Reference Data QC: GigaMUGA
Last updated: 2022-11-08
Checks: 6 1
Knit directory: MUGA_reference_data/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of
the R Markdown file created these results, you’ll want to first commit
it to the Git repo. If you’re still working on the analysis, you can
ignore this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20221108) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 1074529. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: analysis/figure/
Ignored: data/.DS_Store
Untracked files:
Untracked: README.html
Untracked: data/GigaMUGA/FPGM_1-6_FinalReport.txt
Untracked: data/GigaMUGA/FPGM_1-6_FinalReport.zip
Untracked: data/GigaMUGA/GigaMUGA_control_genotypes.txt
Untracked: data/GigaMUGA/GigaMUGA_founder_consensus_genotypes/
Untracked: data/GigaMUGA/GigaMUGA_founder_sample_concordance/
Untracked: data/GigaMUGA/GigaMUGA_founder_sample_dendrogram_genos/
Untracked: data/GigaMUGA/GigaMUGA_founder_sample_genotypes/
Untracked: data/GigaMUGA/GigaMUGA_reference_genotypes/
Untracked: data/GigaMUGA/Marker_QC.RData
Untracked: data/GigaMUGA/bad_samples_markers.RData
Untracked: data/GigaMUGA/sex_check_results.RData
Untracked: output/GigaMUGA/
Unstaged changes:
Modified: analysis/GigaMUGA_Reference_QC.Rmd
Modified: analysis/MUGA_Reference_QC.Rmd
Modified: analysis/MegaMUGA_Reference_QC.Rmd
Deleted: code/GigaMUGA_Reference_QC.Rmd
Deleted: code/GigaMUGA_Reference_QC.html
Deleted: code/MUGA_Reference_QC.Rmd
Deleted: code/MUGA_Reference_QC.html
Deleted: code/MegaMUGA_Reference_QC.Rmd
Deleted: code/MegaMUGA_Reference_QC.html
Modified: data/GigaMUGA/GigaMUGA_BadSamples_BadMarkers.RData
Modified: data/GigaMUGA/GigaMUGA_QC_Results.RData
Modified: data/GigaMUGA/GigaMUGA_SexCheck_Results.RData
Modified: data/GigaMUGA/gm_uwisc_v2.csv
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/GigaMUGA_Reference_QC.Rmd)
and HTML (docs/GigaMUGA_Reference_QC.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 1074529 | sam-widmayer | 2022-11-08 | update markdown formatting for workflowr |
| Rmd | efe8849 | sam-widmayer | 2022-11-08 | initialize workflowr integration |
GigaMUGA Annotations
Loading in data from Sumner analyses
load("data/GigaMUGA/GigaMUGA_QC_Results.RData")
load("data/GigaMUGA/GigaMUGA_SexCheck_Results.RData")
load("data/GigaMUGA/GigaMUGA_BadSamples_BadMarkers.RData")
gm_metadata <- vroom::vroom("data/GigaMUGA/gm_uwisc_v2.csv")Marker QC: Searching for missing genotype calls
no.calls <- control_allele_freqs_df %>%
dplyr::ungroup() %>%
