Peak_Calling
ERM
2024-02-27
Last updated: 2024-02-27
Checks: 7 0
Knit directory: ATAC_learning/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20231016) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 1a8126f. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/Ind1_75DA24h_dedup_peaks.csv
Ignored: data/Ind4_79B24h_dedup_peaks.csv
Ignored: data/Ind4_V24h_fraglength.txt
Ignored: data/Ind4_fragment_files.txt
Ignored: data/Ind4_summary.txt
Ignored: data/aln_run1_results.txt
Ignored: data/anno_ind1_DA24h.RDS
Ignored: data/anno_ind4_V24h.RDS
Ignored: data/ind1_DA24hpeaks.RDS
Ignored: data/ind4_V24hpeaks.RDS
Ignored: data/initial_complete_stats_run1.txt
Ignored: data/multiqc_fastqc_run1.txt
Ignored: data/multiqc_fastqc_run2.txt
Ignored: data/multiqc_genestat_run1.txt
Ignored: data/multiqc_genestat_run2.txt
Ignored: data/trimmed_seq_length.csv
Untracked files:
Untracked: code/just_for_Fun.R
Unstaged changes:
Modified: analysis/Fastqc_results.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/Peak_calling.Rmd) and HTML
(docs/Peak_calling.html) files. If you’ve configured a
remote Git repository (see ?wflow_git_remote), click on the
hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 1a8126f | reneeisnowhere | 2024-02-27 | adding the peak-calling files |
library(tidyverse)
# library(ggsignif)
# library(cowplot)
# library(ggpubr)
# library(scales)
# library(sjmisc)
library(kableExtra)
