Enrichment Overview
Last updated: 2024-01-30
Checks: 7 0
Knit directory: dgrp-starve/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20221101) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version d373edf. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .snakemake/
Ignored: code/methodComp/bglr/err-bglr-f.5381.err
Ignored: code/methodComp/bglr/err-bglr-m.5382.err
Ignored: code/methodComp/m/meth-m.4676.err
Ignored: code/methodComp/m/meth-m.4685.err
Ignored: code/methodComp/method-f.4751.out
Ignored: data/fb/
Ignored: data/snake/
Ignored: snake/.snakemake/
Ignored: snake/GOfile.yaml
Ignored: snake/ReadMe.md
Ignored: snake/code/bayesC/
Ignored: snake/code/misc/
Ignored: snake/customL.Rds
Ignored: snake/data/
Ignored: snake/datafile.yaml
Ignored: snake/dgrp.yaml
Ignored: snake/fig/
Ignored: snake/fullGO.yaml
Ignored: snake/gofig.yaml
Ignored: snake/guide/
Ignored: snake/logs/
Ignored: snake/pbs_smake.pbs
Ignored: snake/pbsmake.sbatch
Ignored: snake/slurm/
Ignored: snake/smake.sbatch
Ignored: snake/snubnose.sbatch
Ignored: snake/zz_lost/
Ignored: zz_lost/
Untracked files:
Untracked: analysis/allotter.R
Untracked: analysis/old_index.Rmd.Rmd
Untracked: analysis/sparseComp.Rmd
Untracked: forester.R
Untracked: malegofind.R
Untracked: moarnotes.R
Untracked: pippinRMDbackup.R
Untracked: snake/code/binner.R
Untracked: snake/code/combine_GO.R
Untracked: snake/code/dataFinGO.R
Untracked: snake/code/datafile.yaml
Untracked: snake/code/filterNcombine_GO.R
Untracked: snake/code/filter_GO.R
Untracked: snake/code/go/
Untracked: snake/code/idTEMP.R
Untracked: snake/code/imstuff.R
Untracked: snake/code/method/bayesHome.R
Untracked: snake/code/method/multiplotGO.Rmd
Untracked: snake/code/scheming.R
Untracked: snake/code/scripts/
Untracked: snake/code/srfile.yaml
Untracked: teno.R
Unstaged changes:
Modified: analysis/Method/BayesC.Rmd
Modified: analysis/bigGO.Rmd
Modified: snake/code/method/bayesGO.R
Modified: snake/code/method/goFish.R
Modified: snake/code/method/varbvs.R
Modified: snake/temp.txt
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/enrich.Rmd) and HTML (docs/enrich.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | d373edf | nklimko | 2024-01-30 | wflow_publish(“analysis/enrich.Rmd”) |
| html | 8e39493 | nklimko | 2024-01-23 | Build site. |
| Rmd | 2683f74 | nklimko | 2024-01-23 | wflow_publish(“analysis/enrich.Rmd”) |
| html | 991d04c | nklimko | 2024-01-23 | Build site. |
| Rmd | 7c20291 | nklimko | 2024-01-23 | wflow_publish(“analysis/enrich.Rmd”) |
| html | 02fdd7e | nklimko | 2024-01-23 | Build site. |
| Rmd | c5835d5 | nklimko | 2024-01-23 | wflow_publish(“analysis/enrich.Rmd”) |
| html | 8f35a08 | nklimko | 2024-01-23 | Build site. |
| Rmd | dd3c95c | nklimko | 2024-01-23 | wflow_publish(“analysis/enrich.Rmd”) |
Dive back in:
We left off looking to modify the selection cutoff for top terms. While various ideas were floated(standard devs, percentiles, flat cutoff), there was not a consistently most useful method for looking at both male and female data. As such, I opted to run a series of flat cutoffs(100, 50, 25) for both complete methods per sex.
