Run_all_analysis
ERM
20230410
Last updated: 2023-04-10
Checks: 7 0
Knit directory: Cardiotoxicity/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230109) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version d3f8cf7. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .RData
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/ACresponse_cluster24h.csv
Ignored: data/Clamp_Summary.csv
Ignored: data/Cormotif_24_k1-5_raw.RDS
Ignored: data/DAtable1.csv
Ignored: data/DDE_reQTL.txt
Ignored: data/DDEresp_list.csv
Ignored: data/DEG_cormotif.RDS
Ignored: data/DF_Plate_Peak.csv
Ignored: data/Da24counts.txt
Ignored: data/Dx24counts.txt
Ignored: data/Dx_reQTL_specific.txt
Ignored: data/Ep24counts.txt
Ignored: data/GOplots.R
Ignored: data/K_cluster
Ignored: data/K_cluster_kisthree.csv
Ignored: data/K_cluster_kistwo.csv
Ignored: data/Mt24counts.txt
Ignored: data/RINsamplelist.txt
Ignored: data/Seonane2019supp1.txt
Ignored: data/TOP2Bi-24hoursGO_analysis.csv
Ignored: data/TR24counts.txt
Ignored: data/Top2biresp_cluster24h.csv
Ignored: data/Viabilitylistfull.csv
Ignored: data/allexpressedgenes.txt
Ignored: data/allgenes.txt
Ignored: data/allmatrix.RDS
Ignored: data/avgLD50.RDS
Ignored: data/backGL.txt