# library(broom)
# library(biomaRt)
library(RColorBrewer)
# library(gprofiler2)
# library(qvalue)
library(ChIPseeker)
library("TxDb.Hsapiens.UCSC.hg38.knownGene")
library("org.Hs.eg.db")drug_pal_fact <- c("#8B006D","#DF707E","#F1B72B", "#3386DD","#707031","#41B333")
txdb <- TxDb.Hsapiens.UCSC.hg38.knownGene
loadFile_peakCall <- function(){
file <- choose.files()
file <- readPeakFile(file, header = FALSE)
return(file)
}
prepGRangeObj <- function(seek_object){
seek_object$Peaks = seek_object$V4
seek_object$level = seek_object$V5
seek_object$V4 = seek_object$V5 = NULL
return(seek_object)
}
TSS = getBioRegion(TxDb=txdb, upstream=3000, downstream=3000, by = "gene",
type = "start_site")
ind4_V24hpeaks <- readRDS("data/ind4_V24hpeaks.RDS")
ind1_DA24hpeaks <- readRDS("data/ind1_DA24hpeaks.RDS")
anno_ind4_V24h <- readRDS("data/anno_ind4_V24h.RDS")
anno_ind1_DA24h <- readRDS("data/anno_ind1_DA24h.RDS")# library(readr)
# > Ind4_summary <- read_table("~/ATAC_downloads/Ind4/Ind4_summary.txt",
# + col_names = FALSE, col_types = cols(X3 = col_skip(),
# + X4 = col_skip(), X5 = col_skip(),
# + X6 = col_skip(), X7 = col_skip(),
# + X8 = col_skip(), X9 = col_skip(),
# + X10 = col_skip(), X11 = col_skip(),
# + X13 = col_skip(), X14 = col_skip(),
# + X17 = col_skip(), X18 = col_skip()))
#
Ind4_summary <- read.csv("data/Ind4_summary.txt", row.names = 1)
Ind4_summary %>%
separate(counts,into=c("counts",NA),sep= " ") %>%
mutate(counts=as.numeric(counts)) name counts
