BMI - Brain Cerebellum
sheng Qian
2021-2-6
Last updated: 2022-02-21
Checks: 6 1
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/ | data |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/ctwas_config.R | code/ctwas_config.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version bbf6737. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .ipynb_checkpoints/
Untracked files:
Untracked: Rplot.png
Untracked: analysis/Glucose_Adipose_Subcutaneous.Rmd
Untracked: analysis/Glucose_Adipose_Visceral_Omentum.Rmd
Untracked: analysis/Splicing_Test.Rmd
Untracked: code/.ipynb_checkpoints/
Untracked: code/AF_out/
Untracked: code/BMI_S_out/
Untracked: code/BMI_out/
Untracked: code/Glucose_out/
Untracked: code/LDL_S_out/
Untracked: code/T2D_out/
Untracked: code/ctwas_config.R
Untracked: code/mapping.R
Untracked: code/out/
Untracked: code/run_AF_analysis.sbatch
Untracked: code/run_AF_analysis.sh
Untracked: code/run_AF_ctwas_rss_LDR.R
Untracked: code/run_BMI_analysis.sbatch
Untracked: code/run_BMI_analysis.sh
Untracked: code/run_BMI_analysis_S.sbatch
Untracked: code/run_BMI_analysis_S.sh
Untracked: code/run_BMI_ctwas_rss_LDR.R
Untracked: code/run_BMI_ctwas_rss_LDR_S.R
Untracked: code/run_Glucose_analysis.sbatch
Untracked: code/run_Glucose_analysis.sh
Untracked: code/run_Glucose_ctwas_rss_LDR.R
Untracked: code/run_LDL_analysis_S.sbatch
Untracked: code/run_LDL_analysis_S.sh
Untracked: code/run_LDL_ctwas_rss_LDR_S.R
Untracked: code/run_T2D_analysis.sbatch
Untracked: code/run_T2D_analysis.sh
Untracked: code/run_T2D_ctwas_rss_LDR.R
Untracked: data/.ipynb_checkpoints/
Untracked: data/AF/
Untracked: data/BMI/
Untracked: data/BMI_S/
Untracked: data/Glucose/
Untracked: data/LDL_S/
