updated_resampling_strategy2
Haider Inam
5/30/2021
Last updated: 2021-06-19
Checks: 7 0
Knit directory: ~/Box/RProjects/pair_con_select/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190211) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 3155461. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.RData
Ignored: analysis/.Rhistory
Ignored: analysis/.Rproj.user/
Ignored: data/depmap_alkati/Data_Raw/CCLE/CCLE_RNAseq_ExonUsageRatio_20180929.gct
Ignored: data/skmel28_sos1_mekq56p_vemurafenib.csv.sb-ea24b981-dvFz4V/
Ignored: data/tcga_brca_expression/
Ignored: data/tcga_luad_expression/
Ignored: data/tcga_skcm_expression/
Ignored: output/alkati_filtercutoff_allfilters.csv
Untracked files:
Untracked: paircon_boxplot_alkati.pdf
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/updated_resampling_strategy2.Rmd) and HTML (docs/updated_resampling_strategy2.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 3155461 | haiderinam | 2021-06-19 | updated method for pairwise comparisons |
| Rmd | 8520286 | haiderinam | 2021-06-14 | Updated website contents on 061421 |
| Rmd | 58c60f1 | haiderinam | 2021-06-10 | 061021 Updates |
# rm(list=ls())
library(knitr)
library(tictoc)
library(workflowr)
library(VennDiagram)Loading required package: gridLoading required package: futile.loggerlibrary(dplyr)Warning: package 'dplyr' was built under R version 4.0.2
Attaching package: 'dplyr'The following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
intersect, setdiff, setequal, unionlibrary(foreach)
library(doParallel)Loading required package: iteratorsLoading required package: parallellibrary(ggplot2)Warning: package 'ggplot2' was built under R version 4.0.2library(reshape2)
library(RColorBrewer)
library(devtools)Loading required package: usethisError in get(genname, envir = envir) : object 'testthat_print' not foundlibrary(ggsignif)
library(plotly)
Attaching package: 'plotly'The following object is masked from 'package:ggplot2':
last_plotThe following object is masked from 'package:stats':
filterThe following object is masked from 'package:graphics':
layoutlibrary(BiocManager)Bioconductor version 3.12 (BiocManager 1.30.10), ?BiocManager::install for helpBioconductor version '3.12' is out-of-date; the current release version '3.13'
is available with R version '4.1'; see https://bioconductor.org/install
Attaching package: 'BiocManager'The following object is masked from 'package:devtools':
install# library(ensembldb) #Loading this with Dplyr commands seems to throw an error in Rmd
# library(EnsDb.Hsapiens.v86) #Loading this with Dplyr commands seems to throw an error in Rmd
source("code/contab_maker.R")
source("code/alldata_compiler.R")
source("code/quadratic_solver.R")
source("code/mut_excl_genes_generator.R")
source("code/mut_excl_genes_datapoints.R")
source("code/simresults_generator.R")
# source("../code/contab_maker.R")
# source("../code/alldata_compiler.R")
# source("../code/quadratic_solver.R")
# source("../code/mut_excl_genes_generator.R")
# source("../code/mut_excl_genes_datapoints.R")
# source("../code/simresults_generator.R")
######################Cleanup for GGPlot2#########################################
cleanup=theme_bw() +
theme(plot.title = element_text(hjust=.5),
panel.grid.major = element_blank(),
panel.grid.major.y = element_blank(),
panel.background = element_blank(),
# axis.line = element_line(color = "black"),
axis.text = element_text(face="bold",color="black",size="11"),
text=element_text(size=11,face="bold"),
axis.title=element_text(face="bold",size="11")) nameposctrl1<-'BRAF'
#Positive control 1
nameposctrl2<-'NRAS'
#Oncogene in Question
namegene<-'ATI'
#Mutation Boolean (Y or N)
mtn<-'N'
#Name Mutation for Positive Ctrl 1
nameposctrl1mt<-'V600E'
#Name of Mutation for Positive Ctrl 2
nameposctrl2mt<-'Q61L'
alldata=read.csv("output/all_data_skcm.csv",sep=",",header=T,stringsAsFactors=F)
# alldata=read.csv("../output/all_data_skcm.csv",sep=",",header=T,stringsAsFactors=F)
head(alldata) X Patid mean_RPKM_1.19 mean_RPKM_20.29 Ratio20.29 mRNA_count BRAF
1 1 TCGA-BF-A1PU 0.62445977 1.24042009 1.986389 948 V600E
2 2 TCGA-BF-A1PV 0.02099345 0.15815619 7.533598 82 NaN
3 3 TCGA-BF-A1PX 0.01752838 0.09612414 5.483914 92 V600E
4 4 TCGA-BF-A1PZ 0.19874434 7.27553619 36.607514 2822 NaN
5 5 TCGA-BF-A1Q0 2.13353636 3.71661801 1.741999 2211 NaN
6 6 TCGA-BF-A3DJ 0.06244694 0.55656239 8.912565 281 V600E
NRAS RSEM_normalized ATI
1 NaN 107.1429 0
2 Q61L 8.9659 0
3 NaN 14.5985 0
4 Q61R 329.0810 1
5 NaN 277.0434 0
6 NaN 35.1542 0Contab downsampler function that the next code chunk uses:
contab_downsampler<- function(contab,prob) {
p11=contab[1,1]
p10=contab[1,2]
p01=contab[2,1]
p00=contab[2,2]
contab_downsampled=contab ###Just making a dummy contingency table
contab_downsampled[1,1]=prob*p11/(p10+p11)
contab_downsampled[1,2]=prob*p10/(p10+p11)
contab_downsampled[2,1]=(1-prob)*p01/(p00+p01)
contab_downsampled[2,2]=(1-prob)*p00/(p00+p01)
contab_downsampled
}mut_excl_genes_generator2=function(cohort_size,incidence,or_pair1,or_pair2){
# or_pair1=.9
# or_pair2=.01
#
# incidence=40
# cohort_size=1000
or_p=or_pair2
or_q=or_pair1
q00=0.05
p00=0.05
###PC1 vs GOI pair###
p_goi=incidence/cohort_size
q10=(1-q00)-p_goi
q11=(p_goi*(1/q00)*or_q*((1-q00)-p_goi))/(1+(1/q00)*or_q*((1-q00)-p_goi))
q01=(p_goi)/(1+(1/q00)*or_q*((1-q00)-p_goi))
###PC1 vs PC2 pair###
p_pc1=q11+q10
p01=(1-p00)-p_pc1
p11=(p_pc1*(1/p00)*or_p*((1-p00)-p_pc1))/(1+((1/p00)*or_p*((1-p00)-p_pc1)))
p10=(p_pc1)/(1+(1/p00)*or_p*((1-p00)-p_pc1))
# gene_pair_1=(as.numeric(c(q11,q10,q01,q00)))
# gene_pair_2=(as.numeric(c(p11,p10,p01,p00)))
gene_pair_1=(as.numeric(c(q11,q01,q10,q00)))
gene_pair_2=(as.numeric(c(p11,p10,p01,p00)))
return(list(gene_pair_1,gene_pair_2))
}
# mut_excl_genes_generator2(500,12,.1,.05)
# mut_excl_genes_generator2(500,36,.1,.05)
# mut_excl_genes_generator(500,12,.1,.05)
# mut_excl_genes_generator(500,36,.1,.05)Another function that takes in a contingency table, and resamples is based on its probabilities on a multinomial distribution. Number of resampling simulations are an input of the function
contab_simulator=function(contab,nsims,cohort_size){
contabs_resampled=rmultinom(nsims, #number of datasets to generate
