Examine topic modeling results with k = 16 topics
Peter Carbonetto
Last updated: 2022-05-10
Checks: 7 0
Knit directory: lps/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(1) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 567870b. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: data/raw_read_counts.csv.gz
Ignored: output/
Untracked files:
Untracked: analysis/temp.R
Untracked: analysis/volcano_plot_k1
Untracked: analysis/volcano_plot_k1+k5
Untracked: analysis/volcano_plot_k10
Untracked: analysis/volcano_plot_k11
Untracked: analysis/volcano_plot_k12
Untracked: analysis/volcano_plot_k13
Untracked: analysis/volcano_plot_k14
Untracked: analysis/volcano_plot_k15
Untracked: analysis/volcano_plot_k16
Untracked: analysis/volcano_plot_k2
Untracked: analysis/volcano_plot_k2+k13
Untracked: analysis/volcano_plot_k3
Untracked: analysis/volcano_plot_k4
Untracked: analysis/volcano_plot_k5
Untracked: analysis/volcano_plot_k6
Untracked: analysis/volcano_plot_k6+k14
Untracked: analysis/volcano_plot_k7
Untracked: analysis/volcano_plot_k8
Untracked: analysis/volcano_plot_k9
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/examine_topic_model_k16.Rmd) and HTML (docs/examine_topic_model_k16.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 567870b | Peter Carbonetto | 2022-05-10 | workflowr::wflow_publish(“examine_topic_model_k16.Rmd”, verbose = TRUE) |
| html | 6dabc4d | Peter Carbonetto | 2022-05-05 | Added another structure plot to examine_topic_model_k16 analysis. |
| Rmd | 1899304 | Peter Carbonetto | 2022-05-05 | workflowr::wflow_publish(“examine_topic_model_k16.Rmd”) |
| html | b5a1eae | Peter Carbonetto | 2022-05-04 | Build site. |
| Rmd | a628c54 | Peter Carbonetto | 2022-05-04 | workflowr::wflow_publish(“examine_topic_model_k16.Rmd”, verbose = TRUE) |
| Rmd | e01a993 | Peter Carbonetto | 2022-05-04 | First build of overview page. |
Add text here giving an overview of this analysis.
Load the packages used in the analysis.
library(data.table)
library(fastTopics)
library(ggplot2)
library(cowplot)
source("../code/lps_data.R")Initialize the sequence of pseudorandom numbers.
set.seed(1)Load the count data.
dat <- read_lps_data("../data/raw_read_counts.csv.gz")
samples <- dat$samples
counts <- dat$countsRemove genes with very low (or no) expression.
j <- which(colSums(counts) > 20)
counts <- counts[,j]Load the results of the topic modeling analysis.
load("../output/fit-lps-k=16.RData")Plot the improvement in the solution over time.
