GO and KEGG pathway analysis
ERM
2024-06-07
Last updated: 2024-06-07
Checks: 7 0
Knit directory: ATAC_learning/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20231016) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 95e0415. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/ARR_SNP_table.csv
Ignored: data/All_merged_peaks.tsv
Ignored: data/CAD_gwas_dataframe.RDS
Ignored: data/Collapsed_expressed_NG_peak_table.csv
Ignored: data/DEG_toplist_sep_n45.RDS
Ignored: data/FRiP_first_run.txt
Ignored: data/Frip_1_reads.csv
Ignored: data/Frip_2_reads.csv
Ignored: data/Frip_3_reads.csv
Ignored: data/Frip_4_reads.csv
Ignored: data/Frip_5_reads.csv
Ignored: data/Frip_6_reads.csv
Ignored: data/GO_KEGG_analysis/
Ignored: data/HF_SNP_table.csv
Ignored: data/Ind1_75DA24h_dedup_peaks.csv
Ignored: data/Ind1_TSS_peaks.RDS
Ignored: data/Ind1_firstfragment_files.txt
Ignored: data/Ind1_fragment_files.txt
Ignored: data/Ind1_peaks_list.RDS
Ignored: data/Ind1_summary.txt
Ignored: data/Ind2_TSS_peaks.RDS
Ignored: data/Ind2_fragment_files.txt
Ignored: data/Ind2_peaks_list.RDS
Ignored: data/Ind2_summary.txt
Ignored: data/Ind3_TSS_peaks.RDS
Ignored: data/Ind3_fragment_files.txt
Ignored: data/Ind3_peaks_list.RDS
Ignored: data/Ind3_summary.txt
Ignored: data/Ind4_79B24h_dedup_peaks.csv
Ignored: data/Ind4_TSS_peaks.RDS
Ignored: data/Ind4_V24h_fraglength.txt
Ignored: data/Ind4_fragment_files.txt
Ignored: data/Ind4_fragment_filesN.txt
Ignored: data/Ind4_peaks_list.RDS
Ignored: data/Ind4_summary.txt
Ignored: data/Ind5_TSS_peaks.RDS
Ignored: data/Ind5_fragment_files.txt
Ignored: data/Ind5_fragment_filesN.txt
Ignored: data/Ind5_peaks_list.RDS
Ignored: data/Ind5_summary.txt
Ignored: data/Ind6_TSS_peaks.RDS
Ignored: data/Ind6_fragment_files.txt
Ignored: data/Ind6_peaks_list.RDS
Ignored: data/Ind6_summary.txt
Ignored: data/Knowles_4.RDS
Ignored: data/Knowles_5.RDS
Ignored: data/Knowles_6.RDS
Ignored: data/MI_gwas.RDS
Ignored: data/SNP_supp_schneider.RDS
Ignored: data/all_TSSE_scores.RDS
Ignored: data/aln_run1_results.txt
Ignored: data/anno_ind1_DA24h.RDS
Ignored: data/anno_ind4_V24h.RDS
Ignored: data/background_n45_he_peaks.RDS
Ignored: data/cardiac_muscle_FRIP.csv
Ignored: data/cardiomyocyte_FRIP.csv
Ignored: data/col_ng_peak.csv
Ignored: data/cormotif_full_4_run.RDS
Ignored: data/cormotif_full_4_run_he.RDS
Ignored: data/cormotif_full_6_run.RDS
Ignored: data/cormotif_full_6_run_he.RDS
Ignored: data/cormotif_probability_45_list.csv
Ignored: data/cormotif_probability_45_list_he.csv
Ignored: data/cormotif_probability_all_6_list.csv
Ignored: data/cormotif_probability_all_6_list_he.csv
Ignored: data/embryo_heart_FRIP.csv
Ignored: data/enhancer_list_ENCFF126UHK.bed
Ignored: data/enhancerdata/
Ignored: data/filt_Peaks_efit2.RDS
Ignored: data/filt_Peaks_efit2_bl.RDS
Ignored: data/filt_Peaks_efit2_n45.RDS
Ignored: data/first_Peaksummarycounts.csv
Ignored: data/first_run_frag_counts.txt
Ignored: data/full_bedfiles/
Ignored: data/gene_ref.csv
Ignored: data/gwas_1_dataframe.RDS
Ignored: data/gwas_2_dataframe.RDS
Ignored: data/gwas_3_dataframe.RDS
Ignored: data/gwas_4_dataframe.RDS
Ignored: data/gwas_5_dataframe.RDS
Ignored: data/high_conf_peak_counts.csv
Ignored: data/high_conf_peak_counts.txt
Ignored: data/high_conf_peaks_bl_counts.txt
Ignored: data/high_conf_peaks_counts.txt
Ignored: data/hits_files/
Ignored: data/ind1_DA24hpeaks.RDS
Ignored: data/ind1_TSSE.RDS
Ignored: data/ind2_TSSE.RDS
Ignored: data/ind3_TSSE.RDS
Ignored: data/ind4_TSSE.RDS
Ignored: data/ind4_V24hpeaks.RDS
Ignored: data/ind5_TSSE.RDS
Ignored: data/ind6_TSSE.RDS
Ignored: data/initial_complete_stats_run1.txt
Ignored: data/left_ventricle_FRIP.csv
Ignored: data/mergedPeads.gff
Ignored: data/mergedPeaks.gff
Ignored: data/motif_list_full
Ignored: data/motif_list_n45
Ignored: data/motif_list_n45.RDS
Ignored: data/multiqc_fastqc_run1.txt
Ignored: data/multiqc_fastqc_run2.txt
Ignored: data/multiqc_genestat_run1.txt
Ignored: data/multiqc_genestat_run2.txt
Ignored: data/my_hc_filt_counts.RDS
Ignored: data/my_hc_filt_counts_n45.RDS
Ignored: data/n45_bedfiles/
Ignored: data/n45_files
Ignored: data/other_papers/
Ignored: data/peakAnnoList_1.RDS
Ignored: data/peakAnnoList_2.RDS
Ignored: data/peakAnnoList_24_full.RDS
Ignored: data/peakAnnoList_24_n45.RDS
Ignored: data/peakAnnoList_3.RDS
Ignored: data/peakAnnoList_3_full.RDS
Ignored: data/peakAnnoList_3_n45.RDS
Ignored: data/peakAnnoList_4.RDS
Ignored: data/peakAnnoList_5.RDS
Ignored: data/peakAnnoList_6.RDS
Ignored: data/peakAnnoList_full_motif.RDS
Ignored: data/peakAnnoList_n45_motif.RDS
Ignored: data/siglist_full.RDS
Ignored: data/siglist_n45.RDS
Ignored: data/test.list.RDS
Ignored: data/testnames.txt
Ignored: data/toplist_6.RDS
Ignored: data/toplist_full.RDS
Ignored: data/toplist_full_DAR_6.RDS
Ignored: data/toplist_n45.RDS
Ignored: data/trimmed_seq_length.csv
Ignored: data/unclassified_full_set_peaks.RDS
Ignored: data/unclassified_n45_set_peaks.RDS
Ignored: data/xstreme/
Ignored: trimmed_Ind1_75DA24h_S7.nodup.splited.bam/
Untracked files:
Untracked: EAR_2_plot.pdf
Untracked: ESR_1_plot.pdf
Untracked: Firstcorr plotATAC.pdf
Untracked: IND1_2_3_6_corrplot.pdf
Untracked: LR_3_plot.pdf
Untracked: NR_1_plot.pdf
Untracked: analysis/my_hc_filt_counts.csv
Untracked: analysis/nucleosome_explore.Rmd
Untracked: code/IGV_snapshot_code.R
Untracked: code/LongDARlist.R
Untracked: code/TSSE.R
Untracked: code/just_for_Fun.R
Untracked: code/toplist_assembly.R
Untracked: lcpm_filtered_corplot.pdf
Untracked: log2cpmfragcount.pdf
Untracked: output/cormotif_probability_45_list.csv
Untracked: output/cormotif_probability_all_6_list.csv
Untracked: splited/
Untracked: trimmed_Ind1_75DA24h_S7.nodup.fragment.size.distribution.pdf
Untracked: trimmed_Ind1_75DA3h_S1.nodup.fragment.size.distribution.pdf
Unstaged changes:
Modified: analysis/CorMotif_data_n45.Rmd
Modified: analysis/Fastqc_results.Rmd
Modified: analysis/Jaspar_motif.Rmd
Modified: analysis/Peak_calling.Rmd
Modified: analysis/Smaller_set_DAR.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/GO_KEGG_analysis.Rmd) and
HTML (docs/GO_KEGG_analysis.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 95e0415 | reneeisnowhere | 2024-06-07 | updates, i think |
| html | eea0976 | reneeisnowhere | 2024-06-03 | Build site. |
| Rmd | 0672654 | reneeisnowhere | 2024-06-03 | updates to gene lists |
| html | 897af23 | reneeisnowhere | 2024-05-28 | Build site. |
| Rmd | 5c6ef83 | reneeisnowhere | 2024-05-28 | updates to KEGG/GO 20kb lists |
| html | d1a3829 | reneeisnowhere | 2024-05-17 | Build site. |
| Rmd | 9fb4f9a | reneeisnowhere | 2024-05-17 | adding in updates from Neargenes and enhncers |
| html | 2d991d7 | reneeisnowhere | 2024-05-03 | Build site. |
| Rmd | f57f2ee | reneeisnowhere | 2024-05-03 | first update |
library(tidyverse)
# library(ggsignif)
# library(cowplot)
# library(ggpubr)
# library(sjmisc)
library(kableExtra)
library(broom)
# library(biomaRt)
library(RColorBrewer)
library(gprofiler2)
# library(qvalue)
library(ChIPseeker)
library("TxDb.Hsapiens.UCSC.hg38.knownGene")
library("org.Hs.eg.db")
# library(ATACseqQC)
library(rtracklayer)
library(edgeR)
library(ggfortify)
library(limma)
library(readr)
library(BiocGenerics)
library(gridExtra)
library(VennDiagram)
library(scales)
# library(ggVennDiagram)
# library(Cormotif)
library(BiocParallel)
library(ggpubr)
# library(devtools)
# # install_github('davetang/bedr')
# library(bedr)
library(biomaRt)
library(clusterProfiler)data loading
peakAnnoList_n45_motif <- readRDS("data/peakAnnoList_n45_motif.RDS")
# list2env(peakAnnoList_n45_motif, envir = .GlobalEnv)
EAR_df <- as.data.frame(peakAnnoList_n45_motif$EAR_n45_gr)
EAR_df_gr <- GRanges(EAR_df)
ESR_df <- as.data.frame(peakAnnoList_n45_motif$ESR_n45_gr)
ESR_df_gr <- GRanges(ESR_df)
LR_df <- as.data.frame(peakAnnoList_n45_motif$LR_n45_gr)
LR_df_gr <- GRanges(LR_df)
NR_df <- as.data.frame(peakAnnoList_n45_motif$NR_n45_gr)
NR_df_gr <- GRanges(NR_df)
exp_neargene_table <- read_delim("data/n45_bedfiles/exp_neargene_table.tsv",
delim = "\t", escape_double = FALSE,
trim_ws = TRUE)
# col_ng_peak <- exp_neargene_table %>%
# group_by(chr, start, end, peakid) %>%
# summarise(NCBI_gene = paste(unique(ENTREZID),collapse=","),
# ensembl_ID= paste(unique(ensembl_id),collapse = ","),
# SYMBOL= paste(unique(SYMBOL),collapse = ",") ,
# dist_to_NG =min(dist_to_NG))
#
EAR_NG_list <- exp_neargene_table %>%
dplyr::filter(peakid %in% EAR_df$id) %>%
dplyr::filter(dist_to_NG<20000) %>%
dplyr::select(ENTREZID) %>%
distinct()
ESR_NG_list <- exp_neargene_table %>%
dplyr::filter(peakid %in% ESR_df$id) %>%
dplyr::filter(dist_to_NG<20000) %>%
dplyr::select(ENTREZID) %>%
distinct()
LR_NG_list <- exp_neargene_table %>%
dplyr::filter(peakid %in% LR_df$id) %>%
dplyr::filter(dist_to_NG<20000) %>%
dplyr::select(ENTREZID) %>%
distinct()
NR_NG_list <- exp_neargene_table %>%
dplyr::filter(peakid %in% NR_df$id) %>%
dplyr::filter(dist_to_NG<20000) %>%
dplyr::select(ENTREZID) %>%
distinct()
background_df <- exp_neargene_table %>%
dplyr::select(ENTREZID) %>%
distinct()
EAR_resgenes <- readRDS("data/GO_KEGG_analysis/EAR_resgenes.RDS")
ESR_resgenes <- readRDS("data/GO_KEGG_analysis/ESR_resgenes.RDS")
LR_resgenes <- readRDS("data/GO_KEGG_analysis/LR_resgenes.RDS")
NR_resgenes <- readRDS("data/GO_KEGG_analysis/NR_resgenes.RDS")
EAR_enh_peaks_20k <- readRDS("data/enhancerdata/EAR_enh_peaks_20k.RDS")
ESR_enh_peaks_20k <- readRDS("data/enhancerdata/ESR_enh_peaks_20k.RDS")
LR_enh_peaks_20k <- readRDS("data/enhancerdata/LR_enh_peaks_20k.RDS")
NR_enh_peaks_20k <- readRDS("data/enhancerdata/NR_enh_peaks_20k.RDS")
EAR_resgenes_20k <- readRDS("data/GO_KEGG_analysis/EAR_resgenes_20k.RDS")
EAR_resgenes_20k_enh <- readRDS("data/GO_KEGG_analysis/EAR_resgenes_20k_enh.RDS")
ESR_resgenes_20k <- readRDS("data/GO_KEGG_analysis/ESR_resgenes_20k.RDS")
ESR_resgenes_20k_enh <- readRDS("data/GO_KEGG_analysis/ESR_resgenes_20k_enh.RDS")
LR_resgenes_20k <- readRDS("data/GO_KEGG_analysis/LR_resgenes_20k.RDS")
LR_resgenes_20k_enh <- readRDS("data/GO_KEGG_analysis/LR_resgenes_20k_enh.RDS")
NR_resgenes_20k <- readRDS("data/GO_KEGG_analysis/NR_resgenes_20k.RDS")
NR_resgenes_20k_enh <- readRDS("data/GO_KEGG_analysis/NR_resgenes_20k_enh.RDS")
### intersection of peaks files
S13Table <- read.csv( "data/other_papers/S13Table_Matthews2024.csv",row.names = 1)
##14021
EAR_RNA <- S13Table %>%
dplyr::filter(MOTIF=="EAR") %>%
dplyr::select(ENTREZID) %>%
mutate(ENTREZID= as.character(ENTREZID))
ESR_RNA <- S13Table %>%
dplyr::filter(MOTIF=="ESR")%>%
dplyr::select(ENTREZID)%>%
mutate(ENTREZID= as.character(ENTREZID))
LR_RNA <- S13Table %>%
dplyr::filter(MOTIF=="LR")%>%
dplyr::select(ENTREZID)%>%
mutate(ENTREZID=as.character(ENTREZID))
NR_RNA <- S13Table %>%
dplyr::filter(MOTIF=="NR")%>%
dplyr::select(ENTREZID)%>%
mutate(ENTREZID= as.character(ENTREZID))
Resp_RNA <- EAR_RNA %>%
rbind(.,ESR_RNA) %>%
rbind(., LR_RNA) %>%
distinct(ENTREZID)
int_EAR_n_RNAresp <- intersect(EAR_NG_list$ENTREZID, Resp_RNA$ENTREZID)
int_ESR_n_RNAresp <- intersect(ESR_NG_list$ENTREZID, Resp_RNA$ENTREZID)
int_LR_n_RNAresp <- intersect(LR_NG_list$ENTREZID, Resp_RNA$ENTREZID)
int_NR_n_RNAresp <- intersect(NR_NG_list$ENTREZID, Resp_RNA$ENTREZID)
EAR_int_resgenes <- readRDS("data/GO_KEGG_analysis/EAR_int_resgenes.RDS")
ESR_int_resgenes <- readRDS("data/GO_KEGG_analysis/ESR_int_resgenes.RDS")
LR_int_resgenes <- readRDS("data/GO_KEGG_analysis/LR_int_resgenes.RDS")EAR response genes
# EAR_resgenes <- gost(query = EAR_NG_list$ENTREZID,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = FALSE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = background_df$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(EAR_resgenes,"data/GO_KEGG_analysis/EAR_resgenes.RDS")
# EARnom <- gostplot(EAR_resgenes, capped = FALSE, interactive = TRUE)
# EARnom
EARnomtable <- EAR_resgenes$result %>%
dplyr::select(c(source, term_id,
term_name,intersection_size,
term_size, p_value))# %>%
EARnomtable %>%
filter(source=="GO:BP", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(.,caption = "GO:BP terms found in EAR neargenes" ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 898 | 1844 | 4.11e-28 |
| GO:BP | GO:0032502 | developmental process | 1806 | 4179 | 6.83e-26 |
| GO:BP | GO:0048856 | anatomical structure development | 1683 | 3867 | 2.86e-25 |
| GO:BP | GO:0007275 | multicellular organism development | 1413 | 3170 | 3.48e-25 |
| GO:BP | GO:0032501 | multicellular organismal process | 1918 | 4504 | 2.88e-24 |
| GO:BP | GO:0048869 | cellular developmental process | 1262 | 2804 | 8.92e-24 |
| GO:BP | GO:0030154 | cell differentiation | 1261 | 2803 | 1.02e-23 |
| GO:BP | GO:0050793 | regulation of developmental process | 809 | 1697 | 7.18e-22 |
| GO:BP | GO:0048731 | system development | 1211 | 2719 | 1.94e-20 |
| GO:BP | GO:0016477 | cell migration | 515 | 1011 | 3.91e-20 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 890 | 1924 | 1.81e-19 |
| GO:BP | GO:0007154 | cell communication | 1686 | 3983 | 1.59e-18 |
| GO:BP | GO:0023052 | signaling | 1660 | 3918 | 2.46e-18 |
| GO:BP | GO:0048699 | generation of neurons | 535 | 1076 | 3.58e-18 |
| GO:BP | GO:0040012 | regulation of locomotion | 380 | 720 | 1.16e-17 |
| GO:BP | GO:0030334 | regulation of cell migration | 357 | 668 | 1.16e-17 |
| GO:BP | GO:2000145 | regulation of cell motility | 371 | 700 | 1.19e-17 |
| GO:BP | GO:0048870 | cell motility | 549 | 1120 | 4.10e-17 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 357 | 673 | 4.96e-17 |
| GO:BP | GO:0007165 | signal transduction | 1534 | 3611 | 5.22e-17 |
| GO:BP | GO:0022008 | neurogenesis | 602 | 1251 | 8.71e-17 |
| GO:BP | GO:0040011 | locomotion | 413 | 805 | 1.25e-16 |
| GO:BP | GO:0072359 | circulatory system development | 438 | 864 | 1.52e-16 |
| GO:BP | GO:0030182 | neuron differentiation | 500 | 1014 | 4.14e-16 |
| GO:BP | GO:0048513 | animal organ development | 901 | 2004 | 1.36e-15 |
| GO:BP | GO:0050789 | regulation of biological process | 2957 | 7516 | 2.09e-15 |
| GO:BP | GO:0009888 | tissue development | 634 | 1348 | 4.41e-15 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 469 | 951 | 4.83e-15 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 452 | 913 | 8.19e-15 |
| GO:BP | GO:0050794 | regulation of cellular process | 2823 | 7153 | 8.19e-15 |
| GO:BP | GO:0065007 | biological regulation | 3028 | 7734 | 1.41e-14 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 517 | 1073 | 2.39e-14 |
| GO:BP | GO:0051716 | cellular response to stimulus | 1900 | 4640 | 4.73e-14 |
| GO:BP | GO:0048468 | cell development | 845 | 1886 | 4.95e-14 |
| GO:BP | GO:0048519 | negative regulation of biological process | 1571 | 3764 | 4.95e-14 |
| GO:BP | GO:0007155 | cell adhesion | 460 | 941 | 5.94e-14 |
| GO:BP | GO:0007399 | nervous system development | 800 | 1775 | 6.41e-14 |
| GO:BP | GO:0009790 | embryo development | 409 | 821 | 6.86e-14 |
| GO:BP | GO:0050896 | response to stimulus | 2170 | 5379 | 1.18e-13 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 1486 | 3550 | 1.59e-13 |
| GO:BP | GO:0035239 | tube morphogenesis | 328 | 638 | 3.14e-13 |
| GO:BP | GO:0048518 | positive regulation of biological process | 1766 | 4301 | 3.91e-13 |
| GO:BP | GO:0035295 | tube development | 391 | 787 | 5.10e-13 |
| GO:BP | GO:0034330 | cell junction organization | 286 | 545 | 7.88e-13 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 411 | 837 | 9.46e-13 |
| GO:BP | GO:0007423 | sensory organ development | 215 | 387 | 1.12e-12 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 1631 | 3954 | 1.57e-12 |
| GO:BP | GO:0010646 | regulation of cell communication | 1054 | 2444 | 1.80e-12 |
| GO:BP | GO:0048666 | neuron development | 408 | 838 | 5.95e-12 |
| GO:BP | GO:0003008 | system process | 532 | 1139 | 7.86e-12 |
| GO:BP | GO:0023051 | regulation of signaling | 1047 | 2438 | 8.93e-12 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 302 | 591 | 9.93e-12 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 309 | 609 | 1.58e-11 |
| GO:BP | GO:0031175 | neuron projection development | 366 | 743 | 1.67e-11 |
| GO:BP | GO:0035556 | intracellular signal transduction | 854 | 1951 | 2.38e-11 |
| GO:BP | GO:0009966 | regulation of signal transduction | 938 | 2166 | 2.52e-11 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 226 | 422 | 3.57e-11 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 599 | 1314 | 4.36e-11 |
| GO:BP | GO:0000902 | cell morphogenesis | 351 | 713 | 5.76e-11 |
| GO:BP | GO:0001501 | skeletal system development | 191 | 347 | 9.68e-11 |
| GO:BP | GO:0001944 | vasculature development | 280 | 552 | 2.24e-10 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 310 | 622 | 2.24e-10 |
| GO:BP | GO:0030900 | forebrain development | 155 | 272 | 3.85e-10 |
| GO:BP | GO:0001568 | blood vessel development | 267 | 525 | 5.12e-10 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 262 | 516 | 1.02e-09 |
| GO:BP | GO:0065008 | regulation of biological quality | 820 | 1893 | 1.15e-09 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 257 | 505 | 1.15e-09 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 784 | 1801 | 1.15e-09 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 191 | 355 | 1.34e-09 |
| GO:BP | GO:0060429 | epithelium development | 377 | 792 | 2.17e-09 |
| GO:BP | GO:0007507 | heart development | 253 | 499 | 2.66e-09 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 231 | 450 | 4.42e-09 |
| GO:BP | GO:0007267 | cell-cell signaling | 494 | 1082 | 4.64e-09 |
| GO:BP | GO:0007417 | central nervous system development | 334 | 694 | 6.43e-09 |
| GO:BP | GO:0048589 | developmental growth | 252 | 501 | 7.88e-09 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 802 | 1862 | 7.88e-09 |
| GO:BP | GO:0048729 | tissue morphogenesis | 231 | 453 | 9.50e-09 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 475 | 1040 | 1.03e-08 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 242 | 479 | 1.06e-08 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 254 | 508 | 1.35e-08 |
| GO:BP | GO:0001503 | ossification | 164 | 302 | 1.35e-08 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 1130 | 2717 | 1.35e-08 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 472 | 1035 | 1.47e-08 |
| GO:BP | GO:0034329 | cell junction assembly | 173 | 323 | 1.74e-08 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 247 | 494 | 2.33e-08 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 494 | 1093 | 2.57e-08 |
| GO:BP | GO:0030030 | cell projection organization | 529 | 1182 | 3.41e-08 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 248 | 498 | 3.42e-08 |
| GO:BP | GO:0014706 | striated muscle tissue development | 122 | 213 | 3.92e-08 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 228 | 452 | 4.03e-08 |
| GO:BP | GO:0060322 | head development | 265 | 539 | 4.21e-08 |
| GO:BP | GO:0070848 | response to growth factor | 268 | 547 | 5.16e-08 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 116 | 201 | 5.44e-08 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 220 | 435 | 5.88e-08 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 541 | 1218 | 7.89e-08 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 338 | 717 | 8.14e-08 |
| GO:BP | GO:0023056 | positive regulation of signaling | 542 | 1221 | 8.33e-08 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 513 | 1150 | 1.06e-07 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 258 | 528 | 1.43e-07 |
| GO:BP | GO:0007420 | brain development | 245 | 498 | 1.69e-07 |
| GO:BP | GO:0061061 | muscle structure development | 261 | 536 | 1.72e-07 |
| GO:BP | GO:0043583 | ear development | 91 | 151 | 1.85e-07 |
| GO:BP | GO:0048880 | sensory system development | 144 | 266 | 1.96e-07 |
| GO:BP | GO:0032989 | cellular anatomical entity morphogenesis | 282 | 588 | 2.55e-07 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 101 | 173 | 2.72e-07 |
| GO:BP | GO:0001525 | angiogenesis | 194 | 381 | 2.81e-07 |
| GO:BP | GO:0001654 | eye development | 142 | 263 | 3.07e-07 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 118 | 210 | 3.07e-07 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 121 | 217 | 3.59e-07 |
| GO:BP | GO:0150063 | visual system development | 142 | 264 | 4.21e-07 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 59 | 88 | 4.53e-07 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 177 | 344 | 4.89e-07 |
| GO:BP | GO:0060284 | regulation of cell development | 273 | 570 | 5.02e-07 |
| GO:BP | GO:0051216 | cartilage development | 87 | 145 | 5.09e-07 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 527 | 1197 | 5.36e-07 |
| GO:BP | GO:0010631 | epithelial cell migration | 132 | 243 | 6.17e-07 |
| GO:BP | GO:0090132 | epithelium migration | 132 | 243 | 6.17e-07 |
| GO:BP | GO:0098609 | cell-cell adhesion | 270 | 564 | 6.20e-07 |
| GO:BP | GO:0040007 | growth | 325 | 697 | 6.25e-07 |
| GO:BP | GO:0001764 | neuron migration | 79 | 129 | 6.61e-07 |
| GO:BP | GO:0060537 | muscle tissue development | 174 | 339 | 7.73e-07 |
| GO:BP | GO:0050808 | synapse organization | 173 | 337 | 8.34e-07 |
| GO:BP | GO:0003013 | circulatory system process | 211 | 426 | 9.64e-07 |
| GO:BP | GO:0008283 | cell population proliferation | 568 | 1306 | 1.06e-06 |
| GO:BP | GO:0042221 | response to chemical | 955 | 2304 | 1.08e-06 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 131 | 243 | 1.22e-06 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 186 | 369 | 1.35e-06 |
| GO:BP | GO:0007409 | axonogenesis | 172 | 337 | 1.51e-06 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 560 | 1289 | 1.56e-06 |
| GO:BP | GO:0048839 | inner ear development | 80 | 133 | 1.56e-06 |
| GO:BP | GO:0061564 | axon development | 187 | 372 | 1.56e-06 |
| GO:BP | GO:0060021 | roof of mouth development | 48 | 69 | 1.71e-06 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 326 | 706 | 1.89e-06 |
| GO:BP | GO:0090130 | tissue migration | 132 | 247 | 2.04e-06 |
| GO:BP | GO:0016358 | dendrite development | 103 | 183 | 2.18e-06 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 105 | 188 | 2.76e-06 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 201 | 408 | 3.29e-06 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 204 | 416 | 4.07e-06 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 195 | 395 | 4.26e-06 |
| GO:BP | GO:0030029 | actin filament-based process | 297 | 640 | 4.36e-06 |
| GO:BP | GO:0044057 | regulation of system process | 190 | 384 | 5.04e-06 |
| GO:BP | GO:0048568 | embryonic organ development | 155 | 303 | 5.70e-06 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 472 | 1077 | 6.07e-06 |
| GO:BP | GO:0048732 | gland development | 157 | 308 | 6.25e-06 |
| GO:BP | GO:0003007 | heart morphogenesis | 117 | 218 | 8.62e-06 |
| GO:BP | GO:0061448 | connective tissue development | 111 | 205 | 9.75e-06 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 85 | 148 | 9.75e-06 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 219 | 456 | 1.01e-05 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 177 | 358 | 1.44e-05 |
| GO:BP | GO:0007389 | pattern specification process | 146 | 286 | 1.52e-05 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 206 | 427 | 1.53e-05 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 732 | 1751 | 1.58e-05 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 177 | 359 | 1.79e-05 |
| GO:BP | GO:0007264 | small GTPase-mediated signal transduction | 190 | 390 | 1.90e-05 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 45 | 67 | 2.05e-05 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 985 | 2415 | 2.05e-05 |
| GO:BP | GO:0043010 | camera-type eye development | 121 | 230 | 2.05e-05 |
| GO:BP | GO:0007411 | axon guidance | 95 | 172 | 2.13e-05 |
| GO:BP | GO:0097485 | neuron projection guidance | 95 | 172 | 2.13e-05 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 635 | 1504 | 2.24e-05 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 103 | 190 | 2.32e-05 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 897 | 2186 | 2.42e-05 |
| GO:BP | GO:0010033 | response to organic substance | 724 | 1736 | 2.71e-05 |
| GO:BP | GO:0021537 | telencephalon development | 105 | 195 | 2.73e-05 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 134 | 261 | 2.87e-05 |
| GO:BP | GO:0050807 | regulation of synapse organization | 95 | 173 | 2.92e-05 |
| GO:BP | GO:0009952 | anterior/posterior pattern specification | 71 | 121 | 2.99e-05 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 122 | 234 | 3.23e-05 |
| GO:BP | GO:0016043 | cellular component organization | 1874 | 4802 | 3.24e-05 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 96 | 176 | 3.77e-05 |
| GO:BP | GO:0001649 | osteoblast differentiation | 95 | 174 | 4.04e-05 |
| GO:BP | GO:0009987 | cellular process | 3975 | 10686 | 4.12e-05 |
| GO:BP | GO:0042692 | muscle cell differentiation | 160 | 323 | 4.12e-05 |
| GO:BP | GO:0050877 | nervous system process | 275 | 600 | 4.21e-05 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 132 | 258 | 4.23e-05 |
| GO:BP | GO:0048588 | developmental cell growth | 97 | 179 | 4.75e-05 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 149 | 298 | 4.85e-05 |
| GO:BP | GO:0008015 | blood circulation | 174 | 357 | 5.05e-05 |
| GO:BP | GO:0050890 | cognition | 112 | 213 | 5.11e-05 |
| GO:BP | GO:0000165 | MAPK cascade | 235 | 504 | 5.53e-05 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 150 | 301 | 5.61e-05 |
| GO:BP | GO:0035265 | organ growth | 73 | 127 | 5.74e-05 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 1939 | 4988 | 5.76e-05 |
| GO:BP | GO:0099536 | synaptic signaling | 226 | 483 | 6.35e-05 |
| GO:BP | GO:0035313 | wound healing, spreading of epidermal cells | 16 | 17 | 6.38e-05 |
| GO:BP | GO:0006468 | protein phosphorylation | 417 | 958 | 7.02e-05 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 1057 | 2622 | 7.62e-05 |
| GO:BP | GO:0045664 | regulation of neuron differentiation | 74 | 130 | 7.84e-05 |
| GO:BP | GO:0045444 | fat cell differentiation | 96 | 179 | 9.34e-05 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 446 | 1034 | 9.54e-05 |
| GO:BP | GO:0048592 | eye morphogenesis | 63 | 107 | 9.78e-05 |
| GO:BP | GO:0051960 | regulation of nervous system development | 165 | 339 | 9.94e-05 |
| GO:BP | GO:0048638 | regulation of developmental growth | 124 | 243 | 9.94e-05 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 373 | 850 | 1.00e-04 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 219 | 469 | 1.06e-04 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 105 | 200 | 1.12e-04 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 252 | 550 | 1.14e-04 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 382 | 874 | 1.18e-04 |
| GO:BP | GO:0071310 | cellular response to organic substance | 548 | 1297 | 1.20e-04 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 240 | 521 | 1.20e-04 |
| GO:BP | GO:0072001 | renal system development | 124 | 244 | 1.25e-04 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 119 | 233 | 1.44e-04 |
| GO:BP | GO:0023057 | negative regulation of signaling | 445 | 1036 | 1.58e-04 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 74 | 132 | 1.58e-04 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 201 | 428 | 1.66e-04 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 217 | 467 | 1.71e-04 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 157 | 323 | 1.82e-04 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 334 | 757 | 1.87e-04 |
| GO:BP | GO:0010720 | positive regulation of cell development | 151 | 309 | 1.89e-04 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 29 | 40 | 1.92e-04 |
| GO:BP | GO:0045637 | regulation of myeloid cell differentiation | 75 | 135 | 2.07e-04 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 147 | 300 | 2.07e-04 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 141 | 286 | 2.13e-04 |
| GO:BP | GO:0003002 | regionalization | 129 | 258 | 2.19e-04 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 61 | 105 | 2.32e-04 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 87 | 162 | 2.32e-04 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 156 | 322 | 2.36e-04 |
| GO:BP | GO:0045165 | cell fate commitment | 82 | 151 | 2.42e-04 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 105 | 203 | 2.45e-04 |
| GO:BP | GO:0007369 | gastrulation | 77 | 140 | 2.47e-04 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 70 | 125 | 2.83e-04 |
| GO:BP | GO:0043009 | chordate embryonic development | 232 | 507 | 2.89e-04 |
| GO:BP | GO:0051049 | regulation of transport | 467 | 1098 | 2.89e-04 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 128 | 257 | 2.92e-04 |
| GO:BP | GO:0035107 | appendage morphogenesis | 63 | 110 | 2.96e-04 |
| GO:BP | GO:0035108 | limb morphogenesis | 63 | 110 | 2.96e-04 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 142 | 290 | 2.96e-04 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 63 | 110 | 2.96e-04 |
| GO:BP | GO:0042592 | homeostatic process | 482 | 1137 | 3.01e-04 |
| GO:BP | GO:0022612 | gland morphogenesis | 55 | 93 | 3.04e-04 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 124 | 248 | 3.09e-04 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 141 | 288 | 3.14e-04 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 315 | 714 | 3.22e-04 |
| GO:BP | GO:0007610 | behavior | 196 | 420 | 3.22e-04 |
| GO:BP | GO:0002063 | chondrocyte development | 19 | 23 | 3.51e-04 |
| GO:BP | GO:0008150 | biological_process | 4156 | 11244 | 3.65e-04 |
| GO:BP | GO:0016049 | cell growth | 180 | 382 | 3.65e-04 |
| GO:BP | GO:0065009 | regulation of molecular function | 544 | 1299 | 3.81e-04 |
| GO:BP | GO:0048562 | embryonic organ morphogenesis | 95 | 182 | 3.84e-04 |
| GO:BP | GO:0099175 | regulation of postsynapse organization | 49 | 81 | 3.87e-04 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 255 | 566 | 3.90e-04 |
| GO:BP | GO:0007611 | learning or memory | 98 | 189 | 3.97e-04 |
| GO:BP | GO:0002761 | regulation of myeloid leukocyte differentiation | 48 | 79 | 3.98e-04 |
| GO:BP | GO:0045667 | regulation of osteoblast differentiation | 57 | 98 | 4.00e-04 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 213 | 463 | 4.07e-04 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 213 | 463 | 4.07e-04 |
| GO:BP | GO:0048813 | dendrite morphogenesis | 63 | 111 | 4.09e-04 |
| GO:BP | GO:0030097 | hemopoiesis | 278 | 624 | 4.34e-04 |
| GO:BP | GO:0003281 | ventricular septum development | 43 | 69 | 4.57e-04 |
| GO:BP | GO:0010633 | negative regulation of epithelial cell migration | 34 | 51 | 4.71e-04 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 938 | 2333 | 4.73e-04 |
| GO:BP | GO:0060041 | retina development in camera-type eye | 60 | 105 | 4.89e-04 |
| GO:BP | GO:0099173 | postsynapse organization | 79 | 147 | 5.18e-04 |
| GO:BP | GO:0060173 | limb development | 74 | 136 | 5.42e-04 |
| GO:BP | GO:0048736 | appendage development | 74 | 136 | 5.42e-04 |
| GO:BP | GO:0051897 | positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 69 | 125 | 5.53e-04 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 57 | 99 | 5.80e-04 |
| GO:BP | GO:0001822 | kidney development | 118 | 237 | 5.92e-04 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 99 | 193 | 6.04e-04 |
| GO:BP | GO:2001222 | regulation of neuron migration | 27 | 38 | 6.25e-04 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 120 | 242 | 6.32e-04 |
| GO:BP | GO:0070372 | regulation of ERK1 and ERK2 cascade | 91 | 175 | 6.56e-04 |
| GO:BP | GO:0008360 | regulation of cell shape | 60 | 106 | 7.00e-04 |
| GO:BP | GO:0060348 | bone development | 78 | 146 | 7.47e-04 |
| GO:BP | GO:0050772 | positive regulation of axonogenesis | 40 | 64 | 7.85e-04 |
| GO:BP | GO:0003279 | cardiac septum development | 58 | 102 | 7.85e-04 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 40 | 64 | 7.85e-04 |
| GO:BP | GO:0021543 | pallium development | 73 | 135 | 7.85e-04 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 51 | 87 | 7.88e-04 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 51 | 87 | 7.88e-04 |
| GO:BP | GO:0006811 | monoatomic ion transport | 314 | 719 | 7.89e-04 |
| GO:BP | GO:0016310 | phosphorylation | 481 | 1145 | 8.12e-04 |
| GO:BP | GO:0048645 | animal organ formation | 34 | 52 | 8.12e-04 |
| GO:BP | GO:0003205 | cardiac chamber development | 80 | 151 | 8.43e-04 |
| GO:BP | GO:0032879 | regulation of localization | 594 | 1439 | 9.38e-04 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 413 | 972 | 9.46e-04 |
| GO:BP | GO:0032924 | activin receptor signaling pathway | 24 | 33 | 9.46e-04 |
| GO:BP | GO:0042476 | odontogenesis | 47 | 79 | 9.46e-04 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 82 | 156 | 9.60e-04 |
| GO:BP | GO:0031399 | regulation of protein modification process | 389 | 911 | 9.67e-04 |
| GO:BP | GO:0002521 | leukocyte differentiation | 173 | 371 | 9.70e-04 |
| GO:BP | GO:0030198 | extracellular matrix organization | 111 | 223 | 9.80e-04 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 122 | 249 | 1.01e-03 |
| GO:BP | GO:0007416 | synapse assembly | 73 | 136 | 1.03e-03 |
| GO:BP | GO:0008038 | neuron recognition | 25 | 35 | 1.03e-03 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 127 | 261 | 1.04e-03 |
| GO:BP | GO:0071407 | cellular response to organic cyclic compound | 176 | 379 | 1.08e-03 |
| GO:BP | GO:1990138 | neuron projection extension | 72 | 134 | 1.10e-03 |
| GO:BP | GO:0070169 | positive regulation of biomineral tissue development | 26 | 37 | 1.11e-03 |
| GO:BP | GO:0141091 | transforming growth factor beta receptor superfamily signaling pathway | 126 | 259 | 1.12e-03 |
| GO:BP | GO:0048663 | neuron fate commitment | 20 | 26 | 1.15e-03 |
| GO:BP | GO:0060485 | mesenchyme development | 120 | 245 | 1.16e-03 |
| GO:BP | GO:0045778 | positive regulation of ossification | 27 | 39 | 1.18e-03 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 260 | 587 | 1.18e-03 |
| GO:BP | GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 50 | 86 | 1.18e-03 |
| GO:BP | GO:0043062 | extracellular structure organization | 111 | 224 | 1.19e-03 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 111 | 224 | 1.19e-03 |
| GO:BP | GO:0022029 | telencephalon cell migration | 28 | 41 | 1.22e-03 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 388 | 911 | 1.22e-03 |
| GO:BP | GO:0007612 | learning | 61 | 110 | 1.23e-03 |
| GO:BP | GO:0021885 | forebrain cell migration | 29 | 43 | 1.26e-03 |
| GO:BP | GO:0048660 | regulation of smooth muscle cell proliferation | 55 | 97 | 1.27e-03 |
| GO:BP | GO:1903522 | regulation of blood circulation | 98 | 194 | 1.27e-03 |
| GO:BP | GO:0045666 | positive regulation of neuron differentiation | 36 | 57 | 1.29e-03 |
| GO:BP | GO:0044319 | wound healing, spreading of cells | 21 | 28 | 1.32e-03 |
| GO:BP | GO:0090505 | epiboly involved in wound healing | 21 | 28 | 1.32e-03 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 267 | 606 | 1.40e-03 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 135 | 282 | 1.49e-03 |
| GO:BP | GO:0060996 | dendritic spine development | 45 | 76 | 1.49e-03 |
| GO:BP | GO:0001823 | mesonephros development | 44 | 74 | 1.57e-03 |
| GO:BP | GO:0001508 | action potential | 57 | 102 | 1.58e-03 |
| GO:BP | GO:0045995 | regulation of embryonic development | 43 | 72 | 1.63e-03 |
| GO:BP | GO:0001657 | ureteric bud development | 43 | 72 | 1.63e-03 |
| GO:BP | GO:0030501 | positive regulation of bone mineralization | 23 | 32 | 1.67e-03 |
| GO:BP | GO:0072073 | kidney epithelium development | 61 | 111 | 1.67e-03 |
| GO:BP | GO:0048593 | camera-type eye morphogenesis | 50 | 87 | 1.67e-03 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 511 | 1231 | 1.67e-03 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 209 | 463 | 1.67e-03 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 209 | 463 | 1.67e-03 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 512 | 1234 | 1.72e-03 |
| GO:BP | GO:0030879 | mammary gland development | 55 | 98 | 1.76e-03 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 49 | 85 | 1.76e-03 |
| GO:BP | GO:0021795 | cerebral cortex cell migration | 24 | 34 | 1.81e-03 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 76 | 145 | 1.84e-03 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 39 | 64 | 1.87e-03 |
| GO:BP | GO:0045216 | cell-cell junction organization | 79 | 152 | 1.92e-03 |
| GO:BP | GO:0033002 | muscle cell proliferation | 79 | 152 | 1.92e-03 |
| GO:BP | GO:0010463 | mesenchymal cell proliferation | 25 | 36 | 1.92e-03 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 104 | 210 | 1.94e-03 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 104 | 210 | 1.94e-03 |
| GO:BP | GO:0040008 | regulation of growth | 208 | 462 | 2.02e-03 |
| GO:BP | GO:0070371 | ERK1 and ERK2 cascade | 97 | 194 | 2.12e-03 |
| GO:BP | GO:0010718 | positive regulation of epithelial to mesenchymal transition | 31 | 48 | 2.21e-03 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 557 | 1354 | 2.22e-03 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 841 | 2100 | 2.23e-03 |
| GO:BP | GO:0072164 | mesonephric tubule development | 43 | 73 | 2.39e-03 |
| GO:BP | GO:0048706 | embryonic skeletal system development | 43 | 73 | 2.39e-03 |
| GO:BP | GO:0072163 | mesonephric epithelium development | 43 | 73 | 2.39e-03 |
| GO:BP | GO:0009611 | response to wounding | 175 | 382 | 2.44e-03 |
| GO:BP | GO:0060419 | heart growth | 42 | 71 | 2.51e-03 |
| GO:BP | GO:0043542 | endothelial cell migration | 88 | 174 | 2.64e-03 |
| GO:BP | GO:0035270 | endocrine system development | 48 | 84 | 2.68e-03 |
| GO:BP | GO:0030001 | metal ion transport | 232 | 524 | 2.76e-03 |
| GO:BP | GO:0031214 | biomineral tissue development | 58 | 106 | 2.76e-03 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 141 | 300 | 2.76e-03 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 53 | 95 | 2.76e-03 |
| GO:BP | GO:0048675 | axon extension | 53 | 95 | 2.76e-03 |
| GO:BP | GO:0090504 | epiboly | 21 | 29 | 2.77e-03 |
| GO:BP | GO:0003231 | cardiac ventricle development | 62 | 115 | 2.81e-03 |
| GO:BP | GO:0003170 | heart valve development | 39 | 65 | 2.83e-03 |
| GO:BP | GO:0060571 | morphogenesis of an epithelial fold | 13 | 15 | 2.91e-03 |
| GO:BP | GO:0045598 | regulation of fat cell differentiation | 52 | 93 | 2.92e-03 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 264 | 605 | 3.00e-03 |
| GO:BP | GO:0007517 | muscle organ development | 127 | 267 | 3.09e-03 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 741 | 1841 | 3.11e-03 |
| GO:BP | GO:0055006 | cardiac cell development | 45 | 78 | 3.12e-03 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 334 | 783 | 3.20e-03 |
| GO:BP | GO:0033993 | response to lipid | 251 | 573 | 3.25e-03 |
| GO:BP | GO:0051179 | localization | 1448 | 3731 | 3.27e-03 |
| GO:BP | GO:0018210 | peptidyl-threonine modification | 44 | 76 | 3.28e-03 |
| GO:BP | GO:0051057 | positive regulation of small GTPase mediated signal transduction | 34 | 55 | 3.34e-03 |
| GO:BP | GO:0002573 | myeloid leukocyte differentiation | 78 | 152 | 3.34e-03 |
| GO:BP | GO:0003179 | heart valve morphogenesis | 34 | 55 | 3.34e-03 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 493 | 1193 | 3.36e-03 |
| GO:BP | GO:1905207 | regulation of cardiocyte differentiation | 17 | 22 | 3.42e-03 |
| GO:BP | GO:0042471 | ear morphogenesis | 43 | 74 | 3.43e-03 |
| GO:BP | GO:1902105 | regulation of leukocyte differentiation | 96 | 194 | 3.44e-03 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 230 | 521 | 3.45e-03 |
| GO:BP | GO:0006812 | monoatomic cation transport | 267 | 614 | 3.47e-03 |
| GO:BP | GO:0050680 | negative regulation of epithelial cell proliferation | 54 | 98 | 3.48e-03 |
| GO:BP | GO:1990090 | cellular response to nerve growth factor stimulus | 27 | 41 | 3.56e-03 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 42 | 72 | 3.56e-03 |
| GO:BP | GO:1990089 | response to nerve growth factor | 27 | 41 | 3.56e-03 |
| GO:BP | GO:0010812 | negative regulation of cell-substrate adhesion | 27 | 41 | 3.56e-03 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 721 | 1791 | 3.65e-03 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 775 | 1934 | 3.77e-03 |
| GO:BP | GO:0032409 | regulation of transporter activity | 76 | 148 | 3.81e-03 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 131 | 278 | 3.83e-03 |
| GO:BP | GO:0003015 | heart process | 100 | 204 | 3.83e-03 |
| GO:BP | GO:0046328 | regulation of JNK cascade | 57 | 105 | 3.83e-03 |
| GO:BP | GO:0001704 | formation of primary germ layer | 52 | 94 | 3.93e-03 |
| GO:BP | GO:0003272 | endocardial cushion formation | 18 | 24 | 3.95e-03 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 85 | 169 | 3.95e-03 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 646 | 1595 | 3.95e-03 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 745 | 1856 | 4.05e-03 |
| GO:BP | GO:0034764 | positive regulation of transmembrane transport | 68 | 130 | 4.06e-03 |
| GO:BP | GO:0035115 | embryonic forelimb morphogenesis | 14 | 17 | 4.09e-03 |
| GO:BP | GO:0048659 | smooth muscle cell proliferation | 55 | 101 | 4.39e-03 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 453 | 1093 | 4.47e-03 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 59 | 110 | 4.47e-03 |
| GO:BP | GO:0010092 | specification of animal organ identity | 19 | 26 | 4.47e-03 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 103 | 212 | 4.57e-03 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 273 | 633 | 5.34e-03 |
| GO:BP | GO:0060323 | head morphogenesis | 21 | 30 | 5.36e-03 |
| GO:BP | GO:0048546 | digestive tract morphogenesis | 21 | 30 | 5.36e-03 |
| GO:BP | GO:0150146 | cell junction disassembly | 15 | 19 | 5.39e-03 |
| GO:BP | GO:0003156 | regulation of animal organ formation | 15 | 19 | 5.39e-03 |
| GO:BP | GO:0035137 | hindlimb morphogenesis | 15 | 19 | 5.39e-03 |
| GO:BP | GO:0035136 | forelimb morphogenesis | 15 | 19 | 5.39e-03 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 186 | 415 | 5.43e-03 |
| GO:BP | GO:0016331 | morphogenesis of embryonic epithelium | 64 | 122 | 5.45e-03 |
| GO:BP | GO:2000630 | positive regulation of miRNA metabolic process | 31 | 50 | 5.59e-03 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 218 | 495 | 5.59e-03 |
| GO:BP | GO:0018107 | peptidyl-threonine phosphorylation | 40 | 69 | 5.73e-03 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 46 | 82 | 5.73e-03 |
| GO:BP | GO:0003197 | endocardial cushion development | 29 | 46 | 5.84e-03 |
| GO:BP | GO:1902895 | positive regulation of miRNA transcription | 28 | 44 | 5.93e-03 |
| GO:BP | GO:0019827 | stem cell population maintenance | 73 | 143 | 5.95e-03 |
| GO:BP | GO:0010596 | negative regulation of endothelial cell migration | 25 | 38 | 5.95e-03 |
| GO:BP | GO:0048013 | ephrin receptor signaling pathway | 25 | 38 | 5.95e-03 |
| GO:BP | GO:0007632 | visual behavior | 26 | 40 | 5.95e-03 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 39 | 67 | 5.95e-03 |
| GO:BP | GO:0031290 | retinal ganglion cell axon guidance | 12 | 14 | 5.95e-03 |
| GO:BP | GO:2000725 | regulation of cardiac muscle cell differentiation | 12 | 14 | 5.95e-03 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 66 | 127 | 6.02e-03 |
| GO:BP | GO:0030856 | regulation of epithelial cell differentiation | 54 | 100 | 6.25e-03 |
| GO:BP | GO:0030282 | bone mineralization | 44 | 78 | 6.42e-03 |
| GO:BP | GO:0014912 | negative regulation of smooth muscle cell migration | 16 | 21 | 6.43e-03 |
| GO:BP | GO:0007492 | endoderm development | 37 | 63 | 6.61e-03 |
| GO:BP | GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 132 | 284 | 6.86e-03 |
| GO:BP | GO:0010586 | miRNA metabolic process | 48 | 87 | 6.89e-03 |
| GO:BP | GO:0110020 | regulation of actomyosin structure organization | 48 | 87 | 6.89e-03 |
| GO:BP | GO:0022898 | regulation of transmembrane transporter activity | 71 | 139 | 6.89e-03 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 241 | 555 | 7.16e-03 |
| GO:BP | GO:0060840 | artery development | 47 | 85 | 7.38e-03 |
| GO:BP | GO:0061337 | cardiac conduction | 47 | 85 | 7.38e-03 |
| GO:BP | GO:0031128 | developmental induction | 17 | 23 | 7.40e-03 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 211 | 480 | 7.52e-03 |
| GO:BP | GO:1901342 | regulation of vasculature development | 96 | 198 | 7.53e-03 |
| GO:BP | GO:0008016 | regulation of heart contraction | 82 | 165 | 7.56e-03 |
| GO:BP | GO:0030099 | myeloid cell differentiation | 135 | 292 | 7.61e-03 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 41 | 72 | 7.62e-03 |
| GO:BP | GO:0048863 | stem cell differentiation | 93 | 191 | 7.66e-03 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 777 | 1951 | 7.69e-03 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 90 | 184 | 7.80e-03 |
| GO:BP | GO:1905208 | negative regulation of cardiocyte differentiation | 10 | 11 | 7.80e-03 |
| GO:BP | GO:0038180 | nerve growth factor signaling pathway | 10 | 11 | 7.80e-03 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 103 | 215 | 7.92e-03 |
| GO:BP | GO:0046777 | protein autophosphorylation | 72 | 142 | 7.98e-03 |
| GO:BP | GO:1903689 | regulation of wound healing, spreading of epidermal cells | 8 | 8 | 7.98e-03 |
| GO:BP | GO:0060993 | kidney morphogenesis | 39 | 68 | 8.53e-03 |
| GO:BP | GO:0060047 | heart contraction | 94 | 194 | 8.66e-03 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 80 | 161 | 8.69e-03 |
| GO:BP | GO:0001952 | regulation of cell-matrix adhesion | 49 | 90 | 8.69e-03 |
| GO:BP | GO:0010464 | regulation of mesenchymal cell proliferation | 19 | 27 | 8.72e-03 |
| GO:BP | GO:0060325 | face morphogenesis | 19 | 27 | 8.72e-03 |
| GO:BP | GO:0060998 | regulation of dendritic spine development | 30 | 49 | 8.73e-03 |
| GO:BP | GO:0042063 | gliogenesis | 114 | 242 | 8.74e-03 |
| GO:BP | GO:0030278 | regulation of ossification | 44 | 79 | 8.76e-03 |
| GO:BP | GO:0007010 | cytoskeleton organization | 470 | 1146 | 8.83e-03 |
| GO:BP | GO:0001570 | vasculogenesis | 38 | 66 | 8.90e-03 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 67 | 131 | 9.03e-03 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 82 | 166 | 9.17e-03 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 48 | 88 | 9.17e-03 |
| GO:BP | GO:0048565 | digestive tract development | 48 | 88 | 9.17e-03 |
| GO:BP | GO:0098727 | maintenance of cell number | 73 | 145 | 9.21e-03 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 338 | 805 | 9.24e-03 |
| GO:BP | GO:0060038 | cardiac muscle cell proliferation | 27 | 43 | 9.39e-03 |
| GO:BP | GO:0006915 | apoptotic process | 558 | 1377 | 9.44e-03 |
| GO:BP | GO:0010171 | body morphogenesis | 26 | 41 | 9.55e-03 |
| GO:BP | GO:0021536 | diencephalon development | 25 | 39 | 9.68e-03 |
| GO:BP | GO:0048511 | rhythmic process | 105 | 221 | 9.69e-03 |
| GO:BP | GO:0008542 | visual learning | 23 | 35 | 9.69e-03 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 69 | 136 | 9.76e-03 |
| GO:BP | GO:0010811 | positive regulation of cell-substrate adhesion | 51 | 95 | 9.95e-03 |
| GO:BP | GO:0051234 | establishment of localization | 1257 | 3240 | 1.01e-02 |
| GO:BP | GO:0008219 | cell death | 575 | 1423 | 1.03e-02 |
| GO:BP | GO:0042472 | inner ear morphogenesis | 35 | 60 | 1.03e-02 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 35 | 60 | 1.03e-02 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 104 | 219 | 1.03e-02 |
| GO:BP | GO:0060349 | bone morphogenesis | 41 | 73 | 1.04e-02 |
| GO:BP | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 58 | 111 | 1.04e-02 |
| GO:BP | GO:0032412 | regulation of monoatomic ion transmembrane transporter activity | 68 | 134 | 1.04e-02 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 91 | 188 | 1.04e-02 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 337 | 804 | 1.05e-02 |
| GO:BP | GO:0021987 | cerebral cortex development | 50 | 93 | 1.05e-02 |
| GO:BP | GO:0051896 | regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 88 | 181 | 1.06e-02 |
| GO:BP | GO:0051147 | regulation of muscle cell differentiation | 61 | 118 | 1.07e-02 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 172 | 386 | 1.10e-02 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 176 | 396 | 1.10e-02 |
| GO:BP | GO:0061572 | actin filament bundle organization | 67 | 132 | 1.12e-02 |
| GO:BP | GO:0043368 | positive T cell selection | 15 | 20 | 1.18e-02 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 128 | 278 | 1.18e-02 |
| GO:BP | GO:0022030 | telencephalon glial cell migration | 15 | 20 | 1.18e-02 |
| GO:BP | GO:0099560 | synaptic membrane adhesion | 15 | 20 | 1.18e-02 |
| GO:BP | GO:0021801 | cerebral cortex radial glia-guided migration | 15 | 20 | 1.18e-02 |
| GO:BP | GO:0086065 | cell communication involved in cardiac conduction | 32 | 54 | 1.18e-02 |
| GO:BP | GO:0042475 | odontogenesis of dentin-containing tooth | 32 | 54 | 1.18e-02 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 121 | 261 | 1.18e-02 |
| GO:BP | GO:0007398 | ectoderm development | 11 | 13 | 1.19e-02 |
| GO:BP | GO:0031032 | actomyosin structure organization | 89 | 184 | 1.19e-02 |
| GO:BP | GO:0006814 | sodium ion transport | 69 | 137 | 1.20e-02 |
| GO:BP | GO:0002040 | sprouting angiogenesis | 52 | 98 | 1.20e-02 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 69 | 137 | 1.20e-02 |
| GO:BP | GO:0007015 | actin filament organization | 160 | 357 | 1.20e-02 |
| GO:BP | GO:0045669 | positive regulation of osteoblast differentiation | 31 | 52 | 1.22e-02 |
| GO:BP | GO:0003012 | muscle system process | 147 | 325 | 1.22e-02 |
| GO:BP | GO:0048878 | chemical homeostasis | 284 | 670 | 1.23e-02 |
| GO:BP | GO:0061035 | regulation of cartilage development | 30 | 50 | 1.28e-02 |
| GO:BP | GO:0051148 | negative regulation of muscle cell differentiation | 30 | 50 | 1.28e-02 |
| GO:BP | GO:0055123 | digestive system development | 51 | 96 | 1.28e-02 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 68 | 135 | 1.28e-02 |
| GO:BP | GO:0003018 | vascular process in circulatory system | 93 | 194 | 1.29e-02 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 654 | 1635 | 1.29e-02 |
| GO:BP | GO:0022409 | positive regulation of cell-cell adhesion | 85 | 175 | 1.29e-02 |
| GO:BP | GO:0003148 | outflow tract septum morphogenesis | 16 | 22 | 1.29e-02 |
| GO:BP | GO:0012501 | programmed cell death | 572 | 1419 | 1.30e-02 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 61 | 119 | 1.34e-02 |
| GO:BP | GO:0032231 | regulation of actin filament bundle assembly | 46 | 85 | 1.35e-02 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 36 | 63 | 1.35e-02 |
| GO:BP | GO:0045446 | endothelial cell differentiation | 46 | 85 | 1.35e-02 |
| GO:BP | GO:0021761 | limbic system development | 46 | 85 | 1.35e-02 |
| GO:BP | GO:0097061 | dendritic spine organization | 36 | 63 | 1.35e-02 |
| GO:BP | GO:0035019 | somatic stem cell population maintenance | 28 | 46 | 1.36e-02 |
| GO:BP | GO:0003407 | neural retina development | 28 | 46 | 1.36e-02 |
| GO:BP | GO:0050954 | sensory perception of mechanical stimulus | 57 | 110 | 1.38e-02 |
| GO:BP | GO:0030516 | regulation of axon extension | 41 | 74 | 1.38e-02 |
| GO:BP | GO:0031644 | regulation of nervous system process | 41 | 74 | 1.38e-02 |
| GO:BP | GO:0017145 | stem cell division | 17 | 24 | 1.38e-02 |
| GO:BP | GO:0086002 | cardiac muscle cell action potential involved in contraction | 27 | 44 | 1.40e-02 |
| GO:BP | GO:0061614 | miRNA transcription | 35 | 61 | 1.42e-02 |
| GO:BP | GO:0060491 | regulation of cell projection assembly | 78 | 159 | 1.43e-02 |
| GO:BP | GO:0042391 | regulation of membrane potential | 129 | 282 | 1.44e-02 |
| GO:BP | GO:0007623 | circadian rhythm | 75 | 152 | 1.44e-02 |
| GO:BP | GO:0086009 | membrane repolarization | 26 | 42 | 1.44e-02 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 189 | 431 | 1.44e-02 |
| GO:BP | GO:0045058 | T cell selection | 18 | 26 | 1.45e-02 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 649 | 1624 | 1.45e-02 |
| GO:BP | GO:0060317 | cardiac epithelial to mesenchymal transition | 19 | 28 | 1.51e-02 |
| GO:BP | GO:0120032 | regulation of plasma membrane bounded cell projection assembly | 77 | 157 | 1.53e-02 |
| GO:BP | GO:0001759 | organ induction | 12 | 15 | 1.53e-02 |
| GO:BP | GO:0038179 | neurotrophin signaling pathway | 23 | 36 | 1.53e-02 |
| GO:BP | GO:0043369 | CD4-positive or CD8-positive, alpha-beta T cell lineage commitment | 12 | 15 | 1.53e-02 |
| GO:BP | GO:0001937 | negative regulation of endothelial cell proliferation | 20 | 30 | 1.53e-02 |
| GO:BP | GO:0032330 | regulation of chondrocyte differentiation | 22 | 34 | 1.54e-02 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 288 | 683 | 1.55e-02 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 65 | 129 | 1.55e-02 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 82 | 169 | 1.57e-02 |
| GO:BP | GO:0046834 | lipid phosphorylation | 9 | 10 | 1.59e-02 |
| GO:BP | GO:0060572 | morphogenesis of an epithelial bud | 9 | 10 | 1.59e-02 |
| GO:BP | GO:0061458 | reproductive system development | 97 | 205 | 1.61e-02 |
| GO:BP | GO:0048754 | branching morphogenesis of an epithelial tube | 58 | 113 | 1.62e-02 |
| GO:BP | GO:0055085 | transmembrane transport | 380 | 921 | 1.64e-02 |
| GO:BP | GO:0061387 | regulation of extent of cell growth | 47 | 88 | 1.64e-02 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 111 | 239 | 1.64e-02 |
| GO:BP | GO:0007254 | JNK cascade | 64 | 127 | 1.67e-02 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 172 | 390 | 1.73e-02 |
| GO:BP | GO:0051492 | regulation of stress fiber assembly | 42 | 77 | 1.74e-02 |
| GO:BP | GO:0048332 | mesoderm morphogenesis | 31 | 53 | 1.74e-02 |
| GO:BP | GO:0045639 | positive regulation of myeloid cell differentiation | 37 | 66 | 1.75e-02 |
| GO:BP | GO:0051153 | regulation of striated muscle cell differentiation | 37 | 66 | 1.75e-02 |
| GO:BP | GO:0001558 | regulation of cell growth | 145 | 323 | 1.77e-02 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 433 | 1060 | 1.79e-02 |
| GO:BP | GO:0042060 | wound healing | 132 | 291 | 1.79e-02 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 634 | 1588 | 1.79e-02 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 56 | 109 | 1.88e-02 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 92 | 194 | 1.92e-02 |
| GO:BP | GO:0051641 | cellular localization | 1022 | 2623 | 1.95e-02 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 271 | 642 | 1.96e-02 |
| GO:BP | GO:0003158 | endothelium development | 52 | 100 | 1.96e-02 |
| GO:BP | GO:0043149 | stress fiber assembly | 48 | 91 | 2.01e-02 |
| GO:BP | GO:0030038 | contractile actin filament bundle assembly | 48 | 91 | 2.01e-02 |
| GO:BP | GO:0045600 | positive regulation of fat cell differentiation | 28 | 47 | 2.02e-02 |
| GO:BP | GO:0030048 | actin filament-based movement | 55 | 107 | 2.02e-02 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 167 | 379 | 2.08e-02 |
| GO:BP | GO:0002360 | T cell lineage commitment | 14 | 19 | 2.09e-02 |
| GO:BP | GO:0021772 | olfactory bulb development | 14 | 19 | 2.09e-02 |
| GO:BP | GO:0060997 | dendritic spine morphogenesis | 27 | 45 | 2.09e-02 |
| GO:BP | GO:0060512 | prostate gland morphogenesis | 14 | 19 | 2.09e-02 |
| GO:BP | GO:2000647 | negative regulation of stem cell proliferation | 14 | 19 | 2.09e-02 |
| GO:BP | GO:0048566 | embryonic digestive tract development | 14 | 19 | 2.09e-02 |
| GO:BP | GO:0060740 | prostate gland epithelium morphogenesis | 14 | 19 | 2.09e-02 |
| GO:BP | GO:0106027 | neuron projection organization | 39 | 71 | 2.10e-02 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 421 | 1031 | 2.10e-02 |
| GO:BP | GO:0007162 | negative regulation of cell adhesion | 88 | 185 | 2.11e-02 |
| GO:BP | GO:0048608 | reproductive structure development | 95 | 202 | 2.18e-02 |
| GO:BP | GO:0048844 | artery morphogenesis | 33 | 58 | 2.22e-02 |
| GO:BP | GO:0072028 | nephron morphogenesis | 33 | 58 | 2.22e-02 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 467 | 1152 | 2.22e-02 |
| GO:BP | GO:0007605 | sensory perception of sound | 50 | 96 | 2.24e-02 |
| GO:BP | GO:0061383 | trabecula morphogenesis | 25 | 41 | 2.26e-02 |
| GO:BP | GO:0086011 | membrane repolarization during action potential | 15 | 21 | 2.27e-02 |
| GO:BP | GO:0021983 | pituitary gland development | 15 | 21 | 2.27e-02 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 46 | 87 | 2.29e-02 |
| GO:BP | GO:0050773 | regulation of dendrite development | 42 | 78 | 2.30e-02 |
| GO:BP | GO:0001763 | morphogenesis of a branching structure | 71 | 145 | 2.31e-02 |
| GO:BP | GO:0003198 | epithelial to mesenchymal transition involved in endocardial cushion formation | 10 | 12 | 2.31e-02 |
| GO:BP | GO:0045176 | apical protein localization | 10 | 12 | 2.31e-02 |
| GO:BP | GO:0002088 | lens development in camera-type eye | 32 | 56 | 2.31e-02 |
| GO:BP | GO:0045670 | regulation of osteoclast differentiation | 24 | 39 | 2.31e-02 |
| GO:BP | GO:0032486 | Rap protein signal transduction | 10 | 12 | 2.31e-02 |
| GO:BP | GO:0014009 | glial cell proliferation | 24 | 39 | 2.31e-02 |
| GO:BP | GO:0048708 | astrocyte differentiation | 32 | 56 | 2.31e-02 |
| GO:BP | GO:0001658 | branching involved in ureteric bud morphogenesis | 24 | 39 | 2.31e-02 |
| GO:BP | GO:0051491 | positive regulation of filopodium assembly | 16 | 23 | 2.38e-02 |
| GO:BP | GO:0043537 | negative regulation of blood vessel endothelial cell migration | 16 | 23 | 2.38e-02 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 604 | 1514 | 2.42e-02 |
| GO:BP | GO:0051963 | regulation of synapse assembly | 41 | 76 | 2.43e-02 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 45 | 85 | 2.43e-02 |
| GO:BP | GO:0001775 | cell activation | 264 | 627 | 2.51e-02 |
| GO:BP | GO:0030857 | negative regulation of epithelial cell differentiation | 20 | 31 | 2.53e-02 |
| GO:BP | GO:0099622 | cardiac muscle cell membrane repolarization | 20 | 31 | 2.53e-02 |
| GO:BP | GO:0060563 | neuroepithelial cell differentiation | 19 | 29 | 2.53e-02 |
| GO:BP | GO:0006810 | transport | 1154 | 2984 | 2.53e-02 |
| GO:BP | GO:0051050 | positive regulation of transport | 242 | 571 | 2.57e-02 |
| GO:BP | GO:0061180 | mammary gland epithelium development | 30 | 52 | 2.59e-02 |
| GO:BP | GO:0001708 | cell fate specification | 30 | 52 | 2.59e-02 |
| GO:BP | GO:0045927 | positive regulation of growth | 87 | 184 | 2.67e-02 |
| GO:BP | GO:0007043 | cell-cell junction assembly | 54 | 106 | 2.70e-02 |
| GO:BP | GO:0002011 | morphogenesis of an epithelial sheet | 29 | 50 | 2.74e-02 |
| GO:BP | GO:0042490 | mechanoreceptor differentiation | 29 | 50 | 2.74e-02 |
| GO:BP | GO:0030199 | collagen fibril organization | 29 | 50 | 2.74e-02 |
| GO:BP | GO:0006936 | muscle contraction | 116 | 255 | 2.85e-02 |
| GO:BP | GO:0035249 | synaptic transmission, glutamatergic | 34 | 61 | 2.87e-02 |
| GO:BP | GO:0008306 | associative learning | 34 | 61 | 2.87e-02 |
| GO:BP | GO:0001656 | metanephros development | 34 | 61 | 2.87e-02 |
| GO:BP | GO:0060113 | inner ear receptor cell differentiation | 28 | 48 | 2.90e-02 |
| GO:BP | GO:0071772 | response to BMP | 59 | 118 | 2.94e-02 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 59 | 118 | 2.94e-02 |
| GO:BP | GO:0048485 | sympathetic nervous system development | 11 | 14 | 2.94e-02 |
| GO:BP | GO:0010842 | retina layer formation | 11 | 14 | 2.94e-02 |
| GO:BP | GO:0002363 | alpha-beta T cell lineage commitment | 11 | 14 | 2.94e-02 |
| GO:BP | GO:0008347 | glial cell migration | 27 | 46 | 3.05e-02 |
| GO:BP | GO:0072171 | mesonephric tubule morphogenesis | 27 | 46 | 3.05e-02 |
| GO:BP | GO:0035725 | sodium ion transmembrane transport | 49 | 95 | 3.05e-02 |
| GO:BP | GO:0043491 | phosphatidylinositol 3-kinase/protein kinase B signal transduction | 99 | 214 | 3.11e-02 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 55 | 109 | 3.16e-02 |
| GO:BP | GO:0098657 | import into cell | 261 | 622 | 3.19e-02 |
| GO:BP | GO:0060324 | face development | 26 | 44 | 3.20e-02 |
| GO:BP | GO:0002042 | cell migration involved in sprouting angiogenesis | 26 | 44 | 3.20e-02 |
| GO:BP | GO:0014033 | neural crest cell differentiation | 41 | 77 | 3.22e-02 |
| GO:BP | GO:0007600 | sensory perception | 114 | 251 | 3.23e-02 |
| GO:BP | GO:0021954 | central nervous system neuron development | 32 | 57 | 3.24e-02 |
| GO:BP | GO:0001954 | positive regulation of cell-matrix adhesion | 25 | 42 | 3.36e-02 |
| GO:BP | GO:0060601 | lateral sprouting from an epithelium | 8 | 9 | 3.36e-02 |
| GO:BP | GO:1901329 | regulation of odontoblast differentiation | 8 | 9 | 3.36e-02 |
| GO:BP | GO:0045986 | negative regulation of smooth muscle contraction | 8 | 9 | 3.36e-02 |
| GO:BP | GO:0008361 | regulation of cell size | 71 | 147 | 3.37e-02 |
| GO:BP | GO:0006935 | chemotaxis | 111 | 244 | 3.39e-02 |
| GO:BP | GO:0007498 | mesoderm development | 44 | 84 | 3.39e-02 |
| GO:BP | GO:0042330 | taxis | 111 | 244 | 3.39e-02 |
| GO:BP | GO:0055008 | cardiac muscle tissue morphogenesis | 31 | 55 | 3.44e-02 |
| GO:BP | GO:0045668 | negative regulation of osteoblast differentiation | 23 | 38 | 3.68e-02 |
| GO:BP | GO:0061001 | regulation of dendritic spine morphogenesis | 22 | 36 | 3.84e-02 |
| GO:BP | GO:0003203 | endocardial cushion morphogenesis | 22 | 36 | 3.84e-02 |
| GO:BP | GO:0048704 | embryonic skeletal system morphogenesis | 29 | 51 | 3.94e-02 |
| GO:BP | GO:0001707 | mesoderm formation | 29 | 51 | 3.94e-02 |
| GO:BP | GO:0051591 | response to cAMP | 34 | 62 | 3.95e-02 |
| GO:BP | GO:0050865 | regulation of cell activation | 152 | 347 | 3.96e-02 |
| GO:BP | GO:0030509 | BMP signaling pathway | 55 | 110 | 3.96e-02 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 100 | 218 | 3.96e-02 |
| GO:BP | GO:0010629 | negative regulation of gene expression | 325 | 790 | 4.01e-02 |
| GO:BP | GO:0045926 | negative regulation of growth | 86 | 184 | 4.02e-02 |
| GO:BP | GO:0002053 | positive regulation of mesenchymal cell proliferation | 14 | 20 | 4.04e-02 |
| GO:BP | GO:0021516 | dorsal spinal cord development | 6 | 6 | 4.04e-02 |
| GO:BP | GO:0048617 | embryonic foregut morphogenesis | 6 | 6 | 4.04e-02 |
| GO:BP | GO:1901289 | succinyl-CoA catabolic process | 6 | 6 | 4.04e-02 |
| GO:BP | GO:0072178 | nephric duct morphogenesis | 6 | 6 | 4.04e-02 |
| GO:BP | GO:0050861 | positive regulation of B cell receptor signaling pathway | 6 | 6 | 4.04e-02 |
| GO:BP | GO:0021814 | cell motility involved in cerebral cortex radial glia guided migration | 6 | 6 | 4.04e-02 |
| GO:BP | GO:0050982 | detection of mechanical stimulus | 20 | 32 | 4.05e-02 |
| GO:BP | GO:0008154 | actin polymerization or depolymerization | 71 | 148 | 4.05e-02 |
| GO:BP | GO:0032233 | positive regulation of actin filament bundle assembly | 28 | 49 | 4.13e-02 |
| GO:BP | GO:0060914 | heart formation | 19 | 30 | 4.15e-02 |
| GO:BP | GO:0033688 | regulation of osteoblast proliferation | 15 | 22 | 4.16e-02 |
| GO:BP | GO:0048661 | positive regulation of smooth muscle cell proliferation | 33 | 60 | 4.16e-02 |
| GO:BP | GO:0021675 | nerve development | 33 | 60 | 4.16e-02 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 57 | 115 | 4.17e-02 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 57 | 115 | 4.17e-02 |
| GO:BP | GO:0072006 | nephron development | 57 | 115 | 4.17e-02 |
| GO:BP | GO:0030510 | regulation of BMP signaling pathway | 41 | 78 | 4.17e-02 |
| GO:BP | GO:0021799 | cerebral cortex radially oriented cell migration | 17 | 26 | 4.18e-02 |
| GO:BP | GO:0071392 | cellular response to estradiol stimulus | 16 | 24 | 4.18e-02 |
| GO:BP | GO:0048048 | embryonic eye morphogenesis | 16 | 24 | 4.18e-02 |
| GO:BP | GO:0003181 | atrioventricular valve morphogenesis | 17 | 26 | 4.18e-02 |
| GO:BP | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway | 16 | 24 | 4.18e-02 |
| GO:BP | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction | 17 | 26 | 4.18e-02 |
| GO:BP | GO:0110110 | positive regulation of animal organ morphogenesis | 17 | 26 | 4.18e-02 |
| GO:BP | GO:0035282 | segmentation | 37 | 69 | 4.18e-02 |
| GO:BP | GO:0033687 | osteoblast proliferation | 17 | 26 | 4.18e-02 |
| GO:BP | GO:0010721 | negative regulation of cell development | 85 | 182 | 4.19e-02 |
| GO:BP | GO:0033135 | regulation of peptidyl-serine phosphorylation | 44 | 85 | 4.30e-02 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 364 | 893 | 4.37e-02 |
| GO:BP | GO:0061333 | renal tubule morphogenesis | 32 | 58 | 4.38e-02 |
| GO:BP | GO:0008543 | fibroblast growth factor receptor signaling pathway | 32 | 58 | 4.38e-02 |
| GO:BP | GO:0071805 | potassium ion transmembrane transport | 59 | 120 | 4.38e-02 |
| GO:BP | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | 56 | 113 | 4.43e-02 |
| GO:BP | GO:0055001 | muscle cell development | 77 | 163 | 4.43e-02 |
| GO:BP | GO:0060675 | ureteric bud morphogenesis | 26 | 45 | 4.56e-02 |
| GO:BP | GO:0035331 | negative regulation of hippo signaling | 9 | 11 | 4.61e-02 |
| GO:BP | GO:0021895 | cerebral cortex neuron differentiation | 9 | 11 | 4.61e-02 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 43 | 83 | 4.62e-02 |
| GO:BP | GO:0072088 | nephron epithelium morphogenesis | 31 | 56 | 4.67e-02 |
| GO:BP | GO:0003229 | ventricular cardiac muscle tissue development | 31 | 56 | 4.67e-02 |
| GO:BP | GO:0046928 | regulation of neurotransmitter secretion | 31 | 56 | 4.67e-02 |
| GO:BP | GO:0002682 | regulation of immune system process | 354 | 868 | 4.70e-02 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 97 | 212 | 4.70e-02 |
| GO:BP | GO:0061326 | renal tubule development | 39 | 74 | 4.75e-02 |
| GO:BP | GO:0048864 | stem cell development | 39 | 74 | 4.75e-02 |
| GO:BP | GO:0030316 | osteoclast differentiation | 35 | 65 | 4.78e-02 |
| GO:BP | GO:0051496 | positive regulation of stress fiber assembly | 25 | 43 | 4.78e-02 |
| GO:BP | GO:0009880 | embryonic pattern specification | 25 | 43 | 4.78e-02 |
| GO:BP | GO:0050679 | positive regulation of epithelial cell proliferation | 68 | 142 | 4.93e-02 |
| GO:BP | GO:0019722 | calcium-mediated signaling | 60 | 123 | 4.97e-02 |
| GO:BP | GO:0072078 | nephron tubule morphogenesis | 30 | 54 | 4.97e-02 |
EARnomtable %>%
filter(source=="KEGG", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "KEGG terms found in EAR neargenes") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| KEGG | KEGG:04510 | Focal adhesion | 97 | 169 | 6.03e-06 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 90 | 162 | 7.47e-05 |
| KEGG | KEGG:04360 | Axon guidance | 84 | 155 | 4.62e-04 |
| KEGG | KEGG:04512 | ECM-receptor interaction | 42 | 66 | 5.12e-04 |
| KEGG | KEGG:04520 | Adherens junction | 49 | 83 | 1.45e-03 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 71 | 133 | 2.44e-03 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 75 | 147 | 6.33e-03 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 111 | 231 | 6.33e-03 |
| KEGG | KEGG:04726 | Serotonergic synapse | 35 | 58 | 6.33e-03 |
| KEGG | KEGG:05200 | Pathways in cancer | 177 | 391 | 6.33e-03 |
| KEGG | KEGG:05222 | Small cell lung cancer | 46 | 82 | 6.46e-03 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 118 | 250 | 7.19e-03 |
| KEGG | KEGG:04014 | Ras signaling pathway | 81 | 164 | 9.91e-03 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 63 | 122 | 9.91e-03 |
| KEGG | KEGG:05412 | Arrhythmogenic right ventricular cardiomyopathy | 38 | 67 | 1.24e-02 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 48 | 90 | 1.47e-02 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 82 | 169 | 1.47e-02 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 79 | 162 | 1.47e-02 |
| KEGG | KEGG:04514 | Cell adhesion molecules | 38 | 69 | 2.10e-02 |
| KEGG | KEGG:04730 | Long-term depression | 24 | 39 | 2.10e-02 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 64 | 129 | 2.22e-02 |
| KEGG | KEGG:04713 | Circadian entrainment | 37 | 68 | 2.80e-02 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 41 | 78 | 3.73e-02 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 53 | 106 | 3.91e-02 |
| KEGG | KEGG:04974 | Protein digestion and absorption | 31 | 56 | 3.97e-02 |
| KEGG | KEGG:04072 | Phospholipase D signaling pathway | 53 | 107 | 4.66e-02 |
EARnomtable %>%
filter(source=="GO:BP", p_value < 0.05) %>%
slice_head(n=10) %>%
mutate(log10_p=(-log(p_value, base=10))) %>%
ggplot(., aes(x=log10_p, y=reorder(term_name, log10_p)))+
geom_col(aes(fill="#F8766D"))+
theme_bw()+
ylab("")+
xlab(paste0("-log10 p-value"))+
guides(fill="none")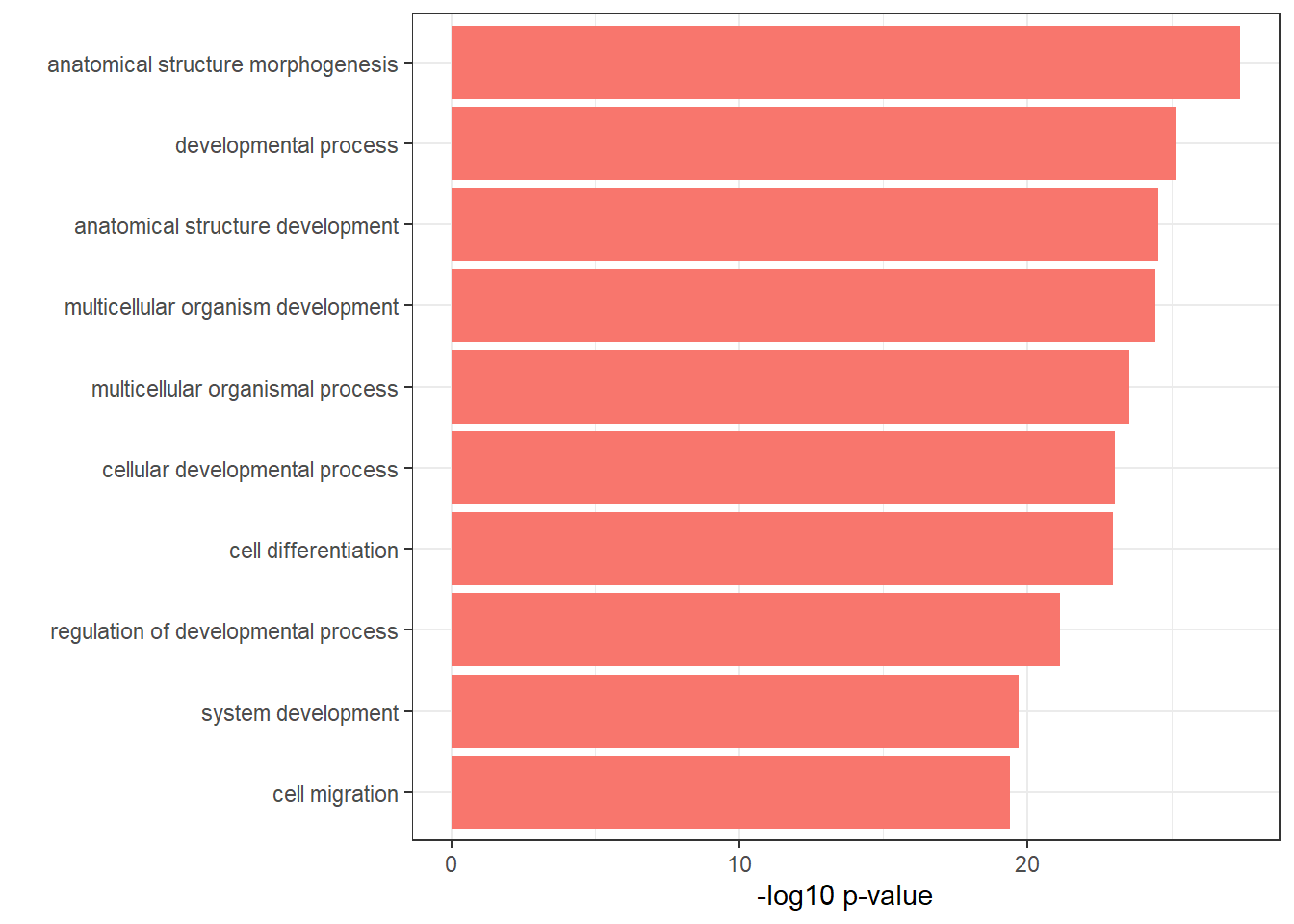
ESR response genes
# ESR_resgenes <- gost(query = ESR_NG_list$ENTREZID,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = FALSE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = background_df$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(ESR_resgenes,"data/GO_KEGG_analysis/ESR_resgenes.RDS")
# ESRnom <- gostplot(ESR_resgenes, capped = FALSE, interactive = TRUE)
# ESRnom
ESRnomtable <- ESR_resgenes$result %>%
dplyr::select(c(source, term_id,
term_name,intersection_size,
term_size, p_value))# %>%
ESRnomtable %>%
filter(source=="GO:BP", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(.,caption = "GO:BP terms found in ESR neargenes" ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0032501 | multicellular organismal process | 2719 | 4504 | 6.19e-51 |
| GO:BP | GO:0048856 | anatomical structure development | 2368 | 3867 | 1.05e-48 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 1237 | 1844 | 1.90e-47 |
| GO:BP | GO:0032502 | developmental process | 2521 | 4179 | 2.35e-45 |
| GO:BP | GO:0007275 | multicellular organism development | 1971 | 3170 | 2.02e-44 |
| GO:BP | GO:0007154 | cell communication | 2411 | 3983 | 2.68e-44 |
| GO:BP | GO:0023052 | signaling | 2374 | 3918 | 7.47e-44 |
| GO:BP | GO:0048731 | system development | 1713 | 2719 | 3.79e-42 |
| GO:BP | GO:0007165 | signal transduction | 2187 | 3611 | 9.13e-39 |
| GO:BP | GO:0030154 | cell differentiation | 1746 | 2803 | 1.06e-38 |
| GO:BP | GO:0048869 | cellular developmental process | 1746 | 2804 | 1.33e-38 |
| GO:BP | GO:0050896 | response to stimulus | 3104 | 5379 | 5.22e-35 |
| GO:BP | GO:0051716 | cellular response to stimulus | 2717 | 4640 | 6.46e-35 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 1237 | 1924 | 5.46e-34 |
| GO:BP | GO:0009888 | tissue development | 904 | 1348 | 2.90e-33 |
| GO:BP | GO:0048513 | animal organ development | 1279 | 2004 | 3.55e-33 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 597 | 837 | 4.28e-32 |
| GO:BP | GO:0007399 | nervous system development | 1136 | 1775 | 1.60e-29 |
| GO:BP | GO:0007155 | cell adhesion | 651 | 941 | 7.22e-29 |
| GO:BP | GO:0010646 | regulation of cell communication | 1503 | 2444 | 4.76e-28 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 1142 | 1801 | 2.61e-27 |
| GO:BP | GO:0023051 | regulation of signaling | 1496 | 2438 | 3.01e-27 |
| GO:BP | GO:0035295 | tube development | 553 | 787 | 7.25e-27 |
| GO:BP | GO:0072359 | circulatory system development | 599 | 864 | 8.41e-27 |
| GO:BP | GO:0016477 | cell migration | 686 | 1011 | 9.57e-27 |
| GO:BP | GO:0048468 | cell development | 1184 | 1886 | 5.73e-26 |
| GO:BP | GO:0050793 | regulation of developmental process | 1076 | 1697 | 1.61e-25 |
| GO:BP | GO:0048870 | cell motility | 745 | 1120 | 2.94e-25 |
| GO:BP | GO:0035239 | tube morphogenesis | 458 | 638 | 3.31e-25 |
| GO:BP | GO:0022008 | neurogenesis | 819 | 1251 | 9.61e-25 |
| GO:BP | GO:0003008 | system process | 752 | 1139 | 4.38e-24 |
| GO:BP | GO:0048729 | tissue morphogenesis | 340 | 453 | 6.87e-24 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 475 | 673 | 1.46e-23 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 692 | 1040 | 2.24e-23 |
| GO:BP | GO:0048699 | generation of neurons | 711 | 1076 | 8.28e-23 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 637 | 951 | 1.83e-22 |
| GO:BP | GO:0030182 | neuron differentiation | 671 | 1014 | 1.21e-21 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 842 | 1314 | 2.89e-21 |
| GO:BP | GO:0007267 | cell-cell signaling | 709 | 1082 | 2.91e-21 |
| GO:BP | GO:0009966 | regulation of signal transduction | 1316 | 2166 | 6.52e-21 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 493 | 717 | 1.33e-20 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 607 | 913 | 3.88e-20 |
| GO:BP | GO:0030334 | regulation of cell migration | 461 | 668 | 1.15e-19 |
| GO:BP | GO:0001944 | vasculature development | 391 | 552 | 1.24e-19 |
| GO:BP | GO:0050789 | regulation of biological process | 4101 | 7516 | 3.07e-19 |
| GO:BP | GO:0040011 | locomotion | 540 | 805 | 5.25e-19 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 1605 | 2717 | 7.46e-19 |
| GO:BP | GO:2000145 | regulation of cell motility | 477 | 700 | 1.07e-18 |
| GO:BP | GO:0001568 | blood vessel development | 372 | 525 | 1.07e-18 |
| GO:BP | GO:0044057 | regulation of system process | 285 | 384 | 1.07e-18 |
| GO:BP | GO:0065007 | biological regulation | 4205 | 7734 | 1.75e-18 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 422 | 609 | 1.86e-18 |
| GO:BP | GO:0040012 | regulation of locomotion | 488 | 720 | 1.89e-18 |
| GO:BP | GO:0060429 | epithelium development | 530 | 792 | 2.43e-18 |
| GO:BP | GO:0034330 | cell junction organization | 383 | 545 | 2.60e-18 |
| GO:BP | GO:0098609 | cell-cell adhesion | 394 | 564 | 3.80e-18 |
| GO:BP | GO:0050794 | regulation of cellular process | 3906 | 7153 | 1.53e-17 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 323 | 450 | 1.64e-17 |
| GO:BP | GO:0030029 | actin filament-based process | 437 | 640 | 2.59e-17 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 272 | 369 | 3.69e-17 |
| GO:BP | GO:0000902 | cell morphogenesis | 478 | 713 | 1.18e-16 |
| GO:BP | GO:0009790 | embryo development | 541 | 821 | 1.22e-16 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 664 | 1035 | 1.36e-16 |
| GO:BP | GO:0035556 | intracellular signal transduction | 1175 | 1951 | 1.58e-16 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 685 | 1073 | 1.95e-16 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 405 | 591 | 2.08e-16 |
| GO:BP | GO:0032989 | cellular anatomical entity morphogenesis | 403 | 588 | 2.42e-16 |
| GO:BP | GO:0003013 | circulatory system process | 305 | 426 | 2.87e-16 |
| GO:BP | GO:0048518 | positive regulation of biological process | 2429 | 4301 | 3.13e-16 |
| GO:BP | GO:0060485 | mesenchyme development | 191 | 245 | 3.48e-16 |
| GO:BP | GO:0048519 | negative regulation of biological process | 2146 | 3764 | 3.74e-16 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 2246 | 3954 | 3.89e-16 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 302 | 422 | 4.42e-16 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 354 | 508 | 5.26e-16 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 2030 | 3550 | 9.49e-16 |
| GO:BP | GO:0070848 | response to growth factor | 376 | 547 | 1.55e-15 |
| GO:BP | GO:0007507 | heart development | 347 | 499 | 1.86e-15 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 1120 | 1862 | 1.96e-15 |
| GO:BP | GO:0008015 | blood circulation | 260 | 357 | 2.56e-15 |
| GO:BP | GO:0065008 | regulation of biological quality | 1136 | 1893 | 2.94e-15 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 749 | 1197 | 4.69e-15 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 722 | 1150 | 6.09e-15 |
| GO:BP | GO:0048666 | neuron development | 544 | 838 | 7.15e-15 |
| GO:BP | GO:0061061 | muscle structure development | 367 | 536 | 9.31e-15 |
| GO:BP | GO:0030030 | cell projection organization | 739 | 1182 | 1.01e-14 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 362 | 528 | 1.09e-14 |
| GO:BP | GO:0031175 | neuron projection development | 488 | 743 | 1.35e-14 |
| GO:BP | GO:0014706 | striated muscle tissue development | 167 | 213 | 1.47e-14 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 158 | 200 | 2.74e-14 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 158 | 201 | 6.30e-14 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 382 | 566 | 6.89e-14 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 676 | 1077 | 6.96e-14 |
| GO:BP | GO:0001525 | angiogenesis | 271 | 381 | 7.21e-14 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 338 | 494 | 1.76e-13 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 315 | 456 | 2.20e-13 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 340 | 498 | 2.30e-13 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 322 | 469 | 3.95e-13 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 460 | 706 | 7.81e-13 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 194 | 261 | 9.34e-13 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 749 | 1218 | 1.35e-12 |
| GO:BP | GO:0023057 | negative regulation of signaling | 647 | 1036 | 1.35e-12 |
| GO:BP | GO:0007423 | sensory organ development | 271 | 387 | 1.52e-12 |
| GO:BP | GO:0023056 | positive regulation of signaling | 750 | 1221 | 1.80e-12 |
| GO:BP | GO:0007010 | cytoskeleton organization | 708 | 1146 | 1.89e-12 |
| GO:BP | GO:0051049 | regulation of transport | 681 | 1098 | 1.98e-12 |
| GO:BP | GO:0008283 | cell population proliferation | 797 | 1306 | 1.99e-12 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 645 | 1034 | 2.00e-12 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 325 | 479 | 3.29e-12 |
| GO:BP | GO:0003012 | muscle system process | 232 | 325 | 4.33e-12 |
| GO:BP | GO:0060537 | muscle tissue development | 240 | 339 | 6.98e-12 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 243 | 344 | 7.18e-12 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 407 | 622 | 1.06e-11 |
| GO:BP | GO:0042221 | response to chemical | 1336 | 2304 | 1.47e-11 |
| GO:BP | GO:0006468 | protein phosphorylation | 598 | 958 | 1.59e-11 |
| GO:BP | GO:0034329 | cell junction assembly | 229 | 323 | 1.99e-11 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 296 | 435 | 2.60e-11 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 337 | 505 | 3.26e-11 |
| GO:BP | GO:0098657 | import into cell | 405 | 622 | 3.52e-11 |
| GO:BP | GO:0007417 | central nervous system development | 446 | 694 | 4.58e-11 |
| GO:BP | GO:0048589 | developmental growth | 334 | 501 | 4.96e-11 |
| GO:BP | GO:0072001 | renal system development | 179 | 244 | 6.49e-11 |
| GO:BP | GO:0010720 | positive regulation of cell development | 219 | 309 | 6.74e-11 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 342 | 516 | 8.22e-11 |
| GO:BP | GO:0048568 | embryonic organ development | 215 | 303 | 8.93e-11 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 725 | 1193 | 1.10e-10 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 777 | 1289 | 1.75e-10 |
| GO:BP | GO:0051960 | regulation of nervous system development | 236 | 339 | 1.76e-10 |
| GO:BP | GO:0060322 | head development | 354 | 539 | 1.98e-10 |
| GO:BP | GO:0072073 | kidney epithelium development | 92 | 111 | 2.23e-10 |
| GO:BP | GO:0030001 | metal ion transport | 345 | 524 | 2.32e-10 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 600 | 972 | 2.48e-10 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 667 | 1093 | 3.12e-10 |
| GO:BP | GO:0001501 | skeletal system development | 240 | 347 | 3.22e-10 |
| GO:BP | GO:0003007 | heart morphogenesis | 161 | 218 | 3.42e-10 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 91 | 110 | 3.49e-10 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 478 | 757 | 3.61e-10 |
| GO:BP | GO:0045165 | cell fate commitment | 118 | 151 | 4.59e-10 |
| GO:BP | GO:0003205 | cardiac chamber development | 118 | 151 | 4.59e-10 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 132 | 173 | 5.78e-10 |
| GO:BP | GO:0007420 | brain development | 328 | 498 | 7.15e-10 |
| GO:BP | GO:0001822 | kidney development | 172 | 237 | 7.36e-10 |
| GO:BP | GO:0032879 | regulation of localization | 855 | 1439 | 9.26e-10 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 209 | 298 | 9.79e-10 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 261 | 386 | 1.63e-09 |
| GO:BP | GO:0001823 | mesonephros development | 65 | 74 | 1.79e-09 |
| GO:BP | GO:0006936 | muscle contraction | 182 | 255 | 1.94e-09 |
| GO:BP | GO:1903522 | regulation of blood circulation | 144 | 194 | 2.29e-09 |
| GO:BP | GO:0003231 | cardiac ventricle development | 93 | 115 | 2.36e-09 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 242 | 355 | 2.45e-09 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 299 | 452 | 2.74e-09 |
| GO:BP | GO:0072164 | mesonephric tubule development | 64 | 73 | 2.95e-09 |
| GO:BP | GO:0072163 | mesonephric epithelium development | 64 | 73 | 2.95e-09 |
| GO:BP | GO:0060284 | regulation of cell development | 367 | 570 | 3.33e-09 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 177 | 248 | 3.47e-09 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 265 | 395 | 3.86e-09 |
| GO:BP | GO:0016310 | phosphorylation | 689 | 1145 | 4.94e-09 |
| GO:BP | GO:0001657 | ureteric bud development | 63 | 72 | 4.94e-09 |
| GO:BP | GO:0016043 | cellular component organization | 2629 | 4802 | 5.18e-09 |
| GO:BP | GO:0051179 | localization | 2071 | 3731 | 5.58e-09 |
| GO:BP | GO:0030900 | forebrain development | 191 | 272 | 5.75e-09 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 272 | 408 | 5.80e-09 |
| GO:BP | GO:0007264 | small GTPase-mediated signal transduction | 261 | 390 | 7.69e-09 |
| GO:BP | GO:0090257 | regulation of muscle system process | 132 | 177 | 8.05e-09 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 208 | 301 | 8.37e-09 |
| GO:BP | GO:0140352 | export from cell | 379 | 594 | 8.42e-09 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 140 | 190 | 9.34e-09 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 59 | 67 | 1.06e-08 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 59 | 67 | 1.06e-08 |
| GO:BP | GO:0007409 | axonogenesis | 229 | 337 | 1.18e-08 |
| GO:BP | GO:0061448 | connective tissue development | 149 | 205 | 1.27e-08 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 196 | 282 | 1.27e-08 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 106 | 137 | 1.32e-08 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 485 | 783 | 1.39e-08 |
| GO:BP | GO:0060047 | heart contraction | 142 | 194 | 1.46e-08 |
| GO:BP | GO:0051216 | cartilage development | 111 | 145 | 1.55e-08 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 275 | 416 | 1.62e-08 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 282 | 428 | 1.62e-08 |
| GO:BP | GO:0048880 | sensory system development | 186 | 266 | 1.62e-08 |
| GO:BP | GO:0042692 | muscle cell differentiation | 220 | 323 | 1.82e-08 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 220 | 323 | 1.82e-08 |
| GO:BP | GO:0061564 | axon development | 249 | 372 | 1.82e-08 |
| GO:BP | GO:0001654 | eye development | 184 | 263 | 1.84e-08 |
| GO:BP | GO:0006810 | transport | 1673 | 2984 | 1.89e-08 |
| GO:BP | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | 90 | 113 | 1.89e-08 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 1015 | 1751 | 2.12e-08 |
| GO:BP | GO:0050808 | synapse organization | 228 | 337 | 2.23e-08 |
| GO:BP | GO:0035265 | organ growth | 99 | 127 | 2.28e-08 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 304 | 467 | 2.31e-08 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 219 | 322 | 2.36e-08 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 335 | 521 | 2.49e-08 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 1480 | 2622 | 2.65e-08 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 73 | 88 | 2.66e-08 |
| GO:BP | GO:0150063 | visual system development | 184 | 264 | 2.85e-08 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 165 | 233 | 3.35e-08 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 301 | 463 | 3.37e-08 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 301 | 463 | 3.37e-08 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 121 | 162 | 3.39e-08 |
| GO:BP | GO:0003018 | vascular process in circulatory system | 141 | 194 | 3.39e-08 |
| GO:BP | GO:0065009 | regulation of molecular function | 768 | 1299 | 3.42e-08 |
| GO:BP | GO:0048863 | stem cell differentiation | 139 | 191 | 3.84e-08 |
| GO:BP | GO:0003015 | heart process | 147 | 204 | 3.96e-08 |
| GO:BP | GO:0060419 | heart growth | 61 | 71 | 4.05e-08 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 240 | 359 | 4.09e-08 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 117 | 156 | 4.09e-08 |
| GO:BP | GO:0040007 | growth | 434 | 697 | 4.15e-08 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 879 | 1504 | 4.15e-08 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 199 | 290 | 4.29e-08 |
| GO:BP | GO:0097485 | neuron projection guidance | 127 | 172 | 4.29e-08 |
| GO:BP | GO:0007411 | axon guidance | 127 | 172 | 4.29e-08 |
| GO:BP | GO:0003279 | cardiac septum development | 82 | 102 | 4.35e-08 |
| GO:BP | GO:0099536 | synaptic signaling | 312 | 483 | 4.35e-08 |
| GO:BP | GO:0090130 | tissue migration | 173 | 247 | 4.71e-08 |
| GO:BP | GO:0043062 | extracellular structure organization | 159 | 224 | 4.73e-08 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 159 | 224 | 4.73e-08 |
| GO:BP | GO:0001775 | cell activation | 394 | 627 | 4.92e-08 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 239 | 358 | 5.11e-08 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 168 | 239 | 5.22e-08 |
| GO:BP | GO:0046903 | secretion | 391 | 622 | 5.28e-08 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 300 | 463 | 5.48e-08 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 300 | 463 | 5.48e-08 |
| GO:BP | GO:0071310 | cellular response to organic substance | 765 | 1297 | 6.20e-08 |
| GO:BP | GO:0045216 | cell-cell junction organization | 114 | 152 | 6.21e-08 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 381 | 605 | 6.21e-08 |
| GO:BP | GO:0030198 | extracellular matrix organization | 158 | 223 | 6.36e-08 |
| GO:BP | GO:0043009 | chordate embryonic development | 325 | 507 | 6.45e-08 |
| GO:BP | GO:0010243 | response to organonitrogen compound | 426 | 685 | 6.91e-08 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 170 | 243 | 7.05e-08 |
| GO:BP | GO:0090132 | epithelium migration | 170 | 243 | 7.05e-08 |
| GO:BP | GO:0010631 | epithelial cell migration | 170 | 243 | 7.05e-08 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 197 | 288 | 7.18e-08 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 179 | 258 | 7.25e-08 |
| GO:BP | GO:0008016 | regulation of heart contraction | 122 | 165 | 7.25e-08 |
| GO:BP | GO:0006897 | endocytosis | 316 | 492 | 7.78e-08 |
| GO:BP | GO:0072006 | nephron development | 90 | 115 | 8.05e-08 |
| GO:BP | GO:0010033 | response to organic substance | 1002 | 1736 | 8.14e-08 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 349 | 550 | 8.55e-08 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 396 | 633 | 9.09e-08 |
| GO:BP | GO:0006816 | calcium ion transport | 181 | 262 | 9.74e-08 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 164 | 234 | 1.07e-07 |
| GO:BP | GO:0006811 | monoatomic ion transport | 444 | 719 | 1.11e-07 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 1364 | 2415 | 1.12e-07 |
| GO:BP | GO:0003208 | cardiac ventricle morphogenesis | 54 | 62 | 1.14e-07 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 190 | 278 | 1.48e-07 |
| GO:BP | GO:0016331 | morphogenesis of embryonic epithelium | 94 | 122 | 1.48e-07 |
| GO:BP | GO:0043010 | camera-type eye development | 161 | 230 | 1.68e-07 |
| GO:BP | GO:0050877 | nervous system process | 376 | 600 | 1.71e-07 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 150 | 212 | 1.77e-07 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 516 | 850 | 1.79e-07 |
| GO:BP | GO:0006915 | apoptotic process | 805 | 1377 | 1.93e-07 |
| GO:BP | GO:0060348 | bone development | 109 | 146 | 2.00e-07 |
| GO:BP | GO:0035148 | tube formation | 95 | 124 | 2.01e-07 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 135 | 188 | 2.28e-07 |
| GO:BP | GO:0061035 | regulation of cartilage development | 45 | 50 | 2.39e-07 |
| GO:BP | GO:0071417 | cellular response to organonitrogen compound | 285 | 442 | 2.61e-07 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 202 | 300 | 3.06e-07 |
| GO:BP | GO:0032940 | secretion by cell | 347 | 551 | 3.08e-07 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 276 | 427 | 3.18e-07 |
| GO:BP | GO:1901698 | response to nitrogen compound | 451 | 736 | 3.30e-07 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 148 | 210 | 3.30e-07 |
| GO:BP | GO:0051234 | establishment of localization | 1795 | 3240 | 3.40e-07 |
| GO:BP | GO:0008219 | cell death | 828 | 1423 | 3.56e-07 |
| GO:BP | GO:0031399 | regulation of protein modification process | 548 | 911 | 3.60e-07 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 367 | 587 | 3.67e-07 |
| GO:BP | GO:0033002 | muscle cell proliferation | 112 | 152 | 4.19e-07 |
| GO:BP | GO:0012501 | programmed cell death | 825 | 1419 | 4.62e-07 |
| GO:BP | GO:0023061 | signal release | 210 | 315 | 5.25e-07 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 647 | 1093 | 5.44e-07 |
| GO:BP | GO:0009605 | response to external stimulus | 900 | 1559 | 5.67e-07 |
| GO:BP | GO:0061138 | morphogenesis of a branching epithelium | 101 | 135 | 5.72e-07 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 90 | 118 | 7.05e-07 |
| GO:BP | GO:0071772 | response to BMP | 90 | 118 | 7.05e-07 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 147 | 210 | 7.29e-07 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 1313 | 2333 | 7.32e-07 |
| GO:BP | GO:0001763 | morphogenesis of a branching structure | 107 | 145 | 7.56e-07 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 95 | 126 | 7.75e-07 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 328 | 521 | 7.90e-07 |
| GO:BP | GO:0061326 | renal tubule development | 61 | 74 | 8.38e-07 |
| GO:BP | GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 191 | 284 | 8.38e-07 |
| GO:BP | GO:0051046 | regulation of secretion | 256 | 396 | 1.02e-06 |
| GO:BP | GO:0072175 | epithelial tube formation | 89 | 117 | 1.03e-06 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 54 | 64 | 1.10e-06 |
| GO:BP | GO:0001503 | ossification | 201 | 302 | 1.24e-06 |
| GO:BP | GO:1901342 | regulation of vasculature development | 139 | 198 | 1.29e-06 |
| GO:BP | GO:0045664 | regulation of neuron differentiation | 97 | 130 | 1.33e-06 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 68 | 85 | 1.35e-06 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 151 | 218 | 1.36e-06 |
| GO:BP | GO:0007162 | negative regulation of cell adhesion | 131 | 185 | 1.38e-06 |
| GO:BP | GO:0060038 | cardiac muscle cell proliferation | 39 | 43 | 1.41e-06 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 146 | 210 | 1.55e-06 |
| GO:BP | GO:0007610 | behavior | 269 | 420 | 1.57e-06 |
| GO:BP | GO:0048562 | embryonic organ morphogenesis | 129 | 182 | 1.58e-06 |
| GO:BP | GO:0042472 | inner ear morphogenesis | 51 | 60 | 1.58e-06 |
| GO:BP | GO:0009611 | response to wounding | 247 | 382 | 1.68e-06 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 98 | 132 | 1.68e-06 |
| GO:BP | GO:0007369 | gastrulation | 103 | 140 | 1.69e-06 |
| GO:BP | GO:0007389 | pattern specification process | 191 | 286 | 1.78e-06 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 150 | 217 | 1.79e-06 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 130 | 184 | 1.86e-06 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 480 | 796 | 1.86e-06 |
| GO:BP | GO:0055085 | transmembrane transport | 549 | 921 | 1.86e-06 |
| GO:BP | GO:0030048 | actin filament-based movement | 82 | 107 | 1.90e-06 |
| GO:BP | GO:0006935 | chemotaxis | 166 | 244 | 1.95e-06 |
| GO:BP | GO:0042330 | taxis | 166 | 244 | 1.95e-06 |
| GO:BP | GO:0003281 | ventricular septum development | 57 | 69 | 1.95e-06 |
| GO:BP | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | 61 | 75 | 2.05e-06 |
| GO:BP | GO:0030510 | regulation of BMP signaling pathway | 63 | 78 | 2.05e-06 |
| GO:BP | GO:0045321 | leukocyte activation | 331 | 530 | 2.06e-06 |
| GO:BP | GO:0046777 | protein autophosphorylation | 104 | 142 | 2.07e-06 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 340 | 546 | 2.08e-06 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 1069 | 1883 | 2.08e-06 |
| GO:BP | GO:0003229 | ventricular cardiac muscle tissue development | 48 | 56 | 2.17e-06 |
| GO:BP | GO:0007015 | actin filament organization | 232 | 357 | 2.19e-06 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 251 | 390 | 2.24e-06 |
| GO:BP | GO:0006812 | monoatomic cation transport | 378 | 614 | 2.31e-06 |
| GO:BP | GO:2000027 | regulation of animal organ morphogenesis | 50 | 59 | 2.45e-06 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 74 | 95 | 2.46e-06 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 484 | 805 | 2.62e-06 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 265 | 415 | 2.69e-06 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 302 | 480 | 2.78e-06 |
| GO:BP | GO:0007517 | muscle organ development | 179 | 267 | 2.87e-06 |
| GO:BP | GO:0048839 | inner ear development | 98 | 133 | 2.92e-06 |
| GO:BP | GO:0060993 | kidney morphogenesis | 56 | 68 | 2.99e-06 |
| GO:BP | GO:0048645 | animal organ formation | 45 | 52 | 2.99e-06 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 542 | 911 | 3.04e-06 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 145 | 210 | 3.05e-06 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 483 | 804 | 3.05e-06 |
| GO:BP | GO:0014033 | neural crest cell differentiation | 62 | 77 | 3.06e-06 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 433 | 714 | 3.17e-06 |
| GO:BP | GO:0051050 | positive regulation of transport | 353 | 571 | 3.29e-06 |
| GO:BP | GO:0043583 | ear development | 109 | 151 | 3.37e-06 |
| GO:BP | GO:0055008 | cardiac muscle tissue morphogenesis | 47 | 55 | 3.45e-06 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 310 | 495 | 3.46e-06 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 1078 | 1904 | 3.53e-06 |
| GO:BP | GO:0001704 | formation of primary germ layer | 73 | 94 | 3.59e-06 |
| GO:BP | GO:0042592 | homeostatic process | 665 | 1137 | 3.62e-06 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 175 | 261 | 3.77e-06 |
| GO:BP | GO:0072028 | nephron morphogenesis | 49 | 58 | 3.81e-06 |
| GO:BP | GO:0061333 | renal tubule morphogenesis | 49 | 58 | 3.81e-06 |
| GO:BP | GO:0048844 | artery morphogenesis | 49 | 58 | 3.81e-06 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 135 | 194 | 3.94e-06 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 69 | 88 | 3.99e-06 |
| GO:BP | GO:0051924 | regulation of calcium ion transport | 110 | 153 | 4.04e-06 |
| GO:BP | GO:0060415 | muscle tissue morphogenesis | 51 | 61 | 4.12e-06 |
| GO:BP | GO:0061337 | cardiac conduction | 67 | 85 | 4.17e-06 |
| GO:BP | GO:0048644 | muscle organ morphogenesis | 53 | 64 | 4.37e-06 |
| GO:BP | GO:0072080 | nephron tubule development | 57 | 70 | 4.61e-06 |
| GO:BP | GO:0042391 | regulation of membrane potential | 187 | 282 | 4.61e-06 |
| GO:BP | GO:0030509 | BMP signaling pathway | 83 | 110 | 4.62e-06 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 2690 | 4988 | 5.01e-06 |
| GO:BP | GO:0022612 | gland morphogenesis | 72 | 93 | 5.24e-06 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 300 | 479 | 5.24e-06 |
| GO:BP | GO:0042060 | wound healing | 192 | 291 | 5.39e-06 |
| GO:BP | GO:0048167 | regulation of synaptic plasticity | 101 | 139 | 5.46e-06 |
| GO:BP | GO:0001952 | regulation of cell-matrix adhesion | 70 | 90 | 5.56e-06 |
| GO:BP | GO:0000165 | MAPK cascade | 314 | 504 | 5.56e-06 |
| GO:BP | GO:0022409 | positive regulation of cell-cell adhesion | 123 | 175 | 5.72e-06 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 68 | 87 | 5.90e-06 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 163 | 242 | 6.11e-06 |
| GO:BP | GO:0072009 | nephron epithelium development | 66 | 84 | 6.22e-06 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 371 | 606 | 6.31e-06 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 36 | 40 | 6.35e-06 |
| GO:BP | GO:0060421 | positive regulation of heart growth | 27 | 28 | 6.52e-06 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 391 | 642 | 6.52e-06 |
| GO:BP | GO:0009987 | cellular process | 5561 | 10686 | 6.84e-06 |
| GO:BP | GO:0021915 | neural tube development | 97 | 133 | 6.88e-06 |
| GO:BP | GO:0090288 | negative regulation of cellular response to growth factor stimulus | 60 | 75 | 6.88e-06 |
| GO:BP | GO:0006941 | striated muscle contraction | 105 | 146 | 7.11e-06 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 80 | 106 | 7.20e-06 |
| GO:BP | GO:0031214 | biomineral tissue development | 80 | 106 | 7.20e-06 |
| GO:BP | GO:0018212 | peptidyl-tyrosine modification | 116 | 164 | 7.20e-06 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 272 | 431 | 7.29e-06 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 252 | 396 | 7.41e-06 |
| GO:BP | GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 67 | 86 | 8.72e-06 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 123 | 176 | 9.02e-06 |
| GO:BP | GO:0072088 | nephron epithelium morphogenesis | 47 | 56 | 9.52e-06 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 86 | 116 | 9.88e-06 |
| GO:BP | GO:0050807 | regulation of synapse organization | 121 | 173 | 1.04e-05 |
| GO:BP | GO:0002521 | leukocyte differentiation | 237 | 371 | 1.07e-05 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 130 | 188 | 1.07e-05 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 196 | 300 | 1.09e-05 |
| GO:BP | GO:1903530 | regulation of secretion by cell | 234 | 366 | 1.14e-05 |
| GO:BP | GO:0141091 | transforming growth factor beta receptor superfamily signaling pathway | 172 | 259 | 1.15e-05 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 483 | 811 | 1.16e-05 |
| GO:BP | GO:0035108 | limb morphogenesis | 82 | 110 | 1.22e-05 |
| GO:BP | GO:0035107 | appendage morphogenesis | 82 | 110 | 1.22e-05 |
| GO:BP | GO:0060420 | regulation of heart growth | 42 | 49 | 1.23e-05 |
| GO:BP | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 108 | 152 | 1.24e-05 |
| GO:BP | GO:0060045 | positive regulation of cardiac muscle cell proliferation | 22 | 22 | 1.24e-05 |
| GO:BP | GO:0003148 | outflow tract septum morphogenesis | 22 | 22 | 1.24e-05 |
| GO:BP | GO:0009612 | response to mechanical stimulus | 108 | 152 | 1.24e-05 |
| GO:BP | GO:0060840 | artery development | 66 | 85 | 1.28e-05 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 412 | 683 | 1.28e-05 |
| GO:BP | GO:0018108 | peptidyl-tyrosine phosphorylation | 114 | 162 | 1.32e-05 |
| GO:BP | GO:0060043 | regulation of cardiac muscle cell proliferation | 29 | 31 | 1.32e-05 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 64 | 82 | 1.35e-05 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 1222 | 2186 | 1.35e-05 |
| GO:BP | GO:1903037 | regulation of leukocyte cell-cell adhesion | 135 | 197 | 1.39e-05 |
| GO:BP | GO:0003179 | heart valve morphogenesis | 46 | 55 | 1.49e-05 |
| GO:BP | GO:0060395 | SMAD protein signal transduction | 60 | 76 | 1.50e-05 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 223 | 348 | 1.58e-05 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 118 | 169 | 1.58e-05 |
| GO:BP | GO:0048588 | developmental cell growth | 124 | 179 | 1.61e-05 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 146 | 216 | 1.71e-05 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 81 | 109 | 1.72e-05 |
| GO:BP | GO:0007498 | mesoderm development | 65 | 84 | 1.90e-05 |
| GO:BP | GO:0032409 | regulation of transporter activity | 105 | 148 | 1.92e-05 |
| GO:BP | GO:0097553 | calcium ion transmembrane import into cytosol | 89 | 122 | 1.92e-05 |
| GO:BP | GO:0050806 | positive regulation of synaptic transmission | 79 | 106 | 1.92e-05 |
| GO:BP | GO:0110110 | positive regulation of animal organ morphogenesis | 25 | 26 | 2.07e-05 |
| GO:BP | GO:0043270 | positive regulation of monoatomic ion transport | 92 | 127 | 2.10e-05 |
| GO:BP | GO:0032330 | regulation of chondrocyte differentiation | 31 | 34 | 2.12e-05 |
| GO:BP | GO:0003161 | cardiac conduction system development | 31 | 34 | 2.12e-05 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 165 | 249 | 2.25e-05 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 240 | 379 | 2.29e-05 |
| GO:BP | GO:1905314 | semi-lunar valve development | 36 | 41 | 2.29e-05 |
| GO:BP | GO:0035924 | cellular response to vascular endothelial growth factor stimulus | 45 | 54 | 2.33e-05 |
| GO:BP | GO:0072078 | nephron tubule morphogenesis | 45 | 54 | 2.33e-05 |
| GO:BP | GO:0014910 | regulation of smooth muscle cell migration | 45 | 54 | 2.33e-05 |
| GO:BP | GO:0010817 | regulation of hormone levels | 205 | 318 | 2.33e-05 |
| GO:BP | GO:0048660 | regulation of smooth muscle cell proliferation | 73 | 97 | 2.61e-05 |
| GO:BP | GO:0048732 | gland development | 199 | 308 | 2.64e-05 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 137 | 202 | 2.70e-05 |
| GO:BP | GO:0046879 | hormone secretion | 137 | 202 | 2.70e-05 |
| GO:BP | GO:0048771 | tissue remodeling | 83 | 113 | 2.73e-05 |
| GO:BP | GO:0048754 | branching morphogenesis of an epithelial tube | 83 | 113 | 2.73e-05 |
| GO:BP | GO:0055010 | ventricular cardiac muscle tissue morphogenesis | 38 | 44 | 2.74e-05 |
| GO:BP | GO:0022898 | regulation of transmembrane transporter activity | 99 | 139 | 2.85e-05 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 186 | 286 | 3.07e-05 |
| GO:BP | GO:0048041 | focal adhesion assembly | 60 | 77 | 3.18e-05 |
| GO:BP | GO:0050865 | regulation of cell activation | 221 | 347 | 3.25e-05 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 117 | 169 | 3.29e-05 |
| GO:BP | GO:0001649 | osteoblast differentiation | 120 | 174 | 3.29e-05 |
| GO:BP | GO:0015833 | peptide transport | 123 | 179 | 3.29e-05 |
| GO:BP | GO:0031644 | regulation of nervous system process | 58 | 74 | 3.33e-05 |
| GO:BP | GO:0048864 | stem cell development | 58 | 74 | 3.33e-05 |
| GO:BP | GO:0042471 | ear morphogenesis | 58 | 74 | 3.33e-05 |
| GO:BP | GO:0046649 | lymphocyte activation | 276 | 444 | 3.36e-05 |
| GO:BP | GO:0048592 | eye morphogenesis | 79 | 107 | 3.41e-05 |
| GO:BP | GO:0055023 | positive regulation of cardiac muscle tissue growth | 24 | 25 | 3.65e-05 |
| GO:BP | GO:0003170 | heart valve development | 52 | 65 | 3.77e-05 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 115 | 166 | 3.77e-05 |
| GO:BP | GO:0035296 | regulation of tube diameter | 75 | 101 | 4.29e-05 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 125 | 183 | 4.29e-05 |
| GO:BP | GO:0035150 | regulation of tube size | 75 | 101 | 4.29e-05 |
| GO:BP | GO:0097746 | blood vessel diameter maintenance | 75 | 101 | 4.29e-05 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 96 | 135 | 4.45e-05 |
| GO:BP | GO:0006813 | potassium ion transport | 96 | 135 | 4.45e-05 |
| GO:BP | GO:0003002 | regionalization | 169 | 258 | 4.65e-05 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 91 | 127 | 4.83e-05 |
| GO:BP | GO:0050878 | regulation of body fluid levels | 152 | 229 | 4.83e-05 |
| GO:BP | GO:0009914 | hormone transport | 139 | 207 | 5.02e-05 |
| GO:BP | GO:0072171 | mesonephric tubule morphogenesis | 39 | 46 | 5.02e-05 |
| GO:BP | GO:0048878 | chemical homeostasis | 401 | 670 | 5.04e-05 |
| GO:BP | GO:0048638 | regulation of developmental growth | 160 | 243 | 5.41e-05 |
| GO:BP | GO:0043491 | phosphatidylinositol 3-kinase/protein kinase B signal transduction | 143 | 214 | 5.43e-05 |
| GO:BP | GO:0071407 | cellular response to organic cyclic compound | 238 | 379 | 5.89e-05 |
| GO:BP | GO:0072091 | regulation of stem cell proliferation | 49 | 61 | 5.89e-05 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 1040 | 1856 | 6.19e-05 |
| GO:BP | GO:0030097 | hemopoiesis | 375 | 624 | 6.22e-05 |
| GO:BP | GO:0060078 | regulation of postsynaptic membrane potential | 60 | 78 | 6.44e-05 |
| GO:BP | GO:0031032 | actomyosin structure organization | 125 | 184 | 6.44e-05 |
| GO:BP | GO:0003272 | endocardial cushion formation | 23 | 24 | 6.53e-05 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 173 | 266 | 6.68e-05 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 442 | 746 | 6.76e-05 |
| GO:BP | GO:0048736 | appendage development | 96 | 136 | 7.25e-05 |
| GO:BP | GO:0060173 | limb development | 96 | 136 | 7.25e-05 |
| GO:BP | GO:0014812 | muscle cell migration | 56 | 72 | 7.43e-05 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 1031 | 1841 | 7.75e-05 |
| GO:BP | GO:0003184 | pulmonary valve morphogenesis | 19 | 19 | 7.75e-05 |
| GO:BP | GO:0014032 | neural crest cell development | 54 | 69 | 7.88e-05 |
| GO:BP | GO:0006939 | smooth muscle contraction | 54 | 69 | 7.88e-05 |
| GO:BP | GO:0060675 | ureteric bud morphogenesis | 38 | 45 | 8.03e-05 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 83 | 115 | 8.09e-05 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 83 | 115 | 8.09e-05 |
| GO:BP | GO:0042110 | T cell activation | 195 | 305 | 8.59e-05 |
| GO:BP | GO:0010718 | positive regulation of epithelial to mesenchymal transition | 40 | 48 | 8.60e-05 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 73 | 99 | 8.60e-05 |
| GO:BP | GO:0071805 | potassium ion transmembrane transport | 86 | 120 | 8.61e-05 |
| GO:BP | GO:0055001 | muscle cell development | 112 | 163 | 8.88e-05 |
| GO:BP | GO:0001707 | mesoderm formation | 42 | 51 | 8.89e-05 |
| GO:BP | GO:0014909 | smooth muscle cell migration | 48 | 60 | 8.89e-05 |
| GO:BP | GO:0072132 | mesenchyme morphogenesis | 42 | 51 | 8.89e-05 |
| GO:BP | GO:0045666 | positive regulation of neuron differentiation | 46 | 57 | 9.01e-05 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 612 | 1060 | 9.15e-05 |
| GO:BP | GO:0070372 | regulation of ERK1 and ERK2 cascade | 119 | 175 | 9.88e-05 |
| GO:BP | GO:0006949 | syncytium formation | 33 | 38 | 1.00e-04 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 101 | 145 | 1.01e-04 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 521 | 893 | 1.02e-04 |
| GO:BP | GO:0008544 | epidermis development | 139 | 209 | 1.04e-04 |
| GO:BP | GO:0008150 | biological_process | 5821 | 11244 | 1.13e-04 |
| GO:BP | GO:0061036 | positive regulation of cartilage development | 22 | 23 | 1.15e-04 |
| GO:BP | GO:0002063 | chondrocyte development | 22 | 23 | 1.15e-04 |
| GO:BP | GO:0070371 | ERK1 and ERK2 cascade | 130 | 194 | 1.16e-04 |
| GO:BP | GO:0060541 | respiratory system development | 111 | 162 | 1.17e-04 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 85 | 119 | 1.18e-04 |
| GO:BP | GO:0045667 | regulation of osteoblast differentiation | 72 | 98 | 1.21e-04 |
| GO:BP | GO:0030010 | establishment of cell polarity | 88 | 124 | 1.24e-04 |
| GO:BP | GO:0055021 | regulation of cardiac muscle tissue growth | 37 | 44 | 1.27e-04 |
| GO:BP | GO:0060976 | coronary vasculature development | 37 | 44 | 1.27e-04 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 595 | 1031 | 1.31e-04 |
| GO:BP | GO:0032412 | regulation of monoatomic ion transmembrane transporter activity | 94 | 134 | 1.31e-04 |
| GO:BP | GO:0046883 | regulation of hormone secretion | 112 | 164 | 1.33e-04 |
| GO:BP | GO:0014855 | striated muscle cell proliferation | 47 | 59 | 1.34e-04 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 135 | 203 | 1.39e-04 |
| GO:BP | GO:0048332 | mesoderm morphogenesis | 43 | 53 | 1.39e-04 |
| GO:BP | GO:0051249 | regulation of lymphocyte activation | 179 | 279 | 1.47e-04 |
| GO:BP | GO:0003158 | endothelium development | 73 | 100 | 1.50e-04 |
| GO:BP | GO:0002790 | peptide secretion | 113 | 166 | 1.52e-04 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 95 | 136 | 1.54e-04 |
| GO:BP | GO:1903039 | positive regulation of leukocyte cell-cell adhesion | 101 | 146 | 1.57e-04 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 1001 | 1791 | 1.61e-04 |
| GO:BP | GO:0003176 | aortic valve development | 32 | 37 | 1.63e-04 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 76 | 105 | 1.63e-04 |
| GO:BP | GO:0050708 | regulation of protein secretion | 120 | 178 | 1.64e-04 |
| GO:BP | GO:0021537 | telencephalon development | 130 | 195 | 1.66e-04 |
| GO:BP | GO:0019722 | calcium-mediated signaling | 87 | 123 | 1.67e-04 |
| GO:BP | GO:0030513 | positive regulation of BMP signaling pathway | 27 | 30 | 1.72e-04 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 102 | 148 | 1.81e-04 |
| GO:BP | GO:0002694 | regulation of leukocyte activation | 198 | 313 | 1.85e-04 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 88 | 125 | 2.00e-04 |
| GO:BP | GO:0048663 | neuron fate commitment | 24 | 26 | 2.04e-04 |
| GO:BP | GO:0010092 | specification of animal organ identity | 24 | 26 | 2.04e-04 |
| GO:BP | GO:0003177 | pulmonary valve development | 21 | 22 | 2.04e-04 |
| GO:BP | GO:0050870 | positive regulation of T cell activation | 94 | 135 | 2.07e-04 |
| GO:BP | GO:0051893 | regulation of focal adhesion assembly | 44 | 55 | 2.09e-04 |
| GO:BP | GO:0003197 | endocardial cushion development | 38 | 46 | 2.09e-04 |
| GO:BP | GO:0090109 | regulation of cell-substrate junction assembly | 44 | 55 | 2.09e-04 |
| GO:BP | GO:0060390 | regulation of SMAD protein signal transduction | 40 | 49 | 2.13e-04 |
| GO:BP | GO:1902105 | regulation of leukocyte differentiation | 129 | 194 | 2.14e-04 |
| GO:BP | GO:0071692 | protein localization to extracellular region | 165 | 256 | 2.18e-04 |
| GO:BP | GO:2000648 | positive regulation of stem cell proliferation | 29 | 33 | 2.21e-04 |
| GO:BP | GO:0003180 | aortic valve morphogenesis | 29 | 33 | 2.21e-04 |
| GO:BP | GO:0061572 | actin filament bundle organization | 92 | 132 | 2.37e-04 |
| GO:BP | GO:0002573 | myeloid leukocyte differentiation | 104 | 152 | 2.37e-04 |
| GO:BP | GO:0070482 | response to oxygen levels | 170 | 265 | 2.39e-04 |
| GO:BP | GO:0048565 | digestive tract development | 65 | 88 | 2.39e-04 |
| GO:BP | GO:0001935 | endothelial cell proliferation | 81 | 114 | 2.51e-04 |
| GO:BP | GO:0036211 | protein modification process | 1263 | 2291 | 2.54e-04 |
| GO:BP | GO:0048659 | smooth muscle cell proliferation | 73 | 101 | 2.55e-04 |
| GO:BP | GO:0060445 | branching involved in salivary gland morphogenesis | 17 | 17 | 2.58e-04 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 84 | 119 | 2.60e-04 |
| GO:BP | GO:0140253 | cell-cell fusion | 31 | 36 | 2.61e-04 |
| GO:BP | GO:0038084 | vascular endothelial growth factor signaling pathway | 31 | 36 | 2.61e-04 |
| GO:BP | GO:0000768 | syncytium formation by plasma membrane fusion | 31 | 36 | 2.61e-04 |
| GO:BP | GO:0010463 | mesenchymal cell proliferation | 31 | 36 | 2.61e-04 |
| GO:BP | GO:0045446 | endothelial cell differentiation | 63 | 85 | 2.66e-04 |
| GO:BP | GO:0007492 | endoderm development | 49 | 63 | 2.68e-04 |
| GO:BP | GO:0007043 | cell-cell junction assembly | 76 | 106 | 2.70e-04 |
| GO:BP | GO:1990138 | neuron projection extension | 93 | 134 | 2.72e-04 |
| GO:BP | GO:0150116 | regulation of cell-substrate junction organization | 47 | 60 | 2.85e-04 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 47 | 60 | 2.85e-04 |
| GO:BP | GO:0002040 | sprouting angiogenesis | 71 | 98 | 2.87e-04 |
| GO:BP | GO:0001658 | branching involved in ureteric bud morphogenesis | 33 | 39 | 2.90e-04 |
| GO:BP | GO:0071774 | response to fibroblast growth factor | 61 | 82 | 2.96e-04 |
| GO:BP | GO:0051283 | negative regulation of sequestering of calcium ion | 66 | 90 | 2.96e-04 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 143 | 219 | 2.97e-04 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 177 | 278 | 2.97e-04 |
| GO:BP | GO:0006954 | inflammatory response | 260 | 425 | 3.08e-04 |
| GO:BP | GO:0032835 | glomerulus development | 43 | 54 | 3.11e-04 |
| GO:BP | GO:0086065 | cell communication involved in cardiac conduction | 43 | 54 | 3.11e-04 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 91 | 131 | 3.11e-04 |
| GO:BP | GO:1903169 | regulation of calcium ion transmembrane transport | 77 | 108 | 3.23e-04 |
| GO:BP | GO:0042476 | odontogenesis | 59 | 79 | 3.27e-04 |
| GO:BP | GO:0044344 | cellular response to fibroblast growth factor stimulus | 59 | 79 | 3.27e-04 |
| GO:BP | GO:0048593 | camera-type eye morphogenesis | 64 | 87 | 3.30e-04 |
| GO:BP | GO:0014020 | primary neural tube formation | 64 | 87 | 3.30e-04 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 64 | 87 | 3.30e-04 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 64 | 87 | 3.30e-04 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 167 | 261 | 3.35e-04 |
| GO:BP | GO:0050863 | regulation of T cell activation | 134 | 204 | 3.45e-04 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 263 | 431 | 3.51e-04 |
| GO:BP | GO:0002064 | epithelial cell development | 101 | 148 | 3.51e-04 |
| GO:BP | GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | 95 | 138 | 3.58e-04 |
| GO:BP | GO:0001841 | neural tube formation | 67 | 92 | 3.63e-04 |
| GO:BP | GO:0010721 | negative regulation of cell development | 121 | 182 | 3.74e-04 |
| GO:BP | GO:0060349 | bone morphogenesis | 55 | 73 | 4.03e-04 |
| GO:BP | GO:0051209 | release of sequestered calcium ion into cytosol | 65 | 89 | 4.13e-04 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 60 | 81 | 4.16e-04 |
| GO:BP | GO:0045860 | positive regulation of protein kinase activity | 122 | 184 | 4.16e-04 |
| GO:BP | GO:0046622 | positive regulation of organ growth | 30 | 35 | 4.16e-04 |
| GO:BP | GO:0030072 | peptide hormone secretion | 109 | 162 | 4.32e-04 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 140 | 215 | 4.32e-04 |
| GO:BP | GO:0045444 | fat cell differentiation | 119 | 179 | 4.33e-04 |
| GO:BP | GO:0098659 | inorganic cation import across plasma membrane | 53 | 70 | 4.40e-04 |
| GO:BP | GO:0016049 | cell growth | 235 | 382 | 4.40e-04 |
| GO:BP | GO:0099565 | chemical synaptic transmission, postsynaptic | 53 | 70 | 4.40e-04 |
| GO:BP | GO:0099587 | inorganic ion import across plasma membrane | 53 | 70 | 4.40e-04 |
| GO:BP | GO:0043616 | keratinocyte proliferation | 32 | 38 | 4.56e-04 |
| GO:BP | GO:0099173 | postsynapse organization | 100 | 147 | 4.61e-04 |
| GO:BP | GO:0050679 | positive regulation of epithelial cell proliferation | 97 | 142 | 4.69e-04 |
| GO:BP | GO:2000463 | positive regulation of excitatory postsynaptic potential | 25 | 28 | 4.70e-04 |
| GO:BP | GO:0098739 | import across plasma membrane | 85 | 122 | 4.70e-04 |
| GO:BP | GO:0030073 | insulin secretion | 94 | 137 | 4.72e-04 |
| GO:BP | GO:0098815 | modulation of excitatory postsynaptic potential | 34 | 41 | 4.78e-04 |
| GO:BP | GO:1905330 | regulation of morphogenesis of an epithelium | 34 | 41 | 4.78e-04 |
| GO:BP | GO:0048010 | vascular endothelial growth factor receptor signaling pathway | 40 | 50 | 4.78e-04 |
| GO:BP | GO:0034332 | adherens junction organization | 36 | 44 | 4.89e-04 |
| GO:BP | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | 66 | 91 | 5.02e-04 |
| GO:BP | GO:0034767 | positive regulation of monoatomic ion transmembrane transport | 66 | 91 | 5.02e-04 |
| GO:BP | GO:0009306 | protein secretion | 161 | 252 | 5.06e-04 |
| GO:BP | GO:0035592 | establishment of protein localization to extracellular region | 161 | 252 | 5.06e-04 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 49 | 64 | 5.24e-04 |
| GO:BP | GO:0016358 | dendrite development | 121 | 183 | 5.26e-04 |
| GO:BP | GO:0030324 | lung development | 98 | 144 | 5.28e-04 |
| GO:BP | GO:0055123 | digestive system development | 69 | 96 | 5.36e-04 |
| GO:BP | GO:0061614 | miRNA transcription | 47 | 61 | 5.69e-04 |
| GO:BP | GO:0042063 | gliogenesis | 155 | 242 | 5.80e-04 |
| GO:BP | GO:0051251 | positive regulation of lymphocyte activation | 122 | 185 | 5.82e-04 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 159 | 249 | 5.83e-04 |
| GO:BP | GO:0042886 | amide transport | 133 | 204 | 6.02e-04 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 378 | 642 | 6.10e-04 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 654 | 1152 | 6.14e-04 |
| GO:BP | GO:1904018 | positive regulation of vasculature development | 81 | 116 | 6.18e-04 |
| GO:BP | GO:0007416 | synapse assembly | 93 | 136 | 6.18e-04 |
| GO:BP | GO:0051051 | negative regulation of transport | 180 | 286 | 6.18e-04 |
| GO:BP | GO:0051235 | maintenance of location | 160 | 251 | 6.25e-04 |
| GO:BP | GO:0032526 | response to retinoic acid | 57 | 77 | 6.51e-04 |
| GO:BP | GO:0001666 | response to hypoxia | 149 | 232 | 6.58e-04 |
| GO:BP | GO:0006898 | receptor-mediated endocytosis | 120 | 182 | 6.71e-04 |
| GO:BP | GO:0030323 | respiratory tube development | 100 | 148 | 6.76e-04 |
| GO:BP | GO:0001708 | cell fate specification | 41 | 52 | 6.91e-04 |
| GO:BP | GO:0001570 | vasculogenesis | 50 | 66 | 6.93e-04 |
| GO:BP | GO:0050796 | regulation of insulin secretion | 79 | 113 | 7.12e-04 |
| GO:BP | GO:0050920 | regulation of chemotaxis | 91 | 133 | 7.15e-04 |
| GO:BP | GO:0030100 | regulation of endocytosis | 128 | 196 | 7.32e-04 |
| GO:BP | GO:0071526 | semaphorin-plexin signaling pathway | 35 | 43 | 7.54e-04 |
| GO:BP | GO:0036293 | response to decreased oxygen levels | 155 | 243 | 7.73e-04 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 247 | 406 | 7.73e-04 |
| GO:BP | GO:0007215 | glutamate receptor signaling pathway | 24 | 27 | 7.79e-04 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 262 | 433 | 7.82e-04 |
| GO:BP | GO:0043542 | endothelial cell migration | 115 | 174 | 8.14e-04 |
| GO:BP | GO:0006836 | neurotransmitter transport | 92 | 135 | 8.15e-04 |
| GO:BP | GO:0002761 | regulation of myeloid leukocyte differentiation | 58 | 79 | 8.16e-04 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 148 | 231 | 8.16e-04 |
| GO:BP | GO:0009410 | response to xenobiotic stimulus | 172 | 273 | 8.17e-04 |
| GO:BP | GO:0034764 | positive regulation of transmembrane transport | 89 | 130 | 8.22e-04 |
| GO:BP | GO:0051897 | positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 86 | 125 | 8.28e-04 |
| GO:BP | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 66 | 92 | 8.36e-04 |
| GO:BP | GO:0051282 | regulation of sequestering of calcium ion | 66 | 92 | 8.36e-04 |
| GO:BP | GO:0001759 | organ induction | 15 | 15 | 8.36e-04 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 126 | 193 | 8.37e-04 |
| GO:BP | GO:0010001 | glial cell differentiation | 119 | 181 | 8.45e-04 |
| GO:BP | GO:0051896 | regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 119 | 181 | 8.45e-04 |
| GO:BP | GO:0060606 | tube closure | 61 | 84 | 8.84e-04 |
| GO:BP | GO:0035270 | endocrine system development | 61 | 84 | 8.84e-04 |
| GO:BP | GO:0010876 | lipid localization | 186 | 298 | 8.84e-04 |
| GO:BP | GO:0043434 | response to peptide hormone | 195 | 314 | 8.99e-04 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 329 | 555 | 9.01e-04 |
| GO:BP | GO:0051963 | regulation of synapse assembly | 56 | 76 | 9.06e-04 |
| GO:BP | GO:1901890 | positive regulation of cell junction assembly | 56 | 76 | 9.06e-04 |
| GO:BP | GO:0060914 | heart formation | 26 | 30 | 9.27e-04 |