cohort_size, #sample size of the total population
c(contab[1,1],
contab[1,2],
contab[2,1],
contab[2,2])) #probability of positive event
contabs_resampled_t=t(contabs_resampled)
colnames(contabs_resampled_t)=c("p11","p10","p01","p00")
contabs_resampled_t
}
# contab_simulator=function(contab,nsims,cohort_size){
# contabs_resampled=rmultinom(nsims, #number of datasets to generate
# 1, #sample size of the total population
# c(contab[1,1],
# contab[1,2],
# contab[2,1],
# contab[2,2])) #probability of positive event
# contabs_resampled_t=t(contabs_resampled)
# colnames(contabs_resampled_t)=c("p11","p10","p01","p00")
# contabs_resampled_t
# }
# contab=pc1pc2_contab_probabilities
# nsims=1000
# cohort_size=cohort_size_curr
# pc1rawpc2_contabs_sims=contab_simulator(pc1pc2_contab_probabilities,1000,cohort_size_curr)# rm(list=ls())
###Not mutation specific generation of counts###
alldata_comp=alldata_compiler(alldata,nameposctrl1,nameposctrl2,namegene,'N',"N/A","N/A")[[2]]
head(alldata_comp) X Patid Positive_Ctrl1 Positive_Ctrl2 genex rndmarray
1 1 TCGA-BF-A1PU 1 0 0 0
2 2 TCGA-BF-A1PV 0 1 0 1
3 3 TCGA-BF-A1PX 1 0 0 0
4 4 TCGA-BF-A1PZ 0 1 1 0
5 5 TCGA-BF-A1Q0 0 0 0 0
6 6 TCGA-BF-A3DJ 1 0 0 0###Calculating Odds ratios and GOI frequencies for the raw data###
cohort_size=length(alldata_comp$Positive_Ctrl1)
pc1pc2_contab_counts=contab_maker(alldata_comp$Positive_Ctrl1,alldata_comp$Positive_Ctrl2,alldata_comp)[2:1, 2:1]
goipc1_contab_counts=contab_maker(alldata_comp$genex,alldata_comp$Positive_Ctrl1,alldata_comp)[2:1, 2:1]
###Had to add the 2:1 bits because the contab maker spits out NN YY whereas we wanted YNYN
goipc2_contab_counts=contab_maker(alldata_comp$genex,alldata_comp$Positive_Ctrl2,alldata_comp)[2:1, 2:1]
# pc1pc2_contab_counts=gene_pair_2_table
# goipc1_contab_counts=contab_maker(alldata_comp$genex,alldata_comp$Positive_Ctrl1,alldata_comp)[2:1, 2:1]
# goipc1_contab_counts=gene_pair_1_table
cohort_size_curr=cohort_size
# goipc2_contab_counts=contab_maker(alldata_comp$genex,alldata_comp$Positive_Ctrl2,alldata_comp)[2:1, 2:1]
pc1pc2_contab_probabilities=pc1pc2_contab_counts/cohort_size_curr
goipc1_contab_probabilities=goipc1_contab_counts/cohort_size_curr
goipc2_contab_probabilities=goipc2_contab_counts/cohort_size_curr
# pc1pc2_contab_probabilities=pc1pc2_contab_counts
# goipc1_contab_probabilities=goipc1_contab_counts
# goipc2_contab_probabilities=goipc2_contab_counts/cohort_size
or_pc1pc2=pc1pc2_contab_probabilities[1,1]*pc1pc2_contab_probabilities[2,2]/(pc1pc2_contab_probabilities[1,2]*pc1pc2_contab_probabilities[2,1])
or_goipc1=goipc1_contab_probabilities[1,1]*goipc1_contab_probabilities[2,2]/(goipc1_contab_probabilities[1,2]*goipc1_contab_probabilities[2,1])
or_goipc2=goipc2_contab_probabilities[1,1]*goipc2_contab_probabilities[2,2]/(goipc2_contab_probabilities[1,2]*goipc2_contab_probabilities[2,1])
goi_freq=goipc1_contab_probabilities[1,1]+goipc1_contab_probabilities[1,2]
goi_freq=.25
# class(goi_freq)
###
###Downsampling PC1 to the probability of GOI without changing ORs###
###The function below converts contingency table data to a new contingency table in which the data is downsampled to the desired frequency, aka the frequency of the GOI in this case###
pc1new_pc2_contab=contab_downsampler(pc1pc2_contab_probabilities,goi_freq)
goinew_pc1_contab=contab_downsampler(goipc1_contab_probabilities,goi_freq)
goinew_pc2_contab=contab_downsampler(goipc2_contab_probabilities,goi_freq)
##original contab:
head(pc1pc2_contab_probabilities) [,1] [,2]
[1,] 0.01424501 0.4985755
[2,] 0.25925926 0.2279202###downsampled contab:
head(pc1new_pc2_contab) [,1] [,2]
[1,] 0.006944444 0.2430556
[2,] 0.399122807 0.3508772pc1rawpc2_contabs_sims=contab_simulator(pc1pc2_contab_probabilities,1000,cohort_size_curr)
pc1pc2_contabs_sims=contab_simulator(pc1new_pc2_contab,1000,cohort_size_curr)
goipc1_contabs_sims=contab_simulator(goinew_pc1_contab,1000,cohort_size_curr)
goipc2_contabs_sims=contab_simulator(goinew_pc2_contab,1000,cohort_size_curr)
# goipc2_contabs_sims=contab_simulator(goinew_pc2_contab,1000,cohort_size)
# head(pc1pc2_contabs_sims) #each row in this dataset is a new contab
pc1rawpc2_contabs_sims=data.frame(pc1rawpc2_contabs_sims)
pc1rawpc2_contabs_sims=pc1rawpc2_contabs_sims%>%
mutate(or=p11*p00/(p10*p01))
pc1pc2_contabs_sims=data.frame(pc1pc2_contabs_sims)
pc1pc2_contabs_sims=pc1pc2_contabs_sims%>%
mutate(or=p11*p00/(p10*p01))
goipc1_contabs_sims=data.frame(goipc1_contabs_sims)
goipc1_contabs_sims=goipc1_contabs_sims%>%
mutate(or=p11*p00/(p10*p01))
goipc2_contabs_sims=data.frame(goipc2_contabs_sims)
goipc2_contabs_sims=goipc2_contabs_sims%>%
mutate(or=p11*p00/(p10*p01))
# goipc2_contabs_sims=data.frame(goipc2_contabs_sims)
# goipc2_contabs_sims=goipc2_contabs_sims%>%
# mutate(or=p11*p00/(p10*p01))
pc1rawpc2_contabs_sims$comparison="pc1rawpc2"
pc1pc2_contabs_sims$comparison="pc1pc2"
goipc1_contabs_sims$comparison="goipc1"
goipc2_contabs_sims$comparison="goipc2"
or_median_raw=quantile(pc1rawpc2_contabs_sims$or,na.rm = T)[3]
or_uq_raw=quantile(pc1rawpc2_contabs_sims$or,na.rm = T)[4]
or_median_downsampled=quantile(pc1pc2_contabs_sims$or,na.rm = T)[3]
or_uq_downsampled=quantile(pc1pc2_contabs_sims$or,na.rm = T)[4]
pc1rawpc2_contabs_sims=pc1rawpc2_contabs_sims%>%
mutate(isgreater_raw_median=case_when(or>or_median_raw~1,
TRUE~0),
isgreater_raw_uq=case_when(or>or_uq_raw~1,
TRUE~0),
isgreater_median=case_when(or>or_median_downsampled~1,
TRUE~0),
isgreater_uq=case_when(or>or_uq_downsampled~1,
TRUE~0)
)
pc1pc2_contabs_sims=pc1pc2_contabs_sims%>%
mutate(isgreater_raw_median=case_when(or>or_median_raw~1,
TRUE~0),
isgreater_raw_uq=case_when(or>or_uq_raw~1,
TRUE~0),
isgreater_median=case_when(or>or_median_downsampled~1,
TRUE~0),
isgreater_uq=case_when(or>or_uq_downsampled~1,
TRUE~0)
)
goipc1_contabs_sims=goipc1_contabs_sims%>%
mutate(isgreater_raw_median=case_when(or>or_median_raw~1,
TRUE~0),
isgreater_raw_uq=case_when(or>or_uq_raw~1,
TRUE~0),
isgreater_median=case_when(or>or_median_downsampled~1,
TRUE~0),
isgreater_uq=case_when(or>or_uq_downsampled~1,
TRUE~0)
)
goipc2_contabs_sims=goipc2_contabs_sims%>%
mutate(isgreater_raw_median=case_when(or>or_median_raw~1,
TRUE~0),