Ignored: data/cormotif_3hk1-8.RDS
Ignored: data/cormotif_ER_respint.txt
Ignored: data/cormotif_ER_respset.txt
Ignored: data/cormotif_LR_respint.txt
Ignored: data/cormotif_LR_respset.txt
Ignored: data/cormotif_NRset.txt
Ignored: data/cormotif_TI_respint.txt
Ignored: data/cormotif_TI_respset.txt
Ignored: data/cormotif_initalK5.RDS
Ignored: data/cormotif_initialK5.RDS
Ignored: data/counts24hours.RDS
Ignored: data/cpmnorm_counts.csv
Ignored: data/dat_cpm.RDS
Ignored: data/data_outline.txt
Ignored: data/efit2results.RDS
Ignored: data/ensgtotal.txt
Ignored: data/filenameonly.txt
Ignored: data/filtered_cpm_counts.csv
Ignored: data/filtermatrix_x.RDS
Ignored: data/folder_05top/
Ignored: data/gene_prob_tran3h.RDS
Ignored: data/gene_probabilityk5.RDS
Ignored: data/individualDRCfile.RDS
Ignored: data/knowles56.GMT
Ignored: data/knowlesGMT.GMT
Ignored: data/mymatrix.RDS
Ignored: data/nonresponse_cluster24h.csv
Ignored: data/norm_LDH.csv
Ignored: data/norm_counts.csv
Ignored: data/plan2plot.png
Ignored: data/raw_counts.csv
Ignored: data/response_cluster24h.csv
Ignored: data/sigVDA24.txt
Ignored: data/sigVDA3.txt
Ignored: data/sigVDX24.txt
Ignored: data/sigVDX3.txt
Ignored: data/sigVEP24.txt
Ignored: data/sigVEP3.txt
