Jaspar_motif_DAR
Renee Matthews
2025-06-16
Last updated: 2025-07-01
Checks: 7 0
Knit directory: ATAC_learning/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20231016) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version be98936. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/H3K27ac_integration_noM.Rmd
Ignored: data/ACresp_SNP_table.csv
Ignored: data/ARR_SNP_table.csv
Ignored: data/All_merged_peaks.tsv
Ignored: data/CAD_gwas_dataframe.RDS
Ignored: data/CTX_SNP_table.csv
Ignored: data/Collapsed_expressed_NG_peak_table.csv
Ignored: data/DEG_toplist_sep_n45.RDS
Ignored: data/FRiP_first_run.txt
Ignored: data/Final_four_data/
Ignored: data/Frip_1_reads.csv
Ignored: data/Frip_2_reads.csv
Ignored: data/Frip_3_reads.csv
Ignored: data/Frip_4_reads.csv
Ignored: data/Frip_5_reads.csv
Ignored: data/Frip_6_reads.csv
Ignored: data/GO_KEGG_analysis/
Ignored: data/HF_SNP_table.csv
Ignored: data/Ind1_75DA24h_dedup_peaks.csv
Ignored: data/Ind1_TSS_peaks.RDS
Ignored: data/Ind1_firstfragment_files.txt
Ignored: data/Ind1_fragment_files.txt
Ignored: data/Ind1_peaks_list.RDS
Ignored: data/Ind1_summary.txt
Ignored: data/Ind2_TSS_peaks.RDS
Ignored: data/Ind2_fragment_files.txt
Ignored: data/Ind2_peaks_list.RDS
Ignored: data/Ind2_summary.txt
Ignored: data/Ind3_TSS_peaks.RDS
Ignored: data/Ind3_fragment_files.txt
Ignored: data/Ind3_peaks_list.RDS
Ignored: data/Ind3_summary.txt
Ignored: data/Ind4_79B24h_dedup_peaks.csv
Ignored: data/Ind4_TSS_peaks.RDS
Ignored: data/Ind4_V24h_fraglength.txt
Ignored: data/Ind4_fragment_files.txt
Ignored: data/Ind4_fragment_filesN.txt
Ignored: data/Ind4_peaks_list.RDS
Ignored: data/Ind4_summary.txt
Ignored: data/Ind5_TSS_peaks.RDS
Ignored: data/Ind5_fragment_files.txt
Ignored: data/Ind5_fragment_filesN.txt
Ignored: data/Ind5_peaks_list.RDS
Ignored: data/Ind5_summary.txt
Ignored: data/Ind6_TSS_peaks.RDS
Ignored: data/Ind6_fragment_files.txt
Ignored: data/Ind6_peaks_list.RDS
Ignored: data/Ind6_summary.txt
Ignored: data/Knowles_4.RDS
Ignored: data/Knowles_5.RDS
Ignored: data/Knowles_6.RDS
Ignored: data/LiSiLTDNRe_TE_df.RDS
Ignored: data/MI_gwas.RDS
Ignored: data/SNP_GWAS_PEAK_MRC_id
Ignored: data/SNP_GWAS_PEAK_MRC_id.csv
Ignored: data/SNP_gene_cat_list.tsv
Ignored: data/SNP_supp_schneider.RDS
Ignored: data/TE_info/
Ignored: data/TFmapnames.RDS
Ignored: data/all_TSSE_scores.RDS
Ignored: data/all_four_filtered_counts.txt
Ignored: data/aln_run1_results.txt
Ignored: data/anno_ind1_DA24h.RDS
Ignored: data/anno_ind4_V24h.RDS
Ignored: data/annotated_gwas_SNPS.csv
Ignored: data/background_n45_he_peaks.RDS
Ignored: data/cardiac_muscle_FRIP.csv
Ignored: data/cardiomyocyte_FRIP.csv
Ignored: data/col_ng_peak.csv
Ignored: data/cormotif_full_4_run.RDS
Ignored: data/cormotif_full_4_run_he.RDS
Ignored: data/cormotif_full_6_run.RDS
Ignored: data/cormotif_full_6_run_he.RDS
Ignored: data/cormotif_probability_45_list.csv
Ignored: data/cormotif_probability_45_list_he.csv
Ignored: data/cormotif_probability_all_6_list.csv
Ignored: data/cormotif_probability_all_6_list_he.csv
Ignored: data/datasave.RDS
Ignored: data/embryo_heart_FRIP.csv
Ignored: data/enhancer_list_ENCFF126UHK.bed
Ignored: data/enhancerdata/
Ignored: data/filt_Peaks_efit2.RDS
Ignored: data/filt_Peaks_efit2_bl.RDS
Ignored: data/filt_Peaks_efit2_n45.RDS
Ignored: data/first_Peaksummarycounts.csv
Ignored: data/first_run_frag_counts.txt
Ignored: data/full_bedfiles/
Ignored: data/gene_ref.csv
Ignored: data/gwas_1_dataframe.RDS
Ignored: data/gwas_2_dataframe.RDS
Ignored: data/gwas_3_dataframe.RDS
Ignored: data/gwas_4_dataframe.RDS
Ignored: data/gwas_5_dataframe.RDS
Ignored: data/high_conf_peak_counts.csv
Ignored: data/high_conf_peak_counts.txt
Ignored: data/high_conf_peaks_bl_counts.txt
Ignored: data/high_conf_peaks_counts.txt
Ignored: data/hits_files/
Ignored: data/hyper_files/
Ignored: data/hypo_files/
Ignored: data/ind1_DA24hpeaks.RDS
Ignored: data/ind1_TSSE.RDS
Ignored: data/ind2_TSSE.RDS
Ignored: data/ind3_TSSE.RDS
Ignored: data/ind4_TSSE.RDS
Ignored: data/ind4_V24hpeaks.RDS
Ignored: data/ind5_TSSE.RDS
Ignored: data/ind6_TSSE.RDS
Ignored: data/initial_complete_stats_run1.txt
Ignored: data/left_ventricle_FRIP.csv
Ignored: data/median_24_lfc.RDS
Ignored: data/median_3_lfc.RDS
Ignored: data/mergedPeads.gff
Ignored: data/mergedPeaks.gff
Ignored: data/motif_list_full
Ignored: data/motif_list_n45
Ignored: data/motif_list_n45.RDS
Ignored: data/multiqc_fastqc_run1.txt
Ignored: data/multiqc_fastqc_run2.txt
Ignored: data/multiqc_genestat_run1.txt
Ignored: data/multiqc_genestat_run2.txt
Ignored: data/my_hc_filt_counts.RDS
Ignored: data/my_hc_filt_counts_n45.RDS
Ignored: data/n45_bedfiles/
Ignored: data/n45_files
Ignored: data/other_papers/
Ignored: data/peakAnnoList_1.RDS
Ignored: data/peakAnnoList_2.RDS
Ignored: data/peakAnnoList_24_full.RDS
Ignored: data/peakAnnoList_24_n45.RDS
Ignored: data/peakAnnoList_3.RDS
Ignored: data/peakAnnoList_3_full.RDS
Ignored: data/peakAnnoList_3_n45.RDS
Ignored: data/peakAnnoList_4.RDS
Ignored: data/peakAnnoList_5.RDS
Ignored: data/peakAnnoList_6.RDS
Ignored: data/peakAnnoList_Eight.RDS
Ignored: data/peakAnnoList_full_motif.RDS
Ignored: data/peakAnnoList_n45_motif.RDS
Ignored: data/siglist_full.RDS
Ignored: data/siglist_n45.RDS
Ignored: data/summarized_peaks_dataframe.txt
Ignored: data/summary_peakIDandReHeat.csv
Ignored: data/test.list.RDS
Ignored: data/testnames.txt
Ignored: data/toplist_6.RDS
Ignored: data/toplist_full.RDS
Ignored: data/toplist_full_DAR_6.RDS
Ignored: data/toplist_n45.RDS
Ignored: data/trimmed_seq_length.csv
Ignored: data/unclassified_full_set_peaks.RDS
Ignored: data/unclassified_n45_set_peaks.RDS
Ignored: data/xstreme/
Untracked files:
Untracked: RNA_seq_integration.Rmd
Untracked: Rplot.pdf
Untracked: Sig_meta
Untracked: analysis/.gitignore
Untracked: analysis/AC_shared_analysis.Rmd
Untracked: analysis/AF_HF_SNP_DAR.Rmd
Untracked: analysis/Cormotif_analysis_testing diff.Rmd
Untracked: analysis/DOX_DAR_heatmap.Rmd
Untracked: analysis/Diagnosis-tmm.Rmd
Untracked: analysis/Expressed_RNA_associations.Rmd
Untracked: analysis/LFC_corr.Rmd
Untracked: analysis/SNP_TAD_peaks.Rmd
Untracked: analysis/SVA.Rmd
Untracked: analysis/Tan2020.Rmd
Untracked: analysis/Top2B_analysis.Rmd
Untracked: analysis/making_master_peaks_list.Rmd
Untracked: analysis/my_hc_filt_counts.csv
Untracked: code/Concatenations_for_export.R
Untracked: code/IGV_snapshot_code.R
Untracked: code/LongDARlist.R
Untracked: code/just_for_Fun.R
Untracked: my_plot.pdf
Untracked: my_plot.png
Untracked: output/cormotif_probability_45_list.csv
Untracked: output/cormotif_probability_all_6_list.csv
Untracked: setup.RData
Unstaged changes:
Modified: ATAC_learning.Rproj
Modified: analysis/AF_HF_SNPs.Rmd
Modified: analysis/Cardiotox_SNPs.Rmd
Modified: analysis/Cormotif_analysis.Rmd
Modified: analysis/DEG_analysis.Rmd
Modified: analysis/H3K27ac_initial_QC.Rmd
Modified: analysis/Jaspar_motif.Rmd
Modified: analysis/Jaspar_motif_ff.Rmd
Modified: analysis/TE_analysis_norm.Rmd
Modified: analysis/final_four_analysis.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/Jaspar_motif_DAR.Rmd) and
HTML (docs/Jaspar_motif_DAR.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | be98936 | reneeisnowhere | 2025-07-01 | updates |
| Rmd | 26eb8eb | reneeisnowhere | 2025-06-23 | adding updates |
library(tidyverse)
library(cowplot)
library(kableExtra)
library(broom)
library(RColorBrewer)
library(ChIPseeker)
library("TxDb.Hsapiens.UCSC.hg38.knownGene")
library("org.Hs.eg.db")
library(rtracklayer)
library(edgeR)
library(ggfortify)
library(limma)
library(readr)
library(BiocGenerics)
library(gridExtra)
library(VennDiagram)
library(scales)
library(Cormotif)
library(BiocParallel)
library(ggpubr)
library(devtools)
library(JASPAR2022)
library(TFBSTools)
library(MotifDb)
library(BSgenome.Hsapiens.UCSC.hg38)
library(data.table)
library(universalmotif)
library(ggseqlogo)
library(motifmatchr)
library(gridExtra)# mrc_palette <- c(
# "EAR_open" = "#F8766D",
# "EAR_close" = "#f6483c",
# "ESR_open" = "#7CAE00",
# "ESR_close" = "#587b00",
# "ESR_C"="grey40",
# "ESR_opcl"="grey40",
# "ESR_D"="tan",
# "ESR_clop"="tan",
# "LR_open" = "#00BFC4",
# "LR_close" = "#008d91",
# "NR" = "#C77CFF"
# )DOX_DAR_3hr data
DOX_DAR_3hr_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DOX_3_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
DOX_DAR_3hr_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DOX_3_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
DOX_DAR_3hr_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DOX_3_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
DOX_DAR_3hr_all <- rbind(DOX_DAR_3hr_sea_disc, DOX_DAR_3hr_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)DOX_DAR_3hr_combo <- DOX_DAR_3hr_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., DOX_DAR_3hr_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ NA_character_
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y) %>%
mutate(mrc_type="DOX_DAR_3hr")
DOX_DAR_3hr_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs in 3 hour DOX-DAR v 3 hour non-DOX DAR") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO | mrc_type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | MA1630.2 (ZNF281) | STREME-2 | 2-CCCCWCCCC | CCCCWCCCC | ZNF281 | 2.3e-83 | 1.82e-83 | 1040 | 33.1 | 24900 | 18.2 | 1.82 | DOX_DAR_3hr |
| 2 | 1 | MA1961.1 | PATZ1 | MA1961.1 | SSGGGGMGGGGS | PATZ1 | 2.48e-66 | 9.82e-67 | 1570 | 50.2 | 47400 | 34.6 | 1.45 | DOX_DAR_3hr |
| 3 | 1 | MA1630.2 (ZNF281) | MEME-1 | GGGWGGGGGSGGGGG | GGGWGGGGGSGGGGG | ZNF281 | 8.82e-64 | 2.33e-64 | 1240 | 39.6 | 34700 | 25.4 | 1.56 | DOX_DAR_3hr |
| 4 | 1 | MA1653.1 | ZNF148 | MA1653.1 | YBCCCCTCCCCC | ZNF148 | 2.61e-61 | 5.16e-62 | 1160 | 37 | 31900 | 23.3 | 1.59 | DOX_DAR_3hr |
| 5 | 1 | MA1653.1 (ZNF148) | STREME-5 | 5-CCCTCCC | CCCTCCC | ZNF148 | 8.33e-60 | 1.32e-60 | 1860 | 59.4 | 60700 | 44.3 | 1.34 | DOX_DAR_3hr |
| 6 | 1 | MA1959.1 | KLF7 | MA1959.1 | DGGGCGGGG | KLF7 | 6.29e-59 | 8.31e-60 | 1650 | 52.7 | 51800 | 37.9 | 1.39 | DOX_DAR_3hr |
| 7 | 1 | MA0740.2 | KLF14 | MA0740.2 | KGGGCGGGG | KLF14 | 2.2e-58 | 2.49e-59 | 1040 | 33.2 | 27900 | 20.4 | 1.63 | DOX_DAR_3hr |
| 8 | 1 | MA0039.4 | KLF4 | MA0039.4 | NNCCCCACCCHN | KLF4 | 8.48e-57 | 8.4e-58 | 1460 | 46.7 | 44500 | 32.5 | 1.44 | DOX_DAR_3hr |
| 9 | 1 | MA0599.1 | KLF5 | MA0599.1 | GCCCCDCCCH | KLF5 | 5.29e-55 | 4.66e-56 | 1200 | 38.4 | 34500 | 25.2 | 1.52 | DOX_DAR_3hr |
| 10 | 1 | MA1522.1 | MAZ | MA1522.1 | BBCCCCTCCCY | MAZ | 9.43e-53 | 7.47e-54 | 1640 | 52.4 | 52600 | 38.4 | 1.37 | DOX_DAR_3hr |
| 11 | 1 | MA0493.2 | KLF1 | MA0493.2 | DGGGYGGGG | KLF1 | 3.83e-52 | 2.76e-53 | 1340 | 42.8 | 40400 | 29.5 | 1.45 | DOX_DAR_3hr |
| 12 | 2 | MA0742.2 (KLF12) | MEME-3 | CCCAGCCC | CCCAGCCC | KLF12 | 1.13e-51 | 7.49e-53 | 1650 | 52.8 | 53300 | 38.9 | 1.36 | DOX_DAR_3hr |
| 13 | 1 | MA0742.2 | KLF12 | MA0742.2 | GGGGCGGGG | KLF12 | 2.27e-49 | 1.38e-50 | 1690 | 54.2 | 55500 | 40.5 | 1.34 | DOX_DAR_3hr |
| 14 | 1 | MA1965.1 | SP5 | MA1965.1 | CBCCTCCCHS | SP5 | 5.78e-47 | 3.27e-48 | 1680 | 53.8 | 55400 | 40.5 | 1.33 | DOX_DAR_3hr |
| 15 | 1 | MA0079.5 | SP1 | MA0079.5 | GGGGCGGGG | SP1 | 1.95e-44 | 1.03e-45 | 1040 | 33.3 | 30000 | 21.9 | 1.52 | DOX_DAR_3hr |
| 16 | 1 | MA1511.2 | KLF10 | MA1511.2 | GGGGCGGGG | KLF10 | 5.2e-43 | 2.58e-44 | 964 | 30.8 | 27300 | 20 | 1.55 | DOX_DAR_3hr |
| 17 | 1 | MA0155.1 (INSM1) | STREME-3 | 3-CCCTGCC | CCCTGCC | INSM1 | 1.08e-42 | 4.97e-44 | 1340 | 42.9 | 42100 | 30.7 | 1.4 | DOX_DAR_3hr |
| 18 | 3 | MA1564.1 | SP9 | MA1564.1 | RCCACGCCCMCY | SP9 | 1.13e-42 | 4.97e-44 | 1590 | 50.9 | 52400 | 38.3 | 1.33 | DOX_DAR_3hr |
| 19 | 1 | MA0741.1 | KLF16 | MA0741.1 | GMCACGCCCCC | KLF16 | 4.01e-42 | 1.67e-43 | 779 | 24.9 | 20700 | 15.1 | 1.65 | DOX_DAR_3hr |
| 20 | 3 | MA0746.2 | SP3 | MA0746.2 | NGCCACGCCCMCH | SP3 | 7.98e-42 | 3.16e-43 | 1110 | 35.5 | 33100 | 24.2 | 1.47 | DOX_DAR_3hr |
| 21 | 1 | MA0516.3 | SP2 | MA0516.3 | GGGGCGGGG | SP2 | 2.71e-41 | 1.02e-42 | 898 | 28.7 | 25100 | 18.4 | 1.57 | DOX_DAR_3hr |
| 22 | 4 | MA1713.1 | ZNF610 | MA1713.1 | SSCGCCGCTCCSSS | ZNF610 | 6.02e-41 | 2.17e-42 | 1910 | 61 | 66400 | 48.5 | 1.26 | DOX_DAR_3hr |
| 23 | 1 | MA0685.2 | SP4 | MA0685.2 | GGGGCGGGG | SP4 | 1.75e-40 | 6.02e-42 | 996 | 31.9 | 28900 | 21.1 | 1.51 | DOX_DAR_3hr |
| 24 | 3 | MA1578.1 | VEZF1 | MA1578.1 | CCCCCCMYDH | VEZF1 | 2.72e-39 | 8.98e-41 | 2070 | 66.4 | 74300 | 54.3 | 1.22 | DOX_DAR_3hr |
| 25 | 5 | MA1548.1 (PLAGL2) | STREME-1 | 1-GGSSCC | GGSSCC | PLAGL2 | 5.98e-35 | 1.89e-36 | 2300 | 73.4 | 85500 | 62.5 | 1.18 | DOX_DAR_3hr |
| 26 | 6 | MA1650.1 | ZBTB14 | MA1650.1 | SSCCGCGCACVS | ZBTB14 | 3.94e-34 | 1.2e-35 | 1910 | 61.2 | 68200 | 49.8 | 1.23 | DOX_DAR_3hr |
| 27 | 3 | MA1107.2 | KLF9 | MA1107.2 | MNGCCACACCCACHYV | KLF9 | 7.26e-32 | 2.13e-33 | 1780 | 56.9 | 62800 | 45.9 | 1.24 | DOX_DAR_3hr |
| 28 | 1 | MA1630.2 | ZNF281 | MA1630.2 | GGGGGAGGGGVV | ZNF281 | 7.97e-32 | 2.26e-33 | 286 | 9.15 | 5560 | 4.06 | 2.26 | DOX_DAR_3hr |
| 29 | 7 | MA1548.1 | PLAGL2 | MA1548.1 | NGGGCCCCCN | PLAGL2 | 5.35e-31 | 1.46e-32 | 1510 | 48.4 | 51600 | 37.7 | 1.29 | DOX_DAR_3hr |
| 30 | 3 | MA0747.1 | SP8 | MA0747.1 | RCCACGCCCMCY | SP8 | 8.35e-31 | 2.2e-32 | 1190 | 38 | 38100 | 27.9 | 1.36 | DOX_DAR_3hr |
| 31 | 8 | MA1976.1 | ZNF320 | MA1976.1 | DGKGGGRCCMRGGGGCCWGTGHVNNNN | ZNF320 | 7.65e-30 | 1.95e-31 | 1680 | 53.9 | 59100 | 43.2 | 1.25 | DOX_DAR_3hr |
| 32 | 9 | MA1596.1 | ZNF460 | MA1596.1 | GCCTCMGCCTCCCRAG | ZNF460 | 4.18e-29 | 1.03e-30 | 1380 | 44.1 | 46300 | 33.8 | 1.3 | DOX_DAR_3hr |
| 33 | 3 | MA0733.1 | EGR4 | MA0733.1 | NHMCGCCCACGCAHWT | EGR4 | 1.77e-28 | 4.24e-30 | 2220 | 70.9 | 83300 | 60.8 | 1.17 | DOX_DAR_3hr |
| 34 | 3 | MA1516.1 | KLF3 | MA1516.1 | GRCCRCGCCCH | KLF3 | 1.05e-27 | 2.45e-29 | 2080 | 66.5 | 77100 | 56.3 | 1.18 | DOX_DAR_3hr |
| 35 | 10 | MA0073.1 (RREB1) | STREME-8 | 8-CACCCCC | CACCCCC | RREB1 | 2.03e-27 | 4.59e-29 | 596 | 19.1 | 16300 | 11.9 | 1.61 | DOX_DAR_3hr |
| 36 | 11 | MA0471.2 | E2F6 | MA0471.2 | VVDGGCGGGAAVV | E2F6 | 8.96e-26 | 1.97e-27 | 2380 | 76 | 91500 | 66.9 | 1.14 | DOX_DAR_3hr |
| 37 | 12 | MA1615.1 | Plagl1 | MA1615.1 | NNCTGGGGCCABN | Plagl1 | 1.33e-25 | 2.85e-27 | 1670 | 53.4 | 59400 | 43.4 | 1.23 | DOX_DAR_3hr |
| 38 | 13 | MA0872.1 | TFAP2A | MA0872.1 | TGCCCYSRGGGCA | TFAP2A | 1.2e-23 | 2.49e-25 | 941 | 30.1 | 29900 | 21.8 | 1.38 | DOX_DAR_3hr |
| 39 | 1 | MA1627.1 | Wt1 | MA1627.1 | YBCCTCCCCCACVB | Wt1 | 1.51e-22 | 3.07e-24 | 532 | 17 | 14700 | 10.8 | 1.58 | DOX_DAR_3hr |
| 40 | 14 | MA0131.2 | HINFP | MA0131.2 | CARCGTCCGCKN | HINFP | 2.39e-22 | 4.74e-24 | 1860 | 59.6 | 68800 | 50.2 | 1.19 | DOX_DAR_3hr |
| 41 | 3 | MA0162.4 | EGR1 | MA0162.4 | VCMCGCCCACGCVS | EGR1 | 3.6e-22 | 6.95e-24 | 1080 | 34.6 | 35700 | 26.1 | 1.32 | DOX_DAR_3hr |
| 42 | 3 | MA1512.1 | KLF11 | MA1512.1 | SCCACGCCCMC | KLF11 | 4.53e-22 | 8.55e-24 | 601 | 19.2 | 17300 | 12.6 | 1.52 | DOX_DAR_3hr |
| 43 | 15 | MA0146.2 | Zfx | MA0146.2 | SSSGCCBVGGCCTS | Zfx | 2.48e-21 | 4.57e-23 | 1370 | 43.9 | 48000 | 35 | 1.25 | DOX_DAR_3hr |
| 44 | 3 | MA1514.1 | KLF17 | MA1514.1 | MMCCACGCACCCMTY | KLF17 | 5.24e-21 | 9.43e-23 | 1780 | 57 | 65600 | 47.9 | 1.19 | DOX_DAR_3hr |
| 45 | 16 | MA0146.2 (Zfx) | STREME-4 | 4-CAGGCC | CAGGCC | Zfx | 7.44e-21 | 1.31e-22 | 1420 | 45.3 | 49900 | 36.4 | 1.24 | DOX_DAR_3hr |
| 46 | 1 | MA1513.1 | KLF15 | MA1513.1 | SCCCCGCCCCS | KLF15 | 2.5e-20 | 4.3e-22 | 973 | 31.1 | 31900 | 23.3 | 1.34 | DOX_DAR_3hr |
| 47 | 17 | MA1712.1 | ZNF454 | MA1712.1 | DGGCTCSSGGCCCYGVKG | ZNF454 | 5.41e-20 | 9.12e-22 | 1750 | 55.9 | 64400 | 47.1 | 1.19 | DOX_DAR_3hr |
| 48 | 48 | MA0528.2 | ZNF263 | MA0528.2 | VNGGGGAGGASG | ZNF263 | 2.82e-19 | 4.66e-21 | 1350 | 43.1 | 47400 | 34.7 | 1.24 | DOX_DAR_3hr |
| 49 | 8 | MA1723.1 | PRDM9 | MA1723.1 | RGDGGGVAGGGRGGVRRMARVARR | PRDM9 | 4.66e-19 | 7.53e-21 | 869 | 27.8 | 28100 | 20.5 | 1.36 | DOX_DAR_3hr |
| 50 | 3 | MA1517.1 | KLF6 | MA1517.1 | NRCCACGCCCH | KLF6 | 1.1e-18 | 1.74e-20 | 1140 | 36.3 | 38800 | 28.4 | 1.28 | DOX_DAR_3hr |
| 51 | 1 | MA0753.2 | ZNF740 | MA0753.2 | CYRCCCCCCCCAC | ZNF740 | 1.33e-18 | 2.06e-20 | 205 | 6.56 | 4290 | 3.13 | 2.1 | DOX_DAR_3hr |
| 52 | 13 | MA0813.1 | TFAP2B | MA0813.1 | TGCCCYBRGGGCA | TFAP2B | 4.92e-18 | 7.49e-20 | 806 | 25.8 | 25900 | 18.9 | 1.37 | DOX_DAR_3hr |
| 53 | 18 | MA1710.1 | ZNF257 | MA1710.1 | VDGAGGCRAGRG | ZNF257 | 1.18e-17 | 1.76e-19 | 1880 | 60.2 | 71100 | 51.9 | 1.16 | DOX_DAR_3hr |
| 54 | 2 | 6-CCCAGC | STREME-6 | 6-CCCAGC | CCCAGC | CCCAGC | 3.68e-17 | 5.39e-19 | 2770 | 88.7 | 113000 | 82.8 | 1.07 | DOX_DAR_3hr |
| 55 | 19 | MA1981.1 | ZNF530 | MA1981.1 | GMARGGMRAGGGGCRGSV | ZNF530 | 1.37e-16 | 1.97e-18 | 672 | 21.5 | 21000 | 15.4 | 1.4 | DOX_DAR_3hr |
| 56 | 20 | MA0814.2 | TFAP2C | MA0814.2 | NNGCCTGAGGCGNN | TFAP2C | 1.47e-16 | 2.08e-18 | 2380 | 76.3 | 94400 | 69 | 1.11 | DOX_DAR_3hr |
| 57 | 3 | MA1515.1 | KLF2 | MA1515.1 | NRCCACRCCCH | KLF2 | 1.91e-16 | 2.66e-18 | 668 | 21.4 | 20900 | 15.3 | 1.4 | DOX_DAR_3hr |
| 58 | 11 | MA1987.1 | ZNF701 | MA1987.1 | GAGCACYAARGGGGGRADDDD | ZNF701 | 2.6e-16 | 3.55e-18 | 2030 | 64.9 | 78000 | 57 | 1.14 | DOX_DAR_3hr |
| 59 | 3 | MA0732.1 | EGR3 | MA0732.1 | HHMCGCCCACGCAHH | EGR3 | 1.85e-15 | 2.48e-17 | 815 | 26.1 | 26800 | 19.6 | 1.33 | DOX_DAR_3hr |
| 60 | 12 | MA1102.2 | CTCFL | MA1102.2 | NSCAGGGGGCGS | CTCFL | 2.22e-15 | 2.94e-17 | 1450 | 46.5 | 53100 | 38.8 | 1.2 | DOX_DAR_3hr |
| 61 | 21 | MA1599.1 | ZNF682 | MA1599.1 | BVGGCYAAGCCCCWNY | ZNF682 | 7.76e-15 | 1.01e-16 | 2400 | 76.9 | 95800 | 70 | 1.1 | DOX_DAR_3hr |
| 62 | 12 | MA1986.1 | ZNF692 | MA1986.1 | BGGGCCCASN | ZNF692 | 1.35e-14 | 1.72e-16 | 1190 | 38.2 | 42400 | 31 | 1.23 | DOX_DAR_3hr |
| 63 | 13 | MA0524.2 | TFAP2C | MA0524.2 | YGCCYBVRGGCA | TFAP2C | 1.97e-14 | 2.48e-16 | 2100 | 67.2 | 82000 | 59.9 | 1.12 | DOX_DAR_3hr |
| 64 | 22 | MA0504.1 | NR2C2 | MA0504.1 | VGRGGTCARAGGTCA | NR2C2 | 9.97e-14 | 1.22e-15 | 1850 | 59.2 | 70900 | 51.8 | 1.14 | DOX_DAR_3hr |
| 65 | 20 | MA1569.1 | TFAP2E | MA1569.1 | YGCCYSRGGCD | TFAP2E | 9.99e-14 | 1.22e-15 | 646 | 20.7 | 20700 | 15.1 | 1.37 | DOX_DAR_3hr |
| 66 | 20 | MA0812.1 | TFAP2B | MA0812.1 | HGCCTSAGGCD | TFAP2B | 1.4e-13 | 1.68e-15 | 2040 | 65.3 | 79500 | 58.1 | 1.12 | DOX_DAR_3hr |
| 67 | 13 | MA0815.1 | TFAP2C | MA0815.1 | TGCCCYSRGGGCA | TFAP2C | 4.7e-13 | 5.56e-15 | 722 | 23.1 | 23800 | 17.4 | 1.33 | DOX_DAR_3hr |
| 68 | 23 | MA0694.1 | ZBTB7B | MA0694.1 | RCGACCACCGAA | ZBTB7B | 6.04e-13 | 7.03e-15 | 819 | 26.2 | 27600 | 20.2 | 1.3 | DOX_DAR_3hr |
| 69 | 23 | MA1990.1 | Gli1 | MA1990.1 | NNRGACCACCCASV | Gli1 | 6.43e-13 | 7.39e-15 | 1760 | 56.1 | 67000 | 48.9 | 1.15 | DOX_DAR_3hr |
| 70 | 24 | MA1528.1 | NFIX | MA1528.1 | STTGGCRNNGTGCCARB | NFIX | 1.25e-12 | 1.42e-14 | 358 | 11.4 | 10200 | 7.42 | 1.55 | DOX_DAR_3hr |
| 71 | 11 | MA1122.1 | TFDP1 | MA1122.1 | VSGCGGGAAVN | TFDP1 | 2.17e-12 | 2.42e-14 | 2690 | 86.1 | 110000 | 80.7 | 1.07 | DOX_DAR_3hr |
| 72 | 13 | MA0811.1 | TFAP2B | MA0811.1 | YGCCCBVRGGCA | TFAP2B | 4.06e-12 | 4.46e-14 | 2120 | 67.9 | 83700 | 61.2 | 1.11 | DOX_DAR_3hr |
| 73 | 25 | MA1721.1 | ZNF93 | MA1721.1 | GGYAGCRGCAGCGGYG | ZNF93 | 1.33e-11 | 1.45e-13 | 1680 | 53.7 | 64100 | 46.8 | 1.15 | DOX_DAR_3hr |
| 74 | 23 | MA0734.3 | Gli2 | MA0734.3 | NRGACCACCCASV | Gli2 | 2.48e-11 | 2.66e-13 | 1090 | 34.9 | 39200 | 28.6 | 1.22 | DOX_DAR_3hr |
| 75 | 13 | MA0810.1 | TFAP2A | MA0810.1 | YGCCCBVRGGCR | TFAP2A | 3.32e-11 | 3.51e-13 | 2570 | 82.2 | 105000 | 76.7 | 1.07 | DOX_DAR_3hr |
| 76 | 26 | MA0751.1 | ZIC4 | MA0751.1 | GRCCCCCCGCKGYGH | ZIC4 | 3.52e-11 | 3.67e-13 | 2450 | 78.2 | 99000 | 72.3 | 1.08 | DOX_DAR_3hr |
| 77 | 27 | MA0830.2 | TCF4 | MA0830.2 | NNGCACCTGCCNN | TCF4 | 7.53e-11 | 7.75e-13 | 2040 | 65.1 | 80200 | 58.6 | 1.11 | DOX_DAR_3hr |
| 78 | 12 | MA1719.1 | ZNF816 | MA1719.1 | DRRKGGGGACATGCHGGG | ZNF816 | 1.37e-10 | 1.39e-12 | 1780 | 57 | 69000 | 50.4 | 1.13 | DOX_DAR_3hr |
| 79 | 27 | MA0506.2 | Nrf1 | MA0506.2 | SNCTGCGCMTGCGCV | Nrf1 | 1.61e-09 | 1.62e-11 | 1420 | 45.3 | 53500 | 39.1 | 1.16 | DOX_DAR_3hr |
| 80 | 24 | MA1643.1 | NFIB | MA1643.1 | NNCCTGGCAYNGTGCCAAGNN | NFIB | 1.66e-09 | 1.64e-11 | 270 | 8.64 | 7570 | 5.53 | 1.57 | DOX_DAR_3hr |
| 81 | 28 | MA0155.1 | INSM1 | MA0155.1 | TGYCWGGGGGCR | INSM1 | 3.46e-09 | 3.39e-11 | 235 | 7.52 | 6390 | 4.67 | 1.62 | DOX_DAR_3hr |
| 82 | 20 | MA0003.4 | TFAP2A | MA0003.4 | NNKGCCTCAGGCNN | TFAP2A | 3.81e-09 | 3.68e-11 | 2660 | 85.2 | 110000 | 80.5 | 1.06 | DOX_DAR_3hr |
| 83 | 27 | MA1631.1 | ASCL1 | MA1631.1 | NNGCACCTGCYNB | ASCL1 | 4.05e-09 | 3.87e-11 | 1340 | 42.7 | 50200 | 36.7 | 1.16 | DOX_DAR_3hr |
| 84 | 29 | MA0130.1 | ZNF354C | MA0130.1 | MTCCAC | ZNF354C | 1.77e-08 | 1.67e-10 | 2340 | 74.9 | 95100 | 69.5 | 1.08 | DOX_DAR_3hr |
| 85 | 30 | 9-GGCWGCC | STREME-9 | 9-GGCWGCC | GGCWGCC | GGCWGCC | 2.54e-08 | 2.37e-10 | 648 | 20.7 | 22200 | 16.2 | 1.28 | DOX_DAR_3hr |
| 86 | 31 | MA0748.2 | YY2 | MA0748.2 | NVATGGCGGCS | YY2 | 2.61e-08 | 2.4e-10 | 2820 | 90.2 | 118000 | 86.2 | 1.05 | DOX_DAR_3hr |
| 87 | 24 | MA0739.1 | Hic1 | MA0739.1 | RTGCCAACY | Hic1 | 2.84e-08 | 2.58e-10 | 1940 | 62.2 | 77100 | 56.4 | 1.1 | DOX_DAR_3hr |
| 88 | 11 | MA0865.2 | E2F8 | MA0865.2 | TTCCCGCCAHWA | E2F8 | 8.4e-08 | 7.56e-10 | 2020 | 64.5 | 80600 | 58.9 | 1.1 | DOX_DAR_3hr |
| 89 | 24 | MA1527.1 | NFIC | MA1527.1 | NTTGGCDNNRTGCCARN | NFIC | 1.13e-07 | 1.01e-09 | 264 | 8.45 | 7680 | 5.61 | 1.51 | DOX_DAR_3hr |
| 90 | 32 | MA1728.1 | ZNF549 | MA1728.1 | NNTGCTGCCCWR | ZNF549 | 1.19e-07 | 1.05e-09 | 2950 | 94.2 | 125000 | 91.2 | 1.03 | DOX_DAR_3hr |
| 91 | 33 | MA1982.1 | ZNF574 | MA1982.1 | VGSCTAGAGMGGCCS | ZNF574 | 2.05e-07 | 1.79e-09 | 1980 | 63.2 | 78900 | 57.7 | 1.1 | DOX_DAR_3hr |
| 92 | 24 | MA0119.1 | NFIC::TLX1 | MA0119.1 | TGGCASSRWGCCAA | NFIC::TLX1 | 1.53e-06 | 1.32e-08 | 295 | 9.44 | 9050 | 6.61 | 1.43 | DOX_DAR_3hr |
| 93 | 8 | MA0163.1 | PLAG1 | MA0163.1 | GGGGCCCWAGGGGG | PLAG1 | 2.79e-06 | 2.38e-08 | 231 | 7.39 | 6760 | 4.94 | 1.5 | DOX_DAR_3hr |
| 94 | 34 | MA1656.1 | ZNF449 | MA1656.1 | NNAAGCCCAACCNN | ZNF449 | 3.04e-06 | 2.56e-08 | 679 | 21.7 | 24100 | 17.6 | 1.24 | DOX_DAR_3hr |
| 95 | 24 | MA0738.1 | HIC2 | MA0738.1 | RTGCCCRSB | HIC2 | 8.79e-06 | 7.33e-08 | 2150 | 68.9 | 87700 | 64.1 | 1.08 | DOX_DAR_3hr |
| 96 | 26 | MA1629.1 | Zic2 | MA1629.1 | NDCACAGCAGGDRG | Zic2 | 2.6e-05 | 2.15e-07 | 2360 | 75.6 | 97400 | 71.2 | 1.06 | DOX_DAR_3hr |
| 97 | 35 | MA1121.1 (TEAD2) | MEME-5 | ACATTCCW | ACATTCCW | TEAD2 | 7.54e-05 | 6.16e-07 | 491 | 15.7 | 17100 | 12.5 | 1.26 | DOX_DAR_3hr |
| 98 | 27 | MA1652.1 | ZKSCAN5 | MA1652.1 | NNRGGARGTGAGRR | ZKSCAN5 | 8.89e-05 | 7.18e-07 | 652 | 20.9 | 23500 | 17.2 | 1.21 | DOX_DAR_3hr |
| 99 | 27 | MA0522.3 | TCF3 | MA0522.3 | NVCACCTGCNN | TCF3 | 9.96e-05 | 7.97e-07 | 1160 | 37 | 44600 | 32.6 | 1.14 | DOX_DAR_3hr |
| 100 | 3 | MA0472.2 | EGR2 | MA0472.2 | MCGCCCACGCA | EGR2 | 0.000118 | 9.34e-07 | 1060 | 33.9 | 40600 | 29.6 | 1.15 | DOX_DAR_3hr |
| 101 | 36 | MA0767.1 | GCM2 | MA0767.1 | BATGCGGGTR | GCM2 | 0.000123 | 9.64e-07 | 2880 | 92.1 | 122000 | 89.3 | 1.03 | DOX_DAR_3hr |
| 102 | 12 | MA1727.1 | ZNF417 | MA1727.1 | VRBVNTGGGCGCCAM | ZNF417 | 2e-04 | 1.55e-06 | 1360 | 43.6 | 53500 | 39.1 | 1.12 | DOX_DAR_3hr |
| 103 | 27 | MA0821.2 | HES5 | MA0821.2 | GRCACGTGYC | HES5 | 0.000288 | 2.22e-06 | 1860 | 59.7 | 75500 | 55.2 | 1.08 | DOX_DAR_3hr |
| 104 | 11 | MA0597.2 | THAP1 | MA0597.2 | VSGCAGGGCASV | THAP1 | 0.000418 | 3.18e-06 | 655 | 21 | 23900 | 17.5 | 1.2 | DOX_DAR_3hr |
| 105 | 37 | MA1979.1 | ZNF416 | MA1979.1 | TATCTGGGCAH | ZNF416 | 0.000491 | 3.7e-06 | 876 | 28 | 33100 | 24.2 | 1.16 | DOX_DAR_3hr |
| 106 | 38 | MA1973.1 | ZKSCAN3 | MA1973.1 | KDKCCCAGGCTAGCCCABNN | ZKSCAN3 | 0.000517 | 3.83e-06 | 1530 | 48.9 | 61000 | 44.6 | 1.1 | DOX_DAR_3hr |
| 107 | 36 | MA0646.1 | GCM1 | MA0646.1 | BATGCGGGTAC | GCM1 | 0.000517 | 3.83e-06 | 2400 | 76.9 | 1e+05 | 73 | 1.05 | DOX_DAR_3hr |
| 108 | 39 | MA1628.1 | Zic1::Zic2 | MA1628.1 | CVCAGCAGGNV | Zic1::Zic2 | 0.000537 | 3.94e-06 | 2550 | 81.5 | 107000 | 77.9 | 1.05 | DOX_DAR_3hr |
| 109 | 40 | MA1637.1 | EBF3 | MA1637.1 | NYCCCAAGGGANN | EBF3 | 0.000676 | 4.91e-06 | 1950 | 62.2 | 79300 | 58 | 1.07 | DOX_DAR_3hr |
| 110 | 41 | MA0159.1 | RARA::RXRA | MA0159.1 | RGKTCANVGRSAGGTCA | RARA::RXRA | 0.001 | 7.22e-06 | 1490 | 47.8 | 59600 | 43.5 | 1.1 | DOX_DAR_3hr |
| 111 | 27 | MA1997.1 | Olig2 | MA1997.1 | NVCAGCTGBN | Olig2 | 0.00299 | 2.13e-05 | 1450 | 46.4 | 58000 | 42.3 | 1.1 | DOX_DAR_3hr |
| 112 | 42 | MA1715.1 | ZNF707 | MA1715.1 | CCCCACTCCTGGTAC | ZNF707 | 0.00355 | 2.51e-05 | 1150 | 36.7 | 44900 | 32.8 | 1.12 | DOX_DAR_3hr |
| 113 | 24 | MA0671.1 | NFIX | MA0671.1 | NNTGCCAAN | NFIX | 0.00388 | 2.72e-05 | 2580 | 82.4 | 108000 | 79.2 | 1.04 | DOX_DAR_3hr |
| 114 | 43 | MA1985.1 | ZNF669 | MA1985.1 | GGGGYGAYGACCRCT | ZNF669 | 0.00518 | 3.6e-05 | 1240 | 39.8 | 49200 | 35.9 | 1.11 | DOX_DAR_3hr |
| 115 | 27 | MA1993.1 | Neurod2 | MA1993.1 | VVCAGCTGBB | Neurod2 | 0.00715 | 4.92e-05 | 1430 | 45.7 | 57300 | 41.8 | 1.09 | DOX_DAR_3hr |
| 116 | 44 | MA1972.1 | ZFP14 | MA1972.1 | GGAGGCHCHGGARBG | ZFP14 | 0.00725 | 4.95e-05 | 948 | 30.3 | 36700 | 26.8 | 1.13 | DOX_DAR_3hr |
| 117 | 45 | MA1964.1 | SMAD2 | MA1964.1 | NBCCAGACDN | SMAD2 | 0.0107 | 7.26e-05 | 614 | 19.6 | 22900 | 16.7 | 1.18 | DOX_DAR_3hr |
| 118 | 40 | MA0154.4 | EBF1 | MA0154.4 | NNTCCCCAGGGGANN | EBF1 | 0.0122 | 8.17e-05 | 2710 | 86.6 | 115000 | 83.9 | 1.03 | DOX_DAR_3hr |
| 119 | 21 | MA0056.2 | MZF1 | MA0056.2 | NNAATCCCCANNN | MZF1 | 0.0136 | 9.03e-05 | 3030 | 96.8 | 130000 | 95.3 | 1.02 | DOX_DAR_3hr |
| 120 | 27 | MA1966.1 | TFAP4::ETV1 | MA1966.1 | RSCGGAAGCAGSTGKN | TFAP4::ETV1 | 0.0169 | 0.000112 | 1890 | 60.5 | 77800 | 56.8 | 1.07 | DOX_DAR_3hr |
| 121 | 27 | MA0633.2 | Twist2 | MA0633.2 | NVCAGCTGBN | Twist2 | 0.0177 | 0.000116 | 1520 | 48.5 | 61300 | 44.8 | 1.08 | DOX_DAR_3hr |
| 122 | 11 | MA1567.2 | Tbx6 | MA1567.2 | NRGGTGTGAANN | Tbx6 | 0.0237 | 0.000154 | 547 | 17.5 | 20300 | 14.8 | 1.18 | DOX_DAR_3hr |
| 123 | 40 | MA1604.1 | Ebf2 | MA1604.1 | NYCCCAAGGGANN | Ebf2 | 0.0252 | 0.000162 | 1980 | 63.4 | 82000 | 59.9 | 1.06 | DOX_DAR_3hr |
| 124 | 27 | MA0649.1 | HEY2 | MA0649.1 | GRCACGTGYC | HEY2 | 0.0306 | 0.000196 | 2100 | 67.2 | 87200 | 63.7 | 1.05 | DOX_DAR_3hr |
| 125 | 11 | MA1583.1 | ZFP57 | MA1583.1 | NNVTTGCCGCANN | ZFP57 | 0.0365 | 0.000232 | 2110 | 67.6 | 87900 | 64.2 | 1.05 | DOX_DAR_3hr |
| 126 | 27 | MA1635.1 | BHLHE22 | MA1635.1 | NVCAGCTGBN | BHLHE22 | 0.0443 | 0.000279 | 1490 | 47.7 | 60500 | 44.2 | 1.08 | DOX_DAR_3hr |
| 127 | 46 | MA0764.3 | ETV4 | MA0764.3 | ACCGGAAGTR | ETV4 | 0.0451 | 0.000281 | 2440 | 78.1 | 103000 | 75 | 1.04 | DOX_DAR_3hr |
| 12 | 47 | MA1979.1 (ZNF416) | STREME-7 | 7-AGAGCCCAG | AGAGCCCAG | ZNF416 | 0.103 | 0.00858 | 2820 | 90.2 | 122000 | 88.8 | 1.02 | DOX_DAR_3hr |
| 13 | 48 | MA0528.2 (ZNF263) | STREME-10 | 10-ATCCTCTCCW | 10-ATCCTCTCCW | ZNF263 | 6.64 | 0.511 | 625 | 20 | 27200 | 19.9 | 1.01 | DOX_DAR_3hr |
| 14 | 49 | MA1107.2 (KLF9) | MEME-4 | ACACACACACACACA | ACACACACACACACA | KLF9 | 10.2 | 0.729 | 7 | 0.22 | 348 | 0.25 | 1 | DOX_DAR_3hr |
| 15 | 50 | MA1125.1 (ZNF384) | MEME-2 | AAAAAAAAAAAAAAA | AAAAAAAAAAAAAAA | ZNF384 | 14.5 | 0.967 | 3 | 0.1 | 301 | 0.22 | 0.58 | DOX_DAR_3hr |
ER_rat <- 1.25
# mrc_type <- "EAR_open"
DOX_DAR_3hr_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
# geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_col(aes(x=log10Evalue))+
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste("DOX_DAR_3hr_combo response peaks(200bp) Enrichment ratio:",ER_rat))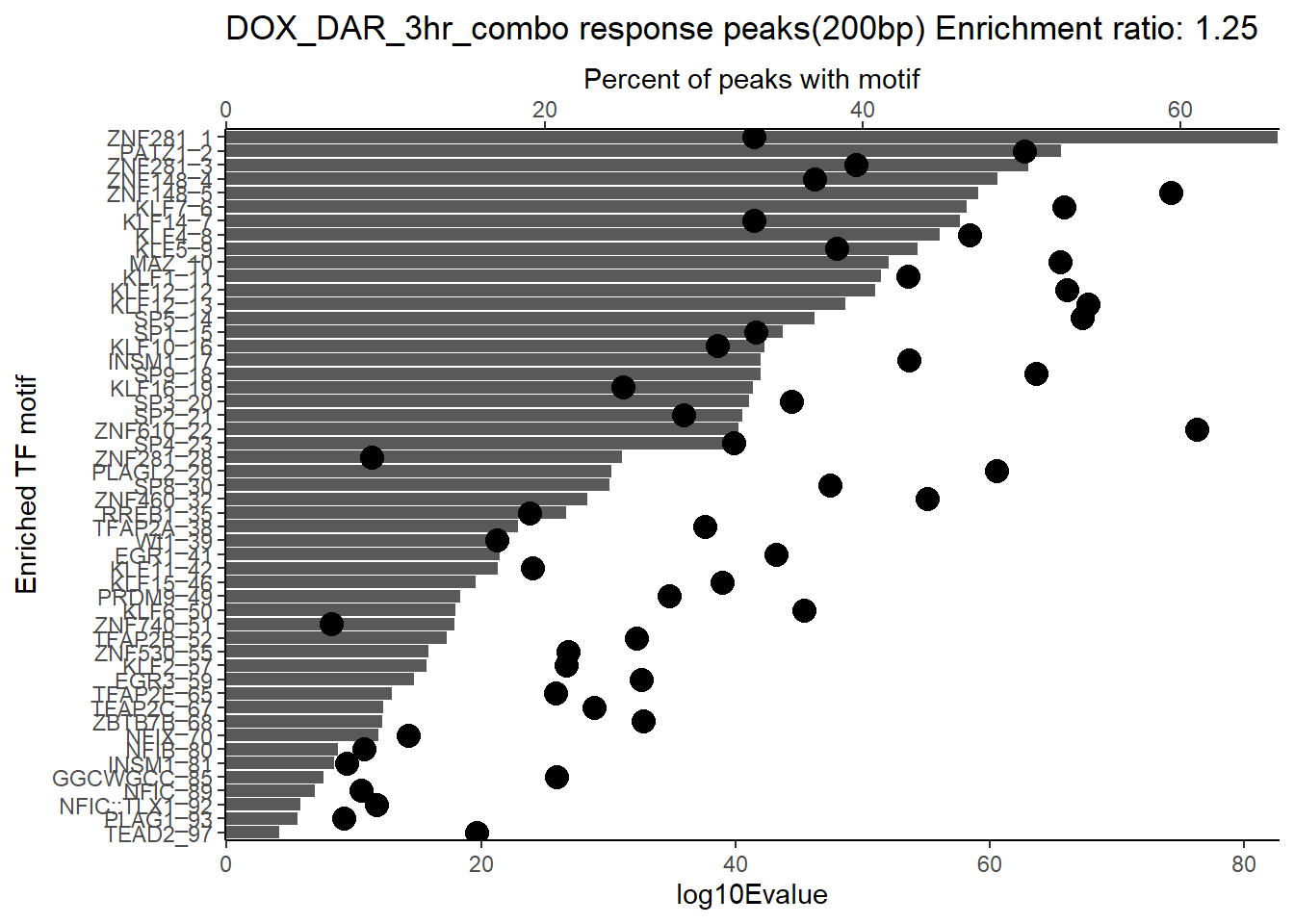
DOX_DAR_3hr_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
slice_head(n=5) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
# geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*.5), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./.5,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste("DOX DAR 3 hour response peaks(200bp) Enrichment ratio:",ER_rat))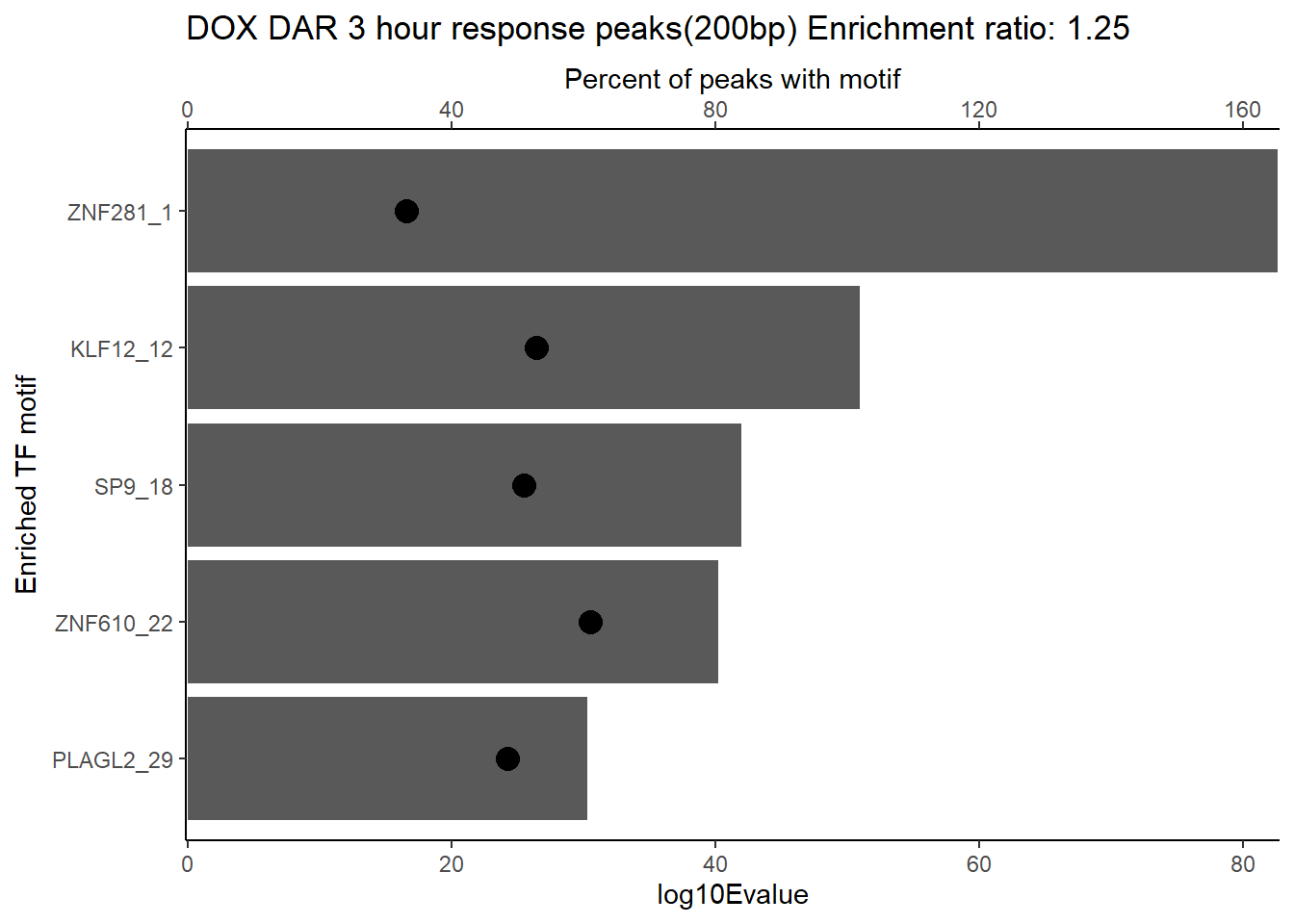
DOX_DAR_24hr data
DOX_DAR_24hr_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DOX_24_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
DOX_DAR_24hr_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DOX_24_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
DOX_DAR_24hr_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DOX_24_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
DOX_DAR_24hr_sea_all <- rbind(DOX_DAR_24hr_sea_disc, DOX_DAR_24hr_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)DOX_DAR_24hr_combo <- DOX_DAR_24hr_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., DOX_DAR_24hr_sea_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ SIM_MOTIF
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y) %>%
mutate(mrc_type="DOX_DAR_24hr")
DOX_DAR_24hr_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs 200 bp DOX_DAR_24hr") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO | mrc_type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | MA1134.1 (FOS::JUNB) | STREME-1 | 1-RATGASTCATY | RATGASTCATY | FOS::JUNB | 0 | 0 | 9050 | 15.5 | 5740 | 7.03 | 2.2 | DOX_DAR_24hr |
| 2 | 1 | MA1134.1 | FOS::JUNB | MA1134.1 | KATGASTCATHN | FOS::JUNB | 0 | 0 | 9320 | 16 | 6130 | 7.51 | 2.13 | DOX_DAR_24hr |
| 3 | 1 | MA1138.1 | FOSL2::JUNB | MA1138.1 | KRTGASTCAT | FOSL2::JUNB | 0 | 0 | 9050 | 15.5 | 5910 | 7.24 | 2.14 | DOX_DAR_24hr |
| 4 | 1 | MA0477.2 | FOSL1 | MA0477.2 | NNATGACTCATNN | FOSL1 | 0 | 0 | 7540 | 12.9 | 4460 | 5.46 | 2.37 | DOX_DAR_24hr |
| 5 | 1 | MA0478.1 (FOSL2) | MEME-2 | MTGASTCAYCH | MTGASTCAYCH | FOSL2 | 0 | 0 | 10100 | 17.4 | 7040 | 8.62 | 2.01 | DOX_DAR_24hr |
| 6 | 1 | MA1144.1 | FOSL2::JUND | MA1144.1 | KATGACTCAT | FOSL2::JUND | 0 | 0 | 8380 | 14.4 | 5290 | 6.47 | 2.22 | DOX_DAR_24hr |
| 7 | 1 | MA0491.2 | JUND | MA0491.2 | NNATGACTCATNN | JUND | 0 | 0 | 9440 | 16.2 | 6370 | 7.8 | 2.08 | DOX_DAR_24hr |
| 8 | 1 | MA1130.1 | FOSL2::JUN | MA1130.1 | NNRTGAGTCAYN | FOSL2::JUN | 0 | 0 | 7650 | 13.1 | 4600 | 5.64 | 2.33 | DOX_DAR_24hr |
| 9 | 1 | MA0490.2 | JUNB | MA0490.2 | NNATGACTCATNN | JUNB | 0 | 0 | 7390 | 12.7 | 4360 | 5.34 | 2.37 | DOX_DAR_24hr |
| 10 | 1 | MA1135.1 | FOSB::JUNB | MA1135.1 | KRTGASTCAT | FOSB::JUNB | 0 | 0 | 8620 | 14.8 | 5570 | 6.82 | 2.17 | DOX_DAR_24hr |
| 11 | 1 | MA0476.1 | FOS | MA0476.1 | DVTGASTCATB | FOS | 0 | 0 | 8860 | 15.2 | 5850 | 7.16 | 2.12 | DOX_DAR_24hr |
| 12 | 1 | MA1633.2 | BACH1 | MA1633.2 | ATGACTCAT | BACH1 | 0 | 0 | 10400 | 17.9 | 7480 | 9.16 | 1.95 | DOX_DAR_24hr |
| 13 | 1 | MA1141.1 | FOS::JUND | MA1141.1 | NKATGAGTCATNN | FOS::JUND | 0 | 0 | 7140 | 12.2 | 4190 | 5.14 | 2.38 | DOX_DAR_24hr |
| 14 | 1 | MA0099.3 | FOS::JUN | MA0099.3 | ATGAGTCAYM | FOS::JUN | 0 | 0 | 7020 | 12 | 4100 | 5.03 | 2.39 | DOX_DAR_24hr |
| 15 | 1 | MA1128.1 | FOSL1::JUN | MA1128.1 | NKATGACTCATNN | FOSL1::JUN | 0 | 0 | 6880 | 11.8 | 4040 | 4.95 | 2.38 | DOX_DAR_24hr |
| 16 | 1 | MA0489.2 | Jun | MA0489.2 | NVTGACTCATNN | Jun | 0 | 0 | 9310 | 16 | 6480 | 7.93 | 2.01 | DOX_DAR_24hr |
| 17 | 1 | MA0462.2 | BATF::JUN | MA0462.2 | DATGACTCATH | BATF::JUN | 0 | 0 | 6230 | 10.7 | 3510 | 4.3 | 2.48 | DOX_DAR_24hr |
| 18 | 1 | MA1137.1 | FOSL1::JUNB | MA1137.1 | NDRTGACTCATNN | FOSL1::JUNB | 0 | 0 | 10600 | 18.1 | 7880 | 9.65 | 1.88 | DOX_DAR_24hr |
| 19 | 1 | MA1634.1 | BATF | MA1634.1 | DATGACTCATH | BATF | 0 | 0 | 6350 | 10.9 | 3630 | 4.44 | 2.45 | DOX_DAR_24hr |
| 20 | 1 | MA0655.1 | JDP2 | MA0655.1 | ATGACTCAT | JDP2 | 0 | 0 | 6090 | 10.4 | 3410 | 4.18 | 2.5 | DOX_DAR_24hr |
| 21 | 1 | MA0835.2 | BATF3 | MA0835.2 | DATGACTCATH | BATF3 | 0 | 0 | 6670 | 11.4 | 3970 | 4.86 | 2.35 | DOX_DAR_24hr |
| 22 | 1 | MA1988.1 | Atf3 | MA1988.1 | NRTGACTCABN | Atf3 | 0 | 0 | 5940 | 10.2 | 3340 | 4.09 | 2.49 | DOX_DAR_24hr |
| 23 | 1 | MA0478.1 | FOSL2 | MA0478.1 | KRRTGASTCAB | FOSL2 | 0 | 0 | 7010 | 12 | 4400 | 5.38 | 2.23 | DOX_DAR_24hr |
| 24 | 1 | MA1132.1 | JUN::JUNB | MA1132.1 | KATGACKCAT | JUN::JUNB | 0 | 0 | 10500 | 18 | 8060 | 9.87 | 1.82 | DOX_DAR_24hr |
| 25 | 1 | MA1142.1 | FOSL1::JUND | MA1142.1 | NRTGACTCAT | FOSL1::JUND | 0 | 0 | 6630 | 11.4 | 4090 | 5.01 | 2.27 | DOX_DAR_24hr |
| 26 | 1 | MA1101.2 | BACH2 | MA1101.2 | DWANCATGASTCATSNTWH | BACH2 | 0 | 0 | 10000 | 17.2 | 7570 | 9.27 | 1.85 | DOX_DAR_24hr |
| 27 | 1 | MA0841.1 | NFE2 | MA0841.1 | VATGACTCATS | NFE2 | 0 | 0 | 5480 | 9.4 | 3060 | 3.74 | 2.51 | DOX_DAR_24hr |
| 28 | 1 | MA1928.1 | BNC2 | MA1928.1 | NDRTGAGTCABN | BNC2 | 0 | 0 | 10800 | 18.5 | 8560 | 10.5 | 1.77 | DOX_DAR_24hr |
| 29 | 1 | MA0476.1 (FOS) | STREME-9 | 9-TCTGASTCA | TCTGASTCA | FOS | 9.64e-253 | 1.63e-254 | 8480 | 14.5 | 7080 | 8.67 | 1.68 | DOX_DAR_24hr |
| 30 | 1 | MA0089.2 | MAFG::NFE2L1 | MA0089.2 | NVNATGACTCAGCADW | MAFG::NFE2L1 | 1.12e-181 | 1.82e-183 | 20400 | 34.9 | 22600 | 27.7 | 1.26 | DOX_DAR_24hr |
| 31 | 1 | MA0496.3 | MAFK | MA0496.3 | NGMTGACTCAGCMNH | MAFK | 7.51e-178 | 1.19e-179 | 8390 | 14.4 | 7670 | 9.39 | 1.53 | DOX_DAR_24hr |
| 32 | 1 | MA0501.1 | MAF::NFE2 | MA0501.1 | ATGACTCAGCANWTY | MAF::NFE2 | 2.72e-145 | 4.15e-147 | 23200 | 39.8 | 27000 | 33.1 | 1.2 | DOX_DAR_24hr |
| 33 | 1 | MA0150.2 | Nfe2l2 | MA0150.2 | CASNATGACTCAGCA | Nfe2l2 | 5.56e-114 | 8.24e-116 | 15900 | 27.2 | 17800 | 21.8 | 1.24 | DOX_DAR_24hr |
| 34 | 2 | MA1970.1 (TRPS1) | STREME-2 | 2-CTTATC | CTTATC | TRPS1 | 2.11e-96 | 3.03e-98 | 41900 | 71.7 | 54300 | 66.4 | 1.08 | DOX_DAR_24hr |
| 35 | 3 | MA0726.1 | VSX2 | MA0726.1 | CTAATTAN | VSX2 | 1.81e-85 | 2.53e-87 | 53200 | 91.1 | 71700 | 87.8 | 1.04 | DOX_DAR_24hr |
| 36 | 3 | MA0720.1 | Shox2 | MA0720.1 | YYAATTAR | Shox2 | 3.17e-85 | 4.31e-87 | 52300 | 89.6 | 70300 | 86.1 | 1.04 | DOX_DAR_24hr |
| 37 | 4 | MA0676.1 | Nr2e1 | MA0676.1 | AAAAGTCAA | Nr2e1 | 3.42e-85 | 4.52e-87 | 50200 | 86 | 67000 | 82.1 | 1.05 | DOX_DAR_24hr |
| 38 | 3 | MA0725.1 | VSX1 | MA0725.1 | YTAATTAN | VSX1 | 3.84e-82 | 4.94e-84 | 52300 | 89.7 | 70400 | 86.2 | 1.04 | DOX_DAR_24hr |
| 39 | 3 | MA0718.1 | RAX | MA0718.1 | RYYAATTARH | RAX | 8e-81 | 1e-82 | 50400 | 86.5 | 67500 | 82.7 | 1.05 | DOX_DAR_24hr |
| 40 | 3 | MA0644.2 | ESX1 | MA0644.2 | YYAATTAN | ESX1 | 3e-79 | 3.67e-81 | 47000 | 80.6 | 62300 | 76.3 | 1.06 | DOX_DAR_24hr |
| 41 | 3 | MA0722.1 | VAX1 | MA0722.1 | YTAATTAH | VAX1 | 2.59e-78 | 3.09e-80 | 51100 | 87.7 | 68600 | 84 | 1.04 | DOX_DAR_24hr |
| 42 | 3 | MA0027.2 | EN1 | MA0027.2 | SYAATTAV | EN1 | 2.45e-76 | 2.85e-78 | 51200 | 87.7 | 68700 | 84.2 | 1.04 | DOX_DAR_24hr |
| 43 | 3 | MA0674.1 | NKX6-1 | MA0674.1 | NTMATTAA | NKX6-1 | 4.36e-76 | 4.95e-78 | 51200 | 87.8 | 68800 | 84.3 | 1.04 | DOX_DAR_24hr |
| 44 | 3 | MA0630.1 | SHOX | MA0630.1 | HTAATTRR | SHOX | 2.51e-74 | 2.79e-76 | 54700 | 93.8 | 74400 | 91.1 | 1.03 | DOX_DAR_24hr |
| 45 | 1 | MA0659.3 | Mafg | MA0659.3 | NWGMTGACTCAGCAN | Mafg | 7.14e-74 | 7.76e-76 | 2870 | 4.92 | 2440 | 2.99 | 1.65 | DOX_DAR_24hr |
| 46 | 3 | MA0723.2 | VAX2 | MA0723.2 | BYAATTAV | VAX2 | 1.97e-73 | 2.09e-75 | 53400 | 91.5 | 72300 | 88.5 | 1.03 | DOX_DAR_24hr |
| 47 | 3 | MA0704.1 | Lhx4 | MA0704.1 | YTAATTAR | Lhx4 | 1.09e-72 | 1.14e-74 | 54100 | 92.8 | 73500 | 90 | 1.03 | DOX_DAR_24hr |
| 48 | 3 | MA0710.1 | NOTO | MA0710.1 | NYTAATTAVN | NOTO | 2.92e-72 | 2.98e-74 | 48100 | 82.5 | 64200 | 78.6 | 1.05 | DOX_DAR_24hr |
| 49 | 3 | MA0879.2 | DLX1 | MA0879.2 | VYAATTAN | DLX1 | 8.12e-70 | 8.11e-72 | 54400 | 93.3 | 74000 | 90.6 | 1.03 | DOX_DAR_24hr |
| 50 | 5 | MA1480.1 | DPRX | MA1480.1 | AGATAATCCN | DPRX | 2.14e-69 | 2.1e-71 | 50600 | 86.7 | 67900 | 83.2 | 1.04 | DOX_DAR_24hr |
| 51 | 3 | MA0882.1 | DLX6 | MA0882.1 | VYAATTAN | DLX6 | 3.49e-69 | 3.35e-71 | 52600 | 90.2 | 71100 | 87.1 | 1.04 | DOX_DAR_24hr |
| 52 | 3 | MA0721.1 | UNCX | MA0721.1 | YYAATTAV | UNCX | 4.2e-69 | 3.95e-71 | 48900 | 83.9 | 65400 | 80.1 | 1.05 | DOX_DAR_24hr |
| 53 | 6 | 7-TMAGMT | STREME-7 | 7-TMAGMT | TMAGMT | TMAGMT | 1.99e-68 | 1.83e-70 | 52900 | 90.6 | 71500 | 87.6 | 1.03 | DOX_DAR_24hr |
| 54 | 1 | MA1640.1 | MEIS2 | MA1640.1 | NNATGATTTATGDNH | MEIS2 | 7.37e-67 | 6.63e-69 | 48500 | 83.2 | 64900 | 79.4 | 1.05 | DOX_DAR_24hr |
| 55 | 1 | MA1639.1 | MEIS1 | MA1639.1 | NHCATMAATCAHN | MEIS1 | 7.45e-67 | 6.63e-69 | 31500 | 54 | 40200 | 49.2 | 1.1 | DOX_DAR_24hr |
| 56 | 3 | MA0716.1 | PRRX1 | MA0716.1 | YYAATTAR | PRRX1 | 1.34e-66 | 1.17e-68 | 55700 | 95.5 | 76200 | 93.3 | 1.02 | DOX_DAR_24hr |
| 57 | 5 | MA0035.4 | GATA1 | MA0035.4 | NHCTAATCTDH | GATA1 | 2.41e-66 | 2.07e-68 | 54000 | 92.6 | 73400 | 89.9 | 1.03 | DOX_DAR_24hr |
| 58 | 3 | MA0793.1 | POU6F2 | MA0793.1 | ASCTMATTAW | POU6F2 | 5.28e-65 | 4.46e-67 | 46900 | 80.4 | 62500 | 76.5 | 1.05 | DOX_DAR_24hr |
| 59 | 7 | MA0635.1 | BARHL2 | MA0635.1 | VHTAAAYGRT | BARHL2 | 1.53e-64 | 1.27e-66 | 52000 | 89.2 | 70300 | 86 | 1.04 | DOX_DAR_24hr |
| 60 | 3 | MA0125.1 | Nobox | MA0125.1 | TAATTRSY | Nobox | 2.23e-64 | 1.82e-66 | 48200 | 82.7 | 64500 | 79 | 1.05 | DOX_DAR_24hr |
| 61 | 3 | MA1481.1 | DRGX | MA1481.1 | YYAATTAN | DRGX | 5.95e-63 | 4.77e-65 | 47600 | 81.6 | 63600 | 77.9 | 1.05 | DOX_DAR_24hr |
| 62 | 3 | MA0707.2 | MNX1 | MA0707.2 | VYRATTAK | MNX1 | 6.1e-62 | 4.82e-64 | 35200 | 60.3 | 45500 | 55.8 | 1.08 | DOX_DAR_24hr |
| 63 | 3 | MA0634.1 | ALX3 | MA0634.1 | YYYAATTANV | ALX3 | 1.84e-61 | 1.43e-63 | 55500 | 95.1 | 75900 | 92.9 | 1.02 | DOX_DAR_24hr |
| 64 | 3 | MA1476.2 | Dlx5 | MA1476.2 | NDGCAATTAGNN | Dlx5 | 2.71e-61 | 2.07e-63 | 49700 | 85.2 | 66800 | 81.8 | 1.04 | DOX_DAR_24hr |
| 65 | 2 | MA1104.2 | GATA6 | MA1104.2 | HWWTCTTATCTNH | GATA6 | 3.95e-61 | 2.97e-63 | 52100 | 89.3 | 70500 | 86.3 | 1.03 | DOX_DAR_24hr |
| 66 | 8 | MA0602.1 | Arid5a | MA0602.1 | SYAATATTGVDANH | Arid5a | 1.16e-60 | 8.62e-63 | 48200 | 82.7 | 64600 | 79.1 | 1.05 | DOX_DAR_24hr |
| 67 | 3 | MA0904.2 | HOXB5 | MA0904.2 | STAATTAS | HOXB5 | 3.62e-60 | 2.65e-62 | 38200 | 65.4 | 49900 | 61.1 | 1.07 | DOX_DAR_24hr |
| 68 | 3 | MA0717.1 | RAX2 | MA0717.1 | YYAATTAV | RAX2 | 1.45e-59 | 1.05e-61 | 47800 | 82 | 64000 | 78.4 | 1.05 | DOX_DAR_24hr |
| 69 | 2 | MA0036.3 | GATA2 | MA0036.3 | NBCTTATCTNH | GATA2 | 2.73e-59 | 1.93e-61 | 43200 | 74 | 57100 | 70 | 1.06 | DOX_DAR_24hr |
| 70 | 3 | MA0628.1 | POU6F1 | MA0628.1 | AYTAATTART | POU6F1 | 5.19e-59 | 3.63e-61 | 44000 | 75.4 | 58400 | 71.5 | 1.06 | DOX_DAR_24hr |
| 71 | 5 | MA0648.1 | GSC | MA0648.1 | VYTAATCCSY | GSC | 2.66e-58 | 1.84e-60 | 48800 | 83.7 | 65500 | 80.2 | 1.04 | DOX_DAR_24hr |
| 72 | 9 | MA0786.1 | POU3F1 | MA0786.1 | WTATGCWAATKW | POU3F1 | 2.78e-58 | 1.89e-60 | 48900 | 83.8 | 65600 | 80.4 | 1.04 | DOX_DAR_24hr |
| 73 | 3 | MA0880.1 | Dlx3 | MA0880.1 | NYAATTAN | Dlx3 | 4.94e-58 | 3.31e-60 | 44800 | 76.8 | 59600 | 73 | 1.05 | DOX_DAR_24hr |
| 74 | 10 | MA0845.1 | FOXB1 | MA0845.1 | WATGTAAATAT | FOXB1 | 6.4e-58 | 4.23e-60 | 49000 | 83.9 | 65700 | 80.5 | 1.04 | DOX_DAR_24hr |
| 75 | 3 | MA0876.1 | BSX | MA0876.1 | SYVATTAV | BSX | 1.85e-57 | 1.21e-59 | 49000 | 84 | 65900 | 80.6 | 1.04 | DOX_DAR_24hr |
| 76 | 3 | MA0881.1 | Dlx4 | MA0881.1 | VYAATTAN | Dlx4 | 1.91e-57 | 1.23e-59 | 45100 | 77.3 | 60000 | 73.5 | 1.05 | DOX_DAR_24hr |
| 77 | 3 | MA1495.1 | HOXA1 | MA1495.1 | STAATTAS | HOXA1 | 2.6e-57 | 1.65e-59 | 29300 | 50.3 | 37500 | 45.9 | 1.1 | DOX_DAR_24hr |
| 78 | 3 | MA1963.1 | SATB1 | MA1963.1 | WWDCTAATAAHWW | SATB1 | 2.71e-56 | 1.7e-58 | 40700 | 69.7 | 53600 | 65.6 | 1.06 | DOX_DAR_24hr |
| 79 | 3 | MA0885.2 | Dlx2 | MA0885.2 | NDGCAATTAGNN | Dlx2 | 2.96e-56 | 1.83e-58 | 52600 | 90.2 | 71300 | 87.4 | 1.03 | DOX_DAR_24hr |
| 80 | 11 | MA0899.1 | HOXA10 | MA0899.1 | DGYMATAAAAH | HOXA10 | 3.09e-56 | 1.89e-58 | 51000 | 87.5 | 68900 | 84.4 | 1.04 | DOX_DAR_24hr |
| 81 | 9 | MA0792.1 | POU5F1B | MA0792.1 | TATGCWAAT | POU5F1B | 4.93e-56 | 2.97e-58 | 53100 | 90.9 | 72100 | 88.3 | 1.03 | DOX_DAR_24hr |
| 82 | 3 | MA0680.2 | Pax7 | MA0680.2 | DWTAATCAATTAWNH | Pax7 | 1.88e-55 | 1.12e-57 | 41200 | 70.7 | 54400 | 66.6 | 1.06 | DOX_DAR_24hr |
| 83 | 3 | MA1549.1 | POU6F1 | MA1549.1 | RTAATGAGVN | POU6F1 | 2.01e-55 | 1.19e-57 | 48000 | 82.3 | 64400 | 78.9 | 1.04 | DOX_DAR_24hr |
| 84 | 12 | MA0084.1 | SRY | MA0084.1 | NWWAACAAT | SRY | 2.42e-55 | 1.41e-57 | 50700 | 87 | 68500 | 83.9 | 1.04 | DOX_DAR_24hr |
| 85 | 13 | 3-ATTCAA | STREME-3 | 3-ATTCAA | ATTCAA | ATTCAA | 4.18e-55 | 2.41e-57 | 51200 | 87.8 | 69200 | 84.8 | 1.04 | DOX_DAR_24hr |
| 86 | 1 | MA1113.2 | PBX2 | MA1113.2 | NHCATAAATCAHN | PBX2 | 1.74e-54 | 9.91e-57 | 35600 | 61 | 46400 | 56.8 | 1.07 | DOX_DAR_24hr |
| 87 | 3 | MA0713.1 | PHOX2A | MA0713.1 | TAATYYAATTA | PHOX2A | 2.91e-54 | 1.64e-56 | 41900 | 71.9 | 55500 | 67.9 | 1.06 | DOX_DAR_24hr |
| 88 | 3 | MA0910.2 | HOXD8 | MA0910.2 | GYMATTAM | HOXD8 | 3.5e-54 | 1.95e-56 | 34400 | 59 | 44700 | 54.8 | 1.08 | DOX_DAR_24hr |
| 89 | 3 | MA1502.1 | HOXB8 | MA1502.1 | GYMATTAM | HOXB8 | 3.83e-54 | 2.11e-56 | 34500 | 59.1 | 44700 | 54.8 | 1.08 | DOX_DAR_24hr |
| 90 | 3 | MA0661.1 | MEOX1 | MA0661.1 | VSTAATTAHC | MEOX1 | 4.87e-54 | 2.65e-56 | 45900 | 78.7 | 61300 | 75 | 1.05 | DOX_DAR_24hr |
| 91 | 14 | MA0790.1 | POU4F1 | MA0790.1 | ATGMATAATTAATG | POU4F1 | 6.37e-54 | 3.42e-56 | 43100 | 73.9 | 57200 | 70 | 1.06 | DOX_DAR_24hr |
| 92 | 15 | MA0498.2 (MEIS1) | STREME-5 | 5-TTGWCAA | TTGWCAA | MEIS1 | 8.1e-54 | 4.31e-56 | 43600 | 74.8 | 58000 | 71 | 1.05 | DOX_DAR_24hr |
| 93 | 3 | MA0890.1 | GBX2 | MA0890.1 | RSYAATTARN | GBX2 | 1.41e-53 | 7.44e-56 | 44300 | 76 | 59000 | 72.2 | 1.05 | DOX_DAR_24hr |
| 94 | 3 | MA0706.1 | MEOX2 | MA0706.1 | DSTAATTAWN | MEOX2 | 1.44e-53 | 7.49e-56 | 46600 | 79.9 | 62400 | 76.4 | 1.05 | DOX_DAR_24hr |
| 95 | 3 | MA0158.2 | HOXA5 | MA0158.2 | GYMATTAS | HOXA5 | 1.52e-53 | 7.84e-56 | 26500 | 45.5 | 33700 | 41.2 | 1.1 | DOX_DAR_24hr |
| 96 | 16 | MA0052.4 | MEF2A | MA0052.4 | DDCTAAAAATAGMHH | MEF2A | 2.12e-53 | 1.08e-55 | 45500 | 77.9 | 60600 | 74.3 | 1.05 | DOX_DAR_24hr |
| 97 | 11 | MA0465.2 | CDX2 | MA0465.2 | NDGCAATAAAWN | CDX2 | 3.04e-52 | 1.53e-54 | 48600 | 83.3 | 65300 | 80 | 1.04 | DOX_DAR_24hr |
| 98 | 3 | MA0068.2 | PAX4 | MA0068.2 | CTAATTAG | PAX4 | 4.9e-52 | 2.45e-54 | 48500 | 83.1 | 65200 | 79.8 | 1.04 | DOX_DAR_24hr |
| 99 | 17 | MA0495.3 | MAFF | MA0495.3 | DWGTCAGCATTTTWHN | MAFF | 8.39e-51 | 4.15e-53 | 49600 | 85.1 | 66900 | 82 | 1.04 | DOX_DAR_24hr |
| 100 | 18 | MA1589.1 (ZNF140) | STREME-8 | 8-ATTGCT | ATTGCT | ZNF140 | 1.33e-50 | 6.52e-53 | 52000 | 89.1 | 70600 | 86.4 | 1.03 | DOX_DAR_24hr |
| 101 | 3 | MA0658.1 | LHX6 | MA0658.1 | RCTAATTARY | LHX6 | 1.89e-50 | 9.16e-53 | 38300 | 65.6 | 50300 | 61.6 | 1.06 | DOX_DAR_24hr |
| 102 | 3 | MA0666.2 | MSX1 | MA0666.2 | SCAATTAR | MSX1 | 2.24e-50 | 1.07e-52 | 42500 | 72.9 | 56400 | 69.1 | 1.05 | DOX_DAR_24hr |
| 103 | 3 | MA0903.1 | HOXB3 | MA0903.1 | NNYMATTARN | HOXB3 | 2.26e-50 | 1.07e-52 | 44000 | 75.4 | 58600 | 71.8 | 1.05 | DOX_DAR_24hr |
| 104 | 3 | MA1505.1 | HOXC8 | MA1505.1 | KTAATTAM | HOXC8 | 2.69e-50 | 1.27e-52 | 45000 | 77.2 | 60100 | 73.6 | 1.05 | DOX_DAR_24hr |
| 105 | 11 | MA0901.2 | HOXB13 | MA0901.2 | WDCCAATAAAAWWW | HOXB13 | 7.59e-50 | 3.54e-52 | 44400 | 76.1 | 59200 | 72.5 | 1.05 | DOX_DAR_24hr |
| 106 | 19 | MA1471.1 | BARX2 | MA1471.1 | NWWAAYMATTAN | BARX2 | 9.88e-50 | 4.56e-52 | 47200 | 80.9 | 63400 | 77.6 | 1.04 | DOX_DAR_24hr |
| 107 | 5 | MA0714.1 | PITX3 | MA0714.1 | NYTAATCCC | PITX3 | 1.39e-49 | 6.35e-52 | 56600 | 96.9 | 77900 | 95.4 | 1.02 | DOX_DAR_24hr |
| 108 | 3 | MA0675.1 | NKX6-2 | MA0675.1 | NYMATTAA | NKX6-2 | 3.69e-49 | 1.67e-51 | 40400 | 69.3 | 53400 | 65.4 | 1.06 | DOX_DAR_24hr |
| 109 | 2 | MA0766.2 | GATA5 | MA0766.2 | SWGATAASRN | GATA5 | 8.18e-49 | 3.67e-51 | 25600 | 43.9 | 32600 | 39.9 | 1.1 | DOX_DAR_24hr |
| 110 | 3 | MA1519.1 | LHX5 | MA1519.1 | STAATTAV | LHX5 | 1.15e-48 | 5.11e-51 | 26300 | 45.1 | 33500 | 41.1 | 1.1 | DOX_DAR_24hr |
| 111 | 9 | MA0785.1 | POU2F1 | MA0785.1 | AWTATGCWAATK | POU2F1 | 1.65e-48 | 7.25e-51 | 48400 | 83 | 65200 | 79.8 | 1.04 | DOX_DAR_24hr |
| 112 | 10 | MA0032.2 | FOXC1 | MA0032.2 | WAWGTAAAYAW | FOXC1 | 4.02e-48 | 1.76e-50 | 48500 | 83.2 | 65400 | 80 | 1.04 | DOX_DAR_24hr |
| 113 | 5 | MA0712.2 | OTX2 | MA0712.2 | NDRGGATTARNN | OTX2 | 1.45e-47 | 6.26e-50 | 53600 | 91.9 | 73100 | 89.5 | 1.03 | DOX_DAR_24hr |
| 114 | 5 | MA0891.1 | GSC2 | MA0891.1 | NYTAATCCBH | GSC2 | 1.53e-47 | 6.55e-50 | 54100 | 92.8 | 73900 | 90.5 | 1.02 | DOX_DAR_24hr |
| 115 | 3 | MA0654.1 | ISX | MA0654.1 | YYAATTAR | ISX | 1.89e-47 | 8.01e-50 | 37200 | 63.8 | 48900 | 59.9 | 1.07 | DOX_DAR_24hr |
| 116 | 3 | MA0888.1 | EVX2 | MA0888.1 | VVTAATTAVB | EVX2 | 1.9e-47 | 8.01e-50 | 47700 | 81.8 | 64200 | 78.6 | 1.04 | DOX_DAR_24hr |
| 117 | 3 | MA0708.2 | MSX2 | MA0708.2 | SCAATTAV | MSX2 | 2.12e-47 | 8.87e-50 | 41100 | 70.4 | 54400 | 66.6 | 1.06 | DOX_DAR_24hr |
| 118 | 10 | MA0041.2 | FOXD3 | MA0041.2 | WAWGHAAATAAACARY | FOXD3 | 1.7e-46 | 7.06e-49 | 49400 | 84.6 | 66700 | 81.6 | 1.04 | DOX_DAR_24hr |
| 119 | 3 | MA0788.1 | POU3F3 | MA0788.1 | WWTATGCWAATTW | POU3F3 | 2.17e-46 | 8.91e-49 | 44700 | 76.6 | 59800 | 73.2 | 1.05 | DOX_DAR_24hr |
| 120 | 3 | MA0902.2 | HOXB2 | MA0902.2 | SYAATTAS | HOXB2 | 2.39e-46 | 9.72e-49 | 26800 | 45.9 | 34300 | 42 | 1.09 | DOX_DAR_24hr |
| 121 | 5 | MA0711.1 | OTX1 | MA0711.1 | HTAATCCS | OTX1 | 2.66e-46 | 1.08e-48 | 49200 | 84.3 | 66400 | 81.3 | 1.04 | DOX_DAR_24hr |
| 122 | 11 | MA0650.3 | Hoxa13 | MA0650.3 | NNCCAATAAAAN | Hoxa13 | 2.93e-46 | 1.18e-48 | 48300 | 82.7 | 65000 | 79.6 | 1.04 | DOX_DAR_24hr |
| 123 | 9 | MA0142.1 | Pou5f1::Sox2 | MA0142.1 | CWTTGTYATGCAAAT | Pou5f1::Sox2 | 3.21e-46 | 1.28e-48 | 46800 | 80.3 | 62900 | 77 | 1.04 | DOX_DAR_24hr |
| 124 | 3 | MA0912.2 | HOXD3 | MA0912.2 | CTAATTAC | HOXD3 | 1.68e-45 | 6.62e-48 | 23600 | 40.4 | 29800 | 36.5 | 1.11 | DOX_DAR_24hr |
| 125 | 14 | MA0791.1 | POU4F3 | MA0791.1 | RTGMATWATTAATGAR | POU4F3 | 3.19e-45 | 1.25e-47 | 38800 | 66.5 | 51200 | 62.8 | 1.06 | DOX_DAR_24hr |
| 126 | 3 | MA0709.1 | Msx3 | MA0709.1 | SCAATTAN | Msx3 | 4.1e-45 | 1.59e-47 | 38700 | 66.4 | 51200 | 62.6 | 1.06 | DOX_DAR_24hr |
| 127 | 10 | MA0031.1 | FOXD1 | MA0031.1 | GTAAACAW | FOXD1 | 2.15e-44 | 8.29e-47 | 41800 | 71.7 | 55700 | 68.2 | 1.05 | DOX_DAR_24hr |
| 128 | 3 | MA0662.1 | MIXL1 | MA0662.1 | NBYAATTAVN | MIXL1 | 3.85e-44 | 1.47e-46 | 42900 | 73.5 | 57200 | 70 | 1.05 | DOX_DAR_24hr |
| 129 | 12 | MA1152.1 | SOX15 | MA1152.1 | CYWTTGTTHW | SOX15 | 6.06e-44 | 2.3e-46 | 51700 | 88.6 | 70300 | 86.1 | 1.03 | DOX_DAR_24hr |
| 130 | 3 | MA0705.1 | Lhx8 | MA0705.1 | CTAATTAV | Lhx8 | 7.01e-44 | 2.64e-46 | 31500 | 54 | 40900 | 50.1 | 1.08 | DOX_DAR_24hr |
| 131 | 3 | MA1498.2 | HOXA7 | MA1498.2 | SYMATTAV | HOXA7 | 1.76e-43 | 6.56e-46 | 54700 | 93.7 | 74900 | 91.7 | 1.02 | DOX_DAR_24hr |
| 132 | 3 | MA0612.2 | EMX1 | MA0612.2 | BTAATTAS | EMX1 | 1.84e-43 | 6.8e-46 | 22800 | 39 | 28800 | 35.3 | 1.11 | DOX_DAR_24hr |
| 133 | 3 | MA0701.2 | LHX9 | MA0701.2 | CYAATTAR | LHX9 | 3.67e-43 | 1.35e-45 | 28700 | 49.1 | 37000 | 45.3 | 1.08 | DOX_DAR_24hr |
| 134 | 7 | MA1608.1 | Isl1 | MA1608.1 | NNCCATTAGNN | Isl1 | 1.83e-42 | 6.69e-45 | 52800 | 90.5 | 71900 | 88.1 | 1.03 | DOX_DAR_24hr |
| 135 | 3 | MA0699.1 | LBX2 | MA0699.1 | NCYAATTARN | LBX2 | 1.94e-42 | 7.05e-45 | 48300 | 82.9 | 65200 | 79.9 | 1.04 | DOX_DAR_24hr |
| 136 | 3 | MA0642.2 | EN2 | MA0642.2 | VSYAATTA | EN2 | 4e-42 | 1.44e-44 | 47300 | 81 | 63600 | 77.9 | 1.04 | DOX_DAR_24hr |
| 137 | 2 | MA0037.4 | Gata3 | MA0037.4 | NHTCTTATCTNH | Gata3 | 4.35e-42 | 1.55e-44 | 54500 | 93.5 | 74700 | 91.5 | 1.02 | DOX_DAR_24hr |
| 138 | 10 | MA0148.4 | FOXA1 | MA0148.4 | WWGTAAACATNN | FOXA1 | 8.65e-42 | 3.07e-44 | 53600 | 91.9 | 73200 | 89.7 | 1.02 | DOX_DAR_24hr |
| 139 | 7 | MA1978.1 | ZNF354A | MA1978.1 | HWAADWATAAATGRAYWAWTT | ZNF354A | 9.29e-42 | 3.27e-44 | 48700 | 83.5 | 65800 | 80.6 | 1.04 | DOX_DAR_24hr |
| 140 | 2 | MA1970.1 | TRPS1 | MA1970.1 | NHTCTTATCTNH | TRPS1 | 9.62e-42 | 3.36e-44 | 34500 | 59.2 | 45300 | 55.4 | 1.07 | DOX_DAR_24hr |
| 141 | 3 | MA0886.1 | EMX2 | MA0886.1 | VBTAATTARB | EMX2 | 2.9e-41 | 1.01e-43 | 28700 | 49.2 | 37100 | 45.4 | 1.08 | DOX_DAR_24hr |
| 142 | 3 | MA0601.1 | Arid3b | MA0601.1 | ATATTAATWAN | Arid3b | 4.24e-41 | 1.46e-43 | 38000 | 65.2 | 50300 | 61.6 | 1.06 | DOX_DAR_24hr |
| 143 | 10 | MA1607.1 | Foxl2 | MA1607.1 | DDWATGTAAACANN | Foxl2 | 1.13e-40 | 3.87e-43 | 52600 | 90.1 | 71700 | 87.8 | 1.03 | DOX_DAR_24hr |
| 144 | 3 | MA0875.1 | BARX1 | MA0875.1 | VCMATTAV | BARX1 | 4.76e-40 | 1.62e-42 | 45000 | 77.2 | 60400 | 74 | 1.04 | DOX_DAR_24hr |
| 145 | 9 | MA1115.1 | POU5F1 | MA1115.1 | WHATGCAAATN | POU5F1 | 4.87e-40 | 1.64e-42 | 52200 | 89.4 | 71100 | 87 | 1.03 | DOX_DAR_24hr |
| 146 | 11 | MA0909.3 | Hoxd13 | MA0909.3 | NNCAATAAAW | Hoxd13 | 6.53e-40 | 2.19e-42 | 37700 | 64.7 | 49900 | 61.1 | 1.06 | DOX_DAR_24hr |
| 147 | 10 | MA0047.3 | FOXA2 | MA0047.3 | RWGTAAACANN | FOXA2 | 7.21e-40 | 2.4e-42 | 53700 | 92 | 73400 | 89.9 | 1.02 | DOX_DAR_24hr |
| 148 | 10 | MA1606.1 | Foxf1 | MA1606.1 | DWRTAAACANN | Foxf1 | 1.23e-39 | 4.08e-42 | 52100 | 89.2 | 70900 | 86.8 | 1.03 | DOX_DAR_24hr |
| 149 | 2 | MA0482.2 | GATA4 | MA0482.2 | NNCCTTATCTNH | GATA4 | 3.07e-39 | 1.01e-41 | 53700 | 92 | 73400 | 89.9 | 1.02 | DOX_DAR_24hr |
| 150 | 3 | MA0892.1 | GSX1 | MA0892.1 | NCYMATTAVN | GSX1 | 8.67e-39 | 2.83e-41 | 46600 | 79.9 | 62800 | 76.9 | 1.04 | DOX_DAR_24hr |
| 151 | 1 | MA0591.1 | Bach1::Mafk | MA0591.1 | RSSATGACTCAGCAB | Bach1::Mafk | 1.1e-38 | 3.58e-41 | 5960 | 10.2 | 6630 | 8.12 | 1.26 | DOX_DAR_24hr |
| 152 | 19 | MA1530.1 | NKX6-3 | MA1530.1 | VYCATTAWW | NKX6-3 | 1.2e-38 | 3.85e-41 | 26700 | 45.7 | 34400 | 42.1 | 1.09 | DOX_DAR_24hr |
| 153 | 20 | MA0679.2 | ONECUT1 | MA0679.2 | WAAAAATCAATAAWWN | ONECUT1 | 1.6e-38 | 5.1e-41 | 36200 | 62.1 | 47800 | 58.6 | 1.06 | DOX_DAR_24hr |
| 154 | 21 | MA0025.2 | NFIL3 | MA0025.2 | NRTTATGYAATNN | NFIL3 | 2.23e-38 | 7.09e-41 | 47300 | 81.1 | 63900 | 78.2 | 1.04 | DOX_DAR_24hr |
| 155 | 12 | MA0143.4 | SOX2 | MA0143.4 | DNACAATGGNN | SOX2 | 3.02e-38 | 9.53e-41 | 50400 | 86.4 | 68400 | 83.8 | 1.03 | DOX_DAR_24hr |
| 156 | 9 | MA0627.2 | POU2F3 | MA0627.2 | NWTATGCAAATNH | POU2F3 | 3.36e-38 | 1.06e-40 | 45200 | 77.4 | 60700 | 74.3 | 1.04 | DOX_DAR_24hr |
| 157 | 12 | MA0868.2 | SOX8 | MA0868.2 | MGAACAATRG | SOX8 | 3.46e-38 | 1.08e-40 | 51300 | 87.9 | 69800 | 85.5 | 1.03 | DOX_DAR_24hr |
| 158 | 3 | MA1497.1 | HOXA6 | MA1497.1 | BGYMATTANN | HOXA6 | 2.7e-37 | 8.36e-40 | 48400 | 82.9 | 65400 | 80.1 | 1.03 | DOX_DAR_24hr |
| 159 | 10 | MA0846.1 | FOXC2 | MA0846.1 | WAWGTAAACAWW | FOXC2 | 3.83e-37 | 1.18e-39 | 47700 | 81.8 | 64500 | 79 | 1.04 | DOX_DAR_24hr |
| 160 | 3 | MA0895.1 | HMBOX1 | MA0895.1 | MYTAGTTAMS | HMBOX1 | 7.43e-37 | 2.27e-39 | 47000 | 80.6 | 63500 | 77.7 | 1.04 | DOX_DAR_24hr |
| 161 | 22 | MA1143.1 | FOSL1::JUND | MA1143.1 | RTGACGTMAY | FOSL1::JUND | 1.69e-36 | 5.14e-39 | 21500 | 36.9 | 27400 | 33.5 | 1.1 | DOX_DAR_24hr |
| 162 | 23 | MA1657.1 | ZNF652 | MA1657.1 | BAAAGRGTTAAN | ZNF652 | 2.38e-36 | 7.17e-39 | 45100 | 77.4 | 60700 | 74.4 | 1.04 | DOX_DAR_24hr |
| 163 | 24 | MA0122.3 | Nkx3-2 | MA0122.3 | WWAAMCACTTAAN | Nkx3-2 | 9.37e-36 | 2.81e-38 | 47100 | 80.7 | 63600 | 77.9 | 1.04 | DOX_DAR_24hr |
| 164 | 17 | MA0117.2 | Mafb | MA0117.2 | AWWNTGCTGACD | Mafb | 1.34e-35 | 4e-38 | 42500 | 72.9 | 56900 | 69.7 | 1.05 | DOX_DAR_24hr |
| 165 | 9 | MA0789.1 | POU3F4 | MA0789.1 | TATGCWAAT | POU3F4 | 1.4e-35 | 4.15e-38 | 41000 | 70.3 | 54800 | 67.1 | 1.05 | DOX_DAR_24hr |
| 166 | 3 | MA0153.2 | HNF1B | MA0153.2 | GTTAATNATTAAY | HNF1B | 1.78e-35 | 5.24e-38 | 26000 | 44.6 | 33600 | 41.1 | 1.08 | DOX_DAR_24hr |
| 167 | 3 | MA1500.1 | HOXB6 | MA1500.1 | BTAATKRC | HOXB6 | 2.06e-35 | 6.04e-38 | 41100 | 70.4 | 54900 | 67.2 | 1.05 | DOX_DAR_24hr |
| 168 | 11 | MA0905.1 | HOXC10 | MA0905.1 | GTCRTAAAAH | HOXC10 | 2.37e-35 | 6.89e-38 | 42900 | 73.5 | 57500 | 70.4 | 1.04 | DOX_DAR_24hr |
| 169 | 25 | MA0769.2 | TCF7 | MA0769.2 | NNCTTTGAWVT | TCF7 | 3.3e-34 | 9.55e-37 | 52200 | 89.4 | 71200 | 87.2 | 1.03 | DOX_DAR_24hr |
| 170 | 22 | MA0018.4 | CREB1 | MA0018.4 | NNRTGATGTCAYN | CREB1 | 3.94e-34 | 1.14e-36 | 52400 | 89.8 | 71600 | 87.7 | 1.02 | DOX_DAR_24hr |
| 171 | 10 | MA1103.2 | FOXK2 | MA1103.2 | DDGTAAACANN | FOXK2 | 4.01e-34 | 1.15e-36 | 55300 | 94.8 | 76100 | 93.2 | 1.02 | DOX_DAR_24hr |
| 172 | 3 | MA1518.2 | Lhx1 | MA1518.2 | NNNTGCTAATTAGCANNN | Lhx1 | 5.09e-34 | 1.45e-36 | 41400 | 71 | 55400 | 67.8 | 1.05 | DOX_DAR_24hr |
| 173 | 3 | MA0787.1 | POU3F2 | MA0787.1 | WTATGCWAATKA | POU3F2 | 5.87e-34 | 1.66e-36 | 40700 | 69.7 | 54300 | 66.5 | 1.05 | DOX_DAR_24hr |
| 174 | 10 | MA1683.1 | FOXA3 | MA1683.1 | DWGTAAACANN | FOXA3 | 5.89e-34 | 1.66e-36 | 49700 | 85.2 | 67500 | 82.6 | 1.03 | DOX_DAR_24hr |
| 175 | 21 | MA0102.4 | CEBPA | MA0102.4 | NDATTGCACAATHN | CEBPA | 6.06e-34 | 1.69e-36 | 48700 | 83.6 | 66100 | 80.9 | 1.03 | DOX_DAR_24hr |
| 176 | 3 | MA0893.2 | GSX2 | MA0893.2 | SYMATTAR | GSX2 | 6.47e-34 | 1.8e-36 | 54200 | 93 | 74400 | 91.1 | 1.02 | DOX_DAR_24hr |
| 177 | 14 | MA0046.2 | HNF1A | MA0046.2 | DRTTAATNATTAACN | HNF1A | 6.67e-34 | 1.84e-36 | 26900 | 46.2 | 34900 | 42.8 | 1.08 | DOX_DAR_24hr |
| 178 | 3 | MA0889.1 | GBX1 | MA0889.1 | VCYAATTASY | GBX1 | 1.28e-33 | 3.52e-36 | 36100 | 61.9 | 47800 | 58.5 | 1.06 | DOX_DAR_24hr |
| 179 | 3 | MA0724.1 | VENTX | MA0724.1 | AMCGATTAR | VENTX | 1.38e-33 | 3.77e-36 | 35000 | 60.1 | 46300 | 56.7 | 1.06 | DOX_DAR_24hr |
| 180 | 10 | MA0481.3 | FOXP1 | MA0481.3 | NDGTAAACANH | FOXP1 | 1.73e-33 | 4.7e-36 | 50600 | 86.8 | 68900 | 84.4 | 1.03 | DOX_DAR_24hr |
| 181 | 5 | MA0682.2 | PITX1 | MA0682.2 | YTAATCCH | PITX1 | 3.02e-33 | 8.15e-36 | 33000 | 56.6 | 43400 | 53.2 | 1.06 | DOX_DAR_24hr |
| 182 | 10 | MA0042.2 | FOXI1 | MA0042.2 | GTAAACA | FOXI1 | 8.25e-32 | 2.22e-34 | 45800 | 78.5 | 61800 | 75.7 | 1.04 | DOX_DAR_24hr |
| 183 | 10 | MA0848.1 | FOXO4 | MA0848.1 | GTAAACA | FOXO4 | 9.62e-32 | 2.57e-34 | 48700 | 83.4 | 66000 | 80.9 | 1.03 | DOX_DAR_24hr |
| 184 | 26 | MA0108.2 | TBP | MA0108.2 | STATAAAWRSVVVBN | TBP | 1.2e-31 | 3.19e-34 | 45600 | 78.2 | 61600 | 75.4 | 1.04 | DOX_DAR_24hr |
| 185 | 10 | MA0033.2 | FOXL1 | MA0033.2 | RTAAACA | FOXL1 | 1.24e-31 | 3.28e-34 | 50500 | 86.5 | 68700 | 84.1 | 1.03 | DOX_DAR_24hr |
| 186 | 3 | MA1501.1 | HOXB7 | MA1501.1 | GGTAATTAVS | HOXB7 | 2.04e-31 | 5.36e-34 | 43600 | 74.7 | 58600 | 71.8 | 1.04 | DOX_DAR_24hr |
| 187 | 2 | MA0140.2 | GATA1::TAL1 | MA0140.2 | MTTATCWSNNNNNHVCAG | GATA1::TAL1 | 2.91e-31 | 7.6e-34 | 20600 | 35.2 | 26200 | 32.1 | 1.1 | DOX_DAR_24hr |
| 188 | 3 | MA0900.2 | HOXA2 | MA0900.2 | STAATTAS | HOXA2 | 3.56e-31 | 9.25e-34 | 19900 | 34.1 | 25400 | 31.1 | 1.1 | DOX_DAR_24hr |
| 189 | 3 | MA1463.1 | ARGFX | MA1463.1 | DCTAATTARM | ARGFX | 2.3e-30 | 5.96e-33 | 27300 | 46.7 | 35500 | 43.5 | 1.07 | DOX_DAR_24hr |
| 190 | 25 | MA0523.1 | TCF7L2 | MA0523.1 | DRASATCAAAGRVA | TCF7L2 | 3.31e-30 | 8.53e-33 | 39100 | 67.1 | 52300 | 64 | 1.05 | DOX_DAR_24hr |
| 191 | 3 | MA0887.1 | EVX1 | MA0887.1 | VVTAATTABS | EVX1 | 6.79e-30 | 1.74e-32 | 55900 | 95.9 | 77200 | 94.5 | 1.01 | DOX_DAR_24hr |
| 192 | 12 | MA1563.2 | SOX18 | MA1563.2 | AACAATDV | SOX18 | 1.02e-29 | 2.61e-32 | 43700 | 74.9 | 58900 | 72.1 | 1.04 | DOX_DAR_24hr |
| 193 | 3 | MA0132.2 | PDX1 | MA0132.2 | VYAATTAR | PDX1 | 1.65e-29 | 4.17e-32 | 30200 | 51.8 | 39700 | 48.6 | 1.07 | DOX_DAR_24hr |
| 194 | 27 | MA0809.2 | TEAD4 | MA0809.2 | NNACATTCCARN | TEAD4 | 7.01e-29 | 1.77e-31 | 46800 | 80.3 | 63400 | 77.7 | 1.03 | DOX_DAR_24hr |
| 195 | 21 | MA1636.1 | CEBPG | MA1636.1 | NDATGATGCAATMHH | CEBPG | 7.3e-29 | 1.83e-31 | 42700 | 73.1 | 57400 | 70.3 | 1.04 | DOX_DAR_24hr |
| 196 | 3 | MA0700.2 | LHX2 | MA0700.2 | DDCCAATTADN | LHX2 | 9.09e-29 | 2.27e-31 | 32400 | 55.5 | 42800 | 52.4 | 1.06 | DOX_DAR_24hr |
| 197 | 11 | MA0898.1 | Hmx3 | MA0898.1 | ASAAGCAATTAAWRRAT | Hmx3 | 1.41e-28 | 3.49e-31 | 37500 | 64.2 | 50000 | 61.2 | 1.05 | DOX_DAR_24hr |
| 198 | 24 | MA0124.2 | Nkx3-1 | MA0124.2 | RCCACTTAA | Nkx3-1 | 1.84e-28 | 4.54e-31 | 46700 | 80.1 | 63300 | 77.5 | 1.03 | DOX_DAR_24hr |
| 199 | 24 | MA0896.1 | Hmx1 | MA0896.1 | VSVAGCAATTAAWGVDB | Hmx1 | 2.72e-28 | 6.68e-31 | 35400 | 60.6 | 47000 | 57.6 | 1.05 | DOX_DAR_24hr |
| 200 | 3 | MA1499.1 | HOXB4 | MA1499.1 | RTMATTAR | HOXB4 | 2.79e-28 | 6.83e-31 | 32400 | 55.5 | 42700 | 52.3 | 1.06 | DOX_DAR_24hr |
| 201 | 28 | MA0498.2 | MEIS1 | MA0498.2 | HTGACAD | MEIS1 | 5.52e-28 | 1.34e-30 | 49700 | 85.3 | 67800 | 83 | 1.03 | DOX_DAR_24hr |
| 202 | 5 | MA0467.2 | Crx | MA0467.2 | NDGGATTANN | Crx | 5.59e-28 | 1.35e-30 | 44500 | 76.2 | 60100 | 73.5 | 1.04 | DOX_DAR_24hr |
| 203 | 12 | MA1120.1 | SOX13 | MA1120.1 | DVACAATGGNN | SOX13 | 9.57e-28 | 2.3e-30 | 50200 | 86 | 68400 | 83.8 | 1.03 | DOX_DAR_24hr |
| 204 | 3 | MA0075.3 | PRRX2 | MA0075.3 | CTAATTAN | PRRX2 | 9.6e-28 | 2.3e-30 | 26300 | 45.1 | 34300 | 42 | 1.07 | DOX_DAR_24hr |
| 205 | 24 | MA0914.1 | ISL2 | MA0914.1 | GCAMTTAR | ISL2 | 1.25e-27 | 2.98e-30 | 53700 | 92 | 73700 | 90.2 | 1.02 | DOX_DAR_24hr |
| 206 | 3 | MA0853.1 | Alx4 | MA0853.1 | CGCRYTAATTARYHNNY | Alx4 | 2.04e-27 | 4.86e-30 | 28000 | 48 | 36700 | 45 | 1.07 | DOX_DAR_24hr |
| 207 | 1 | MA0070.1 | PBX1 | MA0070.1 | HCATCAATCAAW | PBX1 | 3.33e-27 | 7.87e-30 | 25600 | 44 | 33400 | 40.9 | 1.07 | DOX_DAR_24hr |
| 208 | 3 | MA1504.1 | HOXC4 | MA1504.1 | VTMATTAN | HOXC4 | 1.05e-26 | 2.47e-29 | 38700 | 66.3 | 51800 | 63.4 | 1.05 | DOX_DAR_24hr |
| 209 | 12 | MA0077.1 | SOX9 | MA0077.1 | CYATTGTTY | SOX9 | 3.98e-26 | 9.32e-29 | 30400 | 52.1 | 40100 | 49.1 | 1.06 | DOX_DAR_24hr |
| 210 | 12 | MA0514.2 | Sox3 | MA0514.2 | DNACAATGGNN | Sox3 | 6.08e-26 | 1.42e-28 | 50100 | 85.8 | 68300 | 83.7 | 1.03 | DOX_DAR_24hr |
| 211 | 3 | MA0894.1 | HESX1 | MA0894.1 | GHTAATTRRH | HESX1 | 1.19e-25 | 2.75e-28 | 29500 | 50.5 | 38800 | 47.5 | 1.06 | DOX_DAR_24hr |
| 212 | 20 | MA0755.1 | CUX2 | MA0755.1 | TRATCGATAW | CUX2 | 4.54e-25 | 1.05e-27 | 34100 | 58.5 | 45400 | 55.6 | 1.05 | DOX_DAR_24hr |
| 213 | 29 | 4-TAAAGA | STREME-4 | 4-TAAAGA | TAAAGA | TAAAGA | 5.72e-25 | 1.31e-27 | 56700 | 97.3 | 78600 | 96.2 | 1.01 | DOX_DAR_24hr |
| 214 | 30 | MA1489.1 | FOXN3 | MA1489.1 | GTAAACAA | FOXN3 | 1.35e-24 | 3.08e-27 | 43500 | 74.5 | 58800 | 72 | 1.04 | DOX_DAR_24hr |
| 215 | 11 | MA0151.1 | Arid3a | MA0151.1 | ATYAAA | Arid3a | 1.86e-24 | 4.23e-27 | 29300 | 50.2 | 38600 | 47.2 | 1.06 | DOX_DAR_24hr |
| 216 | 10 | MA0157.3 | Foxo3 | MA0157.3 | NDGTAAACARNN | Foxo3 | 1.87e-24 | 4.23e-27 | 54300 | 93 | 74700 | 91.5 | 1.02 | DOX_DAR_24hr |
| 217 | 31 | MA0606.2 | Nfat5 | MA0606.2 | NNATGGAAAAWN | Nfat5 | 1.98e-24 | 4.47e-27 | 55200 | 94.6 | 76100 | 93.2 | 1.02 | DOX_DAR_24hr |
| 218 | 12 | MA1562.1 | SOX14 | MA1562.1 | CCGAACAATG | SOX14 | 2.38e-24 | 5.33e-27 | 42000 | 72 | 56600 | 69.4 | 1.04 | DOX_DAR_24hr |
| 219 | 32 | MA1118.1 | SIX1 | MA1118.1 | GWAACCTGAKM | SIX1 | 4.42e-24 | 9.87e-27 | 54100 | 92.8 | 74500 | 91.2 | 1.02 | DOX_DAR_24hr |
| 220 | 33 | MA1124.1 | ZNF24 | MA1124.1 | CATTCATTCATTC | ZNF24 | 4.9e-24 | 1.09e-26 | 29500 | 50.6 | 39000 | 47.7 | 1.06 | DOX_DAR_24hr |
| 221 | 7 | MA0877.3 | BARHL1 | MA0877.3 | AMCGTTTA | BARHL1 | 2.15e-23 | 4.77e-26 | 49200 | 84.3 | 67100 | 82.2 | 1.03 | DOX_DAR_24hr |
| 222 | 10 | MA0850.1 | FOXP3 | MA0850.1 | RTAAACA | FOXP3 | 2.6e-23 | 5.72e-26 | 51500 | 88.2 | 70500 | 86.3 | 1.02 | DOX_DAR_24hr |
| 223 | 34 | MA1603.1 | Dmrt1 | MA1603.1 | GHTACAAAGTADC | Dmrt1 | 3.03e-23 | 6.66e-26 | 46400 | 79.6 | 63100 | 77.2 | 1.03 | DOX_DAR_24hr |
| 224 | 12 | MA0867.2 | SOX4 | MA0867.2 | RAACAAAGRV | SOX4 | 1.31e-22 | 2.87e-25 | 48500 | 83.1 | 66100 | 80.9 | 1.03 | DOX_DAR_24hr |
| 225 | 3 | MA0618.1 | LBX1 | MA0618.1 | TTAATTAG | LBX1 | 5.91e-22 | 1.28e-24 | 34900 | 59.8 | 46600 | 57.1 | 1.05 | DOX_DAR_24hr |
| 226 | 25 | MA1991.1 | Hnf1A | MA1991.1 | NBCCTTTGATSTBN | Hnf1A | 7.3e-22 | 1.58e-24 | 56800 | 97.3 | 78700 | 96.3 | 1.01 | DOX_DAR_24hr |
| 227 | 3 | MA0681.2 | PHOX2B | MA0681.2 | WWTAATCAAATTAWWW | PHOX2B | 4.4e-21 | 9.49e-24 | 27200 | 46.6 | 35900 | 43.9 | 1.06 | DOX_DAR_24hr |
| 228 | 28 | MA0774.1 | MEIS2 | MA0774.1 | TTGACAGS | MEIS2 | 7.91e-21 | 1.7e-23 | 24700 | 42.4 | 32400 | 39.7 | 1.07 | DOX_DAR_24hr |
| 229 | 16 | MA0497.1 | MEF2C | MA0497.1 | DDDCYAAAAATAGMW | MEF2C | 1.45e-20 | 3.1e-23 | 31500 | 54 | 41900 | 51.4 | 1.05 | DOX_DAR_24hr |
| 230 | 35 | MA0095.3 | Yy1 | MA0095.3 | NNCAAAATGGCN | Yy1 | 2.11e-20 | 4.5e-23 | 55900 | 95.8 | 77300 | 94.7 | 1.01 | DOX_DAR_24hr |
| 231 | 36 | MA0596.1 | SREBF2 | MA0596.1 | RTGGGGTGAY | SREBF2 | 2.96e-20 | 6.27e-23 | 12400 | 21.2 | 15600 | 19.1 | 1.11 | DOX_DAR_24hr |
| 232 | 11 | MA0911.1 | Hoxa11 | MA0911.1 | RGTCGTAAAANT | Hoxa11 | 3.03e-20 | 6.4e-23 | 43700 | 74.8 | 59200 | 72.5 | 1.03 | DOX_DAR_24hr |
| 233 | 11 | MA0878.3 | CDX1 | MA0878.3 | GGYMATAAAA | CDX1 | 4.51e-20 | 9.46e-23 | 25900 | 44.3 | 34100 | 41.7 | 1.06 | DOX_DAR_24hr |
| 234 | 37 | MA1588.1 | ZNF136 | MA1588.1 | RWATTCTTGGTTGRC | ZNF136 | 7.39e-20 | 1.54e-22 | 31000 | 53.1 | 41200 | 50.5 | 1.05 | DOX_DAR_24hr |
| 235 | 10 | MA0852.2 | FOXK1 | MA0852.2 | NRDGTAAACAAGNN | FOXK1 | 8.2e-20 | 1.71e-22 | 53500 | 91.7 | 73700 | 90.2 | 1.02 | DOX_DAR_24hr |
| 236 | 27 | MA0808.1 | TEAD3 | MA0808.1 | RCATTCCW | TEAD3 | 8.23e-20 | 1.71e-22 | 32600 | 55.9 | 43500 | 53.3 | 1.05 | DOX_DAR_24hr |
| 237 | 12 | MA0442.2 | SOX10 | MA0442.2 | NDAACAAAGVN | SOX10 | 1.22e-19 | 2.52e-22 | 53900 | 92.4 | 74300 | 90.9 | 1.02 | DOX_DAR_24hr |
| 238 | 38 | MA1114.1 (PBX3) | STREME-6 | 6-AGTGACA | AGTGACA | PBX3 | 2.11e-19 | 4.34e-22 | 28600 | 49 | 37900 | 46.4 | 1.06 | DOX_DAR_24hr |
| 239 | 20 | MA0754.2 | CUX1 | MA0754.2 | TAATCGATAM | CUX1 | 2.59e-19 | 5.3e-22 | 37500 | 64.3 | 50400 | 61.7 | 1.04 | DOX_DAR_24hr |
| 240 | 5 | MA0883.1 | Dmbx1 | MA0883.1 | WDRWNMGGATTADKNNW | Dmbx1 | 2.82e-19 | 5.75e-22 | 30500 | 52.2 | 40500 | 49.6 | 1.05 | DOX_DAR_24hr |
| 241 | 38 | MA0782.2 | PKNOX1 | MA0782.2 | NNTGAGTGACAGVNN | PKNOX1 | 3.02e-19 | 6.13e-22 | 16200 | 27.8 | 20800 | 25.5 | 1.09 | DOX_DAR_24hr |
| 242 | 39 | MA0106.3 | TP53 | MA0106.3 | RACATGYCCGGRCATGTY | TP53 | 5.74e-19 | 1.16e-21 | 308 | 0.53 | 179 | 0.22 | 2.4 | DOX_DAR_24hr |
| 243 | 3 | MA1577.1 | TLX2 | MA1577.1 | BYAATTAR | TLX2 | 6.55e-19 | 1.32e-21 | 21800 | 37.4 | 28500 | 34.9 | 1.07 | DOX_DAR_24hr |
| 244 | 40 | MA0092.1 | Hand1::Tcf3 | MA0092.1 | BRTCTGGMWT | Hand1::Tcf3 | 7.48e-19 | 1.5e-21 | 53800 | 92.3 | 74200 | 90.8 | 1.02 | DOX_DAR_24hr |
| 245 | 27 | MA0090.3 | TEAD1 | MA0090.3 | NNACATTCCAGSN | TEAD1 | 9.12e-19 | 1.82e-21 | 51200 | 87.7 | 70200 | 86 | 1.02 | DOX_DAR_24hr |
| 246 | 21 | MA0836.2 | CEBPD | MA0836.2 | NRTTGCACAAYHN | CEBPD | 1.51e-18 | 3.01e-21 | 43500 | 74.6 | 59100 | 72.4 | 1.03 | DOX_DAR_24hr |
| 247 | 28 | MA0775.1 | MEIS3 | MA0775.1 | DTGACAGS | MEIS3 | 2.69e-18 | 5.33e-21 | 10000 | 17.2 | 12500 | 15.3 | 1.12 | DOX_DAR_24hr |
| 248 | 32 | MA1119.1 | SIX2 | MA1119.1 | NNCTGAAACCTGATMY | SIX2 | 3.04e-18 | 6e-21 | 50900 | 87.2 | 69800 | 85.4 | 1.02 | DOX_DAR_24hr |
| 249 | 11 | MA0897.1 | Hmx2 | MA0897.1 | ASAAGCAATTAAHVVRT | Hmx2 | 3.93e-18 | 7.72e-21 | 32200 | 55.1 | 43000 | 52.6 | 1.05 | DOX_DAR_24hr |
| 250 | 31 | MA0625.2 | NFATC3 | MA0625.2 | RYGGAAAMH | NFATC3 | 5.27e-18 | 1.03e-20 | 58000 | 99.4 | 80800 | 99 | 1 | DOX_DAR_24hr |
| 251 | 11 | MA0913.2 | HOXD9 | MA0913.2 | GYMATAAAAH | HOXD9 | 9.56e-18 | 1.86e-20 | 33500 | 57.4 | 44800 | 54.9 | 1.05 | DOX_DAR_24hr |
| 252 | 41 | MA1644.1 | NFYC | MA1644.1 | RDCCAATCASN | NFYC | 1.47e-17 | 2.85e-20 | 50900 | 87.3 | 69900 | 85.6 | 1.02 | DOX_DAR_24hr |
| 253 | 21 | MA0043.3 | HLF | MA0043.3 | NNRTTATGCAACHN | HLF | 8.83e-17 | 1.71e-19 | 34400 | 59 | 46200 | 56.6 | 1.04 | DOX_DAR_24hr |
| 254 | 10 | MA0480.2 | Foxo1 | MA0480.2 | NDGTAAACANN | Foxo1 | 1.6e-16 | 3.07e-19 | 56200 | 96.3 | 77900 | 95.3 | 1.01 | DOX_DAR_24hr |
| 255 | 24 | MA0672.1 | NKX2-3 | MA0672.1 | VCCACTTRAV | NKX2-3 | 2.49e-16 | 4.77e-19 | 15400 | 26.3 | 19800 | 24.2 | 1.09 | DOX_DAR_24hr |
| 256 | 42 | MA0626.1 | Npas2 | MA0626.1 | NSCACGTGTN | Npas2 | 3.89e-16 | 7.43e-19 | 28300 | 48.6 | 37700 | 46.2 | 1.05 | DOX_DAR_24hr |
| 257 | 43 | MA0611.2 | Dux | MA0611.2 | NTGATTBAATCAVWNN | Dux | 8.64e-16 | 1.65e-18 | 8840 | 15.2 | 11000 | 13.5 | 1.12 | DOX_DAR_24hr |
| 258 | 44 | MA1105.2 | GRHL2 | MA1105.2 | NRAACAGGTTYN | GRHL2 | 1.61e-15 | 3.05e-18 | 56700 | 97.1 | 78600 | 96.3 | 1.01 | DOX_DAR_24hr |
| 259 | 45 | MA1623.1 | Stat2 | MA1623.1 | RGAAACAGAAASH | Stat2 | 1.94e-15 | 3.67e-18 | 47700 | 81.8 | 65300 | 80 | 1.02 | DOX_DAR_24hr |
| 260 | 41 | MA0060.3 | NFYA | MA0060.3 | RRCCAATCAGM | NFYA | 1.06e-14 | 2e-17 | 45000 | 77.1 | 61400 | 75.2 | 1.03 | DOX_DAR_24hr |
| 261 | 46 | MA1112.2 | NR4A1 | MA1112.2 | NNAAAGGTCANN | NR4A1 | 1.19e-14 | 2.22e-17 | 50600 | 86.8 | 69600 | 85.2 | 1.02 | DOX_DAR_24hr |
| 262 | 47 | MA0691.1 | TFAP4 | MA0691.1 | AWCAGCTGWT | TFAP4 | 1.26e-14 | 2.36e-17 | 4890 | 8.38 | 5840 | 7.16 | 1.17 | DOX_DAR_24hr |
| 263 | 19 | MA1960.1 | MGA::EVX1 | MA1960.1 | RGGTGWTAATKD | MGA::EVX1 | 2.18e-14 | 4.06e-17 | 39200 | 67.1 | 53100 | 65 | 1.03 | DOX_DAR_24hr |
| 264 | 48 | MA1624.1 | Stat5a | MA1624.1 | NTTCCAAGAANH | Stat5a | 2.23e-14 | 4.14e-17 | 49200 | 84.4 | 67500 | 82.7 | 1.02 | DOX_DAR_24hr |
| 265 | 25 | MA0768.2 | Lef1 | MA0768.2 | NNBCCTTTGATSTN | Lef1 | 3.61e-14 | 6.67e-17 | 47600 | 81.6 | 65200 | 79.8 | 1.02 | DOX_DAR_24hr |
| 266 | 48 | MA1625.1 | Stat5b | MA1625.1 | NNNTTCCCAGAANNN | Stat5b | 7.89e-14 | 1.45e-16 | 49900 | 85.5 | 68500 | 83.8 | 1.02 | DOX_DAR_24hr |
| 267 | 49 | MA0093.3 | USF1 | MA0093.3 | NDGTCATGTGACHN | USF1 | 9.5e-14 | 1.74e-16 | 39000 | 66.9 | 52900 | 64.8 | 1.03 | DOX_DAR_24hr |
| 268 | 3 | MA0854.1 | Alx1 | MA0854.1 | CGMRYTAATTARTNMNY | Alx1 | 9.92e-14 | 1.81e-16 | 20200 | 34.6 | 26600 | 32.5 | 1.06 | DOX_DAR_24hr |
| 269 | 5 | MA0719.1 | RHOXF1 | MA0719.1 | NTRATCCN | RHOXF1 | 1.08e-13 | 1.96e-16 | 54900 | 94 | 75900 | 92.9 | 1.01 | DOX_DAR_24hr |
| 270 | 32 | MA2001.1 | Six4 | MA2001.1 | GDAACCTGANH | Six4 | 1.85e-13 | 3.35e-16 | 54300 | 93 | 75000 | 91.9 | 1.01 | DOX_DAR_24hr |
| 271 | 66 | MA0483.1 | Gfi1B | MA0483.1 | AAATCWCWGCH | Gfi1B | 2.37e-13 | 4.29e-16 | 21000 | 36.1 | 27700 | 34 | 1.06 | DOX_DAR_24hr |
| 272 | 24 | MA1645.1 | NKX2-2 | MA1645.1 | NNNCCACTCAANNN | NKX2-2 | 2.45e-13 | 4.4e-16 | 43100 | 73.9 | 58700 | 71.9 | 1.03 | DOX_DAR_24hr |
| 273 | 31 | MA0624.2 | Nfatc1 | MA0624.2 | NVATGGAAAAWN | Nfatc1 | 6.48e-13 | 1.16e-15 | 53700 | 92 | 74100 | 90.8 | 1.01 | DOX_DAR_24hr |
| 274 | 11 | MA0908.1 | HOXD11 | MA0908.1 | RTCGTAAAAH | HOXD11 | 1.16e-12 | 2.06e-15 | 22200 | 38 | 29400 | 36 | 1.06 | DOX_DAR_24hr |
| 275 | 43 | MA0884.2 | DUXA | MA0884.2 | YTRATTWAATYAR | DUXA | 1.18e-12 | 2.09e-15 | 10300 | 17.6 | 13100 | 16 | 1.1 | DOX_DAR_24hr |
| 276 | 10 | MA0851.1 | Foxj3 | MA0851.1 | NDAADGTAAACAAANNM | Foxj3 | 2.32e-12 | 4.12e-15 | 35600 | 61 | 48100 | 58.9 | 1.04 | DOX_DAR_24hr |
| 277 | 24 | MA1994.1 | Nkx2-1 | MA1994.1 | NVCACTTGARHNN | Nkx2-1 | 5.45e-12 | 9.62e-15 | 2990 | 5.12 | 3460 | 4.24 | 1.21 | DOX_DAR_24hr |
| 278 | 24 | MA0673.1 | NKX2-8 | MA0673.1 | VCACTTSAV | NKX2-8 | 7.43e-12 | 1.31e-14 | 26400 | 45.2 | 35300 | 43.2 | 1.05 | DOX_DAR_24hr |
| 279 | 67 | MA0508.3 | PRDM1 | MA0508.3 | YNCTTTCTCTH | PRDM1 | 7.6e-12 | 1.33e-14 | 50500 | 86.6 | 69500 | 85.1 | 1.02 | DOX_DAR_24hr |
| 280 | 66 | MA0038.2 | GFI1 | MA0038.2 | BMAATCACDGCA | GFI1 | 1.65e-11 | 2.88e-14 | 13800 | 23.6 | 17900 | 21.9 | 1.08 | DOX_DAR_24hr |
| 281 | 3 | MA0874.1 | Arx | MA0874.1 | VTBCRYTAATTARTKSW | Arx | 2.12e-11 | 3.69e-14 | 20700 | 35.6 | 27500 | 33.6 | 1.06 | DOX_DAR_24hr |
| 282 | 12 | MA0078.2 | Sox17 | MA0078.2 | NNAGAACAATGGNN | Sox17 | 4.61e-11 | 8e-14 | 27800 | 47.7 | 37300 | 45.7 | 1.04 | DOX_DAR_24hr |
| 283 | 47 | MA1593.1 | ZNF317 | MA1593.1 | WVACAGCAGAYW | ZNF317 | 6.53e-11 | 1.13e-13 | 47800 | 82 | 65600 | 80.4 | 1.02 | DOX_DAR_24hr |
| 284 | 24 | MA0063.2 | NKX2-5 | MA0063.2 | NNCACTCAANN | NKX2-5 | 9.69e-11 | 1.67e-13 | 56400 | 96.6 | 78300 | 95.8 | 1.01 | DOX_DAR_24hr |
| 285 | 45 | MA1709.1 | ZIM3 | MA1709.1 | RVMAACAGAAACCYM | ZIM3 | 1.1e-10 | 1.89e-13 | 40500 | 69.4 | 55100 | 67.5 | 1.03 | DOX_DAR_24hr |
| 286 | 50 | MA1974.1 | ZNF211 | MA1974.1 | HWYATATACCAYRYW | ZNF211 | 1.82e-10 | 3.12e-13 | 28900 | 49.5 | 38800 | 47.5 | 1.04 | DOX_DAR_24hr |
| 287 | 17 | MA0842.2 | NRL | MA0842.2 | AAWWNTGCTGACG | NRL | 2.28e-10 | 3.88e-13 | 4360 | 7.48 | 5290 | 6.48 | 1.15 | DOX_DAR_24hr |
| 288 | 31 | MA0152.2 | Nfatc2 | MA0152.2 | NNRNAATGGAAANWNH | Nfatc2 | 2.35e-10 | 3.99e-13 | 52900 | 90.6 | 73100 | 89.4 | 1.01 | DOX_DAR_24hr |
| 289 | 3 | MA0715.1 | PROP1 | MA0715.1 | TAATYWAATTA | PROP1 | 4.88e-10 | 8.26e-13 | 20900 | 35.9 | 27800 | 34 | 1.05 | DOX_DAR_24hr |
| 290 | 45 | MA0050.3 | Irf1 | MA0050.3 | ANTGAAACTGAAASH | Irf1 | 5.02e-10 | 8.47e-13 | 42000 | 72 | 57300 | 70.2 | 1.02 | DOX_DAR_24hr |
| 291 | 22 | MA1150.1 | RORB | MA1150.1 | AWWTRGGTCAH | RORB | 9.12e-10 | 1.53e-12 | 43900 | 75.2 | 60100 | 73.6 | 1.02 | DOX_DAR_24hr |
| 292 | 3 | MA1496.1 | HOXA4 | MA1496.1 | RTMATTAV | HOXA4 | 3.67e-09 | 6.15e-12 | 57100 | 97.8 | 79400 | 97.3 | 1.01 | DOX_DAR_24hr |
| 293 | 48 | MA0144.2 | STAT3 | MA0144.2 | YTTCYKGGAAD | STAT3 | 5.95e-09 | 9.93e-12 | 34500 | 59.1 | 46800 | 57.3 | 1.03 | DOX_DAR_24hr |
| 294 | 51 | MA0671.1 | NFIX | MA0671.1 | NNTGCCAAN | NFIX | 6.58e-09 | 1.1e-11 | 57700 | 98.8 | 80400 | 98.4 | 1 | DOX_DAR_24hr |
| 295 | 52 | MA0629.1 | Rhox11 | MA0629.1 | ANKNCGCTGTWAWDVRW | Rhox11 | 9.76e-09 | 1.62e-11 | 47300 | 81 | 65000 | 79.6 | 1.02 | DOX_DAR_24hr |
| 296 | 17 | MA1520.1 | MAF | MA1520.1 | NTGCTGASTCAGCAH | MAF | 1.32e-08 | 2.19e-11 | 2340 | 4 | 2720 | 3.33 | 1.2 | DOX_DAR_24hr |
| 297 | 39 | MA0861.1 | TP73 | MA0861.1 | DRCATGTCNNRACAYGYM | TP73 | 3.09e-08 | 5.1e-11 | 1100 | 1.89 | 1170 | 1.44 | 1.31 | DOX_DAR_24hr |
| 298 | 3 | MA1507.1 | HOXD4 | MA1507.1 | VTMATTAD | HOXD4 | 3.5e-08 | 5.74e-11 | 8390 | 14.4 | 10800 | 13.2 | 1.09 | DOX_DAR_24hr |
| 299 | 44 | MA1508.1 | IKZF1 | MA1508.1 | VVAACAGGAARN | IKZF1 | 4.77e-08 | 7.8e-11 | 55600 | 95.3 | 77200 | 94.6 | 1.01 | DOX_DAR_24hr |
| 300 | 22 | MA1139.1 | FOSL2::JUNB | MA1139.1 | DATGACGTCATH | FOSL2::JUNB | 6.75e-08 | 1.1e-10 | 32300 | 55.3 | 43800 | 53.6 | 1.03 | DOX_DAR_24hr |
| 301 | 53 | MA1975.1 | ZNF214 | MA1975.1 | VWTCATCAABGTCCT | ZNF214 | 9.41e-08 | 1.53e-10 | 26000 | 44.6 | 35100 | 42.9 | 1.04 | DOX_DAR_24hr |
| 302 | 22 | MA0488.1 | JUN | MA0488.1 | DDRATGATGTCAT | JUN | 2.67e-07 | 4.32e-10 | 10500 | 17.9 | 13600 | 16.7 | 1.08 | DOX_DAR_24hr |
| 303 | 10 | MA0614.1 | Foxj2 | MA0614.1 | RTAAACAA | Foxj2 | 3.85e-07 | 6.22e-10 | 17000 | 29.2 | 22600 | 27.7 | 1.05 | DOX_DAR_24hr |
| 304 | 41 | MA1523.1 | MSANTD3 | MA1523.1 | STVCACTCAC | MSANTD3 | 5.33e-07 | 8.58e-10 | 56300 | 96.5 | 78300 | 95.9 | 1.01 | DOX_DAR_24hr |
| 305 | 11 | MA1503.1 | HOXB9 | MA1503.1 | GTCGTAAAAH | HOXB9 | 7.77e-07 | 1.25e-09 | 11400 | 19.6 | 14900 | 18.3 | 1.07 | DOX_DAR_24hr |
| 306 | 25 | MA0869.2 | Sox11 | MA0869.2 | NRGAACAAAGVV | Sox11 | 8.22e-07 | 1.31e-09 | 36300 | 62.2 | 49500 | 60.7 | 1.03 | DOX_DAR_24hr |
| 307 | 43 | MA0468.1 | DUX4 | MA0468.1 | TAAYYYAATCA | DUX4 | 8.85e-07 | 1.41e-09 | 5630 | 9.66 | 7120 | 8.72 | 1.11 | DOX_DAR_24hr |
| 308 | 42 | MA0620.3 | MITF | MA0620.3 | NNNDRTCACGTGAYHNNN | MITF | 1.14e-06 | 1.81e-09 | 32600 | 55.9 | 44300 | 54.3 | 1.03 | DOX_DAR_24hr |
| 309 | 16 | MA0773.1 | MEF2D | MA0773.1 | DCTAWAAATAGM | MEF2D | 1.31e-06 | 2.07e-09 | 8090 | 13.9 | 10400 | 12.8 | 1.09 | DOX_DAR_24hr |
| 310 | 27 | MA1121.1 | TEAD2 | MA1121.1 | NYACATTCCWNNS | TEAD2 | 1.6e-06 | 2.52e-09 | 37800 | 64.7 | 51600 | 63.2 | 1.02 | DOX_DAR_24hr |
| 311 | 54 | MA0780.1 | PAX3 | MA0780.1 | TAATYRATTA | PAX3 | 1.64e-06 | 2.58e-09 | 15100 | 25.9 | 20000 | 24.5 | 1.06 | DOX_DAR_24hr |
| 312 | 22 | MA1632.1 | ATF2 | MA1632.1 | NNATGABGTCATN | ATF2 | 1.71e-06 | 2.68e-09 | 19000 | 32.5 | 25300 | 31 | 1.05 | DOX_DAR_24hr |
| 313 | 55 | MA0101.1 | REL | MA0101.1 | BGGRNWTTCC | REL | 2.68e-06 | 4.19e-09 | 53000 | 90.8 | 73400 | 89.9 | 1.01 | DOX_DAR_24hr |
| 314 | 39 | MA0525.2 | TP63 | MA0525.2 | RACATGTYGKGACATGTC | TP63 | 2.86e-06 | 4.46e-09 | 266 | 0.46 | 219 | 0.27 | 1.7 | DOX_DAR_24hr |
| 315 | 41 | MA0502.2 | NFYB | MA0502.2 | CYCATTGGCCVV | NFYB | 4.16e-06 | 6.45e-09 | 3750 | 6.42 | 4640 | 5.68 | 1.13 | DOX_DAR_24hr |
| 316 | 51 | MA0119.1 | NFIC::TLX1 | MA0119.1 | TGGCASSRWGCCAA | NFIC::TLX1 | 7.63e-06 | 1.18e-08 | 4120 | 7.06 | 5150 | 6.3 | 1.12 | DOX_DAR_24hr |
| 317 | 10 | MA0613.1 | FOXG1 | MA0613.1 | RTAAACAW | FOXG1 | 7.98e-06 | 1.23e-08 | 21200 | 36.4 | 28500 | 34.9 | 1.04 | DOX_DAR_24hr |
| 318 | 48 | MA0519.1 | Stat5a::Stat5b | MA0519.1 | DTTTCCMAGAA | Stat5a::Stat5b | 8.54e-06 | 1.31e-08 | 30400 | 52 | 41300 | 50.5 | 1.03 | DOX_DAR_24hr |
| 319 | 3 | MA0702.2 | LMX1A | MA0702.2 | TTAATTAM | LMX1A | 1.4e-05 | 2.14e-08 | 16600 | 28.4 | 22100 | 27.1 | 1.05 | DOX_DAR_24hr |
| 320 | 56 | MA0494.1 | Nr1h3::Rxra | MA0494.1 | TGACCTNNAGTRACCYYDN | Nr1h3::Rxra | 1.63e-05 | 2.5e-08 | 47500 | 81.4 | 65500 | 80.2 | 1.01 | DOX_DAR_24hr |
| 321 | 57 | MA1116.1 | RBPJ | MA1116.1 | BVTGGGAANN | RBPJ | 1.82e-05 | 2.77e-08 | 50500 | 86.5 | 69800 | 85.5 | 1.01 | DOX_DAR_24hr |
| 322 | 58 | MA0693.3 | Vdr | MA0693.3 | NNGAGTTCANN | Vdr | 1.83e-05 | 2.77e-08 | 57000 | 97.8 | 79500 | 97.3 | 1 | DOX_DAR_24hr |
| 323 | 47 | MA0667.1 | MYF6 | MA0667.1 | AACARCTGTT | MYF6 | 2.2e-05 | 3.32e-08 | 14700 | 25.2 | 19500 | 23.9 | 1.05 | DOX_DAR_24hr |
| 324 | 59 | MA1729.1 | ZNF680 | MA1729.1 | GNCCAAGAAGAATDA | ZNF680 | 2.24e-05 | 3.39e-08 | 28800 | 49.4 | 39200 | 48 | 1.03 | DOX_DAR_24hr |
| 325 | 56 | MA1151.1 | RORC | MA1151.1 | NWAWNTRGGTCA | RORC | 2.43e-05 | 3.66e-08 | 55500 | 95.1 | 77100 | 94.4 | 1.01 | DOX_DAR_24hr |
| 326 | 11 | MA1506.1 | HOXD10 | MA1506.1 | RGTCGTAAAAH | HOXD10 | 2.47e-05 | 3.7e-08 | 11800 | 20.3 | 15600 | 19.1 | 1.06 | DOX_DAR_24hr |
| 327 | 60 | MA1478.1 | DMRTA2 | MA1478.1 | AATTGHTACAWT | DMRTA2 | 2.53e-05 | 3.79e-08 | 52100 | 89.4 | 72200 | 88.4 | 1.01 | DOX_DAR_24hr |
| 328 | 21 | MA0837.2 | CEBPE | MA0837.2 | KATTGCGCAATM | CEBPE | 2.87e-05 | 4.28e-08 | 5400 | 9.26 | 6880 | 8.43 | 1.1 | DOX_DAR_24hr |
| 329 | 12 | MA0087.2 | Sox5 | MA0087.2 | VRVAACAATGGNN | Sox5 | 3.68e-05 | 5.48e-08 | 32500 | 55.7 | 44300 | 54.2 | 1.03 | DOX_DAR_24hr |
| 330 | 30 | MA0040.1 | Foxq1 | MA0040.1 | HATTGTTTATW | Foxq1 | 5.15e-05 | 7.64e-08 | 16400 | 28.1 | 21900 | 26.8 | 1.05 | DOX_DAR_24hr |
| 331 | 21 | MA0639.1 | DBP | MA0639.1 | NRTTACGTAAYV | DBP | 7.13e-05 | 1.05e-07 | 14500 | 24.8 | 19200 | 23.6 | 1.05 | DOX_DAR_24hr |
| 332 | 61 | MA1592.1 | ZNF274 | MA1592.1 | NRTRTGAGTTCTCGYN | ZNF274 | 7.45e-05 | 1.1e-07 | 10500 | 18.1 | 13900 | 17 | 1.06 | DOX_DAR_24hr |
| 333 | 22 | MA0492.1 | JUND | MA0492.1 | DDDRATGABGTCATN | JUND | 8e-05 | 1.18e-07 | 9980 | 17.1 | 13100 | 16 | 1.07 | DOX_DAR_24hr |
| 334 | 42 | MA1993.1 | Neurod2 | MA1993.1 | VVCAGCTGBB | Neurod2 | 0.000107 | 1.56e-07 | 7660 | 13.1 | 9960 | 12.2 | 1.08 | DOX_DAR_24hr |
| 335 | 36 | MA0829.2 | SREBF1 | MA0829.2 | DNATCACSTGATNH | SREBF1 | 0.000341 | 4.98e-07 | 6510 | 11.2 | 8430 | 10.3 | 1.08 | DOX_DAR_24hr |
| 336 | 62 | MA0802.1 | TBR1 | MA0802.1 | AGGTGTGAAA | TBR1 | 0.000346 | 5.03e-07 | 36100 | 62 | 49500 | 60.6 | 1.02 | DOX_DAR_24hr |
| 337 | 42 | MA1997.1 | Olig2 | MA1997.1 | NVCAGCTGBN | Olig2 | 0.000388 | 5.63e-07 | 7380 | 12.6 | 9620 | 11.8 | 1.07 | DOX_DAR_24hr |
| 338 | 11 | MA0906.1 | HOXC12 | MA0906.1 | RGTCGTAAAAH | HOXC12 | 0.000449 | 6.49e-07 | 16700 | 28.7 | 22500 | 27.5 | 1.04 | DOX_DAR_24hr |
| 339 | 11 | MA0907.1 | HOXC13 | MA0907.1 | BCTCGTAAAAH | HOXC13 | 0.00048 | 6.92e-07 | 17500 | 30 | 23600 | 28.8 | 1.04 | DOX_DAR_24hr |
| 340 | 27 | MA0114.4 | HNF4A | MA0114.4 | DNCAAAGTCCANN | HNF4A | 0.000493 | 7.1e-07 | 54100 | 92.7 | 75100 | 92 | 1.01 | DOX_DAR_24hr |
| 341 | 16 | MA0660.1 | MEF2B | MA0660.1 | RCTAWAAATAGC | MEF2B | 0.000608 | 8.72e-07 | 8000 | 13.7 | 10500 | 12.8 | 1.07 | DOX_DAR_24hr |
| 342 | 46 | MA1111.1 | NR2F2 | MA1111.1 | NAAAGGTCANR | NR2F2 | 0.000981 | 1.4e-06 | 39300 | 67.3 | 54000 | 66.1 | 1.02 | DOX_DAR_24hr |
| 343 | 17 | MA1521.1 | MAFA | MA1521.1 | NTGCTGASTCAGCAN | MAFA | 0.00142 | 2.03e-06 | 1670 | 2.86 | 2000 | 2.45 | 1.17 | DOX_DAR_24hr |
| 11 | 66 | MA0483.1 (Gfi1B) | MEME-7 | AATCCCAGCWMYTTG | AATCCCAGCWMYTTG | Gfi1B | 0.00173 | 0.000157 | 283 | 0.49 | 289 | 0.35 | 1.37 | DOX_DAR_24hr |
| 344 | 11 | MA0873.1 | HOXD12 | MA0873.1 | RGTCGTAAAAH | HOXD12 | 0.00206 | 2.93e-06 | 12700 | 21.7 | 16900 | 20.7 | 1.05 | DOX_DAR_24hr |
| 345 | 12 | MA0515.1 | Sox6 | MA0515.1 | CCWTTGTYYY | Sox6 | 0.00243 | 3.45e-06 | 12500 | 21.4 | 16600 | 20.4 | 1.05 | DOX_DAR_24hr |
| 346 | 42 | MA0633.2 | Twist2 | MA0633.2 | NVCAGCTGBN | Twist2 | 0.00252 | 3.56e-06 | 7210 | 12.4 | 9440 | 11.6 | 1.07 | DOX_DAR_24hr |
| 347 | 43 | MA1720.1 | ZNF85 | MA1720.1 | DHDGAGATTACWKCAK | ZNF85 | 0.0031 | 4.36e-06 | 12100 | 20.7 | 16100 | 19.7 | 1.05 | DOX_DAR_24hr |
| 348 | 10 | MA0849.1 | FOXO6 | MA0849.1 | GTAAACA | FOXO6 | 0.00363 | 5.1e-06 | 30700 | 52.7 | 42000 | 51.5 | 1.02 | DOX_DAR_24hr |
| 349 | 22 | MA1133.1 | JUN::JUNB | MA1133.1 | KRTGACGTCATN | JUN::JUNB | 0.00368 | 5.15e-06 | 40500 | 69.5 | 55800 | 68.4 | 1.02 | DOX_DAR_24hr |
| 350 | 51 | MA1643.1 | NFIB | MA1643.1 | NNCCTGGCAYNGTGCCAAGNN | NFIB | 0.00447 | 6.25e-06 | 2360 | 4.04 | 2920 | 3.58 | 1.13 | DOX_DAR_24hr |
| 351 | 62 | MA0800.1 | EOMES | MA0800.1 | RAGGTGTGAAAWN | EOMES | 0.0047 | 6.55e-06 | 50800 | 87.1 | 70500 | 86.3 | 1.01 | DOX_DAR_24hr |
| 352 | 63 | MA0761.2 | ETV1 | MA0761.2 | NNACAGGAAGTGNN | ETV1 | 0.00575 | 7.99e-06 | 57200 | 98.1 | 79800 | 97.8 | 1 | DOX_DAR_24hr |
| 353 | 51 | MA1527.1 | NFIC | MA1527.1 | NTTGGCDNNRTGCCARN | NFIC | 0.0086 | 1.19e-05 | 3480 | 5.96 | 4430 | 5.43 | 1.1 | DOX_DAR_24hr |
| 354 | 42 | MA0743.2 | SCRT1 | MA0743.2 | NDWKCAACAGGTGKNN | SCRT1 | 0.0108 | 1.49e-05 | 46300 | 79.3 | 64000 | 78.4 | 1.01 | DOX_DAR_24hr |
| 355 | 3 | MA0135.1 | Lhx3 | MA0135.1 | WAATTAATTAWYY | Lhx3 | 0.0124 | 1.71e-05 | 10100 | 17.4 | 13500 | 16.5 | 1.05 | DOX_DAR_24hr |
| 356 | 64 | MA0631.1 | Six3 | MA0631.1 | DATRGGGTATCAYNWNT | Six3 | 0.0173 | 2.38e-05 | 16100 | 27.7 | 21800 | 26.7 | 1.04 | DOX_DAR_24hr |
| 357 | 49 | MA1524.2 | Msgn1 | MA1524.2 | VRRRACAAATGGTNNN | Msgn1 | 0.0179 | 2.45e-05 | 21500 | 36.9 | 29300 | 35.9 | 1.03 | DOX_DAR_24hr |
| 358 | 3 | MA0683.1 | POU4F2 | MA0683.1 | RTGCATAATTAATGAG | POU4F2 | 0.019 | 2.59e-05 | 3620 | 6.21 | 4640 | 5.68 | 1.09 | DOX_DAR_24hr |
| 359 | 62 | MA1567.2 | Tbx6 | MA1567.2 | NRGGTGTGAANN | Tbx6 | 0.02 | 2.72e-05 | 19500 | 33.5 | 26500 | 32.4 | 1.03 | DOX_DAR_24hr |
| 360 | 22 | MA1129.1 | FOSL1::JUN | MA1129.1 | ATGACGTCAT | FOSL1::JUN | 0.0258 | 3.51e-05 | 36500 | 62.5 | 50200 | 61.4 | 1.02 | DOX_DAR_24hr |
| 361 | 21 | MA0838.1 | CEBPG | MA0838.1 | ATTRCGCAAY | CEBPG | 0.0264 | 3.57e-05 | 18800 | 32.3 | 25500 | 31.3 | 1.03 | DOX_DAR_24hr |
| 362 | 65 | MA0258.2 | ESR2 | MA0258.2 | AGGTCASVNTGMCCY | ESR2 | 0.0281 | 3.8e-05 | 47400 | 81.2 | 65600 | 80.3 | 1.01 | DOX_DAR_24hr |
| 363 | 48 | MA0518.1 | Stat4 | MA0518.1 | YTTCYRGGAARNVR | Stat4 | 0.0291 | 3.92e-05 | 28700 | 49.1 | 39200 | 48.1 | 1.02 | DOX_DAR_24hr |
| 364 | 63 | MA1952.1 | FOXJ2::ELF1 | MA1952.1 | RTAAACMGGAAGTR | FOXJ2::ELF1 | 0.0407 | 5.46e-05 | 52700 | 90.3 | 73200 | 89.7 | 1.01 | DOX_DAR_24hr |
| 365 | 46 | MA0643.1 | Esrrg | MA0643.1 | TCAAGGTCAH | Esrrg | 0.0446 | 5.98e-05 | 21100 | 36.2 | 28700 | 35.2 | 1.03 | DOX_DAR_24hr |
| 12 | 67 | MA0508.3 (PRDM1) | MEME-3 | AVAGAGARAGAGARA | AVAGAGARAGAGARA | PRDM1 | 0.0471 | 0.00393 | 6950 | 11.9 | 9330 | 11.4 | 1.04 | DOX_DAR_24hr |
| 366 | 3 | MA0621.1 | mix-a | MA0621.1 | NDTTAATTAGN | mix-a | 0.0499 | 6.66e-05 | 10400 | 17.8 | 13900 | 17 | 1.05 | DOX_DAR_24hr |
| 13 | 68 | GAGGTTGCAGTGAGC | MEME-4 | GAGGTTGCAGTGAGC | GAGGTTGCAGTGAGC | GAGGTTGCAGTGAGC | 0.34 | 0.0262 | 411 | 0.7 | 500 | 0.61 | 1.15 | DOX_DAR_24hr |
| 14 | 69 | MA1581.1 (ZBTB6) | MEME-8 | CTTGARCCCAGGAGK | CTTGARCCCAGGAGK | ZBTB6 | 0.719 | 0.0514 | 49 | 0.08 | 47 | 0.06 | 1.46 | DOX_DAR_24hr |
| 15 | 70 | MA1107.2 (KLF9) | MEME-5 | ACACACACACACACA | ACACACACACACACA | KLF9 | 1.8 | 0.12 | 16 | 0.03 | 13 | 0.02 | 1.7 | DOX_DAR_24hr |
| 16 | 71 | MA1999.1 (Prdm5) | MEME-6 | ACTGCACTCCAGCCT | ACTGCACTCCAGCCT | Prdm5 | 2.43 | 0.152 | 301 | 0.52 | 386 | 0.47 | 1.09 | DOX_DAR_24hr |
| 17 | 67 | MA1125.1 (ZNF384) | MEME-1 | TTTTTTTTTTTTTTT | TTTTTTTTTTTTTTT | ZNF384 | 4.13 | 0.243 | 4160 | 7.13 | 5740 | 7.03 | 1.01 | DOX_DAR_24hr |
| 18 | 72 | MA1596.1 (ZNF460) | MEME-9 | GGARGCTG | GGARGCTG | ZNF460 | 16 | 0.889 | 959 | 1.64 | 1410 | 1.73 | 0.952 | DOX_DAR_24hr |
ER_rat <- 1.25
mrc_type <- "DOX_DAR_24hr"
DOX_DAR_24hr_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,mrc_type,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks(200bp)Enrichment ratio:",ER_rat))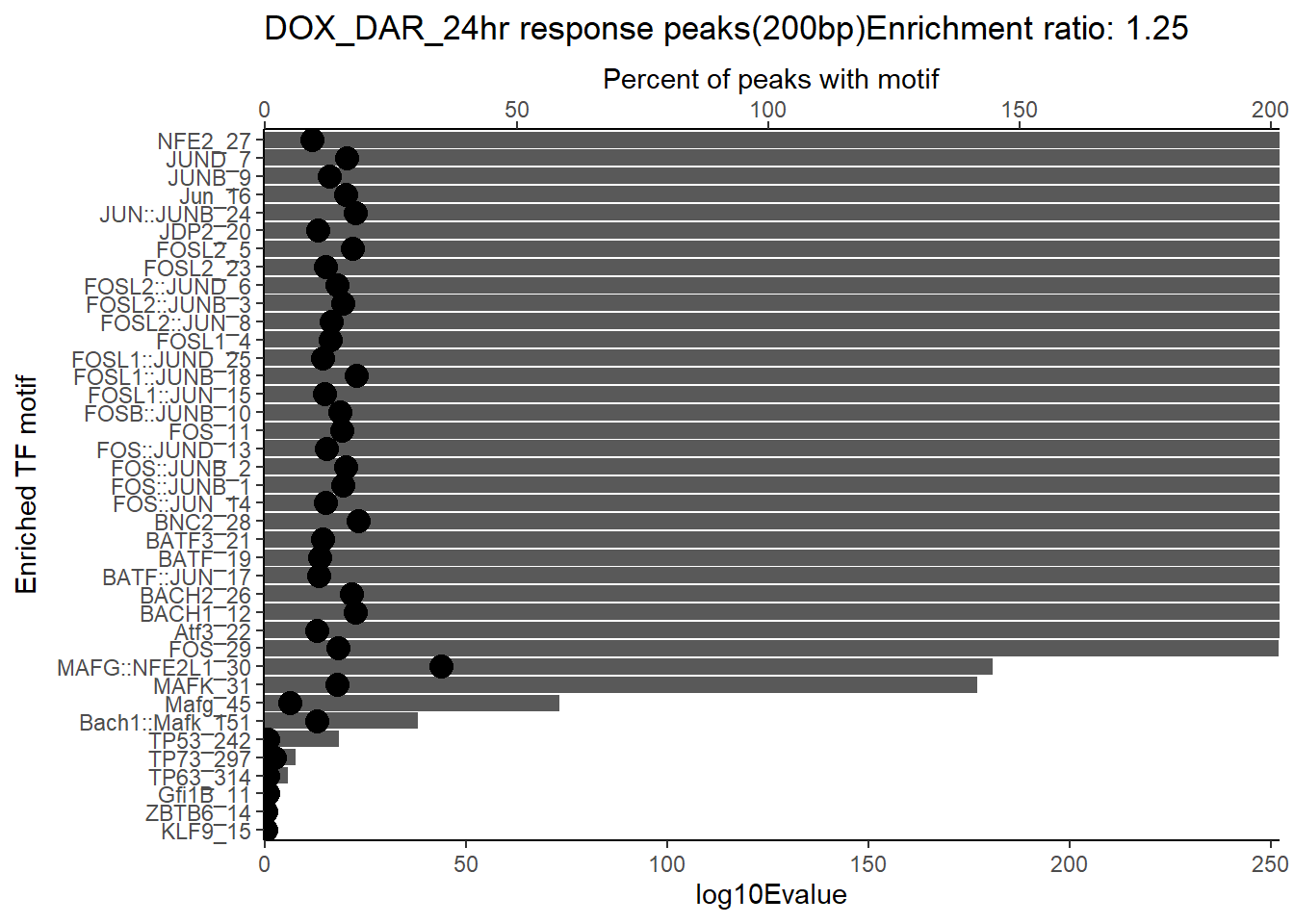
DOX_DAR_24hr_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,mrc_type,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
dplyr::filter(EVALUE.y<0.05) %>%
slice_head(n=5) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*.8), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./.8,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks(200bp)Enrichment ratio:",ER_rat))
DOX_sigup_24hr data
DOX_sigup_24hr_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DOX_24_up_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
DOX_sigup_24hr_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DOX_24_up_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
DOX_sigup_24hr_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DOX_24_up_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
DOX_sigup_24hr_sea_all <- rbind(DOX_sigup_24hr_sea_disc, DOX_sigup_24hr_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)DOX_24_up_combo <-DOX_sigup_24hr_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., DOX_sigup_24hr_sea_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ SIM_MOTIF
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y)
DOX_24_up_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs 200 bp in DOX_24sig up") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | MA1134.1 (FOS::JUNB) | STREME-1 | 1-DATGASTCATH | DATGASTCATH | FOS::JUNB | 0 | 0 | 14000 | 48 | 1740 | 5.97 | 8.03 |
| 2 | 1 | MA1130.1 (FOSL2::JUN) | MEME-1 | RTGACTCAKHH | RTGACTCAKHH | FOSL2::JUN | 0 | 0 | 13200 | 45.3 | 1370 | 4.71 | 9.61 |
| 3 | 1 | MA0476.1 | FOS | MA0476.1 | DVTGASTCATB | FOS | 0 | 0 | 13500 | 46.2 | 1560 | 5.36 | 8.62 |
| 4 | 1 | MA1134.1 | FOS::JUNB | MA1134.1 | KATGASTCATHN | FOS::JUNB | 0 | 0 | 12500 | 42.8 | 1150 | 3.95 | 10.8 |
| 5 | 1 | MA0462.2 (BATF::JUN) | STREME-4 | 4-ATGASTCAT | ATGASTCAT | BATF::JUN | 0 | 0 | 13600 | 46.3 | 1630 | 5.61 | 8.25 |
| 6 | 1 | MA1138.1 | FOSL2::JUNB | MA1138.1 | KRTGASTCAT | FOSL2::JUNB | 0 | 0 | 13300 | 45.5 | 1520 | 5.24 | 8.69 |
| 7 | 1 | MA1135.1 | FOSB::JUNB | MA1135.1 | KRTGASTCAT | FOSB::JUNB | 0 | 0 | 13500 | 46.2 | 1640 | 5.66 | 8.16 |
| 8 | 1 | MA1144.1 | FOSL2::JUND | MA1144.1 | KATGACTCAT | FOSL2::JUND | 0 | 0 | 13200 | 45 | 1510 | 5.2 | 8.66 |
| 9 | 1 | MA1141.1 | FOS::JUND | MA1141.1 | NKATGAGTCATNN | FOS::JUND | 0 | 0 | 12900 | 44 | 1370 | 4.71 | 9.32 |
| 10 | 1 | MA0477.2 | FOSL1 | MA0477.2 | NNATGACTCATNN | FOSL1 | 0 | 0 | 13000 | 44.4 | 1440 | 4.97 | 8.94 |
| 11 | 1 | MA1130.1 | FOSL2::JUN | MA1130.1 | NNRTGAGTCAYN | FOSL2::JUN | 0 | 0 | 12400 | 42.3 | 1180 | 4.05 | 10.4 |
| 12 | 1 | MA0099.3 | FOS::JUN | MA0099.3 | ATGAGTCAYM | FOS::JUN | 0 | 0 | 12700 | 43.3 | 1320 | 4.53 | 9.56 |
| 13 | 1 | MA0462.2 | BATF::JUN | MA0462.2 | DATGACTCATH | BATF::JUN | 0 | 0 | 13200 | 45.2 | 1600 | 5.49 | 8.24 |
| 14 | 1 | MA1633.2 | BACH1 | MA1633.2 | ATGACTCAT | BACH1 | 0 | 0 | 12500 | 42.7 | 1250 | 4.29 | 9.94 |
| 15 | 1 | MA0489.2 | Jun | MA0489.2 | NVTGACTCATNN | Jun | 0 | 0 | 12200 | 41.8 | 1150 | 3.96 | 10.5 |
| 16 | 1 | MA1988.1 | Atf3 | MA1988.1 | NRTGACTCABN | Atf3 | 0 | 0 | 13900 | 47.6 | 2000 | 6.87 | 6.92 |
| 17 | 1 | MA1634.1 | BATF | MA1634.1 | DATGACTCATH | BATF | 0 | 0 | 12400 | 42.3 | 1230 | 4.24 | 9.96 |
| 18 | 1 | MA1128.1 | FOSL1::JUN | MA1128.1 | NKATGACTCATNN | FOSL1::JUN | 0 | 0 | 12800 | 43.6 | 1420 | 4.9 | 8.9 |
| 19 | 1 | MA0835.2 | BATF3 | MA0835.2 | DATGACTCATH | BATF3 | 0 | 0 | 13100 | 44.6 | 1590 | 5.46 | 8.16 |
| 20 | 1 | MA0490.2 | JUNB | MA0490.2 | NNATGACTCATNN | JUNB | 0 | 0 | 13500 | 46.1 | 1860 | 6.4 | 7.2 |
| 21 | 1 | MA0478.1 | FOSL2 | MA0478.1 | KRRTGASTCAB | FOSL2 | 0 | 0 | 12200 | 41.6 | 1230 | 4.24 | 9.8 |
| 22 | 1 | MA0491.2 | JUND | MA0491.2 | NNATGACTCATNN | JUND | 0 | 0 | 12600 | 43 | 1470 | 5.05 | 8.51 |
| 23 | 1 | MA1928.1 | BNC2 | MA1928.1 | NDRTGAGTCABN | BNC2 | 0 | 0 | 12300 | 41.9 | 1310 | 4.51 | 9.28 |
| 24 | 1 | MA1137.1 | FOSL1::JUNB | MA1137.1 | NDRTGACTCATNN | FOSL1::JUNB | 0 | 0 | 12800 | 43.8 | 1820 | 6.24 | 7.02 |
| 25 | 1 | MA0655.1 | JDP2 | MA0655.1 | ATGACTCAT | JDP2 | 0 | 0 | 11500 | 39.3 | 1240 | 4.25 | 9.24 |
| 26 | 1 | MA1142.1 | FOSL1::JUND | MA1142.1 | NRTGACTCAT | FOSL1::JUND | 0 | 0 | 13700 | 46.7 | 2520 | 8.68 | 5.38 |
| 27 | 1 | MA1101.2 | BACH2 | MA1101.2 | DWANCATGASTCATSNTWH | BACH2 | 0 | 0 | 11900 | 40.8 | 1650 | 5.66 | 7.2 |
| 28 | 1 | MA1132.1 | JUN::JUNB | MA1132.1 | KATGACKCAT | JUN::JUNB | 0 | 0 | 12900 | 44 | 2190 | 7.52 | 5.84 |
| 29 | 1 | MA0841.1 | NFE2 | MA0841.1 | VATGACTCATS | NFE2 | 0 | 0 | 11000 | 37.7 | 1390 | 4.77 | 7.9 |
| 30 | 1 | MA0496.3 | MAFK | MA0496.3 | NGMTGACTCAGCMNH | MAFK | 0 | 0 | 10700 | 36.6 | 2760 | 9.48 | 3.86 |
| 31 | 1 | MA0089.2 | MAFG::NFE2L1 | MA0089.2 | NVNATGACTCAGCADW | MAFG::NFE2L1 | 0 | 0 | 10400 | 35.7 | 2600 | 8.94 | 3.99 |
| 32 | 1 | MA0501.1 | MAF::NFE2 | MA0501.1 | ATGACTCAGCANWTY | MAF::NFE2 | 0 | 0 | 12500 | 42.8 | 4380 | 15.1 | 2.84 |
| 33 | 1 | MA0150.2 | Nfe2l2 | MA0150.2 | CASNATGACTCAGCA | Nfe2l2 | 0 | 0 | 13600 | 46.6 | 6140 | 21.1 | 2.2 |
| 34 | 1 | MA0659.3 | Mafg | MA0659.3 | NWGMTGACTCAGCAN | Mafg | 0 | 0 | 10400 | 35.4 | 4240 | 14.6 | 2.43 |
| 35 | 2 | MA0676.1 | Nr2e1 | MA0676.1 | AAAAGTCAA | Nr2e1 | 0 | 0 | 21800 | 74.7 | 15700 | 54.1 | 1.38 |
| 36 | 3 | 20-AWGWCWT | STREME-20 | 20-AWGWCWT | 20-AWGWCWT | AWGWCWT | 0 | 0 | 21800 | 74.6 | 15900 | 54.5 | 1.37 |
| 37 | 4 | MA0791.1 | POU4F3 | MA0791.1 | RTGMATWATTAATGAR | POU4F3 | 0 | 0 | 19200 | 65.7 | 13100 | 44.9 | 1.46 |
| 38 | 4 | MA0790.1 | POU4F1 | MA0790.1 | ATGMATAATTAATG | POU4F1 | 0 | 0 | 21100 | 72.1 | 15200 | 52.2 | 1.38 |
| 39 | 1 | MA1113.2 | PBX2 | MA1113.2 | NHCATAAATCAHN | PBX2 | 0 | 0 | 20100 | 68.5 | 14100 | 48.3 | 1.42 |
| 40 | 5 | MA1978.1 | ZNF354A | MA1978.1 | HWAADWATAAATGRAYWAWTT | ZNF354A | 0 | 0 | 22100 | 75.5 | 16500 | 56.8 | 1.33 |
| 41 | 6 | MA1143.1 | FOSL1::JUND | MA1143.1 | RTGACGTMAY | FOSL1::JUND | 0 | 0 | 22700 | 77.5 | 17200 | 59.1 | 1.31 |
| 42 | 1 | MA1640.1 | MEIS2 | MA1640.1 | NNATGATTTATGDNH | MEIS2 | 0 | 0 | 21100 | 72 | 15400 | 52.9 | 1.36 |
| 43 | 4 | MA0046.2 | HNF1A | MA0046.2 | DRTTAATNATTAACN | HNF1A | 0 | 0 | 17000 | 58 | 11200 | 38.4 | 1.51 |
| 44 | 7 | MA0851.1 (Foxj3) | STREME-2 | 2-AAACAAAWAHAW | AAACAAAWAHAW | Foxj3 | 0 | 0 | 19000 | 64.8 | 13200 | 45.3 | 1.43 |
| 45 | 8 | MA0681.2 | PHOX2B | MA0681.2 | WWTAATCAAATTAWWW | PHOX2B | 0 | 0 | 21000 | 71.9 | 15400 | 53 | 1.36 |
| 46 | 8 | MA0680.2 | Pax7 | MA0680.2 | DWTAATCAATTAWNH | Pax7 | 0 | 0 | 22200 | 75.7 | 16700 | 57.4 | 1.32 |
| 47 | 1 | MA1639.1 | MEIS1 | MA1639.1 | NHCATMAATCAHN | MEIS1 | 0 | 0 | 21500 | 73.4 | 15900 | 54.8 | 1.34 |
| 48 | 9 | MA1588.1 (ZNF136) | STREME-11 | 11-AAGAAT | 11-AAGAAT | ZNF136 | 0 | 0 | 23300 | 79.8 | 18200 | 62.4 | 1.28 |
| 49 | 4 | MA0153.2 | HNF1B | MA0153.2 | GTTAATNATTAAY | HNF1B | 0 | 0 | 16600 | 56.8 | 11000 | 37.7 | 1.51 |
| 50 | 10 | MA0788.1 | POU3F3 | MA0788.1 | WWTATGCWAATTW | POU3F3 | 0 | 0 | 22200 | 75.8 | 17000 | 58.5 | 1.3 |
| 51 | 11 | MA0886.1 | EMX2 | MA0886.1 | VBTAATTARB | EMX2 | 0 | 0 | 23400 | 80 | 18500 | 63.5 | 1.26 |
| 52 | 7 | MA1125.1 (ZNF384) | MEME-2 | TTTTTTTTTTTTTTT | TTTTTTTTTTTTTTT | ZNF384 | 0 | 0 | 16100 | 55.1 | 10700 | 36.9 | 1.49 |
| 53 | 12 | 3-AAWWWWTT | STREME-3 | 3-AAWWWWTT | AAWWWWTT | AAWWWWTT | 0 | 0 | 23200 | 79.2 | 18200 | 62.7 | 1.26 |
| 54 | 13 | MA0679.2 | ONECUT1 | MA0679.2 | WAAAAATCAATAAWWN | ONECUT1 | 0 | 0 | 20100 | 68.6 | 14800 | 50.8 | 1.35 |
| 55 | 10 | MA0786.1 | POU3F1 | MA0786.1 | WTATGCWAATKW | POU3F1 | 0 | 0 | 24100 | 82.4 | 19400 | 66.7 | 1.24 |
| 56 | 14 | MA0846.1 | FOXC2 | MA0846.1 | WAWGTAAACAWW | FOXC2 | 0 | 0 | 23000 | 78.5 | 18100 | 62.1 | 1.26 |
| 57 | 14 | MA0032.2 | FOXC1 | MA0032.2 | WAWGTAAAYAW | FOXC1 | 0 | 0 | 22300 | 76.3 | 17400 | 59.7 | 1.28 |
| 58 | 8 | MA0715.1 | PROP1 | MA0715.1 | TAATYWAATTA | PROP1 | 0 | 0 | 16500 | 56.3 | 11200 | 38.6 | 1.46 |
| 59 | 15 | MA0901.2 | HOXB13 | MA0901.2 | WDCCAATAAAAWWW | HOXB13 | 0 | 0 | 24000 | 81.9 | 19300 | 66.5 | 1.23 |
| 60 | 15 | MA1125.1 | ZNF384 | MA1125.1 | DNWMAAAAAAAA | ZNF384 | 0 | 0 | 17500 | 59.7 | 12200 | 42 | 1.42 |
| 61 | 16 | 18-AAATGA | STREME-18 | 18-AAATGA | 18-AAATGA | AAATGA | 0 | 0 | 20500 | 70 | 15400 | 52.8 | 1.33 |
| 62 | 10 | MA0142.1 | Pou5f1::Sox2 | MA0142.1 | CWTTGTYATGCAAAT | Pou5f1::Sox2 | 0 | 0 | 22300 | 76.3 | 17400 | 59.9 | 1.27 |
| 63 | 10 | MA0787.1 | POU3F2 | MA0787.1 | WTATGCWAATKA | POU3F2 | 0 | 0 | 23100 | 79 | 18400 | 63.2 | 1.25 |
| 64 | 11 | MA0902.2 | HOXB2 | MA0902.2 | SYAATTAS | HOXB2 | 0 | 0 | 22000 | 75 | 17100 | 58.6 | 1.28 |
| 65 | 11 | MA0628.1 | POU6F1 | MA0628.1 | AYTAATTART | POU6F1 | 0 | 0 | 24400 | 83.2 | 19900 | 68.4 | 1.22 |
| 66 | 8 | MA0713.1 | PHOX2A | MA0713.1 | TAATYYAATTA | PHOX2A | 0 | 0 | 20800 | 71.2 | 15900 | 54.5 | 1.31 |
| 67 | 36 | MA0052.4 | MEF2A | MA0052.4 | DDCTAAAAATAGMHH | MEF2A | 0 | 0 | 22700 | 77.7 | 18000 | 61.9 | 1.26 |
| 68 | 14 | MA0845.1 | FOXB1 | MA0845.1 | WATGTAAATAT | FOXB1 | 0 | 0 | 19500 | 66.6 | 14400 | 49.7 | 1.34 |
| 69 | 17 | MA1471.1 | BARX2 | MA1471.1 | NWWAAYMATTAN | BARX2 | 0 | 0 | 22800 | 78.1 | 18100 | 62.4 | 1.25 |
| 70 | 10 | MA0785.1 | POU2F1 | MA0785.1 | AWTATGCWAATK | POU2F1 | 0 | 0 | 22700 | 77.7 | 18000 | 62 | 1.25 |
| 71 | 18 | MA1125.1 (ZNF384) | STREME-5 | 5-AAAAAA | AAAAAA | ZNF384 | 0 | 0 | 20500 | 70.1 | 15500 | 53.4 | 1.31 |
| 72 | 11 | MA1963.1 | SATB1 | MA1963.1 | WWDCTAATAAHWW | SATB1 | 0 | 0 | 20200 | 69.1 | 15300 | 52.6 | 1.31 |
| 73 | 19 | MA1603.1 | Dmrt1 | MA1603.1 | GHTACAAAGTADC | Dmrt1 | 0 | 0 | 24900 | 85 | 20700 | 71 | 1.2 |
| 74 | 14 | MA0041.2 | FOXD3 | MA0041.2 | WAWGHAAATAAACARY | FOXD3 | 0 | 0 | 25000 | 85.6 | 20900 | 72 | 1.19 |
| 75 | 20 | MA0025.2 | NFIL3 | MA0025.2 | NRTTATGYAATNN | NFIL3 | 0 | 0 | 24300 | 83.1 | 20000 | 68.9 | 1.21 |
| 76 | 10 | MA0627.2 | POU2F3 | MA0627.2 | NWTATGCAAATNH | POU2F3 | 0 | 0 | 24900 | 85 | 20700 | 71.3 | 1.19 |
| 77 | 21 | MA0602.1 | Arid5a | MA0602.1 | SYAATATTGVDANH | Arid5a | 0 | 0 | 21800 | 74.4 | 17100 | 58.7 | 1.27 |
| 78 | 14 | MA1607.1 | Foxl2 | MA1607.1 | DDWATGTAAACANN | Foxl2 | 0 | 0 | 23300 | 79.7 | 18900 | 64.9 | 1.23 |
| 79 | 20 | MA1636.1 | CEBPG | MA1636.1 | NDATGATGCAATMHH | CEBPG | 0 | 0 | 22200 | 75.8 | 17600 | 60.4 | 1.25 |
| 80 | 11 | MA1549.1 | POU6F1 | MA1549.1 | RTAATGAGVN | POU6F1 | 0 | 0 | 22900 | 78.1 | 18400 | 63.3 | 1.23 |
| 81 | 22 | 7-AAWWTT | STREME-7 | 7-AAWWTT | AAWWTT | AAWWTT | 0 | 0 | 21500 | 73.4 | 16900 | 58 | 1.26 |
| 82 | 23 | MA0611.2 | Dux | MA0611.2 | NTGATTBAATCAVWNN | Dux | 0 | 0 | 12700 | 43.4 | 8130 | 28 | 1.55 |
| 83 | 11 | MA0722.1 | VAX1 | MA0722.1 | YTAATTAH | VAX1 | 0 | 0 | 23200 | 79.4 | 18900 | 65 | 1.22 |
| 84 | 14 | MA1487.2 | FOXE1 | MA1487.2 | VYTAAAYAAACAAH | FOXE1 | 0 | 0 | 17300 | 59 | 12500 | 42.9 | 1.38 |
| 85 | 11 | MA0793.1 | POU6F2 | MA0793.1 | ASCTMATTAW | POU6F2 | 0 | 0 | 24100 | 82.3 | 19900 | 68.6 | 1.2 |
| 86 | 11 | MA0723.2 | VAX2 | MA0723.2 | BYAATTAV | VAX2 | 0 | 0 | 22200 | 75.7 | 17700 | 60.8 | 1.25 |
| 87 | 11 | MA0706.1 | MEOX2 | MA0706.1 | DSTAATTAWN | MEOX2 | 0 | 0 | 22000 | 75.1 | 17500 | 60.1 | 1.25 |
