data/RINsamplelist.txt
Ignored: data/Seonane2019supp1.txt
Ignored: data/TOP2Bi-24hoursGO_analysis.csv
Ignored: data/TR24counts.txt
Ignored: data/Top2biresp_cluster24h.csv
Ignored: data/Viabilitylistfull.csv
Ignored: data/allexpressedgenes.txt
Ignored: data/allgenes.txt
Ignored: data/allmatrix.RDS
Ignored: data/avgLD50.RDS
Ignored: data/backGL.txt
Ignored: data/cormotif_3hk1-8.RDS
Ignored: data/cormotif_initalK5.RDS
Ignored: data/cormotif_initialK5.RDS
Ignored: data/cormotif_initialall.RDS
Ignored: data/counts24hours.RDS
Ignored: data/cpmnorm_counts.csv
Ignored: data/dat_cpm.RDS
Ignored: data/data_outline.txt
Ignored: data/efit2.RDS
Ignored: data/efit2results.RDS
Ignored: data/ensembl_backup.RDS
Ignored: data/ensgtotal.txt
Ignored: data/filenameonly.txt
Ignored: data/filtered_cpm_counts.csv
Ignored: data/filtered_raw_counts.csv
Ignored: data/filtermatrix_x.RDS
Ignored: data/folder_05top/
Ignored: data/gene_prob_tran3h.RDS
Ignored: data/gene_probabilityk5.RDS
Ignored: data/gostresTop2bi_ER.RDS
Ignored: data/gostresTop2bi_LR
Ignored: data/gostresTop2bi_LR.RDS
Ignored: data/gostresTop2bi_TI.RDS
Ignored: data/gostrescoNR
Ignored: data/heartgenes.csv
Ignored: data/individualDRCfile.RDS
Ignored: data/knowles56.GMT
Ignored: data/knowlesGMT.GMT
Ignored: data/mymatrix.RDS
Ignored: data/nonresponse_cluster24h.csv
Ignored: data/norm_LDH.csv
Ignored: data/norm_counts.csv
Ignored: data/old_sets/
Ignored: data/plan2plot.png
Ignored: data/raw_counts.csv
Ignored: data/response_cluster24h.csv
Ignored: data/sigVDA24.txt
Ignored: data/sigVDA3.txt
Ignored: data/sigVDX24.txt
Ignored: data/sigVDX3.txt
Ignored: data/sigVEP24.txt
Ignored: data/sigVEP3.txt
Ignored: data/sigVMT24.txt
Ignored: data/sigVMT3.txt
Ignored: data/sigVTR24.txt
Ignored: data/sigVTR3.txt
Ignored: data/siglist.RDS
Ignored: data/table3a.omar
Ignored: data/tvl24hour.txt
Ignored: data/tvl24hourw.txt
Ignored: data/venn_code.R
Untracked files:
Untracked: .RDataTmp
Untracked: .RDataTmp1
Untracked: .RDataTmp2
Untracked: analysis/other_analysis.Rmd
Untracked: code/extra_code.R
Untracked: cormotif_probability_genelist.csv
Untracked: output/figure_1.Rmd
Untracked: output/output-old/
Untracked: output/plan48ldh.png
Untracked: output/sequencing_info.txt
Untracked: output/tableNR.csv
Untracked: output/tabletop2Bi_ER.csv
Untracked: output/tabletop2Bi_LR.csv
Untracked: output/tabletop2Bi_TI.csv
Untracked: output/toplistall.csv
Untracked: reneebasecode.R
Unstaged changes:
Modified: analysis/DEG-GO_analysis.Rmd
Modified: analysis/run_all_analysis.Rmd
Modified: code/Cormotifgenelist.R
Modified: code/eQTLcodes.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown
(analysis/Cormotifcluster_analysis.Rmd) and HTML
(docs/Cormotifcluster_analysis.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 6f6aaf4 | reneeisnowhere | 2023-04-18 | new Cormotif analysis with codes |
| html | d0f459b | reneeisnowhere | 2023-04-18 | Build site. |
| Rmd | 67b2a09 | reneeisnowhere | 2023-04-18 | new Cormotif analysis with codes |
| Rmd | a64bbe1 | reneeisnowhere | 2023-04-18 | updated with new cormotif method |
| html | 9e37491 | reneeisnowhere | 2023-04-16 | Build site. |
| Rmd | 6d925a2 | reneeisnowhere | 2023-04-16 | updating cormotif with updated RNAseq counts |
| Rmd | 4e52216 | reneeisnowhere | 2023-03-31 | End of week updates |
| Rmd | c365253 | reneeisnowhere | 2023-03-22 | updated code and plots |
| Rmd | 3a26d52 | reneeisnowhere | 2023-03-22 | Wed poster analysis changes |
| Rmd | a150323 | reneeisnowhere | 2023-03-20 | addingcormotif analysis and go on DEGs |
library(tidyverse)
library(gprofiler2)
library(readr)
library(BiocGenerics)
library(gridExtra)
library(VennDiagram)
library(kableExtra)
library(scales)
library(ggVennDiagram)
library(Cormotif)
library(RColorBrewer)Creation of the data set:
## Fit limma model using code as it is found in the original cormotif code. It has
## only been modified to add names to the matrix of t values, as well as the
## limma fits
limmafit.default <- function(exprs,groupid,compid) {
limmafits <- list()
compnum <- nrow(compid)
genenum <- nrow(exprs)
limmat <- matrix(0,genenum,compnum)
limmas2 <- rep(0,compnum)
limmadf <- rep(0,compnum)
limmav0 <- rep(0,compnum)
limmag1num <- rep(0,compnum)
limmag2num <- rep(0,compnum)
rownames(limmat) <- rownames(exprs)
colnames(limmat) <- rownames(compid)
names(limmas2) <- rownames(compid)
names(limmadf) <- rownames(compid)
names(limmav0) <- rownames(compid)
names(limmag1num) <- rownames(compid)
names(limmag2num) <- rownames(compid)
for(i in 1:compnum) {
selid1 <- which(groupid == compid[i,1])
selid2 <- which(groupid == compid[i,2])
eset <- new("ExpressionSet", exprs=cbind(exprs[,selid1],exprs[,selid2]))
g1num <- length(selid1)
g2num <- length(selid2)
designmat <- cbind(base=rep(1,(g1num+g2num)), delta=c(rep(0,g1num),rep(1,g2num)))
fit <- lmFit(eset,designmat)
fit <- eBayes(fit)
limmat[,i] <- fit$t[,2]
limmas2[i] <- fit$s2.prior
limmadf[i] <- fit$df.prior
limmav0[i] <- fit$var.prior[2]
limmag1num[i] <- g1num
limmag2num[i] <- g2num
limmafits[[i]] <- fit
# log odds
# w<-sqrt(1+fit$var.prior[2]/(1/g1num+1/g2num))
# log(0.99)+dt(fit$t[1,2],g1num+g2num-2+fit$df.prior,log=TRUE)-log(0.01)-dt(fit$t[1,2]/w, g1num+g2num-2+fit$df.prior, log=TRUE)+log(w)
}
names(limmafits) <- rownames(compid)
limmacompnum<-nrow(compid)
result<-list(t = limmat,
v0 = limmav0,
df0 = limmadf,
s20 = limmas2,
g1num = limmag1num,
g2num = limmag2num,
compnum = limmacompnum,
fits = limmafits)
}
limmafit.counts <-
function (exprs, groupid, compid, norm.factor.method = "TMM", voom.normalize.method = "none")
{
limmafits <- list()
compnum <- nrow(compid)
genenum <- nrow(exprs)
limmat <- matrix(NA,genenum,compnum)
limmas2 <- rep(0,compnum)
limmadf <- rep(0,compnum)
limmav0 <- rep(0,compnum)
limmag1num <- rep(0,compnum)
limmag2num <- rep(0,compnum)
rownames(limmat) <- rownames(exprs)
colnames(limmat) <- rownames(compid)
names(limmas2) <- rownames(compid)
names(limmadf) <- rownames(compid)
names(limmav0) <- rownames(compid)
names(limmag1num) <- rownames(compid)
names(limmag2num) <- rownames(compid)
for (i in 1:compnum) {
message(paste("Running limma for comparision",i,"/",compnum))
selid1 <- which(groupid == compid[i, 1])
selid2 <- which(groupid == compid[i, 2])
# make a new count data frame
counts <- cbind(exprs[, selid1], exprs[, selid2])
# remove NAs
not.nas <- which(apply(counts, 1, function(x) !any(is.na(x))) == TRUE)
# runn voom/limma
d <- DGEList(counts[not.nas,])
d <- calcNormFactors(d, method = norm.factor.method)
g1num <- length(selid1)
g2num <- length(selid2)
designmat <- cbind(base = rep(1, (g1num + g2num)), delta = c(rep(0,
g1num), rep(1, g2num)))
y <- voom(d, designmat, normalize.method = voom.normalize.method)
fit <- lmFit(y, designmat)
fit <- eBayes(fit)
limmafits[[i]] <- fit
limmat[not.nas, i] <- fit$t[, 2]
limmas2[i] <- fit$s2.prior
limmadf[i] <- fit$df.prior
limmav0[i] <- fit$var.prior[2]
limmag1num[i] <- g1num
limmag2num[i] <- g2num
}
limmacompnum <- nrow(compid)
names(limmafits) <- rownames(compid)
result <- list(t = limmat,
v0 = limmav0,
df0 = limmadf,
s20 = limmas2,
g1num = limmag1num,
g2num = limmag2num,
compnum = limmacompnum,
fits = limmafits)
}
limmafit.list <-
function (fitlist, cmp.idx=2)
{
compnum <- length(fitlist)
genes <- c()
for (i in 1:compnum) genes <- unique(c(genes, rownames(fitlist[[i]])))
genenum <- length(genes)
limmat <- matrix(NA,genenum,compnum)
limmas2 <- rep(0,compnum)
limmadf <- rep(0,compnum)
limmav0 <- rep(0,compnum)
limmag1num <- rep(0,compnum)
limmag2num <- rep(0,compnum)
rownames(limmat) <- genes
colnames(limmat) <- names(fitlist)
names(limmas2) <- names(fitlist)
names(limmadf) <- names(fitlist)
names(limmav0) <- names(fitlist)
names(limmag1num) <- names(fitlist)
names(limmag2num) <- names(fitlist)
for (i in 1:compnum) {
this.t <- fitlist[[i]]$t[,cmp.idx]
limmat[names(this.t),i] <- this.t
limmas2[i] <- fitlist[[i]]$s2.prior
limmadf[i] <- fitlist[[i]]$df.prior
limmav0[i] <- fitlist[[i]]$var.prior[cmp.idx]
limmag1num[i] <- sum(fitlist[[i]]$design[,cmp.idx]==0)
limmag2num[i] <- sum(fitlist[[i]]$design[,cmp.idx]==1)
}
limmacompnum <- compnum
result <- list(t = limmat,
v0 = limmav0,
df0 = limmadf,
s20 = limmas2,
g1num = limmag1num,
g2num = limmag2num,
compnum = limmacompnum,
fits = limmafits)
}
## Rank genes based on statistics
generank<-function(x) {
xcol<-ncol(x)
xrow<-nrow(x)
result<-matrix(0,xrow,xcol)
z<-(1:1:xrow)
for(i in 1:xcol) {
y<-sort(x[,i],decreasing=TRUE,na.last=TRUE)
result[,i]<-match(x[,i],y)
result[,i]<-order(result[,i])
}
result
}
## Log-likelihood for moderated t under H0
modt.f0.loglike<-function(x,df) {
a<-dt(x, df, log=TRUE)
result<-as.vector(a)
flag<-which(is.na(result)==TRUE)
result[flag]<-0
result
}
## Log-likelihood for moderated t under H1
## param=c(df,g1num,g2num,v0)
modt.f1.loglike<-function(x,param) {
df<-param[1]
g1num<-param[2]
g2num<-param[3]
v0<-param[4]
w<-sqrt(1+v0/(1/g1num+1/g2num))
dt(x/w, df, log=TRUE)-log(w)
a<-dt(x/w, df, log=TRUE)-log(w)
result<-as.vector(a)
flag<-which(is.na(result)==TRUE)
result[flag]<-0
result
}
## Correlation Motif Fit
cmfit.X<-function(x, type, K=1, tol=1e-3, max.iter=100) {
## initialize
xrow <- nrow(x)
xcol <- ncol(x)
loglike0 <- list()
loglike1 <- list()
p <- rep(1, K)/K
q <- matrix(runif(K * xcol), K, xcol)
q[1, ] <- rep(0.01, xcol)
for (i in 1:xcol) {
f0 <- type[[i]][[1]]
f0param <- type[[i]][[2]]
f1 <- type[[i]][[3]]
f1param <- type[[i]][[4]]
loglike0[[i]] <- f0(x[, i], f0param)
loglike1[[i]] <- f1(x[, i], f1param)
}
condlike <- list()
for (i in 1:xcol) {
condlike[[i]] <- matrix(0, xrow, K)
}
loglike.old <- -1e+10
for (i.iter in 1:max.iter) {
if ((i.iter%%50) == 0) {
print(paste("We have run the first ", i.iter, " iterations for K=",
K, sep = ""))
}
err <- tol + 1
clustlike <- matrix(0, xrow, K)
#templike <- matrix(0, xrow, 2)
templike1 <- rep(0, xrow)
templike2 <- rep(0, xrow)
for (j in 1:K) {
for (i in 1:xcol) {
templike1 <- log(q[j, i]) + loglike1[[i]]
templike2 <- log(1 - q[j, i]) + loglike0[[i]]
tempmax <- Rfast::Pmax(templike1, templike2)
templike1 <- exp(templike1 - tempmax)
templike2 <- exp(templike2 - tempmax)
tempsum <- templike1 + templike2
clustlike[, j] <- clustlike[, j] + tempmax +
log(tempsum)
condlike[[i]][, j] <- templike1/tempsum
}
clustlike[, j] <- clustlike[, j] + log(p[j])
}
#tempmax <- apply(clustlike, 1, max)
tempmax <- Rfast::rowMaxs(clustlike, value=TRUE)
for (j in 1:K) {
clustlike[, j] <- exp(clustlike[, j] - tempmax)
}
#tempsum <- apply(clustlike, 1, sum)
tempsum <- Rfast::rowsums(clustlike)
for (j in 1:K) {
clustlike[, j] <- clustlike[, j]/tempsum
}
#p.new <- (apply(clustlike, 2, sum) + 1)/(xrow + K)
p.new <- (Rfast::colsums(clustlike) + 1)/(xrow + K)
q.new <- matrix(0, K, xcol)
for (j in 1:K) {
clustpsum <- sum(clustlike[, j])
for (i in 1:xcol) {
q.new[j, i] <- (sum(clustlike[, j] * condlike[[i]][,
j]) + 1)/(clustpsum + 2)
}
}
err.p <- max(abs(p.new - p)/p)
err.q <- max(abs(q.new - q)/q)
err <- max(err.p, err.q)
loglike.new <- (sum(tempmax + log(tempsum)) + sum(log(p.new)) +
sum(log(q.new) + log(1 - q.new)))/xrow
p <- p.new
q <- q.new
loglike.old <- loglike.new
if (err < tol) {
break
}
}
clustlike <- matrix(0, xrow, K)
for (j in 1:K) {
for (i in 1:xcol) {
templike1 <- log(q[j, i]) + loglike1[[i]]
templike2 <- log(1 - q[j, i]) + loglike0[[i]]
tempmax <- Rfast::Pmax(templike1, templike2)
templike1 <- exp(templike1 - tempmax)
templike2 <- exp(templike2 - tempmax)
tempsum <- templike1 + templike2
clustlike[, j] <- clustlike[, j] + tempmax + log(tempsum)
condlike[[i]][, j] <- templike1/tempsum
}
clustlike[, j] <- clustlike[, j] + log(p[j])
}
#tempmax <- apply(clustlike, 1, max)
tempmax <- Rfast::rowMaxs(clustlike, value=TRUE)
for (j in 1:K) {
clustlike[, j] <- exp(clustlike[, j] - tempmax)
}
#tempsum <- apply(clustlike, 1, sum)
tempsum <- Rfast::rowsums(clustlike)
for (j in 1:K) {
clustlike[, j] <- clustlike[, j]/tempsum
}
p.post <- matrix(0, xrow, xcol)
for (j in 1:K) {
for (i in 1:xcol) {
