Enhancer_files
ERM
2024-10-07
Last updated: 2024-10-07
Checks: 7 0
Knit directory: ATAC_learning/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20231016) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version d926a4a. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/ACresp_SNP_table.csv
Ignored: data/ARR_SNP_table.csv
Ignored: data/All_merged_peaks.tsv
Ignored: data/CAD_gwas_dataframe.RDS
Ignored: data/CTX_SNP_table.csv
Ignored: data/Collapsed_expressed_NG_peak_table.csv
Ignored: data/DEG_toplist_sep_n45.RDS
Ignored: data/FRiP_first_run.txt
Ignored: data/Final_four_data/
Ignored: data/Frip_1_reads.csv
Ignored: data/Frip_2_reads.csv
Ignored: data/Frip_3_reads.csv
Ignored: data/Frip_4_reads.csv
Ignored: data/Frip_5_reads.csv
Ignored: data/Frip_6_reads.csv
Ignored: data/GO_KEGG_analysis/
Ignored: data/HF_SNP_table.csv
Ignored: data/Ind1_75DA24h_dedup_peaks.csv
Ignored: data/Ind1_TSS_peaks.RDS
Ignored: data/Ind1_firstfragment_files.txt
Ignored: data/Ind1_fragment_files.txt
Ignored: data/Ind1_peaks_list.RDS
Ignored: data/Ind1_summary.txt
Ignored: data/Ind2_TSS_peaks.RDS
Ignored: data/Ind2_fragment_files.txt
Ignored: data/Ind2_peaks_list.RDS
Ignored: data/Ind2_summary.txt
Ignored: data/Ind3_TSS_peaks.RDS
Ignored: data/Ind3_fragment_files.txt
Ignored: data/Ind3_peaks_list.RDS
Ignored: data/Ind3_summary.txt
Ignored: data/Ind4_79B24h_dedup_peaks.csv
Ignored: data/Ind4_TSS_peaks.RDS
Ignored: data/Ind4_V24h_fraglength.txt
Ignored: data/Ind4_fragment_files.txt
Ignored: data/Ind4_fragment_filesN.txt
Ignored: data/Ind4_peaks_list.RDS
Ignored: data/Ind4_summary.txt
Ignored: data/Ind5_TSS_peaks.RDS
Ignored: data/Ind5_fragment_files.txt
Ignored: data/Ind5_fragment_filesN.txt
Ignored: data/Ind5_peaks_list.RDS
Ignored: data/Ind5_summary.txt
Ignored: data/Ind6_TSS_peaks.RDS
Ignored: data/Ind6_fragment_files.txt
Ignored: data/Ind6_peaks_list.RDS
Ignored: data/Ind6_summary.txt
Ignored: data/Knowles_4.RDS
Ignored: data/Knowles_5.RDS
Ignored: data/Knowles_6.RDS
Ignored: data/LiSiLTDNRe_TE_df.RDS
Ignored: data/MI_gwas.RDS
Ignored: data/SNP_GWAS_PEAK_MRC_id
Ignored: data/SNP_GWAS_PEAK_MRC_id.csv
Ignored: data/SNP_gene_cat_list.tsv
Ignored: data/SNP_supp_schneider.RDS
Ignored: data/TE_info/
Ignored: data/TFmapnames.RDS
Ignored: data/all_TSSE_scores.RDS
Ignored: data/all_four_filtered_counts.txt
Ignored: data/aln_run1_results.txt
Ignored: data/anno_ind1_DA24h.RDS
Ignored: data/anno_ind4_V24h.RDS
Ignored: data/annotated_gwas_SNPS.csv
Ignored: data/background_n45_he_peaks.RDS
Ignored: data/cardiac_muscle_FRIP.csv
Ignored: data/cardiomyocyte_FRIP.csv
Ignored: data/col_ng_peak.csv
Ignored: data/cormotif_full_4_run.RDS
Ignored: data/cormotif_full_4_run_he.RDS
Ignored: data/cormotif_full_6_run.RDS
Ignored: data/cormotif_full_6_run_he.RDS
Ignored: data/cormotif_probability_45_list.csv
Ignored: data/cormotif_probability_45_list_he.csv
Ignored: data/cormotif_probability_all_6_list.csv
Ignored: data/cormotif_probability_all_6_list_he.csv
Ignored: data/embryo_heart_FRIP.csv
Ignored: data/enhancer_list_ENCFF126UHK.bed
Ignored: data/enhancerdata/
Ignored: data/filt_Peaks_efit2.RDS
Ignored: data/filt_Peaks_efit2_bl.RDS
Ignored: data/filt_Peaks_efit2_n45.RDS
Ignored: data/first_Peaksummarycounts.csv
Ignored: data/first_run_frag_counts.txt
Ignored: data/full_bedfiles/
Ignored: data/gene_ref.csv
Ignored: data/gwas_1_dataframe.RDS
Ignored: data/gwas_2_dataframe.RDS
Ignored: data/gwas_3_dataframe.RDS
Ignored: data/gwas_4_dataframe.RDS
Ignored: data/gwas_5_dataframe.RDS
Ignored: data/high_conf_peak_counts.csv
Ignored: data/high_conf_peak_counts.txt
Ignored: data/high_conf_peaks_bl_counts.txt
Ignored: data/high_conf_peaks_counts.txt
Ignored: data/hits_files/
Ignored: data/hyper_files/
Ignored: data/hypo_files/
Ignored: data/ind1_DA24hpeaks.RDS
Ignored: data/ind1_TSSE.RDS
Ignored: data/ind2_TSSE.RDS
Ignored: data/ind3_TSSE.RDS
Ignored: data/ind4_TSSE.RDS
Ignored: data/ind4_V24hpeaks.RDS
Ignored: data/ind5_TSSE.RDS
Ignored: data/ind6_TSSE.RDS
Ignored: data/initial_complete_stats_run1.txt
Ignored: data/left_ventricle_FRIP.csv
Ignored: data/median_24_lfc.RDS
Ignored: data/median_3_lfc.RDS
Ignored: data/mergedPeads.gff
Ignored: data/mergedPeaks.gff
Ignored: data/motif_list_full
Ignored: data/motif_list_n45
Ignored: data/motif_list_n45.RDS
Ignored: data/multiqc_fastqc_run1.txt
Ignored: data/multiqc_fastqc_run2.txt
Ignored: data/multiqc_genestat_run1.txt
Ignored: data/multiqc_genestat_run2.txt
Ignored: data/my_hc_filt_counts.RDS
Ignored: data/my_hc_filt_counts_n45.RDS
Ignored: data/n45_bedfiles/
Ignored: data/n45_files
Ignored: data/other_papers/
Ignored: data/output_100_MA0835.2.txt
Ignored: data/output_101_MA0018.4.txt
Ignored: data/output_102_MA1102.2.txt
Ignored: data/output_103_MA0162.4.txt
Ignored: data/output_104_MA0598.3.txt
Ignored: data/output_105_MA0473.3.txt
Ignored: data/output_106_MA0640.2.txt
Ignored: data/output_107_MA0761.2.txt
Ignored: data/output_108_MA0477.2.txt
Ignored: data/output_109_MA0062.3.txt
Ignored: data/output_10_MA0076.2.txt
Ignored: data/output_110_MA0035.4.txt
Ignored: data/output_111_MA0482.2.txt
Ignored: data/output_112_MA1104.2.txt
Ignored: data/output_113_MA0490.2.txt
Ignored: data/output_114_MA0491.2.txt
Ignored: data/output_115_MA1107.2.txt
Ignored: data/output_116_MA0496.3.txt
Ignored: data/output_117_MA0620.3.txt
Ignored: data/output_118_MA0500.2.txt
Ignored: data/output_119_MA0089.2.txt
Ignored: data/output_11_MA0258.2.txt
Ignored: data/output_120_MA0782.2.txt
Ignored: data/output_121_MA0090.3.txt
Ignored: data/output_122_MA0809.2.txt
Ignored: data/output_123_MA0093.3.txt
Ignored: data/output_124_MA0609.2.txt
Ignored: data/output_125_MA1684.1.txt
Ignored: data/output_126_MA1928.1.txt
Ignored: data/output_127_MA1929.1.txt
Ignored: data/output_128_MA1930.1.txt
Ignored: data/output_129_MA1934.1.txt
Ignored: data/output_12_MA0150.2.txt
Ignored: data/output_130_MA1940.1.txt
Ignored: data/output_131_MA1942.1.txt
Ignored: data/output_132_MA1944.1.txt
Ignored: data/output_133_MA1951.1.txt
Ignored: data/output_134_MA1959.1.txt
Ignored: data/output_135_MA1964.1.txt
Ignored: data/output_136_MA1970.1.txt
Ignored: data/output_137_MA1988.1.txt
Ignored: data/output_138_MA1992.1.txt
Ignored: data/output_139_MA1993.1.txt
Ignored: data/output_13_MA0140.2.txt
Ignored: data/output_140_MA1995.1.txt
Ignored: data/output_141_MA1997.1.txt
Ignored: data/output_142_MA2002.1.txt
Ignored: data/output_143_MA0037.4.txt
Ignored: data/output_144_MA0156.3.txt
Ignored: data/output_145_MA0474.3.txt
Ignored: data/output_146_MA0489.2.txt
Ignored: data/output_147_MA0493.2.txt
Ignored: data/output_148_MA0516.3.txt
Ignored: data/output_149_MA0521.2.txt
Ignored: data/output_14_MA0591.1.txt
Ignored: data/output_150_MA0633.2.txt
Ignored: data/output_151_MA0659.3.txt
Ignored: data/output_152_MA0685.2.txt
Ignored: data/output_153_MA0697.2.txt
Ignored: data/output_154_MA0740.2.txt
Ignored: data/output_155_MA0742.2.txt
Ignored: data/output_156_MA0764.3.txt
Ignored: data/output_157_MA0831.3.txt
Ignored: data/output_158_MA1472.2.txt
Ignored: data/output_159_MA1483.2.txt
Ignored: data/output_15_MA0595.1.txt
Ignored: data/output_160_MA1511.2.txt
Ignored: data/output_161_MA1633.2.txt
Ignored: data/output_16_MA0596.1.txt
Ignored: data/output_17_MA0599.1.txt
Ignored: data/output_18_MA0603.1.txt
Ignored: data/output_19_MA0645.1.txt
Ignored: data/output_1_MA0029.1.txt
Ignored: data/output_1_Mecom.txt
Ignored: data/output_20_MA0475.2.txt
Ignored: data/output_21_MA0655.1.txt
Ignored: data/output_22_MA0656.1.txt
Ignored: data/output_23_MA0660.1.txt
Ignored: data/output_24_MA0741.1.txt
Ignored: data/output_25_MA0747.1.txt
Ignored: data/output_26_MA0751.1.txt
Ignored: data/output_27_MA0760.1.txt
Ignored: data/output_28_MA0098.3.txt
Ignored: data/output_29_MA0762.1.txt
Ignored: data/output_2_MA0119.1.txt
Ignored: data/output_30_MA0498.2.txt
Ignored: data/output_31_MA0774.1.txt
Ignored: data/output_32_MA0775.1.txt
Ignored: data/output_33_MA0783.1.txt
Ignored: data/output_34_MA0797.1.txt
Ignored: data/output_35_MA0510.2.txt
Ignored: data/output_36_MA0808.1.txt
Ignored: data/output_37_MA0816.1.txt
Ignored: data/output_38_MA0840.1.txt
Ignored: data/output_39_MA0117.2.txt
Ignored: data/output_3_MA0139.1.txt
Ignored: data/output_40_MA0841.1.txt
Ignored: data/output_41_MA0036.3.txt
Ignored: data/output_42_MA1114.1.txt
Ignored: data/output_43_MA1121.1.txt
Ignored: data/output_44_MA0750.2.txt
Ignored: data/output_45_MA0099.3.txt
Ignored: data/output_46_MA1126.1.txt
Ignored: data/output_47_MA1128.1.txt
Ignored: data/output_48_MA1129.1.txt
Ignored: data/output_49_MA1130.1.txt
Ignored: data/output_4_MA0145.2.txt
Ignored: data/output_50_MA1131.1.txt
Ignored: data/output_51_MA1132.1.txt
Ignored: data/output_52_MA1133.1.txt