I first looked at the top terms ordered to look for patterns in the data. While I’m unsure of the significance, the clustering of correlations for top terms shares a shape within sexes across methods. Notably, female data had a sharper increase(negative slope) for top results
load('snake/data/go/50_tables/enrichment.Rdata')
topGrid <- function(data, sex, psize, custom.title, custom.Xlab, custom.Ylab){
plothole <- ggplot(data, aes(x=index, y=cor, label=term))+
geom_point(color=viridis(1, begin=0.5), size=psize)+
theme_minimal() +
labs(x=custom.Xlab, y=custom.Ylab, tag=sex, title=custom.title) +
theme(text=element_text(size=10), plot.tag = element_text(size=15))
return(plothole)
}
gg[[1]] <- topGrid(blupF, 'F', 1, 'Top Female Results: TBLUP', 'Rank', 'Correlation')
gg[[2]] <- topGrid(blupM, 'M', 1, 'Top Male Results: TBLUP', 'Rank', 'Correlation')
gg[[3]] <- topGrid(bayesF, 'F', 1, 'Top Female Results: BayesC', 'Rank', 'Correlation')
gg[[4]] <- topGrid(bayesM, 'M', 1, 'Top Male Results: BayesC', 'Rank', 'Correlation')
plot_grid(gg[[1]], gg[[2]], gg[[3]], gg[[4]], ncol=2)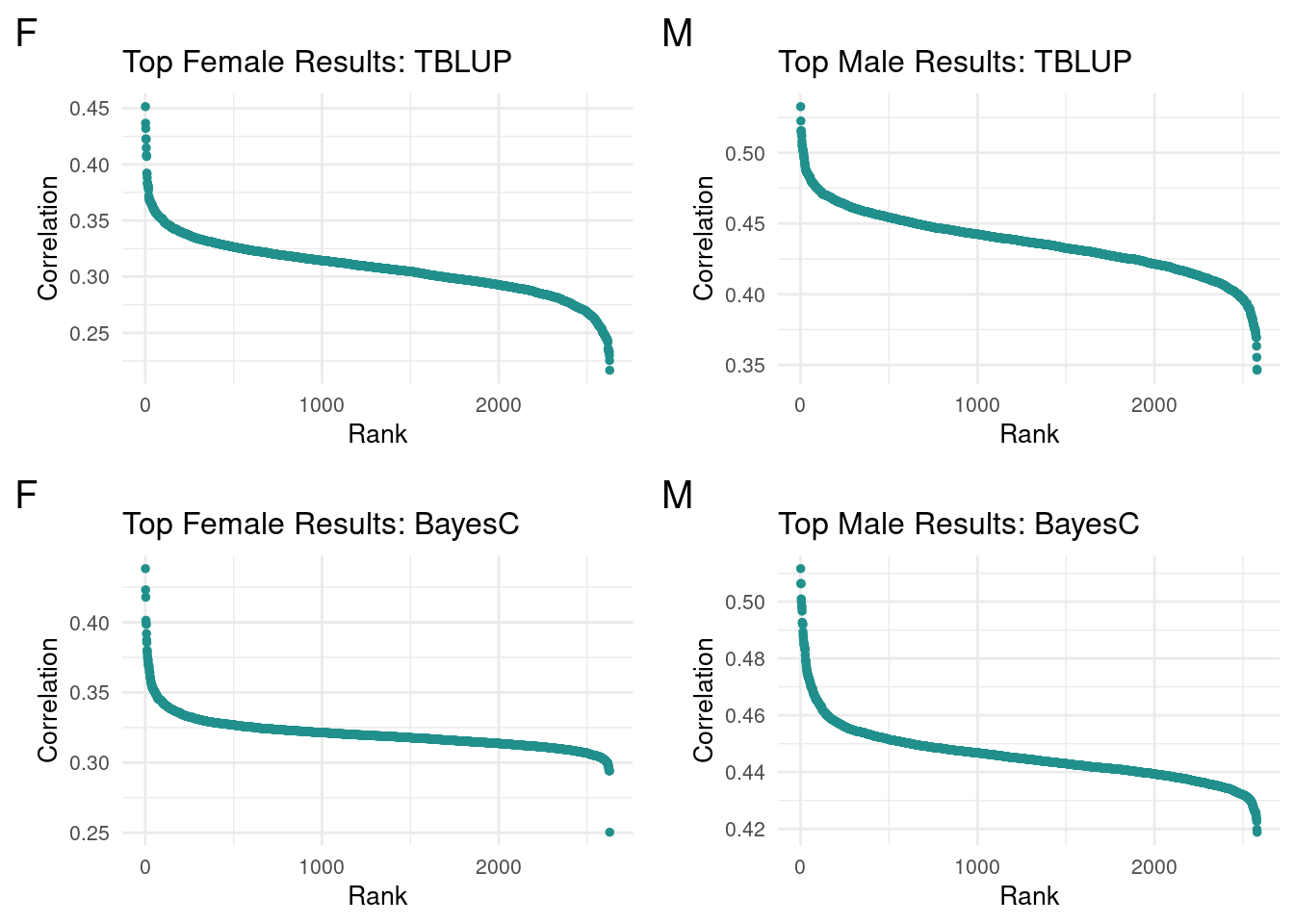
| Version | Author | Date |
|---|---|---|
| 8f35a08 | nklimko | 2024-01-23 |
After this, I found the correlation between the two methods to see how similar generated results are.