p.post[, i] <- p.post[, i] + clustlike[, j] * condlike[[i]][,
j]
}
}
loglike.old <- loglike.old - (sum(log(p)) + sum(log(q) +
log(1 - q)))/xrow
loglike.old <- loglike.old * xrow
result <- list(p.post = p.post, motif.prior = p, motif.q = q,
loglike = loglike.old, clustlike=clustlike, condlike=condlike)
}
## Fit using (0,0,...,0) and (1,1,...,1)
cmfitall<-function(x, type, tol=1e-3, max.iter=100) {
## initialize
xrow<-nrow(x)
xcol<-ncol(x)
loglike0<-list()
loglike1<-list()
p<-0.01
## compute loglikelihood
L0<-matrix(0,xrow,1)
L1<-matrix(0,xrow,1)
for(i in 1:xcol) {
f0<-type[[i]][[1]]
f0param<-type[[i]][[2]]
f1<-type[[i]][[3]]
f1param<-type[[i]][[4]]
loglike0[[i]]<-f0(x[,i],f0param)
loglike1[[i]]<-f1(x[,i],f1param)
L0<-L0+loglike0[[i]]
L1<-L1+loglike1[[i]]
}
## EM algorithm to get MLE of p and q
loglike.old <- -1e10
for(i.iter in 1:max.iter) {
if((i.iter%%50) == 0) {
print(paste("We have run the first ", i.iter, " iterations",sep=""))
}
err<-tol+1
## compute posterior cluster membership
clustlike<-matrix(0,xrow,2)
clustlike[,1]<-log(1-p)+L0
clustlike[,2]<-log(p)+L1
tempmax<-apply(clustlike,1,max)
for(j in 1:2) {
clustlike[,j]<-exp(clustlike[,j]-tempmax)
}
tempsum<-apply(clustlike,1,sum)
## update motif occurrence rate
for(j in 1:2) {
clustlike[,j]<-clustlike[,j]/tempsum
}
p.new<-(sum(clustlike[,2])+1)/(xrow+2)
## evaluate convergence
err<-abs(p.new-p)/p
## evaluate whether the log.likelihood increases
loglike.new<-(sum(tempmax+log(tempsum))+log(p.new)+log(1-p.new))/xrow
loglike.old<-loglike.new
p<-p.new
if(err<tol) {
break;
}
}
## compute posterior p
clustlike<-matrix(0,xrow,2)
clustlike[,1]<-log(1-p)+L0
clustlike[,2]<-log(p)+L1
tempmax<-apply(clustlike,1,max)
for(j in 1:2) {
clustlike[,j]<-exp(clustlike[,j]-tempmax)
}
tempsum<-apply(clustlike,1,sum)
for(j in 1:2) {
clustlike[,j]<-clustlike[,j]/tempsum
}
p.post<-matrix(0,xrow,xcol)
for(i in 1:xcol) {
p.post[,i]<-clustlike[,2]
}
## return
#calculate back loglikelihood
loglike.old<-loglike.old-(log(p)+log(1-p))/xrow
loglike.old<-loglike.old*xrow
result<-list(p.post=p.post, motif.prior=p, loglike=loglike.old)
}
## Fit each dataset separately
cmfitsep<-function(x, type, tol=1e-3, max.iter=100) {
## initialize
xrow<-nrow(x)
xcol<-ncol(x)
loglike0<-list()
loglike1<-list()
p<-0.01*rep(1,xcol)
loglike.final<-rep(0,xcol)
## compute loglikelihood
for(i in 1:xcol) {
f0<-type[[i]][[1]]
f0param<-type[[i]][[2]]
f1<-type[[i]][[3]]
f1param<-type[[i]][[4]]
loglike0[[i]]<-f0(x[,i],f0param)
loglike1[[i]]<-f1(x[,i],f1param)
}
p.post<-matrix(0,xrow,xcol)
## EM algorithm to get MLE of p
for(coli in 1:xcol) {
loglike.old <- -1e10
for(i.iter in 1:max.iter) {
if((i.iter%%50) == 0) {
print(paste("We have run the first ", i.iter, " iterations",sep=""))
}
err<-tol+1
## compute posterior cluster membership
clustlike<-matrix(0,xrow,2)
clustlike[,1]<-log(1-p[coli])+loglike0[[coli]]
clustlike[,2]<-log(p[coli])+loglike1[[coli]]
tempmax<-apply(clustlike,1,max)
for(j in 1:2) {
clustlike[,j]<-exp(clustlike[,j]-tempmax)
}
tempsum<-apply(clustlike,1,sum)
## evaluate whether the log.likelihood increases
loglike.new<-sum(tempmax+log(tempsum))/xrow
## update motif occurrence rate
for(j in 1:2) {
clustlike[,j]<-clustlike[,j]/tempsum
}
p.new<-(sum(clustlike[,2]))/(xrow)
## evaluate convergence
err<-abs(p.new-p[coli])/p[coli]
loglike.old<-loglike.new
p[coli]<-p.new
if(err<tol) {
break;
}
}
## compute posterior p
clustlike<-matrix(0,xrow,2)
clustlike[,1]<-log(1-p[coli])+loglike0[[coli]]
clustlike[,2]<-log(p[coli])+loglike1[[coli]]
tempmax<-apply(clustlike,1,max)
for(j in 1:2) {
clustlike[,j]<-exp(clustlike[,j]-tempmax)
}
tempsum<-apply(clustlike,1,sum)
for(j in 1:2) {
clustlike[,j]<-clustlike[,j]/tempsum
}
p.post[,coli]<-clustlike[,2]
loglike.final[coli]<-loglike.old
}
## return
loglike.final<-loglike.final*xrow
result<-list(p.post=p.post, motif.prior=p, loglike=loglike.final)
}
## Fit the full model
cmfitfull<-function(x, type, tol=1e-3, max.iter=100) {
## initialize
xrow<-nrow(x)
xcol<-ncol(x)
loglike0<-list()
loglike1<-list()
K<-2^xcol
p<-rep(1,K)/K
pattern<-rep(0,xcol)
patid<-matrix(0,K,xcol)
## compute loglikelihood
for(i in 1:xcol) {
f0<-type[[i]][[1]]
f0param<-type[[i]][[2]]
f1<-type[[i]][[3]]
f1param<-type[[i]][[4]]
loglike0[[i]]<-f0(x[,i],f0param)
loglike1[[i]]<-f1(x[,i],f1param)
}
L<-matrix(0,xrow,K)
for(i in 1:K)
{
patid[i,]<-pattern
for(j in 1:xcol) {
if(pattern[j] < 0.5) {
L[,i]<-L[,i]+loglike0[[j]]
} else {
L[,i]<-L[,i]+loglike1[[j]]
}
}
if(i < K) {
pattern[xcol]<-pattern[xcol]+1
j<-xcol
while(pattern[j] > 1) {
pattern[j]<-0
j<-j-1
pattern[j]<-pattern[j]+1
}
}
}
## EM algorithm to get MLE of p and q
loglike.old <- -1e10
for(i.iter in 1:max.iter) {
if((i.iter%%50) == 0) {
print(paste("We have run the first ", i.iter, " iterations",sep=""))
}
err<-tol+1
## compute posterior cluster membership
clustlike<-matrix(0,xrow,K)
for(j in 1:K) {
clustlike[,j]<-log(p[j])+L[,j]
}
tempmax<-apply(clustlike,1,max)
for(j in 1:K) {
clustlike[,j]<-exp(clustlike[,j]-tempmax)
}
tempsum<-apply(clustlike,1,sum)
## update motif occurrence rate
for(j in 1:K) {
clustlike[,j]<-clustlike[,j]/tempsum
}
p.new<-(apply(clustlike,2,sum)+1)/(xrow+K)
## evaluate convergence
err<-max(abs(p.new-p)/p)
## evaluate whether the log.likelihood increases
loglike.new<-(sum(tempmax+log(tempsum))+sum(log(p.new)))/xrow
loglike.old<-loglike.new
p<-p.new
if(err<tol) {
break;
}
}
## compute posterior p
clustlike<-matrix(0,xrow,K)
for(j in 1:K) {
clustlike[,j]<-log(p[j])+L[,j]
}
tempmax<-apply(clustlike,1,max)
for(j in 1:K) {
clustlike[,j]<-exp(clustlike[,j]-tempmax)
}
tempsum<-apply(clustlike,1,sum)
for(j in 1:K) {
clustlike[,j]<-clustlike[,j]/tempsum
}
p.post<-matrix(0,xrow,xcol)
for(j in 1:K) {
for(i in 1:xcol) {
if(patid[j,i] > 0.5) {
p.post[,i]<-p.post[,i]+clustlike[,j]
}
}
}
## return
#calculate back loglikelihood
loglike.old<-loglike.old-sum(log(p))/xrow
loglike.old<-loglike.old*xrow
result<-list(p.post=p.post, motif.prior=p, loglike=loglike.old)
}
generatetype<-function(limfitted)
{
jtype<-list()
df<-limfitted$g1num+limfitted$g2num-2+limfitted$df0
for(j in 1:limfitted$compnum)
{
jtype[[j]]<-list(f0=modt.f0.loglike, f0.param=df[j], f1=modt.f1.loglike, f1.param=c(df[j],limfitted$g1num[j],limfitted$g2num[j],limfitted$v0[j]))
}
jtype
}
cormotiffit <- function(exprs, groupid=NULL, compid=NULL, K=1, tol=1e-3,
max.iter=100, BIC=TRUE, norm.factor.method="TMM",
voom.normalize.method = "none", runtype=c("logCPM","counts","limmafits"), each=3)
{
# first I want to do some typechecking. Input can be either a normalized
# matrix, a count matrix, or a list of limma fits. Dispatch the correct
# limmafit accordingly.
# todo: add some typechecking here
limfitted <- list()
if (runtype=="counts") {
limfitted <- limmafit.counts(exprs,groupid,compid, norm.factor.method, voom.normalize.method)
} else if (runtype=="logCPM") {
limfitted <- limmafit.default(exprs,groupid,compid)
} else if (runtype=="limmafits") {
limfitted <- limmafit.list(exprs)
} else {
stop("runtype must be one of 'logCPM', 'counts', or 'limmafits'")
}
jtype<-generatetype(limfitted)
fitresult<-list()
ks <- rep(K, each = each)
fitresult <- bplapply(1:length(ks), function(i, x, type, ks, tol, max.iter) {
cmfit.X(x, type, K = ks[i], tol = tol, max.iter = max.iter)
}, x=limfitted$t, type=jtype, ks=ks, tol=tol, max.iter=max.iter)
best.fitresults <- list()
for (i in 1:length(K)) {
w.k <- which(ks==K[i])
this.bic <- c()
for (j in w.k) this.bic[j] <- -2 * fitresult[[j]]$loglike + (K[i] - 1 + K[i] * limfitted$compnum) * log(dim(limfitted$t)[1])
w.min <- which(this.bic == min(this.bic, na.rm = TRUE))[1]
best.fitresults[[i]] <- fitresult[[w.min]]
}
fitresult <- best.fitresults
bic <- rep(0, length(K))
aic <- rep(0, length(K))
loglike <- rep(0, length(K))
for (i in 1:length(K)) loglike[i] <- fitresult[[i]]$loglike
for (i in 1:length(K)) bic[i] <- -2 * fitresult[[i]]$loglike + (K[i] - 1 + K[i] * limfitted$compnum) * log(dim(limfitted$t)[1])
for (i in 1:length(K)) aic[i] <- -2 * fitresult[[i]]$loglike + 2 * (K[i] - 1 + K[i] * limfitted$compnum)
if(BIC==TRUE) {
bestflag=which(bic==min(bic))
}
else {
bestflag=which(aic==min(aic))
}
result<-list(bestmotif=fitresult[[bestflag]],bic=cbind(K,bic),
aic=cbind(K,aic),loglike=cbind(K,loglike), allmotifs=fitresult)
}
cormotiffitall<-function(exprs,groupid,compid, tol=1e-3, max.iter=100)
{
limfitted<-limmafit(exprs,groupid,compid)
jtype<-generatetype(limfitted)
fitresult<-cmfitall(limfitted$t,type=jtype,tol=1e-3,max.iter=max.iter)
}
cormotiffitsep<-function(exprs,groupid,compid, tol=1e-3, max.iter=100)
{
limfitted<-limmafit(exprs,groupid,compid)
jtype<-generatetype(limfitted)
fitresult<-cmfitsep(limfitted$t,type=jtype,tol=1e-3,max.iter=max.iter)
}
cormotiffitfull<-function(exprs,groupid,compid, tol=1e-3, max.iter=100)
{
limfitted<-limmafit(exprs,groupid,compid)
jtype<-generatetype(limfitted)
fitresult<-cmfitfull(limfitted$t,type=jtype,tol=1e-3,max.iter=max.iter)
}
plotIC<-function(fitted_cormotif)
{
oldpar<-par(mfrow=c(1,2))
plot(fitted_cormotif$bic[,1], fitted_cormotif$bic[,2], type="b",xlab="Motif Number", ylab="BIC", main="BIC")
plot(fitted_cormotif$aic[,1], fitted_cormotif$aic[,2], type="b",xlab="Motif Number", ylab="AIC", main="AIC")
}
plotMotif<-function(fitted_cormotif,title="")
{
layout(matrix(1:2,ncol=2))
u<-1:dim(fitted_cormotif$bestmotif$motif.q)[2]
v<-1:dim(fitted_cormotif$bestmotif$motif.q)[1]
image(u,v,t(fitted_cormotif$bestmotif$motif.q),
col=gray(seq(from=1,to=0,by=-0.1)),xlab="Study",yaxt = "n",
ylab="Corr. Motifs",main=paste(title,"pattern",sep=" "))
axis(2,at=1:length(v))
for(i in 1:(length(u)+1))
{
abline(v=(i-0.5))
}
for(i in 1:(length(v)+1))
{
abline(h=(i-0.5))
}
Ng=10000
if(is.null(fitted_cormotif$bestmotif$p.post)!=TRUE)
Ng=nrow(fitted_cormotif$bestmotif$p.post)
genecount=floor(fitted_cormotif$bestmotif$motif.p*Ng)
NK=nrow(fitted_cormotif$bestmotif$motif.q)
plot(0,0.7,pch=".",xlim=c(0,1.2),ylim=c(0.75,NK+0.25),
frame.plot=FALSE,axes=FALSE,xlab="No. of genes",ylab="", main=paste(title,"frequency",sep=" "))
segments(0,0.7,fitted_cormotif$bestmotif$motif.p[1],0.7)
rect(0,1:NK-0.3,fitted_cormotif$bestmotif$motif.p,1:NK+0.3,
col="dark grey")
mtext(1:NK,at=1:NK,side=2,cex=0.8)
text(fitted_cormotif$bestmotif$motif.p+0.15,1:NK,
labels=floor(fitted_cormotif$bestmotif$motif.p*Ng))
}library(edgeR)
library(Cormotif)
library(RColorBrewer)
library(BiocParallel)
## read in count file##
design <- read.csv("data/data_outline.txt", row.names = 1)
mymatrix <- readRDS("data/filtermatrix_x.RDS")#should be 14084
x_counts <- mymatrix$counts
indv <- as.factor(rep(c(1,2,3,4,5,6), c(12,12,12,12,12,12)))
time <- rep((rep(c("3h", "24h"), c(6,6))), 6)
time <- ordered(time, levels =c("3h", "24h"))
group <- as.factor(rep((c("1","2","3","4","5","6","7","8","9","10","11","12")),6))
drug <- rep(c("Daunorubicin","Doxorubicin","Epirubicin","Mitoxantrone","Trastuzumab", "Vehicle"),12)
group1 <- interaction(drug,time)
label <- (interaction(substring(drug, 0, 2), indv, time))
colnames(x_counts) <- label
group_fac <- group1
groupid <- as.numeric(group_fac)
compid <- data.frame(c1= c(1,2,3,4,5,7,8,9,10,11), c2 = c( 6,6,6,6,6,12,12,12,12,12))
y_TMM_cpm <- cpm(x_counts, log = TRUE)
colnames(y_TMM_cpm) <- label
y_TMM_cpm
set.seed(12345)
cormotif_initial <- cormotiffit(exprs = y_TMM_cpm,
groupid = groupid,
compid = compid,
K=1:8, max.iter = 500,runtype="logCPM")
gene_prob_tran <- cormotif_initial$bestmotif$p.post
rownames(gene_prob_tran) <- rownames(y_TMM_cpm)
motif_prob <- cormotif_initial$bestmotif$clustlike
rownames(motif_prob) <- rownames(y_TMM_cpm)
write.csv(motif_prob,"cormotif_probability_genelist.csv")cormotif_initial was created after calling corMotif, then running the corMotifcustom.R script. The extra R script enabled me to generate a table containing the likelihood of each gene that belongs to the specific cluster.
After generating the Motifs from 1 to 8, the number of motifs that best fit the data was 4 using the BIC and AIC results below.