| GO:BP | GO:0002791 | regulation of peptide secretion | 90 | 132 | 9.27e-04 |
| GO:BP | GO:0042475 | odontogenesis of dentin-containing tooth | 42 | 54 | 9.55e-04 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 49 | 65 | 9.86e-04 |
| GO:BP | GO:0060079 | excitatory postsynaptic potential | 49 | 65 | 9.86e-04 |
| GO:BP | GO:0033674 | positive regulation of kinase activity | 128 | 197 | 9.95e-04 |
| GO:BP | GO:0051047 | positive regulation of secretion | 132 | 204 | 1.02e-03 |
| GO:BP | GO:0030031 | cell projection assembly | 269 | 447 | 1.03e-03 |
| GO:BP | GO:1901652 | response to peptide | 225 | 368 | 1.03e-03 |
| GO:BP | GO:0032924 | activin receptor signaling pathway | 28 | 33 | 1.04e-03 |
| GO:BP | GO:0003156 | regulation of animal organ formation | 18 | 19 | 1.06e-03 |
| GO:BP | GO:0001764 | neuron migration | 88 | 129 | 1.07e-03 |
| GO:BP | GO:0001755 | neural crest cell migration | 38 | 48 | 1.07e-03 |
| GO:BP | GO:0051058 | negative regulation of small GTPase mediated signal transduction | 38 | 48 | 1.07e-03 |
| GO:BP | GO:0140115 | export across plasma membrane | 38 | 48 | 1.07e-03 |
| GO:BP | GO:0051961 | negative regulation of nervous system development | 76 | 109 | 1.08e-03 |
| GO:BP | GO:0070167 | regulation of biomineral tissue development | 47 | 62 | 1.08e-03 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 101 | 151 | 1.08e-03 |
| GO:BP | GO:0045766 | positive regulation of angiogenesis | 79 | 114 | 1.08e-03 |
| GO:BP | GO:0003203 | endocardial cushion morphogenesis | 30 | 36 | 1.10e-03 |
| GO:BP | GO:0002376 | immune system process | 789 | 1410 | 1.12e-03 |
| GO:BP | GO:0046632 | alpha-beta T cell differentiation | 52 | 70 | 1.13e-03 |
| GO:BP | GO:0046620 | regulation of organ growth | 52 | 70 | 1.13e-03 |
| GO:BP | GO:0040008 | regulation of growth | 277 | 462 | 1.13e-03 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 228 | 374 | 1.17e-03 |
| GO:BP | GO:0001936 | regulation of endothelial cell proliferation | 68 | 96 | 1.18e-03 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 243 | 401 | 1.19e-03 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 263 | 437 | 1.19e-03 |
| GO:BP | GO:0051641 | cellular localization | 1428 | 2623 | 1.19e-03 |
| GO:BP | GO:0007611 | learning or memory | 123 | 189 | 1.20e-03 |
| GO:BP | GO:0001843 | neural tube closure | 60 | 83 | 1.20e-03 |
| GO:BP | GO:0002027 | regulation of heart rate | 60 | 83 | 1.20e-03 |
| GO:BP | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 77 | 111 | 1.24e-03 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 363 | 619 | 1.24e-03 |
| GO:BP | GO:2000181 | negative regulation of blood vessel morphogenesis | 50 | 67 | 1.25e-03 |
| GO:BP | GO:0051588 | regulation of neurotransmitter transport | 50 | 67 | 1.25e-03 |
| GO:BP | GO:1901343 | negative regulation of vasculature development | 50 | 67 | 1.25e-03 |
| GO:BP | GO:0014896 | muscle hypertrophy | 55 | 75 | 1.25e-03 |
| GO:BP | GO:0050867 | positive regulation of cell activation | 131 | 203 | 1.25e-03 |
| GO:BP | GO:0045927 | positive regulation of growth | 120 | 184 | 1.26e-03 |
| GO:BP | GO:0046928 | regulation of neurotransmitter secretion | 43 | 56 | 1.27e-03 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 53 | 72 | 1.42e-03 |
| GO:BP | GO:0014902 | myotube differentiation | 61 | 85 | 1.47e-03 |
| GO:BP | GO:1902414 | protein localization to cell junction | 61 | 85 | 1.47e-03 |
| GO:BP | GO:0060563 | neuroepithelial cell differentiation | 25 | 29 | 1.48e-03 |
| GO:BP | GO:0007520 | myoblast fusion | 25 | 29 | 1.48e-03 |
| GO:BP | GO:0090497 | mesenchymal cell migration | 39 | 50 | 1.49e-03 |
| GO:BP | GO:0023019 | signal transduction involved in regulation of gene expression | 14 | 14 | 1.50e-03 |
| GO:BP | GO:0009991 | response to extracellular stimulus | 229 | 377 | 1.51e-03 |
| GO:BP | GO:0090276 | regulation of peptide hormone secretion | 88 | 130 | 1.57e-03 |
| GO:BP | GO:0046888 | negative regulation of hormone secretion | 37 | 47 | 1.58e-03 |
| GO:BP | GO:0031667 | response to nutrient levels | 216 | 354 | 1.61e-03 |
| GO:BP | GO:1905207 | regulation of cardiocyte differentiation | 20 | 22 | 1.61e-03 |
| GO:BP | GO:0048333 | mesodermal cell differentiation | 20 | 22 | 1.61e-03 |
| GO:BP | GO:1903779 | regulation of cardiac conduction | 20 | 22 | 1.61e-03 |
| GO:BP | GO:1902284 | neuron projection extension involved in neuron projection guidance | 27 | 32 | 1.63e-03 |
| GO:BP | GO:0048846 | axon extension involved in axon guidance | 27 | 32 | 1.63e-03 |
| GO:BP | GO:0086002 | cardiac muscle cell action potential involved in contraction | 35 | 44 | 1.66e-03 |
| GO:BP | GO:0002696 | positive regulation of leukocyte activation | 127 | 197 | 1.67e-03 |
| GO:BP | GO:0032414 | positive regulation of ion transmembrane transporter activity | 44 | 58 | 1.70e-03 |
| GO:BP | GO:0010812 | negative regulation of cell-substrate adhesion | 33 | 41 | 1.71e-03 |
| GO:BP | GO:0006814 | sodium ion transport | 92 | 137 | 1.71e-03 |
| GO:BP | GO:0048168 | regulation of neuronal synaptic plasticity | 31 | 38 | 1.72e-03 |
| GO:BP | GO:0048013 | ephrin receptor signaling pathway | 31 | 38 | 1.72e-03 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 120 | 185 | 1.72e-03 |
| GO:BP | GO:0007613 | memory | 62 | 87 | 1.74e-03 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 54 | 74 | 1.74e-03 |
| GO:BP | GO:0016525 | negative regulation of angiogenesis | 49 | 66 | 1.76e-03 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 106 | 161 | 1.78e-03 |
| GO:BP | GO:0001816 | cytokine production | 264 | 441 | 1.82e-03 |
| GO:BP | GO:0031099 | regeneration | 96 | 144 | 1.83e-03 |
| GO:BP | GO:0050890 | cognition | 136 | 213 | 1.83e-03 |
| GO:BP | GO:0007009 | plasma membrane organization | 74 | 107 | 1.89e-03 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 161 | 257 | 1.91e-03 |
| GO:BP | GO:0043535 | regulation of blood vessel endothelial cell migration | 52 | 71 | 1.97e-03 |
| GO:BP | GO:0090087 | regulation of peptide transport | 90 | 134 | 1.97e-03 |
| GO:BP | GO:0072089 | stem cell proliferation | 60 | 84 | 1.99e-03 |
| GO:BP | GO:0043500 | muscle adaptation | 63 | 89 | 2.08e-03 |
| GO:BP | GO:0010595 | positive regulation of endothelial cell migration | 63 | 89 | 2.08e-03 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 884 | 1595 | 2.08e-03 |
| GO:BP | GO:0006470 | protein dephosphorylation | 115 | 177 | 2.11e-03 |
| GO:BP | GO:0006869 | lipid transport | 163 | 261 | 2.16e-03 |
| GO:BP | GO:0007266 | Rho protein signal transduction | 75 | 109 | 2.18e-03 |
| GO:BP | GO:0045685 | regulation of glial cell differentiation | 38 | 49 | 2.18e-03 |
| GO:BP | GO:1903053 | regulation of extracellular matrix organization | 38 | 49 | 2.18e-03 |
| GO:BP | GO:0099175 | regulation of postsynapse organization | 58 | 81 | 2.28e-03 |
| GO:BP | GO:1904064 | positive regulation of cation transmembrane transport | 58 | 81 | 2.28e-03 |
| GO:BP | GO:0060350 | endochondral bone morphogenesis | 36 | 46 | 2.34e-03 |
| GO:BP | GO:0097120 | receptor localization to synapse | 36 | 46 | 2.34e-03 |
| GO:BP | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway | 24 | 28 | 2.38e-03 |
| GO:BP | GO:0001817 | regulation of cytokine production | 261 | 437 | 2.39e-03 |
| GO:BP | GO:0009952 | anterior/posterior pattern specification | 82 | 121 | 2.41e-03 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 53 | 73 | 2.43e-03 |
| GO:BP | GO:0051208 | sequestering of calcium ion | 67 | 96 | 2.49e-03 |
| GO:BP | GO:0048813 | dendrite morphogenesis | 76 | 111 | 2.49e-03 |
| GO:BP | GO:0035023 | regulation of Rho protein signal transduction | 48 | 65 | 2.50e-03 |
| GO:BP | GO:0010644 | cell communication by electrical coupling | 26 | 31 | 2.56e-03 |
| GO:BP | GO:0071634 | regulation of transforming growth factor beta production | 26 | 31 | 2.56e-03 |
| GO:BP | GO:0050900 | leukocyte migration | 125 | 195 | 2.57e-03 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 110 | 169 | 2.57e-03 |
| GO:BP | GO:0050795 | regulation of behavior | 32 | 40 | 2.58e-03 |
| GO:BP | GO:0055006 | cardiac cell development | 56 | 78 | 2.58e-03 |
| GO:BP | GO:0071868 | cellular response to monoamine stimulus | 28 | 34 | 2.64e-03 |
| GO:BP | GO:0071870 | cellular response to catecholamine stimulus | 28 | 34 | 2.64e-03 |
| GO:BP | GO:0061037 | negative regulation of cartilage development | 19 | 21 | 2.68e-03 |
| GO:BP | GO:0014912 | negative regulation of smooth muscle cell migration | 19 | 21 | 2.68e-03 |
| GO:BP | GO:0046631 | alpha-beta T cell activation | 62 | 88 | 2.81e-03 |
| GO:BP | GO:0043547 | positive regulation of GTPase activity | 77 | 113 | 2.83e-03 |
| GO:BP | GO:0045598 | regulation of fat cell differentiation | 65 | 93 | 2.86e-03 |
| GO:BP | GO:0050680 | negative regulation of epithelial cell proliferation | 68 | 98 | 2.89e-03 |
| GO:BP | GO:0050768 | negative regulation of neurogenesis | 71 | 103 | 2.89e-03 |
| GO:BP | GO:0033993 | response to lipid | 335 | 573 | 2.91e-03 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 689 | 1231 | 2.92e-03 |
| GO:BP | GO:0019935 | cyclic-nucleotide-mediated signaling | 39 | 51 | 2.93e-03 |
| GO:BP | GO:1901653 | cellular response to peptide | 169 | 273 | 3.01e-03 |
| GO:BP | GO:0009725 | response to hormone | 355 | 610 | 3.06e-03 |
| GO:BP | GO:0140747 | regulation of ncRNA transcription | 57 | 80 | 3.10e-03 |
| GO:BP | GO:2001257 | regulation of cation channel activity | 49 | 67 | 3.10e-03 |
| GO:BP | GO:0060977 | coronary vasculature morphogenesis | 16 | 17 | 3.13e-03 |
| GO:BP | GO:0034505 | tooth mineralization | 16 | 17 | 3.13e-03 |
| GO:BP | GO:0003215 | cardiac right ventricle morphogenesis | 16 | 17 | 3.13e-03 |
| GO:BP | GO:0033135 | regulation of peptidyl-serine phosphorylation | 60 | 85 | 3.21e-03 |
| GO:BP | GO:0071396 | cellular response to lipid | 233 | 388 | 3.24e-03 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 166 | 268 | 3.24e-03 |
| GO:BP | GO:0001890 | placenta development | 75 | 110 | 3.25e-03 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 690 | 1234 | 3.30e-03 |
| GO:BP | GO:0032878 | regulation of establishment or maintenance of cell polarity | 21 | 24 | 3.33e-03 |
| GO:BP | GO:0003401 | axis elongation | 21 | 24 | 3.33e-03 |
| GO:BP | GO:1905332 | positive regulation of morphogenesis of an epithelium | 21 | 24 | 3.33e-03 |
| GO:BP | GO:0033036 | macromolecule localization | 1257 | 2309 | 3.34e-03 |
| GO:BP | GO:0019229 | regulation of vasoconstriction | 33 | 42 | 3.70e-03 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 497 | 874 | 3.70e-03 |
| GO:BP | GO:0010927 | cellular component assembly involved in morphogenesis | 73 | 107 | 3.78e-03 |
| GO:BP | GO:0010586 | miRNA metabolic process | 61 | 87 | 3.79e-03 |
| GO:BP | GO:0001508 | action potential | 70 | 102 | 3.81e-03 |
| GO:BP | GO:0120162 | positive regulation of cold-induced thermogenesis | 50 | 69 | 3.81e-03 |
| GO:BP | GO:0099643 | signal release from synapse | 70 | 102 | 3.81e-03 |
| GO:BP | GO:0007269 | neurotransmitter secretion | 70 | 102 | 3.81e-03 |
| GO:BP | GO:0060021 | roof of mouth development | 50 | 69 | 3.81e-03 |
| GO:BP | GO:0043534 | blood vessel endothelial cell migration | 64 | 92 | 3.81e-03 |
| GO:BP | GO:0021536 | diencephalon development | 31 | 39 | 3.87e-03 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 334 | 573 | 3.88e-03 |
| GO:BP | GO:0035249 | synaptic transmission, glutamatergic | 45 | 61 | 3.92e-03 |
| GO:BP | GO:0001656 | metanephros development | 45 | 61 | 3.92e-03 |
| GO:BP | GO:1903532 | positive regulation of secretion by cell | 123 | 193 | 3.92e-03 |
| GO:BP | GO:0048662 | negative regulation of smooth muscle cell proliferation | 29 | 36 | 3.99e-03 |
| GO:BP | GO:0071869 | response to catecholamine | 29 | 36 | 3.99e-03 |
| GO:BP | GO:0071867 | response to monoamine | 29 | 36 | 3.99e-03 |
| GO:BP | GO:0097306 | cellular response to alcohol | 53 | 74 | 4.02e-03 |
| GO:BP | GO:0002792 | negative regulation of peptide secretion | 27 | 33 | 4.05e-03 |
| GO:BP | GO:2000630 | positive regulation of miRNA metabolic process | 38 | 50 | 4.22e-03 |
| GO:BP | GO:0010660 | regulation of muscle cell apoptotic process | 38 | 50 | 4.22e-03 |
| GO:BP | GO:0048008 | platelet-derived growth factor receptor signaling pathway | 38 | 50 | 4.22e-03 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 74 | 109 | 4.24e-03 |
| GO:BP | GO:0051250 | negative regulation of lymphocyte activation | 59 | 84 | 4.31e-03 |
| GO:BP | GO:0051048 | negative regulation of secretion | 71 | 104 | 4.32e-03 |
| GO:BP | GO:0008543 | fibroblast growth factor receptor signaling pathway | 43 | 58 | 4.39e-03 |
| GO:BP | GO:0002028 | regulation of sodium ion transport | 43 | 58 | 4.39e-03 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 286 | 486 | 4.44e-03 |
| GO:BP | GO:1902547 | regulation of cellular response to vascular endothelial growth factor stimulus | 18 | 20 | 4.46e-03 |
| GO:BP | GO:2000116 | regulation of cysteine-type endopeptidase activity | 81 | 121 | 4.50e-03 |
| GO:BP | GO:0002067 | glandular epithelial cell differentiation | 36 | 47 | 4.63e-03 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 46 | 63 | 4.88e-03 |
| GO:BP | GO:0032411 | positive regulation of transporter activity | 46 | 63 | 4.88e-03 |
| GO:BP | GO:0036302 | atrioventricular canal development | 12 | 12 | 4.89e-03 |
| GO:BP | GO:0032332 | positive regulation of chondrocyte differentiation | 12 | 12 | 4.89e-03 |
| GO:BP | GO:0099505 | regulation of presynaptic membrane potential | 12 | 12 | 4.89e-03 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 313 | 536 | 4.89e-03 |
| GO:BP | GO:0008360 | regulation of cell shape | 72 | 106 | 4.89e-03 |
| GO:BP | GO:0003198 | epithelial to mesenchymal transition involved in endocardial cushion formation | 12 | 12 | 4.89e-03 |
| GO:BP | GO:0007405 | neuroblast proliferation | 41 | 55 | 4.89e-03 |
| GO:BP | GO:0045651 | positive regulation of macrophage differentiation | 12 | 12 | 4.89e-03 |
| GO:BP | GO:0010657 | muscle cell apoptotic process | 41 | 55 | 4.89e-03 |
| GO:BP | GO:0038066 | p38MAPK cascade | 34 | 44 | 4.99e-03 |
| GO:BP | GO:0002042 | cell migration involved in sprouting angiogenesis | 34 | 44 | 4.99e-03 |
| GO:BP | GO:1902895 | positive regulation of miRNA transcription | 34 | 44 | 4.99e-03 |
| GO:BP | GO:0034103 | regulation of tissue remodeling | 34 | 44 | 4.99e-03 |
| GO:BP | GO:2000177 | regulation of neural precursor cell proliferation | 49 | 68 | 5.20e-03 |
| GO:BP | GO:0030162 | regulation of proteolysis | 247 | 416 | 5.29e-03 |
| GO:BP | GO:0032331 | negative regulation of chondrocyte differentiation | 15 | 16 | 5.36e-03 |
| GO:BP | GO:0001709 | cell fate determination | 20 | 23 | 5.36e-03 |
| GO:BP | GO:0043537 | negative regulation of blood vessel endothelial cell migration | 20 | 23 | 5.36e-03 |
| GO:BP | GO:0050919 | negative chemotaxis | 32 | 41 | 5.36e-03 |
| GO:BP | GO:0070168 | negative regulation of biomineral tissue development | 15 | 16 | 5.36e-03 |
| GO:BP | GO:0061383 | trabecula morphogenesis | 32 | 41 | 5.36e-03 |
| GO:BP | GO:0031128 | developmental induction | 20 | 23 | 5.36e-03 |
| GO:BP | GO:0014888 | striated muscle adaptation | 32 | 41 | 5.36e-03 |
| GO:BP | GO:0060037 | pharyngeal system development | 20 | 23 | 5.36e-03 |
| GO:BP | GO:0045669 | positive regulation of osteoblast differentiation | 39 | 52 | 5.42e-03 |
| GO:BP | GO:0021675 | nerve development | 44 | 60 | 5.42e-03 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 751 | 1354 | 5.54e-03 |
| GO:BP | GO:0030282 | bone mineralization | 55 | 78 | 5.62e-03 |
| GO:BP | GO:0051480 | regulation of cytosolic calcium ion concentration | 30 | 38 | 5.72e-03 |
| GO:BP | GO:0042752 | regulation of circadian rhythm | 61 | 88 | 5.78e-03 |
| GO:BP | GO:0030099 | myeloid cell differentiation | 178 | 292 | 5.78e-03 |
| GO:BP | GO:0060291 | long-term synaptic potentiation | 47 | 65 | 5.89e-03 |
| GO:BP | GO:0003209 | cardiac atrium morphogenesis | 22 | 26 | 5.90e-03 |
| GO:BP | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction | 22 | 26 | 5.90e-03 |
| GO:BP | GO:0046651 | lymphocyte proliferation | 98 | 151 | 5.93e-03 |
| GO:BP | GO:0086091 | regulation of heart rate by cardiac conduction | 28 | 35 | 5.97e-03 |
| GO:BP | GO:0019933 | cAMP-mediated signaling | 28 | 35 | 5.97e-03 |
| GO:BP | GO:0090278 | negative regulation of peptide hormone secretion | 26 | 32 | 6.11e-03 |
| GO:BP | GO:0010656 | negative regulation of muscle cell apoptotic process | 26 | 32 | 6.11e-03 |
| GO:BP | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules | 24 | 29 | 6.11e-03 |
| GO:BP | GO:0010837 | regulation of keratinocyte proliferation | 26 | 32 | 6.11e-03 |
| GO:BP | GO:0021954 | central nervous system neuron development | 42 | 57 | 6.11e-03 |
| GO:BP | GO:0071604 | transforming growth factor beta production | 26 | 32 | 6.11e-03 |
| GO:BP | GO:0060041 | retina development in camera-type eye | 71 | 105 | 6.30e-03 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 231 | 388 | 6.31e-03 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 200 | 332 | 6.36e-03 |
| GO:BP | GO:0030856 | regulation of epithelial cell differentiation | 68 | 100 | 6.44e-03 |
| GO:BP | GO:0035725 | sodium ion transmembrane transport | 65 | 95 | 6.55e-03 |
| GO:BP | GO:0016311 | dephosphorylation | 144 | 232 | 6.55e-03 |
| GO:BP | GO:0010811 | positive regulation of cell-substrate adhesion | 65 | 95 | 6.55e-03 |
| GO:BP | GO:0050866 | negative regulation of cell activation | 78 | 117 | 6.59e-03 |
| GO:BP | GO:0035019 | somatic stem cell population maintenance | 35 | 46 | 6.59e-03 |
| GO:BP | GO:0006950 | response to stress | 1373 | 2540 | 6.68e-03 |
| GO:BP | GO:0034109 | homotypic cell-cell adhesion | 48 | 67 | 7.09e-03 |
| GO:BP | GO:0001892 | embryonic placenta development | 48 | 67 | 7.09e-03 |
| GO:BP | GO:0030217 | T cell differentiation | 116 | 183 | 7.20e-03 |
| GO:BP | GO:0045665 | negative regulation of neuron differentiation | 33 | 43 | 7.26e-03 |
| GO:BP | GO:0032944 | regulation of mononuclear cell proliferation | 79 | 119 | 7.35e-03 |
| GO:BP | GO:0062009 | secondary palate development | 17 | 19 | 7.35e-03 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 51 | 72 | 7.37e-03 |
| GO:BP | GO:0045995 | regulation of embryonic development | 51 | 72 | 7.37e-03 |
| GO:BP | GO:0002695 | negative regulation of leukocyte activation | 66 | 97 | 7.45e-03 |
| GO:BP | GO:0032943 | mononuclear cell proliferation | 101 | 157 | 7.54e-03 |
| GO:BP | GO:0051279 | regulation of release of sequestered calcium ion into cytosol | 43 | 59 | 7.54e-03 |
| GO:BP | GO:0046849 | bone remodeling | 43 | 59 | 7.54e-03 |
| GO:BP | GO:0045621 | positive regulation of lymphocyte differentiation | 57 | 82 | 7.63e-03 |
| GO:BP | GO:0010633 | negative regulation of epithelial cell migration | 38 | 51 | 7.67e-03 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 175 | 288 | 7.70e-03 |
| GO:BP | GO:0060326 | cell chemotaxis | 98 | 152 | 8.05e-03 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 375 | 654 | 8.47e-03 |
| GO:BP | GO:0062013 | positive regulation of small molecule metabolic process | 67 | 99 | 8.47e-03 |
| GO:BP | GO:0050670 | regulation of lymphocyte proliferation | 77 | 116 | 8.52e-03 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 127 | 203 | 8.58e-03 |
| GO:BP | GO:0021700 | developmental maturation | 127 | 203 | 8.58e-03 |
| GO:BP | GO:0071300 | cellular response to retinoic acid | 36 | 48 | 8.60e-03 |
| GO:BP | GO:0002021 | response to dietary excess | 19 | 22 | 8.64e-03 |
| GO:BP | GO:0097150 | neuronal stem cell population maintenance | 19 | 22 | 8.64e-03 |
| GO:BP | GO:0031623 | receptor internalization | 61 | 89 | 8.77e-03 |
| GO:BP | GO:0045637 | regulation of myeloid cell differentiation | 88 | 135 | 8.85e-03 |
| GO:BP | GO:0070661 | leukocyte proliferation | 111 | 175 | 8.86e-03 |
| GO:BP | GO:0043114 | regulation of vascular permeability | 27 | 34 | 8.99e-03 |
| GO:BP | GO:0003230 | cardiac atrium development | 27 | 34 | 8.99e-03 |
| GO:BP | GO:0060571 | morphogenesis of an epithelial fold | 14 | 15 | 9.15e-03 |
| GO:BP | GO:0014047 | glutamate secretion | 14 | 15 | 9.15e-03 |
| GO:BP | GO:0071636 | positive regulation of transforming growth factor beta production | 14 | 15 | 9.15e-03 |
| GO:BP | GO:0055012 | ventricular cardiac muscle cell differentiation | 14 | 15 | 9.15e-03 |
| GO:BP | GO:0035567 | non-canonical Wnt signaling pathway | 44 | 61 | 9.19e-03 |
| GO:BP | GO:0099072 | regulation of postsynaptic membrane neurotransmitter receptor levels | 44 | 61 | 9.19e-03 |
| GO:BP | GO:0030857 | negative regulation of epithelial cell differentiation | 25 | 31 | 9.28e-03 |
| GO:BP | GO:1900744 | regulation of p38MAPK cascade | 25 | 31 | 9.28e-03 |
| GO:BP | GO:0070663 | regulation of leukocyte proliferation | 85 | 130 | 9.28e-03 |
| GO:BP | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis | 25 | 31 | 9.28e-03 |
| GO:BP | GO:1903351 | cellular response to dopamine | 25 | 31 | 9.28e-03 |
| GO:BP | GO:0060391 | positive regulation of SMAD protein signal transduction | 25 | 31 | 9.28e-03 |
| GO:BP | GO:0002274 | myeloid leukocyte activation | 78 | 118 | 9.29e-03 |
| GO:BP | GO:0009913 | epidermal cell differentiation | 78 | 118 | 9.29e-03 |
| GO:BP | GO:0048483 | autonomic nervous system development | 23 | 28 | 9.40e-03 |
| GO:BP | GO:0048841 | regulation of axon extension involved in axon guidance | 23 | 28 | 9.40e-03 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 175 | 289 | 9.41e-03 |
| GO:BP | GO:0045639 | positive regulation of myeloid cell differentiation | 47 | 66 | 9.59e-03 |
| GO:BP | GO:0022408 | negative regulation of cell-cell adhesion | 75 | 113 | 9.72e-03 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 196 | 327 | 9.73e-03 |
| GO:BP | GO:0051928 | positive regulation of calcium ion transport | 50 | 71 | 9.88e-03 |
| GO:BP | GO:1903038 | negative regulation of leukocyte cell-cell adhesion | 50 | 71 | 9.88e-03 |
| GO:BP | GO:0001819 | positive regulation of cytokine production | 157 | 257 | 9.96e-03 |
| GO:BP | GO:0007229 | integrin-mediated signaling pathway | 59 | 86 | 1.00e-02 |
| GO:BP | GO:0071887 | leukocyte apoptotic process | 53 | 76 | 1.00e-02 |
| GO:BP | GO:0048608 | reproductive structure development | 126 | 202 | 1.03e-02 |
| GO:BP | GO:0045123 | cellular extravasation | 32 | 42 | 1.04e-02 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 214 | 360 | 1.05e-02 |
| GO:BP | GO:0045862 | positive regulation of proteolysis | 150 | 245 | 1.12e-02 |
| GO:BP | GO:0042098 | T cell proliferation | 73 | 110 | 1.13e-02 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 73 | 110 | 1.13e-02 |
| GO:BP | GO:0043502 | regulation of muscle adaptation | 48 | 68 | 1.14e-02 |
| GO:BP | GO:0055002 | striated muscle cell development | 48 | 68 | 1.14e-02 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 211 | 355 | 1.14e-02 |
| GO:BP | GO:0050922 | negative regulation of chemotaxis | 30 | 39 | 1.15e-02 |
| GO:BP | GO:0003014 | renal system process | 51 | 73 | 1.16e-02 |
| GO:BP | GO:0034763 | negative regulation of transmembrane transport | 57 | 83 | 1.16e-02 |
| GO:BP | GO:2001056 | positive regulation of cysteine-type endopeptidase activity | 57 | 83 | 1.16e-02 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 84 | 129 | 1.17e-02 |
| GO:BP | GO:0008217 | regulation of blood pressure | 70 | 105 | 1.17e-02 |
| GO:BP | GO:0042310 | vasoconstriction | 40 | 55 | 1.18e-02 |
| GO:BP | GO:0038094 | Fc-gamma receptor signaling pathway | 16 | 18 | 1.20e-02 |
| GO:BP | GO:0051968 | positive regulation of synaptic transmission, glutamatergic | 16 | 18 | 1.20e-02 |
| GO:BP | GO:1903319 | positive regulation of protein maturation | 16 | 18 | 1.20e-02 |
| GO:BP | GO:0055093 | response to hyperoxia | 16 | 18 | 1.20e-02 |
| GO:BP | GO:0045649 | regulation of macrophage differentiation | 16 | 18 | 1.20e-02 |
| GO:BP | GO:0003228 | atrial cardiac muscle tissue development | 16 | 18 | 1.20e-02 |
| GO:BP | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway | 16 | 18 | 1.20e-02 |
| GO:BP | GO:0002763 | positive regulation of myeloid leukocyte differentiation | 28 | 36 | 1.24e-02 |
| GO:BP | GO:0060443 | mammary gland morphogenesis | 28 | 36 | 1.24e-02 |
| GO:BP | GO:0120032 | regulation of plasma membrane bounded cell projection assembly | 100 | 157 | 1.24e-02 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 242 | 412 | 1.25e-02 |
| GO:BP | GO:0048675 | axon extension | 64 | 95 | 1.25e-02 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 153 | 251 | 1.28e-02 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 153 | 251 | 1.28e-02 |
| GO:BP | GO:0008366 | axon ensheathment | 78 | 119 | 1.30e-02 |
| GO:BP | GO:0007272 | ensheathment of neurons | 78 | 119 | 1.30e-02 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 58 | 85 | 1.31e-02 |
| GO:BP | GO:0032231 | regulation of actin filament bundle assembly | 58 | 85 | 1.31e-02 |
| GO:BP | GO:0060491 | regulation of cell projection assembly | 101 | 159 | 1.32e-02 |
| GO:BP | GO:0022414 | reproductive process | 491 | 874 | 1.36e-02 |
| GO:BP | GO:0021983 | pituitary gland development | 18 | 21 | 1.37e-02 |
| GO:BP | GO:0060384 | innervation | 18 | 21 | 1.37e-02 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 1051 | 1934 | 1.37e-02 |
| GO:BP | GO:0061458 | reproductive system development | 127 | 205 | 1.38e-02 |
| GO:BP | GO:0043412 | macromolecule modification | 1341 | 2489 | 1.39e-02 |
| GO:BP | GO:0001937 | negative regulation of endothelial cell proliferation | 24 | 30 | 1.39e-02 |
| GO:BP | GO:0046676 | negative regulation of insulin secretion | 24 | 30 | 1.39e-02 |
| GO:BP | GO:0048546 | digestive tract morphogenesis | 24 | 30 | 1.39e-02 |
| GO:BP | GO:0048512 | circadian behavior | 24 | 30 | 1.39e-02 |
| GO:BP | GO:0007616 | long-term memory | 24 | 30 | 1.39e-02 |
| GO:BP | GO:0046850 | regulation of bone remodeling | 24 | 30 | 1.39e-02 |
| GO:BP | GO:0050817 | coagulation | 94 | 147 | 1.40e-02 |
| GO:BP | GO:0008361 | regulation of cell size | 94 | 147 | 1.40e-02 |
| GO:BP | GO:1990845 | adaptive thermogenesis | 79 | 121 | 1.41e-02 |
| GO:BP | GO:0071260 | cellular response to mechanical stimulus | 41 | 57 | 1.42e-02 |
| GO:BP | GO:0032147 | activation of protein kinase activity | 41 | 57 | 1.42e-02 |
| GO:BP | GO:0051899 | membrane depolarization | 41 | 57 | 1.42e-02 |
| GO:BP | GO:0014898 | cardiac muscle hypertrophy in response to stress | 20 | 24 | 1.42e-02 |
| GO:BP | GO:0003299 | muscle hypertrophy in response to stress | 20 | 24 | 1.42e-02 |
| GO:BP | GO:1990573 | potassium ion import across plasma membrane | 22 | 27 | 1.42e-02 |
| GO:BP | GO:0010464 | regulation of mesenchymal cell proliferation | 22 | 27 | 1.42e-02 |
| GO:BP | GO:0045124 | regulation of bone resorption | 20 | 24 | 1.42e-02 |
| GO:BP | GO:0002431 | Fc receptor mediated stimulatory signaling pathway | 20 | 24 | 1.42e-02 |
| GO:BP | GO:0031279 | regulation of cyclase activity | 22 | 27 | 1.42e-02 |
| GO:BP | GO:0014887 | cardiac muscle adaptation | 20 | 24 | 1.42e-02 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 224 | 380 | 1.42e-02 |
| GO:BP | GO:0045926 | negative regulation of growth | 115 | 184 | 1.44e-02 |
| GO:BP | GO:0034446 | substrate adhesion-dependent cell spreading | 59 | 87 | 1.46e-02 |
| GO:BP | GO:0030168 | platelet activation | 59 | 87 | 1.46e-02 |
| GO:BP | GO:0030902 | hindbrain development | 69 | 104 | 1.47e-02 |
| GO:BP | GO:0045582 | positive regulation of T cell differentiation | 50 | 72 | 1.52e-02 |
| GO:BP | GO:0021533 | cell differentiation in hindbrain | 13 | 14 | 1.53e-02 |
| GO:BP | GO:0043217 | myelin maintenance | 13 | 14 | 1.53e-02 |
| GO:BP | GO:0010002 | cardioblast differentiation | 13 | 14 | 1.53e-02 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 1137 | 2100 | 1.54e-02 |
| GO:BP | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance | 10 | 10 | 1.54e-02 |
| GO:BP | GO:0072497 | mesenchymal stem cell differentiation | 10 | 10 | 1.54e-02 |
| GO:BP | GO:0045906 | negative regulation of vasoconstriction | 10 | 10 | 1.54e-02 |
| GO:BP | GO:0022410 | circadian sleep/wake cycle process | 10 | 10 | 1.54e-02 |
| GO:BP | GO:0099149 | regulation of postsynaptic neurotransmitter receptor internalization | 10 | 10 | 1.54e-02 |
| GO:BP | GO:1902287 | semaphorin-plexin signaling pathway involved in axon guidance | 10 | 10 | 1.54e-02 |
| GO:BP | GO:0030431 | sleep | 10 | 10 | 1.54e-02 |
| GO:BP | GO:0043549 | regulation of kinase activity | 211 | 357 | 1.59e-02 |
| GO:BP | GO:1903531 | negative regulation of secretion by cell | 63 | 94 | 1.59e-02 |
| GO:BP | GO:0007596 | blood coagulation | 92 | 144 | 1.59e-02 |
| GO:BP | GO:0050918 | positive chemotaxis | 29 | 38 | 1.62e-02 |
| GO:BP | GO:0002066 | columnar/cuboidal epithelial cell development | 29 | 38 | 1.62e-02 |
| GO:BP | GO:0014074 | response to purine-containing compound | 70 | 106 | 1.62e-02 |
| GO:BP | GO:0007200 | phospholipase C-activating G protein-coupled receptor signaling pathway | 29 | 38 | 1.62e-02 |
| GO:BP | GO:0045103 | intermediate filament-based process | 29 | 38 | 1.62e-02 |
| GO:BP | GO:0001706 | endoderm formation | 34 | 46 | 1.67e-02 |
| GO:BP | GO:0033043 | regulation of organelle organization | 532 | 953 | 1.68e-02 |
| GO:BP | GO:0030278 | regulation of ossification | 54 | 79 | 1.72e-02 |
| GO:BP | GO:0010038 | response to metal ion | 151 | 249 | 1.76e-02 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 156 | 258 | 1.76e-02 |
| GO:BP | GO:0007605 | sensory perception of sound | 64 | 96 | 1.77e-02 |
| GO:BP | GO:0008038 | neuron recognition | 27 | 35 | 1.78e-02 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 137 | 224 | 1.78e-02 |
| GO:BP | GO:0042593 | glucose homeostasis | 106 | 169 | 1.80e-02 |
| GO:BP | GO:0051223 | regulation of protein transport | 193 | 325 | 1.81e-02 |
| GO:BP | GO:0042733 | embryonic digit morphogenesis | 37 | 51 | 1.82e-02 |
| GO:BP | GO:0007599 | hemostasis | 94 | 148 | 1.84e-02 |
| GO:BP | GO:0009880 | embryonic pattern specification | 32 | 43 | 1.89e-02 |
| GO:BP | GO:0042129 | regulation of T cell proliferation | 58 | 86 | 1.91e-02 |
| GO:BP | GO:0019725 | cellular homeostasis | 323 | 564 | 1.91e-02 |
| GO:BP | GO:0098884 | postsynaptic neurotransmitter receptor internalization | 15 | 17 | 1.92e-02 |
| GO:BP | GO:0038096 | Fc-gamma receptor signaling pathway involved in phagocytosis | 15 | 17 | 1.92e-02 |
| GO:BP | GO:0051590 | positive regulation of neurotransmitter transport | 15 | 17 | 1.92e-02 |
| GO:BP | GO:0010954 | positive regulation of protein processing | 15 | 17 | 1.92e-02 |
| GO:BP | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis | 15 | 17 | 1.92e-02 |
| GO:BP | GO:0007622 | rhythmic behavior | 25 | 32 | 1.92e-02 |
| GO:BP | GO:0140239 | postsynaptic endocytosis | 15 | 17 | 1.92e-02 |
| GO:BP | GO:0048169 | regulation of long-term neuronal synaptic plasticity | 15 | 17 | 1.92e-02 |
| GO:BP | GO:1903350 | response to dopamine | 25 | 32 | 1.92e-02 |
| GO:BP | GO:0060711 | labyrinthine layer development | 25 | 32 | 1.92e-02 |
| GO:BP | GO:0007612 | learning | 72 | 110 | 1.95e-02 |
| GO:BP | GO:0030879 | mammary gland development | 65 | 98 | 1.95e-02 |
| GO:BP | GO:0001701 | in utero embryonic development | 196 | 331 | 1.98e-02 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 130 | 212 | 1.98e-02 |
| GO:BP | GO:0030239 | myofibril assembly | 46 | 66 | 2.00e-02 |
| GO:BP | GO:0106027 | neuron projection organization | 49 | 71 | 2.00e-02 |
| GO:BP | GO:0035710 | CD4-positive, alpha-beta T cell activation | 46 | 66 | 2.00e-02 |
| GO:BP | GO:0042552 | myelination | 76 | 117 | 2.01e-02 |
| GO:BP | GO:0061384 | heart trabecula morphogenesis | 23 | 29 | 2.04e-02 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 145 | 239 | 2.04e-02 |
| GO:BP | GO:0097106 | postsynaptic density organization | 23 | 29 | 2.04e-02 |
| GO:BP | GO:0042491 | inner ear auditory receptor cell differentiation | 23 | 29 | 2.04e-02 |
| GO:BP | GO:0086019 | cell-cell signaling involved in cardiac conduction | 23 | 29 | 2.04e-02 |
| GO:BP | GO:0031102 | neuron projection regeneration | 35 | 48 | 2.05e-02 |
| GO:BP | GO:0030500 | regulation of bone mineralization | 35 | 48 | 2.05e-02 |
| GO:BP | GO:0048286 | lung alveolus development | 30 | 40 | 2.09e-02 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 88 | 138 | 2.09e-02 |
| GO:BP | GO:0031281 | positive regulation of cyclase activity | 17 | 20 | 2.11e-02 |
| GO:BP | GO:2000114 | regulation of establishment of cell polarity | 17 | 20 | 2.11e-02 |
| GO:BP | GO:0048710 | regulation of astrocyte differentiation | 17 | 20 | 2.11e-02 |
| GO:BP | GO:0048730 | epidermis morphogenesis | 17 | 20 | 2.11e-02 |
| GO:BP | GO:0043588 | skin development | 109 | 175 | 2.12e-02 |
| GO:BP | GO:0048806 | genitalia development | 21 | 26 | 2.13e-02 |
| GO:BP | GO:1903055 | positive regulation of extracellular matrix organization | 21 | 26 | 2.13e-02 |
| GO:BP | GO:0007212 | dopamine receptor signaling pathway | 21 | 26 | 2.13e-02 |
| GO:BP | GO:1902656 | calcium ion import into cytosol | 21 | 26 | 2.13e-02 |
| GO:BP | GO:0010765 | positive regulation of sodium ion transport | 19 | 23 | 2.15e-02 |
| GO:BP | GO:0010827 | regulation of glucose transmembrane transport | 38 | 53 | 2.15e-02 |
| GO:BP | GO:0003416 | endochondral bone growth | 19 | 23 | 2.15e-02 |
| GO:BP | GO:0072012 | glomerulus vasculature development | 19 | 23 | 2.15e-02 |
| GO:BP | GO:1904752 | regulation of vascular associated smooth muscle cell migration | 19 | 23 | 2.15e-02 |
| GO:BP | GO:0043367 | CD4-positive, alpha-beta T cell differentiation | 38 | 53 | 2.15e-02 |
| GO:BP | GO:0045834 | positive regulation of lipid metabolic process | 56 | 83 | 2.15e-02 |
| GO:BP | GO:0045761 | regulation of adenylate cyclase activity | 19 | 23 | 2.15e-02 |
| GO:BP | GO:0019216 | regulation of lipid metabolic process | 132 | 216 | 2.15e-02 |
| GO:BP | GO:0010469 | regulation of signaling receptor activity | 41 | 58 | 2.24e-02 |
| GO:BP | GO:0048706 | embryonic skeletal system development | 50 | 73 | 2.26e-02 |
| GO:BP | GO:0048145 | regulation of fibroblast proliferation | 50 | 73 | 2.26e-02 |
| GO:BP | GO:0097581 | lamellipodium organization | 50 | 73 | 2.26e-02 |
| GO:BP | GO:0033500 | carbohydrate homeostasis | 106 | 170 | 2.27e-02 |
| GO:BP | GO:0097061 | dendritic spine organization | 44 | 63 | 2.27e-02 |
| GO:BP | GO:1903115 | regulation of actin filament-based movement | 28 | 37 | 2.29e-02 |
| GO:BP | GO:0045104 | intermediate filament cytoskeleton organization | 28 | 37 | 2.29e-02 |
| GO:BP | GO:0045747 | positive regulation of Notch signaling pathway | 28 | 37 | 2.29e-02 |
| GO:BP | GO:0010658 | striated muscle cell apoptotic process | 28 | 37 | 2.29e-02 |
| GO:BP | GO:0001818 | negative regulation of cytokine production | 102 | 163 | 2.30e-02 |
| GO:BP | GO:0010761 | fibroblast migration | 36 | 50 | 2.48e-02 |
| GO:BP | GO:0048814 | regulation of dendrite morphogenesis | 36 | 50 | 2.48e-02 |
| GO:BP | GO:0042490 | mechanoreceptor differentiation | 36 | 50 | 2.48e-02 |
| GO:BP | GO:0062012 | regulation of small molecule metabolic process | 140 | 231 | 2.48e-02 |
| GO:BP | GO:1901655 | cellular response to ketone | 54 | 80 | 2.50e-02 |
| GO:BP | GO:0032535 | regulation of cellular component size | 171 | 287 | 2.51e-02 |
| GO:BP | GO:0008104 | protein localization | 1081 | 2000 | 2.51e-02 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 397 | 704 | 2.51e-02 |
| GO:BP | GO:0010662 | regulation of striated muscle cell apoptotic process | 26 | 34 | 2.52e-02 |
| GO:BP | GO:0071895 | odontoblast differentiation | 12 | 13 | 2.52e-02 |
| GO:BP | GO:0010659 | cardiac muscle cell apoptotic process | 26 | 34 | 2.52e-02 |
| GO:BP | GO:0048665 | neuron fate specification | 12 | 13 | 2.52e-02 |
| GO:BP | GO:0099084 | postsynaptic specialization organization | 26 | 34 | 2.52e-02 |
| GO:BP | GO:0046579 | positive regulation of Ras protein signal transduction | 12 | 13 | 2.52e-02 |
| GO:BP | GO:1900153 | positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 12 | 13 | 2.52e-02 |
| GO:BP | GO:0060841 | venous blood vessel development | 12 | 13 | 2.52e-02 |
| GO:BP | GO:0000003 | reproduction | 492 | 882 | 2.52e-02 |
| GO:BP | GO:0030033 | microvillus assembly | 12 | 13 | 2.52e-02 |
| GO:BP | GO:0035315 | hair cell differentiation | 26 | 34 | 2.52e-02 |
| GO:BP | GO:0009954 | proximal/distal pattern formation | 12 | 13 | 2.52e-02 |
| GO:BP | GO:0001956 | positive regulation of neurotransmitter secretion | 12 | 13 | 2.52e-02 |
| GO:BP | GO:0043281 | regulation of cysteine-type endopeptidase activity involved in apoptotic process | 68 | 104 | 2.54e-02 |
| GO:BP | GO:0070509 | calcium ion import | 39 | 55 | 2.54e-02 |
| GO:BP | GO:0003073 | regulation of systemic arterial blood pressure | 39 | 55 | 2.54e-02 |
| GO:BP | GO:0001954 | positive regulation of cell-matrix adhesion | 31 | 42 | 2.58e-02 |
| GO:BP | GO:0048146 | positive regulation of fibroblast proliferation | 31 | 42 | 2.58e-02 |
| GO:BP | GO:0051966 | regulation of synaptic transmission, glutamatergic | 31 | 42 | 2.58e-02 |
| GO:BP | GO:0001938 | positive regulation of endothelial cell proliferation | 48 | 70 | 2.58e-02 |
| GO:BP | GO:0048661 | positive regulation of smooth muscle cell proliferation | 42 | 60 | 2.59e-02 |
| GO:BP | GO:0035418 | protein localization to synapse | 42 | 60 | 2.59e-02 |
| GO:BP | GO:0030316 | osteoclast differentiation | 45 | 65 | 2.59e-02 |
| GO:BP | GO:0001942 | hair follicle development | 45 | 65 | 2.59e-02 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 1055 | 1951 | 2.60e-02 |
| GO:BP | GO:0110020 | regulation of actomyosin structure organization | 58 | 87 | 2.65e-02 |
| GO:BP | GO:0051147 | regulation of muscle cell differentiation | 76 | 118 | 2.65e-02 |
| GO:BP | GO:0050714 | positive regulation of protein secretion | 65 | 99 | 2.67e-02 |
| GO:BP | GO:0046629 | gamma-delta T cell activation | 9 | 9 | 2.68e-02 |
| GO:BP | GO:2000288 | positive regulation of myoblast proliferation | 9 | 9 | 2.68e-02 |
| GO:BP | GO:0002024 | diet induced thermogenesis | 9 | 9 | 2.68e-02 |
| GO:BP | GO:0050802 | circadian sleep/wake cycle, sleep | 9 | 9 | 2.68e-02 |
| GO:BP | GO:0010838 | positive regulation of keratinocyte proliferation | 9 | 9 | 2.68e-02 |
| GO:BP | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration | 9 | 9 | 2.68e-02 |
| GO:BP | GO:0032060 | bleb assembly | 9 | 9 | 2.68e-02 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 540 | 973 | 2.68e-02 |
| GO:BP | GO:2000739 | regulation of mesenchymal stem cell differentiation | 9 | 9 | 2.68e-02 |
| GO:BP | GO:0048845 | venous blood vessel morphogenesis | 9 | 9 | 2.68e-02 |
| GO:BP | GO:0001768 | establishment of T cell polarity | 9 | 9 | 2.68e-02 |
| GO:BP | GO:0032906 | transforming growth factor beta2 production | 9 | 9 | 2.68e-02 |
| GO:BP | GO:0032909 | regulation of transforming growth factor beta2 production | 9 | 9 | 2.68e-02 |
| GO:BP | GO:0051643 | endoplasmic reticulum localization | 9 | 9 | 2.68e-02 |
| GO:BP | GO:0061626 | pharyngeal arch artery morphogenesis | 9 | 9 | 2.68e-02 |
| GO:BP | GO:0010665 | regulation of cardiac muscle cell apoptotic process | 24 | 31 | 2.72e-02 |
| GO:BP | GO:0060688 | regulation of morphogenesis of a branching structure | 24 | 31 | 2.72e-02 |
| GO:BP | GO:0034405 | response to fluid shear stress | 24 | 31 | 2.72e-02 |
| GO:BP | GO:0070664 | negative regulation of leukocyte proliferation | 34 | 47 | 2.74e-02 |
| GO:BP | GO:0014743 | regulation of muscle hypertrophy | 34 | 47 | 2.74e-02 |
| GO:BP | GO:0045600 | positive regulation of fat cell differentiation | 34 | 47 | 2.74e-02 |
| GO:BP | GO:1903317 | regulation of protein maturation | 34 | 47 | 2.74e-02 |
| GO:BP | GO:0007584 | response to nutrient | 73 | 113 | 2.79e-02 |
| GO:BP | GO:0061024 | membrane organization | 354 | 625 | 2.81e-02 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 106 | 171 | 2.82e-02 |
| GO:BP | GO:0045619 | regulation of lymphocyte differentiation | 81 | 127 | 2.85e-02 |
| GO:BP | GO:0042311 | vasodilation | 29 | 39 | 2.86e-02 |
| GO:BP | GO:0051965 | positive regulation of synapse assembly | 29 | 39 | 2.86e-02 |
| GO:BP | GO:0035904 | aorta development | 37 | 52 | 2.87e-02 |
| GO:BP | GO:1904427 | positive regulation of calcium ion transmembrane transport | 37 | 52 | 2.87e-02 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 1085 | 2010 | 2.87e-02 |
| GO:BP | GO:0003171 | atrioventricular valve development | 22 | 28 | 2.92e-02 |
| GO:BP | GO:0022405 | hair cycle process | 46 | 67 | 2.92e-02 |
| GO:BP | GO:0022404 | molting cycle process | 46 | 67 | 2.92e-02 |
| GO:BP | GO:0048713 | regulation of oligodendrocyte differentiation | 22 | 28 | 2.92e-02 |
| GO:BP | GO:0060317 | cardiac epithelial to mesenchymal transition | 22 | 28 | 2.92e-02 |
| GO:BP | GO:0044331 | cell-cell adhesion mediated by cadherin | 22 | 28 | 2.92e-02 |
| GO:BP | GO:0031503 | protein-containing complex localization | 98 | 157 | 2.93e-02 |
| GO:BP | GO:0086014 | atrial cardiac muscle cell action potential | 14 | 16 | 2.99e-02 |
| GO:BP | GO:0086026 | atrial cardiac muscle cell to AV node cell signaling | 14 | 16 | 2.99e-02 |
| GO:BP | GO:0021846 | cell proliferation in forebrain | 14 | 16 | 2.99e-02 |
| GO:BP | GO:0003128 | heart field specification | 14 | 16 | 2.99e-02 |
| GO:BP | GO:0086066 | atrial cardiac muscle cell to AV node cell communication | 14 | 16 | 2.99e-02 |
| GO:BP | GO:1900274 | regulation of phospholipase C activity | 14 | 16 | 2.99e-02 |
| GO:BP | GO:0045762 | positive regulation of adenylate cyclase activity | 14 | 16 | 2.99e-02 |
| GO:BP | GO:0019827 | stem cell population maintenance | 90 | 143 | 3.00e-02 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 535 | 965 | 3.01e-02 |
| GO:BP | GO:1904738 | vascular associated smooth muscle cell migration | 20 | 25 | 3.09e-02 |
| GO:BP | GO:0061437 | renal system vasculature development | 20 | 25 | 3.09e-02 |
| GO:BP | GO:0007435 | salivary gland morphogenesis | 20 | 25 | 3.09e-02 |
| GO:BP | GO:0061440 | kidney vasculature development | 20 | 25 | 3.09e-02 |
| GO:BP | GO:0098703 | calcium ion import across plasma membrane | 20 | 25 | 3.09e-02 |
| GO:BP | GO:0007623 | circadian rhythm | 95 | 152 | 3.15e-02 |
| GO:BP | GO:0030038 | contractile actin filament bundle assembly | 60 | 91 | 3.17e-02 |
| GO:BP | GO:0043149 | stress fiber assembly | 60 | 91 | 3.17e-02 |
| GO:BP | GO:0038179 | neurotrophin signaling pathway | 27 | 36 | 3.17e-02 |
| GO:BP | GO:0035306 | positive regulation of dephosphorylation | 27 | 36 | 3.17e-02 |
| GO:BP | GO:0060512 | prostate gland morphogenesis | 16 | 19 | 3.17e-02 |
| GO:BP | GO:0098810 | neurotransmitter reuptake | 16 | 19 | 3.17e-02 |
| GO:BP | GO:1990806 | ligand-gated ion channel signaling pathway | 16 | 19 | 3.17e-02 |
| GO:BP | GO:0099590 | neurotransmitter receptor internalization | 16 | 19 | 3.17e-02 |
| GO:BP | GO:0060740 | prostate gland epithelium morphogenesis | 16 | 19 | 3.17e-02 |
| GO:BP | GO:1901201 | regulation of extracellular matrix assembly | 16 | 19 | 3.17e-02 |
| GO:BP | GO:0042092 | type 2 immune response | 16 | 19 | 3.17e-02 |
| GO:BP | GO:0003338 | metanephros morphogenesis | 16 | 19 | 3.17e-02 |
| GO:BP | GO:0097529 | myeloid leukocyte migration | 71 | 110 | 3.17e-02 |
| GO:BP | GO:0055075 | potassium ion homeostasis | 18 | 22 | 3.18e-02 |
| GO:BP | GO:0030279 | negative regulation of ossification | 18 | 22 | 3.18e-02 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 191 | 325 | 3.18e-02 |
| GO:BP | GO:1904705 | regulation of vascular associated smooth muscle cell proliferation | 35 | 49 | 3.24e-02 |
| GO:BP | GO:0018107 | peptidyl-threonine phosphorylation | 47 | 69 | 3.28e-02 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 170 | 287 | 3.34e-02 |
| GO:BP | GO:0016239 | positive regulation of macroautophagy | 44 | 64 | 3.34e-02 |
| GO:BP | GO:0050868 | negative regulation of T cell activation | 44 | 64 | 3.34e-02 |
| GO:BP | GO:0050772 | positive regulation of axonogenesis | 44 | 64 | 3.34e-02 |
| GO:BP | GO:0140238 | presynaptic endocytosis | 41 | 59 | 3.36e-02 |
| GO:BP | GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 88 | 140 | 3.41e-02 |
| GO:BP | GO:0106106 | cold-induced thermogenesis | 72 | 112 | 3.42e-02 |
| GO:BP | GO:1903034 | regulation of response to wounding | 76 | 119 | 3.45e-02 |
| GO:BP | GO:0046683 | response to organophosphorus | 61 | 93 | 3.46e-02 |
| GO:BP | GO:0010171 | body morphogenesis | 30 | 41 | 3.49e-02 |
| GO:BP | GO:0045453 | bone resorption | 30 | 41 | 3.49e-02 |
| GO:BP | GO:0007600 | sensory perception | 150 | 251 | 3.50e-02 |
| GO:BP | GO:1902692 | regulation of neuroblast proliferation | 25 | 33 | 3.51e-02 |
| GO:BP | GO:0038093 | Fc receptor signaling pathway | 25 | 33 | 3.51e-02 |
| GO:BP | GO:0018210 | peptidyl-threonine modification | 51 | 76 | 3.58e-02 |
| GO:BP | GO:0060996 | dendritic spine development | 51 | 76 | 3.58e-02 |
| GO:BP | GO:0007219 | Notch signaling pathway | 89 | 142 | 3.63e-02 |
| GO:BP | GO:0030595 | leukocyte chemotaxis | 69 | 107 | 3.65e-02 |
| GO:BP | GO:0032880 | regulation of protein localization | 388 | 691 | 3.70e-02 |
| GO:BP | GO:0001655 | urogenital system development | 33 | 46 | 3.71e-02 |
| GO:BP | GO:0010611 | regulation of cardiac muscle hypertrophy | 33 | 46 | 3.71e-02 |
| GO:BP | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process | 33 | 46 | 3.71e-02 |
| GO:BP | GO:0062014 | negative regulation of small molecule metabolic process | 48 | 71 | 3.71e-02 |
| GO:BP | GO:1904036 | negative regulation of epithelial cell apoptotic process | 33 | 46 | 3.71e-02 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 62 | 95 | 3.78e-02 |
| GO:BP | GO:0051153 | regulation of striated muscle cell differentiation | 45 | 66 | 3.80e-02 |
| GO:BP | GO:0006887 | exocytosis | 142 | 237 | 3.83e-02 |
| GO:BP | GO:0050771 | negative regulation of axonogenesis | 36 | 51 | 3.83e-02 |
| GO:BP | GO:0043266 | regulation of potassium ion transport | 42 | 61 | 3.85e-02 |
| GO:BP | GO:0031668 | cellular response to extracellular stimulus | 132 | 219 | 3.85e-02 |
| GO:BP | GO:0033273 | response to vitamin | 42 | 61 | 3.85e-02 |
| GO:BP | GO:0002088 | lens development in camera-type eye | 39 | 56 | 3.87e-02 |
| GO:BP | GO:1904019 | epithelial cell apoptotic process | 66 | 102 | 3.87e-02 |
| GO:BP | GO:0052652 | cyclic purine nucleotide metabolic process | 23 | 30 | 3.88e-02 |
| GO:BP | GO:0014911 | positive regulation of smooth muscle cell migration | 23 | 30 | 3.88e-02 |
| GO:BP | GO:0001659 | temperature homeostasis | 82 | 130 | 3.93e-02 |
| GO:BP | GO:2001222 | regulation of neuron migration | 28 | 38 | 3.94e-02 |
| GO:BP | GO:0006940 | regulation of smooth muscle contraction | 28 | 38 | 3.94e-02 |
| GO:BP | GO:0045668 | negative regulation of osteoblast differentiation | 28 | 38 | 3.94e-02 |
| GO:BP | GO:0061005 | cell differentiation involved in kidney development | 28 | 38 | 3.94e-02 |
| GO:BP | GO:0052548 | regulation of endopeptidase activity | 95 | 153 | 3.97e-02 |
| GO:BP | GO:0050773 | regulation of dendrite development | 52 | 78 | 3.97e-02 |
| GO:BP | GO:0046942 | carboxylic acid transport | 123 | 203 | 3.98e-02 |
| GO:BP | GO:0072578 | neurotransmitter-gated ion channel clustering | 11 | 12 | 4.07e-02 |
| GO:BP | GO:0045780 | positive regulation of bone resorption | 11 | 12 | 4.07e-02 |
| GO:BP | GO:0048368 | lateral mesoderm development | 11 | 12 | 4.07e-02 |
| GO:BP | GO:0003222 | ventricular trabecula myocardium morphogenesis | 11 | 12 | 4.07e-02 |
| GO:BP | GO:0061154 | endothelial tube morphogenesis | 11 | 12 | 4.07e-02 |
| GO:BP | GO:0003207 | cardiac chamber formation | 11 | 12 | 4.07e-02 |
| GO:BP | GO:0055057 | neuroblast division | 11 | 12 | 4.07e-02 |
| GO:BP | GO:0003159 | morphogenesis of an endothelium | 11 | 12 | 4.07e-02 |
| GO:BP | GO:0030539 | male genitalia development | 11 | 12 | 4.07e-02 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 119 | 196 | 4.12e-02 |
| GO:BP | GO:0001558 | regulation of cell growth | 189 | 323 | 4.13e-02 |
| GO:BP | GO:0038127 | ERBB signaling pathway | 67 | 104 | 4.16e-02 |
| GO:BP | GO:0051651 | maintenance of location in cell | 105 | 171 | 4.16e-02 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 587 | 1067 | 4.18e-02 |
| GO:BP | GO:0120161 | regulation of cold-induced thermogenesis | 71 | 111 | 4.18e-02 |
| GO:BP | GO:1901021 | positive regulation of calcium ion transmembrane transporter activity | 21 | 27 | 4.18e-02 |
| GO:BP | GO:0120033 | negative regulation of plasma membrane bounded cell projection assembly | 21 | 27 | 4.18e-02 |
| GO:BP | GO:0010517 | regulation of phospholipase activity | 21 | 27 | 4.18e-02 |
| GO:BP | GO:0035909 | aorta morphogenesis | 21 | 27 | 4.18e-02 |
| GO:BP | GO:0031333 | negative regulation of protein-containing complex assembly | 75 | 118 | 4.18e-02 |
| GO:BP | GO:0002294 | CD4-positive, alpha-beta T cell differentiation involved in immune response | 31 | 43 | 4.18e-02 |
| GO:BP | GO:0030514 | negative regulation of BMP signaling pathway | 31 | 43 | 4.18e-02 |
| GO:BP | GO:0042093 | T-helper cell differentiation | 31 | 43 | 4.18e-02 |
| GO:BP | GO:0070570 | regulation of neuron projection regeneration | 21 | 27 | 4.18e-02 |
| GO:BP | GO:0032945 | negative regulation of mononuclear cell proliferation | 31 | 43 | 4.18e-02 |
| GO:BP | GO:0002293 | alpha-beta T cell differentiation involved in immune response | 31 | 43 | 4.18e-02 |
| GO:BP | GO:0050672 | negative regulation of lymphocyte proliferation | 31 | 43 | 4.18e-02 |
| GO:BP | GO:0002287 | alpha-beta T cell activation involved in immune response | 31 | 43 | 4.18e-02 |
| GO:BP | GO:0150104 | transport across blood-brain barrier | 46 | 68 | 4.20e-02 |
| GO:BP | GO:0010232 | vascular transport | 46 | 68 | 4.20e-02 |
| GO:BP | GO:0060249 | anatomical structure homeostasis | 101 | 164 | 4.24e-02 |
| GO:BP | GO:0001894 | tissue homeostasis | 101 | 164 | 4.24e-02 |
| GO:BP | GO:0010629 | negative regulation of gene expression | 440 | 790 | 4.24e-02 |
| GO:BP | GO:0060113 | inner ear receptor cell differentiation | 34 | 48 | 4.32e-02 |
| GO:BP | GO:0048488 | synaptic vesicle endocytosis | 40 | 58 | 4.38e-02 |
| GO:BP | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 40 | 58 | 4.38e-02 |
| GO:BP | GO:0007193 | adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway | 26 | 35 | 4.38e-02 |
| GO:BP | GO:0034113 | heterotypic cell-cell adhesion | 26 | 35 | 4.38e-02 |
| GO:BP | GO:0010508 | positive regulation of autophagy | 80 | 127 | 4.43e-02 |
| GO:BP | GO:0098781 | ncRNA transcription | 76 | 120 | 4.47e-02 |
| GO:BP | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway | 19 | 24 | 4.49e-02 |
| GO:BP | GO:0033280 | response to vitamin D | 19 | 24 | 4.49e-02 |
| GO:BP | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling | 19 | 24 | 4.49e-02 |
| GO:BP | GO:0022011 | myelination in peripheral nervous system | 19 | 24 | 4.49e-02 |
| GO:BP | GO:0032292 | peripheral nervous system axon ensheathment | 19 | 24 | 4.49e-02 |
| GO:BP | GO:0001958 | endochondral ossification | 19 | 24 | 4.49e-02 |
| GO:BP | GO:0098868 | bone growth | 19 | 24 | 4.49e-02 |
| GO:BP | GO:0036075 | replacement ossification | 19 | 24 | 4.49e-02 |
| GO:BP | GO:0030534 | adult behavior | 57 | 87 | 4.50e-02 |
| GO:BP | GO:0042303 | molting cycle | 50 | 75 | 4.50e-02 |
| GO:BP | GO:0007204 | positive regulation of cytosolic calcium ion concentration | 50 | 75 | 4.50e-02 |
| GO:BP | GO:0042633 | hair cycle | 50 | 75 | 4.50e-02 |
| GO:BP | GO:0097152 | mesenchymal cell apoptotic process | 8 | 8 | 4.56e-02 |
| GO:BP | GO:0045187 | regulation of circadian sleep/wake cycle, sleep | 8 | 8 | 4.56e-02 |
| GO:BP | GO:0036462 | TRAIL-activated apoptotic signaling pathway | 8 | 8 | 4.56e-02 |
| GO:BP | GO:0071389 | cellular response to mineralocorticoid stimulus | 8 | 8 | 4.56e-02 |
| GO:BP | GO:0040034 | regulation of development, heterochronic | 8 | 8 | 4.56e-02 |
| GO:BP | GO:0060982 | coronary artery morphogenesis | 8 | 8 | 4.56e-02 |
| GO:BP | GO:0048505 | regulation of timing of cell differentiation | 8 | 8 | 4.56e-02 |
| GO:BP | GO:0048715 | negative regulation of oligodendrocyte differentiation | 8 | 8 | 4.56e-02 |
| GO:BP | GO:0003190 | atrioventricular valve formation | 8 | 8 | 4.56e-02 |
| GO:BP | GO:0032674 | regulation of interleukin-5 production | 8 | 8 | 4.56e-02 |
| GO:BP | GO:0060896 | neural plate pattern specification | 8 | 8 | 4.56e-02 |
| GO:BP | GO:0032634 | interleukin-5 production | 8 | 8 | 4.56e-02 |
| GO:BP | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity | 8 | 8 | 4.56e-02 |
| GO:BP | GO:1900116 | extracellular negative regulation of signal transduction | 8 | 8 | 4.56e-02 |
| GO:BP | GO:0099624 | atrial cardiac muscle cell membrane repolarization | 8 | 8 | 4.56e-02 |
| GO:BP | GO:1900115 | extracellular regulation of signal transduction | 8 | 8 | 4.56e-02 |
| GO:BP | GO:0003149 | membranous septum morphogenesis | 8 | 8 | 4.56e-02 |
| GO:BP | GO:0003418 | growth plate cartilage chondrocyte differentiation | 8 | 8 | 4.56e-02 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 314 | 555 | 4.56e-02 |
| GO:BP | GO:0002828 | regulation of type 2 immune response | 13 | 15 | 4.58e-02 |
| GO:BP | GO:0051546 | keratinocyte migration | 13 | 15 | 4.58e-02 |
| GO:BP | GO:1902932 | positive regulation of alcohol biosynthetic process | 13 | 15 | 4.58e-02 |
| GO:BP | GO:0072170 | metanephric tubule development | 13 | 15 | 4.58e-02 |
| GO:BP | GO:0060413 | atrial septum morphogenesis | 13 | 15 | 4.58e-02 |
| GO:BP | GO:1902683 | regulation of receptor localization to synapse | 13 | 15 | 4.58e-02 |
| GO:BP | GO:0070571 | negative regulation of neuron projection regeneration | 13 | 15 | 4.58e-02 |
| GO:BP | GO:0032891 | negative regulation of organic acid transport | 13 | 15 | 4.58e-02 |
| GO:BP | GO:0016079 | synaptic vesicle exocytosis | 47 | 70 | 4.60e-02 |
| GO:BP | GO:1900745 | positive regulation of p38MAPK cascade | 17 | 21 | 4.66e-02 |
| GO:BP | GO:2000178 | negative regulation of neural precursor cell proliferation | 17 | 21 | 4.66e-02 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 879 | 1624 | 4.67e-02 |
| GO:BP | GO:0015849 | organic acid transport | 123 | 204 | 4.71e-02 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 90 | 145 | 4.71e-02 |
| GO:BP | GO:0098727 | maintenance of cell number | 90 | 145 | 4.71e-02 |
| GO:BP | GO:0032869 | cellular response to insulin stimulus | 99 | 161 | 4.71e-02 |
| GO:BP | GO:0032925 | regulation of activin receptor signaling pathway | 15 | 18 | 4.74e-02 |
| GO:BP | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway | 15 | 18 | 4.74e-02 |
| GO:BP | GO:0031069 | hair follicle morphogenesis | 15 | 18 | 4.74e-02 |
| GO:BP | GO:0033032 | regulation of myeloid cell apoptotic process | 15 | 18 | 4.74e-02 |
| GO:BP | GO:0007187 | G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger | 15 | 18 | 4.74e-02 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 178 | 304 | 4.77e-02 |
| GO:BP | GO:0030501 | positive regulation of bone mineralization | 24 | 32 | 4.78e-02 |
| GO:BP | GO:0042220 | response to cocaine | 24 | 32 | 4.78e-02 |
| GO:BP | GO:0062237 | protein localization to postsynapse | 24 | 32 | 4.78e-02 |
| GO:BP | GO:0070884 | regulation of calcineurin-NFAT signaling cascade | 24 | 32 | 4.78e-02 |
| GO:BP | GO:0106056 | regulation of calcineurin-mediated signaling | 24 | 32 | 4.78e-02 |
| GO:BP | GO:1903539 | protein localization to postsynaptic membrane | 24 | 32 | 4.78e-02 |
| GO:BP | GO:0086004 | regulation of cardiac muscle cell contraction | 24 | 32 | 4.78e-02 |
| GO:BP | GO:0050709 | negative regulation of protein secretion | 32 | 45 | 4.80e-02 |
| GO:BP | GO:0002292 | T cell differentiation involved in immune response | 32 | 45 | 4.80e-02 |
| GO:BP | GO:0060071 | Wnt signaling pathway, planar cell polarity pathway | 32 | 45 | 4.80e-02 |
| GO:BP | GO:0007548 | sex differentiation | 109 | 179 | 4.80e-02 |
| GO:BP | GO:0060997 | dendritic spine morphogenesis | 32 | 45 | 4.80e-02 |
| GO:BP | GO:0046486 | glycerolipid metabolic process | 167 | 284 | 4.81e-02 |
| GO:BP | GO:0051492 | regulation of stress fiber assembly | 51 | 77 | 4.85e-02 |
| GO:BP | GO:0010950 | positive regulation of endopeptidase activity | 62 | 96 | 4.88e-02 |
| GO:BP | GO:0070527 | platelet aggregation | 35 | 50 | 4.88e-02 |
| GO:BP | GO:0051145 | smooth muscle cell differentiation | 38 | 55 | 4.88e-02 |
| GO:BP | GO:1901019 | regulation of calcium ion transmembrane transporter activity | 35 | 50 | 4.88e-02 |
| GO:BP | GO:0031016 | pancreas development | 35 | 50 | 4.88e-02 |
| GO:BP | GO:0030041 | actin filament polymerization | 78 | 124 | 4.90e-02 |
| GO:BP | GO:0046718 | symbiont entry into host cell | 66 | 103 | 4.94e-02 |
| GO:BP | GO:0050954 | sensory perception of mechanical stimulus | 70 | 110 | 4.96e-02 |
ESRnomtable %>%
filter(source=="KEGG", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "KEGG terms found in ESR neargenes") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| KEGG | KEGG:04510 | Focal adhesion | 128 | 169 | 4.34e-09 |
| KEGG | KEGG:05200 | Pathways in cancer | 263 | 391 | 4.34e-09 |
| KEGG | KEGG:04020 | Calcium signaling pathway | 123 | 161 | 4.34e-09 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 121 | 162 | 3.73e-08 |
| KEGG | KEGG:04925 | Aldosterone synthesis and secretion | 64 | 75 | 3.73e-08 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 111 | 147 | 5.35e-08 |
| KEGG | KEGG:04360 | Axon guidance | 114 | 155 | 3.91e-07 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 98 | 133 | 3.28e-06 |
| KEGG | KEGG:04911 | Insulin secretion | 51 | 62 | 1.25e-05 |
| KEGG | KEGG:05414 | Dilated cardiomyopathy | 64 | 82 | 1.36e-05 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 114 | 162 | 1.36e-05 |
| KEGG | KEGG:04970 | Salivary secretion | 45 | 54 | 2.12e-05 |
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 90 | 124 | 2.12e-05 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 93 | 129 | 2.14e-05 |
| KEGG | KEGG:04520 | Adherens junction | 64 | 83 | 2.19e-05 |
| KEGG | KEGG:04724 | Glutamatergic synapse | 58 | 74 | 2.54e-05 |
| KEGG | KEGG:04927 | Cortisol synthesis and secretion | 39 | 46 | 3.73e-05 |
| KEGG | KEGG:04014 | Ras signaling pathway | 113 | 164 | 4.85e-05 |
| KEGG | KEGG:04512 | ECM-receptor interaction | 52 | 66 | 5.79e-05 |
| KEGG | KEGG:04971 | Gastric acid secretion | 42 | 51 | 6.00e-05 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 152 | 231 | 6.12e-05 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 163 | 250 | 6.12e-05 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 91 | 129 | 7.77e-05 |
| KEGG | KEGG:04924 | Renin secretion | 41 | 50 | 8.12e-05 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 76 | 105 | 9.35e-05 |
| KEGG | KEGG:05412 | Arrhythmogenic right ventricular cardiomyopathy | 52 | 67 | 9.52e-05 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 114 | 169 | 1.36e-04 |
| KEGG | KEGG:05410 | Hypertrophic cardiomyopathy | 58 | 77 | 1.36e-04 |
| KEGG | KEGG:04934 | Cushing syndrome | 84 | 119 | 1.36e-04 |
| KEGG | KEGG:04972 | Pancreatic secretion | 45 | 57 | 1.50e-04 |
| KEGG | KEGG:04725 | Cholinergic synapse | 61 | 82 | 1.50e-04 |
| KEGG | KEGG:04921 | Oxytocin signaling pathway | 80 | 113 | 1.66e-04 |
| KEGG | KEGG:04974 | Protein digestion and absorption | 44 | 56 | 2.20e-04 |
| KEGG | KEGG:04611 | Platelet activation | 64 | 88 | 2.84e-04 |
| KEGG | KEGG:04713 | Circadian entrainment | 51 | 68 | 4.58e-04 |
| KEGG | KEGG:04670 | Leukocyte transendothelial migration | 56 | 76 | 4.58e-04 |
| KEGG | KEGG:04750 | Inflammatory mediator regulation of TRP channels | 53 | 72 | 7.33e-04 |
| KEGG | KEGG:05224 | Breast cancer | 78 | 113 | 7.33e-04 |
| KEGG | KEGG:04514 | Cell adhesion molecules | 51 | 69 | 8.01e-04 |
| KEGG | KEGG:04923 | Regulation of lipolysis in adipocytes | 33 | 41 | 8.50e-04 |
| KEGG | KEGG:04072 | Phospholipase D signaling pathway | 74 | 107 | 9.33e-04 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 56 | 78 | 1.27e-03 |
| KEGG | KEGG:04961 | Endocrine and other factor-regulated calcium reabsorption | 32 | 40 | 1.27e-03 |
| KEGG | KEGG:05032 | Morphine addiction | 41 | 54 | 1.28e-03 |
| KEGG | KEGG:05226 | Gastric cancer | 77 | 113 | 1.33e-03 |
| KEGG | KEGG:04916 | Melanogenesis | 54 | 75 | 1.37e-03 |
| KEGG | KEGG:04148 | Efferocytosis | 81 | 120 | 1.41e-03 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 91 | 137 | 1.41e-03 |
| KEGG | KEGG:05218 | Melanoma | 42 | 56 | 1.55e-03 |
| KEGG | KEGG:04728 | Dopaminergic synapse | 67 | 97 | 1.66e-03 |
| KEGG | KEGG:04270 | Vascular smooth muscle contraction | 64 | 92 | 1.66e-03 |
| KEGG | KEGG:04726 | Serotonergic synapse | 43 | 58 | 1.91e-03 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 69 | 101 | 2.16e-03 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 81 | 122 | 2.82e-03 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 70 | 104 | 3.70e-03 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 65 | 96 | 4.41e-03 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 154 | 252 | 4.67e-03 |
| KEGG | KEGG:04213 | Longevity regulating pathway - multiple species | 37 | 50 | 4.67e-03 |
| KEGG | KEGG:04929 | GnRH secretion | 37 | 50 | 4.67e-03 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 102 | 160 | 4.67e-03 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 67 | 100 | 5.25e-03 |
| KEGG | KEGG:04915 | Estrogen signaling pathway | 61 | 90 | 5.53e-03 |
| KEGG | KEGG:04727 | GABAergic synapse | 38 | 52 | 5.68e-03 |
| KEGG | KEGG:04976 | Bile secretion | 31 | 41 | 6.23e-03 |
| KEGG | KEGG:04340 | Hedgehog signaling pathway | 36 | 49 | 6.23e-03 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 59 | 87 | 6.23e-03 |
| KEGG | KEGG:04530 | Tight junction | 83 | 129 | 8.32e-03 |
| KEGG | KEGG:04540 | Gap junction | 49 | 71 | 8.54e-03 |
| KEGG | KEGG:04742 | Taste transduction | 17 | 20 | 9.03e-03 |
| KEGG | KEGG:05230 | Central carbon metabolism in cancer | 41 | 58 | 9.57e-03 |
| KEGG | KEGG:04380 | Osteoclast differentiation | 61 | 92 | 1.11e-02 |
| KEGG | KEGG:05214 | Glioma | 45 | 65 | 1.12e-02 |
| KEGG | KEGG:04060 | Cytokine-cytokine receptor interaction | 62 | 94 | 1.21e-02 |
| KEGG | KEGG:04152 | AMPK signaling pathway | 66 | 101 | 1.25e-02 |
| KEGG | KEGG:05146 | Amoebiasis | 43 | 62 | 1.27e-02 |
| KEGG | KEGG:04931 | Insulin resistance | 60 | 91 | 1.37e-02 |
| KEGG | KEGG:05210 | Colorectal cancer | 54 | 81 | 1.50e-02 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 90 | 144 | 1.65e-02 |
| KEGG | KEGG:04720 | Long-term potentiation | 36 | 51 | 1.65e-02 |
| KEGG | KEGG:04922 | Glucagon signaling pathway | 52 | 78 | 1.68e-02 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 59 | 90 | 1.68e-02 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 50 | 75 | 1.96e-02 |
| KEGG | KEGG:05222 | Small cell lung cancer | 54 | 82 | 2.04e-02 |
| KEGG | KEGG:01522 | Endocrine resistance | 54 | 82 | 2.04e-02 |
| KEGG | KEGG:05031 | Amphetamine addiction | 32 | 45 | 2.11e-02 |
| KEGG | KEGG:04071 | Sphingolipid signaling pathway | 66 | 103 | 2.16e-02 |
| KEGG | KEGG:01521 | EGFR tyrosine kinase inhibitor resistance | 48 | 72 | 2.19e-02 |
| KEGG | KEGG:04973 | Carbohydrate digestion and absorption | 22 | 29 | 2.30e-02 |
| KEGG | KEGG:04066 | HIF-1 signaling pathway | 56 | 86 | 2.36e-02 |
| KEGG | KEGG:00000 | KEGG root term | 2855 | 5451 | 2.47e-02 |
| KEGG | KEGG:00051 | Fructose and mannose metabolism | 20 | 26 | 2.47e-02 |
| KEGG | KEGG:04913 | Ovarian steroidogenesis | 20 | 26 | 2.47e-02 |
| KEGG | KEGG:04730 | Long-term depression | 28 | 39 | 2.65e-02 |
| KEGG | KEGG:05213 | Endometrial cancer | 37 | 54 | 2.68e-02 |
| KEGG | KEGG:04666 | Fc gamma R-mediated phagocytosis | 49 | 75 | 3.31e-02 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 40 | 60 | 3.92e-02 |
| KEGG | KEGG:05131 | Shigellosis | 121 | 204 | 3.93e-02 |
| KEGG | KEGG:05207 | Chemical carcinogenesis - receptor activation | 81 | 132 | 3.98e-02 |
| KEGG | KEGG:04625 | C-type lectin receptor signaling pathway | 48 | 74 | 4.08e-02 |
| KEGG | KEGG:00514 | Other types of O-glycan biosynthesis | 25 | 35 | 4.08e-02 |
| KEGG | KEGG:04912 | GnRH signaling pathway | 48 | 74 | 4.08e-02 |
| KEGG | KEGG:04930 | Type II diabetes mellitus | 25 | 35 | 4.08e-02 |
| KEGG | KEGG:04061 | Viral protein interaction with cytokine and cytokine receptor | 15 | 19 | 4.25e-02 |
| KEGG | KEGG:04614 | Renin-angiotensin system | 9 | 10 | 4.35e-02 |
| KEGG | KEGG:05033 | Nicotine addiction | 9 | 10 | 4.35e-02 |
| KEGG | KEGG:00564 | Glycerophospholipid metabolism | 46 | 71 | 4.58e-02 |
| KEGG | KEGG:05030 | Cocaine addiction | 23 | 32 | 4.59e-02 |
| KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 63 | 101 | 4.76e-02 |
| KEGG | KEGG:05231 | Choline metabolism in cancer | 51 | 80 | 4.91e-02 |
ESRnomtable %>%
filter(source=="GO:BP", p_value < 0.05) %>%
slice_head(n=10) %>%
mutate(log10_p=(-log(p_value, base=10))) %>%
ggplot(., aes(x=log10_p, y=reorder(term_name, log10_p)))+
geom_col(fill="#7CAE00")+
theme_bw()+
ylab("")+
xlab(paste0("-log10 p-value"))+
guides(fill="none")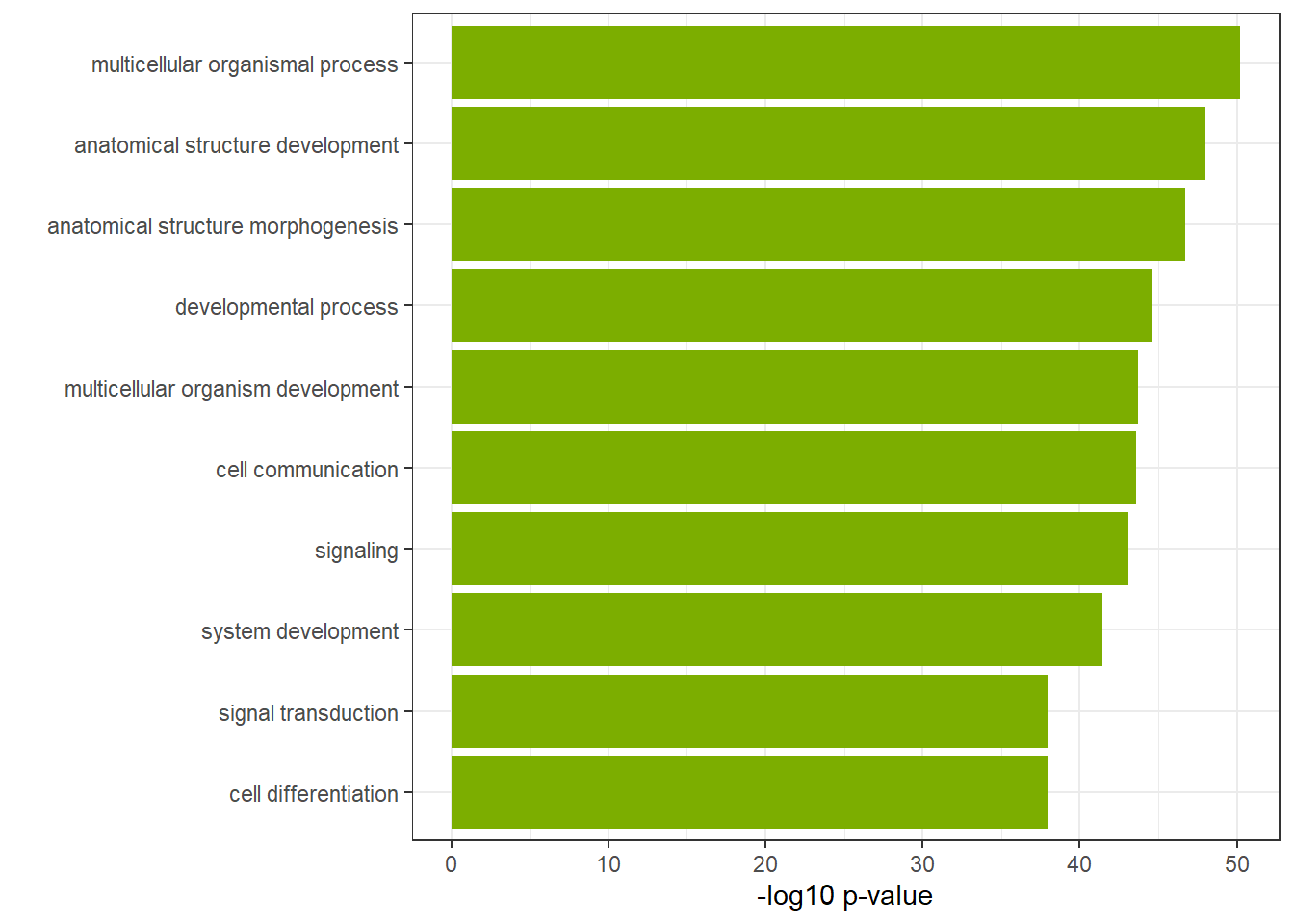
LR response genes
# LR_resgenes <- gost(query = LR_NG_list$ENTREZID,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = FALSE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = background_df$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(LR_resgenes,"data/GO_KEGG_analysis/LR_resgenes.RDS")
# LRnom <- gostplot(LR_resgenes, capped = FALSE, interactive = TRUE)
# LRnom
LRnomtable <- LR_resgenes$result %>%
dplyr::select(c(source, term_id,
term_name,intersection_size,
term_size, p_value))# %>%
LRnomtable %>%
filter(source=="GO:BP", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(.,caption = "GO:BP terms found in LR neargenes" ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0032501 | multicellular organismal process | 3517 | 4504 | 2.85e-37 |
| GO:BP | GO:0048731 | system development | 2194 | 2719 | 1.88e-35 |
| GO:BP | GO:0007154 | cell communication | 3123 | 3983 | 2.81e-34 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 1527 | 1844 | 8.13e-34 |
| GO:BP | GO:0048856 | anatomical structure development | 3035 | 3867 | 8.13e-34 |
| GO:BP | GO:0023052 | signaling | 3073 | 3918 | 8.13e-34 |
| GO:BP | GO:0007275 | multicellular organism development | 2520 | 3170 | 1.90e-33 |
| GO:BP | GO:0032502 | developmental process | 3261 | 4179 | 1.90e-33 |
| GO:BP | GO:0007165 | signal transduction | 2834 | 3611 | 9.26e-31 |
| GO:BP | GO:0050896 | response to stimulus | 4108 | 5379 | 4.91e-29 |
| GO:BP | GO:0051716 | cellular response to stimulus | 3575 | 4640 | 6.17e-29 |
| GO:BP | GO:0007399 | nervous system development | 1449 | 1775 | 5.17e-26 |
| GO:BP | GO:0022008 | neurogenesis | 1048 | 1251 | 8.68e-26 |
| GO:BP | GO:0048869 | cellular developmental process | 2217 | 2804 | 1.25e-25 |
| GO:BP | GO:0030154 | cell differentiation | 2216 | 2803 | 1.35e-25 |
| GO:BP | GO:0048513 | animal organ development | 1616 | 2004 | 1.66e-24 |
| GO:BP | GO:0050793 | regulation of developmental process | 1384 | 1697 | 2.10e-24 |
| GO:BP | GO:0048699 | generation of neurons | 907 | 1076 | 7.45e-24 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 1460 | 1801 | 1.06e-23 |
| GO:BP | GO:0010646 | regulation of cell communication | 1940 | 2444 | 2.20e-23 |
| GO:BP | GO:0030182 | neuron differentiation | 856 | 1014 | 7.74e-23 |
| GO:BP | GO:0023051 | regulation of signaling | 1933 | 2438 | 8.19e-23 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 1547 | 1924 | 2.85e-22 |
| GO:BP | GO:0016477 | cell migration | 848 | 1011 | 7.88e-21 |
| GO:BP | GO:0007155 | cell adhesion | 791 | 941 | 7.79e-20 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 581 | 673 | 1.35e-19 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 2124 | 2717 | 1.94e-19 |
| GO:BP | GO:0009966 | regulation of signal transduction | 1713 | 2166 | 9.59e-19 |
| GO:BP | GO:0048468 | cell development | 1504 | 1886 | 1.08e-18 |
| GO:BP | GO:0048870 | cell motility | 924 | 1120 | 2.51e-18 |
| GO:BP | GO:0009888 | tissue development | 1096 | 1348 | 7.00e-18 |
| GO:BP | GO:0000902 | cell morphogenesis | 607 | 713 | 1.85e-17 |
| GO:BP | GO:0007267 | cell-cell signaling | 892 | 1082 | 1.86e-17 |
| GO:BP | GO:0003008 | system process | 935 | 1139 | 2.58e-17 |
| GO:BP | GO:0040012 | regulation of locomotion | 612 | 720 | 2.58e-17 |
| GO:BP | GO:2000145 | regulation of cell motility | 596 | 700 | 3.50e-17 |
| GO:BP | GO:0072359 | circulatory system development | 723 | 864 | 4.90e-17 |
| GO:BP | GO:0035295 | tube development | 663 | 787 | 6.44e-17 |
| GO:BP | GO:0030334 | regulation of cell migration | 570 | 668 | 7.71e-17 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 760 | 913 | 8.43e-17 |
| GO:BP | GO:0031175 | neuron projection development | 628 | 743 | 1.18e-16 |
| GO:BP | GO:0040011 | locomotion | 676 | 805 | 1.28e-16 |
| GO:BP | GO:0048666 | neuron development | 701 | 838 | 1.92e-16 |
| GO:BP | GO:0035239 | tube morphogenesis | 545 | 638 | 2.70e-16 |
| GO:BP | GO:0032989 | cellular anatomical entity morphogenesis | 505 | 588 | 5.26e-16 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 698 | 837 | 1.02e-15 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 3005 | 3954 | 1.16e-15 |
| GO:BP | GO:0050789 | regulation of biological process | 5544 | 7516 | 1.29e-15 |
| GO:BP | GO:0030030 | cell projection organization | 961 | 1182 | 1.55e-15 |
| GO:BP | GO:0048518 | positive regulation of biological process | 3254 | 4301 | 2.05e-15 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 784 | 951 | 2.95e-15 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 432 | 498 | 3.12e-15 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 935 | 1150 | 4.30e-15 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 603 | 717 | 4.94e-15 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 428 | 494 | 6.88e-15 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 416 | 479 | 7.60e-15 |
| GO:BP | GO:0001568 | blood vessel development | 452 | 525 | 1.08e-14 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 528 | 622 | 1.14e-14 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 874 | 1073 | 1.96e-14 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 517 | 609 | 2.32e-14 |
| GO:BP | GO:0050794 | regulation of cellular process | 5279 | 7153 | 2.55e-14 |
| GO:BP | GO:0001944 | vasculature development | 472 | 552 | 3.18e-14 |
| GO:BP | GO:0065007 | biological regulation | 5684 | 7734 | 8.19e-14 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 845 | 1040 | 2.20e-13 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 1051 | 1314 | 3.49e-13 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 377 | 435 | 4.36e-13 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 840 | 1035 | 4.46e-13 |
| GO:BP | GO:0065008 | regulation of biological quality | 1480 | 1893 | 1.18e-12 |
| GO:BP | GO:0034330 | cell junction organization | 461 | 545 | 3.79e-12 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 1454 | 1862 | 4.53e-12 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 386 | 450 | 6.46e-12 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 496 | 591 | 6.75e-12 |
| GO:BP | GO:0098609 | cell-cell adhesion | 475 | 564 | 6.89e-12 |
| GO:BP | GO:0001503 | ossification | 268 | 302 | 8.52e-12 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 957 | 1197 | 8.53e-12 |
| GO:BP | GO:0048519 | negative regulation of biological process | 2841 | 3764 | 8.92e-12 |
| GO:BP | GO:0048589 | developmental growth | 425 | 501 | 1.43e-11 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 2682 | 3550 | 3.51e-11 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 385 | 452 | 6.08e-11 |
| GO:BP | GO:0072001 | renal system development | 219 | 244 | 1.04e-10 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 428 | 508 | 1.07e-10 |
| GO:BP | GO:0035556 | intracellular signal transduction | 1511 | 1951 | 1.59e-10 |
| GO:BP | GO:0007409 | axonogenesis | 293 | 337 | 2.10e-10 |
| GO:BP | GO:0060429 | epithelium development | 645 | 792 | 2.40e-10 |
| GO:BP | GO:0044057 | regulation of system process | 330 | 384 | 2.44e-10 |
| GO:BP | GO:0007417 | central nervous system development | 570 | 694 | 2.75e-10 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 87 | 88 | 3.70e-10 |
| GO:BP | GO:0001822 | kidney development | 212 | 237 | 5.13e-10 |
| GO:BP | GO:0003013 | circulatory system process | 362 | 426 | 5.59e-10 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 306 | 355 | 6.78e-10 |
| GO:BP | GO:0051960 | regulation of nervous system development | 293 | 339 | 9.50e-10 |
| GO:BP | GO:0061564 | axon development | 319 | 372 | 9.59e-10 |
| GO:BP | GO:0006810 | transport | 2261 | 2984 | 9.59e-10 |
| GO:BP | GO:0051049 | regulation of transport | 873 | 1098 | 1.14e-09 |
| GO:BP | GO:0042221 | response to chemical | 1764 | 2304 | 1.45e-09 |
| GO:BP | GO:0051179 | localization | 2800 | 3731 | 1.60e-09 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 422 | 505 | 1.66e-09 |
| GO:BP | GO:0098657 | import into cell | 512 | 622 | 1.70e-09 |
| GO:BP | GO:0030900 | forebrain development | 239 | 272 | 1.84e-09 |
| GO:BP | GO:0060322 | head development | 448 | 539 | 1.95e-09 |
| GO:BP | GO:0032879 | regulation of localization | 1126 | 1439 | 1.96e-09 |
| GO:BP | GO:0070848 | response to growth factor | 454 | 547 | 2.15e-09 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 307 | 358 | 2.17e-09 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 430 | 516 | 2.26e-09 |
| GO:BP | GO:0001525 | angiogenesis | 325 | 381 | 2.26e-09 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 961 | 1218 | 2.40e-09 |
| GO:BP | GO:0048729 | tissue morphogenesis | 381 | 453 | 2.59e-09 |
| GO:BP | GO:0023056 | positive regulation of signaling | 963 | 1221 | 2.63e-09 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 575 | 706 | 3.23e-09 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 207 | 233 | 3.62e-09 |
| GO:BP | GO:0007423 | sensory organ development | 329 | 387 | 3.96e-09 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 307 | 359 | 4.12e-09 |
| GO:BP | GO:0001501 | skeletal system development | 297 | 347 | 6.89e-09 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 382 | 456 | 6.97e-09 |
| GO:BP | GO:0007420 | brain development | 414 | 498 | 9.84e-09 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 437 | 528 | 1.05e-08 |
| GO:BP | GO:0051234 | establishment of localization | 2438 | 3240 | 1.13e-08 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 228 | 261 | 1.72e-08 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 251 | 290 | 1.77e-08 |
| GO:BP | GO:0061448 | connective tissue development | 183 | 205 | 1.79e-08 |
| GO:BP | GO:0034329 | cell junction assembly | 277 | 323 | 1.82e-08 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 1350 | 1751 | 2.34e-08 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 293 | 344 | 2.79e-08 |
| GO:BP | GO:0008015 | blood circulation | 303 | 357 | 3.22e-08 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 213 | 243 | 3.23e-08 |
| GO:BP | GO:0061061 | muscle structure development | 441 | 536 | 4.28e-08 |
| GO:BP | GO:0071310 | cellular response to organic substance | 1013 | 1297 | 4.30e-08 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 1166 | 1504 | 4.67e-08 |
| GO:BP | GO:0016043 | cellular component organization | 3556 | 4802 | 4.67e-08 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 861 | 1093 | 4.67e-08 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 353 | 422 | 4.79e-08 |
| GO:BP | GO:0006897 | endocytosis | 407 | 492 | 4.79e-08 |
| GO:BP | GO:0007507 | heart development | 412 | 499 | 6.15e-08 |
| GO:BP | GO:0051216 | cartilage development | 133 | 145 | 6.15e-08 |
| GO:BP | GO:0035265 | organ growth | 118 | 127 | 6.64e-08 |
| GO:BP | GO:0030001 | metal ion transport | 431 | 524 | 6.85e-08 |
| GO:BP | GO:0030029 | actin filament-based process | 519 | 640 | 9.17e-08 |
| GO:BP | GO:0006468 | protein phosphorylation | 757 | 958 | 1.97e-07 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 330 | 395 | 2.27e-07 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 340 | 408 | 2.34e-07 |
| GO:BP | GO:0048638 | regulation of developmental growth | 211 | 243 | 2.36e-07 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 141 | 156 | 2.55e-07 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 210 | 242 | 2.87e-07 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 190 | 217 | 3.19e-07 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 184 | 210 | 4.95e-07 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 273 | 323 | 5.14e-07 |
| GO:BP | GO:0016310 | phosphorylation | 894 | 1145 | 5.17e-07 |
| GO:BP | GO:0071417 | cellular response to organonitrogen compound | 365 | 442 | 5.21e-07 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 345 | 416 | 5.21e-07 |
| GO:BP | GO:0043547 | positive regulation of GTPase activity | 105 | 113 | 5.34e-07 |
| GO:BP | GO:0009790 | embryo development | 652 | 821 | 5.82e-07 |
| GO:BP | GO:1903522 | regulation of blood circulation | 171 | 194 | 5.88e-07 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 272 | 322 | 5.89e-07 |
| GO:BP | GO:0045664 | regulation of neuron differentiation | 119 | 130 | 6.08e-07 |
| GO:BP | GO:0009987 | cellular process | 7680 | 10686 | 6.88e-07 |
| GO:BP | GO:0042592 | homeostatic process | 887 | 1137 | 7.63e-07 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 898 | 1152 | 7.94e-07 |
| GO:BP | GO:0060284 | regulation of cell development | 462 | 570 | 7.94e-07 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 308 | 369 | 8.32e-07 |
| GO:BP | GO:0072073 | kidney epithelium development | 103 | 111 | 8.68e-07 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 603 | 757 | 8.81e-07 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 999 | 1289 | 9.23e-07 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 148 | 166 | 9.45e-07 |
| GO:BP | GO:0007264 | small GTPase-mediated signal transduction | 324 | 390 | 9.49e-07 |
| GO:BP | GO:0043010 | camera-type eye development | 199 | 230 | 1.11e-06 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 221 | 258 | 1.20e-06 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 672 | 850 | 1.22e-06 |
| GO:BP | GO:0010243 | response to organonitrogen compound | 548 | 685 | 1.29e-06 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 927 | 1193 | 1.31e-06 |
| GO:BP | GO:0090132 | epithelium migration | 209 | 243 | 1.39e-06 |
| GO:BP | GO:0010631 | epithelial cell migration | 209 | 243 | 1.39e-06 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 133 | 148 | 1.50e-06 |
| GO:BP | GO:0001649 | osteoblast differentiation | 154 | 174 | 1.52e-06 |
| GO:BP | GO:0090130 | tissue migration | 212 | 247 | 1.57e-06 |
| GO:BP | GO:0014706 | striated muscle tissue development | 185 | 213 | 1.76e-06 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 177 | 203 | 1.85e-06 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 382 | 467 | 1.85e-06 |
| GO:BP | GO:0050808 | synapse organization | 282 | 337 | 1.90e-06 |
| GO:BP | GO:0099536 | synaptic signaling | 394 | 483 | 2.08e-06 |
| GO:BP | GO:0040007 | growth | 556 | 697 | 2.08e-06 |
| GO:BP | GO:0023057 | negative regulation of signaling | 809 | 1036 | 2.33e-06 |
| GO:BP | GO:0045165 | cell fate commitment | 135 | 151 | 2.47e-06 |
| GO:BP | GO:0001654 | eye development | 224 | 263 | 2.50e-06 |
| GO:BP | GO:0031214 | biomineral tissue development | 98 | 106 | 2.95e-06 |
| GO:BP | GO:0065009 | regulation of molecular function | 1003 | 1299 | 2.95e-06 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 378 | 463 | 3.20e-06 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 378 | 463 | 3.20e-06 |
| GO:BP | GO:0048880 | sensory system development | 226 | 266 | 3.30e-06 |
| GO:BP | GO:0021537 | telencephalon development | 170 | 195 | 3.35e-06 |
| GO:BP | GO:0008016 | regulation of heart contraction | 146 | 165 | 3.38e-06 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 166 | 190 | 3.38e-06 |
| GO:BP | GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 240 | 284 | 3.39e-06 |
| GO:BP | GO:1901652 | response to peptide | 305 | 368 | 3.54e-06 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 456 | 566 | 3.61e-06 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 351 | 428 | 3.63e-06 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 850 | 1093 | 3.66e-06 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 1968 | 2622 | 3.98e-06 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 806 | 1034 | 4.01e-06 |
| GO:BP | GO:0003007 | heart morphogenesis | 188 | 218 | 4.02e-06 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 433 | 536 | 4.09e-06 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 101 | 110 | 4.40e-06 |
| GO:BP | GO:0150063 | visual system development | 224 | 264 | 4.48e-06 |
| GO:BP | GO:0010720 | positive regulation of cell development | 259 | 309 | 4.57e-06 |
| GO:BP | GO:0030278 | regulation of ossification | 75 | 79 | 4.77e-06 |
| GO:BP | GO:0141091 | transforming growth factor beta receptor superfamily signaling pathway | 220 | 259 | 4.77e-06 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 164 | 188 | 4.82e-06 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 152 | 173 | 4.96e-06 |
| GO:BP | GO:0010033 | response to organic substance | 1322 | 1736 | 5.17e-06 |
| GO:BP | GO:1901698 | response to nitrogen compound | 583 | 736 | 5.60e-06 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 421 | 521 | 5.60e-06 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 759 | 972 | 5.63e-06 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 618 | 783 | 6.01e-06 |
| GO:BP | GO:0006811 | monoatomic ion transport | 570 | 719 | 6.22e-06 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 113 | 125 | 6.45e-06 |
| GO:BP | GO:0006812 | monoatomic cation transport | 491 | 614 | 6.45e-06 |
| GO:BP | GO:0003205 | cardiac chamber development | 134 | 151 | 7.05e-06 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 108 | 119 | 7.29e-06 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 3665 | 4988 | 7.88e-06 |
| GO:BP | GO:0048771 | tissue remodeling | 103 | 113 | 8.09e-06 |
| GO:BP | GO:0008150 | biological_process | 8053 | 11244 | 8.24e-06 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 389 | 480 | 8.92e-06 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 442 | 550 | 9.09e-06 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 376 | 463 | 9.22e-06 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 376 | 463 | 9.22e-06 |
| GO:BP | GO:0060047 | heart contraction | 168 | 194 | 9.64e-06 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 388 | 479 | 1.01e-05 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 317 | 386 | 1.06e-05 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 469 | 587 | 1.45e-05 |
| GO:BP | GO:0007010 | cytoskeleton organization | 885 | 1146 | 1.58e-05 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 173 | 201 | 1.58e-05 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 184 | 215 | 1.61e-05 |
| GO:BP | GO:0097485 | neuron projection guidance | 150 | 172 | 1.61e-05 |
| GO:BP | GO:0007411 | axon guidance | 150 | 172 | 1.61e-05 |
| GO:BP | GO:0043434 | response to peptide hormone | 261 | 314 | 1.62e-05 |
| GO:BP | GO:0035108 | limb morphogenesis | 100 | 110 | 1.62e-05 |
| GO:BP | GO:0035107 | appendage morphogenesis | 100 | 110 | 1.62e-05 |
| GO:BP | GO:0043062 | extracellular structure organization | 191 | 224 | 1.66e-05 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 191 | 224 | 1.66e-05 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 307 | 374 | 1.72e-05 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 187 | 219 | 1.76e-05 |
| GO:BP | GO:0003231 | cardiac ventricle development | 104 | 115 | 1.79e-05 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 250 | 300 | 1.81e-05 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 503 | 633 | 1.84e-05 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 64 | 67 | 1.93e-05 |
| GO:BP | GO:0030198 | extracellular matrix organization | 190 | 223 | 1.95e-05 |
| GO:BP | GO:0050877 | nervous system process | 478 | 600 | 2.01e-05 |
| GO:BP | GO:0007389 | pattern specification process | 239 | 286 | 2.02e-05 |
| GO:BP | GO:0003015 | heart process | 175 | 204 | 2.07e-05 |
| GO:BP | GO:0048568 | embryonic organ development | 252 | 303 | 2.23e-05 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 120 | 135 | 2.28e-05 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 1810 | 2415 | 2.60e-05 |
| GO:BP | GO:0030010 | establishment of cell polarity | 111 | 124 | 2.60e-05 |
| GO:BP | GO:0048588 | developmental cell growth | 155 | 179 | 2.61e-05 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 147 | 169 | 2.79e-05 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 209 | 248 | 2.92e-05 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 398 | 495 | 3.00e-05 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 219 | 261 | 3.14e-05 |
| GO:BP | GO:0048592 | eye morphogenesis | 97 | 107 | 3.16e-05 |
| GO:BP | GO:0035023 | regulation of Rho protein signal transduction | 62 | 65 | 3.31e-05 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 318 | 390 | 3.48e-05 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 232 | 278 | 3.53e-05 |
| GO:BP | GO:0008283 | cell population proliferation | 1000 | 1306 | 3.75e-05 |
| GO:BP | GO:0042391 | regulation of membrane potential | 235 | 282 | 3.76e-05 |
| GO:BP | GO:0090257 | regulation of muscle system process | 153 | 177 | 3.77e-05 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 346 | 427 | 3.77e-05 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 623 | 796 | 3.90e-05 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 1437 | 1904 | 3.96e-05 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 171 | 200 | 4.20e-05 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 76 | 82 | 5.49e-05 |
| GO:BP | GO:0042330 | taxis | 205 | 244 | 5.62e-05 |
| GO:BP | GO:0006935 | chemotaxis | 205 | 244 | 5.62e-05 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 249 | 301 | 5.82e-05 |
| GO:BP | GO:0072006 | nephron development | 103 | 115 | 5.86e-05 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 1420 | 1883 | 6.23e-05 |
| GO:BP | GO:0060537 | muscle tissue development | 278 | 339 | 6.40e-05 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 1640 | 2186 | 6.55e-05 |
| GO:BP | GO:0031399 | regulation of protein modification process | 707 | 911 | 6.56e-05 |
| GO:BP | GO:0003281 | ventricular septum development | 65 | 69 | 6.56e-05 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 80 | 87 | 6.61e-05 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 80 | 87 | 6.61e-05 |
| GO:BP | GO:0045667 | regulation of osteoblast differentiation | 89 | 98 | 7.03e-05 |
| GO:BP | GO:0070167 | regulation of biomineral tissue development | 59 | 62 | 7.48e-05 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 829 | 1077 | 8.19e-05 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 246 | 298 | 9.01e-05 |
| GO:BP | GO:0001823 | mesonephros development | 69 | 74 | 9.01e-05 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 706 | 911 | 9.37e-05 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 1745 | 2333 | 9.39e-05 |
| GO:BP | GO:0046903 | secretion | 491 | 622 | 9.45e-05 |
| GO:BP | GO:0060485 | mesenchyme development | 205 | 245 | 9.67e-05 |
| GO:BP | GO:0003279 | cardiac septum development | 92 | 102 | 9.77e-05 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 184 | 218 | 9.81e-05 |
| GO:BP | GO:0032835 | glomerulus development | 52 | 54 | 1.02e-04 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 335 | 415 | 1.03e-04 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 627 | 805 | 1.08e-04 |
| GO:BP | GO:0055123 | digestive system development | 87 | 96 | 1.11e-04 |
| GO:BP | GO:0072163 | mesonephric epithelium development | 68 | 73 | 1.14e-04 |
| GO:BP | GO:0072164 | mesonephric tubule development | 68 | 73 | 1.14e-04 |
| GO:BP | GO:0000165 | MAPK cascade | 402 | 504 | 1.14e-04 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 626 | 804 | 1.18e-04 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 179 | 212 | 1.25e-04 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 440 | 555 | 1.32e-04 |
| GO:BP | GO:0003012 | muscle system process | 266 | 325 | 1.35e-04 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 86 | 95 | 1.39e-04 |
| GO:BP | GO:0030282 | bone mineralization | 72 | 78 | 1.44e-04 |
| GO:BP | GO:0140352 | export from cell | 469 | 594 | 1.45e-04 |
| GO:BP | GO:0001657 | ureteric bud development | 67 | 72 | 1.46e-04 |
| GO:BP | GO:0007610 | behavior | 338 | 420 | 1.53e-04 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 156 | 183 | 1.62e-04 |
| GO:BP | GO:0014855 | striated muscle cell proliferation | 56 | 59 | 1.66e-04 |
| GO:BP | GO:0001763 | morphogenesis of a branching structure | 126 | 145 | 1.68e-04 |
| GO:BP | GO:0010463 | mesenchymal cell proliferation | 36 | 36 | 1.79e-04 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 346 | 431 | 1.79e-04 |
| GO:BP | GO:0050807 | regulation of synapse organization | 148 | 173 | 1.83e-04 |
| GO:BP | GO:0030100 | regulation of endocytosis | 166 | 196 | 1.84e-04 |
| GO:BP | GO:0007517 | muscle organ development | 221 | 267 | 1.84e-04 |
| GO:BP | GO:0003002 | regionalization | 214 | 258 | 1.92e-04 |
| GO:BP | GO:0048565 | digestive tract development | 80 | 88 | 1.94e-04 |
| GO:BP | GO:0007259 | receptor signaling pathway via JAK-STAT | 80 | 88 | 1.94e-04 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 80 | 88 | 1.94e-04 |
| GO:BP | GO:0048167 | regulation of synaptic plasticity | 121 | 139 | 2.06e-04 |
| GO:BP | GO:0051961 | negative regulation of nervous system development | 97 | 109 | 2.06e-04 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 97 | 109 | 2.06e-04 |
| GO:BP | GO:0009605 | response to external stimulus | 1179 | 1559 | 2.07e-04 |
| GO:BP | GO:1990138 | neuron projection extension | 117 | 134 | 2.10e-04 |
| GO:BP | GO:0001764 | neuron migration | 113 | 129 | 2.13e-04 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 432 | 546 | 2.23e-04 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 319 | 396 | 2.28e-04 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 60 | 64 | 2.33e-04 |
| GO:BP | GO:0046622 | positive regulation of organ growth | 35 | 35 | 2.39e-04 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 150 | 176 | 2.39e-04 |
| GO:BP | GO:0048593 | camera-type eye morphogenesis | 79 | 87 | 2.41e-04 |
| GO:BP | GO:0050768 | negative regulation of neurogenesis | 92 | 103 | 2.42e-04 |
| GO:BP | GO:0007162 | negative regulation of cell adhesion | 157 | 185 | 2.46e-04 |
| GO:BP | GO:0051641 | cellular localization | 1950 | 2623 | 2.46e-04 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 232 | 282 | 2.48e-04 |
| GO:BP | GO:0050920 | regulation of chemotaxis | 116 | 133 | 2.51e-04 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 235 | 286 | 2.58e-04 |
| GO:BP | GO:0033002 | muscle cell proliferation | 131 | 152 | 2.62e-04 |
| GO:BP | GO:1901653 | cellular response to peptide | 225 | 273 | 2.62e-04 |
| GO:BP | GO:0045666 | positive regulation of neuron differentiation | 54 | 57 | 2.73e-04 |
| GO:BP | GO:0055085 | transmembrane transport | 710 | 921 | 2.77e-04 |
| GO:BP | GO:0042063 | gliogenesis | 201 | 242 | 2.88e-04 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 156 | 184 | 2.88e-04 |
| GO:BP | GO:0061035 | regulation of cartilage development | 48 | 50 | 2.96e-04 |
| GO:BP | GO:0007229 | integrin-mediated signaling pathway | 78 | 86 | 2.97e-04 |
| GO:BP | GO:0009725 | response to hormone | 479 | 610 | 3.10e-04 |
| GO:BP | GO:0016331 | morphogenesis of embryonic epithelium | 107 | 122 | 3.12e-04 |
| GO:BP | GO:0032330 | regulation of chondrocyte differentiation | 34 | 34 | 3.19e-04 |
| GO:BP | GO:0097696 | receptor signaling pathway via STAT | 82 | 91 | 3.25e-04 |
| GO:BP | GO:0016358 | dendrite development | 155 | 183 | 3.40e-04 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 176 | 210 | 3.40e-04 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 206 | 249 | 3.70e-04 |
| GO:BP | GO:0060993 | kidney morphogenesis | 63 | 68 | 3.81e-04 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 245 | 300 | 3.88e-04 |
| GO:BP | GO:0010721 | negative regulation of cell development | 154 | 182 | 4.03e-04 |
| GO:BP | GO:0001952 | regulation of cell-matrix adhesion | 81 | 90 | 4.08e-04 |
| GO:BP | GO:0061138 | morphogenesis of a branching epithelium | 117 | 135 | 4.31e-04 |
| GO:BP | GO:0032940 | secretion by cell | 434 | 551 | 4.55e-04 |
| GO:BP | GO:0051057 | positive regulation of small GTPase mediated signal transduction | 52 | 55 | 4.65e-04 |
| GO:BP | GO:0007015 | actin filament organization | 288 | 357 | 4.70e-04 |
| GO:BP | GO:0072009 | nephron epithelium development | 76 | 84 | 4.70e-04 |
| GO:BP | GO:0001656 | metanephros development | 57 | 61 | 4.93e-04 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 532 | 683 | 5.17e-04 |
| GO:BP | GO:0030500 | regulation of bone mineralization | 46 | 48 | 5.17e-04 |
| GO:BP | GO:0003018 | vascular process in circulatory system | 163 | 194 | 5.18e-04 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 145 | 171 | 5.55e-04 |
| GO:BP | GO:1901342 | regulation of vasculature development | 166 | 198 | 5.59e-04 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 88 | 99 | 5.59e-04 |
| GO:BP | GO:0007266 | Rho protein signal transduction | 96 | 109 | 5.76e-04 |
| GO:BP | GO:0001775 | cell activation | 490 | 627 | 5.92e-04 |
| GO:BP | GO:0045216 | cell-cell junction organization | 130 | 152 | 6.10e-04 |
| GO:BP | GO:0031032 | actomyosin structure organization | 155 | 184 | 6.10e-04 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 56 | 60 | 6.33e-04 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 137 | 161 | 6.34e-04 |
| GO:BP | GO:0002521 | leukocyte differentiation | 298 | 371 | 6.34e-04 |
| GO:BP | GO:1903037 | regulation of leukocyte cell-cell adhesion | 165 | 197 | 6.49e-04 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 39 | 40 | 6.49e-04 |
| GO:BP | GO:0051050 | positive regulation of transport | 448 | 571 | 6.53e-04 |
| GO:BP | GO:0048041 | focal adhesion assembly | 70 | 77 | 6.59e-04 |
| GO:BP | GO:0021987 | cerebral cortex development | 83 | 93 | 6.59e-04 |
| GO:BP | GO:0006816 | calcium ion transport | 215 | 262 | 6.66e-04 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 218 | 266 | 6.91e-04 |
| GO:BP | GO:0043542 | endothelial cell migration | 147 | 174 | 7.07e-04 |
| GO:BP | GO:0043583 | ear development | 129 | 151 | 7.16e-04 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 554 | 714 | 7.24e-04 |
| GO:BP | GO:0060419 | heart growth | 65 | 71 | 7.27e-04 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 143 | 169 | 7.61e-04 |
| GO:BP | GO:0060043 | regulation of cardiac muscle cell proliferation | 31 | 31 | 8.05e-04 |
| GO:BP | GO:0018212 | peptidyl-tyrosine modification | 139 | 164 | 8.23e-04 |
| GO:BP | GO:0006898 | receptor-mediated endocytosis | 153 | 182 | 8.31e-04 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 197 | 239 | 8.31e-04 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 280 | 348 | 8.43e-04 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 117 | 136 | 8.45e-04 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 344 | 433 | 8.75e-04 |
| GO:BP | GO:0046620 | regulation of organ growth | 64 | 70 | 9.22e-04 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 310 | 388 | 9.34e-04 |
| GO:BP | GO:0050772 | positive regulation of axonogenesis | 59 | 64 | 1.00e-03 |
| GO:BP | GO:0048645 | animal organ formation | 49 | 52 | 1.00e-03 |
| GO:BP | GO:0021543 | pallium development | 116 | 135 | 1.02e-03 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 162 | 194 | 1.03e-03 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 303 | 379 | 1.03e-03 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 234 | 288 | 1.08e-03 |
| GO:BP | GO:0098900 | regulation of action potential | 37 | 38 | 1.15e-03 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 137 | 162 | 1.15e-03 |
| GO:BP | GO:0018108 | peptidyl-tyrosine phosphorylation | 137 | 162 | 1.15e-03 |
| GO:BP | GO:0061005 | cell differentiation involved in kidney development | 37 | 38 | 1.15e-03 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 76 | 85 | 1.20e-03 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 161 | 193 | 1.20e-03 |
| GO:BP | GO:0032409 | regulation of transporter activity | 126 | 148 | 1.20e-03 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 96 | 110 | 1.23e-03 |
| GO:BP | GO:0043588 | skin development | 147 | 175 | 1.24e-03 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 499 | 642 | 1.27e-03 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 174 | 210 | 1.29e-03 |
| GO:BP | GO:0010633 | negative regulation of epithelial cell migration | 48 | 51 | 1.29e-03 |
| GO:BP | GO:0061326 | renal tubule development | 67 | 74 | 1.29e-03 |
| GO:BP | GO:0021954 | central nervous system neuron development | 53 | 57 | 1.31e-03 |
| GO:BP | GO:0035148 | tube formation | 107 | 124 | 1.33e-03 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 103 | 119 | 1.39e-03 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 190 | 231 | 1.39e-03 |
| GO:BP | GO:0042692 | muscle cell differentiation | 260 | 323 | 1.43e-03 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 1197 | 1595 | 1.44e-03 |
| GO:BP | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules | 29 | 29 | 1.47e-03 |
| GO:BP | GO:0044409 | symbiont entry into host | 95 | 109 | 1.48e-03 |
| GO:BP | GO:0055021 | regulation of cardiac muscle tissue growth | 42 | 44 | 1.49e-03 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 471 | 605 | 1.52e-03 |
| GO:BP | GO:0006814 | sodium ion transport | 117 | 137 | 1.61e-03 |
| GO:BP | GO:0071772 | response to BMP | 102 | 118 | 1.67e-03 |
| GO:BP | GO:0060348 | bone development | 124 | 146 | 1.67e-03 |
| GO:BP | GO:0048008 | platelet-derived growth factor receptor signaling pathway | 47 | 50 | 1.67e-03 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 102 | 118 | 1.67e-03 |
| GO:BP | GO:0050900 | leukocyte migration | 162 | 195 | 1.70e-03 |
| GO:BP | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | 98 | 113 | 1.73e-03 |
| GO:BP | GO:1901343 | negative regulation of vasculature development | 61 | 67 | 1.82e-03 |
| GO:BP | GO:2000181 | negative regulation of blood vessel morphogenesis | 61 | 67 | 1.82e-03 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 61 | 67 | 1.82e-03 |
| GO:BP | GO:0022612 | gland morphogenesis | 82 | 93 | 1.87e-03 |
| GO:BP | GO:0007416 | synapse assembly | 116 | 136 | 1.91e-03 |
| GO:BP | GO:0045665 | negative regulation of neuron differentiation | 41 | 43 | 1.95e-03 |
| GO:BP | GO:0060038 | cardiac muscle cell proliferation | 41 | 43 | 1.95e-03 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 369 | 469 | 1.98e-03 |
| GO:BP | GO:0060421 | positive regulation of heart growth | 28 | 28 | 1.99e-03 |
| GO:BP | GO:0072175 | epithelial tube formation | 101 | 117 | 1.99e-03 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 65 | 72 | 1.99e-03 |
| GO:BP | GO:0061614 | miRNA transcription | 56 | 61 | 1.99e-03 |
| GO:BP | GO:0060420 | regulation of heart growth | 46 | 49 | 2.11e-03 |
| GO:BP | GO:0090109 | regulation of cell-substrate junction assembly | 51 | 55 | 2.11e-03 |
| GO:BP | GO:1903053 | regulation of extracellular matrix organization | 46 | 49 | 2.11e-03 |
| GO:BP | GO:0051893 | regulation of focal adhesion assembly | 51 | 55 | 2.11e-03 |
| GO:BP | GO:0030048 | actin filament-based movement | 93 | 107 | 2.14e-03 |
| GO:BP | GO:0030097 | hemopoiesis | 484 | 624 | 2.20e-03 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 225 | 278 | 2.21e-03 |
| GO:BP | GO:0110020 | regulation of actomyosin structure organization | 77 | 87 | 2.24e-03 |
| GO:BP | GO:0010586 | miRNA metabolic process | 77 | 87 | 2.24e-03 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 302 | 380 | 2.24e-03 |
| GO:BP | GO:0016525 | negative regulation of angiogenesis | 60 | 66 | 2.25e-03 |
| GO:BP | GO:0060541 | respiratory system development | 136 | 162 | 2.30e-03 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 173 | 210 | 2.34e-03 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 156 | 188 | 2.47e-03 |
| GO:BP | GO:0022898 | regulation of transmembrane transporter activity | 118 | 139 | 2.47e-03 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 682 | 893 | 2.49e-03 |
| GO:BP | GO:0150116 | regulation of cell-substrate junction organization | 55 | 60 | 2.49e-03 |
| GO:BP | GO:0002064 | epithelial cell development | 125 | 148 | 2.49e-03 |
| GO:BP | GO:0071407 | cellular response to organic cyclic compound | 301 | 379 | 2.49e-03 |
| GO:BP | GO:0008360 | regulation of cell shape | 92 | 106 | 2.55e-03 |
| GO:BP | GO:0051963 | regulation of synapse assembly | 68 | 76 | 2.59e-03 |
| GO:BP | GO:1901890 | positive regulation of cell junction assembly | 68 | 76 | 2.59e-03 |
| GO:BP | GO:0045663 | positive regulation of myoblast differentiation | 34 | 35 | 2.59e-03 |
| GO:BP | GO:0032412 | regulation of monoatomic ion transmembrane transporter activity | 114 | 134 | 2.62e-03 |
| GO:BP | GO:0098901 | regulation of cardiac muscle cell action potential | 27 | 27 | 2.64e-03 |
| GO:BP | GO:0010464 | regulation of mesenchymal cell proliferation | 27 | 27 | 2.64e-03 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 72 | 81 | 2.66e-03 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 110 | 129 | 2.79e-03 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 59 | 65 | 2.79e-03 |
| GO:BP | GO:0008361 | regulation of cell size | 124 | 147 | 2.92e-03 |
| GO:BP | GO:0030509 | BMP signaling pathway | 95 | 110 | 2.94e-03 |
| GO:BP | GO:0072080 | nephron tubule development | 63 | 70 | 3.04e-03 |
| GO:BP | GO:0046849 | bone remodeling | 54 | 59 | 3.14e-03 |
| GO:BP | GO:0071695 | anatomical structure maturation | 127 | 151 | 3.16e-03 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 505 | 654 | 3.25e-03 |
| GO:BP | GO:0060840 | artery development | 75 | 85 | 3.32e-03 |
| GO:BP | GO:0061337 | cardiac conduction | 75 | 85 | 3.32e-03 |
| GO:BP | GO:0033135 | regulation of peptidyl-serine phosphorylation | 75 | 85 | 3.32e-03 |
| GO:BP | GO:1903039 | positive regulation of leukocyte cell-cell adhesion | 123 | 146 | 3.43e-03 |
| GO:BP | GO:0061572 | actin filament bundle organization | 112 | 132 | 3.69e-03 |
| GO:BP | GO:0036211 | protein modification process | 1696 | 2291 | 3.69e-03 |
| GO:BP | GO:0014013 | regulation of gliogenesis | 62 | 69 | 3.80e-03 |
| GO:BP | GO:0043270 | positive regulation of monoatomic ion transport | 108 | 127 | 3.95e-03 |
| GO:BP | GO:0061333 | renal tubule morphogenesis | 53 | 58 | 3.95e-03 |
| GO:BP | GO:0072028 | nephron morphogenesis | 53 | 58 | 3.95e-03 |
| GO:BP | GO:0048864 | stem cell development | 66 | 74 | 3.95e-03 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 108 | 127 | 3.95e-03 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 66 | 74 | 3.95e-03 |
| GO:BP | GO:0098727 | maintenance of cell number | 122 | 145 | 4.00e-03 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 122 | 145 | 4.00e-03 |
| GO:BP | GO:0031623 | receptor internalization | 78 | 89 | 4.00e-03 |
| GO:BP | GO:0060173 | limb development | 115 | 136 | 4.00e-03 |
| GO:BP | GO:0048736 | appendage development | 115 | 136 | 4.00e-03 |
| GO:BP | GO:0042476 | odontogenesis | 70 | 79 | 4.00e-03 |
| GO:BP | GO:0048878 | chemical homeostasis | 516 | 670 | 4.18e-03 |
| GO:BP | GO:0000003 | reproduction | 672 | 882 | 4.19e-03 |
| GO:BP | GO:0045669 | positive regulation of osteoblast differentiation | 48 | 52 | 4.27e-03 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 111 | 131 | 4.30e-03 |
| GO:BP | GO:0046718 | symbiont entry into host cell | 89 | 103 | 4.40e-03 |
| GO:BP | GO:0060350 | endochondral bone morphogenesis | 43 | 46 | 4.41e-03 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 107 | 126 | 4.64e-03 |
| GO:BP | GO:0030324 | lung development | 121 | 144 | 4.68e-03 |
| GO:BP | GO:0031099 | regeneration | 121 | 144 | 4.68e-03 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 65 | 73 | 4.84e-03 |
| GO:BP | GO:0060349 | bone morphogenesis | 65 | 73 | 4.84e-03 |
| GO:BP | GO:0055023 | positive regulation of cardiac muscle tissue growth | 25 | 25 | 4.84e-03 |
| GO:BP | GO:0055006 | cardiac cell development | 69 | 78 | 4.91e-03 |
| GO:BP | GO:0050890 | cognition | 174 | 213 | 4.91e-03 |
| GO:BP | GO:0030323 | respiratory tube development | 124 | 148 | 5.02e-03 |
| GO:BP | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 127 | 152 | 5.40e-03 |
| GO:BP | GO:0042733 | embryonic digit morphogenesis | 47 | 51 | 5.44e-03 |
| GO:BP | GO:0003208 | cardiac ventricle morphogenesis | 56 | 62 | 5.44e-03 |
| GO:BP | GO:0045778 | positive regulation of ossification | 37 | 39 | 5.46e-03 |
| GO:BP | GO:0019827 | stem cell population maintenance | 120 | 143 | 5.48e-03 |
| GO:BP | GO:0040008 | regulation of growth | 361 | 462 | 5.58e-03 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 780 | 1031 | 5.58e-03 |
| GO:BP | GO:0023061 | signal release | 251 | 315 | 5.62e-03 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 571 | 746 | 5.64e-03 |
| GO:BP | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 80 | 92 | 5.70e-03 |
| GO:BP | GO:0071805 | potassium ion transmembrane transport | 102 | 120 | 5.86e-03 |
| GO:BP | GO:0086004 | regulation of cardiac muscle cell contraction | 31 | 32 | 5.94e-03 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 91 | 106 | 5.94e-03 |
| GO:BP | GO:0035850 | epithelial cell differentiation involved in kidney development | 31 | 32 | 5.94e-03 |
| GO:BP | GO:0007043 | cell-cell junction assembly | 91 | 106 | 5.94e-03 |
| GO:BP | GO:0014033 | neural crest cell differentiation | 68 | 77 | 5.96e-03 |
| GO:BP | GO:0072088 | nephron epithelium morphogenesis | 51 | 56 | 6.14e-03 |
| GO:BP | GO:0048708 | astrocyte differentiation | 51 | 56 | 6.14e-03 |
| GO:BP | GO:0010817 | regulation of hormone levels | 253 | 318 | 6.26e-03 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 443 | 573 | 6.28e-03 |
| GO:BP | GO:0001894 | tissue homeostasis | 136 | 164 | 6.30e-03 |
| GO:BP | GO:0060249 | anatomical structure homeostasis | 136 | 164 | 6.30e-03 |
| GO:BP | GO:0046777 | protein autophosphorylation | 119 | 142 | 6.32e-03 |
| GO:BP | GO:0048839 | inner ear development | 112 | 133 | 6.38e-03 |
| GO:BP | GO:0071875 | adrenergic receptor signaling pathway | 24 | 24 | 6.47e-03 |
| GO:BP | GO:0098868 | bone growth | 24 | 24 | 6.47e-03 |
| GO:BP | GO:0010001 | glial cell differentiation | 149 | 181 | 6.53e-03 |
| GO:BP | GO:0048469 | cell maturation | 94 | 110 | 6.60e-03 |
| GO:BP | GO:0031016 | pancreas development | 46 | 50 | 6.74e-03 |
| GO:BP | GO:2000630 | positive regulation of miRNA metabolic process | 46 | 50 | 6.74e-03 |
| GO:BP | GO:0030038 | contractile actin filament bundle assembly | 79 | 91 | 6.76e-03 |
| GO:BP | GO:0043149 | stress fiber assembly | 79 | 91 | 6.76e-03 |
| GO:BP | GO:0007272 | ensheathment of neurons | 101 | 119 | 6.78e-03 |
| GO:BP | GO:0008366 | axon ensheathment | 101 | 119 | 6.78e-03 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 115 | 137 | 6.81e-03 |
| GO:BP | GO:0010596 | negative regulation of endothelial cell migration | 36 | 38 | 6.91e-03 |
| GO:BP | GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 75 | 86 | 6.95e-03 |
| GO:BP | GO:0060041 | retina development in camera-type eye | 90 | 105 | 6.95e-03 |
| GO:BP | GO:1902895 | positive regulation of miRNA transcription | 41 | 44 | 7.04e-03 |
| GO:BP | GO:0034103 | regulation of tissue remodeling | 41 | 44 | 7.04e-03 |
| GO:BP | GO:0022414 | reproductive process | 664 | 874 | 7.20e-03 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 800 | 1060 | 7.23e-03 |
| GO:BP | GO:0060341 | regulation of cellular localization | 592 | 776 | 7.41e-03 |
| GO:BP | GO:0003179 | heart valve morphogenesis | 50 | 55 | 7.57e-03 |
| GO:BP | GO:0048732 | gland development | 245 | 308 | 7.60e-03 |
| GO:BP | GO:0021955 | central nervous system neuron axonogenesis | 30 | 31 | 7.69e-03 |
| GO:BP | GO:0035725 | sodium ion transmembrane transport | 82 | 95 | 7.72e-03 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 93 | 109 | 7.74e-03 |
| GO:BP | GO:0061458 | reproductive system development | 167 | 205 | 7.87e-03 |
| GO:BP | GO:0008544 | epidermis development | 170 | 209 | 8.09e-03 |
| GO:BP | GO:0021675 | nerve development | 54 | 60 | 8.17e-03 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 229 | 287 | 8.29e-03 |
| GO:BP | GO:0032231 | regulation of actin filament bundle assembly | 74 | 85 | 8.37e-03 |
| GO:BP | GO:0007369 | gastrulation | 117 | 140 | 8.48e-03 |
| GO:BP | GO:0003170 | heart valve development | 58 | 65 | 8.52e-03 |
| GO:BP | GO:0060291 | long-term synaptic potentiation | 58 | 65 | 8.52e-03 |
| GO:BP | GO:0048754 | branching morphogenesis of an epithelial tube | 96 | 113 | 8.54e-03 |
| GO:BP | GO:0051924 | regulation of calcium ion transport | 127 | 153 | 8.56e-03 |
| GO:BP | GO:0061036 | positive regulation of cartilage development | 23 | 23 | 8.56e-03 |
| GO:BP | GO:0003416 | endochondral bone growth | 23 | 23 | 8.56e-03 |
| GO:BP | GO:0002685 | regulation of leukocyte migration | 103 | 122 | 8.56e-03 |
| GO:BP | GO:0002063 | chondrocyte development | 23 | 23 | 8.56e-03 |
| GO:BP | GO:0060037 | pharyngeal system development | 23 | 23 | 8.56e-03 |
| GO:BP | GO:0097553 | calcium ion transmembrane import into cytosol | 103 | 122 | 8.56e-03 |
| GO:BP | GO:0014896 | muscle hypertrophy | 66 | 75 | 8.57e-03 |
| GO:BP | GO:0090288 | negative regulation of cellular response to growth factor stimulus | 66 | 75 | 8.57e-03 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 1366 | 1841 | 8.68e-03 |
| GO:BP | GO:1903115 | regulation of actin filament-based movement | 35 | 37 | 8.77e-03 |
| GO:BP | GO:0002090 | regulation of receptor internalization | 40 | 43 | 8.85e-03 |
| GO:BP | GO:0045321 | leukocyte activation | 410 | 530 | 8.87e-03 |
| GO:BP | GO:0006813 | potassium ion transport | 113 | 135 | 9.08e-03 |
| GO:BP | GO:0050870 | positive regulation of T cell activation | 113 | 135 | 9.08e-03 |
| GO:BP | GO:0051051 | negative regulation of transport | 228 | 286 | 9.08e-03 |
| GO:BP | GO:0042552 | myelination | 99 | 117 | 9.19e-03 |
| GO:BP | GO:0014910 | regulation of smooth muscle cell migration | 49 | 54 | 9.22e-03 |
| GO:BP | GO:0072078 | nephron tubule morphogenesis | 49 | 54 | 9.22e-03 |
| GO:BP | GO:0042475 | odontogenesis of dentin-containing tooth | 49 | 54 | 9.22e-03 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 175 | 216 | 9.37e-03 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 200 | 249 | 9.41e-03 |
| GO:BP | GO:0001659 | temperature homeostasis | 109 | 130 | 9.82e-03 |
| GO:BP | GO:0009612 | response to mechanical stimulus | 126 | 152 | 9.82e-03 |
| GO:BP | GO:0035270 | endocrine system development | 73 | 84 | 9.82e-03 |
| GO:BP | GO:0034764 | positive regulation of transmembrane transport | 109 | 130 | 9.82e-03 |
| GO:BP | GO:0048562 | embryonic organ morphogenesis | 149 | 182 | 9.82e-03 |
| GO:BP | GO:0048546 | digestive tract morphogenesis | 29 | 30 | 9.82e-03 |
| GO:BP | GO:0046850 | regulation of bone remodeling | 29 | 30 | 9.82e-03 |
| GO:BP | GO:0021700 | developmental maturation | 165 | 203 | 9.84e-03 |
| GO:BP | GO:1990845 | adaptive thermogenesis | 102 | 121 | 9.84e-03 |
| GO:BP | GO:0009952 | anterior/posterior pattern specification | 102 | 121 | 9.84e-03 |
| GO:BP | GO:0050867 | positive regulation of cell activation | 165 | 203 | 9.84e-03 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 328 | 420 | 9.87e-03 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 1376 | 1856 | 9.87e-03 |
| GO:BP | GO:0048259 | regulation of receptor-mediated endocytosis | 69 | 79 | 1.00e-02 |
| GO:BP | GO:0050680 | negative regulation of epithelial cell proliferation | 84 | 98 | 1.00e-02 |
| GO:BP | GO:0030031 | cell projection assembly | 348 | 447 | 1.01e-02 |
| GO:BP | GO:0042110 | T cell activation | 242 | 305 | 1.01e-02 |
| GO:BP | GO:0048644 | muscle organ morphogenesis | 57 | 64 | 1.01e-02 |
| GO:BP | GO:0010718 | positive regulation of epithelial to mesenchymal transition | 44 | 48 | 1.02e-02 |
| GO:BP | GO:0014032 | neural crest cell development | 61 | 69 | 1.02e-02 |
| GO:BP | GO:0001755 | neural crest cell migration | 44 | 48 | 1.02e-02 |
| GO:BP | GO:0071396 | cellular response to lipid | 304 | 388 | 1.04e-02 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 98 | 116 | 1.05e-02 |
| GO:BP | GO:0099173 | postsynapse organization | 122 | 147 | 1.05e-02 |
| GO:BP | GO:0045598 | regulation of fat cell differentiation | 80 | 93 | 1.05e-02 |
| GO:BP | GO:0033043 | regulation of organelle organization | 720 | 953 | 1.07e-02 |
| GO:BP | GO:0032880 | regulation of protein localization | 528 | 691 | 1.08e-02 |
| GO:BP | GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | 115 | 138 | 1.09e-02 |
| GO:BP | GO:0031641 | regulation of myelination | 34 | 36 | 1.10e-02 |
| GO:BP | GO:0003203 | endocardial cushion morphogenesis | 34 | 36 | 1.10e-02 |
| GO:BP | GO:0048608 | reproductive structure development | 164 | 202 | 1.10e-02 |
| GO:BP | GO:0033036 | macromolecule localization | 1702 | 2309 | 1.10e-02 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 164 | 202 | 1.10e-02 |
| GO:BP | GO:0003177 | pulmonary valve development | 22 | 22 | 1.11e-02 |
| GO:BP | GO:0060045 | positive regulation of cardiac muscle cell proliferation | 22 | 22 | 1.11e-02 |
| GO:BP | GO:0050921 | positive regulation of chemotaxis | 72 | 83 | 1.15e-02 |
| GO:BP | GO:0050679 | positive regulation of epithelial cell proliferation | 118 | 142 | 1.15e-02 |
| GO:BP | GO:0007611 | learning or memory | 154 | 189 | 1.16e-02 |
| GO:BP | GO:0051235 | maintenance of location | 201 | 251 | 1.16e-02 |
| GO:BP | GO:0006936 | muscle contraction | 204 | 255 | 1.17e-02 |
| GO:BP | GO:0002028 | regulation of sodium ion transport | 52 | 58 | 1.18e-02 |
| GO:BP | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 52 | 58 | 1.18e-02 |
| GO:BP | GO:0021915 | neural tube development | 111 | 133 | 1.18e-02 |
| GO:BP | GO:0045661 | regulation of myoblast differentiation | 52 | 58 | 1.18e-02 |
| GO:BP | GO:0050865 | regulation of cell activation | 273 | 347 | 1.19e-02 |
| GO:BP | GO:0002696 | positive regulation of leukocyte activation | 160 | 197 | 1.21e-02 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 340 | 437 | 1.21e-02 |
| GO:BP | GO:0007492 | endoderm development | 56 | 63 | 1.21e-02 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 56 | 63 | 1.21e-02 |
| GO:BP | GO:0032535 | regulation of cellular component size | 228 | 287 | 1.21e-02 |
| GO:BP | GO:0010232 | vascular transport | 60 | 68 | 1.21e-02 |
| GO:BP | GO:0019915 | lipid storage | 60 | 68 | 1.21e-02 |
| GO:BP | GO:0043502 | regulation of muscle adaptation | 60 | 68 | 1.21e-02 |
| GO:BP | GO:0150104 | transport across blood-brain barrier | 60 | 68 | 1.21e-02 |
| GO:BP | GO:0071675 | regulation of mononuclear cell migration | 60 | 68 | 1.21e-02 |
| GO:BP | GO:0150117 | positive regulation of cell-substrate junction organization | 28 | 29 | 1.24e-02 |
| GO:BP | GO:0060563 | neuroepithelial cell differentiation | 28 | 29 | 1.24e-02 |
| GO:BP | GO:1990776 | response to angiotensin | 28 | 29 | 1.24e-02 |
| GO:BP | GO:0002067 | glandular epithelial cell differentiation | 43 | 47 | 1.24e-02 |
| GO:BP | GO:0014743 | regulation of muscle hypertrophy | 43 | 47 | 1.24e-02 |
| GO:BP | GO:0014015 | positive regulation of gliogenesis | 43 | 47 | 1.24e-02 |
| GO:BP | GO:0045860 | positive regulation of protein kinase activity | 150 | 184 | 1.26e-02 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 75 | 87 | 1.29e-02 |
| GO:BP | GO:0008104 | protein localization | 1478 | 2000 | 1.34e-02 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 427 | 555 | 1.34e-02 |
| GO:BP | GO:1905314 | semi-lunar valve development | 38 | 41 | 1.35e-02 |
| GO:BP | GO:0022409 | positive regulation of cell-cell adhesion | 143 | 175 | 1.35e-02 |
| GO:BP | GO:1905330 | regulation of morphogenesis of an epithelium | 38 | 41 | 1.35e-02 |
| GO:BP | GO:0010171 | body morphogenesis | 38 | 41 | 1.35e-02 |
| GO:BP | GO:0001936 | regulation of endothelial cell proliferation | 82 | 96 | 1.37e-02 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 1485 | 2010 | 1.38e-02 |
| GO:BP | GO:0045444 | fat cell differentiation | 146 | 179 | 1.39e-02 |
| GO:BP | GO:0001935 | endothelial cell proliferation | 96 | 114 | 1.40e-02 |
| GO:BP | GO:0051090 | regulation of DNA-binding transcription factor activity | 193 | 241 | 1.40e-02 |
| GO:BP | GO:0051492 | regulation of stress fiber assembly | 67 | 77 | 1.40e-02 |
| GO:BP | GO:0050863 | regulation of T cell activation | 165 | 204 | 1.40e-02 |
| GO:BP | GO:0051046 | regulation of secretion | 309 | 396 | 1.43e-02 |
| GO:BP | GO:1904892 | regulation of receptor signaling pathway via STAT | 51 | 57 | 1.43e-02 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 63 | 72 | 1.43e-02 |
| GO:BP | GO:0014812 | muscle cell migration | 63 | 72 | 1.43e-02 |
| GO:BP | GO:0008589 | regulation of smoothened signaling pathway | 63 | 72 | 1.43e-02 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 208 | 261 | 1.44e-02 |
| GO:BP | GO:0060384 | innervation | 21 | 21 | 1.45e-02 |
| GO:BP | GO:0042634 | regulation of hair cycle | 21 | 21 | 1.45e-02 |
| GO:BP | GO:0007224 | smoothened signaling pathway | 99 | 118 | 1.50e-02 |
| GO:BP | GO:0035019 | somatic stem cell population maintenance | 42 | 46 | 1.55e-02 |
| GO:BP | GO:0072171 | mesonephric tubule morphogenesis | 42 | 46 | 1.55e-02 |
| GO:BP | GO:0003197 | endocardial cushion development | 42 | 46 | 1.55e-02 |
| GO:BP | GO:0010611 | regulation of cardiac muscle hypertrophy | 42 | 46 | 1.55e-02 |
| GO:BP | GO:0032868 | response to insulin | 164 | 203 | 1.58e-02 |
| GO:BP | GO:0016266 | O-glycan processing | 27 | 28 | 1.60e-02 |
| GO:BP | GO:0048675 | axon extension | 81 | 95 | 1.60e-02 |
| GO:BP | GO:0098739 | import across plasma membrane | 102 | 122 | 1.60e-02 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 1326 | 1791 | 1.60e-02 |
| GO:BP | GO:0038127 | ERBB signaling pathway | 88 | 104 | 1.64e-02 |
| GO:BP | GO:0072132 | mesenchyme morphogenesis | 46 | 51 | 1.68e-02 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 138 | 169 | 1.70e-02 |
| GO:BP | GO:0048286 | lung alveolus development | 37 | 40 | 1.71e-02 |
| GO:BP | GO:0002376 | immune system process | 1050 | 1410 | 1.72e-02 |
| GO:BP | GO:0006954 | inflammatory response | 330 | 425 | 1.72e-02 |
| GO:BP | GO:0006915 | apoptotic process | 1026 | 1377 | 1.74e-02 |
| GO:BP | GO:0003229 | ventricular cardiac muscle tissue development | 50 | 56 | 1.75e-02 |
| GO:BP | GO:0062013 | positive regulation of small molecule metabolic process | 84 | 99 | 1.77e-02 |