dplyr::filter(genotype == "N") %>%
tidyr::pivot_wider(names_from = genotype,
values_from = n) %>%
dplyr::left_join(., gm_metadata) %>%
dplyr::select(marker, chr, bp_grcm39, freq) %>%
dplyr::mutate(chr = as.factor(chr))
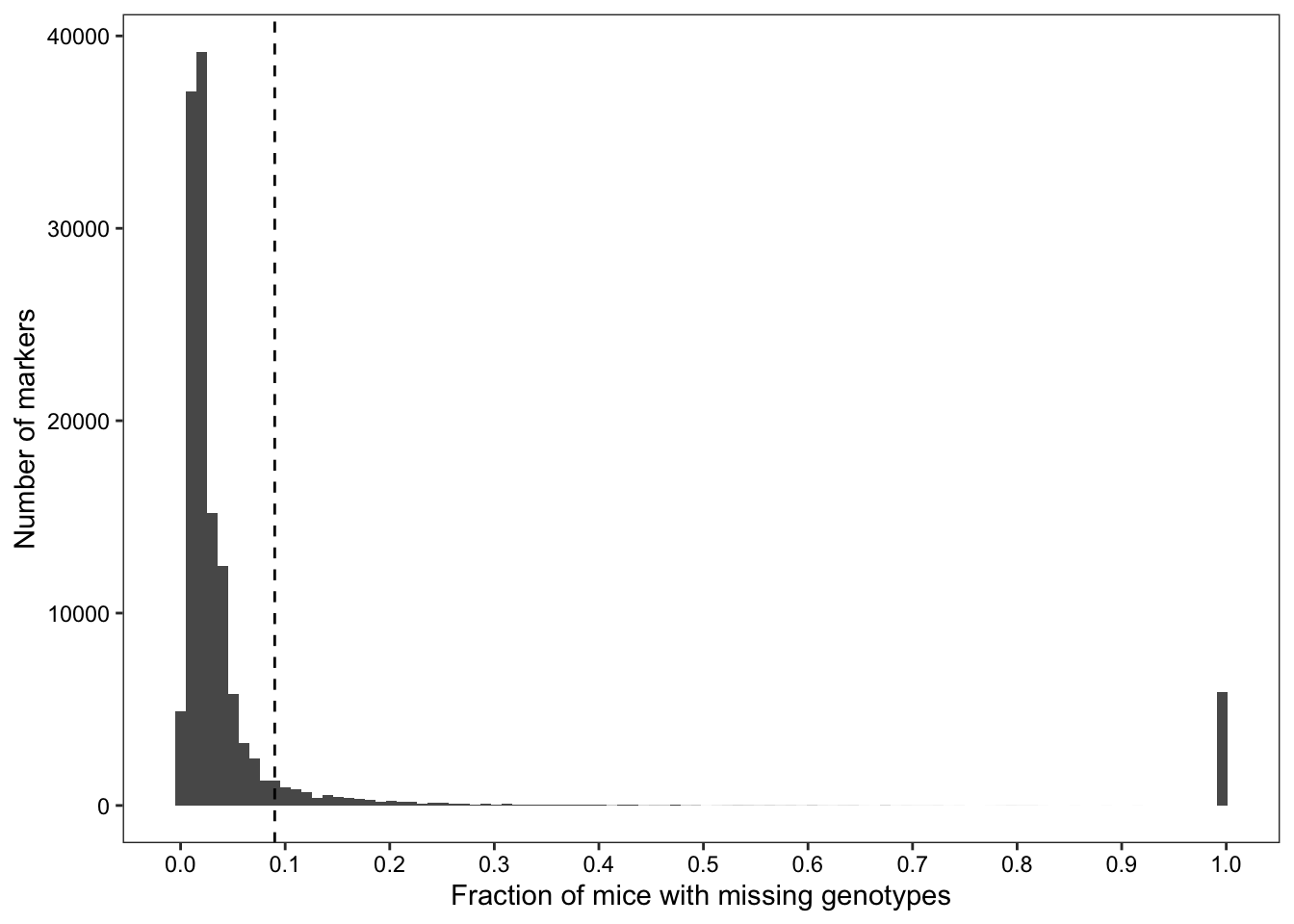
## Identifying markers with missing genotypes at a frequency higher than the 95th percentile of "N" frequencies across all markers
cutoff <- quantile(no.calls$freq, probs = seq(0,1,0.05))[[19]]Of 137337 markers, 135944 failed to genotype at least one sample, and 13348 markers failed to genotype at least 9% of samples.
Sample QC
Searching for samples with poor marker representation
In a similar fashion, we calculated the number of reference samples with missing genotypes. Repeated observations of samples/strains with identical names meant that genotype counts for each marker among them couldn’t be grouped and tallied, so determining no-call frequency occurred column-wise. Mouse over individual samples to see the number of markers with missing genotypes for each sample.
## Interactive plot of the number of missing genotypes for each sample.
bad_sample_cutoff <- quantile(n.calls.strains.df$n.no.calls, probs = seq(0,1,0.05))[20]
sampleQC <- ggplot(n.calls.strains.df,
mapping = aes(x = reorder(sample_id,n.no.calls),
y = n.no.calls,
text = paste("Sample:", sample_id))) +
geom_point() +
QCtheme +
theme(axis.text.x = element_blank(),
axis.ticks.x = element_blank()) +
geom_hline(yintercept = bad_sample_cutoff, linetype = 2) +
labs(x = "Number of mice with missing genotypes",
y = "Number of markers")
ggplotly(sampleQC, tooltip = c("text","y"))Warning: `gather_()` was deprecated in tidyr 1.2.0.
Please use `gather()` instead.Validating sex of reference samples
We next validated the sexes of each sample using sex chromosome probe intensities. We paired up probe intensities, joined available metadata, and filtered down to only markers covering the X and Y chromosomes. Then we flagged markers with high missingness across all samples, as well as samples with high missingness among all markers.The first round of inferring predicted sexes used a rough search of the sample name for expected nomenclature convention, which includes a sex denotation.
## Tabulating the number of samples for each predicted sex
predicted.sex.table <- predicted.sexes %>%
dplyr::group_by(predicted.sex) %>%
dplyr::count() This captured 86 female samples, 130 male samples, leaving 63 samples of unknown predicted sex from nomenclature alone.