isgreater_raw_uq=case_when(or>or_uq_raw~1,
TRUE~0),
isgreater_median=case_when(or>or_median_downsampled~1,
TRUE~0),
isgreater_uq=case_when(or>or_uq_downsampled~1,
TRUE~0)
)
# pc1pc2_contabs_sims=pc1pc2_contabs_sims%>%
# mutate(isgreater=case_when(or>=or_pc1pc2~1,
# TRUE~0))
# goipc1_contabs_sims=goipc1_contabs_sims%>%
# mutate(isgreater=case_when(or>=or_pc1pc2~1,
# TRUE~0))
# goipc2_contabs_sims=goipc2_contabs_sims%>%
# mutate(isgreater=case_when(or>=or_pc1pc2~1,
# TRUE~0))
pc1rawpc2_isgreater_raw_median=sum(pc1rawpc2_contabs_sims$isgreater_raw_median)
pc1rawpc2_isgreater_raw_uq=sum(pc1rawpc2_contabs_sims$isgreater_raw_uq)
pc1rawpc2_isgreater_median=sum(pc1rawpc2_contabs_sims$isgreater_median)
pc1rawpc2_isgreater_uq=sum(pc1rawpc2_contabs_sims$isgreater_uq)
pc1pc2_isgreater_raw_median=sum(pc1pc2_contabs_sims$isgreater_raw_median)
pc1pc2_isgreater_raw_uq=sum(pc1pc2_contabs_sims$isgreater_raw_uq)
pc1pc2_isgreater_median=sum(pc1pc2_contabs_sims$isgreater_median)
pc1pc2_isgreater_uq=sum(pc1pc2_contabs_sims$isgreater_uq)
goipc1_isgreater_raw_median=sum(goipc1_contabs_sims$isgreater_raw_median)
goipc1_isgreater_raw_uq=sum(goipc1_contabs_sims$isgreater_raw_uq)
goipc1_isgreater_median=sum(goipc1_contabs_sims$isgreater_median)
goipc1_isgreater_uq=sum(goipc1_contabs_sims$isgreater_uq)
plotting_df=rbind(pc1pc2_contabs_sims,goipc1_contabs_sims,goipc2_contabs_sims)
# plotting_df=rbind(pc1pc2_contabs_sims,goipc1_contabs_sims)
#
ggplot(plotting_df,aes(x=(or),fill=comparison))+
geom_histogram(bins=40,alpha=0.55,position="identity")+
# geom_histogram(bins=50,alpha=0.55)+
scale_y_continuous(expand=c(0,0),name="Count")+
scale_x_continuous(expand=c(0,0),trans="log10",name="Odds Ratio")+
scale_fill_brewer(palette="Set2")+
# geom_vline(xintercept = or_pc1pc2)+
cleanupWarning: Transformation introduced infinite values in continuous x-axisWarning: Removed 86 rows containing non-finite values (stat_bin).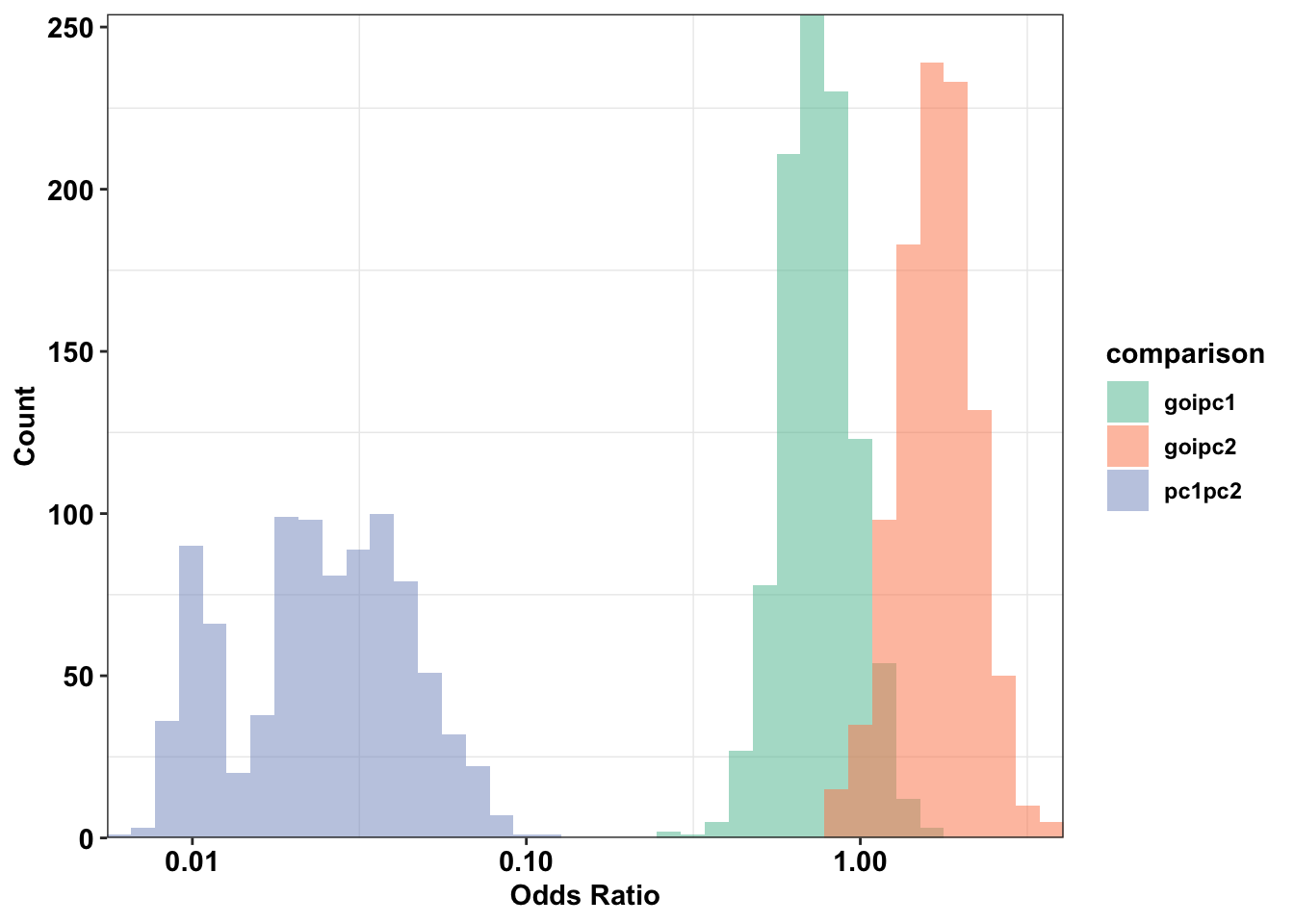
ggplot(plotting_df,aes(y=(or),x=comparison),fill=factor(comparison))+
geom_boxplot()+
scale_y_continuous(name="Odds Ratio",trans="log10")+
scale_x_discrete(name="")+
scale_fill_brewer(palette="Set2")+
geom_hline(yintercept = or_uq_downsampled,linetype="dashed")+
cleanup+
theme(legend.position = "none",
axis.ticks.x = element_blank())Warning: Transformation introduced infinite values in continuous y-axisWarning: Removed 86 rows containing non-finite values (stat_boxplot).
# ggsave("paircon_boxplot.pdf",width = 3,height=2,units="in",useDingbats=F)
ggplot(plotting_df%>%filter(comparison%in%"pc1pc2"),aes(x=or,fill=comparison))+
geom_histogram(bins=50,alpha=0.55,position="identity")+
geom_density(alpha=.6)+
scale_y_continuous(expand=c(0,0),name="Count")+
scale_x_continuous(expand=c(0,0),name="Odds Ratio")+
# geom_vline(xintercept = or_pc1pc2)+
cleanup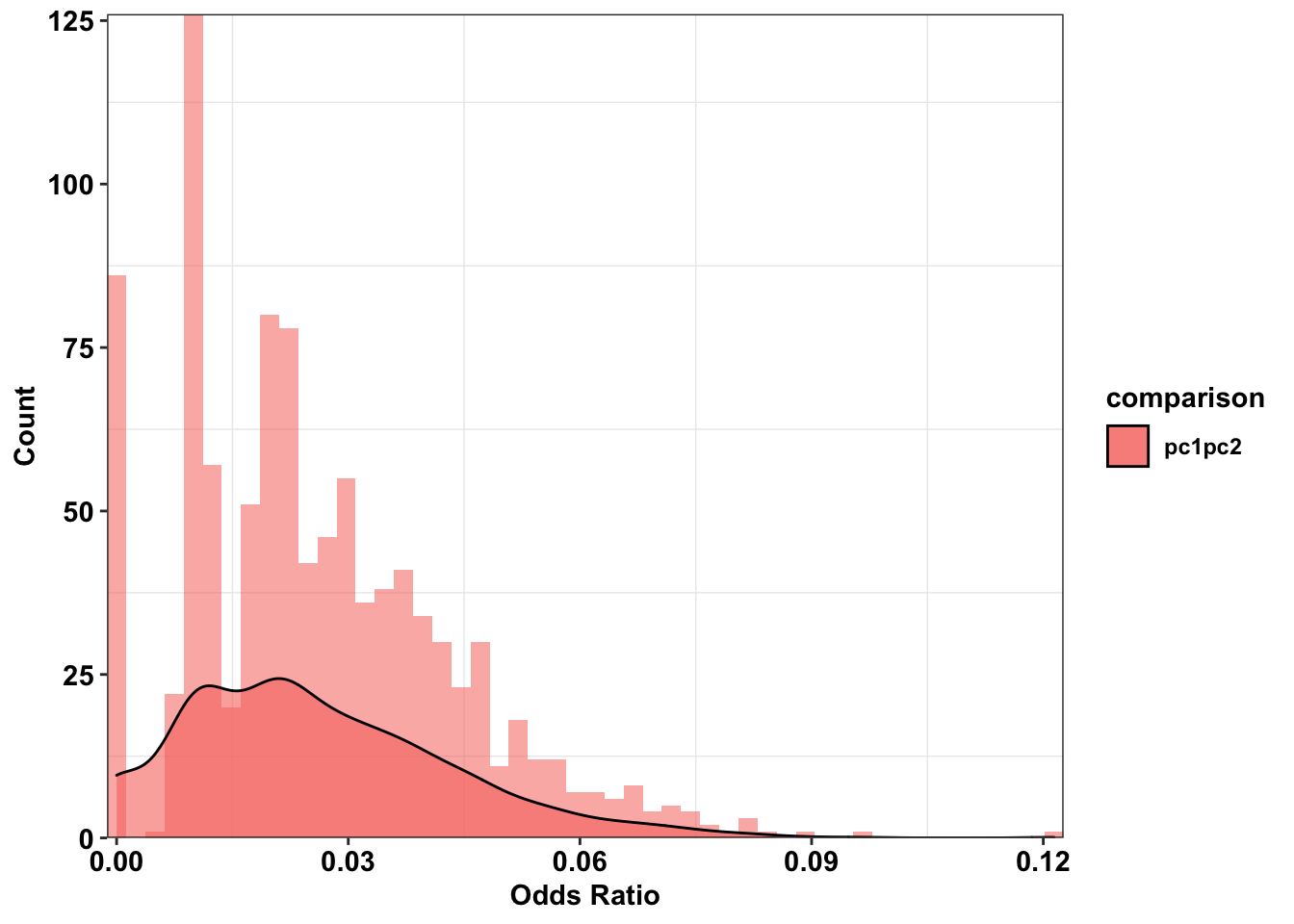
# rm(list=ls())
###Not mutation specific generation of counts###
alldata_comp=alldata_compiler(alldata,nameposctrl1,nameposctrl2,namegene,'N',"N/A","N/A")[[2]]
###Mutation specific
####Decided to not look at mutation specific counts too closely because co-occurrence of brafv600e and nrasq61L is a VERY high bar
# alldata_comp=alldata_compiler(alldata,nameposctrl1,nameposctrl2,namegene,'Y',"V600E","Q61L")[[2]]
head(alldata_comp) X Patid Positive_Ctrl1 Positive_Ctrl2 genex rndmarray
1 1 TCGA-BF-A1PU 1 0 0 0
2 2 TCGA-BF-A1PV 0 1 0 1
3 3 TCGA-BF-A1PX 1 0 0 0
4 4 TCGA-BF-A1PZ 0 1 1 0
5 5 TCGA-BF-A1Q0 0 0 0 0
6 6 TCGA-BF-A3DJ 1 0 0 0###Calculating Odds ratios and GOI frequencies for the raw data###
cohort_size=length(alldata_comp$Positive_Ctrl1)
pc1pc2_contab_counts=contab_maker(alldata_comp$Positive_Ctrl1,alldata_comp$Positive_Ctrl2,alldata_comp)[2:1, 2:1]
pc1pc2_contab_counts=rbind(c(15,25),c(81,230)) ###NRAS ATI
goipc1_contab_counts=contab_maker(alldata_comp$genex,alldata_comp$Positive_Ctrl1,alldata_comp)[2:1, 2:1]
# goipc1_contab_counts=rbind(c(1,110),c(186,365)) ###EGFR KRAS
fisher.test(goipc1_contab_counts)
Fisher's Exact Test for Count Data
data: goipc1_contab_counts
p-value = 0.4071
alternative hypothesis: true odds ratio is not equal to 1
95 percent confidence interval:
0.365048 1.535988
sample estimates:
odds ratio
0.7531399 ###Had to add the 2:1 bits because the contab maker spits out NN YY whereas we wanted YNYN
goipc2_contab_counts=contab_maker(alldata_comp$genex,alldata_comp$Positive_Ctrl2,alldata_comp)[2:1, 2:1]
# pc1pc2_contab_counts=gene_pair_2_table
# goipc1_contab_counts=contab_maker(alldata_comp$genex,alldata_comp$Positive_Ctrl1,alldata_comp)[2:1, 2:1]
# goipc1_contab_counts=gene_pair_1_table
cohort_size_curr=cohort_size
# goipc2_contab_counts=contab_maker(alldata_comp$genex,alldata_comp$Positive_Ctrl2,alldata_comp)[2:1, 2:1]
pc1pc2_contab_probabilities=pc1pc2_contab_counts/cohort_size_curr
goipc1_contab_probabilities=goipc1_contab_counts/cohort_size_curr
goipc2_contab_probabilities=goipc2_contab_counts/cohort_size_curr
# pc1pc2_contab_probabilities=pc1pc2_contab_counts
# goipc1_contab_probabilities=goipc1_contab_counts
# goipc2_contab_probabilities=goipc2_contab_counts/cohort_size
or_pc1pc2=pc1pc2_contab_probabilities[1,1]*pc1pc2_contab_probabilities[2,2]/(pc1pc2_contab_probabilities[1,2]*pc1pc2_contab_probabilities[2,1])
or_goipc1=goipc1_contab_probabilities[1,1]*goipc1_contab_probabilities[2,2]/(goipc1_contab_probabilities[1,2]*goipc1_contab_probabilities[2,1])
or_goipc2=goipc2_contab_probabilities[1,1]*goipc2_contab_probabilities[2,2]/(goipc2_contab_probabilities[1,2]*goipc2_contab_probabilities[2,1])
goi_freq=goipc1_contab_probabilities[1,1]+goipc1_contab_probabilities[1,2]
# goi_freq=.25
# class(goi_freq)
###
###Downsampling PC1 to the probability of GOI without changing ORs###
###The function below converts contingency table data to a new contingency table in which the data is downsampled to the desired frequency, aka the frequency of the GOI in this case###
pc1new_pc2_contab=contab_downsampler(pc1pc2_contab_probabilities,goi_freq)
goinew_pc1_contab=contab_downsampler(goipc1_contab_probabilities,goi_freq)
goinew_pc2_contab=contab_downsampler(goipc2_contab_probabilities,goi_freq)
##original contab:
head(pc1pc2_contab_probabilities) [,1] [,2]
[1,] 0.04273504 0.07122507