Untracked: data/T2D/
Untracked: data/TEST/
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/mashr_sqtl/
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: data/untitled.txt
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/BMI_Brain_Cerebellum.Rmd) and HTML (docs/BMI_Brain_Cerebellum.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | bbf6737 | sq-96 | 2022-02-21 | update |
| html | 91f38fa | sq-96 | 2022-02-13 | Build site. |
| Rmd | eb13ecf | sq-96 | 2022-02-13 | update |
| html | e6bc169 | sq-96 | 2022-02-13 | Build site. |
| Rmd | 87fee8b | sq-96 | 2022-02-13 | update |
Weight QC
#number of imputed weights
nrow(qclist_all)[1] 11531#number of imputed weights by chromosome
table(qclist_all$chr)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
1121 807 665 420 560 646 573 430 448 462 693 623 228 380 382 542
17 18 19 20 21 22
704 176 906 343 127 295 #number of imputed weights without missing variants
sum(qclist_all$nmiss==0)[1] 8840#proportion of imputed weights without missing variants
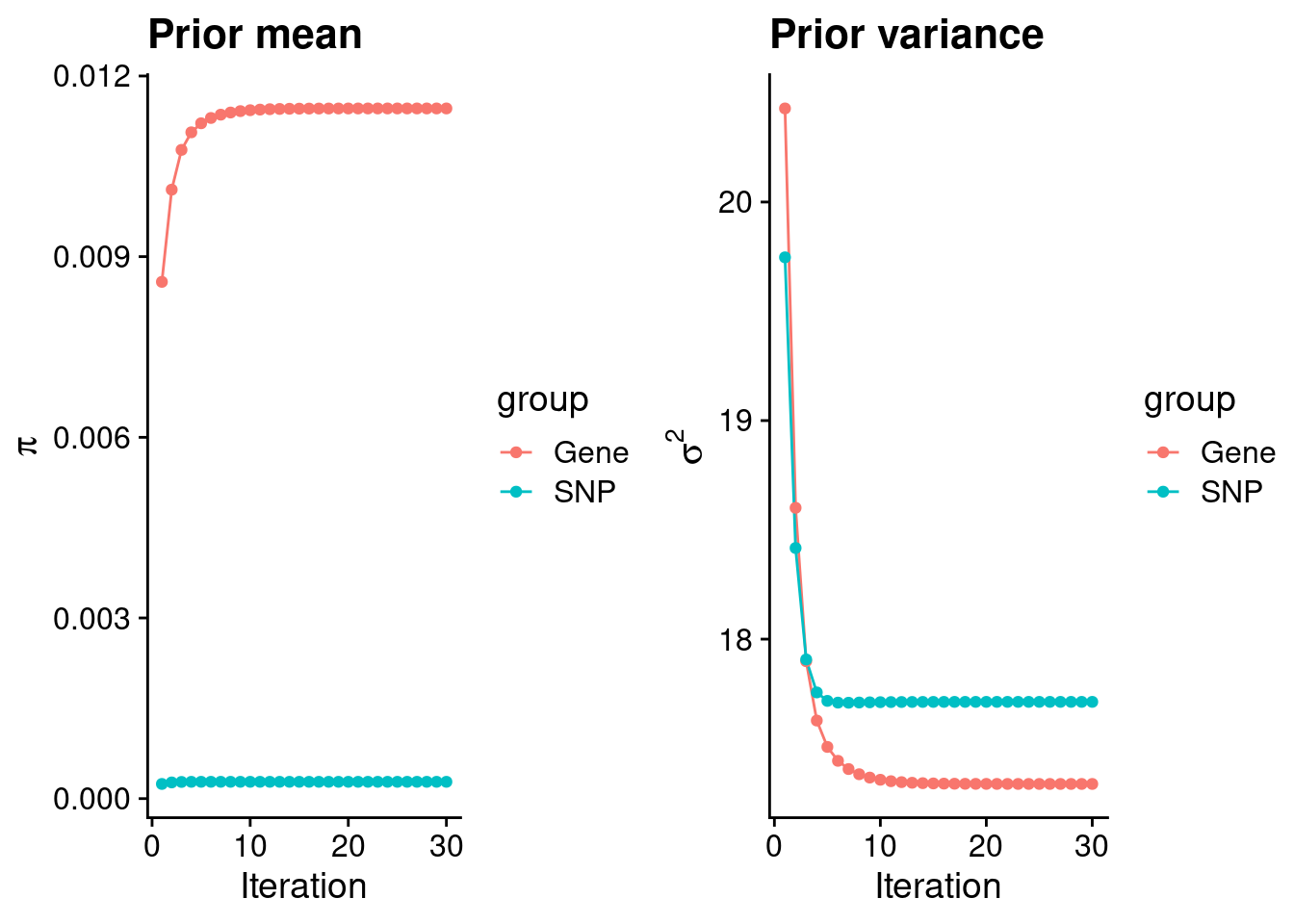
mean(qclist_all$nmiss==0)[1] 0.7666Check convergence of parameters
| Version | Author | Date |
|---|---|---|
| e6bc169 | sq-96 | 2022-02-13 |
#estimated group prior
estimated_group_prior <- group_prior_rec[,ncol(group_prior_rec)]
names(estimated_group_prior) <- c("gene", "snp")
estimated_group_prior["snp"] <- estimated_group_prior["snp"]*thin #adjust parameter to account for thin argument
print(estimated_group_prior) gene snp
0.0114612 0.0002791 #estimated group prior variance
estimated_group_prior_var <- group_prior_var_rec[,ncol(group_prior_var_rec)]
names(estimated_group_prior_var) <- c("gene", "snp")
print(estimated_group_prior_var) gene snp