| 88 | 1 | MA0591.1 | Bach1::Mafk | MA0591.1 | RSSATGACTCAGCAB | Bach1::Mafk | 0 | 0 | 3300 | 11.3 | 931 | 3.2 | 3.52 |
| 89 | 24 | MA1152.1 | SOX15 | MA1152.1 | CYWTTGTTHW | SOX15 | 7.26276499386632e-322 | 7.26276499386632e-322 | 24500 | 83.6 | 20400 | 70.2 | 1.19 |
| 90 | 11 | MA0710.1 | NOTO | MA0710.1 | NYTAATTAVN | NOTO | 4.29837111881884e-321 | 4.29837111881884e-321 | 22700 | 77.6 | 18300 | 63.1 | 1.23 |
| 91 | 24 | MA0084.1 | SRY | MA0084.1 | NWWAACAAT | SRY | 1.12997753860351e-319 | 4.74303020007597e-322 | 20300 | 69.4 | 15700 | 54 | 1.29 |
| 92 | 25 | MA1124.1 | ZNF24 | MA1124.1 | CATTCATTCATTC | ZNF24 | 1.1899999929067e-316 | 5.16002160516598e-319 | 17300 | 59.1 | 12600 | 43.3 | 1.36 |
| 93 | 26 | MA0905.1 | HOXC10 | MA0905.1 | GTCRTAAAAH | HOXC10 | 1.34999999029226e-316 | 5.78002458413216e-319 | 24600 | 84 | 20600 | 70.9 | 1.19 |
| 94 | 10 | MA1115.1 | POU5F1 | MA1115.1 | WHATGCAAATN | POU5F1 | 2.57000000000105e-315 | 1.09000021687027e-317 | 25200 | 86.1 | 21400 | 73.6 | 1.17 |
| 95 | 14 | MA0148.4 | FOXA1 | MA0148.4 | WWGTAAACATNN | FOXA1 | 4.9999999998194e-314 | 2.09999999038868e-316 | 24500 | 83.8 | 20500 | 70.6 | 1.19 |
| 96 | 10 | MA0789.1 | POU3F4 | MA0789.1 | TATGCWAAT | POU3F4 | 7.03999999998287e-313 | 2.92000000169293e-315 | 23700 | 81.1 | 19600 | 67.4 | 1.2 |
| 97 | 26 | MA0908.1 | HOXD11 | MA0908.1 | RTCGTAAAAH | HOXD11 | 9.99999999999997e-311 | 4.12000000001917e-313 | 16400 | 56.2 | 11800 | 40.6 | 1.39 |
| 98 | 15 | MA0650.3 | Hoxa13 | MA0650.3 | NNCCAATAAAAN | Hoxa13 | 1.51e-310 | 6.11999999999634e-313 | 23100 | 79 | 18900 | 65 | 1.22 |
| 99 | 14 | MA0847.3 | FOXD2 | MA0847.3 | SYTAARYAAACA | FOXD2 | 2.11000000000002e-310 | 8.48000000001979e-313 | 20300 | 69.5 | 15800 | 54.3 | 1.28 |
| 100 | 10 | MA0792.1 | POU5F1B | MA0792.1 | TATGCWAAT | POU5F1B | 4.56000000000002e-310 | 1.81999999999849e-312 | 25100 | 85.8 | 21300 | 73.3 | 1.17 |
| 101 | 11 | MA0601.1 | Arid3b | MA0601.1 | ATATTAATWAN | Arid3b | 1.32e-307 | 5.18999999999999e-310 | 19800 | 67.8 | 15300 | 52.5 | 1.29 |
| 102 | 36 | MA0497.1 | MEF2C | MA0497.1 | DDDCYAAAAATAGMW | MEF2C | 1.86e-307 | 7.27000000000001e-310 | 21800 | 74.4 | 17400 | 59.8 | 1.24 |
| 103 | 7 | MA0851.1 | Foxj3 | MA0851.1 | NDAADGTAAACAAANNM | Foxj3 | 6.91e-304 | 2.67e-306 | 20600 | 70.4 | 16100 | 55.4 | 1.27 |
| 104 | 20 | MA0043.3 | HLF | MA0043.3 | NNRTTATGCAACHN | HLF | 4.91e-302 | 1.88e-304 | 21000 | 71.6 | 16500 | 56.9 | 1.26 |
| 105 | 15 | MA0465.2 | CDX2 | MA0465.2 | NDGCAATAAAWN | CDX2 | 2.12e-301 | 8.02e-304 | 23200 | 79.3 | 19100 | 65.6 | 1.21 |
| 106 | 11 | MA1481.1 | DRGX | MA1481.1 | YYAATTAN | DRGX | 7.91e-301 | 2.97e-303 | 21500 | 73.5 | 17100 | 58.9 | 1.25 |
| 107 | 20 | MA0102.4 | CEBPA | MA0102.4 | NDATTGCACAATHN | CEBPA | 2.38e-300 | 8.85e-303 | 19900 | 68.2 | 15500 | 53.1 | 1.28 |
| 108 | 27 | MA0495.3 | MAFF | MA0495.3 | DWGTCAGCATTTTWHN | MAFF | 1.75e-296 | 6.44e-299 | 24300 | 83.1 | 20400 | 70.2 | 1.18 |
| 109 | 28 | MA1657.1 | ZNF652 | MA1657.1 | BAAAGRGTTAAN | ZNF652 | 4.7e-295 | 1.71e-297 | 23400 | 79.9 | 19300 | 66.4 | 1.2 |
| 110 | 26 | MA0911.1 | Hoxa11 | MA0911.1 | RGTCGTAAAANT | Hoxa11 | 7.02e-293 | 2.54e-295 | 24600 | 84.2 | 20800 | 71.6 | 1.18 |
| 111 | 11 | MA0903.1 | HOXB3 | MA0903.1 | NNYMATTARN | HOXB3 | 1.5e-290 | 5.4e-293 | 21000 | 71.7 | 16700 | 57.3 | 1.25 |
| 112 | 14 | MA1489.1 | FOXN3 | MA1489.1 | GTAAACAA | FOXN3 | 2.34e-286 | 8.33e-289 | 22200 | 75.8 | 18000 | 61.8 | 1.22 |
| 113 | 11 | MA0068.2 | PAX4 | MA0068.2 | CTAATTAG | PAX4 | 1.5e-274 | 5.27e-277 | 23500 | 80.2 | 19600 | 67.3 | 1.19 |
| 114 | 29 | 14-ATGWTC | STREME-14 | 14-ATGWTC | 14-ATGWTC | ATGWTC | 1.97e-274 | 6.88e-277 | 21300 | 72.8 | 17100 | 58.8 | 1.24 |
| 115 | 13 | MA0757.1 | ONECUT3 | MA0757.1 | AAAAAATCRATAAH | ONECUT3 | 1.7e-273 | 5.88e-276 | 11600 | 39.7 | 7550 | 26 | 1.53 |
| 116 | 11 | MA0674.1 | NKX6-1 | MA0674.1 | NTMATTAA | NKX6-1 | 2.83e-273 | 9.70999999999999e-276 | 21500 | 73.6 | 17400 | 59.8 | 1.23 |
| 117 | 11 | MA0895.1 | HMBOX1 | MA0895.1 | MYTAGTTAMS | HMBOX1 | 4.02e-273 | 1.37e-275 | 22200 | 75.9 | 18100 | 62.4 | 1.22 |
| 118 | 11 | MA0899.1 | HOXA10 | MA0899.1 | DGYMATAAAAH | HOXA10 | 1.24e-272 | 4.2e-275 | 24900 | 85 | 21200 | 73.1 | 1.16 |
| 119 | 13 | MA0755.1 | CUX2 | MA0755.1 | TRATCGATAW | CUX2 | 6.39e-270 | 2.14e-272 | 22200 | 75.8 | 18100 | 62.3 | 1.22 |
| 120 | 11 | MA0910.2 | HOXD8 | MA0910.2 | GYMATTAM | HOXD8 | 2.7e-268 | 8.94e-271 | 17500 | 59.8 | 13200 | 45.3 | 1.32 |
| 121 | 14 | MA0031.1 | FOXD1 | MA0031.1 | GTAAACAW | FOXD1 | 6.13e-268 | 2.02e-270 | 23200 | 79.1 | 19300 | 66.2 | 1.2 |
| 122 | 14 | MA0047.3 | FOXA2 | MA0047.3 | RWGTAAACANN | FOXA2 | 1.17e-267 | 3.81e-270 | 23400 | 79.9 | 19500 | 67.1 | 1.19 |
| 123 | 30 | MA1623.1 | Stat2 | MA1623.1 | RGAAACAGAAASH | Stat2 | 3.03e-267 | 9.8e-270 | 26000 | 88.9 | 22700 | 78.2 | 1.14 |
| 124 | 11 | MA0658.1 | LHX6 | MA0658.1 | RCTAATTARY | LHX6 | 9.34999999999999e-267 | 3e-269 | 19800 | 67.8 | 15600 | 53.6 | 1.26 |
| 125 | 11 | MA0612.2 | EMX1 | MA0612.2 | BTAATTAS | EMX1 | 9.42e-267 | 3e-269 | 16300 | 55.6 | 12000 | 41.2 | 1.35 |
| 126 | 11 | MA0675.1 | NKX6-2 | MA0675.1 | NYMATTAA | NKX6-2 | 1.97e-266 | 6.22e-269 | 20300 | 69.3 | 16100 | 55.2 | 1.25 |
| 127 | 11 | MA0704.1 | Lhx4 | MA0704.1 | YTAATTAR | Lhx4 | 1.13e-265 | 3.53e-268 | 22000 | 75.1 | 17900 | 61.6 | 1.22 |
| 128 | 31 | MA0606.2 | Nfat5 | MA0606.2 | NNATGGAAAAWN | Nfat5 | 1.65e-263 | 5.12e-266 | 23900 | 81.6 | 20100 | 69.3 | 1.18 |
| 129 | 20 | MA0836.2 | CEBPD | MA0836.2 | NRTTGCACAAYHN | CEBPD | 4.16e-261 | 1.28e-263 | 20500 | 70.1 | 16400 | 56.3 | 1.25 |
| 130 | 30 | MA0050.3 | Irf1 | MA0050.3 | ANTGAAACTGAAASH | Irf1 | 7.16e-258 | 2.19e-260 | 26200 | 89.5 | 23000 | 79.2 | 1.13 |
| 131 | 14 | MA0040.1 | Foxq1 | MA0040.1 | HATTGTTTATW | Foxq1 | 1.62e-257 | 4.93e-260 | 12100 | 41.5 | 8120 | 27.9 | 1.49 |
| 132 | 24 | MA0868.2 | SOX8 | MA0868.2 | MGAACAATRG | SOX8 | 1.02e-256 | 3.08e-259 | 19100 | 65.4 | 14900 | 51.4 | 1.27 |
| 133 | 14 | MA0852.2 | FOXK1 | MA0852.2 | NRDGTAAACAAGNN | FOXK1 | 1.51e-256 | 4.51e-259 | 24600 | 84.1 | 21100 | 72.4 | 1.16 |
| 134 | 11 | MA0158.2 | HOXA5 | MA0158.2 | GYMATTAS | HOXA5 | 1.59e-253 | 4.73e-256 | 17200 | 58.7 | 13000 | 44.6 | 1.32 |
| 135 | 11 | MA1502.1 | HOXB8 | MA1502.1 | GYMATTAM | HOXB8 | 2.12e-251 | 6.26e-254 | 16500 | 56.5 | 12300 | 42.4 | 1.33 |
| 136 | 32 | MA0108.2 | TBP | MA0108.2 | STATAAAWRSVVVBN | TBP | 1.07e-250 | 3.14e-253 | 22900 | 78.1 | 19000 | 65.5 | 1.19 |
| 137 | 14 | MA0157.3 | Foxo3 | MA0157.3 | NDGTAAACARNN | Foxo3 | 1.19e-250 | 3.47e-253 | 25800 | 88.2 | 22600 | 77.7 | 1.14 |
| 138 | 11 | MA0887.1 | EVX1 | MA0887.1 | VVTAATTABS | EVX1 | 3.32e-250 | 9.59e-253 | 22100 | 75.5 | 18200 | 62.5 | 1.21 |
| 139 | 14 | MA1683.1 | FOXA3 | MA1683.1 | DWGTAAACANN | FOXA3 | 4.39e-249 | 1.26e-251 | 26500 | 90.6 | 23500 | 80.9 | 1.12 |
| 140 | 11 | MA1498.2 | HOXA7 | MA1498.2 | SYMATTAV | HOXA7 | 1.5e-248 | 4.25e-251 | 19600 | 67.2 | 15500 | 53.5 | 1.26 |
| 141 | 33 | MA1729.1 | ZNF680 | MA1729.1 | GNCCAAGAAGAATDA | ZNF680 | 4.45e-248 | 1.26e-250 | 18800 | 64.3 | 14700 | 50.4 | 1.27 |
| 142 | 11 | MA1463.1 | ARGFX | MA1463.1 | DCTAATTARM | ARGFX | 1.22e-247 | 3.41e-250 | 16000 | 54.8 | 11900 | 40.9 | 1.34 |
| 143 | 11 | MA1518.2 | Lhx1 | MA1518.2 | NNNTGCTAATTAGCANNN | Lhx1 | 4.03e-247 | 1.12e-249 | 22300 | 76.2 | 18400 | 63.4 | 1.2 |
| 144 | 14 | MA1103.2 | FOXK2 | MA1103.2 | DDGTAAACANN | FOXK2 | 6.55e-247 | 1.81e-249 | 26600 | 90.8 | 23600 | 81.2 | 1.12 |
| 145 | 11 | MA0716.1 | PRRX1 | MA0716.1 | YYAATTAR | PRRX1 | 3.16e-244 | 8.66999999999999e-247 | 21000 | 71.7 | 17000 | 58.5 | 1.23 |
| 146 | 14 | MA1606.1 | Foxf1 | MA1606.1 | DWRTAAACANN | Foxf1 | 8.49e-244 | 2.32e-246 | 26800 | 91.7 | 24000 | 82.5 | 1.11 |
| 147 | 11 | MA0892.1 | GSX1 | MA0892.1 | NCYMATTAVN | GSX1 | 7.05e-243 | 1.91e-245 | 23000 | 78.5 | 19200 | 66.1 | 1.19 |
| 148 | 11 | MA0718.1 | RAX | MA0718.1 | RYYAATTARH | RAX | 2.45e-242 | 6.58e-245 | 24400 | 83.5 | 20900 | 72 | 1.16 |
| 149 | 11 | MA0725.1 | VSX1 | MA0725.1 | YTAATTAN | VSX1 | 6.33e-241 | 1.69e-243 | 16800 | 57.5 | 12700 | 43.8 | 1.31 |
| 150 | 11 | MA0662.1 | MIXL1 | MA0662.1 | NBYAATTAVN | MIXL1 | 2.88e-240 | 7.65e-243 | 21800 | 74.5 | 17900 | 61.6 | 1.21 |
| 151 | 24 | MA0143.4 | SOX2 | MA0143.4 | DNACAATGGNN | SOX2 | 3.02e-240 | 7.97e-243 | 20200 | 69.2 | 16200 | 55.8 | 1.24 |
| 152 | 15 | MA0913.2 | HOXD9 | MA0913.2 | GYMATAAAAH | HOXD9 | 5.16e-240 | 1.35e-242 | 19200 | 65.6 | 15100 | 52.1 | 1.26 |
| 153 | 34 | MA0035.4 | GATA1 | MA0035.4 | NHCTAATCTDH | GATA1 | 7.54e-240 | 1.96e-242 | 22700 | 77.7 | 19000 | 65.4 | 1.19 |
| 154 | 11 | MA1505.1 | HOXC8 | MA1505.1 | KTAATTAM | HOXC8 | 1.45e-239 | 3.74e-242 | 16100 | 55 | 12000 | 41.3 | 1.33 |
| 155 | 11 | MA0132.2 | PDX1 | MA0132.2 | VYAATTAR | PDX1 | 2.49e-239 | 6.4e-242 | 15500 | 52.8 | 11400 | 39.2 | 1.35 |
| 156 | 11 | MA0726.1 | VSX2 | MA0726.1 | CTAATTAN | VSX2 | 6.4e-239 | 1.63e-241 | 24800 | 84.7 | 21400 | 73.6 | 1.15 |
| 157 | 11 | MA0125.1 | Nobox | MA0125.1 | TAATTRSY | Nobox | 4.51e-238 | 1.14e-240 | 21600 | 73.9 | 17800 | 61.1 | 1.21 |
| 158 | 10 | MA1962.1 | POU2F1::SOX2 | MA1962.1 | MATTTRCATMACAATRG | POU2F1::SOX2 | 6.9e-238 | 1.74e-240 | 11500 | 39.2 | 7660 | 26.3 | 1.49 |
| 159 | 11 | MA0654.1 | ISX | MA0654.1 | YYAATTAR | ISX | 2.85e-235 | 7.14e-238 | 20300 | 69.4 | 16400 | 56.2 | 1.23 |
| 160 | 11 | MA0635.1 | BARHL2 | MA0635.1 | VHTAAAYGRT | BARHL2 | 1.72e-234 | 4.28e-237 | 23400 | 80 | 19800 | 68 | 1.17 |
| 161 | 11 | MA0075.3 | PRRX2 | MA0075.3 | CTAATTAN | PRRX2 | 1.53e-233 | 3.78e-236 | 15400 | 52.6 | 11400 | 39 | 1.35 |
| 162 | 14 | MA0613.1 | FOXG1 | MA0613.1 | RTAAACAW | FOXG1 | 1.94e-233 | 4.78e-236 | 14800 | 50.5 | 10800 | 37.1 | 1.36 |
| 163 | 11 | MA0909.3 | Hoxd13 | MA0909.3 | NNCAATAAAW | Hoxd13 | 2.68e-231 | 6.54e-234 | 20400 | 69.6 | 16500 | 56.6 | 1.23 |
| 164 | 11 | MA0888.1 | EVX2 | MA0888.1 | VVTAATTAVB | EVX2 | 2.61e-230 | 6.34e-233 | 25900 | 88.7 | 22900 | 78.8 | 1.13 |
| 165 | 35 | MA1480.1 | DPRX | MA1480.1 | AGATAATCCN | DPRX | 5.04e-230 | 1.22e-232 | 26000 | 88.8 | 23000 | 78.9 | 1.12 |
| 166 | 11 | MA0720.1 | Shox2 | MA0720.1 | YYAATTAR | Shox2 | 1.39e-229 | 3.34e-232 | 19000 | 64.8 | 15000 | 51.5 | 1.26 |
| 167 | 14 | MA0481.3 | FOXP1 | MA0481.3 | NDGTAAACANH | FOXP1 | 1.11e-227 | 2.64e-230 | 27600 | 94.2 | 25100 | 86.3 | 1.09 |
| 168 | 11 | MA0721.1 | UNCX | MA0721.1 | YYAATTAV | UNCX | 1.55e-227 | 3.68e-230 | 22200 | 76 | 18500 | 63.8 | 1.19 |
| 169 | 11 | MA0027.2 | EN1 | MA0027.2 | SYAATTAV | EN1 | 8.04e-227 | 1.89e-229 | 25700 | 88 | 22700 | 77.9 | 1.13 |
| 170 | 11 | MA0700.2 | LHX2 | MA0700.2 | DDCCAATTADN | LHX2 | 1.62e-225 | 3.79e-228 | 21300 | 72.7 | 17500 | 60.1 | 1.21 |
| 171 | 11 | MA0717.1 | RAX2 | MA0717.1 | YYAATTAV | RAX2 | 1.64e-225 | 3.81e-228 | 21700 | 74.3 | 18000 | 61.8 | 1.2 |
| 172 | 11 | MA0882.1 | DLX6 | MA0882.1 | VYAATTAN | DLX6 | 5.62e-225 | 1.3e-227 | 25000 | 85.4 | 21800 | 74.8 | 1.14 |
| 173 | 30 | MA1709.1 | ZIM3 | MA1709.1 | RVMAACAGAAACCYM | ZIM3 | 2.41e-223 | 5.56e-226 | 22400 | 76.6 | 18800 | 64.5 | 1.19 |
| 174 | 20 | MA0833.2 | ATF4 | MA0833.2 | DDATGATGCAATMN | ATF4 | 4.47e-223 | 1.02e-225 | 19800 | 67.6 | 15900 | 54.7 | 1.24 |
| 175 | 35 | MA0682.2 | PITX1 | MA0682.2 | YTAATCCH | PITX1 | 4.34e-221 | 9.86999999999999e-224 | 18700 | 64 | 14800 | 51 | 1.26 |
| 176 | 4 | MA0683.1 | POU4F2 | MA0683.1 | RTGCATAATTAATGAG | POU4F2 | 1.33e-220 | 3.01e-223 | 7240 | 24.7 | 4160 | 14.3 | 1.73 |
| 177 | 11 | MA0661.1 | MEOX1 | MA0661.1 | VSTAATTAHC | MEOX1 | 3.17e-220 | 7.13e-223 | 25200 | 86.3 | 22100 | 76 | 1.14 |
| 178 | 24 | MA0077.1 | SOX9 | MA0077.1 | CYATTGTTY | SOX9 | 3.71e-220 | 8.3e-223 | 22900 | 78.4 | 19400 | 66.6 | 1.18 |
| 179 | 14 | MA0042.2 | FOXI1 | MA0042.2 | GTAAACA | FOXI1 | 8.2e-220 | 1.82e-222 | 25500 | 87.2 | 22500 | 77.2 | 1.13 |
| 180 | 11 | MA0880.1 | Dlx3 | MA0880.1 | NYAATTAN | Dlx3 | 5.42e-219 | 1.2e-221 | 16400 | 56.1 | 12500 | 43 | 1.31 |
| 181 | 11 | MA0634.1 | ALX3 | MA0634.1 | YYYAATTANV | ALX3 | 1.03e-217 | 2.27e-220 | 24700 | 84.3 | 21400 | 73.6 | 1.14 |
| 182 | 17 | MA0780.1 | PAX3 | MA0780.1 | TAATYRATTA | PAX3 | 4.13e-217 | 9.02999999999999e-220 | 9870 | 33.7 | 6400 | 22 | 1.53 |
| 183 | 11 | MA0644.2 | ESX1 | MA0644.2 | YYAATTAN | ESX1 | 6.66e-217 | 1.45e-219 | 21100 | 72.2 | 17400 | 59.8 | 1.21 |
| 184 | 24 | MA0078.2 | Sox17 | MA0078.2 | NNAGAACAATGGNN | Sox17 | 8.35e-215 | 1.81e-217 | 21500 | 73.4 | 17800 | 61.2 | 1.2 |
| 185 | 11 | MA0890.1 | GBX2 | MA0890.1 | RSYAATTARN | GBX2 | 9.25999999999999e-215 | 1.99e-217 | 25800 | 88.1 | 22800 | 78.4 | 1.12 |
| 186 | 11 | MA0881.1 | Dlx4 | MA0881.1 | VYAATTAN | Dlx4 | 9.97e-215 | 2.13e-217 | 22200 | 75.9 | 18600 | 64 | 1.19 |
| 187 | 36 | MA0052.4 (MEF2A) | MEME-4 | WWYTAWAAATA | WWYTAWAAATA | MEF2A | 1.01e-214 | 2.15e-217 | 12700 | 43.4 | 8970 | 30.8 | 1.41 |
| 188 | 11 | MA0724.1 | VENTX | MA0724.1 | AMCGATTAR | VENTX | 1.29e-214 | 2.72e-217 | 18800 | 64.4 | 15000 | 51.6 | 1.25 |
| 189 | 11 | MA1476.2 | Dlx5 | MA1476.2 | NDGCAATTAGNN | Dlx5 | 1.38e-214 | 2.91e-217 | 23800 | 81.4 | 20500 | 70.3 | 1.16 |
| 190 | 11 | MA0874.1 | Arx | MA0874.1 | VTBCRYTAATTARTKSW | Arx | 5.77e-213 | 1.21e-215 | 12900 | 44.2 | 9190 | 31.6 | 1.4 |
| 191 | 11 | MA0876.1 | BSX | MA0876.1 | SYVATTAV | BSX | 7.35e-212 | 1.53e-214 | 23000 | 78.5 | 19500 | 67 | 1.17 |
| 192 | 24 | MA0867.2 | SOX4 | MA0867.2 | RAACAAAGRV | SOX4 | 1.31e-211 | 2.71e-214 | 15200 | 51.8 | 11300 | 39 | 1.33 |
| 193 | 20 | MA0838.1 | CEBPG | MA0838.1 | ATTRCGCAAY | CEBPG | 3.26e-211 | 6.73e-214 | 12400 | 42.3 | 8690 | 29.9 | 1.41 |
| 194 | 11 | MA0648.1 | GSC | MA0648.1 | VYTAATCCSY | GSC | 1.85e-210 | 3.8e-213 | 26100 | 89.4 | 23300 | 80.2 | 1.11 |
| 195 | 11 | MA0630.1 | SHOX | MA0630.1 | HTAATTRR | SHOX | 7.4e-210 | 1.51e-212 | 26800 | 91.6 | 24200 | 83 | 1.1 |
| 196 | 11 | MA0885.2 | Dlx2 | MA0885.2 | NDGCAATTAGNN | Dlx2 | 6.77e-209 | 1.37e-211 | 23800 | 81.3 | 20400 | 70.3 | 1.16 |
| 197 | 13 | MA0754.2 | CUX1 | MA0754.2 | TAATCGATAM | CUX1 | 1.79e-208 | 3.61e-211 | 17600 | 60.2 | 13800 | 47.4 | 1.27 |
| 198 | 11 | MA1497.1 | HOXA6 | MA1497.1 | BGYMATTANN | HOXA6 | 3.04e-208 | 6.11e-211 | 22800 | 77.8 | 19300 | 66.3 | 1.17 |
| 199 | 4 | MA0854.1 | Alx1 | MA0854.1 | CGMRYTAATTARTNMNY | Alx1 | 3.25e-208 | 6.51e-211 | 15100 | 51.5 | 11300 | 38.7 | 1.33 |
| 200 | 26 | MA0907.1 | HOXC13 | MA0907.1 | BCTCGTAAAAH | HOXC13 | 3.76e-208 | 7.49e-211 | 15800 | 54 | 12000 | 41.2 | 1.31 |
| 201 | 11 | MA0891.1 | GSC2 | MA0891.1 | NYTAATCCBH | GSC2 | 1.68e-207 | 3.32e-210 | 23700 | 81.1 | 20400 | 70.2 | 1.16 |
| 202 | 4 | MA0853.1 | Alx4 | MA0853.1 | CGCRYTAATTARYHNNY | Alx4 | 2.46e-207 | 4.86e-210 | 16400 | 55.9 | 12500 | 43.1 | 1.3 |
| 203 | 30 | MA0772.1 | IRF7 | MA0772.1 | HCGAAARYGAAAVT | IRF7 | 1.21e-206 | 2.38e-209 | 11600 | 39.7 | 8030 | 27.6 | 1.44 |
| 204 | 34 | MA1104.2 | GATA6 | MA1104.2 | HWWTCTTATCTNH | GATA6 | 7.69e-205 | 1.5e-207 | 27000 | 92.2 | 24500 | 84.1 | 1.1 |
| 205 | 11 | MA0894.1 | HESX1 | MA0894.1 | GHTAATTRRH | HESX1 | 5.11e-204 | 9.93e-207 | 21400 | 73.2 | 17800 | 61.3 | 1.19 |
| 206 | 37 | MA0898.1 | Hmx3 | MA0898.1 | ASAAGCAATTAAWRRAT | Hmx3 | 6.04e-204 | 1.17e-206 | 20100 | 68.7 | 16400 | 56.4 | 1.22 |
| 207 | 11 | MA0875.1 | BARX1 | MA0875.1 | VCMATTAV | BARX1 | 1.16e-203 | 2.24e-206 | 14400 | 49.4 | 10700 | 36.8 | 1.34 |
| 208 | 14 | MA0848.1 | FOXO4 | MA0848.1 | GTAAACA | FOXO4 | 4.89e-203 | 9.35e-206 | 25500 | 87.3 | 22600 | 77.7 | 1.12 |
| 209 | 19 | MA0442.2 | SOX10 | MA0442.2 | NDAACAAAGVN | SOX10 | 3.28e-202 | 6.25e-205 | 18400 | 63 | 14700 | 50.4 | 1.25 |
| 210 | 24 | MA1120.1 | SOX13 | MA1120.1 | DVACAATGGNN | SOX13 | 1.78e-200 | 3.37e-203 | 17600 | 60.1 | 13800 | 47.5 | 1.26 |
| 211 | 15 | MA0878.3 | CDX1 | MA0878.3 | GGYMATAAAA | CDX1 | 2.57e-200 | 4.86e-203 | 15700 | 53.6 | 12000 | 41.1 | 1.31 |
| 212 | 26 | MA0906.1 | HOXC12 | MA0906.1 | RGTCGTAAAAH | HOXC12 | 8.65999999999999e-200 | 1.63e-202 | 12900 | 44.3 | 9320 | 32.1 | 1.38 |
| 213 | 14 | MA0480.2 | Foxo1 | MA0480.2 | NDGTAAACANN | Foxo1 | 3.37e-199 | 6.29e-202 | 23500 | 80.2 | 20200 | 69.3 | 1.16 |
| 214 | 11 | MA0709.1 | Msx3 | MA0709.1 | SCAATTAN | Msx3 | 1.35e-198 | 2.5e-201 | 21800 | 74.4 | 18200 | 62.7 | 1.19 |
| 215 | 11 | MA1499.1 | HOXB4 | MA1499.1 | RTMATTAR | HOXB4 | 3.12e-196 | 5.78e-199 | 17300 | 59.2 | 13600 | 46.8 | 1.27 |
| 216 | 11 | MA0712.2 | OTX2 | MA0712.2 | NDRGGATTARNN | OTX2 | 1.76e-194 | 3.25e-197 | 26800 | 91.7 | 24300 | 83.6 | 1.1 |
| 217 | 11 | MA1495.1 | HOXA1 | MA1495.1 | STAATTAS | HOXA1 | 4.77e-194 | 8.75e-197 | 12900 | 44.1 | 9320 | 32.1 | 1.37 |
| 218 | 11 | MA0889.1 | GBX1 | MA0889.1 | VCYAATTASY | GBX1 | 3.94e-191 | 7.2e-194 | 20700 | 70.7 | 17200 | 59 | 1.2 |
| 219 | 11 | MA1504.1 | HOXC4 | MA1504.1 | VTMATTAN | HOXC4 | 5.27e-191 | 9.57e-194 | 17600 | 60.2 | 13900 | 48 | 1.26 |
| 220 | 30 | MA1952.1 | FOXJ2::ELF1 | MA1952.1 | RTAAACMGGAAGTR | FOXJ2::ELF1 | 7.28e-191 | 1.32e-193 | 23200 | 79.3 | 19900 | 68.6 | 1.16 |
| 221 | 11 | MA0707.2 | MNX1 | MA0707.2 | VYRATTAK | MNX1 | 1.01e-190 | 1.82e-193 | 17000 | 58 | 13300 | 45.8 | 1.27 |
| 222 | 4 | MA0135.1 | Lhx3 | MA0135.1 | WAATTAATTAWYY | Lhx3 | 2.01e-190 | 3.6e-193 | 7850 | 26.8 | 4860 | 16.7 | 1.6 |
| 223 | 11 | MA1530.1 | NKX6-3 | MA1530.1 | VYCATTAWW | NKX6-3 | 6.27e-190 | 1.12e-192 | 14600 | 50 | 11000 | 37.9 | 1.32 |
| 224 | 14 | MA0849.1 | FOXO6 | MA0849.1 | GTAAACA | FOXO6 | 9.85e-190 | 1.75e-192 | 20900 | 71.5 | 17400 | 59.9 | 1.19 |
| 225 | 14 | MA0614.1 | Foxj2 | MA0614.1 | RTAAACAA | Foxj2 | 6.79e-188 | 1.2e-190 | 18500 | 63.2 | 14900 | 51.2 | 1.24 |
| 226 | 11 | MA1496.1 | HOXA4 | MA1496.1 | RTMATTAV | HOXA4 | 2.02e-187 | 3.55e-190 | 19000 | 65 | 15400 | 53.1 | 1.23 |
| 227 | 6 | MA1126.1 | FOS::JUN | MA1126.1 | DRTGACGTCATHNDTN | FOS::JUN | 3.57e-187 | 6.26e-190 | 24300 | 83 | 21200 | 73 | 1.14 |
| 228 | 38 | MA0152.2 (Nfatc2) | STREME-24 | 24-AAAAGGAAA | 24-AAAAGGAAA | Nfatc2 | 6.08e-187 | 1.06e-189 | 17600 | 60.1 | 14000 | 48 | 1.25 |
| 229 | 11 | MA0879.2 | DLX1 | MA0879.2 | VYAATTAN | DLX1 | 6.98e-187 | 1.21e-189 | 25200 | 86 | 22300 | 76.5 | 1.12 |
| 230 | 39 | MA1974.1 | ZNF211 | MA1974.1 | HWYATATACCAYRYW | ZNF211 | 6.12e-186 | 1.06e-188 | 16500 | 56.5 | 12900 | 44.4 | 1.27 |
| 231 | 37 | MA0897.1 | Hmx2 | MA0897.1 | ASAAGCAATTAAHVVRT | Hmx2 | 1.8e-184 | 3.1e-187 | 19500 | 66.6 | 15900 | 54.8 | 1.22 |
| 232 | 11 | MA1577.1 | TLX2 | MA1577.1 | BYAATTAR | TLX2 | 1.16e-182 | 2e-185 | 12600 | 43 | 9130 | 31.4 | 1.37 |
| 233 | 34 | MA1970.1 | TRPS1 | MA1970.1 | NHTCTTATCTNH | TRPS1 | 2.24e-182 | 3.83e-185 | 26000 | 88.8 | 23300 | 80.2 | 1.11 |
| 234 | 40 | MA1479.1 | DMRTC2 | MA1479.1 | AATTGMTACATT | DMRTC2 | 1.08e-181 | 1.84e-184 | 21000 | 71.7 | 17600 | 60.4 | 1.19 |
| 235 | 24 | MA0514.2 | Sox3 | MA0514.2 | DNACAATGGNN | Sox3 | 1.41e-181 | 2.39e-184 | 16000 | 54.8 | 12500 | 42.9 | 1.28 |
| 236 | 41 | MA0520.1 | Stat6 | MA0520.1 | BDBTTCCWSAGAAVY | Stat6 | 3.63e-181 | 6.12e-184 | 11700 | 40 | 8330 | 28.6 | 1.4 |
| 237 | 42 | MA1562.1 | SOX14 | MA1562.1 | CCGAACAATG | SOX14 | 3.79e-181 | 6.37e-184 | 21100 | 72.2 | 17700 | 61 | 1.18 |
| 238 | 30 | MA0152.2 | Nfatc2 | MA0152.2 | NNRNAATGGAAANWNH | Nfatc2 | 5.01e-181 | 8.38e-184 | 21700 | 74.3 | 18400 | 63.2 | 1.18 |
| 239 | 43 | MA0144.2 | STAT3 | MA0144.2 | YTTCYKGGAAD | STAT3 | 3.53e-180 | 5.87e-183 | 21000 | 71.7 | 17600 | 60.4 | 1.19 |
| 240 | 44 | MA0687.1 | SPIC | MA0687.1 | HAAAAGVGGAAGTA | SPIC | 3.54e-180 | 5.88e-183 | 22900 | 78.3 | 19700 | 67.7 | 1.16 |
| 241 | 11 | MA0618.1 | LBX1 | MA0618.1 | TTAATTAG | LBX1 | 7.59e-180 | 1.25e-182 | 16500 | 56.4 | 12900 | 44.5 | 1.27 |
| 242 | 11 | MA0904.2 | HOXB5 | MA0904.2 | STAATTAS | HOXB5 | 3.14e-178 | 5.16e-181 | 12200 | 41.7 | 8820 | 30.3 | 1.38 |
| 243 | 30 | MA0517.1 | STAT1::STAT2 | MA0517.1 | THAGTTTCRKTTTCY | STAT1::STAT2 | 4.59e-178 | 7.52e-181 | 17300 | 59 | 13700 | 47.1 | 1.25 |
| 244 | 11 | MA0912.2 | HOXD3 | MA0912.2 | CTAATTAC | HOXD3 | 8.6e-178 | 1.4e-180 | 13300 | 45.4 | 9850 | 33.9 | 1.34 |
| 245 | 26 | MA0594.2 | HOXA9 | MA0594.2 | RTCGTWAHNN | HOXA9 | 7.89e-177 | 1.28e-179 | 21800 | 74.5 | 18500 | 63.6 | 1.17 |
| 246 | 11 | MA1500.1 | HOXB6 | MA1500.1 | BTAATKRC | HOXB6 | 4.99e-176 | 8.08e-179 | 19200 | 65.6 | 15700 | 54 | 1.21 |
| 247 | 14 | MA0033.2 | FOXL1 | MA0033.2 | RTAAACA | FOXL1 | 5.51e-176 | 8.88e-179 | 26100 | 89.1 | 23500 | 80.7 | 1.1 |
| 248 | 45 | MA0523.1 | TCF7L2 | MA0523.1 | DRASATCAAAGRVA | TCF7L2 | 1.01e-175 | 1.62e-178 | 22000 | 75.1 | 18700 | 64.3 | 1.17 |
| 249 | 14 | MA0593.1 | FOXP2 | MA0593.1 | RWGTAAACAVR | FOXP2 | 1.82e-174 | 2.91e-177 | 9280 | 31.7 | 6220 | 21.4 | 1.48 |
| 250 | 37 | MA0151.1 | Arid3a | MA0151.1 | ATYAAA | Arid3a | 7.66e-173 | 1.22e-175 | 15700 | 53.6 | 12200 | 41.9 | 1.28 |
| 251 | 11 | MA0642.2 | EN2 | MA0642.2 | VSYAATTA | EN2 | 4.86e-171 | 7.7e-174 | 15000 | 51.4 | 11600 | 39.8 | 1.29 |
| 252 | 14 | MA0850.1 | FOXP3 | MA0850.1 | RTAAACA | FOXP3 | 6.45e-170 | 1.02e-172 | 24100 | 82.4 | 21200 | 72.7 | 1.13 |
| 253 | 11 | MA1608.1 | Isl1 | MA1608.1 | NNCCATTAGNN | Isl1 | 1.21e-169 | 1.9e-172 | 22600 | 77.3 | 19500 | 66.9 | 1.16 |
| 254 | 11 | MA0699.1 | LBX2 | MA0699.1 | NCYAATTARN | LBX2 | 1.24e-169 | 1.94e-172 | 26600 | 90.9 | 24200 | 83.2 | 1.09 |
| 255 | 37 | MA0122.3 | Nkx3-2 | MA0122.3 | WWAAMCACTTAAN | Nkx3-2 | 9.75e-168 | 1.52e-170 | 25300 | 86.6 | 22600 | 77.8 | 1.11 |
| 256 | 46 | MA0629.1 | Rhox11 | MA0629.1 | ANKNCGCTGTWAWDVRW | Rhox11 | 4.11e-167 | 6.38e-170 | 25300 | 86.6 | 22600 | 77.8 | 1.11 |
| 257 | 26 | MA0873.1 | HOXD12 | MA0873.1 | RGTCGTAAAAH | HOXD12 | 2.57e-166 | 3.97e-169 | 11600 | 39.7 | 8380 | 28.8 | 1.38 |
| 258 | 34 | MA0036.3 | GATA2 | MA0036.3 | NBCTTATCTNH | GATA2 | 5.77e-166 | 8.91e-169 | 24900 | 85.3 | 22200 | 76.3 | 1.12 |
| 259 | 11 | MA0900.2 | HOXA2 | MA0900.2 | STAATTAS | HOXA2 | 1.11e-165 | 1.71e-168 | 12200 | 41.8 | 8950 | 30.8 | 1.36 |
| 260 | 34 | MA0037.4 | Gata3 | MA0037.4 | NHTCTTATCTNH | Gata3 | 1.05e-164 | 1.61e-167 | 25400 | 86.7 | 22700 | 78.1 | 1.11 |
| 261 | 11 | MA0893.2 | GSX2 | MA0893.2 | SYMATTAR | GSX2 | 2e-164 | 3.05e-167 | 18300 | 62.5 | 14900 | 51.2 | 1.22 |
| 262 | 11 | MA1519.1 | LHX5 | MA1519.1 | STAATTAV | LHX5 | 5.71e-163 | 8.68e-166 | 14800 | 50.6 | 11400 | 39.3 | 1.29 |
| 263 | 45 | MA0768.2 | Lef1 | MA0768.2 | NNBCCTTTGATSTN | Lef1 | 3.02e-162 | 4.56e-165 | 20100 | 68.6 | 16800 | 57.7 | 1.19 |
| 264 | 11 | MA1507.1 | HOXD4 | MA1507.1 | VTMATTAD | HOXD4 | 3.25e-161 | 4.9e-164 | 12000 | 41.2 | 8820 | 30.4 | 1.36 |
| 265 | 11 | MA0701.2 | LHX9 | MA0701.2 | CYAATTAR | LHX9 | 3.32e-161 | 4.98e-164 | 11100 | 37.8 | 7930 | 27.3 | 1.39 |
| 266 | 47 | MA0069.1 | PAX6 | MA0069.1 | TTCACGCWTSANTK | PAX6 | 1.11e-160 | 1.66e-163 | 24600 | 84.2 | 21800 | 75.1 | 1.12 |
| 267 | 48 | MA0877.3 | BARHL1 | MA0877.3 | AMCGTTTA | BARHL1 | 1.4e-160 | 2.08e-163 | 26300 | 89.8 | 23800 | 81.9 | 1.1 |
| 268 | 45 | MA0769.2 | TCF7 | MA0769.2 | NNCTTTGAWVT | TCF7 | 1.39e-156 | 2.06e-159 | 25600 | 87.7 | 23100 | 79.4 | 1.1 |
| 269 | 23 | MA0884.2 | DUXA | MA0884.2 | YTRATTWAATYAR | DUXA | 1.56e-155 | 2.3e-158 | 6110 | 20.9 | 3670 | 12.6 | 1.65 |
| 270 | 34 | MA0482.2 | GATA4 | MA0482.2 | NNCCTTATCTNH | GATA4 | 3.37e-153 | 4.97e-156 | 26400 | 90.3 | 24100 | 82.8 | 1.09 |
| 271 | 11 | MA0666.2 | MSX1 | MA0666.2 | SCAATTAR | MSX1 | 8.4e-149 | 1.23e-151 | 22900 | 78.4 | 20000 | 68.9 | 1.14 |
| 272 | 6 | MA0843.1 | TEF | MA0843.1 | BRTTACRTAAYM | TEF | 4.2e-148 | 6.14e-151 | 10500 | 35.8 | 7500 | 25.8 | 1.39 |
| 273 | 11 | MA0621.1 | mix-a | MA0621.1 | NDTTAATTAGN | mix-a | 2e-147 | 2.92e-150 | 7860 | 26.9 | 5200 | 17.9 | 1.5 |
| 274 | 35 | MA0714.1 | PITX3 | MA0714.1 | NYTAATCCC | PITX3 | 3.64e-146 | 5.28e-149 | 26400 | 90.1 | 24100 | 82.8 | 1.09 |
| 275 | 49 | MA1588.1 | ZNF136 | MA1588.1 | RWATTCTTGGTTGRC | ZNF136 | 1.26e-145 | 1.83e-148 | 16400 | 55.9 | 13100 | 45.2 | 1.24 |
| 276 | 11 | MA0708.2 | MSX2 | MA0708.2 | SCAATTAV | MSX2 | 2.3e-145 | 3.31e-148 | 18600 | 63.5 | 15400 | 52.9 | 1.2 |
| 277 | 11 | MA0467.2 | Crx | MA0467.2 | NDGGATTANN | Crx | 4.04e-145 | 5.81e-148 | 20200 | 68.9 | 17100 | 58.6 | 1.18 |
| 278 | 14 | MA0030.1 | FOXF2 | MA0030.1 | BNAASGTAAACAAD | FOXF2 | 6.48e-145 | 9.28e-148 | 7380 | 25.2 | 4810 | 16.5 | 1.53 |
| 279 | 50 | MA1119.1 | SIX2 | MA1119.1 | NNCTGAAACCTGATMY | SIX2 | 1.26e-143 | 1.79e-146 | 19400 | 66.4 | 16300 | 56.1 | 1.19 |
| 280 | 51 | 25-GTATAV | STREME-25 | 25-GTATAV | 25-GTATAV | GTATAV | 5.7e-140 | 8.11e-143 | 18300 | 62.5 | 15100 | 52.1 | 1.2 |
| 281 | 50 | MA1118.1 | SIX1 | MA1118.1 | GWAACCTGAKM | SIX1 | 9.75e-138 | 1.38e-140 | 19800 | 67.8 | 16800 | 57.8 | 1.17 |
| 282 | 31 | MA0624.2 | Nfatc1 | MA0624.2 | NVATGGAAAAWN | Nfatc1 | 1.2e-135 | 1.69e-138 | 19600 | 67.1 | 16600 | 57 | 1.18 |
| 283 | 35 | MA0711.1 | OTX1 | MA0711.1 | HTAATCCS | OTX1 | 6.92e-135 | 9.73e-138 | 26000 | 88.8 | 23700 | 81.5 | 1.09 |
| 284 | 11 | MA0705.1 | Lhx8 | MA0705.1 | CTAATTAV | Lhx8 | 3.86e-131 | 5.42e-134 | 15400 | 52.5 | 12300 | 42.3 | 1.24 |
| 285 | 35 | MA0883.1 | Dmbx1 | MA0883.1 | WDRWNMGGATTADKNNW | Dmbx1 | 5.71e-129 | 7.97e-132 | 16300 | 55.8 | 13300 | 45.7 | 1.22 |
| 286 | 24 | MA0515.1 | Sox6 | MA0515.1 | CCWTTGTYYY | Sox6 | 9.11e-127 | 1.27e-129 | 8840 | 30.2 | 6240 | 21.4 | 1.41 |
| 287 | 37 | MA0896.1 | Hmx1 | MA0896.1 | VSVAGCAATTAAWGVDB | Hmx1 | 5.86e-126 | 8.13e-129 | 19500 | 66.8 | 16600 | 57.1 | 1.17 |
| 288 | 6 | MA0639.1 | DBP | MA0639.1 | NRTTACGTAAYV | DBP | 1.38e-125 | 1.91e-128 | 9160 | 31.3 | 6540 | 22.5 | 1.39 |
| 289 | 24 | MA0869.2 | Sox11 | MA0869.2 | NRGAACAAAGVV | Sox11 | 2.56e-123 | 3.52e-126 | 20700 | 70.8 | 17900 | 61.4 | 1.15 |
| 290 | 24 | MA0087.2 | Sox5 | MA0087.2 | VRVAACAATGGNN | Sox5 | 8.06e-123 | 1.11e-125 | 19200 | 65.6 | 16300 | 56 | 1.17 |
| 291 | 52 | MA0678.1 | OLIG2 | MA0678.1 | AMCATATGKT | OLIG2 | 8.15e-123 | 1.11e-125 | 19000 | 64.9 | 16100 | 55.2 | 1.17 |
| 292 | 37 | MA0914.1 | ISL2 | MA0914.1 | GCAMTTAR | ISL2 | 5.15e-120 | 7.02e-123 | 26300 | 89.8 | 24200 | 83.2 | 1.08 |
| 293 | 45 | MA1991.1 | Hnf1A | MA1991.1 | NBCCTTTGATSTBN | Hnf1A | 2.55e-119 | 3.47e-122 | 22400 | 76.6 | 19800 | 68 | 1.13 |
| 294 | 24 | MA1563.2 | SOX18 | MA1563.2 | AACAATDV | SOX18 | 1.11e-118 | 1.5e-121 | 19200 | 65.5 | 16300 | 56 | 1.17 |
| 295 | 27 | MA0117.2 | Mafb | MA0117.2 | AWWNTGCTGACD | Mafb | 3.83e-114 | 5.16e-117 | 24500 | 83.9 | 22200 | 76.3 | 1.1 |
| 296 | 6 | MA1150.1 | RORB | MA1150.1 | AWWTRGGTCAH | RORB | 4.48e-114 | 6.02e-117 | 20800 | 71.2 | 18100 | 62.2 | 1.14 |
| 297 | 53 | MA0693.3 | Vdr | MA0693.3 | NNGAGTTCANN | Vdr | 2.36e-113 | 3.16e-116 | 25400 | 86.9 | 23200 | 79.9 | 1.09 |
| 298 | 27 | MA1520.1 | MAF | MA1520.1 | NTGCTGASTCAGCAH | MAF | 5.04e-112 | 6.73e-115 | 24500 | 83.6 | 22100 | 76 | 1.1 |
| 299 | 11 | MA0702.2 | LMX1A | MA0702.2 | TTAATTAM | LMX1A | 1.22e-111 | 1.63e-114 | 9340 | 31.9 | 6830 | 23.5 | 1.36 |
| 300 | 54 | MA0095.3 | Yy1 | MA0095.3 | NNCAAAATGGCN | Yy1 | 5.55e-111 | 7.36e-114 | 15800 | 54 | 13000 | 44.7 | 1.21 |
| 301 | 26 | MA1506.1 | HOXD10 | MA1506.1 | RGTCGTAAAAH | HOXD10 | 1.81e-110 | 2.39e-113 | 8300 | 28.4 | 5920 | 20.4 | 1.39 |
| 302 | 20 | MA0837.2 | CEBPE | MA0837.2 | KATTGCGCAATM | CEBPE | 6.44e-106 | 8.49e-109 | 3920 | 13.4 | 2270 | 7.8 | 1.72 |
| 303 | 6 | MA1127.1 | FOSB::JUN | MA1127.1 | GATGACGTCAT | FOSB::JUN | 1.29e-105 | 1.7e-108 | 23700 | 80.8 | 21300 | 73.2 | 1.11 |
| 304 | 6 | MA1131.1 | FOSL2::JUN | MA1131.1 | GRTGACGTMAT | FOSL2::JUN | 4.21e-105 | 5.51e-108 | 22800 | 77.8 | 20300 | 69.8 | 1.11 |
| 305 | 11 | MA1501.1 | HOXB7 | MA1501.1 | GGTAATTAVS | HOXB7 | 4.67e-105 | 6.1e-108 | 16700 | 57 | 13900 | 47.9 | 1.19 |
| 306 | 41 | MA1625.1 | Stat5b | MA1625.1 | NNNTTCCCAGAANNN | Stat5b | 7.1e-105 | 9.24e-108 | 22800 | 77.8 | 20300 | 69.8 | 1.11 |
| 307 | 37 | MA0070.1 | PBX1 | MA0070.1 | HCATCAATCAAW | PBX1 | 2.76e-104 | 3.57e-107 | 14400 | 49.2 | 11700 | 40.2 | 1.22 |
| 308 | 40 | MA1707.1 | DMRTA1 | MA1707.1 | AWTTGWTACAWT | DMRTA1 | 3.3e-101 | 4.27e-104 | 18700 | 63.8 | 16000 | 55 | 1.16 |
| 309 | 55 | MA0619.1 | LIN54 | MA0619.1 | DTTTRAATH | LIN54 | 4.46e-101 | 5.75e-104 | 6600 | 22.6 | 4520 | 15.5 | 1.45 |
| 310 | 42 | MA1561.1 | SOX12 | MA1561.1 | ACCGAACAATV | SOX12 | 2.09e-98 | 2.68e-101 | 20100 | 68.8 | 17600 | 60.4 | 1.14 |
| 311 | 26 | MA1503.1 | HOXB9 | MA1503.1 | GTCGTAAAAH | HOXB9 | 3.3e-98 | 4.23e-101 | 5330 | 18.2 | 3470 | 11.9 | 1.53 |
| 312 | 56 | MA0114.4 (HNF4A) | STREME-23 | 23-AAGTCCAW | 23-AAGTCCAW | HNF4A | 2.09e-94 | 2.67e-97 | 17500 | 59.8 | 14900 | 51.2 | 1.17 |
| 313 | 41 | MA0518.1 | Stat4 | MA0518.1 | YTTCYRGGAARNVR | Stat4 | 2.71e-94 | 3.44e-97 | 15900 | 54.3 | 13300 | 45.6 | 1.19 |
| 314 | 40 | MA1478.1 | DMRTA2 | MA1478.1 | AATTGHTACAWT | DMRTA2 | 2.39e-93 | 3.04e-96 | 17300 | 59.2 | 14700 | 50.6 | 1.17 |
| 315 | 23 | MA0468.1 | DUX4 | MA0468.1 | TAAYYYAATCA | DUX4 | 3.89e-92 | 4.92e-95 | 6500 | 22.2 | 4520 | 15.5 | 1.43 |
| 316 | 6 | MA1145.1 | FOSL2::JUND | MA1145.1 | NNRTGACGTCAYHSN | FOSL2::JUND | 5.05e-89 | 6.36e-92 | 21400 | 73.2 | 19000 | 65.4 | 1.12 |
| 317 | 6 | MA1139.1 | FOSL2::JUNB | MA1139.1 | DATGACGTCATH | FOSL2::JUNB | 7.47e-89 | 9.38e-92 | 18400 | 62.9 | 15900 | 54.6 | 1.15 |
| 318 | 26 | MA0651.2 | HOXC11 | MA0651.2 | AGGTCGTAAAAH | HOXC11 | 1.45e-87 | 1.81e-90 | 5380 | 18.4 | 3600 | 12.4 | 1.48 |
| 319 | 72 | MA0106.3 | TP53 | MA0106.3 | RACATGYCCGGRCATGTY | TP53 | 1.85e-86 | 2.3e-89 | 1780 | 6.07 | 788 | 2.71 | 2.24 |
| 320 | 6 | MA1129.1 | FOSL1::JUN | MA1129.1 | ATGACGTCAT | FOSL1::JUN | 5.82e-85 | 7.24e-88 | 20300 | 69.3 | 17900 | 61.5 | 1.13 |
| 321 | 57 | MA1960.1 | MGA::EVX1 | MA1960.1 | RGGTGWTAATKD | MGA::EVX1 | 6.76e-85 | 8.38e-88 | 22200 | 76 | 20000 | 68.7 | 1.11 |
| 322 | 58 | MA1989.1 | Bcl11B | MA1989.1 | NNAAACCACAARNN | Bcl11B | 3.61e-82 | 4.46e-85 | 26700 | 91.4 | 25100 | 86.3 | 1.06 |
| 323 | 52 | MA0827.1 | OLIG3 | MA0827.1 | AMCATATGBY | OLIG3 | 3.9e-82 | 4.8e-85 | 21900 | 74.7 | 19600 | 67.4 | 1.11 |
| 324 | 37 | MA0063.2 | NKX2-5 | MA0063.2 | NNCACTCAANN | NKX2-5 | 2.36e-81 | 2.9e-84 | 27300 | 93.5 | 25900 | 88.9 | 1.05 |
| 325 | 59 | MA1105.2 | GRHL2 | MA1105.2 | NRAACAGGTTYN | GRHL2 | 3.83e-79 | 4.7e-82 | 18100 | 61.8 | 15700 | 54 | 1.14 |
| 326 | 60 | MA1975.1 | ZNF214 | MA1975.1 | VWTCATCAABGTCCT | ZNF214 | 7.12e-79 | 8.69e-82 | 24900 | 85 | 23000 | 79 | 1.08 |
| 327 | 61 | MA0483.1 | Gfi1B | MA0483.1 | AAATCWCWGCH | Gfi1B | 1.28e-78 | 1.56e-81 | 16900 | 57.7 | 14500 | 49.8 | 1.16 |
| 328 | 43 | MA1624.1 | Stat5a | MA1624.1 | NTTCCAAGAANH | Stat5a | 3.79e-78 | 4.6e-81 | 19700 | 67.3 | 17400 | 59.7 | 1.13 |
| 329 | 45 | MA1421.1 | TCF7L1 | MA1421.1 | AAAGATCAAAGG | TCF7L1 | 3.26e-76 | 3.94e-79 | 12800 | 43.9 | 10500 | 36.2 | 1.21 |
| 330 | 15 | MA1473.1 | CDX4 | MA1473.1 | GGYMATAAAAC | CDX4 | 3.73e-76 | 4.5e-79 | 4680 | 16 | 3120 | 10.7 | 1.49 |
| 331 | 30 | MA1953.1 | FOXO1::ELF1 | MA1953.1 | RTMAACAGGAAGTN | FOXO1::ELF1 | 1.28e-75 | 1.53e-78 | 26400 | 90.2 | 24800 | 85.2 | 1.06 |
| 332 | 6 | MA1632.1 | ATF2 | MA1632.1 | NNATGABGTCATN | ATF2 | 1.62e-75 | 1.95e-78 | 21000 | 71.6 | 18700 | 64.4 | 1.11 |
| 333 | 6 | MA1133.1 | JUN::JUNB | MA1133.1 | KRTGACGTCATN | JUN::JUNB | 1.92e-74 | 2.3e-77 | 22800 | 78.1 | 20800 | 71.4 | 1.09 |
| 334 | 43 | MA0463.2 | BCL6 | MA0463.2 | NYGCTTTCKAGGAAYN | BCL6 | 9e-74 | 1.07e-76 | 18300 | 62.6 | 16000 | 55 | 1.14 |
| 335 | 50 | MA2001.1 | Six4 | MA2001.1 | GDAACCTGANH | Six4 | 3.26e-71 | 3.87e-74 | 14800 | 50.6 | 12500 | 43.1 | 1.17 |
| 336 | 44 | MA0508.3 | PRDM1 | MA0508.3 | YNCTTTCTCTH | PRDM1 | 5.72e-71 | 6.78e-74 | 27300 | 93.2 | 25900 | 88.9 | 1.05 |
| 337 | 62 | MA1714.1 | ZNF675 | MA1714.1 | RGGVHYMGGAGGMCAAAATGD | ZNF675 | 1.33e-70 | 1.57e-73 | 18500 | 63.1 | 16200 | 55.7 | 1.13 |
| 338 | 63 | MA0809.2 | TEAD4 | MA0809.2 | NNACATTCCARN | TEAD4 | 9.68e-69 | 1.14e-71 | 27400 | 93.6 | 26000 | 89.4 | 1.05 |
| 339 | 6 | MA0018.4 | CREB1 | MA0018.4 | NNRTGATGTCAYN | CREB1 | 2.78e-68 | 3.27e-71 | 24400 | 83.4 | 22500 | 77.5 | 1.08 |
| 340 | 30 | MA1956.1 | FOXO1::FLI1 | MA1956.1 | GTAAACAGGAAGTK | FOXO1::FLI1 | 3.3e-68 | 3.87e-71 | 6460 | 22.1 | 4740 | 16.3 | 1.36 |
| 341 | 10 | MA0784.2 | POU1F1 | MA0784.2 | ANCTMATTWGCATAWT | POU1F1 | 8.29e-68 | 9.68e-71 | 3190 | 10.9 | 1970 | 6.76 | 1.61 |
| 342 | 20 | MA0466.3 | CEBPB | MA0466.3 | KATTGCGCAATM | CEBPB | 9.09e-67 | 1.06e-69 | 2150 | 7.36 | 1160 | 4.01 | 1.84 |
| 343 | 23 | MA1720.1 | ZNF85 | MA1720.1 | DHDGAGATTACWKCAK | ZNF85 | 2.78e-66 | 3.23e-69 | 20700 | 70.7 | 18600 | 63.8 | 1.11 |
| 344 | 64 | MA1592.1 | ZNF274 | MA1592.1 | NRTRTGAGTTCTCGYN | ZNF274 | 2.79e-66 | 3.23e-69 | 7070 | 24.2 | 5300 | 18.2 | 1.33 |
| 345 | 43 | MA0137.3 | STAT1 | MA0137.3 | TTTCYRGGAAA | STAT1 | 9.63e-66 | 1.11e-68 | 14600 | 50 | 12400 | 42.7 | 1.17 |
| 346 | 53 | MA1534.1 | NR1I3 | MA1534.1 | RTGAACTTT | NR1I3 | 1.86e-65 | 2.14e-68 | 20000 | 68.4 | 17900 | 61.5 | 1.11 |
| 347 | 63 | MA0090.3 | TEAD1 | MA0090.3 | NNACATTCCAGSN | TEAD1 | 8.14e-65 | 9.34e-68 | 26100 | 89.4 | 24600 | 84.5 | 1.06 |
| 348 | 37 | MA1994.1 | Nkx2-1 | MA1994.1 | NVCACTTGARHNN | Nkx2-1 | 4.09e-64 | 4.67e-67 | 27000 | 92.4 | 25700 | 88.2 | 1.05 |
| 349 | 65 | MA1937.1 | ERF::HOXB13 | MA1937.1 | VSCGGAARYNRTWAAN | ERF::HOXB13 | 7.29e-64 | 8.31e-67 | 20200 | 68.9 | 18100 | 62.1 | 1.11 |
| 350 | 6 | MA1136.1 | FOSB::JUNB | MA1136.1 | RTGACGTCAT | FOSB::JUNB | 2.56e-63 | 2.92e-66 | 26400 | 90.1 | 24900 | 85.5 | 1.05 |
| 351 | 44 | MA1949.1 | FLI1::DRGX | MA1949.1 | ACCGGAAGTAATTAT | FLI1::DRGX | 1.63e-62 | 1.85e-65 | 22400 | 76.5 | 20400 | 70.2 | 1.09 |
| 352 | 66 | MA1980.1 | ZNF418 | MA1980.1 | MDGAGGCTAAAAGCA | ZNF418 | 5.03e-62 | 5.68e-65 | 18100 | 62 | 16000 | 55.1 | 1.13 |
| 353 | 67 | MA1151.1 | RORC | MA1151.1 | NWAWNTRGGTCA | RORC | 5.57e-62 | 6.28e-65 | 17300 | 59.2 | 15200 | 52.2 | 1.13 |
| 354 | 68 | MA1112.2 | NR4A1 | MA1112.2 | NNAAAGGTCANN | NR4A1 | 4.66e-59 | 5.24e-62 | 19700 | 67.3 | 17700 | 60.8 | 1.11 |
| 355 | 59 | MA0647.1 | GRHL1 | MA0647.1 | AAAACCGGTTTD | GRHL1 | 6.54e-59 | 7.33e-62 | 11700 | 40 | 9700 | 33.4 | 1.2 |
| 356 | 69 | MA1580.1 | ZBTB32 | MA1580.1 | TGTACAGTAW | ZBTB32 | 6.65e-59 | 7.43e-62 | 7200 | 24.6 | 5520 | 19 | 1.3 |
| 357 | 70 | MA1585.1 | ZKSCAN1 | MA1585.1 | AYAGTAGGTS | ZKSCAN1 | 8.38e-59 | 9.34e-62 | 14400 | 49.3 | 12300 | 42.4 | 1.16 |
| 358 | 71 | MA0479.1 | FOXH1 | MA0479.1 | BNSAATCCACA | FOXH1 | 3.16e-58 | 3.52e-61 | 6440 | 22 | 4830 | 16.6 | 1.32 |
| 359 | 27 | MA1521.1 | MAFA | MA1521.1 | NTGCTGASTCAGCAN | MAFA | 7.31e-58 | 8.11e-61 | 2160 | 7.39 | 1230 | 4.23 | 1.75 |
| 360 | 67 | MA0494.1 | Nr1h3::Rxra | MA0494.1 | TGACCTNNAGTRACCYYDN | Nr1h3::Rxra | 1.42e-55 | 1.57e-58 | 24700 | 84.6 | 23100 | 79.4 | 1.06 |
| 361 | 72 | MA0106.3 (TP53) | STREME-8 | 8-RGRCATGYCY | RGRCATGYCY | TP53 | 2.26e-55 | 2.49e-58 | 1260 | 4.32 | 585 | 2.01 | 2.14 |
| 362 | 72 | MA0861.1 | TP73 | MA0861.1 | DRCATGTCNNRACAYGYM | TP73 | 2.53e-55 | 2.78e-58 | 709 | 2.42 | 229 | 0.79 | 3.07 |
| 363 | 73 | MA1596.1 (ZNF460) | STREME-9 | 9-CCTCAGCCTCCCAAR | CCTCAGCCTCCCAAR | ZNF460 | 4.76e-55 | 5.22e-58 | 823 | 2.81 | 298 | 1.02 | 2.74 |
| 364 | 27 | MA0842.2 | NRL | MA0842.2 | AAWWNTGCTGACG | NRL | 1.13e-54 | 1.24e-57 | 10000 | 34.3 | 8200 | 28.2 | 1.22 |
| 365 | 67 | MA1148.1 | PPARA::RXRA | MA1148.1 | AWNTRGGTYAAAGGTCAN | PPARA::RXRA | 3.71e-53 | 4.05e-56 | 25200 | 86.2 | 23700 | 81.4 | 1.06 |
| 366 | 74 | MA0631.1 | Six3 | MA0631.1 | DATRGGGTATCAYNWNT | Six3 | 7.57e-53 | 8.23e-56 | 14100 | 48.1 | 12100 | 41.7 | 1.16 |
| 367 | 53 | MA1111.1 | NR2F2 | MA1111.1 | NAAAGGTCANR | NR2F2 | 4.4e-52 | 4.77e-55 | 25800 | 88 | 24300 | 83.5 | 1.05 |
| 368 | 58 | MA0684.2 | RUNX3 | MA0684.2 | NHAACCTCAANN | RUNX3 | 1.84e-51 | 1.99e-54 | 26000 | 88.9 | 24600 | 84.5 | 1.05 |
| 369 | 75 | MA1995.1 | Npas4 | MA1995.1 | NNTCGTGACHN | Npas4 | 3.44e-50 | 3.71e-53 | 22800 | 77.8 | 21000 | 72.3 | 1.08 |
| 370 | 76 | MA0782.2 | PKNOX1 | MA0782.2 | NNTGAGTGACAGVNN | PKNOX1 | 1.26e-49 | 1.35e-52 | 21700 | 74.1 | 19900 | 68.4 | 1.08 |
| 371 | 61 | MA0038.2 | GFI1 | MA0038.2 | BMAATCACDGCA | GFI1 | 1.74e-48 | 1.87e-51 | 7390 | 25.3 | 5830 | 20 | 1.26 |
| 372 | 26 | MA0485.2 | HOXC9 | MA0485.2 | GTCGTAAAAH | HOXC9 | 3.58e-48 | 3.84e-51 | 3260 | 11.2 | 2200 | 7.55 | 1.48 |
| 373 | 39 | MA1718.1 | ZNF8 | MA1718.1 | GTATGGATATACCACAKTTT | ZNF8 | 5.75e-48 | 6.13e-51 | 2990 | 10.2 | 1970 | 6.77 | 1.51 |
| 374 | 77 | MA0620.3 | MITF | MA0620.3 | NNNDRTCACGTGAYHNNN | MITF | 2.22e-47 | 2.36e-50 | 13900 | 47.5 | 12000 | 41.4 | 1.15 |
| 375 | 30 | MA1955.1 | FOXO1::ELK3 | MA1955.1 | RWMAACAGGAAGTN | FOXO1::ELK3 | 2.37e-46 | 2.51e-49 | 17800 | 60.9 | 16000 | 54.9 | 1.11 |
| 376 | 78 | MA0029.1 | Mecom | MA0029.1 | AAGAYAAGATAANA | Mecom | 8.21e-45 | 8.69e-48 | 4990 | 17.1 | 3720 | 12.8 | 1.33 |
| 377 | 52 | MA1108.2 | MXI1 | MA1108.2 | NNCACATGBN | MXI1 | 1.55e-44 | 1.64e-47 | 22500 | 76.9 | 20900 | 71.7 | 1.07 |
| 378 | 30 | MA1950.1 | FLI1::FOXI1 | MA1950.1 | RTAAACAGGAARYN | FLI1::FOXI1 | 4.1e-44 | 4.31e-47 | 25700 | 87.8 | 24300 | 83.7 | 1.05 |
| 379 | 43 | MA0519.1 | Stat5a::Stat5b | MA0519.1 | DTTTCCMAGAA | Stat5a::Stat5b | 2e-43 | 2.1e-46 | 14600 | 49.8 | 12800 | 44 | 1.13 |
| 380 | 30 | MA1420.1 | IRF5 | MA1420.1 | CCGAAACCGAAACY | IRF5 | 4.09e-43 | 4.28e-46 | 18500 | 63.3 | 16700 | 57.6 | 1.1 |
| 381 | 30 | MA1508.1 | IKZF1 | MA1508.1 | VVAACAGGAARN | IKZF1 | 4.13e-43 | 4.31e-46 | 21600 | 74 | 20000 | 68.6 | 1.08 |
| 382 | 36 | MA0773.1 | MEF2D | MA0773.1 | DCTAWAAATAGM | MEF2D | 4.82e-43 | 5.03e-46 | 4720 | 16.1 | 3500 | 12 | 1.34 |
| 383 | 79 | MA0525.2 | TP63 | MA0525.2 | RACATGTYGKGACATGTC | TP63 | 4.12e-42 | 4.28e-45 | 452 | 1.54 | 124 | 0.43 | 3.6 |
| 384 | 80 | MA0689.1 | TBX20 | MA0689.1 | WAGGTGTGAAR | TBX20 | 4.25e-41 | 4.41e-44 | 22900 | 78.2 | 21300 | 73.2 | 1.07 |
| 385 | 10 | MA0507.2 | POU2F2 | MA0507.2 | AWTATGCAAATKAG | POU2F2 | 6.94e-40 | 7.17e-43 | 1860 | 6.34 | 1120 | 3.86 | 1.64 |
| 386 | 36 | MA0660.1 | MEF2B | MA0660.1 | RCTAWAAATAGC | MEF2B | 3.18e-39 | 3.28e-42 | 5040 | 17.2 | 3840 | 13.2 | 1.31 |
| 387 | 81 | MA1523.1 | MSANTD3 | MA1523.1 | STVCACTCAC | MSANTD3 | 9.24e-39 | 9.5e-42 | 26600 | 90.9 | 25400 | 87.4 | 1.04 |
| 388 | 52 | MA0461.2 | Atoh1 | MA0461.2 | RMCATATGBY | Atoh1 | 7.71e-38 | 7.91e-41 | 16400 | 56 | 14700 | 50.4 | 1.11 |
| 389 | 35 | MA0719.1 | RHOXF1 | MA0719.1 | NTRATCCN | RHOXF1 | 1.01e-37 | 1.04e-40 | 12000 | 40.9 | 10300 | 35.5 | 1.15 |
| 390 | 30 | MA1947.1 | ETV5::FOXO1 | MA1947.1 | GTMAACAGGAWRY | ETV5::FOXO1 | 3.85e-37 | 3.92e-40 | 26100 | 89.2 | 24900 | 85.6 | 1.04 |
| 391 | 59 | MA0743.2 | SCRT1 | MA0743.2 | NDWKCAACAGGTGKNN | SCRT1 | 1.08e-36 | 1.1e-39 | 15500 | 52.9 | 13800 | 47.4 | 1.11 |
| 392 | 82 | MA0771.1 | HSF4 | MA0771.1 | TTCTAGAABNTTC | HSF4 | 1.55e-36 | 1.57e-39 | 21100 | 72 | 19500 | 67 | 1.07 |
| 393 | 83 | MA0682.2 (PITX1) | STREME-6 | 6-CCTGTAATCC | CCTGTAATCC | PITX1 | 3.61e-36 | 3.66e-39 | 1520 | 5.21 | 893 | 3.07 | 1.7 |
| 394 | 63 | MA1121.1 | TEAD2 | MA1121.1 | NYACATTCCWNNS | TEAD2 | 2.01e-35 | 2.03e-38 | 27900 | 95.5 | 27000 | 93 | 1.03 |
| 395 | 75 | MA1546.1 | PAX3 | MA1546.1 | YSGTCACGSYWATTAN | PAX3 | 4.85e-35 | 4.88e-38 | 17300 | 59 | 15600 | 53.8 | 1.1 |
| 396 | 56 | MA0114.4 | HNF4A | MA0114.4 | DNCAAAGTCCANN | HNF4A | 1.52e-34 | 1.53e-37 | 27200 | 92.8 | 26100 | 89.8 | 1.03 |
| 397 | 84 | MA1515.1 (KLF2) | STREME-22 | 22-GCCACCACGCCCRGC | 22-GCCACCACGCCCRGC | KLF2 | 2.35e-34 | 2.35e-37 | 744 | 2.54 | 332 | 1.14 | 2.22 |
| 398 | 6 | MA0862.1 | GMEB2 | MA0862.1 | TTACGTAA | GMEB2 | 6.72e-34 | 6.73e-37 | 22300 | 76.2 | 20800 | 71.6 | 1.06 |
| 399 | 85 | MA1647.2 | Prdm4 | MA1647.2 | RDNCCTTGAAACYGYYN | Prdm4 | 1.08e-33 | 1.08e-36 | 21900 | 74.9 | 20400 | 70.2 | 1.07 |
| 400 | 52 | MA1524.2 | Msgn1 | MA1524.2 | VRRRACAAATGGTNNN | Msgn1 | 3.68e-33 | 3.66e-36 | 17300 | 59.1 | 15700 | 54 | 1.09 |
| 401 | 13 | MA0756.2 | ONECUT2 | MA0756.2 | CGATCRATA | ONECUT2 | 4.71e-33 | 4.68e-36 | 7040 | 24.1 | 5760 | 19.8 | 1.22 |
| 402 | 59 | MA0744.2 | SCRT2 | MA0744.2 | NNWGCAACAGGTGDNN | SCRT2 | 5e-33 | 4.95e-36 | 25100 | 85.6 | 23800 | 81.8 | 1.05 |
| 403 | 86 | MA1509.1 | IRF6 | MA1509.1 | ACCGAAACT | IRF6 | 6.27e-33 | 6.2e-36 | 10600 | 36.1 | 9080 | 31.2 | 1.16 |
| 404 | 65 | MA1943.1 | ETV2::HOXB13 | MA1943.1 | ACCGGAAMYATWAAM | ETV2::HOXB13 | 7.45e-33 | 7.34e-36 | 7580 | 25.9 | 6260 | 21.5 | 1.2 |
| 405 | 87 | MA0140.2 | GATA1::TAL1 | MA0140.2 | MTTATCWSNNNNNHVCAG | GATA1::TAL1 | 2.37e-32 | 2.33e-35 | 10600 | 36.2 | 9130 | 31.4 | 1.15 |
| 406 | 88 | 10-GARACCCCRTCTC | STREME-10 | 10-GARACCCCRTCTC | 10-GARACCCCRTCTC | GARACCCCRTCTC | 6.96e-32 | 6.83e-35 | 703 | 2.4 | 316 | 1.09 | 2.21 |
| 407 | 30 | MA1954.1 | FOXO1::ELK1 | MA1954.1 | RWMAACAGGAAGTD | FOXO1::ELK1 | 1.05e-31 | 1.02e-34 | 21000 | 71.8 | 19500 | 67.1 | 1.07 |
| 408 | 52 | MA0817.1 | BHLHE23 | MA0817.1 | AAACATATGBTN | BHLHE23 | 1.64e-31 | 1.6e-34 | 2490 | 8.51 | 1720 | 5.91 | 1.44 |
| 409 | 11 | MA1644.1 | NFYC | MA1644.1 | RDCCAATCASN | NFYC | 2.96e-31 | 2.88e-34 | 24700 | 84.5 | 23500 | 80.6 | 1.05 |
| 410 | 40 | MA0610.1 | DMRT3 | MA0610.1 | MATGTATCAAN | DMRT3 | 5.14e-31 | 4.99e-34 | 7760 | 26.5 | 6460 | 22.2 | 1.19 |
| 411 | 30 | MA0080.6 | Spi1 | MA0080.6 | RRAAAGAGGAAGTGGDD | Spi1 | 5.11e-30 | 4.95e-33 | 22000 | 75.1 | 20600 | 70.8 | 1.06 |
| 412 | 35 | MA1547.2 | PITX2 | MA1547.2 | YTAATCCY | PITX2 | 9.24e-29 | 8.93e-32 | 4150 | 14.2 | 3200 | 11 | 1.29 |
| 413 | 73 | MA1596.1 | ZNF460 | MA1596.1 | GCCTCMGCCTCCCRAG | ZNF460 | 1.14e-27 | 1.1e-30 | 533 | 1.82 | 222 | 0.76 | 2.38 |
| 414 | 31 | MA1525.2 | NFATC4 | MA1525.2 | AAYGGAAAMW | NFATC4 | 1.16e-27 | 1.11e-30 | 9030 | 30.9 | 7730 | 26.6 | 1.16 |
| 415 | 52 | MA0826.1 | OLIG1 | MA0826.1 | AMCATATGKT | OLIG1 | 1.56e-27 | 1.5e-30 | 8670 | 29.6 | 7390 | 25.4 | 1.17 |
| 416 | 58 | MA0002.2 | Runx1 | MA0002.2 | BBYTGTGGTTT | Runx1 | 2.15e-27 | 2.06e-30 | 2240 | 7.65 | 1550 | 5.33 | 1.44 |
| 417 | 77 | MA0829.2 | SREBF1 | MA0829.2 | DNATCACSTGATNH | SREBF1 | 2.96e-27 | 2.82e-30 | 5310 | 18.2 | 4270 | 14.7 | 1.24 |
| 418 | 44 | MA1940.1 | ETV2::DRGX | MA1940.1 | RVCGGAAGYAATTA | ETV2::DRGX | 6.3e-27 | 6e-30 | 11300 | 38.7 | 9940 | 34.2 | 1.13 |
| 419 | 56 | MA0484.2 | HNF4G | MA0484.2 | DNCAAAGTCCANN | HNF4G | 6.69e-27 | 6.35e-30 | 26400 | 90.2 | 25400 | 87.2 | 1.03 |
| 420 | 44 | MA1942.1 | ETV2::FOXI1 | MA1942.1 | BGTAAACAGGAAGYR | ETV2::FOXI1 | 1.46e-26 | 1.38e-29 | 20900 | 71.5 | 19500 | 67.2 | 1.06 |
| 421 | 89 | MA1579.1 | ZBTB26 | MA1579.1 | NNCTCCAGAAANNNN | ZBTB26 | 4.23e-26 | 4e-29 | 20100 | 68.7 | 18700 | 64.3 | 1.07 |
| 422 | 90 | MA0101.1 | REL | MA0101.1 | BGGRNWTTCC | REL | 3.75e-25 | 3.54e-28 | 27300 | 93.4 | 26500 | 91 | 1.03 |
| 423 | 6 | MA0840.1 | Creb5 | MA0840.1 | NATGACGTCAYH | Creb5 | 4.35e-25 | 4.09e-28 | 3080 | 10.5 | 2300 | 7.9 | 1.33 |
| 424 | 30 | MA1418.1 | IRF3 | MA1418.1 | NSRRAAMGGAAACCGAAACYR | IRF3 | 4.56e-25 | 4.28e-28 | 2420 | 8.26 | 1730 | 5.95 | 1.39 |
| 425 | 67 | MA0072.1 | RORA | MA0072.1 | NWWAWBTAGGTCAR | RORA | 1.21e-24 | 1.13e-27 | 2540 | 8.67 | 1840 | 6.32 | 1.37 |
| 426 | 52 | MA1618.1 | Ptf1a | MA1618.1 | NNACAGATGTTNN | Ptf1a | 2.79e-24 | 2.61e-27 | 12900 | 44 | 11500 | 39.6 | 1.11 |
| 427 | 91 | 16-AACTCCTGRSCTC | STREME-16 | 16-AACTCCTGRSCTC | 16-AACTCCTGRSCTC | AACTCCTGRSCTC | 4.46e-24 | 4.16e-27 | 474 | 1.62 | 199 | 0.68 | 2.36 |
| 428 | 37 | MA0124.2 | Nkx3-1 | MA0124.2 | RCCACTTAA | Nkx3-1 | 3.36e-23 | 3.13e-26 | 26500 | 90.5 | 25500 | 87.8 | 1.03 |
| 429 | 59 | MA1968.1 | TFCP2 | MA1968.1 | AAACCGGTTY | TFCP2 | 6.84e-23 | 6.35e-26 | 19800 | 67.8 | 18500 | 63.7 | 1.06 |
| 430 | 30 | MA0653.1 | IRF9 | MA0653.1 | AACGAAACCGAAACT | IRF9 | 1e-22 | 9.27e-26 | 489 | 1.67 | 215 | 0.74 | 2.25 |
| 431 | 58 | MA0511.2 | RUNX2 | MA0511.2 | WAACCGCAA | RUNX2 | 1.13e-22 | 1.04e-25 | 1900 | 6.49 | 1320 | 4.53 | 1.43 |
| 432 | 92 | MA0794.1 | PROX1 | MA0794.1 | YAAGACGYCTTA | PROX1 | 1.17e-22 | 1.08e-25 | 6380 | 21.8 | 5340 | 18.3 | 1.19 |
| 433 | 93 | MA0019.1 | Ddit3::Cebpa | MA0019.1 | VNRTGCAATMCC | Ddit3::Cebpa | 3.66e-22 | 3.36e-25 | 10300 | 35.2 | 9060 | 31.2 | 1.13 |
| 434 | 94 | MA1973.1 (ZKSCAN3) | STREME-12 | 12-AGACCAGCCTGGGC | 12-AGACCAGCCTGGGC | ZKSCAN3 | 4.44e-22 | 4.08e-25 | 789 | 2.7 | 432 | 1.49 | 1.81 |
| 435 | 76 | MA0498.2 | MEIS1 | MA0498.2 | HTGACAD | MEIS1 | 6.09e-22 | 5.57e-25 | 24200 | 82.8 | 23100 | 79.4 | 1.04 |
| 436 | 95 | 17-CAGCTACTCRGGAG | STREME-17 | 17-CAGCTACTCRGGAG | 17-CAGCTACTCRGGAG | CAGCTACTCRGGAG | 1.56e-21 | 1.43e-24 | 293 | 1 | 96 | 0.33 | 3.01 |
| 437 | 96 | MA1983.1 | ZNF582 | MA1983.1 | TCTGTTACTTGCAGCCAAAWG | ZNF582 | 1.86e-21 | 1.7e-24 | 6120 | 20.9 | 5120 | 17.6 | 1.19 |
| 438 | 11 | MA0703.2 | LMX1B | MA0703.2 | AWTTTAATTAA | LMX1B | 1.91e-21 | 1.74e-24 | 1570 | 5.36 | 1060 | 3.63 | 1.48 |
| 439 | 97 | MA0596.1 | SREBF2 | MA0596.1 | RTGGGGTGAY | SREBF2 | 2.11e-21 | 1.92e-24 | 8480 | 29 | 7340 | 25.2 | 1.15 |
| 440 | 44 | MA0598.3 | EHF | MA0598.3 | NNCACTTCCTGTTNN | EHF | 8e-21 | 7.24e-24 | 24400 | 83.6 | 23400 | 80.4 | 1.04 |
| 441 | 44 | MA0136.3 | Elf5 | MA0136.3 | RVAAGGAAGTNN | Elf5 | 8.21e-21 | 7.41e-24 | 11300 | 38.7 | 10100 | 34.7 | 1.11 |
| 442 | 87 | MA0766.2 | GATA5 | MA0766.2 | SWGATAASRN | GATA5 | 1.03e-19 | 9.27e-23 | 16400 | 56.1 | 15100 | 52 | 1.08 |
| 443 | 6 | MA0488.1 | JUN | MA0488.1 | DDRATGATGTCAT | JUN | 1.1e-19 | 9.9e-23 | 1520 | 5.19 | 1030 | 3.55 | 1.46 |
| 444 | 82 | MA0486.2 | HSF1 | MA0486.2 | TTCTAGAAYRTTC | HSF1 | 1.76e-18 | 1.58e-21 | 4300 | 14.7 | 3500 | 12 | 1.22 |
| 445 | 77 | MA0093.3 | USF1 | MA0093.3 | NDGTCATGTGACHN | USF1 | 1.37e-17 | 1.22e-20 | 10800 | 36.8 | 9630 | 33.1 | 1.11 |
| 446 | 98 | 13-AAGTGATCCDCC | STREME-13 | 13-AAGTGATCCDCC | 13-AAGTGATCCDCC | AAGTGATCCDCC | 1.46e-17 | 1.31e-20 | 345 | 1.18 | 143 | 0.49 | 2.39 |
| 447 | 99 | 21-AGGCTGCAGTGAGY | STREME-21 | 21-AGGCTGCAGTGAGY | 21-AGGCTGCAGTGAGY | AGGCTGCAGTGAGY | 8.14e-17 | 7.25e-20 | 394 | 1.35 | 179 | 0.62 | 2.18 |
| 448 | 44 | MA0081.2 | SPIB | MA0081.2 | TYTCACTTCCTCTTTY | SPIB | 9.36e-17 | 8.32e-20 | 22900 | 78.2 | 21800 | 75 | 1.04 |
| 449 | 53 | MA1996.1 | Nr1H2 | MA1996.1 | NNRAGGTCANN | Nr1H2 | 1.03e-16 | 9.15e-20 | 17900 | 61.1 | 16700 | 57.4 | 1.06 |
| 450 | 100 | MA0164.1 | Nr2e3 | MA0164.1 | CAAGCTT | Nr2e3 | 2.11e-16 | 1.87e-19 | 3970 | 13.6 | 3240 | 11.2 | 1.22 |
| 451 | 26 | MA1958.1 | HOXD12::ELK1 | MA1958.1 | DRVCGGAAGTCGTAAAD | HOXD12::ELK1 | 1.05e-15 | 9.27e-19 | 1720 | 5.89 | 1250 | 4.3 | 1.37 |
| 452 | 30 | MA0652.1 | IRF8 | MA0652.1 | HCGAAACCGAAACT | IRF8 | 2.98e-15 | 2.62e-18 | 437 | 1.49 | 217 | 0.75 | 2 |
| 453 | 101 | MA1593.1 | ZNF317 | MA1593.1 | WVACAGCAGAYW | ZNF317 | 4.73e-15 | 4.16e-18 | 16800 | 57.3 | 15600 | 53.8 | 1.07 |
| 454 | 30 | MA1935.1 | ERF::FOXI1 | MA1935.1 | RTAAACMGGAARTR | ERF::FOXI1 | 4.76e-15 | 4.17e-18 | 28100 | 96.2 | 27500 | 94.7 | 1.02 |
| 455 | 82 | MA0770.1 | HSF2 | MA0770.1 | TTCTAGAAYRTTC | HSF2 | 5.03e-15 | 4.4e-18 | 2980 | 10.2 | 2370 | 8.15 | 1.25 |
| 456 | 73 | MA1587.1 | ZNF135 | MA1587.1 | CCTCGACCTCCYRR | ZNF135 | 2.35e-14 | 2.05e-17 | 800 | 2.73 | 498 | 1.71 | 1.6 |
| 457 | 77 | MA0626.1 | Npas2 | MA0626.1 | NSCACGTGTN | Npas2 | 7.72e-14 | 6.72e-17 | 15300 | 52.2 | 14200 | 48.8 | 1.07 |
| 458 | 102 | MA1616.1 | Prdm15 | MA1616.1 | NRGAAAACCTGGAVN | Prdm15 | 2.43e-13 | 2.11e-16 | 18900 | 64.5 | 17800 | 61.2 | 1.05 |
| 459 | 30 | MA1419.1 | IRF4 | MA1419.1 | HCGAAACCGAAACYA | IRF4 | 2.57e-13 | 2.23e-16 | 479 | 1.64 | 259 | 0.89 | 1.84 |
| 460 | 103 | MA1998.1 | Prdm14 | MA1998.1 | NDGGTCTCTAWS | Prdm14 | 7.07e-13 | 6.12e-16 | 13100 | 44.9 | 12100 | 41.6 | 1.08 |
| 461 | 52 | MA1123.2 | TWIST1 | MA1123.2 | NNDCCAGATGTBN | TWIST1 | 3.16e-11 | 2.72e-14 | 11200 | 38.2 | 10200 | 35.2 | 1.09 |
| 462 | 68 | MA1541.1 | NR6A1 | MA1541.1 | NYCAAGKTCAAGKTCAN | NR6A1 | 3.43e-11 | 2.96e-14 | 10300 | 35 | 9340 | 32.1 | 1.09 |
| 463 | 53 | MA1110.2 | Nr1H4 | MA1110.2 | NVAGGTCANN | Nr1H4 | 4.17e-11 | 3.59e-14 | 26100 | 89.1 | 25300 | 87.1 | 1.02 |
| 464 | 104 | MA0776.1 | MYBL1 | MA0776.1 | ACCGTTAACSGY | MYBL1 | 4.37e-11 | 3.75e-14 | 7080 | 24.2 | 6280 | 21.6 | 1.12 |
| 465 | 30 | MA0051.1 | IRF2 | MA0051.1 | SGAAAGYGAAASCRWWWM | IRF2 | 5.38e-10 | 4.6e-13 | 381 | 1.3 | 208 | 0.72 | 1.82 |
| 466 | 105 | MA1594.1 | ZNF382 | MA1594.1 | AGGRGAYATCACYACMGAYCCCAC | ZNF382 | 5.98e-10 | 5.11e-13 | 2750 | 9.4 | 2260 | 7.75 | 1.21 |
| 467 | 106 | MA0472.2 (EGR2) | STREME-19 | 19-ACACCCACACAC | 19-ACACCCACACAC | EGR2 | 1.12e-09 | 9.54e-13 | 225 | 0.77 | 99 | 0.34 | 2.25 |
| 468 | 106 | ACACACACACACACA | MEME-3 | ACACACACACACACA | ACACACACACACACA | ACACACACACACACA | 1.28e-09 | 1.09e-12 | 2340 | 8.02 | 1890 | 6.51 | 1.23 |
| 469 | 80 | MA0800.1 | EOMES | MA0800.1 | RAGGTGTGAAAWN | EOMES | 1.76e-09 | 1.49e-12 | 24000 | 82 | 23200 | 79.8 | 1.03 |
| 470 | 30 | MA1946.1 | ETV5::FOXI1 | MA1946.1 | GTAAACAGGAWGY | ETV5::FOXI1 | 1.73e-08 | 1.46e-11 | 5160 | 17.6 | 4540 | 15.6 | 1.13 |
| 471 | 6 | MA0492.1 | JUND | MA0492.1 | DDDRATGABGTCATN | JUND | 2.3e-08 | 1.95e-11 | 1360 | 4.67 | 1040 | 3.58 | 1.3 |
| 472 | 107 | MA0698.1 | ZBTB18 | MA0698.1 | NATCCAGATGTKB | ZBTB18 | 1.29e-07 | 1.08e-10 | 3200 | 10.9 | 2720 | 9.35 | 1.17 |
| 473 | 44 | MA1944.1 | ETV5::DRGX | MA1944.1 | RSMGGAAGYAATTA | ETV5::DRGX | 6.94e-07 | 5.84e-10 | 21600 | 73.7 | 20800 | 71.5 | 1.03 |
| 474 | 106 | MA1493.1 (HES6) | STREME-15 | 15-ACACGTGCACACACA | 15-ACACGTGCACACACA | HES6 | 1.62e-06 | 1.36e-09 | 209 | 0.71 | 104 | 0.36 | 1.99 |
| 475 | 52 | MA0818.2 | BHLHE22 | MA0818.2 | AMCATATGKY | BHLHE22 | 4.01e-06 | 3.36e-09 | 1110 | 3.8 | 855 | 2.94 | 1.29 |
| 476 | 65 | MA1957.1 | HOXB2::ELK1 | MA1957.1 | RCCGGAAGTMRTTA | HOXB2::ELK1 | 9.77e-06 | 8.17e-09 | 9850 | 33.7 | 9160 | 31.5 | 1.07 |
| 477 | 11 | MA0060.3 | NFYA | MA0060.3 | RRCCAATCAGM | NFYA | 1.35e-05 | 1.13e-08 | 22900 | 78.3 | 22200 | 76.4 | 1.03 |
| 478 | 108 | MA0795.1 | SMAD3 | MA0795.1 | YGTCTAGACA | SMAD3 | 1.59e-05 | 1.33e-08 | 5580 | 19.1 | 5040 | 17.3 | 1.1 |
| 479 | 6 | MA0656.1 | JDP2 | MA0656.1 | GATGACGTCAYC | JDP2 | 1.85e-05 | 1.54e-08 | 2560 | 8.76 | 2190 | 7.52 | 1.17 |
| 480 | 44 | MA0761.2 | ETV1 | MA0761.2 | NNACAGGAAGTGNN | ETV1 | 4.08e-05 | 3.38e-08 | 24800 | 84.8 | 24200 | 83.2 | 1.02 |
| 481 | 107 | MA0100.3 | MYB | MA0100.3 | NNCAACTGHH | MYB | 4.19e-05 | 3.47e-08 | 14200 | 48.4 | 13400 | 46.2 | 1.05 |
| 482 | 31 | MA0625.2 | NFATC3 | MA0625.2 | RYGGAAAMH | NFATC3 | 6.03e-05 | 4.97e-08 | 19800 | 67.8 | 19100 | 65.8 | 1.03 |
| 483 | 44 | MA1948.1 | ETV5::HOXA2 | MA1948.1 | RVCGGAWGTMATTA | ETV5::HOXA2 | 6.03e-05 | 4.97e-08 | 12600 | 43 | 11900 | 40.8 | 1.05 |
| 484 | 42 | MA0866.1 | SOX21 | MA0866.1 | AACAATKKYAGTGTT | SOX21 | 0.000111 | 9.13e-08 | 810 | 2.77 | 613 | 2.11 | 1.31 |
| 485 | 108 | MA1602.1 | ZSCAN29 | MA1602.1 | YGTCTACRCNGN | ZSCAN29 | 0.000132 | 1.08e-07 | 2790 | 9.52 | 2420 | 8.31 | 1.15 |
| 486 | 6 | MA0609.2 | CREM | MA0609.2 | NNDGTGACGTCACHNN | CREM | 0.00017 | 1.4e-07 | 25700 | 87.7 | 25100 | 86.3 | 1.02 |
| 487 | 44 | MA1483.2 | ELF2 | MA1483.2 | AAMCCGGAAGTR | ELF2 | 0.000313 | 2.56e-07 | 13700 | 47 | 13100 | 44.9 | 1.05 |
| 488 | 54 | MA1651.1 | ZFP42 | MA1651.1 | NNNHCAARATGGCTGCCNBNN | ZFP42 | 0.000583 | 4.75e-07 | 17800 | 60.9 | 17100 | 58.9 | 1.03 |
| 489 | 44 | MA0062.3 | GABPA | MA0062.3 | NNCACTTCCTGTNN | GABPA | 0.000718 | 5.84e-07 | 7930 | 27.1 | 7370 | 25.4 | 1.07 |
| 490 | 97 | MA0595.1 | SREBF1 | MA0595.1 | VTCACCCCAY | SREBF1 | 0.00107 | 8.73e-07 | 7170 | 24.5 | 6640 | 22.8 | 1.07 |
| 491 | 52 | MA1570.1 | TFAP4 | MA1570.1 | AHCATRTGDT | TFAP4 | 0.00112 | 9.07e-07 | 8780 | 30 | 8210 | 28.2 | 1.06 |
| 492 | 52 | MA1638.1 | HAND2 | MA1638.1 | NVCAGATGNN | HAND2 | 0.00124 | 1.01e-06 | 20300 | 69.4 | 19700 | 67.6 | 1.03 |
| 493 | 6 | MA1951.1 | FOS | MA1951.1 | BGATGACGTCATCR | FOS | 0.00159 | 1.28e-06 | 3300 | 11.3 | 2930 | 10.1 | 1.12 |
| 494 | 67 | MA0071.1 | RORA | MA0071.1 | AWMWAGGTCA | RORA | 0.00459 | 3.7e-06 | 8620 | 29.4 | 8080 | 27.8 | 1.06 |
| 495 | 80 | MA0804.1 | TBX19 | MA0804.1 | DTTMRCACMTAGGTGTGAAW | TBX19 | 0.00483 | 3.89e-06 | 2940 | 10 | 2610 | 8.98 | 1.12 |
| 496 | 109 | MA1977.1 | ZNF324 | MA1977.1 | NNNWRGMAGHYAAGGATGGTT | ZNF324 | 0.00711 | 5.7e-06 | 10900 | 37.4 | 10400 | 35.6 | 1.05 |
| 497 | 52 | MA0607.2 | BHLHA15 | MA0607.2 | ACCATATGGT | BHLHA15 | 0.0108 | 8.68e-06 | 10700 | 36.7 | 10200 | 35 | 1.05 |
| 498 | 110 | MA1717.1 | ZNF784 | MA1717.1 | GTACYTACCGH | ZNF784 | 0.0152 | 1.21e-05 | 5180 | 17.7 | 4780 | 16.4 | 1.08 |
| 499 | 111 | MA1154.1 | ZNF282 | MA1154.1 | CTTTCCCMYAACACGAB | ZNF282 | 0.0182 | 1.45e-05 | 3270 | 11.2 | 2950 | 10.1 | 1.1 |
| 500 | 85 | MA0777.1 | MYBL2 | MA0777.1 | RRCCGTTAAACBGYY | MYBL2 | 0.0252 | 2.01e-05 | 5230 | 17.9 | 4830 | 16.6 | 1.08 |
| 501 | 86 | MA1971.1 | ZBED2 | MA1971.1 | NNMMCGAAACCNNV | ZBED2 | 0.048 | 3.81e-05 | 15300 | 52.4 | 14800 | 50.8 | 1.03 |
ER_rat <- 1.25
DOX_24_up_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( "DOX DAR 24hr sig up response peaks(200bp)Enrichment ratio:",ER_rat))
DOX_24_up_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
slice_head(n=5) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*10), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./10,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( "DOX DAR 24hr sig up response peaks(200bp)Enrichment ratio:",ER_rat))
DOX_sigdown_24hr data
DOX_sigdown_24hr_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DOX_24_down_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
DOX_sigdown_24hr_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DOX_24_down_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
DOX_sigdown_24hr_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DOX_24_down_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
DOX_sigdown_24hr_sea_all <- rbind(DOX_sigdown_24hr_sea_disc, DOX_sigdown_24hr_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)DOX_sigdown_24hr_combo <- DOX_sigdown_24hr_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., DOX_sigdown_24hr_sea_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ SIM_MOTIF
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y)
DOX_sigdown_24hr_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs 200 bp in DOX_sigdown_24hr v non-DOX sigdown 24hour") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | MA1527.1 (NFIC) | STREME-2 | 2-CTGCCAGS | CTGCCAGS | NFIC | 0 | 0 | 21900 | 75.2 | 15800 | 53.9 | 1.39 |
| 2 | 2 | MA1102.2 (CTCFL) | STREME-1 | 1-CRGCCCC | CRGCCCC | CTCFL | 0 | 0 | 15600 | 53.8 | 9740 | 33.3 | 1.61 |
| 3 | 3 | MA1102.2 | CTCFL | MA1102.2 | NSCAGGGGGCGS | CTCFL | 0 | 0 | 18900 | 65.1 | 13200 | 45.1 | 1.44 |
| 4 | 15 | MA1713.1 | ZNF610 | MA1713.1 | SSCGCCGCTCCSSS | ZNF610 | 0 | 0 | 15200 | 52.4 | 9690 | 33.1 | 1.58 |
| 5 | 4 | 9-GGCMCT | STREME-9 | 9-GGCMCT | GGCMCT | GGCMCT | 0 | 0 | 14000 | 48.2 | 8600 | 29.4 | 1.64 |
| 6 | 5 | MA1653.1 (ZNF148) | MEME-1 | GGGGGHGGGGVVGGG | GGGGGHGGGGVVGGG | ZNF148 | 0 | 0 | 20300 | 69.9 | 15000 | 51.2 | 1.36 |
| 7 | 6 | 4-GCCAGCG | STREME-4 | 4-GCCAGCG | GCCAGCG | GCCAGCG | 0 | 0 | 19700 | 67.8 | 14300 | 49 | 1.38 |
| 8 | 7 | MA1548.1 (PLAGL2) | STREME-3 | 3-GKGCMC | GKGCMC | PLAGL2 | 0 | 0 | 12500 | 43 | 7300 | 25 | 1.72 |
| 9 | 8 | MA1548.1 | PLAGL2 | MA1548.1 | NGGGCCCCCN | PLAGL2 | 0 | 0 | 14000 | 48 | 8700 | 29.7 | 1.62 |
| 10 | 5 | MA1653.1 | ZNF148 | MA1653.1 | YBCCCCTCCCCC | ZNF148 | 0 | 0 | 14200 | 49 | 9010 | 30.8 | 1.59 |
| 11 | 5 | MA1961.1 | PATZ1 | MA1961.1 | SSGGGGMGGGGS | PATZ1 | 0 | 0 | 16400 | 56.5 | 11300 | 38.5 | 1.47 |
| 12 | 9 | MA0079.5 | SP1 | MA0079.5 | GGGGCGGGG | SP1 | 0 | 0 | 12300 | 42.4 | 7420 | 25.4 | 1.67 |
| 13 | 10 | MA1522.1 | MAZ | MA1522.1 | BBCCCCTCCCY | MAZ | 0 | 0 | 10900 | 37.6 | 6300 | 21.5 | 1.75 |
| 14 | 9 | MA0685.2 | SP4 | MA0685.2 | GGGGCGGGG | SP4 | 0 | 0 | 13100 | 44.9 | 8240 | 28.2 | 1.59 |
| 15 | 11 | 5-GAGGGC | STREME-5 | 5-GAGGGC | GAGGGC | GAGGGC | 0 | 0 | 24400 | 83.9 | 20300 | 69.3 | 1.21 |
| 16 | 12 | MA0597.2 | THAP1 | MA0597.2 | VSGCAGGGCASV | THAP1 | 0 | 0 | 17100 | 58.9 | 12200 | 41.6 | 1.42 |
| 17 | 13 | MA1653.1 (ZNF148) | STREME-8 | 8-CCCTCC | CCCTCC | ZNF148 | 0 | 0 | 18000 | 61.9 | 13100 | 44.7 | 1.39 |
| 18 | 14 | MA0748.2 (YY2) | STREME-7 | 7-CCGCCA | CCGCCA | YY2 | 0 | 0 | 18800 | 64.8 | 14000 | 47.7 | 1.36 |
| 19 | 15 | MA1713.1 (ZNF610) | STREME-11 | 11-AGCGGC | 11-AGCGGC | ZNF610 | 0 | 0 | 20400 | 70.2 | 15700 | 53.6 | 1.31 |
| 20 | 16 | MA1650.1 | ZBTB14 | MA1650.1 | SSCCGCGCACVS | ZBTB14 | 0 | 0 | 17600 | 60.6 | 12800 | 43.7 | 1.39 |
| 21 | 17 | MA1654.1 (ZNF16) | STREME-6 | 6-GGAGCC | GGAGCC | ZNF16 | 0 | 0 | 23500 | 80.8 | 19300 | 65.9 | 1.23 |
| 22 | 18 | MA0810.1 | TFAP2A | MA0810.1 | YGCCCBVRGGCR | TFAP2A | 0 | 0 | 14000 | 48.2 | 9240 | 31.6 | 1.52 |
| 23 | 19 | MA1712.1 | ZNF454 | MA1712.1 | DGGCTCSSGGCCCYGVKG | ZNF454 | 0 | 0 | 12800 | 43.9 | 8100 | 27.7 | 1.58 |
| 24 | 20 | MA1721.1 | ZNF93 | MA1721.1 | GGYAGCRGCAGCGGYG | ZNF93 | 0 | 0 | 10300 | 35.6 | 5990 | 20.5 | 1.74 |
| 25 | 5 | MA1630.2 | ZNF281 | MA1630.2 | GGGGGAGGGGVV | ZNF281 | 0 | 0 | 7490 | 25.8 | 3700 | 12.6 | 2.04 |
| 26 | 21 | MA1976.1 | ZNF320 | MA1976.1 | DGKGGGRCCMRGGGGCCWGTGHVNNNN | ZNF320 | 0 | 0 | 12200 | 41.9 | 7660 | 26.2 | 1.6 |
| 27 | 22 | MA1981.1 | ZNF530 | MA1981.1 | GMARGGMRAGGGGCRGSV | ZNF530 | 0 | 0 | 17200 | 59.1 | 12400 | 42.5 | 1.39 |
| 28 | 9 | MA0516.3 | SP2 | MA0516.3 | GGGGCGGGG | SP2 | 0 | 0 | 12800 | 44.2 | 8290 | 28.3 | 1.56 |
| 29 | 9 | MA1959.1 | KLF7 | MA1959.1 | DGGGCGGGG | KLF7 | 0 | 0 | 15100 | 51.9 | 10400 | 35.6 | 1.46 |
| 30 | 3 | MA1929.1 | CTCF | MA1929.1 | CTGCAGTKCCNNNNNYNRCCASYAGRKGGCRSWG | CTCF | 0 | 0 | 8630 | 29.7 | 4660 | 15.9 | 1.86 |
| 31 | 23 | MA1728.1 | ZNF549 | MA1728.1 | NNTGCTGCCCWR | ZNF549 | 0 | 0 | 22400 | 77.1 | 18200 | 62.2 | 1.24 |
| 32 | 3 | MA0139.1 | CTCF | MA0139.1 | YGRCCASYAGRKGGCRSYR | CTCF | 0 | 0 | 8260 | 28.4 | 4400 | 15.1 | 1.89 |
| 33 | 24 | MA1615.1 | Plagl1 | MA1615.1 | NNCTGGGGCCABN | Plagl1 | 4.94065645841247e-324 | 4.94065645841247e-324 | 22400 | 76.9 | 18200 | 62.2 | 1.24 |
| 34 | 9 | MA0742.2 | KLF12 | MA0742.2 | GGGGCGGGG | KLF12 | 4.25884586715155e-321 | 1.23516411460312e-322 | 12500 | 43 | 8140 | 27.8 | 1.55 |
| 35 | 9 | MA1513.1 | KLF15 | MA1513.1 | SCCCCGCCCCS | KLF15 | 1.70999998440975e-316 | 2.66000003064469e-318 | 7690 | 26.4 | 4050 | 13.8 | 1.91 |
| 36 | 18 | MA0872.1 | TFAP2A | MA0872.1 | TGCCCYSRGGGCA | TFAP2A | 3.00999999999998e-310 | 4.55999999999834e-312 | 8100 | 27.8 | 4410 | 15.1 | 1.85 |
| 37 | 9 | MA1511.2 | KLF10 | MA1511.2 | GGGGCGGGG | KLF10 | 1.59e-308 | 2.34000000000002e-310 | 13900 | 47.8 | 9520 | 32.5 | 1.47 |
| 38 | 25 | MA1631.1 | ASCL1 | MA1631.1 | NNGCACCTGCYNB | ASCL1 | 2.41e-305 | 3.46e-307 | 20800 | 71.5 | 16600 | 56.6 | 1.26 |
| 39 | 18 | MA0524.2 | TFAP2C | MA0524.2 | YGCCYBVRGGCA | TFAP2C | 3.21e-304 | 4.49e-306 | 12500 | 43.1 | 8280 | 28.3 | 1.52 |
| 40 | 18 | MA0811.1 | TFAP2B | MA0811.1 | YGCCCBVRGGCA | TFAP2B | 1.46e-303 | 1.99e-305 | 12200 | 42.1 | 8020 | 27.4 | 1.54 |
| 41 | 18 | MA0813.1 | TFAP2B | MA0813.1 | TGCCCYBRGGGCA | TFAP2B | 1.63e-302 | 2.16e-304 | 7110 | 24.4 | 3660 | 12.5 | 1.95 |
| 42 | 18 | MA0815.1 | TFAP2C | MA0815.1 | TGCCCYSRGGGCA | TFAP2C | 2.18e-302 | 2.82e-304 | 7060 | 24.3 | 3630 | 12.4 | 1.96 |
| 43 | 9 | MA0740.2 | KLF14 | MA0740.2 | KGGGCGGGG | KLF14 | 7.38e-301 | 9.35999999999999e-303 | 18200 | 62.6 | 13800 | 47.3 | 1.32 |
| 44 | 10 | MA0753.2 | ZNF740 | MA0753.2 | CYRCCCCCCCCAC | ZNF740 | 9.79999999999999e-298 | 1.21e-299 | 10500 | 36.3 | 6540 | 22.3 | 1.62 |
| 45 | 26 | MA1986.1 | ZNF692 | MA1986.1 | BGGGCCCASN | ZNF692 | 1.65e-291 | 2e-293 | 14200 | 48.7 | 9890 | 33.8 | 1.44 |
| 46 | 9 | MA0493.2 | KLF1 | MA0493.2 | DGGGYGGGG | KLF1 | 4.19e-290 | 4.97e-292 | 13200 | 45.4 | 8980 | 30.7 | 1.48 |
| 47 | 27 | MA0751.1 | ZIC4 | MA0751.1 | GRCCCCCCGCKGYGH | ZIC4 | 1.12e-288 | 1.3e-290 | 11600 | 39.8 | 7500 | 25.6 | 1.55 |
| 48 | 28 | MA1643.1 | NFIB | MA1643.1 | NNCCTGGCAYNGTGCCAAGNN | NFIB | 1.52e-288 | 1.73e-290 | 9400 | 32.3 | 5610 | 19.2 | 1.69 |
| 49 | 3 | MA1930.1 | CTCF | MA1930.1 | CTGCAGTKCCNVCHNNYRGCCASYAGRKGGCRAKC | CTCF | 1.62e-285 | 1.8e-287 | 6650 | 22.9 | 3390 | 11.6 | 1.97 |
| 50 | 29 | MA1727.1 | ZNF417 | MA1727.1 | VRBVNTGGGCGCCAM | ZNF417 | 3.3e-283 | 3.6e-285 | 13800 | 47.5 | 9600 | 32.8 | 1.45 |
| 51 | 30 | MA0814.2 | TFAP2C | MA0814.2 | NNGCCTGAGGCGNN | TFAP2C | 1.63e-282 | 1.74e-284 | 20800 | 71.6 | 16800 | 57.4 | 1.25 |
| 52 | 25 | MA0830.2 | TCF4 | MA0830.2 | NNGCACCTGCCNN | TCF4 | 7.65e-282 | 8.02e-284 | 23200 | 79.7 | 19500 | 66.5 | 1.2 |
| 53 | 25 | MA0506.2 | Nrf1 | MA0506.2 | SNCTGCGCMTGCGCV | Nrf1 | 3.74e-278 | 3.85e-280 | 15700 | 54.1 | 11500 | 39.3 | 1.38 |
| 54 | 31 | MA0471.2 | E2F6 | MA0471.2 | VVDGGCGGGAAVV | E2F6 | 1.35e-277 | 1.37e-279 | 10700 | 37 | 6840 | 23.4 | 1.58 |
| 55 | 27 | MA1578.1 | VEZF1 | MA1578.1 | CCCCCCMYDH | VEZF1 | 2.72e-277 | 2.7e-279 | 14500 | 49.7 | 10300 | 35.1 | 1.42 |
| 56 | 31 | MA1122.1 | TFDP1 | MA1122.1 | VSGCGGGAAVN | TFDP1 | 5e-268 | 4.87e-270 | 9240 | 31.8 | 5600 | 19.1 | 1.66 |
| 57 | 18 | MA0812.1 | TFAP2B | MA0812.1 | HGCCTSAGGCD | TFAP2B | 8.21e-268 | 7.85e-270 | 18500 | 63.7 | 14400 | 49.3 | 1.29 |
| 58 | 9 | MA1965.1 | SP5 | MA1965.1 | CBCCTCCCHS | SP5 | 1.94e-263 | 1.83e-265 | 11500 | 39.6 | 7640 | 26.1 | 1.52 |
| 59 | 29 | MA0024.3 | E2F1 | MA0024.3 | TTTGGCGCCAAA | E2F1 | 2.92e-262 | 2.7e-264 | 10700 | 36.7 | 6890 | 23.6 | 1.56 |
| 60 | 25 | MA1628.1 | Zic1::Zic2 | MA1628.1 | CVCAGCAGGNV | Zic1::Zic2 | 1.45e-260 | 1.32e-262 | 15500 | 53.3 | 11400 | 39 | 1.37 |
| 61 | 32 | MA0146.2 | Zfx | MA0146.2 | SSSGCCBVGGCCTS | Zfx | 4.95e-259 | 4.42e-261 | 13500 | 46.2 | 9450 | 32.3 | 1.43 |
| 62 | 25 | MA0697.2 | Zic3 | MA0697.2 | CNCAGCAGGAGNN | Zic3 | 1.81e-258 | 1.6e-260 | 14800 | 51 | 10800 | 36.8 | 1.38 |
| 63 | 33 | MA1982.1 | ZNF574 | MA1982.1 | VGSCTAGAGMGGCCS | ZNF574 | 7.47e-258 | 6.46e-260 | 19500 | 67.1 | 15600 | 53.2 | 1.26 |
| 64 | 28 | MA1528.1 | NFIX | MA1528.1 | STTGGCRNNGTGCCARB | NFIX | 1.79e-249 | 1.53e-251 | 7960 | 27.4 | 4640 | 15.9 | 1.72 |
| 65 | 27 | MA1584.1 | ZIC5 | MA1584.1 | VGACCCCCCGCTGHGM | ZIC5 | 1.12e-247 | 9.39e-250 | 6250 | 21.5 | 3280 | 11.2 | 1.92 |
| 66 | 34 | MA1596.1 | ZNF460 | MA1596.1 | GCCTCMGCCTCCCRAG | ZNF460 | 3.9e-247 | 3.22e-249 | 11800 | 40.7 | 8040 | 27.5 | 1.48 |
| 67 | 28 | MA0671.1 | NFIX | MA0671.1 | NNTGCCAAN | NFIX | 4.14e-246 | 3.37e-248 | 19000 | 65.4 | 15100 | 51.7 | 1.27 |
| 68 | 10 | MA0039.4 | KLF4 | MA0039.4 | NNCCCCACCCHN | KLF4 | 5.94e-245 | 4.76e-247 | 11600 | 39.9 | 7830 | 26.8 | 1.49 |
| 69 | 25 | MA1997.1 | Olig2 | MA1997.1 | NVCAGCTGBN | Olig2 | 1.38e-244 | 1.09e-246 | 9930 | 34.1 | 6350 | 21.7 | 1.57 |
| 70 | 35 | MA1627.1 | Wt1 | MA1627.1 | YBCCTCCCCCACVB | Wt1 | 1.7e-244 | 1.32e-246 | 13500 | 46.4 | 9600 | 32.8 | 1.41 |
| 71 | 25 | MA1648.1 | TCF12 | MA1648.1 | NNCACCTGCNN | TCF12 | 1.6e-243 | 1.23e-245 | 20100 | 69 | 16200 | 55.5 | 1.24 |
| 72 | 9 | MA1564.1 | SP9 | MA1564.1 | RCCACGCCCMCY | SP9 | 2.34e-243 | 1.77e-245 | 16400 | 56.4 | 12400 | 42.5 | 1.33 |
| 73 | 27 | MA0736.1 | GLIS2 | MA0736.1 | GACCCCCCGCRAMG | GLIS2 | 3.87e-242 | 2.89e-244 | 11600 | 39.9 | 7870 | 26.9 | 1.48 |
| 74 | 27 | MA0696.1 | ZIC1 | MA0696.1 | GACCCCCYGCTGTG | ZIC1 | 1.61e-240 | 1.19e-242 | 7210 | 24.8 | 4080 | 14 | 1.78 |
| 75 | 36 | MA1514.1 | KLF17 | MA1514.1 | MMCCACGCACCCMTY | KLF17 | 9.15999999999999e-240 | 6.66e-242 | 16600 | 57.1 | 12700 | 43.4 | 1.32 |
| 76 | 35 | MA0162.4 | EGR1 | MA0162.4 | VCMCGCCCACGCVS | EGR1 | 3.23e-239 | 2.32e-241 | 16900 | 58 | 12900 | 44.3 | 1.31 |
| 77 | 37 | MA1726.1 | ZNF331 | MA1726.1 | NMYTGCAGAGCCCH | ZNF331 | 8.45e-239 | 5.99e-241 | 14600 | 50.2 | 10700 | 36.6 | 1.37 |
| 78 | 38 | MA1599.1 | ZNF682 | MA1599.1 | BVGGCYAAGCCCCWNY | ZNF682 | 4.84e-236 | 3.38e-238 | 12600 | 43.2 | 8810 | 30.1 | 1.44 |
| 79 | 9 | MA0599.1 | KLF5 | MA0599.1 | GCCCCDCCCH | KLF5 | 1.38e-230 | 9.51e-233 | 10700 | 36.8 | 7140 | 24.4 | 1.51 |
| 80 | 39 | MA0738.1 | HIC2 | MA0738.1 | RTGCCCRSB | HIC2 | 1.38e-228 | 9.37e-231 | 14200 | 48.9 | 10400 | 35.6 | 1.37 |
| 81 | 40 | MA0131.2 | HINFP | MA0131.2 | CARCGTCCGCKN | HINFP | 1.27e-227 | 8.57e-230 | 8390 | 28.9 | 5140 | 17.6 | 1.64 |
| 82 | 9 | MA0741.1 | KLF16 | MA0741.1 | GMCACGCCCCC | KLF16 | 2.29e-225 | 1.52e-227 | 5730 | 19.7 | 3000 | 10.2 | 1.92 |
| 83 | 25 | MA0522.3 | TCF3 | MA0522.3 | NVCACCTGCNN | TCF3 | 9.13e-225 | 6e-227 | 20600 | 70.7 | 17000 | 58 | 1.22 |
| 84 | 25 | MA0499.2 | MYOD1 | MA0499.2 | NNGCACCTGTCNB | MYOD1 | 1.23e-224 | 8.01e-227 | 20800 | 71.5 | 17200 | 58.8 | 1.22 |
| 85 | 10 | MA1515.1 | KLF2 | MA1515.1 | NRCCACRCCCH | KLF2 | 2.76e-223 | 1.77e-225 | 21200 | 72.9 | 17700 | 60.4 | 1.21 |
| 86 | 25 | MA1993.1 | Neurod2 | MA1993.1 | VVCAGCTGBB | Neurod2 | 8.95e-221 | 5.67e-223 | 10500 | 36.2 | 7040 | 24.1 | 1.5 |
| 87 | 30 | MA0003.4 | TFAP2A | MA0003.4 | NNKGCCTCAGGCNN | TFAP2A | 6.38e-218 | 4e-220 | 16400 | 56.2 | 12600 | 43.1 | 1.3 |
| 88 | 28 | MA0739.1 | Hic1 | MA0739.1 | RTGCCAACY | Hic1 | 1.82e-217 | 1.13e-219 | 15900 | 54.6 | 12100 | 41.5 | 1.32 |
| 89 | 35 | MA1107.2 | KLF9 | MA1107.2 | MNGCCACACCCACHYV | KLF9 | 1.75e-213 | 1.07e-215 | 21800 | 74.8 | 18400 | 62.8 | 1.19 |
| 90 | 27 | MA1629.1 | Zic2 | MA1629.1 | NDCACAGCAGGDRG | Zic2 | 7.04e-212 | 4.26e-214 | 15900 | 54.6 | 12200 | 41.7 | 1.31 |
| 91 | 41 | MA1719.1 | ZNF816 | MA1719.1 | DRRKGGGGACATGCHGGG | ZNF816 | 4.33e-211 | 2.6e-213 | 18400 | 63.3 | 14800 | 50.5 | 1.25 |
| 92 | 18 | MA1569.1 | TFAP2E | MA1569.1 | YGCCYSRGGCD | TFAP2E | 1.15e-210 | 6.83e-213 | 5340 | 18.4 | 2780 | 9.5 | 1.93 |
| 93 | 9 | MA0747.1 | SP8 | MA0747.1 | RCCACGCCCMCY | SP8 | 3.64e-210 | 2.13e-212 | 14000 | 48.2 | 10400 | 35.5 | 1.36 |
| 94 | 9 | MA1516.1 | KLF3 | MA1516.1 | GRCCRCGCCCH | KLF3 | 5.19e-209 | 3.01e-211 | 14900 | 51.3 | 11300 | 38.5 | 1.33 |
| 95 | 9 | MA0746.2 | SP3 | MA0746.2 | NGCCACGCCCMCH | SP3 | 8.73e-202 | 5.01e-204 | 8420 | 29 | 5340 | 18.3 | 1.59 |
| 96 | 25 | MA0633.2 | Twist2 | MA0633.2 | NVCAGCTGBN | Twist2 | 3.57e-198 | 2.03e-200 | 10600 | 36.3 | 7250 | 24.8 | 1.46 |
| 97 | 42 | MA1637.1 | EBF3 | MA1637.1 | NYCCCAAGGGANN | EBF3 | 2.05e-196 | 1.15e-198 | 16000 | 55.1 | 12500 | 42.6 | 1.29 |
| 98 | 31 | MA0865.2 | E2F8 | MA0865.2 | TTCCCGCCAHWA | E2F8 | 4.15e-194 | 2.31e-196 | 14100 | 48.6 | 10600 | 36.3 | 1.34 |
| 99 | 21 | MA1723.1 | PRDM9 | MA1723.1 | RGDGGGVAGGGRGGVRRMARVARR | PRDM9 | 6.73e-193 | 3.7e-195 | 10900 | 37.4 | 7580 | 25.9 | 1.44 |
| 100 | 25 | MA1635.1 | BHLHE22 | MA1635.1 | NVCAGCTGBN | BHLHE22 | 9.79e-193 | 5.34e-195 | 9950 | 34.2 | 6750 | 23.1 | 1.48 |
| 101 | 3 | MA0155.1 | INSM1 | MA0155.1 | TGYCWGGGGGCR | INSM1 | 1.74e-191 | 9.37e-194 | 8920 | 30.6 | 5850 | 20 | 1.53 |
| 102 | 10 | MA0528.2 | ZNF263 | MA0528.2 | VNGGGGAGGASG | ZNF263 | 7.85e-189 | 4.2e-191 | 17300 | 59.5 | 13900 | 47.4 | 1.26 |
| 103 | 35 | MA0733.1 | EGR4 | MA0733.1 | NHMCGCCCACGCAHWT | EGR4 | 1.08e-188 | 5.71e-191 | 18700 | 64.2 | 15300 | 52.1 | 1.23 |
| 104 | 9 | MA1512.1 | KLF11 | MA1512.1 | SCCACGCCCMC | KLF11 | 1.19e-188 | 6.24e-191 | 12500 | 42.9 | 9100 | 31.1 | 1.38 |
| 105 | 43 | MA0774.1 (MEIS2) | STREME-12 | 12-GACAGCC | 12-GACAGCC | MEIS2 | 4.76e-186 | 2.47e-188 | 11000 | 37.9 | 7770 | 26.6 | 1.43 |
| 106 | 44 | MA1985.1 | ZNF669 | MA1985.1 | GGGGYGAYGACCRCT | ZNF669 | 1.84e-185 | 9.44e-188 | 12500 | 43 | 9170 | 31.3 | 1.37 |
| 107 | 42 | MA0154.4 | EBF1 | MA0154.4 | NNTCCCCAGGGGANN | EBF1 | 1.76e-181 | 8.95e-184 | 15800 | 54.5 | 12400 | 42.5 | 1.28 |
| 108 | 31 | MA1583.1 | ZFP57 | MA1583.1 | NNVTTGCCGCANN | ZFP57 | 1.95e-181 | 9.82e-184 | 8800 | 30.3 | 5820 | 19.9 | 1.52 |
| 109 | 45 | MA1730.1 | ZNF708 | MA1730.1 | NNRGCTGTRCCTCCY | ZNF708 | 8.42e-181 | 4.21e-183 | 21700 | 74.5 | 18600 | 63.5 | 1.17 |
| 110 | 25 | MA0821.2 | HES5 | MA0821.2 | GRCACGTGYC | HES5 | 3.11e-180 | 1.54e-182 | 10400 | 35.8 | 7270 | 24.8 | 1.44 |
| 111 | 25 | MA0104.4 | MYCN | MA0104.4 | VVCCACGTGGBB | MYCN | 4.81e-180 | 2.36e-182 | 13900 | 47.9 | 10600 | 36.1 | 1.33 |
| 112 | 31 | MA1987.1 | ZNF701 | MA1987.1 | GAGCACYAARGGGGGRADDDD | ZNF701 | 3.07e-179 | 1.5e-181 | 10600 | 36.4 | 7420 | 25.4 | 1.43 |
| 113 | 25 | MA0147.3 | MYC | MA0147.3 | NNCCACGTGCNB | MYC | 1.19e-178 | 5.75e-181 | 22500 | 77.5 | 19600 | 66.8 | 1.16 |
| 114 | 46 | MA0748.2 | YY2 | MA0748.2 | NVATGGCGGCS | YY2 | 7.51e-178 | 3.59e-180 | 8520 | 29.3 | 5610 | 19.2 | 1.53 |
| 115 | 47 | MA0759.2 | ELK3 | MA0759.2 | ACCGGAAGTRV | ELK3 | 1.35e-177 | 6.42e-180 | 8660 | 29.8 | 5730 | 19.6 | 1.52 |
| 116 | 48 | MA0734.3 | Gli2 | MA0734.3 | NRGACCACCCASV | Gli2 | 6.05e-173 | 2.85e-175 | 14100 | 48.3 | 10800 | 36.8 | 1.31 |
| 117 | 25 | MA1966.1 | TFAP4::ETV1 | MA1966.1 | RSCGGAAGCAGSTGKN | TFAP4::ETV1 | 7e-171 | 3.26e-173 | 19400 | 66.8 | 16200 | 55.5 | 1.2 |
| 118 | 49 | MA0145.2 | Tfcp2l1 | MA0145.2 | CCAGYYYVADCCRG | Tfcp2l1 | 1.62e-170 | 7.51e-173 | 11700 | 40.2 | 8520 | 29.1 | 1.38 |
| 119 | 50 | 13-CCAGGCCT | STREME-13 | 13-CCAGGCCT | 13-CCAGGCCT | CCAGGCCT | 8.53e-170 | 3.91e-172 | 17400 | 60 | 14200 | 48.4 | 1.24 |
| 120 | 25 | MA1945.1 | ETV5::FIGLA | MA1945.1 | RGCGGAARCAGGTGG | ETV5::FIGLA | 8.8e-167 | 4e-169 | 12200 | 41.9 | 9040 | 30.9 | 1.36 |
| 121 | 51 | MA1656.1 | ZNF449 | MA1656.1 | NNAAGCCCAACCNN | ZNF449 | 2.49e-165 | 1.12e-167 | 16800 | 57.6 | 13500 | 46.2 | 1.25 |
| 122 | 52 | MA1710.1 | ZNF257 | MA1710.1 | VDGAGGCRAGRG | ZNF257 | 4e-161 | 1.79e-163 | 13100 | 45 | 9950 | 34 | 1.32 |
| 123 | 53 | MA0774.1 | MEIS2 | MA0774.1 | TTGACAGS | MEIS2 | 2.04e-159 | 9.03e-162 | 5200 | 17.9 | 2970 | 10.1 | 1.76 |
| 124 | 47 | MA0750.2 | ZBTB7A | MA0750.2 | NVCCGGAAGTGSV | ZBTB7A | 1.09e-158 | 4.78e-161 | 14600 | 50.2 | 11400 | 39.1 | 1.28 |
| 125 | 25 | MA1941.1 | ETV2::FIGLA | MA1941.1 | RSCGGAARCAGSTGBN | ETV2::FIGLA | 3.08e-154 | 1.34e-156 | 21300 | 73.2 | 18400 | 62.9 | 1.16 |
| 126 | 25 | MA1100.2 | ASCL1 | MA1100.2 | VGCAGCTGCN | ASCL1 | 1.78e-152 | 7.71e-155 | 24300 | 83.6 | 21800 | 74.7 | 1.12 |
| 127 | 29 | MA0864.2 | E2F2 | MA0864.2 | RWTTTGGCGCCAWWWY | E2F2 | 5.19e-150 | 2.23e-152 | 9670 | 33.3 | 6860 | 23.5 | 1.42 |
| 128 | 9 | MA1517.1 | KLF6 | MA1517.1 | NRCCACGCCCH | KLF6 | 2.51e-149 | 1.07e-151 | 19100 | 65.7 | 16100 | 55 | 1.19 |
| 129 | 25 | MA1655.1 | ZNF341 | MA1655.1 | NRGAACAGCCNN | ZNF341 | 2.6e-147 | 1.1e-149 | 21300 | 73.2 | 18500 | 63.1 | 1.16 |
| 130 | 35 | MA0732.1 | EGR3 | MA0732.1 | HHMCGCCCACGCAHH | EGR3 | 1.52e-146 | 6.36e-149 | 7160 | 24.6 | 4680 | 16 | 1.54 |
| 131 | 54 | MA0646.1 | GCM1 | MA0646.1 | BATGCGGGTAC | GCM1 | 5.31e-146 | 2.21e-148 | 18900 | 65.1 | 16000 | 54.6 | 1.19 |
| 132 | 25 | MA1472.2 | Bhlha15 | MA1472.2 | NVACAGCTGTBN | Bhlha15 | 5.83e-146 | 2.41e-148 | 16500 | 56.8 | 13500 | 46.1 | 1.23 |
| 133 | 30 | MA1581.1 | ZBTB6 | MA1581.1 | DNSCTTGAGCCNV | ZBTB6 | 6.88e-146 | 2.82e-148 | 16400 | 56.2 | 13300 | 45.5 | 1.24 |
| 134 | 55 | MA1972.1 | ZFP14 | MA1972.1 | GGAGGCHCHGGARBG | ZFP14 | 5.77e-144 | 2.35e-146 | 18200 | 62.6 | 15200 | 52 | 1.2 |
| 135 | 56 | MA0472.2 | EGR2 | MA0472.2 | MCGCCCACGCA | EGR2 | 6.48e-144 | 2.62e-146 | 14400 | 49.4 | 11300 | 38.8 | 1.27 |
| 136 | 25 | MA0823.1 | HEY1 | MA0823.1 | GRCACGTGCC | HEY1 | 7.45e-143 | 2.99e-145 | 12700 | 43.7 | 9760 | 33.4 | 1.31 |
| 137 | 42 | MA1604.1 | Ebf2 | MA1604.1 | NYCCCAAGGGANN | Ebf2 | 3.6e-140 | 1.43e-142 | 20600 | 70.9 | 17800 | 60.9 | 1.16 |
| 138 | 25 | MA2002.1 | Zfp335 | MA2002.1 | VNTCAGGCANH | Zfp335 | 2.14e-136 | 8.47e-139 | 23500 | 80.7 | 21000 | 71.9 | 1.12 |
| 139 | 29 | MA0470.2 | E2F4 | MA0470.2 | TTTTGGCGCCAWWW | E2F4 | 3.27e-136 | 1.28e-138 | 6700 | 23 | 4360 | 14.9 | 1.54 |
| 140 | 34 | MA1587.1 | ZNF135 | MA1587.1 | CCTCGACCTCCYRR | ZNF135 | 1.17e-133 | 4.54e-136 | 14800 | 50.8 | 11900 | 40.6 | 1.25 |
| 141 | 25 | MA0500.2 | MYOG | MA0500.2 | NDRCAGCTGYHN | MYOG | 1.28e-133 | 4.95e-136 | 23700 | 81.6 | 21300 | 72.9 | 1.12 |
| 142 | 57 | MA1979.1 | ZNF416 | MA1979.1 | TATCTGGGCAH | ZNF416 | 1.84e-133 | 7.08e-136 | 22500 | 77.5 | 20000 | 68.4 | 1.13 |
| 143 | 21 | MA0163.1 | PLAG1 | MA0163.1 | GGGGCCCWAGGGGG | PLAG1 | 5.69e-132 | 2.17e-134 | 4330 | 14.9 | 2450 | 8.37 | 1.78 |
| 144 | 58 | MA0527.1 | ZBTB33 | MA0527.1 | BTCTCGCGAGABYTS | ZBTB33 | 2.78e-131 | 1.05e-133 | 7010 | 24.1 | 4670 | 16 | 1.51 |
| 145 | 25 | MA1641.1 | MYF5 | MA1641.1 | NVACAGCTGTBN | MYF5 | 5.81e-130 | 2.18e-132 | 17000 | 58.5 | 14200 | 48.4 | 1.21 |
| 146 | 59 | MA0855.1 (RXRB) | STREME-10 | 10-GACCSC | 10-GACCSC | RXRB | 3.45e-127 | 1.29e-129 | 13300 | 45.9 | 10500 | 36 | 1.27 |
| 147 | 60 | MA1149.1 | RARA::RXRG | MA1149.1 | RRGGTCAHNNNRRGGTCA | RARA::RXRG | 4.25e-125 | 1.57e-127 | 10200 | 35.1 | 7580 | 25.9 | 1.35 |
| 148 | 25 | MA0745.2 | SNAI2 | MA0745.2 | NBGCACCTGTMNY | SNAI2 | 1.03e-123 | 3.79e-126 | 21800 | 74.8 | 19200 | 65.8 | 1.14 |
| 149 | 48 | MA0694.1 | ZBTB7B | MA0694.1 | RCGACCACCGAA | ZBTB7B | 3.41e-123 | 1.25e-125 | 12000 | 41.1 | 9260 | 31.6 | 1.3 |
| 150 | 61 | MA0504.1 | NR2C2 | MA0504.1 | VGRGGTCARAGGTCA | NR2C2 | 3.56e-123 | 1.29e-125 | 10700 | 36.7 | 8050 | 27.5 | 1.33 |
| 151 | 62 | MA1731.1 | ZNF768 | MA1731.1 | YBVCYBRSCCTCTCTGDG | ZNF768 | 1.84e-121 | 6.63e-124 | 12400 | 42.6 | 9670 | 33.1 | 1.29 |
| 152 | 60 | MA1535.1 | NR2C1 | MA1535.1 | NRRGGTCAN | NR2C1 | 4.8e-120 | 1.72e-122 | 23900 | 82.3 | 21700 | 74.3 | 1.11 |
| 153 | 25 | MA0048.2 | NHLH1 | MA0048.2 | CGCAGCTGCK | NHLH1 | 3.93e-116 | 1.4e-118 | 18300 | 62.8 | 15600 | 53.4 | 1.18 |
| 154 | 63 | MA1973.1 | ZKSCAN3 | MA1973.1 | KDKCCCAGGCTAGCCCABNN | ZKSCAN3 | 4.82e-116 | 1.71e-118 | 10200 | 34.9 | 7640 | 26.1 | 1.34 |
| 155 | 53 | MA0138.2 | REST | MA0138.2 | TTCAGCACCATGGACAGCDCC | REST | 9.37e-114 | 3.29e-116 | 6800 | 23.4 | 4640 | 15.8 | 1.47 |
| 156 | 64 | MA1716.1 | ZNF76 | MA1716.1 | NNTWCCCAYAATGCAHYGCGC | ZNF76 | 1.24e-111 | 4.35e-114 | 9430 | 32.4 | 7010 | 24 | 1.35 |
| 157 | 48 | MA1990.1 | Gli1 | MA1990.1 | NNRGACCACCCASV | Gli1 | 8.23e-110 | 2.86e-112 | 26000 | 89.6 | 24300 | 83.2 | 1.08 |
| 158 | 76 | MA0672.1 | NKX2-3 | MA0672.1 | VCCACTTRAV | NKX2-3 | 1.97e-109 | 6.79e-112 | 5920 | 20.4 | 3920 | 13.4 | 1.52 |
| 159 | 25 | MA0649.1 | HEY2 | MA0649.1 | GRCACGTGYC | HEY2 | 3.44e-109 | 1.18e-111 | 11300 | 39 | 8820 | 30.1 | 1.29 |
| 160 | 25 | MA1621.1 | Rbpjl | MA1621.1 | NNVACACCTGTBNN | Rbpjl | 1.07e-104 | 3.64e-107 | 23500 | 80.8 | 21400 | 73.2 | 1.1 |
| 161 | 60 | MA0159.1 | RARA::RXRA | MA0159.1 | RGKTCANVGRSAGGTCA | RARA::RXRA | 2.04e-103 | 6.92e-106 | 14200 | 48.8 | 11700 | 39.8 | 1.23 |
| 162 | 76 | MA2003.1 | NKX2-4 | MA2003.1 | CCACTTSAVV | NKX2-4 | 9.02e-103 | 3.03e-105 | 12600 | 43.4 | 10100 | 34.6 | 1.25 |
| 163 | 65 | MA1715.1 | ZNF707 | MA1715.1 | CCCCACTCCTGGTAC | ZNF707 | 2.99e-101 | 1e-103 | 10600 | 36.3 | 8170 | 27.9 | 1.3 |
| 164 | 25 | MA1938.1 | ERF::NHLH1 | MA1938.1 | VGCAGCTGCCGGAARYN | ERF::NHLH1 | 2.16e-97 | 7.18e-100 | 7980 | 27.4 | 5850 | 20 | 1.37 |
| 165 | 25 | MA1619.1 | Ptf1A | MA1619.1 | NNACAGCTGTNN | Ptf1A | 2.45e-95 | 8.08e-98 | 23600 | 81.2 | 21700 | 74 | 1.1 |
| 166 | 48 | MA0695.1 | ZBTB7C | MA0695.1 | RCGACCACCGAN | ZBTB7C | 5.68e-95 | 1.87e-97 | 17200 | 59.2 | 14800 | 50.6 | 1.17 |
| 167 | 25 | MA0103.3 | ZEB1 | MA0103.3 | SNCACCTGSVN | ZEB1 | 1.03e-94 | 3.35e-97 | 21600 | 74.4 | 19500 | 66.5 | 1.12 |
| 168 | 28 | MA1527.1 | NFIC | MA1527.1 | NTTGGCDNNRTGCCARN | NFIC | 4.23e-91 | 1.37e-93 | 3130 | 10.8 | 1770 | 6.06 | 1.77 |
| 169 | 54 | MA0767.1 | GCM2 | MA0767.1 | BATGCGGGTR | GCM2 | 2.29e-90 | 7.39e-93 | 9880 | 34 | 7670 | 26.2 | 1.3 |
| 170 | 66 | MA1964.1 | SMAD2 | MA1964.1 | NBCCAGACDN | SMAD2 | 9.87e-90 | 3.17e-92 | 11800 | 40.5 | 9480 | 32.4 | 1.25 |
| 171 | 67 | MA0803.1 | TBX15 | MA0803.1 | AGGTGTGA | TBX15 | 1.59e-88 | 5.06e-91 | 23200 | 79.6 | 21200 | 72.5 | 1.1 |
| 172 | 25 | MA1099.2 | HES1 | MA1099.2 | GGCRCGTGBC | HES1 | 1.77e-87 | 5.6e-90 | 4180 | 14.4 | 2640 | 9.03 | 1.59 |
| 173 | 47 | MA0764.3 | ETV4 | MA0764.3 | ACCGGAAGTR | ETV4 | 3.09e-87 | 9.75e-90 | 12900 | 44.5 | 10600 | 36.3 | 1.22 |
| 174 | 25 | MA0521.2 | Tcf12 | MA0521.2 | NNACAGCTGTNN | Tcf12 | 3.75e-86 | 1.18e-88 | 8110 | 27.9 | 6080 | 20.8 | 1.34 |
| 175 | 68 | MA1600.1 | ZNF684 | MA1600.1 | RYACAGTCCACCCCTTKR | ZNF684 | 1.21e-85 | 3.77e-88 | 9640 | 33.2 | 7500 | 25.6 | 1.29 |
| 176 | 69 | MA0632.2 | TCFL5 | MA0632.2 | KCACGCGCMC | TCFL5 | 1.27e-85 | 3.94e-88 | 6560 | 22.6 | 4700 | 16 | 1.4 |
| 177 | 70 | MA0130.1 | ZNF354C | MA0130.1 | MTCCAC | ZNF354C | 4.54e-85 | 1.4e-87 | 27100 | 93.2 | 25900 | 88.5 | 1.05 |
| 178 | 71 | MA1711.1 | ZNF343 | MA1711.1 | CCGCTTCMCCDCGGCMV | ZNF343 | 3.34e-84 | 1.02e-86 | 6490 | 22.3 | 4650 | 15.9 | 1.4 |
| 179 | 72 | MA1575.1 | THRB | MA1575.1 | DTGACCTBRNYVAGGTCAH | THRB | 8.78e-83 | 2.67e-85 | 5700 | 19.6 | 3980 | 13.6 | 1.44 |
| 180 | 53 | MA0775.1 | MEIS3 | MA0775.1 | DTGACAGS | MEIS3 | 3.4e-82 | 1.03e-84 | 3950 | 13.6 | 2500 | 8.53 | 1.59 |
| 181 | 25 | MA0616.2 | HES2 | MA0616.2 | GGCACGTGYC | HES2 | 1.49e-81 | 4.5e-84 | 8620 | 29.6 | 6600 | 22.6 | 1.31 |
| 182 | 25 | MA0816.1 | Ascl2 | MA0816.1 | ARCAGCTGCY | Ascl2 | 5.44e-80 | 1.63e-82 | 18800 | 64.7 | 16600 | 56.9 | 1.14 |
| 183 | 28 | MA0161.2 | NFIC | MA0161.2 | NNCTTGGCANN | NFIC | 8.52e-80 | 2.54e-82 | 15500 | 53.2 | 13200 | 45.2 | 1.18 |
| 184 | 67 | MA1566.2 | TBX3 | MA1566.2 | RAGGTGTGAAV | TBX3 | 1.33e-78 | 3.95e-81 | 22100 | 76 | 20200 | 69 | 1.1 |
| 185 | 67 | MA0806.1 | TBX4 | MA0806.1 | AGGTGTGA | TBX4 | 3.18e-78 | 9.37e-81 | 22800 | 78.5 | 21000 | 71.7 | 1.1 |
| 186 | 28 | MA0119.1 | NFIC::TLX1 | MA0119.1 | TGGCASSRWGCCAA | NFIC::TLX1 | 8.62e-78 | 2.53e-80 | 2820 | 9.7 | 1620 | 5.55 | 1.75 |
| 187 | 25 | MA1485.1 | FERD3L | MA1485.1 | GCRMCAGCTGTYAC | FERD3L | 1.29e-77 | 3.75e-80 | 6040 | 20.8 | 4320 | 14.8 | 1.41 |
| 188 | 25 | MA0807.1 | TBX5 | MA0807.1 | AGGTGTKA | TBX5 | 1.62e-75 | 4.71e-78 | 21800 | 74.8 | 19800 | 67.8 | 1.1 |
| 189 | 25 | MA1620.1 | Ptf1A | MA1620.1 | NVACACCTGTNN | Ptf1A | 2.14e-75 | 6.17e-78 | 25400 | 87.4 | 23900 | 81.8 | 1.07 |
| 190 | 47 | MA0765.3 | ETV5 | MA0765.3 | ACCGGAAGTR | ETV5 | 3.27e-75 | 9.37e-78 | 7000 | 24.1 | 5210 | 17.8 | 1.35 |
| 191 | 53 | MA0498.2 | MEIS1 | MA0498.2 | HTGACAD | MEIS1 | 7.82e-73 | 2.23e-75 | 16200 | 55.6 | 14000 | 48 | 1.16 |
| 192 | 73 | MA0014.3 | PAX5 | MA0014.3 | RRGCGTGACCNN | PAX5 | 8.89e-71 | 2.52e-73 | 3430 | 11.8 | 2160 | 7.39 | 1.6 |
| 193 | 72 | MA1533.1 | NR1I2 | MA1533.1 | DYGAACTSVDTGAACTC | NR1I2 | 1.75e-70 | 4.95e-73 | 11000 | 37.8 | 8970 | 30.6 | 1.23 |
| 194 | 76 | MA0673.1 | NKX2-8 | MA0673.1 | VCACTTSAV | NKX2-8 | 3.2e-70 | 9.01e-73 | 6570 | 22.6 | 4880 | 16.7 | 1.36 |
| 195 | 25 | MA1652.1 | ZKSCAN5 | MA1652.1 | NNRGGARGTGAGRR | ZKSCAN5 | 2.45e-69 | 6.85e-72 | 12200 | 42 | 10200 | 34.8 | 1.21 |
| 196 | 47 | MA1992.1 | Ikzf3 | MA1992.1 | NNRCAGGAAGTGGVN | Ikzf3 | 8.83e-68 | 2.46e-70 | 19700 | 67.7 | 17700 | 60.7 | 1.12 |
| 197 | 25 | MA1558.1 | SNAI1 | MA1558.1 | DRCAGGTGYD | SNAI1 | 9.94e-67 | 2.75e-69 | 17500 | 60.1 | 15500 | 52.9 | 1.14 |
| 198 | 61 | MA0065.2 | Pparg::Rxra | MA0065.2 | STRGGGCARAGGKCA | Pparg::Rxra | 1.74e-66 | 4.78e-69 | 17300 | 59.6 | 15300 | 52.4 | 1.14 |
| 199 | 25 | MA0820.1 | FIGLA | MA0820.1 | WMCACCTGKW | FIGLA | 4.74e-65 | 1.3e-67 | 26100 | 89.6 | 24800 | 84.9 | 1.06 |
| 200 | 47 | MA1933.1 | ELK1::SREBF2 | MA1933.1 | DCCGGAAGTSRCGTGA | ELK1::SREBF2 | 1.17e-64 | 3.19e-67 | 9240 | 31.8 | 7400 | 25.3 | 1.26 |
| 201 | 74 | MA1474.1 | CREB3L4 | MA1474.1 | YGCCACGTCAYC | CREB3L4 | 2.06e-63 | 5.57e-66 | 7930 | 27.3 | 6200 | 21.2 | 1.29 |
| 202 | 25 | MA0668.2 | Neurod2 | MA0668.2 | NNGRACAGATGGYNN | Neurod2 | 2.07e-63 | 5.57e-66 | 20200 | 69.4 | 18400 | 62.7 | 1.11 |
| 203 | 75 | MA1468.1 | ATOH7 | MA1468.1 | AVCATATGBY | ATOH7 | 2.43e-63 | 6.52e-66 | 25900 | 89 | 24600 | 84.1 | 1.06 |
| 204 | 76 | MA1994.1 (Nkx2-1) | STREME-15 | 15-CCACTTGA | 15-CCACTTGA | Nkx2-1 | 3.13e-62 | 8.36e-65 | 3050 | 10.5 | 1920 | 6.56 | 1.6 |
| 205 | 53 | MA1571.1 | TGIF2LX | MA1571.1 | TGACAGCTGTCA | TGIF2LX | 4.17e-62 | 1.11e-64 | 15600 | 53.8 | 13700 | 46.8 | 1.15 |
| 206 | 77 | MA1969.1 | THRA | MA1969.1 | NYTGTGTCCTCABRTGACCTYWBB | THRA | 6.7e-62 | 1.77e-64 | 8820 | 30.3 | 7040 | 24.1 | 1.26 |
| 207 | 25 | MA1106.1 | HIF1A | MA1106.1 | SBACGTGCNN | HIF1A | 8.06e-62 | 2.12e-64 | 11400 | 39.2 | 9510 | 32.5 | 1.21 |
| 208 | 25 | MA0464.2 | BHLHE40 | MA0464.2 | VKCACGTGMC | BHLHE40 | 4.73e-61 | 1.24e-63 | 11800 | 40.6 | 9910 | 33.9 | 1.2 |
| 209 | 25 | MA1560.1 | SOHLH2 | MA1560.1 | YGCACGTGCN | SOHLH2 | 1.44e-60 | 3.75e-63 | 17200 | 59.2 | 15300 | 52.3 | 1.13 |
| 210 | 25 | MA1493.1 | HES6 | MA1493.1 | GGCACGTGTY | HES6 | 1.88e-59 | 4.88e-62 | 15300 | 52.6 | 13400 | 45.7 | 1.15 |
| 211 | 74 | MA0259.1 | ARNT::HIF1A | MA0259.1 | VBACGTGC | ARNT::HIF1A | 1.95e-59 | 5.03e-62 | 10400 | 35.9 | 8610 | 29.4 | 1.22 |
| 212 | 67 | MA0801.1 | MGA | MA0801.1 | AGGTGTGA | MGA | 1.06e-57 | 2.73e-60 | 20100 | 69 | 18300 | 62.5 | 1.1 |
| 213 | 78 | MA0511.2 | RUNX2 | MA0511.2 | WAACCGCAA | RUNX2 | 6.95e-57 | 1.78e-59 | 21100 | 72.6 | 19400 | 66.4 | 1.09 |
| 214 | 79 | MA1545.1 | OVOL2 | MA1545.1 | DDACCGTTABNYN | OVOL2 | 5.31e-56 | 1.35e-58 | 5240 | 18 | 3850 | 13.2 | 1.37 |
| 215 | 61 | MA1574.1 | THRB | MA1574.1 | RRGGTCAAAGGTCAN | THRB | 3.39e-55 | 8.58e-58 | 17100 | 58.8 | 15300 | 52.2 | 1.13 |
| 216 | 25 | MA1559.1 | SNAI3 | MA1559.1 | RRCAGGTGYA | SNAI3 | 6.63e-55 | 1.67e-57 | 17900 | 61.7 | 16100 | 55.2 | 1.12 |
| 217 | 76 | MA1464.1 | ARNT2 | MA1464.1 | GTCACGTGMN | ARNT2 | 3.71e-52 | 9.33e-55 | 8390 | 28.8 | 6780 | 23.2 | 1.24 |
| 218 | 76 | MA0608.1 | Creb3l2 | MA0608.1 | GCCACGTGD | Creb3l2 | 7.01e-52 | 1.75e-54 | 6510 | 22.4 | 5050 | 17.3 | 1.3 |
| 219 | 53 | MA1572.1 | TGIF2LY | MA1572.1 | TGACAGCTGTCA | TGIF2LY | 6.02e-51 | 1.5e-53 | 11300 | 39 | 9610 | 32.9 | 1.19 |
| 220 | 76 | MA0058.3 | MAX | MA0058.3 | AVCACGTGNY | MAX | 6.57e-51 | 1.63e-53 | 17500 | 60.1 | 15700 | 53.8 | 1.12 |
| 221 | 75 | MA0665.1 | MSC | MA0665.1 | AACAGCTGTT | MSC | 5.66e-50 | 1.4e-52 | 13700 | 47 | 11900 | 40.7 | 1.15 |
| 222 | 47 | MA0028.2 | ELK1 | MA0028.2 | ACCGGAAGTR | ELK1 | 7.67e-50 | 1.88e-52 | 12600 | 43.2 | 10800 | 37 | 1.17 |
| 223 | 25 | MA1529.1 | NHLH2 | MA1529.1 | GGGNCGCAGCTGCGYCCC | NHLH2 | 7.85e-50 | 1.92e-52 | 6980 | 24 | 5500 | 18.8 | 1.28 |
| 224 | 80 | MA0778.1 | NFKB2 | MA0778.1 | AGGGGAWTCCCCY | NFKB2 | 7.09e-49 | 1.73e-51 | 5110 | 17.6 | 3830 | 13.1 | 1.34 |
| 225 | 47 | MA0760.1 | ERF | MA0760.1 | ACCGGAAGTR | ERF | 6.92e-48 | 1.68e-50 | 20800 | 71.5 | 19200 | 65.8 | 1.09 |
| 226 | 81 | MA1684.1 | Foxn1 | MA1684.1 | RGAMGC | Foxn1 | 3.74e-47 | 9.03e-50 | 7130 | 24.5 | 5690 | 19.4 | 1.26 |
| 227 | 82 | MA1556.1 | RXRG | MA1556.1 | RRGGTCAYGACCYY | RXRG | 1.55e-46 | 3.71e-49 | 7690 | 26.4 | 6210 | 21.2 | 1.24 |
| 228 | 25 | MA1646.1 | OSR2 | MA1646.1 | NNACAGAAGCNN | OSR2 | 2.39e-46 | 5.7e-49 | 21200 | 73.1 | 19700 | 67.5 | 1.08 |
| 229 | 47 | MA0156.3 | FEV | MA0156.3 | VACCGGAAGTVV | FEV | 4.93e-46 | 1.17e-48 | 7320 | 25.2 | 5880 | 20.1 | 1.25 |
| 230 | 61 | MA1147.1 | NR4A2::RXRA | MA1147.1 | VRGGTCRTTGACCYY | NR4A2::RXRA | 1.62e-44 | 3.85e-47 | 14000 | 48.2 | 12400 | 42.2 | 1.14 |
| 231 | 25 | MA1934.1 | ERF::FIGLA | MA1934.1 | RCCGGAARCASCTGBN | ERF::FIGLA | 4.18e-44 | 9.87e-47 | 3520 | 12.1 | 2490 | 8.51 | 1.42 |
| 232 | 76 | MA0603.1 | Arntl | MA0603.1 | NGTCACGTGH | Arntl | 2.05e-43 | 4.81e-46 | 8620 | 29.7 | 7140 | 24.4 | 1.21 |
| 233 | 83 | MA1531.1 | NR1D1 | MA1531.1 | RGGTYAGTRGGTCAN | NR1D1 | 2.2e-43 | 5.15e-46 | 4940 | 17 | 3750 | 12.8 | 1.33 |
| 234 | 25 | MA0822.1 | HES7 | MA0822.1 | YGGCACGTGCCR | HES7 | 3.85e-42 | 8.96e-45 | 1790 | 6.14 | 1070 | 3.64 | 1.68 |
| 235 | 84 | MA1542.1 | OSR1 | MA1542.1 | HGCTACYGTD | OSR1 | 4.46e-41 | 1.03e-43 | 21100 | 72.7 | 19700 | 67.5 | 1.08 |
| 236 | 85 | MA1554.1 | RFX7 | MA1554.1 | CGTTGCYAY | RFX7 | 8.89e-41 | 2.05e-43 | 25300 | 87.1 | 24300 | 83.1 | 1.05 |
| 237 | 86 | MA0502.2 | NFYB | MA0502.2 | CYCATTGGCCVV | NFYB | 1.36e-40 | 3.12e-43 | 6580 | 22.6 | 5270 | 18 | 1.25 |
| 238 | 31 | MA1565.1 | TBX18 | MA1565.1 | DRAGGTGTGAAR | TBX18 | 2.9e-40 | 6.64e-43 | 10000 | 34.4 | 8530 | 29.2 | 1.18 |
| 239 | 82 | MA1538.1 | NR2F1 | MA1538.1 | RAGGTCANTGACCTY | NR2F1 | 5.4e-39 | 1.23e-41 | 8010 | 27.6 | 6640 | 22.7 | 1.21 |
| 240 | 31 | MA1567.2 | Tbx6 | MA1567.2 | NRGGTGTGAANN | Tbx6 | 5.8e-39 | 1.32e-41 | 16400 | 56.4 | 14900 | 50.9 | 1.11 |
| 241 | 87 | MA1999.1 | Prdm5 | MA1999.1 | CBGTTCTCCATCTNN | Prdm5 | 2.65e-38 | 5.99e-41 | 26800 | 92 | 26000 | 88.8 | 1.04 |
| 242 | 45 | MA1725.1 | ZNF189 | MA1725.1 | NNTGCTGTTCCHB | ZNF189 | 6.23e-37 | 1.4e-39 | 23600 | 81 | 22400 | 76.6 | 1.06 |
| 243 | 60 | MA0258.2 | ESR2 | MA0258.2 | AGGTCASVNTGMCCY | ESR2 | 7.11e-37 | 1.59e-39 | 17700 | 61 | 16300 | 55.6 | 1.1 |
| 244 | 88 | MA0116.1 | Znf423 | MA0116.1 | GSMMCCYARGGKKBM | Znf423 | 7.76e-37 | 1.73e-39 | 1720 | 5.91 | 1050 | 3.6 | 1.64 |
| 245 | 53 | MA0797.1 | TGIF2 | MA0797.1 | TGACAGSTGTCA | TGIF2 | 4.58e-36 | 1.02e-38 | 9350 | 32.1 | 7960 | 27.2 | 1.18 |
| 246 | 28 | MA0670.1 | NFIA | MA0670.1 | NDTGCCAAND | NFIA | 8.96e-36 | 1.99e-38 | 3950 | 13.6 | 2960 | 10.1 | 1.34 |
| 247 | 67 | MA0805.1 | TBX1 | MA0805.1 | AGGTGTGA | TBX1 | 2.53e-35 | 5.59e-38 | 17700 | 60.8 | 16300 | 55.6 | 1.09 |
| 248 | 25 | MA0004.1 | Arnt | MA0004.1 | CACGTG | Arnt | 2.21e-33 | 4.86e-36 | 6540 | 22.5 | 5360 | 18.3 | 1.23 |
| 249 | 25 | MA1109.1 | NEUROD1 | MA1109.1 | NRACAGATGGYNN | NEUROD1 | 2.6e-32 | 5.68e-35 | 27700 | 95.4 | 27200 | 93 | 1.03 |
| 250 | 80 | MA1117.1 | RELB | MA1117.1 | RDATTCCCCNN | RELB | 2.96e-31 | 6.45e-34 | 9340 | 32.1 | 8050 | 27.5 | 1.17 |
| 251 | 53 | MA1114.1 | PBX3 | MA1114.1 | NNNTGAGTGACAGGBNN | PBX3 | 4.1e-31 | 8.9e-34 | 13100 | 45 | 11700 | 40 | 1.12 |
| 252 | 76 | MA0692.1 | TFEB | MA0692.1 | RYCACGTGAC | TFEB | 9.48e-30 | 2.05e-32 | 23800 | 81.9 | 22800 | 78 | 1.05 |
| 253 | 74 | MA1475.1 | CREB3L4 | MA1475.1 | GRTGACGTCAYC | CREB3L4 | 1.93e-29 | 4.16e-32 | 7620 | 26.2 | 6440 | 22 | 1.19 |
| 254 | 47 | MA0076.2 | ELK4 | MA0076.2 | BCRCTTCCGGB | ELK4 | 5.4e-29 | 1.16e-31 | 353 | 1.21 | 109 | 0.37 | 3.24 |
| 255 | 89 | MA0863.1 | MTF1 | MA0863.1 | TTTGCACACGGCAC | MTF1 | 6.04e-29 | 1.29e-31 | 2360 | 8.13 | 1660 | 5.68 | 1.43 |
| 256 | 90 | MA1971.1 | ZBED2 | MA1971.1 | NNMMCGAAACCNNV | ZBED2 | 8.49e-29 | 1.81e-31 | 2440 | 8.38 | 1720 | 5.9 | 1.42 |
| 257 | 67 | MA0690.2 | TBX21 | MA0690.2 | TTTCACACCTT | TBX21 | 3.22e-28 | 6.84e-31 | 20900 | 71.9 | 19800 | 67.5 | 1.07 |
| 258 | 74 | MA0615.1 | Gmeb1 | MA0615.1 | BHBBKKACGTMMNWNNN | Gmeb1 | 1.26e-27 | 2.67e-30 | 1340 | 4.62 | 829 | 2.83 | 1.63 |
| 259 | 48 | MA0737.1 | GLIS3 | MA0737.1 | GACCCCCCACGAAG | GLIS3 | 1.44e-27 | 3.03e-30 | 2280 | 7.85 | 1610 | 5.5 | 1.43 |
| 260 | 86 | MA0060.3 | NFYA | MA0060.3 | RRCCAATCAGM | NFYA | 6.36e-25 | 1.33e-27 | 679 | 2.33 | 340 | 1.16 | 2.01 |
| 261 | 41 | MA0056.2 | MZF1 | MA0056.2 | NNAATCCCCANNN | MZF1 | 3.81e-23 | 7.95e-26 | 25200 | 86.5 | 24400 | 83.4 | 1.04 |
| 262 | 76 | MA0124.2 | Nkx3-1 | MA0124.2 | RCCACTTAA | Nkx3-1 | 1.11e-22 | 2.31e-25 | 1870 | 6.43 | 1310 | 4.47 | 1.44 |
| 263 | 74 | MA0839.1 | CREB3L1 | MA0839.1 | VTGCCACGTCAYCA | CREB3L1 | 1.48e-22 | 3.07e-25 | 7380 | 25.4 | 6360 | 21.7 | 1.17 |
| 264 | 86 | MA1644.1 | NFYC | MA1644.1 | RDCCAATCASN | NFYC | 1.49e-22 | 3.07e-25 | 907 | 3.12 | 525 | 1.79 | 1.74 |
| 265 | 91 | MA1649.1 | ZBTB12 | MA1649.1 | NYCTRGAACHN | ZBTB12 | 3.32e-21 | 6.83e-24 | 21600 | 74.2 | 20600 | 70.5 | 1.05 |
| 266 | 60 | MA0017.2 | NR2F1 | MA0017.2 | CARAGGTCAMVNG | NR2F1 | 4.33e-21 | 8.88e-24 | 7580 | 26.1 | 6580 | 22.5 | 1.16 |
| 267 | 47 | MA1484.1 | ETS2 | MA1484.1 | DACCGGAAGY | ETS2 | 7.39e-21 | 1.51e-23 | 1220 | 4.21 | 791 | 2.7 | 1.56 |
| 268 | 73 | MA0067.2 | PAX2 | MA0067.2 | BCAVTSRAGCGTGACGR | PAX2 | 1.6e-19 | 3.25e-22 | 2160 | 7.44 | 1600 | 5.47 | 1.36 |
| 269 | 74 | MA0636.1 | BHLHE41 | MA0636.1 | RTCACGTGAC | BHLHE41 | 1.61e-19 | 3.26e-22 | 3490 | 12 | 2790 | 9.53 | 1.26 |
| 270 | 75 | MA1967.1 | TFAP4::FLI1 | MA1967.1 | ACCGGAAACAGCTGDY | TFAP4::FLI1 | 2.78e-19 | 5.62e-22 | 2550 | 8.76 | 1940 | 6.63 | 1.32 |
| 271 | 92 | MA1654.1 | ZNF16 | MA1654.1 | AVTRGGGAGCCATGGAAGGTTYT | ZNF16 | 4.98e-19 | 1e-21 | 3440 | 11.8 | 2750 | 9.39 | 1.26 |
| 272 | 93 | MA1597.1 | ZNF528 | MA1597.1 | CCCAGGGAAGCCATTTC | ZNF528 | 6.14e-19 | 1.23e-21 | 6780 | 23.3 | 5860 | 20 | 1.16 |
| 273 | 53 | MA0796.1 | TGIF1 | MA0796.1 | TGACAGCTGTCA | TGIF1 | 7.04e-19 | 1.41e-21 | 944 | 3.25 | 582 | 1.99 | 1.63 |
| 274 | 94 | MA1146.1 | NR1H4::RXRA | MA1146.1 | GAGKTCATTGACCYB | NR1H4::RXRA | 1.06e-18 | 2.1e-21 | 3080 | 10.6 | 2430 | 8.31 | 1.28 |
| 275 | 74 | MA0609.2 | CREM | MA0609.2 | NNDGTGACGTCACHNN | CREM | 4.01e-18 | 7.95e-21 | 13800 | 47.4 | 12700 | 43.5 | 1.09 |
| 276 | 47 | MA0686.1 | SPDEF | MA0686.1 | AMCCGGATGTR | SPDEF | 5.41e-18 | 1.07e-20 | 10900 | 37.4 | 9850 | 33.7 | 1.11 |
| 277 | 95 | MA1651.1 | ZFP42 | MA1651.1 | NNNHCAARATGGCTGCCNBNN | ZFP42 | 1.2e-17 | 2.37e-20 | 3430 | 11.8 | 2760 | 9.43 | 1.25 |
| 278 | 9 | MA0657.1 | KLF13 | MA0657.1 | RTGMCACGCCCCTTTTTG | KLF13 | 1.97e-17 | 3.87e-20 | 2510 | 8.64 | 1940 | 6.62 | 1.3 |
| 279 | 60 | MA1536.1 | NR2C2 | MA1536.1 | RRGGTCAN | NR2C2 | 2.1e-17 | 4.1e-20 | 1170 | 4.01 | 775 | 2.65 | 1.51 |
| 280 | 36 | MA0006.1 | Ahr::Arnt | MA0006.1 | YGCGTG | Ahr::Arnt | 2.12e-17 | 4.13e-20 | 20200 | 69.5 | 19300 | 66 | 1.05 |
| 281 | 103 | MA1573.2 | Thap11 | MA1573.2 | ACTACAABTCCCAG | Thap11 | 3.7e-17 | 7.19e-20 | 15200 | 52.4 | 14200 | 48.6 | 1.08 |
| 282 | 53 | MA0783.1 | PKNOX2 | MA0783.1 | TGACAGGTGTCA | PKNOX2 | 1.21e-16 | 2.33e-19 | 1300 | 4.45 | 890 | 3.04 | 1.46 |
| 283 | 47 | MA0475.2 | FLI1 | MA0475.2 | ACCGGAARTR | FLI1 | 2.09e-16 | 4.02e-19 | 699 | 2.4 | 409 | 1.4 | 1.72 |
| 284 | 76 | MA0664.1 | MLXIPL | MA0664.1 | RTCACGTGAT | MLXIPL | 2.16e-16 | 4.14e-19 | 12100 | 41.7 | 11100 | 38 | 1.1 |
| 285 | 28 | MA0092.1 | Hand1::Tcf3 | MA0092.1 | BRTCTGGMWT | Hand1::Tcf3 | 2.84e-16 | 5.43e-19 | 26400 | 90.9 | 25900 | 88.6 | 1.03 |
| 286 | 76 | MA1994.1 | Nkx2-1 | MA1994.1 | NVCACTTGARHNN | Nkx2-1 | 3.77e-16 | 7.19e-19 | 1410 | 4.86 | 995 | 3.4 | 1.43 |
| 287 | 47 | MA0098.3 | ETS1 | MA0098.3 | ACCGGAARTR | ETS1 | 6.93e-16 | 1.32e-18 | 685 | 2.36 | 402 | 1.37 | 1.71 |
| 288 | 47 | MA0645.1 | ETV6 | MA0645.1 | MSCGGAAGTR | ETV6 | 1.22e-15 | 2.3e-18 | 13000 | 44.7 | 12000 | 41.1 | 1.09 |
| 289 | 85 | MA0799.2 | RFX4 | MA0799.2 | BSGTTGCYWRGCAACSV | RFX4 | 1.73e-15 | 3.26e-18 | 1320 | 4.55 | 927 | 3.17 | 1.44 |
| 290 | 78 | MA0002.2 | Runx1 | MA0002.2 | BBYTGTGGTTT | Runx1 | 3.72e-15 | 7e-18 | 26900 | 92.4 | 26400 | 90.3 | 1.02 |
| 291 | 25 | MA0825.1 | MNT | MA0825.1 | RVCACGTGMH | MNT | 4.02e-15 | 7.53e-18 | 14300 | 49 | 13300 | 45.5 | 1.08 |
| 292 | 61 | MA1537.1 | NR2F1 | MA1537.1 | RRGGTCAAAGGTCAN | NR2F1 | 7.52e-15 | 1.4e-17 | 5130 | 17.6 | 4400 | 15 | 1.17 |
| 293 | 47 | MA0641.1 | ELF4 | MA0641.1 | AACCCGGAAGTR | ELF4 | 7.6e-15 | 1.42e-17 | 4760 | 16.4 | 4050 | 13.8 | 1.18 |
| 294 | 47 | MA0473.3 | ELF1 | MA0473.3 | RDVCAGGAAGTGVN | ELF1 | 8.72e-15 | 1.62e-17 | 18300 | 63 | 17400 | 59.5 | 1.06 |
| 295 | 82 | MA1555.1 | RXRB | MA1555.1 | RRGGTCATGACCYY | RXRB | 5.99e-14 | 1.11e-16 | 12600 | 43.2 | 11700 | 39.9 | 1.08 |
| 296 | 75 | MA1568.1 | TCF21 | MA1568.1 | CACCATATGKYR | TCF21 | 3.95e-13 | 7.27e-16 | 9220 | 31.7 | 8380 | 28.6 | 1.11 |
| 297 | 25 | MA0819.2 | CLOCK | MA0819.2 | NRACACGTGB | CLOCK | 4.64e-13 | 8.52e-16 | 5340 | 18.4 | 4640 | 15.9 | 1.16 |
| 298 | 61 | MA1540.2 | NR5A1 | MA1540.2 | NNRAGTTCAAGGTCANN | NR5A1 | 2.07e-12 | 3.79e-15 | 18200 | 62.6 | 17400 | 59.4 | 1.05 |
| 299 | 80 | MA0107.1 | RELA | MA0107.1 | GGGRATTTCC | RELA | 3.33e-12 | 6.06e-15 | 6710 | 23.1 | 5970 | 20.4 | 1.13 |
| 300 | 96 | MA1121.1 | TEAD2 | MA1121.1 | NYACATTCCWNNS | TEAD2 | 4.02e-12 | 7.3e-15 | 2220 | 7.62 | 1760 | 6 | 1.27 |
| 301 | 96 | MA0808.1 | TEAD3 | MA0808.1 | RCATTCCW | TEAD3 | 5.33e-12 | 9.65e-15 | 4140 | 14.2 | 3530 | 12.1 | 1.18 |
| 302 | 75 | MA1570.1 | TFAP4 | MA1570.1 | AHCATRTGDT | TFAP4 | 6.55e-12 | 1.18e-14 | 19700 | 67.7 | 18900 | 64.7 | 1.05 |