p1 <- plot_progress(fit,x = "timing",y = "loglik",colors = "black",
add.point.every = 10,e = 1e-4) +
guides(color = "none",fill = "none",shape = "none",
linetype = "none",size = "none")
p2 <- plot_progress(fit,x = "timing",y = "res",colors = "black",
add.point.every = 10,e = 1e-4) +
guides(color = "none",fill = "none",shape = "none",
linetype = "none",size = "none")
plot_grid(p1,p2)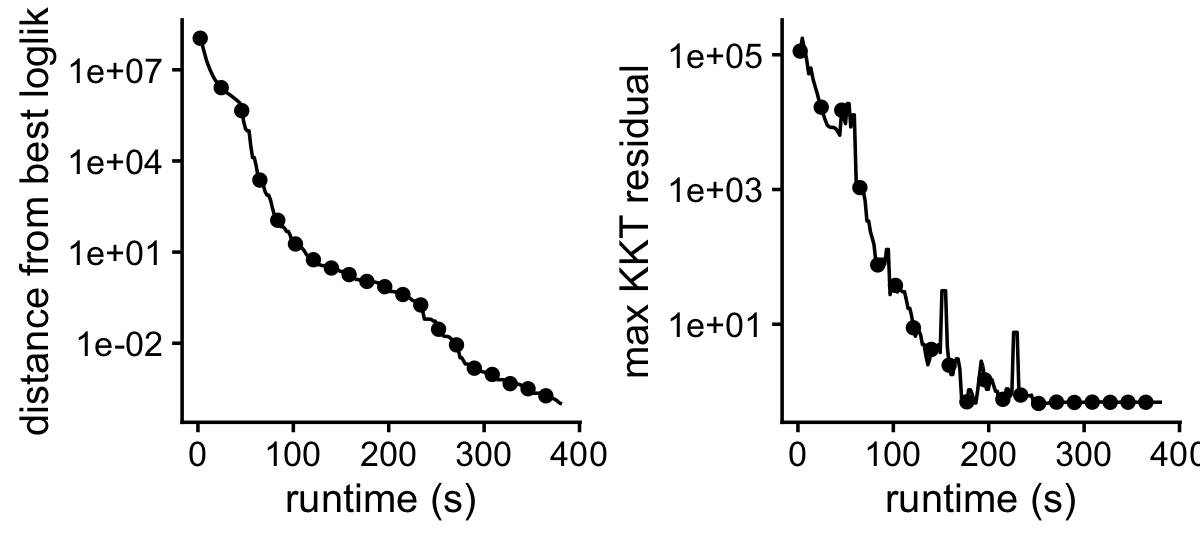
| Version | Author | Date |
|---|---|---|
| b5a1eae | Peter Carbonetto | 2022-05-04 |
Visualize the structure identified in each of the tissues using a Structure plot, in which the samples in each tissue are ordered by time in which the sample was taken:
set.seed(1)
rows <- order(samples$timepoint)
topic_colors <- c("darkblue","dodgerblue","darkorange","forestgreen",
"limegreen","tomato","darkred","olivedrab","magenta",
"darkmagenta","sienna","royalblue","lightskyblue",
"gold","red","cyan")
p <- structure_plot(fit,grouping = samples$tissue,gap = 5,
colors = topic_colors,
topics = c(15,3,4,5,6,7,8,9,10,11,12,13,2,14,1,16),
loadings_order = rows) +
theme(legend.key.height = unit(0.15,"cm"),
legend.text = element_text(size = 7))
print(p)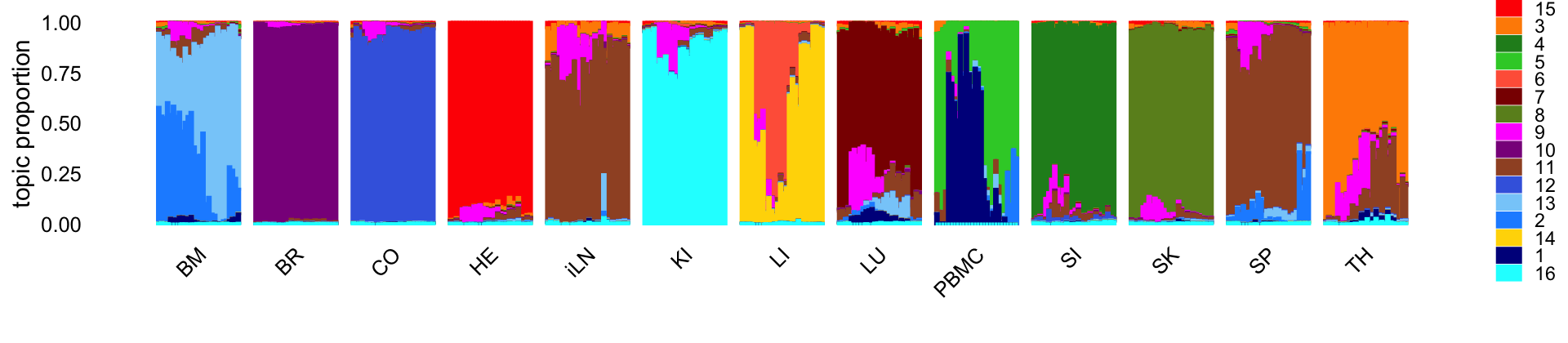
| Version | Author | Date |
|---|---|---|
| 6dabc4d | Peter Carbonetto | 2022-05-05 |
There is a single topic (topic 9, blue in the plot below) that is capturing changes in expression over time across many tissues. Two other topics (topics 1 and 6) show similar patterns, except these patterns are specific to two tissues (PBMC and LI).
