Jaspar_enrichment
ERM
2024-09-26
Last updated: 2024-09-26
Checks: 7 0
Knit directory: ATAC_learning/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20231016) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 6cdc685. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/figure/
Ignored: data/ACresp_SNP_table.csv
Ignored: data/ARR_SNP_table.csv
Ignored: data/All_merged_peaks.tsv
Ignored: data/CAD_gwas_dataframe.RDS
Ignored: data/CTX_SNP_table.csv
Ignored: data/Collapsed_expressed_NG_peak_table.csv
Ignored: data/DEG_toplist_sep_n45.RDS
Ignored: data/FRiP_first_run.txt
Ignored: data/Final_four_data/
Ignored: data/Frip_1_reads.csv
Ignored: data/Frip_2_reads.csv
Ignored: data/Frip_3_reads.csv
Ignored: data/Frip_4_reads.csv
Ignored: data/Frip_5_reads.csv
Ignored: data/Frip_6_reads.csv
Ignored: data/GO_KEGG_analysis/
Ignored: data/HF_SNP_table.csv
Ignored: data/Ind1_75DA24h_dedup_peaks.csv
Ignored: data/Ind1_TSS_peaks.RDS
Ignored: data/Ind1_firstfragment_files.txt
Ignored: data/Ind1_fragment_files.txt
Ignored: data/Ind1_peaks_list.RDS
Ignored: data/Ind1_summary.txt
Ignored: data/Ind2_TSS_peaks.RDS
Ignored: data/Ind2_fragment_files.txt
Ignored: data/Ind2_peaks_list.RDS
Ignored: data/Ind2_summary.txt
Ignored: data/Ind3_TSS_peaks.RDS
Ignored: data/Ind3_fragment_files.txt
Ignored: data/Ind3_peaks_list.RDS
Ignored: data/Ind3_summary.txt
Ignored: data/Ind4_79B24h_dedup_peaks.csv
Ignored: data/Ind4_TSS_peaks.RDS
Ignored: data/Ind4_V24h_fraglength.txt
Ignored: data/Ind4_fragment_files.txt
Ignored: data/Ind4_fragment_filesN.txt
Ignored: data/Ind4_peaks_list.RDS
Ignored: data/Ind4_summary.txt
Ignored: data/Ind5_TSS_peaks.RDS
Ignored: data/Ind5_fragment_files.txt
Ignored: data/Ind5_fragment_filesN.txt
Ignored: data/Ind5_peaks_list.RDS
Ignored: data/Ind5_summary.txt
Ignored: data/Ind6_TSS_peaks.RDS
Ignored: data/Ind6_fragment_files.txt
Ignored: data/Ind6_peaks_list.RDS
Ignored: data/Ind6_summary.txt
Ignored: data/Knowles_4.RDS
Ignored: data/Knowles_5.RDS
Ignored: data/Knowles_6.RDS
Ignored: data/LiSiLTDNRe_TE_df.RDS
Ignored: data/MI_gwas.RDS
Ignored: data/SNP_GWAS_PEAK_MRC_id
Ignored: data/SNP_GWAS_PEAK_MRC_id.csv
Ignored: data/SNP_gene_cat_list.tsv
Ignored: data/SNP_supp_schneider.RDS
Ignored: data/TE_info/
Ignored: data/all_TSSE_scores.RDS
Ignored: data/all_four_filtered_counts.txt
Ignored: data/aln_run1_results.txt
Ignored: data/anno_ind1_DA24h.RDS
Ignored: data/anno_ind4_V24h.RDS
Ignored: data/annotated_gwas_SNPS.csv
Ignored: data/background_n45_he_peaks.RDS
Ignored: data/cardiac_muscle_FRIP.csv
Ignored: data/cardiomyocyte_FRIP.csv
Ignored: data/col_ng_peak.csv
Ignored: data/cormotif_full_4_run.RDS
Ignored: data/cormotif_full_4_run_he.RDS
Ignored: data/cormotif_full_6_run.RDS
Ignored: data/cormotif_full_6_run_he.RDS
Ignored: data/cormotif_probability_45_list.csv
Ignored: data/cormotif_probability_45_list_he.csv
Ignored: data/cormotif_probability_all_6_list.csv
Ignored: data/cormotif_probability_all_6_list_he.csv
Ignored: data/embryo_heart_FRIP.csv
Ignored: data/enhancer_list_ENCFF126UHK.bed
Ignored: data/enhancerdata/
Ignored: data/filt_Peaks_efit2.RDS
Ignored: data/filt_Peaks_efit2_bl.RDS
Ignored: data/filt_Peaks_efit2_n45.RDS
Ignored: data/first_Peaksummarycounts.csv
Ignored: data/first_run_frag_counts.txt
Ignored: data/full_bedfiles/
Ignored: data/gene_ref.csv
Ignored: data/gwas_1_dataframe.RDS
Ignored: data/gwas_2_dataframe.RDS
Ignored: data/gwas_3_dataframe.RDS
Ignored: data/gwas_4_dataframe.RDS
Ignored: data/gwas_5_dataframe.RDS
Ignored: data/high_conf_peak_counts.csv
Ignored: data/high_conf_peak_counts.txt
Ignored: data/high_conf_peaks_bl_counts.txt
Ignored: data/high_conf_peaks_counts.txt
Ignored: data/hits_files/
Ignored: data/hyper_files/
Ignored: data/hypo_files/
Ignored: data/ind1_DA24hpeaks.RDS
Ignored: data/ind1_TSSE.RDS
Ignored: data/ind2_TSSE.RDS
Ignored: data/ind3_TSSE.RDS
Ignored: data/ind4_TSSE.RDS
Ignored: data/ind4_V24hpeaks.RDS
Ignored: data/ind5_TSSE.RDS
Ignored: data/ind6_TSSE.RDS
Ignored: data/initial_complete_stats_run1.txt
Ignored: data/left_ventricle_FRIP.csv
Ignored: data/median_24_lfc.RDS
Ignored: data/median_3_lfc.RDS
Ignored: data/mergedPeads.gff
Ignored: data/mergedPeaks.gff
Ignored: data/motif_list_full
Ignored: data/motif_list_n45
Ignored: data/motif_list_n45.RDS
Ignored: data/multiqc_fastqc_run1.txt
Ignored: data/multiqc_fastqc_run2.txt
Ignored: data/multiqc_genestat_run1.txt
Ignored: data/multiqc_genestat_run2.txt
Ignored: data/my_hc_filt_counts.RDS
Ignored: data/my_hc_filt_counts_n45.RDS
Ignored: data/n45_bedfiles/
Ignored: data/n45_files
Ignored: data/other_papers/
Ignored: data/peakAnnoList_1.RDS
Ignored: data/peakAnnoList_2.RDS
Ignored: data/peakAnnoList_24_full.RDS
Ignored: data/peakAnnoList_24_n45.RDS
Ignored: data/peakAnnoList_3.RDS
Ignored: data/peakAnnoList_3_full.RDS
Ignored: data/peakAnnoList_3_n45.RDS
Ignored: data/peakAnnoList_4.RDS
Ignored: data/peakAnnoList_5.RDS
Ignored: data/peakAnnoList_6.RDS
Ignored: data/peakAnnoList_Eight.RDS
Ignored: data/peakAnnoList_full_motif.RDS
Ignored: data/peakAnnoList_n45_motif.RDS
Ignored: data/siglist_full.RDS
Ignored: data/siglist_n45.RDS
Ignored: data/summary_peakIDandReHeat.csv
Ignored: data/test.list.RDS
Ignored: data/testnames.txt
Ignored: data/toplist_6.RDS
Ignored: data/toplist_full.RDS
Ignored: data/toplist_full_DAR_6.RDS
Ignored: data/toplist_n45.RDS
Ignored: data/trimmed_seq_length.csv
Ignored: data/unclassified_full_set_peaks.RDS
Ignored: data/unclassified_n45_set_peaks.RDS
Ignored: data/xstreme/
Ignored: trimmed_Ind1_75DA24h_S7.nodup.splited.bam/
Untracked files:
Untracked: DOX_DAR_assess.Rmd
Untracked: EAR_2_plot.pdf
Untracked: ESR_1_plot.pdf
Untracked: Firstcorr plotATAC.pdf
Untracked: IND1_2_3_6_corrplot.pdf
Untracked: LR_3_plot.pdf
Untracked: NR_1_plot.pdf
Untracked: analysis/LFC_corr.Rmd
Untracked: analysis/ReHeat_analysis.Rmd
Untracked: analysis/TE_analysis_old.Rmd
Untracked: analysis/my_hc_filt_counts.csv
Untracked: analysis/nucleosome_explore.Rmd
Untracked: code/IGV_snapshot_code.R
Untracked: code/LongDARlist.R
Untracked: code/TSSE.R
Untracked: code/just_for_Fun.R
Untracked: code/toplist_assembly.R
Untracked: lcpm_filtered_corplot.pdf
Untracked: log2cpmfragcount.pdf
Untracked: output/cormotif_probability_45_list.csv
Untracked: output/cormotif_probability_all_6_list.csv
Untracked: splited/
Untracked: trimmed_Ind1_75DA24h_S7.nodup.fragment.size.distribution.pdf
Untracked: trimmed_Ind1_75DA3h_S1.nodup.fragment.size.distribution.pdf
Unstaged changes:
Modified: analysis/CorMotif_data_n45.Rmd
Modified: analysis/Enrichment_motif.Rmd
Modified: analysis/GO_KEGG_analysis.Rmd
Modified: analysis/Peak_calling.Rmd
Modified: analysis/Raodah.Rmd
Modified: analysis/Raodah_mycount.Rmd
Modified: analysis/Smaller_set_DAR.Rmd
Modified: analysis/TE_analysis.Rmd
Modified: analysis/final_four_analysis.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/Jaspar_motif_ff.Rmd) and
HTML (docs/Jaspar_motif_ff.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 6cdc685 | reneeisnowhere | 2024-09-26 | updates with 4 sets of peaks |
library(tidyverse)
# library(ggsignif)
# library(cowplot)
# library(ggpubr)
# library(sjmisc)
library(kableExtra)
library(broom)
# library(biomaRt)
library(RColorBrewer)
# library(gprofiler2)
# library(qvalue)
library(ChIPseeker)
library("TxDb.Hsapiens.UCSC.hg38.knownGene")
library("org.Hs.eg.db")
# library(ATACseqQC)
library(rtracklayer)
library(edgeR)
library(ggfortify)
library(limma)
library(readr)
library(BiocGenerics)
library(gridExtra)
library(VennDiagram)
library(scales)
# library(ggVennDiagram)
library(Cormotif)
library(BiocParallel)
library(ggpubr)
library(devtools)
# install_github('davetang/bedr')
# library(bedr)
library(JASPAR2022)
library(TFBSTools)
library(MotifDb)
library(BSgenome.Hsapiens.UCSC.hg38)# LR_close_xstreme <- read_delim("~/Ward Lab/Cardiotoxicity/ATAC-folder/motif enrichment/meme-ff/LR_close_xstreme/xstreme.tsv",
# delim = "\t", escape_double = FALSE,
# trim_ws = TRUE)
#
#
# saveRDS(EAR_close_xstreme,"data/Final_four_data/xstreme/EAR_close_xstreme.RDS")
# saveRDS(EAR_open_xstreme,"data/Final_four_data/xstreme/EAR_open_xstreme.RDS")
# saveRDS(ESR_open_xstreme,"data/Final_four_data/xstreme/ESR_open_xstreme.RDS")
# saveRDS(ESR_close_xstreme,"data/Final_four_data/xstreme/ESR_close_xstreme.RDS")
# saveRDS(ESR_OC_xstreme,"data/Final_four_data/xstreme/ESR_OC_xstreme.RDS")
# saveRDS(LR_close_xstreme,"data/Final_four_data/xstreme/LR_close_xstreme.RDS")
# saveRDS(LR_open_xstreme,"data/Final_four_data/xstreme/LR_open_xstreme.RDS")
EAR_close_xstreme <-
readRDS("data/Final_four_data/xstreme/EAR_close_xstreme.RDS")%>%
slice_head(n = length(.$ID)-3)
EAR_open_xstreme <-
readRDS("data/Final_four_data/xstreme/EAR_open_xstreme.RDS")%>%
slice_head(n = length(.$ID)-3)
ESR_open_xstreme <-
readRDS("data/Final_four_data/xstreme/ESR_open_xstreme.RDS")%>%
slice_head(n = length(.$ID)-3)
ESR_close_xstreme <-
readRDS("data/Final_four_data/xstreme/ESR_close_xstreme.RDS")%>%
slice_head(n = length(.$ID)-3)
ESR_OC_xstreme <-
readRDS("data/Final_four_data/xstreme/ESR_OC_xstreme.RDS")%>%
slice_head(n = length(.$ID)-3)
LR_close_xstreme <-
readRDS("data/Final_four_data/xstreme/LR_close_xstreme.RDS")%>%
slice_head(n = length(.$ID)-3)
LR_open_xstreme <-
readRDS("data/Final_four_data/xstreme/LR_open_xstreme.RDS")%>%
slice_head(n = length(.$ID)-3)
sea_EAR_open <- readRDS("data/Final_four_data/xstreme/sea_EAR_open.RDS")%>%
slice_head(n = length(.$ID)-3)
sea_EAR_close <- readRDS("data/Final_four_data/xstreme/sea_EAR_close.RDS")%>%
slice_head(n = length(.$ID)-3)
sea_ESR_close <- readRDS("data/Final_four_data/xstreme/sea_ESR_close.RDS")%>%
slice_head(n = length(.$ID)-3)
sea_ESR_open <- readRDS("data/Final_four_data/xstreme/sea_ESR_open.RDS")%>%
slice_head(n = length(.$ID)-3)
sea_ESR_OC <- readRDS("data/Final_four_data/xstreme/sea_ESR_OC.RDS")%>%
slice_head(n = length(.$ID)-3)
sea_LR_open <- readRDS("data/Final_four_data/xstreme/sea_LR_open.RDS")%>%
slice_head(n = length(.$ID)-3)
sea_LR_close <- readRDS("data/Final_four_data/xstreme/sea_LR_close.RDS")%>%
slice_head(n = length(.$ID)-3)
# saveRDS(sea_LR_open,"data/Final_four_data/xstreme/sea_LR_open.RDS")
peakAnnoList_ff_8motif <- readRDS("data/Final_four_data/peakAnnoList_ff_8motif.RDS")
# EAR_df <- as.data.frame(peakAnnoList_n45_motif$EAR_n45_gr)
# EAR_df_gr <- as.GRanges(peakAnnoList_n45_motif$EAR_n45_gr)
#
# ESR_df <- as.data.frame(peakAnnoList_n45_motif$ESR_n45_gr)
# ESR_df_gr <- as.GRanges(peakAnnoList_n45_motif$ESR_n45_gr)
#
# LR_df <- as.data.frame(peakAnnoList_n45_motif$LR_n45_gr)
# LR_df_gr <- as.GRanges(peakAnnoList_n45_motif$LR_n45_gr)
#
# NR_df <- as.data.frame(peakAnnoList_n45_motif$NR_n45_gr)
# NR_df_gr <- as.GRanges(peakAnnoList_n45_motif$NR_n45_gr)
#
# toplistall_RNA <- readRDS("data/other_papers/toplistall_RNA.RDS")
###Because of how I applied the DEG system in RNA-seq analysis, the lFC is opposite of the
###counts. I did trt-veh instead of veh-trt. therefore I need to multiply lfc by -1 to get t
###the right correlation.
# toplistall_RNA <- toplistall_RNA %>%
# mutate(logFC = logFC*(-1))
#
# RNA_expresed_genes <- toplistall_RNA %>%
# # dplyr::filter(adj.P.Val <0.05) %>%
# mutate(expression = if_else(logFC<0,"down","up")) %>%
# dplyr::select(ENTREZID,SYMBOL,expression) %>%
# # dplyr::select(ENTREZID,SYMBOL) %>%
# unique(.)
# RNA_expresed_genes_DE <- toplistall_RNA %>%
# dplyr::filter(adj.P.Val <0.05) %>%
# mutate(expression = if_else(logFC<0,"down","up")) %>%
# dplyr::select(ENTREZID,SYMBOL,expression) %>%
# unique(.) Enrichment all peaks to NR peaks
EAR_open_xstreme%>%
dplyr::select(SIM_MOTIF,ALT_ID, ID,SEA_PVALUE,EVALUE) %>%
arrange(.,EVALUE) %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs in EAR v NR") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| SIM_MOTIF | ALT_ID | ID | SEA_PVALUE | EVALUE |
|---|---|---|---|---|
| MA0028.2 | ELK1 | MA0028.2 | 9.96e-21 | 8.47e-18 |
| MA0759.2 | ELK3 | MA0759.2 | 3.63e-20 | 3.09e-17 |
| MA0764.3 | ETV4 | MA0764.3 | 1.39e-17 | 1.18e-14 |
| MA0765.3 | ETV5 | MA0765.3 | 3e-16 | 2.55e-13 |
| MA0506.2 | Nrf1 | MA0506.2 | 1.8e-15 | 1.53e-12 |
| MA1713.1 | ZNF610 | MA1713.1 | 1.57e-13 | 1.33e-10 |
| MA0748.2 | YY2 | MA0748.2 | 4.05e-13 | 3.44e-10 |
| MA1721.1 | ZNF93 | MA1721.1 | 8.74e-12 | 7.44e-09 |
| MA1583.1 | ZFP57 | MA1583.1 | 9.6e-12 | 8.17e-09 |
| MA1650.1 | ZBTB14 | MA1650.1 | 3.47e-11 | 2.96e-08 |
| MA0645.1 | ETV6 | MA0645.1 | 5.29e-11 | 4.5e-08 |
| MA1651.1 | ZFP42 | MA1651.1 | 2.24e-10 | 1.91e-07 |
| MA0641.1 | ELF4 | MA0641.1 | 5.83e-09 | 4.96e-06 |
| MA1484.1 | ETS2 | MA1484.1 | 9.13e-09 | 7.77e-06 |
| MA1122.1 | TFDP1 | MA1122.1 | 1.08e-08 | 9.17e-06 |
| MA0763.1 | ETV3 | MA0763.1 | 3.48e-08 | 2.96e-05 |
| MA1931.1 | ELK1::HOXA1 | MA1931.1 | 4.49e-08 | 3.82e-05 |
| MA1708.1 | ETV7 | MA1708.1 | 7.67e-08 | 6.52e-05 |
| MA0156.3 | FEV | MA0156.3 | 1.56e-07 | 0.000133 |
| MA1483.2 | ELF2 | MA1483.2 | 1.84e-07 | 0.000157 |
| MA0760.1 | ERF | MA0760.1 | 3.21e-07 | 0.000273 |
| MA0475.2 | FLI1 | MA0475.2 | 3.4e-07 | 0.000289 |
| MA1711.1 | ZNF343 | MA1711.1 | 4.75e-07 | 0.000405 |
| MA0632.2 | TCFL5 | MA0632.2 | 5.58e-07 | 0.000475 |
| MA0024.3 | E2F1 | MA0024.3 | 8.18e-07 | 0.000696 |
| MA0695.1 | ZBTB7C | MA0695.1 | 1.22e-06 | 0.00104 |
| MA0146.2 | Zfx | MA0146.2 | 1.23e-06 | 0.00105 |
| MA0527.1 | ZBTB33 | MA0527.1 | 1.26e-06 | 0.00107 |
| MA0472.2 | EGR2 | MA0472.2 | 1.8e-06 | 0.00153 |
| MA1933.1 | ELK1::SREBF2 | MA1933.1 | 2.38e-06 | 0.00203 |
| MA0615.1 | Gmeb1 | MA0615.1 | 3.63e-06 | 0.00309 |
| MA0750.2 | ZBTB7A | MA0750.2 | 7.24e-06 | 0.00616 |
| MA0098.3 | ETS1 | MA0098.3 | 8.15e-06 | 0.00694 |
| MA0506.2 (Nrf1) | STREME-1 | 1-GCCGGCGCAGCC | 3.9e-12 | 0.00744 |
| MA0131.2 | HINFP | MA0131.2 | 9.26e-06 | 0.00788 |
| MA1727.1 | ZNF417 | MA1727.1 | 1.52e-05 | 0.0129 |
| 2-CGCTTCGHY | STREME-2 | 2-CGCTTCGHY | 8.24e-10 | 0.0157 |
| MA1569.1 | TFAP2E | MA1569.1 | 1.87e-05 | 0.0159 |
| MA0865.2 | E2F8 | MA0865.2 | 2.36e-05 | 0.0201 |
| 3-CGRGCCGCCK | STREME-3 | 3-CGRGCCGCCK | 1.27e-15 | 0.0202 |
| MA1684.1 | Foxn1 | MA1684.1 | 2.46e-05 | 0.0209 |
| MA1721.1 (ZNF93) | STREME-4 | 4-CGGCGGCSGCGSC | 6.87e-12 | 0.0228 |
| MA1099.2 | HES1 | MA1099.2 | 2.77e-05 | 0.0236 |
| MA0006.1 | Ahr::Arnt | MA0006.1 | 3.19e-05 | 0.0272 |
| 5-CGGCCGGCGA | STREME-5 | 5-CGGCCGGCGA | 2.2e-17 | 0.0274 |
| MA1932.1 | ELK1::HOXB13 | MA1932.1 | 3.35e-05 | 0.0285 |
| MA1513.1 | KLF15 | MA1513.1 | 3.47e-05 | 0.0295 |
| 6-CCGGCGCC | STREME-6 | 6-CCGGCGCC | 7.53e-17 | 0.0332 |
| MA0506.2 (Nrf1) | STREME-7 | 7-GCGSGCSCGC | 2.08e-19 | 0.045 |
| MA1107.2 (KLF9) | STREME-8 | 8-ACCACACACACACAA | 0.00786 | 0.0638 |
| 9-CGCCGAACTC | STREME-9 | 9-CGCCGAACTC | 4e-18 | 0.0643 |
| MA0759.2 (ELK3) | STREME-10 | 10-CCGGAAGTGCY | 1.49e-22 | 0.324 |
EAR_close_xstreme %>%
dplyr::select(SIM_MOTIF,ALT_ID, ID,SEA_PVALUE,EVALUE) %>%
arrange(.,EVALUE) %>%
# separate(SIM_MOTIF, into= c("SIM_MOTIF", "NAME"), sep= " ") %>%
# dplyr::filter(., EVALUE<0.05) %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs in EAR close") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "500px")| SIM_MOTIF | ALT_ID | ID | SEA_PVALUE | EVALUE |
|---|---|---|---|---|
| MA0477.2 | FOSL1 | MA0477.2 | 4.1e-93 | 3.55e-90 |
| MA0489.2 | Jun | MA0489.2 | 5.02e-88 | 4.35e-85 |
| MA0490.2 | JUNB | MA0490.2 | 5.4e-87 | 4.68e-84 |
| MA0099.3 | FOS::JUN | MA0099.3 | 2.02e-85 | 1.75e-82 |
| MA1130.1 | FOSL2::JUN | MA1130.1 | 4.88e-85 | 4.23e-82 |
| MA0491.2 | JUND | MA0491.2 | 2.41e-83 | 2.09e-80 |
| MA1141.1 | FOS::JUND | MA1141.1 | 1.24e-82 | 1.08e-79 |
| MA1928.1 | BNC2 | MA1928.1 | 1.69e-82 | 1.47e-79 |
| MA1144.1 | FOSL2::JUND | MA1144.1 | 2.01e-82 | 1.74e-79 |
| MA1634.1 | BATF | MA1634.1 | 3.56e-82 | 3.08e-79 |
| MA0462.2 | BATF::JUN | MA0462.2 | 3.59e-82 | 3.11e-79 |
| MA0835.2 | BATF3 | MA0835.2 | 1.93e-81 | 1.68e-78 |
| MA1633.2 | BACH1 | MA1633.2 | 5.68e-81 | 4.92e-78 |
| MA1138.1 | FOSL2::JUNB | MA1138.1 | 6.58e-80 | 5.7e-77 |
| MA0841.1 | NFE2 | MA0841.1 | 1.94e-77 | 1.68e-74 |
| MA1988.1 | Atf3 | MA1988.1 | 7.55e-77 | 6.54e-74 |
| MA1134.1 | FOS::JUNB | MA1134.1 | 6.56e-74 | 5.69e-71 |
| MA0478.1 | FOSL2 | MA0478.1 | 9.68e-74 | 8.39e-71 |
| MA1101.2 | BACH2 | MA1101.2 | 2.21e-73 | 1.92e-70 |
| MA0476.1 | FOS | MA0476.1 | 2.69e-72 | 2.33e-69 |
| MA0655.1 | JDP2 | MA0655.1 | 6.35e-72 | 5.51e-69 |
| MA1137.1 | FOSL1::JUNB | MA1137.1 | 3.87e-71 | 3.36e-68 |
| MA1128.1 | FOSL1::JUN | MA1128.1 | 1.19e-68 | 1.03e-65 |
| MA1142.1 | FOSL1::JUND | MA1142.1 | 4.84e-67 | 4.2e-64 |
| MA1135.1 | FOSB::JUNB | MA1135.1 | 6.93e-67 | 6.01e-64 |
| MA0501.1 | MAF::NFE2 | MA0501.1 | 3.17e-61 | 2.75e-58 |
| MA0766.2 | GATA5 | MA0766.2 | 2.45e-60 | 2.13e-57 |
| MA0089.2 | MAFG::NFE2L1 | MA0089.2 | 2.62e-59 | 2.27e-56 |
| MA1132.1 | JUN::JUNB | MA1132.1 | 3.14e-59 | 2.72e-56 |
| MA0496.3 | MAFK | MA0496.3 | 1.67e-51 | 1.45e-48 |
| MA1125.1 (ZNF384) | MEME-1 | WTTTTTTTTTTTTTT | 4.13e-10 | 1.5e-48 |
| MA0150.2 | Nfe2l2 | MA0150.2 | 5.26e-51 | 4.56e-48 |
| MA0036.3 | GATA2 | MA0036.3 | 2.16e-47 | 1.87e-44 |
| MA0774.1 | MEIS2 | MA0774.1 | 4.9e-47 | 4.24e-44 |
| MA0018.4 | CREB1 | MA0018.4 | 9.34e-46 | 8.1e-43 |
| MA0037.4 | Gata3 | MA0037.4 | 4.79e-43 | 4.15e-40 |
| MA1970.1 | TRPS1 | MA1970.1 | 1.23e-40 | 1.07e-37 |
| MA0782.2 | PKNOX1 | MA0782.2 | 1.29e-39 | 1.12e-36 |
| MA1652.1 (ZKSCAN5) | MEME-2 | KRRGRWGGVRGDGAG | 3.23e-35 | 1.7e-34 |
| MA1632.1 | ATF2 | MA1632.1 | 2.7e-36 | 2.34e-33 |
| MA1104.2 | GATA6 | MA1104.2 | 1.32e-35 | 1.15e-32 |
| MA0482.2 | GATA4 | MA0482.2 | 1.21e-34 | 1.05e-31 |
| MA1114.1 | PBX3 | MA1114.1 | 5.05e-33 | 4.38e-30 |
| MA0035.4 | GATA1 | MA0035.4 | 6.45e-33 | 5.6e-30 |
| MA0140.2 | GATA1::TAL1 | MA0140.2 | 2.15e-32 | 1.87e-29 |
| MA0775.1 | MEIS3 | MA0775.1 | 5.89e-32 | 5.1e-29 |
| MA0797.1 | TGIF2 | MA0797.1 | 3.88e-31 | 3.37e-28 |
| MA0842.2 | NRL | MA0842.2 | 8.98e-30 | 7.79e-27 |
| MA0612.2 | EMX1 | MA0612.2 | 1.45e-29 | 1.26e-26 |
| GCTGGAGTGCAGTGG | MEME-3 | GCTGGAGTGCAGTGG | 2.2e-22 | 7.7e-26 |
| MA0707.2 | MNX1 | MA0707.2 | 7.41e-28 | 6.42e-25 |
| MA0676.1 | Nr2e1 | MA0676.1 | 9.62e-28 | 8.34e-25 |
| MA1480.1 | DPRX | MA1480.1 | 1.22e-27 | 1.06e-24 |
| MA0829.2 | SREBF1 | MA0829.2 | 5.49e-27 | 4.76e-24 |
| MA0681.2 | PHOX2B | MA0681.2 | 5.89e-26 | 5.1e-23 |
| MA0070.1 | PBX1 | MA0070.1 | 8.7e-26 | 7.55e-23 |
| MA1519.1 | LHX5 | MA1519.1 | 1.77e-25 | 1.54e-22 |
| MA1564.1 | SP9 | MA1564.1 | 3.78e-25 | 3.28e-22 |
| MA1139.1 | FOSL2::JUNB | MA1139.1 | 4.44e-25 | 3.85e-22 |
| MA1495.1 | HOXA1 | MA1495.1 | 8.78e-25 | 7.61e-22 |
| MA0753.2 | ZNF740 | MA0753.2 | 2.48e-24 | 2.15e-21 |
| MA1145.1 | FOSL2::JUND | MA1145.1 | 3.36e-24 | 2.91e-21 |
| MA1653.1 | ZNF148 | MA1653.1 | 4.79e-24 | 4.16e-21 |
| MA0498.2 | MEIS1 | MA0498.2 | 5.18e-24 | 4.49e-21 |
| MA1512.1 | KLF11 | MA1512.1 | 1e-23 | 8.68e-21 |
| MA0596.1 | SREBF2 | MA0596.1 | 1.08e-23 | 9.37e-21 |
| MA0599.1 | KLF5 | MA0599.1 | 1.54e-23 | 1.33e-20 |
| MA0659.3 | Mafg | MA0659.3 | 1.68e-23 | 1.46e-20 |
| MA0595.1 | SREBF1 | MA0595.1 | 1.8e-23 | 1.56e-20 |
| MA0661.1 | MEOX1 | MA0661.1 | 2.31e-23 | 2.01e-20 |
| MA0611.2 | Dux | MA0611.2 | 3.39e-23 | 2.94e-20 |
| MA0492.1 | JUND | MA0492.1 | 4.82e-23 | 4.18e-20 |
| MA0027.2 | EN1 | MA0027.2 | 7.92e-23 | 6.87e-20 |
| MA0039.4 | KLF4 | MA0039.4 | 1.04e-22 | 8.97e-20 |
| MA1110.2 | Nr1H4 | MA1110.2 | 1.85e-22 | 1.6e-19 |
| MA0904.2 | HOXB5 | MA0904.2 | 3.21e-22 | 2.78e-19 |
| MA1593.1 | ZNF317 | MA1593.1 | 5.59e-22 | 4.85e-19 |
| MA0886.1 | EMX2 | MA0886.1 | 6.01e-22 | 5.21e-19 |
| MA1511.2 | KLF10 | MA1511.2 | 8.57e-22 | 7.43e-19 |
| RGRGTCTYRCTCTGT | MEME-6 | RGRGTCTYRCTCTGT | 3.52e-19 | 1.4e-18 |
| MA0742.2 | KLF12 | MA0742.2 | 1.64e-21 | 1.42e-18 |
| GCCTGTAATCCCAGC | MEME-4 | GCCTGTAATCCCAGC | 3.37e-15 | 1.5e-18 |
| MA1107.2 | KLF9 | MA1107.2 | 3.04e-21 | 2.64e-18 |
| MA0881.1 | Dlx4 | MA0881.1 | 4.6e-21 | 3.99e-18 |
| MA0631.1 | Six3 | MA0631.1 | 5.23e-21 | 4.54e-18 |
| MA0709.1 | Msx3 | MA0709.1 | 6.26e-21 | 5.43e-18 |
| MA1959.1 | KLF7 | MA1959.1 | 6.31e-21 | 5.47e-18 |
| MA1571.1 | TGIF2LX | MA1571.1 | 6.63e-21 | 5.75e-18 |
| MA0880.1 | Dlx3 | MA0880.1 | 8.29e-21 | 7.19e-18 |
| MA0666.2 | MSX1 | MA0666.2 | 8.82e-21 | 7.64e-18 |
| MA0705.1 | Lhx8 | MA0705.1 | 1.23e-20 | 1.07e-17 |
| MA1143.1 | FOSL1::JUND | MA1143.1 | 1.28e-20 | 1.11e-17 |
| MA1530.1 | NKX6-3 | MA1530.1 | 2.13e-20 | 1.85e-17 |
| MA1129.1 | FOSL1::JUN | MA1129.1 | 3.06e-20 | 2.65e-17 |
| MA0685.2 | SP4 | MA0685.2 | 5.01e-20 | 4.35e-17 |
| MA0516.3 | SP2 | MA0516.3 | 7.14e-20 | 6.19e-17 |
| MA0132.2 | PDX1 | MA0132.2 | 7.24e-20 | 6.28e-17 |
| MA0882.1 | DLX6 | MA0882.1 | 8.06e-20 | 6.99e-17 |
| MA0741.1 | KLF16 | MA0741.1 | 1.08e-19 | 9.34e-17 |
| MA0680.2 | Pax7 | MA0680.2 | 1.11e-19 | 9.63e-17 |
| MA0628.1 | POU6F1 | MA0628.1 | 1.13e-19 | 9.83e-17 |
| MA0654.1 | ISX | MA0654.1 | 1.22e-19 | 1.06e-16 |
| MA0688.1 | TBX2 | MA0688.1 | 1.26e-19 | 1.09e-16 |
| MA0894.1 | HESX1 | MA0894.1 | 1.7e-19 | 1.47e-16 |
| MA0644.2 | ESX1 | MA0644.2 | 2.36e-19 | 2.05e-16 |
| MA1522.1 | MAZ | MA1522.1 | 2.98e-19 | 2.58e-16 |
| MA0802.1 | TBR1 | MA0802.1 | 4.51e-19 | 3.91e-16 |
| MA0912.2 | HOXD3 | MA0912.2 | 5.43e-19 | 4.71e-16 |
| MA0117.2 | Mafb | MA0117.2 | 1.09e-18 | 9.48e-16 |
| MA0902.2 | HOXB2 | MA0902.2 | 1.23e-18 | 1.07e-15 |
| MA1572.1 | TGIF2LY | MA1572.1 | 1.52e-18 | 1.31e-15 |
| MA0805.1 | TBX1 | MA0805.1 | 2.83e-18 | 2.45e-15 |
| MA1578.1 | VEZF1 | MA1578.1 | 2.95e-18 | 2.56e-15 |
| MA1535.1 | NR2C1 | MA1535.1 | 3.56e-18 | 3.09e-15 |
| MA0889.1 | GBX1 | MA0889.1 | 3.63e-18 | 3.14e-15 |
| MA0708.2 | MSX2 | MA0708.2 | 3.73e-18 | 3.23e-15 |
| MA1720.1 | ZNF85 | MA1720.1 | 4.72e-18 | 4.09e-15 |
| MA1618.1 | Ptf1a | MA1618.1 | 5.6e-18 | 4.85e-15 |
| MA0803.1 | TBX15 | MA0803.1 | 6.04e-18 | 5.24e-15 |
