GO-TBLUP Report
Last updated: 2024-01-16
Checks: 7 0
Knit directory: dgrp-starve/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20221101) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 1aeb6cb. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .snakemake/
Ignored: code/methodComp/bglr/err-bglr-f.5381.err
Ignored: code/methodComp/bglr/err-bglr-m.5382.err
Ignored: code/methodComp/m/meth-m.4676.err
Ignored: code/methodComp/m/meth-m.4685.err
Ignored: code/methodComp/method-f.4751.out
Ignored: data/fb/
Ignored: data/snake/
Ignored: snake/.snakemake/
Ignored: snake/GOfile.yaml
Ignored: snake/ReadMe.md
Ignored: snake/code/bayesC/
Ignored: snake/code/misc/
Ignored: snake/customL.Rds
Ignored: snake/data/
Ignored: snake/datafile.yaml
Ignored: snake/dgrp.yaml
Ignored: snake/fig/
Ignored: snake/fullGO.yaml
Ignored: snake/gofig.yaml
Ignored: snake/guide/
Ignored: snake/logs/
Ignored: snake/slurm/
Ignored: snake/smake.sbatch
Ignored: snake/snubnose.sbatch
Ignored: snake/zz_lost/
Ignored: zz_lost/
Untracked files:
Untracked: analysis/allotter.R
Untracked: analysis/old_index.Rmd.Rmd
Untracked: analysis/sparseComp.Rmd
Untracked: forester.R
Untracked: malegofind.R
Untracked: pippinRMDbackup.R
Untracked: snake/code/binner.R
Untracked: snake/code/combine_GO.R
Untracked: snake/code/dataFinGO.R
Untracked: snake/code/datafile.yaml
Untracked: snake/code/filterNcombine_GO.R
Untracked: snake/code/filter_GO.R
Untracked: snake/code/go/
Untracked: snake/code/idTEMP.R
Untracked: snake/code/imstuff.R
Untracked: snake/code/method/bayesHome.R
Untracked: snake/code/method/multiplotGO.Rmd
Untracked: snake/code/scheming.R
Untracked: snake/code/scripts/
Untracked: snake/code/srfile.yaml
Unstaged changes:
Modified: analysis/Method/BayesC.Rmd
Modified: analysis/bigGO.Rmd
Modified: analysis/goReport.Rmd
Modified: analysis/index.Rmd
Modified: snake/code/method/bayesGO.R
Modified: snake/code/method/goFish.R
Modified: snake/code/method/varbvs.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/blupUp.Rmd) and HTML (docs/blupUp.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 1aeb6cb | nklimko | 2024-01-16 | wflow_publish(“analysis/blupUp.Rmd”) |
| html | 7327d20 | nklimko | 2024-01-16 | Build site. |
| Rmd | 878d371 | nklimko | 2024-01-16 | wflow_publish(“analysis/blupUp.Rmd”) |
#load('snake/data/topTables.Rdata')
options(knitr.kable.NA = '')Summary
GBLUP had the R2 constraints removed from the model to allow each component of the model to maximally explain variance. Overall maximum still 0.8.
While this does remove a degree of certainty, model accuracy not only improved significantly but was able to find similar results to bayesC.