[2,] 0.23076923 0.65527066###downsampled contab:
head(pc1new_pc2_contab) [,1] [,2]
[1,] 0.04273504 0.07122507
[2,] 0.23076923 0.65527066pc1rawpc2_contabs_sims=contab_simulator(pc1pc2_contab_probabilities,1000,cohort_size_curr)
pc1pc2_contabs_sims=contab_simulator(pc1new_pc2_contab,1000,cohort_size_curr)
goipc1_contabs_sims=contab_simulator(goinew_pc1_contab,1000,cohort_size_curr)
goipc2_contabs_sims=contab_simulator(goinew_pc2_contab,1000,cohort_size_curr)
# goipc2_contabs_sims=contab_simulator(goinew_pc2_contab,1000,cohort_size)
# head(pc1pc2_contabs_sims) #each row in this dataset is a new contab
pc1rawpc2_contabs_sims=data.frame(pc1rawpc2_contabs_sims)
pc1rawpc2_contabs_sims=pc1rawpc2_contabs_sims%>%
mutate(or=p11*p00/(p10*p01))
pc1pc2_contabs_sims=data.frame(pc1pc2_contabs_sims)
pc1pc2_contabs_sims=pc1pc2_contabs_sims%>%
mutate(or=p11*p00/(p10*p01))
goipc1_contabs_sims=data.frame(goipc1_contabs_sims)
goipc1_contabs_sims=goipc1_contabs_sims%>%
mutate(or=p11*p00/(p10*p01))
goipc2_contabs_sims=data.frame(goipc2_contabs_sims)
goipc2_contabs_sims=goipc2_contabs_sims%>%
mutate(or=p11*p00/(p10*p01))
# goipc2_contabs_sims=data.frame(goipc2_contabs_sims)
# goipc2_contabs_sims=goipc2_contabs_sims%>%
# mutate(or=p11*p00/(p10*p01))
pc1rawpc2_contabs_sims$comparison="pc1rawpc2"
pc1pc2_contabs_sims$comparison="pc1pc2"
goipc1_contabs_sims$comparison="goipc1"
goipc2_contabs_sims$comparison="goipc2"
or_median_raw=quantile(pc1rawpc2_contabs_sims$or,na.rm = T)[3]
or_uq_raw=quantile(pc1rawpc2_contabs_sims$or,na.rm = T)[4]
or_median_downsampled=quantile(pc1pc2_contabs_sims$or,na.rm = T)[3]
or_uq_downsampled=quantile(pc1pc2_contabs_sims$or,na.rm = T)[4]
pc1rawpc2_contabs_sims=pc1rawpc2_contabs_sims%>%
mutate(isgreater_raw_median=case_when(or>=or_median_raw~1,
TRUE~0),
isgreater_raw_uq=case_when(or>or_uq_raw~1,
TRUE~0),
isgreater_median=case_when(or>or_median_downsampled~1,
TRUE~0),
isgreater_uq=case_when(or>or_uq_downsampled~1,
TRUE~0)
)
pc1pc2_contabs_sims=pc1pc2_contabs_sims%>%
mutate(isgreater_raw_median=case_when(or>or_median_raw~1,
TRUE~0),
isgreater_raw_uq=case_when(or>or_uq_raw~1,
TRUE~0),
isgreater_median=case_when(or>or_median_downsampled~1,
TRUE~0),
isgreater_uq=case_when(or>or_uq_downsampled~1,
TRUE~0)
)
goipc1_contabs_sims=goipc1_contabs_sims%>%
mutate(isgreater_raw_median=case_when(or>or_median_raw~1,
TRUE~0),
isgreater_raw_uq=case_when(or>or_uq_raw~1,
TRUE~0),
isgreater_median=case_when(or>or_median_downsampled~1,
TRUE~0),
isgreater_uq=case_when(or>or_uq_downsampled~1,
TRUE~0)
)
goipc2_contabs_sims=goipc2_contabs_sims%>%
mutate(isgreater_raw_median=case_when(or>or_median_raw~1,
TRUE~0),
isgreater_raw_uq=case_when(or>or_uq_raw~1,
TRUE~0),
isgreater_median=case_when(or>or_median_downsampled~1,
TRUE~0),
isgreater_uq=case_when(or>or_uq_downsampled~1,
TRUE~0)
)
# pc1pc2_contabs_sims=pc1pc2_contabs_sims%>%
# mutate(isgreater=case_when(or>=or_pc1pc2~1,
# TRUE~0))
# goipc1_contabs_sims=goipc1_contabs_sims%>%
# mutate(isgreater=case_when(or>=or_pc1pc2~1,
# TRUE~0))
# goipc2_contabs_sims=goipc2_contabs_sims%>%
# mutate(isgreater=case_when(or>=or_pc1pc2~1,
# TRUE~0))
pc1rawpc2_isgreater_raw_median=sum(pc1rawpc2_contabs_sims$isgreater_raw_median)
pc1rawpc2_isgreater_raw_uq=sum(pc1rawpc2_contabs_sims$isgreater_raw_uq)
pc1rawpc2_isgreater_median=sum(pc1rawpc2_contabs_sims$isgreater_median)
pc1rawpc2_isgreater_uq=sum(pc1rawpc2_contabs_sims$isgreater_uq)
pc1pc2_isgreater_raw_median=sum(pc1pc2_contabs_sims$isgreater_raw_median)
pc1pc2_isgreater_raw_uq=sum(pc1pc2_contabs_sims$isgreater_raw_uq)
pc1pc2_isgreater_median=sum(pc1pc2_contabs_sims$isgreater_median)
pc1pc2_isgreater_uq=sum(pc1pc2_contabs_sims$isgreater_uq)
goipc1_isgreater_raw_median=sum(goipc1_contabs_sims$isgreater_raw_median)
goipc1_isgreater_raw_uq=sum(goipc1_contabs_sims$isgreater_raw_uq)
goipc1_isgreater_median=sum(goipc1_contabs_sims$isgreater_median)
goipc1_isgreater_uq=sum(goipc1_contabs_sims$isgreater_uq)
plotting_df=rbind(pc1pc2_contabs_sims,goipc1_contabs_sims,goipc2_contabs_sims)
# plotting_df=rbind(pc1pc2_contabs_sims,goipc1_contabs_sims)
#
# a=plotting_df%>%filter(comparison%in%"pc1pc2")
ggplot(plotting_df,aes(x=(or),fill=comparison))+
geom_histogram(bins=40,alpha=0.55,position="identity")+
# geom_histogram(bins=50,alpha=0.55)+
scale_y_continuous(expand=c(0,0),name="Count")+
scale_x_continuous(expand=c(0,0),trans="log10",name="Odds Ratio")+
scale_fill_brewer(palette="Set2")+
# geom_vline(xintercept = or_pc1pc2)+
cleanup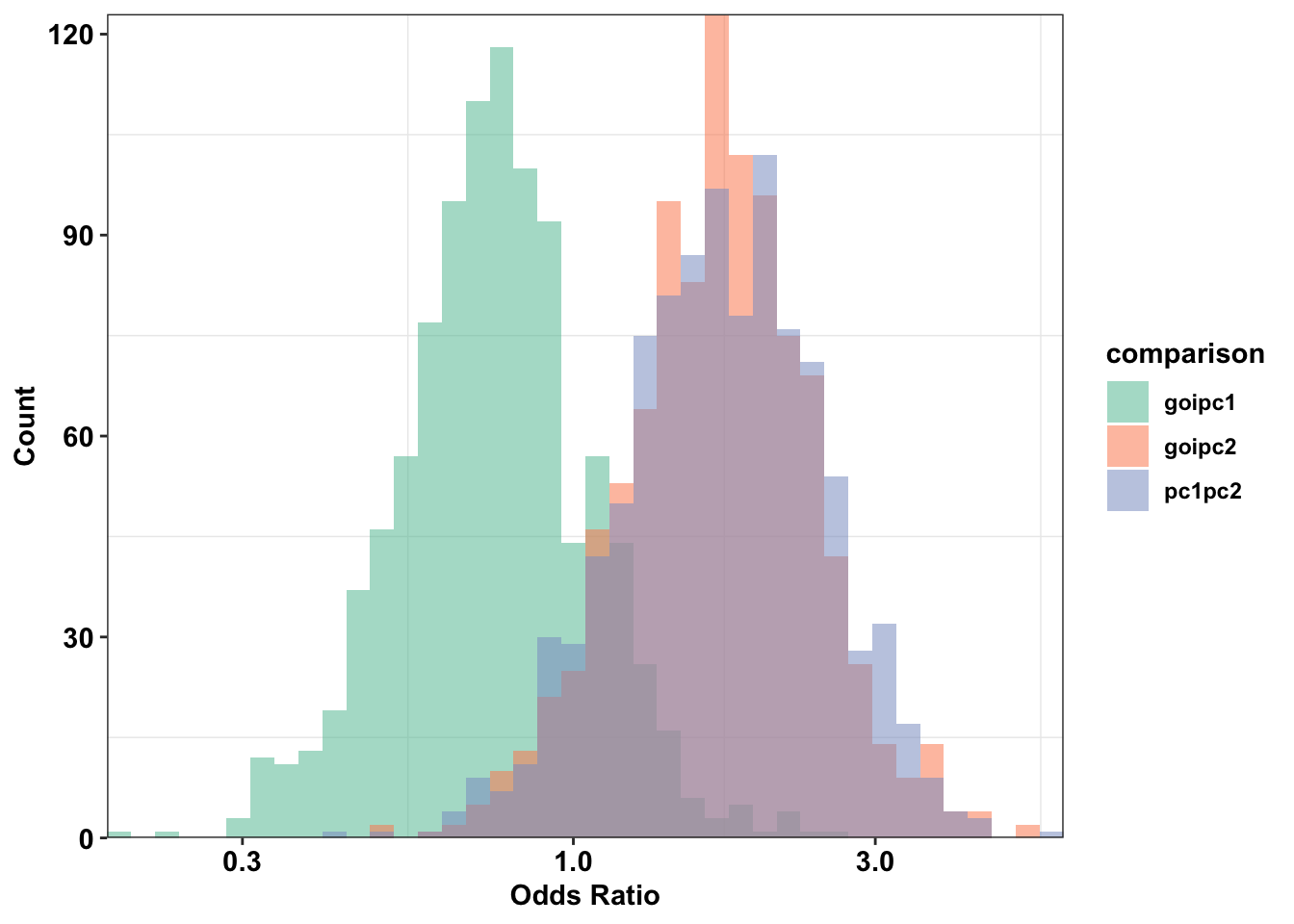
ggplot(plotting_df,aes(y=(or),x=comparison),fill=factor(comparison))+
geom_boxplot()+
scale_y_continuous(name="Odds Ratio",trans="log10")+
scale_x_discrete(name="")+
scale_fill_brewer(palette="Set2")+
geom_hline(yintercept = or_uq_downsampled,linetype="dashed")+
cleanup+
theme(legend.position = "none",
axis.ticks.x = element_blank())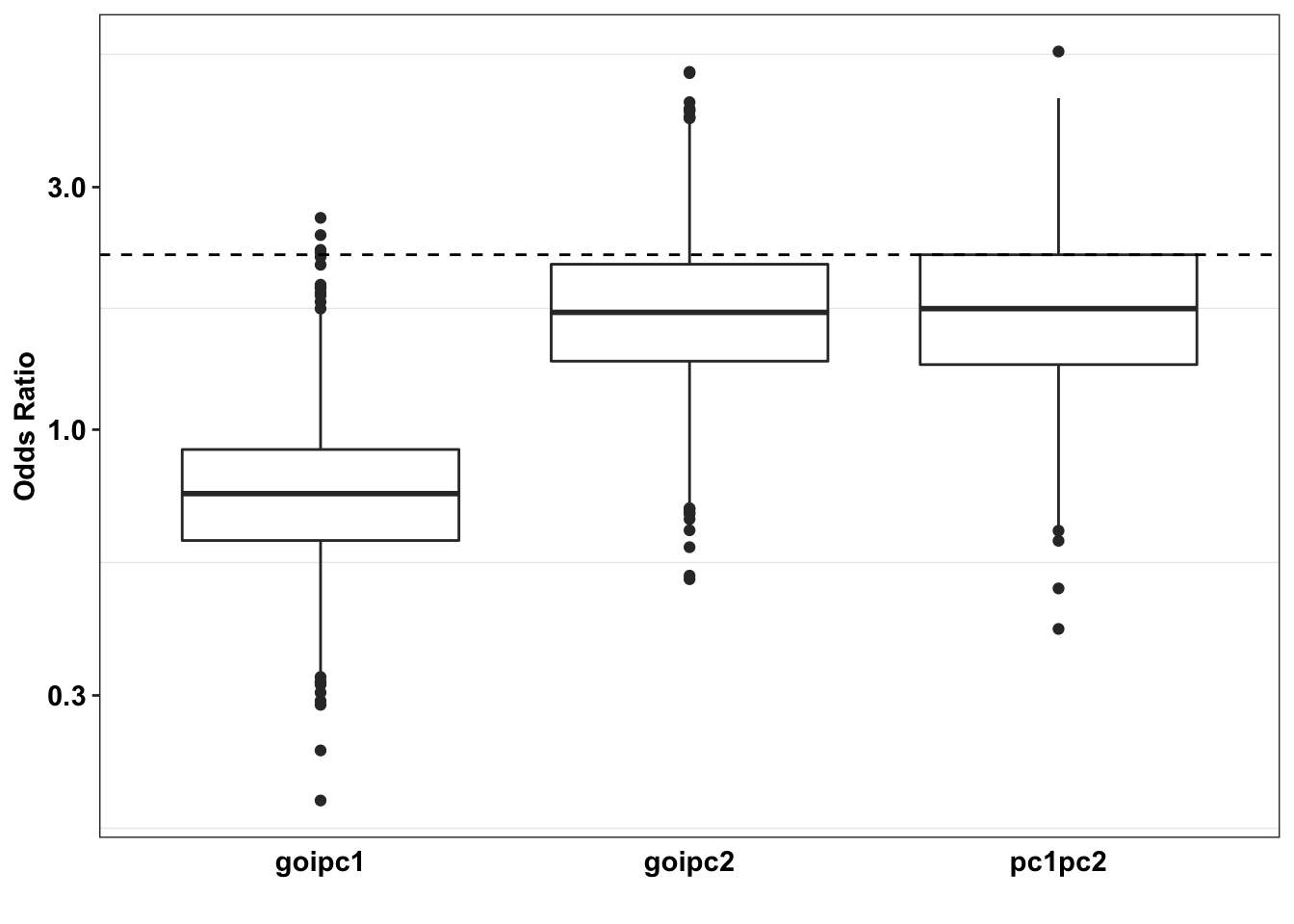
ggsave("paircon_boxplot_alkati.pdf",width = 3,height=2,units="in",useDingbats=F)
ggplot(plotting_df%>%filter(comparison%in%"pc1pc2"),aes(x=or,fill=comparison))+
geom_histogram(bins=50,alpha=0.55,position="identity")+
geom_density(alpha=.6)+
scale_y_continuous(expand=c(0,0),name="Count")+
scale_x_continuous(expand=c(0,0),name="Odds Ratio")+
# geom_vline(xintercept = or_pc1pc2)+
cleanup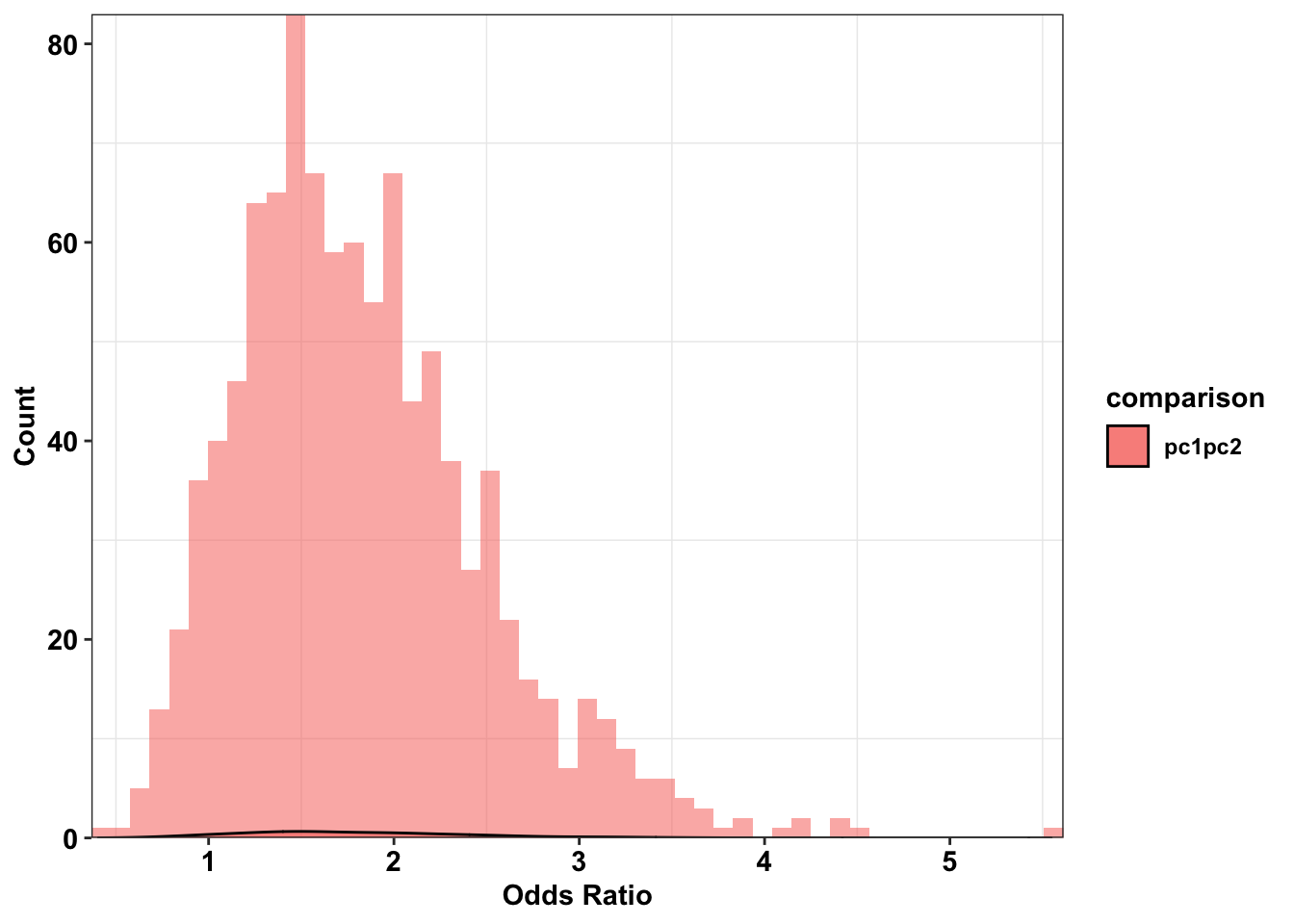
# rm(list=ls())
or_pair1=c(.01,.05,.1,.2,.3,.4,.5,.6,.7,.8,.9,1)
or_pair2=seq(.01,.2,by=.02)
incidence=seq(4,36,by=2)
cohort_size=seq(100,1000,by=200)
# or_pair1=c(.01,.05)
# or_pair2=seq(.01,.1,by=.05)
# incidence=seq(4,36,by=20)
# cohort_size=seq(100,1000,by=400)
# or_pair1=.05
# or_pair2=.01
# incidence=12
# cohort_size=500
# i=1
# j=1
# k=1
# l=1
true_or_vals=or_pair1
cohort_size_vals=cohort_size
gene1_total_vals=incidence
simresults_compiled_alldata=as.list(length(or_pair2)*length(or_pair1)*length(incidence)*length(cohort_size))
simresults_compiled=matrix(,length(or_pair2)*length(or_pair1)*length(incidence)*length(cohort_size),ncol=16)
ct=1
for(j in 1:length(incidence)){
tic()
for(l in 1:length(or_pair2)){
for(k in 1:length(cohort_size_vals)){
for(i in 1:length(true_or_vals)){
gene_pair_1=unlist(mut_excl_genes_generator(cohort_size[k],incidence[j],or_pair1[i],or_pair2[l])[1])
