Chapter 3 - Data Transformation with dplyr
Vebash Naidoo
17/09/2020
Last updated: 2020-10-10
Checks: 7 0
Knit directory: r4ds_book/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20200814) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version bbb6cd2. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rproj.user/
Untracked files:
Untracked: VideoDecodeStats/
Untracked: analysis/images/
Untracked: code_snipp.txt
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/ch3_dplyr.Rmd) and HTML (docs/ch3_dplyr.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | bbb6cd2 | sciencificity | 2020-10-10 | added Ch 4 section |
| html | 226cd16 | sciencificity | 2020-10-10 | Build site. |
| Rmd | ae71e8e | sciencificity | 2020-10-10 | added Ch 4 section |
library(tidyverse)
library(flair)
library(nycflights13)
library(palmerpenguins)
library(gt)
library(skimr)
library(emo)Filter rows
filter() is the verb used to subset rows in a dataframe based on their values - i.e. we take a dataset and we say hey, give me back data that looks like it fulfils this condition(s).
The nycflights13 package has flights for the year 2013 where the departure was from an airport in New York City. You will notice the %>% which we used in the ggplot chapter too. If you can’t recall what it does for now ignore it and just have a look at the data contained in the dataset.
Aside: the pipe into gt() is to print out a neat table 😄.
flights %>%
head(4) %>%
gt()
| year | month | day | dep_time | sched_dep_time | dep_delay | arr_time | sched_arr_time | arr_delay | carrier | flight | tailnum | origin | dest | air_time | distance | hour | minute | time_hour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2013 | 1 | 1 | 517 | 515 | 2 | 830 | 819 | 11 | UA | 1545 | N14228 | EWR | IAH | 227 | 1400 | 5 | 15 | 2013-01-01 05:00:00 |
| 2013 | 1 | 1 | 533 | 529 | 4 | 850 | 830 | 20 | UA | 1714 | N24211 | LGA | IAH | 227 | 1416 | 5 | 29 | 2013-01-01 05:00:00 |
| 2013 | 1 | 1 | 542 | 540 | 2 | 923 | 850 | 33 | AA | 1141 | N619AA | JFK | MIA | 160 | 1089 | 5 | 40 | 2013-01-01 05:00:00 |
| 2013 | 1 | 1 | 544 | 545 | -1 | 1004 | 1022 | -18 | B6 | 725 | N804JB | JFK | BQN | 183 | 1576 | 5 | 45 | 2013-01-01 05:00:00 |
flights %>%
count(origin, sort=TRUE) %>%
gt()| origin | n |
|---|---|
| EWR | 120835 |
| JFK | 111279 |
| LGA | 104662 |
Let’s filter all flights that occurred on Jan 1. The syntax for filter is filter(df, some_condition_s)
filter(flights, month == 1, day == 1)
# A tibble: 842 x 19
year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
<int> <int> <int> <int> <int> <dbl> <int> <int>
1 2013 1 1 517 515 2 830 819
2 2013 1 1 533 529 4 850 830
3 2013 1 1 542 540 2 923 850
4 2013 1 1 544 545 -1 1004 1022
5 2013 1 1 554 600 -6 812 837
6 2013 1 1 554 558 -4 740 728
7 2013 1 1 555 600 -5 913 854
8 2013 1 1 557 600 -3 709 723
9 2013 1 1 557 600 -3 838 846
10 2013 1 1 558 600 -2 753 745
# ... with 832 more rows, and 11 more variables: arr_delay <dbl>,
# carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>,
# air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>
Notice the == for comparison
| sign | meaning |
|---|---|
| > | greater than |
| >= | greater than or equal to |
| < | less than |
| <= | less than or equal to |
| == | equal to |
| != | not equal to |
Okay, truly the {palmerpenguins} data is going to become the new iris! But come on’ who doesn’t love penguins! Just try and keep me away from Boulders Beach in Cape Town when I’m there visiting …
penguins %>%
head(10) %>%
gt()| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | year |
|---|---|---|---|---|---|---|---|
| Adelie | Torgersen | 39.1 | 18.7 | 181 | 3750 | male | 2007 |
| Adelie | Torgersen | 39.5 | 17.4 | 186 | 3800 | female | 2007 |
| Adelie | Torgersen | 40.3 | 18.0 | 195 | 3250 | female | 2007 |
| Adelie | Torgersen | NA | NA | NA | NA | NA | 2007 |
| Adelie | Torgersen | 36.7 | 19.3 | 193 | 3450 | female | 2007 |
| Adelie | Torgersen | 39.3 | 20.6 | 190 | 3650 | male | 2007 |
| Adelie | Torgersen | 38.9 | 17.8 | 181 | 3625 | female | 2007 |
| Adelie | Torgersen | 39.2 | 19.6 | 195 | 4675 | male | 2007 |
| Adelie | Torgersen | 34.1 | 18.1 | 193 | 3475 | NA | 2007 |
| Adelie | Torgersen | 42.0 | 20.2 | 190 | 4250 | NA | 2007 |
Ok, what types of penguins are there?
penguins %>%
count(species, sort=TRUE)# A tibble: 3 x 2
species n
<fct> <int>
1 Adelie 152
2 Gentoo 124
3 Chinstrap 68Let’s filter out Gentoo with bill_depth_mm bigger than 17mm, using some of the operations we learnt.
# lets us save the filter result and
# prints it out ... the extra brackets on the
# outside of the entire code block does that
(gentoo <- filter(penguins, species == 'Gentoo',
bill_depth_mm > 17))
# A tibble: 3 x 8
species island bill_length_mm bill_depth_mm flipper_length_~ body_mass_g sex
<fct> <fct> <dbl> <dbl> <int> <int> <fct>
1 Gentoo Biscoe 44.4 17.3 219 5250 male
2 Gentoo Biscoe 50.8 17.3 228 5600 male
3 Gentoo Biscoe 52.2 17.1 228 5400 male
# ... with 1 more variable: year <int>
Surprising results
Floats are saved a bit differently and hence can cause some unexpected behaviour.
sqrt(2) ^ 2 == 2[1] FALSE1 / 49 * 49 == 1[1] FALSEUse near() to give you if these are indeed nearly same.
near(sqrt(2) ^ 2, 2)
[1] TRUE
near(1 / 49 * 49, 1)
[1] TRUE
Logic
When we explore data we often want to filter data where one condition is true and another is also true. E.g. Which days experienced a thunder storm where a thunder storm was predicted by the Meteorologists.
In R we use logic operators to combine comparisons and get us the data we’re looking for.
| sign | meaning |
|---|---|
| | | Or |
| & | And |
| ! | Not |
Let’s say we want only flights that occurred in Nov and Dec of 2013:
filter(flights, month == 11 | month == 12)
# A tibble: 55,403 x 19
year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
<int> <int> <int> <int> <int> <dbl> <int> <int>
1 2013 11 1 5 2359 6 352 345
2 2013 11 1 35 2250 105 123 2356
3 2013 11 1 455 500 -5 641 651
4 2013 11 1 539 545 -6 856 827
5 2013 11 1 542 545 -3 831 855
6 2013 11 1 549 600 -11 912 923
7 2013 11 1 550 600 -10 705 659
8 2013 11 1 554 600 -6 659 701
9 2013 11 1 554 600 -6 826 827
10 2013 11 1 554 600 -6 749 751
# ... with 55,393 more rows, and 11 more variables: arr_delay <dbl>,
# carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>,
# air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>
We have to repeat the month == part for each comparison.
You may think that you can just say month == (11 | 12) but unfortunately that does not work.
(filtered_fl <- filter(flights, month == (11 | 12)))
# A tibble: 27,004 x 19
year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
<int> <int> <int> <int> <int> <dbl> <int> <int>
1 2013 1 1 517 515 2 830 819
2 2013 1 1 533 529 4 850 830
3 2013 1 1 542 540 2 923 850
4 2013 1 1 544 545 -1 1004 1022
5 2013 1 1 554 600 -6 812 837
6 2013 1 1 554 558 -4 740 728
7 2013 1 1 555 600 -5 913 854
8 2013 1 1 557 600 -3 709 723
9 2013 1 1 557 600 -3 838 846
10 2013 1 1 558 600 -2 753 745
# ... with 26,994 more rows, and 11 more variables: arr_delay <dbl>,
# carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>,
# air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>
Note that the result is unexpected - the tibble returned contains the month == 1 which is January, and should not be in our returned dataset! The (11 | 12) evaluates to TRUE which internally is represented as 1 hence we then look at month == 1. To see this lets load the flights dataframe and filter Jan again. Notice that the number of rows is identical to the number of rows in the filtered filtered_fl which is 27004.
flights %>%
filter(month == 1)# A tibble: 27,004 x 19
year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
<int> <int> <int> <int> <int> <dbl> <int> <int>
1 2013 1 1 517 515 2 830 819
2 2013 1 1 533 529 4 850 830
3 2013 1 1 542 540 2 923 850
4 2013 1 1 544 545 -1 1004 1022
5 2013 1 1 554 600 -6 812 837
6 2013 1 1 554 558 -4 740 728
7 2013 1 1 555 600 -5 913 854
8 2013 1 1 557 600 -3 709 723
9 2013 1 1 557 600 -3 838 846
10 2013 1 1 558 600 -2 753 745
# ... with 26,994 more rows, and 11 more variables: arr_delay <dbl>,
# carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>,
# air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>We can also use the %in% operator instead of repeating the month ==.
filter(flights, month %in% c(11, 12))# A tibble: 55,403 x 19
year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
<int> <int> <int> <int> <int> <dbl> <int> <int>
1 2013 11 1 5 2359 6 352 345
2 2013 11 1 35 2250 105 123 2356
3 2013 11 1 455 500 -5 641 651
4 2013 11 1 539 545 -6 856 827
5 2013 11 1 542 545 -3 831 855
6 2013 11 1 549 600 -11 912 923
7 2013 11 1 550 600 -10 705 659
8 2013 11 1 554 600 -6 659 701
9 2013 11 1 554 600 -6 826 827
10 2013 11 1 554 600 -6 749 751
# ... with 55,393 more rows, and 11 more variables: arr_delay <dbl>,
# carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>,
# air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>If you need the opposite of this i.e. the not in operator it’s a bit more tricky.
You can make your own function for this.
'%!in%' <- function(x,y)!('%in%'(x,y)) # method 1
'%ni%' <- Negate('%in%') # method 2
filter(flights, month %ni% c(11, 12))# A tibble: 281,373 x 19
year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
<int> <int> <int> <int> <int> <dbl> <int> <int>
1 2013 1 1 517 515 2 830 819
2 2013 1 1 533 529 4 850 830
3 2013 1 1 542 540 2 923 850
4 2013 1 1 544 545 -1 1004 1022
5 2013 1 1 554 600 -6 812 837
6 2013 1 1 554 558 -4 740 728
7 2013 1 1 555 600 -5 913 854
8 2013 1 1 557 600 -3 709 723
9 2013 1 1 557 600 -3 838 846
10 2013 1 1 558 600 -2 753 745
# ... with 281,363 more rows, and 11 more variables: arr_delay <dbl>,
# carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>,
# air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>Aside: Some penguin fun …
Let’s see if we can explore our data through filtering.