Ignored: data/output_53_MA1134.1.txt
Ignored: data/output_54_MA1135.1.txt
Ignored: data/output_55_MA1136.1.txt
Ignored: data/output_56_MA1137.1.txt
Ignored: data/output_57_MA1138.1.txt
Ignored: data/output_58_MA1139.1.txt
Ignored: data/output_59_MA1141.1.txt
Ignored: data/output_5_MA0476.1.txt
Ignored: data/output_60_MA1142.1.txt
Ignored: data/output_61_MA1143.1.txt
Ignored: data/output_62_MA1144.1.txt
Ignored: data/output_63_MA1145.1.txt
Ignored: data/output_64_MA1101.2.txt
Ignored: data/output_65_MA1474.1.txt
Ignored: data/output_66_MA1475.1.txt
Ignored: data/output_67_MA1484.1.txt
Ignored: data/output_68_MA1512.1.txt
Ignored: data/output_69_MA1516.1.txt
Ignored: data/output_6_MA0478.1.txt
Ignored: data/output_70_MA1517.1.txt
Ignored: data/output_71_MA1520.1.txt
Ignored: data/output_72_MA1521.1.txt
Ignored: data/output_73_MA1523.1.txt
Ignored: data/output_74_MA1527.1.txt
Ignored: data/output_75_MA1528.1.txt
Ignored: data/output_76_MA1564.1.txt
Ignored: data/output_77_MA1571.1.txt
Ignored: data/output_78_MA1572.1.txt
Ignored: data/output_79_MA1593.1.txt
Ignored: data/output_7_MA0488.1.txt
Ignored: data/output_80_MA1599.1.txt
Ignored: data/output_81_MA1628.1.txt
Ignored: data/output_82_MA1629.1.txt
Ignored: data/output_83_MA1100.2.txt
Ignored: data/output_84_MA0462.2.txt
Ignored: data/output_85_MA0766.2.txt
Ignored: data/output_86_MA1140.2.txt
Ignored: data/output_87_MA0842.2.txt
Ignored: data/output_88_MA0746.2.txt
Ignored: data/output_89_MA1713.1.txt
Ignored: data/output_8_MA0492.1.txt
Ignored: data/output_90_MA1721.1.txt
Ignored: data/output_91_MA1632.1.txt
Ignored: data/output_92_MA1634.1.txt
Ignored: data/output_93_MA1635.1.txt
Ignored: data/output_94_MA1641.1.txt
Ignored: data/output_95_MA1643.1.txt
Ignored: data/output_96_MA1645.1.txt
Ignored: data/output_97_MA1656.1.txt
Ignored: data/output_98_MA1726.1.txt
Ignored: data/output_99_MA1728.1.txt
Ignored: data/output_9_MA0501.1.txt
Ignored: data/peakAnnoList_1.RDS
Ignored: data/peakAnnoList_2.RDS
Ignored: data/peakAnnoList_24_full.RDS
Ignored: data/peakAnnoList_24_n45.RDS
Ignored: data/peakAnnoList_3.RDS
Ignored: data/peakAnnoList_3_full.RDS
Ignored: data/peakAnnoList_3_n45.RDS
Ignored: data/peakAnnoList_4.RDS
Ignored: data/peakAnnoList_5.RDS
Ignored: data/peakAnnoList_6.RDS
Ignored: data/peakAnnoList_Eight.RDS
Ignored: data/peakAnnoList_full_motif.RDS
Ignored: data/peakAnnoList_n45_motif.RDS
Ignored: data/siglist_full.RDS
Ignored: data/siglist_n45.RDS
Ignored: data/summary_peakIDandReHeat.csv
Ignored: data/test.list.RDS
Ignored: data/testnames.txt
Ignored: data/toplist_6.RDS
Ignored: data/toplist_full.RDS
Ignored: data/toplist_full_DAR_6.RDS
Ignored: data/toplist_n45.RDS
Ignored: data/trimmed_seq_length.csv
Ignored: data/unclassified_full_set_peaks.RDS
Ignored: data/unclassified_n45_set_peaks.RDS
Ignored: data/xstreme/
Ignored: trimmed_Ind1_75DA24h_S7.nodup.splited.bam/
Untracked files:
Untracked: Correlationplot_scaled.pdf
Untracked: DOX_DAR_assess.Rmd
Untracked: EAR_2_plot.pdf
Untracked: ESR_1_plot.pdf
Untracked: Firstcorr plotATAC.pdf
Untracked: IND1_2_3_6_corrplot.pdf
Untracked: LR_3_plot.pdf
Untracked: NR_1_plot.pdf
Untracked: analysis/LFC_corr.Rmd
Untracked: analysis/ReHeat_analysis.Rmd
Untracked: analysis/TE_analysis_old.Rmd
Untracked: analysis/my_hc_filt_counts.csv
Untracked: analysis/nucleosome_explore.Rmd
Untracked: code/IGV_snapshot_code.R
Untracked: code/LongDARlist.R
Untracked: code/MRC_clusterlog2cpm.R
Untracked: code/TSSE.R
Untracked: code/just_for_Fun.R
Untracked: code/toplist_assembly.R
Untracked: lcpm_filtered_corplot.pdf
Untracked: log2cpmfragcount.pdf
Untracked: output/cormotif_probability_45_list.csv
Untracked: output/cormotif_probability_all_6_list.csv
Untracked: output_1_Mecom.txt
Untracked: splited/
Untracked: trimmed_Ind1_75DA24h_S7.nodup.fragment.size.distribution.pdf
Untracked: trimmed_Ind1_75DA3h_S1.nodup.fragment.size.distribution.pdf
Unstaged changes:
Modified: analysis/CorMotif_data_n45.Rmd
Modified: analysis/Enrichment_motif.Rmd
Modified: analysis/Jaspar_motif_ff.Rmd
Modified: analysis/Peak_calling.Rmd
Modified: analysis/Raodah.Rmd
Modified: analysis/Smaller_set_DAR.Rmd
Modified: analysis/TE_analysis.Rmd
Modified: analysis/final_four_analysis.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/Enhancer_files_ff.Rmd) and
HTML (docs/Enhancer_files_ff.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | d926a4a | reneeisnowhere | 2024-10-07 | Build site. |
| Rmd | 2132070 | reneeisnowhere | 2024-10-07 | heatmmaps baby! |
| html | 6e3a803 | reneeisnowhere | 2024-09-30 | Build site. |
| Rmd | 74f6474 | reneeisnowhere | 2024-09-30 | updates to code |
| html | e174c87 | reneeisnowhere | 2024-09-23 | Build site. |
| Rmd | c473b77 | reneeisnowhere | 2024-09-23 | adding the updated 8 groups |
| Rmd | a6d16a7 | reneeisnowhere | 2024-09-10 | updates |
####Loading
library(tidyverse)
library(kableExtra)
library(broom)
library(RColorBrewer)
library(ChIPseeker)
library("TxDb.Hsapiens.UCSC.hg38.knownGene")
library("org.Hs.eg.db")
library(rtracklayer)
library(edgeR)
library(ggfortify)
library(limma)
library(readr)
library(BiocGenerics)
library(gridExtra)
library(VennDiagram)
library(scales)
# library(Cormotif)
library(BiocParallel)
library(ggpubr)
library(devtools)
library(JASPAR2022)
library(TFBSTools)
library(MotifDb)
library(BSgenome.Hsapiens.UCSC.hg38)
library(plyranges)
library(genomation)
library(gprofiler2)Data loading
# toplistall_RNA <- readRDS("data/other_papers/toplistall_RNA.RDS")
# S13Table <- read.csv( "data/other_papers/S13Table_Matthews2024.csv",row.names = 1)
# ##14021
#
# EAR_RNA <- S13Table %>%
# dplyr::filter(MOTIF=="EAR") %>%
# dplyr::select(ENTREZID) %>%
# mutate(ENTREZID= as.character(ENTREZID))
# ESR_RNA <- S13Table %>%
# dplyr::filter(MOTIF=="ESR")%>%
# dplyr::select(ENTREZID)%>%
# mutate(ENTREZID= as.character(ENTREZID))
# LR_RNA <- S13Table %>%
# dplyr::filter(MOTIF=="LR")%>%
# dplyr::select(ENTREZID)%>%
# mutate(ENTREZID=as.character(ENTREZID))
# NR_RNA <- S13Table %>%
# dplyr::filter(MOTIF=="NR")%>%
# dplyr::select(ENTREZID)%>%
# mutate(ENTREZID= as.character(ENTREZID))
# exp_neargene_table <- read_delim("data/n45_bedfiles/exp_neargene_table.tsv",
# delim = "\t", escape_double = FALSE,
# trim_ws = TRUE)
TSS_NG_data <- read_delim("data/Final_four_data/TSS_assigned_NG.txt",
delim = "\t", escape_double = FALSE,
trim_ws = TRUE)
col_ng_peak <- read.delim("data/Final_four_data/collapsed_new_peaks.txt")
peakAnnoList_ff_8motif <- readRDS("data/Final_four_data/peakAnnoList_ff_8motif.RDS")
# list2env(peakAnnoList_n45_motif, envir = .GlobalEnv)
# peakAnnoList_3_n45 <- readRDS("data/peakAnnoList_3_n45.RDS")
# peakAnnoList_24_n45<- readRDS("data/peakAnnoList_24_n45.RDS")
all_peak_gr <- as.GRanges(peakAnnoList_ff_8motif$background)
# Joined_tss_neargene <- all_peak_gr %>%
# as.data.frame() %>%
# dplyr::select(seqnames:id,geneId,distanceToTSS) %>%
# dplyr::filter(!grepl("chrX",id)& !grepl("chrY",id))%>%
# dplyr::rename("close_geneId"="geneId") %>%
# mutate(start=start+1) %>%
# left_join(., TSS_NG_data, by=c("id"="peakid", "start"="start","end"="end")) %>%
# dplyr::select(seqnames.x:close_geneId,distanceToTSS, entrezgene_id:dist_to_NG)
EAR_open <- as.data.frame(peakAnnoList_ff_8motif$EAR_open)
EAR_open_gr <- as.GRanges(peakAnnoList_ff_8motif$EAR_open)
EAR_close <- as.data.frame(peakAnnoList_ff_8motif$EAR_close)
EAR_close_gr <- as.GRanges(peakAnnoList_ff_8motif$EAR_close)
ESR_open <- as.data.frame(peakAnnoList_ff_8motif$ESR_open)
ESR_open_gr <- as.GRanges(peakAnnoList_ff_8motif$ESR_open)
ESR_close <- as.data.frame(peakAnnoList_ff_8motif$ESR_close)
ESR_close_gr <- as.GRanges(peakAnnoList_ff_8motif$ESR_close)
ESR_OC <- as.data.frame(peakAnnoList_ff_8motif$ESR_OC)
ESR_OC_gr <- as.GRanges(peakAnnoList_ff_8motif$ESR_OC)
LR_open <- as.data.frame(peakAnnoList_ff_8motif$LR_open)
LR_open_gr <- as.GRanges(peakAnnoList_ff_8motif$LR_open)
LR_close <- as.data.frame(peakAnnoList_ff_8motif$LR_close)
LR_close_gr <- as.GRanges(peakAnnoList_ff_8motif$LR_close)
NR <- as.data.frame(peakAnnoList_ff_8motif$NR)
NR_gr <- as.GRanges(peakAnnoList_ff_8motif$NR)
EAR_open_list_all <- TSS_NG_data %>%
dplyr::filter(Peakid %in% EAR_open$Peakid) %>%
mutate(MRC="EAR_open") %>%
distinct(Peakid,.keep_all = TRUE)
EAR_close_list_all <- TSS_NG_data %>%
dplyr::filter(Peakid %in% EAR_close$Peakid) %>%
mutate(MRC="EAR_close") %>%
distinct(Peakid,.keep_all = TRUE)
ESR_open_list_all <- TSS_NG_data %>%
dplyr::filter(Peakid %in% ESR_open$Peakid) %>%
mutate(MRC="ESR_open")%>%
distinct(Peakid,.keep_all = TRUE)
ESR_close_list_all <- TSS_NG_data %>%
dplyr::filter(Peakid %in% ESR_close$Peakid) %>%
mutate(MRC="ESR_close")%>%
distinct(Peakid,.keep_all = TRUE)
ESR_OC_list_all <- TSS_NG_data %>%
dplyr::filter(Peakid %in% ESR_OC$Peakid) %>%