| GO:BP | GO:0071230 | cellular response to amino acid stimulus | 58 | 66 | 1.77e-02 |
| GO:BP | GO:0060415 | muscle tissue morphogenesis | 54 | 61 | 1.78e-02 |
| GO:BP | GO:1903530 | regulation of secretion by cell | 286 | 366 | 1.81e-02 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 73 | 85 | 1.82e-02 |
| GO:BP | GO:0031667 | response to nutrient levels | 277 | 354 | 1.85e-02 |
| GO:BP | GO:0051251 | positive regulation of lymphocyte activation | 150 | 185 | 1.87e-02 |
| GO:BP | GO:0001704 | formation of primary germ layer | 80 | 94 | 1.90e-02 |
| GO:BP | GO:0140747 | regulation of ncRNA transcription | 69 | 80 | 1.93e-02 |
| GO:BP | GO:0060675 | ureteric bud morphogenesis | 41 | 45 | 1.94e-02 |
| GO:BP | GO:0090183 | regulation of kidney development | 20 | 20 | 1.96e-02 |
| GO:BP | GO:0002053 | positive regulation of mesenchymal cell proliferation | 20 | 20 | 1.96e-02 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 463 | 606 | 2.10e-02 |
| GO:BP | GO:0090497 | mesenchymal cell migration | 45 | 50 | 2.10e-02 |
| GO:BP | GO:0072210 | metanephric nephron development | 26 | 27 | 2.11e-02 |
| GO:BP | GO:1904385 | cellular response to angiotensin | 26 | 27 | 2.11e-02 |
| GO:BP | GO:0060416 | response to growth hormone | 26 | 27 | 2.11e-02 |
| GO:BP | GO:1904646 | cellular response to amyloid-beta | 26 | 27 | 2.11e-02 |
| GO:BP | GO:0055008 | cardiac muscle tissue morphogenesis | 49 | 55 | 2.16e-02 |
| GO:BP | GO:0046578 | regulation of Ras protein signal transduction | 36 | 39 | 2.16e-02 |
| GO:BP | GO:0021536 | diencephalon development | 36 | 39 | 2.16e-02 |
| GO:BP | GO:0046425 | regulation of receptor signaling pathway via JAK-STAT | 49 | 55 | 2.16e-02 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 100 | 120 | 2.17e-02 |
| GO:BP | GO:0014909 | smooth muscle cell migration | 53 | 60 | 2.17e-02 |
| GO:BP | GO:0051301 | cell division | 415 | 541 | 2.17e-02 |
| GO:BP | GO:0046683 | response to organophosphorus | 79 | 93 | 2.24e-02 |
| GO:BP | GO:0001508 | action potential | 86 | 102 | 2.26e-02 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 315 | 406 | 2.34e-02 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 400 | 521 | 2.36e-02 |
| GO:BP | GO:0007548 | sex differentiation | 145 | 179 | 2.36e-02 |
| GO:BP | GO:0030217 | T cell differentiation | 148 | 183 | 2.41e-02 |
| GO:BP | GO:0006638 | neutral lipid metabolic process | 75 | 88 | 2.41e-02 |
| GO:BP | GO:0061387 | regulation of extent of cell growth | 75 | 88 | 2.41e-02 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 239 | 304 | 2.42e-02 |
| GO:BP | GO:0031644 | regulation of nervous system process | 64 | 74 | 2.42e-02 |
| GO:BP | GO:0051649 | establishment of localization in cell | 1161 | 1566 | 2.42e-02 |
| GO:BP | GO:0014074 | response to purine-containing compound | 89 | 106 | 2.43e-02 |
| GO:BP | GO:0008277 | regulation of G protein-coupled receptor signaling pathway | 82 | 97 | 2.44e-02 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 297 | 382 | 2.48e-02 |
| GO:BP | GO:0035264 | multicellular organism growth | 99 | 119 | 2.51e-02 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 109 | 132 | 2.51e-02 |
| GO:BP | GO:0045685 | regulation of glial cell differentiation | 44 | 49 | 2.57e-02 |
| GO:BP | GO:0046942 | carboxylic acid transport | 163 | 203 | 2.57e-02 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 71 | 83 | 2.57e-02 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 56 | 64 | 2.60e-02 |
| GO:BP | GO:0060740 | prostate gland epithelium morphogenesis | 19 | 19 | 2.62e-02 |
| GO:BP | GO:0010878 | cholesterol storage | 19 | 19 | 2.62e-02 |
| GO:BP | GO:0060512 | prostate gland morphogenesis | 19 | 19 | 2.62e-02 |
| GO:BP | GO:0048566 | embryonic digestive tract development | 19 | 19 | 2.62e-02 |
| GO:BP | GO:0071378 | cellular response to growth hormone stimulus | 19 | 19 | 2.62e-02 |
| GO:BP | GO:0003184 | pulmonary valve morphogenesis | 19 | 19 | 2.62e-02 |
| GO:BP | GO:2000027 | regulation of animal organ morphogenesis | 52 | 59 | 2.63e-02 |
| GO:BP | GO:0046503 | glycerolipid catabolic process | 35 | 38 | 2.70e-02 |
| GO:BP | GO:0045668 | negative regulation of osteoblast differentiation | 35 | 38 | 2.70e-02 |
| GO:BP | GO:0002066 | columnar/cuboidal epithelial cell development | 35 | 38 | 2.70e-02 |
| GO:BP | GO:0007200 | phospholipase C-activating G protein-coupled receptor signaling pathway | 35 | 38 | 2.70e-02 |
| GO:BP | GO:0050773 | regulation of dendrite development | 67 | 78 | 2.71e-02 |
| GO:BP | GO:0030510 | regulation of BMP signaling pathway | 67 | 78 | 2.71e-02 |
| GO:BP | GO:0009991 | response to extracellular stimulus | 293 | 377 | 2.71e-02 |
| GO:BP | GO:0010092 | specification of animal organ identity | 25 | 26 | 2.72e-02 |
| GO:BP | GO:0000278 | mitotic cell cycle | 577 | 763 | 2.73e-02 |
| GO:BP | GO:0070371 | ERK1 and ERK2 cascade | 156 | 194 | 2.80e-02 |
| GO:BP | GO:0006639 | acylglycerol metabolic process | 74 | 87 | 2.81e-02 |
| GO:BP | GO:0007613 | memory | 74 | 87 | 2.81e-02 |
| GO:BP | GO:0043009 | chordate embryonic development | 389 | 507 | 2.83e-02 |
| GO:BP | GO:0030501 | positive regulation of bone mineralization | 30 | 32 | 2.85e-02 |
| GO:BP | GO:0071604 | transforming growth factor beta production | 30 | 32 | 2.85e-02 |
| GO:BP | GO:0072583 | clathrin-dependent endocytosis | 39 | 43 | 3.00e-02 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 189 | 238 | 3.02e-02 |
| GO:BP | GO:0044784 | metaphase/anaphase transition of cell cycle | 77 | 91 | 3.08e-02 |
| GO:BP | GO:0034767 | positive regulation of monoatomic ion transmembrane transport | 77 | 91 | 3.08e-02 |
| GO:BP | GO:0006637 | acyl-CoA metabolic process | 55 | 63 | 3.11e-02 |
| GO:BP | GO:0002688 | regulation of leukocyte chemotaxis | 55 | 63 | 3.11e-02 |
| GO:BP | GO:0035383 | thioester metabolic process | 55 | 63 | 3.11e-02 |
| GO:BP | GO:0002694 | regulation of leukocyte activation | 245 | 313 | 3.13e-02 |
| GO:BP | GO:0051058 | negative regulation of small GTPase mediated signal transduction | 43 | 48 | 3.15e-02 |
| GO:BP | GO:0008219 | cell death | 1056 | 1423 | 3.15e-02 |
| GO:BP | GO:0022408 | negative regulation of cell-cell adhesion | 94 | 113 | 3.16e-02 |
| GO:BP | GO:0008543 | fibroblast growth factor receptor signaling pathway | 51 | 58 | 3.17e-02 |
| GO:BP | GO:0033674 | positive regulation of kinase activity | 158 | 197 | 3.17e-02 |
| GO:BP | GO:0048844 | artery morphogenesis | 51 | 58 | 3.17e-02 |
| GO:BP | GO:0012501 | programmed cell death | 1053 | 1419 | 3.20e-02 |
| GO:BP | GO:0032526 | response to retinoic acid | 66 | 77 | 3.20e-02 |
| GO:BP | GO:0033993 | response to lipid | 437 | 573 | 3.31e-02 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 123 | 151 | 3.37e-02 |
| GO:BP | GO:0003176 | aortic valve development | 34 | 37 | 3.39e-02 |
| GO:BP | GO:0070169 | positive regulation of biomineral tissue development | 34 | 37 | 3.39e-02 |
| GO:BP | GO:1902305 | regulation of sodium ion transmembrane transport | 34 | 37 | 3.39e-02 |
| GO:BP | GO:0009611 | response to wounding | 296 | 382 | 3.43e-02 |
| GO:BP | GO:0016049 | cell growth | 296 | 382 | 3.43e-02 |
| GO:BP | GO:0045926 | negative regulation of growth | 148 | 184 | 3.44e-02 |
| GO:BP | GO:1903169 | regulation of calcium ion transmembrane transport | 90 | 108 | 3.45e-02 |
| GO:BP | GO:0035336 | long-chain fatty-acyl-CoA metabolic process | 18 | 18 | 3.49e-02 |
| GO:BP | GO:0010743 | regulation of macrophage derived foam cell differentiation | 18 | 18 | 3.49e-02 |
| GO:BP | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway | 18 | 18 | 3.49e-02 |
| GO:BP | GO:0060396 | growth hormone receptor signaling pathway | 18 | 18 | 3.49e-02 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 310 | 401 | 3.49e-02 |
| GO:BP | GO:0006909 | phagocytosis | 113 | 138 | 3.49e-02 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 113 | 138 | 3.49e-02 |
| GO:BP | GO:0035025 | positive regulation of Rho protein signal transduction | 24 | 25 | 3.50e-02 |
| GO:BP | GO:0099171 | presynaptic modulation of chemical synaptic transmission | 24 | 25 | 3.50e-02 |
| GO:BP | GO:0060351 | cartilage development involved in endochondral bone morphogenesis | 24 | 25 | 3.50e-02 |
| GO:BP | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | 76 | 90 | 3.57e-02 |
| GO:BP | GO:0015849 | organic acid transport | 163 | 204 | 3.57e-02 |
| GO:BP | GO:0007219 | Notch signaling pathway | 116 | 142 | 3.58e-02 |
| GO:BP | GO:1902904 | negative regulation of supramolecular fiber organization | 103 | 125 | 3.58e-02 |
| GO:BP | GO:0071634 | regulation of transforming growth factor beta production | 29 | 31 | 3.59e-02 |
| GO:BP | GO:0045687 | positive regulation of glial cell differentiation | 29 | 31 | 3.59e-02 |
| GO:BP | GO:0030857 | negative regulation of epithelial cell differentiation | 29 | 31 | 3.59e-02 |
| GO:BP | GO:1904645 | response to amyloid-beta | 29 | 31 | 3.59e-02 |
| GO:BP | GO:0060688 | regulation of morphogenesis of a branching structure | 29 | 31 | 3.59e-02 |
| GO:BP | GO:0106106 | cold-induced thermogenesis | 93 | 112 | 3.59e-02 |
| GO:BP | GO:0019058 | viral life cycle | 184 | 232 | 3.64e-02 |
| GO:BP | GO:0051591 | response to cAMP | 54 | 62 | 3.68e-02 |
| GO:BP | GO:0070372 | regulation of ERK1 and ERK2 cascade | 141 | 175 | 3.71e-02 |
| GO:BP | GO:0030705 | cytoskeleton-dependent intracellular transport | 144 | 179 | 3.77e-02 |
| GO:BP | GO:0051899 | membrane depolarization | 50 | 57 | 3.78e-02 |
| GO:BP | GO:0097009 | energy homeostasis | 50 | 57 | 3.78e-02 |
| GO:BP | GO:0032147 | activation of protein kinase activity | 50 | 57 | 3.78e-02 |
| GO:BP | GO:0033555 | multicellular organismal response to stress | 50 | 57 | 3.78e-02 |
| GO:BP | GO:0009953 | dorsal/ventral pattern formation | 42 | 47 | 3.79e-02 |
| GO:BP | GO:0051653 | spindle localization | 46 | 52 | 3.82e-02 |
| GO:BP | GO:0001708 | cell fate specification | 46 | 52 | 3.82e-02 |
| GO:BP | GO:0048863 | stem cell differentiation | 153 | 191 | 3.87e-02 |
| GO:BP | GO:0006629 | lipid metabolic process | 692 | 923 | 3.92e-02 |
| GO:BP | GO:0033045 | regulation of sister chromatid segregation | 82 | 98 | 4.09e-02 |
| GO:BP | GO:0120161 | regulation of cold-induced thermogenesis | 92 | 111 | 4.16e-02 |
| GO:BP | GO:0048813 | dendrite morphogenesis | 92 | 111 | 4.16e-02 |
| GO:BP | GO:0010876 | lipid localization | 233 | 298 | 4.16e-02 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 118 | 145 | 4.17e-02 |
| GO:BP | GO:0038179 | neurotrophin signaling pathway | 33 | 36 | 4.19e-02 |
| GO:BP | GO:0044272 | sulfur compound biosynthetic process | 105 | 128 | 4.24e-02 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 95 | 115 | 4.35e-02 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 95 | 115 | 4.35e-02 |
| GO:BP | GO:0046649 | lymphocyte activation | 341 | 444 | 4.38e-02 |
| GO:BP | GO:0035567 | non-canonical Wnt signaling pathway | 53 | 61 | 4.43e-02 |
| GO:BP | GO:0099072 | regulation of postsynaptic membrane neurotransmitter receptor levels | 53 | 61 | 4.43e-02 |
| GO:BP | GO:0042303 | molting cycle | 64 | 75 | 4.44e-02 |
| GO:BP | GO:0042633 | hair cycle | 64 | 75 | 4.44e-02 |
| GO:BP | GO:0045453 | bone resorption | 37 | 41 | 4.48e-02 |
| GO:BP | GO:0048872 | homeostasis of number of cells | 176 | 222 | 4.48e-02 |
| GO:BP | GO:0021952 | central nervous system projection neuron axonogenesis | 23 | 24 | 4.48e-02 |
| GO:BP | GO:0022029 | telencephalon cell migration | 37 | 41 | 4.48e-02 |
| GO:BP | GO:0010812 | negative regulation of cell-substrate adhesion | 37 | 41 | 4.48e-02 |
| GO:BP | GO:0003272 | endocardial cushion formation | 23 | 24 | 4.48e-02 |
| GO:BP | GO:0051894 | positive regulation of focal adhesion assembly | 23 | 24 | 4.48e-02 |
| GO:BP | GO:0061383 | trabecula morphogenesis | 37 | 41 | 4.48e-02 |
| GO:BP | GO:0043491 | phosphatidylinositol 3-kinase/protein kinase B signal transduction | 170 | 214 | 4.48e-02 |
| GO:BP | GO:0045124 | regulation of bone resorption | 23 | 24 | 4.48e-02 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 185 | 234 | 4.48e-02 |
| GO:BP | GO:2000737 | negative regulation of stem cell differentiation | 23 | 24 | 4.48e-02 |
| GO:BP | GO:0050919 | negative chemotaxis | 37 | 41 | 4.48e-02 |
| GO:BP | GO:0002431 | Fc receptor mediated stimulatory signaling pathway | 23 | 24 | 4.48e-02 |
| GO:BP | GO:0046661 | male sex differentiation | 88 | 106 | 4.50e-02 |
| GO:BP | GO:0001937 | negative regulation of endothelial cell proliferation | 28 | 30 | 4.50e-02 |
| GO:BP | GO:0060914 | heart formation | 28 | 30 | 4.50e-02 |
| GO:BP | GO:0001974 | blood vessel remodeling | 28 | 30 | 4.50e-02 |
| GO:BP | GO:0060323 | head morphogenesis | 28 | 30 | 4.50e-02 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 229 | 293 | 4.51e-02 |
| GO:BP | GO:0016032 | viral process | 255 | 328 | 4.53e-02 |
| GO:BP | GO:0034505 | tooth mineralization | 17 | 17 | 4.53e-02 |
| GO:BP | GO:0009065 | glutamine family amino acid catabolic process | 17 | 17 | 4.53e-02 |
| GO:BP | GO:0003215 | cardiac right ventricle morphogenesis | 17 | 17 | 4.53e-02 |
| GO:BP | GO:0032528 | microvillus organization | 17 | 17 | 4.53e-02 |
| GO:BP | GO:0060977 | coronary vasculature morphogenesis | 17 | 17 | 4.53e-02 |
| GO:BP | GO:0050771 | negative regulation of axonogenesis | 45 | 51 | 4.57e-02 |
| GO:BP | GO:0006865 | amino acid transport | 81 | 97 | 4.64e-02 |
| GO:BP | GO:0006470 | protein dephosphorylation | 142 | 177 | 4.64e-02 |
| GO:BP | GO:0070482 | response to oxygen levels | 208 | 265 | 4.71e-02 |
| GO:BP | GO:0006869 | lipid transport | 205 | 261 | 4.73e-02 |
| GO:BP | GO:0051494 | negative regulation of cytoskeleton organization | 104 | 127 | 4.74e-02 |
| GO:BP | GO:0008593 | regulation of Notch signaling pathway | 67 | 79 | 4.76e-02 |
| GO:BP | GO:0035303 | regulation of dephosphorylation | 67 | 79 | 4.76e-02 |
| GO:BP | GO:0002573 | myeloid leukocyte differentiation | 123 | 152 | 4.81e-02 |
| GO:BP | GO:0006887 | exocytosis | 187 | 237 | 4.84e-02 |
| GO:BP | GO:0001942 | hair follicle development | 56 | 65 | 4.93e-02 |
LRnomtable %>%
filter(source=="KEGG", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "KEGG terms found in LR neargenes") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| KEGG | KEGG:05200 | Pathways in cancer | 328 | 391 | 3.55e-07 |
| KEGG | KEGG:04360 | Axon guidance | 137 | 155 | 2.59e-05 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 142 | 162 | 3.40e-05 |
| KEGG | KEGG:04971 | Gastric acid secretion | 50 | 51 | 4.56e-05 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 118 | 133 | 4.79e-05 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 129 | 147 | 5.16e-05 |
| KEGG | KEGG:04510 | Focal adhesion | 146 | 169 | 7.29e-05 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 140 | 162 | 8.94e-05 |
| KEGG | KEGG:04970 | Salivary secretion | 52 | 54 | 8.94e-05 |
| KEGG | KEGG:04014 | Ras signaling pathway | 141 | 164 | 1.28e-04 |
| KEGG | KEGG:04520 | Adherens junction | 76 | 83 | 1.28e-04 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 113 | 129 | 1.52e-04 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 90 | 101 | 2.61e-04 |
| KEGG | KEGG:04611 | Platelet activation | 79 | 88 | 4.18e-04 |
| KEGG | KEGG:04020 | Calcium signaling pathway | 137 | 161 | 4.18e-04 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 143 | 169 | 4.86e-04 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 135 | 160 | 9.35e-04 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 70 | 78 | 9.35e-04 |
| KEGG | KEGG:04924 | Renin secretion | 47 | 50 | 9.35e-04 |
| KEGG | KEGG:04911 | Insulin secretion | 57 | 62 | 9.35e-04 |
| KEGG | KEGG:05146 | Amoebiasis | 57 | 62 | 9.35e-04 |
| KEGG | KEGG:04512 | ECM-receptor interaction | 60 | 66 | 1.17e-03 |
| KEGG | KEGG:04270 | Vascular smooth muscle contraction | 81 | 92 | 1.17e-03 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 189 | 231 | 1.24e-03 |
| KEGG | KEGG:04925 | Aldosterone synthesis and secretion | 67 | 75 | 1.55e-03 |
| KEGG | KEGG:04916 | Melanogenesis | 67 | 75 | 1.55e-03 |
| KEGG | KEGG:04918 | Thyroid hormone synthesis | 49 | 53 | 1.59e-03 |
| KEGG | KEGG:04927 | Cortisol synthesis and secretion | 43 | 46 | 2.09e-03 |
| KEGG | KEGG:04972 | Pancreatic secretion | 52 | 57 | 2.29e-03 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 202 | 250 | 2.43e-03 |
| KEGG | KEGG:04148 | Efferocytosis | 102 | 120 | 2.50e-03 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 76 | 87 | 2.50e-03 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 109 | 129 | 2.50e-03 |
| KEGG | KEGG:04974 | Protein digestion and absorption | 51 | 56 | 2.53e-03 |
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 105 | 124 | 2.57e-03 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 202 | 252 | 4.72e-03 |
| KEGG | KEGG:04728 | Dopaminergic synapse | 83 | 97 | 5.30e-03 |
| KEGG | KEGG:04713 | Circadian entrainment | 60 | 68 | 5.44e-03 |
| KEGG | KEGG:04976 | Bile secretion | 38 | 41 | 6.05e-03 |
| KEGG | KEGG:04750 | Inflammatory mediator regulation of TRP channels | 63 | 72 | 6.33e-03 |
| KEGG | KEGG:05412 | Arrhythmogenic right ventricular cardiomyopathy | 59 | 67 | 6.33e-03 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 85 | 100 | 6.37e-03 |
| KEGG | KEGG:04961 | Endocrine and other factor-regulated calcium reabsorption | 37 | 40 | 7.11e-03 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 77 | 90 | 7.11e-03 |
| KEGG | KEGG:05417 | Lipid and atherosclerosis | 118 | 143 | 7.35e-03 |
| KEGG | KEGG:04658 | Th1 and Th2 cell differentiation | 45 | 50 | 8.33e-03 |
| KEGG | KEGG:04072 | Phospholipase D signaling pathway | 90 | 107 | 8.33e-03 |
| KEGG | KEGG:04730 | Long-term depression | 36 | 39 | 8.49e-03 |
| KEGG | KEGG:04630 | JAK-STAT signaling pathway | 71 | 83 | 1.00e-02 |
| KEGG | KEGG:04340 | Hedgehog signaling pathway | 44 | 49 | 1.00e-02 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 81 | 96 | 1.09e-02 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 88 | 105 | 1.09e-02 |
| KEGG | KEGG:05418 | Fluid shear stress and atherosclerosis | 91 | 109 | 1.13e-02 |
| KEGG | KEGG:05414 | Dilated cardiomyopathy | 70 | 82 | 1.13e-02 |
| KEGG | KEGG:05167 | Kaposi sarcoma-associated herpesvirus infection | 110 | 134 | 1.28e-02 |
| KEGG | KEGG:04915 | Estrogen signaling pathway | 76 | 90 | 1.34e-02 |
| KEGG | KEGG:04670 | Leukocyte transendothelial migration | 65 | 76 | 1.41e-02 |
| KEGG | KEGG:05135 | Yersinia infection | 89 | 107 | 1.46e-02 |
| KEGG | KEGG:05207 | Chemical carcinogenesis - receptor activation | 108 | 132 | 1.63e-02 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 52 | 60 | 2.00e-02 |
| KEGG | KEGG:04514 | Cell adhesion molecules | 59 | 69 | 2.11e-02 |
| KEGG | KEGG:04921 | Oxytocin signaling pathway | 93 | 113 | 2.12e-02 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 86 | 104 | 2.23e-02 |
| KEGG | KEGG:04725 | Cholinergic synapse | 69 | 82 | 2.25e-02 |
| KEGG | KEGG:04071 | Sphingolipid signaling pathway | 85 | 103 | 2.51e-02 |
| KEGG | KEGG:00000 | KEGG root term | 3931 | 5451 | 2.51e-02 |
| KEGG | KEGG:05210 | Colorectal cancer | 68 | 81 | 2.57e-02 |
| KEGG | KEGG:04726 | Serotonergic synapse | 50 | 58 | 2.69e-02 |
| KEGG | KEGG:04660 | T cell receptor signaling pathway | 70 | 84 | 3.20e-02 |
| KEGG | KEGG:00512 | Mucin type O-glycan biosynthesis | 20 | 21 | 3.56e-02 |
| KEGG | KEGG:03320 | PPAR signaling pathway | 41 | 47 | 3.59e-02 |
| KEGG | KEGG:04625 | C-type lectin receptor signaling pathway | 62 | 74 | 3.77e-02 |
| KEGG | KEGG:04724 | Glutamatergic synapse | 62 | 74 | 3.77e-02 |
| KEGG | KEGG:05170 | Human immunodeficiency virus 1 infection | 119 | 149 | 3.95e-02 |
| KEGG | KEGG:04720 | Long-term potentiation | 44 | 51 | 3.96e-02 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 115 | 144 | 4.31e-02 |
| KEGG | KEGG:04923 | Regulation of lipolysis in adipocytes | 36 | 41 | 4.31e-02 |
| KEGG | KEGG:04742 | Taste transduction | 19 | 20 | 4.32e-02 |
| KEGG | KEGG:05410 | Hypertrophic cardiomyopathy | 64 | 77 | 4.43e-02 |
| KEGG | KEGG:04934 | Cushing syndrome | 96 | 119 | 4.45e-02 |
| KEGG | KEGG:05130 | Pathogenic Escherichia coli infection | 114 | 143 | 4.67e-02 |
| KEGG | KEGG:01521 | EGFR tyrosine kinase inhibitor resistance | 60 | 72 | 4.78e-02 |
| KEGG | KEGG:05032 | Morphine addiction | 46 | 54 | 4.96e-02 |
LRnomtable %>%
filter(source=="GO:BP", p_value < 0.05) %>%
slice_head(n=10) %>%
mutate(log10_p=(-log(p_value, base=10))) %>%
ggplot(., aes(x=log10_p, y=reorder(term_name, log10_p)))+
geom_col(fill="#00BFC4")+
theme_bw()+
ylab("")+
xlab(paste0("-log10 p-value"))+
guides(fill="none")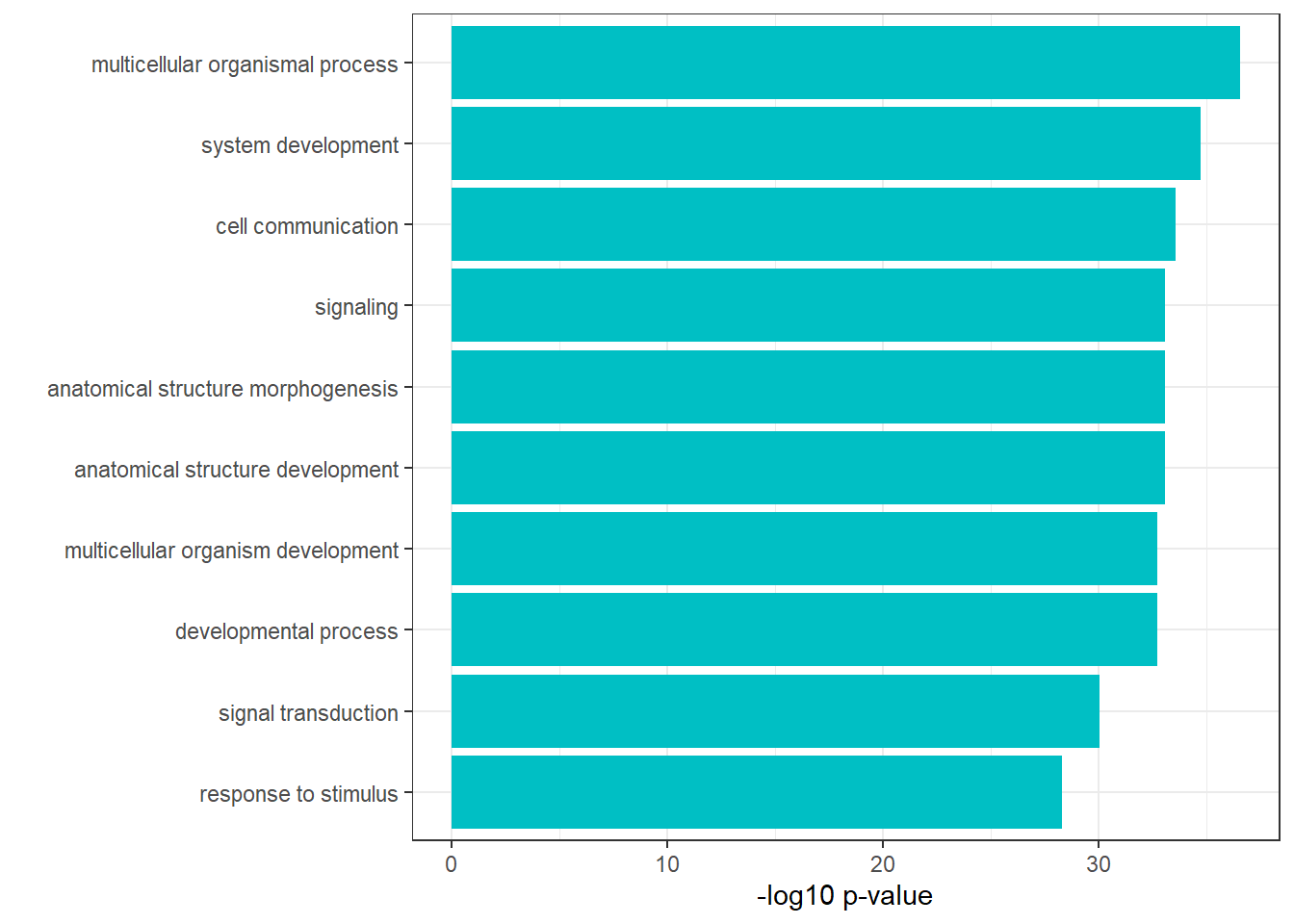
NR response genes
# NR_resgenes <- gost(query = NR_NG_list$ENTREZID,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = FALSE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = background_df$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(NR_resgenes,"data/GO_KEGG_analysis/NR_resgenes.RDS")
NRnom <- gostplot(NR_resgenes, capped = FALSE, interactive = TRUE)
# NRnom
NRnomtable <- NR_resgenes$result %>%
dplyr::select(c(source, term_id,
term_name,intersection_size,
term_size, p_value))# %>%
NRnomtable %>%
filter(source=="GO:BP", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(.,caption = "GO:BP terms found in NR neargenes" ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0007275 | multicellular organism development | 3082 | 3170 | 4.30e-05 |
| GO:BP | GO:0032501 | multicellular organismal process | 4362 | 4504 | 4.30e-05 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 3446 | 3550 | 4.40e-05 |
| GO:BP | GO:0051716 | cellular response to stimulus | 4491 | 4640 | 4.44e-05 |
| GO:BP | GO:0007010 | cytoskeleton organization | 1127 | 1146 | 7.38e-05 |
| GO:BP | GO:0048731 | system development | 2645 | 2719 | 7.49e-05 |
| GO:BP | GO:0048856 | anatomical structure development | 3748 | 3867 | 7.49e-05 |
| GO:BP | GO:0050896 | response to stimulus | 5196 | 5379 | 8.02e-05 |
| GO:BP | GO:0050794 | regulation of cellular process | 6888 | 7153 | 2.38e-04 |
| GO:BP | GO:0065007 | biological regulation | 7441 | 7734 | 3.22e-04 |
| GO:BP | GO:0007154 | cell communication | 3855 | 3983 | 3.67e-04 |
| GO:BP | GO:0048519 | negative regulation of biological process | 3645 | 3764 | 3.67e-04 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 1799 | 1844 | 3.67e-04 |
| GO:BP | GO:0050789 | regulation of biological process | 7232 | 7516 | 3.99e-04 |
| GO:BP | GO:0023052 | signaling | 3792 | 3918 | 4.48e-04 |
| GO:BP | GO:0032502 | developmental process | 4042 | 4179 | 4.62e-04 |
| GO:BP | GO:0008150 | biological_process | 10770 | 11244 | 6.46e-04 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 1815 | 1862 | 6.81e-04 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 1016 | 1035 | 6.93e-04 |
| GO:BP | GO:0050793 | regulation of developmental process | 1655 | 1697 | 1.16e-03 |
| GO:BP | GO:0007165 | signal transduction | 3495 | 3611 | 1.16e-03 |
| GO:BP | GO:0023051 | regulation of signaling | 2368 | 2438 | 1.48e-03 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 2636 | 2717 | 1.58e-03 |
| GO:BP | GO:0009966 | regulation of signal transduction | 2106 | 2166 | 1.65e-03 |
| GO:BP | GO:0007399 | nervous system development | 1729 | 1775 | 2.13e-03 |
| GO:BP | GO:0016477 | cell migration | 991 | 1011 | 2.73e-03 |
| GO:BP | GO:0022008 | neurogenesis | 1223 | 1251 | 2.76e-03 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 601 | 609 | 2.83e-03 |
| GO:BP | GO:0000902 | cell morphogenesis | 702 | 713 | 2.92e-03 |
| GO:BP | GO:0010646 | regulation of cell communication | 2372 | 2444 | 2.95e-03 |
| GO:BP | GO:0030029 | actin filament-based process | 631 | 640 | 3.04e-03 |
| GO:BP | GO:0048518 | positive regulation of biological process | 4153 | 4301 | 3.42e-03 |
| GO:BP | GO:0009888 | tissue development | 1316 | 1348 | 3.77e-03 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 3820 | 3954 | 4.19e-03 |
| GO:BP | GO:0048699 | generation of neurons | 1053 | 1076 | 4.37e-03 |
| GO:BP | GO:0007155 | cell adhesion | 922 | 941 | 6.18e-03 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 821 | 837 | 8.05e-03 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 1169 | 1197 | 8.05e-03 |
| GO:BP | GO:0016043 | cellular component organization | 4630 | 4802 | 8.05e-03 |
| GO:BP | GO:0009987 | cellular process | 10236 | 10686 | 8.78e-03 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 558 | 566 | 8.78e-03 |
| GO:BP | GO:0030182 | neuron differentiation | 992 | 1014 | 8.85e-03 |
| GO:BP | GO:0048513 | animal organ development | 1946 | 2004 | 1.02e-02 |
| GO:BP | GO:0065008 | regulation of biological quality | 1839 | 1893 | 1.13e-02 |
| GO:BP | GO:0035295 | tube development | 772 | 787 | 1.14e-02 |
| GO:BP | GO:0048870 | cell motility | 1094 | 1120 | 1.14e-02 |
| GO:BP | GO:0048869 | cellular developmental process | 2714 | 2804 | 1.27e-02 |
| GO:BP | GO:0030154 | cell differentiation | 2713 | 2803 | 1.27e-02 |
| GO:BP | GO:0007267 | cell-cell signaling | 1057 | 1082 | 1.32e-02 |
| GO:BP | GO:0007015 | actin filament organization | 354 | 357 | 1.44e-02 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 1868 | 1924 | 1.54e-02 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 260 | 261 | 1.76e-02 |
| GO:BP | GO:0048468 | cell development | 1831 | 1886 | 1.89e-02 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 257 | 258 | 1.90e-02 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 1067 | 1093 | 1.90e-02 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 622 | 633 | 2.02e-02 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 298 | 300 | 2.24e-02 |
| GO:BP | GO:0035556 | intracellular signal transduction | 1893 | 1951 | 2.27e-02 |
| GO:BP | GO:0032989 | cellular anatomical entity morphogenesis | 578 | 588 | 2.81e-02 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 4802 | 4988 | 4.57e-02 |
| GO:BP | GO:0023056 | positive regulation of signaling | 1189 | 1221 | 4.59e-02 |
| GO:BP | GO:0006996 | organelle organization | 2721 | 2815 | 4.63e-02 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 1186 | 1218 | 4.77e-02 |
NRnomtable %>%
filter(source=="KEGG", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "KEGG terms found in NR neargenes") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| NA | NA | NA | NA | NA | NA |
| :—— | :——- | :——— | —————–: | ———: | :——- |
Peaks within 20,000 bps of assigned nearest expressed gene, GO and KEGG enrichment
# exp_neargene_table <- read_delim("data/n45_bedfiles/exp_neargene_table.tsv",
# delim = "\t", escape_double = FALSE,
# trim_ws = TRUE)
# col_ng_peak <- exp_neargene_table %>%
# group_by(chr, start, end, peakid) %>%
# summarise(NCBI_gene = paste(unique(ENTREZID),collapse=","),
# ensembl_ID= paste(unique(ensembl_id),collapse = ","),
# SYMBOL= paste(unique(SYMBOL),collapse = ",") ,
# dist_to_NG =min(dist_to_NG)) %>%
# ungroup()
# write.csv(col_ng_peak, "data/Collapsed_expressed_NG_peak_table.csv")
col_ng_peak <- read.csv("data/Collapsed_expressed_NG_peak_table.csv", row.names = 1)
EAR_peak_list_20k <- col_ng_peak %>%
dplyr::filter(dist_to_NG<20000) %>%
dplyr::filter(peakid %in% EAR_df$id)
ESR_peak_list_20k <- col_ng_peak %>%
dplyr::filter(dist_to_NG<20000) %>%
dplyr::filter(peakid %in% ESR_df$id)
LR_peak_list_20k <- col_ng_peak %>%
dplyr::filter(dist_to_NG<20000) %>%
dplyr::filter(peakid %in% LR_df$id)
NR_peak_list_20k <- col_ng_peak %>%
dplyr::filter(dist_to_NG<20000) %>%
dplyr::filter(peakid %in% NR_df$id) EAR 20,000 bp NG
# EAR_resgenes_20k <- gost(query = EAR_peak_list_20k$ENTREZID,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = FALSE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = background_df$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(EAR_resgenes_20k,"data/GO_KEGG_analysis/EAR_resgenes_20k.RDS")
EARnom <- gostplot(EAR_resgenes_20k, capped = FALSE, interactive = TRUE)
# EARnom
EARnomtable_20k <- EAR_resgenes_20k$result %>%
dplyr::select(c(source, term_id,
term_name,intersection_size,
term_size, p_value))# %>%
EARnomtable_20k %>%
filter(source=="GO:BP", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(.,caption = "GO:BP terms found in EAR neargenes_20k" ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 700 | 1844 | 9.84e-20 |
| GO:BP | GO:0048856 | anatomical structure development | 1307 | 3867 | 8.91e-18 |
| GO:BP | GO:0032502 | developmental process | 1397 | 4179 | 1.19e-17 |
| GO:BP | GO:0032501 | multicellular organismal process | 1481 | 4504 | 5.39e-16 |
| GO:BP | GO:0007275 | multicellular organism development | 1086 | 3170 | 8.40e-16 |
| GO:BP | GO:0048731 | system development | 948 | 2719 | 1.49e-15 |
| GO:BP | GO:0048869 | cellular developmental process | 972 | 2804 | 2.68e-15 |
| GO:BP | GO:0050793 | regulation of developmental process | 629 | 1697 | 2.85e-15 |
| GO:BP | GO:0030154 | cell differentiation | 971 | 2803 | 2.85e-15 |
| GO:BP | GO:0050789 | regulation of biological process | 2316 | 7516 | 1.46e-14 |
| GO:BP | GO:0065007 | biological regulation | 2368 | 7734 | 1.72e-13 |
| GO:BP | GO:0050794 | regulation of cellular process | 2207 | 7153 | 3.23e-13 |
| GO:BP | GO:0023052 | signaling | 1286 | 3918 | 9.00e-13 |
| GO:BP | GO:0007154 | cell communication | 1304 | 3983 | 1.18e-12 |
| GO:BP | GO:0048699 | generation of neurons | 414 | 1076 | 4.57e-12 |
| GO:BP | GO:0022008 | neurogenesis | 469 | 1251 | 9.52e-12 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 680 | 1924 | 1.19e-11 |
| GO:BP | GO:0007165 | signal transduction | 1187 | 3611 | 1.30e-11 |
| GO:BP | GO:0072359 | circulatory system development | 340 | 864 | 3.97e-11 |
| GO:BP | GO:0016477 | cell migration | 387 | 1011 | 7.21e-11 |
| GO:BP | GO:0030182 | neuron differentiation | 387 | 1014 | 1.13e-10 |
| GO:BP | GO:0048468 | cell development | 658 | 1886 | 5.47e-10 |
| GO:BP | GO:0048519 | negative regulation of biological process | 1219 | 3764 | 6.07e-10 |
| GO:BP | GO:0050896 | response to stimulus | 1681 | 5379 | 1.37e-09 |
| GO:BP | GO:0007399 | nervous system development | 621 | 1775 | 1.42e-09 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 1153 | 3550 | 1.49e-09 |
| GO:BP | GO:0048870 | cell motility | 415 | 1120 | 1.76e-09 |
| GO:BP | GO:0051716 | cellular response to stimulus | 1468 | 4640 | 2.02e-09 |
| GO:BP | GO:2000145 | regulation of cell motility | 277 | 700 | 3.21e-09 |
| GO:BP | GO:0030334 | regulation of cell migration | 266 | 668 | 3.76e-09 |
| GO:BP | GO:0048666 | neuron development | 322 | 838 | 3.90e-09 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 474 | 1314 | 4.50e-09 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 158 | 355 | 4.59e-09 |
| GO:BP | GO:0034330 | cell junction organization | 224 | 545 | 4.64e-09 |
| GO:BP | GO:0048513 | animal organ development | 687 | 2004 | 4.83e-09 |
| GO:BP | GO:0040012 | regulation of locomotion | 282 | 720 | 6.38e-09 |
| GO:BP | GO:0030029 | actin filament-based process | 255 | 640 | 7.81e-09 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 213 | 516 | 7.92e-09 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 642 | 1862 | 7.92e-09 |
| GO:BP | GO:0031175 | neuron projection development | 289 | 743 | 7.92e-09 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 209 | 505 | 9.01e-09 |
| GO:BP | GO:0035556 | intracellular signal transduction | 668 | 1951 | 1.11e-08 |
| GO:BP | GO:0000902 | cell morphogenesis | 278 | 713 | 1.33e-08 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 243 | 609 | 1.67e-08 |
| GO:BP | GO:0048518 | positive regulation of biological process | 1360 | 4301 | 2.23e-08 |
| GO:BP | GO:0040011 | locomotion | 307 | 805 | 2.26e-08 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 1257 | 3954 | 4.34e-08 |
| GO:BP | GO:0010646 | regulation of cell communication | 809 | 2444 | 1.24e-07 |
| GO:BP | GO:0007155 | cell adhesion | 347 | 941 | 1.33e-07 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 260 | 673 | 1.46e-07 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 337 | 913 | 2.04e-07 |
| GO:BP | GO:0065008 | regulation of biological quality | 641 | 1893 | 2.19e-07 |
| GO:BP | GO:0023051 | regulation of signaling | 804 | 2438 | 2.96e-07 |
| GO:BP | GO:0007507 | heart development | 199 | 499 | 8.15e-07 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 308 | 837 | 1.51e-06 |
| GO:BP | GO:0009966 | regulation of signal transduction | 717 | 2166 | 1.51e-06 |
| GO:BP | GO:0003008 | system process | 403 | 1139 | 1.73e-06 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 91 | 193 | 1.94e-06 |
| GO:BP | GO:0009888 | tissue development | 467 | 1348 | 2.31e-06 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 343 | 951 | 2.39e-06 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 381 | 1073 | 2.73e-06 |
| GO:BP | GO:0009790 | embryo development | 301 | 821 | 3.13e-06 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 218 | 566 | 3.95e-06 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 368 | 1035 | 3.99e-06 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 189 | 479 | 4.32e-06 |
| GO:BP | GO:0035239 | tube morphogenesis | 241 | 638 | 4.86e-06 |
| GO:BP | GO:0016358 | dendrite development | 86 | 183 | 5.11e-06 |
| GO:BP | GO:0030030 | cell projection organization | 413 | 1182 | 5.18e-06 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 403 | 1150 | 5.21e-06 |
| GO:BP | GO:0032989 | cellular anatomical entity morphogenesis | 224 | 588 | 6.50e-06 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 193 | 494 | 6.72e-06 |
| GO:BP | GO:0007423 | sensory organ development | 157 | 387 | 7.32e-06 |
| GO:BP | GO:0007264 | small GTPase-mediated signal transduction | 158 | 390 | 7.32e-06 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 194 | 498 | 7.66e-06 |
| GO:BP | GO:0070848 | response to growth factor | 210 | 547 | 8.12e-06 |
| GO:BP | GO:0008360 | regulation of cell shape | 56 | 106 | 8.39e-06 |
| GO:BP | GO:0035295 | tube development | 287 | 787 | 9.83e-06 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 203 | 528 | 1.15e-05 |
| GO:BP | GO:0048589 | developmental growth | 194 | 501 | 1.21e-05 |
| GO:BP | GO:0016043 | cellular component organization | 1474 | 4802 | 1.71e-05 |
| GO:BP | GO:0001568 | blood vessel development | 201 | 525 | 1.85e-05 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 1525 | 4988 | 2.56e-05 |
| GO:BP | GO:0001944 | vasculature development | 209 | 552 | 2.69e-05 |
| GO:BP | GO:0001501 | skeletal system development | 141 | 347 | 2.70e-05 |
| GO:BP | GO:0009987 | cellular process | 3085 | 10686 | 4.60e-05 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 174 | 450 | 5.74e-05 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 302 | 850 | 6.39e-05 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 409 | 1197 | 8.00e-05 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 272 | 757 | 8.20e-05 |
| GO:BP | GO:0007417 | central nervous system development | 252 | 694 | 8.20e-05 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 863 | 2717 | 9.18e-05 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 259 | 717 | 9.18e-05 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 168 | 435 | 9.18e-05 |
| GO:BP | GO:0001525 | angiogenesis | 150 | 381 | 9.59e-05 |
| GO:BP | GO:0001654 | eye development | 110 | 263 | 9.84e-05 |
| GO:BP | GO:0048880 | sensory system development | 111 | 266 | 9.95e-05 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 591 | 1801 | 9.97e-05 |
| GO:BP | GO:0030900 | forebrain development | 113 | 272 | 1.02e-04 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 436 | 1289 | 1.04e-04 |
| GO:BP | GO:0050808 | synapse organization | 135 | 337 | 1.05e-04 |
| GO:BP | GO:0001764 | neuron migration | 62 | 129 | 1.05e-04 |
| GO:BP | GO:0150063 | visual system development | 110 | 264 | 1.14e-04 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 218 | 591 | 1.14e-04 |
| GO:BP | GO:0034329 | cell junction assembly | 130 | 323 | 1.19e-04 |
| GO:BP | GO:0060322 | head development | 201 | 539 | 1.26e-04 |
| GO:BP | GO:0014706 | striated muscle tissue development | 92 | 213 | 1.31e-04 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 99 | 233 | 1.31e-04 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 87 | 201 | 2.24e-04 |
| GO:BP | GO:0040007 | growth | 250 | 697 | 2.25e-04 |
| GO:BP | GO:0043010 | camera-type eye development | 97 | 230 | 2.40e-04 |
| GO:BP | GO:0007267 | cell-cell signaling | 369 | 1082 | 2.92e-04 |
| GO:BP | GO:0061061 | muscle structure development | 198 | 536 | 2.94e-04 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 106 | 258 | 3.48e-04 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 225 | 622 | 3.54e-04 |
| GO:BP | GO:0060537 | muscle tissue development | 133 | 339 | 3.96e-04 |
| GO:BP | GO:0007420 | brain development | 185 | 498 | 3.96e-04 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 162 | 427 | 3.96e-04 |
| GO:BP | GO:0008150 | biological_process | 3221 | 11244 | 4.26e-04 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 115 | 286 | 4.46e-04 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 160 | 422 | 4.55e-04 |
| GO:BP | GO:0044057 | regulation of system process | 147 | 384 | 5.96e-04 |
| GO:BP | GO:0007010 | cytoskeleton organization | 386 | 1146 | 5.99e-04 |
| GO:BP | GO:0060284 | regulation of cell development | 207 | 570 | 5.99e-04 |
| GO:BP | GO:0007015 | actin filament organization | 138 | 357 | 6.17e-04 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 119 | 300 | 6.28e-04 |
| GO:BP | GO:0042221 | response to chemical | 732 | 2304 | 6.28e-04 |
| GO:BP | GO:0021795 | cerebral cortex cell migration | 22 | 34 | 7.57e-04 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 150 | 395 | 7.57e-04 |
| GO:BP | GO:0006468 | protein phosphorylation | 327 | 958 | 8.77e-04 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 65 | 145 | 8.77e-04 |
| GO:BP | GO:0048588 | developmental cell growth | 77 | 179 | 9.05e-04 |
| GO:BP | GO:0099536 | synaptic signaling | 178 | 483 | 9.22e-04 |
| GO:BP | GO:0050773 | regulation of dendrite development | 40 | 78 | 9.33e-04 |
| GO:BP | GO:0060041 | retina development in camera-type eye | 50 | 105 | 1.18e-03 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 225 | 633 | 1.19e-03 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 694 | 2186 | 1.21e-03 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 153 | 408 | 1.27e-03 |
| GO:BP | GO:0048813 | dendrite morphogenesis | 52 | 111 | 1.39e-03 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 114 | 290 | 1.42e-03 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 34 | 64 | 1.51e-03 |
| GO:BP | GO:0051960 | regulation of nervous system development | 130 | 339 | 1.54e-03 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 95 | 234 | 1.54e-03 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 155 | 416 | 1.60e-03 |
| GO:BP | GO:0000165 | MAPK cascade | 183 | 504 | 1.75e-03 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 65 | 148 | 1.75e-03 |
| GO:BP | GO:0007409 | axonogenesis | 129 | 337 | 1.78e-03 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 184 | 508 | 1.93e-03 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 58 | 129 | 2.00e-03 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 349 | 1040 | 2.00e-03 |
| GO:BP | GO:0061564 | axon development | 140 | 372 | 2.14e-03 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 167 | 456 | 2.22e-03 |
| GO:BP | GO:0048638 | regulation of developmental growth | 97 | 243 | 2.63e-03 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 103 | 261 | 2.65e-03 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 364 | 1093 | 2.69e-03 |
| GO:BP | GO:1990089 | response to nerve growth factor | 24 | 41 | 2.80e-03 |
| GO:BP | GO:1990090 | cellular response to nerve growth factor stimulus | 24 | 41 | 2.80e-03 |
| GO:BP | GO:0051897 | positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 56 | 125 | 2.94e-03 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 69 | 162 | 3.08e-03 |
| GO:BP | GO:0051216 | cartilage development | 63 | 145 | 3.15e-03 |
| GO:BP | GO:0023056 | positive regulation of signaling | 402 | 1221 | 3.17e-03 |
| GO:BP | GO:1990138 | neuron projection extension | 59 | 134 | 3.36e-03 |
| GO:BP | GO:0098609 | cell-cell adhesion | 200 | 564 | 3.42e-03 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 345 | 1034 | 3.42e-03 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 560 | 1751 | 3.42e-03 |
| GO:BP | GO:0003007 | heart morphogenesis | 88 | 218 | 3.42e-03 |
| GO:BP | GO:0016049 | cell growth | 142 | 382 | 3.42e-03 |
| GO:BP | GO:0060021 | roof of mouth development | 35 | 69 | 3.46e-03 |
| GO:BP | GO:0086002 | cardiac muscle cell action potential involved in contraction | 25 | 44 | 3.60e-03 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 169 | 467 | 3.73e-03 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 111 | 288 | 3.96e-03 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 400 | 1218 | 3.96e-03 |
| GO:BP | GO:0030048 | actin filament-based movement | 49 | 107 | 3.96e-03 |
| GO:BP | GO:0048592 | eye morphogenesis | 49 | 107 | 3.96e-03 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 129 | 344 | 4.50e-03 |
| GO:BP | GO:0051049 | regulation of transport | 363 | 1098 | 4.71e-03 |
| GO:BP | GO:0046834 | lipid phosphorylation | 9 | 10 | 4.71e-03 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 167 | 463 | 4.71e-03 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 167 | 463 | 4.71e-03 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 73 | 176 | 4.71e-03 |
| GO:BP | GO:0050807 | regulation of synapse organization | 72 | 173 | 4.71e-03 |
| GO:BP | GO:0023057 | negative regulation of signaling | 344 | 1036 | 4.91e-03 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 77 | 188 | 4.97e-03 |
| GO:BP | GO:0003013 | circulatory system process | 155 | 426 | 5.07e-03 |
| GO:BP | GO:0048511 | rhythmic process | 88 | 221 | 5.40e-03 |
| GO:BP | GO:2001222 | regulation of neuron migration | 22 | 38 | 6.20e-03 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 121 | 322 | 6.27e-03 |
| GO:BP | GO:0022030 | telencephalon glial cell migration | 14 | 20 | 6.55e-03 |
| GO:BP | GO:0021801 | cerebral cortex radial glia-guided migration | 14 | 20 | 6.55e-03 |
| GO:BP | GO:0021885 | forebrain cell migration | 24 | 43 | 6.60e-03 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 113 | 298 | 6.69e-03 |
| GO:BP | GO:0070372 | regulation of ERK1 and ERK2 cascade | 72 | 175 | 6.69e-03 |
| GO:BP | GO:0007623 | circadian rhythm | 64 | 152 | 7.02e-03 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 77 | 190 | 7.02e-03 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 121 | 323 | 7.02e-03 |
| GO:BP | GO:0061448 | connective tissue development | 82 | 205 | 7.26e-03 |
| GO:BP | GO:0021543 | pallium development | 58 | 135 | 7.29e-03 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 86 | 217 | 7.30e-03 |
| GO:BP | GO:0016310 | phosphorylation | 375 | 1145 | 7.58e-03 |
| GO:BP | GO:0022029 | telencephalon cell migration | 23 | 41 | 7.74e-03 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 141 | 386 | 7.74e-03 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 122 | 327 | 7.74e-03 |
| GO:BP | GO:0035265 | organ growth | 55 | 127 | 8.09e-03 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 41 | 88 | 8.13e-03 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 241 | 706 | 8.30e-03 |
| GO:BP | GO:0048729 | tissue morphogenesis | 162 | 453 | 8.56e-03 |
| GO:BP | GO:0046328 | regulation of JNK cascade | 47 | 105 | 9.13e-03 |
| GO:BP | GO:0032879 | regulation of localization | 462 | 1439 | 9.27e-03 |
| GO:BP | GO:0060429 | epithelium development | 267 | 792 | 9.27e-03 |
| GO:BP | GO:0006897 | endocytosis | 174 | 492 | 9.41e-03 |
| GO:BP | GO:0090132 | epithelium migration | 94 | 243 | 9.52e-03 |
| GO:BP | GO:0010631 | epithelial cell migration | 94 | 243 | 9.52e-03 |
| GO:BP | GO:0008154 | actin polymerization or depolymerization | 62 | 148 | 9.52e-03 |
| GO:BP | GO:0060998 | regulation of dendritic spine development | 26 | 49 | 9.52e-03 |
| GO:BP | GO:0021537 | telencephalon development | 78 | 195 | 9.52e-03 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 83 | 210 | 9.64e-03 |
| GO:BP | GO:0051179 | localization | 1129 | 3731 | 9.72e-03 |
| GO:BP | GO:0051641 | cellular localization | 809 | 2623 | 1.02e-02 |
| GO:BP | GO:0001503 | ossification | 113 | 302 | 1.08e-02 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 748 | 2415 | 1.11e-02 |
| GO:BP | GO:0050877 | nervous system process | 207 | 600 | 1.15e-02 |
| GO:BP | GO:0048675 | axon extension | 43 | 95 | 1.16e-02 |
| GO:BP | GO:0010811 | positive regulation of cell-substrate adhesion | 43 | 95 | 1.16e-02 |
| GO:BP | GO:0060996 | dendritic spine development | 36 | 76 | 1.22e-02 |
| GO:BP | GO:0048568 | embryonic organ development | 113 | 303 | 1.22e-02 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 56 | 132 | 1.22e-02 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 40 | 87 | 1.24e-02 |
| GO:BP | GO:0051057 | positive regulation of small GTPase mediated signal transduction | 28 | 55 | 1.28e-02 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 83 | 212 | 1.31e-02 |
| GO:BP | GO:0009952 | anterior/posterior pattern specification | 52 | 121 | 1.34e-02 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 112 | 301 | 1.40e-02 |
| GO:BP | GO:0071407 | cellular response to organic cyclic compound | 137 | 379 | 1.40e-02 |
| GO:BP | GO:0042692 | muscle cell differentiation | 119 | 323 | 1.44e-02 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 160 | 452 | 1.44e-02 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 93 | 243 | 1.44e-02 |
| GO:BP | GO:0030516 | regulation of axon extension | 35 | 74 | 1.45e-02 |
| GO:BP | GO:0007611 | learning or memory | 75 | 189 | 1.51e-02 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 44 | 99 | 1.53e-02 |
| GO:BP | GO:0007398 | ectoderm development | 10 | 13 | 1.57e-02 |
| GO:BP | GO:0001508 | action potential | 45 | 102 | 1.58e-02 |
| GO:BP | GO:0099643 | signal release from synapse | 45 | 102 | 1.58e-02 |
| GO:BP | GO:0007269 | neurotransmitter secretion | 45 | 102 | 1.58e-02 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 153 | 431 | 1.60e-02 |
| GO:BP | GO:0043583 | ear development | 62 | 151 | 1.61e-02 |
| GO:BP | GO:0090130 | tissue migration | 94 | 247 | 1.62e-02 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 46 | 105 | 1.63e-02 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 53 | 125 | 1.64e-02 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 67 | 166 | 1.68e-02 |
| GO:BP | GO:0007411 | axon guidance | 69 | 172 | 1.68e-02 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 190 | 550 | 1.68e-02 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 373 | 1152 | 1.68e-02 |
| GO:BP | GO:0097485 | neuron projection guidance | 69 | 172 | 1.68e-02 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 55 | 131 | 1.69e-02 |
| GO:BP | GO:0045995 | regulation of embryonic development | 34 | 72 | 1.69e-02 |
| GO:BP | GO:0031032 | actomyosin structure organization | 73 | 184 | 1.69e-02 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 31 | 64 | 1.71e-02 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 433 | 1354 | 1.75e-02 |
| GO:BP | GO:0007610 | behavior | 149 | 420 | 1.85e-02 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 65 | 161 | 1.93e-02 |
| GO:BP | GO:0061383 | trabecula morphogenesis | 22 | 41 | 1.95e-02 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 69 | 173 | 1.95e-02 |
| GO:BP | GO:0048617 | embryonic foregut morphogenesis | 6 | 6 | 1.95e-02 |
| GO:BP | GO:0021814 | cell motility involved in cerebral cortex radial glia guided migration | 6 | 6 | 1.95e-02 |
| GO:BP | GO:0065009 | regulation of molecular function | 416 | 1299 | 1.97e-02 |
| GO:BP | GO:0061572 | actin filament bundle organization | 55 | 132 | 2.01e-02 |
| GO:BP | GO:0010033 | response to organic substance | 545 | 1736 | 2.06e-02 |
| GO:BP | GO:0099175 | regulation of postsynapse organization | 37 | 81 | 2.06e-02 |
| GO:BP | GO:0035725 | sodium ion transmembrane transport | 42 | 95 | 2.08e-02 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 42 | 95 | 2.08e-02 |
| GO:BP | GO:0071310 | cellular response to organic substance | 415 | 1297 | 2.11e-02 |
| GO:BP | GO:0086005 | ventricular cardiac muscle cell action potential | 17 | 29 | 2.11e-02 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 139 | 390 | 2.15e-02 |
| GO:BP | GO:0050890 | cognition | 82 | 213 | 2.20e-02 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 803 | 2622 | 2.22e-02 |
| GO:BP | GO:0048593 | camera-type eye morphogenesis | 39 | 87 | 2.27e-02 |
| GO:BP | GO:0007254 | JNK cascade | 53 | 127 | 2.31e-02 |
| GO:BP | GO:0018210 | peptidyl-threonine modification | 35 | 76 | 2.31e-02 |
| GO:BP | GO:0010720 | positive regulation of cell development | 113 | 309 | 2.32e-02 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 200 | 587 | 2.41e-02 |
| GO:BP | GO:0007612 | learning | 47 | 110 | 2.45e-02 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 317 | 972 | 2.45e-02 |
| GO:BP | GO:0021987 | cerebral cortex development | 41 | 93 | 2.47e-02 |
| GO:BP | GO:0120033 | negative regulation of plasma membrane bounded cell projection assembly | 16 | 27 | 2.50e-02 |
| GO:BP | GO:0040008 | regulation of growth | 161 | 462 | 2.57e-02 |
| GO:BP | GO:0032271 | regulation of protein polymerization | 64 | 160 | 2.58e-02 |
| GO:BP | GO:0060419 | heart growth | 33 | 71 | 2.59e-02 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 67 | 169 | 2.59e-02 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 395 | 1234 | 2.62e-02 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 30 | 63 | 2.66e-02 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 394 | 1231 | 2.66e-02 |
| GO:BP | GO:0006811 | monoatomic ion transport | 240 | 719 | 2.66e-02 |
| GO:BP | GO:0032231 | regulation of actin filament bundle assembly | 38 | 85 | 2.67e-02 |
| GO:BP | GO:0001954 | positive regulation of cell-matrix adhesion | 22 | 42 | 2.67e-02 |
| GO:BP | GO:0007517 | muscle organ development | 99 | 267 | 2.72e-02 |
| GO:BP | GO:0030001 | metal ion transport | 180 | 524 | 2.72e-02 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 39 | 88 | 2.78e-02 |
| GO:BP | GO:0006814 | sodium ion transport | 56 | 137 | 2.81e-02 |
| GO:BP | GO:0098657 | import into cell | 210 | 622 | 2.86e-02 |
| GO:BP | GO:0035025 | positive regulation of Rho protein signal transduction | 15 | 25 | 2.90e-02 |
| GO:BP | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 47 | 111 | 2.91e-02 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 205 | 606 | 2.91e-02 |
| GO:BP | GO:0051234 | establishment of localization | 979 | 3240 | 2.92e-02 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 238 | 714 | 2.92e-02 |
| GO:BP | GO:0006898 | receptor-mediated endocytosis | 71 | 182 | 2.93e-02 |
| GO:BP | GO:0018107 | peptidyl-threonine phosphorylation | 32 | 69 | 3.03e-02 |
| GO:BP | GO:0003281 | ventricular septum development | 32 | 69 | 3.03e-02 |
| GO:BP | GO:0050709 | negative regulation of protein secretion | 23 | 45 | 3.04e-02 |
| GO:BP | GO:0019722 | calcium-mediated signaling | 51 | 123 | 3.11e-02 |
| GO:BP | GO:0140352 | export from cell | 201 | 594 | 3.12e-02 |
| GO:BP | GO:0042592 | homeostatic process | 365 | 1137 | 3.12e-02 |
| GO:BP | GO:0048546 | digestive tract morphogenesis | 17 | 30 | 3.13e-02 |
| GO:BP | GO:0060323 | head morphogenesis | 17 | 30 | 3.13e-02 |
| GO:BP | GO:0060065 | uterus development | 10 | 14 | 3.13e-02 |
| GO:BP | GO:0002363 | alpha-beta T cell lineage commitment | 10 | 14 | 3.13e-02 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 21 | 40 | 3.14e-02 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 33 | 72 | 3.17e-02 |
| GO:BP | GO:0086091 | regulation of heart rate by cardiac conduction | 19 | 35 | 3.18e-02 |
| GO:BP | GO:0003012 | muscle system process | 117 | 325 | 3.21e-02 |
| GO:BP | GO:0038203 | TORC2 signaling | 8 | 10 | 3.21e-02 |
| GO:BP | GO:0045176 | apical protein localization | 9 | 12 | 3.25e-02 |
| GO:BP | GO:0032486 | Rap protein signal transduction | 9 | 12 | 3.25e-02 |
| GO:BP | GO:0046339 | diacylglycerol metabolic process | 14 | 23 | 3.30e-02 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 46 | 109 | 3.30e-02 |
| GO:BP | GO:0050772 | positive regulation of axonogenesis | 30 | 64 | 3.30e-02 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 62 | 156 | 3.30e-02 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 648 | 2100 | 3.31e-02 |
| GO:BP | GO:0010718 | positive regulation of epithelial to mesenchymal transition | 24 | 48 | 3.33e-02 |
| GO:BP | GO:0030198 | extracellular matrix organization | 84 | 223 | 3.45e-02 |
| GO:BP | GO:0030832 | regulation of actin filament length | 49 | 118 | 3.49e-02 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 31 | 67 | 3.52e-02 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 110 | 304 | 3.52e-02 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 144 | 412 | 3.57e-02 |
| GO:BP | GO:0030041 | actin filament polymerization | 51 | 124 | 3.57e-02 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 92 | 248 | 3.57e-02 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 52 | 127 | 3.60e-02 |
| GO:BP | GO:0044319 | wound healing, spreading of cells | 16 | 28 | 3.62e-02 |
| GO:BP | GO:0090505 | epiboly involved in wound healing | 16 | 28 | 3.62e-02 |
| GO:BP | GO:0048839 | inner ear development | 54 | 133 | 3.65e-02 |
| GO:BP | GO:0051896 | regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 70 | 181 | 3.72e-02 |
| GO:BP | GO:0006810 | transport | 903 | 2984 | 3.72e-02 |
| GO:BP | GO:0032924 | activin receptor signaling pathway | 18 | 33 | 3.72e-02 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 71 | 184 | 3.72e-02 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 66 | 169 | 3.75e-02 |
| GO:BP | GO:0120032 | regulation of plasma membrane bounded cell projection assembly | 62 | 157 | 3.75e-02 |
| GO:BP | GO:0043062 | extracellular structure organization | 84 | 224 | 3.81e-02 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 84 | 224 | 3.81e-02 |
| GO:BP | GO:0110020 | regulation of actomyosin structure organization | 38 | 87 | 3.82e-02 |
| GO:BP | GO:0010586 | miRNA metabolic process | 38 | 87 | 3.82e-02 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 296 | 911 | 3.83e-02 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 80 | 212 | 3.93e-02 |
| GO:BP | GO:0003407 | neural retina development | 23 | 46 | 3.93e-02 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 598 | 1934 | 4.04e-02 |
| GO:BP | GO:0070371 | ERK1 and ERK2 cascade | 74 | 194 | 4.12e-02 |
| GO:BP | GO:1903522 | regulation of blood circulation | 74 | 194 | 4.12e-02 |
| GO:BP | GO:0003170 | heart valve development | 30 | 65 | 4.12e-02 |
| GO:BP | GO:0010171 | body morphogenesis | 21 | 41 | 4.20e-02 |
| GO:BP | GO:0141091 | transforming growth factor beta receptor superfamily signaling pathway | 95 | 259 | 4.21e-02 |
| GO:BP | GO:0002761 | regulation of myeloid leukocyte differentiation | 35 | 79 | 4.21e-02 |
| GO:BP | GO:0045444 | fat cell differentiation | 69 | 179 | 4.22e-02 |
| GO:BP | GO:0097484 | dendrite extension | 15 | 26 | 4.22e-02 |
| GO:BP | GO:0021799 | cerebral cortex radially oriented cell migration | 15 | 26 | 4.22e-02 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 169 | 495 | 4.28e-02 |
| GO:BP | GO:0008015 | blood circulation | 126 | 357 | 4.29e-02 |
| GO:BP | GO:0002360 | T cell lineage commitment | 12 | 19 | 4.29e-02 |
| GO:BP | GO:0032233 | positive regulation of actin filament bundle assembly | 24 | 49 | 4.29e-02 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 177 | 521 | 4.29e-02 |
| GO:BP | GO:0030833 | regulation of actin filament polymerization | 43 | 102 | 4.30e-02 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 79 | 210 | 4.43e-02 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 284 | 874 | 4.50e-02 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 97 | 266 | 4.53e-02 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 88 | 238 | 4.53e-02 |
| GO:BP | GO:0033036 | macromolecule localization | 706 | 2309 | 4.63e-02 |
| GO:BP | GO:0008064 | regulation of actin polymerization or depolymerization | 48 | 117 | 4.66e-02 |
| GO:BP | GO:0002042 | cell migration involved in sprouting angiogenesis | 22 | 44 | 4.66e-02 |
| GO:BP | GO:0042391 | regulation of membrane potential | 102 | 282 | 4.67e-02 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 70 | 183 | 4.73e-02 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 187 | 555 | 4.74e-02 |
| GO:BP | GO:0003186 | tricuspid valve morphogenesis | 5 | 5 | 4.77e-02 |
| GO:BP | GO:0006836 | neurotransmitter transport | 54 | 135 | 4.77e-02 |
| GO:BP | GO:0061038 | uterus morphogenesis | 5 | 5 | 4.77e-02 |
| GO:BP | GO:0021815 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration | 5 | 5 | 4.77e-02 |
| GO:BP | GO:0097061 | dendritic spine organization | 29 | 63 | 4.77e-02 |
| GO:BP | GO:0035696 | monocyte extravasation | 5 | 5 | 4.77e-02 |
| GO:BP | GO:0048878 | chemical homeostasis | 222 | 670 | 4.77e-02 |
| GO:BP | GO:0045637 | regulation of myeloid cell differentiation | 54 | 135 | 4.77e-02 |
| GO:BP | GO:1905274 | regulation of modification of postsynaptic actin cytoskeleton | 5 | 5 | 4.77e-02 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 55 | 138 | 4.78e-02 |
| GO:BP | GO:0060491 | regulation of cell projection assembly | 62 | 159 | 4.78e-02 |
| GO:BP | GO:0099173 | postsynapse organization | 58 | 147 | 4.79e-02 |
| GO:BP | GO:0035313 | wound healing, spreading of epidermal cells | 11 | 17 | 4.79e-02 |
| GO:BP | GO:0051492 | regulation of stress fiber assembly | 34 | 77 | 4.79e-02 |
| GO:BP | GO:0048048 | embryonic eye morphogenesis | 14 | 24 | 4.84e-02 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 256 | 783 | 4.91e-02 |
| GO:BP | GO:0003179 | heart valve morphogenesis | 26 | 55 | 4.95e-02 |
| GO:BP | GO:0007389 | pattern specification process | 103 | 286 | 4.96e-02 |
EARnomtable_20k %>%
filter(source=="KEGG", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "KEGG terms found in EAR neargenes_20k") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| KEGG | KEGG:04510 | Focal adhesion | 79 | 169 | 5.32e-05 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 73 | 162 | 4.35e-04 |
| KEGG | KEGG:04520 | Adherens junction | 42 | 83 | 1.27e-03 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 58 | 133 | 6.21e-03 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 68 | 162 | 6.21e-03 |
| KEGG | KEGG:04360 | Axon guidance | 65 | 155 | 7.57e-03 |
| KEGG | KEGG:04713 | Circadian entrainment | 33 | 68 | 1.29e-02 |
| KEGG | KEGG:03013 | Nucleocytoplasmic transport | 43 | 96 | 1.40e-02 |
| KEGG | KEGG:05100 | Bacterial invasion of epithelial cells | 34 | 73 | 2.14e-02 |
| KEGG | KEGG:04512 | ECM-receptor interaction | 31 | 66 | 2.78e-02 |
| KEGG | KEGG:04530 | Tight junction | 53 | 129 | 2.94e-02 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 35 | 78 | 3.11e-02 |
| KEGG | KEGG:04720 | Long-term potentiation | 25 | 51 | 3.12e-02 |
| KEGG | KEGG:00561 | Glycerolipid metabolism | 22 | 44 | 4.07e-02 |
| KEGG | KEGG:04730 | Long-term depression | 20 | 39 | 4.12e-02 |
| KEGG | KEGG:04974 | Protein digestion and absorption | 26 | 56 | 4.54e-02 |
| KEGG | KEGG:05412 | Arrhythmogenic right ventricular cardiomyopathy | 30 | 67 | 4.54e-02 |
| KEGG | KEGG:04921 | Oxytocin signaling pathway | 46 | 113 | 4.54e-02 |
| KEGG | KEGG:04742 | Taste transduction | 12 | 20 | 4.54e-02 |
| KEGG | KEGG:05200 | Pathways in cancer | 135 | 391 | 4.54e-02 |
| KEGG | KEGG:04072 | Phospholipase D signaling pathway | 44 | 107 | 4.54e-02 |
EAR peaks overlapping enhancers
# EAR_resgenes_20k_enh <- gost(query = unique(EAR_enh_peaks_20k$NCBI_gene),
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = FALSE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = background_df$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(EAR_resgenes_20k_enh,"data/GO_KEGG_analysis/EAR_resgenes_20k_enh.RDS")
# EARnom <- gostplot(EAR_resgenes_20k_enh, capped = FALSE, interactive = TRUE)
# EARnom
EARnomtable_20k_enh <- EAR_resgenes_20k_enh$result %>%
dplyr::select(c(source, term_id,
term_name,intersection_size,
term_size, p_value))# %>%
EARnomtable_20k_enh %>%
filter(source=="GO:BP", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(.,caption = "GO:BP terms found in EAR enhancer neargenes_20k" ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0050789 | regulation of biological process | 579 | 7516 | 8.40e-13 |
| GO:BP | GO:0050794 | regulation of cellular process | 556 | 7153 | 1.41e-12 |
| GO:BP | GO:0065007 | biological regulation | 587 | 7734 | 5.33e-12 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 344 | 3954 | 1.64e-10 |
| GO:BP | GO:0048518 | positive regulation of biological process | 366 | 4301 | 2.84e-10 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 146 | 1354 | 1.67e-08 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 135 | 1231 | 3.06e-08 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 135 | 1234 | 3.18e-08 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 304 | 3550 | 5.73e-08 |
| GO:BP | GO:0048519 | negative regulation of biological process | 318 | 3764 | 7.00e-08 |
| GO:BP | GO:0032502 | developmental process | 339 | 4179 | 1.68e-06 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 171 | 1791 | 1.77e-06 |
| GO:BP | GO:0050793 | regulation of developmental process | 164 | 1697 | 1.77e-06 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 150 | 1514 | 1.77e-06 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 174 | 1844 | 2.80e-06 |
| GO:BP | GO:0048856 | anatomical structure development | 316 | 3867 | 3.01e-06 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 173 | 1841 | 3.77e-06 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 174 | 1856 | 3.79e-06 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 230 | 2663 | 1.11e-05 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 172 | 1862 | 1.25e-05 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 166 | 1787 | 1.51e-05 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 226 | 2622 | 1.60e-05 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 194 | 2186 | 2.42e-05 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 171 | 1879 | 3.04e-05 |
| GO:BP | GO:0048869 | cellular developmental process | 237 | 2804 | 3.04e-05 |
| GO:BP | GO:0030154 | cell differentiation | 237 | 2803 | 3.04e-05 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 209 | 2410 | 3.31e-05 |
| GO:BP | GO:0032501 | multicellular organismal process | 351 | 4504 | 3.63e-05 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 209 | 2415 | 3.63e-05 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 209 | 2422 | 4.41e-05 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 311 | 3918 | 5.45e-05 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 202 | 2333 | 5.65e-05 |
| GO:BP | GO:0006351 | DNA-templated transcription | 214 | 2510 | 6.90e-05 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 281 | 3486 | 7.41e-05 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 92 | 874 | 8.52e-05 |
| GO:BP | GO:0007275 | multicellular organism development | 259 | 3170 | 8.66e-05 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 240 | 2895 | 8.81e-05 |
| GO:BP | GO:0010468 | regulation of gene expression | 273 | 3379 | 8.99e-05 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 315 | 4015 | 1.10e-04 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 214 | 2533 | 1.10e-04 |
| GO:BP | GO:0040007 | growth | 77 | 697 | 1.10e-04 |
| GO:BP | GO:0048731 | system development | 227 | 2719 | 1.10e-04 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 285 | 3569 | 1.11e-04 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 286 | 3595 | 1.42e-04 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 170 | 1924 | 1.42e-04 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 170 | 1934 | 1.90e-04 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 148 | 1635 | 2.08e-04 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 147 | 1624 | 2.23e-04 |
| GO:BP | GO:0050896 | response to stimulus | 401 | 5379 | 2.36e-04 |
| GO:BP | GO:0072359 | circulatory system development | 89 | 864 | 2.36e-04 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 144 | 1588 | 2.49e-04 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 123 | 1314 | 3.34e-04 |
| GO:BP | GO:0009987 | cellular process | 713 | 10686 | 3.95e-04 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 325 | 4231 | 3.95e-04 |
| GO:BP | GO:0019222 | regulation of metabolic process | 349 | 4603 | 4.36e-04 |
| GO:BP | GO:0008150 | biological_process | 741 | 11244 | 5.30e-04 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 179 | 2100 | 5.42e-04 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 110 | 1157 | 5.42e-04 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 103 | 1067 | 5.71e-04 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 232 | 2867 | 6.18e-04 |
| GO:BP | GO:0010586 | miRNA metabolic process | 18 | 87 | 6.29e-04 |
| GO:BP | GO:0071310 | cellular response to organic substance | 120 | 1297 | 6.70e-04 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 323 | 4233 | 7.16e-04 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 66 | 609 | 8.43e-04 |
| GO:BP | GO:0001568 | blood vessel development | 59 | 525 | 8.47e-04 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 90 | 913 | 9.42e-04 |
| GO:BP | GO:0001944 | vasculature development | 61 | 552 | 9.79e-04 |
| GO:BP | GO:0010033 | response to organic substance | 151 | 1736 | 1.06e-03 |
| GO:BP | GO:0051716 | cellular response to stimulus | 348 | 4640 | 1.11e-03 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 50 | 427 | 1.28e-03 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 143 | 1633 | 1.30e-03 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 93 | 965 | 1.50e-03 |
| GO:BP | GO:0048589 | developmental growth | 56 | 501 | 1.50e-03 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 98 | 1034 | 1.69e-03 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 98 | 1035 | 1.73e-03 |
| GO:BP | GO:0023052 | signaling | 299 | 3918 | 1.79e-03 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 93 | 973 | 1.92e-03 |
| GO:BP | GO:0007154 | cell communication | 303 | 3983 | 1.92e-03 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 232 | 2927 | 2.12e-03 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 232 | 2928 | 2.15e-03 |
| GO:BP | GO:0060419 | heart growth | 15 | 71 | 2.15e-03 |
| GO:BP | GO:0040008 | regulation of growth | 52 | 462 | 2.22e-03 |
| GO:BP | GO:0061061 | muscle structure development | 58 | 536 | 2.42e-03 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 14 | 64 | 2.53e-03 |
| GO:BP | GO:0023057 | negative regulation of signaling | 97 | 1036 | 2.63e-03 |
| GO:BP | GO:0048468 | cell development | 159 | 1886 | 2.63e-03 |
| GO:BP | GO:0016477 | cell migration | 95 | 1011 | 2.67e-03 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 109 | 1197 | 2.67e-03 |
| GO:BP | GO:0016043 | cellular component organization | 355 | 4802 | 2.67e-03 |
| GO:BP | GO:0000165 | MAPK cascade | 55 | 504 | 2.84e-03 |
| GO:BP | GO:0048513 | animal organ development | 167 | 2004 | 2.85e-03 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 163 | 1951 | 3.09e-03 |
| GO:BP | GO:0035556 | intracellular signal transduction | 163 | 1951 | 3.09e-03 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 237 | 3026 | 3.23e-03 |
| GO:BP | GO:0010646 | regulation of cell communication | 197 | 2444 | 3.50e-03 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 50 | 450 | 3.76e-03 |
| GO:BP | GO:0035295 | tube development | 77 | 787 | 3.79e-03 |
| GO:BP | GO:1902895 | positive regulation of miRNA transcription | 11 | 44 | 4.08e-03 |
| GO:BP | GO:0016070 | RNA metabolic process | 258 | 3352 | 4.08e-03 |
| GO:BP | GO:0023051 | regulation of signaling | 196 | 2438 | 4.08e-03 |
| GO:BP | GO:0001525 | angiogenesis | 44 | 381 | 4.08e-03 |
| GO:BP | GO:0016049 | cell growth | 44 | 382 | 4.22e-03 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 365 | 4988 | 4.22e-03 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 13 | 60 | 4.24e-03 |
| GO:BP | GO:0007165 | signal transduction | 275 | 3611 | 4.24e-03 |
| GO:BP | GO:0060644 | mammary gland epithelial cell differentiation | 6 | 13 | 4.24e-03 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 89 | 951 | 4.62e-03 |
| GO:BP | GO:0061614 | miRNA transcription | 13 | 61 | 4.94e-03 |
| GO:BP | GO:0042221 | response to chemical | 186 | 2304 | 4.94e-03 |
| GO:BP | GO:0046328 | regulation of JNK cascade | 18 | 105 | 4.99e-03 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 147 | 1751 | 5.34e-03 |
| GO:BP | GO:0048870 | cell motility | 101 | 1120 | 6.04e-03 |
| GO:BP | GO:0010842 | retina layer formation | 6 | 14 | 6.58e-03 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 14 | 72 | 7.29e-03 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 80 | 850 | 8.39e-03 |
| GO:BP | GO:0033500 | carbohydrate homeostasis | 24 | 170 | 8.62e-03 |
| GO:BP | GO:0001678 | intracellular glucose homeostasis | 18 | 110 | 8.69e-03 |
| GO:BP | GO:0051641 | cellular localization | 206 | 2623 | 8.73e-03 |
| GO:BP | GO:2000145 | regulation of cell motility | 68 | 700 | 1.04e-02 |
| GO:BP | GO:0035239 | tube morphogenesis | 63 | 638 | 1.14e-02 |
| GO:BP | GO:2000630 | positive regulation of miRNA metabolic process | 11 | 50 | 1.15e-02 |
| GO:BP | GO:0048699 | generation of neurons | 96 | 1076 | 1.22e-02 |
| GO:BP | GO:0097240 | chromosome attachment to the nuclear envelope | 3 | 3 | 1.27e-02 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 78 | 837 | 1.29e-02 |
| GO:BP | GO:0040012 | regulation of locomotion | 69 | 720 | 1.32e-02 |
| GO:BP | GO:0006325 | chromatin organization | 60 | 604 | 1.32e-02 |
| GO:BP | GO:0001558 | regulation of cell growth | 37 | 323 | 1.38e-02 |
| GO:BP | GO:0060041 | retina development in camera-type eye | 17 | 105 | 1.38e-02 |
| GO:BP | GO:0030182 | neuron differentiation | 91 | 1014 | 1.38e-02 |
| GO:BP | GO:0061117 | negative regulation of heart growth | 6 | 16 | 1.38e-02 |
| GO:BP | GO:0055022 | negative regulation of cardiac muscle tissue growth | 6 | 16 | 1.38e-02 |
| GO:BP | GO:0042592 | homeostatic process | 100 | 1137 | 1.40e-02 |
| GO:BP | GO:0051179 | localization | 278 | 3731 | 1.50e-02 |
| GO:BP | GO:0035265 | organ growth | 19 | 127 | 1.67e-02 |
| GO:BP | GO:0042593 | glucose homeostasis | 23 | 169 | 1.73e-02 |
| GO:BP | GO:0014706 | striated muscle tissue development | 27 | 213 | 1.79e-02 |
| GO:BP | GO:0030029 | actin filament-based process | 62 | 640 | 1.87e-02 |
| GO:BP | GO:0032879 | regulation of localization | 121 | 1439 | 1.87e-02 |
| GO:BP | GO:0030334 | regulation of cell migration | 64 | 668 | 2.00e-02 |
| GO:BP | GO:0060044 | negative regulation of cardiac muscle cell proliferation | 4 | 7 | 2.09e-02 |
| GO:BP | GO:0060159 | regulation of dopamine receptor signaling pathway | 4 | 7 | 2.09e-02 |
| GO:BP | GO:0071824 | protein-DNA complex organization | 65 | 683 | 2.12e-02 |
| GO:BP | GO:0022008 | neurogenesis | 107 | 1251 | 2.21e-02 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 94 | 1073 | 2.26e-02 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 33 | 286 | 2.27e-02 |
| GO:BP | GO:0045926 | negative regulation of growth | 24 | 184 | 2.27e-02 |
| GO:BP | GO:0040011 | locomotion | 74 | 805 | 2.34e-02 |
| GO:BP | GO:0050772 | positive regulation of axonogenesis | 12 | 64 | 2.37e-02 |
| GO:BP | GO:0009725 | response to hormone | 59 | 610 | 2.46e-02 |
| GO:BP | GO:0033993 | response to lipid | 56 | 573 | 2.61e-02 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 95 | 1093 | 2.61e-02 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 260 | 3495 | 2.61e-02 |
| GO:BP | GO:0097306 | cellular response to alcohol | 13 | 74 | 2.63e-02 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 45 | 435 | 2.65e-02 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 66 | 704 | 2.65e-02 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 86 | 972 | 2.68e-02 |
| GO:BP | GO:0014741 | negative regulation of muscle hypertrophy | 7 | 25 | 2.69e-02 |
| GO:BP | GO:0033036 | macromolecule localization | 180 | 2309 | 2.85e-02 |
| GO:BP | GO:0060537 | muscle tissue development | 37 | 339 | 2.87e-02 |
| GO:BP | GO:0009966 | regulation of signal transduction | 170 | 2166 | 3.01e-02 |
| GO:BP | GO:0048878 | chemical homeostasis | 63 | 670 | 3.12e-02 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 52 | 528 | 3.15e-02 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 25 | 201 | 3.20e-02 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 51 | 516 | 3.22e-02 |
| GO:BP | GO:0006810 | transport | 225 | 2984 | 3.23e-02 |
| GO:BP | GO:0051726 | regulation of cell cycle | 79 | 885 | 3.24e-02 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 207 | 2717 | 3.28e-02 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 38 | 355 | 3.35e-02 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 50 | 505 | 3.37e-02 |
| GO:BP | GO:1903294 | regulation of glutamate secretion, neurotransmission | 3 | 4 | 3.51e-02 |
| GO:BP | GO:0007254 | JNK cascade | 18 | 127 | 3.56e-02 |
| GO:BP | GO:0007015 | actin filament organization | 38 | 357 | 3.64e-02 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 28 | 238 | 3.66e-02 |
| GO:BP | GO:0070848 | response to growth factor | 53 | 547 | 3.81e-02 |
| GO:BP | GO:0007399 | nervous system development | 142 | 1775 | 3.82e-02 |
| GO:BP | GO:0032880 | regulation of protein localization | 64 | 691 | 3.82e-02 |
| GO:BP | GO:0043933 | protein-containing complex organization | 148 | 1863 | 3.91e-02 |
| GO:BP | GO:0048661 | positive regulation of smooth muscle cell proliferation | 11 | 60 | 4.01e-02 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 32 | 287 | 4.01e-02 |
| GO:BP | GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 19 | 140 | 4.17e-02 |
| GO:BP | GO:0006338 | chromatin remodeling | 48 | 486 | 4.17e-02 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 32 | 288 | 4.17e-02 |
| GO:BP | GO:0007266 | Rho protein signal transduction | 16 | 109 | 4.28e-02 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 276 | 3777 | 4.28e-02 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 56 | 591 | 4.40e-02 |
| GO:BP | GO:0030307 | positive regulation of cell growth | 17 | 120 | 4.50e-02 |
| GO:BP | GO:0033002 | muscle cell proliferation | 20 | 152 | 4.50e-02 |
| GO:BP | GO:0140747 | regulation of ncRNA transcription | 13 | 80 | 4.56e-02 |
| GO:BP | GO:0048638 | regulation of developmental growth | 28 | 243 | 4.61e-02 |
| GO:BP | GO:0070372 | regulation of ERK1 and ERK2 cascade | 22 | 175 | 4.78e-02 |
| GO:BP | GO:0007264 | small GTPase-mediated signal transduction | 40 | 390 | 4.91e-02 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 89 | 1040 | 4.91e-02 |
| GO:BP | GO:0048511 | rhythmic process | 26 | 221 | 4.91e-02 |
| GO:BP | GO:0048103 | somatic stem cell division | 4 | 9 | 4.91e-02 |
| GO:BP | GO:0034380 | high-density lipoprotein particle assembly | 4 | 9 | 4.91e-02 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 58 | 622 | 4.98e-02 |
EARnomtable_20k_enh %>%
filter(source=="KEGG", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "KEGG terms found in EAR enhancer neargenes_20k") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| KEGG | KEGG:05203 | Viral carcinogenesis | 22 | 142 | 2.18e-02 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 23 | 162 | 2.93e-02 |
ESR 20,000 bp NG
# ESR_resgenes_20k <- gost(query = ESR_peak_list_20k$ENTREZID,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = FALSE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = background_df$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(ESR_resgenes_20k,"data/GO_KEGG_analysis/ESR_resgenes_20k.RDS")
# ESRnom <- gostplot(ESR_resgenes_20k, capped = FALSE, interactive = TRUE)
# ESRnom
ESRnomtable_20k <- ESR_resgenes_20k$result %>%
dplyr::select(c(source, term_id,
term_name,intersection_size,
term_size, p_value))# %>%
ESRnomtable_20k %>%
filter(source=="GO:BP", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(.,caption = "GO:BP terms found in ESR nESRgenes_20k" ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0032501 | multicellular organismal process | 2259 | 4504 | 1.68e-50 |
| GO:BP | GO:0048856 | anatomical structure development | 1964 | 3867 | 6.68e-46 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 1034 | 1844 | 9.73e-42 |
| GO:BP | GO:0007154 | cell communication | 1992 | 3983 | 9.85e-41 |
| GO:BP | GO:0023052 | signaling | 1962 | 3918 | 2.10e-40 |
| GO:BP | GO:0007275 | multicellular organism development | 1633 | 3170 | 2.57e-40 |
| GO:BP | GO:0032502 | developmental process | 2072 | 4179 | 6.91e-40 |
| GO:BP | GO:0048731 | system development | 1424 | 2719 | 1.37e-38 |
| GO:BP | GO:0050896 | response to stimulus | 2549 | 5379 | 2.99e-33 |
| GO:BP | GO:0030154 | cell differentiation | 1438 | 2803 | 6.60e-33 |
| GO:BP | GO:0007165 | signal transduction | 1794 | 3611 | 6.60e-33 |
| GO:BP | GO:0048869 | cellular developmental process | 1438 | 2804 | 7.71e-33 |
| GO:BP | GO:0051716 | cellular response to stimulus | 2225 | 4640 | 8.80e-31 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 1024 | 1924 | 4.99e-29 |
| GO:BP | GO:0007399 | nervous system development | 953 | 1775 | 1.85e-28 |
| GO:BP | GO:0007155 | cell adhesion | 553 | 941 | 2.52e-27 |
| GO:BP | GO:0048468 | cell development | 998 | 1886 | 5.84e-27 |
| GO:BP | GO:0009888 | tissue development | 746 | 1348 | 1.66e-26 |
| GO:BP | GO:0048513 | animal organ development | 1049 | 2004 | 2.68e-26 |
| GO:BP | GO:0050793 | regulation of developmental process | 899 | 1697 | 5.03e-24 |
| GO:BP | GO:0010646 | regulation of cell communication | 1236 | 2444 | 7.74e-24 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 491 | 837 | 8.49e-24 |
| GO:BP | GO:0023051 | regulation of signaling | 1231 | 2438 | 2.18e-23 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 943 | 1801 | 3.18e-23 |
| GO:BP | GO:0048870 | cell motility | 625 | 1120 | 5.48e-23 |
| GO:BP | GO:0003008 | system process | 633 | 1139 | 1.04e-22 |
| GO:BP | GO:0016477 | cell migration | 571 | 1011 | 1.80e-22 |
| GO:BP | GO:0022008 | neurogenesis | 684 | 1251 | 3.30e-22 |
| GO:BP | GO:0032989 | cellular anatomical entity morphogenesis | 360 | 588 | 1.80e-21 |
| GO:BP | GO:0000902 | cell morphogenesis | 421 | 713 | 5.94e-21 |
| GO:BP | GO:0030029 | actin filament-based process | 384 | 640 | 1.02e-20 |
| GO:BP | GO:0034330 | cell junction organization | 336 | 545 | 1.10e-20 |
| GO:BP | GO:0048699 | generation of neurons | 594 | 1076 | 2.83e-20 |
| GO:BP | GO:0035239 | tube morphogenesis | 381 | 638 | 4.70e-20 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 704 | 1314 | 7.30e-20 |
| GO:BP | GO:0050789 | regulation of biological process | 3343 | 7516 | 8.62e-20 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 365 | 609 | 1.49e-19 |
| GO:BP | GO:0035295 | tube development | 452 | 787 | 2.39e-19 |
| GO:BP | GO:0030182 | neuron differentiation | 560 | 1014 | 4.37e-19 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 416 | 717 | 8.67e-19 |
| GO:BP | GO:0031175 | neuron projection development | 428 | 743 | 1.40e-18 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 392 | 673 | 4.88e-18 |
| GO:BP | GO:0007267 | cell-cell signaling | 588 | 1082 | 4.88e-18 |
| GO:BP | GO:0048666 | neuron development | 472 | 838 | 5.11e-18 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 568 | 1040 | 5.36e-18 |
| GO:BP | GO:0065007 | biological regulation | 3418 | 7734 | 5.47e-18 |
| GO:BP | GO:0072359 | circulatory system development | 483 | 864 | 1.26e-17 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 618 | 1150 | 1.28e-17 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 301 | 494 | 1.95e-17 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 303 | 498 | 1.95e-17 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 337 | 566 | 2.06e-17 |
| GO:BP | GO:0048729 | tissue morphogenesis | 280 | 453 | 2.16e-17 |
| GO:BP | GO:0050794 | regulation of cellular process | 3179 | 7153 | 3.02e-17 |
| GO:BP | GO:0030030 | cell projection organization | 631 | 1182 | 3.34e-17 |
| GO:BP | GO:0044057 | regulation of system process | 243 | 384 | 8.34e-17 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 521 | 951 | 8.81e-17 |
| GO:BP | GO:0009966 | regulation of signal transduction | 1073 | 2166 | 1.71e-16 |
| GO:BP | GO:0065008 | regulation of biological quality | 951 | 1893 | 1.99e-16 |
| GO:BP | GO:0040011 | locomotion | 449 | 805 | 4.21e-16 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 289 | 479 | 6.47e-16 |
| GO:BP | GO:2000145 | regulation of cell motility | 396 | 700 | 2.18e-15 |
| GO:BP | GO:0035556 | intracellular signal transduction | 971 | 1951 | 2.43e-15 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 265 | 435 | 2.80e-15 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 497 | 913 | 2.86e-15 |
| GO:BP | GO:0001568 | blood vessel development | 310 | 525 | 2.86e-15 |
| GO:BP | GO:0030334 | regulation of cell migration | 380 | 668 | 2.87e-15 |
| GO:BP | GO:0048518 | positive regulation of biological process | 1986 | 4301 | 2.87e-15 |
| GO:BP | GO:0001944 | vasculature development | 323 | 552 | 3.52e-15 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 1838 | 3954 | 3.99e-15 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 1306 | 2717 | 4.82e-15 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 929 | 1862 | 5.69e-15 |
| GO:BP | GO:0040012 | regulation of locomotion | 403 | 720 | 1.04e-14 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 551 | 1035 | 1.49e-14 |
| GO:BP | GO:0003013 | circulatory system process | 258 | 426 | 1.85e-14 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 270 | 450 | 1.92e-14 |
| GO:BP | GO:0060485 | mesenchyme development | 164 | 245 | 2.02e-14 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 297 | 505 | 2.52e-14 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 302 | 516 | 3.45e-14 |
| GO:BP | GO:0007010 | cytoskeleton organization | 599 | 1146 | 7.99e-14 |
| GO:BP | GO:0098609 | cell-cell adhesion | 324 | 564 | 1.03e-13 |
| GO:BP | GO:0034329 | cell junction assembly | 203 | 323 | 1.48e-13 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 563 | 1073 | 2.56e-13 |
| GO:BP | GO:0048519 | negative regulation of biological process | 1743 | 3764 | 3.34e-13 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 619 | 1197 | 4.04e-13 |
| GO:BP | GO:0008015 | blood circulation | 219 | 357 | 4.97e-13 |
| GO:BP | GO:0060429 | epithelium development | 430 | 792 | 7.30e-13 |
| GO:BP | GO:0098657 | import into cell | 349 | 622 | 7.61e-13 |
| GO:BP | GO:0030001 | metal ion transport | 301 | 524 | 1.07e-12 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 217 | 355 | 1.10e-12 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 293 | 508 | 1.19e-12 |
| GO:BP | GO:0051049 | regulation of transport | 571 | 1098 | 1.34e-12 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 267 | 456 | 1.60e-12 |
| GO:BP | GO:0003012 | muscle system process | 201 | 325 | 1.96e-12 |
| GO:BP | GO:0070848 | response to growth factor | 310 | 547 | 4.47e-12 |
| GO:BP | GO:0009790 | embryo development | 440 | 821 | 5.10e-12 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 300 | 528 | 7.44e-12 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 1641 | 3550 | 8.41e-12 |
| GO:BP | GO:0007409 | axonogenesis | 205 | 337 | 1.12e-11 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 221 | 369 | 1.22e-11 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 164 | 258 | 1.71e-11 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 329 | 591 | 1.72e-11 |
| GO:BP | GO:0051960 | regulation of nervous system development | 205 | 339 | 2.48e-11 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 133 | 200 | 2.48e-11 |
| GO:BP | GO:0001525 | angiogenesis | 226 | 381 | 2.51e-11 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 74 | 95 | 2.51e-11 |
| GO:BP | GO:0042221 | response to chemical | 1100 | 2304 | 3.45e-11 |
| GO:BP | GO:0007264 | small GTPase-mediated signal transduction | 230 | 390 | 3.54e-11 |
| GO:BP | GO:0061564 | axon development | 221 | 372 | 3.60e-11 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 112 | 162 | 3.60e-11 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 406 | 757 | 4.04e-11 |
| GO:BP | GO:0048589 | developmental growth | 284 | 501 | 4.53e-11 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 245 | 422 | 6.14e-11 |
| GO:BP | GO:0061061 | muscle structure development | 300 | 536 | 7.98e-11 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 69 | 88 | 8.49e-11 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 137 | 210 | 8.88e-11 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 380 | 706 | 1.21e-10 |
| GO:BP | GO:0006936 | muscle contraction | 160 | 255 | 1.42e-10 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 226 | 386 | 1.45e-10 |
| GO:BP | GO:0006816 | calcium ion transport | 163 | 262 | 2.41e-10 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 148 | 233 | 2.42e-10 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 181 | 298 | 3.17e-10 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 613 | 1218 | 3.24e-10 |
| GO:BP | GO:0023056 | positive regulation of signaling | 614 | 1221 | 3.73e-10 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 265 | 469 | 4.14e-10 |
| GO:BP | GO:0023057 | negative regulation of signaling | 529 | 1036 | 6.12e-10 |
| GO:BP | GO:0099536 | synaptic signaling | 271 | 483 | 6.68e-10 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 203 | 344 | 6.95e-10 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 192 | 322 | 7.08e-10 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 106 | 156 | 7.21e-10 |
| GO:BP | GO:0016043 | cellular component organization | 2151 | 4802 | 7.49e-10 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 263 | 467 | 7.71e-10 |
| GO:BP | GO:0006810 | transport | 1383 | 2984 | 7.86e-10 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 527 | 1034 | 9.38e-10 |
| GO:BP | GO:0050808 | synapse organization | 199 | 337 | 9.77e-10 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 192 | 323 | 9.90e-10 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 288 | 521 | 1.34e-09 |
| GO:BP | GO:0006897 | endocytosis | 274 | 492 | 1.43e-09 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 260 | 463 | 1.45e-09 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 260 | 463 | 1.45e-09 |
| GO:BP | GO:0006468 | protein phosphorylation | 491 | 958 | 1.65e-09 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 123 | 190 | 2.31e-09 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 340 | 633 | 2.31e-09 |
| GO:BP | GO:0032879 | regulation of localization | 706 | 1439 | 2.98e-09 |
| GO:BP | GO:0007423 | sensory organ development | 222 | 387 | 2.98e-09 |
| GO:BP | GO:0007507 | heart development | 276 | 499 | 3.11e-09 |
| GO:BP | GO:0048041 | focal adhesion assembly | 60 | 77 | 3.36e-09 |
| GO:BP | GO:0007411 | axon guidance | 113 | 172 | 3.57e-09 |
| GO:BP | GO:0097485 | neuron projection guidance | 113 | 172 | 3.57e-09 |
| GO:BP | GO:0048568 | embryonic organ development | 180 | 303 | 4.25e-09 |
| GO:BP | GO:0010720 | positive regulation of cell development | 183 | 309 | 4.25e-09 |
| GO:BP | GO:0140352 | export from cell | 320 | 594 | 5.44e-09 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 637 | 1289 | 6.34e-09 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 231 | 408 | 6.36e-09 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 147 | 239 | 7.52e-09 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 316 | 587 | 7.84e-09 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 158 | 261 | 8.50e-09 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 593 | 1193 | 9.14e-09 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 224 | 395 | 9.76e-09 |
| GO:BP | GO:0006811 | monoatomic ion transport | 377 | 719 | 1.04e-08 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 234 | 416 | 1.07e-08 |
| GO:BP | GO:0016310 | phosphorylation | 571 | 1145 | 1.07e-08 |
| GO:BP | GO:0008283 | cell population proliferation | 643 | 1306 | 1.09e-08 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 151 | 248 | 1.15e-08 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 437 | 850 | 1.17e-08 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 172 | 290 | 1.24e-08 |
| GO:BP | GO:0014706 | striated muscle tissue development | 133 | 213 | 1.26e-08 |
| GO:BP | GO:0060284 | regulation of cell development | 307 | 570 | 1.26e-08 |
| GO:BP | GO:0007015 | actin filament organization | 205 | 357 | 1.28e-08 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 110 | 169 | 1.32e-08 |
| GO:BP | GO:0051179 | localization | 1689 | 3731 | 1.39e-08 |
| GO:BP | GO:1903522 | regulation of blood circulation | 123 | 194 | 1.44e-08 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 492 | 972 | 1.44e-08 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 271 | 495 | 1.66e-08 |
| GO:BP | GO:0007417 | central nervous system development | 364 | 694 | 1.93e-08 |
| GO:BP | GO:0009605 | response to external stimulus | 753 | 1559 | 2.23e-08 |
| GO:BP | GO:0060322 | head development | 291 | 539 | 2.61e-08 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 263 | 480 | 2.70e-08 |
| GO:BP | GO:0055085 | transmembrane transport | 467 | 921 | 3.09e-08 |
| GO:BP | GO:0060047 | heart contraction | 122 | 194 | 3.48e-08 |
| GO:BP | GO:0050877 | nervous system process | 319 | 600 | 3.57e-08 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 537 | 1077 | 3.74e-08 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 544 | 1093 | 4.02e-08 |
| GO:BP | GO:0046903 | secretion | 329 | 622 | 4.02e-08 |
| GO:BP | GO:0048588 | developmental cell growth | 114 | 179 | 4.13e-08 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 166 | 282 | 5.13e-08 |
| GO:BP | GO:0007420 | brain development | 270 | 498 | 6.22e-08 |
| GO:BP | GO:0006812 | monoatomic cation transport | 324 | 614 | 7.54e-08 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 118 | 188 | 7.54e-08 |
| GO:BP | GO:0097553 | calcium ion transmembrane import into cytosol | 83 | 122 | 8.32e-08 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 108 | 169 | 8.44e-08 |
| GO:BP | GO:0003015 | heart process | 126 | 204 | 8.99e-08 |
| GO:BP | GO:0009611 | response to wounding | 214 | 382 | 8.99e-08 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 129 | 210 | 9.44e-08 |
| GO:BP | GO:0060537 | muscle tissue development | 193 | 339 | 9.98e-08 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 167 | 286 | 1.01e-07 |
| GO:BP | GO:0042391 | regulation of membrane potential | 165 | 282 | 1.01e-07 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 136 | 224 | 1.01e-07 |
| GO:BP | GO:0043062 | extracellular structure organization | 136 | 224 | 1.01e-07 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 327 | 622 | 1.02e-07 |
| GO:BP | GO:0042692 | muscle cell differentiation | 185 | 323 | 1.11e-07 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 124 | 201 | 1.29e-07 |
| GO:BP | GO:0030198 | extracellular matrix organization | 135 | 223 | 1.48e-07 |
| GO:BP | GO:0051234 | establishment of localization | 1471 | 3240 | 1.50e-07 |
| GO:BP | GO:0007610 | behavior | 231 | 420 | 1.78e-07 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 830 | 1751 | 2.03e-07 |
| GO:BP | GO:0030900 | forebrain development | 159 | 272 | 2.10e-07 |
| GO:BP | GO:0003007 | heart morphogenesis | 132 | 218 | 2.13e-07 |
| GO:BP | GO:0090109 | regulation of cell-substrate junction assembly | 44 | 55 | 2.17e-07 |
| GO:BP | GO:0051893 | regulation of focal adhesion assembly | 44 | 55 | 2.17e-07 |
| GO:BP | GO:0150116 | regulation of cell-substrate junction organization | 47 | 60 | 2.28e-07 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 216 | 390 | 2.56e-07 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 278 | 521 | 2.56e-07 |
| GO:BP | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | 77 | 113 | 2.60e-07 |
| GO:BP | GO:0048638 | regulation of developmental growth | 144 | 243 | 3.18e-07 |
| GO:BP | GO:0040007 | growth | 359 | 697 | 3.30e-07 |
| GO:BP | GO:0001952 | regulation of cell-matrix adhesion | 64 | 90 | 3.70e-07 |
| GO:BP | GO:0003018 | vascular process in circulatory system | 119 | 194 | 4.07e-07 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 195 | 348 | 4.15e-07 |
| GO:BP | GO:0023061 | signal release | 179 | 315 | 4.18e-07 |
| GO:BP | GO:0032940 | secretion by cell | 291 | 551 | 4.18e-07 |
| GO:BP | GO:0008016 | regulation of heart contraction | 104 | 165 | 4.41e-07 |
| GO:BP | GO:0001775 | cell activation | 326 | 627 | 4.63e-07 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 172 | 301 | 4.66e-07 |
| GO:BP | GO:0090257 | regulation of muscle system process | 110 | 177 | 5.15e-07 |
| GO:BP | GO:0043009 | chordate embryonic development | 270 | 507 | 5.17e-07 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 397 | 783 | 5.55e-07 |
| GO:BP | GO:0030048 | actin filament-based movement | 73 | 107 | 5.64e-07 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 89 | 137 | 6.08e-07 |
| GO:BP | GO:0065009 | regulation of molecular function | 627 | 1299 | 7.82e-07 |
| GO:BP | GO:0003205 | cardiac chamber development | 96 | 151 | 8.38e-07 |
| GO:BP | GO:0051924 | regulation of calcium ion transport | 97 | 153 | 8.67e-07 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 138 | 234 | 9.29e-07 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 127 | 212 | 9.33e-07 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 74 | 110 | 1.11e-06 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 107 | 173 | 1.13e-06 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 82 | 125 | 1.13e-06 |
| GO:BP | GO:0001823 | mesonephros development | 54 | 74 | 1.19e-06 |
| GO:BP | GO:0006915 | apoptotic process | 660 | 1377 | 1.19e-06 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 715 | 1504 | 1.51e-06 |
| GO:BP | GO:0071310 | cellular response to organic substance | 624 | 1297 | 1.51e-06 |
| GO:BP | GO:0010033 | response to organic substance | 816 | 1736 | 1.62e-06 |
| GO:BP | GO:0006935 | chemotaxis | 142 | 244 | 1.78e-06 |
| GO:BP | GO:0042330 | taxis | 142 | 244 | 1.78e-06 |
| GO:BP | GO:0042060 | wound healing | 165 | 291 | 1.78e-06 |
| GO:BP | GO:0001501 | skeletal system development | 192 | 347 | 1.78e-06 |
| GO:BP | GO:0071417 | cellular response to organonitrogen compound | 237 | 442 | 1.80e-06 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 95 | 151 | 1.98e-06 |
| GO:BP | GO:0072164 | mesonephric tubule development | 53 | 73 | 2.05e-06 |
| GO:BP | GO:0072163 | mesonephric epithelium development | 53 | 73 | 2.05e-06 |
| GO:BP | GO:0001654 | eye development | 151 | 263 | 2.20e-06 |
| GO:BP | GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 161 | 284 | 2.57e-06 |
| GO:BP | GO:0016358 | dendrite development | 111 | 183 | 2.65e-06 |
| GO:BP | GO:1903169 | regulation of calcium ion transmembrane transport | 72 | 108 | 2.81e-06 |
| GO:BP | GO:0048880 | sensory system development | 152 | 266 | 2.98e-06 |
| GO:BP | GO:0150063 | visual system development | 151 | 264 | 3.02e-06 |
| GO:BP | GO:0035265 | organ growth | 82 | 127 | 3.04e-06 |
| GO:BP | GO:0033002 | muscle cell proliferation | 95 | 152 | 3.04e-06 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 206 | 379 | 3.48e-06 |
| GO:BP | GO:0001657 | ureteric bud development | 52 | 72 | 3.52e-06 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 107 | 176 | 3.75e-06 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 2188 | 4988 | 3.78e-06 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 77 | 118 | 3.81e-06 |
| GO:BP | GO:0071772 | response to BMP | 77 | 118 | 3.81e-06 |
| GO:BP | GO:0031032 | actomyosin structure organization | 111 | 184 | 3.82e-06 |
| GO:BP | GO:0008219 | cell death | 676 | 1423 | 4.02e-06 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 1105 | 2415 | 4.06e-06 |
| GO:BP | GO:0001763 | morphogenesis of a branching structure | 91 | 145 | 4.15e-06 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 91 | 145 | 4.15e-06 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 162 | 288 | 4.43e-06 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 81 | 126 | 4.59e-06 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 59 | 85 | 4.65e-06 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 140 | 243 | 4.67e-06 |
| GO:BP | GO:0050807 | regulation of synapse organization | 105 | 173 | 5.31e-06 |
| GO:BP | GO:1990138 | neuron projection extension | 85 | 134 | 5.36e-06 |
| GO:BP | GO:0061448 | connective tissue development | 121 | 205 | 5.48e-06 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 1192 | 2622 | 5.52e-06 |
| GO:BP | GO:0012501 | programmed cell death | 673 | 1419 | 5.62e-06 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 77 | 119 | 6.13e-06 |
| GO:BP | GO:0006941 | striated muscle contraction | 91 | 146 | 6.34e-06 |
| GO:BP | GO:0051050 | positive regulation of transport | 294 | 571 | 6.66e-06 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 310 | 606 | 6.78e-06 |
| GO:BP | GO:0010243 | response to organonitrogen compound | 346 | 685 | 6.80e-06 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 167 | 300 | 6.91e-06 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 126 | 216 | 7.43e-06 |
| GO:BP | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | 53 | 75 | 7.75e-06 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 119 | 202 | 7.75e-06 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 110 | 184 | 7.79e-06 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 112 | 188 | 7.79e-06 |
| GO:BP | GO:0061138 | morphogenesis of a branching epithelium | 85 | 135 | 8.23e-06 |
| GO:BP | GO:0090130 | tissue migration | 141 | 247 | 8.45e-06 |
| GO:BP | GO:0022898 | regulation of transmembrane transporter activity | 87 | 139 | 8.65e-06 |
| GO:BP | GO:0048167 | regulation of synaptic plasticity | 87 | 139 | 8.65e-06 |
| GO:BP | GO:0099173 | postsynapse organization | 91 | 147 | 9.51e-06 |
| GO:BP | GO:0045165 | cell fate commitment | 93 | 151 | 9.85e-06 |
| GO:BP | GO:0051283 | negative regulation of sequestering of calcium ion | 61 | 90 | 1.00e-05 |
| GO:BP | GO:0055001 | muscle cell development | 99 | 163 | 1.06e-05 |
| GO:BP | GO:1905314 | semi-lunar valve development | 33 | 41 | 1.10e-05 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 114 | 193 | 1.10e-05 |
| GO:BP | GO:0072001 | renal system development | 139 | 244 | 1.18e-05 |
| GO:BP | GO:0061572 | actin filament bundle organization | 83 | 132 | 1.20e-05 |
| GO:BP | GO:0072073 | kidney epithelium development | 72 | 111 | 1.24e-05 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 250 | 479 | 1.29e-05 |
| GO:BP | GO:0003231 | cardiac ventricle development | 74 | 115 | 1.36e-05 |
| GO:BP | GO:0032409 | regulation of transporter activity | 91 | 148 | 1.43e-05 |
| GO:BP | GO:0060395 | SMAD protein signal transduction | 53 | 76 | 1.45e-05 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 122 | 210 | 1.45e-05 |
| GO:BP | GO:1901698 | response to nitrogen compound | 367 | 736 | 1.45e-05 |
| GO:BP | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 93 | 152 | 1.45e-05 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 59 | 87 | 1.45e-05 |
| GO:BP | GO:0042472 | inner ear morphogenesis | 44 | 60 | 1.45e-05 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 226 | 428 | 1.46e-05 |
| GO:BP | GO:0010631 | epithelial cell migration | 138 | 243 | 1.58e-05 |
| GO:BP | GO:0090132 | epithelium migration | 138 | 243 | 1.58e-05 |
| GO:BP | GO:0051209 | release of sequestered calcium ion into cytosol | 60 | 89 | 1.58e-05 |
| GO:BP | GO:0045321 | leukocyte activation | 273 | 530 | 1.58e-05 |
| GO:BP | GO:0003179 | heart valve morphogenesis | 41 | 55 | 1.70e-05 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 227 | 431 | 1.75e-05 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 82 | 131 | 1.75e-05 |
| GO:BP | GO:0050878 | regulation of body fluid levels | 131 | 229 | 1.75e-05 |
| GO:BP | GO:0060419 | heart growth | 50 | 71 | 1.83e-05 |
| GO:BP | GO:0030509 | BMP signaling pathway | 71 | 110 | 1.86e-05 |
| GO:BP | GO:0048010 | vascular endothelial growth factor receptor signaling pathway | 38 | 50 | 1.91e-05 |
| GO:BP | GO:0051216 | cartilage development | 89 | 145 | 2.01e-05 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 74 | 116 | 2.12e-05 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 108 | 183 | 2.19e-05 |
| GO:BP | GO:0090288 | negative regulation of cellular response to growth factor stimulus | 52 | 75 | 2.33e-05 |
| GO:BP | GO:0043010 | camera-type eye development | 131 | 230 | 2.38e-05 |
| GO:BP | GO:0051046 | regulation of secretion | 210 | 396 | 2.38e-05 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 59 | 88 | 2.50e-05 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 241 | 463 | 2.53e-05 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 241 | 463 | 2.53e-05 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 1061 | 2333 | 2.77e-05 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 283 | 555 | 2.87e-05 |
| GO:BP | GO:0009987 | cellular process | 4477 | 10686 | 2.87e-05 |
| GO:BP | GO:0051282 | regulation of sequestering of calcium ion | 61 | 92 | 2.87e-05 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 355 | 714 | 2.98e-05 |
| GO:BP | GO:0045216 | cell-cell junction organization | 92 | 152 | 3.05e-05 |
| GO:BP | GO:0042592 | homeostatic process | 543 | 1137 | 3.10e-05 |
| GO:BP | GO:0048878 | chemical homeostasis | 335 | 670 | 3.13e-05 |
| GO:BP | GO:0072006 | nephron development | 73 | 115 | 3.18e-05 |
| GO:BP | GO:0060976 | coronary vasculature development | 34 | 44 | 3.70e-05 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 523 | 1093 | 3.73e-05 |
| GO:BP | GO:0003279 | cardiac septum development | 66 | 102 | 3.83e-05 |
| GO:BP | GO:0021915 | neural tube development | 82 | 133 | 4.02e-05 |
| GO:BP | GO:1901890 | positive regulation of cell junction assembly | 52 | 76 | 4.19e-05 |
| GO:BP | GO:0048844 | artery morphogenesis | 42 | 58 | 4.23e-05 |
| GO:BP | GO:0002521 | leukocyte differentiation | 197 | 371 | 4.38e-05 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 47 | 67 | 4.38e-05 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 47 | 67 | 4.38e-05 |
| GO:BP | GO:2000181 | negative regulation of blood vessel morphogenesis | 47 | 67 | 4.38e-05 |
| GO:BP | GO:0015833 | peptide transport | 105 | 179 | 4.38e-05 |
| GO:BP | GO:1901343 | negative regulation of vasculature development | 47 | 67 | 4.38e-05 |
| GO:BP | GO:0030510 | regulation of BMP signaling pathway | 53 | 78 | 4.52e-05 |
| GO:BP | GO:0060078 | regulation of postsynaptic membrane potential | 53 | 78 | 4.52e-05 |
| GO:BP | GO:0001822 | kidney development | 133 | 237 | 5.47e-05 |
| GO:BP | GO:1901342 | regulation of vasculature development | 114 | 198 | 5.56e-05 |
| GO:BP | GO:0030100 | regulation of endocytosis | 113 | 196 | 5.62e-05 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 112 | 194 | 5.67e-05 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 234 | 452 | 5.77e-05 |
| GO:BP | GO:0032412 | regulation of monoatomic ion transmembrane transporter activity | 82 | 134 | 5.93e-05 |
| GO:BP | GO:0051058 | negative regulation of small GTPase mediated signal transduction | 36 | 48 | 5.93e-05 |
| GO:BP | GO:0055008 | cardiac muscle tissue morphogenesis | 40 | 55 | 6.05e-05 |
| GO:BP | GO:0018212 | peptidyl-tyrosine modification | 97 | 164 | 6.31e-05 |
| GO:BP | GO:0009612 | response to mechanical stimulus | 91 | 152 | 6.31e-05 |
| GO:BP | GO:0048644 | muscle organ morphogenesis | 45 | 64 | 6.39e-05 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 45 | 64 | 6.39e-05 |
| GO:BP | GO:0060038 | cardiac muscle cell proliferation | 33 | 43 | 6.54e-05 |
| GO:BP | GO:0048168 | regulation of neuronal synaptic plasticity | 30 | 38 | 6.72e-05 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 396 | 811 | 6.72e-05 |
| GO:BP | GO:0006949 | syncytium formation | 30 | 38 | 6.72e-05 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 222 | 427 | 6.89e-05 |
| GO:BP | GO:0007389 | pattern specification process | 156 | 286 | 6.89e-05 |
| GO:BP | GO:0048813 | dendrite morphogenesis | 70 | 111 | 6.89e-05 |
| GO:BP | GO:0048771 | tissue remodeling | 71 | 113 | 7.06e-05 |
| GO:BP | GO:0016525 | negative regulation of angiogenesis | 46 | 66 | 7.11e-05 |
| GO:BP | GO:0061035 | regulation of cartilage development | 37 | 50 | 7.12e-05 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 144 | 261 | 7.16e-05 |
| GO:BP | GO:0141091 | transforming growth factor beta receptor superfamily signaling pathway | 143 | 259 | 7.33e-05 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 212 | 406 | 7.75e-05 |
| GO:BP | GO:0007611 | learning or memory | 109 | 189 | 7.84e-05 |
| GO:BP | GO:2000027 | regulation of animal organ morphogenesis | 42 | 59 | 8.04e-05 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 78 | 127 | 8.12e-05 |
| GO:BP | GO:0006813 | potassium ion transport | 82 | 135 | 8.51e-05 |
| GO:BP | GO:0006836 | neurotransmitter transport | 82 | 135 | 8.51e-05 |
| GO:BP | GO:0046632 | alpha-beta T cell differentiation | 48 | 70 | 8.73e-05 |
| GO:BP | GO:0043491 | phosphatidylinositol 3-kinase/protein kinase B signal transduction | 121 | 214 | 8.82e-05 |
| GO:BP | GO:0060415 | muscle tissue morphogenesis | 43 | 61 | 9.17e-05 |
| GO:BP | GO:0060840 | artery development | 56 | 85 | 9.45e-05 |
| GO:BP | GO:0050806 | positive regulation of synaptic transmission | 67 | 106 | 9.62e-05 |
| GO:BP | GO:0046879 | hormone secretion | 115 | 202 | 9.75e-05 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 862 | 1883 | 1.02e-04 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 209 | 401 | 1.04e-04 |
| GO:BP | GO:0031644 | regulation of nervous system process | 50 | 74 | 1.04e-04 |
| GO:BP | GO:0045927 | positive regulation of growth | 106 | 184 | 1.11e-04 |
| GO:BP | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | 59 | 91 | 1.11e-04 |
| GO:BP | GO:0048562 | embryonic organ morphogenesis | 105 | 182 | 1.12e-04 |
| GO:BP | GO:0006898 | receptor-mediated endocytosis | 105 | 182 | 1.12e-04 |
| GO:BP | GO:0071805 | potassium ion transmembrane transport | 74 | 120 | 1.13e-04 |
| GO:BP | GO:0007517 | muscle organ development | 146 | 267 | 1.13e-04 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 122 | 217 | 1.14e-04 |
| GO:BP | GO:0003170 | heart valve development | 45 | 65 | 1.14e-04 |
| GO:BP | GO:0022612 | gland morphogenesis | 60 | 93 | 1.15e-04 |
| GO:BP | GO:0003176 | aortic valve development | 29 | 37 | 1.19e-04 |
| GO:BP | GO:0002790 | peptide secretion | 97 | 166 | 1.19e-04 |
| GO:BP | GO:0035725 | sodium ion transmembrane transport | 61 | 95 | 1.19e-04 |
| GO:BP | GO:0010811 | positive regulation of cell-substrate adhesion | 61 | 95 | 1.19e-04 |
| GO:BP | GO:0018108 | peptidyl-tyrosine phosphorylation | 95 | 162 | 1.20e-04 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 275 | 546 | 1.21e-04 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 870 | 1904 | 1.23e-04 |
| GO:BP | GO:0009914 | hormone transport | 117 | 207 | 1.23e-04 |
| GO:BP | GO:0072028 | nephron morphogenesis | 41 | 58 | 1.31e-04 |
| GO:BP | GO:0051235 | maintenance of location | 138 | 251 | 1.36e-04 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 206 | 396 | 1.37e-04 |
| GO:BP | GO:0006939 | smooth muscle contraction | 47 | 69 | 1.37e-04 |
| GO:BP | GO:0003281 | ventricular septum development | 47 | 69 | 1.37e-04 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 318 | 642 | 1.38e-04 |
| GO:BP | GO:0072132 | mesenchyme morphogenesis | 37 | 51 | 1.41e-04 |
| GO:BP | GO:0048754 | branching morphogenesis of an epithelial tube | 70 | 113 | 1.55e-04 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 118 | 210 | 1.62e-04 |
| GO:BP | GO:0003208 | cardiac ventricle morphogenesis | 43 | 62 | 1.66e-04 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 848 | 1856 | 1.66e-04 |
| GO:BP | GO:0019722 | calcium-mediated signaling | 75 | 123 | 1.67e-04 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 364 | 746 | 1.68e-04 |
| GO:BP | GO:0043270 | positive regulation of monoatomic ion transport | 77 | 127 | 1.69e-04 |
| GO:BP | GO:0016049 | cell growth | 199 | 382 | 1.71e-04 |
| GO:BP | GO:0006814 | sodium ion transport | 82 | 137 | 1.73e-04 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 543 | 1152 | 1.75e-04 |
| GO:BP | GO:0051208 | sequestering of calcium ion | 61 | 96 | 1.87e-04 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 195 | 374 | 1.95e-04 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 188 | 359 | 1.98e-04 |
| GO:BP | GO:0050919 | negative chemotaxis | 31 | 41 | 1.98e-04 |
| GO:BP | GO:0140253 | cell-cell fusion | 28 | 36 | 2.15e-04 |
| GO:BP | GO:0000768 | syncytium formation by plasma membrane fusion | 28 | 36 | 2.15e-04 |
| GO:BP | GO:0060993 | kidney morphogenesis | 46 | 68 | 2.20e-04 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 96 | 166 | 2.28e-04 |
| GO:BP | GO:1902414 | protein localization to cell junction | 55 | 85 | 2.33e-04 |
| GO:BP | GO:0061337 | cardiac conduction | 55 | 85 | 2.33e-04 |
| GO:BP | GO:0098659 | inorganic cation import across plasma membrane | 47 | 70 | 2.37e-04 |
| GO:BP | GO:0099587 | inorganic ion import across plasma membrane | 47 | 70 | 2.37e-04 |
| GO:BP | GO:0099565 | chemical synaptic transmission, postsynaptic | 47 | 70 | 2.37e-04 |
| GO:BP | GO:0031399 | regulation of protein modification process | 436 | 911 | 2.38e-04 |
| GO:BP | GO:0016331 | morphogenesis of embryonic epithelium | 74 | 122 | 2.40e-04 |
| GO:BP | GO:0051279 | regulation of release of sequestered calcium ion into cytosol | 41 | 59 | 2.41e-04 |
| GO:BP | GO:0007613 | memory | 56 | 87 | 2.41e-04 |
| GO:BP | GO:0071526 | semaphorin-plexin signaling pathway | 32 | 43 | 2.42e-04 |
| GO:BP | GO:0060348 | bone development | 86 | 146 | 2.42e-04 |
| GO:BP | GO:0007369 | gastrulation | 83 | 140 | 2.44e-04 |
| GO:BP | GO:0021537 | telencephalon development | 110 | 195 | 2.44e-04 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 187 | 358 | 2.47e-04 |
| GO:BP | GO:0048863 | stem cell differentiation | 108 | 191 | 2.53e-04 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 839 | 1841 | 2.80e-04 |
| GO:BP | GO:0048660 | regulation of smooth muscle cell proliferation | 61 | 97 | 2.87e-04 |
| GO:BP | GO:0003180 | aortic valve morphogenesis | 26 | 33 | 2.90e-04 |
| GO:BP | GO:0060675 | ureteric bud morphogenesis | 33 | 45 | 2.91e-04 |
| GO:BP | GO:0042886 | amide transport | 114 | 204 | 3.01e-04 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 986 | 2186 | 3.07e-04 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 817 | 1791 | 3.13e-04 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 435 | 911 | 3.13e-04 |
| GO:BP | GO:0035023 | regulation of Rho protein signal transduction | 44 | 65 | 3.19e-04 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 67 | 109 | 3.24e-04 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 384 | 796 | 3.28e-04 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 274 | 550 | 3.29e-04 |
| GO:BP | GO:1903530 | regulation of secretion by cell | 190 | 366 | 3.34e-04 |
| GO:BP | GO:0071407 | cellular response to organic cyclic compound | 196 | 379 | 3.34e-04 |
| GO:BP | GO:2001257 | regulation of cation channel activity | 45 | 67 | 3.44e-04 |
| GO:BP | GO:0072088 | nephron epithelium morphogenesis | 39 | 56 | 3.49e-04 |
| GO:BP | GO:0000165 | MAPK cascade | 253 | 504 | 3.50e-04 |
| GO:BP | GO:0050890 | cognition | 118 | 213 | 3.58e-04 |
| GO:BP | GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 55 | 86 | 3.63e-04 |
| GO:BP | GO:0002376 | immune system process | 652 | 1410 | 3.74e-04 |
| GO:BP | GO:0046631 | alpha-beta T cell activation | 56 | 88 | 3.76e-04 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 159 | 300 | 3.86e-04 |
| GO:BP | GO:0008150 | biological_process | 4682 | 11244 | 3.90e-04 |
| GO:BP | GO:0003177 | pulmonary valve development | 19 | 22 | 3.91e-04 |
| GO:BP | GO:0060390 | regulation of SMAD protein signal transduction | 35 | 49 | 3.94e-04 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 212 | 415 | 4.15e-04 |
| GO:BP | GO:0003184 | pulmonary valve morphogenesis | 17 | 19 | 4.23e-04 |
| GO:BP | GO:0030072 | peptide hormone secretion | 93 | 162 | 4.29e-04 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 387 | 805 | 4.34e-04 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 120 | 218 | 4.42e-04 |
| GO:BP | GO:0099643 | signal release from synapse | 63 | 102 | 4.47e-04 |
| GO:BP | GO:0007269 | neurotransmitter secretion | 63 | 102 | 4.47e-04 |
| GO:BP | GO:0071692 | protein localization to extracellular region | 138 | 256 | 4.50e-04 |
| GO:BP | GO:0002573 | myeloid leukocyte differentiation | 88 | 152 | 4.53e-04 |
| GO:BP | GO:0014033 | neural crest cell differentiation | 50 | 77 | 4.63e-04 |
| GO:BP | GO:0035108 | limb morphogenesis | 67 | 110 | 4.70e-04 |
| GO:BP | GO:0035107 | appendage morphogenesis | 67 | 110 | 4.70e-04 |
| GO:BP | GO:0060173 | limb development | 80 | 136 | 4.80e-04 |
| GO:BP | GO:0007416 | synapse assembly | 80 | 136 | 4.80e-04 |
| GO:BP | GO:0048736 | appendage development | 80 | 136 | 4.80e-04 |
| GO:BP | GO:0098739 | import across plasma membrane | 73 | 122 | 4.85e-04 |
| GO:BP | GO:0035148 | tube formation | 74 | 124 | 4.85e-04 |
| GO:BP | GO:0055010 | ventricular cardiac muscle tissue morphogenesis | 32 | 44 | 4.85e-04 |
| GO:BP | GO:0007215 | glutamate receptor signaling pathway | 22 | 27 | 4.95e-04 |
| GO:BP | GO:1904064 | positive regulation of cation transmembrane transport | 52 | 81 | 4.99e-04 |
| GO:BP | GO:0099175 | regulation of postsynapse organization | 52 | 81 | 4.99e-04 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 386 | 804 | 4.99e-04 |
| GO:BP | GO:0006954 | inflammatory response | 216 | 425 | 5.09e-04 |
| GO:BP | GO:0002027 | regulation of heart rate | 53 | 83 | 5.18e-04 |
| GO:BP | GO:0014902 | myotube differentiation | 54 | 85 | 5.38e-04 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 332 | 683 | 5.46e-04 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 319 | 654 | 5.59e-04 |
| GO:BP | GO:0097120 | receptor localization to synapse | 33 | 46 | 5.63e-04 |
| GO:BP | GO:0072171 | mesonephric tubule morphogenesis | 33 | 46 | 5.63e-04 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 485 | 1031 | 6.21e-04 |
| GO:BP | GO:0043583 | ear development | 87 | 151 | 6.21e-04 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 142 | 266 | 6.27e-04 |
| GO:BP | GO:0003272 | endocardial cushion formation | 20 | 24 | 6.27e-04 |
| GO:BP | GO:0001755 | neural crest cell migration | 34 | 48 | 6.48e-04 |
| GO:BP | GO:0010718 | positive regulation of epithelial to mesenchymal transition | 34 | 48 | 6.48e-04 |
| GO:BP | GO:0046649 | lymphocyte activation | 224 | 444 | 6.54e-04 |
| GO:BP | GO:0140238 | presynaptic endocytosis | 40 | 59 | 6.65e-04 |
| GO:BP | GO:0050920 | regulation of chemotaxis | 78 | 133 | 6.65e-04 |
| GO:BP | GO:0042471 | ear morphogenesis | 48 | 74 | 6.65e-04 |
| GO:BP | GO:0051961 | negative regulation of nervous system development | 66 | 109 | 6.65e-04 |
| GO:BP | GO:0048864 | stem cell development | 48 | 74 | 6.65e-04 |
| GO:BP | GO:0010927 | cellular component assembly involved in morphogenesis | 65 | 107 | 6.65e-04 |
| GO:BP | GO:0007266 | Rho protein signal transduction | 66 | 109 | 6.65e-04 |
| GO:BP | GO:0048839 | inner ear development | 78 | 133 | 6.65e-04 |
| GO:BP | GO:0014888 | striated muscle adaptation | 30 | 41 | 6.87e-04 |
| GO:BP | GO:0040008 | regulation of growth | 232 | 462 | 6.87e-04 |
| GO:BP | GO:0061383 | trabecula morphogenesis | 30 | 41 | 6.87e-04 |
| GO:BP | GO:0098815 | modulation of excitatory postsynaptic potential | 30 | 41 | 6.87e-04 |
| GO:BP | GO:0003002 | regionalization | 138 | 258 | 6.91e-04 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 424 | 893 | 6.91e-04 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 497 | 1060 | 7.18e-04 |
| GO:BP | GO:0090497 | mesenchymal cell migration | 35 | 50 | 7.18e-04 |
| GO:BP | GO:0009991 | response to extracellular stimulus | 193 | 377 | 7.52e-04 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 125 | 231 | 7.94e-04 |
| GO:BP | GO:0007498 | mesoderm development | 53 | 84 | 7.97e-04 |
| GO:BP | GO:0048645 | animal organ formation | 36 | 52 | 8.05e-04 |
| GO:BP | GO:0060079 | excitatory postsynaptic potential | 43 | 65 | 8.21e-04 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 147 | 278 | 8.25e-04 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 130 | 242 | 8.75e-04 |
| GO:BP | GO:0072078 | nephron tubule morphogenesis | 37 | 54 | 8.91e-04 |
| GO:BP | GO:0001704 | formation of primary germ layer | 58 | 94 | 8.93e-04 |
| GO:BP | GO:0070372 | regulation of ERK1 and ERK2 cascade | 98 | 175 | 9.11e-04 |
| GO:BP | GO:0110110 | positive regulation of animal organ morphogenesis | 21 | 26 | 9.11e-04 |
| GO:BP | GO:0060043 | regulation of cardiac muscle cell proliferation | 24 | 31 | 9.11e-04 |
| GO:BP | GO:0014032 | neural crest cell development | 45 | 69 | 9.18e-04 |
| GO:BP | GO:0003158 | endothelium development | 61 | 100 | 9.25e-04 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 242 | 486 | 9.29e-04 |
| GO:BP | GO:0034764 | positive regulation of transmembrane transport | 76 | 130 | 9.29e-04 |
| GO:BP | GO:0030010 | establishment of cell polarity | 73 | 124 | 9.41e-04 |
| GO:BP | GO:0008360 | regulation of cell shape | 64 | 106 | 9.41e-04 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 64 | 106 | 9.41e-04 |
| GO:BP | GO:0031214 | biomineral tissue development | 64 | 106 | 9.41e-04 |
| GO:BP | GO:0007612 | learning | 66 | 110 | 9.48e-04 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 217 | 431 | 9.52e-04 |
| GO:BP | GO:0003229 | ventricular cardiac muscle tissue development | 38 | 56 | 9.57e-04 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 47 | 73 | 9.98e-04 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 91 | 161 | 1.03e-03 |
| GO:BP | GO:0061333 | renal tubule morphogenesis | 39 | 58 | 1.04e-03 |
| GO:BP | GO:0048488 | synaptic vesicle endocytosis | 39 | 58 | 1.04e-03 |
| GO:BP | GO:0014896 | muscle hypertrophy | 48 | 75 | 1.04e-03 |
| GO:BP | GO:0008544 | epidermis development | 114 | 209 | 1.04e-03 |
| GO:BP | GO:0002761 | regulation of myeloid leukocyte differentiation | 50 | 79 | 1.11e-03 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 40 | 60 | 1.12e-03 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 51 | 81 | 1.15e-03 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 29 | 40 | 1.16e-03 |
| GO:BP | GO:0050708 | regulation of protein secretion | 99 | 178 | 1.16e-03 |