1 flagstat_first/trimmed_Ind4_79V24h_S12_flagstat.txt 77013488
2 flagstat_noM/trimmed_Ind4_79V24h_S12_noM_flagstat.txt 27439164
3 filt_files/trimmed_Ind4_79V24h_S12.nodup.flagstat.qc 1786687
4 filt_files/trimmed_Ind4_79V24h_S12_fin_flagstat.txt 18870010
5 flagstat_first/trimmed_Ind4_79DA24h_S7_flagstat.txt 83930268
6 flagstat_noM/trimmed_Ind4_79DA24h_S7_noM_flagstat.txt 39561574
7 filt_files/trimmed_Ind4_79DA24h_S7.nodup.flagstat.qc 17862648
8 filt_files/trimmed_Ind4_79DA24h_S7_fin_flagstat.txt 29459906
9 flagstat_first/trimmed_Ind4_79DA3h_S1_flagstat.txt 114610072
10 flagstat_noM/trimmed_Ind4_79DA3h_S1_noM_flagstat.txt 74283554
11 filt_files/trimmed_Ind4_79DA3h_S1.nodup.flagstat.qc 26487126
12 filt_files/trimmed_Ind4_79DA3h_S1_fin_flagstat.txt 55956416
13 flagstat_first/trimmed_Ind4_79DX24h_S8_flagstat.txt 78395382
14 flagstat_noM/trimmed_Ind4_79DX24h_S8_noM_flagstat.txt 25391928
15 filt_files/trimmed_Ind4_79DX24h_S8.nodup.flagstat.qc 11544950
16 filt_files/trimmed_Ind4_79DX24h_S8_fin_flagstat.txt 17146642
17 flagstat_first/trimmed_Ind4_79DX3h_S2_flagstat.txt 77601292
18 flagstat_noM/trimmed_Ind4_79DX3h_S2_noM_flagstat.txt 47116936
19 filt_files/trimmed_Ind4_79DX3h_S2.nodup.flagstat.qc 18908344
20 filt_files/trimmed_Ind4_79DX3h_S2_fin_flagstat.txt 35460994
21 flagstat_first/trimmed_Ind4_79E24h_S9_flagstat.txt 86039200
22 flagstat_noM/trimmed_Ind4_79E24h_S9_noM_flagstat.txt 29965542