| 303 | 97 | MA1601.2 | ZNF75D | MA1601.2 | GTGGGAAAGCCT | ZNF75D | 1.11e-11 | 2e-14 | 13100 | 45.2 | 12300 | 42 | 1.07 |
| 304 | 47 | MA1483.2 | ELF2 | MA1483.2 | AAMCCGGAAGTR | ELF2 | 2.26e-11 | 4.05e-14 | 514 | 1.77 | 303 | 1.04 | 1.7 |
| 305 | 25 | MA1642.1 | NEUROG2 | MA1642.1 | NNVACAGATGGNN | NEUROG2 | 1.21e-10 | 2.16e-13 | 27100 | 93.3 | 26800 | 91.7 | 1.02 |
| 306 | 80 | MA0101.1 | REL | MA0101.1 | BGGRNWTTCC | REL | 1.64e-10 | 2.93e-13 | 3420 | 11.8 | 2900 | 9.91 | 1.19 |
| 307 | 94 | MA1539.1 | NR2F6 | MA1539.1 | RAGGTCANTGACCTY | NR2F6 | 1.89e-10 | 3.35e-13 | 2550 | 8.76 | 2090 | 7.14 | 1.23 |
| 308 | 98 | MA1645.1 | NKX2-2 | MA1645.1 | NNNCCACTCAANNN | NKX2-2 | 1.9e-10 | 3.36e-13 | 3300 | 11.3 | 2780 | 9.51 | 1.19 |
| 309 | 99 | MA1593.1 (ZNF317) | STREME-14 | 14-TTCAGCAGACTTAAA | 14-TTCAGCAGACTTAAA | ZNF317 | 2.52e-10 | 4.44e-13 | 46 | 0.16 | 1 | 0 | 23.6 |
| 310 | 47 | MA0763.1 | ETV3 | MA0763.1 | ACCGGAAGTR | ETV3 | 2.9e-10 | 5.09e-13 | 3570 | 12.3 | 3040 | 10.4 | 1.18 |
| 311 | 92 | MA1977.1 | ZNF324 | MA1977.1 | NNNWRGMAGHYAAGGATGGTT | ZNF324 | 3.42e-10 | 6e-13 | 15600 | 53.5 | 14800 | 50.6 | 1.06 |
| 312 | 47 | MA1708.1 | ETV7 | MA1708.1 | DGSCGGAAGTR | ETV7 | 1.15e-09 | 2.01e-12 | 8500 | 29.2 | 7800 | 26.6 | 1.1 |
| 313 | 25 | MA0626.1 | Npas2 | MA0626.1 | NSCACGTGTN | Npas2 | 3.41e-09 | 5.94e-12 | 5450 | 18.8 | 4850 | 16.6 | 1.13 |
| 314 | 85 | MA1724.1 | Rfx6 | MA1724.1 | HNCCTAGCAACHV | Rfx6 | 4.23e-09 | 7.34e-12 | 28700 | 98.7 | 28700 | 98 | 1.01 |
| 315 | 48 | MA1491.2 | GLI3 | MA1491.2 | RGACCACCCACGWHGH | GLI3 | 8.38e-09 | 1.45e-11 | 1250 | 4.31 | 951 | 3.25 | 1.33 |
| 316 | 25 | MA0640.2 | ELF3 | MA0640.2 | NNCCACTTCCTGNT | ELF3 | 2.23e-08 | 3.85e-11 | 22300 | 76.7 | 21800 | 74.4 | 1.03 |
| 317 | 47 | MA0474.3 | Erg | MA0474.3 | NNACAGGAAGTGVN | Erg | 3.51e-08 | 6.04e-11 | 17200 | 59.1 | 16500 | 56.4 | 1.05 |
| 318 | 80 | MA0105.4 | NFKB1 | MA0105.4 | AGGGGAWTCCCCT | NFKB1 | 4.41e-08 | 7.57e-11 | 1170 | 4.01 | 885 | 3.02 | 1.33 |
| 319 | 100 | MA1984.1 | ZNF667 | MA1984.1 | TTAAGAGCTCAV | ZNF667 | 5.02e-08 | 8.58e-11 | 11800 | 40.4 | 11100 | 37.8 | 1.07 |
| 320 | 47 | MA0062.3 | GABPA | MA0062.3 | NNCACTTCCTGTNN | GABPA | 5.72e-08 | 9.75e-11 | 16800 | 57.6 | 16100 | 55 | 1.05 |
| 321 | 61 | MA0677.1 | Nr2f6 | MA0677.1 | RRGGTCAAAGGTCA | Nr2f6 | 1.22e-07 | 2.08e-10 | 8340 | 28.7 | 7700 | 26.3 | 1.09 |
| 322 | 74 | MA1466.1 | ATF6 | MA1466.1 | TGRTGACGTGGCAN | ATF6 | 3.5e-07 | 5.93e-10 | 1730 | 5.96 | 1410 | 4.81 | 1.24 |
| 323 | 76 | MA0914.1 | ISL2 | MA0914.1 | GCAMTTAR | ISL2 | 3.87e-07 | 6.54e-10 | 1280 | 4.38 | 995 | 3.4 | 1.29 |
| 324 | 47 | MA1931.1 | ELK1::HOXA1 | MA1931.1 | ACCGGAAGTAATTA | ELK1::HOXA1 | 4.45e-07 | 7.48e-10 | 6220 | 21.4 | 5660 | 19.4 | 1.11 |
| 325 | 85 | MA0798.3 | RFX3 | MA0798.3 | CGTTRCCATGGCAACS | RFX3 | 7.68e-07 | 1.29e-09 | 86 | 0.3 | 24 | 0.08 | 3.5 |
| 326 | 96 | MA0809.2 | TEAD4 | MA0809.2 | NNACATTCCARN | TEAD4 | 9.84e-07 | 1.65e-09 | 1840 | 6.34 | 1520 | 5.19 | 1.22 |
| 327 | 85 | MA0509.3 | RFX1 | MA0509.3 | SGTTGCCATGGCAACS | RFX1 | 3.31e-06 | 5.52e-09 | 105 | 0.36 | 37 | 0.13 | 2.81 |
| 328 | 25 | MA1638.1 | HAND2 | MA1638.1 | NVCAGATGNN | HAND2 | 4.92e-06 | 8.17e-09 | 25000 | 85.9 | 24600 | 84.2 | 1.02 |
| 329 | 97 | MA1116.1 | RBPJ | MA1116.1 | BVTGGGAANN | RBPJ | 5.41e-06 | 8.97e-09 | 3560 | 12.2 | 3140 | 10.7 | 1.14 |
| 330 | 101 | MA0505.2 | Nr5A2 | MA0505.2 | NTGACCTTGAMCT | Nr5A2 | 5.93e-06 | 9.8e-09 | 10200 | 35.2 | 9640 | 32.9 | 1.07 |
| 331 | 73 | MA1995.1 | Npas4 | MA1995.1 | NNTCGTGACHN | Npas4 | 6.65e-06 | 1.1e-08 | 2280 | 7.84 | 1940 | 6.62 | 1.18 |
| 332 | 75 | MA0667.1 | MYF6 | MA0667.1 | AACARCTGTT | MYF6 | 7.15e-06 | 1.17e-08 | 9170 | 31.5 | 8600 | 29.4 | 1.07 |
| 333 | 96 | MA0090.3 | TEAD1 | MA0090.3 | NNACATTCCAGSN | TEAD1 | 8.44e-06 | 1.38e-08 | 2190 | 7.52 | 1850 | 6.33 | 1.19 |
| 334 | 25 | MA0100.3 | MYB | MA0100.3 | NNCAACTGHH | MYB | 9.34e-06 | 1.52e-08 | 5320 | 18.3 | 4830 | 16.5 | 1.11 |
| 335 | 102 | MA0111.1 | Spz1 | MA0111.1 | AGGGTWWCAGC | Spz1 | 1.04e-05 | 1.69e-08 | 15200 | 52.3 | 14600 | 50 | 1.05 |
| 336 | 85 | MA0600.2 | RFX2 | MA0600.2 | SGTTGCCATGGYAACS | RFX2 | 1.3e-05 | 2.11e-08 | 122 | 0.42 | 50 | 0.17 | 2.43 |
| 337 | 76 | MA0622.1 | Mlxip | MA0622.1 | BCACGTGK | Mlxip | 2.84e-05 | 4.59e-08 | 829 | 2.85 | 629 | 2.15 | 1.33 |
| 338 | 103 | MA1573.2 (Thap11) | STREME-16 | 16-ACTACATTTCCCAG | 16-ACTACATTTCCCAG | Thap11 | 4.57e-05 | 7.37e-08 | 489 | 1.68 | 339 | 1.16 | 1.45 |
| 339 | 67 | MA0688.1 | TBX2 | MA0688.1 | DAGGTGTGAAA | TBX2 | 0.000179 | 2.88e-07 | 9570 | 32.9 | 9060 | 31 | 1.06 |
| 340 | 25 | MA0059.1 | MAX::MYC | MA0059.1 | RASCACGTGGT | MAX::MYC | 0.000275 | 4.41e-07 | 8560 | 29.4 | 8070 | 27.6 | 1.07 |
| 341 | 74 | MA0844.1 | XBP1 | MA0844.1 | WDKGMCACGTCATC | XBP1 | 0.000304 | 4.86e-07 | 3780 | 13 | 3410 | 11.6 | 1.12 |
| 342 | 25 | MA0832.1 | Tcf21 | MA0832.1 | RYAACAGCTGTTRN | Tcf21 | 0.000371 | 5.91e-07 | 1610 | 5.53 | 1360 | 4.63 | 1.19 |
| 343 | 47 | MA0761.2 | ETV1 | MA0761.2 | NNACAGGAAGTGNN | ETV1 | 0.000499 | 7.93e-07 | 24300 | 83.5 | 24000 | 82 | 1.02 |
| 344 | 77 | MA1576.1 | THRB | MA1576.1 | DTGACCTBACGTGACCTYA | THRB | 0.000504 | 7.98e-07 | 2740 | 9.44 | 2420 | 8.29 | 1.14 |
| 345 | 60 | MA0730.1 | RARA | MA0730.1 | AGGTCAHSYAAAGGTCA | RARA | 0.000547 | 8.65e-07 | 137 | 0.47 | 68 | 0.23 | 2.01 |
| 346 | 98 | MA1523.1 | MSANTD3 | MA1523.1 | STVCACTCAC | MSANTD3 | 0.000679 | 1.07e-06 | 10100 | 34.6 | 9570 | 32.7 | 1.06 |
| 347 | 104 | MA1557.1 | SMAD5 | MA1557.1 | YGTCTAGACA | SMAD5 | 0.00149 | 2.34e-06 | 7010 | 24.1 | 6580 | 22.5 | 1.07 |
| 348 | 75 | MA0691.1 | TFAP4 | MA0691.1 | AWCAGCTGWT | TFAP4 | 0.00156 | 2.44e-06 | 3410 | 11.7 | 3070 | 10.5 | 1.11 |
| 349 | 48 | MA0735.1 | GLIS1 | MA0735.1 | MGACCCCCCACGAWGC | GLIS1 | 0.0021 | 3.28e-06 | 91 | 0.31 | 39 | 0.13 | 2.31 |
| 350 | 25 | MA1467.2 | Atoh1 | MA1467.2 | RVCAGATGGYN | Atoh1 | 0.00267 | 4.17e-06 | 26400 | 90.8 | 26300 | 89.7 | 1.01 |
| 351 | 85 | MA0510.2 | RFX5 | MA0510.2 | SGTTGCCATGGYAACV | RFX5 | 0.006 | 9.32e-06 | 50 | 0.17 | 15 | 0.05 | 3.21 |
| 352 | 105 | MA0083.3 | SRF | MA0083.3 | TGMCCATATATGGKCA | SRF | 0.0296 | 4.59e-05 | 355 | 1.22 | 258 | 0.88 | 1.38 |
| 353 | 77 | MA0860.1 | Rarg | MA0860.1 | AAGGTCAMSARAGGTCA | Rarg | 0.0301 | 4.65e-05 | 93 | 0.32 | 46 | 0.16 | 2.01 |
| 354 | 25 | MA0526.4 | USF2 | MA0526.4 | DRTCACGTGAYH | USF2 | 0.0335 | 5.16e-05 | 292 | 1 | 205 | 0.7 | 1.43 |
| 355 | 106 | MA1998.1 | Prdm14 | MA1998.1 | NDGGTCTCTAWS | Prdm14 | 0.0343 | 5.26e-05 | 24200 | 83.2 | 24000 | 82 | 1.02 |
| 356 | 47 | MA1936.1 | ERF::FOXO1 | MA1936.1 | RTMAACAGGAARBS | ERF::FOXO1 | 0.0396 | 6.06e-05 | 17700 | 60.7 | 17300 | 59.1 | 1.03 |
| 357 | 76 | MA1939.1 | ERF::SREBF2 | MA1939.1 | NSCGGAARTCACGTGAT | ERF::SREBF2 | 0.0489 | 7.48e-05 | 2960 | 10.2 | 2700 | 9.24 | 1.1 |
| 18 | 107 | MA1125.1 (ZNF384) | MEME-2 | AAAAAAAAAAAAAAA | AAAAAAAAAAAAAAA | ZNF384 | 18 | 1 | 173 | 0.59 | 253 | 0.86 | 0.689 |
ER_rat <- 1.25
DOX_sigdown_24hr_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste("DOX_sigdown_24hr response peaks(200bp)Enrichment ratio:",ER_rat))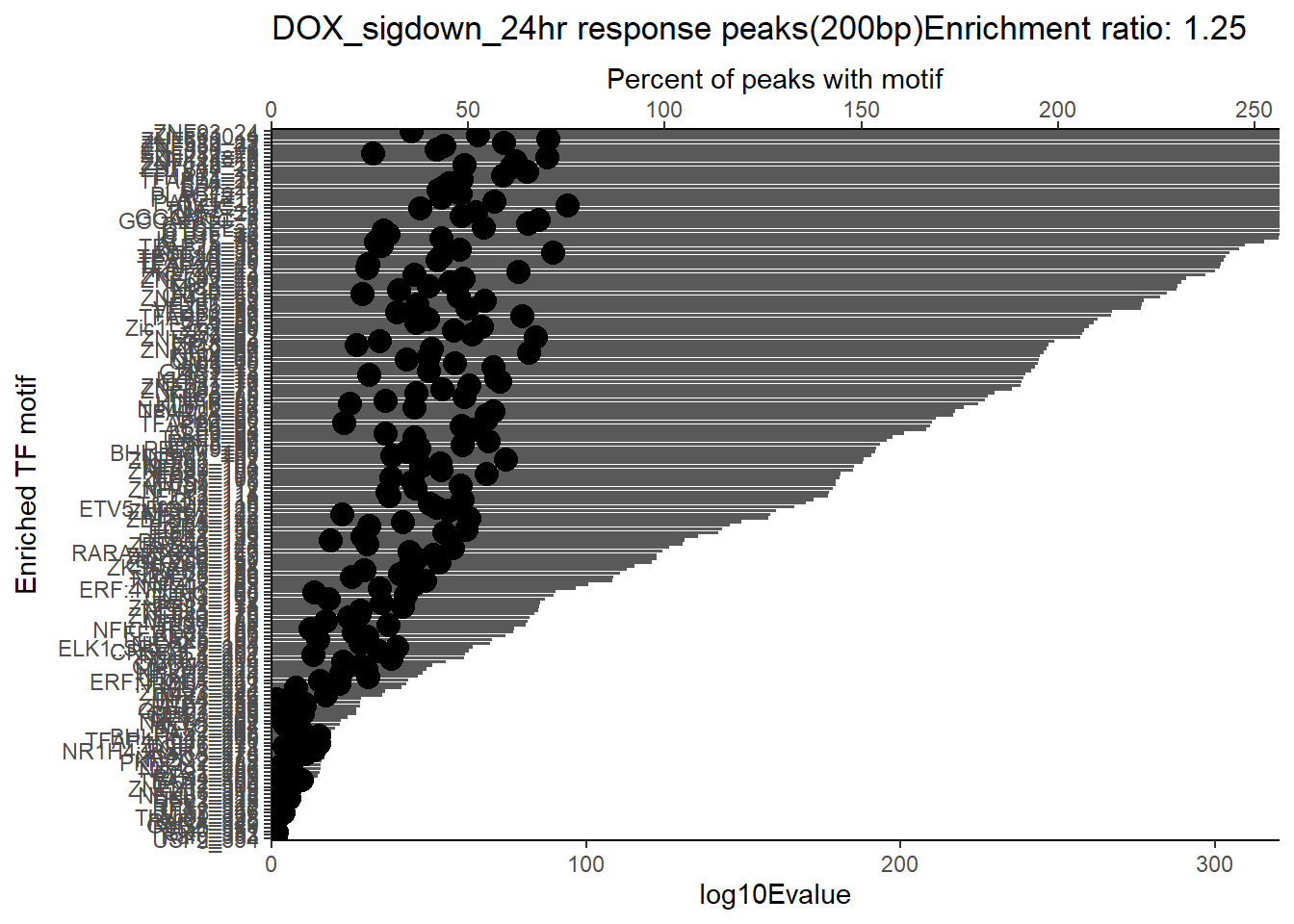
DOX_sigdown_24hr_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
slice_head(n=5) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*1.75), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.75,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste(" DOX_sigdown_24hr response peaks(200bp)Enrichment ratio:",ER_rat))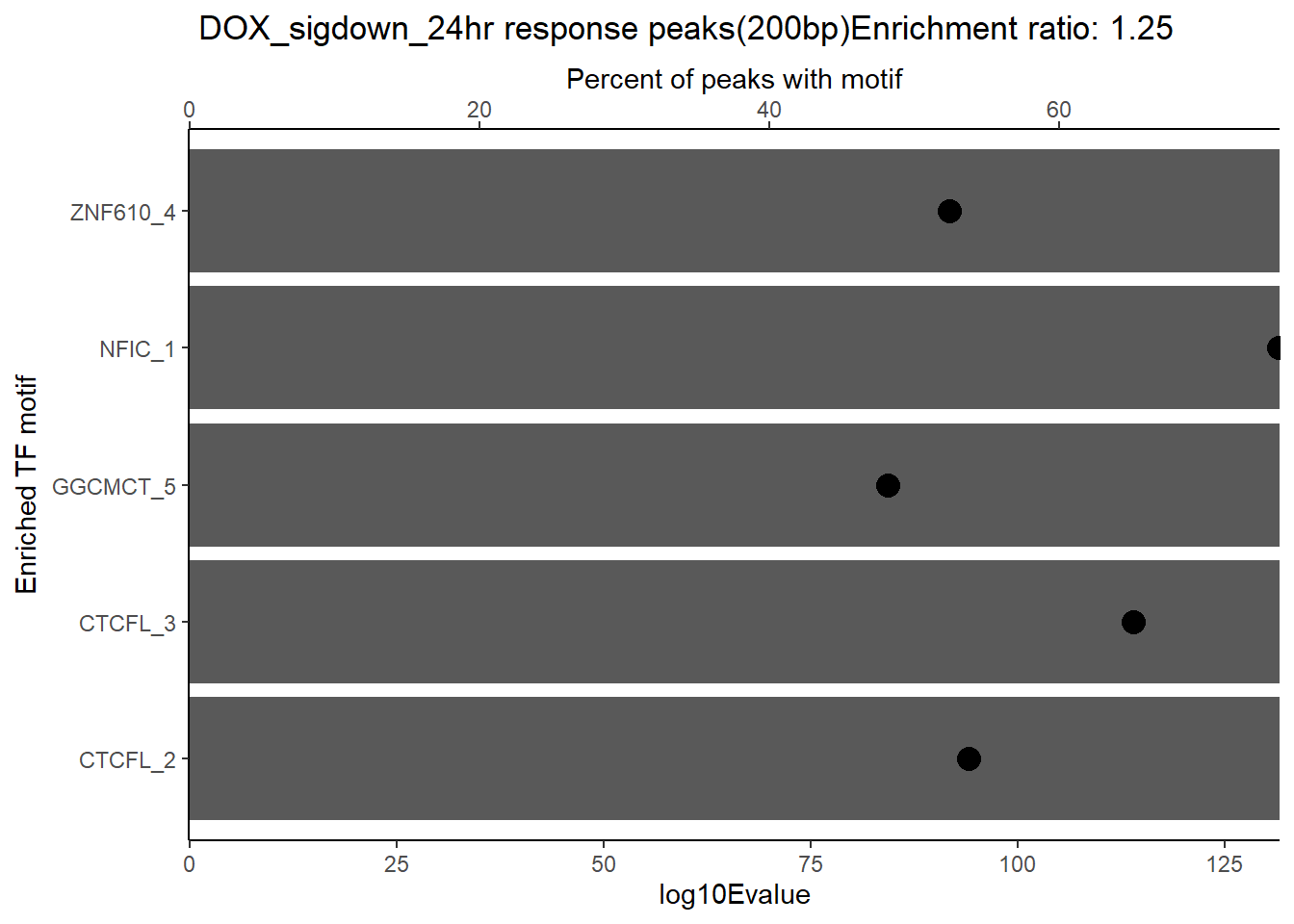
DOX_DAR 24 2kb data
DOX_242kb_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DOX_24_2kb_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
DOX_242kb_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DOX_24_2kb_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
DOX_242kb_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DOX_24_2kb_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
DOX_242kb_sea_all <- rbind(DOX_242kb_sea_disc, DOX_242kb_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)DOX_242kb_combo <- DOX_242kb_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., DOX_242kb_sea_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ SIM_MOTIF
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y)
DOX_242kb_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs 200 bp in DOX_24 2kb") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | MA1138.1 | FOSL2::JUNB | MA1138.1 | KRTGASTCAT | FOSL2::JUNB | 1.84e-120 | 6.35e-121 | 1400 | 11.3 | 889 | 4.33 | 2.61 |
| 2 | 1 | MA1144.1 | FOSL2::JUND | MA1144.1 | KATGACTCAT | FOSL2::JUND | 9.25e-120 | 1.59e-120 | 1300 | 10.5 | 784 | 3.82 | 2.74 |
| 3 | 1 | MA1135.1 | FOSB::JUNB | MA1135.1 | KRTGASTCAT | FOSB::JUNB | 3.61e-119 | 4.14e-120 | 1530 | 12.4 | 1040 | 5.06 | 2.44 |
| 4 | 1 | MA1134.1 (FOS::JUNB) | STREME-1 | 1-GATGASTCATC | GATGASTCATC | FOS::JUNB | 3.12e-117 | 2.68e-118 | 1420 | 11.4 | 919 | 4.48 | 2.55 |
| 5 | 1 | MA1134.1 | FOS::JUNB | MA1134.1 | KATGASTCATHN | FOS::JUNB | 2.2e-116 | 1.51e-117 | 1370 | 11.1 | 878 | 4.28 | 2.59 |
| 6 | 1 | MA1141.1 | FOS::JUND | MA1141.1 | NKATGAGTCATNN | FOS::JUND | 4.1e-115 | 2.35e-116 | 1260 | 10.2 | 769 | 3.75 | 2.72 |
| 7 | 1 | MA0478.1 | FOSL2 | MA0478.1 | KRRTGASTCAB | FOSL2 | 5.56e-114 | 2.73e-115 | 1570 | 12.7 | 1110 | 5.41 | 2.35 |
| 8 | 1 | MA0476.1 | FOS | MA0476.1 | DVTGASTCATB | FOS | 9.92e-111 | 4.27e-112 | 1620 | 13.1 | 1180 | 5.77 | 2.27 |
| 9 | 1 | MA1128.1 | FOSL1::JUN | MA1128.1 | NKATGACTCATNN | FOSL1::JUN | 1.27e-110 | 4.87e-112 | 1330 | 10.7 | 859 | 4.18 | 2.56 |
| 10 | 1 | MA0476.1 (FOS) | MEME-2 | ATGASTCASHN | ATGASTCASHN | FOS | 7.52e-110 | 2.59e-111 | 1100 | 8.86 | 623 | 3.03 | 2.92 |
| 11 | 1 | MA1137.1 | FOSL1::JUNB | MA1137.1 | NDRTGACTCATNN | FOSL1::JUNB | 1.11e-106 | 3.47e-108 | 1250 | 10 | 788 | 3.84 | 2.62 |
| 12 | 1 | MA1633.2 | BACH1 | MA1633.2 | ATGACTCAT | BACH1 | 3.98e-106 | 1.14e-107 | 1780 | 14.3 | 1390 | 6.75 | 2.12 |
| 13 | 1 | MA0489.2 | Jun | MA0489.2 | NVTGACTCATNN | Jun | 9.81e-106 | 2.6e-107 | 1920 | 15.5 | 1560 | 7.61 | 2.03 |
| 14 | 1 | MA1130.1 | FOSL2::JUN | MA1130.1 | NNRTGAGTCAYN | FOSL2::JUN | 1.97e-105 | 4.83e-107 | 1050 | 8.48 | 594 | 2.89 | 2.93 |
| 15 | 1 | MA1634.1 | BATF | MA1634.1 | DATGACTCATH | BATF | 2.97e-103 | 6.82e-105 | 1070 | 8.61 | 619 | 3.02 | 2.85 |
| 16 | 1 | MA0491.2 | JUND | MA0491.2 | NNATGACTCATNN | JUND | 5.54e-103 | 1.19e-104 | 2020 | 16.2 | 1700 | 8.26 | 1.97 |
| 17 | 1 | MA0490.2 | JUNB | MA0490.2 | NNATGACTCATNN | JUNB | 9.74e-103 | 1.87e-104 | 1970 | 15.9 | 1650 | 8.02 | 1.98 |
| 18 | 1 | MA0477.2 | FOSL1 | MA0477.2 | NNATGACTCATNN | FOSL1 | 9.77e-103 | 1.87e-104 | 2100 | 17 | 1810 | 8.81 | 1.93 |
| 19 | 1 | MA0099.3 | FOS::JUN | MA0099.3 | ATGAGTCAYM | FOS::JUN | 1.25e-100 | 2.26e-102 | 975 | 7.86 | 537 | 2.62 | 3 |
| 20 | 1 | MA0462.2 | BATF::JUN | MA0462.2 | DATGACTCATH | BATF::JUN | 2.42e-100 | 4.17e-102 | 1700 | 13.7 | 1330 | 6.48 | 2.11 |
| 21 | 1 | MA1988.1 | Atf3 | MA1988.1 | NRTGACTCABN | Atf3 | 2.69e-100 | 4.41e-102 | 1910 | 15.4 | 1580 | 7.72 | 1.99 |
| 22 | 1 | MA0835.2 | BATF3 | MA0835.2 | DATGACTCATH | BATF3 | 2.17e-97 | 3.4e-99 | 1010 | 8.16 | 587 | 2.86 | 2.85 |
| 23 | 1 | MA1928.1 | BNC2 | MA1928.1 | NDRTGAGTCABN | BNC2 | 3.93e-94 | 5.89e-96 | 1920 | 15.5 | 1640 | 7.99 | 1.94 |
| 24 | 1 | MA1142.1 | FOSL1::JUND | MA1142.1 | NRTGACTCAT | FOSL1::JUND | 4.85e-92 | 6.95e-94 | 1360 | 11 | 991 | 4.83 | 2.28 |
| 25 | 1 | MA0841.1 | NFE2 | MA0841.1 | VATGACTCATS | NFE2 | 8.77e-92 | 1.21e-93 | 1470 | 11.9 | 1120 | 5.43 | 2.18 |
| 26 | 1 | MA1101.2 | BACH2 | MA1101.2 | DWANCATGASTCATSNTWH | BACH2 | 2.03e-87 | 2.69e-89 | 1100 | 8.91 | 729 | 3.55 | 2.51 |
| 27 | 1 | MA1132.1 | JUN::JUNB | MA1132.1 | KATGACKCAT | JUN::JUNB | 1.41e-72 | 1.79e-74 | 2390 | 19.2 | 2420 | 11.8 | 1.63 |
| 28 | 1 | MA0655.1 | JDP2 | MA0655.1 | ATGACTCAT | JDP2 | 2.33e-71 | 2.87e-73 | 3090 | 24.9 | 3410 | 16.6 | 1.5 |
| 29 | 1 | MA0089.2 | MAFG::NFE2L1 | MA0089.2 | NVNATGACTCAGCADW | MAFG::NFE2L1 | 2.72e-53 | 3.22e-55 | 4540 | 36.6 | 5800 | 28.3 | 1.3 |
| 30 | 1 | MA0501.1 | MAF::NFE2 | MA0501.1 | ATGACTCAGCANWTY | MAF::NFE2 | 7.04e-51 | 8.07e-53 | 2330 | 18.8 | 2560 | 12.5 | 1.51 |
| 31 | 1 | MA0496.3 | MAFK | MA0496.3 | NGMTGACTCAGCMNH | MAFK | 2.32e-48 | 2.58e-50 | 1180 | 9.51 | 1060 | 5.14 | 1.85 |
| 32 | 2 | MA0710.1 | NOTO | MA0710.1 | NYTAATTAVN | NOTO | 3.82e-46 | 4.11e-48 | 8210 | 66.2 | 11900 | 58 | 1.14 |
| 33 | 2 | MA0885.2 | Dlx2 | MA0885.2 | NDGCAATTAGNN | Dlx2 | 1.24e-45 | 1.29e-47 | 9230 | 74.4 | 13700 | 66.8 | 1.11 |
| 34 | 3 | 3-AKKMMT | STREME-3 | 3-AKKMMT | AKKMMT | AKKMMT | 9.78e-45 | 9.89e-47 | 5720 | 46.2 | 7810 | 38 | 1.21 |
| 35 | 2 | MA0718.1 | RAX | MA0718.1 | RYYAATTARH | RAX | 1.19e-44 | 1.17e-46 | 8030 | 64.7 | 11600 | 56.7 | 1.14 |
| 36 | 2 | MA1476.2 | Dlx5 | MA1476.2 | NDGCAATTAGNN | Dlx5 | 3.6e-44 | 3.44e-46 | 8720 | 70.3 | 12800 | 62.6 | 1.12 |
| 37 | 2 | MA0644.2 | ESX1 | MA0644.2 | YYAATTAN | ESX1 | 1.38e-43 | 1.28e-45 | 8570 | 69.1 | 12600 | 61.4 | 1.13 |
| 38 | 2 | MA0726.1 | VSX2 | MA0726.1 | CTAATTAN | VSX2 | 2.02e-43 | 1.83e-45 | 8980 | 72.4 | 13300 | 64.9 | 1.12 |
| 39 | 1 | MA0150.2 | Nfe2l2 | MA0150.2 | CASNATGACTCAGCA | Nfe2l2 | 7.32e-43 | 6.46e-45 | 4280 | 34.5 | 5560 | 27.1 | 1.27 |
| 40 | 2 | MA1963.1 | SATB1 | MA1963.1 | WWDCTAATAAHWW | SATB1 | 9.65e-43 | 8.3e-45 | 6960 | 56.1 | 9870 | 48.1 | 1.17 |
| 41 | 4 | MA0142.1 | Pou5f1::Sox2 | MA0142.1 | CWTTGTYATGCAAAT | Pou5f1::Sox2 | 1.45e-42 | 1.22e-44 | 8260 | 66.6 | 12100 | 58.8 | 1.13 |
| 42 | 2 | MA0720.1 | Shox2 | MA0720.1 | YYAATTAR | Shox2 | 1.17e-41 | 9.57e-44 | 8820 | 71.1 | 13100 | 63.6 | 1.12 |
| 43 | 5 | 5-ATTGAG | STREME-5 | 5-ATTGAG | ATTGAG | ATTGAG | 1.93e-41 | 1.54e-43 | 9490 | 76.5 | 14300 | 69.5 | 1.1 |
| 44 | 2 | MA0666.2 | MSX1 | MA0666.2 | SCAATTAR | MSX1 | 8.62e-41 | 6.74e-43 | 6980 | 56.3 | 9940 | 48.4 | 1.16 |
| 45 | 1 | MA1639.1 | MEIS1 | MA1639.1 | NHCATMAATCAHN | MEIS1 | 1.12e-40 | 8.55e-43 | 8730 | 70.4 | 12900 | 63 | 1.12 |
| 46 | 2 | MA0681.2 | PHOX2B | MA0681.2 | WWTAATCAAATTAWWW | PHOX2B | 1.75e-40 | 1.3e-42 | 5650 | 45.6 | 7770 | 37.8 | 1.2 |
| 47 | 2 | MA0654.1 | ISX | MA0654.1 | YYAATTAR | ISX | 1.77e-40 | 1.3e-42 | 6050 | 48.8 | 8410 | 41 | 1.19 |
| 48 | 2 | MA0680.2 | Pax7 | MA0680.2 | DWTAATCAATTAWNH | Pax7 | 2.39e-40 | 1.71e-42 | 6860 | 55.3 | 9760 | 47.5 | 1.16 |
| 49 | 6 | MA0498.2 (MEIS1) | STREME-4 | 4-YTGWCAR | YTGWCAR | MEIS1 | 3.43e-40 | 2.41e-42 | 9260 | 74.6 | 13900 | 67.5 | 1.11 |
| 50 | 2 | MA0628.1 | POU6F1 | MA0628.1 | AYTAATTART | POU6F1 | 4.32e-40 | 2.97e-42 | 6820 | 55 | 9680 | 47.2 | 1.17 |
| 51 | 1 | MA1113.2 | PBX2 | MA1113.2 | NHCATAAATCAHN | PBX2 | 5.11e-40 | 3.45e-42 | 8070 | 65.1 | 11800 | 57.5 | 1.13 |
| 52 | 2 | MA0700.2 | LHX2 | MA0700.2 | DDCCAATTADN | LHX2 | 1.41e-39 | 9.34e-42 | 8000 | 64.5 | 11700 | 56.9 | 1.13 |
| 53 | 7 | MA1104.2 | GATA6 | MA1104.2 | HWWTCTTATCTNH | GATA6 | 1.68e-39 | 1.09e-41 | 6440 | 52 | 9080 | 44.2 | 1.17 |
| 54 | 4 | MA0789.1 | POU3F4 | MA0789.1 | TATGCWAAT | POU3F4 | 2.59e-39 | 1.65e-41 | 7190 | 58 | 10300 | 50.3 | 1.15 |
| 55 | 8 | MA0790.1 | POU4F1 | MA0790.1 | ATGMATAATTAATG | POU4F1 | 2.82e-39 | 1.75e-41 | 7230 | 58.3 | 10400 | 50.6 | 1.15 |
| 56 | 2 | MA0717.1 | RAX2 | MA0717.1 | YYAATTAV | RAX2 | 2.86e-39 | 1.75e-41 | 8370 | 67.5 | 12300 | 60 | 1.12 |
| 57 | 2 | MA0787.1 | POU3F2 | MA0787.1 | WTATGCWAATKA | POU3F2 | 2.89e-39 | 1.75e-41 | 6540 | 52.7 | 9230 | 45 | 1.17 |
| 58 | 2 | MA0027.2 | EN1 | MA0027.2 | SYAATTAV | EN1 | 3.05e-39 | 1.81e-41 | 8540 | 68.9 | 12600 | 61.5 | 1.12 |
| 59 | 2 | MA0704.1 | Lhx4 | MA0704.1 | YTAATTAR | Lhx4 | 3.46e-39 | 2.02e-41 | 8400 | 67.7 | 12400 | 60.3 | 1.12 |
| 60 | 8 | MA0791.1 | POU4F3 | MA0791.1 | RTGMATWATTAATGAR | POU4F3 | 3.97e-39 | 2.28e-41 | 6640 | 53.5 | 9400 | 45.8 | 1.17 |
| 61 | 9 | MA0676.1 | Nr2e1 | MA0676.1 | AAAAGTCAA | Nr2e1 | 2.17e-38 | 1.22e-40 | 9510 | 76.6 | 14300 | 69.9 | 1.1 |
| 62 | 2 | MA0725.1 | VSX1 | MA0725.1 | YTAATTAN | VSX1 | 2.82e-38 | 1.56e-40 | 9700 | 78.2 | 14700 | 71.6 | 1.09 |
| 63 | 10 | MA0896.1 | Hmx1 | MA0896.1 | VSVAGCAATTAAWGVDB | Hmx1 | 4.11e-38 | 2.25e-40 | 6480 | 52.2 | 9160 | 44.6 | 1.17 |
| 64 | 11 | MA0495.3 | MAFF | MA0495.3 | DWGTCAGCATTTTWHN | MAFF | 1.4e-37 | 7.53e-40 | 4940 | 39.8 | 6690 | 32.6 | 1.22 |
| 65 | 2 | MA0889.1 | GBX1 | MA0889.1 | VCYAATTASY | GBX1 | 1.47e-37 | 7.78e-40 | 8010 | 64.5 | 11700 | 57.2 | 1.13 |
| 66 | 2 | MA0722.1 | VAX1 | MA0722.1 | YTAATTAH | VAX1 | 1.83e-37 | 9.52e-40 | 9180 | 74 | 13800 | 67.1 | 1.1 |
| 67 | 2 | MA0886.1 | EMX2 | MA0886.1 | VBTAATTARB | EMX2 | 2.24e-37 | 1.15e-39 | 6530 | 52.6 | 9260 | 45.1 | 1.17 |
| 68 | 1 | MA1640.1 | MEIS2 | MA1640.1 | NNATGATTTATGDNH | MEIS2 | 3.98e-37 | 2.01e-39 | 7220 | 58.2 | 10400 | 50.8 | 1.15 |
| 69 | 7 | MA0036.3 | GATA2 | MA0036.3 | NBCTTATCTNH | GATA2 | 4.85e-37 | 2.42e-39 | 4850 | 39.1 | 6560 | 31.9 | 1.22 |
| 70 | 2 | MA0674.1 | NKX6-1 | MA0674.1 | NTMATTAA | NKX6-1 | 5.14e-37 | 2.52e-39 | 9730 | 78.5 | 14800 | 72 | 1.09 |
| 71 | 4 | MA1115.1 | POU5F1 | MA1115.1 | WHATGCAAATN | POU5F1 | 7.12e-37 | 3.45e-39 | 8190 | 66 | 12100 | 58.8 | 1.12 |
| 72 | 2 | MA0675.1 | NKX6-2 | MA0675.1 | NYMATTAA | NKX6-2 | 8.92e-37 | 4.26e-39 | 9060 | 73 | 13600 | 66.1 | 1.1 |
| 73 | 2 | MA0661.1 | MEOX1 | MA0661.1 | VSTAATTAHC | MEOX1 | 1.59e-36 | 7.49e-39 | 8190 | 66 | 12100 | 58.8 | 1.12 |
| 74 | 2 | MA1519.1 | LHX5 | MA1519.1 | STAATTAV | LHX5 | 2.66e-36 | 1.23e-38 | 3910 | 31.5 | 5100 | 24.9 | 1.27 |
| 75 | 2 | MA0125.1 | Nobox | MA0125.1 | TAATTRSY | Nobox | 3.53e-36 | 1.62e-38 | 7670 | 61.8 | 11200 | 54.5 | 1.13 |
| 76 | 10 | MA0122.3 | Nkx3-2 | MA0122.3 | WWAAMCACTTAAN | Nkx3-2 | 4.01e-36 | 1.82e-38 | 9230 | 74.4 | 13900 | 67.6 | 1.1 |
| 77 | 12 | MA0845.1 | FOXB1 | MA0845.1 | WATGTAAATAT | FOXB1 | 5.62e-36 | 2.51e-38 | 5820 | 46.9 | 8130 | 39.6 | 1.18 |
| 78 | 2 | MA0721.1 | UNCX | MA0721.1 | YYAATTAV | UNCX | 1.17e-35 | 5.18e-38 | 8820 | 71.1 | 13200 | 64.2 | 1.11 |
| 79 | 10 | MA0650.3 | Hoxa13 | MA0650.3 | NNCCAATAAAAN | Hoxa13 | 1.92e-35 | 8.35e-38 | 8640 | 69.6 | 12900 | 62.6 | 1.11 |
| 80 | 13 | MA0025.2 | NFIL3 | MA0025.2 | NRTTATGYAATNN | NFIL3 | 1.97e-35 | 8.46e-38 | 7250 | 58.4 | 10500 | 51.1 | 1.14 |
| 81 | 14 | MA0782.2 | PKNOX1 | MA0782.2 | NNTGAGTGACAGVNN | PKNOX1 | 2.91e-35 | 1.24e-37 | 7690 | 62 | 11200 | 54.7 | 1.13 |
| 82 | 2 | MA0879.2 | DLX1 | MA0879.2 | VYAATTAN | DLX1 | 3.35e-35 | 1.41e-37 | 9640 | 77.7 | 14600 | 71.3 | 1.09 |
| 83 | 10 | MA0041.2 | FOXD3 | MA0041.2 | WAWGHAAATAAACARY | FOXD3 | 3.53e-35 | 1.46e-37 | 8870 | 71.5 | 13300 | 64.6 | 1.11 |
| 84 | 2 | MA1505.1 | HOXC8 | MA1505.1 | KTAATTAM | HOXC8 | 3.66e-35 | 1.5e-37 | 4220 | 34 | 5600 | 27.3 | 1.25 |
| 85 | 15 | MA0052.4 | MEF2A | MA0052.4 | DDCTAAAAATAGMHH | MEF2A | 4.31e-35 | 1.75e-37 | 7850 | 63.3 | 11500 | 56.1 | 1.13 |
| 86 | 2 | MA0887.1 | EVX1 | MA0887.1 | VVTAATTABS | EVX1 | 4.61e-35 | 1.85e-37 | 9260 | 74.6 | 14000 | 68 | 1.1 |
| 87 | 16 | MA0602.1 | Arid5a | MA0602.1 | SYAATATTGVDANH | Arid5a | 4.91e-35 | 1.94e-37 | 8100 | 65.3 | 12000 | 58.2 | 1.12 |
| 88 | 7 | MA0035.4 | GATA1 | MA0035.4 | NHCTAATCTDH | GATA1 | 6.42e-35 | 2.51e-37 | 8220 | 66.2 | 12200 | 59.2 | 1.12 |
| 89 | 2 | MA0903.1 | HOXB3 | MA0903.1 | NNYMATTARN | HOXB3 | 8.52e-35 | 3.29e-37 | 7710 | 62.1 | 11300 | 55 | 1.13 |
| 90 | 10 | MA0679.2 | ONECUT1 | MA0679.2 | WAAAAATCAATAAWWN | ONECUT1 | 9.09e-35 | 3.48e-37 | 6150 | 49.6 | 8690 | 42.3 | 1.17 |
| 91 | 2 | MA0890.1 | GBX2 | MA0890.1 | RSYAATTARN | GBX2 | 1.21e-34 | 4.57e-37 | 8340 | 67.2 | 12400 | 60.2 | 1.12 |
| 92 | 2 | MA0713.1 | PHOX2A | MA0713.1 | TAATYYAATTA | PHOX2A | 1.89e-34 | 7.07e-37 | 7140 | 57.6 | 10300 | 50.4 | 1.14 |
| 93 | 2 | MA0902.2 | HOXB2 | MA0902.2 | SYAATTAS | HOXB2 | 3.4e-34 | 1.26e-36 | 6130 | 49.4 | 8680 | 42.3 | 1.17 |
| 94 | 2 | MA0793.1 | POU6F2 | MA0793.1 | ASCTMATTAW | POU6F2 | 5.25e-34 | 1.92e-36 | 7720 | 62.3 | 11300 | 55.2 | 1.13 |
| 95 | 2 | MA0716.1 | PRRX1 | MA0716.1 | YYAATTAR | PRRX1 | 1.34e-33 | 4.85e-36 | 9050 | 73 | 13600 | 66.4 | 1.1 |
| 96 | 2 | MA1502.1 | HOXB8 | MA1502.1 | GYMATTAM | HOXB8 | 1.41e-33 | 5.04e-36 | 5680 | 45.8 | 7960 | 38.8 | 1.18 |
| 97 | 2 | MA0788.1 | POU3F3 | MA0788.1 | WWTATGCWAATTW | POU3F3 | 1.53e-33 | 5.42e-36 | 6280 | 50.6 | 8930 | 43.5 | 1.16 |
| 98 | 2 | MA0642.2 | EN2 | MA0642.2 | VSYAATTA | EN2 | 2.05e-33 | 7.2e-36 | 7990 | 64.4 | 11800 | 57.5 | 1.12 |
| 99 | 2 | MA0630.1 | SHOX | MA0630.1 | HTAATTRR | SHOX | 2.34e-33 | 8.15e-36 | 9640 | 77.7 | 14700 | 71.5 | 1.09 |
| 100 | 17 | MA0029.1 (Mecom) | STREME-2 | 2-AGATAAT | AGATAAT | Mecom | 2.92e-33 | 1e-35 | 4180 | 33.7 | 5580 | 27.2 | 1.24 |
| 101 | 2 | MA0068.2 | PAX4 | MA0068.2 | CTAATTAG | PAX4 | 3.97e-33 | 1.35e-35 | 6930 | 55.9 | 10000 | 48.8 | 1.15 |
| 102 | 12 | MA0032.2 | FOXC1 | MA0032.2 | WAWGTAAAYAW | FOXC1 | 5.16e-33 | 1.74e-35 | 8280 | 66.8 | 12300 | 59.9 | 1.11 |
| 103 | 18 | MA1562.1 | SOX14 | MA1562.1 | CCGAACAATG | SOX14 | 5.33e-33 | 1.78e-35 | 8700 | 70.1 | 13000 | 63.4 | 1.11 |
| 104 | 2 | MA0648.1 | GSC | MA0648.1 | VYTAATCCSY | GSC | 6.18e-33 | 2.04e-35 | 7490 | 60.4 | 11000 | 53.4 | 1.13 |
| 105 | 10 | MA0898.1 | Hmx3 | MA0898.1 | ASAAGCAATTAAWRRAT | Hmx3 | 6.24e-33 | 2.04e-35 | 5740 | 46.2 | 8060 | 39.2 | 1.18 |
| 106 | 2 | MA0853.1 | Alx4 | MA0853.1 | CGCRYTAATTARYHNNY | Alx4 | 6.47e-33 | 2.1e-35 | 5020 | 40.5 | 6900 | 33.6 | 1.2 |
| 107 | 2 | MA0888.1 | EVX2 | MA0888.1 | VVTAATTAVB | EVX2 | 7.37e-33 | 2.37e-35 | 9000 | 72.6 | 13600 | 66 | 1.1 |
| 108 | 18 | MA0084.1 | SRY | MA0084.1 | NWWAACAAT | SRY | 9.34e-33 | 2.98e-35 | 9540 | 76.9 | 14500 | 70.7 | 1.09 |
| 109 | 10 | MA0909.3 | Hoxd13 | MA0909.3 | NNCAATAAAW | Hoxd13 | 9.91e-33 | 3.13e-35 | 6380 | 51.5 | 9120 | 44.4 | 1.16 |
| 110 | 12 | MA0846.1 | FOXC2 | MA0846.1 | WAWGTAAACAWW | FOXC2 | 1.43e-32 | 4.49e-35 | 7280 | 58.7 | 10600 | 51.7 | 1.14 |
| 111 | 18 | MA0077.1 | SOX9 | MA0077.1 | CYATTGTTY | SOX9 | 1.69e-32 | 5.24e-35 | 6040 | 48.7 | 8560 | 41.7 | 1.17 |
| 112 | 2 | MA1463.1 | ARGFX | MA1463.1 | DCTAATTARM | ARGFX | 2.97e-32 | 9.12e-35 | 4500 | 36.2 | 6090 | 29.7 | 1.22 |
| 113 | 2 | MA0892.1 | GSX1 | MA0892.1 | NCYMATTAVN | GSX1 | 4.42e-32 | 1.34e-34 | 7260 | 58.5 | 10600 | 51.6 | 1.13 |
| 114 | 2 | MA0891.1 | GSC2 | MA0891.1 | NYTAATCCBH | GSC2 | 5.14e-32 | 1.55e-34 | 8930 | 72 | 13500 | 65.5 | 1.1 |
| 115 | 2 | MA0881.1 | Dlx4 | MA0881.1 | VYAATTAN | Dlx4 | 5.43e-32 | 1.62e-34 | 9950 | 80.2 | 15300 | 74.4 | 1.08 |
| 116 | 2 | MA0706.1 | MEOX2 | MA0706.1 | DSTAATTAWN | MEOX2 | 6.17e-32 | 1.82e-34 | 9360 | 75.4 | 14200 | 69.2 | 1.09 |
| 117 | 19 | 10-TAASCA | STREME-10 | 10-TAASCA | 10-TAASCA | TAASCA | 6.19e-32 | 1.82e-34 | 8220 | 66.2 | 12200 | 59.5 | 1.11 |
| 118 | 2 | MA0882.1 | DLX6 | MA0882.1 | VYAATTAN | DLX6 | 6.7e-32 | 1.95e-34 | 10300 | 82.9 | 15900 | 77.4 | 1.07 |
| 119 | 20 | MA0769.2 | TCF7 | MA0769.2 | NNCTTTGAWVT | TCF7 | 6.79e-32 | 1.96e-34 | 9100 | 73.4 | 13800 | 67 | 1.1 |
| 120 | 8 | MA0046.2 | HNF1A | MA0046.2 | DRTTAATNATTAACN | HNF1A | 7.88e-32 | 2.26e-34 | 4600 | 37.1 | 6270 | 30.6 | 1.22 |
| 121 | 10 | MA0901.2 | HOXB13 | MA0901.2 | WDCCAATAAAAWWW | HOXB13 | 8.86e-32 | 2.52e-34 | 6880 | 55.4 | 9950 | 48.5 | 1.14 |
| 122 | 2 | MA1498.2 | HOXA7 | MA1498.2 | SYMATTAV | HOXA7 | 1.01e-31 | 2.86e-34 | 8540 | 68.8 | 12800 | 62.2 | 1.11 |
| 123 | 21 | MA0611.2 | Dux | MA0611.2 | NTGATTBAATCAVWNN | Dux | 1.05e-31 | 2.91e-34 | 3580 | 28.9 | 4680 | 22.8 | 1.27 |
| 124 | 2 | MA0900.2 | HOXA2 | MA0900.2 | STAATTAS | HOXA2 | 1.05e-31 | 2.91e-34 | 3580 | 28.9 | 4680 | 22.8 | 1.27 |
| 125 | 2 | MA1518.2 | Lhx1 | MA1518.2 | NNNTGCTAATTAGCANNN | Lhx1 | 1.5e-31 | 4.12e-34 | 8350 | 67.3 | 12400 | 60.6 | 1.11 |
| 126 | 2 | MA0701.2 | LHX9 | MA0701.2 | CYAATTAR | LHX9 | 2.67e-31 | 7.28e-34 | 3480 | 28.1 | 4540 | 22.1 | 1.27 |
| 127 | 2 | MA0709.1 | Msx3 | MA0709.1 | SCAATTAN | Msx3 | 3.07e-31 | 8.33e-34 | 8800 | 71 | 13200 | 64.5 | 1.1 |
| 128 | 4 | MA0627.2 | POU2F3 | MA0627.2 | NWTATGCAAATNH | POU2F3 | 3.52e-31 | 9.47e-34 | 6000 | 48.4 | 8520 | 41.5 | 1.17 |
| 129 | 18 | MA0078.2 | Sox17 | MA0078.2 | NNAGAACAATGGNN | Sox17 | 3.91e-31 | 1.04e-33 | 8900 | 71.7 | 13400 | 65.3 | 1.1 |
| 130 | 22 | MA1480.1 | DPRX | MA1480.1 | AGATAATCCN | DPRX | 4.09e-31 | 1.08e-33 | 8590 | 69.3 | 12900 | 62.7 | 1.1 |
| 131 | 7 | MA1970.1 | TRPS1 | MA1970.1 | NHTCTTATCTNH | TRPS1 | 4.37e-31 | 1.15e-33 | 4880 | 39.3 | 6720 | 32.7 | 1.2 |
| 132 | 2 | MA0708.2 | MSX2 | MA0708.2 | SCAATTAV | MSX2 | 5.17e-31 | 1.35e-33 | 7120 | 57.4 | 10400 | 50.6 | 1.14 |
| 133 | 2 | MA0854.1 | Alx1 | MA0854.1 | CGMRYTAATTARTNMNY | Alx1 | 6.84e-31 | 1.77e-33 | 4430 | 35.7 | 6020 | 29.3 | 1.22 |
| 134 | 2 | MA1549.1 | POU6F1 | MA1549.1 | RTAATGAGVN | POU6F1 | 7.21e-31 | 1.85e-33 | 5090 | 41 | 7050 | 34.4 | 1.19 |
| 135 | 2 | MA0880.1 | Dlx3 | MA0880.1 | NYAATTAN | Dlx3 | 7.65e-31 | 1.95e-33 | 10400 | 83.8 | 16100 | 78.4 | 1.07 |
| 136 | 2 | MA1471.1 | BARX2 | MA1471.1 | NWWAAYMATTAN | BARX2 | 8.1e-31 | 2.05e-33 | 7120 | 57.4 | 10400 | 50.6 | 1.14 |
| 137 | 8 | MA0153.2 | HNF1B | MA0153.2 | GTTAATNATTAAY | HNF1B | 1.19e-30 | 2.99e-33 | 3770 | 30.4 | 4990 | 24.3 | 1.25 |
| 138 | 7 | MA0037.4 | Gata3 | MA0037.4 | NHTCTTATCTNH | Gata3 | 1.25e-30 | 3.11e-33 | 5800 | 46.7 | 8200 | 40 | 1.17 |
| 139 | 23 | MA0905.1 | HOXC10 | MA0905.1 | GTCRTAAAAH | HOXC10 | 1.38e-30 | 3.41e-33 | 7800 | 62.9 | 11500 | 56.2 | 1.12 |
| 140 | 2 | MA1530.1 | NKX6-3 | MA1530.1 | VYCATTAWW | NKX6-3 | 1.58e-30 | 3.89e-33 | 5860 | 47.2 | 8310 | 40.5 | 1.17 |
| 141 | 4 | MA0792.1 | POU5F1B | MA0792.1 | TATGCWAAT | POU5F1B | 1.84e-30 | 4.49e-33 | 9500 | 76.6 | 14500 | 70.6 | 1.09 |
| 142 | 22 | MA0711.1 | OTX1 | MA0711.1 | HTAATCCS | OTX1 | 2.01e-30 | 4.87e-33 | 8510 | 68.6 | 12800 | 62.1 | 1.1 |
| 143 | 2 | MA0904.2 | HOXB5 | MA0904.2 | STAATTAS | HOXB5 | 2.14e-30 | 5.15e-33 | 6790 | 54.7 | 9840 | 47.9 | 1.14 |
| 144 | 24 | MA1487.2 | FOXE1 | MA1487.2 | VYTAAAYAAACAAH | FOXE1 | 3.1e-30 | 7.4e-33 | 6360 | 51.2 | 9130 | 44.4 | 1.15 |
| 145 | 2 | MA0634.1 | ALX3 | MA0634.1 | YYYAATTANV | ALX3 | 4.71e-30 | 1.12e-32 | 10400 | 83.5 | 16000 | 78.1 | 1.07 |
| 146 | 4 | MA0786.1 | POU3F1 | MA0786.1 | WTATGCWAATKW | POU3F1 | 5.86e-30 | 1.38e-32 | 4530 | 36.5 | 6190 | 30.1 | 1.21 |
| 147 | 2 | MA0699.1 | LBX2 | MA0699.1 | NCYAATTARN | LBX2 | 6.07e-30 | 1.42e-32 | 9160 | 73.8 | 13900 | 67.7 | 1.09 |
| 148 | 2 | MA0612.2 | EMX1 | MA0612.2 | BTAATTAS | EMX1 | 6.99e-30 | 1.63e-32 | 4410 | 35.6 | 6000 | 29.2 | 1.22 |
| 149 | 2 | MA0635.1 | BARHL2 | MA0635.1 | VHTAAAYGRT | BARHL2 | 9.63e-30 | 2.22e-32 | 8600 | 69.4 | 12900 | 63 | 1.1 |
| 150 | 2 | MA0912.2 | HOXD3 | MA0912.2 | CTAATTAC | HOXD3 | 1.02e-29 | 2.34e-32 | 3800 | 30.7 | 5060 | 24.7 | 1.24 |
| 151 | 12 | MA1683.1 | FOXA3 | MA1683.1 | DWGTAAACANN | FOXA3 | 1.34e-29 | 3.05e-32 | 8190 | 66 | 12200 | 59.5 | 1.11 |
| 152 | 2 | MA0662.1 | MIXL1 | MA0662.1 | NBYAATTAVN | MIXL1 | 1.38e-29 | 3.11e-32 | 9770 | 78.7 | 15000 | 73 | 1.08 |
| 153 | 2 | MA0707.2 | MNX1 | MA0707.2 | VYRATTAK | MNX1 | 1.78e-29 | 4.01e-32 | 5830 | 47 | 8280 | 40.4 | 1.16 |
| 154 | 4 | MA0785.1 | POU2F1 | MA0785.1 | AWTATGCWAATK | POU2F1 | 2e-29 | 4.47e-32 | 8460 | 68.2 | 12700 | 61.8 | 1.1 |
| 155 | 2 | MA1608.1 | Isl1 | MA1608.1 | NNCCATTAGNN | Isl1 | 2.58e-29 | 5.73e-32 | 7420 | 59.8 | 10900 | 53.2 | 1.12 |
| 156 | 10 | MA0914.1 | ISL2 | MA0914.1 | GCAMTTAR | ISL2 | 3.7e-29 | 8.15e-32 | 7870 | 63.4 | 11700 | 56.9 | 1.12 |
| 157 | 2 | MA0715.1 | PROP1 | MA0715.1 | TAATYWAATTA | PROP1 | 4.48e-29 | 9.82e-32 | 4180 | 33.7 | 5660 | 27.6 | 1.22 |
| 158 | 12 | MA0148.4 | FOXA1 | MA0148.4 | WWGTAAACATNN | FOXA1 | 4.64e-29 | 1.01e-31 | 9390 | 75.7 | 14300 | 69.7 | 1.09 |
| 159 | 2 | MA0910.2 | HOXD8 | MA0910.2 | GYMATTAM | HOXD8 | 4.68e-29 | 1.01e-31 | 5350 | 43.1 | 7520 | 36.6 | 1.18 |
| 160 | 22 | MA0712.2 | OTX2 | MA0712.2 | NDRGGATTARNN | OTX2 | 5.42e-29 | 1.17e-31 | 7440 | 60 | 11000 | 53.4 | 1.12 |
| 161 | 25 | 7-TCACAG | STREME-7 | 7-TCACAG | TCACAG | TCACAG | 8.74e-29 | 1.87e-31 | 10600 | 85.8 | 16600 | 80.8 | 1.06 |
| 162 | 18 | MA1152.1 | SOX15 | MA1152.1 | CYWTTGTTHW | SOX15 | 9.43e-29 | 2e-31 | 6810 | 54.9 | 9910 | 48.3 | 1.14 |
| 163 | 2 | MA0705.1 | Lhx8 | MA0705.1 | CTAATTAV | Lhx8 | 1.11e-28 | 2.35e-31 | 6020 | 48.5 | 8610 | 42 | 1.16 |
| 164 | 2 | MA1497.1 | HOXA6 | MA1497.1 | BGYMATTANN | HOXA6 | 1.42e-28 | 2.99e-31 | 9730 | 78.5 | 14900 | 72.8 | 1.08 |
| 165 | 13 | MA1636.1 | CEBPG | MA1636.1 | NDATGATGCAATMHH | CEBPG | 2.42e-28 | 5.04e-31 | 8280 | 66.8 | 12400 | 60.4 | 1.11 |
| 166 | 2 | MA0894.1 | HESX1 | MA0894.1 | GHTAATTRRH | HESX1 | 2.79e-28 | 5.78e-31 | 6120 | 49.3 | 8780 | 42.8 | 1.15 |
| 167 | 12 | MA0047.3 | FOXA2 | MA0047.3 | RWGTAAACANN | FOXA2 | 4.52e-28 | 9.32e-31 | 6820 | 55 | 9940 | 48.4 | 1.14 |
| 168 | 18 | MA1563.2 | SOX18 | MA1563.2 | AACAATDV | SOX18 | 5.31e-28 | 1.09e-30 | 7680 | 62 | 11400 | 55.5 | 1.12 |
| 169 | 10 | MA0897.1 | Hmx2 | MA0897.1 | ASAAGCAATTAAHVVRT | Hmx2 | 7.08e-28 | 1.44e-30 | 4680 | 37.8 | 6480 | 31.6 | 1.2 |
| 170 | 26 | MA0108.2 | TBP | MA0108.2 | STATAAAWRSVVVBN | TBP | 8.13e-28 | 1.64e-30 | 6600 | 53.2 | 9580 | 46.6 | 1.14 |
| 171 | 2 | MA1500.1 | HOXB6 | MA1500.1 | BTAATKRC | HOXB6 | 1.04e-27 | 2.08e-30 | 10500 | 84.8 | 16400 | 79.9 | 1.06 |
| 172 | 2 | MA0601.1 | Arid3b | MA0601.1 | ATATTAATWAN | Arid3b | 1.13e-27 | 2.25e-30 | 5210 | 42 | 7330 | 35.7 | 1.18 |
| 173 | 27 | MA1124.1 | ZNF24 | MA1124.1 | CATTCATTCATTC | ZNF24 | 1.97e-27 | 3.92e-30 | 5950 | 48 | 8520 | 41.5 | 1.16 |
| 174 | 2 | MA0158.2 | HOXA5 | MA0158.2 | GYMATTAS | HOXA5 | 2.27e-27 | 4.48e-30 | 4630 | 37.3 | 6400 | 31.2 | 1.2 |
| 175 | 2 | MA1501.1 | HOXB7 | MA1501.1 | GGTAATTAVS | HOXB7 | 3.53e-27 | 6.93e-30 | 7570 | 61 | 11200 | 54.6 | 1.12 |
| 176 | 2 | MA0876.1 | BSX | MA0876.1 | SYVATTAV | BSX | 3.81e-27 | 7.45e-30 | 7060 | 56.9 | 10400 | 50.5 | 1.13 |
| 177 | 28 | MA1644.1 | NFYC | MA1644.1 | RDCCAATCASN | NFYC | 6.07e-27 | 1.18e-29 | 7410 | 59.7 | 11000 | 53.4 | 1.12 |
| 178 | 12 | MA1606.1 | Foxf1 | MA1606.1 | DWRTAAACANN | Foxf1 | 9.74e-27 | 1.88e-29 | 9590 | 77.3 | 14700 | 71.7 | 1.08 |
| 179 | 2 | MA0723.2 | VAX2 | MA0723.2 | BYAATTAV | VAX2 | 1.08e-26 | 2.07e-29 | 10100 | 81.3 | 15600 | 76.1 | 1.07 |
| 180 | 2 | MA1481.1 | DRGX | MA1481.1 | YYAATTAN | DRGX | 1.42e-26 | 2.71e-29 | 3580 | 28.9 | 4780 | 23.3 | 1.24 |
| 181 | 29 | MA0755.1 | CUX2 | MA0755.1 | TRATCGATAW | CUX2 | 1.66e-26 | 3.16e-29 | 7100 | 57.3 | 10500 | 50.9 | 1.12 |
| 182 | 18 | MA0868.2 | SOX8 | MA0868.2 | MGAACAATRG | SOX8 | 2.05e-26 | 3.88e-29 | 8980 | 72.4 | 13700 | 66.5 | 1.09 |
| 183 | 18 | MA0143.4 | SOX2 | MA0143.4 | DNACAATGGNN | SOX2 | 2.43e-26 | 4.57e-29 | 9230 | 74.4 | 14100 | 68.7 | 1.08 |
| 184 | 2 | MA0875.1 | BARX1 | MA0875.1 | VCMATTAV | BARX1 | 2.94e-26 | 5.49e-29 | 7040 | 56.7 | 10300 | 50.4 | 1.13 |
| 185 | 24 | MA0847.3 | FOXD2 | MA0847.3 | SYTAARYAAACA | FOXD2 | 7.62e-26 | 1.42e-28 | 5600 | 45.2 | 8000 | 39 | 1.16 |
| 186 | 30 | MA0018.4 | CREB1 | MA0018.4 | NNRTGATGTCAYN | CREB1 | 1.15e-25 | 2.13e-28 | 5670 | 45.7 | 8110 | 39.5 | 1.16 |
| 187 | 21 | MA1720.1 | ZNF85 | MA1720.1 | DHDGAGATTACWKCAK | ZNF85 | 1.25e-25 | 2.3e-28 | 7160 | 57.7 | 10600 | 51.4 | 1.12 |
| 188 | 15 | MA0497.1 | MEF2C | MA0497.1 | DDDCYAAAAATAGMW | MEF2C | 1.47e-25 | 2.68e-28 | 5380 | 43.3 | 7640 | 37.2 | 1.17 |
| 189 | 31 | MA0809.2 | TEAD4 | MA0809.2 | NNACATTCCARN | TEAD4 | 2.3e-25 | 4.18e-28 | 9700 | 78.2 | 14900 | 72.8 | 1.07 |
| 190 | 2 | MA0658.1 | LHX6 | MA0658.1 | RCTAATTARY | LHX6 | 2.94e-25 | 5.33e-28 | 4720 | 38.1 | 6600 | 32.2 | 1.18 |
| 191 | 2 | MA0075.3 | PRRX2 | MA0075.3 | CTAATTAN | PRRX2 | 3.22e-25 | 5.8e-28 | 3700 | 29.8 | 4980 | 24.3 | 1.23 |
| 192 | 18 | MA1120.1 | SOX13 | MA1120.1 | DVACAATGGNN | SOX13 | 6.22e-25 | 1.11e-27 | 9150 | 73.8 | 14000 | 68.1 | 1.08 |
| 193 | 12 | MA1607.1 | Foxl2 | MA1607.1 | DDWATGTAAACANN | Foxl2 | 6.4e-25 | 1.14e-27 | 7100 | 57.3 | 10500 | 51.1 | 1.12 |
| 194 | 2 | MA1499.1 | HOXB4 | MA1499.1 | RTMATTAR | HOXB4 | 7.53e-25 | 1.33e-27 | 5520 | 44.5 | 7880 | 38.4 | 1.16 |
| 195 | 10 | MA0899.1 | HOXA10 | MA0899.1 | DGYMATAAAAH | HOXA10 | 9.69e-25 | 1.71e-27 | 9520 | 76.8 | 14600 | 71.3 | 1.08 |
| 196 | 32 | MA1974.1 | ZNF211 | MA1974.1 | HWYATATACCAYRYW | ZNF211 | 1.06e-24 | 1.87e-27 | 5020 | 40.4 | 7080 | 34.5 | 1.17 |
| 197 | 33 | MA0483.1 | Gfi1B | MA0483.1 | AAATCWCWGCH | Gfi1B | 2.44e-24 | 4.25e-27 | 4790 | 38.6 | 6720 | 32.8 | 1.18 |
| 198 | 18 | MA0514.2 | Sox3 | MA0514.2 | DNACAATGGNN | Sox3 | 2.65e-24 | 4.6e-27 | 10100 | 81.7 | 15700 | 76.7 | 1.06 |
| 199 | 22 | MA0714.1 | PITX3 | MA0714.1 | NYTAATCCC | PITX3 | 2.74e-24 | 4.73e-27 | 8900 | 71.8 | 13600 | 66.1 | 1.09 |
| 200 | 34 | MA0144.2 | STAT3 | MA0144.2 | YTTCYKGGAAD | STAT3 | 2.75e-24 | 4.73e-27 | 6190 | 49.9 | 8990 | 43.8 | 1.14 |
| 201 | 12 | MA0850.1 | FOXP3 | MA0850.1 | RTAAACA | FOXP3 | 5.6e-24 | 9.59e-27 | 8640 | 69.7 | 13100 | 64 | 1.09 |
| 202 | 35 | MA1657.1 | ZNF652 | MA1657.1 | BAAAGRGTTAAN | ZNF652 | 6.42e-24 | 1.09e-26 | 7730 | 62.3 | 11600 | 56.4 | 1.11 |
| 203 | 13 | MA0102.4 | CEBPA | MA0102.4 | NDATTGCACAATHN | CEBPA | 8.79e-24 | 1.49e-26 | 8900 | 71.8 | 13600 | 66.1 | 1.08 |
| 204 | 36 | MA0877.3 | BARHL1 | MA0877.3 | AMCGTTTA | BARHL1 | 9.22e-24 | 1.56e-26 | 9220 | 74.3 | 14100 | 68.8 | 1.08 |
| 205 | 12 | MA0481.3 | FOXP1 | MA0481.3 | NDGTAAACANH | FOXP1 | 1.35e-23 | 2.27e-26 | 9950 | 80.2 | 15400 | 75.2 | 1.07 |
| 206 | 22 | MA0682.2 | PITX1 | MA0682.2 | YTAATCCH | PITX1 | 1.53e-23 | 2.55e-26 | 5420 | 43.7 | 7750 | 37.8 | 1.16 |
| 207 | 2 | MA1504.1 | HOXC4 | MA1504.1 | VTMATTAN | HOXC4 | 2.14e-23 | 3.56e-26 | 6280 | 50.6 | 9160 | 44.6 | 1.13 |
| 208 | 37 | MA0052.4 (MEF2A) | MEME-1 | WWDKWTTKTWTTTTW | WWDKWTTKTWTTTTW | MEF2A | 4.02e-23 | 6.66e-26 | 4140 | 33.4 | 5730 | 27.9 | 1.2 |
| 209 | 10 | MA0883.1 | Dmbx1 | MA0883.1 | WDRWNMGGATTADKNNW | Dmbx1 | 4.28e-23 | 7.04e-26 | 4810 | 38.8 | 6780 | 33 | 1.17 |
| 210 | 7 | MA0482.2 | GATA4 | MA0482.2 | NNCCTTATCTNH | GATA4 | 8.9e-23 | 1.46e-25 | 2090 | 16.9 | 2600 | 12.7 | 1.33 |
| 211 | 10 | MA0151.1 | Arid3a | MA0151.1 | ATYAAA | Arid3a | 1.03e-22 | 1.68e-25 | 4320 | 34.8 | 6010 | 29.3 | 1.19 |
| 212 | 2 | MA1577.1 | TLX2 | MA1577.1 | BYAATTAR | TLX2 | 1.14e-22 | 1.85e-25 | 3050 | 24.6 | 4040 | 19.7 | 1.25 |
| 213 | 2 | MA0874.1 | Arx | MA0874.1 | VTBCRYTAATTARTKSW | Arx | 1.17e-22 | 1.88e-25 | 3840 | 30.9 | 5260 | 25.6 | 1.21 |
| 214 | 38 | MA0050.3 | Irf1 | MA0050.3 | ANTGAAACTGAAASH | Irf1 | 1.24e-22 | 2e-25 | 8040 | 64.8 | 12100 | 59.1 | 1.1 |
| 215 | 12 | MA0031.1 | FOXD1 | MA0031.1 | GTAAACAW | FOXD1 | 1.74e-22 | 2.78e-25 | 6040 | 48.7 | 8800 | 42.8 | 1.14 |
| 216 | 2 | MA0895.1 | HMBOX1 | MA0895.1 | MYTAGTTAMS | HMBOX1 | 1.92e-22 | 3.05e-25 | 8760 | 70.6 | 13400 | 65.1 | 1.08 |
| 217 | 2 | MA0893.2 | GSX2 | MA0893.2 | SYMATTAR | GSX2 | 3.05e-22 | 4.84e-25 | 6290 | 50.7 | 9210 | 44.9 | 1.13 |
| 218 | 20 | MA0523.1 | TCF7L2 | MA0523.1 | DRASATCAAAGRVA | TCF7L2 | 3.67e-22 | 5.79e-25 | 4400 | 35.5 | 6160 | 30 | 1.18 |
| 219 | 10 | MA0070.1 | PBX1 | MA0070.1 | HCATCAATCAAW | PBX1 | 3.87e-22 | 6.07e-25 | 4640 | 37.4 | 6540 | 31.8 | 1.17 |
| 220 | 2 | MA1495.1 | HOXA1 | MA1495.1 | STAATTAS | HOXA1 | 4.41e-22 | 6.9e-25 | 7000 | 56.4 | 10400 | 50.6 | 1.12 |
| 221 | 31 | MA1121.1 | TEAD2 | MA1121.1 | NYACATTCCWNNS | TEAD2 | 6.66e-22 | 1.04e-24 | 8420 | 67.9 | 12800 | 62.3 | 1.09 |
| 222 | 12 | MA0848.1 | FOXO4 | MA0848.1 | GTAAACA | FOXO4 | 7.01e-22 | 1.09e-24 | 7190 | 57.9 | 10700 | 52.2 | 1.11 |
| 223 | 20 | MA0768.2 | Lef1 | MA0768.2 | NNBCCTTTGATSTN | Lef1 | 8.81e-22 | 1.36e-24 | 10100 | 81.6 | 15800 | 76.9 | 1.06 |
| 224 | 2 | MA0467.2 | Crx | MA0467.2 | NDGGATTANN | Crx | 9.41e-22 | 1.45e-24 | 6110 | 49.2 | 8920 | 43.5 | 1.13 |
| 225 | 12 | MA0157.3 | Foxo3 | MA0157.3 | NDGTAAACARNN | Foxo3 | 1.12e-21 | 1.71e-24 | 10100 | 81.3 | 15700 | 76.6 | 1.06 |
| 226 | 12 | MA1103.2 | FOXK2 | MA1103.2 | DDGTAAACANN | FOXK2 | 1.18e-21 | 1.8e-24 | 9660 | 77.9 | 15000 | 72.9 | 1.07 |
| 227 | 12 | MA0033.2 | FOXL1 | MA0033.2 | RTAAACA | FOXL1 | 1.57e-21 | 2.38e-24 | 9270 | 74.7 | 14300 | 69.5 | 1.07 |
| 228 | 23 | MA0911.1 | Hoxa11 | MA0911.1 | RGTCGTAAAANT | Hoxa11 | 1.59e-21 | 2.39e-24 | 7650 | 61.7 | 11500 | 56 | 1.1 |
| 229 | 12 | MA0480.2 | Foxo1 | MA0480.2 | NDGTAAACANN | Foxo1 | 1.82e-21 | 2.73e-24 | 9550 | 77 | 14800 | 72 | 1.07 |
| 230 | 39 | MA0052.4 (MEF2A) | STREME-9 | 9-AAAATA | AAAATA | MEF2A | 3.55e-21 | 5.31e-24 | 7210 | 58.1 | 10800 | 52.4 | 1.11 |
| 231 | 40 | MA0867.2 | SOX4 | MA0867.2 | RAACAAAGRV | SOX4 | 4.48e-21 | 6.68e-24 | 9340 | 75.3 | 14400 | 70.2 | 1.07 |
| 232 | 2 | MA1960.1 | MGA::EVX1 | MA1960.1 | RGGTGWTAATKD | MGA::EVX1 | 5.29e-21 | 7.84e-24 | 7170 | 57.8 | 10700 | 52.1 | 1.11 |
| 233 | 13 | MA0833.2 | ATF4 | MA0833.2 | DDATGATGCAATMN | ATF4 | 5.33e-21 | 7.87e-24 | 5660 | 45.7 | 8220 | 40 | 1.14 |
| 234 | 40 | MA1603.1 | Dmrt1 | MA1603.1 | GHTACAAAGTADC | Dmrt1 | 5.59e-21 | 8.22e-24 | 5840 | 47.1 | 8500 | 41.4 | 1.14 |
| 235 | 30 | MA1126.1 | FOS::JUN | MA1126.1 | DRTGACGTCATHNDTN | FOS::JUN | 1.18e-20 | 1.72e-23 | 5890 | 47.4 | 8590 | 41.8 | 1.13 |
| 236 | 28 | MA1523.1 | MSANTD3 | MA1523.1 | STVCACTCAC | MSANTD3 | 1.22e-20 | 1.78e-23 | 8900 | 71.8 | 13700 | 66.5 | 1.08 |
| 237 | 41 | MA1593.1 | ZNF317 | MA1593.1 | WVACAGCAGAYW | ZNF317 | 1.24e-20 | 1.81e-23 | 8900 | 71.7 | 13700 | 66.5 | 1.08 |
| 238 | 10 | MA0851.1 | Foxj3 | MA0851.1 | NDAADGTAAACAAANNM | Foxj3 | 1.28e-20 | 1.85e-23 | 5380 | 43.4 | 7760 | 37.8 | 1.15 |
| 239 | 12 | MA0852.2 | FOXK1 | MA0852.2 | NRDGTAAACAAGNN | FOXK1 | 1.72e-20 | 2.47e-23 | 7770 | 62.6 | 11700 | 57.1 | 1.1 |
| 240 | 2 | MA0132.2 | PDX1 | MA0132.2 | VYAATTAR | PDX1 | 2.34e-20 | 3.35e-23 | 3040 | 24.5 | 4080 | 19.9 | 1.23 |
| 241 | 30 | MA0093.3 | USF1 | MA0093.3 | NDGTCATGTGACHN | USF1 | 2.8e-20 | 3.99e-23 | 9580 | 77.3 | 14900 | 72.4 | 1.07 |
| 242 | 12 | MA0042.2 | FOXI1 | MA0042.2 | GTAAACA | FOXI1 | 3.83e-20 | 5.45e-23 | 5930 | 47.8 | 8670 | 42.2 | 1.13 |
| 243 | 13 | MA0043.3 | HLF | MA0043.3 | NNRTTATGCAACHN | HLF | 5.95e-20 | 8.42e-23 | 5400 | 43.5 | 7810 | 38 | 1.14 |
| 244 | 42 | MA1119.1 | SIX2 | MA1119.1 | NNCTGAAACCTGATMY | SIX2 | 6.8e-20 | 9.58e-23 | 7140 | 57.6 | 10700 | 52 | 1.11 |
| 245 | 14 | MA1114.1 | PBX3 | MA1114.1 | NNNTGAGTGACAGGBNN | PBX3 | 9.11e-20 | 1.28e-22 | 4490 | 36.2 | 6360 | 31 | 1.17 |
| 246 | 43 | MA0461.2 | Atoh1 | MA0461.2 | RMCATATGBY | Atoh1 | 1.52e-19 | 2.12e-22 | 9060 | 73 | 14000 | 68 | 1.07 |
| 247 | 13 | MA0836.2 | CEBPD | MA0836.2 | NRTTGCACAAYHN | CEBPD | 1.73e-19 | 2.41e-22 | 7980 | 64.3 | 12100 | 58.9 | 1.09 |
| 248 | 44 | MA0878.3 | CDX1 | MA0878.3 | GGYMATAAAA | CDX1 | 1.91e-19 | 2.64e-22 | 4400 | 35.4 | 6220 | 30.3 | 1.17 |
| 249 | 34 | MA1625.1 | Stat5b | MA1625.1 | NNNTTCCCAGAANNN | Stat5b | 2.93e-19 | 4.05e-22 | 9400 | 75.8 | 14600 | 71 | 1.07 |
| 250 | 31 | MA0808.1 | TEAD3 | MA0808.1 | RCATTCCW | TEAD3 | 3.11e-19 | 4.27e-22 | 3590 | 29 | 4950 | 24.1 | 1.2 |
| 251 | 45 | MA1709.1 | ZIM3 | MA1709.1 | RVMAACAGAAACCYM | ZIM3 | 4.05e-19 | 5.55e-22 | 7060 | 56.9 | 10600 | 51.5 | 1.11 |
| 252 | 12 | MA0614.1 | Foxj2 | MA0614.1 | RTAAACAA | Foxj2 | 4.23e-19 | 5.78e-22 | 5290 | 42.6 | 7650 | 37.3 | 1.14 |
| 253 | 30 | MA1143.1 | FOSL1::JUND | MA1143.1 | RTGACGTMAY | FOSL1::JUND | 4.6e-19 | 6.25e-22 | 7290 | 58.8 | 11000 | 53.4 | 1.1 |
| 254 | 2 | MA1507.1 | HOXD4 | MA1507.1 | VTMATTAD | HOXD4 | 8.33e-19 | 1.13e-21 | 4800 | 38.7 | 6880 | 33.5 | 1.15 |
| 255 | 30 | MA1112.2 | NR4A1 | MA1112.2 | NNAAAGGTCANN | NR4A1 | 1.25e-18 | 1.69e-21 | 9230 | 74.4 | 14300 | 69.5 | 1.07 |
| 256 | 45 | MA1953.1 | FOXO1::ELF1 | MA1953.1 | RTMAACAGGAAGTN | FOXO1::ELF1 | 1.57e-18 | 2.11e-21 | 8090 | 65.2 | 12300 | 60 | 1.09 |
| 257 | 46 | MA1151.1 | RORC | MA1151.1 | NWAWNTRGGTCA | RORC | 2.03e-18 | 2.72e-21 | 9440 | 76.1 | 14600 | 71.4 | 1.07 |
| 258 | 22 | MA0719.1 | RHOXF1 | MA0719.1 | NTRATCCN | RHOXF1 | 4.23e-18 | 5.65e-21 | 8580 | 69.2 | 13200 | 64.2 | 1.08 |
| 259 | 4 | MA1962.1 | POU2F1::SOX2 | MA1962.1 | MATTTRCATMACAATRG | POU2F1::SOX2 | 4.74e-18 | 6.29e-21 | 3240 | 26.1 | 4440 | 21.6 | 1.21 |
| 260 | 31 | MA0090.3 | TEAD1 | MA0090.3 | NNACATTCCAGSN | TEAD1 | 6.36e-18 | 8.42e-21 | 4640 | 37.4 | 6650 | 32.4 | 1.16 |
| 261 | 47 | MA0040.1 (Foxq1) | STREME-6 | 6-AACAAT | AACAAT | Foxq1 | 7.1e-18 | 9.36e-21 | 9990 | 80.5 | 15600 | 76.2 | 1.06 |
| 262 | 14 | MA0498.2 | MEIS1 | MA0498.2 | HTGACAD | MEIS1 | 8.61e-18 | 1.13e-20 | 3980 | 32.1 | 5600 | 27.3 | 1.18 |
| 263 | 34 | MA1624.1 | Stat5a | MA1624.1 | NTTCCAAGAANH | Stat5a | 1.08e-17 | 1.41e-20 | 8680 | 70 | 13300 | 65 | 1.08 |
| 264 | 43 | MA1524.2 | Msgn1 | MA1524.2 | VRRRACAAATGGTNNN | Msgn1 | 1.27e-17 | 1.66e-20 | 6310 | 50.9 | 9370 | 45.7 | 1.11 |
| 265 | 23 | MA0907.1 | HOXC13 | MA0907.1 | BCTCGTAAAAH | HOXC13 | 2.29e-17 | 2.98e-20 | 4590 | 37 | 6570 | 32 | 1.15 |
| 266 | 30 | MA1632.1 | ATF2 | MA1632.1 | NNATGABGTCATN | ATF2 | 2.34e-17 | 3.03e-20 | 3810 | 30.7 | 5340 | 26 | 1.18 |
| 267 | 48 | MA1105.2 | GRHL2 | MA1105.2 | NRAACAGGTTYN | GRHL2 | 3.06e-17 | 3.94e-20 | 8900 | 71.8 | 13700 | 67 | 1.07 |
| 268 | 49 | MA1479.1 | DMRTC2 | MA1479.1 | AATTGMTACATT | DMRTC2 | 3.6e-17 | 4.62e-20 | 6060 | 48.8 | 8960 | 43.6 | 1.12 |
| 269 | 18 | MA0087.2 | Sox5 | MA0087.2 | VRVAACAATGGNN | Sox5 | 3.68e-17 | 4.71e-20 | 5770 | 46.5 | 8490 | 41.4 | 1.12 |
| 270 | 1 | MA0659.3 | Mafg | MA0659.3 | NWGMTGACTCAGCAN | Mafg | 4.6e-17 | 5.86e-20 | 631 | 5.09 | 630 | 3.07 | 1.66 |
| 271 | 20 | MA1991.1 | Hnf1A | MA1991.1 | NBCCTTTGATSTBN | Hnf1A | 5.5e-17 | 6.99e-20 | 10600 | 85.6 | 16800 | 81.8 | 1.05 |
| 272 | 2 | MA0618.1 | LBX1 | MA0618.1 | TTAATTAG | LBX1 | 6.27e-17 | 7.93e-20 | 4870 | 39.3 | 7040 | 34.3 | 1.14 |
| 273 | 45 | MA0152.2 | Nfatc2 | MA0152.2 | NNRNAATGGAAANWNH | Nfatc2 | 1.02e-16 | 1.29e-19 | 8650 | 69.7 | 13300 | 64.9 | 1.07 |
| 274 | 50 | MA0606.2 | Nfat5 | MA0606.2 | NNATGGAAAAWN | Nfat5 | 1.08e-16 | 1.36e-19 | 9500 | 76.5 | 14800 | 72.1 | 1.06 |
| 275 | 2 | MA1496.1 | HOXA4 | MA1496.1 | RTMATTAV | HOXA4 | 1.13e-16 | 1.41e-19 | 6890 | 55.5 | 10300 | 50.4 | 1.1 |
| 276 | 30 | MA0829.2 | SREBF1 | MA0829.2 | DNATCACSTGATNH | SREBF1 | 1.16e-16 | 1.45e-19 | 8490 | 68.4 | 13100 | 63.6 | 1.08 |
| 277 | 51 | MA1588.1 | ZNF136 | MA1588.1 | RWATTCTTGGTTGRC | ZNF136 | 1.28e-16 | 1.59e-19 | 4700 | 37.8 | 6770 | 33 | 1.15 |
| 278 | 10 | MA0629.1 | Rhox11 | MA0629.1 | ANKNCGCTGTWAWDVRW | Rhox11 | 1.39e-16 | 1.72e-19 | 8750 | 70.5 | 13500 | 65.7 | 1.07 |
| 279 | 2 | MA0724.1 | VENTX | MA0724.1 | AMCGATTAR | VENTX | 1.5e-16 | 1.84e-19 | 2650 | 21.4 | 3560 | 17.4 | 1.23 |
| 280 | 10 | MA0465.2 | CDX2 | MA0465.2 | NDGCAATAAAWN | CDX2 | 1.68e-16 | 2.06e-19 | 3960 | 31.9 | 5600 | 27.3 | 1.17 |
| 281 | 52 | MA1978.1 | ZNF354A | MA1978.1 | HWAADWATAAATGRAYWAWTT | ZNF354A | 2.79e-16 | 3.41e-19 | 2060 | 16.6 | 2680 | 13.1 | 1.27 |
| 282 | 7 | MA0766.2 | GATA5 | MA0766.2 | SWGATAASRN | GATA5 | 2.84e-16 | 3.46e-19 | 1520 | 12.2 | 1870 | 9.12 | 1.34 |
| 283 | 46 | MA1150.1 | RORB | MA1150.1 | AWWTRGGTCAH | RORB | 4.08e-16 | 4.97e-19 | 5770 | 46.5 | 8530 | 41.5 | 1.12 |
| 284 | 28 | MA0060.3 | NFYA | MA0060.3 | RRCCAATCAGM | NFYA | 4.44e-16 | 5.37e-19 | 6530 | 52.6 | 9780 | 47.6 | 1.11 |
| 285 | 30 | MA1145.1 | FOSL2::JUND | MA1145.1 | NNRTGACGTCAYHSN | FOSL2::JUND | 7.14e-16 | 8.62e-19 | 8260 | 66.6 | 12700 | 61.8 | 1.08 |
| 286 | 29 | MA0754.2 | CUX1 | MA0754.2 | TAATCGATAM | CUX1 | 2.35e-15 | 2.82e-18 | 3270 | 26.3 | 4540 | 22.1 | 1.19 |
| 287 | 49 | MA1478.1 | DMRTA2 | MA1478.1 | AATTGHTACAWT | DMRTA2 | 3.08e-15 | 3.69e-18 | 7910 | 63.8 | 12100 | 59 | 1.08 |
| 288 | 14 | MA0774.1 | MEIS2 | MA0774.1 | TTGACAGS | MEIS2 | 3.27e-15 | 3.91e-18 | 5610 | 45.2 | 8280 | 40.4 | 1.12 |
| 289 | 53 | MA0069.1 | PAX6 | MA0069.1 | TTCACGCWTSANTK | PAX6 | 3.45e-15 | 4.11e-18 | 8740 | 70.5 | 13500 | 65.9 | 1.07 |
| 290 | 54 | MA1125.1 | ZNF384 | MA1125.1 | DNWMAAAAAAAA | ZNF384 | 5e-15 | 5.93e-18 | 4810 | 38.8 | 7000 | 34.1 | 1.14 |
| 291 | 30 | MA0631.1 | Six3 | MA0631.1 | DATRGGGTATCAYNWNT | Six3 | 5.46e-15 | 6.46e-18 | 4910 | 39.6 | 7160 | 34.9 | 1.13 |
| 292 | 10 | MA0124.2 | Nkx3-1 | MA0124.2 | RCCACTTAA | Nkx3-1 | 6.37e-15 | 7.51e-18 | 5790 | 46.7 | 8600 | 41.9 | 1.12 |
| 293 | 43 | MA1618.1 | Ptf1a | MA1618.1 | NNACAGATGTTNN | Ptf1a | 1.81e-13 | 2.12e-16 | 11100 | 89.8 | 17800 | 86.8 | 1.03 |
| 294 | 38 | MA1623.1 | Stat2 | MA1623.1 | RGAAACAGAAASH | Stat2 | 1.95e-13 | 2.29e-16 | 9950 | 80.2 | 15700 | 76.4 | 1.05 |
| 295 | 12 | MA0849.1 | FOXO6 | MA0849.1 | GTAAACA | FOXO6 | 2.16e-13 | 2.51e-16 | 6260 | 50.5 | 9410 | 45.8 | 1.1 |
| 296 | 55 | MA1989.1 | Bcl11B | MA1989.1 | NNAAACCACAARNN | Bcl11B | 2.43e-13 | 2.83e-16 | 9070 | 73.1 | 14200 | 68.9 | 1.06 |
| 297 | 56 | MA0800.1 | EOMES | MA0800.1 | RAGGTGTGAAAWN | EOMES | 2.96e-13 | 3.43e-16 | 7020 | 56.6 | 10700 | 52 | 1.09 |
| 298 | 23 | MA0906.1 | HOXC12 | MA0906.1 | RGTCGTAAAAH | HOXC12 | 4.23e-13 | 4.88e-16 | 3950 | 31.9 | 5690 | 27.7 | 1.15 |
| 299 | 2 | MA0621.1 | mix-a | MA0621.1 | NDTTAATTAGN | mix-a | 5.25e-13 | 6.04e-16 | 1790 | 14.4 | 2340 | 11.4 | 1.27 |
| 300 | 57 | MA0626.1 | Npas2 | MA0626.1 | NSCACGTGTN | Npas2 | 5.98e-13 | 6.85e-16 | 4740 | 38.2 | 6950 | 33.9 | 1.13 |
| 301 | 58 | MA0494.1 | Nr1h3::Rxra | MA0494.1 | TGACCTNNAGTRACCYYDN | Nr1h3::Rxra | 6.64e-13 | 7.59e-16 | 9230 | 74.4 | 14400 | 70.3 | 1.06 |
| 302 | 50 | MA0624.2 | Nfatc1 | MA0624.2 | NVATGGAAAAWN | Nfatc1 | 9.62e-13 | 1.1e-15 | 7050 | 56.8 | 10700 | 52.4 | 1.09 |
| 303 | 13 | MA0843.1 | TEF | MA0843.1 | BRTTACRTAAYM | TEF | 1.03e-12 | 1.17e-15 | 2620 | 21.1 | 3600 | 17.6 | 1.2 |
| 304 | 31 | MA0114.4 | HNF4A | MA0114.4 | DNCAAAGTCCANN | HNF4A | 1.08e-12 | 1.22e-15 | 8310 | 67 | 12900 | 62.7 | 1.07 |
| 305 | 2 | MA0135.1 | Lhx3 | MA0135.1 | WAATTAATTAWYY | Lhx3 | 1.09e-12 | 1.23e-15 | 2040 | 16.4 | 2720 | 13.2 | 1.24 |
| 306 | 43 | MA0678.1 | OLIG2 | MA0678.1 | AMCATATGKT | OLIG2 | 1.7e-12 | 1.92e-15 | 6030 | 48.6 | 9060 | 44.2 | 1.1 |
| 307 | 59 | MA0095.3 | Yy1 | MA0095.3 | NNCAAAATGGCN | Yy1 | 3.09e-12 | 3.46e-15 | 9650 | 77.8 | 15200 | 74 | 1.05 |
| 308 | 21 | MA0884.2 | DUXA | MA0884.2 | YTRATTWAATYAR | DUXA | 3.1e-12 | 3.47e-15 | 1730 | 13.9 | 2260 | 11 | 1.26 |
| 309 | 60 | MA0508.3 | PRDM1 | MA0508.3 | YNCTTTCTCTH | PRDM1 | 3.6e-12 | 4.01e-15 | 10200 | 82.4 | 16200 | 79 | 1.04 |
| 310 | 30 | MA1139.1 | FOSL2::JUNB | MA1139.1 | DATGACGTCATH | FOSL2::JUNB | 5.53e-12 | 6.14e-15 | 5700 | 46 | 8540 | 41.6 | 1.1 |
| 311 | 78 | MA0502.2 | NFYB | MA0502.2 | CYCATTGGCCVV | NFYB | 9.66e-12 | 1.07e-14 | 869 | 7.01 | 1020 | 4.96 | 1.41 |
| 312 | 61 | MA1729.1 | ZNF680 | MA1729.1 | GNCCAAGAAGAATDA | ZNF680 | 1.64e-11 | 1.81e-14 | 5370 | 43.3 | 8010 | 39 | 1.11 |
| 313 | 62 | MA1647.2 | Prdm4 | MA1647.2 | RDNCCTTGAAACYGYYN | Prdm4 | 1.86e-11 | 2.04e-14 | 8330 | 67.2 | 12900 | 63.1 | 1.07 |
| 314 | 43 | MA0827.1 | OLIG3 | MA0827.1 | AMCATATGBY | OLIG3 | 2.02e-11 | 2.21e-14 | 6530 | 52.7 | 9930 | 48.4 | 1.09 |
| 315 | 2 | MA1975.1 | ZNF214 | MA1975.1 | VWTCATCAABGTCCT | ZNF214 | 3.16e-11 | 3.45e-14 | 5340 | 43.1 | 7980 | 38.9 | 1.11 |
| 316 | 63 | MA1977.1 | ZNF324 | MA1977.1 | NNNWRGMAGHYAAGGATGGTT | ZNF324 | 3.79e-11 | 4.13e-14 | 6400 | 51.6 | 9710 | 47.3 | 1.09 |
| 317 | 29 | MA0757.1 | ONECUT3 | MA0757.1 | AAAAAATCRATAAH | ONECUT3 | 3.96e-11 | 4.3e-14 | 3250 | 26.2 | 4630 | 22.5 | 1.16 |
| 318 | 30 | MA1127.1 | FOSB::JUN | MA1127.1 | GATGACGTCAT | FOSB::JUN | 4.2e-11 | 4.54e-14 | 5000 | 40.4 | 7440 | 36.2 | 1.11 |
| 319 | 18 | MA1561.1 | SOX12 | MA1561.1 | ACCGAACAATV | SOX12 | 5.19e-11 | 5.6e-14 | 8240 | 66.4 | 12800 | 62.4 | 1.06 |
| 320 | 20 | MA0442.2 | SOX10 | MA0442.2 | NDAACAAAGVN | SOX10 | 5.22e-11 | 5.61e-14 | 4070 | 32.8 | 5930 | 28.9 | 1.13 |
| 321 | 42 | MA2001.1 | Six4 | MA2001.1 | GDAACCTGANH | Six4 | 8.13e-11 | 8.71e-14 | 6380 | 51.4 | 9700 | 47.2 | 1.09 |
| 322 | 30 | MA1129.1 | FOSL1::JUN | MA1129.1 | ATGACGTCAT | FOSL1::JUN | 8.47e-11 | 9.05e-14 | 5970 | 48.1 | 9020 | 43.9 | 1.09 |
| 323 | 56 | MA0802.1 | TBR1 | MA0802.1 | AGGTGTGAAA | TBR1 | 8.62e-11 | 9.18e-14 | 9600 | 77.4 | 15100 | 73.8 | 1.05 |
| 324 | 43 | MA1123.2 | TWIST1 | MA1123.2 | NNDCCAGATGTBN | TWIST1 | 1.17e-10 | 1.24e-13 | 7080 | 57.1 | 10900 | 52.9 | 1.08 |
| 325 | 14 | MA0775.1 | MEIS3 | MA0775.1 | DTGACAGS | MEIS3 | 1.17e-10 | 1.24e-13 | 4060 | 32.8 | 5940 | 28.9 | 1.13 |
| 326 | 64 | MA1111.1 | NR2F2 | MA1111.1 | NAAAGGTCANR | NR2F2 | 1.48e-10 | 1.56e-13 | 7840 | 63.2 | 12100 | 59.1 | 1.07 |
| 327 | 56 | MA0689.1 | TBX20 | MA0689.1 | WAGGTGTGAAR | TBX20 | 1.84e-10 | 1.94e-13 | 5770 | 46.5 | 8710 | 42.4 | 1.1 |
| 328 | 31 | MA0484.2 | HNF4G | MA0484.2 | DNCAAAGTCCANN | HNF4G | 2.17e-10 | 2.27e-13 | 8550 | 68.9 | 13300 | 65 | 1.06 |
| 329 | 10 | MA0913.2 | HOXD9 | MA0913.2 | GYMATAAAAH | HOXD9 | 2.17e-10 | 2.27e-13 | 2610 | 21 | 3650 | 17.8 | 1.18 |
| 330 | 45 | MA1950.1 | FLI1::FOXI1 | MA1950.1 | RTAAACAGGAARYN | FLI1::FOXI1 | 2.18e-10 | 2.27e-13 | 8130 | 65.5 | 12600 | 61.6 | 1.06 |
| 331 | 30 | MA0488.1 | JUN | MA0488.1 | DDRATGATGTCAT | JUN | 2.26e-10 | 2.35e-13 | 2010 | 16.2 | 2720 | 13.3 | 1.22 |
| 332 | 48 | MA0743.2 | SCRT1 | MA0743.2 | NDWKCAACAGGTGKNN | SCRT1 | 3.07e-10 | 3.18e-13 | 5380 | 43.4 | 8080 | 39.4 | 1.1 |
| 333 | 22 | MA1547.2 | PITX2 | MA1547.2 | YTAATCCY | PITX2 | 3.69e-10 | 3.81e-13 | 3100 | 25 | 4430 | 21.6 | 1.16 |
| 334 | 46 | MA1148.1 | PPARA::RXRA | MA1148.1 | AWNTRGGTYAAAGGTCAN | PPARA::RXRA | 3.79e-10 | 3.9e-13 | 6940 | 56 | 10700 | 51.9 | 1.08 |
| 335 | 65 | MA0620.3 | MITF | MA0620.3 | NNNDRTCACGTGAYHNNN | MITF | 6.07e-10 | 6.23e-13 | 2070 | 16.7 | 2830 | 13.8 | 1.21 |
| 336 | 66 | MA0673.1 | NKX2-8 | MA0673.1 | VCACTTSAV | NKX2-8 | 6.17e-10 | 6.32e-13 | 9770 | 78.8 | 15500 | 75.4 | 1.05 |
| 337 | 30 | MA1131.1 | FOSL2::JUN | MA1131.1 | GRTGACGTMAT | FOSL2::JUN | 7.3e-10 | 7.45e-13 | 4700 | 37.9 | 6990 | 34 | 1.11 |
| 338 | 2 | MA0702.2 | LMX1A | MA0702.2 | TTAATTAM | LMX1A | 2.95e-09 | 3e-12 | 1890 | 15.2 | 2580 | 12.5 | 1.21 |
| 339 | 67 | MA1571.1 | TGIF2LX | MA1571.1 | TGACAGCTGTCA | TGIF2LX | 3.37e-09 | 3.42e-12 | 4750 | 38.3 | 7100 | 34.6 | 1.11 |
| 340 | 2 | MA0683.1 | POU4F2 | MA0683.1 | RTGCATAATTAATGAG | POU4F2 | 4.91e-09 | 4.97e-12 | 1460 | 11.8 | 1930 | 9.4 | 1.25 |
| 341 | 1 | MA0591.1 | Bach1::Mafk | MA0591.1 | RSSATGACTCAGCAB | Bach1::Mafk | 5.75e-09 | 5.8e-12 | 2560 | 20.6 | 3610 | 17.6 | 1.17 |
| 342 | 56 | MA0804.1 | TBX19 | MA0804.1 | DTTMRCACMTAGGTGTGAAW | TBX19 | 6.38e-09 | 6.42e-12 | 2910 | 23.5 | 4160 | 20.3 | 1.16 |
| 343 | 18 | MA0869.2 | Sox11 | MA0869.2 | NRGAACAAAGVV | Sox11 | 7.45e-09 | 7.47e-12 | 10500 | 84.4 | 16700 | 81.6 | 1.04 |
| 344 | 30 | MA0693.3 | Vdr | MA0693.3 | NNGAGTTCANN | Vdr | 9.42e-09 | 9.42e-12 | 10700 | 86.4 | 17200 | 83.6 | 1.03 |
| 345 | 34 | MA0519.1 | Stat5a::Stat5b | MA0519.1 | DTTTCCMAGAA | Stat5a::Stat5b | 9.9e-09 | 9.87e-12 | 5840 | 47.1 | 8890 | 43.3 | 1.09 |
| 346 | 7 | MA0029.1 | Mecom | MA0029.1 | AAGAYAAGATAANA | Mecom | 1.08e-08 | 1.08e-11 | 1030 | 8.28 | 1300 | 6.31 | 1.31 |
| 347 | 45 | MA1579.1 | ZBTB26 | MA1579.1 | NNCTCCAGAAANNNN | ZBTB26 | 1.71e-08 | 1.69e-11 | 9810 | 79.1 | 15600 | 75.9 | 1.04 |
| 348 | 30 | MA0684.2 | RUNX3 | MA0684.2 | NHAACCTCAANN | RUNX3 | 2.31e-08 | 2.29e-11 | 10100 | 81.1 | 16000 | 78 | 1.04 |
| 349 | 21 | MA0468.1 | DUX4 | MA0468.1 | TAAYYYAATCA | DUX4 | 2.56e-08 | 2.52e-11 | 1280 | 10.3 | 1670 | 8.14 | 1.26 |
| 350 | 68 | MA0092.1 | Hand1::Tcf3 | MA0092.1 | BRTCTGGMWT | Hand1::Tcf3 | 3.24e-08 | 3.19e-11 | 5170 | 41.7 | 7820 | 38.1 | 1.1 |
| 351 | 42 | MA1118.1 | SIX1 | MA1118.1 | GWAACCTGAKM | SIX1 | 3.37e-08 | 3.3e-11 | 3190 | 25.7 | 4630 | 22.6 | 1.14 |
| 352 | 10 | MA0063.2 | NKX2-5 | MA0063.2 | NNCACTCAANN | NKX2-5 | 4.86e-08 | 4.75e-11 | 2680 | 21.6 | 3830 | 18.7 | 1.16 |
| 353 | 64 | MA1541.1 | NR6A1 | MA1541.1 | NYCAAGKTCAAGKTCAN | NR6A1 | 5.92e-08 | 5.77e-11 | 7580 | 61.1 | 11800 | 57.5 | 1.06 |
| 354 | 64 | MA0160.2 | NR4A2 | MA0160.2 | BWAAAGGTCA | NR4A2 | 6.52e-08 | 6.34e-11 | 8930 | 72 | 14100 | 68.6 | 1.05 |
| 355 | 43 | MA1642.1 | NEUROG2 | MA1642.1 | NNVACAGATGGNN | NEUROG2 | 6.92e-08 | 6.7e-11 | 9640 | 77.7 | 15300 | 74.6 | 1.04 |
| 356 | 69 | MA0080.6 | Spi1 | MA0080.6 | RRAAAGAGGAAGTGGDD | Spi1 | 8.77e-08 | 8.48e-11 | 7450 | 60 | 11600 | 56.5 | 1.06 |
| 357 | 30 | MA1136.1 | FOSB::JUNB | MA1136.1 | RTGACGTCAT | FOSB::JUNB | 9.74e-08 | 9.39e-11 | 4810 | 38.8 | 7240 | 35.3 | 1.1 |
| 358 | 84 | MA1714.1 | ZNF675 | MA1714.1 | RGGVHYMGGAGGMCAAAATGD | ZNF675 | 1.22e-07 | 1.17e-10 | 6520 | 52.5 | 10000 | 48.9 | 1.07 |
| 359 | 30 | MA1133.1 | JUN::JUNB | MA1133.1 | KRTGACGTCATN | JUN::JUNB | 1.22e-07 | 1.17e-10 | 9660 | 77.9 | 15400 | 74.8 | 1.04 |
| 360 | 10 | MA1994.1 | Nkx2-1 | MA1994.1 | NVCACTTGARHNN | Nkx2-1 | 1.43e-07 | 1.36e-10 | 2230 | 18 | 3140 | 15.3 | 1.17 |
| 361 | 70 | MA0596.1 | SREBF2 | MA0596.1 | RTGGGGTGAY | SREBF2 | 1.44e-07 | 1.37e-10 | 1120 | 9 | 1450 | 7.07 | 1.27 |
| 362 | 71 | MA0691.1 | TFAP4 | MA0691.1 | AWCAGCTGWT | TFAP4 | 2.23e-07 | 2.12e-10 | 1120 | 9.02 | 1460 | 7.1 | 1.27 |
| 363 | 23 | MA0908.1 | HOXD11 | MA0908.1 | RTCGTAAAAH | HOXD11 | 2.36e-07 | 2.23e-10 | 2380 | 19.2 | 3380 | 16.5 | 1.16 |
| 364 | 72 | MA0671.1 | NFIX | MA0671.1 | NNTGCCAAN | NFIX | 2.53e-07 | 2.39e-10 | 7880 | 63.5 | 12300 | 60.1 | 1.06 |
| 365 | 73 | MA1984.1 | ZNF667 | MA1984.1 | TTAAGAGCTCAV | ZNF667 | 2.61e-07 | 2.46e-10 | 8030 | 64.8 | 12600 | 61.4 | 1.06 |
| 366 | 57 | MA0672.1 | NKX2-3 | MA0672.1 | VCCACTTRAV | NKX2-3 | 2.96e-07 | 2.79e-10 | 10500 | 84.3 | 16800 | 81.6 | 1.03 |
| 367 | 67 | MA0667.1 | MYF6 | MA0667.1 | AACARCTGTT | MYF6 | 3.13e-07 | 2.94e-10 | 3760 | 30.3 | 5570 | 27.1 | 1.12 |
| 368 | 43 | MA1467.2 | Atoh1 | MA1467.2 | RVCAGATGGYN | Atoh1 | 4.1e-07 | 3.83e-10 | 9450 | 76.2 | 15000 | 73.1 | 1.04 |
| 369 | 74 | TGTGTGKGTRTGTGT | MEME-4 | TGTGTGKGTRTGTGT | TGTGTGKGTRTGTGT | TGTGTGKGTRTGTGT | 4.91e-07 | 4.58e-10 | 2770 | 22.4 | 4010 | 19.5 | 1.14 |
| 370 | 43 | MA1108.2 | MXI1 | MA1108.2 | NNCACATGBN | MXI1 | 7.01e-07 | 6.52e-10 | 6460 | 52 | 9980 | 48.6 | 1.07 |
| 371 | 33 | MA0038.2 | GFI1 | MA0038.2 | BMAATCACDGCA | GFI1 | 7.24e-07 | 6.71e-10 | 2030 | 16.4 | 2860 | 13.9 | 1.18 |
| 372 | 24 | MA0040.1 | Foxq1 | MA0040.1 | HATTGTTTATW | Foxq1 | 7.25e-07 | 6.71e-10 | 1610 | 13 | 2200 | 10.7 | 1.21 |
| 373 | 15 | MA0773.1 | MEF2D | MA0773.1 | DCTAWAAATAGM | MEF2D | 8.75e-07 | 8.07e-10 | 856 | 6.9 | 1080 | 5.28 | 1.31 |
| 374 | 30 | MA0492.1 | JUND | MA0492.1 | DDDRATGABGTCATN | JUND | 9.61e-07 | 8.84e-10 | 1630 | 13.2 | 2250 | 10.9 | 1.2 |
| 375 | 13 | MA0838.1 | CEBPG | MA0838.1 | ATTRCGCAAY | CEBPG | 1e-06 | 9.18e-10 | 3970 | 32 | 5920 | 28.9 | 1.11 |
| 376 | 34 | MA0463.2 | BCL6 | MA0463.2 | NYGCTTTCKAGGAAYN | BCL6 | 1.08e-06 | 9.87e-10 | 6540 | 52.7 | 10100 | 49.3 | 1.07 |
| 377 | 75 | MA0479.1 | FOXH1 | MA0479.1 | BNSAATCCACA | FOXH1 | 1.45e-06 | 1.32e-09 | 1650 | 13.3 | 2280 | 11.1 | 1.2 |
| 378 | 66 | MA2003.1 | NKX2-4 | MA2003.1 | CCACTTSAVV | NKX2-4 | 1.72e-06 | 1.57e-09 | 11000 | 88.4 | 17700 | 86.2 | 1.03 |
| 379 | 1 | MA1520.1 | MAF | MA1520.1 | NTGCTGASTCAGCAH | MAF | 1.83e-06 | 1.66e-09 | 4810 | 38.8 | 7300 | 35.6 | 1.09 |
| 380 | 11 | MA0842.2 | NRL | MA0842.2 | AAWWNTGCTGACG | NRL | 2.71e-06 | 2.45e-09 | 1000 | 8.06 | 1300 | 6.36 | 1.27 |
| 381 | 34 | MA1116.1 | RBPJ | MA1116.1 | BVTGGGAANN | RBPJ | 2.82e-06 | 2.55e-09 | 10800 | 87.4 | 17500 | 85.1 | 1.03 |
| 382 | 49 | MA1707.1 | DMRTA1 | MA1707.1 | AWTTGWTACAWT | DMRTA1 | 3.59e-06 | 3.24e-09 | 2570 | 20.7 | 3730 | 18.2 | 1.14 |
| 383 | 45 | MA1508.1 | IKZF1 | MA1508.1 | VVAACAGGAARN | IKZF1 | 4.56e-06 | 4.1e-09 | 7660 | 61.7 | 12000 | 58.6 | 1.05 |
| 384 | 69 | MA1952.1 | FOXJ2::ELF1 | MA1952.1 | RTAAACMGGAAGTR | FOXJ2::ELF1 | 5.23e-06 | 4.68e-09 | 6830 | 55 | 10600 | 51.8 | 1.06 |
| 385 | 62 | MA1724.1 | Rfx6 | MA1724.1 | HNCCTAGCAACHV | Rfx6 | 6.19e-06 | 5.53e-09 | 8440 | 68 | 13300 | 65 | 1.05 |
| 386 | 76 | MA1592.1 | ZNF274 | MA1592.1 | NRTRTGAGTTCTCGYN | ZNF274 | 6.45e-06 | 5.74e-09 | 2790 | 22.5 | 4070 | 19.8 | 1.13 |
| 387 | 77 | MA1937.1 | ERF::HOXB13 | MA1937.1 | VSCGGAARYNRTWAAN | ERF::HOXB13 | 6.46e-06 | 5.74e-09 | 10300 | 83.1 | 16600 | 80.6 | 1.03 |
| 388 | 67 | MA0797.1 | TGIF2 | MA0797.1 | TGACAGSTGTCA | TGIF2 | 6.93e-06 | 6.15e-09 | 873 | 7.04 | 1130 | 5.48 | 1.28 |
| 389 | 78 | MA0502.2 (NFYB) | STREME-8 | 8-CSATTGGCCS | CSATTGGCCS | NFYB | 7.64e-06 | 6.76e-09 | 325 | 2.62 | 348 | 1.7 | 1.55 |
| 390 | 56 | MA0688.1 | TBX2 | MA0688.1 | DAGGTGTGAAA | TBX2 | 7.76e-06 | 6.85e-09 | 5190 | 41.8 | 7940 | 38.7 | 1.08 |
| 391 | 30 | MA0592.3 | ESRRA | MA0592.3 | NBTCAAGGTCAHN | ESRRA | 8.7e-06 | 7.65e-09 | 4250 | 34.3 | 6420 | 31.3 | 1.1 |
| 392 | 34 | MA0518.1 | Stat4 | MA0518.1 | YTTCYRGGAARNVR | Stat4 | 1.26e-05 | 1.11e-08 | 2870 | 23.2 | 4220 | 20.6 | 1.13 |
| 393 | 64 | MA0258.2 | ESR2 | MA0258.2 | AGGTCASVNTGMCCY | ESR2 | 1.63e-05 | 1.43e-08 | 8310 | 67 | 13100 | 64 | 1.05 |
| 394 | 77 | MA1949.1 | FLI1::DRGX | MA1949.1 | ACCGGAAGTAATTAT | FLI1::DRGX | 1.84e-05 | 1.61e-08 | 7220 | 58.2 | 11300 | 55.1 | 1.06 |
| 395 | 30 | MA0609.2 | CREM | MA0609.2 | NNDGTGACGTCACHNN | CREM | 2.02e-05 | 1.76e-08 | 10300 | 83 | 16500 | 80.6 | 1.03 |
| 396 | 43 | MA1570.1 | TFAP4 | MA1570.1 | AHCATRTGDT | TFAP4 | 2.2e-05 | 1.91e-08 | 2630 | 21.2 | 3840 | 18.7 | 1.13 |
| 397 | 64 | MA0141.3 | ESRRB | MA0141.3 | TCAAGGTCAWW | ESRRB | 2.3e-05 | 2e-08 | 3240 | 26.1 | 4810 | 23.4 | 1.11 |
| 398 | 48 | MA0647.1 | GRHL1 | MA0647.1 | AAAACCGGTTTD | GRHL1 | 2.68e-05 | 2.31e-08 | 3910 | 31.5 | 5890 | 28.7 | 1.1 |
| 399 | 69 | MA0136.3 | Elf5 | MA0136.3 | RVAAGGAAGTNN | Elf5 | 2.71e-05 | 2.34e-08 | 8840 | 71.2 | 14000 | 68.4 | 1.04 |
| 400 | 40 | MA0515.1 | Sox6 | MA0515.1 | CCWTTGTYYY | Sox6 | 2.79e-05 | 2.4e-08 | 2370 | 19.1 | 3430 | 16.7 | 1.14 |
| 401 | 15 | MA0660.1 | MEF2B | MA0660.1 | RCTAWAAATAGC | MEF2B | 3.18e-05 | 2.73e-08 | 685 | 5.52 | 864 | 4.21 | 1.31 |
| 402 | 28 | MA2002.1 | Zfp335 | MA2002.1 | VNTCAGGCANH | Zfp335 | 3.52e-05 | 3.01e-08 | 3540 | 28.6 | 5310 | 25.8 | 1.11 |
| 403 | 71 | MA0698.1 | ZBTB18 | MA0698.1 | NATCCAGATGTKB | ZBTB18 | 4.11e-05 | 3.51e-08 | 5580 | 45 | 8620 | 42 | 1.07 |
| 404 | 70 | MA0595.1 | SREBF1 | MA0595.1 | VTCACCCCAY | SREBF1 | 5.3e-05 | 4.51e-08 | 1480 | 12 | 2070 | 10.1 | 1.19 |
| 405 | 48 | MA1968.1 | TFCP2 | MA1968.1 | AAACCGGTTY | TFCP2 | 5.73e-05 | 4.87e-08 | 8950 | 72.2 | 14300 | 69.4 | 1.04 |
| 406 | 69 | MA0687.1 | SPIC | MA0687.1 | HAAAAGVGGAAGTA | SPIC | 6.82e-05 | 5.78e-08 | 8700 | 70.2 | 13800 | 67.4 | 1.04 |
| 407 | 69 | MA0761.2 | ETV1 | MA0761.2 | NNACAGGAAGTGNN | ETV1 | 7.67e-05 | 6.49e-08 | 7590 | 61.2 | 12000 | 58.3 | 1.05 |
| 408 | 13 | MA0639.1 | DBP | MA0639.1 | NRTTACGTAAYV | DBP | 8.35e-05 | 7.04e-08 | 2720 | 21.9 | 4000 | 19.5 | 1.12 |
| 409 | 69 | MA0598.3 | EHF | MA0598.3 | NNCACTTCCTGTTNN | EHF | 9.72e-05 | 8.18e-08 | 7770 | 62.6 | 12300 | 59.7 | 1.05 |
| 410 | 69 | MA0081.2 | SPIB | MA0081.2 | TYTCACTTCCTCTTTY | SPIB | 0.000107 | 8.99e-08 | 3590 | 29 | 5400 | 26.3 | 1.1 |
| 411 | 56 | MA1565.1 | TBX18 | MA1565.1 | DRAGGTGTGAAR | TBX18 | 0.000159 | 1.33e-07 | 5560 | 44.8 | 8620 | 42 | 1.07 |
| 412 | 11 | MA0117.2 | Mafb | MA0117.2 | AWWNTGCTGACD | Mafb | 0.000228 | 1.91e-07 | 480 | 3.87 | 584 | 2.84 | 1.36 |
| 413 | 65 | MA1995.1 | Npas4 | MA1995.1 | NNTCGTGACHN | Npas4 | 0.000278 | 2.32e-07 | 10100 | 81.1 | 16200 | 78.8 | 1.03 |
| 414 | 56 | MA1567.2 | Tbx6 | MA1567.2 | NRGGTGTGAANN | Tbx6 | 0.000393 | 3.27e-07 | 4690 | 37.8 | 7210 | 35.1 | 1.08 |
| 415 | 56 | MA0690.2 | TBX21 | MA0690.2 | TTTCACACCTT | TBX21 | 0.000396 | 3.28e-07 | 6270 | 50.6 | 9810 | 47.8 | 1.06 |
| 416 | 43 | MA1109.1 | NEUROD1 | MA1109.1 | NRACAGATGGYNN | NEUROD1 | 0.000462 | 3.82e-07 | 10700 | 86.2 | 17300 | 84.3 | 1.02 |
| 417 | 2 | MA1546.1 | PAX3 | MA1546.1 | YSGTCACGSYWATTAN | PAX3 | 0.000576 | 4.75e-07 | 5410 | 43.6 | 8400 | 40.9 | 1.07 |
| 418 | 69 | MA0640.2 | ELF3 | MA0640.2 | NNCCACTTCCTGNT | ELF3 | 0.000834 | 6.86e-07 | 9390 | 75.7 | 15000 | 73.3 | 1.03 |
| 419 | 55 | MA0002.2 | Runx1 | MA0002.2 | BBYTGTGGTTT | Runx1 | 0.000889 | 7.3e-07 | 8220 | 66.3 | 13100 | 63.7 | 1.04 |
| 420 | 43 | MA0669.1 | NEUROG2 | MA0669.1 | RACATATGTC | NEUROG2 | 0.00114 | 9.37e-07 | 1550 | 12.5 | 2210 | 10.8 | 1.16 |
| 421 | 48 | MA0744.2 | SCRT2 | MA0744.2 | NNWGCAACAGGTGDNN | SCRT2 | 0.00144 | 1.17e-06 | 2160 | 17.4 | 3180 | 15.5 | 1.13 |
| 422 | 64 | MA1540.2 | NR5A1 | MA1540.2 | NNRAGTTCAAGGTCANN | NR5A1 | 0.00157 | 1.28e-06 | 2140 | 17.3 | 3150 | 15.3 | 1.13 |
| 423 | 43 | MA0668.2 | Neurod2 | MA0668.2 | NNGRACAGATGGYNN | Neurod2 | 0.00189 | 1.53e-06 | 9120 | 73.5 | 14600 | 71.2 | 1.03 |
| 424 | 79 | MA0101.1 | REL | MA0101.1 | BGGRNWTTCC | REL | 0.00233 | 1.89e-06 | 10900 | 87.8 | 17700 | 86 | 1.02 |
| 425 | 43 | MA1638.1 | HAND2 | MA1638.1 | NVCAGATGNN | HAND2 | 0.00249 | 2.02e-06 | 7020 | 56.6 | 11100 | 54 | 1.05 |
| 426 | 62 | MA1649.1 | ZBTB12 | MA1649.1 | NYCTRGAACHN | ZBTB12 | 0.00272 | 2.2e-06 | 5990 | 48.3 | 9390 | 45.7 | 1.06 |
| 427 | 45 | MA1954.1 | FOXO1::ELK1 | MA1954.1 | RWMAACAGGAAGTD | FOXO1::ELK1 | 0.00312 | 2.51e-06 | 3540 | 28.5 | 5390 | 26.2 | 1.09 |
| 428 | 7 | MA0140.2 | GATA1::TAL1 | MA0140.2 | MTTATCWSNNNNNHVCAG | GATA1::TAL1 | 0.00357 | 2.87e-06 | 578 | 4.66 | 749 | 3.65 | 1.28 |
| 429 | 30 | MA1996.1 | Nr1H2 | MA1996.1 | NNRAGGTCANN | Nr1H2 | 0.00373 | 2.99e-06 | 9290 | 74.9 | 14900 | 72.7 | 1.03 |
| 430 | 56 | MA0803.1 | TBX15 | MA0803.1 | AGGTGTGA | TBX15 | 0.00386 | 3.09e-06 | 4670 | 37.6 | 7230 | 35.2 | 1.07 |
| 431 | 29 | MA0780.1 | PAX3 | MA0780.1 | TAATYRATTA | PAX3 | 0.00388 | 3.1e-06 | 893 | 7.2 | 1220 | 5.95 | 1.21 |
| 432 | 1 | MA1521.1 | MAFA | MA1521.1 | NTGCTGASTCAGCAN | MAFA | 0.0042 | 3.34e-06 | 5010 | 40.4 | 7780 | 37.9 | 1.06 |
| 433 | 2 | MA0703.2 | LMX1B | MA0703.2 | AWTTTAATTAA | LMX1B | 0.00598 | 4.75e-06 | 595 | 4.8 | 779 | 3.79 | 1.26 |
| 434 | 43 | MA0607.2 | BHLHA15 | MA0607.2 | ACCATATGGT | BHLHA15 | 0.00614 | 4.87e-06 | 1500 | 12.1 | 2170 | 10.6 | 1.15 |
| 435 | 69 | MA0062.3 | GABPA | MA0062.3 | NNCACTTCCTGTNN | GABPA | 0.00732 | 5.79e-06 | 6680 | 53.8 | 10500 | 51.4 | 1.05 |
| 436 | 77 | MA1940.1 | ETV2::DRGX | MA1940.1 | RVCGGAAGYAATTA | ETV2::DRGX | 0.00896 | 7.07e-06 | 6360 | 51.3 | 10000 | 48.8 | 1.05 |
| 437 | 80 | MA0106.3 | TP53 | MA0106.3 | RACATGYCCGGRCATGTY | TP53 | 0.0098 | 7.72e-06 | 967 | 7.8 | 1340 | 6.55 | 1.19 |
| 438 | 77 | MA1944.1 | ETV5::DRGX | MA1944.1 | RSMGGAAGYAATTA | ETV5::DRGX | 0.012 | 9.44e-06 | 6880 | 55.5 | 10900 | 53.1 | 1.04 |
| 439 | 64 | MA1534.1 | NR1I3 | MA1534.1 | RTGAACTTT | NR1I3 | 0.0135 | 1.06e-05 | 3140 | 25.3 | 4780 | 23.3 | 1.09 |
| 440 | 72 | MA0161.2 | NFIC | MA0161.2 | NNCTTGGCANN | NFIC | 0.0141 | 1.1e-05 | 1150 | 9.27 | 1630 | 7.94 | 1.17 |
| 441 | 81 | MA1717.1 | ZNF784 | MA1717.1 | GTACYTACCGH | ZNF784 | 0.0182 | 1.42e-05 | 9160 | 73.9 | 14700 | 71.8 | 1.03 |
| 442 | 67 | MA1572.1 | TGIF2LY | MA1572.1 | TGACAGCTGTCA | TGIF2LY | 0.0199 | 1.55e-05 | 1030 | 8.3 | 1450 | 7.07 | 1.18 |
| 443 | 82 | MA1117.1 | RELB | MA1117.1 | RDATTCCCCNN | RELB | 0.0208 | 1.62e-05 | 7970 | 64.2 | 12700 | 62 | 1.04 |
| 444 | 69 | MA0474.3 | Erg | MA0474.3 | NNACAGGAAGTGVN | Erg | 0.0211 | 1.64e-05 | 7600 | 61.2 | 12100 | 59 | 1.04 |
| 445 | 44 | MA1473.1 | CDX4 | MA1473.1 | GGYMATAAAAC | CDX4 | 0.0214 | 1.66e-05 | 908 | 7.32 | 1260 | 6.16 | 1.19 |
| 446 | 23 | MA0651.2 | HOXC11 | MA0651.2 | AGGTCGTAAAAH | HOXC11 | 0.0219 | 1.69e-05 | 1760 | 14.2 | 2600 | 12.6 | 1.12 |
| 447 | 80 | MA0861.1 | TP73 | MA0861.1 | DRCATGTCNNRACAYGYM | TP73 | 0.0221 | 1.7e-05 | 1440 | 11.6 | 2090 | 10.2 | 1.14 |
| 448 | 64 | MA0643.1 | Esrrg | MA0643.1 | TCAAGGTCAH | Esrrg | 0.0304 | 2.33e-05 | 7760 | 62.6 | 12400 | 60.4 | 1.04 |
| 449 | 64 | MA0505.2 | Nr5A2 | MA0505.2 | NTGACCTTGAMCT | Nr5A2 | 0.0307 | 2.35e-05 | 2080 | 16.8 | 3100 | 15.1 | 1.11 |
| 14 | 83 | MA1728.1 (ZNF549) | STREME-12 | 12-AGGCAGGAT | 12-AGGCAGGAT | ZNF549 | 0.0478 | 0.00342 | 1570 | 12.6 | 2380 | 11.6 | 1.09 |
| 15 | 84 | MA1596.1 (ZNF460) | MEME-3 | GGGAGGMKGRGGMGG | GGGAGGMKGRGGMGG | ZNF460 | 0.0913 | 0.00609 | 110 | 0.89 | 130 | 0.63 | 1.4 |
| 16 | 85 | 11-CAAGCCCATGGA | STREME-11 | 11-CAAGCCCATGGA | 11-CAAGCCCATGGA | CAAGCCCATGGA | 0.144 | 0.00898 | 1510 | 12.2 | 2320 | 11.3 | 1.08 |
ER_rat <- 1.25
DOX_242kb_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste("DOX_242kb_combo response peaks(200bp)Enrichment ratio:",ER_rat))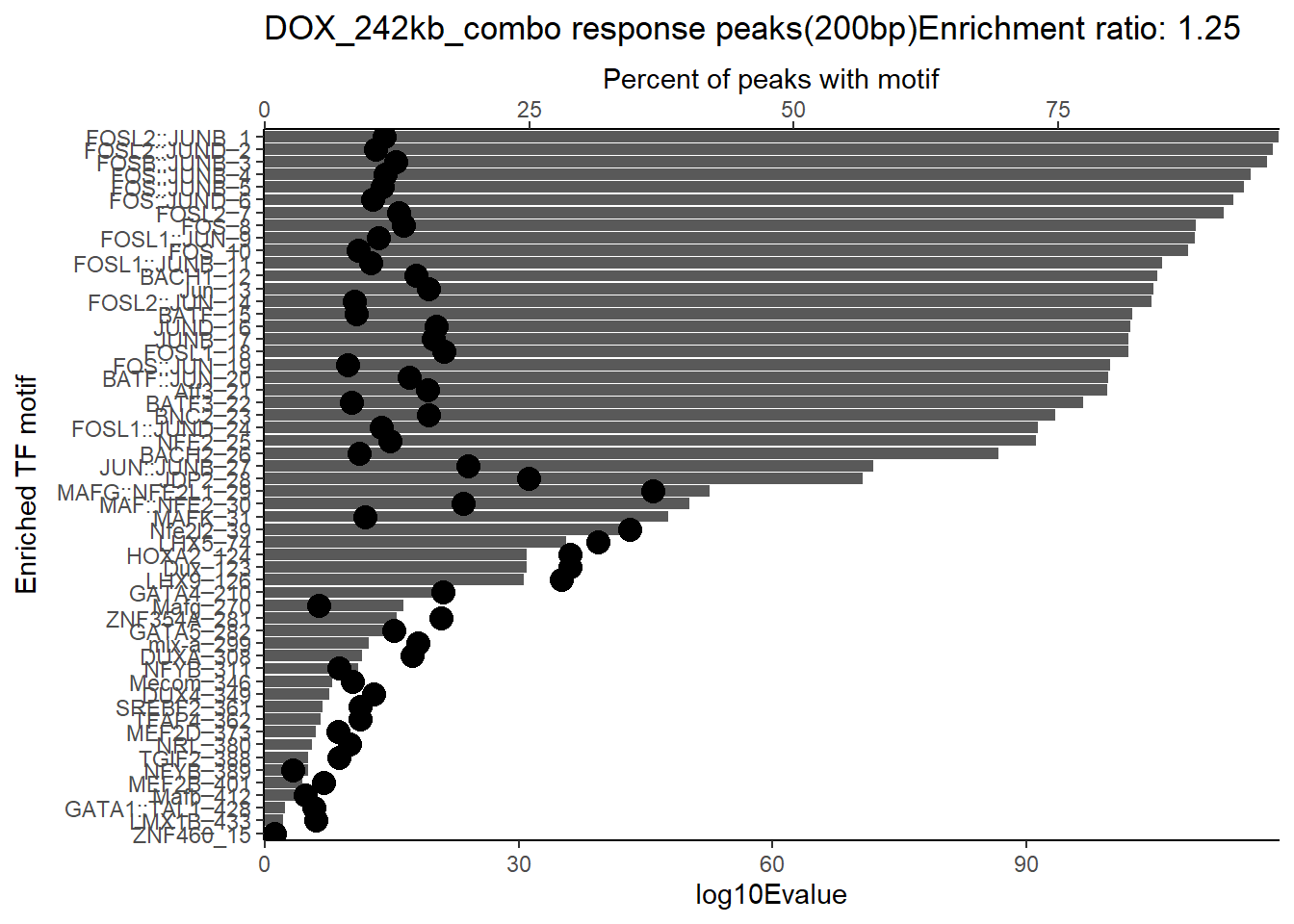
DOX_242kb_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y, TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
slice_head(n=5) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*4), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./4,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( "DOX_24 2kb response peaks(200bp)Enrichment ratio:",ER_rat))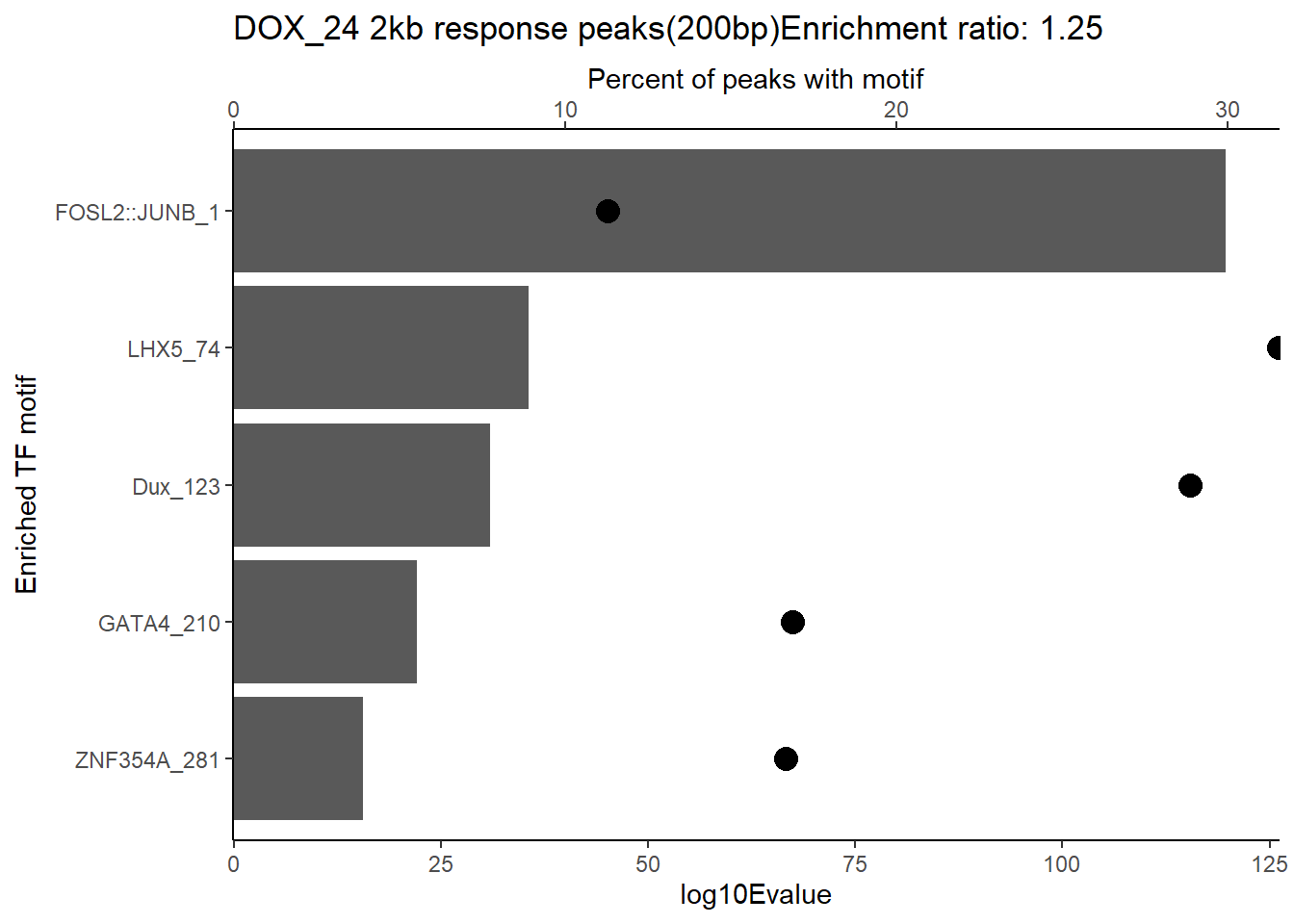
DOX_DAR 24 20kb data
DOX_24_20kb_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DOX_24_20kb_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
DOX_24_20kb_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DOX_24_20kb_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
DOX_24_20kb_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DOX_24_20kb_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
DOX_24_20kb_sea_all <- rbind(DOX_24_20kb_sea_disc, DOX_24_20kb_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)DOX_24_20kb_combo <- DOX_24_20kb_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., DOX_24_20kb_sea_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ SIM_MOTIF
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y)
DOX_24_20kb_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs 200 bp in DOX24 20kb ") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | MA1134.1 (FOS::JUNB) | STREME-1 | 1-RATGASTCATY | RATGASTCATY | FOS::JUNB | 0 | 0 | 6210 | 17.4 | 4170 | 8.15 | 2.14 |
| 2 | 1 | MA1144.1 | FOSL2::JUND | MA1144.1 | KATGACTCAT | FOSL2::JUND | 0 | 0 | 5840 | 16.4 | 3860 | 7.55 | 2.17 |
| 3 | 1 | MA0099.3 | FOS::JUN | MA0099.3 | ATGAGTCAYM | FOS::JUN | 0 | 0 | 6400 | 18 | 4450 | 8.7 | 2.07 |
| 4 | 1 | MA1138.1 | FOSL2::JUNB | MA1138.1 | KRTGASTCAT | FOSL2::JUNB | 0 | 0 | 5770 | 16.2 | 3800 | 7.44 | 2.18 |
| 5 | 1 | MA1134.1 | FOS::JUNB | MA1134.1 | KATGASTCATHN | FOS::JUNB | 0 | 0 | 5270 | 14.8 | 3300 | 6.45 | 2.29 |
| 6 | 1 | MA1130.1 | FOSL2::JUN | MA1130.1 | NNRTGAGTCAYN | FOSL2::JUN | 0 | 0 | 4970 | 14 | 3040 | 5.95 | 2.35 |
| 7 | 1 | MA0477.2 | FOSL1 | MA0477.2 | NNATGACTCATNN | FOSL1 | 0 | 0 | 4470 | 12.6 | 2560 | 5 | 2.51 |
| 8 | 1 | MA1633.2 | BACH1 | MA1633.2 | ATGACTCAT | BACH1 | 0 | 0 | 5540 | 15.6 | 3620 | 7.08 | 2.2 |
| 9 | 1 | MA0491.2 | JUND | MA0491.2 | NNATGACTCATNN | JUND | 0 | 0 | 5050 | 14.2 | 3140 | 6.14 | 2.31 |
| 10 | 1 | MA1141.1 | FOS::JUND | MA1141.1 | NKATGAGTCATNN | FOS::JUND | 0 | 0 | 6780 | 19.1 | 4960 | 9.68 | 1.97 |
| 11 | 1 | MA1128.1 | FOSL1::JUN | MA1128.1 | NKATGACTCATNN | FOSL1::JUN | 0 | 0 | 5990 | 16.8 | 4140 | 8.1 | 2.08 |
| 12 | 1 | MA0476.1 (FOS) | MEME-1 | ATGASTCA | ATGASTCA | FOS | 0 | 0 | 6870 | 19.3 | 5100 | 9.97 | 1.94 |
| 13 | 1 | MA0476.1 | FOS | MA0476.1 | DVTGASTCATB | FOS | 0 | 0 | 6610 | 18.6 | 4820 | 9.42 | 1.97 |
| 14 | 1 | MA1135.1 | FOSB::JUNB | MA1135.1 | KRTGASTCAT | FOSB::JUNB | 5.48906932529625e-321 | 5.48906932529625e-321 | 6930 | 19.5 | 5210 | 10.2 | 1.91 |
| 15 | 1 | MA0490.2 | JUNB | MA0490.2 | NNATGACTCATNN | JUNB | 2.22999987709968e-318 | 6.65012359302318e-320 | 5500 | 15.5 | 3710 | 7.24 | 2.14 |
| 16 | 1 | MA0489.2 | Jun | MA0489.2 | NVTGACTCATNN | Jun | 5.96999999997705e-313 | 1.680000000216e-314 | 6560 | 18.4 | 4860 | 9.5 | 1.94 |
| 17 | 1 | MA1988.1 | Atf3 | MA1988.1 | NRTGACTCABN | Atf3 | 2.35e-309 | 6.19000000000023e-311 | 5230 | 14.7 | 3480 | 6.8 | 2.16 |
| 18 | 1 | MA0655.1 | JDP2 | MA0655.1 | ATGACTCAT | JDP2 | 2.78e-308 | 6.92e-310 | 6060 | 17 | 4350 | 8.49 | 2 |
| 19 | 1 | MA1137.1 | FOSL1::JUNB | MA1137.1 | NDRTGACTCATNN | FOSL1::JUNB | 3.45e-304 | 8.14999999999999e-306 | 5700 | 16 | 3990 | 7.79 | 2.05 |
| 20 | 1 | MA0478.1 | FOSL2 | MA0478.1 | KRRTGASTCAB | FOSL2 | 3.72e-298 | 8.34e-300 | 7320 | 20.6 | 5800 | 11.3 | 1.82 |
| 21 | 1 | MA1142.1 | FOSL1::JUND | MA1142.1 | NRTGACTCAT | FOSL1::JUND | 1.49e-293 | 3.19e-295 | 5790 | 16.3 | 4150 | 8.1 | 2.01 |
| 22 | 1 | MA0835.2 | BATF3 | MA0835.2 | DATGACTCATH | BATF3 | 1.8e-292 | 3.67e-294 | 5750 | 16.2 | 4110 | 8.03 | 2.01 |
| 23 | 1 | MA1634.1 | BATF | MA1634.1 | DATGACTCATH | BATF | 1.2e-289 | 2.35e-291 | 6360 | 17.9 | 4790 | 9.36 | 1.91 |
| 24 | 1 | MA0841.1 | NFE2 | MA0841.1 | VATGACTCATS | NFE2 | 3.97e-286 | 7.43e-288 | 4590 | 12.9 | 2950 | 5.77 | 2.24 |
| 25 | 1 | MA1132.1 | JUN::JUNB | MA1132.1 | KATGACKCAT | JUN::JUNB | 5.85e-285 | 1.05e-286 | 4920 | 13.8 | 3290 | 6.43 | 2.15 |
| 26 | 1 | MA0462.2 | BATF::JUN | MA0462.2 | DATGACTCATH | BATF::JUN | 1.5e-283 | 2.59e-285 | 6730 | 18.9 | 5240 | 10.2 | 1.85 |
| 27 | 1 | MA1101.2 | BACH2 | MA1101.2 | DWANCATGASTCATSNTWH | BACH2 | 1.12e-281 | 1.86e-283 | 5990 | 16.8 | 4440 | 8.68 | 1.94 |
| 28 | 1 | MA1928.1 | BNC2 | MA1928.1 | NDRTGAGTCABN | BNC2 | 9.1e-230 | 1.46e-231 | 4010 | 11.3 | 2670 | 5.21 | 2.16 |
| 29 | 1 | MA0496.3 | MAFK | MA0496.3 | NGMTGACTCAGCMNH | MAFK | 5.7e-154 | 8.81e-156 | 8820 | 24.8 | 8870 | 17.3 | 1.43 |
| 30 | 1 | MA0089.2 | MAFG::NFE2L1 | MA0089.2 | NVNATGACTCAGCADW | MAFG::NFE2L1 | 4.46e-151 | 6.67e-153 | 4670 | 13.1 | 3910 | 7.64 | 1.72 |
| 31 | 1 | MA0501.1 | MAF::NFE2 | MA0501.1 | ATGACTCAGCANWTY | MAF::NFE2 | 1.59e-112 | 2.3e-114 | 13700 | 38.5 | 15900 | 31 | 1.24 |
| 32 | 2 | MA0018.4 (CREB1) | STREME-2 | 2-ATGACW | ATGACW | CREB1 | 3.05e-102 | 4.27e-104 | 26900 | 75.7 | 35300 | 69 | 1.1 |
| 33 | 1 | MA0659.3 | Mafg | MA0659.3 | NWGMTGACTCAGCAN | Mafg | 5.9e-78 | 8.02e-80 | 13800 | 38.8 | 16600 | 32.5 | 1.19 |
| 34 | 3 | MA0718.1 | RAX | MA0718.1 | RYYAATTARH | RAX | 5.22e-73 | 6.89e-75 | 29400 | 82.7 | 39700 | 77.6 | 1.07 |
| 35 | 4 | 3-RATAAG | STREME-3 | 3-RATAAG | RATAAG | RATAAG | 1.84e-72 | 2.36e-74 | 26800 | 75.4 | 35700 | 69.7 | 1.08 |
| 36 | 5 | MA0808.1 (TEAD3) | STREME-4 | 4-ATWCMA | ATWCMA | TEAD3 | 4.13e-71 | 5.15e-73 | 29600 | 83.3 | 40100 | 78.4 | 1.06 |
| 37 | 3 | MA0720.1 | Shox2 | MA0720.1 | YYAATTAR | Shox2 | 3.06e-70 | 3.72e-72 | 29100 | 81.7 | 39200 | 76.6 | 1.07 |
| 38 | 1 | MA0150.2 | Nfe2l2 | MA0150.2 | CASNATGACTCAGCA | Nfe2l2 | 6.91e-70 | 8.16e-72 | 20700 | 58.1 | 26600 | 52 | 1.12 |
| 39 | 3 | MA0725.1 | VSX1 | MA0725.1 | YTAATTAN | VSX1 | 4.64e-69 | 5.33e-71 | 31100 | 87.4 | 42500 | 83.1 | 1.05 |
| 40 | 3 | MA0027.2 | EN1 | MA0027.2 | SYAATTAV | EN1 | 9.31e-69 | 1.04e-70 | 29800 | 83.8 | 40400 | 79 | 1.06 |
| 41 | 1 | MA1639.1 | MEIS1 | MA1639.1 | NHCATMAATCAHN | MEIS1 | 1.61e-66 | 1.76e-68 | 20100 | 56.5 | 25800 | 50.4 | 1.12 |
| 42 | 3 | MA0674.1 | NKX6-1 | MA0674.1 | NTMATTAA | NKX6-1 | 5.16e-66 | 5.52e-68 | 31500 | 88.5 | 43200 | 84.3 | 1.05 |
| 43 | 3 | MA0710.1 | NOTO | MA0710.1 | NYTAATTAVN | NOTO | 3.38e-65 | 3.53e-67 | 27900 | 78.5 | 37500 | 73.4 | 1.07 |
| 44 | 3 | MA0726.1 | VSX2 | MA0726.1 | CTAATTAN | VSX2 | 1.79e-64 | 1.82e-66 | 29500 | 82.9 | 40000 | 78.1 | 1.06 |
| 45 | 1 | MA1113.2 | PBX2 | MA1113.2 | NHCATAAATCAHN | PBX2 | 2.17e-64 | 2.16e-66 | 27800 | 78.1 | 37300 | 72.9 | 1.07 |
| 46 | 3 | MA0704.1 | Lhx4 | MA0704.1 | YTAATTAR | Lhx4 | 3.74e-64 | 3.65e-66 | 26500 | 74.6 | 35400 | 69.2 | 1.08 |
| 47 | 6 | MA0035.4 | GATA1 | MA0035.4 | NHCTAATCTDH | GATA1 | 4.93e-64 | 4.71e-66 | 30000 | 84.3 | 40800 | 79.7 | 1.06 |
| 48 | 3 | MA0716.1 | PRRX1 | MA0716.1 | YYAATTAR | PRRX1 | 5.33e-63 | 4.98e-65 | 26500 | 74.5 | 35400 | 69.2 | 1.08 |
| 49 | 3 | MA0882.1 | DLX6 | MA0882.1 | VYAATTAN | DLX6 | 1.89e-62 | 1.71e-64 | 28000 | 78.8 | 37700 | 73.7 | 1.07 |
| 50 | 7 | MA0676.1 | Nr2e1 | MA0676.1 | AAAAGTCAA | Nr2e1 | 1.91e-62 | 1.71e-64 | 23700 | 66.6 | 31200 | 60.9 | 1.09 |
| 51 | 3 | MA0902.2 | HOXB2 | MA0902.2 | SYAATTAS | HOXB2 | 2.9e-62 | 2.55e-64 | 21800 | 61.2 | 28300 | 55.4 | 1.1 |
| 52 | 3 | MA0717.1 | RAX2 | MA0717.1 | YYAATTAV | RAX2 | 3.04e-61 | 2.63e-63 | 29100 | 81.9 | 39500 | 77.2 | 1.06 |
| 53 | 3 | MA1476.2 | Dlx5 | MA1476.2 | NDGCAATTAGNN | Dlx5 | 5.82e-61 | 4.93e-63 | 30400 | 85.5 | 41500 | 81.1 | 1.05 |
| 54 | 1 | MA1640.1 | MEIS2 | MA1640.1 | NNATGATTTATGDNH | MEIS2 | 7.84e-61 | 6.51e-63 | 26800 | 75.4 | 36000 | 70.3 | 1.07 |
| 55 | 3 | MA0630.1 | SHOX | MA0630.1 | HTAATTRR | SHOX | 9.06e-61 | 7.39e-63 | 27200 | 76.6 | 36600 | 71.5 | 1.07 |
| 56 | 3 | MA1963.1 | SATB1 | MA1963.1 | WWDCTAATAAHWW | SATB1 | 1.79e-60 | 1.43e-62 | 24300 | 68.2 | 32100 | 62.7 | 1.09 |
| 57 | 3 | MA0675.1 | NKX6-2 | MA0675.1 | NYMATTAA | NKX6-2 | 6.14e-60 | 4.84e-62 | 29000 | 81.4 | 39200 | 76.7 | 1.06 |
| 58 | 3 | MA0885.2 | Dlx2 | MA0885.2 | NDGCAATTAGNN | Dlx2 | 5.07e-58 | 3.92e-60 | 29800 | 83.9 | 40700 | 79.5 | 1.06 |
| 59 | 3 | MA0721.1 | UNCX | MA0721.1 | YYAATTAV | UNCX | 2.47e-57 | 1.88e-59 | 27100 | 76 | 36400 | 71.1 | 1.07 |
| 60 | 3 | MA0723.2 | VAX2 | MA0723.2 | BYAATTAV | VAX2 | 2.52e-57 | 1.89e-59 | 26600 | 74.9 | 35700 | 69.8 | 1.07 |
| 61 | 3 | MA0125.1 | Nobox | MA0125.1 | TAATTRSY | Nobox | 2.77e-57 | 2.03e-59 | 29900 | 84.1 | 40800 | 79.7 | 1.05 |
| 62 | 3 | MA0680.2 | Pax7 | MA0680.2 | DWTAATCAATTAWNH | Pax7 | 4.43e-57 | 3.21e-59 | 23600 | 66.3 | 31200 | 60.9 | 1.09 |
| 63 | 8 | MA0845.1 | FOXB1 | MA0845.1 | WATGTAAATAT | FOXB1 | 6.42e-56 | 4.57e-58 | 28200 | 79.4 | 38200 | 74.7 | 1.06 |
| 64 | 3 | MA0876.1 | BSX | MA0876.1 | SYVATTAV | BSX | 2.83e-55 | 1.98e-57 | 29400 | 82.6 | 40000 | 78.2 | 1.06 |
| 65 | 9 | MA1480.1 | DPRX | MA1480.1 | AGATAATCCN | DPRX | 3.06e-55 | 2.11e-57 | 29200 | 82.1 | 39800 | 77.7 | 1.06 |
| 66 | 3 | MA0628.1 | POU6F1 | MA0628.1 | AYTAATTART | POU6F1 | 7.53e-55 | 5.12e-57 | 27700 | 77.9 | 37400 | 73.2 | 1.06 |
| 67 | 10 | MA0602.1 | Arid5a | MA0602.1 | SYAATATTGVDANH | Arid5a | 8.17e-55 | 5.47e-57 | 27900 | 78.5 | 37700 | 73.7 | 1.06 |
| 68 | 3 | MA0886.1 | EMX2 | MA0886.1 | VBTAATTARB | EMX2 | 8.59e-55 | 5.67e-57 | 19900 | 55.9 | 25800 | 50.4 | 1.11 |
| 69 | 3 | MA0648.1 | GSC | MA0648.1 | VYTAATCCSY | GSC | 1.08e-54 | 7.04e-57 | 29700 | 83.4 | 40500 | 79.1 | 1.05 |
| 70 | 3 | MA0713.1 | PHOX2A | MA0713.1 | TAATYYAATTA | PHOX2A | 2.23e-54 | 1.43e-56 | 23900 | 67.1 | 31600 | 61.8 | 1.09 |
| 71 | 11 | MA0786.1 | POU3F1 | MA0786.1 | WTATGCWAATKW | POU3F1 | 3.33e-54 | 2.11e-56 | 27300 | 76.7 | 36800 | 71.9 | 1.07 |
| 72 | 3 | MA1495.1 | HOXA1 | MA1495.1 | STAATTAS | HOXA1 | 4.87e-54 | 3.03e-56 | 16900 | 47.4 | 21500 | 41.9 | 1.13 |
| 73 | 11 | MA0142.1 | Pou5f1::Sox2 | MA0142.1 | CWTTGTYATGCAAAT | Pou5f1::Sox2 | 1.31e-53 | 8.04e-56 | 27300 | 76.7 | 36800 | 71.9 | 1.07 |
| 74 | 3 | MA0880.1 | Dlx3 | MA0880.1 | NYAATTAN | Dlx3 | 1.44e-53 | 8.72e-56 | 23500 | 66 | 31100 | 60.8 | 1.09 |
| 75 | 3 | MA0890.1 | GBX2 | MA0890.1 | RSYAATTARN | GBX2 | 2.17e-53 | 1.3e-55 | 25300 | 71 | 33800 | 66 | 1.08 |
| 76 | 12 | MA0790.1 | POU4F1 | MA0790.1 | ATGMATAATTAATG | POU4F1 | 6.01e-53 | 3.55e-55 | 24600 | 69.1 | 32700 | 64 | 1.08 |
| 77 | 12 | MA0791.1 | POU4F3 | MA0791.1 | RTGMATWATTAATGAR | POU4F3 | 6.95e-53 | 4.05e-55 | 21600 | 60.7 | 28300 | 55.3 | 1.1 |
| 78 | 3 | MA0635.1 | BARHL2 | MA0635.1 | VHTAAAYGRT | BARHL2 | 1.46e-52 | 8.38e-55 | 30000 | 84.2 | 41000 | 80.1 | 1.05 |
| 79 | 6 | MA1104.2 | GATA6 | MA1104.2 | HWWTCTTATCTNH | GATA6 | 2.78e-52 | 1.58e-54 | 21000 | 59 | 27500 | 53.7 | 1.1 |
| 80 | 3 | MA0700.2 | LHX2 | MA0700.2 | DDCCAATTADN | LHX2 | 1.26e-51 | 7.08e-54 | 26200 | 73.8 | 35300 | 68.9 | 1.07 |
| 81 | 8 | MA0041.2 | FOXD3 | MA0041.2 | WAWGHAAATAAACARY | FOXD3 | 1.6e-51 | 8.85e-54 | 27400 | 77 | 37000 | 72.3 | 1.06 |
| 82 | 3 | MA0708.2 | MSX2 | MA0708.2 | SCAATTAV | MSX2 | 3.42e-51 | 1.87e-53 | 28200 | 79.3 | 38300 | 74.8 | 1.06 |
| 83 | 3 | MA0881.1 | Dlx4 | MA0881.1 | VYAATTAN | Dlx4 | 4.9e-51 | 2.65e-53 | 22600 | 63.6 | 29900 | 58.4 | 1.09 |
| 84 | 3 | MA0658.1 | LHX6 | MA0658.1 | RCTAATTARY | LHX6 | 7.43e-51 | 3.97e-53 | 24400 | 68.6 | 32500 | 63.5 | 1.08 |
| 85 | 3 | MA0899.1 | HOXA10 | MA0899.1 | DGYMATAAAAH | HOXA10 | 1.76e-50 | 9.27e-53 | 29500 | 82.8 | 40200 | 78.6 | 1.05 |
| 86 | 3 | MA0709.1 | Msx3 | MA0709.1 | SCAATTAN | Msx3 | 1.99e-50 | 1.04e-52 | 21900 | 61.7 | 28900 | 56.5 | 1.09 |
| 87 | 11 | MA0792.1 | POU5F1B | MA0792.1 | TATGCWAAT | POU5F1B | 1.09e-49 | 5.6e-52 | 28100 | 78.8 | 38100 | 74.4 | 1.06 |
| 88 | 13 | MA0901.2 | HOXB13 | MA0901.2 | WDCCAATAAAAWWW | HOXB13 | 1.28e-49 | 6.55e-52 | 25100 | 70.4 | 33500 | 65.5 | 1.07 |
| 89 | 3 | MA0650.3 | Hoxa13 | MA0650.3 | NNCCAATAAAAN | Hoxa13 | 2.4e-49 | 1.21e-51 | 29000 | 81.4 | 39500 | 77.2 | 1.06 |
| 90 | 8 | MA0032.2 | FOXC1 | MA0032.2 | WAWGTAAAYAW | FOXC1 | 3.74e-49 | 1.87e-51 | 28400 | 80 | 38700 | 75.6 | 1.06 |
| 91 | 3 | MA1502.1 | HOXB8 | MA1502.1 | GYMATTAM | HOXB8 | 4.72e-49 | 2.33e-51 | 19300 | 54.3 | 25100 | 49.1 | 1.11 |
| 92 | 3 | MA0068.2 | PAX4 | MA0068.2 | CTAATTAG | PAX4 | 6.52e-49 | 3.18e-51 | 30800 | 86.5 | 42300 | 82.7 | 1.05 |
| 93 | 3 | MA0891.1 | GSC2 | MA0891.1 | NYTAATCCBH | GSC2 | 9.44e-49 | 4.56e-51 | 25500 | 71.8 | 34300 | 67 | 1.07 |
| 94 | 3 | MA0122.3 | Nkx3-2 | MA0122.3 | WWAAMCACTTAAN | Nkx3-2 | 1.69e-48 | 8.04e-51 | 28700 | 80.7 | 39100 | 76.4 | 1.06 |
| 95 | 3 | MA0910.2 | HOXD8 | MA0910.2 | GYMATTAM | HOXD8 | 2.07e-48 | 9.78e-51 | 19400 | 54.4 | 25200 | 49.2 | 1.11 |
| 96 | 3 | MA0707.2 | MNX1 | MA0707.2 | VYRATTAK | MNX1 | 4.94e-48 | 2.31e-50 | 18900 | 53.2 | 24600 | 48 | 1.11 |
| 97 | 3 | MA1549.1 | POU6F1 | MA1549.1 | RTAATGAGVN | POU6F1 | 1.22e-47 | 5.63e-50 | 19100 | 53.6 | 24800 | 48.4 | 1.11 |
| 98 | 11 | MA0785.1 | POU2F1 | MA0785.1 | AWTATGCWAATK | POU2F1 | 1.39e-47 | 6.36e-50 | 28100 | 79.1 | 38200 | 74.7 | 1.06 |
| 99 | 3 | MA0661.1 | MEOX1 | MA0661.1 | VSTAATTAHC | MEOX1 | 3.19e-47 | 1.45e-49 | 27300 | 76.8 | 37000 | 72.4 | 1.06 |
| 100 | 11 | MA0788.1 | POU3F3 | MA0788.1 | WWTATGCWAATTW | POU3F3 | 4.65e-47 | 2.09e-49 | 25800 | 72.5 | 34700 | 67.8 | 1.07 |
| 101 | 3 | MA0903.1 | HOXB3 | MA0903.1 | NNYMATTARN | HOXB3 | 6.64e-47 | 2.95e-49 | 27500 | 77.3 | 37300 | 72.9 | 1.06 |
| 102 | 3 | MA1498.2 | HOXA7 | MA1498.2 | SYMATTAV | HOXA7 | 1.39e-46 | 6.13e-49 | 26500 | 74.6 | 35800 | 70 | 1.07 |
| 103 | 6 | MA0036.3 | GATA2 | MA0036.3 | NBCTTATCTNH | GATA2 | 1.58e-46 | 6.89e-49 | 16600 | 46.8 | 21400 | 41.7 | 1.12 |
| 104 | 11 | MA0627.2 | POU2F3 | MA0627.2 | NWTATGCAAATNH | POU2F3 | 2.44e-46 | 1.05e-48 | 26200 | 73.6 | 35300 | 69 | 1.07 |
| 105 | 14 | 7-ABTGAG | STREME-7 | 7-ABTGAG | ABTGAG | ABTGAG | 5.21e-46 | 2.23e-48 | 33600 | 94.5 | 47000 | 91.9 | 1.03 |
| 106 | 15 | MA1623.1 (Stat2) | STREME-6 | 6-AACAGA | AACAGA | Stat2 | 5.66e-46 | 2.39e-48 | 32500 | 91.2 | 45100 | 88.1 | 1.04 |
| 107 | 6 | MA0712.2 | OTX2 | MA0712.2 | NDRGGATTARNN | OTX2 | 6.1e-46 | 2.56e-48 | 30800 | 86.6 | 42500 | 83 | 1.04 |
| 108 | 3 | MA0654.1 | ISX | MA0654.1 | YYAATTAR | ISX | 1.78e-45 | 7.38e-48 | 19700 | 55.3 | 25700 | 50.3 | 1.1 |
| 109 | 3 | MA0662.1 | MIXL1 | MA0662.1 | NBYAATTAVN | MIXL1 | 2.1e-45 | 8.65e-48 | 25100 | 70.6 | 33700 | 65.9 | 1.07 |
| 110 | 3 | MA0681.2 | PHOX2B | MA0681.2 | WWTAATCAAATTAWWW | PHOX2B | 3.8e-45 | 1.55e-47 | 20600 | 58 | 27100 | 53 | 1.09 |
| 111 | 3 | MA0887.1 | EVX1 | MA0887.1 | VVTAATTABS | EVX1 | 4.08e-45 | 1.65e-47 | 25900 | 72.7 | 34900 | 68.1 | 1.07 |
| 112 | 16 | MA1978.1 | ZNF354A | MA1978.1 | HWAADWATAAATGRAYWAWTT | ZNF354A | 5.1e-45 | 2.04e-47 | 26500 | 74.6 | 35900 | 70.1 | 1.06 |
| 113 | 17 | MA0143.4 | SOX2 | MA0143.4 | DNACAATGGNN | SOX2 | 6.51e-45 | 2.57e-47 | 30200 | 85 | 41600 | 81.2 | 1.05 |
| 114 | 18 | MA0495.3 | MAFF | MA0495.3 | DWGTCAGCATTTTWHN | MAFF | 6.54e-45 | 2.57e-47 | 29100 | 81.8 | 39800 | 77.7 | 1.05 |
| 115 | 9 | MA0714.1 | PITX3 | MA0714.1 | NYTAATCCC | PITX3 | 1.29e-44 | 5.03e-47 | 33100 | 93 | 46200 | 90.3 | 1.03 |
| 116 | 8 | MA0846.1 | FOXC2 | MA0846.1 | WAWGTAAACAWW | FOXC2 | 2.37e-44 | 9.15e-47 | 29800 | 83.8 | 40900 | 79.9 | 1.05 |
| 117 | 19 | 5-TCACAA | STREME-5 | 5-TCACAA | TCACAA | TCACAA | 2.59e-44 | 9.93e-47 | 31400 | 88.2 | 43400 | 84.8 | 1.04 |
| 118 | 3 | MA0706.1 | MEOX2 | MA0706.1 | DSTAATTAWN | MEOX2 | 5.09e-44 | 1.94e-46 | 19000 | 53.4 | 24800 | 48.5 | 1.1 |
| 119 | 3 | MA0158.2 | HOXA5 | MA0158.2 | GYMATTAS | HOXA5 | 9.12e-44 | 3.44e-46 | 13200 | 37.1 | 16600 | 32.4 | 1.14 |
| 120 | 3 | MA1519.1 | LHX5 | MA1519.1 | STAATTAV | LHX5 | 1.17e-43 | 4.37e-46 | 14400 | 40.5 | 18300 | 35.7 | 1.13 |
| 121 | 20 | MA0084.1 | SRY | MA0084.1 | NWWAACAAT | SRY | 2.08e-43 | 7.7e-46 | 25600 | 71.8 | 34400 | 67.3 | 1.07 |
| 122 | 3 | MA0793.1 | POU6F2 | MA0793.1 | ASCTMATTAW | POU6F2 | 2.48e-43 | 9.1e-46 | 16600 | 46.6 | 21400 | 41.8 | 1.12 |
| 123 | 6 | MA1970.1 | TRPS1 | MA1970.1 | NHTCTTATCTNH | TRPS1 | 3.41e-43 | 1.24e-45 | 18800 | 52.8 | 24500 | 47.9 | 1.1 |
| 124 | 21 | MA0052.4 | MEF2A | MA0052.4 | DDCTAAAAATAGMHH | MEF2A | 5.04e-43 | 1.82e-45 | 22600 | 63.6 | 30100 | 58.8 | 1.08 |
| 125 | 3 | MA1518.2 | Lhx1 | MA1518.2 | NNNTGCTAATTAGCANNN | Lhx1 | 6.24e-43 | 2.24e-45 | 26700 | 75.2 | 36200 | 70.8 | 1.06 |
| 126 | 3 | MA0699.1 | LBX2 | MA0699.1 | NCYAATTARN | LBX2 | 6.84e-43 | 2.43e-45 | 26100 | 73.3 | 35200 | 68.8 | 1.06 |
| 127 | 3 | MA0722.1 | VAX1 | MA0722.1 | YTAATTAH | VAX1 | 2.34e-42 | 8.26e-45 | 19100 | 53.6 | 25000 | 48.8 | 1.1 |
| 128 | 9 | MA0711.1 | OTX1 | MA0711.1 | HTAATCCS | OTX1 | 2.99e-42 | 1.05e-44 | 27700 | 77.9 | 37700 | 73.7 | 1.06 |
| 129 | 11 | MA0789.1 | POU3F4 | MA0789.1 | TATGCWAAT | POU3F4 | 4.92e-42 | 1.71e-44 | 24000 | 67.4 | 32100 | 62.8 | 1.07 |
| 130 | 6 | MA0766.2 | GATA5 | MA0766.2 | SWGATAASRN | GATA5 | 1.25e-41 | 4.32e-44 | 13900 | 39 | 17600 | 34.4 | 1.13 |
| 131 | 3 | MA1481.1 | DRGX | MA1481.1 | YYAATTAN | DRGX | 1.29e-41 | 4.4e-44 | 19600 | 55 | 25700 | 50.2 | 1.1 |
| 132 | 13 | MA0465.2 | CDX2 | MA0465.2 | NDGCAATAAAWN | CDX2 | 1.69e-41 | 5.74e-44 | 25700 | 72.2 | 34700 | 67.8 | 1.07 |
| 133 | 3 | MA0904.2 | HOXB5 | MA0904.2 | STAATTAS | HOXB5 | 1.94e-41 | 6.55e-44 | 12200 | 34.2 | 15200 | 29.7 | 1.15 |
| 134 | 3 | MA0888.1 | EVX2 | MA0888.1 | VVTAATTAVB | EVX2 | 2.18e-41 | 7.28e-44 | 30000 | 84.3 | 41300 | 80.6 | 1.05 |
| 135 | 3 | MA0666.2 | MSX1 | MA0666.2 | SCAATTAR | MSX1 | 4.13e-41 | 1.37e-43 | 23000 | 64.6 | 30700 | 59.9 | 1.08 |
| 136 | 3 | MA1471.1 | BARX2 | MA1471.1 | NWWAAYMATTAN | BARX2 | 4.18e-41 | 1.38e-43 | 26300 | 73.9 | 35600 | 69.6 | 1.06 |
| 137 | 3 | MA0875.1 | BARX1 | MA0875.1 | VCMATTAV | BARX1 | 5.41e-41 | 1.77e-43 | 27300 | 76.8 | 37200 | 72.7 | 1.06 |
| 138 | 3 | MA0634.1 | ALX3 | MA0634.1 | YYYAATTANV | ALX3 | 1.21e-40 | 3.95e-43 | 23200 | 65.3 | 31100 | 60.7 | 1.08 |
| 139 | 3 | MA0892.1 | GSX1 | MA0892.1 | NCYMATTAVN | GSX1 | 1.78e-40 | 5.75e-43 | 27300 | 76.7 | 37100 | 72.5 | 1.06 |
| 140 | 8 | MA0148.4 | FOXA1 | MA0148.4 | WWGTAAACATNN | FOXA1 | 3.71e-40 | 1.19e-42 | 30700 | 86.2 | 42400 | 82.8 | 1.04 |
| 141 | 8 | MA1607.1 | Foxl2 | MA1607.1 | DDWATGTAAACANN | Foxl2 | 4.63e-40 | 1.47e-42 | 30900 | 86.8 | 42700 | 83.4 | 1.04 |
| 142 | 17 | MA1152.1 | SOX15 | MA1152.1 | CYWTTGTTHW | SOX15 | 1.95e-39 | 6.17e-42 | 27300 | 76.8 | 37200 | 72.7 | 1.06 |
| 143 | 6 | MA0037.4 | Gata3 | MA0037.4 | NHTCTTATCTNH | Gata3 | 2.25e-38 | 7.06e-41 | 18700 | 52.5 | 24500 | 47.9 | 1.1 |
| 144 | 3 | MA0879.2 | DLX1 | MA0879.2 | VYAATTAN | DLX1 | 2.71e-38 | 8.45e-41 | 22900 | 64.5 | 30700 | 60 | 1.07 |
| 145 | 3 | MA0705.1 | Lhx8 | MA0705.1 | CTAATTAV | Lhx8 | 3.98e-38 | 1.23e-40 | 17400 | 49 | 22700 | 44.4 | 1.1 |
| 146 | 3 | MA1505.1 | HOXC8 | MA1505.1 | KTAATTAM | HOXC8 | 5.24e-38 | 1.61e-40 | 17600 | 49.5 | 23000 | 44.9 | 1.1 |
| 147 | 3 | MA1530.1 | NKX6-3 | MA1530.1 | VYCATTAWW | NKX6-3 | 9.39e-38 | 2.87e-40 | 21100 | 59.3 | 28000 | 54.7 | 1.08 |
| 148 | 17 | MA0077.1 | SOX9 | MA0077.1 | CYATTGTTY | SOX9 | 2.63e-37 | 7.97e-40 | 32600 | 91.7 | 45600 | 89 | 1.03 |
| 149 | 8 | MA1606.1 | Foxf1 | MA1606.1 | DWRTAAACANN | Foxf1 | 5.79e-37 | 1.74e-39 | 28800 | 80.9 | 39500 | 77.2 | 1.05 |
| 150 | 22 | MA0769.2 | TCF7 | MA0769.2 | NNCTTTGAWVT | TCF7 | 1.35e-36 | 4.04e-39 | 29500 | 83 | 40600 | 79.4 | 1.04 |
| 151 | 3 | MA0701.2 | LHX9 | MA0701.2 | CYAATTAR | LHX9 | 1.52e-36 | 4.53e-39 | 13300 | 37.4 | 16900 | 33.1 | 1.13 |
| 152 | 22 | MA0523.1 | TCF7L2 | MA0523.1 | DRASATCAAAGRVA | TCF7L2 | 6.39e-36 | 1.89e-38 | 20600 | 58 | 27400 | 53.6 | 1.08 |
| 153 | 3 | MA0889.1 | GBX1 | MA0889.1 | VCYAATTASY | GBX1 | 2.34e-35 | 6.85e-38 | 19100 | 53.8 | 25200 | 49.3 | 1.09 |
| 154 | 3 | MA0612.2 | EMX1 | MA0612.2 | BTAATTAS | EMX1 | 2.44e-35 | 7.1e-38 | 14400 | 40.4 | 18400 | 36 | 1.12 |
| 155 | 11 | MA1115.1 | POU5F1 | MA1115.1 | WHATGCAAATN | POU5F1 | 8.59e-35 | 2.47e-37 | 29600 | 83.3 | 40900 | 79.9 | 1.04 |
| 156 | 23 | MA0809.2 | TEAD4 | MA0809.2 | NNACATTCCARN | TEAD4 | 8.59e-35 | 2.47e-37 | 32100 | 90.2 | 44800 | 87.4 | 1.03 |
| 157 | 8 | MA1103.2 | FOXK2 | MA1103.2 | DDGTAAACANN | FOXK2 | 1.08e-34 | 3.08e-37 | 31100 | 87.6 | 43200 | 84.5 | 1.04 |
| 158 | 3 | MA0894.1 | HESX1 | MA0894.1 | GHTAATTRRH | HESX1 | 1.09e-34 | 3.1e-37 | 20000 | 56.2 | 26500 | 51.8 | 1.08 |
| 159 | 8 | MA0848.1 | FOXO4 | MA0848.1 | GTAAACA | FOXO4 | 1.15e-34 | 3.26e-37 | 29100 | 81.9 | 40100 | 78.3 | 1.04 |
| 160 | 24 | MA0108.2 | TBP | MA0108.2 | STATAAAWRSVVVBN | TBP | 1.18e-34 | 3.31e-37 | 28900 | 81.3 | 39800 | 77.7 | 1.05 |
| 161 | 3 | MA0644.2 | ESX1 | MA0644.2 | YYAATTAN | ESX1 | 1.36e-34 | 3.8e-37 | 16100 | 45.3 | 20900 | 40.9 | 1.11 |
| 162 | 3 | MA0075.3 | PRRX2 | MA0075.3 | CTAATTAN | PRRX2 | 1.7e-34 | 4.7e-37 | 14800 | 41.6 | 19100 | 37.3 | 1.12 |
| 163 | 17 | MA0868.2 | SOX8 | MA0868.2 | MGAACAATRG | SOX8 | 2.67e-34 | 7.34e-37 | 30100 | 84.6 | 41600 | 81.3 | 1.04 |
| 164 | 9 | MA0682.2 | PITX1 | MA0682.2 | YTAATCCH | PITX1 | 2.76e-34 | 7.55e-37 | 18900 | 53.1 | 24900 | 48.7 | 1.09 |
| 165 | 17 | MA1120.1 | SOX13 | MA1120.1 | DVACAATGGNN | SOX13 | 3.81e-34 | 1.03e-36 | 29600 | 83.3 | 40900 | 79.9 | 1.04 |
| 166 | 8 | MA0042.2 | FOXI1 | MA0042.2 | GTAAACA | FOXI1 | 3.83e-34 | 1.04e-36 | 26200 | 73.6 | 35600 | 69.6 | 1.06 |
| 167 | 3 | MA0642.2 | EN2 | MA0642.2 | VSYAATTA | EN2 | 5.03e-34 | 1.35e-36 | 25400 | 71.3 | 34400 | 67.3 | 1.06 |
| 168 | 17 | MA1563.2 | SOX18 | MA1563.2 | AACAATDV | SOX18 | 6.49e-34 | 1.73e-36 | 26700 | 75.1 | 36400 | 71.2 | 1.05 |
| 169 | 8 | MA1683.1 | FOXA3 | MA1683.1 | DWGTAAACANN | FOXA3 | 6.74e-34 | 1.79e-36 | 32000 | 89.9 | 44600 | 87.1 | 1.03 |
| 170 | 3 | MA1497.1 | HOXA6 | MA1497.1 | BGYMATTANN | HOXA6 | 1.41e-33 | 3.73e-36 | 27100 | 76.1 | 37000 | 72.3 | 1.05 |
| 171 | 8 | MA0047.3 | FOXA2 | MA0047.3 | RWGTAAACANN | FOXA2 | 3.34e-33 | 8.77e-36 | 28800 | 80.8 | 39600 | 77.3 | 1.05 |
| 172 | 3 | MA0912.2 | HOXD3 | MA0912.2 | CTAATTAC | HOXD3 | 3.92e-33 | 1.02e-35 | 10400 | 29.1 | 12900 | 25.3 | 1.15 |
| 173 | 12 | MA0046.2 | HNF1A | MA0046.2 | DRTTAATNATTAACN | HNF1A | 1.61e-32 | 4.17e-35 | 15600 | 44 | 20300 | 39.7 | 1.11 |
| 174 | 8 | MA0481.3 | FOXP1 | MA0481.3 | NDGTAAACANH | FOXP1 | 1.63e-32 | 4.21e-35 | 28900 | 81.2 | 39800 | 77.7 | 1.04 |
| 175 | 3 | MA0914.1 | ISL2 | MA0914.1 | GCAMTTAR | ISL2 | 2.14e-32 | 5.49e-35 | 29000 | 81.4 | 39900 | 78 | 1.04 |
| 176 | 12 | MA0153.2 | HNF1B | MA0153.2 | GTTAATNATTAAY | HNF1B | 3.14e-32 | 8.01e-35 | 14800 | 41.6 | 19200 | 37.5 | 1.11 |
| 177 | 25 | MA0679.2 | ONECUT1 | MA0679.2 | WAAAAATCAATAAWWN | ONECUT1 | 6.31e-32 | 1.6e-34 | 18800 | 52.8 | 24900 | 48.6 | 1.09 |
| 178 | 8 | MA0033.2 | FOXL1 | MA0033.2 | RTAAACA | FOXL1 | 1.05e-31 | 2.64e-34 | 29900 | 84 | 41300 | 80.8 | 1.04 |
| 179 | 3 | MA0893.2 | GSX2 | MA0893.2 | SYMATTAR | GSX2 | 1.53e-31 | 3.84e-34 | 29200 | 82.1 | 40300 | 78.8 | 1.04 |
| 180 | 3 | MA0900.2 | HOXA2 | MA0900.2 | STAATTAS | HOXA2 | 2.32e-31 | 5.79e-34 | 10600 | 29.9 | 13400 | 26.1 | 1.14 |
| 181 | 3 | MA0896.1 | Hmx1 | MA0896.1 | VSVAGCAATTAAWGVDB | Hmx1 | 3.92e-31 | 9.71e-34 | 18900 | 53.2 | 25100 | 49 | 1.09 |
| 182 | 25 | MA0755.1 | CUX2 | MA0755.1 | TRATCGATAW | CUX2 | 4.15e-31 | 1.02e-33 | 22000 | 62 | 29600 | 57.9 | 1.07 |
| 183 | 1 | MA0591.1 | Bach1::Mafk | MA0591.1 | RSSATGACTCAGCAB | Bach1::Mafk | 6.63e-31 | 1.63e-33 | 1310 | 3.69 | 1170 | 2.29 | 1.61 |
| 184 | 18 | MA0117.2 | Mafb | MA0117.2 | AWWNTGCTGACD | Mafb | 7.55e-31 | 1.84e-33 | 28100 | 79 | 38600 | 75.4 | 1.05 |
| 185 | 3 | MA0124.2 | Nkx3-1 | MA0124.2 | RCCACTTAA | Nkx3-1 | 1.73e-30 | 4.2e-33 | 29800 | 83.8 | 41300 | 80.7 | 1.04 |
| 186 | 26 | MA0018.4 | CREB1 | MA0018.4 | NNRTGATGTCAYN | CREB1 | 1.95e-30 | 4.7e-33 | 26000 | 73.2 | 35500 | 69.4 | 1.05 |
| 187 | 6 | MA0482.2 | GATA4 | MA0482.2 | NNCCTTATCTNH | GATA4 | 2.63e-30 | 6.32e-33 | 13400 | 37.7 | 17300 | 33.8 | 1.12 |
| 188 | 17 | MA0514.2 | Sox3 | MA0514.2 | DNACAATGGNN | Sox3 | 9.88e-30 | 2.36e-32 | 32600 | 91.8 | 45700 | 89.4 | 1.03 |
| 189 | 27 | MA0836.2 | CEBPD | MA0836.2 | NRTTGCACAAYHN | CEBPD | 2.47e-29 | 5.86e-32 | 28000 | 78.7 | 38500 | 75.3 | 1.05 |
| 190 | 28 | MA0905.1 | HOXC10 | MA0905.1 | GTCRTAAAAH | HOXC10 | 4.84e-29 | 1.14e-31 | 22300 | 62.6 | 30000 | 58.6 | 1.07 |
| 191 | 3 | MA0898.1 | Hmx3 | MA0898.1 | ASAAGCAATTAAWRRAT | Hmx3 | 9.72e-29 | 2.28e-31 | 18200 | 51.2 | 24100 | 47.2 | 1.09 |
| 192 | 26 | MA1143.1 | FOSL1::JUND | MA1143.1 | RTGACGTMAY | FOSL1::JUND | 2.67e-28 | 6.23e-31 | 23600 | 66.3 | 32000 | 62.4 | 1.06 |
| 193 | 3 | MA1463.1 | ARGFX | MA1463.1 | DCTAATTARM | ARGFX | 5.75e-28 | 1.34e-30 | 14500 | 40.7 | 18800 | 36.8 | 1.11 |
| 194 | 29 | MA0502.2 (NFYB) | STREME-9 | 9-CCAATG | CCAATG | NFYB | 9.09e-28 | 2.1e-30 | 34300 | 96.3 | 48500 | 94.7 | 1.02 |
| 195 | 3 | MA0895.1 | HMBOX1 | MA0895.1 | MYTAGTTAMS | HMBOX1 | 1.54e-27 | 3.55e-30 | 24700 | 69.5 | 33700 | 65.8 | 1.06 |
| 196 | 30 | MA1489.1 | FOXN3 | MA1489.1 | GTAAACAA | FOXN3 | 1.81e-27 | 4.15e-30 | 24200 | 68 | 32900 | 64.3 | 1.06 |
| 197 | 8 | MA0850.1 | FOXP3 | MA0850.1 | RTAAACA | FOXP3 | 5.82e-27 | 1.32e-29 | 29300 | 82.4 | 40600 | 79.3 | 1.04 |
| 198 | 17 | MA1603.1 | Dmrt1 | MA1603.1 | GHTACAAAGTADC | Dmrt1 | 5.87e-27 | 1.33e-29 | 26000 | 73 | 35500 | 69.4 | 1.05 |
| 199 | 3 | MA1500.1 | HOXB6 | MA1500.1 | BTAATKRC | HOXB6 | 1.16e-26 | 2.62e-29 | 22200 | 62.3 | 30000 | 58.6 | 1.06 |
| 200 | 31 | MA1623.1 | Stat2 | MA1623.1 | RGAAACAGAAASH | Stat2 | 1.94e-26 | 4.36e-29 | 31300 | 87.9 | 43700 | 85.3 | 1.03 |
| 201 | 17 | MA0078.2 | Sox17 | MA0078.2 | NNAGAACAATGGNN | Sox17 | 2.03e-26 | 4.52e-29 | 27300 | 76.7 | 37500 | 73.3 | 1.05 |
| 202 | 32 | MA1657.1 | ZNF652 | MA1657.1 | BAAAGRGTTAAN | ZNF652 | 2.06e-26 | 4.57e-29 | 21500 | 60.5 | 29000 | 56.7 | 1.07 |
| 203 | 22 | MA0768.2 | Lef1 | MA0768.2 | NNBCCTTTGATSTN | Lef1 | 6.1e-26 | 1.35e-28 | 30300 | 85.1 | 42100 | 82.3 | 1.03 |
| 204 | 3 | MA1608.1 | Isl1 | MA1608.1 | NNCCATTAGNN | Isl1 | 6.58e-26 | 1.45e-28 | 22700 | 63.8 | 30700 | 60 | 1.06 |
| 205 | 33 | MA1562.1 | SOX14 | MA1562.1 | CCGAACAATG | SOX14 | 1.6e-25 | 3.51e-28 | 27500 | 77.4 | 37900 | 74.1 | 1.04 |
| 206 | 31 | MA0050.3 | Irf1 | MA0050.3 | ANTGAAACTGAAASH | Irf1 | 2.95e-25 | 6.42e-28 | 30500 | 85.7 | 42400 | 82.9 | 1.03 |
| 207 | 3 | MA0853.1 | Alx4 | MA0853.1 | CGCRYTAATTARYHNNY | Alx4 | 5.29e-25 | 1.15e-27 | 15200 | 42.7 | 19900 | 39 | 1.09 |
| 208 | 8 | MA0852.2 | FOXK1 | MA0852.2 | NRDGTAAACAAGNN | FOXK1 | 1.02e-24 | 2.21e-27 | 32300 | 90.9 | 45300 | 88.6 | 1.03 |
| 209 | 34 | MA0611.2 | Dux | MA0611.2 | NTGATTBAATCAVWNN | Dux | 1.09e-24 | 2.34e-27 | 11300 | 31.8 | 14500 | 28.4 | 1.12 |
| 210 | 3 | MA0467.2 | Crx | MA0467.2 | NDGGATTANN | Crx | 2e-24 | 4.27e-27 | 22200 | 62.3 | 30000 | 58.7 | 1.06 |
| 211 | 35 | MA0774.1 | MEIS2 | MA0774.1 | TTGACAGS | MEIS2 | 2.98e-24 | 6.34e-27 | 33300 | 93.6 | 46900 | 91.6 | 1.02 |
| 212 | 36 | MA1125.1 (ZNF384) | MEME-2 | TTTTTTTTTTTTTTT | TTTTTTTTTTTTTTT | ZNF384 | 3.09e-24 | 6.54e-27 | 22900 | 64.4 | 31100 | 60.8 | 1.06 |
| 213 | 3 | MA0724.1 | VENTX | MA0724.1 | AMCGATTAR | VENTX | 3.46e-24 | 7.29e-27 | 14500 | 40.7 | 19000 | 37.1 | 1.1 |
| 214 | 13 | MA0913.2 | HOXD9 | MA0913.2 | GYMATAAAAH | HOXD9 | 3.91e-24 | 8.2e-27 | 19100 | 53.8 | 25600 | 50.1 | 1.07 |