# Get all penguins where the species is anything
# other than Adelie
filter(penguins, species %ni% ('Adelie'))# A tibble: 192 x 8
species island bill_length_mm bill_depth_mm flipper_length_~ body_mass_g
<fct> <fct> <dbl> <dbl> <int> <int>
1 Gentoo Biscoe 46.1 13.2 211 4500
2 Gentoo Biscoe 50 16.3 230 5700
3 Gentoo Biscoe 48.7 14.1 210 4450
4 Gentoo Biscoe 50 15.2 218 5700
5 Gentoo Biscoe 47.6 14.5 215 5400
6 Gentoo Biscoe 46.5 13.5 210 4550
7 Gentoo Biscoe 45.4 14.6 211 4800
8 Gentoo Biscoe 46.7 15.3 219 5200
9 Gentoo Biscoe 43.3 13.4 209 4400
10 Gentoo Biscoe 46.8 15.4 215 5150
# ... with 182 more rows, and 2 more variables: sex <fct>, year <int>The skim function also works great in conjunction with filter. I am going to use the pipe again which we tackle later in R4DS. For now when you see %>% read it as and then e.g. Take this df and then filter out data that meets the condition and then show me a summary.
df %>%
filter(some_col meets some condition) %>%
summary(avg_col = mean(some_col))
penguins %>%
filter(species == "Chinstrap") %>%
skim()| Name | Piped data |
| Number of rows | 68 |
| Number of columns | 8 |
| _______________________ | |
| Column type frequency: | |
| factor | 3 |
| numeric | 5 |
| ________________________ | |
| Group variables | None |
Variable type: factor
| skim_variable | n_missing | complete_rate | ordered | n_unique | top_counts |
|---|---|---|---|---|---|
| species | 0 | 1 | FALSE | 1 | Chi: 68, Ade: 0, Gen: 0 |
| island | 0 | 1 | FALSE | 1 | Dre: 68, Bis: 0, Tor: 0 |
| sex | 0 | 1 | FALSE | 2 | fem: 34, mal: 34 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| bill_length_mm | 0 | 1 | 48.83 | 3.34 | 40.9 | 46.35 | 49.55 | 51.08 | 58.0 | ▂▇▇▅▁ |
| bill_depth_mm | 0 | 1 | 18.42 | 1.14 | 16.4 | 17.50 | 18.45 | 19.40 | 20.8 | ▅▇▇▆▂ |
| flipper_length_mm | 0 | 1 | 195.82 | 7.13 | 178.0 | 191.00 | 196.00 | 201.00 | 212.0 | ▁▅▇▅▂ |
| body_mass_g | 0 | 1 | 3733.09 | 384.34 | 2700.0 | 3487.50 | 3700.00 | 3950.00 | 4800.0 | ▁▅▇▃▁ |
| year | 0 | 1 | 2007.97 | 0.86 | 2007.0 | 2007.00 | 2008.00 | 2009.00 | 2009.0 | ▇▁▆▁▇ |
Let’s see if the Chinstrap penguins with a body mass above the mean are more likely male / female / equally represented?
The pipe way:
penguins %>%
filter(species == "Chinstrap") %>%
filter(body_mass_g > mean(body_mass_g)) %>%
skim()| Name | Piped data |
| Number of rows | 31 |
| Number of columns | 8 |
| _______________________ | |
| Column type frequency: | |
| factor | 3 |
| numeric | 5 |
| ________________________ | |
| Group variables | None |
Variable type: factor
| skim_variable | n_missing | complete_rate | ordered | n_unique | top_counts |
|---|---|---|---|---|---|
| species | 0 | 1 | FALSE | 1 | Chi: 31, Ade: 0, Gen: 0 |
| island | 0 | 1 | FALSE | 1 | Dre: 31, Bis: 0, Tor: 0 |
| sex | 0 | 1 | FALSE | 2 | mal: 25, fem: 6 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| bill_length_mm | 0 | 1 | 50.38 | 2.30 | 45.2 | 49.25 | 50.6 | 51.80 | 55.8 | ▂▃▇▅▁ |
| bill_depth_mm | 0 | 1 | 19.07 | 0.90 | 16.8 | 18.40 | 19.0 | 19.65 | 20.8 | ▁▅▆▇▂ |
| flipper_length_mm | 0 | 1 | 200.77 | 5.47 | 193.0 | 196.50 | 201.0 | 204.00 | 212.0 | ▇▅▇▃▃ |
| body_mass_g | 0 | 1 | 4055.65 | 267.14 | 3750.0 | 3825.00 | 4000.0 | 4150.00 | 4800.0 | ▇▅▁▂▁ |
| year | 0 | 1 | 2008.00 | 0.86 | 2007.0 | 2007.00 | 2008.0 | 2009.00 | 2009.0 | ▇▁▆▁▇ |
We get the same using the non-piped way.
skim(filter(filter(penguins, species == "Chinstrap"),
body_mass_g > mean(body_mass_g))) | Name | filter(…) |
| Number of rows | 31 |
| Number of columns | 8 |
| _______________________ | |
| Column type frequency: | |
| factor | 3 |
| numeric | 5 |
| ________________________ | |
| Group variables | None |
Variable type: factor
| skim_variable | n_missing | complete_rate | ordered | n_unique | top_counts |
|---|---|---|---|---|---|
| species | 0 | 1 | FALSE | 1 | Chi: 31, Ade: 0, Gen: 0 |
| island | 0 | 1 | FALSE | 1 | Dre: 31, Bis: 0, Tor: 0 |
| sex | 0 | 1 | FALSE | 2 | mal: 25, fem: 6 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| bill_length_mm | 0 | 1 | 50.38 | 2.30 | 45.2 | 49.25 | 50.6 | 51.80 | 55.8 | ▂▃▇▅▁ |
| bill_depth_mm | 0 | 1 | 19.07 | 0.90 | 16.8 | 18.40 | 19.0 | 19.65 | 20.8 | ▁▅▆▇▂ |
| flipper_length_mm | 0 | 1 | 200.77 | 5.47 | 193.0 | 196.50 | 201.0 | 204.00 | 212.0 | ▇▅▇▃▃ |
| body_mass_g | 0 | 1 | 4055.65 | 267.14 | 3750.0 | 3825.00 | 4000.0 | 4150.00 | 4800.0 | ▇▅▁▂▁ |
| year | 0 | 1 | 2008.00 | 0.86 | 2007.0 | 2007.00 | 2008.0 | 2009.00 | 2009.0 | ▇▁▆▁▇ |
It seems there are more male penguins above the mean of body mass.
Exercises
Find all flights that
- Had an arrival delay of two or more hours
filter(flights,
arr_delay >= 120)# A tibble: 10,200 x 19 year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time <int> <int> <int> <int> <int> <dbl> <int> <int> 1 2013 1 1 811 630 101 1047 830 2 2013 1 1 848 1835 853 1001 1950 3 2013 1 1 957 733 144 1056 853 4 2013 1 1 1114 900 134 1447 1222 5 2013 1 1 1505 1310 115 1638 1431 6 2013 1 1 1525 1340 105 1831 1626 7 2013 1 1 1549 1445 64 1912 1656 8 2013 1 1 1558 1359 119 1718 1515 9 2013 1 1 1732 1630 62 2028 1825 10 2013 1 1 1803 1620 103 2008 1750 # ... with 10,190 more rows, and 11 more variables: arr_delay <dbl>, # carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>, # air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>- Flew to Houston (
IAHorHOU)
filter(flights, dest %in% c('IAH', 'HOU'))# A tibble: 9,313 x 19 year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time <int> <int> <int> <int> <int> <dbl> <int> <int> 1 2013 1 1 517 515 2 830 819 2 2013 1 1 533 529 4 850 830 3 2013 1 1 623 627 -4 933 932 4 2013 1 1 728 732 -4 1041 1038 5 2013 1 1 739 739 0 1104 1038 6 2013 1 1 908 908 0 1228 1219 7 2013 1 1 1028 1026 2 1350 1339 8 2013 1 1 1044 1045 -1 1352 1351 9 2013 1 1 1114 900 134 1447 1222 10 2013 1 1 1205 1200 5 1503 1505 # ... with 9,303 more rows, and 11 more variables: arr_delay <dbl>, # carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>, # air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>- Were operated by United, American, or Delta
as_tibble(flights) %>% distinct(carrier) %>% arrange(carrier)# A tibble: 16 x 1 carrier <chr> 1 9E 2 AA 3 AS 4 B6 5 DL 6 EV 7 F9 8 FL 9 HA 10 MQ 11 OO 12 UA 13 US 14 VX 15 WN 16 YV# think these are United = UA, American = AA, Delta = DL filter(flights, carrier %in% c('UA', 'AA', 'DL'))# A tibble: 139,504 x 19 year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time <int> <int> <int> <int> <int> <dbl> <int> <int> 1 2013 1 1 517 515 2 830 819 2 2013 1 1 533 529 4 850 830 3 2013 1 1 542 540 2 923 850 4 2013 1 1 554 600 -6 812 837 5 2013 1 1 554 558 -4 740 728 6 2013 1 1 558 600 -2 753 745 7 2013 1 1 558 600 -2 924 917 8 2013 1 1 558 600 -2 923 937 9 2013 1 1 559 600 -1 941 910 10 2013 1 1 559 600 -1 854 902 # ... with 139,494 more rows, and 11 more variables: arr_delay <dbl>, # carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>, # air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>- Departed in summer (July, August, and September)
filter(flights, month %in% c(7, 8, 9))# A tibble: 86,326 x 19 year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time <int> <int> <int> <int> <int> <dbl> <int> <int> 1 2013 7 1 1 2029 212 236 2359 2 2013 7 1 2 2359 3 344 344 3 2013 7 1 29 2245 104 151 1 4 2013 7 1 43 2130 193 322 14 5 2013 7 1 44 2150 174 300 100 6 2013 7 1 46 2051 235 304 2358 7 2013 7 1 48 2001 287 308 2305 8 2013 7 1 58 2155 183 335 43 9 2013 7 1 100 2146 194 327 30 10 2013 7 1 100 2245 135 337 135 # ... with 86,316 more rows, and 11 more variables: arr_delay <dbl>, # carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>, # air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>- Arrived more than two hours late, but didn’t leave late
filter(flights, arr_delay > 120 & dep_delay <= 0)# A tibble: 29 x 19 year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time <int> <int> <int> <int> <int> <dbl> <int> <int> 1 2013 1 27 1419 1420 -1 1754 1550 2 2013 10 7 1350 1350 0 1736 1526 3 2013 10 7 1357 1359 -2 1858 1654 4 2013 10 16 657 700 -3 1258 1056 5 2013 11 1 658 700 -2 1329 1015 6 2013 3 18 1844 1847 -3 39 2219 7 2013 4 17 1635 1640 -5 2049 1845 8 2013 4 18 558 600 -2 1149 850 9 2013 4 18 655 700 -5 1213 950 10 2013 5 22 1827 1830 -3 2217 2010 # ... with 19 more rows, and 11 more variables: arr_delay <dbl>, carrier <chr>, # flight <int>, tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, # distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>- Were delayed by at least an hour, but made up over 30 minutes in flight
filter(flights, dep_delay >= 60 & arr_delay < (dep_delay - 30))# A tibble: 1,844 x 19 year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time <int> <int> <int> <int> <int> <dbl> <int> <int> 1 2013 1 1 2205 1720 285 46 2040 2 2013 1 1 2326 2130 116 131 18 3 2013 1 3 1503 1221 162 1803 1555 4 2013 1 3 1839 1700 99 2056 1950 5 2013 1 3 1850 1745 65 2148 2120 6 2013 1 3 1941 1759 102 2246 2139 7 2013 1 3 1950 1845 65 2228 2227 8 2013 1 3 2015 1915 60 2135 2111 9 2013 1 3 2257 2000 177 45 2224 10 2013 1 4 1917 1700 137 2135 1950 # ... with 1,834 more rows, and 11 more variables: arr_delay <dbl>, # carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>, # air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>- Departed between midnight and 6am (inclusive)