cormotif_initial <- readRDS("data/cormotif_initialall.RDS")#
plotIC(cormotif_initial)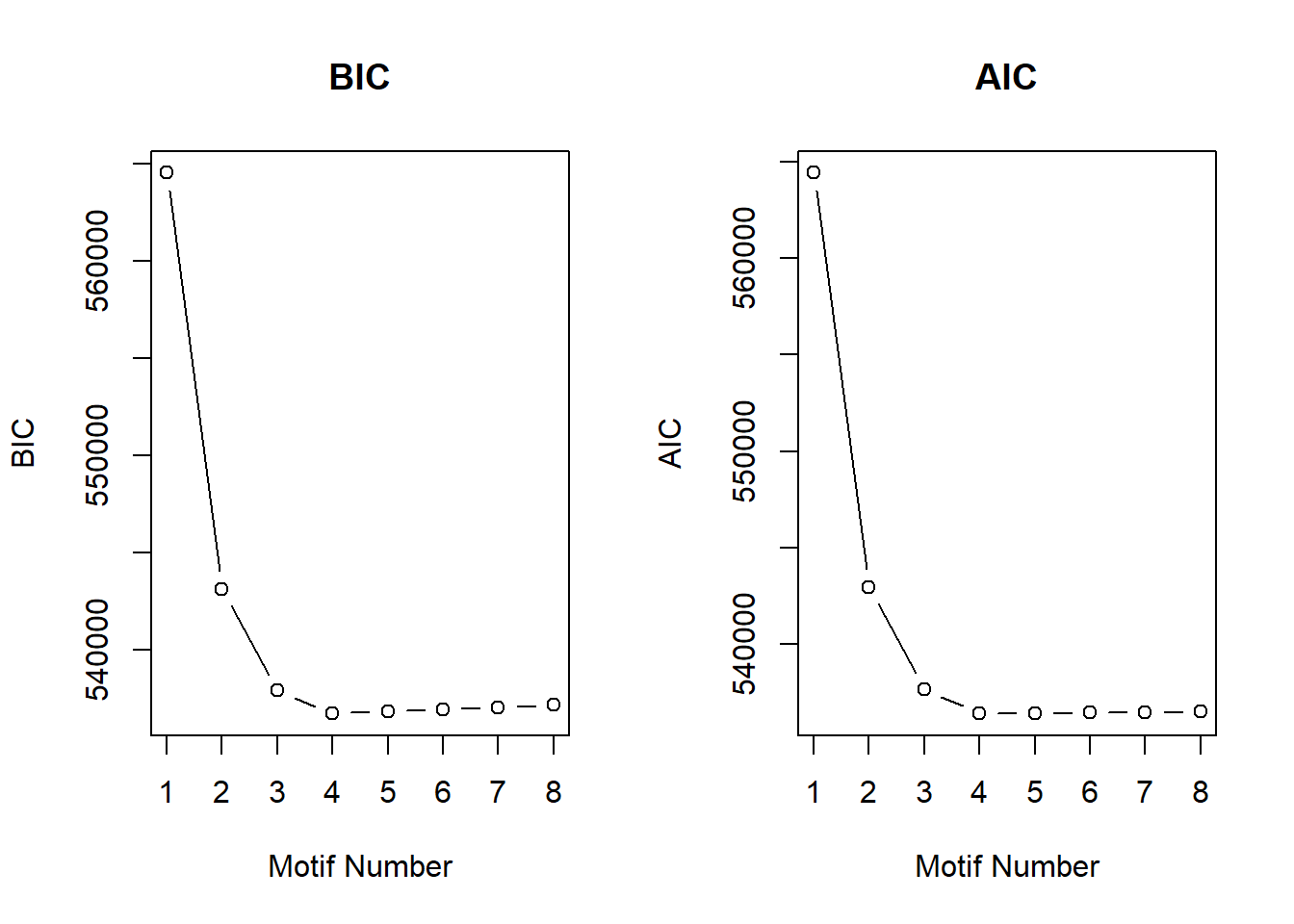
plotMotif(cormotif_initial)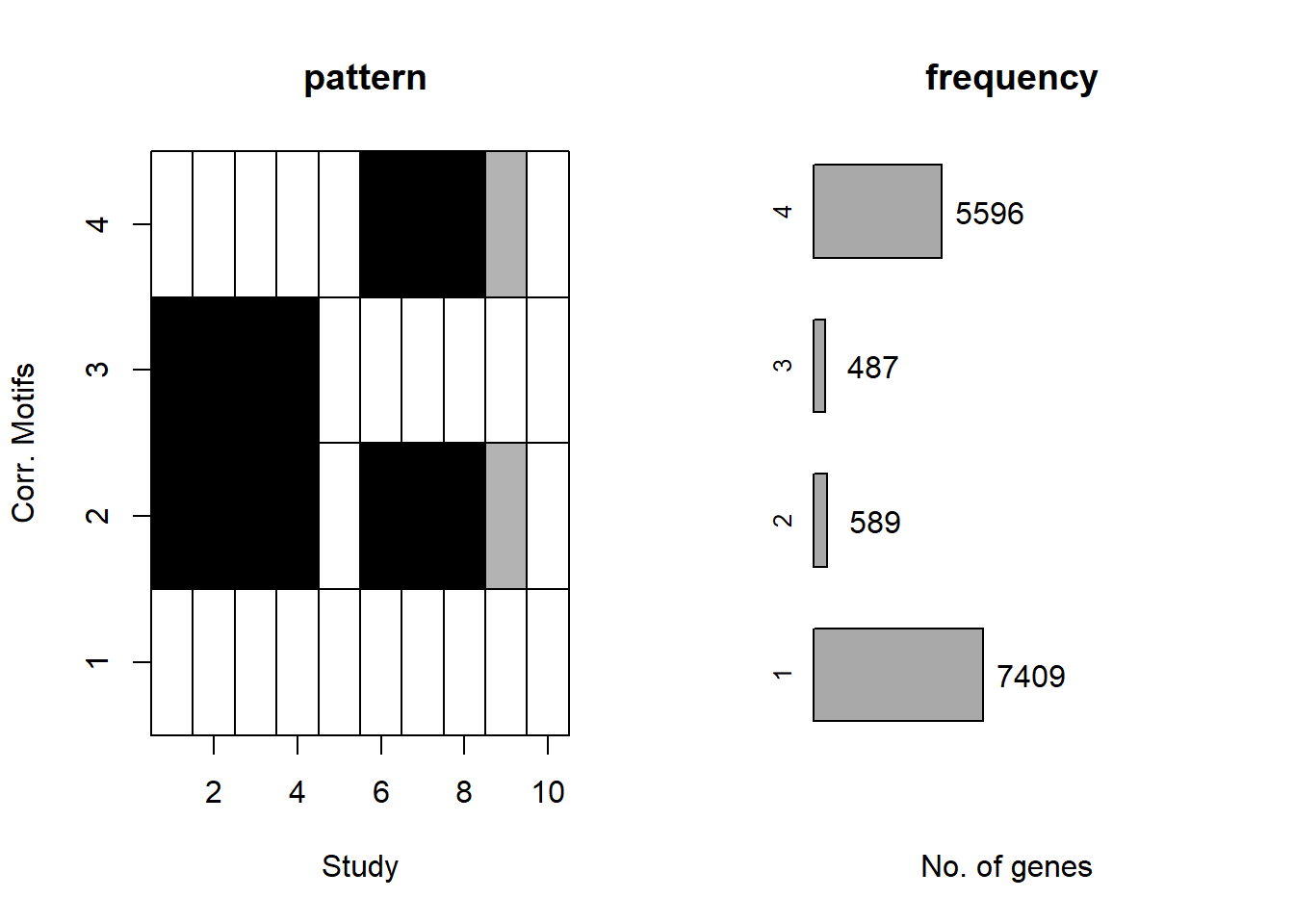
Viewing the motifs, the following groups were named:
Motif 1: No Response (n=7409)
Motif 2: Top2\(\beta\) inhibitor response, Time-independent
- (Time-independent response, n= 589)
Motif 3: Top2\(\beta\) inhibitor response, Early
- (Early response, n= 487)
Motif 4: Top2\(\beta\) inhibitor response, Late
- (Late response, n= 5596)
clust1 <- motif_prob %>%
as.data.frame() %>%
filter(V1>0.5) %>%
rownames
clust2 <- motif_prob %>%
as.data.frame() %>%
filter(V2>0.5) %>%
rownames
clust3 <- motif_prob %>%
as.data.frame() %>%
filter(V3>0.5) %>%
rownames
clust4 <- motif_prob %>%
as.data.frame() %>%
filter(V4>0.5) %>%
rownames
old_clust1 <- rownames(gene_prob_tran[(gene_prob_tran[,1] <0.45 &
gene_prob_tran[,2] <0.45 &
gene_prob_tran[,3] <0.45 &
gene_prob_tran[,4] <0.45 &
gene_prob_tran[,5] <0.45 &
gene_prob_tran[,6] <0.45 &
gene_prob_tran[,7] <0.45 &
gene_prob_tran[,8] <0.45 &
gene_prob_tran[,9] <0.45 &
gene_prob_tran[,10] <0.45),])
length(intersect(old_clust1,clust1))##7358 out of 7362 oldclust1
old_clust2 <- rownames(gene_prob_tran[(gene_prob_tran[,1]> 0.10 &
gene_prob_tran[,2] > 0.10 &
gene_prob_tran[,3] > 0.10 &
gene_prob_tran[,4] > 0.10 &
# gene_prob_tran[,5] < 0.10 &
gene_prob_tran[,6] > 0.10&
gene_prob_tran[,7]> 0.10 &
gene_prob_tran[,8]> 0.10 &
gene_prob_tran[,9]> 0.10),])
# & # gene_prob_tran[,10]< 0.10),])
length(intersect(old_clust2,clust2))##251 out of 432 oldclust2
old_clust3 <- rownames(gene_prob_tran[(gene_prob_tran[,1]>0.55 &
gene_prob_tran[,2] >0.55 &
gene_prob_tran[,3] >0.55 &
gene_prob_tran[,4] >0.25&
gene_prob_tran[,5] <0.90 &
gene_prob_tran[,6] <0.9 &
gene_prob_tran[,7]<0.9&
gene_prob_tran[,8]<0.9 &
gene_prob_tran[,9]<0.9 &
gene_prob_tran[,10]<0.90),])
length(intersect(old_clust3,clust3)) ##414 out of 481 oldclust3
old_clust4 <- rownames(gene_prob_tran[(gene_prob_tran[,1] <0.970 &
gene_prob_tran[,2] <0.97 &
gene_prob_tran[,3] <0.97 &
gene_prob_tran[,4] <0.97 &
gene_prob_tran[,5] <0.9&
gene_prob_tran[,6] >0.55 &
gene_prob_tran[,7] >0.55 &
gene_prob_tran[,8] >0.55 &
gene_prob_tran[,9] >0.05 &
gene_prob_tran[,10] <0.9),])
length(intersect(old_clust4,clust4))##4675 out of 4850 oldclust4
backGL <- read.csv("data/backGL.txt") ##14084
length(setdiff(backGL$ENTREZID,(union(clust1,union(clust2,union(clust3,clust4))))))
##63 genes not used overall same as (14084-7504-528-444-5545)There was overlap between the previous sets and the new sets, so I moved on expecting similar responses in the GO analysis. I did subset out the genes not used overall from the background gene list (rowmeans>0 from log(cpm(count matrix)))
DEG_cormotif <- readRDS("data/DEG_cormotif.RDS")
motif_NR <- DEG_cormotif$motif_NR
motif_TI <- DEG_cormotif$motif_TI
motif_LR <- DEG_cormotif$motif_LR
motif_ER <- DEG_cormotif$motif_ER
backGL <- read.csv("data/backGL.txt")
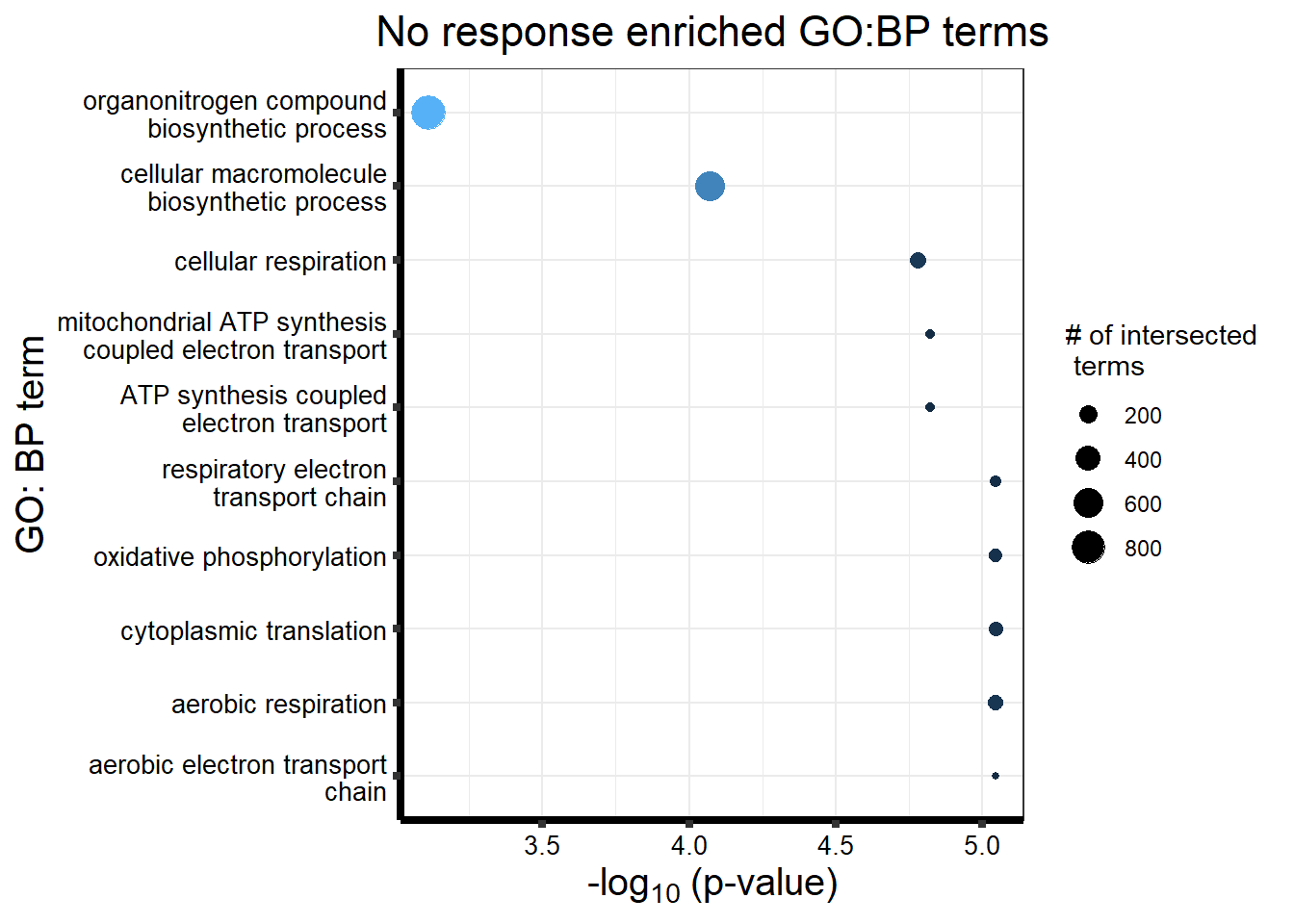
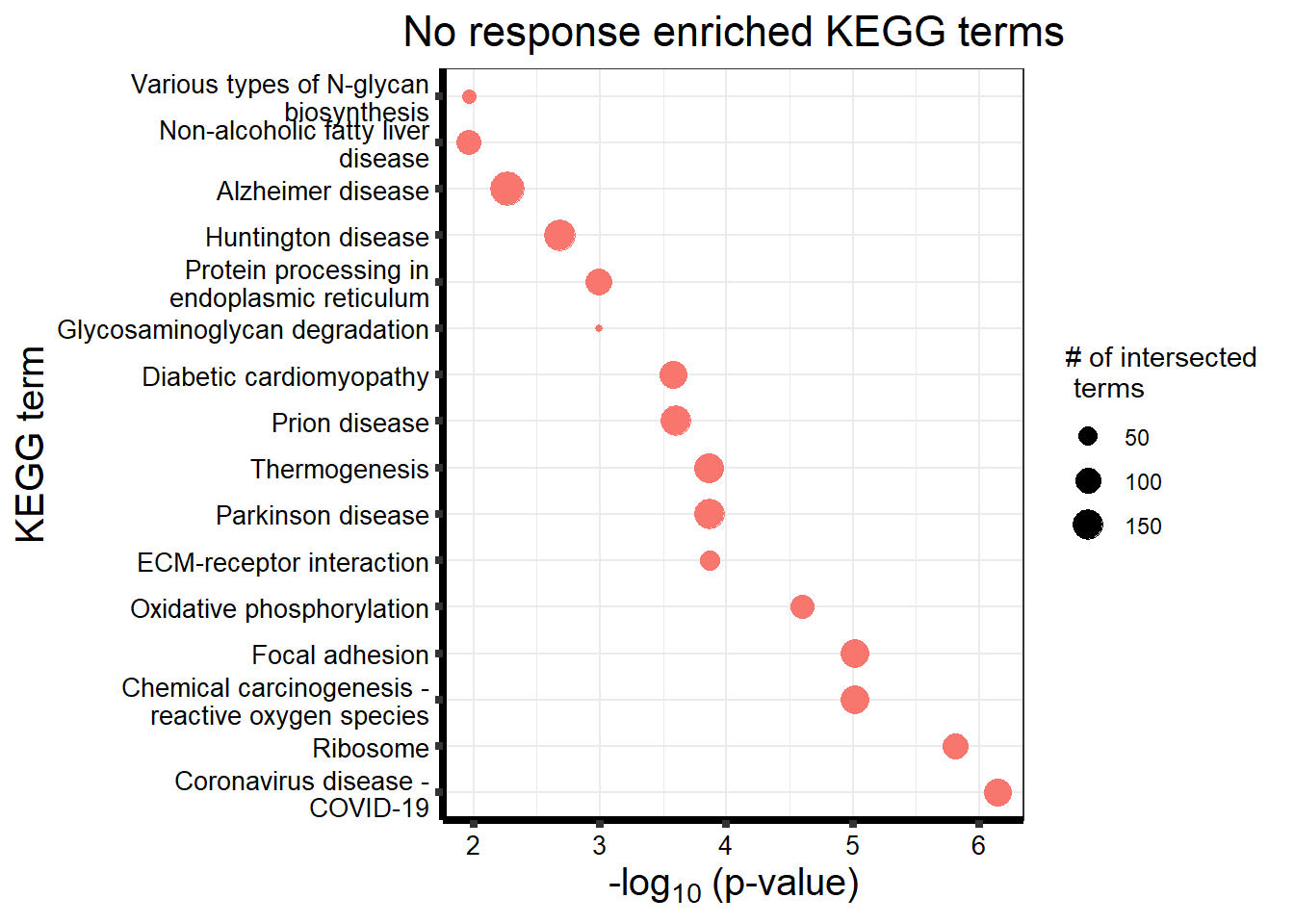
#NRresp <- read_csv("data/cormotif_NRset.txt")No response motif genes
| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0002181 | cytoplasmic translation | 117 | 155 | 9.02e-06 |
| GO:BP | GO:0019646 | aerobic electron transport chain | 63 | 74 | 9.02e-06 |
| GO:BP | GO:0009060 | aerobic respiration | 133 | 180 | 9.02e-06 |
| GO:BP | GO:0006119 | oxidative phosphorylation | 104 | 134 | 9.02e-06 |
| GO:BP | GO:0022904 | respiratory electron transport chain | 83 | 103 | 9.02e-06 |
| GO:BP | GO:0042775 | mitochondrial ATP synthesis coupled electron transport | 68 | 82 | 1.51e-05 |
| GO:BP | GO:0042773 | ATP synthesis coupled electron transport | 68 | 82 | 1.51e-05 |
| GO:BP | GO:0045333 | cellular respiration | 158 | 222 | 1.67e-05 |
| GO:BP | GO:0034645 | cellular macromolecule biosynthetic process | 624 | 1030 | 8.49e-05 |
| GO:BP | GO:1901566 | organonitrogen compound biosynthetic process | 878 | 1499 | 7.73e-04 |
| GO:BP | GO:0009100 | glycoprotein metabolic process | 209 | 317 | 1.00e-03 |
| GO:BP | GO:0022900 | electron transport chain | 102 | 141 | 1.29e-03 |
| GO:BP | GO:1901564 | organonitrogen compound metabolic process | 2774 | 5018 | 1.40e-03 |
| GO:BP | GO:0043603 | amide metabolic process | 597 | 999 | 1.44e-03 |
| GO:BP | GO:0006518 | peptide metabolic process | 481 | 793 | 1.50e-03 |
| GO:BP | GO:0015980 | energy derivation by oxidation of organic compounds | 196 | 298 | 1.91e-03 |
| GO:BP | GO:0070085 | glycosylation | 139 | 204 | 2.73e-03 |
| GO:BP | GO:0033108 | mitochondrial respiratory chain complex assembly | 71 | 94 | 2.73e-03 |
| GO:BP | GO:0006486 | protein glycosylation | 130 | 189 | 2.73e-03 |
| GO:BP | GO:0043413 | macromolecule glycosylation | 130 | 189 | 2.73e-03 |
| GO:BP | GO:0006412 | translation | 393 | 644 | 4.10e-03 |
| GO:BP | GO:0006091 | generation of precursor metabolites and energy | 272 | 432 | 4.10e-03 |
| GO:BP | GO:0022613 | ribonucleoprotein complex biogenesis | 281 | 449 | 5.52e-03 |
| GO:BP | GO:0019538 | protein metabolic process | 2347 | 4252 | 1.54e-02 |
| GO:BP | GO:0015986 | proton motive force-driven ATP synthesis | 51 | 66 | 1.54e-02 |
| GO:BP | GO:0043043 | peptide biosynthetic process | 400 | 664 | 1.54e-02 |
| GO:BP | GO:0043604 | amide biosynthetic process | 458 | 767 | 1.54e-02 |
| GO:BP | GO:0009101 | glycoprotein biosynthetic process | 165 | 254 | 1.87e-02 |
| GO:BP | GO:0006487 | protein N-linked glycosylation | 50 | 65 | 2.02e-02 |
| GO:BP | GO:0006754 | ATP biosynthetic process | 64 | 87 | 2.10e-02 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 529 | 899 | 2.68e-02 |
| GO:BP | GO:0010257 | NADH dehydrogenase complex assembly | 43 | 55 | 3.07e-02 |
| GO:BP | GO:0032981 | mitochondrial respiratory chain complex I assembly | 43 | 55 | 3.07e-02 |
| GO:BP | GO:0010466 | negative regulation of peptidase activity | 101 | 149 | 4.20e-02 |
| GO:BP | GO:0042776 | proton motive force-driven mitochondrial ATP synthesis | 44 | 57 | 4.20e-02 |
| KEGG | KEGG:05171 | Coronavirus disease - COVID-19 | 122 | 162 | 7.18e-07 |
| KEGG | KEGG:03010 | Ribosome | 98 | 127 | 1.54e-06 |
| KEGG | KEGG:04510 | Focal adhesion | 126 | 175 | 9.65e-06 |
| KEGG | KEGG:05208 | Chemical carcinogenesis - reactive oxygen species | 133 | 186 | 9.65e-06 |
| KEGG | KEGG:00190 | Oxidative phosphorylation | 81 | 106 | 2.52e-05 |
| KEGG | KEGG:04512 | ECM-receptor interaction | 55 | 69 | 1.36e-04 |
| KEGG | KEGG:05012 | Parkinson disease | 153 | 226 | 1.36e-04 |
| KEGG | KEGG:04714 | Thermogenesis | 134 | 195 | 1.39e-04 |
| KEGG | KEGG:05020 | Prion disease | 148 | 220 | 2.52e-04 |
| KEGG | KEGG:05415 | Diabetic cardiomyopathy | 117 | 169 | 2.64e-04 |
| KEGG | KEGG:00531 | Glycosaminoglycan degradation | 16 | 16 | 1.02e-03 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 110 | 161 | 1.02e-03 |
| KEGG | KEGG:05016 | Huntington disease | 165 | 256 | 2.07e-03 |
| KEGG | KEGG:05010 | Alzheimer disease | 197 | 315 | 5.40e-03 |
| KEGG | KEGG:00513 | Various types of N-glycan biosynthesis | 29 | 36 | 1.08e-02 |
| KEGG | KEGG:04932 | Non-alcoholic fatty liver disease | 88 | 131 | 1.08e-02 |
| KEGG | KEGG:04142 | Lysosome | 80 | 118 | 1.17e-02 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 36 | 48 | 2.41e-02 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 114 | 179 | 3.22e-02 |
| KEGG | KEGG:01200 | Carbon metabolism | 66 | 98 | 3.80e-02 |
| KEGG | KEGG:03040 | Spliceosome | 86 | 132 | 3.93e-02 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 230 | 385 | 4.28e-02 |
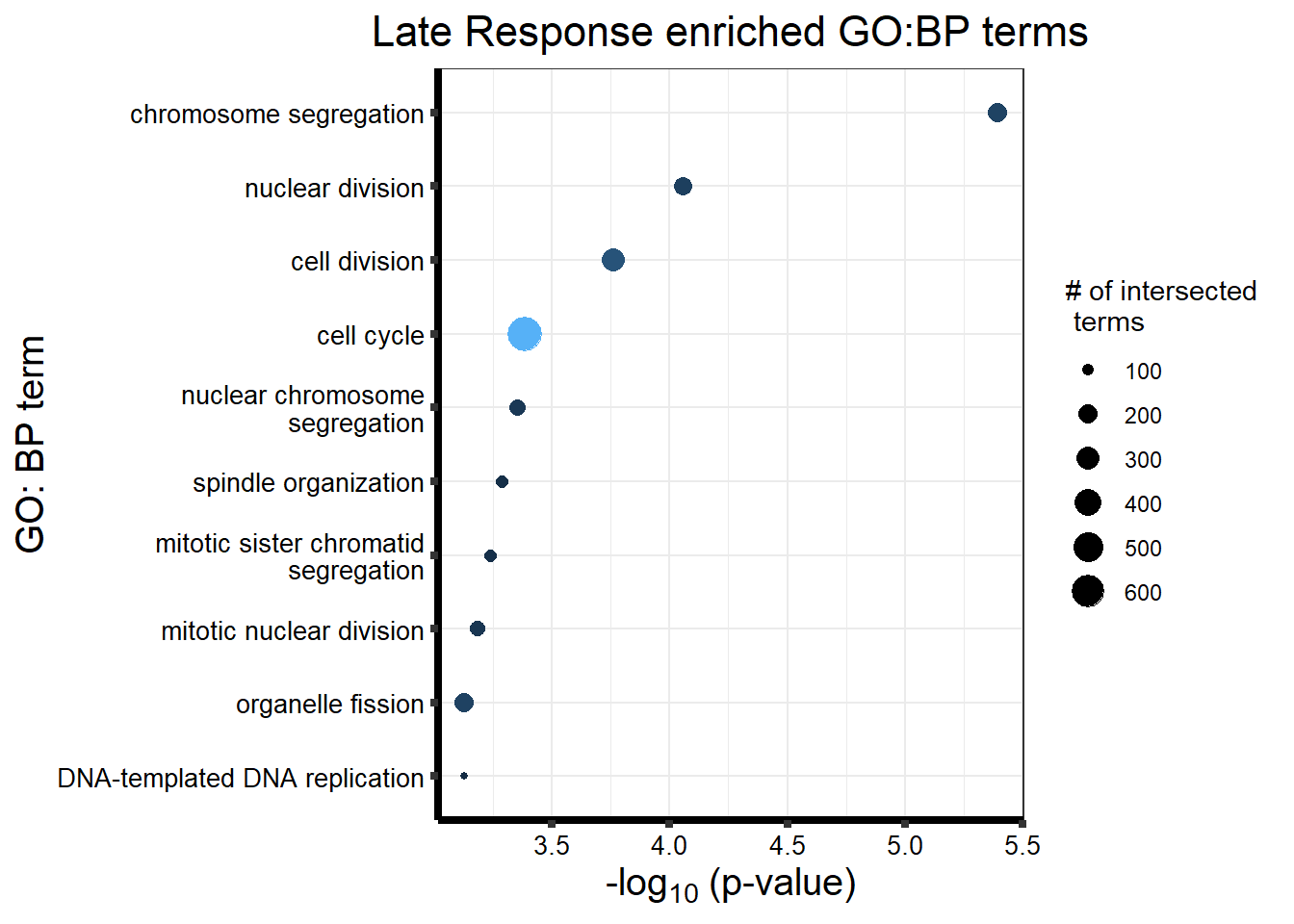
Late response Top2\(\beta\) inhibitor motif genes
| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0007059 | chromosome segregation | 207 | 378 | 4.07e-06 |
| GO:BP | GO:0000280 | nuclear division | 190 | 354 | 8.76e-05 |
| GO:BP | GO:0051301 | cell division | 284 | 567 | 1.73e-04 |
| GO:BP | GO:0007049 | cell cycle | 689 | 1522 | 4.11e-04 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 155 | 287 | 4.42e-04 |
| GO:BP | GO:0007051 | spindle organization | 107 | 186 | 5.13e-04 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 110 | 193 | 5.75e-04 |
| GO:BP | GO:0140014 | mitotic nuclear division | 143 | 264 | 6.50e-04 |
| GO:BP | GO:0006261 | DNA-templated DNA replication | 88 | 149 | 7.42e-04 |
| GO:BP | GO:0048285 | organelle fission | 203 | 397 | 7.42e-04 |
| GO:BP | GO:0061982 | meiosis I cell cycle process | 53 | 80 | 8.85e-04 |
| GO:BP | GO:0022402 | cell cycle process | 495 | 1075 | 9.31e-04 |
| GO:BP | GO:0140013 | meiotic nuclear division | 73 | 121 | 1.68e-03 |
| GO:BP | GO:0000278 | mitotic cell cycle | 390 | 833 | 1.71e-03 |
| GO:BP | GO:0007127 | meiosis I | 50 | 76 | 1.81e-03 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 80 | 136 | 1.85e-03 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 268 | 552 | 2.03e-03 |
| GO:BP | GO:0000819 | sister chromatid segregation | 126 | 234 | 2.07e-03 |
| GO:BP | GO:0045132 | meiotic chromosome segregation | 46 | 69 | 2.09e-03 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 331 | 700 | 2.51e-03 |
| GO:BP | GO:0051276 | chromosome organization | 261 | 543 | 5.74e-03 |
| GO:BP | GO:0007017 | microtubule-based process | 352 | 756 | 6.39e-03 |
| GO:BP | GO:0006260 | DNA replication | 134 | 258 | 9.24e-03 |
| GO:BP | GO:2001251 | negative regulation of chromosome organization | 55 | 90 | 9.83e-03 |
| GO:BP | GO:0070925 | organelle assembly | 370 | 803 | 1.06e-02 |
| GO:BP | GO:0051716 | cellular response to stimulus | 2043 | 4935 | 1.14e-02 |
| GO:BP | GO:0050896 | response to stimulus | 2360 | 5741 | 1.35e-02 |