After predicting the sexes of the vast majority of reference samples, we visualized the average probe intensity among X Chromosome markers for each sample, labeling them by predicted sex. Samples colored black were unabled to have their sex inferred by the sample name, but cluster well with mice for which sex could be inferred. Conversely, some samples’ predicted sex is discordant with X and Y Chromosome marker intensities (i.e. blue samples that cluster with mostly orange samples, and vice versa). Mouse over individual dots to view the sample, as well as whether it was flagged for having many markers with missing genotype information. In many cases, pulling substrings of sample names as the sex of the sample was too sensitive and misclassified samples.
predicted.sex.plot.palettes <- sex.chr.intensities %>%
dplyr::ungroup() %>%
dplyr::distinct(sample_id, predicted.sex, high_missing_sample) %>%
dplyr::mutate(predicted.sex.palette = dplyr::case_when(predicted.sex == "f" ~ "#5856b7",
predicted.sex == "m" ~ "#eeb868",
predicted.sex == "unknown" ~ "black"))
predicted.sex.palette <- predicted.sex.plot.palettes$predicted.sex.palette
names(predicted.sex.palette) <- predicted.sex.plot.palettes$predicted.sex
mean.x.intensities.by.sex.plot <- sex.chr.intensities %>%
dplyr::mutate(high_missing_sample = dplyr::if_else(high_missing_sample == "FLAG",
true = "Bad Samples",
false = "Good Samples")) %>%
ggplot(., mapping = aes(x = mean.x.chr.int,
y = mean.y.int,
colour = predicted.sex,
text = sample_id)) +
geom_point() +
scale_colour_manual(values = predicted.sex.palette) +
facet_grid(.~high_missing_sample) +
QCtheme
ggplotly(mean.x.intensities.by.sex.plot,
tooltip = c("text"))Because the split between inferred sexes of good samples was so distinct, we used k-means clustering to quickly match the clusters to sexed samples and assign or re-assign sexes to samples with unknown or apparently incorrect sex information, respectively. Samples highlighted above were also re-evaluated using strain-specific marker information.
# Prints a table of all samples with an option to view whether a sample had its sex redesignated.
reSexed_samples_table <- reSexed_samples %>%
dplyr::mutate(resexed = predicted.sex != inferred.sex)
reSexed_samples_table %>%
dplyr::select(sample_id, resexed, predicted.sex, inferred.sex) %>%
DT::datatable(., filter = "top",
escape = FALSE)The plot below demonstrates that this clustering technique does a pretty good job at capturing the information we want. Moving forward with sample QC we used the reassigned inferred sexes of the samples.
# Interactive scatter plot of intensities similar to above, but recolors and outlines samples based on redesignated sexes.
reSexed.plot <- ggplot(reSexed_samples_table %>%
dplyr::arrange(inferred.sex),
mapping = aes(x = mean.x.chr.int,
y = mean.y.int,
fill = inferred.sex,
colour = predicted.sex,
text = sample_id,
label = resexed)) +
geom_point(shape = 21,size = 3, alpha = 0.7) +
scale_colour_manual(values = predicted.sex.palette) +
scale_fill_manual(values = predicted.sex.palette) +
QCtheme
ggplotly(reSexed.plot,
tooltip = c("text","label"))Visualizing the corrected sex assignments, we can see two samples that stand out of the clusters: FAM and JAXMP_cookii_TH_BC19281. Because the X and Y probe intensities were supplied to k-means clustering independently in this datasest and the values for each channel were intermediate in this sample, FAM was predicted as female using the y channel and male using the x channel data. Because FAM is not a CC/DO founder or a sample representative of non-musculus species, we assigned this sample to the “unknown” sex category. JAXMP_cookii_TH_BC19281 clustered with both sexes in a similar fashion, but because of genetic divergence between its genome and the mouse reference genome from which probes were designed, the irregular probe intensity data is not unexpected. For this reason, and the Y channel data being much more consistent with female sample, we categorized this sample as female.
Validating reference sample genetic backgrounds
A key component of sample QC for our purposes is knowing that markers that we expect to deliver the consensus genotype (i.e. in a cross) actually provide us the correct strain information and allow us to correctly infer haplotypes.