mutate(MRC="ESR_OC")%>%
distinct(Peakid,.keep_all = TRUE)
LR_open_list_all <- TSS_NG_data %>%
dplyr::filter(Peakid %in% LR_open$Peakid) %>%
mutate(MRC="LR_open")%>%
distinct(Peakid,.keep_all = TRUE)
LR_close_list_all <- TSS_NG_data %>%
dplyr::filter(Peakid %in% LR_close$Peakid) %>%
mutate(MRC="LR_close")%>%
distinct(Peakid,.keep_all = TRUE)
NR_list_all <- TSS_NG_data %>%
dplyr::filter(Peakid %in% NR$Peakid) %>%
mutate(MRC="NR")%>%
distinct(Peakid,.keep_all = TRUE)
ESR_C <- readRDS("data/Final_four_data/ESR_C.RDS")
ESR_D <- readRDS("data/Final_four_data/ESR_D.RDS")
ESRC_list_all <- TSS_NG_data %>%
dplyr::filter(Peakid %in% NR$Peakid) %>%
mutate(MRC="ESR_opcl")%>%
distinct(Peakid,.keep_all = TRUE)
ESRD_list_all <- TSS_NG_data %>%
dplyr::filter(Peakid %in%ESR_D$Peakid) %>%
mutate(MRC="ESR_clop")%>%
distinct(Peakid,.keep_all = TRUE)
# ESR_opcl <-
# ESR_clop <-
peak_list_all_mrc <- col_ng_peak %>%
mutate(mrc=if_else(Peakid %in% EAR_open$Peakid, "EAR_open",
if_else(Peakid %in% EAR_close$Peakid, "EAR_close",
if_else(Peakid %in% ESR_open$Peakid,"ESR_open",
if_else(Peakid %in% ESR_close$Peakid,"ESR_close",
if_else(Peakid %in% ESR_OC$Peakid,"ESR_OC",
if_else(Peakid %in% LR_open$Peakid,"LR_open",
if_else(Peakid %in% LR_close$Peakid,"LR_close",
if_else(Peakid %in% NR$Peakid,"NR","not_mrc"))))))))) %>%
mutate(mrc9=if_else(Peakid %in% EAR_open$Peakid, "EAR_open",
if_else(Peakid %in% EAR_close$Peakid, "EAR_close",
if_else(Peakid %in% ESR_open$Peakid,"ESR_open",
if_else(Peakid %in% ESR_close$Peakid,"ESR_close",
if_else(Peakid %in% ESR_C$Peakid,"ESR_opcl",
if_else(Peakid %in% LR_open$Peakid,"LR_open",
if_else(Peakid %in% LR_close$Peakid,"LR_close",
if_else(Peakid %in% NR$Peakid,"NR",if_else(Peakid %in% ESR_D$Peakid,"ESR_clop","not_mrc"))))))))) )
ESR_C <- readRDS("data/Final_four_data/ESR_C.RDS")
ESR_opcl_gr <- ESR_C %>% GRanges()
ESR_D <- readRDS("data/Final_four_data/ESR_D.RDS")
ESR_clop_gr <- ESR_D %>% GRanges()
EAR_open_NG_2k<- peak_list_all_mrc %>%
dplyr::filter(mrc =="EAR_open") %>%
dplyr::filter(dist_to_NG >-2000&dist_to_NG<2000) %>%
dplyr::select(Peakid, NCBI_gene:SYMBOL,dist_to_NG, mrc) %>%
separate_longer_delim(., cols=NCBI_gene:SYMBOL, delim= ",") %>%
distinct(NCBI_gene,SYMBOL)
EAR_close_NG_2k<- peak_list_all_mrc %>%
dplyr::filter(mrc =="EAR_close") %>%
dplyr::filter(dist_to_NG >-2000&dist_to_NG<2000) %>%
dplyr::select(Peakid, NCBI_gene:SYMBOL,dist_to_NG, mrc) %>%
separate_longer_delim(., cols=NCBI_gene:SYMBOL, delim= ",") %>%
distinct(NCBI_gene,SYMBOL)
ESR_open_NG_2k<- peak_list_all_mrc %>%
dplyr::filter(mrc =="ESR_open") %>%
dplyr::filter(dist_to_NG >-2000&dist_to_NG<2000) %>%
dplyr::select(Peakid, NCBI_gene:SYMBOL,dist_to_NG, mrc) %>%
separate_longer_delim(., cols=NCBI_gene:SYMBOL, delim= ",") %>%
distinct(NCBI_gene,SYMBOL)
ESR_close_NG_2k<- peak_list_all_mrc %>%
dplyr::filter(mrc =="ESR_close") %>%
dplyr::filter(dist_to_NG >-2000&dist_to_NG<2000) %>%
dplyr::select(Peakid, NCBI_gene:SYMBOL,dist_to_NG, mrc) %>%
separate_longer_delim(., cols=NCBI_gene:SYMBOL, delim= ",") %>%
distinct(NCBI_gene,SYMBOL)
ESR_OC_NG_2k<- peak_list_all_mrc %>%
dplyr::filter(mrc =="ESR_OC") %>%
dplyr::filter(dist_to_NG >-2000&dist_to_NG<2000) %>%
dplyr::select(Peakid, NCBI_gene:SYMBOL,dist_to_NG, mrc) %>%
separate_longer_delim(., cols=NCBI_gene:SYMBOL, delim= ",") %>%
distinct(NCBI_gene,SYMBOL)
ESR_opcl_NG_2k<- peak_list_all_mrc %>%
dplyr::filter(mrc9 =="ESR_opcl") %>%
dplyr::filter(dist_to_NG >-2000&dist_to_NG<2000) %>%
dplyr::select(Peakid, NCBI_gene:SYMBOL,dist_to_NG, mrc9) %>%
dplyr::rename("mrc"=mrc9) %>%
separate_longer_delim(., cols=NCBI_gene:SYMBOL, delim= ",") %>%
distinct(NCBI_gene,SYMBOL)
ESR_clop_NG_2k<- peak_list_all_mrc %>%
dplyr::filter(mrc9 =="ESR_clop") %>%
dplyr::filter(dist_to_NG >-2000&dist_to_NG<2000) %>%
dplyr::select(Peakid, NCBI_gene:SYMBOL,dist_to_NG, mrc9) %>%
dplyr::rename("mrc"=mrc9) %>%
separate_longer_delim(., cols=NCBI_gene:SYMBOL, delim= ",") %>%
distinct(NCBI_gene,SYMBOL)
LR_open_NG_2k<- peak_list_all_mrc %>%
dplyr::filter(mrc =="LR_open") %>%
dplyr::filter(dist_to_NG >-2000&dist_to_NG<2000) %>%
dplyr::select(Peakid, NCBI_gene:SYMBOL,dist_to_NG, mrc) %>%
separate_longer_delim(., cols=NCBI_gene:SYMBOL, delim= ",") %>%
distinct(NCBI_gene,SYMBOL)
LR_close_NG_2k<- peak_list_all_mrc %>%
dplyr::filter(mrc =="LR_close") %>%
dplyr::filter(dist_to_NG >-2000&dist_to_NG<2000) %>%
dplyr::select(Peakid, NCBI_gene:SYMBOL,dist_to_NG, mrc) %>%
separate_longer_delim(., cols=NCBI_gene:SYMBOL, delim= ",") %>%
distinct(NCBI_gene,SYMBOL)
NR_NG_2k <- peak_list_all_mrc %>%
dplyr::filter(mrc =="NR") %>%
dplyr::filter(dist_to_NG >-2000&dist_to_NG<2000) %>%
dplyr::select(Peakid, NCBI_gene:SYMBOL,dist_to_NG, mrc) %>%
separate_longer_delim(., cols=NCBI_gene, delim= ",") %>%
separate_longer_delim(., cols=,SYMBOL, delim= ",")
median_24_lfc <- read_csv("data/Final_four_data/median_24_lfc.csv")
median_3_lfc <- read_csv("data/Final_four_data/median_3_lfc.csv")
# saveRDS(peak_list_all_mrc, "data/Final_four_data/Peak_list_all_mrc_NG.RDS")
Top2b_peaks_gr <- readBed("data/n45_bedfiles/TOP2B_CM.bed")Enhancers and Response clusters
First: obtained a list of cis Regulatory Elements from Encode Screen [(https://screen.encodeproject.org/#)]
# BiocManager::install("plyranges")
# enhancers_HLV_46F <- genomation::readBed("C:/Users/renee/Downloads/Supplements folde manuscriptr/ENCODE/heart_left_ventricle_tissue_female_adult_46_years.enhancers.bed")
cREs_HLV_46F <- genomation::readBed("data/enhancerdata/ENCFF867HAD_ENCFF152PBB_ENCFF352YYH_ENCFF252IVK.7group.bed")
# cREs_HLV_53F <- genomation::readBed("data/enhancerdata/ENCFF417JSF_ENCFF651XRK_ENCFF320IPT_ENCFF440RUS.7group.bed")
NR_cREs <- join_overlap_intersect(NR_gr,cREs_HLV_46F)
LR_open_cREs <- join_overlap_intersect(LR_open_gr,cREs_HLV_46F)
LR_close_cREs <- join_overlap_intersect(LR_close_gr,cREs_HLV_46F)
ESR_open_cREs <- join_overlap_intersect(ESR_open_gr,cREs_HLV_46F)
ESR_close_cREs <- join_overlap_intersect(ESR_close_gr,cREs_HLV_46F)
ESR_OC_cREs <- join_overlap_intersect(ESR_OC_gr,cREs_HLV_46F)
ESR_opcl_cREs <- join_overlap_intersect(ESR_opcl_gr, cREs_HLV_46F)
ESR_clop_cREs <- join_overlap_intersect(ESR_clop_gr, cREs_HLV_46F)
EAR_open_cREs <- join_overlap_intersect(EAR_open_gr,cREs_HLV_46F)
EAR_close_cREs <- join_overlap_intersect(EAR_close_gr,cREs_HLV_46F)
### These unique peaks are cREs that contain all types of cREs such as
# [1] "Low-DNase" "DNase-only" "CTCF-only,CTCF-bound"
# [4] "PLS,CTCF-bound" "PLS" "dELS"
# [7] "pELS" "DNase-H3K4me3" "DNase-H3K4me3,CTCF-bound"
# [10] "dELS,CTCF-bound" "pELS,CTCF-bound" #### NOT the exact ones I am interested in.
uni_EAR_open <- EAR_open_cREs %>% as.data.frame() %>% distinct(Peakid)
uni_EAR_close <- EAR_close_cREs %>% as.data.frame() %>% distinct(Peakid)
uni_ESR_open <- ESR_open_cREs%>% as.data.frame() %>% distinct(Peakid)
uni_ESR_close <- ESR_close_cREs%>% as.data.frame() %>% distinct(Peakid)
uni_ESR_OC <- ESR_OC_cREs%>% as.data.frame() %>% distinct(Peakid)
uni_ESR_opcl <- ESR_opcl_cREs%>% as.data.frame() %>% distinct(Peakid)
uni_ESR_clop <- ESR_clop_cREs%>% as.data.frame() %>% distinct(Peakid)
uni_LR_open <- LR_open_cREs%>% as.data.frame() %>% distinct(Peakid)
uni_LR_close <- LR_close_cREs%>% as.data.frame() %>% distinct(Peakid)
uni_NR <- NR_cREs%>% as.data.frame() %>% distinct(Peakid)
allpeak_gr <- peak_list_all_mrc %>%
GRanges(.)
Whole_peaks <- join_overlap_intersect(allpeak_gr, cREs_HLV_46F)
Whole_peaks %>%
as.data.frame() %>%
group_by(blockCount, mrc9) %>% tally %>%
pivot_wider(., id_cols = mrc9, names_from = blockCount, values_from = n) %>%
dplyr::select(mrc9, PLS:'pELS,CTCF-bound') %>%
kable(., caption="Breakdown of peaks overlapping cREs") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 14)| mrc9 | PLS | PLS,CTCF-bound | dELS | dELS,CTCF-bound | pELS | pELS,CTCF-bound |
|---|---|---|---|---|---|---|
| EAR_close | 44 | 9 | 442 | 33 | 97 | 9 |
| EAR_open | 880 | 161 | 137 | 31 | 875 | 124 |
| ESR_close | 294 | 64 | 1588 | 248 | 543 | 106 |
| ESR_opcl | 1 | NA | 8 | 1 | 4 | NA |
| ESR_open | 173 | 33 | 171 | 35 | 302 | 66 |
| LR_close | 833 | 218 | 1620 | 304 | 996 | 225 |
| LR_open | 219 | 37 | 1081 | 116 | 435 | 72 |
| NR | 11375 | 3140 | 6464 | 1717 | 12589 | 2925 |
| not_mrc | 176 | 42 | 339 | 70 | 235 | 47 |
| ESR_clop | NA | NA | 36 | NA | 6 | NA |
Visualization of categories:
First, filtering cRE set by type to include ‘CTCF-only,CTCF-bound’, ‘PLS’, ‘PLS,CTCF-bound’,‘dELS’, ‘dELS,CTCF-bound’, ‘pELS’, ‘pELS,CTCF-bound’.
Second, reclassify (new column) to group the CTCF-bound into their respective groups so only 4 groups, (“CTCF-only”,“PLS”,“dELS”, “pELS”) are created.
Third, left-join the median LFC for both 3 hour and 24 hour data to data set and boxplot by group.