| MA0690.2 | TBX21 | MA0690.2 | 6.13e-18 | 5.31e-15 |
| MA0162.4 | EGR1 | MA0162.4 | 6.18e-18 | 5.36e-15 |
| MA0495.3 | MAFF | MA0495.3 | 6.98e-18 | 6.05e-15 |
| MA1710.1 | ZNF257 | MA1710.1 | 7.54e-18 | 6.54e-15 |
| MA0740.2 | KLF14 | MA0740.2 | 8.03e-18 | 6.96e-15 |
| MA1630.2 | ZNF281 | MA1630.2 | 8.34e-18 | 7.23e-15 |
| MA1996.1 | Nr1H2 | MA1996.1 | 9.66e-18 | 8.37e-15 |
| MA0710.1 | NOTO | MA0710.1 | 1.29e-17 | 1.12e-14 |
| MA0691.1 | TFAP4 | MA0691.1 | 1.48e-17 | 1.28e-14 |
| MA0806.1 | TBX4 | MA0806.1 | 1.54e-17 | 1.33e-14 |
| MA0258.2 | ESR2 | MA0258.2 | 1.9e-17 | 1.65e-14 |
| MA0704.1 | Lhx4 | MA0704.1 | 2.49e-17 | 2.16e-14 |
| MA2003.1 | NKX2-4 | MA2003.1 | 3.82e-17 | 3.31e-14 |
| MA0672.1 | NKX2-3 | MA0672.1 | 3.86e-17 | 3.34e-14 |
| MA0528.2 | ZNF263 | MA0528.2 | 3.92e-17 | 3.4e-14 |
| MA0700.2 | LHX2 | MA0700.2 | 4e-17 | 3.47e-14 |
| MA0093.3 | USF1 | MA0093.3 | 4.5e-17 | 3.9e-14 |
| MA0620.3 | MITF | MA0620.3 | 8.27e-17 | 7.17e-14 |
| MA0879.2 | DLX1 | MA0879.2 | 9.57e-17 | 8.3e-14 |
| MA0746.2 | SP3 | MA0746.2 | 1.15e-16 | 9.98e-14 |
| MA0713.1 | PHOX2A | MA0713.1 | 1.27e-16 | 1.1e-13 |
| MA0726.1 | VSX2 | MA0726.1 | 1.4e-16 | 1.22e-13 |
| MA1723.1 | PRDM9 | MA1723.1 | 1.62e-16 | 1.41e-13 |
| MA0483.1 | Gfi1B | MA0483.1 | 1.69e-16 | 1.47e-13 |
| MA0017.2 | NR2F1 | MA0017.2 | 1.7e-16 | 1.48e-13 |
| MA1119.1 | SIX2 | MA1119.1 | 1.79e-16 | 1.55e-13 |
| MA0014.3 | PAX5 | MA0014.3 | 2.36e-16 | 2.05e-13 |
| MA0800.1 | EOMES | MA0800.1 | 3.49e-16 | 3.02e-13 |
| MA0910.2 | HOXD8 | MA0910.2 | 3.68e-16 | 3.19e-13 |
| MA0717.1 | RAX2 | MA0717.1 | 3.78e-16 | 3.27e-13 |
| MA1566.2 | TBX3 | MA1566.2 | 3.98e-16 | 3.45e-13 |
| MA1146.1 | NR1H4::RXRA | MA1146.1 | 4.31e-16 | 3.74e-13 |
| MA1505.1 | HOXC8 | MA1505.1 | 4.61e-16 | 4e-13 |
| MA1729.1 | ZNF680 | MA1729.1 | 5.1e-16 | 4.42e-13 |
| MA1481.1 | DRGX | MA1481.1 | 5.31e-16 | 4.6e-13 |
| MA0674.1 | NKX6-1 | MA0674.1 | 6.43e-16 | 5.58e-13 |
| MA1108.2 | MXI1 | MA1108.2 | 8.62e-16 | 7.47e-13 |
| MA1960.1 | MGA::EVX1 | MA1960.1 | 1.01e-15 | 8.72e-13 |
| MA0673.1 | NKX2-8 | MA0673.1 | 1.05e-15 | 9.1e-13 |
| MA0591.1 | Bach1::Mafk | MA0591.1 | 1.09e-15 | 9.45e-13 |
| MA1644.1 | NFYC | MA1644.1 | 1.26e-15 | 1.09e-12 |
| MA0747.1 | SP8 | MA0747.1 | 1.32e-15 | 1.14e-12 |
| MA1961.1 | PATZ1 | MA1961.1 | 1.62e-15 | 1.41e-12 |
| MA0900.2 | HOXA2 | MA0900.2 | 1.68e-15 | 1.45e-12 |
| MA1102.2 | CTCFL | MA1102.2 | 1.82e-15 | 1.57e-12 |
| TGTGTGTGTGTGTGT | MEME-5 | TGTGTGTGTGTGTGT | 0.614 | 1.7e-12 |
| MA0719.1 | RHOXF1 | MA0719.1 | 2.01e-15 | 1.74e-12 |
| MA1474.1 | CREB3L4 | MA1474.1 | 2.72e-15 | 2.36e-12 |
| MA0715.1 | PROP1 | MA0715.1 | 3.27e-15 | 2.83e-12 |
| MA1476.2 | Dlx5 | MA1476.2 | 3.54e-15 | 3.07e-12 |
| MA1994.1 | Nkx2-1 | MA1994.1 | 3.83e-15 | 3.32e-12 |
| MA0125.1 | Nobox | MA0125.1 | 3.92e-15 | 3.39e-12 |
| MA1615.1 | Plagl1 | MA1615.1 | 3.95e-15 | 3.42e-12 |
| MA0908.1 | HOXD11 | MA0908.1 | 4.87e-15 | 4.22e-12 |
| MA0875.1 | BARX1 | MA0875.1 | 5.1e-15 | 4.42e-12 |
| MA0883.1 | Dmbx1 | MA0883.1 | 5.28e-15 | 4.58e-12 |
| MA1965.1 | SP5 | MA1965.1 | 5.34e-15 | 4.63e-12 |
| MA0504.1 | NR2C2 | MA0504.1 | 6.76e-15 | 5.86e-12 |
| MA1133.1 | JUN::JUNB | MA1133.1 | 7.53e-15 | 6.53e-12 |
| MA0807.1 | TBX5 | MA0807.1 | 7.87e-15 | 6.82e-12 |
| MA1515.1 | KLF2 | MA1515.1 | 1.05e-14 | 9.08e-12 |
| MA0052.4 | MEF2A | MA0052.4 | 1.26e-14 | 1.09e-11 |
| MA0626.1 | Npas2 | MA0626.1 | 1.92e-14 | 1.67e-11 |
| MA1150.1 | RORB | MA1150.1 | 2.11e-14 | 1.83e-11 |
| MA0725.1 | VSX1 | MA0725.1 | 2.14e-14 | 1.85e-11 |
| MA0079.5 | SP1 | MA0079.5 | 2.2e-14 | 1.91e-11 |
| MA0689.1 | TBX20 | MA0689.1 | 2.46e-14 | 2.13e-11 |
| MA1126.1 | FOS::JUN | MA1126.1 | 2.98e-14 | 2.58e-11 |
| MA0071.1 | RORA | MA0071.1 | 3.03e-14 | 2.63e-11 |
| MA0768.2 | Lef1 | MA0768.2 | 3.04e-14 | 2.64e-11 |
| MA0075.3 | PRRX2 | MA0075.3 | 3.51e-14 | 3.04e-11 |
| MA1147.1 | NR4A2::RXRA | MA1147.1 | 4.48e-14 | 3.88e-11 |
| MA1475.1 | CREB3L4 | MA1475.1 | 4.91e-14 | 4.26e-11 |
| MA0639.1 | DBP | MA0639.1 | 5.29e-14 | 4.58e-11 |
| MA0701.2 | LHX9 | MA0701.2 | 7.45e-14 | 6.46e-11 |
| MA0130.1 | ZNF354C | MA0130.1 | 8.79e-14 | 7.62e-11 |
| MA0833.2 | ATF4 | MA0833.2 | 1.08e-13 | 9.33e-11 |
| MA0601.1 | Arid3b | MA0601.1 | 1.09e-13 | 9.48e-11 |
| MA1514.1 | KLF17 | MA1514.1 | 1.23e-13 | 1.07e-10 |
| MA0642.2 | EN2 | MA0642.2 | 1.25e-13 | 1.09e-10 |
| MA0761.2 | ETV1 | MA0761.2 | 1.27e-13 | 1.1e-10 |
| MA1463.1 | ARGFX | MA1463.1 | 1.42e-13 | 1.23e-10 |
| MA1587.1 | ZNF135 | MA1587.1 | 1.63e-13 | 1.41e-10 |
| MA0063.2 | NKX2-5 | MA0063.2 | 1.63e-13 | 1.41e-10 |
| MA1995.1 | Npas4 | MA1995.1 | 1.7e-13 | 1.48e-10 |
| MA1636.1 | CEBPG | MA1636.1 | 1.99e-13 | 1.73e-10 |
| MA1643.1 | NFIB | MA1643.1 | 2.01e-13 | 1.74e-10 |
| MA1567.2 | Tbx6 | MA1567.2 | 2.04e-13 | 1.77e-10 |
| MA0748.2 | YY2 | MA0748.2 | 2.17e-13 | 1.89e-10 |
| MA0043.3 | HLF | MA0043.3 | 2.24e-13 | 1.94e-10 |
| MA1112.2 | NR4A1 | MA1112.2 | 2.37e-13 | 2.05e-10 |
| MA0714.1 | PITX3 | MA0714.1 | 3.38e-13 | 2.93e-10 |
| MA0500.2 | MYOG | MA0500.2 | 4.01e-13 | 3.48e-10 |
| MA0597.2 | THAP1 | MA0597.2 | 4.51e-13 | 3.91e-10 |
| MA1652.1 | ZKSCAN5 | MA1652.1 | 5e-13 | 4.33e-10 |
| MA0695.1 | ZBTB7C | MA0695.1 | 5.28e-13 | 4.57e-10 |
| MA0060.3 | NFYA | MA0060.3 | 5.6e-13 | 4.86e-10 |
| MA1136.1 | FOSB::JUNB | MA1136.1 | 6.64e-13 | 5.75e-10 |
| MA1972.1 | ZFP14 | MA1972.1 | 7.01e-13 | 6.08e-10 |
| MA0733.1 | EGR4 | MA0733.1 | 7.35e-13 | 6.37e-10 |
| MA0718.1 | RAX | MA0718.1 | 7.65e-13 | 6.63e-10 |
| MA0845.1 | FOXB1 | MA0845.1 | 7.77e-13 | 6.74e-10 |
| MA1650.1 | ZBTB14 | MA1650.1 | 8.36e-13 | 7.24e-10 |
| MA0029.1 | Mecom | MA0029.1 | 8.94e-13 | 7.75e-10 |
| MA0031.1 | FOXD1 | MA0031.1 | 9.74e-13 | 8.45e-10 |
| MA0903.1 | HOXB3 | MA0903.1 | 1.33e-12 | 1.15e-09 |
| MA1577.1 | TLX2 | MA1577.1 | 1.42e-12 | 1.23e-09 |
| MA1547.2 | PITX2 | MA1547.2 | 1.55e-12 | 1.34e-09 |
| MA0801.1 | MGA | MA0801.1 | 1.65e-12 | 1.43e-09 |
| MA1113.2 | PBX2 | MA1113.2 | 1.72e-12 | 1.49e-09 |
| MA1640.1 | MEIS2 | MA1640.1 | 1.86e-12 | 1.61e-09 |
| MA0098.3 | ETS1 | MA0098.3 | 1.89e-12 | 1.64e-09 |
| MA0820.1 | FIGLA | MA0820.1 | 2.43e-12 | 2.11e-09 |
| MA1974.1 | ZNF211 | MA1974.1 | 2.55e-12 | 2.21e-09 |
| MA0141.3 | ESRRB | MA0141.3 | 2.56e-12 | 2.22e-09 |
| MA1639.1 | MEIS1 | MA1639.1 | 2.7e-12 | 2.34e-09 |
| MA0003.4 | TFAP2A | MA0003.4 | 3e-12 | 2.6e-09 |
| MA1520.1 | MAF | MA1520.1 | 3.25e-12 | 2.81e-09 |
| MA0890.1 | GBX2 | MA0890.1 | 3.62e-12 | 3.14e-09 |
| MA0698.1 | ZBTB18 | MA0698.1 | 3.64e-12 | 3.15e-09 |
| MA0124.2 | Nkx3-1 | MA0124.2 | 3.65e-12 | 3.16e-09 |
| MA1536.1 | NR2C2 | MA1536.1 | 3.98e-12 | 3.45e-09 |
| MA0899.1 | HOXA10 | MA0899.1 | 4.23e-12 | 3.67e-09 |
| MA0667.1 | MYF6 | MA0667.1 | 4.32e-12 | 3.74e-09 |
| MA1467.2 | Atoh1 | MA1467.2 | 4.41e-12 | 3.82e-09 |
| MA0142.1 | Pou5f1::Sox2 | MA0142.1 | 4.99e-12 | 4.33e-09 |
| MA0602.1 | Arid5a | MA0602.1 | 5e-12 | 4.34e-09 |
| MA0724.1 | VENTX | MA0724.1 | 5.24e-12 | 4.54e-09 |
| MA0158.2 | HOXA5 | MA0158.2 | 5.27e-12 | 4.57e-09 |
| MA1500.1 | HOXB6 | MA1500.1 | 6.16e-12 | 5.34e-09 |
| MA1523.1 | MSANTD3 | MA1523.1 | 6.68e-12 | 5.79e-09 |
| MA0046.2 | HNF1A | MA0046.2 | 7.07e-12 | 6.13e-09 |
| MA1570.1 | TFAP4 | MA1570.1 | 7.53e-12 | 6.53e-09 |
| MA0769.2 | TCF7 | MA0769.2 | 7.75e-12 | 6.72e-09 |
| MA0523.1 | TCF7L2 | MA0523.1 | 9.12e-12 | 7.9e-09 |
| MA0468.1 | DUX4 | MA0468.1 | 1.01e-11 | 8.71e-09 |
| MA1503.1 | HOXB9 | MA1503.1 | 1.02e-11 | 8.87e-09 |
| MA1621.1 | Rbpjl | MA1621.1 | 1.09e-11 | 9.43e-09 |
| MA1521.1 | MAFA | MA1521.1 | 1.13e-11 | 9.78e-09 |
| MA0658.1 | LHX6 | MA0658.1 | 1.15e-11 | 9.99e-09 |
| MA0656.1 | JDP2 | MA0656.1 | 1.18e-11 | 1.03e-08 |
| MA1986.1 | ZNF692 | MA1986.1 | 1.31e-11 | 1.14e-08 |
| MA0095.3 | Yy1 | MA0095.3 | 1.42e-11 | 1.24e-08 |
| MA0675.1 | NKX6-2 | MA0675.1 | 1.52e-11 | 1.32e-08 |
| MA0621.1 | mix-a | MA0621.1 | 1.57e-11 | 1.37e-08 |
| MA1728.1 | ZNF549 | MA1728.1 | 2.42e-11 | 2.1e-08 |
| MA1991.1 | Hnf1A | MA1991.1 | 2.8e-11 | 2.43e-08 |
| MA0151.1 | Arid3a | MA0151.1 | 2.8e-11 | 2.43e-08 |
| MA0830.2 | TCF4 | MA0830.2 | 2.91e-11 | 2.52e-08 |
| MA1109.1 | NEUROD1 | MA1109.1 | 3.27e-11 | 2.83e-08 |
| MA0684.2 | RUNX3 | MA0684.2 | 3.34e-11 | 2.9e-08 |
| MA0143.4 | SOX2 | MA0143.4 | 3.89e-11 | 3.37e-08 |
| MA1707.1 | DMRTA1 | MA1707.1 | 4.12e-11 | 3.57e-08 |
| MA0682.2 | PITX1 | MA0682.2 | 4.36e-11 | 3.78e-08 |
| MA0788.1 | POU3F3 | MA0788.1 | 4.85e-11 | 4.21e-08 |
| MA1651.1 | ZFP42 | MA1651.1 | 5.08e-11 | 4.4e-08 |
| MA0767.1 | GCM2 | MA0767.1 | 5.18e-11 | 4.49e-08 |
| MA1527.1 | NFIC | MA1527.1 | 5.26e-11 | 4.56e-08 |
| MA0734.3 | Gli2 | MA0734.3 | 5.31e-11 | 4.61e-08 |
| MA0515.1 | Sox6 | MA0515.1 | 5.39e-11 | 4.67e-08 |
| MA0122.3 | Nkx3-2 | MA0122.3 | 5.55e-11 | 4.81e-08 |
| MA0648.1 | GSC | MA0648.1 | 6.15e-11 | 5.33e-08 |
| MA0692.1 | TFEB | MA0692.1 | 6.85e-11 | 5.94e-08 |
| MA0669.1 | NEUROG2 | MA0669.1 | 7.16e-11 | 6.21e-08 |
| MA0751.1 | ZIC4 | MA0751.1 | 8.42e-11 | 7.3e-08 |
| MA0650.3 | Hoxa13 | MA0650.3 | 8.54e-11 | 7.4e-08 |
| MA0793.1 | POU6F2 | MA0793.1 | 9.27e-11 | 8.04e-08 |
| MA1152.1 | SOX15 | MA1152.1 | 1.01e-10 | 8.74e-08 |
| MA1979.1 | ZNF416 | MA1979.1 | 1.01e-10 | 8.8e-08 |
| MA1565.1 | TBX18 | MA1565.1 | 1.04e-10 | 9e-08 |
| MA1990.1 | Gli1 | MA1990.1 | 1.08e-10 | 9.33e-08 |
| MA0119.1 | NFIC::TLX1 | MA0119.1 | 1.09e-10 | 9.45e-08 |
| MA0901.2 | HOXB13 | MA0901.2 | 1.11e-10 | 9.59e-08 |
| MA0892.1 | GSX1 | MA0892.1 | 1.12e-10 | 9.75e-08 |
| MA1710.1 (ZNF257) | MEME-7 | CTGAGGCAGGAGRAT | 3.49e-13 | 1e-07 |
| MA0831.3 | TFE3 | MA0831.3 | 1.54e-10 | 1.33e-07 |
| MA0102.4 | CEBPA | MA0102.4 | 1.6e-10 | 1.39e-07 |
| MA1629.1 | Zic2 | MA1629.1 | 1.64e-10 | 1.42e-07 |
| MA0511.2 | RUNX2 | MA0511.2 | 1.71e-10 | 1.48e-07 |
| MA1501.1 | HOXB7 | MA1501.1 | 1.73e-10 | 1.5e-07 |
| MA1504.1 | HOXC4 | MA1504.1 | 1.97e-10 | 1.71e-07 |
| MA0025.2 | NFIL3 | MA0025.2 | 2.06e-10 | 1.78e-07 |
| MA0664.1 | MLXIPL | MA0664.1 | 2.38e-10 | 2.06e-07 |
| MA0136.3 | Elf5 | MA0136.3 | 2.46e-10 | 2.13e-07 |
| MA0712.2 | OTX2 | MA0712.2 | 2.47e-10 | 2.14e-07 |
| MA1502.1 | HOXB8 | MA1502.1 | 2.58e-10 | 2.24e-07 |
| MA1120.1 | SOX13 | MA1120.1 | 2.69e-10 | 2.33e-07 |
| MA1950.1 | FLI1::FOXI1 | MA1950.1 | 2.74e-10 | 2.38e-07 |
| MA0706.1 | MEOX2 | MA0706.1 | 3.15e-10 | 2.73e-07 |
| MA1468.1 | ATOH7 | MA1468.1 | 3.22e-10 | 2.79e-07 |
| MA0465.2 | CDX2 | MA0465.2 | 3.27e-10 | 2.84e-07 |
| MA1516.1 | KLF3 | MA1516.1 | 3.36e-10 | 2.91e-07 |
| MA0905.1 | HOXC10 | MA0905.1 | 3.38e-10 | 2.93e-07 |
| MA0723.2 | VAX2 | MA0723.2 | 3.5e-10 | 3.03e-07 |
| MA1606.1 | Foxf1 | MA1606.1 | 3.6e-10 | 3.12e-07 |
| MA0789.1 | POU3F4 | MA0789.1 | 4.43e-10 | 3.84e-07 |
| MA0876.1 | BSX | MA0876.1 | 4.68e-10 | 4.05e-07 |
| MA0111.1 | Spz1 | MA0111.1 | 5.04e-10 | 4.37e-07 |
| MA0846.1 | FOXC2 | MA0846.1 | 5.16e-10 | 4.47e-07 |
| MA1713.1 | ZNF610 | MA1713.1 | 5.28e-10 | 4.58e-07 |
| MA0032.2 | FOXC1 | MA0032.2 | 5.47e-10 | 4.75e-07 |
| MA1631.1 | ASCL1 | MA1631.1 | 5.55e-10 | 4.82e-07 |
| MA1645.1 | NKX2-2 | MA1645.1 | 5.58e-10 | 4.84e-07 |
| MA1963.1 | SATB1 | MA1963.1 | 6.15e-10 | 5.33e-07 |
| MA1528.1 | NFIX | MA1528.1 | 6.18e-10 | 5.36e-07 |
| MA0614.1 | Foxj2 | MA0614.1 | 7.14e-10 | 6.19e-07 |
| MA1518.2 | Lhx1 | MA1518.2 | 7.92e-10 | 6.86e-07 |
| MA0623.2 | NEUROG1 | MA0623.2 | 8.33e-10 | 7.22e-07 |
| MA1131.1 | FOSL2::JUN | MA1131.1 | 8.37e-10 | 7.26e-07 |
| MA0711.1 | OTX1 | MA0711.1 | 8.99e-10 | 7.8e-07 |
| MA0693.3 | Vdr | MA0693.3 | 9.27e-10 | 8.04e-07 |
| MA0630.1 | SHOX | MA0630.1 | 9.82e-10 | 8.52e-07 |
| MA0884.2 | DUXA | MA0884.2 | 1.01e-09 | 8.72e-07 |
| MA1549.1 | POU6F1 | MA1549.1 | 1.04e-09 | 9.04e-07 |
| MA0897.1 | Hmx2 | MA0897.1 | 1.11e-09 | 9.62e-07 |
| MA0720.1 | Shox2 | MA0720.1 | 1.17e-09 | 1.02e-06 |
| MA0522.3 | TCF3 | MA0522.3 | 1.2e-09 | 1.04e-06 |
| MA1517.1 | KLF6 | MA1517.1 | 1.25e-09 | 1.09e-06 |
| MA0041.2 | FOXD3 | MA0041.2 | 1.27e-09 | 1.1e-06 |
| MA1574.1 | THRB | MA1574.1 | 1.33e-09 | 1.15e-06 |
| MA0699.1 | LBX2 | MA0699.1 | 1.36e-09 | 1.18e-06 |
| MA1656.1 | ZNF449 | MA1656.1 | 1.38e-09 | 1.19e-06 |
| MA0488.1 | JUN | MA0488.1 | 1.51e-09 | 1.31e-06 |
| MA0603.1 | Arntl | MA0603.1 | 1.56e-09 | 1.35e-06 |
| MA1483.2 | ELF2 | MA1483.2 | 1.66e-09 | 1.44e-06 |
| MA0497.1 | MEF2C | MA0497.1 | 1.76e-09 | 1.53e-06 |
| MA1607.1 | Foxl2 | MA1607.1 | 1.82e-09 | 1.58e-06 |
| MA1628.1 | Zic1::Zic2 | MA1628.1 | 2.04e-09 | 1.77e-06 |
| MA0867.2 | SOX4 | MA0867.2 | 2.08e-09 | 1.8e-06 |
| MA1977.1 | ZNF324 | MA1977.1 | 2.2e-09 | 1.91e-06 |
| MA0745.2 | SNAI2 | MA0745.2 | 2.38e-09 | 2.06e-06 |
| MA0754.2 | CUX1 | MA0754.2 | 2.65e-09 | 2.3e-06 |
| MA1111.1 | NR2F2 | MA1111.1 | 2.65e-09 | 2.3e-06 |
| MA0502.2 | NFYB | MA0502.2 | 2.8e-09 | 2.43e-06 |
| MA0847.3 | FOXD2 | MA0847.3 | 2.95e-09 | 2.55e-06 |
| MA1603.1 | Dmrt1 | MA1603.1 | 2.99e-09 | 2.59e-06 |
| MA0885.2 | Dlx2 | MA0885.2 | 2.99e-09 | 2.59e-06 |
| MA0508.3 | PRDM1 | MA0508.3 | 3.2e-09 | 2.77e-06 |
| MA0808.1 | TEAD3 | MA0808.1 | 3.32e-09 | 2.88e-06 |
| MA1992.1 | Ikzf3 | MA1992.1 | 3.46e-09 | 3e-06 |
| MA0721.1 | UNCX | MA0721.1 | 3.61e-09 | 3.13e-06 |
| MA1498.2 | HOXA7 | MA1498.2 | 3.66e-09 | 3.17e-06 |
| MA1149.1 | RARA::RXRG | MA1149.1 | 3.73e-09 | 3.24e-06 |
| MA1731.1 | ZNF768 | MA1731.1 | 3.92e-09 | 3.4e-06 |
| MA0467.2 | Crx | MA0467.2 | 4.24e-09 | 3.68e-06 |
| MA1620.1 | Ptf1A | MA1620.1 | 4.32e-09 | 3.75e-06 |
| MA0092.1 | Hand1::Tcf3 | MA0092.1 | 4.46e-09 | 3.86e-06 |
| MA0499.2 | MYOD1 | MA0499.2 | 5.09e-09 | 4.41e-06 |
| MA0613.1 | FOXG1 | MA0613.1 | 5.2e-09 | 4.51e-06 |
| MA1944.1 | ETV5::DRGX | MA1944.1 | 5.3e-09 | 4.6e-06 |
| MA0834.1 | ATF7 | MA0834.1 | 5.83e-09 | 5.05e-06 |
| MA0609.2 | CREM | MA0609.2 | 6.61e-09 | 5.73e-06 |
| MA1493.1 | HES6 | MA1493.1 | 6.67e-09 | 5.78e-06 |
| MA0635.1 | BARHL2 | MA0635.1 | 6.84e-09 | 5.93e-06 |
| MA1100.2 | ASCL1 | MA1100.2 | 7.19e-09 | 6.23e-06 |
| MA0750.2 | ZBTB7A | MA0750.2 | 7.22e-09 | 6.26e-06 |
| MA1594.1 | ZNF382 | MA1594.1 | 7.36e-09 | 6.38e-06 |
| MA1127.1 | FOSB::JUN | MA1127.1 | 7.92e-09 | 6.87e-06 |
| MA0814.2 | TFAP2C | MA0814.2 | 8.05e-09 | 6.98e-06 |
| MA0146.2 | Zfx | MA0146.2 | 8.99e-09 | 7.79e-06 |
| MA0042.2 | FOXI1 | MA0042.2 | 9.38e-09 | 8.14e-06 |
| MA1987.1 | ZNF701 | MA1987.1 | 9.42e-09 | 8.17e-06 |
| MA0816.1 | Ascl2 | MA0816.1 | 1.06e-08 | 9.18e-06 |
| MA2002.1 | Zfp335 | MA2002.1 | 1.17e-08 | 1.01e-05 |
| MA0479.1 | FOXH1 | MA0479.1 | 1.25e-08 | 1.09e-05 |
| MA0153.2 | HNF1B | MA0153.2 | 1.26e-08 | 1.1e-05 |
| MA0019.1 | Ddit3::Cebpa | MA0019.1 | 1.28e-08 | 1.11e-05 |
| MA0810.1 | TFAP2A | MA0810.1 | 1.33e-08 | 1.15e-05 |
| MA0716.1 | PRRX1 | MA0716.1 | 1.36e-08 | 1.18e-05 |
| MA0090.3 | TEAD1 | MA0090.3 | 1.43e-08 | 1.24e-05 |
| MA0514.2 | Sox3 | MA0514.2 | 1.64e-08 | 1.42e-05 |
| MA0683.1 | POU4F2 | MA0683.1 | 1.67e-08 | 1.45e-05 |
| MA1478.1 | DMRTA2 | MA1478.1 | 1.97e-08 | 1.71e-05 |
| MA1988.1 (Atf3) | STREME-1 | 1-BVTGASTCABV | 5.65e-86 | 1.97e-05 |
| MA1122.1 | TFDP1 | MA1122.1 | 2.34e-08 | 2.03e-05 |
| MA0913.2 | HOXD9 | MA0913.2 | 2.51e-08 | 2.18e-05 |
| MA1151.1 | RORC | MA1151.1 | 2.53e-08 | 2.19e-05 |
| MA0668.2 | Neurod2 | MA0668.2 | 2.72e-08 | 2.36e-05 |
| MA0471.2 | E2F6 | MA0471.2 | 2.76e-08 | 2.39e-05 |
| MA1558.1 | SNAI1 | MA1558.1 | 3.02e-08 | 2.61e-05 |
| MA0157.3 | Foxo3 | MA0157.3 | 3.25e-08 | 2.81e-05 |
| MA1497.1 | HOXA6 | MA1497.1 | 3.25e-08 | 2.82e-05 |
| MA0645.1 | ETV6 | MA0645.1 | 3.34e-08 | 2.9e-05 |
| MA0677.1 | Nr2f6 | MA0677.1 | 3.61e-08 | 3.13e-05 |
| MA1683.1 | FOXA3 | MA1683.1 | 3.72e-08 | 3.23e-05 |
| MA0006.1 | Ahr::Arnt | MA0006.1 | 4.02e-08 | 3.48e-05 |
| MA0909.3 | Hoxd13 | MA0909.3 | 4.15e-08 | 3.6e-05 |
| MA1581.1 | ZBTB6 | MA1581.1 | 4.36e-08 | 3.78e-05 |
| MA1125.1 | ZNF384 | MA1125.1 | 4.57e-08 | 3.96e-05 |
| MA0481.3 | FOXP1 | MA0481.3 | 4.64e-08 | 4.02e-05 |
| MA0081.2 | SPIB | MA0081.2 | 4.7e-08 | 4.08e-05 |
| MA0145.2 | Tfcp2l1 | MA0145.2 | 4.73e-08 | 4.1e-05 |
| MA0047.3 | FOXA2 | MA0047.3 | 4.81e-08 | 4.17e-05 |
| MA1604.1 | Ebf2 | MA1604.1 | 4.83e-08 | 4.19e-05 |
| MA0838.1 | CEBPG | MA0838.1 | 4.95e-08 | 4.29e-05 |
| MA0722.1 | VAX1 | MA0722.1 | 5.32e-08 | 4.61e-05 |
| MA0600.2 | RFX2 | MA0600.2 | 5.66e-08 | 4.91e-05 |
| MA0744.2 | SCRT2 | MA0744.2 | 5.72e-08 | 4.96e-05 |
| MA0077.1 | SOX9 | MA0077.1 | 6.28e-08 | 5.44e-05 |
| 2-AGATAT | STREME-2 | 2-AGATAT | 2e-33 | 5.78e-05 |
| MA0796.1 | TGIF1 | MA0796.1 | 6.86e-08 | 5.95e-05 |
| MA0080.6 | Spi1 | MA0080.6 | 7.48e-08 | 6.49e-05 |
| MA1619.1 | Ptf1A | MA1619.1 | 7.62e-08 | 6.61e-05 |
| MA1487.2 | FOXE1 | MA1487.2 | 7.82e-08 | 6.78e-05 |
| MA0442.2 | SOX10 | MA0442.2 | 7.9e-08 | 6.85e-05 |
| MA1984.1 | ZNF667 | MA1984.1 | 7.97e-08 | 6.91e-05 |
| MA0868.2 | SOX8 | MA0868.2 | 8.03e-08 | 6.96e-05 |
| MA0103.3 | ZEB1 | MA0103.3 | 8.05e-08 | 6.98e-05 |
| MA1951.1 | FOS | MA1951.1 | 8.62e-08 | 7.47e-05 |
| MA1648.1 | TCF12 | MA1648.1 | 8.66e-08 | 7.51e-05 |
| MA1953.1 | FOXO1::ELF1 | MA1953.1 | 9.49e-08 | 8.23e-05 |
| MA1946.1 | ETV5::FOXI1 | MA1946.1 | 1e-07 | 8.68e-05 |
| MA0859.1 | Rarg | MA0859.1 | 1e-07 | 8.68e-05 |
| MA0914.1 | ISL2 | MA0914.1 | 1.02e-07 | 8.82e-05 |
| MA0809.2 | TEAD4 | MA0809.2 | 1.02e-07 | 8.82e-05 |
| MA1978.1 | ZNF354A | MA1978.1 | 1.07e-07 | 9.26e-05 |
| MA1563.2 | SOX18 | MA1563.2 | 1.17e-07 | 0.000101 |
| MA1954.1 | FOXO1::ELK1 | MA1954.1 | 1.17e-07 | 0.000101 |
| MA0078.2 | Sox17 | MA0078.2 | 1.18e-07 | 0.000102 |
| MA1479.1 | DMRTC2 | MA1479.1 | 1.36e-07 | 0.000118 |
| MA0159.1 | RARA::RXRA | MA0159.1 | 1.54e-07 | 0.000133 |
| MA0634.1 | ALX3 | MA0634.1 | 1.54e-07 | 0.000133 |
| MA0850.1 | FOXP3 | MA0850.1 | 1.58e-07 | 0.000137 |
| MA0649.1 | HEY2 | MA0649.1 | 1.61e-07 | 0.000139 |
| MA1717.1 | ZNF784 | MA1717.1 | 1.65e-07 | 0.000143 |
| MA1641.1 | MYF5 | MA1641.1 | 1.73e-07 | 0.00015 |
| MA0891.1 | GSC2 | MA0891.1 | 1.78e-07 | 0.000155 |
| MA0473.3 | ELF1 | MA0473.3 | 1.8e-07 | 0.000156 |
| MA0598.3 | EHF | MA0598.3 | 1.86e-07 | 0.000161 |
| MA0827.1 | OLIG3 | MA0827.1 | 2.02e-07 | 0.000175 |
| MA1499.1 | HOXB4 | MA1499.1 | 2.18e-07 | 0.000189 |
| MA1638.1 | HAND2 | MA1638.1 | 2.4e-07 | 0.000209 |
| MA0002.2 | Runx1 | MA0002.2 | 2.43e-07 | 0.000211 |
| MA0826.1 | OLIG1 | MA0826.1 | 2.46e-07 | 0.000213 |
| MA0783.1 | PKNOX2 | MA0783.1 | 2.59e-07 | 0.000225 |
| MA0837.2 | CEBPE | MA0837.2 | 2.62e-07 | 0.000228 |
| MA0893.2 | GSX2 | MA0893.2 | 2.64e-07 | 0.000229 |
| MA1532.2 | NR1D2 | MA1532.2 | 3.1e-07 | 0.000269 |
| MA0464.2 | BHLHE40 | MA0464.2 | 3.16e-07 | 0.000274 |
| MA0144.2 | STAT3 | MA0144.2 | 3.18e-07 | 0.000276 |
| MA1148.1 | PPARA::RXRA | MA1148.1 | 3.37e-07 | 0.000292 |
| MA0646.1 | GCM1 | MA0646.1 | 3.38e-07 | 0.000293 |
| MA1421.1 | TCF7L1 | MA1421.1 | 3.42e-07 | 0.000297 |
| MA0480.2 | Foxo1 | MA0480.2 | 3.63e-07 | 0.000315 |
| MA0874.1 | Arx | MA0874.1 | 3.68e-07 | 0.000319 |
| MA0792.1 | POU5F1B | MA0792.1 | 3.72e-07 | 0.000322 |
| MA0524.2 | TFAP2C | MA0524.2 | 3.79e-07 | 0.000329 |
| MA0828.2 | SREBF2 | MA0828.2 | 4.03e-07 | 0.000349 |
| MA0851.1 | Foxj3 | MA0851.1 | 4.42e-07 | 0.000383 |
| MA0033.2 | FOXL1 | MA0033.2 | 4.51e-07 | 0.000391 |
| MA0811.1 | TFAP2B | MA0811.1 | 4.74e-07 | 0.000411 |
| MA0738.1 | HIC2 | MA0738.1 | 4.79e-07 | 0.000415 |
| MA1105.2 | GRHL2 | MA1105.2 | 5.48e-07 | 0.000475 |
| MA0461.2 | Atoh1 | MA0461.2 | 5.78e-07 | 0.000501 |
| MA1507.1 | HOXD4 | MA1507.1 | 6.23e-07 | 0.00054 |
| MA0498.2 (MEIS1) | STREME-3 | 3-STGWCAS | 1.56e-52 | 0.000544 |
| MA0076.2 | ELK4 | MA0076.2 | 6.42e-07 | 0.000557 |
| MA0849.1 | FOXO6 | MA0849.1 | 6.46e-07 | 0.00056 |
| MA1645.1 (NKX2-2) | STREME-4 | 4-CTACTCAG | 4.28e-13 | 0.000563 |
| MA0160.2 | NR4A2 | MA0160.2 | 6.82e-07 | 0.000591 |
| MA1472.2 | Bhlha15 | MA1472.2 | 7.15e-07 | 0.00062 |
| MA1719.1 | ZNF816 | MA1719.1 | 8.24e-07 | 0.000714 |
| MA1534.1 | NR1I3 | MA1534.1 | 8.87e-07 | 0.000769 |
| MA1596.1 | ZNF460 | MA1596.1 | 9.34e-07 | 0.00081 |
| MA1484.1 | ETS2 | MA1484.1 | 9.81e-07 | 0.000851 |
| MA1116.1 | RBPJ | MA1116.1 | 9.84e-07 | 0.000853 |
| MA1124.1 | ZNF24 | MA1124.1 | 9.96e-07 | 0.000864 |
| MA0521.2 | Tcf12 | MA0521.2 | 9.99e-07 | 0.000866 |
| MA0104.4 | MYCN | MA0104.4 | 1e-06 | 0.00087 |
| MA0148.4 | FOXA1 | MA0148.4 | 1.02e-06 | 0.000888 |
| MA1976.1 | ZNF320 | MA1976.1 | 1.07e-06 | 0.000925 |
| MA1637.1 | EBF3 | MA1637.1 | 1.45e-06 | 0.00125 |
| MA0493.2 | KLF1 | MA0493.2 | 1.51e-06 | 0.00131 |
| MA0259.1 | ARNT::HIF1A | MA0259.1 | 1.55e-06 | 0.00134 |
| MA0475.2 | FLI1 | MA0475.2 | 1.55e-06 | 0.00135 |
| MA0755.1 | CUX2 | MA0755.1 | 1.59e-06 | 0.00138 |
| MA1962.1 (POU2F1::SOX2) | STREME-5 | 5-CATCACA | 4.15e-49 | 0.00139 |
| MA1489.1 | FOXN3 | MA1489.1 | 1.9e-06 | 0.00165 |
| MA0887.1 | EVX1 | MA0887.1 | 1.96e-06 | 0.0017 |
| MA0766.2 (GATA5) | STREME-6 | 6-WGATAASRD | 9.24e-59 | 0.00173 |
| MA0679.2 | ONECUT1 | MA0679.2 | 2.01e-06 | 0.00174 |
| MA1721.1 | ZNF93 | MA1721.1 | 2.1e-06 | 0.00182 |
| MA1583.1 | ZFP57 | MA1583.1 | 2.31e-06 | 0.002 |
| MA0087.2 | Sox5 | MA0087.2 | 2.67e-06 | 0.00232 |
| MA0627.2 | POU2F3 | MA0627.2 | 2.92e-06 | 0.00253 |
| MA0494.1 | Nr1h3::Rxra | MA0494.1 | 2.99e-06 | 0.00259 |
| MA0852.2 | FOXK1 | MA0852.2 | 3.43e-06 | 0.00297 |
| MA0791.1 | POU4F3 | MA0791.1 | 3.48e-06 | 0.00302 |
| MA0030.1 | FOXF2 | MA0030.1 | 3.59e-06 | 0.00311 |
| MA0068.2 | PAX4 | MA0068.2 | 3.69e-06 | 0.0032 |
| MA1573.2 | Thap11 | MA1573.2 | 3.95e-06 | 0.00343 |
| MA1471.1 | BARX2 | MA1471.1 | 3.96e-06 | 0.00343 |
| MA0640.2 | ELF3 | MA0640.2 | 4.25e-06 | 0.00369 |
| MA1125.1 (ZNF384) | STREME-7 | 7-GTCTCWAAAAAAAA | 6.53e-18 | 0.00383 |
| MA1552.1 | RARB | MA1552.1 | 4.58e-06 | 0.00397 |
| MA1957.1 | HOXB2::ELK1 | MA1957.1 | 5.26e-06 | 0.00456 |
| MA0038.2 (GFI1) | STREME-8 | 8-CTCACTGCA | 1.06e-34 | 0.00474 |
| MA1642.1 | NEUROG2 | MA1642.1 | 6.16e-06 | 0.00534 |
| MA1624.1 | Stat5a | MA1624.1 | 6.7e-06 | 0.00581 |