| 215 | 3 | MA1504.1 | HOXC4 | MA1504.1 | VTMATTAN | HOXC4 | 5.32e-24 | 1.11e-26 | 21600 | 60.6 | 29100 | 57 | 1.06 |
| 216 | 37 | MA0606.2 | Nfat5 | MA0606.2 | NNATGGAAAAWN | Nfat5 | 6.69e-24 | 1.39e-26 | 29600 | 83.2 | 41100 | 80.4 | 1.04 |
| 217 | 3 | MA0897.1 | Hmx2 | MA0897.1 | ASAAGCAATTAAHVVRT | Hmx2 | 7.77e-24 | 1.61e-26 | 19200 | 54.1 | 25800 | 50.4 | 1.07 |
| 218 | 3 | MA1496.1 | HOXA4 | MA1496.1 | RTMATTAV | HOXA4 | 2.98e-23 | 6.13e-26 | 24300 | 68.2 | 33100 | 64.8 | 1.05 |
| 219 | 38 | MA0151.1 | Arid3a | MA0151.1 | ATYAAA | Arid3a | 3.59e-23 | 7.35e-26 | 15600 | 43.7 | 20600 | 40.2 | 1.09 |
| 220 | 9 | MA0883.1 | Dmbx1 | MA0883.1 | WDRWNMGGATTADKNNW | Dmbx1 | 4.12e-23 | 8.4e-26 | 17300 | 48.5 | 23000 | 44.9 | 1.08 |
| 221 | 3 | MA0909.3 | Hoxd13 | MA0909.3 | NNCAATAAAW | Hoxd13 | 4.21e-23 | 8.55e-26 | 17600 | 49.6 | 23500 | 45.9 | 1.08 |
| 222 | 26 | MA1112.2 | NR4A1 | MA1112.2 | NNAAAGGTCANN | NR4A1 | 6.52e-23 | 1.32e-25 | 29100 | 81.7 | 40300 | 78.8 | 1.04 |
| 223 | 28 | MA0911.1 | Hoxa11 | MA0911.1 | RGTCGTAAAANT | Hoxa11 | 9.02e-23 | 1.82e-25 | 24200 | 68.1 | 33100 | 64.7 | 1.05 |
| 224 | 3 | MA1644.1 | NFYC | MA1644.1 | RDCCAATCASN | NFYC | 1.06e-22 | 2.11e-25 | 28700 | 80.7 | 39800 | 77.8 | 1.04 |
| 225 | 39 | MA0877.3 | BARHL1 | MA0877.3 | AMCGTTTA | BARHL1 | 1.27e-22 | 2.54e-25 | 29100 | 81.9 | 40500 | 79.1 | 1.04 |
| 226 | 28 | MA0908.1 | HOXD11 | MA0908.1 | RTCGTAAAAH | HOXD11 | 2.19e-22 | 4.35e-25 | 14100 | 39.8 | 18600 | 36.3 | 1.1 |
| 227 | 3 | MA0132.2 | PDX1 | MA0132.2 | VYAATTAR | PDX1 | 5.78e-22 | 1.14e-24 | 14600 | 41.1 | 19300 | 37.6 | 1.09 |
| 228 | 11 | MA0787.1 | POU3F2 | MA0787.1 | WTATGCWAATKA | POU3F2 | 1.19e-21 | 2.35e-24 | 19000 | 53.3 | 25500 | 49.8 | 1.07 |
| 229 | 23 | MA0808.1 | TEAD3 | MA0808.1 | RCATTCCW | TEAD3 | 1.44e-21 | 2.82e-24 | 13600 | 38.1 | 17800 | 34.8 | 1.1 |
| 230 | 3 | MA1501.1 | HOXB7 | MA1501.1 | GGTAATTAVS | HOXB7 | 2.61e-21 | 5.09e-24 | 18500 | 51.9 | 24800 | 48.4 | 1.07 |
| 231 | 40 | MA0063.2 | NKX2-5 | MA0063.2 | NNCACTCAANN | NKX2-5 | 2.88e-21 | 5.59e-24 | 27300 | 76.6 | 37700 | 73.6 | 1.04 |
| 232 | 26 | MA1636.1 | CEBPG | MA1636.1 | NDATGATGCAATMHH | CEBPG | 2.91e-21 | 5.62e-24 | 17300 | 48.7 | 23100 | 45.2 | 1.08 |
| 233 | 41 | MA0092.1 | Hand1::Tcf3 | MA0092.1 | BRTCTGGMWT | Hand1::Tcf3 | 4.52e-21 | 8.7e-24 | 32600 | 91.7 | 45900 | 89.7 | 1.02 |
| 234 | 3 | MA1499.1 | HOXB4 | MA1499.1 | RTMATTAR | HOXB4 | 8.96e-21 | 1.72e-23 | 13700 | 38.5 | 18000 | 35.2 | 1.09 |
| 235 | 42 | MA1479.1 | DMRTC2 | MA1479.1 | AATTGMTACATT | DMRTC2 | 1.94e-20 | 3.7e-23 | 26800 | 75.3 | 37000 | 72.3 | 1.04 |
| 236 | 8 | MA0157.3 | Foxo3 | MA0157.3 | NDGTAAACARNN | Foxo3 | 3.49e-20 | 6.64e-23 | 33800 | 95 | 47800 | 93.4 | 1.02 |
| 237 | 9 | MA0719.1 | RHOXF1 | MA0719.1 | NTRATCCN | RHOXF1 | 4.59e-20 | 8.7e-23 | 22600 | 63.7 | 30900 | 60.4 | 1.05 |
| 238 | 13 | MA0878.3 | CDX1 | MA0878.3 | GGYMATAAAA | CDX1 | 5.95e-20 | 1.12e-22 | 13300 | 37.4 | 17500 | 34.2 | 1.09 |
| 239 | 3 | MA1960.1 | MGA::EVX1 | MA1960.1 | RGGTGWTAATKD | MGA::EVX1 | 8.66e-20 | 1.63e-22 | 19300 | 54.4 | 26100 | 51 | 1.07 |
| 240 | 23 | MA0090.3 | TEAD1 | MA0090.3 | NNACATTCCAGSN | TEAD1 | 1.24e-19 | 2.32e-22 | 24400 | 68.6 | 33500 | 65.4 | 1.05 |
| 241 | 43 | MA1624.1 | Stat5a | MA1624.1 | NTTCCAAGAANH | Stat5a | 1.61e-19 | 3e-22 | 29700 | 83.6 | 41500 | 81 | 1.03 |
| 242 | 3 | MA0601.1 | Arid3b | MA0601.1 | ATATTAATWAN | Arid3b | 2.57e-19 | 4.77e-22 | 14800 | 41.6 | 19600 | 38.3 | 1.08 |
| 243 | 44 | MA0626.1 | Npas2 | MA0626.1 | NSCACGTGTN | Npas2 | 2.82e-19 | 5.22e-22 | 16900 | 47.6 | 22700 | 44.3 | 1.07 |
| 244 | 18 | MA0842.2 | NRL | MA0842.2 | AAWWNTGCTGACG | NRL | 2.84e-19 | 5.22e-22 | 9290 | 26.1 | 11900 | 23.3 | 1.12 |
| 245 | 3 | MA0715.1 | PROP1 | MA0715.1 | TAATYWAATTA | PROP1 | 4.39e-19 | 8.04e-22 | 13400 | 37.6 | 17600 | 34.4 | 1.09 |
| 246 | 21 | MA0497.1 | MEF2C | MA0497.1 | DDDCYAAAAATAGMW | MEF2C | 7.19e-19 | 1.31e-21 | 13500 | 38 | 17800 | 34.8 | 1.09 |
| 247 | 35 | MA0782.2 | PKNOX1 | MA0782.2 | NNTGAGTGACAGVNN | PKNOX1 | 1.28e-18 | 2.32e-21 | 12500 | 35.1 | 16400 | 32 | 1.1 |
| 248 | 43 | MA1625.1 | Stat5b | MA1625.1 | NNNTTCCCAGAANNN | Stat5b | 5.15e-18 | 9.31e-21 | 29800 | 83.8 | 41700 | 81.4 | 1.03 |
| 249 | 8 | MA0480.2 | Foxo1 | MA0480.2 | NDGTAAACANN | Foxo1 | 6.54e-18 | 1.18e-20 | 33800 | 94.9 | 47800 | 93.4 | 1.02 |
| 250 | 45 | MA0483.1 | Gfi1B | MA0483.1 | AAATCWCWGCH | Gfi1B | 7.59e-18 | 1.36e-20 | 13100 | 36.9 | 17300 | 33.8 | 1.09 |
| 251 | 26 | MA1131.1 | FOSL2::JUN | MA1131.1 | GRTGACGTMAT | FOSL2::JUN | 9.85e-18 | 1.76e-20 | 25200 | 70.9 | 34800 | 67.9 | 1.04 |
| 252 | 26 | MA0025.2 | NFIL3 | MA0025.2 | NRTTATGYAATNN | NFIL3 | 1.47e-17 | 2.63e-20 | 13800 | 38.8 | 18300 | 35.8 | 1.09 |
| 253 | 46 | MA1588.1 | ZNF136 | MA1588.1 | RWATTCTTGGTTGRC | ZNF136 | 1.76e-17 | 3.12e-20 | 16600 | 46.7 | 22300 | 43.5 | 1.07 |
| 254 | 3 | MA0618.1 | LBX1 | MA0618.1 | TTAATTAG | LBX1 | 2.73e-17 | 4.83e-20 | 16900 | 47.6 | 22700 | 44.4 | 1.07 |
| 255 | 37 | MA0152.2 | Nfatc2 | MA0152.2 | NNRNAATGGAAANWNH | Nfatc2 | 7.32e-17 | 1.29e-19 | 33100 | 92.9 | 46700 | 91.2 | 1.02 |
| 256 | 47 | MA1975.1 | ZNF214 | MA1975.1 | VWTCATCAABGTCCT | ZNF214 | 1.11e-16 | 1.95e-19 | 28400 | 80 | 39600 | 77.4 | 1.03 |
| 257 | 25 | MA0754.2 | CUX1 | MA0754.2 | TAATCGATAM | CUX1 | 1.19e-16 | 2.08e-19 | 17900 | 50.4 | 24200 | 47.3 | 1.07 |
| 258 | 9 | MA1547.2 | PITX2 | MA1547.2 | YTAATCCY | PITX2 | 1.32e-16 | 2.29e-19 | 12000 | 33.8 | 15800 | 30.9 | 1.09 |
| 259 | 8 | MA0031.1 | FOXD1 | MA0031.1 | GTAAACAW | FOXD1 | 1.49e-16 | 2.58e-19 | 15900 | 44.8 | 21300 | 41.7 | 1.07 |
| 260 | 8 | MA0851.1 | Foxj3 | MA0851.1 | NDAADGTAAACAAANNM | Foxj3 | 2.15e-16 | 3.71e-19 | 18200 | 51.2 | 24600 | 48.1 | 1.06 |
| 261 | 48 | MA1118.1 | SIX1 | MA1118.1 | GWAACCTGAKM | SIX1 | 2.27e-16 | 3.91e-19 | 29500 | 83 | 41300 | 80.6 | 1.03 |
| 262 | 31 | MA1709.1 | ZIM3 | MA1709.1 | RVMAACAGAAACCYM | ZIM3 | 6.37e-16 | 1.09e-18 | 23800 | 67 | 32800 | 64.1 | 1.05 |
| 263 | 26 | MA1127.1 | FOSB::JUN | MA1127.1 | GATGACGTCAT | FOSB::JUN | 9.29e-16 | 1.59e-18 | 27300 | 76.6 | 37900 | 74 | 1.04 |
| 264 | 17 | MA0095.3 | Yy1 | MA0095.3 | NNCAAAATGGCN | Yy1 | 1.55e-15 | 2.63e-18 | 33400 | 93.8 | 47200 | 92.2 | 1.02 |
| 265 | 49 | MA1729.1 | ZNF680 | MA1729.1 | GNCCAAGAAGAATDA | ZNF680 | 1.61e-15 | 2.72e-18 | 22500 | 63.4 | 30900 | 60.5 | 1.05 |
| 266 | 50 | MA1974.1 | ZNF211 | MA1974.1 | HWYATATACCAYRYW | ZNF211 | 1.62e-15 | 2.73e-18 | 15100 | 42.5 | 20200 | 39.6 | 1.07 |
| 267 | 40 | MA1523.1 | MSANTD3 | MA1523.1 | STVCACTCAC | MSANTD3 | 1.71e-15 | 2.87e-18 | 30300 | 85.1 | 42400 | 82.9 | 1.03 |
| 268 | 43 | MA0144.2 | STAT3 | MA0144.2 | YTTCYKGGAAD | STAT3 | 1.99e-15 | 3.34e-18 | 20000 | 56.1 | 27200 | 53.2 | 1.06 |
| 269 | 26 | MA0833.2 | ATF4 | MA0833.2 | DDATGATGCAATMN | ATF4 | 2.49e-15 | 4.15e-18 | 16400 | 46.2 | 22100 | 43.3 | 1.07 |
| 270 | 51 | MA1150.1 | RORB | MA1150.1 | AWWTRGGTCAH | RORB | 3.31e-15 | 5.5e-18 | 32400 | 91 | 45700 | 89.2 | 1.02 |
| 271 | 27 | MA0102.4 | CEBPA | MA0102.4 | NDATTGCACAATHN | CEBPA | 3.39e-15 | 5.61e-18 | 16800 | 47.2 | 22600 | 44.2 | 1.07 |
| 272 | 42 | MA1707.1 | DMRTA1 | MA1707.1 | AWTTGWTACAWT | DMRTA1 | 4.1e-15 | 6.77e-18 | 25500 | 71.7 | 35300 | 69 | 1.04 |
| 273 | 40 | MA1645.1 | NKX2-2 | MA1645.1 | NNNCCACTCAANNN | NKX2-2 | 5.53e-15 | 9.08e-18 | 25900 | 72.8 | 35900 | 70.1 | 1.04 |
| 274 | 52 | MA1593.1 | ZNF317 | MA1593.1 | WVACAGCAGAYW | ZNF317 | 7.38e-15 | 1.21e-17 | 33600 | 94.5 | 47600 | 93.1 | 1.02 |
| 275 | 53 | MA1124.1 | ZNF24 | MA1124.1 | CATTCATTCATTC | ZNF24 | 7.94e-15 | 1.3e-17 | 8450 | 23.7 | 10900 | 21.3 | 1.12 |
| 276 | 3 | MA0874.1 | Arx | MA0874.1 | VTBCRYTAATTARTKSW | Arx | 1.37e-14 | 2.23e-17 | 12200 | 34.2 | 16100 | 31.4 | 1.09 |
| 277 | 40 | MA1994.1 | Nkx2-1 | MA1994.1 | NVCACTTGARHNN | Nkx2-1 | 1.94e-14 | 3.14e-17 | 26000 | 73.1 | 36000 | 70.4 | 1.04 |
| 278 | 17 | MA0442.2 | SOX10 | MA0442.2 | NDAACAAAGVN | SOX10 | 2.08e-14 | 3.36e-17 | 26000 | 73 | 36000 | 70.4 | 1.04 |
| 279 | 23 | MA1121.1 | TEAD2 | MA1121.1 | NYACATTCCWNNS | TEAD2 | 5.71e-14 | 9.18e-17 | 24600 | 69.2 | 34000 | 66.5 | 1.04 |
| 280 | 35 | MA0775.1 | MEIS3 | MA0775.1 | DTGACAGS | MEIS3 | 6.18e-14 | 9.9e-17 | 6820 | 19.2 | 8690 | 17 | 1.13 |
| 281 | 22 | MA1991.1 | Hnf1A | MA1991.1 | NBCCTTTGATSTBN | Hnf1A | 8.18e-14 | 1.31e-16 | 17400 | 49 | 23600 | 46.2 | 1.06 |
| 282 | 54 | MA0140.2 | GATA1::TAL1 | MA0140.2 | MTTATCWSNNNNNHVCAG | GATA1::TAL1 | 1.27e-13 | 2.01e-16 | 6640 | 18.6 | 8450 | 16.5 | 1.13 |
| 283 | 26 | MA1632.1 | ATF2 | MA1632.1 | NNATGABGTCATN | ATF2 | 1.52e-13 | 2.41e-16 | 15300 | 42.9 | 20500 | 40.1 | 1.07 |
| 284 | 13 | MA1125.1 | ZNF384 | MA1125.1 | DNWMAAAAAAAA | ZNF384 | 1.79e-13 | 2.83e-16 | 16800 | 47.2 | 22700 | 44.4 | 1.06 |
| 285 | 26 | MA0093.3 | USF1 | MA0093.3 | NDGTCATGTGACHN | USF1 | 1.85e-13 | 2.91e-16 | 21500 | 60.3 | 29400 | 57.5 | 1.05 |
| 286 | 37 | MA0624.2 | Nfatc1 | MA0624.2 | NVATGGAAAAWN | Nfatc1 | 2.32e-13 | 3.64e-16 | 32000 | 90 | 45100 | 88.2 | 1.02 |
| 287 | 26 | MA0498.2 | MEIS1 | MA0498.2 | HTGACAD | MEIS1 | 5.55e-13 | 8.68e-16 | 20800 | 58.4 | 28500 | 55.6 | 1.05 |
| 288 | 48 | MA1119.1 | SIX2 | MA1119.1 | NNCTGAAACCTGATMY | SIX2 | 9.24e-13 | 1.44e-15 | 25000 | 70.3 | 34700 | 67.8 | 1.04 |
| 289 | 3 | MA1577.1 | TLX2 | MA1577.1 | BYAATTAR | TLX2 | 1.29e-12 | 2e-15 | 9040 | 25.4 | 11800 | 23.1 | 1.1 |
| 290 | 28 | MA0906.1 | HOXC12 | MA0906.1 | RGTCGTAAAAH | HOXC12 | 1.71e-12 | 2.65e-15 | 12800 | 36 | 17100 | 33.4 | 1.08 |
| 291 | 31 | MA1508.1 | IKZF1 | MA1508.1 | VVAACAGGAARN | IKZF1 | 2.23e-12 | 3.43e-15 | 35300 | 99.2 | 50500 | 98.7 | 1.01 |
| 292 | 55 | MA0847.3 | FOXD2 | MA0847.3 | SYTAARYAAACA | FOXD2 | 3.4e-12 | 5.22e-15 | 19300 | 54.4 | 26400 | 51.7 | 1.05 |
| 293 | 3 | MA1507.1 | HOXD4 | MA1507.1 | VTMATTAD | HOXD4 | 3.7e-12 | 5.66e-15 | 7900 | 22.2 | 10300 | 20 | 1.11 |
| 294 | 28 | MA0907.1 | HOXC13 | MA0907.1 | BCTCGTAAAAH | HOXC13 | 4.88e-12 | 7.45e-15 | 15600 | 43.8 | 21100 | 41.2 | 1.06 |
| 295 | 26 | MA0829.2 | SREBF1 | MA0829.2 | DNATCACSTGATNH | SREBF1 | 6.15e-12 | 9.35e-15 | 25600 | 72.1 | 35600 | 69.7 | 1.03 |
| 296 | 31 | MA1953.1 | FOXO1::ELF1 | MA1953.1 | RTMAACAGGAAGTN | FOXO1::ELF1 | 1.78e-11 | 2.7e-14 | 26000 | 73.1 | 36200 | 70.8 | 1.03 |
| 297 | 8 | MA0613.1 | FOXG1 | MA0613.1 | RTAAACAW | FOXG1 | 1.82e-11 | 2.75e-14 | 15400 | 43.4 | 20900 | 40.8 | 1.06 |
| 298 | 3 | MA0060.3 | NFYA | MA0060.3 | RRCCAATCAGM | NFYA | 1.94e-11 | 2.92e-14 | 13000 | 36.4 | 17400 | 33.9 | 1.07 |
| 299 | 56 | MA0101.1 | REL | MA0101.1 | BGGRNWTTCC | REL | 2.87e-11 | 4.31e-14 | 32400 | 91.1 | 45800 | 89.5 | 1.02 |
| 300 | 30 | MA1487.2 | FOXE1 | MA1487.2 | VYTAAAYAAACAAH | FOXE1 | 3.26e-11 | 4.88e-14 | 16800 | 47.2 | 22800 | 44.6 | 1.06 |
| 301 | 26 | MA1139.1 | FOSL2::JUNB | MA1139.1 | DATGACGTCATH | FOSL2::JUNB | 3.8e-11 | 5.67e-14 | 19200 | 54 | 26300 | 51.4 | 1.05 |
| 302 | 45 | MA0038.2 | GFI1 | MA0038.2 | BMAATCACDGCA | GFI1 | 3.88e-11 | 5.76e-14 | 7930 | 22.3 | 10300 | 20.2 | 1.1 |
| 303 | 34 | MA0884.2 | DUXA | MA0884.2 | YTRATTWAATYAR | DUXA | 5.97e-11 | 8.84e-14 | 5480 | 15.4 | 6960 | 13.6 | 1.13 |
| 304 | 22 | MA0869.2 | Sox11 | MA0869.2 | NRGAACAAAGVV | Sox11 | 6.61e-11 | 9.76e-14 | 33900 | 95.4 | 48200 | 94.3 | 1.01 |
| 305 | 57 | MA1524.2 | Msgn1 | MA1524.2 | VRRRACAAATGGTNNN | Msgn1 | 8.55e-11 | 1.26e-13 | 21800 | 61.3 | 30100 | 58.8 | 1.04 |
| 306 | 57 | MA0461.2 | Atoh1 | MA0461.2 | RMCATATGBY | Atoh1 | 1.15e-10 | 1.69e-13 | 23300 | 65.4 | 32200 | 63 | 1.04 |
| 307 | 3 | MA0702.2 | LMX1A | MA0702.2 | TTAATTAM | LMX1A | 2.54e-10 | 3.71e-13 | 9310 | 26.2 | 12300 | 24 | 1.09 |
| 308 | 26 | MA0043.3 | HLF | MA0043.3 | NNRTTATGCAACHN | HLF | 3.39e-10 | 4.94e-13 | 10600 | 29.8 | 14100 | 27.5 | 1.08 |
| 309 | 26 | MA0693.3 | Vdr | MA0693.3 | NNGAGTTCANN | Vdr | 3.48e-10 | 5.05e-13 | 29300 | 82.3 | 41200 | 80.4 | 1.02 |
| 310 | 37 | MA0625.2 | NFATC3 | MA0625.2 | RYGGAAAMH | NFATC3 | 3.83e-10 | 5.54e-13 | 35300 | 99.3 | 50600 | 98.9 | 1 |
| 311 | 26 | MA1126.1 | FOS::JUN | MA1126.1 | DRTGACGTCATHNDTN | FOS::JUN | 1.04e-09 | 1.51e-12 | 28500 | 80.2 | 40100 | 78.3 | 1.03 |
| 312 | 28 | MA1503.1 | HOXB9 | MA1503.1 | GTCGTAAAAH | HOXB9 | 1.09e-09 | 1.57e-12 | 6180 | 17.4 | 7980 | 15.6 | 1.12 |
| 313 | 58 | MA1105.2 | GRHL2 | MA1105.2 | NRAACAGGTTYN | GRHL2 | 1.45e-09 | 2.08e-12 | 23600 | 66.2 | 32700 | 63.9 | 1.04 |
| 314 | 59 | MA0479.1 | FOXH1 | MA0479.1 | BNSAATCCACA | FOXH1 | 2.12e-09 | 3.03e-12 | 6870 | 19.3 | 8940 | 17.5 | 1.11 |
| 315 | 35 | MA1114.1 | PBX3 | MA1114.1 | NNNTGAGTGACAGGBNN | PBX3 | 2.49e-09 | 3.55e-12 | 13500 | 38.1 | 18300 | 35.8 | 1.06 |
| 316 | 21 | MA0773.1 | MEF2D | MA0773.1 | DCTAWAAATAGM | MEF2D | 3.97e-09 | 5.63e-12 | 4680 | 13.2 | 5940 | 11.6 | 1.13 |
| 317 | 26 | MA0843.1 | TEF | MA0843.1 | BRTTACRTAAYM | TEF | 5.46e-09 | 7.72e-12 | 9860 | 27.7 | 13100 | 25.6 | 1.08 |
| 318 | 35 | MA0797.1 | TGIF2 | MA0797.1 | TGACAGSTGTCA | TGIF2 | 6.08e-09 | 8.58e-12 | 3900 | 11 | 4890 | 9.56 | 1.15 |
| 319 | 42 | MA1478.1 | DMRTA2 | MA1478.1 | AATTGHTACAWT | DMRTA2 | 6.1e-09 | 8.58e-12 | 25200 | 70.7 | 35100 | 68.6 | 1.03 |
| 320 | 51 | MA1151.1 | RORC | MA1151.1 | NWAWNTRGGTCA | RORC | 6.89e-09 | 9.67e-12 | 34600 | 97.2 | 49300 | 96.3 | 1.01 |
| 321 | 40 | MA0672.1 | NKX2-3 | MA0672.1 | VCCACTTRAV | NKX2-3 | 8.28e-09 | 1.16e-11 | 22400 | 63 | 31100 | 60.7 | 1.04 |
| 322 | 43 | MA0519.1 | Stat5a::Stat5b | MA0519.1 | DTTTCCMAGAA | Stat5a::Stat5b | 8.83e-09 | 1.23e-11 | 18000 | 50.5 | 24700 | 48.2 | 1.05 |
| 323 | 26 | MA0488.1 | JUN | MA0488.1 | DDRATGATGTCAT | JUN | 1.17e-08 | 1.63e-11 | 5350 | 15 | 6870 | 13.4 | 1.12 |
| 324 | 35 | MA1572.1 | TGIF2LY | MA1572.1 | TGACAGCTGTCA | TGIF2LY | 1.42e-08 | 1.96e-11 | 21100 | 59.3 | 29200 | 57.1 | 1.04 |
| 325 | 44 | MA0620.3 | MITF | MA0620.3 | NNNDRTCACGTGAYHNNN | MITF | 2.18e-08 | 3.01e-11 | 22100 | 62 | 30600 | 59.8 | 1.04 |
| 326 | 43 | MA0518.1 | Stat4 | MA0518.1 | YTTCYRGGAARNVR | Stat4 | 2.54e-08 | 3.49e-11 | 18400 | 51.7 | 25300 | 49.4 | 1.05 |
| 327 | 3 | MA0621.1 | mix-a | MA0621.1 | NDTTAATTAGN | mix-a | 2.98e-08 | 4.08e-11 | 7100 | 20 | 9310 | 18.2 | 1.1 |
| 328 | 21 | MA0660.1 | MEF2B | MA0660.1 | RCTAWAAATAGC | MEF2B | 4.11e-08 | 5.62e-11 | 4730 | 13.3 | 6050 | 11.8 | 1.13 |
| 329 | 51 | MA1111.1 | NR2F2 | MA1111.1 | NAAAGGTCANR | NR2F2 | 6.53e-08 | 8.9e-11 | 24300 | 68.3 | 33900 | 66.2 | 1.03 |
| 330 | 60 | MA0595.1 | SREBF1 | MA0595.1 | VTCACCCCAY | SREBF1 | 1.03e-07 | 1.4e-10 | 7590 | 21.3 | 10000 | 19.6 | 1.09 |
| 331 | 26 | MA1136.1 | FOSB::JUNB | MA1136.1 | RTGACGTCAT | FOSB::JUNB | 1.11e-07 | 1.51e-10 | 28200 | 79.1 | 39600 | 77.3 | 1.02 |
| 332 | 26 | MA0631.1 | Six3 | MA0631.1 | DATRGGGTATCAYNWNT | Six3 | 3.49e-07 | 4.71e-10 | 14300 | 40.1 | 19500 | 38.1 | 1.05 |
| 333 | 61 | MA1116.1 | RBPJ | MA1116.1 | BVTGGGAANN | RBPJ | 3.91e-07 | 5.26e-10 | 30500 | 85.8 | 43100 | 84.3 | 1.02 |
| 334 | 30 | MA0040.1 | Foxq1 | MA0040.1 | HATTGTTTATW | Foxq1 | 5.43e-07 | 7.29e-10 | 10900 | 30.6 | 14700 | 28.7 | 1.07 |
| 335 | 8 | MA0849.1 | FOXO6 | MA0849.1 | GTAAACA | FOXO6 | 8.96e-07 | 1.2e-09 | 20100 | 56.4 | 27800 | 54.4 | 1.04 |
| 336 | 11 | MA1962.1 | POU2F1::SOX2 | MA1962.1 | MATTTRCATMACAATRG | POU2F1::SOX2 | 9.1e-07 | 1.22e-09 | 7300 | 20.5 | 9650 | 18.9 | 1.09 |
| 337 | 62 | MA0525.2 | TP63 | MA0525.2 | RACATGTYGKGACATGTC | TP63 | 9.89e-07 | 1.32e-09 | 285 | 0.8 | 243 | 0.47 | 1.69 |
| 338 | 3 | MA0854.1 | Alx1 | MA0854.1 | CGMRYTAATTARTNMNY | Alx1 | 1.05e-06 | 1.39e-09 | 8210 | 23.1 | 10900 | 21.4 | 1.08 |
| 339 | 34 | MA1720.1 | ZNF85 | MA1720.1 | DHDGAGATTACWKCAK | ZNF85 | 1.32e-06 | 1.75e-09 | 17500 | 49.2 | 24100 | 47.1 | 1.04 |
| 340 | 28 | MA0873.1 | HOXD12 | MA0873.1 | RGTCGTAAAAH | HOXD12 | 1.41e-06 | 1.86e-09 | 9680 | 27.2 | 13000 | 25.4 | 1.07 |
| 341 | 60 | MA0596.1 | SREBF2 | MA0596.1 | RTGGGGTGAY | SREBF2 | 1.45e-06 | 1.9e-09 | 2060 | 5.8 | 2500 | 4.89 | 1.19 |
| 342 | 18 | MA1520.1 | MAF | MA1520.1 | NTGCTGASTCAGCAH | MAF | 1.54e-06 | 2.02e-09 | 1620 | 4.55 | 1920 | 3.74 | 1.22 |
| 343 | 63 | MA0689.1 | TBX20 | MA0689.1 | WAGGTGTGAAR | TBX20 | 2.9e-06 | 3.79e-09 | 16100 | 45.2 | 22100 | 43.2 | 1.05 |
| 344 | 63 | MA0800.1 | EOMES | MA0800.1 | RAGGTGTGAAAWN | EOMES | 3.37e-06 | 4.39e-09 | 29900 | 83.9 | 42200 | 82.4 | 1.02 |
| 345 | 64 | MA0682.2 (PITX1) | MEME-3 | CTGGGATTACAGGCR | CTGGGATTACAGGCR | PITX1 | 4.4e-06 | 5.73e-09 | 1100 | 3.1 | 1260 | 2.45 | 1.26 |
| 346 | 51 | MA0494.1 | Nr1h3::Rxra | MA0494.1 | TGACCTNNAGTRACCYYDN | Nr1h3::Rxra | 5.41e-06 | 7e-09 | 24400 | 68.6 | 34200 | 66.8 | 1.03 |
| 347 | 40 | MA0673.1 | NKX2-8 | MA0673.1 | VCACTTSAV | NKX2-8 | 5.42e-06 | 7e-09 | 30700 | 86.4 | 43500 | 85 | 1.02 |
| 348 | 65 | MA0062.3 | GABPA | MA0062.3 | NNCACTTCCTGTNN | GABPA | 6.06e-06 | 7.82e-09 | 35300 | 99.1 | 50500 | 98.7 | 1 |
| 349 | 26 | MA0492.1 | JUND | MA0492.1 | DDDRATGABGTCATN | JUND | 7.53e-06 | 9.68e-09 | 5870 | 16.5 | 7720 | 15.1 | 1.09 |
| 350 | 17 | MA0867.2 | SOX4 | MA0867.2 | RAACAAAGRV | SOX4 | 1.31e-05 | 1.67e-08 | 11700 | 32.9 | 15900 | 31.1 | 1.06 |
| 351 | 23 | MA0484.2 | HNF4G | MA0484.2 | DNCAAAGTCCANN | HNF4G | 1.31e-05 | 1.67e-08 | 11200 | 31.6 | 15300 | 29.8 | 1.06 |
| 352 | 26 | MA1129.1 | FOSL1::JUN | MA1129.1 | ATGACGTCAT | FOSL1::JUN | 1.78e-05 | 2.27e-08 | 31300 | 88 | 44400 | 86.7 | 1.01 |
| 353 | 62 | MA0106.3 | TP53 | MA0106.3 | RACATGYCCGGRCATGTY | TP53 | 1.89e-05 | 2.41e-08 | 127 | 0.36 | 85 | 0.17 | 2.14 |
| 354 | 17 | MA0087.2 | Sox5 | MA0087.2 | VRVAACAATGGNN | Sox5 | 1.93e-05 | 2.45e-08 | 15900 | 44.8 | 22000 | 42.9 | 1.04 |
| 355 | 44 | MA1997.1 | Olig2 | MA1997.1 | NVCAGCTGBN | Olig2 | 2.85e-05 | 3.6e-08 | 5090 | 14.3 | 6670 | 13 | 1.1 |
| 356 | 44 | MA0743.2 | SCRT1 | MA0743.2 | NDWKCAACAGGTGKNN | SCRT1 | 2.87e-05 | 3.61e-08 | 29100 | 81.7 | 41000 | 80.2 | 1.02 |
| 357 | 57 | MA0827.1 | OLIG3 | MA0827.1 | AMCATATGBY | OLIG3 | 3.12e-05 | 3.92e-08 | 27100 | 76.1 | 38100 | 74.5 | 1.02 |
| 358 | 26 | MA0639.1 | DBP | MA0639.1 | NRTTACGTAAYV | DBP | 3.13e-05 | 3.92e-08 | 7090 | 19.9 | 9450 | 18.5 | 1.08 |
| 359 | 29 | MA0502.2 | NFYB | MA0502.2 | CYCATTGGCCVV | NFYB | 3.24e-05 | 4.04e-08 | 8530 | 24 | 11500 | 22.4 | 1.07 |
| 360 | 63 | MA0802.1 | TBR1 | MA0802.1 | AGGTGTGAAA | TBR1 | 3.43e-05 | 4.28e-08 | 28100 | 78.9 | 39600 | 77.3 | 1.02 |
| 361 | 41 | MA1643.1 | NFIB | MA1643.1 | NNCCTGGCAYNGTGCCAAGNN | NFIB | 3.59e-05 | 4.46e-08 | 2290 | 6.44 | 2850 | 5.56 | 1.16 |
| 362 | 27 | MA0838.1 | CEBPG | MA0838.1 | ATTRCGCAAY | CEBPG | 3.64e-05 | 4.51e-08 | 2060 | 5.79 | 2540 | 4.96 | 1.17 |
| 363 | 44 | MA1635.1 | BHLHE22 | MA1635.1 | NVCAGCTGBN | BHLHE22 | 4.38e-05 | 5.41e-08 | 5180 | 14.6 | 6800 | 13.3 | 1.1 |
| 364 | 66 | MA0069.1 | PAX6 | MA0069.1 | TTCACGCWTSANTK | PAX6 | 5.96e-05 | 7.34e-08 | 17300 | 48.7 | 24000 | 46.9 | 1.04 |
| 365 | 67 | MA0080.6 (Spi1) | MEME-5 | CWBYCTYCCTCTSTC | CWBYCTYCCTCTSTC | Spi1 | 8.45e-05 | 1.04e-07 | 14800 | 41.5 | 20300 | 39.7 | 1.04 |
| 366 | 57 | MA1618.1 | Ptf1a | MA1618.1 | NNACAGATGTTNN | Ptf1a | 9.55e-05 | 1.17e-07 | 26600 | 74.6 | 37400 | 73.1 | 1.02 |
| 367 | 23 | MA0114.4 | HNF4A | MA0114.4 | DNCAAAGTCCANN | HNF4A | 9.89e-05 | 1.21e-07 | 15800 | 44.5 | 21900 | 42.7 | 1.04 |
| 368 | 26 | MA0592.3 | ESRRA | MA0592.3 | NBTCAAGGTCAHN | ESRRA | 0.000128 | 1.55e-07 | 32300 | 90.7 | 45900 | 89.6 | 1.01 |
| 369 | 26 | MA1133.1 | JUN::JUNB | MA1133.1 | KRTGACGTCATN | JUN::JUNB | 0.000137 | 1.67e-07 | 24900 | 69.9 | 34900 | 68.2 | 1.02 |
| 370 | 34 | MA0468.1 | DUX4 | MA0468.1 | TAAYYYAATCA | DUX4 | 0.000208 | 2.52e-07 | 3530 | 9.93 | 4560 | 8.92 | 1.11 |
| 371 | 44 | MA0633.2 | Twist2 | MA0633.2 | NVCAGCTGBN | Twist2 | 0.000213 | 2.58e-07 | 4890 | 13.8 | 6440 | 12.6 | 1.09 |
| 372 | 12 | MA0683.1 | POU4F2 | MA0683.1 | RTGCATAATTAATGAG | POU4F2 | 0.00051 | 6.16e-07 | 1980 | 5.57 | 2470 | 4.83 | 1.15 |
| 373 | 26 | MA0629.1 | Rhox11 | MA0629.1 | ANKNCGCTGTWAWDVRW | Rhox11 | 0.000564 | 6.78e-07 | 18100 | 50.8 | 25100 | 49.1 | 1.03 |
| 374 | 26 | MA1996.1 | Nr1H2 | MA1996.1 | NNRAGGTCANN | Nr1H2 | 0.000566 | 6.79e-07 | 33200 | 93.4 | 47400 | 92.6 | 1.01 |
| 375 | 35 | MA1571.1 | TGIF2LX | MA1571.1 | TGACAGCTGTCA | TGIF2LX | 0.00057 | 6.82e-07 | 14600 | 41 | 20100 | 39.4 | 1.04 |
| 376 | 52 | MA0691.1 | TFAP4 | MA0691.1 | AWCAGCTGWT | TFAP4 | 0.000599 | 7.15e-07 | 1240 | 3.48 | 1480 | 2.89 | 1.2 |
| 377 | 26 | MA1145.1 | FOSL2::JUND | MA1145.1 | NNRTGACGTCAYHSN | FOSL2::JUND | 0.000748 | 8.9e-07 | 21700 | 61.1 | 30400 | 59.5 | 1.03 |
| 378 | 68 | MA0029.1 | Mecom | MA0029.1 | AAGAYAAGATAANA | Mecom | 0.000752 | 8.93e-07 | 3100 | 8.72 | 4000 | 7.81 | 1.12 |
| 379 | 69 | MA0687.1 | SPIC | MA0687.1 | HAAAAGVGGAAGTA | SPIC | 0.000899 | 1.06e-06 | 27400 | 76.9 | 38600 | 75.5 | 1.02 |
| 380 | 41 | MA0698.1 | ZBTB18 | MA0698.1 | NATCCAGATGTKB | ZBTB18 | 0.00129 | 1.53e-06 | 11800 | 33.2 | 16200 | 31.7 | 1.05 |
| 381 | 41 | MA1123.2 | TWIST1 | MA1123.2 | NNDCCAGATGTBN | TWIST1 | 0.00195 | 2.3e-06 | 20500 | 57.6 | 28700 | 56 | 1.03 |
| 382 | 70 | MA1989.1 | Bcl11B | MA1989.1 | NNAAACCACAARNN | Bcl11B | 0.00254 | 2.99e-06 | 30800 | 86.4 | 43700 | 85.4 | 1.01 |
| 383 | 71 | MA0160.2 | NR4A2 | MA0160.2 | BWAAAGGTCA | NR4A2 | 0.00327 | 3.83e-06 | 27100 | 76.2 | 38300 | 74.8 | 1.02 |
| 384 | 72 | MA0161.2 | NFIC | MA0161.2 | NNCTTGGCANN | NFIC | 0.00395 | 4.61e-06 | 33100 | 93 | 47200 | 92.2 | 1.01 |
| 385 | 8 | MA0614.1 | Foxj2 | MA0614.1 | RTAAACAA | Foxj2 | 0.00492 | 5.74e-06 | 3820 | 10.7 | 5020 | 9.82 | 1.09 |
| 386 | 18 | MA1521.1 | MAFA | MA1521.1 | NTGCTGASTCAGCAN | MAFA | 0.00615 | 7.14e-06 | 915 | 2.57 | 1080 | 2.12 | 1.21 |
| 387 | 73 | MA0119.1 | NFIC::TLX1 | MA0119.1 | TGGCASSRWGCCAA | NFIC::TLX1 | 0.00662 | 7.68e-06 | 1710 | 4.8 | 2140 | 4.18 | 1.15 |
| 388 | 74 | MA1977.1 | ZNF324 | MA1977.1 | NNNWRGMAGHYAAGGATGGTT | ZNF324 | 0.00668 | 7.72e-06 | 13800 | 38.8 | 19100 | 37.4 | 1.04 |
| 389 | 51 | MA0071.1 | RORA | MA0071.1 | AWMWAGGTCA | RORA | 0.0119 | 1.37e-05 | 9730 | 27.4 | 13300 | 26.1 | 1.05 |
| 390 | 33 | MA1561.1 | SOX12 | MA1561.1 | ACCGAACAATV | SOX12 | 0.0122 | 1.41e-05 | 19500 | 54.7 | 27300 | 53.3 | 1.03 |
| 391 | 17 | MA0515.1 | Sox6 | MA0515.1 | CCWTTGTYYY | Sox6 | 0.0124 | 1.42e-05 | 3140 | 8.83 | 4110 | 8.02 | 1.1 |
| 392 | 63 | MA0688.1 | TBX2 | MA0688.1 | DAGGTGTGAAA | TBX2 | 0.013 | 1.49e-05 | 30900 | 86.8 | 43900 | 85.8 | 1.01 |
| 393 | 75 | MA0508.3 | PRDM1 | MA0508.3 | YNCTTTCTCTH | PRDM1 | 0.015 | 1.71e-05 | 13500 | 38 | 18800 | 36.6 | 1.04 |
| 394 | 76 | MA0520.1 | Stat6 | MA0520.1 | BDBTTCCWSAGAAVY | Stat6 | 0.0187 | 2.13e-05 | 10100 | 28.4 | 13900 | 27.1 | 1.05 |
| 395 | 77 | MA0780.1 | PAX3 | MA0780.1 | TAATYRATTA | PAX3 | 0.0208 | 2.36e-05 | 5620 | 15.8 | 7560 | 14.8 | 1.07 |
| 396 | 65 | MA0640.2 | ELF3 | MA0640.2 | NNCCACTTCCTGNT | ELF3 | 0.0217 | 2.46e-05 | 34100 | 95.9 | 48800 | 95.4 | 1.01 |
| 397 | 51 | MA1148.1 | PPARA::RXRA | MA1148.1 | AWNTRGGTYAAAGGTCAN | PPARA::RXRA | 0.0224 | 2.53e-05 | 24200 | 68 | 34100 | 66.7 | 1.02 |
| 398 | 73 | MA1528.1 | NFIX | MA1528.1 | STTGGCRNNGTGCCARB | NFIX | 0.0252 | 2.84e-05 | 2050 | 5.76 | 2620 | 5.13 | 1.12 |
| 399 | 28 | MA1506.1 | HOXD10 | MA1506.1 | RGTCGTAAAAH | HOXD10 | 0.031 | 3.49e-05 | 2970 | 8.35 | 3890 | 7.61 | 1.1 |
| 400 | 35 | MA0796.1 | TGIF1 | MA0796.1 | TGACAGCTGTCA | TGIF1 | 0.0346 | 3.88e-05 | 1070 | 3 | 1310 | 2.55 | 1.18 |
| 401 | 3 | MA0135.1 | Lhx3 | MA0135.1 | WAATTAATTAWYY | Lhx3 | 0.0355 | 3.97e-05 | 5570 | 15.7 | 7510 | 14.7 | 1.07 |
| 402 | 78 | MA0083.3 | SRF | MA0083.3 | TGMCCATATATGGKCA | SRF | 0.0361 | 4.03e-05 | 455 | 1.28 | 507 | 0.99 | 1.29 |
| 403 | 71 | MA1541.1 | NR6A1 | MA1541.1 | NYCAAGKTCAAGKTCAN | NR6A1 | 0.0365 | 4.06e-05 | 23300 | 65.4 | 32800 | 64.1 | 1.02 |
| 13 | 79 | MA0528.2 (ZNF263) | STREME-10 | 10-CCTCCTCCCACCCC | 10-CCTCCTCCCACCCC | ZNF263 | 0.0368 | 0.00283 | 73 | 0.21 | 64 | 0.13 | 1.64 |
| 14 | 80 | 8-TGCCTGCCAS | STREME-8 | 8-TGCCTGCCAS | TGCCTGCCAS | TGCCTGCCAS | 0.0485 | 0.00346 | 5690 | 16 | 7840 | 15.3 | 1.04 |
| 15 | 79 | MA1596.1 (ZNF460) | MEME-4 | GGRGGCKGRGGCDGG | GGRGGCKGRGGCDGG | ZNF460 | 5.57 | 0.371 | 96 | 0.27 | 131 | 0.26 | 1.06 |
ER_rat <- 1.25
DOX_24_20kb_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,TP.)%>%
arrange(.,EVALUE.y) %>%
slice_head(n=80)%>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste("DOX_24_20kb_combo response peaks(200bp)Enrichment ratio:",ER_rat))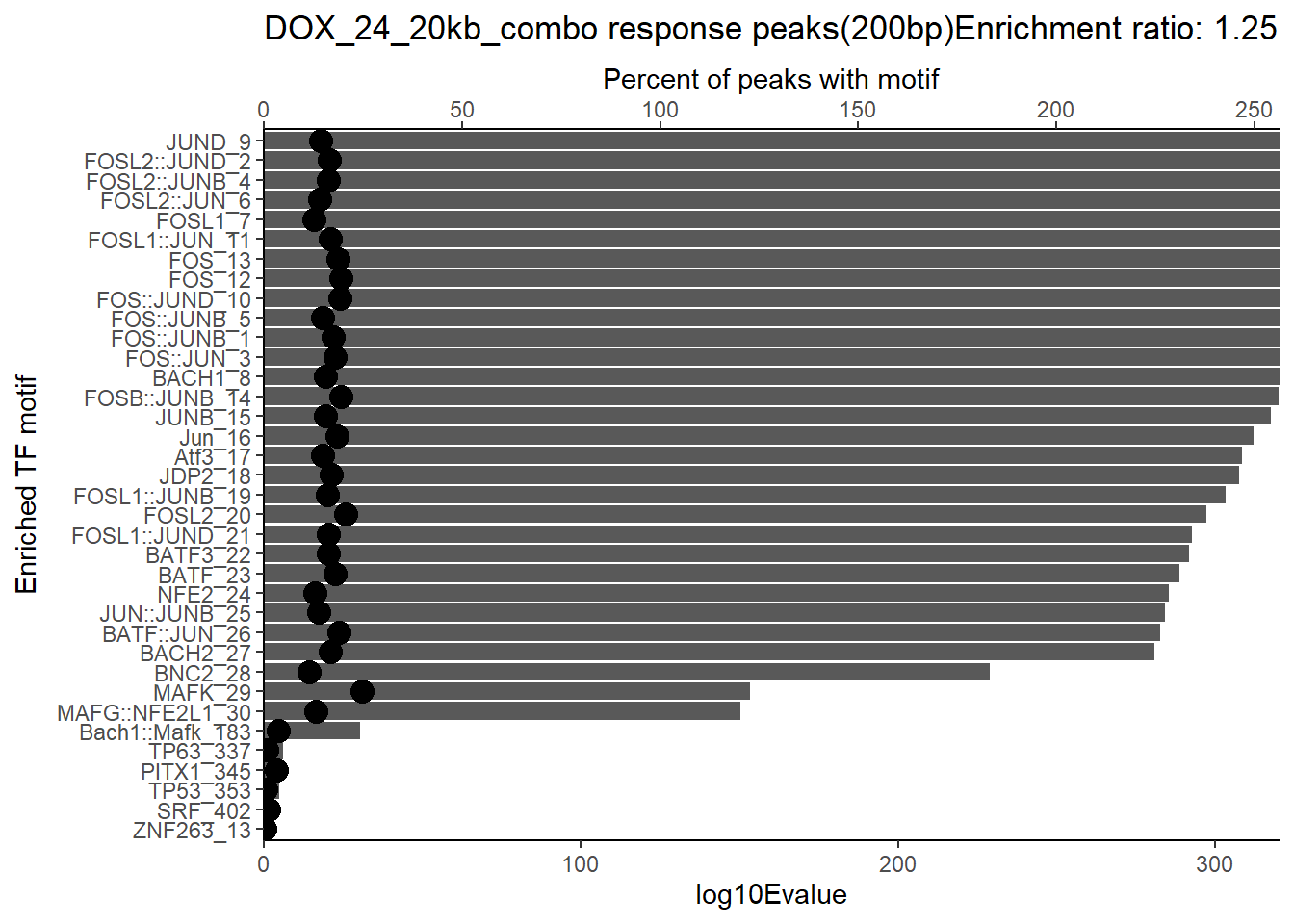
DOX_24_20kb_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue = -log10(if_else(EVALUE.y == 0, 1e-300, EVALUE.y))) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
slice_head(n=5) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*9), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./9,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste("DOX_24_20kb_combo response peaks(200bp)Enrichment ratio:",ER_rat))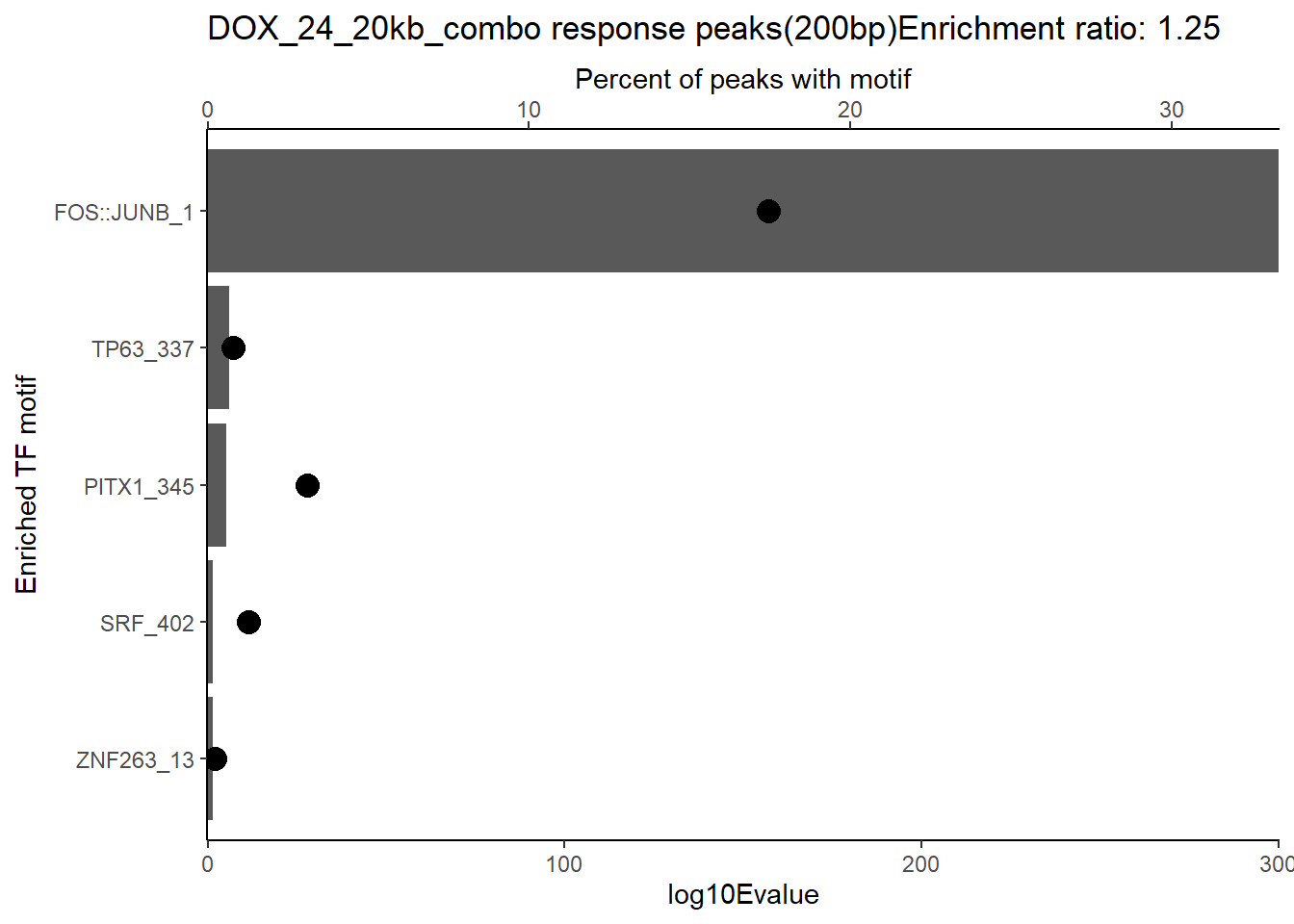 ## Other treaments
## Other treaments
EPI_DAR_3hr data
EPI_DAR_3hr_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/EPI_3_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
EPI_DAR_3hr_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/EPI_3_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
EPI_DAR_3hr_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/EPI_3_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
EPI_DAR_3hr_all <- rbind(EPI_DAR_3hr_sea_disc, EPI_DAR_3hr_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)EPI_DAR_3hr_combo <- EPI_DAR_3hr_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., EPI_DAR_3hr_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ NA_character_
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y) %>%
mutate(mrc_type="EPI_DAR_3hr")
EPI_DAR_3hr_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs in 3 hour EPI-DAR v 3 hour non-EPI DAR") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO | mrc_type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | MA1961.1 (PATZ1) | MEME-1 | CCCVSCCCCGCCCCC | CCCVSCCCCGCCCCC | PATZ1 | 6.82e-286 | 4.49e-286 | 6590 | 51.5 | 44500 | 35 | 1.47 | EPI_DAR_3hr |
| 2 | 1 | MA1511.2 (KLF10) | STREME-1 | 1-CCCCGCCCCCGCC | CCCCGCCCCCGCC | KLF10 | 8.01e-283 | 2.64e-283 | 5800 | 45.3 | 37300 | 29.4 | 1.54 | EPI_DAR_3hr |
| 3 | 1 | MA1961.1 | PATZ1 | MA1961.1 | SSGGGGMGGGGS | PATZ1 | 1.99e-237 | 4.36e-238 | 5020 | 39.2 | 32000 | 25.1 | 1.56 | EPI_DAR_3hr |
| 4 | 2 | MA1713.1 | ZNF610 | MA1713.1 | SSCGCCGCTCCSSS | ZNF610 | 1.35e-233 | 2.22e-234 | 6260 | 48.9 | 43300 | 34 | 1.44 | EPI_DAR_3hr |
| 5 | 1 | MA1653.1 | ZNF148 | MA1653.1 | YBCCCCTCCCCC | ZNF148 | 2.33e-227 | 3.06e-228 | 4560 | 35.6 | 28300 | 22.3 | 1.6 | EPI_DAR_3hr |
| 6 | 1 | MA0079.5 | SP1 | MA0079.5 | GGGGCGGGG | SP1 | 2.78e-220 | 3.05e-221 | 5420 | 42.3 | 36100 | 28.4 | 1.49 | EPI_DAR_3hr |
| 7 | 1 | MA1513.1 | KLF15 | MA1513.1 | SCCCCGCCCCS | KLF15 | 3.87e-219 | 3.64e-220 | 4670 | 36.5 | 29600 | 23.3 | 1.57 | EPI_DAR_3hr |
| 8 | 3 | MA1650.1 | ZBTB14 | MA1650.1 | SSCCGCGCACVS | ZBTB14 | 5.2e-211 | 4.28e-212 | 5310 | 41.4 | 35500 | 27.9 | 1.49 | EPI_DAR_3hr |
| 9 | 1 | MA1522.1 | MAZ | MA1522.1 | BBCCCCTCCCY | MAZ | 2.27e-209 | 1.66e-210 | 5970 | 46.6 | 41500 | 32.6 | 1.43 | EPI_DAR_3hr |
| 10 | 1 | MA0685.2 | SP4 | MA0685.2 | GGGGCGGGG | SP4 | 8.84e-205 | 5.82e-206 | 6400 | 50 | 45800 | 36 | 1.39 | EPI_DAR_3hr |
| 11 | 1 | MA1959.1 | KLF7 | MA1959.1 | DGGGCGGGG | KLF7 | 5.94e-203 | 3.55e-204 | 6590 | 51.4 | 47600 | 37.4 | 1.37 | EPI_DAR_3hr |
| 12 | 1 | MA0742.2 | KLF12 | MA0742.2 | GGGGCGGGG | KLF12 | 4.23e-200 | 2.32e-201 | 6560 | 51.2 | 47400 | 37.3 | 1.37 | EPI_DAR_3hr |
| 13 | 4 | MA0162.4 | EGR1 | MA0162.4 | VCMCGCCCACGCVS | EGR1 | 5.67e-200 | 2.87e-201 | 6950 | 54.3 | 51200 | 40.3 | 1.35 | EPI_DAR_3hr |
| 14 | 5 | 6-CCGGCC | STREME-6 | 6-CCGGCC | CCGGCC | CCGGCC | 6.36e-188 | 2.99e-189 | 5850 | 45.7 | 41300 | 32.5 | 1.41 | EPI_DAR_3hr |
| 15 | 1 | MA0516.3 | SP2 | MA0516.3 | GGGGCGGGG | SP2 | 5.1e-186 | 2.24e-187 | 7020 | 54.8 | 52500 | 41.3 | 1.33 | EPI_DAR_3hr |
| 16 | 1 | MA0740.2 | KLF14 | MA0740.2 | KGGGCGGGG | KLF14 | 1.31e-184 | 5.37e-186 | 6990 | 54.6 | 52300 | 41.1 | 1.33 | EPI_DAR_3hr |
| 17 | 6 | 2-CGGSSCCG | STREME-2 | 2-CGGSSCCG | CGGSSCCG | CGGSSCCG | 4.24e-183 | 1.64e-184 | 6800 | 53.1 | 50500 | 39.7 | 1.34 | EPI_DAR_3hr |
| 18 | 7 | 3-CCGSCGG | STREME-3 | 3-CCGSCGG | CCGSCGG | CCGSCGG | 4.54e-183 | 1.66e-184 | 5860 | 45.8 | 41700 | 32.8 | 1.4 | EPI_DAR_3hr |
| 19 | 1 | MA0753.2 | ZNF740 | MA0753.2 | CYRCCCCCCCCAC | ZNF740 | 6.75e-180 | 2.26e-181 | 6280 | 49 | 45700 | 35.9 | 1.36 | EPI_DAR_3hr |
| 20 | 1 | MA1630.2 | ZNF281 | MA1630.2 | GGGGGAGGGGVV | ZNF281 | 6.86e-180 | 2.26e-181 | 4810 | 37.5 | 32200 | 25.4 | 1.48 | EPI_DAR_3hr |
| 21 | 1 | MA1511.2 | KLF10 | MA1511.2 | GGGGCGGGG | KLF10 | 1.89e-179 | 5.92e-181 | 6800 | 53.1 | 50700 | 39.8 | 1.33 | EPI_DAR_3hr |
| 22 | 4 | MA1564.1 | SP9 | MA1564.1 | RCCACGCCCMCY | SP9 | 5.38e-179 | 1.61e-180 | 6170 | 48.2 | 44700 | 35.2 | 1.37 | EPI_DAR_3hr |
| 23 | 4 | MA1627.1 | Wt1 | MA1627.1 | YBCCTCCCCCACVB | Wt1 | 1.67e-176 | 4.79e-178 | 6470 | 50.5 | 47700 | 37.5 | 1.35 | EPI_DAR_3hr |
| 24 | 1 | MA1515.1 | KLF2 | MA1515.1 | NRCCACRCCCH | KLF2 | 1.84e-170 | 5.04e-172 | 5800 | 45.3 | 41600 | 32.7 | 1.38 | EPI_DAR_3hr |
| 25 | 8 | MA0742.2 (KLF12) | STREME-4 | 4-CGCCCC | CGCCCC | KLF12 | 1.57e-167 | 4.13e-169 | 6920 | 54 | 52300 | 41.2 | 1.31 | EPI_DAR_3hr |
| 26 | 1 | MA0493.2 | KLF1 | MA0493.2 | DGGGYGGGG | KLF1 | 1.5e-164 | 3.8e-166 | 7640 | 59.6 | 59600 | 46.9 | 1.27 | EPI_DAR_3hr |
| 27 | 1 | MA0039.4 | KLF4 | MA0039.4 | NNCCCCACCCHN | KLF4 | 3.43e-162 | 8.35e-164 | 7450 | 58.1 | 57800 | 45.5 | 1.28 | EPI_DAR_3hr |
| 28 | 19 | MA1965.1 | SP5 | MA1965.1 | CBCCTCCCHS | SP5 | 3.73e-161 | 8.77e-163 | 6080 | 47.4 | 44600 | 35.1 | 1.35 | EPI_DAR_3hr |
| 29 | 1 | MA0079.5 (SP1) | STREME-5 | 5-CCCCGC | CCCCGC | SP1 | 1.24e-157 | 2.82e-159 | 5580 | 43.6 | 40200 | 31.6 | 1.38 | EPI_DAR_3hr |
| 30 | 1 | MA0741.1 | KLF16 | MA0741.1 | GMCACGCCCCC | KLF16 | 5.97e-153 | 1.31e-154 | 4690 | 36.6 | 32300 | 25.4 | 1.44 | EPI_DAR_3hr |
| 31 | 9 | MA1976.1 | ZNF320 | MA1976.1 | DGKGGGRCCMRGGGGCCWGTGHVNNNN | ZNF320 | 2.37e-151 | 5.03e-153 | 6960 | 54.3 | 53600 | 42.1 | 1.29 | EPI_DAR_3hr |
| 32 | 10 | MA1517.1 | KLF6 | MA1517.1 | NRCCACGCCCH | KLF6 | 9.66e-151 | 1.99e-152 | 6040 | 47.2 | 44800 | 35.2 | 1.34 | EPI_DAR_3hr |
| 33 | 4 | MA1512.1 | KLF11 | MA1512.1 | SCCACGCCCMC | KLF11 | 2.92e-150 | 5.83e-152 | 7150 | 55.8 | 55500 | 43.6 | 1.28 | EPI_DAR_3hr |
| 34 | 10 | MA0746.2 | SP3 | MA0746.2 | NGCCACGCCCMCH | SP3 | 2.68e-149 | 5.19e-151 | 6860 | 53.6 | 52700 | 41.4 | 1.29 | EPI_DAR_3hr |
| 35 | 11 | MA0506.2 | Nrf1 | MA0506.2 | SNCTGCGCMTGCGCV | Nrf1 | 1.53e-144 | 2.87e-146 | 6310 | 49.3 | 47700 | 37.5 | 1.31 | EPI_DAR_3hr |
| 36 | 12 | MA1122.1 | TFDP1 | MA1122.1 | VSGCGGGAAVN | TFDP1 | 1.41e-143 | 2.57e-145 | 7060 | 55.1 | 55000 | 43.2 | 1.28 | EPI_DAR_3hr |
| 37 | 13 | MA1596.1 | ZNF460 | MA1596.1 | GCCTCMGCCTCCCRAG | ZNF460 | 4.85e-141 | 8.62e-143 | 5310 | 41.4 | 38400 | 30.2 | 1.37 | EPI_DAR_3hr |
| 38 | 4 | MA0733.1 | EGR4 | MA0733.1 | NHMCGCCCACGCAHWT | EGR4 | 1.66e-139 | 2.87e-141 | 8720 | 68.1 | 72000 | 56.6 | 1.2 | EPI_DAR_3hr |
| 39 | 14 | MA1981.1 | ZNF530 | MA1981.1 | GMARGGMRAGGGGCRGSV | ZNF530 | 1.56e-138 | 2.63e-140 | 6960 | 54.3 | 54200 | 42.6 | 1.27 | EPI_DAR_3hr |
| 40 | 4 | MA0732.1 | EGR3 | MA0732.1 | HHMCGCCCACGCAHH | EGR3 | 5.48e-136 | 9.02e-138 | 3480 | 27.2 | 22500 | 17.7 | 1.53 | EPI_DAR_3hr |
| 41 | 15 | MA1712.1 | ZNF454 | MA1712.1 | DGGCTCSSGGCCCYGVKG | ZNF454 | 6.96e-136 | 1.12e-137 | 3390 | 26.4 | 21700 | 17.1 | 1.55 | EPI_DAR_3hr |
| 42 | 4 | MA0747.1 | SP8 | MA0747.1 | RCCACGCCCMCY | SP8 | 8.03e-136 | 1.26e-137 | 5900 | 46 | 44200 | 34.7 | 1.33 | EPI_DAR_3hr |
| 43 | 16 | MA1548.1 | PLAGL2 | MA1548.1 | NGGGCCCCCN | PLAGL2 | 3.23e-135 | 4.94e-137 | 6360 | 49.6 | 48600 | 38.2 | 1.3 | EPI_DAR_3hr |
| 44 | 10 | MA1516.1 | KLF3 | MA1516.1 | GRCCRCGCCCH | KLF3 | 3.11e-134 | 4.66e-136 | 7790 | 60.8 | 62700 | 49.3 | 1.23 | EPI_DAR_3hr |
| 45 | 17 | MA1578.1 | VEZF1 | MA1578.1 | CCCCCCMYDH | VEZF1 | 2.62e-128 | 3.83e-130 | 7740 | 60.4 | 62500 | 49.2 | 1.23 | EPI_DAR_3hr |
| 46 | 18 | MA1986.1 | ZNF692 | MA1986.1 | BGGGCCCASN | ZNF692 | 2.81e-126 | 4.02e-128 | 5210 | 40.7 | 38300 | 30.1 | 1.35 | EPI_DAR_3hr |
| 47 | 12 | MA0471.2 | E2F6 | MA0471.2 | VVDGGCGGGAAVV | E2F6 | 1.7e-125 | 2.38e-127 | 8410 | 65.6 | 69500 | 54.7 | 1.2 | EPI_DAR_3hr |
| 48 | 4 | MA1107.2 | KLF9 | MA1107.2 | MNGCCACACCCACHYV | KLF9 | 2.55e-124 | 3.5e-126 | 5200 | 40.6 | 38300 | 30.1 | 1.35 | EPI_DAR_3hr |
| 49 | 19 | MA1653.1 (ZNF148) | STREME-8 | 8-CCCCTCC | CCCCTCC | ZNF148 | 8.15e-121 | 1.09e-122 | 8020 | 62.6 | 65800 | 51.8 | 1.21 | EPI_DAR_3hr |
| 50 | 20 | MA0131.2 | HINFP | MA0131.2 | CARCGTCCGCKN | HINFP | 6.19e-120 | 8.15e-122 | 7640 | 59.6 | 62000 | 48.7 | 1.22 | EPI_DAR_3hr |
| 51 | 21 | MA0814.2 | TFAP2C | MA0814.2 | NNGCCTGAGGCGNN | TFAP2C | 1.3e-118 | 1.67e-120 | 7780 | 60.7 | 63500 | 49.9 | 1.22 | EPI_DAR_3hr |
| 52 | 4 | MA0472.2 | EGR2 | MA0472.2 | MCGCCCACGCA | EGR2 | 1.41e-114 | 1.79e-116 | 6000 | 46.9 | 46300 | 36.4 | 1.29 | EPI_DAR_3hr |
| 53 | 22 | MA1615.1 | Plagl1 | MA1615.1 | NNCTGGGGCCABN | Plagl1 | 1.8e-114 | 2.23e-116 | 7150 | 55.8 | 57400 | 45.2 | 1.24 | EPI_DAR_3hr |
| 54 | 23 | MA0810.1 | TFAP2A | MA0810.1 | YGCCCBVRGGCR | TFAP2A | 9.38e-112 | 1.14e-113 | 9240 | 72.2 | 79100 | 62.2 | 1.16 | EPI_DAR_3hr |
| 55 | 24 | MA1102.2 | CTCFL | MA1102.2 | NSCAGGGGGCGS | CTCFL | 6.8e-108 | 8.14e-110 | 5070 | 39.6 | 37900 | 29.8 | 1.33 | EPI_DAR_3hr |
| 56 | 25 | MA0146.2 | Zfx | MA0146.2 | SSSGCCBVGGCCTS | Zfx | 3.38e-107 | 3.97e-109 | 5590 | 43.6 | 42800 | 33.6 | 1.3 | EPI_DAR_3hr |
| 57 | 21 | MA0812.1 | TFAP2B | MA0812.1 | HGCCTSAGGCD | TFAP2B | 2.22e-105 | 2.57e-107 | 7200 | 56.2 | 58500 | 46 | 1.22 | EPI_DAR_3hr |
| 58 | 26 | MA0528.2 | ZNF263 | MA0528.2 | VNGGGGAGGASG | ZNF263 | 5.08e-105 | 5.76e-107 | 5460 | 42.6 | 41700 | 32.8 | 1.3 | EPI_DAR_3hr |
| 59 | 27 | MA1721.1 | ZNF93 | MA1721.1 | GGYAGCRGCAGCGGYG | ZNF93 | 9.85e-105 | 1.1e-106 | 7410 | 57.8 | 60600 | 47.6 | 1.21 | EPI_DAR_3hr |
| 60 | 10 | MA1514.1 | KLF17 | MA1514.1 | MMCCACGCACCCMTY | KLF17 | 3.34e-103 | 3.66e-105 | 7530 | 58.8 | 61900 | 48.7 | 1.21 | EPI_DAR_3hr |
| 61 | 9 | MA1723.1 | PRDM9 | MA1723.1 | RGDGGGVAGGGRGGVRRMARVARR | PRDM9 | 3.52e-97 | 3.8e-99 | 6440 | 50.2 | 51500 | 40.5 | 1.24 | EPI_DAR_3hr |
| 62 | 23 | MA0811.1 | TFAP2B | MA0811.1 | YGCCCBVRGGCA | TFAP2B | 8.39e-97 | 8.91e-99 | 9380 | 73.2 | 81400 | 64 | 1.14 | EPI_DAR_3hr |
| 63 | 23 | MA0872.1 | TFAP2A | MA0872.1 | TGCCCYSRGGGCA | TFAP2A | 3.15e-96 | 3.29e-98 | 3800 | 29.6 | 27000 | 21.3 | 1.39 | EPI_DAR_3hr |
| 64 | 28 | MA0597.2 | THAP1 | MA0597.2 | VSGCAGGGCASV | THAP1 | 1.31e-94 | 1.35e-96 | 7970 | 62.2 | 66900 | 52.6 | 1.18 | EPI_DAR_3hr |
| 65 | 29 | MA0865.2 | E2F8 | MA0865.2 | TTCCCGCCAHWA | E2F8 | 1.71e-94 | 1.73e-96 | 7140 | 55.8 | 58600 | 46.1 | 1.21 | EPI_DAR_3hr |
| 66 | 30 | MA0799.2 (RFX4) | STREME-9 | 9-GCCWGGC | GCCWGGC | RFX4 | 3.66e-93 | 3.65e-95 | 5650 | 44.1 | 44200 | 34.8 | 1.27 | EPI_DAR_3hr |
| 67 | 23 | MA0815.1 | TFAP2C | MA0815.1 | TGCCCYSRGGGCA | TFAP2C | 4.25e-93 | 4.17e-95 | 3320 | 25.9 | 23000 | 18.1 | 1.43 | EPI_DAR_3hr |
| 68 | 21 | MA1569.1 | TFAP2E | MA1569.1 | YGCCYSRGGCD | TFAP2E | 7.55e-93 | 7.31e-95 | 2570 | 20 | 16600 | 13.1 | 1.53 | EPI_DAR_3hr |
| 69 | 31 | MA0734.3 | Gli2 | MA0734.3 | NRGACCACCCASV | Gli2 | 4.81e-91 | 4.58e-93 | 8260 | 64.5 | 70100 | 55.1 | 1.17 | EPI_DAR_3hr |
| 70 | 32 | MA1599.1 | ZNF682 | MA1599.1 | BVGGCYAAGCCCCWNY | ZNF682 | 6.8e-91 | 6.4e-93 | 8040 | 62.8 | 67900 | 53.4 | 1.18 | EPI_DAR_3hr |
| 71 | 12 | MA1583.1 | ZFP57 | MA1583.1 | NNVTTGCCGCANN | ZFP57 | 6.53e-90 | 6.05e-92 | 6270 | 49 | 50400 | 39.6 | 1.24 | EPI_DAR_3hr |
| 72 | 23 | MA0813.1 | TFAP2B | MA0813.1 | TGCCCYBRGGGCA | TFAP2B | 3.82e-89 | 3.49e-91 | 3190 | 24.9 | 22100 | 17.4 | 1.43 | EPI_DAR_3hr |
| 73 | 1 | MA0599.1 | KLF5 | MA0599.1 | GCCCCDCCCH | KLF5 | 6.28e-86 | 5.66e-88 | 1880 | 14.6 | 11300 | 8.89 | 1.65 | EPI_DAR_3hr |
| 74 | 17 | MA0736.1 | GLIS2 | MA0736.1 | GACCCCCCGCRAMG | GLIS2 | 1.58e-85 | 1.41e-87 | 5080 | 39.7 | 39300 | 30.9 | 1.28 | EPI_DAR_3hr |
| 75 | 23 | MA0524.2 | TFAP2C | MA0524.2 | YGCCYBVRGGCA | TFAP2C | 2.22e-85 | 1.95e-87 | 3170 | 24.8 | 22100 | 17.4 | 1.42 | EPI_DAR_3hr |
| 76 | 21 | MA0003.4 | TFAP2A | MA0003.4 | NNKGCCTCAGGCNN | TFAP2A | 1.28e-84 | 1.11e-86 | 5190 | 40.5 | 40400 | 31.8 | 1.28 | EPI_DAR_3hr |
| 77 | 33 | MA1648.1 | TCF12 | MA1648.1 | NNCACCTGCNN | TCF12 | 4.76e-82 | 4.07e-84 | 7250 | 56.6 | 60500 | 47.6 | 1.19 | EPI_DAR_3hr |
| 78 | 34 | MA1719.1 | ZNF816 | MA1719.1 | DRRKGGGGACATGCHGGG | ZNF816 | 6.23e-82 | 5.25e-84 | 8220 | 64.2 | 70300 | 55.3 | 1.16 | EPI_DAR_3hr |
| 79 | 33 | MA0522.3 | TCF3 | MA0522.3 | NVCACCTGCNN | TCF3 | 1.69e-81 | 1.41e-83 | 8140 | 63.5 | 69500 | 54.6 | 1.16 | EPI_DAR_3hr |
| 80 | 29 | MA0024.3 | E2F1 | MA0024.3 | TTTGGCGCCAAA | E2F1 | 2.52e-81 | 2.07e-83 | 3850 | 30.1 | 28300 | 22.3 | 1.35 | EPI_DAR_3hr |
| 81 | 35 | MA0751.1 | ZIC4 | MA0751.1 | GRCCCCCCGCKGYGH | ZIC4 | 6.79e-79 | 5.52e-81 | 7860 | 61.3 | 66800 | 52.5 | 1.17 | EPI_DAR_3hr |
| 82 | 33 | MA0823.1 | HEY1 | MA0823.1 | GRCACGTGCC | HEY1 | 4.72e-78 | 3.79e-80 | 5230 | 40.8 | 41200 | 32.4 | 1.26 | EPI_DAR_3hr |
| 83 | 31 | MA1990.1 | Gli1 | MA1990.1 | NNRGACCACCCASV | Gli1 | 9.97e-78 | 7.91e-80 | 7540 | 58.8 | 63700 | 50.1 | 1.18 | EPI_DAR_3hr |
| 84 | 36 | MA0759.2 | ELK3 | MA0759.2 | ACCGGAAGTRV | ELK3 | 6.65e-76 | 5.21e-78 | 4050 | 31.6 | 30400 | 23.9 | 1.32 | EPI_DAR_3hr |
| 85 | 4 | MA1656.1 | ZNF449 | MA1656.1 | NNAAGCCCAACCNN | ZNF449 | 4.32e-75 | 3.35e-77 | 7310 | 57.1 | 61600 | 48.4 | 1.18 | EPI_DAR_3hr |
| 86 | 29 | MA0527.1 | ZBTB33 | MA0527.1 | BTCTCGCGAGABYTS | ZBTB33 | 1.22e-71 | 9.31e-74 | 4620 | 36 | 35900 | 28.2 | 1.28 | EPI_DAR_3hr |
| 87 | 33 | MA0632.2 | TCFL5 | MA0632.2 | KCACGCGCMC | TCFL5 | 4.71e-71 | 3.56e-73 | 3740 | 29.2 | 27900 | 22 | 1.33 | EPI_DAR_3hr |
| 88 | 29 | MA0748.2 | YY2 | MA0748.2 | NVATGGCGGCS | YY2 | 4.11e-70 | 3.07e-72 | 4430 | 34.6 | 34300 | 27 | 1.28 | EPI_DAR_3hr |
| 89 | 33 | MA0830.2 | TCF4 | MA0830.2 | NNGCACCTGCCNN | TCF4 | 2.87e-67 | 2.12e-69 | 6170 | 48.2 | 50900 | 40 | 1.2 | EPI_DAR_3hr |
| 90 | 37 | 7-GCGYCW | STREME-7 | 7-GCGYCW | GCGYCW | GCGYCW | 3.71e-67 | 2.71e-69 | 9940 | 77.6 | 89400 | 70.3 | 1.1 | EPI_DAR_3hr |
| 91 | 38 | MA1985.1 | ZNF669 | MA1985.1 | GGGGYGAYGACCRCT | ZNF669 | 1.03e-65 | 7.48e-68 | 3720 | 29 | 28000 | 22 | 1.32 | EPI_DAR_3hr |
| 92 | 33 | MA1106.1 | HIF1A | MA1106.1 | SBACGTGCNN | HIF1A | 4.16e-65 | 2.97e-67 | 6950 | 54.2 | 58700 | 46.2 | 1.17 | EPI_DAR_3hr |
| 93 | 24 | MA0155.1 | INSM1 | MA0155.1 | TGYCWGGGGGCR | INSM1 | 1.78e-64 | 1.26e-66 | 3910 | 30.5 | 29900 | 23.5 | 1.3 | EPI_DAR_3hr |
| 94 | 39 | MA1727.1 | ZNF417 | MA1727.1 | VRBVNTGGGCGCCAM | ZNF417 | 2.26e-61 | 1.58e-63 | 4670 | 36.4 | 37100 | 29.2 | 1.25 | EPI_DAR_3hr |
| 95 | 26 | MA1987.1 | ZNF701 | MA1987.1 | GAGCACYAARGGGGGRADDDD | ZNF701 | 1.11e-59 | 7.71e-62 | 9000 | 70.2 | 80000 | 62.9 | 1.12 | EPI_DAR_3hr |
| 96 | 13 | MA1587.1 | ZNF135 | MA1587.1 | CCTCGACCTCCYRR | ZNF135 | 3.23e-57 | 2.22e-59 | 6560 | 51.2 | 55500 | 43.6 | 1.17 | EPI_DAR_3hr |
| 97 | 33 | MA1628.1 | Zic1::Zic2 | MA1628.1 | CVCAGCAGGNV | Zic1::Zic2 | 7.75e-57 | 5.26e-59 | 4690 | 36.6 | 37600 | 29.6 | 1.24 | EPI_DAR_3hr |
| 98 | 29 | MA0864.2 | E2F2 | MA0864.2 | RWTTTGGCGCCAWWWY | E2F2 | 1.48e-55 | 9.95e-58 | 4520 | 35.2 | 36100 | 28.4 | 1.24 | EPI_DAR_3hr |
| 99 | 33 | MA1945.1 | ETV5::FIGLA | MA1945.1 | RGCGGAARCAGGTGG | ETV5::FIGLA | 1.53e-55 | 1.02e-57 | 5710 | 44.6 | 47400 | 37.3 | 1.2 | EPI_DAR_3hr |
| 100 | 40 | MA1710.1 | ZNF257 | MA1710.1 | VDGAGGCRAGRG | ZNF257 | 2.73e-55 | 1.79e-57 | 6060 | 47.3 | 50800 | 40 | 1.18 | EPI_DAR_3hr |
| 101 | 33 | MA1997.1 | Olig2 | MA1997.1 | NVCAGCTGBN | Olig2 | 1.36e-54 | 8.88e-57 | 5480 | 42.8 | 45300 | 35.6 | 1.2 | EPI_DAR_3hr |
| 102 | 31 | MA0695.1 | ZBTB7C | MA0695.1 | RCGACCACCGAN | ZBTB7C | 3.77e-54 | 2.43e-56 | 6470 | 50.5 | 54900 | 43.2 | 1.17 | EPI_DAR_3hr |
| 103 | 31 | MA0694.1 | ZBTB7B | MA0694.1 | RCGACCACCGAA | ZBTB7B | 1.8e-53 | 1.15e-55 | 3640 | 28.4 | 28200 | 22.2 | 1.28 | EPI_DAR_3hr |
| 104 | 33 | MA0821.2 | HES5 | MA0821.2 | GRCACGTGYC | HES5 | 1.12e-52 | 7.1e-55 | 9360 | 73.1 | 84400 | 66.4 | 1.1 | EPI_DAR_3hr |
| 105 | 41 | MA1972.1 | ZFP14 | MA1972.1 | GGAGGCHCHGGARBG | ZFP14 | 3.09e-52 | 1.94e-54 | 7320 | 57.1 | 63500 | 49.9 | 1.14 | EPI_DAR_3hr |
| 106 | 42 | MA0615.1 | Gmeb1 | MA0615.1 | BHBBKKACGTMMNWNNN | Gmeb1 | 8.94e-52 | 5.55e-54 | 3460 | 27 | 26600 | 21 | 1.29 | EPI_DAR_3hr |
| 107 | 43 | MA0014.3 | PAX5 | MA0014.3 | RRGCGTGACCNN | PAX5 | 3.52e-51 | 2.17e-53 | 4980 | 38.8 | 40700 | 32 | 1.21 | EPI_DAR_3hr |
| 108 | 33 | MA0633.2 | Twist2 | MA0633.2 | NVCAGCTGBN | Twist2 | 3.96e-50 | 2.41e-52 | 4280 | 33.4 | 34300 | 27 | 1.24 | EPI_DAR_3hr |
| 109 | 33 | MA0147.3 | MYC | MA0147.3 | NNCCACGTGCNB | MYC | 8.81e-50 | 5.32e-52 | 6320 | 49.3 | 53800 | 42.3 | 1.17 | EPI_DAR_3hr |
| 110 | 33 | MA1631.1 | ASCL1 | MA1631.1 | NNGCACCTGCYNB | ASCL1 | 7.43e-49 | 4.45e-51 | 4410 | 34.4 | 35600 | 28 | 1.23 | EPI_DAR_3hr |
| 111 | 33 | MA0616.2 | HES2 | MA0616.2 | GGCACGTGYC | HES2 | 1.09e-48 | 6.45e-51 | 4540 | 35.4 | 36800 | 28.9 | 1.22 | EPI_DAR_3hr |
| 112 | 44 | MA1528.1 | NFIX | MA1528.1 | STTGGCRNNGTGCCARB | NFIX | 3.8e-48 | 2.23e-50 | 3340 | 26.1 | 25800 | 20.3 | 1.29 | EPI_DAR_3hr |
| 113 | 45 | MA0259.1 | ARNT::HIF1A | MA0259.1 | VBACGTGC | ARNT::HIF1A | 6.09e-48 | 3.55e-50 | 4600 | 35.9 | 37400 | 29.4 | 1.22 | EPI_DAR_3hr |
| 114 | 35 | MA0696.1 | ZIC1 | MA0696.1 | GACCCCCYGCTGTG | ZIC1 | 3.23e-47 | 1.86e-49 | 3380 | 26.4 | 26200 | 20.6 | 1.28 | EPI_DAR_3hr |
| 115 | 26 | MA1728.1 | ZNF549 | MA1728.1 | NNTGCTGCCCWR | ZNF549 | 1.53e-46 | 8.74e-49 | 6430 | 50.2 | 55200 | 43.4 | 1.16 | EPI_DAR_3hr |
| 116 | 46 | MA1982.1 | ZNF574 | MA1982.1 | VGSCTAGAGMGGCCS | ZNF574 | 2.7e-45 | 1.53e-47 | 8280 | 64.6 | 73800 | 58 | 1.11 | EPI_DAR_3hr |
| 117 | 33 | MA1099.2 | HES1 | MA1099.2 | GGCRCGTGBC | HES1 | 3.45e-45 | 1.94e-47 | 2470 | 19.3 | 18300 | 14.4 | 1.35 | EPI_DAR_3hr |
| 118 | 33 | MA0104.4 | MYCN | MA0104.4 | VVCCACGTGGBB | MYCN | 3.66e-45 | 2.04e-47 | 8760 | 68.4 | 78800 | 61.9 | 1.1 | EPI_DAR_3hr |
| 119 | 33 | MA1993.1 | Neurod2 | MA1993.1 | VVCAGCTGBB | Neurod2 | 3.72e-45 | 2.06e-47 | 4130 | 32.2 | 33300 | 26.2 | 1.23 | EPI_DAR_3hr |
| 120 | 47 | MA1637.1 | EBF3 | MA1637.1 | NYCCCAAGGGANN | EBF3 | 3.98e-45 | 2.19e-47 | 8690 | 67.8 | 78000 | 61.4 | 1.11 | EPI_DAR_3hr |
| 121 | 36 | MA0750.2 | ZBTB7A | MA0750.2 | NVCCGGAAGTGSV | ZBTB7A | 4.22e-45 | 2.3e-47 | 2460 | 19.2 | 18100 | 14.2 | 1.35 | EPI_DAR_3hr |
| 122 | 33 | MA0103.3 | ZEB1 | MA0103.3 | SNCACCTGSVN | ZEB1 | 9.58e-45 | 5.17e-47 | 7900 | 61.6 | 70000 | 55 | 1.12 | EPI_DAR_3hr |
| 123 | 33 | MA1966.1 | TFAP4::ETV1 | MA1966.1 | RSCGGAAGCAGSTGKN | TFAP4::ETV1 | 4.69e-44 | 2.51e-46 | 2550 | 19.9 | 19000 | 15 | 1.33 | EPI_DAR_3hr |
| 124 | 48 | MA1684.1 | Foxn1 | MA1684.1 | RGAMGC | Foxn1 | 6.31e-44 | 3.35e-46 | 1880 | 14.7 | 13200 | 10.4 | 1.41 | EPI_DAR_3hr |
| 125 | 49 | MA0504.1 | NR2C2 | MA0504.1 | VGRGGTCARAGGTCA | NR2C2 | 8.85e-43 | 4.66e-45 | 7460 | 58.3 | 65800 | 51.7 | 1.13 | EPI_DAR_3hr |
| 126 | 35 | MA1584.1 | ZIC5 | MA1584.1 | VGACCCCCCGCTGHGM | ZIC5 | 9.42e-43 | 4.92e-45 | 1670 | 13 | 11500 | 9.02 | 1.44 | EPI_DAR_3hr |
| 127 | 9 | MA0163.1 | PLAG1 | MA0163.1 | GGGGCCCWAGGGGG | PLAG1 | 2.58e-41 | 1.34e-43 | 1810 | 14.1 | 12800 | 10 | 1.41 | EPI_DAR_3hr |
| 128 | 33 | MA1941.1 | ETV2::FIGLA | MA1941.1 | RSCGGAARCAGSTGBN | ETV2::FIGLA | 1.95e-40 | 1e-42 | 8700 | 67.9 | 78500 | 61.8 | 1.1 | EPI_DAR_3hr |
| 129 | 33 | MA1635.1 | BHLHE22 | MA1635.1 | NVCAGCTGBN | BHLHE22 | 3.16e-40 | 1.61e-42 | 4290 | 33.5 | 35200 | 27.7 | 1.21 | EPI_DAR_3hr |
| 130 | 50 | MA1964.1 | SMAD2 | MA1964.1 | NBCCAGACDN | SMAD2 | 1.14e-38 | 5.79e-41 | 6030 | 47.1 | 52000 | 40.9 | 1.15 | EPI_DAR_3hr |
| 131 | 51 | MA1973.1 | ZKSCAN3 | MA1973.1 | KDKCCCAGGCTAGCCCABNN | ZKSCAN3 | 1.77e-38 | 8.9e-41 | 5000 | 39 | 42100 | 33.1 | 1.18 | EPI_DAR_3hr |
| 132 | 29 | MA0470.2 | E2F4 | MA0470.2 | TTTTGGCGCCAWWW | E2F4 | 2.46e-38 | 1.23e-40 | 2750 | 21.5 | 21200 | 16.7 | 1.29 | EPI_DAR_3hr |
| 133 | 44 | MA1643.1 | NFIB | MA1643.1 | NNCCTGGCAYNGTGCCAAGNN | NFIB | 1.07e-35 | 5.28e-38 | 4620 | 36.1 | 38800 | 30.5 | 1.18 | EPI_DAR_3hr |
| 134 | 52 | MA1726.1 | ZNF331 | MA1726.1 | NMYTGCAGAGCCCH | ZNF331 | 1.35e-35 | 6.64e-38 | 8930 | 69.7 | 81500 | 64 | 1.09 | EPI_DAR_3hr |
| 135 | 47 | MA0154.4 | EBF1 | MA0154.4 | NNTCCCCAGGGGANN | EBF1 | 8.79e-35 | 4.28e-37 | 8880 | 69.4 | 81000 | 63.7 | 1.09 | EPI_DAR_3hr |
| 136 | 53 | MA1474.1 | CREB3L4 | MA1474.1 | YGCCACGTCAYC | CREB3L4 | 1.73e-34 | 8.36e-37 | 3540 | 27.6 | 28700 | 22.5 | 1.23 | EPI_DAR_3hr |
| 137 | 54 | MA0767.1 | GCM2 | MA0767.1 | BATGCGGGTR | GCM2 | 3.51e-34 | 1.69e-36 | 3080 | 24 | 24500 | 19.3 | 1.25 | EPI_DAR_3hr |
| 138 | 36 | MA0765.3 | ETV5 | MA0765.3 | ACCGGAAGTR | ETV5 | 5.32e-34 | 2.54e-36 | 4800 | 37.5 | 40600 | 32 | 1.17 | EPI_DAR_3hr |
| 139 | 47 | MA1604.1 | Ebf2 | MA1604.1 | NYCCCAAGGGANN | Ebf2 | 3.06e-33 | 1.45e-35 | 8870 | 69.2 | 81000 | 63.7 | 1.09 | EPI_DAR_3hr |
| 140 | 36 | MA0764.3 | ETV4 | MA0764.3 | ACCGGAAGTR | ETV4 | 3.38e-33 | 1.59e-35 | 8510 | 66.4 | 77300 | 60.8 | 1.09 | EPI_DAR_3hr |
| 141 | 55 | MA1149.1 | RARA::RXRG | MA1149.1 | RRGGTCAHNNNRRGGTCA | RARA::RXRG | 1.28e-31 | 5.95e-34 | 8820 | 68.8 | 80700 | 63.5 | 1.08 | EPI_DAR_3hr |
| 142 | 54 | MA0646.1 | GCM1 | MA0646.1 | BATGCGGGTAC | GCM1 | 1.35e-30 | 6.24e-33 | 5010 | 39.1 | 43000 | 33.8 | 1.16 | EPI_DAR_3hr |
| 143 | 33 | MA0697.2 | Zic3 | MA0697.2 | CNCAGCAGGAGNN | Zic3 | 6.24e-29 | 2.87e-31 | 10000 | 78 | 93300 | 73.4 | 1.06 | EPI_DAR_3hr |
| 144 | 56 | MA1730.1 | ZNF708 | MA1730.1 | NNRGCTGTRCCTCCY | ZNF708 | 4.68e-28 | 2.14e-30 | 9120 | 71.2 | 84200 | 66.2 | 1.08 | EPI_DAR_3hr |
| 145 | 33 | MA1629.1 | Zic2 | MA1629.1 | NDCACAGCAGGDRG | Zic2 | 1.54e-27 | 7.01e-30 | 10400 | 81.2 | 97800 | 76.9 | 1.06 | EPI_DAR_3hr |
| 146 | 57 | MA1711.1 | ZNF343 | MA1711.1 | CCGCTTCMCCDCGGCMV | ZNF343 | 1.51e-26 | 6.78e-29 | 2800 | 21.9 | 22600 | 17.8 | 1.23 | EPI_DAR_3hr |
| 147 | 45 | MA0006.1 | Ahr::Arnt | MA0006.1 | YGCGTG | Ahr::Arnt | 7.76e-26 | 3.47e-28 | 1790 | 14 | 13600 | 10.7 | 1.31 | EPI_DAR_3hr |
| 148 | 43 | MA0067.2 | PAX2 | MA0067.2 | BCAVTSRAGCGTGACGR | PAX2 | 1.22e-25 | 5.41e-28 | 3090 | 24.1 | 25300 | 19.9 | 1.21 | EPI_DAR_3hr |
| 149 | 24 | MA1929.1 | CTCF | MA1929.1 | CTGCAGTKCCNNNNNYNRCCASYAGRKGGCRSWG | CTCF | 1.14e-24 | 5.02e-27 | 3480 | 27.1 | 29000 | 22.8 | 1.19 | EPI_DAR_3hr |
| 150 | 33 | MA0649.1 | HEY2 | MA0649.1 | GRCACGTGYC | HEY2 | 6.47e-24 | 2.84e-26 | 1950 | 15.2 | 15100 | 11.9 | 1.28 | EPI_DAR_3hr |
| 151 | 58 | MA0511.2 | RUNX2 | MA0511.2 | WAACCGCAA | RUNX2 | 1.58e-23 | 6.87e-26 | 9250 | 72.2 | 86100 | 67.7 | 1.07 | EPI_DAR_3hr |
| 152 | 33 | MA0499.2 | MYOD1 | MA0499.2 | NNGCACCTGTCNB | MYOD1 | 3.81e-23 | 1.65e-25 | 6820 | 53.2 | 61500 | 48.4 | 1.1 | EPI_DAR_3hr |
| 153 | 59 | MA1529.1 | NHLH2 | MA1529.1 | GGGNCGCAGCTGCGYCCC | NHLH2 | 4.65e-23 | 2e-25 | 3050 | 23.8 | 25200 | 19.8 | 1.2 | EPI_DAR_3hr |
| 154 | 60 | MA0738.1 | HIC2 | MA0738.1 | RTGCCCRSB | HIC2 | 2.88e-22 | 1.23e-24 | 11600 | 90.6 | 111000 | 87.6 | 1.03 | EPI_DAR_3hr |
| 155 | 61 | MA0145.2 | Tfcp2l1 | MA0145.2 | CCAGYYYVADCCRG | Tfcp2l1 | 3.08e-22 | 1.31e-24 | 2790 | 21.8 | 22900 | 18 | 1.21 | EPI_DAR_3hr |
| 156 | 62 | MA1971.1 | ZBED2 | MA1971.1 | NNMMCGAAACCNNV | ZBED2 | 3.54e-22 | 1.49e-24 | 702 | 5.48 | 4530 | 3.56 | 1.54 | EPI_DAR_3hr |
| 157 | 63 | MA1979.1 | ZNF416 | MA1979.1 | TATCTGGGCAH | ZNF416 | 2.4e-21 | 1.01e-23 | 8890 | 69.4 | 82600 | 65 | 1.07 | EPI_DAR_3hr |
| 158 | 36 | MA0028.2 | ELK1 | MA0028.2 | ACCGGAAGTR | ELK1 | 3.12e-21 | 1.3e-23 | 5530 | 43.2 | 49100 | 38.6 | 1.12 | EPI_DAR_3hr |
| 159 | 49 | MA1535.1 | NR2C1 | MA1535.1 | NRRGGTCAN | NR2C1 | 5.4e-21 | 2.23e-23 | 10900 | 85.2 | 104000 | 81.8 | 1.04 | EPI_DAR_3hr |
| 160 | 64 | MA1545.1 | OVOL2 | MA1545.1 | DDACCGTTABNYN | OVOL2 | 7.28e-21 | 3e-23 | 1790 | 14 | 14000 | 11 | 1.28 | EPI_DAR_3hr |
| 161 | 65 | MA1731.1 | ZNF768 | MA1731.1 | YBVCYBRSCCTCTCTGDG | ZNF768 | 7.48e-21 | 3.06e-23 | 5560 | 43.4 | 49400 | 38.9 | 1.12 | EPI_DAR_3hr |
| 162 | 33 | MA1100.2 | ASCL1 | MA1100.2 | VGCAGCTGCN | ASCL1 | 2.64e-20 | 1.07e-22 | 8390 | 65.5 | 77700 | 61.1 | 1.07 | EPI_DAR_3hr |
| 163 | 33 | MA2002.1 | Zfp335 | MA2002.1 | VNTCAGGCANH | Zfp335 | 5.17e-20 | 2.09e-22 | 8600 | 67.2 | 79900 | 62.8 | 1.07 | EPI_DAR_3hr |
| 164 | 36 | MA0076.2 | ELK4 | MA0076.2 | BCRCTTCCGGB | ELK4 | 9.58e-20 | 3.84e-22 | 327 | 2.55 | 1740 | 1.36 | 1.88 | EPI_DAR_3hr |
| 165 | 36 | MA1933.1 | ELK1::SREBF2 | MA1933.1 | DCCGGAAGTSRCGTGA | ELK1::SREBF2 | 1.05e-19 | 4.17e-22 | 2540 | 19.8 | 20800 | 16.4 | 1.21 | EPI_DAR_3hr |
| 166 | 33 | MA1934.1 | ERF::FIGLA | MA1934.1 | RCCGGAARCASCTGBN | ERF::FIGLA | 1.53e-19 | 6.08e-22 | 2720 | 21.2 | 22500 | 17.7 | 1.2 | EPI_DAR_3hr |
| 167 | 33 | MA0745.2 | SNAI2 | MA0745.2 | NBGCACCTGTMNY | SNAI2 | 1.68e-19 | 6.61e-22 | 9950 | 77.7 | 93900 | 73.8 | 1.05 | EPI_DAR_3hr |
| 168 | 66 | MA1715.1 | ZNF707 | MA1715.1 | CCCCACTCCTGGTAC | ZNF707 | 1.78e-19 | 6.96e-22 | 5190 | 40.5 | 46000 | 36.2 | 1.12 | EPI_DAR_3hr |
| 169 | 33 | MA0048.2 | NHLH1 | MA0048.2 | CGCAGCTGCK | NHLH1 | 2.85e-19 | 1.11e-21 | 7840 | 61.2 | 72300 | 56.8 | 1.08 | EPI_DAR_3hr |
| 170 | 36 | MA1938.1 | ERF::NHLH1 | MA1938.1 | VGCAGCTGCCGGAARYN | ERF::NHLH1 | 3.35e-19 | 1.3e-21 | 3190 | 24.9 | 26900 | 21.2 | 1.18 | EPI_DAR_3hr |
| 171 | 67 | MA1575.1 | THRB | MA1575.1 | DTGACCTBRNYVAGGTCAH | THRB | 6.07e-19 | 2.34e-21 | 2370 | 18.5 | 19300 | 15.2 | 1.22 | EPI_DAR_3hr |
| 172 | 67 | MA1533.1 | NR1I2 | MA1533.1 | DYGAACTSVDTGAACTC | NR1I2 | 7.69e-19 | 2.94e-21 | 7280 | 56.8 | 66700 | 52.4 | 1.08 | EPI_DAR_3hr |
| 173 | 33 | MA1560.1 | SOHLH2 | MA1560.1 | YGCACGTGCN | SOHLH2 | 6.05e-18 | 2.3e-20 | 1730 | 13.5 | 13700 | 10.8 | 1.26 | EPI_DAR_3hr |
| 174 | 21 | MA1581.1 | ZBTB6 | MA1581.1 | DNSCTTGAGCCNV | ZBTB6 | 4.2e-17 | 1.59e-19 | 10000 | 78.2 | 94900 | 74.6 | 1.05 | EPI_DAR_3hr |
| 175 | 68 | MA0778.1 | NFKB2 | MA0778.1 | AGGGGAWTCCCCY | NFKB2 | 6.17e-17 | 2.32e-19 | 2250 | 17.6 | 18500 | 14.5 | 1.21 | EPI_DAR_3hr |
| 176 | 33 | MA1464.1 | ARNT2 | MA1464.1 | GTCACGTGMN | ARNT2 | 1.27e-16 | 4.74e-19 | 2790 | 21.8 | 23500 | 18.5 | 1.18 | EPI_DAR_3hr |
| 177 | 45 | MA0608.1 | Creb3l2 | MA0608.1 | GCCACGTGD | Creb3l2 | 1.95e-16 | 7.24e-19 | 2450 | 19.2 | 20400 | 16 | 1.19 | EPI_DAR_3hr |
| 178 | 36 | MA1992.1 | Ikzf3 | MA1992.1 | NNRCAGGAAGTGGVN | Ikzf3 | 3.59e-16 | 1.33e-18 | 7410 | 57.9 | 68400 | 53.8 | 1.08 | EPI_DAR_3hr |
| 179 | 33 | MA1621.1 | Rbpjl | MA1621.1 | NNVACACCTGTBNN | Rbpjl | 6.16e-16 | 2.27e-18 | 8990 | 70.1 | 84400 | 66.3 | 1.06 | EPI_DAR_3hr |
| 180 | 53 | MA1475.1 | CREB3L4 | MA1475.1 | GRTGACGTCAYC | CREB3L4 | 8.4e-16 | 3.07e-18 | 6350 | 49.6 | 57900 | 45.5 | 1.09 | EPI_DAR_3hr |
| 181 | 31 | MA0737.1 | GLIS3 | MA0737.1 | GACCCCCCACGAAG | GLIS3 | 8.17e-15 | 2.97e-17 | 962 | 7.51 | 7120 | 5.6 | 1.34 | EPI_DAR_3hr |
| 182 | 69 | MA0803.1 | TBX15 | MA0803.1 | AGGTGTGA | TBX15 | 1.38e-14 | 4.98e-17 | 7110 | 55.5 | 65600 | 51.6 | 1.08 | EPI_DAR_3hr |
| 183 | 33 | MA1652.1 | ZKSCAN5 | MA1652.1 | NNRGGARGTGAGRR | ZKSCAN5 | 2.45e-14 | 8.8e-17 | 1790 | 14 | 14600 | 11.4 | 1.22 | EPI_DAR_3hr |
| 184 | 24 | MA1930.1 | CTCF | MA1930.1 | CTGCAGTKCCNVCHNNYRGCCASYAGRKGGCRAKC | CTCF | 5.12e-14 | 1.83e-16 | 2640 | 20.6 | 22400 | 17.6 | 1.17 | EPI_DAR_3hr |
| 185 | 70 | MA0138.2 | REST | MA0138.2 | TTCAGCACCATGGACAGCDCC | REST | 5.44e-14 | 1.94e-16 | 2080 | 16.2 | 17200 | 13.5 | 1.2 | EPI_DAR_3hr |
| 186 | 45 | MA0004.1 | Arnt | MA0004.1 | CACGTG | Arnt | 1.21e-13 | 4.29e-16 | 2020 | 15.8 | 16700 | 13.1 | 1.2 | EPI_DAR_3hr |
| 187 | 59 | MA1485.1 | FERD3L | MA1485.1 | GCRMCAGCTGTYAC | FERD3L | 1.44e-13 | 5.06e-16 | 2710 | 21.2 | 23100 | 18.2 | 1.16 | EPI_DAR_3hr |
| 188 | 36 | MA0156.3 | FEV | MA0156.3 | VACCGGAAGTVV | FEV | 5.91e-13 | 2.07e-15 | 3500 | 27.3 | 30700 | 24.1 | 1.13 | EPI_DAR_3hr |
| 189 | 33 | MA1493.1 | HES6 | MA1493.1 | GGCACGTGTY | HES6 | 6.75e-13 | 2.35e-15 | 3420 | 26.7 | 29900 | 23.5 | 1.14 | EPI_DAR_3hr |
| 190 | 44 | MA0739.1 | Hic1 | MA0739.1 | RTGCCAACY | Hic1 | 1.04e-12 | 3.6e-15 | 12000 | 94 | 117000 | 92.1 | 1.02 | EPI_DAR_3hr |
| 191 | 36 | MA1708.1 | ETV7 | MA1708.1 | DGSCGGAAGTR | ETV7 | 1.44e-12 | 4.95e-15 | 283 | 2.21 | 1640 | 1.29 | 1.72 | EPI_DAR_3hr |
| 192 | 33 | MA0500.2 | MYOG | MA0500.2 | NDRCAGCTGYHN | MYOG | 2.16e-12 | 7.42e-15 | 6000 | 46.8 | 55000 | 43.2 | 1.08 | EPI_DAR_3hr |
| 193 | 71 | MA1600.1 | ZNF684 | MA1600.1 | RYACAGTCCACCCCTTKR | ZNF684 | 4.14e-12 | 1.41e-14 | 3060 | 23.9 | 26600 | 20.9 | 1.14 | EPI_DAR_3hr |
| 194 | 34 | MA0056.2 | MZF1 | MA0056.2 | NNAATCCCCANNN | MZF1 | 1.58e-11 | 5.36e-14 | 11700 | 91.4 | 114000 | 89.3 | 1.02 | EPI_DAR_3hr |
| 195 | 49 | MA1574.1 | THRB | MA1574.1 | RRGGTCAAAGGTCAN | THRB | 2.12e-11 | 7.14e-14 | 4850 | 37.8 | 43900 | 34.5 | 1.1 | EPI_DAR_3hr |
| 196 | 33 | MA0806.1 | TBX4 | MA0806.1 | AGGTGTGA | TBX4 | 2.17e-11 | 7.27e-14 | 11100 | 86.4 | 107000 | 83.9 | 1.03 | EPI_DAR_3hr |
| 197 | 49 | MA1147.1 | NR4A2::RXRA | MA1147.1 | VRGGTCRTTGACCYY | NR4A2::RXRA | 5.67e-11 | 1.89e-13 | 8510 | 66.4 | 80300 | 63.1 | 1.05 | EPI_DAR_3hr |
| 198 | 33 | MA1619.1 | Ptf1A | MA1619.1 | NNACAGCTGTNN | Ptf1A | 1.7e-10 | 5.66e-13 | 10200 | 79.4 | 97400 | 76.5 | 1.04 | EPI_DAR_3hr |
| 199 | 36 | MA0763.1 | ETV3 | MA0763.1 | ACCGGAAGTR | ETV3 | 2.45e-10 | 8.1e-13 | 361 | 2.82 | 2340 | 1.84 | 1.53 | EPI_DAR_3hr |
| 200 | 69 | MA1565.1 | TBX18 | MA1565.1 | DRAGGTGTGAAR | TBX18 | 2.5e-10 | 8.22e-13 | 5960 | 46.6 | 55000 | 43.2 | 1.08 | EPI_DAR_3hr |
| 201 | 44 | MA0671.1 | NFIX | MA0671.1 | NNTGCCAAN | NFIX | 3.41e-09 | 1.12e-11 | 11400 | 89.2 | 111000 | 87.2 | 1.02 | EPI_DAR_3hr |
| 202 | 45 | MA0822.1 | HES7 | MA0822.1 | YGGCACGTGCCR | HES7 | 5.31e-09 | 1.73e-11 | 528 | 4.12 | 3800 | 2.98 | 1.38 | EPI_DAR_3hr |
| 203 | 33 | MA0464.2 | BHLHE40 | MA0464.2 | VKCACGTGMC | BHLHE40 | 5.52e-09 | 1.79e-11 | 1980 | 15.4 | 16900 | 13.3 | 1.17 | EPI_DAR_3hr |
| 204 | 68 | MA1117.1 | RELB | MA1117.1 | RDATTCCCCNN | RELB | 6.26e-09 | 2.02e-11 | 6990 | 54.5 | 65400 | 51.4 | 1.06 | EPI_DAR_3hr |
| 205 | 72 | MA1556.1 | RXRG | MA1556.1 | RRGGTCAYGACCYY | RXRG | 8.81e-09 | 2.83e-11 | 3360 | 26.2 | 29900 | 23.5 | 1.11 | EPI_DAR_3hr |
| 206 | 69 | MA0801.1 | MGA | MA0801.1 | AGGTGTGA | MGA | 1.14e-08 | 3.63e-11 | 6970 | 54.4 | 65300 | 51.4 | 1.06 | EPI_DAR_3hr |
| 207 | 33 | MA0058.3 | MAX | MA0058.3 | AVCACGTGNY | MAX | 2.6e-08 | 8.26e-11 | 1020 | 7.98 | 8180 | 6.43 | 1.24 | EPI_DAR_3hr |
| 208 | 33 | MA0807.1 | TBX5 | MA0807.1 | AGGTGTKA | TBX5 | 2.63e-08 | 8.33e-11 | 6300 | 49.2 | 58700 | 46.1 | 1.07 | EPI_DAR_3hr |
| 209 | 49 | MA0017.2 | NR2F1 | MA0017.2 | CARAGGTCAMVNG | NR2F1 | 2.67e-08 | 8.39e-11 | 3540 | 27.6 | 31800 | 25 | 1.11 | EPI_DAR_3hr |
| 210 | 24 | MA0139.1 | CTCF | MA0139.1 | YGRCCASYAGRKGGCRSYR | CTCF | 2.76e-08 | 8.66e-11 | 4330 | 33.8 | 39400 | 31 | 1.09 | EPI_DAR_3hr |
| 211 | 33 | MA1559.1 | SNAI3 | MA1559.1 | RRCAGGTGYA | SNAI3 | 6.89e-08 | 2.15e-10 | 7120 | 55.6 | 66900 | 52.6 | 1.06 | EPI_DAR_3hr |
| 212 | 73 | MA1557.1 | SMAD5 | MA1557.1 | YGTCTAGACA | SMAD5 | 6.95e-08 | 2.16e-10 | 6180 | 48.2 | 57600 | 45.3 | 1.07 | EPI_DAR_3hr |
| 213 | 55 | MA0258.2 | ESR2 | MA0258.2 | AGGTCASVNTGMCCY | ESR2 | 9.37e-08 | 2.9e-10 | 7700 | 60.1 | 72700 | 57.2 | 1.05 | EPI_DAR_3hr |
| 214 | 74 | MA0863.1 | MTF1 | MA0863.1 | TTTGCACACGGCAC | MTF1 | 1.05e-07 | 3.22e-10 | 1620 | 12.6 | 13700 | 10.7 | 1.17 | EPI_DAR_3hr |
| 215 | 72 | MA1538.1 | NR2F1 | MA1538.1 | RAGGTCANTGACCTY | NR2F1 | 1.2e-07 | 3.66e-10 | 3240 | 25.3 | 29000 | 22.8 | 1.11 | EPI_DAR_3hr |
| 216 | 31 | MA1491.2 | GLI3 | MA1491.2 | RGACCACCCACGWHGH | GLI3 | 1.21e-07 | 3.68e-10 | 709 | 5.53 | 5460 | 4.29 | 1.29 | EPI_DAR_3hr |
| 217 | 75 | MA0073.1 | RREB1 | MA0073.1 | CCCCMAAMCAMCCMCMMMCV | RREB1 | 2.75e-07 | 8.35e-10 | 365 | 2.85 | 2530 | 1.99 | 1.44 | EPI_DAR_3hr |
| 218 | 36 | MA0686.1 | SPDEF | MA0686.1 | AMCCGGATGTR | SPDEF | 4.14e-07 | 1.25e-09 | 2900 | 22.6 | 25800 | 20.3 | 1.11 | EPI_DAR_3hr |
| 219 | 33 | MA1641.1 | MYF5 | MA1641.1 | NVACAGCTGTBN | MYF5 | 5.16e-07 | 1.55e-09 | 8980 | 70.1 | 85800 | 67.5 | 1.04 | EPI_DAR_3hr |
| 220 | 33 | MA1472.2 | Bhlha15 | MA1472.2 | NVACAGCTGTBN | Bhlha15 | 1.33e-06 | 3.98e-09 | 7600 | 59.3 | 71900 | 56.6 | 1.05 | EPI_DAR_3hr |
| 221 | 33 | MA0603.1 | Arntl | MA0603.1 | NGTCACGTGH | Arntl | 1.37e-06 | 4.09e-09 | 10100 | 78.8 | 97400 | 76.6 | 1.03 | EPI_DAR_3hr |
| 222 | 53 | MA0638.1 | CREB3 | MA0638.1 | VTGCCACGTCAYCR | CREB3 | 2.19e-06 | 6.49e-09 | 1800 | 14.1 | 15600 | 12.2 | 1.15 | EPI_DAR_3hr |
| 223 | 76 | MA0130.1 | ZNF354C | MA0130.1 | MTCCAC | ZNF354C | 5.36e-06 | 1.58e-08 | 11400 | 89.1 | 111000 | 87.4 | 1.02 | EPI_DAR_3hr |
| 224 | 77 | MA1999.1 | Prdm5 | MA1999.1 | CBGTTCTCCATCTNN | Prdm5 | 1.7e-05 | 4.98e-08 | 10600 | 82.8 | 103000 | 80.9 | 1.02 | EPI_DAR_3hr |
| 225 | 36 | MA0760.1 | ERF | MA0760.1 | ACCGGAAGTR | ERF | 2.18e-05 | 6.37e-08 | 4190 | 32.7 | 38600 | 30.4 | 1.08 | EPI_DAR_3hr |
| 226 | 33 | MA0825.1 | MNT | MA0825.1 | RVCACGTGMH | MNT | 2.72e-05 | 7.91e-08 | 1710 | 13.3 | 14900 | 11.7 | 1.14 | EPI_DAR_3hr |
| 227 | 53 | MA0839.1 | CREB3L1 | MA0839.1 | VTGCCACGTCAYCA | CREB3L1 | 3.85e-05 | 1.12e-07 | 1710 | 13.4 | 14900 | 11.7 | 1.14 | EPI_DAR_3hr |
| 228 | 78 | MA1529.1 (NHLH2) | STREME-11 | 11-CAGGGAGGCAGCTS | 11-CAGGGAGGCAGCTS | NHLH2 | 4.37e-05 | 1.26e-07 | 1140 | 8.91 | 9630 | 7.57 | 1.18 | EPI_DAR_3hr |
| 229 | 53 | MA0636.1 | BHLHE41 | MA0636.1 | RTCACGTGAC | BHLHE41 | 7.77e-05 | 2.23e-07 | 1780 | 13.9 | 15600 | 12.2 | 1.13 | EPI_DAR_3hr |
| 230 | 53 | MA0604.1 | Atf1 | MA0604.1 | RTGACGTA | Atf1 | 9.81e-05 | 2.81e-07 | 1490 | 11.6 | 12900 | 10.1 | 1.15 | EPI_DAR_3hr |
| 231 | 43 | MA0779.1 | PAX1 | MA0779.1 | CGTCACGCWTGAVTGMM | PAX1 | 0.000156 | 4.44e-07 | 980 | 7.65 | 8210 | 6.45 | 1.19 | EPI_DAR_3hr |
| 232 | 33 | MA0521.2 | Tcf12 | MA0521.2 | NNACAGCTGTNN | Tcf12 | 0.000276 | 7.82e-07 | 6150 | 48 | 58100 | 45.7 | 1.05 | EPI_DAR_3hr |
| 233 | 45 | MA0622.1 | Mlxip | MA0622.1 | BCACGTGK | Mlxip | 0.000291 | 8.21e-07 | 449 | 3.5 | 3460 | 2.72 | 1.29 | EPI_DAR_3hr |
| 234 | 79 | MA1651.1 | ZFP42 | MA1651.1 | NNNHCAARATGGCTGCCNBNN | ZFP42 | 0.000324 | 9.1e-07 | 391 | 3.05 | 2950 | 2.32 | 1.32 | EPI_DAR_3hr |
| 235 | 80 | MA1468.1 | ATOH7 | MA1468.1 | AVCATATGBY | ATOH7 | 0.000401 | 1.12e-06 | 10600 | 82.6 | 103000 | 80.8 | 1.02 | EPI_DAR_3hr |
| 236 | 53 | MA0844.1 | XBP1 | MA0844.1 | WDKGMCACGTCATC | XBP1 | 0.000432 | 1.21e-06 | 1680 | 13.1 | 14800 | 11.7 | 1.13 | EPI_DAR_3hr |
| 237 | 42 | MA0609.2 | CREM | MA0609.2 | NNDGTGACGTCACHNN | CREM | 0.000709 | 1.97e-06 | 8110 | 63.3 | 77800 | 61.2 | 1.04 | EPI_DAR_3hr |
| 238 | 36 | MA1931.1 | ELK1::HOXA1 | MA1931.1 | ACCGGAAGTAATTA | ELK1::HOXA1 | 0.00157 | 4.33e-06 | 43 | 0.34 | 178 | 0.14 | 2.44 | EPI_DAR_3hr |
| 239 | 53 | MA1969.1 | THRA | MA1969.1 | NYTGTGTCCTCABRTGACCTYWBB | THRA | 0.00187 | 5.16e-06 | 2080 | 16.3 | 18700 | 14.7 | 1.1 | EPI_DAR_3hr |
| 240 | 33 | MA1620.1 | Ptf1A | MA1620.1 | NVACACCTGTNN | Ptf1A | 0.00205 | 5.63e-06 | 7010 | 54.7 | 66900 | 52.6 | 1.04 | EPI_DAR_3hr |
| 241 | 81 | MA1554.1 | RFX7 | MA1554.1 | CGTTGCYAY | RFX7 | 0.00241 | 6.59e-06 | 5970 | 46.6 | 56600 | 44.5 | 1.05 | EPI_DAR_3hr |
| 242 | 44 | MA0119.1 | NFIC::TLX1 | MA0119.1 | TGGCASSRWGCCAA | NFIC::TLX1 | 0.00542 | 1.47e-05 | 834 | 6.51 | 7060 | 5.55 | 1.17 | EPI_DAR_3hr |
| 243 | 42 | MA0862.1 | GMEB2 | MA0862.1 | TTACGTAA | GMEB2 | 0.00561 | 1.52e-05 | 1020 | 7.95 | 8770 | 6.89 | 1.15 | EPI_DAR_3hr |
| 244 | 36 | MA0098.3 | ETS1 | MA0098.3 | ACCGGAARTR | ETS1 | 0.00579 | 1.56e-05 | 6200 | 48.4 | 58900 | 46.3 | 1.04 | EPI_DAR_3hr |
| 245 | 36 | MA1957.1 | HOXB2::ELK1 | MA1957.1 | RCCGGAAGTMRTTA | HOXB2::ELK1 | 0.00773 | 2.08e-05 | 18 | 0.14 | 45 | 0.04 | 4.1 | EPI_DAR_3hr |
| 246 | 33 | MA0820.1 | FIGLA | MA0820.1 | WMCACCTGKW | FIGLA | 0.00871 | 2.33e-05 | 12100 | 94.3 | 119000 | 93.3 | 1.01 | EPI_DAR_3hr |
| 247 | 10 | MA0657.1 | KLF13 | MA0657.1 | RTGMCACGCCCCTTTTTG | KLF13 | 0.00883 | 2.35e-05 | 1010 | 7.86 | 8700 | 6.84 | 1.15 | EPI_DAR_3hr |
| 248 | 82 | MA1716.1 | ZNF76 | MA1716.1 | NNTWCCCAYAATGCAHYGCGC | ZNF76 | 0.0102 | 2.72e-05 | 536 | 4.18 | 4380 | 3.44 | 1.22 | EPI_DAR_3hr |
| 250 | 36 | MA0645.1 | ETV6 | MA0645.1 | MSCGGAAGTR | ETV6 | 0.0104 | 2.73e-05 | 8020 | 62.6 | 77200 | 60.7 | 1.03 | EPI_DAR_3hr |
| 249 | 72 | MA1146.1 | NR1H4::RXRA | MA1146.1 | GAGKTCATTGACCYB | NR1H4::RXRA | 0.0104 | 2.73e-05 | 9640 | 75.3 | 93500 | 73.6 | 1.02 | EPI_DAR_3hr |
| 251 | 36 | MA1484.1 | ETS2 | MA1484.1 | DACCGGAAGY | ETS2 | 0.0123 | 3.24e-05 | 951 | 7.42 | 8200 | 6.44 | 1.15 | EPI_DAR_3hr |
| 252 | 44 | MA1527.1 | NFIC | MA1527.1 | NTTGGCDNNRTGCCARN | NFIC | 0.0134 | 3.5e-05 | 475 | 3.71 | 3840 | 3.02 | 1.23 | EPI_DAR_3hr |
| 253 | 83 | MA0502.2 | NFYB | MA0502.2 | CYCATTGGCCVV | NFYB | 0.019 | 4.95e-05 | 8290 | 64.7 | 80000 | 62.9 | 1.03 | EPI_DAR_3hr |
| 254 | 84 | MA1491.2 (GLI3) | STREME-10 | 10-ACACACACGA | 10-ACACACACGA | GLI3 | 0.0317 | 8.23e-05 | 348 | 2.72 | 2740 | 2.16 | 1.26 | EPI_DAR_3hr |
| 255 | 69 | MA1567.2 | Tbx6 | MA1567.2 | NRGGTGTGAANN | Tbx6 | 0.0322 | 8.32e-05 | 5900 | 46 | 56200 | 44.2 | 1.04 | EPI_DAR_3hr |
| 256 | 53 | MA1466.1 | ATF6 | MA1466.1 | TGRTGACGTGGCAN | ATF6 | 0.041 | 0.000105 | 434 | 3.39 | 3520 | 2.77 | 1.23 | EPI_DAR_3hr |
| 257 | 53 | MA0663.1 | MLX | MA0663.1 | RTCACGTGAT | MLX | 0.047 | 0.00012 | 3580 | 27.9 | 33500 | 26.4 | 1.06 | EPI_DAR_3hr |
| 258 | 36 | MA0475.2 | FLI1 | MA0475.2 | ACCGGAARTR | FLI1 | 0.047 | 0.00012 | 1410 | 11 | 12600 | 9.92 | 1.11 | EPI_DAR_3hr |
| 259 | 85 | MA1531.1 | NR1D1 | MA1531.1 | RGGTYAGTRGGTCAN | NR1D1 | 0.0486 | 0.000124 | 1960 | 15.3 | 17800 | 14 | 1.09 | EPI_DAR_3hr |
| 13 | 86 | MA1125.1 (ZNF384) | MEME-2 | TTTTTTTTTTTTTTT | TTTTTTTTTTTTTTT | ZNF384 | 11.7 | 0.903 | 129 | 1.01 | 1440 | 1.13 | 0.899 | EPI_DAR_3hr |
ER_rat <- 1.25
# mrc_type <- "EAR_open"
EPI_DAR_3hr_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
# geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_col(aes(x=log10Evalue))+
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste("EPI_DAR_3hr_combo response peaks(200bp) Enrichment ratio:",ER_rat))
EPI_DAR_3hr_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
slice_head(n=5) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
# geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*3), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./3,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste("EPI DAR 3 hour response peaks(200bp) Enrichment ratio:",ER_rat))
EPI_DAR_24hr data
EPI_DAR_24hr_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/EPI_24_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
EPI_DAR_24hr_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/EPI_24_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
EPI_DAR_24hr_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/EPI_24_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
EPI_DAR_24hr_sea_all <- rbind(EPI_DAR_24hr_sea_disc, EPI_DAR_24hr_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)EPI_DAR_24hr_combo <- EPI_DAR_24hr_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., EPI_DAR_24hr_sea_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ SIM_MOTIF
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y) %>%
mutate(mrc_type="EPI_DAR_24hr")
EPI_DAR_24hr_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs 200 bp EPI_DAR_24hr") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO | mrc_type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | MA1134.1 (FOS::JUNB) | STREME-1 | 1-DATGASTCATH | DATGASTCATH | FOS::JUNB | 0 | 0 | 8380 | 14 | 4880 | 6.09 | 2.3 | EPI_DAR_24hr |
| 2 | 1 | MA1144.1 | FOSL2::JUND | MA1144.1 | KATGACTCAT | FOSL2::JUND | 0 | 0 | 11100 | 18.5 | 7580 | 9.45 | 1.96 | EPI_DAR_24hr |
| 3 | 1 | MA0491.2 | JUND | MA0491.2 | NNATGACTCATNN | JUND | 0 | 0 | 9740 | 16.3 | 6260 | 7.82 | 2.08 | EPI_DAR_24hr |
| 4 | 1 | MA1138.1 | FOSL2::JUNB | MA1138.1 | KRTGASTCAT | FOSL2::JUNB | 0 | 0 | 10200 | 17 | 6700 | 8.36 | 2.03 | EPI_DAR_24hr |
| 5 | 1 | MA0655.1 (JDP2) | STREME-6 | 6-ATGACTCA | ATGACTCA | JDP2 | 0 | 0 | 9900 | 16.5 | 6480 | 8.09 | 2.05 | EPI_DAR_24hr |
| 6 | 1 | MA1134.1 | FOS::JUNB | MA1134.1 | KATGASTCATHN | FOS::JUNB | 0 | 0 | 10300 | 17.3 | 6940 | 8.66 | 2 | EPI_DAR_24hr |
| 7 | 1 | MA0477.2 | FOSL1 | MA0477.2 | NNATGACTCATNN | FOSL1 | 0 | 0 | 10400 | 17.4 | 7000 | 8.74 | 1.99 | EPI_DAR_24hr |
| 8 | 1 | MA1135.1 | FOSB::JUNB | MA1135.1 | KRTGASTCAT | FOSB::JUNB | 0 | 0 | 9510 | 15.9 | 6120 | 7.64 | 2.08 | EPI_DAR_24hr |
| 9 | 1 | MA1130.1 | FOSL2::JUN | MA1130.1 | NNRTGAGTCAYN | FOSL2::JUN | 0 | 0 | 10500 | 17.5 | 7110 | 8.87 | 1.98 | EPI_DAR_24hr |
| 10 | 1 | MA1141.1 | FOS::JUND | MA1141.1 | NKATGAGTCATNN | FOS::JUND | 0 | 0 | 10500 | 17.6 | 7170 | 8.95 | 1.97 | EPI_DAR_24hr |
| 11 | 1 | MA1128.1 | FOSL1::JUN | MA1128.1 | NKATGACTCATNN | FOSL1::JUN | 0 | 0 | 10800 | 18 | 7400 | 9.23 | 1.95 | EPI_DAR_24hr |
| 12 | 1 | MA0490.2 | JUNB | MA0490.2 | NNATGACTCATNN | JUNB | 0 | 0 | 7780 | 13 | 4520 | 5.64 | 2.31 | EPI_DAR_24hr |
| 13 | 1 | MA1633.2 | BACH1 | MA1633.2 | ATGACTCAT | BACH1 | 0 | 0 | 10500 | 17.5 | 7120 | 8.89 | 1.97 | EPI_DAR_24hr |
| 14 | 1 | MA0099.3 | FOS::JUN | MA0099.3 | ATGAGTCAYM | FOS::JUN | 0 | 0 | 9760 | 16.3 | 6420 | 8.02 | 2.04 | EPI_DAR_24hr |
| 15 | 1 | MA0476.1 | FOS | MA0476.1 | DVTGASTCATB | FOS | 0 | 0 | 11500 | 19.2 | 8160 | 10.2 | 1.88 | EPI_DAR_24hr |
| 16 | 1 | MA1137.1 | FOSL1::JUNB | MA1137.1 | NDRTGACTCATNN | FOSL1::JUNB | 0 | 0 | 11000 | 18.4 | 7800 | 9.73 | 1.89 | EPI_DAR_24hr |
| 17 | 1 | MA0478.1 | FOSL2 | MA0478.1 | KRRTGASTCAB | FOSL2 | 0 | 0 | 11200 | 18.7 | 8000 | 9.98 | 1.88 | EPI_DAR_24hr |
| 18 | 1 | MA0489.2 | Jun | MA0489.2 | NVTGACTCATNN | Jun | 0 | 0 | 11000 | 18.4 | 7860 | 9.8 | 1.88 | EPI_DAR_24hr |
| 19 | 1 | MA0462.2 | BATF::JUN | MA0462.2 | DATGACTCATH | BATF::JUN | 0 | 0 | 8710 | 14.6 | 5570 | 6.95 | 2.09 | EPI_DAR_24hr |
| 20 | 1 | MA1988.1 | Atf3 | MA1988.1 | NRTGACTCABN | Atf3 | 0 | 0 | 7920 | 13.2 | 4830 | 6.02 | 2.2 | EPI_DAR_24hr |
| 21 | 1 | MA0655.1 | JDP2 | MA0655.1 | ATGACTCAT | JDP2 | 0 | 0 | 11100 | 18.5 | 7950 | 9.92 | 1.87 | EPI_DAR_24hr |
| 22 | 1 | MA0476.1 (FOS) | MEME-2 | ATGASTCABHH | ATGASTCABHH | FOS | 0 | 0 | 6700 | 11.2 | 3730 | 4.66 | 2.4 | EPI_DAR_24hr |
| 23 | 1 | MA1634.1 | BATF | MA1634.1 | DATGACTCATH | BATF | 0 | 0 | 6260 | 10.5 | 3400 | 4.25 | 2.46 | EPI_DAR_24hr |
| 24 | 1 | MA0841.1 | NFE2 | MA0841.1 | VATGACTCATS | NFE2 | 0 | 0 | 7840 | 13.1 | 4920 | 6.14 | 2.13 | EPI_DAR_24hr |
| 25 | 1 | MA1928.1 | BNC2 | MA1928.1 | NDRTGAGTCABN | BNC2 | 0 | 0 | 12300 | 20.6 | 9680 | 12.1 | 1.71 | EPI_DAR_24hr |
| 26 | 1 | MA1142.1 | FOSL1::JUND | MA1142.1 | NRTGACTCAT | FOSL1::JUND | 0 | 0 | 6640 | 11.1 | 3930 | 4.9 | 2.26 | EPI_DAR_24hr |
| 27 | 1 | MA0835.2 | BATF3 | MA0835.2 | DATGACTCATH | BATF3 | 0 | 0 | 5550 | 9.28 | 2960 | 3.69 | 2.51 | EPI_DAR_24hr |
| 28 | 1 | MA1101.2 | BACH2 | MA1101.2 | DWANCATGASTCATSNTWH | BACH2 | 0 | 0 | 11800 | 19.8 | 9160 | 11.4 | 1.73 | EPI_DAR_24hr |
| 29 | 1 | MA1132.1 | JUN::JUNB | MA1132.1 | KATGACKCAT | JUN::JUNB | 0 | 0 | 8050 | 13.4 | 5340 | 6.66 | 2.02 | EPI_DAR_24hr |
| 30 | 1 | MA0496.3 | MAFK | MA0496.3 | NGMTGACTCAGCMNH | MAFK | 6.41e-201 | 1.04e-202 | 14400 | 24 | 13900 | 17.4 | 1.38 | EPI_DAR_24hr |
| 31 | 1 | MA0089.2 | MAFG::NFE2L1 | MA0089.2 | NVNATGACTCAGCADW | MAFG::NFE2L1 | 2.12e-189 | 3.33e-191 | 20800 | 34.7 | 21900 | 27.3 | 1.27 | EPI_DAR_24hr |
| 32 | 1 | MA0501.1 | MAF::NFE2 | MA0501.1 | ATGACTCAGCANWTY | MAF::NFE2 | 6.28e-146 | 9.58e-148 | 23500 | 39.3 | 26100 | 32.6 | 1.21 | EPI_DAR_24hr |
| 33 | 1 | MA0150.2 | Nfe2l2 | MA0150.2 | CASNATGACTCAGCA | Nfe2l2 | 1.56e-123 | 2.31e-125 | 14100 | 23.6 | 14700 | 18.4 | 1.29 | EPI_DAR_24hr |
| 34 | 2 | MA0498.2 (MEIS1) | STREME-2 | 2-YTGWCAR | YTGWCAR | MEIS1 | 1.29e-108 | 1.86e-110 | 54700 | 91.3 | 70200 | 87.6 | 1.04 | EPI_DAR_24hr |
| 35 | 1 | MA0659.3 | Mafg | MA0659.3 | NWGMTGACTCAGCAN | Mafg | 3.5e-104 | 4.87e-106 | 6450 | 10.8 | 5920 | 7.39 | 1.46 | EPI_DAR_24hr |
| 36 | 3 | MA0720.1 | Shox2 | MA0720.1 | YYAATTAR | Shox2 | 3.41e-90 | 4.63e-92 | 52900 | 88.3 | 67800 | 84.6 | 1.04 | EPI_DAR_24hr |
| 37 | 3 | MA0722.1 | VAX1 | MA0722.1 | YTAATTAH | VAX1 | 8.43e-86 | 1.11e-87 | 52700 | 88.1 | 67600 | 84.3 | 1.04 | EPI_DAR_24hr |
| 38 | 3 | MA0674.1 | NKX6-1 | MA0674.1 | NTMATTAA | NKX6-1 | 6.19e-84 | 7.95e-86 | 54800 | 91.6 | 70800 | 88.4 | 1.04 | EPI_DAR_24hr |
| 39 | 3 | MA0725.1 | VSX1 | MA0725.1 | YTAATTAN | VSX1 | 5.52e-83 | 6.91e-85 | 53400 | 89.3 | 68700 | 85.8 | 1.04 | EPI_DAR_24hr |
| 40 | 3 | MA0630.1 | SHOX | MA0630.1 | HTAATTRR | SHOX | 5.92e-81 | 7.23e-83 | 52300 | 87.4 | 67100 | 83.8 | 1.04 | EPI_DAR_24hr |
| 41 | 3 | MA0726.1 | VSX2 | MA0726.1 | CTAATTAN | VSX2 | 1.55e-75 | 1.84e-77 | 49700 | 83 | 63400 | 79 | 1.05 | EPI_DAR_24hr |
| 42 | 3 | MA0027.2 | EN1 | MA0027.2 | SYAATTAV | EN1 | 8.19e-75 | 9.52e-77 | 50500 | 84.4 | 64600 | 80.5 | 1.05 | EPI_DAR_24hr |
| 43 | 3 | MA0704.1 | Lhx4 | MA0704.1 | YTAATTAR | Lhx4 | 1.27e-74 | 1.44e-76 | 46000 | 76.8 | 58100 | 72.4 | 1.06 | EPI_DAR_24hr |
| 44 | 4 | MA0676.1 | Nr2e1 | MA0676.1 | AAAAGTCAA | Nr2e1 | 3.89e-73 | 4.31e-75 | 47300 | 79.1 | 60000 | 74.9 | 1.06 | EPI_DAR_24hr |
| 45 | 3 | MA0886.1 | EMX2 | MA0886.1 | VBTAATTARB | EMX2 | 4.52e-73 | 4.9e-75 | 42300 | 70.6 | 52900 | 66 | 1.07 | EPI_DAR_24hr |
| 46 | 5 | MA0084.1 | SRY | MA0084.1 | NWWAACAAT | SRY | 7.77e-73 | 8.24e-75 | 54700 | 91.3 | 70800 | 88.3 | 1.03 | EPI_DAR_24hr |
| 47 | 3 | MA0902.2 | HOXB2 | MA0902.2 | SYAATTAS | HOXB2 | 1.17e-72 | 1.21e-74 | 39400 | 65.8 | 48900 | 61 | 1.08 | EPI_DAR_24hr |
| 48 | 3 | MA0675.1 | NKX6-2 | MA0675.1 | NYMATTAA | NKX6-2 | 1.52e-70 | 1.55e-72 | 49400 | 82.6 | 63100 | 78.8 | 1.05 | EPI_DAR_24hr |
| 49 | 3 | MA0716.1 | PRRX1 | MA0716.1 | YYAATTAR | PRRX1 | 3.78e-70 | 3.76e-72 | 52200 | 87.1 | 67100 | 83.7 | 1.04 | EPI_DAR_24hr |
| 50 | 6 | 3-GAGTTA | STREME-3 | 3-GAGTTA | GAGTTA | GAGTTA | 7.51e-70 | 7.33e-72 | 57900 | 96.7 | 75900 | 94.7 | 1.02 | EPI_DAR_24hr |
| 51 | 1 | MA1113.2 | PBX2 | MA1113.2 | NHCATAAATCAHN | PBX2 | 1.31e-69 | 1.26e-71 | 48900 | 81.7 | 62400 | 77.8 | 1.05 | EPI_DAR_24hr |
| 52 | 3 | MA0125.1 | Nobox | MA0125.1 | TAATTRSY | Nobox | 7.75e-69 | 7.27e-71 | 52100 | 87 | 67000 | 83.6 | 1.04 | EPI_DAR_24hr |
| 53 | 3 | MA0644.2 | ESX1 | MA0644.2 | YYAATTAN | ESX1 | 1.54e-67 | 1.42e-69 | 46400 | 77.6 | 58900 | 73.4 | 1.06 | EPI_DAR_24hr |
| 54 | 3 | MA0879.2 | DLX1 | MA0879.2 | VYAATTAN | DLX1 | 2.69e-67 | 2.43e-69 | 50800 | 84.9 | 65200 | 81.3 | 1.04 | EPI_DAR_24hr |
| 55 | 7 | 10-AATAAG | STREME-10 | 10-AATAAG | 10-AATAAG | AATAAG | 3.32e-67 | 2.95e-69 | 49800 | 83.2 | 63700 | 79.4 | 1.05 | EPI_DAR_24hr |
| 56 | 3 | MA0718.1 | RAX | MA0718.1 | RYYAATTARH | RAX | 4.49e-67 | 3.91e-69 | 46000 | 76.8 | 58200 | 72.6 | 1.06 | EPI_DAR_24hr |
| 57 | 8 | MA1480.1 | DPRX | MA1480.1 | AGATAATCCN | DPRX | 9.58e-66 | 8.2e-68 | 51500 | 86 | 66200 | 82.5 | 1.04 | EPI_DAR_24hr |
| 58 | 3 | MA0721.1 | UNCX | MA0721.1 | YYAATTAV | UNCX | 2.55e-65 | 2.14e-67 | 51300 | 85.7 | 65900 | 82.2 | 1.04 | EPI_DAR_24hr |
| 59 | 3 | MA0717.1 | RAX2 | MA0717.1 | YYAATTAV | RAX2 | 5.83e-65 | 4.82e-67 | 52200 | 87.2 | 67300 | 83.9 | 1.04 | EPI_DAR_24hr |
| 60 | 3 | MA0882.1 | DLX6 | MA0882.1 | VYAATTAN | DLX6 | 1.3e-64 | 1.05e-66 | 47000 | 78.5 | 59700 | 74.5 | 1.05 | EPI_DAR_24hr |
| 61 | 3 | MA0881.1 | Dlx4 | MA0881.1 | VYAATTAN | Dlx4 | 8e-63 | 6.4e-65 | 51600 | 86.1 | 66400 | 82.8 | 1.04 | EPI_DAR_24hr |
| 62 | 1 | MA1639.1 | MEIS1 | MA1639.1 | NHCATMAATCAHN | MEIS1 | 9.09e-62 | 7.16e-64 | 50200 | 83.9 | 64400 | 80.4 | 1.04 | EPI_DAR_24hr |
| 63 | 9 | MA1963.1 | SATB1 | MA1963.1 | WWDCTAATAAHWW | SATB1 | 9.38e-62 | 7.27e-64 | 43300 | 72.4 | 54700 | 68.2 | 1.06 | EPI_DAR_24hr |
| 64 | 3 | MA1476.2 | Dlx5 | MA1476.2 | NDGCAATTAGNN | Dlx5 | 1.16e-61 | 8.86e-64 | 53700 | 89.8 | 69600 | 86.8 | 1.03 | EPI_DAR_24hr |
| 65 | 3 | MA0793.1 | POU6F2 | MA0793.1 | ASCTMATTAW | POU6F2 | 1.21e-61 | 9.06e-64 | 46200 | 77.2 | 58700 | 73.2 | 1.05 | EPI_DAR_24hr |
| 66 | 1 | MA1640.1 | MEIS2 | MA1640.1 | NNATGATTTATGDNH | MEIS2 | 1.25e-61 | 9.23e-64 | 48000 | 80.2 | 61300 | 76.4 | 1.05 | EPI_DAR_24hr |
| 67 | 3 | MA1502.1 | HOXB8 | MA1502.1 | GYMATTAM | HOXB8 | 2.47e-61 | 1.8e-63 | 36700 | 61.4 | 45600 | 56.9 | 1.08 | EPI_DAR_24hr |
| 68 | 3 | MA0634.1 | ALX3 | MA0634.1 | YYYAATTANV | ALX3 | 2.87e-60 | 2.06e-62 | 56200 | 93.9 | 73400 | 91.5 | 1.03 | EPI_DAR_24hr |
| 69 | 3 | MA0068.2 | PAX4 | MA0068.2 | CTAATTAG | PAX4 | 8.14e-60 | 5.76e-62 | 53300 | 89.1 | 69000 | 86.1 | 1.03 | EPI_DAR_24hr |
| 70 | 10 | MA0035.4 | GATA1 | MA0035.4 | NHCTAATCTDH | GATA1 | 1.44e-59 | 1e-61 | 52100 | 87.1 | 67300 | 83.9 | 1.04 | EPI_DAR_24hr |
| 71 | 3 | MA0706.1 | MEOX2 | MA0706.1 | DSTAATTAWN | MEOX2 | 2.42e-59 | 1.66e-61 | 51800 | 86.6 | 66800 | 83.4 | 1.04 | EPI_DAR_24hr |
| 72 | 3 | MA0628.1 | POU6F1 | MA0628.1 | AYTAATTART | POU6F1 | 4.21e-59 | 2.85e-61 | 48300 | 80.8 | 61800 | 77.1 | 1.05 | EPI_DAR_24hr |
| 73 | 3 | MA0880.1 | Dlx3 | MA0880.1 | NYAATTAN | Dlx3 | 1.79e-58 | 1.2e-60 | 53200 | 88.8 | 68800 | 85.9 | 1.03 | EPI_DAR_24hr |
| 74 | 3 | MA0910.2 | HOXD8 | MA0910.2 | GYMATTAM | HOXD8 | 5.33e-58 | 3.52e-60 | 35100 | 58.6 | 43500 | 54.2 | 1.08 | EPI_DAR_24hr |
| 75 | 10 | MA0036.3 | GATA2 | MA0036.3 | NBCTTATCTNH | GATA2 | 3.59e-57 | 2.33e-59 | 53800 | 90 | 69900 | 87.2 | 1.03 | EPI_DAR_24hr |
| 76 | 3 | MA1481.1 | DRGX | MA1481.1 | YYAATTAN | DRGX | 3.81e-57 | 2.45e-59 | 44400 | 74.1 | 56300 | 70.2 | 1.06 | EPI_DAR_24hr |
| 77 | 3 | MA0885.2 | Dlx2 | MA0885.2 | NDGCAATTAGNN | Dlx2 | 9.96e-57 | 6.31e-59 | 53000 | 88.6 | 68600 | 85.6 | 1.03 | EPI_DAR_24hr |
| 78 | 3 | MA0708.2 | MSX2 | MA0708.2 | SCAATTAV | MSX2 | 2.6e-56 | 1.62e-58 | 49300 | 82.4 | 63300 | 78.9 | 1.04 | EPI_DAR_24hr |
| 79 | 11 | MA0899.1 | HOXA10 | MA0899.1 | DGYMATAAAAH | HOXA10 | 4.85e-56 | 2.99e-58 | 53000 | 88.5 | 68600 | 85.5 | 1.03 | EPI_DAR_24hr |
| 80 | 11 | MA0650.3 | Hoxa13 | MA0650.3 | NNCCAATAAAAN | Hoxa13 | 6.22e-56 | 3.79e-58 | 51900 | 86.7 | 67000 | 83.6 | 1.04 | EPI_DAR_24hr |
| 81 | 3 | MA0887.1 | EVX1 | MA0887.1 | VVTAATTABS | EVX1 | 9.03e-55 | 5.44e-57 | 52500 | 87.7 | 67900 | 84.8 | 1.04 | EPI_DAR_24hr |
| 82 | 10 | MA1104.2 | GATA6 | MA1104.2 | HWWTCTTATCTNH | GATA6 | 9.29e-55 | 5.53e-57 | 54900 | 91.7 | 71400 | 89.1 | 1.03 | EPI_DAR_24hr |
| 83 | 3 | MA0648.1 | GSC | MA0648.1 | VYTAATCCSY | GSC | 1.11e-54 | 6.52e-57 | 47400 | 79.3 | 60600 | 75.7 | 1.05 | EPI_DAR_24hr |
| 84 | 12 | MA0845.1 | FOXB1 | MA0845.1 | WATGTAAATAT | FOXB1 | 2.3e-54 | 1.34e-56 | 50100 | 83.7 | 64400 | 80.4 | 1.04 | EPI_DAR_24hr |
| 85 | 3 | MA0903.1 | HOXB3 | MA0903.1 | NNYMATTARN | HOXB3 | 2.38e-54 | 1.37e-56 | 53700 | 89.7 | 69700 | 86.9 | 1.03 | EPI_DAR_24hr |
| 86 | 3 | MA0890.1 | GBX2 | MA0890.1 | RSYAATTARN | GBX2 | 3.23e-54 | 1.83e-56 | 47200 | 78.9 | 60400 | 75.3 | 1.05 | EPI_DAR_24hr |
| 87 | 3 | MA0661.1 | MEOX1 | MA0661.1 | VSTAATTAHC | MEOX1 | 2.49e-53 | 1.4e-55 | 48900 | 81.6 | 62700 | 78.2 | 1.04 | EPI_DAR_24hr |
| 88 | 3 | MA0713.1 | PHOX2A | MA0713.1 | TAATYYAATTA | PHOX2A | 5.49e-53 | 3.05e-55 | 43600 | 72.9 | 55300 | 69 | 1.06 | EPI_DAR_24hr |
| 89 | 3 | MA0710.1 | NOTO | MA0710.1 | NYTAATTAVN | NOTO | 8.67e-53 | 4.75e-55 | 42300 | 70.7 | 53500 | 66.8 | 1.06 | EPI_DAR_24hr |
| 90 | 11 | MA0901.2 | HOXB13 | MA0901.2 | WDCCAATAAAAWWW | HOXB13 | 1.69e-52 | 9.18e-55 | 45200 | 75.5 | 57500 | 71.7 | 1.05 | EPI_DAR_24hr |
| 91 | 3 | MA1505.1 | HOXC8 | MA1505.1 | KTAATTAM | HOXC8 | 4.18e-52 | 2.24e-54 | 39300 | 65.7 | 49400 | 61.6 | 1.07 | EPI_DAR_24hr |
| 92 | 3 | MA0707.2 | MNX1 | MA0707.2 | VYRATTAK | MNX1 | 4.29e-52 | 2.27e-54 | 31000 | 51.8 | 38200 | 47.6 | 1.09 | EPI_DAR_24hr |
| 93 | 3 | MA0889.1 | GBX1 | MA0889.1 | VCYAATTASY | GBX1 | 5.73e-52 | 3e-54 | 44900 | 75.1 | 57200 | 71.3 | 1.05 | EPI_DAR_24hr |
| 94 | 3 | MA0723.2 | VAX2 | MA0723.2 | BYAATTAV | VAX2 | 1.2e-51 | 6.24e-54 | 43100 | 72 | 54600 | 68.1 | 1.06 | EPI_DAR_24hr |
| 95 | 13 | MA0635.1 | BARHL2 | MA0635.1 | VHTAAAYGRT | BARHL2 | 1.76e-51 | 9.03e-54 | 53300 | 89 | 69100 | 86.2 | 1.03 | EPI_DAR_24hr |
| 96 | 3 | MA0658.1 | LHX6 | MA0658.1 | RCTAATTARY | LHX6 | 3.04e-51 | 1.55e-53 | 40500 | 67.7 | 51100 | 63.7 | 1.06 | EPI_DAR_24hr |
| 97 | 14 | MA0786.1 | POU3F1 | MA0786.1 | WTATGCWAATKW | POU3F1 | 1.09e-50 | 5.47e-53 | 49000 | 81.9 | 63000 | 78.6 | 1.04 | EPI_DAR_24hr |
| 98 | 12 | MA1606.1 | Foxf1 | MA1606.1 | DWRTAAACANN | Foxf1 | 1.38e-50 | 6.87e-53 | 54400 | 91 | 70900 | 88.4 | 1.03 | EPI_DAR_24hr |
| 99 | 3 | MA1471.1 | BARX2 | MA1471.1 | NWWAAYMATTAN | BARX2 | 1.65e-50 | 8.13e-53 | 48900 | 81.7 | 62800 | 78.4 | 1.04 | EPI_DAR_24hr |
| 100 | 11 | MA0465.2 | CDX2 | MA0465.2 | NDGCAATAAAWN | CDX2 | 1.73e-50 | 8.43e-53 | 53800 | 89.9 | 69900 | 87.3 | 1.03 | EPI_DAR_24hr |
| 101 | 3 | MA0876.1 | BSX | MA0876.1 | SYVATTAV | BSX | 5.39e-50 | 2.6e-52 | 49900 | 83.4 | 64300 | 80.2 | 1.04 | EPI_DAR_24hr |
| 102 | 12 | MA0041.2 | FOXD3 | MA0041.2 | WAWGHAAATAAACARY | FOXD3 | 7.39e-50 | 3.54e-52 | 49900 | 83.4 | 64300 | 80.3 | 1.04 | EPI_DAR_24hr |
| 103 | 10 | MA1970.1 | TRPS1 | MA1970.1 | NHTCTTATCTNH | TRPS1 | 8.34e-50 | 3.95e-52 | 46900 | 78.4 | 60100 | 75 | 1.05 | EPI_DAR_24hr |
| 104 | 15 | MA0790.1 | POU4F1 | MA0790.1 | ATGMATAATTAATG | POU4F1 | 8.96e-50 | 4.2e-52 | 43900 | 73.4 | 55900 | 69.7 | 1.05 | EPI_DAR_24hr |
| 105 | 16 | 4-TTTSAAA | STREME-4 | 4-TTTSAAA | TTTSAAA | TTTSAAA | 2.3e-49 | 1.07e-51 | 53900 | 90 | 70000 | 87.4 | 1.03 | EPI_DAR_24hr |
| 106 | 3 | MA0680.2 | Pax7 | MA0680.2 | DWTAATCAATTAWNH | Pax7 | 3.23e-49 | 1.49e-51 | 39500 | 66 | 49800 | 62.1 | 1.06 | EPI_DAR_24hr |
| 107 | 3 | MA0666.2 | MSX1 | MA0666.2 | SCAATTAR | MSX1 | 3.81e-49 | 1.74e-51 | 41000 | 68.5 | 51800 | 64.6 | 1.06 | EPI_DAR_24hr |
| 108 | 3 | MA1549.1 | POU6F1 | MA1549.1 | RTAATGAGVN | POU6F1 | 2.26e-48 | 1.01e-50 | 38900 | 64.9 | 48900 | 61 | 1.06 | EPI_DAR_24hr |
| 109 | 3 | MA0709.1 | Msx3 | MA0709.1 | SCAATTAN | Msx3 | 2.26e-48 | 1.01e-50 | 47600 | 79.6 | 61100 | 76.2 | 1.04 | EPI_DAR_24hr |
| 110 | 14 | MA0792.1 | POU5F1B | MA0792.1 | TATGCWAAT | POU5F1B | 2.42e-48 | 1.07e-50 | 50500 | 84.4 | 65200 | 81.4 | 1.04 | EPI_DAR_24hr |
| 111 | 14 | MA0785.1 | POU2F1 | MA0785.1 | AWTATGCWAATK | POU2F1 | 2.77e-48 | 1.22e-50 | 49700 | 83.1 | 64000 | 79.9 | 1.04 | EPI_DAR_24hr |
| 112 | 17 | MA0052.4 | MEF2A | MA0052.4 | DDCTAAAAATAGMHH | MEF2A | 3.84e-48 | 1.67e-50 | 46100 | 77.1 | 59000 | 73.6 | 1.05 | EPI_DAR_24hr |
| 113 | 3 | MA0699.1 | LBX2 | MA0699.1 | NCYAATTARN | LBX2 | 6.88e-48 | 2.97e-50 | 53200 | 89 | 69200 | 86.3 | 1.03 | EPI_DAR_24hr |
| 114 | 3 | MA1498.2 | HOXA7 | MA1498.2 | SYMATTAV | HOXA7 | 1.26e-47 | 5.4e-50 | 46800 | 78.2 | 60000 | 74.8 | 1.05 | EPI_DAR_24hr |
| 115 | 18 | MA0602.1 | Arid5a | MA0602.1 | SYAATATTGVDANH | Arid5a | 1.32e-47 | 5.61e-50 | 49300 | 82.3 | 63400 | 79.1 | 1.04 | EPI_DAR_24hr |
| 116 | 3 | MA0158.2 | HOXA5 | MA0158.2 | GYMATTAS | HOXA5 | 4.85e-47 | 2.04e-49 | 25100 | 42 | 30500 | 38.1 | 1.1 | EPI_DAR_24hr |
| 117 | 3 | MA0700.2 | LHX2 | MA0700.2 | DDCCAATTADN | LHX2 | 1.05e-46 | 4.38e-49 | 48400 | 80.9 | 62200 | 77.6 | 1.04 | EPI_DAR_24hr |
| 118 | 3 | MA0654.1 | ISX | MA0654.1 | YYAATTAR | ISX | 1.22e-46 | 5.02e-49 | 37800 | 63.1 | 47500 | 59.2 | 1.07 | EPI_DAR_24hr |
| 119 | 3 | MA0701.2 | LHX9 | MA0701.2 | CYAATTAR | LHX9 | 1.37e-46 | 5.63e-49 | 29300 | 48.9 | 36000 | 44.9 | 1.09 | EPI_DAR_24hr |
| 120 | 3 | MA0662.1 | MIXL1 | MA0662.1 | NBYAATTAVN | MIXL1 | 1.45e-46 | 5.89e-49 | 48500 | 81.1 | 62400 | 77.8 | 1.04 | EPI_DAR_24hr |
| 121 | 15 | MA0791.1 | POU4F3 | MA0791.1 | RTGMATWATTAATGAR | POU4F3 | 3.13e-46 | 1.26e-48 | 40800 | 68.1 | 51600 | 64.3 | 1.06 | EPI_DAR_24hr |
| 122 | 14 | MA0142.1 | Pou5f1::Sox2 | MA0142.1 | CWTTGTYATGCAAAT | Pou5f1::Sox2 | 4.35e-46 | 1.74e-48 | 47400 | 79.2 | 60800 | 75.8 | 1.04 | EPI_DAR_24hr |
| 123 | 3 | MA1519.1 | LHX5 | MA1519.1 | STAATTAV | LHX5 | 5.33e-46 | 2.11e-48 | 27000 | 45.1 | 33000 | 41.2 | 1.1 | EPI_DAR_24hr |
| 124 | 14 | MA0788.1 | POU3F3 | MA0788.1 | WWTATGCWAATTW | POU3F3 | 8.36e-46 | 3.29e-48 | 46000 | 76.8 | 58800 | 73.4 | 1.05 | EPI_DAR_24hr |
| 125 | 3 | MA0894.1 | HESX1 | MA0894.1 | GHTAATTRRH | HESX1 | 1.81e-45 | 7.06e-48 | 39700 | 66.3 | 50100 | 62.5 | 1.06 | EPI_DAR_24hr |
| 126 | 3 | MA0711.1 | OTX1 | MA0711.1 | HTAATCCS | OTX1 | 4.55e-45 | 1.76e-47 | 53200 | 88.9 | 69200 | 86.4 | 1.03 | EPI_DAR_24hr |
| 127 | 12 | MA0032.2 | FOXC1 | MA0032.2 | WAWGTAAAYAW | FOXC1 | 5.74e-45 | 2.21e-47 | 50100 | 83.7 | 64700 | 80.7 | 1.04 | EPI_DAR_24hr |
| 128 | 3 | MA1495.1 | HOXA1 | MA1495.1 | STAATTAS | HOXA1 | 5.85e-45 | 2.23e-47 | 26300 | 43.9 | 32100 | 40 | 1.1 | EPI_DAR_24hr |
| 129 | 12 | MA0047.3 | FOXA2 | MA0047.3 | RWGTAAACANN | FOXA2 | 7.17e-45 | 2.71e-47 | 53700 | 89.7 | 69900 | 87.2 | 1.03 | EPI_DAR_24hr |
| 130 | 5 | MA1152.1 | SOX15 | MA1152.1 | CYWTTGTTHW | SOX15 | 1.97e-44 | 7.39e-47 | 53600 | 89.5 | 69800 | 87 | 1.03 | EPI_DAR_24hr |
| 131 | 3 | MA1530.1 | NKX6-3 | MA1530.1 | VYCATTAWW | NKX6-3 | 2.9e-44 | 1.08e-46 | 33700 | 56.3 | 42000 | 52.4 | 1.07 | EPI_DAR_24hr |
| 132 | 19 | MA0769.2 | TCF7 | MA0769.2 | NNCTTTGAWVT | TCF7 | 3.2e-44 | 1.18e-46 | 54100 | 90.4 | 70600 | 88 | 1.03 | EPI_DAR_24hr |
| 133 | 20 | MA0495.3 | MAFF | MA0495.3 | DWGTCAGCATTTTWHN | MAFF | 4.19e-44 | 1.54e-46 | 40100 | 67.1 | 50800 | 63.4 | 1.06 | EPI_DAR_24hr |
| 134 | 3 | MA0891.1 | GSC2 | MA0891.1 | NYTAATCCBH | GSC2 | 1.64e-43 | 5.97e-46 | 51700 | 86.4 | 67100 | 83.7 | 1.03 | EPI_DAR_24hr |
| 135 | 3 | MA0888.1 | EVX2 | MA0888.1 | VVTAATTAVB | EVX2 | 1.83e-43 | 6.63e-46 | 48900 | 81.6 | 63000 | 78.6 | 1.04 | EPI_DAR_24hr |
| 136 | 10 | MA0037.4 | Gata3 | MA0037.4 | NHTCTTATCTNH | Gata3 | 1.91e-43 | 6.84e-46 | 50200 | 83.9 | 64900 | 81 | 1.04 | EPI_DAR_24hr |
| 137 | 3 | MA1608.1 | Isl1 | MA1608.1 | NNCCATTAGNN | Isl1 | 2.79e-43 | 9.93e-46 | 52900 | 88.3 | 68700 | 85.7 | 1.03 | EPI_DAR_24hr |
| 138 | 12 | MA0033.2 | FOXL1 | MA0033.2 | RTAAACA | FOXL1 | 1.04e-42 | 3.69e-45 | 52400 | 87.6 | 68100 | 85 | 1.03 | EPI_DAR_24hr |
| 139 | 8 | MA0714.1 | PITX3 | MA0714.1 | NYTAATCCC | PITX3 | 1.57e-42 | 5.52e-45 | 56900 | 95 | 74700 | 93.2 | 1.02 | EPI_DAR_24hr |
| 140 | 12 | MA1607.1 | Foxl2 | MA1607.1 | DDWATGTAAACANN | Foxl2 | 1.67e-42 | 5.81e-45 | 53800 | 89.9 | 70100 | 87.5 | 1.03 | EPI_DAR_24hr |
| 141 | 3 | MA0705.1 | Lhx8 | MA0705.1 | CTAATTAV | Lhx8 | 7.19e-42 | 2.49e-44 | 32800 | 54.8 | 40900 | 51 | 1.07 | EPI_DAR_24hr |
| 142 | 5 | MA0077.1 | SOX9 | MA0077.1 | CYATTGTTY | SOX9 | 7.59e-42 | 2.61e-44 | 50100 | 83.8 | 64800 | 80.9 | 1.04 | EPI_DAR_24hr |
| 143 | 12 | MA0148.4 | FOXA1 | MA0148.4 | WWGTAAACATNN | FOXA1 | 8.48e-42 | 2.89e-44 | 52400 | 87.5 | 68100 | 84.9 | 1.03 | EPI_DAR_24hr |
| 144 | 3 | MA0892.1 | GSX1 | MA0892.1 | NCYMATTAVN | GSX1 | 1.61e-41 | 5.44e-44 | 55000 | 92 | 72000 | 89.8 | 1.02 | EPI_DAR_24hr |
| 145 | 8 | MA0712.2 | OTX2 | MA0712.2 | NDRGGATTARNN | OTX2 | 1.01e-40 | 3.41e-43 | 54100 | 90.3 | 70500 | 88 | 1.03 | EPI_DAR_24hr |
| 146 | 3 | MA1518.2 | Lhx1 | MA1518.2 | NNNTGCTAATTAGCANNN | Lhx1 | 2.46e-40 | 8.22e-43 | 47800 | 79.8 | 61500 | 76.8 | 1.04 | EPI_DAR_24hr |
| 147 | 3 | MA0895.1 | HMBOX1 | MA0895.1 | MYTAGTTAMS | HMBOX1 | 4.24e-40 | 1.41e-42 | 55700 | 93.1 | 73000 | 91 | 1.02 | EPI_DAR_24hr |
| 148 | 11 | MA0905.1 | HOXC10 | MA0905.1 | GTCRTAAAAH | HOXC10 | 4.53e-40 | 1.49e-42 | 43900 | 73.4 | 56100 | 70 | 1.05 | EPI_DAR_24hr |
| 149 | 21 | MA1657.1 | ZNF652 | MA1657.1 | BAAAGRGTTAAN | ZNF652 | 4.8e-40 | 1.57e-42 | 48700 | 81.4 | 62900 | 78.5 | 1.04 | EPI_DAR_24hr |
| 150 | 3 | MA0904.2 | HOXB5 | MA0904.2 | STAATTAS | HOXB5 | 1.03e-39 | 3.34e-42 | 25000 | 41.8 | 30600 | 38.2 | 1.09 | EPI_DAR_24hr |
| 151 | 13 | MA1978.1 | ZNF354A | MA1978.1 | HWAADWATAAATGRAYWAWTT | ZNF354A | 1.19e-39 | 3.84e-42 | 51500 | 86 | 66800 | 83.4 | 1.03 | EPI_DAR_24hr |
| 152 | 12 | MA0481.3 | FOXP1 | MA0481.3 | NDGTAAACANH | FOXP1 | 2.6e-39 | 8.36e-42 | 53600 | 89.6 | 69900 | 87.2 | 1.03 | EPI_DAR_24hr |
| 153 | 22 | MA0679.2 | ONECUT1 | MA0679.2 | WAAAAATCAATAAWWN | ONECUT1 | 3.35e-39 | 1.07e-41 | 37600 | 62.9 | 47500 | 59.3 | 1.06 | EPI_DAR_24hr |
| 154 | 12 | MA0846.1 | FOXC2 | MA0846.1 | WAWGTAAACAWW | FOXC2 | 6.63e-39 | 2.1e-41 | 51500 | 86.1 | 66900 | 83.5 | 1.03 | EPI_DAR_24hr |
| 155 | 12 | MA1683.1 | FOXA3 | MA1683.1 | DWGTAAACANN | FOXA3 | 1.99e-38 | 6.28e-41 | 52000 | 87 | 67700 | 84.4 | 1.03 | EPI_DAR_24hr |
| 156 | 3 | MA1497.1 | HOXA6 | MA1497.1 | BGYMATTANN | HOXA6 | 2.7e-38 | 8.44e-41 | 46700 | 78 | 60000 | 74.9 | 1.04 | EPI_DAR_24hr |
| 157 | 14 | MA0789.1 | POU3F4 | MA0789.1 | TATGCWAAT | POU3F4 | 7.84e-38 | 2.44e-40 | 43300 | 72.4 | 55400 | 69.1 | 1.05 | EPI_DAR_24hr |
| 158 | 14 | MA1115.1 | POU5F1 | MA1115.1 | WHATGCAAATN | POU5F1 | 8.47e-38 | 2.62e-40 | 48000 | 80.2 | 61900 | 77.3 | 1.04 | EPI_DAR_24hr |
| 159 | 3 | MA0875.1 | BARX1 | MA0875.1 | VCMATTAV | BARX1 | 1.49e-37 | 4.59e-40 | 45400 | 75.9 | 58300 | 72.8 | 1.04 | EPI_DAR_24hr |
| 160 | 23 | MA0809.2 | TEAD4 | MA0809.2 | NNACATTCCARN | TEAD4 | 2.03e-37 | 6.2e-40 | 51100 | 85.4 | 66400 | 82.8 | 1.03 | EPI_DAR_24hr |
| 161 | 15 | MA0153.2 | HNF1B | MA0153.2 | GTTAATNATTAAY | HNF1B | 8.05e-37 | 2.44e-39 | 24400 | 40.8 | 29900 | 37.3 | 1.09 | EPI_DAR_24hr |
| 162 | 5 | MA0868.2 | SOX8 | MA0868.2 | MGAACAATRG | SOX8 | 1.04e-35 | 3.13e-38 | 49200 | 82.1 | 63600 | 79.4 | 1.03 | EPI_DAR_24hr |
| 163 | 3 | MA0601.1 | Arid3b | MA0601.1 | ATATTAATWAN | Arid3b | 1.17e-35 | 3.51e-38 | 34400 | 57.4 | 43200 | 53.9 | 1.06 | EPI_DAR_24hr |
| 164 | 5 | MA0143.4 | SOX2 | MA0143.4 | DNACAATGGNN | SOX2 | 1.26e-35 | 3.75e-38 | 52500 | 87.8 | 68400 | 85.4 | 1.03 | EPI_DAR_24hr |
| 165 | 24 | MA0766.2 | GATA5 | MA0766.2 | SWGATAASRN | GATA5 | 1.41e-35 | 4.17e-38 | 26900 | 45 | 33300 | 41.5 | 1.08 | EPI_DAR_24hr |
| 166 | 3 | MA0912.2 | HOXD3 | MA0912.2 | CTAATTAC | HOXD3 | 3.3e-35 | 9.71e-38 | 21900 | 36.6 | 26600 | 33.2 | 1.1 | EPI_DAR_24hr |
| 167 | 3 | MA0612.2 | EMX1 | MA0612.2 | BTAATTAS | EMX1 | 4.26e-35 | 1.24e-37 | 21800 | 36.3 | 26500 | 33 | 1.1 | EPI_DAR_24hr |
| 168 | 1 | MA0591.1 | Bach1::Mafk | MA0591.1 | RSSATGACTCAGCAB | Bach1::Mafk | 8.48e-35 | 2.46e-37 | 2890 | 4.84 | 2780 | 3.46 | 1.4 | EPI_DAR_24hr |
| 169 | 3 | MA0681.2 | PHOX2B | MA0681.2 | WWTAATCAAATTAWWW | PHOX2B | 8.75e-35 | 2.53e-37 | 35700 | 59.7 | 45100 | 56.3 | 1.06 | EPI_DAR_24hr |
| 170 | 25 | MA0108.2 | TBP | MA0108.2 | STATAAAWRSVVVBN | TBP | 1.28e-34 | 3.68e-37 | 50100 | 83.7 | 65000 | 81 | 1.03 | EPI_DAR_24hr |
| 171 | 26 | MA0782.2 | PKNOX1 | MA0782.2 | NNTGAGTGACAGVNN | PKNOX1 | 2.75e-34 | 7.85e-37 | 41900 | 70 | 53600 | 66.8 | 1.05 | EPI_DAR_24hr |
| 172 | 5 | MA1563.2 | SOX18 | MA1563.2 | AACAATDV | SOX18 | 5.78e-34 | 1.64e-36 | 47400 | 79.1 | 61100 | 76.3 | 1.04 | EPI_DAR_24hr |
| 173 | 3 | MA0724.1 | VENTX | MA0724.1 | AMCGATTAR | VENTX | 5.95e-34 | 1.68e-36 | 35800 | 59.8 | 45200 | 56.4 | 1.06 | EPI_DAR_24hr |
| 174 | 27 | MA0122.3 | Nkx3-2 | MA0122.3 | WWAAMCACTTAAN | Nkx3-2 | 7.91e-34 | 2.22e-36 | 46800 | 78.2 | 60300 | 75.3 | 1.04 | EPI_DAR_24hr |
| 175 | 12 | MA0042.2 | FOXI1 | MA0042.2 | GTAAACA | FOXI1 | 8.65e-34 | 2.41e-36 | 50400 | 84.2 | 65400 | 81.6 | 1.03 | EPI_DAR_24hr |
| 176 | 3 | MA0132.2 | PDX1 | MA0132.2 | VYAATTAR | PDX1 | 1.65e-33 | 4.56e-36 | 33000 | 55.1 | 41400 | 51.7 | 1.07 | EPI_DAR_24hr |
| 177 | 12 | MA0848.1 | FOXO4 | MA0848.1 | GTAAACA | FOXO4 | 3.66e-33 | 1.01e-35 | 53600 | 89.5 | 70000 | 87.3 | 1.02 | EPI_DAR_24hr |
| 178 | 12 | MA0031.1 | FOXD1 | MA0031.1 | GTAAACAW | FOXD1 | 3.68e-33 | 1.01e-35 | 35100 | 58.7 | 44300 | 55.3 | 1.06 | EPI_DAR_24hr |
| 179 | 3 | MA1500.1 | HOXB6 | MA1500.1 | BTAATKRC | HOXB6 | 5.33e-33 | 1.45e-35 | 58700 | 98 | 77700 | 97 | 1.01 | EPI_DAR_24hr |
| 180 | 11 | MA0909.3 | Hoxd13 | MA0909.3 | NNCAATAAAW | Hoxd13 | 6.49e-33 | 1.76e-35 | 36200 | 60.4 | 45800 | 57.1 | 1.06 | EPI_DAR_24hr |
| 181 | 15 | MA0046.2 | HNF1A | MA0046.2 | DRTTAATNATTAACN | HNF1A | 1.23e-32 | 3.33e-35 | 24200 | 40.4 | 29800 | 37.2 | 1.09 | EPI_DAR_24hr |
| 182 | 12 | MA1103.2 | FOXK2 | MA1103.2 | DDGTAAACANN | FOXK2 | 6.23e-32 | 1.67e-34 | 53100 | 88.7 | 69300 | 86.5 | 1.03 | EPI_DAR_24hr |
| 183 | 10 | MA0482.2 | GATA4 | MA0482.2 | NNCCTTATCTNH | GATA4 | 1.91e-31 | 5.08e-34 | 50500 | 84.3 | 65600 | 81.8 | 1.03 | EPI_DAR_24hr |
| 184 | 3 | MA0075.3 | PRRX2 | MA0075.3 | CTAATTAN | PRRX2 | 2.95e-31 | 7.83e-34 | 26700 | 44.6 | 33200 | 41.4 | 1.08 | EPI_DAR_24hr |
| 185 | 3 | MA0900.2 | HOXA2 | MA0900.2 | STAATTAS | HOXA2 | 9.4e-31 | 2.48e-33 | 23300 | 39 | 28700 | 35.8 | 1.09 | EPI_DAR_24hr |
| 186 | 14 | MA0787.1 | POU3F2 | MA0787.1 | WTATGCWAATKA | POU3F2 | 2.84e-30 | 7.44e-33 | 41000 | 68.5 | 52500 | 65.5 | 1.05 | EPI_DAR_24hr |
| 187 | 26 | MA0498.2 | MEIS1 | MA0498.2 | HTGACAD | MEIS1 | 9.03e-30 | 2.36e-32 | 52400 | 87.5 | 68400 | 85.3 | 1.03 | EPI_DAR_24hr |
| 188 | 3 | MA0898.1 | Hmx3 | MA0898.1 | ASAAGCAATTAAWRRAT | Hmx3 | 1.19e-29 | 3.1e-32 | 37300 | 62.4 | 47500 | 59.2 | 1.05 | EPI_DAR_24hr |
| 189 | 8 | MA0682.2 | PITX1 | MA0682.2 | YTAATCCH | PITX1 | 2.36e-29 | 6.08e-32 | 26900 | 45 | 33500 | 41.8 | 1.08 | EPI_DAR_24hr |
| 190 | 28 | MA0102.4 | CEBPA | MA0102.4 | NDATTGCACAATHN | CEBPA | 2.95e-29 | 7.58e-32 | 51800 | 86.6 | 67600 | 84.4 | 1.03 | EPI_DAR_24hr |
| 191 | 8 | MA0467.2 | Crx | MA0467.2 | NDGGATTANN | Crx | 3.11e-29 | 7.94e-32 | 42300 | 70.7 | 54300 | 67.8 | 1.04 | EPI_DAR_24hr |
| 192 | 3 | MA1499.1 | HOXB4 | MA1499.1 | RTMATTAR | HOXB4 | 4.82e-29 | 1.23e-31 | 33200 | 55.5 | 42000 | 52.4 | 1.06 | EPI_DAR_24hr |
| 193 | 14 | MA0627.2 | POU2F3 | MA0627.2 | NWTATGCAAATNH | POU2F3 | 7.61e-29 | 1.92e-31 | 43200 | 72.2 | 55600 | 69.4 | 1.04 | EPI_DAR_24hr |
| 194 | 12 | MA0157.3 | Foxo3 | MA0157.3 | NDGTAAACARNN | Foxo3 | 9.22e-29 | 2.32e-31 | 53600 | 89.6 | 70200 | 87.6 | 1.02 | EPI_DAR_24hr |
| 195 | 28 | MA0025.2 | NFIL3 | MA0025.2 | NRTTATGYAATNN | NFIL3 | 1.96e-28 | 4.89e-31 | 45800 | 76.5 | 59100 | 73.8 | 1.04 | EPI_DAR_24hr |
| 196 | 29 | MA0611.2 | Dux | MA0611.2 | NTGATTBAATCAVWNN | Dux | 2.84e-28 | 7.06e-31 | 20000 | 33.3 | 24400 | 30.4 | 1.1 | EPI_DAR_24hr |
| 197 | 22 | MA0070.1 | PBX1 | MA0070.1 | HCATCAATCAAW | PBX1 | 4.05e-28 | 1e-30 | 28000 | 46.8 | 35100 | 43.8 | 1.07 | EPI_DAR_24hr |
| 198 | 5 | MA1562.1 | SOX14 | MA1562.1 | CCGAACAATG | SOX14 | 5.54e-28 | 1.36e-30 | 46800 | 78.2 | 60600 | 75.6 | 1.03 | EPI_DAR_24hr |
| 199 | 5 | MA0514.2 | Sox3 | MA0514.2 | DNACAATGGNN | Sox3 | 1.06e-27 | 2.61e-30 | 55200 | 92.3 | 72600 | 90.5 | 1.02 | EPI_DAR_24hr |
| 200 | 22 | MA0755.1 | CUX2 | MA0755.1 | TRATCGATAW | CUX2 | 1.54e-27 | 3.75e-30 | 39800 | 66.5 | 51000 | 63.6 | 1.05 | EPI_DAR_24hr |
| 201 | 17 | MA0497.1 | MEF2C | MA0497.1 | DDDCYAAAAATAGMW | MEF2C | 2.74e-27 | 6.64e-30 | 39800 | 66.6 | 51000 | 63.6 | 1.05 | EPI_DAR_24hr |
| 202 | 3 | MA0853.1 | Alx4 | MA0853.1 | CGCRYTAATTARYHNNY | Alx4 | 4.19e-27 | 1.01e-29 | 28500 | 47.7 | 35800 | 44.6 | 1.07 | EPI_DAR_24hr |
| 203 | 3 | MA0893.2 | GSX2 | MA0893.2 | SYMATTAR | GSX2 | 5.74e-27 | 1.38e-29 | 56200 | 93.8 | 73900 | 92.3 | 1.02 | EPI_DAR_24hr |
| 204 | 3 | MA0896.1 | Hmx1 | MA0896.1 | VSVAGCAATTAAWGVDB | Hmx1 | 6.61e-27 | 1.58e-29 | 36600 | 61.2 | 46600 | 58.2 | 1.05 | EPI_DAR_24hr |
| 205 | 20 | MA0117.2 | Mafb | MA0117.2 | AWWNTGCTGACD | Mafb | 7.37e-27 | 1.75e-29 | 19100 | 32 | 23400 | 29.2 | 1.1 | EPI_DAR_24hr |
| 206 | 19 | MA0523.1 | TCF7L2 | MA0523.1 | DRASATCAAAGRVA | TCF7L2 | 1.18e-26 | 2.8e-29 | 39300 | 65.7 | 50300 | 62.8 | 1.05 | EPI_DAR_24hr |