Ignored: data/sigVMT24.txt
Ignored: data/sigVMT3.txt
Ignored: data/sigVTR24.txt
Ignored: data/sigVTR3.txt
Ignored: data/table3a.omar
Ignored: data/tvl24hour.txt
Ignored: data/tvl24hourw.txt
Ignored: data/venn_code.R
Untracked files:
Untracked: .RDataTmp
Untracked: .RDataTmp1
Untracked: code/extra_code.R
Untracked: output/output-old/
Untracked: reneebasecode.R
Unstaged changes:
Modified: analysis/DRC_analysis.Rmd
Modified: code/eQTLcodes.R
Deleted: output/Cormotif.svg
Deleted: output/Ctrxn24-3-23.svg
Deleted: output/Ctxnrate3-23.png
Deleted: output/Decay_Slope3-23.svg
Deleted: output/ERmotif5_LFC.svg
Deleted: output/GOBP_motif345.svg
Deleted: output/KEGGmotif_345.svg
Deleted: output/LD503-21-23.png
Deleted: output/LDH_24-3-23.svg
Deleted: output/LDH_243-23.svg
Deleted: output/LFCbytreatment3-25.svg
Deleted: output/LFCmotif4_LR.svg
Deleted: output/LR_RespMoti4.svg
Deleted: output/MeanAmp243-23.svg
Deleted: output/NRmotif1_LFC.svg
Deleted: output/Rise_Slope3-23.svg
Deleted: output/TI_LFC.svg
Deleted: output/TVLcorr3-23.svg
Deleted: output/TropI3-23.svg
Deleted: output/Venn24DEG-3-24.png
Deleted: output/motif1NR_LFC.svg
Deleted: output/motif3TIlfc3-25.svg
Deleted: output/motif4LR3-25LFC.svg