| MA1588.1 | ZNF136 | MA1588.1 | 8.04e-06 | 0.00697 |
| MA0911.1 | Hoxa11 | MA0911.1 | 8.12e-06 | 0.00704 |
| MA0739.1 | Hic1 | MA0739.1 | 8.34e-06 | 0.00723 |
| MA0804.1 | TBX19 | MA0804.1 | 8.67e-06 | 0.00752 |
| MA1542.1 | OSR1 | MA1542.1 | 9.21e-06 | 0.00798 |
| MA0888.1 | EVX2 | MA0888.1 | 9.29e-06 | 0.00805 |
| MA1965.1 (SP5) | STREME-9 | 9-CCTCCCA | 3.96e-31 | 0.00821 |
| MA0662.1 | MIXL1 | MA0662.1 | 9.51e-06 | 0.00824 |
| MA0633.2 | Twist2 | MA0633.2 | 9.67e-06 | 0.00838 |
| MA1555.1 | RXRB | MA1555.1 | 9.81e-06 | 0.0085 |
| MA1625.1 | Stat5b | MA1625.1 | 1.07e-05 | 0.00929 |
| MA1985.1 | ZNF669 | MA1985.1 | 1.08e-05 | 0.00936 |
| MA1496.1 | HOXA4 | MA1496.1 | 1.16e-05 | 0.0101 |
| MA1103.2 | FOXK2 | MA1103.2 | 1.17e-05 | 0.0101 |
| MA1635.1 | BHLHE22 | MA1635.1 | 1.44e-05 | 0.0125 |
| MA0878.3 | CDX1 | MA0878.3 | 1.62e-05 | 0.014 |
| MA1524.2 | Msgn1 | MA1524.2 | 1.76e-05 | 0.0153 |
| MA1712.1 | ZNF454 | MA1712.1 | 1.76e-05 | 0.0153 |
| MA0108.2 | TBP | MA0108.2 | 1.89e-05 | 0.0164 |
| MA0697.2 | Zic3 | MA0697.2 | 1.96e-05 | 0.017 |
| MA1948.1 | ETV5::HOXA2 | MA1948.1 | 1.99e-05 | 0.0172 |
| MA0896.1 | Hmx1 | MA0896.1 | 2e-05 | 0.0173 |
| 10-AGGCAT | STREME-10 | 10-AGGCAT | 7.79e-32 | 0.0178 |
| MA1726.1 | ZNF331 | MA1726.1 | 2.37e-05 | 0.0206 |
| 11-CATCTAA | STREME-11 | 11-CATCTAA | 1.83e-38 | 0.0207 |
| MA0712.2 (OTX2) | STREME-12 | 12-CTGTAATCCCAG | 1.51e-14 | 0.0212 |
| MA0817.1 | BHLHE23 | MA0817.1 | 2.52e-05 | 0.0219 |
| MA1121.1 | TEAD2 | MA1121.1 | 2.57e-05 | 0.0223 |
| MA0038.2 | GFI1 | MA0038.2 | 2.7e-05 | 0.0234 |
| MA0072.1 | RORA | MA0072.1 | 2.94e-05 | 0.0255 |
| MA1999.1 | Prdm5 | MA1999.1 | 3.19e-05 | 0.0276 |
| MA1728.1 (ZNF549) | STREME-13 | 13-CTGGGCAACAGA | 1.14e-09 | 0.0286 |
| MA0131.2 | HINFP | MA0131.2 | 3.32e-05 | 0.0288 |
| MA1983.1 | ZNF582 | MA1983.1 | 3.32e-05 | 0.0288 |
| MA0137.3 | STAT1 | MA0137.3 | 3.42e-05 | 0.0296 |
| MA1506.1 | HOXD10 | MA1506.1 | 3.64e-05 | 0.0315 |
| MA1599.1 | ZNF682 | MA1599.1 | 3.69e-05 | 0.032 |
| MA0058.3 | MAX | MA0058.3 | 3.77e-05 | 0.0327 |
| MA1608.1 | Isl1 | MA1608.1 | 3.84e-05 | 0.0333 |
| MA1710.1 (ZNF257) | STREME-14 | 14-TCTCCTGCCTCARC | 1.25e-08 | 0.0344 |
| MA0466.3 | CEBPB | MA0466.3 | 4.05e-05 | 0.0351 |
| MA1993.1 | Neurod2 | MA1993.1 | 4.48e-05 | 0.0388 |
| 15-CTGAACA | STREME-15 | 15-CTGAACA | 1.13e-15 | 0.0389 |
| MA1997.1 | Olig2 | MA1997.1 | 4.59e-05 | 0.0398 |
| MA1937.1 | ERF::HOXB13 | MA1937.1 | 4.95e-05 | 0.0429 |
| MA0510.2 | RFX5 | MA0510.2 | 5.12e-05 | 0.0444 |
| MA0676.1 (Nr2e1) | STREME-16 | 16-AACAGBCA | 5.29e-41 | 0.045 |
| MA0161.2 | NFIC | MA0161.2 | 5.36e-05 | 0.0465 |
| MA0832.1 | Tcf21 | MA0832.1 | 5.64e-05 | 0.0489 |
| MA0592.3 (ESRRA) | STREME-17 | 17-AGGTCACA | 2.49e-25 | 0.0581 |
| MA1653.1 (ZNF148) | STREME-18 | 18-CMTCCCC | 3.31e-20 | 0.0627 |
| MA1632.1 (ATF2) | STREME-19 | 19-ACCTCAT | 1.13e-29 | 0.071 |
ESR_open_xstreme %>%
dplyr::select(SIM_MOTIF,ALT_ID, ID,SEA_PVALUE,EVALUE) %>%
arrange(.,EVALUE) %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
# separate(SIM_MOTIF, into= c("SIM_MOTIF", "NAME"), sep= " ") %>%
# dplyr::filter(., EVALUE<0.05) %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
kable(., caption = "Enriched motifs in ESR open") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "800px")| SIM_MOTIF | ALT_ID | ID | SEA_PVALUE | EVALUE |
|---|---|---|---|---|
| MA0490.2 | JUNB | MA0490.2 | 4.45e-102 | 3.85e-99 |
| MA0491.2 | JUND | MA0491.2 | 9.18e-95 | 7.95e-92 |
| MA1130.1 | FOSL2::JUN | MA1130.1 | 3.34e-92 | 2.89e-89 |
| MA1142.1 | FOSL1::JUND | MA1142.1 | 1.32e-89 | 1.14e-86 |
| MA0099.3 | FOS::JUN | MA0099.3 | 4.73e-89 | 4.09e-86 |
| MA0477.2 | FOSL1 | MA0477.2 | 2.64e-87 | 2.29e-84 |
| MA1138.1 | FOSL2::JUNB | MA1138.1 | 6.32e-87 | 5.47e-84 |
| MA1141.1 | FOS::JUND | MA1141.1 | 1.41e-85 | 1.22e-82 |
| MA1137.1 | FOSL1::JUNB | MA1137.1 | 2.54e-85 | 2.2e-82 |
| MA1144.1 | FOSL2::JUND | MA1144.1 | 3.81e-85 | 3.3e-82 |
| MA1135.1 | FOSB::JUNB | MA1135.1 | 2.38e-84 | 2.06e-81 |
| MA1988.1 | Atf3 | MA1988.1 | 3.05e-84 | 2.64e-81 |
| MA0655.1 | JDP2 | MA0655.1 | 1.01e-82 | 8.78e-80 |
| MA1134.1 | FOS::JUNB | MA1134.1 | 1.18e-79 | 1.02e-76 |
| MA0841.1 | NFE2 | MA0841.1 | 1.84e-79 | 1.59e-76 |
| MA1633.2 | BACH1 | MA1633.2 | 5.98e-79 | 5.18e-76 |
| MA0476.1 | FOS | MA0476.1 | 1.41e-77 | 1.22e-74 |
| MA1634.1 | BATF | MA1634.1 | 1.49e-76 | 1.29e-73 |
| MA0835.2 | BATF3 | MA0835.2 | 1.76e-75 | 1.52e-72 |
| MA0462.2 | BATF::JUN | MA0462.2 | 1.45e-74 | 1.26e-71 |
| MA0489.2 | Jun | MA0489.2 | 2.18e-71 | 1.89e-68 |
| MA1101.2 | BACH2 | MA1101.2 | 1.26e-69 | 1.09e-66 |
| MA0861.1 | TP73 | MA0861.1 | 4.55e-68 | 3.94e-65 |
| MA1155.1 | ZSCAN4 | MA1155.1 | 9.54e-67 | 8.26e-64 |
| MA0106.3 | TP53 | MA0106.3 | 2.63e-62 | 2.28e-59 |
| MA1128.1 | FOSL1::JUN | MA1128.1 | 2.24e-53 | 1.94e-50 |
| MA1132.1 | JUN::JUNB | MA1132.1 | 1.75e-50 | 1.52e-47 |
| MA1125.1 (ZNF384) | MEME-1 | TWTTTTTTTHTTTTT | 0.186 | 4.3e-44 |
| MA0525.2 | TP63 | MA0525.2 | 4.56e-38 | 3.95e-35 |
| ACACACACACACACA | MEME-2 | ACACACACACACACA | 7.04e-91 | 4.1e-30 |
| MA1108.2 | MXI1 | MA1108.2 | 1.46e-25 | 1.27e-22 |
| MA1547.2 (PITX2) | MEME-3 | WGCTGGGATTACAGG | 0.000211 | 1.5e-22 |
| MA0626.1 | Npas2 | MA0626.1 | 2.64e-24 | 2.29e-21 |
| MA0006.1 | Ahr::Arnt | MA0006.1 | 9.17e-24 | 7.95e-21 |
| MA0819.2 | CLOCK | MA0819.2 | 3.84e-21 | 3.32e-18 |
| MA0089.2 | MAFG::NFE2L1 | MA0089.2 | 5.88e-21 | 5.09e-18 |
| MA0622.1 | Mlxip | MA0622.1 | 6.84e-21 | 5.92e-18 |
| MA0478.1 | FOSL2 | MA0478.1 | 1.4e-18 | 1.21e-15 |
| MA0817.1 | BHLHE23 | MA0817.1 | 1.61e-17 | 1.4e-14 |
| MA1928.1 | BNC2 | MA1928.1 | 9.49e-16 | 8.22e-13 |
| MA0073.1 | RREB1 | MA0073.1 | 1.5e-15 | 1.3e-12 |
| MA0623.2 | NEUROG1 | MA0623.2 | 2.75e-15 | 2.39e-12 |
| MA0496.3 | MAFK | MA0496.3 | 3.55e-15 | 3.07e-12 |
| MA0004.1 | Arnt | MA0004.1 | 2.66e-14 | 2.3e-11 |
| MA1107.2 (KLF9) | STREME-1 | 1-CACACGCACAC | 1.13e-103 | 2.47e-11 |
| MA1718.1 | ZNF8 | MA1718.1 | 3.58e-14 | 3.1e-11 |
| MA1107.2 | KLF9 | MA1107.2 | 3.18e-13 | 2.76e-10 |
| MA1560.1 | SOHLH2 | MA1560.1 | 1.51e-12 | 1.31e-09 |
| MA1974.1 | ZNF211 | MA1974.1 | 2.11e-12 | 1.83e-09 |
| MA0823.1 | HEY1 | MA0823.1 | 2.37e-12 | 2.05e-09 |
| MA0058.3 | MAX | MA0058.3 | 2.8e-12 | 2.42e-09 |
| MA0825.1 | MNT | MA0825.1 | 2.83e-12 | 2.45e-09 |
| 2-ACACACATACAC | STREME-2 | 2-ACACACATACAC | 5.97e-115 | 1.31e-08 |
| 3-CATGCACACACA | STREME-3 | 3-CATGCACACACA | 2.51e-115 | 1.35e-08 |
| 4-CACACAYAYACACAC | STREME-4 | 4-CACACAYAYACACAC | 1.69e-63 | 5.94e-08 |
| MA0818.2 | BHLHE22 | MA0818.2 | 1.35e-10 | 1.17e-07 |
| 5-ACACACACCACAC | STREME-5 | 5-ACACACACCACAC | 1.49e-87 | 2.07e-07 |
| MA1973.1 (ZKSCAN3) | MEME-4 | TGTTGCCCAGGCTGG | 0.0041 | 2.2e-07 |
| MA1990.1 (Gli1) | STREME-6 | 6-CACACACCACACACA | 7.32e-100 | 1.08e-06 |
| MA1107.2 (KLF9) | STREME-7 | 7-ACACACACACAC | 2.27e-82 | 5.35e-06 |
| MA1602.1 | ZSCAN29 | MA1602.1 | 7.62e-09 | 6.6e-06 |
| MA0464.2 | BHLHE40 | MA0464.2 | 7.81e-09 | 6.76e-06 |
| MA0501.1 | MAF::NFE2 | MA0501.1 | 8.08e-09 | 7e-06 |
| MA0827.1 | OLIG3 | MA0827.1 | 2.83e-08 | 2.45e-05 |
| MA0147.3 | MYC | MA0147.3 | 2.92e-08 | 2.53e-05 |
| MA0150.2 | Nfe2l2 | MA0150.2 | 5.04e-08 | 4.37e-05 |
| MA0659.3 | Mafg | MA0659.3 | 5.32e-08 | 4.61e-05 |
| MA0491.2 (JUND) | STREME-8 | 8-NATGASTCATN | 1.31e-84 | 5.98e-05 |
| MA1107.2 (KLF9) | STREME-9 | 9-CACACACACATG | 1.98e-100 | 6.43e-05 |
| MA0259.1 | ARNT::HIF1A | MA0259.1 | 8e-08 | 6.93e-05 |
| MA1464.1 | ARNT2 | MA1464.1 | 2.1e-07 | 0.000182 |
| MA0052.4 | MEF2A | MA0052.4 | 4.84e-07 | 0.000419 |
| MA0821.2 | HES5 | MA0821.2 | 7.8e-07 | 0.000676 |
| MA1718.1 (ZNF8) | STREME-10 | 10-CAYACACCACA | 4.89e-72 | 0.00093 |
| 11-CATACACACG | STREME-11 | 11-CATACACACG | 1.49e-110 | 0.00104 |
| MA0678.1 | OLIG2 | MA0678.1 | 1.38e-06 | 0.0012 |
| MA0608.1 | Creb3l2 | MA0608.1 | 1.47e-06 | 0.00127 |
| MA1155.1 (ZSCAN4) | STREME-12 | 12-CACACACAG | 5.98e-75 | 0.00227 |
| 13-CACACACATA | STREME-13 | 13-CACACACATA | 4.8e-99 | 0.00235 |
| MA0507.2 | POU2F2 | MA0507.2 | 3.02e-06 | 0.00262 |
| MA0789.1 | POU3F4 | MA0789.1 | 3.29e-06 | 0.00285 |
| MA0009.2 | TBXT | MA0009.2 | 3.39e-06 | 0.00294 |
| MA0461.2 | Atoh1 | MA0461.2 | 4.66e-06 | 0.00403 |
| MA0472.2 | EGR2 | MA0472.2 | 5.01e-06 | 0.00433 |
| MA0259.1 (ARNT::HIF1A) | STREME-14 | 14-YABACRTGCMY | 2.41e-50 | 0.00578 |
| MA0616.2 | HES2 | MA0616.2 | 7.38e-06 | 0.00639 |
| MA0649.1 | HEY2 | MA0649.1 | 7.68e-06 | 0.00665 |
| 15-CATGCCCACAC | STREME-15 | 15-CATGCCCACAC | 6.81e-111 | 0.00932 |
| MA1107.2 (KLF9) | STREME-16 | 16-CACACACA | 1.56e-96 | 0.0193 |
| MA1155.1 (ZSCAN4) | STREME-17 | 17-ACACAC | 2.2e-46 | 0.0253 |
| MA1106.1 | HIF1A | MA1106.1 | 3.07e-05 | 0.0266 |
| MA0497.1 | MEF2C | MA0497.1 | 3.48e-05 | 0.0301 |
| MA0059.1 | MAX::MYC | MA0059.1 | 4.43e-05 | 0.0384 |
| MA0801.1 | MGA | MA0801.1 | 4.73e-05 | 0.041 |
| MA0148.4 | FOXA1 | MA0148.4 | 4.77e-05 | 0.0413 |
| MA0698.1 | ZBTB18 | MA0698.1 | 5e-05 | 0.0433 |
| 18-ACACACA | STREME-18 | 18-ACACACA | 7.08e-53 | 0.0481 |
| 19-ACRTGYAC | STREME-19 | 19-ACRTGYAC | 5.15e-56 | 0.0556 |
| MA1108.2 (MXI1) | STREME-20 | 20-CACACATGTG | 4.64e-58 | 0.832 |
| MA1135.1 (FOSB::JUNB) | STREME-21 | 21-ATGASTCAT | 9.62e-57 | 4.74 |
ESR_close_xstreme %>%
dplyr::select(SIM_MOTIF,ALT_ID, ID,SEA_PVALUE,EVALUE) %>%
arrange(.,EVALUE) %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
# separate(SIM_MOTIF, into= c("SIM_MOTIF", "NAME"), sep= " ") %>%
# dplyr::filter(., EVALUE<0.05) %>%
kable(., caption = "Enriched motifs ESR close") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "800px")| SIM_MOTIF | ALT_ID | ID | SEA_PVALUE | EVALUE |
|---|---|---|---|---|
| MA1653.1 | ZNF148 | MA1653.1 | 6.2e-144 | 5.35e-141 |
| MA0493.2 | KLF1 | MA0493.2 | 4.59e-141 | 3.96e-138 |
| MA0685.2 | SP4 | MA0685.2 | 7.2e-138 | 6.2e-135 |
| MA1959.1 | KLF7 | MA1959.1 | 2.94e-133 | 2.53e-130 |
| MA1522.1 | MAZ | MA1522.1 | 9.09e-133 | 7.83e-130 |
| MA1511.2 | KLF10 | MA1511.2 | 2.36e-132 | 2.03e-129 |
| MA0742.2 | KLF12 | MA0742.2 | 2.25e-131 | 1.94e-128 |
| MA1630.2 | ZNF281 | MA1630.2 | 3e-130 | 2.59e-127 |
| MA0741.1 | KLF16 | MA0741.1 | 1.01e-126 | 8.71e-124 |
| MA0740.2 | KLF14 | MA0740.2 | 1.74e-126 | 1.5e-123 |
| MA0079.5 | SP1 | MA0079.5 | 7.28e-122 | 6.28e-119 |
| MA0599.1 | KLF5 | MA0599.1 | 3.49e-120 | 3.01e-117 |
| MA0753.2 | ZNF740 | MA0753.2 | 3.09e-118 | 2.67e-115 |
| MA1713.1 | ZNF610 | MA1713.1 | 5.98e-113 | 5.16e-110 |
| MA0516.3 | SP2 | MA0516.3 | 7.47e-112 | 6.44e-109 |
| MA0146.2 | Zfx | MA0146.2 | 2.93e-109 | 2.52e-106 |
| MA1107.2 | KLF9 | MA1107.2 | 3.95e-109 | 3.4e-106 |
| MA1650.1 | ZBTB14 | MA1650.1 | 2.82e-108 | 2.43e-105 |
| MA1564.1 | SP9 | MA1564.1 | 2.94e-106 | 2.54e-103 |
| MA0746.2 | SP3 | MA0746.2 | 2.58e-105 | 2.23e-102 |
| MA0774.1 | MEIS2 | MA0774.1 | 2.05e-104 | 1.76e-101 |
| MA1961.1 | PATZ1 | MA1961.1 | 3.04e-104 | 2.62e-101 |
| MA1548.1 | PLAGL2 | MA1548.1 | 5.47e-104 | 4.71e-101 |
| MA0039.4 | KLF4 | MA0039.4 | 8.53e-104 | 7.35e-101 |
| MA1578.1 | VEZF1 | MA1578.1 | 5.18e-101 | 4.46e-98 |
| MA1615.1 | Plagl1 | MA1615.1 | 1.02e-99 | 8.76e-97 |
| MA0597.2 | THAP1 | MA0597.2 | 2.47e-98 | 2.13e-95 |
| MA0162.4 | EGR1 | MA0162.4 | 2.74e-97 | 2.37e-94 |
| MA1515.1 | KLF2 | MA1515.1 | 5.49e-95 | 4.73e-92 |
| MA1965.1 | SP5 | MA1965.1 | 1.03e-91 | 8.85e-89 |
| MA0810.1 | TFAP2A | MA0810.1 | 1.16e-91 | 9.95e-89 |
| MA1643.1 | NFIB | MA1643.1 | 1.37e-91 | 1.18e-88 |
| MA1102.2 | CTCFL | MA1102.2 | 1.71e-91 | 1.48e-88 |
| MA1627.1 | Wt1 | MA1627.1 | 8.8e-91 | 7.59e-88 |
| MA1981.1 | ZNF530 | MA1981.1 | 6.2e-89 | 5.34e-86 |
| MA1514.1 | KLF17 | MA1514.1 | 7.01e-85 | 6.04e-82 |
| MA1712.1 | ZNF454 | MA1712.1 | 2.77e-84 | 2.39e-81 |
| MA1986.1 | ZNF692 | MA1986.1 | 1.52e-82 | 1.31e-79 |
| MA0747.1 | SP8 | MA0747.1 | 3.77e-81 | 3.25e-78 |
| MA1599.1 | ZNF682 | MA1599.1 | 3.49e-79 | 3.01e-76 |
| MA1516.1 | KLF3 | MA1516.1 | 6.69e-79 | 5.76e-76 |
| MA1719.1 | ZNF816 | MA1719.1 | 2.01e-77 | 1.73e-74 |
| MA1512.1 | KLF11 | MA1512.1 | 1.99e-76 | 1.72e-73 |
| MA1517.1 | KLF6 | MA1517.1 | 5.35e-75 | 4.61e-72 |
| MA0738.1 | HIC2 | MA0738.1 | 2.64e-74 | 2.28e-71 |
| MA1721.1 | ZNF93 | MA1721.1 | 4.13e-74 | 3.56e-71 |
| MA0528.2 | ZNF263 | MA0528.2 | 2.78e-73 | 2.39e-70 |
| MA1596.1 | ZNF460 | MA1596.1 | 1.17e-71 | 1.01e-68 |
| MA0131.2 | HINFP | MA0131.2 | 1.66e-71 | 1.43e-68 |
| MA1527.1 | NFIC | MA1527.1 | 7.62e-71 | 6.57e-68 |
| MA1976.1 | ZNF320 | MA1976.1 | 1.14e-70 | 9.79e-68 |
| MA1723.1 | PRDM9 | MA1723.1 | 2.5e-70 | 2.15e-67 |
| MA1528.1 | NFIX | MA1528.1 | 4.83e-70 | 4.16e-67 |
| MA1631.1 | ASCL1 | MA1631.1 | 3.19e-68 | 2.75e-65 |
| MA0811.1 | TFAP2B | MA0811.1 | 6.6e-68 | 5.69e-65 |
| MA0775.1 | MEIS3 | MA0775.1 | 1.22e-67 | 1.05e-64 |
| MA0751.1 | ZIC4 | MA0751.1 | 1.75e-67 | 1.51e-64 |
| MA1656.1 | ZNF449 | MA1656.1 | 1.08e-65 | 9.34e-63 |
| MA1710.1 | ZNF257 | MA1710.1 | 1.14e-65 | 9.79e-63 |
| MA0524.2 | TFAP2C | MA0524.2 | 1.67e-65 | 1.44e-62 |
| MA0471.2 | E2F6 | MA0471.2 | 3.59e-65 | 3.09e-62 |
| MA0119.1 | NFIC::TLX1 | MA0119.1 | 3.47e-64 | 2.99e-61 |
| MA0830.2 | TCF4 | MA0830.2 | 3.46e-63 | 2.98e-60 |
| MA0130.1 | ZNF354C | MA0130.1 | 6.66e-61 | 5.74e-58 |
| MA1648.1 | TCF12 | MA1648.1 | 7.57e-57 | 6.53e-54 |
| MA0734.3 | Gli2 | MA0734.3 | 1.23e-56 | 1.06e-53 |
| MA1728.1 | ZNF549 | MA1728.1 | 1.88e-56 | 1.62e-53 |
| MA1635.1 | BHLHE22 | MA1635.1 | 1.05e-55 | 9.03e-53 |
| MA0646.1 | GCM1 | MA0646.1 | 5.21e-55 | 4.49e-52 |
| MA0814.2 | TFAP2C | MA0814.2 | 8.78e-55 | 7.57e-52 |
| MA0506.2 | Nrf1 | MA0506.2 | 1e-54 | 8.66e-52 |
| MA0048.2 | NHLH1 | MA0048.2 | 1.96e-54 | 1.69e-51 |
| MA0733.1 | EGR4 | MA0733.1 | 3.69e-54 | 3.18e-51 |
| MA0504.1 | NR2C2 | MA0504.1 | 7.01e-54 | 6.05e-51 |
| MA0739.1 | Hic1 | MA0739.1 | 1.05e-53 | 9.08e-51 |
| MA1628.1 | Zic1::Zic2 | MA1628.1 | 2.65e-53 | 2.28e-50 |
| MA0003.4 | TFAP2A | MA0003.4 | 8.58e-53 | 7.4e-50 |
| MA0672.1 | NKX2-3 | MA0672.1 | 2.77e-52 | 2.39e-49 |
| MA1997.1 | Olig2 | MA1997.1 | 3.98e-52 | 3.43e-49 |
| MA0633.2 | Twist2 | MA0633.2 | 7.47e-52 | 6.44e-49 |
| MA1535.1 | NR2C1 | MA1535.1 | 8.34e-52 | 7.19e-49 |
| MA0803.1 | TBX15 | MA0803.1 | 1.04e-51 | 9.01e-49 |
| MA1987.1 | ZNF701 | MA1987.1 | 5.77e-51 | 4.97e-48 |
| MA0872.1 | TFAP2A | MA0872.1 | 9.64e-51 | 8.31e-48 |
| MA0697.2 | Zic3 | MA0697.2 | 6.33e-50 | 5.45e-47 |
| MA1982.1 | ZNF574 | MA1982.1 | 7.44e-50 | 6.42e-47 |
| MA1122.1 | TFDP1 | MA1122.1 | 1.5e-49 | 1.29e-46 |
| MA1629.1 | Zic2 | MA1629.1 | 3.47e-48 | 2.99e-45 |
| MA1972.1 | ZFP14 | MA1972.1 | 8.18e-48 | 7.05e-45 |
| MA1979.1 | ZNF416 | MA1979.1 | 2.87e-47 | 2.47e-44 |
| MA1993.1 | Neurod2 | MA1993.1 | 5.19e-47 | 4.48e-44 |
| MA1730.1 | ZNF708 | MA1730.1 | 1.51e-46 | 1.31e-43 |
| MA1985.1 | ZNF669 | MA1985.1 | 1.76e-46 | 1.52e-43 |
| MA0104.4 | MYCN | MA0104.4 | 1.89e-46 | 1.63e-43 |
| MA0694.1 | ZBTB7B | MA0694.1 | 2.22e-46 | 1.91e-43 |
| MA0812.1 | TFAP2B | MA0812.1 | 2.39e-46 | 2.06e-43 |
| MA1973.1 | ZKSCAN3 | MA1973.1 | 4.78e-46 | 4.12e-43 |
| MA0745.2 | SNAI2 | MA0745.2 | 7.74e-46 | 6.67e-43 |
| MA1637.1 | EBF3 | MA1637.1 | 1.64e-45 | 1.42e-42 |
| MA0145.2 | Tfcp2l1 | MA0145.2 | 1.74e-45 | 1.5e-42 |
| MA0155.1 | INSM1 | MA0155.1 | 6.8e-45 | 5.86e-42 |
| MA0807.1 | TBX5 | MA0807.1 | 8.6e-45 | 7.42e-42 |
| MA0500.2 | MYOG | MA0500.2 | 1.11e-44 | 9.6e-42 |
| MA1604.1 | Ebf2 | MA1604.1 | 1.63e-44 | 1.41e-41 |
| MA0258.2 | ESR2 | MA0258.2 | 1.64e-44 | 1.41e-41 |
| MA0816.1 | Ascl2 | MA0816.1 | 1.66e-44 | 1.43e-41 |
| MA0522.3 | TCF3 | MA0522.3 | 1.91e-44 | 1.65e-41 |
| MA0103.3 | ZEB1 | MA0103.3 | 4.7e-44 | 4.05e-41 |
| MA0767.1 | GCM2 | MA0767.1 | 1.34e-43 | 1.15e-40 |
| MA1990.1 | Gli1 | MA1990.1 | 1.72e-43 | 1.48e-40 |
| MA1652.1 | ZKSCAN5 | MA1652.1 | 3.07e-42 | 2.65e-39 |
| MA0159.1 | RARA::RXRA | MA0159.1 | 8.94e-42 | 7.71e-39 |
| MA1641.1 | MYF5 | MA1641.1 | 1.01e-41 | 8.7e-39 |
| MA0736.1 | GLIS2 | MA0736.1 | 3.45e-41 | 2.97e-38 |
| MA0865.2 | E2F8 | MA0865.2 | 4.65e-41 | 4.01e-38 |
| MA0154.4 | EBF1 | MA0154.4 | 1.27e-40 | 1.1e-37 |
| MA1100.2 | ASCL1 | MA1100.2 | 1.36e-40 | 1.17e-37 |
| MA1727.1 | ZNF417 | MA1727.1 | 3.7e-39 | 3.19e-36 |
| MA1472.2 | Bhlha15 | MA1472.2 | 7.02e-39 | 6.05e-36 |
| MA0671.1 | NFIX | MA0671.1 | 1.35e-38 | 1.16e-35 |
| MA1621.1 | Rbpjl | MA1621.1 | 5.14e-38 | 4.43e-35 |
| MA1567.2 | Tbx6 | MA1567.2 | 1.01e-37 | 8.69e-35 |
| MA2003.1 | NKX2-4 | MA2003.1 | 2.1e-35 | 1.81e-32 |
| MA1587.1 | ZNF135 | MA1587.1 | 2.54e-35 | 2.19e-32 |
| MA0820.1 | FIGLA | MA0820.1 | 2.79e-35 | 2.4e-32 |
| MA0649.1 | HEY2 | MA0649.1 | 5.31e-35 | 4.58e-32 |
| MA0805.1 | TBX1 | MA0805.1 | 1.19e-34 | 1.02e-31 |
| MA2002.1 | Zfp335 | MA2002.1 | 1.2e-34 | 1.03e-31 |
| MA0511.2 | RUNX2 | MA0511.2 | 1.26e-34 | 1.09e-31 |
| MA0821.2 | HES5 | MA0821.2 | 2.15e-34 | 1.85e-31 |
| MA1726.1 | ZNF331 | MA1726.1 | 2.25e-34 | 1.94e-31 |
| MA0499.2 | MYOD1 | MA0499.2 | 3.37e-34 | 2.9e-31 |
| MA0065.2 | Pparg::Rxra | MA0065.2 | 9.98e-34 | 8.61e-31 |
| MA0748.2 | YY2 | MA0748.2 | 1.46e-33 | 1.26e-30 |
| MA0695.1 | ZBTB7C | MA0695.1 | 1.77e-33 | 1.52e-30 |
| MA1149.1 | RARA::RXRG | MA1149.1 | 1.98e-33 | 1.71e-30 |
| MA0521.2 | Tcf12 | MA0521.2 | 2.3e-33 | 1.99e-30 |
| MA1731.1 | ZNF768 | MA1731.1 | 2.6e-33 | 2.24e-30 |
| MA0806.1 | TBX4 | MA0806.1 | 5.18e-33 | 4.46e-30 |
| MA0665.1 | MSC | MA0665.1 | 6.34e-33 | 5.46e-30 |
| MA0014.3 | PAX5 | MA0014.3 | 6.58e-33 | 5.67e-30 |
| MA1581.1 | ZBTB6 | MA1581.1 | 1.08e-32 | 9.3e-30 |
| MA0472.2 | EGR2 | MA0472.2 | 2.23e-32 | 1.92e-29 |
| MA1620.1 | Ptf1A | MA1620.1 | 4.74e-32 | 4.08e-29 |
| MA1600.1 | ZNF684 | MA1600.1 | 6.24e-32 | 5.38e-29 |
| MA1574.1 | THRB | MA1574.1 | 1.07e-31 | 9.21e-29 |
| MA0071.1 | RORA | MA0071.1 | 3.04e-31 | 2.62e-28 |
| MA0750.2 | ZBTB7A | MA0750.2 | 3.83e-31 | 3.31e-28 |
| MA1619.1 | Ptf1A | MA1619.1 | 4.72e-31 | 4.07e-28 |
| MA0039.4 (KLF4) | MEME-3 | SWGGSWGGAGK | 1.79e-131 | 6.2e-28 |
| MA0498.2 | MEIS1 | MA0498.2 | 1.4e-30 | 1.21e-27 |
| MA0161.2 | NFIC | MA0161.2 | 2.08e-30 | 1.79e-27 |
| MA1566.2 | TBX3 | MA1566.2 | 4.18e-30 | 3.6e-27 |
| MA0815.1 | TFAP2C | MA0815.1 | 7.71e-30 | 6.65e-27 |
| MA0139.1 | CTCF | MA0139.1 | 8.87e-30 | 7.65e-27 |
| MA0691.1 | TFAP4 | MA0691.1 | 1.05e-29 | 9.09e-27 |
| MA1565.1 | TBX18 | MA1565.1 | 1.08e-29 | 9.33e-27 |
| MA1964.1 | SMAD2 | MA1964.1 | 2.07e-29 | 1.79e-26 |
| MA1583.1 | ZFP57 | MA1583.1 | 2.83e-29 | 2.44e-26 |
| MA1474.1 | CREB3L4 | MA1474.1 | 1.01e-28 | 8.75e-26 |
| MA0006.1 | Ahr::Arnt | MA0006.1 | 1.19e-27 | 1.03e-24 |
| MA1941.1 | ETV2::FIGLA | MA1941.1 | 1.88e-27 | 1.62e-24 |
| MA1966.1 | TFAP4::ETV1 | MA1966.1 | 9.87e-27 | 8.51e-24 |
| MA0783.1 | PKNOX2 | MA0783.1 | 1.05e-26 | 9.05e-24 |
| MA0813.1 | TFAP2B | MA0813.1 | 8.36e-26 | 7.21e-23 |
| MA0670.1 | NFIA | MA0670.1 | 8.95e-26 | 7.71e-23 |
| MA0595.1 | SREBF1 | MA0595.1 | 1.2e-25 | 1.04e-22 |
| MA1109.1 | NEUROD1 | MA1109.1 | 3.23e-25 | 2.78e-22 |
| MA1945.1 | ETV5::FIGLA | MA1945.1 | 3.66e-25 | 3.15e-22 |
| MA0801.1 | MGA | MA0801.1 | 4.2e-25 | 3.62e-22 |
| MA1558.1 | SNAI1 | MA1558.1 | 4.27e-25 | 3.68e-22 |
| MA0664.1 | MLXIPL | MA0664.1 | 6.58e-25 | 5.67e-22 |
| MA0797.1 | TGIF2 | MA0797.1 | 7.82e-25 | 6.74e-22 |
| MA1723.1 (PRDM9) | MEME-1 | RGSWGGGMWGG | 2.77e-87 | 2.4e-21 |
| MA0596.1 | SREBF2 | MA0596.1 | 2.88e-24 | 2.48e-21 |
| MA0017.2 | NR2F1 | MA0017.2 | 4e-24 | 3.45e-21 |
| MA0692.1 | TFEB | MA0692.1 | 4.13e-23 | 3.56e-20 |
| MA0796.1 | TGIF1 | MA0796.1 | 1.5e-21 | 1.3e-18 |
| MA0626.1 | Npas2 | MA0626.1 | 2.31e-21 | 1.99e-18 |
| MA1125.1 (ZNF384) | MEME-2 | AAAAAAAAAAAAAAA | 0.0161 | 3e-18 |
| MA1531.1 | NR1D1 | MA1531.1 | 3.59e-21 | 3.09e-18 |
| MA0668.2 | Neurod2 | MA0668.2 | 5.21e-21 | 4.49e-18 |
| MA1493.1 | HES6 | MA1493.1 | 6.07e-21 | 5.24e-18 |
| MA1929.1 | CTCF | MA1929.1 | 8.88e-21 | 7.66e-18 |
| MA1715.1 | ZNF707 | MA1715.1 | 1.04e-20 | 8.97e-18 |
| MA1571.1 | TGIF2LX | MA1571.1 | 2.06e-20 | 1.77e-17 |
| MA0698.1 | ZBTB18 | MA0698.1 | 2.54e-20 | 2.19e-17 |
| MA0502.2 | NFYB | MA0502.2 | 5.09e-20 | 4.39e-17 |
| MA1114.1 | PBX3 | MA1114.1 | 8.97e-20 | 7.73e-17 |
| MA0632.2 | TCFL5 | MA0632.2 | 1.05e-19 | 9.07e-17 |
| MA1655.1 | ZNF341 | MA1655.1 | 1.06e-19 | 9.15e-17 |
| MA1645.1 | NKX2-2 | MA1645.1 | 5.54e-19 | 4.78e-16 |
| MA0677.1 | Nr2f6 | MA0677.1 | 6.54e-19 | 5.63e-16 |
| MA1108.2 | MXI1 | MA1108.2 | 8.03e-19 | 6.92e-16 |
| MA1572.1 | TGIF2LY | MA1572.1 | 1.12e-18 | 9.69e-16 |
| MA1560.1 | SOHLH2 | MA1560.1 | 1.47e-18 | 1.27e-15 |
| MA1653.1 (ZNF148) | STREME-1 | 1-CCCCWCCCM | 6.38e-172 | 1.51e-15 |
| MA0092.1 | Hand1::Tcf3 | MA0092.1 | 2.46e-18 | 2.12e-15 |
| MA1996.1 | Nr1H2 | MA1996.1 | 2.91e-18 | 2.51e-15 |
| MA0782.2 | PKNOX1 | MA0782.2 | 4.59e-18 | 3.96e-15 |
| MA1569.1 | TFAP2E | MA1569.1 | 4.95e-18 | 4.27e-15 |
| MA0018.4 | CREB1 | MA0018.4 | 2.31e-17 | 1.99e-14 |
| MA0645.1 | ETV6 | MA0645.1 | 2.38e-17 | 2.05e-14 |
| MA0058.3 | MAX | MA0058.3 | 7.59e-17 | 6.54e-14 |
| MA0259.1 | ARNT::HIF1A | MA0259.1 | 1.83e-16 | 1.58e-13 |
| MA1570.1 | TFAP4 | MA1570.1 | 5.71e-16 | 4.92e-13 |
| MA0060.3 | NFYA | MA0060.3 | 5.81e-16 | 5e-13 |
| MA0690.2 | TBX21 | MA0690.2 | 6.68e-16 | 5.76e-13 |
| MA0608.1 | Creb3l2 | MA0608.1 | 8.02e-16 | 6.92e-13 |
| MA1992.1 | Ikzf3 | MA1992.1 | 2.39e-15 | 2.06e-12 |
| MA0667.1 | MYF6 | MA0667.1 | 2.52e-15 | 2.17e-12 |
| MA0831.3 | TFE3 | MA0831.3 | 3.33e-15 | 2.87e-12 |
| MA1642.1 | NEUROG2 | MA1642.1 | 3.46e-15 | 2.98e-12 |
| MA1527.1 (NFIC) | STREME-2 | 2-GSTGNCASC | 1.29e-157 | 4.3e-12 |
| MA1601.2 | ZNF75D | MA1601.2 | 7.58e-15 | 6.54e-12 |
| MA0696.1 | ZIC1 | MA0696.1 | 2e-14 | 1.72e-11 |
| MA0823.1 | HEY1 | MA0823.1 | 2.28e-14 | 1.96e-11 |
| MA1547.2 (PITX2) | MEME-4 | AAAGTGCTGGGATTA | 0.289 | 2.1e-11 |
| MA1153.1 | Smad4 | MA1153.1 | 2.55e-14 | 2.19e-11 |
| MA0766.2 | GATA5 | MA0766.2 | 5.37e-14 | 4.63e-11 |
| MA1716.1 | ZNF76 | MA1716.1 | 5.64e-14 | 4.86e-11 |