| 207 | 5 | MA0078.2 | Sox17 | MA0078.2 | NNAGAACAATGGNN | Sox17 | 1.26e-26 | 2.97e-29 | 52000 | 86.9 | 67900 | 84.7 | 1.03 | EPI_DAR_24hr |
| 208 | 11 | MA0878.3 | CDX1 | MA0878.3 | GGYMATAAAA | CDX1 | 6.53e-26 | 1.53e-28 | 26500 | 44.3 | 33100 | 41.3 | 1.07 | EPI_DAR_24hr |
| 209 | 30 | MA1489.1 | FOXN3 | MA1489.1 | GTAAACAA | FOXN3 | 4.19e-25 | 9.78e-28 | 43400 | 72.5 | 56000 | 69.8 | 1.04 | EPI_DAR_24hr |
| 210 | 19 | MA1991.1 | Hnf1A | MA1991.1 | NBCCTTTGATSTBN | Hnf1A | 6.62e-25 | 1.54e-27 | 54600 | 91.2 | 71700 | 89.5 | 1.02 | EPI_DAR_24hr |
| 211 | 31 | MA0606.2 | Nfat5 | MA0606.2 | NNATGGAAAAWN | Nfat5 | 9.62e-25 | 2.22e-27 | 57400 | 95.9 | 75800 | 94.6 | 1.01 | EPI_DAR_24hr |
| 212 | 32 | MA1124.1 | ZNF24 | MA1124.1 | CATTCATTCATTC | ZNF24 | 4.13e-24 | 9.52e-27 | 34100 | 57 | 43300 | 54.1 | 1.05 | EPI_DAR_24hr |
| 213 | 11 | MA0908.1 | HOXD11 | MA0908.1 | RTCGTAAAAH | HOXD11 | 6.48e-24 | 1.49e-26 | 27400 | 45.8 | 34400 | 42.9 | 1.07 | EPI_DAR_24hr |
| 214 | 33 | MA0018.4 | CREB1 | MA0018.4 | NNRTGATGTCAYN | CREB1 | 7.53e-24 | 1.72e-26 | 44500 | 74.4 | 57600 | 71.8 | 1.04 | EPI_DAR_24hr |
| 215 | 12 | MA0480.2 | Foxo1 | MA0480.2 | NDGTAAACANN | Foxo1 | 1.08e-23 | 2.46e-26 | 56600 | 94.5 | 74600 | 93.1 | 1.01 | EPI_DAR_24hr |
| 216 | 33 | MA1143.1 | FOSL1::JUND | MA1143.1 | RTGACGTMAY | FOSL1::JUND | 1.24e-23 | 2.79e-26 | 14700 | 24.5 | 17700 | 22.1 | 1.11 | EPI_DAR_24hr |
| 217 | 34 | MA1644.1 | NFYC | MA1644.1 | RDCCAATCASN | NFYC | 2.65e-23 | 5.95e-26 | 31300 | 52.3 | 39600 | 49.4 | 1.06 | EPI_DAR_24hr |
| 218 | 28 | MA1636.1 | CEBPG | MA1636.1 | NDATGATGCAATMHH | CEBPG | 2.95e-23 | 6.61e-26 | 46800 | 78.2 | 60700 | 75.8 | 1.03 | EPI_DAR_24hr |
| 219 | 12 | MA0850.1 | FOXP3 | MA0850.1 | RTAAACA | FOXP3 | 3.98e-23 | 8.87e-26 | 57200 | 95.6 | 75600 | 94.3 | 1.01 | EPI_DAR_24hr |
| 220 | 12 | MA0852.2 | FOXK1 | MA0852.2 | NRDGTAAACAAGNN | FOXK1 | 6.97e-23 | 1.55e-25 | 54000 | 90.3 | 70900 | 88.5 | 1.02 | EPI_DAR_24hr |
| 221 | 35 | MA0095.3 | Yy1 | MA0095.3 | NNCAAAATGGCN | Yy1 | 2.88e-22 | 6.36e-25 | 57000 | 95.2 | 75300 | 93.9 | 1.01 | EPI_DAR_24hr |
| 222 | 36 | MA1603.1 | Dmrt1 | MA1603.1 | GHTACAAAGTADC | Dmrt1 | 4.59e-22 | 1.01e-24 | 45000 | 75.2 | 58300 | 72.8 | 1.03 | EPI_DAR_24hr |
| 223 | 37 | MA0502.2 (NFYB) | STREME-7 | 7-CATTGG | CATTGG | NFYB | 7.82e-22 | 1.71e-24 | 30600 | 51.2 | 38800 | 48.4 | 1.06 | EPI_DAR_24hr |
| 224 | 24 | MA0140.2 | GATA1::TAL1 | MA0140.2 | MTTATCWSNNNNNHVCAG | GATA1::TAL1 | 2.08e-21 | 4.53e-24 | 20600 | 34.4 | 25500 | 31.8 | 1.08 | EPI_DAR_24hr |
| 225 | 27 | MA0124.2 | Nkx3-1 | MA0124.2 | RCCACTTAA | Nkx3-1 | 2.33e-21 | 5.04e-24 | 44500 | 74.4 | 57700 | 71.9 | 1.03 | EPI_DAR_24hr |
| 226 | 3 | MA0642.2 | EN2 | MA0642.2 | VSYAATTA | EN2 | 2.53e-21 | 5.45e-24 | 25700 | 42.9 | 32200 | 40.2 | 1.07 | EPI_DAR_24hr |
| 227 | 5 | MA1120.1 | SOX13 | MA1120.1 | DVACAATGGNN | SOX13 | 4.1e-21 | 8.81e-24 | 50000 | 83.5 | 65300 | 81.4 | 1.03 | EPI_DAR_24hr |
| 228 | 11 | MA0913.2 | HOXD9 | MA0913.2 | GYMATAAAAH | HOXD9 | 6e-21 | 1.28e-23 | 30700 | 51.3 | 39000 | 48.6 | 1.06 | EPI_DAR_24hr |
| 229 | 8 | MA0719.1 | RHOXF1 | MA0719.1 | NTRATCCN | RHOXF1 | 1.54e-20 | 3.27e-23 | 52900 | 88.3 | 69400 | 86.5 | 1.02 | EPI_DAR_24hr |
| 230 | 11 | MA0151.1 | Arid3a | MA0151.1 | ATYAAA | Arid3a | 2.06e-20 | 4.37e-23 | 26100 | 43.6 | 32800 | 41 | 1.06 | EPI_DAR_24hr |
| 231 | 19 | MA0768.2 | Lef1 | MA0768.2 | NNBCCTTTGATSTN | Lef1 | 2.69e-20 | 5.69e-23 | 49400 | 82.6 | 64500 | 80.5 | 1.03 | EPI_DAR_24hr |
| 232 | 20 | MA0842.2 | NRL | MA0842.2 | AAWWNTGCTGACG | NRL | 4.45e-20 | 9.36e-23 | 17500 | 29.2 | 21500 | 26.8 | 1.09 | EPI_DAR_24hr |
| 233 | 23 | MA0090.3 | TEAD1 | MA0090.3 | NNACATTCCAGSN | TEAD1 | 6.49e-20 | 1.36e-22 | 51900 | 86.6 | 68000 | 84.8 | 1.02 | EPI_DAR_24hr |
| 234 | 38 | MA1588.1 | ZNF136 | MA1588.1 | RWATTCTTGGTTGRC | ZNF136 | 8.68e-20 | 1.81e-22 | 31700 | 52.9 | 40300 | 50.3 | 1.05 | EPI_DAR_24hr |
| 235 | 23 | MA0808.1 | TEAD3 | MA0808.1 | RCATTCCW | TEAD3 | 1.14e-19 | 2.37e-22 | 34700 | 57.9 | 44300 | 55.3 | 1.05 | EPI_DAR_24hr |
| 236 | 3 | MA0715.1 | PROP1 | MA0715.1 | TAATYWAATTA | PROP1 | 1.17e-19 | 2.42e-22 | 27000 | 45.1 | 34100 | 42.5 | 1.06 | EPI_DAR_24hr |
| 237 | 11 | MA0911.1 | Hoxa11 | MA0911.1 | RGTCGTAAAANT | Hoxa11 | 1.56e-19 | 3.22e-22 | 45500 | 76.1 | 59100 | 73.8 | 1.03 | EPI_DAR_24hr |
| 238 | 26 | MA0774.1 | MEIS2 | MA0774.1 | TTGACAGS | MEIS2 | 2.57e-19 | 5.28e-22 | 28900 | 48.2 | 36600 | 45.7 | 1.06 | EPI_DAR_24hr |
| 239 | 3 | MA0914.1 | ISL2 | MA0914.1 | GCAMTTAR | ISL2 | 2.81e-19 | 5.74e-22 | 44000 | 73.6 | 57100 | 71.3 | 1.03 | EPI_DAR_24hr |
| 240 | 5 | MA0442.2 | SOX10 | MA0442.2 | NDAACAAAGVN | SOX10 | 2.95e-19 | 6e-22 | 52700 | 88.1 | 69200 | 86.4 | 1.02 | EPI_DAR_24hr |
| 241 | 5 | MA0867.2 | SOX4 | MA0867.2 | RAACAAAGRV | SOX4 | 7.13e-19 | 1.44e-21 | 49900 | 83.4 | 65300 | 81.5 | 1.02 | EPI_DAR_24hr |
| 242 | 13 | MA0877.3 | BARHL1 | MA0877.3 | AMCGTTTA | BARHL1 | 1.12e-18 | 2.25e-21 | 51000 | 85.1 | 66700 | 83.3 | 1.02 | EPI_DAR_24hr |
| 243 | 8 | MA0883.1 | Dmbx1 | MA0883.1 | WDRWNMGGATTADKNNW | Dmbx1 | 3.51e-18 | 7.05e-21 | 31700 | 52.9 | 40400 | 50.4 | 1.05 | EPI_DAR_24hr |
| 244 | 12 | MA0614.1 | Foxj2 | MA0614.1 | RTAAACAA | Foxj2 | 4.9e-18 | 9.79e-21 | 19000 | 31.8 | 23600 | 29.5 | 1.08 | EPI_DAR_24hr |
| 245 | 3 | MA0854.1 | Alx1 | MA0854.1 | CGMRYTAATTARTNMNY | Alx1 | 5.23e-18 | 1.04e-20 | 25200 | 42.1 | 31800 | 39.6 | 1.06 | EPI_DAR_24hr |
| 246 | 22 | MA0754.2 | CUX1 | MA0754.2 | TAATCGATAM | CUX1 | 1.21e-17 | 2.41e-20 | 24300 | 40.6 | 30600 | 38.1 | 1.06 | EPI_DAR_24hr |
| 247 | 39 | MA0050.3 | Irf1 | MA0050.3 | ANTGAAACTGAAASH | Irf1 | 1.34e-17 | 2.64e-20 | 48900 | 81.7 | 63900 | 79.7 | 1.02 | EPI_DAR_24hr |
| 248 | 12 | MA0849.1 | FOXO6 | MA0849.1 | GTAAACA | FOXO6 | 2.36e-17 | 4.64e-20 | 5320 | 8.89 | 6040 | 7.53 | 1.18 | EPI_DAR_24hr |
| 249 | 3 | MA1507.1 | HOXD4 | MA1507.1 | VTMATTAD | HOXD4 | 2.67e-17 | 5.23e-20 | 22900 | 38.3 | 28800 | 36 | 1.07 | EPI_DAR_24hr |
| 250 | 3 | MA1504.1 | HOXC4 | MA1504.1 | VTMATTAN | HOXC4 | 3.56e-17 | 6.94e-20 | 24000 | 40.1 | 30200 | 37.7 | 1.06 | EPI_DAR_24hr |
| 251 | 40 | MA1479.1 | DMRTC2 | MA1479.1 | AATTGMTACATT | DMRTC2 | 5.01e-17 | 9.75e-20 | 45000 | 75.2 | 58600 | 73.1 | 1.03 | EPI_DAR_24hr |
| 252 | 41 | MA0483.1 | Gfi1B | MA0483.1 | AAATCWCWGCH | Gfi1B | 5.59e-17 | 1.08e-19 | 24900 | 41.6 | 31400 | 39.2 | 1.06 | EPI_DAR_24hr |
| 253 | 26 | MA0775.1 | MEIS3 | MA0775.1 | DTGACAGS | MEIS3 | 1.56e-16 | 3.01e-19 | 11700 | 19.6 | 14200 | 17.7 | 1.11 | EPI_DAR_24hr |
| 254 | 28 | MA0833.2 | ATF4 | MA0833.2 | DDATGATGCAATMN | ATF4 | 1.72e-16 | 3.3e-19 | 35600 | 59.6 | 45800 | 57.2 | 1.04 | EPI_DAR_24hr |
| 255 | 34 | MA0060.3 | NFYA | MA0060.3 | RRCCAATCAGM | NFYA | 2.32e-16 | 4.44e-19 | 39400 | 65.8 | 50900 | 63.5 | 1.04 | EPI_DAR_24hr |
| 256 | 31 | MA0624.2 | Nfatc1 | MA0624.2 | NVATGGAAAAWN | Nfatc1 | 7.65e-16 | 1.46e-18 | 55300 | 92.4 | 73000 | 91.1 | 1.01 | EPI_DAR_24hr |
| 257 | 3 | MA1960.1 | MGA::EVX1 | MA1960.1 | RGGTGWTAATKD | MGA::EVX1 | 7.7e-16 | 1.46e-18 | 38700 | 64.7 | 50100 | 62.4 | 1.04 | EPI_DAR_24hr |
| 258 | 42 | MA0689.1 | TBX20 | MA0689.1 | WAGGTGTGAAR | TBX20 | 4.41e-15 | 8.34e-18 | 42900 | 71.7 | 55800 | 69.6 | 1.03 | EPI_DAR_24hr |
| 259 | 43 | MA1593.1 | ZNF317 | MA1593.1 | WVACAGCAGAYW | ZNF317 | 5.91e-15 | 1.11e-17 | 54400 | 90.8 | 71700 | 89.4 | 1.02 | EPI_DAR_24hr |
| 260 | 12 | MA0851.1 | Foxj3 | MA0851.1 | NDAADGTAAACAAANNM | Foxj3 | 7.63e-15 | 1.43e-17 | 32300 | 54 | 41500 | 51.7 | 1.04 | EPI_DAR_24hr |
| 261 | 44 | MA1125.1 | ZNF384 | MA1125.1 | DNWMAAAAAAAA | ZNF384 | 1.09e-14 | 2.03e-17 | 29200 | 48.7 | 37200 | 46.4 | 1.05 | EPI_DAR_24hr |
| 262 | 28 | MA0836.2 | CEBPD | MA0836.2 | NRTTGCACAAYHN | CEBPD | 1.11e-14 | 2.06e-17 | 48400 | 80.9 | 63400 | 79.1 | 1.02 | EPI_DAR_24hr |
| 263 | 45 | MA0739.1 (Hic1) | STREME-9 | 9-ATGCCWD | ATGCCWD | Hic1 | 1.41e-14 | 2.62e-17 | 47700 | 79.7 | 62400 | 77.8 | 1.02 | EPI_DAR_24hr |
| 264 | 3 | MA0897.1 | Hmx2 | MA0897.1 | ASAAGCAATTAAHVVRT | Hmx2 | 1.74e-14 | 3.22e-17 | 28600 | 47.8 | 36500 | 45.5 | 1.05 | EPI_DAR_24hr |
| 265 | 46 | MA1118.1 | SIX1 | MA1118.1 | GWAACCTGAKM | SIX1 | 3.73e-14 | 6.86e-17 | 47600 | 79.5 | 62200 | 77.6 | 1.02 | EPI_DAR_24hr |
| 266 | 33 | MA1632.1 | ATF2 | MA1632.1 | NNATGABGTCATN | ATF2 | 6.43e-14 | 1.18e-16 | 41800 | 69.9 | 54400 | 67.8 | 1.03 | EPI_DAR_24hr |
| 267 | 11 | MA0906.1 | HOXC12 | MA0906.1 | RGTCGTAAAAH | HOXC12 | 6.68e-14 | 1.22e-16 | 22300 | 37.2 | 28100 | 35.1 | 1.06 | EPI_DAR_24hr |
| 268 | 47 | MA1105.2 | GRHL2 | MA1105.2 | NRAACAGGTTYN | GRHL2 | 6.97e-14 | 1.27e-16 | 54400 | 90.9 | 71800 | 89.6 | 1.01 | EPI_DAR_24hr |
| 269 | 3 | MA1496.1 | HOXA4 | MA1496.1 | RTMATTAV | HOXA4 | 8.23e-14 | 1.49e-16 | 23400 | 39 | 29500 | 36.9 | 1.06 | EPI_DAR_24hr |
| 270 | 27 | MA0672.1 | NKX2-3 | MA0672.1 | VCCACTTRAV | NKX2-3 | 8.39e-14 | 1.52e-16 | 6690 | 11.2 | 7870 | 9.82 | 1.14 | EPI_DAR_24hr |
| 271 | 46 | MA1119.1 | SIX2 | MA1119.1 | NNCTGAAACCTGATMY | SIX2 | 2.92e-13 | 5.26e-16 | 54900 | 91.8 | 72600 | 90.6 | 1.01 | EPI_DAR_24hr |
| 272 | 3 | MA1463.1 | ARGFX | MA1463.1 | DCTAATTARM | ARGFX | 3.94e-13 | 7.07e-16 | 18300 | 30.6 | 22900 | 28.6 | 1.07 | EPI_DAR_24hr |
| 273 | 27 | MA0063.2 | NKX2-5 | MA0063.2 | NNCACTCAANN | NKX2-5 | 5.91e-13 | 1.06e-15 | 55500 | 92.8 | 73400 | 91.6 | 1.01 | EPI_DAR_24hr |
| 274 | 39 | MA1623.1 | Stat2 | MA1623.1 | RGAAACAGAAASH | Stat2 | 1.48e-12 | 2.63e-15 | 49600 | 82.9 | 65100 | 81.2 | 1.02 | EPI_DAR_24hr |
| 275 | 27 | MA2003.1 | NKX2-4 | MA2003.1 | CCACTTSAVV | NKX2-4 | 1.51e-12 | 2.68e-15 | 8540 | 14.3 | 10300 | 12.8 | 1.11 | EPI_DAR_24hr |
| 276 | 48 | MA0626.1 | Npas2 | MA0626.1 | NSCACGTGTN | Npas2 | 1.66e-12 | 2.93e-15 | 26200 | 43.7 | 33400 | 41.6 | 1.05 | EPI_DAR_24hr |
| 277 | 49 | MA1150.1 | RORB | MA1150.1 | AWWTRGGTCAH | RORB | 2.29e-12 | 4.03e-15 | 42800 | 71.4 | 55700 | 69.5 | 1.03 | EPI_DAR_24hr |
| 278 | 50 | MA1709.1 | ZIM3 | MA1709.1 | RVMAACAGAAACCYM | ZIM3 | 8.17e-12 | 1.43e-14 | 41000 | 68.6 | 53400 | 66.6 | 1.03 | EPI_DAR_24hr |
| 279 | 41 | MA0038.2 | GFI1 | MA0038.2 | BMAATCACDGCA | GFI1 | 9.52e-12 | 1.67e-14 | 13000 | 21.8 | 16100 | 20.1 | 1.08 | EPI_DAR_24hr |
| 280 | 33 | MA1112.2 | NR4A1 | MA1112.2 | NNAAAGGTCANN | NR4A1 | 1.76e-11 | 3.06e-14 | 44500 | 74.3 | 58100 | 72.5 | 1.02 | EPI_DAR_24hr |
| 281 | 51 | MA0106.3 | TP53 | MA0106.3 | RACATGYCCGGRCATGTY | TP53 | 2.45e-11 | 4.25e-14 | 406 | 0.68 | 309 | 0.39 | 1.76 | EPI_DAR_24hr |
| 282 | 34 | MA1523.1 | MSANTD3 | MA1523.1 | STVCACTCAC | MSANTD3 | 3.71e-11 | 6.42e-14 | 58500 | 97.7 | 77800 | 97 | 1.01 | EPI_DAR_24hr |
| 283 | 52 | MA1625.1 | Stat5b | MA1625.1 | NNNTTCCCAGAANNN | Stat5b | 5.13e-11 | 8.84e-14 | 52400 | 87.5 | 69000 | 86.1 | 1.02 | EPI_DAR_24hr |
| 284 | 52 | MA0144.2 | STAT3 | MA0144.2 | YTTCYKGGAAD | STAT3 | 5.38e-11 | 9.25e-14 | 39200 | 65.4 | 50900 | 63.5 | 1.03 | EPI_DAR_24hr |
| 285 | 3 | MA1501.1 | HOXB7 | MA1501.1 | GGTAATTAVS | HOXB7 | 5.88e-11 | 1.01e-13 | 19700 | 32.9 | 24900 | 31 | 1.06 | EPI_DAR_24hr |
| 286 | 33 | MA0093.3 | USF1 | MA0093.3 | NDGTCATGTGACHN | USF1 | 9.8e-11 | 1.67e-13 | 44300 | 74 | 57900 | 72.3 | 1.02 | EPI_DAR_24hr |
| 287 | 53 | MA1975.1 | ZNF214 | MA1975.1 | VWTCATCAABGTCCT | ZNF214 | 1.82e-10 | 3.1e-13 | 45800 | 76.6 | 60100 | 74.9 | 1.02 | EPI_DAR_24hr |
| 288 | 14 | MA1962.1 | POU2F1::SOX2 | MA1962.1 | MATTTRCATMACAATRG | POU2F1::SOX2 | 4.88e-10 | 8.26e-13 | 17800 | 29.7 | 22400 | 28 | 1.06 | EPI_DAR_24hr |
| 289 | 30 | MA1487.2 | FOXE1 | MA1487.2 | VYTAAAYAAACAAH | FOXE1 | 5.57e-10 | 9.4e-13 | 32700 | 54.7 | 42300 | 52.8 | 1.04 | EPI_DAR_24hr |
| 290 | 40 | MA1707.1 | DMRTA1 | MA1707.1 | AWTTGWTACAWT | DMRTA1 | 5.94e-10 | 9.99e-13 | 47300 | 79.1 | 62100 | 77.5 | 1.02 | EPI_DAR_24hr |
| 291 | 28 | MA0043.3 | HLF | MA0043.3 | NNRTTATGCAACHN | HLF | 6.05e-10 | 1.01e-12 | 30600 | 51.1 | 39500 | 49.2 | 1.04 | EPI_DAR_24hr |
| 292 | 49 | MA1111.1 | NR2F2 | MA1111.1 | NAAAGGTCANR | NR2F2 | 7.72e-10 | 1.29e-12 | 56500 | 94.5 | 75000 | 93.6 | 1.01 | EPI_DAR_24hr |
| 293 | 54 | MA0797.1 | TGIF2 | MA0797.1 | TGACAGSTGTCA | TGIF2 | 8.45e-10 | 1.41e-12 | 14500 | 24.3 | 18200 | 22.7 | 1.07 | EPI_DAR_24hr |
| 294 | 47 | MA1508.1 | IKZF1 | MA1508.1 | VVAACAGGAARN | IKZF1 | 9.04e-10 | 1.5e-12 | 59500 | 99.4 | 79400 | 99.1 | 1 | EPI_DAR_24hr |
| 295 | 33 | MA1131.1 | FOSL2::JUN | MA1131.1 | GRTGACGTMAT | FOSL2::JUN | 9.58e-10 | 1.59e-12 | 39700 | 66.2 | 51700 | 64.4 | 1.03 | EPI_DAR_24hr |
| 296 | 55 | MA0691.1 | TFAP4 | MA0691.1 | AWCAGCTGWT | TFAP4 | 1.35e-09 | 2.23e-12 | 12100 | 20.2 | 15000 | 18.7 | 1.08 | EPI_DAR_24hr |
| 297 | 56 | MA0092.1 | Hand1::Tcf3 | MA0092.1 | BRTCTGGMWT | Hand1::Tcf3 | 1.96e-09 | 3.23e-12 | 47100 | 78.7 | 61800 | 77.2 | 1.02 | EPI_DAR_24hr |
| 298 | 57 | MA0508.3 | PRDM1 | MA0508.3 | YNCTTTCTCTH | PRDM1 | 3.43e-09 | 5.62e-12 | 51100 | 85.4 | 67300 | 84 | 1.02 | EPI_DAR_24hr |
| 299 | 58 | MA0596.1 | SREBF2 | MA0596.1 | RTGGGGTGAY | SREBF2 | 3.51e-09 | 5.72e-12 | 6790 | 11.3 | 8170 | 10.2 | 1.11 | EPI_DAR_24hr |
| 300 | 59 | MA1125.1 (ZNF384) | MEME-1 | TTTTTTTTTTTTTTT | TTTTTTTTTTTTTTT | ZNF384 | 4.7e-09 | 7.65e-12 | 29500 | 49.3 | 38000 | 47.4 | 1.04 | EPI_DAR_24hr |
| 301 | 11 | MA0907.1 | HOXC13 | MA0907.1 | BCTCGTAAAAH | HOXC13 | 4.83e-09 | 7.83e-12 | 27300 | 45.7 | 35100 | 43.8 | 1.04 | EPI_DAR_24hr |
| 302 | 8 | MA1547.2 | PITX2 | MA1547.2 | YTAATCCY | PITX2 | 5.24e-09 | 8.47e-12 | 20600 | 34.4 | 26200 | 32.6 | 1.05 | EPI_DAR_24hr |
| 303 | 60 | MA1974.1 | ZNF211 | MA1974.1 | HWYATATACCAYRYW | ZNF211 | 5.56e-09 | 8.95e-12 | 28500 | 47.7 | 36800 | 45.9 | 1.04 | EPI_DAR_24hr |
| 304 | 15 | MA0683.1 | POU4F2 | MA0683.1 | RTGCATAATTAATGAG | POU4F2 | 8.9e-09 | 1.43e-11 | 8230 | 13.8 | 10000 | 12.5 | 1.1 | EPI_DAR_24hr |
| 305 | 31 | MA0152.2 | Nfatc2 | MA0152.2 | NNRNAATGGAAANWNH | Nfatc2 | 1.23e-08 | 1.96e-11 | 53900 | 90 | 71300 | 88.9 | 1.01 | EPI_DAR_24hr |
| 306 | 61 | MA0847.3 | FOXD2 | MA0847.3 | SYTAARYAAACA | FOXD2 | 2.74e-08 | 4.37e-11 | 34300 | 57.3 | 44500 | 55.6 | 1.03 | EPI_DAR_24hr |
| 307 | 5 | MA0515.1 | Sox6 | MA0515.1 | CCWTTGTYYY | Sox6 | 3.62e-08 | 5.75e-11 | 6480 | 10.8 | 7830 | 9.77 | 1.11 | EPI_DAR_24hr |
| 308 | 30 | MA0040.1 | Foxq1 | MA0040.1 | HATTGTTTATW | Foxq1 | 5.61e-08 | 8.88e-11 | 4830 | 8.07 | 5730 | 7.15 | 1.13 | EPI_DAR_24hr |
| 309 | 23 | MA1121.1 | TEAD2 | MA1121.1 | NYACATTCCWNNS | TEAD2 | 1e-07 | 1.59e-10 | 45800 | 76.5 | 60100 | 75 | 1.02 | EPI_DAR_24hr |
| 310 | 3 | MA0874.1 | Arx | MA0874.1 | VTBCRYTAATTARTKSW | Arx | 1.41e-07 | 2.21e-10 | 19200 | 32.1 | 24500 | 30.5 | 1.05 | EPI_DAR_24hr |
| 311 | 55 | MA0829.2 | SREBF1 | MA0829.2 | DNATCACSTGATNH | SREBF1 | 1.99e-07 | 3.11e-10 | 41500 | 69.4 | 54400 | 67.8 | 1.02 | EPI_DAR_24hr |
| 312 | 29 | MA0884.2 | DUXA | MA0884.2 | YTRATTWAATYAR | DUXA | 2.29e-07 | 3.58e-10 | 10400 | 17.3 | 12900 | 16.1 | 1.08 | EPI_DAR_24hr |
| 313 | 49 | MA1151.1 | RORC | MA1151.1 | NWAWNTRGGTCA | RORC | 3.7e-07 | 5.76e-10 | 36700 | 61.4 | 47900 | 59.7 | 1.03 | EPI_DAR_24hr |
| 314 | 58 | MA0595.1 | SREBF1 | MA0595.1 | VTCACCCCAY | SREBF1 | 3.89e-07 | 6.04e-10 | 9910 | 16.6 | 12300 | 15.3 | 1.08 | EPI_DAR_24hr |
| 315 | 19 | MA0869.2 | Sox11 | MA0869.2 | NRGAACAAAGVV | Sox11 | 5.84e-07 | 9.05e-10 | 57400 | 96 | 76400 | 95.3 | 1.01 | EPI_DAR_24hr |
| 316 | 5 | MA0087.2 | Sox5 | MA0087.2 | VRVAACAATGGNN | Sox5 | 6.13e-07 | 9.46e-10 | 43700 | 73.1 | 57400 | 71.6 | 1.02 | EPI_DAR_24hr |
| 317 | 62 | MA0479.1 | FOXH1 | MA0479.1 | BNSAATCCACA | FOXH1 | 1.56e-06 | 2.39e-09 | 8340 | 13.9 | 10300 | 12.8 | 1.08 | EPI_DAR_24hr |
| 318 | 22 | MA0756.2 | ONECUT2 | MA0756.2 | CGATCRATA | ONECUT2 | 1.94e-06 | 2.97e-09 | 13000 | 21.6 | 16300 | 20.4 | 1.06 | EPI_DAR_24hr |
| 319 | 55 | MA0698.1 | ZBTB18 | MA0698.1 | NATCCAGATGTKB | ZBTB18 | 1.96e-06 | 3e-09 | 22700 | 37.9 | 29100 | 36.4 | 1.04 | EPI_DAR_24hr |
| 320 | 63 | MA1729.1 | ZNF680 | MA1729.1 | GNCCAAGAAGAATDA | ZNF680 | 2.25e-06 | 3.43e-09 | 32900 | 54.9 | 42800 | 53.3 | 1.03 | EPI_DAR_24hr |
| 321 | 11 | MA1503.1 | HOXB9 | MA1503.1 | GTCGTAAAAH | HOXB9 | 2.7e-06 | 4.1e-09 | 5210 | 8.7 | 6280 | 7.83 | 1.11 | EPI_DAR_24hr |
| 322 | 52 | MA1624.1 | Stat5a | MA1624.1 | NTTCCAAGAANH | Stat5a | 3.56e-06 | 5.4e-09 | 48100 | 80.4 | 63400 | 79.1 | 1.02 | EPI_DAR_24hr |
| 323 | 54 | MA0667.1 | MYF6 | MA0667.1 | AACARCTGTT | MYF6 | 3.74e-06 | 5.65e-09 | 14700 | 24.6 | 18700 | 23.3 | 1.06 | EPI_DAR_24hr |
| 324 | 22 | MA0757.1 | ONECUT3 | MA0757.1 | AAAAAATCRATAAH | ONECUT3 | 4.32e-06 | 6.5e-09 | 10900 | 18.2 | 13600 | 17 | 1.07 | EPI_DAR_24hr |
| 325 | 33 | MA1127.1 | FOSB::JUN | MA1127.1 | GATGACGTCAT | FOSB::JUN | 4.52e-06 | 6.79e-09 | 43800 | 73.2 | 57500 | 71.8 | 1.02 | EPI_DAR_24hr |
| 326 | 3 | MA0618.1 | LBX1 | MA0618.1 | TTAATTAG | LBX1 | 4.97e-06 | 7.43e-09 | 14100 | 23.6 | 17900 | 22.3 | 1.06 | EPI_DAR_24hr |
| 327 | 64 | MA1592.1 | ZNF274 | MA1592.1 | NRTRTGAGTTCTCGYN | ZNF274 | 6.09e-06 | 9.08e-09 | 13900 | 23.3 | 17600 | 22 | 1.06 | EPI_DAR_24hr |
| 328 | 11 | MA0873.1 | HOXD12 | MA0873.1 | RGTCGTAAAAH | HOXD12 | 7.76e-06 | 1.15e-08 | 11100 | 18.5 | 13900 | 17.3 | 1.07 | EPI_DAR_24hr |
| 329 | 33 | MA0488.1 | JUN | MA0488.1 | DDRATGATGTCAT | JUN | 9.23e-06 | 1.37e-08 | 12000 | 20 | 15100 | 18.8 | 1.06 | EPI_DAR_24hr |
| 330 | 65 | MA0671.1 | NFIX | MA0671.1 | NNTGCCAAN | NFIX | 1.09e-05 | 1.61e-08 | 59700 | 99.7 | 79700 | 99.5 | 1 | EPI_DAR_24hr |
| 331 | 33 | MA0693.3 | Vdr | MA0693.3 | NNGAGTTCANN | Vdr | 2.23e-05 | 3.28e-08 | 58200 | 97.2 | 77500 | 96.7 | 1.01 | EPI_DAR_24hr |
| 332 | 3 | MA0780.1 | PAX3 | MA0780.1 | TAATYRATTA | PAX3 | 2.62e-05 | 3.85e-08 | 15500 | 25.9 | 19800 | 24.7 | 1.05 | EPI_DAR_24hr |
| 333 | 23 | MA0484.2 | HNF4G | MA0484.2 | DNCAAAGTCCANN | HNF4G | 4.16e-05 | 6.1e-08 | 51900 | 86.7 | 68700 | 85.7 | 1.01 | EPI_DAR_24hr |
| 334 | 23 | MA0114.4 | HNF4A | MA0114.4 | DNCAAAGTCCANN | HNF4A | 5e-05 | 7.3e-08 | 54300 | 90.8 | 72100 | 90 | 1.01 | EPI_DAR_24hr |
| 335 | 11 | MA1506.1 | HOXD10 | MA1506.1 | RGTCGTAAAAH | HOXD10 | 5.65e-05 | 8.23e-08 | 10700 | 17.9 | 13500 | 16.8 | 1.06 | EPI_DAR_24hr |
| 336 | 49 | MA0258.2 | ESR2 | MA0258.2 | AGGTCASVNTGMCCY | ESR2 | 6.17e-05 | 8.97e-08 | 52800 | 88.2 | 69900 | 87.3 | 1.01 | EPI_DAR_24hr |
| 337 | 52 | MA0519.1 | Stat5a::Stat5b | MA0519.1 | DTTTCCMAGAA | Stat5a::Stat5b | 6.55e-05 | 9.48e-08 | 30500 | 51 | 39700 | 49.5 | 1.03 | EPI_DAR_24hr |
| 338 | 65 | MA0161.2 | NFIC | MA0161.2 | NNCTTGGCANN | NFIC | 7.78e-05 | 1.12e-07 | 31700 | 53 | 41400 | 51.6 | 1.03 | EPI_DAR_24hr |
| 339 | 42 | MA0802.1 | TBR1 | MA0802.1 | AGGTGTGAAA | TBR1 | 8.06e-05 | 1.16e-07 | 48500 | 81 | 64000 | 79.9 | 1.01 | EPI_DAR_24hr |
| 340 | 37 | MA0502.2 | NFYB | MA0502.2 | CYCATTGGCCVV | NFYB | 0.000122 | 1.75e-07 | 1900 | 3.17 | 2160 | 2.7 | 1.17 | EPI_DAR_24hr |
| 341 | 48 | MA0461.2 | Atoh1 | MA0461.2 | RMCATATGBY | Atoh1 | 0.000128 | 1.83e-07 | 49400 | 82.6 | 65300 | 81.5 | 1.01 | EPI_DAR_24hr |
| 342 | 65 | MA1527.1 | NFIC | MA1527.1 | NTTGGCDNNRTGCCARN | NFIC | 0.000142 | 2.03e-07 | 3840 | 6.42 | 4620 | 5.76 | 1.11 | EPI_DAR_24hr |
| 343 | 27 | MA1994.1 | Nkx2-1 | MA1994.1 | NVCACTTGARHNN | Nkx2-1 | 0.000154 | 2.19e-07 | 23800 | 39.7 | 30800 | 38.4 | 1.03 | EPI_DAR_24hr |
| 344 | 66 | MA0065.2 | Pparg::Rxra | MA0065.2 | STRGGGCARAGGKCA | Pparg::Rxra | 0.000169 | 2.39e-07 | 59300 | 99 | 79100 | 98.7 | 1 | EPI_DAR_24hr |
| 345 | 17 | MA0773.1 | MEF2D | MA0773.1 | DCTAWAAATAGM | MEF2D | 0.000176 | 2.48e-07 | 8030 | 13.4 | 10000 | 12.5 | 1.07 | EPI_DAR_24hr |
| 346 | 54 | MA0669.1 | NEUROG2 | MA0669.1 | RACATATGTC | NEUROG2 | 0.000195 | 2.76e-07 | 21200 | 35.5 | 27400 | 34.2 | 1.04 | EPI_DAR_24hr |
| 347 | 67 | MA0640.2 | ELF3 | MA0640.2 | NNCCACTTCCTGNT | ELF3 | 0.000246 | 3.47e-07 | 59200 | 99 | 79100 | 98.7 | 1 | EPI_DAR_24hr |
| 348 | 52 | MA1579.1 | ZBTB26 | MA1579.1 | NNCTCCAGAAANNNN | ZBTB26 | 0.000251 | 3.52e-07 | 56200 | 93.9 | 74800 | 93.3 | 1.01 | EPI_DAR_24hr |
| 349 | 51 | MA0525.2 | TP63 | MA0525.2 | RACATGTYGKGACATGTC | TP63 | 0.000268 | 3.75e-07 | 196 | 0.33 | 153 | 0.19 | 1.71 | EPI_DAR_24hr |
| 350 | 54 | MA1618.1 | Ptf1a | MA1618.1 | NNACAGATGTTNN | Ptf1a | 0.000304 | 4.25e-07 | 32000 | 53.5 | 41800 | 52.1 | 1.03 | EPI_DAR_24hr |
| 351 | 3 | MA1577.1 | TLX2 | MA1577.1 | BYAATTAR | TLX2 | 0.000356 | 4.95e-07 | 15000 | 25.1 | 19200 | 24 | 1.05 | EPI_DAR_24hr |
| 352 | 40 | MA1478.1 | DMRTA2 | MA1478.1 | AATTGHTACAWT | DMRTA2 | 0.000426 | 5.91e-07 | 51300 | 85.8 | 68000 | 84.8 | 1.01 | EPI_DAR_24hr |
| 353 | 27 | MA1645.1 | NKX2-2 | MA1645.1 | NNNCCACTCAANNN | NKX2-2 | 0.000661 | 9.14e-07 | 53700 | 89.8 | 71300 | 89 | 1.01 | EPI_DAR_24hr |
| 354 | 65 | MA0670.1 | NFIA | MA0670.1 | NDTGCCAAND | NFIA | 0.000703 | 9.7e-07 | 7090 | 11.8 | 8840 | 11 | 1.07 | EPI_DAR_24hr |
| 355 | 50 | MA1953.1 | FOXO1::ELF1 | MA1953.1 | RTMAACAGGAAGTN | FOXO1::ELF1 | 0.000745 | 1.02e-06 | 36200 | 60.5 | 47500 | 59.3 | 1.02 | EPI_DAR_24hr |
| 356 | 48 | MA1108.2 | MXI1 | MA1108.2 | NNCACATGBN | MXI1 | 0.000942 | 1.29e-06 | 37500 | 62.6 | 49200 | 61.4 | 1.02 | EPI_DAR_24hr |
| 357 | 52 | MA1116.1 | RBPJ | MA1116.1 | BVTGGGAANN | RBPJ | 0.00108 | 1.48e-06 | 54600 | 91.2 | 72500 | 90.4 | 1.01 | EPI_DAR_24hr |
| 358 | 42 | MA0800.1 | EOMES | MA0800.1 | RAGGTGTGAAAWN | EOMES | 0.00122 | 1.66e-06 | 46600 | 77.9 | 61600 | 76.8 | 1.01 | EPI_DAR_24hr |
| 359 | 12 | MA0593.1 | FOXP2 | MA0593.1 | RWGTAAACAVR | FOXP2 | 0.00151 | 2.05e-06 | 3460 | 5.78 | 4180 | 5.21 | 1.11 | EPI_DAR_24hr |
| 360 | 51 | MA0861.1 | TP73 | MA0861.1 | DRCATGTCNNRACAYGYM | TP73 | 0.00164 | 2.23e-06 | 278 | 0.46 | 248 | 0.31 | 1.5 | EPI_DAR_24hr |
| 361 | 12 | MA0613.1 | FOXG1 | MA0613.1 | RTAAACAW | FOXG1 | 0.00179 | 2.42e-06 | 7280 | 12.2 | 9100 | 11.4 | 1.07 | EPI_DAR_24hr |
| 362 | 33 | MA1139.1 | FOSL2::JUNB | MA1139.1 | DATGACGTCATH | FOSL2::JUNB | 0.00229 | 3.08e-06 | 32300 | 54 | 42300 | 52.8 | 1.02 | EPI_DAR_24hr |
| 363 | 65 | MA0119.1 | NFIC::TLX1 | MA0119.1 | TGGCASSRWGCCAA | NFIC::TLX1 | 0.00257 | 3.46e-06 | 2000 | 3.34 | 2340 | 2.92 | 1.15 | EPI_DAR_24hr |
| 364 | 31 | MA0625.2 | NFATC3 | MA0625.2 | RYGGAAAMH | NFATC3 | 0.00389 | 5.21e-06 | 58700 | 98.1 | 78300 | 97.7 | 1 | EPI_DAR_24hr |
| 365 | 33 | MA1145.1 | FOSL2::JUND | MA1145.1 | NNRTGACGTCAYHSN | FOSL2::JUND | 0.00417 | 5.57e-06 | 42100 | 70.4 | 55600 | 69.3 | 1.02 | EPI_DAR_24hr |
| 366 | 48 | MA1638.1 | HAND2 | MA1638.1 | NVCAGATGNN | HAND2 | 0.00471 | 6.27e-06 | 9630 | 16.1 | 12200 | 15.2 | 1.06 | EPI_DAR_24hr |
| 367 | 26 | MA1114.1 | PBX3 | MA1114.1 | NNNTGAGTGACAGGBNN | PBX3 | 0.00539 | 7.17e-06 | 5300 | 8.86 | 6570 | 8.2 | 1.08 | EPI_DAR_24hr |
| 11 | 69 | 5-CCTCCCAAAG | STREME-5 | 5-CCTCCCAAAG | CCTCCCAAAG | CCTCCCAAAG | 0.0057 | 0.000518 | 1490 | 2.5 | 1780 | 2.22 | 1.12 | EPI_DAR_24hr |
| 368 | 48 | MA0620.3 | MITF | MA0620.3 | NNNDRTCACGTGAYHNNN | MITF | 0.007 | 9.29e-06 | 6620 | 11.1 | 8280 | 10.3 | 1.07 | EPI_DAR_24hr |
| 369 | 48 | MA0633.2 | Twist2 | MA0633.2 | NVCAGCTGBN | Twist2 | 0.00869 | 1.15e-05 | 8540 | 14.3 | 10800 | 13.5 | 1.06 | EPI_DAR_24hr |
| 370 | 52 | MA0520.1 | Stat6 | MA0520.1 | BDBTTCCWSAGAAVY | Stat6 | 0.00878 | 1.16e-05 | 19700 | 32.9 | 25500 | 31.8 | 1.03 | EPI_DAR_24hr |
| 371 | 66 | MA0160.2 | NR4A2 | MA0160.2 | BWAAAGGTCA | NR4A2 | 0.00929 | 1.22e-05 | 47400 | 79.3 | 62800 | 78.3 | 1.01 | EPI_DAR_24hr |
| 372 | 46 | MA2001.1 | Six4 | MA2001.1 | GDAACCTGANH | Six4 | 0.0106 | 1.38e-05 | 22100 | 36.8 | 28700 | 35.8 | 1.03 | EPI_DAR_24hr |
| 373 | 48 | MA1997.1 | Olig2 | MA1997.1 | NVCAGCTGBN | Olig2 | 0.0106 | 1.38e-05 | 8290 | 13.9 | 10500 | 13.1 | 1.06 | EPI_DAR_24hr |
| 374 | 3 | MA0621.1 | mix-a | MA0621.1 | NDTTAATTAGN | mix-a | 0.0109 | 1.42e-05 | 13400 | 22.4 | 17200 | 21.5 | 1.04 | EPI_DAR_24hr |
| 375 | 11 | MA0485.2 | HOXC9 | MA0485.2 | GTCGTAAAAH | HOXC9 | 0.0114 | 1.49e-05 | 1850 | 3.09 | 2170 | 2.7 | 1.14 | EPI_DAR_24hr |
| 376 | 52 | MA0518.1 | Stat4 | MA0518.1 | YTTCYRGGAARNVR | Stat4 | 0.0142 | 1.84e-05 | 31000 | 51.7 | 40600 | 50.6 | 1.02 | EPI_DAR_24hr |
| 377 | 33 | MA1126.1 | FOS::JUN | MA1126.1 | DRTGACGTCATHNDTN | FOS::JUN | 0.0265 | 3.43e-05 | 43200 | 72.1 | 57000 | 71.1 | 1.01 | EPI_DAR_24hr |
| 12 | 70 | MA1630.2 (ZNF281) | STREME-8 | 8-CCCCTTCCCCCAG | CCCCTTCCCCCAG | ZNF281 | 0.029 | 0.00242 | 593 | 0.99 | 675 | 0.84 | 1.18 | EPI_DAR_24hr |
| 378 | 48 | MA1635.1 | BHLHE22 | MA1635.1 | NVCAGCTGBN | BHLHE22 | 0.0344 | 4.44e-05 | 8410 | 14 | 10700 | 13.3 | 1.06 | EPI_DAR_24hr |
| 379 | 68 | MA0083.3 | SRF | MA0083.3 | TGMCCATATATGGKCA | SRF | 0.0377 | 4.85e-05 | 147 | 0.25 | 121 | 0.15 | 1.62 | EPI_DAR_24hr |
| 380 | 3 | MA0135.1 | Lhx3 | MA0135.1 | WAATTAATTAWYY | Lhx3 | 0.038 | 4.88e-05 | 11700 | 19.6 | 15000 | 18.7 | 1.04 | EPI_DAR_24hr |
| 381 | 48 | MA1993.1 | Neurod2 | MA1993.1 | VVCAGCTGBB | Neurod2 | 0.0437 | 5.59e-05 | 8520 | 14.2 | 10800 | 13.5 | 1.05 | EPI_DAR_24hr |
| 13 | 71 | MA0090.3 (TEAD1) | MEME-4 | GGCTGGAGTGCAGTG | GGCTGGAGTGCAGTG | TEAD1 | 6.23 | 0.48 | 1610 | 2.69 | 2150 | 2.68 | 1.01 | EPI_DAR_24hr |
| 14 | 70 | MA0528.2 (ZNF263) | MEME-3 | GGSBGRGGSWGGRGG | GGSBGRGGSWGGRGG | ZNF263 | 13.7 | 0.975 | 118 | 0.2 | 197 | 0.25 | 0.805 | EPI_DAR_24hr |
ER_rat <- 1.25
mrc_type <- "EPI_DAR_24hr"
EPI_DAR_24hr_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,mrc_type,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks(200bp)Enrichment ratio:",ER_rat))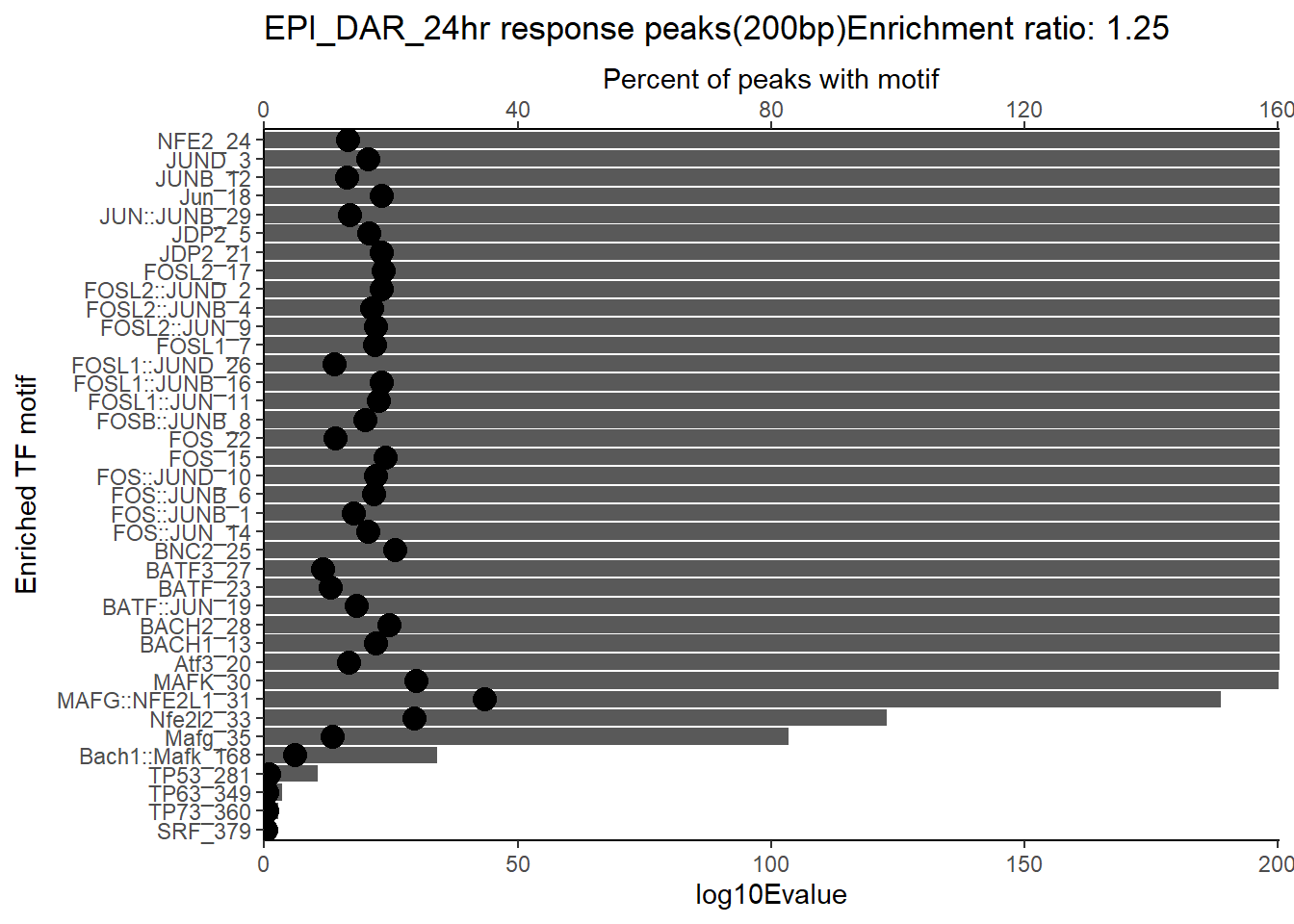
EPI_DAR_24hr_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,mrc_type,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
dplyr::filter(EVALUE.y<0.05) %>%
slice_head(n=5) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*.8), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./.8,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks(200bp)Enrichment ratio:",ER_rat))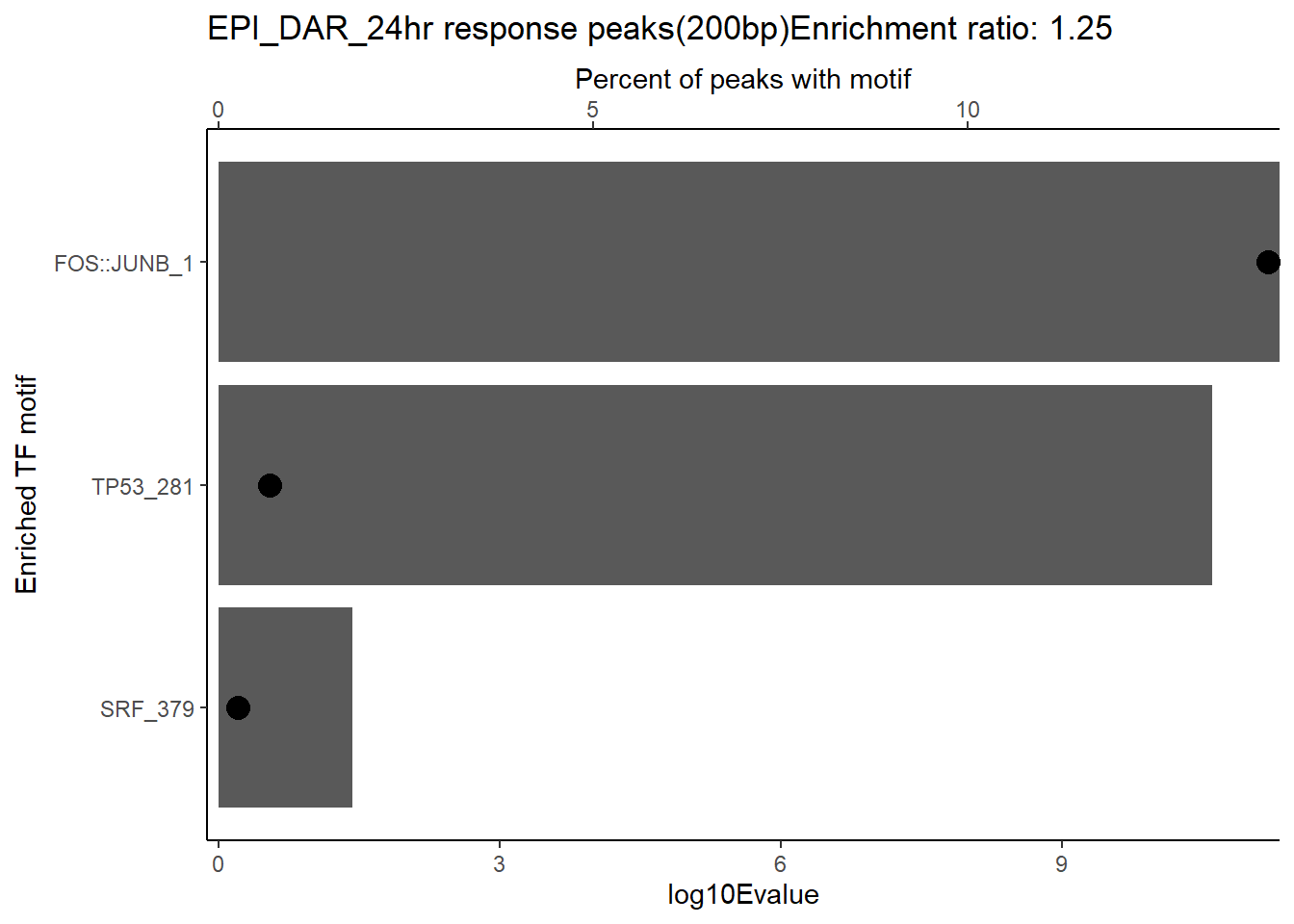
EPI_DAR 24 2kb data
EPI_242kb_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/EPI_24_2kb_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
EPI_242kb_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/EPI_24_2kb_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
EPI_242kb_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/EPI_24_2kb_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
EPI_242kb_sea_all <- rbind(EPI_242kb_sea_disc, EPI_242kb_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)EPI_242kb_combo <- EPI_242kb_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., EPI_242kb_sea_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ SIM_MOTIF
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y)
EPI_242kb_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs 200 bp in EPI_24 2kb") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | MA0491.2 | JUND | MA0491.2 | NNATGACTCATNN | JUND | 5.97e-108 | 2.06e-108 | 1050 | 8.35 | 564 | 2.77 | 3.01 |
| 2 | 1 | MA1141.1 | FOS::JUND | MA1141.1 | NKATGAGTCATNN | FOS::JUND | 1.9e-107 | 3.27e-108 | 1600 | 12.8 | 1150 | 5.64 | 2.26 |
| 3 | 1 | MA0476.1 | FOS | MA0476.1 | DVTGASTCATB | FOS | 2.44e-104 | 2.81e-105 | 1630 | 12.9 | 1190 | 5.85 | 2.21 |
| 4 | 1 | MA1128.1 | FOSL1::JUN | MA1128.1 | NKATGACTCATNN | FOSL1::JUN | 1.12e-103 | 9.65e-105 | 1280 | 10.2 | 816 | 4.01 | 2.54 |
| 5 | 1 | MA0489.2 | Jun | MA0489.2 | NVTGACTCATNN | Jun | 1.68e-102 | 1.16e-103 | 1110 | 8.81 | 644 | 3.16 | 2.78 |
| 6 | 1 | MA0476.1 (FOS) | MEME-4 | TGASTCAT | TGASTCAT | FOS | 2.59e-101 | 1.49e-102 | 1360 | 10.8 | 905 | 4.45 | 2.42 |
| 7 | 1 | MA0477.2 | FOSL1 | MA0477.2 | NNATGACTCATNN | FOSL1 | 3.34e-101 | 1.65e-102 | 959 | 7.62 | 503 | 2.47 | 3.08 |
| 8 | 1 | MA0099.3 | FOS::JUN | MA0099.3 | ATGAGTCAYM | FOS::JUN | 3.95e-101 | 1.71e-102 | 1740 | 13.8 | 1330 | 6.54 | 2.11 |
| 9 | 1 | MA0478.1 | FOSL2 | MA0478.1 | KRRTGASTCAB | FOSL2 | 1.01e-100 | 3.87e-102 | 1830 | 14.5 | 1440 | 7.08 | 2.05 |
| 10 | 1 | MA1144.1 | FOSL2::JUND | MA1144.1 | KATGACTCAT | FOSL2::JUND | 1.24e-100 | 4.29e-102 | 1780 | 14.1 | 1380 | 6.8 | 2.08 |
| 11 | 1 | MA1138.1 | FOSL2::JUNB | MA1138.1 | KRTGASTCAT | FOSL2::JUNB | 3.98e-99 | 1.25e-100 | 2010 | 16 | 1670 | 8.19 | 1.95 |
| 12 | 1 | MA1135.1 | FOSB::JUNB | MA1135.1 | KRTGASTCAT | FOSB::JUNB | 5.42e-99 | 1.56e-100 | 1870 | 14.8 | 1500 | 7.37 | 2.01 |
| 13 | 1 | MA1134.1 | FOS::JUNB | MA1134.1 | KATGASTCATHN | FOS::JUNB | 6.89e-99 | 1.83e-100 | 2070 | 16.4 | 1740 | 8.54 | 1.92 |
| 14 | 1 | MA1634.1 | BATF | MA1634.1 | DATGACTCATH | BATF | 2.18e-98 | 5.38e-100 | 1020 | 8.1 | 572 | 2.81 | 2.88 |
| 15 | 1 | MA1130.1 | FOSL2::JUN | MA1130.1 | NNRTGAGTCAYN | FOSL2::JUN | 2.08e-97 | 4.79e-99 | 1760 | 14 | 1380 | 6.77 | 2.06 |
| 16 | 1 | MA0462.2 | BATF::JUN | MA0462.2 | DATGACTCATH | BATF::JUN | 2.47e-97 | 5.33e-99 | 1070 | 8.47 | 623 | 3.06 | 2.77 |
| 17 | 1 | MA1928.1 | BNC2 | MA1928.1 | NDRTGAGTCABN | BNC2 | 4.24e-96 | 8.6e-98 | 1620 | 12.9 | 1230 | 6.06 | 2.13 |
| 18 | 1 | MA0490.2 | JUNB | MA0490.2 | NNATGACTCATNN | JUNB | 5.28e-96 | 1.01e-97 | 916 | 7.28 | 482 | 2.37 | 3.07 |
| 19 | 1 | MA0477.2 (FOSL1) | STREME-1 | 1-DDDATGASTCATHHH | DDDATGASTCATHHH | FOSL1 | 8.92e-96 | 1.62e-97 | 1850 | 14.7 | 1500 | 7.35 | 2 |
| 20 | 1 | MA0835.2 | BATF3 | MA0835.2 | DATGACTCATH | BATF3 | 2.36e-92 | 4.08e-94 | 1080 | 8.57 | 657 | 3.23 | 2.65 |
| 21 | 1 | MA1633.2 | BACH1 | MA1633.2 | ATGACTCAT | BACH1 | 4.67e-92 | 7.68e-94 | 1610 | 12.8 | 1240 | 6.09 | 2.1 |
| 22 | 1 | MA1137.1 | FOSL1::JUNB | MA1137.1 | NDRTGACTCATNN | FOSL1::JUNB | 6.85e-91 | 1.07e-92 | 1640 | 13.1 | 1290 | 6.33 | 2.06 |
| 23 | 1 | MA1101.2 | BACH2 | MA1101.2 | DWANCATGASTCATSNTWH | BACH2 | 7.85e-86 | 1.18e-87 | 1420 | 11.3 | 1060 | 5.23 | 2.16 |
| 24 | 1 | MA1988.1 | Atf3 | MA1988.1 | NRTGACTCABN | Atf3 | 4.02e-84 | 5.79e-86 | 708 | 5.63 | 333 | 1.64 | 3.43 |
| 25 | 1 | MA0841.1 | NFE2 | MA0841.1 | VATGACTCATS | NFE2 | 2.75e-78 | 3.79e-80 | 1630 | 13 | 1360 | 6.66 | 1.95 |
| 26 | 1 | MA0655.1 | JDP2 | MA0655.1 | ATGACTCAT | JDP2 | 9.01e-74 | 1.2e-75 | 2420 | 19.2 | 2390 | 11.7 | 1.64 |
| 27 | 1 | MA1142.1 | FOSL1::JUND | MA1142.1 | NRTGACTCAT | FOSL1::JUND | 1.52e-70 | 1.94e-72 | 2530 | 20.1 | 2570 | 12.6 | 1.6 |
| 28 | 1 | MA1132.1 | JUN::JUNB | MA1132.1 | KATGACKCAT | JUN::JUNB | 5.19e-62 | 6.4e-64 | 2410 | 19.1 | 2480 | 12.2 | 1.57 |
| 29 | 1 | MA0496.3 | MAFK | MA0496.3 | NGMTGACTCAGCMNH | MAFK | 4.14e-60 | 4.93e-62 | 2930 | 23.3 | 3220 | 15.8 | 1.47 |
| 30 | 1 | MA0089.2 | MAFG::NFE2L1 | MA0089.2 | NVNATGACTCAGCADW | MAFG::NFE2L1 | 2.92e-59 | 3.36e-61 | 2600 | 20.7 | 2770 | 13.6 | 1.52 |
| 31 | 2 | 3-AGKYATSW | STREME-3 | 3-AGKYATSW | AGKYATSW | AGKYATSW | 4.18e-56 | 4.66e-58 | 6660 | 52.9 | 8910 | 43.8 | 1.21 |
| 32 | 3 | MA0704.1 | Lhx4 | MA0704.1 | YTAATTAR | Lhx4 | 1.81e-54 | 1.95e-56 | 6750 | 53.7 | 9090 | 44.6 | 1.2 |
| 33 | 3 | MA0710.1 | NOTO | MA0710.1 | NYTAATTAVN | NOTO | 7.6e-54 | 7.95e-56 | 8170 | 65 | 11400 | 56.2 | 1.16 |
| 34 | 4 | MA1963.1 | SATB1 | MA1963.1 | WWDCTAATAAHWW | SATB1 | 9.1e-54 | 9.24e-56 | 6560 | 52.1 | 8780 | 43.1 | 1.21 |
| 35 | 3 | MA0722.1 | VAX1 | MA0722.1 | YTAATTAH | VAX1 | 2.15e-52 | 2.13e-54 | 8900 | 70.7 | 12700 | 62.3 | 1.13 |
| 36 | 1 | MA1639.1 | MEIS1 | MA1639.1 | NHCATMAATCAHN | MEIS1 | 5.15e-52 | 4.94e-54 | 6590 | 52.3 | 8860 | 43.5 | 1.2 |
| 37 | 5 | MA0882.1 | DLX6 | MA0882.1 | VYAATTAN | DLX6 | 1.48e-50 | 1.38e-52 | 7350 | 58.4 | 10100 | 49.7 | 1.17 |
| 38 | 5 | MA0642.2 (EN2) | STREME-2 | 2-ATTRGM | ATTRGM | EN2 | 1.77e-50 | 1.61e-52 | 6240 | 49.6 | 8340 | 41 | 1.21 |
| 39 | 3 | MA0723.2 | VAX2 | MA0723.2 | BYAATTAV | VAX2 | 3.69e-50 | 3.27e-52 | 7450 | 59.2 | 10300 | 50.6 | 1.17 |
| 40 | 1 | MA0501.1 | MAF::NFE2 | MA0501.1 | ATGACTCAGCANWTY | MAF::NFE2 | 5.78e-50 | 4.99e-52 | 2370 | 18.8 | 2560 | 12.6 | 1.5 |
| 41 | 3 | MA0661.1 | MEOX1 | MA0661.1 | VSTAATTAHC | MEOX1 | 2.04e-49 | 1.72e-51 | 8340 | 66.3 | 11800 | 57.9 | 1.14 |
| 42 | 5 | MA0720.1 | Shox2 | MA0720.1 | YYAATTAR | Shox2 | 6.85e-48 | 5.63e-50 | 6380 | 50.7 | 8600 | 42.2 | 1.2 |
| 43 | 6 | MA0885.2 | Dlx2 | MA0885.2 | NDGCAATTAGNN | Dlx2 | 8.25e-48 | 6.62e-50 | 7690 | 61.1 | 10700 | 52.7 | 1.16 |
| 44 | 7 | MA1113.2 | PBX2 | MA1113.2 | NHCATAAATCAHN | PBX2 | 1.59e-47 | 1.24e-49 | 6380 | 50.7 | 8610 | 42.3 | 1.2 |
| 45 | 8 | 4-TMTSAKA | STREME-4 | 4-TMTSAKA | TMTSAKA | TMTSAKA | 1.61e-47 | 1.24e-49 | 6360 | 50.5 | 8570 | 42.1 | 1.2 |
| 46 | 9 | MA0495.3 | MAFF | MA0495.3 | DWGTCAGCATTTTWHN | MAFF | 2.16e-47 | 1.62e-49 | 6980 | 55.5 | 9580 | 47.1 | 1.18 |
| 47 | 1 | MA0150.2 | Nfe2l2 | MA0150.2 | CASNATGACTCAGCA | Nfe2l2 | 6.77e-47 | 4.97e-49 | 3300 | 26.2 | 3910 | 19.2 | 1.36 |
| 48 | 5 | MA0721.1 | UNCX | MA0721.1 | YYAATTAV | UNCX | 1.3e-46 | 9.36e-49 | 7010 | 55.7 | 9640 | 47.4 | 1.18 |
| 49 | 10 | MA0789.1 | POU3F4 | MA0789.1 | TATGCWAAT | POU3F4 | 1.56e-46 | 1.1e-48 | 6960 | 55.3 | 9560 | 47 | 1.18 |
| 50 | 5 | MA0644.2 | ESX1 | MA0644.2 | YYAATTAN | ESX1 | 1.16e-44 | 7.99e-47 | 6100 | 48.5 | 8220 | 40.4 | 1.2 |
| 51 | 3 | MA0681.2 | PHOX2B | MA0681.2 | WWTAATCAAATTAWWW | PHOX2B | 1.38e-44 | 9.35e-47 | 6090 | 48.4 | 8190 | 40.2 | 1.2 |
| 52 | 6 | MA1476.2 | Dlx5 | MA1476.2 | NDGCAATTAGNN | Dlx5 | 5.46e-44 | 3.63e-46 | 7590 | 60.3 | 10600 | 52.2 | 1.15 |
| 53 | 3 | MA0675.1 | NKX6-2 | MA0675.1 | NYMATTAA | NKX6-2 | 5.69e-44 | 3.71e-46 | 6000 | 47.7 | 8070 | 39.6 | 1.2 |
| 54 | 3 | MA0680.2 | Pax7 | MA0680.2 | DWTAATCAATTAWNH | Pax7 | 7.05e-44 | 4.51e-46 | 6500 | 51.7 | 8860 | 43.6 | 1.19 |
| 55 | 5 | MA0880.1 | Dlx3 | MA0880.1 | NYAATTAN | Dlx3 | 1.54e-43 | 9.69e-46 | 6020 | 47.8 | 8100 | 39.8 | 1.2 |
| 56 | 3 | MA0628.1 | POU6F1 | MA0628.1 | AYTAATTART | POU6F1 | 1.95e-43 | 1.2e-45 | 8460 | 67.2 | 12100 | 59.4 | 1.13 |
| 57 | 5 | MA0650.3 | Hoxa13 | MA0650.3 | NNCCAATAAAAN | Hoxa13 | 2.69e-43 | 1.63e-45 | 8390 | 66.7 | 12000 | 58.9 | 1.13 |
| 58 | 5 | MA0634.1 | ALX3 | MA0634.1 | YYYAATTANV | ALX3 | 2.99e-43 | 1.78e-45 | 8470 | 67.3 | 12100 | 59.6 | 1.13 |
| 59 | 3 | MA0853.1 | Alx4 | MA0853.1 | CGCRYTAATTARYHNNY | Alx4 | 6.26e-43 | 3.66e-45 | 5220 | 41.5 | 6860 | 33.7 | 1.23 |
| 60 | 3 | MA0726.1 | VSX2 | MA0726.1 | CTAATTAN | VSX2 | 7.33e-43 | 4.22e-45 | 10200 | 81.2 | 15200 | 74.5 | 1.09 |
| 61 | 11 | WARAAWAAAAWAAWA | MEME-1 | WARAAWAAAAWAAWA | WARAAWAAAAWAAWA | WARAAWAAAAWAAWA | 1.36e-42 | 7.69e-45 | 6950 | 55.2 | 9610 | 47.2 | 1.17 |
| 62 | 5 | MA0718.1 | RAX | MA0718.1 | RYYAATTARH | RAX | 4.95e-42 | 2.75e-44 | 6070 | 48.2 | 8210 | 40.3 | 1.2 |
| 63 | 12 | MA0052.4 | MEF2A | MA0052.4 | DDCTAAAAATAGMHH | MEF2A | 1.67e-41 | 9.15e-44 | 6750 | 53.6 | 9300 | 45.7 | 1.17 |
| 64 | 3 | MA0793.1 | POU6F2 | MA0793.1 | ASCTMATTAW | POU6F2 | 3.09e-41 | 1.67e-43 | 7250 | 57.6 | 10100 | 49.7 | 1.16 |
| 65 | 13 | MA0077.1 | SOX9 | MA0077.1 | CYATTGTTY | SOX9 | 3.25e-41 | 1.73e-43 | 9290 | 73.9 | 13600 | 66.6 | 1.11 |
| 66 | 5 | MA0713.1 | PHOX2A | MA0713.1 | TAATYYAATTA | PHOX2A | 7.02e-41 | 3.67e-43 | 5360 | 42.6 | 7110 | 34.9 | 1.22 |
| 67 | 5 | MA0700.2 | LHX2 | MA0700.2 | DDCCAATTADN | LHX2 | 1.05e-40 | 5.41e-43 | 7560 | 60.1 | 10600 | 52.3 | 1.15 |
| 68 | 14 | MA0845.1 | FOXB1 | MA0845.1 | WATGTAAATAT | FOXB1 | 1.4e-40 | 7.09e-43 | 5910 | 47 | 7980 | 39.2 | 1.2 |
| 69 | 3 | MA0674.1 | NKX6-1 | MA0674.1 | NTMATTAA | NKX6-1 | 1.87e-40 | 9.38e-43 | 10000 | 79.5 | 14800 | 72.8 | 1.09 |
| 70 | 5 | MA0889.1 | GBX1 | MA0889.1 | VCYAATTASY | GBX1 | 1.98e-40 | 9.75e-43 | 6810 | 54.1 | 9420 | 46.3 | 1.17 |
| 71 | 15 | MA1104.2 | GATA6 | MA1104.2 | HWWTCTTATCTNH | GATA6 | 2.58e-40 | 1.26e-42 | 5060 | 40.3 | 6670 | 32.8 | 1.23 |
| 72 | 3 | MA0886.1 | EMX2 | MA0886.1 | VBTAATTARB | EMX2 | 3.73e-40 | 1.79e-42 | 6000 | 47.7 | 8140 | 40 | 1.19 |
| 73 | 3 | MA1518.2 | Lhx1 | MA1518.2 | NNNTGCTAATTAGCANNN | Lhx1 | 1.49e-39 | 7.04e-42 | 6710 | 53.3 | 9280 | 45.6 | 1.17 |
| 74 | 3 | MA0887.1 | EVX1 | MA0887.1 | VVTAATTABS | EVX1 | 1.75e-39 | 8.1e-42 | 7700 | 61.2 | 10900 | 53.6 | 1.14 |
| 75 | 5 | MA0901.2 | HOXB13 | MA0901.2 | WDCCAATAAAAWWW | HOXB13 | 1.76e-39 | 8.1e-42 | 7310 | 58.1 | 10300 | 50.4 | 1.15 |
| 76 | 16 | MA0676.1 | Nr2e1 | MA0676.1 | AAAAGTCAA | Nr2e1 | 1.79e-39 | 8.11e-42 | 8590 | 68.3 | 12400 | 60.9 | 1.12 |
| 77 | 13 | MA0084.1 | SRY | MA0084.1 | NWWAACAAT | SRY | 1.88e-39 | 8.45e-42 | 9450 | 75.1 | 13900 | 68.1 | 1.1 |
| 78 | 3 | MA0725.1 | VSX1 | MA0725.1 | YTAATTAN | VSX1 | 2.3e-39 | 1.02e-41 | 5390 | 42.8 | 7190 | 35.3 | 1.21 |
| 79 | 3 | MA1495.1 | HOXA1 | MA1495.1 | STAATTAS | HOXA1 | 3.22e-39 | 1.41e-41 | 4910 | 39 | 6450 | 31.7 | 1.23 |
| 80 | 5 | MA0881.1 | Dlx4 | MA0881.1 | VYAATTAN | Dlx4 | 4.9e-39 | 2.12e-41 | 5420 | 43.1 | 7250 | 35.6 | 1.21 |
| 81 | 3 | MA0706.1 | MEOX2 | MA0706.1 | DSTAATTAWN | MEOX2 | 1.24e-38 | 5.3e-41 | 5200 | 41.3 | 6910 | 33.9 | 1.22 |
| 82 | 10 | MA1115.1 | POU5F1 | MA1115.1 | WHATGCAAATN | POU5F1 | 3.5e-38 | 1.47e-40 | 6710 | 53.3 | 9310 | 45.7 | 1.17 |
| 83 | 5 | MA0630.1 | SHOX | MA0630.1 | HTAATTRR | SHOX | 4.65e-38 | 1.93e-40 | 5330 | 42.3 | 7120 | 35 | 1.21 |
| 84 | 3 | MA1530.1 | NKX6-3 | MA1530.1 | VYCATTAWW | NKX6-3 | 5.8e-38 | 2.36e-40 | 5730 | 45.6 | 7760 | 38.1 | 1.2 |
| 85 | 5 | MA0662.1 | MIXL1 | MA0662.1 | NBYAATTAVN | MIXL1 | 5.82e-38 | 2.36e-40 | 6480 | 51.5 | 8950 | 44 | 1.17 |
| 86 | 3 | MA0904.2 | HOXB5 | MA0904.2 | STAATTAS | HOXB5 | 6.45e-38 | 2.59e-40 | 6840 | 54.4 | 9530 | 46.8 | 1.16 |
| 87 | 6 | MA0035.4 | GATA1 | MA0035.4 | NHCTAATCTDH | GATA1 | 9.74e-38 | 3.87e-40 | 5770 | 45.9 | 7820 | 38.4 | 1.19 |
| 88 | 3 | MA0788.1 | POU3F3 | MA0788.1 | WWTATGCWAATTW | POU3F3 | 1.19e-37 | 4.68e-40 | 6310 | 50.2 | 8680 | 42.6 | 1.18 |
| 89 | 3 | MA0903.1 | HOXB3 | MA0903.1 | NNYMATTARN | HOXB3 | 2.33e-37 | 9.05e-40 | 7910 | 62.9 | 11300 | 55.5 | 1.13 |
| 90 | 5 | MA0709.1 | Msx3 | MA0709.1 | SCAATTAN | Msx3 | 4.17e-37 | 1.6e-39 | 5030 | 40 | 6680 | 32.8 | 1.22 |
| 91 | 6 | MA0122.3 | Nkx3-2 | MA0122.3 | WWAAMCACTTAAN | Nkx3-2 | 1.16e-36 | 4.39e-39 | 8350 | 66.3 | 12000 | 59.1 | 1.12 |
| 92 | 1 | MA0659.3 | Mafg | MA0659.3 | NWGMTGACTCAGCAN | Mafg | 1.31e-36 | 4.91e-39 | 5240 | 41.6 | 7010 | 34.4 | 1.21 |
| 93 | 13 | MA1562.1 | SOX14 | MA1562.1 | CCGAACAATG | SOX14 | 1.56e-36 | 5.78e-39 | 7830 | 62.2 | 11200 | 54.9 | 1.13 |
| 94 | 6 | MA1978.1 | ZNF354A | MA1978.1 | HWAADWATAAATGRAYWAWTT | ZNF354A | 1.64e-36 | 6.01e-39 | 8800 | 70 | 12800 | 63 | 1.11 |
| 95 | 10 | MA0792.1 | POU5F1B | MA0792.1 | TATGCWAAT | POU5F1B | 2.46e-36 | 8.95e-39 | 6090 | 48.4 | 8350 | 41 | 1.18 |
| 96 | 3 | MA1463.1 | ARGFX | MA1463.1 | DCTAATTARM | ARGFX | 3.35e-36 | 1.2e-38 | 4670 | 37.1 | 6130 | 30.1 | 1.23 |
| 97 | 14 | MA0032.2 | FOXC1 | MA0032.2 | WAWGTAAAYAW | FOXC1 | 3.47e-36 | 1.23e-38 | 6130 | 48.7 | 8420 | 41.4 | 1.18 |
| 98 | 17 | MA0809.2 | TEAD4 | MA0809.2 | NNACATTCCARN | TEAD4 | 3.75e-36 | 1.32e-38 | 9610 | 76.4 | 14200 | 69.8 | 1.09 |
| 99 | 3 | MA1519.1 | LHX5 | MA1519.1 | STAATTAV | LHX5 | 4.37e-36 | 1.52e-38 | 4100 | 32.6 | 5270 | 25.9 | 1.26 |
| 100 | 3 | MA0910.2 | HOXD8 | MA0910.2 | GYMATTAM | HOXD8 | 6.79e-36 | 2.34e-38 | 5340 | 42.4 | 7170 | 35.2 | 1.2 |
| 101 | 18 | MA0025.2 | NFIL3 | MA0025.2 | NRTTATGYAATNN | NFIL3 | 9.01e-36 | 3.08e-38 | 6930 | 55.1 | 9720 | 47.7 | 1.15 |
| 102 | 3 | MA0701.2 | LHX9 | MA0701.2 | CYAATTAR | LHX9 | 1.54e-35 | 5.21e-38 | 4450 | 35.4 | 5810 | 28.6 | 1.24 |
| 103 | 19 | 7-ATAACA | STREME-7 | 7-ATAACA | ATAACA | ATAACA | 1.84e-35 | 6.18e-38 | 7740 | 61.5 | 11100 | 54.3 | 1.13 |
| 104 | 3 | MA0900.2 | HOXA2 | MA0900.2 | STAATTAS | HOXA2 | 3.25e-35 | 1.08e-37 | 4170 | 33.2 | 5390 | 26.5 | 1.25 |
| 105 | 7 | MA1640.1 | MEIS2 | MA1640.1 | NNATGATTTATGDNH | MEIS2 | 3.27e-35 | 1.08e-37 | 4740 | 37.6 | 6250 | 30.7 | 1.22 |
| 106 | 5 | MA0654.1 | ISX | MA0654.1 | YYAATTAR | ISX | 3.65e-35 | 1.19e-37 | 4860 | 38.6 | 6440 | 31.7 | 1.22 |
| 107 | 3 | MA1502.1 | HOXB8 | MA1502.1 | GYMATTAM | HOXB8 | 5.48e-35 | 1.77e-37 | 5280 | 41.9 | 7100 | 34.9 | 1.2 |
| 108 | 3 | MA0068.2 | PAX4 | MA0068.2 | CTAATTAG | PAX4 | 6.78e-35 | 2.17e-37 | 5220 | 41.5 | 7010 | 34.5 | 1.2 |
| 109 | 6 | MA0899.1 | HOXA10 | MA0899.1 | DGYMATAAAAH | HOXA10 | 6.89e-35 | 2.18e-37 | 8930 | 71 | 13100 | 64.2 | 1.11 |
| 110 | 13 | MA0078.2 | Sox17 | MA0078.2 | NNAGAACAATGGNN | Sox17 | 7.97e-35 | 2.5e-37 | 8840 | 70.2 | 12900 | 63.4 | 1.11 |
| 111 | 15 | MA1970.1 | TRPS1 | MA1970.1 | NHTCTTATCTNH | TRPS1 | 8.6e-35 | 2.68e-37 | 4640 | 36.9 | 6120 | 30.1 | 1.23 |
| 112 | 5 | MA0717.1 | RAX2 | MA0717.1 | YYAATTAV | RAX2 | 9.58e-35 | 2.95e-37 | 5450 | 43.3 | 7370 | 36.2 | 1.2 |
| 113 | 6 | MA1549.1 | POU6F1 | MA1549.1 | RTAATGAGVN | POU6F1 | 1.5e-34 | 4.57e-37 | 5320 | 42.3 | 7180 | 35.3 | 1.2 |
| 114 | 14 | MA1683.1 | FOXA3 | MA1683.1 | DWGTAAACANN | FOXA3 | 1.6e-34 | 4.86e-37 | 8400 | 66.8 | 12200 | 59.8 | 1.12 |
| 115 | 3 | MA0854.1 | Alx1 | MA0854.1 | CGMRYTAATTARTNMNY | Alx1 | 2.6e-34 | 7.81e-37 | 4680 | 37.2 | 6190 | 30.4 | 1.22 |
| 116 | 5 | MA0909.3 | Hoxd13 | MA0909.3 | NNCAATAAAW | Hoxd13 | 3.17e-34 | 9.45e-37 | 6600 | 52.4 | 9210 | 45.3 | 1.16 |
| 117 | 15 | MA0036.3 | GATA2 | MA0036.3 | NBCTTATCTNH | GATA2 | 4.5e-34 | 1.33e-36 | 4990 | 39.7 | 6670 | 32.8 | 1.21 |
| 118 | 5 | MA0602.1 | Arid5a | MA0602.1 | SYAATATTGVDANH | Arid5a | 6.41e-34 | 1.87e-36 | 7200 | 57.2 | 10200 | 50.1 | 1.14 |
| 119 | 6 | MA0712.2 | OTX2 | MA0712.2 | NDRGGATTARNN | OTX2 | 6.44e-34 | 1.87e-36 | 9180 | 73 | 13500 | 66.4 | 1.1 |
| 120 | 3 | MA0892.1 | GSX1 | MA0892.1 | NCYMATTAVN | GSX1 | 1.24e-33 | 3.56e-36 | 8830 | 70.2 | 12900 | 63.4 | 1.11 |
| 121 | 15 | MA0766.2 | GATA5 | MA0766.2 | SWGATAASRN | GATA5 | 1.34e-33 | 3.83e-36 | 4930 | 39.2 | 6580 | 32.3 | 1.21 |
| 122 | 14 | MA1606.1 | Foxf1 | MA1606.1 | DWRTAAACANN | Foxf1 | 2.13e-33 | 6.04e-36 | 10200 | 81.4 | 15400 | 75.6 | 1.08 |
| 123 | 20 | MA0108.2 | TBP | MA0108.2 | STATAAAWRSVVVBN | TBP | 2.77e-33 | 7.76e-36 | 9050 | 71.9 | 13300 | 65.3 | 1.1 |
| 124 | 6 | MA0157.3 | Foxo3 | MA0157.3 | NDGTAAACARNN | Foxo3 | 5.63e-33 | 1.57e-35 | 9540 | 75.8 | 14200 | 69.5 | 1.09 |
| 125 | 5 | MA0699.1 | LBX2 | MA0699.1 | NCYAATTARN | LBX2 | 8.5e-33 | 2.35e-35 | 5030 | 40 | 6750 | 33.2 | 1.2 |
| 126 | 3 | MA0601.1 | Arid3b | MA0601.1 | ATATTAATWAN | Arid3b | 1.04e-32 | 2.84e-35 | 5650 | 44.9 | 7720 | 38 | 1.18 |
| 127 | 6 | MA1608.1 | Isl1 | MA1608.1 | NNCCATTAGNN | Isl1 | 1.05e-32 | 2.84e-35 | 8750 | 69.5 | 12800 | 62.8 | 1.11 |
| 128 | 5 | MA1481.1 | DRGX | MA1481.1 | YYAATTAN | DRGX | 1.51e-32 | 4.06e-35 | 4550 | 36.1 | 6020 | 29.6 | 1.22 |
| 129 | 14 | MA0148.4 | FOXA1 | MA0148.4 | WWGTAAACATNN | FOXA1 | 1.52e-32 | 4.07e-35 | 6680 | 53.1 | 9380 | 46.1 | 1.15 |
| 130 | 21 | MA0905.1 | HOXC10 | MA0905.1 | GTCRTAAAAH | HOXC10 | 1.69e-32 | 4.5e-35 | 7240 | 57.6 | 10300 | 50.6 | 1.14 |
| 131 | 22 | MA0153.2 | HNF1B | MA0153.2 | GTTAATNATTAAY | HNF1B | 2.75e-32 | 7.24e-35 | 3870 | 30.8 | 5000 | 24.6 | 1.25 |
| 132 | 3 | MA1498.2 | HOXA7 | MA1498.2 | SYMATTAV | HOXA7 | 2.93e-32 | 7.67e-35 | 9050 | 71.9 | 13300 | 65.4 | 1.1 |
| 133 | 3 | MA0715.1 | PROP1 | MA0715.1 | TAATYWAATTA | PROP1 | 3.53e-32 | 9.16e-35 | 4570 | 36.3 | 6060 | 29.8 | 1.22 |
| 134 | 5 | MA0716.1 | PRRX1 | MA0716.1 | YYAATTAR | PRRX1 | 3.64e-32 | 9.37e-35 | 4130 | 32.8 | 5380 | 26.4 | 1.24 |
| 135 | 5 | MA0890.1 | GBX2 | MA0890.1 | RSYAATTARN | GBX2 | 3.76e-32 | 9.61e-35 | 5250 | 41.7 | 7110 | 34.9 | 1.19 |
| 136 | 3 | MA1471.1 | BARX2 | MA1471.1 | NWWAAYMATTAN | BARX2 | 4.25e-32 | 1.08e-34 | 8040 | 63.9 | 11600 | 57 | 1.12 |
| 137 | 3 | MA1497.1 | HOXA6 | MA1497.1 | BGYMATTANN | HOXA6 | 8.83e-32 | 2.23e-34 | 6360 | 50.5 | 8870 | 43.6 | 1.16 |
| 138 | 14 | MA0047.3 | FOXA2 | MA0047.3 | RWGTAAACANN | FOXA2 | 1.15e-31 | 2.87e-34 | 9110 | 72.4 | 13400 | 66 | 1.1 |
| 139 | 14 | MA0851.1 | Foxj3 | MA0851.1 | NDAADGTAAACAAANNM | Foxj3 | 1.21e-31 | 3.01e-34 | 6290 | 50 | 8770 | 43.1 | 1.16 |
| 140 | 5 | MA0125.1 | Nobox | MA0125.1 | TAATTRSY | Nobox | 1.3e-31 | 3.2e-34 | 7280 | 57.8 | 10400 | 50.9 | 1.14 |
| 141 | 13 | MA0143.4 | SOX2 | MA0143.4 | DNACAATGGNN | SOX2 | 1.33e-31 | 3.26e-34 | 9730 | 77.4 | 14500 | 71.3 | 1.09 |
| 142 | 23 | MA0711.1 | OTX1 | MA0711.1 | HTAATCCS | OTX1 | 1.39e-31 | 3.39e-34 | 7890 | 62.7 | 11400 | 55.9 | 1.12 |
| 143 | 6 | MA1644.1 | NFYC | MA1644.1 | RDCCAATCASN | NFYC | 1.58e-31 | 3.81e-34 | 5560 | 44.2 | 7610 | 37.4 | 1.18 |
| 144 | 24 | MA1974.1 | ZNF211 | MA1974.1 | HWYATATACCAYRYW | ZNF211 | 6.46e-31 | 1.55e-33 | 5200 | 41.3 | 7060 | 34.7 | 1.19 |
| 145 | 9 | MA0117.2 | Mafb | MA0117.2 | AWWNTGCTGACD | Mafb | 7.71e-31 | 1.84e-33 | 8800 | 69.9 | 12900 | 63.5 | 1.1 |
| 146 | 14 | MA0033.2 | FOXL1 | MA0033.2 | RTAAACA | FOXL1 | 8.41e-31 | 1.99e-33 | 8580 | 68.2 | 12600 | 61.7 | 1.11 |
| 147 | 5 | MA0642.2 | EN2 | MA0642.2 | VSYAATTA | EN2 | 9.29e-31 | 2.18e-33 | 8290 | 65.9 | 12100 | 59.3 | 1.11 |
| 148 | 6 | MA0480.2 | Foxo1 | MA0480.2 | NDGTAAACANN | Foxo1 | 1.13e-30 | 2.63e-33 | 8890 | 70.6 | 13100 | 64.2 | 1.1 |
| 149 | 15 | MA0037.4 | Gata3 | MA0037.4 | NHTCTTATCTNH | Gata3 | 1.34e-30 | 3.11e-33 | 5700 | 45.3 | 7850 | 38.6 | 1.17 |
| 150 | 5 | MA0879.2 | DLX1 | MA0879.2 | VYAATTAN | DLX1 | 2.56e-30 | 5.9e-33 | 4400 | 35 | 5840 | 28.7 | 1.22 |
| 151 | 23 | MA1480.1 | DPRX | MA1480.1 | AGATAATCCN | DPRX | 2.63e-30 | 6.01e-33 | 6070 | 48.3 | 8450 | 41.5 | 1.16 |
| 152 | 3 | MA0912.2 | HOXD3 | MA0912.2 | CTAATTAC | HOXD3 | 2.84e-30 | 6.46e-33 | 3810 | 30.3 | 4940 | 24.3 | 1.25 |
| 153 | 14 | MA0848.1 | FOXO4 | MA0848.1 | GTAAACA | FOXO4 | 2.93e-30 | 6.62e-33 | 8870 | 70.5 | 13100 | 64.2 | 1.1 |
| 154 | 6 | MA0891.1 | GSC2 | MA0891.1 | NYTAATCCBH | GSC2 | 2.99e-30 | 6.71e-33 | 8860 | 70.4 | 13000 | 64 | 1.1 |
| 155 | 6 | MA0465.2 | CDX2 | MA0465.2 | NDGCAATAAAWN | CDX2 | 4.59e-30 | 1.02e-32 | 5370 | 42.7 | 7340 | 36.1 | 1.18 |
| 156 | 18 | MA1636.1 | CEBPG | MA1636.1 | NDATGATGCAATMHH | CEBPG | 5.2e-30 | 1.15e-32 | 8340 | 66.2 | 12200 | 59.7 | 1.11 |
| 157 | 25 | MA0611.2 | Dux | MA0611.2 | NTGATTBAATCAVWNN | Dux | 5.42e-30 | 1.19e-32 | 3660 | 29.1 | 4710 | 23.2 | 1.26 |
| 158 | 14 | MA0481.3 | FOXP1 | MA0481.3 | NDGTAAACANH | FOXP1 | 5.48e-30 | 1.2e-32 | 8100 | 64.4 | 11800 | 57.8 | 1.11 |
| 159 | 3 | MA1500.1 | HOXB6 | MA1500.1 | BTAATKRC | HOXB6 | 7.16e-30 | 1.56e-32 | 10300 | 81.7 | 15500 | 76.2 | 1.07 |
| 160 | 14 | MA1103.2 | FOXK2 | MA1103.2 | DDGTAAACANN | FOXK2 | 7.23e-30 | 1.56e-32 | 7260 | 57.7 | 10400 | 51 | 1.13 |
| 161 | 26 | MA0679.2 | ONECUT1 | MA0679.2 | WAAAAATCAATAAWWN | ONECUT1 | 8.06e-30 | 1.73e-32 | 4590 | 36.5 | 6140 | 30.2 | 1.21 |
| 162 | 26 | MA0755.1 | CUX2 | MA0755.1 | TRATCGATAW | CUX2 | 8.94e-30 | 1.9e-32 | 5760 | 45.8 | 7970 | 39.2 | 1.17 |
| 163 | 13 | MA0514.2 | Sox3 | MA0514.2 | DNACAATGGNN | Sox3 | 9.06e-30 | 1.92e-32 | 9500 | 75.5 | 14100 | 69.5 | 1.09 |
| 164 | 7 | MA0070.1 | PBX1 | MA0070.1 | HCATCAATCAAW | PBX1 | 1.54e-29 | 3.25e-32 | 5180 | 41.2 | 7060 | 34.7 | 1.19 |
| 165 | 3 | MA0707.2 | MNX1 | MA0707.2 | VYRATTAK | MNX1 | 3.61e-29 | 7.56e-32 | 4150 | 33 | 5480 | 26.9 | 1.23 |
| 166 | 14 | MA0031.1 | FOXD1 | MA0031.1 | GTAAACAW | FOXD1 | 4.53e-29 | 9.42e-32 | 6490 | 51.6 | 9150 | 45 | 1.15 |
| 167 | 5 | MA0027.2 | EN1 | MA0027.2 | SYAATTAV | EN1 | 9.68e-29 | 2e-31 | 3490 | 27.7 | 4490 | 22 | 1.26 |
| 168 | 6 | MA0896.1 | Hmx1 | MA0896.1 | VSVAGCAATTAAWGVDB | Hmx1 | 1.01e-28 | 2.07e-31 | 4670 | 37.1 | 6280 | 30.9 | 1.2 |
| 169 | 12 | MA0497.1 | MEF2C | MA0497.1 | DDDCYAAAAATAGMW | MEF2C | 1.02e-28 | 2.09e-31 | 6310 | 50.1 | 8860 | 43.5 | 1.15 |
| 170 | 27 | MA0483.1 | Gfi1B | MA0483.1 | AAATCWCWGCH | Gfi1B | 1.07e-28 | 2.16e-31 | 5760 | 45.8 | 7990 | 39.3 | 1.17 |
| 171 | 28 | MA1487.2 | FOXE1 | MA1487.2 | VYTAAAYAAACAAH | FOXE1 | 1.2e-28 | 2.42e-31 | 6070 | 48.3 | 8490 | 41.7 | 1.16 |
| 172 | 29 | MA0063.2 | NKX2-5 | MA0063.2 | NNCACTCAANN | NKX2-5 | 1.7e-28 | 3.41e-31 | 8200 | 65.2 | 12000 | 58.8 | 1.11 |
| 173 | 14 | MA0846.1 | FOXC2 | MA0846.1 | WAWGTAAACAWW | FOXC2 | 2.44e-28 | 4.87e-31 | 9330 | 74.2 | 13900 | 68.2 | 1.09 |
| 174 | 30 | MA0523.1 | TCF7L2 | MA0523.1 | DRASATCAAAGRVA | TCF7L2 | 2.81e-28 | 5.58e-31 | 7110 | 56.5 | 10200 | 50 | 1.13 |
| 175 | 3 | MA0888.1 | EVX2 | MA0888.1 | VVTAATTAVB | EVX2 | 2.95e-28 | 5.82e-31 | 4920 | 39.1 | 6680 | 32.8 | 1.19 |
| 176 | 28 | MA0847.3 | FOXD2 | MA0847.3 | SYTAARYAAACA | FOXD2 | 3.9e-28 | 7.66e-31 | 6240 | 49.6 | 8770 | 43.1 | 1.15 |
| 177 | 10 | MA0142.1 | Pou5f1::Sox2 | MA0142.1 | CWTTGTYATGCAAAT | Pou5f1::Sox2 | 3.97e-28 | 7.74e-31 | 4770 | 37.9 | 6460 | 31.7 | 1.2 |
| 178 | 14 | MA1607.1 | Foxl2 | MA1607.1 | DDWATGTAAACANN | Foxl2 | 5.68e-28 | 1.1e-30 | 9430 | 75 | 14100 | 69.1 | 1.08 |
| 179 | 18 | MA0836.2 | CEBPD | MA0836.2 | NRTTGCACAAYHN | CEBPD | 1.24e-27 | 2.4e-30 | 9320 | 74 | 13900 | 68.2 | 1.09 |
| 180 | 1 | MA1124.1 | ZNF24 | MA1124.1 | CATTCATTCATTC | ZNF24 | 2.26e-27 | 4.33e-30 | 4820 | 38.3 | 6540 | 32.1 | 1.19 |
| 181 | 30 | MA0769.2 | TCF7 | MA0769.2 | NNCTTTGAWVT | TCF7 | 4.08e-27 | 7.79e-30 | 10100 | 80.1 | 15200 | 74.7 | 1.07 |
| 182 | 26 | MA0754.2 | CUX1 | MA0754.2 | TAATCGATAM | CUX1 | 4.15e-27 | 7.87e-30 | 5710 | 45.4 | 7950 | 39.1 | 1.16 |
| 183 | 6 | MA0648.1 | GSC | MA0648.1 | VYTAATCCSY | GSC | 5.3e-27 | 1e-29 | 4350 | 34.6 | 5830 | 28.6 | 1.21 |
| 184 | 30 | MA0768.2 | Lef1 | MA0768.2 | NNBCCTTTGATSTN | Lef1 | 9.23e-27 | 1.73e-29 | 7970 | 63.3 | 11600 | 57.1 | 1.11 |
| 185 | 6 | MA0635.1 | BARHL2 | MA0635.1 | VHTAAAYGRT | BARHL2 | 1.08e-26 | 2.02e-29 | 8680 | 69 | 12800 | 63 | 1.1 |
| 186 | 6 | MA0897.1 | Hmx2 | MA0897.1 | ASAAGCAATTAAHVVRT | Hmx2 | 2.42e-26 | 4.49e-29 | 4860 | 38.6 | 6620 | 32.5 | 1.19 |
| 187 | 13 | MA1563.2 | SOX18 | MA1563.2 | AACAATDV | SOX18 | 2.45e-26 | 4.53e-29 | 8180 | 65 | 12000 | 58.9 | 1.1 |
| 188 | 15 | MA0482.2 | GATA4 | MA0482.2 | NNCCTTATCTNH | GATA4 | 3.13e-26 | 5.75e-29 | 6330 | 50.3 | 8960 | 44 | 1.14 |
| 189 | 3 | MA0658.1 | LHX6 | MA0658.1 | RCTAATTARY | LHX6 | 3.15e-26 | 5.76e-29 | 4040 | 32.1 | 5370 | 26.4 | 1.22 |
| 190 | 31 | MA1657.1 | ZNF652 | MA1657.1 | BAAAGRGTTAAN | ZNF652 | 3.67e-26 | 6.66e-29 | 6280 | 50 | 8880 | 43.6 | 1.14 |
| 191 | 3 | MA0876.1 | BSX | MA0876.1 | SYVATTAV | BSX | 4e-26 | 7.23e-29 | 7760 | 61.7 | 11300 | 55.5 | 1.11 |
| 192 | 17 | MA0808.1 | TEAD3 | MA0808.1 | RCATTCCW | TEAD3 | 4.29e-26 | 7.71e-29 | 5460 | 43.4 | 7580 | 37.2 | 1.17 |
| 193 | 23 | MA0714.1 | PITX3 | MA0714.1 | NYTAATCCC | PITX3 | 5.7e-26 | 1.02e-28 | 10100 | 80 | 15200 | 74.8 | 1.07 |
| 194 | 23 | MA0682.2 | PITX1 | MA0682.2 | YTAATCCH | PITX1 | 6.71e-26 | 1.19e-28 | 5480 | 43.6 | 7620 | 37.4 | 1.16 |
| 195 | 3 | MA0914.1 | ISL2 | MA0914.1 | GCAMTTAR | ISL2 | 6.78e-26 | 1.2e-28 | 7820 | 62.2 | 11400 | 56 | 1.11 |
| 196 | 3 | MA0787.1 | POU3F2 | MA0787.1 | WTATGCWAATKA | POU3F2 | 7.54e-26 | 1.33e-28 | 4490 | 35.7 | 6060 | 29.8 | 1.2 |
| 197 | 3 | MA1504.1 | HOXC4 | MA1504.1 | VTMATTAN | HOXC4 | 9.19e-26 | 1.61e-28 | 5240 | 41.7 | 7240 | 35.6 | 1.17 |
| 198 | 14 | MA0850.1 | FOXP3 | MA0850.1 | RTAAACA | FOXP3 | 9.44e-26 | 1.65e-28 | 6730 | 53.5 | 9620 | 47.2 | 1.13 |
| 199 | 13 | MA1120.1 | SOX13 | MA1120.1 | DVACAATGGNN | SOX13 | 1.48e-25 | 2.57e-28 | 10900 | 86.9 | 16800 | 82.4 | 1.05 |
| 200 | 3 | MA0791.1 | POU4F3 | MA0791.1 | RTGMATWATTAATGAR | POU4F3 | 1.68e-25 | 2.9e-28 | 4070 | 32.4 | 5430 | 26.7 | 1.21 |
| 201 | 32 | MA0151.1 | Arid3a | MA0151.1 | ATYAAA | Arid3a | 2.67e-25 | 4.58e-28 | 4290 | 34.1 | 5770 | 28.4 | 1.2 |
| 202 | 33 | MA1603.1 | Dmrt1 | MA1603.1 | GHTACAAAGTADC | Dmrt1 | 7e-25 | 1.2e-27 | 8440 | 67.1 | 12400 | 61.2 | 1.1 |
| 203 | 34 | MA1632.1 | ATF2 | MA1632.1 | NNATGABGTCATN | ATF2 | 7.4e-25 | 1.26e-27 | 7810 | 62.1 | 11400 | 56 | 1.11 |
| 204 | 6 | MA0060.3 | NFYA | MA0060.3 | RRCCAATCAGM | NFYA | 8.77e-25 | 1.48e-27 | 5710 | 45.4 | 8000 | 39.3 | 1.15 |
| 205 | 17 | MA0090.3 | TEAD1 | MA0090.3 | NNACATTCCAGSN | TEAD1 | 1.37e-24 | 2.3e-27 | 10600 | 83.9 | 16100 | 79.1 | 1.06 |
| 206 | 9 | MA0842.2 | NRL | MA0842.2 | AAWWNTGCTGACG | NRL | 1.61e-24 | 2.69e-27 | 2810 | 22.3 | 3550 | 17.4 | 1.28 |
| 207 | 18 | MA0102.4 | CEBPA | MA0102.4 | NDATTGCACAATHN | CEBPA | 1.7e-24 | 2.84e-27 | 9460 | 75.2 | 14200 | 69.7 | 1.08 |
| 208 | 5 | MA0666.2 | MSX1 | MA0666.2 | SCAATTAR | MSX1 | 1.9e-24 | 3.16e-27 | 3050 | 24.3 | 3910 | 19.2 | 1.26 |
| 209 | 22 | MA0790.1 | POU4F1 | MA0790.1 | ATGMATAATTAATG | POU4F1 | 2.13e-24 | 3.52e-27 | 3790 | 30.1 | 5020 | 24.7 | 1.22 |
| 210 | 3 | MA0075.3 | PRRX2 | MA0075.3 | CTAATTAN | PRRX2 | 2.96e-24 | 4.86e-27 | 3260 | 25.9 | 4230 | 20.8 | 1.25 |
| 211 | 14 | MA0042.2 | FOXI1 | MA0042.2 | GTAAACA | FOXI1 | 3.54e-24 | 5.79e-27 | 6560 | 52.2 | 9380 | 46.1 | 1.13 |
| 212 | 5 | MA0708.2 | MSX2 | MA0708.2 | SCAATTAV | MSX2 | 4.65e-24 | 7.57e-27 | 3150 | 25 | 4060 | 20 | 1.25 |
| 213 | 35 | MA0788.1 (POU3F3) | STREME-8 | 8-AGCATA | AGCATA | POU3F3 | 8.22e-24 | 1.33e-26 | 11200 | 88.6 | 17200 | 84.5 | 1.05 |
| 214 | 13 | MA1152.1 | SOX15 | MA1152.1 | CYWTTGTTHW | SOX15 | 9.84e-24 | 1.59e-26 | 10900 | 86.5 | 16700 | 82.1 | 1.05 |
| 215 | 3 | MA0158.2 | HOXA5 | MA0158.2 | GYMATTAS | HOXA5 | 1.28e-23 | 2.05e-26 | 3740 | 29.7 | 4960 | 24.4 | 1.22 |
| 216 | 36 | MA1479.1 | DMRTC2 | MA1479.1 | AATTGMTACATT | DMRTC2 | 1.3e-23 | 2.08e-26 | 8800 | 69.9 | 13100 | 64.3 | 1.09 |
| 217 | 17 | MA1121.1 | TEAD2 | MA1121.1 | NYACATTCCWNNS | TEAD2 | 2.05e-23 | 3.26e-26 | 9070 | 72.1 | 13500 | 66.6 | 1.08 |
| 218 | 37 | MA1150.1 | RORB | MA1150.1 | AWWTRGGTCAH | RORB | 2.39e-23 | 3.79e-26 | 9280 | 73.7 | 13900 | 68.3 | 1.08 |
| 219 | 38 | MA1953.1 | FOXO1::ELF1 | MA1953.1 | RTMAACAGGAAGTN | FOXO1::ELF1 | 4.27e-23 | 6.73e-26 | 9720 | 77.3 | 14700 | 72.1 | 1.07 |
| 220 | 39 | MA0144.2 | STAT3 | MA0144.2 | YTTCYKGGAAD | STAT3 | 4.76e-23 | 7.46e-26 | 6910 | 55 | 9980 | 49 | 1.12 |
| 221 | 3 | MA0874.1 | Arx | MA0874.1 | VTBCRYTAATTARTKSW | Arx | 7.47e-23 | 1.17e-25 | 3890 | 30.9 | 5210 | 25.6 | 1.21 |
| 222 | 29 | MA1645.1 | NKX2-2 | MA1645.1 | NNNCCACTCAANNN | NKX2-2 | 9.87e-23 | 1.53e-25 | 8120 | 64.5 | 12000 | 58.8 | 1.1 |
| 223 | 40 | MA0782.2 | PKNOX1 | MA0782.2 | NNTGAGTGACAGVNN | PKNOX1 | 1.31e-22 | 2.03e-25 | 4330 | 34.4 | 5890 | 28.9 | 1.19 |
| 224 | 14 | MA0041.2 | FOXD3 | MA0041.2 | WAWGHAAATAAACARY | FOXD3 | 1.35e-22 | 2.09e-25 | 4410 | 35.1 | 6020 | 29.6 | 1.19 |
| 225 | 37 | MA1151.1 | RORC | MA1151.1 | NWAWNTRGGTCA | RORC | 1.47e-22 | 2.25e-25 | 10500 | 83.8 | 16100 | 79.2 | 1.06 |
| 226 | 30 | MA1991.1 | Hnf1A | MA1991.1 | NBCCTTTGATSTBN | Hnf1A | 1.5e-22 | 2.3e-25 | 6600 | 52.4 | 9470 | 46.5 | 1.13 |
| 227 | 3 | MA0902.2 | HOXB2 | MA0902.2 | SYAATTAS | HOXB2 | 2.59e-22 | 3.94e-25 | 2510 | 19.9 | 3150 | 15.5 | 1.29 |
| 228 | 3 | MA1505.1 | HOXC8 | MA1505.1 | KTAATTAM | HOXC8 | 3.05e-22 | 4.62e-25 | 3030 | 24.1 | 3920 | 19.3 | 1.25 |
| 229 | 14 | MA0852.2 | FOXK1 | MA0852.2 | NRDGTAAACAAGNN | FOXK1 | 3.72e-22 | 5.6e-25 | 9110 | 72.4 | 13600 | 67 | 1.08 |
| 230 | 3 | MA0612.2 | EMX1 | MA0612.2 | BTAATTAS | EMX1 | 3.95e-22 | 5.93e-25 | 2630 | 20.9 | 3330 | 16.4 | 1.28 |
| 231 | 18 | MA0833.2 | ATF4 | MA0833.2 | DDATGATGCAATMN | ATF4 | 1.01e-21 | 1.51e-24 | 5110 | 40.6 | 7120 | 35 | 1.16 |
| 232 | 10 | MA0627.2 | POU2F3 | MA0627.2 | NWTATGCAAATNH | POU2F3 | 1.04e-21 | 1.55e-24 | 4290 | 34.1 | 5850 | 28.7 | 1.19 |
| 233 | 3 | MA1501.1 | HOXB7 | MA1501.1 | GGTAATTAVS | HOXB7 | 1.23e-21 | 1.82e-24 | 4700 | 37.4 | 6490 | 31.9 | 1.17 |
| 234 | 41 | MA1118.1 | SIX1 | MA1118.1 | GWAACCTGAKM | SIX1 | 2.11e-21 | 3.12e-24 | 10000 | 79.7 | 15200 | 74.9 | 1.06 |
| 235 | 3 | MA0132.2 | PDX1 | MA0132.2 | VYAATTAR | PDX1 | 2.17e-21 | 3.19e-24 | 3220 | 25.6 | 4230 | 20.8 | 1.23 |
| 236 | 3 | MA1499.1 | HOXB4 | MA1499.1 | RTMATTAR | HOXB4 | 2.23e-21 | 3.26e-24 | 5200 | 41.3 | 7270 | 35.7 | 1.16 |
| 237 | 13 | MA0868.2 | SOX8 | MA0868.2 | MGAACAATRG | SOX8 | 2.63e-21 | 3.83e-24 | 10600 | 84 | 16200 | 79.6 | 1.06 |
| 238 | 38 | MA1709.1 | ZIM3 | MA1709.1 | RVMAACAGAAACCYM | ZIM3 | 3.07e-21 | 4.45e-24 | 7550 | 60 | 11100 | 54.4 | 1.1 |
| 239 | 10 | MA0785.1 | POU2F1 | MA0785.1 | AWTATGCWAATK | POU2F1 | 3.83e-21 | 5.54e-24 | 4020 | 31.9 | 5440 | 26.7 | 1.19 |
| 240 | 42 | MA0606.2 | Nfat5 | MA0606.2 | NNATGGAAAAWN | Nfat5 | 5.62e-21 | 8.08e-24 | 8290 | 65.9 | 12300 | 60.4 | 1.09 |
| 241 | 43 | MA1524.2 | Msgn1 | MA1524.2 | VRRRACAAATGGTNNN | Msgn1 | 9.95e-21 | 1.43e-23 | 6760 | 53.7 | 9780 | 48.1 | 1.12 |
| 242 | 25 | MA1720.1 | ZNF85 | MA1720.1 | DHDGAGATTACWKCAK | ZNF85 | 2.71e-20 | 3.87e-23 | 7430 | 59 | 10900 | 53.5 | 1.1 |
| 243 | 6 | MA0898.1 | Hmx3 | MA0898.1 | ASAAGCAATTAAWRRAT | Hmx3 | 2.73e-20 | 3.88e-23 | 3830 | 30.4 | 5170 | 25.4 | 1.2 |
| 244 | 3 | MA1577.1 | TLX2 | MA1577.1 | BYAATTAR | TLX2 | 3.02e-20 | 4.28e-23 | 3130 | 24.9 | 4110 | 20.2 | 1.23 |
| 245 | 15 | MA0140.2 | GATA1::TAL1 | MA0140.2 | MTTATCWSNNNNNHVCAG | GATA1::TAL1 | 3.24e-20 | 4.57e-23 | 3280 | 26.1 | 4340 | 21.4 | 1.22 |
| 246 | 44 | 6-CTTTAAA | STREME-6 | 6-CTTTAAA | CTTTAAA | CTTTAAA | 3.86e-20 | 5.42e-23 | 5340 | 42.5 | 7530 | 37 | 1.15 |
| 247 | 34 | MA1126.1 | FOS::JUN | MA1126.1 | DRTGACGTCATHNDTN | FOS::JUN | 3.97e-20 | 5.54e-23 | 5680 | 45.2 | 8070 | 39.7 | 1.14 |
| 248 | 21 | MA0907.1 | HOXC13 | MA0907.1 | BCTCGTAAAAH | HOXC13 | 6.21e-20 | 8.65e-23 | 4910 | 39.1 | 6860 | 33.7 | 1.16 |
| 249 | 45 | MA1112.2 | NR4A1 | MA1112.2 | NNAAAGGTCANN | NR4A1 | 6.78e-20 | 9.4e-23 | 9350 | 74.3 | 14100 | 69.3 | 1.07 |
| 250 | 46 | MA1588.1 | ZNF136 | MA1588.1 | RWATTCTTGGTTGRC | ZNF136 | 6.91e-20 | 9.54e-23 | 4520 | 35.9 | 6240 | 30.7 | 1.17 |
| 251 | 47 | MA0631.1 | Six3 | MA0631.1 | DATRGGGTATCAYNWNT | Six3 | 7.74e-20 | 1.06e-22 | 5120 | 40.7 | 7190 | 35.3 | 1.15 |
| 252 | 23 | MA0467.2 | Crx | MA0467.2 | NDGGATTANN | Crx | 8.41e-20 | 1.15e-22 | 7340 | 58.4 | 10800 | 52.8 | 1.1 |
| 253 | 41 | MA1119.1 | SIX2 | MA1119.1 | NNCTGAAACCTGATMY | SIX2 | 1.43e-19 | 1.95e-22 | 8030 | 63.8 | 11900 | 58.4 | 1.09 |
| 254 | 3 | MA0895.1 | HMBOX1 | MA0895.1 | MYTAGTTAMS | HMBOX1 | 1.54e-19 | 2.1e-22 | 7170 | 57 | 10500 | 51.5 | 1.11 |
| 255 | 36 | MA1707.1 | DMRTA1 | MA1707.1 | AWTTGWTACAWT | DMRTA1 | 1.55e-19 | 2.1e-22 | 7870 | 62.6 | 11600 | 57.2 | 1.09 |
| 256 | 21 | MA0908.1 | HOXD11 | MA0908.1 | RTCGTAAAAH | HOXD11 | 1.6e-19 | 2.16e-22 | 4310 | 34.3 | 5940 | 29.2 | 1.18 |
| 257 | 3 | MA0893.2 | GSX2 | MA0893.2 | SYMATTAR | GSX2 | 2.87e-19 | 3.85e-22 | 11100 | 87.9 | 17100 | 84 | 1.05 |
| 258 | 3 | MA0724.1 | VENTX | MA0724.1 | AMCGATTAR | VENTX | 2.96e-19 | 3.95e-22 | 2580 | 20.5 | 3320 | 16.3 | 1.26 |
| 259 | 23 | MA0883.1 | Dmbx1 | MA0883.1 | WDRWNMGGATTADKNNW | Dmbx1 | 4.14e-19 | 5.52e-22 | 4370 | 34.7 | 6030 | 29.6 | 1.17 |
| 260 | 3 | MA1507.1 | HOXD4 | MA1507.1 | VTMATTAD | HOXD4 | 4.53e-19 | 6.02e-22 | 3590 | 28.6 | 4840 | 23.8 | 1.2 |
| 261 | 6 | MA0124.2 | Nkx3-1 | MA0124.2 | RCCACTTAA | Nkx3-1 | 8.17e-19 | 1.08e-21 | 8620 | 68.5 | 12900 | 63.4 | 1.08 |
| 262 | 34 | MA0018.4 | CREB1 | MA0018.4 | NNRTGATGTCAYN | CREB1 | 1.54e-18 | 2.03e-21 | 10700 | 85.2 | 16500 | 81.2 | 1.05 |
| 263 | 10 | MA1962.1 | POU2F1::SOX2 | MA1962.1 | MATTTRCATMACAATRG | POU2F1::SOX2 | 1.97e-18 | 2.58e-21 | 2970 | 23.6 | 3910 | 19.2 | 1.23 |
| 264 | 29 | MA1523.1 | MSANTD3 | MA1523.1 | STVCACTCAC | MSANTD3 | 1.98e-18 | 2.59e-21 | 9140 | 72.6 | 13800 | 67.7 | 1.07 |
| 265 | 48 | MA0629.1 | Rhox11 | MA0629.1 | ANKNCGCTGTWAWDVRW | Rhox11 | 1.99e-18 | 2.59e-21 | 8960 | 71.2 | 13500 | 66.3 | 1.07 |
| 266 | 43 | MA1618.1 | Ptf1a | MA1618.1 | NNACAGATGTTNN | Ptf1a | 2.28e-18 | 2.96e-21 | 10000 | 79.7 | 15300 | 75.2 | 1.06 |
| 267 | 3 | MA0875.1 | BARX1 | MA0875.1 | VCMATTAV | BARX1 | 2.94e-18 | 3.8e-21 | 2300 | 18.3 | 2920 | 14.3 | 1.27 |
| 268 | 34 | MA1127.1 | FOSB::JUN | MA1127.1 | GATGACGTCAT | FOSB::JUN | 3.84e-18 | 4.94e-21 | 8650 | 68.7 | 13000 | 63.7 | 1.08 |
| 269 | 43 | MA0093.3 | USF1 | MA0093.3 | NDGTCATGTGACHN | USF1 | 4.26e-18 | 5.47e-21 | 9880 | 78.6 | 15100 | 74 | 1.06 |
| 270 | 34 | MA1131.1 | FOSL2::JUN | MA1131.1 | GRTGACGTMAT | FOSL2::JUN | 4.46e-18 | 5.7e-21 | 8420 | 66.9 | 12600 | 61.9 | 1.08 |
| 271 | 22 | MA0046.2 | HNF1A | MA0046.2 | DRTTAATNATTAACN | HNF1A | 4.85e-18 | 6.18e-21 | 2570 | 20.5 | 3330 | 16.4 | 1.25 |
| 272 | 3 | MA0618.1 | LBX1 | MA0618.1 | TTAATTAG | LBX1 | 5.32e-18 | 6.75e-21 | 5180 | 41.2 | 7340 | 36 | 1.14 |
| 273 | 45 | MA0693.3 | Vdr | MA0693.3 | NNGAGTTCANN | Vdr | 6.94e-18 | 8.77e-21 | 9310 | 74 | 14100 | 69.2 | 1.07 |
| 274 | 10 | MA0786.1 | POU3F1 | MA0786.1 | WTATGCWAATKW | POU3F1 | 1.08e-17 | 1.36e-20 | 3400 | 27 | 4580 | 22.5 | 1.2 |
| 275 | 14 | MA0613.1 | FOXG1 | MA0613.1 | RTAAACAW | FOXG1 | 1.8e-17 | 2.26e-20 | 3830 | 30.5 | 5250 | 25.8 | 1.18 |
| 276 | 49 | MA0050.3 | Irf1 | MA0050.3 | ANTGAAACTGAAASH | Irf1 | 2.02e-17 | 2.53e-20 | 9660 | 76.8 | 14700 | 72.2 | 1.06 |
| 277 | 23 | MA0719.1 | RHOXF1 | MA0719.1 | NTRATCCN | RHOXF1 | 2.04e-17 | 2.54e-20 | 8450 | 67.2 | 12700 | 62.2 | 1.08 |
| 278 | 5 | MA0894.1 | HESX1 | MA0894.1 | GHTAATTRRH | HESX1 | 2.82e-17 | 3.5e-20 | 2870 | 22.8 | 3780 | 18.6 | 1.23 |
| 279 | 43 | MA1123.2 | TWIST1 | MA1123.2 | NNDCCAGATGTBN | TWIST1 | 3.34e-17 | 4.14e-20 | 9620 | 76.5 | 14600 | 72 | 1.06 |
| 280 | 30 | MA0442.2 | SOX10 | MA0442.2 | NDAACAAAGVN | SOX10 | 4.41e-17 | 5.44e-20 | 8670 | 68.9 | 13000 | 64 | 1.08 |
| 281 | 43 | MA0620.3 | MITF | MA0620.3 | NNNDRTCACGTGAYHNNN | MITF | 8.57e-17 | 1.05e-19 | 6880 | 54.7 | 10100 | 49.6 | 1.1 |
| 282 | 3 | MA1496.1 | HOXA4 | MA1496.1 | RTMATTAV | HOXA4 | 1.1e-16 | 1.35e-19 | 4540 | 36.1 | 6370 | 31.3 | 1.15 |
| 283 | 14 | MA0849.1 | FOXO6 | MA0849.1 | GTAAACA | FOXO6 | 1.19e-16 | 1.46e-19 | 6400 | 50.8 | 9310 | 45.7 | 1.11 |
| 284 | 13 | MA0867.2 | SOX4 | MA0867.2 | RAACAAAGRV | SOX4 | 1.33e-16 | 1.62e-19 | 7160 | 56.9 | 10600 | 51.9 | 1.1 |
| 285 | 39 | MA1624.1 | Stat5a | MA1624.1 | NTTCCAAGAANH | Stat5a | 1.64e-16 | 1.99e-19 | 9700 | 77.1 | 14800 | 72.7 | 1.06 |
| 286 | 50 | MA1593.1 | ZNF317 | MA1593.1 | WVACAGCAGAYW | ZNF317 | 3.97e-16 | 4.79e-19 | 10600 | 84 | 16300 | 80.1 | 1.05 |
| 287 | 38 | MA1105.2 | GRHL2 | MA1105.2 | NRAACAGGTTYN | GRHL2 | 6.5e-16 | 7.82e-19 | 11000 | 87.2 | 17000 | 83.7 | 1.04 |
| 288 | 51 | MA1980.1 | ZNF418 | MA1980.1 | MDGAGGCTAAAAGCA | ZNF418 | 6.6e-16 | 7.92e-19 | 6560 | 52.2 | 9600 | 47.2 | 1.11 |
| 289 | 18 | MA0043.3 | HLF | MA0043.3 | NNRTTATGCAACHN | HLF | 6.86e-16 | 8.19e-19 | 4080 | 32.4 | 5680 | 27.9 | 1.16 |
| 290 | 38 | MA1623.1 | Stat2 | MA1623.1 | RGAAACAGAAASH | Stat2 | 1.3e-15 | 1.54e-18 | 9550 | 75.9 | 14600 | 71.6 | 1.06 |
| 291 | 6 | MA0913.2 | HOXD9 | MA0913.2 | GYMATAAAAH | HOXD9 | 1.67e-15 | 1.98e-18 | 3540 | 28.1 | 4840 | 23.8 | 1.18 |
| 292 | 40 | MA0774.1 | MEIS2 | MA0774.1 | TTGACAGS | MEIS2 | 1.91e-15 | 2.26e-18 | 7130 | 56.7 | 10500 | 51.8 | 1.09 |
| 293 | 36 | MA1478.1 | DMRTA2 | MA1478.1 | AATTGHTACAWT | DMRTA2 | 2.4e-15 | 2.83e-18 | 8460 | 67.2 | 12700 | 62.5 | 1.07 |
| 294 | 3 | MA0705.1 | Lhx8 | MA0705.1 | CTAATTAV | Lhx8 | 2.55e-15 | 2.99e-18 | 2620 | 20.9 | 3470 | 17 | 1.22 |
| 295 | 29 | MA1114.1 | PBX3 | MA1114.1 | NNNTGAGTGACAGGBNN | PBX3 | 2.59e-15 | 3.03e-18 | 10900 | 86.9 | 17000 | 83.4 | 1.04 |
| 296 | 52 | MA0780.1 | PAX3 | MA0780.1 | TAATYRATTA | PAX3 | 3.01e-15 | 3.51e-18 | 2710 | 21.5 | 3590 | 17.6 | 1.22 |
| 297 | 53 | MA0092.1 | Hand1::Tcf3 | MA0092.1 | BRTCTGGMWT | Hand1::Tcf3 | 4.58e-15 | 5.33e-18 | 11100 | 87.9 | 17200 | 84.6 | 1.04 |
| 298 | 43 | MA0827.1 | OLIG3 | MA0827.1 | AMCATATGBY | OLIG3 | 4.91e-15 | 5.69e-18 | 9210 | 73.2 | 14000 | 68.8 | 1.06 |
| 299 | 54 | MA0800.1 | EOMES | MA0800.1 | RAGGTGTGAAAWN | EOMES | 8.94e-15 | 1.03e-17 | 7170 | 57 | 10600 | 52.2 | 1.09 |
| 300 | 43 | MA0095.3 | Yy1 | MA0095.3 | NNCAAAATGGCN | Yy1 | 8.96e-15 | 1.03e-17 | 11400 | 90.5 | 17800 | 87.4 | 1.03 |
| 301 | 39 | MA1625.1 | Stat5b | MA1625.1 | NNNTTCCCAGAANNN | Stat5b | 9.01e-15 | 1.03e-17 | 9440 | 75 | 14400 | 70.7 | 1.06 |
| 302 | 55 | MA1975.1 | ZNF214 | MA1975.1 | VWTCATCAABGTCCT | ZNF214 | 9.88e-15 | 1.13e-17 | 5560 | 44.2 | 8020 | 39.4 | 1.12 |
| 303 | 56 | MA1125.1 | ZNF384 | MA1125.1 | DNWMAAAAAAAA | ZNF384 | 1.02e-14 | 1.16e-17 | 4450 | 35.3 | 6270 | 30.8 | 1.15 |
| 304 | 40 | MA0498.2 | MEIS1 | MA0498.2 | HTGACAD | MEIS1 | 1.43e-14 | 1.63e-17 | 3860 | 30.6 | 5360 | 26.3 | 1.16 |
| 305 | 26 | MA0757.1 | ONECUT3 | MA0757.1 | AAAAAATCRATAAH | ONECUT3 | 2e-14 | 2.27e-17 | 3320 | 26.4 | 4540 | 22.3 | 1.18 |
| 306 | 34 | MA1139.1 | FOSL2::JUNB | MA1139.1 | DATGACGTCATH | FOSL2::JUNB | 2.88e-14 | 3.25e-17 | 6300 | 50.1 | 9220 | 45.3 | 1.1 |
| 307 | 57 | MA0069.1 | PAX6 | MA0069.1 | TTCACGCWTSANTK | PAX6 | 1.48e-13 | 1.66e-16 | 4670 | 37.1 | 6660 | 32.7 | 1.13 |
| 308 | 34 | MA1145.1 | FOSL2::JUND | MA1145.1 | NNRTGACGTCAYHSN | FOSL2::JUND | 1.67e-13 | 1.87e-16 | 6920 | 55 | 10300 | 50.4 | 1.09 |
| 309 | 3 | MA0702.2 | LMX1A | MA0702.2 | TTAATTAM | LMX1A | 2.54e-13 | 2.83e-16 | 2710 | 21.6 | 3640 | 17.9 | 1.2 |
| 310 | 14 | MA0614.1 | Foxj2 | MA0614.1 | RTAAACAA | Foxj2 | 3.01e-13 | 3.35e-16 | 2980 | 23.7 | 4050 | 19.9 | 1.19 |
| 311 | 21 | MA0911.1 | Hoxa11 | MA0911.1 | RGTCGTAAAANT | Hoxa11 | 3.22e-13 | 3.57e-16 | 4000 | 31.8 | 5620 | 27.6 | 1.15 |
| 312 | 43 | MA0461.2 | Atoh1 | MA0461.2 | RMCATATGBY | Atoh1 | 4e-13 | 4.43e-16 | 10100 | 80.4 | 15600 | 76.7 | 1.05 |
| 313 | 34 | MA1129.1 | FOSL1::JUN | MA1129.1 | ATGACGTCAT | FOSL1::JUN | 6.06e-13 | 6.68e-16 | 8590 | 68.2 | 13000 | 64 | 1.07 |
| 314 | 34 | MA1143.1 | FOSL1::JUND | MA1143.1 | RTGACGTMAY | FOSL1::JUND | 1.06e-12 | 1.16e-15 | 10100 | 80.5 | 15600 | 76.8 | 1.05 |
| 315 | 38 | MA0743.2 | SCRT1 | MA0743.2 | NDWKCAACAGGTGKNN | SCRT1 | 3.07e-12 | 3.36e-15 | 9630 | 76.5 | 14800 | 72.7 | 1.05 |
| 316 | 58 | MA0877.3 | BARHL1 | MA0877.3 | AMCGTTTA | BARHL1 | 3.66e-12 | 4e-15 | 6990 | 55.6 | 10400 | 51.2 | 1.09 |
| 317 | 42 | MA0624.2 | Nfatc1 | MA0624.2 | NVATGGAAAAWN | Nfatc1 | 4.72e-12 | 5.14e-15 | 10500 | 83.5 | 16300 | 80.1 | 1.04 |
| 318 | 38 | MA0744.2 | SCRT2 | MA0744.2 | NNWGCAACAGGTGDNN | SCRT2 | 7.9e-12 | 8.58e-15 | 10800 | 85.9 | 16800 | 82.8 | 1.04 |
| 319 | 40 | MA0775.1 | MEIS3 | MA0775.1 | DTGACAGS | MEIS3 | 8.6e-12 | 9.31e-15 | 2240 | 17.8 | 2970 | 14.6 | 1.22 |
| 320 | 59 | MA0124.2 (Nkx3-1) | STREME-9 | 9-TTAAGT | TTAAGT | Nkx3-1 | 9.73e-12 | 1.05e-14 | 2210 | 17.6 | 2930 | 14.4 | 1.22 |
| 321 | 60 | YWWRAAAWTRKTTTT | MEME-6 | YWWRAAAWTRKTTTT | YWWRAAAWTRKTTTT | YWWRAAAWTRKTTTT | 1.38e-11 | 1.48e-14 | 1760 | 14 | 2260 | 11.1 | 1.26 |
| 322 | 61 | MA1977.1 | ZNF324 | MA1977.1 | NNNWRGMAGHYAAGGATGGTT | ZNF324 | 2.07e-11 | 2.21e-14 | 6660 | 52.9 | 9900 | 48.6 | 1.09 |
| 323 | 12 | MA0773.1 | MEF2D | MA0773.1 | DCTAWAAATAGM | MEF2D | 2.27e-11 | 2.43e-14 | 1160 | 9.23 | 1410 | 6.91 | 1.33 |
| 324 | 3 | MA0621.1 | mix-a | MA0621.1 | NDTTAATTAGN | mix-a | 2.49e-11 | 2.66e-14 | 1780 | 14.1 | 2300 | 11.3 | 1.25 |
| 325 | 37 | MA1148.1 | PPARA::RXRA | MA1148.1 | AWNTRGGTYAAAGGTCAN | PPARA::RXRA | 3.08e-11 | 3.27e-14 | 7000 | 55.6 | 10500 | 51.4 | 1.08 |
| 326 | 54 | MA1567.2 | Tbx6 | MA1567.2 | NRGGTGTGAANN | Tbx6 | 4.98e-11 | 5.28e-14 | 8960 | 71.2 | 13700 | 67.3 | 1.06 |
| 327 | 62 | MA1984.1 | ZNF667 | MA1984.1 | TTAAGAGCTCAV | ZNF667 | 5.31e-11 | 5.61e-14 | 7780 | 61.9 | 11800 | 57.7 | 1.07 |
| 328 | 43 | MA0678.1 | OLIG2 | MA0678.1 | AMCATATGKT | OLIG2 | 6.53e-11 | 6.87e-14 | 3660 | 29.1 | 5160 | 25.4 | 1.15 |
| 329 | 45 | MA1540.2 | NR5A1 | MA1540.2 | NNRAGTTCAAGGTCANN | NR5A1 | 7.16e-11 | 7.52e-14 | 10400 | 82.6 | 16100 | 79.3 | 1.04 |
| 330 | 63 | MA0626.1 | Npas2 | MA0626.1 | NSCACGTGTN | Npas2 | 7.78e-11 | 8.15e-14 | 4310 | 34.2 | 6170 | 30.3 | 1.13 |
| 331 | 64 | MA1579.1 | ZBTB26 | MA1579.1 | NNCTCCAGAAANNNN | ZBTB26 | 8.49e-11 | 8.86e-14 | 9910 | 78.8 | 15300 | 75.3 | 1.05 |
| 332 | 65 | MA0521.2 (Tcf12) | STREME-5 | 5-WYAGCTRW | WYAGCTRW | Tcf12 | 1.16e-10 | 1.2e-13 | 1850 | 14.7 | 2420 | 11.9 | 1.24 |
| 333 | 45 | MA0592.3 | ESRRA | MA0592.3 | NBTCAAGGTCAHN | ESRRA | 1.17e-10 | 1.21e-13 | 7010 | 55.7 | 10500 | 51.6 | 1.08 |
| 334 | 37 | MA0258.2 | ESR2 | MA0258.2 | AGGTCASVNTGMCCY | ESR2 | 1.61e-10 | 1.66e-13 | 9900 | 78.7 | 15300 | 75.2 | 1.05 |
| 335 | 21 | MA0594.2 | HOXA9 | MA0594.2 | RTCGTWAHNN | HOXA9 | 1.7e-10 | 1.76e-13 | 9350 | 74.3 | 14400 | 70.6 | 1.05 |
| 336 | 39 | MA0518.1 | Stat4 | MA0518.1 | YTTCYRGGAARNVR | Stat4 | 2.01e-10 | 2.07e-13 | 5930 | 47.2 | 8760 | 43.1 | 1.09 |
| 337 | 34 | MA1136.1 | FOSB::JUNB | MA1136.1 | RTGACGTCAT | FOSB::JUNB | 2.49e-10 | 2.55e-13 | 9380 | 74.6 | 14400 | 70.9 | 1.05 |
| 338 | 34 | MA0488.1 | JUN | MA0488.1 | DDRATGATGTCAT | JUN | 2.61e-10 | 2.67e-13 | 1810 | 14.4 | 2370 | 11.7 | 1.24 |
| 339 | 34 | MA0596.1 | SREBF2 | MA0596.1 | RTGGGGTGAY | SREBF2 | 2.98e-10 | 3.04e-13 | 2680 | 21.3 | 3680 | 18.1 | 1.18 |
| 340 | 42 | MA0152.2 | Nfatc2 | MA0152.2 | NNRNAATGGAAANWNH | Nfatc2 | 3.34e-10 | 3.39e-13 | 11000 | 87.5 | 17200 | 84.7 | 1.03 |
| 341 | 34 | MA0829.2 | SREBF1 | MA0829.2 | DNATCACSTGATNH | SREBF1 | 3.74e-10 | 3.79e-13 | 7430 | 59 | 11200 | 55 | 1.07 |
| 342 | 66 | MA1647.2 | Prdm4 | MA1647.2 | RDNCCTTGAAACYGYYN | Prdm4 | 3.89e-10 | 3.92e-13 | 7710 | 61.3 | 11700 | 57.3 | 1.07 |
| 343 | 12 | MA0660.1 | MEF2B | MA0660.1 | RCTAWAAATAGC | MEF2B | 5.14e-10 | 5.17e-13 | 1150 | 9.16 | 1420 | 6.98 | 1.31 |
| 344 | 67 | MA1729.1 | ZNF680 | MA1729.1 | GNCCAAGAAGAATDA | ZNF680 | 5.79e-10 | 5.81e-13 | 3610 | 28.7 | 5110 | 25.1 | 1.14 |
| 345 | 39 | MA0137.3 | STAT1 | MA0137.3 | TTTCYRGGAAA | STAT1 | 6.66e-10 | 6.66e-13 | 5010 | 39.8 | 7320 | 36 | 1.11 |
| 346 | 3 | MA0135.1 | Lhx3 | MA0135.1 | WAATTAATTAWYY | Lhx3 | 6.74e-10 | 6.71e-13 | 1910 | 15.2 | 2530 | 12.4 | 1.22 |
| 347 | 54 | MA0802.1 | TBR1 | MA0802.1 | AGGTGTGAAA | TBR1 | 6.75e-10 | 6.71e-13 | 7420 | 58.9 | 11200 | 55 | 1.07 |
| 348 | 68 | MA0502.2 | NFYB | MA0502.2 | CYCATTGGCCVV | NFYB | 9.36e-10 | 9.29e-13 | 3580 | 28.5 | 5080 | 25 | 1.14 |
| 349 | 39 | MA0463.2 | BCL6 | MA0463.2 | NYGCTTTCKAGGAAYN | BCL6 | 1.26e-09 | 1.25e-12 | 6760 | 53.7 | 10100 | 49.8 | 1.08 |
| 350 | 3 | MA0683.1 | POU4F2 | MA0683.1 | RTGCATAATTAATGAG | POU4F2 | 1.4e-09 | 1.38e-12 | 1350 | 10.8 | 1720 | 8.43 | 1.28 |
| 351 | 69 | MA1995.1 | Npas4 | MA1995.1 | NNTCGTGACHN | Npas4 | 1.47e-09 | 1.45e-12 | 10000 | 79.6 | 15500 | 76.4 | 1.04 |
| 352 | 30 | MA0869.2 | Sox11 | MA0869.2 | NRGAACAAAGVV | Sox11 | 1.76e-09 | 1.73e-12 | 8030 | 63.8 | 12200 | 60 | 1.06 |
| 353 | 39 | MA0519.1 | Stat5a::Stat5b | MA0519.1 | DTTTCCMAGAA | Stat5a::Stat5b | 1.77e-09 | 1.73e-12 | 5190 | 41.2 | 7610 | 37.4 | 1.1 |
| 354 | 17 | MA0114.4 | HNF4A | MA0114.4 | DNCAAAGTCCANN | HNF4A | 2.14e-09 | 2.09e-12 | 6120 | 48.6 | 9100 | 44.7 | 1.09 |
| 355 | 66 | MA1724.1 | Rfx6 | MA1724.1 | HNCCTAGCAACHV | Rfx6 | 2.46e-09 | 2.39e-12 | 7400 | 58.8 | 11200 | 55 | 1.07 |
| 356 | 70 | TYTCTCYYTCT | MEME-5 | TYTCTCYYTCT | TYTCTCYYTCT | TYTCTCYYTCT | 3.87e-09 | 3.76e-12 | 3370 | 26.8 | 4780 | 23.5 | 1.14 |
| 357 | 1 | MA0591.1 | Bach1::Mafk | MA0591.1 | RSSATGACTCAGCAB | Bach1::Mafk | 4e-09 | 3.87e-12 | 2610 | 20.7 | 3600 | 17.7 | 1.17 |
| 358 | 13 | MA1561.1 | SOX12 | MA1561.1 | ACCGAACAATV | SOX12 | 5.21e-09 | 5.03e-12 | 4320 | 34.3 | 6250 | 30.7 | 1.12 |
| 359 | 71 | MA1534.1 | NR1I3 | MA1534.1 | RTGAACTTT | NR1I3 | 5.29e-09 | 5.09e-12 | 8530 | 67.8 | 13100 | 64.2 | 1.06 |
| 360 | 37 | MA0494.1 | Nr1h3::Rxra | MA0494.1 | TGACCTNNAGTRACCYYDN | Nr1h3::Rxra | 5.47e-09 | 5.24e-12 | 5280 | 42 | 7780 | 38.2 | 1.1 |
| 361 | 34 | MA0492.1 | JUND | MA0492.1 | DDDRATGABGTCATN | JUND | 6.49e-09 | 6.21e-12 | 1900 | 15.1 | 2540 | 12.5 | 1.21 |
| 362 | 3 | MA0884.2 | DUXA | MA0884.2 | YTRATTWAATYAR | DUXA | 7.88e-09 | 7.52e-12 | 1680 | 13.4 | 2220 | 10.9 | 1.23 |
| 363 | 17 | MA0484.2 | HNF4G | MA0484.2 | DNCAAAGTCCANN | HNF4G | 9.69e-09 | 9.22e-12 | 6890 | 54.8 | 10400 | 51 | 1.07 |
| 364 | 72 | MA0671.1 | NFIX | MA0671.1 | NNTGCCAAN | NFIX | 1.02e-08 | 9.71e-12 | 7560 | 60 | 11500 | 56.3 | 1.07 |
| 365 | 25 | MA0468.1 | DUX4 | MA0468.1 | TAAYYYAATCA | DUX4 | 1.55e-08 | 1.46e-11 | 1640 | 13 | 2160 | 10.6 | 1.23 |
| 366 | 41 | MA2001.1 | Six4 | MA2001.1 | GDAACCTGANH | Six4 | 1.66e-08 | 1.56e-11 | 11400 | 90.8 | 18000 | 88.6 | 1.03 |
| 367 | 29 | MA1994.1 | Nkx2-1 | MA1994.1 | NVCACTTGARHNN | Nkx2-1 | 2.07e-08 | 1.95e-11 | 10900 | 86.7 | 17100 | 84.1 | 1.03 |
| 368 | 73 | MA0508.3 | PRDM1 | MA0508.3 | YNCTTTCTCTH | PRDM1 | 2.56e-08 | 2.41e-11 | 8920 | 70.9 | 13700 | 67.4 | 1.05 |
| 369 | 34 | MA1133.1 | JUN::JUNB | MA1133.1 | KRTGACGTCATN | JUN::JUNB | 2.95e-08 | 2.76e-11 | 11300 | 90.2 | 17900 | 87.8 | 1.03 |
| 370 | 74 | MA1714.1 | ZNF675 | MA1714.1 | RGGVHYMGGAGGMCAAAATGD | ZNF675 | 3.15e-08 | 2.94e-11 | 4950 | 39.3 | 7280 | 35.8 | 1.1 |
| 371 | 28 | MA0040.1 | Foxq1 | MA0040.1 | HATTGTTTATW | Foxq1 | 3.24e-08 | 3.01e-11 | 2030 | 16.1 | 2750 | 13.5 | 1.19 |
| 372 | 1 | MA1520.1 | MAF | MA1520.1 | NTGCTGASTCAGCAH | MAF | 4.3e-08 | 4e-11 | 6170 | 49.1 | 9240 | 45.4 | 1.08 |
| 373 | 15 | MA0029.1 | Mecom | MA0029.1 | AAGAYAAGATAANA | Mecom | 4.75e-08 | 4.4e-11 | 945 | 7.51 | 1160 | 5.7 | 1.32 |
| 374 | 14 | MA0593.1 | FOXP2 | MA0593.1 | RWGTAAACAVR | FOXP2 | 5.46e-08 | 5.04e-11 | 2330 | 18.5 | 3200 | 15.7 | 1.17 |
| 375 | 75 | MA1989.1 | Bcl11B | MA1989.1 | NNAAACCACAARNN | Bcl11B | 6.4e-08 | 5.89e-11 | 11700 | 93.2 | 18600 | 91.2 | 1.02 |
| 376 | 27 | MA0038.2 | GFI1 | MA0038.2 | BMAATCACDGCA | GFI1 | 7.14e-08 | 6.56e-11 | 1320 | 10.5 | 1710 | 8.39 | 1.25 |
| 377 | 72 | MA0161.2 | NFIC | MA0161.2 | NNCTTGGCANN | NFIC | 7.46e-08 | 6.83e-11 | 7920 | 62.9 | 12100 | 59.4 | 1.06 |
| 378 | 3 | MA1960.1 | MGA::EVX1 | MA1960.1 | RGGTGWTAATKD | MGA::EVX1 | 8.77e-08 | 8.01e-11 | 2040 | 16.2 | 2780 | 13.7 | 1.19 |
| 379 | 38 | MA1952.1 | FOXJ2::ELF1 | MA1952.1 | RTAAACMGGAAGTR | FOXJ2::ELF1 | 9.15e-08 | 8.33e-11 | 10300 | 82 | 16100 | 79.1 | 1.04 |
| 380 | 38 | MA1950.1 | FLI1::FOXI1 | MA1950.1 | RTAAACAGGAARYN | FLI1::FOXI1 | 9.71e-08 | 8.82e-11 | 9260 | 73.6 | 14300 | 70.4 | 1.05 |
| 381 | 21 | MA0906.1 | HOXC12 | MA0906.1 | RGTCGTAAAAH | HOXC12 | 1.03e-07 | 9.3e-11 | 2370 | 18.8 | 3280 | 16.1 | 1.17 |
| 382 | 75 | MA0002.2 | Runx1 | MA0002.2 | BBYTGTGGTTT | Runx1 | 1.58e-07 | 1.43e-10 | 11500 | 91.2 | 18100 | 89 | 1.02 |
| 383 | 76 | MA0797.1 | TGIF2 | MA0797.1 | TGACAGSTGTCA | TGIF2 | 1.73e-07 | 1.56e-10 | 3010 | 23.9 | 4270 | 21 | 1.14 |
| 384 | 13 | MA0087.2 | Sox5 | MA0087.2 | VRVAACAATGGNN | Sox5 | 2.27e-07 | 2.05e-10 | 2220 | 17.6 | 3060 | 15 | 1.17 |
| 385 | 38 | MA0081.2 | SPIB | MA0081.2 | TYTCACTTCCTCTTTY | SPIB | 2.74e-07 | 2.46e-10 | 10300 | 81.9 | 16100 | 79.2 | 1.04 |
| 386 | 43 | MA1108.2 | MXI1 | MA1108.2 | NNCACATGBN | MXI1 | 4.45e-07 | 3.98e-10 | 6530 | 51.9 | 9850 | 48.4 | 1.07 |
| 387 | 77 | MA0878.3 | CDX1 | MA0878.3 | GGYMATAAAA | CDX1 | 5.92e-07 | 5.28e-10 | 1500 | 12 | 2000 | 9.82 | 1.22 |
| 388 | 43 | MA1109.1 | NEUROD1 | MA1109.1 | NRACAGATGGYNN | NEUROD1 | 1.08e-06 | 9.62e-10 | 9900 | 78.7 | 15400 | 75.8 | 1.04 |
| 389 | 34 | MA0595.1 | SREBF1 | MA0595.1 | VTCACCCCAY | SREBF1 | 1.7e-06 | 1.51e-09 | 2440 | 19.4 | 3430 | 16.8 | 1.15 |
| 390 | 34 | MA0684.2 | RUNX3 | MA0684.2 | NHAACCTCAANN | RUNX3 | 2.42e-06 | 2.14e-09 | 11200 | 88.8 | 17600 | 86.6 | 1.03 |
| 391 | 18 | MA0639.1 | DBP | MA0639.1 | NRTTACGTAAYV | DBP | 2.86e-06 | 2.52e-09 | 2310 | 18.3 | 3230 | 15.9 | 1.16 |
| 392 | 78 | MA1580.1 | ZBTB32 | MA1580.1 | TGTACAGTAW | ZBTB32 | 3.02e-06 | 2.66e-09 | 2570 | 20.4 | 3630 | 17.8 | 1.14 |
| 393 | 38 | MA1954.1 | FOXO1::ELK1 | MA1954.1 | RWMAACAGGAAGTD | FOXO1::ELK1 | 3.74e-06 | 3.29e-09 | 8600 | 68.4 | 13300 | 65.3 | 1.05 |
| 394 | 79 | MA1998.1 | Prdm14 | MA1998.1 | NDGGTCTCTAWS | Prdm14 | 4.04e-06 | 3.54e-09 | 8800 | 69.9 | 13600 | 66.9 | 1.05 |
| 395 | 80 | MA1968.1 | TFCP2 | MA1968.1 | AAACCGGTTY | TFCP2 | 8.12e-06 | 7.1e-09 | 9060 | 72 | 14100 | 69.1 | 1.04 |
| 396 | 3 | MA1949.1 | FLI1::DRGX | MA1949.1 | ACCGGAAGTAATTAT | FLI1::DRGX | 8.76e-06 | 7.64e-09 | 8500 | 67.6 | 13100 | 64.6 | 1.05 |
| 397 | 29 | MA0673.1 | NKX2-8 | MA0673.1 | VCACTTSAV | NKX2-8 | 8.81e-06 | 7.66e-09 | 10900 | 86.4 | 17100 | 84.1 | 1.03 |
| 398 | 10 | MA0784.2 | POU1F1 | MA0784.2 | ANCTMATTWGCATAWT | POU1F1 | 9.46e-06 | 8.21e-09 | 1290 | 10.2 | 1710 | 8.4 | 1.22 |
| 399 | 38 | MA1955.1 | FOXO1::ELK3 | MA1955.1 | RWMAACAGGAAGTN | FOXO1::ELK3 | 1.04e-05 | 9.03e-09 | 6150 | 48.8 | 9300 | 45.7 | 1.07 |
| 400 | 45 | MA0160.2 | NR4A2 | MA0160.2 | BWAAAGGTCA | NR4A2 | 1.12e-05 | 9.68e-09 | 5460 | 43.4 | 8200 | 40.3 | 1.08 |
| 401 | 54 | MA0689.1 | TBX20 | MA0689.1 | WAGGTGTGAAR | TBX20 | 1.22e-05 | 1.05e-08 | 3200 | 25.5 | 4640 | 22.8 | 1.12 |
| 402 | 45 | MA0065.2 | Pparg::Rxra | MA0065.2 | STRGGGCARAGGKCA | Pparg::Rxra | 1.91e-05 | 1.64e-08 | 5380 | 42.8 | 8080 | 39.7 | 1.08 |
| 403 | 81 | MA0770.1 | HSF2 | MA0770.1 | TTCTAGAAYRTTC | HSF2 | 2.03e-05 | 1.74e-08 | 3280 | 26.1 | 4760 | 23.4 | 1.11 |
| 404 | 38 | MA1508.1 | IKZF1 | MA1508.1 | VVAACAGGAARN | IKZF1 | 2.18e-05 | 1.86e-08 | 7200 | 57.2 | 11000 | 54.2 | 1.06 |
| 405 | 34 | MA0609.2 | CREM | MA0609.2 | NNDGTGACGTCACHNN | CREM | 2.24e-05 | 1.91e-08 | 10700 | 85.3 | 16900 | 83.1 | 1.03 |
| 406 | 82 | MA0520.1 | Stat6 | MA0520.1 | BDBTTCCWSAGAAVY | Stat6 | 2.56e-05 | 2.18e-08 | 2940 | 23.4 | 4240 | 20.8 | 1.12 |
| 407 | 3 | MA1546.1 | PAX3 | MA1546.1 | YSGTCACGSYWATTAN | PAX3 | 2.82e-05 | 2.39e-08 | 5370 | 42.7 | 8070 | 39.7 | 1.08 |
| 408 | 43 | MA0668.2 | Neurod2 | MA0668.2 | NNGRACAGATGGYNN | Neurod2 | 2.89e-05 | 2.45e-08 | 10200 | 80.7 | 15900 | 78.2 | 1.03 |
| 409 | 76 | MA0667.1 | MYF6 | MA0667.1 | AACARCTGTT | MYF6 | 3.62e-05 | 3.06e-08 | 2480 | 19.7 | 3530 | 17.3 | 1.14 |
| 410 | 28 | MA0030.1 | FOXF2 | MA0030.1 | BNAASGTAAACAAD | FOXF2 | 6.27e-05 | 5.28e-08 | 1490 | 11.8 | 2030 | 9.95 | 1.19 |
| 411 | 83 | MA0019.1 | Ddit3::Cebpa | MA0019.1 | VNRTGCAATMCC | Ddit3::Cebpa | 8.14e-05 | 6.84e-08 | 7720 | 61.3 | 11900 | 58.4 | 1.05 |
| 412 | 29 | MA2003.1 | NKX2-4 | MA2003.1 | CCACTTSAVV | NKX2-4 | 8.36e-05 | 7.01e-08 | 10500 | 83.4 | 16500 | 81.1 | 1.03 |
| 413 | 43 | MA1651.1 | ZFP42 | MA1651.1 | NNNHCAARATGGCTGCCNBNN | ZFP42 | 8.65e-05 | 7.23e-08 | 7520 | 59.7 | 11600 | 56.8 | 1.05 |
| 414 | 43 | MA1467.2 | Atoh1 | MA1467.2 | RVCAGATGGYN | Atoh1 | 0.000112 | 9.31e-08 | 11600 | 92.3 | 18400 | 90.6 | 1.02 |
| 415 | 84 | MA1983.1 | ZNF582 | MA1983.1 | TCTGTTACTTGCAGCCAAAWG | ZNF582 | 0.00012 | 9.95e-08 | 2390 | 19 | 3410 | 16.8 | 1.13 |
| 416 | 45 | MA0643.1 | Esrrg | MA0643.1 | TCAAGGTCAH | Esrrg | 0.000123 | 1.02e-07 | 4000 | 31.8 | 5920 | 29.1 | 1.09 |
| 417 | 38 | MA0080.6 | Spi1 | MA0080.6 | RRAAAGAGGAAGTGGDD | Spi1 | 0.000134 | 1.11e-07 | 9140 | 72.6 | 14200 | 70 | 1.04 |
| 418 | 77 | MA1473.1 | CDX4 | MA1473.1 | GGYMATAAAAC | CDX4 | 0.000158 | 1.31e-07 | 997 | 7.92 | 1310 | 6.44 | 1.23 |
| 419 | 37 | MA0072.1 | RORA | MA0072.1 | NWWAWBTAGGTCAR | RORA | 0.000185 | 1.52e-07 | 771 | 6.13 | 982 | 4.82 | 1.27 |
| 420 | 43 | MA1570.1 | TFAP4 | MA1570.1 | AHCATRTGDT | TFAP4 | 0.000222 | 1.83e-07 | 2500 | 19.9 | 3590 | 17.6 | 1.13 |
| 421 | 43 | MA1642.1 | NEUROG2 | MA1642.1 | NNVACAGATGGNN | NEUROG2 | 0.000226 | 1.86e-07 | 11100 | 88.5 | 17600 | 86.6 | 1.02 |
| 422 | 43 | MA0698.1 | ZBTB18 | MA0698.1 | NATCCAGATGTKB | ZBTB18 | 0.000246 | 2.01e-07 | 2750 | 21.9 | 3980 | 19.6 | 1.12 |
| 423 | 43 | MA0607.2 | BHLHA15 | MA0607.2 | ACCATATGGT | BHLHA15 | 0.000255 | 2.08e-07 | 2620 | 20.8 | 3780 | 18.6 | 1.12 |
| 424 | 36 | MA0610.1 | DMRT3 | MA0610.1 | MATGTATCAAN | DMRT3 | 0.00033 | 2.68e-07 | 1920 | 15.3 | 2710 | 13.3 | 1.15 |
| 425 | 80 | MA0647.1 | GRHL1 | MA0647.1 | AAAACCGGTTTD | GRHL1 | 0.000409 | 3.32e-07 | 3680 | 29.3 | 5450 | 26.8 | 1.09 |
| 426 | 37 | MA1111.1 | NR2F2 | MA1111.1 | NAAAGGTCANR | NR2F2 | 0.000488 | 3.96e-07 | 1760 | 14 | 2480 | 12.2 | 1.15 |
| 427 | 65 | MA0691.1 | TFAP4 | MA0691.1 | AWCAGCTGWT | TFAP4 | 0.000599 | 4.84e-07 | 924 | 7.34 | 1220 | 5.98 | 1.23 |
| 428 | 29 | MA0672.1 | NKX2-3 | MA0672.1 | VCCACTTRAV | NKX2-3 | 0.000615 | 4.96e-07 | 10700 | 85 | 16900 | 83 | 1.02 |
| 429 | 54 | MA1565.1 | TBX18 | MA1565.1 | DRAGGTGTGAAR | TBX18 | 0.000632 | 5.09e-07 | 5880 | 46.7 | 8950 | 44 | 1.06 |
| 430 | 85 | MA0619.1 | LIN54 | MA0619.1 | DTTTRAATH | LIN54 | 0.000696 | 5.59e-07 | 1730 | 13.8 | 2430 | 11.9 | 1.15 |
| 431 | 34 | MA1996.1 | Nr1H2 | MA1996.1 | NNRAGGTCANN | Nr1H2 | 0.000704 | 5.64e-07 | 10500 | 83.5 | 16600 | 81.4 | 1.03 |
| 432 | 45 | MA0141.3 | ESRRB | MA0141.3 | TCAAGGTCAWW | ESRRB | 0.000885 | 7.08e-07 | 1210 | 9.63 | 1650 | 8.1 | 1.19 |
| 433 | 45 | MA1541.1 | NR6A1 | MA1541.1 | NYCAAGKTCAAGKTCAN | NR6A1 | 0.0012 | 9.56e-07 | 5920 | 47 | 9030 | 44.4 | 1.06 |
| 434 | 23 | MA1547.2 | PITX2 | MA1547.2 | YTAATCCY | PITX2 | 0.00136 | 1.08e-06 | 1210 | 9.64 | 1660 | 8.14 | 1.19 |
| 435 | 86 | MA0106.3 | TP53 | MA0106.3 | RACATGYCCGGRCATGTY | TP53 | 0.0014 | 1.11e-06 | 1310 | 10.4 | 1800 | 8.84 | 1.18 |
| 436 | 76 | MA1572.1 | TGIF2LY | MA1572.1 | TGACAGCTGTCA | TGIF2LY | 0.00149 | 1.18e-06 | 2760 | 22 | 4030 | 19.8 | 1.11 |
| 437 | 72 | MA1528.1 | NFIX | MA1528.1 | STTGGCRNNGTGCCARB | NFIX | 0.00211 | 1.66e-06 | 799 | 6.35 | 1050 | 5.14 | 1.23 |
| 438 | 5 | MA0479.1 | FOXH1 | MA0479.1 | BNSAATCCACA | FOXH1 | 0.00211 | 1.66e-06 | 616 | 4.9 | 781 | 3.84 | 1.28 |
| 439 | 39 | MA1116.1 | RBPJ | MA1116.1 | BVTGGGAANN | RBPJ | 0.00212 | 1.67e-06 | 9640 | 76.7 | 15100 | 74.4 | 1.03 |
| 440 | 54 | MA0688.1 | TBX2 | MA0688.1 | DAGGTGTGAAA | TBX2 | 0.00222 | 1.74e-06 | 3010 | 23.9 | 4430 | 21.8 | 1.1 |
| 441 | 54 | MA0803.1 | TBX15 | MA0803.1 | AGGTGTGA | TBX15 | 0.00262 | 2.05e-06 | 1440 | 11.4 | 2000 | 9.83 | 1.16 |
| 442 | 43 | MA1638.1 | HAND2 | MA1638.1 | NVCAGATGNN | HAND2 | 0.00328 | 2.56e-06 | 9590 | 76.2 | 15100 | 74 | 1.03 |
| 443 | 54 | MA0801.1 | MGA | MA0801.1 | AGGTGTGA | MGA | 0.00334 | 2.61e-06 | 4300 | 34.2 | 6460 | 31.8 | 1.08 |
| 444 | 38 | MA1937.1 | ERF::HOXB13 | MA1937.1 | VSCGGAARYNRTWAAN | ERF::HOXB13 | 0.00353 | 2.74e-06 | 6940 | 55.2 | 10700 | 52.7 | 1.05 |
| 445 | 72 | MA0119.1 | NFIC::TLX1 | MA0119.1 | TGGCASSRWGCCAA | NFIC::TLX1 | 0.00464 | 3.6e-06 | 768 | 6.1 | 1010 | 4.96 | 1.23 |
| 446 | 37 | MA0071.1 | RORA | MA0071.1 | AWMWAGGTCA | RORA | 0.00511 | 3.95e-06 | 3380 | 26.8 | 5020 | 24.7 | 1.09 |
| 447 | 37 | MA0677.1 | Nr2f6 | MA0677.1 | RRGGTCAAAGGTCA | Nr2f6 | 0.00545 | 4.21e-06 | 5540 | 44 | 8450 | 41.5 | 1.06 |
| 448 | 54 | MA0805.1 | TBX1 | MA0805.1 | AGGTGTGA | TBX1 | 0.00585 | 4.5e-06 | 2000 | 15.9 | 2870 | 14.1 | 1.13 |
| 449 | 43 | MA0818.2 | BHLHE22 | MA0818.2 | AMCATATGKY | BHLHE22 | 0.00585 | 4.5e-06 | 1640 | 13 | 2320 | 11.4 | 1.14 |
| 450 | 45 | MA0505.2 | Nr5A2 | MA0505.2 | NTGACCTTGAMCT | Nr5A2 | 0.00724 | 5.56e-06 | 3360 | 26.7 | 4990 | 24.5 | 1.09 |
| 451 | 54 | MA0807.1 | TBX5 | MA0807.1 | AGGTGTKA | TBX5 | 0.00799 | 6.12e-06 | 1160 | 9.22 | 1600 | 7.86 | 1.17 |
| 452 | 3 | MA1944.1 | ETV5::DRGX | MA1944.1 | RSMGGAAGYAATTA | ETV5::DRGX | 0.00818 | 6.25e-06 | 7660 | 60.9 | 11900 | 58.5 | 1.04 |
| 453 | 38 | MA0062.3 | GABPA | MA0062.3 | NNCACTTCCTGTNN | GABPA | 0.00877 | 6.68e-06 | 10900 | 86.9 | 17300 | 85.2 | 1.02 |
| 454 | 38 | MA0761.2 | ETV1 | MA0761.2 | NNACAGGAAGTGNN | ETV1 | 0.0104 | 7.92e-06 | 10400 | 82.5 | 16400 | 80.7 | 1.02 |
| 14 | 90 | MA1599.1 (ZNF682) | STREME-10 | 10-CCAAGCCCAGGC | 10-CCAAGCCCAGGC | ZNF682 | 0.011 | 0.000783 | 153 | 1.22 | 172 | 0.85 | 1.44 |
| 455 | 38 | MA0136.3 | Elf5 | MA0136.3 | RVAAGGAAGTNN | Elf5 | 0.0111 | 8.45e-06 | 11900 | 94.3 | 19000 | 93.1 | 1.01 |
| 456 | 63 | MA1620.1 | Ptf1A | MA1620.1 | NVACACCTGTNN | Ptf1A | 0.0112 | 8.51e-06 | 10300 | 81.8 | 16300 | 79.9 | 1.02 |
| 457 | 76 | MA0783.1 | PKNOX2 | MA0783.1 | TGACAGGTGTCA | PKNOX2 | 0.0123 | 9.33e-06 | 455 | 3.62 | 566 | 2.78 | 1.3 |
| 458 | 49 | MA0517.1 | STAT1::STAT2 | MA0517.1 | THAGTTTCRKTTTCY | STAT1::STAT2 | 0.0141 | 1.07e-05 | 5670 | 45 | 8690 | 42.7 | 1.05 |
| 459 | 87 | MA0838.1 | CEBPG | MA0838.1 | ATTRCGCAAY | CEBPG | 0.0231 | 1.73e-05 | 986 | 7.84 | 1350 | 6.65 | 1.18 |
| 460 | 88 | MA0101.1 | REL | MA0101.1 | BGGRNWTTCC | REL | 0.0276 | 2.07e-05 | 8770 | 69.7 | 13800 | 67.6 | 1.03 |
| 461 | 63 | MA0006.1 | Ahr::Arnt | MA0006.1 | YGCGTG | Ahr::Arnt | 0.0318 | 2.38e-05 | 10700 | 85.2 | 17000 | 83.5 | 1.02 |
| 462 | 89 | MA1594.1 | ZNF382 | MA1594.1 | AGGRGAYATCACYACMGAYCCCAC | ZNF382 | 0.0331 | 2.47e-05 | 1020 | 8.09 | 1410 | 6.91 | 1.17 |
| 463 | 54 | MA1566.2 | TBX3 | MA1566.2 | RAGGTGTGAAV | TBX3 | 0.0354 | 2.64e-05 | 3060 | 24.3 | 4560 | 22.4 | 1.08 |
| 464 | 38 | MA0473.3 | ELF1 | MA0473.3 | RDVCAGGAAGTGVN | ELF1 | 0.0357 | 2.65e-05 | 10300 | 81.9 | 16300 | 80.1 | 1.02 |
| 465 | 3 | MA0703.2 | LMX1B | MA0703.2 | AWTTTAATTAA | LMX1B | 0.0395 | 2.93e-05 | 524 | 4.16 | 676 | 3.32 | 1.25 |
| 466 | 38 | MA1956.1 | FOXO1::FLI1 | MA1956.1 | GTAAACAGGAAGTK | FOXO1::FLI1 | 0.0431 | 3.19e-05 | 1100 | 8.77 | 1540 | 7.56 | 1.16 |
| 15 | 91 | MA1107.2 (KLF9) | MEME-3 | CACACACACACACAC | CACACACACACACAC | KLF9 | 10.6 | 0.707 | 17 | 0.14 | 29 | 0.14 | 0.971 |
| 16 | 92 | MA1961.1 (PATZ1) | MEME-2 | GGSRGGGGGCGGGGC | GGSRGGGGGCGGGGC | PATZ1 | 12.1 | 0.759 | 9 | 0.07 | 17 | 0.08 | 0.899 |
| 17 | 92 | MA1965.1 (SP5) | MEME-7 | GRGRRGGAGGRRGDG | GRGRRGGAGGRRGDG | SP5 | 17 | 1 | 5830 | 46.4 | 9850 | 48.4 | 0.958 |
ER_rat <- 1.25
EPI_242kb_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste("EPI_242kb_combo response peaks(200bp)Enrichment ratio:",ER_rat))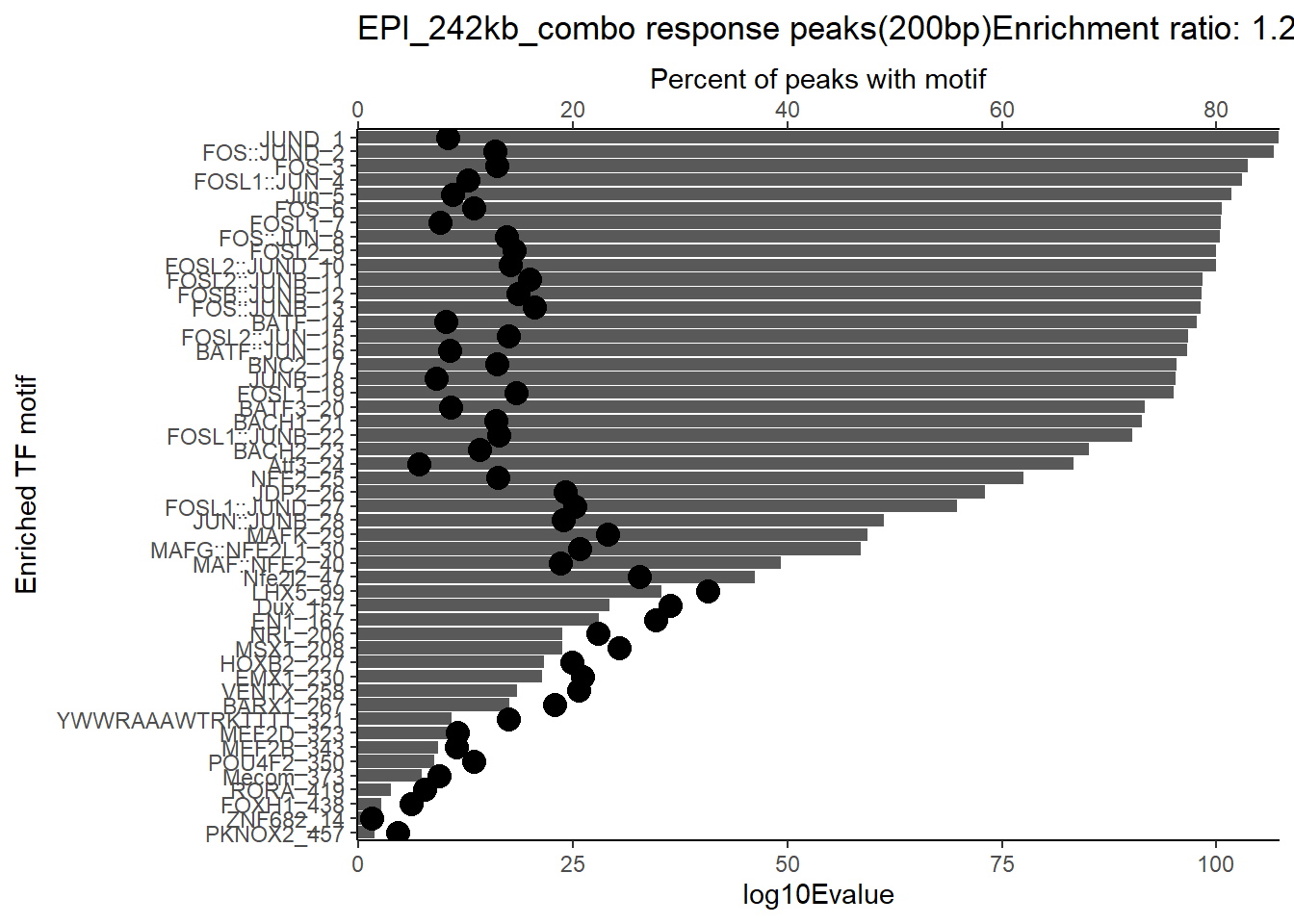
EPI_242kb_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y, TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
slice_head(n=5) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*4), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./4,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( "EPI_24 2kb response peaks(200bp)Enrichment ratio:",ER_rat))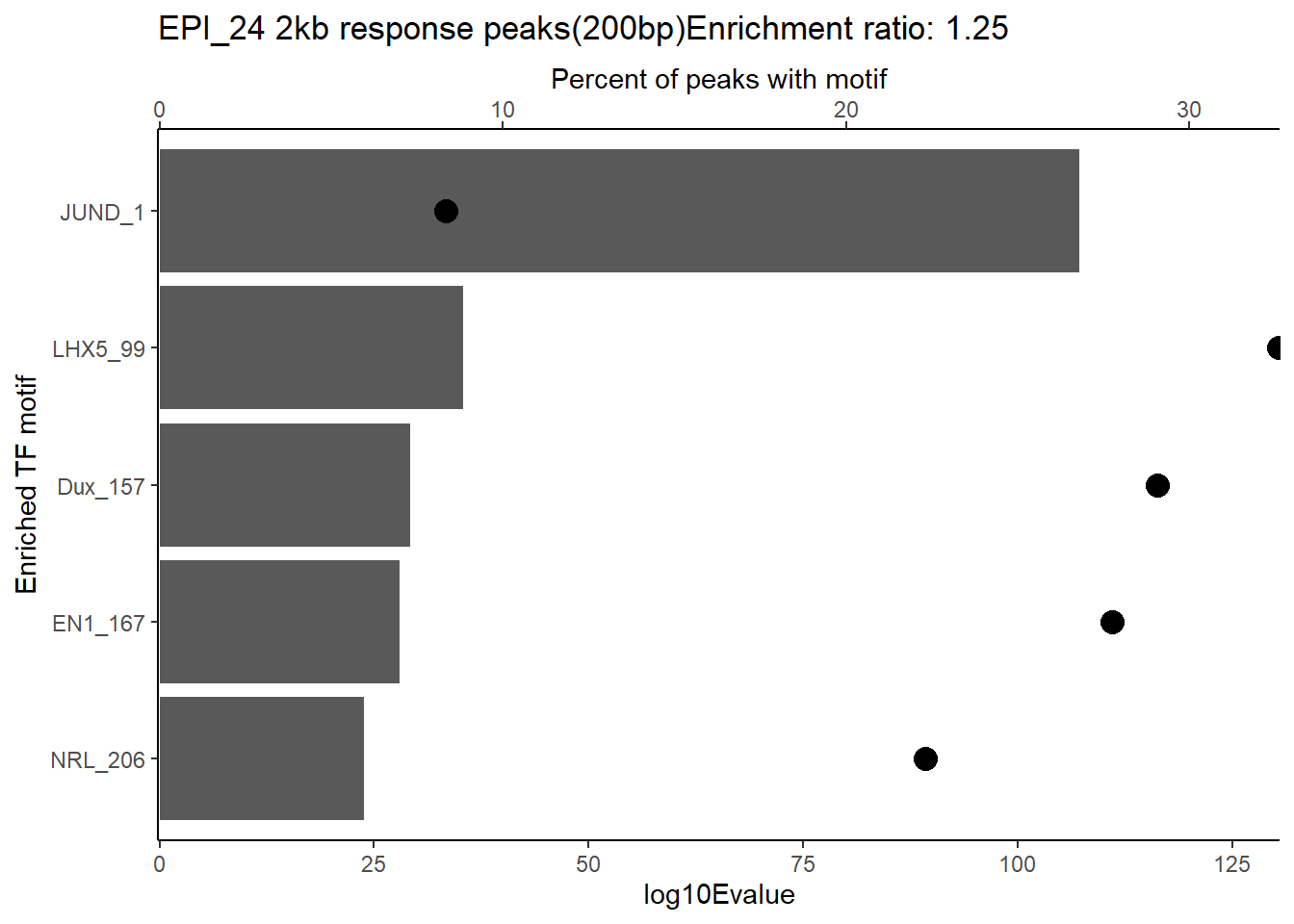
EPI_DAR 24 20kb data
EPI_24_20kb_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/EPI_24_20kb_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
EPI_24_20kb_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/EPI_24_20kb_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
EPI_24_20kb_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/EPI_24_20kb_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
EPI_24_20kb_sea_all <- rbind(EPI_24_20kb_sea_disc, EPI_24_20kb_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)EPI_24_20kb_combo <- EPI_24_20kb_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., EPI_24_20kb_sea_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ SIM_MOTIF
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y)
EPI_24_20kb_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs 200 bp in EPI_24h 20kb") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | MA1134.1 (FOS::JUNB) | STREME-1 | 1-RATGASTCATY | RATGASTCATY | FOS::JUNB | 0 | 0 | 5480 | 15 | 3320 | 6.6 | 2.28 |
| 2 | 1 | MA1138.1 | FOSL2::JUNB | MA1138.1 | KRTGASTCAT | FOSL2::JUNB | 0 | 0 | 6050 | 16.6 | 4020 | 7.98 | 2.08 |
| 3 | 1 | MA1144.1 | FOSL2::JUND | MA1144.1 | KATGACTCAT | FOSL2::JUND | 0 | 0 | 5460 | 15 | 3450 | 6.85 | 2.19 |
| 4 | 1 | MA1134.1 | FOS::JUNB | MA1134.1 | KATGASTCATHN | FOS::JUNB | 1.24998608397835e-320 | 1.06224113855868e-321 | 4810 | 13.2 | 2850 | 5.66 | 2.34 |
| 5 | 1 | MA1135.1 | FOSB::JUNB | MA1135.1 | KRTGASTCAT | FOSB::JUNB | 2.38001302914645e-319 | 2.20007432093107e-320 | 5540 | 15.2 | 3560 | 7.08 | 2.15 |
| 6 | 1 | MA0477.2 | FOSL1 | MA0477.2 | NNATGACTCATNN | FOSL1 | 9.10000244514804e-318 | 7.1199800222182e-319 | 4910 | 13.5 | 2950 | 5.87 | 2.3 |
| 7 | 1 | MA0491.2 | JUND | MA0491.2 | NNATGACTCATNN | JUND | 5.82000000000717e-313 | 3.90000000000735e-314 | 5580 | 15.3 | 3640 | 7.23 | 2.12 |
| 8 | 1 | MA1128.1 | FOSL1::JUN | MA1128.1 | NKATGACTCATNN | FOSL1::JUN | 4.80000000000004e-311 | 2.8200000000019e-312 | 4400 | 12.1 | 2510 | 4.98 | 2.43 |
| 9 | 1 | MA1130.1 | FOSL2::JUN | MA1130.1 | NNRTGAGTCAYN | FOSL2::JUN | 1.03e-309 | 5.36000000000007e-311 | 4430 | 12.2 | 2540 | 5.05 | 2.41 |
| 10 | 1 | MA0099.3 | FOS::JUN | MA0099.3 | ATGAGTCAYM | FOS::JUN | 2.61e-309 | 1.22000000000001e-310 | 4500 | 12.4 | 2610 | 5.18 | 2.38 |
| 11 | 1 | MA1141.1 | FOS::JUND | MA1141.1 | NKATGAGTCATNN | FOS::JUND | 1.05e-304 | 4.35e-306 | 4940 | 13.6 | 3040 | 6.05 | 2.24 |
| 12 | 1 | MA0489.2 | Jun | MA0489.2 | NVTGACTCATNN | Jun | 1.11e-304 | 4.35e-306 | 5450 | 15 | 3550 | 7.05 | 2.12 |
| 13 | 1 | MA0476.1 | FOS | MA0476.1 | DVTGASTCATB | FOS | 4.73e-304 | 1.71e-305 | 4350 | 12 | 2490 | 4.95 | 2.41 |
| 14 | 1 | MA1633.2 | BACH1 | MA1633.2 | ATGACTCAT | BACH1 | 3.87e-303 | 1.3e-304 | 4780 | 13.1 | 2910 | 5.77 | 2.28 |
| 15 | 1 | MA0490.2 | JUNB | MA0490.2 | NNATGACTCATNN | JUNB | 1.34e-301 | 4.18e-303 | 5420 | 14.9 | 3540 | 7.02 | 2.12 |
| 16 | 1 | MA0476.1 (FOS) | MEME-2 | ATGASTCASH | ATGASTCASH | FOS | 2.27e-295 | 6.66e-297 | 4640 | 12.8 | 2810 | 5.59 | 2.28 |
| 17 | 1 | MA1634.1 | BATF | MA1634.1 | DATGACTCATH | BATF | 1.68e-293 | 4.62e-295 | 5150 | 14.1 | 3310 | 6.57 | 2.15 |
| 18 | 1 | MA1137.1 | FOSL1::JUNB | MA1137.1 | NDRTGACTCATNN | FOSL1::JUNB | 1.48e-290 | 3.85e-292 | 5220 | 14.3 | 3400 | 6.75 | 2.12 |
| 19 | 1 | MA0462.2 | BATF::JUN | MA0462.2 | DATGACTCATH | BATF::JUN | 4.54e-289 | 1.12e-290 | 4890 | 13.4 | 3080 | 6.13 | 2.19 |
| 20 | 1 | MA1988.1 | Atf3 | MA1988.1 | NRTGACTCABN | Atf3 | 8.72999999999999e-288 | 2.05e-289 | 4530 | 12.4 | 2740 | 5.45 | 2.28 |
| 21 | 1 | MA0655.1 | JDP2 | MA0655.1 | ATGACTCAT | JDP2 | 2.06e-284 | 4.6e-286 | 6460 | 17.8 | 4710 | 9.35 | 1.9 |
| 22 | 1 | MA0478.1 | FOSL2 | MA0478.1 | KRRTGASTCAB | FOSL2 | 6.12e-280 | 1.31e-281 | 5160 | 14.2 | 3400 | 6.75 | 2.1 |
| 23 | 1 | MA1142.1 | FOSL1::JUND | MA1142.1 | NRTGACTCAT | FOSL1::JUND | 2.84e-273 | 5.79e-275 | 5480 | 15.1 | 3760 | 7.46 | 2.02 |
| 24 | 1 | MA1928.1 | BNC2 | MA1928.1 | NDRTGAGTCABN | BNC2 | 6.07e-270 | 1.19e-271 | 6050 | 16.6 | 4370 | 8.67 | 1.92 |
| 25 | 1 | MA0835.2 | BATF3 | MA0835.2 | DATGACTCATH | BATF3 | 2.35e-264 | 4.41e-266 | 4060 | 11.2 | 2420 | 4.8 | 2.33 |
| 26 | 1 | MA1132.1 | JUN::JUNB | MA1132.1 | KATGACKCAT | JUN::JUNB | 5.41e-262 | 9.76e-264 | 4870 | 13.4 | 3210 | 6.37 | 2.1 |
| 27 | 1 | MA1101.2 | BACH2 | MA1101.2 | DWANCATGASTCATSNTWH | BACH2 | 1.61e-260 | 2.8e-262 | 6040 | 16.6 | 4420 | 8.77 | 1.89 |
| 28 | 1 | MA0841.1 | NFE2 | MA0841.1 | VATGACTCATS | NFE2 | 1.87e-245 | 3.13e-247 | 5130 | 14.1 | 3560 | 7.08 | 1.99 |
| 29 | 1 | MA0089.2 | MAFG::NFE2L1 | MA0089.2 | NVNATGACTCAGCADW | MAFG::NFE2L1 | 5.17e-154 | 8.36e-156 | 6800 | 18.7 | 6090 | 12.1 | 1.54 |
| 30 | 1 | MA0496.3 | MAFK | MA0496.3 | NGMTGACTCAGCMNH | MAFK | 1.57e-137 | 2.45e-139 | 5360 | 14.7 | 4610 | 9.15 | 1.61 |
| 31 | 1 | MA0501.1 | MAF::NFE2 | MA0501.1 | ATGACTCAGCANWTY | MAF::NFE2 | 1.43e-126 | 2.17e-128 | 8160 | 22.4 | 8000 | 15.9 | 1.41 |
| 32 | 1 | MA0150.2 | Nfe2l2 | MA0150.2 | CASNATGACTCAGCA | Nfe2l2 | 9.41e-90 | 1.38e-91 | 8360 | 23 | 8740 | 17.4 | 1.32 |
| 33 | 2 | MA1991.1 (Hnf1A) | STREME-8 | 8-TCAAAG | TCAAAG | Hnf1A | 7.75e-78 | 1.1e-79 | 32700 | 89.7 | 43000 | 85.4 | 1.05 |
| 34 | 1 | MA1137.1 (FOSL1::JUNB) | STREME-2 | 2-ABTCAKM | ABTCAKM | FOSL1::JUNB | 7.34e-76 | 1.01e-77 | 24600 | 67.5 | 30900 | 61.4 | 1.1 |
| 35 | 1 | MA0659.3 | Mafg | MA0659.3 | NWGMTGACTCAGCAN | Mafg | 7.98e-76 | 1.07e-77 | 4670 | 12.8 | 4450 | 8.84 | 1.45 |
| 36 | 3 | MA0720.1 | Shox2 | MA0720.1 | YYAATTAR | Shox2 | 2.16e-74 | 2.82e-76 | 31100 | 85.5 | 40600 | 80.7 | 1.06 |
| 37 | 3 | MA0723.2 | VAX2 | MA0723.2 | BYAATTAV | VAX2 | 3.3e-73 | 4.18e-75 | 30900 | 85 | 40400 | 80.2 | 1.06 |
| 38 | 4 | MA0084.1 | SRY | MA0084.1 | NWWAACAAT | SRY | 2.58e-71 | 3.18e-73 | 32800 | 90 | 43200 | 85.9 | 1.05 |
| 39 | 3 | MA0027.2 | EN1 | MA0027.2 | SYAATTAV | EN1 | 4.92e-71 | 5.92e-73 | 31100 | 85.5 | 40700 | 80.8 | 1.06 |
| 40 | 5 | 9-ATNAGG | STREME-9 | 9-ATNAGG | ATNAGG | ATNAGG | 6.67e-71 | 7.82e-73 | 32100 | 88.1 | 42100 | 83.7 | 1.05 |
| 41 | 3 | MA0718.1 | RAX | MA0718.1 | RYYAATTARH | RAX | 2.42e-70 | 2.77e-72 | 29300 | 80.4 | 37900 | 75.3 | 1.07 |
| 42 | 3 | MA0725.1 | VSX1 | MA0725.1 | YTAATTAN | VSX1 | 2.84e-68 | 3.18e-70 | 32700 | 89.9 | 43200 | 85.9 | 1.05 |
| 43 | 1 | MA1639.1 | MEIS1 | MA1639.1 | NHCATMAATCAHN | MEIS1 | 3.66e-68 | 3.99e-70 | 25200 | 69.1 | 31900 | 63.3 | 1.09 |
| 44 | 6 | MA0676.1 | Nr2e1 | MA0676.1 | AAAAGTCAA | Nr2e1 | 4.08e-67 | 4.35e-69 | 24300 | 66.7 | 30700 | 60.9 | 1.1 |
| 45 | 7 | MA1963.1 | SATB1 | MA1963.1 | WWDCTAATAAHWW | SATB1 | 4.74e-67 | 4.94e-69 | 24300 | 66.7 | 30700 | 60.9 | 1.1 |