set.seed(1)
topic_colors <- c("gold","darkorange","dodgerblue","white")
fit2 <- poisson2multinom(fit)
fit2 <- merge_topics(fit2,paste0("k",setdiff(1:16,c(1,6,9))))
p <- structure_plot(fit2,grouping = samples$tissue,gap = 5,
colors = topic_colors,topics = c(4,1:3),
loadings_order = rows)
print(p)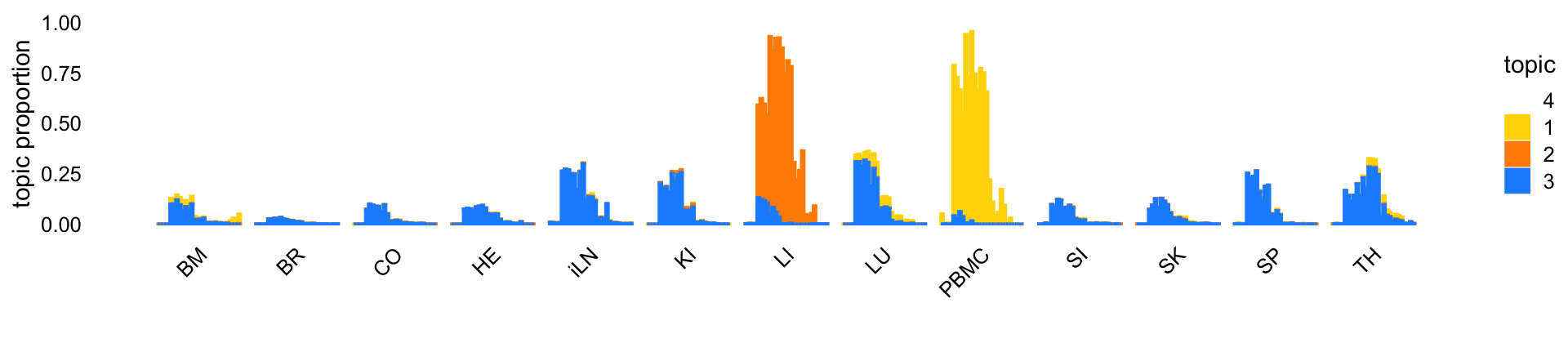
| Version | Author | Date |
|---|---|---|
| 6dabc4d | Peter Carbonetto | 2022-05-05 |
These volcano plots summarize the results of the DE analysis for topics capturing different tissues (and topic 9, which is capturing changes in expression at different time points):
topics <- colnames(de_merged$z)
p <- vector("list",13)
names(p) <- topics
for (k in topics) {
p[[k]] <- volcano_plot(de_merged,k = k,ymax = 500) +
guides(fill = "none")
volcano_plotly(de_merged,k = k,ymax = 500,
file = paste("volcano_plot",k,sep = "_"))