Deleted: output/motif5ER-LFC-3-25.svg
Deleted: output/nolegendLDH.svg
Deleted: output/resultsigVDA24.csv
Deleted: output/tropI_24-3-23.svg
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/run_all_analysis.Rmd) and
HTML (docs/run_all_analysis.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | d3f8cf7 | reneeisnowhere | 2023-04-10 | update push of new data |
| html | f0a75e1 | reneeisnowhere | 2023-04-10 | Build site. |
| Rmd | 8ca4c7e | reneeisnowhere | 2023-04-10 | first rmd commit |
| Rmd | 2e69969 | reneeisnowhere | 2023-04-10 | adding data |
| Rmd | 0f1f1da | reneeisnowhere | 2023-04-10 | final run analysis |
This starts the documentation of the RNA-seq cardiotoxicity analysis for my manuscript
library(edgeR)#
library(limma)#
library(RColorBrewer)
library(mixOmics)
library(gridExtra)#
library(reshape2)#
#library(devtools)
library(AnnotationHub)
library(tidyverse)
library(scales)
library(biomaRt)#
library(Homo.sapiens)
library(cowplot)#
library(ggrepel)#
library(corrplot)
library(Hmisc) ###now we add genenames to the geneid###
geneid <- rownames(mymatrix) ### pulls the names we have in the counts file
genes <- select(Homo.sapiens, keys=geneid, columns=c("SYMBOL"),
keytype="ENTREZID")