| GO:BP | GO:0061036 | positive regulation of cartilage development | 19 | 23 | 1.16e-03 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 184 | 360 | 1.20e-03 |
| GO:BP | GO:0009306 | protein secretion | 134 | 252 | 1.20e-03 |
| GO:BP | GO:0035592 | establishment of protein localization to extracellular region | 134 | 252 | 1.20e-03 |
| GO:BP | GO:0030097 | hemopoiesis | 303 | 624 | 1.22e-03 |
| GO:BP | GO:0034446 | substrate adhesion-dependent cell spreading | 54 | 87 | 1.22e-03 |
| GO:BP | GO:0030073 | insulin secretion | 79 | 137 | 1.22e-03 |
| GO:BP | GO:0060421 | positive regulation of heart growth | 22 | 28 | 1.22e-03 |
| GO:BP | GO:2000463 | positive regulation of excitatory postsynaptic potential | 22 | 28 | 1.22e-03 |
| GO:BP | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway | 22 | 28 | 1.22e-03 |
| GO:BP | GO:0048732 | gland development | 160 | 308 | 1.22e-03 |
| GO:BP | GO:0050900 | leukocyte migration | 107 | 195 | 1.23e-03 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 42 | 64 | 1.25e-03 |
| GO:BP | GO:0034767 | positive regulation of monoatomic ion transmembrane transport | 56 | 91 | 1.25e-03 |
| GO:BP | GO:0019935 | cyclic-nucleotide-mediated signaling | 35 | 51 | 1.26e-03 |
| GO:BP | GO:0050771 | negative regulation of axonogenesis | 35 | 51 | 1.26e-03 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 146 | 278 | 1.26e-03 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 75 | 129 | 1.26e-03 |
| GO:BP | GO:0001707 | mesoderm formation | 35 | 51 | 1.26e-03 |
| GO:BP | GO:0048675 | axon extension | 58 | 95 | 1.27e-03 |
| GO:BP | GO:0031667 | response to nutrient levels | 181 | 354 | 1.30e-03 |
| GO:BP | GO:0035296 | regulation of tube diameter | 61 | 101 | 1.30e-03 |
| GO:BP | GO:0097746 | blood vessel diameter maintenance | 61 | 101 | 1.30e-03 |
| GO:BP | GO:0035150 | regulation of tube size | 61 | 101 | 1.30e-03 |
| GO:BP | GO:0048659 | smooth muscle cell proliferation | 61 | 101 | 1.30e-03 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 68 | 115 | 1.30e-03 |
| GO:BP | GO:0050768 | negative regulation of neurogenesis | 62 | 103 | 1.30e-03 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 68 | 115 | 1.30e-03 |
| GO:BP | GO:0032535 | regulation of cellular component size | 150 | 287 | 1.35e-03 |
| GO:BP | GO:0055002 | striated muscle cell development | 44 | 68 | 1.35e-03 |
| GO:BP | GO:0048332 | mesoderm morphogenesis | 36 | 53 | 1.36e-03 |
| GO:BP | GO:0007162 | negative regulation of cell adhesion | 102 | 185 | 1.36e-03 |
| GO:BP | GO:0051896 | regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 100 | 181 | 1.43e-03 |
| GO:BP | GO:0030031 | cell projection assembly | 223 | 447 | 1.45e-03 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 118 | 219 | 1.47e-03 |
| GO:BP | GO:0002064 | epithelial cell development | 84 | 148 | 1.51e-03 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 47 | 74 | 1.52e-03 |
| GO:BP | GO:0061326 | renal tubule development | 47 | 74 | 1.52e-03 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 181 | 355 | 1.54e-03 |
| GO:BP | GO:0010817 | regulation of hormone levels | 164 | 318 | 1.54e-03 |
| GO:BP | GO:0031099 | regeneration | 82 | 144 | 1.55e-03 |
| GO:BP | GO:0030513 | positive regulation of BMP signaling pathway | 23 | 30 | 1.55e-03 |
| GO:BP | GO:0060996 | dendritic spine development | 48 | 76 | 1.56e-03 |
| GO:BP | GO:0046777 | protein autophosphorylation | 81 | 142 | 1.57e-03 |
| GO:BP | GO:0070371 | ERK1 and ERK2 cascade | 106 | 194 | 1.58e-03 |
| GO:BP | GO:0043217 | myelin maintenance | 13 | 14 | 1.59e-03 |
| GO:BP | GO:0010954 | positive regulation of protein processing | 15 | 17 | 1.59e-03 |
| GO:BP | GO:0060977 | coronary vasculature morphogenesis | 15 | 17 | 1.59e-03 |
| GO:BP | GO:0055023 | positive regulation of cardiac muscle tissue growth | 20 | 25 | 1.60e-03 |
| GO:BP | GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | 79 | 138 | 1.60e-03 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 168 | 327 | 1.62e-03 |
| GO:BP | GO:0002791 | regulation of peptide secretion | 76 | 132 | 1.67e-03 |
| GO:BP | GO:0014855 | striated muscle cell proliferation | 39 | 59 | 1.67e-03 |
| GO:BP | GO:0072009 | nephron epithelium development | 52 | 84 | 1.70e-03 |
| GO:BP | GO:0061387 | regulation of extent of cell growth | 54 | 88 | 1.76e-03 |
| GO:BP | GO:0061614 | miRNA transcription | 40 | 61 | 1.78e-03 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 216 | 433 | 1.81e-03 |
| GO:BP | GO:0140115 | export across plasma membrane | 33 | 48 | 1.81e-03 |
| GO:BP | GO:0010721 | negative regulation of cell development | 100 | 182 | 1.83e-03 |
| GO:BP | GO:0031503 | protein-containing complex localization | 88 | 157 | 1.83e-03 |
| GO:BP | GO:0007043 | cell-cell junction assembly | 63 | 106 | 1.83e-03 |
| GO:BP | GO:0038127 | ERBB signaling pathway | 62 | 104 | 1.83e-03 |
| GO:BP | GO:0001658 | branching involved in ureteric bud morphogenesis | 28 | 39 | 1.85e-03 |
| GO:BP | GO:0050922 | negative regulation of chemotaxis | 28 | 39 | 1.85e-03 |
| GO:BP | GO:0097061 | dendritic spine organization | 41 | 63 | 1.85e-03 |
| GO:BP | GO:0007492 | endoderm development | 41 | 63 | 1.85e-03 |
| GO:BP | GO:0070482 | response to oxygen levels | 139 | 265 | 1.85e-03 |
| GO:BP | GO:1902284 | neuron projection extension involved in neuron projection guidance | 24 | 32 | 1.90e-03 |
| GO:BP | GO:0048846 | axon extension involved in axon guidance | 24 | 32 | 1.90e-03 |
| GO:BP | GO:0008361 | regulation of cell size | 83 | 147 | 2.00e-03 |
| GO:BP | GO:0003148 | outflow tract septum morphogenesis | 18 | 22 | 2.09e-03 |
| GO:BP | GO:0060045 | positive regulation of cardiac muscle cell proliferation | 18 | 22 | 2.09e-03 |
| GO:BP | GO:1903779 | regulation of cardiac conduction | 18 | 22 | 2.09e-03 |
| GO:BP | GO:1905330 | regulation of morphogenesis of an epithelium | 29 | 41 | 2.12e-03 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 206 | 412 | 2.14e-03 |
| GO:BP | GO:0106027 | neuron projection organization | 45 | 71 | 2.17e-03 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 77 | 135 | 2.22e-03 |
| GO:BP | GO:0046883 | regulation of hormone secretion | 91 | 164 | 2.24e-03 |
| GO:BP | GO:0014910 | regulation of smooth muscle cell migration | 36 | 54 | 2.29e-03 |
| GO:BP | GO:0060541 | respiratory system development | 90 | 162 | 2.29e-03 |
| GO:BP | GO:0071868 | cellular response to monoamine stimulus | 25 | 34 | 2.29e-03 |
| GO:BP | GO:0071870 | cellular response to catecholamine stimulus | 25 | 34 | 2.29e-03 |
| GO:BP | GO:0001764 | neuron migration | 74 | 129 | 2.30e-03 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 217 | 437 | 2.37e-03 |
| GO:BP | GO:0072175 | epithelial tube formation | 68 | 117 | 2.48e-03 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 115 | 215 | 2.49e-03 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 52 | 85 | 2.50e-03 |
| GO:BP | GO:0032231 | regulation of actin filament bundle assembly | 52 | 85 | 2.50e-03 |
| GO:BP | GO:0045446 | endothelial cell differentiation | 52 | 85 | 2.50e-03 |
| GO:BP | GO:0050865 | regulation of cell activation | 176 | 347 | 2.52e-03 |
| GO:BP | GO:0014020 | primary neural tube formation | 53 | 87 | 2.52e-03 |
| GO:BP | GO:0043500 | muscle adaptation | 54 | 89 | 2.54e-03 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 64 | 109 | 2.54e-03 |
| GO:BP | GO:0007009 | plasma membrane organization | 63 | 107 | 2.55e-03 |
| GO:BP | GO:0008217 | regulation of blood pressure | 62 | 105 | 2.56e-03 |
| GO:BP | GO:0002028 | regulation of sodium ion transport | 38 | 58 | 2.56e-03 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 57 | 95 | 2.56e-03 |
| GO:BP | GO:1902105 | regulation of leukocyte differentiation | 105 | 194 | 2.58e-03 |
| GO:BP | GO:0007520 | myoblast fusion | 22 | 29 | 2.65e-03 |
| GO:BP | GO:0150117 | positive regulation of cell-substrate junction organization | 22 | 29 | 2.65e-03 |
| GO:BP | GO:0003203 | endocardial cushion morphogenesis | 26 | 36 | 2.65e-03 |
| GO:BP | GO:0010463 | mesenchymal cell proliferation | 26 | 36 | 2.65e-03 |
| GO:BP | GO:0051703 | biological process involved in intraspecies interaction between organisms | 22 | 29 | 2.65e-03 |
| GO:BP | GO:0061384 | heart trabecula morphogenesis | 22 | 29 | 2.65e-03 |
| GO:BP | GO:0035176 | social behavior | 22 | 29 | 2.65e-03 |
| GO:BP | GO:0071867 | response to monoamine | 26 | 36 | 2.65e-03 |
| GO:BP | GO:0071869 | response to catecholamine | 26 | 36 | 2.65e-03 |
| GO:BP | GO:0035418 | protein localization to synapse | 39 | 60 | 2.65e-03 |
| GO:BP | GO:0014909 | smooth muscle cell migration | 39 | 60 | 2.65e-03 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 93 | 169 | 2.71e-03 |
| GO:BP | GO:0032878 | regulation of establishment or maintenance of cell polarity | 19 | 24 | 2.85e-03 |
| GO:BP | GO:0003299 | muscle hypertrophy in response to stress | 19 | 24 | 2.85e-03 |
| GO:BP | GO:0014898 | cardiac muscle hypertrophy in response to stress | 19 | 24 | 2.85e-03 |
| GO:BP | GO:0014887 | cardiac muscle adaptation | 19 | 24 | 2.85e-03 |
| GO:BP | GO:0021700 | developmental maturation | 109 | 203 | 2.88e-03 |
| GO:BP | GO:0050772 | positive regulation of axonogenesis | 41 | 64 | 2.89e-03 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 77 | 136 | 2.89e-03 |
| GO:BP | GO:0090087 | regulation of peptide transport | 76 | 134 | 2.94e-03 |
| GO:BP | GO:0030239 | myofibril assembly | 42 | 66 | 2.99e-03 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 136 | 261 | 2.99e-03 |
| GO:BP | GO:0051641 | cellular localization | 1157 | 2623 | 3.01e-03 |
| GO:BP | GO:0090276 | regulation of peptide hormone secretion | 74 | 130 | 3.04e-03 |
| GO:BP | GO:0060420 | regulation of heart growth | 33 | 49 | 3.05e-03 |
| GO:BP | GO:0070168 | negative regulation of biomineral tissue development | 14 | 16 | 3.05e-03 |
| GO:BP | GO:0072080 | nephron tubule development | 44 | 70 | 3.17e-03 |
| GO:BP | GO:0014812 | muscle cell migration | 45 | 72 | 3.25e-03 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 45 | 72 | 3.25e-03 |
| GO:BP | GO:0002274 | myeloid leukocyte activation | 68 | 118 | 3.31e-03 |
| GO:BP | GO:0051963 | regulation of synapse assembly | 47 | 76 | 3.38e-03 |
| GO:BP | GO:0050773 | regulation of dendrite development | 48 | 78 | 3.44e-03 |
| GO:BP | GO:0050817 | coagulation | 82 | 147 | 3.47e-03 |
| GO:BP | GO:0001508 | action potential | 60 | 102 | 3.57e-03 |
| GO:BP | GO:0007173 | epidermal growth factor receptor signaling pathway | 52 | 86 | 3.58e-03 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 307 | 642 | 3.58e-03 |
| GO:BP | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 55 | 92 | 3.60e-03 |
| GO:BP | GO:0001841 | neural tube formation | 55 | 92 | 3.60e-03 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 130 | 249 | 3.60e-03 |
| GO:BP | GO:0010657 | muscle cell apoptotic process | 36 | 55 | 3.69e-03 |
| GO:BP | GO:1902656 | calcium ion import into cytosol | 20 | 26 | 3.70e-03 |
| GO:BP | GO:1901653 | cellular response to peptide | 141 | 273 | 3.75e-03 |
| GO:BP | GO:0032924 | activin receptor signaling pathway | 24 | 33 | 3.82e-03 |
| GO:BP | GO:0019725 | cellular homeostasis | 272 | 564 | 4.09e-03 |
| GO:BP | GO:0086002 | cardiac muscle cell action potential involved in contraction | 30 | 44 | 4.16e-03 |
| GO:BP | GO:0034103 | regulation of tissue remodeling | 30 | 44 | 4.16e-03 |
| GO:BP | GO:0055021 | regulation of cardiac muscle tissue growth | 30 | 44 | 4.16e-03 |
| GO:BP | GO:0099072 | regulation of postsynaptic membrane neurotransmitter receptor levels | 39 | 61 | 4.23e-03 |
| GO:BP | GO:0022409 | positive regulation of cell-cell adhesion | 95 | 175 | 4.23e-03 |
| GO:BP | GO:0051897 | positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 71 | 125 | 4.28e-03 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 290 | 605 | 4.28e-03 |
| GO:BP | GO:0002694 | regulation of leukocyte activation | 159 | 313 | 4.33e-03 |
| GO:BP | GO:0019933 | cAMP-mediated signaling | 25 | 35 | 4.41e-03 |
| GO:BP | GO:0009952 | anterior/posterior pattern specification | 69 | 121 | 4.41e-03 |
| GO:BP | GO:0046622 | positive regulation of organ growth | 25 | 35 | 4.41e-03 |
| GO:BP | GO:2000116 | regulation of cysteine-type endopeptidase activity | 69 | 121 | 4.41e-03 |
| GO:BP | GO:0051651 | maintenance of location in cell | 93 | 171 | 4.44e-03 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 68 | 119 | 4.46e-03 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 147 | 287 | 4.46e-03 |
| GO:BP | GO:0030316 | osteoclast differentiation | 41 | 65 | 4.46e-03 |
| GO:BP | GO:0003197 | endocardial cushion development | 31 | 46 | 4.47e-03 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 82 | 148 | 4.48e-03 |
| GO:BP | GO:0030323 | respiratory tube development | 82 | 148 | 4.48e-03 |
| GO:BP | GO:0007599 | hemostasis | 82 | 148 | 4.48e-03 |
| GO:BP | GO:0036465 | synaptic vesicle recycling | 42 | 67 | 4.55e-03 |
| GO:BP | GO:0051588 | regulation of neurotransmitter transport | 42 | 67 | 4.55e-03 |
| GO:BP | GO:0003171 | atrioventricular valve development | 21 | 28 | 4.55e-03 |
| GO:BP | GO:0048841 | regulation of axon extension involved in axon guidance | 21 | 28 | 4.55e-03 |
| GO:BP | GO:0050796 | regulation of insulin secretion | 65 | 113 | 4.61e-03 |
| GO:BP | GO:0043547 | positive regulation of GTPase activity | 65 | 113 | 4.61e-03 |
| GO:BP | GO:0018107 | peptidyl-threonine phosphorylation | 43 | 69 | 4.64e-03 |
| GO:BP | GO:0007596 | blood coagulation | 80 | 144 | 4.66e-03 |
| GO:BP | GO:0030324 | lung development | 80 | 144 | 4.66e-03 |
| GO:BP | GO:0031102 | neuron projection regeneration | 32 | 48 | 4.75e-03 |
| GO:BP | GO:0048592 | eye morphogenesis | 62 | 107 | 4.77e-03 |
| GO:BP | GO:0097581 | lamellipodium organization | 45 | 73 | 4.79e-03 |
| GO:BP | GO:0042110 | T cell activation | 155 | 305 | 4.84e-03 |
| GO:BP | GO:1903319 | positive regulation of protein maturation | 15 | 18 | 4.86e-03 |
| GO:BP | GO:0042063 | gliogenesis | 126 | 242 | 4.89e-03 |
| GO:BP | GO:0030517 | negative regulation of axon extension | 26 | 37 | 4.89e-03 |
| GO:BP | GO:0010658 | striated muscle cell apoptotic process | 26 | 37 | 4.89e-03 |
| GO:BP | GO:0010660 | regulation of muscle cell apoptotic process | 33 | 50 | 4.97e-03 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 52 | 87 | 4.97e-03 |
| GO:BP | GO:1901019 | regulation of calcium ion transmembrane transporter activity | 33 | 50 | 4.97e-03 |
| GO:BP | GO:0048814 | regulation of dendrite morphogenesis | 33 | 50 | 4.97e-03 |
| GO:BP | GO:0031623 | receptor internalization | 53 | 89 | 4.97e-03 |
| GO:BP | GO:0030038 | contractile actin filament bundle assembly | 54 | 91 | 4.97e-03 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 112 | 212 | 4.97e-03 |
| GO:BP | GO:0033135 | regulation of peptidyl-serine phosphorylation | 51 | 85 | 4.97e-03 |
| GO:BP | GO:0043149 | stress fiber assembly | 54 | 91 | 4.97e-03 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 52 | 87 | 4.97e-03 |
| GO:BP | GO:0010765 | positive regulation of sodium ion transport | 18 | 23 | 4.99e-03 |
| GO:BP | GO:0048668 | collateral sprouting | 18 | 23 | 4.99e-03 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 74 | 132 | 5.22e-03 |
| GO:BP | GO:0032906 | transforming growth factor beta2 production | 9 | 9 | 5.23e-03 |
| GO:BP | GO:0032909 | regulation of transforming growth factor beta2 production | 9 | 9 | 5.23e-03 |
| GO:BP | GO:1900024 | regulation of substrate adhesion-dependent cell spreading | 34 | 52 | 5.24e-03 |
| GO:BP | GO:1904427 | positive regulation of calcium ion transmembrane transport | 34 | 52 | 5.24e-03 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 129 | 249 | 5.34e-03 |
| GO:BP | GO:0035924 | cellular response to vascular endothelial growth factor stimulus | 35 | 54 | 5.50e-03 |
| GO:BP | GO:0086065 | cell communication involved in cardiac conduction | 35 | 54 | 5.50e-03 |
| GO:BP | GO:0001666 | response to hypoxia | 121 | 232 | 5.67e-03 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 295 | 619 | 5.73e-03 |
| GO:BP | GO:0046928 | regulation of neurotransmitter secretion | 36 | 56 | 5.75e-03 |
| GO:BP | GO:0051251 | positive regulation of lymphocyte activation | 99 | 185 | 5.84e-03 |
| GO:BP | GO:0010812 | negative regulation of cell-substrate adhesion | 28 | 41 | 5.92e-03 |
| GO:BP | GO:0008543 | fibroblast growth factor receptor signaling pathway | 37 | 58 | 5.96e-03 |
| GO:BP | GO:0032414 | positive regulation of ion transmembrane transporter activity | 37 | 58 | 5.96e-03 |
| GO:BP | GO:0001935 | endothelial cell proliferation | 65 | 114 | 6.14e-03 |
| GO:BP | GO:0010001 | glial cell differentiation | 97 | 181 | 6.19e-03 |
| GO:BP | GO:0098703 | calcium ion import across plasma membrane | 19 | 25 | 6.39e-03 |
| GO:BP | GO:0072578 | neurotransmitter-gated ion channel clustering | 11 | 12 | 6.42e-03 |
| GO:BP | GO:0003222 | ventricular trabecula myocardium morphogenesis | 11 | 12 | 6.42e-03 |
| GO:BP | GO:0009880 | embryonic pattern specification | 29 | 43 | 6.42e-03 |
| GO:BP | GO:0032332 | positive regulation of chondrocyte differentiation | 11 | 12 | 6.42e-03 |
| GO:BP | GO:0001701 | in utero embryonic development | 166 | 331 | 6.51e-03 |
| GO:BP | GO:0001570 | vasculogenesis | 41 | 66 | 6.61e-03 |
| GO:BP | GO:0002040 | sprouting angiogenesis | 57 | 98 | 6.81e-03 |
| GO:BP | GO:0046620 | regulation of organ growth | 43 | 70 | 6.82e-03 |
| GO:BP | GO:0016079 | synaptic vesicle exocytosis | 43 | 70 | 6.82e-03 |
| GO:BP | GO:0055123 | digestive system development | 56 | 96 | 6.85e-03 |
| GO:BP | GO:0060997 | dendritic spine morphogenesis | 30 | 45 | 6.87e-03 |
| GO:BP | GO:0030516 | regulation of axon extension | 45 | 74 | 6.96e-03 |
| GO:BP | GO:0048565 | digestive tract development | 52 | 88 | 7.03e-03 |
| GO:BP | GO:0050867 | positive regulation of cell activation | 107 | 203 | 7.03e-03 |
| GO:BP | GO:0010659 | cardiac muscle cell apoptotic process | 24 | 34 | 7.03e-03 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 560 | 1231 | 7.03e-03 |
| GO:BP | GO:0010662 | regulation of striated muscle cell apoptotic process | 24 | 34 | 7.03e-03 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 107 | 203 | 7.03e-03 |
| GO:BP | GO:0060606 | tube closure | 50 | 84 | 7.03e-03 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 49 | 82 | 7.03e-03 |
| GO:BP | GO:0043114 | regulation of vascular permeability | 24 | 34 | 7.03e-03 |
| GO:BP | GO:0140747 | regulation of ncRNA transcription | 48 | 80 | 7.03e-03 |
| GO:BP | GO:0032330 | regulation of chondrocyte differentiation | 24 | 34 | 7.03e-03 |
| GO:BP | GO:0003230 | cardiac atrium development | 24 | 34 | 7.03e-03 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 137 | 268 | 7.13e-03 |
| GO:BP | GO:0051249 | regulation of lymphocyte activation | 142 | 279 | 7.24e-03 |
| GO:BP | GO:0045860 | positive regulation of protein kinase activity | 98 | 184 | 7.44e-03 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 561 | 1234 | 7.44e-03 |
| GO:BP | GO:1903053 | regulation of extracellular matrix organization | 32 | 49 | 7.64e-03 |
| GO:BP | GO:1901021 | positive regulation of calcium ion transmembrane transporter activity | 20 | 27 | 7.65e-03 |
| GO:BP | GO:0070570 | regulation of neuron projection regeneration | 20 | 27 | 7.65e-03 |
| GO:BP | GO:0031279 | regulation of cyclase activity | 20 | 27 | 7.65e-03 |
| GO:BP | GO:0002696 | positive regulation of leukocyte activation | 104 | 197 | 7.69e-03 |
| GO:BP | GO:1903037 | regulation of leukocyte cell-cell adhesion | 104 | 197 | 7.69e-03 |
| GO:BP | GO:0038084 | vascular endothelial growth factor signaling pathway | 25 | 36 | 7.83e-03 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 612 | 1354 | 8.15e-03 |
| GO:BP | GO:0043367 | CD4-positive, alpha-beta T cell differentiation | 34 | 53 | 8.32e-03 |
| GO:BP | GO:0060491 | regulation of cell projection assembly | 86 | 159 | 8.32e-03 |
| GO:BP | GO:0120032 | regulation of plasma membrane bounded cell projection assembly | 85 | 157 | 8.56e-03 |
| GO:BP | GO:0003073 | regulation of systemic arterial blood pressure | 35 | 55 | 8.63e-03 |
| GO:BP | GO:0036293 | response to decreased oxygen levels | 125 | 243 | 8.83e-03 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 60 | 105 | 8.84e-03 |
| GO:BP | GO:0021954 | central nervous system neuron development | 36 | 57 | 8.88e-03 |
| GO:BP | GO:0051899 | membrane depolarization | 36 | 57 | 8.88e-03 |
| GO:BP | GO:0140239 | postsynaptic endocytosis | 14 | 17 | 8.95e-03 |
| GO:BP | GO:0048169 | regulation of long-term neuronal synaptic plasticity | 14 | 17 | 8.95e-03 |
| GO:BP | GO:0002433 | immune response-regulating cell surface receptor signaling pathway involved in phagocytosis | 14 | 17 | 8.95e-03 |
| GO:BP | GO:0098884 | postsynaptic neurotransmitter receptor internalization | 14 | 17 | 8.95e-03 |
| GO:BP | GO:0051590 | positive regulation of neurotransmitter transport | 14 | 17 | 8.95e-03 |
| GO:BP | GO:0060445 | branching involved in salivary gland morphogenesis | 14 | 17 | 8.95e-03 |
| GO:BP | GO:0038096 | Fc-gamma receptor signaling pathway involved in phagocytosis | 14 | 17 | 8.95e-03 |
| GO:BP | GO:0097106 | postsynaptic density organization | 21 | 29 | 8.96e-03 |
| GO:BP | GO:0062013 | positive regulation of small molecule metabolic process | 57 | 99 | 9.15e-03 |
| GO:BP | GO:0035249 | synaptic transmission, glutamatergic | 38 | 61 | 9.23e-03 |
| GO:BP | GO:0072091 | regulation of stem cell proliferation | 38 | 61 | 9.23e-03 |
| GO:BP | GO:0035567 | non-canonical Wnt signaling pathway | 38 | 61 | 9.23e-03 |
| GO:BP | GO:0006950 | response to stress | 1114 | 2540 | 9.26e-03 |
| GO:BP | GO:0050795 | regulation of behavior | 27 | 40 | 9.26e-03 |
| GO:BP | GO:0032411 | positive regulation of transporter activity | 39 | 63 | 9.37e-03 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 39 | 63 | 9.37e-03 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 40 | 65 | 9.52e-03 |
| GO:BP | GO:0006887 | exocytosis | 122 | 237 | 9.60e-03 |
| GO:BP | GO:0010595 | positive regulation of endothelial cell migration | 52 | 89 | 9.60e-03 |
| GO:BP | GO:0001892 | embryonic placenta development | 41 | 67 | 9.61e-03 |
| GO:BP | GO:0030534 | adult behavior | 51 | 87 | 9.64e-03 |
| GO:BP | GO:0048593 | camera-type eye morphogenesis | 51 | 87 | 9.64e-03 |
| GO:BP | GO:0110020 | regulation of actomyosin structure organization | 51 | 87 | 9.64e-03 |
| GO:BP | GO:0001843 | neural tube closure | 49 | 83 | 9.76e-03 |
| GO:BP | GO:0003014 | renal system process | 44 | 73 | 9.79e-03 |
| GO:BP | GO:0051492 | regulation of stress fiber assembly | 46 | 77 | 9.80e-03 |
| GO:BP | GO:0044344 | cellular response to fibroblast growth factor stimulus | 47 | 79 | 9.80e-03 |
| GO:BP | GO:0001954 | positive regulation of cell-matrix adhesion | 28 | 42 | 9.84e-03 |
| GO:BP | GO:0019229 | regulation of vasoconstriction | 28 | 42 | 9.84e-03 |
| GO:BP | GO:0043434 | response to peptide hormone | 157 | 314 | 1.00e-02 |
| GO:BP | GO:0010644 | cell communication by electrical coupling | 22 | 31 | 1.01e-02 |
| GO:BP | GO:0010665 | regulation of cardiac muscle cell apoptotic process | 22 | 31 | 1.01e-02 |
| GO:BP | GO:0030307 | positive regulation of cell growth | 67 | 120 | 1.01e-02 |
| GO:BP | GO:1903351 | cellular response to dopamine | 22 | 31 | 1.01e-02 |
| GO:BP | GO:1902895 | positive regulation of miRNA transcription | 29 | 44 | 1.04e-02 |
| GO:BP | GO:0034332 | adherens junction organization | 29 | 44 | 1.04e-02 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 165 | 332 | 1.05e-02 |
| GO:BP | GO:0051894 | positive regulation of focal adhesion assembly | 18 | 24 | 1.08e-02 |
| GO:BP | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling | 18 | 24 | 1.08e-02 |
| GO:BP | GO:0045124 | regulation of bone resorption | 18 | 24 | 1.08e-02 |
| GO:BP | GO:0060350 | endochondral bone morphogenesis | 30 | 46 | 1.10e-02 |
| GO:BP | GO:0001890 | placenta development | 62 | 110 | 1.11e-02 |
| GO:BP | GO:0097529 | myeloid leukocyte migration | 62 | 110 | 1.11e-02 |
| GO:BP | GO:0098969 | neurotransmitter receptor transport to postsynaptic membrane | 12 | 14 | 1.11e-02 |
| GO:BP | GO:0003190 | atrioventricular valve formation | 8 | 8 | 1.11e-02 |
| GO:BP | GO:0032634 | interleukin-5 production | 8 | 8 | 1.11e-02 |
| GO:BP | GO:1903540 | establishment of protein localization to postsynaptic membrane | 12 | 14 | 1.11e-02 |
| GO:BP | GO:0060896 | neural plate pattern specification | 8 | 8 | 1.11e-02 |
| GO:BP | GO:0060982 | coronary artery morphogenesis | 8 | 8 | 1.11e-02 |
| GO:BP | GO:0003418 | growth plate cartilage chondrocyte differentiation | 8 | 8 | 1.11e-02 |
| GO:BP | GO:0032674 | regulation of interleukin-5 production | 8 | 8 | 1.11e-02 |
| GO:BP | GO:0051051 | negative regulation of transport | 144 | 286 | 1.11e-02 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 122 | 238 | 1.13e-02 |
| GO:BP | GO:0009410 | response to xenobiotic stimulus | 138 | 273 | 1.14e-02 |
| GO:BP | GO:0060326 | cell chemotaxis | 82 | 152 | 1.14e-02 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 152 | 304 | 1.17e-02 |
| GO:BP | GO:2000630 | positive regulation of miRNA metabolic process | 32 | 50 | 1.18e-02 |
| GO:BP | GO:1990806 | ligand-gated ion channel signaling pathway | 15 | 19 | 1.20e-02 |
| GO:BP | GO:0099637 | neurotransmitter receptor transport | 15 | 19 | 1.20e-02 |
| GO:BP | GO:0099590 | neurotransmitter receptor internalization | 15 | 19 | 1.20e-02 |
| GO:BP | GO:0098810 | neurotransmitter reuptake | 15 | 19 | 1.20e-02 |
| GO:BP | GO:0009725 | response to hormone | 288 | 610 | 1.21e-02 |
| GO:BP | GO:0001708 | cell fate specification | 33 | 52 | 1.22e-02 |
| GO:BP | GO:0008038 | neuron recognition | 24 | 35 | 1.23e-02 |
| GO:BP | GO:0007605 | sensory perception of sound | 55 | 96 | 1.25e-02 |
| GO:BP | GO:0060143 | positive regulation of syncytium formation by plasma membrane fusion | 10 | 11 | 1.27e-02 |
| GO:BP | GO:0051923 | sulfation | 10 | 11 | 1.27e-02 |
| GO:BP | GO:0021670 | lateral ventricle development | 10 | 11 | 1.27e-02 |
| GO:BP | GO:0045055 | regulated exocytosis | 78 | 144 | 1.27e-02 |
| GO:BP | GO:0003209 | cardiac atrium morphogenesis | 19 | 26 | 1.27e-02 |
| GO:BP | GO:0003181 | atrioventricular valve morphogenesis | 19 | 26 | 1.27e-02 |
| GO:BP | GO:1903055 | positive regulation of extracellular matrix organization | 19 | 26 | 1.27e-02 |
| GO:BP | GO:0032941 | secretion by tissue | 19 | 26 | 1.27e-02 |
| GO:BP | GO:0051047 | positive regulation of secretion | 106 | 204 | 1.29e-02 |
| GO:BP | GO:0010469 | regulation of signaling receptor activity | 36 | 58 | 1.30e-02 |
| GO:BP | GO:0043542 | endothelial cell migration | 92 | 174 | 1.33e-02 |
| GO:BP | GO:0008366 | axon ensheathment | 66 | 119 | 1.33e-02 |
| GO:BP | GO:0007272 | ensheathment of neurons | 66 | 119 | 1.33e-02 |
| GO:BP | GO:1902305 | regulation of sodium ion transmembrane transport | 25 | 37 | 1.33e-02 |
| GO:BP | GO:0098661 | inorganic anion transmembrane transport | 38 | 62 | 1.34e-02 |
| GO:BP | GO:0070167 | regulation of biomineral tissue development | 38 | 62 | 1.34e-02 |
| GO:BP | GO:0071774 | response to fibroblast growth factor | 48 | 82 | 1.34e-02 |
| GO:BP | GO:0150104 | transport across blood-brain barrier | 41 | 68 | 1.36e-02 |
| GO:BP | GO:0071887 | leukocyte apoptotic process | 45 | 76 | 1.36e-02 |
| GO:BP | GO:0018210 | peptidyl-threonine modification | 45 | 76 | 1.36e-02 |
| GO:BP | GO:0097191 | extrinsic apoptotic signaling pathway | 91 | 172 | 1.36e-02 |
| GO:BP | GO:0010232 | vascular transport | 41 | 68 | 1.36e-02 |
| GO:BP | GO:0042552 | myelination | 65 | 117 | 1.36e-02 |
| GO:BP | GO:0035710 | CD4-positive, alpha-beta T cell activation | 40 | 66 | 1.36e-02 |
| GO:BP | GO:0097306 | cellular response to alcohol | 44 | 74 | 1.36e-02 |
| GO:BP | GO:0046578 | regulation of Ras protein signal transduction | 26 | 39 | 1.42e-02 |
| GO:BP | GO:0051965 | positive regulation of synapse assembly | 26 | 39 | 1.42e-02 |
| GO:BP | GO:0052548 | regulation of endopeptidase activity | 82 | 153 | 1.42e-02 |
| GO:BP | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 62 | 111 | 1.44e-02 |
| GO:BP | GO:0048713 | regulation of oligodendrocyte differentiation | 20 | 28 | 1.45e-02 |
| GO:BP | GO:0030705 | cytoskeleton-dependent intracellular transport | 94 | 179 | 1.54e-02 |
| GO:BP | GO:0060041 | retina development in camera-type eye | 59 | 105 | 1.54e-02 |
| GO:BP | GO:0032814 | regulation of natural killer cell activation | 13 | 16 | 1.63e-02 |
| GO:BP | GO:0021846 | cell proliferation in forebrain | 13 | 16 | 1.63e-02 |
| GO:BP | GO:0030041 | actin filament polymerization | 68 | 124 | 1.64e-02 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 56 | 99 | 1.64e-02 |
| GO:BP | GO:0007616 | long-term memory | 21 | 30 | 1.64e-02 |
| GO:BP | GO:0046850 | regulation of bone remodeling | 21 | 30 | 1.64e-02 |
| GO:BP | GO:0001937 | negative regulation of endothelial cell proliferation | 21 | 30 | 1.64e-02 |
| GO:BP | GO:0048512 | circadian behavior | 21 | 30 | 1.64e-02 |
| GO:BP | GO:0060914 | heart formation | 21 | 30 | 1.64e-02 |
| GO:BP | GO:0008277 | regulation of G protein-coupled receptor signaling pathway | 55 | 97 | 1.66e-02 |
| GO:BP | GO:0010038 | response to metal ion | 126 | 249 | 1.67e-02 |
| GO:BP | GO:0045926 | negative regulation of growth | 96 | 184 | 1.77e-02 |
| GO:BP | GO:0045685 | regulation of glial cell differentiation | 31 | 49 | 1.77e-02 |
| GO:BP | GO:0060711 | labyrinthine layer development | 22 | 32 | 1.81e-02 |
| GO:BP | GO:0062237 | protein localization to postsynapse | 22 | 32 | 1.81e-02 |
| GO:BP | GO:1903539 | protein localization to postsynaptic membrane | 22 | 32 | 1.81e-02 |
| GO:BP | GO:0010656 | negative regulation of muscle cell apoptotic process | 22 | 32 | 1.81e-02 |
| GO:BP | GO:1903350 | response to dopamine | 22 | 32 | 1.81e-02 |
| GO:BP | GO:0007622 | rhythmic behavior | 22 | 32 | 1.81e-02 |
| GO:BP | GO:0030168 | platelet activation | 50 | 87 | 1.81e-02 |
| GO:BP | GO:0010586 | miRNA metabolic process | 50 | 87 | 1.81e-02 |
| GO:BP | GO:0010876 | lipid localization | 148 | 298 | 1.81e-02 |
| GO:BP | GO:0010664 | negative regulation of striated muscle cell apoptotic process | 17 | 23 | 1.83e-02 |
| GO:BP | GO:0048843 | negative regulation of axon extension involved in axon guidance | 17 | 23 | 1.83e-02 |
| GO:BP | GO:0002063 | chondrocyte development | 17 | 23 | 1.83e-02 |
| GO:BP | GO:0060037 | pharyngeal system development | 17 | 23 | 1.83e-02 |
| GO:BP | GO:0045761 | regulation of adenylate cyclase activity | 17 | 23 | 1.83e-02 |
| GO:BP | GO:0045834 | positive regulation of lipid metabolic process | 48 | 83 | 1.83e-02 |
| GO:BP | GO:0034763 | negative regulation of transmembrane transport | 48 | 83 | 1.83e-02 |
| GO:BP | GO:0046847 | filopodium assembly | 33 | 53 | 1.84e-02 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 270 | 573 | 1.86e-02 |
| GO:BP | GO:0070509 | calcium ion import | 34 | 55 | 1.87e-02 |
| GO:BP | GO:0042476 | odontogenesis | 46 | 79 | 1.87e-02 |
| GO:BP | GO:0043393 | regulation of protein binding | 62 | 112 | 1.88e-02 |
| GO:BP | GO:1901184 | regulation of ERBB signaling pathway | 35 | 57 | 1.89e-02 |
| GO:BP | GO:0071260 | cellular response to mechanical stimulus | 35 | 57 | 1.89e-02 |
| GO:BP | GO:0046849 | bone remodeling | 36 | 59 | 1.91e-02 |
| GO:BP | GO:0048145 | regulation of fibroblast proliferation | 43 | 73 | 1.91e-02 |
| GO:BP | GO:0043535 | regulation of blood vessel endothelial cell migration | 42 | 71 | 1.92e-02 |
| GO:BP | GO:0051928 | positive regulation of calcium ion transport | 42 | 71 | 1.92e-02 |
| GO:BP | GO:0099084 | postsynaptic specialization organization | 23 | 34 | 1.94e-02 |
| GO:BP | GO:0051592 | response to calcium ion | 60 | 108 | 1.96e-02 |
| GO:BP | GO:0006869 | lipid transport | 131 | 261 | 1.96e-02 |
| GO:BP | GO:1903039 | positive regulation of leukocyte cell-cell adhesion | 78 | 146 | 1.98e-02 |
| GO:BP | GO:1901652 | response to peptide | 179 | 368 | 2.05e-02 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 709 | 1595 | 2.05e-02 |
| GO:BP | GO:0030902 | hindbrain development | 58 | 104 | 2.05e-02 |
| GO:BP | GO:0022607 | cellular component assembly | 1009 | 2305 | 2.05e-02 |
| GO:BP | GO:2001259 | positive regulation of cation channel activity | 24 | 36 | 2.07e-02 |
| GO:BP | GO:0048662 | negative regulation of smooth muscle cell proliferation | 24 | 36 | 2.07e-02 |
| GO:BP | GO:1902904 | negative regulation of supramolecular fiber organization | 68 | 125 | 2.07e-02 |
| GO:BP | GO:0002763 | positive regulation of myeloid leukocyte differentiation | 24 | 36 | 2.07e-02 |
| GO:BP | GO:1904019 | epithelial cell apoptotic process | 57 | 102 | 2.09e-02 |
| GO:BP | GO:0001956 | positive regulation of neurotransmitter secretion | 11 | 13 | 2.09e-02 |
| GO:BP | GO:0046321 | positive regulation of fatty acid oxidation | 11 | 13 | 2.09e-02 |
| GO:BP | GO:0046579 | positive regulation of Ras protein signal transduction | 11 | 13 | 2.09e-02 |
| GO:BP | GO:0060351 | cartilage development involved in endochondral bone morphogenesis | 18 | 25 | 2.10e-02 |
| GO:BP | GO:0032232 | negative regulation of actin filament bundle assembly | 18 | 25 | 2.10e-02 |
| GO:BP | GO:0038094 | Fc-gamma receptor signaling pathway | 14 | 18 | 2.11e-02 |
| GO:BP | GO:0032925 | regulation of activin receptor signaling pathway | 14 | 18 | 2.11e-02 |
| GO:BP | GO:0051968 | positive regulation of synaptic transmission, glutamatergic | 14 | 18 | 2.11e-02 |
| GO:BP | GO:0043113 | receptor clustering | 25 | 38 | 2.18e-02 |
| GO:BP | GO:0030217 | T cell differentiation | 95 | 183 | 2.18e-02 |
| GO:BP | GO:0045103 | intermediate filament-based process | 25 | 38 | 2.18e-02 |
| GO:BP | GO:0051480 | regulation of cytosolic calcium ion concentration | 25 | 38 | 2.18e-02 |
| GO:BP | GO:0061024 | membrane organization | 292 | 625 | 2.20e-02 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 74 | 138 | 2.20e-02 |
| GO:BP | GO:0072523 | purine-containing compound catabolic process | 65 | 119 | 2.22e-02 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 400 | 874 | 2.24e-02 |
| GO:BP | GO:0001816 | cytokine production | 211 | 441 | 2.30e-02 |
| GO:BP | GO:0021591 | ventricular system development | 19 | 27 | 2.38e-02 |
| GO:BP | GO:0120033 | negative regulation of plasma membrane bounded cell projection assembly | 19 | 27 | 2.38e-02 |
| GO:BP | GO:0010464 | regulation of mesenchymal cell proliferation | 19 | 27 | 2.38e-02 |
| GO:BP | GO:0031334 | positive regulation of protein-containing complex assembly | 79 | 149 | 2.38e-02 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 261 | 555 | 2.42e-02 |
| GO:BP | GO:0002682 | regulation of immune system process | 397 | 868 | 2.44e-02 |
| GO:BP | GO:0001817 | regulation of cytokine production | 209 | 437 | 2.45e-02 |
| GO:BP | GO:0072089 | stem cell proliferation | 48 | 84 | 2.46e-02 |
| GO:BP | GO:0009261 | ribonucleotide catabolic process | 61 | 111 | 2.46e-02 |
| GO:BP | GO:0070613 | regulation of protein processing | 28 | 44 | 2.46e-02 |
| GO:BP | GO:0045664 | regulation of neuron differentiation | 70 | 130 | 2.47e-02 |
| GO:BP | GO:0046129 | purine ribonucleoside biosynthetic process | 9 | 10 | 2.49e-02 |
| GO:BP | GO:0002093 | auditory receptor cell morphogenesis | 9 | 10 | 2.49e-02 |
| GO:BP | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway | 9 | 10 | 2.49e-02 |
| GO:BP | GO:0030431 | sleep | 9 | 10 | 2.49e-02 |
| GO:BP | GO:0042451 | purine nucleoside biosynthetic process | 9 | 10 | 2.49e-02 |
| GO:BP | GO:0042455 | ribonucleoside biosynthetic process | 9 | 10 | 2.49e-02 |
| GO:BP | GO:0045906 | negative regulation of vasoconstriction | 9 | 10 | 2.49e-02 |
| GO:BP | GO:0071625 | vocalization behavior | 9 | 10 | 2.49e-02 |
| GO:BP | GO:0099149 | regulation of postsynaptic neurotransmitter receptor internalization | 9 | 10 | 2.49e-02 |
| GO:BP | GO:0022410 | circadian sleep/wake cycle process | 9 | 10 | 2.49e-02 |
| GO:BP | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process | 15 | 20 | 2.60e-02 |
| GO:BP | GO:0035024 | negative regulation of Rho protein signal transduction | 15 | 20 | 2.60e-02 |
| GO:BP | GO:0031281 | positive regulation of cyclase activity | 15 | 20 | 2.60e-02 |
| GO:BP | GO:2000114 | regulation of establishment of cell polarity | 15 | 20 | 2.60e-02 |
| GO:BP | GO:0086005 | ventricular cardiac muscle cell action potential | 20 | 29 | 2.60e-02 |
| GO:BP | GO:0086019 | cell-cell signaling involved in cardiac conduction | 20 | 29 | 2.60e-02 |
| GO:BP | GO:0010761 | fibroblast migration | 31 | 50 | 2.61e-02 |
| GO:BP | GO:0051606 | detection of stimulus | 58 | 105 | 2.62e-02 |
| GO:BP | GO:0048709 | oligodendrocyte differentiation | 42 | 72 | 2.63e-02 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 42 | 72 | 2.63e-02 |
| GO:BP | GO:0001558 | regulation of cell growth | 158 | 323 | 2.63e-02 |
| GO:BP | GO:1904063 | negative regulation of cation transmembrane transport | 32 | 52 | 2.64e-02 |
| GO:BP | GO:0042475 | odontogenesis of dentin-containing tooth | 33 | 54 | 2.66e-02 |
| GO:BP | GO:0032835 | glomerulus development | 33 | 54 | 2.66e-02 |
| GO:BP | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 35 | 58 | 2.70e-02 |
| GO:BP | GO:0010565 | regulation of cellular ketone metabolic process | 55 | 99 | 2.81e-02 |
| GO:BP | GO:0070296 | sarcoplasmic reticulum calcium ion transport | 21 | 31 | 2.81e-02 |
| GO:BP | GO:0060391 | positive regulation of SMAD protein signal transduction | 21 | 31 | 2.81e-02 |
| GO:BP | GO:0021955 | central nervous system neuron axonogenesis | 21 | 31 | 2.81e-02 |
| GO:BP | GO:0050863 | regulation of T cell activation | 104 | 204 | 2.82e-02 |
| GO:BP | GO:0009913 | epidermal cell differentiation | 64 | 118 | 2.89e-02 |
| GO:BP | GO:0014047 | glutamate secretion | 12 | 15 | 2.89e-02 |
| GO:BP | GO:1900242 | regulation of synaptic vesicle endocytosis | 12 | 15 | 2.89e-02 |
| GO:BP | GO:0098877 | neurotransmitter receptor transport to plasma membrane | 12 | 15 | 2.89e-02 |
| GO:BP | GO:0001759 | organ induction | 12 | 15 | 2.89e-02 |
| GO:BP | GO:0003188 | heart valve formation | 12 | 15 | 2.89e-02 |
| GO:BP | GO:1902932 | positive regulation of alcohol biosynthetic process | 12 | 15 | 2.89e-02 |
| GO:BP | GO:0060571 | morphogenesis of an epithelial fold | 12 | 15 | 2.89e-02 |
| GO:BP | GO:0097091 | synaptic vesicle clustering | 12 | 15 | 2.89e-02 |
| GO:BP | GO:0009166 | nucleotide catabolic process | 72 | 135 | 2.89e-02 |
| GO:BP | GO:0070571 | negative regulation of neuron projection regeneration | 12 | 15 | 2.89e-02 |
| GO:BP | GO:0050870 | positive regulation of T cell activation | 72 | 135 | 2.89e-02 |
| GO:BP | GO:0046128 | purine ribonucleoside metabolic process | 12 | 15 | 2.89e-02 |
| GO:BP | GO:0090103 | cochlea morphogenesis | 12 | 15 | 2.89e-02 |
| GO:BP | GO:0033993 | response to lipid | 268 | 573 | 2.92e-02 |
| GO:BP | GO:0002684 | positive regulation of immune system process | 271 | 580 | 2.96e-02 |
| GO:BP | GO:0008154 | actin polymerization or depolymerization | 78 | 148 | 2.98e-02 |
| GO:BP | GO:0046635 | positive regulation of alpha-beta T cell activation | 22 | 33 | 2.98e-02 |
| GO:BP | GO:0060343 | trabecula formation | 16 | 22 | 3.01e-02 |
| GO:BP | GO:0002021 | response to dietary excess | 16 | 22 | 3.01e-02 |
| GO:BP | GO:0030279 | negative regulation of ossification | 16 | 22 | 3.01e-02 |
| GO:BP | GO:0048499 | synaptic vesicle membrane organization | 16 | 22 | 3.01e-02 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 95 | 185 | 3.13e-02 |
| GO:BP | GO:0086091 | regulation of heart rate by cardiac conduction | 23 | 35 | 3.16e-02 |
| GO:BP | GO:0015698 | inorganic anion transport | 48 | 85 | 3.24e-02 |
| GO:BP | GO:2001056 | positive regulation of cysteine-type endopeptidase activity | 47 | 83 | 3.30e-02 |
| GO:BP | GO:0015800 | acidic amino acid transport | 24 | 37 | 3.31e-02 |
| GO:BP | GO:0045104 | intermediate filament cytoskeleton organization | 24 | 37 | 3.31e-02 |
| GO:BP | GO:0046637 | regulation of alpha-beta T cell differentiation | 24 | 37 | 3.31e-02 |
| GO:BP | GO:0014074 | response to purine-containing compound | 58 | 106 | 3.36e-02 |
| GO:BP | GO:0009154 | purine ribonucleotide catabolic process | 58 | 106 | 3.36e-02 |
| GO:BP | GO:0022011 | myelination in peripheral nervous system | 17 | 24 | 3.43e-02 |
| GO:BP | GO:0048679 | regulation of axon regeneration | 17 | 24 | 3.43e-02 |
| GO:BP | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway | 17 | 24 | 3.43e-02 |
| GO:BP | GO:0032292 | peripheral nervous system axon ensheathment | 17 | 24 | 3.43e-02 |
| GO:BP | GO:1905332 | positive regulation of morphogenesis of an epithelium | 17 | 24 | 3.43e-02 |
| GO:BP | GO:0002431 | Fc receptor mediated stimulatory signaling pathway | 17 | 24 | 3.43e-02 |
| GO:BP | GO:0060074 | synapse maturation | 17 | 24 | 3.43e-02 |
| GO:BP | GO:0043281 | regulation of cysteine-type endopeptidase activity involved in apoptotic process | 57 | 104 | 3.43e-02 |
| GO:BP | GO:0046942 | carboxylic acid transport | 103 | 203 | 3.43e-02 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 186 | 388 | 3.44e-02 |
| GO:BP | GO:0006909 | phagocytosis | 73 | 138 | 3.46e-02 |
| GO:BP | GO:0007204 | positive regulation of cytosolic calcium ion concentration | 43 | 75 | 3.51e-02 |
| GO:BP | GO:1990845 | adaptive thermogenesis | 65 | 121 | 3.51e-02 |
| GO:BP | GO:0045453 | bone resorption | 26 | 41 | 3.51e-02 |
| GO:BP | GO:0031103 | axon regeneration | 26 | 41 | 3.51e-02 |
| GO:BP | GO:0030833 | regulation of actin filament polymerization | 56 | 102 | 3.51e-02 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 251 | 536 | 3.53e-02 |
| GO:BP | GO:0060349 | bone morphogenesis | 42 | 73 | 3.55e-02 |
| GO:BP | GO:0042093 | T-helper cell differentiation | 27 | 43 | 3.59e-02 |
| GO:BP | GO:1903034 | regulation of response to wounding | 64 | 119 | 3.59e-02 |
| GO:BP | GO:0002294 | CD4-positive, alpha-beta T cell differentiation involved in immune response | 27 | 43 | 3.59e-02 |
| GO:BP | GO:0002293 | alpha-beta T cell differentiation involved in immune response | 27 | 43 | 3.59e-02 |
| GO:BP | GO:0045214 | sarcomere organization | 27 | 43 | 3.59e-02 |
| GO:BP | GO:0002287 | alpha-beta T cell activation involved in immune response | 27 | 43 | 3.59e-02 |
| GO:BP | GO:0060392 | negative regulation of SMAD protein signal transduction | 13 | 17 | 3.65e-02 |
| GO:BP | GO:0072673 | lamellipodium morphogenesis | 13 | 17 | 3.65e-02 |
| GO:BP | GO:0048670 | regulation of collateral sprouting | 13 | 17 | 3.65e-02 |
| GO:BP | GO:0042100 | B cell proliferation | 28 | 45 | 3.65e-02 |
| GO:BP | GO:0061323 | cell proliferation involved in heart morphogenesis | 13 | 17 | 3.65e-02 |
| GO:BP | GO:2000136 | regulation of cell proliferation involved in heart morphogenesis | 13 | 17 | 3.65e-02 |
| GO:BP | GO:0043549 | regulation of kinase activity | 172 | 357 | 3.65e-02 |
| GO:BP | GO:0050680 | negative regulation of epithelial cell proliferation | 54 | 98 | 3.66e-02 |
| GO:BP | GO:0030099 | myeloid cell differentiation | 143 | 292 | 3.66e-02 |
| GO:BP | GO:0002067 | glandular epithelial cell differentiation | 29 | 47 | 3.69e-02 |
| GO:BP | GO:1903317 | regulation of protein maturation | 29 | 47 | 3.69e-02 |
| GO:BP | GO:0060291 | long-term synaptic potentiation | 38 | 65 | 3.69e-02 |
| GO:BP | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 18 | 26 | 3.72e-02 |
| GO:BP | GO:0048678 | response to axon injury | 36 | 61 | 3.72e-02 |
| GO:BP | GO:0010092 | specification of animal organ identity | 18 | 26 | 3.72e-02 |
| GO:BP | GO:0060998 | regulation of dendritic spine development | 30 | 49 | 3.72e-02 |
| GO:BP | GO:0033674 | positive regulation of kinase activity | 100 | 197 | 3.72e-02 |
| GO:BP | GO:0032092 | positive regulation of protein binding | 30 | 49 | 3.72e-02 |
| GO:BP | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction | 18 | 26 | 3.72e-02 |
| GO:BP | GO:0007212 | dopamine receptor signaling pathway | 18 | 26 | 3.72e-02 |
| GO:BP | GO:0001936 | regulation of endothelial cell proliferation | 53 | 96 | 3.72e-02 |
| GO:BP | GO:0071621 | granulocyte chemotaxis | 30 | 49 | 3.72e-02 |
| GO:BP | GO:0032271 | regulation of protein polymerization | 83 | 160 | 3.72e-02 |
| GO:BP | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway | 31 | 51 | 3.72e-02 |
| GO:BP | GO:0043954 | cellular component maintenance | 34 | 57 | 3.73e-02 |
| GO:BP | GO:0042310 | vasoconstriction | 33 | 55 | 3.73e-02 |
| GO:BP | GO:0042116 | macrophage activation | 33 | 55 | 3.73e-02 |
| GO:BP | GO:0007589 | body fluid secretion | 34 | 57 | 3.73e-02 |
| GO:BP | GO:0030032 | lamellipodium assembly | 33 | 55 | 3.73e-02 |
| GO:BP | GO:0036211 | protein modification process | 998 | 2291 | 3.75e-02 |
| GO:BP | GO:0036302 | atrioventricular canal development | 10 | 12 | 3.76e-02 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 119 | 239 | 3.76e-02 |
| GO:BP | GO:0045651 | positive regulation of macrophage differentiation | 10 | 12 | 3.76e-02 |
| GO:BP | GO:0061154 | endothelial tube morphogenesis | 10 | 12 | 3.76e-02 |
| GO:BP | GO:0033043 | regulation of organelle organization | 431 | 953 | 3.76e-02 |
| GO:BP | GO:0048368 | lateral mesoderm development | 10 | 12 | 3.76e-02 |
| GO:BP | GO:0071871 | response to epinephrine | 10 | 12 | 3.76e-02 |
| GO:BP | GO:0045780 | positive regulation of bone resorption | 10 | 12 | 3.76e-02 |
| GO:BP | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis | 10 | 12 | 3.76e-02 |
| GO:BP | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis | 10 | 12 | 3.76e-02 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 182 | 380 | 3.76e-02 |
| GO:BP | GO:0003159 | morphogenesis of an endothelium | 10 | 12 | 3.76e-02 |
| GO:BP | GO:0099505 | regulation of presynaptic membrane potential | 10 | 12 | 3.76e-02 |
| GO:BP | GO:0032516 | positive regulation of phosphoprotein phosphatase activity | 10 | 12 | 3.76e-02 |
| GO:BP | GO:0006955 | immune response | 394 | 867 | 3.77e-02 |
| GO:BP | GO:0022414 | reproductive process | 397 | 874 | 3.77e-02 |
| GO:BP | GO:0007584 | response to nutrient | 61 | 113 | 3.79e-02 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 141 | 288 | 3.81e-02 |
| GO:BP | GO:0043534 | blood vessel endothelial cell migration | 51 | 92 | 3.82e-02 |
| GO:BP | GO:1903532 | positive regulation of secretion by cell | 98 | 193 | 3.90e-02 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 127 | 257 | 3.91e-02 |
| GO:BP | GO:0001819 | positive regulation of cytokine production | 127 | 257 | 3.91e-02 |
| GO:BP | GO:0015849 | organic acid transport | 103 | 204 | 3.92e-02 |
| GO:BP | GO:0033036 | macromolecule localization | 1005 | 2309 | 3.96e-02 |
| GO:BP | GO:1903514 | release of sequestered calcium ion into cytosol by endoplasmic reticulum | 19 | 28 | 3.96e-02 |
| GO:BP | GO:0048483 | autonomic nervous system development | 19 | 28 | 3.96e-02 |
| GO:BP | GO:0014808 | release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 19 | 28 | 3.96e-02 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 112 | 224 | 4.04e-02 |
| GO:BP | GO:0050905 | neuromuscular process | 58 | 107 | 4.11e-02 |
| GO:BP | GO:0030595 | leukocyte chemotaxis | 58 | 107 | 4.11e-02 |
| GO:BP | GO:0001503 | ossification | 147 | 302 | 4.13e-02 |
| GO:BP | GO:0035270 | endocrine system development | 47 | 84 | 4.19e-02 |
| GO:BP | GO:0003338 | metanephros morphogenesis | 14 | 19 | 4.23e-02 |
| GO:BP | GO:0019934 | cGMP-mediated signaling | 14 | 19 | 4.23e-02 |
| GO:BP | GO:0042278 | purine nucleoside metabolic process | 14 | 19 | 4.23e-02 |
| GO:BP | GO:1901201 | regulation of extracellular matrix assembly | 14 | 19 | 4.23e-02 |
| GO:BP | GO:0010878 | cholesterol storage | 14 | 19 | 4.23e-02 |
| GO:BP | GO:0042092 | type 2 immune response | 14 | 19 | 4.23e-02 |
| GO:BP | GO:0003156 | regulation of animal organ formation | 14 | 19 | 4.23e-02 |
| GO:BP | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion | 14 | 19 | 4.23e-02 |
| GO:BP | GO:0052652 | cyclic purine nucleotide metabolic process | 20 | 30 | 4.23e-02 |
| GO:BP | GO:0062009 | secondary palate development | 14 | 19 | 4.23e-02 |
| GO:BP | GO:0045621 | positive regulation of lymphocyte differentiation | 46 | 82 | 4.25e-02 |
| GO:BP | GO:0007600 | sensory perception | 124 | 251 | 4.29e-02 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 141 | 289 | 4.36e-02 |
| GO:BP | GO:0045637 | regulation of myeloid cell differentiation | 71 | 135 | 4.36e-02 |
| GO:BP | GO:0042113 | B cell activation | 83 | 161 | 4.42e-02 |
| GO:BP | GO:0032869 | cellular response to insulin stimulus | 83 | 161 | 4.42e-02 |
| GO:BP | GO:0055006 | cardiac cell development | 44 | 78 | 4.42e-02 |
| GO:BP | GO:0030282 | bone mineralization | 44 | 78 | 4.42e-02 |
| GO:BP | GO:0031333 | negative regulation of protein-containing complex assembly | 63 | 118 | 4.47e-02 |
| GO:BP | GO:0050714 | positive regulation of protein secretion | 54 | 99 | 4.56e-02 |
| GO:BP | GO:1904018 | positive regulation of vasculature development | 62 | 116 | 4.61e-02 |
| GO:BP | GO:0035272 | exocrine system development | 22 | 34 | 4.63e-02 |
| GO:BP | GO:0030888 | regulation of B cell proliferation | 22 | 34 | 4.63e-02 |
| GO:BP | GO:0090102 | cochlea development | 22 | 34 | 4.63e-02 |
| GO:BP | GO:0060999 | positive regulation of dendritic spine development | 22 | 34 | 4.63e-02 |
| GO:BP | GO:0043666 | regulation of phosphoprotein phosphatase activity | 22 | 34 | 4.63e-02 |
| GO:BP | GO:0003161 | cardiac conduction system development | 22 | 34 | 4.63e-02 |
| GO:BP | GO:0071711 | basement membrane organization | 22 | 34 | 4.63e-02 |
| GO:BP | GO:0002793 | positive regulation of peptide secretion | 41 | 72 | 4.67e-02 |
| GO:BP | GO:0097475 | motor neuron migration | 8 | 9 | 4.69e-02 |
| GO:BP | GO:0003162 | atrioventricular node development | 8 | 9 | 4.69e-02 |
| GO:BP | GO:0050802 | circadian sleep/wake cycle, sleep | 8 | 9 | 4.69e-02 |
| GO:BP | GO:0071361 | cellular response to ethanol | 8 | 9 | 4.69e-02 |
| GO:BP | GO:0060088 | auditory receptor cell stereocilium organization | 8 | 9 | 4.69e-02 |
| GO:BP | GO:0061626 | pharyngeal arch artery morphogenesis | 8 | 9 | 4.69e-02 |
| GO:BP | GO:0001768 | establishment of T cell polarity | 8 | 9 | 4.69e-02 |
| GO:BP | GO:0038065 | collagen-activated signaling pathway | 8 | 9 | 4.69e-02 |
| GO:BP | GO:0030162 | regulation of proteolysis | 197 | 416 | 4.69e-02 |
| GO:BP | GO:0006195 | purine nucleotide catabolic process | 61 | 114 | 4.69e-02 |
| GO:BP | GO:0045766 | positive regulation of angiogenesis | 61 | 114 | 4.69e-02 |
| GO:BP | GO:0045862 | positive regulation of proteolysis | 121 | 245 | 4.69e-02 |
| GO:BP | GO:0030593 | neutrophil chemotaxis | 23 | 36 | 4.75e-02 |
| GO:BP | GO:0061001 | regulation of dendritic spine morphogenesis | 23 | 36 | 4.75e-02 |
| GO:BP | GO:0060443 | mammary gland morphogenesis | 23 | 36 | 4.75e-02 |
| GO:BP | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway | 6 | 6 | 4.75e-02 |
| GO:BP | GO:0023041 | neuronal signal transduction | 6 | 6 | 4.75e-02 |
| GO:BP | GO:0014912 | negative regulation of smooth muscle cell migration | 15 | 21 | 4.75e-02 |
| GO:BP | GO:0032754 | positive regulation of interleukin-5 production | 6 | 6 | 4.75e-02 |
| GO:BP | GO:0031223 | auditory behavior | 6 | 6 | 4.75e-02 |
| GO:BP | GO:1903421 | regulation of synaptic vesicle recycling | 15 | 21 | 4.75e-02 |
| GO:BP | GO:0072173 | metanephric tubule morphogenesis | 6 | 6 | 4.75e-02 |
| GO:BP | GO:1903903 | regulation of establishment of T cell polarity | 6 | 6 | 4.75e-02 |
| GO:BP | GO:0010737 | protein kinase A signaling | 15 | 21 | 4.75e-02 |
| GO:BP | GO:0060710 | chorio-allantoic fusion | 6 | 6 | 4.75e-02 |
| GO:BP | GO:0061037 | negative regulation of cartilage development | 15 | 21 | 4.75e-02 |
| GO:BP | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis | 6 | 6 | 4.75e-02 |
| GO:BP | GO:0099545 | trans-synaptic signaling by trans-synaptic complex | 6 | 6 | 4.75e-02 |
| GO:BP | GO:0051639 | actin filament network formation | 6 | 6 | 4.75e-02 |
| GO:BP | GO:0050679 | positive regulation of epithelial cell proliferation | 74 | 142 | 4.75e-02 |
| GO:BP | GO:2000192 | negative regulation of fatty acid transport | 6 | 6 | 4.75e-02 |
| GO:BP | GO:0014031 | mesenchymal cell development | 6 | 6 | 4.75e-02 |
| GO:BP | GO:0050861 | positive regulation of B cell receptor signaling pathway | 6 | 6 | 4.75e-02 |
| GO:BP | GO:0060897 | neural plate regionalization | 6 | 6 | 4.75e-02 |
| GO:BP | GO:0003175 | tricuspid valve development | 6 | 6 | 4.75e-02 |
| GO:BP | GO:0043502 | regulation of muscle adaptation | 39 | 68 | 4.75e-02 |
| GO:BP | GO:0098976 | excitatory chemical synaptic transmission | 6 | 6 | 4.75e-02 |
| GO:BP | GO:0003415 | chondrocyte hypertrophy | 6 | 6 | 4.75e-02 |
| GO:BP | GO:0001771 | immunological synapse formation | 6 | 6 | 4.75e-02 |
| GO:BP | GO:0045639 | positive regulation of myeloid cell differentiation | 38 | 66 | 4.77e-02 |
| GO:BP | GO:0051098 | regulation of binding | 101 | 201 | 4.78e-02 |
| GO:BP | GO:0048013 | ephrin receptor signaling pathway | 24 | 38 | 4.78e-02 |
| GO:BP | GO:0051494 | negative regulation of cytoskeleton organization | 67 | 127 | 4.78e-02 |
| GO:BP | GO:2001222 | regulation of neuron migration | 24 | 38 | 4.78e-02 |
| GO:BP | GO:2000117 | negative regulation of cysteine-type endopeptidase activity | 24 | 38 | 4.78e-02 |
| GO:BP | GO:0019217 | regulation of fatty acid metabolic process | 37 | 64 | 4.82e-02 |
| GO:BP | GO:0038166 | angiotensin-activated signaling pathway | 11 | 14 | 4.84e-02 |
| GO:BP | GO:0060142 | regulation of syncytium formation by plasma membrane fusion | 11 | 14 | 4.84e-02 |
| GO:BP | GO:0010763 | positive regulation of fibroblast migration | 11 | 14 | 4.84e-02 |
| GO:BP | GO:0002252 | immune effector process | 145 | 299 | 4.84e-02 |
| GO:BP | GO:1901292 | nucleoside phosphate catabolic process | 73 | 140 | 4.84e-02 |
| GO:BP | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway | 11 | 14 | 4.84e-02 |
| GO:BP | GO:0050954 | sensory perception of mechanical stimulus | 59 | 110 | 4.84e-02 |
| GO:BP | GO:0010885 | regulation of cholesterol storage | 11 | 14 | 4.84e-02 |
| GO:BP | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter | 11 | 14 | 4.84e-02 |
| GO:BP | GO:0006835 | dicarboxylic acid transport | 35 | 60 | 4.91e-02 |
| GO:BP | GO:0048146 | positive regulation of fibroblast proliferation | 26 | 42 | 4.93e-02 |
| GO:BP | GO:0045123 | cellular extravasation | 26 | 42 | 4.93e-02 |
| GO:BP | GO:0051489 | regulation of filopodium assembly | 26 | 42 | 4.93e-02 |
| GO:BP | GO:0048144 | fibroblast proliferation | 49 | 89 | 4.98e-02 |
| GO:BP | GO:0002042 | cell migration involved in sprouting angiogenesis | 27 | 44 | 4.98e-02 |
| GO:BP | GO:0071695 | anatomical structure maturation | 78 | 151 | 4.99e-02 |
ESRnomtable_20k %>%
filter(source=="KEGG", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "KEGG terms found in ESR nESRgenes_20k") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| KEGG | KEGG:04510 | Focal adhesion | 113 | 169 | 2.81e-09 |
| KEGG | KEGG:04020 | Calcium signaling pathway | 107 | 161 | 5.90e-09 |
| KEGG | KEGG:04360 | Axon guidance | 104 | 155 | 5.90e-09 |
| KEGG | KEGG:05200 | Pathways in cancer | 223 | 391 | 5.98e-09 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 98 | 147 | 1.47e-08 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 106 | 162 | 1.47e-08 |
| KEGG | KEGG:04925 | Aldosterone synthesis and secretion | 57 | 75 | 3.12e-08 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 105 | 162 | 3.12e-08 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 84 | 129 | 8.83e-07 |
| KEGG | KEGG:04724 | Glutamatergic synapse | 54 | 74 | 8.83e-07 |
| KEGG | KEGG:05414 | Dilated cardiomyopathy | 58 | 82 | 1.48e-06 |
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 80 | 124 | 2.88e-06 |
| KEGG | KEGG:04921 | Oxytocin signaling pathway | 74 | 113 | 2.99e-06 |
| KEGG | KEGG:04911 | Insulin secretion | 46 | 62 | 2.99e-06 |
| KEGG | KEGG:04270 | Vascular smooth muscle contraction | 62 | 92 | 6.32e-06 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 83 | 133 | 1.05e-05 |
| KEGG | KEGG:04924 | Renin secretion | 38 | 50 | 1.06e-05 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 101 | 169 | 1.16e-05 |
| KEGG | KEGG:05410 | Hypertrophic cardiomyopathy | 53 | 77 | 1.18e-05 |
| KEGG | KEGG:04713 | Circadian entrainment | 48 | 68 | 1.18e-05 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 68 | 105 | 1.18e-05 |
| KEGG | KEGG:04611 | Platelet activation | 59 | 88 | 1.18e-05 |
| KEGG | KEGG:04927 | Cortisol synthesis and secretion | 35 | 46 | 2.12e-05 |
| KEGG | KEGG:04512 | ECM-receptor interaction | 46 | 66 | 3.24e-05 |
| KEGG | KEGG:04750 | Inflammatory mediator regulation of TRP channels | 49 | 72 | 4.35e-05 |
| KEGG | KEGG:04014 | Ras signaling pathway | 96 | 164 | 5.46e-05 |
| KEGG | KEGG:04726 | Serotonergic synapse | 41 | 58 | 5.82e-05 |
| KEGG | KEGG:04725 | Cholinergic synapse | 54 | 82 | 5.84e-05 |
| KEGG | KEGG:04971 | Gastric acid secretion | 37 | 51 | 5.94e-05 |
| KEGG | KEGG:04934 | Cushing syndrome | 73 | 119 | 6.66e-05 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 137 | 250 | 7.18e-05 |
| KEGG | KEGG:04520 | Adherens junction | 54 | 83 | 8.80e-05 |
| KEGG | KEGG:04970 | Salivary secretion | 38 | 54 | 1.26e-04 |
| KEGG | KEGG:04961 | Endocrine and other factor-regulated calcium reabsorption | 30 | 40 | 1.36e-04 |
| KEGG | KEGG:04974 | Protein digestion and absorption | 39 | 56 | 1.36e-04 |
| KEGG | KEGG:04728 | Dopaminergic synapse | 60 | 97 | 2.51e-04 |
| KEGG | KEGG:04972 | Pancreatic secretion | 39 | 57 | 2.51e-04 |
| KEGG | KEGG:04072 | Phospholipase D signaling pathway | 65 | 107 | 2.61e-04 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 76 | 129 | 2.61e-04 |
| KEGG | KEGG:04670 | Leukocyte transendothelial migration | 49 | 76 | 2.63e-04 |
| KEGG | KEGG:04929 | GnRH secretion | 35 | 50 | 2.70e-04 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 125 | 231 | 2.92e-04 |
| KEGG | KEGG:05032 | Morphine addiction | 37 | 54 | 3.17e-04 |
| KEGG | KEGG:05412 | Arrhythmogenic right ventricular cardiomyopathy | 44 | 67 | 3.17e-04 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 82 | 144 | 6.05e-04 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 49 | 78 | 6.17e-04 |
| KEGG | KEGG:04915 | Estrogen signaling pathway | 55 | 90 | 6.81e-04 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 60 | 100 | 6.92e-04 |
| KEGG | KEGG:04730 | Long-term depression | 28 | 39 | 6.92e-04 |
| KEGG | KEGG:04530 | Tight junction | 74 | 129 | 8.61e-04 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 89 | 160 | 8.61e-04 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 132 | 252 | 1.05e-03 |
| KEGG | KEGG:04148 | Efferocytosis | 69 | 120 | 1.24e-03 |
| KEGG | KEGG:04066 | HIF-1 signaling pathway | 52 | 86 | 1.31e-03 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 61 | 104 | 1.31e-03 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 57 | 96 | 1.31e-03 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 77 | 137 | 1.38e-03 |
| KEGG | KEGG:04976 | Bile secretion | 28 | 41 | 2.24e-03 |
| KEGG | KEGG:04923 | Regulation of lipolysis in adipocytes | 28 | 41 | 2.24e-03 |
| KEGG | KEGG:05146 | Amoebiasis | 39 | 62 | 2.38e-03 |
| KEGG | KEGG:05031 | Amphetamine addiction | 30 | 45 | 2.57e-03 |
| KEGG | KEGG:04720 | Long-term potentiation | 33 | 51 | 3.03e-03 |
| KEGG | KEGG:05226 | Gastric cancer | 64 | 113 | 3.09e-03 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 51 | 87 | 3.68e-03 |
| KEGG | KEGG:04916 | Melanogenesis | 45 | 75 | 3.70e-03 |
| KEGG | KEGG:04814 | Motor proteins | 77 | 141 | 3.89e-03 |
| KEGG | KEGG:00051 | Fructose and mannose metabolism | 19 | 26 | 4.90e-03 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 37 | 60 | 5.06e-03 |
| KEGG | KEGG:04912 | GnRH signaling pathway | 44 | 74 | 5.12e-03 |
| KEGG | KEGG:04922 | Glucagon signaling pathway | 46 | 78 | 5.12e-03 |
| KEGG | KEGG:05224 | Breast cancer | 63 | 113 | 5.26e-03 |
| KEGG | KEGG:04370 | VEGF signaling pathway | 30 | 47 | 6.43e-03 |
| KEGG | KEGG:05100 | Bacterial invasion of epithelial cells | 43 | 73 | 6.96e-03 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 44 | 75 | 6.96e-03 |
| KEGG | KEGG:04666 | Fc gamma R-mediated phagocytosis | 44 | 75 | 6.96e-03 |
| KEGG | KEGG:05214 | Glioma | 39 | 65 | 6.96e-03 |
| KEGG | KEGG:04540 | Gap junction | 42 | 71 | 6.96e-03 |
| KEGG | KEGG:00000 | KEGG root term | 2317 | 5451 | 7.40e-03 |
| KEGG | KEGG:04740 | Olfactory transduction | 18 | 25 | 7.53e-03 |
| KEGG | KEGG:01522 | Endocrine resistance | 47 | 82 | 9.04e-03 |
| KEGG | KEGG:04727 | GABAergic synapse | 32 | 52 | 9.64e-03 |
| KEGG | KEGG:05230 | Central carbon metabolism in cancer | 35 | 58 | 9.77e-03 |
| KEGG | KEGG:04152 | AMPK signaling pathway | 56 | 101 | 9.84e-03 |
| KEGG | KEGG:05017 | Spinocerebellar ataxia | 65 | 120 | 9.89e-03 |
| KEGG | KEGG:05131 | Shigellosis | 104 | 204 | 9.89e-03 |
| KEGG | KEGG:04930 | Type II diabetes mellitus | 23 | 35 | 1.12e-02 |
| KEGG | KEGG:04721 | Synaptic vesicle cycle | 30 | 49 | 1.35e-02 |
| KEGG | KEGG:05207 | Chemical carcinogenesis - receptor activation | 70 | 132 | 1.35e-02 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 61 | 113 | 1.40e-02 |
| KEGG | KEGG:04380 | Osteoclast differentiation | 51 | 92 | 1.40e-02 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 50 | 90 | 1.42e-02 |
| KEGG | KEGG:05135 | Yersinia infection | 58 | 107 | 1.48e-02 |
| KEGG | KEGG:05231 | Choline metabolism in cancer | 45 | 80 | 1.57e-02 |
| KEGG | KEGG:05030 | Cocaine addiction | 21 | 32 | 1.60e-02 |
| KEGG | KEGG:01521 | EGFR tyrosine kinase inhibitor resistance | 41 | 72 | 1.68e-02 |
| KEGG | KEGG:00052 | Galactose metabolism | 15 | 21 | 1.71e-02 |
| KEGG | KEGG:00512 | Mucin type O-glycan biosynthesis | 15 | 21 | 1.71e-02 |
| KEGG | KEGG:04931 | Insulin resistance | 50 | 91 | 1.77e-02 |
| KEGG | KEGG:04070 | Phosphatidylinositol signaling system | 46 | 83 | 1.96e-02 |
| KEGG | KEGG:04060 | Cytokine-cytokine receptor interaction | 51 | 94 | 2.24e-02 |
| KEGG | KEGG:04514 | Cell adhesion molecules | 39 | 69 | 2.29e-02 |
| KEGG | KEGG:04973 | Carbohydrate digestion and absorption | 19 | 29 | 2.30e-02 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 64 | 122 | 2.32e-02 |
| KEGG | KEGG:04071 | Sphingolipid signaling pathway | 55 | 103 | 2.46e-02 |
| KEGG | KEGG:04650 | Natural killer cell mediated cytotoxicity | 31 | 53 | 2.54e-02 |
| KEGG | KEGG:04625 | C-type lectin receptor signaling pathway | 41 | 74 | 2.85e-02 |
| KEGG | KEGG:05211 | Renal cell carcinoma | 35 | 62 | 3.26e-02 |
| KEGG | KEGG:04960 | Aldosterone-regulated sodium reabsorption | 16 | 24 | 3.26e-02 |
| KEGG | KEGG:05210 | Colorectal cancer | 44 | 81 | 3.35e-02 |
| KEGG | KEGG:04913 | Ovarian steroidogenesis | 17 | 26 | 3.35e-02 |
| KEGG | KEGG:05218 | Melanoma | 32 | 56 | 3.39e-02 |
| KEGG | KEGG:05418 | Fluid shear stress and atherosclerosis | 57 | 109 | 3.45e-02 |
| KEGG | KEGG:04062 | Chemokine signaling pathway | 57 | 109 | 3.45e-02 |
| KEGG | KEGG:04213 | Longevity regulating pathway - multiple species | 29 | 50 | 3.48e-02 |
| KEGG | KEGG:05212 | Pancreatic cancer | 39 | 71 | 3.73e-02 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 66 | 129 | 3.73e-02 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 53 | 101 | 3.86e-02 |
| KEGG | KEGG:05170 | Human immunodeficiency virus 1 infection | 75 | 149 | 3.88e-02 |
| KEGG | KEGG:04260 | Cardiac muscle contraction | 35 | 63 | 4.04e-02 |
| KEGG | KEGG:05222 | Small cell lung cancer | 44 | 82 | 4.04e-02 |
| KEGG | KEGG:05167 | Kaposi sarcoma-associated herpesvirus infection | 68 | 134 | 4.07e-02 |
| KEGG | KEGG:04614 | Renin-angiotensin system | 8 | 10 | 4.18e-02 |
| KEGG | KEGG:05033 | Nicotine addiction | 8 | 10 | 4.18e-02 |
| KEGG | KEGG:04340 | Hedgehog signaling pathway | 28 | 49 | 4.61e-02 |
| KEGG | KEGG:05142 | Chagas disease | 39 | 72 | 4.61e-02 |
| KEGG | KEGG:03320 | PPAR signaling pathway | 27 | 47 | 4.67e-02 |
| KEGG | KEGG:04392 | Hippo signaling pathway - multiple species | 16 | 25 | 4.75e-02 |
| KEGG | KEGG:05219 | Bladder cancer | 21 | 35 | 4.97e-02 |
ESR peaks overlapping enhancers
# ESR_resgenes_20k_enh <- gost(query = unique(ESR_enh_peaks_20k$NCBI_gene),
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = FALSE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = background_df$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(ESR_resgenes_20k_enh,"data/GO_KEGG_analysis/ESR_resgenes_20k_enh.RDS")
# ESRnom <- gostplot(ESR_resgenes_20k_enh, capped = FALSE, interactive = TRUE)
# ESRnom
ESRnomtable_20k_enh <- ESR_resgenes_20k_enh$result %>%
dplyr::select(c(source, term_id,
term_name,intersection_size,
term_size, p_value))# %>%
ESRnomtable_20k_enh %>%
filter(source=="GO:BP", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(.,caption = "GO:BP terms found in ESR neargenes_20k" ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0032501 | multicellular organismal process | 623 | 4504 | 2.35e-19 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 318 | 1924 | 1.69e-17 |
| GO:BP | GO:0030154 | cell differentiation | 415 | 2803 | 1.63e-15 |
| GO:BP | GO:0048856 | anatomical structure development | 536 | 3867 | 1.63e-15 |
| GO:BP | GO:0048869 | cellular developmental process | 415 | 2804 | 1.63e-15 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 299 | 1844 | 2.56e-15 |
| GO:BP | GO:0032502 | developmental process | 567 | 4179 | 4.03e-15 |
| GO:BP | GO:0007154 | cell communication | 533 | 3983 | 2.17e-12 |
| GO:BP | GO:0050896 | response to stimulus | 680 | 5379 | 3.74e-12 |
| GO:BP | GO:0072359 | circulatory system development | 161 | 864 | 4.11e-12 |
| GO:BP | GO:0048731 | system development | 389 | 2719 | 5.72e-12 |
| GO:BP | GO:0007275 | multicellular organism development | 440 | 3170 | 5.72e-12 |
| GO:BP | GO:0023052 | signaling | 523 | 3918 | 5.72e-12 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 156 | 837 | 8.14e-12 |
| GO:BP | GO:0050789 | regulation of biological process | 891 | 7516 | 9.21e-12 |
| GO:BP | GO:0003013 | circulatory system process | 96 | 426 | 1.43e-11 |
| GO:BP | GO:0008015 | blood circulation | 85 | 357 | 1.64e-11 |
| GO:BP | GO:0065007 | biological regulation | 908 | 7734 | 5.15e-11 |
| GO:BP | GO:0003008 | system process | 193 | 1139 | 5.57e-11 |
| GO:BP | GO:0007165 | signal transduction | 483 | 3611 | 6.96e-11 |
| GO:BP | GO:0030029 | actin filament-based process | 125 | 640 | 9.97e-11 |
| GO:BP | GO:0050793 | regulation of developmental process | 260 | 1697 | 2.48e-10 |
| GO:BP | GO:0061061 | muscle structure development | 109 | 536 | 2.48e-10 |
| GO:BP | GO:0048468 | cell development | 282 | 1886 | 3.66e-10 |
| GO:BP | GO:0060047 | heart contraction | 54 | 194 | 9.51e-10 |
| GO:BP | GO:0051716 | cellular response to stimulus | 587 | 4640 | 1.20e-09 |
| GO:BP | GO:0050794 | regulation of cellular process | 842 | 7153 | 2.34e-09 |
| GO:BP | GO:0048518 | positive regulation of biological process | 547 | 4301 | 5.59e-09 |
| GO:BP | GO:0003015 | heart process | 54 | 204 | 6.73e-09 |
| GO:BP | GO:0009888 | tissue development | 211 | 1348 | 6.73e-09 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 192 | 1197 | 7.02e-09 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 508 | 3954 | 9.71e-09 |
| GO:BP | GO:0048519 | negative regulation of biological process | 487 | 3764 | 1.11e-08 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 170 | 1035 | 1.58e-08 |
| GO:BP | GO:0008016 | regulation of heart contraction | 46 | 165 | 2.64e-08 |
| GO:BP | GO:0016477 | cell migration | 165 | 1011 | 4.95e-08 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 106 | 566 | 5.58e-08 |
| GO:BP | GO:0003012 | muscle system process | 71 | 325 | 6.70e-08 |
| GO:BP | GO:0035239 | tube morphogenesis | 115 | 638 | 9.94e-08 |
| GO:BP | GO:0040011 | locomotion | 137 | 805 | 1.11e-07 |
| GO:BP | GO:0042692 | muscle cell differentiation | 70 | 323 | 1.17e-07 |
| GO:BP | GO:2000145 | regulation of cell motility | 123 | 700 | 1.17e-07 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 457 | 3550 | 1.22e-07 |
| GO:BP | GO:0044057 | regulation of system process | 79 | 384 | 1.22e-07 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 259 | 1801 | 1.39e-07 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 91 | 469 | 1.39e-07 |
| GO:BP | GO:0035295 | tube development | 134 | 787 | 1.44e-07 |
| GO:BP | GO:0055001 | muscle cell development | 44 | 163 | 1.50e-07 |
| GO:BP | GO:0040012 | regulation of locomotion | 125 | 720 | 1.56e-07 |
| GO:BP | GO:0030334 | regulation of cell migration | 118 | 668 | 1.62e-07 |
| GO:BP | GO:1903522 | regulation of blood circulation | 49 | 194 | 1.90e-07 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 55 | 234 | 3.18e-07 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 109 | 609 | 3.24e-07 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 87 | 450 | 3.38e-07 |
| GO:BP | GO:0030048 | actin filament-based movement | 33 | 107 | 4.41e-07 |
| GO:BP | GO:0008283 | cell population proliferation | 197 | 1306 | 4.49e-07 |
| GO:BP | GO:0001568 | blood vessel development | 97 | 525 | 4.54e-07 |
| GO:BP | GO:0048870 | cell motility | 174 | 1120 | 4.71e-07 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 29 | 87 | 5.05e-07 |
| GO:BP | GO:0048513 | animal organ development | 279 | 2004 | 5.79e-07 |
| GO:BP | GO:0007507 | heart development | 93 | 499 | 5.79e-07 |
| GO:BP | GO:0001944 | vasculature development | 100 | 552 | 6.93e-07 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 163 | 1040 | 7.78e-07 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 34 | 116 | 9.47e-07 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 121 | 717 | 1.26e-06 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 115 | 673 | 1.45e-06 |
| GO:BP | GO:0060537 | muscle tissue development | 69 | 339 | 1.53e-06 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 108 | 622 | 1.68e-06 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 166 | 1077 | 1.71e-06 |
| GO:BP | GO:0007155 | cell adhesion | 149 | 941 | 1.73e-06 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 150 | 951 | 1.98e-06 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 165 | 1073 | 2.15e-06 |
| GO:BP | GO:0006936 | muscle contraction | 56 | 255 | 2.18e-06 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 356 | 2717 | 2.43e-06 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 92 | 508 | 2.45e-06 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 69 | 344 | 2.50e-06 |
| GO:BP | GO:0000902 | cell morphogenesis | 119 | 713 | 2.72e-06 |
| GO:BP | GO:0014706 | striated muscle tissue development | 49 | 213 | 3.34e-06 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 47 | 201 | 3.68e-06 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 54 | 248 | 4.82e-06 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 116 | 706 | 9.39e-06 |
| GO:BP | GO:0006941 | striated muscle contraction | 37 | 146 | 1.14e-05 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 106 | 633 | 1.29e-05 |
| GO:BP | GO:0023051 | regulation of signaling | 320 | 2438 | 1.32e-05 |
| GO:BP | GO:0001525 | angiogenesis | 72 | 381 | 1.38e-05 |
| GO:BP | GO:0065009 | regulation of molecular function | 188 | 1299 | 1.64e-05 |
| GO:BP | GO:0010646 | regulation of cell communication | 320 | 2444 | 1.64e-05 |
| GO:BP | GO:0003007 | heart morphogenesis | 48 | 218 | 1.70e-05 |
| GO:BP | GO:0090130 | tissue migration | 52 | 247 | 2.38e-05 |
| GO:BP | GO:0070848 | response to growth factor | 93 | 547 | 3.56e-05 |
| GO:BP | GO:0002027 | regulation of heart rate | 25 | 83 | 3.56e-05 |
| GO:BP | GO:0098657 | import into cell | 102 | 622 | 5.64e-05 |
| GO:BP | GO:0061337 | cardiac conduction | 25 | 85 | 5.74e-05 |
| GO:BP | GO:0065008 | regulation of biological quality | 254 | 1893 | 6.06e-05 |
| GO:BP | GO:0071417 | cellular response to organonitrogen compound | 78 | 442 | 6.81e-05 |
| GO:BP | GO:0009605 | response to external stimulus | 215 | 1559 | 7.27e-05 |
| GO:BP | GO:0090132 | epithelium migration | 50 | 243 | 7.54e-05 |
| GO:BP | GO:0010631 | epithelial cell migration | 50 | 243 | 7.54e-05 |
| GO:BP | GO:0032989 | cellular anatomical entity morphogenesis | 97 | 588 | 7.57e-05 |
| GO:BP | GO:0031032 | actomyosin structure organization | 41 | 184 | 8.32e-05 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 89 | 528 | 8.37e-05 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 137 | 913 | 1.13e-04 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 82 | 479 | 1.17e-04 |
| GO:BP | GO:0048568 | embryonic organ development | 58 | 303 | 1.20e-04 |
| GO:BP | GO:0042221 | response to chemical | 298 | 2304 | 1.33e-04 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 41 | 188 | 1.41e-04 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 78 | 452 | 1.48e-04 |
| GO:BP | GO:0009966 | regulation of signal transduction | 282 | 2166 | 1.54e-04 |
| GO:BP | GO:0010243 | response to organonitrogen compound | 108 | 685 | 1.54e-04 |
| GO:BP | GO:0071526 | semaphorin-plexin signaling pathway | 16 | 43 | 1.64e-04 |
| GO:BP | GO:0003205 | cardiac chamber development | 35 | 151 | 1.76e-04 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 300 | 2333 | 1.91e-04 |
| GO:BP | GO:0003231 | cardiac ventricle development | 29 | 115 | 2.10e-04 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 123 | 811 | 2.13e-04 |
| GO:BP | GO:0007517 | muscle organ development | 52 | 267 | 2.20e-04 |
| GO:BP | GO:0071310 | cellular response to organic substance | 181 | 1297 | 2.45e-04 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 28 | 110 | 2.45e-04 |
| GO:BP | GO:0035556 | intracellular signal transduction | 256 | 1951 | 2.67e-04 |
| GO:BP | GO:0048729 | tissue morphogenesis | 77 | 453 | 2.78e-04 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 71 | 408 | 2.81e-04 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 19 | 60 | 2.85e-04 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 233 | 1751 | 2.86e-04 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 72 | 416 | 2.87e-04 |
| GO:BP | GO:0030239 | myofibril assembly | 20 | 66 | 3.35e-04 |
| GO:BP | GO:0061614 | miRNA transcription | 19 | 61 | 3.63e-04 |
| GO:BP | GO:0007010 | cytoskeleton organization | 162 | 1146 | 3.76e-04 |
| GO:BP | GO:0014888 | striated muscle adaptation | 15 | 41 | 3.76e-04 |
| GO:BP | GO:0090257 | regulation of muscle system process | 38 | 177 | 4.07e-04 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 94 | 591 | 4.20e-04 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 329 | 2622 | 4.43e-04 |
| GO:BP | GO:1901698 | response to nitrogen compound | 112 | 736 | 4.49e-04 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 181 | 1314 | 4.90e-04 |
| GO:BP | GO:0055002 | striated muscle cell development | 20 | 68 | 5.16e-04 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 68 | 395 | 5.83e-04 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 40 | 194 | 6.23e-04 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 19 | 64 | 7.16e-04 |
| GO:BP | GO:0048644 | muscle organ morphogenesis | 19 | 64 | 7.16e-04 |
| GO:BP | GO:0070482 | response to oxygen levels | 50 | 265 | 7.16e-04 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 107 | 704 | 7.20e-04 |
| GO:BP | GO:0010720 | positive regulation of cell development | 56 | 309 | 7.42e-04 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 304 | 2415 | 8.12e-04 |
| GO:BP | GO:1901342 | regulation of vasculature development | 40 | 198 | 9.82e-04 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 153 | 1093 | 1.06e-03 |
| GO:BP | GO:0043500 | muscle adaptation | 23 | 89 | 1.10e-03 |
| GO:BP | GO:0006915 | apoptotic process | 186 | 1377 | 1.10e-03 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 29 | 126 | 1.10e-03 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 20 | 72 | 1.19e-03 |
| GO:BP | GO:0060415 | muscle tissue morphogenesis | 18 | 61 | 1.23e-03 |
| GO:BP | GO:0022008 | neurogenesis | 171 | 1251 | 1.24e-03 |
| GO:BP | GO:0007264 | small GTPase-mediated signal transduction | 66 | 390 | 1.24e-03 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 46 | 243 | 1.30e-03 |
| GO:BP | GO:0003018 | vascular process in circulatory system | 39 | 194 | 1.31e-03 |
| GO:BP | GO:0048589 | developmental growth | 80 | 501 | 1.50e-03 |
| GO:BP | GO:0010033 | response to organic substance | 226 | 1736 | 1.50e-03 |
| GO:BP | GO:0014902 | myotube differentiation | 22 | 85 | 1.51e-03 |
| GO:BP | GO:0071260 | cellular response to mechanical stimulus | 17 | 57 | 1.68e-03 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 239 | 1856 | 1.76e-03 |
| GO:BP | GO:0007015 | actin filament organization | 61 | 357 | 1.77e-03 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 18 | 63 | 1.86e-03 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 116 | 796 | 1.86e-03 |
| GO:BP | GO:1902894 | negative regulation of miRNA transcription | 10 | 23 | 1.89e-03 |
| GO:BP | GO:0042092 | type 2 immune response | 9 | 19 | 2.02e-03 |
| GO:BP | GO:0014074 | response to purine-containing compound | 25 | 106 | 2.31e-03 |
| GO:BP | GO:0008360 | regulation of cell shape | 25 | 106 | 2.31e-03 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 181 | 1354 | 2.33e-03 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 38 | 193 | 2.44e-03 |
| GO:BP | GO:0036293 | response to decreased oxygen levels | 45 | 243 | 2.51e-03 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 236 | 1841 | 2.52e-03 |
| GO:BP | GO:0012501 | programmed cell death | 188 | 1419 | 2.69e-03 |
| GO:BP | GO:0009790 | embryo development | 118 | 821 | 2.76e-03 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 212 | 1633 | 2.99e-03 |
| GO:BP | GO:0034330 | cell junction organization | 84 | 545 | 3.09e-03 |
| GO:BP | GO:0008219 | cell death | 188 | 1423 | 3.09e-03 |
| GO:BP | GO:0009725 | response to hormone | 92 | 610 | 3.16e-03 |
| GO:BP | GO:0086005 | ventricular cardiac muscle cell action potential | 11 | 29 | 3.26e-03 |
| GO:BP | GO:0016525 | negative regulation of angiogenesis | 18 | 66 | 3.30e-03 |
| GO:BP | GO:0001666 | response to hypoxia | 43 | 232 | 3.37e-03 |
| GO:BP | GO:0035272 | exocrine system development | 12 | 34 | 3.45e-03 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 161 | 1193 | 3.63e-03 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 237 | 1862 | 3.63e-03 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 229 | 1791 | 3.67e-03 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 68 | 422 | 3.73e-03 |
| GO:BP | GO:0032879 | regulation of localization | 189 | 1439 | 3.87e-03 |
| GO:BP | GO:2000629 | negative regulation of miRNA metabolic process | 10 | 25 | 3.87e-03 |
| GO:BP | GO:1901343 | negative regulation of vasculature development | 18 | 67 | 3.87e-03 |
| GO:BP | GO:2000181 | negative regulation of blood vessel morphogenesis | 18 | 67 | 3.87e-03 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 135 | 973 | 3.94e-03 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 146 | 1067 | 3.95e-03 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 165 | 1231 | 3.96e-03 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 134 | 965 | 3.96e-03 |
| GO:BP | GO:0048699 | generation of neurons | 147 | 1076 | 3.96e-03 |
| GO:BP | GO:0051960 | regulation of nervous system development | 57 | 339 | 4.09e-03 |
| GO:BP | GO:0007162 | negative regulation of cell adhesion | 36 | 185 | 4.21e-03 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 72 | 456 | 4.22e-03 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 165 | 1234 | 4.34e-03 |
| GO:BP | GO:0086091 | regulation of heart rate by cardiac conduction | 12 | 35 | 4.34e-03 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 69 | 433 | 4.34e-03 |
| GO:BP | GO:0023057 | negative regulation of signaling | 142 | 1036 | 4.34e-03 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 156 | 1157 | 4.46e-03 |
| GO:BP | GO:0060284 | regulation of cell development | 86 | 570 | 4.67e-03 |
| GO:BP | GO:0051049 | regulation of transport | 149 | 1098 | 4.67e-03 |
| GO:BP | GO:0007399 | nervous system development | 226 | 1775 | 4.81e-03 |
| GO:BP | GO:0007267 | cell-cell signaling | 147 | 1082 | 4.88e-03 |
| GO:BP | GO:0032944 | regulation of mononuclear cell proliferation | 26 | 119 | 5.19e-03 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 20 | 81 | 5.26e-03 |
| GO:BP | GO:0030001 | metal ion transport | 80 | 524 | 5.27e-03 |
| GO:BP | GO:0090288 | negative regulation of cellular response to growth factor stimulus | 19 | 75 | 5.28e-03 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 50 | 290 | 5.28e-03 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 51 | 298 | 5.43e-03 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 47 | 268 | 5.43e-03 |
| GO:BP | GO:0043542 | endothelial cell migration | 34 | 174 | 5.43e-03 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 141 | 1034 | 5.43e-03 |
| GO:BP | GO:0038084 | vascular endothelial growth factor signaling pathway | 12 | 36 | 5.43e-03 |
| GO:BP | GO:0040007 | growth | 101 | 697 | 5.54e-03 |
| GO:BP | GO:0010927 | cellular component assembly involved in morphogenesis | 24 | 107 | 5.74e-03 |
| GO:BP | GO:0098915 | membrane repolarization during ventricular cardiac muscle cell action potential | 6 | 10 | 5.82e-03 |
| GO:BP | GO:1902287 | semaphorin-plexin signaling pathway involved in axon guidance | 6 | 10 | 5.82e-03 |
| GO:BP | GO:0036363 | transforming growth factor beta activation | 6 | 10 | 5.82e-03 |
| GO:BP | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance | 6 | 10 | 5.82e-03 |
| GO:BP | GO:0045064 | T-helper 2 cell differentiation | 6 | 10 | 5.82e-03 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 27 | 127 | 5.83e-03 |
| GO:BP | GO:0099625 | ventricular cardiac muscle cell membrane repolarization | 9 | 22 | 5.97e-03 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 261 | 2100 | 6.08e-03 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 51 | 300 | 6.08e-03 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 77 | 504 | 6.28e-03 |
| GO:BP | GO:0030182 | neuron differentiation | 138 | 1014 | 6.61e-03 |
| GO:BP | GO:1990573 | potassium ion import across plasma membrane | 10 | 27 | 6.76e-03 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 270 | 2186 | 6.85e-03 |
| GO:BP | GO:0048846 | axon extension involved in axon guidance | 11 | 32 | 6.94e-03 |
| GO:BP | GO:1902284 | neuron projection extension involved in neuron projection guidance | 11 | 32 | 6.94e-03 |
| GO:BP | GO:0098739 | import across plasma membrane | 26 | 122 | 7.02e-03 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 28 | 136 | 7.51e-03 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 45 | 258 | 7.63e-03 |
| GO:BP | GO:0086065 | cell communication involved in cardiac conduction | 15 | 54 | 7.78e-03 |
| GO:BP | GO:0006897 | endocytosis | 75 | 492 | 7.79e-03 |
| GO:BP | GO:0030510 | regulation of BMP signaling pathway | 19 | 78 | 8.05e-03 |
| GO:BP | GO:0070663 | regulation of leukocyte proliferation | 27 | 130 | 8.14e-03 |
| GO:BP | GO:0001817 | regulation of cytokine production | 68 | 437 | 8.20e-03 |
| GO:BP | GO:0030224 | monocyte differentiation | 9 | 23 | 8.26e-03 |
| GO:BP | GO:0002026 | regulation of the force of heart contraction | 9 | 23 | 8.26e-03 |
| GO:BP | GO:0045444 | fat cell differentiation | 34 | 179 | 8.34e-03 |
| GO:BP | GO:0048588 | developmental cell growth | 34 | 179 | 8.34e-03 |
| GO:BP | GO:0045668 | negative regulation of osteoblast differentiation | 12 | 38 | 8.48e-03 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 123 | 893 | 8.49e-03 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 110 | 783 | 8.49e-03 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 139 | 1031 | 8.77e-03 |
| GO:BP | GO:0048841 | regulation of axon extension involved in axon guidance | 10 | 28 | 8.79e-03 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 42 | 238 | 8.82e-03 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 241 | 1934 | 8.82e-03 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 67 | 431 | 8.90e-03 |
| GO:BP | GO:0055008 | cardiac muscle tissue morphogenesis | 15 | 55 | 9.02e-03 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 132 | 972 | 9.02e-03 |
| GO:BP | GO:0042592 | homeostatic process | 151 | 1137 | 9.32e-03 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 17 | 67 | 9.32e-03 |
| GO:BP | GO:0060976 | coronary vasculature development | 13 | 44 | 9.36e-03 |
| GO:BP | GO:0086002 | cardiac muscle cell action potential involved in contraction | 13 | 44 | 9.36e-03 |
| GO:BP | GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 48 | 284 | 9.36e-03 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 26 | 125 | 9.36e-03 |
| GO:BP | GO:0043501 | skeletal muscle adaptation | 8 | 19 | 9.36e-03 |
| GO:BP | GO:0055010 | ventricular cardiac muscle tissue morphogenesis | 13 | 44 | 9.36e-03 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 38 | 210 | 9.66e-03 |
| GO:BP | GO:0001816 | cytokine production | 68 | 441 | 9.72e-03 |
| GO:BP | GO:0014733 | regulation of skeletal muscle adaptation | 6 | 11 | 9.75e-03 |
| GO:BP | GO:1902307 | positive regulation of sodium ion transmembrane transport | 6 | 11 | 9.75e-03 |
| GO:BP | GO:0048010 | vascular endothelial growth factor receptor signaling pathway | 14 | 50 | 1.01e-02 |
| GO:BP | GO:0140747 | regulation of ncRNA transcription | 19 | 80 | 1.01e-02 |
| GO:BP | GO:0002828 | regulation of type 2 immune response | 7 | 15 | 1.02e-02 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 41 | 233 | 1.02e-02 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 50 | 301 | 1.03e-02 |
| GO:BP | GO:0003229 | ventricular cardiac muscle tissue development | 15 | 56 | 1.04e-02 |
| GO:BP | GO:0046683 | response to organophosphorus | 21 | 93 | 1.04e-02 |
| GO:BP | GO:0003208 | cardiac ventricle morphogenesis | 16 | 62 | 1.06e-02 |
| GO:BP | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling | 9 | 24 | 1.06e-02 |
| GO:BP | GO:0010586 | miRNA metabolic process | 20 | 87 | 1.10e-02 |
| GO:BP | GO:0007009 | plasma membrane organization | 23 | 107 | 1.18e-02 |
| GO:BP | GO:0014896 | muscle hypertrophy | 18 | 75 | 1.21e-02 |
| GO:BP | GO:0002376 | immune system process | 181 | 1410 | 1.23e-02 |
| GO:BP | GO:0048638 | regulation of developmental growth | 42 | 243 | 1.24e-02 |
| GO:BP | GO:0002286 | T cell activation involved in immune response | 15 | 57 | 1.24e-02 |
| GO:BP | GO:0016049 | cell growth | 60 | 382 | 1.24e-02 |
| GO:BP | GO:0050873 | brown fat cell differentiation | 12 | 40 | 1.25e-02 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 141 | 1060 | 1.25e-02 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 43 | 251 | 1.27e-02 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 43 | 251 | 1.27e-02 |
| GO:BP | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity | 5 | 8 | 1.30e-02 |
| GO:BP | GO:0141091 | transforming growth factor beta receptor superfamily signaling pathway | 44 | 259 | 1.31e-02 |
| GO:BP | GO:0006935 | chemotaxis | 42 | 244 | 1.31e-02 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 47 | 282 | 1.31e-02 |
| GO:BP | GO:0042330 | taxis | 42 | 244 | 1.31e-02 |
| GO:BP | GO:0006468 | protein phosphorylation | 129 | 958 | 1.31e-02 |
| GO:BP | GO:0035725 | sodium ion transmembrane transport | 21 | 95 | 1.32e-02 |
| GO:BP | GO:1901490 | regulation of lymphangiogenesis | 4 | 5 | 1.33e-02 |
| GO:BP | GO:0009987 | cellular process | 1129 | 10686 | 1.35e-02 |
| GO:BP | GO:0060485 | mesenchyme development | 42 | 245 | 1.38e-02 |
| GO:BP | GO:0099587 | inorganic ion import across plasma membrane | 17 | 70 | 1.38e-02 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 38 | 215 | 1.38e-02 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 119 | 874 | 1.38e-02 |
| GO:BP | GO:0098659 | inorganic cation import across plasma membrane | 17 | 70 | 1.38e-02 |
| GO:BP | GO:0003279 | cardiac septum development | 22 | 102 | 1.38e-02 |
| GO:BP | GO:0045321 | leukocyte activation | 78 | 530 | 1.39e-02 |
| GO:BP | GO:0030198 | extracellular matrix organization | 39 | 223 | 1.42e-02 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 74 | 498 | 1.44e-02 |
| GO:BP | GO:0001843 | neural tube closure | 19 | 83 | 1.45e-02 |
| GO:BP | GO:0050670 | regulation of lymphocyte proliferation | 24 | 116 | 1.45e-02 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 40 | 231 | 1.47e-02 |
| GO:BP | GO:0045165 | cell fate commitment | 29 | 151 | 1.51e-02 |
| GO:BP | GO:0043062 | extracellular structure organization | 39 | 224 | 1.52e-02 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 39 | 224 | 1.52e-02 |
| GO:BP | GO:0060419 | heart growth | 17 | 71 | 1.56e-02 |
| GO:BP | GO:0071407 | cellular response to organic cyclic compound | 59 | 379 | 1.56e-02 |
| GO:BP | GO:1903038 | negative regulation of leukocyte cell-cell adhesion | 17 | 71 | 1.56e-02 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 59 | 379 | 1.56e-02 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 38 | 217 | 1.56e-02 |
| GO:BP | GO:0070664 | negative regulation of leukocyte proliferation | 13 | 47 | 1.56e-02 |
| GO:BP | GO:0048662 | negative regulation of smooth muscle cell proliferation | 11 | 36 | 1.61e-02 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 16 | 65 | 1.61e-02 |
| GO:BP | GO:0035148 | tube formation | 25 | 124 | 1.62e-02 |
| GO:BP | GO:0061564 | axon development | 58 | 372 | 1.62e-02 |
| GO:BP | GO:0060606 | tube closure | 19 | 84 | 1.62e-02 |
| GO:BP | GO:0014912 | negative regulation of smooth muscle cell migration | 8 | 21 | 1.70e-02 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 190 | 1504 | 1.71e-02 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 73 | 494 | 1.72e-02 |
| GO:BP | GO:0040008 | regulation of growth | 69 | 462 | 1.76e-02 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 191 | 1514 | 1.76e-02 |
| GO:BP | GO:0031333 | negative regulation of protein-containing complex assembly | 24 | 118 | 1.76e-02 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 26 | 132 | 1.78e-02 |
| GO:BP | GO:0010721 | negative regulation of cell development | 33 | 182 | 1.78e-02 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 38 | 219 | 1.78e-02 |
| GO:BP | GO:0014812 | muscle cell migration | 17 | 72 | 1.78e-02 |
| GO:BP | GO:0002040 | sprouting angiogenesis | 21 | 98 | 1.79e-02 |
| GO:BP | GO:0001819 | positive regulation of cytokine production | 43 | 257 | 1.80e-02 |
| GO:BP | GO:0001570 | vasculogenesis | 16 | 66 | 1.83e-02 |
| GO:BP | GO:0035710 | CD4-positive, alpha-beta T cell activation | 16 | 66 | 1.83e-02 |
| GO:BP | GO:0051058 | negative regulation of small GTPase mediated signal transduction | 13 | 48 | 1.83e-02 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 22 | 105 | 1.84e-02 |
| GO:BP | GO:0035924 | cellular response to vascular endothelial growth factor stimulus | 14 | 54 | 1.85e-02 |
| GO:BP | GO:0046323 | glucose import | 14 | 54 | 1.85e-02 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 34 | 190 | 1.85e-02 |
| GO:BP | GO:0032869 | cellular response to insulin stimulus | 30 | 161 | 1.86e-02 |
| GO:BP | GO:1902305 | regulation of sodium ion transmembrane transport | 11 | 37 | 1.93e-02 |
| GO:BP | GO:0021915 | neural tube development | 26 | 133 | 1.93e-02 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 17 | 73 | 2.00e-02 |
| GO:BP | GO:0030903 | notochord development | 7 | 17 | 2.00e-02 |
| GO:BP | GO:0010954 | positive regulation of protein processing | 7 | 17 | 2.00e-02 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 30 | 162 | 2.03e-02 |
| GO:BP | GO:0042129 | regulation of T cell proliferation | 19 | 86 | 2.03e-02 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 86 | 606 | 2.03e-02 |
| GO:BP | GO:0007409 | axonogenesis | 53 | 337 | 2.03e-02 |
| GO:BP | GO:0002293 | alpha-beta T cell differentiation involved in immune response | 12 | 43 | 2.04e-02 |
| GO:BP | GO:0002287 | alpha-beta T cell activation involved in immune response | 12 | 43 | 2.04e-02 |
| GO:BP | GO:0042093 | T-helper cell differentiation | 12 | 43 | 2.04e-02 |
| GO:BP | GO:0043086 | negative regulation of catalytic activity | 48 | 298 | 2.04e-02 |
| GO:BP | GO:0045214 | sarcomere organization | 12 | 43 | 2.04e-02 |
| GO:BP | GO:0002294 | CD4-positive, alpha-beta T cell differentiation involved in immune response | 12 | 43 | 2.04e-02 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 16 | 67 | 2.04e-02 |
| GO:BP | GO:0071805 | potassium ion transmembrane transport | 24 | 120 | 2.06e-02 |
| GO:BP | GO:0043434 | response to peptide hormone | 50 | 314 | 2.07e-02 |
| GO:BP | GO:0035265 | organ growth | 25 | 127 | 2.07e-02 |
| GO:BP | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity | 5 | 9 | 2.12e-02 |
| GO:BP | GO:0007405 | neuroblast proliferation | 14 | 55 | 2.12e-02 |
| GO:BP | GO:1901388 | regulation of transforming growth factor beta activation | 5 | 9 | 2.12e-02 |
| GO:BP | GO:0032673 | regulation of interleukin-4 production | 6 | 13 | 2.19e-02 |
| GO:BP | GO:1903729 | regulation of plasma membrane organization | 6 | 13 | 2.19e-02 |
| GO:BP | GO:0032633 | interleukin-4 production | 6 | 13 | 2.19e-02 |
| GO:BP | GO:0048666 | neuron development | 113 | 838 | 2.21e-02 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 17 | 74 | 2.22e-02 |
| GO:BP | GO:0014020 | primary neural tube formation | 19 | 87 | 2.22e-02 |
| GO:BP | GO:0006811 | monoatomic ion transport | 99 | 719 | 2.24e-02 |
| GO:BP | GO:1901491 | negative regulation of lymphangiogenesis | 3 | 3 | 2.24e-02 |
| GO:BP | GO:0086047 | membrane depolarization during Purkinje myocyte cell action potential | 3 | 3 | 2.24e-02 |
| GO:BP | GO:0045760 | positive regulation of action potential | 3 | 3 | 2.24e-02 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 31 | 171 | 2.24e-02 |
| GO:BP | GO:0006949 | syncytium formation | 11 | 38 | 2.25e-02 |
| GO:BP | GO:0043502 | regulation of muscle adaptation | 16 | 68 | 2.30e-02 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 54 | 348 | 2.32e-02 |
| GO:BP | GO:0001775 | cell activation | 88 | 627 | 2.32e-02 |
| GO:BP | GO:0032943 | mononuclear cell proliferation | 29 | 157 | 2.34e-02 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 40 | 239 | 2.35e-02 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 87 | 619 | 2.35e-02 |
| GO:BP | GO:0048878 | chemical homeostasis | 93 | 670 | 2.39e-02 |
| GO:BP | GO:0097485 | neuron projection guidance | 31 | 172 | 2.41e-02 |
| GO:BP | GO:0007411 | axon guidance | 31 | 172 | 2.41e-02 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 23 | 115 | 2.42e-02 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 23 | 115 | 2.42e-02 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 72 | 495 | 2.42e-02 |
| GO:BP | GO:0061387 | regulation of extent of cell growth | 19 | 88 | 2.43e-02 |
| GO:BP | GO:0046631 | alpha-beta T cell activation | 19 | 88 | 2.43e-02 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 20 | 95 | 2.54e-02 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 36 | 210 | 2.54e-02 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 75 | 521 | 2.54e-02 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 103 | 757 | 2.54e-02 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 36 | 210 | 2.54e-02 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 46 | 287 | 2.57e-02 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 70 | 480 | 2.57e-02 |
| GO:BP | GO:0003281 | ventricular septum development | 16 | 69 | 2.57e-02 |
| GO:BP | GO:0001508 | action potential | 21 | 102 | 2.57e-02 |
| GO:BP | GO:1903319 | positive regulation of protein maturation | 7 | 18 | 2.59e-02 |
| GO:BP | GO:0006814 | sodium ion transport | 26 | 137 | 2.60e-02 |
| GO:BP | GO:0032868 | response to insulin | 35 | 203 | 2.61e-02 |
| GO:BP | GO:0009266 | response to temperature stimulus | 23 | 116 | 2.62e-02 |
| GO:BP | GO:0006812 | monoatomic cation transport | 86 | 614 | 2.63e-02 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 148 | 1150 | 2.63e-02 |
| GO:BP | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis | 4 | 6 | 2.66e-02 |
| GO:BP | GO:0048843 | negative regulation of axon extension involved in axon guidance | 8 | 23 | 2.67e-02 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 237 | 1951 | 2.67e-02 |
| GO:BP | GO:0006950 | response to stress | 301 | 2540 | 2.67e-02 |
| GO:BP | GO:0010765 | positive regulation of sodium ion transport | 8 | 23 | 2.67e-02 |
| GO:BP | GO:0009612 | response to mechanical stimulus | 28 | 152 | 2.71e-02 |
| GO:BP | GO:0002573 | myeloid leukocyte differentiation | 28 | 152 | 2.71e-02 |
| GO:BP | GO:0002292 | T cell differentiation involved in immune response | 12 | 45 | 2.72e-02 |
| GO:BP | GO:0051346 | negative regulation of hydrolase activity | 23 | 117 | 2.87e-02 |
| GO:BP | GO:0072175 | epithelial tube formation | 23 | 117 | 2.87e-02 |
| GO:BP | GO:1903037 | regulation of leukocyte cell-cell adhesion | 34 | 197 | 2.88e-02 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 89 | 642 | 2.90e-02 |
| GO:BP | GO:0009991 | response to extracellular stimulus | 57 | 377 | 2.90e-02 |
| GO:BP | GO:0016310 | phosphorylation | 147 | 1145 | 2.92e-02 |
| GO:BP | GO:0043114 | regulation of vascular permeability | 10 | 34 | 2.94e-02 |
| GO:BP | GO:0070661 | leukocyte proliferation | 31 | 175 | 2.94e-02 |
| GO:BP | GO:0050868 | negative regulation of T cell activation | 15 | 64 | 3.02e-02 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 15 | 64 | 3.02e-02 |
| GO:BP | GO:0010885 | regulation of cholesterol storage | 6 | 14 | 3.03e-02 |
| GO:BP | GO:0050996 | positive regulation of lipid catabolic process | 6 | 14 | 3.03e-02 |
| GO:BP | GO:0031667 | response to nutrient levels | 54 | 354 | 3.06e-02 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 32 | 183 | 3.06e-02 |
| GO:BP | GO:0046324 | regulation of glucose import | 11 | 40 | 3.11e-02 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 23 | 118 | 3.11e-02 |
| GO:BP | GO:0002028 | regulation of sodium ion transport | 14 | 58 | 3.11e-02 |
| GO:BP | GO:0071772 | response to BMP | 23 | 118 | 3.11e-02 |
| GO:BP | GO:0002521 | leukocyte differentiation | 56 | 371 | 3.19e-02 |
| GO:BP | GO:0001501 | skeletal system development | 53 | 347 | 3.21e-02 |
| GO:BP | GO:0019222 | regulation of metabolic process | 518 | 4603 | 3.21e-02 |
| GO:BP | GO:0033275 | actin-myosin filament sliding | 5 | 10 | 3.22e-02 |
| GO:BP | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis | 5 | 10 | 3.22e-02 |
| GO:BP | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway | 5 | 10 | 3.22e-02 |
| GO:BP | GO:0010832 | negative regulation of myotube differentiation | 5 | 10 | 3.22e-02 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 30 | 169 | 3.22e-02 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 218 | 1787 | 3.22e-02 |
| GO:BP | GO:0045926 | negative regulation of growth | 32 | 184 | 3.23e-02 |
| GO:BP | GO:0055085 | transmembrane transport | 121 | 921 | 3.23e-02 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 32 | 184 | 3.23e-02 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 82 | 587 | 3.31e-02 |
| GO:BP | GO:0060429 | epithelium development | 106 | 792 | 3.31e-02 |
| GO:BP | GO:0086010 | membrane depolarization during action potential | 8 | 24 | 3.31e-02 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 42 | 261 | 3.31e-02 |
| GO:BP | GO:0014887 | cardiac muscle adaptation | 8 | 24 | 3.31e-02 |
| GO:BP | GO:0071398 | cellular response to fatty acid | 8 | 24 | 3.31e-02 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 42 | 261 | 3.31e-02 |
| GO:BP | GO:0014898 | cardiac muscle hypertrophy in response to stress | 8 | 24 | 3.31e-02 |
| GO:BP | GO:0003299 | muscle hypertrophy in response to stress | 8 | 24 | 3.31e-02 |
| GO:BP | GO:0055006 | cardiac cell development | 17 | 78 | 3.33e-02 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 27 | 148 | 3.43e-02 |
| GO:BP | GO:0070228 | regulation of lymphocyte apoptotic process | 10 | 35 | 3.46e-02 |
| GO:BP | GO:0050994 | regulation of lipid catabolic process | 10 | 35 | 3.46e-02 |
| GO:BP | GO:0060840 | artery development | 18 | 85 | 3.47e-02 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 18 | 85 | 3.47e-02 |
| GO:BP | GO:0006816 | calcium ion transport | 42 | 262 | 3.51e-02 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 228 | 1883 | 3.52e-02 |
| GO:BP | GO:0001657 | ureteric bud development | 16 | 72 | 3.53e-02 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 44 | 278 | 3.53e-02 |
| GO:BP | GO:0001841 | neural tube formation | 19 | 92 | 3.53e-02 |
| GO:BP | GO:0098609 | cell-cell adhesion | 79 | 564 | 3.53e-02 |
| GO:BP | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 19 | 92 | 3.53e-02 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 16 | 72 | 3.53e-02 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 44 | 278 | 3.53e-02 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 21 | 106 | 3.62e-02 |
| GO:BP | GO:0030097 | hemopoiesis | 86 | 624 | 3.64e-02 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 140 | 1093 | 3.68e-02 |
| GO:BP | GO:0002761 | regulation of myeloid leukocyte differentiation | 17 | 79 | 3.70e-02 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 230 | 1904 | 3.70e-02 |
| GO:BP | GO:0018212 | peptidyl-tyrosine modification | 29 | 164 | 3.74e-02 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 154 | 1218 | 3.75e-02 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 50 | 327 | 3.80e-02 |
| GO:BP | GO:0014904 | myotube cell development | 9 | 30 | 3.80e-02 |
| GO:BP | GO:0006813 | potassium ion transport | 25 | 135 | 3.81e-02 |
| GO:BP | GO:0014909 | smooth muscle cell migration | 14 | 60 | 3.94e-02 |
| GO:BP | GO:0072164 | mesonephric tubule development | 16 | 73 | 3.97e-02 |
| GO:BP | GO:0072163 | mesonephric epithelium development | 16 | 73 | 3.97e-02 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 36 | 218 | 4.06e-02 |
| GO:BP | GO:0023056 | positive regulation of signaling | 154 | 1221 | 4.07e-02 |
| GO:BP | GO:0071320 | cellular response to cAMP | 10 | 36 | 4.09e-02 |
| GO:BP | GO:0042475 | odontogenesis of dentin-containing tooth | 13 | 54 | 4.09e-02 |
| GO:BP | GO:0085029 | extracellular matrix assembly | 10 | 36 | 4.09e-02 |
| GO:BP | GO:0042130 | negative regulation of T cell proliferation | 10 | 36 | 4.09e-02 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 55 | 369 | 4.10e-02 |
| GO:BP | GO:0007435 | salivary gland morphogenesis | 8 | 25 | 4.11e-02 |
| GO:BP | GO:0009410 | response to xenobiotic stimulus | 43 | 273 | 4.13e-02 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 32 | 188 | 4.15e-02 |
| GO:BP | GO:0010718 | positive regulation of epithelial to mesenchymal transition | 12 | 48 | 4.16e-02 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 63 | 435 | 4.17e-02 |
| GO:BP | GO:0086009 | membrane repolarization | 11 | 42 | 4.17e-02 |
| GO:BP | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity | 7 | 20 | 4.23e-02 |
| GO:BP | GO:0046651 | lymphocyte proliferation | 27 | 151 | 4.23e-02 |
| GO:BP | GO:1902547 | regulation of cellular response to vascular endothelial growth factor stimulus | 7 | 20 | 4.23e-02 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 23 | 122 | 4.23e-02 |
| GO:BP | GO:0030308 | negative regulation of cell growth | 26 | 144 | 4.35e-02 |
| GO:BP | GO:0042391 | regulation of membrane potential | 44 | 282 | 4.37e-02 |
| GO:BP | GO:0001823 | mesonephros development | 16 | 74 | 4.41e-02 |
| GO:BP | GO:0010644 | cell communication by electrical coupling | 9 | 31 | 4.51e-02 |
| GO:BP | GO:0045657 | positive regulation of monocyte differentiation | 4 | 7 | 4.51e-02 |
| GO:BP | GO:0044351 | macropinocytosis | 4 | 7 | 4.51e-02 |
| GO:BP | GO:0042060 | wound healing | 45 | 291 | 4.51e-02 |
| GO:BP | GO:1900373 | positive regulation of purine nucleotide biosynthetic process | 4 | 7 | 4.51e-02 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 23 | 123 | 4.51e-02 |
| GO:BP | GO:0010830 | regulation of myotube differentiation | 9 | 31 | 4.51e-02 |
| GO:BP | GO:1903351 | cellular response to dopamine | 9 | 31 | 4.51e-02 |
| GO:BP | GO:0002830 | positive regulation of type 2 immune response | 4 | 7 | 4.51e-02 |
| GO:BP | GO:0032753 | positive regulation of interleukin-4 production | 4 | 7 | 4.51e-02 |
| GO:BP | GO:0001558 | regulation of cell growth | 49 | 323 | 4.51e-02 |
| GO:BP | GO:0014883 | transition between fast and slow fiber | 4 | 7 | 4.51e-02 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 68 | 479 | 4.51e-02 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 53 | 355 | 4.51e-02 |
| GO:BP | GO:0070296 | sarcoplasmic reticulum calcium ion transport | 9 | 31 | 4.51e-02 |
| GO:BP | GO:0045628 | regulation of T-helper 2 cell differentiation | 4 | 7 | 4.51e-02 |
| GO:BP | GO:0030810 | positive regulation of nucleotide biosynthetic process | 4 | 7 | 4.51e-02 |
| GO:BP | GO:0071634 | regulation of transforming growth factor beta production | 9 | 31 | 4.51e-02 |
| GO:BP | GO:0008150 | biological_process | 1175 | 11244 | 4.51e-02 |
| GO:BP | GO:0099622 | cardiac muscle cell membrane repolarization | 9 | 31 | 4.51e-02 |
| GO:BP | GO:0030030 | cell projection organization | 149 | 1182 | 4.51e-02 |
| GO:BP | GO:0090189 | regulation of branching involved in ureteric bud morphogenesis | 5 | 11 | 4.52e-02 |
| GO:BP | GO:0021783 | preganglionic parasympathetic fiber development | 5 | 11 | 4.52e-02 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 18 | 88 | 4.53e-02 |
| GO:BP | GO:0031175 | neuron projection development | 99 | 743 | 4.54e-02 |
| GO:BP | GO:0150104 | transport across blood-brain barrier | 15 | 68 | 4.56e-02 |
| GO:BP | GO:0009894 | regulation of catabolic process | 109 | 830 | 4.56e-02 |
| GO:BP | GO:0010232 | vascular transport | 15 | 68 | 4.56e-02 |
| GO:BP | GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | 25 | 138 | 4.67e-02 |
| GO:BP | GO:0050672 | negative regulation of lymphocyte proliferation | 11 | 43 | 4.72e-02 |
| GO:BP | GO:0032945 | negative regulation of mononuclear cell proliferation | 11 | 43 | 4.72e-02 |
| GO:BP | GO:0051591 | response to cAMP | 14 | 62 | 4.90e-02 |
| GO:BP | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 8 | 26 | 4.94e-02 |
| GO:BP | GO:0007431 | salivary gland development | 8 | 26 | 4.94e-02 |
| GO:BP | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction | 8 | 26 | 4.94e-02 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 476 | 4233 | 4.98e-02 |
ESRnomtable_20k_enh %>%
filter(source=="KEGG", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "KEGG terms found in ESR enhancer neargenes_20k") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 37 | 124 | 3.15e-07 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 37 | 129 | 5.30e-07 |
| KEGG | KEGG:04925 | Aldosterone synthesis and secretion | 25 | 75 | 4.82e-06 |
| KEGG | KEGG:05414 | Dilated cardiomyopathy | 26 | 82 | 6.21e-06 |
| KEGG | KEGG:05200 | Pathways in cancer | 73 | 391 | 1.30e-05 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 35 | 147 | 6.89e-05 |
| KEGG | KEGG:05410 | Hypertrophic cardiomyopathy | 22 | 77 | 2.45e-04 |
| KEGG | KEGG:04976 | Bile secretion | 15 | 41 | 2.45e-04 |
| KEGG | KEGG:04931 | Insulin resistance | 24 | 91 | 3.33e-04 |
| KEGG | KEGG:04921 | Oxytocin signaling pathway | 27 | 113 | 6.15e-04 |
| KEGG | KEGG:04020 | Calcium signaling pathway | 34 | 161 | 7.81e-04 |
| KEGG | KEGG:04360 | Axon guidance | 33 | 155 | 7.81e-04 |
| KEGG | KEGG:04927 | Cortisol synthesis and secretion | 15 | 46 | 7.81e-04 |
| KEGG | KEGG:04725 | Cholinergic synapse | 21 | 82 | 1.14e-03 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 24 | 100 | 1.14e-03 |
| KEGG | KEGG:05203 | Viral carcinogenesis | 30 | 142 | 1.66e-03 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 33 | 164 | 1.93e-03 |
| KEGG | KEGG:04911 | Insulin secretion | 17 | 62 | 1.94e-03 |
| KEGG | KEGG:04971 | Gastric acid secretion | 15 | 51 | 1.95e-03 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 19 | 78 | 3.91e-03 |
| KEGG | KEGG:04934 | Cushing syndrome | 25 | 119 | 5.26e-03 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 20 | 87 | 5.63e-03 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 31 | 162 | 5.79e-03 |
| KEGG | KEGG:04260 | Cardiac muscle contraction | 16 | 63 | 5.98e-03 |
| KEGG | KEGG:04740 | Olfactory transduction | 9 | 25 | 6.49e-03 |
| KEGG | KEGG:04014 | Ras signaling pathway | 31 | 164 | 6.49e-03 |
| KEGG | KEGG:03320 | PPAR signaling pathway | 13 | 47 | 7.20e-03 |
| KEGG | KEGG:04915 | Estrogen signaling pathway | 20 | 90 | 7.20e-03 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 22 | 104 | 7.64e-03 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 30 | 160 | 7.77e-03 |
| KEGG | KEGG:04066 | HIF-1 signaling pathway | 19 | 86 | 9.32e-03 |
| KEGG | KEGG:05030 | Cocaine addiction | 10 | 32 | 9.39e-03 |
| KEGG | KEGG:04713 | Circadian entrainment | 16 | 68 | 1.07e-02 |
| KEGG | KEGG:04520 | Adherens junction | 18 | 83 | 1.44e-02 |
| KEGG | KEGG:04728 | Dopaminergic synapse | 20 | 97 | 1.51e-02 |
| KEGG | KEGG:04922 | Glucagon signaling pathway | 17 | 78 | 1.71e-02 |
| KEGG | KEGG:04923 | Regulation of lipolysis in adipocytes | 11 | 41 | 1.75e-02 |
| KEGG | KEGG:04814 | Motor proteins | 26 | 141 | 1.75e-02 |
| KEGG | KEGG:04380 | Osteoclast differentiation | 19 | 92 | 1.75e-02 |
| KEGG | KEGG:00000 | KEGG root term | 604 | 5451 | 1.96e-02 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 40 | 250 | 2.10e-02 |
| KEGG | KEGG:04916 | Melanogenesis | 16 | 75 | 2.48e-02 |
| KEGG | KEGG:04913 | Ovarian steroidogenesis | 8 | 26 | 2.51e-02 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 19 | 96 | 2.53e-02 |
| KEGG | KEGG:04670 | Leukocyte transendothelial migration | 16 | 76 | 2.66e-02 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 24 | 133 | 2.76e-02 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 20 | 105 | 2.98e-02 |
| KEGG | KEGG:05031 | Amphetamine addiction | 11 | 45 | 3.12e-02 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 21 | 113 | 3.13e-02 |
| KEGG | KEGG:04750 | Inflammatory mediator regulation of TRP channels | 15 | 72 | 3.48e-02 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 25 | 144 | 3.51e-02 |
| KEGG | KEGG:04961 | Endocrine and other factor-regulated calcium reabsorption | 10 | 40 | 3.57e-02 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 13 | 60 | 4.01e-02 |
| KEGG | KEGG:05034 | Alcoholism | 19 | 102 | 4.02e-02 |
| KEGG | KEGG:04510 | Focal adhesion | 28 | 169 | 4.02e-02 |
| KEGG | KEGG:04970 | Salivary secretion | 12 | 54 | 4.08e-02 |
| KEGG | KEGG:05032 | Morphine addiction | 12 | 54 | 4.08e-02 |
| KEGG | KEGG:05211 | Renal cell carcinoma | 13 | 62 | 4.79e-02 |
| KEGG | KEGG:05146 | Amoebiasis | 13 | 62 | 4.79e-02 |
| KEGG | KEGG:04721 | Synaptic vesicle cycle | 11 | 49 | 4.91e-02 |
LR 20,000 bp NG
# LR_resgenes_20k <- gost(query = LR_peak_list_20k$ENTREZID,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = FALSE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = background_df$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(LR_resgenes_20k,"data/GO_KEGG_analysis/LR_resgenes_20k.RDS")
# LRnom <- gostplot(LR_resgenes_20k, capped = FALSE, interactive = TRUE)
# LRnom
LRnomtable_20k <- LR_resgenes_20k$result %>%
dplyr::select(c(source, term_id,
term_name,intersection_size,
term_size, p_value))# %>%
LRnomtable_20k %>%
filter(source=="GO:BP", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(.,caption = "GO:BP terms found in LR nESRgenes_20k" ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0032501 | multicellular organismal process | 3247 | 4504 | 2.82e-47 |
| GO:BP | GO:0048856 | anatomical structure development | 2795 | 3867 | 2.23e-39 |
| GO:BP | GO:0048731 | system development | 2022 | 2719 | 3.44e-38 |
| GO:BP | GO:0007275 | multicellular organism development | 2324 | 3170 | 7.17e-38 |
| GO:BP | GO:0032502 | developmental process | 2991 | 4179 | 1.92e-37 |
| GO:BP | GO:0007154 | cell communication | 2854 | 3983 | 1.21e-35 |
| GO:BP | GO:0023052 | signaling | 2810 | 3918 | 1.94e-35 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 1401 | 1844 | 6.31e-32 |
| GO:BP | GO:0007165 | signal transduction | 2587 | 3611 | 6.46e-31 |
| GO:BP | GO:0050896 | response to stimulus | 3737 | 5379 | 2.12e-30 |
| GO:BP | GO:0048869 | cellular developmental process | 2041 | 2804 | 6.29e-29 |
| GO:BP | GO:0030154 | cell differentiation | 2040 | 2803 | 6.90e-29 |
| GO:BP | GO:0051716 | cellular response to stimulus | 3243 | 4640 | 2.39e-27 |
| GO:BP | GO:0007399 | nervous system development | 1335 | 1775 | 8.03e-27 |
| GO:BP | GO:0022008 | neurogenesis | 963 | 1251 | 4.06e-24 |
| GO:BP | GO:0010646 | regulation of cell communication | 1772 | 2444 | 9.29e-23 |
| GO:BP | GO:0048699 | generation of neurons | 834 | 1076 | 3.77e-22 |
| GO:BP | GO:0023051 | regulation of signaling | 1765 | 2438 | 3.77e-22 |
| GO:BP | GO:0048513 | animal organ development | 1472 | 2004 | 4.86e-22 |
| GO:BP | GO:0050793 | regulation of developmental process | 1263 | 1697 | 5.10e-22 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 1416 | 1924 | 1.17e-21 |
| GO:BP | GO:0048468 | cell development | 1386 | 1886 | 1.13e-20 |
| GO:BP | GO:0030182 | neuron differentiation | 785 | 1014 | 1.66e-20 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 1327 | 1801 | 2.11e-20 |
| GO:BP | GO:0050789 | regulation of biological process | 5035 | 7516 | 3.13e-20 |
| GO:BP | GO:0031175 | neuron projection development | 592 | 743 | 3.28e-20 |
| GO:BP | GO:0007155 | cell adhesion | 732 | 941 | 4.87e-20 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 1940 | 2717 | 5.30e-20 |
| GO:BP | GO:0048666 | neuron development | 656 | 838 | 5.56e-19 |
| GO:BP | GO:0000902 | cell morphogenesis | 567 | 713 | 5.79e-19 |
| GO:BP | GO:0065007 | biological regulation | 5159 | 7734 | 2.58e-18 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 872 | 1150 | 2.65e-18 |
| GO:BP | GO:0030030 | cell projection organization | 894 | 1182 | 2.85e-18 |
| GO:BP | GO:0003008 | system process | 864 | 1139 | 3.17e-18 |
| GO:BP | GO:0032989 | cellular anatomical entity morphogenesis | 475 | 588 | 3.28e-18 |
| GO:BP | GO:0050794 | regulation of cellular process | 4791 | 7153 | 6.38e-18 |
| GO:BP | GO:0009966 | regulation of signal transduction | 1557 | 2166 | 4.58e-17 |
| GO:BP | GO:0016477 | cell migration | 770 | 1011 | 7.96e-17 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 406 | 498 | 9.87e-17 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 392 | 479 | 1.11e-16 |
| GO:BP | GO:0048518 | positive regulation of biological process | 2959 | 4301 | 1.11e-16 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 402 | 494 | 2.61e-16 |
| GO:BP | GO:0048870 | cell motility | 843 | 1120 | 3.51e-16 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 2728 | 3954 | 7.52e-16 |
| GO:BP | GO:0007267 | cell-cell signaling | 815 | 1082 | 9.81e-16 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 357 | 435 | 1.66e-15 |
| GO:BP | GO:0065008 | regulation of biological quality | 1364 | 1893 | 3.54e-15 |
| GO:BP | GO:0009888 | tissue development | 993 | 1348 | 1.17e-14 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 969 | 1314 | 1.78e-14 |
| GO:BP | GO:0034330 | cell junction organization | 433 | 545 | 3.80e-14 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 691 | 913 | 6.65e-14 |
| GO:BP | GO:0040012 | regulation of locomotion | 556 | 720 | 8.00e-14 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 403 | 505 | 1.10e-13 |
| GO:BP | GO:0040011 | locomotion | 615 | 805 | 1.10e-13 |
| GO:BP | GO:0072359 | circulatory system development | 656 | 864 | 1.12e-13 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 294 | 355 | 1.12e-13 |
| GO:BP | GO:2000145 | regulation of cell motility | 541 | 700 | 1.42e-13 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 409 | 516 | 4.84e-13 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 551 | 717 | 5.29e-13 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 520 | 673 | 5.64e-13 |
| GO:BP | GO:0061564 | axon development | 304 | 372 | 1.22e-12 |
| GO:BP | GO:0001568 | blood vessel development | 414 | 525 | 1.43e-12 |
| GO:BP | GO:0030334 | regulation of cell migration | 515 | 668 | 1.53e-12 |
| GO:BP | GO:0007409 | axonogenesis | 278 | 337 | 1.71e-12 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 473 | 609 | 2.07e-12 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 794 | 1073 | 3.38e-12 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 631 | 837 | 4.65e-12 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 770 | 1040 | 7.02e-12 |
| GO:BP | GO:0098657 | import into cell | 480 | 622 | 9.09e-12 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 708 | 951 | 1.15e-11 |
| GO:BP | GO:0035556 | intracellular signal transduction | 1383 | 1951 | 1.15e-11 |
| GO:BP | GO:0001944 | vasculature development | 430 | 552 | 1.40e-11 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 1323 | 1862 | 1.45e-11 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 764 | 1035 | 2.55e-11 |
| GO:BP | GO:0035295 | tube development | 593 | 787 | 3.20e-11 |
| GO:BP | GO:0003013 | circulatory system process | 339 | 426 | 3.72e-11 |
| GO:BP | GO:0098609 | cell-cell adhesion | 437 | 564 | 3.81e-11 |
| GO:BP | GO:0030029 | actin filament-based process | 490 | 640 | 4.81e-11 |
| GO:BP | GO:0044057 | regulation of system process | 308 | 384 | 7.16e-11 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 197 | 233 | 1.09e-10 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 454 | 591 | 1.44e-10 |
| GO:BP | GO:0048519 | negative regulation of biological process | 2565 | 3764 | 1.53e-10 |
| GO:BP | GO:0006897 | endocytosis | 384 | 492 | 1.68e-10 |
| GO:BP | GO:0035239 | tube morphogenesis | 486 | 638 | 2.45e-10 |
| GO:BP | GO:0006810 | transport | 2054 | 2984 | 3.55e-10 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 474 | 622 | 4.11e-10 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 868 | 1197 | 4.54e-10 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 2422 | 3550 | 4.64e-10 |
| GO:BP | GO:0032879 | regulation of localization | 1031 | 1439 | 4.65e-10 |
| GO:BP | GO:0016043 | cellular component organization | 3229 | 4802 | 7.98e-10 |
| GO:BP | GO:0051179 | localization | 2537 | 3731 | 9.27e-10 |
| GO:BP | GO:0051049 | regulation of transport | 798 | 1098 | 1.70e-09 |
| GO:BP | GO:0007264 | small GTPase-mediated signal transduction | 308 | 390 | 2.21e-09 |
| GO:BP | GO:0048589 | developmental growth | 386 | 501 | 3.35e-09 |
| GO:BP | GO:0030001 | metal ion transport | 402 | 524 | 3.65e-09 |
| GO:BP | GO:0009987 | cellular process | 6915 | 10686 | 5.43e-09 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 349 | 450 | 6.96e-09 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 430 | 566 | 8.49e-09 |
| GO:BP | GO:0023056 | positive regulation of signaling | 877 | 1221 | 9.65e-09 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 875 | 1218 | 9.65e-09 |