flights %>% # notice that some flights were schedule to leave befor 00h00 but # were delayed and hence left after 00h00 filter(dep_time > 0 & dep_time < 100)# A tibble: 881 x 19 year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time <int> <int> <int> <int> <int> <dbl> <int> <int> 1 2013 1 2 42 2359 43 518 442 2 2013 1 3 32 2359 33 504 442 3 2013 1 3 50 2145 185 203 2311 4 2013 1 4 25 2359 26 505 442 5 2013 1 5 14 2359 15 503 445 6 2013 1 5 37 2230 127 341 131 7 2013 1 6 16 2359 17 451 442 8 2013 1 7 49 2359 50 531 444 9 2013 1 9 2 2359 3 432 444 10 2013 1 9 8 2359 9 432 437 # ... with 871 more rows, and 11 more variables: arr_delay <dbl>, # carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>, # air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>flights %>% # are any flights exactly at 24h00? filter(dep_time == 2400)# A tibble: 29 x 19 year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time <int> <int> <int> <int> <int> <dbl> <int> <int> 1 2013 10 30 2400 2359 1 327 337 2 2013 11 27 2400 2359 1 515 445 3 2013 12 5 2400 2359 1 427 440 4 2013 12 9 2400 2359 1 432 440 5 2013 12 9 2400 2250 70 59 2356 6 2013 12 13 2400 2359 1 432 440 7 2013 12 19 2400 2359 1 434 440 8 2013 12 29 2400 1700 420 302 2025 9 2013 2 7 2400 2359 1 432 436 10 2013 2 7 2400 2359 1 443 444 # ... with 19 more rows, and 11 more variables: arr_delay <dbl>, carrier <chr>, # flight <int>, tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, # distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>filter(flights, (dep_time >= 0 & dep_time <= 600) | (dep_time == 2400))# A tibble: 9,373 x 19 year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time <int> <int> <int> <int> <int> <dbl> <int> <int> 1 2013 1 1 517 515 2 830 819 2 2013 1 1 533 529 4 850 830 3 2013 1 1 542 540 2 923 850 4 2013 1 1 544 545 -1 1004 1022 5 2013 1 1 554 600 -6 812 837 6 2013 1 1 554 558 -4 740 728 7 2013 1 1 555 600 -5 913 854 8 2013 1 1 557 600 -3 709 723 9 2013 1 1 557 600 -3 838 846 10 2013 1 1 558 600 -2 753 745 # ... with 9,363 more rows, and 11 more variables: arr_delay <dbl>, # carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>, # air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>Another useful dplyr filtering helper is
between(). What does it do? Can you use it to simplify the code needed to answer the previous challenges?# Departed in summer (July, August, and September) filter(flights, between(month,7,9))# A tibble: 86,326 x 19 year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time <int> <int> <int> <int> <int> <dbl> <int> <int> 1 2013 7 1 1 2029 212 236 2359 2 2013 7 1 2 2359 3 344 344 3 2013 7 1 29 2245 104 151 1 4 2013 7 1 43 2130 193 322 14 5 2013 7 1 44 2150 174 300 100 6 2013 7 1 46 2051 235 304 2358 7 2013 7 1 48 2001 287 308 2305 8 2013 7 1 58 2155 183 335 43 9 2013 7 1 100 2146 194 327 30 10 2013 7 1 100 2245 135 337 135 # ... with 86,316 more rows, and 11 more variables: arr_delay <dbl>, # carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>, # air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm># Departed between midnight and 6am (inclusive) filter(flights, between(dep_time, 0, 600) | (dep_time == 2400))# A tibble: 9,373 x 19 year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time <int> <int> <int> <int> <int> <dbl> <int> <int> 1 2013 1 1 517 515 2 830 819 2 2013 1 1 533 529 4 850 830 3 2013 1 1 542 540 2 923 850 4 2013 1 1 544 545 -1 1004 1022 5 2013 1 1 554 600 -6 812 837 6 2013 1 1 554 558 -4 740 728 7 2013 1 1 555 600 -5 913 854 8 2013 1 1 557 600 -3 709 723 9 2013 1 1 557 600 -3 838 846 10 2013 1 1 558 600 -2 753 745 # ... with 9,363 more rows, and 11 more variables: arr_delay <dbl>, # carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>, # air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>How many flights have a missing
dep_time? What other variables are missing? What might these rows represent?filter(flights, is.na(dep_time))# A tibble: 8,255 x 19 year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time <int> <int> <int> <int> <int> <dbl> <int> <int> 1 2013 1 1 NA 1630 NA NA 1815 2 2013 1 1 NA 1935 NA NA 2240 3 2013 1 1 NA 1500 NA NA 1825 4 2013 1 1 NA 600 NA NA 901 5 2013 1 2 NA 1540 NA NA 1747 6 2013 1 2 NA 1620 NA NA 1746 7 2013 1 2 NA 1355 NA NA 1459 8 2013 1 2 NA 1420 NA NA 1644 9 2013 1 2 NA 1321 NA NA 1536 10 2013 1 2 NA 1545 NA NA 1910 # ... with 8,245 more rows, and 11 more variables: arr_delay <dbl>, # carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>, # air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>skim(flights) # Great summary function from skimrData summary Name flights Number of rows 336776 Number of columns 19 _______________________ Column type frequency: character 4 numeric 14 POSIXct 1 ________________________ Group variables None Variable type: character
skim_variable n_missing complete_rate min max empty n_unique whitespace carrier 0 1.00 2 2 0 16 0 tailnum 2512 0.99 5 6 0 4043 0 origin 0 1.00 3 3 0 3 0 dest 0 1.00 3 3 0 105 0 Variable type: numeric
skim_variable n_missing complete_rate mean sd p0 p25 p50 p75 p100 hist year 0 1.00 2013.00 0.00 2013 2013 2013 2013 2013 ▁▁▇▁▁ month 0 1.00 6.55 3.41 1 4 7 10 12 ▇▆▆▆▇ day 0 1.00 15.71 8.77 1 8 16 23 31 ▇▇▇▇▆ dep_time 8255 0.98 1349.11 488.28 1 907 1401 1744 2400 ▁▇▆▇▃ sched_dep_time 0 1.00 1344.25 467.34 106 906 1359 1729 2359 ▁▇▇▇▃ dep_delay 8255 0.98 12.64 40.21 -43 -5 -2 11 1301 ▇▁▁▁▁ arr_time 8713 0.97 1502.05 533.26 1 1104 1535 1940 2400 ▁▃▇▇▇ sched_arr_time 0 1.00 1536.38 497.46 1 1124 1556 1945 2359 ▁▃▇▇▇ arr_delay 9430 0.97 6.90 44.63 -86 -17 -5 14 1272 ▇▁▁▁▁ flight 0 1.00 1971.92 1632.47 1 553 1496 3465 8500 ▇▃▃▁▁ air_time 9430 0.97 150.69 93.69 20 82 129 192 695 ▇▂▂▁▁ distance 0 1.00 1039.91 733.23 17 502 872 1389 4983 ▇▃▂▁▁ hour 0 1.00 13.18 4.66 1 9 13 17 23 ▁▇▇▇▅ minute 0 1.00 26.23 19.30 0 8 29 44 59 ▇▃▆▃▅ Variable type: POSIXct
skim_variable n_missing complete_rate min max median n_unique time_hour 0 1 2013-01-01 05:00:00 2013-12-31 23:00:00 2013-07-03 10:00:00 6936 Why is
NA ^ 0not missing? Why isNA | TRUEnot missing? Why isFALSE & NAnot missing? Can you figure out the general rule? (NA * 0is a tricky counterexample!)# Anything to the power 0, is 1 NA ^ 0[1] 1# Anything OR TRUE, is still TRUE NA | TRUE[1] TRUE# FALSE and anything, is FALSE FALSE & NA[1] FALSE# Not everything * 0 is 0; e.g sqrt(-2) * 0 is NaN NA * 0[1] NA
Check out Suzan Baert’s great dplyr tutorial on filter
Arrange
arrange() changes the order of the observations based on the columns you provide and the order you want it arranged. The syntax is arrange(df, cols_to_order_by) E.g. arrange(df, col1, col2) says “Hey, take this df and arrange the observations in alphabetical/increasing numeric order of col1 and col2.” col2 comes in when there are ties in col1.
We use desc() or - when we want to order the observations in reverse for a column.
arrange(flights, desc(arr_delay),
desc(dep_delay),
day, month, year) %>%
head(10) %>%
gt()| year | month | day | dep_time | sched_dep_time | dep_delay | arr_time | sched_arr_time | arr_delay | carrier | flight | tailnum | origin | dest | air_time | distance | hour | minute | time_hour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2013 | 1 | 9 | 641 | 900 | 1301 | 1242 | 1530 | 1272 | HA | 51 | N384HA | JFK | HNL | 640 | 4983 | 9 | 0 | 2013-01-09 09:00:00 |
| 2013 | 6 | 15 | 1432 | 1935 | 1137 | 1607 | 2120 | 1127 | MQ | 3535 | N504MQ | JFK | CMH | 74 | 483 | 19 | 35 | 2013-06-15 19:00:00 |
| 2013 | 1 | 10 | 1121 | 1635 | 1126 | 1239 | 1810 | 1109 | MQ | 3695 | N517MQ | EWR | ORD | 111 | 719 | 16 | 35 | 2013-01-10 16:00:00 |
| 2013 | 9 | 20 | 1139 | 1845 | 1014 | 1457 | 2210 | 1007 | AA | 177 | N338AA | JFK | SFO | 354 | 2586 | 18 | 45 | 2013-09-20 18:00:00 |
| 2013 | 7 | 22 | 845 | 1600 | 1005 | 1044 | 1815 | 989 | MQ | 3075 | N665MQ | JFK | CVG | 96 | 589 | 16 | 0 | 2013-07-22 16:00:00 |
| 2013 | 4 | 10 | 1100 | 1900 | 960 | 1342 | 2211 | 931 | DL | 2391 | N959DL | JFK | TPA | 139 | 1005 | 19 | 0 | 2013-04-10 19:00:00 |
| 2013 | 3 | 17 | 2321 | 810 | 911 | 135 | 1020 | 915 | DL | 2119 | N927DA | LGA | MSP | 167 | 1020 | 8 | 10 | 2013-03-17 08:00:00 |
| 2013 | 7 | 22 | 2257 | 759 | 898 | 121 | 1026 | 895 | DL | 2047 | N6716C | LGA | ATL | 109 | 762 | 7 | 59 | 2013-07-22 07:00:00 |
| 2013 | 12 | 5 | 756 | 1700 | 896 | 1058 | 2020 | 878 | AA | 172 | N5DMAA | EWR | MIA | 149 | 1085 | 17 | 0 | 2013-12-05 17:00:00 |
| 2013 | 5 | 3 | 1133 | 2055 | 878 | 1250 | 2215 | 875 | MQ | 3744 | N523MQ | EWR | ORD | 112 | 719 | 20 | 55 | 2013-05-03 20:00:00 |
Using the - sign instead of desc() yields the same results.
