analysis_congreso_forestal
ajpelu
2022-01-13
Last updated: 2022-01-14
Checks: 7 0
Knit directory:
dronveg_alcontar/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210923) was run prior to running the code in the R Markdown file.
Setting a seed ensures that any results that rely on randomness, e.g.
subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version a4923cc. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the
analysis have been committed to Git prior to generating the results (you can
use wflow_publish or wflow_git_commit). workflowr only
checks the R Markdown file, but you know if there are other scripts or data
files that it depends on. Below is the status of the Git repository when the
results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Untracked files:
Untracked: analysis/ecology-letters.csl
Untracked: analysis/references.bib
Unstaged changes:
Modified: analysis/compara_drone_campo.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made
to the R Markdown (analysis/analysis_congreso_forestal.Rmd) and HTML (docs/analysis_congreso_forestal.html)
files. If you’ve configured a remote Git repository (see
?wflow_git_remote), click on the hyperlinks in the table below to
view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | a4923cc | ajpelu | 2022-01-14 | add analysis congreso |
0.1 Prepara Datos
knitr::opts_chunk$set(echo = FALSE,
warning = FALSE,
message = FALSE,
fig.width=10, fig.height=7)Usamos datos de cobertura vegetal de las parcelas de campo (cob.campo) y datos derivados de dron (cob.dron).
De los datos de dron, utilizamos el método de estimación denominado COBERTURA (ver análisis preliminar)
Los datos de campo corresponden al muestreo realizado el 19/05/2021.
El vuelo del dron se realizó el día 21/05/2021.
Los rangos de cobertura se han reclasifiaco de acuerdo a:
| RANGO_INFOCA | Nombre | Cobertura |
|---|---|---|
| 1 | “Matorral claro” | “<25%” |
| 2 | “Matorral medio” | “25-50%” |
| 3 | “Espartal denso” | “>75%” |
| 4 | “Aulagar denso” | “>75%” |
0.2 Correlación General
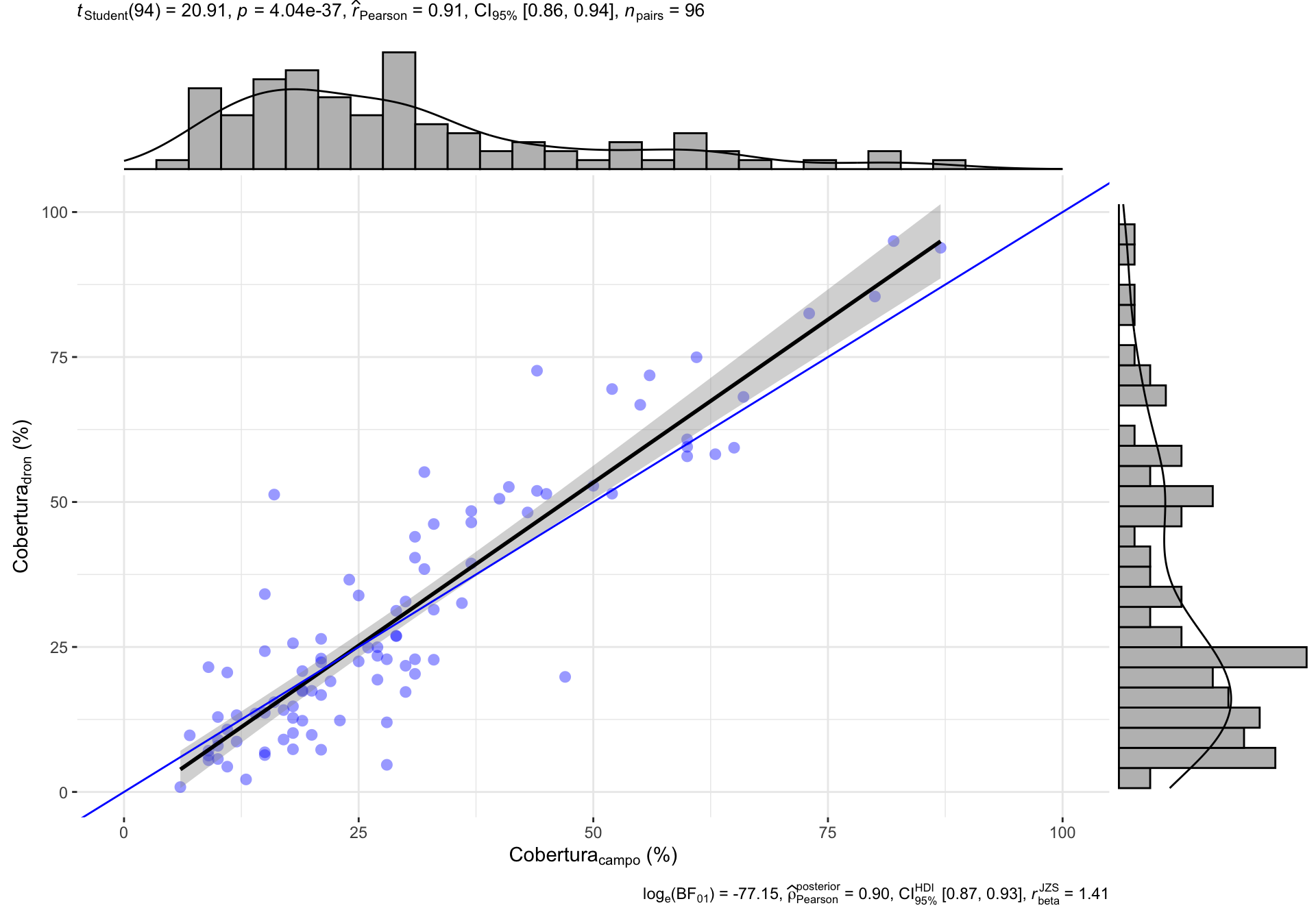
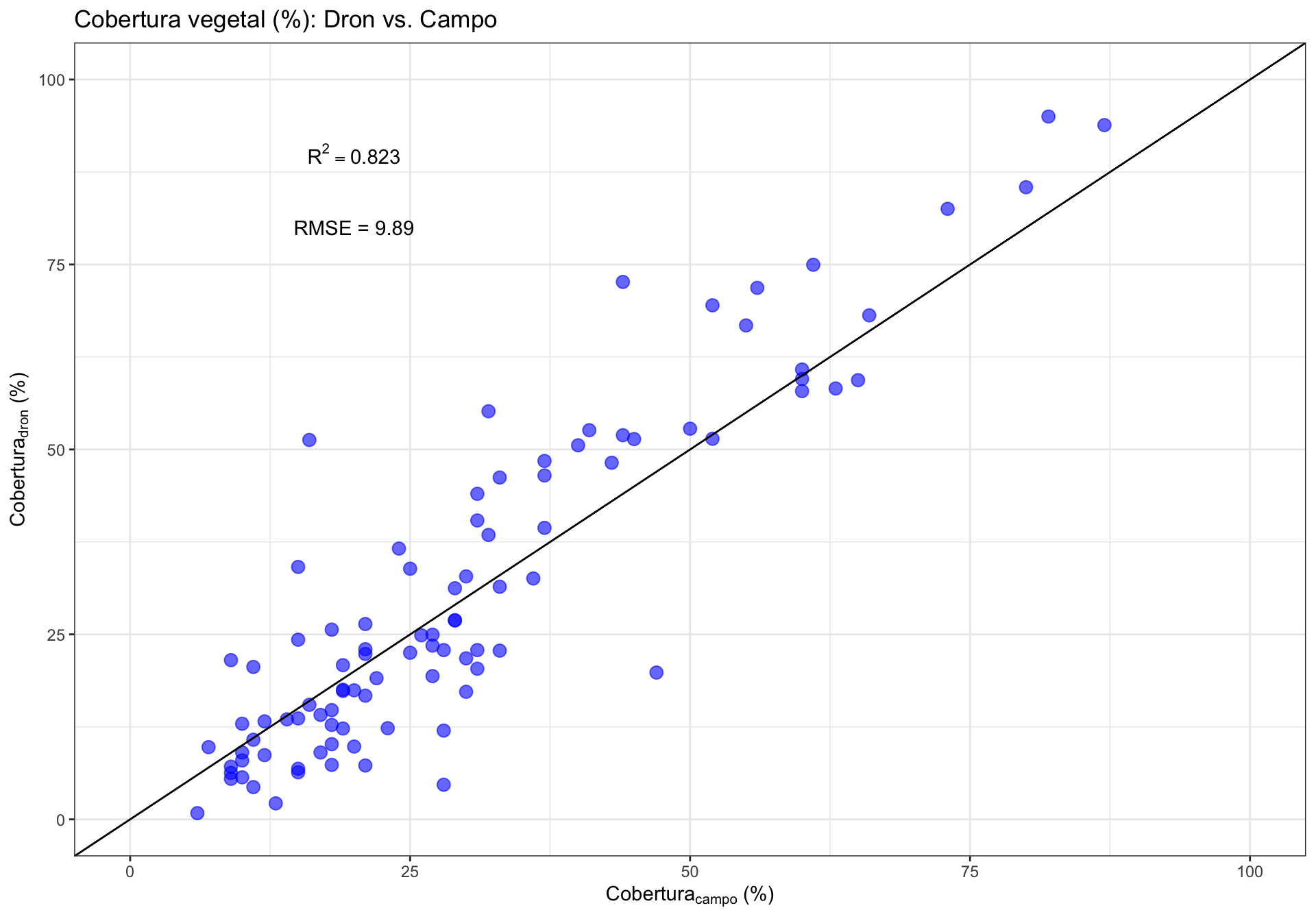
0.3 Correlación por Rangos