#function
partMake <- function(data, sex, nullInt, upperCutoff, lowerCutoff, psize, custom.title, custom.Xlab, custom.Ylab){
plothole <- ggplot(data, aes(x=term, y=cor, label=term))+
geom_point(color=viridis(1, begin=0.5), size=psize)+
geom_text(aes(label=ifelse(cor>upperCutoff, as.character(term),'')), hjust=0, size=2, angle=0)+
geom_text(aes(label=ifelse(cor<lowerCutoff, as.character(term),'')), hjust=1, size=2, angle=90)+
geom_hline(yintercept = nullInt) +
theme_minimal() +
labs(x=custom.Xlab, y=custom.Ylab, tag=sex, title=custom.title) +
theme(text=element_text(size=10), plot.tag = element_text(size=15))
return(plothole)
}
#data
load('snake/data/go/50_tables/saveTables.Rdata')
#Cutoff selection
sigFactor <- 3
sdf <- sd(unlist(allDataF[,2]))
meanf <- mean(unlist(allDataF[,2]))
cutoffF <- meanf + sigFactor*sdf
sdm <- sd(unlist(allDataM[,2]))
meanm <- mean(unlist(allDataM[,2]))
cutoffM <- meanm + sigFactor*sdm
#graphs
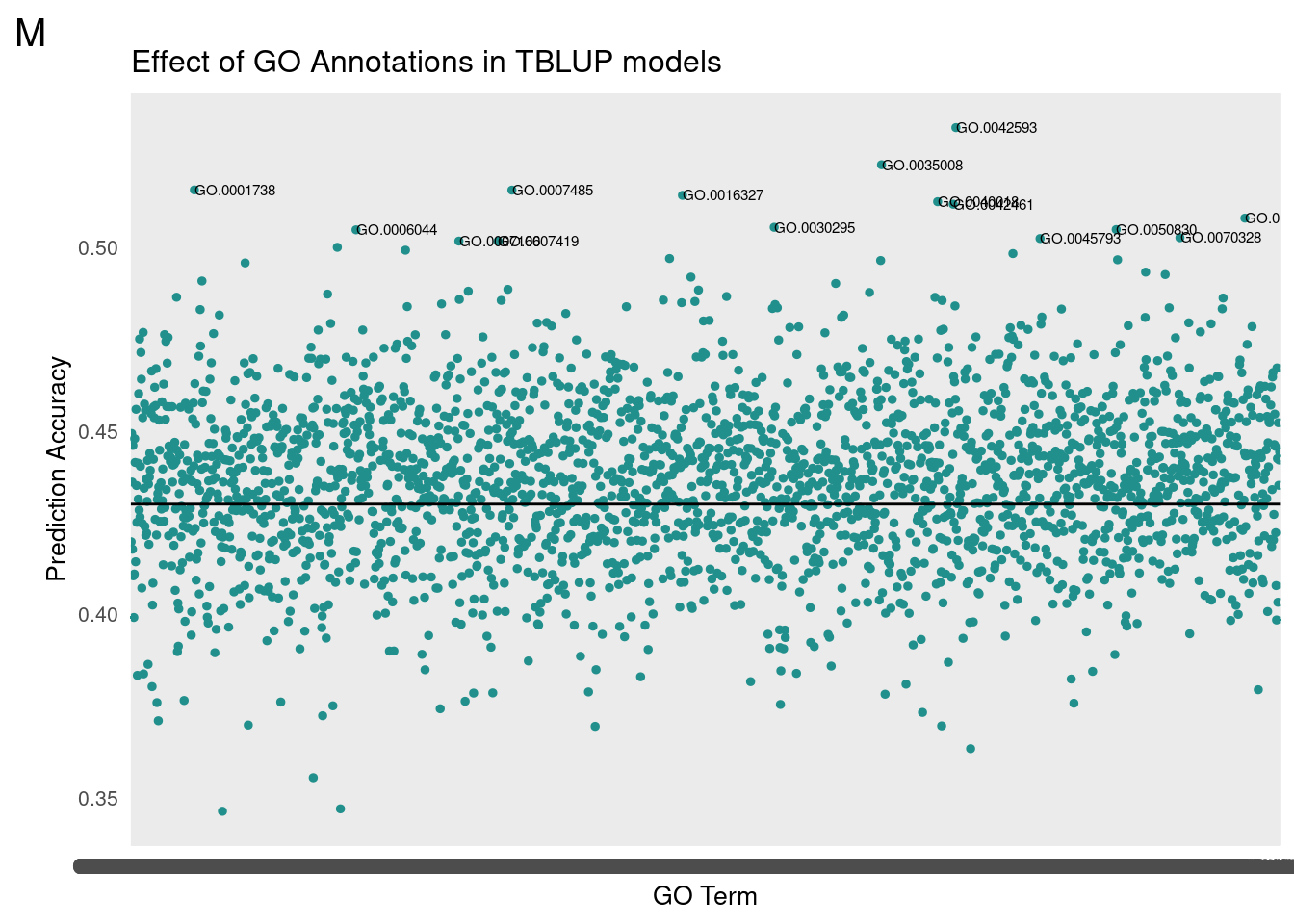
gg[[1]] <- partMake(allDataF, 'F', 0.31, cutoffF, 0.2, 1, 'Effect of GO Annotations in TBLUP models', 'GO Term', 'Prediction Accuracy')
gg[[2]] <- partMake(allDataM, 'M', 0.43, cutoffM, 0.2, 1, 'Effect of GO Annotations in TBLUP models', 'GO Term', 'Prediction Accuracy')
subF <- allDataF[which(cor>cutoffF),]
subM <- allDataM[which(cor>cutoffM),]
subF <- subF[order(-cor),]
subM <- subM[order(-cor),]
#write ordered GO terms to table file for enrichment purposes
cat(unlist(subF[,1]), sep = '\n', file='snake/data/go/50_tables/topHitsF.txt')
cat(unlist(subM[,1]), sep = '\n', file='snake/data/go/50_tables/topHitsM.txt')
cat(unlist(subF[,1]), sep = '\n', file='snake/code/go/enrichment/blup/f/topHitsF.txt')
cat(unlist(subM[,1]), sep = '\n', file='snake/code/go/enrichment/blup/m/topHitsM.txt')
topBlupSoloF <- readRDS('snake/code/go/enrichment/blup/f/finalData.Rds')