23 filt_files/trimmed_Ind4_79E24h_S9.nodup.flagstat.qc 12495788
24 filt_files/trimmed_Ind4_79E24h_S9_fin_flagstat.txt 21004354
25 flagstat_first/trimmed_Ind4_79E3h_S3_flagstat.txt 86993222
26 flagstat_noM/trimmed_Ind4_79E3h_S3_noM_flagstat.txt 46372712
27 filt_files/trimmed_Ind4_79E3h_S3.nodup.flagstat.qc 19109612
28 filt_files/trimmed_Ind4_79E3h_S3_fin_flagstat.txt 34578548
29 flagstat_first/trimmed_Ind4_79M24h_S10_flagstat.txt 82061002
30 flagstat_noM/trimmed_Ind4_79M24h_S10_noM_flagstat.txt 27148372
31 filt_files/trimmed_Ind4_79M24h_S10.nodup.flagstat.qc 9894182
32 filt_files/trimmed_Ind4_79M24h_S10_fin_flagstat.txt 19085838
33 flagstat_first/trimmed_Ind4_79M3h_S4_flagstat.txt 83929214
34 flagstat_noM/trimmed_Ind4_79M3h_S4_noM_flagstat.txt 48985626
35 filt_files/trimmed_Ind4_79M3h_S4.nodup.flagstat.qc 16384910
36 filt_files/trimmed_Ind4_79M3h_S4_fin_flagstat.txt 36937534
37 flagstat_first/trimmed_Ind4_79T24h_S11_flagstat.txt 90875858
38 flagstat_noM/trimmed_Ind4_79T24h_S11_noM_flagstat.txt 31347532
39 filt_files/trimmed_Ind4_79T24h_S11.nodup.flagstat.qc 8948490
40 filt_files/trimmed_Ind4_79T24h_S11_fin_flagstat.txt 21789834
41 flagstat_first/trimmed_Ind4_79T3h_S5_flagstat.txt 106856444
42 flagstat_noM/trimmed_Ind4_79T3h_S5_noM_flagstat.txt 65690664
43 filt_files/trimmed_Ind4_79T3h_S5.nodup.flagstat.qc 28930176
44 filt_files/trimmed_Ind4_79T3h_S5_fin_flagstat.txt 49669024
45 flagstat_first/trimmed_Ind4_79V24h_S12_flagstat.txt 77013488