| GO:BP | GO:0032465 | regulation of cytokinesis | 49 | 79 | 1.35e-02 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 86 | 156 | 1.45e-02 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 110 | 208 | 1.45e-02 |
| GO:BP | GO:0045839 | negative regulation of mitotic nuclear division | 36 | 54 | 1.49e-02 |
| GO:BP | GO:2000816 | negative regulation of mitotic sister chromatid separation | 32 | 47 | 1.89e-02 |
| GO:BP | GO:0033048 | negative regulation of mitotic sister chromatid segregation | 32 | 47 | 1.89e-02 |
| GO:BP | GO:0046474 | glycerophospholipid biosynthetic process | 101 | 190 | 1.89e-02 |
| GO:BP | GO:0033046 | negative regulation of sister chromatid segregation | 32 | 47 | 1.89e-02 |
| GO:BP | GO:0008654 | phospholipid biosynthetic process | 122 | 236 | 1.89e-02 |
| GO:BP | GO:0045017 | glycerolipid biosynthetic process | 114 | 219 | 2.09e-02 |
| GO:BP | GO:0006996 | organelle organization | 1253 | 2971 | 2.23e-02 |
| GO:BP | GO:0051784 | negative regulation of nuclear division | 37 | 57 | 2.23e-02 |
| GO:BP | GO:0071174 | mitotic spindle checkpoint signaling | 30 | 44 | 2.59e-02 |
| GO:BP | GO:0071173 | spindle assembly checkpoint signaling | 30 | 44 | 2.59e-02 |
| GO:BP | GO:0007094 | mitotic spindle assembly checkpoint signaling | 30 | 44 | 2.59e-02 |
| GO:BP | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition | 31 | 46 | 2.72e-02 |
| GO:BP | GO:1905819 | negative regulation of chromosome separation | 32 | 48 | 2.76e-02 |
| GO:BP | GO:0051985 | negative regulation of chromosome segregation | 32 | 48 | 2.76e-02 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 75 | 136 | 2.79e-02 |
| GO:BP | GO:0006270 | DNA replication initiation | 25 | 35 | 2.79e-02 |
| GO:BP | GO:0033047 | regulation of mitotic sister chromatid segregation | 34 | 52 | 2.85e-02 |
| GO:BP | GO:0051304 | chromosome separation | 45 | 74 | 3.10e-02 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 95 | 180 | 3.10e-02 |
| GO:BP | GO:0007052 | mitotic spindle organization | 71 | 128 | 3.10e-02 |
| GO:BP | GO:0021537 | telencephalon development | 103 | 198 | 3.39e-02 |
| GO:BP | GO:0051225 | spindle assembly | 66 | 118 | 3.52e-02 |
| GO:BP | GO:0031577 | spindle checkpoint signaling | 30 | 45 | 3.79e-02 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 163 | 334 | 3.84e-02 |
| GO:BP | GO:1902100 | negative regulation of metaphase/anaphase transition of cell cycle | 31 | 47 | 3.84e-02 |
| GO:BP | GO:1901653 | cellular response to peptide | 139 | 280 | 4.26e-02 |
| GO:BP | GO:1905818 | regulation of chromosome separation | 42 | 69 | 4.33e-02 |
| GO:BP | GO:0051256 | mitotic spindle midzone assembly | 9 | 9 | 4.43e-02 |
| GO:BP | GO:0090407 | organophosphate biosynthetic process | 241 | 517 | 4.81e-02 |
| GO:BP | GO:0010889 | regulation of sequestering of triglyceride | 11 | 12 | 4.81e-02 |
| GO:BP | GO:0019692 | deoxyribose phosphate metabolic process | 26 | 38 | 4.81e-02 |
| GO:BP | GO:0051255 | spindle midzone assembly | 11 | 12 | 4.81e-02 |
| GO:BP | GO:0009262 | deoxyribonucleotide metabolic process | 26 | 38 | 4.81e-02 |
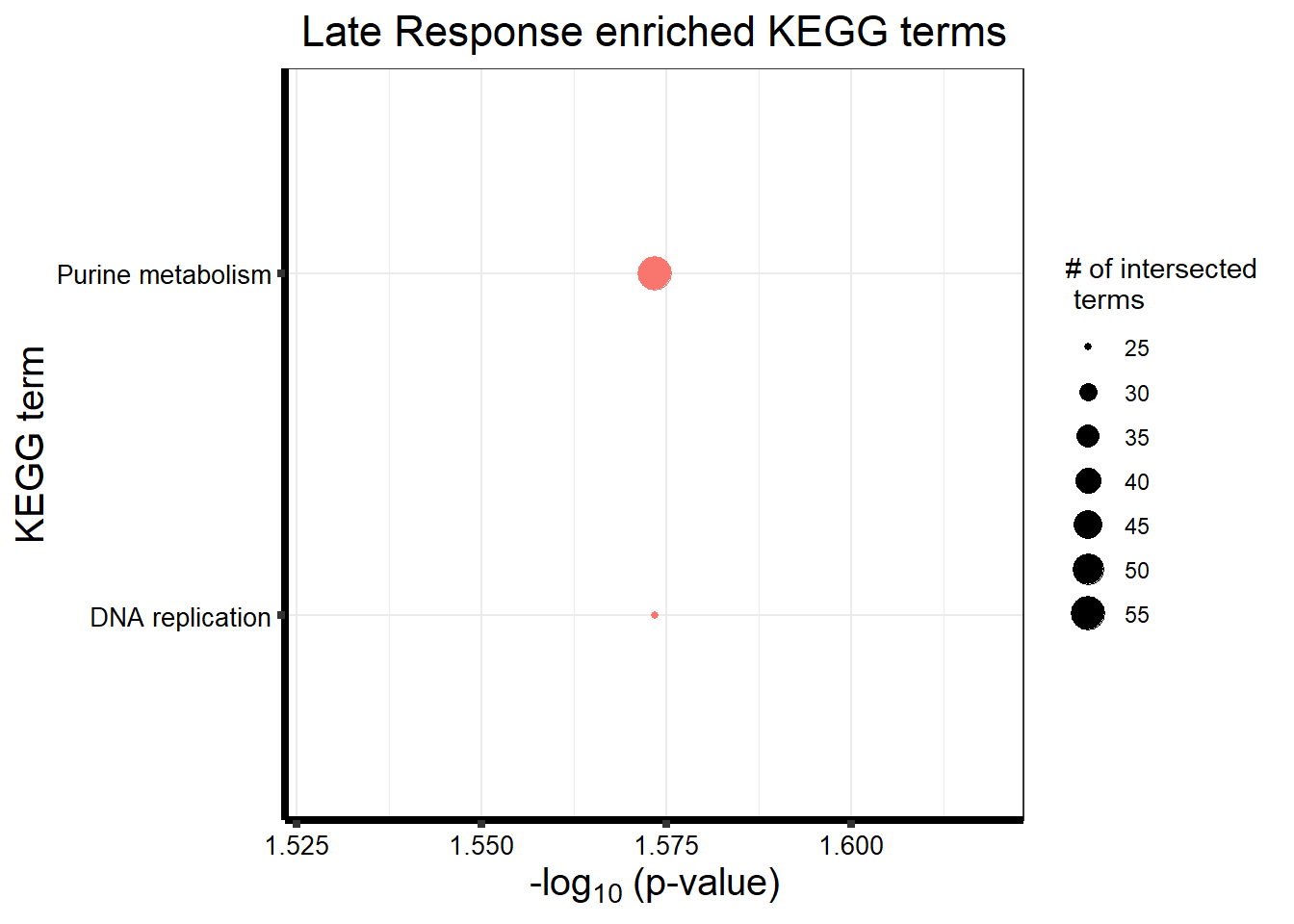
| KEGG | KEGG:00230 | Purine metabolism | 56 | 97 | 2.67e-02 |
| KEGG | KEGG:03030 | DNA replication | 25 | 35 | 2.67e-02 |
Early Response Top2\(\beta\) inhibitor motif genes
| query | significant | p_value | term_size | query_size | intersection_size | precision | recall | term_id | source | term_name | effective_domain_size | source_order | parents |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| query_1 | TRUE | 0.0000000 | 2683 | 428 | 2.16e+02 | 0.5046729 | 0.0805069 | GO:0006351 | GO:BP | DNA-templated transcription | 13587 | 2189 | GO:0097659 |
| query_1 | TRUE | 0.0000000 | 2684 | 428 | 2.16e+02 | 0.5046729 | 0.0804769 | GO:0097659 | GO:BP | nucleic acid-templated transcription | 13587 | 19903 | GO:0010467, GO:0032774 |
| query_1 | TRUE | 0.0000000 | 2714 | 428 | 2.16e+02 | 0.5046729 | 0.0795873 | GO:0032774 | GO:BP | RNA biosynthetic process | 13587 | 8281 | GO:0009059, GO:0016070, GO:0034654 |
| query_1 | TRUE | 0.0000000 | 1991 | 428 | 1.83e+02 | 0.4275701 | 0.0919136 | GO:0006366 | GO:BP | transcription by RNA polymerase II | 13587 | 2202 | GO:0006351 |
| query_1 | TRUE | 0.0000000 | 2581 | 428 | 2.09e+02 | 0.4883178 | 0.0809764 | GO:1903506 | GO:BP | regulation of nucleic acid-templated transcription | 13587 | 24286 | GO:0097659, GO:2001141 |
| query_1 | TRUE | 0.0000000 | 3105 | 428 | 2.31e+02 | 0.5397196 | 0.0743961 | GO:0019219 | GO:BP | regulation of nucleobase-containing compound metabolic process | 13587 | 5932 | GO:0006139, GO:0031323, GO:0051171, GO:0080090 |
| query_1 | TRUE | 0.0000000 | 2579 | 428 | 2.09e+02 | 0.4883178 | 0.0810392 | GO:0006355 | GO:BP | regulation of DNA-templated transcription | 13587 | 2193 | GO:0006351, GO:0010468, GO:1903506 |
| query_1 | TRUE | 0.0000000 | 1915 | 428 | 1.78e+02 | 0.4158879 | 0.0929504 | GO:0006357 | GO:BP | regulation of transcription by RNA polymerase II | 13587 | 2195 | GO:0006355, GO:0006366 |
| query_1 | TRUE | 0.0000000 | 2598 | 428 | 2.09e+02 | 0.4883178 | 0.0804465 | GO:2001141 | GO:BP | regulation of RNA biosynthetic process | 13587 | 27809 | GO:0010556, GO:0031326, GO:0032774, GO:0051252 |
| query_1 | TRUE | 0.0000000 | 2849 | 428 | 2.19e+02 | 0.5116822 | 0.0768691 | GO:0051252 | GO:BP | regulation of RNA metabolic process | 13587 | 14451 | GO:0016070, GO:0019219, GO:0060255 |
| query_1 | TRUE | 0.0000000 | 4047 | 428 | 2.60e+02 | 0.6074766 | 0.0642451 | GO:0090304 | GO:BP | nucleic acid metabolic process | 13587 | 19342 | GO:0006139, GO:0043170 |
| query_1 | TRUE | 0.0000000 | 3015 | 428 | 2.19e+02 | 0.5116822 | 0.0726368 | GO:0010556 | GO:BP | regulation of macromolecule biosynthetic process | 13587 | 4326 | GO:0009059, GO:0009889, GO:0060255 |
| query_1 | TRUE | 0.0000000 | 3593 | 428 | 2.41e+02 | 0.5630841 | 0.0670749 | GO:0016070 | GO:BP | RNA metabolic process | 13587 | 5254 | GO:0090304 |
| query_1 | TRUE | 0.0000000 | 4305 | 428 | 2.66e+02 | 0.6214953 | 0.0617886 | GO:0031323 | GO:BP | regulation of cellular metabolic process | 13587 | 7549 | GO:0019222, GO:0044237, GO:0050794 |
| query_1 | TRUE | 0.0000000 | 3083 | 428 | 2.19e+02 | 0.5116822 | 0.0710347 | GO:0034654 | GO:BP | nucleobase-containing compound biosynthetic process | 13587 | 9218 | GO:0006139, GO:0018130, GO:0019438, GO:0044271, GO:1901362 |
| query_1 | TRUE | 0.0000000 | 3123 | 428 | 2.20e+02 | 0.5140187 | 0.0704451 | GO:0031326 | GO:BP | regulation of cellular biosynthetic process | 13587 | 7552 | GO:0009889, GO:0031323, GO:0044249 |
| query_1 | TRUE | 0.0000000 | 3149 | 428 | 2.20e+02 | 0.5140187 | 0.0698634 | GO:0019438 | GO:BP | aromatic compound biosynthetic process | 13587 | 6132 | GO:0006725, GO:0044249 |
| query_1 | TRUE | 0.0000000 | 3148 | 428 | 2.20e+02 | 0.5140187 | 0.0698856 | GO:0018130 | GO:BP | heterocycle biosynthetic process | 13587 | 5537 | GO:0044249, GO:0046483 |
| query_1 | TRUE | 0.0000000 | 3174 | 428 | 2.20e+02 | 0.5140187 | 0.0693132 | GO:0009889 | GO:BP | regulation of biosynthetic process | 13587 | 3843 | GO:0009058, GO:0019222 |
| query_1 | TRUE | 0.0000000 | 3253 | 428 | 2.21e+02 | 0.5163551 | 0.0679373 | GO:1901362 | GO:BP | organic cyclic compound biosynthetic process | 13587 | 22499 | GO:1901360, GO:1901576 |
| query_1 | TRUE | 0.0000000 | 3583 | 428 | 2.33e+02 | 0.5443925 | 0.0650293 | GO:0010468 | GO:BP | regulation of gene expression | 13587 | 4271 | GO:0010467, GO:0060255 |
| query_1 | TRUE | 0.0000000 | 4440 | 428 | 2.63e+02 | 0.6144860 | 0.0592342 | GO:0080090 | GO:BP | regulation of primary metabolic process | 13587 | 18924 | GO:0019222, GO:0044238 |
| query_1 | TRUE | 0.0000000 | 4330 | 428 | 2.59e+02 | 0.6051402 | 0.0598152 | GO:0051171 | GO:BP | regulation of nitrogen compound metabolic process | 13587 | 14399 | GO:0006807, GO:0019222 |
| query_1 | TRUE | 0.0000000 | 3730 | 428 | 2.36e+02 | 0.5514019 | 0.0632708 | GO:0009059 | GO:BP | macromolecule biosynthetic process | 13587 | 3327 | GO:0043170, GO:1901576 |
| query_1 | TRUE | 0.0000000 | 4503 | 428 | 2.63e+02 | 0.6144860 | 0.0584055 | GO:0006139 | GO:BP | nucleobase-containing compound metabolic process | 13587 | 2036 | GO:0006725, GO:0034641, GO:0044238, GO:0046483, GO:1901360 |
| query_1 | TRUE | 0.0000000 | 4640 | 428 | 2.65e+02 | 0.6191589 | 0.0571121 | GO:0060255 | GO:BP | regulation of macromolecule metabolic process | 13587 | 15399 | GO:0019222, GO:0043170 |
| query_1 | TRUE | 0.0000000 | 4616 | 428 | 2.64e+02 | 0.6168224 | 0.0571924 | GO:0046483 | GO:BP | heterocycle metabolic process | 13587 | 12974 | GO:0044237 |
| query_1 | TRUE | 0.0000000 | 4646 | 428 | 2.64e+02 | 0.6168224 | 0.0568231 | GO:0006725 | GO:BP | cellular aromatic compound metabolic process | 13587 | 2497 | GO:0044237 |
| query_1 | TRUE | 0.0000000 | 3758 | 428 | 2.31e+02 | 0.5397196 | 0.0614689 | GO:0044271 | GO:BP | cellular nitrogen compound biosynthetic process | 13587 | 11656 | GO:0034641, GO:0044249 |
| query_1 | TRUE | 0.0000000 | 4790 | 428 | 2.66e+02 | 0.6214953 | 0.0555324 | GO:1901360 | GO:BP | organic cyclic compound metabolic process | 13587 | 22497 | GO:0071704 |
| query_1 | TRUE | 0.0000000 | 4572 | 428 | 2.58e+02 | 0.6028037 | 0.0564304 | GO:0010467 | GO:BP | gene expression | 13587 | 4270 | GO:0043170 |
| query_1 | TRUE | 0.0000000 | 5039 | 428 | 2.73e+02 | 0.6378505 | 0.0541774 | GO:0019222 | GO:BP | regulation of metabolic process | 13587 | 5935 | GO:0008152, GO:0050789 |
| query_1 | TRUE | 0.0000000 | 4991 | 428 | 2.68e+02 | 0.6261682 | 0.0536967 | GO:0034641 | GO:BP | cellular nitrogen compound metabolic process | 13587 | 9210 | GO:0006807, GO:0044237 |
| query_1 | TRUE | 0.0000000 | 4465 | 428 | 2.49e+02 | 0.5817757 | 0.0557671 | GO:0044249 | GO:BP | cellular biosynthetic process | 13587 | 11645 | GO:0009058, GO:0044237 |