# Count up samples for each founder and resulting cross and display a table
# Dam names = row names; Sire name = column names
founder_sample_table <- founderSamples %>%
dplyr::group_by(dam,sire) %>%
dplyr::count() %>%
tidyr::pivot_wider(names_from = sire, values_from = n)
DT::datatable(founder_sample_table, escape = FALSE,
options = list(columnDefs = list(list(width = '20%', targets = c(8)))))From the table, we can see that all possible pairwise combinations of CC/DO founder strains are represented, with the exception of (NZO/HILtJxCAST/EiJ)F1 and (NZO/HILtJxPWK/PhJ)F1. These missing samples could be interesting; these two crosses have been previously noted as “reproductively incompatible” in the literature. We constructed a rough dendrogram from good marker genotypes to determine whether samples cluster according to known relationships among founder strains. Edge colors represent rough clustering into six groups - three of which contain samples derived from wild-derived founder strains and their F1 hybrids with other founder strains.
recoded_wide_geno_files <- list.files("data/GigaMUGA/GigaMUGA_founder_sample_dendrogram_genos//", pattern = "gm_widegenos_chr")
recoded_wide_sample_genos_list <- list()
for(i in 1:length(recoded_wide_geno_files)){
recoded_wide_sample_genos_list[[i]] <- fst::read.fst(path = paste0("data/GigaMUGA/GigaMUGA_founder_sample_dendrogram_genos/",recoded_wide_geno_files[[i]]))
}
recoded_wide_sample_genos <- Reduce(dplyr::bind_cols,recoded_wide_sample_genos_list)
sample_id_cols <- recoded_wide_sample_genos %>%
dplyr::select(contains("sample_id"))
recoded_wide_sample_genos_nosamps <- recoded_wide_sample_genos %>%
dplyr::select(-contains("sample_id"))
rownames(recoded_wide_sample_genos_nosamps) <- sample_id_cols[,1]
# Scaling the genotype matrix, then calculating euclidean distance between all samples
dd <- dist(scale(recoded_wide_sample_genos_nosamps), method = "euclidean")
hc <- hclust(dd, method = "ward.D2")
# Plotting sample distances as a dendrogram
dend_colors = c("slateblue", # classical strains
"blue",
qtl2::CCcolors[6:8]) # official colors for wild-derived CC/DO founder strains
clus8 = cutree(hc, 5)
plot(as.phylo(hc), type = "f", cex = 0.5, tip.color = dend_colors[clus8],
no.margin = T, label.offset = 1, edge.width = 0.5)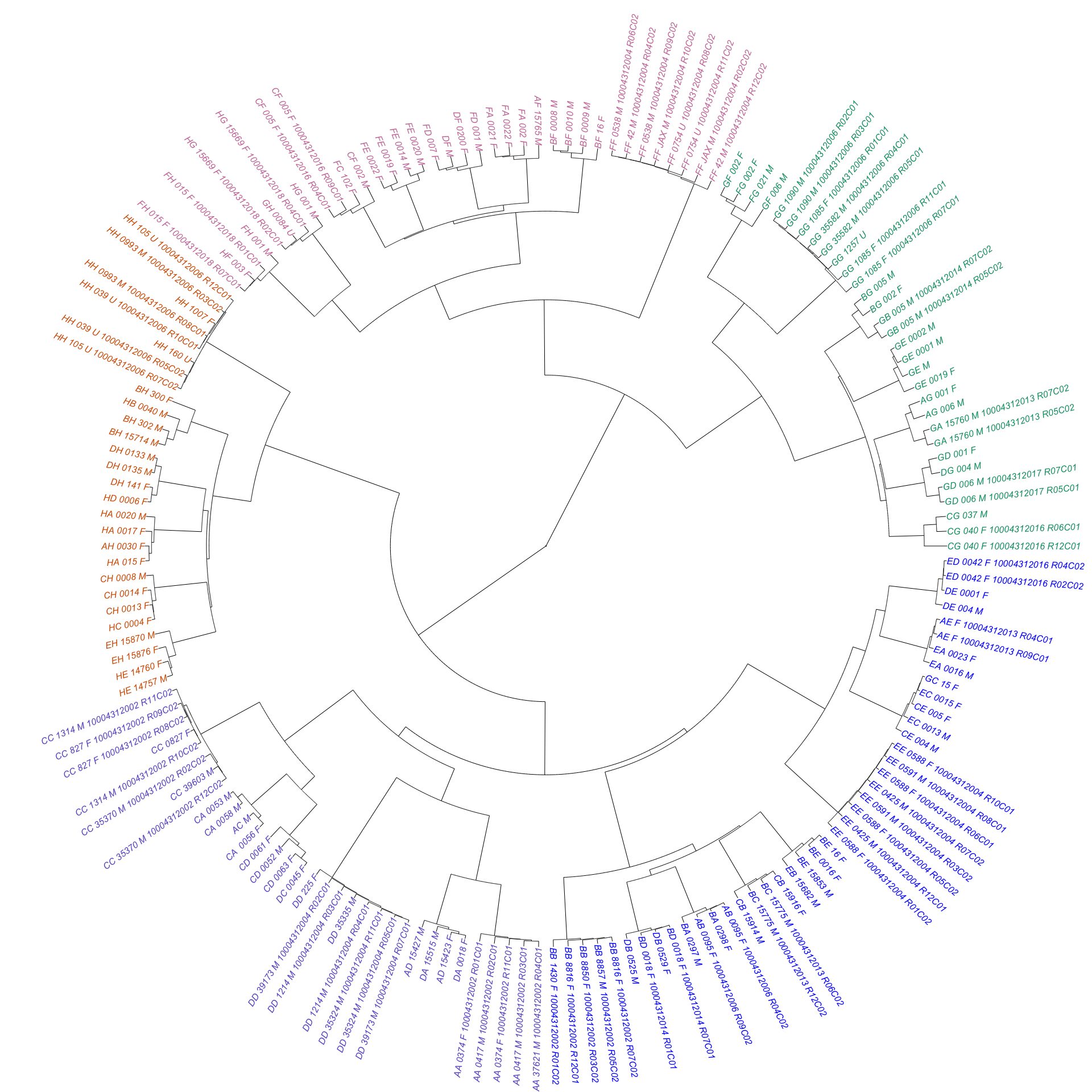
From here we curated a set of genotypes for each CC/DO founder strain that were fixed across replicate samples from that strain. The genotypes for intersecting markers between two strains were combined (“crossed”) to form predicted genotypes for each F1 hybrid of CC/DO founders. Then, the genotypes of each CC/DO founder F1 hybrid were compared directly to what was predicted, and the concordance shown below is the proportion