arrange(flights, -arr_delay,
-dep_delay,
day, month, year) %>%
head(10) %>%
gt()
| year | month | day | dep_time | sched_dep_time | dep_delay | arr_time | sched_arr_time | arr_delay | carrier | flight | tailnum | origin | dest | air_time | distance | hour | minute | time_hour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2013 | 1 | 9 | 641 | 900 | 1301 | 1242 | 1530 | 1272 | HA | 51 | N384HA | JFK | HNL | 640 | 4983 | 9 | 0 | 2013-01-09 09:00:00 |
| 2013 | 6 | 15 | 1432 | 1935 | 1137 | 1607 | 2120 | 1127 | MQ | 3535 | N504MQ | JFK | CMH | 74 | 483 | 19 | 35 | 2013-06-15 19:00:00 |
| 2013 | 1 | 10 | 1121 | 1635 | 1126 | 1239 | 1810 | 1109 | MQ | 3695 | N517MQ | EWR | ORD | 111 | 719 | 16 | 35 | 2013-01-10 16:00:00 |
| 2013 | 9 | 20 | 1139 | 1845 | 1014 | 1457 | 2210 | 1007 | AA | 177 | N338AA | JFK | SFO | 354 | 2586 | 18 | 45 | 2013-09-20 18:00:00 |
| 2013 | 7 | 22 | 845 | 1600 | 1005 | 1044 | 1815 | 989 | MQ | 3075 | N665MQ | JFK | CVG | 96 | 589 | 16 | 0 | 2013-07-22 16:00:00 |
| 2013 | 4 | 10 | 1100 | 1900 | 960 | 1342 | 2211 | 931 | DL | 2391 | N959DL | JFK | TPA | 139 | 1005 | 19 | 0 | 2013-04-10 19:00:00 |
| 2013 | 3 | 17 | 2321 | 810 | 911 | 135 | 1020 | 915 | DL | 2119 | N927DA | LGA | MSP | 167 | 1020 | 8 | 10 | 2013-03-17 08:00:00 |
| 2013 | 7 | 22 | 2257 | 759 | 898 | 121 | 1026 | 895 | DL | 2047 | N6716C | LGA | ATL | 109 | 762 | 7 | 59 | 2013-07-22 07:00:00 |
| 2013 | 12 | 5 | 756 | 1700 | 896 | 1058 | 2020 | 878 | AA | 172 | N5DMAA | EWR | MIA | 149 | 1085 | 17 | 0 | 2013-12-05 17:00:00 |
| 2013 | 5 | 3 | 1133 | 2055 | 878 | 1250 | 2215 | 875 | MQ | 3744 | N523MQ | EWR | ORD | 112 | 719 | 20 | 55 | 2013-05-03 20:00:00 |
Exercises
How could you use
arrange()to sort all missing values to the start? Answer found here. (Hint: useis.na()).flights %>% # arrange by highest row sum of NA's # observations with most NAs float to the # top of the df returned arrange(desc(rowSums(is.na(.))))# A tibble: 336,776 x 19 year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time <int> <int> <int> <int> <int> <dbl> <int> <int> 1 2013 1 2 NA 1545 NA NA 1910 2 2013 1 2 NA 1601 NA NA 1735 3 2013 1 3 NA 857 NA NA 1209 4 2013 1 3 NA 645 NA NA 952 5 2013 1 4 NA 845 NA NA 1015 6 2013 1 4 NA 1830 NA NA 2044 7 2013 1 5 NA 840 NA NA 1001 8 2013 1 7 NA 820 NA NA 958 9 2013 1 8 NA 1645 NA NA 1838 10 2013 1 9 NA 755 NA NA 1012 # ... with 336,766 more rows, and 11 more variables: arr_delay <dbl>, # carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>, # air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>Sort
flightsto find the most delayed flights. Find the flights that left earliest.arrange(flights, -dep_delay, dep_time)# A tibble: 336,776 x 19 year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time <int> <int> <int> <int> <int> <dbl> <int> <int> 1 2013 1 9 641 900 1301 1242 1530 2 2013 6 15 1432 1935 1137 1607 2120 3 2013 1 10 1121 1635 1126 1239 1810 4 2013 9 20 1139 1845 1014 1457 2210 5 2013 7 22 845 1600 1005 1044 1815 6 2013 4 10 1100 1900 960 1342 2211 7 2013 3 17 2321 810 911 135 1020 8 2013 6 27 959 1900 899 1236 2226 9 2013 7 22 2257 759 898 121 1026 10 2013 12 5 756 1700 896 1058 2020 # ... with 336,766 more rows, and 11 more variables: arr_delay <dbl>, # carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>, # air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>Sort
flightsto find the fastest (highest speed) flights.arrange(flights, desc(distance/air_time))# A tibble: 336,776 x 19 year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time <int> <int> <int> <int> <int> <dbl> <int> <int> 1 2013 5 25 1709 1700 9 1923 1937 2 2013 7 2 1558 1513 45 1745 1719 3 2013 5 13 2040 2025 15 2225 2226 4 2013 3 23 1914 1910 4 2045 2043 5 2013 1 12 1559 1600 -1 1849 1917 6 2013 11 17 650 655 -5 1059 1150 7 2013 2 21 2355 2358 -3 412 438 8 2013 11 17 759 800 -1 1212 1255 9 2013 11 16 2003 1925 38 17 36 10 2013 11 16 2349 2359 -10 402 440 # ... with 336,766 more rows, and 11 more variables: arr_delay <dbl>, # carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>, # air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>Which flights travelled the farthest? Which travelled the shortest?
- farthest
# farthest
arrange(flights, desc(distance))# A tibble: 336,776 x 19
year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
<int> <int> <int> <int> <int> <dbl> <int> <int>
1 2013 1 1 857 900 -3 1516 1530
2 2013 1 2 909 900 9 1525 1530
3 2013 1 3 914 900 14 1504 1530
4 2013 1 4 900 900 0 1516 1530
5 2013 1 5 858 900 -2 1519 1530
6 2013 1 6 1019 900 79 1558 1530
7 2013 1 7 1042 900 102 1620 1530
8 2013 1 8 901 900 1 1504 1530
9 2013 1 9 641 900 1301 1242 1530
10 2013 1 10 859 900 -1 1449 1530
# ... with 336,766 more rows, and 11 more variables: arr_delay <dbl>,
# carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>,
# air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>- shortest (assuming shortest distance here in context above)
arrange(flights, distance)# A tibble: 336,776 x 19
year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
<int> <int> <int> <int> <int> <dbl> <int> <int>
1 2013 7 27 NA 106 NA NA 245
2 2013 1 3 2127 2129 -2 2222 2224
3 2013 1 4 1240 1200 40 1333 1306
4 2013 1 4 1829 1615 134 1937 1721
5 2013 1 4 2128 2129 -1 2218 2224
6 2013 1 5 1155 1200 -5 1241 1306
7 2013 1 6 2125 2129 -4 2224 2224
8 2013 1 7 2124 2129 -5 2212 2224
9 2013 1 8 2127 2130 -3 2304 2225
10 2013 1 9 2126 2129 -3 2217 2224
# ... with 336,766 more rows, and 11 more variables: arr_delay <dbl>,
# carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>,
# air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>Select columns
select() allows you to choose a subset of columns / variables in your data.
Let’s try out some using the penguins data.
as_tibble(names(penguins_raw) ) %>%
gt() %>%
tab_options(
heading.title.font.size = "small",
table.font.size = "small"
)| value |
|---|
| studyName |
| Sample Number |
| Species |
| Region |
| Island |
| Stage |
| Individual ID |
| Clutch Completion |
| Date Egg |
| Culmen Length (mm) |
| Culmen Depth (mm) |
| Flipper Length (mm) |
| Body Mass (g) |
| Sex |
| Delta 15 N (o/oo) |
| Delta 13 C (o/oo) |
| Comments |
- Select columns by name
select(penguins_raw, "Sample Number",
Island, Sex)
# A tibble: 344 x 3
`Sample Number` Island Sex
<dbl> <chr> <chr>
1 1 Torgersen MALE
2 2 Torgersen FEMALE
3 3 Torgersen FEMALE
4 4 Torgersen <NA>
5 5 Torgersen FEMALE
6 6 Torgersen MALE
7 7 Torgersen FEMALE
8 8 Torgersen MALE
9 9 Torgersen <NA>
10 10 Torgersen <NA>
# ... with 334 more rows
- Select columns in a range
select(penguins_raw,
"Sample Number":"Individual ID")
# A tibble: 344 x 6
`Sample Number` Species Region Island Stage `Individual ID`
<dbl> <chr> <chr> <chr> <chr> <chr>
1 1 Adelie Penguin (Py~ Anvers Torger~ Adult, 1 ~ N1A1
2 2 Adelie Penguin (Py~ Anvers Torger~ Adult, 1 ~ N1A2
3 3 Adelie Penguin (Py~ Anvers Torger~ Adult, 1 ~ N2A1
4 4 Adelie Penguin (Py~ Anvers Torger~ Adult, 1 ~ N2A2
5 5 Adelie Penguin (Py~ Anvers Torger~ Adult, 1 ~ N3A1
6 6 Adelie Penguin (Py~ Anvers Torger~ Adult, 1 ~ N3A2
7 7 Adelie Penguin (Py~ Anvers Torger~ Adult, 1 ~ N4A1
8 8 Adelie Penguin (Py~ Anvers Torger~ Adult, 1 ~ N4A2
9 9 Adelie Penguin (Py~ Anvers Torger~ Adult, 1 ~ N5A1
10 10 Adelie Penguin (Py~ Anvers Torger~ Adult, 1 ~ N5A2
# ... with 334 more rows
- Select everything except for
select(penguins_raw, -Comments,
-(Region:"Date Egg"),
-ends_with("(o/oo)"))
# A tibble: 344 x 8
studyName `Sample Number` Species `Culmen Length ~ `Culmen Depth (~
<chr> <dbl> <chr> <dbl> <dbl>
1 PAL0708 1 Adelie~ 39.1 18.7
2 PAL0708 2 Adelie~ 39.5 17.4
3 PAL0708 3 Adelie~ 40.3 18
4 PAL0708 4 Adelie~ NA NA
5 PAL0708 5 Adelie~ 36.7 19.3
6 PAL0708 6 Adelie~ 39.3 20.6
7 PAL0708 7 Adelie~ 38.9 17.8
8 PAL0708 8 Adelie~ 39.2 19.6
9 PAL0708 9 Adelie~ 34.1 18.1
10 PAL0708 10 Adelie~ 42 20.2
# ... with 334 more rows, and 3 more variables: `Flipper Length (mm)` <dbl>,
# `Body Mass (g)` <dbl>, Sex <chr>
Select using helper functions (we saw one above)
starts_with("abc"): matches names that begin with “abc”.ends_with("xyz"): matches names that end with “xyz”.contains("ijk"): matches names that contain “ijk”.matches("(.)\\1"): selects variables that match a regular expression. This one matches any variables that contain repeated characters. You’ll learn more about regular expressions in [strings].num_range("x", 1:3): matchesx1,x2andx3.