| GO:BP | GO:0042221 | response to chemical | 1596 | 2304 | 1.28e-08 |
| GO:BP | GO:0008015 | blood circulation | 282 | 357 | 1.41e-08 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 624 | 850 | 2.01e-08 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 388 | 508 | 2.12e-08 |
| GO:BP | GO:0008150 | biological_process | 7248 | 11244 | 2.12e-08 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 351 | 456 | 3.01e-08 |
| GO:BP | GO:0051234 | establishment of localization | 2205 | 3240 | 3.03e-08 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 1062 | 1504 | 5.39e-08 |
| GO:BP | GO:0007010 | cytoskeleton organization | 822 | 1146 | 5.91e-08 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 473 | 633 | 5.91e-08 |
| GO:BP | GO:0071417 | cellular response to organonitrogen compound | 340 | 442 | 6.37e-08 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 347 | 452 | 6.39e-08 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 81 | 88 | 7.39e-08 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 1225 | 1751 | 7.72e-08 |
| GO:BP | GO:0034329 | cell junction assembly | 255 | 323 | 1.10e-07 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 824 | 1152 | 1.30e-07 |
| GO:BP | GO:0007417 | central nervous system development | 513 | 694 | 1.59e-07 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 356 | 467 | 1.62e-07 |
| GO:BP | GO:0099536 | synaptic signaling | 367 | 483 | 1.79e-07 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 439 | 587 | 1.92e-07 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 254 | 323 | 2.29e-07 |
| GO:BP | GO:0006811 | monoatomic ion transport | 529 | 719 | 2.71e-07 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 253 | 322 | 2.81e-07 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 782 | 1093 | 3.16e-07 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 352 | 463 | 3.27e-07 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 352 | 463 | 3.27e-07 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 392 | 521 | 3.95e-07 |
| GO:BP | GO:0016310 | phosphorylation | 816 | 1145 | 4.44e-07 |
| GO:BP | GO:0051960 | regulation of nervous system development | 264 | 339 | 5.96e-07 |
| GO:BP | GO:0071310 | cellular response to organic substance | 916 | 1297 | 8.30e-07 |
| GO:BP | GO:0042391 | regulation of membrane potential | 223 | 282 | 8.39e-07 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 312 | 408 | 8.56e-07 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 108 | 125 | 8.81e-07 |
| GO:BP | GO:0050808 | synapse organization | 262 | 337 | 8.81e-07 |
| GO:BP | GO:0001501 | skeletal system development | 269 | 347 | 9.14e-07 |
| GO:BP | GO:0001503 | ossification | 237 | 302 | 9.90e-07 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 910 | 1289 | 1.00e-06 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 361 | 479 | 1.10e-06 |
| GO:BP | GO:0070848 | response to growth factor | 408 | 547 | 1.11e-06 |
| GO:BP | GO:0006812 | monoatomic cation transport | 454 | 614 | 1.15e-06 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 317 | 416 | 1.18e-06 |
| GO:BP | GO:0061448 | connective tissue development | 167 | 205 | 1.18e-06 |
| GO:BP | GO:0061061 | muscle structure development | 400 | 536 | 1.36e-06 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 302 | 395 | 1.41e-06 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 225 | 286 | 1.49e-06 |
| GO:BP | GO:0030900 | forebrain development | 215 | 272 | 1.49e-06 |
| GO:BP | GO:0001525 | angiogenesis | 292 | 381 | 1.57e-06 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 287 | 374 | 1.66e-06 |
| GO:BP | GO:0007507 | heart development | 374 | 499 | 1.68e-06 |
| GO:BP | GO:0006468 | protein phosphorylation | 686 | 958 | 2.16e-06 |
| GO:BP | GO:0048771 | tissue remodeling | 98 | 113 | 2.51e-06 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 360 | 480 | 2.67e-06 |
| GO:BP | GO:0051216 | cartilage development | 122 | 145 | 2.67e-06 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 227 | 290 | 2.67e-06 |
| GO:BP | GO:1903522 | regulation of blood circulation | 158 | 194 | 2.75e-06 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 3312 | 4988 | 2.76e-06 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 204 | 258 | 3.07e-06 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 297 | 390 | 3.39e-06 |
| GO:BP | GO:0048638 | regulation of developmental growth | 193 | 243 | 3.74e-06 |
| GO:BP | GO:0060322 | head development | 400 | 539 | 3.74e-06 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 137 | 166 | 4.43e-06 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 392 | 528 | 4.66e-06 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 274 | 358 | 4.90e-06 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 548 | 757 | 5.19e-06 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 369 | 495 | 5.31e-06 |
| GO:BP | GO:0010243 | response to organonitrogen compound | 499 | 685 | 5.42e-06 |
| GO:BP | GO:0007015 | actin filament organization | 273 | 357 | 5.82e-06 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 513 | 706 | 6.07e-06 |
| GO:BP | GO:0097485 | neuron projection guidance | 141 | 172 | 6.24e-06 |
| GO:BP | GO:0007411 | axon guidance | 141 | 172 | 6.24e-06 |
| GO:BP | GO:0060047 | heart contraction | 157 | 194 | 6.32e-06 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 274 | 359 | 7.32e-06 |
| GO:BP | GO:0003015 | heart process | 164 | 204 | 7.73e-06 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 231 | 298 | 7.99e-06 |
| GO:BP | GO:0003012 | muscle system process | 250 | 325 | 8.03e-06 |
| GO:BP | GO:0140352 | export from cell | 436 | 594 | 8.12e-06 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 839 | 1193 | 9.34e-06 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 564 | 783 | 9.57e-06 |
| GO:BP | GO:0060429 | epithelium development | 570 | 792 | 9.84e-06 |
| GO:BP | GO:0030010 | establishment of cell polarity | 105 | 124 | 1.05e-05 |
| GO:BP | GO:0014706 | striated muscle tissue development | 170 | 213 | 1.12e-05 |
| GO:BP | GO:0009790 | embryo development | 589 | 821 | 1.23e-05 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 1308 | 1904 | 1.24e-05 |
| GO:BP | GO:0010720 | positive regulation of cell development | 238 | 309 | 1.27e-05 |
| GO:BP | GO:0046903 | secretion | 454 | 622 | 1.31e-05 |
| GO:BP | GO:0008016 | regulation of heart contraction | 135 | 165 | 1.34e-05 |
| GO:BP | GO:0035265 | organ growth | 107 | 127 | 1.34e-05 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 204 | 261 | 1.34e-05 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 101 | 119 | 1.36e-05 |
| GO:BP | GO:0048729 | tissue morphogenesis | 338 | 453 | 1.41e-05 |
| GO:BP | GO:1901698 | response to nitrogen compound | 531 | 736 | 1.50e-05 |
| GO:BP | GO:0060284 | regulation of cell development | 418 | 570 | 1.64e-05 |
| GO:BP | GO:0040007 | growth | 504 | 697 | 1.93e-05 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 770 | 1093 | 1.93e-05 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 1292 | 1883 | 2.12e-05 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 286 | 379 | 2.12e-05 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 177 | 224 | 2.12e-05 |
| GO:BP | GO:0043062 | extracellular structure organization | 177 | 224 | 2.12e-05 |
| GO:BP | GO:0032409 | regulation of transporter activity | 122 | 148 | 2.19e-05 |
| GO:BP | GO:0007420 | brain development | 368 | 498 | 2.25e-05 |
| GO:BP | GO:0090257 | regulation of muscle system process | 143 | 177 | 2.47e-05 |
| GO:BP | GO:0031032 | actomyosin structure organization | 148 | 184 | 2.56e-05 |
| GO:BP | GO:0030198 | extracellular matrix organization | 176 | 223 | 2.56e-05 |
| GO:BP | GO:1901652 | response to peptide | 278 | 368 | 2.56e-05 |
| GO:BP | GO:0023057 | negative regulation of signaling | 731 | 1036 | 2.56e-05 |
| GO:BP | GO:0050877 | nervous system process | 437 | 600 | 3.04e-05 |
| GO:BP | GO:0022898 | regulation of transmembrane transporter activity | 115 | 139 | 3.12e-05 |
| GO:BP | GO:0016358 | dendrite development | 147 | 183 | 3.23e-05 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 230 | 300 | 3.47e-05 |
| GO:BP | GO:1901653 | cellular response to peptide | 211 | 273 | 3.50e-05 |
| GO:BP | GO:0048588 | developmental cell growth | 144 | 179 | 3.51e-05 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 569 | 796 | 3.79e-05 |
| GO:BP | GO:0010033 | response to organic substance | 1193 | 1736 | 3.81e-05 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 82 | 95 | 3.82e-05 |
| GO:BP | GO:0006816 | calcium ion transport | 203 | 262 | 4.05e-05 |
| GO:BP | GO:0032412 | regulation of monoatomic ion transmembrane transporter activity | 111 | 134 | 4.17e-05 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 217 | 282 | 4.26e-05 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 728 | 1034 | 4.32e-05 |
| GO:BP | GO:0050772 | positive regulation of axonogenesis | 58 | 64 | 4.56e-05 |
| GO:BP | GO:0007423 | sensory organ development | 290 | 387 | 4.70e-05 |
| GO:BP | GO:0072001 | renal system development | 190 | 244 | 4.80e-05 |
| GO:BP | GO:0055085 | transmembrane transport | 652 | 921 | 5.07e-05 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 220 | 287 | 5.83e-05 |
| GO:BP | GO:0065009 | regulation of molecular function | 903 | 1299 | 5.98e-05 |
| GO:BP | GO:0032940 | secretion by cell | 402 | 551 | 5.98e-05 |
| GO:BP | GO:0043547 | positive regulation of GTPase activity | 95 | 113 | 6.60e-05 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 1769 | 2622 | 7.36e-05 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 165 | 210 | 8.86e-05 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 76 | 88 | 8.86e-05 |
| GO:BP | GO:0030100 | regulation of endocytosis | 155 | 196 | 8.93e-05 |
| GO:BP | GO:0018212 | peptidyl-tyrosine modification | 132 | 164 | 8.96e-05 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 259 | 344 | 8.96e-05 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 288 | 386 | 9.24e-05 |
| GO:BP | GO:0009605 | response to external stimulus | 1073 | 1559 | 9.26e-05 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 229 | 301 | 9.26e-05 |
| GO:BP | GO:0042592 | homeostatic process | 794 | 1137 | 9.38e-05 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 684 | 972 | 9.94e-05 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 126 | 156 | 1.05e-04 |
| GO:BP | GO:0032535 | regulation of cellular component size | 219 | 287 | 1.05e-04 |
| GO:BP | GO:0045216 | cell-cell junction organization | 123 | 152 | 1.14e-04 |
| GO:BP | GO:0030048 | actin filament-based movement | 90 | 107 | 1.14e-04 |
| GO:BP | GO:0001654 | eye development | 202 | 263 | 1.16e-04 |
| GO:BP | GO:0048880 | sensory system development | 204 | 266 | 1.23e-04 |
| GO:BP | GO:0051641 | cellular localization | 1767 | 2623 | 1.23e-04 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 120 | 148 | 1.23e-04 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 390 | 536 | 1.25e-04 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 158 | 201 | 1.30e-04 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 312 | 422 | 1.32e-04 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 187 | 242 | 1.32e-04 |
| GO:BP | GO:0018108 | peptidyl-tyrosine phosphorylation | 130 | 162 | 1.35e-04 |
| GO:BP | GO:0035023 | regulation of Rho protein signal transduction | 58 | 65 | 1.40e-04 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 150 | 190 | 1.44e-04 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 179 | 231 | 1.54e-04 |
| GO:BP | GO:0043434 | response to peptide hormone | 237 | 314 | 1.64e-04 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 164 | 210 | 1.70e-04 |
| GO:BP | GO:0042330 | taxis | 188 | 244 | 1.70e-04 |
| GO:BP | GO:0006935 | chemotaxis | 188 | 244 | 1.70e-04 |
| GO:BP | GO:0150063 | visual system development | 202 | 264 | 1.74e-04 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 469 | 654 | 1.76e-04 |
| GO:BP | GO:0001822 | kidney development | 183 | 237 | 1.76e-04 |
| GO:BP | GO:0048167 | regulation of synaptic plasticity | 113 | 139 | 1.77e-04 |
| GO:BP | GO:0043010 | camera-type eye development | 178 | 230 | 1.83e-04 |
| GO:BP | GO:0061337 | cardiac conduction | 73 | 85 | 1.96e-04 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 641 | 911 | 1.98e-04 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 170 | 219 | 2.14e-04 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 167 | 215 | 2.41e-04 |
| GO:BP | GO:1990138 | neuron projection extension | 109 | 134 | 2.43e-04 |
| GO:BP | GO:0003018 | vascular process in circulatory system | 152 | 194 | 2.61e-04 |
| GO:BP | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 78 | 92 | 2.61e-04 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 318 | 433 | 2.63e-04 |
| GO:BP | GO:0009725 | response to hormone | 438 | 610 | 2.89e-04 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 134 | 169 | 2.95e-04 |
| GO:BP | GO:0045165 | cell fate commitment | 121 | 151 | 3.13e-04 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 105 | 129 | 3.32e-04 |
| GO:BP | GO:0001764 | neuron migration | 105 | 129 | 3.32e-04 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 158 | 203 | 3.37e-04 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 397 | 550 | 3.39e-04 |
| GO:BP | GO:0006898 | receptor-mediated endocytosis | 143 | 182 | 3.60e-04 |
| GO:BP | GO:0035108 | limb morphogenesis | 91 | 110 | 3.63e-04 |
| GO:BP | GO:0035107 | appendage morphogenesis | 91 | 110 | 3.63e-04 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 186 | 243 | 3.70e-04 |
| GO:BP | GO:0061572 | actin filament bundle organization | 107 | 132 | 3.83e-04 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 183 | 239 | 4.18e-04 |
| GO:BP | GO:0021537 | telencephalon development | 152 | 195 | 4.19e-04 |
| GO:BP | GO:0060348 | bone development | 117 | 146 | 4.23e-04 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 147 | 188 | 4.27e-04 |
| GO:BP | GO:0007610 | behavior | 308 | 420 | 4.27e-04 |
| GO:BP | GO:0001952 | regulation of cell-matrix adhesion | 76 | 90 | 4.40e-04 |
| GO:BP | GO:0007266 | Rho protein signal transduction | 90 | 109 | 4.62e-04 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 106 | 131 | 4.80e-04 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 337 | 463 | 4.89e-04 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 337 | 463 | 4.89e-04 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 182 | 238 | 4.92e-04 |
| GO:BP | GO:0060537 | muscle tissue development | 252 | 339 | 5.20e-04 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 393 | 546 | 5.51e-04 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 1624 | 2415 | 5.60e-04 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 399 | 555 | 5.66e-04 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 143 | 183 | 5.75e-04 |
| GO:BP | GO:0048041 | focal adhesion assembly | 66 | 77 | 5.84e-04 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 128 | 162 | 5.96e-04 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 150 | 193 | 6.05e-04 |
| GO:BP | GO:0038127 | ERBB signaling pathway | 86 | 104 | 6.18e-04 |
| GO:BP | GO:0021955 | central nervous system neuron axonogenesis | 30 | 31 | 6.27e-04 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 272 | 369 | 6.93e-04 |
| GO:BP | GO:0008361 | regulation of cell size | 117 | 147 | 7.27e-04 |
| GO:BP | GO:0050900 | leukocyte migration | 151 | 195 | 7.99e-04 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 303 | 415 | 8.53e-04 |
| GO:BP | GO:0046849 | bone remodeling | 52 | 59 | 8.81e-04 |
| GO:BP | GO:0001649 | osteoblast differentiation | 136 | 174 | 8.81e-04 |
| GO:BP | GO:0035725 | sodium ion transmembrane transport | 79 | 95 | 8.92e-04 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 311 | 427 | 9.14e-04 |
| GO:BP | GO:0008360 | regulation of cell shape | 87 | 106 | 9.57e-04 |
| GO:BP | GO:0110020 | regulation of actomyosin structure organization | 73 | 87 | 9.68e-04 |
| GO:BP | GO:0001775 | cell activation | 446 | 627 | 9.73e-04 |
| GO:BP | GO:0090130 | tissue migration | 187 | 247 | 1.03e-03 |
| GO:BP | GO:0050807 | regulation of synapse organization | 135 | 173 | 1.07e-03 |
| GO:BP | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 120 | 152 | 1.12e-03 |
| GO:BP | GO:0090132 | epithelium migration | 184 | 243 | 1.16e-03 |
| GO:BP | GO:0010631 | epithelial cell migration | 184 | 243 | 1.16e-03 |
| GO:BP | GO:0051494 | negative regulation of cytoskeleton organization | 102 | 127 | 1.17e-03 |
| GO:BP | GO:0002063 | chondrocyte development | 23 | 23 | 1.19e-03 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 313 | 431 | 1.23e-03 |
| GO:BP | GO:0007229 | integrin-mediated signaling pathway | 72 | 86 | 1.25e-03 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 226 | 304 | 1.31e-03 |
| GO:BP | GO:0006814 | sodium ion transport | 109 | 137 | 1.36e-03 |
| GO:BP | GO:0006936 | muscle contraction | 192 | 255 | 1.39e-03 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 162 | 212 | 1.42e-03 |
| GO:BP | GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 212 | 284 | 1.44e-03 |
| GO:BP | GO:0000165 | MAPK cascade | 362 | 504 | 1.46e-03 |
| GO:BP | GO:0021987 | cerebral cortex development | 77 | 93 | 1.46e-03 |
| GO:BP | GO:0034103 | regulation of tissue remodeling | 40 | 44 | 1.46e-03 |
| GO:BP | GO:0008283 | cell population proliferation | 895 | 1306 | 1.46e-03 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 1470 | 2186 | 1.47e-03 |
| GO:BP | GO:0050920 | regulation of chemotaxis | 106 | 133 | 1.47e-03 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 196 | 261 | 1.49e-03 |
| GO:BP | GO:0031623 | receptor internalization | 74 | 89 | 1.52e-03 |
| GO:BP | GO:0051050 | positive regulation of transport | 407 | 571 | 1.52e-03 |
| GO:BP | GO:0009611 | response to wounding | 279 | 382 | 1.56e-03 |
| GO:BP | GO:0021954 | central nervous system neuron development | 50 | 57 | 1.56e-03 |
| GO:BP | GO:0032231 | regulation of actin filament bundle assembly | 71 | 85 | 1.58e-03 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 142 | 184 | 1.71e-03 |
| GO:BP | GO:0048592 | eye morphogenesis | 87 | 107 | 1.76e-03 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 165 | 217 | 1.86e-03 |
| GO:BP | GO:0141091 | transforming growth factor beta receptor superfamily signaling pathway | 194 | 259 | 2.06e-03 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 136 | 176 | 2.19e-03 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 52 | 60 | 2.23e-03 |
| GO:BP | GO:0006835 | dicarboxylic acid transport | 52 | 60 | 2.23e-03 |
| GO:BP | GO:0150116 | regulation of cell-substrate junction organization | 52 | 60 | 2.23e-03 |
| GO:BP | GO:0098900 | regulation of action potential | 35 | 38 | 2.23e-03 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 114 | 145 | 2.24e-03 |
| GO:BP | GO:0010232 | vascular transport | 58 | 68 | 2.27e-03 |
| GO:BP | GO:0043502 | regulation of muscle adaptation | 58 | 68 | 2.27e-03 |
| GO:BP | GO:0150104 | transport across blood-brain barrier | 58 | 68 | 2.27e-03 |
| GO:BP | GO:0043149 | stress fiber assembly | 75 | 91 | 2.33e-03 |
| GO:BP | GO:0030038 | contractile actin filament bundle assembly | 75 | 91 | 2.33e-03 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 72 | 87 | 2.49e-03 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 88 | 109 | 2.55e-03 |
| GO:BP | GO:0071695 | anatomical structure maturation | 118 | 151 | 2.67e-03 |
| GO:BP | GO:0003205 | cardiac chamber development | 118 | 151 | 2.67e-03 |
| GO:BP | GO:0003007 | heart morphogenesis | 165 | 218 | 2.72e-03 |
| GO:BP | GO:0048259 | regulation of receptor-mediated endocytosis | 66 | 79 | 2.74e-03 |
| GO:BP | GO:0030278 | regulation of ossification | 66 | 79 | 2.74e-03 |
| GO:BP | GO:0051893 | regulation of focal adhesion assembly | 48 | 55 | 2.80e-03 |
| GO:BP | GO:0051057 | positive regulation of small GTPase mediated signal transduction | 48 | 55 | 2.80e-03 |
| GO:BP | GO:0090109 | regulation of cell-substrate junction assembly | 48 | 55 | 2.80e-03 |
| GO:BP | GO:0098901 | regulation of cardiac muscle cell action potential | 26 | 27 | 2.81e-03 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 287 | 396 | 2.91e-03 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 54 | 63 | 2.96e-03 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 95 | 119 | 2.96e-03 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 198 | 266 | 3.05e-03 |
| GO:BP | GO:1902305 | regulation of sodium ion transmembrane transport | 34 | 37 | 3.06e-03 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 561 | 805 | 3.08e-03 |
| GO:BP | GO:0042692 | muscle cell differentiation | 237 | 323 | 3.24e-03 |
| GO:BP | GO:0097553 | calcium ion transmembrane import into cytosol | 97 | 122 | 3.32e-03 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 560 | 804 | 3.39e-03 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 159 | 210 | 3.40e-03 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 152 | 200 | 3.45e-03 |
| GO:BP | GO:0048008 | platelet-derived growth factor receptor signaling pathway | 44 | 50 | 3.50e-03 |
| GO:BP | GO:0050890 | cognition | 161 | 213 | 3.55e-03 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 62 | 74 | 3.63e-03 |
| GO:BP | GO:1902904 | negative regulation of supramolecular fiber organization | 99 | 125 | 3.68e-03 |
| GO:BP | GO:0002028 | regulation of sodium ion transport | 50 | 58 | 3.84e-03 |
| GO:BP | GO:0060485 | mesenchyme development | 183 | 245 | 3.90e-03 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 297 | 412 | 4.02e-03 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 106 | 135 | 4.03e-03 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 130 | 169 | 4.10e-03 |
| GO:BP | GO:0010817 | regulation of hormone levels | 233 | 318 | 4.14e-03 |
| GO:BP | GO:0050768 | negative regulation of neurogenesis | 83 | 103 | 4.27e-03 |
| GO:BP | GO:0051051 | negative regulation of transport | 211 | 286 | 4.31e-03 |
| GO:BP | GO:0002521 | leukocyte differentiation | 269 | 371 | 4.31e-03 |
| GO:BP | GO:0050730 | regulation of peptidyl-tyrosine phosphorylation | 108 | 138 | 4.37e-03 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 108 | 138 | 4.37e-03 |
| GO:BP | GO:0006909 | phagocytosis | 108 | 138 | 4.37e-03 |
| GO:BP | GO:0048469 | cell maturation | 88 | 110 | 4.37e-03 |
| GO:BP | GO:0051492 | regulation of stress fiber assembly | 64 | 77 | 4.39e-03 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 1559 | 2333 | 4.39e-03 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 80 | 99 | 4.60e-03 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 61 | 73 | 4.61e-03 |
| GO:BP | GO:0007259 | receptor signaling pathway via JAK-STAT | 72 | 88 | 4.62e-03 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 143 | 188 | 4.83e-03 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 253 | 348 | 4.95e-03 |
| GO:BP | GO:0099072 | regulation of postsynaptic membrane neurotransmitter receptor levels | 52 | 61 | 4.98e-03 |
| GO:BP | GO:0061614 | miRNA transcription | 52 | 61 | 4.98e-03 |
| GO:BP | GO:0036211 | protein modification process | 1531 | 2291 | 4.98e-03 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 124 | 161 | 5.05e-03 |
| GO:BP | GO:0031099 | regeneration | 112 | 144 | 5.09e-03 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 131 | 171 | 5.09e-03 |
| GO:BP | GO:0001508 | action potential | 82 | 102 | 5.19e-03 |
| GO:BP | GO:0045664 | regulation of neuron differentiation | 102 | 130 | 5.33e-03 |
| GO:BP | GO:0034764 | positive regulation of transmembrane transport | 102 | 130 | 5.33e-03 |
| GO:BP | GO:1903530 | regulation of secretion by cell | 265 | 366 | 5.34e-03 |
| GO:BP | GO:0097696 | receptor signaling pathway via STAT | 74 | 91 | 5.34e-03 |
| GO:BP | GO:0099173 | postsynapse organization | 114 | 147 | 5.45e-03 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 71 | 87 | 5.77e-03 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 71 | 87 | 5.77e-03 |
| GO:BP | GO:0046622 | positive regulation of organ growth | 32 | 35 | 5.77e-03 |
| GO:BP | GO:0042063 | gliogenesis | 180 | 242 | 5.95e-03 |
| GO:BP | GO:0008366 | axon ensheathment | 94 | 119 | 5.95e-03 |
| GO:BP | GO:0007272 | ensheathment of neurons | 94 | 119 | 5.95e-03 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 738 | 1077 | 5.96e-03 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 498 | 714 | 5.99e-03 |
| GO:BP | GO:0051046 | regulation of secretion | 285 | 396 | 5.99e-03 |
| GO:BP | GO:0023061 | signal release | 230 | 315 | 5.99e-03 |
| GO:BP | GO:0030500 | regulation of bone mineralization | 42 | 48 | 6.06e-03 |
| GO:BP | GO:0046850 | regulation of bone remodeling | 28 | 30 | 6.06e-03 |
| GO:BP | GO:0030097 | hemopoiesis | 438 | 624 | 6.06e-03 |
| GO:BP | GO:0048736 | appendage development | 106 | 136 | 6.18e-03 |
| GO:BP | GO:0060173 | limb development | 106 | 136 | 6.18e-03 |
| GO:BP | GO:0060341 | regulation of cellular localization | 539 | 776 | 6.18e-03 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 132 | 173 | 6.26e-03 |
| GO:BP | GO:0002376 | immune system process | 956 | 1410 | 6.44e-03 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 159 | 212 | 6.73e-03 |
| GO:BP | GO:0031399 | regulation of protein modification process | 628 | 911 | 6.73e-03 |
| GO:BP | GO:0014896 | muscle hypertrophy | 62 | 75 | 6.97e-03 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 129 | 169 | 7.06e-03 |
| GO:BP | GO:0007173 | epidermal growth factor receptor signaling pathway | 70 | 86 | 7.13e-03 |
| GO:BP | GO:0071407 | cellular response to organic cyclic compound | 273 | 379 | 7.15e-03 |
| GO:BP | GO:0048813 | dendrite morphogenesis | 88 | 111 | 7.16e-03 |
| GO:BP | GO:0046777 | protein autophosphorylation | 110 | 142 | 7.19e-03 |
| GO:BP | GO:0010721 | negative regulation of cell development | 138 | 182 | 7.31e-03 |
| GO:BP | GO:0021543 | pallium development | 105 | 135 | 7.45e-03 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 211 | 288 | 7.49e-03 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 1076 | 1595 | 7.49e-03 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 312 | 437 | 7.49e-03 |
| GO:BP | GO:0007162 | negative regulation of cell adhesion | 140 | 185 | 7.64e-03 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 112 | 145 | 7.67e-03 |
| GO:BP | GO:0071711 | basement membrane organization | 31 | 34 | 7.74e-03 |
| GO:BP | GO:0032330 | regulation of chondrocyte differentiation | 31 | 34 | 7.74e-03 |
| GO:BP | GO:1990845 | adaptive thermogenesis | 95 | 121 | 7.74e-03 |
| GO:BP | GO:0033043 | regulation of organelle organization | 655 | 953 | 7.85e-03 |
| GO:BP | GO:0014743 | regulation of muscle hypertrophy | 41 | 47 | 7.91e-03 |
| GO:BP | GO:0098868 | bone growth | 23 | 24 | 7.95e-03 |
| GO:BP | GO:0021952 | central nervous system projection neuron axonogenesis | 23 | 24 | 7.95e-03 |
| GO:BP | GO:0050773 | regulation of dendrite development | 64 | 78 | 8.09e-03 |
| GO:BP | GO:0030282 | bone mineralization | 64 | 78 | 8.09e-03 |
| GO:BP | GO:0010743 | regulation of macrophage derived foam cell differentiation | 18 | 18 | 8.15e-03 |
| GO:BP | GO:1990776 | response to angiotensin | 27 | 29 | 8.32e-03 |
| GO:BP | GO:0150117 | positive regulation of cell-substrate junction organization | 27 | 29 | 8.32e-03 |
| GO:BP | GO:0033036 | macromolecule localization | 1539 | 2309 | 8.54e-03 |
| GO:BP | GO:0042552 | myelination | 92 | 117 | 8.54e-03 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 87 | 110 | 8.58e-03 |
| GO:BP | GO:0043270 | positive regulation of monoatomic ion transport | 99 | 127 | 9.13e-03 |
| GO:BP | GO:0060249 | anatomical structure homeostasis | 125 | 164 | 9.15e-03 |
| GO:BP | GO:0046620 | regulation of organ growth | 58 | 70 | 9.15e-03 |
| GO:BP | GO:0001894 | tissue homeostasis | 125 | 164 | 9.15e-03 |
| GO:BP | GO:0030031 | cell projection assembly | 318 | 447 | 9.45e-03 |
| GO:BP | GO:0007188 | adenylate cyclase-modulating G protein-coupled receptor signaling pathway | 89 | 113 | 9.46e-03 |
| GO:BP | GO:0031214 | biomineral tissue development | 84 | 106 | 9.47e-03 |
| GO:BP | GO:0007043 | cell-cell junction assembly | 84 | 106 | 9.47e-03 |
| GO:BP | GO:0046942 | carboxylic acid transport | 152 | 203 | 9.60e-03 |
| GO:BP | GO:0007517 | muscle organ development | 196 | 267 | 9.60e-03 |
| GO:BP | GO:0040008 | regulation of growth | 328 | 462 | 9.60e-03 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 183 | 248 | 9.60e-03 |
| GO:BP | GO:0120032 | regulation of plasma membrane bounded cell projection assembly | 120 | 157 | 9.60e-03 |
| GO:BP | GO:0001659 | temperature homeostasis | 101 | 130 | 9.71e-03 |
| GO:BP | GO:0061387 | regulation of extent of cell growth | 71 | 88 | 9.90e-03 |
| GO:BP | GO:0032271 | regulation of protein polymerization | 122 | 160 | 1.02e-02 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 91 | 116 | 1.02e-02 |
| GO:BP | GO:0048675 | axon extension | 76 | 95 | 1.02e-02 |
| GO:BP | GO:0010611 | regulation of cardiac muscle hypertrophy | 40 | 46 | 1.03e-02 |
| GO:BP | GO:0060350 | endochondral bone morphogenesis | 40 | 46 | 1.03e-02 |
| GO:BP | GO:0051961 | negative regulation of nervous system development | 86 | 109 | 1.03e-02 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 390 | 555 | 1.03e-02 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 424 | 606 | 1.03e-02 |
| GO:BP | GO:0086065 | cell communication involved in cardiac conduction | 46 | 54 | 1.04e-02 |
| GO:BP | GO:0051924 | regulation of calcium ion transport | 117 | 153 | 1.07e-02 |
| GO:BP | GO:0060349 | bone morphogenesis | 60 | 73 | 1.07e-02 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 98 | 126 | 1.07e-02 |
| GO:BP | GO:0007611 | learning or memory | 142 | 189 | 1.11e-02 |
| GO:BP | GO:0003416 | endochondral bone growth | 22 | 23 | 1.11e-02 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 162 | 218 | 1.12e-02 |
| GO:BP | GO:0016266 | O-glycan processing | 26 | 28 | 1.14e-02 |
| GO:BP | GO:0006887 | exocytosis | 175 | 237 | 1.16e-02 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 203 | 278 | 1.18e-02 |
| GO:BP | GO:0032528 | microvillus organization | 17 | 17 | 1.18e-02 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 1343 | 2010 | 1.18e-02 |
| GO:BP | GO:0060491 | regulation of cell projection assembly | 121 | 159 | 1.18e-02 |
| GO:BP | GO:0015800 | acidic amino acid transport | 33 | 37 | 1.19e-02 |
| GO:BP | GO:0002685 | regulation of leukocyte migration | 95 | 122 | 1.19e-02 |
| GO:BP | GO:0098739 | import across plasma membrane | 95 | 122 | 1.19e-02 |
| GO:BP | GO:0060291 | long-term synaptic potentiation | 54 | 65 | 1.19e-02 |
| GO:BP | GO:0032272 | negative regulation of protein polymerization | 54 | 65 | 1.19e-02 |
| GO:BP | GO:0007613 | memory | 70 | 87 | 1.20e-02 |
| GO:BP | GO:0097191 | extrinsic apoptotic signaling pathway | 130 | 172 | 1.21e-02 |
| GO:BP | GO:0003231 | cardiac ventricle development | 90 | 115 | 1.21e-02 |
| GO:BP | GO:1901342 | regulation of vasculature development | 148 | 198 | 1.22e-02 |
| GO:BP | GO:1901890 | positive regulation of cell junction assembly | 62 | 76 | 1.23e-02 |
| GO:BP | GO:0033002 | muscle cell proliferation | 116 | 152 | 1.24e-02 |
| GO:BP | GO:0048678 | response to axon injury | 51 | 61 | 1.25e-02 |
| GO:BP | GO:0008104 | protein localization | 1336 | 2000 | 1.26e-02 |
| GO:BP | GO:0031103 | axon regeneration | 36 | 41 | 1.27e-02 |
| GO:BP | GO:0051899 | membrane depolarization | 48 | 57 | 1.30e-02 |
| GO:BP | GO:0050921 | positive regulation of chemotaxis | 67 | 83 | 1.30e-02 |
| GO:BP | GO:0015849 | organic acid transport | 152 | 204 | 1.30e-02 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 613 | 893 | 1.31e-02 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 59 | 72 | 1.32e-02 |
| GO:BP | GO:1903053 | regulation of extracellular matrix organization | 42 | 49 | 1.34e-02 |
| GO:BP | GO:0006954 | inflammatory response | 302 | 425 | 1.35e-02 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 304 | 428 | 1.35e-02 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 145 | 194 | 1.35e-02 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 191 | 261 | 1.35e-02 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 474 | 683 | 1.37e-02 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 277 | 388 | 1.39e-02 |
| GO:BP | GO:0045927 | positive regulation of growth | 138 | 184 | 1.41e-02 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 115 | 151 | 1.46e-02 |
| GO:BP | GO:0010876 | lipid localization | 216 | 298 | 1.47e-02 |
| GO:BP | GO:0032880 | regulation of protein localization | 479 | 691 | 1.48e-02 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 53 | 64 | 1.50e-02 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 53 | 64 | 1.50e-02 |
| GO:BP | GO:1904646 | cellular response to amyloid-beta | 25 | 27 | 1.55e-02 |
| GO:BP | GO:1904385 | cellular response to angiotensin | 25 | 27 | 1.55e-02 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 98 | 127 | 1.63e-02 |
| GO:BP | GO:0060419 | heart growth | 58 | 71 | 1.66e-02 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 217 | 300 | 1.70e-02 |
| GO:BP | GO:0071805 | potassium ion transmembrane transport | 93 | 120 | 1.70e-02 |
| GO:BP | GO:0048568 | embryonic organ development | 219 | 303 | 1.71e-02 |
| GO:BP | GO:0051653 | spindle localization | 44 | 52 | 1.71e-02 |
| GO:BP | GO:0021846 | cell proliferation in forebrain | 16 | 16 | 1.71e-02 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 202 | 278 | 1.72e-02 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 150 | 202 | 1.74e-02 |
| GO:BP | GO:0051058 | negative regulation of small GTPase mediated signal transduction | 41 | 48 | 1.74e-02 |
| GO:BP | GO:0031102 | neuron projection regeneration | 41 | 48 | 1.74e-02 |
| GO:BP | GO:0055006 | cardiac cell development | 63 | 78 | 1.75e-02 |
| GO:BP | GO:0055001 | muscle cell development | 123 | 163 | 1.76e-02 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 68 | 85 | 1.81e-02 |
| GO:BP | GO:1904645 | response to amyloid-beta | 28 | 31 | 1.90e-02 |
| GO:BP | GO:0032411 | positive regulation of transporter activity | 52 | 63 | 1.90e-02 |
| GO:BP | GO:0031644 | regulation of nervous system process | 60 | 74 | 1.91e-02 |
| GO:BP | GO:0048864 | stem cell development | 60 | 74 | 1.91e-02 |
| GO:BP | GO:0016049 | cell growth | 272 | 382 | 1.92e-02 |
| GO:BP | GO:0000003 | reproduction | 604 | 882 | 1.92e-02 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 65 | 81 | 1.99e-02 |
| GO:BP | GO:0003279 | cardiac septum development | 80 | 102 | 2.00e-02 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 290 | 409 | 2.01e-02 |
| GO:BP | GO:0014855 | striated muscle cell proliferation | 49 | 59 | 2.01e-02 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 75 | 95 | 2.02e-02 |
| GO:BP | GO:0043588 | skin development | 131 | 175 | 2.07e-02 |
| GO:BP | GO:0106106 | cold-induced thermogenesis | 87 | 112 | 2.10e-02 |
| GO:BP | GO:0008038 | neuron recognition | 31 | 35 | 2.11e-02 |
| GO:BP | GO:0045663 | positive regulation of myoblast differentiation | 31 | 35 | 2.11e-02 |
| GO:BP | GO:0021799 | cerebral cortex radially oriented cell migration | 24 | 26 | 2.16e-02 |
| GO:BP | GO:0051235 | maintenance of location | 183 | 251 | 2.20e-02 |
| GO:BP | GO:0045667 | regulation of osteoblast differentiation | 77 | 98 | 2.22e-02 |
| GO:BP | GO:0046578 | regulation of Ras protein signal transduction | 34 | 39 | 2.23e-02 |
| GO:BP | GO:0034767 | positive regulation of monoatomic ion transmembrane transport | 72 | 91 | 2.24e-02 |
| GO:BP | GO:0045665 | negative regulation of neuron differentiation | 37 | 43 | 2.27e-02 |
| GO:BP | GO:0002090 | regulation of receptor internalization | 37 | 43 | 2.27e-02 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 159 | 216 | 2.27e-02 |
| GO:BP | GO:0006813 | potassium ion transport | 103 | 135 | 2.29e-02 |
| GO:BP | GO:1903169 | regulation of calcium ion transmembrane transport | 84 | 108 | 2.34e-02 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 270 | 380 | 2.37e-02 |
| GO:BP | GO:0070167 | regulation of biomineral tissue development | 51 | 62 | 2.39e-02 |
| GO:BP | GO:0044272 | sulfur compound biosynthetic process | 98 | 128 | 2.44e-02 |
| GO:BP | GO:0010586 | miRNA metabolic process | 69 | 87 | 2.47e-02 |
| GO:BP | GO:0048593 | camera-type eye morphogenesis | 69 | 87 | 2.47e-02 |
| GO:BP | GO:1904375 | regulation of protein localization to cell periphery | 86 | 111 | 2.52e-02 |
| GO:BP | GO:0120161 | regulation of cold-induced thermogenesis | 86 | 111 | 2.52e-02 |
| GO:BP | GO:0032414 | positive regulation of ion transmembrane transporter activity | 48 | 58 | 2.55e-02 |
| GO:BP | GO:0001974 | blood vessel remodeling | 27 | 30 | 2.57e-02 |
| GO:BP | GO:1904707 | positive regulation of vascular associated smooth muscle cell proliferation | 27 | 30 | 2.57e-02 |
| GO:BP | GO:0120162 | positive regulation of cold-induced thermogenesis | 56 | 69 | 2.57e-02 |
| GO:BP | GO:0046486 | glycerolipid metabolic process | 205 | 284 | 2.57e-02 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 171 | 234 | 2.57e-02 |
| GO:BP | GO:0071396 | cellular response to lipid | 275 | 388 | 2.66e-02 |
| GO:BP | GO:0006950 | response to stress | 1680 | 2540 | 2.66e-02 |
| GO:BP | GO:0014910 | regulation of smooth muscle cell migration | 45 | 54 | 2.69e-02 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 66 | 83 | 2.72e-02 |
| GO:BP | GO:0051648 | vesicle localization | 140 | 189 | 2.80e-02 |
| GO:BP | GO:0010927 | cellular component assembly involved in morphogenesis | 83 | 107 | 2.80e-02 |
| GO:BP | GO:0031667 | response to nutrient levels | 252 | 354 | 2.80e-02 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 329 | 469 | 2.81e-02 |
| GO:BP | GO:0021795 | cerebral cortex cell migration | 30 | 34 | 2.81e-02 |
| GO:BP | GO:2001236 | regulation of extrinsic apoptotic signaling pathway | 90 | 117 | 2.86e-02 |
| GO:BP | GO:0006629 | lipid metabolic process | 629 | 923 | 2.89e-02 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 58 | 72 | 2.93e-02 |
| GO:BP | GO:0046503 | glycerolipid catabolic process | 33 | 38 | 2.93e-02 |
| GO:BP | GO:1904407 | positive regulation of nitric oxide metabolic process | 23 | 25 | 2.94e-02 |
| GO:BP | GO:0060351 | cartilage development involved in endochondral bone morphogenesis | 23 | 25 | 2.94e-02 |
| GO:BP | GO:0001954 | positive regulation of cell-matrix adhesion | 36 | 42 | 2.94e-02 |
| GO:BP | GO:0022612 | gland morphogenesis | 73 | 93 | 2.94e-02 |
| GO:BP | GO:0033993 | response to lipid | 398 | 573 | 2.94e-02 |
| GO:BP | GO:0030705 | cytoskeleton-dependent intracellular transport | 133 | 179 | 2.94e-02 |
| GO:BP | GO:0006869 | lipid transport | 189 | 261 | 2.96e-02 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 85 | 110 | 2.97e-02 |
| GO:BP | GO:0051043 | regulation of membrane protein ectodomain proteolysis | 19 | 20 | 3.09e-02 |
| GO:BP | GO:0071675 | regulation of mononuclear cell migration | 55 | 68 | 3.18e-02 |
| GO:BP | GO:0019915 | lipid storage | 55 | 68 | 3.18e-02 |
| GO:BP | GO:0007584 | response to nutrient | 87 | 113 | 3.18e-02 |
| GO:BP | GO:0055123 | digestive system development | 75 | 96 | 3.20e-02 |
| GO:BP | GO:0006941 | striated muscle contraction | 110 | 146 | 3.22e-02 |
| GO:BP | GO:0009914 | hormone transport | 152 | 207 | 3.22e-02 |
| GO:BP | GO:0007189 | adenylate cyclase-activating G protein-coupled receptor signaling pathway | 60 | 75 | 3.27e-02 |
| GO:BP | GO:0045321 | leukocyte activation | 369 | 530 | 3.29e-02 |
| GO:BP | GO:0007416 | synapse assembly | 103 | 136 | 3.31e-02 |
| GO:BP | GO:0046661 | male sex differentiation | 82 | 106 | 3.31e-02 |
| GO:BP | GO:0050806 | positive regulation of synaptic transmission | 82 | 106 | 3.31e-02 |
| GO:BP | GO:0022414 | reproductive process | 596 | 874 | 3.38e-02 |
| GO:BP | GO:2000649 | regulation of sodium ion transmembrane transporter activity | 26 | 29 | 3.43e-02 |
| GO:BP | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules | 26 | 29 | 3.43e-02 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 443 | 642 | 3.48e-02 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 84 | 109 | 3.55e-02 |
| GO:BP | GO:1904705 | regulation of vascular associated smooth muscle cell proliferation | 41 | 49 | 3.56e-02 |
| GO:BP | GO:1903078 | positive regulation of protein localization to plasma membrane | 41 | 49 | 3.56e-02 |
| GO:BP | GO:0060998 | regulation of dendritic spine development | 41 | 49 | 3.56e-02 |
| GO:BP | GO:0060420 | regulation of heart growth | 41 | 49 | 3.56e-02 |
| GO:BP | GO:0106027 | neuron projection organization | 57 | 71 | 3.57e-02 |
| GO:BP | GO:0021700 | developmental maturation | 149 | 203 | 3.58e-02 |
| GO:BP | GO:0008037 | cell recognition | 62 | 78 | 3.62e-02 |
| GO:BP | GO:0040019 | positive regulation of embryonic development | 14 | 14 | 3.63e-02 |
| GO:BP | GO:0086069 | bundle of His cell to Purkinje myocyte communication | 14 | 14 | 3.63e-02 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 183 | 253 | 3.64e-02 |
| GO:BP | GO:0042100 | B cell proliferation | 38 | 45 | 3.68e-02 |
| GO:BP | GO:0007260 | tyrosine phosphorylation of STAT protein | 29 | 33 | 3.68e-02 |
| GO:BP | GO:0015711 | organic anion transport | 185 | 256 | 3.68e-02 |
| GO:BP | GO:1904377 | positive regulation of protein localization to cell periphery | 49 | 60 | 3.68e-02 |
| GO:BP | GO:0001763 | morphogenesis of a branching structure | 109 | 145 | 3.71e-02 |
| GO:BP | GO:0045453 | bone resorption | 35 | 41 | 3.76e-02 |
| GO:BP | GO:0010171 | body morphogenesis | 35 | 41 | 3.76e-02 |
| GO:BP | GO:1903115 | regulation of actin filament-based movement | 32 | 37 | 3.77e-02 |
| GO:BP | GO:0010811 | positive regulation of cell-substrate adhesion | 74 | 95 | 3.79e-02 |
| GO:BP | GO:0050870 | positive regulation of T cell activation | 102 | 135 | 3.82e-02 |
| GO:BP | GO:0008154 | actin polymerization or depolymerization | 111 | 148 | 3.84e-02 |
| GO:BP | GO:0002064 | epithelial cell development | 111 | 148 | 3.84e-02 |
| GO:BP | GO:1901343 | negative regulation of vasculature development | 54 | 67 | 3.85e-02 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 54 | 67 | 3.85e-02 |
| GO:BP | GO:2000181 | negative regulation of blood vessel morphogenesis | 54 | 67 | 3.85e-02 |
| GO:BP | GO:1903076 | regulation of protein localization to plasma membrane | 69 | 88 | 3.87e-02 |
| GO:BP | GO:0006638 | neutral lipid metabolic process | 69 | 88 | 3.87e-02 |
| GO:BP | GO:0008217 | regulation of blood pressure | 81 | 105 | 3.87e-02 |
| GO:BP | GO:0060041 | retina development in camera-type eye | 81 | 105 | 3.87e-02 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 88 | 115 | 3.89e-02 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 88 | 115 | 3.89e-02 |
| GO:BP | GO:0001823 | mesonephros development | 59 | 74 | 3.89e-02 |
| GO:BP | GO:0030516 | regulation of axon extension | 59 | 74 | 3.89e-02 |
| GO:BP | GO:0003229 | ventricular cardiac muscle tissue development | 46 | 56 | 3.89e-02 |
| GO:BP | GO:0051894 | positive regulation of focal adhesion assembly | 22 | 24 | 3.92e-02 |
| GO:BP | GO:0045429 | positive regulation of nitric oxide biosynthetic process | 22 | 24 | 3.92e-02 |
| GO:BP | GO:0002431 | Fc receptor mediated stimulatory signaling pathway | 22 | 24 | 3.92e-02 |
| GO:BP | GO:0045124 | regulation of bone resorption | 22 | 24 | 3.92e-02 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 180 | 249 | 3.95e-02 |
| GO:BP | GO:0046879 | hormone secretion | 148 | 202 | 3.97e-02 |
| GO:BP | GO:0007389 | pattern specification process | 205 | 286 | 3.99e-02 |
| GO:BP | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | 71 | 91 | 4.16e-02 |
| GO:BP | GO:0071378 | cellular response to growth hormone stimulus | 18 | 19 | 4.23e-02 |
| GO:BP | GO:0051650 | establishment of vesicle localization | 128 | 173 | 4.23e-02 |
| GO:BP | GO:0035270 | endocrine system development | 66 | 84 | 4.26e-02 |
| GO:BP | GO:0002684 | positive regulation of immune system process | 401 | 580 | 4.27e-02 |
| GO:BP | GO:1901879 | regulation of protein depolymerization | 56 | 70 | 4.28e-02 |
| GO:BP | GO:0014033 | neural crest cell differentiation | 61 | 77 | 4.30e-02 |
| GO:BP | GO:0072073 | kidney epithelium development | 85 | 111 | 4.33e-02 |
| GO:BP | GO:0001755 | neural crest cell migration | 40 | 48 | 4.41e-02 |
| GO:BP | GO:0051293 | establishment of spindle localization | 40 | 48 | 4.41e-02 |
| GO:BP | GO:0015833 | peptide transport | 132 | 179 | 4.44e-02 |
| GO:BP | GO:0060421 | positive regulation of heart growth | 25 | 28 | 4.49e-02 |
| GO:BP | GO:0030041 | actin filament polymerization | 94 | 124 | 4.51e-02 |
| GO:BP | GO:0042060 | wound healing | 208 | 291 | 4.55e-02 |
| GO:BP | GO:0030073 | insulin secretion | 103 | 137 | 4.57e-02 |
| GO:BP | GO:0055021 | regulation of cardiac muscle tissue growth | 37 | 44 | 4.60e-02 |
| GO:BP | GO:0006639 | acylglycerol metabolic process | 68 | 87 | 4.60e-02 |
| GO:BP | GO:1902895 | positive regulation of miRNA transcription | 37 | 44 | 4.60e-02 |
| GO:BP | GO:0086002 | cardiac muscle cell action potential involved in contraction | 37 | 44 | 4.60e-02 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 396 | 573 | 4.63e-02 |
| GO:BP | GO:0051668 | localization within membrane | 392 | 567 | 4.66e-02 |
| GO:BP | GO:0071230 | cellular response to amino acid stimulus | 53 | 66 | 4.66e-02 |
| GO:BP | GO:0016525 | negative regulation of angiogenesis | 53 | 66 | 4.66e-02 |
| GO:BP | GO:0007369 | gastrulation | 105 | 140 | 4.71e-02 |
| GO:BP | GO:0072164 | mesonephric tubule development | 58 | 73 | 4.71e-02 |
| GO:BP | GO:0072163 | mesonephric epithelium development | 58 | 73 | 4.71e-02 |
| GO:BP | GO:0042509 | regulation of tyrosine phosphorylation of STAT protein | 28 | 32 | 4.71e-02 |
| GO:BP | GO:0086004 | regulation of cardiac muscle cell contraction | 28 | 32 | 4.71e-02 |
| GO:BP | GO:0098927 | vesicle-mediated transport between endosomal compartments | 34 | 40 | 4.72e-02 |
| GO:BP | GO:0008277 | regulation of G protein-coupled receptor signaling pathway | 75 | 97 | 4.72e-02 |
| GO:BP | GO:0006865 | amino acid transport | 75 | 97 | 4.72e-02 |
| GO:BP | GO:0038179 | neurotrophin signaling pathway | 31 | 36 | 4.77e-02 |
| GO:BP | GO:0031641 | regulation of myelination | 31 | 36 | 4.77e-02 |
| GO:BP | GO:0003073 | regulation of systemic arterial blood pressure | 45 | 55 | 4.79e-02 |
| GO:BP | GO:0042246 | tissue regeneration | 45 | 55 | 4.79e-02 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 232 | 327 | 4.79e-02 |
| GO:BP | GO:0002682 | regulation of immune system process | 590 | 868 | 4.85e-02 |
LRnomtable_20k %>%
filter(source=="KEGG", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "KEGG terms found in LR nESRgenes_20k") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| KEGG | KEGG:04360 | Axon guidance | 130 | 155 | 5.82e-06 |
| KEGG | KEGG:04971 | Gastric acid secretion | 49 | 51 | 6.86e-06 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 109 | 129 | 8.01e-06 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 133 | 162 | 8.01e-06 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 122 | 147 | 8.01e-06 |
| KEGG | KEGG:04520 | Adherens junction | 74 | 83 | 8.01e-06 |
| KEGG | KEGG:04270 | Vascular smooth muscle contraction | 81 | 92 | 8.01e-06 |
| KEGG | KEGG:04020 | Calcium signaling pathway | 132 | 161 | 9.13e-06 |
| KEGG | KEGG:04911 | Insulin secretion | 57 | 62 | 9.72e-06 |
| KEGG | KEGG:04014 | Ras signaling pathway | 134 | 164 | 9.72e-06 |
| KEGG | KEGG:04925 | Aldosterone synthesis and secretion | 67 | 75 | 1.26e-05 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 137 | 169 | 1.34e-05 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 69 | 78 | 1.74e-05 |
| KEGG | KEGG:04510 | Focal adhesion | 136 | 169 | 2.83e-05 |
| KEGG | KEGG:04611 | Platelet activation | 76 | 88 | 3.64e-05 |
| KEGG | KEGG:04974 | Protein digestion and absorption | 51 | 56 | 5.62e-05 |
| KEGG | KEGG:04927 | Cortisol synthesis and secretion | 43 | 46 | 5.69e-05 |
| KEGG | KEGG:05200 | Pathways in cancer | 290 | 391 | 6.90e-05 |
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 102 | 124 | 6.90e-05 |
| KEGG | KEGG:04970 | Salivary secretion | 49 | 54 | 9.33e-05 |
| KEGG | KEGG:05146 | Amoebiasis | 55 | 62 | 1.27e-04 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 177 | 231 | 1.88e-04 |
| KEGG | KEGG:04750 | Inflammatory mediator regulation of TRP channels | 62 | 72 | 2.63e-04 |
| KEGG | KEGG:04713 | Circadian entrainment | 59 | 68 | 2.63e-04 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 126 | 160 | 3.03e-04 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 189 | 250 | 3.54e-04 |
| KEGG | KEGG:04961 | Endocrine and other factor-regulated calcium reabsorption | 37 | 40 | 3.64e-04 |
| KEGG | KEGG:04512 | ECM-receptor interaction | 57 | 66 | 4.11e-04 |
| KEGG | KEGG:04918 | Thyroid hormone synthesis | 47 | 53 | 4.46e-04 |
| KEGG | KEGG:04972 | Pancreatic secretion | 50 | 57 | 4.65e-04 |
| KEGG | KEGG:04730 | Long-term depression | 36 | 39 | 4.65e-04 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 126 | 162 | 6.91e-04 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 72 | 87 | 7.24e-04 |
| KEGG | KEGG:05412 | Arrhythmogenic right ventricular cardiomyopathy | 57 | 67 | 8.62e-04 |
| KEGG | KEGG:04728 | Dopaminergic synapse | 79 | 97 | 9.25e-04 |
| KEGG | KEGG:04924 | Renin secretion | 44 | 50 | 9.91e-04 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 81 | 100 | 1.02e-03 |
| KEGG | KEGG:04976 | Bile secretion | 37 | 41 | 1.02e-03 |
| KEGG | KEGG:04148 | Efferocytosis | 95 | 120 | 1.37e-03 |
| KEGG | KEGG:04915 | Estrogen signaling pathway | 73 | 90 | 1.91e-03 |
| KEGG | KEGG:05135 | Yersinia infection | 85 | 107 | 2.20e-03 |
| KEGG | KEGG:05414 | Dilated cardiomyopathy | 67 | 82 | 2.20e-03 |
| KEGG | KEGG:04072 | Phospholipase D signaling pathway | 85 | 107 | 2.20e-03 |
| KEGG | KEGG:04720 | Long-term potentiation | 44 | 51 | 2.26e-03 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 77 | 96 | 2.30e-03 |
| KEGG | KEGG:04726 | Serotonergic synapse | 49 | 58 | 2.90e-03 |
| KEGG | KEGG:05417 | Lipid and atherosclerosis | 110 | 143 | 2.96e-03 |
| KEGG | KEGG:04670 | Leukocyte transendothelial migration | 62 | 76 | 3.51e-03 |
| KEGG | KEGG:00000 | KEGG root term | 3552 | 5451 | 3.54e-03 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 82 | 104 | 3.80e-03 |
| KEGG | KEGG:04725 | Cholinergic synapse | 66 | 82 | 4.54e-03 |
| KEGG | KEGG:04921 | Oxytocin signaling pathway | 88 | 113 | 4.91e-03 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 102 | 133 | 5.10e-03 |
| KEGG | KEGG:04724 | Glutamatergic synapse | 60 | 74 | 5.24e-03 |
| KEGG | KEGG:05410 | Hypertrophic cardiomyopathy | 62 | 77 | 5.99e-03 |
| KEGG | KEGG:04144 | Endocytosis | 159 | 216 | 6.22e-03 |
| KEGG | KEGG:04530 | Tight junction | 98 | 129 | 1.04e-02 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 49 | 60 | 1.07e-02 |
| KEGG | KEGG:05207 | Chemical carcinogenesis - receptor activation | 100 | 132 | 1.07e-02 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 81 | 105 | 1.11e-02 |
| KEGG | KEGG:04625 | C-type lectin receptor signaling pathway | 59 | 74 | 1.11e-02 |
| KEGG | KEGG:04960 | Aldosterone-regulated sodium reabsorption | 22 | 24 | 1.11e-02 |
| KEGG | KEGG:04912 | GnRH signaling pathway | 59 | 74 | 1.11e-02 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 78 | 101 | 1.19e-02 |
| KEGG | KEGG:04660 | T cell receptor signaling pathway | 66 | 84 | 1.19e-02 |
| KEGG | KEGG:05231 | Choline metabolism in cancer | 63 | 80 | 1.30e-02 |
| KEGG | KEGG:05100 | Bacterial invasion of epithelial cells | 58 | 73 | 1.30e-02 |
| KEGG | KEGG:04070 | Phosphatidylinositol signaling system | 65 | 83 | 1.41e-02 |
| KEGG | KEGG:04514 | Cell adhesion molecules | 55 | 69 | 1.42e-02 |
| KEGG | KEGG:04071 | Sphingolipid signaling pathway | 79 | 103 | 1.46e-02 |
| KEGG | KEGG:03320 | PPAR signaling pathway | 39 | 47 | 1.49e-02 |
| KEGG | KEGG:01521 | EGFR tyrosine kinase inhibitor resistance | 57 | 72 | 1.54e-02 |
| KEGG | KEGG:05032 | Morphine addiction | 44 | 54 | 1.61e-02 |
| KEGG | KEGG:04916 | Melanogenesis | 59 | 75 | 1.64e-02 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 59 | 75 | 1.64e-02 |
| KEGG | KEGG:00564 | Glycerophospholipid metabolism | 56 | 71 | 1.84e-02 |
| KEGG | KEGG:05170 | Human immunodeficiency virus 1 infection | 110 | 149 | 2.20e-02 |
| KEGG | KEGG:04630 | JAK-STAT signaling pathway | 64 | 83 | 2.57e-02 |
| KEGG | KEGG:05418 | Fluid shear stress and atherosclerosis | 82 | 109 | 2.68e-02 |
| KEGG | KEGG:04062 | Chemokine signaling pathway | 82 | 109 | 2.68e-02 |
| KEGG | KEGG:00512 | Mucin type O-glycan biosynthesis | 19 | 21 | 2.68e-02 |
| KEGG | KEGG:05167 | Kaposi sarcoma-associated herpesvirus infection | 99 | 134 | 3.01e-02 |
| KEGG | KEGG:05130 | Pathogenic Escherichia coli infection | 105 | 143 | 3.23e-02 |
| KEGG | KEGG:05210 | Colorectal cancer | 62 | 81 | 3.50e-02 |
| KEGG | KEGG:04722 | Neurotrophin signaling pathway | 78 | 104 | 3.50e-02 |
| KEGG | KEGG:04742 | Taste transduction | 18 | 20 | 3.50e-02 |
| KEGG | KEGG:04931 | Insulin resistance | 69 | 91 | 3.50e-02 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 95 | 129 | 3.79e-02 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 178 | 252 | 3.97e-02 |
| KEGG | KEGG:01212 | Fatty acid metabolism | 37 | 46 | 4.08e-02 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 79 | 106 | 4.10e-02 |
| KEGG | KEGG:00514 | Other types of O-glycan biosynthesis | 29 | 35 | 4.12e-02 |
| KEGG | KEGG:04930 | Type II diabetes mellitus | 29 | 35 | 4.12e-02 |
| KEGG | KEGG:00280 | Valine, leucine and isoleucine degradation | 34 | 42 | 4.33e-02 |
| KEGG | KEGG:04664 | Fc epsilon RI signaling pathway | 34 | 42 | 4.33e-02 |
| KEGG | KEGG:04723 | Retrograde endocannabinoid signaling | 69 | 92 | 4.64e-02 |
| KEGG | KEGG:04380 | Osteoclast differentiation | 69 | 92 | 4.64e-02 |
| KEGG | KEGG:01040 | Biosynthesis of unsaturated fatty acids | 20 | 23 | 4.64e-02 |
| KEGG | KEGG:04727 | GABAergic synapse | 41 | 52 | 4.67e-02 |
| KEGG | KEGG:04662 | B cell receptor signaling pathway | 41 | 52 | 4.67e-02 |
| KEGG | KEGG:05031 | Amphetamine addiction | 36 | 45 | 4.68e-02 |
| KEGG | KEGG:05131 | Shigellosis | 145 | 204 | 4.69e-02 |
| KEGG | KEGG:04211 | Longevity regulating pathway | 57 | 75 | 4.95e-02 |
| KEGG | KEGG:04152 | AMPK signaling pathway | 75 | 101 | 4.95e-02 |
| KEGG | KEGG:04666 | Fc gamma R-mediated phagocytosis | 57 | 75 | 4.95e-02 |
LR peaks overlapping enhancers
# LR_resgenes_20k_enh <- gost(query = unique(LR_enh_peaks_20k$NCBI_gene),
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = FALSE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = background_df$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(LR_resgenes_20k_enh,"data/GO_KEGG_analysis/LR_resgenes_20k_enh.RDS")
# LRnom <- gostplot(LR_resgenes_20k_enh, capped = FALSE, interactive = TRUE)
# LRnom
LRnomtable_20k_enh <- LR_resgenes_20k_enh$result %>%
dplyr::select(c(source, term_id,
term_name,intersection_size,
term_size, p_value))# %>%
LRnomtable_20k_enh %>%
filter(source=="GO:BP", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(.,caption = "GO:BP terms found in LR neargenes_20k" ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0050896 | response to stimulus | 1012 | 5379 | 1.25e-17 |
| GO:BP | GO:0007154 | cell communication | 789 | 3983 | 1.25e-17 |
| GO:BP | GO:0023052 | signaling | 778 | 3918 | 1.25e-17 |
| GO:BP | GO:0048856 | anatomical structure development | 768 | 3867 | 2.10e-17 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 434 | 1924 | 4.48e-17 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 418 | 1844 | 9.43e-17 |
| GO:BP | GO:0032501 | multicellular organismal process | 864 | 4504 | 3.39e-16 |
| GO:BP | GO:0007165 | signal transduction | 718 | 3611 | 4.68e-16 |
| GO:BP | GO:0007275 | multicellular organism development | 641 | 3170 | 2.41e-15 |
| GO:BP | GO:0051716 | cellular response to stimulus | 878 | 4640 | 7.34e-15 |
| GO:BP | GO:0032502 | developmental process | 803 | 4179 | 1.11e-14 |
| GO:BP | GO:0030029 | actin filament-based process | 176 | 640 | 2.32e-13 |
| GO:BP | GO:0072359 | circulatory system development | 220 | 864 | 4.21e-13 |
| GO:BP | GO:0048731 | system development | 553 | 2719 | 4.37e-13 |
| GO:BP | GO:0048519 | negative regulation of biological process | 725 | 3764 | 7.59e-13 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 690 | 3550 | 7.59e-13 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 390 | 1801 | 2.55e-12 |
| GO:BP | GO:0050789 | regulation of biological process | 1301 | 7516 | 3.24e-12 |
| GO:BP | GO:0010646 | regulation of cell communication | 501 | 2444 | 3.51e-12 |
| GO:BP | GO:0007155 | cell adhesion | 231 | 941 | 3.82e-12 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 547 | 2717 | 3.82e-12 |
| GO:BP | GO:0048513 | animal organ development | 424 | 2004 | 4.73e-12 |
| GO:BP | GO:0061061 | muscle structure development | 149 | 536 | 1.04e-11 |
| GO:BP | GO:0023051 | regulation of signaling | 495 | 2438 | 2.67e-11 |
| GO:BP | GO:0065007 | biological regulation | 1326 | 7734 | 3.75e-11 |
| GO:BP | GO:0016477 | cell migration | 240 | 1011 | 4.70e-11 |
| GO:BP | GO:0009966 | regulation of signal transduction | 446 | 2166 | 6.97e-11 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 86 | 258 | 8.53e-11 |
| GO:BP | GO:0009888 | tissue development | 301 | 1348 | 8.79e-11 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 79 | 234 | 3.46e-10 |
| GO:BP | GO:0030154 | cell differentiation | 550 | 2803 | 3.46e-10 |
| GO:BP | GO:0048869 | cellular developmental process | 550 | 2804 | 3.55e-10 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 61 | 162 | 6.44e-10 |
| GO:BP | GO:0048870 | cell motility | 255 | 1120 | 7.91e-10 |
| GO:BP | GO:0044057 | regulation of system process | 111 | 384 | 1.08e-09 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 148 | 566 | 1.56e-09 |
| GO:BP | GO:0050793 | regulation of developmental process | 357 | 1697 | 1.58e-09 |
| GO:BP | GO:0048518 | positive regulation of biological process | 788 | 4301 | 2.69e-09 |
| GO:BP | GO:0050794 | regulation of cellular process | 1227 | 7153 | 3.11e-09 |
| GO:BP | GO:0003013 | circulatory system process | 118 | 426 | 4.29e-09 |
| GO:BP | GO:0042692 | muscle cell differentiation | 96 | 323 | 4.89e-09 |
| GO:BP | GO:0008015 | blood circulation | 103 | 357 | 6.19e-09 |
| GO:BP | GO:0003012 | muscle system process | 96 | 325 | 6.85e-09 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 133 | 508 | 1.45e-08 |
| GO:BP | GO:0040011 | locomotion | 190 | 805 | 2.19e-08 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 173 | 717 | 2.53e-08 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 725 | 3954 | 2.89e-08 |
| GO:BP | GO:0060537 | muscle tissue development | 97 | 339 | 3.42e-08 |
| GO:BP | GO:0001568 | blood vessel development | 135 | 525 | 3.51e-08 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 195 | 837 | 3.57e-08 |
| GO:BP | GO:2000145 | regulation of cell motility | 169 | 700 | 3.62e-08 |
| GO:BP | GO:0001944 | vasculature development | 140 | 552 | 4.20e-08 |
| GO:BP | GO:0003008 | system process | 250 | 1139 | 4.52e-08 |
| GO:BP | GO:0060047 | heart contraction | 64 | 194 | 8.25e-08 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 218 | 972 | 9.89e-08 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 258 | 1193 | 9.89e-08 |
| GO:BP | GO:0040012 | regulation of locomotion | 171 | 720 | 9.89e-08 |
| GO:BP | GO:0030334 | regulation of cell migration | 161 | 668 | 1.01e-07 |
| GO:BP | GO:0003015 | heart process | 66 | 204 | 1.02e-07 |
| GO:BP | GO:0009987 | cellular process | 1718 | 10686 | 1.43e-07 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 381 | 1904 | 1.64e-07 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 377 | 1883 | 1.90e-07 |
| GO:BP | GO:0035239 | tube morphogenesis | 154 | 638 | 2.01e-07 |
| GO:BP | GO:0023057 | negative regulation of signaling | 228 | 1036 | 2.02e-07 |
| GO:BP | GO:0090257 | regulation of muscle system process | 59 | 177 | 2.08e-07 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 117 | 450 | 2.21e-07 |
| GO:BP | GO:0014706 | striated muscle tissue development | 67 | 213 | 2.52e-07 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 227 | 1035 | 2.86e-07 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 256 | 1197 | 2.95e-07 |
| GO:BP | GO:0008150 | biological_process | 1789 | 11244 | 2.95e-07 |
| GO:BP | GO:0035295 | tube development | 181 | 787 | 3.49e-07 |
| GO:BP | GO:0006941 | striated muscle contraction | 51 | 146 | 3.97e-07 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 233 | 1073 | 3.97e-07 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 226 | 1034 | 3.97e-07 |
| GO:BP | GO:0055001 | muscle cell development | 55 | 163 | 3.97e-07 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 149 | 622 | 5.42e-07 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 94 | 344 | 5.59e-07 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 275 | 1314 | 5.84e-07 |
| GO:BP | GO:0030048 | actin filament-based movement | 41 | 107 | 5.84e-07 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 43 | 116 | 7.97e-07 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 202 | 913 | 9.93e-07 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 84 | 300 | 1.03e-06 |
| GO:BP | GO:0001952 | regulation of cell-matrix adhesion | 36 | 90 | 1.26e-06 |
| GO:BP | GO:0043502 | regulation of muscle adaptation | 30 | 68 | 1.40e-06 |
| GO:BP | GO:0008016 | regulation of heart contraction | 54 | 165 | 1.61e-06 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 35 | 87 | 1.62e-06 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 62 | 201 | 1.79e-06 |
| GO:BP | GO:0090130 | tissue migration | 72 | 247 | 1.82e-06 |
| GO:BP | GO:0010631 | epithelial cell migration | 71 | 243 | 2.01e-06 |
| GO:BP | GO:0090132 | epithelium migration | 71 | 243 | 2.01e-06 |
| GO:BP | GO:0065008 | regulation of biological quality | 371 | 1893 | 2.51e-06 |
| GO:BP | GO:0032879 | regulation of localization | 293 | 1439 | 2.57e-06 |
| GO:BP | GO:1903522 | regulation of blood circulation | 60 | 194 | 2.57e-06 |
| GO:BP | GO:0043500 | muscle adaptation | 35 | 89 | 2.96e-06 |
| GO:BP | GO:0035556 | intracellular signal transduction | 380 | 1951 | 3.24e-06 |
| GO:BP | GO:0007507 | heart development | 122 | 499 | 3.38e-06 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 54 | 169 | 3.55e-06 |
| GO:BP | GO:0043542 | endothelial cell migration | 55 | 174 | 4.00e-06 |
| GO:BP | GO:0048468 | cell development | 368 | 1886 | 4.48e-06 |
| GO:BP | GO:0065009 | regulation of molecular function | 267 | 1299 | 4.64e-06 |
| GO:BP | GO:0031032 | actomyosin structure organization | 57 | 184 | 4.89e-06 |
| GO:BP | GO:0051049 | regulation of transport | 231 | 1098 | 5.92e-06 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 146 | 633 | 7.57e-06 |
| GO:BP | GO:0009605 | response to external stimulus | 310 | 1559 | 9.93e-06 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 159 | 706 | 1.01e-05 |
| GO:BP | GO:0001525 | angiogenesis | 97 | 381 | 1.05e-05 |
| GO:BP | GO:0032409 | regulation of transporter activity | 48 | 148 | 1.09e-05 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 361 | 1862 | 1.17e-05 |
| GO:BP | GO:0055006 | cardiac cell development | 31 | 78 | 1.17e-05 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 27 | 63 | 1.18e-05 |
| GO:BP | GO:0007162 | negative regulation of cell adhesion | 56 | 185 | 1.39e-05 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 35 | 95 | 1.68e-05 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 29 | 72 | 2.03e-05 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 67 | 239 | 2.22e-05 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 217 | 1040 | 2.67e-05 |
| GO:BP | GO:0034330 | cell junction organization | 127 | 545 | 2.67e-05 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 37 | 105 | 2.70e-05 |
| GO:BP | GO:0009611 | response to wounding | 95 | 382 | 4.06e-05 |
| GO:BP | GO:0010243 | response to organonitrogen compound | 152 | 685 | 4.46e-05 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 222 | 1077 | 4.88e-05 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 78 | 298 | 4.92e-05 |
| GO:BP | GO:0007010 | cytoskeleton organization | 234 | 1146 | 4.98e-05 |
| GO:BP | GO:0042221 | response to chemical | 430 | 2304 | 5.06e-05 |
| GO:BP | GO:0098657 | import into cell | 140 | 622 | 5.25e-05 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 55 | 188 | 5.37e-05 |
| GO:BP | GO:0150116 | regulation of cell-substrate junction organization | 25 | 60 | 5.67e-05 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 26 | 64 | 6.13e-05 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 149 | 673 | 6.19e-05 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 70 | 261 | 6.57e-05 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 137 | 609 | 6.82e-05 |
| GO:BP | GO:0097553 | calcium ion transmembrane import into cytosol | 40 | 122 | 7.12e-05 |
| GO:BP | GO:0030198 | extracellular matrix organization | 62 | 223 | 7.19e-05 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 74 | 282 | 7.77e-05 |
| GO:BP | GO:0043062 | extracellular structure organization | 62 | 224 | 8.30e-05 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 62 | 224 | 8.30e-05 |
| GO:BP | GO:0061337 | cardiac conduction | 31 | 85 | 8.66e-05 |
| GO:BP | GO:0014902 | myotube differentiation | 31 | 85 | 8.66e-05 |
| GO:BP | GO:0008283 | cell population proliferation | 260 | 1306 | 8.68e-05 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 107 | 452 | 8.70e-05 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 42 | 132 | 8.87e-05 |
| GO:BP | GO:0002040 | sprouting angiogenesis | 34 | 98 | 9.96e-05 |
| GO:BP | GO:0006936 | muscle contraction | 68 | 255 | 1.06e-04 |
| GO:BP | GO:0051283 | negative regulation of sequestering of calcium ion | 32 | 90 | 1.07e-04 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 54 | 188 | 1.10e-04 |
| GO:BP | GO:0009725 | response to hormone | 136 | 610 | 1.12e-04 |
| GO:BP | GO:0007015 | actin filament organization | 88 | 357 | 1.27e-04 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 107 | 456 | 1.28e-04 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 197 | 951 | 1.28e-04 |
| GO:BP | GO:0030038 | contractile actin filament bundle assembly | 32 | 91 | 1.35e-04 |
| GO:BP | GO:0043149 | stress fiber assembly | 32 | 91 | 1.35e-04 |
| GO:BP | GO:0007517 | muscle organ development | 70 | 267 | 1.37e-04 |
| GO:BP | GO:0042391 | regulation of membrane potential | 73 | 282 | 1.38e-04 |
| GO:BP | GO:0022898 | regulation of transmembrane transporter activity | 43 | 139 | 1.41e-04 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 102 | 431 | 1.41e-04 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 66 | 248 | 1.49e-04 |
| GO:BP | GO:1900024 | regulation of substrate adhesion-dependent cell spreading | 22 | 52 | 1.54e-04 |
| GO:BP | GO:0043534 | blood vessel endothelial cell migration | 32 | 92 | 1.67e-04 |
| GO:BP | GO:0051282 | regulation of sequestering of calcium ion | 32 | 92 | 1.67e-04 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 58 | 210 | 1.67e-04 |
| GO:BP | GO:0016310 | phosphorylation | 230 | 1145 | 1.71e-04 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 31 | 88 | 1.73e-04 |
| GO:BP | GO:0071310 | cellular response to organic substance | 256 | 1297 | 1.80e-04 |
| GO:BP | GO:0043270 | positive regulation of monoatomic ion transport | 40 | 127 | 1.80e-04 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 42 | 136 | 1.80e-04 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 131 | 591 | 2.11e-04 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 58 | 212 | 2.19e-04 |
| GO:BP | GO:0051209 | release of sequestered calcium ion into cytosol | 31 | 89 | 2.19e-04 |
| GO:BP | GO:0051899 | membrane depolarization | 23 | 57 | 2.26e-04 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 29 | 81 | 2.35e-04 |
| GO:BP | GO:0048041 | focal adhesion assembly | 28 | 77 | 2.40e-04 |
| GO:BP | GO:0006816 | calcium ion transport | 68 | 262 | 2.45e-04 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 64 | 243 | 2.75e-04 |
| GO:BP | GO:0032412 | regulation of monoatomic ion transmembrane transporter activity | 41 | 134 | 2.88e-04 |
| GO:BP | GO:0086065 | cell communication involved in cardiac conduction | 22 | 54 | 2.92e-04 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 59 | 219 | 2.92e-04 |
| GO:BP | GO:0002028 | regulation of sodium ion transport | 23 | 58 | 3.06e-04 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 331 | 1751 | 3.13e-04 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 57 | 210 | 3.22e-04 |
| GO:BP | GO:0010811 | positive regulation of cell-substrate adhesion | 32 | 95 | 3.23e-04 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 58 | 215 | 3.25e-04 |
| GO:BP | GO:0000902 | cell morphogenesis | 152 | 713 | 3.30e-04 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 112 | 495 | 3.67e-04 |
| GO:BP | GO:1901652 | response to peptide | 88 | 368 | 3.75e-04 |
| GO:BP | GO:0002027 | regulation of heart rate | 29 | 83 | 3.75e-04 |
| GO:BP | GO:0071417 | cellular response to organonitrogen compound | 102 | 442 | 3.80e-04 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 118 | 528 | 3.80e-04 |
| GO:BP | GO:0090109 | regulation of cell-substrate junction assembly | 22 | 55 | 3.86e-04 |
| GO:BP | GO:0051893 | regulation of focal adhesion assembly | 22 | 55 | 3.86e-04 |
| GO:BP | GO:0051235 | maintenance of location | 65 | 251 | 3.87e-04 |
| GO:BP | GO:0051208 | sequestering of calcium ion | 32 | 96 | 3.90e-04 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 109 | 480 | 3.90e-04 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 93 | 395 | 3.92e-04 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 39 | 127 | 3.94e-04 |
| GO:BP | GO:0006897 | endocytosis | 111 | 492 | 4.32e-04 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 218 | 1093 | 4.32e-04 |
| GO:BP | GO:1902305 | regulation of sodium ion transmembrane transport | 17 | 37 | 4.41e-04 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 61 | 233 | 4.85e-04 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 95 | 408 | 4.92e-04 |
| GO:BP | GO:1901698 | response to nitrogen compound | 155 | 736 | 4.92e-04 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 75 | 304 | 5.04e-04 |
| GO:BP | GO:0010812 | negative regulation of cell-substrate adhesion | 18 | 41 | 5.16e-04 |
| GO:BP | GO:0086010 | membrane depolarization during action potential | 13 | 24 | 5.16e-04 |
| GO:BP | GO:0046486 | glycerolipid metabolic process | 71 | 284 | 5.24e-04 |
| GO:BP | GO:0001508 | action potential | 33 | 102 | 5.41e-04 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 99 | 431 | 5.69e-04 |
| GO:BP | GO:0032231 | regulation of actin filament bundle assembly | 29 | 85 | 5.69e-04 |
| GO:BP | GO:0061448 | connective tissue development | 55 | 205 | 5.78e-04 |
| GO:BP | GO:0051492 | regulation of stress fiber assembly | 27 | 77 | 6.23e-04 |
| GO:BP | GO:1903053 | regulation of extracellular matrix organization | 20 | 49 | 6.23e-04 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 96 | 416 | 6.23e-04 |
| GO:BP | GO:0010644 | cell communication by electrical coupling | 15 | 31 | 6.29e-04 |
| GO:BP | GO:0070296 | sarcoplasmic reticulum calcium ion transport | 15 | 31 | 6.29e-04 |
| GO:BP | GO:0051924 | regulation of calcium ion transport | 44 | 153 | 6.33e-04 |
| GO:BP | GO:0006470 | protein dephosphorylation | 49 | 177 | 6.61e-04 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 99 | 433 | 6.68e-04 |
| GO:BP | GO:0070848 | response to growth factor | 120 | 547 | 6.85e-04 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 250 | 1289 | 6.88e-04 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 308 | 1633 | 6.95e-04 |
| GO:BP | GO:0014808 | release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 14 | 28 | 7.29e-04 |
| GO:BP | GO:1903514 | release of sequestered calcium ion into cytosol by endoplasmic reticulum | 14 | 28 | 7.29e-04 |
| GO:BP | GO:0010033 | response to organic substance | 325 | 1736 | 7.30e-04 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 57 | 217 | 7.46e-04 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 115 | 521 | 7.47e-04 |
| GO:BP | GO:0010762 | regulation of fibroblast migration | 16 | 35 | 7.65e-04 |
| GO:BP | GO:0051050 | positive regulation of transport | 124 | 571 | 7.86e-04 |
| GO:BP | GO:0043434 | response to peptide hormone | 76 | 314 | 8.03e-04 |
| GO:BP | GO:0007399 | nervous system development | 331 | 1775 | 8.22e-04 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 130 | 605 | 8.35e-04 |
| GO:BP | GO:0034446 | substrate adhesion-dependent cell spreading | 29 | 87 | 8.44e-04 |
| GO:BP | GO:0110020 | regulation of actomyosin structure organization | 29 | 87 | 8.44e-04 |
| GO:BP | GO:0006810 | transport | 528 | 2984 | 8.99e-04 |
| GO:BP | GO:0019722 | calcium-mediated signaling | 37 | 123 | 9.22e-04 |
| GO:BP | GO:0001935 | endothelial cell proliferation | 35 | 114 | 9.31e-04 |
| GO:BP | GO:0030001 | metal ion transport | 115 | 524 | 9.31e-04 |
| GO:BP | GO:0016043 | cellular component organization | 815 | 4802 | 1.00e-03 |
| GO:BP | GO:0040007 | growth | 146 | 697 | 1.00e-03 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 73 | 301 | 1.03e-03 |
| GO:BP | GO:0051928 | positive regulation of calcium ion transport | 25 | 71 | 1.04e-03 |
| GO:BP | GO:2001257 | regulation of cation channel activity | 24 | 67 | 1.08e-03 |
| GO:BP | GO:0010595 | positive regulation of endothelial cell migration | 29 | 89 | 1.31e-03 |
| GO:BP | GO:1903055 | positive regulation of extracellular matrix organization | 13 | 26 | 1.32e-03 |
| GO:BP | GO:0048384 | retinoic acid receptor signaling pathway | 13 | 26 | 1.32e-03 |
| GO:BP | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 13 | 26 | 1.32e-03 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 104 | 469 | 1.40e-03 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 164 | 804 | 1.40e-03 |
| GO:BP | GO:1904064 | positive regulation of cation transmembrane transport | 27 | 81 | 1.49e-03 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 164 | 805 | 1.49e-03 |
| GO:BP | GO:0006650 | glycerophospholipid metabolic process | 58 | 228 | 1.55e-03 |
| GO:BP | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway | 9 | 14 | 1.59e-03 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 38 | 131 | 1.64e-03 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 110 | 504 | 1.66e-03 |
| GO:BP | GO:0010649 | regulation of cell communication by electrical coupling | 10 | 17 | 1.71e-03 |
| GO:BP | GO:1902105 | regulation of leukocyte differentiation | 51 | 194 | 1.73e-03 |
| GO:BP | GO:0006468 | protein phosphorylation | 190 | 958 | 1.83e-03 |
| GO:BP | GO:0042060 | wound healing | 70 | 291 | 1.84e-03 |
| GO:BP | GO:0061572 | actin filament bundle organization | 38 | 132 | 1.92e-03 |
| GO:BP | GO:0043010 | camera-type eye development | 58 | 230 | 1.95e-03 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 105 | 479 | 1.99e-03 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 171 | 850 | 1.99e-03 |
| GO:BP | GO:0002521 | leukocyte differentiation | 85 | 371 | 2.00e-03 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 36 | 123 | 2.00e-03 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 69 | 287 | 2.06e-03 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 46 | 171 | 2.10e-03 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 282 | 1504 | 2.18e-03 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 159 | 783 | 2.18e-03 |
| GO:BP | GO:0006950 | response to stress | 452 | 2540 | 2.26e-03 |
| GO:BP | GO:0006812 | monoatomic cation transport | 129 | 614 | 2.28e-03 |
| GO:BP | GO:0001954 | positive regulation of cell-matrix adhesion | 17 | 42 | 2.40e-03 |
| GO:BP | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling | 12 | 24 | 2.40e-03 |
| GO:BP | GO:0002761 | regulation of myeloid leukocyte differentiation | 26 | 79 | 2.41e-03 |
| GO:BP | GO:0016311 | dephosphorylation | 58 | 232 | 2.41e-03 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 124 | 587 | 2.41e-03 |
| GO:BP | GO:0022008 | neurogenesis | 239 | 1251 | 2.43e-03 |
| GO:BP | GO:0001654 | eye development | 64 | 263 | 2.46e-03 |
| GO:BP | GO:0001501 | skeletal system development | 80 | 347 | 2.48e-03 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 28 | 88 | 2.49e-03 |
| GO:BP | GO:0048660 | regulation of smooth muscle cell proliferation | 30 | 97 | 2.51e-03 |
| GO:BP | GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 68 | 284 | 2.51e-03 |
| GO:BP | GO:0006915 | apoptotic process | 260 | 1377 | 2.55e-03 |
| GO:BP | GO:0045655 | regulation of monocyte differentiation | 8 | 12 | 2.66e-03 |
| GO:BP | GO:1900025 | negative regulation of substrate adhesion-dependent cell spreading | 8 | 12 | 2.66e-03 |
| GO:BP | GO:0043535 | regulation of blood vessel endothelial cell migration | 24 | 71 | 2.67e-03 |
| GO:BP | GO:0086012 | membrane depolarization during cardiac muscle cell action potential | 11 | 21 | 2.67e-03 |
| GO:BP | GO:0150063 | visual system development | 64 | 264 | 2.68e-03 |
| GO:BP | GO:0048592 | eye morphogenesis | 32 | 107 | 2.95e-03 |
| GO:BP | GO:1990845 | adaptive thermogenesis | 35 | 121 | 2.98e-03 |
| GO:BP | GO:1905709 | negative regulation of membrane permeability | 9 | 15 | 2.98e-03 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 177 | 893 | 3.06e-03 |
| GO:BP | GO:0051279 | regulation of release of sequestered calcium ion into cytosol | 21 | 59 | 3.09e-03 |
| GO:BP | GO:0051216 | cartilage development | 40 | 145 | 3.10e-03 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 36 | 126 | 3.11e-03 |
| GO:BP | GO:0006629 | lipid metabolic process | 182 | 923 | 3.21e-03 |
| GO:BP | GO:0048880 | sensory system development | 64 | 266 | 3.28e-03 |
| GO:BP | GO:0098609 | cell-cell adhesion | 119 | 564 | 3.28e-03 |
| GO:BP | GO:0051346 | negative regulation of hydrolase activity | 34 | 117 | 3.28e-03 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 211 | 1093 | 3.29e-03 |
| GO:BP | GO:0060284 | regulation of cell development | 120 | 570 | 3.35e-03 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 53 | 210 | 3.38e-03 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 35 | 122 | 3.43e-03 |
| GO:BP | GO:0032989 | cellular anatomical entity morphogenesis | 123 | 588 | 3.64e-03 |
| GO:BP | GO:0048771 | tissue remodeling | 33 | 113 | 3.64e-03 |
| GO:BP | GO:0000768 | syncytium formation by plasma membrane fusion | 15 | 36 | 3.80e-03 |
| GO:BP | GO:0140253 | cell-cell fusion | 15 | 36 | 3.80e-03 |
| GO:BP | GO:0085029 | extracellular matrix assembly | 15 | 36 | 3.80e-03 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 21 | 60 | 3.85e-03 |
| GO:BP | GO:0007229 | integrin-mediated signaling pathway | 27 | 86 | 3.85e-03 |
| GO:BP | GO:0035725 | sodium ion transmembrane transport | 29 | 95 | 3.85e-03 |
| GO:BP | GO:0051179 | localization | 639 | 3731 | 4.07e-03 |
| GO:BP | GO:0033002 | muscle cell proliferation | 41 | 152 | 4.11e-03 |
| GO:BP | GO:0030097 | hemopoiesis | 129 | 624 | 4.12e-03 |
| GO:BP | GO:0051234 | establishment of localization | 561 | 3240 | 4.12e-03 |
| GO:BP | GO:0086002 | cardiac muscle cell action potential involved in contraction | 17 | 44 | 4.12e-03 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 70 | 300 | 4.12e-03 |
| GO:BP | GO:0034767 | positive regulation of monoatomic ion transmembrane transport | 28 | 91 | 4.20e-03 |
| GO:BP | GO:0007520 | myoblast fusion | 13 | 29 | 4.22e-03 |
| GO:BP | GO:2000649 | regulation of sodium ion transmembrane transporter activity | 13 | 29 | 4.22e-03 |
| GO:BP | GO:0150117 | positive regulation of cell-substrate junction organization | 13 | 29 | 4.22e-03 |
| GO:BP | GO:0036293 | response to decreased oxygen levels | 59 | 243 | 4.25e-03 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 159 | 796 | 4.26e-03 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 231 | 1218 | 4.41e-03 |
| GO:BP | GO:0048593 | camera-type eye morphogenesis | 27 | 87 | 4.57e-03 |
| GO:BP | GO:0002376 | immune system process | 263 | 1410 | 4.67e-03 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 49 | 193 | 4.76e-03 |
| GO:BP | GO:0061614 | miRNA transcription | 21 | 61 | 4.76e-03 |
| GO:BP | GO:0070482 | response to oxygen levels | 63 | 265 | 4.78e-03 |
| GO:BP | GO:0042592 | homeostatic process | 217 | 1137 | 4.78e-03 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 24 | 74 | 4.80e-03 |
| GO:BP | GO:0048659 | smooth muscle cell proliferation | 30 | 101 | 4.80e-03 |
| GO:BP | GO:1901184 | regulation of ERBB signaling pathway | 20 | 57 | 4.95e-03 |
| GO:BP | GO:0023056 | positive regulation of signaling | 231 | 1221 | 4.99e-03 |
| GO:BP | GO:0014874 | response to stimulus involved in regulation of muscle adaptation | 8 | 13 | 5.08e-03 |
| GO:BP | GO:0008360 | regulation of cell shape | 31 | 106 | 5.08e-03 |
| GO:BP | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction | 12 | 26 | 5.14e-03 |
| GO:BP | GO:0012501 | programmed cell death | 264 | 1419 | 5.20e-03 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 111 | 527 | 5.24e-03 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 49 | 194 | 5.25e-03 |
| GO:BP | GO:0043536 | positive regulation of blood vessel endothelial cell migration | 17 | 45 | 5.25e-03 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 376 | 2100 | 5.37e-03 |
| GO:BP | GO:0030030 | cell projection organization | 224 | 1182 | 5.54e-03 |
| GO:BP | GO:0007264 | small GTPase-mediated signal transduction | 86 | 390 | 5.64e-03 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 90 | 412 | 5.70e-03 |
| GO:BP | GO:0014896 | muscle hypertrophy | 24 | 75 | 5.80e-03 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 56 | 231 | 5.97e-03 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 61 | 257 | 5.97e-03 |
| GO:BP | GO:0008219 | cell death | 264 | 1423 | 6.07e-03 |
| GO:BP | GO:1901653 | cellular response to peptide | 64 | 273 | 6.11e-03 |
| GO:BP | GO:0030224 | monocyte differentiation | 11 | 23 | 6.15e-03 |
| GO:BP | GO:1902894 | negative regulation of miRNA transcription | 11 | 23 | 6.15e-03 |
| GO:BP | GO:0006644 | phospholipid metabolic process | 67 | 289 | 6.15e-03 |
| GO:BP | GO:0002026 | regulation of the force of heart contraction | 11 | 23 | 6.15e-03 |
| GO:BP | GO:0009790 | embryo development | 162 | 821 | 6.25e-03 |
| GO:BP | GO:1901019 | regulation of calcium ion transmembrane transporter activity | 18 | 50 | 6.67e-03 |
| GO:BP | GO:0051651 | maintenance of location in cell | 44 | 171 | 6.68e-03 |
| GO:BP | GO:0045668 | negative regulation of osteoblast differentiation | 15 | 38 | 6.69e-03 |
| GO:BP | GO:0006949 | syncytium formation | 15 | 38 | 6.69e-03 |
| GO:BP | GO:0035265 | organ growth | 35 | 127 | 6.82e-03 |
| GO:BP | GO:1903169 | regulation of calcium ion transmembrane transport | 31 | 108 | 6.83e-03 |
| GO:BP | GO:0014894 | response to denervation involved in regulation of muscle adaptation | 6 | 8 | 6.83e-03 |
| GO:BP | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation | 6 | 8 | 6.83e-03 |
| GO:BP | GO:0006811 | monoatomic ion transport | 144 | 719 | 6.85e-03 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 218 | 1152 | 6.92e-03 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 459 | 2622 | 7.03e-03 |
| GO:BP | GO:0032411 | positive regulation of transporter activity | 21 | 63 | 7.08e-03 |
| GO:BP | GO:0001558 | regulation of cell growth | 73 | 323 | 7.21e-03 |
| GO:BP | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process | 10 | 20 | 7.22e-03 |
| GO:BP | GO:1901021 | positive regulation of calcium ion transmembrane transporter activity | 12 | 27 | 7.25e-03 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 115 | 555 | 7.26e-03 |
| GO:BP | GO:0038127 | ERBB signaling pathway | 30 | 104 | 7.53e-03 |
| GO:BP | GO:0048878 | chemical homeostasis | 135 | 670 | 7.93e-03 |
| GO:BP | GO:0090559 | regulation of membrane permeability | 19 | 55 | 7.98e-03 |
| GO:BP | GO:1901342 | regulation of vasculature development | 49 | 198 | 7.98e-03 |
| GO:BP | GO:0008154 | actin polymerization or depolymerization | 39 | 148 | 8.26e-03 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 44 | 173 | 8.32e-03 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 152 | 769 | 8.41e-03 |
| GO:BP | GO:0040008 | regulation of growth | 98 | 462 | 8.42e-03 |
| GO:BP | GO:0014743 | regulation of muscle hypertrophy | 17 | 47 | 8.62e-03 |
| GO:BP | GO:0030010 | establishment of cell polarity | 34 | 124 | 8.68e-03 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 23 | 73 | 9.07e-03 |
| GO:BP | GO:0048661 | positive regulation of smooth muscle cell proliferation | 20 | 60 | 9.37e-03 |
| GO:BP | GO:0001936 | regulation of endothelial cell proliferation | 28 | 96 | 9.37e-03 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 32 | 115 | 9.45e-03 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 32 | 115 | 9.45e-03 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 77 | 348 | 9.45e-03 |
| GO:BP | GO:1990849 | vacuolar localization | 19 | 56 | 9.96e-03 |
| GO:BP | GO:0032418 | lysosome localization | 19 | 56 | 9.96e-03 |
| GO:BP | GO:1902904 | negative regulation of supramolecular fiber organization | 34 | 125 | 9.96e-03 |
| GO:BP | GO:0034764 | positive regulation of transmembrane transport | 35 | 130 | 1.02e-02 |
| GO:BP | GO:1903670 | regulation of sprouting angiogenesis | 12 | 28 | 1.03e-02 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 52 | 216 | 1.04e-02 |
| GO:BP | GO:1904427 | positive regulation of calcium ion transmembrane transport | 18 | 52 | 1.05e-02 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 216 | 1150 | 1.05e-02 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 21 | 65 | 1.06e-02 |
| GO:BP | GO:0001666 | response to hypoxia | 55 | 232 | 1.07e-02 |
| GO:BP | GO:0009991 | response to extracellular stimulus | 82 | 377 | 1.07e-02 |
| GO:BP | GO:0045834 | positive regulation of lipid metabolic process | 25 | 83 | 1.07e-02 |
| GO:BP | GO:0032787 | monocarboxylic acid metabolic process | 89 | 416 | 1.10e-02 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 829 | 4988 | 1.13e-02 |
| GO:BP | GO:0044255 | cellular lipid metabolic process | 138 | 694 | 1.13e-02 |
| GO:BP | GO:0141091 | transforming growth factor beta receptor superfamily signaling pathway | 60 | 259 | 1.13e-02 |
| GO:BP | GO:0003254 | regulation of membrane depolarization | 14 | 36 | 1.13e-02 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 56 | 238 | 1.14e-02 |
| GO:BP | GO:0045165 | cell fate commitment | 39 | 151 | 1.18e-02 |
| GO:BP | GO:0003205 | cardiac chamber development | 39 | 151 | 1.18e-02 |
| GO:BP | GO:0048589 | developmental growth | 104 | 501 | 1.18e-02 |
| GO:BP | GO:0007267 | cell-cell signaling | 204 | 1082 | 1.20e-02 |
| GO:BP | GO:0106106 | cold-induced thermogenesis | 31 | 112 | 1.21e-02 |
| GO:BP | GO:1903589 | positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 4 | 4 | 1.22e-02 |
| GO:BP | GO:0008064 | regulation of actin polymerization or depolymerization | 32 | 117 | 1.24e-02 |
| GO:BP | GO:0051494 | negative regulation of cytoskeleton organization | 34 | 127 | 1.29e-02 |
| GO:BP | GO:2000629 | negative regulation of miRNA metabolic process | 11 | 25 | 1.29e-02 |
| GO:BP | GO:0006658 | phosphatidylserine metabolic process | 9 | 18 | 1.29e-02 |
| GO:BP | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis | 9 | 18 | 1.29e-02 |
| GO:BP | GO:0060402 | calcium ion transport into cytosol | 9 | 18 | 1.29e-02 |
| GO:BP | GO:0043304 | regulation of mast cell degranulation | 9 | 18 | 1.29e-02 |
| GO:BP | GO:0010743 | regulation of macrophage derived foam cell differentiation | 9 | 18 | 1.29e-02 |
| GO:BP | GO:0006814 | sodium ion transport | 36 | 137 | 1.30e-02 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 195 | 1031 | 1.31e-02 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 347 | 1951 | 1.34e-02 |
| GO:BP | GO:0032233 | positive regulation of actin filament bundle assembly | 17 | 49 | 1.35e-02 |
| GO:BP | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 24 | 80 | 1.37e-02 |
| GO:BP | GO:0086005 | ventricular cardiac muscle cell action potential | 12 | 29 | 1.39e-02 |
| GO:BP | GO:0006096 | glycolytic process | 22 | 71 | 1.39e-02 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 344 | 1934 | 1.39e-02 |
| GO:BP | GO:0030832 | regulation of actin filament length | 32 | 118 | 1.40e-02 |
| GO:BP | GO:0031667 | response to nutrient levels | 77 | 354 | 1.45e-02 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 63 | 278 | 1.45e-02 |
| GO:BP | GO:0006909 | phagocytosis | 36 | 138 | 1.46e-02 |
| GO:BP | GO:0032414 | positive regulation of ion transmembrane transporter activity | 19 | 58 | 1.47e-02 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 65 | 289 | 1.47e-02 |
| GO:BP | GO:0016049 | cell growth | 82 | 382 | 1.48e-02 |
| GO:BP | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity | 6 | 9 | 1.48e-02 |
| GO:BP | GO:0014745 | negative regulation of muscle adaptation | 6 | 9 | 1.48e-02 |
| GO:BP | GO:0043306 | positive regulation of mast cell degranulation | 6 | 9 | 1.48e-02 |
| GO:BP | GO:0014870 | response to muscle inactivity | 6 | 9 | 1.48e-02 |
| GO:BP | GO:0019222 | regulation of metabolic process | 767 | 4603 | 1.52e-02 |
| GO:BP | GO:0007266 | Rho protein signal transduction | 30 | 109 | 1.52e-02 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 50 | 210 | 1.54e-02 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 288 | 1595 | 1.55e-02 |
| GO:BP | GO:0071871 | response to epinephrine | 7 | 12 | 1.56e-02 |
| GO:BP | GO:0034329 | cell junction assembly | 71 | 323 | 1.64e-02 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 22 | 72 | 1.65e-02 |
| GO:BP | GO:0010761 | fibroblast migration | 17 | 50 | 1.67e-02 |
| GO:BP | GO:0048008 | platelet-derived growth factor receptor signaling pathway | 17 | 50 | 1.67e-02 |
| GO:BP | GO:0001775 | cell activation | 125 | 627 | 1.67e-02 |
| GO:BP | GO:0009179 | purine ribonucleoside diphosphate metabolic process | 25 | 86 | 1.68e-02 |
| GO:BP | GO:0009135 | purine nucleoside diphosphate metabolic process | 25 | 86 | 1.68e-02 |
| GO:BP | GO:0007173 | epidermal growth factor receptor signaling pathway | 25 | 86 | 1.68e-02 |
| GO:BP | GO:1905952 | regulation of lipid localization | 30 | 110 | 1.74e-02 |
| GO:BP | GO:0046902 | regulation of mitochondrial membrane permeability | 16 | 46 | 1.75e-02 |
| GO:BP | GO:0050654 | chondroitin sulfate proteoglycan metabolic process | 11 | 26 | 1.75e-02 |
| GO:BP | GO:0071407 | cellular response to organic cyclic compound | 81 | 379 | 1.75e-02 |
| GO:BP | GO:0010611 | regulation of cardiac muscle hypertrophy | 16 | 46 | 1.75e-02 |
| GO:BP | GO:0045637 | regulation of myeloid cell differentiation | 35 | 135 | 1.82e-02 |
| GO:BP | GO:0031529 | ruffle organization | 15 | 42 | 1.82e-02 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 80 | 374 | 1.82e-02 |
| GO:BP | GO:0086009 | membrane repolarization | 15 | 42 | 1.82e-02 |
| GO:BP | GO:0030099 | myeloid cell differentiation | 65 | 292 | 1.83e-02 |
| GO:BP | GO:0001974 | blood vessel remodeling | 12 | 30 | 1.85e-02 |
| GO:BP | GO:0098900 | regulation of action potential | 14 | 38 | 1.87e-02 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 147 | 757 | 1.87e-02 |
| GO:BP | GO:0010662 | regulation of striated muscle cell apoptotic process | 13 | 34 | 1.88e-02 |
| GO:BP | GO:1901201 | regulation of extracellular matrix assembly | 9 | 19 | 1.90e-02 |
| GO:BP | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion | 9 | 19 | 1.90e-02 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 48 | 202 | 1.91e-02 |
| GO:BP | GO:0045017 | glycerolipid biosynthetic process | 48 | 202 | 1.91e-02 |
| GO:BP | GO:0046032 | ADP catabolic process | 22 | 73 | 1.91e-02 |
| GO:BP | GO:0120161 | regulation of cold-induced thermogenesis | 30 | 111 | 1.95e-02 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 156 | 811 | 1.98e-02 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 134 | 683 | 2.02e-02 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 198 | 1060 | 2.02e-02 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 64 | 288 | 2.03e-02 |
| GO:BP | GO:0007423 | sensory organ development | 82 | 387 | 2.04e-02 |
| GO:BP | GO:0046031 | ADP metabolic process | 23 | 78 | 2.04e-02 |
| GO:BP | GO:0120162 | positive regulation of cold-induced thermogenesis | 21 | 69 | 2.11e-02 |
| GO:BP | GO:0014909 | smooth muscle cell migration | 19 | 60 | 2.11e-02 |
| GO:BP | GO:0030833 | regulation of actin filament polymerization | 28 | 102 | 2.15e-02 |
| GO:BP | GO:0034763 | negative regulation of transmembrane transport | 24 | 83 | 2.15e-02 |
| GO:BP | GO:0003416 | endochondral bone growth | 10 | 23 | 2.19e-02 |
| GO:BP | GO:0010765 | positive regulation of sodium ion transport | 10 | 23 | 2.19e-02 |
| GO:BP | GO:0030162 | regulation of proteolysis | 87 | 416 | 2.19e-02 |
| GO:BP | GO:0002573 | myeloid leukocyte differentiation | 38 | 152 | 2.24e-02 |
| GO:BP | GO:0019725 | cellular homeostasis | 113 | 564 | 2.25e-02 |
| GO:BP | GO:0051496 | positive regulation of stress fiber assembly | 15 | 43 | 2.27e-02 |
| GO:BP | GO:0032272 | negative regulation of protein polymerization | 20 | 65 | 2.31e-02 |
| GO:BP | GO:0035023 | regulation of Rho protein signal transduction | 20 | 65 | 2.31e-02 |
| GO:BP | GO:0048699 | generation of neurons | 200 | 1076 | 2.36e-02 |
| GO:BP | GO:0062012 | regulation of small molecule metabolic process | 53 | 231 | 2.42e-02 |
| GO:BP | GO:0045657 | positive regulation of monocyte differentiation | 5 | 7 | 2.49e-02 |
| GO:BP | GO:0002064 | epithelial cell development | 37 | 148 | 2.54e-02 |
| GO:BP | GO:0031399 | regulation of protein modification process | 172 | 911 | 2.54e-02 |
| GO:BP | GO:0007584 | response to nutrient | 30 | 113 | 2.54e-02 |
| GO:BP | GO:1905475 | regulation of protein localization to membrane | 36 | 143 | 2.54e-02 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 35 | 138 | 2.56e-02 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 181 | 965 | 2.59e-02 |
| GO:BP | GO:0035795 | negative regulation of mitochondrial membrane permeability | 7 | 13 | 2.61e-02 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 122 | 619 | 2.65e-02 |
| GO:BP | GO:0009137 | purine nucleoside diphosphate catabolic process | 22 | 75 | 2.65e-02 |
| GO:BP | GO:0019364 | pyridine nucleotide catabolic process | 22 | 75 | 2.65e-02 |
| GO:BP | GO:0009181 | purine ribonucleoside diphosphate catabolic process | 22 | 75 | 2.65e-02 |
| GO:BP | GO:0006082 | organic acid metabolic process | 123 | 625 | 2.68e-02 |
| GO:BP | GO:0071872 | cellular response to epinephrine stimulus | 6 | 10 | 2.76e-02 |
| GO:BP | GO:0032530 | regulation of microvillus organization | 6 | 10 | 2.76e-02 |
| GO:BP | GO:0033275 | actin-myosin filament sliding | 6 | 10 | 2.76e-02 |
| GO:BP | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity | 9 | 20 | 2.76e-02 |
| GO:BP | GO:1903671 | negative regulation of sprouting angiogenesis | 6 | 10 | 2.76e-02 |
| GO:BP | GO:0031099 | regeneration | 36 | 144 | 2.81e-02 |
| GO:BP | GO:0070613 | regulation of protein processing | 15 | 44 | 2.81e-02 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 56 | 249 | 2.81e-02 |
| GO:BP | GO:0002042 | cell migration involved in sprouting angiogenesis | 15 | 44 | 2.81e-02 |
| GO:BP | GO:0010876 | lipid localization | 65 | 298 | 2.82e-02 |
| GO:BP | GO:0043086 | negative regulation of catalytic activity | 65 | 298 | 2.82e-02 |
| GO:BP | GO:0030041 | actin filament polymerization | 32 | 124 | 2.84e-02 |
| GO:BP | GO:0043409 | negative regulation of MAPK cascade | 32 | 124 | 2.84e-02 |
| GO:BP | GO:0007519 | skeletal muscle tissue development | 31 | 119 | 2.84e-02 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 24 | 85 | 2.86e-02 |
| GO:BP | GO:0032271 | regulation of protein polymerization | 39 | 160 | 3.00e-02 |
| GO:BP | GO:0045923 | positive regulation of fatty acid metabolic process | 10 | 24 | 3.01e-02 |
| GO:BP | GO:0098868 | bone growth | 10 | 24 | 3.01e-02 |
| GO:BP | GO:0002763 | positive regulation of myeloid leukocyte differentiation | 13 | 36 | 3.06e-02 |
| GO:BP | GO:0048662 | negative regulation of smooth muscle cell proliferation | 13 | 36 | 3.06e-02 |
| GO:BP | GO:2001259 | positive regulation of cation channel activity | 13 | 36 | 3.06e-02 |
| GO:BP | GO:0006869 | lipid transport | 58 | 261 | 3.06e-02 |
| GO:BP | GO:0060429 | epithelium development | 151 | 792 | 3.09e-02 |
| GO:BP | GO:0050878 | regulation of body fluid levels | 52 | 229 | 3.15e-02 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 29 | 110 | 3.17e-02 |
| GO:BP | GO:0071805 | potassium ion transmembrane transport | 31 | 120 | 3.20e-02 |
| GO:BP | GO:1904705 | regulation of vascular associated smooth muscle cell proliferation | 16 | 49 | 3.21e-02 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 61 | 278 | 3.23e-02 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 119 | 606 | 3.24e-02 |
| GO:BP | GO:0019752 | carboxylic acid metabolic process | 119 | 606 | 3.24e-02 |
| GO:BP | GO:0048844 | artery morphogenesis | 18 | 58 | 3.27e-02 |
| GO:BP | GO:0010955 | negative regulation of protein processing | 8 | 17 | 3.34e-02 |
| GO:BP | GO:0030206 | chondroitin sulfate biosynthetic process | 8 | 17 | 3.34e-02 |
| GO:BP | GO:1903318 | negative regulation of protein maturation | 8 | 17 | 3.34e-02 |
| GO:BP | GO:0034383 | low-density lipoprotein particle clearance | 8 | 17 | 3.34e-02 |
| GO:BP | GO:0006090 | pyruvate metabolic process | 25 | 91 | 3.38e-02 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 83 | 401 | 3.41e-02 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 125 | 642 | 3.41e-02 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 89 | 435 | 3.42e-02 |
| GO:BP | GO:0009185 | ribonucleoside diphosphate metabolic process | 26 | 96 | 3.44e-02 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 181 | 973 | 3.47e-02 |
| GO:BP | GO:0097191 | extrinsic apoptotic signaling pathway | 41 | 172 | 3.47e-02 |
| GO:BP | GO:0051258 | protein polymerization | 51 | 225 | 3.48e-02 |
| GO:BP | GO:0009132 | nucleoside diphosphate metabolic process | 27 | 101 | 3.48e-02 |
| GO:BP | GO:0002262 | myeloid cell homeostasis | 34 | 136 | 3.48e-02 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 669 | 4015 | 3.51e-02 |
| GO:BP | GO:0014910 | regulation of smooth muscle cell migration | 17 | 54 | 3.53e-02 |
| GO:BP | GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 30 | 116 | 3.54e-02 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 43 | 183 | 3.63e-02 |
| GO:BP | GO:0031638 | zymogen activation | 14 | 41 | 3.63e-02 |
| GO:BP | GO:0055085 | transmembrane transport | 172 | 921 | 3.69e-02 |
| GO:BP | GO:0045216 | cell-cell junction organization | 37 | 152 | 3.72e-02 |
| GO:BP | GO:0019915 | lipid storage | 20 | 68 | 3.72e-02 |
| GO:BP | GO:0055002 | striated muscle cell development | 20 | 68 | 3.72e-02 |
| GO:BP | GO:0043436 | oxoacid metabolic process | 121 | 621 | 3.78e-02 |
| GO:BP | GO:0010658 | striated muscle cell apoptotic process | 13 | 37 | 3.81e-02 |
| GO:BP | GO:1903115 | regulation of actin filament-based movement | 13 | 37 | 3.81e-02 |
| GO:BP | GO:0010660 | regulation of muscle cell apoptotic process | 16 | 50 | 3.82e-02 |
| GO:BP | GO:0015918 | sterol transport | 25 | 92 | 3.82e-02 |
| GO:BP | GO:0070373 | negative regulation of ERK1 and ERK2 cascade | 16 | 50 | 3.82e-02 |
| GO:BP | GO:0035914 | skeletal muscle cell differentiation | 16 | 50 | 3.82e-02 |
| GO:BP | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis | 4 | 5 | 3.83e-02 |
| GO:BP | GO:1904261 | positive regulation of basement membrane assembly involved in embryonic body morphogenesis | 4 | 5 | 3.83e-02 |
| GO:BP | GO:2001197 | basement membrane assembly involved in embryonic body morphogenesis | 4 | 5 | 3.83e-02 |
| GO:BP | GO:2000669 | negative regulation of dendritic cell apoptotic process | 4 | 5 | 3.83e-02 |
| GO:BP | GO:1904978 | regulation of endosome organization | 4 | 5 | 3.83e-02 |
| GO:BP | GO:0021960 | anterior commissure morphogenesis | 4 | 5 | 3.83e-02 |
| GO:BP | GO:0046898 | response to cycloheximide | 4 | 5 | 3.83e-02 |
| GO:BP | GO:1902267 | regulation of polyamine transmembrane transport | 4 | 5 | 3.83e-02 |
| GO:BP | GO:0097190 | apoptotic signaling pathway | 94 | 466 | 3.85e-02 |
| GO:BP | GO:0006790 | sulfur compound metabolic process | 52 | 232 | 3.88e-02 |
| GO:BP | GO:0010927 | cellular component assembly involved in morphogenesis | 28 | 107 | 3.89e-02 |
| GO:BP | GO:0032232 | negative regulation of actin filament bundle assembly | 10 | 25 | 3.90e-02 |
| GO:BP | GO:1901203 | positive regulation of extracellular matrix assembly | 7 | 14 | 3.90e-02 |
| GO:BP | GO:0060142 | regulation of syncytium formation by plasma membrane fusion | 7 | 14 | 3.90e-02 |
| GO:BP | GO:1904738 | vascular associated smooth muscle cell migration | 10 | 25 | 3.90e-02 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 46 | 200 | 3.90e-02 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 402 | 2333 | 3.95e-02 |
| GO:BP | GO:0072526 | pyridine-containing compound catabolic process | 22 | 78 | 3.96e-02 |
| GO:BP | GO:0009191 | ribonucleoside diphosphate catabolic process | 22 | 78 | 3.96e-02 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 19 | 64 | 4.01e-02 |
| GO:BP | GO:1904036 | negative regulation of epithelial cell apoptotic process | 15 | 46 | 4.05e-02 |
| GO:BP | GO:0030182 | neuron differentiation | 187 | 1014 | 4.07e-02 |
| GO:BP | GO:0010657 | muscle cell apoptotic process | 17 | 55 | 4.12e-02 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 86 | 422 | 4.17e-02 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 378 | 2186 | 4.24e-02 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 87 | 428 | 4.25e-02 |
| GO:BP | GO:0060538 | skeletal muscle organ development | 32 | 128 | 4.29e-02 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 124 | 642 | 4.34e-02 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 53 | 239 | 4.36e-02 |
| GO:BP | GO:0022408 | negative regulation of cell-cell adhesion | 29 | 113 | 4.36e-02 |
| GO:BP | GO:1905954 | positive regulation of lipid localization | 21 | 74 | 4.42e-02 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 83 | 406 | 4.42e-02 |
| GO:BP | GO:0014854 | response to inactivity | 6 | 11 | 4.46e-02 |
| GO:BP | GO:0014733 | regulation of skeletal muscle adaptation | 6 | 11 | 4.46e-02 |
| GO:BP | GO:1901739 | regulation of myoblast fusion | 6 | 11 | 4.46e-02 |
| GO:BP | GO:0010172 | embryonic body morphogenesis | 6 | 11 | 4.46e-02 |
| GO:BP | GO:0021783 | preganglionic parasympathetic fiber development | 6 | 11 | 4.46e-02 |
| GO:BP | GO:1990874 | vascular associated smooth muscle cell proliferation | 16 | 51 | 4.47e-02 |
| GO:BP | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway | 16 | 51 | 4.47e-02 |
| GO:BP | GO:0030278 | regulation of ossification | 22 | 79 | 4.54e-02 |
| GO:BP | GO:0048666 | neuron development | 157 | 838 | 4.56e-02 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 62 | 289 | 4.56e-02 |
| GO:BP | GO:1901028 | regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 8 | 18 | 4.61e-02 |
| GO:BP | GO:0055093 | response to hyperoxia | 8 | 18 | 4.61e-02 |
| GO:BP | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway | 5 | 8 | 4.66e-02 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 414 | 2415 | 4.66e-02 |
| GO:BP | GO:0045861 | negative regulation of proteolysis | 32 | 129 | 4.73e-02 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 50 | 224 | 4.76e-02 |
| GO:BP | GO:0046434 | organophosphate catabolic process | 41 | 176 | 4.81e-02 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 30 | 119 | 4.81e-02 |
| GO:BP | GO:0062013 | positive regulation of small molecule metabolic process | 26 | 99 | 4.83e-02 |
| GO:BP | GO:0045766 | positive regulation of angiogenesis | 29 | 114 | 4.83e-02 |
| GO:BP | GO:0003229 | ventricular cardiac muscle tissue development | 17 | 56 | 4.83e-02 |
| GO:BP | GO:0002067 | glandular epithelial cell differentiation | 15 | 47 | 4.83e-02 |
| GO:BP | GO:0097006 | regulation of plasma lipoprotein particle levels | 15 | 47 | 4.83e-02 |
| GO:BP | GO:1903317 | regulation of protein maturation | 15 | 47 | 4.83e-02 |
| GO:BP | GO:0001938 | positive regulation of endothelial cell proliferation | 20 | 70 | 4.84e-02 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 107 | 546 | 4.91e-02 |
| GO:BP | GO:0001945 | lymph vessel development | 9 | 22 | 4.91e-02 |
| GO:BP | GO:1900027 | regulation of ruffle assembly | 9 | 22 | 4.91e-02 |
| GO:BP | GO:1903779 | regulation of cardiac conduction | 9 | 22 | 4.91e-02 |
| GO:BP | GO:0030279 | negative regulation of ossification | 9 | 22 | 4.91e-02 |
| GO:BP | GO:0033688 | regulation of osteoblast proliferation | 9 | 22 | 4.91e-02 |
| GO:BP | GO:0051497 | negative regulation of stress fiber assembly | 9 | 22 | 4.91e-02 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 35 | 145 | 4.92e-02 |
| GO:BP | GO:0032373 | positive regulation of sterol transport | 11 | 30 | 4.94e-02 |
| GO:BP | GO:0032376 | positive regulation of cholesterol transport | 11 | 30 | 4.94e-02 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 39 | 166 | 4.96e-02 |
| GO:BP | GO:1904035 | regulation of epithelial cell apoptotic process | 21 | 75 | 4.96e-02 |
| GO:BP | GO:0090288 | negative regulation of cellular response to growth factor stimulus | 21 | 75 | 4.96e-02 |
LRnomtable_20k_enh %>%
filter(source=="KEGG", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "KEGG terms found in LR enhancer neargenes_20k") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 43 | 124 | 2.31e-05 |
| KEGG | KEGG:05414 | Dilated cardiomyopathy | 32 | 82 | 2.50e-05 |
| KEGG | KEGG:05410 | Hypertrophic cardiomyopathy | 30 | 77 | 4.24e-05 |
| KEGG | KEGG:03320 | PPAR signaling pathway | 21 | 47 | 1.29e-04 |
| KEGG | KEGG:04510 | Focal adhesion | 48 | 169 | 4.37e-04 |
| KEGG | KEGG:05412 | Arrhythmogenic right ventricular cardiomyopathy | 25 | 67 | 4.37e-04 |
| KEGG | KEGG:04925 | Aldosterone synthesis and secretion | 27 | 75 | 4.37e-04 |
| KEGG | KEGG:04971 | Gastric acid secretion | 20 | 51 | 1.23e-03 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 63 | 252 | 1.35e-03 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 26 | 78 | 1.74e-03 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 62 | 250 | 1.74e-03 |
| KEGG | KEGG:05200 | Pathways in cancer | 88 | 391 | 2.40e-03 |
| KEGG | KEGG:04260 | Cardiac muscle contraction | 22 | 63 | 2.47e-03 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 31 | 104 | 3.08e-03 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 31 | 105 | 3.49e-03 |
| KEGG | KEGG:04512 | ECM-receptor interaction | 22 | 66 | 4.36e-03 |
| KEGG | KEGG:04927 | Cortisol synthesis and secretion | 17 | 46 | 5.32e-03 |
| KEGG | KEGG:04974 | Protein digestion and absorption | 19 | 56 | 8.00e-03 |
| KEGG | KEGG:04973 | Carbohydrate digestion and absorption | 12 | 29 | 1.11e-02 |
| KEGG | KEGG:04911 | Insulin secretion | 20 | 62 | 1.11e-02 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 34 | 129 | 1.28e-02 |
| KEGG | KEGG:04148 | Efferocytosis | 32 | 120 | 1.28e-02 |
| KEGG | KEGG:04931 | Insulin resistance | 26 | 91 | 1.28e-02 |
| KEGG | KEGG:04360 | Axon guidance | 39 | 155 | 1.28e-02 |
| KEGG | KEGG:05215 | Prostate cancer | 24 | 82 | 1.28e-02 |
| KEGG | KEGG:04020 | Calcium signaling pathway | 40 | 161 | 1.33e-02 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 40 | 162 | 1.41e-02 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 40 | 162 | 1.41e-02 |
| KEGG | KEGG:05203 | Viral carcinogenesis | 36 | 142 | 1.42e-02 |
| KEGG | KEGG:04972 | Pancreatic secretion | 18 | 57 | 1.74e-02 |
| KEGG | KEGG:05146 | Amoebiasis | 19 | 62 | 1.82e-02 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 39 | 160 | 1.82e-02 |
| KEGG | KEGG:00000 | KEGG root term | 893 | 5451 | 1.83e-02 |
| KEGG | KEGG:04750 | Inflammatory mediator regulation of TRP channels | 21 | 72 | 2.00e-02 |
| KEGG | KEGG:05222 | Small cell lung cancer | 23 | 82 | 2.15e-02 |
| KEGG | KEGG:04713 | Circadian entrainment | 20 | 68 | 2.15e-02 |
| KEGG | KEGG:00330 | Arginine and proline metabolism | 12 | 33 | 2.22e-02 |
| KEGG | KEGG:04740 | Olfactory transduction | 10 | 25 | 2.22e-02 |
| KEGG | KEGG:04520 | Adherens junction | 23 | 83 | 2.29e-02 |
| KEGG | KEGG:04611 | Platelet activation | 24 | 88 | 2.30e-02 |
| KEGG | KEGG:05131 | Shigellosis | 46 | 204 | 3.02e-02 |
| KEGG | KEGG:04330 | Notch signaling pathway | 16 | 52 | 3.02e-02 |
| KEGG | KEGG:04930 | Type II diabetes mellitus | 12 | 35 | 3.33e-02 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 35 | 147 | 3.33e-02 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 23 | 87 | 3.76e-02 |
| KEGG | KEGG:04961 | Endocrine and other factor-regulated calcium reabsorption | 13 | 40 | 3.76e-02 |
| KEGG | KEGG:05418 | Fluid shear stress and atherosclerosis | 27 | 109 | 4.52e-02 |
| KEGG | KEGG:04976 | Bile secretion | 13 | 41 | 4.52e-02 |
| KEGG | KEGG:04934 | Cushing syndrome | 29 | 119 | 4.52e-02 |
| KEGG | KEGG:00532 | Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate | 8 | 20 | 4.54e-02 |
NR 20,000 bp NG
# NR_resgenes_20k <- gost(query = NR_peak_list_20k$ENTREZID,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = FALSE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = background_df$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(NR_resgenes_20k,"data/GO_KEGG_analysis/NR_resgenes_20k.RDS")
# NRnom <- gostplot(NR_resgenes_20k, capped = FALSE, interactive = TRUE)
# NRnom
NRnomtable_20k <- NR_resgenes_20k$result %>%
dplyr::select(c(source, term_id,
term_name,intersection_size,
term_size, p_value))# %>%
NRnomtable_20k %>%
filter(source=="GO:BP", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(.,caption = "GO:BP terms found in NR nESRgenes_20k" ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0032501 | multicellular organismal process | 4317 | 4504 | 9.74e-06 |
| GO:BP | GO:0007275 | multicellular organism development | 3050 | 3170 | 1.83e-05 |
| GO:BP | GO:0048856 | anatomical structure development | 3708 | 3867 | 6.42e-05 |
| GO:BP | GO:0048731 | system development | 2618 | 2719 | 7.09e-05 |
| GO:BP | GO:0008150 | biological_process | 10636 | 11244 | 8.31e-05 |
| GO:BP | GO:0032502 | developmental process | 4001 | 4179 | 8.31e-05 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 3406 | 3550 | 8.31e-05 |
| GO:BP | GO:0051716 | cellular response to stimulus | 4437 | 4640 | 8.31e-05 |
| GO:BP | GO:0050794 | regulation of cellular process | 6806 | 7153 | 1.22e-04 |
| GO:BP | GO:0050896 | response to stimulus | 5133 | 5379 | 1.72e-04 |
| GO:BP | GO:0065007 | biological regulation | 7350 | 7734 | 2.63e-04 |
| GO:BP | GO:0050789 | regulation of biological process | 7144 | 7516 | 3.70e-04 |
| GO:BP | GO:0007154 | cell communication | 3811 | 3983 | 3.83e-04 |
| GO:BP | GO:0023052 | signaling | 3749 | 3918 | 4.40e-04 |
| GO:BP | GO:0007010 | cytoskeleton organization | 1113 | 1146 | 8.70e-04 |
| GO:BP | GO:0007399 | nervous system development | 1713 | 1775 | 9.59e-04 |
| GO:BP | GO:0009987 | cellular process | 10111 | 10686 | 9.59e-04 |
| GO:BP | GO:0007165 | signal transduction | 3456 | 3611 | 9.61e-04 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 1778 | 1844 | 1.25e-03 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 1006 | 1035 | 1.25e-03 |
| GO:BP | GO:0022008 | neurogenesis | 1212 | 1251 | 1.74e-03 |
| GO:BP | GO:0048519 | negative regulation of biological process | 3599 | 3764 | 1.74e-03 |
| GO:BP | GO:0048869 | cellular developmental process | 2688 | 2804 | 2.76e-03 |
| GO:BP | GO:0007015 | actin filament organization | 353 | 357 | 2.76e-03 |
| GO:BP | GO:0016043 | cellular component organization | 4579 | 4802 | 2.76e-03 |
| GO:BP | GO:0030154 | cell differentiation | 2687 | 2803 | 2.76e-03 |
| GO:BP | GO:0030029 | actin filament-based process | 626 | 640 | 2.76e-03 |
| GO:BP | GO:0048518 | positive regulation of biological process | 4105 | 4301 | 2.96e-03 |
| GO:BP | GO:0023051 | regulation of signaling | 2340 | 2438 | 3.51e-03 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 3776 | 3954 | 3.89e-03 |
| GO:BP | GO:0048699 | generation of neurons | 1043 | 1076 | 4.39e-03 |
| GO:BP | GO:0050793 | regulation of developmental process | 1635 | 1697 | 4.59e-03 |
| GO:BP | GO:0035556 | intracellular signal transduction | 1876 | 1951 | 5.84e-03 |
| GO:BP | GO:0009888 | tissue development | 1302 | 1348 | 5.84e-03 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 1158 | 1197 | 5.84e-03 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 297 | 300 | 6.20e-03 |
| GO:BP | GO:0010646 | regulation of cell communication | 2344 | 2444 | 6.20e-03 |
| GO:BP | GO:0030182 | neuron differentiation | 983 | 1014 | 6.20e-03 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 618 | 633 | 6.72e-03 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 259 | 261 | 7.37e-03 |
| GO:BP | GO:0006996 | organelle organization | 2695 | 2815 | 8.01e-03 |
| GO:BP | GO:0009966 | regulation of signal transduction | 2079 | 2166 | 8.77e-03 |
| GO:BP | GO:0048468 | cell development | 1813 | 1886 | 8.77e-03 |
| GO:BP | GO:0048513 | animal organ development | 1925 | 2004 | 9.14e-03 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 1790 | 1862 | 9.19e-03 |
| GO:BP | GO:0007267 | cell-cell signaling | 1047 | 1082 | 9.96e-03 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 553 | 566 | 1.03e-02 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 425 | 433 | 1.03e-02 |
| GO:BP | GO:0000902 | cell morphogenesis | 694 | 713 | 1.03e-02 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 2601 | 2717 | 1.03e-02 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 4749 | 4988 | 1.03e-02 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 166 | 166 | 1.22e-02 |
| GO:BP | GO:0016477 | cell migration | 978 | 1011 | 1.97e-02 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 353 | 359 | 2.16e-02 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 352 | 358 | 2.22e-02 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 404 | 412 | 2.31e-02 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 1110 | 1150 | 2.63e-02 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 2312 | 2415 | 2.73e-02 |
| GO:BP | GO:0030030 | cell projection organization | 1140 | 1182 | 3.40e-02 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 1845 | 1924 | 4.34e-02 |
NRnomtable_20k %>%
filter(source=="KEGG", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "KEGG terms found in NR nESRgenes_20k") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| NA | NA | NA | NA | NA | NA |
| :—— | :——- | :——— | —————–: | ———: | :——- |
NR peaks overlapping enhancers
# NR_resgenes_20k_enh <- gost(query = unique(NR_enh_peaks_20k$NCBI_gene),
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = FALSE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = background_df$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(NR_resgenes_20k_enh,"data/GO_KEGG_analysis/NR_resgenes_20k_enh.RDS")
# NRnom <- gostplot(NR_resgenes_20k_enh, capped = FALSE, interactive = TRUE)
# NRnom
NRnomtable_20k_enh <- NR_resgenes_20k_enh$result %>%
dplyr::select(c(source, term_id,
term_name,intersection_size,
term_size, p_value))# %>%
NRnomtable_20k_enh %>%
filter(source=="GO:BP", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(.,caption = "GO:BP terms found in NR neargenes_20k" ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0065007 | biological regulation | 4230 | 7734 | 9.66e-28 |
| GO:BP | GO:0050789 | regulation of biological process | 4120 | 7516 | 1.29e-27 |
| GO:BP | GO:0050794 | regulation of cellular process | 3931 | 7153 | 3.23e-26 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 2068 | 3550 | 5.64e-24 |
| GO:BP | GO:0007165 | signal transduction | 2097 | 3611 | 1.83e-23 |
| GO:BP | GO:0048519 | negative regulation of biological process | 2176 | 3764 | 3.12e-23 |
| GO:BP | GO:0051716 | cellular response to stimulus | 2632 | 4640 | 5.29e-23 |
| GO:BP | GO:0050896 | response to stimulus | 3012 | 5379 | 6.75e-23 |
| GO:BP | GO:0008150 | biological_process | 5864 | 11244 | 8.77e-23 |
| GO:BP | GO:0023052 | signaling | 2243 | 3918 | 5.28e-21 |
| GO:BP | GO:0048518 | positive regulation of biological process | 2442 | 4301 | 7.27e-21 |
| GO:BP | GO:0007154 | cell communication | 2275 | 3983 | 9.78e-21 |
| GO:BP | GO:0019222 | regulation of metabolic process | 2585 | 4603 | 9.97e-19 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 1539 | 2622 | 4.31e-18 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 2243 | 3954 | 4.41e-18 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 1588 | 2717 | 8.05e-18 |
| GO:BP | GO:0071310 | cellular response to organic substance | 813 | 1297 | 1.07e-17 |
| GO:BP | GO:0009987 | cellular process | 5587 | 10686 | 1.45e-17 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 1063 | 1751 | 2.03e-17 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 997 | 1633 | 3.36e-17 |
| GO:BP | GO:0010033 | response to organic substance | 1053 | 1736 | 4.29e-17 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 2378 | 4231 | 8.17e-17 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 840 | 1354 | 1.08e-16 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 752 | 1197 | 1.19e-16 |
| GO:BP | GO:0009966 | regulation of signal transduction | 1284 | 2166 | 1.42e-16 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 2263 | 4015 | 1.63e-16 |
| GO:BP | GO:0042221 | response to chemical | 1358 | 2304 | 1.63e-16 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 1247 | 2100 | 2.22e-16 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 1549 | 2663 | 2.59e-16 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 1416 | 2415 | 2.82e-16 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 1165 | 1951 | 3.39e-16 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 1292 | 2186 | 3.63e-16 |
| GO:BP | GO:0030029 | actin filament-based process | 431 | 640 | 3.91e-16 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 658 | 1035 | 4.03e-16 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 1155 | 1934 | 4.21e-16 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 2208 | 3918 | 5.56e-16 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 617 | 965 | 8.15e-16 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 2370 | 4233 | 1.00e-15 |
| GO:BP | GO:0035556 | intracellular signal transduction | 1162 | 1951 | 1.00e-15 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 621 | 973 | 1.04e-15 |
| GO:BP | GO:0032502 | developmental process | 2340 | 4179 | 1.77e-15 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 673 | 1067 | 1.89e-15 |