| query_1 | TRUE | 0.0000000 | 4537 | 428 | 2.49e+02 | 0.5817757 | 0.0548821 | GO:1901576 | GO:BP | organic substance biosynthetic process | 13587 | 22687 | GO:0009058, GO:0071704 |
| query_1 | TRUE | 0.0000000 | 4595 | 428 | 2.49e+02 | 0.5817757 | 0.0541893 | GO:0009058 | GO:BP | biosynthetic process | 13587 | 3326 | GO:0008152 |
| query_1 | TRUE | 0.0000000 | 7057 | 428 | 3.22e+02 | 0.7523364 | 0.0456285 | GO:0043170 | GO:BP | macromolecule metabolic process | 13587 | 11211 | GO:0071704 |
| query_1 | TRUE | 0.0000000 | 7579 | 428 | 3.29e+02 | 0.7686916 | 0.0434094 | GO:0050794 | GO:BP | regulation of cellular process | 13587 | 14091 | GO:0009987, GO:0050789 |
| query_1 | TRUE | 0.0000000 | 1304 | 428 | 1.05e+02 | 0.2453271 | 0.0805215 | GO:1903508 | GO:BP | positive regulation of nucleic acid-templated transcription | 13587 | 24288 | GO:0097659, GO:1902680, GO:1903506 |
| query_1 | TRUE | 0.0000000 | 1304 | 428 | 1.05e+02 | 0.2453271 | 0.0805215 | GO:0045893 | GO:BP | positive regulation of DNA-templated transcription | 13587 | 12467 | GO:0006351, GO:0006355, GO:1903508 |
| query_1 | TRUE | 0.0000000 | 1431 | 428 | 1.11e+02 | 0.2593458 | 0.0775681 | GO:0051254 | GO:BP | positive regulation of RNA metabolic process | 13587 | 14453 | GO:0010604, GO:0016070, GO:0045935, GO:0051252 |
| query_1 | TRUE | 0.0000000 | 1311 | 428 | 1.05e+02 | 0.2453271 | 0.0800915 | GO:1902680 | GO:BP | positive regulation of RNA biosynthetic process | 13587 | 23603 | GO:0010557, GO:0031328, GO:0032774, GO:0051254, GO:2001141 |
| query_1 | TRUE | 0.0000000 | 7503 | 428 | 3.25e+02 | 0.7593458 | 0.0433160 | GO:0044237 | GO:BP | cellular metabolic process | 13587 | 11638 | GO:0008152, GO:0009987 |
| query_1 | TRUE | 0.0000000 | 1600 | 428 | 1.16e+02 | 0.2710280 | 0.0725000 | GO:0045935 | GO:BP | positive regulation of nucleobase-containing compound metabolic process | 13587 | 12505 | GO:0006139, GO:0019219, GO:0031325, GO:0051173 |
| query_1 | TRUE | 0.0000000 | 7474 | 428 | 3.21e+02 | 0.7500000 | 0.0429489 | GO:0006807 | GO:BP | nitrogen compound metabolic process | 13587 | 2557 | GO:0008152 |
| query_1 | TRUE | 0.0000000 | 7992 | 428 | 3.35e+02 | 0.7827103 | 0.0419169 | GO:0050789 | GO:BP | regulation of biological process | 13587 | 14087 | GO:0008150, GO:0065007 |
| query_1 | TRUE | 0.0000000 | 7800 | 428 | 3.29e+02 | 0.7686916 | 0.0421795 | GO:0044238 | GO:BP | primary metabolic process | 13587 | 11639 | GO:0008152 |
| query_1 | TRUE | 0.0000000 | 8348 | 428 | 3.44e+02 | 0.8037383 | 0.0412075 | GO:0065007 | GO:BP | biological regulation | 13587 | 16927 | GO:0008150 |
| query_1 | TRUE | 0.0000000 | 1499 | 428 | 1.09e+02 | 0.2546729 | 0.0727151 | GO:0010557 | GO:BP | positive regulation of macromolecule biosynthetic process | 13587 | 4327 | GO:0009059, GO:0009891, GO:0010556, GO:0010604 |
| query_1 | TRUE | 0.0000000 | 1009 | 428 | 8.50e+01 | 0.1985981 | 0.0842418 | GO:0045892 | GO:BP | negative regulation of DNA-templated transcription | 13587 | 12466 | GO:0006351, GO:0006355, GO:1903507 |
| query_1 | TRUE | 0.0000000 | 1011 | 428 | 8.50e+01 | 0.1985981 | 0.0840752 | GO:1903507 | GO:BP | negative regulation of nucleic acid-templated transcription | 13587 | 24287 | GO:0097659, GO:1902679, GO:1903506 |
| query_1 | TRUE | 0.0000000 | 1020 | 428 | 8.50e+01 | 0.1985981 | 0.0833333 | GO:1902679 | GO:BP | negative regulation of RNA biosynthetic process | 13587 | 23602 | GO:0010558, GO:0031327, GO:0032774, GO:0051253, GO:2001141 |
| query_1 | TRUE | 0.0000000 | 1215 | 428 | 9.40e+01 | 0.2196262 | 0.0773663 | GO:0045934 | GO:BP | negative regulation of nucleobase-containing compound metabolic process | 13587 | 12504 | GO:0006139, GO:0019219, GO:0031324, GO:0051172 |
| query_1 | TRUE | 0.0000000 | 1115 | 428 | 8.90e+01 | 0.2079439 | 0.0798206 | GO:0051253 | GO:BP | negative regulation of RNA metabolic process | 13587 | 14452 | GO:0010605, GO:0016070, GO:0045934, GO:0051252 |
| query_1 | TRUE | 0.0000000 | 925 | 428 | 7.90e+01 | 0.1845794 | 0.0854054 | GO:0045944 | GO:BP | positive regulation of transcription by RNA polymerase II | 13587 | 12513 | GO:0006357, GO:0006366, GO:0045893 |
| query_1 | TRUE | 0.0000000 | 1571 | 428 | 1.09e+02 | 0.2546729 | 0.0693826 | GO:0031328 | GO:BP | positive regulation of cellular biosynthetic process | 13587 | 7554 | GO:0009891, GO:0031325, GO:0031326, GO:0044249 |
| query_1 | TRUE | 0.0000000 | 8150 | 428 | 3.34e+02 | 0.7803738 | 0.0409816 | GO:0071704 | GO:BP | organic substance metabolic process | 13587 | 17953 | GO:0008152 |
| query_1 | TRUE | 0.0000000 | 1598 | 428 | 1.09e+02 | 0.2546729 | 0.0682103 | GO:0009891 | GO:BP | positive regulation of biosynthetic process | 13587 | 3845 | GO:0009058, GO:0009889, GO:0009893 |
| query_1 | TRUE | 0.0000000 | 1780 | 428 | 1.16e+02 | 0.2710280 | 0.0651685 | GO:0031324 | GO:BP | negative regulation of cellular metabolic process | 13587 | 7550 | GO:0009892, GO:0031323, GO:0044237, GO:0048523 |
| query_1 | TRUE | 0.0000000 | 1206 | 428 | 9.00e+01 | 0.2102804 | 0.0746269 | GO:0010558 | GO:BP | negative regulation of macromolecule biosynthetic process | 13587 | 4328 | GO:0009059, GO:0009890, GO:0010556, GO:0010605 |
| query_1 | TRUE | 0.0000000 | 735 | 428 | 6.50e+01 | 0.1518692 | 0.0884354 | GO:0000122 | GO:BP | negative regulation of transcription by RNA polymerase II | 13587 | 51 | GO:0006357, GO:0006366, GO:0045892 |
| query_1 | TRUE | 0.0000000 | 2373 | 428 | 1.38e+02 | 0.3224299 | 0.0581542 | GO:0031325 | GO:BP | positive regulation of cellular metabolic process | 13587 | 7551 | GO:0009893, GO:0031323, GO:0044237, GO:0048522 |
| query_1 | TRUE | 0.0000000 | 1249 | 428 | 9.00e+01 | 0.2102804 | 0.0720576 | GO:0031327 | GO:BP | negative regulation of cellular biosynthetic process | 13587 | 7553 | GO:0009890, GO:0031324, GO:0031326, GO:0044249 |
| query_1 | TRUE | 0.0000000 | 2425 | 428 | 1.40e+02 | 0.3271028 | 0.0577320 | GO:0051173 | GO:BP | positive regulation of nitrogen compound metabolic process | 13587 | 14401 | GO:0006807, GO:0009893, GO:0051171 |
| query_1 | TRUE | 0.0000000 | 1273 | 428 | 9.00e+01 | 0.2102804 | 0.0706991 | GO:0009890 | GO:BP | negative regulation of biosynthetic process | 13587 | 3844 | GO:0009058, GO:0009889, GO:0009892 |
| query_1 | TRUE | 0.0000000 | 2648 | 428 | 1.47e+02 | 0.3434579 | 0.0555136 | GO:0010604 | GO:BP | positive regulation of macromolecule metabolic process | 13587 | 4369 | GO:0009893, GO:0043170, GO:0060255 |
| query_1 | TRUE | 0.0000000 | 8501 | 428 | 3.36e+02 | 0.7850467 | 0.0395248 | GO:0008152 | GO:BP | metabolic process | 13587 | 3213 | GO:0008150 |
| query_1 | TRUE | 0.0000000 | 2306 | 428 | 1.30e+02 | 0.3037383 | 0.0563747 | GO:0009892 | GO:BP | negative regulation of metabolic process | 13587 | 3846 | GO:0008152, GO:0019222, GO:0048519 |
| query_1 | TRUE | 0.0000000 | 1866 | 428 | 1.12e+02 | 0.2616822 | 0.0600214 | GO:0051172 | GO:BP | negative regulation of nitrogen compound metabolic process | 13587 | 14400 | GO:0006807, GO:0009892, GO:0051171 |
| query_1 | TRUE | 0.0000000 | 2146 | 428 | 1.23e+02 | 0.2873832 | 0.0573159 | GO:0010605 | GO:BP | negative regulation of macromolecule metabolic process | 13587 | 4370 | GO:0009892, GO:0043170, GO:0060255 |
| query_1 | TRUE | 0.0000000 | 2884 | 428 | 1.49e+02 | 0.3481308 | 0.0516644 | GO:0009893 | GO:BP | positive regulation of metabolic process | 13587 | 3847 | GO:0008152, GO:0019222, GO:0048518 |
| query_1 | TRUE | 0.0000000 | 3628 | 428 | 1.75e+02 | 0.4088785 | 0.0482359 | GO:0048523 | GO:BP | negative regulation of cellular process | 13587 | 13627 | GO:0009987, GO:0048519, GO:0050794 |
| query_1 | TRUE | 0.0000000 | 4035 | 428 | 1.87e+02 | 0.4369159 | 0.0463445 | GO:0048519 | GO:BP | negative regulation of biological process | 13587 | 13623 | GO:0008150, GO:0050789 |
| query_1 | TRUE | 0.0000001 | 536 | 428 | 4.50e+01 | 0.1051402 | 0.0839552 | GO:0006325 | GO:BP | chromatin organization | 13587 | 2182 | GO:0016043 |
| query_1 | TRUE | 0.0000027 | 309 | 428 | 3.00e+01 | 0.0700935 | 0.0970874 | GO:0006338 | GO:BP | chromatin remodeling | 13587 | 2186 | GO:0006325 |
| query_1 | TRUE | 0.0000562 | 4148 | 428 | 1.77e+02 | 0.4135514 | 0.0426712 | GO:0048522 | GO:BP | positive regulation of cellular process | 13587 | 13626 | GO:0009987, GO:0048518, GO:0050794 |
| query_1 | TRUE | 0.0001845 | 4577 | 428 | 1.89e+02 | 0.4415888 | 0.0412934 | GO:0048518 | GO:BP | positive regulation of biological process | 13587 | 13622 | GO:0008150, GO:0050789 |
| query_1 | TRUE | 0.0002647 | 424 | 428 | 3.20e+01 | 0.0747664 | 0.0754717 | GO:0016570 | GO:BP | histone modification | 13587 | 5415 | GO:0036211 |
| query_1 | TRUE | 0.0011631 | 12 | 428 | 5.00e+00 | 0.0116822 | 0.4166667 | GO:0010452 | GO:BP | histone H3-K36 methylation | 13587 | 4257 | GO:0034968 |
| query_1 | TRUE | 0.0013697 | 1829 | 428 | 8.80e+01 | 0.2056075 | 0.0481137 | GO:0050793 | GO:BP | regulation of developmental process | 13587 | 14090 | GO:0032502, GO:0050789 |
| query_1 | TRUE | 0.0013866 | 120 | 428 | 1.40e+01 | 0.0327103 | 0.1166667 | GO:0018022 | GO:BP | peptidyl-lysine methylation | 13587 | 5494 | GO:0006479, GO:0018205 |
| query_1 | TRUE | 0.0015606 | 224 | 428 | 2.00e+01 | 0.0467290 | 0.0892857 | GO:0006354 | GO:BP | DNA-templated transcription elongation | 13587 | 2192 | GO:0006351, GO:0032774 |
| query_1 | TRUE | 0.0017063 | 107 | 428 | 1.30e+01 | 0.0303738 | 0.1214953 | GO:0034968 | GO:BP | histone lysine methylation | 13587 | 9262 | GO:0016571, GO:0018022 |
| query_1 | TRUE | 0.0020798 | 141 | 428 | 1.50e+01 | 0.0350467 | 0.1063830 | GO:0016571 | GO:BP | histone methylation | 13587 | 5416 | GO:0006479, GO:0016570 |
| query_1 | TRUE | 0.0055369 | 367 | 428 | 2.60e+01 | 0.0607477 | 0.0708447 | GO:0018205 | GO:BP | peptidyl-lysine modification | 13587 | 5601 | GO:0018193 |