of markers of each individual that match this prediction. In order to generate a consensus call for each CC/DO founder, we had to determine whether individual samples for a given founder were correctly typed and, if the marker failed, eliminate it so that consensus could be formed. Below is an example of what that might look like for an individual marker. Note how when at least one sample for a given strain is not assigned one of the two expected alleles, the consensus is an “NA”.
chr_concordance_files <- list.files("data/GigaMUGA/GigaMUGA_founder_sample_concordance/")
chr_concordance_list <- list()
for(i in 1:length(chr_concordance_files)){
chr_concordance_list[[i]] <- fst::read.fst(path = paste0("data/GigaMUGA/GigaMUGA_founder_sample_concordance/",chr_concordance_files[[i]]))
}
founder_concordance_df <- Reduce(dplyr::full_join, chr_concordance_list)
founder_concordance_df_3 <- founder_concordance_df %>%
dplyr::group_by(sample, alt_chr, dam, sire, inferred.sex) %>%
dplyr::summarise(total.MATCH = sum(MATCH),
total.NO_MATCH = sum(`NO MATCH`),
concordance = total.MATCH/(total.MATCH+total.NO_MATCH))
auto_concordance <- founder_concordance_df_3 %>%
dplyr::filter(alt_chr == "Autosome")
X_concordance <- founder_concordance_df_3 %>%
dplyr::filter(alt_chr == "X")
founder_palette <- qtl2::CCcolors
names(founder_palette) <- unique(founderSamples$dam)
auto_concordance_plot <- ggplot(auto_concordance, mapping = aes(x = dam,
y = concordance,
color = dam,
fill = sire,
label = sample,
text = concordance)) +
geom_jitter(shape = 21, width = 0.25) +
scale_fill_manual(values = founder_palette) +
scale_colour_manual(values = founder_palette) +
# ylim(c(0.5,1.05)) +
labs(x = "Dam Founder Strain",
y = "Autosomal Genotype Concordance") +
facet_grid(.~inferred.sex) +
QCtheme +
theme(axis.text.x = element_text(angle = 45, hjust = 1, vjust = 1))
ggplotly(auto_concordance_plot, tooltip = c("label", "text"))X_concordance_plot <- ggplot(X_concordance, mapping = aes(x = dam,
y = concordance,
color = dam,
fill = sire,
label = sample,
text = concordance)) +
geom_jitter(shape = 21, width = 0.25) +
scale_fill_manual(values = founder_palette) +
scale_colour_manual(values = founder_palette) +
# ylim(c(0.5,1.05)) +
labs(x = "Dam Founder Strain",
y = "X Chromosome Genotype Concordance") +
facet_grid(.~inferred.sex) +
QCtheme +
theme(axis.text.x = element_text(angle = 45, hjust = 1, vjust = 1))
ggplotly(X_concordance_plot, tooltip = c("label","text"))Writing reference files
The list of output file types following QC is as follows:
Genotype file; rows = markers, columns = samples
Probe intensity file; rows = markers, columns = samples
Sample metadata; columns = sample, strain, sex
Each file type is generated for:
All good samples
Eight CC/DO founder strains