17.34 17.71 #report sample size
print(sample_size)[1] 336107#report group size
group_size <- c(nrow(ctwas_gene_res), n_snps)
print(group_size)[1] 11531 7535010#estimated group PVE
estimated_group_pve <- estimated_group_prior_var*estimated_group_prior*group_size/sample_size #check PVE calculation
names(estimated_group_pve) <- c("gene", "snp")
print(estimated_group_pve) gene snp
0.006817 0.110820 #compare sum(PIP*mu2/sample_size) with above PVE calculation
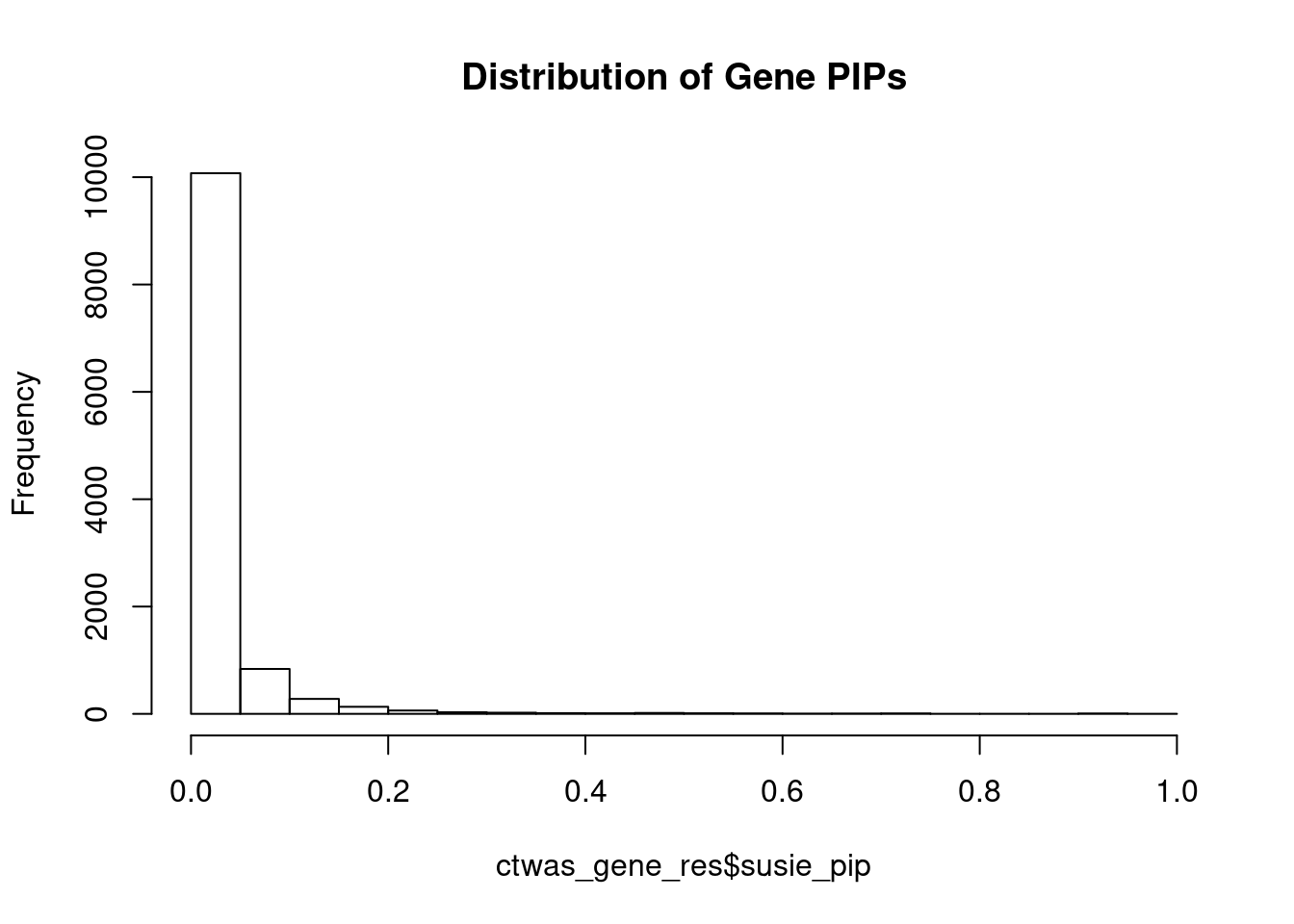
c(sum(ctwas_gene_res$PVE),sum(ctwas_snp_res$PVE))[1] 0.08778 17.62682Genes with highest PIPs
genename region_tag susie_pip mu2 PVE z num_eqtl
3434 CCND2 12_4 0.9754 28.48 8.266e-05 -5.120 1
983 PIK3C3 18_23 0.9392 51.84 1.449e-04 6.828 2
8986 C1QTNF4 11_29 0.9284 1287.45 3.556e-03 11.152 2
507 KCNH2 7_93 0.9260 43.04 1.186e-04 6.515 2
4444 TRAF3 14_54 0.9110 60.27 1.634e-04 -8.170 1
12533 ETV5 3_114 0.9061 94.53 2.549e-04 9.862 1
5033 DCAF7 17_37 0.9007 28.31 7.587e-05 5.437 1
1797 PPP1R16B 20_23 0.9001 21.05 5.638e-05 -4.129 1
8020 CASP7 10_71 0.8958 24.20 6.450e-05 4.584 1
9598 ZBTB41 1_98 0.8825 1744.49 4.580e-03 4.618 1
6041 ECE2 3_113 0.8424 29.53 7.402e-05 -5.315 1
13701 RP11-823E8.3 12_54 0.7583 102.48 2.312e-04 -6.438 1
10915 ZKSCAN5 7_61 0.7330 52.16 1.138e-04 7.133 1
7609 SERPINI1 3_103 0.7283 21.23 4.600e-05 -4.173 2
3223 EDEM3 1_92 0.7278 28.50 6.171e-05 5.238 1
13885 PRICKLE4 6_32 0.7231 23.69 5.097e-05 -4.797 1
12931 RP11-218E20.3 14_20 0.7194 21.32 4.563e-05 -3.497 2
13700 NOL12 22_15 0.7137 28.48 6.046e-05 -4.159 1
6995 DYRK1A 21_18 0.7102 21.12 4.462e-05 -4.006 1
11862 TEX40 11_36 0.7099 30.73 6.492e-05 -5.495 1Genes with largest effect sizes