| query_1 | TRUE | 0.0064837 | 4 | 428 | 3.00e+00 | 0.0070093 | 0.7500000 | GO:0097676 | GO:BP | histone H3-K36 dimethylation | 13587 | 19904 | GO:0010452, GO:0018027 |
| query_1 | TRUE | 0.0083303 | 178 | 428 | 1.60e+01 | 0.0373832 | 0.0898876 | GO:0006479 | GO:BP | protein methylation | 13587 | 2280 | GO:0008213, GO:0043414 |
| query_1 | TRUE | 0.0083303 | 178 | 428 | 1.60e+01 | 0.0373832 | 0.0898876 | GO:0008213 | GO:BP | protein alkylation | 13587 | 3228 | GO:0036211 |
| query_1 | TRUE | 0.0083303 | 789 | 428 | 4.40e+01 | 0.1028037 | 0.0557668 | GO:0006974 | GO:BP | cellular response to DNA damage stimulus | 13587 | 2675 | GO:0033554 |
| query_1 | TRUE | 0.0094433 | 18 | 428 | 5.00e+00 | 0.0116822 | 0.2777778 | GO:0006607 | GO:BP | NLS-bearing protein import into nucleus | 13587 | 2391 | GO:0006606 |
| query_1 | TRUE | 0.0099108 | 845 | 428 | 4.60e+01 | 0.1074766 | 0.0544379 | GO:0060429 | GO:BP | epithelium development | 13587 | 15559 | GO:0009888 |
| query_1 | TRUE | 0.0110510 | 657 | 428 | 3.80e+01 | 0.0887850 | 0.0578387 | GO:0016071 | GO:BP | mRNA metabolic process | 13587 | 5255 | GO:0016070 |
| query_1 | TRUE | 0.0147308 | 84 | 428 | 1.00e+01 | 0.0233645 | 0.1190476 | GO:0045814 | GO:BP | negative regulation of gene expression, epigenetic | 13587 | 12410 | GO:0010629, GO:0040029 |
| query_1 | TRUE | 0.0151756 | 227 | 428 | 1.80e+01 | 0.0420561 | 0.0792952 | GO:0030522 | GO:BP | intracellular receptor signaling pathway | 13587 | 7237 | GO:0007165 |
| query_1 | TRUE | 0.0227542 | 879 | 428 | 4.60e+01 | 0.1074766 | 0.0523322 | GO:0072359 | GO:BP | circulatory system development | 13587 | 18474 | GO:0048731 |
| query_1 | TRUE | 0.0231340 | 177 | 428 | 1.50e+01 | 0.0350467 | 0.0847458 | GO:0090596 | GO:BP | sensory organ morphogenesis | 13587 | 19524 | GO:0007423, GO:0009887 |
| query_1 | TRUE | 0.0235158 | 277 | 428 | 2.00e+01 | 0.0467290 | 0.0722022 | GO:1903706 | GO:BP | regulation of hemopoiesis | 13587 | 24477 | GO:0002682, GO:0030097, GO:0060284, GO:2000026 |
| query_1 | TRUE | 0.0235158 | 197 | 428 | 1.60e+01 | 0.0373832 | 0.0812183 | GO:1902105 | GO:BP | regulation of leukocyte differentiation | 13587 | 23159 | GO:0002521, GO:1903706 |
| query_1 | TRUE | 0.0235158 | 74 | 428 | 9.00e+00 | 0.0210280 | 0.1216216 | GO:2000736 | GO:BP | regulation of stem cell differentiation | 13587 | 27440 | GO:0045595, GO:0048863 |
| query_1 | TRUE | 0.0235158 | 141 | 428 | 1.30e+01 | 0.0303738 | 0.0921986 | GO:0040029 | GO:BP | epigenetic regulation of gene expression | 13587 | 10536 | GO:0006325, GO:0010468 |
| query_1 | TRUE | 0.0263267 | 6 | 428 | 3.00e+00 | 0.0070093 | 0.5000000 | GO:0086023 | GO:BP | adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process | 13587 | 19024 | GO:0071880, GO:0086103 |
| query_1 | TRUE | 0.0278172 | 76 | 428 | 9.00e+00 | 0.0210280 | 0.1184211 | GO:0031507 | GO:BP | heterochromatin formation | 13587 | 7631 | GO:0045814, GO:0070828 |
| query_1 | TRUE | 0.0306558 | 164 | 428 | 1.40e+01 | 0.0327103 | 0.0853659 | GO:0007623 | GO:BP | circadian rhythm | 13587 | 3168 | GO:0048511 |
| query_1 | TRUE | 0.0318091 | 531 | 428 | 3.10e+01 | 0.0724299 | 0.0583804 | GO:0043009 | GO:BP | chordate embryonic development | 13587 | 11117 | GO:0009792 |
| query_1 | TRUE | 0.0360655 | 36 | 428 | 6.00e+00 | 0.0140187 | 0.1666667 | GO:1902275 | GO:BP | regulation of chromatin organization | 13587 | 23312 | GO:0006325, GO:0051128 |
| query_1 | TRUE | 0.0398125 | 150 | 428 | 1.30e+01 | 0.0303738 | 0.0866667 | GO:0019827 | GO:BP | stem cell population maintenance | 13587 | 6416 | GO:0032501, GO:0098727 |
| query_1 | TRUE | 0.0417637 | 7 | 428 | 3.00e+00 | 0.0070093 | 0.4285714 | GO:1900246 | GO:BP | positive regulation of RIG-I signaling pathway | 13587 | 21551 | GO:0039529, GO:0039535, GO:0062208 |
| query_1 | TRUE | 0.0417637 | 2 | 428 | 2.00e+00 | 0.0046729 | 1.0000000 | GO:0032242 | GO:BP | regulation of nucleoside transport | 13587 | 7889 | GO:0015858, GO:0032239 |
| query_1 | TRUE | 0.0417637 | 2 | 428 | 2.00e+00 | 0.0046729 | 1.0000000 | GO:1901898 | GO:BP | negative regulation of relaxation of cardiac muscle | 13587 | 22972 | GO:0055119, GO:1901078, GO:1901897 |
| query_1 | TRUE | 0.0420232 | 15 | 428 | 4.00e+00 | 0.0093458 | 0.2666667 | GO:0032239 | GO:BP | regulation of nucleobase-containing compound transport | 13587 | 7886 | GO:0015931, GO:0051049 |
| query_1 | TRUE | 0.0429893 | 152 | 428 | 1.30e+01 | 0.0303738 | 0.0855263 | GO:0098727 | GO:BP | maintenance of cell number | 13587 | 20042 | GO:0032502 |
| query_1 | TRUE | 0.0468968 | 1415 | 428 | 6.50e+01 | 0.1518692 | 0.0459364 | GO:0009888 | GO:BP | tissue development | 13587 | 3842 | GO:0048856 |
| query_1 | TRUE | 0.0473282 | 154 | 428 | 1.30e+01 | 0.0303738 | 0.0844156 | GO:0045165 | GO:BP | cell fate commitment | 13587 | 12071 | GO:0030154, GO:0048869 |
| query_1 | TRUE | 0.0473282 | 83 | 428 | 9.00e+00 | 0.0210280 | 0.1084337 | GO:0070828 | GO:BP | heterochromatin organization | 13587 | 17383 | GO:0006338 |
| query_1 | TRUE | 0.0475458 | 501 | 428 | 2.90e+01 | 0.0677570 | 0.0578842 | GO:0048729 | GO:BP | tissue morphogenesis | 13587 | 13809 | GO:0009653, GO:0009888 |
| query_1 | TRUE | 0.0475458 | 234 | 428 | 1.70e+01 | 0.0397196 | 0.0726496 | GO:0048511 | GO:BP | rhythmic process | 13587 | 13617 | GO:0008150 |
| query_1 | TRUE | 0.0475458 | 548 | 428 | 3.10e+01 | 0.0724299 | 0.0565693 | GO:0009792 | GO:BP | embryo development ending in birth or egg hatching | 13587 | 3773 | GO:0009790 |
| query_1 | TRUE | 0.0477912 | 118 | 428 | 1.10e+01 | 0.0257009 | 0.0932203 | GO:1902107 | GO:BP | positive regulation of leukocyte differentiation | 13587 | 23161 | GO:0002521, GO:1902105, GO:1903708 |
| query_1 | TRUE | 0.0477912 | 118 | 428 | 1.10e+01 | 0.0257009 | 0.0932203 | GO:1903708 | GO:BP | positive regulation of hemopoiesis | 13587 | 24479 | GO:0002684, GO:0010720, GO:0030097, GO:0051240, GO:1903706 |
| query_1 | TRUE | 0.0477912 | 502 | 428 | 2.90e+01 | 0.0677570 | 0.0577689 | GO:0007507 | GO:BP | heart development | 13587 | 3080 | GO:0048513, GO:0072359 |
| query_1 | TRUE | 0.0481745 | 39 | 428 | 6.00e+00 | 0.0140187 | 0.1538462 | GO:2001222 | GO:BP | regulation of neuron migration | 13587 | 27870 | GO:0001764, GO:0030334 |
| query_1 | TRUE | 0.0481745 | 871 | 428 | 4.40e+01 | 0.1028037 | 0.0505166 | GO:0009790 | GO:BP | embryo development | 13587 | 3771 | GO:0007275 |
| query_1 | TRUE | 0.0000000 | 415 | 428 | 4.50e+01 | 0.1051402 | 0.1084337 | KEGG:05168 | KEGG | Herpes simplex virus 1 infection | 13587 | 442 | KEGG:00000 |
| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0006351 | DNA-templated transcription | 216 | 2683 | 4.83e-44 |
| GO:BP | GO:0097659 | nucleic acid-templated transcription | 216 | 2684 | 4.83e-44 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 216 | 2714 | 1.67e-43 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 183 | 1991 | 1.67e-43 |
| GO:BP | GO:1903506 | regulation of nucleic acid-templated transcription | 209 | 2581 | 6.11e-43 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 231 | 3105 | 6.11e-43 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 209 | 2579 | 6.11e-43 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 178 | 1915 | 6.11e-43 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 209 | 2598 | 1.31e-42 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 219 | 2849 | 3.56e-42 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 260 | 4047 | 8.30e-39 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 219 | 3015 | 4.49e-38 |
| GO:BP | GO:0016070 | RNA metabolic process | 241 | 3593 | 1.21e-37 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 266 | 4305 | 4.72e-37 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 219 | 3083 | 1.48e-36 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 220 | 3123 | 3.12e-36 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 220 | 3149 | 1.09e-35 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 220 | 3148 | 1.09e-35 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 220 | 3174 | 3.81e-35 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 221 | 3253 | 5.63e-34 |
| GO:BP | GO:0010468 | regulation of gene expression | 233 | 3583 | 1.42e-33 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 263 | 4440 | 5.67e-33 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 259 | 4330 | 6.53e-33 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 236 | 3730 | 2.90e-32 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 263 | 4503 | 7.26e-32 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 265 | 4640 | 1.81e-30 |
| GO:BP | GO:0046483 | heterocycle metabolic process | 264 | 4616 | 2.15e-30 |
| GO:BP | GO:0006725 | cellular aromatic compound metabolic process | 264 | 4646 | 6.97e-30 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 231 | 3758 | 3.33e-29 |
| GO:BP | GO:1901360 | organic cyclic compound metabolic process | 266 | 4790 | 1.86e-28 |
| GO:BP | GO:0010467 | gene expression | 258 | 4572 | 3.44e-28 |
| GO:BP | GO:0019222 | regulation of metabolic process | 273 | 5039 | 7.88e-28 |
| GO:BP | GO:0034641 | cellular nitrogen compound metabolic process | 268 | 4991 | 3.45e-26 |
| GO:BP | GO:0044249 | cellular biosynthetic process | 249 | 4465 | 1.14e-25 |
| GO:BP | GO:1901576 | organic substance biosynthetic process | 249 | 4537 | 1.61e-24 |
| GO:BP | GO:0009058 | biosynthetic process | 249 | 4595 | 1.28e-23 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 322 | 7057 | 9.57e-22 |
| GO:BP | GO:0050794 | regulation of cellular process | 329 | 7579 | 2.78e-18 |