genes <- genes[!duplicated(genes$ENTREZID),]
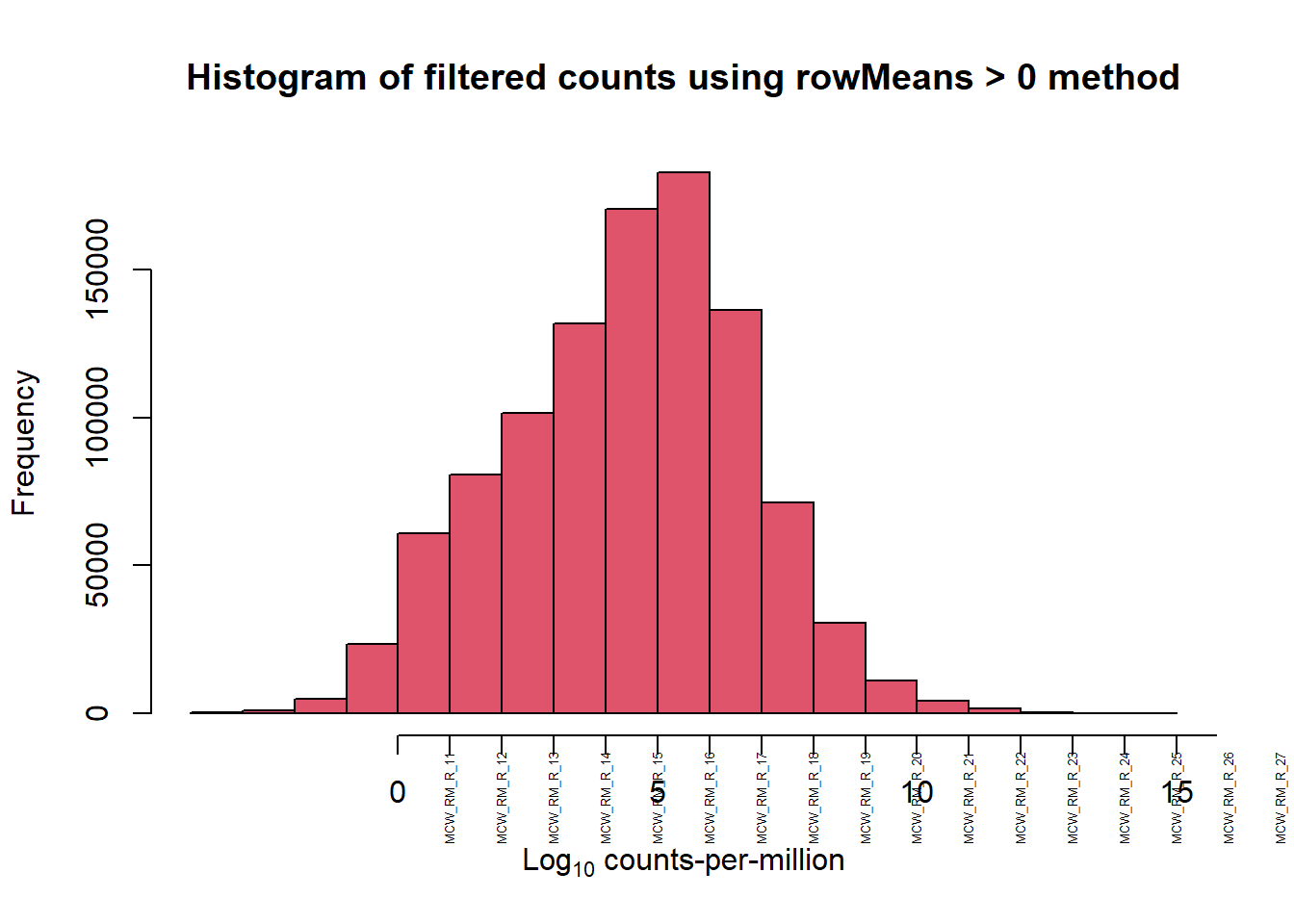
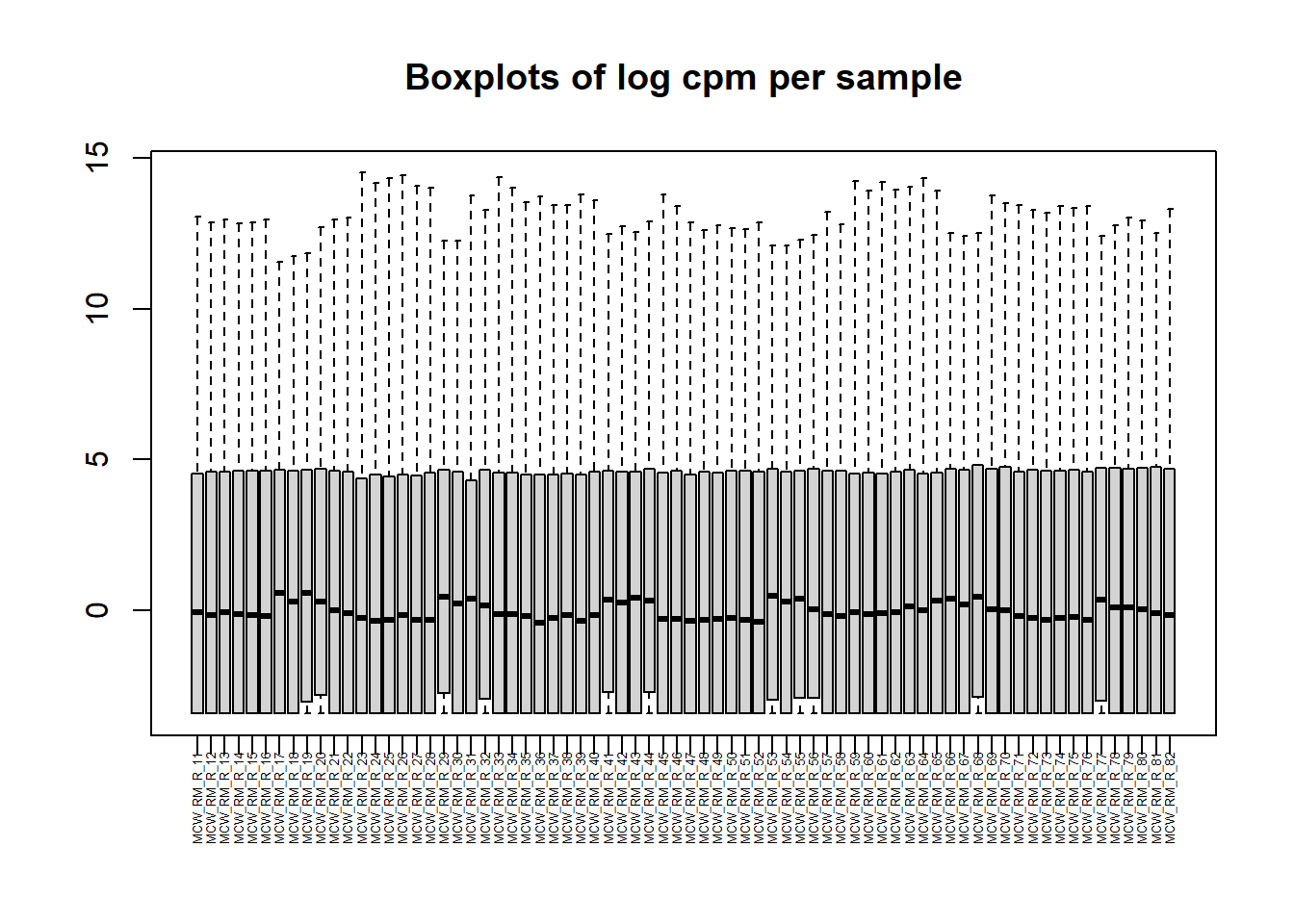
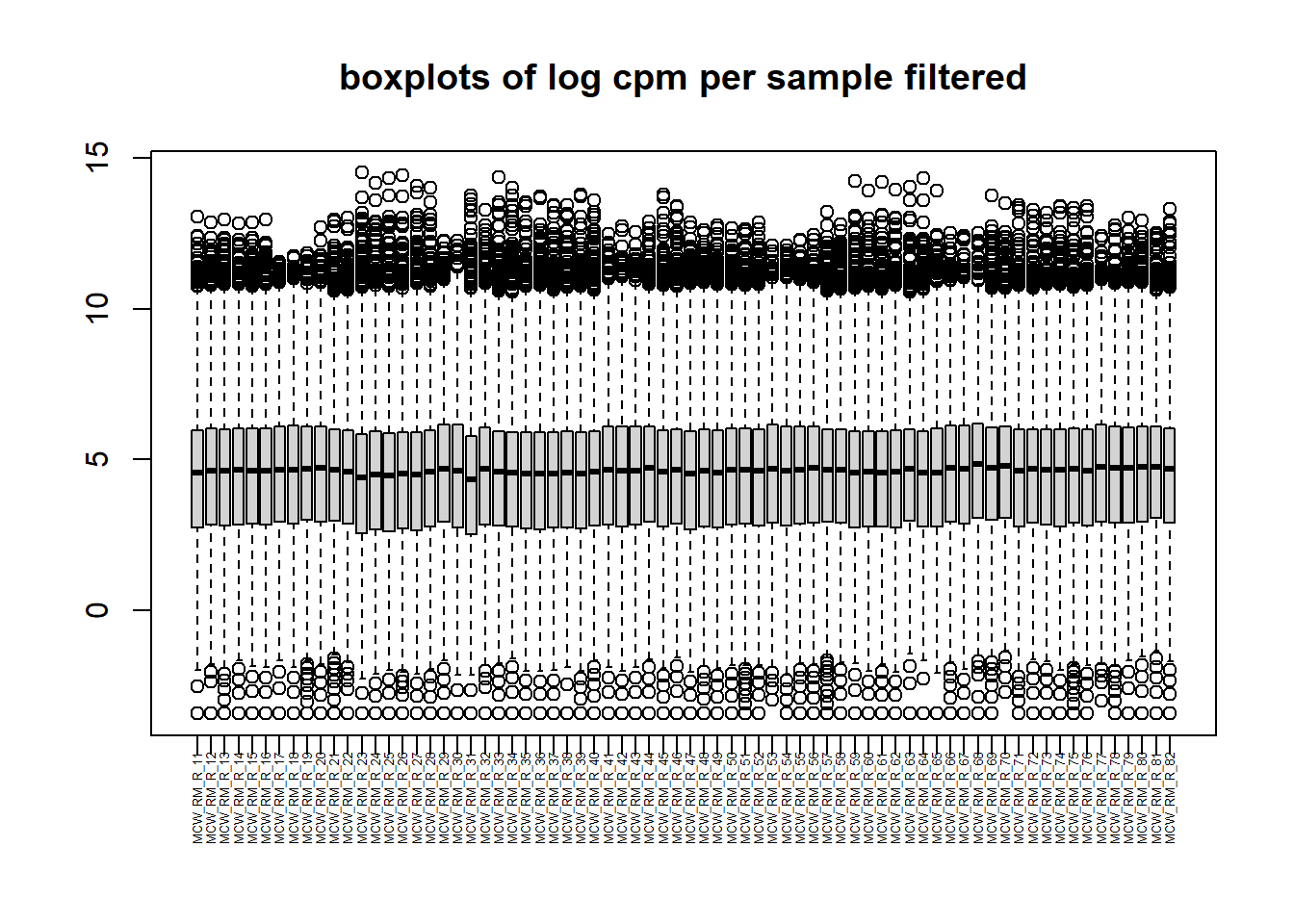
mymatrix$genes <- genes[1] 28395 72[1] 14086 72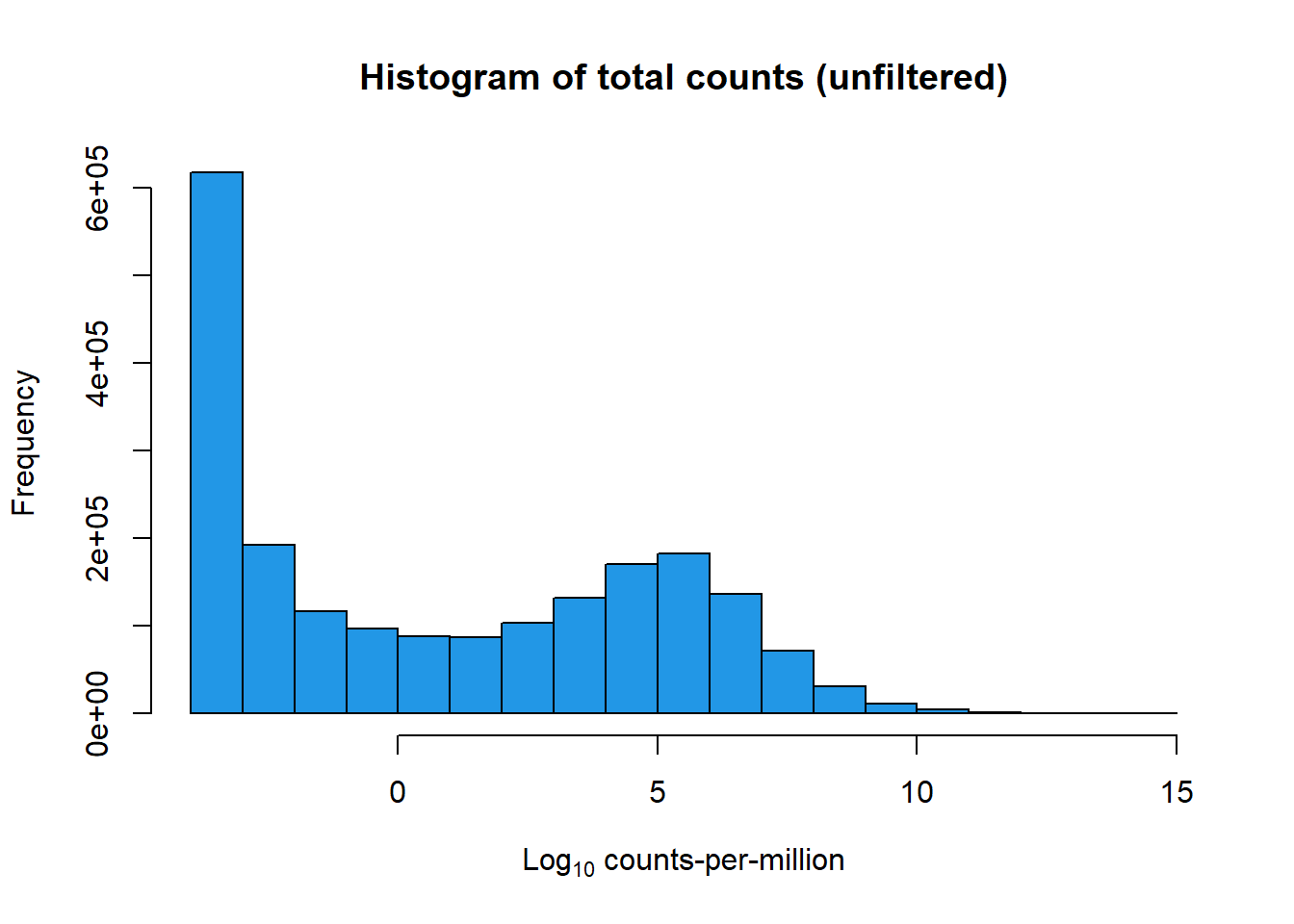
### Vehicle 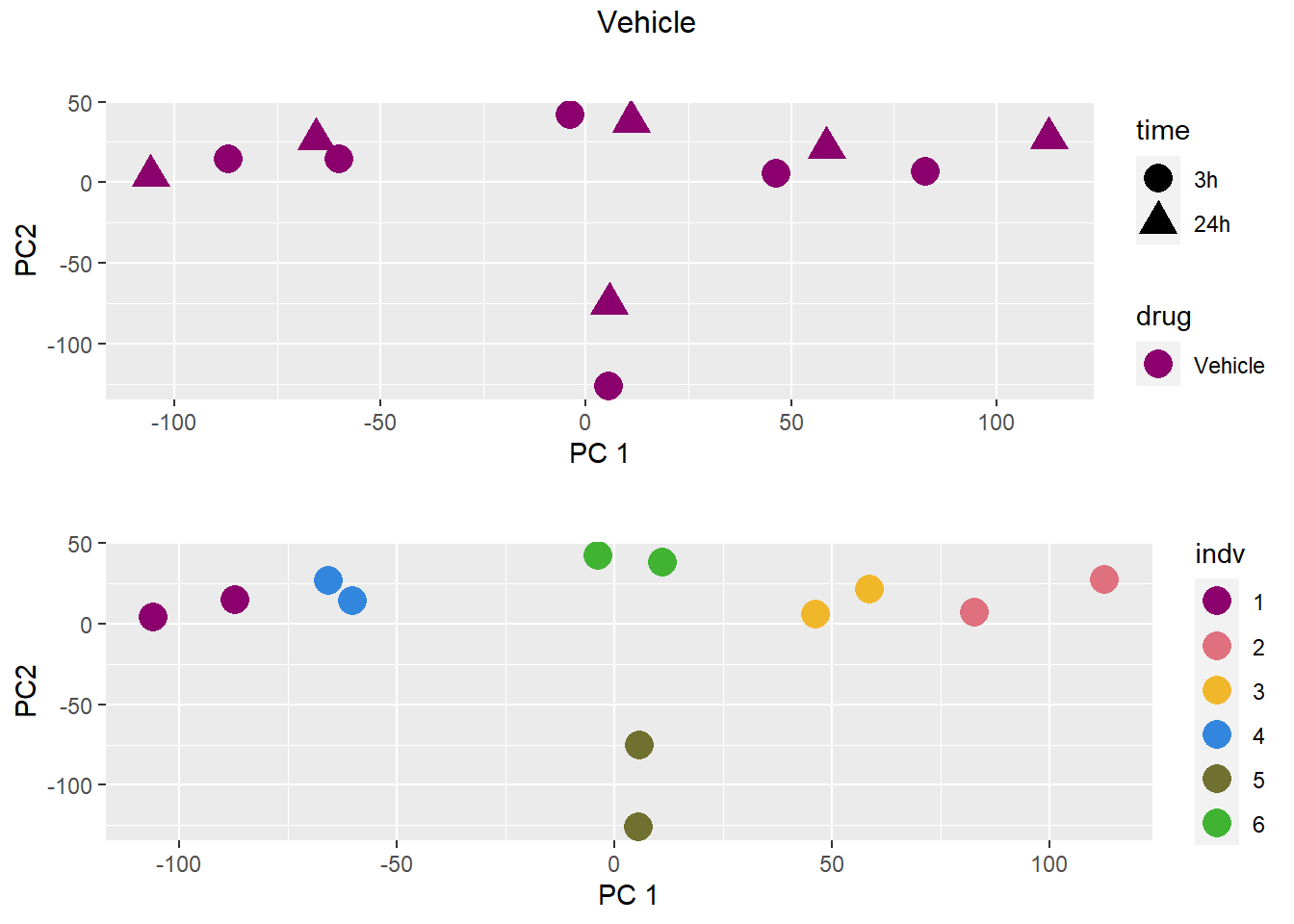
### Daunorubicin 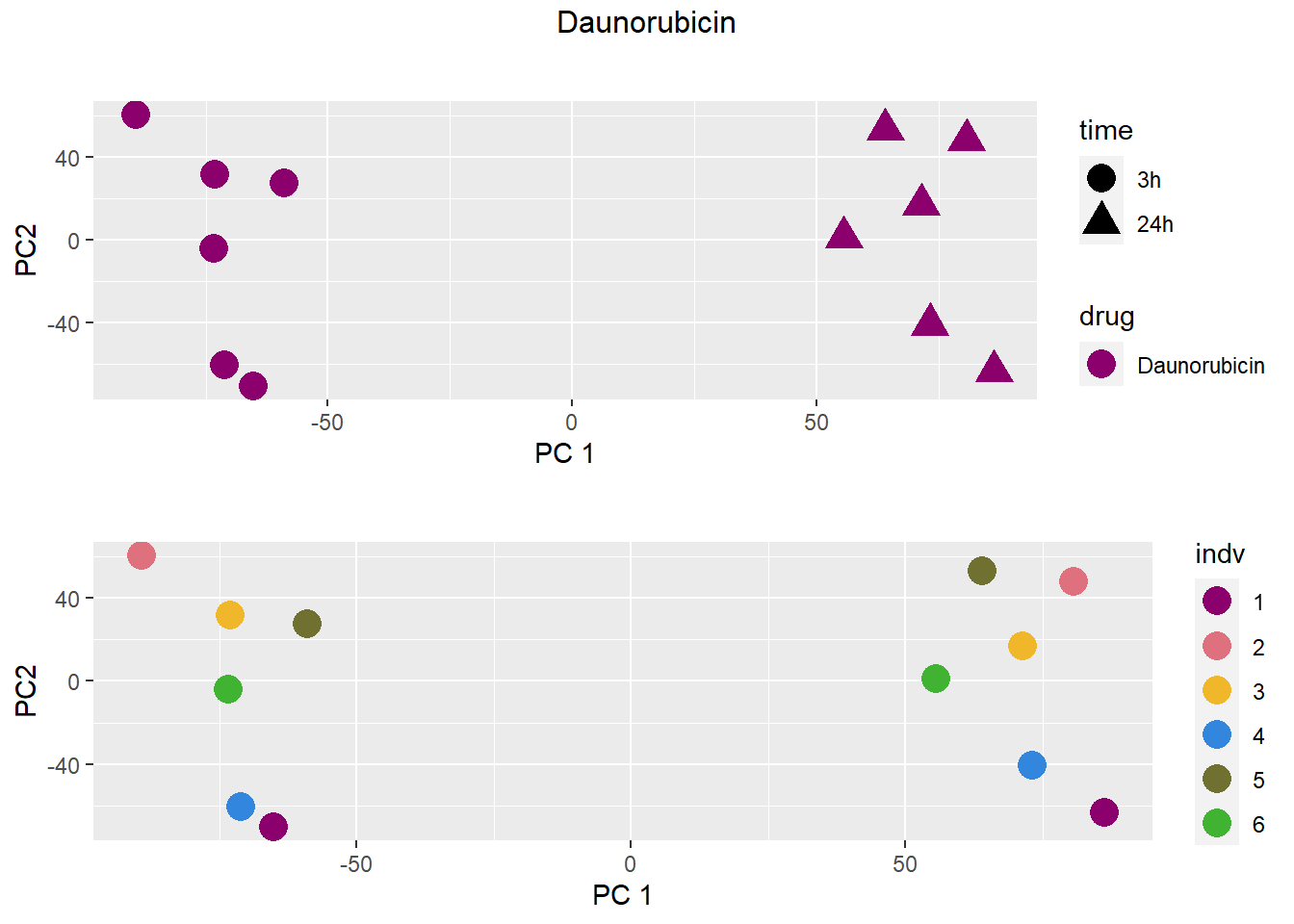
### Doxorubicin 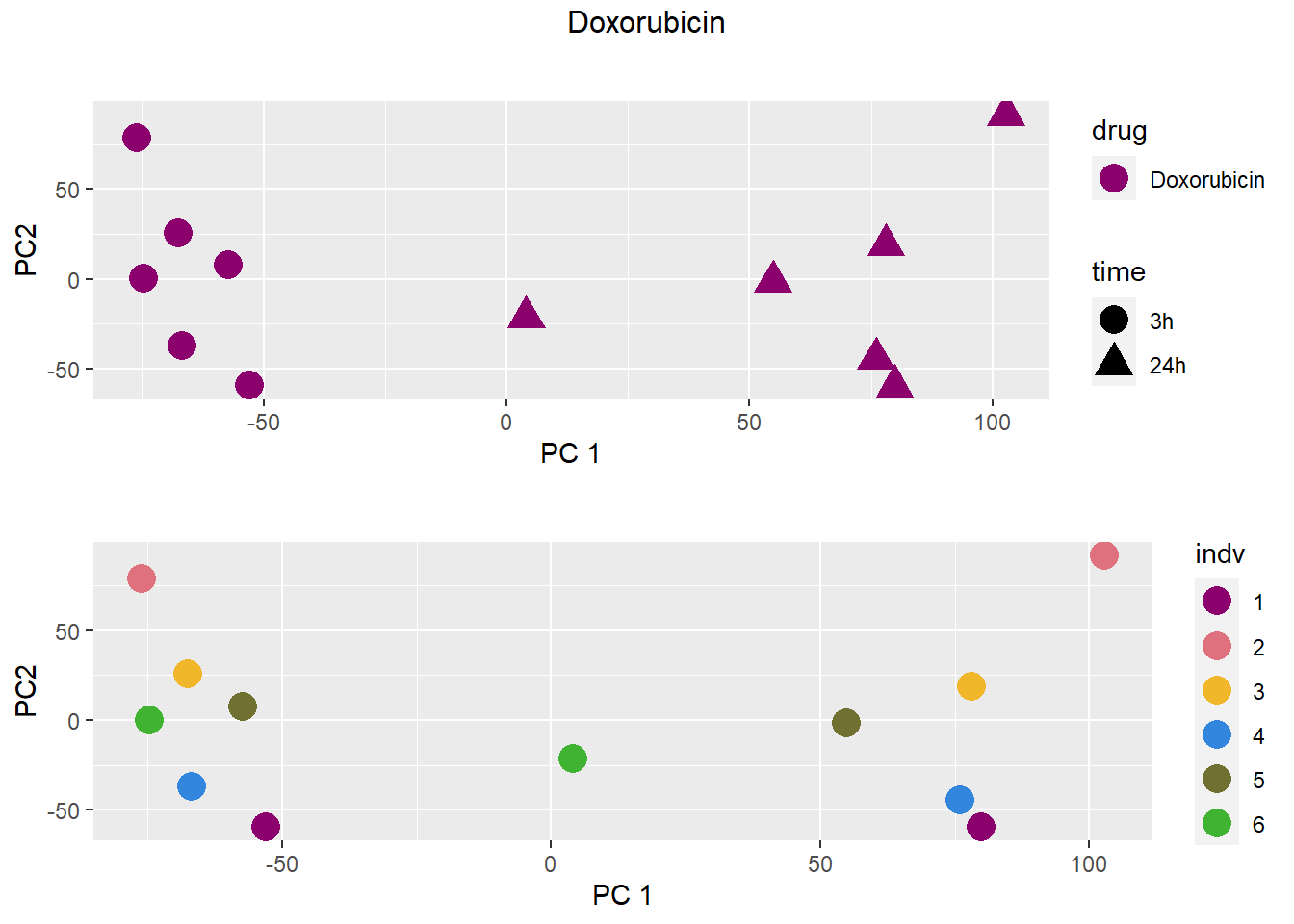
### Epirubicin 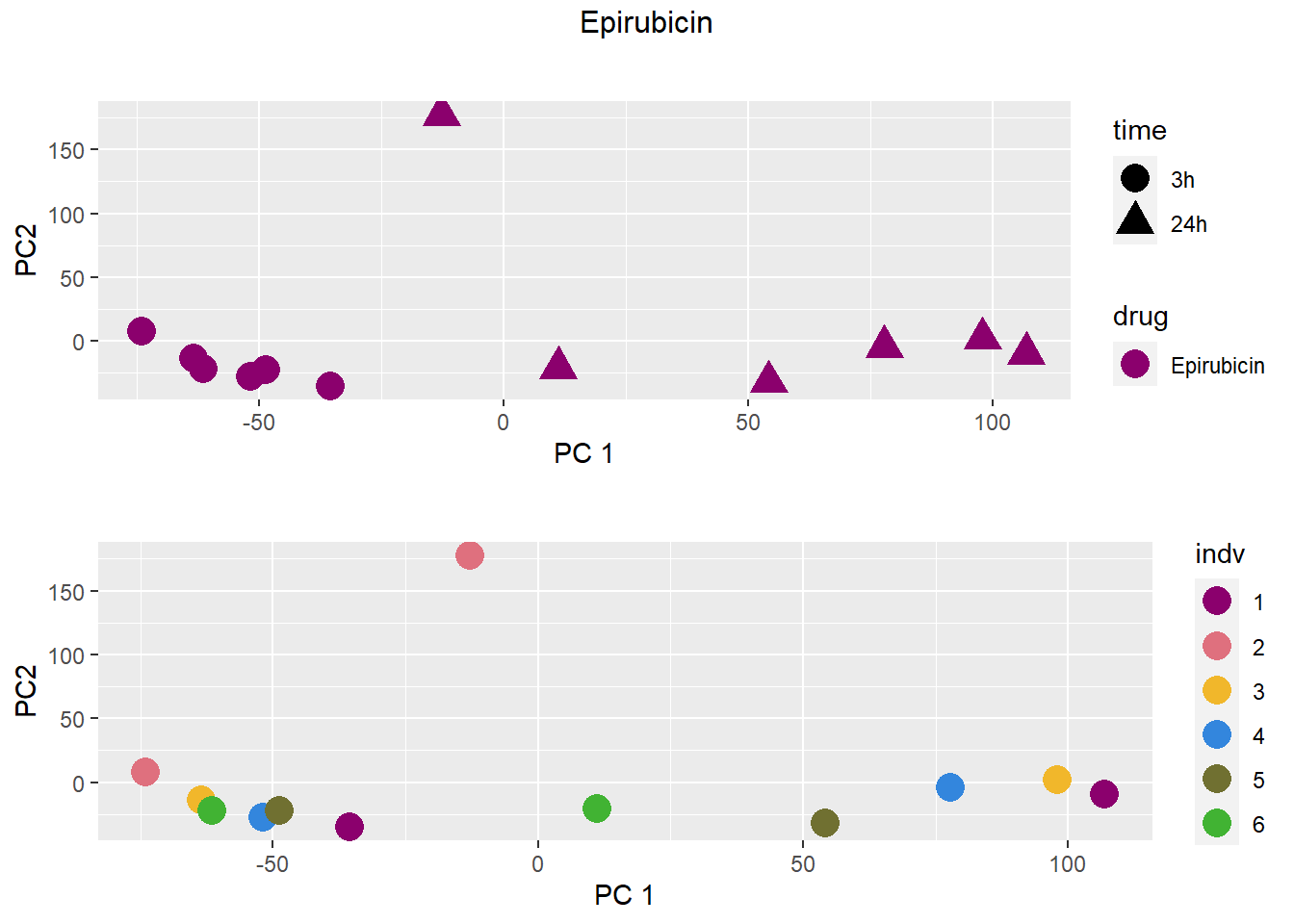
### Mitoxantrone 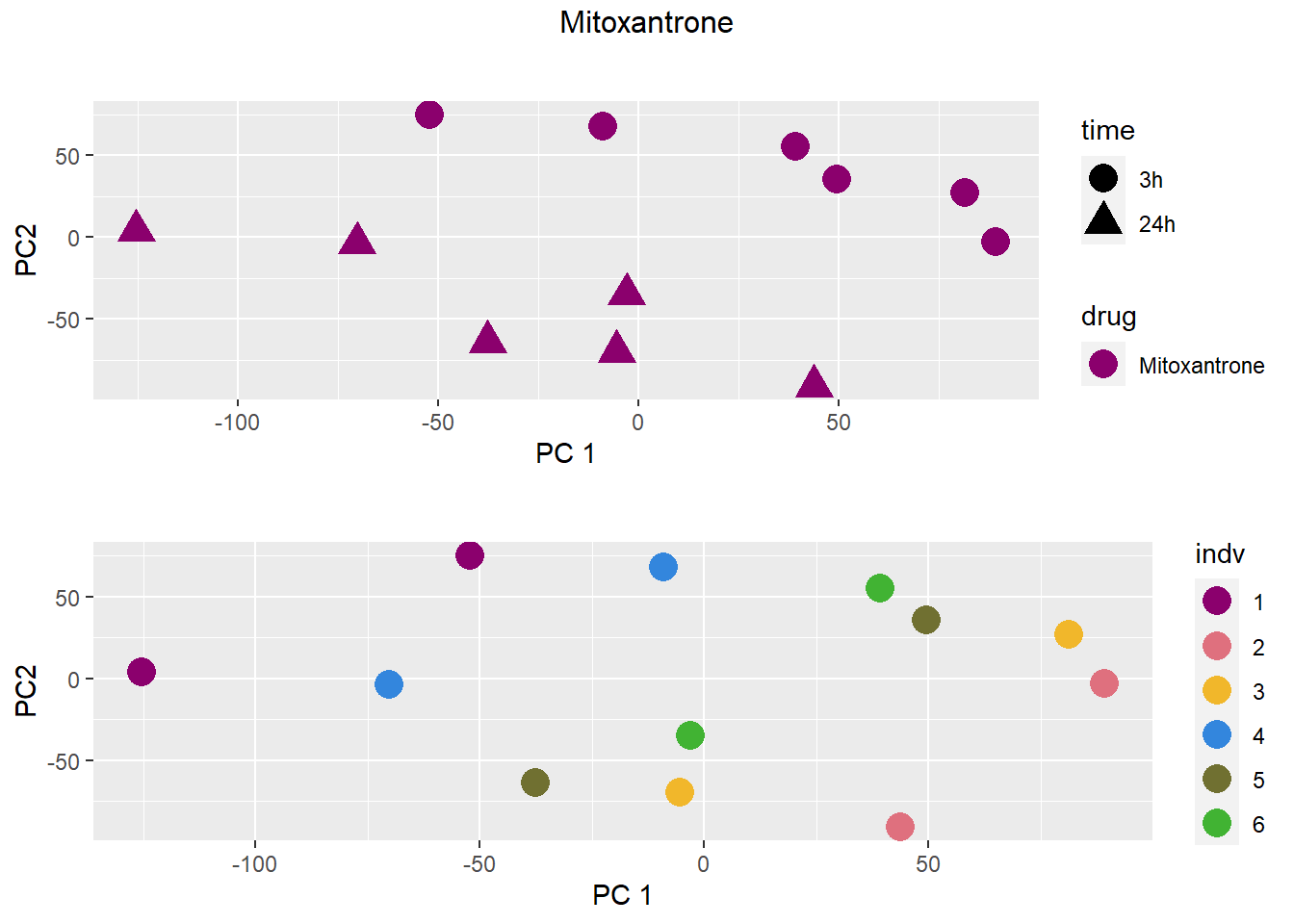
### Trastuzumab 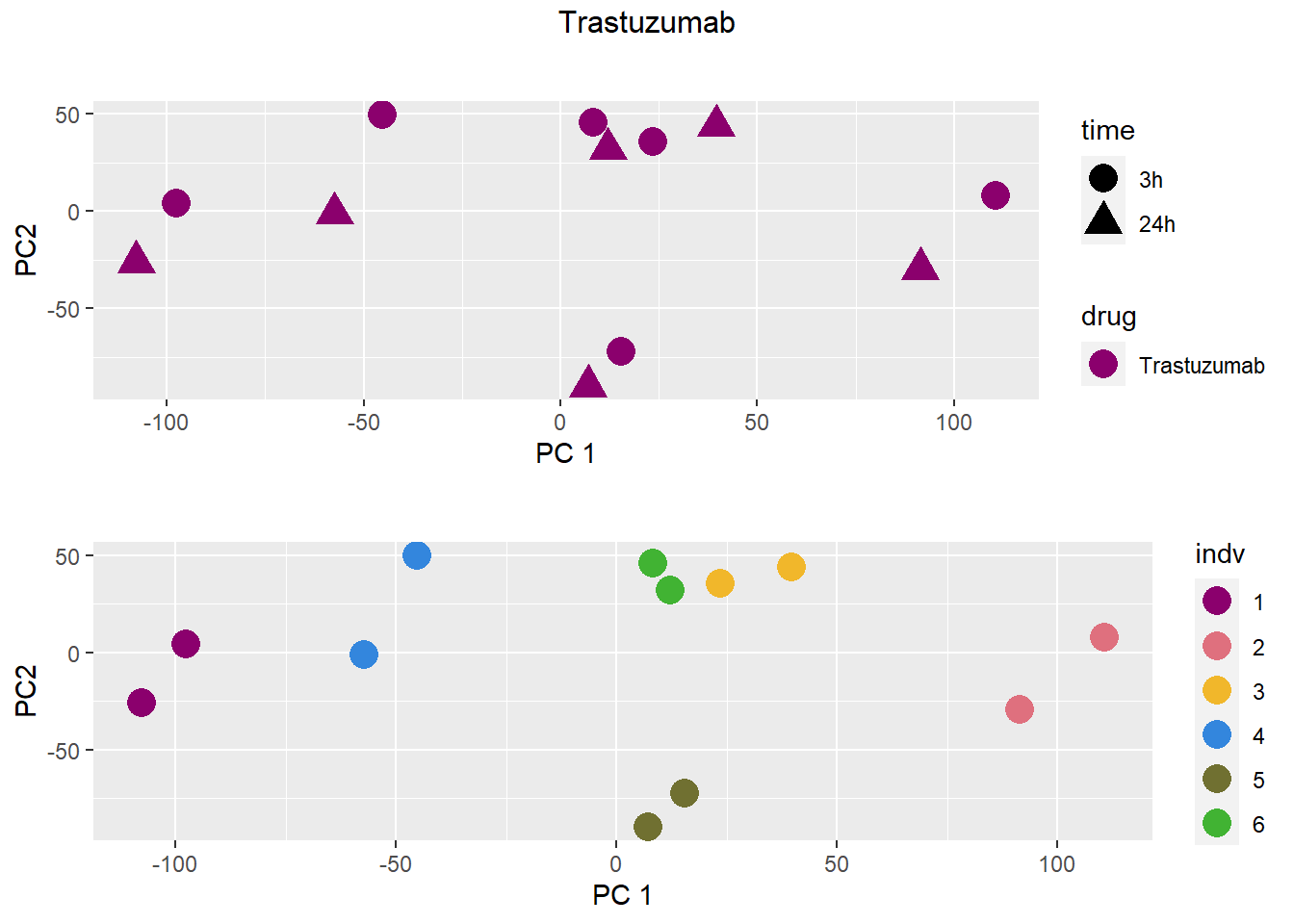
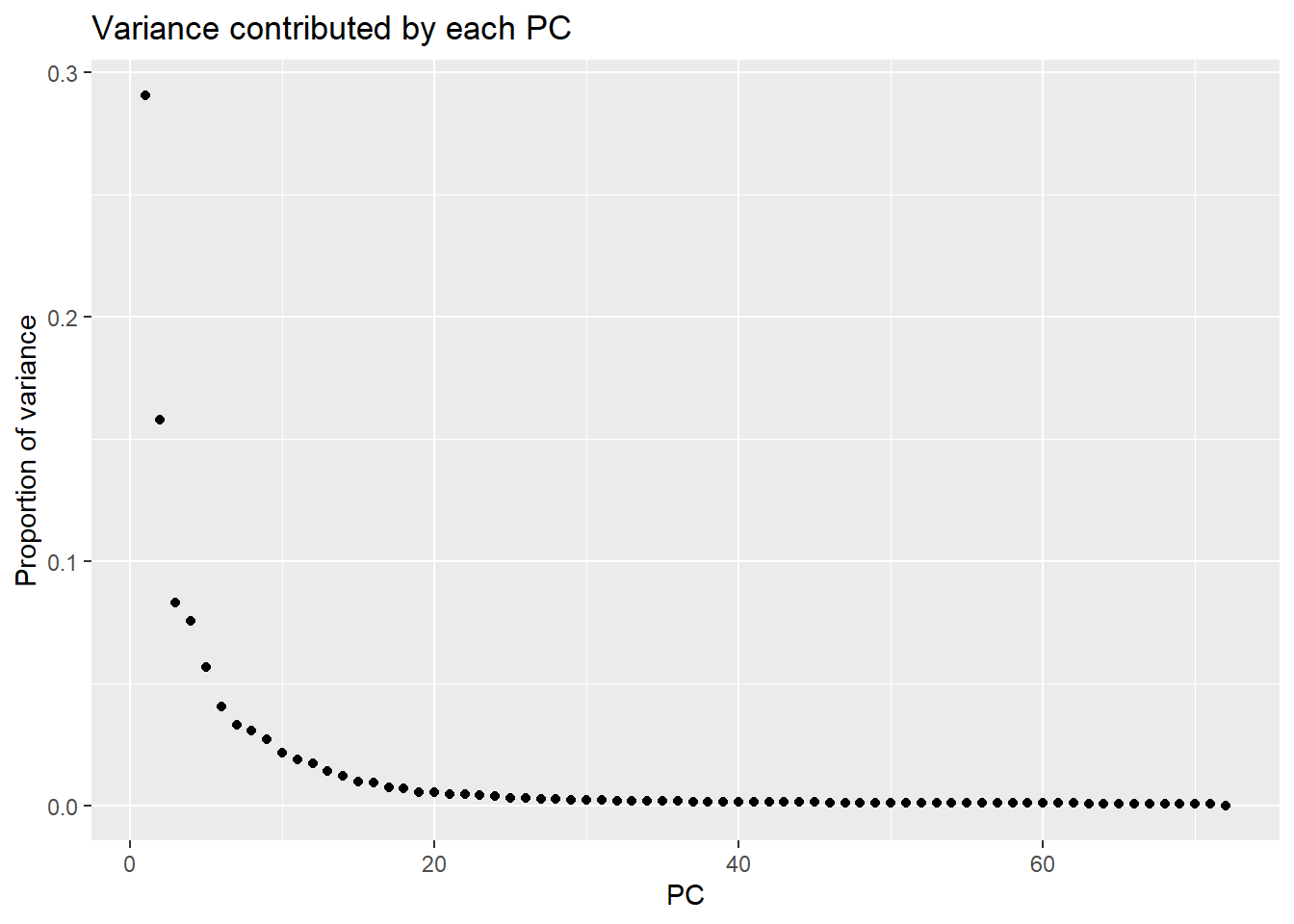
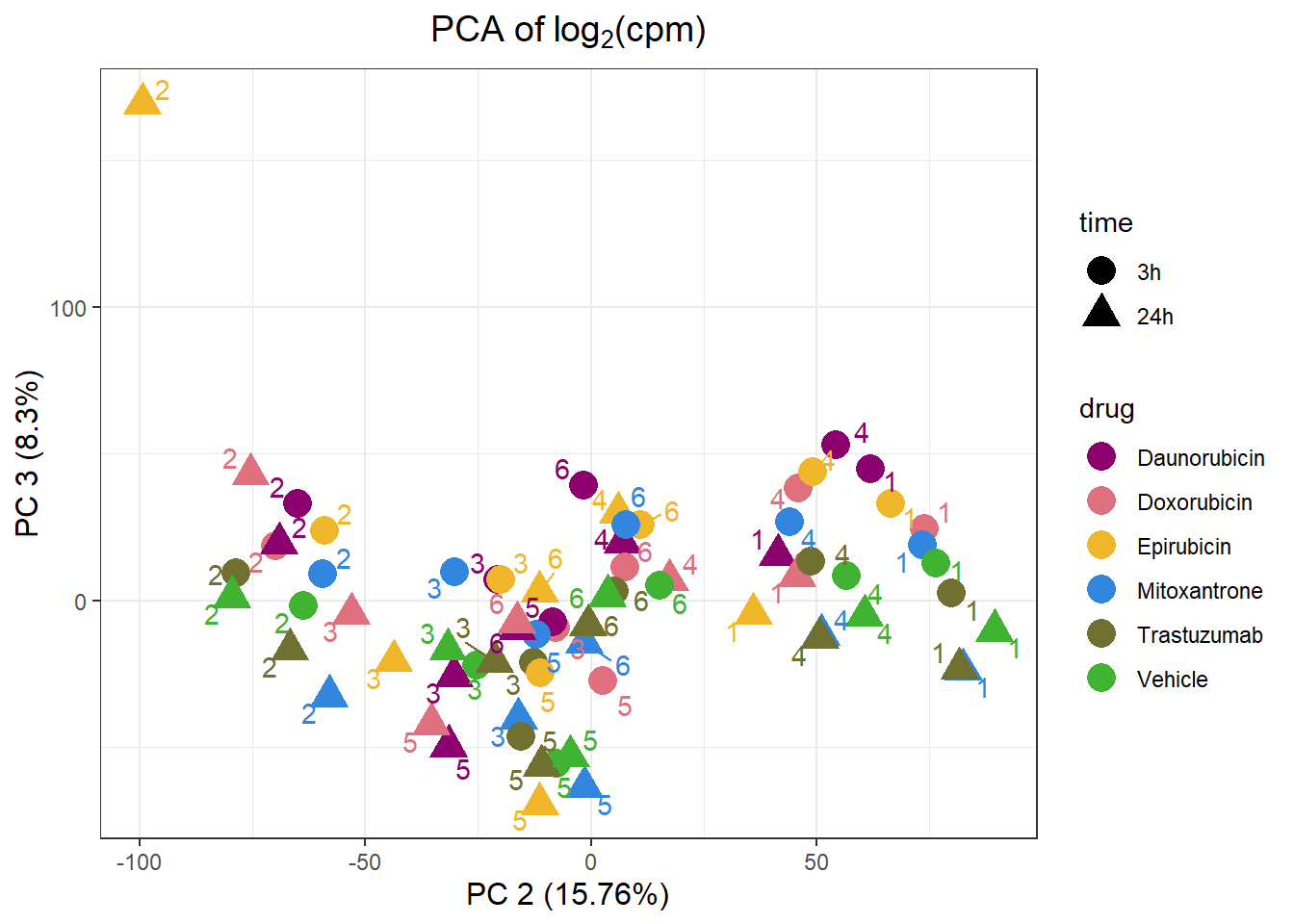
samplenames indv drug time RIN group PC1 PC2
Da.1.3h MCW_RM_R_11 1 Daunorubicin 3h 9.3 1 -18.53902 62.11914
Do.1.3h MCW_RM_R_12 1 Doxorubicin 3h 9.8 2 -12.37874 73.96424
Ep.1.3h MCW_RM_R_13 1 Epirubicin 3h 9.8 3 -11.19226 66.47554
Mi.1.3h MCW_RM_R_14 1 Mitoxantrone 3h 10 4 -10.24538 73.46900
Tr.1.3h MCW_RM_R_15 1 Trastuzumab 3h 9.6 5 -12.21578 79.97646
Ve.1.3h MCW_RM_R_16 1 Vehicle 3h 9.9 6 -15.01396 76.61739
PC3 PC4 PC5 PC6
Da.1.3h 44.793907 -4.229684 25.33474 -35.96203
Do.1.3h 24.565270 -8.522490 -19.86738 -19.00810