gene_pair_1_table=rbind(c(gene_pair_1[1],gene_pair_1[2]),c(gene_pair_1[3],gene_pair_1[4])) ###make sure these are the right indices
gene_pair_2=unlist(mut_excl_genes_generator(cohort_size[k],incidence[j],or_pair1[i],or_pair2[l])[2])
gene_pair_2_table=rbind(c(gene_pair_2[1],gene_pair_2[2]),c(gene_pair_2[3],gene_pair_2[4]))
# alldata_1=mut_excl_genes_datapoints(gene_pair_1)
#
# alldata_2=mut_excl_genes_datapoints(gene_pair_2)
# alldata_comp_1=alldata_compiler(alldata_1,"gene2","gene3","gene1",'N',"N/A","N/A")[[2]]
#
# genex_replication_prop_1=alldata_compiler(alldata_1,"gene2","gene3","gene1",'N',"N/A","N/A")[[1]]
# alldata_comp_2=alldata_compiler(alldata_2,"gene2","gene3","gene1",'N',"N/A","N/A")[[2]]
# genex_replication_prop_2=alldata_compiler(alldata_2,"gene2","gene3","gene1",'N',"N/A","N/A")[[1]]
###Calculating Odds ratios and GOI frequencies for the raw data###
# cohort_size_curr=length(alldata_comp$Positive_Ctrl1)
cohort_size_curr=cohort_size[k]
# pc1pc2_contab_counts=contab_maker(alldata_comp$Positive_Ctrl1,alldata_comp$Positive_Ctrl2,alldata_comp)[2:1, 2:1]
pc1pc2_contab_counts=gene_pair_2_table
# goipc1_contab_counts=contab_maker(alldata_comp$genex,alldata_comp$Positive_Ctrl1,alldata_comp)[2:1, 2:1]
goipc1_contab_counts=gene_pair_1_table
# goipc2_contab_counts=contab_maker(alldata_comp$genex,alldata_comp$Positive_Ctrl2,alldata_comp)[2:1, 2:1]
# pc1pc2_contab_probabilities=pc1pc2_contab_counts/cohort_size_curr
# goipc1_contab_probabilities=goipc1_contab_counts/cohort_size_curr
pc1pc2_contab_probabilities=pc1pc2_contab_counts
goipc1_contab_probabilities=goipc1_contab_counts
# goipc2_contab_probabilities=goipc2_contab_counts/cohort_size
or_pc1pc2=pc1pc2_contab_probabilities[1,1]*pc1pc2_contab_probabilities[2,2]/(pc1pc2_contab_probabilities[1,2]*pc1pc2_contab_probabilities[2,1])
or_goipc1=goipc1_contab_probabilities[1,1]*goipc1_contab_probabilities[2,2]/(goipc1_contab_probabilities[1,2]*goipc1_contab_probabilities[2,1])
# or_goipc2=goipc2_contab_probabilities[1,1]*goipc2_contab_probabilities[2,2]/(goipc2_contab_probabilities[1,2]*goipc2_contab_probabilities[2,1])
goi_freq=goipc1_contab_probabilities[1,1]+goipc1_contab_probabilities[1,2]
# goi_freq=.01
# class(goi_freq)
###
###Downsampling PC1 to the probability of GOI without changing ORs###
###The function below converts contingency table data to a new contingency table in which the data is downsampled to the desired frequency, aka the frequency of the GOI in this case###
pc1new_pc2_contab=contab_downsampler(pc1pc2_contab_probabilities,goi_freq)
goinew_pc1_contab=contab_downsampler(goipc1_contab_probabilities,goi_freq)
# goinew_pc2_contab=contab_downsampler(goipc2_contab_probabilities,goi_freq)
###original contab:
# head(pc1pc2_contab_probabilities)
###downsampled contab:
# head(pc1new_pc2_contab)
pc1rawpc2_contabs_sims=contab_simulator(pc1pc2_contab_probabilities,1000,cohort_size_curr)
pc1pc2_contabs_sims=contab_simulator(pc1new_pc2_contab,1000,cohort_size_curr)
goipc1_contabs_sims=contab_simulator(goinew_pc1_contab,1000,cohort_size_curr)
# goipc2_contabs_sims=contab_simulator(goinew_pc2_contab,1000,cohort_size)
# head(pc1pc2_contabs_sims) #each row in this dataset is a new contab
pc1rawpc2_contabs_sims=data.frame(pc1rawpc2_contabs_sims)
pc1rawpc2_contabs_sims=pc1rawpc2_contabs_sims%>%
mutate(or=p11*p00/(p10*p01))
pc1pc2_contabs_sims=data.frame(pc1pc2_contabs_sims)
pc1pc2_contabs_sims=pc1pc2_contabs_sims%>%
mutate(or=p11*p00/(p10*p01))
goipc1_contabs_sims=data.frame(goipc1_contabs_sims)
goipc1_contabs_sims=goipc1_contabs_sims%>%
mutate(or=p11*p00/(p10*p01))
# goipc2_contabs_sims=data.frame(goipc2_contabs_sims)
# goipc2_contabs_sims=goipc2_contabs_sims%>%
# mutate(or=p11*p00/(p10*p01))
pc1rawpc2_contabs_sims$comparison="pc1rawpc2"
pc1pc2_contabs_sims$comparison="pc1pc2"
goipc1_contabs_sims$comparison="goipc1"
# goipc2_contabs_sims$comparison="goipc2"
or_median_raw=quantile(pc1rawpc2_contabs_sims$or,na.rm = T)[3]
or_uq_raw=quantile(pc1rawpc2_contabs_sims$or,na.rm = T)[4]
or_median_downsampled=quantile(pc1pc2_contabs_sims$or,na.rm = T)[3]
or_uq_downsampled=quantile(pc1pc2_contabs_sims$or,na.rm = T)[4]
# pc1rawpc2_contabs_sims=pc1rawpc2_contabs_sims%>%
# mutate(isgreater_raw_median=case_when(or>=or_median_raw~1,
# TRUE~0),
# isgreater_raw_uq=case_when(or>=or_uq_raw~1,
# TRUE~0),
# isgreater_median=case_when(or>=or_median_downsampled~1,
# TRUE~0),
# isgreater_uq=case_when(or>=or_uq_downsampled~1,
# TRUE~0)
# )
# pc1pc2_contabs_sims=pc1pc2_contabs_sims%>%
# mutate(isgreater_raw_median=case_when(or>=or_median_raw~1,
# TRUE~0),
# isgreater_raw_uq=case_when(or>=or_uq_raw~1,
# TRUE~0),
# isgreater_median=case_when(or>=or_median_downsampled~1,
# TRUE~0),
# isgreater_uq=case_when(or>=or_uq_downsampled~1,
# TRUE~0)
# )
# goipc1_contabs_sims=goipc1_contabs_sims%>%
# mutate(isgreater_raw_median=case_when(or>=or_median_raw~1,
# TRUE~0),
# isgreater_raw_uq=case_when(or>=or_uq_raw~1,
# TRUE~0),
# isgreater_median=case_when(or>=or_median_downsampled~1,
# TRUE~0),
# isgreater_uq=case_when(or>=or_uq_downsampled~1,
# TRUE~0)
# )
pc1rawpc2_contabs_sims=pc1rawpc2_contabs_sims%>%
mutate(isgreater_raw_median=case_when(or>=or_median_raw~1,
TRUE~0),
isgreater_raw_uq=case_when(or>or_uq_raw~1,
TRUE~0),
isgreater_median=case_when(or>or_median_downsampled~1,
TRUE~0),
isgreater_uq=case_when(or>or_uq_downsampled~1,
TRUE~0)
)
pc1pc2_contabs_sims=pc1pc2_contabs_sims%>%
mutate(isgreater_raw_median=case_when(or>=or_median_raw~1,
TRUE~0),
isgreater_raw_uq=case_when(or>or_uq_raw~1,
TRUE~0),
isgreater_median=case_when(or>or_median_downsampled~1,
TRUE~0),
isgreater_uq=case_when(or>or_uq_downsampled~1,
TRUE~0)
)
goipc1_contabs_sims=goipc1_contabs_sims%>%
mutate(isgreater_raw_median=case_when(or>or_median_raw~1,
TRUE~0),
isgreater_raw_uq=case_when(or>or_uq_raw~1,
TRUE~0),
isgreater_median=case_when(or>or_median_downsampled~1,
TRUE~0),
isgreater_uq=case_when(or>or_uq_downsampled~1,
TRUE~0)
)
# pc1pc2_contabs_sims=pc1pc2_contabs_sims%>%
# mutate(isgreater=case_when(or>=or_pc1pc2~1,
# TRUE~0))
# goipc1_contabs_sims=goipc1_contabs_sims%>%
# mutate(isgreater=case_when(or>=or_pc1pc2~1,
# TRUE~0))
# goipc2_contabs_sims=goipc2_contabs_sims%>%
# mutate(isgreater=case_when(or>=or_pc1pc2~1,
# TRUE~0))
pc1rawpc2_isgreater_raw_median=sum(pc1rawpc2_contabs_sims$isgreater_raw_median)