}
# This version of bslib is designed to work with shiny version 1.6.0 or higher.
# Warning: `arrange_()` was deprecated in dplyr 0.7.0.
# Please use `arrange()` instead.
# See vignette('programming') for more help
# This warning is displayed once every 8 hours.
# Call `lifecycle::last_warnings()` to see where this warning was generated.
# Warning: Ignoring 290 observations
# Warning: Ignoring 222 observations
# Warning: Ignoring 234 observations
# Warning: Ignoring 207 observations
# Warning: Ignoring 107 observations
# Warning: Ignoring 228 observations
# Warning: Ignoring 267 observations
# Warning: Ignoring 227 observations
# Warning: Ignoring 244 observations
# Warning: Ignoring 240 observations
# Warning: Ignoring 187 observations
# Warning: Ignoring 251 observations
# Warning: Ignoring 246 observations
do.call("plot_grid",c(p,list(ncol = 3,nrow = 5)))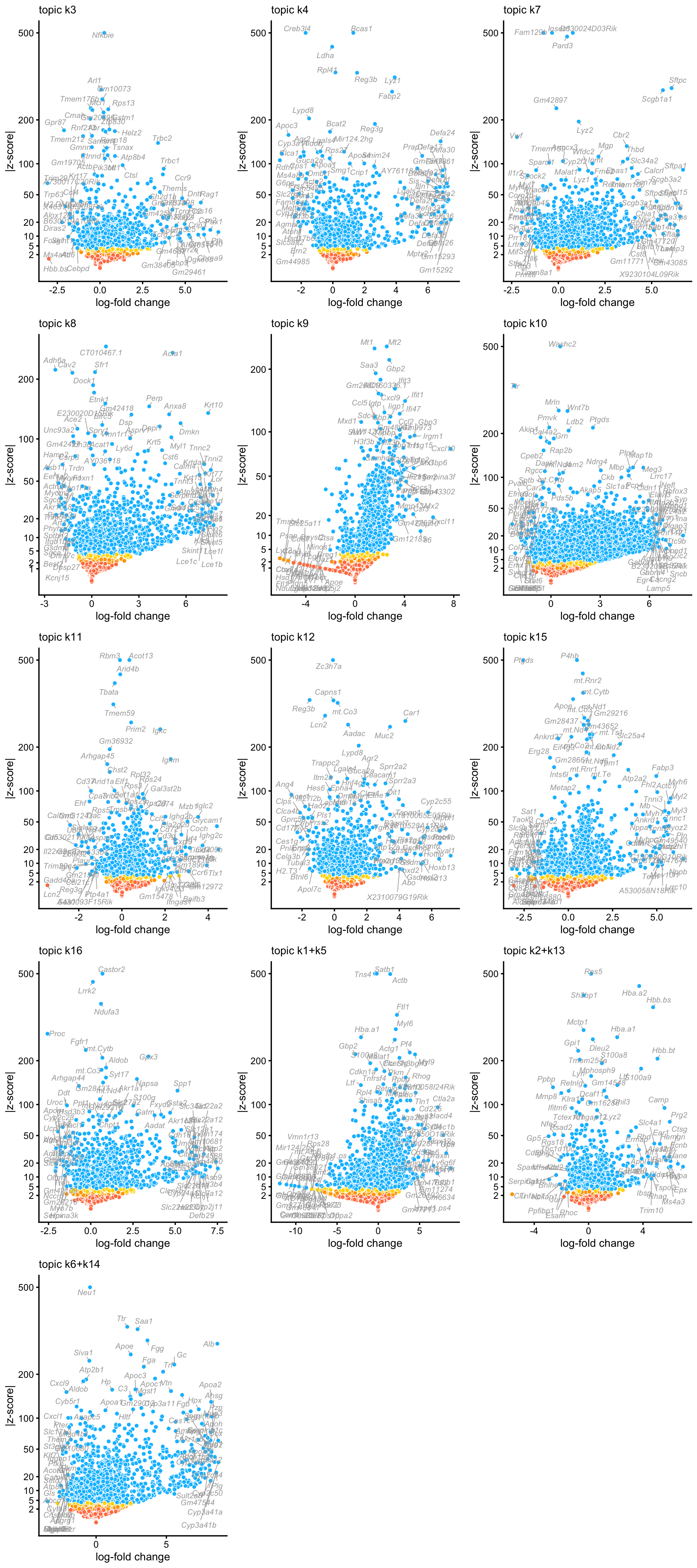
These volcano plots summarize the results of the DE analysis for topics capturing expression patterns within tissues:
topics <- c("k1","k2","k5","k6","k13","k14")
p <- vector("list",6)
names(p) <- topics
for (k in topics) {
p[[k]] <- volcano_plot(de,k = k,ymax = 300) +
guides(fill = "none")