Ep.1.3h 33.103318 -9.146883 18.17321 -43.12911
Mi.1.3h 19.048074 -14.540036 -9.00047 -24.33769
Tr.1.3h 2.646249 -17.004856 -34.18428 -11.83222
Ve.1.3h 12.636907 -4.140009 -39.84065 -17.16481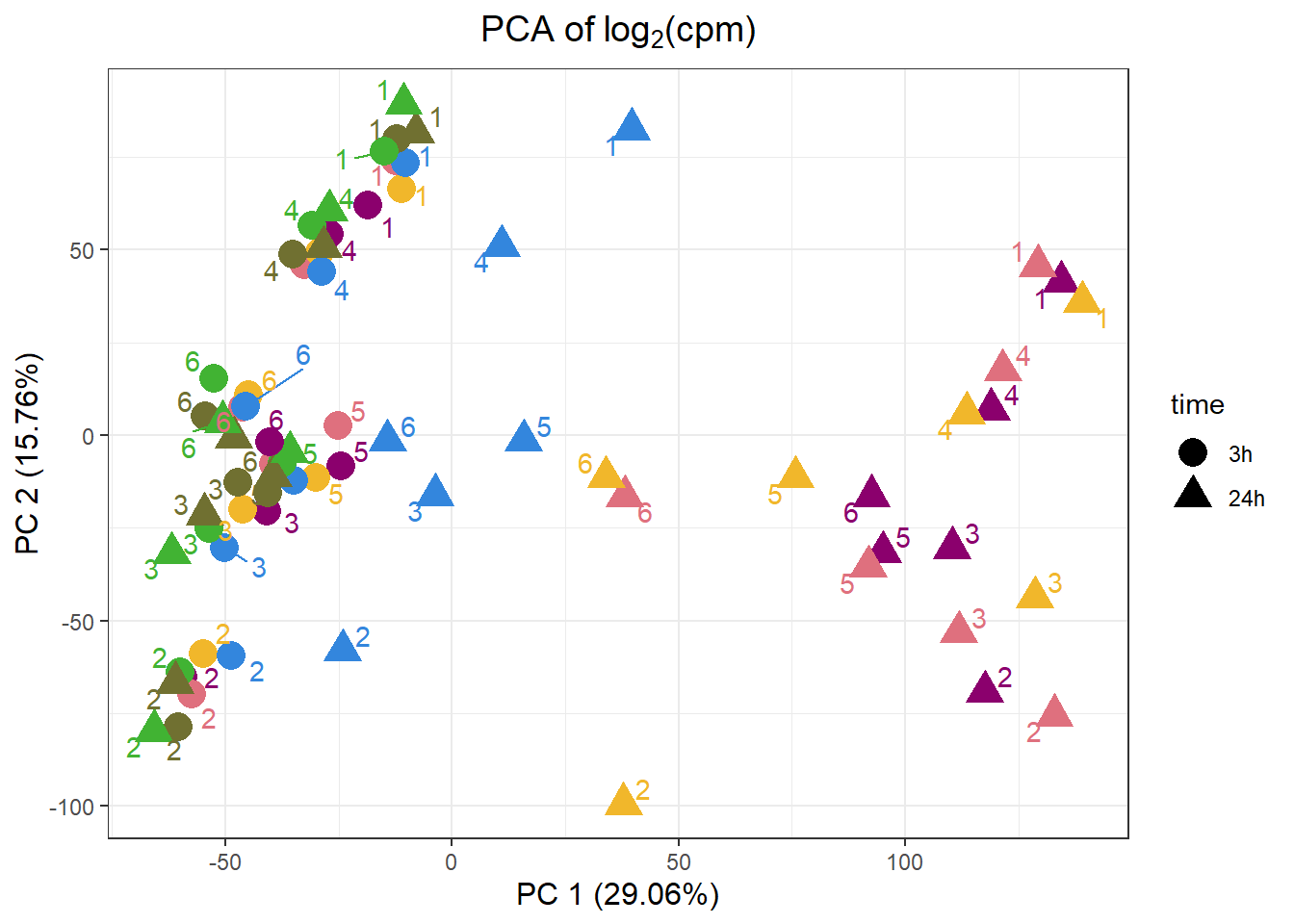
entrezgene_id ensembl_gene_id hgnc_symbol
1 88 ENSG00000077522 ACTN2
2 153 ENSG00000043591 ADRB1
3 154 ENSG00000169252 ADRB2
4 651 ENSG00000152785 BMP3
5 775 ENSG00000285479 CACNA1C
6 100874369 ENSG00000285479 CACNA1C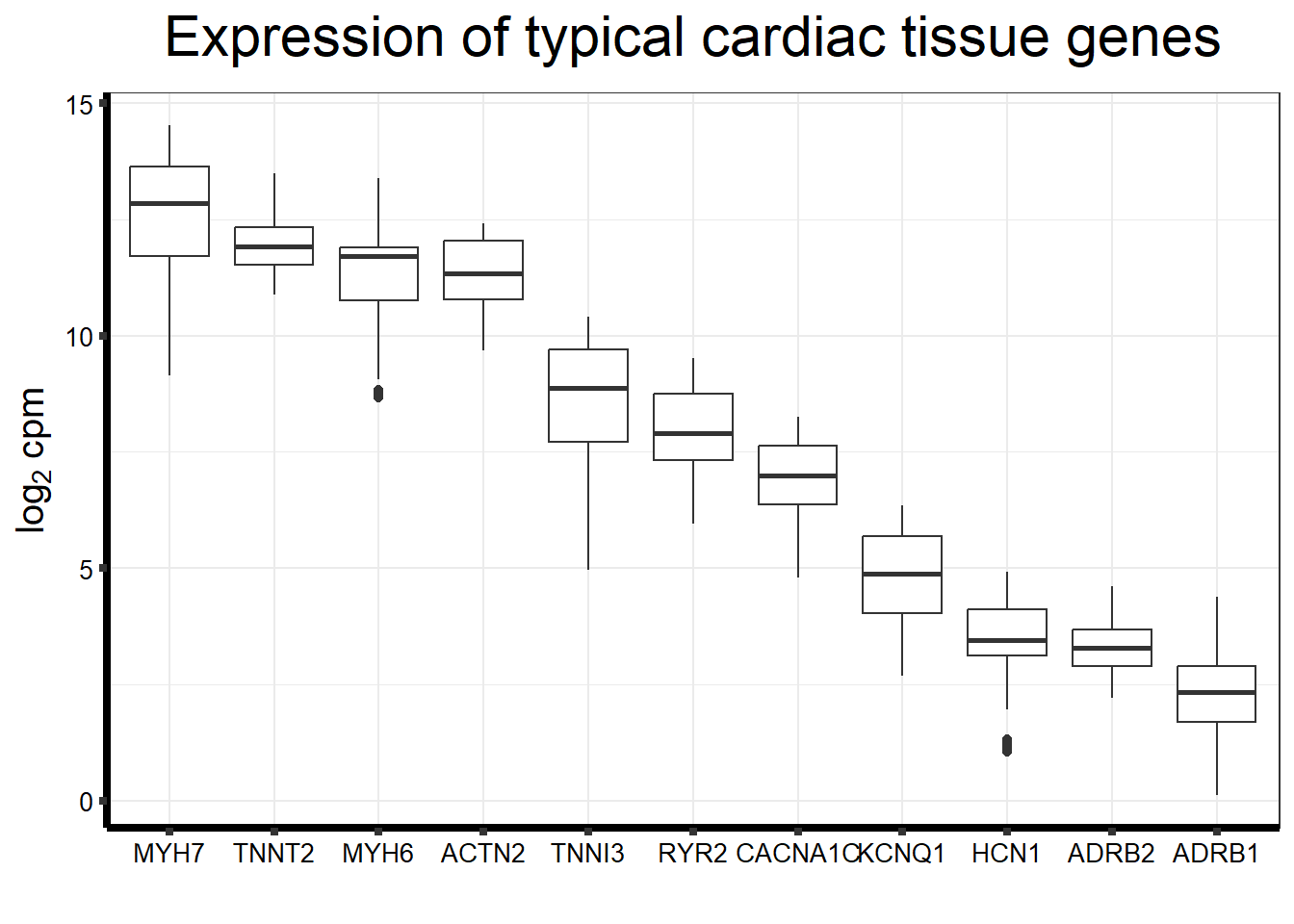
##correlation heatmap of counts matrix
R version 4.2.2 (2022-10-31 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19044)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] Hmisc_4.8-0
[2] Formula_1.2-5
[3] survival_3.5-3
[4] corrplot_0.92
[5] ggrepel_0.9.3
[6] cowplot_1.1.1
[7] Homo.sapiens_1.3.1
[8] TxDb.Hsapiens.UCSC.hg19.knownGene_3.2.2
[9] org.Hs.eg.db_3.15.0
[10] GO.db_3.15.0
[11] OrganismDbi_1.38.1
[12] GenomicFeatures_1.48.4
[13] GenomicRanges_1.48.0
[14] GenomeInfoDb_1.32.4
[15] AnnotationDbi_1.58.0
[16] IRanges_2.30.1
[17] S4Vectors_0.34.0
[18] Biobase_2.56.0
[19] biomaRt_2.52.0
[20] scales_1.2.1
[21] lubridate_1.9.2
[22] forcats_1.0.0
[23] stringr_1.5.0
[24] dplyr_1.1.0
[25] purrr_1.0.1
[26] readr_2.1.4
[27] tidyr_1.3.0