| MA0147.3 | MYC | MA0147.3 | 7.11e-14 | 6.13e-11 |
| MA0673.1 | NKX2-8 | MA0673.1 | 7.25e-14 | 6.25e-11 |
| MA0163.1 | PLAG1 | MA0163.1 | 1.19e-13 | 1.02e-10 |
| MA0124.2 | Nkx3-1 | MA0124.2 | 1.75e-13 | 1.51e-10 |
| MA0464.2 | BHLHE40 | MA0464.2 | 3.78e-13 | 3.26e-10 |
| MA0688.1 | TBX2 | MA0688.1 | 4.23e-13 | 3.65e-10 |
| MA1513.1 (KLF15) | STREME-3 | 3-CCAGCCC | 6.85e-130 | 3.93e-10 |
| MA0669.1 | NEUROG2 | MA0669.1 | 4.82e-13 | 4.15e-10 |
| MA1559.1 | SNAI3 | MA1559.1 | 5.04e-13 | 4.34e-10 |
| MA0829.2 | SREBF1 | MA0829.2 | 8.25e-13 | 7.11e-10 |
| MA1533.1 | NR1I2 | MA1533.1 | 1.12e-12 | 9.66e-10 |
| MA1485.1 | FERD3L | MA1485.1 | 1.76e-12 | 1.51e-09 |
| MA0056.2 | MZF1 | MA0056.2 | 2.27e-12 | 1.95e-09 |
| MA0825.1 | MNT | MA0825.1 | 2.91e-12 | 2.5e-09 |
| MA1468.1 | ATOH7 | MA1468.1 | 6.91e-12 | 5.95e-09 |
| MA1618.1 | Ptf1a | MA1618.1 | 1.17e-11 | 1.01e-08 |
| MA0802.1 | TBR1 | MA0802.1 | 1.47e-11 | 1.27e-08 |
| MA1537.1 | NR2F1 | MA1537.1 | 1.58e-11 | 1.36e-08 |
| MA1554.1 | RFX7 | MA1554.1 | 1.72e-11 | 1.48e-08 |
| MA0093.3 | USF1 | MA0093.3 | 2.53e-11 | 2.18e-08 |
| MA0764.3 | ETV4 | MA0764.3 | 3.16e-11 | 2.73e-08 |
| MA0073.1 | RREB1 | MA0073.1 | 3.89e-11 | 3.35e-08 |
| MA1542.1 | OSR1 | MA1542.1 | 4.23e-11 | 3.65e-08 |
| MA0505.2 | Nr5A2 | MA0505.2 | 6.86e-11 | 5.92e-08 |
| MA0488.1 | JUN | MA0488.1 | 1.43e-10 | 1.23e-07 |
| MA0689.1 | TBX20 | MA0689.1 | 3.54e-10 | 3.06e-07 |
| MA1475.1 | CREB3L4 | MA1475.1 | 3.75e-10 | 3.24e-07 |
| MA1523.1 | MSANTD3 | MA1523.1 | 3.88e-10 | 3.35e-07 |
| MA1638.1 | HAND2 | MA1638.1 | 4.06e-10 | 3.5e-07 |
| MA1467.2 | Atoh1 | MA1467.2 | 6.08e-10 | 5.24e-07 |
| MA1123.2 | TWIST1 | MA1123.2 | 6.42e-10 | 5.54e-07 |
| MA1979.1 (ZNF416) | STREME-4 | 4-CTGGGCC | 4.99e-119 | 6.62e-07 |
| MA1629.1 (Zic2) | STREME-5 | 5-CACAGCAG | 8.19e-87 | 7.02e-07 |
| MA1540.2 | NR5A1 | MA1540.2 | 8.29e-10 | 7.15e-07 |
| MA1724.1 | Rfx6 | MA1724.1 | 1.14e-09 | 9.84e-07 |
| MA1644.1 | NFYC | MA1644.1 | 1.4e-09 | 1.21e-06 |
| MA0607.2 | BHLHA15 | MA0607.2 | 1.72e-09 | 1.48e-06 |
| MA1708.1 | ETV7 | MA1708.1 | 1.79e-09 | 1.55e-06 |
| MA0492.1 | JUND | MA0492.1 | 1.85e-09 | 1.6e-06 |
| MA0111.1 | Spz1 | MA0111.1 | 2.25e-09 | 1.94e-06 |
| MA0091.1 | TAL1::TCF3 | MA0091.1 | 3.09e-09 | 2.66e-06 |
| MA0737.1 | GLIS3 | MA0737.1 | 3.29e-09 | 2.84e-06 |
| MA0832.1 | Tcf21 | MA0832.1 | 3.8e-09 | 3.28e-06 |
| MA0002.2 | Runx1 | MA0002.2 | 4.22e-09 | 3.64e-06 |
| MA0141.3 | ESRRB | MA0141.3 | 4.5e-09 | 3.88e-06 |
| MA1593.1 | ZNF317 | MA1593.1 | 5.51e-09 | 4.75e-06 |
| MA0528.2 (ZNF263) | STREME-6 | 6-CCTCCC | 2.51e-109 | 5.06e-06 |
| MA0036.3 | GATA2 | MA0036.3 | 6.15e-09 | 5.3e-06 |
| MA1725.1 | ZNF189 | MA1725.1 | 7.96e-09 | 6.86e-06 |
| MA0871.2 | TFEC | MA0871.2 | 8.52e-09 | 7.34e-06 |
| MA1099.2 | HES1 | MA1099.2 | 1.62e-08 | 1.4e-05 |
| MA1556.1 | RXRG | MA1556.1 | 1.97e-08 | 1.7e-05 |
| MA0643.1 | Esrrg | MA0643.1 | 1.98e-08 | 1.71e-05 |
| MA1596.1 (ZNF460) | MEME-5 | GGGAGGCYGAGGCAG | 0.106 | 1.8e-05 |
| MA0609.2 | CREM | MA0609.2 | 2.1e-08 | 1.81e-05 |
| MA1116.1 | RBPJ | MA1116.1 | 2.21e-08 | 1.91e-05 |
| MA1984.1 | ZNF667 | MA1984.1 | 2.48e-08 | 2.14e-05 |
| MA1573.2 | Thap11 | MA1573.2 | 2.77e-08 | 2.39e-05 |
| MA1557.1 | SMAD5 | MA1557.1 | 3.22e-08 | 2.77e-05 |
| MA0623.2 | NEUROG1 | MA0623.2 | 5.37e-08 | 4.63e-05 |
| MA1110.2 | Nr1H4 | MA1110.2 | 6.05e-08 | 5.21e-05 |
| MA1711.1 | ZNF343 | MA1711.1 | 7.9e-08 | 6.81e-05 |
| MA1111.1 | NR2F2 | MA1111.1 | 8.49e-08 | 7.32e-05 |
| MA1513.1 | KLF15 | MA1513.1 | 9.13e-08 | 7.87e-05 |
| MA1117.1 | RELB | MA1117.1 | 9.8e-08 | 8.45e-05 |
| MA0473.3 | ELF1 | MA0473.3 | 1.15e-07 | 9.94e-05 |
| MA1529.1 | NHLH2 | MA1529.1 | 2.3e-07 | 0.000198 |
| MA0062.3 | GABPA | MA0062.3 | 2.84e-07 | 0.000245 |
| MA1555.1 | RXRB | MA1555.1 | 2.94e-07 | 0.000254 |
| MA1106.1 | HIF1A | MA1106.1 | 3.5e-07 | 0.000302 |
| 7-GCCWGGC | STREME-7 | 7-GCCWGGC | 2.19e-109 | 0.000332 |
| MA0818.2 | BHLHE22 | MA0818.2 | 4.55e-07 | 0.000392 |
| MA0640.2 | ELF3 | MA0640.2 | 4.62e-07 | 0.000398 |
| MA1967.1 | TFAP4::FLI1 | MA1967.1 | 4.63e-07 | 0.000399 |
| MA1495.1 | HOXA1 | MA1495.1 | 5.4e-07 | 0.000465 |
| MA1999.1 | Prdm5 | MA1999.1 | 5.41e-07 | 0.000466 |
| MA0482.2 | GATA4 | MA0482.2 | 6.25e-07 | 0.000539 |
| MA0904.2 | HOXB5 | MA0904.2 | 6.79e-07 | 0.000586 |
| MA0732.1 | EGR3 | MA0732.1 | 1.06e-06 | 0.000912 |
| MA1983.1 (ZNF582) | STREME-8 | 8-GCAGCC | 9.53e-48 | 0.000988 |
| MA1584.1 | ZIC5 | MA1584.1 | 1.35e-06 | 0.00116 |
| MA1969.1 | THRA | MA1969.1 | 1.64e-06 | 0.00141 |
| MA0138.2 | REST | MA0138.2 | 1.83e-06 | 0.00158 |
| MA1717.1 | ZNF784 | MA1717.1 | 2.86e-06 | 0.00246 |
| MA1998.1 | Prdm14 | MA1998.1 | 3.19e-06 | 0.00275 |
| MA0782.2 (PKNOX1) | STREME-9 | 9-AGTGMCAGT | 2.25e-30 | 0.00305 |
| MA1722.1 (ZSCAN31) | STREME-10 | 10-CAGGGC | 5.47e-95 | 0.00309 |
| MA0592.3 | ESRRA | MA0592.3 | 3.94e-06 | 0.0034 |
| MA0474.3 | Erg | MA0474.3 | 4.12e-06 | 0.00355 |
| MA0063.2 | NKX2-5 | MA0063.2 | 6.6e-06 | 0.00569 |
| MA1988.1 (Atf3) | MEME-6 | CAGGCRTGAGCCACY | 0.000708 | 0.0057 |
| MA0003.4 (TFAP2A) | STREME-11 | 11-CCCTCAG | 3.8e-70 | 0.00615 |
| MA0860.1 | Rarg | MA0860.1 | 9.4e-06 | 0.00811 |
| MA0707.2 | MNX1 | MA0707.2 | 9.6e-06 | 0.00828 |
| MA0616.2 | HES2 | MA0616.2 | 1.23e-05 | 0.0106 |
| MA1532.2 | NR1D2 | MA1532.2 | 1.38e-05 | 0.0119 |
| MA1632.1 | ATF2 | MA1632.1 | 1.4e-05 | 0.0121 |
| MA0778.1 | NFKB2 | MA0778.1 | 1.46e-05 | 0.0126 |
| MA0800.1 | EOMES | MA0800.1 | 1.7e-05 | 0.0146 |
| MA1654.1 | ZNF16 | MA1654.1 | 1.83e-05 | 0.0158 |
| MA1944.1 | ETV5::DRGX | MA1944.1 | 2.15e-05 | 0.0185 |
| MA1102.2 (CTCFL) | STREME-12 | 12-AGGGGG | 2.31e-85 | 0.029 |
| MA0842.2 | NRL | MA0842.2 | 4.36e-05 | 0.0376 |
| MA0730.1 | RARA | MA0730.1 | 4.39e-05 | 0.0379 |
| MA0603.1 | Arntl | MA0603.1 | 4.81e-05 | 0.0415 |
| MA1964.1 (SMAD2) | STREME-13 | 13-CAGACAG | 5.44e-67 | 1.32 |
| MA1994.1 (Nkx2-1) | STREME-14 | 14-MCACTTGA | 7.24e-16 | 2.55 |
| 15-CAGAGG | STREME-15 | 15-CAGAGG | 5.52e-49 | 4.3 |
ESR_OC_xstreme %>%
dplyr::select(SIM_MOTIF,ALT_ID, ID,SEA_PVALUE,EVALUE) %>%
arrange(.,EVALUE) %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
# separate(SIM_MOTIF, into= c("SIM_MOTIF", "NAME"), sep= " ") %>%
# dplyr::filter(., EVALUE<0.05) %>%
kable(., caption = "Enriched motifs in ESR OC") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "800px")| SIM_MOTIF | ALT_ID | ID | SEA_PVALUE | EVALUE |
|---|---|---|---|---|
| MA0462.2 | BATF::JUN | MA0462.2 | 7.25e-193 | 6.23e-190 |
| MA1988.1 | Atf3 | MA1988.1 | 1.02e-188 | 8.76e-186 |
| MA1144.1 | FOSL2::JUND | MA1144.1 | 5.1e-183 | 4.39e-180 |
| MA0489.2 | Jun | MA0489.2 | 1.23e-182 | 1.06e-179 |
| MA0477.2 | FOSL1 | MA0477.2 | 3.36e-181 | 2.88e-178 |
| MA1633.2 | BACH1 | MA1633.2 | 4.37e-181 | 3.76e-178 |
| MA0099.3 | FOS::JUN | MA0099.3 | 3.39e-180 | 2.91e-177 |
| MA1141.1 | FOS::JUND | MA1141.1 | 7.05e-180 | 6.06e-177 |
| MA0835.2 | BATF3 | MA0835.2 | 1.29e-179 | 1.11e-176 |
| MA1134.1 | FOS::JUNB | MA1134.1 | 4.84e-179 | 4.16e-176 |
| MA0476.1 | FOS | MA0476.1 | 8.53e-179 | 7.33e-176 |
| MA1138.1 | FOSL2::JUNB | MA1138.1 | 1.59e-176 | 1.37e-173 |
| MA1128.1 | FOSL1::JUN | MA1128.1 | 1.07e-175 | 9.22e-173 |
| MA1928.1 | BNC2 | MA1928.1 | 5.14e-174 | 4.42e-171 |
| MA1130.1 | FOSL2::JUN | MA1130.1 | 3.9e-173 | 3.35e-170 |
| MA0478.1 | FOSL2 | MA0478.1 | 7.14e-171 | 6.14e-168 |
| MA0490.2 | JUNB | MA0490.2 | 4.43e-170 | 3.81e-167 |
| MA0491.2 | JUND | MA0491.2 | 1.93e-167 | 1.66e-164 |
| MA1101.2 | BACH2 | MA1101.2 | 6.18e-166 | 5.31e-163 |
| MA1135.1 | FOSB::JUNB | MA1135.1 | 1.5e-164 | 1.29e-161 |
| MA1137.1 | FOSL1::JUNB | MA1137.1 | 2.27e-160 | 1.95e-157 |
| MA1142.1 | FOSL1::JUND | MA1142.1 | 2.5e-155 | 2.15e-152 |
| MA1634.1 | BATF | MA1634.1 | 5.11e-145 | 4.39e-142 |
| MA1132.1 | JUN::JUNB | MA1132.1 | 1e-141 | 8.6e-139 |
| MA0841.1 | NFE2 | MA0841.1 | 8.01e-139 | 6.87e-136 |
| MA0655.1 | JDP2 | MA0655.1 | 1.12e-122 | 9.65e-120 |
| MA0089.2 | MAFG::NFE2L1 | MA0089.2 | 5.75e-97 | 4.94e-94 |
| MA0496.3 | MAFK | MA0496.3 | 1.38e-91 | 1.18e-88 |
| MA0501.1 | MAF::NFE2 | MA0501.1 | 3.58e-74 | 3.07e-71 |
| MA1138.1 (FOSL2::JUNB) | MEME-1 | ATGASTCA | 3.52e-182 | 8.1e-65 |
| MA0150.2 | Nfe2l2 | MA0150.2 | 9.42e-66 | 8.09e-63 |
| MA1125.1 (ZNF384) | MEME-2 | TTTTTTTTTTTTTTT | 1.19e-23 | 5.1e-61 |
| MA0659.3 | Mafg | MA0659.3 | 2.92e-58 | 2.51e-55 |
| MA1639.1 | MEIS1 | MA1639.1 | 2.08e-49 | 1.79e-46 |
| MA0679.2 | ONECUT1 | MA0679.2 | 1.43e-45 | 1.23e-42 |
| MA0611.2 | Dux | MA0611.2 | 5.67e-45 | 4.87e-42 |
| MA1113.2 | PBX2 | MA1113.2 | 1.18e-41 | 1.02e-38 |
| MA0680.2 | Pax7 | MA0680.2 | 1.69e-39 | 1.45e-36 |
| MA0158.2 | HOXA5 | MA0158.2 | 1.42e-36 | 1.22e-33 |
| MA0046.2 | HNF1A | MA0046.2 | 1.45e-36 | 1.24e-33 |
| MA0681.2 | PHOX2B | MA0681.2 | 7.83e-36 | 6.73e-33 |
| MA0910.2 | HOXD8 | MA0910.2 | 9.39e-36 | 8.06e-33 |
| MA0706.1 | MEOX2 | MA0706.1 | 2.32e-35 | 1.99e-32 |
| MA0846.1 | FOXC2 | MA0846.1 | 1.11e-34 | 9.54e-32 |
| MA1502.1 | HOXB8 | MA1502.1 | 1.94e-34 | 1.67e-31 |
| MA0902.2 | HOXB2 | MA0902.2 | 1.03e-33 | 8.82e-31 |
| MA0755.1 | CUX2 | MA0755.1 | 6.57e-33 | 5.64e-30 |
| MA0788.1 | POU3F3 | MA0788.1 | 1.51e-32 | 1.3e-29 |
| MA0032.2 | FOXC1 | MA0032.2 | 1.58e-32 | 1.36e-29 |
| MA0886.1 | EMX2 | MA0886.1 | 3.4e-32 | 2.92e-29 |
| MA0041.2 | FOXD3 | MA0041.2 | 3.41e-32 | 2.93e-29 |
| MA1505.1 | HOXC8 | MA1505.1 | 6.5e-32 | 5.59e-29 |
| MA1640.1 | MEIS2 | MA1640.1 | 1.46e-31 | 1.26e-28 |
| MA0791.1 | POU4F3 | MA0791.1 | 2.24e-31 | 1.93e-28 |
| MA0845.1 | FOXB1 | MA0845.1 | 3.86e-31 | 3.32e-28 |
| MA0785.1 | POU2F1 | MA0785.1 | 6.65e-31 | 5.71e-28 |
| MA0495.3 | MAFF | MA0495.3 | 1.09e-30 | 9.38e-28 |
| MA1124.1 | ZNF24 | MA1124.1 | 1.18e-30 | 1.01e-27 |
| MA0786.1 | POU3F1 | MA0786.1 | 2.02e-30 | 1.73e-27 |
| MA1495.1 | HOXA1 | MA1495.1 | 3.16e-30 | 2.71e-27 |
| MA0851.1 | Foxj3 | MA0851.1 | 6.65e-30 | 5.72e-27 |
| MA0833.2 | ATF4 | MA0833.2 | 9.82e-30 | 8.43e-27 |
| MA0757.1 | ONECUT3 | MA0757.1 | 9.96e-30 | 8.56e-27 |
| MA0715.1 | PROP1 | MA0715.1 | 1.11e-29 | 9.52e-27 |
| MA0766.2 | GATA5 | MA0766.2 | 2.5e-29 | 2.15e-26 |
| MA1636.1 | CEBPG | MA1636.1 | 3.48e-29 | 2.99e-26 |
| MA0713.1 | PHOX2A | MA0713.1 | 3.5e-29 | 3e-26 |
| MA0675.1 | NKX6-2 | MA0675.1 | 3.87e-29 | 3.33e-26 |
| MA0612.2 | EMX1 | MA0612.2 | 4.47e-29 | 3.84e-26 |
| MA0707.2 | MNX1 | MA0707.2 | 1.12e-28 | 9.58e-26 |
| MA0613.1 | FOXG1 | MA0613.1 | 1.54e-28 | 1.32e-25 |
| MA0787.1 | POU3F2 | MA0787.1 | 2.11e-28 | 1.82e-25 |
| MA0661.1 | MEOX1 | MA0661.1 | 3.47e-28 | 2.98e-25 |
| MA0628.1 | POU6F1 | MA0628.1 | 5.59e-28 | 4.8e-25 |
| MA1134.1 (FOS::JUNB) | STREME-1 | 1-DATGASTCATH | 9.03999999999999e-197 | 7.09e-25 |
| MA0722.1 | VAX1 | MA0722.1 | 1.31e-27 | 1.13e-24 |
| MA1143.1 | FOSL1::JUND | MA1143.1 | 1.98e-27 | 1.7e-24 |
| MA1978.1 | ZNF354A | MA1978.1 | 2.14e-27 | 1.84e-24 |
| MA0682.2 (PITX1) | MEME-3 | GCCTGTAATCCCAGC | 3.15e-22 | 2e-24 |
| MA1519.1 | LHX5 | MA1519.1 | 2.43e-27 | 2.09e-24 |
| MA0912.2 | HOXD3 | MA0912.2 | 3.76e-27 | 3.23e-24 |
| MA0602.1 | Arid5a | MA0602.1 | 8.52e-27 | 7.32e-24 |
| MA1480.1 | DPRX | MA1480.1 | 9.24e-27 | 7.94e-24 |
| MA0676.1 | Nr2e1 | MA0676.1 | 9.64e-27 | 8.28e-24 |
| MA0674.1 | NKX6-1 | MA0674.1 | 1.26e-26 | 1.09e-23 |
| MA0148.4 | FOXA1 | MA0148.4 | 1.38e-26 | 1.18e-23 |
| MA0132.2 | PDX1 | MA0132.2 | 1.54e-26 | 1.32e-23 |
| MA1963.1 | SATB1 | MA1963.1 | 1.64e-26 | 1.41e-23 |
| MA0888.1 | EVX2 | MA0888.1 | 2.75e-26 | 2.36e-23 |
| MA0142.1 | Pou5f1::Sox2 | MA0142.1 | 3.59e-26 | 3.08e-23 |
| MA0792.1 | POU5F1B | MA0792.1 | 3.74e-26 | 3.21e-23 |
| MA1481.1 | DRGX | MA1481.1 | 7.83e-26 | 6.72e-23 |
| MA0630.1 | SHOX | MA0630.1 | 1.51e-25 | 1.3e-22 |
| MA0683.1 | POU4F2 | MA0683.1 | 2.4e-25 | 2.06e-22 |
| MA1487.2 | FOXE1 | MA1487.2 | 2.63e-25 | 2.26e-22 |
| MA0701.2 | LHX9 | MA0701.2 | 2.63e-25 | 2.26e-22 |
| MA0153.2 | HNF1B | MA0153.2 | 2.81e-25 | 2.41e-22 |
| MA0724.1 | VENTX | MA0724.1 | 2.99e-25 | 2.57e-22 |
| MA0650.3 | Hoxa13 | MA0650.3 | 3.19e-25 | 2.74e-22 |
| MA1115.1 | POU5F1 | MA1115.1 | 3.32e-25 | 2.86e-22 |
| MA1962.1 | POU2F1::SOX2 | MA1962.1 | 4.21e-25 | 3.61e-22 |
| MA0789.1 | POU3F4 | MA0789.1 | 4.4e-25 | 3.78e-22 |
| MA0025.2 | NFIL3 | MA0025.2 | 5.77e-25 | 4.95e-22 |
| MA0908.1 | HOXD11 | MA0908.1 | 7.82e-25 | 6.71e-22 |
| MA0847.3 | FOXD2 | MA0847.3 | 1e-24 | 8.61e-22 |
| MA0078.2 | Sox17 | MA0078.2 | 1.07e-24 | 9.19e-22 |
| MA0035.4 | GATA1 | MA0035.4 | 1.39e-24 | 1.2e-21 |
| MA0601.1 | Arid3b | MA0601.1 | 1.51e-24 | 1.29e-21 |
| MA0700.2 | LHX2 | MA0700.2 | 1.77e-24 | 1.52e-21 |
| MA0075.3 | PRRX2 | MA0075.3 | 3.23e-24 | 2.77e-21 |
| MA0790.1 | POU4F1 | MA0790.1 | 3.37e-24 | 2.89e-21 |
| MA0043.3 | HLF | MA0043.3 | 4.11e-24 | 3.53e-21 |
| MA0037.4 | Gata3 | MA0037.4 | 4.68e-24 | 4.02e-21 |
| MA0627.2 | POU2F3 | MA0627.2 | 5.29e-24 | 4.54e-21 |
| MA1497.1 | HOXA6 | MA1497.1 | 7.64e-24 | 6.56e-21 |
| MA0754.2 | CUX1 | MA0754.2 | 9.06e-24 | 7.78e-21 |
| MA0903.1 | HOXB3 | MA0903.1 | 1.35e-23 | 1.16e-20 |
| MA1577.1 | TLX2 | MA1577.1 | 1.69e-23 | 1.45e-20 |
| MA0614.1 | Foxj2 | MA0614.1 | 1.7e-23 | 1.46e-20 |
| MA0084.1 | SRY | MA0084.1 | 2.3e-23 | 1.98e-20 |
| MA0644.2 | ESX1 | MA0644.2 | 2.62e-23 | 2.25e-20 |
| MA0900.2 | HOXA2 | MA0900.2 | 3.69e-23 | 3.17e-20 |
| MA0031.1 | FOXD1 | MA0031.1 | 4.05e-23 | 3.48e-20 |
| MA0718.1 | RAX | MA0718.1 | 7.96e-23 | 6.84e-20 |
| MA0887.1 | EVX1 | MA0887.1 | 8.53e-23 | 7.33e-20 |
| MA1471.1 | BARX2 | MA1471.1 | 1.08e-22 | 9.29e-20 |
| MA0102.4 | CEBPA | MA0102.4 | 1.47e-22 | 1.26e-19 |
| MA0904.2 | HOXB5 | MA0904.2 | 1.53e-22 | 1.31e-19 |
| MA0793.1 | POU6F2 | MA0793.1 | 1.63e-22 | 1.4e-19 |
| MA1104.2 | GATA6 | MA1104.2 | 1.71e-22 | 1.47e-19 |
| MA0654.1 | ISX | MA0654.1 | 1.85e-22 | 1.59e-19 |
| MA0854.1 | Alx1 | MA0854.1 | 2.06e-22 | 1.77e-19 |
| MA1606.1 | Foxf1 | MA1606.1 | 2.59e-22 | 2.22e-19 |
| MA1125.1 | ZNF384 | MA1125.1 | 2.67e-22 | 2.3e-19 |
| MA0052.4 | MEF2A | MA0052.4 | 2.83e-22 | 2.43e-19 |
| MA0658.1 | LHX6 | MA0658.1 | 3.37e-22 | 2.89e-19 |
| MA0890.1 | GBX2 | MA0890.1 | 3.61e-22 | 3.1e-19 |
| MA0726.1 | VSX2 | MA0726.1 | 4.78e-22 | 4.11e-19 |
| MA0704.1 | Lhx4 | MA0704.1 | 5.24e-22 | 4.5e-19 |
| MA0682.2 | PITX1 | MA0682.2 | 5.99e-22 | 5.14e-19 |
| MA0648.1 | GSC | MA0648.1 | 6.19e-22 | 5.32e-19 |
| MA0135.1 | Lhx3 | MA0135.1 | 1.02e-21 | 8.72e-19 |
| MA0723.2 | VAX2 | MA0723.2 | 1.09e-21 | 9.4e-19 |
| MA0716.1 | PRRX1 | MA0716.1 | 1.65e-21 | 1.42e-18 |
| ACTGCACTCCAGCCT | MEME-4 | ACTGCACTCCAGCCT | 7.09e-21 | 1.5e-18 |
| MA1683.1 | FOXA3 | MA1683.1 | 2.55e-21 | 2.19e-18 |
| MA0465.2 | CDX2 | MA0465.2 | 2.59e-21 | 2.22e-18 |
| MA0909.3 | Hoxd13 | MA0909.3 | 2.63e-21 | 2.26e-18 |
| MA1530.1 | NKX6-3 | MA1530.1 | 2.87e-21 | 2.46e-18 |
| MA0906.1 | HOXC12 | MA0906.1 | 3.55e-21 | 3.05e-18 |
| MA1463.1 | ARGFX | MA1463.1 | 3.65e-21 | 3.14e-18 |
| MA0874.1 | Arx | MA0874.1 | 3.81e-21 | 3.27e-18 |
| MA0710.1 | NOTO | MA0710.1 | 4.33e-21 | 3.72e-18 |
| MA0875.1 | BARX1 | MA0875.1 | 4.8e-21 | 4.12e-18 |
| MA0523.1 | TCF7L2 | MA0523.1 | 4.85e-21 | 4.17e-18 |
| MA1476.2 | Dlx5 | MA1476.2 | 5.13e-21 | 4.41e-18 |
| MA0040.1 | Foxq1 | MA0040.1 | 6.17e-21 | 5.3e-18 |
| MA0880.1 | Dlx3 | MA0880.1 | 6.26e-21 | 5.37e-18 |
| MA0068.2 | PAX4 | MA0068.2 | 6.64e-21 | 5.7e-18 |
| MA0894.1 | HESX1 | MA0894.1 | 7.22e-21 | 6.2e-18 |
| MA0885.2 | Dlx2 | MA0885.2 | 8.7e-21 | 7.47e-18 |
| MA0666.2 | MSX1 | MA0666.2 | 8.94e-21 | 7.68e-18 |
| MA0047.3 | FOXA2 | MA0047.3 | 9.26e-21 | 7.96e-18 |
| MA0882.1 | DLX6 | MA0882.1 | 1.08e-20 | 9.27e-18 |
| MA0699.1 | LBX2 | MA0699.1 | 1.34e-20 | 1.15e-17 |
| MA0725.1 | VSX1 | MA0725.1 | 1.66e-20 | 1.43e-17 |
| MA0709.1 | Msx3 | MA0709.1 | 1.94e-20 | 1.66e-17 |
| MA0717.1 | RAX2 | MA0717.1 | 3.79e-20 | 3.26e-17 |
| MA0712.2 | OTX2 | MA0712.2 | 7.12e-20 | 6.11e-17 |
| MA0836.2 | CEBPD | MA0836.2 | 7.65e-20 | 6.58e-17 |
| MA0867.2 | SOX4 | MA0867.2 | 8.39e-20 | 7.21e-17 |
| MA0780.1 | PAX3 | MA0780.1 | 9.82e-20 | 8.44e-17 |
| MA0853.1 | Alx4 | MA0853.1 | 1.04e-19 | 8.89e-17 |
| MA0898.1 | Hmx3 | MA0898.1 | 1.17e-19 | 1.01e-16 |
| MA1489.1 | FOXN3 | MA1489.1 | 1.2e-19 | 1.03e-16 |
| MA1991.1 | Hnf1A | MA1991.1 | 1.48e-19 | 1.27e-16 |
| MA0036.3 | GATA2 | MA0036.3 | 1.58e-19 | 1.35e-16 |
| MA1607.1 | Foxl2 | MA1607.1 | 1.62e-19 | 1.39e-16 |
| MA0884.2 | DUXA | MA0884.2 | 1.69e-19 | 1.45e-16 |
| MA0881.1 | Dlx4 | MA0881.1 | 2.33e-19 | 2.01e-16 |
| MA0720.1 | Shox2 | MA0720.1 | 2.34e-19 | 2.01e-16 |
| MA0027.2 | EN1 | MA0027.2 | 2.87e-19 | 2.46e-16 |
| MA0892.1 | GSX1 | MA0892.1 | 3.9e-19 | 3.35e-16 |
| MA0913.2 | HOXD9 | MA0913.2 | 4.31e-19 | 3.7e-16 |
| MA1139.1 | FOSL2::JUNB | MA1139.1 | 5.57e-19 | 4.79e-16 |
| MA1103.2 | FOXK2 | MA1103.2 | 5.69e-19 | 4.88e-16 |
| MA1632.1 | ATF2 | MA1632.1 | 5.8e-19 | 4.98e-16 |
| MA0829.2 | SREBF1 | MA0829.2 | 6.15e-19 | 5.28e-16 |
| MA1729.1 | ZNF680 | MA1729.1 | 7.3e-19 | 6.27e-16 |
| MA0705.1 | Lhx8 | MA0705.1 | 8.4e-19 | 7.22e-16 |
| MA0889.1 | GBX1 | MA0889.1 | 9.59e-19 | 8.23e-16 |
| MA1974.1 | ZNF211 | MA1974.1 | 1.01e-18 | 8.64e-16 |
| MA1131.1 | FOSL2::JUN | MA1131.1 | 1.26e-18 | 1.08e-15 |
| MA0702.2 | LMX1A | MA0702.2 | 1.36e-18 | 1.17e-15 |
| MA0621.1 | mix-a | MA0621.1 | 1.54e-18 | 1.33e-15 |
| MA1518.2 | Lhx1 | MA1518.2 | 2.01e-18 | 1.72e-15 |
| MA0721.1 | UNCX | MA0721.1 | 2.3e-18 | 1.98e-15 |
| MA0635.1 | BARHL2 | MA0635.1 | 2.33e-18 | 2e-15 |
| MA0849.1 | FOXO6 | MA0849.1 | 2.92e-18 | 2.51e-15 |
| MA1421.1 | TCF7L1 | MA1421.1 | 3.31e-18 | 2.85e-15 |
| MA0018.4 | CREB1 | MA0018.4 | 3.35e-18 | 2.88e-15 |
| MA0639.1 | DBP | MA0639.1 | 4.93e-18 | 4.23e-15 |
| MA0896.1 | Hmx1 | MA0896.1 | 5.79e-18 | 4.98e-15 |
| MA0662.1 | MIXL1 | MA0662.1 | 6.51e-18 | 5.59e-15 |
| MA1496.1 | HOXA4 | MA1496.1 | 7.15e-18 | 6.14e-15 |
| MA0030.1 | FOXF2 | MA0030.1 | 7.22e-18 | 6.2e-15 |
| MA0497.1 | MEF2C | MA0497.1 | 9.77e-18 | 8.39e-15 |
| MA0879.2 | DLX1 | MA0879.2 | 9.98e-18 | 8.58e-15 |
| MA1970.1 | TRPS1 | MA1970.1 | 1.14e-17 | 9.77e-15 |
| MA0895.1 | HMBOX1 | MA0895.1 | 1.19e-17 | 1.02e-14 |
| MA0070.1 | PBX1 | MA0070.1 | 1.2e-17 | 1.03e-14 |
| MA0768.2 | Lef1 | MA0768.2 | 1.21e-17 | 1.04e-14 |
| MA0077.1 | SOX9 | MA0077.1 | 1.3e-17 | 1.11e-14 |
| MA1960.1 | MGA::EVX1 | MA1960.1 | 1.59e-17 | 1.37e-14 |
| MA0482.2 | GATA4 | MA0482.2 | 1.6e-17 | 1.37e-14 |
| MA1603.1 | Dmrt1 | MA1603.1 | 1.65e-17 | 1.42e-14 |
| MA1500.1 | HOXB6 | MA1500.1 | 1.74e-17 | 1.5e-14 |
| MA1152.1 | SOX15 | MA1152.1 | 1.84e-17 | 1.58e-14 |
| MA1145.1 | FOSL2::JUND | MA1145.1 | 2.72e-17 | 2.33e-14 |
| MA1126.1 | FOS::JUN | MA1126.1 | 3.05e-17 | 2.62e-14 |
| MA0911.1 | Hoxa11 | MA0911.1 | 3.42e-17 | 2.94e-14 |
| MA0634.1 | ALX3 | MA0634.1 | 3.44e-17 | 2.96e-14 |
| MA1499.1 | HOXB4 | MA1499.1 | 3.85e-17 | 3.3e-14 |
| MA0852.2 | FOXK1 | MA0852.2 | 3.91e-17 | 3.36e-14 |
| MA0143.4 | SOX2 | MA0143.4 | 4.56e-17 | 3.91e-14 |
| MA1549.1 | POU6F1 | MA1549.1 | 6.52e-17 | 5.6e-14 |
| MA0901.2 | HOXB13 | MA0901.2 | 1.02e-16 | 8.73e-14 |
| MA0848.1 | FOXO4 | MA0848.1 | 1.12e-16 | 9.61e-14 |
| MA0117.2 | Mafb | MA0117.2 | 1.21e-16 | 1.04e-13 |
| MA0784.2 | POU1F1 | MA0784.2 | 1.26e-16 | 1.08e-13 |
| MA0483.1 | Gfi1B | MA0483.1 | 1.3e-16 | 1.12e-13 |
| MA0481.3 | FOXP1 | MA0481.3 | 1.33e-16 | 1.15e-13 |
| MA0042.2 | FOXI1 | MA0042.2 | 1.7e-16 | 1.46e-13 |
| MA0708.2 | MSX2 | MA0708.2 | 1.74e-16 | 1.5e-13 |
| MA0593.1 | FOXP2 | MA0593.1 | 1.76e-16 | 1.51e-13 |
| MA0891.1 | GSC2 | MA0891.1 | 1.9e-16 | 1.63e-13 |
| MA0642.2 | EN2 | MA0642.2 | 2.29e-16 | 1.96e-13 |
| MA1720.1 | ZNF85 | MA1720.1 | 2.31e-16 | 1.98e-13 |
| MA0842.2 | NRL | MA0842.2 | 2.68e-16 | 2.3e-13 |
| MA1501.1 | HOXB7 | MA1501.1 | 2.96e-16 | 2.54e-13 |
| MA0782.2 | PKNOX1 | MA0782.2 | 3.12e-16 | 2.68e-13 |
| MA0876.1 | BSX | MA0876.1 | 3.28e-16 | 2.82e-13 |
| MA0618.1 | LBX1 | MA0618.1 | 6.67e-16 | 5.73e-13 |
| MA0897.1 | Hmx2 | MA0897.1 | 1.04e-15 | 8.91e-13 |
| MA0899.1 | HOXA10 | MA0899.1 | 1.13e-15 | 9.68e-13 |
| MA1507.1 | HOXD4 | MA1507.1 | 1.2e-15 | 1.04e-12 |
| MA1608.1 | Isl1 | MA1608.1 | 1.39e-15 | 1.2e-12 |
| MA1588.1 | ZNF136 | MA1588.1 | 1.41e-15 | 1.21e-12 |
| MA0905.1 | HOXC10 | MA0905.1 | 1.51e-15 | 1.29e-12 |
| MA1127.1 | FOSB::JUN | MA1127.1 | 1.58e-15 | 1.36e-12 |
| MA0838.1 | CEBPG | MA0838.1 | 2.16e-15 | 1.85e-12 |
| MA0689.1 | TBX20 | MA0689.1 | 2.38e-15 | 2.04e-12 |
| MA0591.1 | Bach1::Mafk | MA0591.1 | 2.52e-15 | 2.17e-12 |
| MA0492.1 | JUND | MA0492.1 | 6.19e-15 | 5.32e-12 |
| MA1657.1 | ZNF652 | MA1657.1 | 1.01e-14 | 8.67e-12 |
| MA1707.1 | DMRTA1 | MA1707.1 | 1.1e-14 | 9.44e-12 |
| MA0711.1 | OTX1 | MA0711.1 | 1.39e-14 | 1.2e-11 |
| MA1498.2 | HOXA7 | MA1498.2 | 1.44e-14 | 1.23e-11 |
| MA1111.1 | NR2F2 | MA1111.1 | 1.74e-14 | 1.49e-11 |
| MA0850.1 | FOXP3 | MA0850.1 | 1.76e-14 | 1.51e-11 |
| MA0620.3 | MITF | MA0620.3 | 1.92e-14 | 1.65e-11 |
| MA0514.2 | Sox3 | MA0514.2 | 2.39e-14 | 2.05e-11 |
| MA0033.2 | FOXL1 | MA0033.2 | 3.98e-14 | 3.42e-11 |
| MA0442.2 | SOX10 | MA0442.2 | 6.92e-14 | 5.95e-11 |
| MA0461.2 | Atoh1 | MA0461.2 | 7.42e-14 | 6.37e-11 |
| MA0038.2 | GFI1 | MA0038.2 | 7.98e-14 | 6.86e-11 |
| MA0108.2 | TBP | MA0108.2 | 9.02e-14 | 7.75e-11 |
| MA1114.1 | PBX3 | MA1114.1 | 1.49e-13 | 1.28e-10 |
| MA1645.1 | NKX2-2 | MA1645.1 | 1.8e-13 | 1.55e-10 |
| MA0479.1 | FOXH1 | MA0479.1 | 1.89e-13 | 1.63e-10 |
| MA0093.3 | USF1 | MA0093.3 | 2.07e-13 | 1.78e-10 |
| MA0029.1 | Mecom | MA0029.1 | 2.1e-13 | 1.8e-10 |
| MA0488.1 | JUN | MA0488.1 | 2.54e-13 | 2.18e-10 |
| MA0157.3 | Foxo3 | MA0157.3 | 4.85e-13 | 4.17e-10 |
| MA1504.1 | HOXC4 | MA1504.1 | 5.64e-13 | 4.84e-10 |
| MA0873.1 | HOXD12 | MA0873.1 | 8.97e-13 | 7.71e-10 |
| MA1133.1 | JUN::JUNB | MA1133.1 | 1.11e-12 | 9.5e-10 |
| MA1129.1 | FOSL1::JUN | MA1129.1 | 1.19e-12 | 1.02e-09 |
| MA1120.1 | SOX13 | MA1120.1 | 1.27e-12 | 1.09e-09 |
| MA0714.1 | PITX3 | MA0714.1 | 2.54e-12 | 2.18e-09 |