| GO:BP | GO:0010468 | regulation of gene expression | 1921 | 3379 | 2.41e-15 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 1412 | 2422 | 4.20e-15 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 1405 | 2410 | 5.14e-15 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 1362 | 2333 | 9.91e-15 |
| GO:BP | GO:0048856 | anatomical structure development | 2172 | 3867 | 9.91e-15 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 917 | 1514 | 1.50e-14 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 957 | 1588 | 1.92e-14 |
| GO:BP | GO:0010646 | regulation of cell communication | 1419 | 2444 | 2.31e-14 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 761 | 1234 | 2.75e-14 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 1969 | 3486 | 3.09e-14 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 759 | 1231 | 3.09e-14 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 381 | 566 | 3.09e-14 |
| GO:BP | GO:0006351 | DNA-templated transcription | 1452 | 2510 | 4.74e-14 |
| GO:BP | GO:1901698 | response to nitrogen compound | 479 | 736 | 4.74e-14 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 1464 | 2533 | 5.13e-14 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 974 | 1624 | 5.40e-14 |
| GO:BP | GO:0023051 | regulation of signaling | 1413 | 2438 | 5.59e-14 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 1070 | 1801 | 6.07e-14 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 716 | 1157 | 6.93e-14 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 979 | 1635 | 7.85e-14 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 1092 | 1844 | 1.00e-13 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 2021 | 3595 | 1.35e-13 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 1653 | 2895 | 1.37e-13 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 2007 | 3569 | 1.50e-13 |
| GO:BP | GO:0010243 | response to organonitrogen compound | 447 | 685 | 2.19e-13 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 1059 | 1791 | 5.37e-13 |
| GO:BP | GO:0050793 | regulation of developmental process | 1008 | 1697 | 5.37e-13 |
| GO:BP | GO:0048869 | cellular developmental process | 1600 | 2804 | 6.75e-13 |
| GO:BP | GO:0030154 | cell differentiation | 1599 | 2803 | 7.81e-13 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 1055 | 1787 | 1.08e-12 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 1091 | 1856 | 1.78e-12 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 662 | 1073 | 2.23e-12 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 1082 | 1841 | 2.47e-12 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 323 | 479 | 3.53e-12 |
| GO:BP | GO:0036211 | protein modification process | 1322 | 2291 | 4.14e-12 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 505 | 796 | 4.89e-12 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 793 | 1314 | 5.91e-12 |
| GO:BP | GO:0071417 | cellular response to organonitrogen compound | 300 | 442 | 8.21e-12 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 670 | 1093 | 9.42e-12 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 777 | 1289 | 1.57e-11 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 1097 | 1879 | 1.83e-11 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 1654 | 2927 | 2.01e-11 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 1705 | 3026 | 2.77e-11 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 1621 | 2867 | 3.05e-11 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 1652 | 2928 | 4.27e-11 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 303 | 452 | 6.27e-11 |
| GO:BP | GO:0009725 | response to hormone | 394 | 610 | 8.60e-11 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 407 | 633 | 9.37e-11 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 720 | 1193 | 9.39e-11 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 291 | 433 | 1.11e-10 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 597 | 972 | 1.38e-10 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 400 | 622 | 1.38e-10 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 542 | 874 | 1.63e-10 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 446 | 704 | 1.96e-10 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 453 | 717 | 2.37e-10 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 1113 | 1924 | 3.01e-10 |
| GO:BP | GO:0007275 | multicellular organism development | 1772 | 3170 | 3.07e-10 |
| GO:BP | GO:0032501 | multicellular organismal process | 2464 | 4504 | 3.29e-10 |
| GO:BP | GO:0007015 | actin filament organization | 244 | 357 | 5.05e-10 |
| GO:BP | GO:0023057 | negative regulation of signaling | 629 | 1036 | 6.35e-10 |
| GO:BP | GO:0043434 | response to peptide hormone | 218 | 314 | 6.60e-10 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 884 | 1504 | 8.18e-10 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 627 | 1034 | 9.27e-10 |
| GO:BP | GO:1901652 | response to peptide | 249 | 368 | 1.52e-09 |
| GO:BP | GO:0016477 | cell migration | 612 | 1011 | 2.57e-09 |
| GO:BP | GO:0072359 | circulatory system development | 529 | 864 | 5.47e-09 |
| GO:BP | GO:0007010 | cytoskeleton organization | 684 | 1146 | 6.28e-09 |
| GO:BP | GO:0001944 | vasculature development | 353 | 552 | 7.66e-09 |
| GO:BP | GO:0008152 | metabolic process | 4177 | 7921 | 1.78e-08 |
| GO:BP | GO:0044271 | cellular nitrogen compound biosynthetic process | 1926 | 3495 | 1.88e-08 |
| GO:BP | GO:0009605 | response to external stimulus | 903 | 1559 | 3.62e-08 |
| GO:BP | GO:0032868 | response to insulin | 146 | 203 | 3.94e-08 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 325 | 508 | 4.00e-08 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 163 | 231 | 4.09e-08 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 295 | 456 | 4.54e-08 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 1086 | 1904 | 5.64e-08 |
| GO:BP | GO:0001568 | blood vessel development | 334 | 525 | 5.72e-08 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 371 | 591 | 6.96e-08 |
| GO:BP | GO:0009888 | tissue development | 787 | 1348 | 7.67e-08 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 238 | 359 | 7.75e-08 |
| GO:BP | GO:0048513 | animal organ development | 1138 | 2004 | 7.75e-08 |
| GO:BP | GO:0048731 | system development | 1515 | 2719 | 8.04e-08 |
| GO:BP | GO:0002376 | immune system process | 820 | 1410 | 8.73e-08 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 1073 | 1883 | 9.32e-08 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 237 | 358 | 9.96e-08 |
| GO:BP | GO:1901653 | cellular response to peptide | 187 | 273 | 9.97e-08 |
| GO:BP | GO:0044238 | primary metabolic process | 3816 | 7216 | 1.09e-07 |
| GO:BP | GO:0071407 | cellular response to organic cyclic compound | 249 | 379 | 1.11e-07 |
| GO:BP | GO:0030334 | regulation of cell migration | 413 | 668 | 1.20e-07 |
| GO:BP | GO:0065009 | regulation of molecular function | 759 | 1299 | 1.22e-07 |
| GO:BP | GO:0019538 | protein metabolic process | 1982 | 3619 | 1.27e-07 |
| GO:BP | GO:0065008 | regulation of biological quality | 1077 | 1893 | 1.32e-07 |
| GO:BP | GO:0016310 | phosphorylation | 675 | 1145 | 1.45e-07 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 202 | 300 | 1.95e-07 |
| GO:BP | GO:0043412 | macromolecule modification | 1390 | 2489 | 2.29e-07 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 916 | 1595 | 2.43e-07 |
| GO:BP | GO:0051179 | localization | 2037 | 3731 | 2.47e-07 |
| GO:BP | GO:0061061 | muscle structure development | 337 | 536 | 2.89e-07 |
| GO:BP | GO:0071704 | organic substance metabolic process | 3992 | 7578 | 3.25e-07 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 277 | 431 | 3.43e-07 |
| GO:BP | GO:0006810 | transport | 1647 | 2984 | 3.85e-07 |
| GO:BP | GO:0007155 | cell adhesion | 561 | 941 | 3.97e-07 |
| GO:BP | GO:0008283 | cell population proliferation | 759 | 1306 | 4.22e-07 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 3474 | 6550 | 4.26e-07 |
| GO:BP | GO:0030097 | hemopoiesis | 385 | 624 | 5.69e-07 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 503 | 837 | 5.69e-07 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 276 | 431 | 5.89e-07 |
| GO:BP | GO:0035295 | tube development | 475 | 787 | 6.84e-07 |
| GO:BP | GO:0040011 | locomotion | 484 | 805 | 1.01e-06 |
| GO:BP | GO:2000145 | regulation of cell motility | 426 | 700 | 1.01e-06 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 543 | 913 | 1.10e-06 |
| GO:BP | GO:0046907 | intracellular transport | 677 | 1160 | 1.21e-06 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 632 | 1077 | 1.22e-06 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 165 | 242 | 1.31e-06 |
| GO:BP | GO:0006468 | protein phosphorylation | 567 | 958 | 1.34e-06 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 373 | 606 | 1.36e-06 |
| GO:BP | GO:0040012 | regulation of locomotion | 436 | 720 | 1.55e-06 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 639 | 1093 | 2.16e-06 |
| GO:BP | GO:0048468 | cell development | 1063 | 1886 | 2.24e-06 |
| GO:BP | GO:0048870 | cell motility | 653 | 1120 | 2.65e-06 |
| GO:BP | GO:0042592 | homeostatic process | 662 | 1137 | 2.81e-06 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 373 | 609 | 2.89e-06 |
| GO:BP | GO:0016070 | RNA metabolic process | 1829 | 3352 | 3.05e-06 |
| GO:BP | GO:0006807 | nitrogen compound metabolic process | 3646 | 6912 | 3.19e-06 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 609 | 1040 | 3.29e-06 |
| GO:BP | GO:0016567 | protein ubiquitination | 377 | 617 | 3.47e-06 |
| GO:BP | GO:0032869 | cellular response to insulin stimulus | 115 | 161 | 3.66e-06 |
| GO:BP | GO:0006915 | apoptotic process | 790 | 1377 | 3.84e-06 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 459 | 766 | 3.92e-06 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 283 | 450 | 4.25e-06 |
| GO:BP | GO:0006950 | response to stress | 1404 | 2540 | 4.76e-06 |
| GO:BP | GO:0043687 | post-translational protein modification | 483 | 811 | 5.09e-06 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 313 | 504 | 5.16e-06 |
| GO:BP | GO:0012501 | programmed cell death | 811 | 1419 | 5.87e-06 |
| GO:BP | GO:0044237 | cellular metabolic process | 3716 | 7058 | 5.91e-06 |
| GO:BP | GO:0030099 | myeloid cell differentiation | 192 | 292 | 5.94e-06 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 369 | 605 | 6.01e-06 |
| GO:BP | GO:0031399 | regulation of protein modification process | 536 | 911 | 8.98e-06 |
| GO:BP | GO:0002684 | positive regulation of immune system process | 354 | 580 | 9.88e-06 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 139 | 203 | 1.01e-05 |
| GO:BP | GO:0016043 | cellular component organization | 2569 | 4802 | 1.02e-05 |
| GO:BP | GO:0033993 | response to lipid | 350 | 573 | 1.02e-05 |
| GO:BP | GO:0035239 | tube morphogenesis | 386 | 638 | 1.03e-05 |
| GO:BP | GO:0008219 | cell death | 811 | 1423 | 1.06e-05 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 173 | 261 | 1.07e-05 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 173 | 261 | 1.07e-05 |
| GO:BP | GO:1901564 | organonitrogen compound metabolic process | 2339 | 4354 | 1.09e-05 |
| GO:BP | GO:0009894 | regulation of catabolic process | 490 | 830 | 1.73e-05 |
| GO:BP | GO:0060284 | regulation of cell development | 347 | 570 | 1.78e-05 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 416 | 695 | 1.78e-05 |
| GO:BP | GO:0042692 | muscle cell differentiation | 208 | 323 | 1.78e-05 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 249 | 395 | 1.78e-05 |
| GO:BP | GO:0051234 | establishment of localization | 1762 | 3240 | 1.81e-05 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 476 | 805 | 1.92e-05 |
| GO:BP | GO:0070848 | response to growth factor | 334 | 547 | 1.94e-05 |
| GO:BP | GO:0044249 | cellular biosynthetic process | 2897 | 5453 | 2.03e-05 |
| GO:BP | GO:0007507 | heart development | 307 | 499 | 2.17e-05 |
| GO:BP | GO:1901576 | organic substance biosynthetic process | 2925 | 5509 | 2.20e-05 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 475 | 804 | 2.23e-05 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 258 | 412 | 2.35e-05 |
| GO:BP | GO:0023056 | positive regulation of signaling | 700 | 1221 | 2.41e-05 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 167 | 253 | 2.44e-05 |
| GO:BP | GO:0010629 | negative regulation of gene expression | 467 | 790 | 2.51e-05 |
| GO:BP | GO:0044419 | biological process involved in interspecies interaction between organisms | 544 | 932 | 2.66e-05 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 1040 | 1862 | 2.78e-05 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 554 | 951 | 2.91e-05 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 196 | 304 | 3.18e-05 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 208 | 325 | 3.21e-05 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 209 | 327 | 3.46e-05 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 697 | 1218 | 3.54e-05 |
| GO:BP | GO:0009058 | biosynthetic process | 2941 | 5547 | 3.81e-05 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 2574 | 4827 | 3.83e-05 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 596 | 1031 | 4.20e-05 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 521 | 893 | 4.89e-05 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 611 | 1060 | 5.07e-05 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 321 | 528 | 5.32e-05 |
| GO:BP | GO:0002682 | regulation of immune system process | 507 | 868 | 5.65e-05 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 660 | 1152 | 5.65e-05 |
| GO:BP | GO:0034097 | response to cytokine | 354 | 588 | 5.67e-05 |
| GO:BP | GO:0080134 | regulation of response to stress | 539 | 928 | 6.55e-05 |
| GO:BP | GO:0009991 | response to extracellular stimulus | 236 | 377 | 6.93e-05 |
| GO:BP | GO:0050776 | regulation of immune response | 292 | 477 | 6.98e-05 |
| GO:BP | GO:0001525 | angiogenesis | 238 | 381 | 7.84e-05 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 154 | 234 | 8.08e-05 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 253 | 408 | 8.51e-05 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 134 | 200 | 8.90e-05 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 318 | 525 | 9.14e-05 |
| GO:BP | GO:0000902 | cell morphogenesis | 421 | 713 | 1.01e-04 |
| GO:BP | GO:0050778 | positive regulation of immune response | 235 | 377 | 1.12e-04 |
| GO:BP | GO:0071396 | cellular response to lipid | 241 | 388 | 1.22e-04 |
| GO:BP | GO:0007264 | small GTPase-mediated signal transduction | 242 | 390 | 1.30e-04 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 216 | 344 | 1.36e-04 |
| GO:BP | GO:0070936 | protein K48-linked ubiquitination | 57 | 74 | 1.36e-04 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 184 | 288 | 1.50e-04 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 527 | 911 | 1.57e-04 |
| GO:BP | GO:0061448 | connective tissue development | 136 | 205 | 1.63e-04 |
| GO:BP | GO:0010467 | gene expression | 2372 | 4449 | 1.64e-04 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 256 | 416 | 1.65e-04 |
| GO:BP | GO:0006897 | endocytosis | 298 | 492 | 1.85e-04 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 171 | 266 | 1.87e-04 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 443 | 757 | 1.94e-04 |
| GO:BP | GO:0033554 | cellular response to stress | 805 | 1433 | 1.96e-04 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 457 | 783 | 1.99e-04 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 184 | 289 | 2.00e-04 |
| GO:BP | GO:0032879 | regulation of localization | 808 | 1439 | 2.03e-04 |
| GO:BP | GO:0006955 | immune response | 502 | 867 | 2.27e-04 |
| GO:BP | GO:0006470 | protein dephosphorylation | 119 | 177 | 2.36e-04 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 419 | 714 | 2.36e-04 |
| GO:BP | GO:0016311 | dephosphorylation | 151 | 232 | 2.38e-04 |
| GO:BP | GO:0048872 | homeostasis of number of cells | 145 | 222 | 2.74e-04 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 95 | 137 | 2.87e-04 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 46 | 58 | 2.88e-04 |
| GO:BP | GO:0031667 | response to nutrient levels | 220 | 354 | 2.89e-04 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 414 | 706 | 2.91e-04 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 284 | 469 | 3.01e-04 |
| GO:BP | GO:0060485 | mesenchyme development | 158 | 245 | 3.12e-04 |
| GO:BP | GO:0098657 | import into cell | 368 | 622 | 3.13e-04 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 171 | 268 | 3.40e-04 |
| GO:BP | GO:0051641 | cellular localization | 1426 | 2623 | 3.50e-04 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 63 | 85 | 3.55e-04 |
| GO:BP | GO:0014902 | myotube differentiation | 63 | 85 | 3.55e-04 |
| GO:BP | GO:0034101 | erythrocyte homeostasis | 79 | 111 | 3.58e-04 |
| GO:BP | GO:0007162 | negative regulation of cell adhesion | 123 | 185 | 3.63e-04 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 234 | 380 | 3.66e-04 |
| GO:BP | GO:0003012 | muscle system process | 203 | 325 | 3.91e-04 |
| GO:BP | GO:0009896 | positive regulation of catabolic process | 265 | 436 | 3.95e-04 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 320 | 536 | 4.27e-04 |
| GO:BP | GO:0007520 | myoblast fusion | 26 | 29 | 4.50e-04 |
| GO:BP | GO:0032271 | regulation of protein polymerization | 108 | 160 | 4.50e-04 |
| GO:BP | GO:0031032 | actomyosin structure organization | 122 | 184 | 4.70e-04 |
| GO:BP | GO:0010631 | epithelial cell migration | 156 | 243 | 4.88e-04 |
| GO:BP | GO:0090132 | epithelium migration | 156 | 243 | 4.88e-04 |
| GO:BP | GO:0045444 | fat cell differentiation | 119 | 179 | 4.92e-04 |
| GO:BP | GO:0003170 | heart valve development | 50 | 65 | 5.13e-04 |
| GO:BP | GO:0002253 | activation of immune response | 182 | 289 | 5.35e-04 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 127 | 193 | 5.37e-04 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 138 | 212 | 5.37e-04 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 150 | 233 | 5.61e-04 |
| GO:BP | GO:0030218 | erythrocyte differentiation | 74 | 104 | 6.58e-04 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 2638 | 4988 | 6.67e-04 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 433 | 746 | 6.67e-04 |
| GO:BP | GO:0010921 | regulation of phosphatase activity | 36 | 44 | 7.55e-04 |
| GO:BP | GO:0002262 | myeloid cell homeostasis | 93 | 136 | 7.81e-04 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 188 | 301 | 7.81e-04 |
| GO:BP | GO:0008360 | regulation of cell shape | 75 | 106 | 7.81e-04 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 54 | 72 | 8.11e-04 |
| GO:BP | GO:0090257 | regulation of muscle system process | 117 | 177 | 8.26e-04 |
| GO:BP | GO:0030323 | respiratory tube development | 100 | 148 | 8.30e-04 |
| GO:BP | GO:0002521 | leukocyte differentiation | 227 | 371 | 8.49e-04 |
| GO:BP | GO:0009607 | response to biotic stimulus | 472 | 820 | 8.65e-04 |
| GO:BP | GO:0008286 | insulin receptor signaling pathway | 70 | 98 | 8.65e-04 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 398 | 683 | 8.70e-04 |
| GO:BP | GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 178 | 284 | 9.37e-04 |
| GO:BP | GO:0090130 | tissue migration | 157 | 247 | 9.47e-04 |
| GO:BP | GO:0031668 | cellular response to extracellular stimulus | 141 | 219 | 9.47e-04 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 108 | 162 | 9.48e-04 |
| GO:BP | GO:0003179 | heart valve morphogenesis | 43 | 55 | 9.59e-04 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 328 | 555 | 9.74e-04 |
| GO:BP | GO:0046649 | lymphocyte activation | 267 | 444 | 1.00e-03 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 236 | 388 | 1.01e-03 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 55 | 74 | 1.01e-03 |
| GO:BP | GO:0035966 | response to topologically incorrect protein | 89 | 130 | 1.03e-03 |
| GO:BP | GO:0045646 | regulation of erythrocyte differentiation | 29 | 34 | 1.03e-03 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 163 | 258 | 1.05e-03 |
| GO:BP | GO:0010586 | miRNA metabolic process | 63 | 87 | 1.06e-03 |
| GO:BP | GO:0051707 | response to other organism | 457 | 794 | 1.13e-03 |
| GO:BP | GO:0043207 | response to external biotic stimulus | 457 | 794 | 1.13e-03 |
| GO:BP | GO:0048878 | chemical homeostasis | 390 | 670 | 1.14e-03 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 110 | 166 | 1.15e-03 |
| GO:BP | GO:0051494 | negative regulation of cytoskeleton organization | 87 | 127 | 1.18e-03 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 46 | 60 | 1.18e-03 |
| GO:BP | GO:0008064 | regulation of actin polymerization or depolymerization | 81 | 117 | 1.21e-03 |
| GO:BP | GO:0030324 | lung development | 97 | 144 | 1.23e-03 |
| GO:BP | GO:0006996 | organelle organization | 1518 | 2815 | 1.23e-03 |
| GO:BP | GO:0036293 | response to decreased oxygen levels | 154 | 243 | 1.33e-03 |
| GO:BP | GO:0055001 | muscle cell development | 108 | 163 | 1.33e-03 |
| GO:BP | GO:0001775 | cell activation | 366 | 627 | 1.41e-03 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 76 | 109 | 1.41e-03 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 54 | 73 | 1.42e-03 |
| GO:BP | GO:0002764 | immune response-regulating signaling pathway | 168 | 268 | 1.43e-03 |
| GO:BP | GO:0052547 | regulation of peptidase activity | 113 | 172 | 1.53e-03 |
| GO:BP | GO:0008154 | actin polymerization or depolymerization | 99 | 148 | 1.53e-03 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 140 | 219 | 1.56e-03 |
| GO:BP | GO:0051049 | regulation of transport | 618 | 1098 | 1.65e-03 |
| GO:BP | GO:0002757 | immune response-activating signaling pathway | 162 | 258 | 1.69e-03 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 380 | 654 | 1.70e-03 |
| GO:BP | GO:1901201 | regulation of extracellular matrix assembly | 18 | 19 | 1.74e-03 |
| GO:BP | GO:0014896 | muscle hypertrophy | 55 | 75 | 1.78e-03 |
| GO:BP | GO:0034109 | homotypic cell-cell adhesion | 50 | 67 | 1.80e-03 |
| GO:BP | GO:0030832 | regulation of actin filament length | 81 | 118 | 1.85e-03 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 263 | 440 | 1.90e-03 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 173 | 278 | 1.91e-03 |
| GO:BP | GO:0140747 | regulation of ncRNA transcription | 58 | 80 | 1.91e-03 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 373 | 642 | 1.96e-03 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 156 | 248 | 1.97e-03 |
| GO:BP | GO:0071383 | cellular response to steroid hormone stimulus | 105 | 159 | 1.97e-03 |
| GO:BP | GO:0048384 | retinoic acid receptor signaling pathway | 23 | 26 | 2.02e-03 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 161 | 257 | 2.04e-03 |
| GO:BP | GO:0033036 | macromolecule localization | 1252 | 2309 | 2.17e-03 |
| GO:BP | GO:0141091 | transforming growth factor beta receptor superfamily signaling pathway | 162 | 259 | 2.17e-03 |
| GO:BP | GO:0006325 | chromatin organization | 352 | 604 | 2.21e-03 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 137 | 215 | 2.23e-03 |
| GO:BP | GO:0061614 | miRNA transcription | 46 | 61 | 2.23e-03 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 126 | 196 | 2.40e-03 |
| GO:BP | GO:0035967 | cellular response to topologically incorrect protein | 62 | 87 | 2.40e-03 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 389 | 673 | 2.43e-03 |
| GO:BP | GO:0030048 | actin filament-based movement | 74 | 107 | 2.51e-03 |
| GO:BP | GO:0001570 | vasculogenesis | 49 | 66 | 2.54e-03 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 156 | 249 | 2.55e-03 |
| GO:BP | GO:0035303 | regulation of dephosphorylation | 57 | 79 | 2.63e-03 |
| GO:BP | GO:0000165 | MAPK cascade | 297 | 504 | 2.68e-03 |
| GO:BP | GO:0051235 | maintenance of location | 157 | 251 | 2.70e-03 |
| GO:BP | GO:0009611 | response to wounding | 230 | 382 | 2.74e-03 |
| GO:BP | GO:0006338 | chromatin remodeling | 287 | 486 | 2.84e-03 |
| GO:BP | GO:0043433 | negative regulation of DNA-binding transcription factor activity | 72 | 104 | 2.90e-03 |
| GO:BP | GO:0032535 | regulation of cellular component size | 177 | 287 | 2.99e-03 |
| GO:BP | GO:1903902 | positive regulation of viral life cycle | 17 | 18 | 3.00e-03 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 150 | 239 | 3.00e-03 |
| GO:BP | GO:0001666 | response to hypoxia | 146 | 232 | 3.05e-03 |
| GO:BP | GO:0051649 | establishment of localization in cell | 862 | 1566 | 3.07e-03 |
| GO:BP | GO:1990845 | adaptive thermogenesis | 82 | 121 | 3.07e-03 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 228 | 379 | 3.07e-03 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 92 | 138 | 3.07e-03 |
| GO:BP | GO:0009952 | anterior/posterior pattern specification | 82 | 121 | 3.07e-03 |
| GO:BP | GO:0003007 | heart morphogenesis | 138 | 218 | 3.10e-03 |
| GO:BP | GO:0098609 | cell-cell adhesion | 329 | 564 | 3.23e-03 |
| GO:BP | GO:0120161 | regulation of cold-induced thermogenesis | 76 | 111 | 3.23e-03 |
| GO:BP | GO:0032989 | cellular anatomical entity morphogenesis | 342 | 588 | 3.23e-03 |
| GO:BP | GO:0010720 | positive regulation of cell development | 189 | 309 | 3.30e-03 |
| GO:BP | GO:0031329 | regulation of cellular catabolic process | 324 | 555 | 3.33e-03 |
| GO:BP | GO:0097581 | lamellipodium organization | 53 | 73 | 3.37e-03 |
| GO:BP | GO:0060341 | regulation of cellular localization | 443 | 776 | 3.47e-03 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 90 | 135 | 3.53e-03 |
| GO:BP | GO:0009790 | embryo development | 467 | 821 | 3.55e-03 |
| GO:BP | GO:0045598 | regulation of fat cell differentiation | 65 | 93 | 3.85e-03 |
| GO:BP | GO:1902904 | negative regulation of supramolecular fiber organization | 84 | 125 | 3.86e-03 |
| GO:BP | GO:0070482 | response to oxygen levels | 164 | 265 | 3.93e-03 |
| GO:BP | GO:0000302 | response to reactive oxygen species | 102 | 156 | 4.01e-03 |
| GO:BP | GO:0003176 | aortic valve development | 30 | 37 | 4.01e-03 |
| GO:BP | GO:0060429 | epithelium development | 451 | 792 | 4.01e-03 |
| GO:BP | GO:0043666 | regulation of phosphoprotein phosphatase activity | 28 | 34 | 4.05e-03 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 156 | 251 | 4.16e-03 |
| GO:BP | GO:0003205 | cardiac chamber development | 99 | 151 | 4.25e-03 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 171 | 278 | 4.40e-03 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 1092 | 2010 | 4.40e-03 |
| GO:BP | GO:0006886 | intracellular protein transport | 342 | 590 | 4.41e-03 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 323 | 555 | 4.48e-03 |
| GO:BP | GO:0052548 | regulation of endopeptidase activity | 100 | 153 | 4.64e-03 |
| GO:BP | GO:0009056 | catabolic process | 1052 | 1934 | 4.69e-03 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 2004 | 3777 | 4.75e-03 |
| GO:BP | GO:0031331 | positive regulation of cellular catabolic process | 168 | 273 | 4.76e-03 |
| GO:BP | GO:0043393 | regulation of protein binding | 76 | 112 | 4.76e-03 |
| GO:BP | GO:0106106 | cold-induced thermogenesis | 76 | 112 | 4.76e-03 |
| GO:BP | GO:0042060 | wound healing | 178 | 291 | 4.85e-03 |
| GO:BP | GO:0051090 | regulation of DNA-binding transcription factor activity | 150 | 241 | 4.87e-03 |
| GO:BP | GO:0030833 | regulation of actin filament polymerization | 70 | 102 | 4.98e-03 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 242 | 407 | 4.98e-03 |
| GO:BP | GO:0030903 | notochord development | 16 | 17 | 5.05e-03 |
| GO:BP | GO:0045321 | leukocyte activation | 309 | 530 | 5.05e-03 |
| GO:BP | GO:0060612 | adipose tissue development | 35 | 45 | 5.05e-03 |
| GO:BP | GO:0060541 | respiratory system development | 105 | 162 | 5.07e-03 |
| GO:BP | GO:0006986 | response to unfolded protein | 80 | 119 | 5.10e-03 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 117 | 183 | 5.16e-03 |
| GO:BP | GO:2000737 | negative regulation of stem cell differentiation | 21 | 24 | 5.20e-03 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 2212 | 4185 | 5.27e-03 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 110 | 171 | 5.55e-03 |
| GO:BP | GO:0032103 | positive regulation of response to external stimulus | 207 | 344 | 5.57e-03 |
| GO:BP | GO:0050851 | antigen receptor-mediated signaling pathway | 71 | 104 | 5.61e-03 |
| GO:BP | GO:0036294 | cellular response to decreased oxygen levels | 81 | 121 | 5.68e-03 |
| GO:BP | GO:0010464 | regulation of mesenchymal cell proliferation | 23 | 27 | 5.79e-03 |
| GO:BP | GO:0035886 | vascular associated smooth muscle cell differentiation | 23 | 27 | 5.79e-03 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 140 | 224 | 5.82e-03 |
| GO:BP | GO:0043124 | negative regulation of canonical NF-kappaB signal transduction | 29 | 36 | 5.94e-03 |
| GO:BP | GO:0000768 | syncytium formation by plasma membrane fusion | 29 | 36 | 5.94e-03 |
| GO:BP | GO:0010463 | mesenchymal cell proliferation | 29 | 36 | 5.94e-03 |
| GO:BP | GO:0140253 | cell-cell fusion | 29 | 36 | 5.94e-03 |
| GO:BP | GO:0048008 | platelet-derived growth factor receptor signaling pathway | 38 | 50 | 6.05e-03 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 317 | 546 | 6.06e-03 |
| GO:BP | GO:0000209 | protein polyubiquitination | 141 | 226 | 6.11e-03 |
| GO:BP | GO:0045637 | regulation of myeloid cell differentiation | 89 | 135 | 6.23e-03 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 319 | 550 | 6.36e-03 |
| GO:BP | GO:0097696 | receptor signaling pathway via STAT | 63 | 91 | 6.68e-03 |
| GO:BP | GO:0008104 | protein localization | 1084 | 2000 | 6.74e-03 |
| GO:BP | GO:0007584 | response to nutrient | 76 | 113 | 6.83e-03 |
| GO:BP | GO:0030032 | lamellipodium assembly | 41 | 55 | 6.97e-03 |
| GO:BP | GO:0048193 | Golgi vesicle transport | 162 | 264 | 6.97e-03 |
| GO:BP | GO:0061572 | actin filament bundle organization | 87 | 132 | 7.27e-03 |
| GO:BP | GO:0043244 | regulation of protein-containing complex disassembly | 70 | 103 | 7.32e-03 |
| GO:BP | GO:0002768 | immune response-regulating cell surface receptor signaling pathway | 106 | 165 | 7.36e-03 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 300 | 516 | 7.59e-03 |
| GO:BP | GO:0055006 | cardiac cell development | 55 | 78 | 7.68e-03 |
| GO:BP | GO:0003013 | circulatory system process | 251 | 426 | 7.76e-03 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 150 | 243 | 7.78e-03 |
| GO:BP | GO:0046483 | heterocycle metabolic process | 2265 | 4295 | 7.81e-03 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 103 | 160 | 7.87e-03 |
| GO:BP | GO:1905314 | semi-lunar valve development | 32 | 41 | 7.90e-03 |
| GO:BP | GO:0010952 | positive regulation of peptidase activity | 71 | 105 | 8.22e-03 |
| GO:BP | GO:1903580 | positive regulation of ATP metabolic process | 20 | 23 | 8.34e-03 |
| GO:BP | GO:1902894 | negative regulation of miRNA transcription | 20 | 23 | 8.34e-03 |
| GO:BP | GO:0002429 | immune response-activating cell surface receptor signaling pathway | 100 | 155 | 8.42e-03 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 147 | 238 | 8.42e-03 |
| GO:BP | GO:0006949 | syncytium formation | 30 | 38 | 8.45e-03 |
| GO:BP | GO:0071474 | cellular hyperosmotic response | 15 | 16 | 8.50e-03 |
| GO:BP | GO:1901879 | regulation of protein depolymerization | 50 | 70 | 8.52e-03 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 65 | 95 | 8.61e-03 |
| GO:BP | GO:0090288 | negative regulation of cellular response to growth factor stimulus | 53 | 75 | 8.75e-03 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 86 | 131 | 9.19e-03 |
| GO:BP | GO:0034614 | cellular response to reactive oxygen species | 79 | 119 | 9.31e-03 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 131 | 210 | 9.50e-03 |
| GO:BP | GO:0045216 | cell-cell junction organization | 98 | 152 | 9.72e-03 |
| GO:BP | GO:0007623 | circadian rhythm | 98 | 152 | 9.72e-03 |
| GO:BP | GO:0033002 | muscle cell proliferation | 98 | 152 | 9.72e-03 |
| GO:BP | GO:0071456 | cellular response to hypoxia | 76 | 114 | 9.74e-03 |
| GO:BP | GO:0046822 | regulation of nucleocytoplasmic transport | 66 | 97 | 9.74e-03 |
| GO:BP | GO:0043086 | negative regulation of catalytic activity | 180 | 298 | 9.76e-03 |
| GO:BP | GO:0030182 | neuron differentiation | 565 | 1014 | 9.82e-03 |
| GO:BP | GO:0098542 | defense response to other organism | 347 | 605 | 9.93e-03 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 293 | 505 | 1.01e-02 |
| GO:BP | GO:0044409 | symbiont entry into host | 73 | 109 | 1.01e-02 |
| GO:BP | GO:2000116 | regulation of cysteine-type endopeptidase activity | 80 | 121 | 1.02e-02 |
| GO:BP | GO:0009267 | cellular response to starvation | 95 | 147 | 1.03e-02 |
| GO:BP | GO:0040007 | growth | 396 | 697 | 1.08e-02 |
| GO:BP | GO:0046777 | protein autophosphorylation | 92 | 142 | 1.11e-02 |
| GO:BP | GO:0043242 | negative regulation of protein-containing complex disassembly | 46 | 64 | 1.12e-02 |
| GO:BP | GO:0051258 | protein polymerization | 139 | 225 | 1.14e-02 |
| GO:BP | GO:1901360 | organic cyclic compound metabolic process | 2346 | 4460 | 1.14e-02 |
| GO:BP | GO:0070935 | 3’-UTR-mediated mRNA stabilization | 17 | 19 | 1.15e-02 |
| GO:BP | GO:0120162 | positive regulation of cold-induced thermogenesis | 49 | 69 | 1.15e-02 |
| GO:BP | GO:0001659 | temperature homeostasis | 85 | 130 | 1.15e-02 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 31 | 40 | 1.15e-02 |
| GO:BP | GO:0043500 | muscle adaptation | 61 | 89 | 1.15e-02 |
| GO:BP | GO:0022008 | neurogenesis | 689 | 1251 | 1.18e-02 |
| GO:BP | GO:0007249 | canonical NF-kappaB signal transduction | 140 | 227 | 1.18e-02 |
| GO:BP | GO:1901880 | negative regulation of protein depolymerization | 41 | 56 | 1.20e-02 |
| GO:BP | GO:0060537 | muscle tissue development | 202 | 339 | 1.20e-02 |
| GO:BP | GO:0051058 | negative regulation of small GTPase mediated signal transduction | 36 | 48 | 1.22e-02 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 250 | 427 | 1.23e-02 |
| GO:BP | GO:0030866 | cortical actin cytoskeleton organization | 29 | 37 | 1.24e-02 |
| GO:BP | GO:0051261 | protein depolymerization | 65 | 96 | 1.26e-02 |
| GO:BP | GO:0048568 | embryonic organ development | 182 | 303 | 1.28e-02 |
| GO:BP | GO:0098781 | ncRNA transcription | 79 | 120 | 1.29e-02 |
| GO:BP | GO:0045639 | positive regulation of myeloid cell differentiation | 47 | 66 | 1.32e-02 |
| GO:BP | GO:0042110 | T cell activation | 183 | 305 | 1.33e-02 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 83 | 127 | 1.33e-02 |
| GO:BP | GO:0042594 | response to starvation | 111 | 176 | 1.34e-02 |
| GO:BP | GO:0010035 | response to inorganic substance | 223 | 378 | 1.34e-02 |
| GO:BP | GO:0046718 | symbiont entry into host cell | 69 | 103 | 1.35e-02 |
| GO:BP | GO:0030865 | cortical cytoskeleton organization | 34 | 45 | 1.35e-02 |
| GO:BP | GO:0003231 | cardiac ventricle development | 76 | 115 | 1.35e-02 |
| GO:BP | GO:0002831 | regulation of response to biotic stimulus | 184 | 307 | 1.36e-02 |
| GO:BP | GO:1900744 | regulation of p38MAPK cascade | 25 | 31 | 1.37e-02 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 134 | 217 | 1.38e-02 |
| GO:BP | GO:0045981 | positive regulation of nucleotide metabolic process | 21 | 25 | 1.38e-02 |
| GO:BP | GO:0075294 | positive regulation by symbiont of entry into host | 11 | 11 | 1.38e-02 |
| GO:BP | GO:2000629 | negative regulation of miRNA metabolic process | 21 | 25 | 1.38e-02 |
| GO:BP | GO:0046598 | positive regulation of viral entry into host cell | 11 | 11 | 1.38e-02 |
| GO:BP | GO:1900544 | positive regulation of purine nucleotide metabolic process | 21 | 25 | 1.38e-02 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 125 | 201 | 1.38e-02 |
| GO:BP | GO:0060317 | cardiac epithelial to mesenchymal transition | 23 | 28 | 1.39e-02 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 73 | 110 | 1.40e-02 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 269 | 463 | 1.41e-02 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 269 | 463 | 1.41e-02 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 130 | 210 | 1.41e-02 |
| GO:BP | GO:0022411 | cellular component disassembly | 250 | 428 | 1.41e-02 |
| GO:BP | GO:0009755 | hormone-mediated signaling pathway | 88 | 136 | 1.43e-02 |
| GO:BP | GO:0007259 | receptor signaling pathway via JAK-STAT | 60 | 88 | 1.47e-02 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 60 | 88 | 1.47e-02 |
| GO:BP | GO:0006952 | defense response | 512 | 918 | 1.49e-02 |
| GO:BP | GO:2000630 | positive regulation of miRNA metabolic process | 37 | 50 | 1.49e-02 |
| GO:BP | GO:0048511 | rhythmic process | 136 | 221 | 1.49e-02 |
| GO:BP | GO:0071824 | protein-DNA complex organization | 387 | 683 | 1.49e-02 |
| GO:BP | GO:0030041 | actin filament polymerization | 81 | 124 | 1.49e-02 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 210 | 355 | 1.49e-02 |
| GO:BP | GO:0019915 | lipid storage | 48 | 68 | 1.50e-02 |
| GO:BP | GO:0043502 | regulation of muscle adaptation | 48 | 68 | 1.50e-02 |
| GO:BP | GO:0003158 | endothelium development | 67 | 100 | 1.51e-02 |
| GO:BP | GO:0030856 | regulation of epithelial cell differentiation | 67 | 100 | 1.51e-02 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 93 | 145 | 1.54e-02 |
| GO:BP | GO:0031347 | regulation of defense response | 261 | 449 | 1.56e-02 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 281 | 486 | 1.57e-02 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 345 | 605 | 1.58e-02 |
| GO:BP | GO:0051145 | smooth muscle cell differentiation | 40 | 55 | 1.59e-02 |
| GO:BP | GO:0016032 | viral process | 195 | 328 | 1.59e-02 |
| GO:BP | GO:0007009 | plasma membrane organization | 71 | 107 | 1.59e-02 |
| GO:BP | GO:0001952 | regulation of cell-matrix adhesion | 61 | 90 | 1.63e-02 |
| GO:BP | GO:0071453 | cellular response to oxygen levels | 86 | 133 | 1.63e-02 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 174 | 290 | 1.65e-02 |
| GO:BP | GO:0071702 | organic substance transport | 933 | 1722 | 1.68e-02 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 134 | 218 | 1.69e-02 |
| GO:BP | GO:0006725 | cellular aromatic compound metabolic process | 2267 | 4313 | 1.69e-02 |
| GO:BP | GO:0006900 | vesicle budding from membrane | 52 | 75 | 1.74e-02 |
| GO:BP | GO:0085029 | extracellular matrix assembly | 28 | 36 | 1.74e-02 |
| GO:BP | GO:0048660 | regulation of smooth muscle cell proliferation | 65 | 97 | 1.75e-02 |
| GO:BP | GO:0050878 | regulation of body fluid levels | 140 | 229 | 1.78e-02 |
| GO:BP | GO:0006979 | response to oxidative stress | 182 | 305 | 1.80e-02 |
| GO:BP | GO:0001501 | skeletal system development | 205 | 347 | 1.81e-02 |
| GO:BP | GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 76 | 116 | 1.81e-02 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 598 | 1083 | 1.82e-02 |
| GO:BP | GO:1902895 | positive regulation of miRNA transcription | 33 | 44 | 1.85e-02 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 80 | 123 | 1.85e-02 |
| GO:BP | GO:0038066 | p38MAPK cascade | 33 | 44 | 1.85e-02 |
| GO:BP | GO:0003180 | aortic valve morphogenesis | 26 | 33 | 1.87e-02 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 237 | 406 | 1.88e-02 |
| GO:BP | GO:0006402 | mRNA catabolic process | 127 | 206 | 1.89e-02 |
| GO:BP | GO:0010608 | post-transcriptional regulation of gene expression | 264 | 456 | 1.94e-02 |
| GO:BP | GO:0003208 | cardiac ventricle morphogenesis | 44 | 62 | 1.97e-02 |
| GO:BP | GO:0033043 | regulation of organelle organization | 529 | 953 | 1.98e-02 |
| GO:BP | GO:1903828 | negative regulation of protein localization | 101 | 160 | 1.98e-02 |
| GO:BP | GO:0061013 | regulation of mRNA catabolic process | 101 | 160 | 1.98e-02 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 93 | 146 | 2.02e-02 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 234 | 401 | 2.05e-02 |
| GO:BP | GO:1904385 | cellular response to angiotensin | 22 | 27 | 2.05e-02 |
| GO:BP | GO:0051098 | regulation of binding | 124 | 201 | 2.05e-02 |
| GO:BP | GO:0043122 | regulation of canonical NF-kappaB signal transduction | 124 | 201 | 2.05e-02 |
| GO:BP | GO:0072538 | T-helper 17 type immune response | 22 | 27 | 2.05e-02 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 242 | 416 | 2.13e-02 |
| GO:BP | GO:0097190 | apoptotic signaling pathway | 269 | 466 | 2.16e-02 |
| GO:BP | GO:0051651 | maintenance of location in cell | 107 | 171 | 2.19e-02 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 284 | 494 | 2.22e-02 |
| GO:BP | GO:0048732 | gland development | 183 | 308 | 2.23e-02 |
| GO:BP | GO:0010950 | positive regulation of endopeptidase activity | 64 | 96 | 2.23e-02 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 172 | 288 | 2.23e-02 |
| GO:BP | GO:0051169 | nuclear transport | 172 | 288 | 2.23e-02 |
| GO:BP | GO:0031397 | negative regulation of protein ubiquitination | 42 | 59 | 2.25e-02 |
| GO:BP | GO:0035304 | regulation of protein dephosphorylation | 42 | 59 | 2.25e-02 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 75 | 115 | 2.26e-02 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 75 | 115 | 2.26e-02 |
| GO:BP | GO:1901203 | positive regulation of extracellular matrix assembly | 13 | 14 | 2.28e-02 |
| GO:BP | GO:1903321 | negative regulation of protein modification by small protein conjugation or removal | 48 | 69 | 2.28e-02 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 286 | 498 | 2.28e-02 |
| GO:BP | GO:0034260 | negative regulation of GTPase activity | 13 | 14 | 2.28e-02 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 252 | 435 | 2.28e-02 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 79 | 122 | 2.28e-02 |
| GO:BP | GO:0007267 | cell-cell signaling | 596 | 1082 | 2.35e-02 |
| GO:BP | GO:0034976 | response to endoplasmic reticulum stress | 113 | 182 | 2.35e-02 |
| GO:BP | GO:0044030 | regulation of DNA methylation | 10 | 10 | 2.35e-02 |
| GO:BP | GO:2000479 | regulation of cAMP-dependent protein kinase activity | 10 | 10 | 2.35e-02 |
| GO:BP | GO:0034599 | cellular response to oxidative stress | 123 | 200 | 2.43e-02 |
| GO:BP | GO:0045667 | regulation of osteoblast differentiation | 65 | 98 | 2.44e-02 |
| GO:BP | GO:0051346 | negative regulation of hydrolase activity | 76 | 117 | 2.44e-02 |
| GO:BP | GO:0042113 | B cell activation | 101 | 161 | 2.54e-02 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 124 | 202 | 2.55e-02 |
| GO:BP | GO:0048708 | astrocyte differentiation | 40 | 56 | 2.57e-02 |
| GO:BP | GO:0007517 | muscle organ development | 160 | 267 | 2.57e-02 |
| GO:BP | GO:0030514 | negative regulation of BMP signaling pathway | 32 | 43 | 2.57e-02 |
| GO:BP | GO:0007369 | gastrulation | 89 | 140 | 2.66e-02 |
| GO:BP | GO:0050920 | regulation of chemotaxis | 85 | 133 | 2.67e-02 |
| GO:BP | GO:0048589 | developmental growth | 287 | 501 | 2.67e-02 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 59 | 88 | 2.68e-02 |
| GO:BP | GO:0032386 | regulation of intracellular transport | 135 | 222 | 2.68e-02 |
| GO:BP | GO:0002833 | positive regulation of response to biotic stimulus | 130 | 213 | 2.68e-02 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 116 | 188 | 2.73e-02 |
| GO:BP | GO:1903037 | regulation of leukocyte cell-cell adhesion | 121 | 197 | 2.77e-02 |
| GO:BP | GO:0034641 | cellular nitrogen compound metabolic process | 2432 | 4646 | 2.77e-02 |
| GO:BP | GO:0050852 | T cell receptor signaling pathway | 56 | 83 | 2.77e-02 |
| GO:BP | GO:0030162 | regulation of proteolysis | 241 | 416 | 2.77e-02 |
| GO:BP | GO:0008015 | blood circulation | 209 | 357 | 2.77e-02 |
| GO:BP | GO:0043549 | regulation of kinase activity | 209 | 357 | 2.77e-02 |
| GO:BP | GO:0030042 | actin filament depolymerization | 35 | 48 | 2.77e-02 |
| GO:BP | GO:0010718 | positive regulation of epithelial to mesenchymal transition | 35 | 48 | 2.77e-02 |
| GO:BP | GO:0010811 | positive regulation of cell-substrate adhesion | 63 | 95 | 2.79e-02 |
| GO:BP | GO:0071219 | cellular response to molecule of bacterial origin | 74 | 114 | 2.79e-02 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 452 | 811 | 2.80e-02 |
| GO:BP | GO:0003215 | cardiac right ventricle morphogenesis | 15 | 17 | 2.80e-02 |
| GO:BP | GO:0009166 | nucleotide catabolic process | 86 | 135 | 2.81e-02 |
| GO:BP | GO:0007409 | axonogenesis | 198 | 337 | 2.84e-02 |
| GO:BP | GO:0034620 | cellular response to unfolded protein | 53 | 78 | 2.84e-02 |
| GO:BP | GO:0035305 | negative regulation of dephosphorylation | 23 | 29 | 2.86e-02 |
| GO:BP | GO:1990776 | response to angiotensin | 23 | 29 | 2.86e-02 |
| GO:BP | GO:0043542 | endothelial cell migration | 108 | 174 | 2.86e-02 |
| GO:BP | GO:0003279 | cardiac septum development | 67 | 102 | 2.87e-02 |
| GO:BP | GO:0045814 | negative regulation of gene expression, epigenetic | 67 | 102 | 2.87e-02 |
| GO:BP | GO:0048524 | positive regulation of viral process | 38 | 53 | 2.87e-02 |
| GO:BP | GO:0043401 | steroid hormone mediated signaling pathway | 71 | 109 | 2.95e-02 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 176 | 297 | 2.96e-02 |
| GO:BP | GO:0032880 | regulation of protein localization | 388 | 691 | 2.96e-02 |
| GO:BP | GO:0048710 | regulation of astrocyte differentiation | 17 | 20 | 3.03e-02 |
| GO:BP | GO:0045446 | endothelial cell differentiation | 57 | 85 | 3.03e-02 |
| GO:BP | GO:0001783 | B cell apoptotic process | 17 | 20 | 3.03e-02 |
| GO:BP | GO:0045088 | regulation of innate immune response | 149 | 248 | 3.03e-02 |
| GO:BP | GO:0002053 | positive regulation of mesenchymal cell proliferation | 17 | 20 | 3.03e-02 |
| GO:BP | GO:0019221 | cytokine-mediated signaling pathway | 160 | 268 | 3.03e-02 |
| GO:BP | GO:0046700 | heterocycle catabolic process | 253 | 439 | 3.05e-02 |
| GO:BP | GO:0090077 | foam cell differentiation | 19 | 23 | 3.06e-02 |
| GO:BP | GO:0010742 | macrophage derived foam cell differentiation | 19 | 23 | 3.06e-02 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 105 | 169 | 3.07e-02 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 105 | 169 | 3.07e-02 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 297 | 521 | 3.11e-02 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 472 | 850 | 3.11e-02 |
| GO:BP | GO:0019725 | cellular homeostasis | 320 | 564 | 3.12e-02 |
| GO:BP | GO:1902305 | regulation of sodium ion transmembrane transport | 28 | 37 | 3.13e-02 |
| GO:BP | GO:0009261 | ribonucleotide catabolic process | 72 | 111 | 3.16e-02 |
| GO:BP | GO:0019058 | viral life cycle | 140 | 232 | 3.18e-02 |
| GO:BP | GO:0031333 | negative regulation of protein-containing complex assembly | 76 | 118 | 3.18e-02 |
| GO:BP | GO:0030100 | regulation of endocytosis | 120 | 196 | 3.19e-02 |
| GO:BP | GO:1901575 | organic substance catabolic process | 845 | 1562 | 3.20e-02 |
| GO:BP | GO:0070527 | platelet aggregation | 36 | 50 | 3.25e-02 |
| GO:BP | GO:0010660 | regulation of muscle cell apoptotic process | 36 | 50 | 3.25e-02 |
| GO:BP | GO:0097193 | intrinsic apoptotic signaling pathway | 152 | 254 | 3.33e-02 |
| GO:BP | GO:0006611 | protein export from nucleus | 39 | 55 | 3.35e-02 |
| GO:BP | GO:0009154 | purine ribonucleotide catabolic process | 69 | 106 | 3.36e-02 |
| GO:BP | GO:0043547 | positive regulation of GTPase activity | 73 | 113 | 3.41e-02 |
| GO:BP | GO:0010256 | endomembrane system organization | 273 | 477 | 3.41e-02 |
| GO:BP | GO:0043009 | chordate embryonic development | 289 | 507 | 3.50e-02 |
| GO:BP | GO:0044403 | biological process involved in symbiotic interaction | 127 | 209 | 3.50e-02 |
| GO:BP | GO:0022409 | positive regulation of cell-cell adhesion | 108 | 175 | 3.57e-02 |
| GO:BP | GO:0048659 | smooth muscle cell proliferation | 66 | 101 | 3.58e-02 |
| GO:BP | GO:0001101 | response to acid chemical | 66 | 101 | 3.58e-02 |
| GO:BP | GO:0019439 | aromatic compound catabolic process | 255 | 444 | 3.64e-02 |
| GO:BP | GO:0031623 | receptor internalization | 59 | 89 | 3.67e-02 |
| GO:BP | GO:0048385 | regulation of retinoic acid receptor signaling pathway | 12 | 13 | 3.67e-02 |
| GO:BP | GO:0060841 | venous blood vessel development | 12 | 13 | 3.67e-02 |
| GO:BP | GO:1903729 | regulation of plasma membrane organization | 12 | 13 | 3.67e-02 |
| GO:BP | GO:0140546 | defense response to symbiont | 315 | 556 | 3.67e-02 |
| GO:BP | GO:0003174 | mitral valve development | 12 | 13 | 3.67e-02 |
| GO:BP | GO:0045600 | positive regulation of fat cell differentiation | 34 | 47 | 3.70e-02 |
| GO:BP | GO:0014743 | regulation of muscle hypertrophy | 34 | 47 | 3.70e-02 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 49 | 72 | 3.71e-02 |
| GO:BP | GO:0071229 | cellular response to acid chemical | 49 | 72 | 3.71e-02 |
| GO:BP | GO:0019318 | hexose metabolic process | 109 | 177 | 3.71e-02 |
| GO:BP | GO:0061564 | axon development | 216 | 372 | 3.72e-02 |
| GO:BP | GO:0032984 | protein-containing complex disassembly | 129 | 213 | 3.75e-02 |
| GO:BP | GO:0014706 | striated muscle tissue development | 129 | 213 | 3.75e-02 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 100 | 161 | 3.75e-02 |
| GO:BP | GO:0031349 | positive regulation of defense response | 156 | 262 | 3.76e-02 |
| GO:BP | GO:0030857 | negative regulation of epithelial cell differentiation | 24 | 31 | 3.77e-02 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 46 | 67 | 3.78e-02 |
| GO:BP | GO:0072089 | stem cell proliferation | 56 | 84 | 3.79e-02 |
| GO:BP | GO:1904063 | negative regulation of cation transmembrane transport | 37 | 52 | 3.79e-02 |
| GO:BP | GO:0048545 | response to steroid hormone | 140 | 233 | 3.80e-02 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 96 | 154 | 3.84e-02 |
| GO:BP | GO:0035108 | limb morphogenesis | 71 | 110 | 3.87e-02 |
| GO:BP | GO:0035107 | appendage morphogenesis | 71 | 110 | 3.87e-02 |
| GO:BP | GO:0042330 | taxis | 146 | 244 | 3.87e-02 |
| GO:BP | GO:0006935 | chemotaxis | 146 | 244 | 3.87e-02 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 239 | 415 | 3.92e-02 |
| GO:BP | GO:0030163 | protein catabolic process | 440 | 792 | 3.93e-02 |
| GO:BP | GO:0060397 | growth hormone receptor signaling pathway via JAK-STAT | 9 | 9 | 3.93e-02 |
| GO:BP | GO:0090336 | positive regulation of brown fat cell differentiation | 9 | 9 | 3.93e-02 |
| GO:BP | GO:0030038 | contractile actin filament bundle assembly | 60 | 91 | 3.93e-02 |
| GO:BP | GO:0048845 | venous blood vessel morphogenesis | 9 | 9 | 3.93e-02 |
| GO:BP | GO:0043149 | stress fiber assembly | 60 | 91 | 3.93e-02 |
| GO:BP | GO:1901298 | regulation of hydrogen peroxide-mediated programmed cell death | 9 | 9 | 3.93e-02 |
| GO:BP | GO:0001765 | membrane raft assembly | 9 | 9 | 3.93e-02 |
| GO:BP | GO:1901292 | nucleoside phosphate catabolic process | 88 | 140 | 3.97e-02 |
| GO:BP | GO:0033673 | negative regulation of kinase activity | 84 | 133 | 4.03e-02 |
| GO:BP | GO:0003171 | atrioventricular valve development | 22 | 28 | 4.03e-02 |
| GO:BP | GO:0098876 | vesicle-mediated transport to the plasma membrane | 84 | 133 | 4.03e-02 |
| GO:BP | GO:0010591 | regulation of lamellipodium assembly | 22 | 28 | 4.03e-02 |
| GO:BP | GO:0032456 | endocytic recycling | 50 | 74 | 4.06e-02 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 68 | 105 | 4.07e-02 |
| GO:BP | GO:1905039 | carboxylic acid transmembrane transport | 68 | 105 | 4.07e-02 |
| GO:BP | GO:0072523 | purine-containing compound catabolic process | 76 | 119 | 4.10e-02 |
| GO:BP | GO:0034332 | adherens junction organization | 32 | 44 | 4.11e-02 |
| GO:BP | GO:0030217 | T cell differentiation | 112 | 183 | 4.16e-02 |
| GO:BP | GO:0071901 | negative regulation of protein serine/threonine kinase activity | 47 | 69 | 4.19e-02 |
| GO:BP | GO:0032273 | positive regulation of protein polymerization | 47 | 69 | 4.19e-02 |
| GO:BP | GO:0044270 | cellular nitrogen compound catabolic process | 250 | 436 | 4.19e-02 |
| GO:BP | GO:0048644 | muscle organ morphogenesis | 44 | 64 | 4.24e-02 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 273 | 479 | 4.24e-02 |
| GO:BP | GO:0048662 | negative regulation of smooth muscle cell proliferation | 27 | 36 | 4.24e-02 |
| GO:BP | GO:0048699 | generation of neurons | 589 | 1076 | 4.24e-02 |
| GO:BP | GO:1902176 | negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 20 | 25 | 4.24e-02 |
| GO:BP | GO:0072111 | cell proliferation involved in kidney development | 14 | 16 | 4.24e-02 |
| GO:BP | GO:0038095 | Fc-epsilon receptor signaling pathway | 14 | 16 | 4.24e-02 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 103 | 167 | 4.24e-02 |
| GO:BP | GO:0032092 | positive regulation of protein binding | 35 | 49 | 4.24e-02 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 360 | 642 | 4.24e-02 |
| GO:BP | GO:0071320 | cellular response to cAMP | 27 | 36 | 4.24e-02 |
| GO:BP | GO:0046323 | glucose import | 38 | 54 | 4.29e-02 |
| GO:BP | GO:0005996 | monosaccharide metabolic process | 118 | 194 | 4.29e-02 |
| GO:BP | GO:0048729 | tissue morphogenesis | 259 | 453 | 4.29e-02 |
| GO:BP | GO:0014910 | regulation of smooth muscle cell migration | 38 | 54 | 4.29e-02 |
| GO:BP | GO:0034766 | negative regulation of monoatomic ion transmembrane transport | 41 | 59 | 4.29e-02 |
| GO:BP | GO:0006195 | purine nucleotide catabolic process | 73 | 114 | 4.34e-02 |
| GO:BP | GO:0006006 | glucose metabolic process | 90 | 144 | 4.36e-02 |
| GO:BP | GO:0035767 | endothelial cell chemotaxis | 18 | 22 | 4.42e-02 |
| GO:BP | GO:0018210 | peptidyl-threonine modification | 51 | 76 | 4.45e-02 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 169 | 287 | 4.53e-02 |
| GO:BP | GO:0045862 | positive regulation of proteolysis | 146 | 245 | 4.54e-02 |
| GO:BP | GO:0019722 | calcium-mediated signaling | 78 | 123 | 4.62e-02 |
| GO:BP | GO:0051701 | biological process involved in interaction with host | 91 | 146 | 4.62e-02 |
| GO:BP | GO:1901873 | regulation of post-translational protein modification | 125 | 207 | 4.63e-02 |
| GO:BP | GO:0055085 | transmembrane transport | 507 | 921 | 4.64e-02 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 74 | 116 | 4.65e-02 |
| GO:BP | GO:0001889 | liver development | 70 | 109 | 4.65e-02 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 55 | 83 | 4.67e-02 |
| GO:BP | GO:2001056 | positive regulation of cysteine-type endopeptidase activity | 55 | 83 | 4.67e-02 |
| GO:BP | GO:0001503 | ossification | 177 | 302 | 4.76e-02 |
| GO:BP | GO:0043933 | protein-containing complex organization | 998 | 1863 | 4.82e-02 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 83 | 132 | 4.82e-02 |
| GO:BP | GO:0060350 | endochondral bone morphogenesis | 33 | 46 | 4.83e-02 |
| GO:BP | GO:0010611 | regulation of cardiac muscle hypertrophy | 33 | 46 | 4.83e-02 |
| GO:BP | GO:0010721 | negative regulation of cell development | 111 | 182 | 4.84e-02 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 92 | 148 | 4.84e-02 |
| GO:BP | GO:0060415 | muscle tissue morphogenesis | 42 | 61 | 4.86e-02 |
| GO:BP | GO:0072091 | regulation of stem cell proliferation | 42 | 61 | 4.86e-02 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 79 | 125 | 4.88e-02 |
| GO:BP | GO:0030510 | regulation of BMP signaling pathway | 52 | 78 | 4.88e-02 |
| GO:BP | GO:0030518 | intracellular steroid hormone receptor signaling pathway | 63 | 97 | 4.91e-02 |
| GO:BP | GO:0071772 | response to BMP | 75 | 118 | 4.92e-02 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 75 | 118 | 4.92e-02 |
| GO:BP | GO:0043281 | regulation of cysteine-type endopeptidase activity involved in apoptotic process | 67 | 104 | 4.94e-02 |
| GO:BP | GO:0002520 | immune system development | 88 | 141 | 4.94e-02 |
| GO:BP | GO:0002218 | activation of innate immune response | 102 | 166 | 4.97e-02 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 299 | 529 | 4.98e-02 |
NRnomtable_20k_enh %>%
filter(source=="KEGG", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "KEGG terms found in NR enhancer neargenes_20k") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| KEGG | KEGG:00000 | KEGG root term | 2946 | 5451 | 2.10e-09 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 51 | 60 | 4.20e-06 |
| KEGG | KEGG:05200 | Pathways in cancer | 249 | 391 | 9.31e-06 |
| KEGG | KEGG:04510 | Focal adhesion | 114 | 169 | 5.20e-04 |
| KEGG | KEGG:04380 | Osteoclast differentiation | 67 | 92 | 7.50e-04 |
| KEGG | KEGG:04072 | Phospholipase D signaling pathway | 76 | 107 | 7.50e-04 |
| KEGG | KEGG:04931 | Insulin resistance | 66 | 91 | 7.86e-04 |
| KEGG | KEGG:04148 | Efferocytosis | 83 | 120 | 1.15e-03 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 88 | 129 | 1.18e-03 |
| KEGG | KEGG:05135 | Yersinia infection | 75 | 107 | 1.18e-03 |
| KEGG | KEGG:05100 | Bacterial invasion of epithelial cells | 54 | 73 | 1.18e-03 |
| KEGG | KEGG:04630 | JAK-STAT signaling pathway | 60 | 83 | 1.30e-03 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 78 | 113 | 1.32e-03 |
| KEGG | KEGG:05131 | Shigellosis | 131 | 204 | 1.32e-03 |
| KEGG | KEGG:04658 | Th1 and Th2 cell differentiation | 39 | 50 | 1.45e-03 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 70 | 101 | 2.19e-03 |
| KEGG | KEGG:04917 | Prolactin signaling pathway | 41 | 54 | 2.34e-03 |
| KEGG | KEGG:04520 | Adherens junction | 59 | 83 | 2.34e-03 |
| KEGG | KEGG:05167 | Kaposi sarcoma-associated herpesvirus infection | 89 | 134 | 2.65e-03 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 61 | 87 | 2.78e-03 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 105 | 162 | 2.78e-03 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 88 | 133 | 3.05e-03 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 66 | 96 | 3.46e-03 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 103 | 160 | 4.09e-03 |
| KEGG | KEGG:05231 | Choline metabolism in cancer | 56 | 80 | 4.20e-03 |
| KEGG | KEGG:05130 | Pathogenic Escherichia coli infection | 93 | 143 | 4.20e-03 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 62 | 90 | 4.20e-03 |
| KEGG | KEGG:04062 | Chemokine signaling pathway | 73 | 109 | 4.75e-03 |
| KEGG | KEGG:04722 | Neurotrophin signaling pathway | 70 | 104 | 4.81e-03 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 89 | 137 | 5.21e-03 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 93 | 144 | 5.21e-03 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 142 | 231 | 5.57e-03 |
| KEGG | KEGG:04066 | HIF-1 signaling pathway | 59 | 86 | 5.57e-03 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 103 | 162 | 5.63e-03 |
| KEGG | KEGG:04934 | Cushing syndrome | 78 | 119 | 6.70e-03 |
| KEGG | KEGG:04662 | B cell receptor signaling pathway | 38 | 52 | 7.71e-03 |
| KEGG | KEGG:05132 | Salmonella infection | 124 | 201 | 8.78e-03 |
| KEGG | KEGG:04152 | AMPK signaling pathway | 67 | 101 | 8.92e-03 |
| KEGG | KEGG:04625 | C-type lectin receptor signaling pathway | 51 | 74 | 9.41e-03 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 66 | 100 | 1.13e-02 |
| KEGG | KEGG:05170 | Human immunodeficiency virus 1 infection | 94 | 149 | 1.16e-02 |
| KEGG | KEGG:04360 | Axon guidance | 97 | 155 | 1.28e-02 |
| KEGG | KEGG:04657 | IL-17 signaling pathway | 39 | 55 | 1.28e-02 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 105 | 169 | 1.28e-02 |
| KEGG | KEGG:05226 | Gastric cancer | 73 | 113 | 1.28e-02 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 102 | 164 | 1.28e-02 |
| KEGG | KEGG:03083 | Polycomb repressive complex | 48 | 70 | 1.28e-02 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 150 | 250 | 1.28e-02 |
| KEGG | KEGG:04217 | Necroptosis | 65 | 99 | 1.28e-02 |
| KEGG | KEGG:04664 | Fc epsilon RI signaling pathway | 31 | 42 | 1.29e-02 |
| KEGG | KEGG:05417 | Lipid and atherosclerosis | 90 | 143 | 1.29e-02 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 49 | 72 | 1.35e-02 |
| KEGG | KEGG:04660 | T cell receptor signaling pathway | 56 | 84 | 1.37e-02 |
| KEGG | KEGG:04730 | Long-term depression | 29 | 39 | 1.38e-02 |
| KEGG | KEGG:03082 | ATP-dependent chromatin remodeling | 60 | 91 | 1.39e-02 |
| KEGG | KEGG:04340 | Hedgehog signaling pathway | 35 | 49 | 1.47e-02 |
| KEGG | KEGG:04920 | Adipocytokine signaling pathway | 35 | 49 | 1.47e-02 |
| KEGG | KEGG:05210 | Colorectal cancer | 54 | 81 | 1.47e-02 |
| KEGG | KEGG:04392 | Hippo signaling pathway - multiple species | 20 | 25 | 1.47e-02 |
| KEGG | KEGG:04144 | Endocytosis | 130 | 216 | 1.58e-02 |
| KEGG | KEGG:04014 | Ras signaling pathway | 101 | 164 | 1.64e-02 |
| KEGG | KEGG:04921 | Oxytocin signaling pathway | 72 | 113 | 1.81e-02 |
| KEGG | KEGG:05224 | Breast cancer | 72 | 113 | 1.81e-02 |
| KEGG | KEGG:04218 | Cellular senescence | 85 | 136 | 1.81e-02 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 50 | 75 | 1.81e-02 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 77 | 122 | 1.81e-02 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 81 | 129 | 1.81e-02 |
| KEGG | KEGG:04666 | Fc gamma R-mediated phagocytosis | 50 | 75 | 1.81e-02 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 92 | 149 | 1.96e-02 |
| KEGG | KEGG:05145 | Toxoplasmosis | 48 | 72 | 2.11e-02 |
| KEGG | KEGG:05211 | Renal cell carcinoma | 42 | 62 | 2.30e-02 |
| KEGG | KEGG:04330 | Notch signaling pathway | 36 | 52 | 2.34e-02 |
| KEGG | KEGG:04710 | Circadian rhythm | 22 | 29 | 2.34e-02 |
| KEGG | KEGG:04611 | Platelet activation | 57 | 88 | 2.34e-02 |
| KEGG | KEGG:05418 | Fluid shear stress and atherosclerosis | 69 | 109 | 2.34e-02 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 80 | 129 | 2.65e-02 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 67 | 106 | 2.67e-02 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 66 | 105 | 3.35e-02 |
| KEGG | KEGG:04979 | Cholesterol metabolism | 24 | 33 | 3.63e-02 |
| KEGG | KEGG:01521 | EGFR tyrosine kinase inhibitor resistance | 47 | 72 | 3.66e-02 |
| KEGG | KEGG:05412 | Arrhythmogenic right ventricular cardiomyopathy | 44 | 67 | 3.90e-02 |
| KEGG | KEGG:04915 | Estrogen signaling pathway | 57 | 90 | 4.26e-02 |
| KEGG | KEGG:04070 | Phosphatidylinositol signaling system | 53 | 83 | 4.26e-02 |
| KEGG | KEGG:05161 | Hepatitis B | 77 | 126 | 4.51e-02 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 50 | 78 | 4.51e-02 |
| KEGG | KEGG:05212 | Pancreatic cancer | 46 | 71 | 4.51e-02 |
intersection of EAR_ATAC and RNA-response genes
EAR
# EAR_int_resgenes <- gost(query = int_EAR_n_RNAresp,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = FALSE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = background_df$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(EAR_int_resgenes,"data/GO_KEGG_analysis/EAR_int_resgenes.RDS")
EAR_int_restable <- EAR_int_resgenes$result %>%
dplyr::select(c(source, term_id,
term_name,intersection_size,
term_size, p_value))# %>%
EAR_int_restable %>%
filter(source=="GO:BP", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(.,caption = "GO:BP terms found in EAR neargenes intersected with RNA response genes" ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0050794 | regulation of cellular process | 1092 | 7153 | 2.38e-13 |
| GO:BP | GO:0065007 | biological regulation | 1165 | 7734 | 2.38e-13 |
| GO:BP | GO:0050789 | regulation of biological process | 1139 | 7516 | 2.38e-13 |
| GO:BP | GO:0032502 | developmental process | 689 | 4179 | 1.08e-11 |
| GO:BP | GO:0050793 | regulation of developmental process | 326 | 1697 | 2.76e-11 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 348 | 1844 | 3.19e-11 |
| GO:BP | GO:0007275 | multicellular organism development | 542 | 3170 | 5.67e-11 |
| GO:BP | GO:0048856 | anatomical structure development | 637 | 3867 | 2.06e-10 |
| GO:BP | GO:0048519 | negative regulation of biological process | 621 | 3764 | 3.20e-10 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 591 | 3550 | 3.20e-10 |
| GO:BP | GO:0048731 | system development | 471 | 2719 | 5.16e-10 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 353 | 1924 | 6.07e-10 |
| GO:BP | GO:0072359 | circulatory system development | 186 | 864 | 6.09e-10 |
| GO:BP | GO:0023052 | signaling | 640 | 3918 | 6.14e-10 |
| GO:BP | GO:0007154 | cell communication | 648 | 3983 | 8.57e-10 |
| GO:BP | GO:0048869 | cellular developmental process | 481 | 2804 | 9.61e-10 |
| GO:BP | GO:0030154 | cell differentiation | 480 | 2803 | 1.30e-09 |
| GO:BP | GO:0007165 | signal transduction | 592 | 3611 | 3.68e-09 |
| GO:BP | GO:0032501 | multicellular organismal process | 715 | 4504 | 3.92e-09 |
| GO:BP | GO:0051716 | cellular response to stimulus | 732 | 4640 | 6.49e-09 |
| GO:BP | GO:0048518 | positive regulation of biological process | 681 | 4301 | 3.47e-08 |
| GO:BP | GO:0014706 | striated muscle tissue development | 63 | 213 | 6.41e-08 |
| GO:BP | GO:0048513 | animal organ development | 353 | 2004 | 7.86e-08 |
| GO:BP | GO:0050896 | response to stimulus | 823 | 5379 | 1.02e-07 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 629 | 3954 | 1.34e-07 |
| GO:BP | GO:0048589 | developmental growth | 114 | 501 | 3.77e-07 |
| GO:BP | GO:0009888 | tissue development | 250 | 1348 | 4.61e-07 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 58 | 201 | 7.51e-07 |
| GO:BP | GO:0007507 | heart development | 111 | 499 | 2.18e-06 |
| GO:BP | GO:0022008 | neurogenesis | 229 | 1251 | 7.39e-06 |
| GO:BP | GO:0035556 | intracellular signal transduction | 333 | 1951 | 8.53e-06 |
| GO:BP | GO:0007417 | central nervous system development | 141 | 694 | 9.28e-06 |
| GO:BP | GO:0048468 | cell development | 323 | 1886 | 9.64e-06 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 129 | 622 | 1.00e-05 |
| GO:BP | GO:0048699 | generation of neurons | 200 | 1076 | 1.71e-05 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 174 | 913 | 2.23e-05 |
| GO:BP | GO:0030334 | regulation of cell migration | 135 | 668 | 2.23e-05 |
| GO:BP | GO:2000145 | regulation of cell motility | 140 | 700 | 2.38e-05 |
| GO:BP | GO:0016477 | cell migration | 189 | 1011 | 2.40e-05 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 135 | 673 | 3.15e-05 |
| GO:BP | GO:0060537 | muscle tissue development | 79 | 339 | 3.15e-05 |
| GO:BP | GO:0040012 | regulation of locomotion | 142 | 720 | 4.17e-05 |
| GO:BP | GO:0007399 | nervous system development | 302 | 1775 | 4.48e-05 |
| GO:BP | GO:0040007 | growth | 138 | 697 | 4.72e-05 |
| GO:BP | GO:0045444 | fat cell differentiation | 49 | 179 | 4.81e-05 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 233 | 1314 | 5.41e-05 |
| GO:BP | GO:0030900 | forebrain development | 66 | 272 | 6.19e-05 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 123 | 609 | 6.75e-05 |
| GO:BP | GO:0060419 | heart growth | 26 | 71 | 7.54e-05 |
| GO:BP | GO:0040011 | locomotion | 154 | 805 | 7.54e-05 |
| GO:BP | GO:0060322 | head development | 111 | 539 | 8.85e-05 |
| GO:BP | GO:0035239 | tube morphogenesis | 127 | 638 | 9.12e-05 |
| GO:BP | GO:0007420 | brain development | 104 | 498 | 9.83e-05 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 24 | 64 | 1.16e-04 |
| GO:BP | GO:0035295 | tube development | 150 | 787 | 1.23e-04 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 79 | 355 | 1.68e-04 |
| GO:BP | GO:0030182 | neuron differentiation | 184 | 1014 | 1.99e-04 |
| GO:BP | GO:0048638 | regulation of developmental growth | 59 | 243 | 2.03e-04 |
| GO:BP | GO:0009790 | embryo development | 154 | 821 | 2.16e-04 |
| GO:BP | GO:0030029 | actin filament-based process | 125 | 640 | 2.73e-04 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 192 | 1073 | 2.83e-04 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 135 | 706 | 3.29e-04 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 173 | 951 | 3.29e-04 |
| GO:BP | GO:0010646 | regulation of cell communication | 391 | 2444 | 3.37e-04 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 94 | 452 | 3.37e-04 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 233 | 1354 | 4.01e-04 |
| GO:BP | GO:0048870 | cell motility | 198 | 1120 | 4.05e-04 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 214 | 1231 | 5.12e-04 |
| GO:BP | GO:0001568 | blood vessel development | 105 | 525 | 5.41e-04 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 214 | 1234 | 5.91e-04 |
| GO:BP | GO:0001944 | vasculature development | 109 | 552 | 6.36e-04 |
| GO:BP | GO:0009966 | regulation of signal transduction | 349 | 2166 | 6.57e-04 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 351 | 2186 | 8.32e-04 |
| GO:BP | GO:0016358 | dendrite development | 46 | 183 | 8.32e-04 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 153 | 837 | 8.32e-04 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 315 | 1934 | 8.32e-04 |
| GO:BP | GO:0048666 | neuron development | 153 | 838 | 8.72e-04 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 412 | 2622 | 9.21e-04 |
| GO:BP | GO:0023051 | regulation of signaling | 386 | 2438 | 9.33e-04 |
| GO:BP | GO:0021537 | telencephalon development | 48 | 195 | 9.33e-04 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 304 | 1862 | 9.38e-04 |
| GO:BP | GO:0008360 | regulation of cell shape | 31 | 106 | 9.48e-04 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 371 | 2333 | 9.48e-04 |
| GO:BP | GO:0007423 | sensory organ development | 81 | 387 | 9.98e-04 |
| GO:BP | GO:0035265 | organ growth | 35 | 127 | 1.08e-03 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 100 | 505 | 1.14e-03 |
| GO:BP | GO:0044057 | regulation of system process | 80 | 384 | 1.30e-03 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 263 | 1588 | 1.37e-03 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 381 | 2415 | 1.39e-03 |
| GO:BP | GO:0001764 | neuron migration | 35 | 129 | 1.48e-03 |
| GO:BP | GO:0048568 | embryonic organ development | 66 | 303 | 1.60e-03 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 101 | 516 | 1.60e-03 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 269 | 1635 | 1.69e-03 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 90 | 450 | 1.87e-03 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 314 | 1951 | 1.87e-03 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 267 | 1624 | 1.87e-03 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 335 | 2100 | 1.95e-03 |
| GO:BP | GO:0031175 | neuron projection development | 136 | 743 | 1.97e-03 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 421 | 2717 | 2.32e-03 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 23 | 72 | 2.32e-03 |
| GO:BP | GO:0086065 | cell communication involved in cardiac conduction | 19 | 54 | 2.50e-03 |
| GO:BP | GO:0065008 | regulation of biological quality | 304 | 1893 | 2.92e-03 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 21 | 64 | 3.18e-03 |
| GO:BP | GO:0045598 | regulation of fat cell differentiation | 27 | 93 | 3.21e-03 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 298 | 1856 | 3.51e-03 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 29 | 105 | 4.58e-03 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 45 | 193 | 5.46e-03 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 178 | 1040 | 5.54e-03 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 287 | 1791 | 5.60e-03 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 294 | 1841 | 5.76e-03 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 247 | 1514 | 5.93e-03 |
| GO:BP | GO:0048729 | tissue morphogenesis | 88 | 453 | 5.99e-03 |
| GO:BP | GO:0007010 | cytoskeleton organization | 193 | 1146 | 6.67e-03 |
| GO:BP | GO:0060420 | regulation of heart growth | 17 | 49 | 7.05e-03 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 20 | 63 | 7.07e-03 |
| GO:BP | GO:0000902 | cell morphogenesis | 128 | 713 | 7.07e-03 |
| GO:BP | GO:0060284 | regulation of cell development | 106 | 570 | 7.07e-03 |
| GO:BP | GO:0043583 | ear development | 37 | 151 | 7.07e-03 |
| GO:BP | GO:0003205 | cardiac chamber development | 37 | 151 | 7.07e-03 |
| GO:BP | GO:0086004 | regulation of cardiac muscle cell contraction | 13 | 32 | 7.07e-03 |
| GO:BP | GO:0048588 | developmental cell growth | 42 | 179 | 7.15e-03 |
| GO:BP | GO:0042221 | response to chemical | 358 | 2304 | 7.50e-03 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 194 | 1157 | 7.50e-03 |
| GO:BP | GO:0050773 | regulation of dendrite development | 23 | 78 | 7.50e-03 |
| GO:BP | GO:0055006 | cardiac cell development | 23 | 78 | 7.50e-03 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 27 | 99 | 8.64e-03 |
| GO:BP | GO:0006468 | protein phosphorylation | 164 | 958 | 8.90e-03 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 22 | 74 | 8.90e-03 |
| GO:BP | GO:0034329 | cell junction assembly | 66 | 323 | 8.90e-03 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 82 | 422 | 8.90e-03 |
| GO:BP | GO:0016043 | cellular component organization | 698 | 4802 | 8.92e-03 |
| GO:BP | GO:0009952 | anterior/posterior pattern specification | 31 | 121 | 9.65e-03 |
| GO:BP | GO:0034330 | cell junction organization | 101 | 545 | 1.03e-02 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 151 | 874 | 1.03e-02 |
| GO:BP | GO:0014896 | muscle hypertrophy | 22 | 75 | 1.06e-02 |
| GO:BP | GO:0003008 | system process | 190 | 1139 | 1.08e-02 |
| GO:BP | GO:0003007 | heart morphogenesis | 48 | 218 | 1.10e-02 |
| GO:BP | GO:0048839 | inner ear development | 33 | 133 | 1.11e-02 |
| GO:BP | GO:0045668 | negative regulation of osteoblast differentiation | 14 | 38 | 1.14e-02 |
| GO:BP | GO:0065009 | regulation of molecular function | 213 | 1299 | 1.16e-02 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 179 | 1067 | 1.21e-02 |
| GO:BP | GO:1903243 | negative regulation of cardiac muscle hypertrophy in response to stress | 5 | 6 | 1.21e-02 |
| GO:BP | GO:0010616 | negative regulation of cardiac muscle adaptation | 5 | 6 | 1.21e-02 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 52 | 243 | 1.23e-02 |
| GO:BP | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 11 | 26 | 1.30e-02 |
| GO:BP | GO:0060038 | cardiac muscle cell proliferation | 15 | 43 | 1.31e-02 |
| GO:BP | GO:0016310 | phosphorylation | 190 | 1145 | 1.33e-02 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 24 | 87 | 1.41e-02 |
| GO:BP | GO:0043367 | CD4-positive, alpha-beta T cell differentiation | 17 | 53 | 1.60e-02 |
| GO:BP | GO:0086091 | regulation of heart rate by cardiac conduction | 13 | 35 | 1.63e-02 |
| GO:BP | GO:0075732 | viral penetration into host nucleus | 4 | 4 | 1.64e-02 |
| GO:BP | GO:0014745 | negative regulation of muscle adaptation | 6 | 9 | 1.65e-02 |
| GO:BP | GO:0055021 | regulation of cardiac muscle tissue growth | 15 | 44 | 1.66e-02 |
| GO:BP | GO:0086002 | cardiac muscle cell action potential involved in contraction | 15 | 44 | 1.66e-02 |
| GO:BP | GO:0060429 | epithelium development | 137 | 792 | 1.67e-02 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 173 | 1035 | 1.67e-02 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 47 | 217 | 1.67e-02 |
| GO:BP | GO:0003013 | circulatory system process | 81 | 426 | 1.67e-02 |
| GO:BP | GO:0060043 | regulation of cardiac muscle cell proliferation | 12 | 31 | 1.67e-02 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 21 | 73 | 1.67e-02 |
| GO:BP | GO:0003231 | cardiac ventricle development | 29 | 115 | 1.70e-02 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 259 | 1633 | 1.78e-02 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 32 | 132 | 1.82e-02 |
| GO:BP | GO:0043010 | camera-type eye development | 49 | 230 | 1.82e-02 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 74 | 384 | 1.88e-02 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 42 | 190 | 2.06e-02 |
| GO:BP | GO:0086066 | atrial cardiac muscle cell to AV node cell communication | 8 | 16 | 2.08e-02 |
| GO:BP | GO:0086026 | atrial cardiac muscle cell to AV node cell signaling | 8 | 16 | 2.08e-02 |
| GO:BP | GO:0086014 | atrial cardiac muscle cell action potential | 8 | 16 | 2.08e-02 |
| GO:BP | GO:0040008 | regulation of growth | 86 | 462 | 2.08e-02 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 102 | 566 | 2.13e-02 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 34 | 145 | 2.23e-02 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 60 | 300 | 2.33e-02 |
| GO:BP | GO:0014808 | release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 11 | 28 | 2.33e-02 |
| GO:BP | GO:1903514 | release of sequestered calcium ion into cytosol by endoplasmic reticulum | 11 | 28 | 2.33e-02 |
| GO:BP | GO:0051171 | regulation of nitrogen compound metabolic process | 573 | 3918 | 2.35e-02 |
| GO:BP | GO:0003012 | muscle system process | 64 | 325 | 2.36e-02 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 45 | 210 | 2.50e-02 |
| GO:BP | GO:0001508 | action potential | 26 | 102 | 2.53e-02 |
| GO:BP | GO:1903115 | regulation of actin filament-based movement | 13 | 37 | 2.53e-02 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 615 | 4233 | 2.53e-02 |
| GO:BP | GO:0009214 | cyclic nucleotide catabolic process | 7 | 13 | 2.54e-02 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 161 | 965 | 2.54e-02 |
| GO:BP | GO:0051960 | regulation of nervous system development | 66 | 339 | 2.58e-02 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 58 | 290 | 2.72e-02 |
| GO:BP | GO:0007416 | synapse assembly | 32 | 136 | 2.81e-02 |
| GO:BP | GO:0048880 | sensory system development | 54 | 266 | 2.83e-02 |
| GO:BP | GO:0008283 | cell population proliferation | 210 | 1306 | 2.87e-02 |
| GO:BP | GO:0001501 | skeletal system development | 67 | 347 | 2.89e-02 |
| GO:BP | GO:0046834 | lipid phosphorylation | 6 | 10 | 2.93e-02 |
| GO:BP | GO:0045600 | positive regulation of fat cell differentiation | 15 | 47 | 2.98e-02 |
| GO:BP | GO:0090257 | regulation of muscle system process | 39 | 177 | 2.98e-02 |
| GO:BP | GO:0086019 | cell-cell signaling involved in cardiac conduction | 11 | 29 | 2.98e-02 |
| GO:BP | GO:0070848 | response to growth factor | 98 | 547 | 2.98e-02 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 27 | 109 | 3.00e-02 |
| GO:BP | GO:2001222 | regulation of neuron migration | 13 | 38 | 3.13e-02 |
| GO:BP | GO:0014741 | negative regulation of muscle hypertrophy | 10 | 25 | 3.14e-02 |
| GO:BP | GO:0099536 | synaptic signaling | 88 | 483 | 3.14e-02 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 613 | 4231 | 3.16e-02 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 207 | 1289 | 3.16e-02 |
| GO:BP | GO:0007267 | cell-cell signaling | 177 | 1082 | 3.24e-02 |
| GO:BP | GO:0001525 | angiogenesis | 72 | 381 | 3.24e-02 |
| GO:BP | GO:0061061 | muscle structure development | 96 | 536 | 3.24e-02 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 27 | 110 | 3.30e-02 |
| GO:BP | GO:1990830 | cellular response to leukemia inhibitory factor | 22 | 83 | 3.30e-02 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 161 | 973 | 3.30e-02 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 133 | 783 | 3.33e-02 |
| GO:BP | GO:0001654 | eye development | 53 | 263 | 3.40e-02 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 38 | 173 | 3.42e-02 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 74 | 395 | 3.44e-02 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 28 | 116 | 3.45e-02 |
| GO:BP | GO:0008015 | blood circulation | 68 | 357 | 3.45e-02 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 121 | 704 | 3.53e-02 |
| GO:BP | GO:0150063 | visual system development | 53 | 264 | 3.62e-02 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 85 | 467 | 3.64e-02 |
| GO:BP | GO:0001503 | ossification | 59 | 302 | 3.85e-02 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 93 | 521 | 4.06e-02 |
| GO:BP | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction | 10 | 26 | 4.07e-02 |
| GO:BP | GO:0070372 | regulation of ERK1 and ERK2 cascade | 38 | 175 | 4.07e-02 |
| GO:BP | GO:0019222 | regulation of metabolic process | 661 | 4603 | 4.07e-02 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 94 | 528 | 4.09e-02 |
| GO:BP | GO:1990823 | response to leukemia inhibitory factor | 22 | 85 | 4.12e-02 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 712 | 4988 | 4.12e-02 |
| GO:BP | GO:0048644 | muscle organ morphogenesis | 18 | 64 | 4.12e-02 |
| GO:BP | GO:0061448 | connective tissue development | 43 | 205 | 4.12e-02 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 84 | 463 | 4.12e-02 |
| GO:BP | GO:0014855 | striated muscle cell proliferation | 17 | 59 | 4.12e-02 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 30 | 129 | 4.12e-02 |
| GO:BP | GO:0061337 | cardiac conduction | 22 | 85 | 4.12e-02 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 84 | 463 | 4.12e-02 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 22 | 85 | 4.12e-02 |
| GO:BP | GO:0021543 | pallium development | 31 | 135 | 4.15e-02 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 271 | 1751 | 4.15e-02 |
| GO:BP | GO:0045637 | regulation of myeloid cell differentiation | 31 | 135 | 4.15e-02 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 276 | 1787 | 4.16e-02 |
| GO:BP | GO:0030048 | actin filament-based movement | 26 | 107 | 4.16e-02 |
| GO:BP | GO:0010629 | negative regulation of gene expression | 133 | 790 | 4.16e-02 |
| GO:BP | GO:0007389 | pattern specification process | 56 | 286 | 4.39e-02 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 192 | 1197 | 4.40e-02 |
| GO:BP | GO:0044773 | mitotic DNA damage checkpoint signaling | 20 | 75 | 4.40e-02 |
| GO:BP | GO:0043584 | nose development | 6 | 11 | 4.42e-02 |
| GO:BP | GO:0002251 | organ or tissue specific immune response | 6 | 11 | 4.42e-02 |
| GO:BP | GO:0070296 | sarcoplasmic reticulum calcium ion transport | 11 | 31 | 4.42e-02 |
| GO:BP | GO:0046620 | regulation of organ growth | 19 | 70 | 4.42e-02 |
| GO:BP | GO:0071878 | negative regulation of adenylate cyclase-activating adrenergic receptor signaling pathway | 4 | 5 | 4.42e-02 |
| GO:BP | GO:0016079 | synaptic vesicle exocytosis | 19 | 70 | 4.42e-02 |
| GO:BP | GO:0086067 | AV node cell to bundle of His cell communication | 6 | 11 | 4.42e-02 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 28 | 119 | 4.42e-02 |
| GO:BP | GO:0061644 | protein localization to CENP-A containing chromatin | 4 | 5 | 4.42e-02 |
| GO:BP | GO:0086070 | SA node cell to atrial cardiac muscle cell communication | 6 | 11 | 4.42e-02 |
| GO:BP | GO:0010644 | cell communication by electrical coupling | 11 | 31 | 4.42e-02 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 103 | 591 | 4.43e-02 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 18 | 65 | 4.49e-02 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 397 | 2663 | 4.49e-02 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 75 | 408 | 4.53e-02 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 65 | 344 | 4.53e-02 |
| GO:BP | GO:0055008 | cardiac muscle tissue morphogenesis | 16 | 55 | 4.57e-02 |
| GO:BP | GO:0021591 | ventricular system development | 10 | 27 | 4.84e-02 |
| GO:BP | GO:0043009 | chordate embryonic development | 90 | 507 | 4.84e-02 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 76 | 416 | 4.94e-02 |
EAR_int_restable %>%
filter(source=="KEGG") %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "KEGG terms found in EAR neargenes intersected with RNA response genes") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| KEGG | KEGG:03013 | Nucleocytoplasmic transport | 23 | 96 | 8.45e-01 |
| KEGG | KEGG:03083 | Polycomb repressive complex | 17 | 70 | 8.45e-01 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 33 | 162 | 8.45e-01 |
| KEGG | KEGG:04730 | Long-term depression | 10 | 39 | 9.42e-01 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 24 | 122 | 9.42e-01 |
| KEGG | KEGG:04713 | Circadian entrainment | 15 | 68 | 9.42e-01 |
| KEGG | KEGG:05033 | Nicotine addiction | 4 | 10 | 9.42e-01 |
| KEGG | KEGG:04720 | Long-term potentiation | 12 | 51 | 9.42e-01 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 27 | 133 | 9.42e-01 |
| KEGG | KEGG:00592 | alpha-Linolenic acid metabolism | 4 | 10 | 9.42e-01 |
| KEGG | KEGG:04070 | Phosphatidylinositol signaling system | 18 | 83 | 9.42e-01 |
| KEGG | KEGG:04014 | Ras signaling pathway | 30 | 164 | 9.49e-01 |
| KEGG | KEGG:00564 | Glycerophospholipid metabolism | 15 | 71 | 9.49e-01 |
| KEGG | KEGG:05034 | Alcoholism | 20 | 102 | 9.49e-01 |
| KEGG | KEGG:04925 | Aldosterone synthesis and secretion | 11 | 75 | 1.00e+00 |
| KEGG | KEGG:04924 | Renin secretion | 7 | 50 | 1.00e+00 |
| KEGG | KEGG:04923 | Regulation of lipolysis in adipocytes | 3 | 41 | 1.00e+00 |
| KEGG | KEGG:04922 | Glucagon signaling pathway | 10 | 78 | 1.00e+00 |
| KEGG | KEGG:05410 | Hypertrophic cardiomyopathy | 9 | 77 | 1.00e+00 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 7 | 100 | 1.00e+00 |
| KEGG | KEGG:04921 | Oxytocin signaling pathway | 19 | 113 | 1.00e+00 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 17 | 105 | 1.00e+00 |
| KEGG | KEGG:04918 | Thyroid hormone synthesis | 5 | 53 | 1.00e+00 |
| KEGG | KEGG:04917 | Prolactin signaling pathway | 6 | 54 | 1.00e+00 |
| KEGG | KEGG:04916 | Melanogenesis | 11 | 75 | 1.00e+00 |
| KEGG | KEGG:04915 | Estrogen signaling pathway | 8 | 90 | 1.00e+00 |
| KEGG | KEGG:04914 | Progesterone-mediated oocyte maturation | 8 | 82 | 1.00e+00 |
| KEGG | KEGG:04913 | Ovarian steroidogenesis | 2 | 26 | 1.00e+00 |
| KEGG | KEGG:04920 | Adipocytokine signaling pathway | 9 | 49 | 1.00e+00 |
| KEGG | KEGG:04927 | Cortisol synthesis and secretion | 6 | 46 | 1.00e+00 |
| KEGG | KEGG:05226 | Gastric cancer | 20 | 113 | 1.00e+00 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 13 | 78 | 1.00e+00 |
| KEGG | KEGG:05323 | Rheumatoid arthritis | 4 | 38 | 1.00e+00 |
| KEGG | KEGG:04966 | Collecting duct acid secretion | 2 | 18 | 1.00e+00 |
| KEGG | KEGG:04964 | Proximal tubule bicarbonate reclamation | 1 | 18 | 1.00e+00 |
| KEGG | KEGG:04962 | Vasopressin-regulated water reabsorption | 1 | 37 | 1.00e+00 |
| KEGG | KEGG:04961 | Endocrine and other factor-regulated calcium reabsorption | 6 | 40 | 1.00e+00 |
| KEGG | KEGG:04960 | Aldosterone-regulated sodium reabsorption | 3 | 24 | 1.00e+00 |
| KEGG | KEGG:04940 | Type I diabetes mellitus | 2 | 7 | 1.00e+00 |
| KEGG | KEGG:04936 | Alcoholic liver disease | 14 | 91 | 1.00e+00 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 14 | 96 | 1.00e+00 |
| KEGG | KEGG:04934 | Cushing syndrome | 20 | 119 | 1.00e+00 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 12 | 87 | 1.00e+00 |
| KEGG | KEGG:04932 | Non-alcoholic fatty liver disease | 12 | 126 | 1.00e+00 |
| KEGG | KEGG:04931 | Insulin resistance | 14 | 91 | 1.00e+00 |
| KEGG | KEGG:04930 | Type II diabetes mellitus | 5 | 35 | 1.00e+00 |
| KEGG | KEGG:04929 | GnRH secretion | 6 | 50 | 1.00e+00 |
| KEGG | KEGG:04912 | GnRH signaling pathway | 9 | 74 | 1.00e+00 |
| KEGG | KEGG:04911 | Insulin secretion | 11 | 62 | 1.00e+00 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 17 | 113 | 1.00e+00 |
| KEGG | KEGG:04814 | Motor proteins | 13 | 141 | 1.00e+00 |
| KEGG | KEGG:04670 | Leukocyte transendothelial migration | 8 | 76 | 1.00e+00 |
| KEGG | KEGG:04668 | TNF signaling pathway | 10 | 83 | 1.00e+00 |
| KEGG | KEGG:04666 | Fc gamma R-mediated phagocytosis | 10 | 75 | 1.00e+00 |
| KEGG | KEGG:04664 | Fc epsilon RI signaling pathway | 2 | 42 | 1.00e+00 |
| KEGG | KEGG:04662 | B cell receptor signaling pathway | 3 | 52 | 1.00e+00 |
| KEGG | KEGG:04660 | T cell receptor signaling pathway | 6 | 84 | 1.00e+00 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 11 | 60 | 1.00e+00 |
| KEGG | KEGG:04658 | Th1 and Th2 cell differentiation | 7 | 50 | 1.00e+00 |
| KEGG | KEGG:04657 | IL-17 signaling pathway | 6 | 55 | 1.00e+00 |
| KEGG | KEGG:04650 | Natural killer cell mediated cytotoxicity | 5 | 53 | 1.00e+00 |
| KEGG | KEGG:04640 | Hematopoietic cell lineage | 5 | 28 | 1.00e+00 |
| KEGG | KEGG:04630 | JAK-STAT signaling pathway | 10 | 83 | 1.00e+00 |
| KEGG | KEGG:04625 | C-type lectin receptor signaling pathway | 11 | 74 | 1.00e+00 |
| KEGG | KEGG:05416 | Viral myocarditis | 4 | 36 | 1.00e+00 |
| KEGG | KEGG:04623 | Cytosolic DNA-sensing pathway | 4 | 46 | 1.00e+00 |
| KEGG | KEGG:04672 | Intestinal immune network for IgA production | 1 | 10 | 1.00e+00 |
| KEGG | KEGG:04970 | Salivary secretion | 9 | 54 | 1.00e+00 |
| KEGG | KEGG:04710 | Circadian rhythm | 6 | 29 | 1.00e+00 |
| KEGG | KEGG:04714 | Thermogenesis | 21 | 185 | 1.00e+00 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 18 | 169 | 1.00e+00 |
| KEGG | KEGG:04750 | Inflammatory mediator regulation of TRP channels | 12 | 72 | 1.00e+00 |
| KEGG | KEGG:04744 | Phototransduction | 3 | 12 | 1.00e+00 |
| KEGG | KEGG:04742 | Taste transduction | 5 | 20 | 1.00e+00 |
| KEGG | KEGG:04740 | Olfactory transduction | 6 | 25 | 1.00e+00 |
| KEGG | KEGG:05412 | Arrhythmogenic right ventricular cardiomyopathy | 11 | 67 | 1.00e+00 |
| KEGG | KEGG:04728 | Dopaminergic synapse | 13 | 97 | 1.00e+00 |
| KEGG | KEGG:04727 | GABAergic synapse | 6 | 52 | 1.00e+00 |
| KEGG | KEGG:04726 | Serotonergic synapse | 10 | 58 | 1.00e+00 |
| KEGG | KEGG:04725 | Cholinergic synapse | 11 | 82 | 1.00e+00 |
| KEGG | KEGG:04724 | Glutamatergic synapse | 10 | 74 | 1.00e+00 |
| KEGG | KEGG:04723 | Retrograde endocannabinoid signaling | 12 | 92 | 1.00e+00 |
| KEGG | KEGG:04722 | Neurotrophin signaling pathway | 12 | 104 | 1.00e+00 |
| KEGG | KEGG:04721 | Synaptic vesicle cycle | 10 | 49 | 1.00e+00 |
| KEGG | KEGG:05414 | Dilated cardiomyopathy | 8 | 82 | 1.00e+00 |
| KEGG | KEGG:05415 | Diabetic cardiomyopathy | 16 | 158 | 1.00e+00 |
| KEGG | KEGG:04971 | Gastric acid secretion | 8 | 51 | 1.00e+00 |
| KEGG | KEGG:04972 | Pancreatic secretion | 9 | 57 | 1.00e+00 |
| KEGG | KEGG:04973 | Carbohydrate digestion and absorption | 1 | 29 | 1.00e+00 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 23 | 162 | 1.00e+00 |
| KEGG | KEGG:05203 | Viral carcinogenesis | 20 | 142 | 1.00e+00 |
| KEGG | KEGG:05231 | Choline metabolism in cancer | 13 | 80 | 1.00e+00 |
| KEGG | KEGG:05200 | Pathways in cancer | 50 | 391 | 1.00e+00 |
| KEGG | KEGG:05171 | Coronavirus disease - COVID-19 | 9 | 149 | 1.00e+00 |
| KEGG | KEGG:05170 | Human immunodeficiency virus 1 infection | 17 | 149 | 1.00e+00 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 15 | 136 | 1.00e+00 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 24 | 388 | 1.00e+00 |
| KEGG | KEGG:05167 | Kaposi sarcoma-associated herpesvirus infection | 20 | 134 | 1.00e+00 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 21 | 164 | 1.00e+00 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 28 | 252 | 1.00e+00 |
| KEGG | KEGG:05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 8 | 65 | 1.00e+00 |
| KEGG | KEGG:05164 | Influenza A | 12 | 95 | 1.00e+00 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 21 | 160 | 1.00e+00 |
| KEGG | KEGG:05162 | Measles | 8 | 85 | 1.00e+00 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 20 | 144 | 1.00e+00 |
| KEGG | KEGG:05321 | Inflammatory bowel disease | 4 | 20 | 1.00e+00 |
| KEGG | KEGG:05207 | Chemical carcinogenesis - receptor activation | 17 | 132 | 1.00e+00 |
| KEGG | KEGG:05210 | Colorectal cancer | 13 | 81 | 1.00e+00 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 19 | 137 | 1.00e+00 |
| KEGG | KEGG:05224 | Breast cancer | 16 | 113 | 1.00e+00 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 10 | 64 | 1.00e+00 |
| KEGG | KEGG:05222 | Small cell lung cancer | 8 | 82 | 1.00e+00 |
| KEGG | KEGG:05221 | Acute myeloid leukemia | 10 | 52 | 1.00e+00 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 10 | 72 | 1.00e+00 |
| KEGG | KEGG:05219 | Bladder cancer | 4 | 35 | 1.00e+00 |
| KEGG | KEGG:05218 | Melanoma | 9 | 56 | 1.00e+00 |
| KEGG | KEGG:05217 | Basal cell carcinoma | 8 | 48 | 1.00e+00 |
| KEGG | KEGG:05216 | Thyroid cancer | 6 | 33 | 1.00e+00 |
| KEGG | KEGG:05215 | Prostate cancer | 10 | 82 | 1.00e+00 |
| KEGG | KEGG:05214 | Glioma | 10 | 65 | 1.00e+00 |
| KEGG | KEGG:05213 | Endometrial cancer | 8 | 54 | 1.00e+00 |
| KEGG | KEGG:05212 | Pancreatic cancer | 12 | 71 | 1.00e+00 |
| KEGG | KEGG:05211 | Renal cell carcinoma | 9 | 62 | 1.00e+00 |
| KEGG | KEGG:05208 | Chemical carcinogenesis - reactive oxygen species | 10 | 173 | 1.00e+00 |
| KEGG | KEGG:05230 | Central carbon metabolism in cancer | 2 | 58 | 1.00e+00 |
| KEGG | KEGG:05161 | Hepatitis B | 14 | 126 | 1.00e+00 |
| KEGG | KEGG:05152 | Tuberculosis | 9 | 101 | 1.00e+00 |
| KEGG | KEGG:05030 | Cocaine addiction | 3 | 32 | 1.00e+00 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 37 | 364 | 1.00e+00 |
| KEGG | KEGG:05020 | Prion disease | 17 | 204 | 1.00e+00 |
| KEGG | KEGG:05017 | Spinocerebellar ataxia | 15 | 120 | 1.00e+00 |
| KEGG | KEGG:05016 | Huntington disease | 20 | 246 | 1.00e+00 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 34 | 291 | 1.00e+00 |
| KEGG | KEGG:05012 | Parkinson disease | 19 | 211 | 1.00e+00 |
| KEGG | KEGG:05010 | Alzheimer disease | 27 | 296 | 1.00e+00 |
| KEGG | KEGG:04979 | Cholesterol metabolism | 4 | 33 | 1.00e+00 |
| KEGG | KEGG:04978 | Mineral absorption | 5 | 38 | 1.00e+00 |
| KEGG | KEGG:04622 | RIG-I-like receptor signaling pathway | 4 | 44 | 1.00e+00 |
| KEGG | KEGG:04977 | Vitamin digestion and absorption | 3 | 13 | 1.00e+00 |
| KEGG | KEGG:04976 | Bile secretion | 6 | 41 | 1.00e+00 |
| KEGG | KEGG:04975 | Fat digestion and absorption | 2 | 18 | 1.00e+00 |
| KEGG | KEGG:04974 | Protein digestion and absorption | 9 | 56 | 1.00e+00 |
| KEGG | KEGG:05031 | Amphetamine addiction | 8 | 45 | 1.00e+00 |
| KEGG | KEGG:05160 | Hepatitis C | 16 | 110 | 1.00e+00 |
| KEGG | KEGG:05032 | Morphine addiction | 11 | 54 | 1.00e+00 |
| KEGG | KEGG:05100 | Bacterial invasion of epithelial cells | 10 | 73 | 1.00e+00 |
| KEGG | KEGG:05150 | Staphylococcus aureus infection | 1 | 13 | 1.00e+00 |
| KEGG | KEGG:05146 | Amoebiasis | 1 | 62 | 1.00e+00 |
| KEGG | KEGG:05145 | Toxoplasmosis | 7 | 72 | 1.00e+00 |
| KEGG | KEGG:05144 | Malaria | 1 | 20 | 1.00e+00 |
| KEGG | KEGG:05143 | African trypanosomiasis | 1 | 16 | 1.00e+00 |
| KEGG | KEGG:05142 | Chagas disease | 4 | 72 | 1.00e+00 |
| KEGG | KEGG:05140 | Leishmaniasis | 4 | 38 | 1.00e+00 |
| KEGG | KEGG:05135 | Yersinia infection | 12 | 107 | 1.00e+00 |
| KEGG | KEGG:05134 | Legionellosis | 3 | 34 | 1.00e+00 |
| KEGG | KEGG:05133 | Pertussis | 4 | 42 | 1.00e+00 |
| KEGG | KEGG:05132 | Salmonella infection | 16 | 201 | 1.00e+00 |
| KEGG | KEGG:05131 | Shigellosis | 20 | 204 | 1.00e+00 |
| KEGG | KEGG:05130 | Pathogenic Escherichia coli infection | 15 | 143 | 1.00e+00 |
| KEGG | KEGG:05120 | Epithelial cell signaling in Helicobacter pylori infection | 5 | 55 | 1.00e+00 |
| KEGG | KEGG:05110 | Vibrio cholerae infection | 5 | 41 | 1.00e+00 |
| KEGG | KEGG:05322 | Systemic lupus erythematosus | 8 | 44 | 1.00e+00 |
| KEGG | KEGG:04621 | NOD-like receptor signaling pathway | 8 | 109 | 1.00e+00 |
| KEGG | KEGG:04611 | Platelet activation | 13 | 88 | 1.00e+00 |
| KEGG | KEGG:04614 | Renin-angiotensin system | 1 | 10 | 1.00e+00 |
| KEGG | KEGG:00760 | Nicotinate and nicotinamide metabolism | 3 | 28 | 1.00e+00 |
| KEGG | KEGG:00750 | Vitamin B6 metabolism | 1 | 5 | 1.00e+00 |
| KEGG | KEGG:00730 | Thiamine metabolism | 2 | 10 | 1.00e+00 |
| KEGG | KEGG:00670 | One carbon pool by folate | 2 | 18 | 1.00e+00 |
| KEGG | KEGG:00650 | Butanoate metabolism | 2 | 17 | 1.00e+00 |
| KEGG | KEGG:00640 | Propanoate metabolism | 2 | 27 | 1.00e+00 |
| KEGG | KEGG:00630 | Glyoxylate and dicarboxylate metabolism | 2 | 25 | 1.00e+00 |
| KEGG | KEGG:00620 | Pyruvate metabolism | 2 | 31 | 1.00e+00 |
| KEGG | KEGG:00601 | Glycosphingolipid biosynthesis - lacto and neolacto series | 1 | 14 | 1.00e+00 |
| KEGG | KEGG:00600 | Sphingolipid metabolism | 7 | 42 | 1.00e+00 |
| KEGG | KEGG:00591 | Linoleic acid metabolism | 2 | 7 | 1.00e+00 |
| KEGG | KEGG:00590 | Arachidonic acid metabolism | 3 | 25 | 1.00e+00 |
| KEGG | KEGG:00565 | Ether lipid metabolism | 4 | 28 | 1.00e+00 |
| KEGG | KEGG:00563 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 2 | 23 | 1.00e+00 |
| KEGG | KEGG:00562 | Inositol phosphate metabolism | 7 | 60 | 1.00e+00 |
| KEGG | KEGG:00770 | Pantothenate and CoA biosynthesis | 3 | 15 | 1.00e+00 |
| KEGG | KEGG:00785 | Lipoic acid metabolism | 1 | 16 | 1.00e+00 |
| KEGG | KEGG:00790 | Folate biosynthesis | 1 | 18 | 1.00e+00 |
| KEGG | KEGG:00830 | Retinol metabolism | 1 | 14 | 1.00e+00 |
| KEGG | KEGG:01524 | Platinum drug resistance | 5 | 60 | 1.00e+00 |
| KEGG | KEGG:01523 | Antifolate resistance | 4 | 22 | 1.00e+00 |
| KEGG | KEGG:01522 | Endocrine resistance | 9 | 82 | 1.00e+00 |
| KEGG | KEGG:01521 | EGFR tyrosine kinase inhibitor resistance | 14 | 72 | 1.00e+00 |
| KEGG | KEGG:01250 | Biosynthesis of nucleotide sugars | 3 | 34 | 1.00e+00 |
| KEGG | KEGG:01240 | Biosynthesis of cofactors | 11 | 109 | 1.00e+00 |
| KEGG | KEGG:01232 | Nucleotide metabolism | 7 | 65 | 1.00e+00 |
| KEGG | KEGG:00561 | Glycerolipid metabolism | 10 | 44 | 1.00e+00 |
| KEGG | KEGG:01230 | Biosynthesis of amino acids | 5 | 52 | 1.00e+00 |
| KEGG | KEGG:01200 | Carbon metabolism | 6 | 88 | 1.00e+00 |
| KEGG | KEGG:01100 | Metabolic pathways | 119 | 1077 | 1.00e+00 |
| KEGG | KEGG:01040 | Biosynthesis of unsaturated fatty acids | 6 | 23 | 1.00e+00 |
| KEGG | KEGG:00983 | Drug metabolism - other enzymes | 2 | 40 | 1.00e+00 |
| KEGG | KEGG:00970 | Aminoacyl-tRNA biosynthesis | 5 | 42 | 1.00e+00 |
| KEGG | KEGG:00910 | Nitrogen metabolism | 1 | 8 | 1.00e+00 |
| KEGG | KEGG:00900 | Terpenoid backbone biosynthesis | 2 | 21 | 1.00e+00 |
| KEGG | KEGG:01212 | Fatty acid metabolism | 10 | 46 | 1.00e+00 |
| KEGG | KEGG:00534 | Glycosaminoglycan biosynthesis - heparan sulfate / heparin | 4 | 19 | 1.00e+00 |
| KEGG | KEGG:00532 | Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate | 2 | 20 | 1.00e+00 |
| KEGG | KEGG:00520 | Amino sugar and nucleotide sugar metabolism | 3 | 44 | 1.00e+00 |
| KEGG | KEGG:00240 | Pyrimidine metabolism | 3 | 44 | 1.00e+00 |
| KEGG | KEGG:00230 | Purine metabolism | 18 | 93 | 1.00e+00 |
| KEGG | KEGG:00220 | Arginine biosynthesis | 1 | 15 | 1.00e+00 |
| KEGG | KEGG:00190 | Oxidative phosphorylation | 6 | 99 | 1.00e+00 |
| KEGG | KEGG:00130 | Ubiquinone and other terpenoid-quinone biosynthesis | 1 | 9 | 1.00e+00 |
| KEGG | KEGG:00071 | Fatty acid degradation | 5 | 31 | 1.00e+00 |
| KEGG | KEGG:00062 | Fatty acid elongation | 3 | 22 | 1.00e+00 |
| KEGG | KEGG:00250 | Alanine, aspartate and glutamate metabolism | 2 | 28 | 1.00e+00 |
| KEGG | KEGG:00061 | Fatty acid biosynthesis | 2 | 9 | 1.00e+00 |
| KEGG | KEGG:00052 | Galactose metabolism | 2 | 21 | 1.00e+00 |
| KEGG | KEGG:00051 | Fructose and mannose metabolism | 2 | 26 | 1.00e+00 |
| KEGG | KEGG:00040 | Pentose and glucuronate interconversions | 1 | 11 | 1.00e+00 |
| KEGG | KEGG:00030 | Pentose phosphate pathway | 4 | 20 | 1.00e+00 |
| KEGG | KEGG:00020 | Citrate cycle (TCA cycle) | 2 | 26 | 1.00e+00 |
| KEGG | KEGG:00010 | Glycolysis / Gluconeogenesis | 3 | 41 | 1.00e+00 |
| KEGG | KEGG:00000 | KEGG root term | 713 | 5451 | 1.00e+00 |
| KEGG | KEGG:00053 | Ascorbate and aldarate metabolism | 1 | 8 | 1.00e+00 |
| KEGG | KEGG:02010 | ABC transporters | 4 | 24 | 1.00e+00 |
| KEGG | KEGG:00260 | Glycine, serine and threonine metabolism | 1 | 26 | 1.00e+00 |
| KEGG | KEGG:00280 | Valine, leucine and isoleucine degradation | 4 | 42 | 1.00e+00 |
| KEGG | KEGG:00515 | Mannose type O-glycan biosynthesis | 1 | 19 | 1.00e+00 |
| KEGG | KEGG:00514 | Other types of O-glycan biosynthesis | 4 | 35 | 1.00e+00 |
| KEGG | KEGG:00513 | Various types of N-glycan biosynthesis | 2 | 37 | 1.00e+00 |
| KEGG | KEGG:00512 | Mucin type O-glycan biosynthesis | 2 | 21 | 1.00e+00 |
| KEGG | KEGG:00511 | Other glycan degradation | 1 | 14 | 1.00e+00 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 4 | 49 | 1.00e+00 |
| KEGG | KEGG:00500 | Starch and sucrose metabolism | 2 | 22 | 1.00e+00 |
| KEGG | KEGG:00270 | Cysteine and methionine metabolism | 1 | 37 | 1.00e+00 |
| KEGG | KEGG:00480 | Glutathione metabolism | 2 | 39 | 1.00e+00 |
| KEGG | KEGG:00450 | Selenocompound metabolism | 2 | 14 | 1.00e+00 |
| KEGG | KEGG:00440 | Phosphonate and phosphinate metabolism | 1 | 5 | 1.00e+00 |
| KEGG | KEGG:00410 | beta-Alanine metabolism | 3 | 22 | 1.00e+00 |
| KEGG | KEGG:00380 | Tryptophan metabolism | 1 | 21 | 1.00e+00 |
| KEGG | KEGG:00340 | Histidine metabolism | 1 | 10 | 1.00e+00 |
| KEGG | KEGG:00330 | Arginine and proline metabolism | 5 | 33 | 1.00e+00 |
| KEGG | KEGG:00310 | Lysine degradation | 10 | 56 | 1.00e+00 |
| KEGG | KEGG:00470 | D-Amino acid metabolism | 1 | 4 | 1.00e+00 |
| KEGG | KEGG:04620 | Toll-like receptor signaling pathway | 6 | 60 | 1.00e+00 |
| KEGG | KEGG:03008 | Ribosome biogenesis in eukaryotes | 4 | 65 | 1.00e+00 |
| KEGG | KEGG:03015 | mRNA surveillance pathway | 7 | 80 | 1.00e+00 |
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 20 | 124 | 1.00e+00 |
| KEGG | KEGG:04260 | Cardiac muscle contraction | 7 | 63 | 1.00e+00 |
| KEGG | KEGG:04218 | Cellular senescence | 23 | 136 | 1.00e+00 |
| KEGG | KEGG:04217 | Necroptosis | 10 | 99 | 1.00e+00 |
| KEGG | KEGG:04216 | Ferroptosis | 6 | 31 | 1.00e+00 |
| KEGG | KEGG:04213 | Longevity regulating pathway - multiple species | 6 | 50 | 1.00e+00 |
| KEGG | KEGG:04211 | Longevity regulating pathway | 9 | 75 | 1.00e+00 |
| KEGG | KEGG:04210 | Apoptosis | 11 | 112 | 1.00e+00 |
| KEGG | KEGG:04152 | AMPK signaling pathway | 17 | 101 | 1.00e+00 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 24 | 250 | 1.00e+00 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 16 | 129 | 1.00e+00 |
| KEGG | KEGG:04148 | Efferocytosis | 17 | 120 | 1.00e+00 |
| KEGG | KEGG:04146 | Peroxisome | 5 | 66 | 1.00e+00 |
| KEGG | KEGG:04145 | Phagosome | 6 | 84 | 1.00e+00 |
| KEGG | KEGG:04144 | Endocytosis | 27 | 216 | 1.00e+00 |
| KEGG | KEGG:04270 | Vascular smooth muscle contraction | 13 | 92 | 1.00e+00 |
| KEGG | KEGG:04330 | Notch signaling pathway | 10 | 52 | 1.00e+00 |
| KEGG | KEGG:04340 | Hedgehog signaling pathway | 5 | 49 | 1.00e+00 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 16 | 90 | 1.00e+00 |
| KEGG | KEGG:04613 | Neutrophil extracellular trap formation | 14 | 89 | 1.00e+00 |
| KEGG | KEGG:04612 | Antigen processing and presentation | 3 | 26 | 1.00e+00 |
| KEGG | KEGG:04610 | Complement and coagulation cascades | 2 | 35 | 1.00e+00 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 19 | 101 | 1.00e+00 |
| KEGG | KEGG:04540 | Gap junction | 8 | 71 | 1.00e+00 |
| KEGG | KEGG:04530 | Tight junction | 21 | 129 | 1.00e+00 |
| KEGG | KEGG:04520 | Adherens junction | 15 | 83 | 1.00e+00 |
| KEGG | KEGG:04142 | Lysosome | 6 | 109 | 1.00e+00 |
| KEGG | KEGG:04514 | Cell adhesion molecules | 12 | 69 | 1.00e+00 |
| KEGG | KEGG:04510 | Focal adhesion | 18 | 169 | 1.00e+00 |
| KEGG | KEGG:04392 | Hippo signaling pathway - multiple species | 5 | 25 | 1.00e+00 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 24 | 129 | 1.00e+00 |
| KEGG | KEGG:04380 | Osteoclast differentiation | 13 | 92 | 1.00e+00 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 10 | 104 | 1.00e+00 |
| KEGG | KEGG:04370 | VEGF signaling pathway | 3 | 47 | 1.00e+00 |
| KEGG | KEGG:04360 | Axon guidance | 26 | 155 | 1.00e+00 |
| KEGG | KEGG:04512 | ECM-receptor interaction | 4 | 66 | 1.00e+00 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 12 | 149 | 1.00e+00 |
| KEGG | KEGG:04140 | Autophagy - animal | 14 | 149 | 1.00e+00 |
| KEGG | KEGG:04137 | Mitophagy - animal | 10 | 94 | 1.00e+00 |
| KEGG | KEGG:03460 | Fanconi anemia pathway | 5 | 46 | 1.00e+00 |
| KEGG | KEGG:03450 | Non-homologous end-joining | 2 | 12 | 1.00e+00 |
| KEGG | KEGG:03440 | Homologous recombination | 9 | 38 | 1.00e+00 |
| KEGG | KEGG:03430 | Mismatch repair | 4 | 22 | 1.00e+00 |
| KEGG | KEGG:03420 | Nucleotide excision repair | 5 | 54 | 1.00e+00 |
| KEGG | KEGG:03410 | Base excision repair | 5 | 44 | 1.00e+00 |
| KEGG | KEGG:03320 | PPAR signaling pathway | 10 | 47 | 1.00e+00 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 36 | 231 | 1.00e+00 |
| KEGG | KEGG:03265 | Virion - Ebolavirus and Lyssavirus | 2 | 5 | 1.00e+00 |
| KEGG | KEGG:03082 | ATP-dependent chromatin remodeling | 16 | 91 | 1.00e+00 |
| KEGG | KEGG:03060 | Protein export | 3 | 28 | 1.00e+00 |
| KEGG | KEGG:03050 | Proteasome | 3 | 40 | 1.00e+00 |
| KEGG | KEGG:03040 | Spliceosome | 8 | 124 | 1.00e+00 |
| KEGG | KEGG:03030 | DNA replication | 5 | 34 | 1.00e+00 |
| KEGG | KEGG:03022 | Basal transcription factors | 4 | 33 | 1.00e+00 |
| KEGG | KEGG:03018 | RNA degradation | 6 | 67 | 1.00e+00 |
| KEGG | KEGG:03250 | Viral life cycle - HIV-1 | 6 | 54 | 1.00e+00 |
| KEGG | KEGG:03010 | Ribosome | 4 | 121 | 1.00e+00 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 13 | 75 | 1.00e+00 |
| KEGG | KEGG:04020 | Calcium signaling pathway | 26 | 161 | 1.00e+00 |
| KEGG | KEGG:04136 | Autophagy - other | 3 | 29 | 1.00e+00 |
| KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 20 | 127 | 1.00e+00 |
| KEGG | KEGG:04115 | p53 signaling pathway | 7 | 66 | 1.00e+00 |
| KEGG | KEGG:04114 | Oocyte meiosis | 12 | 103 | 1.00e+00 |
| KEGG | KEGG:04110 | Cell cycle | 18 | 146 | 1.00e+00 |
| KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 13 | 101 | 1.00e+00 |
| KEGG | KEGG:04072 | Phospholipase D signaling pathway | 18 | 107 | 1.00e+00 |
| KEGG | KEGG:05417 | Lipid and atherosclerosis | 19 | 143 | 1.00e+00 |
| KEGG | KEGG:04071 | Sphingolipid signaling pathway | 12 | 103 | 1.00e+00 |
| KEGG | KEGG:04066 | HIF-1 signaling pathway | 13 | 86 | 1.00e+00 |
| KEGG | KEGG:04064 | NF-kappa B signaling pathway | 9 | 61 | 1.00e+00 |
| KEGG | KEGG:04062 | Chemokine signaling pathway | 13 | 109 | 1.00e+00 |
| KEGG | KEGG:04061 | Viral protein interaction with cytokine and cytokine receptor | 2 | 19 | 1.00e+00 |
| KEGG | KEGG:04060 | Cytokine-cytokine receptor interaction | 8 | 94 | 1.00e+00 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 24 | 147 | 1.00e+00 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 20 | 129 | 1.00e+00 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 19 | 106 | 1.00e+00 |
| KEGG | KEGG:05418 | Fluid shear stress and atherosclerosis | 8 | 109 | 1.00e+00 |
EAR_int_restable %>%
filter(source=="GO:BP", p_value < 0.05) %>%
slice_head(n=10) %>%
mutate(log10_p=(-log(p_value, base=10))) %>%
ggplot(., aes(x=log10_p, y=reorder(term_name, log10_p)))+
geom_col(aes(fill="#F8766D"))+
theme_bw()+
ylab("")+
xlab(paste0("-log10 p-value"))+
guides(fill="none")+
theme(axis.text.y = element_text(color="black"))+
ggtitle("Intersection EAR")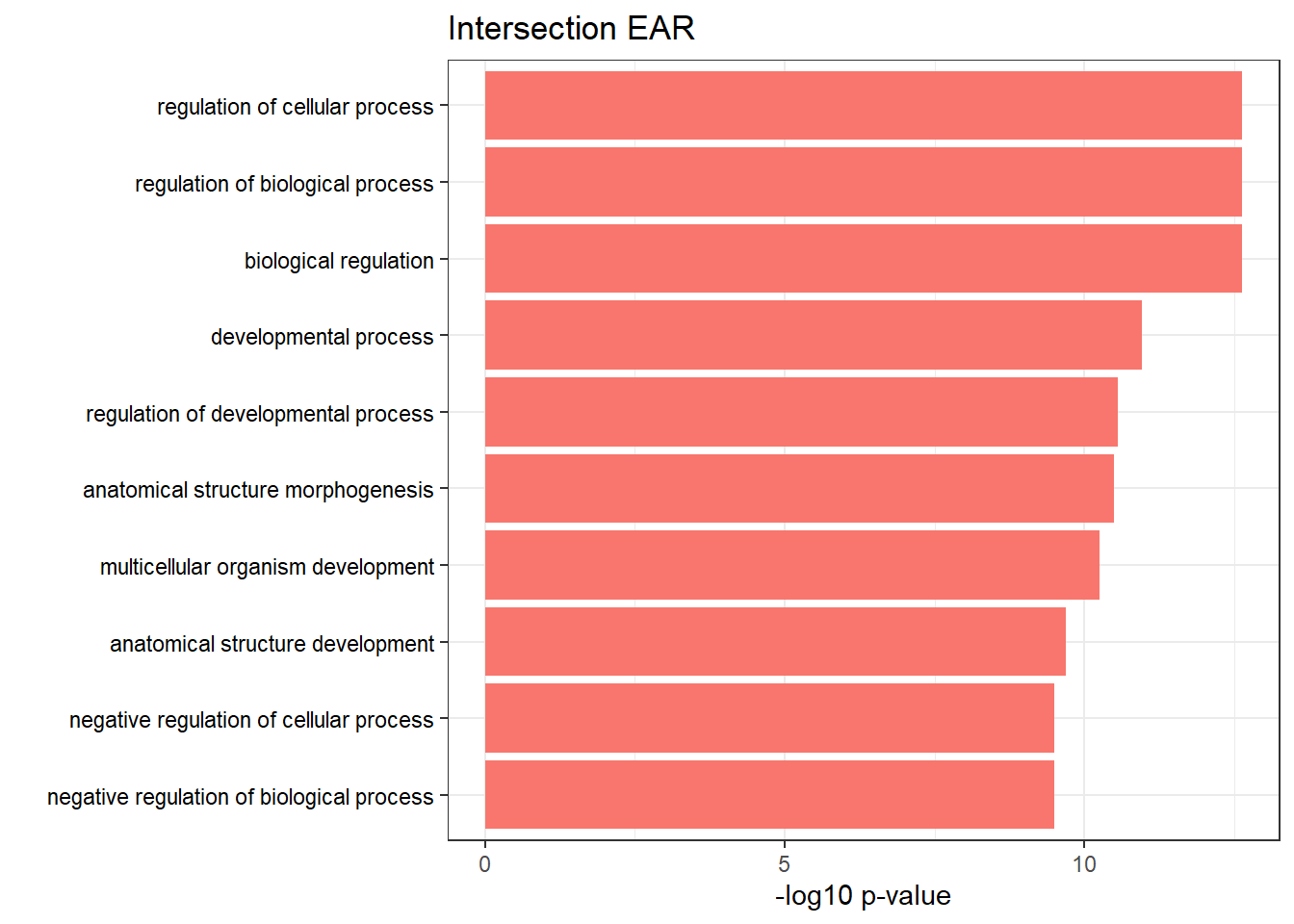
ESR
# ESR_int_resgenes <- gost(query = int_ESR_n_RNAresp,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = FALSE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = background_df$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(ESR_int_resgenes,"data/GO_KEGG_analysis/ESR_int_resgenes.RDS")
ESR_int_restable <- ESR_int_resgenes$result %>%
dplyr::select(c(source, term_id,
term_name,intersection_size,
term_size, p_value))# %>%
ESR_int_restable %>%
filter(source=="GO:BP", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(.,caption = "GO:BP terms found in ESR neargenes intersected with RNA response genes" ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0032501 | multicellular organismal process | 1141 | 4504 | 9.64e-21 |
| GO:BP | GO:0007275 | multicellular organism development | 845 | 3170 | 7.84e-20 |
| GO:BP | GO:0048856 | anatomical structure development | 995 | 3867 | 2.24e-19 |
| GO:BP | GO:0030154 | cell differentiation | 755 | 2803 | 1.65e-18 |
| GO:BP | GO:0048869 | cellular developmental process | 755 | 2804 | 1.65e-18 |
| GO:BP | GO:0032502 | developmental process | 1056 | 4179 | 1.90e-18 |
| GO:BP | GO:0048731 | system development | 733 | 2719 | 4.74e-18 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 526 | 1844 | 7.62e-17 |
| GO:BP | GO:0007154 | cell communication | 1004 | 3983 | 9.23e-17 |
| GO:BP | GO:0050896 | response to stimulus | 1298 | 5379 | 1.04e-16 |
| GO:BP | GO:0023052 | signaling | 986 | 3918 | 4.09e-16 |
| GO:BP | GO:0051716 | cellular response to stimulus | 1136 | 4640 | 1.80e-15 |
| GO:BP | GO:0009888 | tissue development | 399 | 1348 | 6.29e-15 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 535 | 1924 | 6.66e-15 |
| GO:BP | GO:0007165 | signal transduction | 909 | 3611 | 2.40e-14 |
| GO:BP | GO:0050794 | regulation of cellular process | 1638 | 7153 | 1.22e-13 |
| GO:BP | GO:0048468 | cell development | 519 | 1886 | 1.86e-13 |
| GO:BP | GO:0048513 | animal organ development | 546 | 2004 | 1.94e-13 |
| GO:BP | GO:0050789 | regulation of biological process | 1705 | 7516 | 6.05e-13 |
| GO:BP | GO:0050793 | regulation of developmental process | 471 | 1697 | 1.17e-12 |
| GO:BP | GO:0065007 | biological regulation | 1742 | 7734 | 4.66e-12 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 261 | 837 | 6.04e-12 |
| GO:BP | GO:0007399 | nervous system development | 485 | 1775 | 7.08e-12 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 308 | 1040 | 3.45e-11 |
| GO:BP | GO:0022008 | neurogenesis | 356 | 1251 | 1.39e-10 |
| GO:BP | GO:0044057 | regulation of system process | 138 | 384 | 1.96e-10 |
| GO:BP | GO:0072359 | circulatory system development | 261 | 864 | 2.83e-10 |
| GO:BP | GO:0010646 | regulation of cell communication | 627 | 2444 | 3.19e-10 |
| GO:BP | GO:0035239 | tube morphogenesis | 202 | 638 | 1.22e-09 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 951 | 3954 | 1.22e-09 |
| GO:BP | GO:0034330 | cell junction organization | 178 | 545 | 1.29e-09 |
| GO:BP | GO:0023051 | regulation of signaling | 621 | 2438 | 1.64e-09 |
| GO:BP | GO:0035295 | tube development | 237 | 787 | 4.07e-09 |
| GO:BP | GO:0048699 | generation of neurons | 307 | 1076 | 4.07e-09 |
| GO:BP | GO:0048518 | positive regulation of biological process | 1019 | 4301 | 5.72e-09 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 267 | 913 | 5.72e-09 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 208 | 673 | 5.72e-09 |
| GO:BP | GO:0048519 | negative regulation of biological process | 905 | 3764 | 5.74e-09 |
| GO:BP | GO:0003008 | system process | 321 | 1139 | 5.76e-09 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 276 | 951 | 5.82e-09 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 305 | 1073 | 6.41e-09 |
| GO:BP | GO:0048729 | tissue morphogenesis | 150 | 453 | 1.63e-08 |
| GO:BP | GO:0014706 | striated muscle tissue development | 84 | 213 | 2.40e-08 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 853 | 3550 | 3.57e-08 |
| GO:BP | GO:0030182 | neuron differentiation | 287 | 1014 | 4.07e-08 |
| GO:BP | GO:0009966 | regulation of signal transduction | 550 | 2166 | 5.46e-08 |
| GO:BP | GO:0030029 | actin filament-based process | 195 | 640 | 7.42e-08 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 466 | 1801 | 1.20e-07 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 95 | 261 | 2.15e-07 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 667 | 2717 | 2.15e-07 |
| GO:BP | GO:0048589 | developmental growth | 158 | 501 | 2.42e-07 |
| GO:BP | GO:0007155 | cell adhesion | 265 | 941 | 3.29e-07 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 180 | 591 | 3.38e-07 |
| GO:BP | GO:0060429 | epithelium development | 229 | 792 | 3.72e-07 |
| GO:BP | GO:0060537 | muscle tissue development | 115 | 339 | 4.53e-07 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 77 | 201 | 5.26e-07 |
| GO:BP | GO:0016477 | cell migration | 280 | 1011 | 6.19e-07 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 207 | 706 | 6.21e-07 |
| GO:BP | GO:0031175 | neuron projection development | 216 | 743 | 6.21e-07 |
| GO:BP | GO:0060485 | mesenchyme development | 89 | 245 | 6.61e-07 |
| GO:BP | GO:0048870 | cell motility | 305 | 1120 | 7.20e-07 |
| GO:BP | GO:0035556 | intracellular signal transduction | 494 | 1951 | 7.20e-07 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 186 | 622 | 7.34e-07 |
| GO:BP | GO:0007267 | cell-cell signaling | 296 | 1082 | 7.38e-07 |
| GO:BP | GO:0009790 | embryo development | 234 | 821 | 8.30e-07 |
| GO:BP | GO:0048666 | neuron development | 237 | 838 | 1.37e-06 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 158 | 516 | 1.83e-06 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 471 | 1862 | 1.92e-06 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 155 | 505 | 2.03e-06 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 347 | 1314 | 2.03e-06 |
| GO:BP | GO:0007423 | sensory organ development | 125 | 387 | 2.06e-06 |
| GO:BP | GO:0048568 | embryonic organ development | 103 | 303 | 2.09e-06 |
| GO:BP | GO:0032989 | cellular anatomical entity morphogenesis | 175 | 588 | 2.70e-06 |
| GO:BP | GO:0001944 | vasculature development | 166 | 552 | 2.85e-06 |
| GO:BP | GO:0008283 | cell population proliferation | 344 | 1306 | 2.99e-06 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 116 | 355 | 3.09e-06 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 308 | 1150 | 3.09e-06 |
| GO:BP | GO:0007010 | cytoskeleton organization | 307 | 1146 | 3.12e-06 |
| GO:BP | GO:0030900 | forebrain development | 94 | 272 | 3.27e-06 |
| GO:BP | GO:0003012 | muscle system process | 108 | 325 | 3.35e-06 |
| GO:BP | GO:0060322 | head development | 162 | 539 | 4.04e-06 |
| GO:BP | GO:0007507 | heart development | 152 | 499 | 4.15e-06 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 119 | 369 | 4.34e-06 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 179 | 609 | 4.34e-06 |
| GO:BP | GO:0030030 | cell projection organization | 314 | 1182 | 4.84e-06 |
| GO:BP | GO:0060419 | heart growth | 35 | 71 | 5.56e-06 |
| GO:BP | GO:0090257 | regulation of muscle system process | 67 | 177 | 6.13e-06 |
| GO:BP | GO:0001568 | blood vessel development | 157 | 525 | 8.75e-06 |
| GO:BP | GO:0007417 | central nervous system development | 198 | 694 | 9.01e-06 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 167 | 566 | 9.01e-06 |
| GO:BP | GO:0035265 | organ growth | 52 | 127 | 9.67e-06 |
| GO:BP | GO:0008016 | regulation of heart contraction | 63 | 165 | 9.92e-06 |
| GO:BP | GO:0050808 | synapse organization | 109 | 337 | 1.23e-05 |
| GO:BP | GO:0021537 | telencephalon development | 71 | 195 | 1.40e-05 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 277 | 1035 | 1.49e-05 |
| GO:BP | GO:0007420 | brain development | 149 | 498 | 1.68e-05 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 130 | 422 | 1.69e-05 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 137 | 450 | 1.69e-05 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 284 | 1067 | 1.69e-05 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 202 | 717 | 1.69e-05 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 286 | 1077 | 1.85e-05 |
| GO:BP | GO:0061061 | muscle structure development | 158 | 536 | 1.90e-05 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 151 | 508 | 2.01e-05 |
| GO:BP | GO:0060284 | regulation of cell development | 166 | 570 | 2.17e-05 |
| GO:BP | GO:0034329 | cell junction assembly | 104 | 323 | 2.71e-05 |
| GO:BP | GO:0065008 | regulation of biological quality | 468 | 1893 | 3.07e-05 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 140 | 467 | 3.15e-05 |
| GO:BP | GO:0010720 | positive regulation of cell development | 100 | 309 | 3.40e-05 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 136 | 452 | 3.63e-05 |
| GO:BP | GO:0000902 | cell morphogenesis | 199 | 713 | 3.91e-05 |
| GO:BP | GO:0008015 | blood circulation | 112 | 357 | 3.91e-05 |
| GO:BP | GO:0046632 | alpha-beta T cell differentiation | 33 | 70 | 3.91e-05 |
| GO:BP | GO:0006915 | apoptotic process | 352 | 1377 | 3.91e-05 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 31 | 64 | 4.01e-05 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 311 | 1197 | 4.15e-05 |
| GO:BP | GO:0098609 | cell-cell adhesion | 163 | 564 | 4.27e-05 |
| GO:BP | GO:0043009 | chordate embryonic development | 149 | 507 | 4.53e-05 |
| GO:BP | GO:0003013 | circulatory system process | 129 | 426 | 4.56e-05 |
| GO:BP | GO:0099536 | synaptic signaling | 143 | 483 | 4.74e-05 |
| GO:BP | GO:0040011 | locomotion | 220 | 805 | 4.79e-05 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 138 | 463 | 4.79e-05 |
| GO:BP | GO:0009605 | response to external stimulus | 392 | 1559 | 4.79e-05 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 138 | 463 | 4.79e-05 |
| GO:BP | GO:0032879 | regulation of localization | 365 | 1439 | 4.91e-05 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 139 | 469 | 6.06e-05 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 624 | 2622 | 6.06e-05 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 561 | 2333 | 6.76e-05 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 70 | 200 | 7.00e-05 |
| GO:BP | GO:0051960 | regulation of nervous system development | 106 | 339 | 8.29e-05 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 151 | 521 | 8.43e-05 |
| GO:BP | GO:0040007 | growth | 193 | 697 | 8.88e-05 |
| GO:BP | GO:0060047 | heart contraction | 68 | 194 | 9.11e-05 |
| GO:BP | GO:0030334 | regulation of cell migration | 186 | 668 | 9.28e-05 |
| GO:BP | GO:0048638 | regulation of developmental growth | 81 | 243 | 9.41e-05 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 49 | 126 | 9.47e-05 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 257 | 973 | 9.75e-05 |
| GO:BP | GO:0006936 | muscle contraction | 84 | 255 | 1.02e-04 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 255 | 965 | 1.02e-04 |
| GO:BP | GO:0055001 | muscle cell development | 59 | 163 | 1.33e-04 |
| GO:BP | GO:0008219 | cell death | 358 | 1423 | 1.34e-04 |
| GO:BP | GO:0043010 | camera-type eye development | 77 | 230 | 1.35e-04 |
| GO:BP | GO:0012501 | programmed cell death | 357 | 1419 | 1.36e-04 |
| GO:BP | GO:0002521 | leukocyte differentiation | 113 | 371 | 1.39e-04 |
| GO:BP | GO:0003015 | heart process | 70 | 204 | 1.42e-04 |
| GO:BP | GO:0048880 | sensory system development | 86 | 266 | 1.72e-04 |
| GO:BP | GO:0016043 | cellular component organization | 1079 | 4802 | 1.78e-04 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 404 | 1635 | 1.84e-04 |
| GO:BP | GO:0001654 | eye development | 85 | 263 | 1.95e-04 |
| GO:BP | GO:0046631 | alpha-beta T cell activation | 37 | 88 | 1.96e-04 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 100 | 322 | 2.00e-04 |
| GO:BP | GO:0061337 | cardiac conduction | 36 | 85 | 2.14e-04 |
| GO:BP | GO:0003205 | cardiac chamber development | 55 | 151 | 2.15e-04 |
| GO:BP | GO:0040012 | regulation of locomotion | 196 | 720 | 2.16e-04 |
| GO:BP | GO:0150063 | visual system development | 85 | 264 | 2.22e-04 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 100 | 323 | 2.25e-04 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 326 | 1289 | 2.28e-04 |
| GO:BP | GO:2000145 | regulation of cell motility | 191 | 700 | 2.40e-04 |
| GO:BP | GO:0042221 | response to chemical | 549 | 2304 | 2.50e-04 |
| GO:BP | GO:0033002 | muscle cell proliferation | 55 | 152 | 2.59e-04 |
| GO:BP | GO:0016310 | phosphorylation | 293 | 1145 | 2.66e-04 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 400 | 1624 | 2.67e-04 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 295 | 1157 | 3.35e-04 |
| GO:BP | GO:1903522 | regulation of blood circulation | 66 | 194 | 3.37e-04 |