| 46 | 3 | MA0886.1 | EMX2 | MA0886.1 | VBTAATTARB | EMX2 | 8.66e-67 | 8.83e-69 | 23100 | 63.5 | 29000 | 57.5 | 1.1 |
| 47 | 3 | MA0902.2 | HOXB2 | MA0902.2 | SYAATTAS | HOXB2 | 2.65e-66 | 2.64e-68 | 22300 | 61.4 | 27900 | 55.4 | 1.11 |
| 48 | 3 | MA0726.1 | VSX2 | MA0726.1 | CTAATTAN | VSX2 | 2.13e-64 | 2.08e-66 | 32600 | 89.4 | 43000 | 85.5 | 1.05 |
| 49 | 3 | MA0704.1 | Lhx4 | MA0704.1 | YTAATTAR | Lhx4 | 4.31e-64 | 4.12e-66 | 31500 | 86.6 | 41400 | 82.3 | 1.05 |
| 50 | 1 | MA1113.2 | PBX2 | MA1113.2 | NHCATAAATCAHN | PBX2 | 6.29e-63 | 5.91e-65 | 28300 | 77.8 | 36600 | 72.8 | 1.07 |
| 51 | 3 | MA0710.1 | NOTO | MA0710.1 | NYTAATTAVN | NOTO | 4.18e-62 | 3.85e-64 | 25700 | 70.5 | 32700 | 65 | 1.08 |
| 52 | 3 | MA0644.2 | ESX1 | MA0644.2 | YYAATTAN | ESX1 | 1.11e-61 | 1e-63 | 24900 | 68.3 | 31600 | 62.8 | 1.09 |
| 53 | 8 | MA0142.1 | Pou5f1::Sox2 | MA0142.1 | CWTTGTYATGCAAAT | Pou5f1::Sox2 | 2.24e-61 | 1.98e-63 | 27700 | 76.1 | 35700 | 70.9 | 1.07 |
| 54 | 3 | MA0674.1 | NKX6-1 | MA0674.1 | NTMATTAA | NKX6-1 | 1.04e-60 | 8.99e-63 | 33000 | 90.5 | 43700 | 86.9 | 1.04 |
| 55 | 1 | MA1640.1 | MEIS2 | MA1640.1 | NNATGATTTATGDNH | MEIS2 | 2.65e-59 | 2.26e-61 | 29600 | 81.3 | 38600 | 76.6 | 1.06 |
| 56 | 3 | MA0634.1 | ALX3 | MA0634.1 | YYYAATTANV | ALX3 | 4.88e-59 | 4.09e-61 | 31600 | 86.9 | 41700 | 82.8 | 1.05 |
| 57 | 8 | MA0786.1 | POU3F1 | MA0786.1 | WTATGCWAATKW | POU3F1 | 5.71e-59 | 4.7e-61 | 28900 | 79.5 | 37600 | 74.7 | 1.06 |
| 58 | 3 | MA0680.2 | Pax7 | MA0680.2 | DWTAATCAATTAWNH | Pax7 | 1.68e-58 | 1.36e-60 | 24300 | 66.7 | 30800 | 61.3 | 1.09 |
| 59 | 9 | MA0650.3 | Hoxa13 | MA0650.3 | NNCCAATAAAAN | Hoxa13 | 1.82e-58 | 1.45e-60 | 29900 | 82.1 | 39000 | 77.5 | 1.06 |
| 60 | 10 | MA1104.2 | GATA6 | MA1104.2 | HWWTCTTATCTNH | GATA6 | 1.96e-58 | 1.53e-60 | 27800 | 76.4 | 36000 | 71.4 | 1.07 |
| 61 | 8 | MA0788.1 | POU3F3 | MA0788.1 | WWTATGCWAATTW | POU3F3 | 2.74e-58 | 2.11e-60 | 26600 | 73 | 34200 | 67.9 | 1.08 |
| 62 | 10 | MA0035.4 | GATA1 | MA0035.4 | NHCTAATCTDH | GATA1 | 4.3e-58 | 3.25e-60 | 28600 | 78.5 | 37100 | 73.7 | 1.07 |
| 63 | 8 | MA0785.1 | POU2F1 | MA0785.1 | AWTATGCWAATK | POU2F1 | 5.47e-58 | 4.08e-60 | 28600 | 78.7 | 37200 | 73.9 | 1.07 |
| 64 | 3 | MA0713.1 | PHOX2A | MA0713.1 | TAATYYAATTA | PHOX2A | 1.96e-57 | 1.43e-59 | 25400 | 69.7 | 32400 | 64.4 | 1.08 |
| 65 | 3 | MA0628.1 | POU6F1 | MA0628.1 | AYTAATTART | POU6F1 | 2.21e-57 | 1.59e-59 | 28900 | 79.3 | 37500 | 74.5 | 1.06 |
| 66 | 3 | MA0879.2 | DLX1 | MA0879.2 | VYAATTAN | DLX1 | 4.58e-57 | 3.25e-59 | 33600 | 92.4 | 44900 | 89.2 | 1.04 |
| 67 | 3 | MA0890.1 | GBX2 | MA0890.1 | RSYAATTARN | GBX2 | 4.65e-57 | 3.25e-59 | 27500 | 75.6 | 35600 | 70.6 | 1.07 |
| 68 | 3 | MA1476.2 | Dlx5 | MA1476.2 | NDGCAATTAGNN | Dlx5 | 7.13e-57 | 4.92e-59 | 31500 | 86.4 | 41500 | 82.4 | 1.05 |
| 69 | 3 | MA0904.2 | HOXB5 | MA0904.2 | STAATTAS | HOXB5 | 9.09e-57 | 6.18e-59 | 20800 | 57.1 | 26000 | 51.6 | 1.11 |
| 70 | 11 | MA0041.2 | FOXD3 | MA0041.2 | WAWGHAAATAAACARY | FOXD3 | 1.58e-56 | 1.06e-58 | 29200 | 80.1 | 38000 | 75.4 | 1.06 |
| 71 | 11 | MA0845.1 | FOXB1 | MA0845.1 | WATGTAAATAT | FOXB1 | 2.47e-56 | 1.63e-58 | 28000 | 76.8 | 36200 | 71.9 | 1.07 |
| 72 | 3 | MA0654.1 | ISX | MA0654.1 | YYAATTAR | ISX | 4.62e-56 | 3.01e-58 | 21500 | 59 | 26900 | 53.5 | 1.1 |
| 73 | 3 | MA0889.1 | GBX1 | MA0889.1 | VCYAATTASY | GBX1 | 1.19e-55 | 7.64e-58 | 26200 | 71.8 | 33600 | 66.7 | 1.08 |
| 74 | 3 | MA1481.1 | DRGX | MA1481.1 | YYAATTAN | DRGX | 1.44e-55 | 9.12e-58 | 29100 | 80 | 37900 | 75.3 | 1.06 |
| 75 | 3 | MA0675.1 | NKX6-2 | MA0675.1 | NYMATTAA | NKX6-2 | 2.2e-55 | 1.37e-57 | 33200 | 91.1 | 44100 | 87.7 | 1.04 |
| 76 | 3 | MA0882.1 | DLX6 | MA0882.1 | VYAATTAN | DLX6 | 2.21e-55 | 1.37e-57 | 33900 | 93 | 45300 | 89.9 | 1.03 |
| 77 | 3 | MA1502.1 | HOXB8 | MA1502.1 | GYMATTAM | HOXB8 | 8.27e-55 | 5.04e-57 | 19400 | 53.2 | 24000 | 47.7 | 1.12 |
| 78 | 12 | MA0052.4 | MEF2A | MA0052.4 | DDCTAAAAATAGMHH | MEF2A | 1.19e-54 | 7.17e-57 | 26100 | 71.6 | 33500 | 66.5 | 1.08 |
| 79 | 8 | MA0789.1 | POU3F4 | MA0789.1 | TATGCWAAT | POU3F4 | 2.47e-54 | 1.47e-56 | 24700 | 68 | 31600 | 62.8 | 1.08 |
| 80 | 3 | MA0125.1 | Nobox | MA0125.1 | TAATTRSY | Nobox | 3.65e-54 | 2.14e-56 | 30300 | 83.1 | 39700 | 78.8 | 1.05 |
| 81 | 13 | MA1480.1 | DPRX | MA1480.1 | AGATAATCCN | DPRX | 4.12e-54 | 2.39e-56 | 29300 | 80.4 | 38200 | 75.9 | 1.06 |
| 82 | 9 | MA0465.2 | CDX2 | MA0465.2 | NDGCAATAAAWN | CDX2 | 4.97e-54 | 2.85e-56 | 29300 | 80.4 | 38200 | 75.9 | 1.06 |
| 83 | 3 | MA0722.1 | VAX1 | MA0722.1 | YTAATTAH | VAX1 | 6.26e-54 | 3.54e-56 | 33100 | 91 | 44100 | 87.6 | 1.04 |
| 84 | 14 | MA0635.1 | BARHL2 | MA0635.1 | VHTAAAYGRT | BARHL2 | 8.09e-54 | 4.52e-56 | 31300 | 86 | 41300 | 82 | 1.05 |
| 85 | 9 | MA0901.2 | HOXB13 | MA0901.2 | WDCCAATAAAAWWW | HOXB13 | 8.43e-54 | 4.66e-56 | 25400 | 69.8 | 32500 | 64.6 | 1.08 |
| 86 | 9 | MA0899.1 | HOXA10 | MA0899.1 | DGYMATAAAAH | HOXA10 | 1.71e-53 | 9.32e-56 | 25300 | 69.4 | 32400 | 64.3 | 1.08 |
| 87 | 3 | MA0793.1 | POU6F2 | MA0793.1 | ASCTMATTAW | POU6F2 | 2.02e-53 | 1.08e-55 | 28000 | 76.8 | 36300 | 72.1 | 1.07 |
| 88 | 3 | MA0158.2 | HOXA5 | MA0158.2 | GYMATTAS | HOXA5 | 2.03e-53 | 1.08e-55 | 17200 | 47.1 | 21000 | 41.7 | 1.13 |
| 89 | 8 | MA0787.1 | POU3F2 | MA0787.1 | WTATGCWAATKA | POU3F2 | 2.61e-53 | 1.38e-55 | 25100 | 69 | 32200 | 63.9 | 1.08 |
| 90 | 3 | MA0721.1 | UNCX | MA0721.1 | YYAATTAV | UNCX | 5.57e-53 | 2.9e-55 | 32500 | 89.4 | 43200 | 85.8 | 1.04 |
| 91 | 3 | MA0876.1 | BSX | MA0876.1 | SYVATTAV | BSX | 1.57e-52 | 8.1e-55 | 30000 | 82.4 | 39300 | 78.2 | 1.05 |
| 92 | 3 | MA1495.1 | HOXA1 | MA1495.1 | STAATTAS | HOXA1 | 2.96e-52 | 1.51e-54 | 16400 | 45.2 | 20100 | 39.9 | 1.13 |
| 93 | 3 | MA1505.1 | HOXC8 | MA1505.1 | KTAATTAM | HOXC8 | 3.44e-52 | 1.74e-54 | 32300 | 88.8 | 42900 | 85.1 | 1.04 |
| 94 | 3 | MA0707.2 | MNX1 | MA0707.2 | VYRATTAK | MNX1 | 5.13e-52 | 2.56e-54 | 19100 | 52.5 | 23800 | 47.2 | 1.11 |
| 95 | 3 | MA0709.1 | Msx3 | MA0709.1 | SCAATTAN | Msx3 | 6.43e-52 | 3.18e-54 | 28800 | 79.1 | 37600 | 74.6 | 1.06 |
| 96 | 3 | MA0885.2 | Dlx2 | MA0885.2 | NDGCAATTAGNN | Dlx2 | 1.54e-51 | 7.53e-54 | 32400 | 89.1 | 43100 | 85.6 | 1.04 |
| 97 | 3 | MA0708.2 | MSX2 | MA0708.2 | SCAATTAV | MSX2 | 1.91e-51 | 9.23e-54 | 27900 | 76.6 | 36200 | 71.9 | 1.06 |
| 98 | 3 | MA0601.1 | Arid3b | MA0601.1 | ATATTAATWAN | Arid3b | 4.98e-51 | 2.38e-53 | 21600 | 59.2 | 27200 | 54 | 1.1 |
| 99 | 3 | MA0658.1 | LHX6 | MA0658.1 | RCTAATTARY | LHX6 | 7.17e-51 | 3.4e-53 | 22500 | 61.8 | 28500 | 56.6 | 1.09 |
| 100 | 10 | MA0036.3 | GATA2 | MA0036.3 | NBCTTATCTNH | GATA2 | 1.37e-50 | 6.42e-53 | 30400 | 83.6 | 40000 | 79.5 | 1.05 |
| 101 | 15 | MA0791.1 | POU4F3 | MA0791.1 | RTGMATWATTAATGAR | POU4F3 | 1.76e-50 | 8.16e-53 | 21800 | 60 | 27600 | 54.8 | 1.1 |
| 102 | 3 | MA0716.1 | PRRX1 | MA0716.1 | YYAATTAR | PRRX1 | 2.63e-50 | 1.21e-52 | 33900 | 93.2 | 45500 | 90.3 | 1.03 |
| 103 | 3 | MA0701.2 | LHX9 | MA0701.2 | CYAATTAR | LHX9 | 3.9e-50 | 1.78e-52 | 15200 | 41.7 | 18400 | 36.6 | 1.14 |
| 104 | 3 | MA0700.2 | LHX2 | MA0700.2 | DDCCAATTADN | LHX2 | 5.47e-50 | 2.47e-52 | 28400 | 78 | 37000 | 73.5 | 1.06 |
| 105 | 8 | MA0792.1 | POU5F1B | MA0792.1 | TATGCWAAT | POU5F1B | 5.64e-50 | 2.52e-52 | 31900 | 87.8 | 42300 | 84.1 | 1.04 |
| 106 | 16 | MA0018.4 (CREB1) | STREME-3 | 3-ATGWCAT | ATGWCAT | CREB1 | 8.62e-50 | 3.82e-52 | 34600 | 94.9 | 46500 | 92.4 | 1.03 |
| 107 | 11 | MA1606.1 | Foxf1 | MA1606.1 | DWRTAAACANN | Foxf1 | 8.8e-50 | 3.86e-52 | 32600 | 89.6 | 43400 | 86.2 | 1.04 |
| 108 | 3 | MA0648.1 | GSC | MA0648.1 | VYTAATCCSY | GSC | 1.33e-49 | 5.77e-52 | 28900 | 79.3 | 37700 | 74.9 | 1.06 |
| 109 | 3 | MA0666.2 | MSX1 | MA0666.2 | SCAATTAR | MSX1 | 2.24e-49 | 9.64e-52 | 24100 | 66.3 | 30800 | 61.2 | 1.08 |
| 110 | 17 | MA0495.3 | MAFF | MA0495.3 | DWGTCAGCATTTTWHN | MAFF | 2.78e-49 | 1.18e-51 | 31100 | 85.5 | 41100 | 81.6 | 1.05 |
| 111 | 3 | MA0910.2 | HOXD8 | MA0910.2 | GYMATTAM | HOXD8 | 6.8e-49 | 2.87e-51 | 17200 | 47.4 | 21300 | 42.2 | 1.12 |
| 112 | 15 | MA0790.1 | POU4F1 | MA0790.1 | ATGMATAATTAATG | POU4F1 | 1.33e-48 | 5.57e-51 | 22100 | 60.7 | 28000 | 55.6 | 1.09 |
| 113 | 11 | MA0032.2 | FOXC1 | MA0032.2 | WAWGTAAAYAW | FOXC1 | 1.68e-47 | 6.96e-50 | 29200 | 80.3 | 38300 | 76.1 | 1.06 |
| 114 | 3 | MA1497.1 | HOXA6 | MA1497.1 | BGYMATTANN | HOXA6 | 1.7e-47 | 6.98e-50 | 31100 | 85.4 | 41100 | 81.6 | 1.05 |
| 115 | 8 | MA0627.2 | POU2F3 | MA0627.2 | NWTATGCAAATNH | POU2F3 | 1.92e-47 | 7.85e-50 | 26700 | 73.2 | 34500 | 68.6 | 1.07 |
| 116 | 3 | MA0681.2 | PHOX2B | MA0681.2 | WWTAATCAAATTAWWW | PHOX2B | 2.46e-47 | 9.95e-50 | 21000 | 57.6 | 26400 | 52.5 | 1.1 |
| 117 | 3 | MA0630.1 | SHOX | MA0630.1 | HTAATTRR | SHOX | 3.63e-47 | 1.45e-49 | 34300 | 94.3 | 46100 | 91.6 | 1.03 |
| 118 | 18 | MA0769.2 | TCF7 | MA0769.2 | NNCTTTGAWVT | TCF7 | 3.64e-47 | 1.45e-49 | 34000 | 93.5 | 45700 | 90.8 | 1.03 |
| 119 | 3 | MA1549.1 | POU6F1 | MA1549.1 | RTAATGAGVN | POU6F1 | 4.91e-47 | 1.93e-49 | 30100 | 82.6 | 39600 | 78.6 | 1.05 |
| 120 | 3 | MA0661.1 | MEOX1 | MA0661.1 | VSTAATTAHC | MEOX1 | 5.54e-47 | 2.17e-49 | 31100 | 85.4 | 41100 | 81.6 | 1.05 |
| 121 | 10 | MA1970.1 | TRPS1 | MA1970.1 | NHTCTTATCTNH | TRPS1 | 5.83e-47 | 2.26e-49 | 30000 | 82.4 | 39500 | 78.4 | 1.05 |
| 122 | 3 | MA1518.2 | Lhx1 | MA1518.2 | NNNTGCTAATTAGCANNN | Lhx1 | 5.87e-47 | 2.26e-49 | 27300 | 74.9 | 35400 | 70.3 | 1.06 |
| 123 | 3 | MA0068.2 | PAX4 | MA0068.2 | CTAATTAG | PAX4 | 7.26e-47 | 2.77e-49 | 30900 | 84.9 | 40800 | 81.1 | 1.05 |
| 124 | 3 | MA1471.1 | BARX2 | MA1471.1 | NWWAAYMATTAN | BARX2 | 8.49e-47 | 3.21e-49 | 28000 | 77 | 36500 | 72.5 | 1.06 |
| 125 | 3 | MA0912.2 | HOXD3 | MA0912.2 | CTAATTAC | HOXD3 | 3.33e-46 | 1.25e-48 | 13700 | 37.5 | 16500 | 32.7 | 1.15 |
| 126 | 19 | MA0905.1 | HOXC10 | MA0905.1 | GTCRTAAAAH | HOXC10 | 4.32e-46 | 1.61e-48 | 24500 | 67.3 | 31400 | 62.5 | 1.08 |
| 127 | 3 | MA0894.1 | HESX1 | MA0894.1 | GHTAATTRRH | HESX1 | 1.53e-45 | 5.65e-48 | 22600 | 62.2 | 28800 | 57.3 | 1.09 |
| 128 | 3 | MA0706.1 | MEOX2 | MA0706.1 | DSTAATTAWN | MEOX2 | 6.1e-45 | 2.24e-47 | 31800 | 87.4 | 42200 | 83.9 | 1.04 |
| 129 | 4 | MA0143.4 | SOX2 | MA0143.4 | DNACAATGGNN | SOX2 | 1.26e-44 | 4.59e-47 | 30600 | 84 | 40400 | 80.2 | 1.05 |
| 130 | 3 | MA0642.2 | EN2 | MA0642.2 | VSYAATTA | EN2 | 3.87e-44 | 1.4e-46 | 28800 | 79.1 | 37700 | 74.9 | 1.06 |
| 131 | 3 | MA1519.1 | LHX5 | MA1519.1 | STAATTAV | LHX5 | 7.61e-44 | 2.73e-46 | 14500 | 39.8 | 17600 | 35 | 1.14 |
| 132 | 20 | MA1657.1 | ZNF652 | MA1657.1 | BAAAGRGTTAAN | ZNF652 | 8.5e-44 | 3.02e-46 | 30300 | 83.3 | 40000 | 79.4 | 1.05 |
| 133 | 21 | MA0122.3 | Nkx3-2 | MA0122.3 | WWAAMCACTTAAN | Nkx3-2 | 9.85e-44 | 3.47e-46 | 27400 | 75.2 | 35600 | 70.8 | 1.06 |
| 134 | 11 | MA0846.1 | FOXC2 | MA0846.1 | WAWGTAAACAWW | FOXC2 | 1.12e-43 | 3.93e-46 | 30200 | 82.8 | 39700 | 79 | 1.05 |
| 135 | 11 | MA1978.1 | ZNF354A | MA1978.1 | HWAADWATAAATGRAYWAWTT | ZNF354A | 1.51e-43 | 5.25e-46 | 25100 | 69 | 32400 | 64.3 | 1.07 |
| 136 | 3 | MA0711.1 | OTX1 | MA0711.1 | HTAATCCS | OTX1 | 2.14e-43 | 7.38e-46 | 31400 | 86.3 | 41600 | 82.7 | 1.04 |
| 137 | 8 | MA1115.1 | POU5F1 | MA1115.1 | WHATGCAAATN | POU5F1 | 7.01e-43 | 2.4e-45 | 31400 | 86.2 | 41600 | 82.6 | 1.04 |
| 138 | 3 | MA0903.1 | HOXB3 | MA0903.1 | NNYMATTARN | HOXB3 | 7.34e-43 | 2.5e-45 | 31900 | 87.6 | 42400 | 84.2 | 1.04 |
| 139 | 22 | MA0047.3 | FOXA2 | MA0047.3 | RWGTAAACANN | FOXA2 | 3e-42 | 1.01e-44 | 32000 | 87.8 | 42500 | 84.4 | 1.04 |
| 140 | 23 | MA0499.2 (MYOD1) | STREME-5 | 5-AGACAGG | AGACAGG | MYOD1 | 6.66e-42 | 2.23e-44 | 27900 | 76.6 | 36500 | 72.4 | 1.06 |
| 141 | 3 | MA0892.1 | GSX1 | MA0892.1 | NCYMATTAVN | GSX1 | 8.32e-42 | 2.77e-44 | 30600 | 84 | 40400 | 80.3 | 1.05 |
| 142 | 3 | MA0705.1 | Lhx8 | MA0705.1 | CTAATTAV | Lhx8 | 1.06e-41 | 3.52e-44 | 19700 | 54.2 | 24900 | 49.4 | 1.1 |
| 143 | 22 | MA0148.4 | FOXA1 | MA0148.4 | WWGTAAACATNN | FOXA1 | 1.32e-41 | 4.33e-44 | 31500 | 86.5 | 41800 | 83.1 | 1.04 |
| 144 | 4 | MA1152.1 | SOX15 | MA1152.1 | CYWTTGTTHW | SOX15 | 5e-41 | 1.63e-43 | 33500 | 91.9 | 44900 | 89.1 | 1.03 |
| 145 | 22 | MA0031.1 | FOXD1 | MA0031.1 | GTAAACAW | FOXD1 | 6.59e-41 | 2.13e-43 | 21900 | 60 | 27900 | 55.4 | 1.09 |
| 146 | 11 | MA1607.1 | Foxl2 | MA1607.1 | DDWATGTAAACANN | Foxl2 | 7.51e-41 | 2.41e-43 | 31500 | 86.5 | 41800 | 83.1 | 1.04 |
| 147 | 22 | MA1683.1 | FOXA3 | MA1683.1 | DWGTAAACANN | FOXA3 | 2.5e-40 | 7.99e-43 | 30300 | 83.2 | 40000 | 79.4 | 1.05 |
| 148 | 3 | MA1530.1 | NKX6-3 | MA1530.1 | VYCATTAWW | NKX6-3 | 4.47e-40 | 1.42e-42 | 19800 | 54.5 | 25100 | 49.8 | 1.09 |
| 149 | 10 | MA0037.4 | Gata3 | MA0037.4 | NHTCTTATCTNH | Gata3 | 7.08e-40 | 2.23e-42 | 31700 | 87.2 | 42200 | 83.8 | 1.04 |
| 150 | 4 | MA0077.1 | SOX9 | MA0077.1 | CYATTGTTY | SOX9 | 3.74e-39 | 1.17e-41 | 34900 | 95.8 | 47200 | 93.8 | 1.02 |
| 151 | 3 | MA0875.1 | BARX1 | MA0875.1 | VCMATTAV | BARX1 | 8.22e-39 | 2.55e-41 | 27900 | 76.7 | 36600 | 72.6 | 1.06 |
| 152 | 3 | MA0075.3 | PRRX2 | MA0075.3 | CTAATTAN | PRRX2 | 3.82e-38 | 1.18e-40 | 14900 | 40.9 | 18300 | 36.4 | 1.12 |
| 153 | 24 | MA0108.2 | TBP | MA0108.2 | STATAAAWRSVVVBN | TBP | 6.16e-38 | 1.89e-40 | 29100 | 79.9 | 38300 | 76.1 | 1.05 |
| 154 | 15 | MA0153.2 | HNF1B | MA0153.2 | GTTAATNATTAAY | HNF1B | 9.18e-38 | 2.8e-40 | 15200 | 41.7 | 18700 | 37.2 | 1.12 |
| 155 | 3 | MA0612.2 | EMX1 | MA0612.2 | BTAATTAS | EMX1 | 1.17e-37 | 3.55e-40 | 11600 | 31.9 | 14000 | 27.7 | 1.15 |
| 156 | 25 | MA0895.1 | HMBOX1 | MA0895.1 | MYTAGTTAMS | HMBOX1 | 1.76e-37 | 5.31e-40 | 32400 | 88.9 | 43200 | 85.8 | 1.04 |
| 157 | 3 | MA0900.2 | HOXA2 | MA0900.2 | STAATTAS | HOXA2 | 2.58e-37 | 7.71e-40 | 14000 | 38.6 | 17200 | 34.2 | 1.13 |
| 158 | 26 | MA0025.2 | NFIL3 | MA0025.2 | NRTTATGYAATNN | NFIL3 | 3.07e-37 | 9.11e-40 | 28900 | 79.2 | 38000 | 75.4 | 1.05 |
| 159 | 10 | MA0766.2 | GATA5 | MA0766.2 | SWGATAASRN | GATA5 | 4.31e-37 | 1.27e-39 | 17200 | 47.3 | 21500 | 42.8 | 1.11 |
| 160 | 3 | MA0717.1 | RAX2 | MA0717.1 | YYAATTAV | RAX2 | 6.66e-37 | 1.95e-39 | 34800 | 95.5 | 47000 | 93.4 | 1.02 |
| 161 | 3 | MA0891.1 | GSC2 | MA0891.1 | NYTAATCCBH | GSC2 | 9.95e-37 | 2.9e-39 | 33200 | 91.2 | 44500 | 88.5 | 1.03 |
| 162 | 22 | MA0481.3 | FOXP1 | MA0481.3 | NDGTAAACANH | FOXP1 | 1.22e-36 | 3.52e-39 | 28600 | 78.5 | 37600 | 74.7 | 1.05 |
| 163 | 3 | MA1463.1 | ARGFX | MA1463.1 | DCTAATTARM | ARGFX | 1.29e-36 | 3.71e-39 | 15900 | 43.6 | 19700 | 39.2 | 1.11 |
| 164 | 11 | MA0157.3 | Foxo3 | MA0157.3 | NDGTAAACARNN | Foxo3 | 1.68e-36 | 4.81e-39 | 31800 | 87.3 | 42400 | 84.2 | 1.04 |
| 165 | 13 | MA0682.2 | PITX1 | MA0682.2 | YTAATCCH | PITX1 | 2.44e-36 | 6.93e-39 | 19000 | 52 | 23900 | 47.6 | 1.09 |
| 166 | 15 | MA0046.2 | HNF1A | MA0046.2 | DRTTAATNATTAACN | HNF1A | 2.45e-36 | 6.93e-39 | 15500 | 42.5 | 19200 | 38.1 | 1.12 |
| 167 | 4 | MA1563.2 | SOX18 | MA1563.2 | AACAATDV | SOX18 | 3.7e-36 | 1.04e-38 | 27600 | 75.9 | 36200 | 72 | 1.05 |
| 168 | 27 | MA0602.1 | Arid5a | MA0602.1 | SYAATATTGVDANH | Arid5a | 4.41e-36 | 1.23e-38 | 22800 | 62.6 | 29300 | 58.2 | 1.08 |
| 169 | 3 | MA1608.1 | Isl1 | MA1608.1 | NNCCATTAGNN | Isl1 | 5.1e-36 | 1.42e-38 | 33700 | 92.7 | 45400 | 90.2 | 1.03 |
| 170 | 3 | MA0898.1 | Hmx3 | MA0898.1 | ASAAGCAATTAAWRRAT | Hmx3 | 6.69e-36 | 1.85e-38 | 21600 | 59.4 | 27700 | 55 | 1.08 |
| 171 | 12 | MA0497.1 | MEF2C | MA0497.1 | DDDCYAAAAATAGMW | MEF2C | 7.79e-36 | 2.14e-38 | 22900 | 62.9 | 29400 | 58.5 | 1.07 |
| 172 | 28 | MA0809.2 | TEAD4 | MA0809.2 | NNACATTCCARN | TEAD4 | 9.33e-36 | 2.55e-38 | 27600 | 75.9 | 36200 | 72 | 1.05 |
| 173 | 22 | MA0033.2 | FOXL1 | MA0033.2 | RTAAACA | FOXL1 | 1.15e-35 | 3.12e-38 | 29600 | 81.4 | 39200 | 77.8 | 1.05 |
| 174 | 4 | MA0868.2 | SOX8 | MA0868.2 | MGAACAATRG | SOX8 | 1.22e-35 | 3.3e-38 | 33200 | 91.2 | 44500 | 88.4 | 1.03 |
| 175 | 13 | MA0712.2 | OTX2 | MA0712.2 | NDRGGATTARNN | OTX2 | 1.48e-35 | 3.97e-38 | 26500 | 72.8 | 34600 | 68.7 | 1.06 |
| 176 | 3 | MA0662.1 | MIXL1 | MA0662.1 | NBYAATTAVN | MIXL1 | 3.59e-35 | 9.58e-38 | 34600 | 95.1 | 46800 | 93 | 1.02 |
| 177 | 4 | MA1120.1 | SOX13 | MA1120.1 | DVACAATGGNN | SOX13 | 1.29e-34 | 3.43e-37 | 30600 | 84.1 | 40700 | 80.8 | 1.04 |
| 178 | 9 | MA0909.3 | Hoxd13 | MA0909.3 | NNCAATAAAW | Hoxd13 | 2.39e-34 | 6.31e-37 | 22100 | 60.7 | 28400 | 56.4 | 1.08 |
| 179 | 29 | 7-AGAATA | STREME-7 | 7-AGAATA | AGAATA | AGAATA | 5e-34 | 1.31e-36 | 32800 | 90.1 | 44000 | 87.4 | 1.03 |
| 180 | 22 | MA0042.2 | FOXI1 | MA0042.2 | GTAAACA | FOXI1 | 5.6e-34 | 1.46e-36 | 26200 | 72 | 34300 | 68 | 1.06 |
| 181 | 30 | MA0018.4 | CREB1 | MA0018.4 | NNRTGATGTCAYN | CREB1 | 1.09e-33 | 2.83e-36 | 28100 | 77.1 | 36900 | 73.3 | 1.05 |
| 182 | 22 | MA1103.2 | FOXK2 | MA1103.2 | DDGTAAACANN | FOXK2 | 1.84e-33 | 4.75e-36 | 34000 | 93.4 | 45900 | 91.1 | 1.03 |
| 183 | 3 | MA0888.1 | EVX2 | MA0888.1 | VVTAATTAVB | EVX2 | 2.16e-33 | 5.53e-36 | 33000 | 90.5 | 44200 | 87.8 | 1.03 |
| 184 | 31 | MA1125.1 (ZNF384) | MEME-1 | THTTTTTKTWTTTTT | THTTTTTKTWTTTTT | ZNF384 | 3.33e-33 | 8.48e-36 | 22600 | 62 | 29100 | 57.8 | 1.07 |
| 185 | 4 | MA0514.2 | Sox3 | MA0514.2 | DNACAATGGNN | Sox3 | 4.04e-33 | 1.02e-35 | 32100 | 88.2 | 43000 | 85.3 | 1.03 |
| 186 | 22 | MA0848.1 | FOXO4 | MA0848.1 | GTAAACA | FOXO4 | 4.17e-33 | 1.05e-35 | 31300 | 86 | 41700 | 82.8 | 1.04 |
| 187 | 4 | MA0078.2 | Sox17 | MA0078.2 | NNAGAACAATGGNN | Sox17 | 8.89e-33 | 2.23e-35 | 30900 | 84.9 | 41100 | 81.7 | 1.04 |
| 188 | 26 | MA0102.4 | CEBPA | MA0102.4 | NDATTGCACAATHN | CEBPA | 1.04e-32 | 2.59e-35 | 30000 | 82.3 | 39700 | 79 | 1.04 |
| 189 | 3 | MA0881.1 | Dlx4 | MA0881.1 | VYAATTAN | Dlx4 | 1.06e-32 | 2.64e-35 | 35200 | 96.6 | 47800 | 94.9 | 1.02 |
| 190 | 3 | MA0914.1 | ISL2 | MA0914.1 | GCAMTTAR | ISL2 | 1.28e-32 | 3.15e-35 | 30500 | 83.7 | 40500 | 80.4 | 1.04 |
| 191 | 4 | MA1562.1 | SOX14 | MA1562.1 | CCGAACAATG | SOX14 | 1.66e-32 | 4.09e-35 | 27200 | 74.7 | 35700 | 70.9 | 1.05 |
| 192 | 3 | MA1498.2 | HOXA7 | MA1498.2 | SYMATTAV | HOXA7 | 1.84e-32 | 4.48e-35 | 34300 | 94.3 | 46400 | 92.1 | 1.02 |
| 193 | 18 | MA0523.1 | TCF7L2 | MA0523.1 | DRASATCAAAGRVA | TCF7L2 | 2.31e-32 | 5.62e-35 | 22800 | 62.6 | 29400 | 58.5 | 1.07 |
| 194 | 3 | MA0893.2 | GSX2 | MA0893.2 | SYMATTAR | GSX2 | 3.24e-32 | 7.83e-35 | 30700 | 84.4 | 40800 | 81.1 | 1.04 |
| 195 | 3 | MA0132.2 | PDX1 | MA0132.2 | VYAATTAR | PDX1 | 1.26e-31 | 3.03e-34 | 20100 | 55.2 | 25700 | 51.1 | 1.08 |
| 196 | 17 | MA0842.2 | NRL | MA0842.2 | AAWWNTGCTGACG | NRL | 3.06e-31 | 7.32e-34 | 14900 | 41 | 18600 | 36.9 | 1.11 |
| 197 | 13 | MA0467.2 | Crx | MA0467.2 | NDGGATTANN | Crx | 1.31e-30 | 3.12e-33 | 26500 | 72.7 | 34700 | 68.9 | 1.05 |
| 198 | 3 | MA0854.1 | Alx1 | MA0854.1 | CGMRYTAATTARTNMNY | Alx1 | 1.33e-30 | 3.15e-33 | 15000 | 41.3 | 18800 | 37.3 | 1.11 |
| 199 | 32 | MA0755.1 | CUX2 | MA0755.1 | TRATCGATAW | CUX2 | 1.55e-30 | 3.65e-33 | 17100 | 46.9 | 21500 | 42.8 | 1.1 |
| 200 | 32 | MA0679.2 | ONECUT1 | MA0679.2 | WAAAAATCAATAAWWN | ONECUT1 | 1.64e-30 | 3.84e-33 | 14800 | 40.6 | 18400 | 36.6 | 1.11 |
| 201 | 33 | MA0070.1 | PBX1 | MA0070.1 | HCATCAATCAAW | PBX1 | 5.35e-30 | 1.25e-32 | 15500 | 42.6 | 19400 | 38.6 | 1.1 |
| 202 | 3 | MA0618.1 | LBX1 | MA0618.1 | TTAATTAG | LBX1 | 5.94e-30 | 1.38e-32 | 19800 | 54.3 | 25300 | 50.2 | 1.08 |
| 203 | 28 | MA0090.3 | TEAD1 | MA0090.3 | NNACATTCCAGSN | TEAD1 | 1.36e-29 | 3.15e-32 | 29300 | 80.6 | 38900 | 77.2 | 1.04 |
| 204 | 26 | MA1636.1 | CEBPG | MA1636.1 | NDATGATGCAATMHH | CEBPG | 4.52e-28 | 1.04e-30 | 23100 | 63.6 | 30100 | 59.7 | 1.06 |
| 205 | 34 | MA0151.1 | Arid3a | MA0151.1 | ATYAAA | Arid3a | 4.71e-28 | 1.08e-30 | 15000 | 41.1 | 18700 | 37.2 | 1.1 |
| 206 | 1 | MA0591.1 | Bach1::Mafk | MA0591.1 | RSSATGACTCAGCAB | Bach1::Mafk | 6.68e-28 | 1.52e-30 | 2740 | 7.54 | 2820 | 5.59 | 1.35 |
| 207 | 35 | MA0483.1 | Gfi1B | MA0483.1 | AAATCWCWGCH | Gfi1B | 1.56e-27 | 3.54e-30 | 20100 | 55.3 | 25900 | 51.4 | 1.08 |
| 208 | 11 | MA0852.2 | FOXK1 | MA0852.2 | NRDGTAAACAAGNN | FOXK1 | 2.71e-27 | 6.11e-30 | 32300 | 88.8 | 43400 | 86.2 | 1.03 |
| 209 | 9 | MA0878.3 | CDX1 | MA0878.3 | GGYMATAAAA | CDX1 | 3.01e-27 | 6.77e-30 | 14300 | 39.4 | 17900 | 35.6 | 1.11 |
| 210 | 3 | MA1499.1 | HOXB4 | MA1499.1 | RTMATTAR | HOXB4 | 4.48e-27 | 1e-29 | 15800 | 43.4 | 19900 | 39.6 | 1.1 |
| 211 | 21 | MA0124.2 | Nkx3-1 | MA0124.2 | RCCACTTAA | Nkx3-1 | 5.92e-27 | 1.32e-29 | 27700 | 76.2 | 36600 | 72.8 | 1.05 |
| 212 | 26 | MA0836.2 | CEBPD | MA0836.2 | NRTTGCACAAYHN | CEBPD | 9.6e-27 | 2.13e-29 | 28300 | 77.6 | 37400 | 74.3 | 1.04 |
| 213 | 3 | MA0880.1 | Dlx3 | MA0880.1 | NYAATTAN | Dlx3 | 1.61e-26 | 3.55e-29 | 35200 | 96.8 | 48000 | 95.3 | 1.02 |
| 214 | 3 | MA0724.1 | VENTX | MA0724.1 | AMCGATTAR | VENTX | 3.14e-26 | 6.89e-29 | 12500 | 34.3 | 15400 | 30.7 | 1.12 |
| 215 | 17 | MA0117.2 | Mafb | MA0117.2 | AWWNTGCTGACD | Mafb | 4.65e-26 | 1.01e-28 | 32300 | 88.8 | 43400 | 86.2 | 1.03 |
| 216 | 3 | MA1500.1 | HOXB6 | MA1500.1 | BTAATKRC | HOXB6 | 5.6e-26 | 1.22e-28 | 35300 | 97 | 48100 | 95.5 | 1.02 |
| 217 | 13 | MA0714.1 | PITX3 | MA0714.1 | NYTAATCCC | PITX3 | 5.89e-26 | 1.27e-28 | 35300 | 96.9 | 48100 | 95.5 | 1.02 |
| 218 | 3 | MA0853.1 | Alx4 | MA0853.1 | CGCRYTAATTARYHNNY | Alx4 | 1.18e-24 | 2.54e-27 | 13900 | 38.1 | 17400 | 34.5 | 1.1 |
| 219 | 22 | MA1489.1 | FOXN3 | MA1489.1 | GTAAACAA | FOXN3 | 1.57e-24 | 3.36e-27 | 23100 | 63.4 | 30100 | 59.8 | 1.06 |
| 220 | 36 | MA1623.1 | Stat2 | MA1623.1 | RGAAACAGAAASH | Stat2 | 2.28e-24 | 4.86e-27 | 32000 | 88 | 43000 | 85.5 | 1.03 |
| 221 | 3 | MA1577.1 | TLX2 | MA1577.1 | BYAATTAR | TLX2 | 2.41e-24 | 5.11e-27 | 12100 | 33.2 | 15000 | 29.8 | 1.12 |
| 222 | 37 | MA0611.2 | Dux | MA0611.2 | NTGATTBAATCAVWNN | Dux | 6.11e-24 | 1.29e-26 | 11700 | 32.2 | 14500 | 28.8 | 1.12 |
| 223 | 18 | MA1991.1 | Hnf1A | MA1991.1 | NBCCTTTGATSTBN | Hnf1A | 2.26e-23 | 4.76e-26 | 35100 | 96.4 | 47800 | 94.9 | 1.02 |
| 224 | 22 | MA0480.2 | Foxo1 | MA0480.2 | NDGTAAACANN | Foxo1 | 2.66e-23 | 5.58e-26 | 34600 | 95 | 47000 | 93.3 | 1.02 |
| 225 | 38 | MA0606.2 | Nfat5 | MA0606.2 | NNATGGAAAAWN | Nfat5 | 4.78e-23 | 9.97e-26 | 31300 | 85.9 | 42000 | 83.3 | 1.03 |
| 226 | 22 | MA0850.1 | FOXP3 | MA0850.1 | RTAAACA | FOXP3 | 7.55e-23 | 1.57e-25 | 33900 | 93.2 | 45900 | 91.2 | 1.02 |
| 227 | 39 | MA1603.1 | Dmrt1 | MA1603.1 | GHTACAAAGTADC | Dmrt1 | 9.66e-23 | 2e-25 | 22800 | 62.7 | 29800 | 59.2 | 1.06 |
| 228 | 40 | MA0782.2 | PKNOX1 | MA0782.2 | NNTGAGTGACAGVNN | PKNOX1 | 1.17e-22 | 2.41e-25 | 16400 | 45.2 | 21000 | 41.6 | 1.09 |
| 229 | 13 | MA0719.1 | RHOXF1 | MA0719.1 | NTRATCCN | RHOXF1 | 1.41e-22 | 2.89e-25 | 30800 | 84.5 | 41200 | 81.8 | 1.03 |
| 230 | 28 | MA0808.1 | TEAD3 | MA0808.1 | RCATTCCW | TEAD3 | 4.39e-22 | 8.95e-25 | 15400 | 42.2 | 19500 | 38.8 | 1.09 |
| 231 | 32 | MA0754.2 | CUX1 | MA0754.2 | TAATCGATAM | CUX1 | 5.44e-22 | 1.1e-24 | 16700 | 45.8 | 21300 | 42.3 | 1.08 |
| 232 | 30 | MA1632.1 | ATF2 | MA1632.1 | NNATGABGTCATN | ATF2 | 1.65e-21 | 3.33e-24 | 24700 | 67.9 | 32500 | 64.6 | 1.05 |
| 233 | 41 | MA0095.3 | Yy1 | MA0095.3 | NNCAAAATGGCN | Yy1 | 1.74e-21 | 3.5e-24 | 33700 | 92.7 | 45700 | 90.8 | 1.02 |
| 234 | 3 | MA1504.1 | HOXC4 | MA1504.1 | VTMATTAN | HOXC4 | 3.73e-21 | 7.48e-24 | 17200 | 47.3 | 22100 | 43.9 | 1.08 |
| 235 | 3 | MA1507.1 | HOXD4 | MA1507.1 | VTMATTAD | HOXD4 | 4.37e-21 | 8.72e-24 | 11500 | 31.6 | 14300 | 28.4 | 1.11 |
| 236 | 13 | MA0883.1 | Dmbx1 | MA0883.1 | WDRWNMGGATTADKNNW | Dmbx1 | 6.37e-21 | 1.27e-23 | 18500 | 50.8 | 23900 | 47.4 | 1.07 |
| 237 | 42 | MA1974.1 | ZNF211 | MA1974.1 | HWYATATACCAYRYW | ZNF211 | 7.63e-21 | 1.51e-23 | 16900 | 46.3 | 21600 | 42.9 | 1.08 |
| 238 | 3 | MA0715.1 | PROP1 | MA0715.1 | TAATYWAATTA | PROP1 | 7.68e-21 | 1.51e-23 | 14200 | 39 | 18000 | 35.7 | 1.09 |
| 239 | 26 | MA0043.3 | HLF | MA0043.3 | NNRTTATGCAACHN | HLF | 1.18e-20 | 2.32e-23 | 22400 | 61.5 | 29300 | 58.2 | 1.06 |
| 240 | 3 | MA1496.1 | HOXA4 | MA1496.1 | RTMATTAV | HOXA4 | 1.48e-20 | 2.89e-23 | 15100 | 41.4 | 19200 | 38.1 | 1.09 |
| 241 | 36 | MA0050.3 | Irf1 | MA0050.3 | ANTGAAACTGAAASH | Irf1 | 2.62e-20 | 5.1e-23 | 26900 | 73.9 | 35700 | 70.8 | 1.04 |
| 242 | 3 | MA1501.1 | HOXB7 | MA1501.1 | GGTAATTAVS | HOXB7 | 6.07e-20 | 1.18e-22 | 28900 | 79.3 | 38500 | 76.5 | 1.04 |
| 243 | 26 | MA0833.2 | ATF4 | MA0833.2 | DDATGATGCAATMN | ATF4 | 8.63e-20 | 1.67e-22 | 19400 | 53.3 | 25100 | 49.9 | 1.07 |
| 244 | 43 | MA1118.1 | SIX1 | MA1118.1 | GWAACCTGAKM | SIX1 | 1.42e-19 | 2.73e-22 | 29900 | 82.2 | 40100 | 79.6 | 1.03 |
| 245 | 44 | MA0626.1 | Npas2 | MA0626.1 | NSCACGTGTN | Npas2 | 2.71e-19 | 5.2e-22 | 17300 | 47.4 | 22200 | 44.1 | 1.07 |
| 246 | 40 | MA0774.1 | MEIS2 | MA0774.1 | TTGACAGS | MEIS2 | 7.32e-19 | 1.4e-21 | 5800 | 15.9 | 6850 | 13.6 | 1.17 |
| 247 | 3 | MA0699.1 | LBX2 | MA0699.1 | NCYAATTARN | LBX2 | 1.03e-18 | 1.96e-21 | 35100 | 96.4 | 47800 | 95 | 1.01 |
| 248 | 3 | MA1960.1 | MGA::EVX1 | MA1960.1 | RGGTGWTAATKD | MGA::EVX1 | 1.23e-18 | 2.33e-21 | 21200 | 58.2 | 27700 | 54.9 | 1.06 |
| 249 | 22 | MA0614.1 | Foxj2 | MA0614.1 | RTAAACAA | Foxj2 | 2.09e-18 | 3.95e-21 | 18800 | 51.6 | 24400 | 48.4 | 1.07 |
| 250 | 30 | MA0498.2 | MEIS1 | MA0498.2 | HTGACAD | MEIS1 | 2.51e-18 | 4.7e-21 | 14800 | 40.7 | 18900 | 37.6 | 1.08 |
| 251 | 4 | MA0867.2 | SOX4 | MA0867.2 | RAACAAAGRV | SOX4 | 3.7e-18 | 6.91e-21 | 27800 | 76.4 | 37100 | 73.7 | 1.04 |
| 252 | 19 | MA0911.1 | Hoxa11 | MA0911.1 | RGTCGTAAAANT | Hoxa11 | 4.07e-18 | 7.58e-21 | 17100 | 46.8 | 22000 | 43.6 | 1.07 |
| 253 | 10 | MA0482.2 | GATA4 | MA0482.2 | NNCCTTATCTNH | GATA4 | 5.32e-18 | 9.86e-21 | 10900 | 30.1 | 13700 | 27.2 | 1.11 |
| 254 | 45 | MA0144.2 | STAT3 | MA0144.2 | YTTCYKGGAAD | STAT3 | 7.51e-18 | 1.39e-20 | 22700 | 62.5 | 29900 | 59.4 | 1.05 |
| 255 | 30 | MA1131.1 | FOSL2::JUN | MA1131.1 | GRTGACGTMAT | FOSL2::JUN | 1.43e-17 | 2.62e-20 | 24900 | 68.5 | 33000 | 65.5 | 1.05 |
| 256 | 46 | MA1479.1 | DMRTC2 | MA1479.1 | AATTGMTACATT | DMRTC2 | 1.7e-17 | 3.12e-20 | 26900 | 73.9 | 35800 | 71.1 | 1.04 |
| 257 | 11 | MA1125.1 | ZNF384 | MA1125.1 | DNWMAAAAAAAA | ZNF384 | 3.65e-17 | 6.66e-20 | 17200 | 47.2 | 22200 | 44 | 1.07 |
| 258 | 4 | MA0087.2 | Sox5 | MA0087.2 | VRVAACAATGGNN | Sox5 | 4.41e-17 | 8.02e-20 | 26400 | 72.5 | 35100 | 69.7 | 1.04 |
| 259 | 3 | MA0874.1 | Arx | MA0874.1 | VTBCRYTAATTARTKSW | Arx | 4.91e-17 | 8.9e-20 | 12400 | 34 | 15700 | 31.1 | 1.09 |
| 260 | 47 | MA1150.1 | RORB | MA1150.1 | AWWTRGGTCAH | RORB | 2.2e-16 | 3.98e-19 | 34900 | 95.8 | 47500 | 94.4 | 1.01 |
| 261 | 11 | MA0851.1 | Foxj3 | MA0851.1 | NDAADGTAAACAAANNM | Foxj3 | 2.27e-16 | 4.07e-19 | 17500 | 48 | 22600 | 44.9 | 1.07 |
| 262 | 3 | MA0887.1 | EVX1 | MA0887.1 | VVTAATTABS | EVX1 | 3.44e-16 | 6.16e-19 | 34800 | 95.6 | 47400 | 94.2 | 1.01 |
| 263 | 8 | MA1962.1 | POU2F1::SOX2 | MA1962.1 | MATTTRCATMACAATRG | POU2F1::SOX2 | 3.63e-16 | 6.47e-19 | 10400 | 28.7 | 13100 | 26 | 1.1 |
| 264 | 11 | MA0847.3 | FOXD2 | MA0847.3 | SYTAARYAAACA | FOXD2 | 3.82e-16 | 6.78e-19 | 20800 | 57.2 | 27300 | 54.2 | 1.06 |
| 265 | 48 | MA1593.1 | ZNF317 | MA1593.1 | WVACAGCAGAYW | ZNF317 | 1.04e-15 | 1.84e-18 | 32400 | 88.9 | 43800 | 86.9 | 1.02 |
| 266 | 49 | MA1523.1 | MSANTD3 | MA1523.1 | STVCACTCAC | MSANTD3 | 1.05e-15 | 1.85e-18 | 31000 | 85.1 | 41700 | 82.8 | 1.03 |
| 267 | 46 | MA1707.1 | DMRTA1 | MA1707.1 | AWTTGWTACAWT | DMRTA1 | 2.03e-15 | 3.56e-18 | 27700 | 76 | 37000 | 73.4 | 1.04 |
| 268 | 40 | MA1114.1 | PBX3 | MA1114.1 | NNNTGAGTGACAGGBNN | PBX3 | 6.06e-15 | 1.06e-17 | 21000 | 57.7 | 27600 | 54.8 | 1.05 |
| 269 | 43 | MA1119.1 | SIX2 | MA1119.1 | NNCTGAAACCTGATMY | SIX2 | 7.31e-15 | 1.27e-17 | 31800 | 87.4 | 43000 | 85.4 | 1.02 |
| 270 | 19 | MA0907.1 | HOXC13 | MA0907.1 | BCTCGTAAAAH | HOXC13 | 7.6e-15 | 1.32e-17 | 16900 | 46.5 | 21900 | 43.6 | 1.07 |
| 271 | 19 | MA0906.1 | HOXC12 | MA0906.1 | RGTCGTAAAAH | HOXC12 | 2.69e-14 | 4.64e-17 | 12800 | 35 | 16300 | 32.3 | 1.08 |
| 272 | 10 | MA0140.2 | GATA1::TAL1 | MA0140.2 | MTTATCWSNNNNNHVCAG | GATA1::TAL1 | 2.69e-14 | 4.64e-17 | 10700 | 29.4 | 13500 | 26.8 | 1.1 |
| 273 | 36 | MA0152.2 | Nfatc2 | MA0152.2 | NNRNAATGGAAANWNH | Nfatc2 | 6.76e-14 | 1.16e-16 | 33800 | 93 | 46000 | 91.4 | 1.02 |
| 274 | 50 | MA0092.1 | Hand1::Tcf3 | MA0092.1 | BRTCTGGMWT | Hand1::Tcf3 | 7.85e-14 | 1.34e-16 | 18400 | 50.5 | 24000 | 47.6 | 1.06 |
| 275 | 30 | MA1127.1 | FOSB::JUN | MA1127.1 | GATGACGTCAT | FOSB::JUN | 1.23e-13 | 2.09e-16 | 27000 | 74.2 | 36100 | 71.7 | 1.03 |
| 276 | 30 | MA0093.3 | USF1 | MA0093.3 | NDGTCATGTGACHN | USF1 | 3.22e-13 | 5.48e-16 | 21200 | 58.2 | 27900 | 55.4 | 1.05 |
| 277 | 38 | MA0624.2 | Nfatc1 | MA0624.2 | NVATGGAAAAWN | Nfatc1 | 3.29e-13 | 5.58e-16 | 33000 | 90.7 | 44800 | 89 | 1.02 |
| 278 | 28 | MA1121.1 | TEAD2 | MA1121.1 | NYACATTCCWNNS | TEAD2 | 4.64e-13 | 7.84e-16 | 24300 | 66.8 | 32300 | 64.2 | 1.04 |
| 279 | 51 | MA1524.2 | Msgn1 | MA1524.2 | VRRRACAAATGGTNNN | Msgn1 | 5.09e-13 | 8.56e-16 | 21200 | 58.3 | 28000 | 55.5 | 1.05 |
| 280 | 18 | MA0768.2 | Lef1 | MA0768.2 | NNBCCTTTGATSTN | Lef1 | 6.13e-13 | 1.03e-15 | 15800 | 43.4 | 20500 | 40.7 | 1.07 |
| 281 | 3 | MA0896.1 | Hmx1 | MA0896.1 | VSVAGCAATTAAWGVDB | Hmx1 | 2.4e-12 | 4.01e-15 | 9450 | 26 | 11900 | 23.6 | 1.1 |
| 282 | 30 | MA1112.2 | NR4A1 | MA1112.2 | NNAAAGGTCANN | NR4A1 | 2.55e-12 | 4.24e-15 | 23700 | 65 | 31400 | 62.4 | 1.04 |
| 283 | 9 | MA1645.1 | NKX2-2 | MA1645.1 | NNNCCACTCAANNN | NKX2-2 | 2.57e-12 | 4.26e-15 | 33100 | 90.9 | 45000 | 89.3 | 1.02 |
| 284 | 52 | MA1588.1 | ZNF136 | MA1588.1 | RWATTCTTGGTTGRC | ZNF136 | 8.44e-12 | 1.39e-14 | 11400 | 31.4 | 14600 | 29 | 1.08 |
| 285 | 4 | MA0442.2 | SOX10 | MA0442.2 | NDAACAAAGVN | SOX10 | 1.88e-11 | 3.09e-14 | 18300 | 50.1 | 23900 | 47.6 | 1.05 |
| 286 | 30 | MA1139.1 | FOSL2::JUNB | MA1139.1 | DATGACGTCATH | FOSL2::JUNB | 4.63e-11 | 7.6e-14 | 19700 | 54.2 | 26000 | 51.7 | 1.05 |
| 287 | 36 | MA1953.1 | FOXO1::ELF1 | MA1953.1 | RTMAACAGGAAGTN | FOXO1::ELF1 | 5.4e-11 | 8.82e-14 | 31900 | 87.6 | 43300 | 85.9 | 1.02 |
| 288 | 22 | MA0849.1 | FOXO6 | MA0849.1 | GTAAACA | FOXO6 | 6.09e-11 | 9.93e-14 | 2880 | 7.9 | 3310 | 6.58 | 1.2 |
| 289 | 21 | MA0063.2 | NKX2-5 | MA0063.2 | NNCACTCAANN | NKX2-5 | 8.29e-11 | 1.35e-13 | 35500 | 97.4 | 48600 | 96.5 | 1.01 |
| 290 | 37 | MA0884.2 | DUXA | MA0884.2 | YTRATTWAATYAR | DUXA | 8.61e-11 | 1.39e-13 | 5960 | 16.4 | 7320 | 14.6 | 1.13 |
| 291 | 53 | MA0596.1 | SREBF2 | MA0596.1 | RTGGGGTGAY | SREBF2 | 8.75e-11 | 1.41e-13 | 7110 | 19.5 | 8840 | 17.6 | 1.11 |
| 292 | 12 | MA0773.1 | MEF2D | MA0773.1 | DCTAWAAATAGM | MEF2D | 1.07e-10 | 1.71e-13 | 4610 | 12.7 | 5560 | 11 | 1.15 |
| 293 | 54 | MA1975.1 | ZNF214 | MA1975.1 | VWTCATCAABGTCCT | ZNF214 | 1.08e-10 | 1.74e-13 | 22500 | 61.7 | 29800 | 59.2 | 1.04 |
| 294 | 45 | MA1624.1 | Stat5a | MA1624.1 | NTTCCAAGAANH | Stat5a | 1.26e-10 | 2.01e-13 | 28900 | 79.4 | 39000 | 77.4 | 1.03 |
| 295 | 30 | MA1111.1 | NR2F2 | MA1111.1 | NAAAGGTCANR | NR2F2 | 1.56e-10 | 2.47e-13 | 31200 | 85.8 | 42300 | 84 | 1.02 |
| 296 | 35 | MA0038.2 | GFI1 | MA0038.2 | BMAATCACDGCA | GFI1 | 2.1e-10 | 3.33e-13 | 6610 | 18.2 | 8200 | 16.3 | 1.12 |
| 297 | 15 | MA0683.1 | POU4F2 | MA0683.1 | RTGCATAATTAATGAG | POU4F2 | 4.32e-10 | 6.83e-13 | 6170 | 17 | 7630 | 15.2 | 1.12 |
| 298 | 51 | MA0620.3 | MITF | MA0620.3 | NNNDRTCACGTGAYHNNN | MITF | 9.29e-10 | 1.46e-12 | 18500 | 50.9 | 24400 | 48.5 | 1.05 |
| 299 | 55 | MA0508.3 | PRDM1 | MA0508.3 | YNCTTTCTCTH | PRDM1 | 1.72e-09 | 2.69e-12 | 28400 | 77.9 | 38200 | 75.9 | 1.03 |
| 300 | 9 | MA1644.1 | NFYC | MA1644.1 | RDCCAATCASN | NFYC | 2.14e-09 | 3.35e-12 | 7840 | 21.5 | 9870 | 19.6 | 1.1 |
| 301 | 19 | MA0908.1 | HOXD11 | MA0908.1 | RTCGTAAAAH | HOXD11 | 2.17e-09 | 3.39e-12 | 4940 | 13.6 | 6030 | 12 | 1.13 |
| 302 | 36 | MA1709.1 | ZIM3 | MA1709.1 | RVMAACAGAAACCYM | ZIM3 | 2.48e-09 | 3.85e-12 | 19900 | 54.6 | 26300 | 52.3 | 1.05 |
| 303 | 30 | MA0488.1 | JUN | MA0488.1 | DDRATGATGTCAT | JUN | 2.75e-09 | 4.26e-12 | 6060 | 16.6 | 7510 | 14.9 | 1.12 |
| 304 | 37 | MA1720.1 | ZNF85 | MA1720.1 | DHDGAGATTACWKCAK | ZNF85 | 2.84e-09 | 4.38e-12 | 22100 | 60.8 | 29400 | 58.4 | 1.04 |
| 305 | 46 | MA1478.1 | DMRTA2 | MA1478.1 | AATTGHTACAWT | DMRTA2 | 3.47e-09 | 5.34e-12 | 32300 | 88.6 | 43800 | 87.1 | 1.02 |
| 306 | 51 | MA0698.1 | ZBTB18 | MA0698.1 | NATCCAGATGTKB | ZBTB18 | 4.5e-09 | 6.9e-12 | 12300 | 33.9 | 16000 | 31.7 | 1.07 |
| 307 | 4 | MA0515.1 | Sox6 | MA0515.1 | CCWTTGTYYY | Sox6 | 6.64e-09 | 1.01e-11 | 4270 | 11.7 | 5180 | 10.3 | 1.14 |
| 308 | 56 | MA0479.1 | FOXH1 | MA0479.1 | BNSAATCCACA | FOXH1 | 9.19e-09 | 1.4e-11 | 3880 | 10.6 | 4670 | 9.27 | 1.15 |
| 309 | 30 | MA1143.1 | FOSL1::JUND | MA1143.1 | RTGACGTMAY | FOSL1::JUND | 1.46e-08 | 2.22e-11 | 33100 | 91 | 45200 | 89.7 | 1.02 |
| 310 | 47 | MA1151.1 | RORC | MA1151.1 | NWAWNTRGGTCA | RORC | 1.61e-08 | 2.43e-11 | 33100 | 91 | 45100 | 89.6 | 1.02 |
| 311 | 32 | MA0757.1 | ONECUT3 | MA0757.1 | AAAAAATCRATAAH | ONECUT3 | 1.63e-08 | 2.46e-11 | 10200 | 28 | 13100 | 26 | 1.08 |
| 312 | 21 | MA1994.1 | Nkx2-1 | MA1994.1 | NVCACTTGARHNN | Nkx2-1 | 2.68e-08 | 4.02e-11 | 34500 | 94.8 | 47200 | 93.8 | 1.01 |
| 313 | 57 | MA0161.2 (NFIC) | STREME-4 | 4-TGCCTGGCACA | TGCCTGGCACA | NFIC | 2.84e-08 | 4.26e-11 | 267 | 0.73 | 201 | 0.4 | 1.83 |
| 314 | 43 | MA2001.1 | Six4 | MA2001.1 | GDAACCTGANH | Six4 | 3.97e-08 | 5.94e-11 | 35400 | 97.3 | 48600 | 96.5 | 1.01 |
| 315 | 30 | MA0631.1 | Six3 | MA0631.1 | DATRGGGTATCAYNWNT | Six3 | 5.63e-08 | 8.39e-11 | 14700 | 40.4 | 19200 | 38.2 | 1.06 |
| 316 | 58 | MA0691.1 | TFAP4 | MA0691.1 | AWCAGCTGWT | TFAP4 | 1.14e-07 | 1.7e-10 | 6440 | 17.7 | 8090 | 16.1 | 1.1 |
| 317 | 59 | MA1643.1 | NFIB | MA1643.1 | NNCCTGGCAYNGTGCCAAGNN | NFIB | 1.6e-07 | 2.36e-10 | 1700 | 4.66 | 1910 | 3.79 | 1.23 |
| 318 | 60 | MA1561.1 | SOX12 | MA1561.1 | ACCGAACAATV | SOX12 | 2.12e-07 | 3.12e-10 | 25900 | 71.2 | 34800 | 69.2 | 1.03 |
| 319 | 19 | MA1503.1 | HOXB9 | MA1503.1 | GTCGTAAAAH | HOXB9 | 2.19e-07 | 3.22e-10 | 2290 | 6.29 | 2660 | 5.29 | 1.19 |
| 320 | 22 | MA0040.1 | Foxq1 | MA0040.1 | HATTGTTTATW | Foxq1 | 2.29e-07 | 3.36e-10 | 6320 | 17.4 | 7940 | 15.8 | 1.1 |
| 321 | 61 | MA0060.3 | NFYA | MA0060.3 | RRCCAATCAGM | NFYA | 2.62e-07 | 3.83e-10 | 6040 | 16.6 | 7570 | 15 | 1.1 |
| 322 | 3 | MA0468.1 | DUX4 | MA0468.1 | TAAYYYAATCA | DUX4 | 3.67e-07 | 5.35e-10 | 6020 | 16.5 | 7550 | 15 | 1.1 |
| 323 | 61 | MA0502.2 | NFYB | MA0502.2 | CYCATTGGCCVV | NFYB | 4.22e-07 | 6.13e-10 | 1390 | 3.83 | 1540 | 3.06 | 1.25 |
| 324 | 51 | MA0678.1 | OLIG2 | MA0678.1 | AMCATATGKT | OLIG2 | 5.4e-07 | 7.83e-10 | 20700 | 56.8 | 27500 | 54.7 | 1.04 |
| 325 | 40 | MA0775.1 | MEIS3 | MA0775.1 | DTGACAGS | MEIS3 | 5.54e-07 | 8e-10 | 1800 | 4.94 | 2050 | 4.08 | 1.21 |
| 326 | 62 | MA1980.1 | ZNF418 | MA1980.1 | MDGAGGCTAAAAGCA | ZNF418 | 6.14e-07 | 8.84e-10 | 20000 | 54.9 | 26600 | 52.8 | 1.04 |
| 327 | 51 | MA0669.1 | NEUROG2 | MA0669.1 | RACATATGTC | NEUROG2 | 7.73e-07 | 1.11e-09 | 13200 | 36.2 | 17200 | 34.2 | 1.06 |
| 328 | 3 | MA0897.1 | Hmx2 | MA0897.1 | ASAAGCAATTAAHVVRT | Hmx2 | 7.85e-07 | 1.12e-09 | 7270 | 20 | 9240 | 18.4 | 1.09 |
| 329 | 9 | MA1473.1 | CDX4 | MA1473.1 | GGYMATAAAAC | CDX4 | 9.45e-07 | 1.35e-09 | 2960 | 8.12 | 3540 | 7.03 | 1.15 |
| 330 | 22 | MA0593.1 | FOXP2 | MA0593.1 | RWGTAAACAVR | FOXP2 | 1.48e-06 | 2.11e-09 | 8660 | 23.8 | 11100 | 22.1 | 1.08 |
| 331 | 19 | MA1506.1 | HOXD10 | MA1506.1 | RGTCGTAAAAH | HOXD10 | 1.69e-06 | 2.39e-09 | 5450 | 15 | 6820 | 13.6 | 1.1 |
| 332 | 30 | MA0693.3 | Vdr | MA0693.3 | NNGAGTTCANN | Vdr | 2.23e-06 | 3.16e-09 | 35500 | 97.6 | 48800 | 97 | 1.01 |
| 333 | 51 | MA0461.2 | Atoh1 | MA0461.2 | RMCATATGBY | Atoh1 | 2.56e-06 | 3.6e-09 | 12600 | 34.7 | 16500 | 32.8 | 1.06 |
| 334 | 28 | MA0114.4 | HNF4A | MA0114.4 | DNCAAAGTCCANN | HNF4A | 2.77e-06 | 3.89e-09 | 35100 | 96.5 | 48200 | 95.7 | 1.01 |
| 335 | 53 | MA0595.1 | SREBF1 | MA0595.1 | VTCACCCCAY | SREBF1 | 2.9e-06 | 4.07e-09 | 7500 | 20.6 | 9570 | 19 | 1.08 |
| 336 | 59 | MA0119.1 | NFIC::TLX1 | MA0119.1 | TGGCASSRWGCCAA | NFIC::TLX1 | 4.52e-06 | 6.32e-09 | 1570 | 4.32 | 1790 | 3.56 | 1.22 |
| 337 | 63 | MA1124.1 | ZNF24 | MA1124.1 | CATTCATTCATTC | ZNF24 | 7.2e-06 | 1e-08 | 3650 | 10 | 4470 | 8.88 | 1.13 |
| 338 | 28 | MA0484.2 | HNF4G | MA0484.2 | DNCAAAGTCCANN | HNF4G | 9.22e-06 | 1.28e-08 | 35100 | 96.4 | 48200 | 95.6 | 1.01 |
| 339 | 3 | MA0135.1 | Lhx3 | MA0135.1 | WAATTAATTAWYY | Lhx3 | 1.17e-05 | 1.62e-08 | 6460 | 17.7 | 8200 | 16.3 | 1.09 |
| 340 | 10 | MA0029.1 | Mecom | MA0029.1 | AAGAYAAGATAANA | Mecom | 1.38e-05 | 1.9e-08 | 4860 | 13.3 | 6080 | 12.1 | 1.11 |
| 341 | 18 | MA0869.2 | Sox11 | MA0869.2 | NRGAACAAAGVV | Sox11 | 1.4e-05 | 1.93e-08 | 16700 | 45.8 | 22100 | 43.9 | 1.04 |
| 342 | 30 | MA1145.1 | FOSL2::JUND | MA1145.1 | NNRTGACGTCAYHSN | FOSL2::JUND | 1.82e-05 | 2.5e-08 | 25200 | 69.2 | 33900 | 67.4 | 1.03 |
| 343 | 30 | MA0492.1 | JUND | MA0492.1 | DDDRATGABGTCATN | JUND | 2.65e-05 | 3.62e-08 | 6370 | 17.5 | 8100 | 16.1 | 1.09 |
| 344 | 45 | MA0518.1 | Stat4 | MA0518.1 | YTTCYRGGAARNVR | Stat4 | 3.16e-05 | 4.31e-08 | 19000 | 52.2 | 25300 | 50.3 | 1.04 |
| 345 | 64 | MA0797.1 | TGIF2 | MA0797.1 | TGACAGSTGTCA | TGIF2 | 5.35e-05 | 7.27e-08 | 3690 | 10.1 | 4560 | 9.05 | 1.12 |
| 346 | 30 | MA0829.2 | SREBF1 | MA0829.2 | DNATCACSTGATNH | SREBF1 | 6.05e-05 | 8.21e-08 | 17000 | 46.8 | 22600 | 45 | 1.04 |
| 347 | 30 | MA1133.1 | JUN::JUNB | MA1133.1 | KRTGACGTCATN | JUN::JUNB | 8.03e-05 | 1.09e-07 | 22300 | 61.2 | 29900 | 59.5 | 1.03 |
| 348 | 59 | MA1527.1 | NFIC | MA1527.1 | NTTGGCDNNRTGCCARN | NFIC | 9.65e-05 | 1.3e-07 | 1700 | 4.67 | 1990 | 3.95 | 1.18 |
| 349 | 30 | MA1136.1 | FOSB::JUNB | MA1136.1 | RTGACGTCAT | FOSB::JUNB | 0.000105 | 1.41e-07 | 23400 | 64.2 | 31400 | 62.5 | 1.03 |
| 350 | 47 | MA0494.1 | Nr1h3::Rxra | MA0494.1 | TGACCTNNAGTRACCYYDN | Nr1h3::Rxra | 0.000113 | 1.52e-07 | 25100 | 68.9 | 33800 | 67.2 | 1.02 |
| 351 | 65 | MA1989.1 | Bcl11B | MA1989.1 | NNAAACCACAARNN | Bcl11B | 0.000116 | 1.56e-07 | 27100 | 74.5 | 36700 | 72.9 | 1.02 |
| 352 | 36 | MA1508.1 | IKZF1 | MA1508.1 | VVAACAGGAARN | IKZF1 | 0.000132 | 1.76e-07 | 31300 | 86 | 42700 | 84.7 | 1.01 |
| 353 | 9 | MA0913.2 | HOXD9 | MA0913.2 | GYMATAAAAH | HOXD9 | 0.00014 | 1.87e-07 | 3060 | 8.42 | 3760 | 7.47 | 1.13 |
| 354 | 30 | MA1126.1 | FOS::JUN | MA1126.1 | DRTGACGTCATHNDTN | FOS::JUN | 0.000164 | 2.17e-07 | 33200 | 91.3 | 45400 | 90.3 | 1.01 |
| 355 | 66 | MA0106.3 | TP53 | MA0106.3 | RACATGYCCGGRCATGTY | TP53 | 0.000165 | 2.18e-07 | 278 | 0.76 | 246 | 0.49 | 1.56 |
| 356 | 67 | MA0101.1 | REL | MA0101.1 | BGGRNWTTCC | REL | 0.000174 | 2.29e-07 | 32000 | 87.9 | 43700 | 86.7 | 1.01 |
| 357 | 68 | MA0136.3 | Elf5 | MA0136.3 | RVAAGGAAGTNN | Elf5 | 0.000217 | 2.85e-07 | 35800 | 98.4 | 49300 | 97.9 | 1 |
| 358 | 30 | MA1129.1 | FOSL1::JUN | MA1129.1 | ATGACGTCAT | FOSL1::JUN | 0.000362 | 4.75e-07 | 23700 | 65.1 | 31900 | 63.4 | 1.03 |
| 359 | 3 | MA0784.2 | POU1F1 | MA0784.2 | ANCTMATTWGCATAWT | POU1F1 | 0.000631 | 8.24e-07 | 4870 | 13.4 | 6170 | 12.3 | 1.09 |
| 360 | 36 | MA1952.1 | FOXJ2::ELF1 | MA1952.1 | RTAAACMGGAAGTR | FOXJ2::ELF1 | 0.000663 | 8.63e-07 | 32400 | 88.9 | 44200 | 87.8 | 1.01 |
| 361 | 51 | MA1618.1 | Ptf1a | MA1618.1 | NNACAGATGTTNN | Ptf1a | 0.00077 | 1e-06 | 34500 | 94.8 | 47400 | 94.1 | 1.01 |
| 362 | 51 | MA0744.2 | SCRT2 | MA0744.2 | NNWGCAACAGGTGDNN | SCRT2 | 0.000789 | 1.02e-06 | 31800 | 87.4 | 43400 | 86.3 | 1.01 |
| 363 | 51 | MA0827.1 | OLIG3 | MA0827.1 | AMCATATGBY | OLIG3 | 0.000845 | 1.09e-06 | 24700 | 67.8 | 33300 | 66.2 | 1.02 |
| 364 | 32 | MA0780.1 | PAX3 | MA0780.1 | TAATYRATTA | PAX3 | 0.000922 | 1.19e-06 | 4810 | 13.2 | 6110 | 12.1 | 1.09 |
| 13 | 74 | TTGAGSYCAGGAGTT | MEME-6 | TTGAGSYCAGGAGTT | TTGAGSYCAGGAGTT | TTGAGSYCAGGAGTT | 0.0011 | 8.47e-05 | 683 | 1.88 | 773 | 1.54 | 1.22 |
| 365 | 22 | MA0030.1 | FOXF2 | MA0030.1 | BNAASGTAAACAAD | FOXF2 | 0.00119 | 1.53e-06 | 4580 | 12.6 | 5800 | 11.5 | 1.09 |
| 366 | 59 | MA1528.1 | NFIX | MA1528.1 | STTGGCRNNGTGCCARB | NFIX | 0.00142 | 1.81e-06 | 2880 | 7.91 | 3560 | 7.07 | 1.12 |
| 367 | 69 | MA1592.1 | ZNF274 | MA1592.1 | NRTRTGAGTTCTCGYN | ZNF274 | 0.00311 | 3.98e-06 | 7670 | 21.1 | 9980 | 19.8 | 1.06 |
| 368 | 70 | MA0520.1 | Stat6 | MA0520.1 | BDBTTCCWSAGAAVY | Stat6 | 0.00419 | 5.34e-06 | 10100 | 27.8 | 13300 | 26.5 | 1.05 |
| 369 | 51 | MA0826.1 | OLIG1 | MA0826.1 | AMCATATGKT | OLIG1 | 0.00524 | 6.67e-06 | 8720 | 24 | 11400 | 22.7 | 1.06 |
| 370 | 26 | MA0639.1 | DBP | MA0639.1 | NRTTACGTAAYV | DBP | 0.00547 | 6.93e-06 | 6230 | 17.1 | 8050 | 16 | 1.07 |
| 371 | 22 | MA0613.1 | FOXG1 | MA0613.1 | RTAAACAW | FOXG1 | 0.00572 | 7.23e-06 | 5560 | 15.3 | 7160 | 14.2 | 1.07 |
| 372 | 30 | MA0592.3 | ESRRA | MA0592.3 | NBTCAAGGTCAHN | ESRRA | 0.00604 | 7.62e-06 | 33000 | 90.6 | 45200 | 89.7 | 1.01 |
| 373 | 58 | MA0633.2 | Twist2 | MA0633.2 | NVCAGCTGBN | Twist2 | 0.00714 | 8.98e-06 | 5010 | 13.8 | 6420 | 12.8 | 1.08 |
| 374 | 18 | MA1421.1 | TCF7L1 | MA1421.1 | AAAGATCAAAGG | TCF7L1 | 0.00734 | 9.21e-06 | 3390 | 9.31 | 4260 | 8.47 | 1.1 |
| 375 | 14 | MA0877.3 | BARHL1 | MA0877.3 | AMCGTTTA | BARHL1 | 0.0129 | 1.61e-05 | 8070 | 22.2 | 10600 | 21 | 1.06 |
| 376 | 36 | MA0517.1 | STAT1::STAT2 | MA0517.1 | THAGTTTCRKTTTCY | STAT1::STAT2 | 0.0136 | 1.7e-05 | 17200 | 47.1 | 23000 | 45.7 | 1.03 |
| 377 | 57 | MA0161.2 | NFIC | MA0161.2 | NNCTTGGCANN | NFIC | 0.0164 | 2.04e-05 | 35200 | 96.6 | 48400 | 96.1 | 1.01 |
| 378 | 11 | MA1487.2 | FOXE1 | MA1487.2 | VYTAAAYAAACAAH | FOXE1 | 0.0174 | 2.17e-05 | 553 | 1.52 | 600 | 1.19 | 1.27 |
| 379 | 71 | MA0484.2 (HNF4G) | STREME-6 | 6-CAAAGTGCTGGG | CAAAGTGCTGGG | HNF4G | 0.0182 | 2.25e-05 | 301 | 0.83 | 297 | 0.59 | 1.4 |
| 380 | 30 | MA1996.1 | Nr1H2 | MA1996.1 | NNRAGGTCANN | Nr1H2 | 0.0195 | 2.41e-05 | 34900 | 96 | 48000 | 95.4 | 1.01 |
| 381 | 30 | MA0629.1 | Rhox11 | MA0629.1 | ANKNCGCTGTWAWDVRW | Rhox11 | 0.0204 | 2.51e-05 | 14800 | 40.7 | 19800 | 39.4 | 1.03 |
| 382 | 72 | MA0682.2 (PITX1) | MEME-4 | WGCTGGGATTACAGG | WGCTGGGATTACAGG | PITX1 | 0.023 | 2.83e-05 | 900 | 2.47 | 1040 | 2.06 | 1.2 |
| 383 | 58 | MA0667.1 | MYF6 | MA0667.1 | AACARCTGTT | MYF6 | 0.0357 | 4.37e-05 | 5340 | 14.7 | 6910 | 13.7 | 1.07 |
| 384 | 19 | MA0485.2 | HOXC9 | MA0485.2 | GTCGTAAAAH | HOXC9 | 0.0361 | 4.41e-05 | 3250 | 8.94 | 4120 | 8.18 | 1.09 |
| 385 | 45 | MA1625.1 | Stat5b | MA1625.1 | NNNTTCCCAGAANNN | Stat5b | 0.0364 | 4.44e-05 | 20200 | 55.6 | 27300 | 54.3 | 1.02 |
| 386 | 66 | MA0525.2 | TP63 | MA0525.2 | RACATGTYGKGACATGTC | TP63 | 0.0376 | 4.57e-05 | 275 | 0.76 | 271 | 0.54 | 1.4 |
| 387 | 73 | MA0069.1 | PAX6 | MA0069.1 | TTCACGCWTSANTK | PAX6 | 0.0385 | 4.66e-05 | 27900 | 76.6 | 38000 | 75.4 | 1.02 |
| 14 | 75 | MA0090.3 (TEAD1) | MEME-5 | CTGCACTCCAGCCTG | CTGCACTCCAGCCTG | TEAD1 | 0.105 | 0.00752 | 3540 | 9.72 | 4640 | 9.21 | 1.05 |
| 15 | 76 | MA1596.1 (ZNF460) | MEME-7 | RGAGGCWGAGGSAGG | RGAGGCWGAGGSAGG | ZNF460 | 0.329 | 0.0219 | 74 | 0.2 | 72 | 0.14 | 1.42 |
| 16 | 76 | MA1653.1 (ZNF148) | MEME-3 | CCCDSCCCCKCCYCC | CCCDSCCCCKCCYCC | ZNF148 | 5.87 | 0.367 | 142 | 0.39 | 188 | 0.37 | 1.05 |
ER_rat <- 1.25
EPI_24_20kb_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,TP.)%>%
arrange(.,EVALUE.y) %>%
slice_head(n=80)%>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste("EPI_24_20kb_combo response peaks(200bp)Enrichment ratio:",ER_rat))
EPI_24_20kb_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue = -log10(if_else(EVALUE.y == 0, 1e-300, EVALUE.y))) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
slice_head(n=5) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*9), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./9,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste("EPI_24_20kb_combo response peaks(200bp)Enrichment ratio:",ER_rat)) ### DNR_DAR_3hr data
### DNR_DAR_3hr data
DNR_DAR_3hr_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DNR_3_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
DNR_DAR_3hr_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DNR_3_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
DNR_DAR_3hr_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DNR_3_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
DNR_DAR_3hr_all <- rbind(DNR_DAR_3hr_sea_disc, DNR_DAR_3hr_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)DNR_DAR_3hr_combo <- DNR_DAR_3hr_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., DNR_DAR_3hr_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ NA_character_
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y) %>%
mutate(mrc_type="DNR_DAR_3hr")
DNR_DAR_3hr_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs in 3 hour DNR-DAR v 3 hour non-DNR DAR") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO | mrc_type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | MA0079.5 (SP1) | MEME-1 | CSCSSSCSCCGSCSC | CSCSSSCSCCGSCSC | SP1 | 0 | 0 | 9080 | 44.4 | 30800 | 25.8 | 1.72 | DNR_DAR_3hr |
| 2 | 1 | MA0162.4 (EGR1) | STREME-1 | 1-CCGCCCCCGCCCCS | CCGCCCCCGCCCCS | EGR1 | 0 | 0 | 6270 | 30.6 | 17900 | 14.9 | 2.05 | DNR_DAR_3hr |
| 3 | 2 | MA1513.1 | KLF15 | MA1513.1 | SCCCCGCCCCS | KLF15 | 0 | 0 | 7060 | 34.5 | 22400 | 18.8 | 1.84 | DNR_DAR_3hr |
| 4 | 2 | MA1713.1 | ZNF610 | MA1713.1 | SSCGCCGCTCCSSS | ZNF610 | 0 | 0 | 6710 | 32.8 | 21000 | 17.6 | 1.87 | DNR_DAR_3hr |
| 5 | 2 | MA1653.1 | ZNF148 | MA1653.1 | YBCCCCTCCCCC | ZNF148 | 0 | 0 | 8030 | 39.2 | 27400 | 22.9 | 1.71 | DNR_DAR_3hr |
| 6 | 2 | MA1961.1 | PATZ1 | MA1961.1 | SSGGGGMGGGGS | PATZ1 | 0 | 0 | 10400 | 50.7 | 39700 | 33.2 | 1.53 | DNR_DAR_3hr |
| 7 | 3 | MA0162.4 | EGR1 | MA0162.4 | VCMCGCCCACGCVS | EGR1 | 0 | 0 | 7890 | 38.5 | 27000 | 22.6 | 1.71 | DNR_DAR_3hr |
| 8 | 3 | MA1650.1 | ZBTB14 | MA1650.1 | SSCCGCGCACVS | ZBTB14 | 0 | 0 | 6870 | 33.6 | 22300 | 18.6 | 1.8 | DNR_DAR_3hr |
| 9 | 2 | MA0079.5 | SP1 | MA0079.5 | GGGGCGGGG | SP1 | 0 | 0 | 8930 | 43.6 | 32700 | 27.4 | 1.6 | DNR_DAR_3hr |
| 10 | 4 | 2-CSCGSG | STREME-2 | 2-CSCGSG | CSCGSG | CSCGSG | 0 | 0 | 10500 | 51.5 | 41800 | 34.9 | 1.47 | DNR_DAR_3hr |
| 11 | 2 | MA1522.1 | MAZ | MA1522.1 | BBCCCCTCCCY | MAZ | 0 | 0 | 7460 | 36.5 | 25800 | 21.6 | 1.69 | DNR_DAR_3hr |
| 12 | 5 | MA1713.1 (ZNF610) | STREME-4 | 4-CGCTCCCC | CGCTCCCC | ZNF610 | 0 | 0 | 11000 | 53.5 | 44100 | 36.9 | 1.45 | DNR_DAR_3hr |
| 13 | 2 | MA1959.1 | KLF7 | MA1959.1 | DGGGCGGGG | KLF7 | 0 | 0 | 12200 | 59.8 | 51700 | 43.3 | 1.38 | DNR_DAR_3hr |
| 14 | 2 | MA1564.1 | SP9 | MA1564.1 | RCCACGCCCMCY | SP9 | 0 | 0 | 8980 | 43.9 | 33900 | 28.4 | 1.54 | DNR_DAR_3hr |
| 15 | 2 | MA1515.1 | KLF2 | MA1515.1 | NRCCACRCCCH | KLF2 | 0 | 0 | 11400 | 55.8 | 47300 | 39.6 | 1.41 | DNR_DAR_3hr |
| 16 | 6 | 6-CCCCSA | STREME-6 | 6-CCCCSA | CCCCSA | CCCCSA | 0 | 0 | 9640 | 47.1 | 37500 | 31.4 | 1.5 | DNR_DAR_3hr |
| 17 | 2 | MA0742.2 | KLF12 | MA0742.2 | GGGGCGGGG | KLF12 | 0 | 0 | 11500 | 56.4 | 48100 | 40.3 | 1.4 | DNR_DAR_3hr |
| 18 | 2 | MA0685.2 | SP4 | MA0685.2 | GGGGCGGGG | SP4 | 0 | 0 | 8640 | 42.2 | 32400 | 27.1 | 1.56 | DNR_DAR_3hr |
| 19 | 7 | MA0506.2 | Nrf1 | MA0506.2 | SNCTGCGCMTGCGCV | Nrf1 | 0 | 0 | 6330 | 30.9 | 21200 | 17.7 | 1.75 | DNR_DAR_3hr |
| 20 | 8 | MA1712.1 | ZNF454 | MA1712.1 | DGGCTCSSGGCCCYGVKG | ZNF454 | 0 | 0 | 8800 | 43 | 33500 | 28 | 1.53 | DNR_DAR_3hr |
| 21 | 9 | 14-CKSSMG | STREME-14 | 14-CKSSMG | 14-CKSSMG | CKSSMG | 0 | 0 | 7020 | 34.3 | 24600 | 20.6 | 1.67 | DNR_DAR_3hr |
| 22 | 3 | MA1627.1 | Wt1 | MA1627.1 | YBCCTCCCCCACVB | Wt1 | 0 | 0 | 10200 | 50 | 41400 | 34.6 | 1.44 | DNR_DAR_3hr |
| 23 | 10 | 3-RGCCGS | STREME-3 | 3-RGCCGS | RGCCGS | RGCCGS | 0 | 0 | 7730 | 37.8 | 28200 | 23.6 | 1.6 | DNR_DAR_3hr |
| 24 | 2 | MA1511.2 | KLF10 | MA1511.2 | GGGGCGGGG | KLF10 | 0 | 0 | 11700 | 57.4 | 49900 | 41.8 | 1.37 | DNR_DAR_3hr |
| 25 | 2 | MA0039.4 | KLF4 | MA0039.4 | NNCCCCACCCHN | KLF4 | 0 | 0 | 11000 | 53.7 | 45700 | 38.2 | 1.41 | DNR_DAR_3hr |
| 26 | 3 | MA0733.1 | EGR4 | MA0733.1 | NHMCGCCCACGCAHWT | EGR4 | 0 | 0 | 8980 | 43.9 | 34800 | 29.1 | 1.51 | DNR_DAR_3hr |
| 27 | 2 | MA0740.2 | KLF14 | MA0740.2 | KGGGCGGGG | KLF14 | 0 | 0 | 12000 | 58.7 | 51800 | 43.3 | 1.35 | DNR_DAR_3hr |
| 28 | 11 | MA1122.1 | TFDP1 | MA1122.1 | VSGCGGGAAVN | TFDP1 | 0 | 0 | 8520 | 41.6 | 32600 | 27.2 | 1.53 | DNR_DAR_3hr |
| 29 | 2 | MA0516.3 | SP2 | MA0516.3 | GGGGCGGGG | SP2 | 0 | 0 | 11700 | 56.9 | 49900 | 41.8 | 1.36 | DNR_DAR_3hr |
| 30 | 2 | MA0753.2 | ZNF740 | MA0753.2 | CYRCCCCCCCCAC | ZNF740 | 0 | 0 | 7760 | 37.9 | 28800 | 24.1 | 1.57 | DNR_DAR_3hr |
| 31 | 2 | MA0493.2 | KLF1 | MA0493.2 | DGGGYGGGG | KLF1 | 0 | 0 | 11800 | 57.7 | 50900 | 42.6 | 1.36 | DNR_DAR_3hr |
| 32 | 2 | MA0471.2 | E2F6 | MA0471.2 | VVDGGCGGGAAVV | E2F6 | 0 | 0 | 7740 | 37.8 | 28700 | 24 | 1.57 | DNR_DAR_3hr |
| 33 | 12 | MA1596.1 (ZNF460) | STREME-5 | 5-GCGGAGG | GCGGAGG | ZNF460 | 0 | 0 | 7720 | 37.7 | 28700 | 24 | 1.57 | DNR_DAR_3hr |
| 34 | 13 | MA1721.1 | ZNF93 | MA1721.1 | GGYAGCRGCAGCGGYG | ZNF93 | 0 | 0 | 8120 | 39.7 | 30900 | 25.9 | 1.53 | DNR_DAR_3hr |
| 35 | 14 | MA1596.1 | ZNF460 | MA1596.1 | GCCTCMGCCTCCCRAG | ZNF460 | 0 | 0 | 9180 | 44.9 | 36500 | 30.6 | 1.47 | DNR_DAR_3hr |
| 36 | 3 | MA0732.1 | EGR3 | MA0732.1 | HHMCGCCCACGCAHH | EGR3 | 0 | 0 | 6130 | 30 | 21100 | 17.6 | 1.7 | DNR_DAR_3hr |
| 37 | 2 | MA1630.2 | ZNF281 | MA1630.2 | GGGGGAGGGGVV | ZNF281 | 0 | 0 | 9040 | 44.2 | 35800 | 30 | 1.47 | DNR_DAR_3hr |
| 38 | 2 | MA1965.1 | SP5 | MA1965.1 | CBCCTCCCHS | SP5 | 0 | 0 | 10200 | 50.1 | 42400 | 35.5 | 1.41 | DNR_DAR_3hr |
| 39 | 15 | MA0814.2 | TFAP2C | MA0814.2 | NNGCCTGAGGCGNN | TFAP2C | 0 | 0 | 9510 | 46.5 | 38500 | 32.2 | 1.44 | DNR_DAR_3hr |
| 40 | 79 | MA1107.2 | KLF9 | MA1107.2 | MNGCCACACCCACHYV | KLF9 | 0 | 0 | 10900 | 53.5 | 46400 | 38.8 | 1.38 | DNR_DAR_3hr |
| 41 | 2 | MA1517.1 | KLF6 | MA1517.1 | NRCCACGCCCH | KLF6 | 0 | 0 | 8960 | 43.8 | 35500 | 29.7 | 1.47 | DNR_DAR_3hr |
| 42 | 16 | MA0810.1 | TFAP2A | MA0810.1 | YGCCCBVRGGCR | TFAP2A | 0 | 0 | 7290 | 35.6 | 26900 | 22.5 | 1.58 | DNR_DAR_3hr |
| 43 | 2 | MA0741.1 | KLF16 | MA0741.1 | GMCACGCCCCC | KLF16 | 0 | 0 | 10200 | 49.7 | 42100 | 35.2 | 1.41 | DNR_DAR_3hr |
| 44 | 17 | MA1976.1 | ZNF320 | MA1976.1 | DGKGGGRCCMRGGGGCCWGTGHVNNNN | ZNF320 | 0 | 0 | 7780 | 38 | 29400 | 24.6 | 1.55 | DNR_DAR_3hr |
| 45 | 18 | MA0506.2 (Nrf1) | STREME-16 | 16-CWGCGS | 16-CWGCGS | Nrf1 | 0 | 0 | 8090 | 39.5 | 31100 | 26 | 1.52 | DNR_DAR_3hr |
| 46 | 3 | MA0472.2 | EGR2 | MA0472.2 | MCGCCCACGCA | EGR2 | 2.9e-308 | 3.90999999999998e-310 | 8920 | 43.6 | 35900 | 30 | 1.45 | DNR_DAR_3hr |
| 47 | 19 | MA1986.1 | ZNF692 | MA1986.1 | BGGGCCCASN | ZNF692 | 3.05e-304 | 4.03e-306 | 8260 | 40.4 | 32500 | 27.2 | 1.49 | DNR_DAR_3hr |
| 48 | 2 | MA1512.1 | KLF11 | MA1512.1 | SCCACGCCCMC | KLF11 | 1.46e-302 | 1.89e-304 | 9480 | 46.3 | 39000 | 32.6 | 1.42 | DNR_DAR_3hr |
| 49 | 2 | MA0746.2 | SP3 | MA0746.2 | NGCCACGCCCMCH | SP3 | 2.76e-297 | 3.49e-299 | 8300 | 40.5 | 32800 | 27.5 | 1.48 | DNR_DAR_3hr |
| 50 | 20 | MA1981.1 | ZNF530 | MA1981.1 | GMARGGMRAGGGGCRGSV | ZNF530 | 3.47e-296 | 4.31e-298 | 10400 | 50.6 | 44000 | 36.8 | 1.37 | DNR_DAR_3hr |
| 51 | 21 | MA0131.2 | HINFP | MA0131.2 | CARCGTCCGCKN | HINFP | 9.8e-292 | 1.19e-293 | 10400 | 51 | 44600 | 37.3 | 1.37 | DNR_DAR_3hr |
| 52 | 22 | MA1102.2 | CTCFL | MA1102.2 | NSCAGGGGGCGS | CTCFL | 7.74e-291 | 9.23999999999999e-293 | 10300 | 50.2 | 43700 | 36.5 | 1.37 | DNR_DAR_3hr |
| 53 | 16 | MA0811.1 | TFAP2B | MA0811.1 | YGCCCBVRGGCA | TFAP2B | 3.65e-284 | 4.28e-286 | 9870 | 48.2 | 41600 | 34.8 | 1.38 | DNR_DAR_3hr |
| 54 | 23 | MA0865.2 | E2F8 | MA0865.2 | TTCCCGCCAHWA | E2F8 | 7.26e-282 | 8.35e-284 | 6420 | 31.4 | 23600 | 19.7 | 1.59 | DNR_DAR_3hr |
| 55 | 16 | MA0524.2 | TFAP2C | MA0524.2 | YGCCYBVRGGCA | TFAP2C | 4.7e-280 | 5.3e-282 | 9240 | 45.2 | 38300 | 32.1 | 1.41 | DNR_DAR_3hr |
| 56 | 2 | MA1516.1 | KLF3 | MA1516.1 | GRCCRCGCCCH | KLF3 | 3.83e-278 | 4.25e-280 | 10000 | 48.9 | 42600 | 35.6 | 1.37 | DNR_DAR_3hr |
| 57 | 2 | MA0747.1 | SP8 | MA0747.1 | RCCACGCCCMCY | SP8 | 1.42e-274 | 1.55e-276 | 9730 | 47.6 | 41100 | 34.4 | 1.38 | DNR_DAR_3hr |
| 58 | 24 | MA1548.1 | PLAGL2 | MA1548.1 | NGGGCCCCCN | PLAGL2 | 2.09e-271 | 2.24e-273 | 8820 | 43.1 | 36300 | 30.4 | 1.42 | DNR_DAR_3hr |
| 59 | 25 | MA1514.1 | KLF17 | MA1514.1 | MMCCACGCACCCMTY | KLF17 | 1.15e-261 | 1.21e-263 | 11400 | 55.6 | 50800 | 42.5 | 1.31 | DNR_DAR_3hr |
| 60 | 26 | MA0146.2 | Zfx | MA0146.2 | SSSGCCBVGGCCTS | Zfx | 5.6e-259 | 5.8e-261 | 8420 | 41.2 | 34500 | 28.8 | 1.43 | DNR_DAR_3hr |
| 61 | 27 | MA1615.1 | Plagl1 | MA1615.1 | NNCTGGGGCCABN | Plagl1 | 1.71e-257 | 1.74e-259 | 9980 | 48.8 | 43000 | 36 | 1.36 | DNR_DAR_3hr |
| 62 | 23 | MA0024.3 | E2F1 | MA0024.3 | TTTGGCGCCAAA | E2F1 | 4.32e-254 | 4.32e-256 | 6630 | 32.4 | 25300 | 21.2 | 1.53 | DNR_DAR_3hr |
| 63 | 28 | MA1599.1 | ZNF682 | MA1599.1 | BVGGCYAAGCCCCWNY | ZNF682 | 1.6e-242 | 1.58e-244 | 10100 | 49.6 | 44400 | 37.1 | 1.34 | DNR_DAR_3hr |
| 64 | 29 | MA1712.1 (ZNF454) | STREME-10 | 10-CCAGGGCC | 10-CCAGGGCC | ZNF454 | 2e-237 | 1.94e-239 | 9450 | 46.2 | 40600 | 34 | 1.36 | DNR_DAR_3hr |
| 65 | 23 | MA0527.1 | ZBTB33 | MA0527.1 | BTCTCGCGAGABYTS | ZBTB33 | 1.53e-235 | 1.46e-237 | 5910 | 28.9 | 22100 | 18.5 | 1.56 | DNR_DAR_3hr |
| 66 | 15 | MA0812.1 | TFAP2B | MA0812.1 | HGCCTSAGGCD | TFAP2B | 1.03e-232 | 9.70999999999999e-235 | 7700 | 37.6 | 31300 | 26.2 | 1.43 | DNR_DAR_3hr |
| 67 | 30 | MA1578.1 | VEZF1 | MA1578.1 | CCCCCCMYDH | VEZF1 | 7.94e-229 | 7.36e-231 | 6660 | 32.6 | 26100 | 21.8 | 1.49 | DNR_DAR_3hr |
| 68 | 31 | MA0597.2 | THAP1 | MA0597.2 | VSGCAGGGCASV | THAP1 | 1.22e-225 | 1.11e-227 | 11200 | 54.9 | 51100 | 42.7 | 1.28 | DNR_DAR_3hr |
| 69 | 15 | MA1569.1 | TFAP2E | MA1569.1 | YGCCYSRGGCD | TFAP2E | 1.06e-224 | 9.5e-227 | 4640 | 22.7 | 16200 | 13.6 | 1.67 | DNR_DAR_3hr |
| 70 | 11 | MA1583.1 | ZFP57 | MA1583.1 | NNVTTGCCGCANN | ZFP57 | 2.73e-224 | 2.42e-226 | 4700 | 23 | 16500 | 13.8 | 1.66 | DNR_DAR_3hr |
| 71 | 32 | MA1517.1 (KLF6) | STREME-11 | 11-CACSCMC | 11-CACSCMC | KLF6 | 7.63e-223 | 6.67e-225 | 13800 | 67.4 | 66500 | 55.6 | 1.21 | DNR_DAR_3hr |
| 72 | 33 | MA0734.3 | Gli2 | MA0734.3 | NRGACCACCCASV | Gli2 | 7.4e-218 | 6.38e-220 | 10600 | 51.7 | 47600 | 39.8 | 1.3 | DNR_DAR_3hr |
| 73 | 17 | MA1723.1 | PRDM9 | MA1723.1 | RGDGGGVAGGGRGGVRRMARVARR | PRDM9 | 2.12e-215 | 1.8e-217 | 10400 | 50.8 | 46500 | 38.9 | 1.3 | DNR_DAR_3hr |
| 74 | 34 | MA0823.1 | HEY1 | MA0823.1 | GRCACGTGCC | HEY1 | 3.04e-214 | 2.55e-216 | 10400 | 50.8 | 46600 | 39 | 1.3 | DNR_DAR_3hr |
| 75 | 2 | MA0528.2 | ZNF263 | MA0528.2 | VNGGGGAGGASG | ZNF263 | 3.28e-213 | 2.72e-215 | 6510 | 31.8 | 25700 | 21.5 | 1.48 | DNR_DAR_3hr |
| 76 | 35 | MA1727.1 | ZNF417 | MA1727.1 | VRBVNTGGGCGCCAM | ZNF417 | 3e-212 | 2.45e-214 | 6980 | 34.1 | 28100 | 23.5 | 1.45 | DNR_DAR_3hr |
| 77 | 23 | MA0748.2 | YY2 | MA0748.2 | NVATGGCGGCS | YY2 | 1.61e-210 | 1.3e-212 | 8360 | 40.8 | 35500 | 29.7 | 1.38 | DNR_DAR_3hr |
| 78 | 36 | MA0751.1 | ZIC4 | MA0751.1 | GRCCCCCCGCKGYGH | ZIC4 | 9.18e-210 | 7.31e-212 | 11000 | 53.8 | 50300 | 42.1 | 1.28 | DNR_DAR_3hr |
| 79 | 34 | MA0632.2 | TCFL5 | MA0632.2 | KCACGCGCMC | TCFL5 | 4.96e-206 | 3.9e-208 | 5060 | 24.7 | 18700 | 15.6 | 1.58 | DNR_DAR_3hr |
| 80 | 15 | MA0003.4 | TFAP2A | MA0003.4 | NNKGCCTCAGGCNN | TFAP2A | 6.52e-205 | 5.06e-207 | 12000 | 58.8 | 56400 | 47.2 | 1.25 | DNR_DAR_3hr |
| 81 | 16 | MA0872.1 | TFAP2A | MA0872.1 | TGCCCYSRGGGCA | TFAP2A | 7.14e-202 | 5.47e-204 | 6000 | 29.3 | 23400 | 19.6 | 1.5 | DNR_DAR_3hr |
| 82 | 37 | MA0759.2 | ELK3 | MA0759.2 | ACCGGAAGTRV | ELK3 | 2.9e-199 | 2.2e-201 | 6350 | 31 | 25300 | 21.1 | 1.47 | DNR_DAR_3hr |
| 83 | 16 | MA0815.1 | TFAP2C | MA0815.1 | TGCCCYSRGGGCA | TFAP2C | 7.93e-196 | 5.93e-198 | 5060 | 24.7 | 18900 | 15.8 | 1.57 | DNR_DAR_3hr |
| 84 | 30 | MA0736.1 | GLIS2 | MA0736.1 | GACCCCCCGCRAMG | GLIS2 | 9.25e-195 | 6.84e-197 | 6740 | 32.9 | 27400 | 22.9 | 1.44 | DNR_DAR_3hr |
| 85 | 38 | MA1985.1 | ZNF669 | MA1985.1 | GGGGYGAYGACCRCT | ZNF669 | 4.62e-193 | 3.38e-195 | 8120 | 39.7 | 34700 | 29.1 | 1.37 | DNR_DAR_3hr |
| 86 | 16 | MA0813.1 | TFAP2B | MA0813.1 | TGCCCYBRGGGCA | TFAP2B | 6.53e-192 | 4.71e-194 | 5490 | 26.8 | 21100 | 17.7 | 1.52 | DNR_DAR_3hr |
| 87 | 34 | MA0821.2 | HES5 | MA0821.2 | GRCACGTGYC | HES5 | 2.82e-191 | 2.01e-193 | 6180 | 30.2 | 24600 | 20.6 | 1.47 | DNR_DAR_3hr |
| 88 | 2 | MA0599.1 | KLF5 | MA0599.1 | GCCCCDCCCH | KLF5 | 1.05e-189 | 7.38e-192 | 6230 | 30.4 | 24900 | 20.8 | 1.46 | DNR_DAR_3hr |
| 89 | 39 | MA0014.3 | PAX5 | MA0014.3 | RRGCGTGACCNN | PAX5 | 2.42e-187 | 1.69e-189 | 9120 | 44.6 | 40400 | 33.8 | 1.32 | DNR_DAR_3hr |
| 90 | 34 | MA1631.1 | ASCL1 | MA1631.1 | NNGCACCTGCYNB | ASCL1 | 1.58e-183 | 1.09e-185 | 13000 | 63.7 | 63100 | 52.8 | 1.21 | DNR_DAR_3hr |
| 91 | 40 | MA0615.1 | Gmeb1 | MA0615.1 | BHBBKKACGTMMNWNNN | Gmeb1 | 7.13e-180 | 4.86e-182 | 6890 | 33.7 | 28600 | 24 | 1.41 | DNR_DAR_3hr |
| 92 | 36 | MA1584.1 | ZIC5 | MA1584.1 | VGACCCCCCGCTGHGM | ZIC5 | 2.48e-177 | 1.67e-179 | 4910 | 24 | 18600 | 15.6 | 1.54 | DNR_DAR_3hr |
| 93 | 34 | MA0522.3 | TCF3 | MA0522.3 | NVCACCTGCNN | TCF3 | 6.92e-175 | 4.62e-177 | 14800 | 72.3 | 74300 | 62.2 | 1.16 | DNR_DAR_3hr |
| 94 | 41 | MA1966.1 | TFAP4::ETV1 | MA1966.1 | RSCGGAAGCAGSTGKN | TFAP4::ETV1 | 1.86e-174 | 1.23e-176 | 4870 | 23.8 | 18500 | 15.5 | 1.54 | DNR_DAR_3hr |
| 95 | 23 | MA0470.2 | E2F4 | MA0470.2 | TTTTGGCGCCAWWW | E2F4 | 1.39e-173 | 9.06e-176 | 5340 | 26.1 | 20800 | 17.4 | 1.5 | DNR_DAR_3hr |
| 96 | 37 | MA0750.2 | ZBTB7A | MA0750.2 | NVCCGGAAGTGSV | ZBTB7A | 1.71e-173 | 1.11e-175 | 4850 | 23.7 | 18400 | 15.4 | 1.54 | DNR_DAR_3hr |
| 97 | 23 | MA0864.2 | E2F2 | MA0864.2 | RWTTTGGCGCCAWWWY | E2F2 | 4.97e-167 | 3.18e-169 | 4240 | 20.7 | 15600 | 13 | 1.59 | DNR_DAR_3hr |
| 98 | 36 | MA0696.1 | ZIC1 | MA0696.1 | GACCCCCYGCTGTG | ZIC1 | 3.33e-163 | 2.11e-165 | 6010 | 29.4 | 24500 | 20.5 | 1.43 | DNR_DAR_3hr |
| 99 | 30 | MA1987.1 | ZNF701 | MA1987.1 | GAGCACYAARGGGGGRADDDD | ZNF701 | 5.94e-163 | 3.73e-165 | 11900 | 58.4 | 57400 | 48 | 1.22 | DNR_DAR_3hr |
| 100 | 33 | MA1990.1 | Gli1 | MA1990.1 | NNRGACCACCCASV | Gli1 | 2.31e-162 | 1.43e-164 | 14200 | 69.5 | 71200 | 59.5 | 1.17 | DNR_DAR_3hr |
| 101 | 41 | MA1997.1 | Olig2 | MA1997.1 | NVCAGCTGBN | Olig2 | 3.29e-162 | 2.02e-164 | 9250 | 45.2 | 42000 | 35.1 | 1.29 | DNR_DAR_3hr |
| 102 | 42 | MA1719.1 | ZNF816 | MA1719.1 | DRRKGGGGACATGCHGGG | ZNF816 | 4.02e-161 | 2.44e-163 | 14100 | 68.7 | 70200 | 58.7 | 1.17 | DNR_DAR_3hr |
| 103 | 34 | MA1106.1 | HIF1A | MA1106.1 | SBACGTGCNN | HIF1A | 1.3e-155 | 7.82e-158 | 7960 | 38.9 | 35100 | 29.4 | 1.32 | DNR_DAR_3hr |
| 104 | 41 | MA1941.1 | ETV2::FIGLA | MA1941.1 | RSCGGAARCAGSTGBN | ETV2::FIGLA | 8.4e-155 | 5.02e-157 | 6580 | 32.1 | 27800 | 23.2 | 1.38 | DNR_DAR_3hr |
| 105 | 34 | MA1648.1 | TCF12 | MA1648.1 | NNCACCTGCNN | TCF12 | 1.46e-154 | 8.65e-157 | 16600 | 81.3 | 86800 | 72.6 | 1.12 | DNR_DAR_3hr |
| 106 | 33 | MA0694.1 | ZBTB7B | MA0694.1 | RCGACCACCGAA | ZBTB7B | 1.36e-153 | 7.96e-156 | 9970 | 48.7 | 46300 | 38.8 | 1.26 | DNR_DAR_3hr |
| 107 | 43 | MA0146.2 (Zfx) | STREME-9 | 9-CCCAGGCC | CCCAGGCC | Zfx | 2.87e-153 | 1.67e-155 | 6900 | 33.7 | 29500 | 24.7 | 1.37 | DNR_DAR_3hr |
| 108 | 34 | MA1945.1 | ETV5::FIGLA | MA1945.1 | RGCGGAARCAGGTGG | ETV5::FIGLA | 8.6e-152 | 4.95e-154 | 10000 | 49.1 | 46800 | 39.2 | 1.25 | DNR_DAR_3hr |
| 109 | 34 | MA0830.2 | TCF4 | MA0830.2 | NNGCACCTGCCNN | TCF4 | 1.59e-151 | 9.03e-154 | 16300 | 79.7 | 84800 | 70.9 | 1.12 | DNR_DAR_3hr |
| 110 | 34 | MA0147.3 | MYC | MA0147.3 | NNCCACGTGCNB | MYC | 1.79e-147 | 1.01e-149 | 12800 | 62.4 | 62900 | 52.6 | 1.19 | DNR_DAR_3hr |
| 111 | 34 | MA1099.2 | HES1 | MA1099.2 | GGCRCGTGBC | HES1 | 1e-144 | 5.59e-147 | 4050 | 19.8 | 15200 | 12.7 | 1.55 | DNR_DAR_3hr |
| 112 | 34 | MA0616.2 | HES2 | MA0616.2 | GGCACGTGYC | HES2 | 2.11e-144 | 1.17e-146 | 8500 | 41.5 | 38500 | 32.2 | 1.29 | DNR_DAR_3hr |
| 113 | 44 | MA1628.1 | Zic1::Zic2 | MA1628.1 | CVCAGCAGGNV | Zic1::Zic2 | 2.87e-144 | 1.58e-146 | 8570 | 41.9 | 38800 | 32.5 | 1.29 | DNR_DAR_3hr |
| 114 | 22 | MA1929.1 | CTCF | MA1929.1 | CTGCAGTKCCNNNNNYNRCCASYAGRKGGCRSWG | CTCF | 1.93e-140 | 1.05e-142 | 9480 | 46.3 | 44000 | 36.8 | 1.26 | DNR_DAR_3hr |
| 115 | 33 | MA0695.1 | ZBTB7C | MA0695.1 | RCGACCACCGAN | ZBTB7C | 4.24e-139 | 2.29e-141 | 9940 | 48.6 | 46700 | 39.1 | 1.24 | DNR_DAR_3hr |
| 116 | 14 | MA1587.1 | ZNF135 | MA1587.1 | CCTCGACCTCCYRR | ZNF135 | 6.87e-137 | 3.68e-139 | 10300 | 50.1 | 48600 | 40.7 | 1.23 | DNR_DAR_3hr |
| 117 | 41 | MA1993.1 | Neurod2 | MA1993.1 | VVCAGCTGBB | Neurod2 | 4.96e-136 | 2.63e-138 | 8940 | 43.7 | 41200 | 34.5 | 1.27 | DNR_DAR_3hr |
| 118 | 22 | MA0155.1 | INSM1 | MA0155.1 | TGYCWGGGGGCR | INSM1 | 2.63e-135 | 1.38e-137 | 6030 | 29.5 | 25500 | 21.4 | 1.38 | DNR_DAR_3hr |
| 119 | 41 | MA0633.2 | Twist2 | MA0633.2 | NVCAGCTGBN | Twist2 | 4.28e-132 | 2.23e-134 | 8390 | 41 | 38400 | 32.1 | 1.28 | DNR_DAR_3hr |
| 120 | 41 | MA1684.1 | Foxn1 | MA1684.1 | RGAMGC | Foxn1 | 2.37e-127 | 1.23e-129 | 3080 | 15 | 11000 | 9.23 | 1.63 | DNR_DAR_3hr |
| 121 | 45 | MA1522.1 (MAZ) | STREME-8 | 8-CCCCCCTC | CCCCCCTC | MAZ | 1.28e-119 | 6.54e-122 | 6480 | 31.7 | 28500 | 23.8 | 1.33 | DNR_DAR_3hr |
| 122 | 34 | MA0104.4 | MYCN | MA0104.4 | VVCCACGTGGBB | MYCN | 2.61e-119 | 1.33e-121 | 13400 | 65.6 | 68100 | 57 | 1.15 | DNR_DAR_3hr |
| 123 | 46 | MA1710.1 | ZNF257 | MA1710.1 | VDGAGGCRAGRG | ZNF257 | 5.37e-118 | 2.71e-120 | 10400 | 50.8 | 50200 | 42 | 1.21 | DNR_DAR_3hr |
| 124 | 47 | MA1637.1 | EBF3 | MA1637.1 | NYCCCAAGGGANN | EBF3 | 1.16e-116 | 5.79e-119 | 13300 | 65.2 | 67600 | 56.6 | 1.15 | DNR_DAR_3hr |
| 125 | 34 | MA0649.1 | HEY2 | MA0649.1 | GRCACGTGYC | HEY2 | 2.22e-116 | 1.1e-118 | 5490 | 26.8 | 23300 | 19.5 | 1.37 | DNR_DAR_3hr |
| 126 | 48 | MA0767.1 | GCM2 | MA0767.1 | BATGCGGGTR | GCM2 | 1.12e-115 | 5.51e-118 | 10400 | 50.7 | 50200 | 42 | 1.21 | DNR_DAR_3hr |
| 127 | 17 | MA1930.1 | CTCF | MA1930.1 | CTGCAGTKCCNVCHNNYRGCCASYAGRKGGCRAKC | CTCF | 2.79e-113 | 1.36e-115 | 7140 | 34.9 | 32200 | 27 | 1.29 | DNR_DAR_3hr |
| 128 | 49 | MA1982.1 | ZNF574 | MA1982.1 | VGSCTAGAGMGGCCS | ZNF574 | 3.76e-113 | 1.82e-115 | 8860 | 43.3 | 41700 | 34.9 | 1.24 | DNR_DAR_3hr |
| 129 | 50 | MA1973.1 | ZKSCAN3 | MA1973.1 | KDKCCCAGGCTAGCCCABNN | ZKSCAN3 | 2.51e-112 | 1.21e-114 | 9790 | 47.8 | 47000 | 39.3 | 1.22 | DNR_DAR_3hr |
| 130 | 34 | MA1474.1 | CREB3L4 | MA1474.1 | YGCCACGTCAYC | CREB3L4 | 2.08e-109 | 9.91e-112 | 6700 | 32.7 | 30000 | 25.1 | 1.3 | DNR_DAR_3hr |
| 131 | 51 | MA1528.1 | NFIX | MA1528.1 | STTGGCRNNGTGCCARB | NFIX | 3.5e-109 | 1.66e-111 | 5440 | 26.6 | 23300 | 19.5 | 1.36 | DNR_DAR_3hr |
| 132 | 3 | MA1656.1 | ZNF449 | MA1656.1 | NNAAGCCCAACCNN | ZNF449 | 4.2e-108 | 1.98e-110 | 15700 | 76.6 | 82500 | 69 | 1.11 | DNR_DAR_3hr |
| 133 | 52 | MA1728.1 | ZNF549 | MA1728.1 | NNTGCTGCCCWR | ZNF549 | 7.5e-108 | 3.5e-110 | 8190 | 40 | 38200 | 32 | 1.25 | DNR_DAR_3hr |
| 134 | 39 | MA0067.2 | PAX2 | MA0067.2 | BCAVTSRAGCGTGACGR | PAX2 | 6.62e-106 | 3.07e-108 | 7170 | 35 | 32700 | 27.3 | 1.28 | DNR_DAR_3hr |
| 135 | 53 | MA0145.2 | Tfcp2l1 | MA0145.2 | CCAGYYYVADCCRG | Tfcp2l1 | 5.02e-103 | 2.31e-105 | 10600 | 51.6 | 51800 | 43.3 | 1.19 | DNR_DAR_3hr |
| 136 | 36 | MA0697.2 | Zic3 | MA0697.2 | CNCAGCAGGAGNN | Zic3 | 1.49e-102 | 6.82e-105 | 9540 | 46.6 | 46000 | 38.5 | 1.21 | DNR_DAR_3hr |
| 137 | 54 | MA1972.1 | ZFP14 | MA1972.1 | GGAGGCHCHGGARBG | ZFP14 | 2.8e-102 | 1.27e-104 | 12300 | 59.9 | 61800 | 51.7 | 1.16 | DNR_DAR_3hr |
| 138 | 55 | MA1716.1 | ZNF76 | MA1716.1 | NNTWCCCAYAATGCAHYGCGC | ZNF76 | 8.11e-102 | 3.65e-104 | 6900 | 33.7 | 31400 | 26.2 | 1.28 | DNR_DAR_3hr |
| 139 | 25 | MA0006.1 | Ahr::Arnt | MA0006.1 | YGCGTG | Ahr::Arnt | 1.55e-101 | 6.91e-104 | 3620 | 17.7 | 14300 | 12 | 1.48 | DNR_DAR_3hr |
| 140 | 34 | MA0103.3 | ZEB1 | MA0103.3 | SNCACCTGSVN | ZEB1 | 1.81e-101 | 8.03e-104 | 13500 | 65.9 | 69200 | 57.9 | 1.14 | DNR_DAR_3hr |
| 141 | 34 | MA0259.1 | ARNT::HIF1A | MA0259.1 | VBACGTGC | ARNT::HIF1A | 8.71e-100 | 3.83e-102 | 5020 | 24.5 | 21500 | 18 | 1.36 | DNR_DAR_3hr |
| 142 | 56 | MA1730.1 | ZNF708 | MA1730.1 | NNRGCTGTRCCTCCY | ZNF708 | 6.49e-99 | 2.84e-101 | 11000 | 53.8 | 54700 | 45.7 | 1.18 | DNR_DAR_3hr |
| 143 | 57 | MA0504.1 | NR2C2 | MA0504.1 | VGRGGTCARAGGTCA | NR2C2 | 1.6e-98 | 6.95e-101 | 9670 | 47.2 | 46900 | 39.2 | 1.2 | DNR_DAR_3hr |
| 144 | 37 | MA0765.3 | ETV5 | MA0765.3 | ACCGGAAGTR | ETV5 | 3.33e-97 | 1.43e-99 | 6480 | 31.6 | 29300 | 24.5 | 1.29 | DNR_DAR_3hr |
| 145 | 58 | MA1149.1 | RARA::RXRG | MA1149.1 | RRGGTCAHNNNRRGGTCA | RARA::RXRG | 1.45e-95 | 6.21e-98 | 12700 | 62.1 | 64800 | 54.2 | 1.15 | DNR_DAR_3hr |
| 146 | 34 | MA1560.1 | SOHLH2 | MA1560.1 | YGCACGTGCN | SOHLH2 | 2.04e-93 | 8.66e-96 | 8550 | 41.8 | 40900 | 34.2 | 1.22 | DNR_DAR_3hr |
| 147 | 51 | MA1643.1 | NFIB | MA1643.1 | NNCCTGGCAYNGTGCCAAGNN | NFIB | 2.31e-93 | 9.74e-96 | 7850 | 38.4 | 37000 | 30.9 | 1.24 | DNR_DAR_3hr |
| 148 | 37 | MA0764.3 | ETV4 | MA0764.3 | ACCGGAAGTR | ETV4 | 1.4e-92 | 5.89e-95 | 5830 | 28.5 | 26000 | 21.8 | 1.31 | DNR_DAR_3hr |
| 149 | 47 | MA0154.4 | EBF1 | MA0154.4 | NNTCCCCAGGGGANN | EBF1 | 2.43e-88 | 1.01e-90 | 9530 | 46.6 | 46600 | 39 | 1.19 | DNR_DAR_3hr |
| 150 | 41 | MA1635.1 | BHLHE22 | MA1635.1 | NVCAGCTGBN | BHLHE22 | 1.31e-86 | 5.42e-89 | 9030 | 44.1 | 43900 | 36.7 | 1.2 | DNR_DAR_3hr |
| 151 | 36 | MA1629.1 | Zic2 | MA1629.1 | NDCACAGCAGGDRG | Zic2 | 8.35e-85 | 3.43e-87 | 13200 | 64.3 | 68100 | 57 | 1.13 | DNR_DAR_3hr |
| 152 | 59 | MA1964.1 | SMAD2 | MA1964.1 | NBCCAGACDN | SMAD2 | 4.03e-84 | 1.65e-86 | 12900 | 63.1 | 66700 | 55.8 | 1.13 | DNR_DAR_3hr |
| 153 | 17 | MA0163.1 | PLAG1 | MA0163.1 | GGGGCCCWAGGGGG | PLAG1 | 4.51e-82 | 1.83e-84 | 3080 | 15 | 12200 | 10.2 | 1.47 | DNR_DAR_3hr |
| 154 | 47 | MA1604.1 | Ebf2 | MA1604.1 | NYCCCAAGGGANN | Ebf2 | 1.67e-79 | 6.72e-82 | 14600 | 71.4 | 77200 | 64.6 | 1.11 | DNR_DAR_3hr |
| 155 | 60 | MA1726.1 | ZNF331 | MA1726.1 | NMYTGCAGAGCCCH | ZNF331 | 4.55e-79 | 1.82e-81 | 10200 | 49.9 | 51100 | 42.7 | 1.17 | DNR_DAR_3hr |
| 156 | 34 | MA0464.2 | BHLHE40 | MA0464.2 | VKCACGTGMC | BHLHE40 | 7.8e-79 | 3.1e-81 | 9490 | 46.4 | 46900 | 39.2 | 1.18 | DNR_DAR_3hr |
| 157 | 48 | MA0646.1 | GCM1 | MA0646.1 | BATGCGGGTAC | GCM1 | 3.75e-78 | 1.48e-80 | 6570 | 32.1 | 30700 | 25.6 | 1.25 | DNR_DAR_3hr |
| 158 | 34 | MA1464.1 | ARNT2 | MA1464.1 | GTCACGTGMN | ARNT2 | 4.21e-77 | 1.65e-79 | 8130 | 39.8 | 39300 | 32.9 | 1.21 | DNR_DAR_3hr |
| 159 | 61 | MA1971.1 | ZBED2 | MA1971.1 | NNMMCGAAACCNNV | ZBED2 | 2.7e-75 | 1.05e-77 | 1980 | 9.69 | 7180 | 6 | 1.61 | DNR_DAR_3hr |
| 160 | 15 | MA1581.1 | ZBTB6 | MA1581.1 | DNSCTTGAGCCNV | ZBTB6 | 1.26e-73 | 4.88e-76 | 10300 | 50.2 | 51700 | 43.2 | 1.16 | DNR_DAR_3hr |
| 161 | 62 | MA1711.1 | ZNF343 | MA1711.1 | CCGCTTCMCCDCGGCMV | ZNF343 | 1.05e-71 | 4.06e-74 | 3630 | 17.8 | 15400 | 12.9 | 1.38 | DNR_DAR_3hr |
| 162 | 34 | MA0499.2 | MYOD1 | MA0499.2 | NNGCACCTGTCNB | MYOD1 | 4.7e-71 | 1.8e-73 | 15500 | 75.9 | 83300 | 69.7 | 1.09 | DNR_DAR_3hr |
| 163 | 63 | MA1575.1 | THRB | MA1575.1 | DTGACCTBRNYVAGGTCAH | THRB | 4.12e-70 | 1.57e-72 | 4100 | 20 | 17800 | 14.9 | 1.34 | DNR_DAR_3hr |
| 164 | 22 | MA0139.1 | CTCF | MA0139.1 | YGRCCASYAGRKGGCRSYR | CTCF | 3.79e-68 | 1.44e-70 | 11700 | 57 | 60200 | 50.3 | 1.13 | DNR_DAR_3hr |
| 165 | 64 | MA1545.1 | OVOL2 | MA1545.1 | DDACCGTTABNYN | OVOL2 | 6.33e-68 | 2.38e-70 | 3750 | 18.3 | 16100 | 13.5 | 1.36 | DNR_DAR_3hr |
| 166 | 63 | MA1533.1 | NR1I2 | MA1533.1 | DYGAACTSVDTGAACTC | NR1I2 | 8.61e-66 | 3.22e-68 | 11900 | 58.4 | 61900 | 51.8 | 1.13 | DNR_DAR_3hr |
| 167 | 37 | MA0028.2 | ELK1 | MA0028.2 | ACCGGAAGTR | ELK1 | 9.22e-64 | 3.43e-66 | 7990 | 39.1 | 39300 | 32.8 | 1.19 | DNR_DAR_3hr |
| 168 | 41 | MA0048.2 | NHLH1 | MA0048.2 | CGCAGCTGCK | NHLH1 | 1.04e-61 | 3.86e-64 | 13200 | 64.6 | 69800 | 58.4 | 1.11 | DNR_DAR_3hr |
| 169 | 65 | MA0511.2 | RUNX2 | MA0511.2 | WAACCGCAA | RUNX2 | 2.39e-61 | 8.76e-64 | 15200 | 74.4 | 82100 | 68.6 | 1.08 | DNR_DAR_3hr |
| 170 | 66 | MA1979.1 | ZNF416 | MA1979.1 | TATCTGGGCAH | ZNF416 | 1.09e-60 | 3.98e-63 | 15600 | 76.1 | 84100 | 70.4 | 1.08 | DNR_DAR_3hr |
| 171 | 67 | MA1652.1 | ZKSCAN5 | MA1652.1 | NNRGGARGTGAGRR | ZKSCAN5 | 2.47e-59 | 8.95e-62 | 12200 | 59.5 | 63600 | 53.2 | 1.12 | DNR_DAR_3hr |
| 172 | 68 | MA1475.1 | CREB3L4 | MA1475.1 | GRTGACGTCAYC | CREB3L4 | 1.21e-55 | 4.37e-58 | 3830 | 18.7 | 17100 | 14.3 | 1.31 | DNR_DAR_3hr |
| 173 | 37 | MA1933.1 | ELK1::SREBF2 | MA1933.1 | DCCGGAAGTSRCGTGA | ELK1::SREBF2 | 9.6e-55 | 3.44e-57 | 1700 | 8.29 | 6380 | 5.33 | 1.56 | DNR_DAR_3hr |
| 174 | 69 | MA1731.1 | ZNF768 | MA1731.1 | YBVCYBRSCCTCTCTGDG | ZNF768 | 4.47e-54 | 1.59e-56 | 10100 | 49.2 | 51600 | 43.2 | 1.14 | DNR_DAR_3hr |
| 175 | 37 | MA0156.3 | FEV | MA0156.3 | VACCGGAAGTVV | FEV | 4.32e-51 | 1.53e-53 | 6540 | 32 | 31900 | 26.7 | 1.2 | DNR_DAR_3hr |
| 176 | 41 | MA1529.1 | NHLH2 | MA1529.1 | GGGNCGCAGCTGCGYCCC | NHLH2 | 4.6e-51 | 1.62e-53 | 5140 | 25.1 | 24300 | 20.3 | 1.24 | DNR_DAR_3hr |
| 177 | 70 | MA0138.2 | REST | MA0138.2 | TTCAGCACCATGGACAGCDCC | REST | 1.91e-49 | 6.7e-52 | 4220 | 20.6 | 19400 | 16.2 | 1.27 | DNR_DAR_3hr |
| 178 | 34 | MA1934.1 | ERF::FIGLA | MA1934.1 | RCCGGAARCASCTGBN | ERF::FIGLA | 4.36e-49 | 1.52e-51 | 3000 | 14.7 | 13000 | 10.9 | 1.34 | DNR_DAR_3hr |
| 179 | 34 | MA1100.2 | ASCL1 | MA1100.2 | VGCAGCTGCN | ASCL1 | 1.04e-48 | 3.59e-51 | 17400 | 85.1 | 96600 | 80.8 | 1.05 | DNR_DAR_3hr |
| 180 | 71 | MA2002.1 | Zfp335 | MA2002.1 | VNTCAGGCANH | Zfp335 | 9.11e-48 | 3.14e-50 | 7850 | 38.3 | 39400 | 33 | 1.16 | DNR_DAR_3hr |
| 181 | 41 | MA1485.1 | FERD3L | MA1485.1 | GCRMCAGCTGTYAC | FERD3L | 5.03e-47 | 1.73e-49 | 4670 | 22.8 | 21900 | 18.4 | 1.24 | DNR_DAR_3hr |
| 182 | 72 | MA1557.1 | SMAD5 | MA1557.1 | YGTCTAGACA | SMAD5 | 5.49e-46 | 1.87e-48 | 11300 | 55 | 59100 | 49.5 | 1.11 | DNR_DAR_3hr |
| 183 | 73 | MA0738.1 | HIC2 | MA0738.1 | RTGCCCRSB | HIC2 | 1.42e-45 | 4.81e-48 | 3910 | 19.1 | 17900 | 15 | 1.27 | DNR_DAR_3hr |
| 184 | 74 | MA1108.2 (MXI1) | STREME-7 | 7-CACACATG | CACACATG | MXI1 | 1.03e-43 | 3.48e-46 | 1430 | 6.97 | 5410 | 4.52 | 1.54 | DNR_DAR_3hr |
| 185 | 58 | MA0159.1 | RARA::RXRA | MA0159.1 | RGKTCANVGRSAGGTCA | RARA::RXRA | 1.21e-41 | 4.06e-44 | 7630 | 37.3 | 38600 | 32.3 | 1.16 | DNR_DAR_3hr |
| 186 | 34 | MA1493.1 | HES6 | MA1493.1 | GGCACGTGTY | HES6 | 2.22e-41 | 7.4e-44 | 13600 | 66.7 | 73700 | 61.6 | 1.08 | DNR_DAR_3hr |
| 187 | 75 | MA0778.1 | NFKB2 | MA0778.1 | AGGGGAWTCCCCY | NFKB2 | 6.44e-41 | 2.14e-43 | 3540 | 17.3 | 16200 | 13.6 | 1.28 | DNR_DAR_3hr |
| 188 | 34 | MA0608.1 | Creb3l2 | MA0608.1 | GCCACGTGD | Creb3l2 | 1.07e-40 | 3.54e-43 | 2480 | 12.1 | 10700 | 8.95 | 1.35 | DNR_DAR_3hr |
| 189 | 76 | MA1715.1 | ZNF707 | MA1715.1 | CCCCACTCCTGGTAC | ZNF707 | 4.26e-40 | 1.4e-42 | 7990 | 39.1 | 40700 | 34.1 | 1.15 | DNR_DAR_3hr |
| 190 | 34 | MA0822.1 | HES7 | MA0822.1 | YGGCACGTGCCR | HES7 | 9.7e-40 | 3.17e-42 | 1380 | 6.74 | 5300 | 4.43 | 1.52 | DNR_DAR_3hr |
| 191 | 34 | MA0745.2 | SNAI2 | MA0745.2 | NBGCACCTGTMNY | SNAI2 | 1.32e-39 | 4.3e-42 | 16800 | 82.1 | 93100 | 77.9 | 1.05 | DNR_DAR_3hr |
| 192 | 37 | MA0076.2 | ELK4 | MA0076.2 | BCRCTTCCGGB | ELK4 | 7.8e-39 | 2.52e-41 | 893 | 4.36 | 3060 | 2.56 | 1.7 | DNR_DAR_3hr |
| 193 | 34 | MA0004.1 | Arnt | MA0004.1 | CACGTG | Arnt | 5.68e-38 | 1.83e-40 | 4180 | 20.4 | 19800 | 16.5 | 1.23 | DNR_DAR_3hr |
| 194 | 77 | MA0863.1 | MTF1 | MA0863.1 | TTTGCACACGGCAC | MTF1 | 9.45e-37 | 3.02e-39 | 2860 | 14 | 12900 | 10.8 | 1.3 | DNR_DAR_3hr |
| 195 | 57 | MA1147.1 | NR4A2::RXRA | MA1147.1 | VRGGTCRTTGACCYY | NR4A2::RXRA | 1.19e-36 | 3.8e-39 | 9720 | 47.5 | 50900 | 42.6 | 1.12 | DNR_DAR_3hr |
| 196 | 41 | MA0500.2 | MYOG | MA0500.2 | NDRCAGCTGYHN | MYOG | 2.06e-36 | 6.52e-39 | 12800 | 62.7 | 69200 | 57.9 | 1.08 | DNR_DAR_3hr |
| 197 | 34 | MA0603.1 | Arntl | MA0603.1 | NGTCACGTGH | Arntl | 4.13e-36 | 1.3e-38 | 6120 | 29.9 | 30500 | 25.5 | 1.17 | DNR_DAR_3hr |
| 198 | 78 | MA0807.1 | TBX5 | MA0807.1 | AGGTGTKA | TBX5 | 4.45e-35 | 1.4e-37 | 13200 | 64.6 | 71600 | 59.9 | 1.08 | DNR_DAR_3hr |
| 199 | 30 | MA0737.1 | GLIS3 | MA0737.1 | GACCCCCCACGAAG | GLIS3 | 5.47e-35 | 1.71e-37 | 1700 | 8.29 | 7000 | 5.85 | 1.42 | DNR_DAR_3hr |
| 200 | 37 | MA1938.1 | ERF::NHLH1 | MA1938.1 | VGCAGCTGCCGGAARYN | ERF::NHLH1 | 1.54e-34 | 4.77e-37 | 5470 | 26.7 | 27000 | 22.6 | 1.18 | DNR_DAR_3hr |
| 201 | 34 | MA0058.3 | MAX | MA0058.3 | AVCACGTGNY | MAX | 4.26e-34 | 1.31e-36 | 13100 | 64 | 70900 | 59.3 | 1.08 | DNR_DAR_3hr |
| 202 | 34 | MA1621.1 | Rbpjl | MA1621.1 | NNVACACCTGTBNN | Rbpjl | 1.27e-32 | 3.91e-35 | 17800 | 87.1 | 1e+05 | 83.8 | 1.04 | DNR_DAR_3hr |
| 203 | 79 | MA1107.2 (KLF9) | STREME-13 | 13-CCACACACA | 13-CCACACACA | KLF9 | 4.65e-32 | 1.42e-34 | 4130 | 20.2 | 19900 | 16.6 | 1.21 | DNR_DAR_3hr |
| 204 | 68 | MA0604.1 | Atf1 | MA0604.1 | RTGACGTA | Atf1 | 8.09e-32 | 2.46e-34 | 3670 | 17.9 | 17400 | 14.5 | 1.23 | DNR_DAR_3hr |
| 205 | 39 | MA1995.1 | Npas4 | MA1995.1 | NNTCGTGACHN | Npas4 | 2.53e-31 | 7.67e-34 | 8210 | 40.1 | 42600 | 35.7 | 1.12 | DNR_DAR_3hr |
| 206 | 42 | MA0056.2 | MZF1 | MA0056.2 | NNAATCCCCANNN | MZF1 | 8.14e-31 | 2.45e-33 | 16100 | 78.7 | 89400 | 74.8 | 1.05 | DNR_DAR_3hr |
| 207 | 51 | MA0739.1 | Hic1 | MA0739.1 | RTGCCAACY | Hic1 | 1.91e-30 | 5.72e-33 | 9020 | 44.1 | 47400 | 39.6 | 1.11 | DNR_DAR_3hr |
| 208 | 80 | MA1554.1 | RFX7 | MA1554.1 | CGTTGCYAY | RFX7 | 2.67e-30 | 7.97e-33 | 11700 | 57.2 | 62900 | 52.6 | 1.09 | DNR_DAR_3hr |
| 209 | 37 | MA1992.1 | Ikzf3 | MA1992.1 | NNRCAGGAAGTGGVN | Ikzf3 | 7.47e-30 | 2.22e-32 | 16300 | 79.8 | 90900 | 76.1 | 1.05 | DNR_DAR_3hr |
| 210 | 37 | MA0475.2 | FLI1 | MA0475.2 | ACCGGAARTR | FLI1 | 1.89e-29 | 5.58e-32 | 465 | 2.27 | 1400 | 1.17 | 1.95 | DNR_DAR_3hr |
| 211 | 78 | MA0801.1 | MGA | MA0801.1 | AGGTGTGA | MGA | 3.27e-29 | 9.62e-32 | 17300 | 84.6 | 97100 | 81.2 | 1.04 | DNR_DAR_3hr |
| 212 | 81 | MA1538.1 | NR2F1 | MA1538.1 | RAGGTCANTGACCTY | NR2F1 | 7.51e-29 | 2.2e-31 | 5430 | 26.5 | 27200 | 22.8 | 1.17 | DNR_DAR_3hr |
| 213 | 40 | MA0609.2 | CREM | MA0609.2 | NNDGTGACGTCACHNN | CREM | 1.74e-28 | 5.07e-31 | 5150 | 25.2 | 25700 | 21.5 | 1.17 | DNR_DAR_3hr |
| 214 | 37 | MA0763.1 | ETV3 | MA0763.1 | ACCGGAAGTR | ETV3 | 6.22e-28 | 1.8e-30 | 403 | 1.97 | 1160 | 0.97 | 2.02 | DNR_DAR_3hr |
| 215 | 57 | MA1574.1 | THRB | MA1574.1 | RRGGTCAAAGGTCAN | THRB | 6.47e-28 | 1.87e-30 | 7890 | 38.6 | 41100 | 34.4 | 1.12 | DNR_DAR_3hr |
| 216 | 82 | MA1600.1 | ZNF684 | MA1600.1 | RYACAGTCCACCCCTTKR | ZNF684 | 6.73e-28 | 1.93e-30 | 6070 | 29.7 | 30800 | 25.8 | 1.15 | DNR_DAR_3hr |
| 217 | 41 | MA1619.1 | Ptf1A | MA1619.1 | NNACAGCTGTNN | Ptf1A | 1.05e-27 | 3e-30 | 17800 | 87 | 1e+05 | 84 | 1.04 | DNR_DAR_3hr |
| 218 | 56 | MA1655.1 | ZNF341 | MA1655.1 | NRGAACAGCCNN | ZNF341 | 1.25e-27 | 3.55e-30 | 8110 | 39.6 | 42400 | 35.5 | 1.12 | DNR_DAR_3hr |
| 219 | 78 | MA0803.1 | TBX15 | MA0803.1 | AGGTGTGA | TBX15 | 3.87e-27 | 1.1e-29 | 12200 | 59.4 | 65900 | 55.1 | 1.08 | DNR_DAR_3hr |
| 220 | 37 | MA0686.1 | SPDEF | MA0686.1 | AMCCGGATGTR | SPDEF | 3.88e-26 | 1.1e-28 | 6050 | 29.5 | 30800 | 25.8 | 1.15 | DNR_DAR_3hr |
| 221 | 41 | MA0521.2 | Tcf12 | MA0521.2 | NNACAGCTGTNN | Tcf12 | 2.55e-24 | 7.15e-27 | 18200 | 89.2 | 103000 | 86.5 | 1.03 | DNR_DAR_3hr |
| 222 | 81 | MA1556.1 | RXRG | MA1556.1 | RRGGTCAYGACCYY | RXRG | 4.96e-24 | 1.39e-26 | 5130 | 25 | 25900 | 21.6 | 1.16 | DNR_DAR_3hr |
| 223 | 57 | MA1536.1 | NR2C2 | MA1536.1 | RRGGTCAN | NR2C2 | 8.12e-24 | 2.26e-26 | 754 | 3.68 | 2810 | 2.35 | 1.57 | DNR_DAR_3hr |
| 224 | 57 | MA1535.1 | NR2C1 | MA1535.1 | NRRGGTCAN | NR2C1 | 8.4e-24 | 2.33e-26 | 9920 | 48.5 | 53100 | 44.5 | 1.09 | DNR_DAR_3hr |
| 225 | 83 | MA1999.1 | Prdm5 | MA1999.1 | CBGTTCTCCATCTNN | Prdm5 | 4.23e-23 | 1.17e-25 | 13000 | 63.4 | 71100 | 59.5 | 1.07 | DNR_DAR_3hr |
| 226 | 37 | MA0641.1 | ELF4 | MA0641.1 | AACCCGGAAGTR | ELF4 | 4.67e-23 | 1.28e-25 | 4430 | 21.6 | 22100 | 18.5 | 1.17 | DNR_DAR_3hr |
| 227 | 75 | MA1117.1 | RELB | MA1117.1 | RDATTCCCCNN | RELB | 2.34e-22 | 6.41e-25 | 13900 | 68 | 76800 | 64.3 | 1.06 | DNR_DAR_3hr |
| 228 | 84 | MA0130.1 | ZNF354C | MA0130.1 | MTCCAC | ZNF354C | 3e-22 | 8.16e-25 | 14900 | 72.7 | 82600 | 69.1 | 1.05 | DNR_DAR_3hr |
| 229 | 34 | MA0663.1 | MLX | MA0663.1 | RTCACGTGAT | MLX | 5.39e-22 | 1.46e-24 | 6240 | 30.5 | 32300 | 27 | 1.13 | DNR_DAR_3hr |
| 230 | 57 | MA0065.2 | Pparg::Rxra | MA0065.2 | STRGGGCARAGGKCA | Pparg::Rxra | 8.97e-22 | 2.42e-24 | 9870 | 48.2 | 53000 | 44.4 | 1.09 | DNR_DAR_3hr |
| 231 | 37 | MA1484.1 | ETS2 | MA1484.1 | DACCGGAAGY | ETS2 | 1.14e-21 | 3.06e-24 | 311 | 1.52 | 892 | 0.75 | 2.04 | DNR_DAR_3hr |
| 232 | 37 | MA1708.1 | ETV7 | MA1708.1 | DGSCGGAAGTR | ETV7 | 4.91e-21 | 1.31e-23 | 926 | 4.52 | 3710 | 3.11 | 1.46 | DNR_DAR_3hr |
| 233 | 85 | MA0073.1 | RREB1 | MA0073.1 | CCCCMAAMCAMCCMCMMMCV | RREB1 | 1.24e-20 | 3.3e-23 | 1520 | 7.44 | 6710 | 5.61 | 1.33 | DNR_DAR_3hr |
| 234 | 80 | MA0799.2 | RFX4 | MA0799.2 | BSGTTGCYWRGCAACSV | RFX4 | 2.95e-20 | 7.83e-23 | 1230 | 6.02 | 5250 | 4.4 | 1.37 | DNR_DAR_3hr |
| 235 | 34 | MA0636.1 | BHLHE41 | MA0636.1 | RTCACGTGAC | BHLHE41 | 3.85e-20 | 1.02e-22 | 3050 | 14.9 | 14800 | 12.4 | 1.2 | DNR_DAR_3hr |
| 236 | 51 | MA0671.1 | NFIX | MA0671.1 | NNTGCCAAN | NFIX | 4.66e-20 | 1.22e-22 | 18100 | 88.2 | 102000 | 85.7 | 1.03 | DNR_DAR_3hr |
| 237 | 44 | MA1472.2 | Bhlha15 | MA1472.2 | NVACAGCTGTBN | Bhlha15 | 1.11e-19 | 2.9e-22 | 15600 | 76.2 | 87200 | 73 | 1.04 | DNR_DAR_3hr |
| 238 | 79 | MA1491.2 | GLI3 | MA1491.2 | RGACCACCCACGWHGH | GLI3 | 4.85e-19 | 1.26e-21 | 1150 | 5.64 | 4910 | 4.11 | 1.37 | DNR_DAR_3hr |
| 239 | 44 | MA1641.1 | MYF5 | MA1641.1 | NVACAGCTGTBN | MYF5 | 9.62e-18 | 2.5e-20 | 14300 | 69.6 | 79300 | 66.4 | 1.05 | DNR_DAR_3hr |
| 240 | 57 | MA0017.2 | NR2F1 | MA0017.2 | CARAGGTCAMVNG | NR2F1 | 6.35e-17 | 1.64e-19 | 5680 | 27.7 | 29600 | 24.7 | 1.12 | DNR_DAR_3hr |
| 241 | 72 | MA1602.1 | ZSCAN29 | MA1602.1 | YGTCTACRCNGN | ZSCAN29 | 1.14e-16 | 2.93e-19 | 6190 | 30.3 | 32500 | 27.2 | 1.11 | DNR_DAR_3hr |
| 242 | 34 | MA1558.1 | SNAI1 | MA1558.1 | DRCAGGTGYD | SNAI1 | 9.43e-16 | 2.42e-18 | 13800 | 67.5 | 77000 | 64.4 | 1.05 | DNR_DAR_3hr |
| 243 | 81 | MA1555.1 | RXRB | MA1555.1 | RRGGTCATGACCYY | RXRB | 1.5e-15 | 3.82e-18 | 5430 | 26.5 | 28300 | 23.7 | 1.12 | DNR_DAR_3hr |
| 244 | 78 | MA1565.1 | TBX18 | MA1565.1 | DRAGGTGTGAAR | TBX18 | 2.12e-15 | 5.39e-18 | 17600 | 86 | 1e+05 | 83.7 | 1.03 | DNR_DAR_3hr |
| 245 | 41 | MA1468.1 | ATOH7 | MA1468.1 | AVCATATGBY | ATOH7 | 2.32e-15 | 5.88e-18 | 17900 | 87.5 | 102000 | 85.3 | 1.03 | DNR_DAR_3hr |
| 246 | 37 | MA0645.1 | ETV6 | MA0645.1 | MSCGGAAGTR | ETV6 | 4.08e-15 | 1.03e-17 | 5750 | 28.1 | 30200 | 25.2 | 1.11 | DNR_DAR_3hr |
| 247 | 34 | MA1620.1 | Ptf1A | MA1620.1 | NVACACCTGTNN | Ptf1A | 1.57e-14 | 3.95e-17 | 18100 | 88.6 | 103000 | 86.5 | 1.02 | DNR_DAR_3hr |
| 248 | 34 | MA0819.2 | CLOCK | MA0819.2 | NRACACGTGB | CLOCK | 2.23e-14 | 5.58e-17 | 4610 | 22.5 | 23800 | 19.9 | 1.13 | DNR_DAR_3hr |
| 249 | 58 | MA0258.2 | ESR2 | MA0258.2 | AGGTCASVNTGMCCY | ESR2 | 5.37e-14 | 1.34e-16 | 11200 | 54.9 | 61800 | 51.7 | 1.06 | DNR_DAR_3hr |
| 250 | 79 | MA1107.2 (KLF9) | STREME-12 | 12-CACACACACAC | 12-CACACACACAC | KLF9 | 6.4e-14 | 1.59e-16 | 855 | 4.18 | 3630 | 3.04 | 1.38 | DNR_DAR_3hr |
| 251 | 37 | MA0760.1 | ERF | MA0760.1 | ACCGGAAGTR | ERF | 1.06e-13 | 2.63e-16 | 432 | 2.11 | 1590 | 1.33 | 1.59 | DNR_DAR_3hr |
| 252 | 68 | MA1969.1 | THRA | MA1969.1 | NYTGTGTCCTCABRTGACCTYWBB | THRA | 9.95e-13 | 2.45e-15 | 4450 | 21.8 | 23100 | 19.4 | 1.12 | DNR_DAR_3hr |
| 253 | 51 | MA1527.1 | NFIC | MA1527.1 | NTTGGCDNNRTGCCARN | NFIC | 1.11e-12 | 2.73e-15 | 1640 | 8.03 | 7770 | 6.5 | 1.24 | DNR_DAR_3hr |
| 254 | 86 | MA1521.1 | MAFA | MA1521.1 | NTGCTGASTCAGCAN | MAFA | 6.65e-12 | 1.63e-14 | 10800 | 52.9 | 59800 | 50 | 1.06 | DNR_DAR_3hr |
| 255 | 81 | MA1552.1 | RARB | MA1552.1 | RAGGTCATGACCTY | RARB | 9.62e-12 | 2.34e-14 | 4100 | 20.1 | 21300 | 17.8 | 1.13 | DNR_DAR_3hr |
| 256 | 87 | MA0161.2 | NFIC | MA0161.2 | NNCTTGGCANN | NFIC | 1.03e-11 | 2.5e-14 | 14300 | 69.8 | 80200 | 67.1 | 1.04 | DNR_DAR_3hr |
| 257 | 78 | MA1566.2 | TBX3 | MA1566.2 | RAGGTGTGAAV | TBX3 | 1.93e-11 | 4.65e-14 | 15200 | 74.2 | 85600 | 71.6 | 1.04 | DNR_DAR_3hr |
| 258 | 68 | MA0638.1 | CREB3 | MA0638.1 | VTGCCACGTCAYCR | CREB3 | 2.36e-11 | 5.68e-14 | 4520 | 22.1 | 23700 | 19.8 | 1.12 | DNR_DAR_3hr |
| 259 | 34 | MA1559.1 | SNAI3 | MA1559.1 | RRCAGGTGYA | SNAI3 | 7.12e-11 | 1.71e-13 | 10500 | 51.5 | 58300 | 48.7 | 1.06 | DNR_DAR_3hr |
| 260 | 40 | MA0862.1 | GMEB2 | MA0862.1 | TTACGTAA | GMEB2 | 8.3e-11 | 1.98e-13 | 4380 | 21.4 | 22900 | 19.2 | 1.12 | DNR_DAR_3hr |
| 261 | 88 | MA1651.1 | ZFP42 | MA1651.1 | NNNHCAARATGGCTGCCNBNN | ZFP42 | 9.77e-11 | 2.32e-13 | 290 | 1.42 | 1020 | 0.85 | 1.67 | DNR_DAR_3hr |
| 262 | 68 | MA0844.1 | XBP1 | MA0844.1 | WDKGMCACGTCATC | XBP1 | 1.59e-10 | 3.76e-13 | 909 | 4.44 | 4050 | 3.39 | 1.31 | DNR_DAR_3hr |
| 263 | 78 | MA0806.1 | TBX4 | MA0806.1 | AGGTGTGA | TBX4 | 4.59e-10 | 1.08e-12 | 20100 | 98 | 116000 | 97.2 | 1.01 | DNR_DAR_3hr |
| 264 | 37 | MA1931.1 | ELK1::HOXA1 | MA1931.1 | ACCGGAAGTAATTA | ELK1::HOXA1 | 2.41e-09 | 5.67e-12 | 510 | 2.49 | 2100 | 1.76 | 1.42 | DNR_DAR_3hr |
| 265 | 79 | 15-CACACACCACACA | STREME-15 | 15-CACACACCACACA | 15-CACACACCACACA | CACACACCACACA | 3.04e-09 | 7.12e-12 | 1330 | 6.48 | 6310 | 5.28 | 1.23 | DNR_DAR_3hr |
| 266 | 87 | MA0502.2 | NFYB | MA0502.2 | CYCATTGGCCVV | NFYB | 3.8e-09 | 8.87e-12 | 13300 | 65.1 | 74800 | 62.6 | 1.04 | DNR_DAR_3hr |
| 267 | 68 | MA0839.1 | CREB3L1 | MA0839.1 | VTGCCACGTCAYCA | CREB3L1 | 1.98e-08 | 4.59e-11 | 777 | 3.8 | 3480 | 2.91 | 1.3 | DNR_DAR_3hr |
| 268 | 41 | MA0668.2 | Neurod2 | MA0668.2 | NNGRACAGATGGYNN | Neurod2 | 3.13e-08 | 7.24e-11 | 17400 | 84.9 | 99300 | 83 | 1.02 | DNR_DAR_3hr |
| 269 | 34 | MA0825.1 | MNT | MA0825.1 | RVCACGTGMH | MNT | 6.16e-08 | 1.42e-10 | 5790 | 28.3 | 31200 | 26.1 | 1.08 | DNR_DAR_3hr |
| 270 | 68 | MA1466.1 | ATF6 | MA1466.1 | TGRTGACGTGGCAN | ATF6 | 2.75e-07 | 6.32e-10 | 642 | 3.14 | 2850 | 2.38 | 1.32 | DNR_DAR_3hr |
| 271 | 37 | MA0098.3 | ETS1 | MA0098.3 | ACCGGAARTR | ETS1 | 3.75e-07 | 8.59e-10 | 14600 | 71.3 | 82700 | 69.1 | 1.03 | DNR_DAR_3hr |
| 272 | 89 | MA1542.1 | OSR1 | MA1542.1 | HGCTACYGTD | OSR1 | 1.35e-06 | 3.08e-09 | 15900 | 77.6 | 90500 | 75.7 | 1.03 | DNR_DAR_3hr |
| 273 | 51 | MA0119.1 | NFIC::TLX1 | MA0119.1 | TGGCASSRWGCCAA | NFIC::TLX1 | 2.34e-06 | 5.33e-09 | 1360 | 6.64 | 6680 | 5.59 | 1.19 | DNR_DAR_3hr |
| 274 | 75 | MA0101.1 | REL | MA0101.1 | BGGRNWTTCC | REL | 2.57e-06 | 5.81e-09 | 1470 | 7.16 | 7260 | 6.08 | 1.18 | DNR_DAR_3hr |
| 275 | 90 | MA1114.1 | PBX3 | MA1114.1 | NNNTGAGTGACAGGBNN | PBX3 | 4.28e-06 | 9.67e-09 | 18800 | 91.9 | 108000 | 90.6 | 1.01 | DNR_DAR_3hr |
| 276 | 34 | MA0820.1 | FIGLA | MA0820.1 | WMCACCTGKW | FIGLA | 4.97e-06 | 1.12e-08 | 19500 | 95 | 112000 | 94.1 | 1.01 | DNR_DAR_3hr |
| 277 | 44 | MA1646.1 | OSR2 | MA1646.1 | NNACAGAAGCNN | OSR2 | 5.42e-06 | 1.21e-08 | 15800 | 77 | 89900 | 75.2 | 1.02 | DNR_DAR_3hr |
| 278 | 78 | MA0805.1 | TBX1 | MA0805.1 | AGGTGTGA | TBX1 | 6.08e-06 | 1.36e-08 | 15300 | 74.8 | 87200 | 72.9 | 1.03 | DNR_DAR_3hr |
| 279 | 56 | MA1725.1 | ZNF189 | MA1725.1 | NNTGCTGTTCCHB | ZNF189 | 1.07e-05 | 2.39e-08 | 14900 | 72.8 | 84700 | 70.9 | 1.03 | DNR_DAR_3hr |
| 280 | 41 | MA1638.1 | HAND2 | MA1638.1 | NVCAGATGNN | HAND2 | 1.15e-05 | 2.56e-08 | 6820 | 33.3 | 37500 | 31.4 | 1.06 | DNR_DAR_3hr |
| 281 | 78 | MA1571.1 | TGIF2LX | MA1571.1 | TGACAGCTGTCA | TGIF2LX | 1.57e-05 | 3.47e-08 | 14400 | 70.2 | 81600 | 68.2 | 1.03 | DNR_DAR_3hr |
| 282 | 34 | MA0059.1 | MAX::MYC | MA0059.1 | RASCACGTGGT | MAX::MYC | 3.55e-05 | 7.81e-08 | 1020 | 4.96 | 4930 | 4.12 | 1.2 | DNR_DAR_3hr |
| 283 | 34 | MA0622.1 | Mlxip | MA0622.1 | BCACGTGK | Mlxip | 5.29e-05 | 1.16e-07 | 791 | 3.87 | 3750 | 3.14 | 1.23 | DNR_DAR_3hr |
| 284 | 57 | MA1537.1 | NR2F1 | MA1537.1 | RRGGTCAAAGGTCAN | NR2F1 | 0.000251 | 5.49e-07 | 3110 | 15.2 | 16600 | 13.9 | 1.1 | DNR_DAR_3hr |
| 285 | 78 | MA1572.1 | TGIF2LY | MA1572.1 | TGACAGCTGTCA | TGIF2LY | 0.000286 | 6.23e-07 | 14000 | 68.2 | 79400 | 66.4 | 1.03 | DNR_DAR_3hr |
| 286 | 37 | MA0473.3 | ELF1 | MA0473.3 | RDVCAGGAAGTGVN | ELF1 | 0.000463 | 1e-06 | 17300 | 84.7 | 99600 | 83.3 | 1.02 | DNR_DAR_3hr |
| 287 | 91 | MA1531.1 | NR1D1 | MA1531.1 | RGGTYAGTRGGTCAN | NR1D1 | 0.000556 | 1.2e-06 | 2700 | 13.2 | 14300 | 12 | 1.1 | DNR_DAR_3hr |
| 288 | 88 | MA0095.3 | Yy1 | MA0095.3 | NNCAAAATGGCN | Yy1 | 0.000758 | 1.63e-06 | 32 | 0.16 | 59 | 0.05 | 3.21 | DNR_DAR_3hr |
| 289 | 37 | MA1957.1 | HOXB2::ELK1 | MA1957.1 | RCCGGAAGTMRTTA | HOXB2::ELK1 | 0.00155 | 3.33e-06 | 447 | 2.18 | 2040 | 1.71 | 1.28 | DNR_DAR_3hr |
| 290 | 2 | MA0657.1 | KLF13 | MA0657.1 | RTGMCACGCCCCTTTTTG | KLF13 | 0.00226 | 4.84e-06 | 260 | 1.27 | 1100 | 0.92 | 1.39 | DNR_DAR_3hr |
| 291 | 80 | MA0510.2 | RFX5 | MA0510.2 | SGTTGCCATGGYAACV | RFX5 | 0.0027 | 5.75e-06 | 42 | 0.21 | 99 | 0.08 | 2.51 | DNR_DAR_3hr |
| 292 | 41 | MA1109.1 | NEUROD1 | MA1109.1 | NRACAGATGGYNN | NEUROD1 | 0.0043 | 9.15e-06 | 18700 | 91.3 | 108000 | 90.3 | 1.01 | DNR_DAR_3hr |
| 293 | 75 | MA0107.1 | RELA | MA0107.1 | GGGRATTTCC | RELA | 0.00736 | 1.56e-05 | 4520 | 22.1 | 24800 | 20.8 | 1.06 | DNR_DAR_3hr |
| 294 | 11 | MA1116.1 | RBPJ | MA1116.1 | BVTGGGAANN | RBPJ | 0.0101 | 2.12e-05 | 403 | 1.97 | 1860 | 1.55 | 1.27 | DNR_DAR_3hr |
| 295 | 34 | MA0626.1 | Npas2 | MA0626.1 | NSCACGTGTN | Npas2 | 0.0101 | 2.13e-05 | 362 | 1.77 | 1640 | 1.38 | 1.29 | DNR_DAR_3hr |
| 296 | 78 | MA1567.2 | Tbx6 | MA1567.2 | NRGGTGTGAANN | Tbx6 | 0.0107 | 2.25e-05 | 20200 | 98.5 | 117000 | 98 | 1 | DNR_DAR_3hr |
| 297 | 80 | MA0798.3 | RFX3 | MA0798.3 | CGTTRCCATGGCAACS | RFX3 | 0.0143 | 2.99e-05 | 34 | 0.17 | 78 | 0.07 | 2.59 | DNR_DAR_3hr |
| 298 | 81 | MA1553.1 | RARG | MA1553.1 | RAGGTCATGACCTY | RARG | 0.0172 | 3.58e-05 | 3330 | 16.3 | 18100 | 15.2 | 1.07 | DNR_DAR_3hr |
| 299 | 80 | MA0509.3 | RFX1 | MA0509.3 | SGTTGCCATGGCAACS | RFX1 | 0.0245 | 5.09e-05 | 42 | 0.21 | 111 | 0.09 | 2.24 | DNR_DAR_3hr |
| 300 | 37 | MA1935.1 | ERF::FOXI1 | MA1935.1 | RTAAACMGGAARTR | ERF::FOXI1 | 0.0262 | 5.42e-05 | 65 | 0.32 | 206 | 0.17 | 1.86 | DNR_DAR_3hr |
| 301 | 34 | MA0526.4 | USF2 | MA0526.4 | DRTCACGTGAYH | USF2 | 0.0297 | 6.13e-05 | 111 | 0.54 | 414 | 0.35 | 1.58 | DNR_DAR_3hr |
| 302 | 81 | MA1146.1 | NR1H4::RXRA | MA1146.1 | GAGKTCATTGACCYB | NR1H4::RXRA | 0.0331 | 6.81e-05 | 577 | 2.82 | 2810 | 2.35 | 1.2 | DNR_DAR_3hr |
| 303 | 75 | MA0105.4 | NFKB1 | MA0105.4 | AGGGGAWTCCCCT | NFKB1 | 0.0399 | 8.17e-05 | 255 | 1.25 | 1130 | 0.94 | 1.33 | DNR_DAR_3hr |
| 304 | 34 | MA0831.3 | TFE3 | MA0831.3 | GTCACGTGAC | TFE3 | 0.0459 | 9.37e-05 | 48 | 0.23 | 139 | 0.12 | 2.04 | DNR_DAR_3hr |
| 305 | 92 | MA1573.2 | Thap11 | MA1573.2 | ACTACAABTCCCAG | Thap11 | 0.0466 | 9.49e-05 | 6880 | 33.6 | 38500 | 32.2 | 1.04 | DNR_DAR_3hr |
| 306 | 37 | MA0762.1 | ETV2 | MA0762.1 | AACCGGAAATR | ETV2 | 0.0471 | 9.56e-05 | 34 | 0.17 | 84 | 0.07 | 2.41 | DNR_DAR_3hr |
ER_rat <- 1.25
# mrc_type <- "EAR_open"
DNR_DAR_3hr_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
# geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_col(aes(x=log10Evalue))+
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste("DNR_DAR_3hr_combo response peaks(200bp) Enrichment ratio:",ER_rat))
DNR_DAR_3hr_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
slice_head(n=5) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
# geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*.5), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./.5,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste("DNR DAR 3 hour response peaks(200bp) Enrichment ratio:",ER_rat))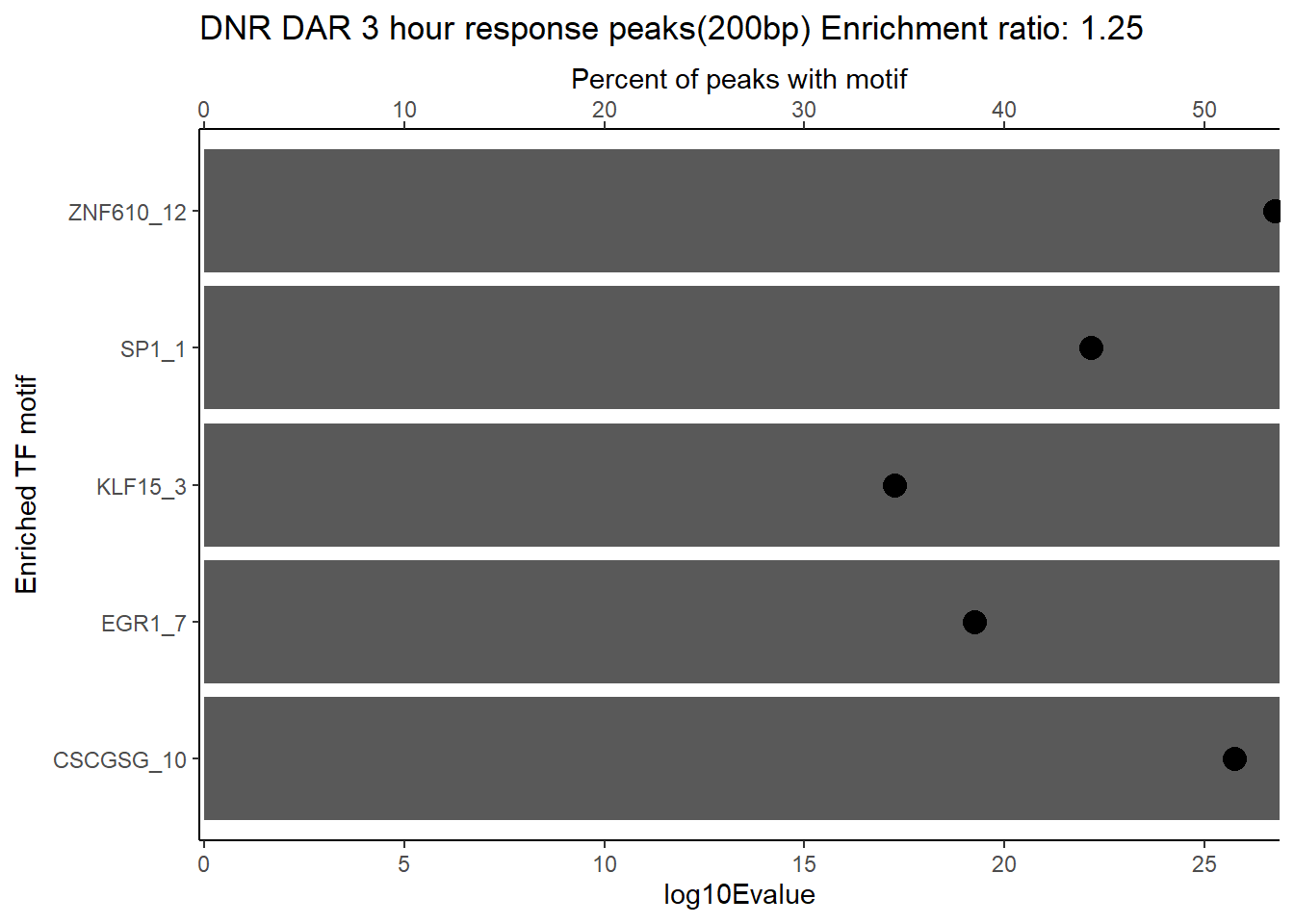
DNR_DAR_24hr data
DNR_DAR_24hr_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DNR_24_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
DNR_DAR_24hr_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DNR_24_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
DNR_DAR_24hr_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DNR_24_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
DNR_DAR_24hr_sea_all <- rbind(DNR_DAR_24hr_sea_disc, DNR_DAR_24hr_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)DNR_DAR_24hr_combo <- DNR_DAR_24hr_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., DNR_DAR_24hr_sea_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ SIM_MOTIF
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y) %>%
mutate(mrc_type="DNR_DAR_24hr")
DNR_DAR_24hr_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs 200 bp DNR_DAR_24hr") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO | mrc_type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | MA0477.2 (FOSL1) | STREME-1 | 1-NRATGASTCATYN | NRATGASTCATYN | FOSL1 | 0 | 0 | 9200 | 12.8 | 4200 | 6.17 | 2.07 | DNR_DAR_24hr |
| 2 | 1 | MA0099.3 | FOS::JUN | MA0099.3 | ATGAGTCAYM | FOS::JUN | 0 | 0 | 8840 | 12.3 | 3960 | 5.82 | 2.11 | DNR_DAR_24hr |
| 3 | 1 | MA1144.1 | FOSL2::JUND | MA1144.1 | KATGACTCAT | FOSL2::JUND | 0 | 0 | 8670 | 12 | 3920 | 5.77 | 2.09 | DNR_DAR_24hr |
| 4 | 1 | MA0477.2 | FOSL1 | MA0477.2 | NNATGACTCATNN | FOSL1 | 0 | 0 | 7810 | 10.8 | 3360 | 4.94 | 2.19 | DNR_DAR_24hr |
| 5 | 1 | MA1134.1 | FOS::JUNB | MA1134.1 | KATGASTCATHN | FOS::JUNB | 0 | 0 | 8350 | 11.6 | 3720 | 5.47 | 2.12 | DNR_DAR_24hr |
| 6 | 1 | MA1135.1 | FOSB::JUNB | MA1135.1 | KRTGASTCAT | FOSB::JUNB | 0 | 0 | 9410 | 13.1 | 4460 | 6.56 | 1.99 | DNR_DAR_24hr |
| 7 | 1 | MA1141.1 | FOS::JUND | MA1141.1 | NKATGAGTCATNN | FOS::JUND | 0 | 0 | 8800 | 12.2 | 4050 | 5.96 | 2.05 | DNR_DAR_24hr |
| 8 | 1 | MA1130.1 | FOSL2::JUN | MA1130.1 | NNRTGAGTCAYN | FOSL2::JUN | 0 | 0 | 9570 | 13.3 | 4590 | 6.75 | 1.97 | DNR_DAR_24hr |
| 9 | 1 | MA1138.1 | FOSL2::JUNB | MA1138.1 | KRTGASTCAT | FOSL2::JUNB | 0 | 0 | 10900 | 15.1 | 5510 | 8.1 | 1.86 | DNR_DAR_24hr |
| 10 | 1 | MA1128.1 | FOSL1::JUN | MA1128.1 | NKATGACTCATNN | FOSL1::JUN | 0 | 0 | 7620 | 10.6 | 3270 | 4.81 | 2.2 | DNR_DAR_24hr |
| 11 | 1 | MA0490.2 | JUNB | MA0490.2 | NNATGACTCATNN | JUNB | 0 | 0 | 8970 | 12.4 | 4210 | 6.19 | 2.01 | DNR_DAR_24hr |
| 12 | 1 | MA0491.2 | JUND | MA0491.2 | NNATGACTCATNN | JUND | 0 | 0 | 6790 | 9.44 | 2760 | 4.06 | 2.33 | DNR_DAR_24hr |
| 13 | 1 | MA0476.1 | FOS | MA0476.1 | DVTGASTCATB | FOS | 0 | 0 | 9160 | 12.7 | 4360 | 6.42 | 1.98 | DNR_DAR_24hr |
| 14 | 1 | MA0655.1 (JDP2) | STREME-2 | 2-ATGAATCA | ATGAATCA | JDP2 | 0 | 0 | 10400 | 14.5 | 5280 | 7.77 | 1.86 | DNR_DAR_24hr |
| 15 | 1 | MA1633.2 | BACH1 | MA1633.2 | ATGACTCAT | BACH1 | 0 | 0 | 11700 | 16.3 | 6280 | 9.24 | 1.76 | DNR_DAR_24hr |
| 16 | 1 | MA1137.1 | FOSL1::JUNB | MA1137.1 | NDRTGACTCATNN | FOSL1::JUNB | 0 | 0 | 8130 | 11.3 | 3750 | 5.51 | 2.05 | DNR_DAR_24hr |
| 17 | 1 | MA1988.1 | Atf3 | MA1988.1 | NRTGACTCABN | Atf3 | 1.28457067918724e-322 | 1.28457067918724e-322 | 6960 | 9.66 | 3010 | 4.43 | 2.18 | DNR_DAR_24hr |
| 18 | 1 | MA0476.1 (FOS) | MEME-2 | ATGAGTCA | ATGAGTCA | FOS | 1.90000000007959e-314 | 6.62000001534372e-316 | 11100 | 15.4 | 5970 | 8.78 | 1.75 | DNR_DAR_24hr |
| 19 | 1 | MA0841.1 | NFE2 | MA0841.1 | VATGACTCATS | NFE2 | 1.66000000000002e-310 | 5.47000000000005e-312 | 7610 | 10.6 | 3510 | 5.16 | 2.05 | DNR_DAR_24hr |
| 20 | 1 | MA0489.2 | Jun | MA0489.2 | NVTGACTCATNN | Jun | 4.16999999999997e-310 | 1.31000000000011e-311 | 9380 | 13 | 4760 | 6.99 | 1.86 | DNR_DAR_24hr |
| 21 | 1 | MA1142.1 | FOSL1::JUND | MA1142.1 | NRTGACTCAT | FOSL1::JUND | 6.17000000000002e-310 | 1.84000000000024e-311 | 11100 | 15.5 | 6040 | 8.89 | 1.74 | DNR_DAR_24hr |
| 22 | 1 | MA1634.1 | BATF | MA1634.1 | DATGACTCATH | BATF | 1.37e-309 | 3.89999999999994e-311 | 9980 | 13.9 | 5200 | 7.64 | 1.81 | DNR_DAR_24hr |
| 23 | 1 | MA0462.2 | BATF::JUN | MA0462.2 | DATGACTCATH | BATF::JUN | 3.63e-301 | 9.9e-303 | 10400 | 14.5 | 5560 | 8.18 | 1.77 | DNR_DAR_24hr |
| 24 | 1 | MA1101.2 | BACH2 | MA1101.2 | DWANCATGASTCATSNTWH | BACH2 | 5.49e-297 | 1.43e-298 | 7920 | 11 | 3790 | 5.58 | 1.97 | DNR_DAR_24hr |