| MA0480.2 | Foxo1 | MA0480.2 | 2.63e-12 | 2.26e-09 |
| MA0878.3 | CDX1 | MA0878.3 | 2.65e-12 | 2.27e-09 |
| MA0893.2 | GSX2 | MA0893.2 | 2.74e-12 | 2.36e-09 |
| MA1479.1 | DMRTC2 | MA1479.1 | 4.1e-12 | 3.52e-09 |
| MA0087.2 | Sox5 | MA0087.2 | 4.18e-12 | 3.59e-09 |
| MA0515.1 | Sox6 | MA0515.1 | 4.32e-12 | 3.71e-09 |
| MA0907.1 | HOXC13 | MA0907.1 | 4.84e-12 | 4.16e-09 |
| MA1975.1 | ZNF214 | MA1975.1 | 6.59e-12 | 5.66e-09 |
| MA0144.2 | STAT3 | MA0144.2 | 6.72e-12 | 5.77e-09 |
| MA0467.2 | Crx | MA0467.2 | 7.67e-12 | 6.58e-09 |
| MA0808.1 | TEAD3 | MA0808.1 | 7.8e-12 | 6.7e-09 |
| MA1644.1 | NFYC | MA1644.1 | 1e-11 | 8.62e-09 |
| MA1562.1 | SOX14 | MA1562.1 | 1.29e-11 | 1.11e-08 |
| MA2001.1 | Six4 | MA2001.1 | 1.39e-11 | 1.2e-08 |
| MA0517.1 | STAT1::STAT2 | MA0517.1 | 1.62e-11 | 1.39e-08 |
| MA1593.1 | ZNF317 | MA1593.1 | 1.84e-11 | 1.58e-08 |
| MA0152.2 | Nfatc2 | MA0152.2 | 2.21e-11 | 1.9e-08 |
| MA0596.1 | SREBF2 | MA0596.1 | 2.68e-11 | 2.3e-08 |
| MA0883.1 | Dmbx1 | MA0883.1 | 3.43e-11 | 2.95e-08 |
| MA1478.1 | DMRTA2 | MA1478.1 | 3.91e-11 | 3.36e-08 |
| MA1996.1 | Nr1H2 | MA1996.1 | 3.97e-11 | 3.41e-08 |
| MA0498.2 | MEIS1 | MA0498.2 | 4.75e-11 | 4.08e-08 |
| MA0914.1 | ISL2 | MA0914.1 | 6.05e-11 | 5.19e-08 |
| MA0151.1 | Arid3a | MA0151.1 | 8.28e-11 | 7.11e-08 |
| MA1709.1 | ZIM3 | MA1709.1 | 9.99e-11 | 8.58e-08 |
| MA0684.2 | RUNX3 | MA0684.2 | 1.4e-10 | 1.2e-07 |
| MA0629.1 | Rhox11 | MA0629.1 | 2.23e-10 | 1.91e-07 |
| MA1596.1 (ZNF460) | MEME-5 | RCCTCRGCCTCCCA | 2.48e-12 | 2.1e-07 |
| MA0095.3 | Yy1 | MA0095.3 | 2.63e-10 | 2.26e-07 |
| MA0804.1 | TBX19 | MA0804.1 | 2.88e-10 | 2.47e-07 |
| MA0703.2 | LMX1B | MA0703.2 | 3.11e-10 | 2.67e-07 |
| MA0800.1 | EOMES | MA0800.1 | 3.43e-10 | 2.95e-07 |
| MA1995.1 | Npas4 | MA1995.1 | 3.56e-10 | 3.06e-07 |
| MA0125.1 | Nobox | MA0125.1 | 3.76e-10 | 3.23e-07 |
| MA1136.1 | FOSB::JUNB | MA1136.1 | 4.02e-10 | 3.45e-07 |
| MA0594.2 | HOXA9 | MA0594.2 | 5.19e-10 | 4.46e-07 |
| MA0595.1 | SREBF1 | MA0595.1 | 5.58e-10 | 4.79e-07 |
| MA1148.1 | PPARA::RXRA | MA1148.1 | 6.16e-10 | 5.3e-07 |
| MA1503.1 | HOXB9 | MA1503.1 | 6.52e-10 | 5.6e-07 |
| MA0050.3 | Irf1 | MA0050.3 | 6.68e-10 | 5.74e-07 |
| MA1714.1 | ZNF675 | MA1714.1 | 7.59e-10 | 6.52e-07 |
| MA1534.1 | NR1I3 | MA1534.1 | 1.05e-09 | 8.98e-07 |
| MA1118.1 | SIX1 | MA1118.1 | 1.07e-09 | 9.17e-07 |
| MA0494.1 | Nr1h3::Rxra | MA0494.1 | 1.08e-09 | 9.27e-07 |
| MA1119.1 | SIX2 | MA1119.1 | 1.14e-09 | 9.78e-07 |
| MA0869.2 | Sox11 | MA0869.2 | 1.46e-09 | 1.25e-06 |
| MA1105.2 | GRHL2 | MA1105.2 | 1.96e-09 | 1.68e-06 |
| MA1520.1 | MAF | MA1520.1 | 2.29e-09 | 1.97e-06 |
| MA0769.2 | TCF7 | MA0769.2 | 2.55e-09 | 2.19e-06 |
| MA1110.2 | Nr1H4 | MA1110.2 | 2.87e-09 | 2.46e-06 |
| MA1570.1 | TFAP4 | MA1570.1 | 2.9e-09 | 2.49e-06 |
| MA1523.1 | MSANTD3 | MA1523.1 | 6.12e-09 | 5.25e-06 |
| MA1540.2 | NR5A1 | MA1540.2 | 7.72e-09 | 6.63e-06 |
| MA0063.2 | NKX2-5 | MA0063.2 | 9.87e-09 | 8.48e-06 |
| MA1112.2 | NR4A1 | MA1112.2 | 1.66e-08 | 1.43e-05 |
| MA0831.3 | TFE3 | MA0831.3 | 2.45e-08 | 2.11e-05 |
| MA0756.2 | ONECUT2 | MA0756.2 | 2.64e-08 | 2.26e-05 |
| MA0688.1 | TBX2 | MA0688.1 | 3.73e-08 | 3.21e-05 |
| MA0606.2 | Nfat5 | MA0606.2 | 4.61e-08 | 3.96e-05 |
| MA1506.1 | HOXD10 | MA1506.1 | 4.64e-08 | 3.99e-05 |
| MA1718.1 | ZNF8 | MA1718.1 | 4.84e-08 | 4.16e-05 |
| MA1977.1 | ZNF324 | MA1977.1 | 4.97e-08 | 4.27e-05 |
| MA0802.1 | TBR1 | MA0802.1 | 5.19e-08 | 4.45e-05 |
| MA0140.2 | GATA1::TAL1 | MA0140.2 | 6.47e-08 | 5.56e-05 |
| MA0258.2 | ESR2 | MA0258.2 | 6.74e-08 | 5.79e-05 |
| MA1536.1 | NR2C2 | MA1536.1 | 6.9e-08 | 5.92e-05 |
| MA0677.1 | Nr2f6 | MA0677.1 | 8.02e-08 | 6.89e-05 |
| MA1420.1 | IRF5 | MA1420.1 | 8.06e-08 | 6.92e-05 |
| MA0840.1 | Creb5 | MA0840.1 | 8.3e-08 | 7.13e-05 |
| MA1984.1 | ZNF667 | MA1984.1 | 1.03e-07 | 8.88e-05 |
| MA0485.2 | HOXC9 | MA0485.2 | 1.09e-07 | 9.33e-05 |
| MA0744.2 | SCRT2 | MA0744.2 | 1.12e-07 | 9.6e-05 |
| MA0060.3 | NFYA | MA0060.3 | 1.13e-07 | 9.68e-05 |
| MA1968.1 | TFCP2 | MA1968.1 | 1.2e-07 | 0.000103 |
| MA1473.1 | CDX4 | MA1473.1 | 1.22e-07 | 0.000105 |
| MA0673.1 | NKX2-8 | MA0673.1 | 1.69e-07 | 0.000145 |
| MA0124.2 | Nkx3-1 | MA0124.2 | 2.01e-07 | 0.000173 |
| MA0141.3 | ESRRB | MA0141.3 | 2.2e-07 | 0.000189 |
| MA0122.3 | Nkx3-2 | MA0122.3 | 2.46e-07 | 0.000211 |
| MA1994.1 | Nkx2-1 | MA1994.1 | 2.55e-07 | 0.000219 |
| MA0592.3 | ESRRA | MA0592.3 | 3.67e-07 | 0.000315 |
| MA1651.1 | ZFP42 | MA1651.1 | 3.92e-07 | 0.000337 |
| MA1561.1 | SOX12 | MA1561.1 | 4.28e-07 | 0.000368 |
| MA0691.1 | TFAP4 | MA0691.1 | 4.43e-07 | 0.000381 |
| MA1521.1 | MAFA | MA1521.1 | 4.53e-07 | 0.000389 |
| MA1147.1 | NR4A2::RXRA | MA1147.1 | 5.23e-07 | 0.000449 |
| MA0624.2 | Nfatc1 | MA0624.2 | 5.97e-07 | 0.000513 |
| MA0868.2 | SOX8 | MA0868.2 | 6.46e-07 | 0.000555 |
| MA0809.2 | TEAD4 | MA0809.2 | 6.95e-07 | 0.000597 |
| MA0743.2 | SCRT1 | MA0743.2 | 7.49e-07 | 0.000643 |
| MA1647.2 | Prdm4 | MA1647.2 | 7.6e-07 | 0.000653 |
| MA1731.1 | ZNF768 | MA1731.1 | 7.76e-07 | 0.000667 |
| MA0670.1 | NFIA | MA0670.1 | 7.94e-07 | 0.000682 |
| MA0687.1 | SPIC | MA0687.1 | 8.22e-07 | 0.000706 |
| MA0672.1 | NKX2-3 | MA0672.1 | 1.04e-06 | 0.000895 |
| MA1524.2 | Msgn1 | MA1524.2 | 1.12e-06 | 0.000959 |
| MA1979.1 | ZNF416 | MA1979.1 | 1.21e-06 | 0.00104 |
| MA0610.1 | DMRT3 | MA0610.1 | 1.6e-06 | 0.00138 |
| MA1623.1 | Stat2 | MA1623.1 | 1.7e-06 | 0.00146 |
| MA1652.1 | ZKSCAN5 | MA1652.1 | 1.81e-06 | 0.00155 |
| MA0669.1 | NEUROG2 | MA0669.1 | 1.94e-06 | 0.00167 |
| MA1956.1 | FOXO1::FLI1 | MA1956.1 | 1.98e-06 | 0.0017 |
| MA0619.1 | LIN54 | MA0619.1 | 2.26e-06 | 0.00194 |
| MA0719.1 | RHOXF1 | MA0719.1 | 2.29e-06 | 0.00197 |
| MA1574.1 | THRB | MA1574.1 | 2.33e-06 | 0.002 |
| MA1940.1 | ETV2::DRGX | MA1940.1 | 2.35e-06 | 0.00202 |
| MA0826.1 | OLIG1 | MA0826.1 | 2.59e-06 | 0.00222 |
| MA0817.1 | BHLHE23 | MA0817.1 | 2.68e-06 | 0.0023 |
| MA1150.1 | RORB | MA1150.1 | 3.2e-06 | 0.00275 |
| MA0827.1 | OLIG3 | MA0827.1 | 3.31e-06 | 0.00284 |
| MA0160.2 | NR4A2 | MA0160.2 | 3.51e-06 | 0.00302 |
| MA0065.2 | Pparg::Rxra | MA0065.2 | 3.7e-06 | 0.00317 |
| MA1937.1 | ERF::HOXB13 | MA1937.1 | 3.72e-06 | 0.00319 |
| MA0664.1 | MLXIPL | MA0664.1 | 4.1e-06 | 0.00352 |
| MA1567.2 | Tbx6 | MA1567.2 | 4.25e-06 | 0.00365 |
| MA0072.1 | RORA | MA0072.1 | 5.16e-06 | 0.00443 |
| MA0660.1 | MEF2B | MA0660.1 | 5.31e-06 | 0.00456 |
| MA1563.2 | SOX18 | MA1563.2 | 5.34e-06 | 0.00458 |
| MA0690.2 | TBX21 | MA0690.2 | 5.96e-06 | 0.00512 |
| MA0014.3 | PAX5 | MA0014.3 | 6.23e-06 | 0.00535 |
| MA0711.1 (OTX1) | STREME-2 | 2-ATAATC | 5.98e-38 | 0.0055 |
| MA0518.1 | Stat4 | MA0518.1 | 6.82e-06 | 0.00586 |
| MA0130.1 | ZNF354C | MA0130.1 | 6.97e-06 | 0.00599 |
| MA0651.2 | HOXC11 | MA0651.2 | 7.18e-06 | 0.00617 |
| MA1998.1 | Prdm14 | MA1998.1 | 7.49e-06 | 0.00643 |
| MA0643.1 | Esrrg | MA0643.1 | 7.94e-06 | 0.00682 |
| MA0681.2 (PHOX2B) | STREME-3 | 3-TCAAAT | 7.9e-18 | 0.00808 |
| MA0017.2 | NR2F1 | MA0017.2 | 9.69e-06 | 0.00832 |
| MA0071.1 | RORA | MA0071.1 | 1.09e-05 | 0.00939 |
| MA0631.1 | Six3 | MA0631.1 | 1.15e-05 | 0.00986 |
| MA0626.1 | Npas2 | MA0626.1 | 1.23e-05 | 0.0106 |
| MA0114.4 | HNF4A | MA0114.4 | 1.25e-05 | 0.0107 |
| MA0137.3 | STAT1 | MA0137.3 | 1.27e-05 | 0.0109 |
| MA1646.1 | OSR2 | MA1646.1 | 1.4e-05 | 0.012 |
| MA0825.1 | MNT | MA0825.1 | 1.51e-05 | 0.013 |
| MA0069.1 | PAX6 | MA0069.1 | 1.59e-05 | 0.0136 |
| MA1618.1 | Ptf1a | MA1618.1 | 1.61e-05 | 0.0138 |
| MA0507.2 | POU2F2 | MA0507.2 | 1.69e-05 | 0.0145 |
| MA0056.2 | MZF1 | MA0056.2 | 1.72e-05 | 0.0147 |
| 4-ATCTAA | STREME-4 | 4-ATCTAA | 3.39e-22 | 0.0181 |
| MA1949.1 | FLI1::DRGX | MA1949.1 | 2.13e-05 | 0.0183 |
| MA1990.1 | Gli1 | MA1990.1 | 2.19e-05 | 0.0188 |
| MA1146.1 | NR1H4::RXRA | MA1146.1 | 2.22e-05 | 0.019 |
| MA0090.3 | TEAD1 | MA0090.3 | 2.28e-05 | 0.0196 |
| MA0006.1 | Ahr::Arnt | MA0006.1 | 2.42e-05 | 0.0208 |
| MA0843.1 | TEF | MA0843.1 | 2.43e-05 | 0.0209 |
| MA1108.2 | MXI1 | MA1108.2 | 2.51e-05 | 0.0216 |
| MA1625.1 | Stat5b | MA1625.1 | 2.67e-05 | 0.023 |
| MA0058.3 | MAX | MA0058.3 | 2.95e-05 | 0.0253 |
| MA0505.2 | Nr5A2 | MA0505.2 | 3.01e-05 | 0.0258 |
| MA1957.1 | HOXB2::ELK1 | MA1957.1 | 3.14e-05 | 0.0269 |
| MA0036.3 (GATA2) | STREME-5 | 5-RDAGATAA | 2.61e-50 | 0.0287 |
| MA0484.2 | HNF4G | MA0484.2 | 3.54e-05 | 0.0304 |
| 6-ATATCA | STREME-6 | 6-ATATCA | 1.86e-35 | 0.0316 |
| MA2003.1 | NKX2-4 | MA2003.1 | 3.81e-05 | 0.0327 |
| MA0774.1 | MEIS2 | MA0774.1 | 4.16e-05 | 0.0358 |
| MA1555.1 (RXRB) | STREME-7 | 7-CATGAC | 3.14e-40 | 0.0361 |
| MA0775.1 | MEIS3 | MA0775.1 | 4.8e-05 | 0.0412 |
| MA0776.1 | MYBL1 | MA0776.1 | 5.27e-05 | 0.0452 |
| MA1643.1 (NFIB) | MEME-6 | GCCACCRYGCCYRGC | 6.27e-21 | 0.046 |
| MA1596.1 (ZNF460) | STREME-8 | 8-GCCTCAGCCTCCCA | 4.05e-11 | 0.0472 |
| MA0609.2 | CREM | MA0609.2 | 5.73e-05 | 0.0492 |
| MA1564.1 | SP9 | MA1564.1 | 5.78e-05 | 0.0497 |
| 9-KTCAAT | STREME-9 | 9-KTCAAT | 9e-23 | 0.0617 |
| MA1489.1 (FOXN3) | STREME-10 | 10-AAACAAA | 1.86e-33 | 0.106 |
| 11-AGTGAA | STREME-11 | 11-AGTGAA | 8.85e-20 | 0.122 |
| MA0790.1 (POU4F1) | STREME-12 | 12-ATKMAT | 4.42e-36 | 0.231 |
LR_open_xstreme %>%
dplyr::select(SIM_MOTIF,ALT_ID, ID,SEA_PVALUE,EVALUE) %>%
arrange(.,EVALUE) %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
# separate(SIM_MOTIF, into= c("SIM_MOTIF", "NAME"), sep= " ") %>%
# dplyr::filter(., EVALUE<0.05) %>%
kable(., caption = "Enriched motifs in LR_open") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "800px")| SIM_MOTIF | ALT_ID | ID | SEA_PVALUE | EVALUE |
|---|---|---|---|---|
| MA1134.1 (FOS::JUNB) | STREME-1 | 1-DATGASTCATH | 3.3e-305 | 5.25e-305 |
| MA1125.1 (ZNF384) | MEME-1 | AAAWAAAAAAAAAAA | 4.1e-229 | 6e-73 |
| MA0477.2 (FOSL1) | MEME-2 | TGAGTCAT | 4.52e-305 | 2.4e-70 |
| MA1634.1 (BATF) | STREME-2 | 2-NTGASTCAN | 3.82e-305 | 4.73e-53 |
| 3-ATRTAYAT | STREME-3 | 3-ATRTAYAT | 6.9e-303 | 5.97e-38 |
| 4-TGTAATC | STREME-4 | 4-TGTAATC | 5.17e-269 | 3.02e-32 |
| MA0817.1 (BHLHE23) | STREME-5 | 5-AAWCATAT | 4.04e-305 | 3.83e-30 |
| MA1632.1 (ATF2) | STREME-6 | 6-AAGTSATCH | 4.27e-305 | 1.96e-28 |
| 7-AVTABT | STREME-7 | 7-AVTABT | 1.59e-265 | 1.82e-24 |
| 8-WCATGW | STREME-8 | 8-WCATGW | 1.55e-305 | 3.13e-22 |
| 9-AAYWRTT | STREME-9 | 9-AAYWRTT | 6.27e-173 | 4.43e-22 |
| 10-TRTAYA | STREME-10 | 10-TRTAYA | 5.08e-291 | 1.68e-18 |
| MA1973.1 (ZKSCAN3) | MEME-3 | CCAGCCTGGGYRACA | 3.92e-118 | 4.3e-17 |
| MA1107.2 (KLF9) | MEME-4 | GTGTGTGTGTGTGTG | 4.84e-43 | 8.3e-14 |
| 11-TAAGAMA | STREME-11 | 11-TAAGAMA | 9.35e-305 | 4.66e-13 |
| MA0041.2 (FOXD3) | MEME-5 | AAAWGMAAAWWAAAA | 7.97e-173 | 1.4e-09 |
| MA1596.1 (ZNF460) | MEME-6 | TTGGGAGGCYGAGGY | 6.14e-119 | 6.3e-07 |
| MA0682.2 (PITX1) | MEME-7 | CCTGTAATCCCAGCA | 4.02e-129 | 7.9e-06 |
LR_close_xstreme %>%
dplyr::select(SIM_MOTIF,ALT_ID, ID,SEA_PVALUE,EVALUE) %>%
arrange(.,EVALUE) %>%
dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3)))) %>%
# separate(SIM_MOTIF, into= c("SIM_MOTIF", "NAME"), sep= " ") %>%
# dplyr::filter(., EVALUE<0.05) %>%
kable(., caption = "Enriched motifs in LR close") %>%
kable_paper("striped", full_width = TRUE) %>%
kable_styling(full_width = FALSE, font_size = 16) %>%
scroll_box(height = "800px")| SIM_MOTIF | ALT_ID | ID | SEA_PVALUE | EVALUE |
|---|---|---|---|---|
| MA0774.1 | MEIS2 | MA0774.1 | 5.29e-115 | 4.56e-112 |
| MA0775.1 | MEIS3 | MA0775.1 | 8.3e-91 | 7.15e-88 |
| MA0672.1 | NKX2-3 | MA0672.1 | 6.29e-87 | 5.41e-84 |
| MA0673.1 | NKX2-8 | MA0673.1 | 1.93e-72 | 1.66e-69 |
| MA2003.1 | NKX2-4 | MA2003.1 | 1.05e-65 | 9.04e-63 |
| MA0498.2 | MEIS1 | MA0498.2 | 4.09e-56 | 3.52e-53 |
| MA0797.1 | TGIF2 | MA0797.1 | 6.65e-55 | 5.72e-52 |
| MA0738.1 | HIC2 | MA0738.1 | 1.11e-53 | 9.53e-51 |
| MA0739.1 | Hic1 | MA0739.1 | 1.74e-53 | 1.5e-50 |
| MA0766.2 | GATA5 | MA0766.2 | 1.49e-51 | 1.28e-48 |
| MA0670.1 | NFIA | MA0670.1 | 3.94e-50 | 3.4e-47 |
| MA0626.1 | Npas2 | MA0626.1 | 1.06e-49 | 9.12e-47 |
| MA1994.1 | Nkx2-1 | MA1994.1 | 2.02e-49 | 1.74e-46 |
| MA0671.1 | NFIX | MA0671.1 | 1.37e-44 | 1.18e-41 |
| MA1615.1 | Plagl1 | MA1615.1 | 7.4e-43 | 6.37e-40 |
| MA0161.2 | NFIC | MA0161.2 | 1.95e-42 | 1.68e-39 |
| MA0783.1 | PKNOX2 | MA0783.1 | 5.38e-40 | 4.63e-37 |
| MA1108.2 | MXI1 | MA1108.2 | 4.6e-38 | 3.96e-35 |
| MA0809.2 | TEAD4 | MA0809.2 | 4.89e-38 | 4.21e-35 |
| MA1643.1 | NFIB | MA1643.1 | 5.06e-38 | 4.35e-35 |
| MA0092.1 | Hand1::Tcf3 | MA0092.1 | 8.33e-36 | 7.17e-33 |
| MA0669.1 | NEUROG2 | MA0669.1 | 1.14e-35 | 9.85e-33 |
| MA0036.3 | GATA2 | MA0036.3 | 5.26e-35 | 4.53e-32 |
| MA1641.1 | MYF5 | MA1641.1 | 8.45e-35 | 7.27e-32 |
| MA1986.1 | ZNF692 | MA1986.1 | 1.79e-34 | 1.54e-31 |
| MA1571.1 | TGIF2LX | MA1571.1 | 5.51e-34 | 4.74e-31 |
| MA0499.2 | MYOD1 | MA0499.2 | 2.26e-33 | 1.94e-30 |
| MA1631.1 | ASCL1 | MA1631.1 | 2.86e-33 | 2.46e-30 |
| MA1979.1 | ZNF416 | MA1979.1 | 6.24e-33 | 5.37e-30 |
| MA0090.3 | TEAD1 | MA0090.3 | 1.63e-32 | 1.4e-29 |
| MA0140.2 | GATA1::TAL1 | MA0140.2 | 4.39e-32 | 3.78e-29 |
| MA0623.2 | NEUROG1 | MA0623.2 | 5.56e-32 | 4.79e-29 |
| MA0808.1 | TEAD3 | MA0808.1 | 2.62e-31 | 2.26e-28 |
| MA0667.1 | MYF6 | MA0667.1 | 1.49e-30 | 1.28e-27 |
| MA0633.2 | Twist2 | MA0633.2 | 4.03e-30 | 3.47e-27 |
| MA0830.2 | TCF4 | MA0830.2 | 4.34e-30 | 3.73e-27 |
| MA0607.2 | BHLHA15 | MA0607.2 | 8.9e-30 | 7.66e-27 |
| MA1997.1 | Olig2 | MA1997.1 | 1.37e-29 | 1.18e-26 |
| MA0796.1 | TGIF1 | MA0796.1 | 1.63e-29 | 1.4e-26 |
| MA0597.2 | THAP1 | MA0597.2 | 2.64e-29 | 2.27e-26 |
| MA0060.3 | NFYA | MA0060.3 | 4.78e-29 | 4.12e-26 |
| MA0741.1 | KLF16 | MA0741.1 | 6.95e-29 | 5.99e-26 |
| MA1472.2 | Bhlha15 | MA1472.2 | 1.34e-28 | 1.16e-25 |
| MA1656.1 | ZNF449 | MA1656.1 | 2.13e-28 | 1.83e-25 |
| MA1730.1 | ZNF708 | MA1730.1 | 2.16e-28 | 1.86e-25 |
| MA1645.1 | NKX2-2 | MA1645.1 | 2.42e-28 | 2.08e-25 |
| MA1993.1 | Neurod2 | MA1993.1 | 2.52e-28 | 2.17e-25 |
| MA0826.1 | OLIG1 | MA0826.1 | 2.38e-27 | 2.05e-24 |
| MA1102.2 | CTCFL | MA1102.2 | 2.84e-27 | 2.44e-24 |
| MA1599.1 | ZNF682 | MA1599.1 | 1.36e-26 | 1.17e-23 |
| MA0039.4 | KLF4 | MA0039.4 | 2.42e-26 | 2.08e-23 |
| MA1719.1 | ZNF816 | MA1719.1 | 7.92e-26 | 6.82e-23 |
| MA1982.1 | ZNF574 | MA1982.1 | 8.83e-26 | 7.61e-23 |
| MA0070.1 | PBX1 | MA0070.1 | 1.73e-25 | 1.49e-22 |
| MA0698.1 | ZBTB18 | MA0698.1 | 2.64e-25 | 2.27e-22 |
| MA0048.2 | NHLH1 | MA0048.2 | 2.93e-25 | 2.53e-22 |
| MA0063.2 | NKX2-5 | MA0063.2 | 3.71e-25 | 3.19e-22 |
| MA1572.1 | TGIF2LY | MA1572.1 | 3.77e-25 | 3.24e-22 |
| MA1635.1 | BHLHE22 | MA1635.1 | 4.31e-25 | 3.71e-22 |
| MA1726.1 | ZNF331 | MA1726.1 | 8.16e-25 | 7.02e-22 |
| MA0817.1 | BHLHE23 | MA0817.1 | 1.14e-24 | 9.81e-22 |
| MA0810.1 | TFAP2A | MA0810.1 | 1.35e-24 | 1.16e-21 |
| MA0816.1 | Ascl2 | MA0816.1 | 1.41e-24 | 1.21e-21 |
| MA1468.1 | ATOH7 | MA1468.1 | 1.63e-24 | 1.41e-21 |
| MA1125.1 (ZNF384) | MEME-1 | AAAWAAAAAWAAAAA | 0.788 | 1.5e-21 |
| MA1650.1 | ZBTB14 | MA1650.1 | 4e-24 | 3.44e-21 |
| MA0122.3 | Nkx3-2 | MA0122.3 | 6.35e-24 | 5.47e-21 |
| MA1712.1 | ZNF454 | MA1712.1 | 1.31e-23 | 1.13e-20 |
| MA1629.1 | Zic2 | MA1629.1 | 3.23e-23 | 2.78e-20 |
| MA1578.1 | VEZF1 | MA1578.1 | 3.52e-23 | 3.03e-20 |
| MA1515.1 | KLF2 | MA1515.1 | 5.16e-23 | 4.45e-20 |
| MA1619.1 | Ptf1A | MA1619.1 | 1.18e-22 | 1.01e-19 |
| MA0697.2 | Zic3 | MA0697.2 | 1.18e-22 | 1.02e-19 |
| MA1620.1 | Ptf1A | MA1620.1 | 1.43e-22 | 1.23e-19 |
| MA1107.2 | KLF9 | MA1107.2 | 1.99e-22 | 1.71e-19 |
| MA1558.1 | SNAI1 | MA1558.1 | 2.94e-22 | 2.54e-19 |
| MA1621.1 | Rbpjl | MA1621.1 | 3.8e-22 | 3.27e-19 |
| MA0502.2 | NFYB | MA0502.2 | 5.14e-22 | 4.43e-19 |
| MA0914.1 | ISL2 | MA0914.1 | 5.86e-22 | 5.05e-19 |
| MA0521.2 | Tcf12 | MA0521.2 | 8.3e-22 | 7.14e-19 |
| MA0818.2 | BHLHE22 | MA0818.2 | 1.13e-21 | 9.74e-19 |
| MA1566.2 | TBX3 | MA1566.2 | 1.21e-21 | 1.04e-18 |
| MA1974.1 | ZNF211 | MA1974.1 | 2.12e-21 | 1.82e-18 |
| MA0130.1 | ZNF354C | MA0130.1 | 2.54e-21 | 2.19e-18 |
| MA0745.2 | SNAI2 | MA0745.2 | 2.84e-21 | 2.44e-18 |
| MA0747.1 | SP8 | MA0747.1 | 5.54e-21 | 4.77e-18 |
| MA0811.1 | TFAP2B | MA0811.1 | 6.98e-21 | 6.01e-18 |
| MA0018.4 | CREB1 | MA0018.4 | 7.28e-21 | 6.27e-18 |
| MA0691.1 | TFAP4 | MA0691.1 | 8.67e-21 | 7.46e-18 |
| MA1581.1 | ZBTB6 | MA1581.1 | 9.52e-21 | 8.2e-18 |
| MA0147.3 | MYC | MA0147.3 | 9.61e-21 | 8.27e-18 |
| MA0678.1 | OLIG2 | MA0678.1 | 1.08e-20 | 9.27e-18 |
| MA1644.1 | NFYC | MA1644.1 | 1.83e-20 | 1.57e-17 |
| MA0821.2 | HES5 | MA0821.2 | 1.85e-20 | 1.59e-17 |
| MA1655.1 | ZNF341 | MA1655.1 | 2.11e-20 | 1.81e-17 |
| MA1100.2 | ASCL1 | MA1100.2 | 2.12e-20 | 1.83e-17 |
| MA0734.3 | Gli2 | MA0734.3 | 2.66e-20 | 2.29e-17 |
| MA1570.1 | TFAP4 | MA1570.1 | 2.97e-20 | 2.56e-17 |
| MA0103.3 | ZEB1 | MA0103.3 | 4.69e-20 | 4.04e-17 |
| MA1153.1 | Smad4 | MA1153.1 | 4.85e-20 | 4.18e-17 |
| MA1959.1 | KLF7 | MA1959.1 | 5.09e-20 | 4.38e-17 |
| MA1564.1 | SP9 | MA1564.1 | 8.28e-20 | 7.13e-17 |
| MA1653.1 | ZNF148 | MA1653.1 | 1.25e-19 | 1.07e-16 |
| MA1623.1 (Stat2) | MEME-2 | TYYYTYTYTYTCTYT | 0.105 | 1.5e-16 |
| MA1567.2 | Tbx6 | MA1567.2 | 1.8e-19 | 1.55e-16 |
| MA1628.1 | Zic1::Zic2 | MA1628.1 | 1.97e-19 | 1.69e-16 |
| MA0471.2 | E2F6 | MA0471.2 | 2.74e-19 | 2.36e-16 |
| MA0003.4 | TFAP2A | MA0003.4 | 5.17e-19 | 4.45e-16 |
| MA0500.2 | MYOG | MA0500.2 | 6.44e-19 | 5.54e-16 |
| MA1563.2 | SOX18 | MA1563.2 | 8.58e-19 | 7.39e-16 |
| MA1121.1 | TEAD2 | MA1121.1 | 9.7e-19 | 8.35e-16 |
| MA1648.1 | TCF12 | MA1648.1 | 1.17e-18 | 1.01e-15 |
| MA0802.1 | TBR1 | MA0802.1 | 1.29e-18 | 1.11e-15 |
| MA1112.2 | NR4A1 | MA1112.2 | 1.58e-18 | 1.36e-15 |
| MA0524.2 | TFAP2C | MA0524.2 | 1.87e-18 | 1.61e-15 |
| MA1547.2 | PITX2 | MA1547.2 | 1.96e-18 | 1.69e-15 |
| MA0820.1 | FIGLA | MA0820.1 | 2.7e-18 | 2.33e-15 |
| MA0709.1 | Msx3 | MA0709.1 | 3.64e-18 | 3.13e-15 |
| MA0789.1 | POU3F4 | MA0789.1 | 4e-18 | 3.44e-15 |
| MA1123.2 | TWIST1 | MA1123.2 | 5.22e-18 | 4.49e-15 |
| MA0688.1 | TBX2 | MA0688.1 | 5.24e-18 | 4.52e-15 |
| MA1524.2 | Msgn1 | MA1524.2 | 5.38e-18 | 4.63e-15 |
| MA0807.1 | TBX5 | MA0807.1 | 7.68e-18 | 6.61e-15 |
| MA1981.1 | ZNF530 | MA1981.1 | 8.06e-18 | 6.94e-15 |
| MA1990.1 | Gli1 | MA1990.1 | 8.18e-18 | 7.05e-15 |
| MA0814.2 | TFAP2C | MA0814.2 | 2.03e-17 | 1.75e-14 |
| MA1109.1 | NEUROD1 | MA1109.1 | 2.29e-17 | 1.97e-14 |
| MA0896.1 | Hmx1 | MA0896.1 | 2.91e-17 | 2.51e-14 |
| MA0119.1 | NFIC::TLX1 | MA0119.1 | 2.94e-17 | 2.53e-14 |
| MA0006.1 | Ahr::Arnt | MA0006.1 | 3.23e-17 | 2.78e-14 |
| MA1618.1 | Ptf1a | MA1618.1 | 4.14e-17 | 3.57e-14 |
| MA0033.2 | FOXL1 | MA0033.2 | 4.32e-17 | 3.72e-14 |
| MA1724.1 | Rfx6 | MA1724.1 | 4.47e-17 | 3.84e-14 |
| MA0792.1 | POU5F1B | MA0792.1 | 5.36e-17 | 4.62e-14 |
| MA0827.1 | OLIG3 | MA0827.1 | 6.64e-17 | 5.71e-14 |
| MA1122.1 | TFDP1 | MA1122.1 | 7.23e-17 | 6.22e-14 |
| MA0654.1 | ISX | MA0654.1 | 8.65e-17 | 7.45e-14 |
| MA0091.1 | TAL1::TCF3 | MA0091.1 | 1.33e-16 | 1.14e-13 |
| MA0800.1 | EOMES | MA0800.1 | 1.48e-16 | 1.27e-13 |
| MA0493.2 | KLF1 | MA0493.2 | 1.56e-16 | 1.35e-13 |
| MA0646.1 | GCM1 | MA0646.1 | 1.71e-16 | 1.47e-13 |
| MA0803.1 | TBX15 | MA0803.1 | 1.99e-16 | 1.71e-13 |
| MA0146.2 | Zfx | MA0146.2 | 2e-16 | 1.72e-13 |
| MA0812.1 | TFAP2B | MA0812.1 | 2.2e-16 | 1.9e-13 |
| MA0154.4 | EBF1 | MA0154.4 | 2.24e-16 | 1.93e-13 |
| MA0258.2 | ESR2 | MA0258.2 | 4.47e-16 | 3.85e-13 |
| MA0666.2 | MSX1 | MA0666.2 | 6.21e-16 | 5.35e-13 |
| MA0665.1 | MSC | MA0665.1 | 8.28e-16 | 7.13e-13 |
| MA1723.1 | PRDM9 | MA1723.1 | 8.32e-16 | 7.16e-13 |
| MA0912.2 | HOXD3 | MA0912.2 | 1.1e-15 | 9.47e-13 |
| MA1593.1 | ZNF317 | MA1593.1 | 1.13e-15 | 9.72e-13 |
| MA0751.1 | ZIC4 | MA0751.1 | 1.2e-15 | 1.03e-12 |
| MA0482.2 | GATA4 | MA0482.2 | 2.4e-15 | 2.06e-12 |
| MA1522.1 | MAZ | MA1522.1 | 2.44e-15 | 2.1e-12 |
| MA1514.1 | KLF17 | MA1514.1 | 2.47e-15 | 2.13e-12 |
| MA0664.1 | MLXIPL | MA0664.1 | 2.78e-15 | 2.39e-12 |
| MA0145.2 | Tfcp2l1 | MA0145.2 | 3.16e-15 | 2.72e-12 |
| MA0596.1 | SREBF2 | MA0596.1 | 3.26e-15 | 2.81e-12 |
| MA1500.1 | HOXB6 | MA1500.1 | 3.88e-15 | 3.34e-12 |
| MA2002.1 | Zfp335 | MA2002.1 | 3.95e-15 | 3.4e-12 |
| MA0904.2 | HOXB5 | MA0904.2 | 4.22e-15 | 3.63e-12 |
| MA0124.2 | Nkx3-1 | MA0124.2 | 4.97e-15 | 4.28e-12 |
| MA1731.1 | ZNF768 | MA1731.1 | 6.41e-15 | 5.52e-12 |
| MA0767.1 | GCM2 | MA0767.1 | 6.54e-15 | 5.63e-12 |
| MA1527.1 | NFIC | MA1527.1 | 7.09e-15 | 6.11e-12 |
| MA1638.1 | HAND2 | MA1638.1 | 7.23e-15 | 6.22e-12 |
| MA0782.2 | PKNOX1 | MA0782.2 | 1.04e-14 | 8.94e-12 |
| MA0806.1 | TBX4 | MA0806.1 | 1.17e-14 | 1.01e-11 |
| MA0158.2 | HOXA5 | MA0158.2 | 1.24e-14 | 1.07e-11 |
| MA0674.1 | NKX6-1 | MA0674.1 | 1.26e-14 | 1.08e-11 |
| MA1637.1 | EBF3 | MA1637.1 | 1.85e-14 | 1.59e-11 |
| MA0668.2 | Neurod2 | MA0668.2 | 2.27e-14 | 1.95e-11 |
| MA1627.1 | Wt1 | MA1627.1 | 3.08e-14 | 2.65e-11 |
| MA0825.1 | MNT | MA0825.1 | 3.13e-14 | 2.7e-11 |
| MA1493.1 | HES6 | MA1493.1 | 3.55e-14 | 3.05e-11 |
| MA0522.3 | TCF3 | MA0522.3 | 3.65e-14 | 3.15e-11 |
| MA0644.2 | ESX1 | MA0644.2 | 4.07e-14 | 3.5e-11 |
| MA0523.1 | TCF7L2 | MA0523.1 | 5.07e-14 | 4.36e-11 |
| MA0461.2 | Atoh1 | MA0461.2 | 5.95e-14 | 5.13e-11 |
| MA0724.1 | VENTX | MA0724.1 | 6.2e-14 | 5.33e-11 |
| MA0052.4 | MEF2A | MA0052.4 | 6.27e-14 | 5.4e-11 |
| MA1151.1 | RORC | MA1151.1 | 6.29e-14 | 5.42e-11 |
| MA0618.1 | LBX1 | MA0618.1 | 6.61e-14 | 5.69e-11 |
| MA0160.2 | NR4A2 | MA0160.2 | 6.67e-14 | 5.74e-11 |
| MA1479.1 | DMRTC2 | MA1479.1 | 7.02e-14 | 6.04e-11 |
| MA0601.1 | Arid3b | MA0601.1 | 7.44e-14 | 6.41e-11 |
| MA1499.1 | HOXB4 | MA1499.1 | 7.75e-14 | 6.67e-11 |
| MA0614.1 | Foxj2 | MA0614.1 | 8.34e-14 | 7.18e-11 |
| MA1505.1 | HOXC8 | MA1505.1 | 8.7e-14 | 7.49e-11 |
| MA0701.2 | LHX9 | MA0701.2 | 8.72e-14 | 7.51e-11 |
| MA1560.1 | SOHLH2 | MA1560.1 | 1.01e-13 | 8.73e-11 |
| MA0071.1 | RORA | MA0071.1 | 1.06e-13 | 9.11e-11 |
| MA0058.3 | MAX | MA0058.3 | 1.18e-13 | 1.01e-10 |