select(penguins_raw,
# cols starting with "Delta"
starts_with("Delta"))
# A tibble: 344 x 2
`Delta 15 N (o/oo)` `Delta 13 C (o/oo)`
<dbl> <dbl>
1 NA NA
2 8.95 -24.7
3 8.37 -25.3
4 NA NA
5 8.77 -25.3
6 8.66 -25.3
7 9.19 -25.2
8 9.46 -24.9
9 NA NA
10 9.13 -25.1
# ... with 334 more rows
select(penguins_raw,
# all cols except those ending with "(o/oo)"
-ends_with("(o/oo)"))
# A tibble: 344 x 15
studyName `Sample Number` Species Region Island Stage `Individual ID`
<chr> <dbl> <chr> <chr> <chr> <chr> <chr>
1 PAL0708 1 Adelie~ Anvers Torge~ Adul~ N1A1
2 PAL0708 2 Adelie~ Anvers Torge~ Adul~ N1A2
3 PAL0708 3 Adelie~ Anvers Torge~ Adul~ N2A1
4 PAL0708 4 Adelie~ Anvers Torge~ Adul~ N2A2
5 PAL0708 5 Adelie~ Anvers Torge~ Adul~ N3A1
6 PAL0708 6 Adelie~ Anvers Torge~ Adul~ N3A2
7 PAL0708 7 Adelie~ Anvers Torge~ Adul~ N4A1
8 PAL0708 8 Adelie~ Anvers Torge~ Adul~ N4A2
9 PAL0708 9 Adelie~ Anvers Torge~ Adul~ N5A1
10 PAL0708 10 Adelie~ Anvers Torge~ Adul~ N5A2
# ... with 334 more rows, and 8 more variables: `Clutch Completion` <chr>,
# `Date Egg` <date>, `Culmen Length (mm)` <dbl>, `Culmen Depth (mm)` <dbl>,
# `Flipper Length (mm)` <dbl>, `Body Mass (g)` <dbl>, Sex <chr>,
# Comments <chr>
select(penguins_raw,
# all columns except those with mm
-contains("(mm)"))
# A tibble: 344 x 14
studyName `Sample Number` Species Region Island Stage `Individual ID`
<chr> <dbl> <chr> <chr> <chr> <chr> <chr>
1 PAL0708 1 Adelie~ Anvers Torge~ Adul~ N1A1
2 PAL0708 2 Adelie~ Anvers Torge~ Adul~ N1A2
3 PAL0708 3 Adelie~ Anvers Torge~ Adul~ N2A1
4 PAL0708 4 Adelie~ Anvers Torge~ Adul~ N2A2
5 PAL0708 5 Adelie~ Anvers Torge~ Adul~ N3A1
6 PAL0708 6 Adelie~ Anvers Torge~ Adul~ N3A2
7 PAL0708 7 Adelie~ Anvers Torge~ Adul~ N4A1
8 PAL0708 8 Adelie~ Anvers Torge~ Adul~ N4A2
9 PAL0708 9 Adelie~ Anvers Torge~ Adul~ N5A1
10 PAL0708 10 Adelie~ Anvers Torge~ Adul~ N5A2
# ... with 334 more rows, and 7 more variables: `Clutch Completion` <chr>,
# `Date Egg` <date>, `Body Mass (g)` <dbl>, Sex <chr>, `Delta 15 N
# (o/oo)` <dbl>, `Delta 13 C (o/oo)` <dbl>, Comments <chr>
select(penguins_raw,
# any cols having brackets
# with letters or / inside
matches("\\([a-zA-Z/]+\\)"))
# A tibble: 344 x 6
`Culmen Length ~ `Culmen Depth (~ `Flipper Length~ `Body Mass (g)`
<dbl> <dbl> <dbl> <dbl>
1 39.1 18.7 181 3750
2 39.5 17.4 186 3800
3 40.3 18 195 3250
4 NA NA NA NA
5 36.7 19.3 193 3450
6 39.3 20.6 190 3650
7 38.9 17.8 181 3625
8 39.2 19.6 195 4675
9 34.1 18.1 193 3475
10 42 20.2 190 4250
# ... with 334 more rows, and 2 more variables: `Delta 15 N (o/oo)` <dbl>,
# `Delta 13 C (o/oo)` <dbl>
- Use
select()to reorder columns.
# move Species, Region, Island upfront them put
# in the rest
select(penguins_raw, Species, Region, Island,
everything())
# A tibble: 344 x 17
Species Region Island studyName `Sample Number` Stage `Individual ID`
<chr> <chr> <chr> <chr> <dbl> <chr> <chr>
1 Adelie~ Anvers Torge~ PAL0708 1 Adul~ N1A1
2 Adelie~ Anvers Torge~ PAL0708 2 Adul~ N1A2
3 Adelie~ Anvers Torge~ PAL0708 3 Adul~ N2A1
4 Adelie~ Anvers Torge~ PAL0708 4 Adul~ N2A2
5 Adelie~ Anvers Torge~ PAL0708 5 Adul~ N3A1
6 Adelie~ Anvers Torge~ PAL0708 6 Adul~ N3A2
7 Adelie~ Anvers Torge~ PAL0708 7 Adul~ N4A1
8 Adelie~ Anvers Torge~ PAL0708 8 Adul~ N4A2
9 Adelie~ Anvers Torge~ PAL0708 9 Adul~ N5A1
10 Adelie~ Anvers Torge~ PAL0708 10 Adul~ N5A2
# ... with 334 more rows, and 10 more variables: `Clutch Completion` <chr>,
# `Date Egg` <date>, `Culmen Length (mm)` <dbl>, `Culmen Depth (mm)` <dbl>,
# `Flipper Length (mm)` <dbl>, `Body Mass (g)` <dbl>, Sex <chr>, `Delta 15 N
# (o/oo)` <dbl>, `Delta 13 C (o/oo)` <dbl>, Comments <chr>
Exercises
Brainstorm as many ways as possible to select
dep_time,dep_delay,arr_time, andarr_delayfromflights.# by name select(flights, dep_time, dep_delay, arr_time, arr_delay)# A tibble: 336,776 x 4 dep_time dep_delay arr_time arr_delay <int> <dbl> <int> <dbl> 1 517 2 830 11 2 533 4 850 20 3 542 2 923 33 4 544 -1 1004 -18 5 554 -6 812 -25 6 554 -4 740 12 7 555 -5 913 19 8 557 -3 709 -14 9 557 -3 838 -8 10 558 -2 753 8 # ... with 336,766 more rows# range select(flights, dep_time:arr_delay, -sched_dep_time, -sched_arr_time)# A tibble: 336,776 x 4 dep_time dep_delay arr_time arr_delay <int> <dbl> <int> <dbl> 1 517 2 830 11 2 533 4 850 20 3 542 2 923 33 4 544 -1 1004 -18 5 554 -6 812 -25 6 554 -4 740 12 7 555 -5 913 19 8 557 -3 709 -14 9 557 -3 838 -8 10 558 -2 753 8 # ... with 336,766 more rows# except for select(flights, -(year:day), -sched_dep_time, -sched_arr_time, -(carrier:time_hour))# A tibble: 336,776 x 4 dep_time dep_delay arr_time arr_delay <int> <dbl> <int> <dbl> 1 517 2 830 11 2 533 4 850 20 3 542 2 923 33 4 544 -1 1004 -18 5 554 -6 812 -25 6 554 -4 740 12 7 555 -5 913 19 8 557 -3 709 -14 9 557 -3 838 -8 10 558 -2 753 8 # ... with 336,766 more rows# starts with select(flights, starts_with("dep"), starts_with("arr"))# A tibble: 336,776 x 4 dep_time dep_delay arr_time arr_delay <int> <dbl> <int> <dbl> 1 517 2 830 11 2 533 4 850 20 3 542 2 923 33 4 544 -1 1004 -18 5 554 -6 812 -25 6 554 -4 740 12 7 555 -5 913 19 8 557 -3 709 -14 9 557 -3 838 -8 10 558 -2 753 8 # ... with 336,766 more rows# ends with select(flights, ends_with("time"), ends_with("delay"), -sched_dep_time, -sched_arr_time, -air_time)# A tibble: 336,776 x 4 dep_time arr_time dep_delay arr_delay <int> <int> <dbl> <dbl> 1 517 830 2 11 2 533 850 4 20 3 542 923 2 33 4 544 1004 -1 -18 5 554 812 -6 -25 6 554 740 -4 12 7 555 913 -5 19 8 557 709 -3 -14 9 557 838 -3 -8 10 558 753 -2 8 # ... with 336,766 more rows# contains select(flights, contains("dep_"), contains("arr_"), -sched_dep_time, -sched_arr_time)# A tibble: 336,776 x 4 dep_time dep_delay arr_time arr_delay <int> <dbl> <int> <dbl> 1 517 2 830 11 2 533 4 850 20 3 542 2 923 33 4 544 -1 1004 -18 5 554 -6 812 -25 6 554 -4 740 12 7 555 -5 913 19 8 557 -3 709 -14 9 557 -3 838 -8 10 558 -2 753 8 # ... with 336,766 more rows# matches select(flights, matches("^(dep|arr)_(delay|time)"))# A tibble: 336,776 x 4 dep_time dep_delay arr_time arr_delay <int> <dbl> <int> <dbl> 1 517 2 830 11 2 533 4 850 20 3 542 2 923 33 4 544 -1 1004 -18 5 554 -6 812 -25 6 554 -4 740 12 7 555 -5 913 19 8 557 -3 709 -14 9 557 -3 838 -8 10 558 -2 753 8 # ... with 336,766 more rowsWhat happens if you include the name of a variable multiple times in a
select()call?Only one instance is included. If you want both variables you have to rename both variables.
select(penguins, species, island,
ends_with("mm"),
species,
body_mass_g:year)# A tibble: 344 x 8 species island bill_length_mm bill_depth_mm flipper_length_~ body_mass_g <fct> <fct> <dbl> <dbl> <int> <int> 1 Adelie Torge~ 39.1 18.7 181 3750 2 Adelie Torge~ 39.5 17.4 186 3800 3 Adelie Torge~ 40.3 18 195 3250 4 Adelie Torge~ NA NA NA NA 5 Adelie Torge~ 36.7 19.3 193 3450 6 Adelie Torge~ 39.3 20.6 190 3650 7 Adelie Torge~ 38.9 17.8 181 3625 8 Adelie Torge~ 39.2 19.6 195 4675 9 Adelie Torge~ 34.1 18.1 193 3475 10 Adelie Torge~ 42 20.2 190 4250 # ... with 334 more rows, and 2 more variables: sex <fct>, year <int>select(penguins, species = species, island,
ends_with("mm"),
rep_species = species,
body_mass_g:year)# A tibble: 344 x 9 species island bill_length_mm bill_depth_mm flipper_length_~ rep_species <fct> <fct> <dbl> <dbl> <int> <fct> 1 Adelie Torge~ 39.1 18.7 181 Adelie 2 Adelie Torge~ 39.5 17.4 186 Adelie 3 Adelie Torge~ 40.3 18 195 Adelie 4 Adelie Torge~ NA NA NA Adelie 5 Adelie Torge~ 36.7 19.3 193 Adelie 6 Adelie Torge~ 39.3 20.6 190 Adelie 7 Adelie Torge~ 38.9 17.8 181 Adelie 8 Adelie Torge~ 39.2 19.6 195 Adelie 9 Adelie Torge~ 34.1 18.1 193 Adelie 10 Adelie Torge~ 42 20.2 190 Adelie # ... with 334 more rows, and 3 more variables: body_mass_g <int>, sex <fct>, # year <int>What does the
one_of()function do? Why might it be helpful in conjunction with this vector?vars <- c("year", "month", "day", "dep_delay", "arr_delay")It selects a column if it forms a part of the vector.
select(flights, one_of(vars))# A tibble: 336,776 x 5 year month day dep_delay arr_delay <int> <int> <int> <dbl> <dbl> 1 2013 1 1 2 11 2 2013 1 1 4 20 3 2013 1 1 2 33 4 2013 1 1 -1 -18 5 2013 1 1 -6 -25 6 2013 1 1 -4 12 7 2013 1 1 -5 19 8 2013 1 1 -3 -14 9 2013 1 1 -3 -8 10 2013 1 1 -2 8 # ... with 336,766 more rowsDoes the result of running the following code surprise you? How do the select helpers deal with case by default? How can you change that default?