pc1rawpc2_isgreater_raw_uq=sum(pc1rawpc2_contabs_sims$isgreater_raw_uq)
pc1rawpc2_isgreater_median=sum(pc1rawpc2_contabs_sims$isgreater_median)
pc1rawpc2_isgreater_uq=sum(pc1rawpc2_contabs_sims$isgreater_uq)
pc1pc2_isgreater_raw_median=sum(pc1pc2_contabs_sims$isgreater_raw_median)
pc1pc2_isgreater_raw_uq=sum(pc1pc2_contabs_sims$isgreater_raw_uq)
pc1pc2_isgreater_median=sum(pc1pc2_contabs_sims$isgreater_median)
pc1pc2_isgreater_uq=sum(pc1pc2_contabs_sims$isgreater_uq)
goipc1_isgreater_raw_median=sum(goipc1_contabs_sims$isgreater_raw_median)
goipc1_isgreater_raw_uq=sum(goipc1_contabs_sims$isgreater_raw_uq)
goipc1_isgreater_median=sum(goipc1_contabs_sims$isgreater_median)
goipc1_isgreater_uq=sum(goipc1_contabs_sims$isgreater_uq)
# pc1rawpc2_isgreater=sum(pc1rawpc2_contabs_sims$isgreater)
# pc1pc2_isgreater=sum(pc1pc2_contabs_sims$isgreater)
# goipc1_isgreater=sum(goipc1_contabs_sims$isgreater)
simresults=c(cohort_size[k],
incidence[j],
or_pair1[i],
or_pair2[l],
pc1rawpc2_isgreater_raw_median,
pc1rawpc2_isgreater_raw_uq,
pc1rawpc2_isgreater_median,
pc1rawpc2_isgreater_uq,
pc1pc2_isgreater_raw_median,
pc1pc2_isgreater_raw_uq,
pc1pc2_isgreater_median,
pc1pc2_isgreater_uq,
goipc1_isgreater_raw_median,
goipc1_isgreater_raw_uq,
goipc1_isgreater_median,
goipc1_isgreater_uq)
simresults_alldata=c(cohort_size[k],
incidence[j],
or_pair1[i],
or_pair2[l],
list(goipc1_contabs_sims$or),
list(pc1pc2_contabs_sims$or))
######Comment out this section to not gather all the 1,000 ORs for each simulation####
simresults_compiled_alldata[[ct]]=simresults_alldata
# a=simresults_compiled_alldata[[1]]
simresults_compiled[ct,]=simresults
ct=ct+1
}
}
}
toc()
}9.846 sec elapsed
9.035 sec elapsed
9.122 sec elapsed
9.083 sec elapsed
9.072 sec elapsed
9.452 sec elapsed
9.196 sec elapsed
9.349 sec elapsed
9.575 sec elapsed
9.264 sec elapsed
9.143 sec elapsed
9.076 sec elapsed
8.912 sec elapsed
9.049 sec elapsed
9.209 sec elapsed
9.019 sec elapsed
9.234 sec elapsedsimresults_compiled=data.frame(simresults_compiled)
colnames(simresults_compiled)=c("cohort_size",
"incidence",
"or1",
"or2",
"pc1rawpc2_isgreater_raw_median",
"pc1rawpc2_isgreater_raw_uq",
"pc1rawpc2_isgreater_median",
"pc1rawpc2_isgreater_uq",
"pc1pc2_isgreater_raw_median",
"pc1pc2_isgreater_raw_uq",
"pc1pc2_isgreater_median",
"pc1pc2_isgreater_uq",
"goipc1_isgreater_raw_median",
"goipc1_isgreater_raw_uq",
"goipc1_isgreater_median",
"goipc1_isgreater_uq")
simresults_compiled$delta_median=simresults_compiled$goipc1_isgreater_raw_median-simresults_compiled$goipc1_isgreater_median
simresults_compiled$delta_uq=simresults_compiled$goipc1_isgreater_raw_uq-simresults_compiled$goipc1_isgreater_uq
simresults_compiled=simresults_compiled%>%
mutate(fp_corrected_95=case_when(
goipc1_isgreater_raw_uq<950~-1,
(goipc1_isgreater_raw_uq>=950&(goipc1_isgreater_raw_uq-delta_uq)<=950)~1,
TRUE~0),
fp_corrected_99=case_when(
goipc1_isgreater_raw_uq<990~-1,
(goipc1_isgreater_raw_uq>=990&(goipc1_isgreater_raw_uq-delta_uq)<=990)~1,
TRUE~0))
# simresults_compiled$goipc1_isgreater_percent=simresults_compiled$goipc1_isgreater*100/1000
simresults_subset=simresults_compiled%>%filter(cohort_size%in%500)
ggplot(simresults_compiled%>%filter(or1<=.4),aes(x=incidence,y=or2))+geom_tile(aes(fill=goipc1_isgreater_median/10))+facet_grid(cohort_size~or1)+scale_fill_gradient2(low ="red" ,mid ="white",midpoint=50,high ="blue",name="Score")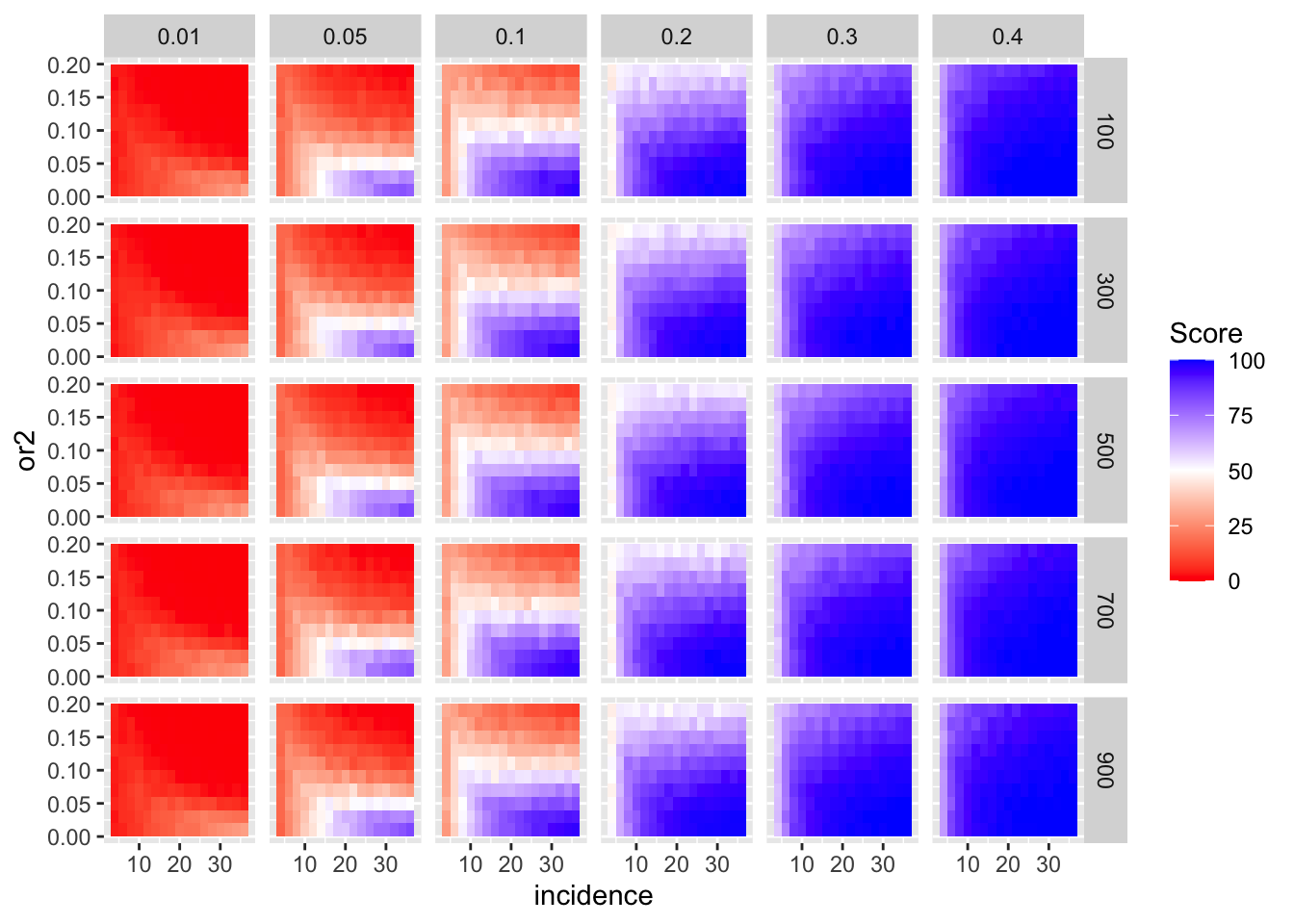
ggplot(simresults_compiled%>%filter(or1<=.4),aes(x=incidence,y=or2))+geom_tile(aes(fill=goipc1_isgreater_raw_median/10))+facet_grid(cohort_size~or1)+scale_fill_gradient2(low ="red" ,mid ="white",midpoint=50,high ="blue",name="Percent succesful trials")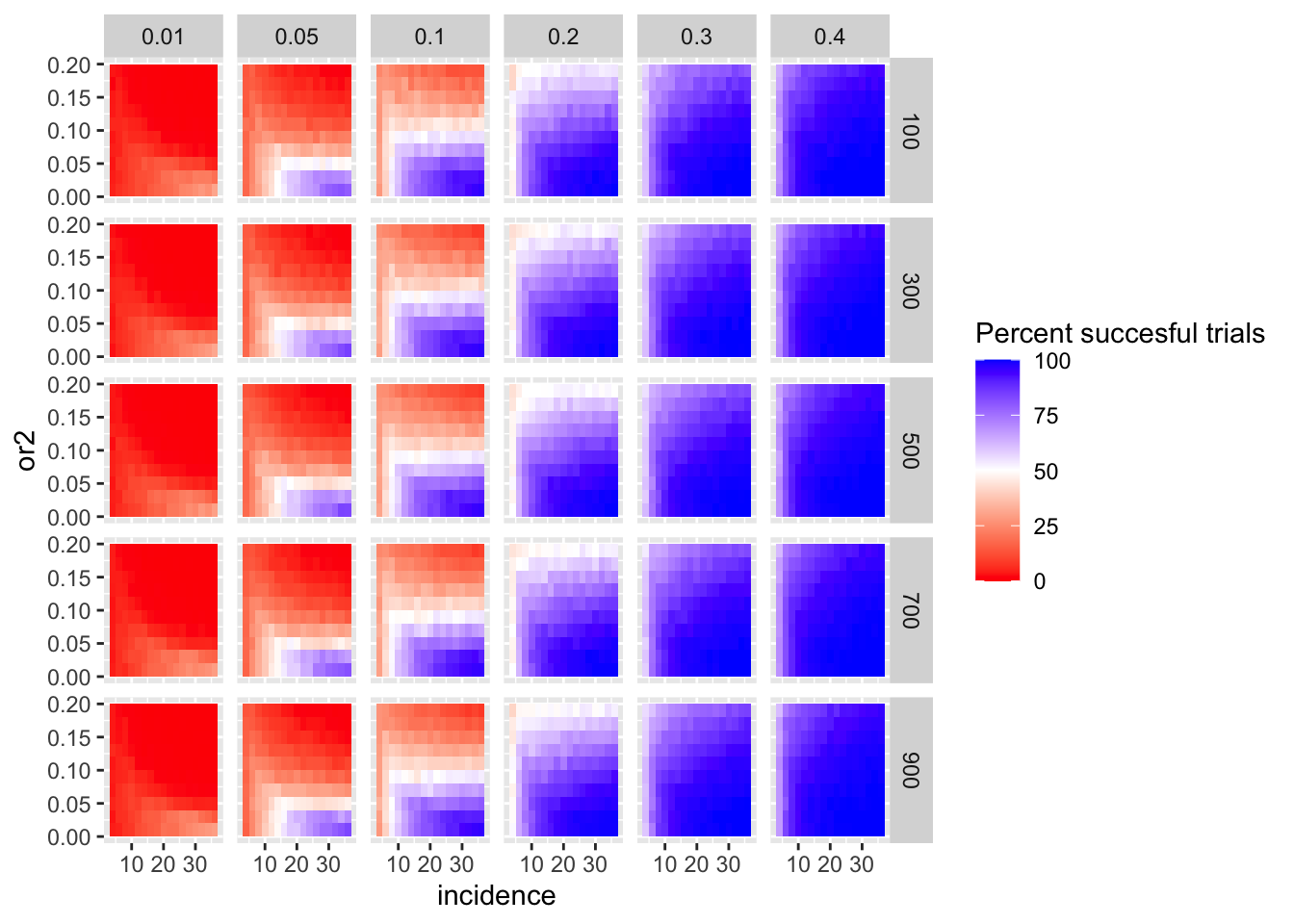
ggplot(simresults_compiled%>%filter(or1<=.4),aes(x=incidence,y=or2))+geom_tile(aes(fill=goipc1_isgreater_uq/10))+facet_grid(cohort_size~or1)+scale_fill_gradient2(low ="red" ,mid ="white",midpoint=50,high ="blue",name="Percent succesful trials")
ggplot(simresults_compiled%>%filter(or1<=.4),aes(x=incidence,y=or2))+geom_tile(aes(fill=goipc1_isgreater_raw_uq/10))+facet_grid(cohort_size~or1)+scale_fill_gradient2(low ="red" ,mid ="white",midpoint=50,high ="blue",name="Percent succesful trials")
ggplot(simresults_compiled%>%filter(or1<=.7,or1>=.2),aes(x=incidence,y=or2))+geom_tile(aes(fill=delta_median))+facet_grid(cohort_size~or1)+scale_fill_gradient2(low ="red" ,mid ="white",high ="blue",name="Delta bw raw and downsampled")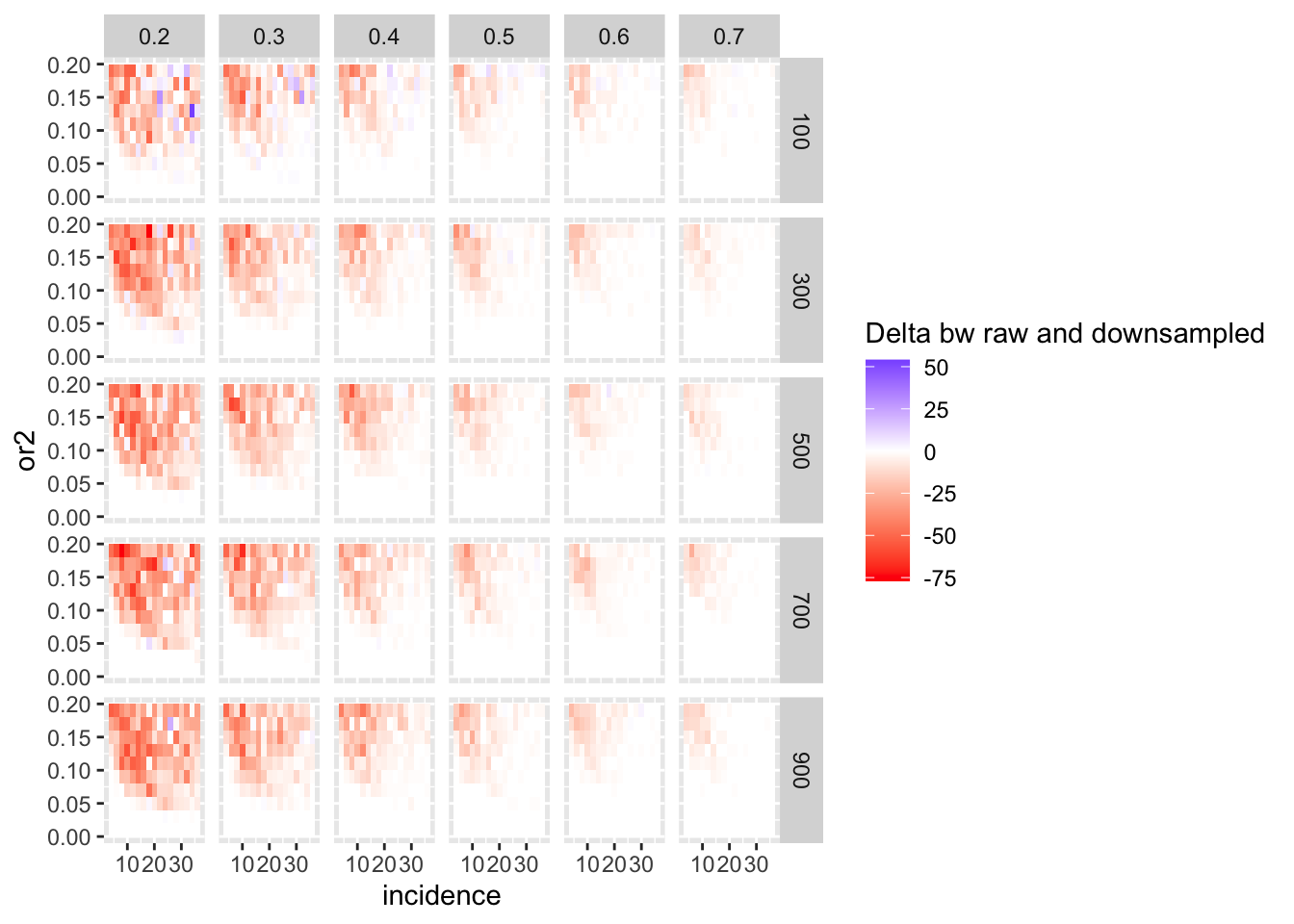
ggplot(simresults_compiled%>%filter(or1<=.7,or1>=.2),aes(x=incidence,y=or2))+geom_tile(aes(fill=delta_uq))+facet_grid(cohort_size~or1)+scale_fill_gradient2(low ="red" ,mid ="white",high ="blue",name="Delta bw raw and downsampled")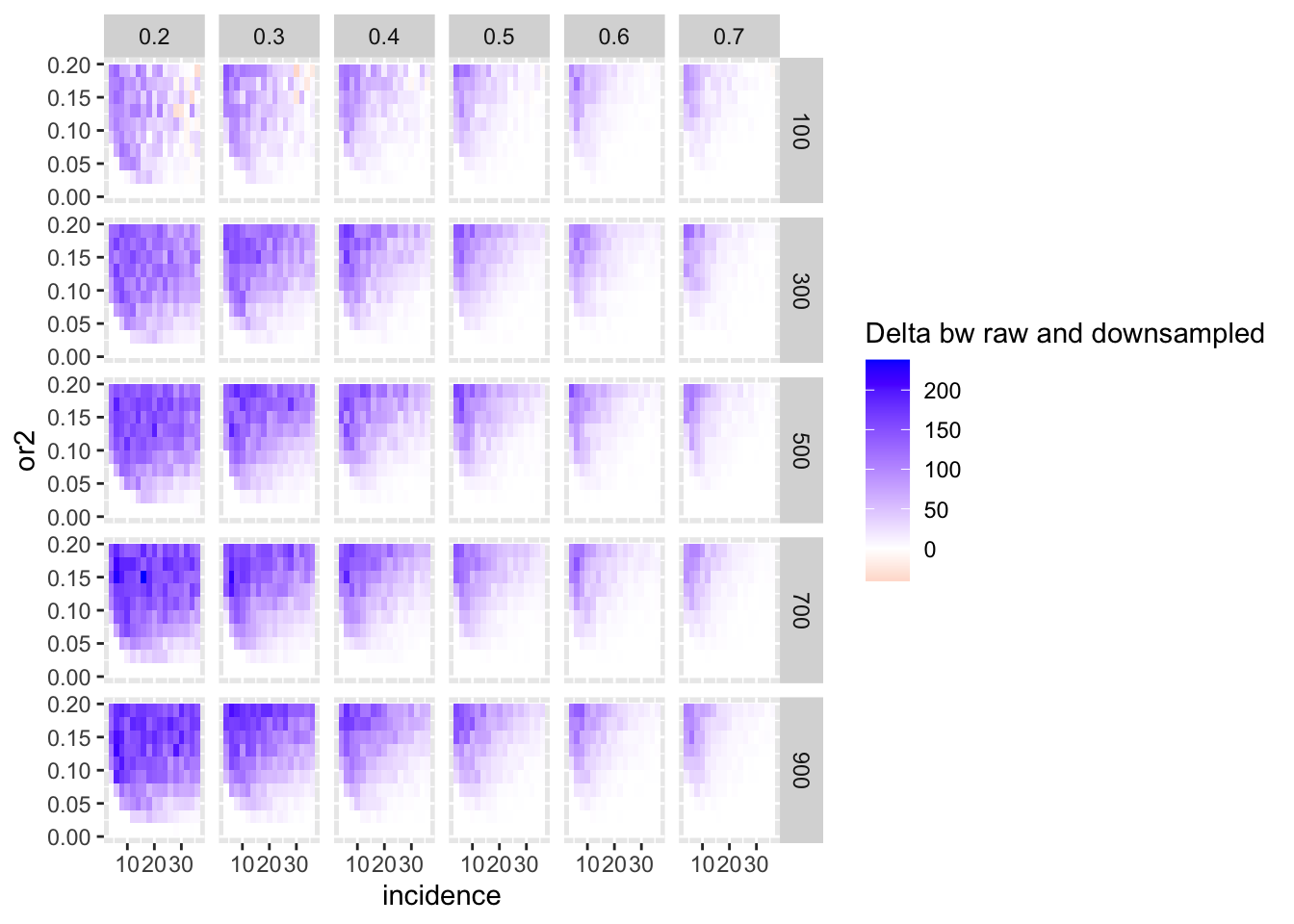
ggplot(simresults_compiled%>%filter(or1<=.7,or1>=.2),aes(x=incidence,y=or2))+geom_tile(color="black",aes(fill=fp_corrected_95))+facet_grid(cohort_size~or1)+scale_fill_gradient2(low ="red" ,mid ="white",high ="blue",name="Regions where downsampling helps")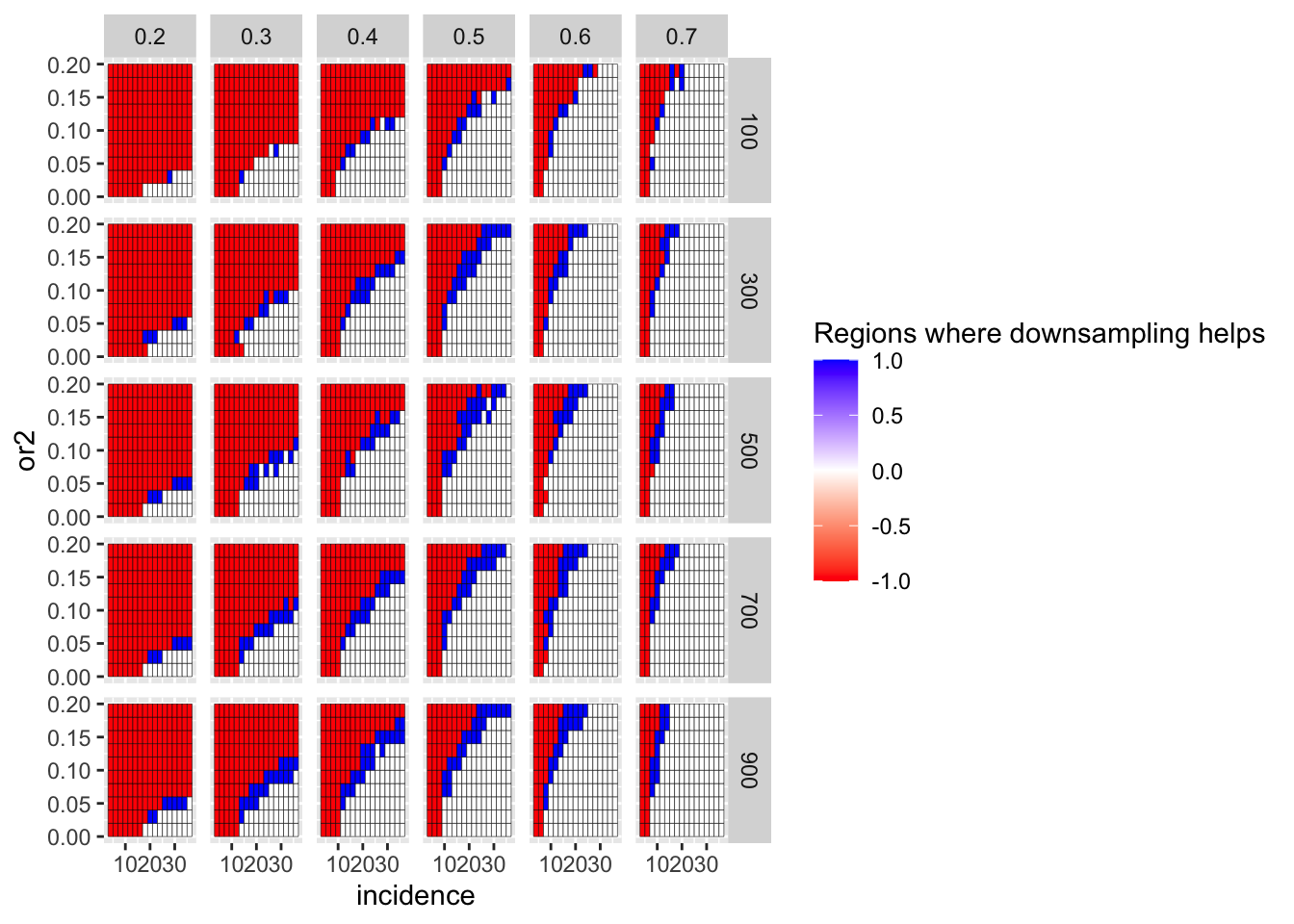
ggplot(simresults_compiled%>%filter(or1<=.7,or1>=.2),aes(x=incidence,y=or2))+geom_tile(color="black",aes(fill=fp_corrected_99))+facet_grid(cohort_size~or1)+scale_fill_gradient2(low ="red" ,mid ="white",high ="blue",name="Regions where downsampling helps")
ggplot(simresults_compiled%>%filter(or1<=.4),aes(x=incidence,y=or2))+geom_tile(aes(fill=goipc1_isgreater_median/10))+facet_grid(cohort_size~or1)+scale_fill_gradient2(low ="red" ,mid ="white",midpoint=50,high ="blue",name="Score")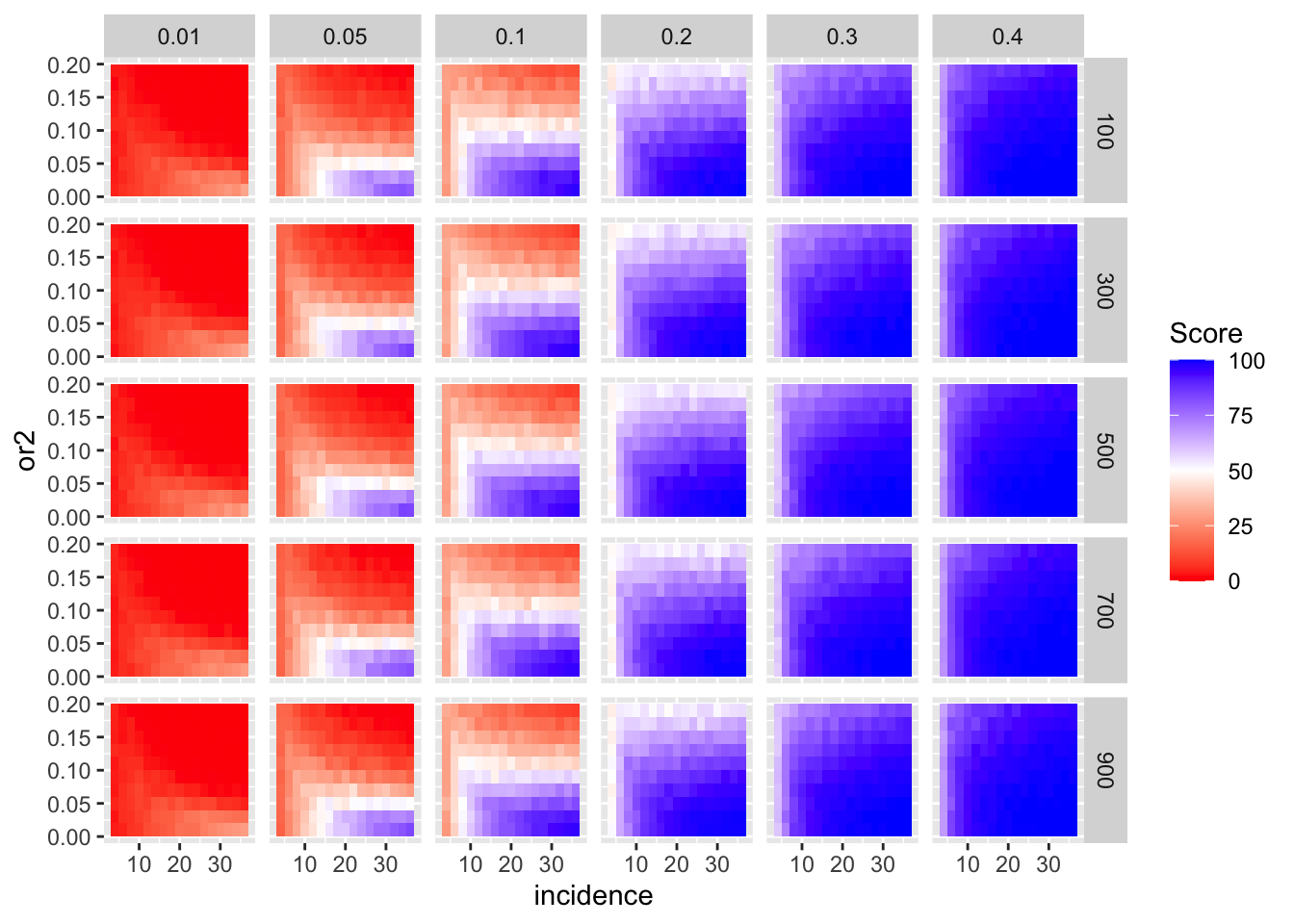
ggplot(simresults_compiled%>%filter(or1<=.4,cohort_size%in%500),aes(x=incidence,y=or2))+geom_tile(aes(fill=goipc1_isgreater_median/10))+facet_wrap(~or1)+scale_fill_gradient2(low ="red" ,mid ="white",midpoint=50,high ="blue",name="Score")
ggplot(simresults_compiled%>%filter(or1<=.4,cohort_size%in%500),aes(x=incidence,y=or2))+geom_tile(aes(fill=goipc1_isgreater_median/10))+facet_wrap(~or1,ncol=6)+scale_fill_gradient2(low ="red" ,mid ="white",midpoint=50,high ="blue",name="Score")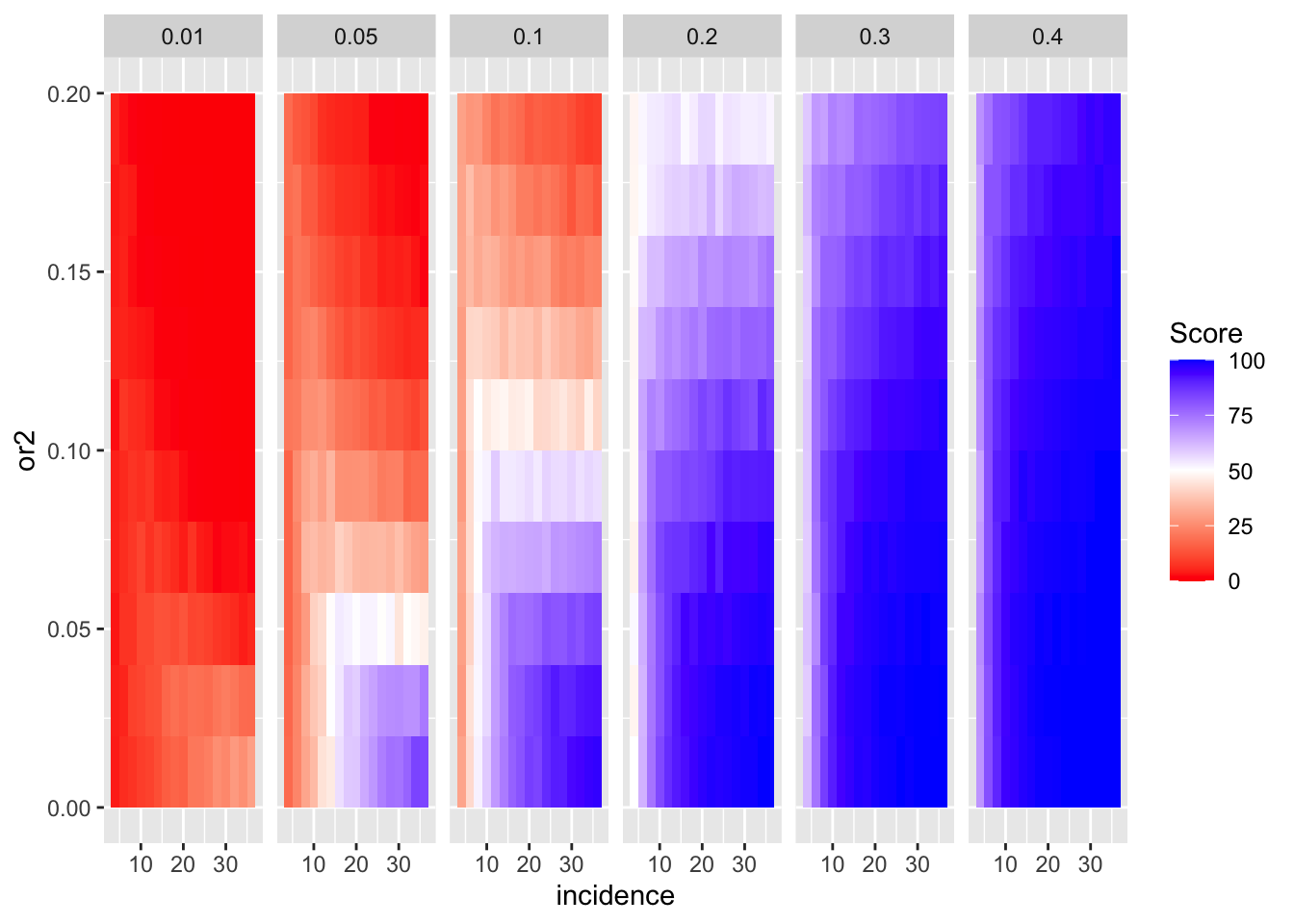