46 flagstat_noM/trimmed_Ind4_79V24h_S12_noM_flagstat.txt 27439164
47 filt_files/trimmed_Ind4_79V24h_S12.nodup.flagstat.qc 1786687
48 filt_files/trimmed_Ind4_79V24h_S12_fin_flagstat.txt 18870010
49 flagstat_first/trimmed_Ind4_79V3h_S6_flagstat.txt 74863328
50 flagstat_noM/trimmed_Ind4_79V3h_S6_noM_flagstat.txt 52535548
51 filt_files/trimmed_Ind4_79V3h_S6.nodup.flagstat.qc 25112288
52 filt_files/trimmed_Ind4_79V3h_S6_fin_flagstat.txt 40497298
mapped
1 75962835 + 0 mapped (98.64% : N/A)
2 26415467 + 0 mapped (96.27% : N/A)
3 1786687 + 0 mapped (100.00% : N/A)
4 18870010 + 0 mapped (100.00% : N/A)
5 83140985 + 0 mapped (99.06% : N/A)
6 38811928 + 0 mapped (98.11% : N/A)
7 17862648 + 0 mapped (100.00% : N/A)
8 29459906 + 0 mapped (100.00% : N/A)
9 112679708 + 0 mapped (98.32% : N/A)
10 72386250 + 0 mapped (97.45% : N/A)
11 26487126 + 0 mapped (100.00% : N/A)
12 55956416 + 0 mapped (100.00% : N/A)
13 77692597 + 0 mapped (99.10% : N/A)
14 24733417 + 0 mapped (97.41% : N/A)
15 11544950 + 0 mapped (100.00% : N/A)
16 17146642 + 0 mapped (100.00% : N/A)
17 76058132 + 0 mapped (98.01% : N/A)
18 45593581 + 0 mapped (96.77% : N/A)
19 18908344 + 0 mapped (100.00% : N/A)
20 35460994 + 0 mapped (100.00% : N/A)
21 85324746 + 0 mapped (99.17% : N/A)
22 29300484 + 0 mapped (97.78% : N/A)
23 12495788 + 0 mapped (100.00% : N/A)
24 21004354 + 0 mapped (100.00% : N/A)
25 85671962 + 0 mapped (98.48% : N/A)
26 45084280 + 0 mapped (97.22% : N/A)