select(flights, contains("TIME"))The default is
ignore.case = TRUEhence this behaviour. If you want to find the precise thing you’re looking for addignore.case = FALSEin your helper function.select(flights, contains("TIME"))# A tibble: 336,776 x 6 dep_time sched_dep_time arr_time sched_arr_time air_time time_hour <int> <int> <int> <int> <dbl> <dttm> 1 517 515 830 819 227 2013-01-01 05:00:00 2 533 529 850 830 227 2013-01-01 05:00:00 3 542 540 923 850 160 2013-01-01 05:00:00 4 544 545 1004 1022 183 2013-01-01 05:00:00 5 554 600 812 837 116 2013-01-01 06:00:00 6 554 558 740 728 150 2013-01-01 05:00:00 7 555 600 913 854 158 2013-01-01 06:00:00 8 557 600 709 723 53 2013-01-01 06:00:00 9 557 600 838 846 140 2013-01-01 06:00:00 10 558 600 753 745 138 2013-01-01 06:00:00 # ... with 336,766 more rowsselect(flights,
contains("TIME",
ignore.case = FALSE))# A tibble: 336,776 x 0
Mutate
mutate() is the verb used to create new variables in your dataframe based on other variables in your dataset.
flights_sml <- select(flights,
year:day,
ends_with("delay"),
distance,
air_time
)
mutate(flights_sml,
gain = dep_delay - arr_delay,
speed = distance / air_time * 60,
hours = air_time / 60,
gain_per_hour = gain / hours
)
# A tibble: 336,776 x 11
year month day dep_delay arr_delay distance air_time gain speed hours
<int> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 2013 1 1 2 11 1400 227 -9 370. 3.78
2 2013 1 1 4 20 1416 227 -16 374. 3.78
3 2013 1 1 2 33 1089 160 -31 408. 2.67
4 2013 1 1 -1 -18 1576 183 17 517. 3.05
5 2013 1 1 -6 -25 762 116 19 394. 1.93
6 2013 1 1 -4 12 719 150 -16 288. 2.5
7 2013 1 1 -5 19 1065 158 -24 404. 2.63
8 2013 1 1 -3 -14 229 53 11 259. 0.883
9 2013 1 1 -3 -8 944 140 5 405. 2.33
10 2013 1 1 -2 8 733 138 -10 319. 2.3
# ... with 336,766 more rows, and 1 more variable: gain_per_hour <dbl>
To keep the new variables only, use transmute().
transmute(flights,
gain = dep_delay - arr_delay,
speed = distance / air_time * 60,
hours = air_time / 60,
gain_per_hour = gain / hours
)
# A tibble: 336,776 x 4
gain speed hours gain_per_hour
<dbl> <dbl> <dbl> <dbl>
1 -9 370. 3.78 -2.38
2 -16 374. 3.78 -4.23
3 -31 408. 2.67 -11.6
4 17 517. 3.05 5.57
5 19 394. 1.93 9.83
6 -16 288. 2.5 -6.4
7 -24 404. 2.63 -9.11
8 11 259. 0.883 12.5
9 5 405. 2.33 2.14
10 -10 319. 2.3 -4.35
# ... with 336,766 more rows
There are many operations you can use inside mutate() and transmute(), the key is that the function is vectorisable.
Modular arithmetic: %/% (integer division) and %% (remainder) are handy tools because it allows you to break integers up into pieces. For example we can compute hour and minute from dep_time using:
transmute(flights,
dep_time,
hour = dep_time %/% 100,
minute = dep_time %% 100
)# A tibble: 336,776 x 3
dep_time hour minute
<int> <dbl> <dbl>
1 517 5 17
2 533 5 33
3 542 5 42
4 544 5 44
5 554 5 54
6 554 5 54
7 555 5 55
8 557 5 57
9 557 5 57
10 558 5 58
# ... with 336,766 more rowsExercises
Currently
dep_timeandsched_dep_timeare convenient to look at, but hard to compute with because they’re not really continuous numbers. Convert them to a more convenient representation of number of minutes since midnight.flights %>%
select(dep_time,
sched_dep_time, time_hour)# A tibble: 336,776 x 3 dep_time sched_dep_time time_hour <int> <int> <dttm> 1 517 515 2013-01-01 05:00:00 2 533 529 2013-01-01 05:00:00 3 542 540 2013-01-01 05:00:00 4 544 545 2013-01-01 05:00:00 5 554 600 2013-01-01 06:00:00 6 554 558 2013-01-01 05:00:00 7 555 600 2013-01-01 06:00:00 8 557 600 2013-01-01 06:00:00 9 557 600 2013-01-01 06:00:00 10 558 600 2013-01-01 06:00:00 # ... with 336,766 more rowsmutate(flights,
minutes_since_00 =
# get int hr and mult by 60 to
# get hours converted to minutes.
(dep_time %/% 100) * 60 +
(dep_time %% 100)) # add minutes# A tibble: 336,776 x 20 year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time <int> <int> <int> <int> <int> <dbl> <int> <int> 1 2013 1 1 517 515 2 830 819 2 2013 1 1 533 529 4 850 830 3 2013 1 1 542 540 2 923 850 4 2013 1 1 544 545 -1 1004 1022 5 2013 1 1 554 600 -6 812 837 6 2013 1 1 554 558 -4 740 728 7 2013 1 1 555 600 -5 913 854 8 2013 1 1 557 600 -3 709 723 9 2013 1 1 557 600 -3 838 846 10 2013 1 1 558 600 -2 753 745 # ... with 336,766 more rows, and 12 more variables: arr_delay <dbl>, # carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>, # air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>, # minutes_since_00 <dbl>flights %>%
select(dep_time,
sched_dep_time, time_hour) %>%
filter(dep_time == 2400 | dep_time == 0) %>%
mutate(minutes_since_00 =
# get int hr and mult by 60 to
# get hours converted to minutes.
(dep_time %/% 100) * 60 +
(dep_time %% 100)) # add minutes# A tibble: 29 x 4 dep_time sched_dep_time time_hour minutes_since_00 <int> <int> <dttm> <dbl> 1 2400 2359 2013-10-30 23:00:00 1440 2 2400 2359 2013-11-27 23:00:00 1440 3 2400 2359 2013-12-05 23:00:00 1440 4 2400 2359 2013-12-09 23:00:00 1440 5 2400 2250 2013-12-09 22:00:00 1440 6 2400 2359 2013-12-13 23:00:00 1440 7 2400 2359 2013-12-19 23:00:00 1440 8 2400 1700 2013-12-29 17:00:00 1440 9 2400 2359 2013-02-07 23:00:00 1440 10 2400 2359 2013-02-07 23:00:00 1440 # ... with 19 more rowsWe see that the flights have midnight flights as 2400 and there’s none that are marked as 0. So we can change all 2400 to 0. We will learn the
if_else()construct later on, for now it follows the syntaxif_else(condition, do_this_if_condition_met_true, do_this_otherwise)flights_mut2 <- mutate(flights,
# if dep_time == 2400 (i.e. midnights)
# convert it to 0, else we keep the orig
# dep_time
dep_time = if_else(dep_time == 2400,
as.integer(0),
as.integer(dep_time)),
sched_dep_time =
if_else(sched_dep_time == 2400,
as.integer(0),
as.integer(sched_dep_time)),
minutes_dep_time =
(dep_time %/% 100) * 60 +
(dep_time %% 100),
minutes_sched_dep_time =
(sched_dep_time %/% 100) * 60 +
(sched_dep_time %% 100))
flights_mut2 %>%
select(dep_time, sched_dep_time,
minutes_dep_time,
minutes_sched_dep_time) %>%
head(10)# A tibble: 10 x 4 dep_time sched_dep_time minutes_dep_time minutes_sched_dep_time <int> <int> <dbl> <dbl> 1 517 515 317 315 2 533 529 333 329 3 542 540 342 340 4 544 545 344 345 5 554 600 354 360 6 554 558 354 358 7 555 600 355 360 8 557 600 357 360 9 557 600 357 360 10 558 600 358 360flights_mut2 %>%
select(dep_time, sched_dep_time,
minutes_dep_time,
minutes_sched_dep_time) %>%
filter(dep_time == 0 |
sched_dep_time == 0) %>%
head(10)# A tibble: 10 x 4 dep_time sched_dep_time minutes_dep_time minutes_sched_dep_time <int> <int> <dbl> <dbl> 1 0 2359 0 1439 2 0 2359 0 1439 3 0 2359 0 1439 4 0 2359 0 1439 5 0 2250 0 1370 6 0 2359 0 1439 7 0 2359 0 1439 8 0 1700 0 1020 9 0 2359 0 1439 10 0 2359 0 1439Compare
air_timewitharr_time - dep_time. What do you expect to see? What do you see? What do you need to do to fix it?flights %>% select(air_time, arr_time, dep_time) %>% mutate(diff_time = arr_time - dep_time)# A tibble: 336,776 x 4 air_time arr_time dep_time diff_time <dbl> <int> <int> <int> 1 227 830 517 313 2 227 850 533 317 3 160 923 542 381 4 183 1004 544 460 5 116 812 554 258 6 150 740 554 186 7 158 913 555 358 8 53 709 557 152 9 140 838 557 281 10 138 753 558 195 # ... with 336,766 more rowsWe see that just taking the difference between these times is misleading. Maybe we need to do the same as before, convert these to minutes since midnight before taking the difference? 🤷
flights_mut2 <- mutate(flights_mut2, arr_time = if_else(arr_time == 2400, as.integer(0), as.integer(arr_time)), minutes_arr_time = (arr_time %/% 100) * 60 + (arr_time %% 100), diff_time = minutes_arr_time - minutes_dep_time, diff_in_metrics = air_time - diff_time ) flights_mut2 %>% select(origin, dest, air_time, arr_time, dep_time, minutes_arr_time, minutes_dep_time, diff_time, diff_in_metrics) %>% head(10) %>% gt()origin dest air_time arr_time dep_time minutes_arr_time minutes_dep_time diff_time diff_in_metrics EWR IAH 227 830 517 510 317 193 34 LGA IAH 227 850 533 530 333 197 30 JFK MIA 160 923 542 563 342 221 -61 JFK BQN 183 1004 544 604 344 260 -77 LGA ATL 116 812 554 492 354 138 -22 EWR ORD 150 740 554 460 354 106 44 EWR FLL 158 913 555 553 355 198 -40 LGA IAD 53 709 557 429 357 72 -19 JFK MCO 140 838 557 518 357 161 -21 LGA ORD 138 753 558 473 358 115 23
Not quite, we see. So what else could be the difference? It could be that the arrival time is the local time at the destination. For example the time difference between New York and Houston is 1 hour. Houston is 1 hour behind New York. But upon checking the first couple some airports share the same timezone so I am a bit perplexed to tell the truth. E.g. the 3rd entry and the 4th entry, upon googling, suggests these are the same timezones? 😕
Compare
dep_time,sched_dep_time, anddep_delay. How would you expect those three numbers to be related?I would expect that:
dep_delay = dep_time - sched_dep_time
flights_mut3 <- mutate(flights_mut2, dep_delay_test = minutes_dep_time - minutes_sched_dep_time) %>% select(dep_time, sched_dep_time, minutes_dep_time, minutes_sched_dep_time, dep_delay, dep_delay_test) flights_mut3# A tibble: 336,776 x 6 dep_time sched_dep_time minutes_dep_time minutes_sched_d~ dep_delay <int> <int> <dbl> <dbl> <dbl> 1 517 515 317 315 2 2 533 529 333 329 4 3 542 540 342 340 2 4 544 545 344 345 -1 5 554 600 354 360 -6 6 554 558 354 358 -4 7 555 600 355 360 -5 8 557 600 357 360 -3 9 557 600 357 360 -3 10 558 600 358 360 -2 # ... with 336,766 more rows, and 1 more variable: dep_delay_test <dbl>flights_mut3 %>% filter(dep_delay != dep_delay_test)# A tibble: 1,236 x 6 dep_time sched_dep_time minutes_dep_time minutes_sched_d~ dep_delay <int> <int> <dbl> <dbl> <dbl> 1 848 1835 528 1115 853 2 42 2359 42 1439 43 3 126 2250 86 1370 156 4 32 2359 32 1439 33 5 50 2145 50 1305 185 6 235 2359 155 1439 156 7 25 2359 25 1439 26 8 106 2245 66 1365 141 9 14 2359 14 1439 15 10 37 2230 37 1350 127 # ... with 1,226 more rows, and 1 more variable: dep_delay_test <dbl>This does seem to be the case for most flights except for flights that were delayed overnight. To cater for these we’d need to work out whether the flight departed on the same day. If they departed on different days we would find how many minutes left to midnight, and then subtract the scheduled dep time from this and add to the departure time to get the delay.