| GO:BP | GO:1903508 | positive regulation of nucleic acid-templated transcription | 105 | 1304 | 4.26e-18 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 105 | 1304 | 4.26e-18 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 111 | 1431 | 4.68e-18 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 105 | 1311 | 6.07e-18 |
| GO:BP | GO:0044237 | cellular metabolic process | 325 | 7503 | 1.55e-17 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 116 | 1600 | 1.05e-16 |
| GO:BP | GO:0006807 | nitrogen compound metabolic process | 321 | 7474 | 3.03e-16 |
| GO:BP | GO:0050789 | regulation of biological process | 335 | 7992 | 4.68e-16 |
| GO:BP | GO:0044238 | primary metabolic process | 329 | 7800 | 8.56e-16 |
| GO:BP | GO:0065007 | biological regulation | 344 | 8348 | 8.56e-16 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 109 | 1499 | 1.23e-15 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 85 | 1009 | 2.00e-15 |
| GO:BP | GO:1903507 | negative regulation of nucleic acid-templated transcription | 85 | 1011 | 2.21e-15 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 85 | 1020 | 3.73e-15 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 94 | 1215 | 7.17e-15 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 89 | 1115 | 8.33e-15 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 79 | 925 | 1.42e-14 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 109 | 1571 | 3.23e-14 |
| GO:BP | GO:0071704 | organic substance metabolic process | 334 | 8150 | 5.29e-14 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 109 | 1598 | 1.04e-13 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 116 | 1780 | 2.57e-13 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 90 | 1206 | 3.13e-13 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 65 | 735 | 1.83e-12 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 138 | 2373 | 2.39e-12 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 90 | 1249 | 2.44e-12 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 140 | 2425 | 2.46e-12 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 90 | 1273 | 7.26e-12 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 147 | 2648 | 1.02e-11 |
| GO:BP | GO:0008152 | metabolic process | 336 | 8501 | 3.09e-11 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 130 | 2306 | 1.80e-10 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 112 | 1866 | 2.11e-10 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 123 | 2146 | 2.90e-10 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 149 | 2884 | 2.01e-09 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 175 | 3628 | 4.49e-09 |
| GO:BP | GO:0048519 | negative regulation of biological process | 187 | 4035 | 2.05e-08 |
| GO:BP | GO:0006325 | chromatin organization | 45 | 536 | 1.13e-07 |
| GO:BP | GO:0006338 | chromatin remodeling | 30 | 309 | 2.71e-06 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 177 | 4148 | 5.62e-05 |
| GO:BP | GO:0048518 | positive regulation of biological process | 189 | 4577 | 1.85e-04 |
| GO:BP | GO:0016570 | histone modification | 32 | 424 | 2.65e-04 |
| GO:BP | GO:0010452 | histone H3-K36 methylation | 5 | 12 | 1.16e-03 |
| GO:BP | GO:0050793 | regulation of developmental process | 88 | 1829 | 1.37e-03 |
| GO:BP | GO:0018022 | peptidyl-lysine methylation | 14 | 120 | 1.39e-03 |
| GO:BP | GO:0006354 | DNA-templated transcription elongation | 20 | 224 | 1.56e-03 |
| GO:BP | GO:0034968 | histone lysine methylation | 13 | 107 | 1.71e-03 |
| GO:BP | GO:0016571 | histone methylation | 15 | 141 | 2.08e-03 |
| GO:BP | GO:0018205 | peptidyl-lysine modification | 26 | 367 | 5.54e-03 |
| GO:BP | GO:0097676 | histone H3-K36 dimethylation | 3 | 4 | 6.48e-03 |
| GO:BP | GO:0006479 | protein methylation | 16 | 178 | 8.33e-03 |
| GO:BP | GO:0008213 | protein alkylation | 16 | 178 | 8.33e-03 |
| GO:BP | GO:0006974 | cellular response to DNA damage stimulus | 44 | 789 | 8.33e-03 |
| GO:BP | GO:0006607 | NLS-bearing protein import into nucleus | 5 | 18 | 9.44e-03 |
| GO:BP | GO:0060429 | epithelium development | 46 | 845 | 9.91e-03 |
| GO:BP | GO:0016071 | mRNA metabolic process | 38 | 657 | 1.11e-02 |
| GO:BP | GO:0045814 | negative regulation of gene expression, epigenetic | 10 | 84 | 1.47e-02 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 18 | 227 | 1.52e-02 |
| GO:BP | GO:0072359 | circulatory system development | 46 | 879 | 2.28e-02 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 15 | 177 | 2.31e-02 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 20 | 277 | 2.35e-02 |
| GO:BP | GO:1902105 | regulation of leukocyte differentiation | 16 | 197 | 2.35e-02 |
| GO:BP | GO:2000736 | regulation of stem cell differentiation | 9 | 74 | 2.35e-02 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 13 | 141 | 2.35e-02 |
| GO:BP | GO:0086023 | adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process | 3 | 6 | 2.63e-02 |
| GO:BP | GO:0031507 | heterochromatin formation | 9 | 76 | 2.78e-02 |
| GO:BP | GO:0007623 | circadian rhythm | 14 | 164 | 3.07e-02 |
| GO:BP | GO:0043009 | chordate embryonic development | 31 | 531 | 3.18e-02 |
| GO:BP | GO:1902275 | regulation of chromatin organization | 6 | 36 | 3.61e-02 |
| GO:BP | GO:0019827 | stem cell population maintenance | 13 | 150 | 3.98e-02 |
| GO:BP | GO:1900246 | positive regulation of RIG-I signaling pathway | 3 | 7 | 4.18e-02 |
| GO:BP | GO:0032242 | regulation of nucleoside transport | 2 | 2 | 4.18e-02 |
| GO:BP | GO:1901898 | negative regulation of relaxation of cardiac muscle | 2 | 2 | 4.18e-02 |
| GO:BP | GO:0032239 | regulation of nucleobase-containing compound transport | 4 | 15 | 4.20e-02 |
| GO:BP | GO:0098727 | maintenance of cell number | 13 | 152 | 4.30e-02 |
| GO:BP | GO:0009888 | tissue development | 65 | 1415 | 4.69e-02 |
| GO:BP | GO:0045165 | cell fate commitment | 13 | 154 | 4.73e-02 |
| GO:BP | GO:0070828 | heterochromatin organization | 9 | 83 | 4.73e-02 |
| GO:BP | GO:0048729 | tissue morphogenesis | 29 | 501 | 4.75e-02 |
| GO:BP | GO:0048511 | rhythmic process | 17 | 234 | 4.75e-02 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 31 | 548 | 4.75e-02 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 11 | 118 | 4.78e-02 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 11 | 118 | 4.78e-02 |
| GO:BP | GO:0007507 | heart development | 29 | 502 | 4.78e-02 |
| GO:BP | GO:2001222 | regulation of neuron migration | 6 | 39 | 4.82e-02 |
| GO:BP | GO:0009790 | embryo development | 44 | 871 | 4.82e-02 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 45 | 415 | 6.65e-11 |
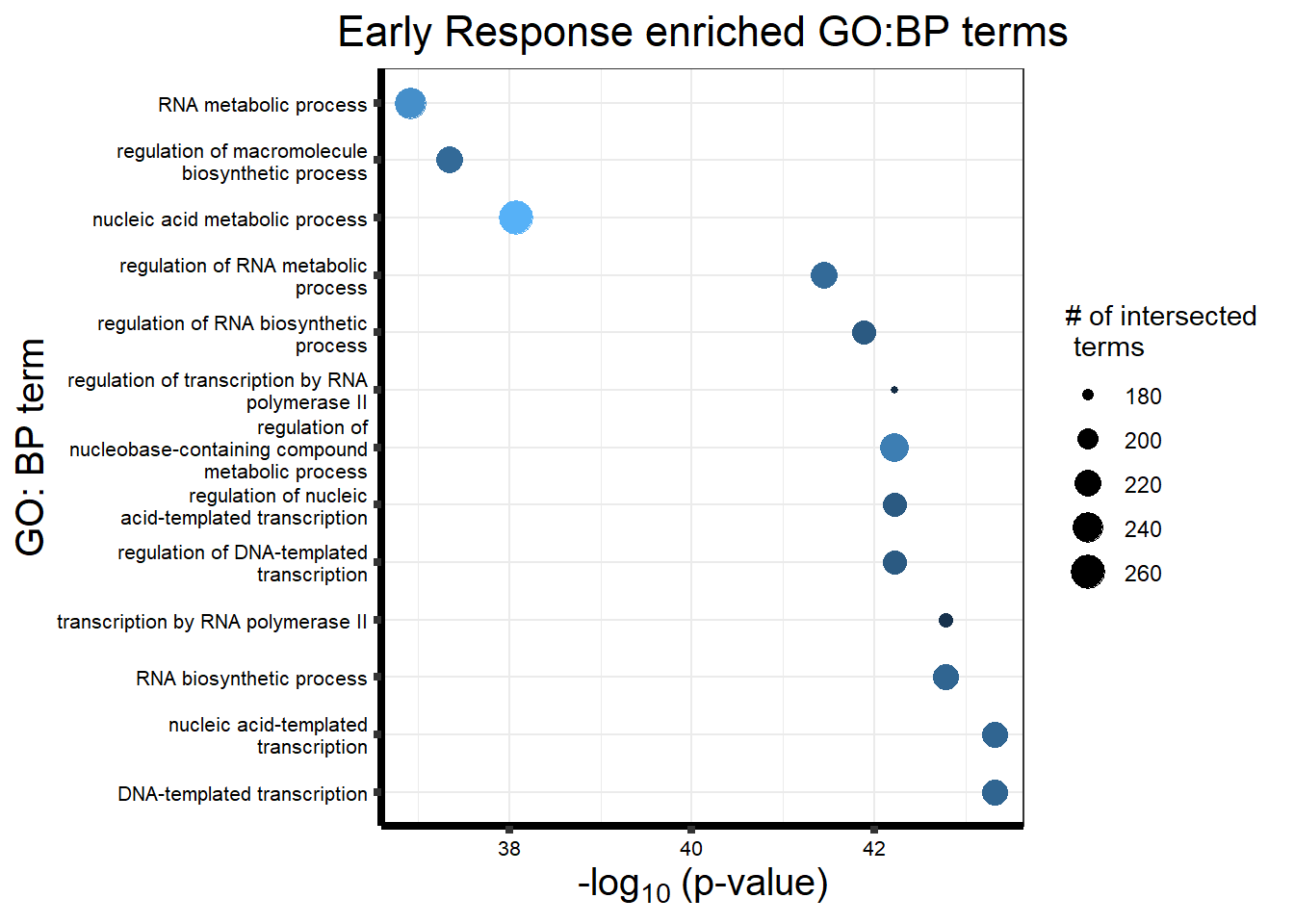
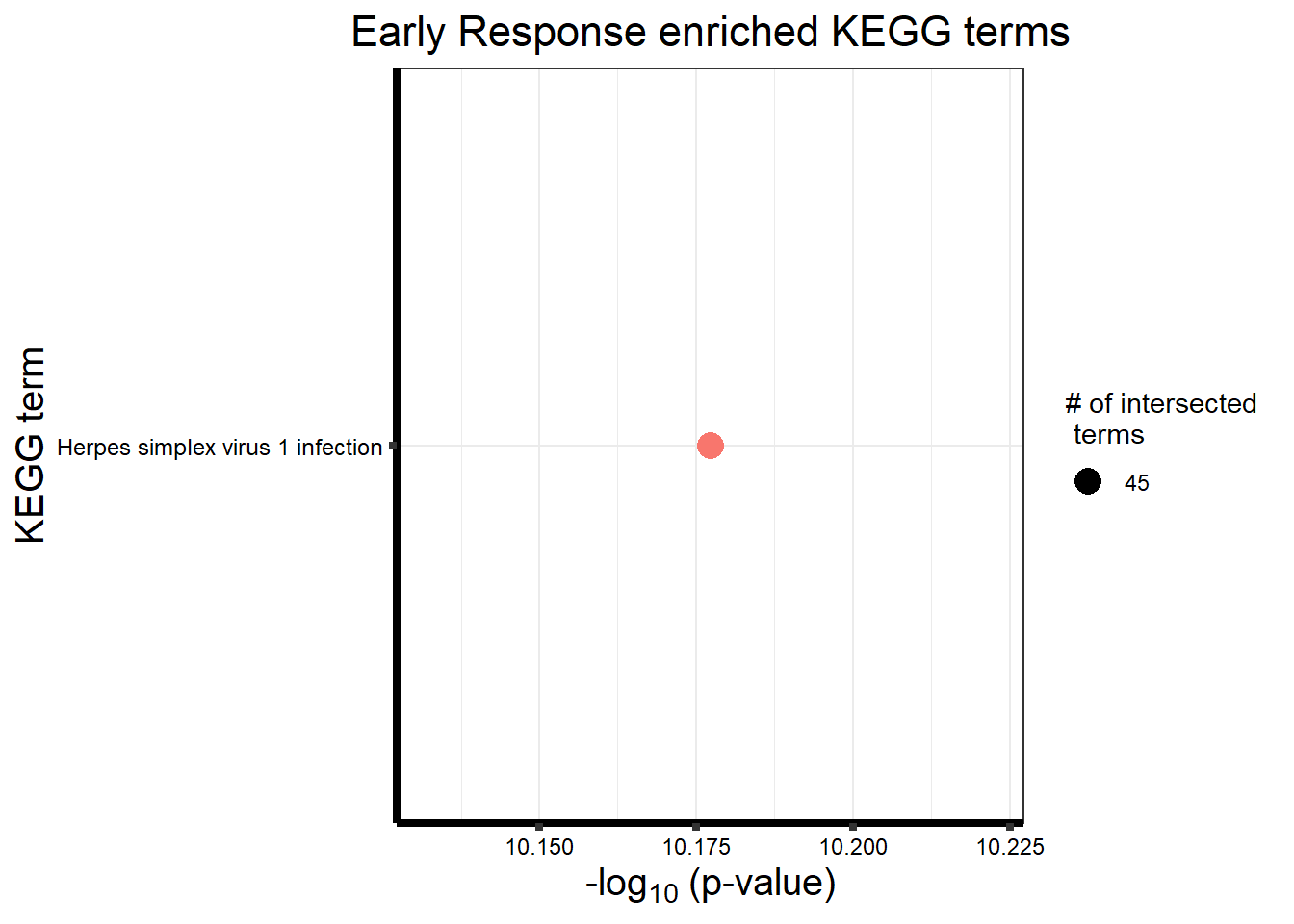
Time-independent Top2\(\beta\) inhibitor motif genes
| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 178 | 1915 | 4.27e-29 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 180 | 1991 | 3.00e-28 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 217 | 2849 | 2.60e-25 |
| GO:BP | GO:1903506 | regulation of nucleic acid-templated transcription | 204 | 2581 | 2.60e-25 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 204 | 2579 | 2.60e-25 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 204 | 2598 | 4.68e-25 |
| GO:BP | GO:0097659 | nucleic acid-templated transcription | 206 | 2684 | 3.74e-24 |
| GO:BP | GO:0006351 | DNA-templated transcription | 206 | 2683 | 3.74e-24 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 206 | 2714 | 1.52e-23 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 224 | 3105 | 2.64e-23 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 213 | 3015 | 1.79e-20 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 216 | 3123 | 1.09e-19 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 257 | 4047 | 1.14e-19 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 218 | 3174 | 1.29e-19 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 213 | 3083 | 2.52e-19 |
| GO:BP | GO:0010468 | regulation of gene expression | 235 | 3583 | 4.29e-19 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 215 | 3149 | 5.36e-19 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 215 | 3148 | 5.36e-19 |
| GO:BP | GO:0016070 | RNA metabolic process | 235 | 3593 | 5.36e-19 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 217 | 3253 | 5.69e-18 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 260 | 4330 | 1.50e-16 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 263 | 4440 | 5.17e-16 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 271 | 4640 | 6.15e-16 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 255 | 4305 | 3.04e-15 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 261 | 4503 | 1.65e-14 |
| GO:BP | GO:0046483 | heterocycle metabolic process | 264 | 4616 | 5.52e-14 |
| GO:BP | GO:0019222 | regulation of metabolic process | 281 | 5039 | 6.07e-14 |
| GO:BP | GO:0006725 | cellular aromatic compound metabolic process | 264 | 4646 | 1.27e-13 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 225 | 3730 | 1.70e-13 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 225 | 3758 | 4.06e-13 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 97 | 1115 | 7.31e-13 |
| GO:BP | GO:1901360 | organic cyclic compound metabolic process | 267 | 4790 | 8.90e-13 |
| GO:BP | GO:0010467 | gene expression | 256 | 4572 | 3.76e-12 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 89 | 1009 | 5.07e-12 |
| GO:BP | GO:1903507 | negative regulation of nucleic acid-templated transcription | 89 | 1011 | 5.53e-12 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 89 | 1020 | 9.04e-12 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 72 | 735 | 1.15e-11 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 99 | 1215 | 1.89e-11 |