topBlupSoloM <- readRDS('snake/code/go/enrichment/blup/m/finalData.Rds')Surface Level / Sanity Check / Congruence / On course
Comparison of top 20 terms from both BayesC and TBLUP yields familiar results: 11 of top 20 match for females, 10 of top 20 match for males
This suggests the models are both accurate and able to detect GO terms of interest, even with delimited R2.
Females
kable(finF, caption = 'Female BayesC/BLUP Comparison', "simple")| BayesC_Cor | TBLUP_Cor | Term | BayesC_Rank | TBLUP_Rank |
|---|---|---|---|---|
| 0.4382497 | 0.4319931 | GO.0045819 | 1 | 3 |
| 0.4231229 | 0.3881740 | GO.0033500 | 2 | 11 |
| 0.4178735 | 0.4514600 | GO.0055088 | 3 | 1 |
| 0.4014416 | 0.4147260 | GO.0042675 | 4 | 6 |
| 0.3984704 | 0.4366905 | GO.0008586 | 6 | 2 |
| 0.3919625 | 0.3811383 | GO.0016042 | 7 | 15 |
| 0.3873729 | 0.3805084 | GO.0007368 | 8 | 17 |
| 0.3799043 | 0.3833308 | GO.0006644 | 10 | 13 |
| 0.3777651 | 0.3805090 | GO.0046488 | 12 | 16 |
| 0.3736580 | 0.4223885 | GO.0017056 | 14 | 5 |
| 0.3691317 | 0.3786734 | GO.0061883 | 19 | 18 |
Males
kable(finM, caption = 'Male BayesC/BLUP Comparison', "simple")| BayesC_Cor | TBLUP_Cor | Term | BayesC_Rank | TBLUP_Rank |
|---|---|---|---|---|
| 0.5116379 | 0.5225415 | GO.0035008 | 1 | 2 |
| 0.5064311 | 0.5079819 | GO.0140042 | 2 | 8 |
| 0.5062906 | 0.5156219 | GO.0007485 | 3 | 4 |
| 0.4984272 | 0.5117790 | GO.0042461 | 6 | 7 |
| 0.4978650 | 0.5141982 | GO.0016327 | 8 | 5 |
| 0.4924000 | 0.5048001 | GO.0006044 | 11 | 11 |
| 0.4923077 | 0.5124686 | GO.0040018 | 12 | 6 |
| 0.4918994 | 0.5326720 | GO.0042593 | 13 | 1 |
| 0.4882313 | 0.5156778 | GO.0001738 | 15 | 3 |
| 0.4866070 | 0.4983168 | GO.0045196 | 17 | 18 |
Overall Results
For GO-TBLUP, I filtered top terms that were 3 standard deviations above the mean for each sex.