print(cor(blupF$cor, bayesF$cor))[1] 0.9490421Female Overall
print(cor(blupF[1:200,cor], bayesF[1:200, cor]))[1] 0.9937145Female Top 200
print(cor(blupM$cor, bayesM$cor))[1] 0.9598015Male Overall
print(cor(blupM[1:200,cor], bayesM[1:200, cor]))[1] 0.9967979Male Top 100
Moving past this, I wanted to assess the effect of term count on enrichment
load("snake/data/go/50_tables/enrich/kables.Rdata")
percentModder <- function(dataKable, custom.caption){
dataKable[,5] <- dataKable[,5]*2
dataKable[,8] <- dataKable[,8]*2
colnames(dataKable) <- rep(c('Flybase Gene', 'Percent', 'Gene'), 3)
kabled <- kable(dataKable, caption=custom.caption, "simple", header = c('Top 100 GO Terms' = 3, 'Top 50 GO Terms' = 3, 'Top 25 GO Terms' = 3))
return(kabled)
}
bayesF_KableMod <- percentModder(bayesF_Kable, 'Female BayesC')
print(bayesF_KableMod)
Table: Female BayesC
Flybase Gene Percent Gene Flybase Gene Percent Gene Flybase Gene Percent Gene
------------- -------- ---------- ------------- -------- -------- ------------- -------- --------
FBgn0262738 15 norpA FBgn0025595 18 AkhR FBgn0025595 16 AkhR
FBgn0003731 13 Egfr FBgn0003731 18 Egfr FBgn0000575 12 emc
FBgn0004635 13 rho FBgn0003205 18 Ras85D FBgn0004552 8 Akh
FBgn0003205 10 Ras85D FBgn0004635 18 rho FBgn0283499 8 InR
FBgn0025595 9 AkhR FBgn0000575 14 emc FBgn0000490 8 dpp
FBgn0003310 8 NULL FBgn0262738 14 norpA FBgn0010303 6 hep
FBgn0000575 7 emc FBgn0004552 10 Akh FBgn0015279 6 Pi3K92E
FBgn0015795 7 Rab7 FBgn0015279 10 Pi3K92E FBgn0033799 6 GLaz
FBgn0283499 6 InR FBgn0283499 10 InR FBgn0036449 6 bmm
FBgn0039114 6 Lsd-1 FBgn0003310 10 NULL FBgn0003731 6 Egfr
FBgn0004552 5 Akh FBgn0035586 8 CG10671 FBgn0003205 6 Ras85D
FBgn0015279 5 Pi3K92E FBgn0026252 8 msk FBgn0003463 6 sog
FBgn0035586 5 CG10671 FBgn0000490 8 dpp FBgn0003719 6 tld
FBgn0003463 5 sog FBgn0261648 8 salm FBgn0262738 6 norpA
FBgn0003719 5 tld FBgn0010303 6 hep
FBgn0036545 5 GXIVsPLA2 FBgn0036046 6 Ilp2
FBgn0039655 5 CG14507 FBgn0024248 6 chico
FBgn0003118 5 pnt FBgn0033799 6 GLaz
FBgn0005672 5 spi FBgn0036449 6 bmm
FBgn0036046 4 Ilp2 FBgn0036260 6 Rh7
FBgn0003651 4 svp FBgn0003463 6 sog
FBgn0010379 4 Akt1 FBgn0003719 6 tld
FBgn0024248 4 chico FBgn0003984 6 vn
FBgn0036449 4 bmm FBgn0038197 6 foxo
FBgn0026252 4 msk FBgn0039114 6 Lsd-1
FBgn0036260 4 Rh7 FBgn0003218 6 rdgB
FBgn0000490 4 dpp FBgn0026207 6 mbo
FBgn0003984 4 vn FBgn0027537 6 Nup93-1
FBgn0261648 4 salm FBgn0031078 6 Nup205
FBgn0029720 4 CG3009 FBgn0033737 6 Nup54
FBgn0030013 4 GIIIspla2 FBgn0033766 6 CG8771
FBgn0033170 4 sPLA2 FBgn0038274 6 Nup93-2
FBgn0050503 4 CG30503 FBgn0061200 6 Nup153
FBgn0250862 4 CG42237 FBgn0003256 6 rl
FBgn0003218 4 rdgB FBgn0034140 6 Lst
FBgn0030608 4 Lsd-2 FBgn0015795 6 Rab7
FBgn0035206 4 CG9186 FBgn0002940 6 ninaE
FBgn0040336 4 Seipin FBgn0003295 6 ru
FBgn0026207 4 mbo
FBgn0004435 4 Galphaq
FBgn0000257 4 car
FBgn0000482 4 dor
FBgn0003256 4 rl
FBgn0035871 4 BI-1