Fourth, rbind two more data frames to help with ggplot visualization
PLS= promoter like sequences pELS= proximal enhancer like sequences dELS=distal enhancer like sequences
EAR open
all_EAR_open_list <-
peak_list_all_mrc %>%
dplyr::filter(mrc=="EAR_open") %>%
mutate(type="all_EAR_open") %>%
dplyr::select(Peakid,mrc,type) %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
notCRE_EAR_open_list <- peak_list_all_mrc %>%
dplyr::filter(mrc=="EAR_open") %>%
dplyr::filter(!Peakid %in% uni_EAR_open$Peakid) %>%
mutate(type="not_cRE_peaks") %>%
dplyr::select(Peakid,mrc,type) %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
EAR_open_cRE_list <- EAR_open_cREs %>%
as.data.frame() %>%
dplyr::filter(blockCount=="CTCF-only,CTCF-bound"|
blockCount =="PLS"|
blockCount =="PLS,CTCF-bound"|
blockCount =="dELS"|
blockCount =="dELS,CTCF-bound"|
blockCount =="pELS"|
blockCount =="pELS,CTCF-bound") %>%
mutate(type=if_else(blockCount=="CTCF-only,CTCF-bound","CTCF-only", if_else(grepl("PLS",blockCount),"Promoter-like", if_else(grepl("pELS",blockCount),"Proximal enhancer-like", "Distal enhancer-like"),blockCount))) %>%
dplyr::select(Peakid,type) %>%
mutate(mrc="EAR_open") %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
EAR_open_cRE_list %>%
rbind(notCRE_EAR_open_list) %>%
rbind(all_EAR_open_list) %>%
ggplot(., aes(y=type, x=lfc,group=type))+
geom_boxplot() +
scale_fill_discrete()+
ggtitle(paste(" 24 hour EAR_open\n n = ",length(unique(EAR_open_cRE_list$Peakid))," out of ",peakAnnoList_ff_8motif$EAR_open@peakNum, " total peaks (",sprintf("%.1f",(length(unique(EAR_open_cRE_list$Peakid))/peakAnnoList_ff_8motif$EAR_open@peakNum*100)),"%)"))+
# facet_wrap(~direction)+
theme_bw()#+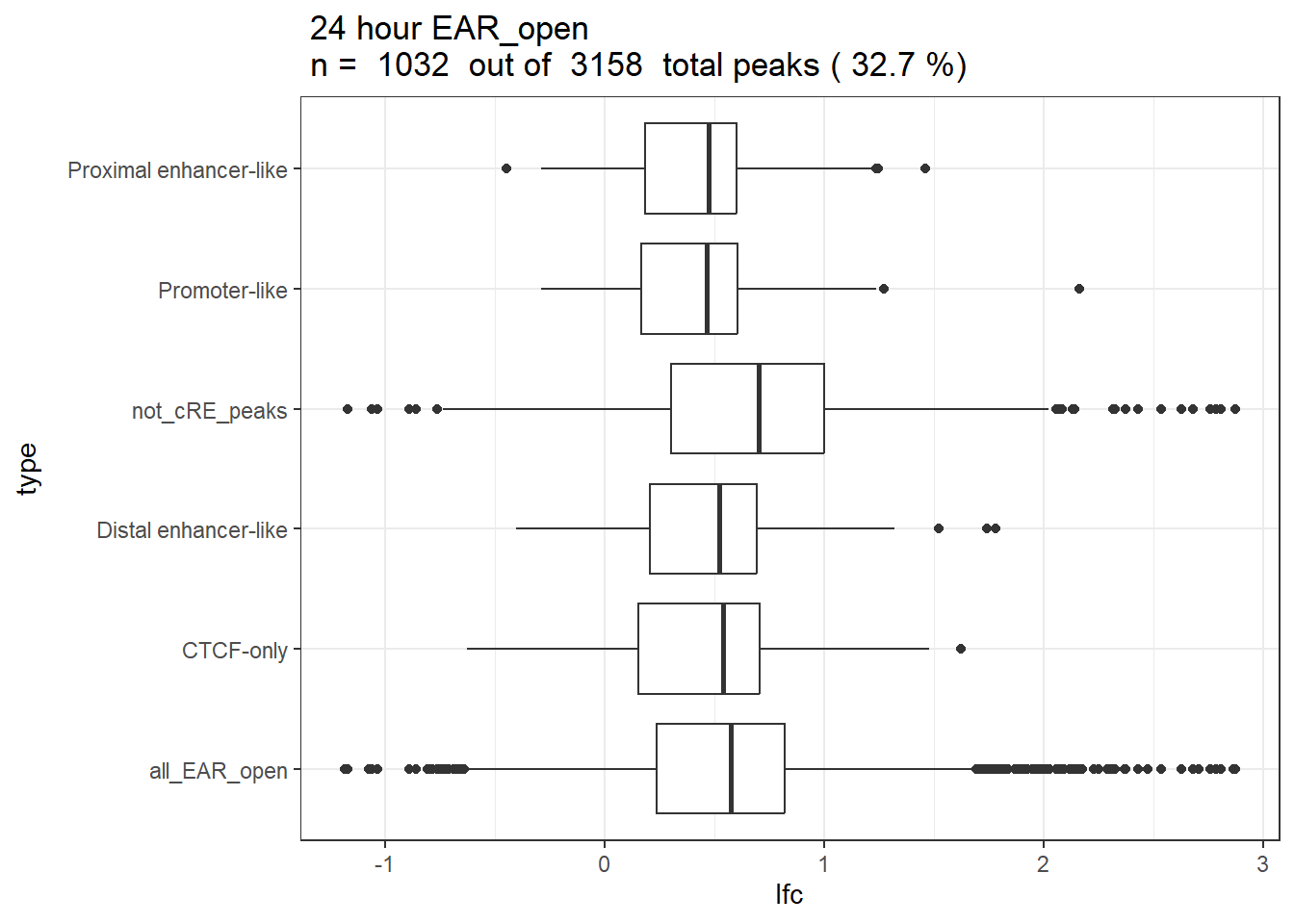
# xlim(-4,4)
EAR_open_complete_list <- EAR_open_cRE_list %>%
rbind(notCRE_EAR_open_list) %>%
rbind(all_EAR_open_list)EAR close
all_EAR_close_list <-
peak_list_all_mrc %>%
dplyr::filter(mrc=="EAR_close") %>%
mutate(type="all_EAR_close") %>%
dplyr::select(Peakid,mrc,type) %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
notCRE_EAR_close_list <- peak_list_all_mrc %>%
dplyr::filter(mrc=="EAR_close") %>%
dplyr::filter(!Peakid %in% uni_EAR_close$Peakid) %>%
mutate(type="not_cRE_peaks") %>%
dplyr::select(Peakid,mrc,type) %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
EAR_close_cRE_list <- EAR_close_cREs %>%
as.data.frame() %>%
dplyr::filter(blockCount=="CTCF-only,CTCF-bound"|
blockCount =="PLS"|
blockCount =="PLS,CTCF-bound"|
blockCount =="dELS"|
blockCount =="dELS,CTCF-bound"|
blockCount =="pELS"|
blockCount =="pELS,CTCF-bound") %>%
mutate(type=if_else(blockCount=="CTCF-only,CTCF-bound","CTCF-only", if_else(grepl("PLS",blockCount),"Promoter-like", if_else(grepl("pELS",blockCount),"Proximal enhancer-like", "Distal enhancer-like"),blockCount))) %>%
dplyr::select(Peakid,type) %>%
mutate(mrc="EAR_close") %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
EAR_close_cRE_list %>%
rbind(notCRE_EAR_close_list) %>%
rbind(all_EAR_close_list) %>%
ggplot(., aes(y=type, x=lfc,group=type))+
geom_boxplot() +
scale_fill_discrete()+
ggtitle(paste0(" 24 hour EAR_close\n n = ",length(unique(EAR_close_cRE_list$Peakid))," out of ", peakAnnoList_ff_8motif$EAR_close@peakNum, " total peaks (",sprintf("%.1f",length(unique(EAR_close_cRE_list$Peakid))/ peakAnnoList_ff_8motif$EAR_close@peakNum*100),"%)"))+
# facet_wrap(~direction)+
theme_bw()
| Version | Author | Date |
|---|---|---|
| d926a4a | reneeisnowhere | 2024-10-07 |
# xlim(-4,4)
EAR_close_complete_list <- EAR_close_cRE_list %>%
rbind(notCRE_EAR_close_list) %>%
rbind(all_EAR_close_list)ESR close
all_ESR_close_list <-
peak_list_all_mrc %>%
dplyr::filter(mrc=="ESR_close") %>%
mutate(type="all_ESR_close") %>%
dplyr::select(Peakid,mrc,type) %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
notCRE_ESR_close_list <- peak_list_all_mrc %>%
dplyr::filter(mrc=="ESR_close") %>%
dplyr::filter(!Peakid %in% uni_ESR_close$Peakid) %>%
mutate(type="not_cRE_peaks") %>%
dplyr::select(Peakid,mrc,type) %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
ESR_close_cRE_list <- ESR_close_cREs %>%
as.data.frame() %>%
dplyr::filter(blockCount=="CTCF-only,CTCF-bound"|
blockCount =="PLS"|
blockCount =="PLS,CTCF-bound"|
blockCount =="dELS"|
blockCount =="dELS,CTCF-bound"|
blockCount =="pELS"|
blockCount =="pELS,CTCF-bound") %>%
mutate(type=if_else(blockCount=="CTCF-only,CTCF-bound","CTCF-only", if_else(grepl("PLS",blockCount),"Promoter-like", if_else(grepl("pELS",blockCount),"Proximal enhancer-like", "Distal enhancer-like"),blockCount))) %>%
dplyr::select(Peakid,type) %>%
mutate(mrc="ESR_close") %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
ESR_close_cRE_list %>%
rbind(notCRE_ESR_close_list) %>%
rbind(all_ESR_close_list) %>%
ggplot(., aes(y=type, x=lfc,group=type))+
geom_boxplot() +
scale_fill_discrete()+
ggtitle(paste0(" 24 hour ESR_close\n n = ",length(unique(ESR_close_cRE_list$Peakid))," out of ", peakAnnoList_ff_8motif$ESR_close@peakNum, " total peaks (",sprintf("%.1f",length(unique(ESR_close_cRE_list$Peakid))/ peakAnnoList_ff_8motif$ESR_close@peakNum*100),"%)"))+
# facet_wrap(~direction)+
theme_bw()
| Version | Author | Date |
|---|---|---|
| d926a4a | reneeisnowhere | 2024-10-07 |
# xlim(-4,4)
ESR_close_complete_list <- ESR_close_cRE_list %>%
rbind(notCRE_ESR_close_list) %>%
rbind(all_ESR_close_list)ESR open
all_ESR_open_list <-
peak_list_all_mrc %>%
dplyr::filter(mrc=="ESR_open") %>%
mutate(type="all_ESR_open") %>%
dplyr::select(Peakid,mrc,type) %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
notCRE_ESR_open_list <- peak_list_all_mrc %>%
dplyr::filter(mrc=="ESR_open") %>%
dplyr::filter(!Peakid %in% uni_ESR_open$Peakid) %>%
mutate(type="not_cRE_peaks") %>%
dplyr::select(Peakid,mrc,type) %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
ESR_open_cRE_list <- ESR_open_cREs %>%
as.data.frame() %>%
dplyr::filter(blockCount=="CTCF-only,CTCF-bound"|
blockCount =="PLS"|
blockCount =="PLS,CTCF-bound"|
blockCount =="dELS"|
blockCount =="dELS,CTCF-bound"|
blockCount =="pELS"|
blockCount =="pELS,CTCF-bound") %>%
mutate(type=if_else(blockCount=="CTCF-only,CTCF-bound","CTCF-only", if_else(grepl("PLS",blockCount),"Promoter-like", if_else(grepl("pELS",blockCount),"Proximal enhancer-like", "Distal enhancer-like"),blockCount))) %>%
dplyr::select(Peakid,type) %>%
mutate(mrc="ESR_open") %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
ESR_open_cRE_list %>%
rbind(notCRE_ESR_open_list) %>%
rbind(all_ESR_open_list) %>%
ggplot(., aes(y=type, x=lfc,group=type))+
geom_boxplot() +
scale_fill_discrete()+
ggtitle(paste0(" 24 hour ESR_open\n n = ",length(unique(ESR_open_cRE_list$Peakid))," out of ", peakAnnoList_ff_8motif$ESR_open@peakNum, " total peaks (",sprintf("%.1f",length(unique(ESR_open_cRE_list$Peakid))/ peakAnnoList_ff_8motif$ESR_open@peakNum*100),"%)"))+
# facet_wrap(~direction)+
theme_bw()
| Version | Author | Date |
|---|---|---|
| d926a4a | reneeisnowhere | 2024-10-07 |
# xlim(-4,4)
ESR_open_complete_list <- ESR_open_cRE_list %>%
rbind(notCRE_ESR_open_list) %>%
rbind(all_ESR_open_list)ESR OC
all_ESR_OC_list <-
peak_list_all_mrc %>%
dplyr::filter(mrc=="ESR_OC") %>%
# mutate(mrc=mrc9) %>%
mutate(type="all_ESR_OC") %>%
dplyr::select(Peakid,mrc,type) %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
notCRE_ESR_OC_list <- peak_list_all_mrc %>%
dplyr::filter(mrc=="ESR_OC") %>%
dplyr::filter(!Peakid %in% uni_ESR_OC$Peakid) %>%
mutate(type="not_cRE_peaks") %>%
dplyr::select(Peakid,mrc,type) %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
ESR_OC_cRE_list <- ESR_OC_cREs %>%
as.data.frame() %>%
dplyr::filter(blockCount=="CTCF-only,CTCF-bound"|
blockCount =="PLS"|
blockCount =="PLS,CTCF-bound"|
blockCount =="dELS"|
blockCount =="dELS,CTCF-bound"|
blockCount =="pELS"|
blockCount =="pELS,CTCF-bound") %>%
mutate(type=if_else(blockCount=="CTCF-only,CTCF-bound","CTCF-only", if_else(grepl("PLS",blockCount),"Promoter-like", if_else(grepl("pELS",blockCount),"Proximal enhancer-like", "Distal enhancer-like"),blockCount))) %>%
dplyr::select(Peakid,type) %>%
mutate(mrc="ESR_OC") %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
ESR_OC_cRE_list %>%
rbind(notCRE_ESR_OC_list) %>%
rbind(all_ESR_OC_list) %>%
ggplot(., aes(y=type, x=lfc,group=type))+
geom_boxplot() +
scale_fill_discrete()+
ggtitle(paste0(" 24 hour ESR_OC\n n = ",length(unique(ESR_OC_cRE_list$Peakid))," out of ", peakAnnoList_ff_8motif$ESR_OC@peakNum, " total peaks (",sprintf("%.1f",length(unique(ESR_OC_cRE_list$Peakid))/ peakAnnoList_ff_8motif$ESR_OC@peakNum*100),"%)"))#+
| Version | Author | Date |
|---|---|---|
| d926a4a | reneeisnowhere | 2024-10-07 |
# facet_wrap(~direction)+
# theme_bw()+
# xlim(-4,4)
ESR_OC_complete_list <- ESR_OC_cRE_list %>%
rbind(notCRE_ESR_OC_list) %>%
rbind(all_ESR_OC_list)ESR open/close