ggplot(simresults_compiled%>%filter(or1%in%c(.05,0.1,.5),cohort_size%in%500),aes(x=incidence,y=or2))+geom_tile(color="black",aes(fill=goipc1_isgreater_median/10))+facet_wrap(~or1,ncol=6)+scale_fill_gradient2(low ="red" ,mid ="white",midpoint=50,high ="blue",name="Score")+scale_x_continuous(expand = c(0,0),name="Incidence")+
scale_y_continuous(expand = c(0,0),name="Odds ratio of PC1vsPC2")+ggtitle("Odds ratio of GOIvsPC1")+
theme(plot.title = element_text(hjust = 0.5))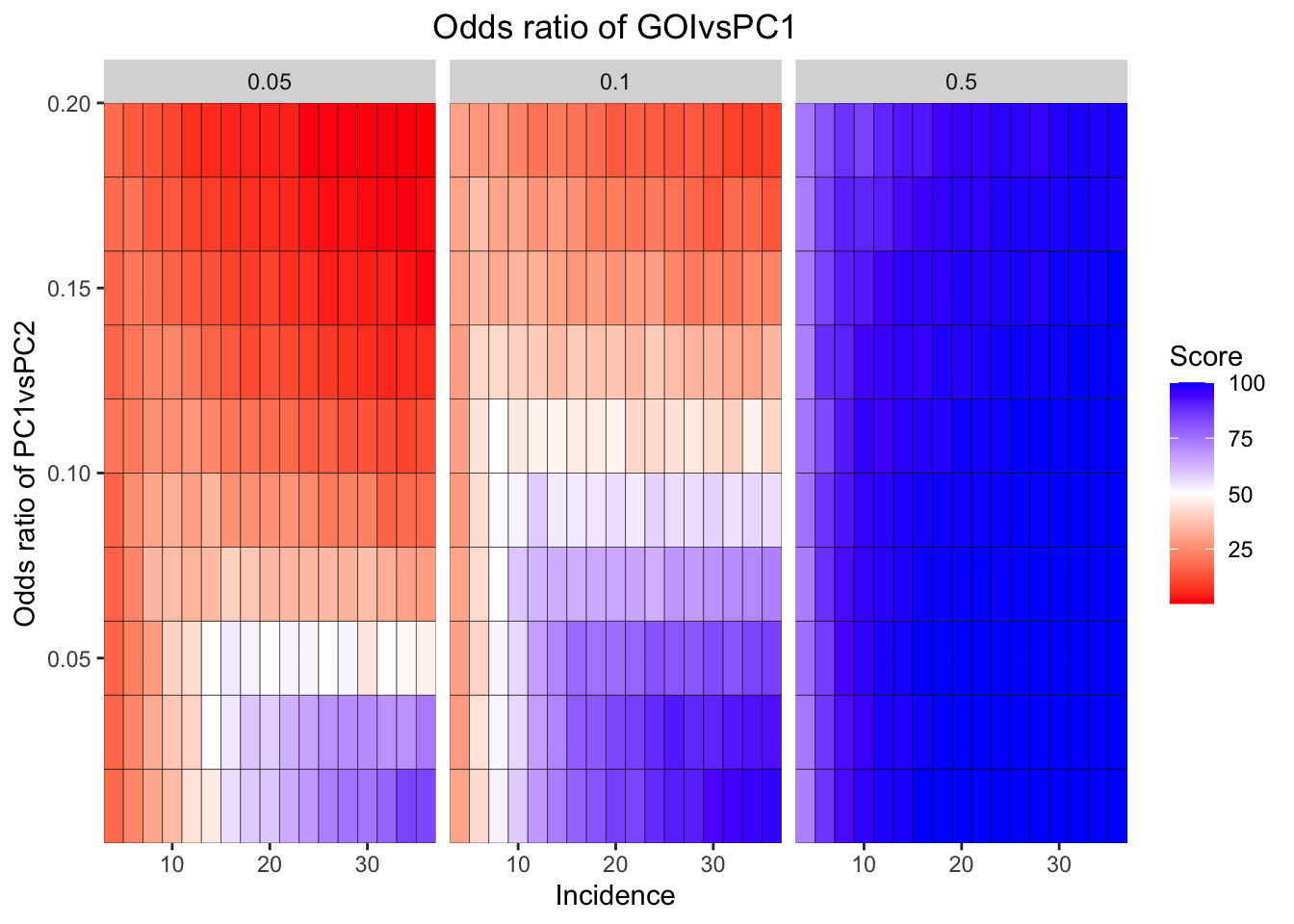
# ggsave("score_heatmap.pdf",width=8,heigh=3,units="in",useDingbats=F)
ggplot(simresults_compiled%>%filter(or1%in%c(.05,0.1,.5),cohort_size%in%c(100,500,900)),aes(x=incidence,y=or2))+geom_tile(color="black",aes(fill=goipc1_isgreater_median/10))+facet_grid(cohort_size~or1)+scale_fill_gradient2(low ="red" ,mid ="white",midpoint=50,high ="blue",name="Score")+scale_x_continuous(expand = c(0,0),name="Incidence")+
scale_y_continuous(expand = c(0,0),name="Odds ratio of PC1vsPC2")+ggtitle("Odds ratio of GOIvsPC1")+
theme(plot.title = element_text(hjust = 0.5))
# ggsave("score_heatmap_supplement.pdf",width=8,heigh=7,units="in",useDingbats=F)
ggplot(simresults_compiled%>%filter(or2%in%.05,cohort_size%in%c(500)),aes(x=factor(or1),y=goipc1_isgreater_median/10))+geom_boxplot()+scale_x_discrete(name="Odds ratio of PC1vsPC2")+
scale_y_continuous(name="Score")+
theme(plot.title = element_text(hjust = 0.5))+cleanup
# ggsave("score_plot.pdf",width=8,heigh=3,units="in",useDingbats=F)
ggplot(simresults_compiled%>%filter(incidence%in%c(6,16,26,36),cohort_size%in%c(100,300,500,900)),aes(x=factor(or1),y=or2))+geom_tile(color="black",aes(fill=goipc1_isgreater_uq/10))+scale_fill_gradient2(low ="red" ,mid ="white",midpoint=50,high ="blue",name="Score")+scale_x_discrete(expand = c(0,0),name="Odds ratio of GOI and PC1")+facet_grid(cohort_size~incidence)+
scale_y_continuous(expand = c(0,0),name="Odds ratio of PC1 and PC2")+theme(plot.title = element_text(hjust = 0.5))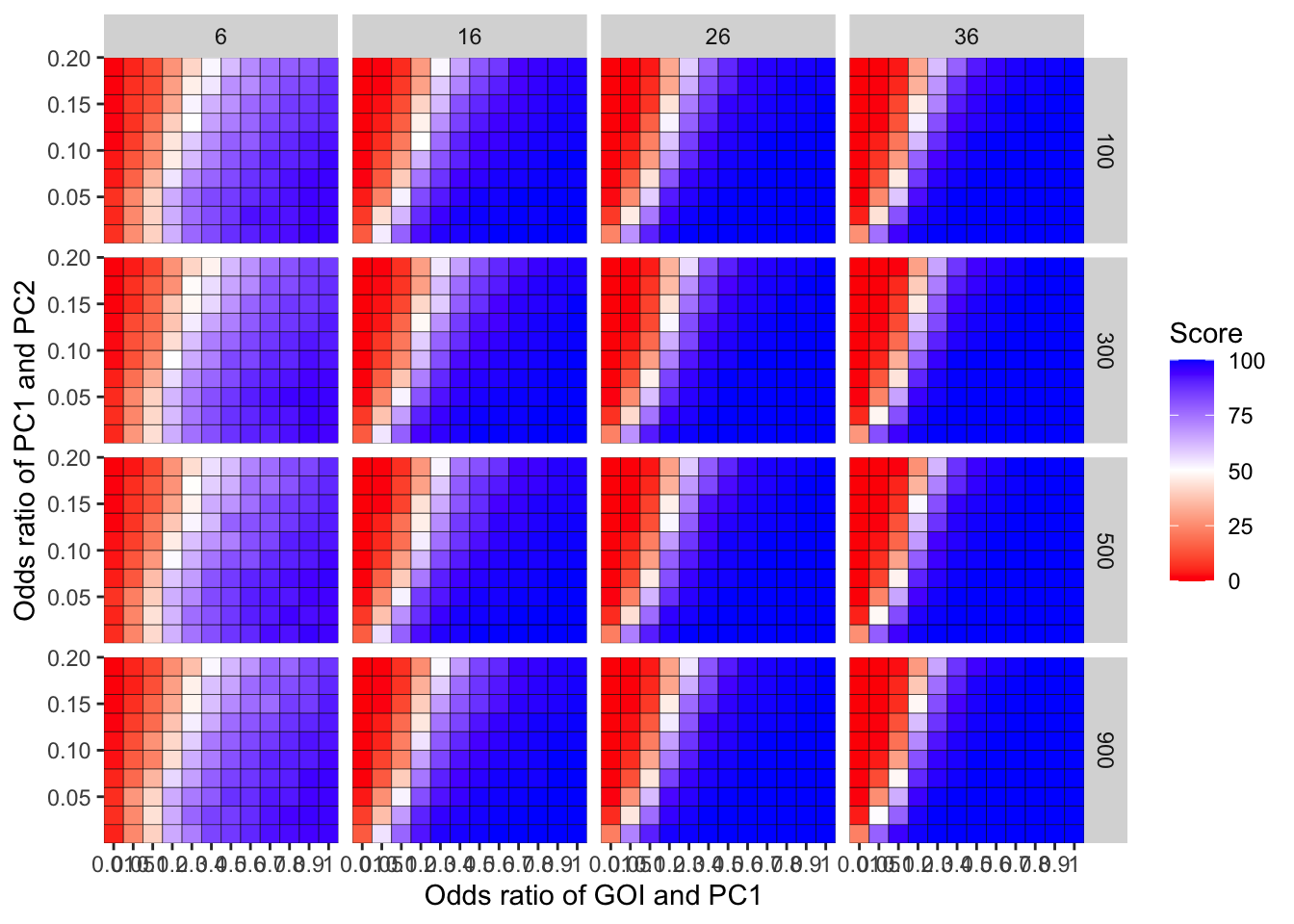
ggplot(simresults_compiled%>%filter(incidence%in%c(6,16,26,36),cohort_size%in%500),aes(x=factor(or1),y=or2))+geom_tile(color="black",aes(fill=goipc1_isgreater_uq/10))+scale_fill_gradient2(low ="red" ,mid ="white",midpoint=50,high ="blue",name="Score")+scale_x_discrete(expand = c(0,0),name="Odds ratio of GOI and PC1")+facet_wrap(~incidence,ncol=4)+
scale_y_continuous(expand = c(0,0),name="Odds ratio of PC1 and PC2")+theme(plot.title = element_text(hjust = 0.5),axis.text.x=element_text(angle=90,hjust=.5,vjust=.5),axis.text=element_text(face="bold",size="9",color="black"))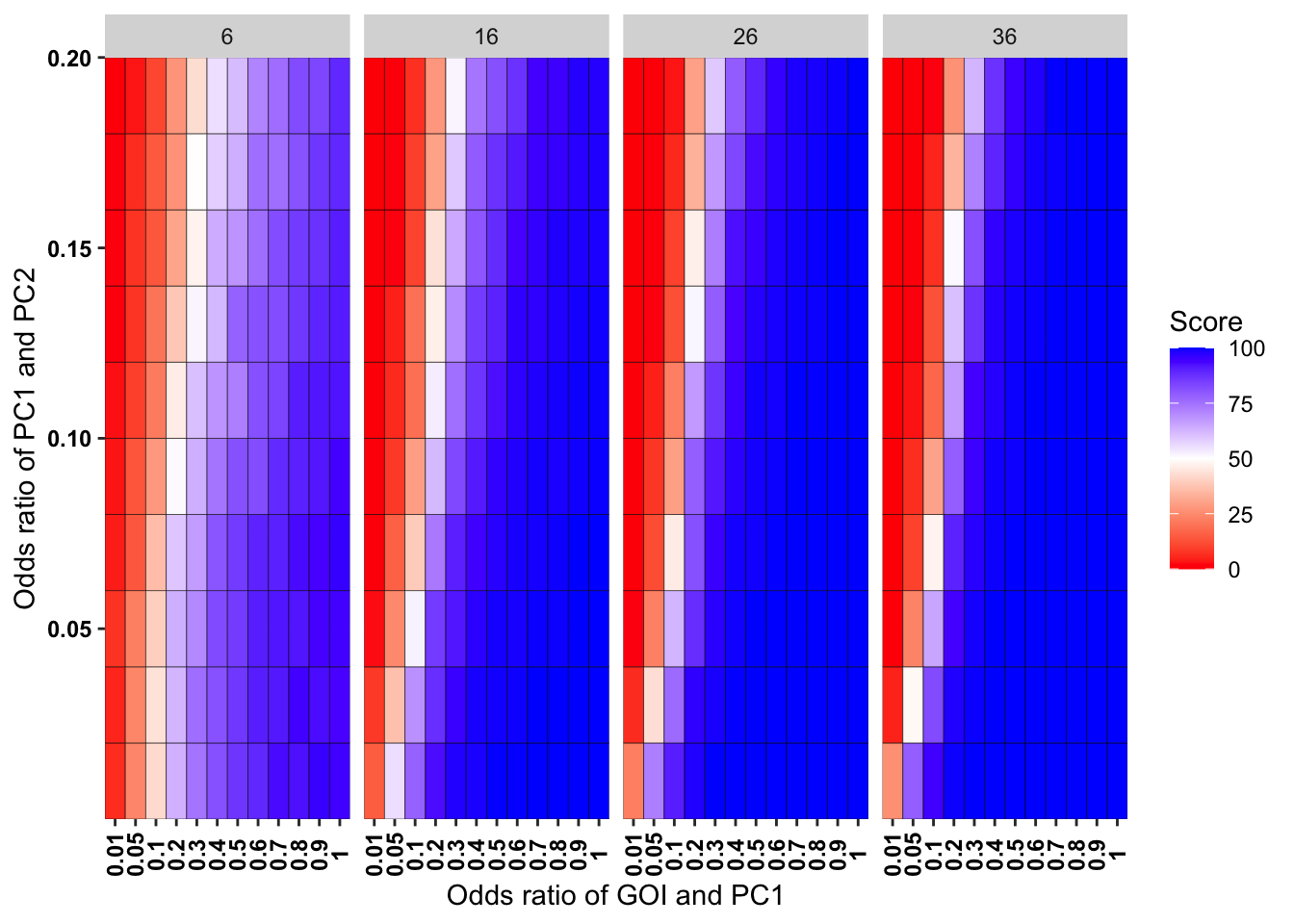
# ggsave("score_heatmap_bestoption1.pdf",width=8,heigh=2.5,units="in",useDingbats=F)
# sort(unique(simresults_compiled$or2))
ggplot(simresults_compiled%>%filter(cohort_size%in%500),aes(x=factor(or1),y=or2))+geom_tile(color="black",aes(fill=goipc1_isgreater_median/10))+scale_fill_gradient2(low ="red" ,mid ="white",midpoint=50,high ="blue",name="Score")+scale_x_discrete(expand = c(0,0),name="Odds ratio of GOI and PC1")+
scale_y_continuous(expand = c(0,0),name="Odds ratio of PC1 and PC2")+theme(plot.title = element_text(hjust = 0.5))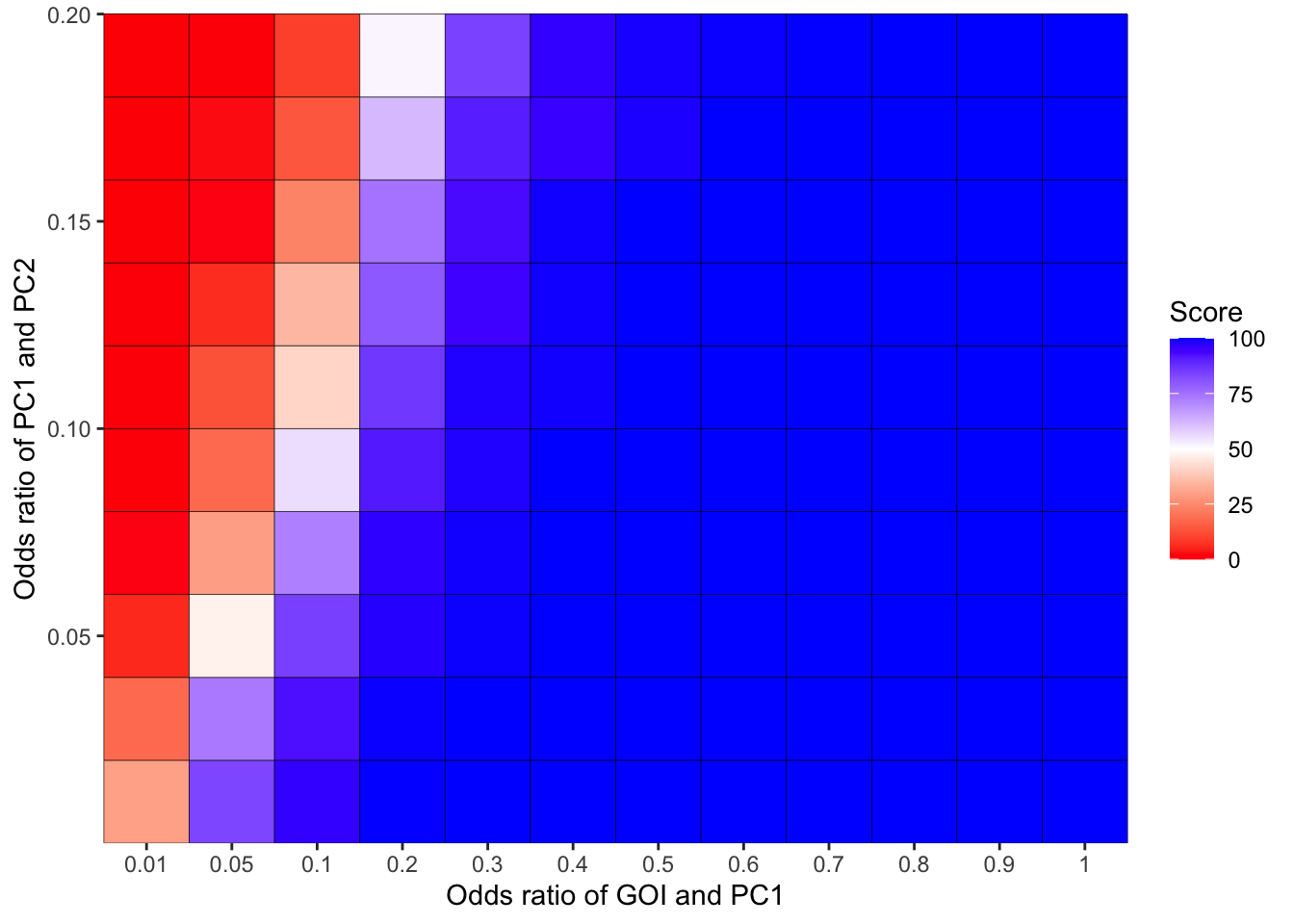
# ggsave("score_heatmap_bestoption.pdf",width=4,heigh=3,units="in",useDingbats=F)
ggplot(simresults_compiled%>%filter(or1%in%.3,or2%in%.09),aes(x=incidence/cohort_size,y=goipc1_isgreater_median/10))+geom_point()
ggplot(simresults_compiled%>%filter(or1%in%.1,or2%in%.05,cohort_size%in%500),aes(x=incidence,y=goipc1_isgreater_median/10))+geom_point()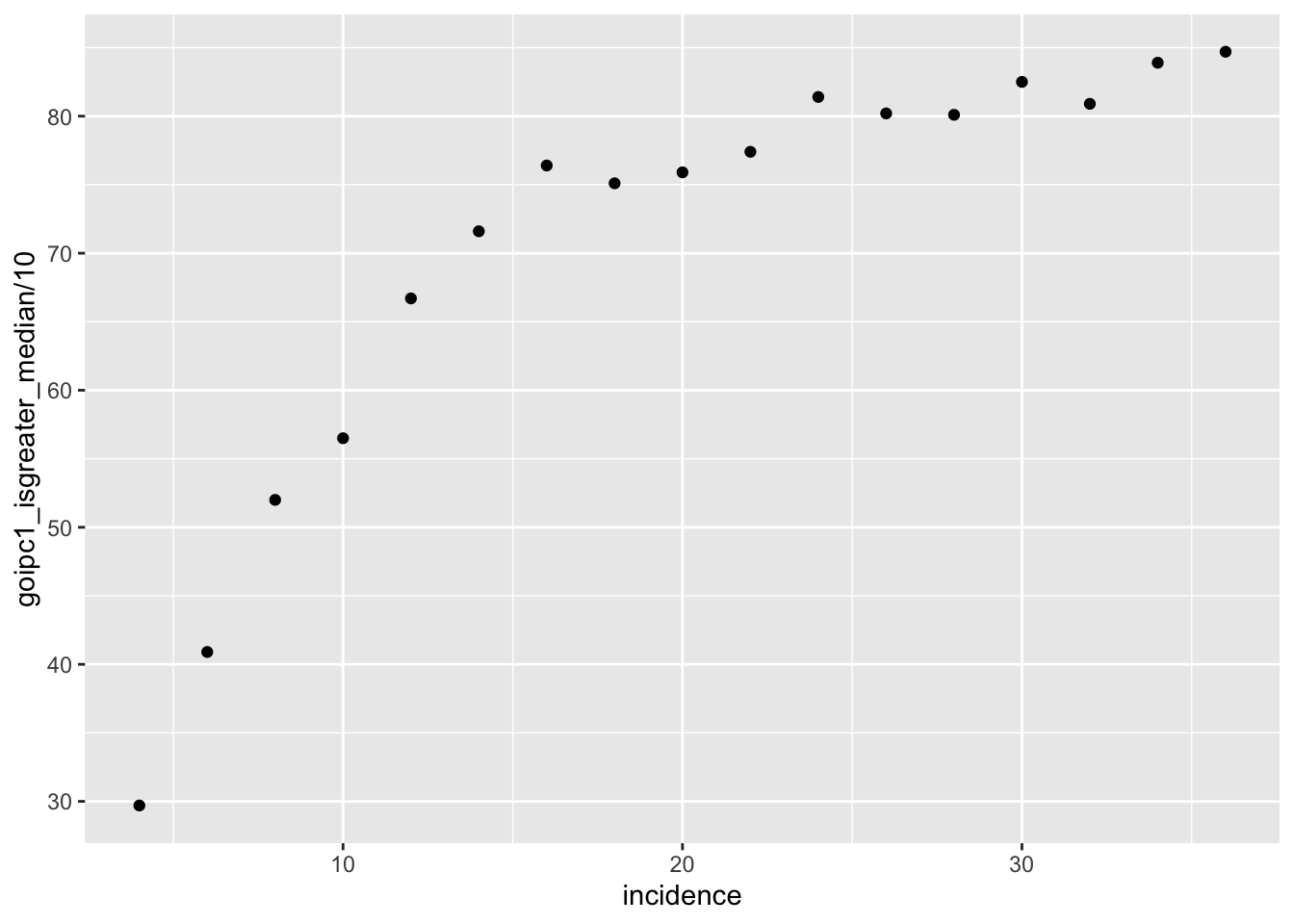
# scale_fill_gradient2(low ="red" ,mid ="white",midpoint=50,high ="blue",name="Score")+scale_x_discrete(expand = c(0,0),name="Odds ratio of GOI and PC1")+
# scale_y_continuous(expand = c(0,0),name="Odds ratio of PC1 and PC2")+theme(plot.title = element_text(hjust = 0.5))
# or1%in%c(.05,0.1,.5),cohort_size%in%c(100,500,900)
ggplot(simresults_compiled%>%filter(cohort_size%in%c(100,300,500)),aes(x=incidence,y=or2))+geom_tile(color="black",aes(fill=goipc1_isgreater_median/10))+facet_grid(cohort_size~or1)+scale_fill_gradient2(low ="red" ,mid ="white",midpoint=50,high ="blue",name="Score")+scale_x_continuous(expand = c(0,0),name="Incidence")+
scale_y_continuous(expand = c(0,0),name="Odds ratio of PC1vsPC2")+ggtitle("Odds ratio of GOIvsPC1")+
theme(plot.title = element_text(hjust = 0.5)) Choosing plots for putting in paper
Choosing plots for putting in paper
ggplot(simresults_compiled%>%filter(cohort_size%in%500),aes(x=factor(or1),y=or2))+geom_tile(color="black",aes(fill=goipc1_isgreater_uq/10))+scale_fill_gradient2(low ="red" ,mid ="white",midpoint=50,high ="blue",name="Score")+scale_x_discrete(expand = c(0,0),name="Odds ratio of GOI and PC1")+
scale_y_continuous(expand = c(0,0),name="Odds ratio of PC1 and PC2")+theme(plot.title = element_text(hjust = 0.5))+theme(plot.title = element_text(hjust = 0.5),axis.text.x=element_text(angle=90,hjust=.5,vjust=.5),axis.text=element_text(face="bold",size="9",color="black"))
# ggsave("score_heatmap_bestoption.pdf",width=4,heigh=3,units="in",useDingbats=F)
ggplot(simresults_compiled%>%filter(or1%in%c(.05,.3,.6),incidence%in%c(4,8,12,16,20,24,28,32,36),cohort_size%in%500),aes(x=factor(incidence),y=or2))+geom_tile(color="black",aes(fill=goipc1_isgreater_median/10))+facet_wrap(~or1,ncol=6)+scale_fill_gradient2(low ="red" ,mid ="white",midpoint=50,high ="blue",name="Score")+scale_x_discrete(expand = c(0,0),name="Incidence")+
scale_y_continuous(expand = c(0,0),name="Odds ratio of PC1 and PC2")+theme(strip.text=element_blank(),plot.title = element_text(hjust = 0.5),axis.text.x=element_text(angle=90,hjust=.5,vjust=.5),axis.text=element_text(face="bold",size="9",color="black"),legend.position = "none")
# ggsave("score_heatmap_bestoption1.pdf",width=6,heigh=2.5,units="in",useDingbats=F)
ggplot(simresults_compiled%>%filter(or1%in%c(.1,.2,.5),incidence%in%c(4,8,12,16,20,24,28,32,36),cohort_size%in%500),aes(x=factor(incidence),y=or2))+geom_tile(color="black",aes(fill=goipc1_isgreater_raw_median/10))+facet_wrap(~or1,ncol=6)+scale_fill_gradient2(low ="red" ,mid ="white",midpoint=50,high ="blue",name="Score")+scale_x_discrete(expand = c(0,0),name="Incidence")+