We then translated the top GO terms into human readable categories to assess our findings. Below are the top ten ordered by correlation.
Female
Top Terms
id: GO:0055088 name: lipid homeostasis – id: GO:0008586 name: imaginal disc-derived wing vein morphogenesis – id: GO:0045819 name: positive regulation of glycogen catabolic process – id: GO:0043066 name: negative regulation of apoptotic process – id: GO:0017056 name: structural constituent of nuclear pore – id: GO:0042675 name: compound eye cone cell differentiation – id: GO:0035556 name: intracellular signal transduction – id: GO:0006606 name: protein import into nucleus – id: GO:0005524 name: ATP binding – id: GO:0000281 name: mitotic cytokinesis – id: GO:0033500 name: carbohydrate homeostasis – id: GO:0042749 name: regulation of circadian sleep/wake cycle – id: GO:0006644 name: phospholipid metabolic process – id: GO:0004672 name: protein kinase activity – id: GO:0016042 name: lipid catabolic process – id: GO:0046488 name: phosphatidylinositol metabolic process – id: GO:0007368 name: determination of left/right symmetry
Male
Top Terms
id: GO:0042593 name: glucose homeostasis – id: GO:0035008 name: positive regulation of melanization defense response – id: GO:0001738 name: morphogenesis of a polarized epithelium – id: GO:0007485 name: imaginal disc-derived male genitalia development – id: GO:0016327 name: apicolateral plasma membrane – id: GO:0040018 name: positive regulation of multicellular organism growth – id: GO:0042461 name: photoreceptor cell development – id: GO:0140042 name: lipid droplet formation – id: GO:0030295 name: protein kinase activator activity – id: GO:0050830 name: defense response to Gram-positive bacterium – id: GO:0006044 name: N-acetylglucosamine metabolic process – id: GO:0070328 name: triglyceride homeostasis – id: GO:0045793 name: positive regulation of cell size – id: GO:0007166 name: cell surface receptor signaling pathway – id: GO:0007419 name: ventral cord development
Post Processing
Beyond this, we took the models to determine if certain genes were enriched in the GO terms of interest. From the selected terms, we pooled the associated genes and totaled gene occurrence.
Females involved 81 unique genes at least 3 times across top terms. Of these, 7 were present 4 or more times.
Males had a significantly lower number of genes involved than expected. Only 7 genes were involved at least 3 times across top terms. This may suggest that the selection criteria is too stringent for males despite males having a higher base prediction accuracy.
After establishing unique genes, we translated the FlyBase gene codes to human-readable genes.