all_ESR_opcl_list <-
peak_list_all_mrc %>%
dplyr::filter(mrc9=="ESR_opcl") %>%
mutate(mrc=mrc9) %>%
mutate(type="all_ESR_opcl") %>%
dplyr::select(Peakid,mrc,type) %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
notCRE_ESR_opcl_list <- peak_list_all_mrc %>%
dplyr::filter(mrc9=="ESR_opcl") %>%
mutate(mrc=mrc9) %>%
dplyr::filter(!Peakid %in% uni_ESR_opcl$Peakid) %>%
mutate(type="not_cRE_peaks") %>%
dplyr::select(Peakid,mrc,type) %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
ESR_opcl_cRE_list <- ESR_opcl_cREs %>%
as.data.frame() %>%
dplyr::filter(blockCount=="CTCF-only,CTCF-bound"|
blockCount =="PLS"|
blockCount =="PLS,CTCF-bound"|
blockCount =="dELS"|
blockCount =="dELS,CTCF-bound"|
blockCount =="pELS"|
blockCount =="pELS,CTCF-bound") %>%
mutate(type=if_else(blockCount=="CTCF-only,CTCF-bound","CTCF-only", if_else(grepl("PLS",blockCount),"Promoter-like", if_else(grepl("pELS",blockCount),"Proximal enhancer-like", "Distal enhancer-like"),blockCount))) %>%
dplyr::select(Peakid,type) %>%
mutate(mrc="ESR_opcl") %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
ESR_opcl_cRE_list %>%
rbind(notCRE_ESR_opcl_list) %>%
rbind(all_ESR_opcl_list) %>%
ggplot(., aes(y=type, x=lfc,group=type))+
geom_boxplot() +
scale_fill_discrete()+
ggtitle(paste0(" 24 hour ESR_opcl\n n = ",length(unique(ESR_opcl_cRE_list$Peakid))," out of ", length(ESR_C$Peakid), " total peaks (",sprintf("%.1f",length(unique(ESR_opcl_cRE_list$Peakid))/ length(ESR_C$Peakid)*100),"%)"))+
# facet_wrap(~direction)+
theme_bw()
| Version | Author | Date |
|---|---|---|
| d926a4a | reneeisnowhere | 2024-10-07 |
# xlim(-4,4)
ESR_opcl_complete_list <- ESR_opcl_cRE_list %>%
rbind(notCRE_ESR_opcl_list) %>%
rbind(all_ESR_opcl_list)ESR close/open
all_ESR_clop_list <-
peak_list_all_mrc %>%
dplyr::filter(mrc9=="ESR_clop") %>%
mutate(mrc=mrc9) %>%
mutate(type="all_ESR_clop") %>%
dplyr::select(Peakid,mrc,type) %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
notCRE_ESR_clop_list <- peak_list_all_mrc %>%
dplyr::filter(mrc9=="ESR_clop") %>%
mutate(mrc=mrc9) %>%
dplyr::filter(!Peakid %in% uni_ESR_clop$Peakid) %>%
mutate(type="not_cRE_peaks") %>%
dplyr::select(Peakid,mrc,type) %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
ESR_clop_cRE_list <- ESR_clop_cREs %>%
as.data.frame() %>%
dplyr::filter(blockCount=="CTCF-only,CTCF-bound"|
blockCount =="PLS"|
blockCount =="PLS,CTCF-bound"|
blockCount =="dELS"|
blockCount =="dELS,CTCF-bound"|
blockCount =="pELS"|
blockCount =="pELS,CTCF-bound") %>%
mutate(type=if_else(blockCount=="CTCF-only,CTCF-bound","CTCF-only", if_else(grepl("PLS",blockCount),"Promoter-like", if_else(grepl("pELS",blockCount),"Proximal enhancer-like", "Distal enhancer-like"),blockCount))) %>%
dplyr::select(Peakid,type) %>%
mutate(mrc="ESR_clop") %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
ESR_clop_cRE_list %>%
rbind(notCRE_ESR_clop_list) %>%
rbind(all_ESR_clop_list) %>%
ggplot(., aes(y=type, x=lfc,group=type))+
geom_boxplot() +
scale_fill_discrete()+
ggtitle(paste0(" 24 hour ESR_clop\n n = ",length(unique(ESR_clop_cRE_list$Peakid))," out of ", length(ESR_C$Peakid), " total peaks (",sprintf("%.1f",length(unique(ESR_clop_cRE_list$Peakid))/ length(ESR_C$Peakid)*100),"%)"))+
# facet_wrap(~direction)+
theme_bw()
| Version | Author | Date |
|---|---|---|
| d926a4a | reneeisnowhere | 2024-10-07 |
# xlim(-4,4)
ESR_clop_complete_list <- ESR_clop_cRE_list %>%
rbind(notCRE_ESR_clop_list) %>%
rbind(all_ESR_clop_list)LR open
all_LR_open_list <-
peak_list_all_mrc %>%
dplyr::filter(mrc=="LR_open") %>%
mutate(type="all_LR_open") %>%
dplyr::select(Peakid,mrc,type) %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
notCRE_LR_open_list <- peak_list_all_mrc %>%
dplyr::filter(mrc=="LR_open") %>%
dplyr::filter(!Peakid %in% uni_LR_open$Peakid) %>%
mutate(type="not_cRE_peaks") %>%
dplyr::select(Peakid,mrc,type) %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
LR_open_cRE_list <- LR_open_cREs %>%
as.data.frame() %>%
dplyr::filter(blockCount=="CTCF-only,CTCF-bound"|
blockCount =="PLS"|
blockCount =="PLS,CTCF-bound"|
blockCount =="dELS"|
blockCount =="dELS,CTCF-bound"|
blockCount =="pELS"|
blockCount =="pELS,CTCF-bound") %>%
mutate(type=if_else(blockCount=="CTCF-only,CTCF-bound","CTCF-only", if_else(grepl("PLS",blockCount),"Promoter-like", if_else(grepl("pELS",blockCount),"Proximal enhancer-like", "Distal enhancer-like"),blockCount))) %>%
dplyr::select(Peakid,type) %>%
mutate(mrc="LR_open") %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
LR_open_cRE_list %>%
rbind(notCRE_LR_open_list) %>%
rbind(all_LR_open_list) %>%
ggplot(., aes(y=type, x=lfc,group=type))+
geom_boxplot() +
scale_fill_discrete()+
ggtitle(paste0(" 24 hour LR_open\n n = ",length(unique(LR_open_cRE_list$Peakid))," out of ", peakAnnoList_ff_8motif$LR_open@peakNum, " total peaks (",sprintf("%.1f",length(unique(LR_open_cRE_list$Peakid))/ peakAnnoList_ff_8motif$LR_open@peakNum*100),"%)"))+
# facet_wrap(~direction)+
theme_bw()
| Version | Author | Date |
|---|---|---|
| d926a4a | reneeisnowhere | 2024-10-07 |
# xlim(-4,4)
LR_open_complete_list <- LR_open_cRE_list %>%
rbind(notCRE_LR_open_list) %>%
rbind(all_LR_open_list)
# LR_openLR close
all_LR_close_list <-
peak_list_all_mrc %>%
dplyr::filter(mrc=="LR_close") %>%
mutate(type="all_LR_close") %>%
dplyr::select(Peakid,mrc,type) %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
notCRE_LR_close_list <- peak_list_all_mrc %>%
dplyr::filter(mrc=="LR_close") %>%
dplyr::filter(!Peakid %in% uni_LR_close$Peakid) %>%
mutate(type="not_cRE_peaks") %>%
dplyr::select(Peakid,mrc,type) %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
LR_close_cRE_list <- LR_close_cREs %>%
as.data.frame() %>%
dplyr::filter(blockCount=="CTCF-only,CTCF-bound"|
blockCount =="PLS"|
blockCount =="PLS,CTCF-bound"|
blockCount =="dELS"|
blockCount =="dELS,CTCF-bound"|
blockCount =="pELS"|
blockCount =="pELS,CTCF-bound") %>%
mutate(type=if_else(blockCount=="CTCF-only,CTCF-bound","CTCF-only", if_else(grepl("PLS",blockCount),"Promoter-like", if_else(grepl("pELS",blockCount),"Proximal enhancer-like", "Distal enhancer-like"),blockCount))) %>%
dplyr::select(Peakid,type) %>%
mutate(mrc="LR_close") %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
LR_close_cRE_list %>%
rbind(notCRE_LR_close_list) %>%
rbind(all_LR_close_list) %>%
ggplot(., aes(y=type, x=lfc,group=type))+
geom_boxplot() +
scale_fill_discrete()+
ggtitle(paste0(" 24 hour LR_close\n n = ",length(unique(LR_close_cRE_list$Peakid))," out of ", peakAnnoList_ff_8motif$LR_close@peakNum, " total peaks (",sprintf("%.1f",length(unique(LR_close_cRE_list$Peakid))/ peakAnnoList_ff_8motif$LR_close@peakNum*100),"%)"))+
# facet_wrap(~direction)+
theme_bw()
| Version | Author | Date |
|---|---|---|
| d926a4a | reneeisnowhere | 2024-10-07 |
# xlim(-4,4)
LR_close_complete_list <- LR_close_cRE_list %>%
rbind(notCRE_LR_close_list) %>%
rbind(all_LR_close_list)No response
all_NR_list <-
peak_list_all_mrc %>%
dplyr::filter(mrc=="NR") %>%
mutate(type="all_NR") %>%
dplyr::select(Peakid,mrc,type) %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
notCRE_NR_list <- peak_list_all_mrc %>%
dplyr::filter(mrc=="NR") %>%
dplyr::filter(!Peakid %in% uni_NR$Peakid) %>%
mutate(type="not_cRE_peaks") %>%
dplyr::select(Peakid,mrc,type) %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
NR_cRE_list <- NR_cREs %>%
as.data.frame() %>%
dplyr::filter(blockCount=="CTCF-only,CTCF-bound"|
blockCount =="PLS"|
blockCount =="PLS,CTCF-bound"|
blockCount =="dELS"|
blockCount =="dELS,CTCF-bound"|
blockCount =="pELS"|
blockCount =="pELS,CTCF-bound") %>%
mutate(type=if_else(blockCount=="CTCF-only,CTCF-bound","CTCF-only", if_else(grepl("PLS",blockCount),"Promoter-like", if_else(grepl("pELS",blockCount),"Proximal enhancer-like", "Distal enhancer-like"),blockCount))) %>%
dplyr::select(Peakid,type) %>%
mutate(mrc="NR") %>%
left_join(.,(median_24_lfc %>% ungroup%>%
dplyr::select(peak,med_24h_lfc)),by = c("Peakid"="peak")) %>%
left_join(.,(median_3_lfc %>%ungroup %>%
dplyr::select(peak,med_3h_lfc)),by = c("Peakid"="peak")) %>%
pivot_longer(cols = c("med_24h_lfc","med_3h_lfc"), names_to = "time", values_to = "lfc")
NR_cRE_list %>%
rbind(notCRE_NR_list) %>%
rbind(all_NR_list) %>%
ggplot(., aes(y=type, x=lfc,group=type))+
geom_boxplot() +
scale_fill_discrete()+
ggtitle(paste0(" 24 hour NR\n n = ",length(unique(NR_cRE_list$Peakid))," out of ", peakAnnoList_ff_8motif$NR@peakNum, " total peaks (",sprintf("%.1f",length(unique(NR_cRE_list$Peakid))/ peakAnnoList_ff_8motif$NR@peakNum*100),"%)"))+
# facet_wrap(~direction)+
theme_bw()
| Version | Author | Date |
|---|---|---|
| d926a4a | reneeisnowhere | 2024-10-07 |
# xlim(-4,4)
NR_complete_list <- NR_cRE_list %>%
rbind(notCRE_NR_list) %>%
rbind(all_NR_list)GO analysis of neargenes from Peaks that are within 2kb of Neargene TSS.