bad_samples <- high.n.samples %>%
dplyr::filter(!sample_id %in% c("JAXMP_caroli_UNK_BC21325",
"JAXMP_minu_KE_BC24327",
"JAXMP_minu_KE_BC24339",
"JAXMP_pahari_TH_BC21314"))
#############################################
## Write Reference Genotypes - All Samples ##
#############################################
founder_sample_geno_files <- c(as.list(list.files(path = "data/GigaMUGA/GigaMUGA_founder_sample_genotypes/", pattern = "gm_founder_genos_chr_")[1:19]),
"M",
"NA",
list.files(path = "data/GigaMUGA/GigaMUGA_founder_sample_genotypes/", pattern = "gm_founder_genos_chr_")[20],
"Y")
nonfounder_sample_geno_files <- as.list(list.files(path = "data/GigaMUGA/GigaMUGA_reference_genotypes/", pattern = "gm_genos_chr_"))
future::plan(multisession, workers = 16)
make_chunks <- furrr:::make_chunks
make_chunks(n_x = length(founder_sample_geno_files), n_workers = 16)[[1]]
[1] 1 2
[[2]]
[1] 3
[[3]]
[1] 4 5
[[4]]
[1] 6
[[5]]
[1] 7
[[6]]
[1] 8 9
[[7]]
[1] 10
[[8]]
[1] 11
[[9]]
[1] 12 13
[[10]]
[1] 14
[[11]]
[1] 15 16
[[12]]
[1] 17
[[13]]
[1] 18
[[14]]
[1] 19 20
[[15]]
[1] 21
[[16]]
[1] 22 23reference_genos_list <- furrr::future_map2(.x = founder_sample_geno_files,
.y = nonfounder_sample_geno_files,
.f = writeReferenceGenotypes,
.options = furrr_options(seed = TRUE)) %>%
purrr::discard(., is.character)[1] "NO REFERENCE GENOTYPES"
[1] "NO REFERENCE GENOTYPES"# All reference genotypes
reference_genos_df <- Reduce(dplyr::bind_rows, reference_genos_list)
###############################################
## Write Reference Intensities - All Samples ##
###############################################
reference_intensities_list <- furrr::future_map(.x = nonfounder_sample_geno_files,
.f = writeReferenceIntensities,
.options = furrr_options(seed = TRUE))[1] "NO REFERENCE INTENSITIES"
[1] "NO REFERENCE INTENSITIES"reference_X_intensities_df <- purrr::transpose(reference_intensities_list)[[1]] %>%
purrr::keep(., is.data.frame) %>%
Reduce(dplyr::bind_rows, .)Warning: Element 21 must be length 2, not 1Warning: Element 23 must be length 2, not 1reference_Y_intensities_df <- purrr::transpose(reference_intensities_list)[[2]] %>%
purrr::keep(., is.data.frame) %>%
Reduce(dplyr::bind_rows, .)Warning: Element 21 must be length 2, not 1
Warning: Element 23 must be length 2, not 1#################################################
## Write Consensus Genotypes - Founder Strains ##
#################################################
founder_consensus_files <- list.files(path = "data/GigaMUGA/GigaMUGA_founder_consensus_genotypes/", pattern = "gm_founder_consensus_chr_")
founder_consensus_genos <- furrr::future_map(.x = founder_consensus_files,
.f = writeFounderConsensusGenotypes,
.options = furrr_options(seed = TRUE)) %>%
Reduce(dplyr::bind_rows, .)