| GO:BP | GO:0048167 | regulation of synaptic plasticity | 51 | 139 | 3.52e-04 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 391 | 1588 | 3.53e-04 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 450 | 1856 | 3.53e-04 |
| GO:BP | GO:0046637 | regulation of alpha-beta T cell differentiation | 20 | 37 | 3.62e-04 |
| GO:BP | GO:0042692 | muscle cell differentiation | 99 | 323 | 3.76e-04 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 203 | 757 | 3.83e-04 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 76 | 233 | 3.83e-04 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 85 | 268 | 3.83e-04 |
| GO:BP | GO:0031644 | regulation of nervous system process | 32 | 74 | 3.83e-04 |
| GO:BP | GO:0001823 | mesonephros development | 32 | 74 | 3.83e-04 |
| GO:BP | GO:0060038 | cardiac muscle cell proliferation | 22 | 43 | 3.94e-04 |
| GO:BP | GO:0099173 | postsynapse organization | 53 | 147 | 3.95e-04 |
| GO:BP | GO:0060420 | regulation of heart growth | 24 | 49 | 4.00e-04 |
| GO:BP | GO:0003231 | cardiac ventricle development | 44 | 115 | 4.06e-04 |
| GO:BP | GO:0006468 | protein phosphorylation | 249 | 958 | 4.06e-04 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 93 | 301 | 4.65e-04 |
| GO:BP | GO:1990823 | response to leukemia inhibitory factor | 35 | 85 | 5.22e-04 |
| GO:BP | GO:0051049 | regulation of transport | 280 | 1098 | 5.26e-04 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 141 | 498 | 5.27e-04 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 208 | 783 | 5.27e-04 |
| GO:BP | GO:0014855 | striated muscle cell proliferation | 27 | 59 | 5.27e-04 |
| GO:BP | GO:0001657 | ureteric bud development | 31 | 72 | 5.52e-04 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 148 | 528 | 5.65e-04 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 569 | 2415 | 5.76e-04 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 42 | 110 | 6.50e-04 |
| GO:BP | GO:0042592 | homeostatic process | 288 | 1137 | 6.56e-04 |
| GO:BP | GO:0006941 | striated muscle contraction | 52 | 146 | 6.58e-04 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 306 | 1218 | 6.90e-04 |
| GO:BP | GO:0070848 | response to growth factor | 152 | 547 | 6.93e-04 |
| GO:BP | GO:0003007 | heart morphogenesis | 71 | 218 | 7.15e-04 |
| GO:BP | GO:0072163 | mesonephric epithelium development | 31 | 73 | 7.27e-04 |
| GO:BP | GO:1990830 | cellular response to leukemia inhibitory factor | 34 | 83 | 7.27e-04 |
| GO:BP | GO:0042472 | inner ear morphogenesis | 27 | 60 | 7.27e-04 |
| GO:BP | GO:0072164 | mesonephric tubule development | 31 | 73 | 7.27e-04 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 125 | 435 | 7.30e-04 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 75 | 234 | 7.56e-04 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 113 | 386 | 7.56e-04 |
| GO:BP | GO:0051179 | localization | 847 | 3731 | 7.80e-04 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 139 | 494 | 7.82e-04 |
| GO:BP | GO:0046634 | regulation of alpha-beta T cell activation | 25 | 54 | 7.86e-04 |
| GO:BP | GO:0050877 | nervous system process | 164 | 600 | 8.14e-04 |
| GO:BP | GO:0045165 | cell fate commitment | 53 | 151 | 8.14e-04 |
| GO:BP | GO:0016331 | morphogenesis of embryonic epithelium | 45 | 122 | 8.28e-04 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 86 | 278 | 8.28e-04 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 89 | 290 | 8.31e-04 |
| GO:BP | GO:0048644 | muscle organ morphogenesis | 28 | 64 | 9.04e-04 |
| GO:BP | GO:0001501 | skeletal system development | 103 | 347 | 9.04e-04 |
| GO:BP | GO:0060421 | positive regulation of heart growth | 16 | 28 | 9.73e-04 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 442 | 1841 | 9.80e-04 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 102 | 344 | 1.03e-03 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 63 | 190 | 1.04e-03 |
| GO:BP | GO:0001763 | morphogenesis of a branching structure | 51 | 145 | 1.04e-03 |
| GO:BP | GO:1902105 | regulation of leukocyte differentiation | 64 | 194 | 1.06e-03 |
| GO:BP | GO:0009991 | response to extracellular stimulus | 110 | 377 | 1.06e-03 |
| GO:BP | GO:0060043 | regulation of cardiac muscle cell proliferation | 17 | 31 | 1.08e-03 |
| GO:BP | GO:0055008 | cardiac muscle tissue morphogenesis | 25 | 55 | 1.08e-03 |
| GO:BP | GO:0030217 | T cell differentiation | 61 | 183 | 1.13e-03 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 78 | 249 | 1.16e-03 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 430 | 1791 | 1.24e-03 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 59 | 176 | 1.25e-03 |
| GO:BP | GO:0023056 | positive regulation of signaling | 304 | 1221 | 1.35e-03 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 134 | 479 | 1.35e-03 |
| GO:BP | GO:0007389 | pattern specification process | 87 | 286 | 1.36e-03 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 57 | 169 | 1.37e-03 |
| GO:BP | GO:0050807 | regulation of synapse organization | 58 | 173 | 1.42e-03 |
| GO:BP | GO:0065009 | regulation of molecular function | 321 | 1299 | 1.47e-03 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 170 | 633 | 1.47e-03 |
| GO:BP | GO:0014032 | neural crest cell development | 29 | 69 | 1.47e-03 |
| GO:BP | GO:0030048 | actin filament-based movement | 40 | 107 | 1.49e-03 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 196 | 746 | 1.55e-03 |
| GO:BP | GO:0043367 | CD4-positive, alpha-beta T cell differentiation | 24 | 53 | 1.58e-03 |
| GO:BP | GO:0097106 | postsynaptic density organization | 16 | 29 | 1.58e-03 |
| GO:BP | GO:0048839 | inner ear development | 47 | 133 | 1.71e-03 |
| GO:BP | GO:0086004 | regulation of cardiac muscle cell contraction | 17 | 32 | 1.72e-03 |
| GO:BP | GO:0048562 | embryonic organ morphogenesis | 60 | 182 | 1.75e-03 |
| GO:BP | GO:0055021 | regulation of cardiac muscle tissue growth | 21 | 44 | 1.80e-03 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 34 | 87 | 1.92e-03 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 43 | 119 | 1.92e-03 |
| GO:BP | GO:0046620 | regulation of organ growth | 29 | 70 | 1.93e-03 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 50 | 145 | 2.00e-03 |
| GO:BP | GO:0016358 | dendrite development | 60 | 183 | 2.05e-03 |
| GO:BP | GO:0014033 | neural crest cell differentiation | 31 | 77 | 2.05e-03 |
| GO:BP | GO:0048863 | stem cell differentiation | 62 | 191 | 2.14e-03 |
| GO:BP | GO:0086065 | cell communication involved in cardiac conduction | 24 | 54 | 2.18e-03 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 84 | 278 | 2.20e-03 |
| GO:BP | GO:1901021 | positive regulation of calcium ion transmembrane transporter activity | 15 | 27 | 2.35e-03 |
| GO:BP | GO:0003338 | metanephros morphogenesis | 12 | 19 | 2.37e-03 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 32 | 81 | 2.37e-03 |
| GO:BP | GO:0048565 | digestive tract development | 34 | 88 | 2.40e-03 |
| GO:BP | GO:0048754 | branching morphogenesis of an epithelial tube | 41 | 113 | 2.43e-03 |
| GO:BP | GO:0048771 | tissue remodeling | 41 | 113 | 2.43e-03 |
| GO:BP | GO:0030001 | metal ion transport | 143 | 524 | 2.45e-03 |
| GO:BP | GO:0035148 | tube formation | 44 | 124 | 2.45e-03 |
| GO:BP | GO:0060415 | muscle tissue morphogenesis | 26 | 61 | 2.49e-03 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 57 | 173 | 2.53e-03 |
| GO:BP | GO:0046635 | positive regulation of alpha-beta T cell activation | 17 | 33 | 2.63e-03 |
| GO:BP | GO:0060443 | mammary gland morphogenesis | 18 | 36 | 2.68e-03 |
| GO:BP | GO:0071310 | cellular response to organic substance | 318 | 1297 | 2.69e-03 |
| GO:BP | GO:0099545 | trans-synaptic signaling by trans-synaptic complex | 6 | 6 | 2.71e-03 |
| GO:BP | GO:0030097 | hemopoiesis | 166 | 624 | 2.74e-03 |
| GO:BP | GO:0043583 | ear development | 51 | 151 | 2.88e-03 |
| GO:BP | GO:0055123 | digestive system development | 36 | 96 | 2.92e-03 |
| GO:BP | GO:0060045 | positive regulation of cardiac muscle cell proliferation | 13 | 22 | 2.92e-03 |
| GO:BP | GO:0061448 | connective tissue development | 65 | 205 | 2.93e-03 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 27 | 65 | 2.98e-03 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 245 | 972 | 3.04e-03 |
| GO:BP | GO:0010721 | negative regulation of cell development | 59 | 182 | 3.05e-03 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 78 | 257 | 3.11e-03 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 272 | 1093 | 3.12e-03 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 184 | 704 | 3.14e-03 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 29 | 72 | 3.15e-03 |
| GO:BP | GO:0030903 | notochord development | 11 | 17 | 3.23e-03 |
| GO:BP | GO:0072073 | kidney epithelium development | 40 | 111 | 3.26e-03 |
| GO:BP | GO:0061564 | axon development | 106 | 372 | 3.28e-03 |
| GO:BP | GO:0055023 | positive regulation of cardiac muscle tissue growth | 14 | 25 | 3.28e-03 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 294 | 1193 | 3.28e-03 |
| GO:BP | GO:0060603 | mammary gland duct morphogenesis | 14 | 25 | 3.28e-03 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 88 | 298 | 3.28e-03 |
| GO:BP | GO:0097120 | receptor localization to synapse | 21 | 46 | 3.41e-03 |
| GO:BP | GO:0060350 | endochondral bone morphogenesis | 21 | 46 | 3.41e-03 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 41 | 115 | 3.41e-03 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 41 | 115 | 3.41e-03 |
| GO:BP | GO:0001525 | angiogenesis | 108 | 381 | 3.47e-03 |
| GO:BP | GO:0071887 | leukocyte apoptotic process | 30 | 76 | 3.55e-03 |
| GO:BP | GO:0048713 | regulation of oligodendrocyte differentiation | 15 | 28 | 3.56e-03 |
| GO:BP | GO:0002027 | regulation of heart rate | 32 | 83 | 3.57e-03 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 149 | 555 | 3.62e-03 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 510 | 2186 | 3.62e-03 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 141 | 521 | 3.68e-03 |
| GO:BP | GO:0010644 | cell communication by electrical coupling | 16 | 31 | 3.70e-03 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 329 | 1354 | 3.71e-03 |
| GO:BP | GO:0007015 | actin filament organization | 102 | 357 | 3.71e-03 |
| GO:BP | GO:0099084 | postsynaptic specialization organization | 17 | 34 | 3.71e-03 |
| GO:BP | GO:1903115 | regulation of actin filament-based movement | 18 | 37 | 3.71e-03 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 119 | 428 | 3.71e-03 |
| GO:BP | GO:0045321 | leukocyte activation | 143 | 530 | 3.75e-03 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 29 | 73 | 3.84e-03 |
| GO:BP | GO:0031503 | protein-containing complex localization | 52 | 157 | 3.84e-03 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 59 | 184 | 3.84e-03 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 38 | 105 | 3.97e-03 |
| GO:BP | GO:0006950 | response to stress | 585 | 2540 | 4.12e-03 |
| GO:BP | GO:0031667 | response to nutrient levels | 101 | 354 | 4.14e-03 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 66 | 212 | 4.15e-03 |
| GO:BP | GO:0061138 | morphogenesis of a branching epithelium | 46 | 135 | 4.30e-03 |
| GO:BP | GO:0042110 | T cell activation | 89 | 305 | 4.33e-03 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 415 | 1751 | 4.34e-03 |
| GO:BP | GO:0001775 | cell activation | 165 | 627 | 4.43e-03 |
| GO:BP | GO:1903037 | regulation of leukocyte cell-cell adhesion | 62 | 197 | 4.60e-03 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 125 | 456 | 4.67e-03 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 27 | 67 | 4.73e-03 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 38 | 106 | 4.81e-03 |
| GO:BP | GO:0048864 | stem cell development | 29 | 74 | 4.84e-03 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 29 | 74 | 4.84e-03 |
| GO:BP | GO:0042471 | ear morphogenesis | 29 | 74 | 4.84e-03 |
| GO:BP | GO:0099175 | regulation of postsynapse organization | 31 | 81 | 4.85e-03 |
| GO:BP | GO:0046649 | lymphocyte activation | 122 | 444 | 4.90e-03 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 84 | 286 | 5.01e-03 |
| GO:BP | GO:0046638 | positive regulation of alpha-beta T cell differentiation | 14 | 26 | 5.03e-03 |
| GO:BP | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction | 14 | 26 | 5.03e-03 |
| GO:BP | GO:0110110 | positive regulation of animal organ morphogenesis | 14 | 26 | 5.03e-03 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 26 | 64 | 5.15e-03 |
| GO:BP | GO:0023057 | negative regulation of signaling | 257 | 1036 | 5.15e-03 |
| GO:BP | GO:0006816 | calcium ion transport | 78 | 262 | 5.18e-03 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 65 | 210 | 5.24e-03 |
| GO:BP | GO:0007159 | leukocyte cell-cell adhesion | 67 | 218 | 5.25e-03 |
| GO:BP | GO:1902742 | apoptotic process involved in development | 15 | 29 | 5.25e-03 |
| GO:BP | GO:0046622 | positive regulation of organ growth | 17 | 35 | 5.29e-03 |
| GO:BP | GO:0055006 | cardiac cell development | 30 | 78 | 5.34e-03 |
| GO:BP | GO:0048588 | developmental cell growth | 57 | 179 | 5.55e-03 |
| GO:BP | GO:0072132 | mesenchyme morphogenesis | 22 | 51 | 5.59e-03 |
| GO:BP | GO:0021915 | neural tube development | 45 | 133 | 5.59e-03 |
| GO:BP | GO:0030033 | microvillus assembly | 9 | 13 | 5.69e-03 |
| GO:BP | GO:0007517 | muscle organ development | 79 | 267 | 5.69e-03 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 51 | 156 | 5.70e-03 |
| GO:BP | GO:0007409 | axonogenesis | 96 | 337 | 5.77e-03 |
| GO:BP | GO:0071417 | cellular response to organonitrogen compound | 121 | 442 | 5.79e-03 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 256 | 1034 | 5.80e-03 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 73 | 243 | 5.83e-03 |
| GO:BP | GO:0031668 | cellular response to extracellular stimulus | 67 | 219 | 5.84e-03 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 31 | 82 | 5.84e-03 |
| GO:BP | GO:0014896 | muscle hypertrophy | 29 | 75 | 5.87e-03 |
| GO:BP | GO:0051058 | negative regulation of small GTPase mediated signal transduction | 21 | 48 | 5.92e-03 |
| GO:BP | GO:0042594 | response to starvation | 56 | 176 | 6.18e-03 |
| GO:BP | GO:0050863 | regulation of T cell activation | 63 | 204 | 6.53e-03 |
| GO:BP | GO:0072001 | renal system development | 73 | 244 | 6.53e-03 |
| GO:BP | GO:0042330 | taxis | 73 | 244 | 6.53e-03 |
| GO:BP | GO:0006935 | chemotaxis | 73 | 244 | 6.53e-03 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 488 | 2100 | 6.53e-03 |
| GO:BP | GO:0032944 | regulation of mononuclear cell proliferation | 41 | 119 | 6.53e-03 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 452 | 1934 | 6.93e-03 |
| GO:BP | GO:0060348 | bone development | 48 | 146 | 7.12e-03 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 40 | 116 | 7.48e-03 |
| GO:BP | GO:0042391 | regulation of membrane potential | 82 | 282 | 7.65e-03 |
| GO:BP | GO:0003180 | aortic valve morphogenesis | 16 | 33 | 7.77e-03 |
| GO:BP | GO:0086026 | atrial cardiac muscle cell to AV node cell signaling | 10 | 16 | 7.96e-03 |
| GO:BP | GO:0086066 | atrial cardiac muscle cell to AV node cell communication | 10 | 16 | 7.96e-03 |
| GO:BP | GO:0086014 | atrial cardiac muscle cell action potential | 10 | 16 | 7.96e-03 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 38 | 109 | 8.05e-03 |
| GO:BP | GO:0035710 | CD4-positive, alpha-beta T cell activation | 26 | 66 | 8.26e-03 |
| GO:BP | GO:0001570 | vasculogenesis | 26 | 66 | 8.26e-03 |
| GO:BP | GO:0060349 | bone morphogenesis | 28 | 73 | 8.31e-03 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 49 | 151 | 8.47e-03 |
| GO:BP | GO:0003279 | cardiac septum development | 36 | 102 | 8.64e-03 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 44 | 132 | 8.68e-03 |
| GO:BP | GO:0010033 | response to organic substance | 408 | 1736 | 8.79e-03 |
| GO:BP | GO:0072175 | epithelial tube formation | 40 | 117 | 8.81e-03 |
| GO:BP | GO:0007416 | synapse assembly | 45 | 136 | 8.87e-03 |
| GO:BP | GO:0060173 | limb development | 45 | 136 | 8.87e-03 |
| GO:BP | GO:0048736 | appendage development | 45 | 136 | 8.87e-03 |
| GO:BP | GO:0031214 | biomineral tissue development | 37 | 106 | 9.05e-03 |
| GO:BP | GO:0002287 | alpha-beta T cell activation involved in immune response | 19 | 43 | 9.22e-03 |
| GO:BP | GO:0002294 | CD4-positive, alpha-beta T cell differentiation involved in immune response | 19 | 43 | 9.22e-03 |
| GO:BP | GO:0002293 | alpha-beta T cell differentiation involved in immune response | 19 | 43 | 9.22e-03 |
| GO:BP | GO:0042093 | T-helper cell differentiation | 19 | 43 | 9.22e-03 |
| GO:BP | GO:0035108 | limb morphogenesis | 38 | 110 | 9.45e-03 |
| GO:BP | GO:0035107 | appendage morphogenesis | 38 | 110 | 9.45e-03 |
| GO:BP | GO:0002573 | myeloid leukocyte differentiation | 49 | 152 | 9.62e-03 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 35 | 99 | 9.62e-03 |
| GO:BP | GO:0045216 | cell-cell junction organization | 49 | 152 | 9.62e-03 |
| GO:BP | GO:0001764 | neuron migration | 43 | 129 | 9.67e-03 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 128 | 479 | 1.01e-02 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 73 | 248 | 1.01e-02 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 1091 | 4988 | 1.01e-02 |
| GO:BP | GO:2000181 | negative regulation of blood vessel morphogenesis | 26 | 67 | 1.02e-02 |
| GO:BP | GO:1901343 | negative regulation of vasculature development | 26 | 67 | 1.02e-02 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 26 | 67 | 1.02e-02 |
| GO:BP | GO:0010629 | negative regulation of gene expression | 199 | 790 | 1.03e-02 |
| GO:BP | GO:0061035 | regulation of cartilage development | 21 | 50 | 1.03e-02 |
| GO:BP | GO:0003148 | outflow tract septum morphogenesis | 12 | 22 | 1.04e-02 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 60 | 196 | 1.04e-02 |
| GO:BP | GO:0001816 | cytokine production | 119 | 441 | 1.04e-02 |
| GO:BP | GO:1903779 | regulation of cardiac conduction | 12 | 22 | 1.04e-02 |
| GO:BP | GO:0007264 | small GTPase-mediated signal transduction | 107 | 390 | 1.04e-02 |
| GO:BP | GO:0001817 | regulation of cytokine production | 118 | 437 | 1.06e-02 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 31 | 85 | 1.06e-02 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 453 | 1951 | 1.09e-02 |
| GO:BP | GO:0060351 | cartilage development involved in endochondral bone morphogenesis | 13 | 25 | 1.09e-02 |
| GO:BP | GO:0021954 | central nervous system neuron development | 23 | 57 | 1.09e-02 |
| GO:BP | GO:0070296 | sarcoplasmic reticulum calcium ion transport | 15 | 31 | 1.10e-02 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 934 | 4233 | 1.10e-02 |
| GO:BP | GO:0048483 | autonomic nervous system development | 14 | 28 | 1.11e-02 |
| GO:BP | GO:1903038 | negative regulation of leukocyte cell-cell adhesion | 27 | 71 | 1.12e-02 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 297 | 1234 | 1.18e-02 |
| GO:BP | GO:0045621 | positive regulation of lymphocyte differentiation | 30 | 82 | 1.19e-02 |
| GO:BP | GO:0019222 | regulation of metabolic process | 1010 | 4603 | 1.21e-02 |
| GO:BP | GO:0055010 | ventricular cardiac muscle tissue morphogenesis | 19 | 44 | 1.21e-02 |
| GO:BP | GO:0022612 | gland morphogenesis | 33 | 93 | 1.21e-02 |
| GO:BP | GO:0045580 | regulation of T cell differentiation | 37 | 108 | 1.24e-02 |
| GO:BP | GO:0042063 | gliogenesis | 71 | 242 | 1.25e-02 |
| GO:BP | GO:0051249 | regulation of lymphocyte activation | 80 | 279 | 1.25e-02 |
| GO:BP | GO:0045619 | regulation of lymphocyte differentiation | 42 | 127 | 1.25e-02 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 217 | 874 | 1.25e-02 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 296 | 1231 | 1.25e-02 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 116 | 431 | 1.28e-02 |
| GO:BP | GO:0050919 | negative chemotaxis | 18 | 41 | 1.30e-02 |
| GO:BP | GO:0006811 | monoatomic ion transport | 182 | 719 | 1.30e-02 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 59 | 194 | 1.31e-02 |
| GO:BP | GO:1901342 | regulation of vasculature development | 60 | 198 | 1.31e-02 |
| GO:BP | GO:0050771 | negative regulation of axonogenesis | 21 | 51 | 1.32e-02 |
| GO:BP | GO:0050670 | regulation of lymphocyte proliferation | 39 | 116 | 1.32e-02 |
| GO:BP | GO:0051235 | maintenance of location | 73 | 251 | 1.34e-02 |
| GO:BP | GO:0051283 | negative regulation of sequestering of calcium ion | 32 | 90 | 1.35e-02 |
| GO:BP | GO:0045582 | positive regulation of T cell differentiation | 27 | 72 | 1.37e-02 |
| GO:BP | GO:0048709 | oligodendrocyte differentiation | 27 | 72 | 1.37e-02 |
| GO:BP | GO:0009267 | cellular response to starvation | 47 | 147 | 1.37e-02 |
| GO:BP | GO:0050865 | regulation of cell activation | 96 | 347 | 1.37e-02 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 70 | 239 | 1.37e-02 |
| GO:BP | GO:0098900 | regulation of action potential | 17 | 38 | 1.37e-02 |
| GO:BP | GO:0030316 | osteoclast differentiation | 25 | 65 | 1.38e-02 |
| GO:BP | GO:0022409 | positive regulation of cell-cell adhesion | 54 | 175 | 1.46e-02 |
| GO:BP | GO:0045444 | fat cell differentiation | 55 | 179 | 1.46e-02 |
| GO:BP | GO:0048486 | parasympathetic nervous system development | 8 | 12 | 1.50e-02 |
| GO:BP | GO:0060561 | apoptotic process involved in morphogenesis | 11 | 20 | 1.50e-02 |
| GO:BP | GO:0032332 | positive regulation of chondrocyte differentiation | 8 | 12 | 1.50e-02 |
| GO:BP | GO:0030879 | mammary gland development | 34 | 98 | 1.50e-02 |
| GO:BP | GO:0014020 | primary neural tube formation | 31 | 87 | 1.51e-02 |
| GO:BP | GO:0007613 | memory | 31 | 87 | 1.51e-02 |
| GO:BP | GO:0003179 | heart valve morphogenesis | 22 | 55 | 1.51e-02 |
| GO:BP | GO:0003281 | ventricular septum development | 26 | 69 | 1.52e-02 |
| GO:BP | GO:0062237 | protein localization to postsynapse | 15 | 32 | 1.52e-02 |
| GO:BP | GO:1903539 | protein localization to postsynaptic membrane | 15 | 32 | 1.52e-02 |
| GO:BP | GO:0050866 | negative regulation of cell activation | 39 | 117 | 1.52e-02 |
| GO:BP | GO:0070167 | regulation of biomineral tissue development | 24 | 62 | 1.53e-02 |
| GO:BP | GO:0002292 | T cell differentiation involved in immune response | 19 | 45 | 1.56e-02 |
| GO:BP | GO:0086019 | cell-cell signaling involved in cardiac conduction | 14 | 29 | 1.57e-02 |
| GO:BP | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 13 | 26 | 1.59e-02 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 41 | 125 | 1.61e-02 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 436 | 1883 | 1.61e-02 |
| GO:BP | GO:0050806 | positive regulation of synaptic transmission | 36 | 106 | 1.61e-02 |
| GO:BP | GO:0048878 | chemical homeostasis | 170 | 670 | 1.62e-02 |
| GO:BP | GO:0061180 | mammary gland epithelium development | 21 | 52 | 1.66e-02 |
| GO:BP | GO:0048645 | animal organ formation | 21 | 52 | 1.66e-02 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 33 | 95 | 1.67e-02 |
| GO:BP | GO:0035270 | endocrine system development | 30 | 84 | 1.68e-02 |
| GO:BP | GO:0072089 | stem cell proliferation | 30 | 84 | 1.68e-02 |
| GO:BP | GO:0060606 | tube closure | 30 | 84 | 1.68e-02 |
| GO:BP | GO:0051250 | negative regulation of lymphocyte activation | 30 | 84 | 1.68e-02 |
| GO:BP | GO:0016525 | negative regulation of angiogenesis | 25 | 66 | 1.68e-02 |
| GO:BP | GO:0032943 | mononuclear cell proliferation | 49 | 157 | 1.80e-02 |
| GO:BP | GO:0051641 | cellular localization | 593 | 2623 | 1.80e-02 |
| GO:BP | GO:0010001 | glial cell differentiation | 55 | 181 | 1.83e-02 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 84 | 300 | 1.83e-02 |
| GO:BP | GO:0007162 | negative regulation of cell adhesion | 56 | 185 | 1.83e-02 |
| GO:BP | GO:0051251 | positive regulation of lymphocyte activation | 56 | 185 | 1.83e-02 |
| GO:BP | GO:0040008 | regulation of growth | 122 | 462 | 1.83e-02 |
| GO:BP | GO:0006810 | transport | 669 | 2984 | 1.84e-02 |
| GO:BP | GO:0099565 | chemical synaptic transmission, postsynaptic | 26 | 70 | 1.84e-02 |
| GO:BP | GO:0002376 | immune system process | 333 | 1410 | 1.84e-02 |
| GO:BP | GO:0003417 | growth plate cartilage development | 9 | 15 | 1.86e-02 |
| GO:BP | GO:0001780 | neutrophil homeostasis | 9 | 15 | 1.86e-02 |
| GO:BP | GO:0003229 | ventricular cardiac muscle tissue development | 22 | 56 | 1.86e-02 |
| GO:BP | GO:0002694 | regulation of leukocyte activation | 87 | 313 | 1.86e-02 |
| GO:BP | GO:0070663 | regulation of leukocyte proliferation | 42 | 130 | 1.86e-02 |
| GO:BP | GO:0001841 | neural tube formation | 32 | 92 | 1.86e-02 |
| GO:BP | GO:0010927 | cellular component assembly involved in morphogenesis | 36 | 107 | 1.86e-02 |
| GO:BP | GO:0051282 | regulation of sequestering of calcium ion | 32 | 92 | 1.86e-02 |
| GO:BP | GO:0072170 | metanephric tubule development | 9 | 15 | 1.86e-02 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 24 | 63 | 1.86e-02 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 601 | 2663 | 1.88e-02 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 126 | 480 | 1.88e-02 |
| GO:BP | GO:0031641 | regulation of myelination | 16 | 36 | 1.92e-02 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 62 | 210 | 1.95e-02 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 60 | 202 | 1.98e-02 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 30 | 85 | 1.98e-02 |
| GO:BP | GO:0071872 | cellular response to epinephrine stimulus | 7 | 10 | 1.98e-02 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 50 | 162 | 2.00e-02 |
| GO:BP | GO:0001649 | osteoblast differentiation | 53 | 174 | 2.01e-02 |
| GO:BP | GO:0010799 | regulation of peptidyl-threonine phosphorylation | 15 | 33 | 2.05e-02 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 95 | 348 | 2.05e-02 |
| GO:BP | GO:0030902 | hindbrain development | 35 | 104 | 2.08e-02 |
| GO:BP | GO:0051209 | release of sequestered calcium ion into cytosol | 31 | 89 | 2.08e-02 |
| GO:BP | GO:0030282 | bone mineralization | 28 | 78 | 2.10e-02 |
| GO:BP | GO:0060444 | branching involved in mammary gland duct morphogenesis | 10 | 18 | 2.11e-02 |
| GO:BP | GO:0031646 | positive regulation of nervous system process | 10 | 18 | 2.11e-02 |
| GO:BP | GO:0050870 | positive regulation of T cell activation | 43 | 135 | 2.15e-02 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 150 | 587 | 2.16e-02 |
| GO:BP | GO:0071526 | semaphorin-plexin signaling pathway | 18 | 43 | 2.16e-02 |
| GO:BP | GO:0045598 | regulation of fat cell differentiation | 32 | 93 | 2.17e-02 |
| GO:BP | GO:0043370 | regulation of CD4-positive, alpha-beta T cell differentiation | 13 | 27 | 2.22e-02 |
| GO:BP | GO:0031279 | regulation of cyclase activity | 13 | 27 | 2.22e-02 |
| GO:BP | GO:0098901 | regulation of cardiac muscle cell action potential | 13 | 27 | 2.22e-02 |
| GO:BP | GO:0021591 | ventricular system development | 13 | 27 | 2.22e-02 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 55 | 183 | 2.24e-02 |
| GO:BP | GO:0001818 | negative regulation of cytokine production | 50 | 163 | 2.24e-02 |
| GO:BP | GO:0070372 | regulation of ERK1 and ERK2 cascade | 53 | 175 | 2.24e-02 |
| GO:BP | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling | 12 | 24 | 2.25e-02 |
| GO:BP | GO:0050868 | negative regulation of T cell activation | 24 | 64 | 2.26e-02 |
| GO:BP | GO:0090497 | mesenchymal cell migration | 20 | 50 | 2.26e-02 |
| GO:BP | GO:1901019 | regulation of calcium ion transmembrane transporter activity | 20 | 50 | 2.26e-02 |
| GO:BP | GO:0043954 | cellular component maintenance | 22 | 57 | 2.29e-02 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 438 | 1904 | 2.35e-02 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 138 | 536 | 2.39e-02 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 108 | 406 | 2.46e-02 |
| GO:BP | GO:0055002 | striated muscle cell development | 25 | 68 | 2.47e-02 |
| GO:BP | GO:0001822 | kidney development | 68 | 237 | 2.47e-02 |
| GO:BP | GO:0031099 | regeneration | 45 | 144 | 2.49e-02 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 56 | 188 | 2.49e-02 |
| GO:BP | GO:0031032 | actomyosin structure organization | 55 | 184 | 2.49e-02 |
| GO:BP | GO:0002761 | regulation of myeloid leukocyte differentiation | 28 | 79 | 2.49e-02 |
| GO:BP | GO:0045927 | positive regulation of growth | 55 | 184 | 2.49e-02 |
| GO:BP | GO:0007411 | axon guidance | 52 | 172 | 2.49e-02 |
| GO:BP | GO:0009612 | response to mechanical stimulus | 47 | 152 | 2.49e-02 |
| GO:BP | GO:0003002 | regionalization | 73 | 258 | 2.49e-02 |
| GO:BP | GO:0097485 | neuron projection guidance | 52 | 172 | 2.49e-02 |
| GO:BP | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 47 | 152 | 2.49e-02 |
| GO:BP | GO:0002067 | glandular epithelial cell differentiation | 19 | 47 | 2.49e-02 |
| GO:BP | GO:0003176 | aortic valve development | 16 | 37 | 2.52e-02 |
| GO:BP | GO:1900116 | extracellular negative regulation of signal transduction | 6 | 8 | 2.52e-02 |
| GO:BP | GO:0106120 | positive regulation of sterol biosynthetic process | 6 | 8 | 2.52e-02 |
| GO:BP | GO:0045542 | positive regulation of cholesterol biosynthetic process | 6 | 8 | 2.52e-02 |
| GO:BP | GO:0032634 | interleukin-5 production | 6 | 8 | 2.52e-02 |
| GO:BP | GO:0032674 | regulation of interleukin-5 production | 6 | 8 | 2.52e-02 |
| GO:BP | GO:0003418 | growth plate cartilage chondrocyte differentiation | 6 | 8 | 2.52e-02 |
| GO:BP | GO:1900115 | extracellular regulation of signal transduction | 6 | 8 | 2.52e-02 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 140 | 546 | 2.52e-02 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 63 | 217 | 2.55e-02 |
| GO:BP | GO:0001843 | neural tube closure | 29 | 83 | 2.58e-02 |
| GO:BP | GO:0009952 | anterior/posterior pattern specification | 39 | 121 | 2.58e-02 |
| GO:BP | GO:1905874 | regulation of postsynaptic density organization | 8 | 13 | 2.58e-02 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 26 | 72 | 2.58e-02 |
| GO:BP | GO:0002696 | positive regulation of leukocyte activation | 58 | 197 | 2.68e-02 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 196 | 796 | 2.68e-02 |
| GO:BP | GO:0003230 | cardiac atrium development | 15 | 34 | 2.68e-02 |
| GO:BP | GO:0003161 | cardiac conduction system development | 15 | 34 | 2.68e-02 |
| GO:BP | GO:0032330 | regulation of chondrocyte differentiation | 15 | 34 | 2.68e-02 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 43 | 137 | 2.68e-02 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 351 | 1504 | 2.68e-02 |
| GO:BP | GO:0010586 | miRNA metabolic process | 30 | 87 | 2.68e-02 |
| GO:BP | GO:0003170 | heart valve development | 24 | 65 | 2.68e-02 |
| GO:BP | GO:0034103 | regulation of tissue remodeling | 18 | 44 | 2.68e-02 |
| GO:BP | GO:0051216 | cartilage development | 45 | 145 | 2.72e-02 |
| GO:BP | GO:0060996 | dendritic spine development | 27 | 76 | 2.72e-02 |
| GO:BP | GO:0060395 | SMAD protein signal transduction | 27 | 76 | 2.72e-02 |
| GO:BP | GO:0048844 | artery morphogenesis | 22 | 58 | 2.75e-02 |
| GO:BP | GO:0019935 | cyclic-nucleotide-mediated signaling | 20 | 51 | 2.76e-02 |
| GO:BP | GO:0006812 | monoatomic cation transport | 155 | 614 | 2.78e-02 |
| GO:BP | GO:0045687 | positive regulation of glial cell differentiation | 14 | 31 | 2.85e-02 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 61 | 210 | 2.85e-02 |
| GO:BP | GO:0140747 | regulation of ncRNA transcription | 28 | 80 | 2.88e-02 |
| GO:BP | GO:0001503 | ossification | 83 | 302 | 2.92e-02 |
| GO:BP | GO:0010468 | regulation of gene expression | 748 | 3379 | 2.92e-02 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 105 | 396 | 2.92e-02 |
| GO:BP | GO:0097553 | calcium ion transmembrane import into cytosol | 39 | 122 | 2.92e-02 |
| GO:BP | GO:1905314 | semi-lunar valve development | 17 | 41 | 2.93e-02 |
| GO:BP | GO:0098815 | modulation of excitatory postsynaptic potential | 17 | 41 | 2.93e-02 |
| GO:BP | GO:0072273 | metanephric nephron morphogenesis | 9 | 16 | 2.93e-02 |
| GO:BP | GO:0010893 | positive regulation of steroid biosynthetic process | 9 | 16 | 2.93e-02 |
| GO:BP | GO:0032814 | regulation of natural killer cell activation | 9 | 16 | 2.93e-02 |
| GO:BP | GO:0048339 | paraxial mesoderm development | 9 | 16 | 2.93e-02 |
| GO:BP | GO:0072207 | metanephric epithelium development | 9 | 16 | 2.93e-02 |
| GO:BP | GO:2000463 | positive regulation of excitatory postsynaptic potential | 13 | 28 | 2.94e-02 |
| GO:BP | GO:0032898 | neurotrophin production | 4 | 4 | 2.94e-02 |
| GO:BP | GO:0098910 | regulation of atrial cardiac muscle cell action potential | 4 | 4 | 2.94e-02 |
| GO:BP | GO:0003171 | atrioventricular valve development | 13 | 28 | 2.94e-02 |
| GO:BP | GO:0003208 | cardiac ventricle morphogenesis | 23 | 62 | 2.94e-02 |
| GO:BP | GO:0014808 | release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 13 | 28 | 2.94e-02 |
| GO:BP | GO:1903514 | release of sequestered calcium ion into cytosol by endoplasmic reticulum | 13 | 28 | 2.94e-02 |
| GO:BP | GO:0001659 | temperature homeostasis | 41 | 130 | 2.94e-02 |
| GO:BP | GO:0110030 | regulation of G2/MI transition of meiotic cell cycle | 4 | 4 | 2.94e-02 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 217 | 893 | 2.94e-02 |
| GO:BP | GO:1904045 | cellular response to aldosterone | 4 | 4 | 2.94e-02 |
| GO:BP | GO:0061026 | cardiac muscle tissue regeneration | 5 | 6 | 2.94e-02 |
| GO:BP | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway | 5 | 6 | 2.94e-02 |
| GO:BP | GO:1903243 | negative regulation of cardiac muscle hypertrophy in response to stress | 5 | 6 | 2.94e-02 |
| GO:BP | GO:0010616 | negative regulation of cardiac muscle adaptation | 5 | 6 | 2.94e-02 |
| GO:BP | GO:0030432 | peristalsis | 5 | 6 | 2.94e-02 |
| GO:BP | GO:0006166 | purine ribonucleoside salvage | 5 | 6 | 2.94e-02 |
| GO:BP | GO:0072173 | metanephric tubule morphogenesis | 5 | 6 | 2.94e-02 |
| GO:BP | GO:0050768 | negative regulation of neurogenesis | 34 | 103 | 2.95e-02 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 106 | 401 | 2.95e-02 |
| GO:BP | GO:1903039 | positive regulation of leukocyte cell-cell adhesion | 45 | 146 | 2.96e-02 |
| GO:BP | GO:0048145 | regulation of fibroblast proliferation | 26 | 73 | 2.96e-02 |
| GO:BP | GO:0001755 | neural crest cell migration | 19 | 48 | 2.96e-02 |
| GO:BP | GO:0051145 | smooth muscle cell differentiation | 21 | 55 | 2.96e-02 |
| GO:BP | GO:2000629 | negative regulation of miRNA metabolic process | 12 | 25 | 3.00e-02 |
| GO:BP | GO:0003184 | pulmonary valve morphogenesis | 10 | 19 | 3.01e-02 |
| GO:BP | GO:0019934 | cGMP-mediated signaling | 10 | 19 | 3.01e-02 |
| GO:BP | GO:0099637 | neurotransmitter receptor transport | 10 | 19 | 3.01e-02 |
| GO:BP | GO:0099558 | maintenance of synapse structure | 11 | 22 | 3.03e-02 |
| GO:BP | GO:0048499 | synaptic vesicle membrane organization | 11 | 22 | 3.03e-02 |
| GO:BP | GO:0002021 | response to dietary excess | 11 | 22 | 3.03e-02 |
| GO:BP | GO:0009119 | ribonucleoside metabolic process | 11 | 22 | 3.03e-02 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 787 | 3569 | 3.03e-02 |
| GO:BP | GO:0050867 | positive regulation of cell activation | 59 | 203 | 3.07e-02 |
| GO:BP | GO:0045668 | negative regulation of osteoblast differentiation | 16 | 38 | 3.08e-02 |
| GO:BP | GO:1902106 | negative regulation of leukocyte differentiation | 24 | 66 | 3.10e-02 |
| GO:BP | GO:0072006 | nephron development | 37 | 115 | 3.10e-02 |
| GO:BP | GO:0051234 | establishment of localization | 718 | 3240 | 3.11e-02 |
| GO:BP | GO:0007272 | ensheathment of neurons | 38 | 119 | 3.14e-02 |
| GO:BP | GO:0008366 | axon ensheathment | 38 | 119 | 3.14e-02 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 207 | 850 | 3.15e-02 |
| GO:BP | GO:0051208 | sequestering of calcium ion | 32 | 96 | 3.19e-02 |
| GO:BP | GO:0048732 | gland development | 84 | 308 | 3.19e-02 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 145 | 573 | 3.19e-02 |
| GO:BP | GO:0007548 | sex differentiation | 53 | 179 | 3.21e-02 |
| GO:BP | GO:2000027 | regulation of animal organ morphogenesis | 22 | 59 | 3.21e-02 |
| GO:BP | GO:0070661 | leukocyte proliferation | 52 | 175 | 3.22e-02 |
| GO:BP | GO:1903312 | negative regulation of mRNA metabolic process | 28 | 81 | 3.22e-02 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 41 | 131 | 3.22e-02 |
| GO:BP | GO:0045637 | regulation of myeloid cell differentiation | 42 | 135 | 3.24e-02 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 769 | 3486 | 3.26e-02 |
| GO:BP | GO:0098659 | inorganic cation import across plasma membrane | 25 | 70 | 3.26e-02 |
| GO:BP | GO:0099587 | inorganic ion import across plasma membrane | 25 | 70 | 3.26e-02 |
| GO:BP | GO:0046651 | lymphocyte proliferation | 46 | 151 | 3.26e-02 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 103 | 390 | 3.31e-02 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 96 | 360 | 3.35e-02 |
| GO:BP | GO:0060840 | artery development | 29 | 85 | 3.35e-02 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 163 | 654 | 3.36e-02 |
| GO:BP | GO:1903169 | regulation of calcium ion transmembrane transport | 35 | 108 | 3.41e-02 |
| GO:BP | GO:0021670 | lateral ventricle development | 7 | 11 | 3.41e-02 |
| GO:BP | GO:0072077 | renal vesicle morphogenesis | 7 | 11 | 3.41e-02 |
| GO:BP | GO:0035435 | phosphate ion transmembrane transport | 7 | 11 | 3.41e-02 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 127 | 495 | 3.41e-02 |
| GO:BP | GO:0021783 | preganglionic parasympathetic fiber development | 7 | 11 | 3.41e-02 |
| GO:BP | GO:0030852 | regulation of granulocyte differentiation | 7 | 11 | 3.41e-02 |
| GO:BP | GO:1904044 | response to aldosterone | 7 | 11 | 3.41e-02 |
| GO:BP | GO:0060033 | anatomical structure regression | 7 | 11 | 3.41e-02 |
| GO:BP | GO:0032411 | positive regulation of transporter activity | 23 | 63 | 3.41e-02 |
| GO:BP | GO:0009611 | response to wounding | 101 | 382 | 3.43e-02 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 545 | 2422 | 3.43e-02 |
| GO:BP | GO:0043500 | muscle adaptation | 30 | 89 | 3.43e-02 |
| GO:BP | GO:0014070 | response to organic cyclic compound | 152 | 606 | 3.51e-02 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 791 | 3595 | 3.51e-02 |
| GO:BP | GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 78 | 284 | 3.51e-02 |
| GO:BP | GO:0010656 | negative regulation of muscle cell apoptotic process | 14 | 32 | 3.56e-02 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 923 | 4231 | 3.57e-02 |
| GO:BP | GO:0060078 | regulation of postsynaptic membrane potential | 27 | 78 | 3.58e-02 |
| GO:BP | GO:0045685 | regulation of glial cell differentiation | 19 | 49 | 3.58e-02 |
| GO:BP | GO:1901698 | response to nitrogen compound | 181 | 736 | 3.58e-02 |
| GO:BP | GO:0061572 | actin filament bundle organization | 41 | 132 | 3.59e-02 |
| GO:BP | GO:0032409 | regulation of transporter activity | 45 | 148 | 3.62e-02 |
| GO:BP | GO:0002695 | negative regulation of leukocyte activation | 32 | 97 | 3.63e-02 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 542 | 2410 | 3.64e-02 |
| GO:BP | GO:0071407 | cellular response to organic cyclic compound | 100 | 379 | 3.76e-02 |
| GO:BP | GO:0060041 | retina development in camera-type eye | 34 | 105 | 3.81e-02 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 22 | 60 | 3.85e-02 |
| GO:BP | GO:0051961 | negative regulation of nervous system development | 35 | 109 | 3.88e-02 |
| GO:BP | GO:0042129 | regulation of T cell proliferation | 29 | 86 | 3.90e-02 |
| GO:BP | GO:0014009 | glial cell proliferation | 16 | 39 | 3.90e-02 |
| GO:BP | GO:0046578 | regulation of Ras protein signal transduction | 16 | 39 | 3.90e-02 |
| GO:BP | GO:0022408 | negative regulation of cell-cell adhesion | 36 | 113 | 3.93e-02 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 376 | 1633 | 3.95e-02 |
| GO:BP | GO:2000106 | regulation of leukocyte apoptotic process | 20 | 53 | 3.97e-02 |
| GO:BP | GO:0042552 | myelination | 37 | 117 | 3.97e-02 |
| GO:BP | GO:0046847 | filopodium assembly | 20 | 53 | 3.97e-02 |
| GO:BP | GO:0003197 | endocardial cushion development | 18 | 46 | 3.97e-02 |
| GO:BP | GO:1903540 | establishment of protein localization to postsynaptic membrane | 8 | 14 | 3.97e-02 |
| GO:BP | GO:0016202 | regulation of striated muscle tissue development | 8 | 14 | 3.97e-02 |
| GO:BP | GO:1990845 | adaptive thermogenesis | 38 | 121 | 3.97e-02 |
| GO:BP | GO:0048485 | sympathetic nervous system development | 8 | 14 | 3.97e-02 |
| GO:BP | GO:0010885 | regulation of cholesterol storage | 8 | 14 | 3.97e-02 |
| GO:BP | GO:0043217 | myelin maintenance | 8 | 14 | 3.97e-02 |
| GO:BP | GO:0098969 | neurotransmitter receptor transport to postsynaptic membrane | 8 | 14 | 3.97e-02 |
| GO:BP | GO:0072234 | metanephric nephron tubule development | 8 | 14 | 3.97e-02 |
| GO:BP | GO:0098657 | import into cell | 155 | 622 | 4.00e-02 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 40 | 129 | 4.01e-02 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 119 | 463 | 4.04e-02 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 119 | 463 | 4.04e-02 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 108 | 415 | 4.05e-02 |
| GO:BP | GO:0070371 | ERK1 and ERK2 cascade | 56 | 194 | 4.13e-02 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 77 | 282 | 4.13e-02 |
| GO:BP | GO:0044342 | type B pancreatic cell proliferation | 11 | 23 | 4.13e-02 |
| GO:BP | GO:0003283 | atrial septum development | 11 | 23 | 4.13e-02 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 133 | 525 | 4.13e-02 |
| GO:BP | GO:0061036 | positive regulation of cartilage development | 11 | 23 | 4.13e-02 |
| GO:BP | GO:0010664 | negative regulation of striated muscle cell apoptotic process | 11 | 23 | 4.13e-02 |
| GO:BP | GO:0030278 | regulation of ossification | 27 | 79 | 4.13e-02 |
| GO:BP | GO:0098698 | postsynaptic specialization assembly | 11 | 23 | 4.13e-02 |
| GO:BP | GO:0051491 | positive regulation of filopodium assembly | 11 | 23 | 4.13e-02 |
| GO:BP | GO:1902894 | negative regulation of miRNA transcription | 11 | 23 | 4.13e-02 |
| GO:BP | GO:0045761 | regulation of adenylate cyclase activity | 11 | 23 | 4.13e-02 |
| GO:BP | GO:0048668 | collateral sprouting | 11 | 23 | 4.13e-02 |
| GO:BP | GO:0045667 | regulation of osteoblast differentiation | 32 | 98 | 4.13e-02 |
| GO:BP | GO:0003416 | endochondral bone growth | 11 | 23 | 4.13e-02 |
| GO:BP | GO:0003215 | cardiac right ventricle morphogenesis | 9 | 17 | 4.15e-02 |
| GO:BP | GO:0060392 | negative regulation of SMAD protein signal transduction | 9 | 17 | 4.15e-02 |
| GO:BP | GO:0097107 | postsynaptic density assembly | 9 | 17 | 4.15e-02 |
| GO:BP | GO:0003203 | endocardial cushion morphogenesis | 15 | 36 | 4.15e-02 |
| GO:BP | GO:0032528 | microvillus organization | 9 | 17 | 4.15e-02 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 94 | 355 | 4.15e-02 |
| GO:BP | GO:0048670 | regulation of collateral sprouting | 9 | 17 | 4.15e-02 |
| GO:BP | GO:0060575 | intestinal epithelial cell differentiation | 9 | 17 | 4.15e-02 |
| GO:BP | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process | 10 | 20 | 4.16e-02 |
| GO:BP | GO:0035024 | negative regulation of Rho protein signal transduction | 10 | 20 | 4.16e-02 |
| GO:BP | GO:0043502 | regulation of muscle adaptation | 24 | 68 | 4.16e-02 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 124 | 486 | 4.16e-02 |
| GO:BP | GO:0099560 | synaptic membrane adhesion | 10 | 20 | 4.16e-02 |
| GO:BP | GO:0032946 | positive regulation of mononuclear cell proliferation | 24 | 68 | 4.16e-02 |
| GO:BP | GO:0007043 | cell-cell junction assembly | 34 | 106 | 4.16e-02 |
| GO:BP | GO:0002286 | T cell activation involved in immune response | 21 | 57 | 4.16e-02 |
| GO:BP | GO:0001819 | positive regulation of cytokine production | 71 | 257 | 4.16e-02 |
| GO:BP | GO:0014074 | response to purine-containing compound | 34 | 106 | 4.16e-02 |
| GO:BP | GO:1902373 | negative regulation of mRNA catabolic process | 21 | 57 | 4.16e-02 |
| GO:BP | GO:0060993 | kidney morphogenesis | 24 | 68 | 4.16e-02 |
| GO:BP | GO:2001056 | positive regulation of cysteine-type endopeptidase activity | 28 | 83 | 4.18e-02 |
| GO:BP | GO:0045834 | positive regulation of lipid metabolic process | 28 | 83 | 4.18e-02 |
| GO:BP | GO:0042490 | mechanoreceptor differentiation | 19 | 50 | 4.23e-02 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 245 | 1031 | 4.23e-02 |
| GO:BP | GO:0010660 | regulation of muscle cell apoptotic process | 19 | 50 | 4.23e-02 |
| GO:BP | GO:0031016 | pancreas development | 19 | 50 | 4.23e-02 |
| GO:BP | GO:0009880 | embryonic pattern specification | 17 | 43 | 4.24e-02 |
| GO:BP | GO:0046660 | female sex differentiation | 29 | 87 | 4.29e-02 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 159 | 642 | 4.29e-02 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 29 | 87 | 4.29e-02 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 29 | 87 | 4.29e-02 |
| GO:BP | GO:0048593 | camera-type eye morphogenesis | 29 | 87 | 4.29e-02 |
| GO:BP | GO:0051050 | positive regulation of transport | 143 | 571 | 4.38e-02 |
| GO:BP | GO:0061614 | miRNA transcription | 22 | 61 | 4.40e-02 |
| GO:BP | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules | 30 | 91 | 4.41e-02 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 61 | 216 | 4.48e-02 |
| GO:BP | GO:0140352 | export from cell | 148 | 594 | 4.53e-02 |
| GO:BP | GO:0003162 | atrioventricular node development | 6 | 9 | 4.53e-02 |
| GO:BP | GO:0072189 | ureter development | 6 | 9 | 4.53e-02 |
| GO:BP | GO:0014745 | negative regulation of muscle adaptation | 6 | 9 | 4.53e-02 |
| GO:BP | GO:0086027 | AV node cell to bundle of His cell signaling | 6 | 9 | 4.53e-02 |
| GO:BP | GO:0048671 | negative regulation of collateral sprouting | 6 | 9 | 4.53e-02 |
| GO:BP | GO:0042693 | muscle cell fate commitment | 6 | 9 | 4.53e-02 |
| GO:BP | GO:0048340 | paraxial mesoderm morphogenesis | 6 | 9 | 4.53e-02 |
| GO:BP | GO:0086016 | AV node cell action potential | 6 | 9 | 4.53e-02 |
| GO:BP | GO:0046854 | phosphatidylinositol phosphate biosynthetic process | 20 | 54 | 4.60e-02 |
| GO:BP | GO:0060079 | excitatory postsynaptic potential | 23 | 65 | 4.67e-02 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 349 | 1514 | 4.70e-02 |
| GO:BP | GO:0045600 | positive regulation of fat cell differentiation | 18 | 47 | 4.72e-02 |
| GO:BP | GO:0014015 | positive regulation of gliogenesis | 18 | 47 | 4.72e-02 |
| GO:BP | GO:0060491 | regulation of cell projection assembly | 47 | 159 | 4.77e-02 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 67 | 242 | 4.83e-02 |
| GO:BP | GO:0099068 | postsynapse assembly | 13 | 30 | 4.84e-02 |
| GO:BP | GO:0046850 | regulation of bone remodeling | 13 | 30 | 4.84e-02 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 37 | 119 | 4.85e-02 |
| GO:BP | GO:0141091 | transforming growth factor beta receptor superfamily signaling pathway | 71 | 259 | 4.86e-02 |
| GO:BP | GO:0022898 | regulation of transmembrane transporter activity | 42 | 139 | 4.88e-02 |
| GO:BP | GO:0060021 | roof of mouth development | 24 | 69 | 4.89e-02 |
| GO:BP | GO:0006939 | smooth muscle contraction | 24 | 69 | 4.89e-02 |
| GO:BP | GO:0010243 | response to organonitrogen compound | 168 | 685 | 4.89e-02 |
| GO:BP | GO:0032414 | positive regulation of ion transmembrane transporter activity | 21 | 58 | 4.93e-02 |
| GO:BP | GO:0072028 | nephron morphogenesis | 21 | 58 | 4.93e-02 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 29 | 88 | 4.96e-02 |
ESR_int_restable %>%
filter(source=="KEGG") %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "KEGG terms found in ESR neargenes intersected with RNA response genes") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| KEGG | KEGG:04020 | Calcium signaling pathway | 51 | 161 | 5.13e-02 |
| KEGG | KEGG:04360 | Axon guidance | 50 | 155 | 5.13e-02 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 43 | 129 | 5.13e-02 |
| KEGG | KEGG:04934 | Cushing syndrome | 39 | 119 | 7.61e-02 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 46 | 147 | 7.61e-02 |
| KEGG | KEGG:04750 | Inflammatory mediator regulation of TRP channels | 26 | 72 | 7.98e-02 |
| KEGG | KEGG:04070 | Phosphatidylinositol signaling system | 28 | 83 | 1.16e-01 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 40 | 129 | 1.16e-01 |
| KEGG | KEGG:04924 | Renin secretion | 19 | 50 | 1.16e-01 |
| KEGG | KEGG:05410 | Hypertrophic cardiomyopathy | 25 | 77 | 1.28e-01 |
| KEGG | KEGG:04148 | Efferocytosis | 37 | 120 | 1.28e-01 |
| KEGG | KEGG:04911 | Insulin secretion | 22 | 62 | 1.28e-01 |
| KEGG | KEGG:00230 | Purine metabolism | 29 | 93 | 1.28e-01 |
| KEGG | KEGG:03250 | Viral life cycle - HIV-1 | 19 | 54 | 1.28e-01 |
| KEGG | KEGG:04740 | Olfactory transduction | 11 | 25 | 1.28e-01 |
| KEGG | KEGG:05226 | Gastric cancer | 34 | 113 | 1.28e-01 |
| KEGG | KEGG:05224 | Breast cancer | 35 | 113 | 1.28e-01 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 33 | 105 | 1.28e-01 |
| KEGG | KEGG:04814 | Motor proteins | 41 | 141 | 1.28e-01 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 47 | 162 | 1.28e-01 |
| KEGG | KEGG:04730 | Long-term depression | 15 | 39 | 1.28e-01 |
| KEGG | KEGG:04927 | Cortisol synthesis and secretion | 17 | 46 | 1.28e-01 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 42 | 144 | 1.28e-01 |
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 37 | 124 | 1.28e-01 |
| KEGG | KEGG:05414 | Dilated cardiomyopathy | 26 | 82 | 1.34e-01 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 36 | 122 | 1.34e-01 |
| KEGG | KEGG:04260 | Cardiac muscle contraction | 21 | 63 | 1.36e-01 |
| KEGG | KEGG:04925 | Aldosterone synthesis and secretion | 24 | 75 | 1.36e-01 |
| KEGG | KEGG:05412 | Arrhythmogenic right ventricular cardiomyopathy | 22 | 67 | 1.36e-01 |
| KEGG | KEGG:04976 | Bile secretion | 15 | 41 | 1.36e-01 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 20 | 60 | 1.41e-01 |
| KEGG | KEGG:04611 | Platelet activation | 27 | 88 | 1.46e-01 |
| KEGG | KEGG:01521 | EGFR tyrosine kinase inhibitor resistance | 23 | 72 | 1.46e-01 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 38 | 133 | 1.46e-01 |
| KEGG | KEGG:04014 | Ras signaling pathway | 45 | 164 | 1.68e-01 |
| KEGG | KEGG:05032 | Morphine addiction | 18 | 54 | 1.68e-01 |
| KEGG | KEGG:04724 | Glutamatergic synapse | 23 | 74 | 1.78e-01 |
| KEGG | KEGG:03320 | PPAR signaling pathway | 16 | 47 | 1.80e-01 |
| KEGG | KEGG:03430 | Mismatch repair | 9 | 22 | 1.92e-01 |
| KEGG | KEGG:05164 | Influenza A | 28 | 95 | 1.92e-01 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 36 | 129 | 1.98e-01 |
| KEGG | KEGG:05217 | Basal cell carcinoma | 16 | 48 | 1.98e-01 |
| KEGG | KEGG:00562 | Inositol phosphate metabolism | 19 | 60 | 1.98e-01 |
| KEGG | KEGG:05218 | Melanoma | 18 | 56 | 1.98e-01 |
| KEGG | KEGG:04913 | Ovarian steroidogenesis | 10 | 26 | 1.98e-01 |
| KEGG | KEGG:04950 | Maturity onset diabetes of the young | 3 | 4 | 2.08e-01 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 29 | 101 | 2.08e-01 |
| KEGG | KEGG:04972 | Pancreatic secretion | 18 | 57 | 2.18e-01 |
| KEGG | KEGG:05034 | Alcoholism | 29 | 102 | 2.22e-01 |
| KEGG | KEGG:04658 | Th1 and Th2 cell differentiation | 16 | 50 | 2.37e-01 |
| KEGG | KEGG:04115 | p53 signaling pathway | 20 | 66 | 2.37e-01 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 23 | 78 | 2.37e-01 |
| KEGG | KEGG:04640 | Hematopoietic cell lineage | 10 | 28 | 2.52e-01 |
| KEGG | KEGG:04921 | Oxytocin signaling pathway | 31 | 113 | 2.52e-01 |
| KEGG | KEGG:04971 | Gastric acid secretion | 16 | 51 | 2.52e-01 |
| KEGG | KEGG:04960 | Aldosterone-regulated sodium reabsorption | 9 | 24 | 2.52e-01 |
| KEGG | KEGG:04380 | Osteoclast differentiation | 26 | 92 | 2.52e-01 |
| KEGG | KEGG:04540 | Gap junction | 21 | 71 | 2.52e-01 |
| KEGG | KEGG:04916 | Melanogenesis | 22 | 75 | 2.52e-01 |
| KEGG | KEGG:04713 | Circadian entrainment | 20 | 68 | 2.75e-01 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 19 | 64 | 2.75e-01 |
| KEGG | KEGG:04330 | Notch signaling pathway | 16 | 52 | 2.76e-01 |
| KEGG | KEGG:04392 | Hippo signaling pathway - multiple species | 9 | 25 | 2.80e-01 |
| KEGG | KEGG:04721 | Synaptic vesicle cycle | 15 | 49 | 2.95e-01 |
| KEGG | KEGG:00910 | Nitrogen metabolism | 4 | 8 | 2.95e-01 |
| KEGG | KEGG:04725 | Cholinergic synapse | 23 | 82 | 2.95e-01 |
| KEGG | KEGG:04923 | Regulation of lipolysis in adipocytes | 13 | 41 | 2.95e-01 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 36 | 137 | 2.95e-01 |
| KEGG | KEGG:05110 | Vibrio cholerae infection | 13 | 41 | 2.95e-01 |
| KEGG | KEGG:04929 | GnRH secretion | 15 | 50 | 3.02e-01 |
| KEGG | KEGG:04726 | Serotonergic synapse | 17 | 58 | 3.02e-01 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 26 | 96 | 3.02e-01 |
| KEGG | KEGG:00000 | KEGG root term | 1145 | 5451 | 3.02e-01 |
| KEGG | KEGG:05213 | Endometrial cancer | 16 | 54 | 3.02e-01 |
| KEGG | KEGG:03083 | Polycomb repressive complex | 20 | 70 | 3.02e-01 |
| KEGG | KEGG:05200 | Pathways in cancer | 92 | 391 | 3.02e-01 |
| KEGG | KEGG:04970 | Salivary secretion | 16 | 54 | 3.02e-01 |
| KEGG | KEGG:04931 | Insulin resistance | 25 | 91 | 3.02e-01 |
| KEGG | KEGG:04061 | Viral protein interaction with cytokine and cytokine receptor | 7 | 19 | 3.05e-01 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 41 | 162 | 3.05e-01 |
| KEGG | KEGG:04270 | Vascular smooth muscle contraction | 25 | 92 | 3.05e-01 |
| KEGG | KEGG:00564 | Glycerophospholipid metabolism | 20 | 71 | 3.05e-01 |
| KEGG | KEGG:04720 | Long-term potentiation | 15 | 51 | 3.29e-01 |
| KEGG | KEGG:04613 | Neutrophil extracellular trap formation | 24 | 89 | 3.30e-01 |
| KEGG | KEGG:04930 | Type II diabetes mellitus | 11 | 35 | 3.33e-01 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 56 | 231 | 3.33e-01 |
| KEGG | KEGG:04060 | Cytokine-cytokine receptor interaction | 25 | 94 | 3.42e-01 |
| KEGG | KEGG:04530 | Tight junction | 33 | 129 | 3.42e-01 |
| KEGG | KEGG:05167 | Kaposi sarcoma-associated herpesvirus infection | 34 | 134 | 3.48e-01 |
| KEGG | KEGG:05221 | Acute myeloid leukemia | 15 | 52 | 3.48e-01 |
| KEGG | KEGG:04742 | Taste transduction | 7 | 20 | 3.48e-01 |
| KEGG | KEGG:04210 | Apoptosis | 29 | 112 | 3.48e-01 |
| KEGG | KEGG:04961 | Endocrine and other factor-regulated calcium reabsorption | 12 | 40 | 3.53e-01 |
| KEGG | KEGG:05214 | Glioma | 18 | 65 | 3.53e-01 |
| KEGG | KEGG:05030 | Cocaine addiction | 10 | 32 | 3.55e-01 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 29 | 113 | 3.60e-01 |
| KEGG | KEGG:04625 | C-type lectin receptor signaling pathway | 20 | 74 | 3.61e-01 |
| KEGG | KEGG:04650 | Natural killer cell mediated cytotoxicity | 15 | 53 | 3.68e-01 |
| KEGG | KEGG:05211 | Renal cell carcinoma | 17 | 62 | 3.85e-01 |
| KEGG | KEGG:04152 | AMPK signaling pathway | 26 | 101 | 3.85e-01 |
| KEGG | KEGG:04728 | Dopaminergic synapse | 25 | 97 | 3.92e-01 |
| KEGG | KEGG:05216 | Thyroid cancer | 10 | 33 | 3.92e-01 |
| KEGG | KEGG:04917 | Prolactin signaling pathway | 15 | 54 | 3.97e-01 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 39 | 160 | 4.03e-01 |
| KEGG | KEGG:04072 | Phospholipase D signaling pathway | 27 | 107 | 4.21e-01 |
| KEGG | KEGG:05210 | Colorectal cancer | 21 | 81 | 4.27e-01 |
| KEGG | KEGG:04114 | Oocyte meiosis | 26 | 103 | 4.27e-01 |
| KEGG | KEGG:04071 | Sphingolipid signaling pathway | 26 | 103 | 4.27e-01 |
| KEGG | KEGG:04966 | Collecting duct acid secretion | 6 | 18 | 4.39e-01 |
| KEGG | KEGG:00410 | beta-Alanine metabolism | 7 | 22 | 4.39e-01 |
| KEGG | KEGG:04964 | Proximal tubule bicarbonate reclamation | 6 | 18 | 4.39e-01 |
| KEGG | KEGG:04370 | VEGF signaling pathway | 13 | 47 | 4.41e-01 |
| KEGG | KEGG:01522 | Endocrine resistance | 21 | 82 | 4.48e-01 |
| KEGG | KEGG:04974 | Protein digestion and absorption | 15 | 56 | 4.54e-01 |
| KEGG | KEGG:04922 | Glucagon signaling pathway | 20 | 78 | 4.54e-01 |
| KEGG | KEGG:00591 | Linoleic acid metabolism | 3 | 7 | 4.54e-01 |
| KEGG | KEGG:04727 | GABAergic synapse | 14 | 52 | 4.56e-01 |
| KEGG | KEGG:05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 17 | 65 | 4.56e-01 |
| KEGG | KEGG:04520 | Adherens junction | 21 | 83 | 4.65e-01 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 58 | 252 | 4.65e-01 |
| KEGG | KEGG:05160 | Hepatitis C | 27 | 110 | 4.65e-01 |
| KEGG | KEGG:03410 | Base excision repair | 12 | 44 | 4.74e-01 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 19 | 75 | 4.85e-01 |
| KEGG | KEGG:05417 | Lipid and atherosclerosis | 34 | 143 | 4.85e-01 |
| KEGG | KEGG:05130 | Pathogenic Escherichia coli infection | 34 | 143 | 4.85e-01 |
| KEGG | KEGG:05017 | Spinocerebellar ataxia | 29 | 120 | 4.85e-01 |
| KEGG | KEGG:05146 | Amoebiasis | 16 | 62 | 4.85e-01 |
| KEGG | KEGG:05212 | Pancreatic cancer | 18 | 71 | 4.88e-01 |
| KEGG | KEGG:00470 | D-Amino acid metabolism | 2 | 4 | 4.93e-01 |
| KEGG | KEGG:05031 | Amphetamine addiction | 12 | 45 | 5.01e-01 |
| KEGG | KEGG:05142 | Chagas disease | 18 | 72 | 5.07e-01 |
| KEGG | KEGG:04915 | Estrogen signaling pathway | 22 | 90 | 5.07e-01 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 22 | 90 | 5.07e-01 |
| KEGG | KEGG:05161 | Hepatitis B | 30 | 126 | 5.07e-01 |
| KEGG | KEGG:04217 | Necroptosis | 24 | 99 | 5.07e-01 |
| KEGG | KEGG:03260 | Virion - Human immunodeficiency virus | 1 | 1 | 5.09e-01 |
| KEGG | KEGG:04213 | Longevity regulating pathway - multiple species | 13 | 50 | 5.09e-01 |
| KEGG | KEGG:05321 | Inflammatory bowel disease | 6 | 20 | 5.14e-01 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 38 | 164 | 5.16e-01 |
| KEGG | KEGG:04914 | Progesterone-mediated oocyte maturation | 20 | 82 | 5.19e-01 |
| KEGG | KEGG:03450 | Non-homologous end-joining | 4 | 12 | 5.19e-01 |
| KEGG | KEGG:05222 | Small cell lung cancer | 20 | 82 | 5.19e-01 |
| KEGG | KEGG:05120 | Epithelial cell signaling in Helicobacter pylori infection | 14 | 55 | 5.19e-01 |
| KEGG | KEGG:04973 | Carbohydrate digestion and absorption | 8 | 29 | 5.29e-01 |
| KEGG | KEGG:04136 | Autophagy - other | 8 | 29 | 5.29e-01 |
| KEGG | KEGG:05152 | Tuberculosis | 24 | 101 | 5.39e-01 |
| KEGG | KEGG:04978 | Mineral absorption | 10 | 38 | 5.41e-01 |
| KEGG | KEGG:04912 | GnRH signaling pathway | 18 | 74 | 5.46e-01 |
| KEGG | KEGG:03030 | DNA replication | 9 | 34 | 5.58e-01 |
| KEGG | KEGG:04660 | T cell receptor signaling pathway | 20 | 84 | 5.72e-01 |
| KEGG | KEGG:04145 | Phagosome | 20 | 84 | 5.72e-01 |
| KEGG | KEGG:04140 | Autophagy - animal | 34 | 149 | 5.75e-01 |
| KEGG | KEGG:04977 | Vitamin digestion and absorption | 4 | 13 | 5.89e-01 |
| KEGG | KEGG:00750 | Vitamin B6 metabolism | 2 | 5 | 5.94e-01 |
| KEGG | KEGG:03265 | Virion - Ebolavirus and Lyssavirus | 2 | 5 | 5.94e-01 |
| KEGG | KEGG:00061 | Fatty acid biosynthesis | 3 | 9 | 5.94e-01 |
| KEGG | KEGG:05207 | Chemical carcinogenesis - receptor activation | 30 | 132 | 6.09e-01 |
| KEGG | KEGG:00500 | Starch and sucrose metabolism | 6 | 22 | 6.09e-01 |
| KEGG | KEGG:04216 | Ferroptosis | 8 | 31 | 6.15e-01 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 17 | 72 | 6.15e-01 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 20 | 87 | 6.54e-01 |
| KEGG | KEGG:05100 | Bacterial invasion of epithelial cells | 17 | 73 | 6.54e-01 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 24 | 106 | 6.57e-01 |
| KEGG | KEGG:00601 | Glycosphingolipid biosynthesis - lacto and neolacto series | 4 | 14 | 6.57e-01 |
| KEGG | KEGG:04514 | Cell adhesion molecules | 16 | 69 | 6.57e-01 |
| KEGG | KEGG:04144 | Endocytosis | 47 | 216 | 6.57e-01 |
| KEGG | KEGG:04630 | JAK-STAT signaling pathway | 19 | 83 | 6.57e-01 |
| KEGG | KEGG:01524 | Platinum drug resistance | 14 | 60 | 6.57e-01 |
| KEGG | KEGG:05033 | Nicotine addiction | 3 | 10 | 6.57e-01 |
| KEGG | KEGG:04614 | Renin-angiotensin system | 3 | 10 | 6.57e-01 |
| KEGG | KEGG:00592 | alpha-Linolenic acid metabolism | 3 | 10 | 6.57e-01 |
| KEGG | KEGG:04668 | TNF signaling pathway | 19 | 83 | 6.57e-01 |
| KEGG | KEGG:04672 | Intestinal immune network for IgA production | 3 | 10 | 6.57e-01 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 54 | 250 | 6.61e-01 |
| KEGG | KEGG:00515 | Mannose type O-glycan biosynthesis | 5 | 19 | 6.63e-01 |
| KEGG | KEGG:00534 | Glycosaminoglycan biosynthesis - heparan sulfate / heparin | 5 | 19 | 6.63e-01 |
| KEGG | KEGG:04218 | Cellular senescence | 30 | 136 | 6.63e-01 |
| KEGG | KEGG:04211 | Longevity regulating pathway | 17 | 75 | 6.74e-01 |
| KEGG | KEGG:04666 | Fc gamma R-mediated phagocytosis | 17 | 75 | 6.74e-01 |
| KEGG | KEGG:05203 | Viral carcinogenesis | 31 | 142 | 6.75e-01 |
| KEGG | KEGG:05330 | Allograft rejection | 1 | 2 | 6.75e-01 |
| KEGG | KEGG:05323 | Rheumatoid arthritis | 9 | 38 | 6.75e-01 |
| KEGG | KEGG:05332 | Graft-versus-host disease | 1 | 2 | 6.75e-01 |
| KEGG | KEGG:00770 | Pantothenate and CoA biosynthesis | 4 | 15 | 6.75e-01 |
| KEGG | KEGG:02010 | ABC transporters | 6 | 24 | 6.75e-01 |
| KEGG | KEGG:04062 | Chemokine signaling pathway | 24 | 109 | 6.80e-01 |
| KEGG | KEGG:00532 | Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate | 5 | 20 | 7.05e-01 |
| KEGG | KEGG:04936 | Alcoholic liver disease | 20 | 91 | 7.11e-01 |
| KEGG | KEGG:05230 | Central carbon metabolism in cancer | 13 | 58 | 7.19e-01 |
| KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 22 | 101 | 7.19e-01 |
| KEGG | KEGG:05215 | Prostate cancer | 18 | 82 | 7.21e-01 |
| KEGG | KEGG:05322 | Systemic lupus erythematosus | 10 | 44 | 7.22e-01 |
| KEGG | KEGG:04920 | Adipocytokine signaling pathway | 11 | 49 | 7.31e-01 |
| KEGG | KEGG:05143 | African trypanosomiasis | 4 | 16 | 7.33e-01 |
| KEGG | KEGG:00514 | Other types of O-glycan biosynthesis | 8 | 35 | 7.35e-01 |
| KEGG | KEGG:03420 | Nucleotide excision repair | 12 | 54 | 7.35e-01 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 29 | 136 | 7.35e-01 |
| KEGG | KEGG:00052 | Galactose metabolism | 5 | 21 | 7.42e-01 |
| KEGG | KEGG:04940 | Type I diabetes mellitus | 2 | 7 | 7.43e-01 |
| KEGG | KEGG:04744 | Phototransduction | 3 | 12 | 7.76e-01 |
| KEGG | KEGG:04064 | NF-kappa B signaling pathway | 13 | 61 | 8.11e-01 |
| KEGG | KEGG:04670 | Leukocyte transendothelial migration | 16 | 76 | 8.11e-01 |
| KEGG | KEGG:05170 | Human immunodeficiency virus 1 infection | 31 | 149 | 8.11e-01 |
| KEGG | KEGG:04066 | HIF-1 signaling pathway | 18 | 86 | 8.11e-01 |
| KEGG | KEGG:05320 | Autoimmune thyroid disease | 1 | 3 | 8.11e-01 |
| KEGG | KEGG:04662 | B cell receptor signaling pathway | 11 | 52 | 8.11e-01 |
| KEGG | KEGG:00600 | Sphingolipid metabolism | 9 | 42 | 8.11e-01 |
| KEGG | KEGG:05131 | Shigellosis | 42 | 204 | 8.11e-01 |
| KEGG | KEGG:00290 | Valine, leucine and isoleucine biosynthesis | 1 | 3 | 8.11e-01 |
| KEGG | KEGG:00270 | Cysteine and methionine metabolism | 8 | 37 | 8.11e-01 |
| KEGG | KEGG:05340 | Primary immunodeficiency | 2 | 8 | 8.20e-01 |
| KEGG | KEGG:01040 | Biosynthesis of unsaturated fatty acids | 5 | 23 | 8.31e-01 |
| KEGG | KEGG:05135 | Yersinia infection | 22 | 107 | 8.31e-01 |
| KEGG | KEGG:05140 | Leishmaniasis | 8 | 38 | 8.33e-01 |
| KEGG | KEGG:04918 | Thyroid hormone synthesis | 11 | 53 | 8.36e-01 |
| KEGG | KEGG:00830 | Retinol metabolism | 3 | 14 | 8.93e-01 |
| KEGG | KEGG:04620 | Toll-like receptor signaling pathway | 12 | 60 | 9.11e-01 |
| KEGG | KEGG:00524 | Neomycin, kanamycin and gentamicin biosynthesis | 1 | 4 | 9.35e-01 |
| KEGG | KEGG:00220 | Arginine biosynthesis | 3 | 15 | 9.59e-01 |
| KEGG | KEGG:05416 | Viral myocarditis | 7 | 36 | 9.62e-01 |
| KEGG | KEGG:04130 | SNARE interactions in vesicular transport | 6 | 31 | 9.67e-01 |
| KEGG | KEGG:00051 | Fructose and mannose metabolism | 5 | 26 | 9.67e-01 |
| KEGG | KEGG:00730 | Thiamine metabolism | 2 | 10 | 9.67e-01 |
| KEGG | KEGG:00340 | Histidine metabolism | 2 | 10 | 9.67e-01 |
| KEGG | KEGG:00970 | Aminoacyl-tRNA biosynthesis | 8 | 42 | 9.70e-01 |
| KEGG | KEGG:00512 | Mucin type O-glycan biosynthesis | 4 | 21 | 9.70e-01 |
| KEGG | KEGG:05133 | Pertussis | 8 | 42 | 9.70e-01 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 20 | 104 | 9.76e-01 |
| KEGG | KEGG:05132 | Salmonella infection | 39 | 201 | 9.86e-01 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 19 | 100 | 9.89e-01 |
| KEGG | KEGG:04110 | Cell cycle | 28 | 146 | 9.89e-01 |
| KEGG | KEGG:00640 | Propanoate metabolism | 5 | 27 | 9.89e-01 |
| KEGG | KEGG:05162 | Measles | 16 | 85 | 9.94e-01 |
| KEGG | KEGG:00120 | Primary bile acid biosynthesis | 2 | 11 | 9.94e-01 |
| KEGG | KEGG:01232 | Nucleotide metabolism | 12 | 65 | 9.94e-01 |
| KEGG | KEGG:05231 | Choline metabolism in cancer | 15 | 80 | 9.94e-01 |
| KEGG | KEGG:03082 | ATP-dependent chromatin remodeling | 17 | 91 | 9.94e-01 |
| KEGG | KEGG:04340 | Hedgehog signaling pathway | 9 | 49 | 9.94e-01 |
| KEGG | KEGG:03013 | Nucleocytoplasmic transport | 18 | 96 | 9.94e-01 |
| KEGG | KEGG:00565 | Ether lipid metabolism | 5 | 28 | 9.98e-01 |
| KEGG | KEGG:04512 | ECM-receptor interaction | 9 | 66 | 1.00e+00 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 32 | 169 | 1.00e+00 |
| KEGG | KEGG:04962 | Vasopressin-regulated water reabsorption | 5 | 37 | 1.00e+00 |
| KEGG | KEGG:05208 | Chemical carcinogenesis - reactive oxygen species | 18 | 173 | 1.00e+00 |
| KEGG | KEGG:05171 | Coronavirus disease - COVID-19 | 18 | 149 | 1.00e+00 |
| KEGG | KEGG:05415 | Diabetic cardiomyopathy | 25 | 158 | 1.00e+00 |
| KEGG | KEGG:04510 | Focal adhesion | 28 | 169 | 1.00e+00 |
| KEGG | KEGG:04932 | Non-alcoholic fatty liver disease | 17 | 126 | 1.00e+00 |
| KEGG | KEGG:04975 | Fat digestion and absorption | 2 | 18 | 1.00e+00 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 65 | 364 | 1.00e+00 |
| KEGG | KEGG:04979 | Cholesterol metabolism | 5 | 33 | 1.00e+00 |
| KEGG | KEGG:04714 | Thermogenesis | 24 | 185 | 1.00e+00 |
| KEGG | KEGG:05016 | Huntington disease | 40 | 246 | 1.00e+00 |
| KEGG | KEGG:04710 | Circadian rhythm | 5 | 29 | 1.00e+00 |
| KEGG | KEGG:04722 | Neurotrophin signaling pathway | 19 | 104 | 1.00e+00 |
| KEGG | KEGG:04723 | Retrograde endocannabinoid signaling | 16 | 92 | 1.00e+00 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 47 | 291 | 1.00e+00 |
| KEGG | KEGG:04664 | Fc epsilon RI signaling pathway | 7 | 42 | 1.00e+00 |
| KEGG | KEGG:05012 | Parkinson disease | 31 | 211 | 1.00e+00 |
| KEGG | KEGG:05010 | Alzheimer disease | 54 | 296 | 1.00e+00 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 41 | 388 | 1.00e+00 |
| KEGG | KEGG:04657 | IL-17 signaling pathway | 9 | 55 | 1.00e+00 |
| KEGG | KEGG:04623 | Cytosolic DNA-sensing pathway | 5 | 46 | 1.00e+00 |
| KEGG | KEGG:05144 | Malaria | 3 | 20 | 1.00e+00 |
| KEGG | KEGG:04622 | RIG-I-like receptor signaling pathway | 5 | 44 | 1.00e+00 |
| KEGG | KEGG:04621 | NOD-like receptor signaling pathway | 18 | 109 | 1.00e+00 |
| KEGG | KEGG:04612 | Antigen processing and presentation | 2 | 26 | 1.00e+00 |
| KEGG | KEGG:05145 | Toxoplasmosis | 9 | 72 | 1.00e+00 |
| KEGG | KEGG:04610 | Complement and coagulation cascades | 6 | 35 | 1.00e+00 |
| KEGG | KEGG:05150 | Staphylococcus aureus infection | 1 | 13 | 1.00e+00 |
| KEGG | KEGG:05020 | Prion disease | 29 | 204 | 1.00e+00 |
| KEGG | KEGG:05134 | Legionellosis | 3 | 34 | 1.00e+00 |
| KEGG | KEGG:05219 | Bladder cancer | 6 | 35 | 1.00e+00 |
| KEGG | KEGG:04146 | Peroxisome | 10 | 66 | 1.00e+00 |
| KEGG | KEGG:05418 | Fluid shear stress and atherosclerosis | 20 | 109 | 1.00e+00 |
| KEGG | KEGG:00860 | Porphyrin metabolism | 1 | 18 | 1.00e+00 |
| KEGG | KEGG:00900 | Terpenoid backbone biosynthesis | 2 | 21 | 1.00e+00 |
| KEGG | KEGG:00140 | Steroid hormone biosynthesis | 2 | 16 | 1.00e+00 |
| KEGG | KEGG:00920 | Sulfur metabolism | 1 | 10 | 1.00e+00 |
| KEGG | KEGG:00100 | Steroid biosynthesis | 1 | 15 | 1.00e+00 |
| KEGG | KEGG:00983 | Drug metabolism - other enzymes | 6 | 40 | 1.00e+00 |
| KEGG | KEGG:00071 | Fatty acid degradation | 2 | 31 | 1.00e+00 |
| KEGG | KEGG:01100 | Metabolic pathways | 195 | 1077 | 1.00e+00 |
| KEGG | KEGG:00062 | Fatty acid elongation | 2 | 22 | 1.00e+00 |
| KEGG | KEGG:00040 | Pentose and glucuronate interconversions | 1 | 11 | 1.00e+00 |
| KEGG | KEGG:01200 | Carbon metabolism | 11 | 88 | 1.00e+00 |
| KEGG | KEGG:00030 | Pentose phosphate pathway | 3 | 20 | 1.00e+00 |
| KEGG | KEGG:01210 | 2-Oxocarboxylic acid metabolism | 2 | 26 | 1.00e+00 |
| KEGG | KEGG:01212 | Fatty acid metabolism | 6 | 46 | 1.00e+00 |
| KEGG | KEGG:00430 | Taurine and hypotaurine metabolism | 1 | 8 | 1.00e+00 |
| KEGG | KEGG:01240 | Biosynthesis of cofactors | 16 | 109 | 1.00e+00 |
| KEGG | KEGG:01250 | Biosynthesis of nucleotide sugars | 5 | 34 | 1.00e+00 |
| KEGG | KEGG:00020 | Citrate cycle (TCA cycle) | 1 | 26 | 1.00e+00 |
| KEGG | KEGG:01523 | Antifolate resistance | 2 | 22 | 1.00e+00 |
| KEGG | KEGG:03008 | Ribosome biogenesis in eukaryotes | 5 | 65 | 1.00e+00 |
| KEGG | KEGG:03010 | Ribosome | 7 | 121 | 1.00e+00 |
| KEGG | KEGG:00790 | Folate biosynthesis | 2 | 18 | 1.00e+00 |
| KEGG | KEGG:00785 | Lipoic acid metabolism | 2 | 16 | 1.00e+00 |
| KEGG | KEGG:00190 | Oxidative phosphorylation | 6 | 99 | 1.00e+00 |
| KEGG | KEGG:00760 | Nicotinate and nicotinamide metabolism | 4 | 28 | 1.00e+00 |
| KEGG | KEGG:00480 | Glutathione metabolism | 3 | 39 | 1.00e+00 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 5 | 49 | 1.00e+00 |
| KEGG | KEGG:00511 | Other glycan degradation | 1 | 14 | 1.00e+00 |
| KEGG | KEGG:00513 | Various types of N-glycan biosynthesis | 4 | 37 | 1.00e+00 |
| KEGG | KEGG:00520 | Amino sugar and nucleotide sugar metabolism | 5 | 44 | 1.00e+00 |
| KEGG | KEGG:00330 | Arginine and proline metabolism | 5 | 33 | 1.00e+00 |
| KEGG | KEGG:00533 | Glycosaminoglycan biosynthesis - keratan sulfate | 2 | 13 | 1.00e+00 |
| KEGG | KEGG:00310 | Lysine degradation | 10 | 56 | 1.00e+00 |
| KEGG | KEGG:00561 | Glycerolipid metabolism | 7 | 44 | 1.00e+00 |
| KEGG | KEGG:00563 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 2 | 23 | 1.00e+00 |
| KEGG | KEGG:03015 | mRNA surveillance pathway | 13 | 80 | 1.00e+00 |
| KEGG | KEGG:00590 | Arachidonic acid metabolism | 4 | 25 | 1.00e+00 |
| KEGG | KEGG:00603 | Glycosphingolipid biosynthesis - globo and isoglobo series | 1 | 8 | 1.00e+00 |
| KEGG | KEGG:00604 | Glycosphingolipid biosynthesis - ganglio series | 1 | 12 | 1.00e+00 |
| KEGG | KEGG:00620 | Pyruvate metabolism | 4 | 31 | 1.00e+00 |
| KEGG | KEGG:00630 | Glyoxylate and dicarboxylate metabolism | 2 | 25 | 1.00e+00 |
| KEGG | KEGG:00260 | Glycine, serine and threonine metabolism | 3 | 26 | 1.00e+00 |
| KEGG | KEGG:00250 | Alanine, aspartate and glutamate metabolism | 3 | 28 | 1.00e+00 |
| KEGG | KEGG:00650 | Butanoate metabolism | 2 | 17 | 1.00e+00 |
| KEGG | KEGG:00240 | Pyrimidine metabolism | 5 | 44 | 1.00e+00 |
| KEGG | KEGG:00670 | One carbon pool by folate | 1 | 18 | 1.00e+00 |
| KEGG | KEGG:00740 | Riboflavin metabolism | 1 | 6 | 1.00e+00 |
| KEGG | KEGG:00280 | Valine, leucine and isoleucine degradation | 4 | 42 | 1.00e+00 |
| KEGG | KEGG:03018 | RNA degradation | 6 | 67 | 1.00e+00 |
| KEGG | KEGG:01230 | Biosynthesis of amino acids | 6 | 52 | 1.00e+00 |
| KEGG | KEGG:03022 | Basal transcription factors | 5 | 33 | 1.00e+00 |
| KEGG | KEGG:04215 | Apoptosis - multiple species | 5 | 29 | 1.00e+00 |
| KEGG | KEGG:04142 | Lysosome | 15 | 109 | 1.00e+00 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 21 | 149 | 1.00e+00 |
| KEGG | KEGG:04137 | Mitophagy - animal | 15 | 94 | 1.00e+00 |
| KEGG | KEGG:04122 | Sulfur relay system | 1 | 8 | 1.00e+00 |
| KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 23 | 127 | 1.00e+00 |
| KEGG | KEGG:03020 | RNA polymerase | 4 | 33 | 1.00e+00 |
| KEGG | KEGG:03460 | Fanconi anemia pathway | 8 | 46 | 1.00e+00 |
| KEGG | KEGG:03440 | Homologous recombination | 6 | 38 | 1.00e+00 |
| KEGG | KEGG:00450 | Selenocompound metabolism | 2 | 14 | 1.00e+00 |
| KEGG | KEGG:03060 | Protein export | 2 | 28 | 1.00e+00 |
| KEGG | KEGG:00010 | Glycolysis / Gluconeogenesis | 6 | 41 | 1.00e+00 |
| KEGG | KEGG:03266 | Virion - Herpesvirus | 1 | 6 | 1.00e+00 |
| KEGG | KEGG:03040 | Spliceosome | 10 | 124 | 1.00e+00 |
| KEGG | KEGG:03050 | Proteasome | 5 | 40 | 1.00e+00 |
ESR_int_restable %>%
filter(source=="GO:BP", p_value < 0.05) %>%
slice_head(n=10) %>%
mutate(log10_p=(-log(p_value, base=10))) %>%
ggplot(., aes(x=log10_p, y=reorder(term_name, log10_p)))+
geom_col(fill="#7CAE00")+
theme_bw()+
ylab("")+
xlab(paste0("-log10 p-value"))+
guides(fill="none")+
theme(axis.text.y = element_text(color="black"))+
ggtitle("Intersection ESR")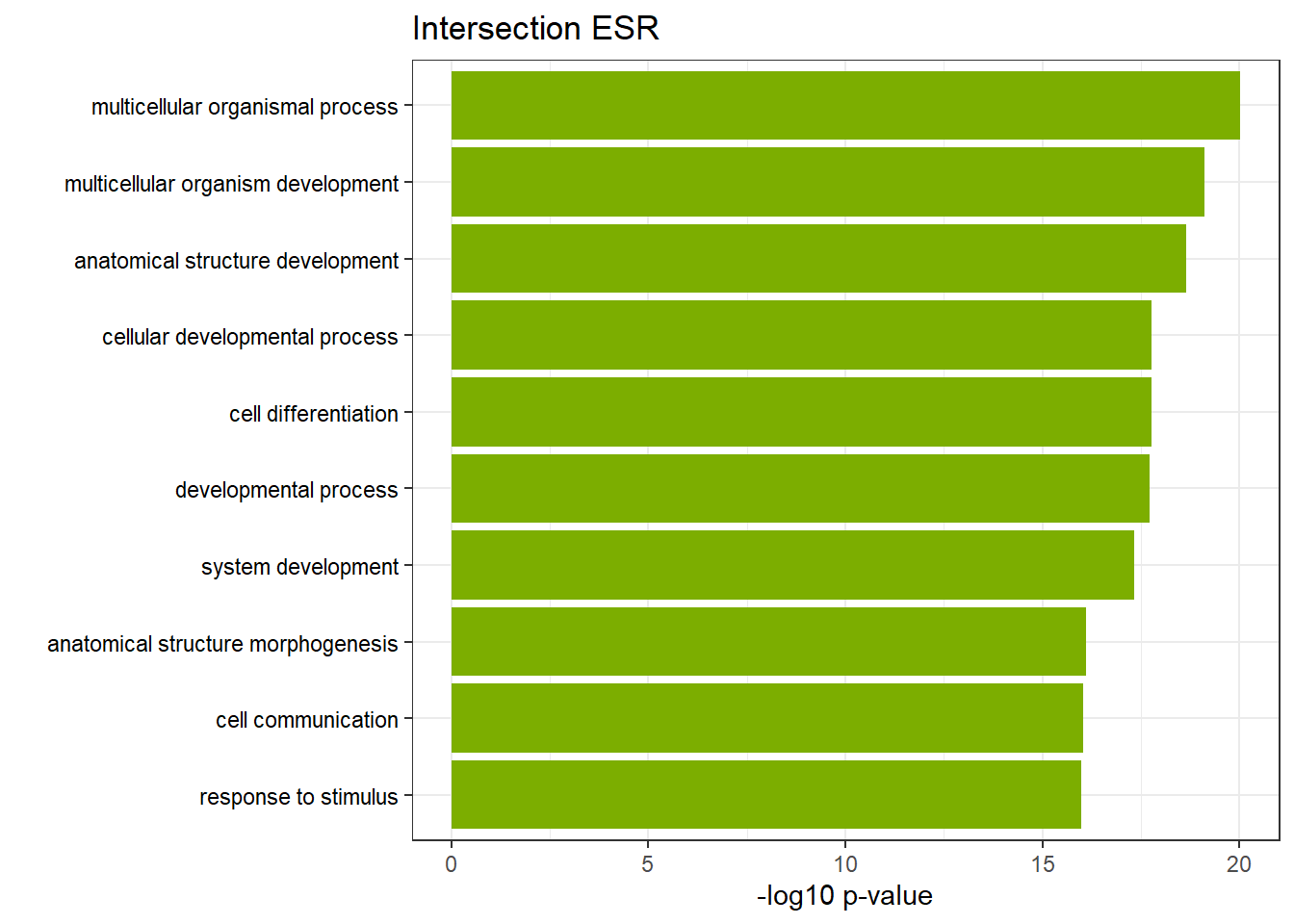
LR
# LR_int_resgenes <- gost(query = int_LR_n_RNAresp,
# organism = "hsapiens",
# significant = FALSE,
# ordered_query = FALSE,
# domain_scope = "custom",
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# custom_bg = background_df$ENTREZID,
# sources=c("GO:BP","KEGG"))
# saveRDS(LR_int_resgenes,"data/GO_KEGG_analysis/LR_int_resgenes.RDS")
LR_int_restable <- LR_int_resgenes$result %>%
dplyr::select(c(source, term_id,
term_name,intersection_size,
term_size, p_value))# %>%
LR_int_restable %>%
filter(source=="GO:BP", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(.,caption = "GO:BP terms found in LR neargenes intersected with RNA response genes" ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0051716 | cellular response to stimulus | 1660 | 4640 | 5.46e-18 |
| GO:BP | GO:0050896 | response to stimulus | 1879 | 5379 | 1.95e-16 |
| GO:BP | GO:0007154 | cell communication | 1439 | 3983 | 1.95e-16 |
| GO:BP | GO:0023052 | signaling | 1413 | 3918 | 1.02e-15 |
| GO:BP | GO:0007165 | signal transduction | 1313 | 3611 | 1.40e-15 |
| GO:BP | GO:0032501 | multicellular organismal process | 1596 | 4504 | 2.20e-15 |
| GO:BP | GO:0050794 | regulation of cellular process | 2401 | 7153 | 6.11e-14 |
| GO:BP | GO:0007275 | multicellular organism development | 1153 | 3170 | 4.23e-13 |
| GO:BP | GO:0032502 | developmental process | 1477 | 4179 | 4.23e-13 |
| GO:BP | GO:0048869 | cellular developmental process | 1034 | 2804 | 4.23e-13 |
| GO:BP | GO:0050789 | regulation of biological process | 2502 | 7516 | 4.50e-13 |
| GO:BP | GO:0030154 | cell differentiation | 1033 | 2803 | 4.50e-13 |
| GO:BP | GO:0048518 | positive regulation of biological process | 1514 | 4301 | 4.52e-13 |
| GO:BP | GO:0065007 | biological regulation | 2565 | 7734 | 7.27e-13 |
| GO:BP | GO:0048731 | system development | 1002 | 2719 | 1.21e-12 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 1400 | 3954 | 1.34e-12 |
| GO:BP | GO:0048856 | anatomical structure development | 1372 | 3867 | 1.39e-12 |
| GO:BP | GO:0048519 | negative regulation of biological process | 1335 | 3764 | 5.10e-12 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 1265 | 3550 | 8.05e-12 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 703 | 1844 | 2.67e-11 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 727 | 1924 | 7.75e-11 |
| GO:BP | GO:0010646 | regulation of cell communication | 895 | 2444 | 3.36e-10 |
| GO:BP | GO:0048513 | animal organ development | 749 | 2004 | 4.17e-10 |
| GO:BP | GO:0048468 | cell development | 709 | 1886 | 5.16e-10 |
| GO:BP | GO:0050793 | regulation of developmental process | 644 | 1697 | 9.53e-10 |
| GO:BP | GO:0023051 | regulation of signaling | 888 | 2438 | 1.58e-09 |
| GO:BP | GO:0009966 | regulation of signal transduction | 798 | 2166 | 1.88e-09 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 978 | 2717 | 2.01e-09 |
| GO:BP | GO:0009888 | tissue development | 522 | 1348 | 4.03e-09 |
| GO:BP | GO:0035556 | intracellular signal transduction | 724 | 1951 | 4.67e-09 |
| GO:BP | GO:0072359 | circulatory system development | 352 | 864 | 1.31e-08 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 669 | 1801 | 2.75e-08 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 502 | 1314 | 1.05e-07 |
| GO:BP | GO:0048589 | developmental growth | 218 | 501 | 1.10e-07 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 364 | 913 | 1.28e-07 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 336 | 837 | 2.52e-07 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 406 | 1040 | 2.70e-07 |
| GO:BP | GO:0008283 | cell population proliferation | 495 | 1306 | 4.69e-07 |
| GO:BP | GO:0007399 | nervous system development | 651 | 1775 | 5.48e-07 |
| GO:BP | GO:0022008 | neurogenesis | 475 | 1251 | 7.62e-07 |
| GO:BP | GO:0014706 | striated muscle tissue development | 106 | 213 | 8.14e-07 |
| GO:BP | GO:0030900 | forebrain development | 128 | 272 | 1.71e-06 |
| GO:BP | GO:0044057 | regulation of system process | 170 | 384 | 1.80e-06 |
| GO:BP | GO:0048699 | generation of neurons | 411 | 1076 | 4.17e-06 |
| GO:BP | GO:0003008 | system process | 432 | 1139 | 4.63e-06 |
| GO:BP | GO:0007507 | heart development | 210 | 499 | 4.94e-06 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 98 | 201 | 1.03e-05 |
| GO:BP | GO:0060419 | heart growth | 44 | 71 | 1.03e-05 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 201 | 479 | 1.22e-05 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 408 | 1077 | 1.28e-05 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 240 | 591 | 1.70e-05 |
| GO:BP | GO:0071417 | cellular response to organonitrogen compound | 187 | 442 | 1.70e-05 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 478 | 1289 | 1.84e-05 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 405 | 1073 | 2.16e-05 |
| GO:BP | GO:0060322 | head development | 221 | 539 | 2.18e-05 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 268 | 673 | 2.18e-05 |
| GO:BP | GO:0040007 | growth | 276 | 697 | 2.49e-05 |
| GO:BP | GO:0035295 | tube development | 307 | 787 | 2.54e-05 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 911 | 2622 | 2.86e-05 |
| GO:BP | GO:0003013 | circulatory system process | 180 | 426 | 2.86e-05 |
| GO:BP | GO:0035239 | tube morphogenesis | 255 | 638 | 2.86e-05 |
| GO:BP | GO:0021537 | telencephalon development | 94 | 195 | 2.90e-05 |
| GO:BP | GO:0060537 | muscle tissue development | 148 | 339 | 3.33e-05 |
| GO:BP | GO:0007417 | central nervous system development | 274 | 694 | 3.35e-05 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 429 | 1150 | 3.49e-05 |
| GO:BP | GO:0007267 | cell-cell signaling | 406 | 1082 | 3.64e-05 |
| GO:BP | GO:0007010 | cytoskeleton organization | 427 | 1146 | 4.24e-05 |
| GO:BP | GO:0030030 | cell projection organization | 439 | 1182 | 4.25e-05 |
| GO:BP | GO:0008015 | blood circulation | 154 | 357 | 4.29e-05 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 207 | 505 | 4.46e-05 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 361 | 951 | 4.46e-05 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 248 | 622 | 4.56e-05 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 626 | 1751 | 5.38e-05 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 662 | 1862 | 5.59e-05 |
| GO:BP | GO:0035265 | organ growth | 66 | 127 | 5.59e-05 |
| GO:BP | GO:0006950 | response to stress | 881 | 2540 | 5.69e-05 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 210 | 516 | 6.22e-05 |
| GO:BP | GO:0048638 | regulation of developmental growth | 111 | 243 | 6.22e-05 |
| GO:BP | GO:0030182 | neuron differentiation | 381 | 1014 | 6.36e-05 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 276 | 706 | 6.58e-05 |
| GO:BP | GO:0034330 | cell junction organization | 220 | 545 | 6.58e-05 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 39 | 64 | 6.61e-05 |
| GO:BP | GO:0060420 | regulation of heart growth | 32 | 49 | 7.47e-05 |
| GO:BP | GO:0042221 | response to chemical | 802 | 2304 | 1.12e-04 |
| GO:BP | GO:0033002 | muscle cell proliferation | 75 | 152 | 1.18e-04 |
| GO:BP | GO:0016477 | cell migration | 378 | 1011 | 1.18e-04 |
| GO:BP | GO:0007420 | brain development | 202 | 498 | 1.22e-04 |
| GO:BP | GO:0001568 | blood vessel development | 211 | 525 | 1.51e-04 |
| GO:BP | GO:0014855 | striated muscle cell proliferation | 36 | 59 | 1.60e-04 |
| GO:BP | GO:0032879 | regulation of localization | 519 | 1439 | 1.65e-04 |
| GO:BP | GO:0001944 | vasculature development | 220 | 552 | 1.77e-04 |
| GO:BP | GO:0031325 | positive regulation of cellular metabolic process | 809 | 2333 | 1.79e-04 |
| GO:BP | GO:0045165 | cell fate commitment | 74 | 151 | 1.79e-04 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 190 | 467 | 1.87e-04 |
| GO:BP | GO:0090257 | regulation of muscle system process | 84 | 177 | 1.91e-04 |
| GO:BP | GO:1901653 | cellular response to peptide | 120 | 273 | 2.05e-04 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 384 | 1035 | 2.06e-04 |
| GO:BP | GO:0071310 | cellular response to organic substance | 471 | 1297 | 2.13e-04 |
| GO:BP | GO:0006915 | apoptotic process | 497 | 1377 | 2.34e-04 |
| GO:BP | GO:0008016 | regulation of heart contraction | 79 | 165 | 2.34e-04 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 303 | 796 | 2.34e-04 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 276 | 717 | 2.34e-04 |
| GO:BP | GO:0016310 | phosphorylation | 420 | 1145 | 2.43e-04 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 115 | 261 | 2.68e-04 |
| GO:BP | GO:0060429 | epithelium development | 301 | 792 | 2.80e-04 |
| GO:BP | GO:0031175 | neuron projection development | 284 | 743 | 3.14e-04 |
| GO:BP | GO:0003012 | muscle system process | 138 | 325 | 3.23e-04 |
| GO:BP | GO:0065008 | regulation of biological quality | 664 | 1893 | 3.23e-04 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 187 | 463 | 3.38e-04 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 187 | 463 | 3.38e-04 |
| GO:BP | GO:0099536 | synaptic signaling | 194 | 483 | 3.38e-04 |
| GO:BP | GO:0046620 | regulation of organ growth | 40 | 70 | 3.51e-04 |
| GO:BP | GO:0009790 | embryo development | 310 | 821 | 3.51e-04 |
| GO:BP | GO:0030029 | actin filament-based process | 248 | 640 | 3.94e-04 |
| GO:BP | GO:0048729 | tissue morphogenesis | 183 | 453 | 4.05e-04 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 435 | 1197 | 4.30e-04 |
| GO:BP | GO:0048666 | neuron development | 315 | 838 | 4.40e-04 |
| GO:BP | GO:0012501 | programmed cell death | 508 | 1419 | 4.41e-04 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 148 | 355 | 4.43e-04 |
| GO:BP | GO:0008219 | cell death | 509 | 1423 | 4.73e-04 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 391 | 1067 | 5.37e-04 |
| GO:BP | GO:0055021 | regulation of cardiac muscle tissue growth | 28 | 44 | 5.54e-04 |
| GO:BP | GO:0040011 | locomotion | 303 | 805 | 5.65e-04 |
| GO:BP | GO:1901652 | response to peptide | 152 | 368 | 5.89e-04 |
| GO:BP | GO:0040012 | regulation of locomotion | 274 | 720 | 5.96e-04 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 103 | 233 | 6.10e-04 |
| GO:BP | GO:0048870 | cell motility | 408 | 1120 | 6.15e-04 |
| GO:BP | GO:2000145 | regulation of cell motility | 267 | 700 | 6.26e-04 |
| GO:BP | GO:0030334 | regulation of cell migration | 256 | 668 | 6.43e-04 |
| GO:BP | GO:0001503 | ossification | 128 | 302 | 6.69e-04 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 235 | 609 | 8.60e-04 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 419 | 1157 | 8.60e-04 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 170 | 422 | 9.25e-04 |
| GO:BP | GO:1901698 | response to nitrogen compound | 278 | 736 | 9.32e-04 |
| GO:BP | GO:0045444 | fat cell differentiation | 82 | 179 | 9.74e-04 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 826 | 2415 | 9.89e-04 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 357 | 972 | 9.90e-04 |
| GO:BP | GO:0033554 | cellular response to stress | 509 | 1433 | 9.96e-04 |
| GO:BP | GO:0006468 | protein phosphorylation | 352 | 958 | 1.08e-03 |
| GO:BP | GO:0016043 | cellular component organization | 1575 | 4802 | 1.13e-03 |
| GO:BP | GO:0003015 | heart process | 91 | 204 | 1.19e-03 |
| GO:BP | GO:0035108 | limb morphogenesis | 55 | 110 | 1.19e-03 |
| GO:BP | GO:0035107 | appendage morphogenesis | 55 | 110 | 1.19e-03 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 158 | 390 | 1.21e-03 |
| GO:BP | GO:0030097 | hemopoiesis | 239 | 624 | 1.21e-03 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 654 | 1883 | 1.23e-03 |
| GO:BP | GO:0060047 | heart contraction | 87 | 194 | 1.36e-03 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 437 | 1218 | 1.40e-03 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 150 | 369 | 1.54e-03 |
| GO:BP | GO:0014745 | negative regulation of muscle adaptation | 9 | 9 | 1.74e-03 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 659 | 1904 | 1.77e-03 |
| GO:BP | GO:0023056 | positive regulation of signaling | 437 | 1221 | 1.77e-03 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 34 | 60 | 1.77e-03 |
| GO:BP | GO:0042592 | homeostatic process | 409 | 1137 | 1.94e-03 |
| GO:BP | GO:0002521 | leukocyte differentiation | 150 | 371 | 2.07e-03 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 427 | 1193 | 2.16e-03 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 282 | 757 | 2.30e-03 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 178 | 452 | 2.31e-03 |
| GO:BP | GO:1903522 | regulation of blood circulation | 86 | 194 | 2.37e-03 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 642 | 1856 | 2.39e-03 |
| GO:BP | GO:0010033 | response to organic substance | 603 | 1736 | 2.54e-03 |
| GO:BP | GO:0061614 | miRNA transcription | 34 | 61 | 2.63e-03 |
| GO:BP | GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | 179 | 456 | 2.63e-03 |
| GO:BP | GO:0086065 | cell communication involved in cardiac conduction | 31 | 54 | 2.63e-03 |
| GO:BP | GO:0010243 | response to organonitrogen compound | 257 | 685 | 2.81e-03 |
| GO:BP | GO:0023057 | negative regulation of signaling | 374 | 1036 | 2.90e-03 |
| GO:BP | GO:0048568 | embryonic organ development | 125 | 303 | 3.01e-03 |
| GO:BP | GO:0086004 | regulation of cardiac muscle cell contraction | 21 | 32 | 3.04e-03 |
| GO:BP | GO:0001764 | neuron migration | 61 | 129 | 3.10e-03 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 373 | 1034 | 3.10e-03 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 183 | 469 | 3.10e-03 |
| GO:BP | GO:0035148 | tube formation | 59 | 124 | 3.24e-03 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 350 | 965 | 3.24e-03 |
| GO:BP | GO:0051173 | positive regulation of nitrogen compound metabolic process | 746 | 2186 | 3.28e-03 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 57 | 119 | 3.37e-03 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 392 | 1093 | 3.41e-03 |
| GO:BP | GO:0007423 | sensory organ development | 154 | 387 | 3.74e-03 |
| GO:BP | GO:0016331 | morphogenesis of embryonic epithelium | 58 | 122 | 3.74e-03 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 352 | 973 | 3.76e-03 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 49 | 99 | 3.78e-03 |
| GO:BP | GO:0007155 | cell adhesion | 341 | 941 | 4.18e-03 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 38 | 72 | 4.28e-03 |
| GO:BP | GO:0007049 | cell cycle | 503 | 1436 | 4.37e-03 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 525 | 1504 | 4.41e-03 |
| GO:BP | GO:0034329 | cell junction assembly | 131 | 323 | 4.60e-03 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 567 | 1635 | 4.77e-03 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 98 | 231 | 4.94e-03 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 663 | 1934 | 4.94e-03 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 175 | 450 | 4.98e-03 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 716 | 2100 | 5.01e-03 |
| GO:BP | GO:0048167 | regulation of synaptic plasticity | 64 | 139 | 5.01e-03 |
| GO:BP | GO:0061337 | cardiac conduction | 43 | 85 | 5.16e-03 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 668 | 1951 | 5.29e-03 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 34 | 63 | 5.30e-03 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 390 | 1093 | 5.30e-03 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 551 | 1588 | 5.45e-03 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 111 | 268 | 5.49e-03 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 244 | 654 | 5.56e-03 |
| GO:BP | GO:0060043 | regulation of cardiac muscle cell proliferation | 20 | 31 | 5.58e-03 |
| GO:BP | GO:0045598 | regulation of fat cell differentiation | 46 | 93 | 5.77e-03 |
| GO:BP | GO:0030282 | bone mineralization | 40 | 78 | 5.84e-03 |
| GO:BP | GO:0031328 | positive regulation of cellular biosynthetic process | 632 | 1841 | 5.87e-03 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 565 | 1633 | 5.92e-03 |
| GO:BP | GO:0043434 | response to peptide hormone | 127 | 314 | 6.11e-03 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 56 | 119 | 6.11e-03 |
| GO:BP | GO:0043010 | camera-type eye development | 97 | 230 | 6.38e-03 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 210 | 555 | 6.39e-03 |
| GO:BP | GO:0032989 | cellular anatomical entity morphogenesis | 221 | 588 | 6.72e-03 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 260 | 704 | 6.72e-03 |
| GO:BP | GO:0000902 | cell morphogenesis | 263 | 713 | 6.72e-03 |
| GO:BP | GO:0014856 | skeletal muscle cell proliferation | 13 | 17 | 6.72e-03 |
| GO:BP | GO:2000629 | negative regulation of miRNA metabolic process | 17 | 25 | 6.72e-03 |
| GO:BP | GO:0014741 | negative regulation of muscle hypertrophy | 17 | 25 | 6.72e-03 |
| GO:BP | GO:0061049 | cell growth involved in cardiac muscle cell development | 14 | 19 | 6.93e-03 |
| GO:BP | GO:0072175 | epithelial tube formation | 55 | 117 | 6.93e-03 |
| GO:BP | GO:0003338 | metanephros morphogenesis | 14 | 19 | 6.93e-03 |
| GO:BP | GO:0003301 | physiological cardiac muscle hypertrophy | 14 | 19 | 6.93e-03 |
| GO:BP | GO:1902894 | negative regulation of miRNA transcription | 16 | 23 | 6.93e-03 |
| GO:BP | GO:0003298 | physiological muscle hypertrophy | 14 | 19 | 6.93e-03 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 561 | 1624 | 6.97e-03 |
| GO:BP | GO:0042100 | B cell proliferation | 26 | 45 | 7.00e-03 |
| GO:BP | GO:0061180 | mammary gland epithelium development | 29 | 52 | 7.00e-03 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 34 | 64 | 7.00e-03 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 368 | 1031 | 7.12e-03 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 286 | 783 | 7.12e-03 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 38 | 74 | 7.39e-03 |
| GO:BP | GO:0060173 | limb development | 62 | 136 | 7.46e-03 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 213 | 566 | 7.46e-03 |
| GO:BP | GO:0032147 | activation of protein kinase activity | 31 | 57 | 7.46e-03 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 1384 | 4233 | 7.46e-03 |
| GO:BP | GO:0048736 | appendage development | 62 | 136 | 7.46e-03 |
| GO:BP | GO:0002682 | regulation of immune system process | 314 | 868 | 7.46e-03 |
| GO:BP | GO:0010586 | miRNA metabolic process | 43 | 87 | 8.33e-03 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 43 | 87 | 8.33e-03 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 43 | 87 | 8.33e-03 |
| GO:BP | GO:0050808 | synapse organization | 134 | 337 | 8.58e-03 |
| GO:BP | GO:0007186 | G protein-coupled receptor signaling pathway | 151 | 386 | 8.62e-03 |
| GO:BP | GO:0048771 | tissue remodeling | 53 | 113 | 8.82e-03 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 129 | 323 | 8.87e-03 |
| GO:BP | GO:0007389 | pattern specification process | 116 | 286 | 8.87e-03 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 613 | 1791 | 9.14e-03 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 202 | 536 | 9.43e-03 |
| GO:BP | GO:0061061 | muscle structure development | 202 | 536 | 9.43e-03 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 150 | 384 | 9.46e-03 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 141 | 358 | 9.46e-03 |
| GO:BP | GO:0002376 | immune system process | 490 | 1410 | 9.47e-03 |
| GO:BP | GO:0009605 | response to external stimulus | 538 | 1559 | 9.67e-03 |
| GO:BP | GO:0014896 | muscle hypertrophy | 38 | 75 | 9.67e-03 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 166 | 431 | 9.99e-03 |
| GO:BP | GO:0031347 | regulation of defense response | 172 | 449 | 1.06e-02 |
| GO:BP | GO:0060421 | positive regulation of heart growth | 18 | 28 | 1.06e-02 |
| GO:BP | GO:0046661 | male sex differentiation | 50 | 106 | 1.06e-02 |
| GO:BP | GO:0198738 | cell-cell signaling by wnt | 141 | 359 | 1.06e-02 |
| GO:BP | GO:0060008 | Sertoli cell differentiation | 10 | 12 | 1.08e-02 |
| GO:BP | GO:0072087 | renal vesicle development | 10 | 12 | 1.08e-02 |
| GO:BP | GO:0042391 | regulation of membrane potential | 114 | 282 | 1.10e-02 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 128 | 322 | 1.10e-02 |
| GO:BP | GO:0043502 | regulation of muscle adaptation | 35 | 68 | 1.10e-02 |
| GO:BP | GO:1903115 | regulation of actin filament-based movement | 22 | 37 | 1.10e-02 |
| GO:BP | GO:0001775 | cell activation | 232 | 627 | 1.10e-02 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 37 | 73 | 1.10e-02 |
| GO:BP | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction | 17 | 26 | 1.12e-02 |
| GO:BP | GO:0032943 | mononuclear cell proliferation | 69 | 157 | 1.12e-02 |
| GO:BP | GO:0007015 | actin filament organization | 140 | 357 | 1.15e-02 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 164 | 427 | 1.19e-02 |
| GO:BP | GO:0071875 | adrenergic receptor signaling pathway | 16 | 24 | 1.20e-02 |
| GO:BP | GO:0010614 | negative regulation of cardiac muscle hypertrophy | 16 | 24 | 1.20e-02 |
| GO:BP | GO:0022402 | cell cycle process | 363 | 1024 | 1.21e-02 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 120 | 300 | 1.21e-02 |
| GO:BP | GO:0042692 | muscle cell differentiation | 128 | 323 | 1.22e-02 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 102 | 249 | 1.22e-02 |
| GO:BP | GO:0006271 | DNA strand elongation involved in DNA replication | 11 | 14 | 1.22e-02 |
| GO:BP | GO:0061117 | negative regulation of heart growth | 12 | 16 | 1.29e-02 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 431 | 1234 | 1.29e-02 |
| GO:BP | GO:0055022 | negative regulation of cardiac muscle tissue growth | 12 | 16 | 1.29e-02 |
| GO:BP | GO:0086014 | atrial cardiac muscle cell action potential | 12 | 16 | 1.29e-02 |
| GO:BP | GO:0021879 | forebrain neuron differentiation | 14 | 20 | 1.29e-02 |
| GO:BP | GO:0002683 | negative regulation of immune system process | 118 | 295 | 1.29e-02 |
| GO:BP | GO:0086066 | atrial cardiac muscle cell to AV node cell communication | 12 | 16 | 1.29e-02 |
| GO:BP | GO:0019222 | regulation of metabolic process | 1494 | 4603 | 1.29e-02 |
| GO:BP | GO:0007264 | small GTPase-mediated signal transduction | 151 | 390 | 1.29e-02 |
| GO:BP | GO:0086026 | atrial cardiac muscle cell to AV node cell signaling | 12 | 16 | 1.29e-02 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 430 | 1231 | 1.29e-02 |
| GO:BP | GO:0045017 | glycerolipid biosynthetic process | 85 | 202 | 1.29e-02 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 112 | 278 | 1.30e-02 |
| GO:BP | GO:0001501 | skeletal system development | 136 | 347 | 1.33e-02 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 374 | 1060 | 1.37e-02 |
| GO:BP | GO:0030278 | regulation of ossification | 39 | 79 | 1.38e-02 |
| GO:BP | GO:0045600 | positive regulation of fat cell differentiation | 26 | 47 | 1.40e-02 |
| GO:BP | GO:0001823 | mesonephros development | 37 | 74 | 1.40e-02 |
| GO:BP | GO:0006974 | DNA damage response | 270 | 745 | 1.49e-02 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 42 | 87 | 1.51e-02 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 469 | 1354 | 1.52e-02 |
| GO:BP | GO:0040008 | regulation of growth | 175 | 462 | 1.52e-02 |
| GO:BP | GO:0045321 | leukocyte activation | 198 | 530 | 1.53e-02 |
| GO:BP | GO:0021954 | central nervous system neuron development | 30 | 57 | 1.58e-02 |
| GO:BP | GO:0003205 | cardiac chamber development | 66 | 151 | 1.60e-02 |
| GO:BP | GO:0098900 | regulation of action potential | 22 | 38 | 1.61e-02 |
| GO:BP | GO:0046474 | glycerophospholipid biosynthetic process | 74 | 173 | 1.61e-02 |
| GO:BP | GO:0043009 | chordate embryonic development | 190 | 507 | 1.61e-02 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 74 | 173 | 1.61e-02 |
| GO:BP | GO:0001657 | ureteric bud development | 36 | 72 | 1.61e-02 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 36 | 72 | 1.61e-02 |
| GO:BP | GO:0030851 | granulocyte differentiation | 18 | 29 | 1.64e-02 |
| GO:BP | GO:0097106 | postsynaptic density organization | 18 | 29 | 1.64e-02 |
| GO:BP | GO:0086019 | cell-cell signaling involved in cardiac conduction | 18 | 29 | 1.64e-02 |
| GO:BP | GO:0021543 | pallium development | 60 | 135 | 1.64e-02 |
| GO:BP | GO:0001654 | eye development | 106 | 263 | 1.68e-02 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 41 | 85 | 1.70e-02 |
| GO:BP | GO:0030879 | mammary gland development | 46 | 98 | 1.71e-02 |
| GO:BP | GO:0048880 | sensory system development | 107 | 266 | 1.71e-02 |
| GO:BP | GO:0003007 | heart morphogenesis | 90 | 218 | 1.72e-02 |
| GO:BP | GO:0007548 | sex differentiation | 76 | 179 | 1.72e-02 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 49 | 106 | 1.74e-02 |
| GO:BP | GO:0140747 | regulation of ncRNA transcription | 39 | 80 | 1.74e-02 |
| GO:BP | GO:0031214 | biomineral tissue development | 49 | 106 | 1.74e-02 |
| GO:BP | GO:0001659 | temperature homeostasis | 58 | 130 | 1.74e-02 |
| GO:BP | GO:0021987 | cerebral cortex development | 44 | 93 | 1.76e-02 |
| GO:BP | GO:0021872 | forebrain generation of neurons | 17 | 27 | 1.76e-02 |
| GO:BP | GO:0060038 | cardiac muscle cell proliferation | 24 | 43 | 1.76e-02 |
| GO:BP | GO:0060443 | mammary gland morphogenesis | 21 | 36 | 1.76e-02 |
| GO:BP | GO:0042246 | tissue regeneration | 29 | 55 | 1.76e-02 |
| GO:BP | GO:0000278 | mitotic cell cycle | 275 | 763 | 1.76e-02 |
| GO:BP | GO:0060416 | response to growth hormone | 17 | 27 | 1.76e-02 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 118 | 298 | 1.78e-02 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 149 | 388 | 1.82e-02 |
| GO:BP | GO:0001649 | osteoblast differentiation | 74 | 174 | 1.83e-02 |
| GO:BP | GO:0150063 | visual system development | 106 | 264 | 1.85e-02 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 194 | 521 | 1.89e-02 |
| GO:BP | GO:0030500 | regulation of bone mineralization | 26 | 48 | 1.90e-02 |
| GO:BP | GO:0051960 | regulation of nervous system development | 132 | 339 | 1.90e-02 |
| GO:BP | GO:1904407 | positive regulation of nitric oxide metabolic process | 16 | 25 | 1.92e-02 |
| GO:BP | GO:0060603 | mammary gland duct morphogenesis | 16 | 25 | 1.92e-02 |
| GO:BP | GO:0033043 | regulation of organelle organization | 337 | 953 | 1.95e-02 |
| GO:BP | GO:0055006 | cardiac cell development | 38 | 78 | 1.95e-02 |
| GO:BP | GO:0003161 | cardiac conduction system development | 20 | 34 | 1.97e-02 |
| GO:BP | GO:0072163 | mesonephric epithelium development | 36 | 73 | 2.00e-02 |
| GO:BP | GO:0072164 | mesonephric tubule development | 36 | 73 | 2.00e-02 |
| GO:BP | GO:0046621 | negative regulation of organ growth | 15 | 23 | 2.10e-02 |
| GO:BP | GO:0048592 | eye morphogenesis | 49 | 107 | 2.10e-02 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 236 | 648 | 2.13e-02 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 231 | 633 | 2.13e-02 |
| GO:BP | GO:0070661 | leukocyte proliferation | 74 | 175 | 2.13e-02 |
| GO:BP | GO:0051179 | localization | 1218 | 3731 | 2.17e-02 |
| GO:BP | GO:0072077 | renal vesicle morphogenesis | 9 | 11 | 2.19e-02 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 63 | 145 | 2.22e-02 |
| GO:BP | GO:1990874 | vascular associated smooth muscle cell proliferation | 27 | 51 | 2.29e-02 |
| GO:BP | GO:0019935 | cyclic-nucleotide-mediated signaling | 27 | 51 | 2.29e-02 |
| GO:BP | GO:0006955 | immune response | 308 | 867 | 2.29e-02 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 310 | 874 | 2.45e-02 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 287 | 804 | 2.45e-02 |
| GO:BP | GO:0014841 | skeletal muscle satellite cell proliferation | 10 | 13 | 2.45e-02 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 195 | 527 | 2.46e-02 |
| GO:BP | GO:0034103 | regulation of tissue remodeling | 24 | 44 | 2.50e-02 |
| GO:BP | GO:0086002 | cardiac muscle cell action potential involved in contraction | 24 | 44 | 2.50e-02 |
| GO:BP | GO:0060575 | intestinal epithelial cell differentiation | 12 | 17 | 2.50e-02 |
| GO:BP | GO:0099545 | trans-synaptic signaling by trans-synaptic complex | 6 | 6 | 2.53e-02 |
| GO:BP | GO:0030010 | establishment of cell polarity | 55 | 124 | 2.53e-02 |
| GO:BP | GO:0010616 | negative regulation of cardiac muscle adaptation | 6 | 6 | 2.53e-02 |
| GO:BP | GO:1903243 | negative regulation of cardiac muscle hypertrophy in response to stress | 6 | 6 | 2.53e-02 |
| GO:BP | GO:0003166 | bundle of His development | 6 | 6 | 2.53e-02 |
| GO:BP | GO:0009987 | cellular process | 3327 | 10686 | 2.58e-02 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 287 | 805 | 2.60e-02 |
| GO:BP | GO:0048593 | camera-type eye morphogenesis | 41 | 87 | 2.61e-02 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 63 | 146 | 2.61e-02 |
| GO:BP | GO:1904705 | regulation of vascular associated smooth muscle cell proliferation | 26 | 49 | 2.61e-02 |
| GO:BP | GO:0060284 | regulation of cell development | 209 | 570 | 2.68e-02 |
| GO:BP | GO:0032944 | regulation of mononuclear cell proliferation | 53 | 119 | 2.70e-02 |
| GO:BP | GO:0055001 | muscle cell development | 69 | 163 | 2.85e-02 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 192 | 520 | 2.89e-02 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 164 | 437 | 2.93e-02 |
| GO:BP | GO:0045667 | regulation of osteoblast differentiation | 45 | 98 | 2.93e-02 |
| GO:BP | GO:0046622 | positive regulation of organ growth | 20 | 35 | 2.96e-02 |
| GO:BP | GO:0031099 | regeneration | 62 | 144 | 2.98e-02 |
| GO:BP | GO:1902117 | positive regulation of organelle assembly | 35 | 72 | 2.98e-02 |
| GO:BP | GO:0014743 | regulation of muscle hypertrophy | 25 | 47 | 3.02e-02 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 246 | 683 | 3.07e-02 |
| GO:BP | GO:0070167 | regulation of biomineral tissue development | 31 | 62 | 3.13e-02 |
| GO:BP | GO:0110110 | positive regulation of animal organ morphogenesis | 16 | 26 | 3.13e-02 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 49 | 109 | 3.13e-02 |
| GO:BP | GO:0002286 | T cell activation involved in immune response | 29 | 57 | 3.13e-02 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 220 | 605 | 3.14e-02 |
| GO:BP | GO:0008654 | phospholipid biosynthetic process | 88 | 217 | 3.14e-02 |
| GO:BP | GO:0030031 | cell projection assembly | 167 | 447 | 3.23e-02 |
| GO:BP | GO:0070372 | regulation of ERK1 and ERK2 cascade | 73 | 175 | 3.24e-02 |
| GO:BP | GO:0046632 | alpha-beta T cell differentiation | 34 | 70 | 3.45e-02 |
| GO:BP | GO:0007017 | microtubule-based process | 257 | 718 | 3.48e-02 |
| GO:BP | GO:0002027 | regulation of heart rate | 39 | 83 | 3.48e-02 |
| GO:BP | GO:0045429 | positive regulation of nitric oxide biosynthetic process | 15 | 24 | 3.50e-02 |
| GO:BP | GO:0086010 | membrane depolarization during action potential | 15 | 24 | 3.50e-02 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 32 | 65 | 3.52e-02 |
| GO:BP | GO:0060291 | long-term synaptic potentiation | 32 | 65 | 3.52e-02 |
| GO:BP | GO:0006936 | muscle contraction | 101 | 255 | 3.54e-02 |
| GO:BP | GO:0030048 | actin filament-based movement | 48 | 107 | 3.57e-02 |
| GO:BP | GO:0046486 | glycerolipid metabolic process | 111 | 284 | 3.59e-02 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 266 | 746 | 3.59e-02 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 55 | 126 | 3.59e-02 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 1603 | 4988 | 3.59e-02 |
| GO:BP | GO:0060491 | regulation of cell projection assembly | 67 | 159 | 3.59e-02 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 63 | 148 | 3.62e-02 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 300 | 850 | 3.68e-02 |
| GO:BP | GO:0072001 | renal system development | 97 | 244 | 3.71e-02 |
| GO:BP | GO:2001222 | regulation of neuron migration | 21 | 38 | 3.71e-02 |
| GO:BP | GO:0120034 | positive regulation of plasma membrane bounded cell projection assembly | 40 | 86 | 3.72e-02 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 49 | 110 | 3.74e-02 |
| GO:BP | GO:0046651 | lymphocyte proliferation | 64 | 151 | 3.74e-02 |
| GO:BP | GO:0010644 | cell communication by electrical coupling | 18 | 31 | 3.76e-02 |
| GO:BP | GO:0050890 | cognition | 86 | 213 | 3.89e-02 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 57 | 132 | 3.93e-02 |
| GO:BP | GO:0003093 | regulation of glomerular filtration | 7 | 8 | 3.98e-02 |
| GO:BP | GO:0014842 | regulation of skeletal muscle satellite cell proliferation | 7 | 8 | 3.98e-02 |
| GO:BP | GO:0001977 | renal system process involved in regulation of blood volume | 7 | 8 | 3.98e-02 |
| GO:BP | GO:0120032 | regulation of plasma membrane bounded cell projection assembly | 66 | 157 | 4.02e-02 |
| GO:BP | GO:0031668 | cellular response to extracellular stimulus | 88 | 219 | 4.04e-02 |
| GO:BP | GO:0065009 | regulation of molecular function | 445 | 1299 | 4.04e-02 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 514 | 1514 | 4.04e-02 |
| GO:BP | GO:0043367 | CD4-positive, alpha-beta T cell differentiation | 27 | 53 | 4.10e-02 |
| GO:BP | GO:0061448 | connective tissue development | 83 | 205 | 4.11e-02 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 231 | 642 | 4.11e-02 |
| GO:BP | GO:0046649 | lymphocyte activation | 165 | 444 | 4.11e-02 |
| GO:BP | GO:0051049 | regulation of transport | 380 | 1098 | 4.11e-02 |
| GO:BP | GO:0003094 | glomerular filtration | 13 | 20 | 4.12e-02 |
| GO:BP | GO:0050776 | regulation of immune response | 176 | 477 | 4.19e-02 |
| GO:BP | GO:0048863 | stem cell differentiation | 78 | 191 | 4.19e-02 |
| GO:BP | GO:0000165 | MAPK cascade | 185 | 504 | 4.22e-02 |
| GO:BP | GO:0098609 | cell-cell adhesion | 205 | 564 | 4.22e-02 |
| GO:BP | GO:0002573 | myeloid leukocyte differentiation | 64 | 152 | 4.33e-02 |
| GO:BP | GO:0045216 | cell-cell junction organization | 64 | 152 | 4.33e-02 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 93 | 234 | 4.36e-02 |
| GO:BP | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway | 12 | 18 | 4.46e-02 |
| GO:BP | GO:0031646 | positive regulation of nervous system process | 12 | 18 | 4.46e-02 |
| GO:BP | GO:0010171 | body morphogenesis | 22 | 41 | 4.48e-02 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 143 | 380 | 4.54e-02 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 43 | 95 | 4.57e-02 |
| GO:BP | GO:0010611 | regulation of cardiac muscle hypertrophy | 24 | 46 | 4.65e-02 |
| GO:BP | GO:0014857 | regulation of skeletal muscle cell proliferation | 8 | 10 | 4.68e-02 |
| GO:BP | GO:0031644 | regulation of nervous system process | 35 | 74 | 4.68e-02 |
| GO:BP | GO:1904646 | cellular response to amyloid-beta | 16 | 27 | 4.74e-02 |
| GO:BP | GO:0099084 | postsynaptic specialization organization | 19 | 34 | 4.74e-02 |
| GO:BP | GO:0098901 | regulation of cardiac muscle cell action potential | 16 | 27 | 4.74e-02 |
| GO:BP | GO:0072273 | metanephric nephron morphogenesis | 11 | 16 | 4.74e-02 |
| GO:BP | GO:0007043 | cell-cell junction assembly | 47 | 106 | 4.74e-02 |
| GO:BP | GO:1901021 | positive regulation of calcium ion transmembrane transporter activity | 16 | 27 | 4.74e-02 |
| GO:BP | GO:0031279 | regulation of cyclase activity | 16 | 27 | 4.74e-02 |
| GO:BP | GO:0002262 | myeloid cell homeostasis | 58 | 136 | 4.74e-02 |
| GO:BP | GO:0001817 | regulation of cytokine production | 162 | 437 | 4.76e-02 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 38 | 82 | 4.76e-02 |
| GO:BP | GO:0051283 | negative regulation of sequestering of calcium ion | 41 | 90 | 4.81e-02 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 111 | 287 | 4.83e-02 |
| GO:BP | GO:0010889 | regulation of sequestering of triglyceride | 9 | 12 | 4.85e-02 |
| GO:BP | GO:0098792 | xenophagy | 9 | 12 | 4.85e-02 |
| GO:BP | GO:0016202 | regulation of striated muscle tissue development | 10 | 14 | 4.85e-02 |
| GO:BP | GO:0035721 | intraciliary retrograde transport | 10 | 14 | 4.85e-02 |
LR_int_restable %>%
filter(source=="KEGG") %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "KEGG terms found in LR neargenes intersected with RNA response genes") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| KEGG | KEGG:05167 | Kaposi sarcoma-associated herpesvirus infection | 54 | 134 | 1.79e-01 |
| KEGG | KEGG:00564 | Glycerophospholipid metabolism | 33 | 71 | 1.79e-01 |
| KEGG | KEGG:04911 | Insulin secretion | 29 | 62 | 1.79e-01 |
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 50 | 124 | 1.79e-01 |
| KEGG | KEGG:04927 | Cortisol synthesis and secretion | 24 | 46 | 1.79e-01 |
| KEGG | KEGG:01521 | EGFR tyrosine kinase inhibitor resistance | 32 | 72 | 1.79e-01 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 42 | 101 | 1.79e-01 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 34 | 78 | 1.79e-01 |
| KEGG | KEGG:04923 | Regulation of lipolysis in adipocytes | 20 | 41 | 1.79e-01 |
| KEGG | KEGG:03030 | DNA replication | 17 | 34 | 1.79e-01 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 40 | 96 | 1.79e-01 |
| KEGG | KEGG:04360 | Axon guidance | 61 | 155 | 1.79e-01 |
| KEGG | KEGG:00230 | Purine metabolism | 41 | 93 | 1.79e-01 |
| KEGG | KEGG:04976 | Bile secretion | 21 | 41 | 1.79e-01 |
| KEGG | KEGG:04750 | Inflammatory mediator regulation of TRP channels | 34 | 72 | 1.79e-01 |
| KEGG | KEGG:05221 | Acute myeloid leukemia | 24 | 52 | 1.79e-01 |
| KEGG | KEGG:04148 | Efferocytosis | 50 | 120 | 1.79e-01 |
| KEGG | KEGG:04070 | Phosphatidylinositol signaling system | 39 | 83 | 1.79e-01 |
| KEGG | KEGG:05164 | Influenza A | 41 | 95 | 1.79e-01 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 46 | 106 | 1.79e-01 |
| KEGG | KEGG:05032 | Morphine addiction | 25 | 54 | 1.79e-01 |
| KEGG | KEGG:04020 | Calcium signaling pathway | 63 | 161 | 1.79e-01 |
| KEGG | KEGG:04925 | Aldosterone synthesis and secretion | 33 | 75 | 1.79e-01 |
| KEGG | KEGG:05217 | Basal cell carcinoma | 23 | 48 | 1.79e-01 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 59 | 147 | 1.79e-01 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 64 | 162 | 1.79e-01 |
| KEGG | KEGG:05161 | Hepatitis B | 52 | 126 | 1.79e-01 |
| KEGG | KEGG:04611 | Platelet activation | 37 | 88 | 1.84e-01 |
| KEGG | KEGG:04924 | Renin secretion | 23 | 50 | 1.87e-01 |
| KEGG | KEGG:04668 | TNF signaling pathway | 35 | 83 | 1.93e-01 |
| KEGG | KEGG:00770 | Pantothenate and CoA biosynthesis | 9 | 15 | 1.96e-01 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 51 | 129 | 1.97e-01 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 51 | 129 | 1.97e-01 |
| KEGG | KEGG:04014 | Ras signaling pathway | 63 | 164 | 1.97e-01 |
| KEGG | KEGG:04970 | Salivary secretion | 24 | 54 | 2.18e-01 |
| KEGG | KEGG:04931 | Insulin resistance | 37 | 91 | 2.24e-01 |
| KEGG | KEGG:00000 | KEGG root term | 1718 | 5451 | 2.24e-01 |
| KEGG | KEGG:04727 | GABAergic synapse | 23 | 52 | 2.24e-01 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 26 | 60 | 2.24e-01 |
| KEGG | KEGG:04110 | Cell cycle | 56 | 146 | 2.24e-01 |
| KEGG | KEGG:03410 | Base excision repair | 20 | 44 | 2.24e-01 |
| KEGG | KEGG:00562 | Inositol phosphate metabolism | 26 | 60 | 2.24e-01 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 62 | 164 | 2.25e-01 |
| KEGG | KEGG:04814 | Motor proteins | 54 | 141 | 2.25e-01 |
| KEGG | KEGG:05142 | Chagas disease | 30 | 72 | 2.25e-01 |
| KEGG | KEGG:04730 | Long-term depression | 18 | 39 | 2.25e-01 |
| KEGG | KEGG:04913 | Ovarian steroidogenesis | 13 | 26 | 2.25e-01 |
| KEGG | KEGG:05152 | Tuberculosis | 40 | 101 | 2.41e-01 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 84 | 231 | 2.41e-01 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 60 | 160 | 2.46e-01 |
| KEGG | KEGG:04934 | Cushing syndrome | 46 | 119 | 2.46e-01 |
| KEGG | KEGG:00830 | Retinol metabolism | 8 | 14 | 2.46e-01 |
| KEGG | KEGG:04971 | Gastric acid secretion | 22 | 51 | 2.52e-01 |
| KEGG | KEGG:04725 | Cholinergic synapse | 33 | 82 | 2.52e-01 |
| KEGG | KEGG:05417 | Lipid and atherosclerosis | 54 | 143 | 2.52e-01 |
| KEGG | KEGG:03250 | Viral life cycle - HIV-1 | 23 | 54 | 2.52e-01 |
| KEGG | KEGG:04713 | Circadian entrainment | 28 | 68 | 2.52e-01 |
| KEGG | KEGG:04625 | C-type lectin receptor signaling pathway | 30 | 74 | 2.59e-01 |
| KEGG | KEGG:05207 | Chemical carcinogenesis - receptor activation | 50 | 132 | 2.59e-01 |
| KEGG | KEGG:04978 | Mineral absorption | 17 | 38 | 2.68e-01 |
| KEGG | KEGG:04210 | Apoptosis | 43 | 112 | 2.69e-01 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 50 | 133 | 2.76e-01 |
| KEGG | KEGG:04380 | Osteoclast differentiation | 36 | 92 | 2.76e-01 |
| KEGG | KEGG:05216 | Thyroid cancer | 15 | 33 | 2.79e-01 |
| KEGG | KEGG:04064 | NF-kappa B signaling pathway | 25 | 61 | 2.89e-01 |
| KEGG | KEGG:05226 | Gastric cancer | 43 | 113 | 2.89e-01 |
| KEGG | KEGG:00220 | Arginine biosynthesis | 8 | 15 | 2.90e-01 |
| KEGG | KEGG:04914 | Progesterone-mediated oocyte maturation | 32 | 82 | 3.20e-01 |
| KEGG | KEGG:00910 | Nitrogen metabolism | 5 | 8 | 3.20e-01 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 53 | 144 | 3.20e-01 |
| KEGG | KEGG:04724 | Glutamatergic synapse | 29 | 74 | 3.51e-01 |
| KEGG | KEGG:05213 | Endometrial cancer | 22 | 54 | 3.53e-01 |
| KEGG | KEGG:05200 | Pathways in cancer | 133 | 391 | 3.62e-01 |
| KEGG | KEGG:01524 | Platinum drug resistance | 24 | 60 | 3.62e-01 |
| KEGG | KEGG:04115 | p53 signaling pathway | 26 | 66 | 3.63e-01 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 45 | 122 | 3.63e-01 |
| KEGG | KEGG:05170 | Human immunodeficiency virus 1 infection | 54 | 149 | 3.63e-01 |
| KEGG | KEGG:04211 | Longevity regulating pathway | 29 | 75 | 3.70e-01 |
| KEGG | KEGG:04960 | Aldosterone-regulated sodium reabsorption | 11 | 24 | 3.70e-01 |
| KEGG | KEGG:04950 | Maturity onset diabetes of the young | 3 | 4 | 3.80e-01 |
| KEGG | KEGG:00470 | D-Amino acid metabolism | 3 | 4 | 3.80e-01 |
| KEGG | KEGG:05412 | Arrhythmogenic right ventricular cardiomyopathy | 26 | 67 | 3.94e-01 |
| KEGG | KEGG:04061 | Viral protein interaction with cytokine and cytokine receptor | 9 | 19 | 3.94e-01 |
| KEGG | KEGG:00561 | Glycerolipid metabolism | 18 | 44 | 3.95e-01 |
| KEGG | KEGG:03320 | PPAR signaling pathway | 19 | 47 | 4.01e-01 |
| KEGG | KEGG:04936 | Alcoholic liver disease | 34 | 91 | 4.03e-01 |
| KEGG | KEGG:04728 | Dopaminergic synapse | 36 | 97 | 4.03e-01 |
| KEGG | KEGG:04918 | Thyroid hormone synthesis | 21 | 53 | 4.05e-01 |
| KEGG | KEGG:05218 | Melanoma | 22 | 56 | 4.08e-01 |
| KEGG | KEGG:04740 | Olfactory transduction | 11 | 25 | 4.17e-01 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 41 | 113 | 4.17e-01 |
| KEGG | KEGG:04140 | Autophagy - animal | 53 | 149 | 4.17e-01 |
| KEGG | KEGG:04152 | AMPK signaling pathway | 37 | 101 | 4.17e-01 |
| KEGG | KEGG:04540 | Gap junction | 27 | 71 | 4.17e-01 |
| KEGG | KEGG:04630 | JAK-STAT signaling pathway | 31 | 83 | 4.17e-01 |
| KEGG | KEGG:04216 | Ferroptosis | 13 | 31 | 4.40e-01 |
| KEGG | KEGG:04972 | Pancreatic secretion | 22 | 57 | 4.40e-01 |
| KEGG | KEGG:04915 | Estrogen signaling pathway | 33 | 90 | 4.43e-01 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 32 | 87 | 4.43e-01 |
| KEGG | KEGG:05210 | Colorectal cancer | 30 | 81 | 4.43e-01 |
| KEGG | KEGG:04742 | Taste transduction | 9 | 20 | 4.43e-01 |
| KEGG | KEGG:04940 | Type I diabetes mellitus | 4 | 7 | 4.54e-01 |
| KEGG | KEGG:01040 | Biosynthesis of unsaturated fatty acids | 10 | 23 | 4.54e-01 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 24 | 64 | 4.77e-01 |
| KEGG | KEGG:05215 | Prostate cancer | 30 | 82 | 4.77e-01 |
| KEGG | KEGG:04726 | Serotonergic synapse | 22 | 58 | 4.77e-01 |
| KEGG | KEGG:05030 | Cocaine addiction | 13 | 32 | 4.87e-01 |
| KEGG | KEGG:04930 | Type II diabetes mellitus | 14 | 35 | 4.87e-01 |
| KEGG | KEGG:05224 | Breast cancer | 40 | 113 | 4.87e-01 |
| KEGG | KEGG:04964 | Proximal tubule bicarbonate reclamation | 8 | 18 | 4.87e-01 |
| KEGG | KEGG:04966 | Collecting duct acid secretion | 8 | 18 | 4.87e-01 |
| KEGG | KEGG:03440 | Homologous recombination | 15 | 38 | 4.87e-01 |
| KEGG | KEGG:04072 | Phospholipase D signaling pathway | 38 | 107 | 4.88e-01 |
| KEGG | KEGG:04658 | Th1 and Th2 cell differentiation | 19 | 50 | 4.97e-01 |
| KEGG | KEGG:05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 24 | 65 | 5.00e-01 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 45 | 129 | 5.02e-01 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 37 | 105 | 5.24e-01 |
| KEGG | KEGG:05133 | Pertussis | 16 | 42 | 5.32e-01 |
| KEGG | KEGG:04916 | Melanogenesis | 27 | 75 | 5.32e-01 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 27 | 75 | 5.32e-01 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 32 | 90 | 5.32e-01 |
| KEGG | KEGG:00480 | Glutathione metabolism | 15 | 39 | 5.32e-01 |
| KEGG | KEGG:04917 | Prolactin signaling pathway | 20 | 54 | 5.33e-01 |
| KEGG | KEGG:05130 | Pathogenic Escherichia coli infection | 49 | 143 | 5.42e-01 |
| KEGG | KEGG:04071 | Sphingolipid signaling pathway | 36 | 103 | 5.45e-01 |
| KEGG | KEGG:03430 | Mismatch repair | 9 | 22 | 5.64e-01 |
| KEGG | KEGG:05160 | Hepatitis C | 38 | 110 | 5.67e-01 |
| KEGG | KEGG:04392 | Hippo signaling pathway - multiple species | 10 | 25 | 5.67e-01 |
| KEGG | KEGG:04640 | Hematopoietic cell lineage | 11 | 28 | 5.67e-01 |
| KEGG | KEGG:03083 | Polycomb repressive complex | 25 | 70 | 5.67e-01 |
| KEGG | KEGG:04962 | Vasopressin-regulated water reabsorption | 14 | 37 | 5.71e-01 |
| KEGG | KEGG:04961 | Endocrine and other factor-regulated calcium reabsorption | 15 | 40 | 5.71e-01 |
| KEGG | KEGG:04530 | Tight junction | 44 | 129 | 5.73e-01 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 83 | 252 | 5.92e-01 |
| KEGG | KEGG:04912 | GnRH signaling pathway | 26 | 74 | 6.01e-01 |
| KEGG | KEGG:04217 | Necroptosis | 34 | 99 | 6.14e-01 |
| KEGG | KEGG:04974 | Protein digestion and absorption | 20 | 56 | 6.14e-01 |
| KEGG | KEGG:00650 | Butanoate metabolism | 7 | 17 | 6.14e-01 |
| KEGG | KEGG:05321 | Inflammatory bowel disease | 8 | 20 | 6.14e-01 |
| KEGG | KEGG:00030 | Pentose phosphate pathway | 8 | 20 | 6.14e-01 |
| KEGG | KEGG:04136 | Autophagy - other | 11 | 29 | 6.18e-01 |
| KEGG | KEGG:05323 | Rheumatoid arthritis | 14 | 38 | 6.18e-01 |
| KEGG | KEGG:05418 | Fluid shear stress and atherosclerosis | 37 | 109 | 6.18e-01 |
| KEGG | KEGG:04922 | Glucagon signaling pathway | 27 | 78 | 6.19e-01 |
| KEGG | KEGG:04514 | Cell adhesion molecules | 24 | 69 | 6.32e-01 |
| KEGG | KEGG:04060 | Cytokine-cytokine receptor interaction | 32 | 94 | 6.37e-01 |
| KEGG | KEGG:04620 | Toll-like receptor signaling pathway | 21 | 60 | 6.41e-01 |
| KEGG | KEGG:01522 | Endocrine resistance | 28 | 82 | 6.47e-01 |
| KEGG | KEGG:05414 | Dilated cardiomyopathy | 28 | 82 | 6.47e-01 |
| KEGG | KEGG:05162 | Measles | 29 | 85 | 6.47e-01 |