background_NGs <- col_ng_peak %>%
dplyr::select (Peakid,NCBI_gene:dist_to_NG) %>%
# dplyr::filter (dist_to_NG >-2000&dist_to_NG <2000) %>%
distinct(SYMBOL)
EAR_openNG <- EAR_open_list_all %>%
dplyr::select (Peakid,SYMBOL,dist_to_NG) %>%
dplyr::filter (dist_to_NG >-2000&dist_to_NG <2000) %>%
distinct(SYMBOL)
EAR_closeNG <- EAR_close_list_all %>%
dplyr::select (Peakid,SYMBOL,dist_to_NG) %>%
dplyr::filter (dist_to_NG >-2000&dist_to_NG <2000) %>%
distinct(SYMBOL)
ESR_openNG <- ESR_open_list_all %>%
dplyr::select (Peakid,SYMBOL,dist_to_NG) %>%
dplyr::filter (dist_to_NG >-2000&dist_to_NG <2000) %>%
distinct(SYMBOL)
ESR_closeNG <- ESR_close_list_all %>%
dplyr::select (Peakid,SYMBOL,dist_to_NG) %>%
dplyr::filter (dist_to_NG >-2000&dist_to_NG <2000) %>%
distinct(SYMBOL)
ESR_OCNG <- ESR_OC_list_all %>%
dplyr::select (Peakid,SYMBOL,dist_to_NG) %>%
dplyr::filter (dist_to_NG >-2000&dist_to_NG <2000) %>%
distinct(SYMBOL)
LR_openNG <- LR_open_list_all %>%
dplyr::select (Peakid,SYMBOL,dist_to_NG) %>%
dplyr::filter (dist_to_NG >-2000&dist_to_NG <2000) %>%
distinct(SYMBOL)
LR_closeNG <- LR_close_list_all %>%
dplyr::select (Peakid,SYMBOL,dist_to_NG) %>%
dplyr::filter (dist_to_NG >-2000&dist_to_NG <2000) %>%
distinct(SYMBOL)
NR_NG <- NR_list_all %>%
dplyr::select (Peakid,SYMBOL,dist_to_NG) %>%
dplyr::filter (dist_to_NG >-2000&dist_to_NG <2000) %>%
distinct(SYMBOL)scale_fill_mrc <- function(...){
ggplot2:::manual_scale(
'fill',
values = setNames(c("#F8766D","#f6483c","#7CAE00","#587b00","#6a9500","cornflowerblue","grey60", "#00BFC4","#008d91", "#C77CFF"), c("EAR_open","EAR_close","ESR_open","ESR_close", "ESR_OC","ESR_opcl","ESR_clop","LR_open","LR_close","NR")),
...
)
}summarizing plots:
ggplot_combo_df <-
rbind(EAR_open_complete_list,
EAR_close_complete_list,
ESR_open_complete_list,
ESR_close_complete_list,
ESR_OC_complete_list,
LR_open_complete_list,
LR_close_complete_list,
NR_complete_list)
ggplot_combo_df %>%
mutate(time=factor(time, levels= c("med_3h_lfc", "med_24h_lfc"), labels=c("3 hours","24 hours"))) %>%
mutate(type= if_else(type=="Promoter-like",type,
if_else(type =="Proximal enhancer-like",type,if_else(type=="Distal enhancer-like", type,if_else(type=="CTCF-only",type,if_else(type=="not_cRE_peaks", type, "all")))))) %>%
mutate(type=factor(type, levels = c("Promoter-like","Proximal enhancer-like","Distal enhancer-like","CTCF-only","not_cRE_peaks","all"))) %>%
ggplot(., aes(y=type, x=lfc, fill=mrc))+
geom_boxplot()+
# geom_pointrange(aes(xmin=min_val, xmax=max_val),position=position_dodge(width =.7), width=.3,size=0.5,shape=15, fatten=8)+
geom_vline(xintercept = 0, linetype=2)+
facet_wrap(~time) +
theme_classic()+
scale_fill_mrc()
| Version | Author | Date |
|---|---|---|
| d926a4a | reneeisnowhere | 2024-10-07 |
ggplot_combo_df_9 <-
rbind(EAR_open_complete_list,
EAR_close_complete_list,
ESR_open_complete_list,
ESR_close_complete_list,
ESR_opcl_complete_list,
ESR_clop_complete_list,
LR_open_complete_list,
LR_close_complete_list,
NR_complete_list)
ggplot_combo_df_9 %>%
mutate(time=factor(time, levels= c("med_3h_lfc", "med_24h_lfc"), labels=c("3 hours","24 hours"))) %>%
mutate(type= if_else(type=="Promoter-like",type,
if_else(type =="Proximal enhancer-like",type,if_else(type=="Distal enhancer-like", type,if_else(type=="CTCF-only",type,if_else(type=="not_cRE_peaks", type, "all")))))) %>%
mutate(type=factor(type, levels = c("Promoter-like","Proximal enhancer-like","Distal enhancer-like","CTCF-only","not_cRE_peaks","all"))) %>%
ggplot(., aes(y=type, x=lfc, fill=mrc))+
geom_boxplot()+
# geom_pointrange(aes(xmin=min_val, xmax=max_val),position=position_dodge(width =.7), width=.3,size=0.5,shape=15, fatten=8)+
geom_vline(xintercept = 0, linetype=2)+
facet_wrap(~time) +
theme_classic()+
scale_fill_mrc()
| Version | Author | Date |
|---|---|---|
| d926a4a | reneeisnowhere | 2024-10-07 |
keep_cRE_names <- c("CTCF-only,CTCF-bound" ,"PLS,CTCF-bound","PLS","dELS,CTCF-bound", "pELS","pELS,CTCF-bound","dELS")
is_cRE <- Whole_peaks %>%
as.data.frame() %>%
dplyr::filter(blockCount %in% keep_cRE_names) %>%
distinct(Peakid,blockCount)
is_CTCF <- Whole_peaks %>%
as.data.frame() %>%
dplyr::filter(blockCount == "CTCF-only,CTCF-bound") %>%
distinct(Peakid,blockCount)
is_dELS <- Whole_peaks %>%
as.data.frame() %>%
dplyr::filter(blockCount == "dELS,CTCF-bound"|blockCount == "dELS") %>%
distinct(Peakid,blockCount)
is_pELS <- Whole_peaks %>%
as.data.frame() %>%
dplyr::filter(blockCount == "pELS,CTCF-bound"|blockCount == "pELS") %>%
distinct(Peakid,blockCount)
is_PLS <- Whole_peaks %>%
as.data.frame() %>%
dplyr::filter(blockCount == "PLS,CTCF-bound"|blockCount == "PLS") %>%
distinct(Peakid,blockCount)
CRE_summary <- col_ng_peak %>%
mutate(cRE_status=if_else(Peakid %in% is_cRE$Peakid,"cRE_peak","not_cRE_peak")) %>%
mutate(CTCF_status=if_else(Peakid %in% is_CTCF$Peakid,"CTCF_peak","not_CTCF_peak")) %>%
mutate(dELS_status=if_else(Peakid %in% is_dELS$Peakid,"dELS_peak","not_dELS_peak")) %>%
mutate(pELS_status=if_else(Peakid %in% is_pELS$Peakid,"pELS_peak","not_pELS_peak")) %>%
mutate(PLS_status=if_else(Peakid %in% is_PLS$Peakid,"PLS_peak","not_PLS_peak")) %>%
mutate(mrc=if_else(Peakid %in% EAR_open$Peakid, "EAR_open",
if_else(Peakid %in% EAR_close$Peakid, "EAR_close",
if_else(Peakid %in% ESR_open$Peakid,"ESR_open",
if_else(Peakid %in% ESR_close$Peakid,"ESR_close",
if_else(Peakid %in% ESR_C$Peakid,"ESR_opcl",
if_else(Peakid %in% LR_open$Peakid,"LR_open",
if_else(Peakid %in% LR_close$Peakid,"LR_close",
if_else(Peakid %in% NR$Peakid,"NR",if_else(Peakid %in% ESR_D$Peakid,"ESR_clop","not_mrc"))))))))) )
cRE_mat<- CRE_summary %>%
dplyr::filter(mrc != "not_mrc") %>%
group_by(cRE_status, mrc) %>%
tally %>%
mutate(mrc= factor(mrc, levels = c("EAR_open","EAR_close","ESR_open", "ESR_close", "ESR_opcl", "ESR_clop","LR_open","LR_close","NR"))) %>%
pivot_wider(id_cols = mrc, names_from = cRE_status,values_from = n) %>%
column_to_rownames("mrc") %>%
na.omit(.) %>%
as.matrix(.)
CTCF_mat<- CRE_summary %>%
dplyr::filter(mrc != "not_mrc") %>%
group_by(CTCF_status, mrc) %>%
tally %>%
mutate(mrc= factor(mrc, levels = c("EAR_open","EAR_close","ESR_open", "ESR_close", "ESR_opcl", "ESR_clop","LR_open","LR_close","NR"))) %>%
pivot_wider(id_cols = mrc, names_from = CTCF_status,values_from = n) %>%
column_to_rownames("mrc") %>%
na.omit(.) %>%
as.matrix(.)
dELS_mat<- CRE_summary %>%
dplyr::filter(mrc != "not_mrc") %>%
group_by(dELS_status, mrc) %>%
tally %>%
mutate(mrc= factor(mrc, levels = c("EAR_open","EAR_close","ESR_open", "ESR_close", "ESR_opcl", "ESR_clop","LR_open","LR_close","NR"))) %>%
pivot_wider(id_cols = mrc, names_from = dELS_status,values_from = n) %>%
column_to_rownames("mrc") %>%
na.omit(.) %>%
as.matrix(.)
pELS_mat<- CRE_summary %>%
dplyr::filter(mrc != "not_mrc") %>%
group_by(pELS_status, mrc) %>%
tally %>%
mutate(mrc= factor(mrc, levels = c("EAR_open","EAR_close","ESR_open", "ESR_close", "ESR_opcl", "ESR_clop","LR_open","LR_close","NR"))) %>%
pivot_wider(id_cols = mrc, names_from = pELS_status,values_from = n) %>%
column_to_rownames("mrc") %>%
na.omit(.) %>%
as.matrix(.)