Females
kable(topBlupSoloF, caption = 'GO-TBLUP Genes', "simple")| flybase | count | gene |
|---|---|---|
| FBgn0010379 | 5 | Akt1 |
| FBgn0003731 | 5 | Egfr |
| FBgn0025595 | 4 | AkhR |
| FBgn0262103 | 4 | Sik3 |
| FBgn0283499 | 4 | InR |
| FBgn0020386 | 4 | Pdk1 |
| FBgn0028484 | 4 | Ack |
| FBgn0004552 | 3 | Akh |
| FBgn0035039 | 3 | Adck |
| FBgn0261984 | 3 | Ire1 |
| FBgn0283472 | 3 | S6k |
| FBgn0000575 | 3 | emc |
| FBgn0004635 | 3 | rho |
| FBgn0026252 | 3 | msk |
| FBgn0035142 | 3 | Hipk |
| FBgn0003169 | 3 | put |
| FBgn0011300 | 3 | babo |
| FBgn0260945 | 3 | Atg1 |
| FBgn0002413 | 3 | dco |
| FBgn0032006 | 3 | Pvr |
| FBgn0003091 | 3 | Pkc53E |
| FBgn0003093 | 3 | Pkc98E |
| FBgn0003256 | 3 | rl |
| FBgn0003502 | 3 | Btk29A |
| FBgn0003744 | 3 | trc |
| FBgn0004784 | 3 | inaC |
| FBgn0004864 | 3 | hop |
| FBgn0010197 | 3 | Gyc32E |
| FBgn0010441 | 3 | pll |
| FBgn0011817 | 3 | nmo |
| FBgn0013987 | 3 | MAPk-Ak2 |
| FBgn0015765 | 3 | p38a |
| FBgn0017581 | 3 | Lk6 |
| FBgn0020621 | 3 | Pkn |
| FBgn0023169 | 3 | AMPKalpha |
| FBgn0024846 | 3 | p38b |
| FBgn0025625 | 3 | Sik2 |
| FBgn0025743 | 3 | mbt |
| FBgn0026063 | 3 | KP78b |
| FBgn0026064 | 3 | KP78a |
| FBgn0027497 | 3 | Madm |
| FBgn0028741 | 3 | fab1 |
| FBgn0031299 | 3 | CG4629 |
| FBgn0031784 | 3 | CG9222 |
| FBgn0033915 | 3 | CG8485 |
| FBgn0034568 | 3 | CG3216 |
| FBgn0034950 | 3 | Pask |
| FBgn0036368 | 3 | CG10738 |
| FBgn0036544 | 3 | sff |
| FBgn0037098 | 3 | Wnk |
| FBgn0038167 | 3 | Lkb1 |
| FBgn0038630 | 3 | CG14305 |
| FBgn0039083 | 3 | CG10177 |
| FBgn0040056 | 3 | CG17698 |
| FBgn0044826 | 3 | Pak3 |
| FBgn0046706 | 3 | Haspin |
| FBgn0051183 | 3 | CG31183 |
| FBgn0052666 | 3 | Drak |
| FBgn0052703 | 3 | Erk7 |
| FBgn0052944 | 3 | CG32944 |
| FBgn0085386 | 3 | CG34357 |
| FBgn0259712 | 3 | CG42366 |
| FBgn0260399 | 3 | gwl |
| FBgn0260934 | 3 | par-1 |
| FBgn0261278 | 3 | grp |
| FBgn0261360 | 3 | CG42637 |
| FBgn0261387 | 3 | CG17528 |
| FBgn0261456 | 3 | hpo |
| FBgn0261854 | 3 | aPKC |
| FBgn0262738 | 3 | norpA |
| FBgn0262866 | 3 | S6kII |
| FBgn0263395 | 3 | hppy |
| FBgn0266136 | 3 | Gyc76C |
| FBgn0267339 | 3 | p38c |
| FBgn0267390 | 3 | dop |
| FBgn0267698 | 3 | Pak |
| FBgn0283657 | 3 | Tlk |
| FBgn0002466 | 3 | sti |
| FBgn0010303 | 3 | hep |
| FBgn0024227 | 3 | aurB |
| FBgn0026181 | 3 | Rok |
Males
kable(topBlupSoloM, caption = 'GO-TBLUP Genes', "simple")| flybase | count | gene |
|---|---|---|
| FBgn0036046 | 4 | Ilp2 |
| FBgn0086687 | 4 | Desat1 |
| FBgn0283499 | 4 | InR |
| FBgn0020386 | 4 | Pdk1 |
| FBgn0024248 | 3 | chico |