[28] tibble_3.1.8
[29] tidyverse_2.0.0
[30] AnnotationHub_3.4.0
[31] BiocFileCache_2.4.0
[32] dbplyr_2.3.1
[33] BiocGenerics_0.42.0
[34] reshape2_1.4.4
[35] gridExtra_2.3
[36] mixOmics_6.20.0
[37] ggplot2_3.4.1
[38] lattice_0.20-45
[39] MASS_7.3-58.2
[40] RColorBrewer_1.1-3
[41] edgeR_3.38.4
[42] limma_3.52.4
[43] workflowr_1.7.0
loaded via a namespace (and not attached):
[1] backports_1.4.1 plyr_1.8.8
[3] igraph_1.4.1 splines_4.2.2
[5] BiocParallel_1.30.4 digest_0.6.31
[7] htmltools_0.5.4 fansi_1.0.4
[9] checkmate_2.1.0 magrittr_2.0.3
[11] memoise_2.0.1 cluster_2.1.4
[13] tzdb_0.3.0 Biostrings_2.64.1
[15] matrixStats_0.63.0 rARPACK_0.11-0
[17] timechange_0.2.0 prettyunits_1.1.1
[19] jpeg_0.1-10 colorspace_2.1-0
[21] blob_1.2.3 rappdirs_0.3.3
[23] xfun_0.37 callr_3.7.3
[25] crayon_1.5.2 RCurl_1.98-1.10
[27] jsonlite_1.8.4 graph_1.74.0
[29] glue_1.6.2 gtable_0.3.1
[31] zlibbioc_1.42.0 XVector_0.36.0
[33] DelayedArray_0.22.0 DBI_1.1.3
[35] Rcpp_1.0.10 htmlTable_2.4.1
[37] xtable_1.8-4 progress_1.2.2
[39] foreign_0.8-84 bit_4.0.5
[41] htmlwidgets_1.6.1 httr_1.4.5
[43] ellipsis_0.3.2 farver_2.1.1
[45] pkgconfig_2.0.3 XML_3.99-0.13
[47] nnet_7.3-18 deldir_1.0-6
[49] sass_0.4.5 locfit_1.5-9.7