27 19109612 + 0 mapped (100.00% : N/A)
28 34578548 + 0 mapped (100.00% : N/A)
29 81424543 + 0 mapped (99.22% : N/A)
30 26562054 + 0 mapped (97.84% : N/A)
31 9894182 + 0 mapped (100.00% : N/A)
32 19085838 + 0 mapped (100.00% : N/A)
33 82368935 + 0 mapped (98.14% : N/A)
34 47454435 + 0 mapped (96.87% : N/A)
35 16384910 + 0 mapped (100.00% : N/A)
36 36937534 + 0 mapped (100.00% : N/A)
37 89680567 + 0 mapped (98.68% : N/A)
38 30204755 + 0 mapped (96.35% : N/A)
39 8948490 + 0 mapped (100.00% : N/A)
40 21789834 + 0 mapped (100.00% : N/A)
41 105027081 + 0 mapped (98.29% : N/A)
42 63898507 + 0 mapped (97.27% : N/A)
43 28930176 + 0 mapped (100.00% : N/A)
44 49669024 + 0 mapped (100.00% : N/A)
45 75962835 + 0 mapped (98.64% : N/A)
46 26415467 + 0 mapped (96.27% : N/A)
47 1786687 + 0 mapped (100.00% : N/A)
48 18870010 + 0 mapped (100.00% : N/A)
49 73508067 + 0 mapped (98.19% : N/A)
50 51196256 + 0 mapped (97.45% : N/A)
51 25112288 + 0 mapped (100.00% : N/A)
52 40497298 + 0 mapped (100.00% : N/A)# plotAvgProf(anno_ind4_V24h,xlim=c(-3000,3000))
plotAnnoBar(anno_ind4_V24h, main = "Genomic Feature Distribution")+ ggtitle("Ind4 VEH 24 hour")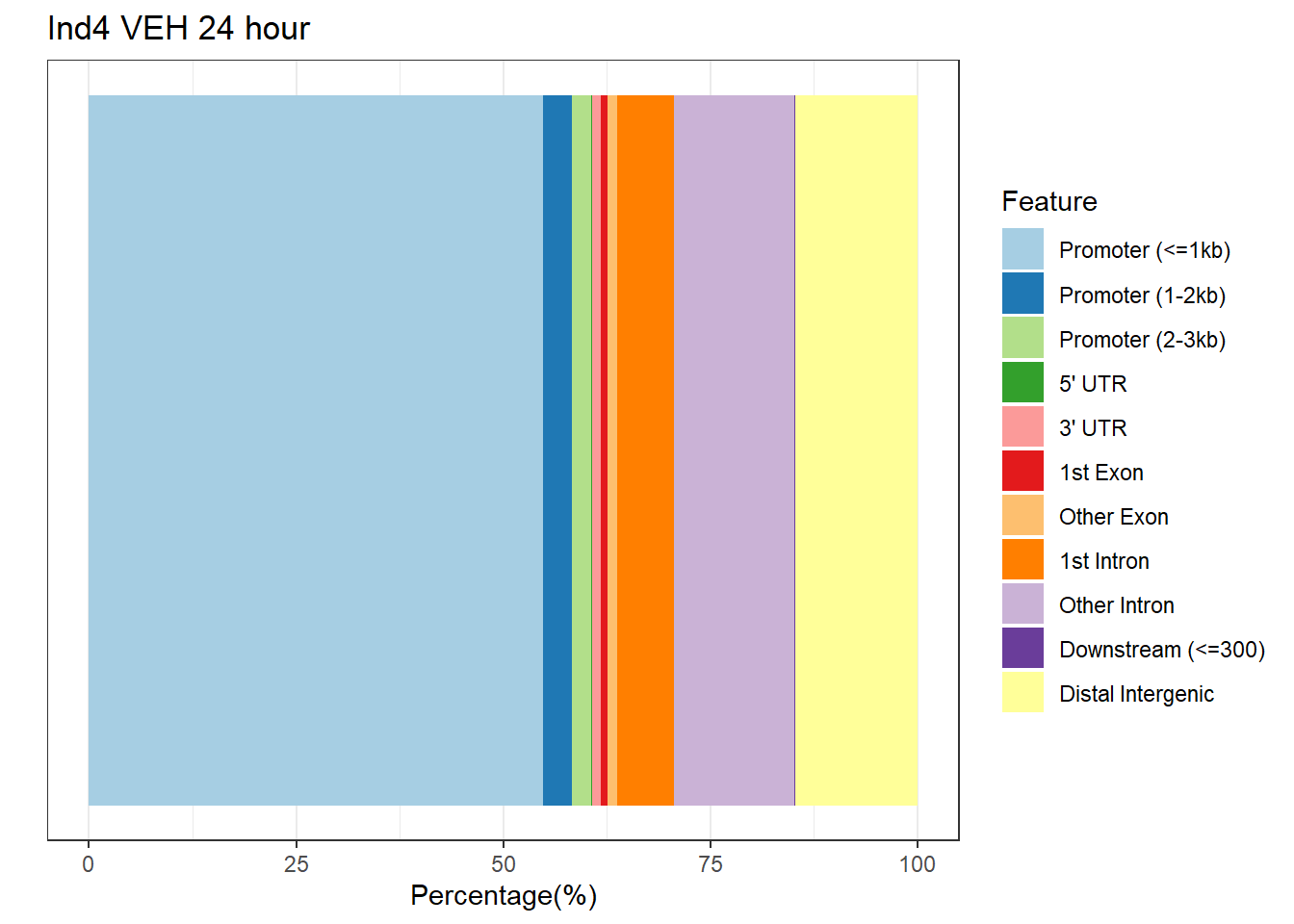
plotAnnoBar(anno_ind1_DA24h, main = "Genomic Feature Distribution")+ ggtitle("Ind1 DNR 24 hour")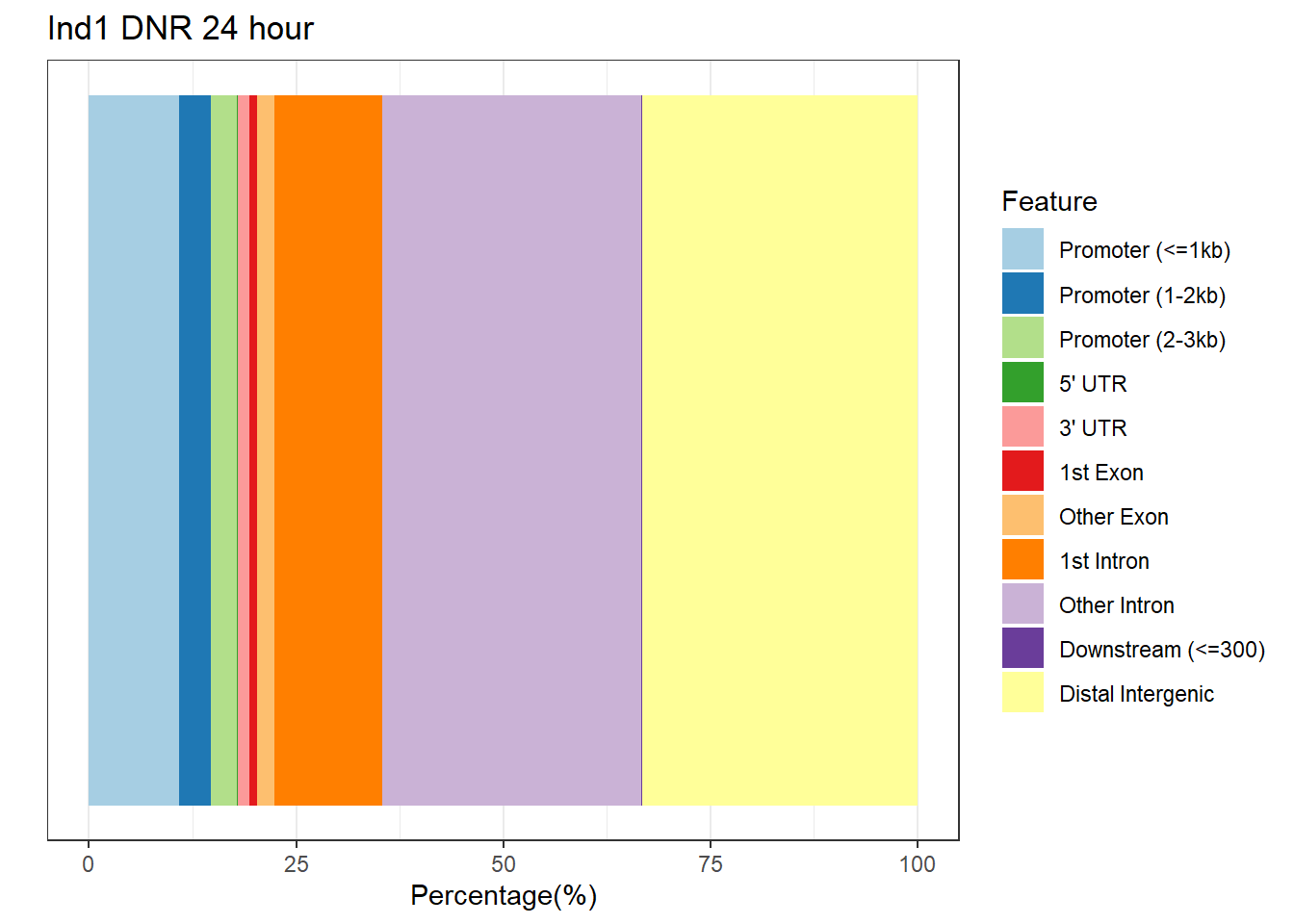
ind4_V24hpeaks_gr <- prepGRangeObj(ind4_V24hpeaks)
ind1_DA24hpeaks_gr <- prepGRangeObj((ind1_DA24hpeaks))