| KEGG | KEGG:00600 | Sphingolipid metabolism | 15 | 42 | 6.59e-01 |
| KEGG | KEGG:03450 | Non-homologous end-joining | 5 | 12 | 6.62e-01 |
| KEGG | KEGG:04744 | Phototransduction | 5 | 12 | 6.62e-01 |
| KEGG | KEGG:04218 | Cellular senescence | 45 | 136 | 6.62e-01 |
| KEGG | KEGG:05120 | Epithelial cell signaling in Helicobacter pylori infection | 19 | 55 | 6.83e-01 |
| KEGG | KEGG:00232 | Caffeine metabolism | 1 | 1 | 6.83e-01 |
| KEGG | KEGG:03260 | Virion - Human immunodeficiency virus | 1 | 1 | 6.83e-01 |
| KEGG | KEGG:05410 | Hypertrophic cardiomyopathy | 26 | 77 | 6.87e-01 |
| KEGG | KEGG:05212 | Pancreatic cancer | 24 | 71 | 6.96e-01 |
| KEGG | KEGG:05211 | Renal cell carcinoma | 21 | 62 | 6.96e-01 |
| KEGG | KEGG:04621 | NOD-like receptor signaling pathway | 36 | 109 | 6.96e-01 |
| KEGG | KEGG:01232 | Nucleotide metabolism | 22 | 65 | 6.96e-01 |
| KEGG | KEGG:05146 | Amoebiasis | 21 | 62 | 6.96e-01 |
| KEGG | KEGG:04660 | T cell receptor signaling pathway | 28 | 84 | 6.96e-01 |
| KEGG | KEGG:04114 | Oocyte meiosis | 34 | 103 | 6.96e-01 |
| KEGG | KEGG:00270 | Cysteine and methionine metabolism | 13 | 37 | 6.96e-01 |
| KEGG | KEGG:04672 | Intestinal immune network for IgA production | 4 | 10 | 6.99e-01 |
| KEGG | KEGG:00250 | Alanine, aspartate and glutamate metabolism | 10 | 28 | 6.99e-01 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 33 | 100 | 6.99e-01 |
| KEGG | KEGG:00592 | alpha-Linolenic acid metabolism | 4 | 10 | 6.99e-01 |
| KEGG | KEGG:00591 | Linoleic acid metabolism | 3 | 7 | 6.99e-01 |
| KEGG | KEGG:04921 | Oxytocin signaling pathway | 37 | 113 | 6.99e-01 |
| KEGG | KEGG:04213 | Longevity regulating pathway - multiple species | 17 | 50 | 6.99e-01 |
| KEGG | KEGG:05143 | African trypanosomiasis | 6 | 16 | 6.99e-01 |
| KEGG | KEGG:00534 | Glycosaminoglycan biosynthesis - heparan sulfate / heparin | 7 | 19 | 6.99e-01 |
| KEGG | KEGG:05203 | Viral carcinogenesis | 46 | 142 | 6.99e-01 |
| KEGG | KEGG:00240 | Pyrimidine metabolism | 15 | 44 | 6.99e-01 |
| KEGG | KEGG:00310 | Lysine degradation | 19 | 56 | 6.99e-01 |
| KEGG | KEGG:04650 | Natural killer cell mediated cytotoxicity | 18 | 53 | 6.99e-01 |
| KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 33 | 101 | 6.99e-01 |
| KEGG | KEGG:04977 | Vitamin digestion and absorption | 5 | 13 | 6.99e-01 |
| KEGG | KEGG:00515 | Mannose type O-glycan biosynthesis | 7 | 19 | 6.99e-01 |
| KEGG | KEGG:00410 | beta-Alanine metabolism | 8 | 22 | 6.99e-01 |
| KEGG | KEGG:05140 | Leishmaniasis | 13 | 38 | 7.05e-01 |
| KEGG | KEGG:04723 | Retrograde endocannabinoid signaling | 30 | 92 | 7.07e-01 |
| KEGG | KEGG:04215 | Apoptosis - multiple species | 10 | 29 | 7.18e-01 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 79 | 250 | 7.18e-01 |
| KEGG | KEGG:04710 | Circadian rhythm | 10 | 29 | 7.18e-01 |
| KEGG | KEGG:04973 | Carbohydrate digestion and absorption | 10 | 29 | 7.18e-01 |
| KEGG | KEGG:04520 | Adherens junction | 27 | 83 | 7.18e-01 |
| KEGG | KEGG:05231 | Choline metabolism in cancer | 26 | 80 | 7.24e-01 |
| KEGG | KEGG:04144 | Endocytosis | 68 | 216 | 7.52e-01 |
| KEGG | KEGG:00330 | Arginine and proline metabolism | 11 | 33 | 7.62e-01 |
| KEGG | KEGG:05214 | Glioma | 21 | 65 | 7.62e-01 |
| KEGG | KEGG:04721 | Synaptic vesicle cycle | 16 | 49 | 7.62e-01 |
| KEGG | KEGG:04137 | Mitophagy - animal | 30 | 94 | 7.62e-01 |
| KEGG | KEGG:04330 | Notch signaling pathway | 17 | 52 | 7.62e-01 |
| KEGG | KEGG:05132 | Salmonella infection | 63 | 201 | 7.64e-01 |
| KEGG | KEGG:00601 | Glycosphingolipid biosynthesis - lacto and neolacto series | 5 | 14 | 7.64e-01 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 23 | 72 | 7.74e-01 |
| KEGG | KEGG:01100 | Metabolic pathways | 332 | 1077 | 7.74e-01 |
| KEGG | KEGG:02010 | ABC transporters | 8 | 24 | 7.81e-01 |
| KEGG | KEGG:05034 | Alcoholism | 32 | 102 | 7.81e-01 |
| KEGG | KEGG:04270 | Vascular smooth muscle contraction | 29 | 92 | 7.81e-01 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 43 | 137 | 7.81e-01 |
| KEGG | KEGG:04670 | Leukocyte transendothelial migration | 24 | 76 | 7.81e-01 |
| KEGG | KEGG:04260 | Cardiac muscle contraction | 20 | 63 | 7.81e-01 |
| KEGG | KEGG:00983 | Drug metabolism - other enzymes | 13 | 40 | 7.81e-01 |
| KEGG | KEGG:04929 | GnRH secretion | 16 | 50 | 7.81e-01 |
| KEGG | KEGG:05340 | Primary immunodeficiency | 3 | 8 | 7.81e-01 |
| KEGG | KEGG:03013 | Nucleocytoplasmic transport | 30 | 96 | 7.89e-01 |
| KEGG | KEGG:00440 | Phosphonate and phosphinate metabolism | 2 | 5 | 7.89e-01 |
| KEGG | KEGG:03265 | Virion - Ebolavirus and Lyssavirus | 2 | 5 | 7.89e-01 |
| KEGG | KEGG:04062 | Chemokine signaling pathway | 34 | 109 | 7.89e-01 |
| KEGG | KEGG:04720 | Long-term potentiation | 16 | 51 | 8.16e-01 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 42 | 136 | 8.16e-01 |
| KEGG | KEGG:03267 | Virion - Adenovirus | 1 | 2 | 8.26e-01 |
| KEGG | KEGG:00400 | Phenylalanine, tyrosine and tryptophan biosynthesis | 1 | 2 | 8.26e-01 |
| KEGG | KEGG:05135 | Yersinia infection | 33 | 107 | 8.26e-01 |
| KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 39 | 127 | 8.30e-01 |
| KEGG | KEGG:05031 | Amphetamine addiction | 14 | 45 | 8.30e-01 |
| KEGG | KEGG:00062 | Fatty acid elongation | 7 | 22 | 8.30e-01 |
| KEGG | KEGG:00500 | Starch and sucrose metabolism | 7 | 22 | 8.30e-01 |
| KEGG | KEGG:04920 | Adipocytokine signaling pathway | 15 | 49 | 8.32e-01 |
| KEGG | KEGG:04666 | Fc gamma R-mediated phagocytosis | 23 | 75 | 8.32e-01 |
| KEGG | KEGG:05145 | Toxoplasmosis | 22 | 72 | 8.32e-01 |
| KEGG | KEGG:04340 | Hedgehog signaling pathway | 15 | 49 | 8.32e-01 |
| KEGG | KEGG:00280 | Valine, leucine and isoleucine degradation | 13 | 42 | 8.32e-01 |
| KEGG | KEGG:05222 | Small cell lung cancer | 25 | 82 | 8.32e-01 |
| KEGG | KEGG:00061 | Fatty acid biosynthesis | 3 | 9 | 8.32e-01 |
| KEGG | KEGG:04662 | B cell receptor signaling pathway | 16 | 52 | 8.32e-01 |
| KEGG | KEGG:01230 | Biosynthesis of amino acids | 16 | 52 | 8.32e-01 |
| KEGG | KEGG:01210 | 2-Oxocarboxylic acid metabolism | 8 | 26 | 8.47e-01 |
| KEGG | KEGG:04066 | HIF-1 signaling pathway | 26 | 86 | 8.47e-01 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 49 | 162 | 8.47e-01 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 51 | 169 | 8.53e-01 |
| KEGG | KEGG:00533 | Glycosaminoglycan biosynthesis - keratan sulfate | 4 | 13 | 8.75e-01 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 31 | 104 | 8.75e-01 |
| KEGG | KEGG:05144 | Malaria | 6 | 20 | 8.75e-01 |
| KEGG | KEGG:04370 | VEGF signaling pathway | 14 | 47 | 8.75e-01 |
| KEGG | KEGG:04145 | Phagosome | 25 | 84 | 8.75e-01 |
| KEGG | KEGG:05150 | Staphylococcus aureus infection | 4 | 13 | 8.75e-01 |
| KEGG | KEGG:03420 | Nucleotide excision repair | 16 | 54 | 8.79e-01 |
| KEGG | KEGG:04622 | RIG-I-like receptor signaling pathway | 13 | 44 | 8.82e-01 |
| KEGG | KEGG:00730 | Thiamine metabolism | 3 | 10 | 8.87e-01 |
| KEGG | KEGG:00920 | Sulfur metabolism | 3 | 10 | 8.87e-01 |
| KEGG | KEGG:05033 | Nicotine addiction | 3 | 10 | 8.87e-01 |
| KEGG | KEGG:05110 | Vibrio cholerae infection | 12 | 41 | 8.87e-01 |
| KEGG | KEGG:04614 | Renin-angiotensin system | 3 | 10 | 8.87e-01 |
| KEGG | KEGG:04130 | SNARE interactions in vesicular transport | 9 | 31 | 8.95e-01 |
| KEGG | KEGG:04932 | Non-alcoholic fatty liver disease | 37 | 126 | 9.03e-01 |
| KEGG | KEGG:00360 | Phenylalanine metabolism | 1 | 3 | 9.03e-01 |
| KEGG | KEGG:00290 | Valine, leucine and isoleucine biosynthesis | 1 | 3 | 9.03e-01 |
| KEGG | KEGG:04664 | Fc epsilon RI signaling pathway | 12 | 42 | 9.03e-01 |
| KEGG | KEGG:00565 | Ether lipid metabolism | 8 | 28 | 9.03e-01 |
| KEGG | KEGG:05017 | Spinocerebellar ataxia | 35 | 120 | 9.03e-01 |
| KEGG | KEGG:00450 | Selenocompound metabolism | 4 | 14 | 9.03e-01 |
| KEGG | KEGG:04146 | Peroxisome | 19 | 66 | 9.03e-01 |
| KEGG | KEGG:00900 | Terpenoid backbone biosynthesis | 6 | 21 | 9.03e-01 |
| KEGG | KEGG:04722 | Neurotrophin signaling pathway | 30 | 104 | 9.12e-01 |
| KEGG | KEGG:01212 | Fatty acid metabolism | 13 | 46 | 9.12e-01 |
| KEGG | KEGG:00860 | Porphyrin metabolism | 5 | 18 | 9.12e-01 |
| KEGG | KEGG:03460 | Fanconi anemia pathway | 13 | 46 | 9.12e-01 |
| KEGG | KEGG:00350 | Tyrosine metabolism | 3 | 11 | 9.22e-01 |
| KEGG | KEGG:05416 | Viral myocarditis | 10 | 36 | 9.22e-01 |
| KEGG | KEGG:03082 | ATP-dependent chromatin remodeling | 26 | 91 | 9.22e-01 |
| KEGG | KEGG:03022 | Basal transcription factors | 9 | 33 | 9.36e-01 |
| KEGG | KEGG:00100 | Steroid biosynthesis | 4 | 15 | 9.36e-01 |
| KEGG | KEGG:04979 | Cholesterol metabolism | 9 | 33 | 9.36e-01 |
| KEGG | KEGG:04613 | Neutrophil extracellular trap formation | 25 | 89 | 9.47e-01 |
| KEGG | KEGG:00053 | Ascorbate and aldarate metabolism | 2 | 8 | 9.57e-01 |
| KEGG | KEGG:00563 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 6 | 23 | 9.57e-01 |
| KEGG | KEGG:04122 | Sulfur relay system | 2 | 8 | 9.57e-01 |
| KEGG | KEGG:00430 | Taurine and hypotaurine metabolism | 2 | 8 | 9.57e-01 |
| KEGG | KEGG:04657 | IL-17 signaling pathway | 15 | 55 | 9.57e-01 |
| KEGG | KEGG:00980 | Metabolism of xenobiotics by cytochrome P450 | 6 | 23 | 9.57e-01 |
| KEGG | KEGG:00524 | Neomycin, kanamycin and gentamicin biosynthesis | 1 | 4 | 9.57e-01 |
| KEGG | KEGG:05100 | Bacterial invasion of epithelial cells | 20 | 73 | 9.57e-01 |
| KEGG | KEGG:00640 | Propanoate metabolism | 7 | 27 | 9.57e-01 |
| KEGG | KEGG:00785 | Lipoic acid metabolism | 4 | 16 | 9.57e-01 |
| KEGG | KEGG:00140 | Steroid hormone biosynthesis | 4 | 16 | 9.57e-01 |
| KEGG | KEGG:00620 | Pyruvate metabolism | 8 | 31 | 9.62e-01 |
| KEGG | KEGG:00532 | Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate | 5 | 20 | 9.62e-01 |
| KEGG | KEGG:04610 | Complement and coagulation cascades | 9 | 35 | 9.70e-01 |
| KEGG | KEGG:00512 | Mucin type O-glycan biosynthesis | 5 | 21 | 9.88e-01 |
| KEGG | KEGG:05131 | Shigellosis | 57 | 204 | 9.88e-01 |
| KEGG | KEGG:00052 | Galactose metabolism | 5 | 21 | 9.88e-01 |
| KEGG | KEGG:00380 | Tryptophan metabolism | 5 | 21 | 9.88e-01 |
| KEGG | KEGG:05230 | Central carbon metabolism in cancer | 15 | 58 | 9.88e-01 |
| KEGG | KEGG:00590 | Arachidonic acid metabolism | 6 | 25 | 9.88e-01 |
| KEGG | KEGG:00630 | Glyoxylate and dicarboxylate metabolism | 6 | 25 | 9.88e-01 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 93 | 364 | 1.00e+00 |
| KEGG | KEGG:05020 | Prion disease | 48 | 204 | 1.00e+00 |
| KEGG | KEGG:05016 | Huntington disease | 63 | 246 | 1.00e+00 |
| KEGG | KEGG:05012 | Parkinson disease | 49 | 211 | 1.00e+00 |
| KEGG | KEGG:05010 | Alzheimer disease | 72 | 296 | 1.00e+00 |
| KEGG | KEGG:04612 | Antigen processing and presentation | 5 | 26 | 1.00e+00 |
| KEGG | KEGG:00603 | Glycosphingolipid biosynthesis - globo and isoglobo series | 1 | 8 | 1.00e+00 |
| KEGG | KEGG:00604 | Glycosphingolipid biosynthesis - ganglio series | 2 | 12 | 1.00e+00 |
| KEGG | KEGG:04975 | Fat digestion and absorption | 4 | 18 | 1.00e+00 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 76 | 291 | 1.00e+00 |
| KEGG | KEGG:00520 | Amino sugar and nucleotide sugar metabolism | 8 | 44 | 1.00e+00 |
| KEGG | KEGG:00760 | Nicotinate and nicotinamide metabolism | 6 | 28 | 1.00e+00 |
| KEGG | KEGG:00790 | Folate biosynthesis | 3 | 18 | 1.00e+00 |
| KEGG | KEGG:00970 | Aminoacyl-tRNA biosynthesis | 7 | 42 | 1.00e+00 |
| KEGG | KEGG:00982 | Drug metabolism - cytochrome P450 | 3 | 16 | 1.00e+00 |
| KEGG | KEGG:01200 | Carbon metabolism | 22 | 88 | 1.00e+00 |
| KEGG | KEGG:01240 | Biosynthesis of cofactors | 28 | 109 | 1.00e+00 |
| KEGG | KEGG:01250 | Biosynthesis of nucleotide sugars | 5 | 34 | 1.00e+00 |
| KEGG | KEGG:01523 | Antifolate resistance | 4 | 22 | 1.00e+00 |
| KEGG | KEGG:00740 | Riboflavin metabolism | 1 | 6 | 1.00e+00 |
| KEGG | KEGG:00750 | Vitamin B6 metabolism | 1 | 5 | 1.00e+00 |
| KEGG | KEGG:00513 | Various types of N-glycan biosynthesis | 5 | 37 | 1.00e+00 |
| KEGG | KEGG:03008 | Ribosome biogenesis in eukaryotes | 10 | 65 | 1.00e+00 |
| KEGG | KEGG:05415 | Diabetic cardiomyopathy | 40 | 158 | 1.00e+00 |
| KEGG | KEGG:05322 | Systemic lupus erythematosus | 5 | 44 | 1.00e+00 |
| KEGG | KEGG:04510 | Focal adhesion | 34 | 169 | 1.00e+00 |
| KEGG | KEGG:04512 | ECM-receptor interaction | 11 | 66 | 1.00e+00 |
| KEGG | KEGG:05219 | Bladder cancer | 6 | 35 | 1.00e+00 |
| KEGG | KEGG:05208 | Chemical carcinogenesis - reactive oxygen species | 40 | 173 | 1.00e+00 |
| KEGG | KEGG:00010 | Glycolysis / Gluconeogenesis | 9 | 41 | 1.00e+00 |
| KEGG | KEGG:05204 | Chemical carcinogenesis - DNA adducts | 4 | 20 | 1.00e+00 |
| KEGG | KEGG:00020 | Citrate cycle (TCA cycle) | 6 | 26 | 1.00e+00 |
| KEGG | KEGG:00040 | Pentose and glucuronate interconversions | 1 | 11 | 1.00e+00 |
| KEGG | KEGG:05171 | Coronavirus disease - COVID-19 | 30 | 149 | 1.00e+00 |
| KEGG | KEGG:00051 | Fructose and mannose metabolism | 3 | 26 | 1.00e+00 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 78 | 388 | 1.00e+00 |
| KEGG | KEGG:00071 | Fatty acid degradation | 7 | 31 | 1.00e+00 |
| KEGG | KEGG:00120 | Primary bile acid biosynthesis | 1 | 11 | 1.00e+00 |
| KEGG | KEGG:00190 | Oxidative phosphorylation | 17 | 99 | 1.00e+00 |
| KEGG | KEGG:00260 | Glycine, serine and threonine metabolism | 5 | 26 | 1.00e+00 |
| KEGG | KEGG:05134 | Legionellosis | 7 | 34 | 1.00e+00 |
| KEGG | KEGG:00340 | Histidine metabolism | 2 | 10 | 1.00e+00 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 8 | 49 | 1.00e+00 |
| KEGG | KEGG:00511 | Other glycan degradation | 2 | 14 | 1.00e+00 |
| KEGG | KEGG:00514 | Other types of O-glycan biosynthesis | 8 | 35 | 1.00e+00 |
| KEGG | KEGG:03010 | Ribosome | 15 | 121 | 1.00e+00 |
| KEGG | KEGG:00670 | One carbon pool by folate | 3 | 18 | 1.00e+00 |
| KEGG | KEGG:03018 | RNA degradation | 17 | 67 | 1.00e+00 |
| KEGG | KEGG:03015 | mRNA surveillance pathway | 17 | 80 | 1.00e+00 |
| KEGG | KEGG:04714 | Thermogenesis | 45 | 185 | 1.00e+00 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 27 | 149 | 1.00e+00 |
| KEGG | KEGG:04142 | Lysosome | 26 | 109 | 1.00e+00 |
| KEGG | KEGG:03266 | Virion - Herpesvirus | 1 | 6 | 1.00e+00 |
| KEGG | KEGG:03060 | Protein export | 5 | 28 | 1.00e+00 |
| KEGG | KEGG:04623 | Cytosolic DNA-sensing pathway | 10 | 46 | 1.00e+00 |
| KEGG | KEGG:03050 | Proteasome | 6 | 40 | 1.00e+00 |
| KEGG | KEGG:03040 | Spliceosome | 14 | 124 | 1.00e+00 |
| KEGG | KEGG:03020 | RNA polymerase | 6 | 33 | 1.00e+00 |
# intersect(unique(Resp_RNA$ENTREZID), unique(NR_peak_list_20k$ENTREZID)))
LR_int_restable %>%
filter(source=="GO:BP", p_value < 0.05) %>%
slice_head(n=10) %>%
mutate(log10_p=(-log(p_value, base=10))) %>%
ggplot(., aes(x=log10_p, y=reorder(term_name, log10_p)))+
geom_col(fill="#00BFC4")+
theme_bw()+
ylab("")+
xlab(paste0("-log10 p-value"))+
guides(fill="none")+
theme(axis.text.y = element_text(color="black"))+
ggtitle("Intersection LR")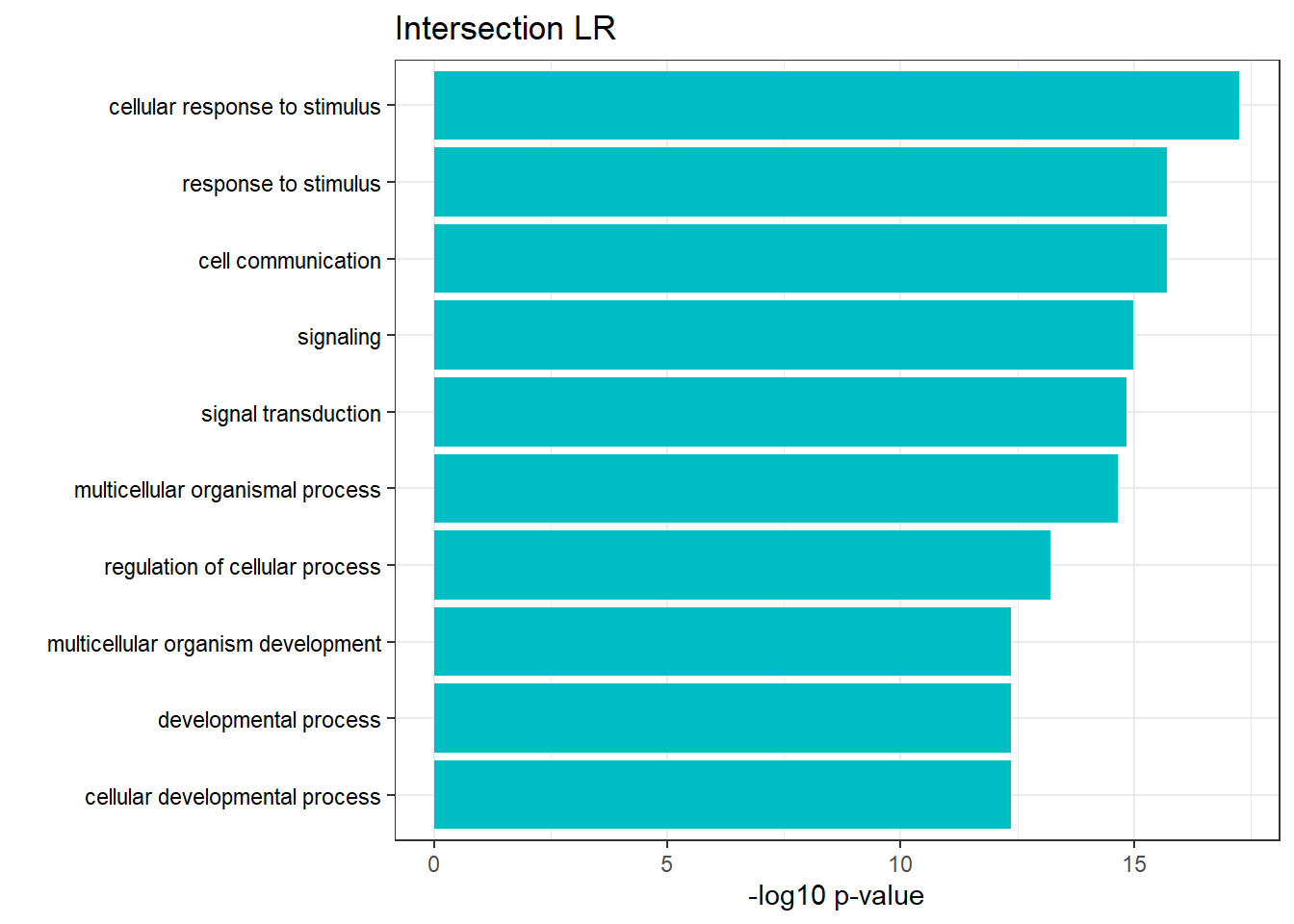 #### NR
#### NR
NR_int_resgenes <- gost(query = int_NR_n_RNAresp,
organism = "hsapiens",
significant = FALSE,
ordered_query = FALSE,
domain_scope = "custom",
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
custom_bg = background_df$ENTREZID,
sources=c("GO:BP","KEGG"))
saveRDS(NR_int_resgenes,"data/GO_KEGG_analysis/NR_int_resgenes.RDS")
NR_int_restable <- NR_int_resgenes$result %>%
dplyr::select(c(source, term_id,
term_name,intersection_size,
term_size, p_value))# %>%
NR_int_restable %>%
filter(source=="GO:BP", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(.,caption = "GO:BP terms found in NR neargenes intersected with RNA response genes" ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0007049 | cell cycle | 749 | 1436 | 3.20e-07 |
| GO:BP | GO:0007059 | chromosome segregation | 214 | 351 | 4.26e-07 |
| GO:BP | GO:0022402 | cell cycle process | 548 | 1024 | 4.67e-07 |
| GO:BP | GO:0050794 | regulation of cellular process | 3320 | 7153 | 5.99e-07 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 1709 | 3550 | 6.52e-06 |
| GO:BP | GO:0050789 | regulation of biological process | 3463 | 7516 | 1.01e-05 |
| GO:BP | GO:0065007 | biological regulation | 3558 | 7734 | 1.01e-05 |
| GO:BP | GO:0000280 | nuclear division | 198 | 335 | 1.77e-05 |
| GO:BP | GO:0051716 | cellular response to stimulus | 2192 | 4640 | 1.77e-05 |
| GO:BP | GO:0006974 | DNA damage response | 400 | 745 | 3.39e-05 |
| GO:BP | GO:0048519 | negative regulation of biological process | 1793 | 3764 | 5.18e-05 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 155 | 256 | 5.75e-05 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 595 | 1157 | 5.77e-05 |
| GO:BP | GO:0000278 | mitotic cell cycle | 406 | 763 | 7.44e-05 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 349 | 648 | 1.19e-04 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 108 | 170 | 1.49e-04 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 285 | 520 | 2.02e-04 |
| GO:BP | GO:0000819 | sister chromatid segregation | 127 | 207 | 2.12e-04 |
| GO:BP | GO:0061982 | meiosis I cell cycle process | 57 | 79 | 2.24e-04 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 547 | 1067 | 2.30e-04 |
| GO:BP | GO:0051276 | chromosome organization | 277 | 506 | 2.60e-04 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 321 | 596 | 2.60e-04 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 94 | 146 | 2.60e-04 |
| GO:BP | GO:0033554 | cellular response to stress | 717 | 1433 | 3.22e-04 |
| GO:BP | GO:0007051 | spindle organization | 110 | 177 | 3.75e-04 |
| GO:BP | GO:0051726 | regulation of cell cycle | 457 | 885 | 6.27e-04 |
| GO:BP | GO:0050896 | response to stimulus | 2496 | 5379 | 6.31e-04 |
| GO:BP | GO:0048285 | organelle fission | 211 | 377 | 6.31e-04 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 1384 | 2895 | 6.31e-04 |
| GO:BP | GO:0140013 | meiotic nuclear division | 78 | 119 | 6.51e-04 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 494 | 965 | 6.88e-04 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 86 | 134 | 6.88e-04 |
| GO:BP | GO:0007127 | meiosis I | 52 | 73 | 7.96e-04 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 875 | 1787 | 1.08e-03 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 496 | 973 | 1.11e-03 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 1275 | 2663 | 1.11e-03 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 368 | 704 | 1.12e-03 |
| GO:BP | GO:0140014 | mitotic nuclear division | 141 | 241 | 1.12e-03 |
| GO:BP | GO:0021537 | telencephalon development | 117 | 195 | 1.33e-03 |
| GO:BP | GO:0051301 | cell division | 289 | 541 | 1.33e-03 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 1774 | 3777 | 1.88e-03 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 1156 | 2410 | 2.17e-03 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 913 | 1879 | 2.28e-03 |
| GO:BP | GO:0007010 | cytoskeleton organization | 574 | 1146 | 2.28e-03 |
| GO:BP | GO:0006260 | DNA replication | 142 | 246 | 2.36e-03 |
| GO:BP | GO:0007017 | microtubule-based process | 372 | 718 | 2.41e-03 |
| GO:BP | GO:0030900 | forebrain development | 155 | 272 | 2.41e-03 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 1160 | 2422 | 2.56e-03 |
| GO:BP | GO:0040007 | growth | 361 | 697 | 3.12e-03 |
| GO:BP | GO:0031323 | regulation of cellular metabolic process | 1974 | 4233 | 3.12e-03 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 777 | 1588 | 3.27e-03 |
| GO:BP | GO:0006351 | DNA-templated transcription | 1198 | 2510 | 3.50e-03 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 797 | 1635 | 4.54e-03 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 1207 | 2533 | 4.54e-03 |
| GO:BP | GO:0006281 | DNA repair | 269 | 508 | 4.84e-03 |
| GO:BP | GO:0051321 | meiotic cell cycle | 107 | 181 | 5.94e-03 |
| GO:BP | GO:0031327 | negative regulation of cellular biosynthetic process | 790 | 1624 | 6.69e-03 |
| GO:BP | GO:0033043 | regulation of organelle organization | 478 | 953 | 8.28e-03 |
| GO:BP | GO:0048589 | developmental growth | 264 | 501 | 8.28e-03 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 177 | 322 | 8.28e-03 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 81 | 132 | 8.28e-03 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 1842 | 3954 | 8.36e-03 |
| GO:BP | GO:0007052 | mitotic spindle organization | 73 | 117 | 8.52e-03 |
| GO:BP | GO:0048518 | positive regulation of biological process | 1996 | 4301 | 9.30e-03 |
| GO:BP | GO:0008283 | cell population proliferation | 641 | 1306 | 1.04e-02 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 1351 | 2867 | 1.31e-02 |
| GO:BP | GO:0031324 | negative regulation of cellular metabolic process | 928 | 1934 | 1.31e-02 |
| GO:BP | GO:0007154 | cell communication | 1851 | 3983 | 1.42e-02 |
| GO:BP | GO:1990830 | cellular response to leukemia inhibitory factor | 54 | 83 | 1.50e-02 |
| GO:BP | GO:1990823 | response to leukemia inhibitory factor | 55 | 85 | 1.59e-02 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 104 | 179 | 1.60e-02 |
| GO:BP | GO:0019222 | regulation of metabolic process | 2126 | 4603 | 1.62e-02 |
| GO:BP | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore | 18 | 21 | 1.63e-02 |
| GO:BP | GO:0018130 | heterocycle biosynthetic process | 1376 | 2927 | 1.63e-02 |
| GO:BP | GO:0007165 | signal transduction | 1683 | 3611 | 1.63e-02 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 934 | 1951 | 1.63e-02 |
| GO:BP | GO:0006261 | DNA-templated DNA replication | 88 | 148 | 1.63e-02 |
| GO:BP | GO:0019438 | aromatic compound biosynthetic process | 1376 | 2928 | 1.70e-02 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 1959 | 4231 | 1.83e-02 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 148 | 268 | 1.89e-02 |
| GO:BP | GO:0060322 | head development | 279 | 539 | 1.95e-02 |
| GO:BP | GO:0051303 | establishment of chromosome localization | 62 | 99 | 2.05e-02 |
| GO:BP | GO:0010468 | regulation of gene expression | 1577 | 3379 | 2.07e-02 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 73 | 120 | 2.07e-02 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 158 | 289 | 2.09e-02 |
| GO:BP | GO:0050000 | chromosome localization | 66 | 107 | 2.33e-02 |
| GO:BP | GO:0045444 | fat cell differentiation | 103 | 179 | 2.33e-02 |
| GO:BP | GO:1902894 | negative regulation of miRNA transcription | 19 | 23 | 2.33e-02 |
| GO:BP | GO:0051225 | spindle assembly | 68 | 111 | 2.42e-02 |
| GO:BP | GO:0051310 | metaphase chromosome alignment | 59 | 94 | 2.46e-02 |
| GO:BP | GO:0006259 | DNA metabolic process | 416 | 832 | 2.48e-02 |
| GO:BP | GO:0023052 | signaling | 1816 | 3918 | 2.59e-02 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 998 | 2100 | 2.71e-02 |
| GO:BP | GO:0035556 | intracellular signal transduction | 930 | 1951 | 2.74e-02 |
| GO:BP | GO:1901362 | organic cyclic compound biosynthetic process | 1416 | 3026 | 2.74e-02 |
| GO:BP | GO:0051172 | negative regulation of nitrogen compound metabolic process | 785 | 1633 | 2.74e-02 |
| GO:BP | GO:2001251 | negative regulation of chromosome organization | 54 | 85 | 2.74e-02 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 1622 | 3486 | 2.78e-02 |
| GO:BP | GO:2000629 | negative regulation of miRNA metabolic process | 20 | 25 | 3.17e-02 |
| GO:BP | GO:0009262 | deoxyribonucleotide metabolic process | 26 | 35 | 3.22e-02 |
| GO:BP | GO:0019692 | deoxyribose phosphate metabolic process | 26 | 35 | 3.22e-02 |
| GO:BP | GO:0007275 | multicellular organism development | 1479 | 3170 | 3.28e-02 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 888 | 1862 | 3.42e-02 |
| GO:BP | GO:0009888 | tissue development | 653 | 1348 | 3.42e-02 |
| GO:BP | GO:0007420 | brain development | 257 | 498 | 3.49e-02 |
| GO:BP | GO:0031326 | regulation of cellular biosynthetic process | 1657 | 3569 | 3.53e-02 |
| GO:BP | GO:0072359 | circulatory system development | 429 | 864 | 3.54e-02 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 1668 | 3595 | 3.79e-02 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 915 | 1924 | 4.01e-02 |
| GO:BP | GO:0086004 | regulation of cardiac muscle cell contraction | 24 | 32 | 4.01e-02 |
| GO:BP | GO:0003338 | metanephros morphogenesis | 16 | 19 | 4.17e-02 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 114 | 204 | 4.20e-02 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 71 | 119 | 4.26e-02 |
| GO:BP | GO:0051299 | centrosome separation | 12 | 13 | 4.26e-02 |
| GO:BP | GO:0007098 | centrosome cycle | 72 | 121 | 4.26e-02 |
| GO:BP | GO:0006996 | organelle organization | 1317 | 2815 | 4.26e-02 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 142 | 261 | 4.26e-02 |
| GO:BP | GO:0007389 | pattern specification process | 154 | 286 | 4.64e-02 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 1929 | 4185 | 4.66e-02 |
| GO:BP | GO:0009394 | 2’-deoxyribonucleotide metabolic process | 25 | 34 | 4.66e-02 |
| GO:BP | GO:0007417 | central nervous system development | 348 | 694 | 4.75e-02 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 526 | 1077 | 4.75e-02 |
| GO:BP | GO:0007507 | heart development | 256 | 499 | 4.86e-02 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 111 | 199 | 4.93e-02 |
NR_int_restable %>%
filter(source=="KEGG") %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "KEGG terms found in NR neargenes intersected with RNA response genes") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| KEGG | KEGG:04115 | p53 signaling pathway | 44 | 66 | 5.84e-02 |
| KEGG | KEGG:04110 | Cell cycle | 85 | 146 | 6.34e-02 |
| KEGG | KEGG:05217 | Basal cell carcinoma | 32 | 48 | 1.12e-01 |
| KEGG | KEGG:00230 | Purine metabolism | 56 | 93 | 1.12e-01 |
| KEGG | KEGG:03410 | Base excision repair | 29 | 44 | 1.95e-01 |
| KEGG | KEGG:04070 | Phosphatidylinositol signaling system | 48 | 83 | 3.92e-01 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 88 | 164 | 3.92e-01 |
| KEGG | KEGG:01524 | Platinum drug resistance | 36 | 60 | 3.99e-01 |
| KEGG | KEGG:03030 | DNA replication | 22 | 34 | 4.50e-01 |
| KEGG | KEGG:05224 | Breast cancer | 62 | 113 | 4.50e-01 |
| KEGG | KEGG:05221 | Acute myeloid leukemia | 31 | 52 | 5.38e-01 |
| KEGG | KEGG:03430 | Mismatch repair | 15 | 22 | 5.66e-01 |
| KEGG | KEGG:05226 | Gastric cancer | 60 | 113 | 7.66e-01 |
| KEGG | KEGG:00562 | Inositol phosphate metabolism | 34 | 60 | 7.66e-01 |
| KEGG | KEGG:03083 | Polycomb repressive complex | 39 | 70 | 7.66e-01 |
| KEGG | KEGG:03460 | Fanconi anemia pathway | 26 | 46 | 8.05e-01 |
| KEGG | KEGG:04978 | Mineral absorption | 22 | 38 | 8.05e-01 |
| KEGG | KEGG:00564 | Glycerophospholipid metabolism | 39 | 71 | 8.05e-01 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 53 | 101 | 8.05e-01 |
| KEGG | KEGG:05034 | Alcoholism | 54 | 102 | 8.05e-01 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 63 | 122 | 8.05e-01 |
| KEGG | KEGG:03250 | Viral life cycle - HIV-1 | 30 | 54 | 8.05e-01 |
| KEGG | KEGG:03440 | Homologous recombination | 22 | 38 | 8.05e-01 |
| KEGG | KEGG:04966 | Collecting duct acid secretion | 12 | 18 | 8.05e-01 |
| KEGG | KEGG:04914 | Progesterone-mediated oocyte maturation | 44 | 82 | 8.05e-01 |
| KEGG | KEGG:04814 | Motor proteins | 72 | 141 | 8.05e-01 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 55 | 106 | 8.06e-01 |
| KEGG | KEGG:04934 | Cushing syndrome | 61 | 119 | 8.07e-01 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 34 | 64 | 8.07e-01 |
| KEGG | KEGG:05216 | Thyroid cancer | 19 | 33 | 8.07e-01 |
| KEGG | KEGG:04217 | Necroptosis | 51 | 99 | 8.07e-01 |
| KEGG | KEGG:00910 | Nitrogen metabolism | 6 | 8 | 8.07e-01 |
| KEGG | KEGG:01521 | EGFR tyrosine kinase inhibitor resistance | 38 | 72 | 8.07e-01 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 72 | 144 | 8.07e-01 |
| KEGG | KEGG:04216 | Ferroptosis | 18 | 31 | 8.07e-01 |
| KEGG | KEGG:04148 | Efferocytosis | 61 | 120 | 8.07e-01 |
| KEGG | KEGG:04146 | Peroxisome | 35 | 66 | 8.07e-01 |
| KEGG | KEGG:04750 | Inflammatory mediator regulation of TRP channels | 38 | 72 | 8.07e-01 |
| KEGG | KEGG:04923 | Regulation of lipolysis in adipocytes | 23 | 41 | 8.07e-01 |
| KEGG | KEGG:05218 | Melanoma | 30 | 56 | 8.13e-01 |
| KEGG | KEGG:03420 | Nucleotide excision repair | 29 | 54 | 8.13e-01 |
| KEGG | KEGG:05213 | Endometrial cancer | 29 | 54 | 8.13e-01 |
| KEGG | KEGG:05161 | Hepatitis B | 63 | 126 | 8.15e-01 |
| KEGG | KEGG:00830 | Retinol metabolism | 9 | 14 | 8.15e-01 |
| KEGG | KEGG:04927 | Cortisol synthesis and secretion | 25 | 46 | 8.15e-01 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 64 | 129 | 8.82e-01 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 67 | 137 | 9.57e-01 |
| KEGG | KEGG:00480 | Glutathione metabolism | 21 | 39 | 9.57e-01 |
| KEGG | KEGG:05210 | Colorectal cancer | 41 | 81 | 9.57e-01 |
| KEGG | KEGG:04668 | TNF signaling pathway | 42 | 83 | 9.57e-01 |
| KEGG | KEGG:04940 | Type I diabetes mellitus | 5 | 7 | 9.57e-01 |
| KEGG | KEGG:04976 | Bile secretion | 22 | 41 | 9.57e-01 |
| KEGG | KEGG:05032 | Morphine addiction | 28 | 54 | 9.80e-01 |
| KEGG | KEGG:00770 | Pantothenate and CoA biosynthesis | 9 | 15 | 9.80e-01 |
| KEGG | KEGG:00515 | Mannose type O-glycan biosynthesis | 11 | 19 | 9.80e-01 |
| KEGG | KEGG:00220 | Arginine biosynthesis | 9 | 15 | 9.80e-01 |
| KEGG | KEGG:04210 | Apoptosis | 55 | 112 | 9.80e-01 |
| KEGG | KEGG:05203 | Viral carcinogenesis | 69 | 142 | 9.80e-01 |
| KEGG | KEGG:03060 | Protein export | 7 | 28 | 1.00e+00 |
| KEGG | KEGG:00330 | Arginine and proline metabolism | 16 | 33 | 1.00e+00 |
| KEGG | KEGG:00591 | Linoleic acid metabolism | 3 | 7 | 1.00e+00 |
| KEGG | KEGG:04014 | Ras signaling pathway | 71 | 164 | 1.00e+00 |
| KEGG | KEGG:00340 | Histidine metabolism | 4 | 10 | 1.00e+00 |
| KEGG | KEGG:01523 | Antifolate resistance | 9 | 22 | 1.00e+00 |
| KEGG | KEGG:00524 | Neomycin, kanamycin and gentamicin biosynthesis | 1 | 4 | 1.00e+00 |
| KEGG | KEGG:00350 | Tyrosine metabolism | 6 | 11 | 1.00e+00 |
| KEGG | KEGG:00514 | Other types of O-glycan biosynthesis | 8 | 35 | 1.00e+00 |
| KEGG | KEGG:00513 | Various types of N-glycan biosynthesis | 6 | 37 | 1.00e+00 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 34 | 75 | 1.00e+00 |
| KEGG | KEGG:00532 | Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate | 7 | 20 | 1.00e+00 |
| KEGG | KEGG:03008 | Ribosome biogenesis in eukaryotes | 28 | 65 | 1.00e+00 |
| KEGG | KEGG:00650 | Butanoate metabolism | 7 | 17 | 1.00e+00 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 105 | 231 | 1.00e+00 |
| KEGG | KEGG:00512 | Mucin type O-glycan biosynthesis | 5 | 21 | 1.00e+00 |
| KEGG | KEGG:00860 | Porphyrin metabolism | 9 | 18 | 1.00e+00 |
| KEGG | KEGG:02010 | ABC transporters | 12 | 24 | 1.00e+00 |
| KEGG | KEGG:00520 | Amino sugar and nucleotide sugar metabolism | 14 | 44 | 1.00e+00 |
| KEGG | KEGG:00604 | Glycosphingolipid biosynthesis - ganglio series | 3 | 12 | 1.00e+00 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 72 | 162 | 1.00e+00 |
| KEGG | KEGG:00290 | Valine, leucine and isoleucine biosynthesis | 1 | 3 | 1.00e+00 |
| KEGG | KEGG:00601 | Glycosphingolipid biosynthesis - lacto and neolacto series | 6 | 14 | 1.00e+00 |
| KEGG | KEGG:01250 | Biosynthesis of nucleotide sugars | 10 | 34 | 1.00e+00 |
| KEGG | KEGG:01240 | Biosynthesis of cofactors | 45 | 109 | 1.00e+00 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 67 | 147 | 1.00e+00 |
| KEGG | KEGG:00563 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 10 | 23 | 1.00e+00 |
| KEGG | KEGG:00603 | Glycosphingolipid biosynthesis - globo and isoglobo series | 1 | 8 | 1.00e+00 |
| KEGG | KEGG:00310 | Lysine degradation | 20 | 56 | 1.00e+00 |
| KEGG | KEGG:01212 | Fatty acid metabolism | 15 | 46 | 1.00e+00 |
| KEGG | KEGG:00750 | Vitamin B6 metabolism | 3 | 5 | 1.00e+00 |
| KEGG | KEGG:00760 | Nicotinate and nicotinamide metabolism | 11 | 28 | 1.00e+00 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 58 | 129 | 1.00e+00 |
| KEGG | KEGG:00785 | Lipoic acid metabolism | 6 | 16 | 1.00e+00 |
| KEGG | KEGG:01232 | Nucleotide metabolism | 31 | 65 | 1.00e+00 |
| KEGG | KEGG:01230 | Biosynthesis of amino acids | 21 | 52 | 1.00e+00 |
| KEGG | KEGG:00790 | Folate biosynthesis | 5 | 18 | 1.00e+00 |
| KEGG | KEGG:01522 | Endocrine resistance | 38 | 82 | 1.00e+00 |
| KEGG | KEGG:04020 | Calcium signaling pathway | 70 | 161 | 1.00e+00 |
| KEGG | KEGG:00600 | Sphingolipid metabolism | 20 | 42 | 1.00e+00 |
| KEGG | KEGG:00640 | Propanoate metabolism | 8 | 27 | 1.00e+00 |
| KEGG | KEGG:03010 | Ribosome | 25 | 121 | 1.00e+00 |
| KEGG | KEGG:01210 | 2-Oxocarboxylic acid metabolism | 10 | 26 | 1.00e+00 |
| KEGG | KEGG:03020 | RNA polymerase | 12 | 33 | 1.00e+00 |
| KEGG | KEGG:01040 | Biosynthesis of unsaturated fatty acids | 10 | 23 | 1.00e+00 |
| KEGG | KEGG:03022 | Basal transcription factors | 16 | 33 | 1.00e+00 |
| KEGG | KEGG:03266 | Virion - Herpesvirus | 1 | 6 | 1.00e+00 |
| KEGG | KEGG:00983 | Drug metabolism - other enzymes | 15 | 40 | 1.00e+00 |
| KEGG | KEGG:00450 | Selenocompound metabolism | 6 | 14 | 1.00e+00 |
| KEGG | KEGG:00590 | Arachidonic acid metabolism | 8 | 25 | 1.00e+00 |
| KEGG | KEGG:00565 | Ether lipid metabolism | 10 | 28 | 1.00e+00 |
| KEGG | KEGG:00630 | Glyoxylate and dicarboxylate metabolism | 6 | 25 | 1.00e+00 |
| KEGG | KEGG:00982 | Drug metabolism - cytochrome P450 | 5 | 16 | 1.00e+00 |
| KEGG | KEGG:00592 | alpha-Linolenic acid metabolism | 4 | 10 | 1.00e+00 |
| KEGG | KEGG:03265 | Virion - Ebolavirus and Lyssavirus | 3 | 5 | 1.00e+00 |
| KEGG | KEGG:00534 | Glycosaminoglycan biosynthesis - heparan sulfate / heparin | 10 | 19 | 1.00e+00 |
| KEGG | KEGG:03040 | Spliceosome | 39 | 124 | 1.00e+00 |
| KEGG | KEGG:00980 | Metabolism of xenobiotics by cytochrome P450 | 8 | 23 | 1.00e+00 |
| KEGG | KEGG:00970 | Aminoacyl-tRNA biosynthesis | 19 | 42 | 1.00e+00 |
| KEGG | KEGG:00500 | Starch and sucrose metabolism | 8 | 22 | 1.00e+00 |
| KEGG | KEGG:03050 | Proteasome | 11 | 40 | 1.00e+00 |
| KEGG | KEGG:03082 | ATP-dependent chromatin remodeling | 43 | 91 | 1.00e+00 |
| KEGG | KEGG:00533 | Glycosaminoglycan biosynthesis - keratan sulfate | 4 | 13 | 1.00e+00 |
| KEGG | KEGG:00620 | Pyruvate metabolism | 9 | 31 | 1.00e+00 |
| KEGG | KEGG:01100 | Metabolic pathways | 445 | 1077 | 1.00e+00 |
| KEGG | KEGG:00440 | Phosphonate and phosphinate metabolism | 2 | 5 | 1.00e+00 |
| KEGG | KEGG:03013 | Nucleocytoplasmic transport | 44 | 96 | 1.00e+00 |
| KEGG | KEGG:00740 | Riboflavin metabolism | 3 | 6 | 1.00e+00 |
| KEGG | KEGG:03450 | Non-homologous end-joining | 6 | 12 | 1.00e+00 |
| KEGG | KEGG:00360 | Phenylalanine metabolism | 2 | 3 | 1.00e+00 |
| KEGG | KEGG:00900 | Terpenoid backbone biosynthesis | 6 | 21 | 1.00e+00 |
| KEGG | KEGG:04060 | Cytokine-cytokine receptor interaction | 40 | 94 | 1.00e+00 |
| KEGG | KEGG:00511 | Other glycan degradation | 2 | 14 | 1.00e+00 |
| KEGG | KEGG:00380 | Tryptophan metabolism | 3 | 21 | 1.00e+00 |
| KEGG | KEGG:03015 | mRNA surveillance pathway | 27 | 80 | 1.00e+00 |
| KEGG | KEGG:00670 | One carbon pool by folate | 7 | 18 | 1.00e+00 |
| KEGG | KEGG:00920 | Sulfur metabolism | 3 | 10 | 1.00e+00 |
| KEGG | KEGG:03018 | RNA degradation | 26 | 67 | 1.00e+00 |
| KEGG | KEGG:00730 | Thiamine metabolism | 4 | 10 | 1.00e+00 |
| KEGG | KEGG:00410 | beta-Alanine metabolism | 10 | 22 | 1.00e+00 |
| KEGG | KEGG:01200 | Carbon metabolism | 26 | 88 | 1.00e+00 |
| KEGG | KEGG:03320 | PPAR signaling pathway | 23 | 47 | 1.00e+00 |
| KEGG | KEGG:03267 | Virion - Adenovirus | 1 | 2 | 1.00e+00 |
| KEGG | KEGG:00430 | Taurine and hypotaurine metabolism | 3 | 8 | 1.00e+00 |
| KEGG | KEGG:00561 | Glycerolipid metabolism | 21 | 44 | 1.00e+00 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 11 | 49 | 1.00e+00 |
| KEGG | KEGG:00400 | Phenylalanine, tyrosine and tryptophan biosynthesis | 2 | 2 | 1.00e+00 |
| KEGG | KEGG:00470 | D-Amino acid metabolism | 2 | 4 | 1.00e+00 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 27 | 60 | 1.00e+00 |
| KEGG | KEGG:04062 | Chemokine signaling pathway | 41 | 109 | 1.00e+00 |
| KEGG | KEGG:04979 | Cholesterol metabolism | 11 | 33 | 1.00e+00 |
| KEGG | KEGG:05010 | Alzheimer disease | 102 | 296 | 1.00e+00 |
| KEGG | KEGG:05012 | Parkinson disease | 62 | 211 | 1.00e+00 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 113 | 291 | 1.00e+00 |
| KEGG | KEGG:05016 | Huntington disease | 82 | 246 | 1.00e+00 |
| KEGG | KEGG:05017 | Spinocerebellar ataxia | 46 | 120 | 1.00e+00 |
| KEGG | KEGG:05020 | Prion disease | 63 | 204 | 1.00e+00 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 137 | 364 | 1.00e+00 |
| KEGG | KEGG:05030 | Cocaine addiction | 16 | 32 | 1.00e+00 |
| KEGG | KEGG:05031 | Amphetamine addiction | 19 | 45 | 1.00e+00 |
| KEGG | KEGG:05033 | Nicotine addiction | 4 | 10 | 1.00e+00 |
| KEGG | KEGG:05100 | Bacterial invasion of epithelial cells | 26 | 73 | 1.00e+00 |
| KEGG | KEGG:05110 | Vibrio cholerae infection | 18 | 41 | 1.00e+00 |
| KEGG | KEGG:05120 | Epithelial cell signaling in Helicobacter pylori infection | 24 | 55 | 1.00e+00 |
| KEGG | KEGG:05130 | Pathogenic Escherichia coli infection | 59 | 143 | 1.00e+00 |
| KEGG | KEGG:05131 | Shigellosis | 78 | 204 | 1.00e+00 |
| KEGG | KEGG:05132 | Salmonella infection | 82 | 201 | 1.00e+00 |
| KEGG | KEGG:05133 | Pertussis | 15 | 42 | 1.00e+00 |
| KEGG | KEGG:05134 | Legionellosis | 8 | 34 | 1.00e+00 |
| KEGG | KEGG:04977 | Vitamin digestion and absorption | 6 | 13 | 1.00e+00 |
| KEGG | KEGG:05135 | Yersinia infection | 38 | 107 | 1.00e+00 |
| KEGG | KEGG:04975 | Fat digestion and absorption | 6 | 18 | 1.00e+00 |
| KEGG | KEGG:04973 | Carbohydrate digestion and absorption | 12 | 29 | 1.00e+00 |
| KEGG | KEGG:04924 | Renin secretion | 25 | 50 | 1.00e+00 |
| KEGG | KEGG:04925 | Aldosterone synthesis and secretion | 33 | 75 | 1.00e+00 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 36 | 100 | 1.00e+00 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 36 | 78 | 1.00e+00 |
| KEGG | KEGG:04929 | GnRH secretion | 20 | 50 | 1.00e+00 |
| KEGG | KEGG:04930 | Type II diabetes mellitus | 18 | 35 | 1.00e+00 |
| KEGG | KEGG:04931 | Insulin resistance | 45 | 91 | 1.00e+00 |
| KEGG | KEGG:04932 | Non-alcoholic fatty liver disease | 39 | 126 | 1.00e+00 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 36 | 87 | 1.00e+00 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 47 | 96 | 1.00e+00 |
| KEGG | KEGG:04936 | Alcoholic liver disease | 43 | 91 | 1.00e+00 |
| KEGG | KEGG:04950 | Maturity onset diabetes of the young | 2 | 4 | 1.00e+00 |
| KEGG | KEGG:04960 | Aldosterone-regulated sodium reabsorption | 13 | 24 | 1.00e+00 |
| KEGG | KEGG:04961 | Endocrine and other factor-regulated calcium reabsorption | 15 | 40 | 1.00e+00 |
| KEGG | KEGG:04962 | Vasopressin-regulated water reabsorption | 14 | 37 | 1.00e+00 |
| KEGG | KEGG:04964 | Proximal tubule bicarbonate reclamation | 9 | 18 | 1.00e+00 |
| KEGG | KEGG:04970 | Salivary secretion | 23 | 54 | 1.00e+00 |
| KEGG | KEGG:04971 | Gastric acid secretion | 22 | 51 | 1.00e+00 |
| KEGG | KEGG:04972 | Pancreatic secretion | 25 | 57 | 1.00e+00 |
| KEGG | KEGG:04974 | Protein digestion and absorption | 21 | 56 | 1.00e+00 |
| KEGG | KEGG:04922 | Glucagon signaling pathway | 29 | 78 | 1.00e+00 |
| KEGG | KEGG:05140 | Leishmaniasis | 13 | 38 | 1.00e+00 |
| KEGG | KEGG:05143 | African trypanosomiasis | 6 | 16 | 1.00e+00 |
| KEGG | KEGG:05215 | Prostate cancer | 38 | 82 | 1.00e+00 |
| KEGG | KEGG:05219 | Bladder cancer | 10 | 35 | 1.00e+00 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 34 | 72 | 1.00e+00 |
| KEGG | KEGG:05222 | Small cell lung cancer | 35 | 82 | 1.00e+00 |
| KEGG | KEGG:05230 | Central carbon metabolism in cancer | 19 | 58 | 1.00e+00 |
| KEGG | KEGG:05231 | Choline metabolism in cancer | 32 | 80 | 1.00e+00 |
| KEGG | KEGG:05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 28 | 65 | 1.00e+00 |
| KEGG | KEGG:05320 | Autoimmune thyroid disease | 1 | 3 | 1.00e+00 |
| KEGG | KEGG:05321 | Inflammatory bowel disease | 8 | 20 | 1.00e+00 |
| KEGG | KEGG:05322 | Systemic lupus erythematosus | 21 | 44 | 1.00e+00 |
| KEGG | KEGG:05323 | Rheumatoid arthritis | 18 | 38 | 1.00e+00 |
| KEGG | KEGG:05330 | Allograft rejection | 1 | 2 | 1.00e+00 |
| KEGG | KEGG:05332 | Graft-versus-host disease | 1 | 2 | 1.00e+00 |
| KEGG | KEGG:05340 | Primary immunodeficiency | 3 | 8 | 1.00e+00 |
| KEGG | KEGG:05410 | Hypertrophic cardiomyopathy | 30 | 77 | 1.00e+00 |
| KEGG | KEGG:05412 | Arrhythmogenic right ventricular cardiomyopathy | 29 | 67 | 1.00e+00 |
| KEGG | KEGG:05414 | Dilated cardiomyopathy | 31 | 82 | 1.00e+00 |
| KEGG | KEGG:05415 | Diabetic cardiomyopathy | 48 | 158 | 1.00e+00 |
| KEGG | KEGG:05416 | Viral myocarditis | 12 | 36 | 1.00e+00 |
| KEGG | KEGG:05214 | Glioma | 28 | 65 | 1.00e+00 |
| KEGG | KEGG:05142 | Chagas disease | 31 | 72 | 1.00e+00 |
| KEGG | KEGG:05212 | Pancreatic cancer | 35 | 71 | 1.00e+00 |
| KEGG | KEGG:05208 | Chemical carcinogenesis - reactive oxygen species | 46 | 173 | 1.00e+00 |
| KEGG | KEGG:05144 | Malaria | 6 | 20 | 1.00e+00 |
| KEGG | KEGG:05145 | Toxoplasmosis | 24 | 72 | 1.00e+00 |
| KEGG | KEGG:05146 | Amoebiasis | 23 | 62 | 1.00e+00 |
| KEGG | KEGG:05150 | Staphylococcus aureus infection | 4 | 13 | 1.00e+00 |
| KEGG | KEGG:05152 | Tuberculosis | 43 | 101 | 1.00e+00 |
| KEGG | KEGG:05160 | Hepatitis C | 50 | 110 | 1.00e+00 |
| KEGG | KEGG:05162 | Measles | 37 | 85 | 1.00e+00 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 68 | 160 | 1.00e+00 |
| KEGG | KEGG:05164 | Influenza A | 46 | 95 | 1.00e+00 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 107 | 252 | 1.00e+00 |
| KEGG | KEGG:05167 | Kaposi sarcoma-associated herpesvirus infection | 61 | 134 | 1.00e+00 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 177 | 388 | 1.00e+00 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 60 | 136 | 1.00e+00 |
| KEGG | KEGG:05170 | Human immunodeficiency virus 1 infection | 66 | 149 | 1.00e+00 |
| KEGG | KEGG:05171 | Coronavirus disease - COVID-19 | 34 | 149 | 1.00e+00 |
| KEGG | KEGG:05200 | Pathways in cancer | 174 | 391 | 1.00e+00 |
| KEGG | KEGG:05204 | Chemical carcinogenesis - DNA adducts | 6 | 20 | 1.00e+00 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 63 | 162 | 1.00e+00 |
| KEGG | KEGG:05207 | Chemical carcinogenesis - receptor activation | 59 | 132 | 1.00e+00 |
| KEGG | KEGG:05211 | Renal cell carcinoma | 25 | 62 | 1.00e+00 |
| KEGG | KEGG:04061 | Viral protein interaction with cytokine and cytokine receptor | 7 | 19 | 1.00e+00 |
| KEGG | KEGG:04921 | Oxytocin signaling pathway | 44 | 113 | 1.00e+00 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 50 | 105 | 1.00e+00 |
| KEGG | KEGG:04260 | Cardiac muscle contraction | 22 | 63 | 1.00e+00 |
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 55 | 124 | 1.00e+00 |
| KEGG | KEGG:04270 | Vascular smooth muscle contraction | 31 | 92 | 1.00e+00 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 64 | 133 | 1.00e+00 |
| KEGG | KEGG:04330 | Notch signaling pathway | 25 | 52 | 1.00e+00 |
| KEGG | KEGG:04340 | Hedgehog signaling pathway | 20 | 49 | 1.00e+00 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 41 | 90 | 1.00e+00 |
| KEGG | KEGG:04360 | Axon guidance | 68 | 155 | 1.00e+00 |
| KEGG | KEGG:04370 | VEGF signaling pathway | 18 | 47 | 1.00e+00 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 38 | 104 | 1.00e+00 |
| KEGG | KEGG:04380 | Osteoclast differentiation | 43 | 92 | 1.00e+00 |
| KEGG | KEGG:04392 | Hippo signaling pathway - multiple species | 13 | 25 | 1.00e+00 |
| KEGG | KEGG:04510 | Focal adhesion | 48 | 169 | 1.00e+00 |
| KEGG | KEGG:04512 | ECM-receptor interaction | 13 | 66 | 1.00e+00 |
| KEGG | KEGG:04514 | Cell adhesion molecules | 24 | 69 | 1.00e+00 |
| KEGG | KEGG:04520 | Adherens junction | 31 | 83 | 1.00e+00 |
| KEGG | KEGG:04530 | Tight junction | 52 | 129 | 1.00e+00 |
| KEGG | KEGG:04540 | Gap junction | 34 | 71 | 1.00e+00 |
| KEGG | KEGG:04610 | Complement and coagulation cascades | 9 | 35 | 1.00e+00 |
| KEGG | KEGG:04218 | Cellular senescence | 64 | 136 | 1.00e+00 |
| KEGG | KEGG:04611 | Platelet activation | 42 | 88 | 1.00e+00 |
| KEGG | KEGG:04215 | Apoptosis - multiple species | 14 | 29 | 1.00e+00 |
| KEGG | KEGG:04211 | Longevity regulating pathway | 35 | 75 | 1.00e+00 |
| KEGG | KEGG:04064 | NF-kappa B signaling pathway | 29 | 61 | 1.00e+00 |
| KEGG | KEGG:04066 | HIF-1 signaling pathway | 28 | 86 | 1.00e+00 |
| KEGG | KEGG:04071 | Sphingolipid signaling pathway | 43 | 103 | 1.00e+00 |
| KEGG | KEGG:04072 | Phospholipase D signaling pathway | 47 | 107 | 1.00e+00 |
| KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 42 | 101 | 1.00e+00 |
| KEGG | KEGG:04114 | Oocyte meiosis | 49 | 103 | 1.00e+00 |
| KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 51 | 127 | 1.00e+00 |
| KEGG | KEGG:04122 | Sulfur relay system | 3 | 8 | 1.00e+00 |
| KEGG | KEGG:04130 | SNARE interactions in vesicular transport | 14 | 31 | 1.00e+00 |
| KEGG | KEGG:04136 | Autophagy - other | 14 | 29 | 1.00e+00 |
| KEGG | KEGG:04137 | Mitophagy - animal | 36 | 94 | 1.00e+00 |
| KEGG | KEGG:04140 | Autophagy - animal | 70 | 149 | 1.00e+00 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 46 | 149 | 1.00e+00 |
| KEGG | KEGG:04142 | Lysosome | 32 | 109 | 1.00e+00 |
| KEGG | KEGG:04144 | Endocytosis | 92 | 216 | 1.00e+00 |
| KEGG | KEGG:04145 | Phagosome | 34 | 84 | 1.00e+00 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 60 | 129 | 1.00e+00 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 97 | 250 | 1.00e+00 |
| KEGG | KEGG:04152 | AMPK signaling pathway | 47 | 101 | 1.00e+00 |
| KEGG | KEGG:04213 | Longevity regulating pathway - multiple species | 22 | 50 | 1.00e+00 |
| KEGG | KEGG:04920 | Adipocytokine signaling pathway | 20 | 49 | 1.00e+00 |
| KEGG | KEGG:04612 | Antigen processing and presentation | 8 | 26 | 1.00e+00 |
| KEGG | KEGG:04614 | Renin-angiotensin system | 2 | 10 | 1.00e+00 |
| KEGG | KEGG:04723 | Retrograde endocannabinoid signaling | 32 | 92 | 1.00e+00 |
| KEGG | KEGG:04724 | Glutamatergic synapse | 34 | 74 | 1.00e+00 |
| KEGG | KEGG:04725 | Cholinergic synapse | 38 | 82 | 1.00e+00 |
| KEGG | KEGG:04726 | Serotonergic synapse | 26 | 58 | 1.00e+00 |
| KEGG | KEGG:04727 | GABAergic synapse | 26 | 52 | 1.00e+00 |
| KEGG | KEGG:04728 | Dopaminergic synapse | 37 | 97 | 1.00e+00 |
| KEGG | KEGG:04730 | Long-term depression | 19 | 39 | 1.00e+00 |
| KEGG | KEGG:04740 | Olfactory transduction | 11 | 25 | 1.00e+00 |
| KEGG | KEGG:04742 | Taste transduction | 10 | 20 | 1.00e+00 |
| KEGG | KEGG:04744 | Phototransduction | 6 | 12 | 1.00e+00 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 58 | 169 | 1.00e+00 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 53 | 113 | 1.00e+00 |
| KEGG | KEGG:04911 | Insulin secretion | 29 | 62 | 1.00e+00 |
| KEGG | KEGG:04912 | GnRH signaling pathway | 29 | 74 | 1.00e+00 |
| KEGG | KEGG:04913 | Ovarian steroidogenesis | 14 | 26 | 1.00e+00 |
| KEGG | KEGG:04915 | Estrogen signaling pathway | 36 | 90 | 1.00e+00 |
| KEGG | KEGG:04916 | Melanogenesis | 32 | 75 | 1.00e+00 |
| KEGG | KEGG:04917 | Prolactin signaling pathway | 26 | 54 | 1.00e+00 |
| KEGG | KEGG:04918 | Thyroid hormone synthesis | 23 | 53 | 1.00e+00 |
| KEGG | KEGG:04722 | Neurotrophin signaling pathway | 37 | 104 | 1.00e+00 |
| KEGG | KEGG:04613 | Neutrophil extracellular trap formation | 40 | 89 | 1.00e+00 |
| KEGG | KEGG:04721 | Synaptic vesicle cycle | 23 | 49 | 1.00e+00 |
| KEGG | KEGG:04714 | Thermogenesis | 54 | 185 | 1.00e+00 |
| KEGG | KEGG:04620 | Toll-like receptor signaling pathway | 24 | 60 | 1.00e+00 |
| KEGG | KEGG:04621 | NOD-like receptor signaling pathway | 41 | 109 | 1.00e+00 |
| KEGG | KEGG:04622 | RIG-I-like receptor signaling pathway | 14 | 44 | 1.00e+00 |
| KEGG | KEGG:04623 | Cytosolic DNA-sensing pathway | 17 | 46 | 1.00e+00 |
| KEGG | KEGG:04625 | C-type lectin receptor signaling pathway | 34 | 74 | 1.00e+00 |
| KEGG | KEGG:04630 | JAK-STAT signaling pathway | 37 | 83 | 1.00e+00 |
| KEGG | KEGG:04640 | Hematopoietic cell lineage | 13 | 28 | 1.00e+00 |
| KEGG | KEGG:04650 | Natural killer cell mediated cytotoxicity | 23 | 53 | 1.00e+00 |
| KEGG | KEGG:04657 | IL-17 signaling pathway | 21 | 55 | 1.00e+00 |
| KEGG | KEGG:04658 | Th1 and Th2 cell differentiation | 23 | 50 | 1.00e+00 |
| KEGG | KEGG:00280 | Valine, leucine and isoleucine degradation | 14 | 42 | 1.00e+00 |
| KEGG | KEGG:04660 | T cell receptor signaling pathway | 32 | 84 | 1.00e+00 |
| KEGG | KEGG:04662 | B cell receptor signaling pathway | 19 | 52 | 1.00e+00 |
| KEGG | KEGG:04664 | Fc epsilon RI signaling pathway | 14 | 42 | 1.00e+00 |
| KEGG | KEGG:04666 | Fc gamma R-mediated phagocytosis | 29 | 75 | 1.00e+00 |
| KEGG | KEGG:04670 | Leukocyte transendothelial migration | 26 | 76 | 1.00e+00 |
| KEGG | KEGG:04672 | Intestinal immune network for IgA production | 3 | 10 | 1.00e+00 |
| KEGG | KEGG:04710 | Circadian rhythm | 13 | 29 | 1.00e+00 |
| KEGG | KEGG:04713 | Circadian entrainment | 31 | 68 | 1.00e+00 |
| KEGG | KEGG:04720 | Long-term potentiation | 21 | 51 | 1.00e+00 |
| KEGG | KEGG:00270 | Cysteine and methionine metabolism | 17 | 37 | 1.00e+00 |
| KEGG | KEGG:05418 | Fluid shear stress and atherosclerosis | 41 | 109 | 1.00e+00 |
| KEGG | KEGG:00250 | Alanine, aspartate and glutamate metabolism | 11 | 28 | 1.00e+00 |
| KEGG | KEGG:05417 | Lipid and atherosclerosis | 59 | 143 | 1.00e+00 |
| KEGG | KEGG:00260 | Glycine, serine and threonine metabolism | 7 | 26 | 1.00e+00 |
| KEGG | KEGG:00000 | KEGG root term | 2405 | 5451 | 1.00e+00 |
| KEGG | KEGG:00020 | Citrate cycle (TCA cycle) | 7 | 26 | 1.00e+00 |
| KEGG | KEGG:00030 | Pentose phosphate pathway | 9 | 20 | 1.00e+00 |
| KEGG | KEGG:00040 | Pentose and glucuronate interconversions | 2 | 11 | 1.00e+00 |
| KEGG | KEGG:00051 | Fructose and mannose metabolism | 6 | 26 | 1.00e+00 |
| KEGG | KEGG:00052 | Galactose metabolism | 7 | 21 | 1.00e+00 |
| KEGG | KEGG:00053 | Ascorbate and aldarate metabolism | 2 | 8 | 1.00e+00 |
| KEGG | KEGG:00061 | Fatty acid biosynthesis | 3 | 9 | 1.00e+00 |
| KEGG | KEGG:00010 | Glycolysis / Gluconeogenesis | 10 | 41 | 1.00e+00 |
| KEGG | KEGG:00071 | Fatty acid degradation | 8 | 31 | 1.00e+00 |
| KEGG | KEGG:00100 | Steroid biosynthesis | 7 | 15 | 1.00e+00 |
| KEGG | KEGG:00120 | Primary bile acid biosynthesis | 4 | 11 | 1.00e+00 |
| KEGG | KEGG:00130 | Ubiquinone and other terpenoid-quinone biosynthesis | 1 | 9 | 1.00e+00 |
| KEGG | KEGG:00140 | Steroid hormone biosynthesis | 5 | 16 | 1.00e+00 |
| KEGG | KEGG:00190 | Oxidative phosphorylation | 22 | 99 | 1.00e+00 |
| KEGG | KEGG:00240 | Pyrimidine metabolism | 21 | 44 | 1.00e+00 |
| KEGG | KEGG:00062 | Fatty acid elongation | 7 | 22 | 1.00e+00 |
| KEGG | KEGG:00232 | Caffeine metabolism | 1 | 1 | 1.00e+00 |
# intersect(unique(Resp_RNA$ENTREZID), unique(NR_peak_list_20k$ENTREZID)))
NR_int_restable %>%
filter(source=="GO:BP", p_value < 0.05) %>%
slice_head(n=10) %>%
mutate(log10_p=(-log(p_value, base=10))) %>%
ggplot(., aes(x=log10_p, y=reorder(term_name, log10_p)))+
geom_col(fill="#C77CFF")+
theme_bw()+
ylab("")+
xlab(paste0("-log10 p-value"))+
guides(fill="none")+
theme(axis.text.y = element_text(color="black"))+
ggtitle("Intersection NR")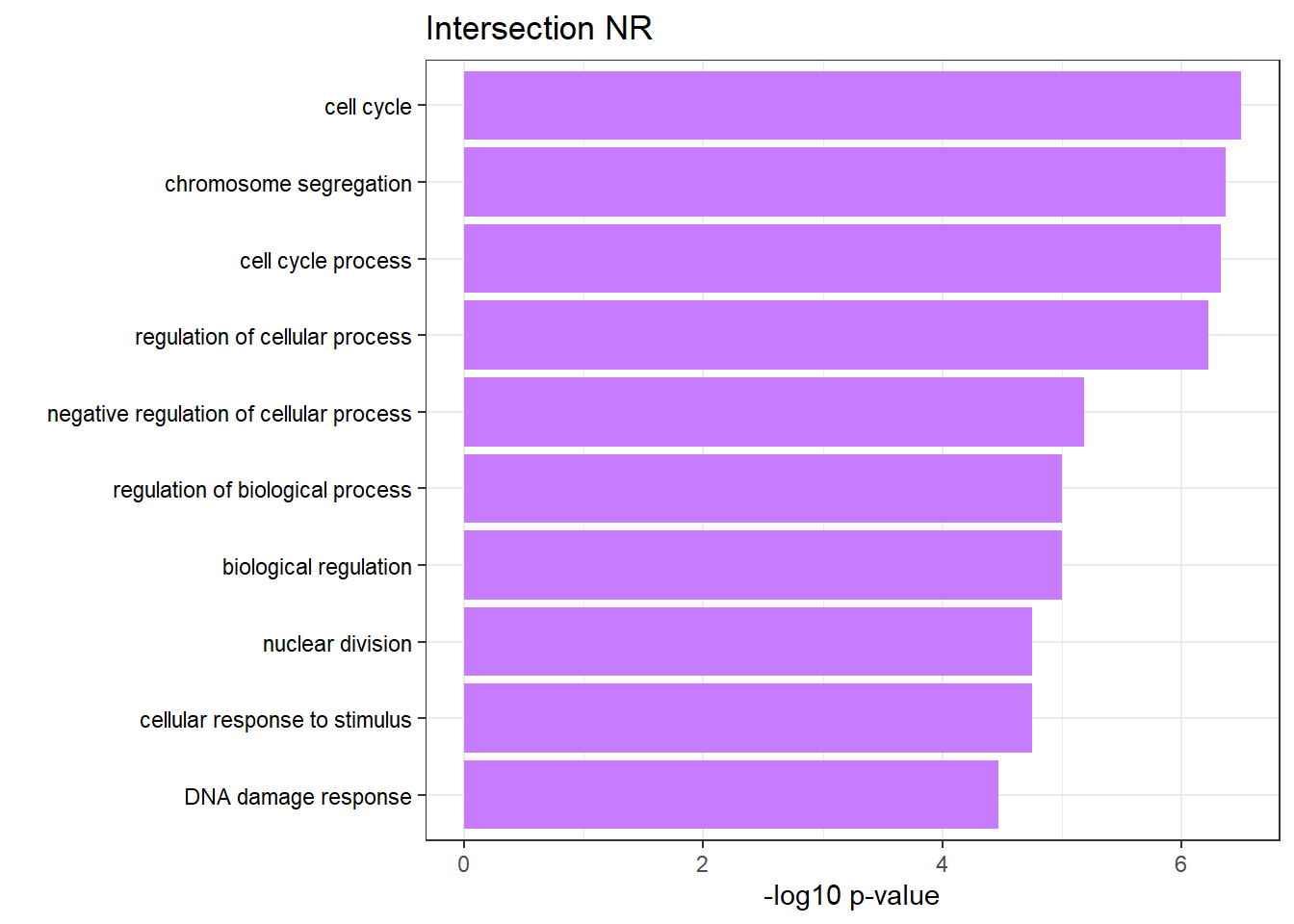
not intersected NR
not_NR_int_resgenes <- gost(query = setdiff(NR_NG_list$ENTREZID, Resp_RNA$ENTREZID),
organism = "hsapiens",
significant = FALSE,
ordered_query = FALSE,
domain_scope = "custom",
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
custom_bg = background_df$ENTREZID,
sources=c("GO:BP","KEGG"))
saveRDS(not_NR_int_resgenes,"data/GO_KEGG_analysis/not_NR_int_resgenes.RDS")
not_NR_int_restable <- not_NR_int_resgenes$result %>%
dplyr::select(c(source, term_id,
term_name,intersection_size,
term_size, p_value))# %>%
not_NR_int_restable %>%
filter(source=="GO:BP", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(.,caption = "GO:BP terms found in NR neargenes intersected with RNA response genes" ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:1901564 | organonitrogen compound metabolic process | 2339 | 4354 | 5.62e-04 |
| GO:BP | GO:0002181 | cytoplasmic translation | 99 | 138 | 1.51e-03 |
| GO:BP | GO:0019538 | protein metabolic process | 1946 | 3619 | 2.62e-03 |
| GO:BP | GO:0009060 | aerobic respiration | 108 | 156 | 2.62e-03 |
| GO:BP | GO:0022904 | respiratory electron transport chain | 73 | 98 | 2.62e-03 |
| GO:BP | GO:0019646 | aerobic electron transport chain | 55 | 70 | 2.62e-03 |
| GO:BP | GO:0006119 | oxidative phosphorylation | 82 | 114 | 2.64e-03 |
| GO:BP | GO:1901566 | organonitrogen compound biosynthetic process | 772 | 1369 | 2.64e-03 |
| GO:BP | GO:0045333 | cellular respiration | 132 | 198 | 2.64e-03 |
| GO:BP | GO:0042775 | mitochondrial ATP synthesis coupled electron transport | 59 | 77 | 2.64e-03 |
| GO:BP | GO:0042773 | ATP synthesis coupled electron transport | 59 | 77 | 2.64e-03 |
| GO:BP | GO:0043603 | amide metabolic process | 535 | 928 | 2.85e-03 |
| GO:BP | GO:0009100 | glycoprotein metabolic process | 179 | 285 | 1.32e-02 |
| GO:BP | GO:0006518 | peptide metabolic process | 425 | 734 | 1.43e-02 |
| GO:BP | GO:0006412 | translation | 347 | 590 | 1.43e-02 |
| GO:BP | GO:0043413 | macromolecule glycosylation | 116 | 176 | 1.53e-02 |
| GO:BP | GO:0006486 | protein glycosylation | 116 | 176 | 1.53e-02 |
| GO:BP | GO:0022900 | electron transport chain | 90 | 132 | 1.69e-02 |
| GO:BP | GO:0070085 | glycosylation | 122 | 187 | 1.74e-02 |
| GO:BP | GO:0006487 | protein N-linked glycosylation | 47 | 62 | 2.23e-02 |
| GO:BP | GO:0043604 | amide biosynthetic process | 406 | 705 | 3.07e-02 |
| GO:BP | GO:0043043 | peptide biosynthetic process | 354 | 609 | 3.17e-02 |
not_NR_int_restable %>%
filter(source=="KEGG") %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "KEGG terms found in NR neargenes intersected with RNA response genes") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| KEGG | KEGG:04510 | Focal adhesion | 117 | 169 | 8.46e-05 |
| KEGG | KEGG:05171 | Coronavirus disease - COVID-19 | 105 | 149 | 8.46e-05 |
| KEGG | KEGG:03010 | Ribosome | 87 | 121 | 1.34e-04 |
| KEGG | KEGG:04512 | ECM-receptor interaction | 51 | 66 | 5.89e-04 |
| KEGG | KEGG:05208 | Chemical carcinogenesis - reactive oxygen species | 115 | 173 | 9.19e-04 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 100 | 149 | 1.51e-03 |
| KEGG | KEGG:00190 | Oxidative phosphorylation | 70 | 99 | 1.59e-03 |
| KEGG | KEGG:04714 | Thermogenesis | 119 | 185 | 3.85e-03 |
| KEGG | KEGG:05012 | Parkinson disease | 133 | 211 | 5.40e-03 |
| KEGG | KEGG:05020 | Prion disease | 128 | 204 | 8.56e-03 |
| KEGG | KEGG:00513 | Various types of N-glycan biosynthesis | 29 | 37 | 1.45e-02 |
| KEGG | KEGG:00531 | Glycosaminoglycan degradation | 14 | 15 | 1.61e-02 |
| KEGG | KEGG:05016 | Huntington disease | 149 | 246 | 2.20e-02 |
| KEGG | KEGG:05010 | Alzheimer disease | 176 | 296 | 2.54e-02 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 105 | 169 | 2.86e-02 |
| KEGG | KEGG:04142 | Lysosome | 71 | 109 | 2.86e-02 |
| KEGG | KEGG:05415 | Diabetic cardiomyopathy | 99 | 158 | 2.86e-02 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 35 | 49 | 4.48e-02 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 210 | 364 | 5.42e-02 |
| KEGG | KEGG:01100 | Metabolic pathways | 582 | 1077 | 1.25e-01 |
| KEGG | KEGG:00511 | Other glycan degradation | 12 | 14 | 1.25e-01 |
| KEGG | KEGG:00514 | Other types of O-glycan biosynthesis | 25 | 35 | 1.35e-01 |
| KEGG | KEGG:05219 | Bladder cancer | 25 | 35 | 1.35e-01 |
| KEGG | KEGG:01200 | Carbon metabolism | 56 | 88 | 1.35e-01 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 145 | 250 | 1.35e-01 |
| KEGG | KEGG:04932 | Non-alcoholic fatty liver disease | 77 | 126 | 1.37e-01 |
| KEGG | KEGG:05100 | Bacterial invasion of epithelial cells | 47 | 73 | 1.47e-01 |
| KEGG | KEGG:00520 | Amino sugar and nucleotide sugar metabolism | 30 | 44 | 1.65e-01 |
| KEGG | KEGG:01250 | Biosynthesis of nucleotide sugars | 24 | 34 | 1.69e-01 |
| KEGG | KEGG:00051 | Fructose and mannose metabolism | 19 | 26 | 1.84e-01 |
| KEGG | KEGG:00010 | Glycolysis / Gluconeogenesis | 28 | 41 | 1.84e-01 |
| KEGG | KEGG:00620 | Pyruvate metabolism | 22 | 31 | 1.84e-01 |
| KEGG | KEGG:03015 | mRNA surveillance pathway | 50 | 80 | 2.12e-01 |
| KEGG | KEGG:05414 | Dilated cardiomyopathy | 51 | 82 | 2.16e-01 |
| KEGG | KEGG:04530 | Tight junction | 77 | 129 | 2.16e-01 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 63 | 104 | 2.29e-01 |
| KEGG | KEGG:04066 | HIF-1 signaling pathway | 53 | 86 | 2.29e-01 |
| KEGG | KEGG:04270 | Vascular smooth muscle contraction | 56 | 92 | 2.51e-01 |
| KEGG | KEGG:04520 | Adherens junction | 51 | 83 | 2.51e-01 |
| KEGG | KEGG:04723 | Retrograde endocannabinoid signaling | 56 | 92 | 2.51e-01 |
| KEGG | KEGG:04670 | Leukocyte transendothelial migration | 47 | 76 | 2.58e-01 |
| KEGG | KEGG:05134 | Legionellosis | 23 | 34 | 2.66e-01 |
| KEGG | KEGG:05135 | Yersinia infection | 64 | 107 | 2.66e-01 |
| KEGG | KEGG:00040 | Pentose and glucuronate interconversions | 9 | 11 | 2.81e-01 |
| KEGG | KEGG:00603 | Glycosphingolipid biosynthesis - globo and isoglobo series | 7 | 8 | 2.93e-01 |
| KEGG | KEGG:00071 | Fatty acid degradation | 21 | 31 | 2.99e-01 |
| KEGG | KEGG:05410 | Hypertrophic cardiomyopathy | 47 | 77 | 2.99e-01 |
| KEGG | KEGG:00380 | Tryptophan metabolism | 15 | 21 | 3.03e-01 |
| KEGG | KEGG:00512 | Mucin type O-glycan biosynthesis | 15 | 21 | 3.03e-01 |
| KEGG | KEGG:00020 | Citrate cycle (TCA cycle) | 18 | 26 | 3.03e-01 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 93 | 162 | 3.05e-01 |
| KEGG | KEGG:03060 | Protein export | 19 | 28 | 3.27e-01 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 59 | 100 | 3.28e-01 |
| KEGG | KEGG:04514 | Cell adhesion molecules | 42 | 69 | 3.28e-01 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 161 | 291 | 3.28e-01 |
| KEGG | KEGG:00280 | Valine, leucine and isoleucine degradation | 27 | 42 | 3.28e-01 |
| KEGG | KEGG:00790 | Folate biosynthesis | 13 | 18 | 3.28e-01 |
| KEGG | KEGG:04921 | Oxytocin signaling pathway | 66 | 113 | 3.30e-01 |
| KEGG | KEGG:05146 | Amoebiasis | 38 | 62 | 3.37e-01 |
| KEGG | KEGG:01212 | Fatty acid metabolism | 29 | 46 | 3.45e-01 |
| KEGG | KEGG:00640 | Propanoate metabolism | 18 | 27 | 3.88e-01 |
| KEGG | KEGG:04260 | Cardiac muscle contraction | 38 | 63 | 4.10e-01 |
| KEGG | KEGG:05416 | Viral myocarditis | 23 | 36 | 4.10e-01 |
| KEGG | KEGG:05132 | Salmonella infection | 112 | 201 | 4.10e-01 |
| KEGG | KEGG:04912 | GnRH signaling pathway | 44 | 74 | 4.10e-01 |
| KEGG | KEGG:00604 | Glycosphingolipid biosynthesis - ganglio series | 9 | 12 | 4.16e-01 |
| KEGG | KEGG:00310 | Lysine degradation | 34 | 56 | 4.21e-01 |
| KEGG | KEGG:05230 | Central carbon metabolism in cancer | 35 | 58 | 4.29e-01 |
| KEGG | KEGG:04722 | Neurotrophin signaling pathway | 60 | 104 | 4.29e-01 |
| KEGG | KEGG:03050 | Proteasome | 25 | 40 | 4.31e-01 |
| KEGG | KEGG:05131 | Shigellosis | 113 | 204 | 4.38e-01 |
| KEGG | KEGG:04612 | Antigen processing and presentation | 17 | 26 | 4.50e-01 |
| KEGG | KEGG:00130 | Ubiquinone and other terpenoid-quinone biosynthesis | 7 | 9 | 4.58e-01 |
| KEGG | KEGG:04610 | Complement and coagulation cascades | 22 | 35 | 4.61e-01 |
| KEGG | KEGG:00900 | Terpenoid backbone biosynthesis | 14 | 21 | 4.71e-01 |
| KEGG | KEGG:03040 | Spliceosome | 70 | 124 | 4.71e-01 |
| KEGG | KEGG:00000 | KEGG root term | 2785 | 5451 | 4.71e-01 |
| KEGG | KEGG:00052 | Galactose metabolism | 14 | 21 | 4.71e-01 |
| KEGG | KEGG:05418 | Fluid shear stress and atherosclerosis | 62 | 109 | 4.71e-01 |
| KEGG | KEGG:05145 | Toxoplasmosis | 42 | 72 | 4.89e-01 |
| KEGG | KEGG:00140 | Steroid hormone biosynthesis | 11 | 16 | 4.89e-01 |
| KEGG | KEGG:03266 | Virion - Herpesvirus | 5 | 6 | 4.92e-01 |
| KEGG | KEGG:04144 | Endocytosis | 118 | 216 | 5.15e-01 |
| KEGG | KEGG:05412 | Arrhythmogenic right ventricular cardiomyopathy | 39 | 67 | 5.20e-01 |
| KEGG | KEGG:05231 | Choline metabolism in cancer | 46 | 80 | 5.20e-01 |
| KEGG | KEGG:04974 | Protein digestion and absorption | 33 | 56 | 5.24e-01 |
| KEGG | KEGG:00780 | Biotin metabolism | 3 | 3 | 5.24e-01 |
| KEGG | KEGG:04660 | T cell receptor signaling pathway | 48 | 84 | 5.27e-01 |
| KEGG | KEGG:04720 | Long-term potentiation | 30 | 51 | 5.30e-01 |
| KEGG | KEGG:05130 | Pathogenic Escherichia coli infection | 79 | 143 | 5.30e-01 |
| KEGG | KEGG:05204 | Chemical carcinogenesis - DNA adducts | 13 | 20 | 5.30e-01 |
| KEGG | KEGG:04961 | Endocrine and other factor-regulated calcium reabsorption | 24 | 40 | 5.30e-01 |
| KEGG | KEGG:00533 | Glycosaminoglycan biosynthesis - keratan sulfate | 9 | 13 | 5.30e-01 |
| KEGG | KEGG:04062 | Chemokine signaling pathway | 61 | 109 | 5.30e-01 |
| KEGG | KEGG:05144 | Malaria | 13 | 20 | 5.30e-01 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 136 | 252 | 5.30e-01 |
| KEGG | KEGG:05200 | Pathways in cancer | 208 | 391 | 5.30e-01 |
| KEGG | KEGG:04666 | Fc gamma R-mediated phagocytosis | 43 | 75 | 5.30e-01 |
| KEGG | KEGG:00053 | Ascorbate and aldarate metabolism | 6 | 8 | 5.39e-01 |
| KEGG | KEGG:04664 | Fc epsilon RI signaling pathway | 25 | 42 | 5.43e-01 |
| KEGG | KEGG:04657 | IL-17 signaling pathway | 32 | 55 | 5.45e-01 |
| KEGG | KEGG:04622 | RIG-I-like receptor signaling pathway | 26 | 44 | 5.51e-01 |
| KEGG | KEGG:04979 | Cholesterol metabolism | 20 | 33 | 5.51e-01 |
| KEGG | KEGG:04728 | Dopaminergic synapse | 54 | 97 | 5.84e-01 |
| KEGG | KEGG:05417 | Lipid and atherosclerosis | 78 | 143 | 5.84e-01 |
| KEGG | KEGG:04929 | GnRH secretion | 29 | 50 | 5.84e-01 |
| KEGG | KEGG:03018 | RNA degradation | 38 | 67 | 5.84e-01 |
| KEGG | KEGG:04145 | Phagosome | 47 | 84 | 5.84e-01 |
| KEGG | KEGG:00260 | Glycine, serine and threonine metabolism | 16 | 26 | 5.84e-01 |
| KEGG | KEGG:04614 | Renin-angiotensin system | 7 | 10 | 5.84e-01 |
| KEGG | KEGG:05017 | Spinocerebellar ataxia | 66 | 120 | 5.84e-01 |
| KEGG | KEGG:04071 | Sphingolipid signaling pathway | 57 | 103 | 5.84e-01 |
| KEGG | KEGG:04915 | Estrogen signaling pathway | 50 | 90 | 6.06e-01 |
| KEGG | KEGG:04360 | Axon guidance | 84 | 155 | 6.08e-01 |
| KEGG | KEGG:04137 | Mitophagy - animal | 52 | 94 | 6.14e-01 |
| KEGG | KEGG:05211 | Renal cell carcinoma | 35 | 62 | 6.30e-01 |
| KEGG | KEGG:04340 | Hedgehog signaling pathway | 28 | 49 | 6.40e-01 |
| KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 69 | 127 | 6.40e-01 |
| KEGG | KEGG:00980 | Metabolism of xenobiotics by cytochrome P450 | 14 | 23 | 6.40e-01 |
| KEGG | KEGG:00630 | Glyoxylate and dicarboxylate metabolism | 15 | 25 | 6.66e-01 |
| KEGG | KEGG:00982 | Drug metabolism - cytochrome P450 | 10 | 16 | 6.99e-01 |
| KEGG | KEGG:04072 | Phospholipase D signaling pathway | 58 | 107 | 7.19e-01 |
| KEGG | KEGG:01240 | Biosynthesis of cofactors | 59 | 109 | 7.20e-01 |
| KEGG | KEGG:04975 | Fat digestion and absorption | 11 | 18 | 7.22e-01 |
| KEGG | KEGG:03271 | Virion - Rotavirus | 2 | 2 | 7.23e-01 |
| KEGG | KEGG:03020 | RNA polymerase | 19 | 33 | 7.28e-01 |
| KEGG | KEGG:04060 | Cytokine-cytokine receptor interaction | 51 | 94 | 7.28e-01 |
| KEGG | KEGG:00061 | Fatty acid biosynthesis | 6 | 9 | 7.28e-01 |
| KEGG | KEGG:04962 | Vasopressin-regulated water reabsorption | 21 | 37 | 7.41e-01 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 85 | 160 | 7.41e-01 |
| KEGG | KEGG:00062 | Fatty acid elongation | 13 | 22 | 7.41e-01 |
| KEGG | KEGG:00500 | Starch and sucrose metabolism | 13 | 22 | 7.41e-01 |
| KEGG | KEGG:04726 | Serotonergic synapse | 32 | 58 | 7.41e-01 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 47 | 87 | 7.55e-01 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 121 | 231 | 7.55e-01 |
| KEGG | KEGG:05110 | Vibrio cholerae infection | 23 | 41 | 7.55e-01 |
| KEGG | KEGG:04370 | VEGF signaling pathway | 26 | 47 | 7.55e-01 |
| KEGG | KEGG:04713 | Circadian entrainment | 37 | 68 | 7.55e-01 |
| KEGG | KEGG:00760 | Nicotinate and nicotinamide metabolism | 16 | 28 | 7.55e-01 |
| KEGG | KEGG:05150 | Staphylococcus aureus infection | 8 | 13 | 7.55e-01 |
| KEGG | KEGG:00565 | Ether lipid metabolism | 16 | 28 | 7.55e-01 |
| KEGG | KEGG:00250 | Alanine, aspartate and glutamate metabolism | 16 | 28 | 7.55e-01 |
| KEGG | KEGG:04650 | Natural killer cell mediated cytotoxicity | 29 | 53 | 7.64e-01 |
| KEGG | KEGG:04922 | Glucagon signaling pathway | 42 | 78 | 7.64e-01 |
| KEGG | KEGG:04971 | Gastric acid secretion | 28 | 51 | 7.64e-01 |
| KEGG | KEGG:04621 | NOD-like receptor signaling pathway | 58 | 109 | 7.64e-01 |
| KEGG | KEGG:00524 | Neomycin, kanamycin and gentamicin biosynthesis | 3 | 4 | 7.71e-01 |
| KEGG | KEGG:04972 | Pancreatic secretion | 31 | 57 | 7.72e-01 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 48 | 90 | 7.83e-01 |
| KEGG | KEGG:05140 | Leishmaniasis | 21 | 38 | 7.83e-01 |
| KEGG | KEGG:04014 | Ras signaling pathway | 86 | 164 | 7.83e-01 |
| KEGG | KEGG:03008 | Ribosome biogenesis in eukaryotes | 35 | 65 | 7.83e-01 |
| KEGG | KEGG:05133 | Pertussis | 23 | 42 | 7.95e-01 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 40 | 75 | 8.01e-01 |
| KEGG | KEGG:04916 | Melanogenesis | 40 | 75 | 8.01e-01 |
| KEGG | KEGG:00563 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 13 | 23 | 8.07e-01 |
| KEGG | KEGG:04662 | B cell receptor signaling pathway | 28 | 52 | 8.09e-01 |
| KEGG | KEGG:04630 | JAK-STAT signaling pathway | 44 | 83 | 8.09e-01 |
| KEGG | KEGG:00590 | Arachidonic acid metabolism | 14 | 25 | 8.09e-01 |
| KEGG | KEGG:04970 | Salivary secretion | 29 | 54 | 8.12e-01 |
| KEGG | KEGG:04710 | Circadian rhythm | 16 | 29 | 8.24e-01 |
| KEGG | KEGG:00340 | Histidine metabolism | 6 | 10 | 8.50e-01 |
| KEGG | KEGG:04724 | Glutamatergic synapse | 39 | 74 | 8.55e-01 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 41 | 78 | 8.59e-01 |
| KEGG | KEGG:05222 | Small cell lung cancer | 43 | 82 | 8.62e-01 |
| KEGG | KEGG:00601 | Glycosphingolipid biosynthesis - lacto and neolacto series | 8 | 14 | 8.73e-01 |
| KEGG | KEGG:05120 | Epithelial cell signaling in Helicobacter pylori infection | 29 | 55 | 8.87e-01 |
| KEGG | KEGG:04742 | Taste transduction | 11 | 20 | 8.87e-01 |
| KEGG | KEGG:00670 | One carbon pool by folate | 10 | 18 | 8.87e-01 |
| KEGG | KEGG:00532 | Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate | 11 | 20 | 8.87e-01 |
| KEGG | KEGG:01523 | Antifolate resistance | 12 | 22 | 8.87e-01 |
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 64 | 124 | 8.87e-01 |
| KEGG | KEGG:05321 | Inflammatory bowel disease | 11 | 20 | 8.87e-01 |
| KEGG | KEGG:01210 | 2-Oxocarboxylic acid metabolism | 14 | 26 | 8.88e-01 |
| KEGG | KEGG:04925 | Aldosterone synthesis and secretion | 39 | 75 | 8.88e-01 |
| KEGG | KEGG:04540 | Gap junction | 37 | 71 | 8.88e-01 |
| KEGG | KEGG:04918 | Thyroid hormone synthesis | 27 | 53 | 9.02e-01 |
| KEGG | KEGG:01230 | Biosynthesis of amino acids | 27 | 52 | 9.02e-01 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 67 | 133 | 9.02e-01 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 53 | 105 | 9.02e-01 |
| KEGG | KEGG:04920 | Adipocytokine signaling pathway | 25 | 49 | 9.02e-01 |
| KEGG | KEGG:01040 | Biosynthesis of unsaturated fatty acids | 12 | 23 | 9.02e-01 |
| KEGG | KEGG:00120 | Primary bile acid biosynthesis | 6 | 11 | 9.02e-01 |
| KEGG | KEGG:04973 | Carbohydrate digestion and absorption | 15 | 29 | 9.02e-01 |
| KEGG | KEGG:00440 | Phosphonate and phosphinate metabolism | 3 | 5 | 9.02e-01 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 31 | 60 | 9.02e-01 |
| KEGG | KEGG:05031 | Amphetamine addiction | 23 | 45 | 9.02e-01 |
| KEGG | KEGG:04623 | Cytosolic DNA-sensing pathway | 24 | 46 | 9.02e-01 |
| KEGG | KEGG:05214 | Glioma | 33 | 65 | 9.02e-01 |
| KEGG | KEGG:00290 | Valine, leucine and isoleucine biosynthesis | 2 | 3 | 9.02e-01 |
| KEGG | KEGG:03013 | Nucleocytoplasmic transport | 49 | 96 | 9.02e-01 |
| KEGG | KEGG:04977 | Vitamin digestion and absorption | 7 | 13 | 9.02e-01 |
| KEGG | KEGG:04215 | Apoptosis - multiple species | 15 | 29 | 9.02e-01 |
| KEGG | KEGG:04061 | Viral protein interaction with cytokine and cytokine receptor | 10 | 19 | 9.02e-01 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 74 | 147 | 9.02e-01 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 65 | 129 | 9.02e-01 |
| KEGG | KEGG:04020 | Calcium signaling pathway | 82 | 161 | 9.02e-01 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 83 | 162 | 9.02e-01 |
| KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 51 | 101 | 9.02e-01 |
| KEGG | KEGG:04911 | Insulin secretion | 32 | 62 | 9.02e-01 |
| KEGG | KEGG:05170 | Human immunodeficiency virus 1 infection | 76 | 149 | 9.02e-01 |
| KEGG | KEGG:04136 | Autophagy - other | 15 | 29 | 9.02e-01 |
| KEGG | KEGG:00100 | Steroid biosynthesis | 8 | 15 | 9.02e-01 |
| KEGG | KEGG:04140 | Autophagy - animal | 75 | 149 | 9.02e-01 |
| KEGG | KEGG:05320 | Autoimmune thyroid disease | 2 | 3 | 9.02e-01 |
| KEGG | KEGG:00600 | Sphingolipid metabolism | 22 | 42 | 9.02e-01 |
| KEGG | KEGG:05310 | Asthma | 1 | 1 | 9.02e-01 |
| KEGG | KEGG:04213 | Longevity regulating pathway - multiple species | 26 | 50 | 9.02e-01 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 65 | 129 | 9.02e-01 |
| KEGG | KEGG:03022 | Basal transcription factors | 17 | 33 | 9.02e-01 |
| KEGG | KEGG:04152 | AMPK signaling pathway | 51 | 101 | 9.02e-01 |
| KEGG | KEGG:04730 | Long-term depression | 20 | 39 | 9.02e-01 |
| KEGG | KEGG:04740 | Olfactory transduction | 13 | 25 | 9.02e-01 |
| KEGG | KEGG:03264 | Virion - Flavivirus | 1 | 1 | 9.02e-01 |
| KEGG | KEGG:04611 | Platelet activation | 43 | 88 | 9.26e-01 |
| KEGG | KEGG:04727 | GABAergic synapse | 26 | 52 | 9.26e-01 |
| KEGG | KEGG:04725 | Cholinergic synapse | 40 | 82 | 9.26e-01 |
| KEGG | KEGG:00561 | Glycerolipid metabolism | 22 | 44 | 9.26e-01 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 67 | 136 | 9.26e-01 |
| KEGG | KEGG:05142 | Chagas disease | 36 | 72 | 9.26e-01 |
| KEGG | KEGG:04640 | Hematopoietic cell lineage | 14 | 28 | 9.26e-01 |
| KEGG | KEGG:05032 | Morphine addiction | 27 | 54 | 9.26e-01 |
| KEGG | KEGG:05340 | Primary immunodeficiency | 4 | 8 | 9.26e-01 |
| KEGG | KEGG:04672 | Intestinal immune network for IgA production | 5 | 10 | 9.26e-01 |
| KEGG | KEGG:00450 | Selenocompound metabolism | 7 | 14 | 9.26e-01 |
| KEGG | KEGG:00270 | Cysteine and methionine metabolism | 18 | 37 | 9.26e-01 |
| KEGG | KEGG:00430 | Taurine and hypotaurine metabolism | 4 | 8 | 9.26e-01 |
| KEGG | KEGG:04721 | Synaptic vesicle cycle | 24 | 49 | 9.26e-01 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 36 | 72 | 9.26e-01 |
| KEGG | KEGG:05160 | Hepatitis C | 55 | 110 | 9.26e-01 |
| KEGG | KEGG:00330 | Arginine and proline metabolism | 16 | 33 | 9.26e-01 |
| KEGG | KEGG:05152 | Tuberculosis | 50 | 101 | 9.26e-01 |
| KEGG | KEGG:00030 | Pentose phosphate pathway | 10 | 20 | 9.26e-01 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 56 | 113 | 9.26e-01 |
| KEGG | KEGG:00410 | beta-Alanine metabolism | 11 | 22 | 9.26e-01 |
| KEGG | KEGG:05143 | African trypanosomiasis | 8 | 16 | 9.26e-01 |
| KEGG | KEGG:00785 | Lipoic acid metabolism | 8 | 16 | 9.26e-01 |
| KEGG | KEGG:05162 | Measles | 42 | 85 | 9.26e-01 |
| KEGG | KEGG:04930 | Type II diabetes mellitus | 17 | 35 | 9.26e-01 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 72 | 144 | 9.26e-01 |
| KEGG | KEGG:02010 | ABC transporters | 12 | 24 | 9.26e-01 |
| KEGG | KEGG:04211 | Longevity regulating pathway | 37 | 75 | 9.26e-01 |
| KEGG | KEGG:04122 | Sulfur relay system | 4 | 8 | 9.26e-01 |
| KEGG | KEGG:01522 | Endocrine resistance | 40 | 82 | 9.26e-01 |
| KEGG | KEGG:00730 | Thiamine metabolism | 5 | 10 | 9.26e-01 |
| KEGG | KEGG:04148 | Efferocytosis | 59 | 120 | 9.26e-01 |
| KEGG | KEGG:03082 | ATP-dependent chromatin remodeling | 45 | 91 | 9.26e-01 |
| KEGG | KEGG:05207 | Chemical carcinogenesis - receptor activation | 66 | 132 | 9.26e-01 |
| KEGG | KEGG:00592 | alpha-Linolenic acid metabolism | 5 | 10 | 9.26e-01 |
| KEGG | KEGG:00860 | Porphyrin metabolism | 9 | 18 | 9.26e-01 |
| KEGG | KEGG:04218 | Cellular senescence | 67 | 136 | 9.26e-01 |
| KEGG | KEGG:05215 | Prostate cancer | 40 | 82 | 9.26e-01 |
| KEGG | KEGG:03450 | Non-homologous end-joining | 6 | 12 | 9.26e-01 |
| KEGG | KEGG:00740 | Riboflavin metabolism | 3 | 6 | 9.29e-01 |
| KEGG | KEGG:04392 | Hippo signaling pathway - multiple species | 12 | 25 | 9.35e-01 |
| KEGG | KEGG:04620 | Toll-like receptor signaling pathway | 29 | 60 | 9.38e-01 |
| KEGG | KEGG:04330 | Notch signaling pathway | 25 | 52 | 9.46e-01 |
| KEGG | KEGG:00650 | Butanoate metabolism | 8 | 17 | 9.57e-01 |
| KEGG | KEGG:05167 | Kaposi sarcoma-associated herpesvirus infection | 65 | 134 | 9.57e-01 |
| KEGG | KEGG:05323 | Rheumatoid arthritis | 18 | 38 | 9.57e-01 |
| KEGG | KEGG:00983 | Drug metabolism - other enzymes | 19 | 40 | 9.57e-01 |
| KEGG | KEGG:00970 | Aminoacyl-tRNA biosynthesis | 20 | 42 | 9.57e-01 |
| KEGG | KEGG:01232 | Nucleotide metabolism | 31 | 65 | 9.62e-01 |
| KEGG | KEGG:05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 31 | 65 | 9.62e-01 |
| KEGG | KEGG:04064 | NF-kappa B signaling pathway | 29 | 61 | 9.64e-01 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 46 | 96 | 9.68e-01 |
| KEGG | KEGG:03320 | PPAR signaling pathway | 22 | 47 | 9.74e-01 |
| KEGG | KEGG:04913 | Ovarian steroidogenesis | 12 | 26 | 9.74e-01 |
| KEGG | KEGG:04960 | Aldosterone-regulated sodium reabsorption | 11 | 24 | 9.74e-01 |
| KEGG | KEGG:04934 | Cushing syndrome | 57 | 119 | 9.74e-01 |
| KEGG | KEGG:00350 | Tyrosine metabolism | 5 | 11 | 9.74e-01 |
| KEGG | KEGG:05332 | Graft-versus-host disease | 1 | 2 | 9.77e-01 |
| KEGG | KEGG:05330 | Allograft rejection | 1 | 2 | 9.77e-01 |
| KEGG | KEGG:03267 | Virion - Adenovirus | 1 | 2 | 9.77e-01 |
| KEGG | KEGG:04917 | Prolactin signaling pathway | 25 | 54 | 9.83e-01 |
| KEGG | KEGG:04924 | Renin secretion | 23 | 50 | 9.83e-01 |
| KEGG | KEGG:05210 | Colorectal cancer | 38 | 81 | 9.83e-01 |
| KEGG | KEGG:04927 | Cortisol synthesis and secretion | 21 | 46 | 9.83e-01 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 65 | 137 | 9.83e-01 |
| KEGG | KEGG:05212 | Pancreatic cancer | 33 | 71 | 9.83e-01 |
| KEGG | KEGG:04658 | Th1 and Th2 cell differentiation | 23 | 50 | 9.83e-01 |
| KEGG | KEGG:04936 | Alcoholic liver disease | 43 | 91 | 9.83e-01 |
| KEGG | KEGG:00591 | Linoleic acid metabolism | 3 | 7 | 9.83e-01 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 61 | 129 | 9.83e-01 |
| KEGG | KEGG:00240 | Pyrimidine metabolism | 20 | 44 | 9.83e-01 |
| KEGG | KEGG:04964 | Proximal tubule bicarbonate reclamation | 8 | 18 | 9.83e-01 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 181 | 388 | 9.99e-01 |
| KEGG | KEGG:05322 | Systemic lupus erythematosus | 15 | 44 | 9.99e-01 |
| KEGG | KEGG:05226 | Gastric cancer | 49 | 113 | 9.99e-01 |
| KEGG | KEGG:05217 | Basal cell carcinoma | 16 | 48 | 9.99e-01 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 52 | 122 | 9.99e-01 |
| KEGG | KEGG:05161 | Hepatitis B | 56 | 126 | 9.99e-01 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 28 | 64 | 9.99e-01 |
| KEGG | KEGG:05203 | Viral carcinogenesis | 64 | 142 | 9.99e-01 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 70 | 164 | 9.99e-01 |
| KEGG | KEGG:05224 | Breast cancer | 46 | 113 | 9.99e-01 |
| KEGG | KEGG:05216 | Thyroid cancer | 14 | 33 | 9.99e-01 |
| KEGG | KEGG:05218 | Melanoma | 23 | 56 | 9.99e-01 |
| KEGG | KEGG:05221 | Acute myeloid leukemia | 18 | 52 | 9.99e-01 |
| KEGG | KEGG:05164 | Influenza A | 43 | 95 | 9.99e-01 |
| KEGG | KEGG:04914 | Progesterone-mediated oocyte maturation | 30 | 82 | 9.99e-01 |
| KEGG | KEGG:05213 | Endometrial cancer | 23 | 54 | 9.99e-01 |
| KEGG | KEGG:04976 | Bile secretion | 18 | 41 | 9.99e-01 |
| KEGG | KEGG:04978 | Mineral absorption | 16 | 38 | 9.99e-01 |
| KEGG | KEGG:04114 | Oocyte meiosis | 45 | 103 | 9.99e-01 |
| KEGG | KEGG:04115 | p53 signaling pathway | 21 | 66 | 9.99e-01 |
| KEGG | KEGG:00564 | Glycerophospholipid metabolism | 27 | 71 | 9.99e-01 |
| KEGG | KEGG:00562 | Inositol phosphate metabolism | 25 | 60 | 9.99e-01 |
| KEGG | KEGG:04130 | SNARE interactions in vesicular transport | 13 | 31 | 9.99e-01 |
| KEGG | KEGG:04110 | Cell cycle | 55 | 146 | 9.99e-01 |
| KEGG | KEGG:04146 | Peroxisome | 29 | 66 | 9.99e-01 |
| KEGG | KEGG:00515 | Mannose type O-glycan biosynthesis | 8 | 19 | 9.99e-01 |
| KEGG | KEGG:05033 | Nicotine addiction | 4 | 10 | 9.99e-01 |
| KEGG | KEGG:05034 | Alcoholism | 37 | 102 | 9.99e-01 |
| KEGG | KEGG:00480 | Glutathione metabolism | 17 | 39 | 9.99e-01 |
| KEGG | KEGG:00470 | D-Amino acid metabolism | 1 | 4 | 9.99e-01 |
| KEGG | KEGG:00360 | Phenylalanine metabolism | 1 | 3 | 9.99e-01 |
| KEGG | KEGG:00230 | Purine metabolism | 36 | 93 | 9.99e-01 |
| KEGG | KEGG:00534 | Glycosaminoglycan biosynthesis - heparan sulfate / heparin | 8 | 19 | 9.99e-01 |
| KEGG | KEGG:00750 | Vitamin B6 metabolism | 2 | 5 | 9.99e-01 |
| KEGG | KEGG:04070 | Phosphatidylinositol signaling system | 34 | 83 | 9.99e-01 |
| KEGG | KEGG:00770 | Pantothenate and CoA biosynthesis | 6 | 15 | 9.99e-01 |
| KEGG | KEGG:03250 | Viral life cycle - HIV-1 | 21 | 54 | 9.99e-01 |
| KEGG | KEGG:03265 | Virion - Ebolavirus and Lyssavirus | 2 | 5 | 9.99e-01 |
| KEGG | KEGG:04950 | Maturity onset diabetes of the young | 1 | 4 | 9.99e-01 |
| KEGG | KEGG:03030 | DNA replication | 9 | 34 | 9.99e-01 |
| KEGG | KEGG:03410 | Base excision repair | 13 | 44 | 9.99e-01 |
| KEGG | KEGG:03420 | Nucleotide excision repair | 23 | 54 | 9.99e-01 |
| KEGG | KEGG:03430 | Mismatch repair | 6 | 22 | 9.99e-01 |
| KEGG | KEGG:03440 | Homologous recombination | 15 | 38 | 9.99e-01 |
| KEGG | KEGG:01524 | Platinum drug resistance | 21 | 60 | 9.99e-01 |
| KEGG | KEGG:01521 | EGFR tyrosine kinase inhibitor resistance | 31 | 72 | 9.99e-01 |
| KEGG | KEGG:03460 | Fanconi anemia pathway | 18 | 46 | 9.99e-01 |
| KEGG | KEGG:04940 | Type I diabetes mellitus | 2 | 7 | 9.99e-01 |
| KEGG | KEGG:04966 | Collecting duct acid secretion | 5 | 18 | 9.99e-01 |
| KEGG | KEGG:00920 | Sulfur metabolism | 4 | 10 | 9.99e-01 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 46 | 106 | 9.99e-01 |
| KEGG | KEGG:00910 | Nitrogen metabolism | 2 | 8 | 9.99e-01 |
| KEGG | KEGG:00830 | Retinol metabolism | 3 | 14 | 9.99e-01 |
| KEGG | KEGG:00220 | Arginine biosynthesis | 5 | 15 | 9.99e-01 |
| KEGG | KEGG:04210 | Apoptosis | 49 | 112 | 9.99e-01 |
| KEGG | KEGG:05030 | Cocaine addiction | 13 | 32 | 9.99e-01 |
| KEGG | KEGG:03083 | Polycomb repressive complex | 31 | 70 | 9.99e-01 |
| KEGG | KEGG:04380 | Osteoclast differentiation | 42 | 92 | 9.99e-01 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 43 | 101 | 9.99e-01 |
| KEGG | KEGG:04923 | Regulation of lipolysis in adipocytes | 14 | 41 | 9.99e-01 |
| KEGG | KEGG:04216 | Ferroptosis | 12 | 31 | 9.99e-01 |
| KEGG | KEGG:04613 | Neutrophil extracellular trap formation | 37 | 89 | 9.99e-01 |
| KEGG | KEGG:04625 | C-type lectin receptor signaling pathway | 34 | 74 | 9.99e-01 |
| KEGG | KEGG:04668 | TNF signaling pathway | 34 | 83 | 9.99e-01 |
| KEGG | KEGG:04744 | Phototransduction | 5 | 12 | 9.99e-01 |
| KEGG | KEGG:04931 | Insulin resistance | 41 | 91 | 9.99e-01 |
| KEGG | KEGG:04217 | Necroptosis | 41 | 99 | 9.99e-01 |
| KEGG | KEGG:04750 | Inflammatory mediator regulation of TRP channels | 31 | 72 | 9.99e-01 |
| KEGG | KEGG:04814 | Motor proteins | 65 | 141 | 9.99e-01 |
# intersect(unique(Resp_RNA$ENTREZID), unique(NR_peak_list_20k$ENTREZID)))
not_NR_int_restable %>%
filter(source=="GO:BP", p_value < 0.05) %>%
slice_head(n=10) %>%
mutate(log10_p=(-log(p_value, base=10))) %>%
ggplot(., aes(x=log10_p, y=reorder(term_name, log10_p)))+
geom_col(fill="#C77CFF")+
theme_bw()+
ylab("")+
xlab(paste0("-log10 p-value"))+
guides(fill="none")+
theme(axis.text.y = element_text(color="black"))+
ggtitle("not Intersection NR")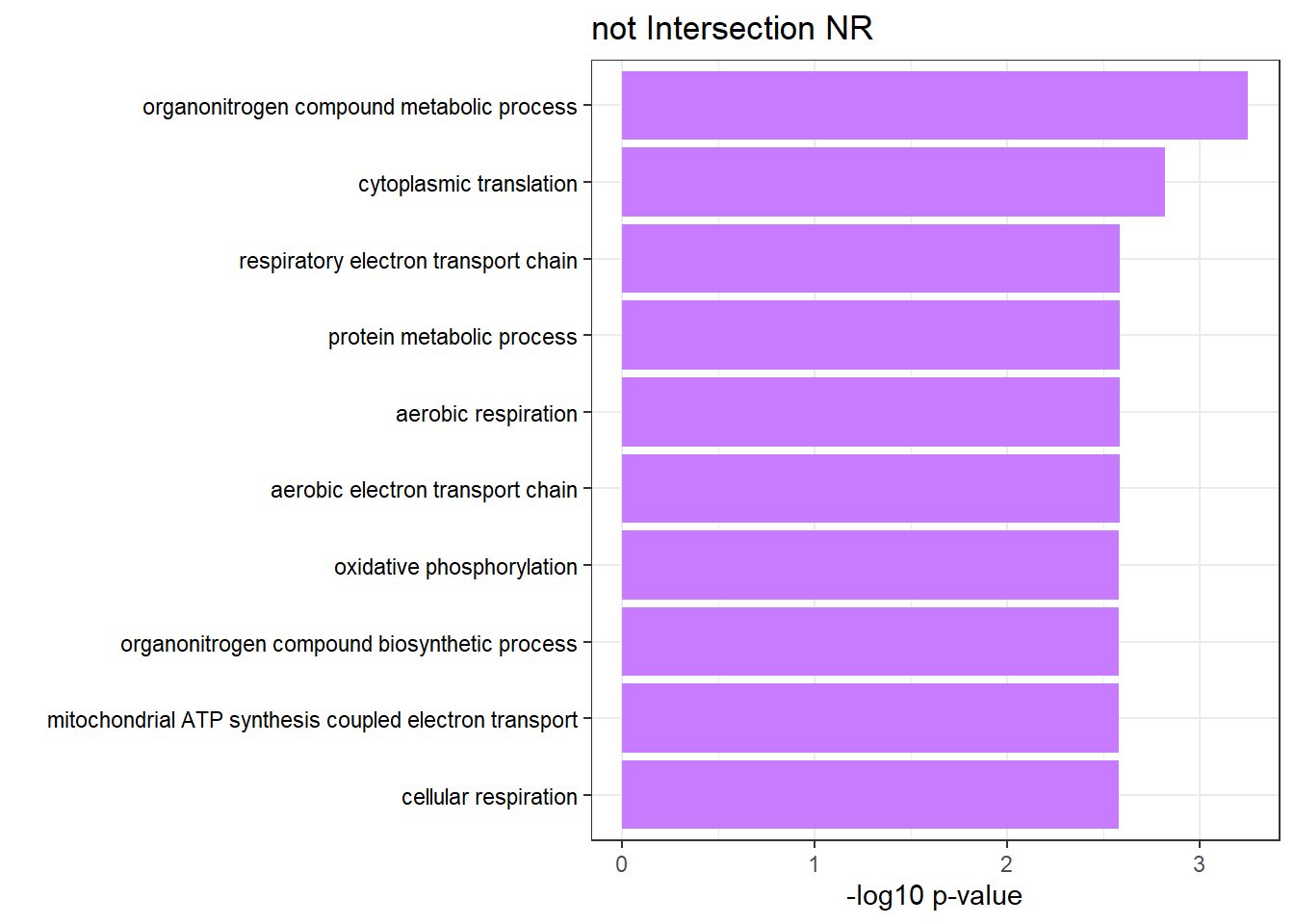
not intersected LR
not_LR_int_resgenes <- gost(query =setdiff(LR_NG_list$ENTREZID, Resp_RNA$ENTREZID),
organism = "hsapiens",
significant = FALSE,
ordered_query = FALSE,
domain_scope = "custom",
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
custom_bg = background_df$ENTREZID,
sources=c("GO:BP","KEGG"))
saveRDS(not_LR_int_resgenes,"data/GO_KEGG_analysis/not_LR_int_resgenes.RDS")
not_LR_int_restable <- not_LR_int_resgenes$result %>%
dplyr::select(c(source, term_id,
term_name,intersection_size,
term_size, p_value))# %>%
not_LR_int_restable %>%
filter(source=="GO:BP", p_value < 0.05) %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(.,caption = "GO:BP terms found in LR neargenes intersected with RNA response genes" ) %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0032501 | multicellular organismal process | 1659 | 4504 | 2.80e-07 |
| GO:BP | GO:0048856 | anatomical structure development | 1430 | 3867 | 3.35e-06 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 220 | 479 | 3.69e-06 |
| GO:BP | GO:0006897 | endocytosis | 226 | 492 | 3.69e-06 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 226 | 494 | 3.69e-06 |
| GO:BP | GO:0098657 | import into cell | 276 | 622 | 3.69e-06 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 226 | 498 | 6.96e-06 |
| GO:BP | GO:0007155 | cell adhesion | 393 | 941 | 7.01e-06 |
| GO:BP | GO:0048731 | system development | 1026 | 2719 | 7.01e-06 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 200 | 435 | 1.10e-05 |
| GO:BP | GO:0000902 | cell morphogenesis | 306 | 713 | 1.23e-05 |
| GO:BP | GO:0032502 | developmental process | 1523 | 4179 | 1.34e-05 |
| GO:BP | GO:0007275 | multicellular organism development | 1177 | 3170 | 1.70e-05 |
| GO:BP | GO:0007399 | nervous system development | 689 | 1775 | 2.38e-05 |
| GO:BP | GO:0032989 | cellular anatomical entity morphogenesis | 256 | 588 | 3.31e-05 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 113 | 224 | 3.39e-05 |
| GO:BP | GO:0030198 | extracellular matrix organization | 112 | 223 | 4.94e-05 |
| GO:BP | GO:0043062 | extracellular structure organization | 112 | 224 | 6.33e-05 |
| GO:BP | GO:0031175 | neuron projection development | 311 | 743 | 1.04e-04 |
| GO:BP | GO:0007409 | axonogenesis | 156 | 337 | 1.37e-04 |
| GO:BP | GO:0048666 | neuron development | 345 | 838 | 1.45e-04 |
| GO:BP | GO:0006810 | transport | 1101 | 2984 | 1.71e-04 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 704 | 1844 | 2.01e-04 |
| GO:BP | GO:0030182 | neuron differentiation | 408 | 1014 | 2.01e-04 |
| GO:BP | GO:0061564 | axon development | 168 | 372 | 2.81e-04 |
| GO:BP | GO:0022008 | neurogenesis | 492 | 1251 | 2.95e-04 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 86 | 169 | 4.87e-04 |
| GO:BP | GO:0048699 | generation of neurons | 427 | 1076 | 5.40e-04 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 454 | 1152 | 5.78e-04 |
| GO:BP | GO:0007229 | integrin-mediated signaling pathway | 49 | 86 | 1.61e-03 |
| GO:BP | GO:0023052 | signaling | 1402 | 3918 | 3.97e-03 |
| GO:BP | GO:0030030 | cell projection organization | 458 | 1182 | 3.97e-03 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 446 | 1150 | 4.34e-03 |
| GO:BP | GO:0051234 | establishment of localization | 1171 | 3240 | 4.34e-03 |
| GO:BP | GO:0048870 | cell motility | 435 | 1120 | 4.59e-03 |
| GO:BP | GO:0098609 | cell-cell adhesion | 233 | 564 | 5.86e-03 |
| GO:BP | GO:0065005 | protein-lipid complex assembly | 16 | 20 | 6.70e-03 |
| GO:BP | GO:0007154 | cell communication | 1420 | 3983 | 6.77e-03 |
| GO:BP | GO:0097485 | neuron projection guidance | 83 | 172 | 6.81e-03 |
| GO:BP | GO:0007411 | axon guidance | 83 | 172 | 6.81e-03 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 48 | 88 | 7.20e-03 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 182 | 431 | 9.59e-03 |
| GO:BP | GO:0071711 | basement membrane organization | 23 | 34 | 9.89e-03 |
| GO:BP | GO:0008150 | biological_process | 3787 | 11244 | 9.89e-03 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 116 | 258 | 1.04e-02 |
| GO:BP | GO:0030001 | metal ion transport | 216 | 524 | 1.08e-02 |
| GO:BP | GO:0030154 | cell differentiation | 1015 | 2803 | 1.08e-02 |
| GO:BP | GO:0006898 | receptor-mediated endocytosis | 86 | 182 | 1.08e-02 |
| GO:BP | GO:0016477 | cell migration | 392 | 1011 | 1.08e-02 |
| GO:BP | GO:0048869 | cellular developmental process | 1015 | 2804 | 1.13e-02 |
| GO:BP | GO:0065008 | regulation of biological quality | 701 | 1893 | 1.19e-02 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 50 | 95 | 1.36e-02 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 47 | 88 | 1.36e-02 |
| GO:BP | GO:0003008 | system process | 435 | 1139 | 1.86e-02 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 237 | 587 | 1.94e-02 |
| GO:BP | GO:0051049 | regulation of transport | 420 | 1098 | 2.04e-02 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 158 | 374 | 2.20e-02 |
| GO:BP | GO:0030100 | regulation of endocytosis | 90 | 196 | 2.25e-02 |
| GO:BP | GO:0006812 | monoatomic cation transport | 246 | 614 | 2.40e-02 |
| GO:BP | GO:0034377 | plasma lipoprotein particle assembly | 14 | 18 | 2.43e-02 |
| GO:BP | GO:0051179 | localization | 1324 | 3731 | 2.58e-02 |
| GO:BP | GO:0006811 | monoatomic ion transport | 283 | 719 | 3.10e-02 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 201 | 495 | 3.91e-02 |
| GO:BP | GO:0043413 | macromolecule glycosylation | 81 | 176 | 3.91e-02 |
| GO:BP | GO:0006486 | protein glycosylation | 81 | 176 | 3.91e-02 |
| GO:BP | GO:0055085 | transmembrane transport | 354 | 921 | 4.07e-02 |
| GO:BP | GO:0023051 | regulation of signaling | 881 | 2438 | 4.12e-02 |
| GO:BP | GO:0030199 | collagen fibril organization | 29 | 50 | 4.13e-02 |
| GO:BP | GO:0040012 | regulation of locomotion | 282 | 720 | 4.19e-02 |
| GO:BP | GO:0009987 | cellular process | 3604 | 10686 | 4.28e-02 |
| GO:BP | GO:0007165 | signal transduction | 1279 | 3611 | 4.46e-02 |
| GO:BP | GO:0040011 | locomotion | 312 | 805 | 4.46e-02 |
| GO:BP | GO:0045663 | positive regulation of myoblast differentiation | 22 | 35 | 4.46e-02 |
| GO:BP | GO:0007267 | cell-cell signaling | 410 | 1082 | 4.62e-02 |
| GO:BP | GO:0090109 | regulation of cell-substrate junction assembly | 31 | 55 | 4.64e-02 |
| GO:BP | GO:0051893 | regulation of focal adhesion assembly | 31 | 55 | 4.64e-02 |
| GO:BP | GO:2000145 | regulation of cell motility | 274 | 700 | 4.79e-02 |
| GO:BP | GO:0072337 | modified amino acid transport | 19 | 29 | 4.88e-02 |
| GO:BP | GO:0010646 | regulation of cell communication | 881 | 2444 | 4.88e-02 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 660 | 1801 | 4.88e-02 |
| GO:BP | GO:0051016 | barbed-end actin filament capping | 14 | 19 | 4.88e-02 |
not_LR_int_restable %>%
filter(source=="KEGG") %>%
mutate_at(.vars = 6, .funs = scientific_format()) %>%
kable(., caption = "KEGG terms found in LR neargenes intersected with RNA response genes") %>%
kable_paper("striped", full_width = FALSE) %>%
kable_styling(
full_width = FALSE,
position = "left",
bootstrap_options = c("striped", "hover")
) %>%
scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| KEGG | KEGG:04510 | Focal adhesion | 103 | 169 | 2.97e-11 |
| KEGG | KEGG:04512 | ECM-receptor interaction | 46 | 66 | 2.18e-07 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 87 | 169 | 6.40e-05 |
| KEGG | KEGG:04270 | Vascular smooth muscle contraction | 53 | 92 | 9.91e-05 |
| KEGG | KEGG:04520 | Adherens junction | 47 | 83 | 6.01e-04 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 77 | 162 | 5.28e-03 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 111 | 250 | 5.77e-03 |
| KEGG | KEGG:05146 | Amoebiasis | 34 | 62 | 1.48e-02 |
| KEGG | KEGG:04720 | Long-term potentiation | 29 | 51 | 1.58e-02 |
| KEGG | KEGG:04974 | Protein digestion and absorption | 31 | 56 | 1.68e-02 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 51 | 104 | 1.68e-02 |
| KEGG | KEGG:05135 | Yersinia infection | 52 | 107 | 1.68e-02 |
| KEGG | KEGG:05100 | Bacterial invasion of epithelial cells | 38 | 73 | 1.68e-02 |
| KEGG | KEGG:00514 | Other types of O-glycan biosynthesis | 21 | 35 | 2.44e-02 |
| KEGG | KEGG:04360 | Axon guidance | 70 | 155 | 2.63e-02 |
| KEGG | KEGG:05200 | Pathways in cancer | 158 | 391 | 2.82e-02 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 48 | 100 | 2.83e-02 |
| KEGG | KEGG:00512 | Mucin type O-glycan biosynthesis | 14 | 21 | 2.95e-02 |
| KEGG | KEGG:05131 | Shigellosis | 88 | 204 | 2.95e-02 |
| KEGG | KEGG:04670 | Leukocyte transendothelial migration | 38 | 76 | 2.95e-02 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 59 | 129 | 3.07e-02 |
| KEGG | KEGG:05414 | Dilated cardiomyopathy | 40 | 82 | 3.65e-02 |
| KEGG | KEGG:04014 | Ras signaling pathway | 72 | 164 | 3.70e-02 |
| KEGG | KEGG:04971 | Gastric acid secretion | 27 | 51 | 3.82e-02 |
| KEGG | KEGG:00051 | Fructose and mannose metabolism | 16 | 26 | 3.82e-02 |
| KEGG | KEGG:04921 | Oxytocin signaling pathway | 52 | 113 | 3.82e-02 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 65 | 147 | 4.04e-02 |
| KEGG | KEGG:04144 | Endocytosis | 91 | 216 | 4.07e-02 |
| KEGG | KEGG:00604 | Glycosphingolipid biosynthesis - ganglio series | 9 | 12 | 4.10e-02 |
| KEGG | KEGG:04722 | Neurotrophin signaling pathway | 48 | 104 | 4.10e-02 |
| KEGG | KEGG:04020 | Calcium signaling pathway | 70 | 161 | 4.10e-02 |
| KEGG | KEGG:04961 | Endocrine and other factor-regulated calcium reabsorption | 22 | 40 | 4.10e-02 |
| KEGG | KEGG:05410 | Hypertrophic cardiomyopathy | 37 | 77 | 4.80e-02 |
| KEGG | KEGG:01212 | Fatty acid metabolism | 24 | 46 | 5.94e-02 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 69 | 162 | 7.05e-02 |
| KEGG | KEGG:04664 | Fc epsilon RI signaling pathway | 22 | 42 | 7.18e-02 |
| KEGG | KEGG:04660 | T cell receptor signaling pathway | 39 | 84 | 7.18e-02 |
| KEGG | KEGG:04728 | Dopaminergic synapse | 44 | 97 | 7.23e-02 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 40 | 87 | 7.47e-02 |
| KEGG | KEGG:05412 | Arrhythmogenic right ventricular cardiomyopathy | 32 | 67 | 7.49e-02 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 94 | 231 | 7.49e-02 |
| KEGG | KEGG:04972 | Pancreatic secretion | 28 | 57 | 7.49e-02 |
| KEGG | KEGG:05031 | Amphetamine addiction | 23 | 45 | 7.61e-02 |
| KEGG | KEGG:05231 | Choline metabolism in cancer | 37 | 80 | 7.72e-02 |
| KEGG | KEGG:04925 | Aldosterone synthesis and secretion | 35 | 75 | 7.72e-02 |
| KEGG | KEGG:00071 | Fatty acid degradation | 17 | 31 | 7.79e-02 |
| KEGG | KEGG:05416 | Viral myocarditis | 19 | 36 | 7.84e-02 |
| KEGG | KEGG:04713 | Circadian entrainment | 32 | 68 | 7.84e-02 |
| KEGG | KEGG:04062 | Chemokine signaling pathway | 48 | 109 | 7.84e-02 |
| KEGG | KEGG:04726 | Serotonergic synapse | 28 | 58 | 7.84e-02 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 63 | 149 | 7.84e-02 |
| KEGG | KEGG:04918 | Thyroid hormone synthesis | 26 | 53 | 7.84e-02 |
| KEGG | KEGG:04072 | Phospholipase D signaling pathway | 47 | 107 | 8.21e-02 |
| KEGG | KEGG:04530 | Tight junction | 55 | 129 | 9.29e-02 |
| KEGG | KEGG:04912 | GnRH signaling pathway | 34 | 74 | 9.29e-02 |
| KEGG | KEGG:04662 | B cell receptor signaling pathway | 25 | 52 | 9.63e-02 |
| KEGG | KEGG:04370 | VEGF signaling pathway | 23 | 47 | 9.63e-02 |
| KEGG | KEGG:03266 | Virion - Herpesvirus | 5 | 6 | 9.63e-02 |
| KEGG | KEGG:00280 | Valine, leucine and isoleucine degradation | 21 | 42 | 9.63e-02 |
| KEGG | KEGG:04611 | Platelet activation | 39 | 88 | 9.63e-02 |
| KEGG | KEGG:04915 | Estrogen signaling pathway | 40 | 90 | 9.63e-02 |
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 53 | 124 | 9.63e-02 |
| KEGG | KEGG:00513 | Various types of N-glycan biosynthesis | 19 | 37 | 9.63e-02 |
| KEGG | KEGG:04142 | Lysosome | 47 | 109 | 9.63e-02 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 66 | 160 | 9.63e-02 |
| KEGG | KEGG:04666 | Fc gamma R-mediated phagocytosis | 34 | 75 | 9.63e-02 |
| KEGG | KEGG:04911 | Insulin secretion | 29 | 62 | 9.63e-02 |
| KEGG | KEGG:05219 | Bladder cancer | 18 | 35 | 9.74e-02 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 35 | 78 | 1.01e-01 |
| KEGG | KEGG:04929 | GnRH secretion | 24 | 50 | 1.01e-01 |
| KEGG | KEGG:04979 | Cholesterol metabolism | 17 | 33 | 1.08e-01 |
| KEGG | KEGG:04723 | Retrograde endocannabinoid signaling | 40 | 92 | 1.17e-01 |
| KEGG | KEGG:04514 | Cell adhesion molecules | 31 | 69 | 1.29e-01 |
| KEGG | KEGG:04970 | Salivary secretion | 25 | 54 | 1.41e-01 |
| KEGG | KEGG:04340 | Hedgehog signaling pathway | 23 | 49 | 1.42e-01 |
| KEGG | KEGG:00062 | Fatty acid elongation | 12 | 22 | 1.43e-01 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 33 | 75 | 1.44e-01 |
| KEGG | KEGG:00640 | Propanoate metabolism | 14 | 27 | 1.52e-01 |
| KEGG | KEGG:04066 | HIF-1 signaling pathway | 37 | 86 | 1.53e-01 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 44 | 105 | 1.59e-01 |
| KEGG | KEGG:04071 | Sphingolipid signaling pathway | 43 | 103 | 1.76e-01 |
| KEGG | KEGG:05230 | Central carbon metabolism in cancer | 26 | 58 | 1.77e-01 |
| KEGG | KEGG:04724 | Glutamatergic synapse | 32 | 74 | 1.84e-01 |
| KEGG | KEGG:05418 | Fluid shear stress and atherosclerosis | 45 | 109 | 1.86e-01 |
| KEGG | KEGG:04916 | Melanogenesis | 32 | 75 | 2.16e-01 |
| KEGG | KEGG:05211 | Renal cell carcinoma | 27 | 62 | 2.25e-01 |
| KEGG | KEGG:04920 | Adipocytokine signaling pathway | 22 | 49 | 2.28e-01 |
| KEGG | KEGG:00520 | Amino sugar and nucleotide sugar metabolism | 20 | 44 | 2.36e-01 |
| KEGG | KEGG:05415 | Diabetic cardiomyopathy | 62 | 158 | 2.39e-01 |
| KEGG | KEGG:04730 | Long-term depression | 18 | 39 | 2.39e-01 |
| KEGG | KEGG:05222 | Small cell lung cancer | 34 | 82 | 2.61e-01 |
| KEGG | KEGG:04725 | Cholinergic synapse | 34 | 82 | 2.61e-01 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 95 | 252 | 2.61e-01 |
| KEGG | KEGG:04924 | Renin secretion | 22 | 50 | 2.63e-01 |
| KEGG | KEGG:04614 | Renin-angiotensin system | 6 | 10 | 2.68e-01 |
| KEGG | KEGG:05130 | Pathogenic Escherichia coli infection | 56 | 143 | 2.68e-01 |
| KEGG | KEGG:05417 | Lipid and atherosclerosis | 56 | 143 | 2.68e-01 |
| KEGG | KEGG:04650 | Natural killer cell mediated cytotoxicity | 23 | 53 | 2.71e-01 |
| KEGG | KEGG:00000 | KEGG root term | 1843 | 5451 | 2.79e-01 |
| KEGG | KEGG:04930 | Type II diabetes mellitus | 16 | 35 | 2.84e-01 |
| KEGG | KEGG:04630 | JAK-STAT signaling pathway | 34 | 83 | 2.84e-01 |
| KEGG | KEGG:00531 | Glycosaminoglycan degradation | 8 | 15 | 2.89e-01 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 52 | 133 | 2.89e-01 |
| KEGG | KEGG:04922 | Glucagon signaling pathway | 32 | 78 | 2.93e-01 |
| KEGG | KEGG:04742 | Taste transduction | 10 | 20 | 2.94e-01 |
| KEGG | KEGG:04927 | Cortisol synthesis and secretion | 20 | 46 | 3.04e-01 |
| KEGG | KEGG:04621 | NOD-like receptor signaling pathway | 43 | 109 | 3.16e-01 |
| KEGG | KEGG:00565 | Ether lipid metabolism | 13 | 28 | 3.19e-01 |
| KEGG | KEGG:04977 | Vitamin digestion and absorption | 7 | 13 | 3.19e-01 |
| KEGG | KEGG:00533 | Glycosaminoglycan biosynthesis - keratan sulfate | 7 | 13 | 3.19e-01 |
| KEGG | KEGG:04975 | Fat digestion and absorption | 9 | 18 | 3.27e-01 |
| KEGG | KEGG:05207 | Chemical carcinogenesis - receptor activation | 51 | 132 | 3.27e-01 |
| KEGG | KEGG:05170 | Human immunodeficiency virus 1 infection | 57 | 149 | 3.29e-01 |
| KEGG | KEGG:03271 | Virion - Rotavirus | 2 | 2 | 3.30e-01 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 38 | 96 | 3.30e-01 |
| KEGG | KEGG:00524 | Neomycin, kanamycin and gentamicin biosynthesis | 3 | 4 | 3.30e-01 |
| KEGG | KEGG:03320 | PPAR signaling pathway | 20 | 47 | 3.35e-01 |
| KEGG | KEGG:00020 | Citrate cycle (TCA cycle) | 12 | 26 | 3.41e-01 |
| KEGG | KEGG:05132 | Salmonella infection | 75 | 201 | 3.41e-01 |
| KEGG | KEGG:00052 | Galactose metabolism | 10 | 21 | 3.44e-01 |
| KEGG | KEGG:00040 | Pentose and glucuronate interconversions | 6 | 11 | 3.44e-01 |
| KEGG | KEGG:00380 | Tryptophan metabolism | 10 | 21 | 3.44e-01 |
| KEGG | KEGG:05145 | Toxoplasmosis | 29 | 72 | 3.47e-01 |
| KEGG | KEGG:04973 | Carbohydrate digestion and absorption | 13 | 29 | 3.56e-01 |
| KEGG | KEGG:04710 | Circadian rhythm | 13 | 29 | 3.56e-01 |
| KEGG | KEGG:04114 | Oocyte meiosis | 40 | 103 | 3.59e-01 |
| KEGG | KEGG:00310 | Lysine degradation | 23 | 56 | 3.59e-01 |
| KEGG | KEGG:04148 | Efferocytosis | 46 | 120 | 3.59e-01 |
| KEGG | KEGG:05210 | Colorectal cancer | 32 | 81 | 3.62e-01 |
| KEGG | KEGG:04960 | Aldosterone-regulated sodium reabsorption | 11 | 24 | 3.62e-01 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 35 | 90 | 3.89e-01 |
| KEGG | KEGG:05214 | Glioma | 26 | 65 | 3.89e-01 |
| KEGG | KEGG:04610 | Complement and coagulation cascades | 15 | 35 | 3.89e-01 |
| KEGG | KEGG:04659 | Th17 cell differentiation | 24 | 60 | 4.08e-01 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 20 | 49 | 4.08e-01 |
| KEGG | KEGG:04625 | C-type lectin receptor signaling pathway | 29 | 74 | 4.08e-01 |
| KEGG | KEGG:01200 | Carbon metabolism | 34 | 88 | 4.08e-01 |
| KEGG | KEGG:05208 | Chemical carcinogenesis - reactive oxygen species | 64 | 173 | 4.08e-01 |
| KEGG | KEGG:00330 | Arginine and proline metabolism | 14 | 33 | 4.24e-01 |
| KEGG | KEGG:00591 | Linoleic acid metabolism | 4 | 7 | 4.24e-01 |
| KEGG | KEGG:04657 | IL-17 signaling pathway | 22 | 55 | 4.27e-01 |
| KEGG | KEGG:05017 | Spinocerebellar ataxia | 45 | 120 | 4.35e-01 |
| KEGG | KEGG:04750 | Inflammatory mediator regulation of TRP channels | 28 | 72 | 4.37e-01 |
| KEGG | KEGG:05020 | Prion disease | 74 | 204 | 4.45e-01 |
| KEGG | KEGG:00532 | Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate | 9 | 20 | 4.45e-01 |
| KEGG | KEGG:03060 | Protein export | 12 | 28 | 4.45e-01 |
| KEGG | KEGG:04658 | Th1 and Th2 cell differentiation | 20 | 50 | 4.45e-01 |
| KEGG | KEGG:04152 | AMPK signaling pathway | 38 | 101 | 4.56e-01 |
| KEGG | KEGG:01100 | Metabolic pathways | 370 | 1077 | 4.56e-01 |
| KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 47 | 127 | 4.62e-01 |
| KEGG | KEGG:01040 | Biosynthesis of unsaturated fatty acids | 10 | 23 | 4.62e-01 |
| KEGG | KEGG:00592 | alpha-Linolenic acid metabolism | 5 | 10 | 4.66e-01 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 42 | 113 | 4.66e-01 |
| KEGG | KEGG:00730 | Thiamine metabolism | 5 | 10 | 4.66e-01 |
| KEGG | KEGG:01250 | Biosynthesis of nucleotide sugars | 14 | 34 | 4.66e-01 |
| KEGG | KEGG:04714 | Thermogenesis | 67 | 185 | 4.66e-01 |
| KEGG | KEGG:05010 | Alzheimer disease | 105 | 296 | 4.66e-01 |
| KEGG | KEGG:05215 | Prostate cancer | 31 | 82 | 4.71e-01 |
| KEGG | KEGG:04962 | Vasopressin-regulated water reabsorption | 15 | 37 | 4.71e-01 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 128 | 364 | 4.71e-01 |
| KEGG | KEGG:00670 | One carbon pool by folate | 8 | 18 | 4.71e-01 |
| KEGG | KEGG:03015 | mRNA surveillance pathway | 30 | 80 | 5.07e-01 |
| KEGG | KEGG:04260 | Cardiac muscle contraction | 24 | 63 | 5.09e-01 |
| KEGG | KEGG:05012 | Parkinson disease | 75 | 211 | 5.26e-01 |
| KEGG | KEGG:04211 | Longevity regulating pathway | 28 | 75 | 5.30e-01 |
| KEGG | KEGG:05143 | African trypanosomiasis | 7 | 16 | 5.30e-01 |
| KEGG | KEGG:00780 | Biotin metabolism | 2 | 3 | 5.30e-01 |
| KEGG | KEGG:00430 | Taurine and hypotaurine metabolism | 4 | 8 | 5.30e-01 |
| KEGG | KEGG:00603 | Glycosphingolipid biosynthesis - globo and isoglobo series | 4 | 8 | 5.30e-01 |
| KEGG | KEGG:04976 | Bile secretion | 16 | 41 | 5.31e-01 |
| KEGG | KEGG:04218 | Cellular senescence | 49 | 136 | 5.34e-01 |
| KEGG | KEGG:00561 | Glycerolipid metabolism | 17 | 44 | 5.35e-01 |
| KEGG | KEGG:04622 | RIG-I-like receptor signaling pathway | 17 | 44 | 5.35e-01 |
| KEGG | KEGG:00534 | Glycosaminoglycan biosynthesis - heparan sulfate / heparin | 8 | 19 | 5.47e-01 |
| KEGG | KEGG:03022 | Basal transcription factors | 13 | 33 | 5.51e-01 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 49 | 137 | 5.60e-01 |
| KEGG | KEGG:00500 | Starch and sucrose metabolism | 9 | 22 | 5.60e-01 |
| KEGG | KEGG:00410 | beta-Alanine metabolism | 9 | 22 | 5.60e-01 |
| KEGG | KEGG:04932 | Non-alcoholic fatty liver disease | 45 | 126 | 5.74e-01 |
| KEGG | KEGG:01232 | Nucleotide metabolism | 24 | 65 | 5.74e-01 |
| KEGG | KEGG:04740 | Olfactory transduction | 10 | 25 | 5.74e-01 |
| KEGG | KEGG:04640 | Hematopoietic cell lineage | 11 | 28 | 5.78e-01 |
| KEGG | KEGG:05206 | MicroRNAs in cancer | 51 | 144 | 5.78e-01 |
| KEGG | KEGG:00250 | Alanine, aspartate and glutamate metabolism | 11 | 28 | 5.78e-01 |
| KEGG | KEGG:00511 | Other glycan degradation | 6 | 14 | 5.78e-01 |
| KEGG | KEGG:05032 | Morphine addiction | 20 | 54 | 5.86e-01 |
| KEGG | KEGG:05213 | Endometrial cancer | 20 | 54 | 5.86e-01 |
| KEGG | KEGG:00740 | Riboflavin metabolism | 3 | 6 | 5.86e-01 |
| KEGG | KEGG:04380 | Osteoclast differentiation | 33 | 92 | 5.96e-01 |
| KEGG | KEGG:05203 | Viral carcinogenesis | 50 | 142 | 6.00e-01 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 36 | 101 | 6.01e-01 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 26 | 72 | 6.09e-01 |
| KEGG | KEGG:04934 | Cushing syndrome | 42 | 119 | 6.12e-01 |
| KEGG | KEGG:00061 | Fatty acid biosynthesis | 4 | 9 | 6.20e-01 |
| KEGG | KEGG:04727 | GABAergic synapse | 19 | 52 | 6.20e-01 |
| KEGG | KEGG:01210 | 2-Oxocarboxylic acid metabolism | 10 | 26 | 6.20e-01 |
| KEGG | KEGG:05120 | Epithelial cell signaling in Helicobacter pylori infection | 20 | 55 | 6.23e-01 |
| KEGG | KEGG:04936 | Alcoholic liver disease | 32 | 91 | 6.51e-01 |
| KEGG | KEGG:04931 | Insulin resistance | 32 | 91 | 6.51e-01 |
| KEGG | KEGG:03082 | ATP-dependent chromatin remodeling | 32 | 91 | 6.51e-01 |
| KEGG | KEGG:04914 | Progesterone-mediated oocyte maturation | 29 | 82 | 6.51e-01 |
| KEGG | KEGG:01522 | Endocrine resistance | 29 | 82 | 6.51e-01 |
| KEGG | KEGG:04213 | Longevity regulating pathway - multiple species | 18 | 50 | 6.59e-01 |
| KEGG | KEGG:05016 | Huntington disease | 84 | 246 | 6.65e-01 |
| KEGG | KEGG:04540 | Gap junction | 25 | 71 | 6.69e-01 |
| KEGG | KEGG:05212 | Pancreatic cancer | 25 | 71 | 6.69e-01 |
| KEGG | KEGG:05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 23 | 65 | 6.69e-01 |
| KEGG | KEGG:05216 | Thyroid cancer | 12 | 33 | 6.86e-01 |
| KEGG | KEGG:00600 | Sphingolipid metabolism | 15 | 42 | 6.96e-01 |
| KEGG | KEGG:04145 | Phagosome | 29 | 84 | 7.09e-01 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 22 | 64 | 7.09e-01 |
| KEGG | KEGG:00785 | Lipoic acid metabolism | 6 | 16 | 7.09e-01 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 44 | 129 | 7.09e-01 |
| KEGG | KEGG:00340 | Histidine metabolism | 4 | 10 | 7.09e-01 |
| KEGG | KEGG:04814 | Motor proteins | 48 | 141 | 7.09e-01 |
| KEGG | KEGG:04721 | Synaptic vesicle cycle | 17 | 49 | 7.09e-01 |
| KEGG | KEGG:04060 | Cytokine-cytokine receptor interaction | 32 | 94 | 7.09e-01 |
| KEGG | KEGG:05033 | Nicotine addiction | 4 | 10 | 7.09e-01 |
| KEGG | KEGG:04137 | Mitophagy - animal | 32 | 94 | 7.09e-01 |
| KEGG | KEGG:04064 | NF-kappa B signaling pathway | 21 | 61 | 7.09e-01 |
| KEGG | KEGG:00920 | Sulfur metabolism | 4 | 10 | 7.09e-01 |
| KEGG | KEGG:00982 | Drug metabolism - cytochrome P450 | 6 | 16 | 7.09e-01 |
| KEGG | KEGG:04130 | SNARE interactions in vesicular transport | 11 | 31 | 7.09e-01 |
| KEGG | KEGG:01523 | Antifolate resistance | 8 | 22 | 7.09e-01 |
| KEGG | KEGG:00630 | Glyoxylate and dicarboxylate metabolism | 9 | 25 | 7.09e-01 |
| KEGG | KEGG:01521 | EGFR tyrosine kinase inhibitor resistance | 25 | 72 | 7.09e-01 |
| KEGG | KEGG:00590 | Arachidonic acid metabolism | 9 | 25 | 7.09e-01 |
| KEGG | KEGG:00620 | Pyruvate metabolism | 11 | 31 | 7.09e-01 |
| KEGG | KEGG:05167 | Kaposi sarcoma-associated herpesvirus infection | 45 | 134 | 7.40e-01 |
| KEGG | KEGG:04140 | Autophagy - animal | 50 | 149 | 7.40e-01 |
| KEGG | KEGG:05160 | Hepatitis C | 37 | 110 | 7.41e-01 |
| KEGG | KEGG:00240 | Pyrimidine metabolism | 15 | 44 | 7.49e-01 |
| KEGG | KEGG:00980 | Metabolism of xenobiotics by cytochrome P450 | 8 | 23 | 7.49e-01 |
| KEGG | KEGG:05144 | Malaria | 7 | 20 | 7.49e-01 |
| KEGG | KEGG:05030 | Cocaine addiction | 11 | 32 | 7.49e-01 |
| KEGG | KEGG:04913 | Ovarian steroidogenesis | 9 | 26 | 7.49e-01 |
| KEGG | KEGG:00260 | Glycine, serine and threonine metabolism | 9 | 26 | 7.49e-01 |
| KEGG | KEGG:04978 | Mineral absorption | 13 | 38 | 7.49e-01 |
| KEGG | KEGG:00563 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 8 | 23 | 7.49e-01 |
| KEGG | KEGG:00650 | Butanoate metabolism | 6 | 17 | 7.49e-01 |
| KEGG | KEGG:00450 | Selenocompound metabolism | 5 | 14 | 7.50e-01 |
| KEGG | KEGG:00120 | Primary bile acid biosynthesis | 4 | 11 | 7.52e-01 |
| KEGG | KEGG:04620 | Toll-like receptor signaling pathway | 20 | 60 | 7.56e-01 |
| KEGG | KEGG:05142 | Chagas disease | 24 | 72 | 7.56e-01 |
| KEGG | KEGG:03265 | Virion - Ebolavirus and Lyssavirus | 2 | 5 | 7.56e-01 |
| KEGG | KEGG:00053 | Ascorbate and aldarate metabolism | 3 | 8 | 7.56e-01 |
| KEGG | KEGG:00440 | Phosphonate and phosphinate metabolism | 2 | 5 | 7.56e-01 |
| KEGG | KEGG:04917 | Prolactin signaling pathway | 18 | 54 | 7.57e-01 |
| KEGG | KEGG:05133 | Pertussis | 14 | 42 | 7.66e-01 |
| KEGG | KEGG:03267 | Virion - Adenovirus | 1 | 2 | 7.72e-01 |
| KEGG | KEGG:03083 | Polycomb repressive complex | 23 | 70 | 7.84e-01 |
| KEGG | KEGG:02010 | ABC transporters | 8 | 24 | 7.86e-01 |
| KEGG | KEGG:05152 | Tuberculosis | 33 | 101 | 7.95e-01 |
| KEGG | KEGG:04613 | Neutrophil extracellular trap formation | 29 | 89 | 8.01e-01 |
| KEGG | KEGG:00100 | Steroid biosynthesis | 5 | 15 | 8.03e-01 |
| KEGG | KEGG:04668 | TNF signaling pathway | 27 | 83 | 8.03e-01 |
| KEGG | KEGG:00983 | Drug metabolism - other enzymes | 13 | 40 | 8.05e-01 |
| KEGG | KEGG:00564 | Glycerophospholipid metabolism | 23 | 71 | 8.07e-01 |
| KEGG | KEGG:05134 | Legionellosis | 11 | 34 | 8.08e-01 |
| KEGG | KEGG:04744 | Phototransduction | 4 | 12 | 8.08e-01 |
| KEGG | KEGG:00190 | Oxidative phosphorylation | 32 | 99 | 8.09e-01 |
| KEGG | KEGG:05218 | Melanoma | 18 | 56 | 8.15e-01 |
| KEGG | KEGG:00130 | Ubiquinone and other terpenoid-quinone biosynthesis | 3 | 9 | 8.20e-01 |
| KEGG | KEGG:04392 | Hippo signaling pathway - multiple species | 8 | 25 | 8.22e-01 |
| KEGG | KEGG:05171 | Coronavirus disease - COVID-19 | 48 | 149 | 8.22e-01 |
| KEGG | KEGG:04923 | Regulation of lipolysis in adipocytes | 13 | 41 | 8.26e-01 |
| KEGG | KEGG:05110 | Vibrio cholerae infection | 13 | 41 | 8.26e-01 |
| KEGG | KEGG:00010 | Glycolysis / Gluconeogenesis | 13 | 41 | 8.26e-01 |
| KEGG | KEGG:05162 | Measles | 27 | 85 | 8.37e-01 |
| KEGG | KEGG:00140 | Steroid hormone biosynthesis | 5 | 16 | 8.46e-01 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 41 | 129 | 8.46e-01 |
| KEGG | KEGG:04080 | Neuroactive ligand-receptor interaction | 32 | 101 | 8.46e-01 |
| KEGG | KEGG:04215 | Apoptosis - multiple species | 9 | 29 | 8.46e-01 |
| KEGG | KEGG:03018 | RNA degradation | 21 | 67 | 8.48e-01 |
| KEGG | KEGG:05150 | Staphylococcus aureus infection | 4 | 13 | 8.53e-01 |
| KEGG | KEGG:04070 | Phosphatidylinositol signaling system | 26 | 83 | 8.55e-01 |
| KEGG | KEGG:00360 | Phenylalanine metabolism | 1 | 3 | 8.64e-01 |
| KEGG | KEGG:05320 | Autoimmune thyroid disease | 1 | 3 | 8.64e-01 |
| KEGG | KEGG:00290 | Valine, leucine and isoleucine biosynthesis | 1 | 3 | 8.64e-01 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 33 | 106 | 8.64e-01 |
| KEGG | KEGG:00230 | Purine metabolism | 29 | 93 | 8.64e-01 |
| KEGG | KEGG:00030 | Pentose phosphate pathway | 6 | 20 | 8.64e-01 |
| KEGG | KEGG:05321 | Inflammatory bowel disease | 6 | 20 | 8.64e-01 |
| KEGG | KEGG:05204 | Chemical carcinogenesis - DNA adducts | 6 | 20 | 8.64e-01 |
| KEGG | KEGG:04146 | Peroxisome | 20 | 66 | 8.93e-01 |
| KEGG | KEGG:00601 | Glycosphingolipid biosynthesis - lacto and neolacto series | 4 | 14 | 8.95e-01 |
| KEGG | KEGG:05140 | Leishmaniasis | 11 | 38 | 9.19e-01 |
| KEGG | KEGG:00350 | Tyrosine metabolism | 3 | 11 | 9.19e-01 |
| KEGG | KEGG:04964 | Proximal tubule bicarbonate reclamation | 5 | 18 | 9.19e-01 |
| KEGG | KEGG:00760 | Nicotinate and nicotinamide metabolism | 8 | 28 | 9.19e-01 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 91 | 291 | 9.24e-01 |
| KEGG | KEGG:04623 | Cytosolic DNA-sensing pathway | 13 | 46 | 9.26e-01 |
| KEGG | KEGG:00770 | Pantothenate and CoA biosynthesis | 4 | 15 | 9.26e-01 |
| KEGG | KEGG:00470 | D-Amino acid metabolism | 1 | 4 | 9.26e-01 |
| KEGG | KEGG:00515 | Mannose type O-glycan biosynthesis | 5 | 19 | 9.26e-01 |
| KEGG | KEGG:05226 | Gastric cancer | 34 | 113 | 9.26e-01 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 50 | 164 | 9.26e-01 |
| KEGG | KEGG:04612 | Antigen processing and presentation | 7 | 26 | 9.26e-01 |
| KEGG | KEGG:05164 | Influenza A | 28 | 95 | 9.26e-01 |
| KEGG | KEGG:03040 | Spliceosome | 37 | 124 | 9.26e-01 |
| KEGG | KEGG:04950 | Maturity onset diabetes of the young | 1 | 4 | 9.26e-01 |
| KEGG | KEGG:04136 | Autophagy - other | 8 | 29 | 9.26e-01 |
| KEGG | KEGG:04061 | Viral protein interaction with cytokine and cytokine receptor | 5 | 19 | 9.26e-01 |
| KEGG | KEGG:00970 | Aminoacyl-tRNA biosynthesis | 12 | 42 | 9.26e-01 |
| KEGG | KEGG:00480 | Glutathione metabolism | 11 | 39 | 9.26e-01 |
| KEGG | KEGG:03020 | RNA polymerase | 9 | 33 | 9.29e-01 |
| KEGG | KEGG:03013 | Nucleocytoplasmic transport | 28 | 96 | 9.32e-01 |
| KEGG | KEGG:04210 | Apoptosis | 33 | 112 | 9.32e-01 |
| KEGG | KEGG:00562 | Inositol phosphate metabolism | 17 | 60 | 9.32e-01 |
| KEGG | KEGG:05202 | Transcriptional misregulation in cancer | 36 | 122 | 9.33e-01 |
| KEGG | KEGG:05161 | Hepatitis B | 37 | 126 | 9.43e-01 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 40 | 136 | 9.46e-01 |
| KEGG | KEGG:03008 | Ribosome biogenesis in eukaryotes | 18 | 65 | 9.50e-01 |
| KEGG | KEGG:05323 | Rheumatoid arthritis | 10 | 38 | 9.50e-01 |
| KEGG | KEGG:04216 | Ferroptosis | 8 | 31 | 9.50e-01 |
| KEGG | KEGG:05221 | Acute myeloid leukemia | 14 | 52 | 9.56e-01 |
| KEGG | KEGG:00900 | Terpenoid backbone biosynthesis | 5 | 21 | 9.63e-01 |
| KEGG | KEGG:05224 | Breast cancer | 32 | 113 | 9.70e-01 |
| KEGG | KEGG:00790 | Folate biosynthesis | 4 | 18 | 9.73e-01 |
| KEGG | KEGG:03050 | Proteasome | 10 | 40 | 9.73e-01 |
| KEGG | KEGG:00860 | Porphyrin metabolism | 4 | 18 | 9.73e-01 |
| KEGG | KEGG:00830 | Retinol metabolism | 3 | 14 | 9.73e-01 |
| KEGG | KEGG:03250 | Viral life cycle - HIV-1 | 14 | 54 | 9.73e-01 |
| KEGG | KEGG:05034 | Alcoholism | 28 | 102 | 9.77e-01 |
| KEGG | KEGG:00270 | Cysteine and methionine metabolism | 9 | 37 | 9.77e-01 |
| KEGG | KEGG:01230 | Biosynthesis of amino acids | 13 | 52 | 9.80e-01 |
| KEGG | KEGG:04330 | Notch signaling pathway | 13 | 52 | 9.80e-01 |
| KEGG | KEGG:00220 | Arginine biosynthesis | 3 | 15 | 9.80e-01 |
| KEGG | KEGG:03440 | Homologous recombination | 9 | 38 | 9.81e-01 |
| KEGG | KEGG:03450 | Non-homologous end-joining | 2 | 12 | 9.93e-01 |
| KEGG | KEGG:01240 | Biosynthesis of cofactors | 29 | 109 | 9.93e-01 |
| KEGG | KEGG:03420 | Nucleotide excision repair | 13 | 54 | 9.93e-01 |
| KEGG | KEGG:04217 | Necroptosis | 25 | 99 | 9.96e-01 |
| KEGG | KEGG:01524 | Platinum drug resistance | 14 | 60 | 9.96e-01 |
| KEGG | KEGG:03430 | Mismatch repair | 4 | 22 | 9.96e-01 |
| KEGG | KEGG:04122 | Sulfur relay system | 1 | 8 | 9.96e-01 |
| KEGG | KEGG:05340 | Primary immunodeficiency | 1 | 8 | 9.96e-01 |
| KEGG | KEGG:00910 | Nitrogen metabolism | 1 | 8 | 9.96e-01 |
| KEGG | KEGG:03460 | Fanconi anemia pathway | 10 | 46 | 9.97e-01 |
| KEGG | KEGG:04966 | Collecting duct acid secretion | 2 | 18 | 1.00e+00 |
| KEGG | KEGG:05322 | Systemic lupus erythematosus | 5 | 44 | 1.00e+00 |
| KEGG | KEGG:04110 | Cell cycle | 38 | 146 | 1.00e+00 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 80 | 388 | 1.00e+00 |
| KEGG | KEGG:04115 | p53 signaling pathway | 10 | 66 | 1.00e+00 |
| KEGG | KEGG:05217 | Basal cell carcinoma | 8 | 48 | 1.00e+00 |
| KEGG | KEGG:03010 | Ribosome | 27 | 121 | 1.00e+00 |
| KEGG | KEGG:04672 | Intestinal immune network for IgA production | 1 | 10 | 1.00e+00 |
| KEGG | KEGG:03030 | DNA replication | 4 | 34 | 1.00e+00 |
| KEGG | KEGG:03410 | Base excision repair | 4 | 44 | 1.00e+00 |
# intersect(unique(Resp_RNA$ENTREZID), unique(NR_peak_list_20k$ENTREZID)))
not_LR_int_restable %>%
filter(source=="GO:BP", p_value < 0.05) %>%
slice_head(n=10) %>%
mutate(log10_p=(-log(p_value, base=10))) %>%
ggplot(., aes(x=log10_p, y=reorder(term_name, log10_p)))+
geom_col(fill="#00BFC4")+
theme_bw()+
ylab("")+
xlab(paste0("-log10 p-value"))+
guides(fill="none")+
theme(axis.text.y = element_text(color="black"))+
ggtitle("not Intersection LR")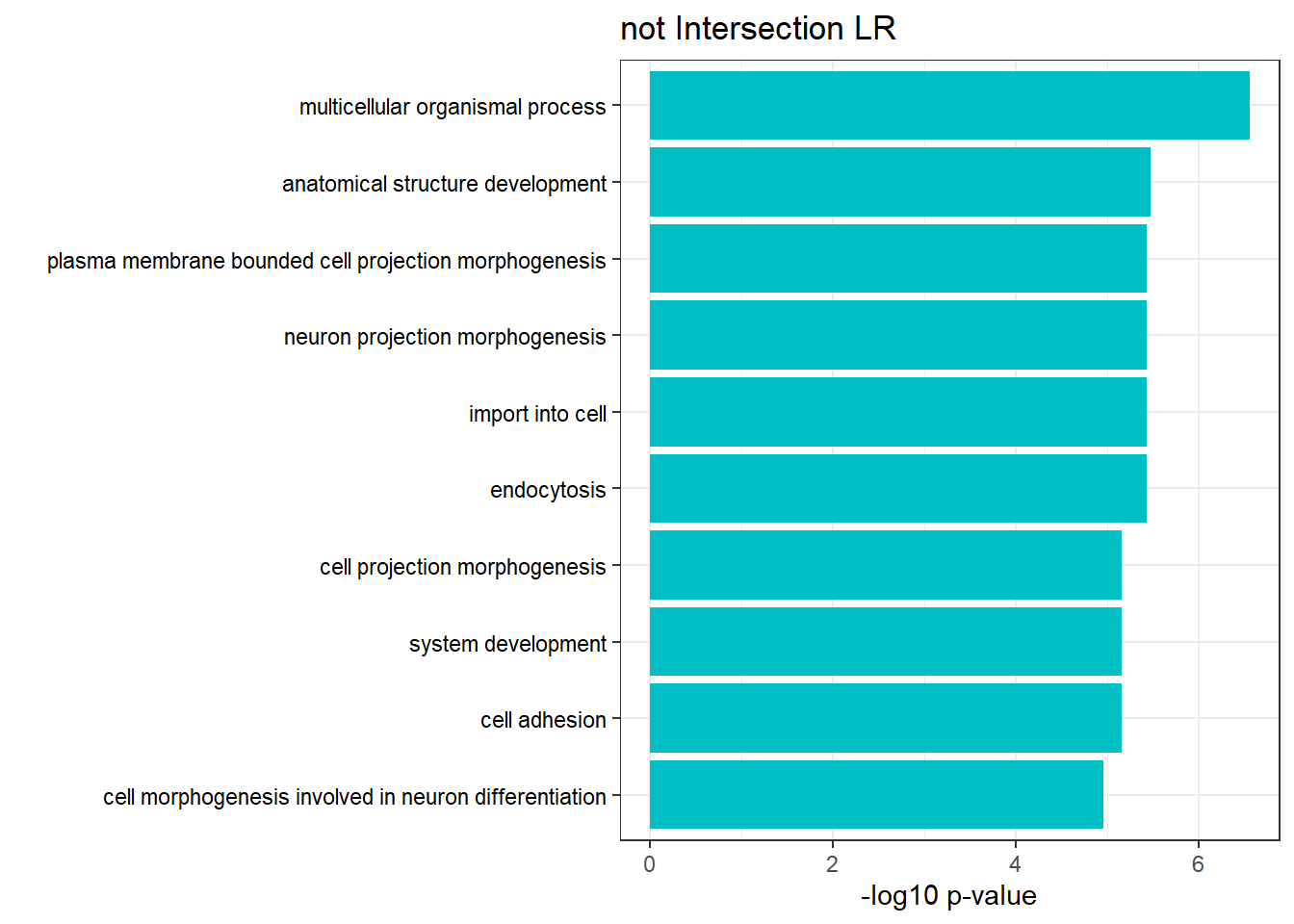
# library(clusterProfiler)
# genes_go = list("EAR"=int_EAR_n_RNAresp,"ESR"=int_ESR_n_RNAresp, "LR"=int_LR_n_RNAresp)
# names(genes) = sub("_", "\n", names(genes))
compEAR <- enrichGO(int_EAR_n_RNAresp,
universe = background_df$ENTREZID,
ont = "BP",
pvalueCutoff = 0.05,
OrgDb='org.Hs.eg.db',
pAdjustMethod = "BH")
# dotplot(compKEGG, showCategory = 15, title = "GO Enrichment Analysis")
compESR <- enrichGO(int_ESR_n_RNAresp,
universe = background_df$ENTREZID,
ont = "BP",
pvalueCutoff = 0.05,
OrgDb='org.Hs.eg.db',
pAdjustMethod = "BH")
# dotplot(compESR, showCategory = 15, title = "GO Enrichment Analysis")
compLR <- enrichGO(int_LR_n_RNAresp,
universe = background_df$ENTREZID,
ont = "BP",
pvalueCutoff = 0.05,
OrgDb='org.Hs.eg.db',
pAdjustMethod = "BH")
barplot(compLR,x="p.adjust", showCategory = 15, title = "GO Enrichment Analysis for LR")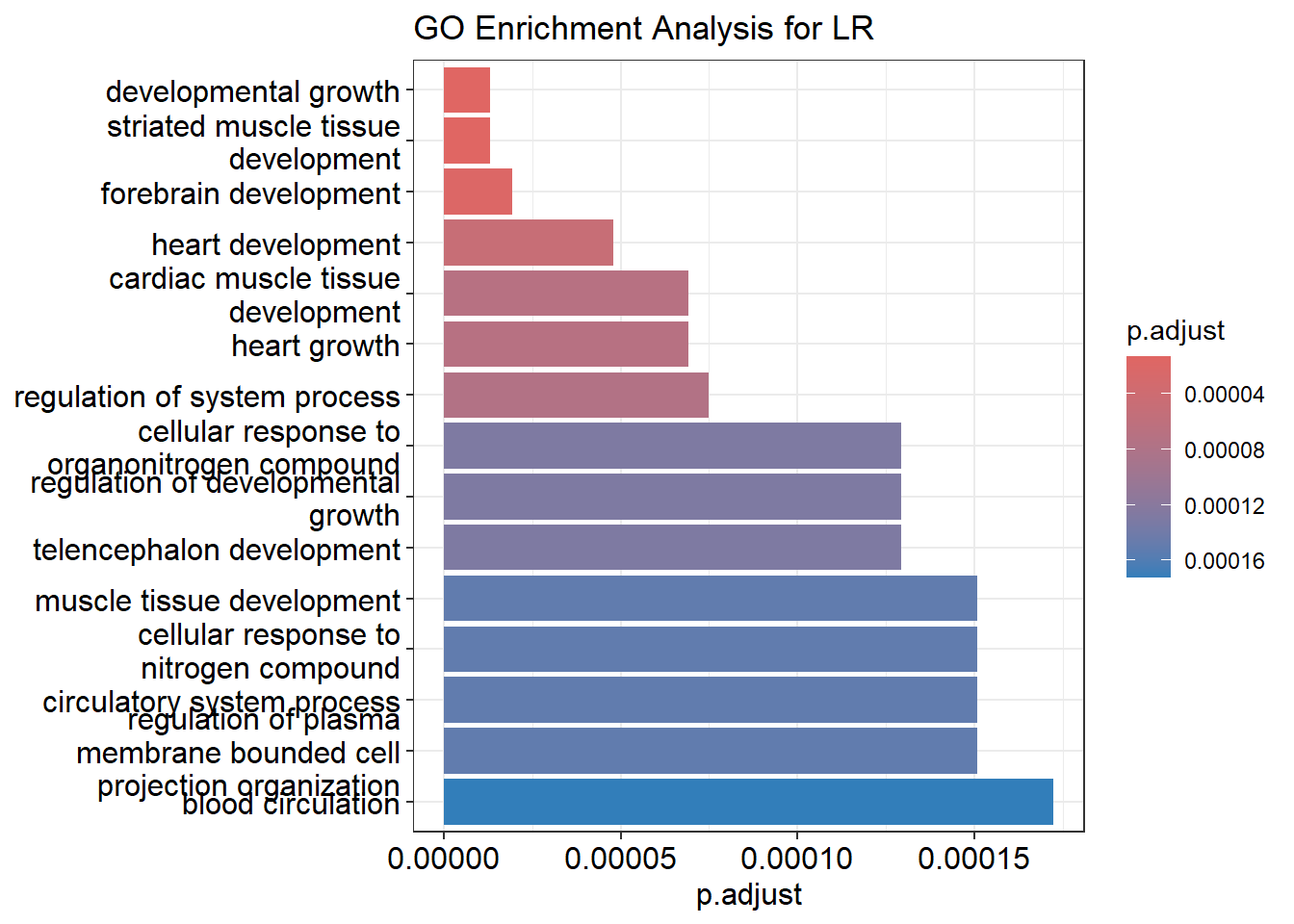
| Version | Author | Date |
|---|---|---|
| eea0976 | reneeisnowhere | 2024-06-03 |
barplot(compEAR,x="p.adjust", showCategory = 15, title = "GO Enrichment Analysis for EAR")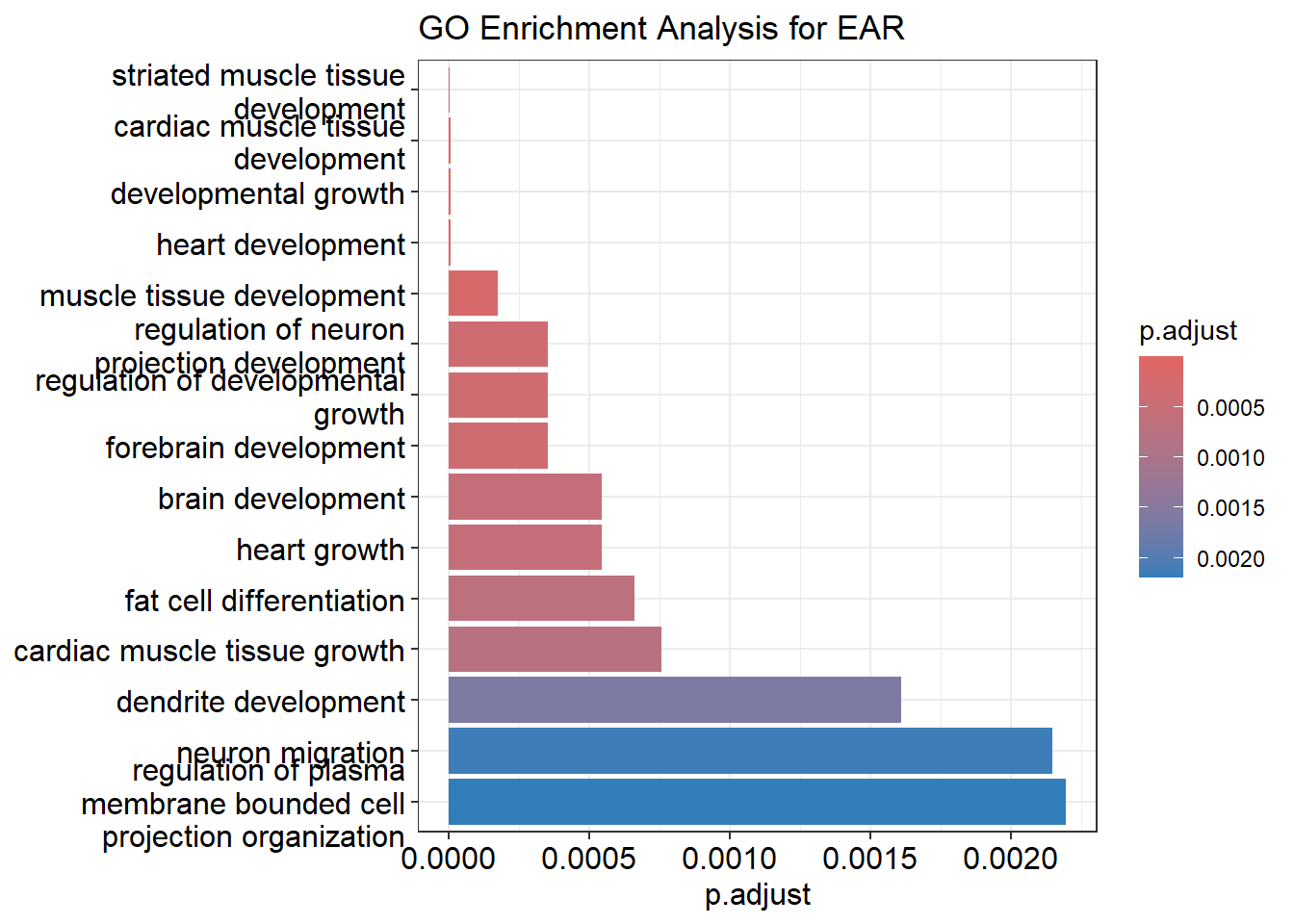
| Version | Author | Date |
|---|---|---|
| eea0976 | reneeisnowhere | 2024-06-03 |
barplot(compESR,x="p.adjust", showCategory = 15, title = "GO Enrichment Analysis for ESR")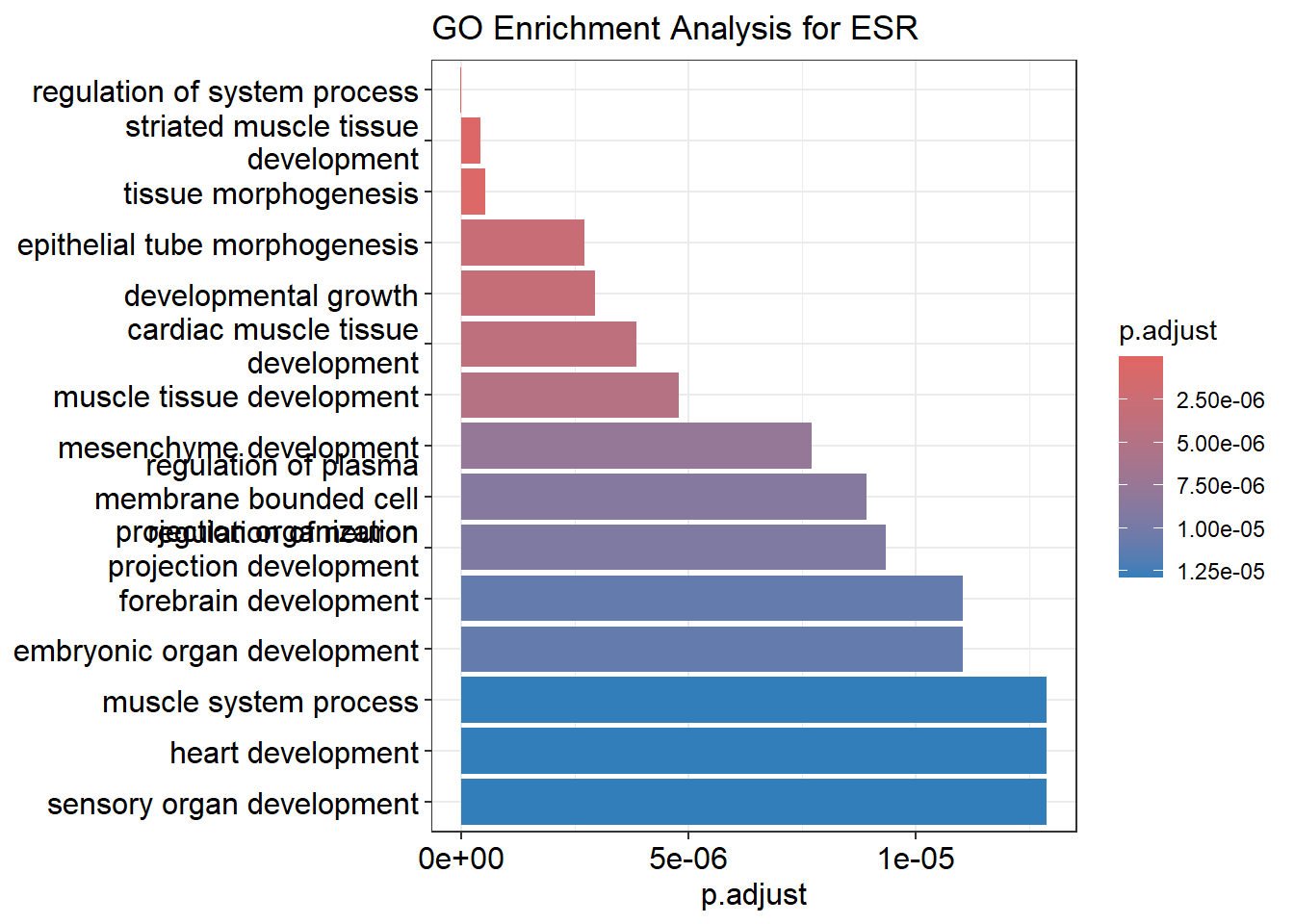
| Version | Author | Date |
|---|---|---|
| eea0976 | reneeisnowhere | 2024-06-03 |
# goplot(compLR)
compEARx <- setReadable(compEAR, 'org.Hs.eg.db', 'ENTREZID')
compESRx <- setReadable(compESR, 'org.Hs.eg.db', 'ENTREZID')
compLRx <- setReadable(compLR, 'org.Hs.eg.db', 'ENTREZID')cnetplot(compEARx)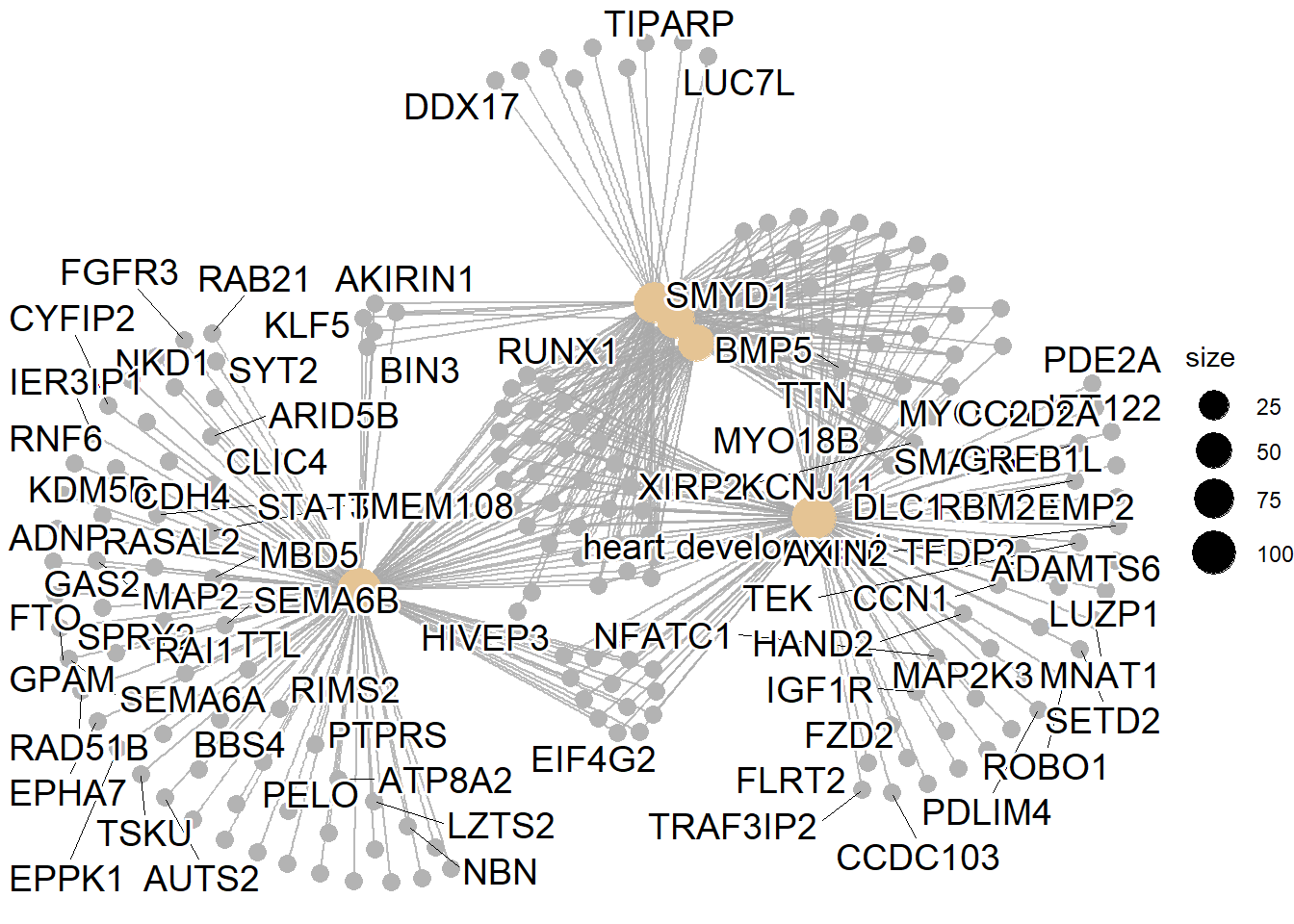
| Version | Author | Date |
|---|---|---|
| eea0976 | reneeisnowhere | 2024-06-03 |
cnetplot(compESRx)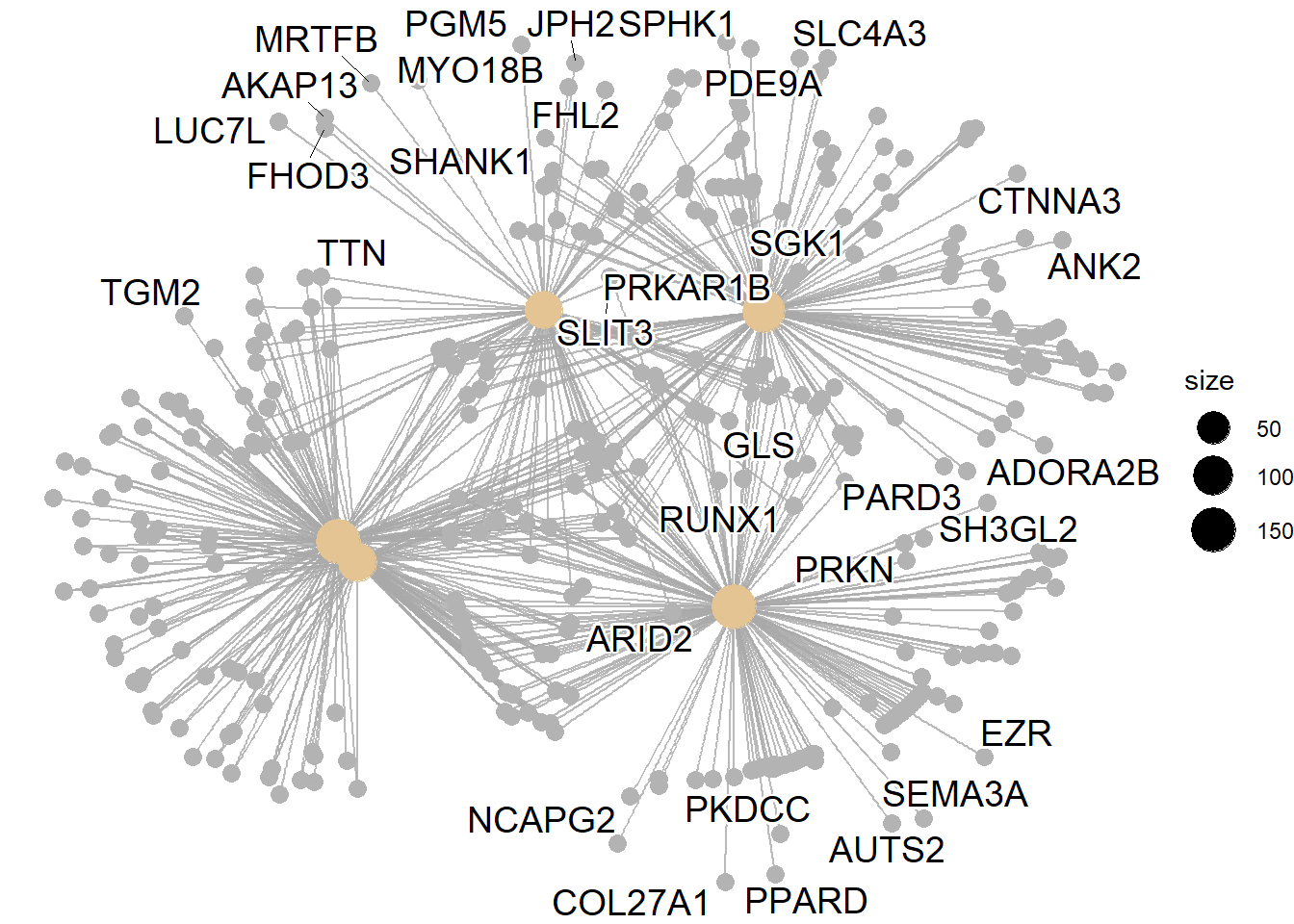
| Version | Author | Date |
|---|---|---|
| eea0976 | reneeisnowhere | 2024-06-03 |
cnetplot(compLRx, node_label="all")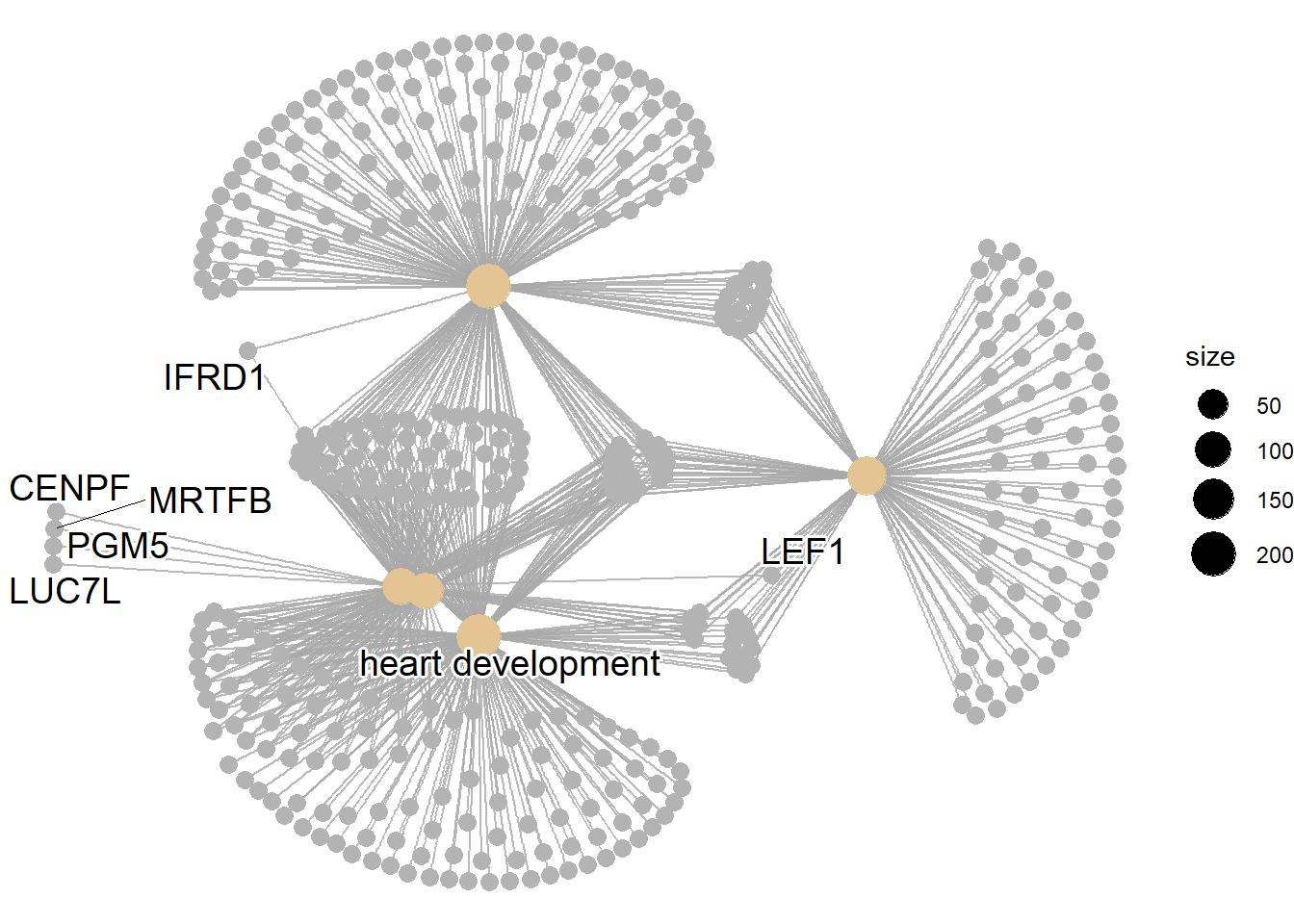
| Version | Author | Date |
|---|---|---|
| eea0976 | reneeisnowhere | 2024-06-03 |
# upsetplot(compLRx)
sessionInfo()R version 4.3.1 (2023-06-16 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19045)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
time zone: America/Chicago
tzcode source: internal
attached base packages:
[1] grid stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] clusterProfiler_4.10.1
[2] biomaRt_2.58.2
[3] ggpubr_0.6.0
[4] BiocParallel_1.36.0
[5] scales_1.3.0
[6] VennDiagram_1.7.3
[7] futile.logger_1.4.3
[8] gridExtra_2.3
[9] ggfortify_0.4.17
[10] edgeR_4.0.16
[11] limma_3.58.1
[12] rtracklayer_1.62.0
[13] org.Hs.eg.db_3.18.0
[14] TxDb.Hsapiens.UCSC.hg38.knownGene_3.18.0
[15] GenomicFeatures_1.54.4
[16] AnnotationDbi_1.64.1
[17] Biobase_2.62.0
[18] GenomicRanges_1.54.1
[19] GenomeInfoDb_1.38.8
[20] IRanges_2.36.0
[21] S4Vectors_0.40.2
[22] BiocGenerics_0.48.1
[23] ChIPseeker_1.38.0
[24] gprofiler2_0.2.3
[25] RColorBrewer_1.1-3
[26] broom_1.0.6
[27] kableExtra_1.4.0
[28] lubridate_1.9.3
[29] forcats_1.0.0
[30] stringr_1.5.1
[31] dplyr_1.1.4
[32] purrr_1.0.2
[33] readr_2.1.5
[34] tidyr_1.3.1
[35] tibble_3.2.1
[36] ggplot2_3.5.1
[37] tidyverse_2.0.0
[38] workflowr_1.7.1
loaded via a namespace (and not attached):
[1] fs_1.6.4
[2] matrixStats_1.3.0
[3] bitops_1.0-7
[4] enrichplot_1.22.0
[5] HDO.db_0.99.1
[6] httr_1.4.7
[7] tools_4.3.1
[8] backports_1.5.0
[9] utf8_1.2.4
[10] R6_2.5.1
[11] lazyeval_0.2.2
[12] withr_3.0.0
[13] prettyunits_1.2.0
[14] cli_3.6.2
[15] formatR_1.14
[16] scatterpie_0.2.2
[17] labeling_0.4.3
[18] sass_0.4.9
[19] Rsamtools_2.18.0
[20] systemfonts_1.1.0
[21] yulab.utils_0.1.4
[22] gson_0.1.0
[23] DOSE_3.28.2
[24] svglite_2.1.3
[25] plotrix_3.8-4
[26] rstudioapi_0.16.0
[27] RSQLite_2.3.7
[28] generics_0.1.3
[29] gridGraphics_0.5-1
[30] TxDb.Hsapiens.UCSC.hg19.knownGene_3.2.2
[31] BiocIO_1.12.0
[32] crosstalk_1.2.1
[33] vroom_1.6.5
[34] gtools_3.9.5
[35] car_3.1-2
[36] GO.db_3.18.0
[37] Matrix_1.6-5
[38] fansi_1.0.6
[39] abind_1.4-5
[40] lifecycle_1.0.4
[41] whisker_0.4.1
[42] yaml_2.3.8
[43] carData_3.0-5
[44] SummarizedExperiment_1.32.0
[45] gplots_3.1.3.1
[46] qvalue_2.34.0
[47] SparseArray_1.2.4
[48] BiocFileCache_2.10.2
[49] blob_1.2.4
[50] promises_1.3.0
[51] crayon_1.5.2
[52] lattice_0.22-6
[53] cowplot_1.1.3
[54] KEGGREST_1.42.0
[55] pillar_1.9.0
[56] knitr_1.47
[57] fgsea_1.28.0
[58] rjson_0.2.21
[59] boot_1.3-30
[60] codetools_0.2-20
[61] fastmatch_1.1-4
[62] glue_1.7.0
[63] getPass_0.2-4
[64] ggfun_0.1.5
[65] data.table_1.15.4
[66] vctrs_0.6.5
[67] png_0.1-8
[68] treeio_1.26.0
[69] gtable_0.3.5
[70] cachem_1.1.0
[71] xfun_0.44
[72] S4Arrays_1.2.1
[73] mime_0.12
[74] tidygraph_1.3.1
[75] statmod_1.5.0
[76] interactiveDisplayBase_1.40.0
[77] nlme_3.1-164
[78] ggtree_3.10.1
[79] bit64_4.0.5
[80] progress_1.2.3
[81] filelock_1.0.3
[82] rprojroot_2.0.4
[83] bslib_0.7.0
[84] KernSmooth_2.23-24
[85] colorspace_2.1-0
[86] DBI_1.2.3
[87] tidyselect_1.2.1
[88] processx_3.8.4
[89] bit_4.0.5
[90] compiler_4.3.1
[91] curl_5.2.1
[92] git2r_0.33.0
[93] xml2_1.3.6
[94] DelayedArray_0.28.0
[95] plotly_4.10.4
[96] shadowtext_0.1.3
[97] caTools_1.18.2
[98] callr_3.7.6
[99] rappdirs_0.3.3
[100] digest_0.6.35
[101] rmarkdown_2.27
[102] XVector_0.42.0
[103] htmltools_0.5.8.1
[104] pkgconfig_2.0.3
[105] MatrixGenerics_1.14.0
[106] highr_0.11
[107] dbplyr_2.5.0
[108] fastmap_1.2.0
[109] rlang_1.1.3
[110] htmlwidgets_1.6.4
[111] shiny_1.8.1.1
[112] farver_2.1.2
[113] jquerylib_0.1.4
[114] jsonlite_1.8.8
[115] GOSemSim_2.28.1
[116] RCurl_1.98-1.14
[117] magrittr_2.0.3
[118] GenomeInfoDbData_1.2.11
[119] ggplotify_0.1.2
[120] patchwork_1.2.0
[121] munsell_0.5.1
[122] Rcpp_1.0.12
[123] ape_5.8
[124] viridis_0.6.5
[125] stringi_1.8.4
[126] ggraph_2.2.1
[127] zlibbioc_1.48.2
[128] MASS_7.3-60.0.1
[129] AnnotationHub_3.10.1
[130] plyr_1.8.9
[131] parallel_4.3.1
[132] ggrepel_0.9.5
[133] Biostrings_2.70.3
[134] graphlayouts_1.1.1
[135] splines_4.3.1
[136] hms_1.1.3
[137] locfit_1.5-9.9
[138] ps_1.7.6
[139] igraph_2.0.3
[140] ggsignif_0.6.4
[141] reshape2_1.4.4
[142] futile.options_1.0.1
[143] BiocVersion_3.18.1
[144] XML_3.99-0.16.1
[145] evaluate_0.23
[146] lambda.r_1.2.4
[147] BiocManager_1.30.23
[148] tzdb_0.4.0
[149] tweenr_2.0.3
[150] httpuv_1.6.15
[151] polyclip_1.10-6
[152] ggforce_0.4.2
[153] xtable_1.8-4
[154] restfulr_0.0.15
[155] tidytree_0.4.6
[156] rstatix_0.7.2
[157] later_1.3.2
[158] viridisLite_0.4.2
[159] aplot_0.2.2
[160] memoise_2.0.1
[161] GenomicAlignments_1.38.2
[162] timechange_0.3.0