| FBgn0261873 | 3 | sdt |
| FBgn0037874 | 3 | Tctp |
Comparison
allSolo <- cbind(topBlupSoloF[1:7], topBlupSoloM)
names(allSolo) <- c('Female Gene', 'Count', 'Name', 'Male Gene', 'Count', 'Name')
kable(allSolo, caption = 'GO-TBLUP Gene Comparison', "simple")| Female Gene | Count | Name | Male Gene | Count | Name |
|---|---|---|---|---|---|
| FBgn0010379 | 5 | Akt1 | FBgn0036046 | 4 | Ilp2 |
| FBgn0003731 | 5 | Egfr | FBgn0086687 | 4 | Desat1 |
| FBgn0025595 | 4 | AkhR | FBgn0283499 | 4 | InR |
| FBgn0262103 | 4 | Sik3 | FBgn0020386 | 4 | Pdk1 |
| FBgn0283499 | 4 | InR | FBgn0024248 | 3 | chico |
| FBgn0020386 | 4 | Pdk1 | FBgn0261873 | 3 | sdt |
| FBgn0028484 | 4 | Ack | FBgn0037874 | 3 | Tctp |
| Looking at bot | h sexes | togethe | r, the only tw | o genes | that are found in both rankings are InR and Pdk1. Coincidentally, both are significantly involved genes for both sexes. |
Intuitively, both are heavily involved in carbohydrate modification activity.
sessionInfo()R version 4.1.2 (2021-11-01)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Rocky Linux 8.5 (Green Obsidian)
Matrix products: default
BLAS/LAPACK: /opt/ohpc/pub/libs/gnu9/openblas/0.3.7/lib/libopenblasp-r0.3.7.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] DT_0.31 kableExtra_1.3.4 knitr_1.43 reshape2_1.4.4
[5] melt_1.10.0 ggcorrplot_0.1.4.1 lubridate_1.9.3 forcats_1.0.0
[9] stringr_1.5.0 purrr_1.0.1 readr_2.1.4 tidyr_1.3.0
[13] tibble_3.2.1 tidyverse_2.0.0 scales_1.2.1 viridis_0.6.4
[17] viridisLite_0.4.2 qqman_0.1.9 cowplot_1.1.1 ggplot2_3.4.4
[21] data.table_1.14.8 dplyr_1.1.3 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] httr_1.4.7 sass_0.4.7 jsonlite_1.8.7 bslib_0.5.0
[5] getPass_0.2-2 highr_0.10 yaml_2.3.7 pillar_1.9.0
[9] glue_1.6.2 digest_0.6.33 promises_1.2.0.1 rvest_1.0.3
[13] colorspace_2.1-0 htmltools_0.5.5 httpuv_1.6.12 plyr_1.8.9
[17] pkgconfig_2.0.3 calibrate_1.7.7 webshot_0.5.5 processx_3.8.2
[21] svglite_2.1.2 whisker_0.4.1 later_1.3.1 tzdb_0.4.0
[25] timechange_0.2.0 git2r_0.32.0 generics_0.1.3 farver_2.1.1
[29] cachem_1.0.8 withr_2.5.0 cli_3.6.1 magrittr_2.0.3
[33] evaluate_0.21 ps_1.7.5 fs_1.6.3 fansi_1.0.4
[37] MASS_7.3-60 xml2_1.3.3 tools_4.1.2 hms_1.1.3
[41] lifecycle_1.0.3 munsell_0.5.0 callr_3.7.3 compiler_4.1.2
[45] jquerylib_0.1.4 systemfonts_1.0.5 rlang_1.1.1 grid_4.1.2
[49] rstudioapi_0.15.0 htmlwidgets_1.6.2 labeling_0.4.3 rmarkdown_2.23
[53] gtable_0.3.4 R6_2.5.1 gridExtra_2.3 fastmap_1.1.1
[57] utf8_1.2.3 rprojroot_2.0.3 stringi_1.7.12 Rcpp_1.0.11
[61] vctrs_0.6.4 tidyselect_1.2.0 xfun_0.39