scale_y_continuous(expand = c(0,0),name="Odds ratio of PC1 and PC2")+theme(strip.text=element_blank(),plot.title = element_text(hjust = 0.5),axis.text.x=element_text(angle=90,hjust=.5,vjust=.5),axis.text=element_text(face="bold",size="9",color="black"),legend.position = "none")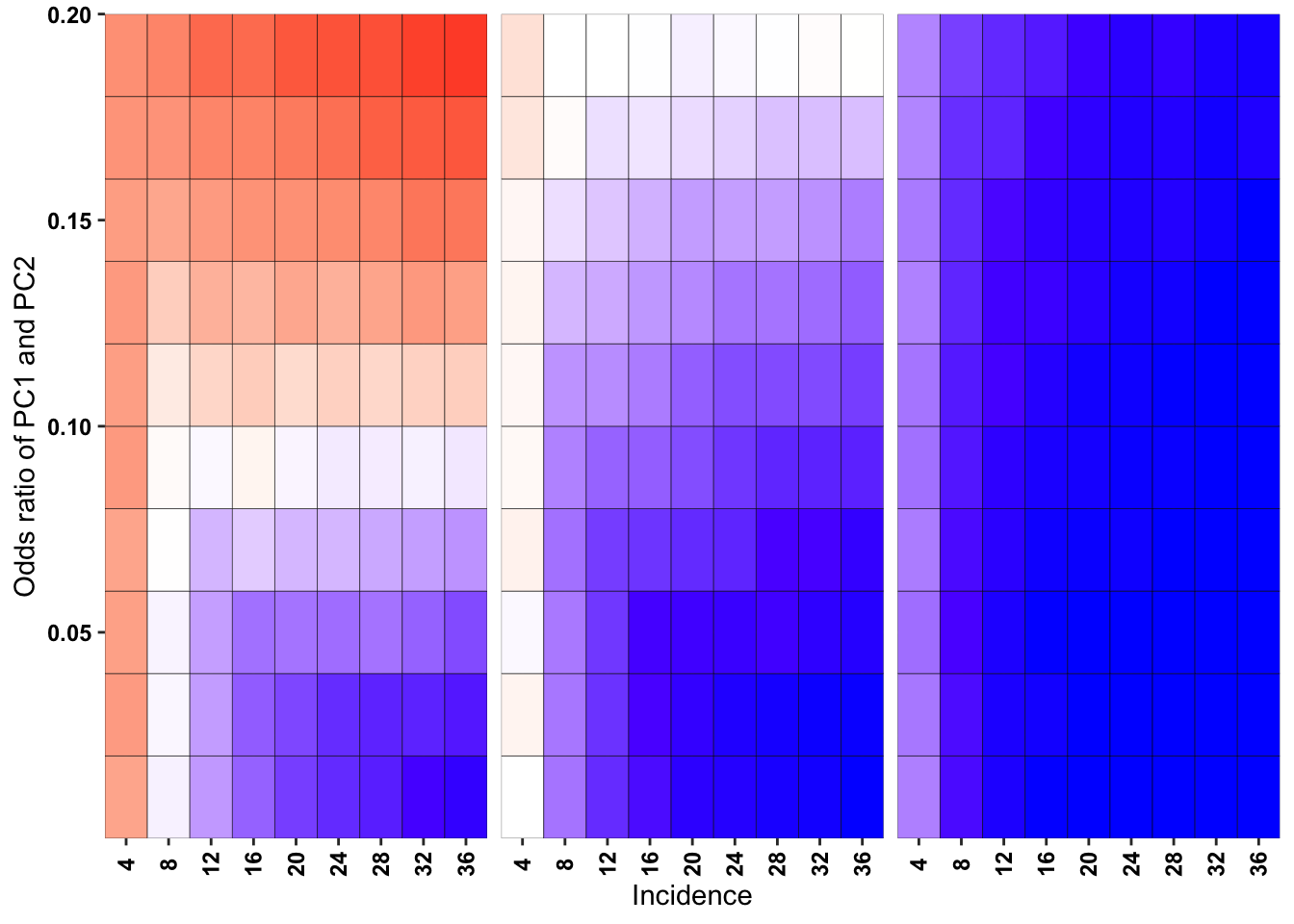
# ggsave("score_heatmap_bestoption1.pdf",width=6,heigh=2.5,units="in",useDingbats=F)
ggplot(simresults_compiled%>%filter(cohort_size%in%500,incidence%in%c(4,8,12,16,20,24,28,32,36)),aes(x=factor(incidence),y=or2))+geom_tile(color="black",aes(fill=goipc1_isgreater_raw_median/10))+facet_wrap(~or1,ncol=4)+scale_fill_gradient2(low ="red" ,mid ="white",midpoint=50,high ="blue",name="Score")+scale_x_discrete(expand = c(0,0),name="Incidence")+
scale_y_continuous(expand = c(0,0),name="Odds ratio of PC1 and PC2")+theme(plot.title = element_text(hjust = 0.5),axis.text.x=element_text(angle=90,hjust=.5,vjust=.5),axis.text=element_text(face="bold",size="9",color="black"),legend.position = "none")
# ggsave("score_heatmap_bestoption1_supplement.pdf",width=6,heigh=6,units="in",useDingbats=F)
ggplot(simresults_compiled%>%filter(or1%in%c(.05,.1,.5),cohort_size%in%c(100,300,500)),aes(x=incidence,y=or2))+geom_tile(color="black",aes(fill=goipc1_isgreater_median/10))+facet_grid(cohort_size~or1)+scale_fill_gradient2(low ="red" ,mid ="white",midpoint=50,high ="blue",name="Score")+scale_x_discrete(expand = c(0,0),name="Incidence")+
scale_y_continuous(expand = c(0,0),name="Odds ratio of PC1 and PC2")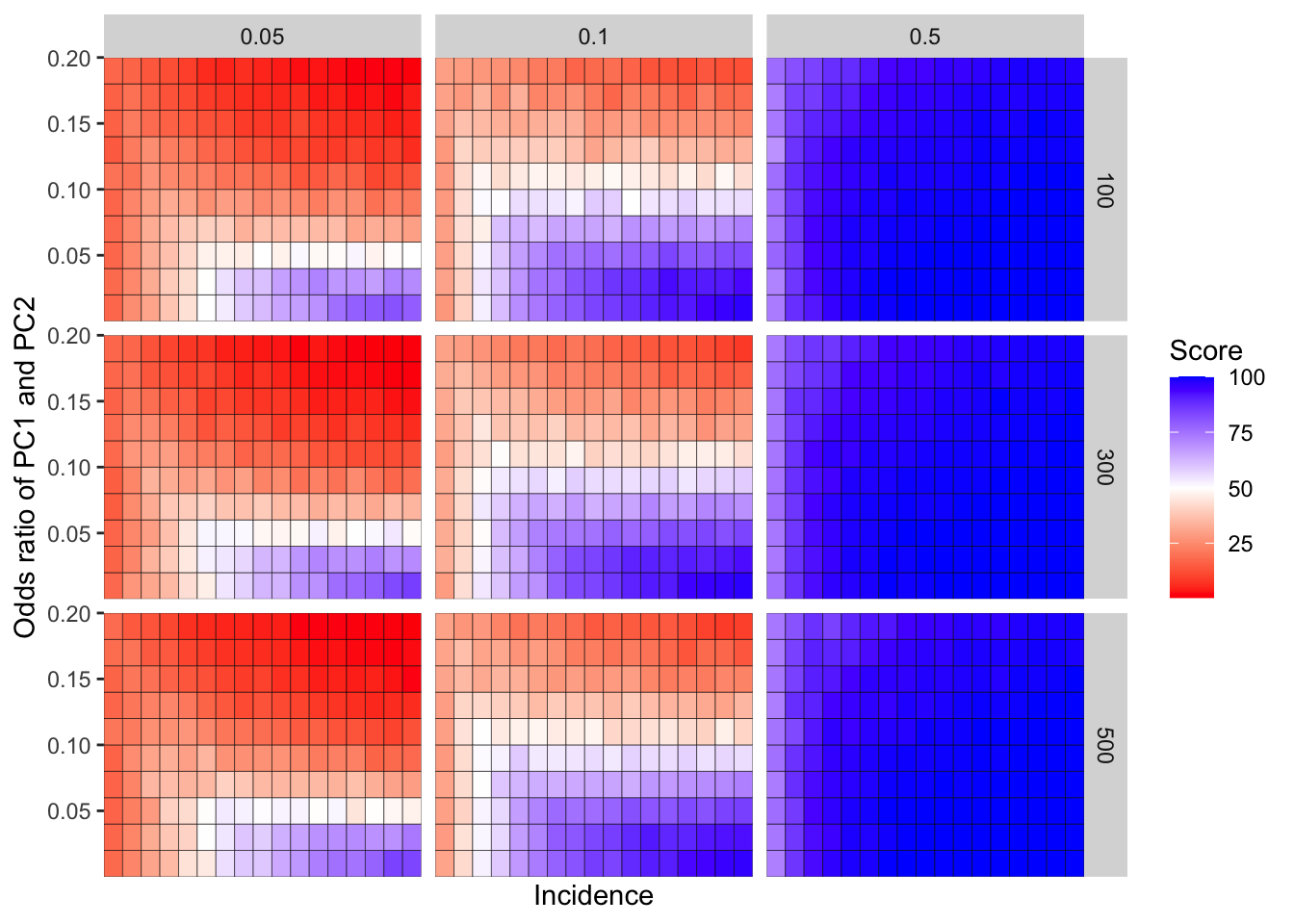
# a=simresults_compiled%>%filter(cohort_size%in%500,or1%in%.7,or2%in%.05,incidence%in%)# simresults_subset=simresults_compiled%>%filter(or2%in%0.01,or1%in%c(0.01,.1,1))
simresults_subset=unlist(simresults_compiled_alldata)
simresults_unlisted=data.frame(unlist(lapply(simresults_compiled_alldata,'[[',1)),
unlist(lapply(simresults_compiled_alldata,'[[',2)),
unlist(lapply(simresults_compiled_alldata,'[[',3)),
unlist(lapply(simresults_compiled_alldata,'[[',4)))
simresults_unlisted$list1=(lapply(simresults_compiled_alldata,'[[',5))
simresults_unlisted$list2=(lapply(simresults_compiled_alldata,'[[',6))
colnames(simresults_unlisted)=c("cohort_size","incidence","or1","or2","or1_list","or2_list")
simresults_unlisted=simresults_unlisted%>%filter(or2%in%0.01,or1%in%c(0.05,.1,1),cohort_size%in%500,incidence%in%c(4,8,12,16,20))
# library(reshape2)
a=simresults_unlisted%>%filter(incidence%in%8,or2%in%0.01,or1%in%.05)
# b=unnest(a)
median(b$or2_list)>median(b$or1_list)
library(tidyr)
simresults_melted=unnest(simresults_unlisted)
simresults_melted2=melt(simresults_melted,
id.vars = c("cohort_size","or1","or2","incidence"),
measure.vars =c("or1_list","or2_list"),
variable.name = "Comparison",
value.name = "OR"
)
ggplot(simresults_melted2,aes(x=factor(incidence),y=OR,fill=Comparison))+facet_wrap(~or1)+geom_boxplot()+scale_y_continuous(trans="log2")+cleanup
plotly=ggplot(simresults_melted2,aes(x=factor(incidence),y=OR,fill=Comparison))+facet_wrap(~or1)+geom_boxplot()+cleanup
ggplotly(plotly)
simresults_unlisted=data.frame(unlist(lapply(simresults_compiled_alldata,'[[',1)),
unlist(lapply(simresults_compiled_alldata,'[[',2)),
unlist(lapply(simresults_compiled_alldata,'[[',3)),
unlist(lapply(simresults_compiled_alldata,'[[',4)))
simresults_unlisted$list1=(lapply(simresults_compiled_alldata,'[[',5))
simresults_unlisted$list2=(lapply(simresults_compiled_alldata,'[[',6))
colnames(simresults_unlisted)=c("cohort_size","incidence","or1","or2","or1_list","or2_list")
simresults_unlisted=simresults_unlisted%>%filter(or1%in%.5,or2%in%c(0.05,.11,.19),cohort_size%in%500,incidence%in%c(4,8,12,16,20))
simresults_melted=unnest(simresults_unlisted)
simresults_melted2=melt(simresults_melted,
id.vars = c("cohort_size","or1","or2","incidence"),
measure.vars =c("or1_list","or2_list"),
variable.name = "Comparison",
value.name = "OR"
)
ggplot(simresults_melted2,aes(x=factor(incidence),y=OR,fill=Comparison))+facet_wrap(~or2)+geom_boxplot()+scale_y_continuous(trans="log2")+cleanup
plotly=ggplot(simresults_melted2,aes(x=factor(incidence),y=OR,fill=Comparison))+facet_wrap(~or2)+geom_boxplot()+cleanup
ggplotly(plotly)
# sort(unique(simresults_unlisted$or2))
#I used the following website to make sure that my Gaussian elimination was working properly
# https://www.emathhelp.net/calculators/linear-algebra/gauss-jordan-elimination-calculator/?i=%5B%5B1%2C1%2C0%2C0%2C.4836%5D%2C%5B1%2C1%2C1%2C1%2C1%5D%2C%5B0%2C1%2C0%2C-1%2C0%5D%2C%5B1%2C0%2C-0.01%2C0%2C0%5D%5D&reduced=on
sessionInfo()R version 4.0.0 (2020-04-24)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRblas.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] parallel grid stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] BiocManager_1.30.10 plotly_4.9.2.1 ggsignif_0.6.0
[4] devtools_2.3.0 usethis_1.6.1 RColorBrewer_1.1-2
[7] reshape2_1.4.4 ggplot2_3.3.3 doParallel_1.0.15
[10] iterators_1.0.12 foreach_1.5.0 dplyr_1.0.6
[13] VennDiagram_1.6.20 futile.logger_1.4.3 tictoc_1.0
[16] knitr_1.28 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rcpp_1.0.4.6 tidyr_1.1.3 prettyunits_1.1.1
[4] ps_1.3.3 assertthat_0.2.1 rprojroot_1.3-2
[7] digest_0.6.25 utf8_1.1.4 R6_2.4.1
[10] plyr_1.8.6 futile.options_1.0.1 backports_1.1.7
[13] evaluate_0.14 httr_1.4.2 pillar_1.6.1
[16] rlang_0.4.11 lazyeval_0.2.2 data.table_1.12.8
[19] whisker_0.4 callr_3.7.0 rmarkdown_2.8
[22] labeling_0.3 desc_1.2.0 stringr_1.4.0
[25] htmlwidgets_1.5.1 munsell_0.5.0 compiler_4.0.0
[28] httpuv_1.5.2 xfun_0.22 pkgconfig_2.0.3
[31] pkgbuild_1.0.8 htmltools_0.4.0 tidyselect_1.1.0
[34] tibble_3.1.2 codetools_0.2-16 viridisLite_0.3.0
[37] fansi_0.4.1 crayon_1.4.1 withr_2.4.2
[40] later_1.0.0 jsonlite_1.7.2 gtable_0.3.0
[43] lifecycle_1.0.0 DBI_1.1.0 git2r_0.27.1
[46] magrittr_2.0.1 formatR_1.7 scales_1.1.1
[49] cli_2.5.0 stringi_1.4.6 farver_2.0.3
[52] fs_1.4.1 promises_1.1.0 remotes_2.1.1
[55] testthat_2.3.2 ellipsis_0.3.2 generics_0.0.2
[58] vctrs_0.3.8 lambda.r_1.2.4 tools_4.0.0
[61] glue_1.4.1 purrr_0.3.4 processx_3.5.2
[64] pkgload_1.0.2 yaml_2.2.1 colorspace_1.4-1
[67] sessioninfo_1.1.1 memoise_1.1.0