PLS_mat<- CRE_summary %>%
dplyr::filter(mrc != "not_mrc") %>%
group_by(PLS_status, mrc) %>%
tally %>%
mutate(mrc= factor(mrc, levels = c("EAR_open","EAR_close","ESR_open", "ESR_close", "ESR_opcl", "ESR_clop","LR_open","LR_close","NR"))) %>%
pivot_wider(id_cols = mrc, names_from = PLS_status,values_from = n) %>%
column_to_rownames("mrc") %>%
na.omit(.) %>%
as.matrix(.)chi square results
matrix_list_cre <- list("PLS"=PLS_mat, "dELS"=dELS_mat, "pELS"=pELS_mat,"CTCF"= CTCF_mat,"All cREs"= cRE_mat)
results_cre <- data.frame(Matrix_Name = character(),
Row_Compared = character(),
Chi_Square_Statistic = numeric(),
P_Value = numeric(),
stringsAsFactors = FALSE)
names_list_cre <- names(matrix_list_cre)
# Loop through each matrix in the list
for (matrix_name in names_list_cre) {
current_matrix <- matrix_list_cre[[matrix_name]]
n_rows <- nrow(current_matrix)
# Print matrix details for debugging
cat("Processing Matrix:", matrix_name, "\n")
print(current_matrix)
# Loop through each row of the current matrix (except the last row)
for (i in 1:(n_rows - 1)) {
# Print row details for debugging
cat("Comparing row", i, "with last row:\n")
print(current_matrix[i, ])
print(current_matrix[n_rows, ])
# Perform chi-square test between row i and the last row
test_result <- tryCatch({
chisq.test(rbind(current_matrix[i, ], current_matrix[n_rows, ]))
}, error = function(e) {
cat("Error in chi-square test:", e$message, "\n")
return(NULL)
})
# Only store the result if test_result is valid (i.e., not NULL)
if (!is.null(test_result)) {
results_cre <- rbind(results_cre, data.frame(Matrix_Name = matrix_name,
Row_Compared = rownames(current_matrix)[i],
Chi_Square_Statistic = test_result$statistic,
P_Value = test_result$p.value))
}
}
}Processing Matrix: PLS
PLS_peak not_PLS_peak
EAR_close 48 4248
EAR_open 697 2461
ESR_close 288 9061
ESR_opcl 1 98
ESR_open 158 5114
LR_close 789 13283
LR_open 219 28318
NR 9778 76553
Comparing row 1 with last row:
PLS_peak not_PLS_peak
48 4248
PLS_peak not_PLS_peak
9778 76553
Comparing row 2 with last row:
PLS_peak not_PLS_peak
697 2461
PLS_peak not_PLS_peak
9778 76553
Comparing row 3 with last row:
PLS_peak not_PLS_peak
288 9061
PLS_peak not_PLS_peak
9778 76553
Comparing row 4 with last row:
PLS_peak not_PLS_peak
1 98
PLS_peak not_PLS_peak
9778 76553
Comparing row 5 with last row:
PLS_peak not_PLS_peak
158 5114
PLS_peak not_PLS_peak
9778 76553
Comparing row 6 with last row:
PLS_peak not_PLS_peak
789 13283
PLS_peak not_PLS_peak
9778 76553
Comparing row 7 with last row:
PLS_peak not_PLS_peak
219 28318
PLS_peak not_PLS_peak
9778 76553
Processing Matrix: dELS
dELS_peak not_dELS_peak
EAR_close 346 3950
EAR_open 125 3033
ESR_clop 31 1030
ESR_close 1354 7995
ESR_opcl 8 91
ESR_open 163 5109
LR_close 1478 12594
LR_open 955 27582
NR 5680 80651
Comparing row 1 with last row:
dELS_peak not_dELS_peak
346 3950
dELS_peak not_dELS_peak
5680 80651
Comparing row 2 with last row:
dELS_peak not_dELS_peak
125 3033
dELS_peak not_dELS_peak
5680 80651
Comparing row 3 with last row:
dELS_peak not_dELS_peak
31 1030
dELS_peak not_dELS_peak
5680 80651
Comparing row 4 with last row:
dELS_peak not_dELS_peak
1354 7995
dELS_peak not_dELS_peak
5680 80651
Comparing row 5 with last row:
dELS_peak not_dELS_peak
8 91
dELS_peak not_dELS_peak
5680 80651
Comparing row 6 with last row:
dELS_peak not_dELS_peak
163 5109
dELS_peak not_dELS_peak
5680 80651
Comparing row 7 with last row:
dELS_peak not_dELS_peak
1478 12594
dELS_peak not_dELS_peak
5680 80651
Comparing row 8 with last row:
dELS_peak not_dELS_peak
955 27582
dELS_peak not_dELS_peak
5680 80651
Processing Matrix: pELS
not_pELS_peak pELS_peak
EAR_close 4214 82
EAR_open 2590 568
ESR_clop 1056 5
ESR_close 8887 462
ESR_opcl 95 4
ESR_open 5058 214
LR_close 13295 777
LR_open 28189 348
NR 78113 8218
Comparing row 1 with last row:
not_pELS_peak pELS_peak
4214 82
not_pELS_peak pELS_peak
78113 8218
Comparing row 2 with last row:
not_pELS_peak pELS_peak
2590 568
not_pELS_peak pELS_peak
78113 8218
Comparing row 3 with last row:
not_pELS_peak pELS_peak
1056 5
not_pELS_peak pELS_peak
78113 8218
Comparing row 4 with last row:
not_pELS_peak pELS_peak
8887 462
not_pELS_peak pELS_peak
78113 8218
Comparing row 5 with last row:
not_pELS_peak pELS_peak
95 4
not_pELS_peak pELS_peak
78113 8218
Comparing row 6 with last row:
not_pELS_peak pELS_peak
5058 214
not_pELS_peak pELS_peak
78113 8218
Comparing row 7 with last row:
not_pELS_peak pELS_peak
13295 777
not_pELS_peak pELS_peak
78113 8218
Comparing row 8 with last row:
not_pELS_peak pELS_peak
28189 348
not_pELS_peak pELS_peak
78113 8218
Processing Matrix: CTCF
CTCF_peak not_CTCF_peak
EAR_close 101 4195
EAR_open 97 3061
ESR_close 377 8972
ESR_opcl 1 98
ESR_open 64 5208
LR_close 592 13480
LR_open 219 28318
NR 5403 80928
Comparing row 1 with last row:
CTCF_peak not_CTCF_peak
101 4195
CTCF_peak not_CTCF_peak
5403 80928
Comparing row 2 with last row:
CTCF_peak not_CTCF_peak
97 3061
CTCF_peak not_CTCF_peak
5403 80928
Comparing row 3 with last row:
CTCF_peak not_CTCF_peak
377 8972
CTCF_peak not_CTCF_peak
5403 80928
Comparing row 4 with last row:
CTCF_peak not_CTCF_peak
1 98
CTCF_peak not_CTCF_peak
5403 80928
Comparing row 5 with last row:
CTCF_peak not_CTCF_peak
64 5208
CTCF_peak not_CTCF_peak
5403 80928
Comparing row 6 with last row:
CTCF_peak not_CTCF_peak
592 13480
CTCF_peak not_CTCF_peak
5403 80928
Comparing row 7 with last row:
CTCF_peak not_CTCF_peak
219 28318
CTCF_peak not_CTCF_peak
5403 80928
Processing Matrix: All cREs
cRE_peak not_cRE_peak
EAR_close 564 3732
EAR_open 1032 2126
ESR_clop 36 1025
ESR_close 2324 7025
ESR_opcl 13 86
ESR_open 512 4760
LR_close 3128 10944
LR_open 1637 26900
NR 22534 63797
Comparing row 1 with last row:
cRE_peak not_cRE_peak
564 3732
cRE_peak not_cRE_peak
22534 63797
Comparing row 2 with last row:
cRE_peak not_cRE_peak
1032 2126
cRE_peak not_cRE_peak
22534 63797
Comparing row 3 with last row:
cRE_peak not_cRE_peak
36 1025
cRE_peak not_cRE_peak
22534 63797
Comparing row 4 with last row:
cRE_peak not_cRE_peak
2324 7025
cRE_peak not_cRE_peak
22534 63797
Comparing row 5 with last row:
cRE_peak not_cRE_peak
13 86
cRE_peak not_cRE_peak
22534 63797
Comparing row 6 with last row:
cRE_peak not_cRE_peak
512 4760
cRE_peak not_cRE_peak
22534 63797
Comparing row 7 with last row:
cRE_peak not_cRE_peak
3128 10944
cRE_peak not_cRE_peak
22534 63797
Comparing row 8 with last row:
cRE_peak not_cRE_peak
1637 26900
cRE_peak not_cRE_peak
22534 63797 # Print the resulting dataframe
print(results_cre) Matrix_Name Row_Compared Chi_Square_Statistic P_Value
X-squared PLS EAR_close 440.1586065 9.998327e-98
X-squared1 PLS EAR_open 339.2781306 9.163922e-76
X-squared2 PLS ESR_close 608.3795659 2.518823e-134
X-squared3 PLS ESR_opcl 9.4848736 2.071729e-03
X-squared4 PLS ESR_open 355.5913847 2.567814e-79
X-squared5 PLS LR_close 419.6860256 2.856187e-93
X-squared6 PLS LR_open 3008.0307322 0.000000e+00
X-squared7 dELS EAR_close 14.1010187 1.732499e-04
X-squared8 dELS EAR_open 34.0740951 5.305287e-09
X-squared9 dELS ESR_clop 22.3614218 2.258657e-06
X-squared10 dELS ESR_close 772.4658531 5.231320e-170
X-squared11 dELS ESR_opcl 0.1595127 6.896056e-01
X-squared12 dELS ESR_open 100.6132894 1.118150e-23
X-squared13 dELS LR_close 280.7696601 5.103577e-63
X-squared14 dELS LR_open 411.2476997 1.961260e-91
X-squared15 pELS EAR_close 283.9817228 1.018430e-63
X-squared16 pELS EAR_open 245.7142389 2.232575e-55
X-squared17 pELS ESR_clop 99.6034670 1.861782e-23
X-squared18 pELS ESR_close 213.7174103 2.122586e-48
X-squared19 pELS ESR_opcl 2.8411858 9.187639e-02
X-squared20 pELS ESR_open 176.5759605 2.710603e-40
X-squared21 pELS LR_close 236.5801977 2.189762e-53
X-squared22 pELS LR_open 2139.6174210 0.000000e+00
X-squared23 CTCF EAR_close 108.8501702 1.750287e-25
X-squared24 CTCF EAR_open 53.0905639 3.185189e-13
X-squared25 CTCF ESR_close 73.2456334 1.144782e-17
X-squared26 CTCF ESR_opcl 3.7947687 5.141298e-02
X-squared27 CTCF ESR_open 224.3993326 9.927024e-51
X-squared28 CTCF LR_close 90.3394922 2.006082e-21
X-squared29 CTCF LR_open 1388.0944201 8.119789e-304
X-squared30 All cREs EAR_close 362.0022835 1.031801e-80
X-squared31 All cREs EAR_open 67.5954503 2.007302e-16
X-squared32 All cREs ESR_clop 280.9720486 4.610746e-63
X-squared33 All cREs ESR_close 6.7192904 9.537557e-03
X-squared34 All cREs ESR_opcl 7.9684063 4.760083e-03
X-squared35 All cREs ESR_open 708.0084840 5.422764e-156
X-squared36 All cREs LR_close 95.2051750 1.716410e-22
X-squared37 All cREs LR_open 5352.7289387 0.000000e+00odds ratio results
results_or_cre <- data.frame(Matrix_Name = character(),
Row_Compared = character(),
Odds_Ratio = numeric(),
Lower_CI = numeric(),
Upper_CI = numeric(),
P_Value = numeric(),
stringsAsFactors = FALSE)
# Loop through each matrix in the list
for (matrix_name in names(matrix_list_cre)) {
current_matrix <- matrix_list_cre[[matrix_name]]
n_rows <- nrow(current_matrix)
# Loop through each row of the current matrix (except the last row)
for (i in 1:(n_rows - 1)) {
# Perform odds ratio test between row i and the last row using epitools
test_result <- tryCatch({
contingency_table <- rbind(current_matrix[i, ], current_matrix[n_rows, ])
# Check if any row in the contingency table contains only zeros
if (any(rowSums(contingency_table) == 0)) {
stop("Contingency table contains empty rows.")
}
oddsratio_result <- oddsratio(contingency_table)
# Ensure the oddsratio result has at least 2 rows
if (nrow(oddsratio_result$measure) < 2) {
stop("oddsratio result does not have enough data.")