flights_mut2 %>% filter(dep_delay < 0) %>% arrange((dep_delay)) %>% head(4) %>% gt()year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time arr_delay carrier flight tailnum origin dest air_time distance hour minute time_hour minutes_dep_time minutes_sched_dep_time minutes_arr_time diff_time diff_in_metrics 2013 12 7 2040 2123 -43 40 2352 48 B6 97 N592JB JFK DEN 265 1626 21 23 2013-12-07 21:00:00 1240 1283 40 -1200 1465 2013 2 3 2022 2055 -33 2240 2338 -58 DL 1715 N612DL LGA MSY 162 1183 20 55 2013-02-03 20:00:00 1222 1255 1360 138 24 2013 11 10 1408 1440 -32 1549 1559 -10 EV 5713 N825AS LGA IAD 52 229 14 40 2013-11-10 14:00:00 848 880 949 101 -49 2013 1 11 1900 1930 -30 2233 2243 -10 DL 1435 N934DL LGA TPA 139 1010 19 30 2013-01-11 19:00:00 1140 1170 1353 213 -74 flights_mut3 <- mutate(flights_mut2, dep_delay_test = if_else( (minutes_dep_time >= minutes_sched_dep_time) | (minutes_dep_time - minutes_sched_dep_time ) > -45, minutes_dep_time - minutes_sched_dep_time, 24*60 - minutes_sched_dep_time + minutes_dep_time)) %>% select(dep_time, sched_dep_time, minutes_dep_time, minutes_sched_dep_time, dep_delay, dep_delay_test) flights_mut3# A tibble: 336,776 x 6 dep_time sched_dep_time minutes_dep_time minutes_sched_d~ dep_delay <int> <int> <dbl> <dbl> <dbl> 1 517 515 317 315 2 2 533 529 333 329 4 3 542 540 342 340 2 4 544 545 344 345 -1 5 554 600 354 360 -6 6 554 558 354 358 -4 7 555 600 355 360 -5 8 557 600 357 360 -3 9 557 600 357 360 -3 10 558 600 358 360 -2 # ... with 336,766 more rows, and 1 more variable: dep_delay_test <dbl>flights_mut3 %>% filter(dep_delay != dep_delay_test) %>% head(10) %>% gt()dep_time sched_dep_time minutes_dep_time minutes_sched_dep_time dep_delay dep_delay_test
The above checks the biggest difference for those flights that had dep_delay < 0. Note: This is a bit hacky. Best approach is to compare dates but both the year-month-day and time_hour reflect the same date.Find the 10 most delayed flights using a ranking function. How do you want to handle ties? Carefully read the documentation for
min_rank().mutate(flights, rank = min_rank(desc(dep_delay))) %>% arrange(rank) %>% head(10) %>% gt()year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time arr_delay carrier flight tailnum origin dest air_time distance hour minute time_hour rank 2013 1 9 641 900 1301 1242 1530 1272 HA 51 N384HA JFK HNL 640 4983 9 0 2013-01-09 09:00:00 1 2013 6 15 1432 1935 1137 1607 2120 1127 MQ 3535 N504MQ JFK CMH 74 483 19 35 2013-06-15 19:00:00 2 2013 1 10 1121 1635 1126 1239 1810 1109 MQ 3695 N517MQ EWR ORD 111 719 16 35 2013-01-10 16:00:00 3 2013 9 20 1139 1845 1014 1457 2210 1007 AA 177 N338AA JFK SFO 354 2586 18 45 2013-09-20 18:00:00 4 2013 7 22 845 1600 1005 1044 1815 989 MQ 3075 N665MQ JFK CVG 96 589 16 0 2013-07-22 16:00:00 5 2013 4 10 1100 1900 960 1342 2211 931 DL 2391 N959DL JFK TPA 139 1005 19 0 2013-04-10 19:00:00 6 2013 3 17 2321 810 911 135 1020 915 DL 2119 N927DA LGA MSP 167 1020 8 10 2013-03-17 08:00:00 7 2013 6 27 959 1900 899 1236 2226 850 DL 2007 N3762Y JFK PDX 313 2454 19 0 2013-06-27 19:00:00 8 2013 7 22 2257 759 898 121 1026 895 DL 2047 N6716C LGA ATL 109 762 7 59 2013-07-22 07:00:00 9 2013 12 5 756 1700 896 1058 2020 878 AA 172 N5DMAA EWR MIA 149 1085 17 0 2013-12-05 17:00:00 10 What does
1:3 + 1:10return? Why?1:3 + 1:10[1] 2 4 6 5 7 9 8 10 12 11The vector that is shorter gets recycled.
So essentially it is outputting:
df <- data.frame(a = c(rep(1:3,3), 1), b = 1:10) df %>% mutate(c = a + b) %>% gt()a b c 1 1 2 2 2 4 3 3 6 1 4 5 2 5 7 3 6 9 1 7 8 2 8 10 3 9 12 1 10 11 What trigonometric functions does R provide?
It provides pretty much all the trig functions. Here is a list.
Grouped summaries with summarise()/summarize()
If you need to find out the maximum of a certain variable, or the mean, median, minimum etc. you use summarise(). na.rm = TRUE means exclude the NA values then get the summary stat. I added a simple example in the code below. Notice that when we have NAs in our dataset and then we take a mean, the result is NA. If there are NAs in the input, there are NAs in the output. The na.rm = TRUE helps us by ignoring the NA values, and getting the summary stat for the non NA values here.
summarise(flights, delay = mean(dep_delay, na.rm = TRUE))# A tibble: 1 x 1
delay
<dbl>
1 12.6summarise(flights,
n = n(),
biggest_dep_delay = max(dep_delay, na.rm = TRUE),
biggest_arr_delay = max(arr_delay, na.rm = TRUE))# A tibble: 1 x 3
n biggest_dep_delay biggest_arr_delay
<int> <dbl> <dbl>
1 336776 1301 1272# Why use na.rm?
test = c(3, 3, 6, 6, NA, NA)
mean(test)[1] NAmean(test, na.rm = TRUE)[1] 4.5Did you see that when we used mean(test, na.rm = TRUE) we got a result of 4.5, which is \((3 + 3+ 6 + 6)/4\). I.e. the NA values are removed and the count that forms the denominator is also updated.
Summary stats are most often valuable when we look across groups. For example, let’s say we wanted to know the median bill length of penguins based on species and sex. We first group by species and sex since we’re interested to see how the stats change based on species and whether the penguin is male/female. When I ran it the first time there were some species that had sex == NA, so I removed the NA values from the penguin dataset before grouping it.
by_species_sex <- group_by(drop_na(penguins), species, sex)
summarise(by_species_sex,
med_bill_length_mm = median(bill_length_mm, na.rm = TRUE),
med_bill_depth_mm = median(bill_depth_mm, na.rm = TRUE),
med_flipper_length_mm = median(flipper_length_mm, na.rm = TRUE),
med_body_mass_g = median(body_mass_g, na.rm = TRUE)) %>%
gt()| sex | med_bill_length_mm | med_bill_depth_mm | med_flipper_length_mm | med_body_mass_g |
|---|---|---|---|---|
| Adelie | ||||
| female | 37.00 | 17.60 | 188.0 | 3400 |
| male | 40.60 | 18.90 | 193.0 | 4000 |
| Chinstrap | ||||
| female | 46.30 | 17.65 | 192.0 | 3550 |
| male | 50.95 | 19.30 | 200.5 | 3950 |
| Gentoo | ||||
| female | 45.50 | 14.25 | 212.0 | 4700 |
| male | 49.50 | 15.70 | 221.0 | 5500 |
The pipe: %>%
The book uses an example with flights, here we add a new example with the penguins data set. As pointed out in the book, it is a bit tedious, and arguably doesn’t flow very well, or read particularly well the first way.
Hence the pipe! The pipe is more inuitive for me, and as discussed previously can be read as as then.