| MA0708.2 | MSX2 | MA0708.2 | 1.22e-13 | 1.05e-10 |
| MA1517.1 | KLF6 | MA1517.1 | 1.37e-13 | 1.18e-10 |
| MA0649.1 | HEY2 | MA0649.1 | 1.48e-13 | 1.27e-10 |
| MA0065.2 | Pparg::Rxra | MA0065.2 | 1.71e-13 | 1.47e-10 |
| MA1710.1 | ZNF257 | MA1710.1 | 1.76e-13 | 1.52e-10 |
| MA0151.1 | Arid3a | MA0151.1 | 2.33e-13 | 2.01e-10 |
| MA1502.1 | HOXB8 | MA1502.1 | 3.05e-13 | 2.62e-10 |
| MA0093.3 | USF1 | MA0093.3 | 3.34e-13 | 2.88e-10 |
| MA0876.1 | BSX | MA0876.1 | 4.6e-13 | 3.96e-10 |
| MA0805.1 | TBX1 | MA0805.1 | 6.3e-13 | 5.42e-10 |
| MA0889.1 | GBX1 | MA0889.1 | 6.86e-13 | 5.9e-10 |
| MA1720.1 | ZNF85 | MA1720.1 | 1.03e-12 | 8.9e-10 |
| MA0707.2 | MNX1 | MA0707.2 | 1.06e-12 | 9.09e-10 |
| MA1642.1 | NEUROG2 | MA1642.1 | 1.22e-12 | 1.05e-09 |
| MA1977.1 (ZNF324) | MEME-3 | MHTBCCWGCMTBCYH | 1.25e-21 | 1.1e-09 |
| MA0881.1 | Dlx4 | MA0881.1 | 1.28e-12 | 1.11e-09 |
| MA1573.2 | Thap11 | MA1573.2 | 1.68e-12 | 1.45e-09 |
| MA0910.2 | HOXD8 | MA0910.2 | 1.9e-12 | 1.64e-09 |
| MA0865.2 | E2F8 | MA0865.2 | 2.08e-12 | 1.79e-09 |
| MA1579.1 | ZBTB26 | MA1579.1 | 2.23e-12 | 1.92e-09 |
| MA0900.2 | HOXA2 | MA0900.2 | 2.36e-12 | 2.03e-09 |
| MA1963.1 | SATB1 | MA1963.1 | 2.37e-12 | 2.04e-09 |
| MA0690.2 | TBX21 | MA0690.2 | 2.74e-12 | 2.36e-09 |
| MA0125.1 | Nobox | MA0125.1 | 2.91e-12 | 2.51e-09 |
| MA0692.1 | TFEB | MA0692.1 | 4.08e-12 | 3.51e-09 |
| MA1115.1 | POU5F1 | MA1115.1 | 4.48e-12 | 3.86e-09 |
| MA1583.1 | ZFP57 | MA1583.1 | 5.18e-12 | 4.46e-09 |
| MA1476.2 | Dlx5 | MA1476.2 | 6.99e-12 | 6.02e-09 |
| MA1987.1 | ZNF701 | MA1987.1 | 7.1e-12 | 6.12e-09 |
| MA0842.2 | NRL | MA0842.2 | 7.38e-12 | 6.35e-09 |
| MA0141.3 | ESRRB | MA0141.3 | 8.17e-12 | 7.04e-09 |
| MA0610.1 | DMRT3 | MA0610.1 | 8.34e-12 | 7.18e-09 |
| MA0259.1 | ARNT::HIF1A | MA0259.1 | 8.42e-12 | 7.25e-09 |
| MA0704.1 | Lhx4 | MA0704.1 | 8.85e-12 | 7.62e-09 |
| MA1707.1 | DMRTA1 | MA1707.1 | 1.14e-11 | 9.83e-09 |
| MA1976.1 | ZNF320 | MA1976.1 | 1.14e-11 | 9.85e-09 |
| MA1104.2 | GATA6 | MA1104.2 | 1.26e-11 | 1.09e-08 |
| MA0630.1 | SHOX | MA0630.1 | 1.31e-11 | 1.13e-08 |
| MA1149.1 | RARA::RXRG | MA1149.1 | 1.35e-11 | 1.16e-08 |
| MA1511.2 | KLF10 | MA1511.2 | 1.35e-11 | 1.16e-08 |
| MA0788.1 | POU3F3 | MA0788.1 | 1.71e-11 | 1.47e-08 |
| MA1966.1 | TFAP4::ETV1 | MA1966.1 | 1.84e-11 | 1.58e-08 |
| MA1652.1 | ZKSCAN5 | MA1652.1 | 2.1e-11 | 1.81e-08 |
| MA1972.1 | ZFP14 | MA1972.1 | 2.19e-11 | 1.89e-08 |
| MA0511.2 | RUNX2 | MA0511.2 | 2.37e-11 | 2.04e-08 |
| MA0132.2 | PDX1 | MA0132.2 | 2.38e-11 | 2.05e-08 |
| MA1523.1 | MSANTD3 | MA1523.1 | 2.48e-11 | 2.14e-08 |
| MA0627.2 | POU2F3 | MA0627.2 | 2.6e-11 | 2.24e-08 |
| MA0670.1 (NFIA) | STREME-1 | 1-RTGCCA | 4.29e-106 | 2.52e-08 |
| MA0902.2 | HOXB2 | MA0902.2 | 2.94e-11 | 2.53e-08 |
| MA0786.1 | POU3F1 | MA0786.1 | 3.3e-11 | 2.84e-08 |
| MA0722.1 | VAX1 | MA0722.1 | 3.45e-11 | 2.97e-08 |
| MA0675.1 | NKX6-2 | MA0675.1 | 3.85e-11 | 3.31e-08 |
| MA0634.1 | ALX3 | MA0634.1 | 4.15e-11 | 3.57e-08 |
| MA0700.2 | LHX2 | MA0700.2 | 4.4e-11 | 3.79e-08 |
| MA1535.1 | NR2C1 | MA1535.1 | 5.1e-11 | 4.39e-08 |
| MA0631.1 | Six3 | MA0631.1 | 5.63e-11 | 4.84e-08 |
| MA1941.1 | ETV2::FIGLA | MA1941.1 | 6.07e-11 | 5.22e-08 |
| MA0612.2 | EMX1 | MA0612.2 | 6.39e-11 | 5.5e-08 |
| MA1507.1 | HOXD4 | MA1507.1 | 7.32e-11 | 6.3e-08 |
| MA0035.4 | GATA1 | MA0035.4 | 7.38e-11 | 6.36e-08 |
| MA1463.1 | ARGFX | MA1463.1 | 7.57e-11 | 6.52e-08 |
| MA1528.1 | NFIX | MA1528.1 | 7.68e-11 | 6.61e-08 |
| MA0689.1 | TBX20 | MA0689.1 | 8.13e-11 | 7e-08 |
| MA1604.1 | Ebf2 | MA1604.1 | 8.7e-11 | 7.49e-08 |
| MA0726.1 | VSX2 | MA0726.1 | 8.95e-11 | 7.71e-08 |
| MA0743.2 | SCRT1 | MA0743.2 | 9.1e-11 | 7.84e-08 |
| MA0899.1 | HOXA10 | MA0899.1 | 1.23e-10 | 1.06e-07 |
| MA0592.3 | ESRRA | MA0592.3 | 1.33e-10 | 1.15e-07 |
| MA1632.1 | ATF2 | MA1632.1 | 1.42e-10 | 1.22e-07 |
| MA0753.2 | ZNF740 | MA0753.2 | 1.46e-10 | 1.25e-07 |
| MA1984.1 | ZNF667 | MA1984.1 | 1.94e-10 | 1.67e-07 |
| MA0720.1 | Shox2 | MA0720.1 | 2.44e-10 | 2.1e-07 |
| MA0832.1 | Tcf21 | MA0832.1 | 2.46e-10 | 2.12e-07 |
| MA1721.1 | ZNF93 | MA1721.1 | 2.81e-10 | 2.42e-07 |
| MA1519.1 | LHX5 | MA1519.1 | 3.44e-10 | 2.96e-07 |
| MA0084.1 | SRY | MA0084.1 | 3.47e-10 | 2.99e-07 |
| MA1651.1 | ZFP42 | MA1651.1 | 3.5e-10 | 3.01e-07 |
| MA0479.1 | FOXH1 | MA0479.1 | 3.5e-10 | 3.01e-07 |
| MA0650.3 | Hoxa13 | MA0650.3 | 4.36e-10 | 3.75e-07 |
| MA1973.1 | ZKSCAN3 | MA1973.1 | 4.85e-10 | 4.18e-07 |
| MA1512.1 | KLF11 | MA1512.1 | 5.46e-10 | 4.7e-07 |
| MA0885.2 | Dlx2 | MA0885.2 | 5.47e-10 | 4.71e-07 |
| MA1975.1 | ZNF214 | MA1975.1 | 5.5e-10 | 4.74e-07 |
| MA1649.1 | ZBTB12 | MA1649.1 | 5.75e-10 | 4.95e-07 |
| MA1646.1 | OSR2 | MA1646.1 | 6.84e-10 | 5.89e-07 |
| MA1504.1 | HOXC4 | MA1504.1 | 7.11e-10 | 6.12e-07 |
| MA0695.1 | ZBTB7C | MA0695.1 | 7.6e-10 | 6.54e-07 |
| MA1540.2 | NR5A1 | MA1540.2 | 9.07e-10 | 7.81e-07 |
| MA0027.2 | EN1 | MA0027.2 | 9.36e-10 | 8.06e-07 |
| MA0075.3 | PRRX2 | MA0075.3 | 1.04e-09 | 8.95e-07 |
| MA1480.1 | DPRX | MA1480.1 | 1.06e-09 | 9.14e-07 |
| MA1562.1 | SOX14 | MA1562.1 | 1.07e-09 | 9.23e-07 |
| MA1978.1 | ZNF354A | MA1978.1 | 1.14e-09 | 9.85e-07 |
| MA0909.3 | Hoxd13 | MA0909.3 | 1.21e-09 | 1.05e-06 |
| MA0079.5 | SP1 | MA0079.5 | 1.39e-09 | 1.2e-06 |
| MA0031.1 | FOXD1 | MA0031.1 | 1.49e-09 | 1.28e-06 |
| MA0787.1 | POU3F2 | MA0787.1 | 1.49e-09 | 1.28e-06 |
| MA0783.1 (PKNOX2) | STREME-2 | 2-CCTGTCA | 7.9e-136 | 1.43e-06 |
| MA0117.2 | Mafb | MA0117.2 | 2.03e-09 | 1.75e-06 |
| MA0740.2 | KLF14 | MA0740.2 | 2.19e-09 | 1.88e-06 |
| MA0505.2 | Nr5A2 | MA0505.2 | 2.28e-09 | 1.96e-06 |
| MA0901.2 | HOXB13 | MA0901.2 | 2.74e-09 | 2.36e-06 |
| MA1487.2 | FOXE1 | MA1487.2 | 2.77e-09 | 2.38e-06 |
| MA0143.4 | SOX2 | MA0143.4 | 3.63e-09 | 3.12e-06 |
| MA0712.2 | OTX2 | MA0712.2 | 3.72e-09 | 3.2e-06 |
| MA0716.1 | PRRX1 | MA0716.1 | 3.72e-09 | 3.2e-06 |
| MA0897.1 | Hmx2 | MA0897.1 | 3.84e-09 | 3.31e-06 |
| MA0890.1 | GBX2 | MA0890.1 | 3.94e-09 | 3.39e-06 |
| MA0483.1 | Gfi1B | MA0483.1 | 4.02e-09 | 3.47e-06 |
| MA0719.1 | RHOXF1 | MA0719.1 | 4.46e-09 | 3.84e-06 |
| MA0713.1 | PHOX2A | MA0713.1 | 4.69e-09 | 4.04e-06 |
| MA0725.1 | VSX1 | MA0725.1 | 4.81e-09 | 4.14e-06 |
| MA1111.1 | NR2F2 | MA1111.1 | 5.25e-09 | 4.52e-06 |
| MA0602.1 | Arid5a | MA0602.1 | 5.52e-09 | 4.75e-06 |
| MA0014.3 | PAX5 | MA0014.3 | 5.52e-09 | 4.75e-06 |
| MA1559.1 | SNAI3 | MA1559.1 | 5.88e-09 | 5.06e-06 |
| MA0886.1 | EMX2 | MA0886.1 | 5.94e-09 | 5.11e-06 |
| MA1467.2 | Atoh1 | MA1467.2 | 6.04e-09 | 5.2e-06 |
| MA0705.1 | Lhx8 | MA0705.1 | 6.13e-09 | 5.28e-06 |
| MA0660.1 | MEF2B | MA0660.1 | 6.29e-09 | 5.41e-06 |
| MA0880.1 | Dlx3 | MA0880.1 | 6.36e-09 | 5.48e-06 |
| MA0898.1 | Hmx3 | MA0898.1 | 6.42e-09 | 5.53e-06 |
| MA1548.1 | PLAGL2 | MA1548.1 | 6.57e-09 | 5.65e-06 |
| MA0845.1 | FOXB1 | MA0845.1 | 6.6e-09 | 5.68e-06 |
| MA0077.1 | SOX9 | MA0077.1 | 7.4e-09 | 6.37e-06 |
| MA0078.2 | Sox17 | MA0078.2 | 7.58e-09 | 6.53e-06 |
| MA1574.1 | THRB | MA1574.1 | 7.76e-09 | 6.68e-06 |
| MA0790.1 | POU4F1 | MA0790.1 | 7.94e-09 | 6.84e-06 |
| MA0025.2 | NFIL3 | MA0025.2 | 7.96e-09 | 6.86e-06 |
| MA1119.1 | SIX2 | MA1119.1 | 8.09e-09 | 6.97e-06 |
| MA1518.2 | Lhx1 | MA1518.2 | 8.13e-09 | 7e-06 |
| MA0744.2 | SCRT2 | MA0744.2 | 8.16e-09 | 7.03e-06 |
| MA1114.1 | PBX3 | MA1114.1 | 8.33e-09 | 7.17e-06 |
| MA0801.1 | MGA | MA0801.1 | 8.4e-09 | 7.23e-06 |
| MA1991.1 | Hnf1A | MA1991.1 | 1.09e-08 | 9.42e-06 |
| MA0710.1 | NOTO | MA0710.1 | 1.13e-08 | 9.77e-06 |
| MA1561.1 | SOX12 | MA1561.1 | 1.25e-08 | 1.07e-05 |
| MA0492.1 | JUND | MA0492.1 | 1.4e-08 | 1.21e-05 |
| MA0111.1 | Spz1 | MA0111.1 | 1.46e-08 | 1.26e-05 |
| MA1601.2 | ZNF75D | MA1601.2 | 1.46e-08 | 1.26e-05 |
| MA0723.2 | VAX2 | MA0723.2 | 1.49e-08 | 1.28e-05 |
| MA1713.1 | ZNF610 | MA1713.1 | 1.52e-08 | 1.31e-05 |
| MA0037.4 | Gata3 | MA0037.4 | 1.95e-08 | 1.68e-05 |
| MA1536.1 | NR2C2 | MA1536.1 | 2.01e-08 | 1.73e-05 |
| MA0162.4 | EGR1 | MA0162.4 | 2.04e-08 | 1.75e-05 |
| MA0718.1 | RAX | MA0718.1 | 2.05e-08 | 1.77e-05 |
| MA1580.1 | ZBTB32 | MA1580.1 | 2.12e-08 | 1.83e-05 |
| MA1603.1 | Dmrt1 | MA1603.1 | 2.37e-08 | 2.04e-05 |
| MA0785.1 | POU2F1 | MA0785.1 | 2.67e-08 | 2.3e-05 |
| MA1989.1 | Bcl11B | MA1989.1 | 3.02e-08 | 2.6e-05 |
| MA0750.2 | ZBTB7A | MA0750.2 | 3.1e-08 | 2.67e-05 |
| MA0599.1 | KLF5 | MA0599.1 | 3.12e-08 | 2.69e-05 |
| MA0662.1 | MIXL1 | MA0662.1 | 3.76e-08 | 3.24e-05 |
| MA0108.2 | TBP | MA0108.2 | 3.99e-08 | 3.43e-05 |
| MA1148.1 | PPARA::RXRA | MA1148.1 | 4.48e-08 | 3.86e-05 |
| MA0442.2 | SOX10 | MA0442.2 | 4.96e-08 | 4.27e-05 |
| MA0494.1 | Nr1h3::Rxra | MA0494.1 | 5.61e-08 | 4.83e-05 |
| MA0748.2 | YY2 | MA0748.2 | 6.79e-08 | 5.84e-05 |
| MA1996.1 | Nr1H2 | MA1996.1 | 7.2e-08 | 6.2e-05 |
| MA1998.1 | Prdm14 | MA1998.1 | 7.23e-08 | 6.22e-05 |
| MA0857.1 | Rarb | MA0857.1 | 7.28e-08 | 6.27e-05 |
| MA0609.2 | CREM | MA0609.2 | 7.3e-08 | 6.28e-05 |
| MA0756.2 | ONECUT2 | MA0756.2 | 7.49e-08 | 6.45e-05 |
| MA1481.1 | DRGX | MA1481.1 | 7.57e-08 | 6.52e-05 |
| MA0087.2 | Sox5 | MA0087.2 | 7.86e-08 | 6.77e-05 |
| MA0504.1 | NR2C2 | MA0504.1 | 9.06e-08 | 7.8e-05 |
| MA0791.1 | POU4F3 | MA0791.1 | 9.17e-08 | 7.89e-05 |
| MA0882.1 | DLX6 | MA0882.1 | 9.4e-08 | 8.09e-05 |
| MA1531.1 | NR1D1 | MA1531.1 | 1.06e-07 | 9.16e-05 |
| MA0528.2 | ZNF263 | MA0528.2 | 1.19e-07 | 0.000102 |
| MA0860.1 | Rarg | MA0860.1 | 1.34e-07 | 0.000115 |
| MA0642.2 | EN2 | MA0642.2 | 1.4e-07 | 0.00012 |
| MA1624.1 | Stat5a | MA1624.1 | 1.61e-07 | 0.000138 |
| MA0104.4 | MYCN | MA0104.4 | 1.62e-07 | 0.00014 |
| MA1501.1 | HOXB7 | MA1501.1 | 1.84e-07 | 0.000159 |
| MA1715.1 | ZNF707 | MA1715.1 | 1.85e-07 | 0.000159 |
| MA1683.1 | FOXA3 | MA1683.1 | 1.86e-07 | 0.00016 |
| MA1970.1 | TRPS1 | MA1970.1 | 1.86e-07 | 0.000161 |
| MA0833.2 | ATF4 | MA0833.2 | 1.91e-07 | 0.000164 |
| MA0516.3 | SP2 | MA0516.3 | 1.97e-07 | 0.00017 |
| MA0488.1 | JUN | MA0488.1 | 1.98e-07 | 0.00017 |
| MA1474.1 | CREB3L4 | MA1474.1 | 2e-07 | 0.000172 |
| MA0843.1 | TEF | MA0843.1 | 2.07e-07 | 0.000178 |
| MA0714.1 | PITX3 | MA0714.1 | 2.07e-07 | 0.000178 |
| MA1968.1 | TFCP2 | MA1968.1 | 2.07e-07 | 0.000178 |
| MA0643.1 | Esrrg | MA0643.1 | 2.15e-07 | 0.000185 |
| MA0102.4 | CEBPA | MA0102.4 | 2.29e-07 | 0.000197 |
| MA1568.1 | TCF21 | MA1568.1 | 2.46e-07 | 0.000212 |
| MA1516.1 | KLF3 | MA1516.1 | 2.97e-07 | 0.000255 |
| MA0661.1 | MEOX1 | MA0661.1 | 3.27e-07 | 0.000281 |
| MA1727.1 | ZNF417 | MA1727.1 | 3.48e-07 | 3e-04 |
| MA0465.2 | CDX2 | MA0465.2 | 3.58e-07 | 0.000309 |
| MA1594.1 | ZNF382 | MA1594.1 | 3.8e-07 | 0.000327 |
| MA0083.3 | SRF | MA0083.3 | 3.92e-07 | 0.000337 |
| MA1541.1 | NR6A1 | MA1541.1 | 4.05e-07 | 0.000348 |
| MA0866.1 | SOX21 | MA0866.1 | 4.18e-07 | 0.00036 |
| MA0159.1 | RARA::RXRA | MA0159.1 | 4.64e-07 | 0.000399 |
| MA1557.1 | SMAD5 | MA1557.1 | 4.77e-07 | 0.000411 |
| MA0009.2 | TBXT | MA0009.2 | 4.94e-07 | 0.000425 |
| MA0514.2 | Sox3 | MA0514.2 | 4.99e-07 | 0.00043 |
| MA0905.1 | HOXC10 | MA0905.1 | 5.32e-07 | 0.000458 |
| 3-ACAKAGCA | STREME-3 | 3-ACAKAGCA | 4.72e-56 | 0.000462 |
| MA1929.1 | CTCF | MA1929.1 | 5.47e-07 | 0.000471 |
| MA1960.1 | MGA::EVX1 | MA1960.1 | 5.54e-07 | 0.000477 |
| MA0846.1 | FOXC2 | MA0846.1 | 6.72e-07 | 0.000578 |
| MA0002.2 | Runx1 | MA0002.2 | 7.49e-07 | 0.000645 |
| 4-CACACAG | STREME-4 | 4-CACACAG | 3.15e-70 | 0.000855 |
| MA1421.1 | TCF7L1 | MA1421.1 | 1.06e-06 | 0.000915 |
| MA0717.1 | RAX2 | MA0717.1 | 1.14e-06 | 0.000984 |
| MA0868.2 | SOX8 | MA0868.2 | 1.19e-06 | 0.00102 |
| MA0645.1 | ETV6 | MA0645.1 | 1.22e-06 | 0.00105 |
| MA1992.1 | Ikzf3 | MA1992.1 | 1.23e-06 | 0.00106 |
| MA0851.1 | Foxj3 | MA0851.1 | 1.32e-06 | 0.00114 |
| MA1152.1 | SOX15 | MA1152.1 | 1.36e-06 | 0.00117 |
| MA0043.3 | HLF | MA0043.3 | 1.4e-06 | 0.0012 |
| MA0848.1 | FOXO4 | MA0848.1 | 1.41e-06 | 0.00121 |
| MA1964.1 | SMAD2 | MA1964.1 | 1.5e-06 | 0.0013 |
| MA1497.1 | HOXA6 | MA1497.1 | 1.76e-06 | 0.00152 |
| MA0721.1 | UNCX | MA0721.1 | 1.85e-06 | 0.00159 |
| MA0090.3 (TEAD1) | STREME-5 | 5-CATTCCA | 2.35e-45 | 0.00181 |
| MA1965.1 | SP5 | MA1965.1 | 2.64e-06 | 0.00228 |
| MA0875.1 | BARX1 | MA0875.1 | 2.69e-06 | 0.00232 |
| 6-CTCTCAG | STREME-6 | 6-CTCTCAG | 1.13e-32 | 0.00237 |
| MA0595.1 | SREBF1 | MA0595.1 | 3.01e-06 | 0.00259 |
| MA0761.2 | ETV1 | MA0761.2 | 3.36e-06 | 0.00289 |
| MA0041.2 | FOXD3 | MA0041.2 | 3.93e-06 | 0.00339 |
| MA0628.1 | POU6F1 | MA0628.1 | 4.12e-06 | 0.00354 |
| MA0467.2 | Crx | MA0467.2 | 4.29e-06 | 0.00369 |
| MA1120.1 | SOX13 | MA1120.1 | 4.35e-06 | 0.00374 |
| MA1636.1 | CEBPG | MA1636.1 | 4.63e-06 | 0.00399 |
| MA0032.2 | FOXC1 | MA0032.2 | 4.68e-06 | 0.00403 |
| 7-ATGGGAG | STREME-7 | 7-ATGGGAG | 6.39e-31 | 0.00463 |
| MA0763.1 | ETV3 | MA0763.1 | 5.55e-06 | 0.00478 |
| MA1977.1 | ZNF324 | MA1977.1 | 5.67e-06 | 0.00488 |
| MA0148.4 | FOXA1 | MA0148.4 | 6.05e-06 | 0.00521 |
| MA0495.3 | MAFF | MA0495.3 | 6.09e-06 | 0.00524 |
| MA1534.1 | NR1I3 | MA1534.1 | 6.48e-06 | 0.00558 |
| MA0635.1 | BARHL2 | MA0635.1 | 6.78e-06 | 0.00584 |
| MA0743.2 (SCRT1) | STREME-8 | 8-STGTTGA | 5.54e-53 | 0.00597 |
| MA0679.2 | ONECUT1 | MA0679.2 | 7.21e-06 | 0.00621 |
| MA0620.3 | MITF | MA0620.3 | 8.56e-06 | 0.00737 |
| MA0831.3 | TFE3 | MA0831.3 | 8.75e-06 | 0.00753 |
| MA1554.1 | RFX7 | MA1554.1 | 8.83e-06 | 0.0076 |
| MA1475.1 | CREB3L4 | MA1475.1 | 9e-06 | 0.00775 |
| MA0685.2 | SP4 | MA0685.2 | 9.15e-06 | 0.00788 |
| MA1969.1 | THRA | MA1969.1 | 9.47e-06 | 0.00815 |
| MA0042.2 | FOXI1 | MA0042.2 | 9.5e-06 | 0.00818 |
| MA0680.2 | Pax7 | MA0680.2 | 9.63e-06 | 0.00829 |
| MA0847.3 | FOXD2 | MA0847.3 | 9.74e-06 | 0.00839 |
| MA0733.1 | EGR4 | MA0733.1 | 1.05e-05 | 0.00908 |
| MA0849.1 | FOXO6 | MA0849.1 | 1.07e-05 | 0.00918 |
| MA0858.1 | Rarb | MA0858.1 | 1.07e-05 | 0.0092 |
| MA0804.1 | TBX19 | MA0804.1 | 1.11e-05 | 0.00959 |
| MA0136.3 | Elf5 | MA0136.3 | 1.14e-05 | 0.00983 |
| MA0497.1 | MEF2C | MA0497.1 | 1.25e-05 | 0.0108 |
| MA0017.2 | NR2F1 | MA0017.2 | 1.29e-05 | 0.0111 |
| MA0518.1 | Stat4 | MA0518.1 | 1.3e-05 | 0.0112 |
| MA0769.2 | TCF7 | MA0769.2 | 1.31e-05 | 0.0113 |
| MA0891.1 | GSC2 | MA0891.1 | 1.44e-05 | 0.0124 |
| MA0059.1 | MAX::MYC | MA0059.1 | 1.51e-05 | 0.013 |
| MA1983.1 | ZNF582 | MA1983.1 | 1.55e-05 | 0.0133 |
| 9-CCCAGC | STREME-9 | 9-CCCAGC | 7.4e-41 | 0.0144 |
| 10-CCAGAGT | STREME-10 | 10-CCAGAGT | 2.61e-49 | 0.0146 |
| MA0682.2 | PITX1 | MA0682.2 | 1.73e-05 | 0.0149 |
| MA0888.1 | EVX2 | MA0888.1 | 1.76e-05 | 0.0151 |
| MA1606.1 | Foxf1 | MA1606.1 | 1.8e-05 | 0.0155 |
| MA0887.1 | EVX1 | MA0887.1 | 1.81e-05 | 0.0156 |
| MA0047.3 | FOXA2 | MA0047.3 | 2.04e-05 | 0.0175 |
| MA0711.1 | OTX1 | MA0711.1 | 2.05e-05 | 0.0177 |
| MA1110.2 | Nr1H4 | MA1110.2 | 2.14e-05 | 0.0184 |
| MA1549.1 | POU6F1 | MA1549.1 | 2.21e-05 | 0.0191 |
| MA0892.1 | GSX1 | MA0892.1 | 2.22e-05 | 0.0191 |
| MA1537.1 | NR2F1 | MA1537.1 | 2.27e-05 | 0.0195 |
| MA1116.1 | RBPJ | MA1116.1 | 2.37e-05 | 0.0204 |
| MA1718.1 | ZNF8 | MA1718.1 | 2.56e-05 | 0.0221 |
| MA1129.1 | FOSL1::JUN | MA1129.1 | 2.57e-05 | 0.0221 |
| MA0684.2 | RUNX3 | MA0684.2 | 2.63e-05 | 0.0227 |
| MA1640.1 | MEIS2 | MA1640.1 | 2.69e-05 | 0.0232 |
| MA0867.2 | SOX4 | MA0867.2 | 2.77e-05 | 0.0238 |
| MA1127.1 | FOSB::JUN | MA1127.1 | 3.06e-05 | 0.0263 |
| MA0699.1 | LBX2 | MA0699.1 | 3.15e-05 | 0.0271 |
| MA0629.1 | Rhox11 | MA0629.1 | 3.35e-05 | 0.0289 |
| MA0879.2 | DLX1 | MA0879.2 | 3.57e-05 | 0.0307 |
| MA0114.4 | HNF4A | MA0114.4 | 3.58e-05 | 0.0308 |
| MA0613.1 | FOXG1 | MA0613.1 | 3.85e-05 | 0.0331 |
| MA0871.2 | TFEC | MA0871.2 | 4.22e-05 | 0.0364 |
| MA1994.1 (Nkx2-1) | STREME-11 | 11-SCACTTGA | 5.27e-91 | 0.0368 |
| MA0693.3 | Vdr | MA0693.3 | 4.42e-05 | 0.0381 |
| MA0683.1 (POU4F2) | STREME-12 | 12-TATGCA | 4.59e-51 | 0.0405 |
| 13-CTGTAAG | STREME-13 | 13-CTGTAAG | 4.07e-30 | 0.0425 |
| MA1117.1 | RELB | MA1117.1 | 5.58e-05 | 0.0481 |
| MA1108.2 (MXI1) | STREME-14 | 14-ATGTGGT | 1.43e-47 | 0.0537 |
| 15-ATAGCTG | STREME-15 | 15-ATAGCTG | 1.52e-35 | 0.0634 |
| 16-TATCAG | STREME-16 | 16-TATCAG | 6.58e-66 | 0.0657 |
| MA0597.2 (THAP1) | STREME-17 | 17-CAGGGC | 2.77e-30 | 0.0865 |
### selecting just the list of names and checking for expressionAll enriched motifs found
This area was for all motifs found in Xstreme analysis using NR peaks as background. #### EAR ##### EAR_open
mrc_palette <- c(
"EAR_open" = "#F8766D",
"EAR_close" = "#f6483c",
"ESR_open" = "#7CAE00",
"ESR_close" = "#587b00",
"ESR_OC" = "#6a9500",
"LR_open" = "#00BFC4",
"LR_close" = "#008d91",
"NR" = "#C77CFF"
)
spec_dataframe <-
EAR_open_xstreme%>%
mutate(CONSENSUS=gsub('[[:digit:]]+-', '', CONSENSUS)) %>%
dplyr::filter(EVALUE<0.05) %>%
left_join(., (sea_EAR_open %>%
dplyr::rename("SEA_PVALUE"=PVALUE) %>%
dplyr::select(RANK,ID:SEA_PVALUE)),
by= c("RANK"="RANK", "ALT_ID"="ALT_ID", "CONSENSUS"="CONSENSUS", "ID"="ID", "SEA_PVALUE"="SEA_PVALUE")) %>%
separate(SIM_MOTIF, into= c("SIM_MOTIF", "NAME"), sep= " ") %>%
mutate(motif_name= gsub("[()]","",NAME), mrc="EAR_open") %>%
mutate(motif_name=
if_else(is.na(motif_name)&str_detect(SIM_MOTIF,"^M"),ALT_ID,
if_else(is.na(motif_name),ID,motif_name))) %>%
# errat_EARo <-
rbind(EAR_close_xstreme%>%
mutate(CONSENSUS=gsub('[[:digit:]]+-', '', CONSENSUS)) %>%
dplyr::filter(EVALUE<0.05) %>%
left_join(., (sea_EAR_close %>%
dplyr::rename("SEA_PVALUE"=PVALUE) %>%
dplyr::select(RANK,ID:SEA_PVALUE)),
by= c("RANK"="RANK", "ALT_ID"="ALT_ID", "CONSENSUS"="CONSENSUS", "ID"="ID", "SEA_PVALUE"="SEA_PVALUE")) %>%
separate(SIM_MOTIF, into= c("SIM_MOTIF", "NAME"), sep= " ") %>%
mutate(motif_name= gsub("[()]","",NAME), mrc="EAR_close") %>%
mutate(motif_name=
if_else(is.na(motif_name)&str_detect(SIM_MOTIF,"^M"),ALT_ID,
if_else(is.na(motif_name),ID,motif_name)))) %>%
# errat_EARo <-
rbind(ESR_open_xstreme%>%
mutate(CONSENSUS=gsub('[[:digit:]]+-', '', CONSENSUS)) %>%
dplyr::filter(EVALUE<0.05) %>%
left_join(., (sea_ESR_open %>%
dplyr::rename("SEA_PVALUE"=PVALUE) %>%
dplyr::select(RANK,ID:SEA_PVALUE)),
by= c("RANK"="RANK", "ALT_ID"="ALT_ID", "CONSENSUS"="CONSENSUS", "ID"="ID", "SEA_PVALUE"="SEA_PVALUE")) %>%
separate(SIM_MOTIF, into= c("SIM_MOTIF", "NAME"), sep= " ") %>%
mutate(motif_name= gsub("[()]","",NAME), mrc="ESR_open") %>%
mutate(motif_name=
if_else(is.na(motif_name)&str_detect(SIM_MOTIF,"^M"),ALT_ID,
if_else(is.na(motif_name),ID,motif_name)))) %>%
# errat_EARo <-
rbind(ESR_close_xstreme%>%
mutate(CONSENSUS=gsub('[[:digit:]]+-', '', CONSENSUS)) %>%
dplyr::filter(EVALUE<0.05) %>%
left_join(., (sea_ESR_close %>%
dplyr::rename("SEA_PVALUE"=PVALUE) %>%
dplyr::select(RANK,ID:SEA_PVALUE)),
by= c("RANK"="RANK", "ALT_ID"="ALT_ID", "CONSENSUS"="CONSENSUS", "ID"="ID", "SEA_PVALUE"="SEA_PVALUE")) %>%
separate(SIM_MOTIF, into= c("SIM_MOTIF", "NAME"), sep= " ") %>%
mutate(motif_name= gsub("[()]","",NAME), mrc="ESR_close") %>%
mutate(motif_name=
if_else(is.na(motif_name)&str_detect(SIM_MOTIF,"^M"),ALT_ID,
if_else(is.na(motif_name),ID,motif_name)))) %>%
# errat_EARo <-
rbind(ESR_OC_xstreme%>%
mutate(CONSENSUS=gsub('[[:digit:]]+-', '', CONSENSUS)) %>%
dplyr::filter(EVALUE<0.05) %>%
left_join(., (sea_ESR_OC %>%
dplyr::rename("SEA_PVALUE"=PVALUE) %>%
dplyr::select(RANK,ID:SEA_PVALUE)),
by= c("RANK"="RANK", "ALT_ID"="ALT_ID", "CONSENSUS"="CONSENSUS", "ID"="ID", "SEA_PVALUE"="SEA_PVALUE")) %>%
separate(SIM_MOTIF, into= c("SIM_MOTIF", "NAME"), sep= " ") %>%
mutate(motif_name= gsub("[()]","",NAME), mrc="ESR_OC") %>%
mutate(motif_name=
if_else(is.na(motif_name)&str_detect(SIM_MOTIF,"^M"),ALT_ID,
if_else(is.na(motif_name),ID,motif_name)))) %>%
###rbind break#####
rbind(LR_open_xstreme%>%
dplyr::select(SEED_MOTIF:MOTIF_URL) %>%
dplyr::select(!"SEA_PVALUE") %>%
rownames_to_column("RANK") %>%
mutate(CONSENSUS=gsub('[[:digit:]]+-', '', CONSENSUS)) %>%
dplyr::filter(EVALUE<0.05) %>%
left_join(., (sea_LR_open %>%
dplyr::rename("SEA_PVALUE"=PVALUE) %>%
dplyr::select(RANK,ID:SEA_PVALUE)),
by= c("RANK"="RANK","ALT_ID"="ALT_ID", "CONSENSUS"="CONSENSUS", "ID"="ID")) %>%
separate(SIM_MOTIF, into= c("SIM_MOTIF", "NAME"), sep= " ") %>%
mutate(motif_name= gsub("[()]","",NAME), mrc="LR_open") %>%
mutate(motif_name=
if_else(is.na(motif_name)&str_detect(SIM_MOTIF,"^M"),ALT_ID,
if_else(is.na(motif_name),ID,motif_name)))) %>%
### rbind break
rbind(LR_close_xstreme%>%
mutate(CONSENSUS=gsub('[[:digit:]]+-', '', CONSENSUS)) %>%
dplyr::filter(EVALUE<0.05) %>%
left_join(., (sea_LR_close %>%
dplyr::rename("SEA_PVALUE"=PVALUE) %>%
dplyr::select(RANK,ID:SEA_PVALUE)),
by= c("RANK"="RANK", "ALT_ID"="ALT_ID", "CONSENSUS"="CONSENSUS", "ID"="ID", "SEA_PVALUE"="SEA_PVALUE")) %>%
separate(SIM_MOTIF, into= c("SIM_MOTIF", "NAME"), sep= " ") %>%
mutate(motif_name= gsub("[()]","",NAME), mrc="LR_close") %>%
mutate(motif_name=
if_else(is.na(motif_name)&str_detect(SIM_MOTIF,"^M"),ALT_ID,
if_else(is.na(motif_name),ID,motif_name))))
###plotting
spec_dataframe %>%
ggplot(.,aes(x=mrc,y=ENR_RATIO,fill=mrc))+
geom_boxplot()+
theme_classic()+
ylab("Enrichment ratio")+
ggtitle("Enrichment ratio values across MRC")+
scale_fill_manual(values=mrc_palette)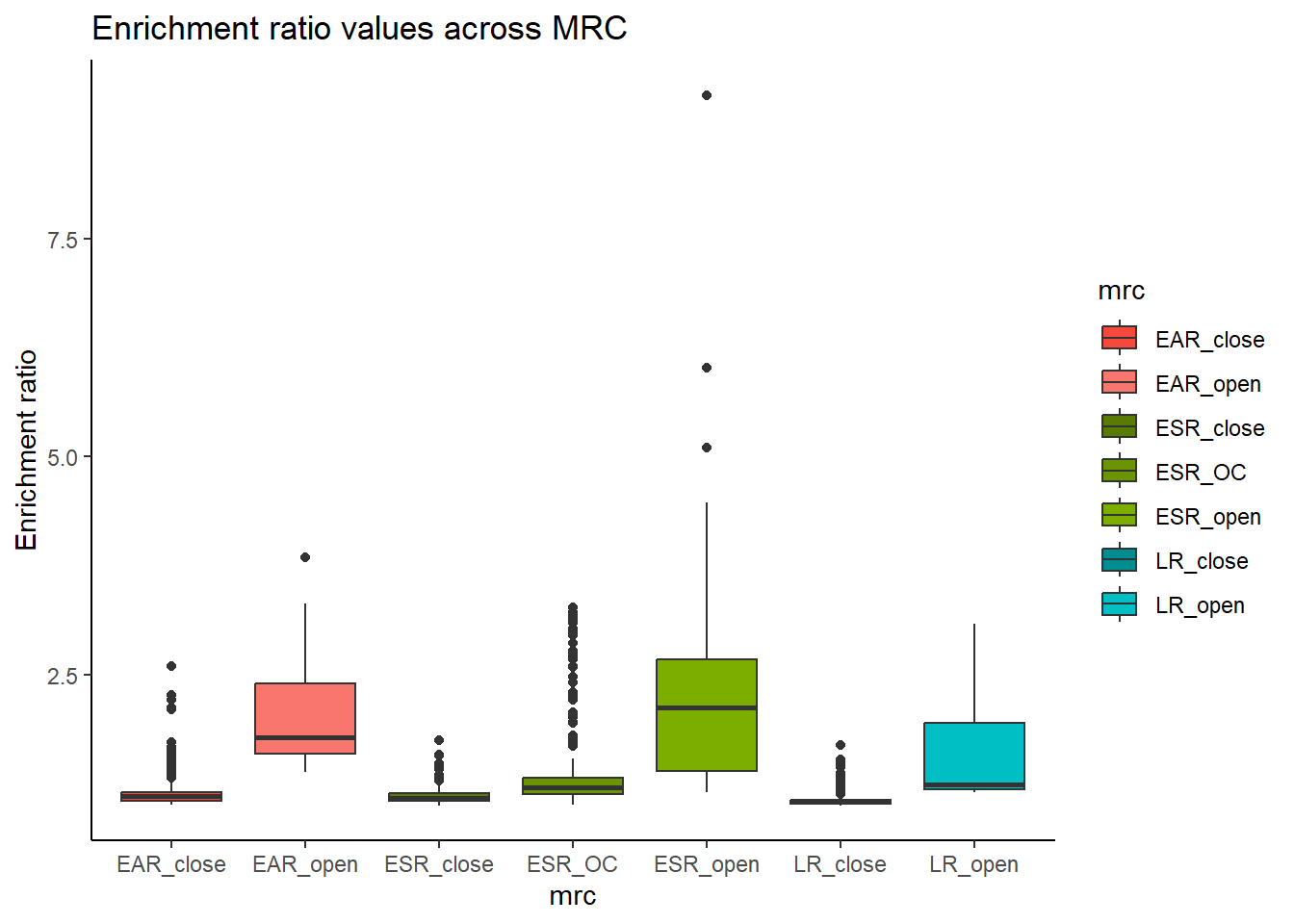
spec_dataframe %>%
ggplot(., aes(x=ENR_RATIO, fill = mrc))+
geom_density(aes(alpha=0.4)) +
theme_classic()+
xlab("Enrichment ratio")+
ggtitle("Enrichment ratio histogram MRC")+
scale_fill_manual(values=mrc_palette)+
coord_cartesian(xlim=c(1,5))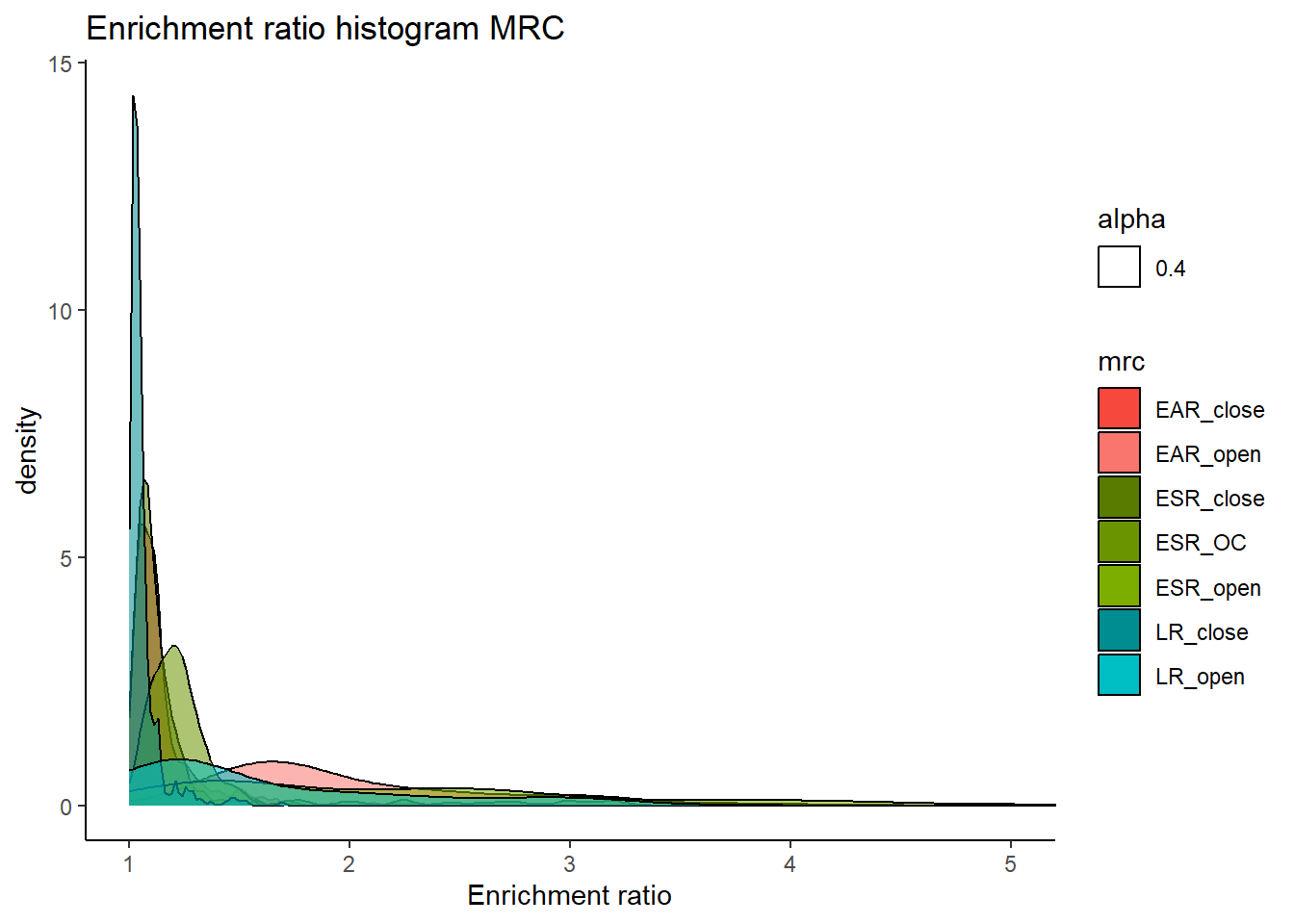
spec_dataframe %>%
# dplyr::filter(mrc=="LR_open") %>%
ggplot(., aes(x=mrc, y=-log(base=10,EVALUE), color=mrc))+
geom_jitter()+
theme_classic()+
ylab(expression("-log[10] Evalue"))+
ggtitle("Significant values across MRC motifs")+
scale_color_manual(values=mrc_palette)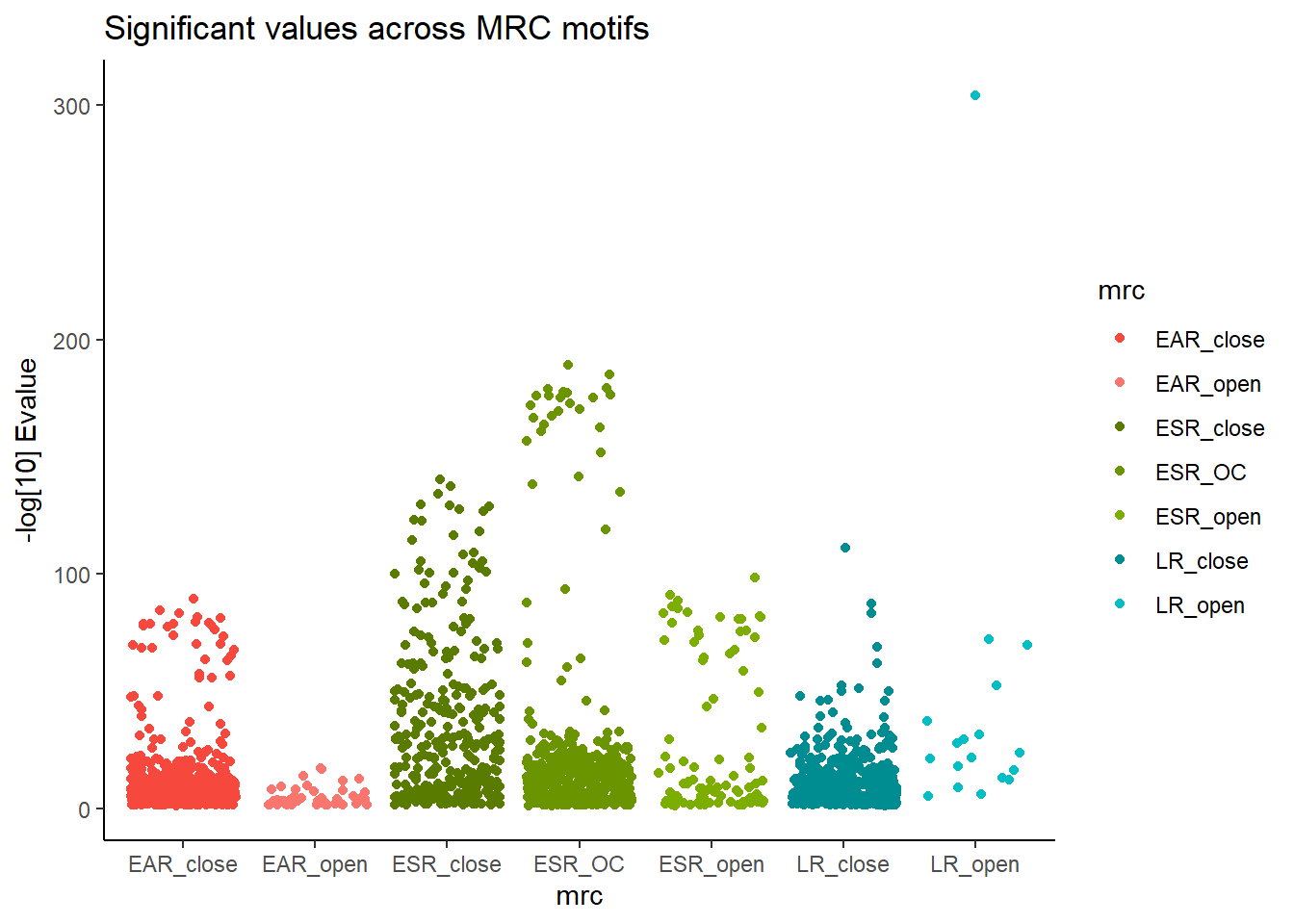
The enrichment ratio plot let me know where (sort of) to cut the plots to find the highest enrichment ratios by MRC. I am going to use 1.75 for most of them for now.