}
list(oddsratio = oddsratio_result, p.value = oddsratio_result$p.value[2,"chi.square"])
}, error = function(e) {
cat("Error in odds ratio test for row", i, "in matrix", matrix_name, ":", e$message, "\n")
return(NULL)
})
# Only store the result if test_result is valid (i.e., not NULL)
if (!is.null(test_result)) {
or_value <- test_result$oddsratio$measure[2, "estimate"]
lower_ci <- test_result$oddsratio$measure[2, "lower"]
upper_ci <- test_result$oddsratio$measure[2, "upper"]
p_value <- test_result$oddsratio$p.value[2,"chi.square"]
# Check if the values are numeric and valid (not NA)
if (!is.na(or_value) && !is.na(lower_ci) && !is.na(upper_ci) && !is.na(p_value)) {
# Store the results in the dataframe
results_or_cre <- rbind(results_or_cre, data.frame(Matrix_Name = matrix_name,
Row_Compared = rownames(current_matrix)[i],
Odds_Ratio = or_value,
Lower_CI = lower_ci,
Upper_CI = upper_ci,
P_Value = p_value))
}
}
}
}Error in odds ratio test for row 1 in matrix PLS : could not find function "oddsratio"
Error in odds ratio test for row 2 in matrix PLS : could not find function "oddsratio"
Error in odds ratio test for row 3 in matrix PLS : could not find function "oddsratio"
Error in odds ratio test for row 4 in matrix PLS : could not find function "oddsratio"
Error in odds ratio test for row 5 in matrix PLS : could not find function "oddsratio"
Error in odds ratio test for row 6 in matrix PLS : could not find function "oddsratio"
Error in odds ratio test for row 7 in matrix PLS : could not find function "oddsratio"
Error in odds ratio test for row 1 in matrix dELS : could not find function "oddsratio"
Error in odds ratio test for row 2 in matrix dELS : could not find function "oddsratio"
Error in odds ratio test for row 3 in matrix dELS : could not find function "oddsratio"
Error in odds ratio test for row 4 in matrix dELS : could not find function "oddsratio"
Error in odds ratio test for row 5 in matrix dELS : could not find function "oddsratio"
Error in odds ratio test for row 6 in matrix dELS : could not find function "oddsratio"
Error in odds ratio test for row 7 in matrix dELS : could not find function "oddsratio"
Error in odds ratio test for row 8 in matrix dELS : could not find function "oddsratio"
Error in odds ratio test for row 1 in matrix pELS : could not find function "oddsratio"
Error in odds ratio test for row 2 in matrix pELS : could not find function "oddsratio"
Error in odds ratio test for row 3 in matrix pELS : could not find function "oddsratio"
Error in odds ratio test for row 4 in matrix pELS : could not find function "oddsratio"
Error in odds ratio test for row 5 in matrix pELS : could not find function "oddsratio"
Error in odds ratio test for row 6 in matrix pELS : could not find function "oddsratio"
Error in odds ratio test for row 7 in matrix pELS : could not find function "oddsratio"
Error in odds ratio test for row 8 in matrix pELS : could not find function "oddsratio"
Error in odds ratio test for row 1 in matrix CTCF : could not find function "oddsratio"
Error in odds ratio test for row 2 in matrix CTCF : could not find function "oddsratio"
Error in odds ratio test for row 3 in matrix CTCF : could not find function "oddsratio"
Error in odds ratio test for row 4 in matrix CTCF : could not find function "oddsratio"
Error in odds ratio test for row 5 in matrix CTCF : could not find function "oddsratio"
Error in odds ratio test for row 6 in matrix CTCF : could not find function "oddsratio"
Error in odds ratio test for row 7 in matrix CTCF : could not find function "oddsratio"
Error in odds ratio test for row 1 in matrix All cREs : could not find function "oddsratio"
Error in odds ratio test for row 2 in matrix All cREs : could not find function "oddsratio"
Error in odds ratio test for row 3 in matrix All cREs : could not find function "oddsratio"
Error in odds ratio test for row 4 in matrix All cREs : could not find function "oddsratio"
Error in odds ratio test for row 5 in matrix All cREs : could not find function "oddsratio"
Error in odds ratio test for row 6 in matrix All cREs : could not find function "oddsratio"
Error in odds ratio test for row 7 in matrix All cREs : could not find function "oddsratio"
Error in odds ratio test for row 8 in matrix All cREs : could not find function "oddsratio" # Print the resulting dataframe
print(results_or_cre)[1] Matrix_Name Row_Compared Odds_Ratio Lower_CI Upper_CI
[6] P_Value
<0 rows> (or 0-length row.names)library(circlize)
col_fun_cre = colorRamp2(c(0,6), c("white", "purple3"))
sig_mat_cre <- results_or_cre %>%
as.data.frame() %>%
dplyr::select( Matrix_Name,Row_Compared,P_Value) %>%
pivot_wider(., id_cols = Matrix_Name, names_from = Row_Compared, values_from = P_Value) %>%
column_to_rownames("Matrix_Name") %>%
as.matrix()
results_or_cre %>%
as.data.frame() %>%
dplyr::select( Matrix_Name,Row_Compared,Odds_Ratio) %>%
pivot_wider(., id_cols = Matrix_Name, names_from = Row_Compared, values_from = Odds_Ratio) %>%
column_to_rownames("Matrix_Name") %>%
as.matrix() %>%
ComplexHeatmap::Heatmap(. ,col = col_fun_cre,
cluster_rows=FALSE,
cluster_columns=FALSE,
column_names_side = "top",
column_names_rot = 45,
cell_fun = function(j, i, x, y, width, height, fill) {if (!is.na(sig_mat_cre[i, j]) && sig_mat_cre[i, j] < 0.05)
grid.text("*", x, y, gp = gpar(fontsize = 20))})
| Version | Author | Date |
|---|---|---|
| d926a4a | reneeisnowhere | 2024-10-07 |
Top2b
to be determined…. ingnore for now
Counts of peaks-
sessionInfo()R version 4.4.1 (2024-06-14 ucrt)
Platform: x86_64-w64-mingw32/x64
Running under: Windows 10 x64 (build 19045)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
time zone: America/Chicago
tzcode source: internal
attached base packages:
[1] grid stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] circlize_0.4.16
[2] gprofiler2_0.2.3
[3] genomation_1.36.0
[4] plyranges_1.24.0
[5] BSgenome.Hsapiens.UCSC.hg38_1.4.5
[6] BSgenome_1.72.0
[7] BiocIO_1.14.0
[8] MotifDb_1.46.0
[9] Biostrings_2.72.1
[10] XVector_0.44.0
[11] TFBSTools_1.42.0
[12] JASPAR2022_0.99.8
[13] BiocFileCache_2.12.0
[14] dbplyr_2.5.0
[15] devtools_2.4.5
[16] usethis_3.0.0
[17] ggpubr_0.6.0
[18] BiocParallel_1.38.0
[19] scales_1.3.0
[20] VennDiagram_1.7.3
[21] futile.logger_1.4.3
[22] gridExtra_2.3
[23] ggfortify_0.4.17
[24] edgeR_4.2.1
[25] limma_3.60.4
[26] rtracklayer_1.64.0
[27] org.Hs.eg.db_3.19.1
[28] TxDb.Hsapiens.UCSC.hg38.knownGene_3.18.0
[29] GenomicFeatures_1.56.0
[30] AnnotationDbi_1.66.0
[31] Biobase_2.64.0
[32] GenomicRanges_1.56.1
[33] GenomeInfoDb_1.40.1
[34] IRanges_2.38.1
[35] S4Vectors_0.42.1
[36] BiocGenerics_0.50.0
[37] ChIPseeker_1.40.0
[38] RColorBrewer_1.1-3
[39] broom_1.0.7
[40] kableExtra_1.4.0
[41] lubridate_1.9.3
[42] forcats_1.0.0
[43] stringr_1.5.1
[44] dplyr_1.1.4
[45] purrr_1.0.2
[46] readr_2.1.5
[47] tidyr_1.3.1
[48] tibble_3.2.1
[49] ggplot2_3.5.1
[50] tidyverse_2.0.0
[51] workflowr_1.7.1
loaded via a namespace (and not attached):
[1] R.methodsS3_1.8.2
[2] vroom_1.6.5
[3] urlchecker_1.0.1
[4] poweRlaw_0.80.0
[5] vctrs_0.6.5
[6] digest_0.6.37
[7] png_0.1-8
[8] shape_1.4.6.1
[9] git2r_0.33.0
[10] ggrepel_0.9.6
[11] magick_2.8.5
[12] MASS_7.3-61
[13] reshape2_1.4.4
[14] httpuv_1.6.15
[15] foreach_1.5.2
[16] qvalue_2.36.0
[17] withr_3.0.1
[18] xfun_0.47
[19] ggfun_0.1.6
[20] ellipsis_0.3.2
[21] memoise_2.0.1
[22] profvis_0.4.0
[23] systemfonts_1.1.0
[24] tidytree_0.4.6
[25] GlobalOptions_0.1.2
[26] gtools_3.9.5
[27] R.oo_1.26.0
[28] Formula_1.2-5
[29] KEGGREST_1.44.1
[30] promises_1.3.0
[31] httr_1.4.7
[32] rstatix_0.7.2
[33] restfulr_0.0.15
[34] ps_1.8.0
[35] rstudioapi_0.16.0
[36] UCSC.utils_1.0.0
[37] miniUI_0.1.1.1
[38] generics_0.1.3
[39] DOSE_3.30.5
[40] processx_3.8.4
[41] curl_5.2.3
[42] zlibbioc_1.50.0
[43] ggraph_2.2.1
[44] polyclip_1.10-7
[45] GenomeInfoDbData_1.2.12
[46] SparseArray_1.4.8
[47] xtable_1.8-4
[48] pracma_2.4.4
[49] doParallel_1.0.17
[50] evaluate_1.0.0
[51] S4Arrays_1.4.1
[52] hms_1.1.3
[53] colorspace_2.1-1
[54] filelock_1.0.3
[55] magrittr_2.0.3
[56] later_1.3.2
[57] viridis_0.6.5
[58] ggtree_3.12.0
[59] lattice_0.22-6
[60] getPass_0.2-4
[61] XML_3.99-0.17
[62] shadowtext_0.1.4
[63] cowplot_1.1.3
[64] matrixStats_1.4.1
[65] pillar_1.9.0
[66] nlme_3.1-166
[67] iterators_1.0.14
[68] pwalign_1.0.0
[69] gridBase_0.4-7
[70] caTools_1.18.3
[71] compiler_4.4.1
[72] stringi_1.8.4
[73] SummarizedExperiment_1.34.0
[74] GenomicAlignments_1.40.0
[75] plyr_1.8.9
[76] crayon_1.5.3
[77] abind_1.4-8
[78] gridGraphics_0.5-1
[79] locfit_1.5-9.10
[80] graphlayouts_1.2.0
[81] bit_4.5.0
[82] fastmatch_1.1-4
[83] whisker_0.4.1
[84] codetools_0.2-20
[85] bslib_0.8.0
[86] TxDb.Hsapiens.UCSC.hg19.knownGene_3.2.2
[87] GetoptLong_1.0.5
[88] plotly_4.10.4
[89] mime_0.12
[90] splines_4.4.1
[91] Rcpp_1.0.13
[92] knitr_1.48
[93] blob_1.2.4
[94] utf8_1.2.4
[95] clue_0.3-65
[96] seqLogo_1.70.0
[97] fs_1.6.4
[98] pkgbuild_1.4.4
[99] ggsignif_0.6.4
[100] ggplotify_0.1.2
[101] Matrix_1.7-0
[102] callr_3.7.6
[103] statmod_1.5.0
[104] tzdb_0.4.0
[105] svglite_2.1.3
[106] tweenr_2.0.3
[107] pkgconfig_2.0.3
[108] tools_4.4.1
[109] cachem_1.1.0
[110] RSQLite_2.3.7
[111] viridisLite_0.4.2
[112] DBI_1.2.3
[113] splitstackshape_1.4.8
[114] impute_1.78.0
[115] fastmap_1.2.0
[116] rmarkdown_2.28
[117] Rsamtools_2.20.0
[118] sass_0.4.9
[119] patchwork_1.3.0
[120] carData_3.0-5
[121] farver_2.1.2
[122] tidygraph_1.3.1
[123] scatterpie_0.2.4
[124] yaml_2.3.10
[125] MatrixGenerics_1.16.0
[126] cli_3.6.3
[127] lifecycle_1.0.4
[128] lambda.r_1.2.4
[129] sessioninfo_1.2.2
[130] backports_1.5.0
[131] annotate_1.82.0
[132] timechange_0.3.0
[133] gtable_0.3.5
[134] rjson_0.2.23
[135] parallel_4.4.1
[136] ape_5.8
[137] jsonlite_1.8.9
[138] bitops_1.0-8
[139] bit64_4.5.2
[140] yulab.utils_0.1.7
[141] CNEr_1.40.0
[142] futile.options_1.0.1
[143] jquerylib_0.1.4
[144] highr_0.11
[145] GOSemSim_2.30.2
[146] R.utils_2.12.3
[147] lazyeval_0.2.2
[148] shiny_1.9.1
[149] htmltools_0.5.8.1
[150] enrichplot_1.24.4
[151] GO.db_3.19.1
[152] rappdirs_0.3.3
[153] formatR_1.14
[154] glue_1.8.0
[155] TFMPvalue_0.0.9
[156] httr2_1.0.5
[157] RCurl_1.98-1.16
[158] rprojroot_2.0.4
[159] treeio_1.28.0
[160] boot_1.3-31
[161] igraph_2.0.3
[162] R6_2.5.1
[163] gplots_3.1.3.1
[164] labeling_0.4.3
[165] cluster_2.1.6
[166] pkgload_1.4.0
[167] aplot_0.2.3
[168] DirichletMultinomial_1.46.0
[169] DelayedArray_0.30.1
[170] tidyselect_1.2.1
[171] plotrix_3.8-4
[172] ggforce_0.4.2
[173] xml2_1.3.6
[174] car_3.1-3
[175] seqPattern_1.36.0
[176] munsell_0.5.1
[177] KernSmooth_2.23-24
[178] data.table_1.16.0
[179] htmlwidgets_1.6.4
[180] fgsea_1.30.0
[181] ComplexHeatmap_2.20.0
[182] rlang_1.1.4
[183] remotes_2.5.0
[184] fansi_1.0.6