[51] utf8_1.2.3 labeling_0.4.2
[53] tidyselect_1.2.0 rlang_1.0.6
[55] later_1.3.0 munsell_0.5.0
[57] BiocVersion_3.15.2 tools_4.2.2
[59] cachem_1.0.7 cli_3.6.0
[61] generics_0.1.3 RSQLite_2.3.0
[63] evaluate_0.20 fastmap_1.1.1
[65] yaml_2.3.7 processx_3.8.0
[67] knitr_1.42 bit64_4.0.5
[69] fs_1.6.1 KEGGREST_1.36.3
[71] RBGL_1.72.0 whisker_0.4.1
[73] mime_0.12 xml2_1.3.3
[75] compiler_4.2.2 rstudioapi_0.14
[77] filelock_1.0.2 curl_5.0.0
[79] png_0.1-8 interactiveDisplayBase_1.34.0
[81] bslib_0.4.2 stringi_1.7.12
[83] highr_0.10 ps_1.7.2
[85] RSpectra_0.16-1 Matrix_1.5-3
[87] vctrs_0.5.2 pillar_1.8.1
[89] lifecycle_1.0.3 BiocManager_1.30.20
[91] jquerylib_0.1.4 data.table_1.14.8
[93] bitops_1.0-7 corpcor_1.6.10
[95] httpuv_1.6.9 rtracklayer_1.56.1
[97] latticeExtra_0.6-30 R6_2.5.1
[99] BiocIO_1.6.0 promises_1.2.0.1
[101] codetools_0.2-19 SummarizedExperiment_1.26.1
[103] rprojroot_2.0.3 rjson_0.2.21
[105] withr_2.5.0 GenomicAlignments_1.32.1
[107] Rsamtools_2.12.0 GenomeInfoDbData_1.2.8
[109] parallel_4.2.2 hms_1.1.2
[111] rpart_4.1.19 grid_4.2.2
[113] rmarkdown_2.20 MatrixGenerics_1.8.1
[115] git2r_0.31.0 getPass_0.2-2
[117] base64enc_0.1-3 shiny_1.7.4
[119] ellipse_0.4.3 interp_1.1-3
[121] restfulr_0.0.15