####################################
## Write Founder Mean Intensities ##
####################################
founder_samples_strains <- founderSamples %>%
dplyr::filter(bg == "INBRED") %>%
dplyr::distinct(sample_id, dam) %>%
dplyr::rename(strain = dam) # if inbred, dam == strain
founder_mean_x_ints <- reference_X_intensities_df %>%
dplyr::select(marker,founder_samples_strains$sample_id) %>%
tidyr::pivot_longer(-marker, names_to = "sample_id", values_to = "X") %>%
dplyr::left_join(., founder_samples_strains) %>%
dplyr::filter(!is.na(X)) %>%
dplyr::group_by(marker, strain) %>%
dplyr::summarise(mean_x = mean(X)) %>%
tidyr::pivot_wider(names_from = strain, values_from = mean_x) %>%
dplyr::filter(marker %in% founder_consensus_genos$marker)
founder_mean_y_ints <- reference_Y_intensities_df %>%
dplyr::select(marker,founder_samples_strains$sample_id) %>%
tidyr::pivot_longer(-marker, names_to = "sample_id", values_to = "Y") %>%
dplyr::left_join(., founder_samples_strains) %>%
dplyr::filter(!is.na(Y)) %>%
dplyr::group_by(marker, strain) %>%
dplyr::summarise(mean_y = mean(Y)) %>%
tidyr::pivot_wider(names_from = strain, values_from = mean_y) %>%
dplyr::filter(marker %in% founder_consensus_genos$marker)
###################################
## Write Founder Sample Metadata ##
###################################
founder_metadata <- founderSamples %>%
dplyr::select(-bg) %>%
dplyr::rename(original_sex = predicted.sex,
correct_sex = inferred.sex,
sample = sample_id) %>%
dplyr::mutate(strain = dplyr::if_else(dam != sire, true = paste0("(",dam,"x",sire,")F1"), false = dam)) %>%
dplyr::mutate(dam = case_when(dam == "A/J" ~ "A",
dam == "C57BL/6J" ~ "B",
dam == "129S1/SvImJ" ~ "C",
dam == "NOD/ShiLtJ" ~ "D",
dam == "NZO/HILtJ" ~ "E",
dam == "CAST/EiJ" ~ "F",
dam == "PWK/PhJ" ~ "G",
dam == "WSB/EiJ" ~ "H"),
sire = case_when(sire == "A/J" ~ "A",
sire == "C57BL/6J" ~ "B",
sire == "129S1/SvImJ" ~ "C",
sire == "NOD/ShiLtJ" ~ "D",
sire == "NZO/HILtJ" ~ "E",
sire == "CAST/EiJ" ~ "F",
sire == "PWK/PhJ" ~ "G",
sire == "WSB/EiJ" ~ "H")) %>%
tidyr::unite("letter", dam:sire, sep = "") %>%
dplyr::ungroup() %>%
dplyr::select(sample, strain, correct_sex, original_sex, letter)
reference_meta_nostrain <- predicted.sexes %>%
dplyr::filter(sample_id %in% colnames(reference_genos_df)[-1]) %>%
dplyr::left_join(., reSexed_samples %>%
dplyr::select(sample_id, inferred.sex)) %>%
dplyr::rename(sample = sample_id,
original_sex = predicted.sex,
correct_sex = inferred.sex) %>%
dplyr::left_join(., founder_metadata %>%
dplyr::select(sample, strain))
unassigned_strains <- reference_meta_nostrain %>%
dplyr::filter(is.na(strain))
new_strains <- purrr::map(unassigned_strains$sample, findStrain) %>%
unlist()
unassigned_strains$strain <- new_strains
reference_sample_metadata <- unassigned_strains %>%
dplyr::mutate(correct_sex = dplyr::if_else(is.na(correct_sex), true = "unknown", false = correct_sex),
mouse.id.1 = stringr::str_sub(strain, -1),
strain = dplyr::if_else(mouse.id.1 %in% c("m","f"),
true = str_sub(string = strain, 1, nchar(strain)-1),
false = strain)) %>%
dplyr::select(-mouse.id.1) %>%
dplyr::mutate(strain = dplyr::case_when(strain == "JAXMP_do" ~ "JAXMP_dom",
strain == "JAXMP_" ~ "JAXMP_minu",
strain == "PANCEVO/EiJ31581" ~ "PANCEVO/EiJ",
str_detect(string = strain, pattern = "MWN") ~ "MWN",
str_detect(string = strain, pattern = "PAE") ~ "PAE",
str_detect(string = strain, pattern = "JAXMP_pahari") ~ "JAXMP_pahari",
str_detect(string = strain, pattern = "JAXMP_spt") ~ "JAXMP_spt",
str_detect(string = strain, pattern = "JAXMP_cooki") ~ "JAXMP_cooki",
str_detect(string = strain, pattern = "JAXMP_cptak") ~ "JAXMP_cptak",
str_detect(string = strain, pattern = "JAXMP_caroli") ~ "JAXMP_caroli",
str_detect(string = strain, pattern = "JAXMP_hort") ~ "JAXMP_hort",
TRUE ~ strain)) %>%