| GO:BP | GO:0034641 | cellular nitrogen compound metabolic process | 270 | 4991 | 2.48e-11 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 79 | 925 | 7.99e-10 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 93 | 1206 | 2.62e-09 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 95 | 1249 | 3.21e-09 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 96 | 1273 | 3.90e-09 |
| GO:BP | GO:0044249 | cellular biosynthetic process | 240 | 4465 | 5.35e-09 |
| GO:BP | GO:1901576 | organic substance biosynthetic process | 242 | 4537 | 9.24e-09 |
| GO:BP | GO:0050789 | regulation of biological process | 373 | 7992 | 1.06e-08 |
| GO:BP | GO:0009058 | biosynthetic process | 244 | 4595 | 1.10e-08 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 339 | 7057 | 1.24e-08 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 95 | 1304 | 2.89e-08 |
| GO:BP | GO:1903508 | positive regulation of nucleic acid-templated transcription | 95 | 1304 | 2.89e-08 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 95 | 1311 | 3.78e-08 |
| GO:BP | GO:0050794 | regulation of cellular process | 355 | 7579 | 6.74e-08 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 99 | 1431 | 1.79e-07 |
| GO:BP | GO:0065007 | biological regulation | 380 | 8348 | 2.54e-07 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 100 | 1499 | 9.63e-07 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 113 | 1780 | 1.33e-06 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 103 | 1571 | 1.33e-06 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 104 | 1598 | 1.60e-06 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 104 | 1600 | 1.68e-06 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 139 | 2373 | 2.94e-06 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 141 | 2425 | 3.53e-06 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 127 | 2146 | 7.92e-06 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 114 | 1866 | 8.40e-06 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 149 | 2648 | 9.95e-06 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 159 | 2884 | 1.16e-05 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 133 | 2306 | 1.52e-05 |
| GO:BP | GO:0006807 | nitrogen compound metabolic process | 339 | 7474 | 3.21e-05 |
| GO:BP | GO:0044238 | primary metabolic process | 350 | 7800 | 4.85e-05 |
| GO:BP | GO:0071704 | organic substance metabolic process | 360 | 8150 | 1.64e-04 |
| GO:BP | GO:0003007 | heart morphogenesis | 23 | 212 | 4.45e-04 |
| GO:BP | GO:0044237 | cellular metabolic process | 334 | 7503 | 4.56e-04 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 72 | 1134 | 7.05e-04 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 180 | 3628 | 1.47e-03 |
| GO:BP | GO:0048645 | animal organ formation | 10 | 52 | 1.52e-03 |
| GO:BP | GO:0009952 | anterior/posterior pattern specification | 16 | 126 | 1.55e-03 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 45 | 619 | 1.55e-03 |
| GO:BP | GO:0140467 | integrated stress response signaling | 8 | 34 | 2.11e-03 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 11 | 67 | 2.67e-03 |
| GO:BP | GO:0008152 | metabolic process | 364 | 8501 | 5.44e-03 |
| GO:BP | GO:0060914 | heart formation | 7 | 30 | 6.77e-03 |
| GO:BP | GO:0007389 | pattern specification process | 26 | 305 | 6.79e-03 |
| GO:BP | GO:0035914 | skeletal muscle cell differentiation | 9 | 52 | 8.36e-03 |
| GO:BP | GO:0014706 | striated muscle tissue development | 20 | 210 | 9.46e-03 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 59 | 958 | 9.56e-03 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 35 | 478 | 9.88e-03 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 61 | 1007 | 1.13e-02 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 19 | 198 | 1.19e-02 |
| GO:BP | GO:0021546 | rhombomere development | 3 | 4 | 1.24e-02 |
| GO:BP | GO:0048518 | positive regulation of biological process | 212 | 4577 | 1.37e-02 |
| GO:BP | GO:0060537 | muscle tissue development | 27 | 340 | 1.46e-02 |
| GO:BP | GO:0048519 | negative regulation of biological process | 190 | 4035 | 1.48e-02 |
| GO:BP | GO:0043009 | chordate embryonic development | 37 | 531 | 1.60e-02 |
| GO:BP | GO:0003002 | regionalization | 23 | 272 | 1.64e-02 |
| GO:BP | GO:0051726 | regulation of cell cycle | 57 | 950 | 2.16e-02 |
| GO:BP | GO:0001756 | somitogenesis | 8 | 48 | 2.22e-02 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 105 | 2023 | 2.26e-02 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 20 | 227 | 2.27e-02 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 193 | 4148 | 2.34e-02 |
| GO:BP | GO:0002293 | alpha-beta T cell differentiation involved in immune response | 7 | 38 | 2.53e-02 |
| GO:BP | GO:0042093 | T-helper cell differentiation | 7 | 38 | 2.53e-02 |
| GO:BP | GO:0002287 | alpha-beta T cell activation involved in immune response | 7 | 38 | 2.53e-02 |
| GO:BP | GO:0002294 | CD4-positive, alpha-beta T cell differentiation involved in immune response | 7 | 38 | 2.53e-02 |
| GO:BP | GO:0035282 | segmentation | 10 | 75 | 2.59e-02 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 37 | 548 | 2.59e-02 |
| GO:BP | GO:0045598 | regulation of fat cell differentiation | 12 | 103 | 2.60e-02 |
| GO:BP | GO:0050793 | regulation of developmental process | 96 | 1829 | 2.61e-02 |
| GO:BP | GO:0048483 | autonomic nervous system development | 6 | 28 | 2.61e-02 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 8 | 51 | 2.98e-02 |
| GO:BP | GO:0007049 | cell cycle | 82 | 1522 | 3.18e-02 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 7 | 40 | 3.20e-02 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 20 | 236 | 3.20e-02 |
| GO:BP | GO:0002292 | T cell differentiation involved in immune response | 7 | 40 | 3.20e-02 |
| GO:BP | GO:0060429 | epithelium development | 51 | 845 | 3.20e-02 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 46 | 741 | 3.21e-02 |
| GO:BP | GO:0061614 | miRNA transcription | 8 | 52 | 3.21e-02 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 42 | 659 | 3.22e-02 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 9 | 65 | 3.28e-02 |
| GO:BP | GO:0060284 | regulation of cell development | 39 | 600 | 3.37e-02 |
| GO:BP | GO:0048486 | parasympathetic nervous system development | 4 | 12 | 3.42e-02 |
| GO:BP | GO:0048729 | tissue morphogenesis | 34 | 501 | 3.42e-02 |
| GO:BP | GO:0045444 | fat cell differentiation | 17 | 187 | 3.42e-02 |
| GO:BP | GO:0007507 | heart development | 34 | 502 | 3.48e-02 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 9 | 66 | 3.50e-02 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 12 | 109 | 3.68e-02 |
| GO:BP | GO:0043954 | cellular component maintenance | 8 | 54 | 3.78e-02 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 22 | 277 | 3.78e-02 |
| GO:BP | GO:0003197 | endocardial cushion development | 7 | 42 | 3.81e-02 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 64 | 1141 | 4.12e-02 |
| GO:BP | GO:0060840 | artery development | 10 | 82 | 4.25e-02 |
| GO:BP | GO:1900744 | regulation of p38MAPK cascade | 6 | 32 | 4.55e-02 |
| GO:BP | GO:0003281 | ventricular septum development | 9 | 69 | 4.55e-02 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 47 | 781 | 4.71e-02 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 21 | 265 | 4.78e-02 |
| GO:BP | GO:0060038 | cardiac muscle cell proliferation | 7 | 44 | 4.80e-02 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 47 | 415 | 3.76e-09 |
| KEGG | KEGG:04115 | p53 signaling pathway | 11 | 65 | 3.17e-03 |
| KEGG | KEGG:05217 | Basal cell carcinoma | 9 | 49 | 5.76e-03 |
| KEGG | KEGG:05224 | Breast cancer | 14 | 117 | 5.91e-03 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 16 | 145 | 5.91e-03 |
| KEGG | KEGG:05226 | Gastric cancer | 13 | 117 | 1.76e-02 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 13 | 128 | 3.52e-02 |
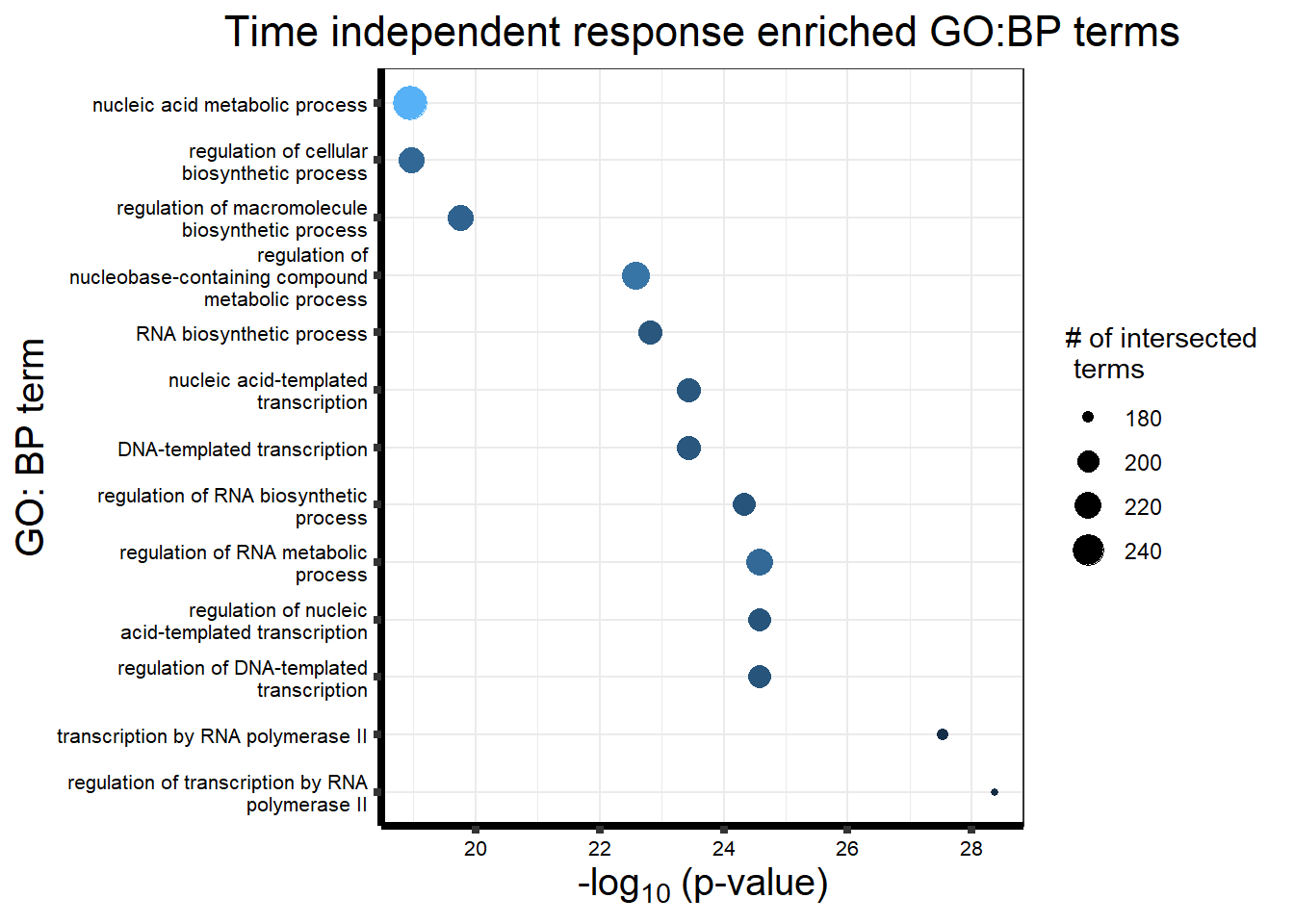
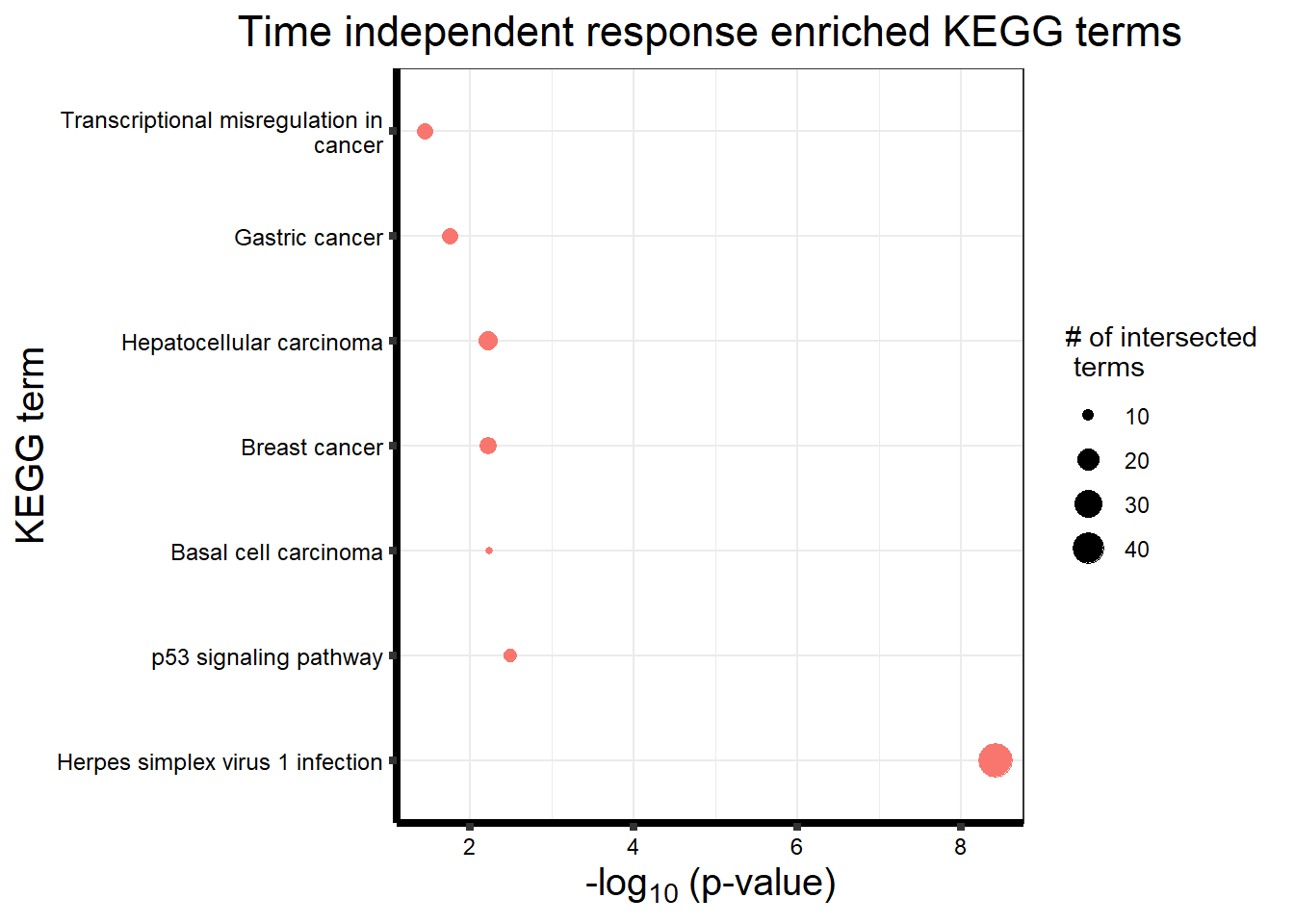
long go frame
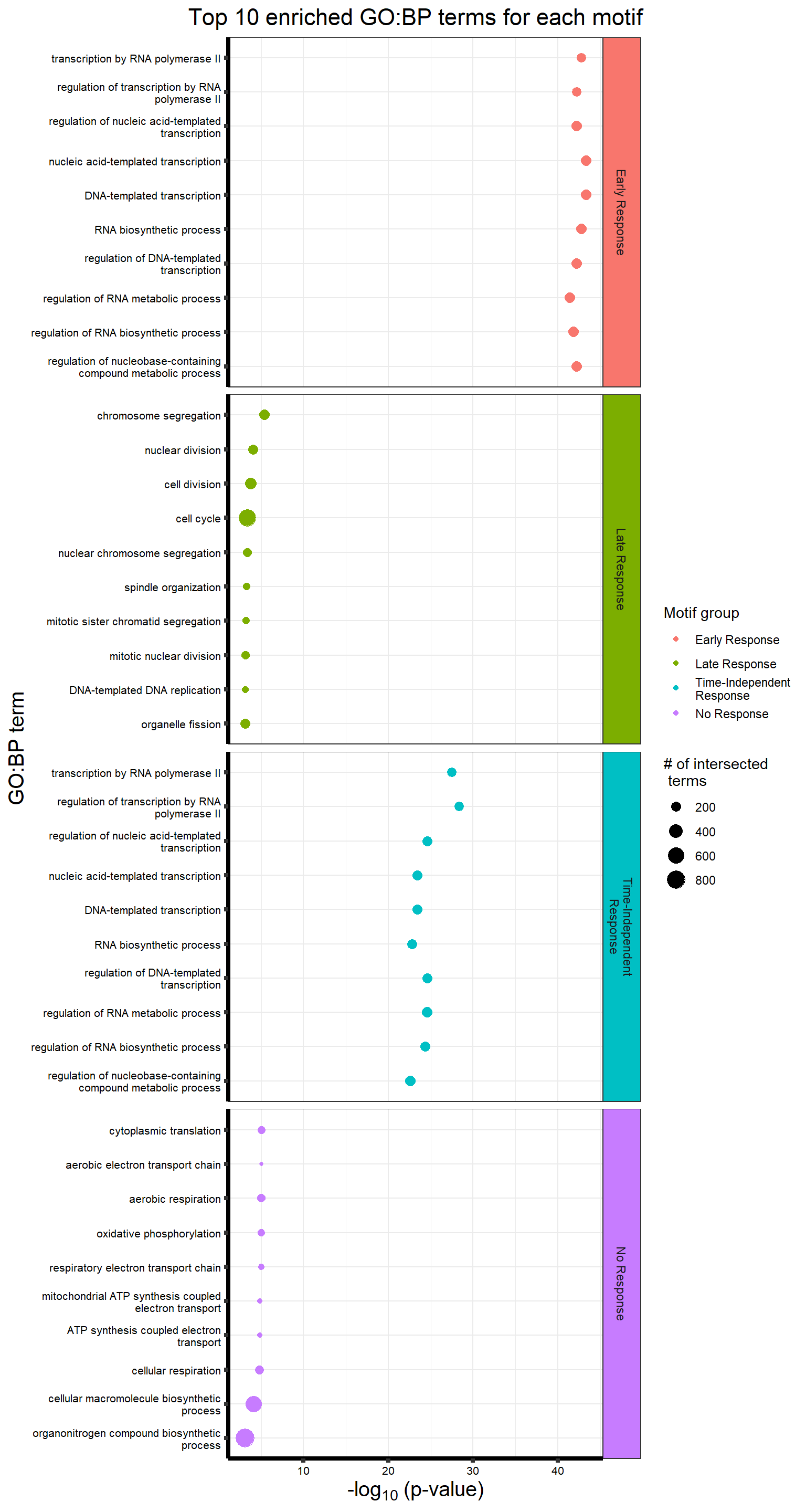
###long go kegg break
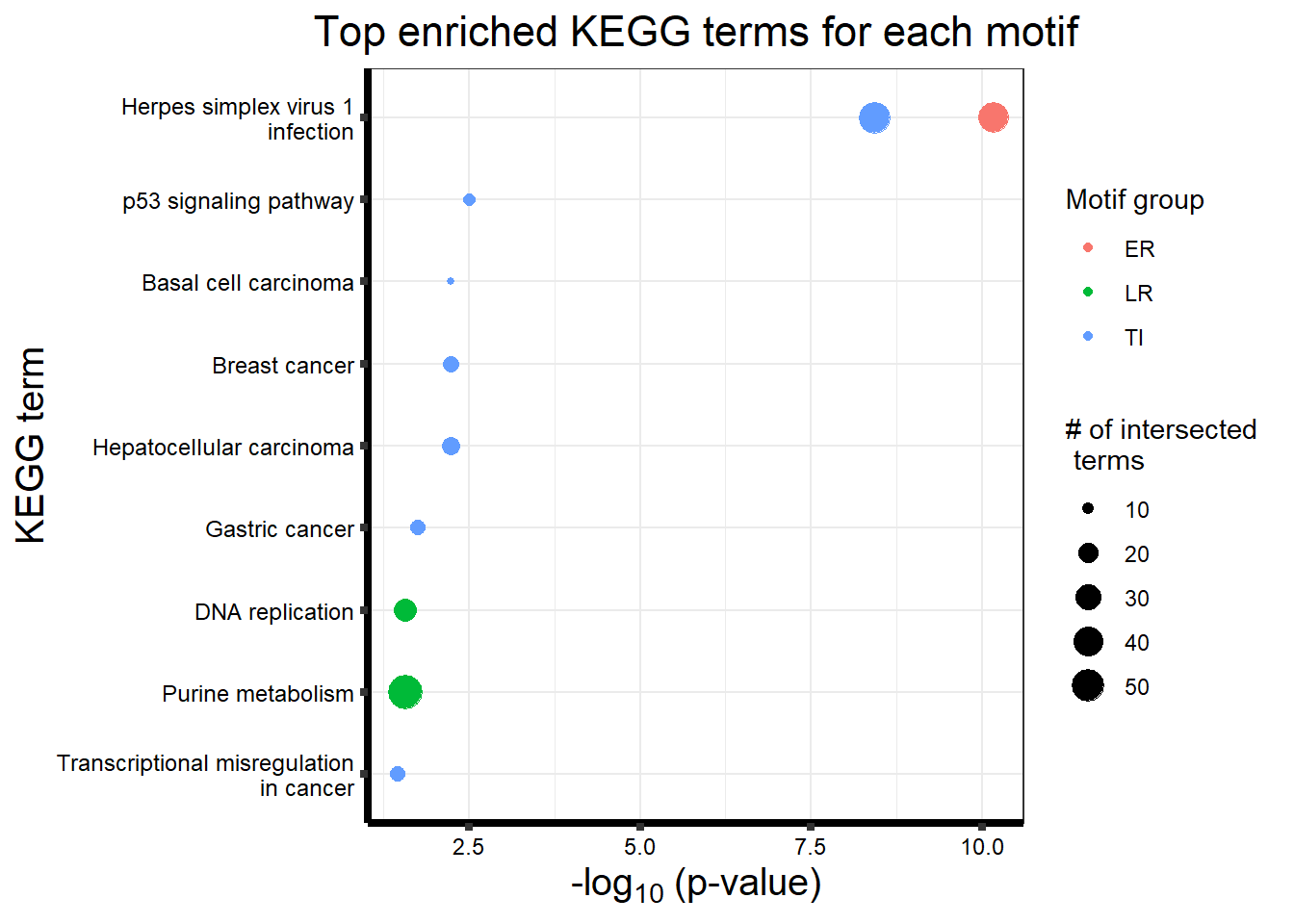
sessionInfo()R version 4.2.2 (2022-10-31 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19044)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
attached base packages:
[1] grid stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] RColorBrewer_1.1-3 Cormotif_1.42.0 limma_3.52.4
[4] affy_1.74.0 Biobase_2.56.0 ggVennDiagram_1.2.2
[7] scales_1.2.1 kableExtra_1.3.4 VennDiagram_1.7.3
[10] futile.logger_1.4.3 gridExtra_2.3 BiocGenerics_0.42.0
[13] gprofiler2_0.2.1 lubridate_1.9.2 forcats_1.0.0
[16] stringr_1.5.0 dplyr_1.1.0 purrr_1.0.1
[19] readr_2.1.4 tidyr_1.3.0 tibble_3.1.8
[22] ggplot2_3.4.1 tidyverse_2.0.0 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] fs_1.6.1 webshot_0.5.4 httr_1.4.5
[4] rprojroot_2.0.3 tools_4.2.2 bslib_0.4.2
[7] utf8_1.2.3 R6_2.5.1 affyio_1.66.0
[10] lazyeval_0.2.2 colorspace_2.1-0 withr_2.5.0
[13] tidyselect_1.2.0 processx_3.8.0 compiler_4.2.2
[16] git2r_0.31.0 preprocessCore_1.58.0 cli_3.6.0
[19] rvest_1.0.3 formatR_1.14 xml2_1.3.3
[22] plotly_4.10.1 labeling_0.4.2 sass_0.4.5
[25] callr_3.7.3 systemfonts_1.0.4 digest_0.6.31
[28] rmarkdown_2.20 svglite_2.1.1 pkgconfig_2.0.3
[31] htmltools_0.5.4 fastmap_1.1.1 highr_0.10
[34] htmlwidgets_1.6.1 rlang_1.0.6 rstudioapi_0.14
[37] shiny_1.7.4 farver_2.1.1 jquerylib_0.1.4
[40] generics_0.1.3 jsonlite_1.8.4 crosstalk_1.2.0
[43] magrittr_2.0.3 Rcpp_1.0.10 munsell_0.5.0
[46] fansi_1.0.4 lifecycle_1.0.3 stringi_1.7.12
[49] whisker_0.4.1 yaml_2.3.7 zlibbioc_1.42.0
[52] promises_1.2.0.1 hms_1.1.2 knitr_1.42
[55] ps_1.7.2 pillar_1.8.1 futile.options_1.0.1
[58] glue_1.6.2 evaluate_0.20 getPass_0.2-2
[61] lambda.r_1.2.4 data.table_1.14.8 BiocManager_1.30.20
[64] vctrs_0.5.2 tzdb_0.3.0 httpuv_1.6.9
[67] gtable_0.3.1 cachem_1.0.7 xfun_0.37
[70] mime_0.12 xtable_1.8-4 later_1.3.0
[73] RVenn_1.1.0 viridisLite_0.4.1 timechange_0.2.0
[76] ellipsis_0.3.2