ER_rat <- 1.5
mrc_type <- "EAR_open"
spec_dataframe %>%
dplyr::filter(mrc==mrc_type) %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER, SITES,SIM_MOTIF,ALT_ID, ID,EVALUE, TP:'FP%', motif_name)%>%
arrange(.,EVALUE) %>%
mutate(log10Evalue= log(EVALUE, base = 10)*(-1)) %>%
mutate(motif_name=if_else(motif_name=="ELK3",paste(motif_name, RANK, sep="_"), if_else(motif_name=="ZNF93",paste(motif_name, RANK, sep="_"), if_else(motif_name=="Nrf1",paste(motif_name, RANK,sep="_"),motif_name))))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP%`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks Enrichment ratio:",ER_rat))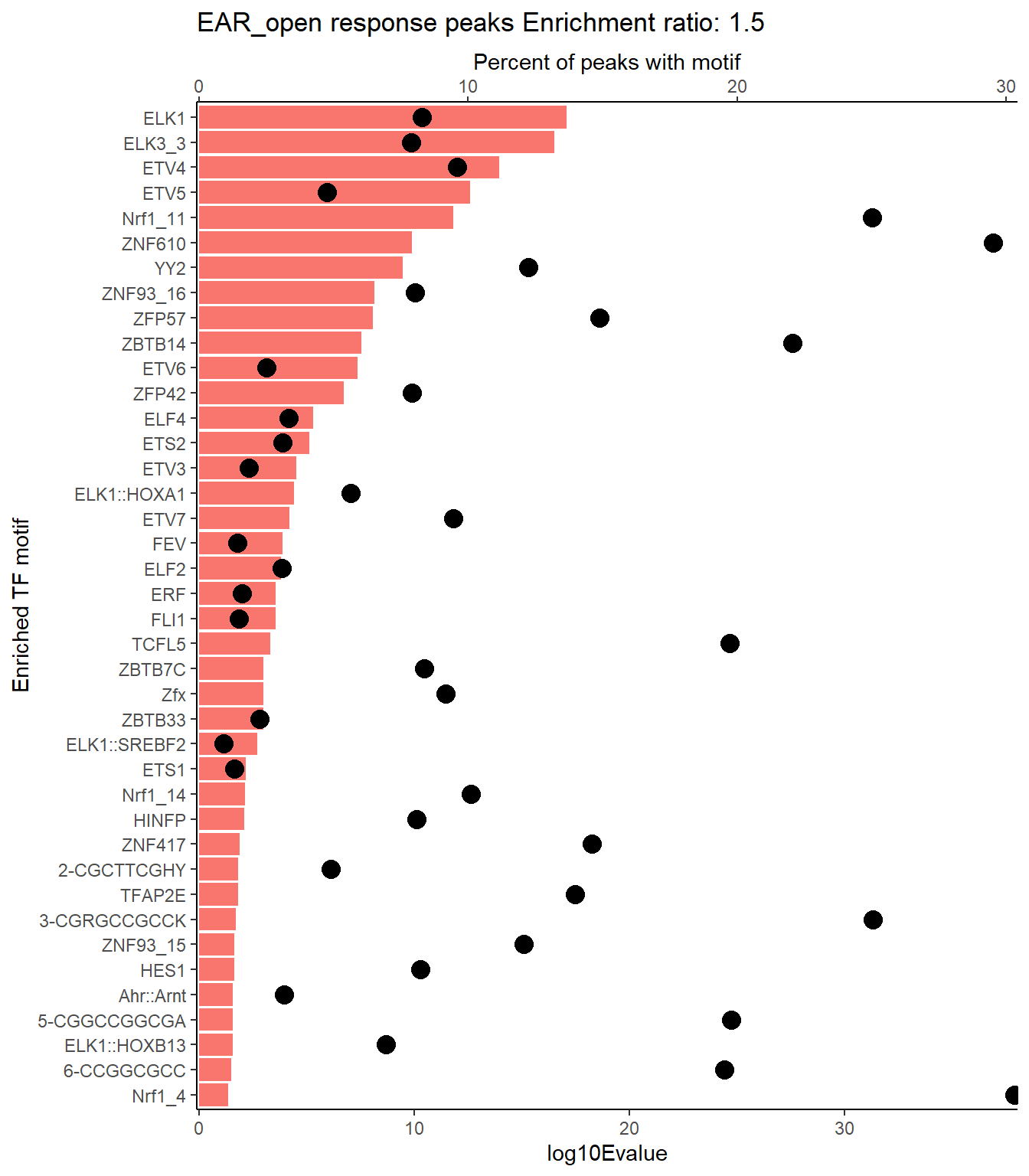
spec_dataframe %>%
dplyr::filter(mrc==mrc_type) %>%
dplyr::filter(ENR_RATIO>1.4) %>%
dplyr::select(RANK,CLUSTER, SITES,SIM_MOTIF,ALT_ID, ID,EVALUE, TP:'FP%', motif_name)%>%
arrange(.,EVALUE) %>%
mutate(log10Evalue= log(EVALUE, base = 10)*(-1)) %>%
mutate(motif_name=if_else(motif_name=="ELK3",paste(motif_name, RANK, sep="_"), if_else(motif_name=="ZNF93",paste(motif_name, RANK, sep="_"), if_else(motif_name=="Nrf1",paste(motif_name, RANK,sep="_"),motif_name))))%>%
distinct(CLUSTER,.keep_all = TRUE) %>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP%`/2.5), size =4)+
# geom_line(aes(x=`TP%`,y= motif_name, group=log10Evalue))+
scale_x_continuous(expand=c (0,.25),sec.axis = sec_axis(transform= ~.*2.5,name="Percent of peaks with motif"))+
# geom_text(aes())
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks Enrichment ratio:",ER_rat," merged clusters"))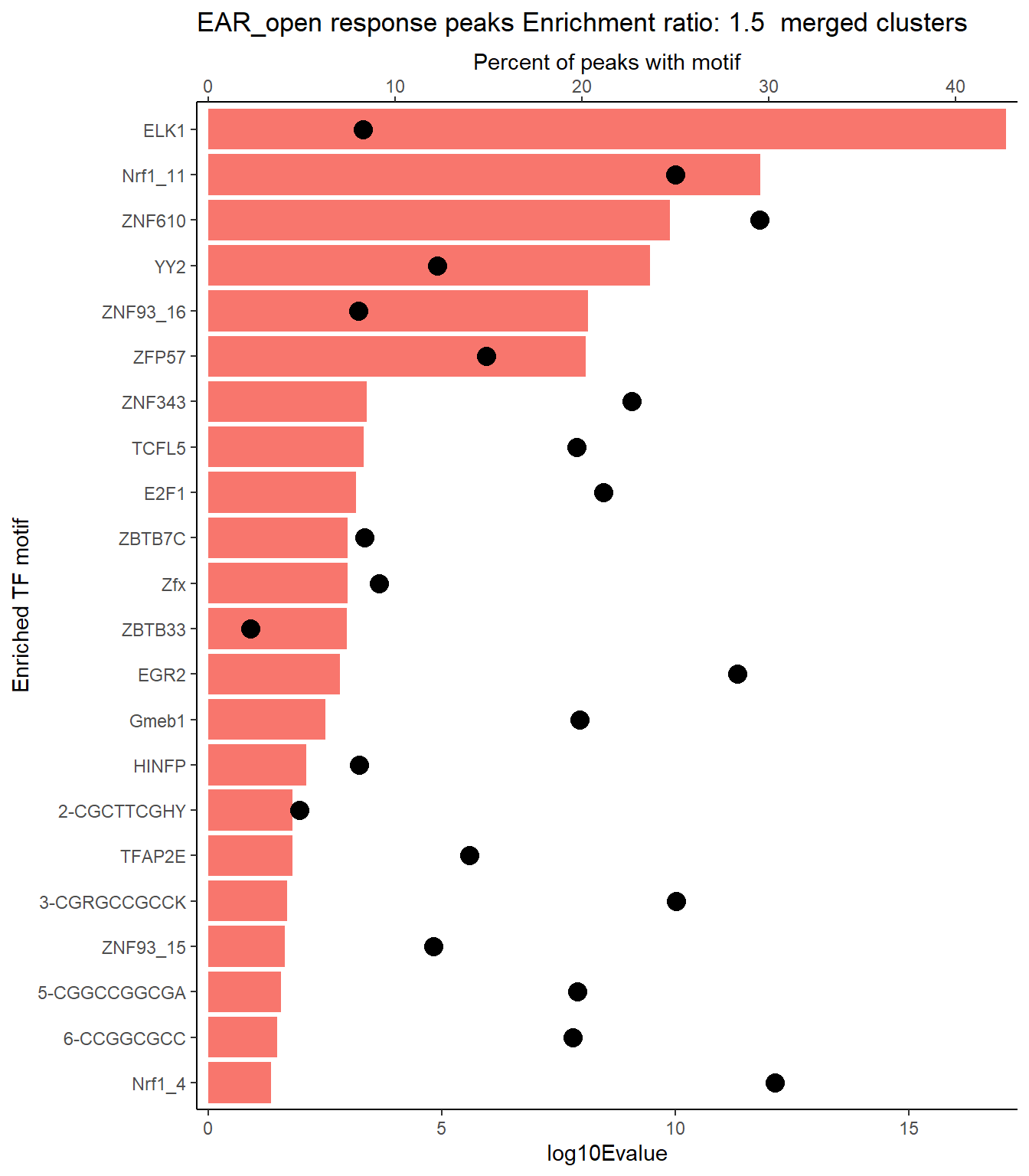
EAR_close
ER_rat <- 1.5
mrc_type <- "EAR_close"
spec_dataframe %>%
dplyr::filter(mrc==mrc_type) %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER, SITES,SIM_MOTIF,ALT_ID, ID,EVALUE, TP:'FP%', motif_name)%>%
arrange(.,EVALUE) %>%
mutate(log10Evalue= log(EVALUE, base = 10)*(-1)) %>%
mutate(motif_name=if_else(motif_name=="MEIS1",paste(motif_name, RANK, sep="_"), if_else(motif_name=="ZNF384",paste(motif_name, RANK, sep="_"), if_else(motif_name=="ZNF257",paste(motif_name, RANK,sep="_"),motif_name))))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP%`*1.25), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./1.25,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks Enrichment ratio:",ER_rat))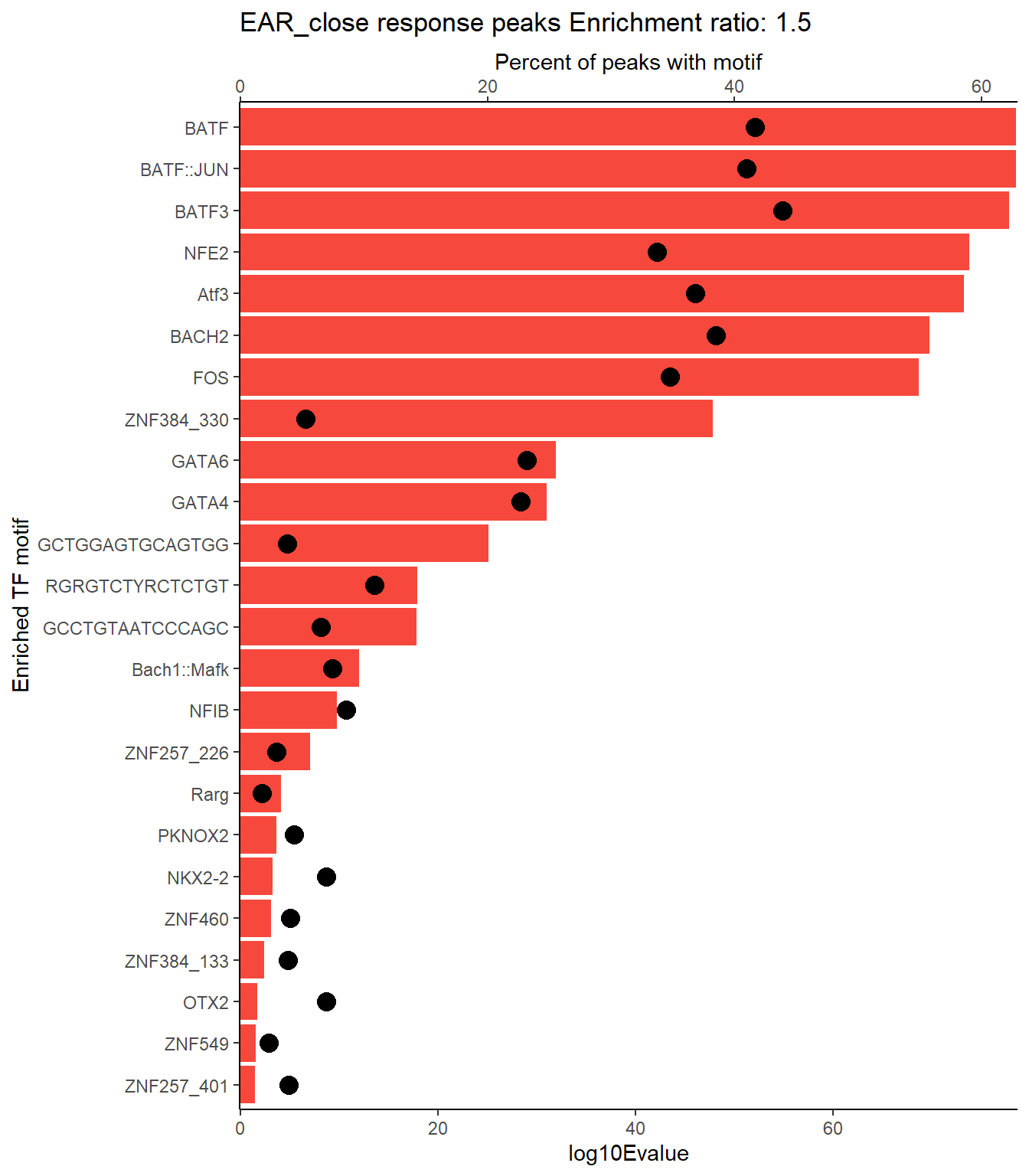
spec_dataframe %>%
dplyr::filter(mrc==mrc_type) %>%
dplyr::filter(ENR_RATIO>1.4) %>%
dplyr::select(RANK,CLUSTER, SITES,SIM_MOTIF,ALT_ID, ID,EVALUE, TP:'FP%', motif_name)%>%
arrange(.,EVALUE) %>%
mutate(log10Evalue= log(EVALUE, base = 10)*(-1)) %>%
mutate(motif_name=if_else(motif_name=="MEIS1",paste(motif_name, RANK, sep="_"), if_else(motif_name=="ZNF384",paste(motif_name, RANK, sep="_"), if_else(motif_name=="ZNF257",paste(motif_name, RANK,sep="_"),motif_name))))%>%
distinct(CLUSTER,.keep_all = TRUE) %>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP%`/2.5), size =4)+
# geom_line(aes(x=`TP%`,y= motif_name, group=log10Evalue))+
scale_x_continuous(expand=c (0,.25),sec.axis = sec_axis(transform= ~.*2.5,name="Percent of peaks with motif"))+
# geom_text(aes())
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks Enrichment ratio:",ER_rat," merged clusters"))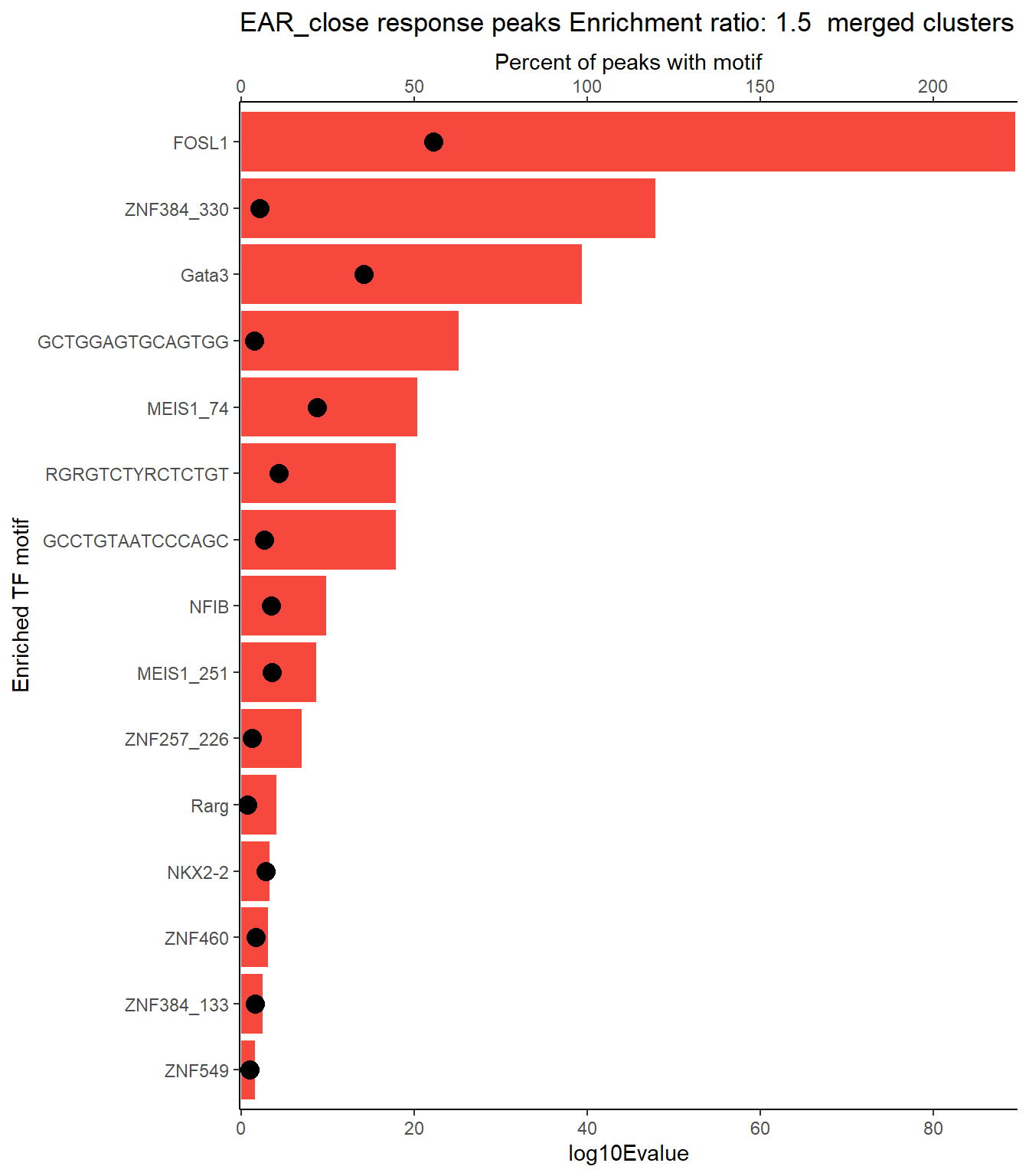
ESR
ESR_open
ER_rat <- 1.5
mrc_type <- "ESR_open"
spec_dataframe %>%
dplyr::filter(mrc=="ESR_open") %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER, SITES,SIM_MOTIF,ALT_ID, ID,EVALUE, TP:'FP%', motif_name)%>%
arrange(.,EVALUE) %>%
mutate(log10Evalue= log(EVALUE, base = 10)*(-1)) %>%
mutate(motif_name=if_else(motif_name=="KLF9",paste(motif_name, RANK, sep="_"), if_else(motif_name=="^ZN*",paste(motif_name, RANK, sep="_"), if_else(motif_name=="ZSCAN4",paste(motif_name, RANK,sep="_"),motif_name))))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette["ESR_open"]) +
geom_point(aes(x=`TP%`*4), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./4,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks Enrichment ratio:",ER_rat))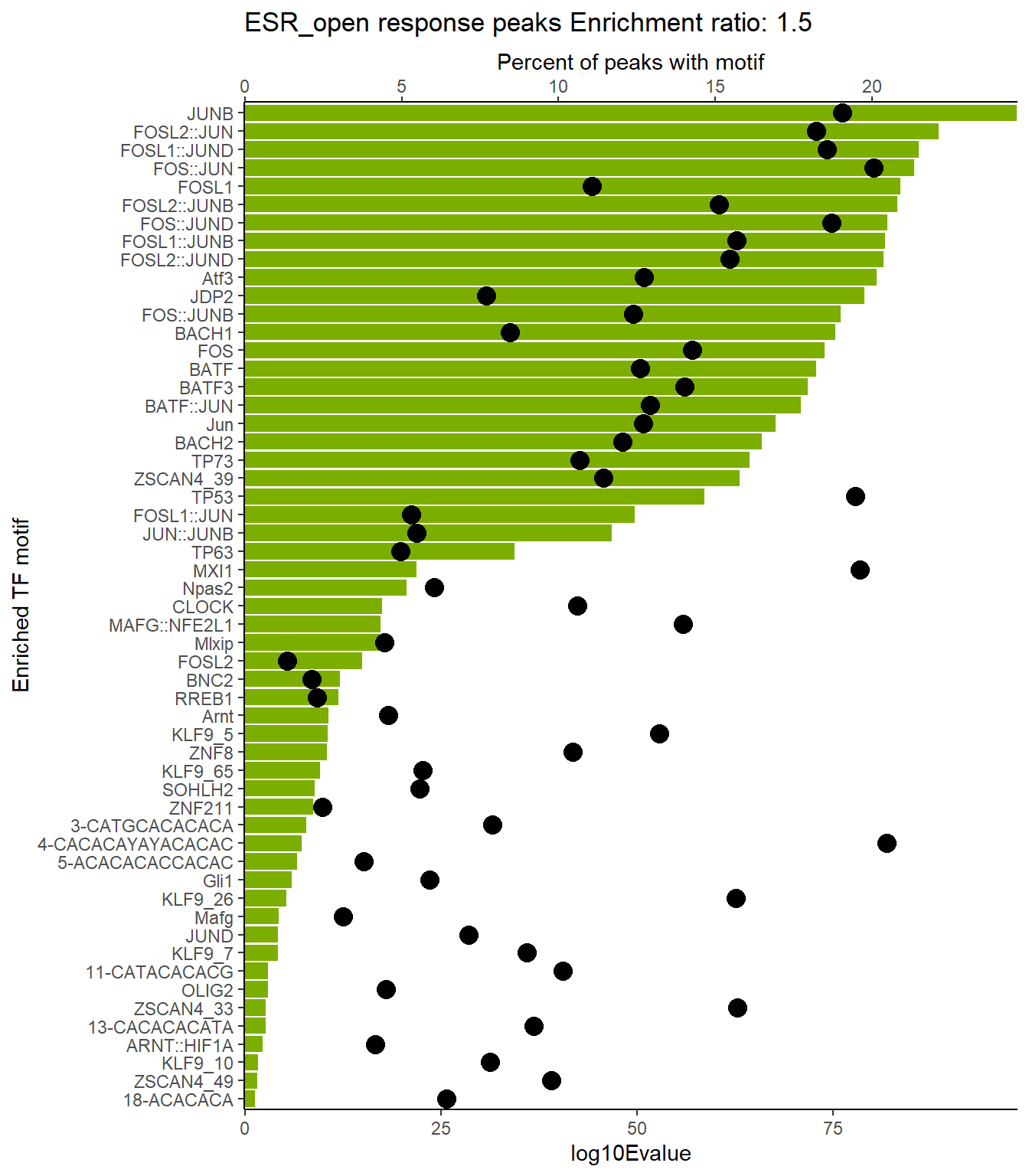
spec_dataframe %>%
dplyr::filter(mrc=="ESR_open") %>%
dplyr::filter(ENR_RATIO>1.4) %>%
dplyr::select(RANK,CLUSTER, SITES,SIM_MOTIF,ALT_ID, ID,EVALUE, TP:'FP%', motif_name)%>%
arrange(.,EVALUE) %>%
mutate(log10Evalue= log(EVALUE, base = 10)*(-1)) %>%
mutate(motif_name=if_else(motif_name=="KLF9",paste(motif_name, RANK, sep="_"), if_else(motif_name=="^ZN*",paste(motif_name, RANK, sep="_"), if_else(motif_name=="ZSCAN4",paste(motif_name, RANK,sep="_"),motif_name))))%>%
distinct(CLUSTER,.keep_all = TRUE) %>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette["ESR_open"]) +
geom_point(aes(x=`TP%`*4), size =4)+
# geom_line(aes(x=`TP%`,y= motif_name, group=log10Evalue))+
scale_x_continuous(expand=c (0,.25),sec.axis = sec_axis(transform= ~./4,name="Percent of peaks with motif"))+
# geom_text(aes())
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks Enrichment ratio:",ER_rat," merged motif clusters"))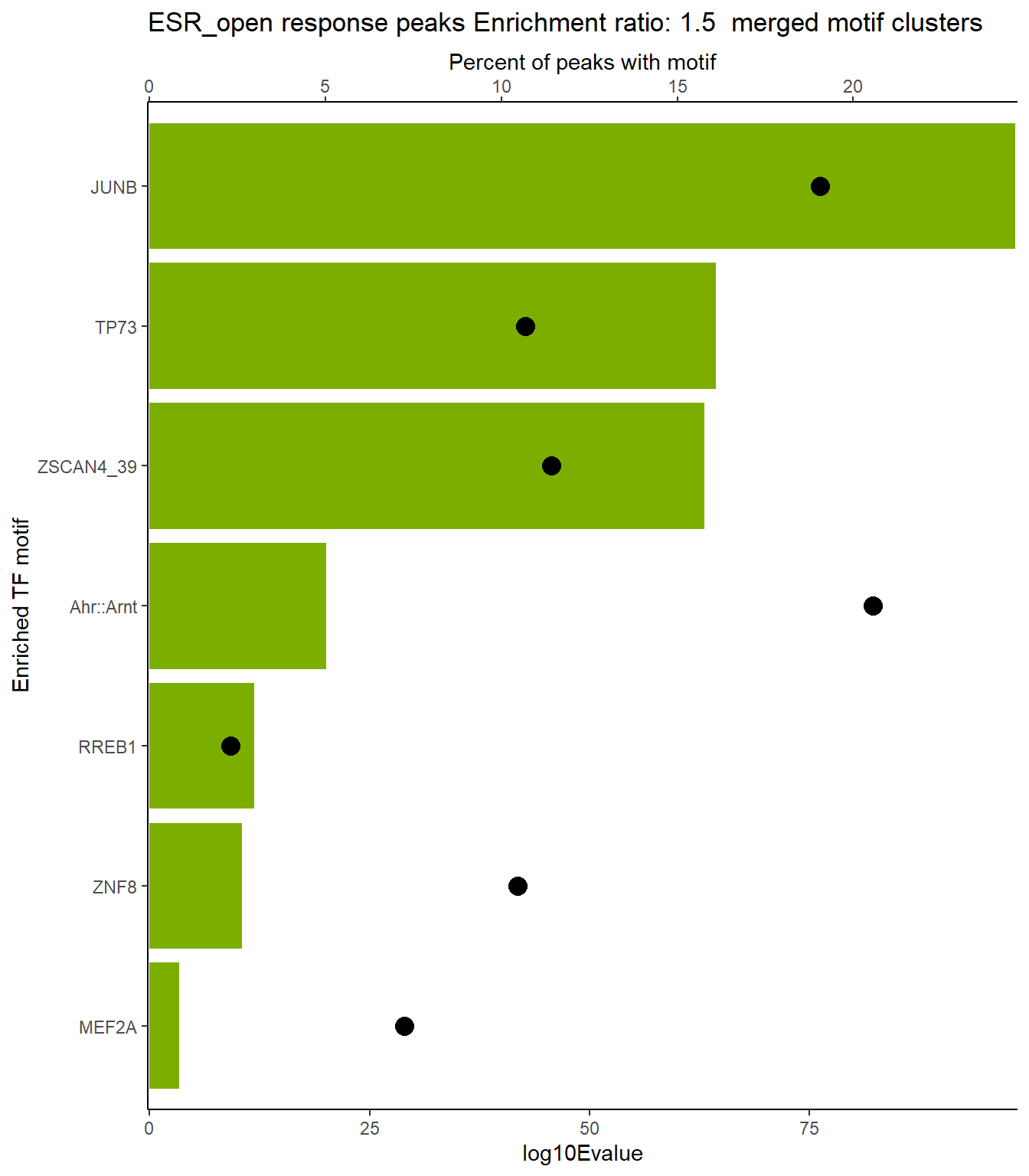
ESR_close
ER_rat <- 1.25
mrc_type <- "ESR_close"
spec_dataframe %>%
dplyr::filter(mrc==mrc_type) %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER, SITES,SIM_MOTIF,ALT_ID, ID,EVALUE, TP:'FP%', motif_name)%>%
arrange(.,EVALUE) %>%
mutate(log10Evalue= log(EVALUE, base = 10)*(-1)) %>%
mutate(motif_name=if_else(motif_name=="KLF9",paste(motif_name, RANK, sep="_"), if_else(motif_name=="^ZN*",paste(motif_name, RANK, sep="_"), if_else(motif_name=="ZSCAN4",paste(motif_name, RANK,sep="_"),motif_name))))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP%`*4), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./4,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks Enrichment ratio:",ER_rat))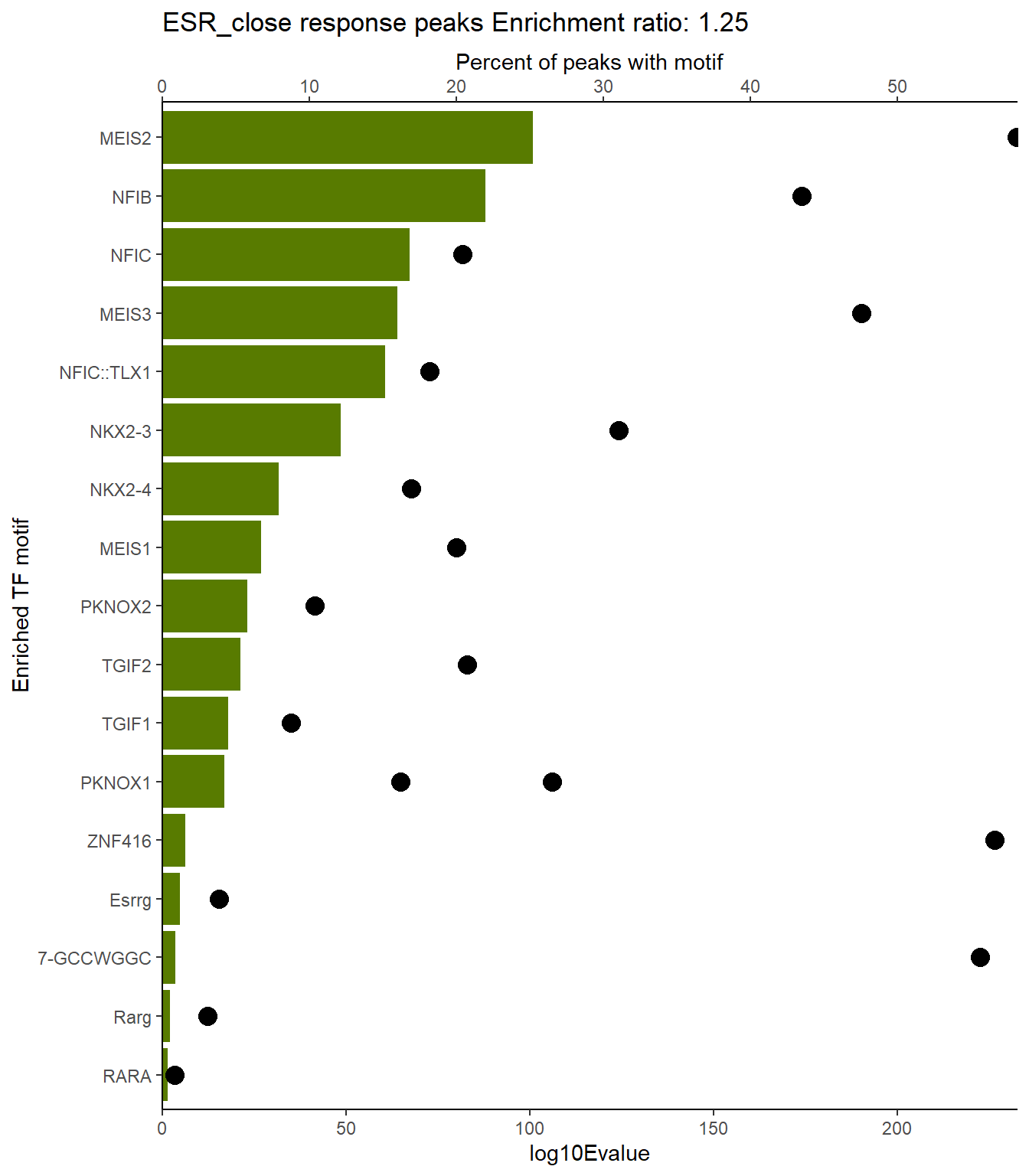
spec_dataframe %>%
dplyr::filter(mrc==mrc_type) %>%
dplyr::filter(ENR_RATIO>1.4) %>%
dplyr::select(RANK,CLUSTER, SITES,SIM_MOTIF,ALT_ID, ID,EVALUE, TP:'FP%', motif_name)%>%
arrange(.,EVALUE) %>%
mutate(log10Evalue= log(EVALUE, base = 10)*(-1)) %>%
mutate(motif_name=if_else(motif_name=="KLF9",paste(motif_name, RANK, sep="_"), if_else(motif_name=="^ZN*",paste(motif_name, RANK, sep="_"), if_else(motif_name=="ZSCAN4",paste(motif_name, RANK,sep="_"),motif_name))))%>%
distinct(CLUSTER,.keep_all = TRUE) %>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP%`*4), size =4)+
# geom_line(aes(x=`TP%`,y= motif_name, group=log10Evalue))+
scale_x_continuous(expand=c (0,.25),sec.axis = sec_axis(transform= ~./4,name="Percent of peaks with motif"))+
# geom_text(aes())
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks Enrichment ratio:",ER_rat," merged motif clusters"))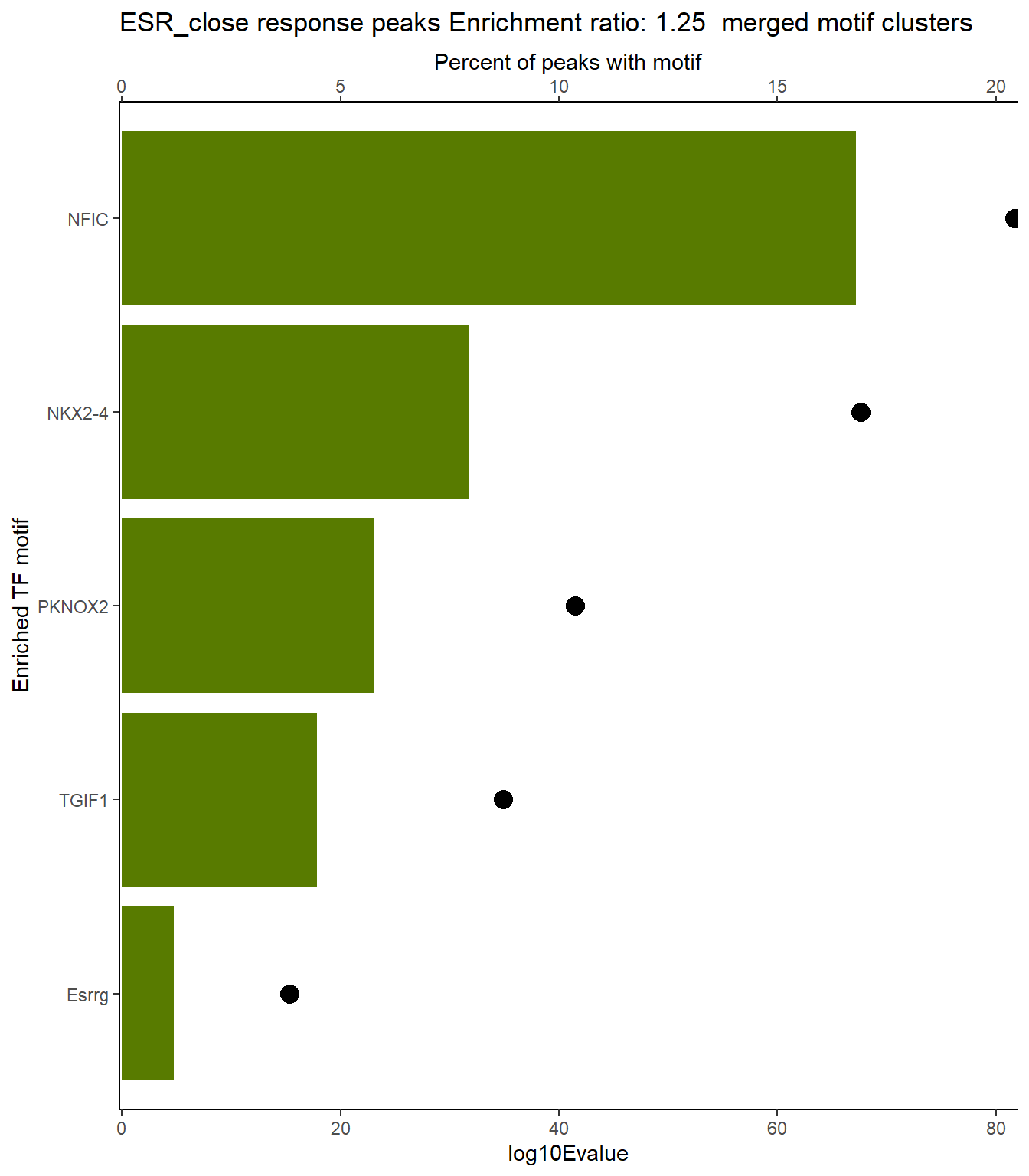
ESR_OC
ER_rat <- 2
mrc_type <- "ESR_OC"
spec_dataframe %>%
dplyr::filter(mrc==mrc_type) %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER, SITES,SIM_MOTIF,ALT_ID, ID,EVALUE, TP:'FP%', motif_name)%>%
arrange(.,EVALUE) %>%
mutate(log10Evalue= log(EVALUE, base = 10)*(-1)) %>%
mutate(motif_name=if_else(motif_name=="KLF9",paste(motif_name, RANK, sep="_"), if_else(motif_name=="^Z*",paste(motif_name, RANK, sep="_"), if_else(motif_name=="FOSL2::JUNB",paste(motif_name, RANK,sep="_"),motif_name))))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP%`*4), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./4,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks Enrichment ratio:",ER_rat))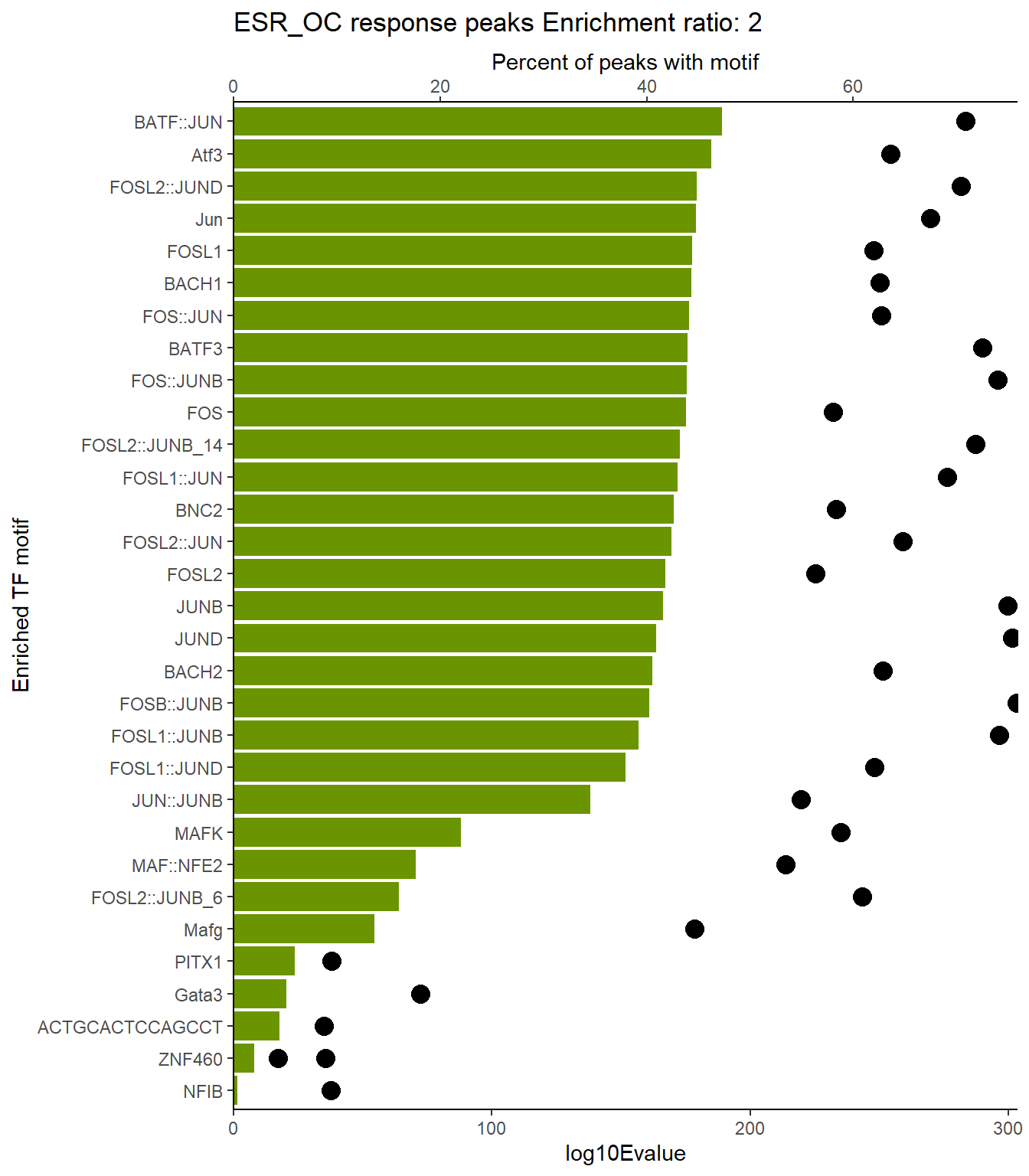
spec_dataframe %>%
dplyr::filter(mrc==mrc_type) %>%
dplyr::filter(ENR_RATIO>1.4) %>%
dplyr::select(RANK,CLUSTER, SITES,SIM_MOTIF,ALT_ID, ID,EVALUE, TP:'FP%', motif_name)%>%
arrange(.,EVALUE) %>%
mutate(log10Evalue= log(EVALUE, base = 10)*(-1)) %>%
mutate(motif_name=if_else(motif_name=="KLF9",paste(motif_name, RANK, sep="_"), if_else(motif_name=="^Z*",paste(motif_name, RANK, sep="_"), if_else(motif_name=="FOSL2::JUNB",paste(motif_name, RANK,sep="_"),motif_name))))%>%
distinct(CLUSTER,.keep_all = TRUE) %>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP%`*4), size =4)+
# geom_line(aes(x=`TP%`,y= motif_name, group=log10Evalue))+
scale_x_continuous(expand=c (0,.25),sec.axis = sec_axis(transform= ~./4,name="Percent of peaks with motif"))+
# geom_text(aes())
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks Enrichment ratio:",ER_rat," merged motif clusters"))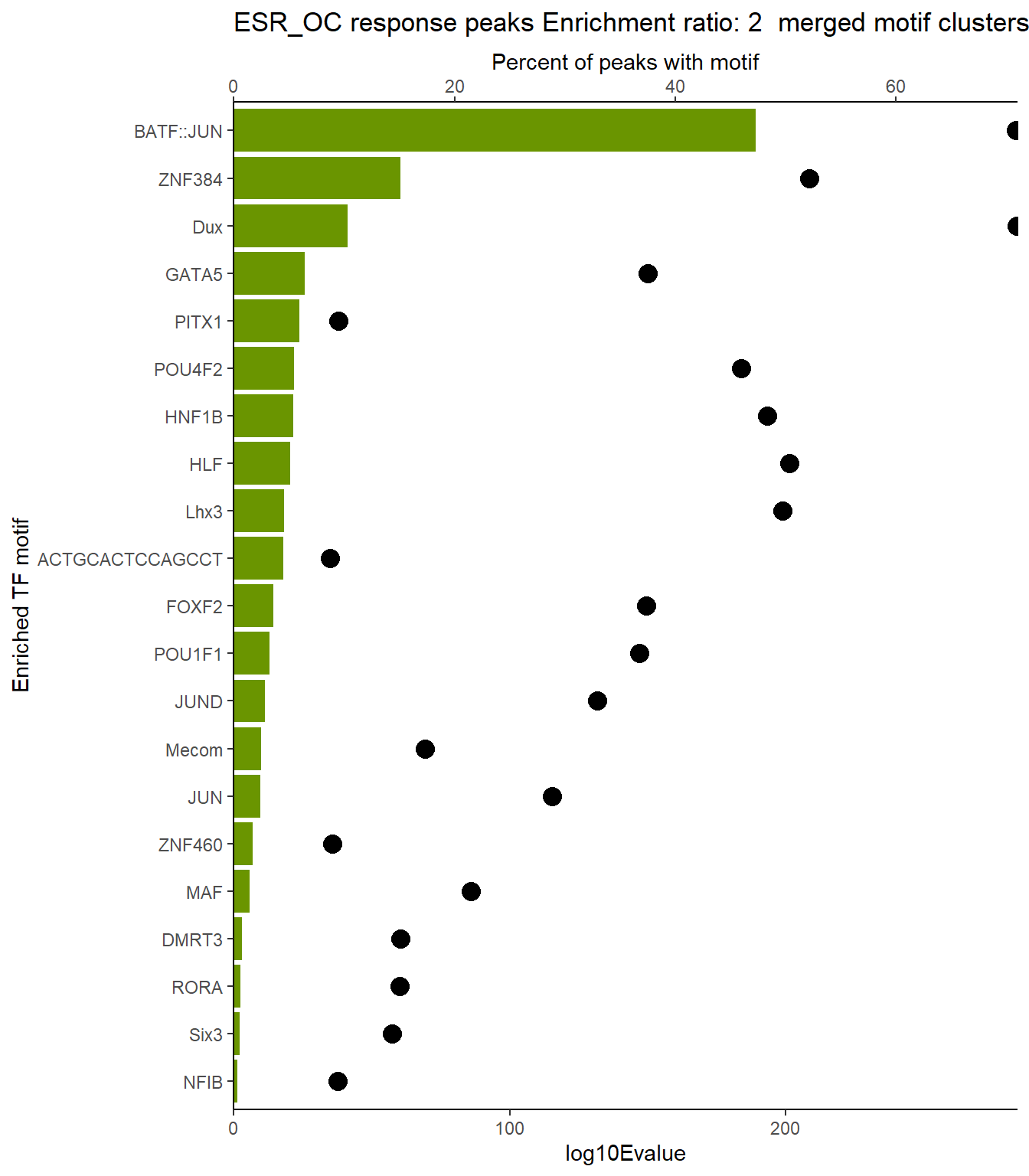
LR
LR_open
ER_rat <- 1
mrc_type <- "LR_open"
spec_dataframe %>%
dplyr::filter(mrc==mrc_type) %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER, SITES,SIM_MOTIF,ALT_ID, ID,EVALUE, TP:'FP%', motif_name)%>%
arrange(.,EVALUE) %>%
mutate(log10Evalue= log(EVALUE, base = 10)*(-1)) %>%
mutate(motif_name=if_else(motif_name=="KLF9",paste(motif_name, RANK, sep="_"), if_else(motif_name=="^R*",paste(motif_name,RANK, sep="_"), if_else(motif_name=="PKNOX2",paste(motif_name, RANK,sep="_"),motif_name))))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP%`*4), size =4)+
scale_x_continuous(expand=c (0,.125),sec.axis = sec_axis(transform= ~./4,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks Enrichment ratio:",ER_rat))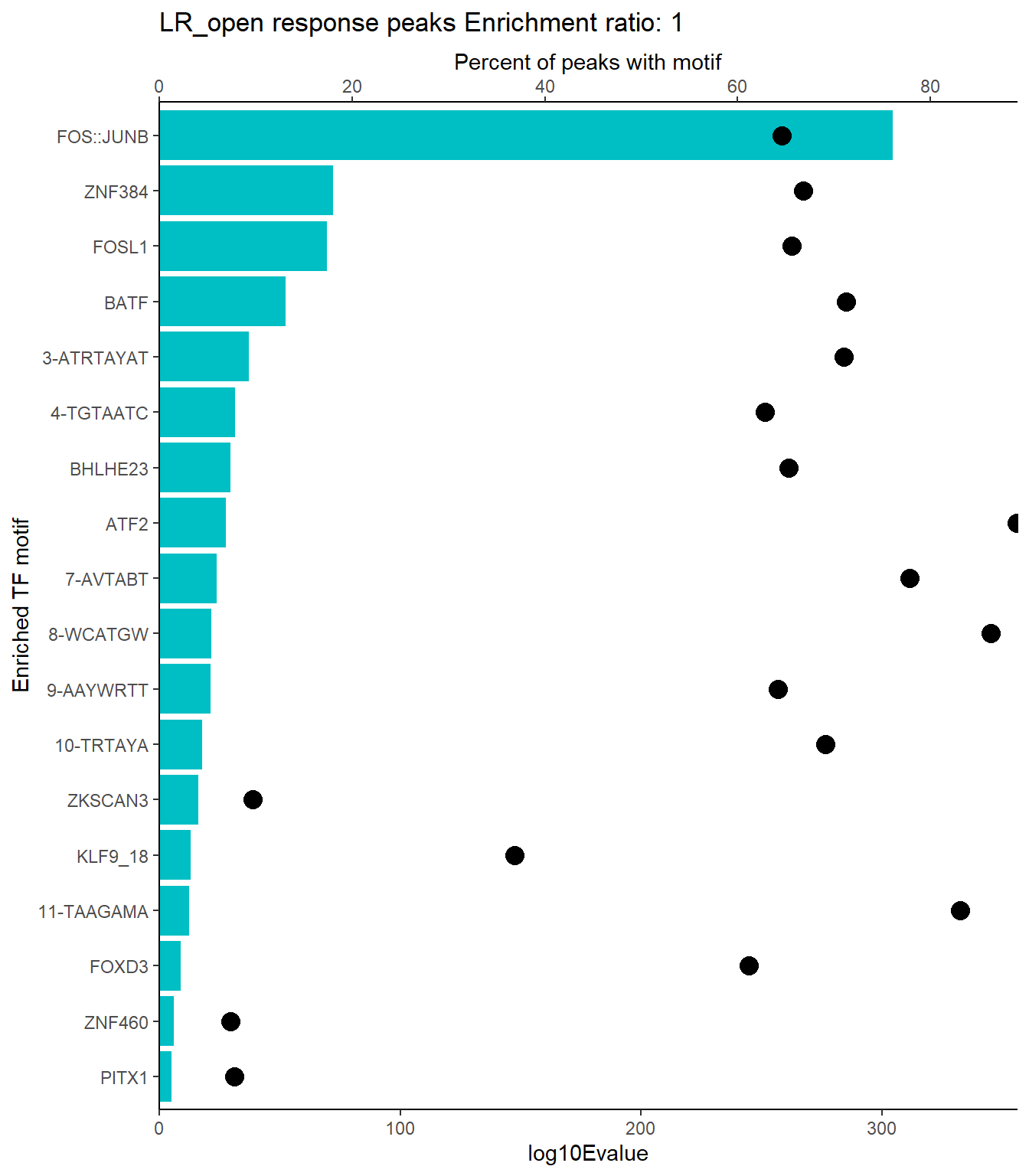
spec_dataframe %>%
dplyr::filter(mrc==mrc_type) %>%
dplyr::filter(ENR_RATIO>ER_rat) %>%
dplyr::select(RANK,CLUSTER, SITES,SIM_MOTIF,ALT_ID, ID,EVALUE, TP:'FP%', motif_name)%>%
arrange(.,EVALUE) %>%
mutate(log10Evalue= log(EVALUE, base = 10)*(-1)) %>%
mutate(motif_name=if_else(motif_name=="KLF9",paste(motif_name, RANK, sep="_"), if_else(motif_name=="^R*",paste(motif_name, RANK, sep="_"), if_else(motif_name=="PKNOX2",paste(motif_name, RANK,sep="_"),motif_name))))%>%
distinct(CLUSTER,.keep_all = TRUE) %>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette[mrc_type]) +
geom_point(aes(x=`TP%`*4), size =4)+
# geom_line(aes(x=`TP%`,y= motif_name, group=log10Evalue))+
scale_x_continuous(expand=c (0,.25),sec.axis = sec_axis(transform= ~./4,name="Percent of peaks with motif"))+
# geom_text(aes())
theme_classic()+
ylab("Enriched TF motif")+
ggtitle(paste( mrc_type,"response peaks Enrichment ratio:",ER_rat," merged motif clusters"))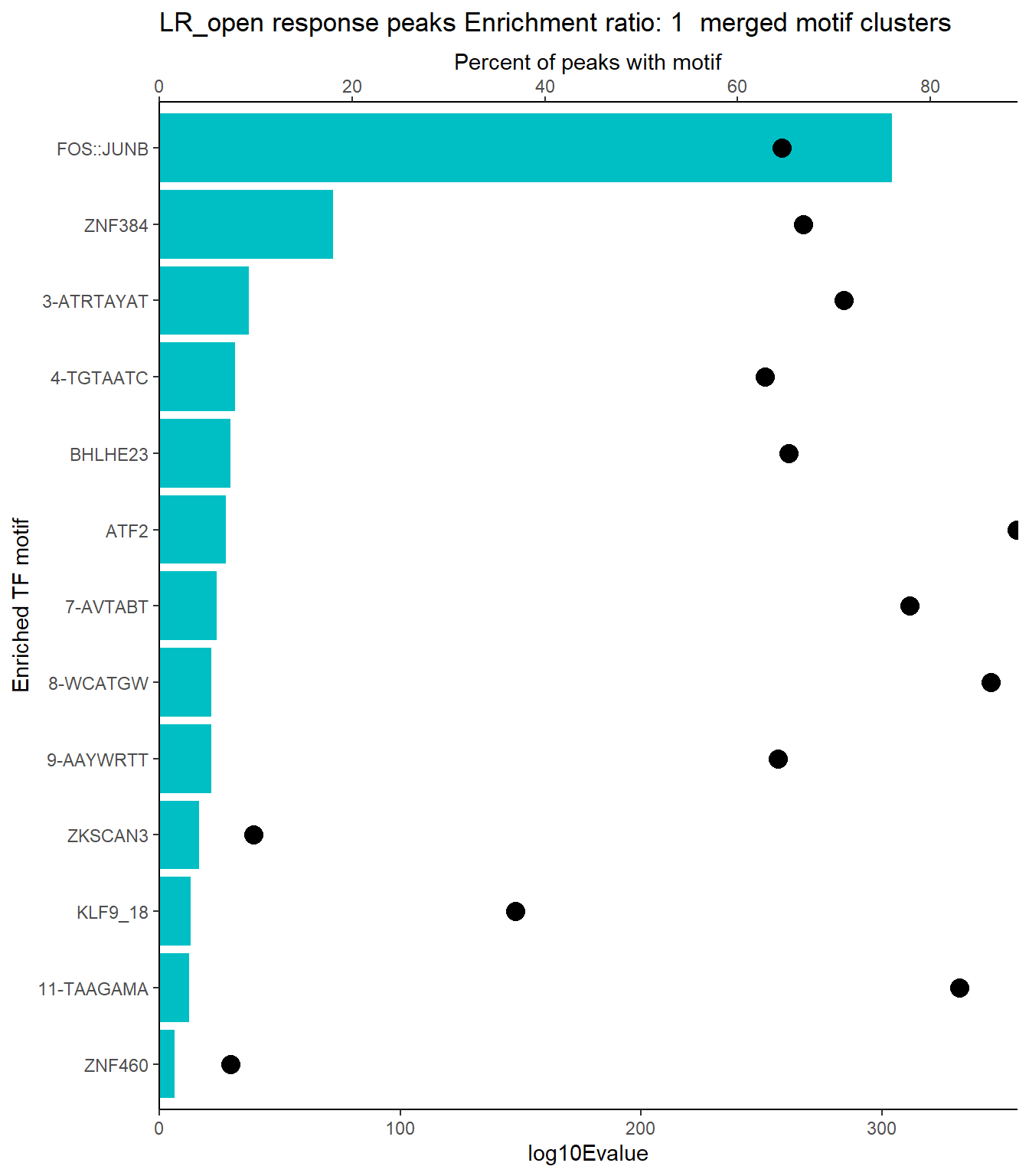
# spec_dataframe %>%
# dplyr::filter(mrc==mrc_type) %>%
# dplyr::filter(ENR_RATIO>ER_rat) #%>%
# dplyr::select(RANK,CLUSTER, SITES,SIM_MOTIF,ALT_ID, ID,EVALUE, TP:'FP%', motif_name)LR_open_xstreme%>%
mutate(CONSENSUS=gsub('[[:digit:]]+-', '', CONSENSUS)) %>%
left_join(., sea_LR_open, by= c("ALT_ID"="ALT_ID","CONSENSUS"="CONSENSUS", "ID"="ID"))%>%
# dplyr::filter(ENR_RATIO>1.25) %>%
dplyr::select(RANK.y,CLUSTER, SITES,SIM_MOTIF,ALT_ID, ID,EVALUE.x, TP:'FP%')%>%
separate(SIM_MOTIF, into= c("SIM_MOTIF", "NAME"), sep= " ") %>%
arrange(.,EVALUE.x) %>%
mutate(log10Evalue= log(EVALUE.x, base = 10)*(-1)) %>%
mutate(motif_name= gsub("[()]","",NAME), mrc="LR_open") %>%
mutate(motif_name=if_else(is.na(motif_name)&str_detect(SIM_MOTIF,"^M"),
ALT_ID, if_else(is.na(motif_name),ID, if_else(motif_name=="Rarb",paste(motif_name, RANK.y, sep="_"), if_else(motif_name=="PKNOX2",paste(motif_name, RANK.y, sep="_"),motif_name)))))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette["LR_open"]) +
geom_point(aes(x=`TP%`*4), size =4)+
scale_x_continuous(expand=c (0,.5),sec.axis = sec_axis(transform= ~./4,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle("Late response open peaks")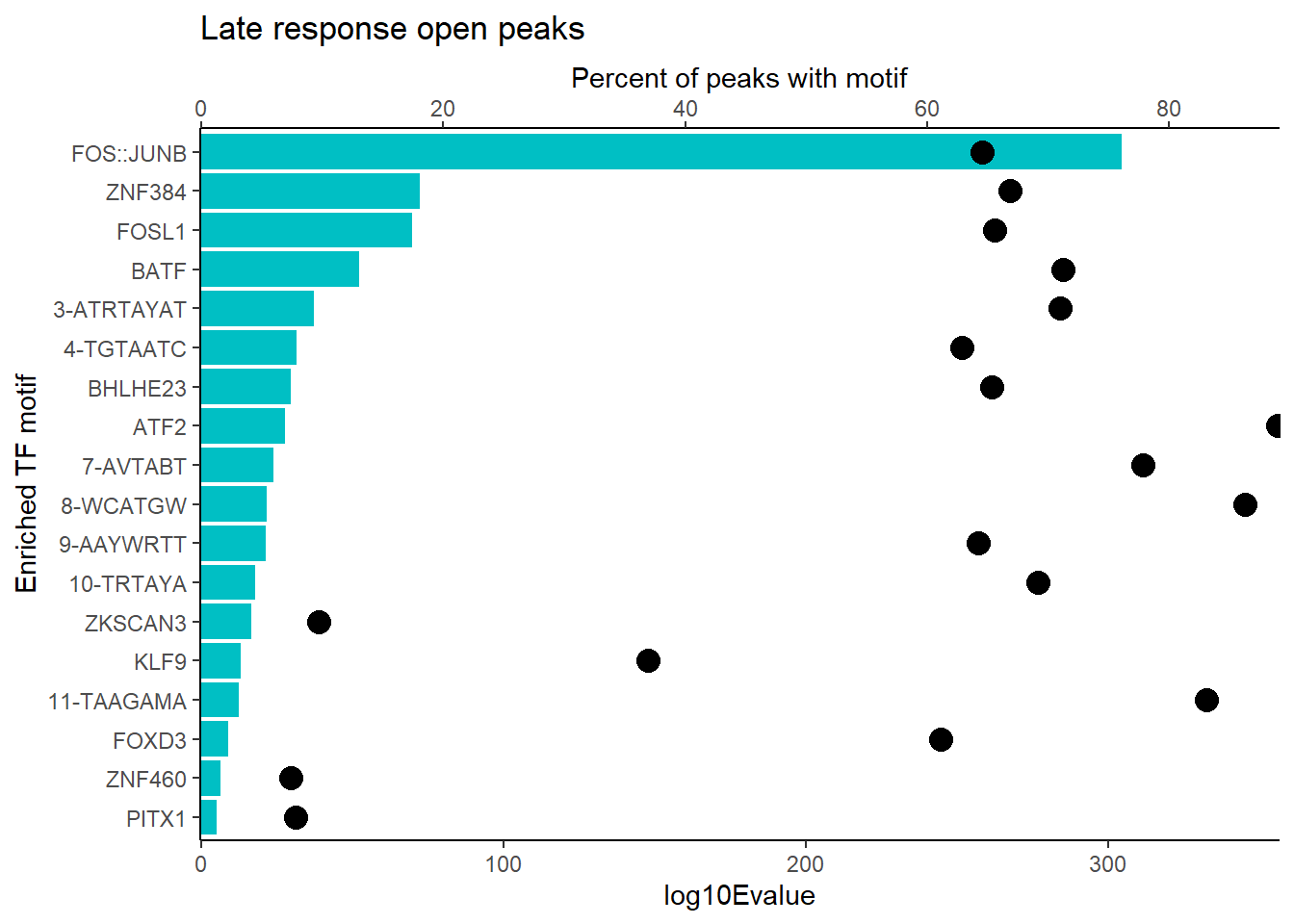
LR_open_xstreme%>%
mutate(CONSENSUS=gsub('[[:digit:]]+-', '', CONSENSUS)) %>%
left_join(., sea_LR_open, by= c("ALT_ID"="ALT_ID","CONSENSUS"="CONSENSUS", "ID"="ID")) %>%
# dplyr::filter(ENR_RATIO>1.25) %>%
dplyr::select(RANK.y,CLUSTER, SITES,SIM_MOTIF,ALT_ID, ID,EVALUE.x, TP:'FP%')%>%
distinct(CLUSTER,.keep_all = TRUE) %>%
separate(SIM_MOTIF, into= c("SIM_MOTIF", "NAME"), sep= " ") %>%
arrange(.,EVALUE.x) %>%
# dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3))))#%>%
mutate(log10Evalue= log(EVALUE.x, base = 10)*(-1)) %>%
mutate(motif_name= gsub("[()]","",NAME), mrc="LR_open") %>%
# mutate(RowGroup=cumsum(ALT_ID != 0)) %>%
mutate(motif_name=if_else(is.na(motif_name)&str_detect(SIM_MOTIF,"^M"),
ALT_ID, if_else(is.na(motif_name),ID, if_else(motif_name=="FOSL2::JUNB",paste(motif_name, RANK.y, sep="_"), if_else(motif_name=="ZNF460",paste(motif_name, RANK.y, sep="_"),motif_name)))))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette["LR_open"]) +
geom_point(aes(x=`TP%`*4), size =4)+
# geom_line(aes(x=`TP%`,y= motif_name, group=log10Evalue))+
scale_x_continuous(expand=c (0,1),sec.axis = sec_axis(transform= ~./4,name="Percent of peaks with motif"))+
# geom_text(aes())
theme_classic()+
ylab("Enriched TF motif")+
ggtitle("Late response open peaks")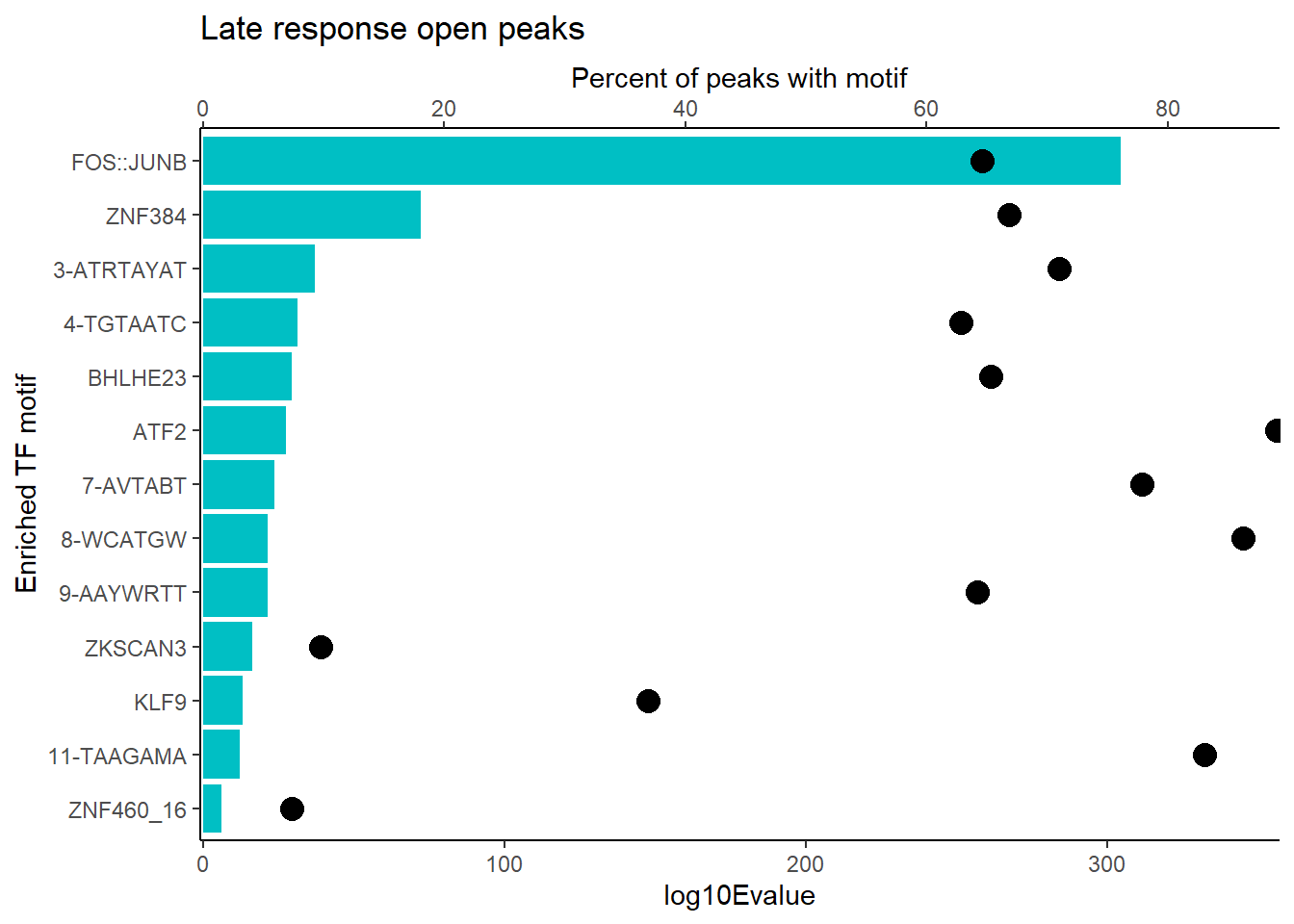
LR_close
LR_close_xstreme%>%
mutate(CONSENSUS=gsub('[[:digit:]]+-', '', CONSENSUS)) %>%
left_join(., sea_LR_close, by= c("RANK"="RANK","ALT_ID"="ALT_ID","CONSENSUS"="CONSENSUS", "ID"="ID"))%>%
dplyr::filter(ENR_RATIO>1.25) %>%
dplyr::select(RANK,CLUSTER, SITES,SIM_MOTIF,ALT_ID, ID,EVALUE.x, TP:'FP%')%>%
separate(SIM_MOTIF, into= c("SIM_MOTIF", "NAME"), sep= " ") %>%
arrange(.,EVALUE.x) %>%
mutate(log10Evalue= log(EVALUE.x, base = 10)*(-1)) %>%
mutate(motif_name= gsub("[()]","",NAME), mrc="LR_close") %>%
mutate(motif_name=if_else(is.na(motif_name)&str_detect(SIM_MOTIF,"^M"),
ALT_ID, if_else(is.na(motif_name),ID, if_else(motif_name=="Rarb",paste(motif_name, RANK, sep="_"), if_else(motif_name=="PKNOX2",paste(motif_name, RANK, sep="_"),motif_name)))))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette["LR_close"]) +
geom_point(aes(x=`TP%`*4), size =4)+
scale_x_continuous(expand=c (0,.5),sec.axis = sec_axis(transform= ~./4,name="Percent of peaks with motif"))+
theme_classic()+
ylab("Enriched TF motif")+
ggtitle("Late response close peaks")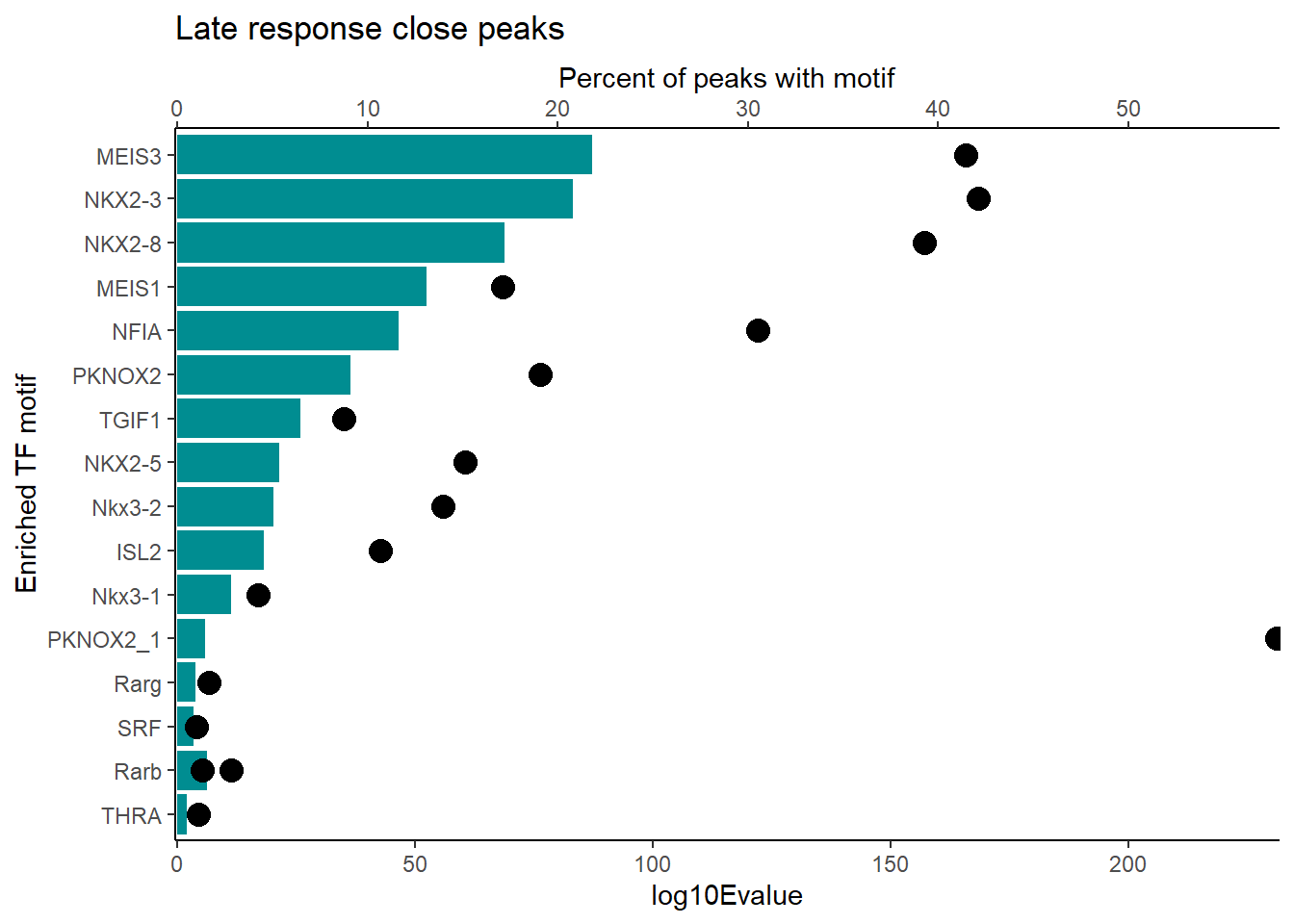
LR_close_xstreme%>%
mutate(CONSENSUS=gsub('[[:digit:]]+-', '', CONSENSUS)) %>%
left_join(., sea_LR_close, by= c("RANK"="RANK","ALT_ID"="ALT_ID","CONSENSUS"="CONSENSUS", "ID"="ID")) %>%
dplyr::filter(ENR_RATIO>1.25) %>%
dplyr::select(RANK,CLUSTER, SITES,SIM_MOTIF,ALT_ID, ID,EVALUE.x, TP:'FP%')%>%
distinct(CLUSTER,.keep_all = TRUE) %>%
separate(SIM_MOTIF, into= c("SIM_MOTIF", "NAME"), sep= " ") %>%
arrange(.,EVALUE.x) %>%
# dplyr::mutate_if(is.numeric, funs(as.character(signif(., 3))))#%>%
mutate(log10Evalue= log(EVALUE.x, base = 10)*(-1)) %>%
mutate(motif_name= gsub("[()]","",NAME), mrc="LR_close") %>%
# mutate(RowGroup=cumsum(ALT_ID != 0)) %>%
mutate(motif_name=if_else(is.na(motif_name)&str_detect(SIM_MOTIF,"^M"),
ALT_ID,
if_else(is.na(motif_name),ID, if_else(motif_name =="FOSL2::JUNB",paste(motif_name, RANK, sep="_"), if_else(motif_name =="ZNF460",paste(motif_name, RANK, sep="_"),motif_name)))))%>%
ggplot(., aes (y= reorder(motif_name,log10Evalue))) +
geom_col(aes(x=log10Evalue),fill=mrc_palette["LR_close"]) +
geom_point(aes(x=`TP%`*4), size =4)+
# geom_line(aes(x=`TP%`,y= motif_name, group=log10Evalue))+
scale_x_continuous(expand=c (0,1),sec.axis = sec_axis(transform= ~./4,name="Percent of peaks with motif"))+
# geom_text(aes())
theme_classic()+
ylab("Enriched TF motif")+
ggtitle("Late response close peaks")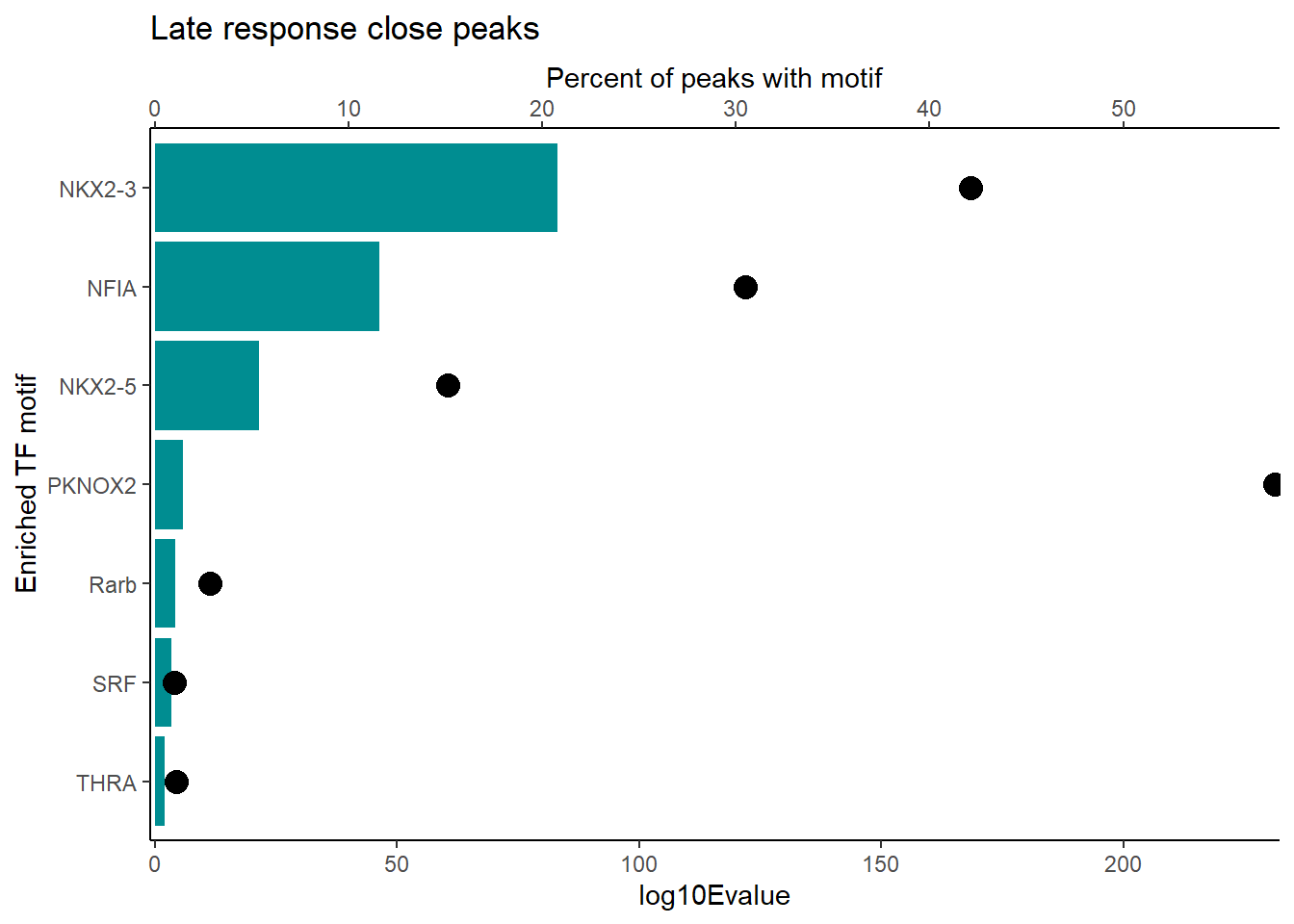
motif and peak section (incomplete)
NR_gr <- import("data/Final_four_data/meme_bed/NR_df.bed")
EAR_open_gr <- import("data/Final_four_data/meme_bed/EAR_open.bed")
EAR_close_gr <- import("data/Final_four_data/meme_bed/EAR_close.bed")
ESR_open_gr <- import("data/Final_four_data/meme_bed/ESR_open.bed")
ESR_OC_gr <- import("data/Final_four_data/meme_bed/ESR_OC.bed")
ESR_close_gr <- import("data/Final_four_data/meme_bed/ESR_close.bed")
LR_open_gr <- import("data/Final_four_data/meme_bed/LR_open.bed")
LR_close_gr <- import("data/Final_four_data/meme_bed/LR_close.bed")
EAR_open_resized <- resize(EAR_open_gr, width = 1000,fix='center')
ESR_open_resized <- resize(ESR_open_gr, width = 1000,fix='center')
LR_open_resized <- resize(LR_open_gr, width = 1000,fix='center')
NR_resized <- resize(NR_gr, width = 1000,fix='center')
EAR_close_resized <- resize(EAR_close_gr, width = 1000,fix='center')
ESR_close_resized <- resize(ESR_close_gr, width = 1000,fix='center')
LR_close_resized <- resize(LR_close_gr, width = 1000,fix='center')
ESR_OC_resized <- resize(ESR_OC_gr, width = 1000,fix='center')
BiocManager::install('PWMEnrich')
seq_list <- list(EAR_open_resized=EAR_open_resized,EAR_close_resized=EAR_close_resized,ESR_open_resized=ESR_open_resized,ESR_close_resized=ESR_close_resized,ESR_OC_resized=ESR_OC_resized,LR_open_resized=LR_open_resized,LR_close_resized=LR_close_resized,NR_resized=NR_resized)
# seq_all <- lapply(seq_list, getSeq, x=BSgenome.Hsapiens.UCSC.hg38)
# saveRDS(seq_all, "data/Final_four_data/sequencing_R_object8_motif.RDS")
seq_all <- readRDS("data/Final_four_data/sequencing_R_object8_motif.RDS")NRF1
#
# motifs_NRF1 = query(query(MotifDb, 'Hsapiens'), 'NRF1')
# NRF1_motif <- motifs_NRF1$`Hsapiens-jaspar2018-NRF1-MA0506.1`
# #
# #
# NRF1_pwm = PWMatrix(
# ID = 'NRF1',
# profileMatrix = NRF1_motif)
#
# list_nrf1 = lapply(unlist(seq_all) , FUN=searchSeq, x=NRF1_pwm, min.score="80%", strand="*")
#
# nrf1storage <- map_dfr(list_nrf1, ~as.data.frame(.x), .id="id")
# saveRDS(nrf1storage, "data/Final_four_data/nrf1_motif_8group.RDS")
nrf1_motif_8group <- readRDS("data/Final_four_data/nrf1_motif_8group.RDS")
nrf1_motif_8group %>%
mutate(id= gsub("_resized","",id)) %>%
# dplyr::filter(id =="EAR_open"|id == "NR"|id=="EAR_close") %>%
mutate(position=start-500) %>%
ggplot(.,aes(position, color = id))+
geom_density(trim = FALSE,size=1.5) +
theme_bw() +
scale_color_manual(values=mrc_palette)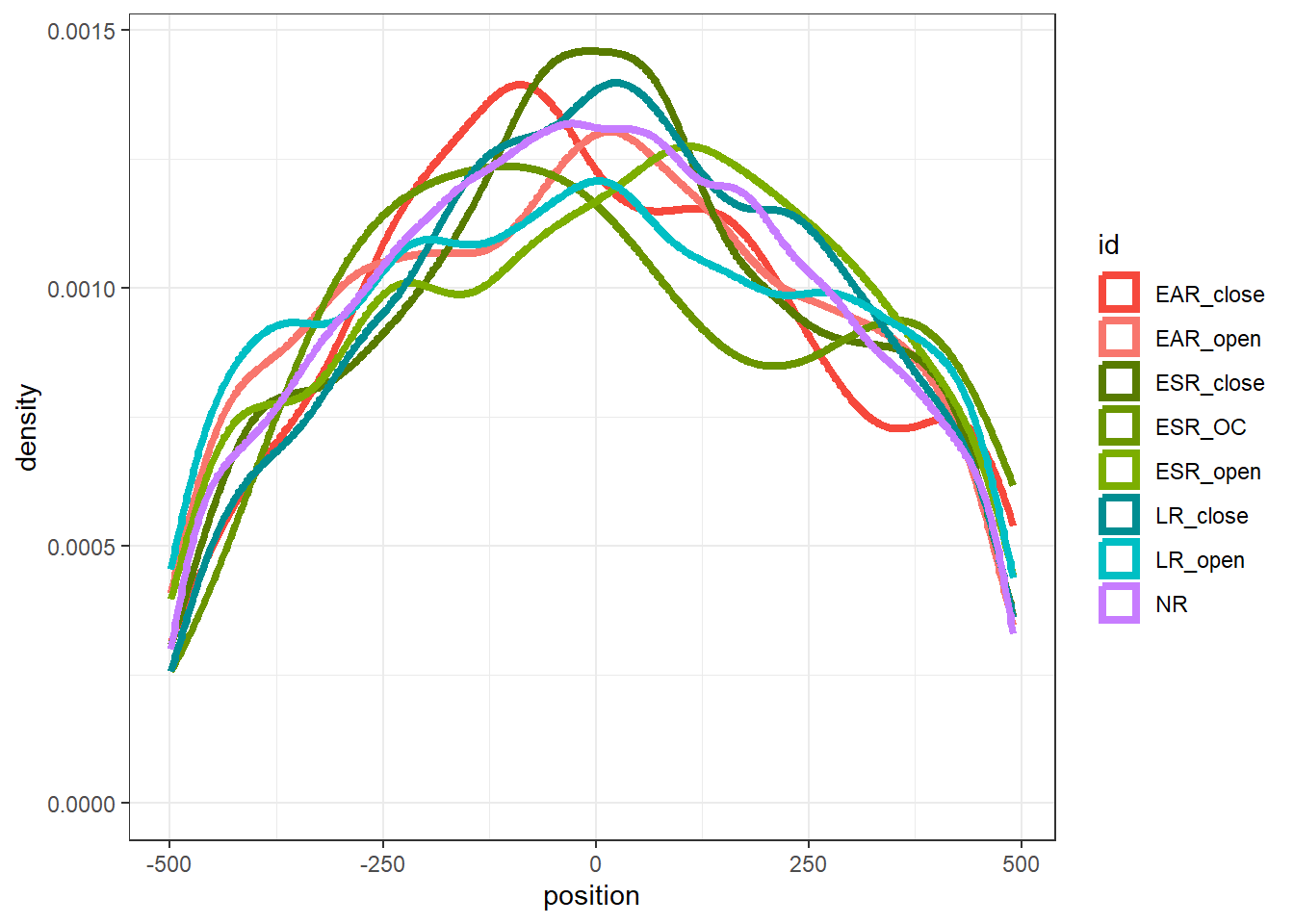
BACH2
ZNF384
enriched in EAR, ESR, LR
# motifs_ZNF384 = query(query(MotifDb, 'Hsapiens'), 'ZNF384')
# ZNF384_motif <- motifs_ZNF384$`Hsapiens-jaspar2022-ZNF384-MA1125.1`
# #
# #
# ZNF384_pwm = PWMatrix(
# ID = 'ZNF384',
# profileMatrix = ZNF384_motif)
# znf384_test = lapply(unlist(seq_all) , FUN=searchSeq, x=BACH2_pwm, min.score="80%", strand="*")
#
# storage <- lapply(list_test, relScore)
# str(storage)
# storage %>%
# map(as_tibble) %>%
# reduce(bind_rows)
# znf384_storage <- map_dfr(znf384_test, ~as.data.frame(.x), .id="id")
# saveRDS(znf384_storage, "data/Final_four_data/znf384_motif_8group.RDS")
znf384_motif_8group <- readRDS("data/Final_four_data/znf384_motif_8group.RDS")
znf384_motif_8group %>%
mutate(id= gsub("_resized","",id)) %>%
# dplyr::filter(id =="EAR_open"|id == "NR"|id=="EAR_close") %>%
# dplyr::filter(relScore>0.9) %>%
mutate(position=start-500) %>%
ggplot(.,aes(position, color = id))+
geom_density(trim = FALSE,size=1.5) +
theme_bw() +
scale_color_manual(values=mrc_palette)+
ggtitle("ZNF384 motif")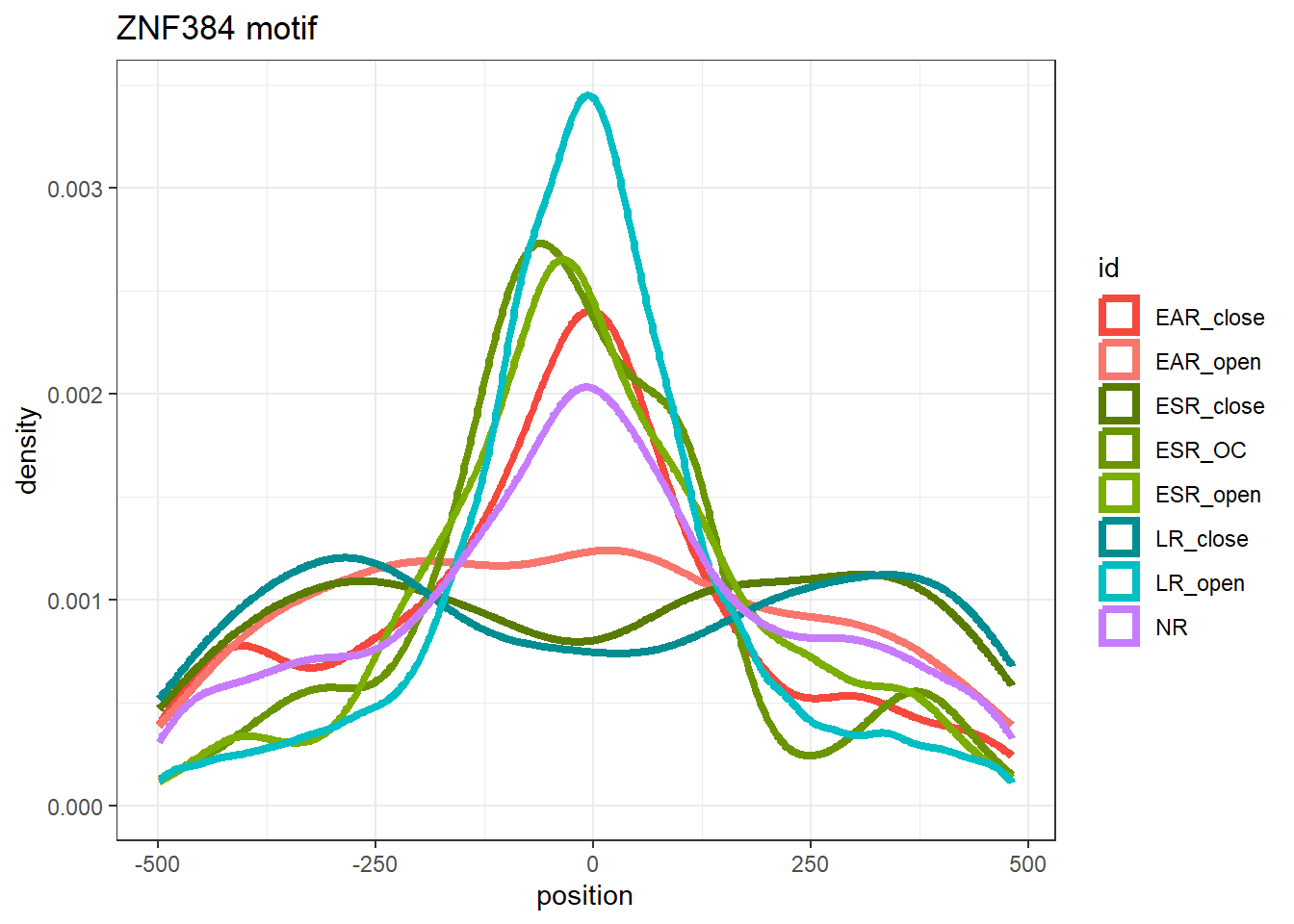
sessionInfo()R version 4.4.1 (2024-06-14 ucrt)
Platform: x86_64-w64-mingw32/x64
Running under: Windows 10 x64 (build 19045)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
time zone: America/Chicago
tzcode source: internal
attached base packages:
[1] grid stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] BSgenome.Hsapiens.UCSC.hg38_1.4.5
[2] BSgenome_1.72.0
[3] BiocIO_1.14.0
[4] MotifDb_1.46.0
[5] Biostrings_2.72.1
[6] XVector_0.44.0
[7] TFBSTools_1.42.0
[8] JASPAR2022_0.99.8
[9] BiocFileCache_2.12.0
[10] dbplyr_2.5.0
[11] devtools_2.4.5
[12] usethis_3.0.0
[13] ggpubr_0.6.0
[14] BiocParallel_1.38.0
[15] Cormotif_1.50.0
[16] affy_1.82.0
[17] scales_1.3.0
[18] VennDiagram_1.7.3
[19] futile.logger_1.4.3
[20] gridExtra_2.3
[21] ggfortify_0.4.17
[22] edgeR_4.2.1
[23] limma_3.60.4
[24] rtracklayer_1.64.0
[25] org.Hs.eg.db_3.19.1
[26] TxDb.Hsapiens.UCSC.hg38.knownGene_3.18.0
[27] GenomicFeatures_1.56.0
[28] AnnotationDbi_1.66.0
[29] Biobase_2.64.0
[30] GenomicRanges_1.56.1
[31] GenomeInfoDb_1.40.1
[32] IRanges_2.38.1
[33] S4Vectors_0.42.1
[34] BiocGenerics_0.50.0
[35] ChIPseeker_1.40.0
[36] RColorBrewer_1.1-3
[37] broom_1.0.6
[38] kableExtra_1.4.0
[39] lubridate_1.9.3
[40] forcats_1.0.0
[41] stringr_1.5.1
[42] dplyr_1.1.4
[43] purrr_1.0.2
[44] readr_2.1.5
[45] tidyr_1.3.1
[46] tibble_3.2.1
[47] ggplot2_3.5.1
[48] tidyverse_2.0.0
[49] workflowr_1.7.1
loaded via a namespace (and not attached):
[1] fs_1.6.4
[2] matrixStats_1.4.1
[3] bitops_1.0-8
[4] DirichletMultinomial_1.46.0
[5] enrichplot_1.24.4
[6] httr_1.4.7
[7] profvis_0.3.8
[8] tools_4.4.1
[9] backports_1.5.0
[10] utf8_1.2.4
[11] R6_2.5.1
[12] lazyeval_0.2.2
[13] urlchecker_1.0.1
[14] withr_3.0.1
[15] preprocessCore_1.66.0
[16] cli_3.6.3
[17] formatR_1.14
[18] scatterpie_0.2.4
[19] labeling_0.4.3
[20] sass_0.4.9
[21] Rsamtools_2.20.0
[22] systemfonts_1.1.0
[23] yulab.utils_0.1.7
[24] DOSE_3.30.5
[25] svglite_2.1.3
[26] R.utils_2.12.3
[27] sessioninfo_1.2.2
[28] plotrix_3.8-4
[29] rstudioapi_0.16.0
[30] RSQLite_2.3.7
[31] generics_0.1.3
[32] gridGraphics_0.5-1
[33] TxDb.Hsapiens.UCSC.hg19.knownGene_3.2.2
[34] gtools_3.9.5
[35] car_3.1-2
[36] GO.db_3.19.1
[37] Matrix_1.7-0
[38] fansi_1.0.6
[39] abind_1.4-8
[40] R.methodsS3_1.8.2
[41] lifecycle_1.0.4
[42] whisker_0.4.1
[43] yaml_2.3.10
[44] carData_3.0-5
[45] SummarizedExperiment_1.34.0
[46] gplots_3.1.3.1
[47] qvalue_2.36.0
[48] SparseArray_1.4.8
[49] blob_1.2.4
[50] promises_1.3.0
[51] pwalign_1.0.0
[52] crayon_1.5.3
[53] miniUI_0.1.1.1
[54] lattice_0.22-6
[55] cowplot_1.1.3
[56] annotate_1.82.0
[57] KEGGREST_1.44.1
[58] pillar_1.9.0
[59] knitr_1.48
[60] fgsea_1.30.0
[61] rjson_0.2.23
[62] boot_1.3-31
[63] codetools_0.2-20
[64] fastmatch_1.1-4
[65] glue_1.7.0
[66] getPass_0.2-4
[67] ggfun_0.1.6
[68] remotes_2.5.0
[69] data.table_1.16.0
[70] vctrs_0.6.5
[71] png_0.1-8
[72] treeio_1.28.0
[73] poweRlaw_0.80.0
[74] gtable_0.3.5
[75] cachem_1.1.0
[76] xfun_0.47
[77] mime_0.12
[78] S4Arrays_1.4.1
[79] tidygraph_1.3.1
[80] pracma_2.4.4
[81] statmod_1.5.0
[82] ellipsis_0.3.2
[83] nlme_3.1-166
[84] ggtree_3.12.0
[85] bit64_4.0.5
[86] filelock_1.0.3
[87] rprojroot_2.0.4
[88] bslib_0.8.0
[89] affyio_1.74.0
[90] KernSmooth_2.23-24
[91] splitstackshape_1.4.8
[92] seqLogo_1.70.0
[93] colorspace_2.1-1
[94] DBI_1.2.3
[95] tidyselect_1.2.1
[96] processx_3.8.4
[97] bit_4.0.5
[98] compiler_4.4.1
[99] curl_5.2.2
[100] git2r_0.33.0
[101] httr2_1.0.4
[102] xml2_1.3.6
[103] DelayedArray_0.30.1
[104] shadowtext_0.1.4
[105] caTools_1.18.3
[106] callr_3.7.6
[107] rappdirs_0.3.3
[108] digest_0.6.37
[109] rmarkdown_2.28
[110] htmltools_0.5.8.1
[111] pkgconfig_2.0.3
[112] MatrixGenerics_1.16.0
[113] highr_0.11
[114] fastmap_1.2.0
[115] htmlwidgets_1.6.4
[116] rlang_1.1.4
[117] UCSC.utils_1.0.0
[118] shiny_1.9.1
[119] farver_2.1.2
[120] jquerylib_0.1.4
[121] jsonlite_1.8.8
[122] GOSemSim_2.30.2
[123] R.oo_1.26.0
[124] RCurl_1.98-1.16
[125] magrittr_2.0.3
[126] GenomeInfoDbData_1.2.12
[127] ggplotify_0.1.2
[128] patchwork_1.3.0
[129] munsell_0.5.1
[130] Rcpp_1.0.13
[131] ape_5.8
[132] viridis_0.6.5
[133] stringi_1.8.4
[134] ggraph_2.2.1
[135] zlibbioc_1.50.0
[136] MASS_7.3-61
[137] pkgbuild_1.4.4
[138] plyr_1.8.9
[139] parallel_4.4.1
[140] ggrepel_0.9.6
[141] CNEr_1.40.0
[142] graphlayouts_1.1.1
[143] splines_4.4.1
[144] hms_1.1.3
[145] locfit_1.5-9.10
[146] ps_1.8.0
[147] igraph_2.0.3
[148] ggsignif_0.6.4
[149] pkgload_1.4.0
[150] reshape2_1.4.4
[151] TFMPvalue_0.0.9
[152] futile.options_1.0.1
[153] XML_3.99-0.17
[154] evaluate_1.0.0
[155] lambda.r_1.2.4
[156] BiocManager_1.30.25
[157] tzdb_0.4.0
[158] tweenr_2.0.3
[159] httpuv_1.6.15
[160] polyclip_1.10-7
[161] ggforce_0.4.2
[162] xtable_1.8-4
[163] restfulr_0.0.15
[164] tidytree_0.4.6
[165] rstatix_0.7.2
[166] later_1.3.2
[167] viridisLite_0.4.2
[168] aplot_0.2.3
[169] memoise_2.0.1
[170] GenomicAlignments_1.40.0
[171] timechange_0.3.0