| 25 | 1 | MA0655.1 | JDP2 | MA0655.1 | ATGACTCAT | JDP2 | 4.28e-296 | 1.07e-297 | 4550 | 6.32 | 1600 | 2.36 | 2.68 | DNR_DAR_24hr |
| 26 | 1 | MA1132.1 | JUN::JUNB | MA1132.1 | KATGACKCAT | JUN::JUNB | 2.82e-292 | 6.81e-294 | 11000 | 15.3 | 6090 | 8.95 | 1.71 | DNR_DAR_24hr |
| 27 | 1 | MA0835.2 | BATF3 | MA0835.2 | DATGACTCATH | BATF3 | 6.82e-277 | 1.58e-278 | 10500 | 14.6 | 5800 | 8.53 | 1.71 | DNR_DAR_24hr |
| 28 | 1 | MA0478.1 | FOSL2 | MA0478.1 | KRRTGASTCAB | FOSL2 | 4.8e-275 | 1.08e-276 | 12900 | 17.9 | 7600 | 11.2 | 1.6 | DNR_DAR_24hr |
| 29 | 1 | MA1928.1 | BNC2 | MA1928.1 | NDRTGAGTCABN | BNC2 | 3.7e-218 | 8.02e-220 | 14100 | 19.6 | 9050 | 13.3 | 1.47 | DNR_DAR_24hr |
| 30 | 1 | MA0089.2 | MAFG::NFE2L1 | MA0089.2 | NVNATGACTCAGCADW | MAFG::NFE2L1 | 5.72e-124 | 1.2e-125 | 15600 | 21.6 | 11300 | 16.6 | 1.3 | DNR_DAR_24hr |
| 31 | 1 | MA0496.3 | MAFK | MA0496.3 | NGMTGACTCAGCMNH | MAFK | 5.36e-116 | 1.09e-117 | 18700 | 26 | 14100 | 20.7 | 1.25 | DNR_DAR_24hr |
| 32 | 1 | MA0501.1 | MAF::NFE2 | MA0501.1 | ATGACTCAGCANWTY | MAF::NFE2 | 1.23e-99 | 2.4e-101 | 12500 | 17.3 | 8980 | 13.2 | 1.31 | DNR_DAR_24hr |
| 33 | 1 | MA0659.3 | Mafg | MA0659.3 | NWGMTGACTCAGCAN | Mafg | 8.94e-74 | 1.7e-75 | 7040 | 9.78 | 4790 | 7.04 | 1.39 | DNR_DAR_24hr |
| 34 | 2 | MA1990.1 (Gli1) | STREME-3 | 3-ACACACCACACACAC | ACACACCACACACAC | Gli1 | 2.1e-71 | 3.87e-73 | 468 | 0.65 | 61 | 0.09 | 7.15 | DNR_DAR_24hr |
| 35 | 2 | 6-ACACACACACCACAC | STREME-6 | 6-ACACACACACCACAC | ACACACACACCACAC | ACACACACACCACAC | 3.04e-64 | 5.46e-66 | 494 | 0.69 | 84 | 0.12 | 5.5 | DNR_DAR_24hr |
| 36 | 1 | MA0150.2 | Nfe2l2 | MA0150.2 | CASNATGACTCAGCA | Nfe2l2 | 1.48e-53 | 2.59e-55 | 25200 | 35 | 21100 | 31.1 | 1.13 | DNR_DAR_24hr |
| 37 | 3 | 9-ACACATRCACAC | STREME-9 | 9-ACACATRCACAC | ACACATRCACAC | ACACATRCACAC | 1.34e-43 | 2.27e-45 | 714 | 0.99 | 253 | 0.37 | 2.66 | DNR_DAR_24hr |
| 38 | 3 | MA0522.3 (TCF3) | STREME-10 | 10-ACACACATGCACD | 10-ACACACATGCACD | TCF3 | 2.82e-41 | 4.65e-43 | 706 | 0.98 | 257 | 0.38 | 2.59 | DNR_DAR_24hr |
| 39 | 2 | MA0073.1 (RREB1) | STREME-7 | 7-CACACACCACACC | CACACACCACACC | RREB1 | 5.63e-39 | 9.06e-41 | 666 | 0.93 | 242 | 0.36 | 2.59 | DNR_DAR_24hr |
| 40 | 2 | MA1107.2 (KLF9) | STREME-11 | 11-CACACACACATAM | 11-CACACACACATAM | KLF9 | 3.52e-35 | 5.52e-37 | 2940 | 4.08 | 1920 | 2.83 | 1.44 | DNR_DAR_24hr |
| 41 | 4 | MA0766.2 (GATA5) | STREME-13 | 13-CWGATA | 13-CWGATA | GATA5 | 1.61e-34 | 2.46e-36 | 48500 | 67.4 | 43600 | 64.1 | 1.05 | DNR_DAR_24hr |
| 42 | 1 | MA0591.1 | Bach1::Mafk | MA0591.1 | RSSATGACTCAGCAB | Bach1::Mafk | 1.02e-31 | 1.52e-33 | 3910 | 5.43 | 2750 | 4.04 | 1.34 | DNR_DAR_24hr |
| 43 | 3 | ACACACACACACACA | MEME-5 | ACACACACACACACA | ACACACACACACACA | ACACACACACACACA | 3.75e-30 | 5.47e-32 | 1900 | 2.64 | 1170 | 1.71 | 1.54 | DNR_DAR_24hr |
| 44 | 5 | MA0904.2 | HOXB5 | MA0904.2 | STAATTAS | HOXB5 | 4.38e-27 | 6.25e-29 | 40400 | 56.1 | 36100 | 53.1 | 1.06 | DNR_DAR_24hr |
| 45 | 1 | MA1639.1 | MEIS1 | MA1639.1 | NHCATMAATCAHN | MEIS1 | 2.66e-26 | 3.7e-28 | 28200 | 39.2 | 24700 | 36.3 | 1.08 | DNR_DAR_24hr |
| 46 | 5 | MA0158.2 | HOXA5 | MA0158.2 | GYMATTAS | HOXA5 | 6.67e-24 | 9.1e-26 | 31000 | 43 | 27300 | 40.2 | 1.07 | DNR_DAR_24hr |
| 47 | 5 | MA1495.1 | HOXA1 | MA1495.1 | STAATTAS | HOXA1 | 1.73e-23 | 2.31e-25 | 33200 | 46.1 | 29400 | 43.2 | 1.07 | DNR_DAR_24hr |
| 48 | 6 | MA0106.3 (TP53) | STREME-4 | 4-CATGTCCA | CATGTCCA | TP53 | 6.8e-23 | 8.89e-25 | 5880 | 8.17 | 4560 | 6.7 | 1.22 | DNR_DAR_24hr |
| 49 | 7 | MA0106.3 | TP53 | MA0106.3 | RACATGYCCGGRCATGTY | TP53 | 9.99e-23 | 1.28e-24 | 1180 | 1.64 | 686 | 1.01 | 1.63 | DNR_DAR_24hr |
| 50 | 5 | MA0902.2 | HOXB2 | MA0902.2 | SYAATTAS | HOXB2 | 1.26e-22 | 1.58e-24 | 45300 | 63 | 41000 | 60.2 | 1.04 | DNR_DAR_24hr |
| 51 | 5 | MA0707.2 | MNX1 | MA0707.2 | VYRATTAK | MNX1 | 1.81e-22 | 2.22e-24 | 37100 | 51.5 | 33200 | 48.8 | 1.06 | DNR_DAR_24hr |
| 52 | 5 | MA0705.1 | Lhx8 | MA0705.1 | CTAATTAV | Lhx8 | 1.83e-20 | 2.21e-22 | 42000 | 58.3 | 37900 | 55.7 | 1.05 | DNR_DAR_24hr |
| 53 | 8 | MA1155.1 | ZSCAN4 | MA1155.1 | TGCACACACTGAAAA | ZSCAN4 | 2.81e-20 | 3.33e-22 | 1600 | 2.23 | 1030 | 1.51 | 1.47 | DNR_DAR_24hr |
| 54 | 5 | MA0886.1 | EMX2 | MA0886.1 | VBTAATTARB | EMX2 | 1.09e-19 | 1.27e-21 | 49700 | 69.1 | 45300 | 66.6 | 1.04 | DNR_DAR_24hr |
| 55 | 9 | MA0626.1 | Npas2 | MA0626.1 | NSCACGTGTN | Npas2 | 6.17e-19 | 7.04e-21 | 27400 | 38 | 24200 | 35.6 | 1.07 | DNR_DAR_24hr |
| 56 | 5 | MA1519.1 | LHX5 | MA1519.1 | STAATTAV | LHX5 | 1.06e-16 | 1.18e-18 | 30800 | 42.8 | 27500 | 40.5 | 1.06 | DNR_DAR_24hr |
| 57 | 5 | MA1502.1 | HOXB8 | MA1502.1 | GYMATTAM | HOXB8 | 5.96e-16 | 6.56e-18 | 43100 | 59.9 | 39100 | 57.6 | 1.04 | DNR_DAR_24hr |
| 58 | 10 | MA0774.1 (MEIS2) | STREME-5 | 5-CTGTCARCAS | CTGTCARCAS | MEIS2 | 7.4e-16 | 8.01e-18 | 1780 | 2.48 | 1220 | 1.8 | 1.38 | DNR_DAR_24hr |
| 59 | 9 | MA1108.2 | MXI1 | MA1108.2 | NNCACATGBN | MXI1 | 1.33e-15 | 1.41e-17 | 8460 | 11.8 | 7000 | 10.3 | 1.14 | DNR_DAR_24hr |
| 60 | 5 | MA0710.1 | NOTO | MA0710.1 | NYTAATTAVN | NOTO | 1.49e-15 | 1.56e-17 | 57000 | 79.2 | 52500 | 77.3 | 1.02 | DNR_DAR_24hr |
| 61 | 11 | MA1143.1 | FOSL1::JUND | MA1143.1 | RTGACGTMAY | FOSL1::JUND | 4.86e-15 | 5e-17 | 32100 | 44.6 | 28800 | 42.3 | 1.05 | DNR_DAR_24hr |
| 62 | 1 | MA1113.2 | PBX2 | MA1113.2 | NHCATAAATCAHN | PBX2 | 6.85e-15 | 6.93e-17 | 39700 | 55.1 | 35900 | 52.8 | 1.04 | DNR_DAR_24hr |
| 63 | 5 | MA0701.2 | LHX9 | MA0701.2 | CYAATTAR | LHX9 | 3.96e-14 | 3.94e-16 | 27900 | 38.8 | 24900 | 36.6 | 1.06 | DNR_DAR_24hr |
| 64 | 12 | MA1606.1 | Foxf1 | MA1606.1 | DWRTAAACANN | Foxf1 | 2.28e-13 | 2.24e-15 | 63800 | 88.6 | 59300 | 87.2 | 1.02 | DNR_DAR_24hr |
| 65 | 13 | MA0093.3 | USF1 | MA0093.3 | NDGTCATGTGACHN | USF1 | 4.65e-13 | 4.49e-15 | 53800 | 74.7 | 49500 | 72.8 | 1.03 | DNR_DAR_24hr |
| 66 | 7 | MA0861.1 | TP73 | MA0861.1 | DRCATGTCNNRACAYGYM | TP73 | 1.17e-12 | 1.12e-14 | 15400 | 21.4 | 13400 | 19.7 | 1.09 | DNR_DAR_24hr |
| 67 | 12 | MA1103.2 | FOXK2 | MA1103.2 | DDGTAAACANN | FOXK2 | 9.72e-12 | 9.1e-14 | 67100 | 93.2 | 62600 | 92.1 | 1.01 | DNR_DAR_24hr |
| 68 | 14 | MA0596.1 | SREBF2 | MA0596.1 | RTGGGGTGAY | SREBF2 | 1.12e-11 | 1.04e-13 | 17400 | 24.2 | 15300 | 22.4 | 1.08 | DNR_DAR_24hr |
| 69 | 5 | MA0612.2 | EMX1 | MA0612.2 | BTAATTAS | EMX1 | 1.19e-11 | 1.08e-13 | 31000 | 43 | 27900 | 41 | 1.05 | DNR_DAR_24hr |
| 70 | 12 | MA0033.2 | FOXL1 | MA0033.2 | RTAAACA | FOXL1 | 1.29e-11 | 1.15e-13 | 61100 | 84.8 | 56700 | 83.3 | 1.02 | DNR_DAR_24hr |
| 71 | 15 | MA0786.1 | POU3F1 | MA0786.1 | WTATGCWAATKW | POU3F1 | 2.05e-11 | 1.81e-13 | 59400 | 82.6 | 55100 | 81 | 1.02 | DNR_DAR_24hr |
| 72 | 12 | MA0148.4 | FOXA1 | MA0148.4 | WWGTAAACATNN | FOXA1 | 2.15e-11 | 1.87e-13 | 60900 | 84.6 | 56500 | 83.1 | 1.02 | DNR_DAR_24hr |
| 73 | 16 | MA0676.1 | Nr2e1 | MA0676.1 | AAAAGTCAA | Nr2e1 | 3.29e-11 | 2.83e-13 | 30200 | 42 | 27200 | 40 | 1.05 | DNR_DAR_24hr |
| 74 | 5 | MA0912.2 | HOXD3 | MA0912.2 | CTAATTAC | HOXD3 | 6.01e-11 | 5.1e-13 | 21100 | 29.3 | 18700 | 27.5 | 1.06 | DNR_DAR_24hr |
| 75 | 10 | MA0498.2 | MEIS1 | MA0498.2 | HTGACAD | MEIS1 | 7.52e-11 | 6.3e-13 | 60800 | 84.5 | 56500 | 83 | 1.02 | DNR_DAR_24hr |
| 76 | 12 | MA0047.3 | FOXA2 | MA0047.3 | RWGTAAACANN | FOXA2 | 8.42e-11 | 6.95e-13 | 66100 | 91.7 | 61600 | 90.6 | 1.01 | DNR_DAR_24hr |
| 77 | 10 | MA0117.2 | Mafb | MA0117.2 | AWWNTGCTGACD | Mafb | 1.18e-10 | 9.59e-13 | 11700 | 16.3 | 10100 | 14.8 | 1.1 | DNR_DAR_24hr |
| 78 | 7 | MA0525.2 | TP63 | MA0525.2 | RACATGTYGKGACATGTC | TP63 | 1.96e-10 | 1.57e-12 | 681 | 0.95 | 412 | 0.61 | 1.56 | DNR_DAR_24hr |
| 79 | 17 | MA0782.2 | PKNOX1 | MA0782.2 | NNTGAGTGACAGVNN | PKNOX1 | 2.06e-10 | 1.63e-12 | 50800 | 70.6 | 46800 | 68.8 | 1.03 | DNR_DAR_24hr |
| 80 | 9 | MA1570.1 | TFAP4 | MA1570.1 | AHCATRTGDT | TFAP4 | 2.44e-10 | 1.91e-12 | 23300 | 32.3 | 20800 | 30.5 | 1.06 | DNR_DAR_24hr |
| 81 | 1 | MA1640.1 | MEIS2 | MA1640.1 | NNATGATTTATGDNH | MEIS2 | 2.68e-10 | 2.06e-12 | 58000 | 80.6 | 53800 | 79.1 | 1.02 | DNR_DAR_24hr |
| 82 | 15 | MA1115.1 | POU5F1 | MA1115.1 | WHATGCAAATN | POU5F1 | 2.7e-10 | 2.06e-12 | 58700 | 81.5 | 54400 | 80 | 1.02 | DNR_DAR_24hr |
| 83 | 5 | MA0644.2 | ESX1 | MA0644.2 | YYAATTAN | ESX1 | 3.04e-10 | 2.3e-12 | 55600 | 77.3 | 51400 | 75.6 | 1.02 | DNR_DAR_24hr |
| 84 | 12 | MA0032.2 | FOXC1 | MA0032.2 | WAWGTAAAYAW | FOXC1 | 3.36e-10 | 2.51e-12 | 58100 | 80.7 | 53800 | 79.2 | 1.02 | DNR_DAR_24hr |
| 85 | 18 | MA0122.3 | Nkx3-2 | MA0122.3 | WWAAMCACTTAAN | Nkx3-2 | 6.93e-10 | 5.12e-12 | 57400 | 79.8 | 53200 | 78.2 | 1.02 | DNR_DAR_24hr |
| 86 | 15 | MA0792.1 | POU5F1B | MA0792.1 | TATGCWAAT | POU5F1B | 1.09e-09 | 7.95e-12 | 63300 | 87.9 | 58900 | 86.6 | 1.01 | DNR_DAR_24hr |
| 87 | 12 | MA0042.2 | FOXI1 | MA0042.2 | GTAAACA | FOXI1 | 1.41e-09 | 1.02e-11 | 54900 | 76.3 | 50800 | 74.7 | 1.02 | DNR_DAR_24hr |
| 88 | 12 | MA1683.1 | FOXA3 | MA1683.1 | DWGTAAACANN | FOXA3 | 1.76e-09 | 1.26e-11 | 64300 | 89.4 | 60000 | 88.2 | 1.01 | DNR_DAR_24hr |
| 89 | 12 | MA0481.3 | FOXP1 | MA0481.3 | NDGTAAACANH | FOXP1 | 2.7e-09 | 1.9e-11 | 67700 | 94 | 63300 | 93.1 | 1.01 | DNR_DAR_24hr |
| 90 | 12 | MA0848.1 | FOXO4 | MA0848.1 | GTAAACA | FOXO4 | 4.07e-09 | 2.84e-11 | 60900 | 84.6 | 56600 | 83.2 | 1.02 | DNR_DAR_24hr |
| 91 | 5 | MA0675.1 | NKX6-2 | MA0675.1 | NYMATTAA | NKX6-2 | 5.32e-09 | 3.67e-11 | 64200 | 89.2 | 59900 | 88 | 1.01 | DNR_DAR_24hr |
| 92 | 5 | MA0027.2 | EN1 | MA0027.2 | SYAATTAV | EN1 | 9.72e-09 | 6.63e-11 | 47100 | 65.4 | 43300 | 63.7 | 1.03 | DNR_DAR_24hr |
| 93 | 9 | MA0620.3 | MITF | MA0620.3 | NNNDRTCACGTGAYHNNN | MITF | 1.41e-08 | 9.5e-11 | 41500 | 57.6 | 38000 | 55.9 | 1.03 | DNR_DAR_24hr |
| 94 | 19 | MA1479.1 | DMRTC2 | MA1479.1 | AATTGMTACATT | DMRTC2 | 2.06e-08 | 1.37e-10 | 53800 | 74.7 | 49700 | 73.1 | 1.02 | DNR_DAR_24hr |
| 95 | 5 | MA0708.2 | MSX2 | MA0708.2 | SCAATTAV | MSX2 | 3.01e-08 | 1.99e-10 | 58400 | 81.1 | 54200 | 79.7 | 1.02 | DNR_DAR_24hr |
| 96 | 4 | MA0766.2 | GATA5 | MA0766.2 | SWGATAASRN | GATA5 | 3.24e-08 | 2.12e-10 | 21100 | 29.2 | 18800 | 27.7 | 1.06 | DNR_DAR_24hr |
| 97 | 20 | MA0502.2 | NFYB | MA0502.2 | CYCATTGGCCVV | NFYB | 4.07e-08 | 2.64e-10 | 4560 | 6.34 | 3750 | 5.52 | 1.15 | DNR_DAR_24hr |
| 98 | 21 | MA0036.3 | GATA2 | MA0036.3 | NBCTTATCTNH | GATA2 | 4.44e-08 | 2.84e-10 | 6630 | 9.2 | 5600 | 8.23 | 1.12 | DNR_DAR_24hr |
| 99 | 5 | MA0910.2 | HOXD8 | MA0910.2 | GYMATTAM | HOXD8 | 5.92e-08 | 3.75e-10 | 26800 | 37.2 | 24200 | 35.6 | 1.05 | DNR_DAR_24hr |
| 100 | 5 | MA0879.2 | DLX1 | MA0879.2 | VYAATTAN | DLX1 | 6.89e-08 | 4.33e-10 | 67400 | 93.6 | 63100 | 92.7 | 1.01 | DNR_DAR_24hr |
| 101 | 5 | MA0718.1 | RAX | MA0718.1 | RYYAATTARH | RAX | 7.03e-08 | 4.37e-10 | 61400 | 85.3 | 57200 | 84.1 | 1.01 | DNR_DAR_24hr |
| 102 | 15 | MA0142.1 | Pou5f1::Sox2 | MA0142.1 | CWTTGTYATGCAAAT | Pou5f1::Sox2 | 8.32e-08 | 5.12e-10 | 55700 | 77.3 | 51600 | 75.9 | 1.02 | DNR_DAR_24hr |
| 103 | 4 | MA0140.2 | GATA1::TAL1 | MA0140.2 | MTTATCWSNNNNNHVCAG | GATA1::TAL1 | 2.79e-07 | 1.7e-09 | 23100 | 32.1 | 20800 | 30.6 | 1.05 | DNR_DAR_24hr |
| 104 | 5 | MA1505.1 | HOXC8 | MA1505.1 | KTAATTAM | HOXC8 | 3.17e-07 | 1.91e-09 | 64600 | 89.7 | 60300 | 88.7 | 1.01 | DNR_DAR_24hr |
| 105 | 15 | MA0785.1 | POU2F1 | MA0785.1 | AWTATGCWAATK | POU2F1 | 3.74e-07 | 2.24e-09 | 57900 | 80.4 | 53800 | 79 | 1.02 | DNR_DAR_24hr |
| 106 | 12 | MA1607.1 | Foxl2 | MA1607.1 | DDWATGTAAACANN | Foxl2 | 4.46e-07 | 2.64e-09 | 63600 | 88.4 | 59400 | 87.3 | 1.01 | DNR_DAR_24hr |
| 107 | 5 | MA1530.1 | NKX6-3 | MA1530.1 | VYCATTAWW | NKX6-3 | 5.51e-07 | 3.23e-09 | 13900 | 19.3 | 12300 | 18 | 1.07 | DNR_DAR_24hr |
| 108 | 5 | MA0630.1 | SHOX | MA0630.1 | HTAATTRR | SHOX | 6.08e-07 | 3.53e-09 | 62700 | 87 | 58400 | 85.9 | 1.01 | DNR_DAR_24hr |
| 109 | 23 | MA0611.2 | Dux | MA0611.2 | NTGATTBAATCAVWNN | Dux | 7.5e-07 | 4.32e-09 | 11700 | 16.2 | 10200 | 15 | 1.08 | DNR_DAR_24hr |
| 110 | 17 | MA0063.2 | NKX2-5 | MA0063.2 | NNCACTCAANN | NKX2-5 | 8.7e-07 | 4.96e-09 | 49000 | 68.1 | 45300 | 66.6 | 1.02 | DNR_DAR_24hr |
| 111 | 12 | MA0849.1 | FOXO6 | MA0849.1 | GTAAACA | FOXO6 | 9.85e-07 | 5.57e-09 | 23700 | 32.9 | 21300 | 31.4 | 1.05 | DNR_DAR_24hr |
| 112 | 12 | MA0845.1 | FOXB1 | MA0845.1 | WATGTAAATAT | FOXB1 | 1.04e-06 | 5.81e-09 | 37800 | 52.6 | 34700 | 51 | 1.03 | DNR_DAR_24hr |
| 113 | 10 | MA0774.1 | MEIS2 | MA0774.1 | TTGACAGS | MEIS2 | 1.11e-06 | 6.14e-09 | 29300 | 40.7 | 26600 | 39.1 | 1.04 | DNR_DAR_24hr |
| 114 | 21 | MA0037.4 | Gata3 | MA0037.4 | NHTCTTATCTNH | Gata3 | 1.23e-06 | 6.78e-09 | 8510 | 11.8 | 7360 | 10.8 | 1.09 | DNR_DAR_24hr |
| 115 | 5 | MA0704.1 | Lhx4 | MA0704.1 | YTAATTAR | Lhx4 | 1.47e-06 | 8.02e-09 | 60800 | 84.5 | 56700 | 83.4 | 1.01 | DNR_DAR_24hr |
| 116 | 1 | MA1645.1 | NKX2-2 | MA1645.1 | NNNCCACTCAANNN | NKX2-2 | 2.02e-06 | 1.1e-08 | 56700 | 78.8 | 52700 | 77.5 | 1.02 | DNR_DAR_24hr |
| 117 | 12 | MA0846.1 | FOXC2 | MA0846.1 | WAWGTAAACAWW | FOXC2 | 2.09e-06 | 1.12e-08 | 49200 | 68.3 | 45400 | 66.8 | 1.02 | DNR_DAR_24hr |
| 118 | 22 | MA0084.1 | SRY | MA0084.1 | NWWAACAAT | SRY | 2.23e-06 | 1.19e-08 | 65900 | 91.5 | 61600 | 90.6 | 1.01 | DNR_DAR_24hr |
| 119 | 12 | MA0480.2 | Foxo1 | MA0480.2 | NDGTAAACANN | Foxo1 | 2.28e-06 | 1.2e-08 | 67100 | 93.2 | 62800 | 92.3 | 1.01 | DNR_DAR_24hr |
| 120 | 17 | MA1523.1 | MSANTD3 | MA1523.1 | STVCACTCAC | MSANTD3 | 2.39e-06 | 1.25e-08 | 61000 | 84.7 | 56800 | 83.6 | 1.01 | DNR_DAR_24hr |
| 121 | 5 | MA0720.1 | Shox2 | MA0720.1 | YYAATTAR | Shox2 | 3.05e-06 | 1.58e-08 | 46000 | 63.9 | 42400 | 62.4 | 1.02 | DNR_DAR_24hr |
| 122 | 9 | MA0672.1 | NKX2-3 | MA0672.1 | VCCACTTRAV | NKX2-3 | 3.57e-06 | 1.84e-08 | 7920 | 11 | 6840 | 10 | 1.09 | DNR_DAR_24hr |
| 123 | 20 | MA1644.1 | NFYC | MA1644.1 | RDCCAATCASN | NFYC | 3.62e-06 | 1.85e-08 | 53200 | 73.9 | 49400 | 72.6 | 1.02 | DNR_DAR_24hr |
| 124 | 23 | MA1720.1 (ZNF85) | STREME-8 | 8-TGATGAAATCTCAT | TGATGAAATCTCAT | ZNF85 | 5.09e-06 | 2.58e-08 | 232 | 0.32 | 116 | 0.17 | 1.88 | DNR_DAR_24hr |
| 125 | 18 | MA1978.1 | ZNF354A | MA1978.1 | HWAADWATAAATGRAYWAWTT | ZNF354A | 5.85e-06 | 2.94e-08 | 60200 | 83.6 | 56100 | 82.5 | 1.01 | DNR_DAR_24hr |
| 126 | 15 | MA0789.1 | POU3F4 | MA0789.1 | TATGCWAAT | POU3F4 | 7.36e-06 | 3.67e-08 | 47400 | 65.8 | 43800 | 64.4 | 1.02 | DNR_DAR_24hr |
| 127 | 5 | MA0654.1 | ISX | MA0654.1 | YYAATTAR | ISX | 8.5e-06 | 4.2e-08 | 43800 | 60.8 | 40400 | 59.3 | 1.02 | DNR_DAR_24hr |
| 128 | 24 | MA1573.2 | Thap11 | MA1573.2 | ACTACAABTCCCAG | Thap11 | 9.57e-06 | 4.69e-08 | 251 | 0.35 | 131 | 0.19 | 1.8 | DNR_DAR_24hr |
| 129 | 25 | 12-CCMATKGG | STREME-12 | 12-CCMATKGG | 12-CCMATKGG | CCMATKGG | 9.78e-06 | 4.76e-08 | 510 | 0.71 | 325 | 0.48 | 1.48 | DNR_DAR_24hr |
| 130 | 21 | MA1970.1 | TRPS1 | MA1970.1 | NHTCTTATCTNH | TRPS1 | 1.22e-05 | 5.88e-08 | 14000 | 19.5 | 12500 | 18.3 | 1.06 | DNR_DAR_24hr |
| 131 | 14 | MA0595.1 | SREBF1 | MA0595.1 | VTCACCCCAY | SREBF1 | 1.47e-05 | 7.02e-08 | 13700 | 19 | 12200 | 17.9 | 1.06 | DNR_DAR_24hr |
| 132 | 26 | MA0650.3 | Hoxa13 | MA0650.3 | NNCCAATAAAAN | Hoxa13 | 1.49e-05 | 7.09e-08 | 63500 | 88.2 | 59300 | 87.2 | 1.01 | DNR_DAR_24hr |
| 133 | 12 | MA0157.3 | Foxo3 | MA0157.3 | NDGTAAACARNN | Foxo3 | 1.57e-05 | 7.37e-08 | 68000 | 94.4 | 63800 | 93.8 | 1.01 | DNR_DAR_24hr |
| 134 | 10 | MA0842.2 | NRL | MA0842.2 | AAWWNTGCTGACG | NRL | 1.57e-05 | 7.37e-08 | 10600 | 14.8 | 9350 | 13.8 | 1.07 | DNR_DAR_24hr |
| 135 | 27 | MA0803.1 | TBX15 | MA0803.1 | AGGTGTGA | TBX15 | 2.09e-05 | 9.72e-08 | 32600 | 45.3 | 29800 | 43.8 | 1.03 | DNR_DAR_24hr |
| 136 | 27 | MA0689.1 | TBX20 | MA0689.1 | WAGGTGTGAAR | TBX20 | 2.2e-05 | 1.02e-07 | 44800 | 62.2 | 41400 | 60.8 | 1.02 | DNR_DAR_24hr |
| 137 | 5 | MA1960.1 | MGA::EVX1 | MA1960.1 | RGGTGWTAATKD | MGA::EVX1 | 2.51e-05 | 1.15e-07 | 53300 | 74.1 | 49500 | 72.8 | 1.02 | DNR_DAR_24hr |
| 138 | 17 | MA1114.1 | PBX3 | MA1114.1 | NNNTGAGTGACAGGBNN | PBX3 | 2.59e-05 | 1.18e-07 | 29900 | 41.6 | 27300 | 40.1 | 1.04 | DNR_DAR_24hr |
| 139 | 15 | MA0627.2 | POU2F3 | MA0627.2 | NWTATGCAAATNH | POU2F3 | 2.6e-05 | 1.18e-07 | 52000 | 72.3 | 48300 | 71 | 1.02 | DNR_DAR_24hr |
| 140 | 12 | MA0614.1 | Foxj2 | MA0614.1 | RTAAACAA | Foxj2 | 4.31e-05 | 1.93e-07 | 39500 | 54.9 | 36400 | 53.5 | 1.03 | DNR_DAR_24hr |
| 141 | 17 | MA1124.1 | ZNF24 | MA1124.1 | CATTCATTCATTC | ZNF24 | 4.53e-05 | 2.01e-07 | 5130 | 7.13 | 4360 | 6.41 | 1.11 | DNR_DAR_24hr |
| 142 | 5 | MA1481.1 | DRGX | MA1481.1 | YYAATTAN | DRGX | 5.65e-05 | 2.5e-07 | 40900 | 56.9 | 37700 | 55.5 | 1.03 | DNR_DAR_24hr |
| 143 | 5 | MA0709.1 | Msx3 | MA0709.1 | SCAATTAN | Msx3 | 5.85e-05 | 2.57e-07 | 42300 | 58.7 | 39000 | 57.3 | 1.02 | DNR_DAR_24hr |
| 144 | 21 | MA0482.2 | GATA4 | MA0482.2 | NNCCTTATCTNH | GATA4 | 5.92e-05 | 2.58e-07 | 3950 | 5.48 | 3300 | 4.86 | 1.13 | DNR_DAR_24hr |
| 145 | 28 | MA0073.1 | RREB1 | MA0073.1 | CCCCMAAMCAMCCMCMMMCV | RREB1 | 6.42e-05 | 2.78e-07 | 439 | 0.61 | 278 | 0.41 | 1.49 | DNR_DAR_24hr |
| 146 | 5 | MA0661.1 | MEOX1 | MA0661.1 | VSTAATTAHC | MEOX1 | 7.88e-05 | 3.39e-07 | 62600 | 87 | 58500 | 86.1 | 1.01 | DNR_DAR_24hr |
| 147 | 9 | MA0831.3 | TFE3 | MA0831.3 | GTCACGTGAC | TFE3 | 8.54e-05 | 3.65e-07 | 8260 | 11.5 | 7200 | 10.6 | 1.08 | DNR_DAR_24hr |
| 148 | 29 | MA0698.1 | ZBTB18 | MA0698.1 | NATCCAGATGTKB | ZBTB18 | 0.000108 | 4.57e-07 | 25500 | 35.4 | 23200 | 34.1 | 1.04 | DNR_DAR_24hr |
| 149 | 18 | MA0914.1 | ISL2 | MA0914.1 | GCAMTTAR | ISL2 | 0.000135 | 5.69e-07 | 66600 | 92.6 | 62500 | 91.8 | 1.01 | DNR_DAR_24hr |
| 150 | 30 | MA0090.3 | TEAD1 | MA0090.3 | NNACATTCCAGSN | TEAD1 | 0.000148 | 6.19e-07 | 58200 | 80.8 | 54200 | 79.7 | 1.01 | DNR_DAR_24hr |
| 151 | 12 | MA0031.1 | FOXD1 | MA0031.1 | GTAAACAW | FOXD1 | 0.000152 | 6.3e-07 | 49800 | 69.2 | 46200 | 68 | 1.02 | DNR_DAR_24hr |
| 152 | 11 | MA0018.4 | CREB1 | MA0018.4 | NNRTGATGTCAYN | CREB1 | 0.000155 | 6.41e-07 | 23800 | 33 | 21600 | 31.8 | 1.04 | DNR_DAR_24hr |
| 153 | 18 | MA2003.1 | NKX2-4 | MA2003.1 | CCACTTSAVV | NKX2-4 | 0.000159 | 6.53e-07 | 8440 | 11.7 | 7390 | 10.9 | 1.08 | DNR_DAR_24hr |
| 154 | 28 | MA1107.2 | KLF9 | MA1107.2 | MNGCCACACCCACHYV | KLF9 | 0.000173 | 7.04e-07 | 10500 | 14.5 | 9250 | 13.6 | 1.07 | DNR_DAR_24hr |
| 155 | 21 | MA0035.4 | GATA1 | MA0035.4 | NHCTAATCTDH | GATA1 | 0.000178 | 7.19e-07 | 36600 | 50.8 | 33600 | 49.4 | 1.03 | DNR_DAR_24hr |
| 156 | 27 | MA0802.1 | TBR1 | MA0802.1 | AGGTGTGAAA | TBR1 | 0.000231 | 9.28e-07 | 45800 | 63.6 | 42300 | 62.2 | 1.02 | DNR_DAR_24hr |
| 157 | 5 | MA0666.2 | MSX1 | MA0666.2 | SCAATTAR | MSX1 | 0.000253 | 1.01e-06 | 44500 | 61.8 | 41100 | 60.5 | 1.02 | DNR_DAR_24hr |
| 158 | 21 | MA1104.2 | GATA6 | MA1104.2 | HWWTCTTATCTNH | GATA6 | 0.00026 | 1.03e-06 | 16100 | 22.3 | 14400 | 21.2 | 1.05 | DNR_DAR_24hr |
| 159 | 31 | MA0790.1 | POU4F1 | MA0790.1 | ATGMATAATTAATG | POU4F1 | 0.000272 | 1.07e-06 | 28200 | 39.2 | 25800 | 37.9 | 1.03 | DNR_DAR_24hr |
| 160 | 5 | MA0880.1 | Dlx3 | MA0880.1 | NYAATTAN | Dlx3 | 0.000282 | 1.11e-06 | 68600 | 95.3 | 64400 | 94.7 | 1.01 | DNR_DAR_24hr |
| 161 | 5 | MA0882.1 | DLX6 | MA0882.1 | VYAATTAN | DLX6 | 0.000404 | 1.57e-06 | 67800 | 94.1 | 63600 | 93.5 | 1.01 | DNR_DAR_24hr |
| 162 | 10 | MA0495.3 | MAFF | MA0495.3 | DWGTCAGCATTTTWHN | MAFF | 0.000416 | 1.61e-06 | 60900 | 84.6 | 56900 | 83.6 | 1.01 | DNR_DAR_24hr |
| 163 | 5 | MA0887.1 | EVX1 | MA0887.1 | VVTAATTABS | EVX1 | 0.00048 | 1.85e-06 | 55400 | 76.9 | 51600 | 75.8 | 1.01 | DNR_DAR_24hr |
| 164 | 5 | MA0726.1 | VSX2 | MA0726.1 | CTAATTAN | VSX2 | 0.000485 | 1.85e-06 | 52500 | 72.9 | 48800 | 71.7 | 1.02 | DNR_DAR_24hr |
| 165 | 32 | MA1562.1 | SOX14 | MA1562.1 | CCGAACAATG | SOX14 | 0.000505 | 1.92e-06 | 52000 | 72.3 | 48400 | 71.1 | 1.02 | DNR_DAR_24hr |
| 166 | 5 | MA0706.1 | MEOX2 | MA0706.1 | DSTAATTAWN | MEOX2 | 0.000523 | 1.98e-06 | 63800 | 88.7 | 59700 | 87.8 | 1.01 | DNR_DAR_24hr |
| 167 | 9 | MA0819.2 | CLOCK | MA0819.2 | NRACACGTGB | CLOCK | 0.000604 | 2.27e-06 | 8540 | 11.9 | 7510 | 11 | 1.07 | DNR_DAR_24hr |
| 168 | 5 | MA0723.2 | VAX2 | MA0723.2 | BYAATTAV | VAX2 | 0.000627 | 2.34e-06 | 69100 | 95.9 | 64900 | 95.4 | 1.01 | DNR_DAR_24hr |
| 169 | 5 | MA0700.2 | LHX2 | MA0700.2 | DDCCAATTADN | LHX2 | 0.000705 | 2.62e-06 | 53800 | 74.7 | 50000 | 73.6 | 1.02 | DNR_DAR_24hr |
| 170 | 12 | MA0851.1 | Foxj3 | MA0851.1 | NDAADGTAAACAAANNM | Foxj3 | 0.000854 | 3.15e-06 | 42000 | 58.3 | 38800 | 57.1 | 1.02 | DNR_DAR_24hr |
| 171 | 33 | MA1974.1 | ZNF211 | MA1974.1 | HWYATATACCAYRYW | ZNF211 | 0.000913 | 3.35e-06 | 1890 | 2.62 | 1520 | 2.23 | 1.18 | DNR_DAR_24hr |
| 172 | 5 | MA0716.1 | PRRX1 | MA0716.1 | YYAATTAR | PRRX1 | 0.00101 | 3.69e-06 | 60600 | 84.1 | 56600 | 83.2 | 1.01 | DNR_DAR_24hr |
| 173 | 31 | MA0791.1 | POU4F3 | MA0791.1 | RTGMATWATTAATGAR | POU4F3 | 0.00103 | 3.73e-06 | 33100 | 46 | 30400 | 44.7 | 1.03 | DNR_DAR_24hr |
| 174 | 5 | MA0674.1 | NKX6-1 | MA0674.1 | NTMATTAA | NKX6-1 | 0.00134 | 4.84e-06 | 62600 | 86.9 | 58500 | 86.1 | 1.01 | DNR_DAR_24hr |
| 175 | 12 | MA0613.1 | FOXG1 | MA0613.1 | RTAAACAW | FOXG1 | 0.00138 | 4.95e-06 | 36000 | 50 | 33200 | 48.8 | 1.03 | DNR_DAR_24hr |
| 176 | 5 | MA0725.1 | VSX1 | MA0725.1 | YTAATTAN | VSX1 | 0.00147 | 5.22e-06 | 59900 | 83.2 | 55900 | 82.2 | 1.01 | DNR_DAR_24hr |
| 177 | 22 | MA0077.1 | SOX9 | MA0077.1 | CYATTGTTY | SOX9 | 0.00147 | 5.22e-06 | 68000 | 94.4 | 63800 | 93.8 | 1.01 | DNR_DAR_24hr |
| 178 | 18 | MA0124.2 | Nkx3-1 | MA0124.2 | RCCACTTAA | Nkx3-1 | 0.00153 | 5.4e-06 | 28300 | 39.4 | 26000 | 38.2 | 1.03 | DNR_DAR_24hr |
| 179 | 27 | MA0806.1 | TBX4 | MA0806.1 | AGGTGTGA | TBX4 | 0.00174 | 6.1e-06 | 8570 | 11.9 | 7560 | 11.1 | 1.07 | DNR_DAR_24hr |
| 180 | 27 | MA0807.1 | TBX5 | MA0807.1 | AGGTGTKA | TBX5 | 0.00237 | 8.25e-06 | 7750 | 10.8 | 6820 | 10 | 1.07 | DNR_DAR_24hr |
| 181 | 18 | MA0673.1 | NKX2-8 | MA0673.1 | VCACTTSAV | NKX2-8 | 0.0025 | 8.68e-06 | 12400 | 17.2 | 11100 | 16.3 | 1.06 | DNR_DAR_24hr |
| 182 | 9 | MA0692.1 | TFEB | MA0692.1 | RYCACGTGAC | TFEB | 0.00287 | 9.89e-06 | 6010 | 8.35 | 5230 | 7.69 | 1.09 | DNR_DAR_24hr |
| 183 | 34 | MA0606.2 | Nfat5 | MA0606.2 | NNATGGAAAAWN | Nfat5 | 0.00295 | 1.01e-05 | 366 | 0.51 | 238 | 0.35 | 1.45 | DNR_DAR_24hr |
| 184 | 5 | MA1500.1 | HOXB6 | MA1500.1 | BTAATKRC | HOXB6 | 0.00299 | 1.02e-05 | 59700 | 83 | 55800 | 82 | 1.01 | DNR_DAR_24hr |
| 185 | 12 | MA0852.2 | FOXK1 | MA0852.2 | NRDGTAAACAAGNN | FOXK1 | 0.00346 | 1.17e-05 | 54200 | 75.3 | 50500 | 74.2 | 1.01 | DNR_DAR_24hr |
| 186 | 5 | MA1476.2 | Dlx5 | MA1476.2 | NDGCAATTAGNN | Dlx5 | 0.00391 | 1.32e-05 | 63700 | 88.4 | 59600 | 87.7 | 1.01 | DNR_DAR_24hr |
| 187 | 5 | MA0648.1 | GSC | MA0648.1 | VYTAATCCSY | GSC | 0.00565 | 1.9e-05 | 55300 | 76.8 | 51600 | 75.8 | 1.01 | DNR_DAR_24hr |
| 188 | 5 | MA0892.1 | GSX1 | MA0892.1 | NCYMATTAVN | GSX1 | 0.00636 | 2.12e-05 | 68900 | 95.6 | 64700 | 95.2 | 1.01 | DNR_DAR_24hr |
| 189 | 27 | MA0805.1 | TBX1 | MA0805.1 | AGGTGTGA | TBX1 | 0.0064 | 2.13e-05 | 24700 | 34.4 | 22600 | 33.3 | 1.03 | DNR_DAR_24hr |
| 190 | 5 | MA0890.1 | GBX2 | MA0890.1 | RSYAATTARN | GBX2 | 0.00758 | 2.5e-05 | 54500 | 75.7 | 50800 | 74.7 | 1.01 | DNR_DAR_24hr |
| 191 | 18 | MA0885.2 | Dlx2 | MA0885.2 | NDGCAATTAGNN | Dlx2 | 0.0079 | 2.59e-05 | 65200 | 90.5 | 61100 | 89.8 | 1.01 | DNR_DAR_24hr |
| 192 | 11 | MA0488.1 | JUN | MA0488.1 | DDRATGATGTCAT | JUN | 0.00826 | 2.7e-05 | 13800 | 19.2 | 12500 | 18.3 | 1.05 | DNR_DAR_24hr |
| 193 | 26 | MA0899.1 | HOXA10 | MA0899.1 | DGYMATAAAAH | HOXA10 | 0.00837 | 2.72e-05 | 60400 | 83.9 | 56500 | 83 | 1.01 | DNR_DAR_24hr |
| 194 | 9 | MA1994.1 | Nkx2-1 | MA1994.1 | NVCACTTGARHNN | Nkx2-1 | 0.00905 | 2.93e-05 | 55800 | 77.4 | 52000 | 76.5 | 1.01 | DNR_DAR_24hr |
| 195 | 13 | MA0829.2 | SREBF1 | MA0829.2 | DNATCACSTGATNH | SREBF1 | 0.00991 | 3.19e-05 | 8360 | 11.6 | 7410 | 10.9 | 1.07 | DNR_DAR_24hr |
| 196 | 5 | MA0721.1 | UNCX | MA0721.1 | YYAATTAV | UNCX | 0.0101 | 3.23e-05 | 55200 | 76.6 | 51500 | 75.7 | 1.01 | DNR_DAR_24hr |
| 197 | 22 | MA0143.4 | SOX2 | MA0143.4 | DNACAATGGNN | SOX2 | 0.0119 | 3.79e-05 | 63800 | 88.6 | 59700 | 87.8 | 1.01 | DNR_DAR_24hr |
| 198 | 22 | MA1152.1 | SOX15 | MA1152.1 | CYWTTGTTHW | SOX15 | 0.012 | 3.82e-05 | 67600 | 93.9 | 63500 | 93.4 | 1.01 | DNR_DAR_24hr |
| 199 | 5 | MA1496.1 | HOXA4 | MA1496.1 | RTMATTAV | HOXA4 | 0.013 | 4.1e-05 | 57600 | 80 | 53800 | 79.1 | 1.01 | DNR_DAR_24hr |
| 200 | 12 | MA0850.1 | FOXP3 | MA0850.1 | RTAAACA | FOXP3 | 0.0154 | 4.83e-05 | 63500 | 88.1 | 59400 | 87.4 | 1.01 | DNR_DAR_24hr |
| 201 | 5 | MA1549.1 | POU6F1 | MA1549.1 | RTAATGAGVN | POU6F1 | 0.0159 | 4.98e-05 | 22000 | 30.6 | 20100 | 29.6 | 1.03 | DNR_DAR_24hr |
| 202 | 5 | MA0888.1 | EVX2 | MA0888.1 | VVTAATTAVB | EVX2 | 0.0172 | 5.33e-05 | 60900 | 84.6 | 57000 | 83.8 | 1.01 | DNR_DAR_24hr |
| 203 | 31 | MA0153.2 | HNF1B | MA0153.2 | GTTAATNATTAAY | HNF1B | 0.0178 | 5.52e-05 | 17700 | 24.6 | 16100 | 23.6 | 1.04 | DNR_DAR_24hr |
| 204 | 5 | MA0889.1 | GBX1 | MA0889.1 | VCYAATTASY | GBX1 | 0.0208 | 6.4e-05 | 51400 | 71.4 | 47900 | 70.4 | 1.01 | DNR_DAR_24hr |
| 205 | 12 | MA0041.2 | FOXD3 | MA0041.2 | WAWGHAAATAAACARY | FOXD3 | 0.0211 | 6.47e-05 | 46900 | 65.2 | 43600 | 64.1 | 1.02 | DNR_DAR_24hr |
| 206 | 9 | MA1620.1 | Ptf1A | MA1620.1 | NVACACCTGTNN | Ptf1A | 0.0233 | 7.1e-05 | 43100 | 59.8 | 40000 | 58.8 | 1.02 | DNR_DAR_24hr |
| 207 | 9 | MA0664.1 | MLXIPL | MA0664.1 | RTCACGTGAT | MLXIPL | 0.0246 | 7.45e-05 | 18000 | 24.9 | 16300 | 24 | 1.04 | DNR_DAR_24hr |
| 208 | 10 | MA0775.1 | MEIS3 | MA0775.1 | DTGACAGS | MEIS3 | 0.0261 | 7.87e-05 | 2490 | 3.46 | 2090 | 3.07 | 1.12 | DNR_DAR_24hr |
| 209 | 12 | MA0030.1 | FOXF2 | MA0030.1 | BNAASGTAAACAAD | FOXF2 | 0.028 | 8.41e-05 | 7950 | 11 | 7060 | 10.4 | 1.06 | DNR_DAR_24hr |
| 210 | 5 | MA0793.1 | POU6F2 | MA0793.1 | ASCTMATTAW | POU6F2 | 0.04 | 0.00012 | 32700 | 45.5 | 30200 | 44.4 | 1.02 | DNR_DAR_24hr |
| 16 | 35 | MA1125.1 (ZNF384) | MEME-1 | AAAAAAAAAAAAAAA | AAAAAAAAAAAAAAA | ZNF384 | 8.9 | 0.556 | 2650 | 3.68 | 2500 | 3.68 | 1 | DNR_DAR_24hr |
| 17 | 36 | MA1723.1 (PRDM9) | MEME-4 | CYCYYCCTCCYTCYC | CYCYYCCTCCYTCYC | PRDM9 | 16.5 | 0.968 | 6 | 0.01 | 10 | 0.01 | 0.601 | DNR_DAR_24hr |
| 18 | 36 | MA1961.1 (PATZ1) | MEME-3 | GSGGSSGGGGCGGGG | GSGGSSGGGGCGGGG | PATZ1 | 17.9 | 0.996 | 55 | 0.08 | 81 | 0.12 | 0.645 | DNR_DAR_24hr |
ER_rat <- 1.25
mrc_type <- "DNR_DAR_24hr"
DNR_DAR_24hr_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,mrc_type,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks(200bp)Enrichment ratio:",ER_rat))
DNR_DAR_24hr_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,mrc_type,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
dplyr::filter(EVALUE.y<0.05) %>%
slice_head(n=5) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*.8), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./.8,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks(200bp)Enrichment ratio:",ER_rat))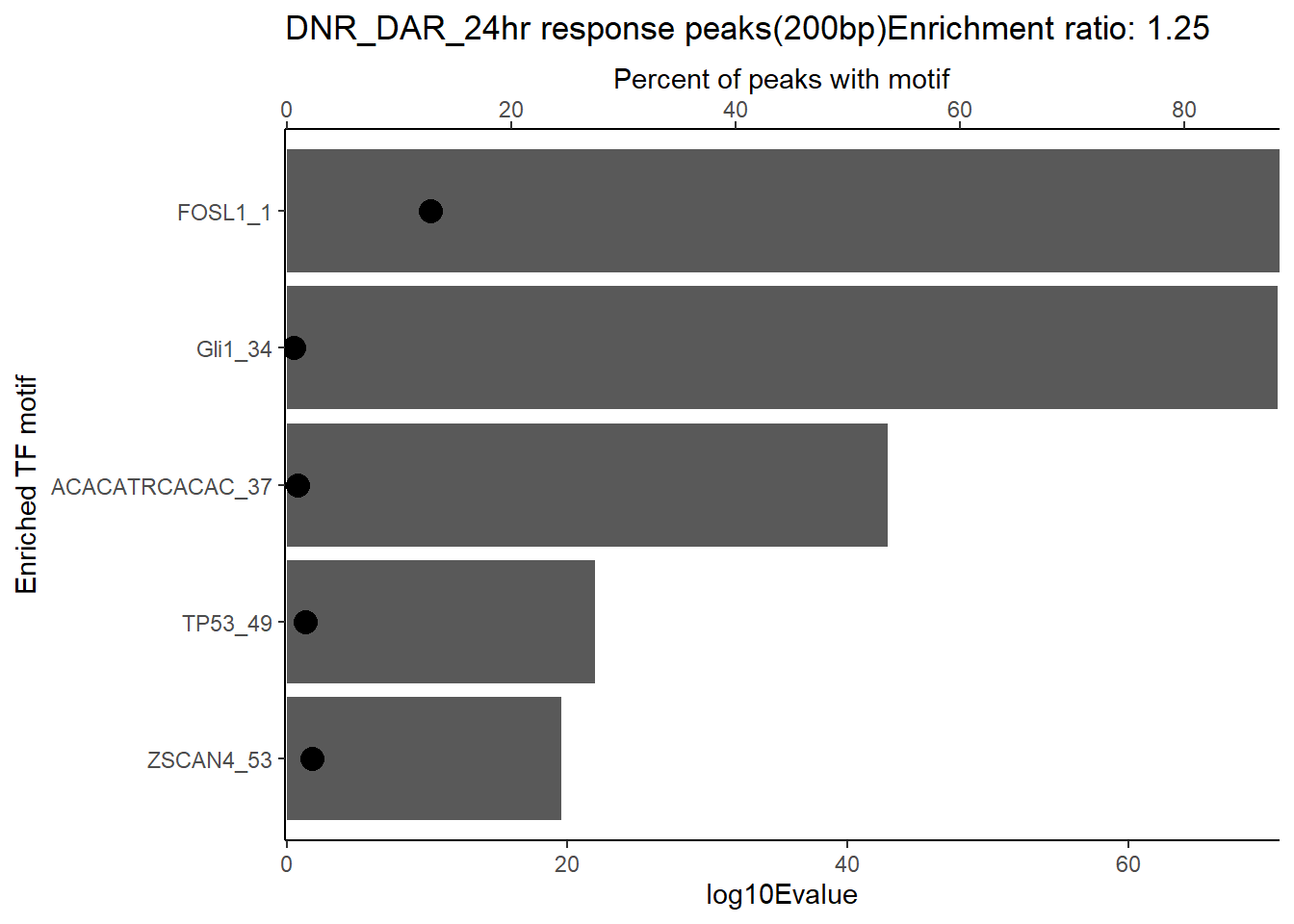
DNR_DAR 24 2kb data
DNR_242kb_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DNR_24_2kb_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
DNR_242kb_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DNR_24_2kb_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
DNR_242kb_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DNR_24_2kb_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
DNR_242kb_sea_all <- rbind(DNR_242kb_sea_disc, DNR_242kb_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)DNR_242kb_combo <- DNR_242kb_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., DNR_242kb_sea_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ SIM_MOTIF
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y)
DNR_242kb_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs 200 bp in DNR_24 2kb") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | MA1134.1 (FOS::JUNB) | STREME-1 | 1-DATGASTCATH | DATGASTCATH | FOS::JUNB | 2.11e-78 | 9.15e-79 | 1020 | 6.3 | 362 | 2.16 | 2.91 |
| 2 | 1 | MA0099.3 | FOS::JUN | MA0099.3 | ATGAGTCAYM | FOS::JUN | 6.33e-71 | 1.38e-71 | 1380 | 8.5 | 630 | 3.76 | 2.26 |
| 3 | 1 | MA1130.1 | FOSL2::JUN | MA1130.1 | NNRTGAGTCAYN | FOSL2::JUN | 2.42e-68 | 3.51e-69 | 1240 | 7.63 | 543 | 3.24 | 2.35 |
| 4 | 1 | MA1988.1 | Atf3 | MA1988.1 | NRTGACTCABN | Atf3 | 8.3e-67 | 9.02e-68 | 1500 | 9.25 | 738 | 4.4 | 2.1 |
| 5 | 1 | MA1128.1 | FOSL1::JUN | MA1128.1 | NKATGACTCATNN | FOSL1::JUN | 3.82e-65 | 3.32e-66 | 961 | 5.94 | 371 | 2.21 | 2.68 |
| 6 | 1 | MA0462.2 | BATF::JUN | MA0462.2 | DATGACTCATH | BATF::JUN | 7.69e-65 | 5.57e-66 | 1020 | 6.29 | 409 | 2.44 | 2.57 |
| 7 | 1 | MA1634.1 | BATF | MA1634.1 | DATGACTCATH | BATF | 3.05e-64 | 1.9e-65 | 1020 | 6.33 | 416 | 2.48 | 2.55 |
| 8 | 1 | MA1141.1 | FOS::JUND | MA1141.1 | NKATGAGTCATNN | FOS::JUND | 1.13e-63 | 6.15e-65 | 911 | 5.63 | 344 | 2.05 | 2.74 |
| 9 | 1 | MA0489.2 | Jun | MA0489.2 | NVTGACTCATNN | Jun | 2.85e-63 | 1.38e-64 | 1140 | 7.06 | 501 | 2.99 | 2.36 |
| 10 | 1 | MA0491.2 | JUND | MA0491.2 | NNATGACTCATNN | JUND | 1.24e-61 | 5.38e-63 | 1200 | 7.44 | 551 | 3.29 | 2.26 |
| 11 | 1 | MA0835.2 | BATF3 | MA0835.2 | DATGACTCATH | BATF3 | 3.25e-61 | 1.28e-62 | 1010 | 6.26 | 420 | 2.51 | 2.49 |
| 12 | 1 | MA0477.2 (FOSL1) | MEME-6 | TGAGTCAT | TGAGTCAT | FOSL1 | 1.58e-59 | 5.73e-61 | 1270 | 7.83 | 606 | 3.62 | 2.16 |
| 13 | 1 | MA1137.1 | FOSL1::JUNB | MA1137.1 | NDRTGACTCATNN | FOSL1::JUNB | 1.41e-58 | 4.7e-60 | 1090 | 6.75 | 486 | 2.9 | 2.32 |
| 14 | 1 | MA0490.2 | JUNB | MA0490.2 | NNATGACTCATNN | JUNB | 3.01e-58 | 9.34e-60 | 837 | 5.17 | 316 | 1.89 | 2.74 |
| 15 | 1 | MA1134.1 | FOS::JUNB | MA1134.1 | KATGASTCATHN | FOS::JUNB | 7.97e-58 | 2.31e-59 | 715 | 4.42 | 241 | 1.44 | 3.06 |
| 16 | 1 | MA0477.2 | FOSL1 | MA0477.2 | NNATGACTCATNN | FOSL1 | 2.37e-57 | 6.44e-59 | 796 | 4.92 | 293 | 1.75 | 2.81 |
| 17 | 1 | MA1633.2 | BACH1 | MA1633.2 | ATGACTCAT | BACH1 | 6.46e-57 | 1.65e-58 | 874 | 5.4 | 345 | 2.06 | 2.62 |
| 18 | 1 | MA0476.1 | FOS | MA0476.1 | DVTGASTCATB | FOS | 1.3e-55 | 3.13e-57 | 750 | 4.64 | 270 | 1.61 | 2.87 |
| 19 | 1 | MA1138.1 | FOSL2::JUNB | MA1138.1 | KRTGASTCAT | FOSL2::JUNB | 2.63e-53 | 6.02e-55 | 699 | 4.32 | 246 | 1.47 | 2.94 |
| 20 | 1 | MA1101.2 | BACH2 | MA1101.2 | DWANCATGASTCATSNTWH | BACH2 | 5.48e-52 | 1.19e-53 | 977 | 6.04 | 435 | 2.6 | 2.32 |
| 21 | 1 | MA1135.1 | FOSB::JUNB | MA1135.1 | KRTGASTCAT | FOSB::JUNB | 1.21e-50 | 2.5e-52 | 713 | 4.41 | 264 | 1.58 | 2.79 |
| 22 | 1 | MA0841.1 | NFE2 | MA0841.1 | VATGACTCATS | NFE2 | 4.44e-50 | 8.77e-52 | 1140 | 7.03 | 559 | 3.34 | 2.11 |
| 23 | 1 | MA0655.1 | JDP2 | MA0655.1 | ATGACTCAT | JDP2 | 5.29e-50 | 9.99e-52 | 785 | 4.85 | 313 | 1.87 | 2.59 |
| 24 | 1 | MA0478.1 | FOSL2 | MA0478.1 | KRRTGASTCAB | FOSL2 | 2.65e-49 | 4.8e-51 | 755 | 4.67 | 296 | 1.77 | 2.64 |
| 25 | 1 | MA1144.1 | FOSL2::JUND | MA1144.1 | KATGACTCAT | FOSL2::JUND | 8.4e-49 | 1.46e-50 | 660 | 4.08 | 237 | 1.41 | 2.88 |
| 26 | 1 | MA1142.1 | FOSL1::JUND | MA1142.1 | NRTGACTCAT | FOSL1::JUND | 4.52e-43 | 7.55e-45 | 848 | 5.24 | 385 | 2.3 | 2.28 |
| 27 | 1 | MA1132.1 | JUN::JUNB | MA1132.1 | KATGACKCAT | JUN::JUNB | 2.45e-37 | 3.94e-39 | 584 | 3.61 | 230 | 1.37 | 2.62 |
| 28 | 1 | 3-AGACATM | STREME-3 | 3-AGACATM | AGACATM | AGACATM | 4.44e-29 | 6.9e-31 | 3880 | 24 | 3130 | 18.7 | 1.28 |
| 29 | 1 | MA1928.1 | BNC2 | MA1928.1 | NDRTGAGTCABN | BNC2 | 8.36e-28 | 1.25e-29 | 448 | 2.77 | 178 | 1.06 | 2.6 |
| 30 | 1 | MA0089.2 | MAFG::NFE2L1 | MA0089.2 | NVNATGACTCAGCADW | MAFG::NFE2L1 | 8.29e-27 | 1.2e-28 | 1420 | 8.78 | 935 | 5.58 | 1.57 |
| 31 | 1 | MA0496.3 | MAFK | MA0496.3 | NGMTGACTCAGCMNH | MAFK | 1.82e-25 | 2.55e-27 | 3240 | 20 | 2580 | 15.4 | 1.3 |
| 32 | 2 | MA0502.2 (NFYB) | STREME-4 | 4-CATTGGC | CATTGGC | NFYB | 3.72e-20 | 5.05e-22 | 6400 | 39.5 | 5750 | 34.3 | 1.15 |
| 33 | 1 | MA0150.2 | Nfe2l2 | MA0150.2 | CASNATGACTCAGCA | Nfe2l2 | 1.02e-19 | 1.34e-21 | 4410 | 27.3 | 3790 | 22.6 | 1.21 |
| 34 | 1 | MA0501.1 | MAF::NFE2 | MA0501.1 | ATGACTCAGCANWTY | MAF::NFE2 | 5.04e-19 | 6.45e-21 | 3400 | 21 | 2830 | 16.9 | 1.24 |
| 35 | 3 | MA0705.1 | Lhx8 | MA0705.1 | CTAATTAV | Lhx8 | 8.08e-19 | 1e-20 | 7600 | 47 | 7000 | 41.8 | 1.12 |
| 36 | 3 | MA0904.2 | HOXB5 | MA0904.2 | STAATTAS | HOXB5 | 8.36e-18 | 1.01e-19 | 8680 | 53.7 | 8140 | 48.6 | 1.11 |
| 37 | 4 | 7-CATGTCA | STREME-7 | 7-CATGTCA | CATGTCA | CATGTCA | 1.28e-17 | 1.5e-19 | 8090 | 50 | 7530 | 45 | 1.11 |
| 38 | 3 | MA1530.1 | NKX6-3 | MA1530.1 | VYCATTAWW | NKX6-3 | 1.09e-16 | 1.25e-18 | 7520 | 46.5 | 6970 | 41.6 | 1.12 |
| 39 | 3 | MA1519.1 | LHX5 | MA1519.1 | STAATTAV | LHX5 | 6.75e-16 | 7.52e-18 | 4830 | 29.8 | 4270 | 25.5 | 1.17 |
| 40 | 5 | MA0774.1 (MEIS2) | STREME-2 | 2-CTGTCA | CTGTCA | MEIS2 | 1.55e-15 | 1.68e-17 | 11900 | 73.5 | 11600 | 69.2 | 1.06 |
| 41 | 3 | MA1495.1 | HOXA1 | MA1495.1 | STAATTAS | HOXA1 | 2.42e-15 | 2.57e-17 | 7320 | 45.3 | 6790 | 40.5 | 1.12 |
| 42 | 2 | MA1644.1 | NFYC | MA1644.1 | RDCCAATCASN | NFYC | 3.37e-15 | 3.49e-17 | 9710 | 60 | 9270 | 55.3 | 1.08 |
| 43 | 3 | MA0707.2 | MNX1 | MA0707.2 | VYRATTAK | MNX1 | 1.3e-14 | 1.32e-16 | 6810 | 42.1 | 6290 | 37.6 | 1.12 |
| 44 | 6 | MA1107.2 (KLF9) | STREME-8 | 8-GTGTGTGTGYATG | GTGTGTGTGYATG | KLF9 | 1.78e-14 | 1.76e-16 | 308 | 1.9 | 140 | 0.84 | 2.27 |
| 45 | 5 | MA0782.2 | PKNOX1 | MA0782.2 | NNTGAGTGACAGVNN | PKNOX1 | 2.08e-14 | 2.01e-16 | 7740 | 47.9 | 7250 | 43.2 | 1.11 |
| 46 | 7 | MA1990.1 (Gli1) | STREME-5 | 5-ACACACCACACAC | ACACACCACACAC | Gli1 | 3.05e-14 | 2.88e-16 | 124 | 0.77 | 27 | 0.16 | 4.62 |
| 47 | 3 | MA0900.2 | HOXA2 | MA0900.2 | STAATTAS | HOXA2 | 3.81e-13 | 3.52e-15 | 4480 | 27.7 | 3980 | 23.8 | 1.16 |
| 48 | 3 | MA0661.1 | MEOX1 | MA0661.1 | VSTAATTAHC | MEOX1 | 4.6e-13 | 4.17e-15 | 5770 | 35.7 | 5280 | 31.5 | 1.13 |
| 49 | 8 | MA0620.3 | MITF | MA0620.3 | NNNDRTCACGTGAYHNNN | MITF | 5.47e-13 | 4.85e-15 | 8140 | 50.3 | 7690 | 45.9 | 1.1 |
| 50 | 1 | MA0659.3 | Mafg | MA0659.3 | NWGMTGACTCAGCAN | Mafg | 1.04e-12 | 9.02e-15 | 8700 | 53.8 | 8280 | 49.4 | 1.09 |
| 51 | 3 | MA0666.2 | MSX1 | MA0666.2 | SCAATTAR | MSX1 | 2.2e-12 | 1.87e-14 | 9450 | 58.4 | 9070 | 54.1 | 1.08 |
| 52 | 3 | MA0027.2 | EN1 | MA0027.2 | SYAATTAV | EN1 | 2.55e-12 | 2.13e-14 | 9580 | 59.2 | 9210 | 55 | 1.08 |
| 53 | 3 | MA0706.1 | MEOX2 | MA0706.1 | DSTAATTAWN | MEOX2 | 3.06e-12 | 2.51e-14 | 7960 | 49.2 | 7520 | 44.9 | 1.1 |
| 54 | 8 | MA0093.3 | USF1 | MA0093.3 | NDGTCATGTGACHN | USF1 | 5.32e-12 | 4.28e-14 | 10600 | 65.6 | 10300 | 61.5 | 1.07 |
| 55 | 9 | MA0077.1 | SOX9 | MA0077.1 | CYATTGTTY | SOX9 | 7.03e-12 | 5.56e-14 | 8060 | 49.8 | 7640 | 45.6 | 1.09 |
| 56 | 3 | MA0704.1 | Lhx4 | MA0704.1 | YTAATTAR | Lhx4 | 7.27e-12 | 5.65e-14 | 9370 | 57.9 | 9000 | 53.7 | 1.08 |
| 57 | 2 | MA0060.3 | NFYA | MA0060.3 | RRCCAATCAGM | NFYA | 2e-11 | 1.53e-13 | 9410 | 58.2 | 9050 | 54 | 1.08 |
| 58 | 3 | MA0630.1 | SHOX | MA0630.1 | HTAATTRR | SHOX | 2.72e-11 | 2.04e-13 | 8960 | 55.4 | 8590 | 51.2 | 1.08 |
| 59 | 3 | MA1502.1 | HOXB8 | MA1502.1 | GYMATTAM | HOXB8 | 3.23e-11 | 2.38e-13 | 7030 | 43.5 | 6600 | 39.4 | 1.1 |
| 60 | 3 | MA1549.1 | POU6F1 | MA1549.1 | RTAATGAGVN | POU6F1 | 4.74e-11 | 3.4e-13 | 6910 | 42.7 | 6480 | 38.7 | 1.1 |
| 61 | 10 | MA0142.1 | Pou5f1::Sox2 | MA0142.1 | CWTTGTYATGCAAAT | Pou5f1::Sox2 | 4.81e-11 | 3.4e-13 | 10100 | 62.5 | 9800 | 58.5 | 1.07 |
| 62 | 3 | MA0902.2 | HOXB2 | MA0902.2 | SYAATTAS | HOXB2 | 4.85e-11 | 3.4e-13 | 6420 | 39.7 | 5990 | 35.7 | 1.11 |
| 63 | 3 | MA0718.1 | RAX | MA0718.1 | RYYAATTARH | RAX | 5.03e-11 | 3.47e-13 | 8820 | 54.6 | 8460 | 50.5 | 1.08 |
| 64 | 3 | MA0726.1 | VSX2 | MA0726.1 | CTAATTAN | VSX2 | 7.83e-11 | 5.26e-13 | 8890 | 54.9 | 8530 | 50.9 | 1.08 |
| 65 | 3 | MA1481.1 | DRGX | MA1481.1 | YYAATTAN | DRGX | 7.86e-11 | 5.26e-13 | 7510 | 46.4 | 7100 | 42.4 | 1.1 |
| 66 | 3 | MA0725.1 | VSX1 | MA0725.1 | YTAATTAN | VSX1 | 9.17e-11 | 6.04e-13 | 5970 | 36.9 | 5540 | 33 | 1.12 |
| 67 | 8 | MA0626.1 | Npas2 | MA0626.1 | NSCACGTGTN | Npas2 | 1.23e-10 | 8.01e-13 | 5470 | 33.8 | 5040 | 30 | 1.12 |
| 68 | 3 | MA0888.1 | EVX2 | MA0888.1 | VVTAATTAVB | EVX2 | 1.44e-10 | 9.22e-13 | 8540 | 52.8 | 8170 | 48.8 | 1.08 |
| 69 | 3 | MA1476.2 | Dlx5 | MA1476.2 | NDGCAATTAGNN | Dlx5 | 1.79e-10 | 1.12e-12 | 9750 | 60.2 | 9440 | 56.3 | 1.07 |
| 70 | 3 | MA0882.1 | DLX6 | MA0882.1 | VYAATTAN | DLX6 | 1.97e-10 | 1.23e-12 | 7920 | 49 | 7540 | 45 | 1.09 |
| 71 | 1 | MA1639.1 | MEIS1 | MA1639.1 | NHCATMAATCAHN | MEIS1 | 2.28e-10 | 1.4e-12 | 3710 | 23 | 3300 | 19.7 | 1.17 |
| 72 | 3 | MA0710.1 | NOTO | MA0710.1 | NYTAATTAVN | NOTO | 2.48e-10 | 1.5e-12 | 7390 | 45.7 | 6990 | 41.7 | 1.09 |
| 73 | 3 | MA0879.2 | DLX1 | MA0879.2 | VYAATTAN | DLX1 | 2.94e-10 | 1.75e-12 | 8590 | 53.1 | 8240 | 49.2 | 1.08 |
| 74 | 11 | MA0063.2 | NKX2-5 | MA0063.2 | NNCACTCAANN | NKX2-5 | 3.57e-10 | 2.1e-12 | 10600 | 65.3 | 10300 | 61.5 | 1.06 |
| 75 | 3 | MA0912.2 | HOXD3 | MA0912.2 | CTAATTAC | HOXD3 | 4.54e-10 | 2.63e-12 | 4140 | 25.6 | 3730 | 22.2 | 1.15 |
| 76 | 3 | MA0709.1 | Msx3 | MA0709.1 | SCAATTAN | Msx3 | 4.89e-10 | 2.8e-12 | 7670 | 47.4 | 7290 | 43.5 | 1.09 |
| 77 | 3 | MA0886.1 | EMX2 | MA0886.1 | VBTAATTARB | EMX2 | 5.01e-10 | 2.83e-12 | 4730 | 29.2 | 4310 | 25.7 | 1.14 |
| 78 | 3 | MA0708.2 | MSX2 | MA0708.2 | SCAATTAV | MSX2 | 5.14e-10 | 2.87e-12 | 10600 | 65.3 | 10300 | 61.5 | 1.06 |
| 79 | 3 | MA0644.2 | ESX1 | MA0644.2 | YYAATTAN | ESX1 | 6.78e-10 | 3.71e-12 | 5280 | 32.6 | 4860 | 29 | 1.12 |
| 80 | 3 | MA0722.1 | VAX1 | MA0722.1 | YTAATTAH | VAX1 | 6.83e-10 | 3.71e-12 | 7480 | 46.2 | 7100 | 42.4 | 1.09 |
| 81 | 12 | 6-CYACAMHCAH | STREME-6 | 6-CYACAMHCAH | CYACAMHCAH | CYACAMHCAH | 8.61e-10 | 4.62e-12 | 3180 | 19.7 | 2800 | 16.7 | 1.18 |
| 82 | 3 | MA0658.1 | LHX6 | MA0658.1 | RCTAATTARY | LHX6 | 9.25e-10 | 4.91e-12 | 7600 | 46.9 | 7220 | 43.1 | 1.09 |
| 83 | 3 | MA0889.1 | GBX1 | MA0889.1 | VCYAATTASY | GBX1 | 1.04e-09 | 5.44e-12 | 8320 | 51.4 | 7960 | 47.5 | 1.08 |
| 84 | 3 | MA0885.2 | Dlx2 | MA0885.2 | NDGCAATTAGNN | Dlx2 | 1.14e-09 | 5.91e-12 | 9200 | 56.9 | 8890 | 53 | 1.07 |
| 85 | 3 | MA0853.1 | Alx4 | MA0853.1 | CGCRYTAATTARYHNNY | Alx4 | 1.64e-09 | 8.37e-12 | 5930 | 36.7 | 5530 | 33 | 1.11 |
| 86 | 8 | MA0018.4 | CREB1 | MA0018.4 | NNRTGATGTCAYN | CREB1 | 1.66e-09 | 8.39e-12 | 9140 | 56.5 | 8820 | 52.6 | 1.07 |
| 87 | 3 | MA0887.1 | EVX1 | MA0887.1 | VVTAATTABS | EVX1 | 1.7e-09 | 8.5e-12 | 7840 | 48.5 | 7480 | 44.6 | 1.09 |
| 88 | 9 | MA1562.1 | SOX14 | MA1562.1 | CCGAACAATG | SOX14 | 3.16e-09 | 1.56e-11 | 7050 | 43.6 | 6670 | 39.8 | 1.09 |
| 89 | 3 | MA1500.1 | HOXB6 | MA1500.1 | BTAATKRC | HOXB6 | 3.35e-09 | 1.64e-11 | 12400 | 76.6 | 12300 | 73.3 | 1.04 |
| 90 | 3 | MA1518.2 | Lhx1 | MA1518.2 | NNNTGCTAATTAGCANNN | Lhx1 | 4.39e-09 | 2.12e-11 | 7760 | 48 | 7410 | 44.2 | 1.08 |
| 91 | 6 | MA1107.2 (KLF9) | MEME-4 | TGTGTGTGTGTRTGT | TGTGTGTGTGTRTGT | KLF9 | 4.93e-09 | 2.36e-11 | 942 | 5.82 | 702 | 4.19 | 1.39 |
| 92 | 13 | MA0070.1 | PBX1 | MA0070.1 | HCATCAATCAAW | PBX1 | 6.49e-09 | 3.07e-11 | 6230 | 38.5 | 5850 | 34.9 | 1.1 |
| 93 | 3 | MA0854.1 | Alx1 | MA0854.1 | CGMRYTAATTARTNMNY | Alx1 | 7.32e-09 | 3.42e-11 | 5180 | 32 | 4800 | 28.6 | 1.12 |
| 94 | 3 | MA0720.1 | Shox2 | MA0720.1 | YYAATTAR | Shox2 | 7.71e-09 | 3.56e-11 | 10200 | 63.3 | 10000 | 59.7 | 1.06 |
| 95 | 3 | MA0675.1 | NKX6-2 | MA0675.1 | NYMATTAA | NKX6-2 | 9.15e-09 | 4.19e-11 | 9300 | 57.5 | 9020 | 53.8 | 1.07 |
| 96 | 3 | MA0642.2 | EN2 | MA0642.2 | VSYAATTA | EN2 | 1.15e-08 | 5.2e-11 | 9280 | 57.4 | 9000 | 53.7 | 1.07 |
| 97 | 16 | MA0746.2 | SP3 | MA0746.2 | NGCCACGCCCMCH | SP3 | 1.17e-08 | 5.26e-11 | 4010 | 24.8 | 3630 | 21.7 | 1.14 |
| 98 | 14 | MA0845.1 | FOXB1 | MA0845.1 | WATGTAAATAT | FOXB1 | 1.37e-08 | 6.08e-11 | 10800 | 67 | 10600 | 63.6 | 1.05 |
| 99 | 15 | MA0842.2 | NRL | MA0842.2 | AAWWNTGCTGACG | NRL | 1.46e-08 | 6.4e-11 | 2960 | 18.3 | 2610 | 15.6 | 1.18 |
| 100 | 3 | MA1505.1 | HOXC8 | MA1505.1 | KTAATTAM | HOXC8 | 1.49e-08 | 6.47e-11 | 11500 | 71.3 | 11400 | 67.9 | 1.05 |
| 101 | 3 | MA0701.2 | LHX9 | MA0701.2 | CYAATTAR | LHX9 | 1.87e-08 | 8.04e-11 | 3380 | 20.9 | 3020 | 18 | 1.16 |
| 102 | 16 | MA0742.2 (KLF12) | STREME-9 | 9-AGGCCCCGCCC | AGGCCCCGCCC | KLF12 | 1.96e-08 | 8.35e-11 | 1200 | 7.43 | 944 | 5.63 | 1.32 |
| 103 | 3 | MA0654.1 | ISX | MA0654.1 | YYAATTAR | ISX | 2.3e-08 | 9.71e-11 | 6610 | 40.9 | 6260 | 37.3 | 1.09 |
| 104 | 3 | MA0158.2 | HOXA5 | MA0158.2 | GYMATTAS | HOXA5 | 2.54e-08 | 1.06e-10 | 4540 | 28.1 | 4170 | 24.9 | 1.13 |
| 105 | 1 | MA1113.2 | PBX2 | MA1113.2 | NHCATAAATCAHN | PBX2 | 3.27e-08 | 1.35e-10 | 6470 | 40 | 6110 | 36.5 | 1.1 |
| 106 | 3 | MA0893.2 | GSX2 | MA0893.2 | SYMATTAR | GSX2 | 3.41e-08 | 1.4e-10 | 5360 | 33.1 | 4990 | 29.8 | 1.11 |
| 107 | 3 | MA0881.1 | Dlx4 | MA0881.1 | VYAATTAN | Dlx4 | 3.49e-08 | 1.42e-10 | 6920 | 42.8 | 6580 | 39.2 | 1.09 |
| 108 | 11 | MA1645.1 | NKX2-2 | MA1645.1 | NNNCCACTCAANNN | NKX2-2 | 4.31e-08 | 1.74e-10 | 15400 | 95.1 | 15700 | 93.4 | 1.02 |
| 109 | 3 | MA0723.2 | VAX2 | MA0723.2 | BYAATTAV | VAX2 | 5.33e-08 | 2.13e-10 | 8370 | 51.8 | 8080 | 48.2 | 1.07 |
| 110 | 3 | MA0721.1 | UNCX | MA0721.1 | YYAATTAV | UNCX | 6.45e-08 | 2.55e-10 | 9660 | 59.7 | 9420 | 56.2 | 1.06 |
| 111 | 16 | MA1107.2 | KLF9 | MA1107.2 | MNGCCACACCCACHYV | KLF9 | 7.52e-08 | 2.94e-10 | 1310 | 8.08 | 1050 | 6.27 | 1.29 |
| 112 | 3 | MA0717.1 | RAX2 | MA0717.1 | YYAATTAV | RAX2 | 7.61e-08 | 2.95e-10 | 9040 | 55.9 | 8780 | 52.4 | 1.07 |
| 113 | 3 | MA0700.2 | LHX2 | MA0700.2 | DDCCAATTADN | LHX2 | 8.78e-08 | 3.38e-10 | 8640 | 53.4 | 8360 | 49.9 | 1.07 |
| 114 | 3 | MA1463.1 | ARGFX | MA1463.1 | DCTAATTARM | ARGFX | 8.9e-08 | 3.39e-10 | 5310 | 32.8 | 4960 | 29.6 | 1.11 |
| 115 | 3 | MA0713.1 | PHOX2A | MA0713.1 | TAATYYAATTA | PHOX2A | 9.99e-08 | 3.78e-10 | 6670 | 41.2 | 6330 | 37.8 | 1.09 |
| 116 | 3 | MA0890.1 | GBX2 | MA0890.1 | RSYAATTARN | GBX2 | 1.02e-07 | 3.82e-10 | 4950 | 30.6 | 4590 | 27.4 | 1.12 |
| 117 | 14 | MA0042.2 | FOXI1 | MA0042.2 | GTAAACA | FOXI1 | 1.53e-07 | 5.68e-10 | 8720 | 53.9 | 8460 | 50.5 | 1.07 |
| 118 | 17 | MA1124.1 | ZNF24 | MA1124.1 | CATTCATTCATTC | ZNF24 | 2.45e-07 | 9.04e-10 | 4850 | 30 | 4500 | 26.9 | 1.11 |
| 119 | 3 | MA0793.1 | POU6F2 | MA0793.1 | ASCTMATTAW | POU6F2 | 2.51e-07 | 9.17e-10 | 7070 | 43.7 | 6760 | 40.3 | 1.08 |
| 120 | 5 | MA1114.1 | PBX3 | MA1114.1 | NNNTGAGTGACAGGBNN | PBX3 | 2.81e-07 | 1.02e-09 | 4930 | 30.5 | 4590 | 27.4 | 1.11 |
| 121 | 14 | MA0032.2 | FOXC1 | MA0032.2 | WAWGTAAAYAW | FOXC1 | 2.84e-07 | 1.02e-09 | 10300 | 63.8 | 10100 | 60.5 | 1.05 |
| 122 | 8 | MA1126.1 | FOS::JUN | MA1126.1 | DRTGACGTCATHNDTN | FOS::JUN | 3.45e-07 | 1.23e-09 | 7170 | 44.3 | 6860 | 41 | 1.08 |
| 123 | 10 | MA1115.1 | POU5F1 | MA1115.1 | WHATGCAAATN | POU5F1 | 3.64e-07 | 1.29e-09 | 5580 | 34.5 | 5240 | 31.3 | 1.1 |
| 124 | 3 | MA0680.2 | Pax7 | MA0680.2 | DWTAATCAATTAWNH | Pax7 | 3.93e-07 | 1.36e-09 | 8150 | 50.4 | 7880 | 47 | 1.07 |
| 125 | 18 | MA0483.1 | Gfi1B | MA0483.1 | AAATCWCWGCH | Gfi1B | 3.93e-07 | 1.36e-09 | 3410 | 21 | 3080 | 18.4 | 1.15 |
| 126 | 11 | MA0122.3 | Nkx3-2 | MA0122.3 | WWAAMCACTTAAN | Nkx3-2 | 3.95e-07 | 1.36e-09 | 6750 | 41.7 | 6430 | 38.4 | 1.09 |
| 127 | 19 | MA1963.1 | SATB1 | MA1963.1 | WWDCTAATAAHWW | SATB1 | 4.66e-07 | 1.6e-09 | 8560 | 52.9 | 8300 | 49.5 | 1.07 |
| 128 | 20 | MA1960.1 | MGA::EVX1 | MA1960.1 | RGGTGWTAATKD | MGA::EVX1 | 5.7e-07 | 1.94e-09 | 10300 | 63.6 | 10100 | 60.4 | 1.05 |
| 129 | 14 | MA1683.1 | FOXA3 | MA1683.1 | DWGTAAACANN | FOXA3 | 6.15e-07 | 2.07e-09 | 9410 | 58.2 | 9190 | 54.8 | 1.06 |
| 130 | 3 | MA0612.2 | EMX1 | MA0612.2 | BTAATTAS | EMX1 | 6.99e-07 | 2.34e-09 | 5120 | 31.6 | 4790 | 28.6 | 1.11 |
| 131 | 3 | MA0634.1 | ALX3 | MA0634.1 | YYYAATTANV | ALX3 | 7.55e-07 | 2.51e-09 | 8580 | 53 | 8330 | 49.7 | 1.07 |
| 132 | 21 | MA0611.2 | Dux | MA0611.2 | NTGATTBAATCAVWNN | Dux | 8.19e-07 | 2.7e-09 | 3760 | 23.2 | 3440 | 20.5 | 1.13 |
| 133 | 9 | MA0143.4 | SOX2 | MA0143.4 | DNACAATGGNN | SOX2 | 1.2e-06 | 3.94e-09 | 11000 | 68.2 | 10900 | 65.1 | 1.05 |
| 134 | 3 | MA0681.2 | PHOX2B | MA0681.2 | WWTAATCAAATTAWWW | PHOX2B | 1.24e-06 | 4.04e-09 | 7220 | 44.6 | 6930 | 41.4 | 1.08 |
| 135 | 22 | MA1974.1 | ZNF211 | MA1974.1 | HWYATATACCAYRYW | ZNF211 | 1.42e-06 | 4.57e-09 | 4180 | 25.8 | 3860 | 23 | 1.12 |
| 136 | 15 | MA0495.3 | MAFF | MA0495.3 | DWGTCAGCATTTTWHN | MAFF | 1.73e-06 | 5.53e-09 | 5770 | 35.6 | 5460 | 32.6 | 1.09 |
| 137 | 3 | MA0880.1 | Dlx3 | MA0880.1 | NYAATTAN | Dlx3 | 1.95e-06 | 6.2e-09 | 6230 | 38.5 | 5930 | 35.4 | 1.09 |
| 138 | 14 | MA0846.1 | FOXC2 | MA0846.1 | WAWGTAAACAWW | FOXC2 | 2.01e-06 | 6.34e-09 | 8370 | 51.7 | 8120 | 48.5 | 1.07 |
| 139 | 11 | MA1523.1 | MSANTD3 | MA1523.1 | STVCACTCAC | MSANTD3 | 2.07e-06 | 6.46e-09 | 13800 | 85.6 | 13900 | 83.2 | 1.03 |
| 140 | 3 | MA0699.1 | LBX2 | MA0699.1 | NCYAATTARN | LBX2 | 2.27e-06 | 7.04e-09 | 10000 | 61.9 | 9840 | 58.7 | 1.05 |
| 141 | 1 | MA1640.1 | MEIS2 | MA1640.1 | NNATGATTTATGDNH | MEIS2 | 2.61e-06 | 8.05e-09 | 6320 | 39.1 | 6030 | 36 | 1.09 |
| 142 | 3 | MA0662.1 | MIXL1 | MA0662.1 | NBYAATTAVN | MIXL1 | 3.03e-06 | 9.26e-09 | 10200 | 63 | 10000 | 59.9 | 1.05 |
| 143 | 10 | MA0785.1 | POU2F1 | MA0785.1 | AWTATGCWAATK | POU2F1 | 3.8e-06 | 1.16e-08 | 9190 | 56.8 | 8990 | 53.6 | 1.06 |
| 144 | 14 | MA0614.1 | Foxj2 | MA0614.1 | RTAAACAA | Foxj2 | 4.44e-06 | 1.34e-08 | 6620 | 40.9 | 6340 | 37.8 | 1.08 |
| 145 | 23 | MA0676.1 | Nr2e1 | MA0676.1 | AAAAGTCAA | Nr2e1 | 5.04e-06 | 1.51e-08 | 6850 | 42.3 | 6570 | 39.2 | 1.08 |
| 146 | 3 | MA0075.3 | PRRX2 | MA0075.3 | CTAATTAN | PRRX2 | 6.48e-06 | 1.93e-08 | 4570 | 28.2 | 4270 | 25.5 | 1.11 |
| 147 | 3 | MA0648.1 | GSC | MA0648.1 | VYTAATCCSY | GSC | 9.84e-06 | 2.91e-08 | 7300 | 45.1 | 7050 | 42 | 1.07 |
| 148 | 14 | MA0848.1 | FOXO4 | MA0848.1 | GTAAACA | FOXO4 | 1.01e-05 | 2.96e-08 | 9860 | 61 | 9710 | 57.9 | 1.05 |
| 149 | 13 | MA0679.2 | ONECUT1 | MA0679.2 | WAAAAATCAATAAWWN | ONECUT1 | 1.21e-05 | 3.54e-08 | 7460 | 46.1 | 7220 | 43.1 | 1.07 |
| 150 | 14 | MA0481.3 | FOXP1 | MA0481.3 | NDGTAAACANH | FOXP1 | 1.43e-05 | 4.16e-08 | 8940 | 55.3 | 8750 | 52.2 | 1.06 |
| 151 | 24 | MA1572.1 | TGIF2LY | MA1572.1 | TGACAGCTGTCA | TGIF2LY | 1.48e-05 | 4.25e-08 | 10200 | 63.3 | 10100 | 60.4 | 1.05 |
| 152 | 25 | MA0025.2 | NFIL3 | MA0025.2 | NRTTATGYAATNN | NFIL3 | 1.52e-05 | 4.35e-08 | 8110 | 50.1 | 7890 | 47.1 | 1.06 |
| 153 | 8 | MA1145.1 | FOSL2::JUND | MA1145.1 | NNRTGACGTCAYHSN | FOSL2::JUND | 1.69e-05 | 4.79e-08 | 5420 | 33.5 | 5140 | 30.6 | 1.09 |
| 154 | 16 | MA1512.1 | KLF11 | MA1512.1 | SCCACGCCCMC | KLF11 | 1.9e-05 | 5.38e-08 | 2790 | 17.3 | 2520 | 15 | 1.15 |
| 155 | 19 | MA0035.4 | GATA1 | MA0035.4 | NHCTAATCTDH | GATA1 | 2.14e-05 | 5.99e-08 | 3330 | 20.6 | 3050 | 18.2 | 1.13 |
| 156 | 19 | MA0712.2 | OTX2 | MA0712.2 | NDRGGATTARNN | OTX2 | 2.24e-05 | 6.24e-08 | 11900 | 73.8 | 11900 | 71.1 | 1.04 |
| 157 | 26 | MA1155.1 | ZSCAN4 | MA1155.1 | TGCACACACTGAAAA | ZSCAN4 | 2.44e-05 | 6.76e-08 | 344 | 2.13 | 225 | 1.34 | 1.58 |
| 158 | 27 | MA0899.1 | HOXA10 | MA0899.1 | DGYMATAAAAH | HOXA10 | 2.87e-05 | 7.88e-08 | 8500 | 52.5 | 8300 | 49.5 | 1.06 |
| 159 | 28 | MA1707.1 | DMRTA1 | MA1707.1 | AWTTGWTACAWT | DMRTA1 | 3e-05 | 8.2e-08 | 9940 | 61.4 | 9800 | 58.5 | 1.05 |
| 160 | 3 | MA0894.1 | HESX1 | MA0894.1 | GHTAATTRRH | HESX1 | 3.05e-05 | 8.29e-08 | 6570 | 40.6 | 6320 | 37.7 | 1.08 |
| 161 | 9 | MA1120.1 | SOX13 | MA1120.1 | DVACAATGGNN | SOX13 | 3.39e-05 | 9.16e-08 | 7190 | 44.4 | 6960 | 41.5 | 1.07 |
| 162 | 29 | MA0461.2 | Atoh1 | MA0461.2 | RMCATATGBY | Atoh1 | 3.61e-05 | 9.68e-08 | 12200 | 75.5 | 12200 | 72.9 | 1.04 |
| 163 | 30 | MA0847.3 | FOXD2 | MA0847.3 | SYTAARYAAACA | FOXD2 | 3.81e-05 | 1.02e-07 | 7300 | 45.1 | 7070 | 42.2 | 1.07 |
| 164 | 31 | MA0790.1 | POU4F1 | MA0790.1 | ATGMATAATTAATG | POU4F1 | 3.9e-05 | 1.03e-07 | 4840 | 29.9 | 4560 | 27.2 | 1.1 |
| 165 | 32 | MA0861.1 | TP73 | MA0861.1 | DRCATGTCNNRACAYGYM | TP73 | 4.12e-05 | 1.09e-07 | 689 | 4.26 | 527 | 3.14 | 1.35 |
| 166 | 30 | MA1487.2 | FOXE1 | MA1487.2 | VYTAAAYAAACAAH | FOXE1 | 4.27e-05 | 1.1e-07 | 7650 | 47.3 | 7430 | 44.3 | 1.07 |
| 167 | 24 | MA1571.1 | TGIF2LX | MA1571.1 | TGACAGCTGTCA | TGIF2LX | 4.28e-05 | 1.1e-07 | 9190 | 56.8 | 9020 | 53.8 | 1.05 |
| 168 | 3 | MA0788.1 | POU3F3 | MA0788.1 | WWTATGCWAATTW | POU3F3 | 4.28e-05 | 1.1e-07 | 7740 | 47.9 | 7530 | 44.9 | 1.07 |
| 169 | 33 | MA0754.2 | CUX1 | MA0754.2 | TAATCGATAM | CUX1 | 4.29e-05 | 1.1e-07 | 5750 | 35.6 | 5490 | 32.8 | 1.09 |
| 170 | 3 | MA0716.1 | PRRX1 | MA0716.1 | YYAATTAR | PRRX1 | 4.43e-05 | 1.13e-07 | 10300 | 63.5 | 10200 | 60.6 | 1.05 |
| 171 | 8 | MA1632.1 | ATF2 | MA1632.1 | NNATGABGTCATN | ATF2 | 4.57e-05 | 1.16e-07 | 2930 | 18.1 | 2670 | 15.9 | 1.14 |
| 172 | 16 | MA0493.2 | KLF1 | MA0493.2 | DGGGYGGGG | KLF1 | 4.94e-05 | 1.25e-07 | 1380 | 8.51 | 1160 | 6.95 | 1.23 |
| 173 | 14 | MA1606.1 | Foxf1 | MA1606.1 | DWRTAAACANN | Foxf1 | 5.24e-05 | 1.32e-07 | 11000 | 68.3 | 11000 | 65.5 | 1.04 |
| 174 | 33 | MA0755.1 | CUX2 | MA0755.1 | TRATCGATAW | CUX2 | 5.33e-05 | 1.33e-07 | 4560 | 28.2 | 4290 | 25.6 | 1.1 |
| 175 | 13 | MA0896.1 | Hmx1 | MA0896.1 | VSVAGCAATTAAWGVDB | Hmx1 | 5.42e-05 | 1.35e-07 | 5570 | 34.4 | 5310 | 31.7 | 1.09 |
| 176 | 34 | MA1978.1 | ZNF354A | MA1978.1 | HWAADWATAAATGRAYWAWTT | ZNF354A | 5.58e-05 | 1.38e-07 | 6300 | 38.9 | 6050 | 36.1 | 1.08 |
| 177 | 35 | MA1480.1 | DPRX | MA1480.1 | AGATAATCCN | DPRX | 5.63e-05 | 1.38e-07 | 7560 | 46.7 | 7340 | 43.8 | 1.07 |
| 178 | 8 | MA1133.1 | JUN::JUNB | MA1133.1 | KRTGACGTCATN | JUN::JUNB | 5.98e-05 | 1.46e-07 | 6240 | 38.6 | 5990 | 35.8 | 1.08 |
| 179 | 24 | MA0797.1 | TGIF2 | MA0797.1 | TGACAGSTGTCA | TGIF2 | 6.62e-05 | 1.61e-07 | 4510 | 27.9 | 4240 | 25.3 | 1.1 |
| 180 | 14 | MA0041.2 | FOXD3 | MA0041.2 | WAWGHAAATAAACARY | FOXD3 | 6.65e-05 | 1.61e-07 | 6760 | 41.8 | 6520 | 38.9 | 1.07 |
| 181 | 14 | MA0033.2 | FOXL1 | MA0033.2 | RTAAACA | FOXL1 | 6.79e-05 | 1.63e-07 | 5800 | 35.8 | 5540 | 33.1 | 1.08 |
| 182 | 3 | MA0628.1 | POU6F1 | MA0628.1 | AYTAATTART | POU6F1 | 8.45e-05 | 2.02e-07 | 10400 | 64.4 | 10300 | 61.6 | 1.05 |
| 183 | 36 | MA0069.1 | PAX6 | MA0069.1 | TTCACGCWTSANTK | PAX6 | 0.000103 | 2.45e-07 | 6240 | 38.6 | 6000 | 35.8 | 1.08 |
| 184 | 19 | MA1608.1 | Isl1 | MA1608.1 | NNCCATTAGNN | Isl1 | 0.000104 | 2.45e-07 | 7020 | 43.4 | 6800 | 40.6 | 1.07 |
| 185 | 8 | MA1129.1 | FOSL1::JUN | MA1129.1 | ATGACGTCAT | FOSL1::JUN | 0.000107 | 2.51e-07 | 13000 | 80.1 | 13000 | 77.7 | 1.03 |
| 186 | 13 | MA0897.1 | Hmx2 | MA0897.1 | ASAAGCAATTAAHVVRT | Hmx2 | 0.000119 | 2.78e-07 | 5540 | 34.2 | 5290 | 31.6 | 1.08 |
| 187 | 15 | MA0117.2 | Mafb | MA0117.2 | AWWNTGCTGACD | Mafb | 0.000128 | 2.98e-07 | 1610 | 9.96 | 1400 | 8.32 | 1.2 |
| 188 | 8 | MA1524.2 | Msgn1 | MA1524.2 | VRRRACAAATGGTNNN | Msgn1 | 0.000165 | 3.82e-07 | 7740 | 47.8 | 7550 | 45.1 | 1.06 |
| 189 | 19 | MA0766.2 | GATA5 | MA0766.2 | SWGATAASRN | GATA5 | 0.000167 | 3.85e-07 | 1010 | 6.24 | 829 | 4.95 | 1.26 |
| 190 | 3 | MA0791.1 | POU4F3 | MA0791.1 | RTGMATWATTAATGAR | POU4F3 | 0.000184 | 4.21e-07 | 4140 | 25.6 | 3890 | 23.2 | 1.1 |
| 191 | 3 | MA0903.1 | HOXB3 | MA0903.1 | NNYMATTARN | HOXB3 | 0.000195 | 4.43e-07 | 9090 | 56.2 | 8950 | 53.4 | 1.05 |
| 192 | 37 | MA0808.1 | TEAD3 | MA0808.1 | RCATTCCW | TEAD3 | 0.000237 | 5.36e-07 | 6290 | 38.9 | 6070 | 36.2 | 1.07 |
| 193 | 3 | MA0674.1 | NKX6-1 | MA0674.1 | NTMATTAA | NKX6-1 | 0.00024 | 5.41e-07 | 11200 | 69.4 | 11200 | 66.8 | 1.04 |
| 194 | 9 | MA0084.1 | SRY | MA0084.1 | NWWAACAAT | SRY | 0.000286 | 6.4e-07 | 12600 | 78 | 12700 | 75.6 | 1.03 |
| 195 | 8 | MA1618.1 | Ptf1a | MA1618.1 | NNACAGATGTTNN | Ptf1a | 0.000287 | 6.4e-07 | 14400 | 89 | 14600 | 87.2 | 1.02 |
| 196 | 8 | MA0609.2 | CREM | MA0609.2 | NNDGTGACGTCACHNN | CREM | 0.000308 | 6.82e-07 | 7550 | 46.7 | 7360 | 43.9 | 1.06 |
| 197 | 28 | MA1478.1 | DMRTA2 | MA1478.1 | AATTGHTACAWT | DMRTA2 | 0.000409 | 9.02e-07 | 7100 | 43.9 | 6900 | 41.2 | 1.06 |
| 198 | 38 | MA0596.1 | SREBF2 | MA0596.1 | RTGGGGTGAY | SREBF2 | 0.000554 | 1.22e-06 | 1220 | 7.51 | 1030 | 6.16 | 1.22 |
| 199 | 3 | MA1501.1 | HOXB7 | MA1501.1 | GGTAATTAVS | HOXB7 | 0.000569 | 1.24e-06 | 7510 | 46.4 | 7340 | 43.8 | 1.06 |
| 200 | 3 | MA0786.1 | POU3F1 | MA0786.1 | WTATGCWAATKW | POU3F1 | 0.000683 | 1.48e-06 | 6030 | 37.3 | 5820 | 34.7 | 1.07 |
| 201 | 10 | MA0789.1 | POU3F4 | MA0789.1 | TATGCWAAT | POU3F4 | 0.000709 | 1.53e-06 | 5890 | 36.4 | 5680 | 33.9 | 1.07 |
| 202 | 9 | MA0868.2 | SOX8 | MA0868.2 | MGAACAATRG | SOX8 | 0.000744 | 1.6e-06 | 10100 | 62.4 | 10000 | 59.8 | 1.04 |
| 203 | 5 | MA0775.1 | MEIS3 | MA0775.1 | DTGACAGS | MEIS3 | 0.000749 | 1.6e-06 | 4800 | 29.6 | 4570 | 27.3 | 1.09 |
| 204 | 10 | MA0627.2 | POU2F3 | MA0627.2 | NWTATGCAAATNH | POU2F3 | 0.00075 | 1.6e-06 | 6330 | 39.1 | 6130 | 36.6 | 1.07 |
| 205 | 10 | MA0792.1 | POU5F1B | MA0792.1 | TATGCWAAT | POU5F1B | 0.000792 | 1.68e-06 | 5430 | 33.6 | 5210 | 31.1 | 1.08 |
| 206 | 39 | MA1151.1 | RORC | MA1151.1 | NWAWNTRGGTCA | RORC | 0.00087 | 1.84e-06 | 10900 | 67.6 | 10900 | 65.1 | 1.04 |
| 207 | 1 | MA1633.2 (BACH1) | MEME-5 | TGRCTCATGCCTGTA | TGRCTCATGCCTGTA | BACH1 | 0.000894 | 1.88e-06 | 565 | 3.49 | 434 | 2.59 | 1.35 |
| 208 | 3 | MA0618.1 | LBX1 | MA0618.1 | TTAATTAG | LBX1 | 0.000907 | 1.89e-06 | 7530 | 46.5 | 7360 | 43.9 | 1.06 |
| 209 | 3 | MA0874.1 | Arx | MA0874.1 | VTBCRYTAATTARTKSW | Arx | 0.000911 | 1.89e-06 | 4240 | 26.2 | 4010 | 24 | 1.09 |
| 210 | 40 | MA0803.1 | TBX15 | MA0803.1 | AGGTGTGA | TBX15 | 0.0011 | 2.28e-06 | 1010 | 6.23 | 843 | 5.03 | 1.24 |
| 211 | 9 | MA0514.2 | Sox3 | MA0514.2 | DNACAATGGNN | Sox3 | 0.00114 | 2.34e-06 | 6730 | 41.6 | 6550 | 39.1 | 1.07 |
| 212 | 3 | MA0787.1 | POU3F2 | MA0787.1 | WTATGCWAATKA | POU3F2 | 0.0012 | 2.45e-06 | 6690 | 41.4 | 6510 | 38.8 | 1.07 |
| 213 | 19 | MA0909.3 | Hoxd13 | MA0909.3 | NNCAATAAAW | Hoxd13 | 0.00127 | 2.58e-06 | 7840 | 48.4 | 7680 | 45.9 | 1.06 |
| 214 | 32 | MA0106.3 | TP53 | MA0106.3 | RACATGYCCGGRCATGTY | TP53 | 0.00137 | 2.77e-06 | 166 | 1.03 | 95 | 0.57 | 1.8 |
| 215 | 9 | MA1561.1 | SOX12 | MA1561.1 | ACCGAACAATV | SOX12 | 0.00146 | 2.94e-06 | 10500 | 65 | 10500 | 62.5 | 1.04 |
| 216 | 25 | MA0043.3 | HLF | MA0043.3 | NNRTTATGCAACHN | HLF | 0.00147 | 2.95e-06 | 8050 | 49.8 | 7910 | 47.2 | 1.05 |
| 217 | 14 | MA0148.4 | FOXA1 | MA0148.4 | WWGTAAACATNN | FOXA1 | 0.00153 | 3.06e-06 | 5640 | 34.9 | 5440 | 32.4 | 1.07 |
| 218 | 20 | MA1567.2 | Tbx6 | MA1567.2 | NRGGTGTGAANN | Tbx6 | 0.0017 | 3.39e-06 | 6900 | 42.7 | 6730 | 40.2 | 1.06 |
| 219 | 35 | MA0467.2 | Crx | MA0467.2 | NDGGATTANN | Crx | 0.00202 | 4.02e-06 | 11200 | 69.3 | 11200 | 67 | 1.04 |
| 220 | 27 | MA0905.1 | HOXC10 | MA0905.1 | GTCRTAAAAH | HOXC10 | 0.00203 | 4.02e-06 | 7980 | 49.4 | 7850 | 46.8 | 1.05 |
| 221 | 3 | MA1498.2 | HOXA7 | MA1498.2 | SYMATTAV | HOXA7 | 0.00222 | 4.37e-06 | 8990 | 55.6 | 8890 | 53.1 | 1.05 |
| 222 | 14 | MA0047.3 | FOXA2 | MA0047.3 | RWGTAAACANN | FOXA2 | 0.00234 | 4.59e-06 | 5160 | 31.9 | 4960 | 29.6 | 1.08 |
| 223 | 8 | MA0831.3 | TFE3 | MA0831.3 | GTCACGTGAC | TFE3 | 0.00239 | 4.65e-06 | 2680 | 16.6 | 2470 | 14.7 | 1.12 |
| 224 | 8 | MA1995.1 | Npas4 | MA1995.1 | NNTCGTGACHN | Npas4 | 0.0024 | 4.67e-06 | 11800 | 72.7 | 11800 | 70.4 | 1.03 |
| 225 | 37 | MA0090.3 | TEAD1 | MA0090.3 | NNACATTCCAGSN | TEAD1 | 0.00249 | 4.81e-06 | 5960 | 36.9 | 5770 | 34.5 | 1.07 |
| 226 | 41 | MA1603.1 | Dmrt1 | MA1603.1 | GHTACAAAGTADC | Dmrt1 | 0.00281 | 5.4e-06 | 10000 | 61.9 | 9960 | 59.5 | 1.04 |
| 227 | 15 | MA1520.1 | MAF | MA1520.1 | NTGCTGASTCAGCAH | MAF | 0.00334 | 6.39e-06 | 7780 | 48.1 | 7650 | 45.6 | 1.05 |
| 228 | 40 | MA0805.1 | TBX1 | MA0805.1 | AGGTGTGA | TBX1 | 0.00373 | 7.12e-06 | 795 | 4.91 | 654 | 3.9 | 1.26 |
| 229 | 16 | MA1564.1 | SP9 | MA1564.1 | RCCACGCCCMCY | SP9 | 0.00384 | 7.29e-06 | 1470 | 9.1 | 1300 | 7.73 | 1.18 |
| 230 | 8 | MA1108.2 | MXI1 | MA1108.2 | NNCACATGBN | MXI1 | 0.00399 | 7.54e-06 | 623 | 3.85 | 496 | 2.96 | 1.3 |
| 231 | 40 | MA0806.1 | TBX4 | MA0806.1 | AGGTGTGA | TBX4 | 0.00413 | 7.78e-06 | 1390 | 8.59 | 1220 | 7.26 | 1.18 |
| 232 | 3 | MA0892.1 | GSX1 | MA0892.1 | NCYMATTAVN | GSX1 | 0.00428 | 8.02e-06 | 10800 | 67 | 10800 | 64.6 | 1.04 |
| 233 | 3 | MA0683.1 | POU4F2 | MA0683.1 | RTGCATAATTAATGAG | POU4F2 | 0.00559 | 1.04e-05 | 2130 | 13.2 | 1940 | 11.6 | 1.14 |
| 234 | 42 | MA1588.1 | ZNF136 | MA1588.1 | RWATTCTTGGTTGRC | ZNF136 | 0.00599 | 1.11e-05 | 4880 | 30.2 | 4690 | 28 | 1.08 |
| 235 | 40 | MA0689.1 | TBX20 | MA0689.1 | WAGGTGTGAAR | TBX20 | 0.00702 | 1.3e-05 | 2240 | 13.9 | 2060 | 12.3 | 1.13 |
| 236 | 43 | MA1953.1 | FOXO1::ELF1 | MA1953.1 | RTMAACAGGAAGTN | FOXO1::ELF1 | 0.00776 | 1.43e-05 | 10200 | 63.3 | 10200 | 61 | 1.04 |
| 237 | 44 | MA1489.1 | FOXN3 | MA1489.1 | GTAAACAA | FOXN3 | 0.00841 | 1.54e-05 | 7110 | 44 | 6980 | 41.6 | 1.06 |
| 238 | 14 | MA0480.2 | Foxo1 | MA0480.2 | NDGTAAACANN | Foxo1 | 0.00884 | 1.61e-05 | 3110 | 19.2 | 2910 | 17.4 | 1.1 |
| 239 | 3 | MA1507.1 | HOXD4 | MA1507.1 | VTMATTAD | HOXD4 | 0.00968 | 1.76e-05 | 3360 | 20.8 | 3160 | 18.9 | 1.1 |
| 240 | 45 | MA1989.1 | Bcl11B | MA1989.1 | NNAAACCACAARNN | Bcl11B | 0.0105 | 1.91e-05 | 14600 | 90.4 | 14900 | 88.9 | 1.02 |
| 241 | 3 | MA1504.1 | HOXC4 | MA1504.1 | VTMATTAN | HOXC4 | 0.0109 | 1.97e-05 | 3400 | 21 | 3200 | 19.1 | 1.1 |
| 242 | 3 | MA1497.1 | HOXA6 | MA1497.1 | BGYMATTANN | HOXA6 | 0.0111 | 1.99e-05 | 6500 | 40.2 | 6350 | 37.9 | 1.06 |
| 243 | 16 | MA0742.2 | KLF12 | MA0742.2 | GGGGCGGGG | KLF12 | 0.0118 | 2.11e-05 | 1190 | 7.36 | 1040 | 6.19 | 1.19 |
| 244 | 14 | MA0031.1 | FOXD1 | MA0031.1 | GTAAACAW | FOXD1 | 0.0121 | 2.16e-05 | 8810 | 54.5 | 8740 | 52.2 | 1.04 |
| 15 | 55 | MA0679.2 (ONECUT1) | MEME-1 | WAAAAAWAMAWAAAW | WAAAAAWAMAWAAAW | ONECUT1 | 0.0125 | 0.000831 | 6200 | 38.3 | 6130 | 36.6 | 1.05 |
| 245 | 9 | MA1962.1 | POU2F1::SOX2 | MA1962.1 | MATTTRCATMACAATRG | POU2F1::SOX2 | 0.0131 | 2.33e-05 | 3770 | 23.3 | 3580 | 21.4 | 1.09 |
| 246 | 46 | MA0698.1 | ZBTB18 | MA0698.1 | NATCCAGATGTKB | ZBTB18 | 0.0132 | 2.33e-05 | 6220 | 38.5 | 6070 | 36.2 | 1.06 |
| 247 | 40 | MA0801.1 | MGA | MA0801.1 | AGGTGTGA | MGA | 0.0133 | 2.34e-05 | 533 | 3.29 | 422 | 2.52 | 1.31 |
| 248 | 19 | MA0036.3 | GATA2 | MA0036.3 | NBCTTATCTNH | GATA2 | 0.0136 | 2.38e-05 | 1070 | 6.61 | 923 | 5.51 | 1.2 |
| 249 | 3 | MA0132.2 | PDX1 | MA0132.2 | VYAATTAR | PDX1 | 0.0154 | 2.68e-05 | 3590 | 22.2 | 3400 | 20.3 | 1.09 |
| 250 | 16 | MA1517.1 | KLF6 | MA1517.1 | NRCCACGCCCH | KLF6 | 0.0158 | 2.74e-05 | 3250 | 20.1 | 3060 | 18.3 | 1.1 |
| 251 | 18 | MA0038.2 | GFI1 | MA0038.2 | BMAATCACDGCA | GFI1 | 0.0162 | 2.81e-05 | 2780 | 17.2 | 2600 | 15.5 | 1.11 |
| 252 | 3 | MA1471.1 | BARX2 | MA1471.1 | NWWAAYMATTAN | BARX2 | 0.0167 | 2.88e-05 | 9840 | 60.8 | 9820 | 58.6 | 1.04 |
| 253 | 47 | MA1576.1 | THRB | MA1576.1 | DTGACCTBACGTGACCTYA | THRB | 0.0169 | 2.9e-05 | 1480 | 9.17 | 1320 | 7.9 | 1.16 |
| 254 | 16 | MA0599.1 | KLF5 | MA0599.1 | GCCCCDCCCH | KLF5 | 0.0174 | 2.98e-05 | 5480 | 33.9 | 5320 | 31.8 | 1.07 |
| 255 | 16 | MA1515.1 | KLF2 | MA1515.1 | NRCCACRCCCH | KLF2 | 0.0178 | 3.03e-05 | 4550 | 28.1 | 4370 | 26.1 | 1.08 |
| 256 | 48 | MA1975.1 | ZNF214 | MA1975.1 | VWTCATCAABGTCCT | ZNF214 | 0.0218 | 3.71e-05 | 11800 | 72.8 | 11900 | 70.8 | 1.03 |
| 257 | 3 | MA0784.2 | POU1F1 | MA0784.2 | ANCTMATTWGCATAWT | POU1F1 | 0.025 | 4.23e-05 | 1470 | 9.07 | 1310 | 7.84 | 1.16 |
| 258 | 29 | MA0678.1 | OLIG2 | MA0678.1 | AMCATATGKT | OLIG2 | 0.0254 | 4.27e-05 | 3340 | 20.6 | 3160 | 18.9 | 1.09 |
| 259 | 8 | MA1994.1 | Nkx2-1 | MA1994.1 | NVCACTTGARHNN | Nkx2-1 | 0.0268 | 4.49e-05 | 14600 | 90 | 14800 | 88.6 | 1.02 |
| 260 | 49 | MA1980.1 | ZNF418 | MA1980.1 | MDGAGGCTAAAAGCA | ZNF418 | 0.0272 | 4.55e-05 | 7490 | 46.3 | 7390 | 44.1 | 1.05 |
| 261 | 8 | MA0492.1 | JUND | MA0492.1 | DDDRATGABGTCATN | JUND | 0.0303 | 5.04e-05 | 1890 | 11.7 | 1730 | 10.3 | 1.13 |
| 262 | 50 | MA1984.1 | ZNF667 | MA1984.1 | TTAAGAGCTCAV | ZNF667 | 0.0316 | 5.24e-05 | 13600 | 84.1 | 13800 | 82.5 | 1.02 |
| 263 | 40 | MA0800.1 | EOMES | MA0800.1 | RAGGTGTGAAAWN | EOMES | 0.0344 | 5.68e-05 | 1600 | 9.92 | 1450 | 8.65 | 1.15 |
| 264 | 51 | MA0002.2 (Runx1) | STREME-10 | 10-CAACCAGAGTG | 10-CAACCAGAGTG | Runx1 | 0.0347 | 5.71e-05 | 9200 | 56.8 | 9160 | 54.7 | 1.04 |
| 265 | 9 | MA1152.1 | SOX15 | MA1152.1 | CYWTTGTTHW | SOX15 | 0.0368 | 6.03e-05 | 7840 | 48.5 | 7760 | 46.3 | 1.05 |
| 266 | 3 | MA0891.1 | GSC2 | MA0891.1 | NYTAATCCBH | GSC2 | 0.0391 | 6.39e-05 | 11300 | 70.1 | 11400 | 68.1 | 1.03 |
| 267 | 52 | MA0050.3 (Irf1) | MEME-7 | ADRDRNRSAVAVWGA | ADRDRNRSAVAVWGA | Irf1 | 0.0396 | 6.44e-05 | 6550 | 40.5 | 6430 | 38.4 | 1.06 |
| 268 | 3 | MA0702.2 | LMX1A | MA0702.2 | TTAATTAM | LMX1A | 0.0407 | 6.6e-05 | 2970 | 18.3 | 2800 | 16.7 | 1.1 |
| 269 | 1 | MA0591.1 | Bach1::Mafk | MA0591.1 | RSSATGACTCAGCAB | Bach1::Mafk | 0.0453 | 7.32e-05 | 136 | 0.84 | 82 | 0.49 | 1.71 |
| 270 | 9 | MA0867.2 | SOX4 | MA0867.2 | RAACAAAGRV | SOX4 | 0.0478 | 7.7e-05 | 11900 | 73.3 | 12000 | 71.3 | 1.03 |
| 271 | 53 | MA0108.2 | TBP | MA0108.2 | STATAAAWRSVVVBN | TBP | 0.0486 | 7.78e-05 | 7270 | 45 | 7180 | 42.8 | 1.05 |
| 272 | 54 | MA1991.1 | Hnf1A | MA1991.1 | NBCCTTTGATSTBN | Hnf1A | 0.0487 | 7.78e-05 | 5480 | 33.9 | 5340 | 31.9 | 1.06 |
| 16 | 56 | MA1596.1 (ZNF460) | MEME-3 | GGGAGGDGGAGGVDG | GGGAGGDGGAGGVDG | ZNF460 | 4.91 | 0.307 | 10 | 0.06 | 7 | 0.04 | 1.42 |
| 17 | 16 | MA1961.1 (PATZ1) | MEME-2 | GGSGGCGGGGSSGGG | GGSGGCGGGGSSGGG | PATZ1 | 16.9 | 0.996 | 363 | 2.24 | 452 | 2.7 | 0.832 |
ER_rat <- 1.25
DNR_242kb_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste("DNR_242kb_combo response peaks(200bp)Enrichment ratio:",ER_rat))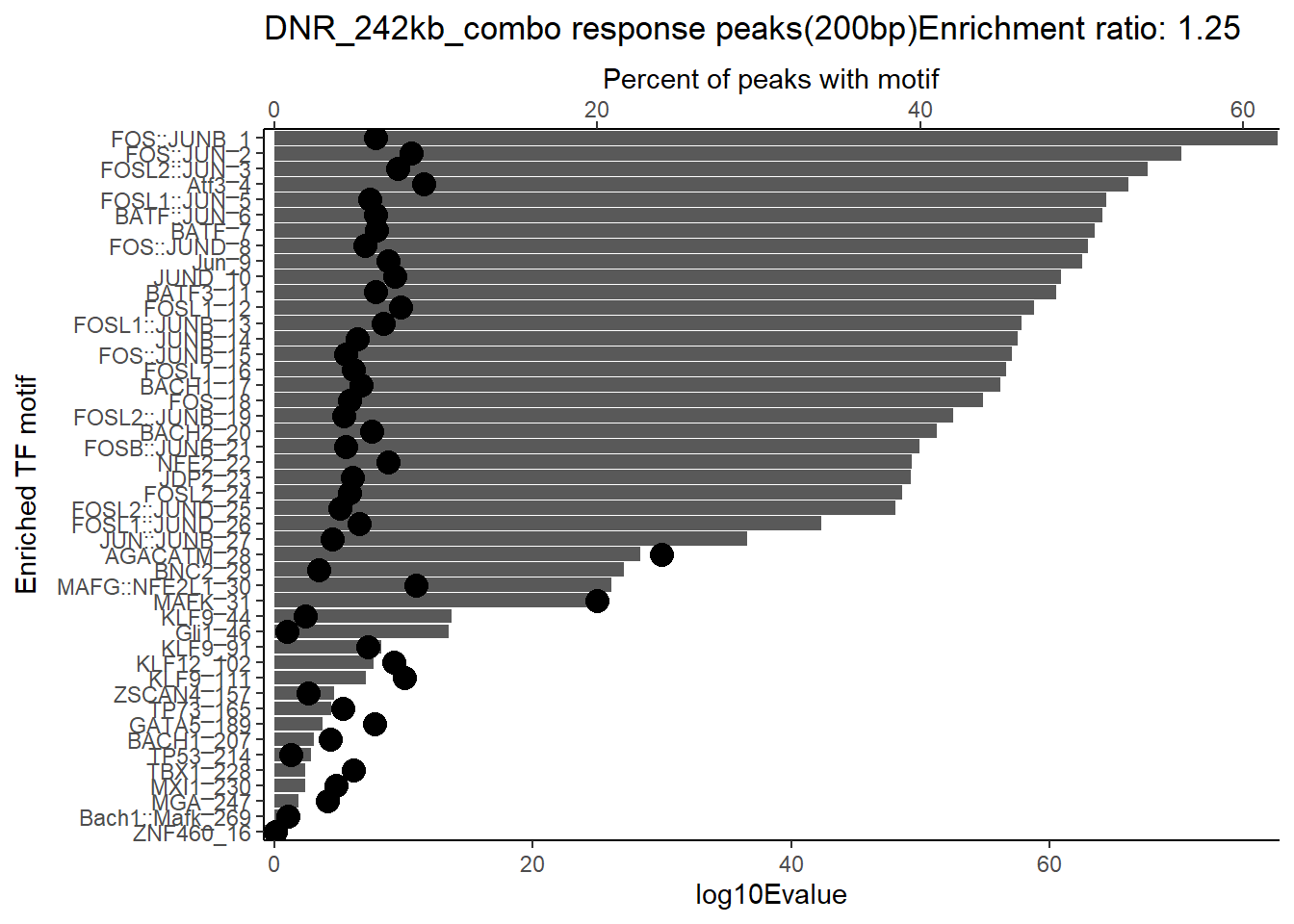
DNR_242kb_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y, TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
slice_head(n=5) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*4), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./4,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( "DNR_24 2kb response peaks(200bp)Enrichment ratio:",ER_rat))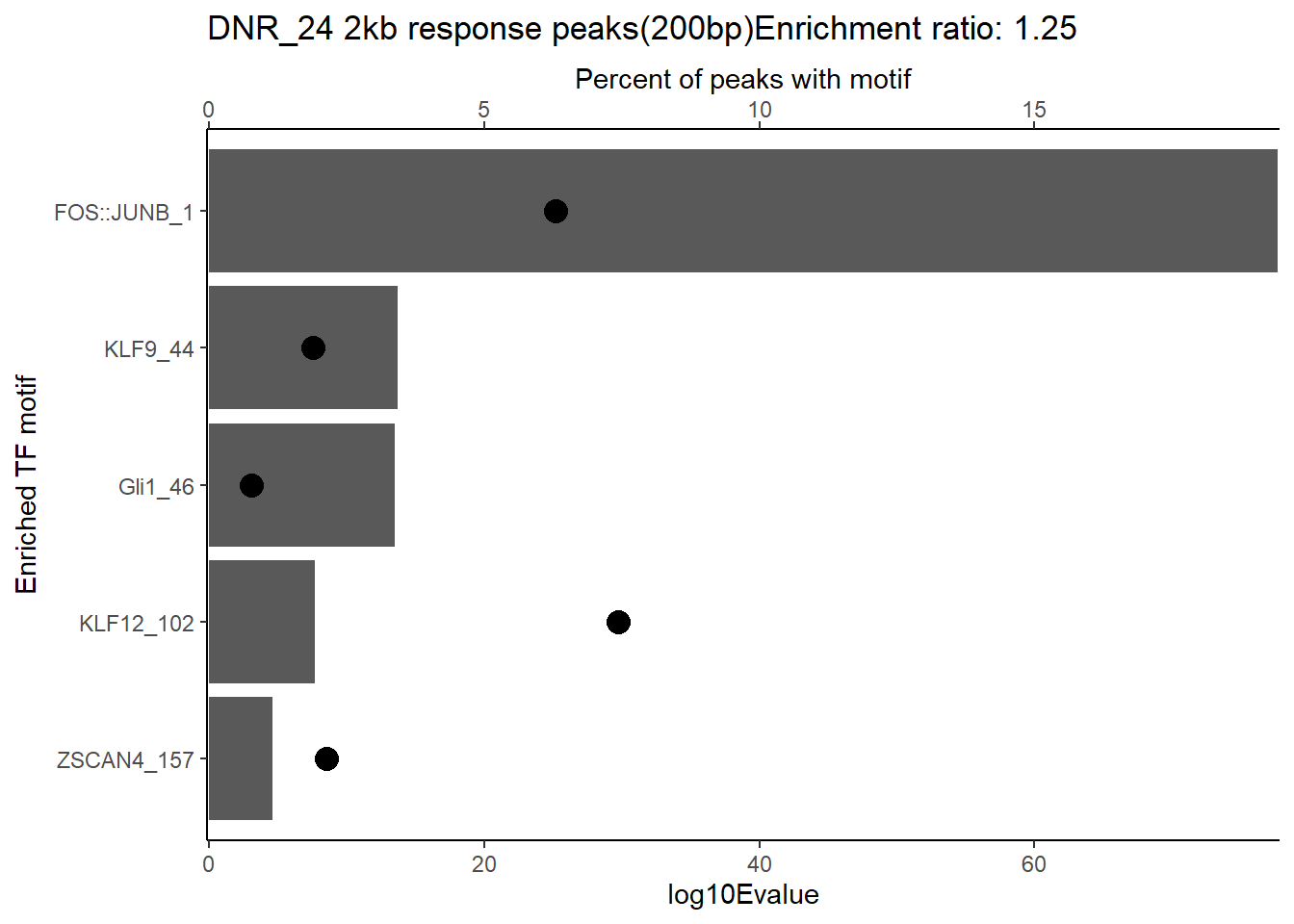
DNR_DAR 24 20kb data
DNR_24_20kb_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DNR_24_20kb_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
DNR_24_20kb_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DNR_24_20kb_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
DNR_24_20kb_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/DNR_24_20kb_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
DNR_24_20kb_sea_all <- rbind(DNR_24_20kb_sea_disc, DNR_24_20kb_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)DNR_24_20kb_combo <- DNR_24_20kb_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., DNR_24_20kb_sea_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ SIM_MOTIF
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y)
DNR_24_20kb_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs 200 bp in DNR 24h 20kb") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | MA1134.1 (FOS::JUNB) | STREME-1 | 1-RATGASTCATY | RATGASTCATY | FOS::JUNB | 1.07e-255 | 6.3e-256 | 4120 | 9.28 | 1530 | 3.63 | 2.56 |
| 2 | 1 | MA1134.1 | FOS::JUNB | MA1134.1 | KATGASTCATHN | FOS::JUNB | 7.89e-246 | 2.32e-246 | 5300 | 11.9 | 2340 | 5.53 | 2.16 |
| 3 | 1 | MA1135.1 | FOSB::JUNB | MA1135.1 | KRTGASTCAT | FOSB::JUNB | 7.37e-244 | 1.44e-244 | 5710 | 12.8 | 2630 | 6.21 | 2.07 |
| 4 | 1 | MA0099.3 | FOS::JUN | MA0099.3 | ATGAGTCAYM | FOS::JUN | 5.01e-243 | 7.36e-244 | 4760 | 10.7 | 2000 | 4.72 | 2.27 |
| 5 | 1 | MA1144.1 | FOSL2::JUND | MA1144.1 | KATGACTCAT | FOSL2::JUND | 5.84e-242 | 6.87e-243 | 5800 | 13.1 | 2700 | 6.39 | 2.04 |
| 6 | 1 | MA1138.1 | FOSL2::JUNB | MA1138.1 | KRTGASTCAT | FOSL2::JUNB | 2.96e-239 | 2.9e-240 | 5660 | 12.7 | 2620 | 6.19 | 2.06 |
| 7 | 1 | MA0655.1 (JDP2) | MEME-2 | ATGASTCA | ATGASTCA | JDP2 | 7.98e-237 | 6.71e-238 | 5960 | 13.4 | 2840 | 6.7 | 2 |
| 8 | 1 | MA1130.1 | FOSL2::JUN | MA1130.1 | NNRTGAGTCAYN | FOSL2::JUN | 3.8e-229 | 2.8e-230 | 4600 | 10.3 | 1940 | 4.59 | 2.25 |
| 9 | 1 | MA1633.2 | BACH1 | MA1633.2 | ATGACTCAT | BACH1 | 3.38e-226 | 2.21e-227 | 5920 | 13.3 | 2870 | 6.78 | 1.97 |
| 10 | 1 | MA1141.1 | FOS::JUND | MA1141.1 | NKATGAGTCATNN | FOS::JUND | 5.53e-226 | 3.25e-227 | 4650 | 10.5 | 1990 | 4.71 | 2.22 |
| 11 | 1 | MA0491.2 | JUND | MA0491.2 | NNATGACTCATNN | JUND | 2.3e-224 | 1.23e-225 | 3810 | 8.58 | 1460 | 3.45 | 2.48 |
| 12 | 1 | MA0489.2 | Jun | MA0489.2 | NVTGACTCATNN | Jun | 3.64e-220 | 1.79e-221 | 6120 | 13.8 | 3040 | 7.18 | 1.92 |
| 13 | 1 | MA0490.2 | JUNB | MA0490.2 | NNATGACTCATNN | JUNB | 1.06e-217 | 4.79e-219 | 3700 | 8.33 | 1420 | 3.35 | 2.49 |
| 14 | 1 | MA0476.1 | FOS | MA0476.1 | DVTGASTCATB | FOS | 9.53999999999999e-213 | 4.01e-214 | 6530 | 14.7 | 3380 | 7.98 | 1.84 |
| 15 | 1 | MA1988.1 | Atf3 | MA1988.1 | NRTGACTCABN | Atf3 | 1.6e-211 | 6.27e-213 | 4130 | 9.29 | 1720 | 4.05 | 2.29 |
| 16 | 1 | MA1137.1 | FOSL1::JUNB | MA1137.1 | NDRTGACTCATNN | FOSL1::JUNB | 4.27e-211 | 1.57e-212 | 4420 | 9.96 | 1910 | 4.52 | 2.2 |
| 17 | 1 | MA0462.2 | BATF::JUN | MA0462.2 | DATGACTCATH | BATF::JUN | 1.43e-209 | 4.97e-211 | 4220 | 9.5 | 1780 | 4.21 | 2.25 |
| 18 | 1 | MA1634.1 | BATF | MA1634.1 | DATGACTCATH | BATF | 1.19e-206 | 3.88e-208 | 3590 | 8.08 | 1390 | 3.29 | 2.46 |
| 19 | 1 | MA0478.1 | FOSL2 | MA0478.1 | KRRTGASTCAB | FOSL2 | 4.85e-205 | 1.5e-206 | 5900 | 13.3 | 2960 | 7.01 | 1.89 |
| 20 | 1 | MA1128.1 | FOSL1::JUN | MA1128.1 | NKATGACTCATNN | FOSL1::JUN | 7.93e-204 | 2.33e-205 | 3290 | 7.41 | 1220 | 2.88 | 2.58 |
| 21 | 1 | MA0835.2 | BATF3 | MA0835.2 | DATGACTCATH | BATF3 | 7.52e-202 | 2.11e-203 | 4050 | 9.11 | 1700 | 4.03 | 2.26 |
| 22 | 1 | MA0477.2 | FOSL1 | MA0477.2 | NNATGACTCATNN | FOSL1 | 3.97e-198 | 1.06e-199 | 3040 | 6.83 | 1080 | 2.55 | 2.67 |
| 23 | 1 | MA0655.1 | JDP2 | MA0655.1 | ATGACTCAT | JDP2 | 5.7e-197 | 1.46e-198 | 5380 | 12.1 | 2640 | 6.24 | 1.94 |
| 24 | 1 | MA1142.1 | FOSL1::JUND | MA1142.1 | NRTGACTCAT | FOSL1::JUND | 7.18e-186 | 1.76e-187 | 3610 | 8.13 | 1490 | 3.52 | 2.31 |
| 25 | 1 | MA1101.2 | BACH2 | MA1101.2 | DWANCATGASTCATSNTWH | BACH2 | 1.05e-178 | 2.47e-180 | 5340 | 12 | 2710 | 6.4 | 1.88 |
| 26 | 1 | MA1928.1 | BNC2 | MA1928.1 | NDRTGAGTCABN | BNC2 | 3.7e-169 | 8.37e-171 | 6080 | 13.7 | 3310 | 7.82 | 1.75 |
| 27 | 1 | MA0841.1 | NFE2 | MA0841.1 | VATGACTCATS | NFE2 | 8.74e-169 | 1.9e-170 | 7040 | 15.8 | 4040 | 9.55 | 1.66 |
| 28 | 1 | MA1132.1 | JUN::JUNB | MA1132.1 | KATGACKCAT | JUN::JUNB | 1.41e-128 | 2.97e-130 | 1910 | 4.3 | 659 | 1.56 | 2.76 |
| 29 | 1 | MA0496.3 | MAFK | MA0496.3 | NGMTGACTCAGCMNH | MAFK | 2.98e-80 | 6.05e-82 | 10100 | 22.8 | 7430 | 17.6 | 1.3 |
| 30 | 1 | MA0501.1 | MAF::NFE2 | MA0501.1 | ATGACTCAGCANWTY | MAF::NFE2 | 2.4e-71 | 4.7e-73 | 9320 | 21 | 6840 | 16.2 | 1.3 |
| 31 | 1 | MA0089.2 | MAFG::NFE2L1 | MA0089.2 | NVNATGACTCAGCADW | MAFG::NFE2L1 | 1.57e-64 | 2.99e-66 | 3060 | 6.88 | 1780 | 4.2 | 1.64 |
| 32 | 1 | MA0659.3 | Mafg | MA0659.3 | NWGMTGACTCAGCAN | Mafg | 3.63e-44 | 6.67e-46 | 3610 | 8.13 | 2390 | 5.66 | 1.44 |
| 33 | 2 | 6-ACATACACACAM | STREME-6 | 6-ACATACACACAM | ACATACACACAM | ACATACACACAM | 1.55e-41 | 2.76e-43 | 1020 | 2.29 | 458 | 1.08 | 2.12 |
| 34 | 1 | MA0150.2 | Nfe2l2 | MA0150.2 | CASNATGACTCAGCA | Nfe2l2 | 7.34e-41 | 1.27e-42 | 14700 | 33 | 12100 | 28.6 | 1.15 |
| 35 | 2 | 4-ACACACATACA | STREME-4 | 4-ACACACATACA | ACACACATACA | ACACACATACA | 2.47e-34 | 4.15e-36 | 1070 | 2.41 | 533 | 1.26 | 1.91 |
| 36 | 2 | TGTGTGTGTGT | MEME-4 | TGTGTGTGTGT | TGTGTGTGTGT | TGTGTGTGTGT | 1.55e-31 | 2.53e-33 | 1900 | 4.28 | 1170 | 2.76 | 1.55 |
| 37 | 2 | MA1107.2 (KLF9) | STREME-2 | 2-CACACACACAC | CACACACACAC | KLF9 | 5.05e-29 | 8.03e-31 | 1670 | 3.76 | 1010 | 2.39 | 1.57 |
| 38 | 3 | MA1990.1 (Gli1) | STREME-8 | 8-CACACACCACACAYA | CACACACCACACAYA | Gli1 | 1.71e-28 | 2.65e-30 | 169 | 0.38 | 17 | 0.04 | 8.99 |
| 39 | 2 | MA1155.1 (ZSCAN4) | STREME-5 | 5-TGCACACACACA | TGCACACACACA | ZSCAN4 | 1.25e-25 | 1.89e-27 | 1360 | 3.06 | 803 | 1.9 | 1.61 |
| 40 | 4 | 3-ATGTGTG | STREME-3 | 3-ATGTGTG | ATGTGTG | ATGTGTG | 5.08e-24 | 7.48e-26 | 29600 | 66.6 | 26700 | 63.1 | 1.05 |
| 41 | 5 | MA0707.2 | MNX1 | MA0707.2 | VYRATTAK | MNX1 | 1.62e-22 | 2.33e-24 | 24100 | 54.2 | 21400 | 50.7 | 1.07 |
| 42 | 5 | MA0904.2 | HOXB5 | MA0904.2 | STAATTAS | HOXB5 | 4.47e-22 | 6.27e-24 | 24400 | 55 | 21800 | 51.5 | 1.07 |
| 43 | 5 | MA1495.1 | HOXA1 | MA1495.1 | STAATTAS | HOXA1 | 1.24e-20 | 1.7e-22 | 24100 | 54.2 | 21500 | 50.8 | 1.07 |
| 44 | 5 | MA1519.1 | LHX5 | MA1519.1 | STAATTAV | LHX5 | 3.89e-20 | 5.2e-22 | 18400 | 41.4 | 16100 | 38.1 | 1.09 |
| 45 | 5 | MA0158.2 | HOXA5 | MA0158.2 | GYMATTAS | HOXA5 | 6e-20 | 7.85e-22 | 20300 | 45.7 | 17900 | 42.4 | 1.08 |
| 46 | 6 | MA0626.1 | Npas2 | MA0626.1 | NSCACGTGTN | Npas2 | 2.66e-18 | 3.4e-20 | 18500 | 41.6 | 16300 | 38.5 | 1.08 |
| 47 | 1 | MA0591.1 | Bach1::Mafk | MA0591.1 | RSSATGACTCAGCAB | Bach1::Mafk | 6.06e-18 | 7.59e-20 | 6670 | 15 | 5420 | 12.8 | 1.17 |
| 48 | 5 | MA0886.1 | EMX2 | MA0886.1 | VBTAATTARB | EMX2 | 1.47e-17 | 1.81e-19 | 28500 | 64.2 | 25900 | 61.2 | 1.05 |
| 49 | 5 | MA0705.1 | Lhx8 | MA0705.1 | CTAATTAV | Lhx8 | 6.22e-16 | 7.47e-18 | 24700 | 55.5 | 22200 | 52.6 | 1.06 |
| 50 | 5 | MA0661.1 | MEOX1 | MA0661.1 | VSTAATTAHC | MEOX1 | 4.86e-15 | 5.72e-17 | 19000 | 42.7 | 16800 | 39.8 | 1.07 |
| 51 | 7 | MA1155.1 | ZSCAN4 | MA1155.1 | TGCACACACTGAAAA | ZSCAN4 | 2.91e-14 | 3.36e-16 | 1040 | 2.33 | 656 | 1.55 | 1.5 |
| 52 | 5 | MA0912.2 | HOXD3 | MA0912.2 | CTAATTAC | HOXD3 | 7.43e-14 | 8.41e-16 | 15000 | 33.9 | 13200 | 31.2 | 1.08 |
| 53 | 8 | MA0041.2 | FOXD3 | MA0041.2 | WAWGHAAATAAACARY | FOXD3 | 1.09e-13 | 1.21e-15 | 33100 | 74.6 | 30500 | 72.1 | 1.03 |
| 54 | 8 | MA1606.1 | Foxf1 | MA1606.1 | DWRTAAACANN | Foxf1 | 2.55e-13 | 2.75e-15 | 39500 | 88.8 | 36800 | 87.1 | 1.02 |
| 55 | 5 | MA0902.2 | HOXB2 | MA0902.2 | SYAATTAS | HOXB2 | 2.57e-13 | 2.75e-15 | 16000 | 36.1 | 14200 | 33.5 | 1.08 |
| 56 | 9 | MA1115.1 | POU5F1 | MA1115.1 | WHATGCAAATN | POU5F1 | 6.76e-13 | 7.1e-15 | 34200 | 77 | 31600 | 74.7 | 1.03 |
| 57 | 5 | MA0612.2 | EMX1 | MA0612.2 | BTAATTAS | EMX1 | 1.74e-12 | 1.79e-14 | 14500 | 32.7 | 12800 | 30.2 | 1.08 |
| 58 | 8 | MA0846.1 | FOXC2 | MA0846.1 | WAWGTAAACAWW | FOXC2 | 2.02e-12 | 2.05e-14 | 36600 | 82.4 | 34000 | 80.4 | 1.03 |
| 59 | 5 | MA0910.2 | HOXD8 | MA0910.2 | GYMATTAM | HOXD8 | 2.27e-12 | 2.27e-14 | 23000 | 51.9 | 20800 | 49.2 | 1.05 |
| 60 | 10 | MA1143.1 | FOSL1::JUND | MA1143.1 | RTGACGTMAY | FOSL1::JUND | 2.4e-12 | 2.35e-14 | 16000 | 36 | 14200 | 33.5 | 1.08 |
| 61 | 1 | MA1639.1 | MEIS1 | MA1639.1 | NHCATMAATCAHN | MEIS1 | 3.26e-12 | 3.14e-14 | 33900 | 76.3 | 31300 | 74 | 1.03 |
| 62 | 9 | MA0792.1 | POU5F1B | MA0792.1 | TATGCWAAT | POU5F1B | 3.4e-12 | 3.23e-14 | 35600 | 80.1 | 33000 | 77.9 | 1.03 |
| 63 | 5 | MA0706.1 | MEOX2 | MA0706.1 | DSTAATTAWN | MEOX2 | 5.85e-12 | 5.46e-14 | 25400 | 57.2 | 23100 | 54.6 | 1.05 |
| 64 | 5 | MA0027.2 | EN1 | MA0027.2 | SYAATTAV | EN1 | 6.26e-12 | 5.75e-14 | 32400 | 72.8 | 29800 | 70.5 | 1.03 |
| 65 | 11 | MA0502.2 (NFYB) | STREME-9 | 9-CRGCCAATGG | CRGCCAATGG | NFYB | 8.75e-12 | 7.91e-14 | 286 | 0.64 | 123 | 0.29 | 2.2 |
| 66 | 13 | MA0766.2 | GATA5 | MA0766.2 | SWGATAASRN | GATA5 | 8.88e-12 | 7.91e-14 | 7680 | 17.3 | 6500 | 15.4 | 1.13 |
| 67 | 5 | MA1502.1 | HOXB8 | MA1502.1 | GYMATTAM | HOXB8 | 9.3e-12 | 8.16e-14 | 22800 | 51.4 | 20700 | 48.8 | 1.05 |
| 68 | 8 | MA0845.1 | FOXB1 | MA0845.1 | WATGTAAATAT | FOXB1 | 1.31e-11 | 1.14e-13 | 34200 | 77 | 31600 | 74.8 | 1.03 |
| 69 | 6 | MA0093.3 | USF1 | MA0093.3 | NDGTCATGTGACHN | USF1 | 1.33e-11 | 1.14e-13 | 35700 | 80.4 | 33100 | 78.3 | 1.03 |
| 70 | 12 | MA0676.1 | Nr2e1 | MA0676.1 | AAAAGTCAA | Nr2e1 | 1.66e-11 | 1.39e-13 | 12600 | 28.3 | 11000 | 26 | 1.09 |
| 71 | 13 | MA0036.3 (GATA2) | STREME-10 | 10-WGATAAG | 10-WGATAAG | GATA2 | 2.08e-11 | 1.73e-13 | 28900 | 65 | 26500 | 62.5 | 1.04 |
| 72 | 11 | MA0502.2 | NFYB | MA0502.2 | CYCATTGGCCVV | NFYB | 2.59e-11 | 2.12e-13 | 2650 | 5.97 | 2040 | 4.81 | 1.24 |
| 73 | 14 | MA0106.3 | TP53 | MA0106.3 | RACATGYCCGGRCATGTY | TP53 | 2.7e-11 | 2.18e-13 | 5100 | 11.5 | 4190 | 9.9 | 1.16 |
| 74 | 5 | MA1530.1 | NKX6-3 | MA1530.1 | VYCATTAWW | NKX6-3 | 5.68e-11 | 4.52e-13 | 23700 | 53.2 | 21500 | 50.7 | 1.05 |
| 75 | 9 | MA0142.1 | Pou5f1::Sox2 | MA0142.1 | CWTTGTYATGCAAAT | Pou5f1::Sox2 | 6.16e-11 | 4.83e-13 | 31500 | 70.9 | 29000 | 68.6 | 1.03 |
| 76 | 15 | MA0148.4 | FOXA1 | MA0148.4 | WWGTAAACATNN | FOXA1 | 6.52e-11 | 5.05e-13 | 36800 | 82.8 | 34200 | 80.8 | 1.02 |
| 77 | 1 | MA1640.1 | MEIS2 | MA1640.1 | NNATGATTTATGDNH | MEIS2 | 7.83e-11 | 5.98e-13 | 35500 | 79.8 | 32900 | 77.8 | 1.03 |
| 78 | 9 | MA0786.1 | POU3F1 | MA0786.1 | WTATGCWAATKW | POU3F1 | 1.16e-10 | 8.72e-13 | 34200 | 77 | 31700 | 74.8 | 1.03 |
| 79 | 1 | MA1113.2 | PBX2 | MA1113.2 | NHCATAAATCAHN | PBX2 | 1.64e-10 | 1.22e-12 | 30300 | 68.2 | 27900 | 65.9 | 1.04 |
| 80 | 9 | MA0785.1 | POU2F1 | MA0785.1 | AWTATGCWAATK | POU2F1 | 2.42e-10 | 1.78e-12 | 32100 | 72.3 | 29700 | 70.1 | 1.03 |
| 81 | 15 | MA0848.1 | FOXO4 | MA0848.1 | GTAAACA | FOXO4 | 2.62e-10 | 1.9e-12 | 36200 | 81.5 | 33600 | 79.5 | 1.02 |
| 82 | 9 | MA0789.1 | POU3F4 | MA0789.1 | TATGCWAAT | POU3F4 | 2.89e-10 | 2.07e-12 | 29200 | 65.7 | 26800 | 63.4 | 1.04 |
| 83 | 8 | MA1683.1 | FOXA3 | MA1683.1 | DWGTAAACANN | FOXA3 | 3.57e-10 | 2.53e-12 | 34400 | 77.3 | 31800 | 75.3 | 1.03 |
| 84 | 5 | MA0879.2 | DLX1 | MA0879.2 | VYAATTAN | DLX1 | 3.78e-10 | 2.65e-12 | 34000 | 76.4 | 31400 | 74.3 | 1.03 |
| 85 | 5 | MA1505.1 | HOXC8 | MA1505.1 | KTAATTAM | HOXC8 | 6.16e-10 | 4.26e-12 | 33700 | 75.9 | 31200 | 73.8 | 1.03 |
| 86 | 15 | MA0033.2 | FOXL1 | MA0033.2 | RTAAACA | FOXL1 | 9.73e-10 | 6.66e-12 | 35800 | 80.4 | 33200 | 78.5 | 1.02 |
| 87 | 9 | MA0787.1 | POU3F2 | MA0787.1 | WTATGCWAATKA | POU3F2 | 9.93e-10 | 6.72e-12 | 29500 | 66.5 | 27200 | 64.2 | 1.04 |
| 88 | 5 | MA0900.2 | HOXA2 | MA0900.2 | STAATTAS | HOXA2 | 1.14e-09 | 7.6e-12 | 16400 | 36.8 | 14600 | 34.6 | 1.07 |
| 89 | 5 | MA0675.1 | NKX6-2 | MA0675.1 | NYMATTAA | NKX6-2 | 1.5e-09 | 9.93e-12 | 32300 | 72.7 | 29900 | 70.6 | 1.03 |
| 90 | 6 | MA1108.2 | MXI1 | MA1108.2 | NNCACATGBN | MXI1 | 1.55e-09 | 1.01e-11 | 19300 | 43.4 | 17400 | 41.1 | 1.06 |
| 91 | 8 | MA0481.3 | FOXP1 | MA0481.3 | NDGTAAACANH | FOXP1 | 1.63e-09 | 1.06e-11 | 38100 | 85.8 | 35600 | 84.1 | 1.02 |
| 92 | 8 | MA0047.3 | FOXA2 | MA0047.3 | RWGTAAACANN | FOXA2 | 2.26e-09 | 1.45e-11 | 39400 | 88.6 | 36900 | 87.1 | 1.02 |
| 93 | 23 | MA0498.2 | MEIS1 | MA0498.2 | HTGACAD | MEIS1 | 4.81e-09 | 3.04e-11 | 35500 | 79.8 | 33000 | 78 | 1.02 |
| 94 | 5 | MA0701.2 | LHX9 | MA0701.2 | CYAATTAR | LHX9 | 5.96e-09 | 3.73e-11 | 13200 | 29.8 | 11700 | 27.7 | 1.08 |
| 95 | 8 | MA1103.2 | FOXK2 | MA1103.2 | DDGTAAACANN | FOXK2 | 6.94e-09 | 4.3e-11 | 41000 | 92.3 | 38500 | 91 | 1.01 |
| 96 | 5 | MA0644.2 | ESX1 | MA0644.2 | YYAATTAN | ESX1 | 7.47e-09 | 4.58e-11 | 26900 | 60.6 | 24700 | 58.3 | 1.04 |
| 97 | 9 | MA0627.2 | POU2F3 | MA0627.2 | NWTATGCAAATNH | POU2F3 | 1.62e-08 | 9.8e-11 | 31900 | 71.7 | 29500 | 69.7 | 1.03 |
| 98 | 16 | MA1479.1 | DMRTC2 | MA1479.1 | AATTGMTACATT | DMRTC2 | 2e-08 | 1.2e-10 | 34600 | 77.9 | 32200 | 76 | 1.02 |
| 99 | 5 | MA0708.2 | MSX2 | MA0708.2 | SCAATTAV | MSX2 | 2.37e-08 | 1.41e-10 | 30900 | 69.6 | 28600 | 67.6 | 1.03 |
| 100 | 9 | MA0788.1 | POU3F3 | MA0788.1 | WWTATGCWAATTW | POU3F3 | 2.56e-08 | 1.5e-10 | 31300 | 70.4 | 28900 | 68.4 | 1.03 |
| 101 | 8 | MA1978.1 | ZNF354A | MA1978.1 | HWAADWATAAATGRAYWAWTT | ZNF354A | 2.63e-08 | 1.53e-10 | 32700 | 73.5 | 30300 | 71.5 | 1.03 |
| 102 | 5 | MA1549.1 | POU6F1 | MA1549.1 | RTAATGAGVN | POU6F1 | 4.01e-08 | 2.31e-10 | 25800 | 58.2 | 23700 | 56 | 1.04 |
| 103 | 5 | MA0666.2 | MSX1 | MA0666.2 | SCAATTAR | MSX1 | 4.56e-08 | 2.61e-10 | 28700 | 64.5 | 26400 | 62.4 | 1.03 |
| 104 | 17 | MA1120.1 | SOX13 | MA1120.1 | DVACAATGGNN | SOX13 | 4.92e-08 | 2.78e-10 | 38400 | 86.3 | 35900 | 84.8 | 1.02 |
| 105 | 5 | MA0882.1 | DLX6 | MA0882.1 | VYAATTAN | DLX6 | 5.95e-08 | 3.34e-10 | 38400 | 86.3 | 35900 | 84.8 | 1.02 |
| 106 | 18 | MA0791.1 | POU4F3 | MA0791.1 | RTGMATWATTAATGAR | POU4F3 | 6.01e-08 | 3.34e-10 | 26400 | 59.4 | 24200 | 57.3 | 1.04 |
| 107 | 5 | MA0718.1 | RAX | MA0718.1 | RYYAATTARH | RAX | 6.68e-08 | 3.67e-10 | 36200 | 81.4 | 33700 | 79.7 | 1.02 |
| 108 | 23 | MA0117.2 | Mafb | MA0117.2 | AWWNTGCTGACD | Mafb | 9.29e-08 | 5.06e-10 | 34500 | 77.5 | 32000 | 75.7 | 1.02 |
| 109 | 23 | MA0842.2 | NRL | MA0842.2 | AAWWNTGCTGACG | NRL | 9.86e-08 | 5.32e-10 | 6930 | 15.6 | 5950 | 14 | 1.11 |
| 110 | 10 | MA0018.4 | CREB1 | MA0018.4 | NNRTGATGTCAYN | CREB1 | 1.04e-07 | 5.59e-10 | 19200 | 43.3 | 17400 | 41.2 | 1.05 |
| 111 | 5 | MA0887.1 | EVX1 | MA0887.1 | VVTAATTABS | EVX1 | 1.48e-07 | 7.85e-10 | 30000 | 67.6 | 27700 | 65.6 | 1.03 |
| 112 | 5 | MA0709.1 | Msx3 | MA0709.1 | SCAATTAN | Msx3 | 1.82e-07 | 9.58e-10 | 23600 | 53 | 21500 | 50.9 | 1.04 |
| 113 | 5 | MA0722.1 | VAX1 | MA0722.1 | YTAATTAH | VAX1 | 2.48e-07 | 1.29e-09 | 25100 | 56.6 | 23100 | 54.5 | 1.04 |
| 114 | 5 | MA0654.1 | ISX | MA0654.1 | YYAATTAR | ISX | 3.63e-07 | 1.87e-09 | 23900 | 53.9 | 21900 | 51.8 | 1.04 |
| 115 | 5 | MA0880.1 | Dlx3 | MA0880.1 | NYAATTAN | Dlx3 | 4.06e-07 | 2.08e-09 | 24900 | 56.1 | 22900 | 54 | 1.04 |
| 116 | 5 | MA0710.1 | NOTO | MA0710.1 | NYTAATTAVN | NOTO | 6.04e-07 | 3.06e-09 | 24900 | 56.1 | 22900 | 54.1 | 1.04 |
| 117 | 19 | MA0611.2 | Dux | MA0611.2 | NTGATTBAATCAVWNN | Dux | 7.78e-07 | 3.91e-09 | 13600 | 30.6 | 12200 | 28.7 | 1.06 |
| 118 | 5 | MA1476.2 | Dlx5 | MA1476.2 | NDGCAATTAGNN | Dlx5 | 8.72e-07 | 4.35e-09 | 37600 | 84.6 | 35200 | 83.1 | 1.02 |
| 119 | 20 | MA0797.1 | TGIF2 | MA0797.1 | TGACAGSTGTCA | TGIF2 | 8.86e-07 | 4.38e-09 | 13600 | 30.7 | 12200 | 28.8 | 1.06 |
| 120 | 21 | MA0090.3 | TEAD1 | MA0090.3 | NNACATTCCAGSN | TEAD1 | 1.05e-06 | 5.15e-09 | 33000 | 74.2 | 30600 | 72.4 | 1.02 |
| 121 | 5 | MA0704.1 | Lhx4 | MA0704.1 | YTAATTAR | Lhx4 | 1.21e-06 | 5.87e-09 | 37200 | 83.6 | 34700 | 82.1 | 1.02 |
| 122 | 5 | MA0700.2 | LHX2 | MA0700.2 | DDCCAATTADN | LHX2 | 1.38e-06 | 6.62e-09 | 33900 | 76.2 | 31500 | 74.5 | 1.02 |
| 123 | 15 | MA0042.2 | FOXI1 | MA0042.2 | GTAAACA | FOXI1 | 1.38e-06 | 6.62e-09 | 35200 | 79.2 | 32800 | 77.6 | 1.02 |
| 124 | 22 | MA1124.1 | ZNF24 | MA1124.1 | CATTCATTCATTC | ZNF24 | 2.21e-06 | 1.05e-08 | 22700 | 51.2 | 20800 | 49.2 | 1.04 |
| 125 | 5 | MA0885.2 | Dlx2 | MA0885.2 | NDGCAATTAGNN | Dlx2 | 3.34e-06 | 1.57e-08 | 36800 | 82.9 | 34400 | 81.4 | 1.02 |
| 126 | 5 | MA0658.1 | LHX6 | MA0658.1 | RCTAATTARY | LHX6 | 3.35e-06 | 1.57e-08 | 18700 | 42.2 | 17000 | 40.2 | 1.05 |
| 127 | 5 | MA0889.1 | GBX1 | MA0889.1 | VCYAATTASY | GBX1 | 4.57e-06 | 2.12e-08 | 18400 | 41.3 | 16700 | 39.4 | 1.05 |
| 128 | 23 | MA0495.3 (MAFF) | STREME-7 | 7-CTGTCAGCACT | CTGTCAGCACT | MAFF | 4.65e-06 | 2.14e-08 | 1140 | 2.57 | 843 | 1.99 | 1.29 |
| 129 | 24 | MA1114.1 | PBX3 | MA1114.1 | NNNTGAGTGACAGGBNN | PBX3 | 4.69e-06 | 2.14e-08 | 14500 | 32.6 | 13000 | 30.8 | 1.06 |
| 130 | 5 | MA0793.1 | POU6F2 | MA0793.1 | ASCTMATTAW | POU6F2 | 4.88e-06 | 2.21e-08 | 23800 | 53.6 | 21900 | 51.7 | 1.04 |
| 131 | 5 | MA1960.1 | MGA::EVX1 | MA1960.1 | RGGTGWTAATKD | MGA::EVX1 | 4.95e-06 | 2.22e-08 | 30100 | 67.6 | 27800 | 65.8 | 1.03 |
| 132 | 8 | MA0032.2 | FOXC1 | MA0032.2 | WAWGTAAAYAW | FOXC1 | 6.47e-06 | 2.88e-08 | 27300 | 61.5 | 25200 | 59.6 | 1.03 |
| 133 | 18 | MA0790.1 | POU4F1 | MA0790.1 | ATGMATAATTAATG | POU4F1 | 6.66e-06 | 2.95e-08 | 27800 | 62.5 | 25700 | 60.6 | 1.03 |
| 134 | 8 | MA0480.2 | Foxo1 | MA0480.2 | NDGTAAACANN | Foxo1 | 7e-06 | 3.07e-08 | 40300 | 90.7 | 37900 | 89.5 | 1.01 |
| 135 | 25 | MA0755.1 | CUX2 | MA0755.1 | TRATCGATAW | CUX2 | 7.1e-06 | 3.09e-08 | 24000 | 53.9 | 22000 | 52 | 1.04 |
| 136 | 5 | MA0890.1 | GBX2 | MA0890.1 | RSYAATTARN | GBX2 | 7.27e-06 | 3.15e-08 | 23800 | 53.5 | 21800 | 51.6 | 1.04 |
| 137 | 14 | MA0861.1 | TP73 | MA0861.1 | DRCATGTCNNRACAYGYM | TP73 | 7.8e-06 | 3.35e-08 | 7720 | 17.4 | 6740 | 15.9 | 1.09 |
| 138 | 20 | MA0774.1 | MEIS2 | MA0774.1 | TTGACAGS | MEIS2 | 7.9e-06 | 3.37e-08 | 4620 | 10.4 | 3920 | 9.27 | 1.12 |
| 139 | 6 | MA1618.1 | Ptf1a | MA1618.1 | NNACAGATGTTNN | Ptf1a | 9.02e-06 | 3.82e-08 | 36900 | 83.1 | 34600 | 81.7 | 1.02 |
| 140 | 6 | MA1570.1 | TFAP4 | MA1570.1 | AHCATRTGDT | TFAP4 | 9.34e-06 | 3.92e-08 | 15400 | 34.6 | 13900 | 32.8 | 1.05 |
| 141 | 5 | MA0488.1 | JUN | MA0488.1 | DDRATGATGTCAT | JUN | 1.01e-05 | 4.21e-08 | 7520 | 16.9 | 6570 | 15.5 | 1.09 |
| 142 | 8 | MA1607.1 | Foxl2 | MA1607.1 | DDWATGTAAACANN | Foxl2 | 1.06e-05 | 4.4e-08 | 37700 | 84.8 | 35300 | 83.4 | 1.02 |
| 143 | 38 | MA1107.2 | KLF9 | MA1107.2 | MNGCCACACCCACHYV | KLF9 | 1.1e-05 | 4.52e-08 | 1990 | 4.48 | 1580 | 3.73 | 1.2 |
| 144 | 26 | MA0805.1 | TBX1 | MA0805.1 | AGGTGTGA | TBX1 | 1.15e-05 | 4.7e-08 | 2260 | 5.09 | 1810 | 4.29 | 1.19 |
| 145 | 5 | MA0723.2 | VAX2 | MA0723.2 | BYAATTAV | VAX2 | 1.25e-05 | 5.08e-08 | 38500 | 86.7 | 36100 | 85.4 | 1.02 |
| 146 | 6 | MA0871.2 | TFEC | MA0871.2 | NVCACGTGACN | TFEC | 1.61e-05 | 6.5e-08 | 10600 | 23.9 | 9430 | 22.3 | 1.07 |
| 147 | 5 | MA0075.3 | PRRX2 | MA0075.3 | CTAATTAN | PRRX2 | 2.05e-05 | 8.2e-08 | 17600 | 39.7 | 16000 | 37.9 | 1.05 |
| 148 | 24 | MA0063.2 | NKX2-5 | MA0063.2 | NNCACTCAANN | NKX2-5 | 2.18e-05 | 8.68e-08 | 41000 | 92.2 | 38600 | 91.2 | 1.01 |
| 149 | 6 | MA0461.2 | Atoh1 | MA0461.2 | RMCATATGBY | Atoh1 | 4.03e-05 | 1.59e-07 | 30900 | 69.5 | 28700 | 67.8 | 1.02 |
| 150 | 17 | MA0514.2 | Sox3 | MA0514.2 | DNACAATGGNN | Sox3 | 4.17e-05 | 1.64e-07 | 40100 | 90.2 | 37700 | 89 | 1.01 |
| 151 | 5 | MA0699.1 | LBX2 | MA0699.1 | NCYAATTARN | LBX2 | 4.48e-05 | 1.74e-07 | 21300 | 48 | 19500 | 46.2 | 1.04 |
| 152 | 14 | MA0525.2 | TP63 | MA0525.2 | RACATGTYGKGACATGTC | TP63 | 4.56e-05 | 1.76e-07 | 404 | 0.91 | 252 | 0.6 | 1.52 |
| 153 | 27 | MA0060.3 | NFYA | MA0060.3 | RRCCAATCAGM | NFYA | 4.8e-05 | 1.85e-07 | 39400 | 88.8 | 37100 | 87.6 | 1.01 |
| 154 | 5 | MA1480.1 | DPRX | MA1480.1 | AGATAATCCN | DPRX | 5.72e-05 | 2.18e-07 | 35500 | 79.8 | 33200 | 78.4 | 1.02 |
| 155 | 8 | MA0851.1 | Foxj3 | MA0851.1 | NDAADGTAAACAAANNM | Foxj3 | 6.97e-05 | 2.65e-07 | 24900 | 56 | 22900 | 54.2 | 1.03 |
| 156 | 5 | MA0881.1 | Dlx4 | MA0881.1 | VYAATTAN | Dlx4 | 8.13e-05 | 3.07e-07 | 38400 | 86.4 | 36000 | 85.1 | 1.01 |
| 157 | 5 | MA1499.1 | HOXB4 | MA1499.1 | RTMATTAR | HOXB4 | 8.71e-05 | 3.26e-07 | 17100 | 38.5 | 15600 | 36.8 | 1.05 |
| 158 | 24 | MA0782.2 | PKNOX1 | MA0782.2 | NNTGAGTGACAGVNN | PKNOX1 | 9.04e-05 | 3.36e-07 | 9480 | 21.3 | 8420 | 19.9 | 1.07 |
| 159 | 5 | MA1481.1 | DRGX | MA1481.1 | YYAATTAN | DRGX | 0.000114 | 4.22e-07 | 34600 | 78 | 32400 | 76.5 | 1.02 |
| 160 | 5 | MA0720.1 | Shox2 | MA0720.1 | YYAATTAR | Shox2 | 0.000129 | 4.73e-07 | 16600 | 37.3 | 15100 | 35.6 | 1.05 |
| 161 | 5 | MA1496.1 | HOXA4 | MA1496.1 | RTMATTAV | HOXA4 | 0.000157 | 5.74e-07 | 27400 | 61.7 | 25400 | 60 | 1.03 |
| 162 | 13 | MA0036.3 | GATA2 | MA0036.3 | NBCTTATCTNH | GATA2 | 0.000198 | 7.19e-07 | 24100 | 54.2 | 22200 | 52.5 | 1.03 |
| 163 | 5 | MA1500.1 | HOXB6 | MA1500.1 | BTAATKRC | HOXB6 | 0.000202 | 7.3e-07 | 29400 | 66.2 | 27300 | 64.6 | 1.03 |
| 164 | 28 | MA0596.1 | SREBF2 | MA0596.1 | RTGGGGTGAY | SREBF2 | 0.00033 | 1.18e-06 | 5520 | 12.4 | 4800 | 11.3 | 1.1 |
| 165 | 15 | MA0613.1 | FOXG1 | MA0613.1 | RTAAACAW | FOXG1 | 0.000345 | 1.23e-06 | 18400 | 41.5 | 16900 | 39.9 | 1.04 |
| 166 | 17 | MA0143.4 | SOX2 | MA0143.4 | DNACAATGGNN | SOX2 | 0.000355 | 1.26e-06 | 41600 | 93.5 | 39200 | 92.7 | 1.01 |
| 167 | 17 | MA1563.2 | SOX18 | MA1563.2 | AACAATDV | SOX18 | 0.00038 | 1.34e-06 | 31900 | 71.8 | 29800 | 70.3 | 1.02 |
| 168 | 5 | MA0680.2 | Pax7 | MA0680.2 | DWTAATCAATTAWNH | Pax7 | 0.000392 | 1.37e-06 | 26500 | 59.6 | 24500 | 57.9 | 1.03 |
| 169 | 5 | MA0888.1 | EVX2 | MA0888.1 | VVTAATTAVB | EVX2 | 0.000392 | 1.37e-06 | 29800 | 67.1 | 27700 | 65.5 | 1.02 |
| 170 | 23 | MA0495.3 | MAFF | MA0495.3 | DWGTCAGCATTTTWHN | MAFF | 0.000413 | 1.43e-06 | 37000 | 83.4 | 34700 | 82.1 | 1.02 |
| 171 | 5 | MA0892.1 | GSX1 | MA0892.1 | NCYMATTAVN | GSX1 | 0.000427 | 1.47e-06 | 43200 | 97.2 | 40900 | 96.6 | 1.01 |
| 172 | 27 | MA0035.4 | GATA1 | MA0035.4 | NHCTAATCTDH | GATA1 | 0.000514 | 1.76e-06 | 19800 | 44.6 | 18200 | 42.9 | 1.04 |
| 173 | 5 | MA0716.1 | PRRX1 | MA0716.1 | YYAATTAR | PRRX1 | 0.000542 | 1.84e-06 | 37900 | 85.3 | 35600 | 84.1 | 1.01 |
| 174 | 17 | MA0077.1 | SOX9 | MA0077.1 | CYATTGTTY | SOX9 | 0.00069 | 2.33e-06 | 41100 | 92.5 | 38800 | 91.6 | 1.01 |
| 175 | 17 | MA1562.1 | SOX14 | MA1562.1 | CCGAACAATG | SOX14 | 0.000693 | 2.33e-06 | 25200 | 56.8 | 23300 | 55.2 | 1.03 |
| 176 | 26 | MA0803.1 | TBX15 | MA0803.1 | AGGTGTGA | TBX15 | 0.000781 | 2.61e-06 | 29600 | 66.6 | 27500 | 65.1 | 1.02 |
| 177 | 5 | MA0628.1 | POU6F1 | MA0628.1 | AYTAATTART | POU6F1 | 0.000872 | 2.9e-06 | 27700 | 62.4 | 25700 | 60.8 | 1.03 |
| 178 | 20 | MA0691.1 | TFAP4 | MA0691.1 | AWCAGCTGWT | TFAP4 | 0.00109 | 3.61e-06 | 4140 | 9.3 | 3550 | 8.39 | 1.11 |
| 179 | 5 | MA0630.1 | SHOX | MA0630.1 | HTAATTRR | SHOX | 0.00119 | 3.9e-06 | 40800 | 91.8 | 38500 | 90.9 | 1.01 |
| 180 | 29 | MA0073.1 | RREB1 | MA0073.1 | CCCCMAAMCAMCCMCMMMCV | RREB1 | 0.00135 | 4.41e-06 | 856 | 1.93 | 640 | 1.51 | 1.27 |
| 181 | 1 | MA0070.1 | PBX1 | MA0070.1 | HCATCAATCAAW | PBX1 | 0.00162 | 5.27e-06 | 15700 | 35.2 | 14300 | 33.7 | 1.04 |
| 182 | 8 | MA0897.1 | Hmx2 | MA0897.1 | ASAAGCAATTAAHVVRT | Hmx2 | 0.0019 | 6.14e-06 | 24100 | 54.2 | 22300 | 52.6 | 1.03 |
| 183 | 30 | MA1573.2 | Thap11 | MA1573.2 | ACTACAABTCCCAG | Thap11 | 0.0024 | 7.72e-06 | 140 | 0.32 | 69 | 0.16 | 1.92 |
| 184 | 8 | MA0157.3 | Foxo3 | MA0157.3 | NDGTAAACARNN | Foxo3 | 0.00291 | 9.32e-06 | 42200 | 95.1 | 39900 | 94.4 | 1.01 |
| 185 | 31 | MA0698.1 | ZBTB18 | MA0698.1 | NATCCAGATGTKB | ZBTB18 | 0.00339 | 1.08e-05 | 10500 | 23.6 | 9430 | 22.3 | 1.06 |
| 186 | 18 | MA0153.2 | HNF1B | MA0153.2 | GTTAATNATTAAY | HNF1B | 0.00369 | 1.17e-05 | 17400 | 39.2 | 15900 | 37.7 | 1.04 |
| 187 | 15 | MA0031.1 | FOXD1 | MA0031.1 | GTAAACAW | FOXD1 | 0.00389 | 1.22e-05 | 28800 | 64.9 | 26800 | 63.4 | 1.02 |
| 188 | 17 | MA1152.1 | SOX15 | MA1152.1 | CYWTTGTTHW | SOX15 | 0.00393 | 1.23e-05 | 36400 | 81.8 | 34100 | 80.7 | 1.01 |
| 189 | 24 | MA1645.1 | NKX2-2 | MA1645.1 | NNNCCACTCAANNN | NKX2-2 | 0.00434 | 1.35e-05 | 7120 | 16 | 6320 | 14.9 | 1.07 |
| 190 | 31 | MA0818.2 | BHLHE22 | MA0818.2 | AMCATATGKY | BHLHE22 | 0.00443 | 1.37e-05 | 9680 | 21.8 | 8690 | 20.6 | 1.06 |
| 13 | 34 | MA0682.2 (PITX1) | MEME-3 | GCCTGTAATCCCAGC | GCCTGTAATCCCAGC | PITX1 | 0.00449 | 0.000345 | 1360 | 3.06 | 1130 | 2.66 | 1.15 |
| 191 | 18 | MA0046.2 | HNF1A | MA0046.2 | DRTTAATNATTAACN | HNF1A | 0.00527 | 1.62e-05 | 18400 | 41.4 | 16900 | 40 | 1.04 |
| 192 | 5 | MA1608.1 | Isl1 | MA1608.1 | NNCCATTAGNN | Isl1 | 0.00606 | 1.86e-05 | 40700 | 91.6 | 38400 | 90.8 | 1.01 |
| 193 | 32 | MA1974.1 | ZNF211 | MA1974.1 | HWYATATACCAYRYW | ZNF211 | 0.00653 | 1.99e-05 | 11200 | 25.2 | 10100 | 24 | 1.05 |
| 194 | 13 | MA1104.2 | GATA6 | MA1104.2 | HWWTCTTATCTNH | GATA6 | 0.00671 | 2.04e-05 | 21900 | 49.2 | 20200 | 47.7 | 1.03 |
| 195 | 15 | MA0849.1 | FOXO6 | MA0849.1 | GTAAACA | FOXO6 | 0.00813 | 2.45e-05 | 24400 | 54.9 | 22600 | 53.4 | 1.03 |
| 196 | 28 | MA0595.1 | SREBF1 | MA0595.1 | VTCACCCCAY | SREBF1 | 0.00886 | 2.66e-05 | 2580 | 5.81 | 2180 | 5.15 | 1.13 |
| 197 | 6 | MA0004.1 | Arnt | MA0004.1 | CACGTG | Arnt | 0.00929 | 2.78e-05 | 752 | 1.69 | 566 | 1.34 | 1.26 |
| 198 | 33 | MA0052.4 | MEF2A | MA0052.4 | DDCTAAAAATAGMHH | MEF2A | 0.0142 | 4.2e-05 | 33800 | 76 | 31600 | 74.8 | 1.02 |
| 199 | 26 | MA0689.1 | TBX20 | MA0689.1 | WAGGTGTGAAR | TBX20 | 0.0142 | 4.21e-05 | 6840 | 15.4 | 6080 | 14.4 | 1.07 |
| 200 | 20 | MA1572.1 | TGIF2LY | MA1572.1 | TGACAGCTGTCA | TGIF2LY | 0.0145 | 4.27e-05 | 36200 | 81.6 | 34000 | 80.5 | 1.01 |
| 201 | 5 | MA0713.1 | PHOX2A | MA0713.1 | TAATYYAATTA | PHOX2A | 0.0174 | 5.09e-05 | 28200 | 63.4 | 26200 | 62 | 1.02 |
| 202 | 5 | MA0914.1 | ISL2 | MA0914.1 | GCAMTTAR | ISL2 | 0.0199 | 5.81e-05 | 30700 | 69.1 | 28700 | 67.8 | 1.02 |
| 203 | 26 | MA1567.2 | Tbx6 | MA1567.2 | NRGGTGTGAANN | Tbx6 | 0.0203 | 5.89e-05 | 5620 | 12.7 | 4970 | 11.8 | 1.08 |
| 204 | 5 | MA0674.1 | NKX6-1 | MA0674.1 | NTMATTAA | NKX6-1 | 0.0224 | 6.45e-05 | 29000 | 65.3 | 27100 | 64 | 1.02 |
| 205 | 15 | MA0850.1 | FOXP3 | MA0850.1 | RTAAACA | FOXP3 | 0.0245 | 7.03e-05 | 30100 | 67.8 | 28100 | 66.5 | 1.02 |
| 206 | 27 | MA1644.1 | NFYC | MA1644.1 | RDCCAATCASN | NFYC | 0.0248 | 7.07e-05 | 39800 | 89.5 | 37500 | 88.6 | 1.01 |
| 207 | 9 | MA0025.2 | NFIL3 | MA0025.2 | NRTTATGYAATNN | NFIL3 | 0.0252 | 7.16e-05 | 18100 | 40.8 | 16700 | 39.5 | 1.03 |
| 208 | 26 | MA0800.1 | EOMES | MA0800.1 | RAGGTGTGAAAWN | EOMES | 0.0298 | 8.43e-05 | 37500 | 84.3 | 35200 | 83.3 | 1.01 |
| 209 | 26 | MA0802.1 | TBR1 | MA0802.1 | AGGTGTGAAA | TBR1 | 0.0323 | 9.1e-05 | 38000 | 85.5 | 35800 | 84.5 | 1.01 |
| 210 | 5 | MA0642.2 | EN2 | MA0642.2 | VSYAATTA | EN2 | 0.0422 | 0.000118 | 29400 | 66.2 | 27500 | 65 | 1.02 |
| 14 | 35 | MA1973.1 (ZKSCAN3) | MEME-6 | TGCCCAGGCTG | TGCCCAGGCTG | ZKSCAN3 | 2.1 | 0.15 | 532 | 1.2 | 470 | 1.11 | 1.08 |
| 15 | 36 | MA0471.2 (E2F6) | MEME-7 | DGAGGARGGAR | DGAGGARGGAR | E2F6 | 3.33 | 0.222 | 14000 | 31.4 | 13200 | 31.2 | 1.01 |
| 16 | 37 | MA1125.1 (ZNF384) | MEME-1 | AAAAAAAAAAAADWA | AAAAAAAAAAAADWA | ZNF384 | 3.75 | 0.235 | 1610 | 3.62 | 1490 | 3.52 | 1.03 |
| 17 | 38 | MA1961.1 (PATZ1) | MEME-5 | GGGRGGCGGRGGCGG | GGGRGGCGGRGGCGG | PATZ1 | 17 | 1 | 9820 | 22.1 | 10400 | 24.5 | 0.902 |
ER_rat <- 1.25
DNR_24_20kb_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,TP.)%>%
arrange(.,EVALUE.y) %>%
slice_head(n=80)%>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste("DNR_24_20kb_combo response peaks(200bp)Enrichment ratio:",ER_rat))
DNR_24_20kb_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue = -log10(if_else(EVALUE.y == 0, 1e-300, EVALUE.y))) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
slice_head(n=7) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*9), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./9,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste("DNR_24_20kb_combo response peaks(200bp)Enrichment ratio:",ER_rat))
MTX_DAR_3hr data
MTX_DAR_3hr_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/MTX_3_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
MTX_DAR_3hr_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/MTX_3_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
MTX_DAR_3hr_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/MTX_3_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
MTX_DAR_3hr_all <- rbind(MTX_DAR_3hr_sea_disc, MTX_DAR_3hr_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)MTX_DAR_3hr_combo <- MTX_DAR_3hr_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., MTX_DAR_3hr_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ NA_character_
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y) %>%
mutate(mrc_type="MTX_DAR_3hr")
MTX_DAR_3hr_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs in 3 hour MTX-DAR v 3 hour non-MTX DAR") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO | mrc_type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | MA1513.1 | KLF15 | MA1513.1 | SCCCCGCCCCS | KLF15 | 3.29e-13 | 2.66e-13 | 204 | 28.2 | 22500 | 16.2 | 1.75 | MTX_DAR_3hr |
| 2 | 2 | MA1513.1 (KLF15) | STREME-1 | 1-GCGGGCC | GCGGGCC | KLF15 | 2.67e-12 | 1.08e-12 | 303 | 41.8 | 39300 | 28.2 | 1.49 | MTX_DAR_3hr |
| 3 | 3 | MA1650.1 (ZBTB14) | STREME-3 | 3-CCGCGCBCCCCSS | CCGCGCBCCCCSS | ZBTB14 | 8.67e-11 | 2.34e-11 | 83 | 11.5 | 6460 | 4.64 | 2.5 | MTX_DAR_3hr |
| 4 | 4 | MA1650.1 (ZBTB14) | STREME-6 | 6-CCGCGMA | CCGCGMA | ZBTB14 | 1.49e-10 | 2.83e-11 | 309 | 42.7 | 41500 | 29.8 | 1.43 | MTX_DAR_3hr |
| 5 | 5 | 2-CCGGCGCK | STREME-2 | 2-CCGGCGCK | CCGGCGCK | CCGGCGCK | 1.9e-10 | 2.83e-11 | 119 | 16.4 | 11300 | 8.1 | 2.04 | MTX_DAR_3hr |
| 6 | 1 | MA0742.2 | KLF12 | MA0742.2 | GGGGCGGGG | KLF12 | 2.1e-10 | 2.83e-11 | 423 | 58.4 | 62600 | 44.9 | 1.3 | MTX_DAR_3hr |