FBgn0052350 4 Vps11
FBgn0285911 4 NULL
FBgn0015754 4 Lis-1
FBgn0004647 4 NULL
FBgn0002940 4 ninaE
FBgn0003295 4 ru
FBgn0020386 4 Pdk1
FBgn0262451 4 ban
FBgn0004784 4 inaC
FBgn0003169 3 put
FBgn0010303 3 hep
FBgn0028717 3 Lnk
FBgn0010051 3 Itp-r83A
FBgn0030607 3 dob
FBgn0033226 3 CG1882
FBgn0033799 3 GLaz
FBgn0262103 3 Sik3
FBgn0002576 3 lz
FBgn0004885 3 tok
FBgn0050418 3 nord
FBgn0039152 3 Root
FBgn0038197 3 foxo
FBgn0028741 3 fab1
FBgn0029870 3 Marf
FBgn0052703 3 Erk7
FBgn0027537 3 Nup93-1
FBgn0031078 3 Nup205
FBgn0033737 3 Nup54
FBgn0033766 3 CG8771
FBgn0038274 3 Nup93-2
FBgn0061200 3 Nup153
FBgn0261549 3 rdgA
FBgn0002566 3 lt
FBgn0034140 3 Lst
FBgn0038659 3 EndoA
FBgn0086676 3 spin
FBgn0015721 3 ktub
FBgn0086687 3 Desat1
FBgn0014010 3 Rab5
FBgn0025680 3 cry
FBgn0038167 3 Lkb1
FBgn0004611 3 Plc21C
FBgn0001263 3 inaD
FBgn0052699 3 LPCAT
FBgn0265048 3 cv-d if(0){
bayesF_KableMod <- percentModder(bayesF_Kable)
bayesM_KableMod <- percentModder(bayesM_Kable)
blupF_KableMod <- percentModder(blupF_Kable )
blupM_KableMod <- percentModder(blupM_Kable )
kable(bayesF_KableMod, caption="Female BayesC", "simple")
kable(bayesM_KableMod, caption="Male BayesC", "simple")
kable(blupF_KableMod, caption="Female TBLUP", "simple")
kable(blupM_KableMod, caption="Male TBLUP", "simple")
}
sessionInfo()R version 4.1.2 (2021-11-01)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Rocky Linux 8.5 (Green Obsidian)
Matrix products: default
BLAS/LAPACK: /opt/ohpc/pub/libs/gnu9/openblas/0.3.7/lib/libopenblasp-r0.3.7.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] DT_0.31 kableExtra_1.3.4 knitr_1.43 reshape2_1.4.4
[5] melt_1.10.0 ggcorrplot_0.1.4.1 lubridate_1.9.3 forcats_1.0.0
[9] stringr_1.5.0 purrr_1.0.1 readr_2.1.4 tidyr_1.3.0
[13] tibble_3.2.1 tidyverse_2.0.0 scales_1.2.1 viridis_0.6.4
[17] viridisLite_0.4.2 qqman_0.1.9 cowplot_1.1.1 ggplot2_3.4.4
[21] data.table_1.14.8 dplyr_1.1.3 workflowr_1.7.1
loaded via a namespace (and not attached):
[1] httr_1.4.7 sass_0.4.7 jsonlite_1.8.7 bslib_0.5.0
[5] getPass_0.2-2 highr_0.10 yaml_2.3.7 pillar_1.9.0
[9] glue_1.6.2 digest_0.6.33 promises_1.2.0.1 rvest_1.0.3
[13] colorspace_2.1-0 htmltools_0.5.5 httpuv_1.6.12 plyr_1.8.9
[17] pkgconfig_2.0.3 calibrate_1.7.7 webshot_0.5.5 processx_3.8.2
[21] svglite_2.1.2 whisker_0.4.1 later_1.3.1 tzdb_0.4.0
[25] timechange_0.2.0 git2r_0.32.0 generics_0.1.3 farver_2.1.1
[29] cachem_1.0.8 withr_2.5.0 cli_3.6.1 magrittr_2.0.3
[33] evaluate_0.21 ps_1.7.5 fs_1.6.3 fansi_1.0.4
[37] MASS_7.3-60 xml2_1.3.3 tools_4.1.2 hms_1.1.3
[41] lifecycle_1.0.3 munsell_0.5.0 callr_3.7.3 compiler_4.1.2
[45] jquerylib_0.1.4 systemfonts_1.0.5 rlang_1.1.1 grid_4.1.2
[49] rstudioapi_0.15.0 htmlwidgets_1.6.2 labeling_0.4.3 rmarkdown_2.23
[53] gtable_0.3.4 R6_2.5.1 gridExtra_2.3 fastmap_1.1.1
[57] utf8_1.2.3 rprojroot_2.0.3 stringi_1.7.12 Rcpp_1.0.11
[61] vctrs_0.6.4 tidyselect_1.2.0 xfun_0.39