Explorar como varía la correlación en los diferentes rangos de cobertura
Computar el RMSE, y el RMSE normalizado. El RMSE es dependiente de la escala, por lo que necesitaríamos normalizar para poder comparar entre las clases de cobertura.
| Rango de cobertura | RMSE | min | max | norm. RMSE % |
|---|---|---|---|---|
| Aulagar denso (>75%) | 12.69 | 10 | 61 | 24.89 |
| Espartal denso (>75%) | 7.63 | 9 | 87 | 9.78 |
| Matorral claro (<25%) | 7.30 | 6 | 28 | 33.18 |
| Matorral medio (25-50%) | 10.89 | 9 | 63 | 20.16 |
- Generamos las ecuaciones para la gráfica
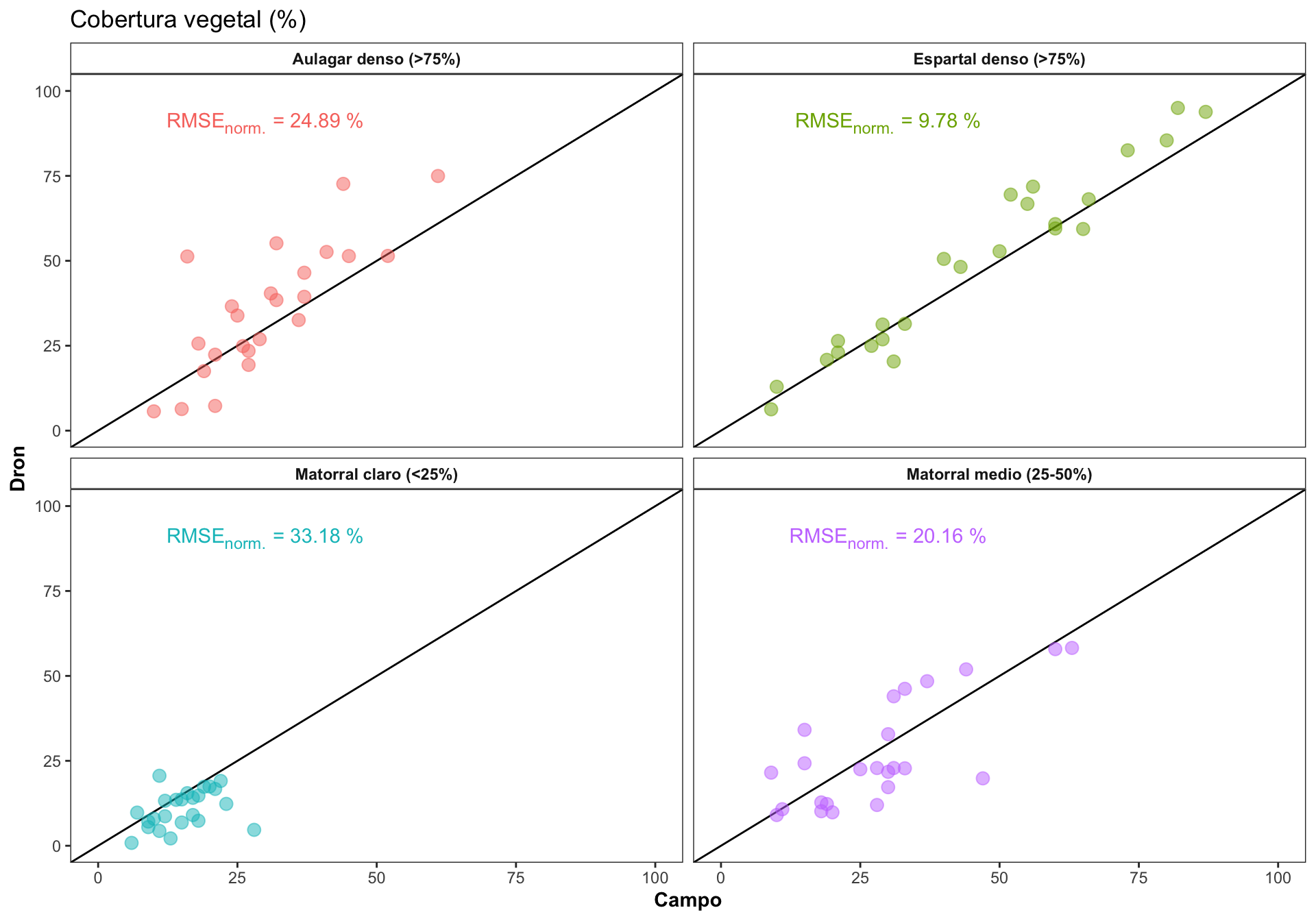
0.4 Influencia de otras variables en la Variación de la correlación
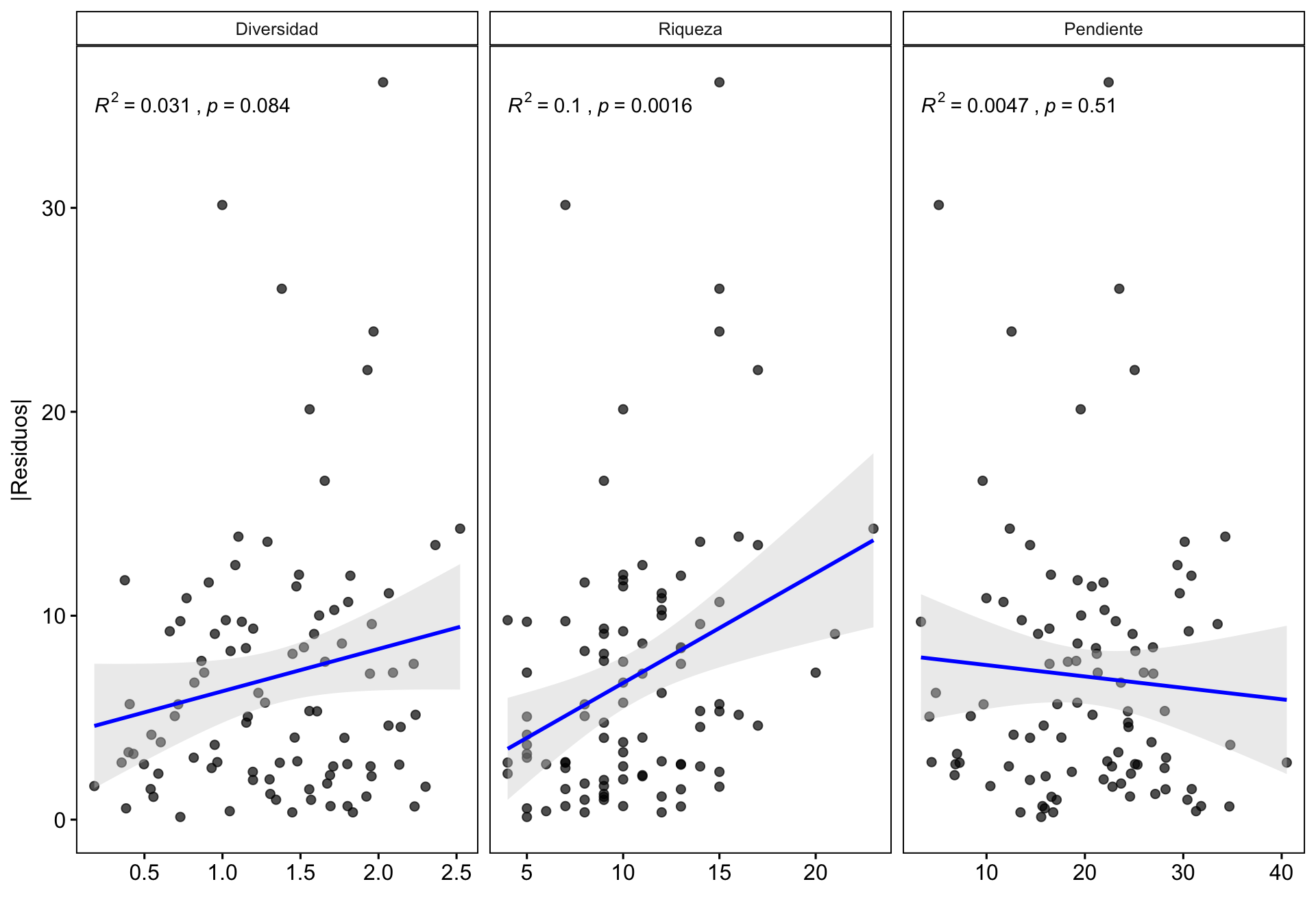
¿Existe alguna relación entre la correlación y otras variables? Podría interesarnos explorar cómo otras variables podrían influir en la correlación dron-campo, por ejemplo la riqueza o la pendiente. Se pueden utilizar varios enfoques (análisis exploratorio, residuos, etc.). En nuestro caso utilizamos la correlación entre los residuos de la correlación y las diferentes variables.