Epi_list <- GRangesList(ind1_DA24hpeaks_gr, ind4_V24hpeaks_gr)
##plotting the TSS average window (making an overlap of each using Epi_list as list holder)
Epi_list_tagMatrix = lapply(Epi_list, getTagMatrix, windows = TSS)>> preparing start_site regions by gene... 2024-02-27 3:04:00 PM
>> preparing tag matrix... 2024-02-27 3:04:00 PM
>> preparing start_site regions by gene... 2024-02-27 3:04:14 PM
>> preparing tag matrix... 2024-02-27 3:04:14 PM plotAvgProf(Epi_list_tagMatrix, xlim=c(-3000, 3000), ylab = "Count Frequency")>> plotting figure... 2024-02-27 3:04:23 PM 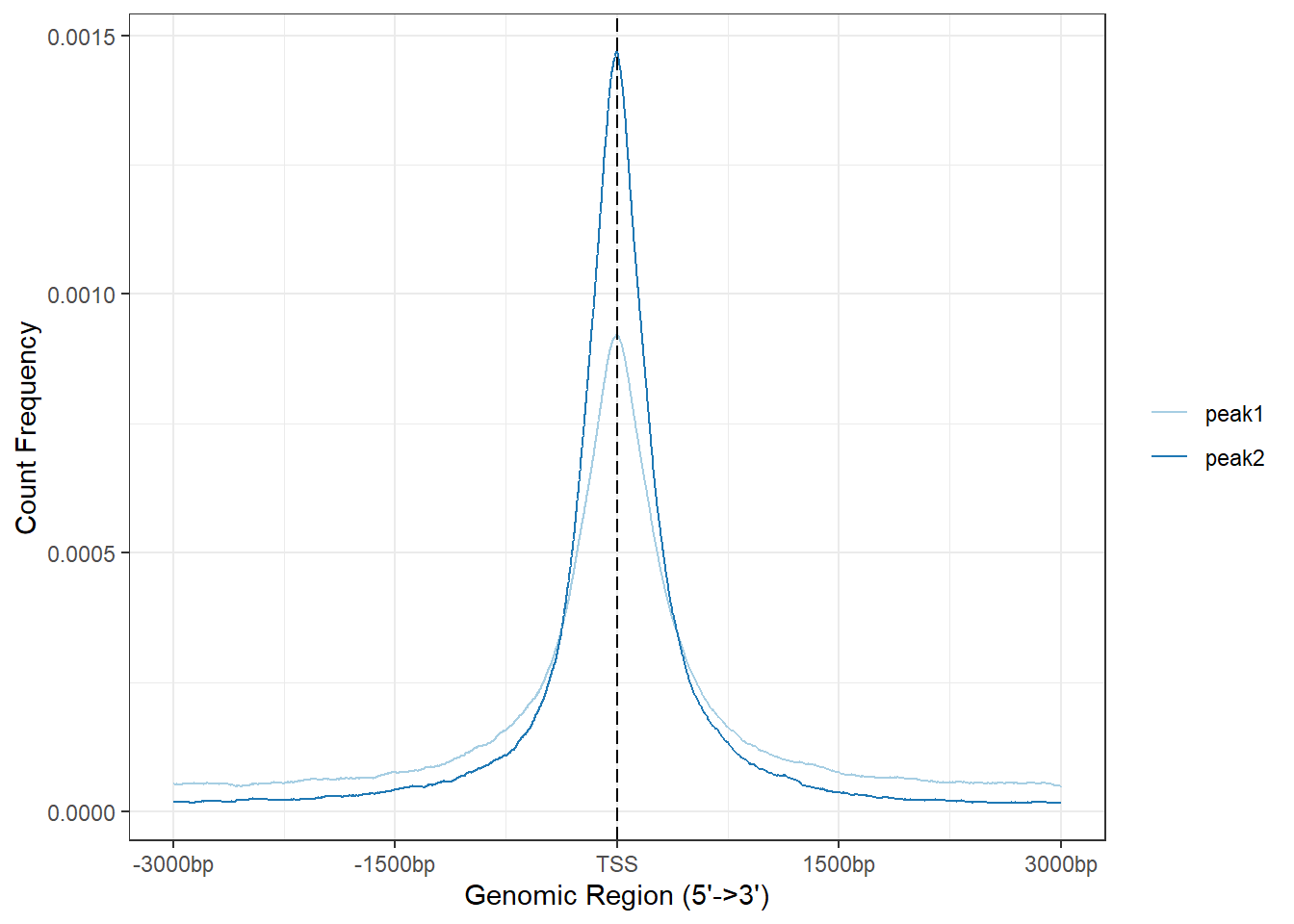
#plotPeakProf(Epi_list_tagMatrix, facet = "none", conf = 0.95)Ind4 3 hour and 24 hour fragment sizes
Ind4_frag_files <- read.csv("data/Ind4_fragment_files.txt", row.names = 1)
Ind4_frag_files %>%
dplyr::filter(time =="3h") %>%
ggplot(., aes(y=counts, x=frag_size, group=trt))+
geom_line(aes(col=trt, alpha = 0.5, linewidth=1 ))+
ggtitle("Individual 4\n3 hour fragment sizes")+
theme_classic()+
scale_color_manual(values=drug_pal_fact)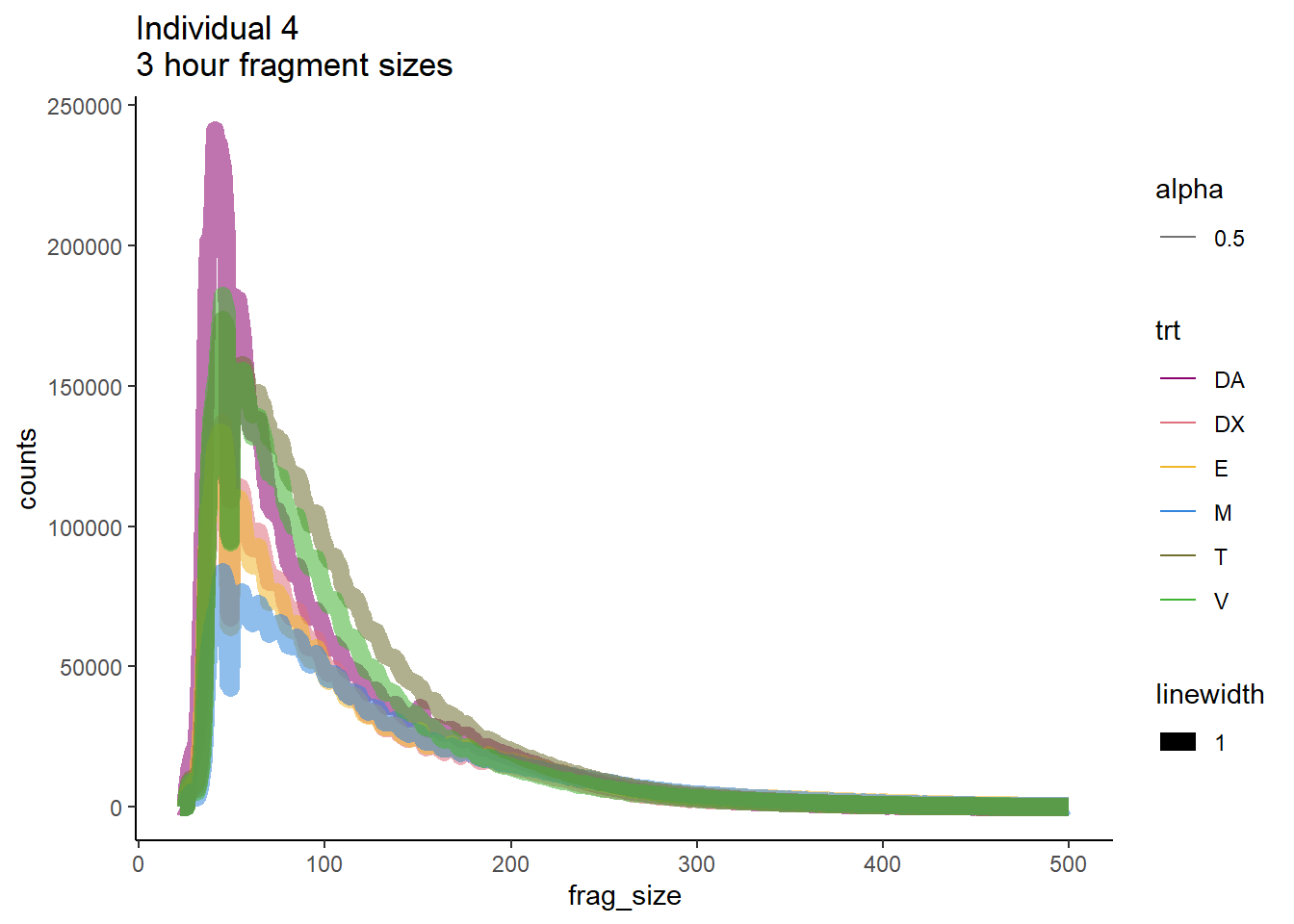
Ind4_frag_files %>%
dplyr::filter(time =="24h") %>%
ggplot(., aes(y=counts, x=frag_size, group=trt))+
geom_line(aes(col=trt, alpha = 0.5, linewidth=1 ))+
ggtitle("Individual 4\n24 hour fragment sizes")+
theme_classic()+
scale_color_manual(values=drug_pal_fact)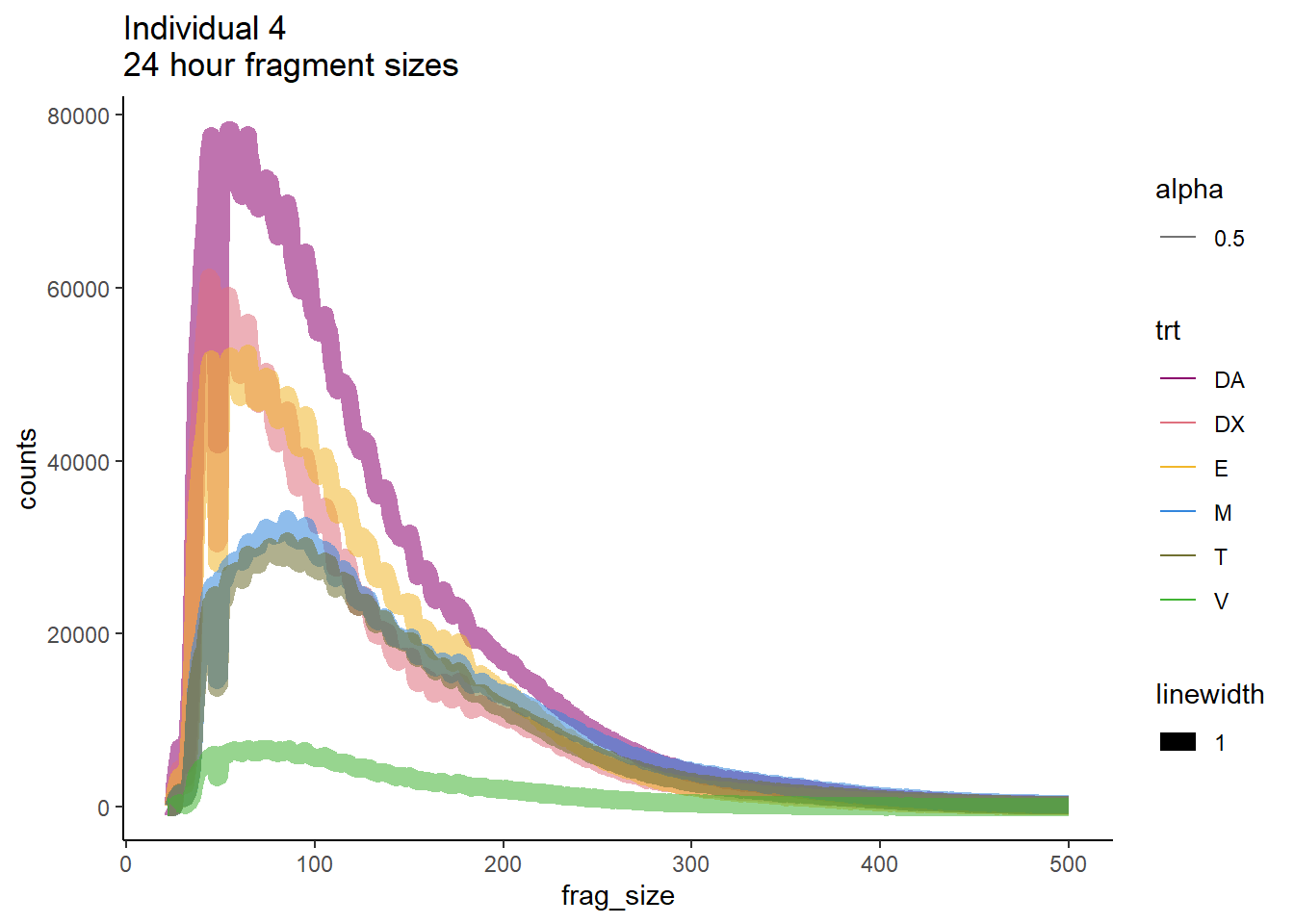
So, fragment lengths are not so great. Lots of noise.