More onerous way
by_species <- group_by(drop_na(penguins), species, sex)
measures <- summarise(by_species,
count = n(),
med_bill_length_mm = median(bill_length_mm, na.rm = TRUE),
med_bill_depth_mm = median(bill_depth_mm, na.rm = TRUE),
med_flipper_length_mm = median(flipper_length_mm, na.rm = TRUE),
med_body_mass_g = median(body_mass_g, na.rm = TRUE)
)
ggplot(data = measures, mapping = aes(x = med_body_mass_g,
y = med_flipper_length_mm)) +
geom_point(aes(size = count), alpha = 1/3) 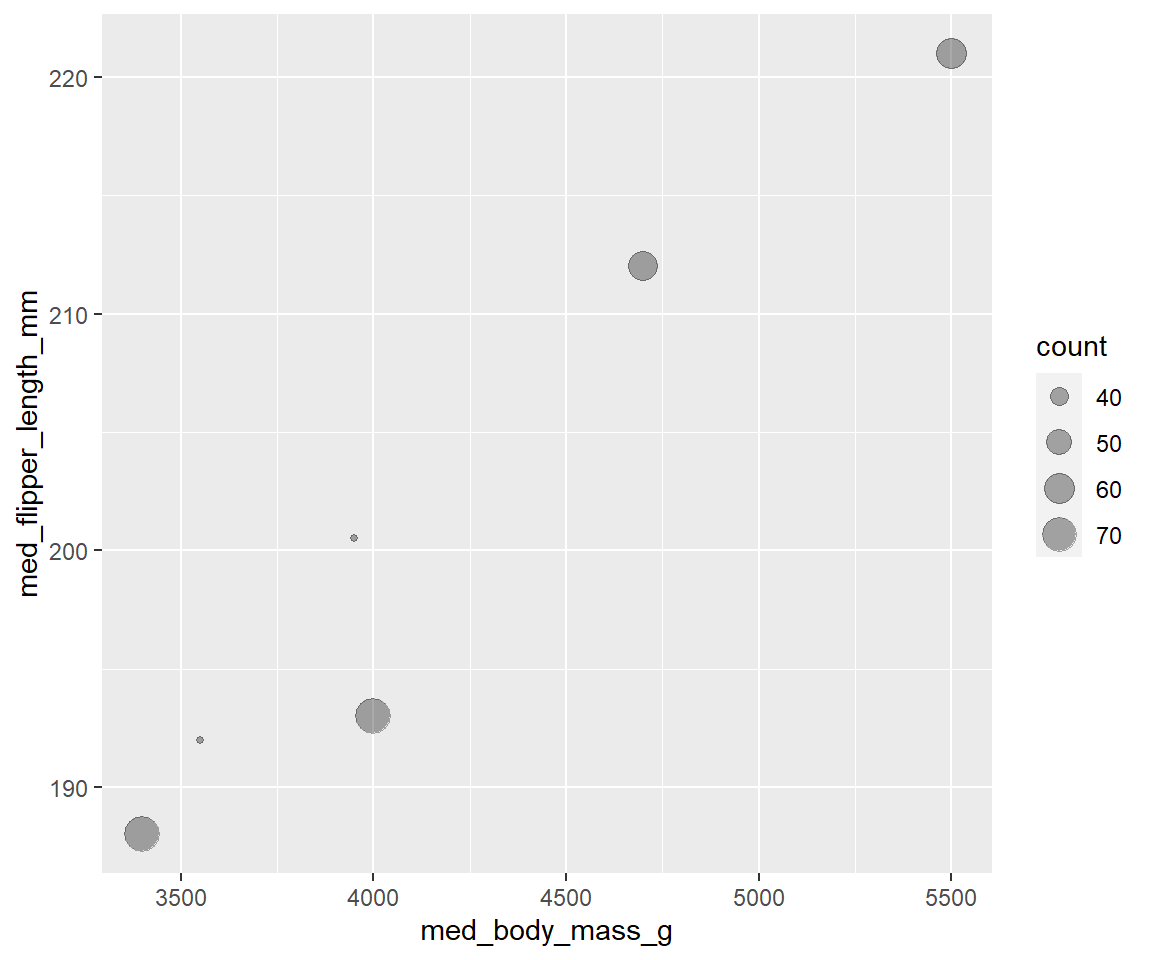
Using the pipe %>%
penguins %>%
drop_na() %>%
group_by(species, sex) %>%
summarise(count = n(),
med_bill_length_mm = median(bill_length_mm, na.rm = TRUE),
med_bill_depth_mm = median(bill_depth_mm, na.rm = TRUE),
med_flipper_length_mm = median(flipper_length_mm, na.rm = TRUE),
med_body_mass_g = median(body_mass_g, na.rm = TRUE)) %>%
ggplot(mapping = aes(x = med_body_mass_g,
y = med_flipper_length_mm)) +
geom_point(aes(size = count), alpha = 1/3) 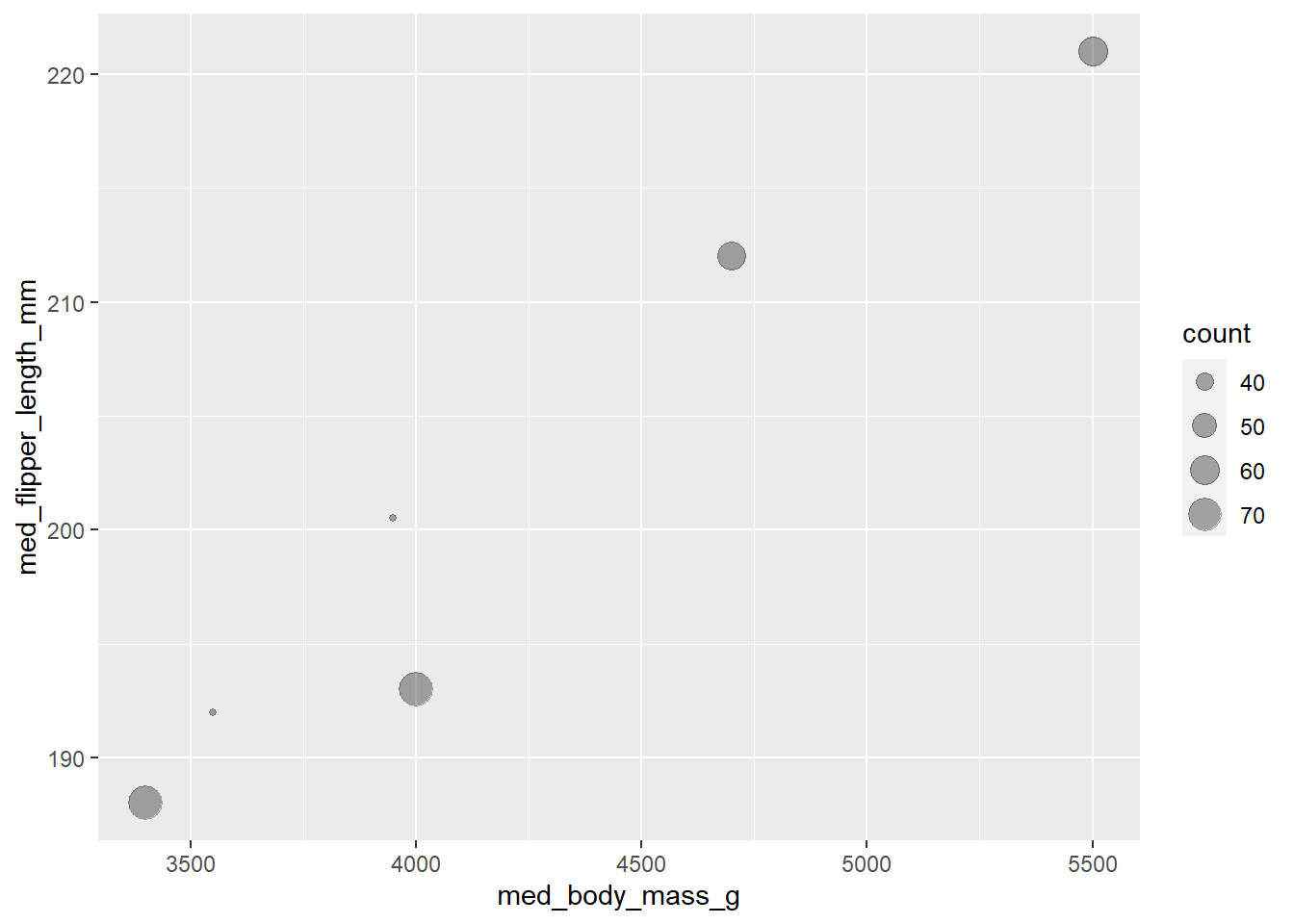
Counts
n()which produces a count and,sum(is.na(some_variable))gives you the NA sum of data.
penguins %>%
group_by(species) %>%
summarise(count = n())# A tibble: 3 x 2
species count
<fct> <int>
1 Adelie 152
2 Chinstrap 68
3 Gentoo 124# how many missing data in certain characteristics?
penguins %>%
select(species, body_mass_g) %>%
group_by(species) %>%
summarise(count = sum(is.na(body_mass_g)))# A tibble: 3 x 2
species count
<fct> <int>
1 Adelie 1
2 Chinstrap 0
3 Gentoo 1# how many missing data in certain characteristics?
penguins %>%
select(species, bill_length_mm) %>%
group_by(species) %>%
summarise(count = sum(is.na(bill_length_mm)))# A tibble: 3 x 2
species count
<fct> <int>
1 Adelie 1
2 Chinstrap 0
3 Gentoo 1Useful summary Functions
Measures of Location (mean, median etc.)
- Median: `median(x) is the value of x where 50% of the variable x lies above that value, and 50% of the variable x lies below that value.
not_cancelled <- flights %>%
filter(!is.na(dep_delay),
!is.na(arr_delay))
not_cancelled %>%
group_by(year, month, day) %>%
summarise(
# avg_delay
avg_delay1 = mean(arr_delay),
# avg positive delay - filter our only positive arrival delay
avg_delay2 = mean(arr_delay[arr_delay > 0])
)# A tibble: 365 x 5
# Groups: year, month [12]
year month day avg_delay1 avg_delay2
<int> <int> <int> <dbl> <dbl>
1 2013 1 1 12.7 32.5
2 2013 1 2 12.7 32.0
3 2013 1 3 5.73 27.7
4 2013 1 4 -1.93 28.3
5 2013 1 5 -1.53 22.6
6 2013 1 6 4.24 24.4
7 2013 1 7 -4.95 27.8
8 2013 1 8 -3.23 20.8
9 2013 1 9 -0.264 25.6
10 2013 1 10 -5.90 27.3
# ... with 355 more rowsMeasures of spread (sd, IQR etc.)
- Standard deviation: sd() measures the spread of a variable, the larger the sd the more dispersed the data.
- Inter Quartile Range: IQR() measures the spread in the middle - i.e. it measures the spread in the middle 50% of the variable. The measure is useful if there are outliers. \(IQR = Q{3} - Q{1}\).
- Median Absolute Devaition: mad() measures how far each point is from the median of the variable in absolute terms.
- Need the Mean Absolute Deviation: That can be found in the DescTools package.
not_cancelled %>%
group_by(dest) %>%
summarise(dist_sd = sd(distance),
dist_iqr = IQR(distance),
dist_mad = mad(distance)) %>%
arrange(desc(dist_sd))# A tibble: 104 x 4
dest dist_sd dist_iqr dist_mad
<chr> <dbl> <dbl> <dbl>
1 EGE 10.5 21 1.48
2 SAN 10.4 21 0
3 SFO 10.2 21 0
4 HNL 10.0 20 0
5 SEA 9.98 20 0
6 LAS 9.91 21 0
7 PDX 9.87 20 0
8 PHX 9.86 20 0
9 LAX 9.66 21 0
10 IND 9.46 20 0
# ... with 94 more rows
sessionInfo()R version 3.6.3 (2020-02-29)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 18363)
Matrix products: default
locale:
[1] LC_COLLATE=English_South Africa.1252 LC_CTYPE=English_South Africa.1252
[3] LC_MONETARY=English_South Africa.1252 LC_NUMERIC=C
[5] LC_TIME=English_South Africa.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] emo_0.0.0.9000 skimr_2.1.1 gt_0.2.2
[4] palmerpenguins_0.1.0 nycflights13_1.0.1 flair_0.0.2
[7] forcats_0.5.0 stringr_1.4.0 dplyr_1.0.0
[10] purrr_0.3.4 readr_1.3.1 tidyr_1.1.0
[13] tibble_3.0.3 ggplot2_3.3.0 tidyverse_1.3.0
[16] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rcpp_1.0.4.6 lubridate_1.7.8 lattice_0.20-38 utf8_1.1.4
[5] assertthat_0.2.1 rprojroot_1.3-2 digest_0.6.25 R6_2.4.1
[9] cellranger_1.1.0 repr_1.1.0 backports_1.1.6 reprex_0.3.0
[13] evaluate_0.14 highr_0.8 httr_1.4.2 pillar_1.4.6
[17] rlang_0.4.7 readxl_1.3.1 rstudioapi_0.11 whisker_0.4
[21] checkmate_2.0.0 rmarkdown_2.1 labeling_0.3 munsell_0.5.0
[25] broom_0.5.6 compiler_3.6.3 httpuv_1.5.2 modelr_0.1.6
[29] xfun_0.13 pkgconfig_2.0.3 base64enc_0.1-3 htmltools_0.5.0
[33] tidyselect_1.1.0 fansi_0.4.1 crayon_1.3.4 dbplyr_1.4.3
[37] withr_2.2.0 later_1.0.0 grid_3.6.3 nlme_3.1-144
[41] jsonlite_1.7.0 gtable_0.3.0 lifecycle_0.2.0 DBI_1.1.0
[45] git2r_0.26.1 magrittr_1.5 scales_1.1.0 cli_2.0.2
[49] stringi_1.4.6 farver_2.0.3 fs_1.4.1 promises_1.1.0
[53] xml2_1.3.2 ellipsis_0.3.1 generics_0.0.2 vctrs_0.3.2
[57] tools_3.6.3 glue_1.4.1 hms_0.5.3 yaml_2.2.1
[61] colorspace_1.4-1 rvest_0.3.5 knitr_1.28 haven_2.2.0
[65] sass_0.2.0