- Calculamos los residuos y los residuos absolutos
m <- lm(cob.dron ~ cob.campo, data=df)
df <- df %>% modelr::add_residuals(m) %>%
mutate(resid.abs = abs(resid))
dfres <- df %>% dplyr::select(coverclass, Diversidad = shannon, Riqueza = rich, Pendiente = slope, resid, resid.abs) %>%
pivot_longer(cols = c("Diversidad", "Riqueza", "Pendiente")) %>%
mutate(variable = fct_relevel(name, c("Diversidad", "Riqueza", "Pendiente")))- Hacemos gráfico de las tres variables
R version 4.0.2 (2020-06-22)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Catalina 10.15.3
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRblas.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] modelr_0.1.8 ggpubr_0.4.0 ggtext_0.1.1 kableExtra_1.3.1
[5] Metrics_0.1.4 ggstatsplot_0.7.2 readxl_1.3.1 here_1.0.1
[9] forcats_0.5.1 stringr_1.4.0 dplyr_1.0.6 purrr_0.3.4
[13] readr_1.4.0 tidyr_1.1.3 tibble_3.1.2 ggplot2_3.3.5
[17] tidyverse_1.3.1 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] pairwiseComparisons_3.1.3 backports_1.2.1
[3] plyr_1.8.6 splines_4.0.2