sessionInfo()R version 4.3.1 (2023-06-16 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19045)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
time zone: America/Chicago
tzcode source: internal
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] org.Hs.eg.db_3.17.0
[2] TxDb.Hsapiens.UCSC.hg38.knownGene_3.17.0
[3] GenomicFeatures_1.52.2
[4] AnnotationDbi_1.62.2
[5] Biobase_2.60.0
[6] GenomicRanges_1.52.1
[7] GenomeInfoDb_1.36.4
[8] IRanges_2.34.1
[9] S4Vectors_0.38.2
[10] BiocGenerics_0.46.0
[11] ChIPseeker_1.36.0
[12] RColorBrewer_1.1-3
[13] kableExtra_1.4.0
[14] lubridate_1.9.3
[15] forcats_1.0.0
[16] stringr_1.5.1
[17] dplyr_1.1.4
[18] purrr_1.0.2
[19] readr_2.1.5
[20] tidyr_1.3.1
[21] tibble_3.2.1
[22] ggplot2_3.4.4
[23] tidyverse_2.0.0
[24] workflowr_1.7.1
loaded via a namespace (and not attached):
[1] splines_4.3.1
[2] later_1.3.2
[3] BiocIO_1.10.0
[4] bitops_1.0-7
[5] ggplotify_0.1.2
[6] filelock_1.0.3
[7] polyclip_1.10-6
[8] XML_3.99-0.16.1
[9] lifecycle_1.0.4
[10] rprojroot_2.0.4
[11] processx_3.8.3
[12] lattice_0.22-5
[13] MASS_7.3-60.0.1
[14] magrittr_2.0.3
[15] sass_0.4.8
[16] rmarkdown_2.25
[17] plotrix_3.8-4
[18] jquerylib_0.1.4
[19] yaml_2.3.8
[20] httpuv_1.6.14
[21] cowplot_1.1.3
[22] DBI_1.2.2
[23] abind_1.4-5
[24] zlibbioc_1.46.0
[25] ggraph_2.1.0
[26] RCurl_1.98-1.14
[27] yulab.utils_0.1.4
[28] tweenr_2.0.2
[29] rappdirs_0.3.3
[30] git2r_0.33.0
[31] GenomeInfoDbData_1.2.10
[32] enrichplot_1.20.3
[33] ggrepel_0.9.5
[34] tidytree_0.4.6
[35] svglite_2.1.3
[36] codetools_0.2-19
[37] DelayedArray_0.26.7
[38] DOSE_3.26.2
[39] xml2_1.3.6
[40] ggforce_0.4.2
[41] tidyselect_1.2.0
[42] aplot_0.2.2
[43] farver_2.1.1
[44] viridis_0.6.5
[45] matrixStats_1.2.0
[46] BiocFileCache_2.8.0
[47] GenomicAlignments_1.36.0
[48] jsonlite_1.8.8
[49] tidygraph_1.3.1
[50] systemfonts_1.0.5
[51] tools_4.3.1
[52] progress_1.2.3
[53] treeio_1.24.3
[54] TxDb.Hsapiens.UCSC.hg19.knownGene_3.2.2
[55] Rcpp_1.0.12
[56] glue_1.7.0
[57] gridExtra_2.3
[58] xfun_0.42
[59] qvalue_2.32.0
[60] MatrixGenerics_1.12.3
[61] withr_3.0.0
[62] fastmap_1.1.1
[63] boot_1.3-29
[64] fansi_1.0.6
[65] caTools_1.18.2
[66] callr_3.7.5
[67] digest_0.6.34
[68] timechange_0.3.0
[69] R6_2.5.1
[70] gridGraphics_0.5-1
[71] colorspace_2.1-0
[72] GO.db_3.17.0
[73] gtools_3.9.5
[74] biomaRt_2.56.1
[75] RSQLite_2.3.5
[76] utf8_1.2.4
[77] generics_0.1.3
[78] data.table_1.15.0
[79] rtracklayer_1.60.1
[80] prettyunits_1.2.0
[81] graphlayouts_1.1.0
[82] httr_1.4.7
[83] S4Arrays_1.0.6
[84] scatterpie_0.2.1
[85] whisker_0.4.1
[86] pkgconfig_2.0.3
[87] gtable_0.3.4
[88] blob_1.2.4
[89] XVector_0.40.0
[90] shadowtext_0.1.3
[91] htmltools_0.5.7
[92] fgsea_1.26.0
[93] scales_1.3.0
[94] png_0.1-8
[95] ggfun_0.1.4
[96] knitr_1.45
[97] rstudioapi_0.15.0
[98] tzdb_0.4.0
[99] reshape2_1.4.4
[100] rjson_0.2.21
[101] nlme_3.1-164
[102] curl_5.2.0
[103] cachem_1.0.8
[104] KernSmooth_2.23-22
[105] parallel_4.3.1
[106] HDO.db_0.99.1
[107] restfulr_0.0.15
[108] pillar_1.9.0
[109] grid_4.3.1
[110] vctrs_0.6.5
[111] gplots_3.1.3.1
[112] promises_1.2.1
[113] dbplyr_2.4.0
[114] evaluate_0.23
[115] cli_3.6.2
[116] compiler_4.3.1
[117] Rsamtools_2.16.0
[118] rlang_1.1.3
[119] crayon_1.5.2
[120] labeling_0.4.3
[121] ps_1.7.6
[122] getPass_0.2-4
[123] plyr_1.8.9
[124] fs_1.6.3
[125] stringi_1.8.3
[126] viridisLite_0.4.2
[127] BiocParallel_1.34.2
[128] munsell_0.5.0
[129] Biostrings_2.68.1
[130] lazyeval_0.2.2
[131] GOSemSim_2.26.1
[132] Matrix_1.6-5
[133] hms_1.1.3
[134] patchwork_1.2.0
[135] bit64_4.0.5
[136] KEGGREST_1.40.1
[137] highr_0.10
[138] SummarizedExperiment_1.30.2
[139] igraph_2.0.2
[140] memoise_2.0.1
[141] bslib_0.6.1
[142] ggtree_3.8.2
[143] fastmatch_1.1-4
[144] bit_4.0.5
[145] ape_5.7-1