dplyr::bind_rows(.,reference_meta_nostrain %>%
dplyr::filter(!sample %in% unassigned_strains$sample)) %>%
dplyr::filter(sample %in% colnames(reference_genos_df)[-1]) %>%
dplyr::mutate(correct_sex = dplyr::case_when(sample == "FAM" ~ "unknown",
sample == "JAXMP_cookii_TH_BC19281" ~ "female",
TRUE ~ correct_sex)) %>%
dplyr::distinct()
###################
## WRITING FILES ##
###################
if(file.exists("output/GigaMUGA/GigaMUGA_genotypes.csv.gz") == "FALSE"){
dir.create("output/GigaMUGA")
write.csv(reference_genos_df, file = "output/GigaMUGA/GigaMUGA_genotypes.csv")
system("gzip output/GigaMUGA/GigaMUGA_genotypes.csv")
}
if(file.exists("output/GigaMUGA/GigaMUGA_x_intensities.csv.gz") == "FALSE"){
write.csv(reference_X_intensities_df, file = "output/GigaMUGA/GigaMUGA_x_intensities.csv")
system("gzip output/GigaMUGA/GigaMUGA_x_intensities.csv")
}
if(file.exists("output/GigaMUGA/GigaMUGA_y_intensities.csv.gz") == "FALSE"){
write.csv(reference_Y_intensities_df, file = "output/GigaMUGA/GigaMUGA_y_intensities.csv")
system("gzip output/GigaMUGA/GigaMUGA_y_intensities.csv")
}
if(file.exists("output/GigaMUGA/GigaMUGA_sample_metadata.csv") == "FALSE"){
write.csv(reference_sample_metadata, file = "output/GigaMUGA/GigaMUGA_sample_metadata.csv")
}
if(file.exists("output/GigaMUGA/GigaMUGA_founder_consensus_genotypes.csv.gz") == "FALSE"){
write.csv(founder_consensus_genos,
file = "output/GigaMUGA/GigaMUGA_founder_consensus_genotypes.csv")
system("gzip output/GigaMUGA/GigaMUGA_founder_consensus_genotypes.csv")
}
if(file.exists("output/GigaMUGA/GigaMUGA_founder_mean_x_intensities.csv.gz") == "FALSE"){
write.csv(founder_mean_x_ints, file = "output/GigaMUGA/GigaMUGA_founder_mean_x_intensities.csv")
system("gzip output/GigaMUGA/GigaMUGA_founder_mean_x_intensities.csv")
}
if(file.exists("output/GigaMUGA/GigaMUGA_founder_mean_y_intensities.csv.gz") == "FALSE"){
write.csv(founder_mean_y_ints, file = "output/GigaMUGA/GigaMUGA_founder_mean_y_intensities.csv")
system("gzip output/GigaMUGA/GigaMUGA_founder_mean_y_intensities.csv")
}
if(file.exists("output/GigaMUGA/GigaMUGA_founder_metadata.csv") == "FALSE"){
write.csv(founder_metadata, file = "output/GigaMUGA/GigaMUGA_founder_metadata.csv")
}
sessionInfo()R version 4.2.1 (2022-06-23)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Big Sur ... 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] parallel stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] fstcore_0.9.12 vroom_1.6.0 tictoc_1.1 RColorBrewer_1.1-3
[5] ape_5.6-2 progress_1.2.2 plotly_4.10.0 ggplot2_3.3.6
[9] DT_0.25 magrittr_2.0.3 qtl2_0.28 furrr_0.3.1
[13] future_1.28.0 purrr_0.3.4 data.table_1.14.2 stringr_1.4.1
[17] tidyr_1.2.1 dplyr_1.0.10 fst_0.9.8 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] httr_1.4.4 sass_0.4.2 bit64_4.0.5 jsonlite_1.8.0
[5] viridisLite_0.4.1 bslib_0.4.0 assertthat_0.2.1 getPass_0.2-2
[9] highr_0.9 blob_1.2.3 yaml_2.3.5 globals_0.16.1
[13] pillar_1.8.1 RSQLite_2.2.17 lattice_0.20-45 glue_1.6.2
[17] digest_0.6.29 promises_1.2.0.1 colorspace_2.0-3 htmltools_0.5.3
[21] httpuv_1.6.6 pkgconfig_2.0.3 listenv_0.8.0 scales_1.2.1
[25] processx_3.7.0 whisker_0.4 later_1.3.0 tzdb_0.3.0
[29] git2r_0.30.1 tibble_3.1.8 farver_2.1.1 generics_0.1.3
[33] ellipsis_0.3.2 cachem_1.0.6 withr_2.5.0 lazyeval_0.2.2
[37] cli_3.4.0 crayon_1.5.1 memoise_2.0.1 evaluate_0.16
[41] ps_1.7.1 fs_1.5.2 fansi_1.0.3 parallelly_1.32.1
[45] nlme_3.1-157 tools_4.2.1 prettyunits_1.1.1 hms_1.1.2
[49] lifecycle_1.0.2 munsell_0.5.0 callr_3.7.2 compiler_4.2.1
[53] jquerylib_0.1.4 rlang_1.0.5 grid_4.2.1 rstudioapi_0.14
[57] htmlwidgets_1.5.4 crosstalk_1.2.0 labeling_0.4.2 rmarkdown_2.16
[61] gtable_0.3.1 codetools_0.2-18 DBI_1.1.3 R6_2.5.1
[65] knitr_1.40 fastmap_1.1.0 bit_4.0.4 utf8_1.2.2
[69] rprojroot_2.0.3 stringi_1.7.8 Rcpp_1.0.9 vctrs_0.4.1
[73] tidyselect_1.1.2 xfun_0.33