[5] gmp_0.6-2 kSamples_1.2-9
[7] ipmisc_5.0.2 TH.data_1.0-10
[9] digest_0.6.27 SuppDists_1.1-9.5
[11] htmltools_0.5.2 fansi_0.4.2
[13] magrittr_2.0.1 memoise_2.0.0
[15] paletteer_1.3.0 openxlsx_4.2.3
[17] sandwich_3.0-0 colorspace_2.0-2
[19] rvest_1.0.0 ggrepel_0.9.1
[21] haven_2.3.1 xfun_0.23
[23] callr_3.7.0 crayon_1.4.1
[25] jsonlite_1.7.2 zeallot_0.1.0
[27] survival_3.2-7 zoo_1.8-8
[29] glue_1.4.2 gtable_0.3.0
[31] emmeans_1.5.4 webshot_0.5.2
[33] MatrixModels_0.4-1 statsExpressions_1.1.0
[35] car_3.0-10 Rmpfr_0.8-2
[37] abind_1.4-5 scales_1.1.1.9000
[39] mvtnorm_1.1-1 DBI_1.1.1
[41] rstatix_0.6.0 PMCMRplus_1.9.0
[43] miniUI_0.1.1.1 Rcpp_1.0.7
[45] viridisLite_0.4.0 xtable_1.8-4
[47] performance_0.8.0 gridtext_0.1.4
[49] foreign_0.8-81 httr_1.4.2
[51] ellipsis_0.3.2 farver_2.1.0
[53] pkgconfig_2.0.3 reshape_0.8.8
[55] multcompView_0.1-8 sass_0.3.1
[57] dbplyr_2.1.1 utf8_1.1.4
[59] labeling_0.4.2 tidyselect_1.1.1
[61] rlang_0.4.12 later_1.1.0.1
[63] ggcorrplot_0.1.3 effectsize_0.4.5
[65] munsell_0.5.0 cellranger_1.1.0
[67] tools_4.0.2 cachem_1.0.4
[69] cli_2.5.0 generics_0.1.0