| 7 | 6 | MA1653.1 (ZNF148) | STREME-4 | 4-CTMCCS | CTMCCS | ZNF148 | 2.8e-10 | 3.24e-11 | 529 | 73.1 | 83900 | 60.2 | 1.21 | MTX_DAR_3hr |
| 8 | 3 | MA1961.1 (PATZ1) | MEME-2 | CSCSCCCCKSCSCGS | CSCSCCCCKSCSCGS | PATZ1 | 9.69e-10 | 9.82e-11 | 170 | 23.5 | 19100 | 13.7 | 1.72 | MTX_DAR_3hr |
| 9 | 1 | MA1653.1 | ZNF148 | MA1653.1 | YBCCCCTCCCCC | ZNF148 | 1.69e-09 | 1.52e-10 | 385 | 53.2 | 56100 | 40.3 | 1.32 | MTX_DAR_3hr |
| 10 | 1 | MA1959.1 | KLF7 | MA1959.1 | DGGGCGGGG | KLF7 | 3.27e-09 | 2.65e-10 | 363 | 50.1 | 52300 | 37.5 | 1.34 | MTX_DAR_3hr |
| 11 | 7 | MA1615.1 | Plagl1 | MA1615.1 | NNCTGGGGCCABN | Plagl1 | 4.18e-09 | 3.08e-10 | 392 | 54.1 | 57700 | 41.5 | 1.31 | MTX_DAR_3hr |
| 12 | 1 | MA1961.1 | PATZ1 | MA1961.1 | SSGGGGMGGGGS | PATZ1 | 3.43e-08 | 2.32e-09 | 116 | 16 | 11800 | 8.48 | 1.9 | MTX_DAR_3hr |
| 13 | 1 | MA0516.3 | SP2 | MA0516.3 | GGGGCGGGG | SP2 | 9.28e-08 | 5.78e-09 | 125 | 17.3 | 13300 | 9.56 | 1.82 | MTX_DAR_3hr |
| 14 | 1 | MA1965.1 | SP5 | MA1965.1 | CBCCTCCCHS | SP5 | 1.12e-07 | 6.49e-09 | 429 | 59.2 | 66100 | 47.4 | 1.25 | MTX_DAR_3hr |
| 15 | 1 | MA1713.1 | ZNF610 | MA1713.1 | SSCGCCGCTCCSSS | ZNF610 | 1.63e-07 | 8.83e-09 | 175 | 24.2 | 21100 | 15.2 | 1.6 | MTX_DAR_3hr |
| 16 | 8 | MA0527.1 | ZBTB33 | MA0527.1 | BTCTCGCGAGABYTS | ZBTB33 | 3.4e-07 | 1.7e-08 | 128 | 17.7 | 14100 | 10.1 | 1.76 | MTX_DAR_3hr |
| 17 | 7 | MA1986.1 | ZNF692 | MA1986.1 | BGGGCCCASN | ZNF692 | 3.58e-07 | 1.7e-08 | 425 | 58.7 | 65800 | 47.2 | 1.24 | MTX_DAR_3hr |
| 18 | 1 | MA1522.1 | MAZ | MA1522.1 | BBCCCCTCCCY | MAZ | 4.1e-07 | 1.84e-08 | 330 | 45.6 | 48000 | 34.5 | 1.32 | MTX_DAR_3hr |
| 19 | 9 | MA0733.1 | EGR4 | MA0733.1 | NHMCGCCCACGCAHWT | EGR4 | 5.68e-07 | 2.42e-08 | 399 | 55.1 | 61000 | 43.8 | 1.26 | MTX_DAR_3hr |
| 20 | 1 | MA1511.2 | KLF10 | MA1511.2 | GGGGCGGGG | KLF10 | 9.02e-07 | 3.65e-08 | 311 | 43 | 44900 | 32.2 | 1.34 | MTX_DAR_3hr |
| 21 | 10 | MA0750.2 | ZBTB7A | MA0750.2 | NVCCGGAAGTGSV | ZBTB7A | 1.44e-06 | 5.57e-08 | 439 | 60.6 | 69100 | 49.6 | 1.22 | MTX_DAR_3hr |
| 22 | 1 | MA0493.2 | KLF1 | MA0493.2 | DGGGYGGGG | KLF1 | 1.52e-06 | 5.6e-08 | 338 | 46.7 | 50000 | 35.9 | 1.3 | MTX_DAR_3hr |
| 23 | 3 | MA1961.1 (PATZ1) | STREME-5 | 5-CGCCCGCGSCGCCSG | CGCCCGCGSCGCCSG | PATZ1 | 1.72e-06 | 6.06e-08 | 24 | 3.31 | 1010 | 0.73 | 4.74 | MTX_DAR_3hr |
| 24 | 9 | MA0162.4 | EGR1 | MA0162.4 | VCMCGCCCACGCVS | EGR1 | 1.99e-06 | 6.72e-08 | 111 | 15.3 | 11900 | 8.56 | 1.8 | MTX_DAR_3hr |
| 25 | 1 | MA0685.2 | SP4 | MA0685.2 | GGGGCGGGG | SP4 | 2.08e-06 | 6.73e-08 | 308 | 42.5 | 44700 | 32.1 | 1.33 | MTX_DAR_3hr |
| 26 | 1 | MA0740.2 | KLF14 | MA0740.2 | KGGGCGGGG | KLF14 | 2.48e-06 | 7.69e-08 | 254 | 35.1 | 35200 | 25.2 | 1.39 | MTX_DAR_3hr |
| 27 | 11 | MA1721.1 | ZNF93 | MA1721.1 | GGYAGCRGCAGCGGYG | ZNF93 | 2.56e-06 | 7.69e-08 | 93 | 12.8 | 9380 | 6.74 | 1.92 | MTX_DAR_3hr |
| 28 | 3 | MA1650.1 | ZBTB14 | MA1650.1 | SSCCGCGCACVS | ZBTB14 | 2.77e-06 | 8.03e-08 | 270 | 37.3 | 38000 | 27.3 | 1.37 | MTX_DAR_3hr |
| 29 | 1 | MA0079.5 | SP1 | MA0079.5 | GGGGCGGGG | SP1 | 4.58e-06 | 1.28e-07 | 277 | 38.3 | 39400 | 28.3 | 1.35 | MTX_DAR_3hr |
| 30 | 12 | MA0823.1 | HEY1 | MA0823.1 | GRCACGTGCC | HEY1 | 5.55e-06 | 1.5e-07 | 130 | 18 | 15000 | 10.8 | 1.67 | MTX_DAR_3hr |
| 31 | 7 | MA1548.1 | PLAGL2 | MA1548.1 | NGGGCCCCCN | PLAGL2 | 5.89e-06 | 1.54e-07 | 248 | 34.2 | 34400 | 24.7 | 1.39 | MTX_DAR_3hr |
| 32 | 13 | MA1985.1 | ZNF669 | MA1985.1 | GGGGYGAYGACCRCT | ZNF669 | 1.1e-05 | 2.78e-07 | 325 | 44.9 | 48400 | 34.8 | 1.29 | MTX_DAR_3hr |
| 33 | 14 | MA1710.1 | ZNF257 | MA1710.1 | VDGAGGCRAGRG | ZNF257 | 1.61e-05 | 3.94e-07 | 501 | 69.2 | 82500 | 59.2 | 1.17 | MTX_DAR_3hr |
| 34 | 15 | MA1712.1 | ZNF454 | MA1712.1 | DGGCTCSSGGCCCYGVKG | ZNF454 | 1.85e-05 | 4.41e-07 | 445 | 61.5 | 71400 | 51.2 | 1.2 | MTX_DAR_3hr |
| 35 | 16 | MA1982.1 | ZNF574 | MA1982.1 | VGSCTAGAGMGGCCS | ZNF574 | 8.19e-05 | 1.9e-06 | 496 | 68.5 | 82200 | 59 | 1.16 | MTX_DAR_3hr |
| 36 | 1 | MA1630.2 | ZNF281 | MA1630.2 | GGGGGAGGGGVV | ZNF281 | 8.82e-05 | 1.98e-06 | 272 | 37.6 | 39700 | 28.5 | 1.32 | MTX_DAR_3hr |
| 37 | 7 | MA1583.1 | ZFP57 | MA1583.1 | NNVTTGCCGCANN | ZFP57 | 9.77e-05 | 2.14e-06 | 164 | 22.6 | 21300 | 15.3 | 1.49 | MTX_DAR_3hr |
| 38 | 17 | MA1723.1 | PRDM9 | MA1723.1 | RGDGGGVAGGGRGGVRRMARVARR | PRDM9 | 0.000119 | 2.53e-06 | 187 | 25.8 | 25100 | 18 | 1.44 | MTX_DAR_3hr |
| 39 | 12 | MA0522.3 | TCF3 | MA0522.3 | NVCACCTGCNN | TCF3 | 0.000129 | 2.64e-06 | 348 | 48.1 | 53800 | 38.6 | 1.25 | MTX_DAR_3hr |
| 40 | 18 | MA0812.1 | TFAP2B | MA0812.1 | HGCCTSAGGCD | TFAP2B | 0.00013 | 2.64e-06 | 129 | 17.8 | 15700 | 11.3 | 1.59 | MTX_DAR_3hr |
| 41 | 19 | MA0872.1 | TFAP2A | MA0872.1 | TGCCCYSRGGGCA | TFAP2A | 0.000147 | 2.91e-06 | 219 | 30.2 | 30700 | 22 | 1.38 | MTX_DAR_3hr |
| 42 | 19 | MA0815.1 | TFAP2C | MA0815.1 | TGCCCYSRGGGCA | TFAP2C | 0.000174 | 3.36e-06 | 192 | 26.5 | 26100 | 18.8 | 1.42 | MTX_DAR_3hr |
| 43 | 20 | MA0024.3 | E2F1 | MA0024.3 | TTTGGCGCCAAA | E2F1 | 0.000193 | 3.64e-06 | 179 | 24.7 | 24000 | 17.2 | 1.44 | MTX_DAR_3hr |
| 44 | 21 | MA1981.1 | ZNF530 | MA1981.1 | GMARGGMRAGGGGCRGSV | ZNF530 | 0.000221 | 4.06e-06 | 329 | 45.4 | 50500 | 36.2 | 1.26 | MTX_DAR_3hr |
| 45 | 20 | MA0470.2 | E2F4 | MA0470.2 | TTTTGGCGCCAWWW | E2F4 | 0.000247 | 4.4e-06 | 116 | 16 | 13900 | 9.96 | 1.62 | MTX_DAR_3hr |
| 46 | 12 | MA1648.1 | TCF12 | MA1648.1 | NNCACCTGCNN | TCF12 | 0.00025 | 4.4e-06 | 501 | 69.2 | 83800 | 60.2 | 1.15 | MTX_DAR_3hr |
| 47 | 7 | MA1102.2 | CTCFL | MA1102.2 | NSCAGGGGGCGS | CTCFL | 0.000302 | 5.2e-06 | 282 | 39 | 42100 | 30.2 | 1.29 | MTX_DAR_3hr |
| 48 | 9 | MA0472.2 | EGR2 | MA0472.2 | MCGCCCACGCA | EGR2 | 0.000314 | 5.3e-06 | 225 | 31.1 | 32000 | 23 | 1.36 | MTX_DAR_3hr |
| 49 | 22 | MA1584.1 | ZIC5 | MA1584.1 | VGACCCCCCGCTGHGM | ZIC5 | 0.000323 | 5.35e-06 | 166 | 22.9 | 22000 | 15.8 | 1.46 | MTX_DAR_3hr |
| 50 | 23 | MA1987.1 | ZNF701 | MA1987.1 | GAGCACYAARGGGGGRADDDD | ZNF701 | 0.000371 | 6e-06 | 513 | 70.9 | 86400 | 62 | 1.14 | MTX_DAR_3hr |
| 51 | 24 | MA1972.1 | ZFP14 | MA1972.1 | GGAGGCHCHGGARBG | ZFP14 | 0.000719 | 1.14e-05 | 539 | 74.4 | 92100 | 66.1 | 1.13 | MTX_DAR_3hr |
| 52 | 19 | MA0813.1 | TFAP2B | MA0813.1 | TGCCCYBRGGGCA | TFAP2B | 0.000729 | 1.14e-05 | 188 | 26 | 26000 | 18.7 | 1.4 | MTX_DAR_3hr |
| 53 | 10 | MA0814.2 | TFAP2C | MA0814.2 | NNGCCTGAGGCGNN | TFAP2C | 0.000896 | 1.37e-05 | 386 | 53.3 | 61900 | 44.4 | 1.2 | MTX_DAR_3hr |
| 54 | 10 | MA0131.2 | HINFP | MA0131.2 | CARCGTCCGCKN | HINFP | 0.00097 | 1.46e-05 | 400 | 55.2 | 64600 | 46.4 | 1.19 | MTX_DAR_3hr |
| 55 | 9 | MA1990.1 | Gli1 | MA1990.1 | NNRGACCACCCASV | Gli1 | 0.00132 | 1.94e-05 | 302 | 41.7 | 46400 | 33.3 | 1.25 | MTX_DAR_3hr |
| 56 | 1 | MA1516.1 | KLF3 | MA1516.1 | GRCCRCGCCCH | KLF3 | 0.00173 | 2.5e-05 | 281 | 38.8 | 42700 | 30.7 | 1.27 | MTX_DAR_3hr |
| 57 | 10 | MA1966.1 | TFAP4::ETV1 | MA1966.1 | RSCGGAAGCAGSTGKN | TFAP4::ETV1 | 0.00206 | 2.92e-05 | 114 | 15.8 | 14200 | 10.2 | 1.56 | MTX_DAR_3hr |
| 58 | 1 | MA1564.1 | SP9 | MA1564.1 | RCCACGCCCMCY | SP9 | 0.00215 | 3e-05 | 587 | 81.1 | 103000 | 73.8 | 1.1 | MTX_DAR_3hr |
| 59 | 23 | MA0528.2 | ZNF263 | MA0528.2 | VNGGGGAGGASG | ZNF263 | 0.0029 | 3.98e-05 | 471 | 65.1 | 79100 | 56.8 | 1.15 | MTX_DAR_3hr |
| 60 | 18 | MA0810.1 | TFAP2A | MA0810.1 | YGCCCBVRGGCR | TFAP2A | 0.00324 | 4.38e-05 | 270 | 37.3 | 41000 | 29.5 | 1.27 | MTX_DAR_3hr |
| 61 | 25 | MA0633.2 | Twist2 | MA0633.2 | NVCAGCTGBN | Twist2 | 0.0041 | 5.45e-05 | 364 | 50.3 | 58500 | 42 | 1.2 | MTX_DAR_3hr |
| 62 | 1 | MA0746.2 | SP3 | MA0746.2 | NGCCACGCCCMCH | SP3 | 0.00469 | 6.13e-05 | 133 | 18.4 | 17500 | 12.6 | 1.47 | MTX_DAR_3hr |
| 63 | 26 | MA1599.1 | ZNF682 | MA1599.1 | BVGGCYAAGCCCCWNY | ZNF682 | 0.00482 | 6.2e-05 | 505 | 69.8 | 86200 | 61.9 | 1.13 | MTX_DAR_3hr |
| 64 | 1 | MA0039.4 | KLF4 | MA0039.4 | NNCCCCACCCHN | KLF4 | 0.00495 | 6.27e-05 | 134 | 18.5 | 17700 | 12.7 | 1.47 | MTX_DAR_3hr |
| 65 | 25 | MA1993.1 | Neurod2 | MA1993.1 | VVCAGCTGBB | Neurod2 | 0.00672 | 8.25e-05 | 356 | 49.2 | 57300 | 41.1 | 1.2 | MTX_DAR_3hr |
| 66 | 20 | MA1727.1 | ZNF417 | MA1727.1 | VRBVNTGGGCGCCAM | ZNF417 | 0.00672 | 8.25e-05 | 314 | 43.4 | 49400 | 35.5 | 1.22 | MTX_DAR_3hr |
| 67 | 20 | MA0864.2 | E2F2 | MA0864.2 | RWTTTGGCGCCAWWWY | E2F2 | 0.00701 | 8.48e-05 | 212 | 29.3 | 31100 | 22.3 | 1.32 | MTX_DAR_3hr |
| 68 | 12 | MA1631.1 | ASCL1 | MA1631.1 | NNGCACCTGCYNB | ASCL1 | 0.00744 | 8.86e-05 | 602 | 83.2 | 107000 | 76.5 | 1.09 | MTX_DAR_3hr |
| 69 | 27 | MA1979.1 | ZNF416 | MA1979.1 | TATCTGGGCAH | ZNF416 | 0.00767 | 9.01e-05 | 599 | 82.7 | 106000 | 76.1 | 1.09 | MTX_DAR_3hr |
| 70 | 28 | MA0146.2 | Zfx | MA0146.2 | SSSGCCBVGGCCTS | Zfx | 0.00801 | 9.22e-05 | 304 | 42 | 47700 | 34.2 | 1.23 | MTX_DAR_3hr |
| 71 | 29 | MA1643.1 | NFIB | MA1643.1 | NNCCTGGCAYNGTGCCAAGNN | NFIB | 0.00808 | 9.22e-05 | 269 | 37.2 | 41300 | 29.7 | 1.26 | MTX_DAR_3hr |
| 72 | 12 | MA0632.2 | TCFL5 | MA0632.2 | KCACGCGCMC | TCFL5 | 0.0099 | 0.000111 | 285 | 39.4 | 44300 | 31.8 | 1.24 | MTX_DAR_3hr |
| 73 | 1 | MA0741.1 | KLF16 | MA0741.1 | GMCACGCCCCC | KLF16 | 0.0103 | 0.000115 | 585 | 80.8 | 103000 | 74 | 1.09 | MTX_DAR_3hr |
| 74 | 7 | MA1719.1 | ZNF816 | MA1719.1 | DRRKGGGGACATGCHGGG | ZNF816 | 0.0116 | 0.000126 | 475 | 65.6 | 80600 | 57.9 | 1.13 | MTX_DAR_3hr |
| 75 | 30 | MA0694.1 | ZBTB7B | MA0694.1 | RCGACCACCGAA | ZBTB7B | 0.0118 | 0.000126 | 349 | 48.2 | 56300 | 40.4 | 1.19 | MTX_DAR_3hr |
| 76 | 30 | MA1587.1 | ZNF135 | MA1587.1 | CCTCGACCTCCYRR | ZNF135 | 0.0118 | 0.000126 | 366 | 50.6 | 59500 | 42.7 | 1.19 | MTX_DAR_3hr |
| 77 | 30 | MA0734.3 | Gli2 | MA0734.3 | NRGACCACCCASV | Gli2 | 0.0121 | 0.000127 | 389 | 53.7 | 63900 | 45.9 | 1.17 | MTX_DAR_3hr |
| 78 | 19 | MA0524.2 | TFAP2C | MA0524.2 | YGCCYBVRGGCA | TFAP2C | 0.0148 | 0.000154 | 107 | 14.8 | 13700 | 9.83 | 1.51 | MTX_DAR_3hr |
| 79 | 12 | MA1099.2 | HES1 | MA1099.2 | GGCRCGTGBC | HES1 | 0.0153 | 0.000157 | 58 | 8.01 | 6200 | 4.45 | 1.83 | MTX_DAR_3hr |
| 80 | 10 | MA0759.2 | ELK3 | MA0759.2 | ACCGGAAGTRV | ELK3 | 0.0172 | 0.000174 | 158 | 21.8 | 22200 | 15.9 | 1.38 | MTX_DAR_3hr |
| 81 | 31 | MA0597.2 | THAP1 | MA0597.2 | VSGCAGGGCASV | THAP1 | 0.0204 | 0.000204 | 189 | 26.1 | 27600 | 19.8 | 1.32 | MTX_DAR_3hr |
| 82 | 1 | MA0599.1 | KLF5 | MA0599.1 | GCCCCDCCCH | KLF5 | 0.0213 | 0.00021 | 223 | 30.8 | 33600 | 24.1 | 1.28 | MTX_DAR_3hr |
| 83 | 25 | MA1997.1 | Olig2 | MA1997.1 | NVCAGCTGBN | Olig2 | 0.0238 | 0.000233 | 354 | 48.9 | 57600 | 41.4 | 1.18 | MTX_DAR_3hr |
| 84 | 32 | 7-AAGCCGCCCTG | STREME-7 | 7-AAGCCGCCCTG | AAGCCGCCCTG | AAGCCGCCCTG | 0.0266 | 0.000257 | 197 | 27.2 | 29100 | 20.9 | 1.31 | MTX_DAR_3hr |
| 85 | 33 | MA1684.1 | Foxn1 | MA1684.1 | RGAMGC | Foxn1 | 0.035 | 0.000333 | 74 | 10.2 | 8790 | 6.31 | 1.64 | MTX_DAR_3hr |
| 9 | 34 | MA1721.1 (ZNF93) | MEME-1 | CCCSCSSCCGCCGCC | CCCSCSSCCGCCGCC | ZNF93 | 0.338 | 0.0375 | 6 | 0.83 | 466 | 0.33 | 2.88 | MTX_DAR_3hr |
ER_rat <- 1.25
# mrc_type <- "EAR_open"
MTX_DAR_3hr_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
# geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_col(aes(x=log10Evalue))+
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste("MTX_DAR_3hr_combo response peaks(200bp) Enrichment ratio:",ER_rat))
MTX_DAR_3hr_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
slice_head(n=5) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
# geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*.5), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./.5,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste("MTX DAR 3 hour response peaks(200bp) Enrichment ratio:",ER_rat))
MTX_DAR_24hr data
MTX_DAR_24hr_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/MTX_24_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
MTX_DAR_24hr_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/MTX_24_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
MTX_DAR_24hr_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/MTX_24_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
MTX_DAR_24hr_sea_all <- rbind(MTX_DAR_24hr_sea_disc, MTX_DAR_24hr_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)MTX_DAR_24hr_combo <- MTX_DAR_24hr_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., MTX_DAR_24hr_sea_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ SIM_MOTIF
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y) %>%
mutate(mrc_type="MTX_DAR_24hr")
MTX_DAR_24hr_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs 200 bp MTX_DAR_24hr") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO | mrc_type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | MA0477.2 (FOSL1) | STREME-1 | 1-GATGAGTCAY | GATGAGTCAY | FOSL1 | 6.82e-110 | 4.56e-110 | 3560 | 16.3 | 12700 | 10.8 | 1.52 | MTX_DAR_24hr |
| 2 | 1 | MA1138.1 | FOSL2::JUNB | MA1138.1 | KRTGASTCAT | FOSL2::JUNB | 4.22e-99 | 1.41e-99 | 3810 | 17.5 | 14200 | 12 | 1.46 | MTX_DAR_24hr |
| 3 | 1 | MA1135.1 | FOSB::JUNB | MA1135.1 | KRTGASTCAT | FOSB::JUNB | 5.59e-96 | 1.25e-96 | 4190 | 19.2 | 16000 | 13.6 | 1.42 | MTX_DAR_24hr |
| 4 | 1 | MA1130.1 | FOSL2::JUN | MA1130.1 | NNRTGAGTCAYN | FOSL2::JUN | 9.33e-96 | 1.56e-96 | 3600 | 16.5 | 13300 | 11.3 | 1.47 | MTX_DAR_24hr |
| 5 | 1 | MA1141.1 | FOS::JUND | MA1141.1 | NKATGAGTCATNN | FOS::JUND | 1.96e-95 | 2.62e-96 | 3840 | 17.6 | 14400 | 12.2 | 1.44 | MTX_DAR_24hr |
| 6 | 1 | MA0478.1 (FOSL2) | MEME-1 | MTGAGTCAYY | MTGAGTCAYY | FOSL2 | 3.62e-93 | 3.52e-94 | 3790 | 17.4 | 14200 | 12.1 | 1.44 | MTX_DAR_24hr |
| 7 | 1 | MA1144.1 | FOSL2::JUND | MA1144.1 | KATGACTCAT | FOSL2::JUND | 3.68e-93 | 3.52e-94 | 4270 | 19.6 | 16500 | 14 | 1.4 | MTX_DAR_24hr |
| 8 | 1 | MA0099.3 | FOS::JUN | MA0099.3 | ATGAGTCAYM | FOS::JUN | 1.12e-92 | 9.33e-94 | 3580 | 16.4 | 13300 | 11.3 | 1.46 | MTX_DAR_24hr |
| 9 | 1 | MA1128.1 | FOSL1::JUN | MA1128.1 | NKATGACTCATNN | FOSL1::JUN | 1.35e-92 | 1e-93 | 4140 | 19 | 15900 | 13.4 | 1.41 | MTX_DAR_24hr |
| 10 | 1 | MA1134.1 | FOS::JUNB | MA1134.1 | KATGASTCATHN | FOS::JUNB | 5.58e-91 | 3.73e-92 | 4300 | 19.7 | 16700 | 14.1 | 1.39 | MTX_DAR_24hr |
| 11 | 1 | MA1633.2 | BACH1 | MA1633.2 | ATGACTCAT | BACH1 | 1.14e-88 | 6.95e-90 | 3700 | 17 | 14000 | 11.8 | 1.43 | MTX_DAR_24hr |
| 12 | 1 | MA0476.1 (FOS) | STREME-2 | 2-VTGASTCAB | VTGASTCAB | FOS | 3.57e-88 | 1.99e-89 | 3890 | 17.8 | 14900 | 12.6 | 1.42 | MTX_DAR_24hr |
| 13 | 1 | MA0655.1 | JDP2 | MA0655.1 | ATGACTCAT | JDP2 | 1.88e-86 | 9.65e-88 | 4440 | 20.4 | 17500 | 14.8 | 1.37 | MTX_DAR_24hr |
| 14 | 1 | MA0489.2 | Jun | MA0489.2 | NVTGACTCATNN | Jun | 5.21e-85 | 2.49e-86 | 4600 | 21.1 | 18300 | 15.5 | 1.36 | MTX_DAR_24hr |
| 15 | 1 | MA0476.1 | FOS | MA0476.1 | DVTGASTCATB | FOS | 7.1e-85 | 3.16e-86 | 3720 | 17 | 14200 | 12 | 1.42 | MTX_DAR_24hr |
| 16 | 1 | MA1137.1 | FOSL1::JUNB | MA1137.1 | NDRTGACTCATNN | FOSL1::JUNB | 1.9e-83 | 7.96e-85 | 3810 | 17.5 | 14700 | 12.4 | 1.41 | MTX_DAR_24hr |
| 17 | 1 | MA0490.2 | JUNB | MA0490.2 | NNATGACTCATNN | JUNB | 1.01e-82 | 3.95e-84 | 3700 | 16.9 | 14100 | 12 | 1.42 | MTX_DAR_24hr |
| 18 | 1 | MA1988.1 | Atf3 | MA1988.1 | NRTGACTCABN | Atf3 | 4.55e-81 | 1.69e-82 | 4120 | 18.9 | 16200 | 13.7 | 1.38 | MTX_DAR_24hr |
| 19 | 1 | MA0477.2 | FOSL1 | MA0477.2 | NNATGACTCATNN | FOSL1 | 7.63e-81 | 2.69e-82 | 4890 | 22.4 | 19900 | 16.8 | 1.33 | MTX_DAR_24hr |
| 20 | 1 | MA1928.1 | BNC2 | MA1928.1 | NDRTGAGTCABN | BNC2 | 1.03e-80 | 3.44e-82 | 4270 | 19.6 | 16900 | 14.3 | 1.37 | MTX_DAR_24hr |
| 21 | 1 | MA0491.2 | JUND | MA0491.2 | NNATGACTCATNN | JUND | 4.07e-80 | 1.3e-81 | 4690 | 21.5 | 18900 | 16 | 1.34 | MTX_DAR_24hr |
| 22 | 1 | MA0478.1 | FOSL2 | MA0478.1 | KRRTGASTCAB | FOSL2 | 4.32e-78 | 1.31e-79 | 4500 | 20.6 | 18100 | 15.3 | 1.35 | MTX_DAR_24hr |
| 23 | 1 | MA1132.1 | JUN::JUNB | MA1132.1 | KATGACKCAT | JUN::JUNB | 6.03e-76 | 1.75e-77 | 4300 | 19.7 | 17200 | 14.6 | 1.35 | MTX_DAR_24hr |
| 24 | 1 | MA0835.2 | BATF3 | MA0835.2 | DATGACTCATH | BATF3 | 5.28e-74 | 1.47e-75 | 4570 | 20.9 | 18600 | 15.7 | 1.33 | MTX_DAR_24hr |
| 25 | 1 | MA1142.1 | FOSL1::JUND | MA1142.1 | NRTGACTCAT | FOSL1::JUND | 1.49e-73 | 3.99e-75 | 3600 | 16.5 | 14000 | 11.8 | 1.39 | MTX_DAR_24hr |
| 26 | 2 | MA1513.1 (KLF15) | STREME-3 | 3-GCCCCGCCC | GCCCCGCCC | KLF15 | 9.43e-73 | 2.42e-74 | 1290 | 5.92 | 3800 | 3.21 | 1.84 | MTX_DAR_24hr |
| 27 | 1 | MA0841.1 | NFE2 | MA0841.1 | VATGACTCATS | NFE2 | 1.02e-72 | 2.52e-74 | 4120 | 18.9 | 16500 | 14 | 1.35 | MTX_DAR_24hr |
| 28 | 1 | MA0462.2 | BATF::JUN | MA0462.2 | DATGACTCATH | BATF::JUN | 2.57e-72 | 6.14e-74 | 4560 | 20.9 | 18600 | 15.8 | 1.33 | MTX_DAR_24hr |
| 29 | 1 | MA1634.1 | BATF | MA1634.1 | DATGACTCATH | BATF | 2.56e-70 | 5.91e-72 | 4580 | 21 | 18800 | 15.9 | 1.32 | MTX_DAR_24hr |
| 30 | 1 | MA1101.2 | BACH2 | MA1101.2 | DWANCATGASTCATSNTWH | BACH2 | 8.54e-61 | 1.9e-62 | 4910 | 22.5 | 20800 | 17.6 | 1.28 | MTX_DAR_24hr |
| 31 | 3 | MA1985.1 (ZNF669) | STREME-5 | 5-CYGTCRNCRSC | CYGTCRNCRSC | ZNF669 | 1.9e-54 | 4.11e-56 | 6370 | 29.2 | 28400 | 24 | 1.21 | MTX_DAR_24hr |
| 32 | 2 | MA1513.1 | KLF15 | MA1513.1 | SCCCCGCCCCS | KLF15 | 1.52e-51 | 3.18e-53 | 1140 | 5.22 | 3580 | 3.03 | 1.72 | MTX_DAR_24hr |
| 33 | 4 | MA0502.2 (NFYB) | STREME-9 | 9-CGATTGGC | CGATTGGC | NFYB | 3.15e-45 | 6.39e-47 | 961 | 4.4 | 2980 | 2.52 | 1.75 | MTX_DAR_24hr |
| 34 | 2 | MA1564.1 | SP9 | MA1564.1 | RCCACGCCCMCY | SP9 | 1.71e-44 | 3.36e-46 | 3090 | 14.1 | 12600 | 10.7 | 1.32 | MTX_DAR_24hr |
| 35 | 2 | MA0742.2 (KLF12) | STREME-11 | 11-CCCGGCC | 11-CCCGGCC | KLF12 | 1.26e-43 | 2.41e-45 | 3530 | 16.2 | 14800 | 12.5 | 1.29 | MTX_DAR_24hr |
| 36 | 5 | MA0162.4 | EGR1 | MA0162.4 | VCMCGCCCACGCVS | EGR1 | 4.32e-40 | 8.03e-42 | 3050 | 14 | 12600 | 10.7 | 1.31 | MTX_DAR_24hr |
| 37 | 6 | 6-CCGSSSCGG | STREME-6 | 6-CCGSSSCGG | CCGSSSCGG | CCGSSSCGG | 2.17e-39 | 3.92e-41 | 1940 | 8.88 | 7430 | 6.29 | 1.41 | MTX_DAR_24hr |
| 38 | 2 | MA0746.2 | SP3 | MA0746.2 | NGCCACGCCCMCH | SP3 | 2.82e-39 | 4.96e-41 | 3700 | 17 | 15900 | 13.4 | 1.26 | MTX_DAR_24hr |
| 39 | 2 | MA1961.1 | PATZ1 | MA1961.1 | SSGGGGMGGGGS | PATZ1 | 1.92e-38 | 3.29e-40 | 3180 | 14.6 | 13300 | 11.3 | 1.29 | MTX_DAR_24hr |
| 40 | 7 | 10-CGGCSGCCG | STREME-10 | 10-CGGCSGCCG | 10-CGGCSGCCG | CGGCSGCCG | 4.56e-38 | 7.62e-40 | 1620 | 7.41 | 6000 | 5.08 | 1.46 | MTX_DAR_24hr |
| 41 | 2 | MA0741.1 | KLF16 | MA0741.1 | GMCACGCCCCC | KLF16 | 2.65e-36 | 4.32e-38 | 2870 | 13.2 | 11900 | 10.1 | 1.3 | MTX_DAR_24hr |
| 42 | 22 | MA1713.1 | ZNF610 | MA1713.1 | SSCGCCGCTCCSSS | ZNF610 | 9.97e-36 | 1.59e-37 | 3240 | 14.9 | 13800 | 11.7 | 1.27 | MTX_DAR_24hr |
| 43 | 2 | MA0742.2 | KLF12 | MA0742.2 | GGGGCGGGG | KLF12 | 1.11e-35 | 1.73e-37 | 2610 | 12 | 10700 | 9.08 | 1.32 | MTX_DAR_24hr |
| 44 | 8 | 12-CGGGCC | STREME-12 | 12-CGGGCC | 12-CGGGCC | CGGGCC | 3.75e-35 | 5.7e-37 | 3240 | 14.9 | 13800 | 11.7 | 1.27 | MTX_DAR_24hr |
| 45 | 9 | MA0516.3 (SP2) | MEME-3 | GCGGCGGSGSSSGCG | GCGGCGGSGSSSGCG | SP2 | 1.37e-34 | 2.03e-36 | 1580 | 7.24 | 5950 | 5.03 | 1.44 | MTX_DAR_24hr |
| 46 | 2 | MA1512.1 | KLF11 | MA1512.1 | SCCACGCCCMC | KLF11 | 3.57e-34 | 5.18e-36 | 4100 | 18.8 | 18100 | 15.3 | 1.23 | MTX_DAR_24hr |
| 47 | 10 | MA0146.2 | Zfx | MA0146.2 | SSSGCCBVGGCCTS | Zfx | 5.13e-33 | 7.29e-35 | 6760 | 31 | 31700 | 26.8 | 1.15 | MTX_DAR_24hr |
| 48 | 2 | MA1959.1 | KLF7 | MA1959.1 | DGGGCGGGG | KLF7 | 6.51e-32 | 9.06e-34 | 2840 | 13 | 12000 | 10.2 | 1.28 | MTX_DAR_24hr |
| 49 | 11 | MA1599.1 | ZNF682 | MA1599.1 | BVGGCYAAGCCCCWNY | ZNF682 | 7.02e-32 | 9.58e-34 | 5020 | 23 | 22900 | 19.3 | 1.19 | MTX_DAR_24hr |
| 50 | 2 | MA0685.2 | SP4 | MA0685.2 | GGGGCGGGG | SP4 | 9.96e-32 | 1.33e-33 | 2410 | 11 | 9950 | 8.42 | 1.31 | MTX_DAR_24hr |
| 51 | 5 | MA0733.1 | EGR4 | MA0733.1 | NHMCGCCCACGCAHWT | EGR4 | 1.23e-31 | 1.61e-33 | 5570 | 25.5 | 25700 | 21.7 | 1.17 | MTX_DAR_24hr |
| 52 | 1 | MA0496.3 | MAFK | MA0496.3 | NGMTGACTCAGCMNH | MAFK | 3.49e-31 | 4.49e-33 | 5200 | 23.8 | 23800 | 20.2 | 1.18 | MTX_DAR_24hr |
| 53 | 2 | MA1653.1 | ZNF148 | MA1653.1 | YBCCCCTCCCCC | ZNF148 | 3.89e-31 | 4.9e-33 | 3020 | 13.8 | 12900 | 10.9 | 1.27 | MTX_DAR_24hr |
| 54 | 12 | MA0506.2 | Nrf1 | MA0506.2 | SNCTGCGCMTGCGCV | Nrf1 | 5.33e-31 | 6.6e-33 | 3020 | 13.8 | 12900 | 10.9 | 1.27 | MTX_DAR_24hr |
| 55 | 2 | MA0079.5 | SP1 | MA0079.5 | GGGGCGGGG | SP1 | 1.81e-30 | 2.2e-32 | 2890 | 13.2 | 12300 | 10.4 | 1.27 | MTX_DAR_24hr |
| 56 | 2 | MA0599.1 | KLF5 | MA0599.1 | GCCCCDCCCH | KLF5 | 1.86e-30 | 2.22e-32 | 2660 | 12.2 | 11200 | 9.47 | 1.29 | MTX_DAR_24hr |
| 57 | 1 | MA0089.2 | MAFG::NFE2L1 | MA0089.2 | NVNATGACTCAGCADW | MAFG::NFE2L1 | 4.69e-29 | 5.51e-31 | 4300 | 19.7 | 19400 | 16.4 | 1.2 | MTX_DAR_24hr |
| 58 | 2 | MA0516.3 | SP2 | MA0516.3 | GGGGCGGGG | SP2 | 1.55e-28 | 1.79e-30 | 4290 | 19.7 | 19400 | 16.4 | 1.2 | MTX_DAR_24hr |
| 59 | 4 | MA0060.3 | NFYA | MA0060.3 | RRCCAATCAGM | NFYA | 1.74e-27 | 1.97e-29 | 3270 | 15 | 14300 | 12.1 | 1.23 | MTX_DAR_24hr |
| 60 | 2 | MA1511.2 | KLF10 | MA1511.2 | GGGGCGGGG | KLF10 | 2.98e-27 | 3.32e-29 | 4570 | 20.9 | 20900 | 17.6 | 1.19 | MTX_DAR_24hr |
| 61 | 13 | MA1122.1 | TFDP1 | MA1122.1 | VSGCGGGAAVN | TFDP1 | 5.29e-27 | 5.8e-29 | 2510 | 11.5 | 10600 | 9 | 1.28 | MTX_DAR_24hr |
| 62 | 2 | MA0740.2 | KLF14 | MA0740.2 | KGGGCGGGG | KLF14 | 1.88e-26 | 2.03e-28 | 5520 | 25.3 | 25800 | 21.8 | 1.16 | MTX_DAR_24hr |
| 63 | 4 | MA0502.2 | NFYB | MA0502.2 | CYCATTGGCCVV | NFYB | 6.74e-25 | 7.15e-27 | 2630 | 12 | 11300 | 9.6 | 1.26 | MTX_DAR_24hr |
| 64 | 2 | MA0039.4 | KLF4 | MA0039.4 | NNCCCCACCCHN | KLF4 | 1.06e-24 | 1.11e-26 | 3630 | 16.6 | 16300 | 13.8 | 1.21 | MTX_DAR_24hr |
| 65 | 1 | MA1114.1 | PBX3 | MA1114.1 | NNNTGAGTGACAGGBNN | PBX3 | 2.38e-24 | 2.44e-26 | 10900 | 50 | 54400 | 46 | 1.09 | MTX_DAR_24hr |
| 66 | 14 | MA1712.1 | ZNF454 | MA1712.1 | DGGCTCSSGGCCCYGVKG | ZNF454 | 2.34e-23 | 2.37e-25 | 7150 | 32.8 | 34500 | 29.2 | 1.12 | MTX_DAR_24hr |
| 67 | 2 | MA1515.1 | KLF2 | MA1515.1 | NRCCACRCCCH | KLF2 | 3.2e-23 | 3.19e-25 | 3940 | 18.1 | 18000 | 15.2 | 1.19 | MTX_DAR_24hr |
| 68 | 2 | MA1517.1 | KLF6 | MA1517.1 | NRCCACGCCCH | KLF6 | 5.11e-23 | 5.02e-25 | 4750 | 21.8 | 22100 | 18.7 | 1.16 | MTX_DAR_24hr |
| 69 | 5 | MA0732.1 | EGR3 | MA0732.1 | HHMCGCCCACGCAHH | EGR3 | 1.17e-22 | 1.13e-24 | 2970 | 13.6 | 13100 | 11.1 | 1.22 | MTX_DAR_24hr |
| 70 | 2 | MA0493.2 | KLF1 | MA0493.2 | DGGGYGGGG | KLF1 | 1.8e-22 | 1.72e-24 | 3540 | 16.2 | 16000 | 13.6 | 1.2 | MTX_DAR_24hr |
| 71 | 5 | MA1650.1 | ZBTB14 | MA1650.1 | SSCCGCGCACVS | ZBTB14 | 3.06e-22 | 2.89e-24 | 3130 | 14.3 | 13900 | 11.8 | 1.21 | MTX_DAR_24hr |
| 72 | 15 | MA1721.1 | ZNF93 | MA1721.1 | GGYAGCRGCAGCGGYG | ZNF93 | 3.89e-22 | 3.62e-24 | 2440 | 11.2 | 10500 | 8.93 | 1.25 | MTX_DAR_24hr |
| 73 | 16 | MA1615.1 | Plagl1 | MA1615.1 | NNCTGGGGCCABN | Plagl1 | 7.75e-22 | 7.1e-24 | 5340 | 24.4 | 25200 | 21.3 | 1.15 | MTX_DAR_24hr |
| 74 | 16 | MA1986.1 | ZNF692 | MA1986.1 | BGGGCCCASN | ZNF692 | 3.59e-21 | 3.24e-23 | 5250 | 24 | 24800 | 21 | 1.15 | MTX_DAR_24hr |
| 75 | 2 | MA1630.2 | ZNF281 | MA1630.2 | GGGGGAGGGGVV | ZNF281 | 1.27e-20 | 1.13e-22 | 1730 | 7.95 | 7210 | 6.1 | 1.3 | MTX_DAR_24hr |
| 76 | 2 | MA0747.1 | SP8 | MA0747.1 | RCCACGCCCMCY | SP8 | 1.69e-20 | 1.48e-22 | 5590 | 25.6 | 26600 | 22.5 | 1.14 | MTX_DAR_24hr |
| 77 | 1 | MA0501.1 | MAF::NFE2 | MA0501.1 | ATGACTCAGCANWTY | MAF::NFE2 | 1.02e-19 | 8.83e-22 | 3010 | 13.8 | 13500 | 11.4 | 1.21 | MTX_DAR_24hr |
| 78 | 2 | MA1522.1 | MAZ | MA1522.1 | BBCCCCTCCCY | MAZ | 2.07e-18 | 1.78e-20 | 7820 | 35.8 | 38400 | 32.5 | 1.1 | MTX_DAR_24hr |
| 79 | 17 | MA1155.1 (ZSCAN4) | STREME-7 | 7-ACATGCACACACGT | ACATGCACACACGT | ZSCAN4 | 3.27e-18 | 2.77e-20 | 355 | 1.63 | 1050 | 0.89 | 1.83 | MTX_DAR_24hr |
| 80 | 18 | MA0748.2 | YY2 | MA0748.2 | NVATGGCGGCS | YY2 | 5.34e-18 | 4.47e-20 | 1510 | 6.94 | 6280 | 5.32 | 1.31 | MTX_DAR_24hr |
| 81 | 19 | MA0774.1 | MEIS2 | MA0774.1 | TTGACAGS | MEIS2 | 1.14e-17 | 9.4e-20 | 1830 | 8.37 | 7800 | 6.6 | 1.27 | MTX_DAR_24hr |
| 82 | 20 | MA1976.1 | ZNF320 | MA1976.1 | DGKGGGRCCMRGGGGCCWGTGHVNNNN | ZNF320 | 2.02e-17 | 1.65e-19 | 5240 | 24 | 25100 | 21.2 | 1.13 | MTX_DAR_24hr |
| 83 | 1 | MA0659.3 | Mafg | MA0659.3 | NWGMTGACTCAGCAN | Mafg | 1.19e-16 | 9.6e-19 | 5780 | 26.5 | 27900 | 23.6 | 1.12 | MTX_DAR_24hr |
| 84 | 1 | MA0150.2 | Nfe2l2 | MA0150.2 | CASNATGACTCAGCA | Nfe2l2 | 1.24e-16 | 9.91e-19 | 8210 | 37.6 | 40700 | 34.4 | 1.09 | MTX_DAR_24hr |
| 85 | 1 | MA0782.2 | PKNOX1 | MA0782.2 | NNTGAGTGACAGVNN | PKNOX1 | 1.45e-16 | 1.14e-18 | 12500 | 57.1 | 63600 | 53.8 | 1.06 | MTX_DAR_24hr |
| 86 | 21 | MA1573.2 | Thap11 | MA1573.2 | ACTACAABTCCCAG | Thap11 | 1.53e-16 | 1.19e-18 | 99 | 0.45 | 155 | 0.13 | 3.47 | MTX_DAR_24hr |
| 87 | 22 | MA1653.1 (ZNF148) | STREME-4 | 4-CCCCTCCCC | CCCCTCCCC | ZNF148 | 1.75e-16 | 1.34e-18 | 2580 | 11.8 | 11600 | 9.79 | 1.21 | MTX_DAR_24hr |
| 88 | 5 | MA1107.2 | KLF9 | MA1107.2 | MNGCCACACCCACHYV | KLF9 | 2.29e-16 | 1.74e-18 | 17600 | 80.5 | 91900 | 77.8 | 1.03 | MTX_DAR_24hr |
| 89 | 23 | MA1569.1 | TFAP2E | MA1569.1 | YGCCYSRGGCD | TFAP2E | 6.14e-16 | 4.62e-18 | 3740 | 17.1 | 17500 | 14.8 | 1.16 | MTX_DAR_24hr |
| 90 | 24 | MA0821.2 | HES5 | MA0821.2 | GRCACGTGYC | HES5 | 1.65e-15 | 1.23e-17 | 4260 | 19.5 | 20200 | 17.1 | 1.14 | MTX_DAR_24hr |
| 91 | 25 | MA1643.1 | NFIB | MA1643.1 | NNCCTGGCAYNGTGCCAAGNN | NFIB | 1.79e-15 | 1.32e-17 | 5720 | 26.2 | 27700 | 23.4 | 1.12 | MTX_DAR_24hr |
| 92 | 26 | MA1548.1 | PLAGL2 | MA1548.1 | NGGGCCCCCN | PLAGL2 | 1.99e-15 | 1.44e-17 | 7530 | 34.5 | 37200 | 31.5 | 1.1 | MTX_DAR_24hr |
| 93 | 24 | MA0632.2 | TCFL5 | MA0632.2 | KCACGCGCMC | TCFL5 | 2.97e-15 | 2.14e-17 | 998 | 4.57 | 3970 | 3.36 | 1.36 | MTX_DAR_24hr |
| 94 | 24 | MA0823.1 | HEY1 | MA0823.1 | GRCACGTGCC | HEY1 | 1.4e-14 | 9.97e-17 | 2930 | 13.4 | 13400 | 11.4 | 1.18 | MTX_DAR_24hr |
| 95 | 27 | MA0527.1 | ZBTB33 | MA0527.1 | BTCTCGCGAGABYTS | ZBTB33 | 2.05e-14 | 1.44e-16 | 1180 | 5.4 | 4850 | 4.1 | 1.32 | MTX_DAR_24hr |
| 96 | 23 | MA0814.2 | TFAP2C | MA0814.2 | NNGCCTGAGGCGNN | TFAP2C | 2.27e-14 | 1.58e-16 | 10100 | 46.2 | 51000 | 43.2 | 1.07 | MTX_DAR_24hr |
| 97 | 24 | MA0603.1 | Arntl | MA0603.1 | NGTCACGTGH | Arntl | 5.78e-14 | 3.98e-16 | 14900 | 68.2 | 77300 | 65.4 | 1.04 | MTX_DAR_24hr |
| 98 | 13 | MA0865.2 | E2F8 | MA0865.2 | TTCCCGCCAHWA | E2F8 | 7.34e-14 | 5.01e-16 | 2750 | 12.6 | 12600 | 10.7 | 1.18 | MTX_DAR_24hr |
| 99 | 5 | MA1656.1 | ZNF449 | MA1656.1 | NNAAGCCCAACCNN | ZNF449 | 1.03e-13 | 6.97e-16 | 9700 | 44.4 | 49000 | 41.4 | 1.07 | MTX_DAR_24hr |
| 100 | 5 | MA0472.2 | EGR2 | MA0472.2 | MCGCCCACGCA | EGR2 | 2.93e-13 | 1.96e-15 | 9230 | 42.3 | 46500 | 39.4 | 1.07 | MTX_DAR_24hr |
| 101 | 13 | MA0471.2 | E2F6 | MA0471.2 | VVDGGCGGGAAVV | E2F6 | 2.99e-13 | 1.98e-15 | 655 | 3 | 2460 | 2.09 | 1.44 | MTX_DAR_24hr |
| 102 | 28 | MA1995.1 | Npas4 | MA1995.1 | NNTCGTGACHN | Npas4 | 3.77e-13 | 2.47e-15 | 11800 | 54 | 60400 | 51.1 | 1.06 | MTX_DAR_24hr |
| 103 | 29 | MA1514.1 | KLF17 | MA1514.1 | MMCCACGCACCCMTY | KLF17 | 6.59e-13 | 4.28e-15 | 8920 | 40.9 | 44900 | 38 | 1.08 | MTX_DAR_24hr |
| 104 | 22 | MA1981.1 | ZNF530 | MA1981.1 | GMARGGMRAGGGGCRGSV | ZNF530 | 1.33e-12 | 8.56e-15 | 2940 | 13.5 | 13700 | 11.6 | 1.17 | MTX_DAR_24hr |
| 105 | 30 | MA0615.1 | Gmeb1 | MA0615.1 | BHBBKKACGTMMNWNNN | Gmeb1 | 2.48e-12 | 1.58e-14 | 1970 | 9.03 | 8820 | 7.46 | 1.21 | MTX_DAR_24hr |
| 106 | 23 | MA0812.1 | TFAP2B | MA0812.1 | HGCCTSAGGCD | TFAP2B | 5.25e-12 | 3.31e-14 | 8610 | 39.4 | 43400 | 36.7 | 1.08 | MTX_DAR_24hr |
| 107 | 31 | MA0067.2 | PAX2 | MA0067.2 | BCAVTSRAGCGTGACGR | PAX2 | 9.01e-12 | 5.63e-14 | 5190 | 23.8 | 25300 | 21.4 | 1.11 | MTX_DAR_24hr |
| 108 | 32 | MA1596.1 | ZNF460 | MA1596.1 | GCCTCMGCCTCCCRAG | ZNF460 | 1.03e-11 | 6.39e-14 | 4730 | 21.7 | 23000 | 19.4 | 1.12 | MTX_DAR_24hr |
| 109 | 24 | MA0464.2 | BHLHE40 | MA0464.2 | VKCACGTGMC | BHLHE40 | 1.04e-11 | 6.4e-14 | 8190 | 37.5 | 41200 | 34.8 | 1.08 | MTX_DAR_24hr |
| 110 | 28 | MA0014.3 | PAX5 | MA0014.3 | RRGCGTGACCNN | PAX5 | 1.72e-11 | 1.05e-13 | 1450 | 6.64 | 6310 | 5.34 | 1.25 | MTX_DAR_24hr |
| 111 | 13 | MA1583.1 | ZFP57 | MA1583.1 | NNVTTGCCGCANN | ZFP57 | 3.2e-11 | 1.93e-13 | 4780 | 21.9 | 23200 | 19.7 | 1.11 | MTX_DAR_24hr |
| 112 | 19 | MA1572.1 | TGIF2LY | MA1572.1 | TGACAGCTGTCA | TGIF2LY | 5.41e-11 | 3.23e-13 | 13700 | 62.6 | 70800 | 59.9 | 1.04 | MTX_DAR_24hr |
| 113 | 33 | MA1964.1 | SMAD2 | MA1964.1 | NBCCAGACDN | SMAD2 | 7.07e-11 | 4.19e-13 | 14900 | 68.4 | 77800 | 65.8 | 1.04 | MTX_DAR_24hr |
| 114 | 34 | MA0803.1 | TBX15 | MA0803.1 | AGGTGTGA | TBX15 | 8.2e-11 | 4.81e-13 | 17300 | 79.4 | 91200 | 77.2 | 1.03 | MTX_DAR_24hr |
| 115 | 3 | MA1985.1 | ZNF669 | MA1985.1 | GGGGYGAYGACCRCT | ZNF669 | 1.38e-10 | 7.99e-13 | 1040 | 4.76 | 4360 | 3.69 | 1.29 | MTX_DAR_24hr |
| 116 | 25 | MA1528.1 | NFIX | MA1528.1 | STTGGCRNNGTGCCARB | NFIX | 1.39e-10 | 8e-13 | 3050 | 14 | 14400 | 12.2 | 1.15 | MTX_DAR_24hr |
| 117 | 35 | MA1557.1 | SMAD5 | MA1557.1 | YGTCTAGACA | SMAD5 | 1.61e-10 | 9.2e-13 | 11000 | 50.4 | 56400 | 47.7 | 1.06 | MTX_DAR_24hr |
| 118 | 22 | MA1627.1 | Wt1 | MA1627.1 | YBCCTCCCCCACVB | Wt1 | 3.22e-10 | 1.83e-12 | 3710 | 17 | 17800 | 15.1 | 1.13 | MTX_DAR_24hr |
| 119 | 24 | MA0616.2 | HES2 | MA0616.2 | GGCACGTGYC | HES2 | 5.41e-10 | 3.04e-12 | 1800 | 8.24 | 8120 | 6.87 | 1.2 | MTX_DAR_24hr |
| 120 | 36 | MA1997.1 | Olig2 | MA1997.1 | NVCAGCTGBN | Olig2 | 5.9e-10 | 3.29e-12 | 11200 | 51.3 | 57600 | 48.7 | 1.05 | MTX_DAR_24hr |
| 121 | 17 | MA1124.1 (ZNF24) | STREME-8 | 8-CACACATGCATGC | CACACATGCATGC | ZNF24 | 9.63e-10 | 5.32e-12 | 456 | 2.09 | 1680 | 1.42 | 1.47 | MTX_DAR_24hr |
| 122 | 22 | MA1965.1 | SP5 | MA1965.1 | CBCCTCCCHS | SP5 | 1.32e-09 | 7.23e-12 | 8640 | 39.6 | 43800 | 37.1 | 1.07 | MTX_DAR_24hr |
| 123 | 37 | MA0767.1 | GCM2 | MA0767.1 | BATGCGGGTR | GCM2 | 1.36e-09 | 7.39e-12 | 9600 | 44 | 49000 | 41.4 | 1.06 | MTX_DAR_24hr |
| 124 | 2 | MA0753.2 | ZNF740 | MA0753.2 | CYRCCCCCCCCAC | ZNF740 | 1.46e-09 | 7.87e-12 | 3510 | 16.1 | 16800 | 14.2 | 1.13 | MTX_DAR_24hr |
| 125 | 38 | MA0759.2 | ELK3 | MA0759.2 | ACCGGAAGTRV | ELK3 | 2.07e-09 | 1.11e-11 | 2650 | 12.1 | 12400 | 10.5 | 1.15 | MTX_DAR_24hr |
| 126 | 39 | MA0131.2 | HINFP | MA0131.2 | CARCGTCCGCKN | HINFP | 2.93e-09 | 1.56e-11 | 925 | 4.24 | 3890 | 3.29 | 1.29 | MTX_DAR_24hr |
| 127 | 19 | MA1571.1 | TGIF2LX | MA1571.1 | TGACAGCTGTCA | TGIF2LX | 8.84e-09 | 4.65e-11 | 11000 | 50.3 | 56500 | 47.8 | 1.05 | MTX_DAR_24hr |
| 128 | 40 | MA0813.1 | TFAP2B | MA0813.1 | TGCCCYBRGGGCA | TFAP2B | 1.33e-08 | 6.96e-11 | 1770 | 8.13 | 8100 | 6.85 | 1.19 | MTX_DAR_24hr |
| 129 | 40 | MA0872.1 | TFAP2A | MA0872.1 | TGCCCYSRGGGCA | TFAP2A | 1.64e-08 | 8.51e-11 | 1420 | 6.48 | 6320 | 5.35 | 1.21 | MTX_DAR_24hr |
| 130 | 41 | MA0734.3 | Gli2 | MA0734.3 | NRGACCACCCASV | Gli2 | 1.7e-08 | 8.72e-11 | 13800 | 63.3 | 72000 | 60.9 | 1.04 | MTX_DAR_24hr |
| 131 | 38 | MA0750.2 | ZBTB7A | MA0750.2 | NVCCGGAAGTGSV | ZBTB7A | 1.83e-08 | 9.33e-11 | 901 | 4.13 | 3810 | 3.23 | 1.28 | MTX_DAR_24hr |
| 132 | 41 | MA1990.1 | Gli1 | MA1990.1 | NNRGACCACCCASV | Gli1 | 1.86e-08 | 9.44e-11 | 12800 | 58.7 | 66500 | 56.3 | 1.04 | MTX_DAR_24hr |
| 133 | 2 | MA1516.1 | KLF3 | MA1516.1 | GRCCRCGCCCH | KLF3 | 2.52e-08 | 1.27e-10 | 986 | 4.52 | 4230 | 3.58 | 1.26 | MTX_DAR_24hr |
| 134 | 24 | MA1464.1 | ARNT2 | MA1464.1 | GTCACGTGMN | ARNT2 | 2.8e-08 | 1.4e-10 | 7270 | 33.3 | 36700 | 31.1 | 1.07 | MTX_DAR_24hr |
| 135 | 24 | MA1099.2 | HES1 | MA1099.2 | GGCRCGTGBC | HES1 | 7.68e-08 | 3.8e-10 | 649 | 2.97 | 2640 | 2.24 | 1.33 | MTX_DAR_24hr |
| 136 | 42 | MA1973.1 | ZKSCAN3 | MA1973.1 | KDKCCCAGGCTAGCCCABNN | ZKSCAN3 | 1.27e-07 | 6.24e-10 | 9430 | 43.2 | 48400 | 40.9 | 1.06 | MTX_DAR_24hr |
| 137 | 38 | MA0764.3 | ETV4 | MA0764.3 | ACCGGAAGTR | ETV4 | 2.26e-07 | 1.1e-09 | 266 | 1.22 | 917 | 0.78 | 1.57 | MTX_DAR_24hr |
| 138 | 23 | MA0003.4 | TFAP2A | MA0003.4 | NNKGCCTCAGGCNN | TFAP2A | 2.27e-07 | 1.1e-09 | 18000 | 82.5 | 95400 | 80.7 | 1.02 | MTX_DAR_24hr |
| 139 | 43 | MA1155.1 | ZSCAN4 | MA1155.1 | TGCACACACTGAAAA | ZSCAN4 | 3.73e-07 | 1.8e-09 | 315 | 1.44 | 1140 | 0.96 | 1.5 | MTX_DAR_24hr |
| 140 | 36 | MA0633.2 | Twist2 | MA0633.2 | NVCAGCTGBN | Twist2 | 3.89e-07 | 1.86e-09 | 11500 | 52.6 | 59500 | 50.3 | 1.04 | MTX_DAR_24hr |
| 141 | 24 | MA0499.2 | MYOD1 | MA0499.2 | NNGCACCTGTCNB | MYOD1 | 4.77e-07 | 2.26e-09 | 20600 | 94.6 | 110000 | 93.5 | 1.01 | MTX_DAR_24hr |
| 142 | 30 | MA1475.1 | CREB3L4 | MA1475.1 | GRTGACGTCAYC | CREB3L4 | 1.19e-06 | 5.61e-09 | 10900 | 50.1 | 56600 | 47.9 | 1.05 | MTX_DAR_24hr |
| 143 | 4 | MA1644.1 | NFYC | MA1644.1 | RDCCAATCASN | NFYC | 1.24e-06 | 5.8e-09 | 38 | 0.17 | 52 | 0.04 | 3.98 | MTX_DAR_24hr |
| 144 | 19 | MA0498.2 | MEIS1 | MA0498.2 | HTGACAD | MEIS1 | 1.25e-06 | 5.81e-09 | 14800 | 68 | 77900 | 65.9 | 1.03 | MTX_DAR_24hr |
| 145 | 24 | MA0608.1 | Creb3l2 | MA0608.1 | GCCACGTGD | Creb3l2 | 1.46e-06 | 6.74e-09 | 5810 | 26.6 | 29200 | 24.7 | 1.08 | MTX_DAR_24hr |
| 146 | 24 | MA0692.1 | TFEB | MA0692.1 | RYCACGTGAC | TFEB | 1.86e-06 | 8.52e-09 | 8830 | 40.5 | 45300 | 38.4 | 1.06 | MTX_DAR_24hr |
| 147 | 22 | MA0528.2 | ZNF263 | MA0528.2 | VNGGGGAGGASG | ZNF263 | 2.51e-06 | 1.14e-08 | 6170 | 28.3 | 31100 | 26.4 | 1.07 | MTX_DAR_24hr |
| 148 | 24 | MA0147.3 | MYC | MA0147.3 | NNCCACGTGCNB | MYC | 2.89e-06 | 1.31e-08 | 16600 | 76.1 | 87800 | 74.3 | 1.02 | MTX_DAR_24hr |
| 149 | 38 | MA0076.2 | ELK4 | MA0076.2 | BCRCTTCCGGB | ELK4 | 3.11e-06 | 1.39e-08 | 197 | 0.9 | 651 | 0.55 | 1.64 | MTX_DAR_24hr |
| 150 | 24 | MA1106.1 | HIF1A | MA1106.1 | SBACGTGCNN | HIF1A | 3.86e-06 | 1.72e-08 | 4540 | 20.8 | 22600 | 19.1 | 1.09 | MTX_DAR_24hr |
| 151 | 40 | MA0815.1 | TFAP2C | MA0815.1 | TGCCCYSRGGGCA | TFAP2C | 4.97e-06 | 2.2e-08 | 1150 | 5.26 | 5160 | 4.36 | 1.21 | MTX_DAR_24hr |
| 152 | 19 | MA0775.1 | MEIS3 | MA0775.1 | DTGACAGS | MEIS3 | 5.65e-06 | 2.48e-08 | 10500 | 47.9 | 54100 | 45.8 | 1.05 | MTX_DAR_24hr |
| 153 | 44 | MA1730.1 | ZNF708 | MA1730.1 | NNRGCTGTRCCTCCY | ZNF708 | 6.61e-06 | 2.89e-08 | 11900 | 54.6 | 62000 | 52.5 | 1.04 | MTX_DAR_24hr |
| 154 | 45 | MA1727.1 | ZNF417 | MA1727.1 | VRBVNTGGGCGCCAM | ZNF417 | 2.25e-05 | 9.77e-08 | 4930 | 22.6 | 24700 | 20.9 | 1.08 | MTX_DAR_24hr |
| 155 | 1 | MA1143.1 | FOSL1::JUND | MA1143.1 | RTGACGTMAY | FOSL1::JUND | 3.22e-05 | 1.39e-07 | 13000 | 59.4 | 67900 | 57.5 | 1.03 | MTX_DAR_24hr |
| 156 | 36 | MA1628.1 | Zic1::Zic2 | MA1628.1 | CVCAGCAGGNV | Zic1::Zic2 | 3.29e-05 | 1.41e-07 | 9550 | 43.8 | 49400 | 41.8 | 1.05 | MTX_DAR_24hr |
| 157 | 36 | MA1966.1 | TFAP4::ETV1 | MA1966.1 | RSCGGAAGCAGSTGKN | TFAP4::ETV1 | 5.39e-05 | 2.3e-07 | 4150 | 19 | 20700 | 17.5 | 1.09 | MTX_DAR_24hr |
| 158 | 30 | MA0609.2 | CREM | MA0609.2 | NNDGTGACGTCACHNN | CREM | 5.44e-05 | 2.3e-07 | 13800 | 63.1 | 72300 | 61.2 | 1.03 | MTX_DAR_24hr |
| 159 | 24 | MA0093.3 | USF1 | MA0093.3 | NDGTCATGTGACHN | USF1 | 6.13e-05 | 2.58e-07 | 14200 | 64.9 | 74500 | 63.1 | 1.03 | MTX_DAR_24hr |
| 160 | 23 | MA0810.1 | TFAP2A | MA0810.1 | YGCCCBVRGGCR | TFAP2A | 0.000129 | 5.39e-07 | 502 | 2.3 | 2100 | 1.77 | 1.3 | MTX_DAR_24hr |
| 161 | 38 | MA0028.2 | ELK1 | MA0028.2 | ACCGGAAGTR | ELK1 | 0.000142 | 5.88e-07 | 362 | 1.66 | 1440 | 1.22 | 1.36 | MTX_DAR_24hr |
| 162 | 38 | MA0156.3 | FEV | MA0156.3 | VACCGGAAGTVV | FEV | 0.00018 | 7.45e-07 | 443 | 2.03 | 1820 | 1.54 | 1.32 | MTX_DAR_24hr |
| 163 | 37 | MA0646.1 | GCM1 | MA0646.1 | BATGCGGGTAC | GCM1 | 0.000252 | 1.03e-06 | 4020 | 18.4 | 20100 | 17 | 1.08 | MTX_DAR_24hr |
| 164 | 36 | MA1638.1 | HAND2 | MA1638.1 | NVCAGATGNN | HAND2 | 0.000256 | 1.04e-06 | 11000 | 50.3 | 57300 | 48.4 | 1.04 | MTX_DAR_24hr |
| 165 | 45 | MA0024.3 | E2F1 | MA0024.3 | TTTGGCGCCAAA | E2F1 | 0.00036 | 1.46e-06 | 2640 | 12.1 | 12900 | 10.9 | 1.11 | MTX_DAR_24hr |
| 166 | 38 | MA0645.1 | ETV6 | MA0645.1 | MSCGGAAGTR | ETV6 | 0.000377 | 1.52e-06 | 154 | 0.71 | 519 | 0.44 | 1.61 | MTX_DAR_24hr |
| 167 | 20 | MA1723.1 | PRDM9 | MA1723.1 | RGDGGGVAGGGRGGVRRMARVARR | PRDM9 | 0.000555 | 2.22e-06 | 11900 | 54.7 | 62500 | 52.9 | 1.03 | MTX_DAR_24hr |
| 168 | 1 | MA0591.1 | Bach1::Mafk | MA0591.1 | RSSATGACTCAGCAB | Bach1::Mafk | 0.00102 | 4.08e-06 | 1280 | 5.86 | 6000 | 5.07 | 1.16 | MTX_DAR_24hr |
| 169 | 46 | MA1149.1 | RARA::RXRG | MA1149.1 | RRGGTCAHNNNRRGGTCA | RARA::RXRG | 0.00126 | 4.98e-06 | 6380 | 29.2 | 32700 | 27.7 | 1.06 | MTX_DAR_24hr |
| 170 | 47 | MA0155.1 | INSM1 | MA0155.1 | TGYCWGGGGGCR | INSM1 | 0.00135 | 5.31e-06 | 3490 | 16 | 17400 | 14.8 | 1.08 | MTX_DAR_24hr |
| 171 | 38 | MA1708.1 | ETV7 | MA1708.1 | DGSCGGAAGTR | ETV7 | 0.00222 | 8.68e-06 | 397 | 1.82 | 1660 | 1.4 | 1.3 | MTX_DAR_24hr |
| 172 | 24 | MA0649.1 | HEY2 | MA0649.1 | GRCACGTGYC | HEY2 | 0.00245 | 9.51e-06 | 15400 | 70.6 | 81600 | 69.1 | 1.02 | MTX_DAR_24hr |
| 173 | 48 | MA1711.1 | ZNF343 | MA1711.1 | CCGCTTCMCCDCGGCMV | ZNF343 | 0.00286 | 1.1e-05 | 2540 | 11.7 | 12500 | 10.6 | 1.1 | MTX_DAR_24hr |
| 174 | 23 | MA0524.2 | TFAP2C | MA0524.2 | YGCCYBVRGGCA | TFAP2C | 0.00304 | 1.17e-05 | 624 | 2.86 | 2760 | 2.34 | 1.22 | MTX_DAR_24hr |
| 175 | 38 | MA0098.3 | ETS1 | MA0098.3 | ACCGGAARTR | ETS1 | 0.004 | 1.53e-05 | 413 | 1.89 | 1740 | 1.48 | 1.28 | MTX_DAR_24hr |
| 176 | 45 | MA0864.2 | E2F2 | MA0864.2 | RWTTTGGCGCCAWWWY | E2F2 | 0.00553 | 2.1e-05 | 3250 | 14.9 | 16300 | 13.8 | 1.08 | MTX_DAR_24hr |
| 177 | 24 | MA0620.3 | MITF | MA0620.3 | NNNDRTCACGTGAYHNNN | MITF | 0.00564 | 2.13e-05 | 13900 | 63.9 | 73600 | 62.3 | 1.02 | MTX_DAR_24hr |
| 178 | 38 | MA0765.3 | ETV5 | MA0765.3 | ACCGGAAGTR | ETV5 | 0.00567 | 2.13e-05 | 1860 | 8.52 | 9040 | 7.65 | 1.11 | MTX_DAR_24hr |
| 179 | 20 | MA0163.1 | PLAG1 | MA0163.1 | GGGGCCCWAGGGGG | PLAG1 | 0.00653 | 2.44e-05 | 2410 | 11 | 11900 | 10 | 1.1 | MTX_DAR_24hr |
| 180 | 24 | MA0058.3 | MAX | MA0058.3 | AVCACGTGNY | MAX | 0.00927 | 3.45e-05 | 10500 | 48.3 | 55300 | 46.8 | 1.03 | MTX_DAR_24hr |
| 181 | 24 | MA0004.1 | Arnt | MA0004.1 | CACGTG | Arnt | 0.00971 | 3.59e-05 | 3820 | 17.5 | 19300 | 16.4 | 1.07 | MTX_DAR_24hr |
| 182 | 24 | MA0831.3 | TFE3 | MA0831.3 | GTCACGTGAC | TFE3 | 0.00976 | 3.59e-05 | 3990 | 18.3 | 20200 | 17.1 | 1.07 | MTX_DAR_24hr |
| 183 | 1 | MA1521.1 | MAFA | MA1521.1 | NTGCTGASTCAGCAN | MAFA | 0.0119 | 4.34e-05 | 18100 | 82.7 | 96400 | 81.6 | 1.01 | MTX_DAR_24hr |
| 184 | 24 | MA0259.1 | ARNT::HIF1A | MA0259.1 | VBACGTGC | ARNT::HIF1A | 0.0119 | 4.34e-05 | 1610 | 7.37 | 7790 | 6.59 | 1.12 | MTX_DAR_24hr |
| 185 | 34 | MA1566.2 | TBX3 | MA1566.2 | RAGGTGTGAAV | TBX3 | 0.0127 | 4.58e-05 | 5400 | 24.7 | 27700 | 23.4 | 1.06 | MTX_DAR_24hr |
| 186 | 49 | MA1716.1 | ZNF76 | MA1716.1 | NNTWCCCAYAATGCAHYGCGC | ZNF76 | 0.0132 | 4.74e-05 | 137 | 0.63 | 486 | 0.41 | 1.53 | MTX_DAR_24hr |
| 187 | 50 | MA0656.1 | JDP2 | MA0656.1 | GATGACGTCAYC | JDP2 | 0.0139 | 4.96e-05 | 90 | 0.41 | 286 | 0.24 | 1.72 | MTX_DAR_24hr |
| 188 | 24 | MA1493.1 | HES6 | MA1493.1 | GGCACGTGTY | HES6 | 0.0161 | 5.73e-05 | 8320 | 38.1 | 43300 | 36.7 | 1.04 | MTX_DAR_24hr |
| 189 | 51 | MA1982.1 | ZNF574 | MA1982.1 | VGSCTAGAGMGGCCS | ZNF574 | 0.0171 | 6.05e-05 | 598 | 2.74 | 2680 | 2.27 | 1.21 | MTX_DAR_24hr |
| 190 | 38 | MA0475.2 | FLI1 | MA0475.2 | ACCGGAARTR | FLI1 | 0.0203 | 7.13e-05 | 341 | 1.56 | 1440 | 1.22 | 1.29 | MTX_DAR_24hr |
| 191 | 52 | MA0705.1 | Lhx8 | MA0705.1 | CTAATTAV | Lhx8 | 0.0217 | 7.61e-05 | 12100 | 55.6 | 64000 | 54.1 | 1.03 | MTX_DAR_24hr |
| 192 | 24 | MA0819.2 | CLOCK | MA0819.2 | NRACACGTGB | CLOCK | 0.0232 | 8.07e-05 | 7740 | 35.5 | 40200 | 34 | 1.04 | MTX_DAR_24hr |
| 193 | 24 | MA0664.1 | MLXIPL | MA0664.1 | RTCACGTGAT | MLXIPL | 0.0233 | 8.09e-05 | 12100 | 55.6 | 63900 | 54.1 | 1.03 | MTX_DAR_24hr |
| 194 | 53 | MA1971.1 | ZBED2 | MA1971.1 | NNMMCGAAACCNNV | ZBED2 | 0.0252 | 8.68e-05 | 1020 | 4.67 | 4810 | 4.07 | 1.15 | MTX_DAR_24hr |
| 195 | 54 | MA1645.1 | NKX2-2 | MA1645.1 | NNNCCACTCAANNN | NKX2-2 | 0.0259 | 8.88e-05 | 5090 | 23.3 | 26100 | 22.1 | 1.06 | MTX_DAR_24hr |
| 196 | 24 | MA1560.1 | SOHLH2 | MA1560.1 | YGCACGTGCN | SOHLH2 | 0.0277 | 9.45e-05 | 457 | 2.09 | 2000 | 1.7 | 1.24 | MTX_DAR_24hr |
| 197 | 36 | MA0697.2 | Zic3 | MA0697.2 | CNCAGCAGGAGNN | Zic3 | 0.0291 | 9.88e-05 | 12300 | 56.5 | 65000 | 55 | 1.03 | MTX_DAR_24hr |
| 198 | 24 | MA1474.1 | CREB3L4 | MA1474.1 | YGCCACGTCAYC | CREB3L4 | 0.03 | 0.000101 | 152 | 0.7 | 563 | 0.48 | 1.47 | MTX_DAR_24hr |
| 199 | 55 | MA1710.1 | ZNF257 | MA1710.1 | VDGAGGCRAGRG | ZNF257 | 0.0357 | 0.00012 | 13900 | 63.8 | 73700 | 62.4 | 1.02 | MTX_DAR_24hr |
| 200 | 38 | MA1484.1 | ETS2 | MA1484.1 | DACCGGAAGY | ETS2 | 0.0367 | 0.000123 | 246 | 1.13 | 998 | 0.84 | 1.34 | MTX_DAR_24hr |
| 201 | 32 | MA1587.1 | ZNF135 | MA1587.1 | CCTCGACCTCCYRR | ZNF135 | 0.038 | 0.000126 | 9650 | 44.2 | 50500 | 42.8 | 1.03 | MTX_DAR_24hr |
| 202 | 56 | MA1731.1 | ZNF768 | MA1731.1 | YBVCYBRSCCTCTCTGDG | ZNF768 | 0.0487 | 0.000161 | 4240 | 19.4 | 21600 | 18.3 | 1.06 | MTX_DAR_24hr |
| 15 | 57 | MA1125.1 (ZNF384) | MEME-2 | AAAAAAAAAAAAAAA | AAAAAAAAAAAAAAA | ZNF384 | 15 | 1 | 1700 | 7.78 | 10700 | 9.03 | 0.863 | MTX_DAR_24hr |
ER_rat <- 1.25
mrc_type <- "MTX_DAR_24hr"
MTX_DAR_24hr_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,mrc_type,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks(200bp)Enrichment ratio:",ER_rat))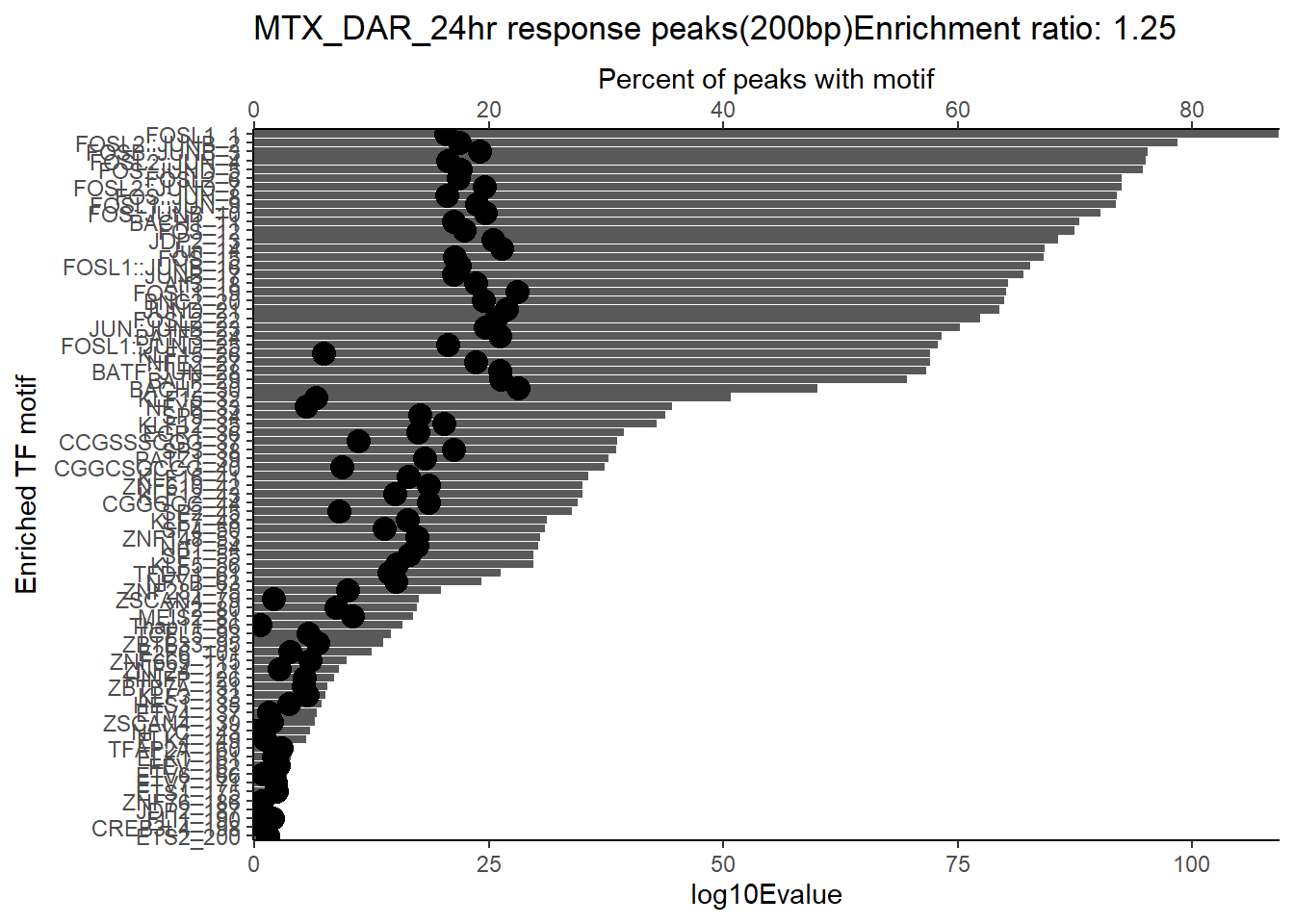
MTX_DAR_24hr_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,mrc_type,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
dplyr::filter(EVALUE.y<0.05) %>%
slice_head(n=5) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*.8), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./.8,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks(200bp)Enrichment ratio:",ER_rat))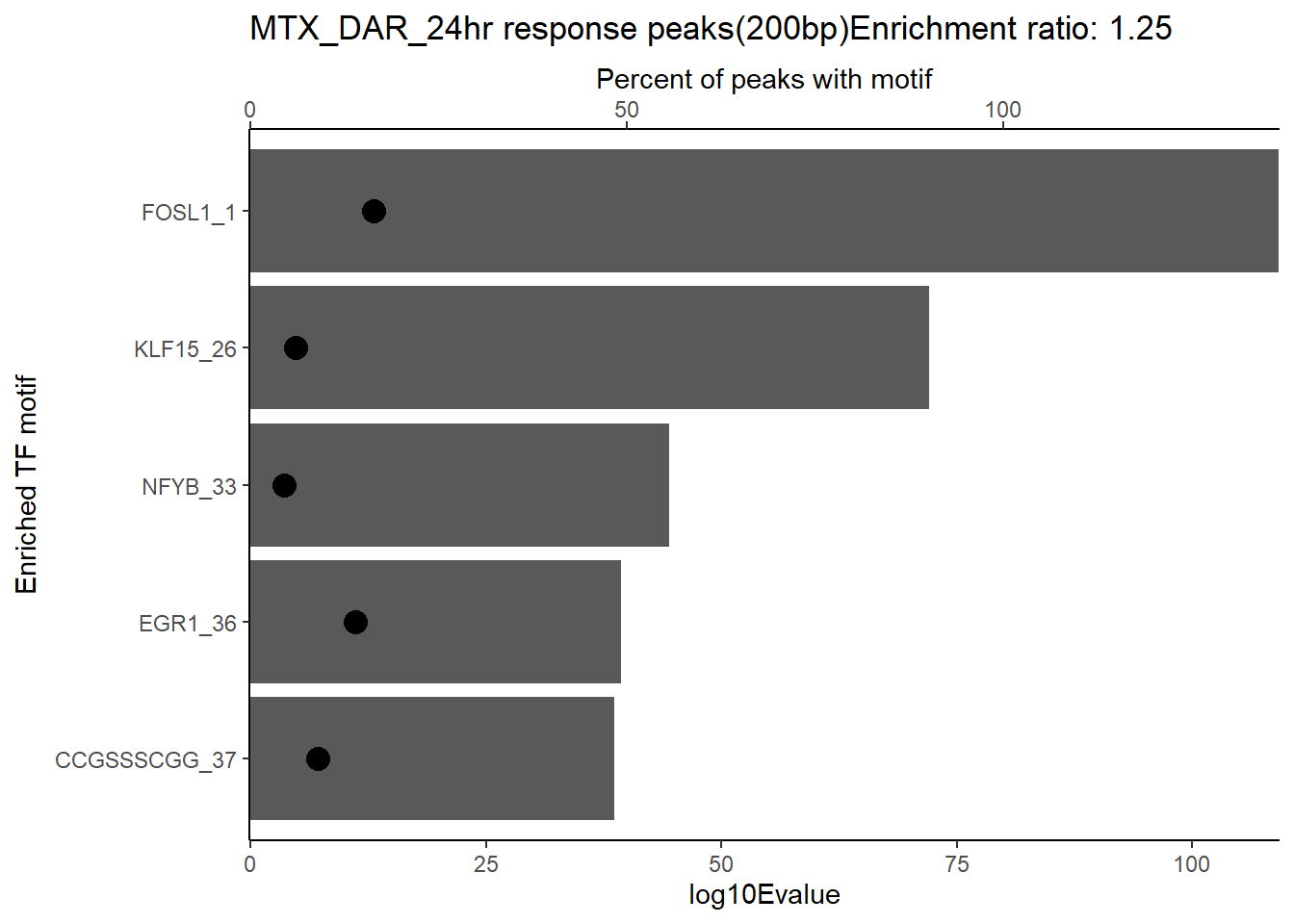 ### MTX_DAR 24 2kb data
### MTX_DAR 24 2kb data
MTX_242kb_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/MTX_24_2kb_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
MTX_242kb_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/MTX_24_2kb_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
MTX_242kb_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/MTX_24_2kb_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
MTX_242kb_sea_all <- rbind(MTX_242kb_sea_disc, MTX_242kb_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)MTX_242kb_combo <- MTX_242kb_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., MTX_242kb_sea_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ SIM_MOTIF
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y)
MTX_242kb_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs 200 bp in MTX_24 2kb") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | MA0060.3 (NFYA) | STREME-2 | 2-YSATTGGYY | YSATTGGYY | NFYA | 3.44e-70 | 2.66e-70 | 486 | 8.15 | 737 | 2.73 | 2.98 |
| 2 | 2 | MA0516.3 (SP2) | STREME-1 | 1-GCCCCGCCCC | GCCCCGCCCC | SP2 | 1.91e-57 | 7.39e-58 | 998 | 16.7 | 2470 | 9.14 | 1.83 |
| 3 | 1 | MA0502.2 | NFYB | MA0502.2 | CYCATTGGCCVV | NFYB | 3.22e-44 | 8.3e-45 | 279 | 4.68 | 387 | 1.43 | 3.26 |
| 4 | 3 | 3-CGCGCCGG | STREME-3 | 3-CGCGCCGG | CGCGCCGG | CGCGCCGG | 7.94e-43 | 1.54e-43 | 2960 | 49.5 | 10700 | 39.5 | 1.25 |
| 5 | 4 | MA0146.2 (Zfx) | STREME-7 | 7-AGGCCGG | AGGCCGG | Zfx | 1.8e-40 | 2.78e-41 | 2310 | 38.7 | 7940 | 29.4 | 1.31 |
| 6 | 2 | MA0516.3 | SP2 | MA0516.3 | GGGGCGGGG | SP2 | 4.47e-40 | 5.77e-41 | 1390 | 23.3 | 4220 | 15.7 | 1.49 |
| 7 | 2 | MA1513.1 | KLF15 | MA1513.1 | SCCCCGCCCCS | KLF15 | 1.15e-37 | 1.27e-38 | 1390 | 23.2 | 4270 | 15.8 | 1.47 |
| 8 | 2 | MA0742.2 | KLF12 | MA0742.2 | GGGGCGGGG | KLF12 | 3.24e-35 | 3.14e-36 | 1430 | 24 | 4520 | 16.7 | 1.44 |
| 9 | 2 | MA0740.2 | KLF14 | MA0740.2 | KGGGCGGGG | KLF14 | 1.69e-33 | 1.45e-34 | 944 | 15.8 | 2680 | 9.95 | 1.59 |
| 10 | 2 | MA1961.1 (PATZ1) | MEME-1 | CCCCGCCSCCSCCSC | CCCCGCCSCCSCCSC | PATZ1 | 1.08e-32 | 8.36e-34 | 3210 | 53.8 | 12100 | 44.9 | 1.2 |
| 11 | 2 | MA1512.1 | KLF11 | MA1512.1 | SCCACGCCCMC | KLF11 | 8.39e-32 | 5.91e-33 | 3030 | 50.8 | 11400 | 42.1 | 1.21 |
| 12 | 2 | MA1511.2 | KLF10 | MA1511.2 | GGGGCGGGG | KLF10 | 1.74e-31 | 1.12e-32 | 1980 | 33.2 | 6840 | 25.4 | 1.31 |
| 13 | 2 | MA1959.1 | KLF7 | MA1959.1 | DGGGCGGGG | KLF7 | 2.32e-31 | 1.38e-32 | 1140 | 19.2 | 3480 | 12.9 | 1.49 |
| 14 | 2 | MA0599.1 | KLF5 | MA0599.1 | GCCCCDCCCH | KLF5 | 5.88e-30 | 3.25e-31 | 1170 | 19.7 | 3630 | 13.5 | 1.46 |
| 15 | 2 | MA0685.2 | SP4 | MA0685.2 | GGGGCGGGG | SP4 | 3.23e-29 | 1.67e-30 | 3030 | 50.7 | 11400 | 42.4 | 1.2 |
| 16 | 2 | MA0746.2 | SP3 | MA0746.2 | NGCCACGCCCMCH | SP3 | 1.26e-28 | 6.11e-30 | 2790 | 46.8 | 10400 | 38.6 | 1.21 |
| 17 | 2 | MA1515.1 | KLF2 | MA1515.1 | NRCCACRCCCH | KLF2 | 1.18e-26 | 5.38e-28 | 3270 | 54.8 | 12600 | 46.8 | 1.17 |
| 18 | 2 | MA0079.5 | SP1 | MA0079.5 | GGGGCGGGG | SP1 | 4.56e-24 | 1.96e-25 | 912 | 15.3 | 2770 | 10.3 | 1.49 |
| 19 | 2 | MA1564.1 | SP9 | MA1564.1 | RCCACGCCCMCY | SP9 | 5.17e-24 | 2.11e-25 | 3290 | 55.2 | 12800 | 47.5 | 1.16 |
| 20 | 2 | MA1961.1 | PATZ1 | MA1961.1 | SSGGGGMGGGGS | PATZ1 | 6.62e-23 | 2.56e-24 | 3240 | 54.3 | 12600 | 46.9 | 1.16 |
| 21 | 2 | MA0493.2 | KLF1 | MA0493.2 | DGGGYGGGG | KLF1 | 9.66e-23 | 3.49e-24 | 3920 | 65.8 | 15800 | 58.5 | 1.12 |
| 22 | 1 | MA1644.1 | NFYC | MA1644.1 | RDCCAATCASN | NFYC | 9.92e-23 | 3.49e-24 | 235 | 3.94 | 440 | 1.63 | 2.42 |
| 23 | 5 | 9-SGCGCS | STREME-9 | 9-SGCGCS | SGCGCS | SGCGCS | 2.01e-21 | 6.76e-23 | 3570 | 59.9 | 14200 | 52.7 | 1.14 |
| 24 | 6 | MA1713.1 | ZNF610 | MA1713.1 | SSCGCCGCTCCSSS | ZNF610 | 4.32e-21 | 1.39e-22 | 2450 | 41.1 | 9200 | 34.1 | 1.2 |
| 25 | 7 | MA0162.4 | EGR1 | MA0162.4 | VCMCGCCCACGCVS | EGR1 | 7.75e-21 | 2.4e-22 | 3530 | 59.2 | 14000 | 52 | 1.14 |
| 26 | 1 | MA0060.3 | NFYA | MA0060.3 | RRCCAATCAGM | NFYA | 1.47e-20 | 4.38e-22 | 168 | 2.82 | 272 | 1.01 | 2.8 |
| 27 | 2 | MA1517.1 | KLF6 | MA1517.1 | NRCCACGCCCH | KLF6 | 2.58e-19 | 7.41e-21 | 2960 | 49.7 | 11500 | 42.8 | 1.16 |
| 28 | 2 | MA1522.1 | MAZ | MA1522.1 | BBCCCCTCCCY | MAZ | 4.96e-19 | 1.37e-20 | 3210 | 53.8 | 12700 | 46.9 | 1.15 |
| 29 | 2 | MA1653.1 | ZNF148 | MA1653.1 | YBCCCCTCCCCC | ZNF148 | 1.73e-17 | 4.63e-19 | 4070 | 68.2 | 16700 | 62 | 1.1 |
| 30 | 7 | MA1107.2 | KLF9 | MA1107.2 | MNGCCACACCCACHYV | KLF9 | 1.9e-17 | 4.91e-19 | 4160 | 69.8 | 17100 | 63.6 | 1.1 |
| 31 | 2 | MA0039.4 | KLF4 | MA0039.4 | NNCCCCACCCHN | KLF4 | 5.98e-17 | 1.49e-18 | 4320 | 72.4 | 17900 | 66.4 | 1.09 |
| 32 | 8 | 4-CGGCCC | STREME-4 | 4-CGGCCC | CGGCCC | CGGCCC | 1.43e-16 | 3.46e-18 | 4230 | 70.9 | 17500 | 64.9 | 1.09 |
| 33 | 1 | MA0502.2 (NFYB) | STREME-10 | 10-ATTGGCC | 10-ATTGGCC | NFYB | 5.56e-16 | 1.31e-17 | 102 | 1.71 | 136 | 0.5 | 3.4 |
| 34 | 9 | MA1965.1 | SP5 | MA1965.1 | CBCCTCCCHS | SP5 | 8.53e-15 | 1.94e-16 | 3960 | 66.4 | 16300 | 60.5 | 1.1 |
| 35 | 2 | MA0741.1 | KLF16 | MA0741.1 | GMCACGCCCCC | KLF16 | 1.2e-14 | 2.67e-16 | 3620 | 60.7 | 14700 | 54.7 | 1.11 |
| 36 | 11 | MA1573.2 | Thap11 | MA1573.2 | ACTACAABTCCCAG | Thap11 | 2.36e-14 | 5.07e-16 | 94 | 1.58 | 127 | 0.47 | 3.35 |
| 37 | 10 | 12-CCGSCGG | STREME-12 | 12-CCGSCGG | 12-CCGSCGG | CCGSCGG | 2.92e-13 | 6.12e-15 | 4150 | 69.5 | 17300 | 64 | 1.09 |
| 38 | 11 | MA1573.2 (Thap11) | STREME-11 | 11-ACTACAAYTCCCAGV | 11-ACTACAAYTCCCAGV | Thap11 | 6.29e-13 | 1.28e-14 | 152 | 2.55 | 297 | 1.1 | 2.32 |
| 39 | 12 | MA1938.1 (ERF::NHLH1) | STREME-8 | 8-GCCGGA | GCCGGA | ERF::NHLH1 | 1.8e-12 | 3.58e-14 | 4120 | 69.1 | 17200 | 63.8 | 1.08 |
| 40 | 7 | MA0733.1 | EGR4 | MA0733.1 | NHMCGCCCACGCAHWT | EGR4 | 1.87e-12 | 3.63e-14 | 3650 | 61.2 | 15000 | 55.6 | 1.1 |
| 41 | 13 | MA1721.1 | ZNF93 | MA1721.1 | GGYAGCRGCAGCGGYG | ZNF93 | 5.47e-12 | 1.03e-13 | 4240 | 71.1 | 17800 | 65.9 | 1.08 |
| 42 | 2 | MA1516.1 | KLF3 | MA1516.1 | GRCCRCGCCCH | KLF3 | 6.62e-12 | 1.22e-13 | 2410 | 40.4 | 9470 | 35.1 | 1.15 |
| 43 | 14 | MA1122.1 | TFDP1 | MA1122.1 | VSGCGGGAAVN | TFDP1 | 7.43e-12 | 1.34e-13 | 1560 | 26.2 | 5820 | 21.6 | 1.22 |
| 44 | 2 | MA0747.1 | SP8 | MA0747.1 | RCCACGCCCMCY | SP8 | 1.17e-11 | 2.05e-13 | 4120 | 69.1 | 17200 | 63.9 | 1.08 |
| 45 | 15 | MA1548.1 | PLAGL2 | MA1548.1 | NGGGCCCCCN | PLAGL2 | 1.63e-11 | 2.8e-13 | 3760 | 63 | 15600 | 57.7 | 1.09 |
| 46 | 16 | MA1986.1 | ZNF692 | MA1986.1 | BGGGCCCASN | ZNF692 | 1.94e-11 | 3.26e-13 | 3250 | 54.5 | 13200 | 49.1 | 1.11 |
| 47 | 17 | MA1599.1 | ZNF682 | MA1599.1 | BVGGCYAAGCCCCWNY | ZNF682 | 6.03e-11 | 9.94e-13 | 1980 | 33.1 | 7620 | 28.3 | 1.17 |
| 48 | 18 | MA1712.1 | ZNF454 | MA1712.1 | DGGCTCSSGGCCCYGVKG | ZNF454 | 1.4e-10 | 2.26e-12 | 2930 | 49.2 | 11900 | 44 | 1.12 |
| 49 | 19 | MA0759.2 | ELK3 | MA0759.2 | ACCGGAAGTRV | ELK3 | 1.65e-10 | 2.61e-12 | 884 | 14.8 | 3060 | 11.4 | 1.31 |
| 50 | 20 | MA0753.2 | ZNF740 | MA0753.2 | CYRCCCCCCCCAC | ZNF740 | 2.63e-10 | 4.07e-12 | 3540 | 59.3 | 14600 | 54.2 | 1.09 |
| 51 | 7 | MA0472.2 | EGR2 | MA0472.2 | MCGCCCACGCA | EGR2 | 4.97e-10 | 7.55e-12 | 3590 | 60.1 | 14900 | 55.1 | 1.09 |
| 52 | 7 | MA1650.1 | ZBTB14 | MA1650.1 | SSCCGCGCACVS | ZBTB14 | 5.22e-10 | 7.78e-12 | 1490 | 25 | 5600 | 20.8 | 1.2 |
| 53 | 9 | MA1596.1 | ZNF460 | MA1596.1 | GCCTCMGCCTCCCRAG | ZNF460 | 7.01e-10 | 1.02e-11 | 2830 | 47.5 | 11400 | 42.4 | 1.12 |
| 54 | 2 | MA1630.2 | ZNF281 | MA1630.2 | GGGGGAGGGGVV | ZNF281 | 1.5e-09 | 2.15e-11 | 894 | 15 | 3140 | 11.6 | 1.29 |
| 55 | 21 | MA0024.3 | E2F1 | MA0024.3 | TTTGGCGCCAAA | E2F1 | 3.28e-09 | 4.62e-11 | 2960 | 49.7 | 12100 | 44.8 | 1.11 |
| 56 | 22 | MA0146.2 | Zfx | MA0146.2 | SSSGCCBVGGCCTS | Zfx | 8.7e-09 | 1.2e-10 | 370 | 6.2 | 1110 | 4.12 | 1.51 |
| 57 | 23 | MA0632.2 | TCFL5 | MA0632.2 | KCACGCGCMC | TCFL5 | 8.9e-09 | 1.21e-10 | 2210 | 37.1 | 8770 | 32.5 | 1.14 |
| 58 | 24 | MA0527.1 | ZBTB33 | MA0527.1 | BTCTCGCGAGABYTS | ZBTB33 | 1.06e-08 | 1.41e-10 | 2470 | 41.4 | 9900 | 36.7 | 1.13 |
| 59 | 19 | MA0131.2 | HINFP | MA0131.2 | CARCGTCCGCKN | HINFP | 1.19e-08 | 1.56e-10 | 2230 | 37.4 | 8870 | 32.9 | 1.14 |
| 60 | 14 | MA0865.2 | E2F8 | MA0865.2 | TTCCCGCCAHWA | E2F8 | 1.61e-08 | 2.08e-10 | 3440 | 57.7 | 14300 | 53 | 1.09 |
| 61 | 19 | MA0765.3 | ETV5 | MA0765.3 | ACCGGAAGTR | ETV5 | 3.21e-08 | 4.01e-10 | 778 | 13 | 2720 | 10.1 | 1.29 |
| 62 | 25 | MA1134.1 (FOS::JUNB) | STREME-6 | 6-KATGAGTCATCM | KATGAGTCATCM | FOS::JUNB | 3.21e-08 | 4.01e-10 | 272 | 4.56 | 767 | 2.84 | 1.61 |
| 63 | 26 | MA1569.1 | TFAP2E | MA1569.1 | YGCCYSRGGCD | TFAP2E | 8.59e-08 | 1.06e-09 | 1910 | 32 | 7500 | 27.8 | 1.15 |
| 64 | 25 | MA1583.1 | ZFP57 | MA1583.1 | NNVTTGCCGCANN | ZFP57 | 1.05e-07 | 1.27e-09 | 4680 | 78.5 | 20100 | 74.7 | 1.05 |
| 65 | 27 | MA0872.1 | TFAP2A | MA0872.1 | TGCCCYSRGGGCA | TFAP2A | 1.1e-07 | 1.31e-09 | 2220 | 37.1 | 8850 | 32.8 | 1.13 |
| 66 | 25 | MA0489.2 | Jun | MA0489.2 | NVTGACTCATNN | Jun | 1.52e-07 | 1.78e-09 | 335 | 5.62 | 1010 | 3.76 | 1.5 |
| 67 | 26 | MA0814.2 | TFAP2C | MA0814.2 | NNGCCTGAGGCGNN | TFAP2C | 1.79e-07 | 2.06e-09 | 4590 | 77 | 19700 | 73 | 1.05 |
| 68 | 19 | MA0764.3 | ETV4 | MA0764.3 | ACCGGAAGTR | ETV4 | 2e-07 | 2.27e-09 | 779 | 13.1 | 2760 | 10.2 | 1.28 |
| 69 | 19 | MA0076.2 | ELK4 | MA0076.2 | BCRCTTCCGGB | ELK4 | 2.06e-07 | 2.32e-09 | 258 | 4.32 | 734 | 2.72 | 1.59 |
| 70 | 28 | MA1102.2 | CTCFL | MA1102.2 | NSCAGGGGGCGS | CTCFL | 4.37e-07 | 4.83e-09 | 5000 | 83.8 | 21700 | 80.4 | 1.04 |
| 71 | 26 | MA0810.1 | TFAP2A | MA0810.1 | YGCCCBVRGGCR | TFAP2A | 7.41e-07 | 8.09e-09 | 4630 | 77.6 | 19900 | 73.9 | 1.05 |
| 72 | 29 | MA1985.1 | ZNF669 | MA1985.1 | GGGGYGAYGACCRCT | ZNF669 | 8.2e-07 | 8.83e-09 | 3710 | 62.2 | 15600 | 58 | 1.07 |
| 73 | 21 | MA0470.2 | E2F4 | MA0470.2 | TTTTGGCGCCAWWW | E2F4 | 8.73e-07 | 9.27e-09 | 1780 | 29.8 | 6990 | 25.9 | 1.15 |
| 74 | 7 | MA1627.1 | Wt1 | MA1627.1 | YBCCTCCCCCACVB | Wt1 | 1.51e-06 | 1.57e-08 | 4810 | 80.7 | 20800 | 77.2 | 1.05 |
| 75 | 25 | MA0476.1 | FOS | MA0476.1 | DVTGASTCATB | FOS | 1.52e-06 | 1.57e-08 | 354 | 5.93 | 1110 | 4.12 | 1.44 |
| 76 | 40 | MA0506.2 | Nrf1 | MA0506.2 | SNCTGCGCMTGCGCV | Nrf1 | 2.48e-06 | 2.53e-08 | 1000 | 16.8 | 3730 | 13.8 | 1.22 |
| 77 | 21 | MA1615.1 | Plagl1 | MA1615.1 | NNCTGGGGCCABN | Plagl1 | 3.27e-06 | 3.29e-08 | 3470 | 58.1 | 14600 | 54 | 1.08 |
| 78 | 14 | MA0471.2 | E2F6 | MA0471.2 | VVDGGCGGGAAVV | E2F6 | 3.71e-06 | 3.68e-08 | 1360 | 22.7 | 5220 | 19.4 | 1.17 |
| 79 | 30 | MA1545.1 | OVOL2 | MA1545.1 | DDACCGTTABNYN | OVOL2 | 4.39e-06 | 4.31e-08 | 931 | 15.6 | 3440 | 12.8 | 1.22 |
| 80 | 26 | MA0811.1 | TFAP2B | MA0811.1 | YGCCCBVRGGCA | TFAP2B | 5.33e-06 | 5.16e-08 | 4160 | 69.7 | 17800 | 65.9 | 1.06 |
| 81 | 31 | MA1976.1 | ZNF320 | MA1976.1 | DGKGGGRCCMRGGGGCCWGTGHVNNNN | ZNF320 | 7.46e-06 | 7.13e-08 | 1720 | 28.8 | 6820 | 25.3 | 1.14 |
| 82 | 32 | MA0067.2 | PAX2 | MA0067.2 | BCAVTSRAGCGTGACGR | PAX2 | 1.04e-05 | 9.81e-08 | 1910 | 32 | 7650 | 28.4 | 1.13 |
| 83 | 26 | MA0812.1 | TFAP2B | MA0812.1 | HGCCTSAGGCD | TFAP2B | 1.15e-05 | 1.07e-07 | 3440 | 57.6 | 14500 | 53.6 | 1.07 |
| 84 | 33 | MA0615.1 | Gmeb1 | MA0615.1 | BHBBKKACGTMMNWNNN | Gmeb1 | 1.96e-05 | 1.81e-07 | 1810 | 30.4 | 7240 | 26.9 | 1.13 |
| 85 | 25 | MA0491.2 | JUND | MA0491.2 | NNATGACTCATNN | JUND | 3.39e-05 | 3.09e-07 | 275 | 4.61 | 850 | 3.15 | 1.47 |
| 86 | 34 | MA0821.2 | HES5 | MA0821.2 | GRCACGTGYC | HES5 | 3.54e-05 | 3.19e-07 | 1820 | 30.4 | 7280 | 27 | 1.13 |
| 87 | 20 | MA1578.1 | VEZF1 | MA1578.1 | CCCCCCMYDH | VEZF1 | 4.05e-05 | 3.61e-07 | 4540 | 76.1 | 19600 | 72.8 | 1.05 |
| 88 | 26 | MA0003.4 | TFAP2A | MA0003.4 | NNKGCCTCAGGCNN | TFAP2A | 4.31e-05 | 3.79e-07 | 4700 | 78.8 | 20400 | 75.6 | 1.04 |
| 89 | 25 | MA0462.2 | BATF::JUN | MA0462.2 | DATGACTCATH | BATF::JUN | 4.93e-05 | 4.29e-07 | 429 | 7.19 | 1450 | 5.37 | 1.34 |
| 90 | 25 | MA1634.1 | BATF | MA1634.1 | DATGACTCATH | BATF | 5.23e-05 | 4.5e-07 | 446 | 7.48 | 1520 | 5.62 | 1.33 |
| 91 | 35 | MA0748.2 | YY2 | MA0748.2 | NVATGGCGGCS | YY2 | 7.09e-05 | 6.04e-07 | 5000 | 83.8 | 21800 | 81 | 1.04 |
| 92 | 26 | MA0524.2 | TFAP2C | MA0524.2 | YGCCYBVRGGCA | TFAP2C | 7.49e-05 | 6.31e-07 | 2800 | 47 | 11700 | 43.2 | 1.09 |
| 93 | 36 | MA0751.1 | ZIC4 | MA0751.1 | GRCCCCCCGCKGYGH | ZIC4 | 8.51e-05 | 7.09e-07 | 4110 | 68.9 | 17600 | 65.4 | 1.05 |
| 94 | 34 | MA0823.1 | HEY1 | MA0823.1 | GRCACGTGCC | HEY1 | 0.00014 | 1.15e-06 | 2320 | 38.8 | 9520 | 35.3 | 1.1 |
| 95 | 27 | MA0815.1 | TFAP2C | MA0815.1 | TGCCCYSRGGGCA | TFAP2C | 0.000156 | 1.27e-06 | 1580 | 26.5 | 6300 | 23.4 | 1.13 |
| 96 | 25 | MA0841.1 | NFE2 | MA0841.1 | VATGACTCATS | NFE2 | 0.000168 | 1.36e-06 | 647 | 10.8 | 2350 | 8.71 | 1.25 |
| 97 | 33 | MA0604.1 | Atf1 | MA0604.1 | RTGACGTA | Atf1 | 0.000192 | 1.53e-06 | 2270 | 38 | 9320 | 34.5 | 1.1 |
| 98 | 25 | MA1988.1 | Atf3 | MA1988.1 | NRTGACTCABN | Atf3 | 0.000195 | 1.54e-06 | 163 | 2.73 | 457 | 1.69 | 1.62 |
| 99 | 34 | MA0259.1 | ARNT::HIF1A | MA0259.1 | VBACGTGC | ARNT::HIF1A | 2e-04 | 1.57e-06 | 1860 | 31.3 | 7540 | 28 | 1.12 |
| 100 | 27 | MA0813.1 | TFAP2B | MA0813.1 | TGCCCYBRGGGCA | TFAP2B | 0.000229 | 1.77e-06 | 1700 | 28.5 | 6840 | 25.4 | 1.13 |
| 101 | 37 | MA0705.1 | Lhx8 | MA0705.1 | CTAATTAV | Lhx8 | 0.000279 | 2.14e-06 | 902 | 15.1 | 3420 | 12.7 | 1.19 |
| 102 | 25 | MA0835.2 | BATF3 | MA0835.2 | DATGACTCATH | BATF3 | 0.000287 | 2.17e-06 | 405 | 6.79 | 1380 | 5.12 | 1.33 |
| 103 | 34 | MA1464.1 | ARNT2 | MA1464.1 | GTCACGTGMN | ARNT2 | 0.000289 | 2.17e-06 | 1890 | 31.7 | 7670 | 28.4 | 1.11 |
| 104 | 38 | MA1514.1 | KLF17 | MA1514.1 | MMCCACGCACCCMTY | KLF17 | 0.00044 | 3.28e-06 | 3940 | 66 | 16900 | 62.6 | 1.05 |
| 105 | 25 | MA0490.2 | JUNB | MA0490.2 | NNATGACTCATNN | JUNB | 0.000881 | 6.5e-06 | 246 | 4.12 | 780 | 2.89 | 1.43 |
| 106 | 34 | MA0616.2 | HES2 | MA0616.2 | GGCACGTGYC | HES2 | 0.00127 | 9.26e-06 | 2200 | 36.8 | 9060 | 33.6 | 1.1 |
| 107 | 21 | MA1727.1 | ZNF417 | MA1727.1 | VRBVNTGGGCGCCAM | ZNF417 | 0.00134 | 9.69e-06 | 3340 | 56 | 14200 | 52.7 | 1.06 |
| 108 | 33 | MA1475.1 | CREB3L4 | MA1475.1 | GRTGACGTCAYC | CREB3L4 | 0.00174 | 1.25e-05 | 3090 | 51.8 | 13100 | 48.5 | 1.07 |
| 109 | 36 | MA0736.1 | GLIS2 | MA0736.1 | GACCCCCCGCRAMG | GLIS2 | 0.002 | 1.42e-05 | 1980 | 33.2 | 8120 | 30.1 | 1.1 |
| 110 | 19 | MA0750.2 | ZBTB7A | MA0750.2 | NVCCGGAAGTGSV | ZBTB7A | 0.00214 | 1.51e-05 | 375 | 6.29 | 1300 | 4.81 | 1.31 |
| 111 | 21 | MA0864.2 | E2F2 | MA0864.2 | RWTTTGGCGCCAWWWY | E2F2 | 0.00317 | 2.21e-05 | 2030 | 34 | 8360 | 31 | 1.1 |
| 112 | 19 | MA0028.2 | ELK1 | MA0028.2 | ACCGGAAGTR | ELK1 | 0.00372 | 2.58e-05 | 296 | 4.96 | 993 | 3.68 | 1.35 |
| 113 | 25 | MA0477.2 | FOSL1 | MA0477.2 | NNATGACTCATNN | FOSL1 | 0.00395 | 2.71e-05 | 178 | 2.98 | 542 | 2.01 | 1.49 |
| 114 | 23 | MA1099.2 | HES1 | MA1099.2 | GGCRCGTGBC | HES1 | 0.00487 | 3.31e-05 | 2450 | 41.1 | 10300 | 38.1 | 1.08 |
| 115 | 25 | MA0655.1 | JDP2 | MA0655.1 | ATGACTCAT | JDP2 | 0.00682 | 4.59e-05 | 127 | 2.13 | 361 | 1.34 | 1.6 |
| 116 | 39 | MA1990.1 | Gli1 | MA1990.1 | NNRGACCACCCASV | Gli1 | 0.00833 | 5.56e-05 | 3160 | 52.9 | 13400 | 49.8 | 1.06 |
| 117 | 19 | MA0156.3 | FEV | MA0156.3 | VACCGGAAGTVV | FEV | 0.0095 | 6.29e-05 | 995 | 16.7 | 3910 | 14.5 | 1.15 |
| 118 | 40 | MA0506.2 (Nrf1) | STREME-5 | 5-GCATGCACAC | GCATGCACAC | Nrf1 | 0.0116 | 7.64e-05 | 18 | 0.3 | 18 | 0.07 | 4.52 |
| 119 | 32 | MA0014.3 | PAX5 | MA0014.3 | RRGCGTGACCNN | PAX5 | 0.02 | 0.000131 | 618 | 10.4 | 2340 | 8.66 | 1.2 |
| 120 | 19 | MA1484.1 | ETS2 | MA1484.1 | DACCGGAAGY | ETS2 | 0.0208 | 0.000134 | 264 | 4.43 | 894 | 3.31 | 1.34 |
| 121 | 7 | MA1656.1 | ZNF449 | MA1656.1 | NNAAGCCCAACCNN | ZNF449 | 0.0281 | 0.00018 | 4370 | 73.2 | 19000 | 70.6 | 1.04 |
| 122 | 41 | MA1951.1 | FOS | MA1951.1 | BGATGACGTCATCR | FOS | 0.032 | 0.000204 | 347 | 5.82 | 1230 | 4.57 | 1.28 |
| 123 | 19 | MA0475.2 | FLI1 | MA0475.2 | ACCGGAARTR | FLI1 | 0.0338 | 0.000213 | 158 | 2.65 | 492 | 1.82 | 1.46 |
| 124 | 37 | MA1495.1 | HOXA1 | MA1495.1 | STAATTAS | HOXA1 | 0.0419 | 0.000262 | 2900 | 48.7 | 12400 | 45.9 | 1.06 |
| 125 | 41 | MA0840.1 | Creb5 | MA0840.1 | NATGACGTCAYH | Creb5 | 0.0462 | 0.000286 | 56 | 0.94 | 131 | 0.49 | 1.95 |
| 126 | 36 | MA1584.1 | ZIC5 | MA1584.1 | VGACCCCCCGCTGHGM | ZIC5 | 0.0488 | 3e-04 | 1490 | 25 | 6110 | 22.6 | 1.1 |
| 14 | 42 | MA1107.2 (KLF9) | MEME-3 | TGTGTGTGYGTGTGT | TGTGTGTGYGTGTGT | KLF9 | 4.06 | 0.29 | 141 | 2.36 | 600 | 2.22 | 1.07 |
| 15 | 43 | MA1125.1 (ZNF384) | MEME-2 | AAAAWRAAAAAAAAA | AAAAWRAAAAAAAAA | ZNF384 | 15 | 1 | 0 | 0 | 8 | 0.03 | 0.502 |
ER_rat <- 1.25
MTX_242kb_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste("MTX_242kb_combo response peaks(200bp)Enrichment ratio:",ER_rat))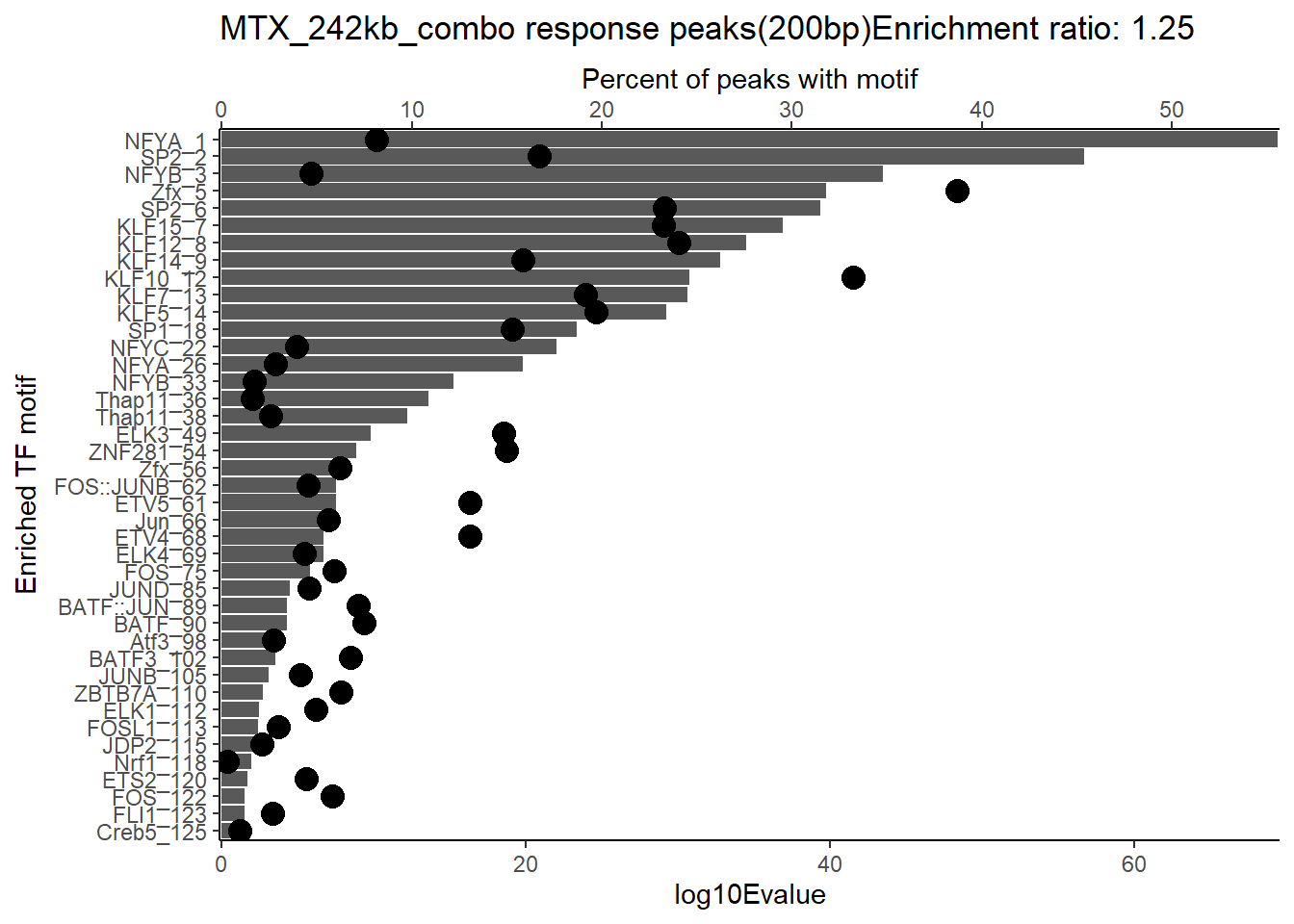
MTX_242kb_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y, TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
slice_head(n=5) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*4), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./4,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( "MTX_24 2kb response peaks(200bp)Enrichment ratio:",ER_rat))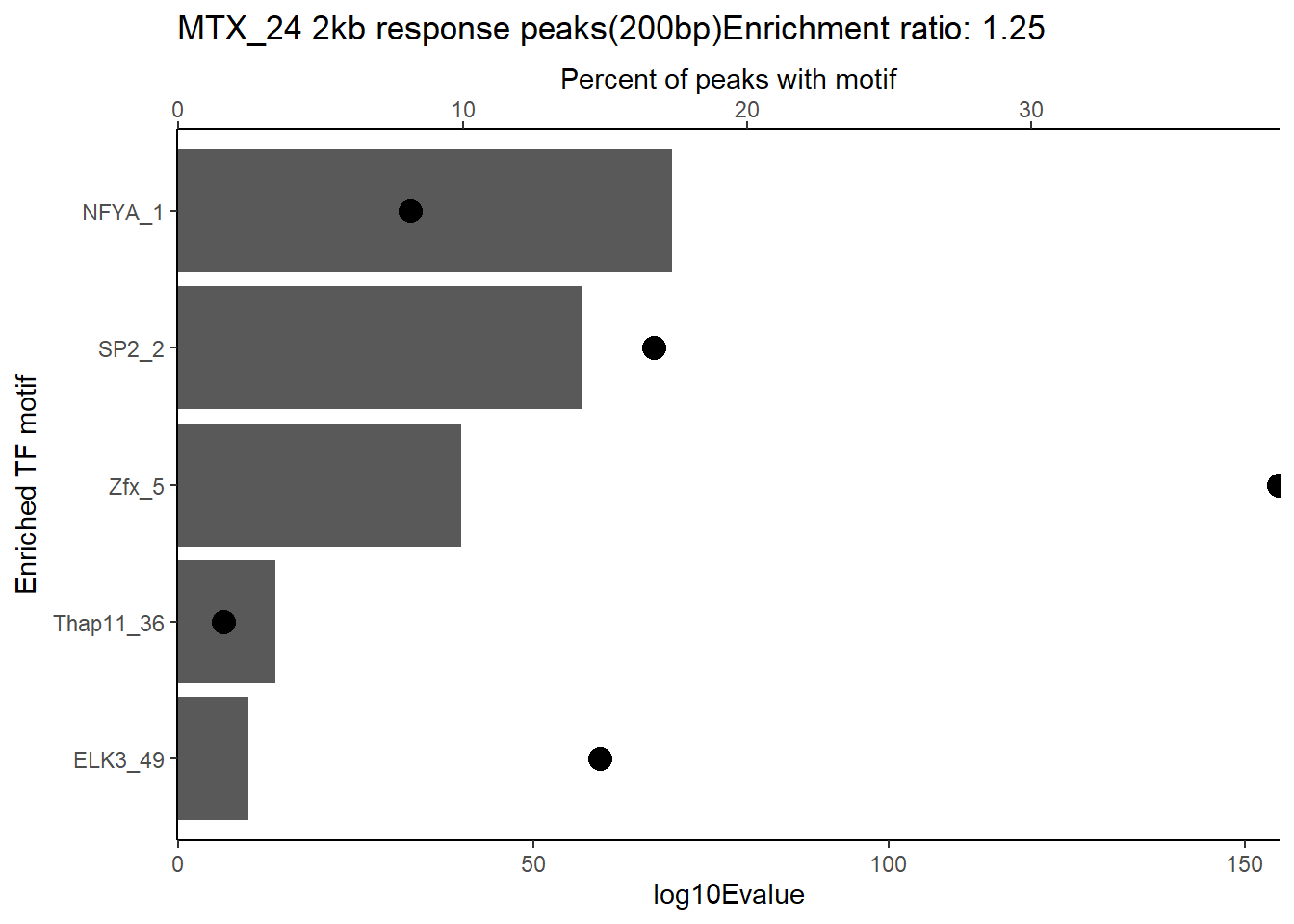
MTX_DAR 24 20kb data
MTX_24_20kb_sea_disc <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/MTX_24_20kb_xstreme/sea_disc_out/sea.tsv") %>% mutate(source="disc") %>% slice_head(n = length(.$ID)-3)
MTX_24_20kb_sea_known <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/MTX_24_20kb_xstreme/sea_out/sea.tsv") %>% mutate(source="known") %>% slice_head(n = length(.$ID)-3)
MTX_24_20kb_xstreme <- read.delim("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/MTX_24_20kb_xstreme/xstreme.tsv") %>% slice_head(n = length(.$ID)-3)
MTX_24_20kb_sea_all <- rbind(MTX_24_20kb_sea_disc, MTX_24_20kb_sea_known) %>%
arrange(ID, desc(source == "known")) %>%
distinct(ID, .keep_all = TRUE)MTX_24_20kb_combo <- MTX_24_20kb_xstreme %>%
dplyr::select(SEED_MOTIF:EVALUE_ACC,SIM_MOTIF) %>%
left_join(., MTX_24_20kb_sea_all, by=c("ID"="ID","ALT_ID"="ALT_ID")) %>%
mutate(motif_name=case_when(
str_detect(SIM_MOTIF, "\\(") ~ str_extract(SIM_MOTIF, "(?<=\\().+?(?=\\))"),
str_detect(SIM_MOTIF, "^MA\\d+\\.\\d+") ~ ALT_ID,
str_detect(SIM_MOTIF, "^\\d+-") ~ str_replace(SIM_MOTIF, "^\\d+-", ""),
TRUE ~ SIM_MOTIF
)) %>%
dplyr::select(RANK,CLUSTER,SIM_MOTIF,ALT_ID, ID,CONSENSUS.x,motif_name,EVALUE.y,QVALUE,TP:ENR_RATIO) %>%
arrange(.,EVALUE.y)
MTX_24_20kb_combo %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs 200 bp in MTX v VEH") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| RANK | CLUSTER | SIM_MOTIF | ALT_ID | ID | CONSENSUS.x | motif_name | EVALUE.y | QVALUE | TP | TP. | FP | FP. | ENR_RATIO |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | MA1513.1 (KLF15) | STREME-1 | 1-GCCCCGCCC | GCCCCGCCC | KLF15 | 8.57e-69 | 5.97e-69 | 1450 | 10.4 | 4390 | 6.02 | 1.73 |
| 2 | 2 | MA0060.3 (NFYA) | STREME-3 | 3-CYGATTGGYYRSN | CYGATTGGYYRSN | NFYA | 2.14e-56 | 7.46e-57 | 1030 | 7.45 | 2980 | 4.09 | 1.82 |
| 3 | 1 | MA0516.3 | SP2 | MA0516.3 | GGGGCGGGG | SP2 | 6.53e-55 | 1.52e-55 | 2080 | 15 | 7400 | 10.2 | 1.47 |
| 4 | 3 | 9-CCGVSBCGG | STREME-9 | 9-CCGVSBCGG | CCGVSBCGG | CCGVSBCGG | 1.53e-52 | 2.67e-53 | 2950 | 21.2 | 11400 | 15.7 | 1.35 |
| 5 | 4 | MA0079.5 (SP1) | MEME-3 | CSCCGCCSCSSCCSC | CSCCGCCSCSSCCSC | SP1 | 6.19e-52 | 8.63e-53 | 2720 | 19.6 | 10400 | 14.2 | 1.37 |
| 6 | 1 | MA1513.1 | KLF15 | MA1513.1 | SCCCCGCCCCS | KLF15 | 8.57e-51 | 9.96e-52 | 1630 | 11.7 | 5550 | 7.61 | 1.54 |
| 7 | 2 | MA0502.2 | NFYB | MA0502.2 | CYCATTGGCCVV | NFYB | 1.05e-49 | 1.04e-50 | 943 | 6.79 | 2740 | 3.76 | 1.81 |
| 8 | 1 | MA1564.1 | SP9 | MA1564.1 | RCCACGCCCMCY | SP9 | 1.39e-46 | 1.21e-47 | 3150 | 22.7 | 12600 | 17.3 | 1.31 |
| 9 | 1 | MA0685.2 | SP4 | MA0685.2 | GGGGCGGGG | SP4 | 1.83e-46 | 1.42e-47 | 1110 | 7.98 | 3470 | 4.76 | 1.68 |
| 10 | 1 | MA0746.2 | SP3 | MA0746.2 | NGCCACGCCCMCH | SP3 | 2.38e-46 | 1.66e-47 | 1860 | 13.4 | 6710 | 9.21 | 1.46 |
| 11 | 5 | MA0733.1 (EGR4) | STREME-4 | 4-CCGCCCGCG | CCGCCCGCG | EGR4 | 1.11e-45 | 7.01e-47 | 3160 | 22.8 | 12700 | 17.4 | 1.31 |
| 12 | 6 | MA0476.1 (FOS) | STREME-2 | 2-GATGASTCA | GATGASTCA | FOS | 8.95e-45 | 5.2e-46 | 1570 | 11.3 | 5480 | 7.52 | 1.51 |
| 13 | 6 | MA1128.1 | FOSL1::JUN | MA1128.1 | NKATGACTCATNN | FOSL1::JUN | 1.46e-44 | 7.83e-46 | 1820 | 13.1 | 6580 | 9.03 | 1.46 |
| 14 | 6 | MA0477.2 | FOSL1 | MA0477.2 | NNATGACTCATNN | FOSL1 | 4.16e-44 | 2.07e-45 | 1810 | 13 | 6530 | 8.96 | 1.46 |
| 15 | 6 | MA1928.1 | BNC2 | MA1928.1 | NDRTGAGTCABN | BNC2 | 1.52e-43 | 7.06e-45 | 2060 | 14.9 | 7680 | 10.6 | 1.41 |
| 16 | 1 | MA1961.1 | PATZ1 | MA1961.1 | SSGGGGMGGGGS | PATZ1 | 3.3e-43 | 1.44e-44 | 2020 | 14.6 | 7520 | 10.3 | 1.41 |
| 17 | 6 | MA0099.3 | FOS::JUN | MA0099.3 | ATGAGTCAYM | FOS::JUN | 5e-43 | 2.05e-44 | 1680 | 12.1 | 5990 | 8.22 | 1.47 |
| 18 | 6 | MA1633.2 | BACH1 | MA1633.2 | ATGACTCAT | BACH1 | 6.87e-42 | 2.66e-43 | 2650 | 19.1 | 10500 | 14.3 | 1.33 |
| 19 | 6 | MA1141.1 | FOS::JUND | MA1141.1 | NKATGAGTCATNN | FOS::JUND | 9.53e-42 | 3.5e-43 | 1710 | 12.3 | 6140 | 8.43 | 1.46 |
| 20 | 6 | MA1137.1 | FOSL1::JUNB | MA1137.1 | NDRTGACTCATNN | FOSL1::JUNB | 6.5e-41 | 2.27e-42 | 2740 | 19.7 | 10900 | 14.9 | 1.32 |
| 21 | 6 | MA1135.1 | FOSB::JUNB | MA1135.1 | KRTGASTCAT | FOSB::JUNB | 7.56e-41 | 2.51e-42 | 1760 | 12.7 | 6400 | 8.79 | 1.44 |
| 22 | 1 | MA0740.2 | KLF14 | MA0740.2 | KGGGCGGGG | KLF14 | 9.17e-40 | 2.91e-41 | 1180 | 8.52 | 3930 | 5.39 | 1.58 |
| 23 | 1 | MA1653.1 | ZNF148 | MA1653.1 | YBCCCCTCCCCC | ZNF148 | 1.22e-39 | 3.69e-41 | 3330 | 24 | 13700 | 18.8 | 1.27 |
| 24 | 1 | MA1959.1 | KLF7 | MA1959.1 | DGGGCGGGG | KLF7 | 1.78e-39 | 5.16e-41 | 1470 | 10.6 | 5150 | 7.07 | 1.49 |
| 25 | 1 | MA0742.2 | KLF12 | MA0742.2 | GGGGCGGGG | KLF12 | 3.3e-39 | 9.2e-41 | 986 | 7.1 | 3120 | 4.28 | 1.66 |
| 26 | 6 | MA0491.2 | JUND | MA0491.2 | NNATGACTCATNN | JUND | 3.59e-39 | 9.64e-41 | 2700 | 19.4 | 10800 | 14.8 | 1.32 |
| 27 | 6 | MA0462.2 | BATF::JUN | MA0462.2 | DATGACTCATH | BATF::JUN | 9.37e-39 | 2.42e-40 | 2170 | 15.6 | 8310 | 11.4 | 1.37 |
| 28 | 6 | MA1634.1 | BATF | MA1634.1 | DATGACTCATH | BATF | 3.32e-37 | 8.06e-39 | 2100 | 15.2 | 8080 | 11.1 | 1.37 |
| 29 | 6 | MA0478.1 | FOSL2 | MA0478.1 | KRRTGASTCAB | FOSL2 | 3.35e-37 | 8.06e-39 | 1610 | 11.6 | 5830 | 8.01 | 1.45 |
| 30 | 6 | MA1130.1 | FOSL2::JUN | MA1130.1 | NNRTGAGTCAYN | FOSL2::JUN | 5.03e-37 | 1.17e-38 | 1490 | 10.7 | 5340 | 7.32 | 1.47 |
| 31 | 6 | MA0835.2 | BATF3 | MA0835.2 | DATGACTCATH | BATF3 | 8.8e-37 | 1.98e-38 | 2070 | 14.9 | 7950 | 10.9 | 1.37 |
| 32 | 1 | MA0741.1 | KLF16 | MA0741.1 | GMCACGCCCCC | KLF16 | 2.5e-36 | 5.45e-38 | 2900 | 20.9 | 11800 | 16.2 | 1.29 |
| 33 | 1 | MA1511.2 | KLF10 | MA1511.2 | GGGGCGGGG | KLF10 | 3.84e-36 | 8.11e-38 | 1110 | 8 | 3720 | 5.1 | 1.57 |
| 34 | 7 | MA1522.1 | MAZ | MA1522.1 | BBCCCCTCCCY | MAZ | 2.06e-35 | 4.23e-37 | 3090 | 22.3 | 12800 | 17.6 | 1.27 |
| 35 | 6 | MA1132.1 | JUN::JUNB | MA1132.1 | KATGACKCAT | JUN::JUNB | 8.32e-35 | 1.66e-36 | 2560 | 18.4 | 10300 | 14.1 | 1.31 |
| 36 | 8 | MA1982.1 (ZNF574) | STREME-5 | 5-SGGCCS | SGGCCS | ZNF574 | 1.16e-34 | 2.24e-36 | 3100 | 22.3 | 12800 | 17.6 | 1.27 |
| 37 | 2 | MA0060.3 | NFYA | MA0060.3 | RRCCAATCAGM | NFYA | 4.31e-34 | 8.12e-36 | 490 | 3.53 | 1270 | 1.74 | 2.03 |
| 38 | 6 | MA0490.2 | JUNB | MA0490.2 | NNATGACTCATNN | JUNB | 4.4e-33 | 8.06e-35 | 2830 | 20.4 | 11600 | 16 | 1.28 |
| 39 | 1 | MA1512.1 | KLF11 | MA1512.1 | SCCACGCCCMC | KLF11 | 6.74e-33 | 1.21e-34 | 5160 | 37.1 | 23100 | 31.7 | 1.17 |
| 40 | 9 | MA1122.1 | TFDP1 | MA1122.1 | VSGCGGGAAVN | TFDP1 | 1.2e-32 | 2.09e-34 | 3400 | 24.5 | 14400 | 19.7 | 1.24 |
| 41 | 7 | MA1713.1 | ZNF610 | MA1713.1 | SSCGCCGCTCCSSS | ZNF610 | 1.81e-32 | 3.07e-34 | 2330 | 16.8 | 9300 | 12.8 | 1.31 |
| 42 | 6 | MA1101.2 | BACH2 | MA1101.2 | DWANCATGASTCATSNTWH | BACH2 | 3.56e-32 | 5.91e-34 | 2580 | 18.6 | 10500 | 14.4 | 1.29 |
| 43 | 1 | MA0079.5 | SP1 | MA0079.5 | GGGGCGGGG | SP1 | 5.09e-32 | 8.25e-34 | 692 | 4.98 | 2080 | 2.86 | 1.75 |
| 44 | 10 | MA0162.4 | EGR1 | MA0162.4 | VCMCGCCCACGCVS | EGR1 | 1.14e-31 | 1.8e-33 | 4140 | 29.8 | 18100 | 24.8 | 1.2 |
| 45 | 11 | 14-MGGSCCK | STREME-14 | 14-MGGSCCK | 14-MGGSCCK | MGGSCCK | 2.45e-31 | 3.8e-33 | 8030 | 57.8 | 38000 | 52.2 | 1.11 |
| 46 | 6 | MA0476.1 | FOS | MA0476.1 | DVTGASTCATB | FOS | 3.57e-31 | 5.41e-33 | 1110 | 7.99 | 3840 | 5.27 | 1.52 |
| 47 | 2 | MA1644.1 | NFYC | MA1644.1 | RDCCAATCASN | NFYC | 1.84e-30 | 2.73e-32 | 566 | 4.08 | 1620 | 2.22 | 1.84 |
| 48 | 1 | MA0493.2 | KLF1 | MA0493.2 | DGGGYGGGG | KLF1 | 3.49e-30 | 5.07e-32 | 1960 | 14.1 | 7700 | 10.6 | 1.34 |
| 49 | 12 | MA1712.1 | ZNF454 | MA1712.1 | DGGCTCSSGGCCCYGVKG | ZNF454 | 1.76e-29 | 2.5e-31 | 5370 | 38.7 | 24400 | 33.4 | 1.16 |
| 50 | 13 | MA0146.2 | Zfx | MA0146.2 | SSSGCCBVGGCCTS | Zfx | 1.84e-29 | 2.56e-31 | 6220 | 44.8 | 28700 | 39.4 | 1.14 |
| 51 | 10 | MA1107.2 | KLF9 | MA1107.2 | MNGCCACACCCACHYV | KLF9 | 1.01e-28 | 1.39e-30 | 5580 | 40.2 | 25500 | 34.9 | 1.15 |
| 52 | 14 | MA0506.2 | Nrf1 | MA0506.2 | SNCTGCGCMTGCGCV | Nrf1 | 1.28e-28 | 1.72e-30 | 4810 | 34.6 | 21600 | 29.6 | 1.17 |
| 53 | 1 | MA0039.4 | KLF4 | MA0039.4 | NNCCCCACCCHN | KLF4 | 1.09e-27 | 1.43e-29 | 2320 | 16.7 | 9450 | 13 | 1.29 |
| 54 | 2 | MA0502.2 (NFYB) | STREME-8 | 8-CCATTGGC | CCATTGGC | NFYB | 1.32e-27 | 1.71e-29 | 371 | 2.67 | 929 | 1.27 | 2.1 |
| 55 | 15 | MA0814.2 | TFAP2C | MA0814.2 | NNGCCTGAGGCGNN | TFAP2C | 6.84e-27 | 8.67e-29 | 5850 | 42.1 | 27000 | 37 | 1.14 |
| 56 | 1 | MA0599.1 | KLF5 | MA0599.1 | GCCCCDCCCH | KLF5 | 1.16e-26 | 1.44e-28 | 528 | 3.8 | 1540 | 2.11 | 1.81 |
| 57 | 6 | MA0489.2 | Jun | MA0489.2 | NVTGACTCATNN | Jun | 3.53e-26 | 4.31e-28 | 1020 | 7.35 | 3590 | 4.93 | 1.49 |
| 58 | 16 | MA1986.1 | ZNF692 | MA1986.1 | BGGGCCCASN | ZNF692 | 1.51e-24 | 1.81e-26 | 4380 | 31.6 | 19700 | 27 | 1.17 |
| 59 | 4 | MA1721.1 | ZNF93 | MA1721.1 | GGYAGCRGCAGCGGYG | ZNF93 | 2.01e-24 | 2.38e-26 | 2520 | 18.1 | 10500 | 14.5 | 1.25 |
| 60 | 10 | MA0733.1 | EGR4 | MA0733.1 | NHMCGCCCACGCAHWT | EGR4 | 2.4e-24 | 2.79e-26 | 3440 | 24.7 | 15000 | 20.6 | 1.2 |
| 61 | 1 | MA1517.1 | KLF6 | MA1517.1 | NRCCACGCCCH | KLF6 | 4.95e-24 | 5.66e-26 | 6640 | 47.8 | 31300 | 42.9 | 1.11 |
| 62 | 10 | MA0732.1 | EGR3 | MA0732.1 | HHMCGCCCACGCAHH | EGR3 | 9.87e-24 | 1.11e-25 | 2620 | 18.9 | 11100 | 15.2 | 1.24 |
| 63 | 17 | MA1599.1 | ZNF682 | MA1599.1 | BVGGCYAAGCCCCWNY | ZNF682 | 2.57e-23 | 2.85e-25 | 2580 | 18.6 | 10900 | 15 | 1.24 |
| 64 | 10 | MA0472.2 | EGR2 | MA0472.2 | MCGCCCACGCA | EGR2 | 2.67e-23 | 2.91e-25 | 2730 | 19.7 | 11700 | 16 | 1.23 |
| 65 | 6 | MA1142.1 | FOSL1::JUND | MA1142.1 | NRTGACTCAT | FOSL1::JUND | 7.05e-23 | 7.56e-25 | 1160 | 8.32 | 4290 | 5.88 | 1.41 |
| 66 | 18 | 13-CACCSCG | STREME-13 | 13-CACCSCG | 13-CACCSCG | CACCSCG | 1.27e-22 | 1.34e-24 | 1780 | 12.8 | 7130 | 9.78 | 1.31 |
| 67 | 19 | MA1548.1 | PLAGL2 | MA1548.1 | NGGGCCCCCN | PLAGL2 | 2.85e-22 | 2.96e-24 | 6880 | 49.5 | 32600 | 44.8 | 1.11 |
| 68 | 1 | MA1515.1 | KLF2 | MA1515.1 | NRCCACRCCCH | KLF2 | 6.95e-22 | 7.12e-24 | 1580 | 11.4 | 6270 | 8.61 | 1.33 |
| 69 | 10 | MA1650.1 | ZBTB14 | MA1650.1 | SSCCGCGCACVS | ZBTB14 | 1.59e-21 | 1.6e-23 | 2010 | 14.5 | 8290 | 11.4 | 1.27 |
| 70 | 20 | MA0748.2 | YY2 | MA0748.2 | NVATGGCGGCS | YY2 | 2.04e-20 | 2.03e-22 | 1200 | 8.67 | 4590 | 6.3 | 1.38 |
| 71 | 1 | MA0747.1 | SP8 | MA0747.1 | RCCACGCCCMCY | SP8 | 3.93e-20 | 3.86e-22 | 1500 | 10.8 | 5940 | 8.15 | 1.32 |
| 72 | 9 | MA0865.2 | E2F8 | MA0865.2 | TTCCCGCCAHWA | E2F8 | 1.67e-19 | 1.62e-21 | 4420 | 31.9 | 20200 | 27.8 | 1.15 |
| 73 | 21 | MA1976.1 | ZNF320 | MA1976.1 | DGKGGGRCCMRGGGGCCWGTGHVNNNN | ZNF320 | 1.92e-19 | 1.84e-21 | 3690 | 26.6 | 16600 | 22.7 | 1.17 |
| 74 | 1 | MA0753.2 | ZNF740 | MA0753.2 | CYRCCCCCCCCAC | ZNF740 | 6.54e-19 | 6.16e-21 | 5250 | 37.8 | 24500 | 33.6 | 1.13 |
| 75 | 22 | MA0774.1 (MEIS2) | STREME-12 | 12-CCTGTCAGC | 12-CCTGTCAGC | MEIS2 | 1.01e-18 | 9.42e-21 | 1340 | 9.62 | 5260 | 7.22 | 1.33 |
| 76 | 23 | MA0632.2 | TCFL5 | MA0632.2 | KCACGCGCMC | TCFL5 | 1.04e-18 | 9.5e-21 | 973 | 7.01 | 3620 | 4.96 | 1.41 |
| 77 | 6 | MA1144.1 | FOSL2::JUND | MA1144.1 | KATGACTCAT | FOSL2::JUND | 5.78e-18 | 5.23e-20 | 743 | 5.35 | 2630 | 3.61 | 1.48 |
| 78 | 6 | MA0841.1 | NFE2 | MA0841.1 | VATGACTCATS | NFE2 | 9.31e-18 | 8.32e-20 | 763 | 5.49 | 2720 | 3.74 | 1.47 |
| 79 | 24 | MA0014.3 | PAX5 | MA0014.3 | RRGCGTGACCNN | PAX5 | 8.3e-17 | 7.32e-19 | 2650 | 19.1 | 11600 | 15.9 | 1.2 |
| 80 | 9 | MA0471.2 | E2F6 | MA0471.2 | VVDGGCGGGAAVV | E2F6 | 8.52e-17 | 7.42e-19 | 3190 | 23 | 14300 | 19.6 | 1.17 |
| 81 | 25 | MA1573.2 (Thap11) | STREME-6 | 6-ACTACAACTCCC | ACTACAACTCCC | Thap11 | 9.98e-17 | 8.59e-19 | 53 | 0.38 | 39 | 0.05 | 7.08 |
| 82 | 6 | MA1138.1 | FOSL2::JUNB | MA1138.1 | KRTGASTCAT | FOSL2::JUNB | 3.59e-16 | 3.05e-18 | 700 | 5.04 | 2500 | 3.43 | 1.47 |
| 83 | 6 | MA0496.3 | MAFK | MA0496.3 | NGMTGACTCAGCMNH | MAFK | 4.56e-16 | 3.83e-18 | 2250 | 16.2 | 9710 | 13.3 | 1.22 |
| 84 | 6 | MA0655.1 | JDP2 | MA0655.1 | ATGACTCAT | JDP2 | 4.81e-16 | 3.99e-18 | 720 | 5.19 | 2590 | 3.55 | 1.46 |
| 85 | 6 | MA0089.2 | MAFG::NFE2L1 | MA0089.2 | NVNATGACTCAGCADW | MAFG::NFE2L1 | 5.45e-16 | 4.47e-18 | 2100 | 15.2 | 9020 | 12.4 | 1.23 |
| 86 | 26 | MA1985.1 | ZNF669 | MA1985.1 | GGGGYGAYGACCRCT | ZNF669 | 5.6e-16 | 4.54e-18 | 5410 | 39 | 25500 | 35 | 1.11 |
| 87 | 27 | MA1615.1 | Plagl1 | MA1615.1 | NNCTGGGGCCABN | Plagl1 | 1.14e-15 | 9.15e-18 | 7150 | 51.5 | 34600 | 47.5 | 1.09 |
| 88 | 28 | MA0131.2 | HINFP | MA0131.2 | CARCGTCCGCKN | HINFP | 7.64e-15 | 6.05e-17 | 2000 | 14.4 | 8570 | 11.8 | 1.22 |
| 89 | 29 | MA0810.1 | TFAP2A | MA0810.1 | YGCCCBVRGGCR | TFAP2A | 1.48e-14 | 1.16e-16 | 6540 | 47.1 | 31500 | 43.2 | 1.09 |
| 90 | 29 | MA0811.1 | TFAP2B | MA0811.1 | YGCCCBVRGGCA | TFAP2B | 2.83e-14 | 2.19e-16 | 6440 | 46.3 | 31000 | 42.5 | 1.09 |
| 91 | 23 | MA1099.2 | HES1 | MA1099.2 | GGCRCGTGBC | HES1 | 8.93e-14 | 6.84e-16 | 1050 | 7.56 | 4150 | 5.7 | 1.33 |
| 92 | 29 | MA0872.1 | TFAP2A | MA0872.1 | TGCCCYSRGGGCA | TFAP2A | 2.79e-13 | 2.11e-15 | 3770 | 27.2 | 17400 | 23.9 | 1.14 |
| 93 | 1 | MA1630.2 | ZNF281 | MA1630.2 | GGGGGAGGGGVV | ZNF281 | 2.81e-13 | 2.11e-15 | 713 | 5.13 | 2650 | 3.63 | 1.41 |
| 94 | 29 | MA0524.2 | TFAP2C | MA0524.2 | YGCCYBVRGGCA | TFAP2C | 3.31e-13 | 2.45e-15 | 6060 | 43.7 | 29100 | 40 | 1.09 |
| 95 | 30 | MA0464.2 | BHLHE40 | MA0464.2 | VKCACGTGMC | BHLHE40 | 8.66e-13 | 6.36e-15 | 5240 | 37.7 | 24900 | 34.2 | 1.1 |
| 96 | 25 | MA1573.2 | Thap11 | MA1573.2 | ACTACAABTCCCAG | Thap11 | 3.46e-12 | 2.52e-14 | 196 | 1.41 | 516 | 0.71 | 2 |
| 97 | 6 | MA0501.1 | MAF::NFE2 | MA0501.1 | ATGACTCAGCANWTY | MAF::NFE2 | 4.34e-12 | 3.12e-14 | 1750 | 12.6 | 7520 | 10.3 | 1.22 |
| 98 | 31 | 10-ACATGCACACACGT | STREME-10 | 10-ACATGCACACACGT | 10-ACATGCACACACGT | ACATGCACACACGT | 4.92e-12 | 3.5e-14 | 102 | 0.73 | 194 | 0.27 | 2.77 |
| 99 | 32 | MA1596.1 | ZNF460 | MA1596.1 | GCCTCMGCCTCCCRAG | ZNF460 | 7.58e-12 | 5.34e-14 | 3450 | 24.9 | 15900 | 21.9 | 1.14 |
| 100 | 6 | MA1134.1 | FOS::JUNB | MA1134.1 | KATGASTCATHN | FOS::JUNB | 8.03e-12 | 5.6e-14 | 483 | 3.48 | 1690 | 2.32 | 1.5 |
| 101 | 29 | MA0813.1 | TFAP2B | MA0813.1 | TGCCCYBRGGGCA | TFAP2B | 8.33e-12 | 5.75e-14 | 3280 | 23.6 | 15100 | 20.7 | 1.14 |
| 102 | 33 | MA0527.1 | ZBTB33 | MA0527.1 | BTCTCGCGAGABYTS | ZBTB33 | 1e-11 | 6.84e-14 | 832 | 5.99 | 3240 | 4.45 | 1.35 |
| 103 | 34 | MA1108.2 (MXI1) | STREME-11 | 11-ACACACATGTGCA | 11-ACACACATGTGCA | MXI1 | 1.48e-11 | 1e-13 | 80 | 0.58 | 132 | 0.18 | 3.2 |
| 104 | 35 | MA1643.1 | NFIB | MA1643.1 | NNCCTGGCAYNGTGCCAAGNN | NFIB | 2.69e-11 | 1.81e-13 | 4960 | 35.7 | 23600 | 32.4 | 1.1 |
| 105 | 6 | MA1988.1 | Atf3 | MA1988.1 | NRTGACTCABN | Atf3 | 4.23e-11 | 2.81e-13 | 495 | 3.56 | 1760 | 2.42 | 1.48 |
| 106 | 10 | MA1627.1 | Wt1 | MA1627.1 | YBCCTCCCCCACVB | Wt1 | 5.31e-11 | 3.49e-13 | 12100 | 87.2 | 61800 | 84.8 | 1.03 |
| 107 | 6 | MA0659.3 | Mafg | MA0659.3 | NWGMTGACTCAGCAN | Mafg | 1.66e-10 | 1.08e-12 | 4810 | 34.6 | 22900 | 31.5 | 1.1 |
| 108 | 29 | MA0815.1 | TFAP2C | MA0815.1 | TGCCCYSRGGGCA | TFAP2C | 9.26e-10 | 5.98e-12 | 1510 | 10.9 | 6520 | 8.95 | 1.22 |
| 109 | 15 | MA0812.1 | TFAP2B | MA0812.1 | HGCCTSAGGCD | TFAP2B | 1.25e-09 | 8.02e-12 | 8700 | 62.7 | 43400 | 59.5 | 1.05 |
| 110 | 36 | MA1514.1 | KLF17 | MA1514.1 | MMCCACGCACCCMTY | KLF17 | 1.61e-09 | 1.02e-11 | 4900 | 35.3 | 23500 | 32.2 | 1.09 |
| 111 | 30 | MA0616.2 | HES2 | MA0616.2 | GGCACGTGYC | HES2 | 2.27e-09 | 1.42e-11 | 1170 | 8.43 | 4920 | 6.75 | 1.25 |
| 112 | 15 | MA0003.4 | TFAP2A | MA0003.4 | NNKGCCTCAGGCNN | TFAP2A | 2.68e-09 | 1.67e-11 | 11800 | 85.2 | 60400 | 82.9 | 1.03 |
| 113 | 6 | MA1114.1 | PBX3 | MA1114.1 | NNNTGAGTGACAGGBNN | PBX3 | 2.71e-09 | 1.67e-11 | 9400 | 67.7 | 47100 | 64.7 | 1.05 |
| 114 | 30 | MA0608.1 | Creb3l2 | MA0608.1 | GCCACGTGD | Creb3l2 | 2.99e-09 | 1.83e-11 | 4460 | 32.2 | 21300 | 29.2 | 1.1 |
| 115 | 30 | MA1631.1 | ASCL1 | MA1631.1 | NNGCACCTGCYNB | ASCL1 | 3.33e-09 | 2.02e-11 | 11400 | 82.3 | 58200 | 79.8 | 1.03 |
| 116 | 31 | MA1155.1 (ZSCAN4) | STREME-7 | 7-GTGTGTGCATGYGTG | GTGTGTGCATGYGTG | ZSCAN4 | 3.5e-09 | 2.1e-11 | 96 | 0.69 | 203 | 0.28 | 2.49 |
| 117 | 37 | MA0734.3 | Gli2 | MA0734.3 | NRGACCACCCASV | Gli2 | 8.06e-09 | 4.8e-11 | 9050 | 65.2 | 45300 | 62.2 | 1.05 |
| 118 | 10 | MA1656.1 | ZNF449 | MA1656.1 | NNAAGCCCAACCNN | ZNF449 | 1.7e-08 | 1e-10 | 7680 | 55.3 | 38100 | 52.3 | 1.06 |
| 119 | 27 | MA1719.1 | ZNF816 | MA1719.1 | DRRKGGGGACATGCHGGG | ZNF816 | 3.43e-08 | 2.01e-10 | 8540 | 61.5 | 42700 | 58.6 | 1.05 |
| 120 | 15 | MA1569.1 | TFAP2E | MA1569.1 | YGCCYSRGGCD | TFAP2E | 3.83e-08 | 2.22e-10 | 1020 | 7.37 | 4300 | 5.9 | 1.25 |
| 121 | 30 | MA0830.2 | TCF4 | MA0830.2 | NNGCACCTGCCNN | TCF4 | 4.89e-08 | 2.82e-10 | 10700 | 76.8 | 54100 | 74.2 | 1.03 |
| 122 | 38 | MA1557.1 | SMAD5 | MA1557.1 | YGTCTAGACA | SMAD5 | 5.21e-08 | 2.98e-10 | 4150 | 29.9 | 19800 | 27.2 | 1.1 |
| 123 | 22 | MA0774.1 | MEIS2 | MA0774.1 | TTGACAGS | MEIS2 | 7.56e-08 | 4.29e-10 | 2670 | 19.2 | 12400 | 17 | 1.13 |
| 124 | 39 | MA0864.2 | E2F2 | MA0864.2 | RWTTTGGCGCCAWWWY | E2F2 | 8.03e-08 | 4.51e-10 | 4360 | 31.4 | 20900 | 28.7 | 1.09 |
| 125 | 40 | MA1966.1 | TFAP4::ETV1 | MA1966.1 | RSCGGAAGCAGSTGKN | TFAP4::ETV1 | 9.07e-08 | 5.06e-10 | 6280 | 45.2 | 30800 | 42.3 | 1.07 |
| 126 | 40 | MA0076.2 | ELK4 | MA0076.2 | BCRCTTCCGGB | ELK4 | 9.93e-08 | 5.49e-10 | 367 | 2.64 | 1310 | 1.8 | 1.47 |
| 127 | 7 | MA0528.2 | ZNF263 | MA0528.2 | VNGGGGAGGASG | ZNF263 | 5.17e-07 | 2.84e-09 | 7320 | 52.8 | 36400 | 49.9 | 1.06 |
| 128 | 30 | MA0649.1 | HEY2 | MA0649.1 | GRCACGTGYC | HEY2 | 5.95e-07 | 3.24e-09 | 3550 | 25.6 | 16900 | 23.2 | 1.1 |
| 129 | 30 | MA0147.3 | MYC | MA0147.3 | NNCCACGTGCNB | MYC | 6.28e-07 | 3.4e-09 | 11100 | 80.1 | 56700 | 77.8 | 1.03 |
| 130 | 24 | MA0067.2 | PAX2 | MA0067.2 | BCAVTSRAGCGTGACGR | PAX2 | 7.94e-07 | 4.26e-09 | 4330 | 31.2 | 20900 | 28.7 | 1.09 |
| 131 | 41 | MA0767.1 | GCM2 | MA0767.1 | BATGCGGGTR | GCM2 | 8.71e-07 | 4.6e-09 | 5700 | 41 | 27900 | 38.3 | 1.07 |
| 132 | 39 | MA1727.1 | ZNF417 | MA1727.1 | VRBVNTGGGCGCCAM | ZNF417 | 8.71e-07 | 4.6e-09 | 3370 | 24.3 | 16000 | 21.9 | 1.11 |
| 133 | 30 | MA0821.2 | HES5 | MA0821.2 | GRCACGTGYC | HES5 | 9.4e-07 | 4.93e-09 | 722 | 5.2 | 2950 | 4.05 | 1.29 |
| 134 | 40 | MA0764.3 | ETV4 | MA0764.3 | ACCGGAAGTR | ETV4 | 1.06e-06 | 5.53e-09 | 246 | 1.77 | 822 | 1.13 | 1.57 |
| 135 | 30 | MA0823.1 | HEY1 | MA0823.1 | GRCACGTGCC | HEY1 | 1.37e-06 | 7.08e-09 | 1290 | 9.28 | 5650 | 7.76 | 1.2 |
| 136 | 30 | MA0603.1 | Arntl | MA0603.1 | NGTCACGTGH | Arntl | 1.53e-06 | 7.85e-09 | 10500 | 75.8 | 53500 | 73.4 | 1.03 |
| 137 | 42 | MA1964.1 | SMAD2 | MA1964.1 | NBCCAGACDN | SMAD2 | 1.57e-06 | 8e-09 | 9780 | 70.4 | 49500 | 67.9 | 1.04 |
| 138 | 30 | MA1106.1 | HIF1A | MA1106.1 | SBACGTGCNN | HIF1A | 4.05e-06 | 2.05e-08 | 3140 | 22.6 | 14900 | 20.4 | 1.11 |
| 139 | 30 | MA1464.1 | ARNT2 | MA1464.1 | GTCACGTGMN | ARNT2 | 5.25e-06 | 2.63e-08 | 3400 | 24.5 | 16200 | 22.3 | 1.1 |
| 140 | 43 | MA1684.1 | Foxn1 | MA1684.1 | RGAMGC | Foxn1 | 5.72e-06 | 2.85e-08 | 2230 | 16 | 10300 | 14.2 | 1.13 |
| 141 | 30 | MA1474.1 | CREB3L4 | MA1474.1 | YGCCACGTCAYC | CREB3L4 | 5.99e-06 | 2.96e-08 | 9250 | 66.6 | 46700 | 64.1 | 1.04 |
| 142 | 7 | MA1578.1 | VEZF1 | MA1578.1 | CCCCCCMYDH | VEZF1 | 8.96e-06 | 4.4e-08 | 1960 | 14.2 | 9030 | 12.4 | 1.14 |
| 143 | 44 | MA0028.2 | ELK1 | MA0028.2 | ACCGGAAGTR | ELK1 | 1.13e-05 | 5.51e-08 | 182 | 1.31 | 582 | 0.8 | 1.65 |
| 144 | 39 | MA0024.3 | E2F1 | MA0024.3 | TTTGGCGCCAAA | E2F1 | 1.15e-05 | 5.57e-08 | 3240 | 23.3 | 15400 | 21.2 | 1.1 |
| 145 | 6 | MA1521.1 | MAFA | MA1521.1 | NTGCTGASTCAGCAN | MAFA | 1.17e-05 | 5.6e-08 | 4410 | 31.7 | 21400 | 29.4 | 1.08 |
| 146 | 24 | MA1995.1 | Npas4 | MA1995.1 | NNTCGTGACHN | Npas4 | 2.58e-05 | 1.23e-07 | 4500 | 32.4 | 21900 | 30 | 1.08 |
| 147 | 44 | MA0765.3 | ETV5 | MA0765.3 | ACCGGAAGTR | ETV5 | 2.96e-05 | 1.4e-07 | 184 | 1.33 | 600 | 0.82 | 1.62 |
| 148 | 45 | MA1710.1 | ZNF257 | MA1710.1 | VDGAGGCRAGRG | ZNF257 | 4.4e-05 | 2.07e-07 | 4500 | 32.4 | 21900 | 30.1 | 1.08 |
| 149 | 46 | MA0597.2 | THAP1 | MA0597.2 | VSGCAGGGCASV | THAP1 | 5.45e-05 | 2.55e-07 | 10100 | 72.9 | 51500 | 70.7 | 1.03 |
| 150 | 39 | MA0470.2 | E2F4 | MA0470.2 | TTTTGGCGCCAWWW | E2F4 | 9.12e-05 | 4.24e-07 | 1950 | 14 | 9050 | 12.4 | 1.13 |
| 151 | 1 | MA1516.1 | KLF3 | MA1516.1 | GRCCRCGCCCH | KLF3 | 9.28e-05 | 4.29e-07 | 144 | 1.04 | 448 | 0.61 | 1.69 |
| 152 | 30 | MA0822.1 | HES7 | MA0822.1 | YGGCACGTGCCR | HES7 | 0.000115 | 5.28e-07 | 793 | 5.71 | 3400 | 4.66 | 1.23 |
| 153 | 40 | MA0633.2 | Twist2 | MA0633.2 | NVCAGCTGBN | Twist2 | 0.000141 | 6.41e-07 | 4020 | 28.9 | 19500 | 26.8 | 1.08 |
| 154 | 44 | MA0759.2 | ELK3 | MA0759.2 | ACCGGAAGTRV | ELK3 | 0.000166 | 7.51e-07 | 186 | 1.34 | 627 | 0.86 | 1.56 |
| 155 | 40 | MA1997.1 | Olig2 | MA1997.1 | NVCAGCTGBN | Olig2 | 0.00023 | 1.03e-06 | 7730 | 55.7 | 38900 | 53.4 | 1.04 |
| 156 | 30 | MA0844.1 | XBP1 | MA0844.1 | WDKGMCACGTCATC | XBP1 | 0.000261 | 1.17e-06 | 4620 | 33.2 | 22700 | 31.1 | 1.07 |
| 157 | 30 | MA1934.1 | ERF::FIGLA | MA1934.1 | RCCGGAARCASCTGBN | ERF::FIGLA | 0.000309 | 1.37e-06 | 2770 | 20 | 13200 | 18.2 | 1.1 |
| 158 | 37 | MA1990.1 | Gli1 | MA1990.1 | NNRGACCACCCASV | Gli1 | 0.000352 | 1.55e-06 | 5140 | 37 | 25400 | 34.8 | 1.06 |
| 159 | 47 | MA1982.1 | ZNF574 | MA1982.1 | VGSCTAGAGMGGCCS | ZNF574 | 0.000374 | 1.64e-06 | 7150 | 51.5 | 35800 | 49.2 | 1.05 |
| 160 | 40 | MA1941.1 | ETV2::FIGLA | MA1941.1 | RSCGGAARCAGSTGBN | ETV2::FIGLA | 0.000419 | 1.82e-06 | 5860 | 42.2 | 29100 | 40 | 1.06 |
| 161 | 48 | MA1730.1 | ZNF708 | MA1730.1 | NNRGCTGTRCCTCCY | ZNF708 | 0.000426 | 1.85e-06 | 7290 | 52.5 | 36600 | 50.2 | 1.05 |
| 162 | 22 | MA0775.1 | MEIS3 | MA0775.1 | DTGACAGS | MEIS3 | 0.000489 | 2.1e-06 | 6010 | 43.3 | 29900 | 41.1 | 1.05 |
| 163 | 30 | MA0692.1 | TFEB | MA0692.1 | RYCACGTGAC | TFEB | 0.000506 | 2.16e-06 | 11700 | 84.2 | 60100 | 82.5 | 1.02 |
| 164 | 6 | MA0782.2 | PKNOX1 | MA0782.2 | NNTGAGTGACAGVNN | PKNOX1 | 0.000525 | 2.23e-06 | 9390 | 67.6 | 47700 | 65.5 | 1.03 |
| 165 | 39 | MA0615.1 | Gmeb1 | MA0615.1 | BHBBKKACGTMMNWNNN | Gmeb1 | 0.000681 | 2.88e-06 | 635 | 4.57 | 2690 | 3.7 | 1.24 |
| 166 | 22 | MA0498.2 | MEIS1 | MA0498.2 | HTGACAD | MEIS1 | 0.000717 | 3.01e-06 | 7520 | 54.1 | 37800 | 51.9 | 1.04 |
| 167 | 30 | MA1493.1 | HES6 | MA1493.1 | GGCACGTGTY | HES6 | 0.000745 | 3.11e-06 | 3580 | 25.8 | 17400 | 23.9 | 1.08 |
| 168 | 30 | MA1560.1 | SOHLH2 | MA1560.1 | YGCACGTGCN | SOHLH2 | 0.000797 | 3.31e-06 | 269 | 1.94 | 1010 | 1.38 | 1.41 |
| 169 | 30 | MA0004.1 | Arnt | MA0004.1 | CACGTG | Arnt | 0.000897 | 3.7e-06 | 2180 | 15.7 | 10300 | 14.2 | 1.11 |
| 170 | 27 | MA1102.2 | CTCFL | MA1102.2 | NSCAGGGGGCGS | CTCFL | 0.00121 | 4.98e-06 | 6740 | 48.5 | 33800 | 46.4 | 1.05 |
| 171 | 30 | MA0499.2 | MYOD1 | MA0499.2 | NNGCACCTGTCNB | MYOD1 | 0.00126 | 5.15e-06 | 11300 | 81.6 | 58200 | 79.9 | 1.02 |
| 172 | 37 | MA0695.1 | ZBTB7C | MA0695.1 | RCGACCACCGAN | ZBTB7C | 0.00181 | 7.32e-06 | 1550 | 11.2 | 7180 | 9.85 | 1.13 |
| 173 | 44 | MA1933.1 | ELK1::SREBF2 | MA1933.1 | DCCGGAAGTSRCGTGA | ELK1::SREBF2 | 0.00246 | 9.92e-06 | 2010 | 14.5 | 9510 | 13 | 1.11 |
| 174 | 40 | MA0763.1 | ETV3 | MA0763.1 | ACCGGAAGTR | ETV3 | 0.00277 | 1.11e-05 | 239 | 1.72 | 893 | 1.23 | 1.41 |
| 175 | 44 | MA0156.3 | FEV | MA0156.3 | VACCGGAAGTVV | FEV | 0.00285 | 1.13e-05 | 238 | 1.71 | 889 | 1.22 | 1.41 |
| 176 | 49 | MA0155.1 | INSM1 | MA0155.1 | TGYCWGGGGGCR | INSM1 | 0.00321 | 1.27e-05 | 2430 | 17.5 | 11600 | 15.9 | 1.1 |
| 177 | 37 | MA0694.1 | ZBTB7B | MA0694.1 | RCGACCACCGAA | ZBTB7B | 0.00518 | 2.04e-05 | 3320 | 23.9 | 16200 | 22.2 | 1.08 |
| 178 | 21 | MA1723.1 | PRDM9 | MA1723.1 | RGDGGGVAGGGRGGVRRMARVARR | PRDM9 | 0.00551 | 2.16e-05 | 3830 | 27.6 | 18800 | 25.8 | 1.07 |
| 179 | 50 | MA1981.1 | ZNF530 | MA1981.1 | GMARGGMRAGGGGCRGSV | ZNF530 | 0.00557 | 2.17e-05 | 393 | 2.83 | 1610 | 2.2 | 1.29 |
| 180 | 21 | MA0163.1 | PLAG1 | MA0163.1 | GGGGCCCWAGGGGG | PLAG1 | 0.00771 | 2.99e-05 | 1360 | 9.82 | 6320 | 8.68 | 1.13 |
| 181 | 51 | MA1584.1 | ZIC5 | MA1584.1 | VGACCCCCCGCTGHGM | ZIC5 | 0.00788 | 3.03e-05 | 1420 | 10.2 | 6600 | 9.05 | 1.13 |
| 182 | 40 | MA0645.1 | ETV6 | MA0645.1 | MSCGGAAGTR | ETV6 | 0.0118 | 4.54e-05 | 151 | 1.09 | 529 | 0.73 | 1.5 |
| 183 | 15 | MA1581.1 | ZBTB6 | MA1581.1 | DNSCTTGAGCCNV | ZBTB6 | 0.0122 | 4.64e-05 | 4470 | 32.2 | 22100 | 30.4 | 1.06 |
| 184 | 40 | MA1484.1 | ETS2 | MA1484.1 | DACCGGAAGY | ETS2 | 0.0137 | 5.18e-05 | 215 | 1.55 | 811 | 1.11 | 1.4 |
| 185 | 30 | MA0058.3 | MAX | MA0058.3 | AVCACGTGNY | MAX | 0.0193 | 7.29e-05 | 8610 | 62 | 43800 | 60.1 | 1.03 |
| 186 | 52 | MA0863.1 | MTF1 | MA0863.1 | TTTGCACACGGCAC | MTF1 | 0.024 | 9.01e-05 | 1540 | 11 | 7220 | 9.91 | 1.12 |
| 187 | 15 | MA1629.1 | Zic2 | MA1629.1 | NDCACAGCAGGDRG | Zic2 | 0.0279 | 0.000104 | 12300 | 88.5 | 63600 | 87.3 | 1.01 |
| 188 | 30 | MA0103.3 | ZEB1 | MA0103.3 | SNCACCTGSVN | ZEB1 | 0.0279 | 0.000104 | 12500 | 90.3 | 65000 | 89.2 | 1.01 |
| 16 | 6 | MA1633.2 (BACH1) | MEME-2 | TGRCTCAYGCCTGTA | TGRCTCAYGCCTGTA | BACH1 | 15.4 | 0.961 | 168 | 1.21 | 959 | 1.32 | 0.924 |
| 17 | 53 | MA1125.1 (ZNF384) | MEME-1 | WAAAAAAAAAAAAAA | WAAAAAAAAAAAAAA | ZNF384 | 16.9 | 0.995 | 22 | 0.16 | 159 | 0.22 | 0.754 |
| 18 | 54 | MA0114.4 (HNF4A) | MEME-4 | CCAAAGTGCTGGGAT | CCAAAGTGCTGGGAT | HNF4A | 18 | 1 | 542 | 3.9 | 3360 | 4.62 | 0.846 |
ER_rat <- 1.25
MTX_24_20kb_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,TP.)%>%
arrange(.,EVALUE.y) %>%
slice_head(n=80)%>%
mutate(log10Evalue= log(EVALUE.y, base = 10)*(-1)) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste("MTX_24_20kb_combo response peaks(200bp)Enrichment ratio:",ER_rat))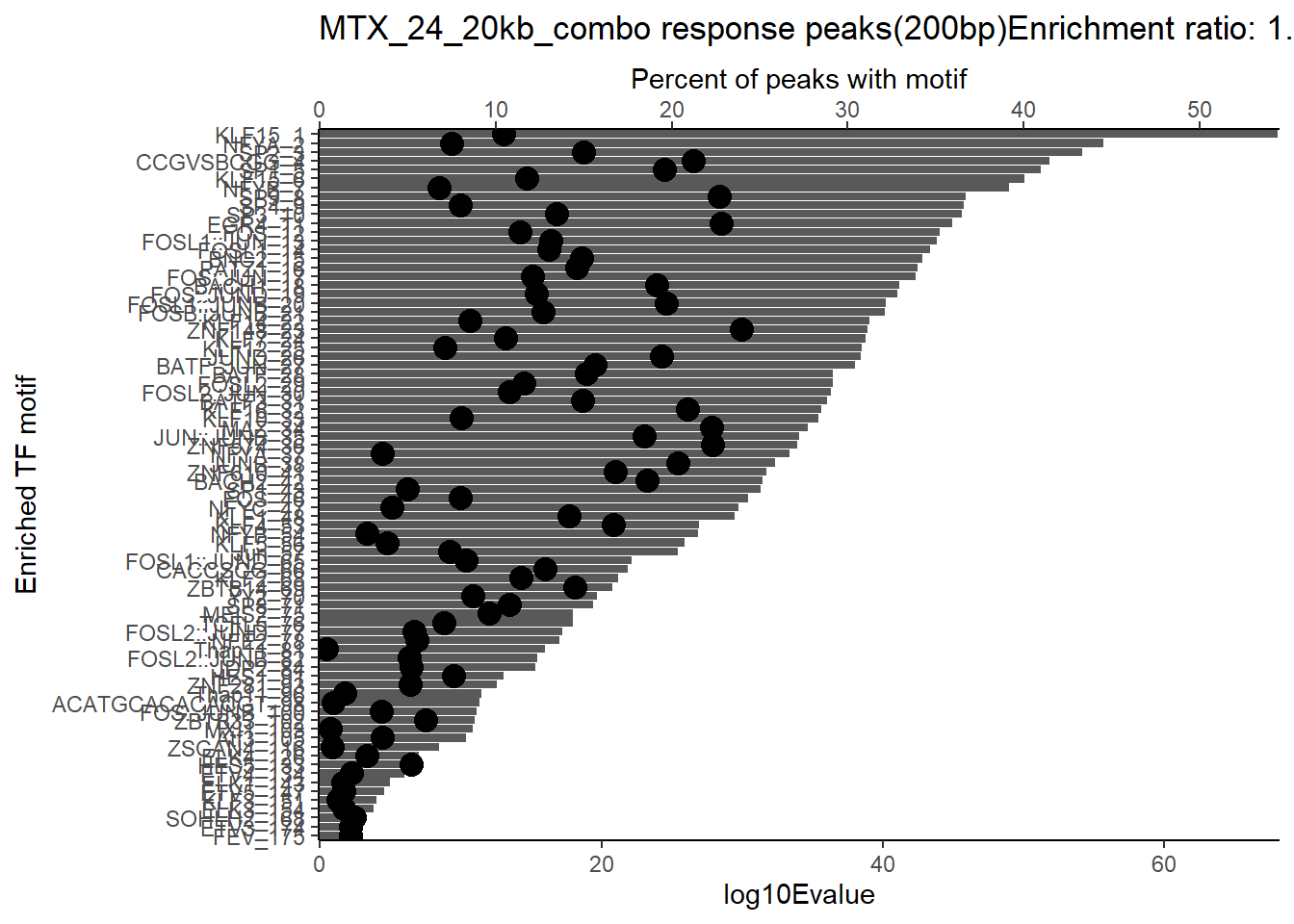
MTX_24_20kb_combo %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER,motif_name,SIM_MOTIF,ALT_ID, ID,EVALUE.y,TP.)%>%
arrange(.,EVALUE.y) %>%
mutate(log10Evalue = -log10(if_else(EVALUE.y == 0, 1e-300, EVALUE.y))) %>%
distinct(CLUSTER,.keep_all = TRUE) %>%
slice_head(n=5) %>%
mutate(motif_name=if_else(str_starts(motif_name,"^Z*"),paste(motif_name, RANK, sep="_"), if_else(motif_name=="KLF9",paste(motif_name, RANK,sep="_"),motif_name)))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue)) +
geom_point(aes(x=`TP.`*9), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./9,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste("MTX_24_20kb_combo response peaks(200bp)Enrichment ratio:",ER_rat))
meme_motifs_EAR_open <- read_meme("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/EAR_open_xstreme/xstreme.txt")
meme_motifs_ESR_open <- read_meme("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/ESR_open_xstreme/xstreme.txt")
meme_motifs_LR_open <- read_meme("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/LR_open_xstreme/xstreme.txt")
meme_motifs_EAR_close <- read_meme("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/EAR_close_xstreme/xstreme.txt")
meme_motifs_ESR_close <- read_meme("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/ESR_close_xstreme/xstreme.txt")
meme_motifs_LR_close <- read_meme("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/LR_close_xstreme/xstreme.txt")
meme_motifs_ESR_opcl <- read_meme("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/ESR_opcl_xstreme/xstreme.txt")
meme_motifs_ESR_clop <- read_meme("C:/Users/renee/ATAC_folder/ATAC_meme_data/new_analysis/ESR_clop_xstreme/xstreme.txt")making table of significant
full_motifs_dataframe <- bind_rows(DOX_DAR_3hr_combo,
DOX_DAR_24hr_combo,
)
# for_supplemental <-
full_motifs_dataframe %>%
dplyr::select(CLUSTER:EVALUE.y, ENR_RATIO,mrc_type) %>%
dplyr::filter(ENR_RATIO>1.25) %>%
# dplyr::filter(EVALUE.y<0.05) %>%
group_by(mrc_type,CLUSTER) %>%
summarize(ID=paste(unique(ID), collapse = ";"),
ALT_ID=paste(unique(ALT_ID), collapse = ";"),
motif_name=paste(unique(motif_name),collapse="; "),
mrc=unique(mrc_type),
sig_val=paste0(min(EVALUE.y),"-",max(EVALUE.y)),
order_val=min(EVALUE.y)) %>%
arrange(mrc,order_val) %>%
DT::datatable(.)
# write_delim(for_supplemental,"data/Final_four_data/re_analysis/motif_cluster_dataframe.txt",delim="\t")
# for_supplemental
# full_motifs_dataframe
full_motifs_dataframe %>%
dplyr::select(CLUSTER:EVALUE.y, ENR_RATIO,mrc_type) %>%
dplyr::filter(ENR_RATIO>1.25) %>%
dplyr::filter(mrc_type=="DOX_DAR_24hr")%>%
group_by(mrc_type,CLUSTER) %>%
summarize(ID=paste(unique(ID), collapse = ";"),
ALT_ID=paste(unique(ALT_ID), collapse = ";"),
motif_name=paste(unique(motif_name),collapse="; "),
mrc=unique(mrc_type),
sig_val=paste0(min(EVALUE.y),"-",max(EVALUE.y)),
order_val=min(EVALUE.y)) %>%
dplyr::select(motif_name,sig_val) %>%
DT::datatable(.)
sessionInfo()R version 4.4.2 (2024-10-31 ucrt)
Platform: x86_64-w64-mingw32/x64
Running under: Windows 11 x64 (build 26100)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
time zone: America/Chicago
tzcode source: internal
attached base packages:
[1] grid stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] motifmatchr_1.28.0
[2] ggseqlogo_0.2
[3] universalmotif_1.24.2
[4] data.table_1.17.6
[5] BSgenome.Hsapiens.UCSC.hg38_1.4.5
[6] BSgenome_1.74.0
[7] BiocIO_1.16.0
[8] MotifDb_1.48.0
[9] Biostrings_2.74.1
[10] XVector_0.46.0
[11] TFBSTools_1.44.0
[12] JASPAR2022_0.99.8
[13] BiocFileCache_2.14.0
[14] dbplyr_2.5.0
[15] devtools_2.4.5
[16] usethis_3.1.0
[17] ggpubr_0.6.0
[18] BiocParallel_1.40.2
[19] Cormotif_1.52.0
[20] affy_1.84.0
[21] scales_1.4.0
[22] VennDiagram_1.7.3
[23] futile.logger_1.4.3
[24] gridExtra_2.3
[25] ggfortify_0.4.17
[26] edgeR_4.4.2
[27] limma_3.62.2
[28] rtracklayer_1.66.0
[29] org.Hs.eg.db_3.20.0
[30] TxDb.Hsapiens.UCSC.hg38.knownGene_3.20.0
[31] GenomicFeatures_1.58.0
[32] AnnotationDbi_1.68.0
[33] Biobase_2.66.0
[34] GenomicRanges_1.58.0
[35] GenomeInfoDb_1.42.3
[36] IRanges_2.40.1
[37] S4Vectors_0.44.0
[38] BiocGenerics_0.52.0
[39] ChIPseeker_1.42.1
[40] RColorBrewer_1.1-3
[41] broom_1.0.8
[42] kableExtra_1.4.0
[43] cowplot_1.1.3
[44] lubridate_1.9.4
[45] forcats_1.0.0
[46] stringr_1.5.1
[47] dplyr_1.1.4
[48] purrr_1.0.4
[49] readr_2.1.5
[50] tidyr_1.3.1
[51] tibble_3.3.0
[52] ggplot2_3.5.2
[53] tidyverse_2.0.0
[54] workflowr_1.7.1
loaded via a namespace (and not attached):
[1] fs_1.6.6
[2] matrixStats_1.5.0
[3] bitops_1.0-9
[4] DirichletMultinomial_1.48.0
[5] enrichplot_1.26.6
[6] httr_1.4.7
[7] profvis_0.4.0
[8] tools_4.4.2
[9] backports_1.5.0
[10] R6_2.6.1
[11] lazyeval_0.2.2
[12] urlchecker_1.0.1
[13] withr_3.0.2
[14] preprocessCore_1.68.0
[15] cli_3.6.5
[16] textshaping_1.0.1
[17] formatR_1.14
[18] labeling_0.4.3
[19] sass_0.4.10
[20] Rsamtools_2.22.0
[21] systemfonts_1.2.3
[22] yulab.utils_0.2.0
[23] DOSE_4.0.1
[24] svglite_2.2.1
[25] R.utils_2.13.0
[26] dichromat_2.0-0.1
[27] sessioninfo_1.2.3
[28] plotrix_3.8-4
[29] rstudioapi_0.17.1
[30] RSQLite_2.4.1
[31] generics_0.1.4
[32] gridGraphics_0.5-1
[33] TxDb.Hsapiens.UCSC.hg19.knownGene_3.2.2
[34] gtools_3.9.5
[35] car_3.1-3
[36] GO.db_3.20.0
[37] Matrix_1.7-3
[38] abind_1.4-8
[39] R.methodsS3_1.8.2
[40] lifecycle_1.0.4
[41] whisker_0.4.1
[42] yaml_2.3.10
[43] carData_3.0-5
[44] SummarizedExperiment_1.36.0
[45] gplots_3.2.0
[46] qvalue_2.38.0
[47] SparseArray_1.6.2
[48] blob_1.2.4
[49] promises_1.3.3
[50] pwalign_1.2.0
[51] crayon_1.5.3
[52] miniUI_0.1.2
[53] ggtangle_0.0.6
[54] lattice_0.22-7
[55] annotate_1.84.0
[56] KEGGREST_1.46.0
[57] pillar_1.10.2
[58] knitr_1.50
[59] fgsea_1.32.4
[60] rjson_0.2.23
[61] boot_1.3-31
[62] codetools_0.2-20
[63] fastmatch_1.1-6
[64] glue_1.8.0
[65] getPass_0.2-4
[66] ggfun_0.1.9
[67] remotes_2.5.0
[68] vctrs_0.6.5
[69] png_0.1-8
[70] treeio_1.30.0
[71] poweRlaw_1.0.0
[72] gtable_0.3.6
[73] cachem_1.1.0
[74] xfun_0.52
[75] S4Arrays_1.6.0
[76] mime_0.13
[77] statmod_1.5.0
[78] ellipsis_0.3.2
[79] nlme_3.1-168
[80] ggtree_3.14.0
[81] bit64_4.6.0-1
[82] filelock_1.0.3
[83] rprojroot_2.0.4
[84] bslib_0.9.0
[85] affyio_1.76.0
[86] KernSmooth_2.23-26
[87] splitstackshape_1.4.8
[88] seqLogo_1.72.0
[89] colorspace_2.1-1
[90] DBI_1.2.3
[91] tidyselect_1.2.1
[92] processx_3.8.6
[93] bit_4.6.0
[94] compiler_4.4.2
[95] curl_6.4.0
[96] git2r_0.36.2
[97] xml2_1.3.8
[98] DelayedArray_0.32.0
[99] caTools_1.18.3
[100] callr_3.7.6
[101] digest_0.6.37
[102] rmarkdown_2.29
[103] htmltools_0.5.8.1
[104] pkgconfig_2.0.3
[105] MatrixGenerics_1.18.1
[106] fastmap_1.2.0
[107] rlang_1.1.6
[108] htmlwidgets_1.6.4
[109] UCSC.utils_1.2.0
[110] shiny_1.11.0
[111] farver_2.1.2
[112] jquerylib_0.1.4
[113] jsonlite_2.0.0
[114] GOSemSim_2.32.0
[115] R.oo_1.27.1
[116] RCurl_1.98-1.17
[117] magrittr_2.0.3
[118] Formula_1.2-5
[119] GenomeInfoDbData_1.2.13
[120] ggplotify_0.1.2
[121] patchwork_1.3.1
[122] Rcpp_1.0.14
[123] ape_5.8-1
[124] stringi_1.8.7
[125] MASS_7.3-65
[126] zlibbioc_1.52.0
[127] plyr_1.8.9
[128] pkgbuild_1.4.8
[129] parallel_4.4.2
[130] ggrepel_0.9.6
[131] CNEr_1.42.0
[132] splines_4.4.2
[133] hms_1.1.3
[134] locfit_1.5-9.12
[135] ps_1.9.1
[136] igraph_2.1.4
[137] ggsignif_0.6.4
[138] reshape2_1.4.4
[139] pkgload_1.4.0
[140] TFMPvalue_0.0.9
[141] futile.options_1.0.1
[142] XML_3.99-0.18
[143] evaluate_1.0.4
[144] lambda.r_1.2.4
[145] BiocManager_1.30.26
[146] tzdb_0.5.0
[147] httpuv_1.6.16
[148] xtable_1.8-4
[149] restfulr_0.0.15
[150] tidytree_0.4.6
[151] rstatix_0.7.2
[152] later_1.4.2
[153] viridisLite_0.4.2
[154] aplot_0.2.7
[155] memoise_2.0.1
[156] GenomicAlignments_1.42.0
[157] timechange_0.3.0