volcano_plotly(de,k = k,ymax = 300,
file = paste("volcano_plot",k,sep = "_"))
}
# Warning: Ignoring 113 observations
# Warning: Ignoring 67 observations
# Warning: Ignoring 114 observations
# Warning: Ignoring 107 observations
# Warning: Ignoring 115 observations
# Warning: Ignoring 121 observations
do.call("plot_grid",c(p,list(ncol = 3,nrow = 2)))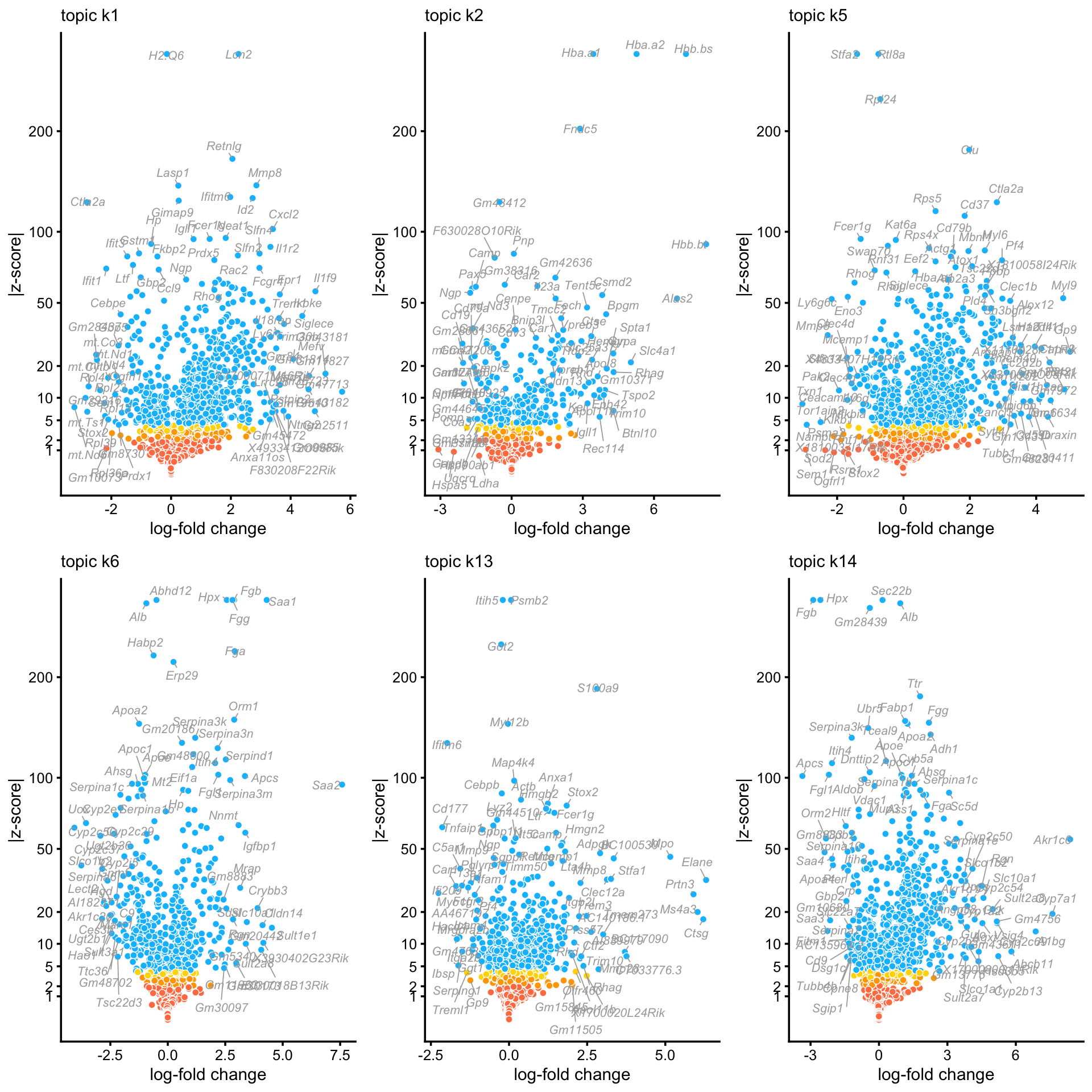
sessionInfo()
# R version 3.6.2 (2019-12-12)
# Platform: x86_64-apple-darwin15.6.0 (64-bit)
# Running under: macOS Catalina 10.15.7
#
# Matrix products: default
# BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
# LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
#
# locale:
# [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
#
# attached base packages:
# [1] stats graphics grDevices utils datasets methods base
#
# other attached packages:
# [1] cowplot_1.0.0 ggplot2_3.3.5 fastTopics_0.6-117 data.table_1.12.8
#
# loaded via a namespace (and not attached):
# [1] mcmc_0.9-6 fs_1.5.2 progress_1.2.2 httr_1.4.2
# [5] rprojroot_1.3-2 tools_3.6.2 backports_1.1.5 bslib_0.3.1
# [9] utf8_1.1.4 R6_2.4.1 irlba_2.3.3 uwot_0.1.10
# [13] DBI_1.1.0 lazyeval_0.2.2 colorspace_1.4-1 