[71] broom_0.7.9 evaluate_0.14
[73] fastmap_1.1.0 BWStest_0.2.2
[75] yaml_2.2.1 rematch2_2.1.2
[77] processx_3.5.1 knitr_1.31
[79] fs_1.5.0 zip_2.1.1
[81] nlme_3.1-152 WRS2_1.1-1
[83] pbapply_1.4-3 mime_0.10
[85] whisker_0.4 ggExtra_0.9
[87] xml2_1.3.2 correlation_0.6.1
[89] compiler_4.0.2 rstudioapi_0.13
[91] curl_4.3 ggsignif_0.6.0
[93] reprex_2.0.0 bslib_0.2.4
[95] stringi_1.7.4 highr_0.8
[97] ps_1.5.0 parameters_0.14.0
[99] lattice_0.20-41 Matrix_1.3-2
[101] markdown_1.1 vctrs_0.3.8
[103] pillar_1.6.1 lifecycle_1.0.1
[105] mc2d_0.1-18 jquerylib_0.1.3
[107] estimability_1.3 data.table_1.14.0
[109] insight_0.14.4 httpuv_1.5.5
[111] patchwork_1.1.1 R6_2.5.1
[113] bookdown_0.21.6 promises_1.2.0.1
[115] rio_0.5.16 BayesFactor_0.9.12-4.2
[117] codetools_0.2-18 MASS_7.3-53
[119] gtools_3.8.2 assertthat_0.2.1
[121] rprojroot_2.0.2 withr_2.4.1
[123] multcomp_1.4-16 mgcv_1.8-33
[125] bayestestR_0.9.0 parallel_4.0.2
[127] hms_1.0.0 grid_4.0.2
[129] coda_0.19-4 rmarkdown_2.8
[131] carData_3.0-4 git2r_0.28.0
[133] getPass_0.2-2 shiny_1.6.0
[135] lubridate_1.7.10