genename region_tag susie_pip mu2 PVE z num_eqtl
135 NADK 1_1 0.000e+00 34223 0.000e+00 4.859 2
9678 STX19 3_59 0.000e+00 31351 0.000e+00 -5.060 1
10427 GSAP 7_49 3.331e-16 31262 3.098e-17 5.260 1
2201 PIK3R2 19_14 0.000e+00 28047 0.000e+00 5.621 1
12651 CTD-3074O7.2 11_37 6.961e-08 26961 5.584e-09 -4.561 2
12665 RP11-757G1.6 11_38 2.704e-01 24015 1.932e-02 4.314 2
5499 MFAP1 15_16 0.000e+00 23944 0.000e+00 4.303 1
11029 MRPL21 11_38 1.278e-03 23927 9.101e-05 4.379 1
4902 HEY2 6_84 0.000e+00 23615 0.000e+00 3.066 1
756 MAPK6 15_21 7.398e-03 23519 5.176e-04 -4.662 1
8147 LEO1 15_21 5.343e-04 23367 3.714e-05 4.647 1
13664 LINC02019 3_35 1.112e-07 22719 7.513e-09 -4.362 2
4212 TMOD2 15_21 0.000e+00 22290 0.000e+00 4.403 1
5505 LYSMD2 15_21 0.000e+00 22290 0.000e+00 4.403 1
1379 WDR76 15_16 0.000e+00 21871 0.000e+00 4.420 2
11904 CKMT1A 15_16 0.000e+00 21445 0.000e+00 4.130 1
3034 CISH 3_35 0.000e+00 20422 0.000e+00 -3.799 1
10708 DPYD 1_60 0.000e+00 19375 0.000e+00 -2.963 2
3033 HEMK1 3_35 0.000e+00 19267 0.000e+00 -4.682 1
13533 U91328.19 6_20 0.000e+00 18947 0.000e+00 -5.327 2Genes with highest PVE
genename region_tag susie_pip mu2 PVE z num_eqtl
12665 RP11-757G1.6 11_38 0.270376 24015.26 0.0193187 4.314 2
6352 CELF1 11_29 0.300033 13975.32 0.0124754 -3.558 1
2658 PTPMT1 11_29 0.300033 13975.32 0.0124754 -3.558 1
276 CPS1 2_124 0.529443 4711.27 0.0074213 -3.535 1
6638 PANK1 10_57 0.320041 6099.70 0.0058081 -3.857 1
9598 ZBTB41 1_98 0.882510 1744.49 0.0045805 4.618 1
8986 C1QTNF4 11_29 0.928384 1287.45 0.0035562 11.152 2
756 MAPK6 15_21 0.007398 23518.62 0.0005176 -4.662 1
10898 AFAP1 4_9 0.244594 587.90 0.0004278 4.142 2
12533 ETV5 3_114 0.906113 94.53 0.0002549 9.862 1