withr_2.5.0
# [17] tidyselect_1.1.1 prettyunits_1.1.1 compiler_3.6.2 git2r_0.29.0
# [21] quantreg_5.54 SparseM_1.78 plotly_4.9.2 labeling_0.3
# [25] sass_0.4.0 scales_1.1.0 SQUAREM_2017.10-1 quadprog_1.5-8
# [29] pbapply_1.5-1 mixsqp_0.3-46 stringr_1.4.0 digest_0.6.23
# [33] rmarkdown_2.11 R.utils_2.11.0 MCMCpack_1.4-5 pkgconfig_2.0.3
# [37] htmltools_0.5.2 fastmap_1.1.0 invgamma_1.1 highr_0.8
# [41] htmlwidgets_1.5.1 rlang_0.4.11 shiny_1.4.0 jquerylib_0.1.4
# [45] generics_0.0.2 farver_2.0.1 jsonlite_1.7.2 crosstalk_1.0.0
# [49] dplyr_1.0.7 R.oo_1.24.0 magrittr_2.0.1 Matrix_1.2-18
# [53] Rcpp_1.0.7 munsell_0.5.0 fansi_0.4.0 lifecycle_1.0.0
# [57] R.methodsS3_1.8.1 stringi_1.4.3 whisker_0.4 yaml_2.2.0
# [61] MASS_7.3-51.4 Rtsne_0.15 grid_3.6.2 parallel_3.6.2
# [65] promises_1.1.0 ggrepel_0.9.1 crayon_1.4.1 lattice_0.20-38
# [69] hms_1.1.0 knitr_1.37 pillar_1.6.2 glue_1.4.2
# [73] evaluate_0.14 RcppParallel_5.1.5 vctrs_0.3.8 httpuv_1.5.2
# [77] MatrixModels_0.4-1 gtable_0.3.0 purrr_0.3.4 tidyr_1.1.3
# [81] assertthat_0.2.1 ashr_2.2-54 xfun_0.29 mime_0.8
# [85] xtable_1.8-4 coda_0.19-3 later_1.0.0 viridisLite_0.3.0
# [89] truncnorm_1.0-8 tibble_3.1.3 workflowr_1.7.0 ellipsis_0.3.2