11901 VPS52 6_28 0.677229 124.40 0.0002507 1.606 1
11712 NDUFS3 11_29 0.059984 1353.72 0.0002416 -10.874 1
13701 RP11-823E8.3 12_54 0.758347 102.48 0.0002312 -6.438 1
4444 TRAF3 14_54 0.911008 60.27 0.0001634 -8.170 1
983 PIK3C3 18_23 0.939184 51.84 0.0001449 6.828 2
507 KCNH2 7_93 0.926029 43.04 0.0001186 6.515 2
9411 NUPR1 16_23 0.606521 63.68 0.0001149 -10.468 2
10915 ZKSCAN5 7_61 0.732954 52.16 0.0001138 7.133 1
5638 C18orf8 18_12 0.596521 56.76 0.0001007 7.506 2
13896 DHRS11 17_22 0.545531 61.62 0.0001000 -8.128 1Genes with largest z scores
genename region_tag susie_pip mu2 PVE z num_eqtl
34 RBM6 3_35 1.402e-03 914.63 3.816e-06 12.536 1
9289 KCTD13 16_24 1.258e-01 109.37 4.093e-05 -11.491 1
7735 MST1R 3_35 1.838e-10 233.55 1.277e-13 -11.458 2
8986 C1QTNF4 11_29 9.284e-01 1287.45 3.556e-03 11.152 2
7729 RNF123 3_35 1.686e-11 829.60 4.161e-14 -10.957 1
1860 MAPK3 16_24 2.536e-02 97.55 7.360e-06 10.880 1
11712 NDUFS3 11_29 5.998e-02 1353.72 2.416e-04 -10.874 1
9411 NUPR1 16_23 6.065e-01 63.68 1.149e-04 -10.468 2
12230 NPIPB7 16_23 5.871e-02 62.12 1.085e-05 10.429 1
8623 INO80E 16_24 4.239e-02 86.81 1.095e-05 10.393 2
10945 C6orf106 6_28 4.877e-05 118.65 1.722e-08 -10.264 1
640 UHRF1BP1 6_28 1.556e-05 97.69 4.523e-09 10.203 2
12533 ETV5 3_114 9.061e-01 94.53 2.549e-04 9.862 1
1952 BCKDK 16_24 1.729e-02 67.73 3.484e-06 -9.556 2
7733 CAMKV 3_35 0.000e+00 1461.86 0.000e+00 -9.545 2
2608 MTCH2 11_29 3.575e-14 508.58 5.409e-17 -9.514 1
10920 FAM180B 11_29 1.743e-14 504.82 2.618e-17 -9.432 1
1953 KAT8 16_24 1.836e-02 63.60 3.473e-06 -9.181 2
8987 NEGR1 1_46 6.023e-01 44.67 8.005e-05 -8.928 1
10248 APOBR 16_23 9.618e-03 41.38 1.184e-06 -8.735 1Comparing z scores and PIPs
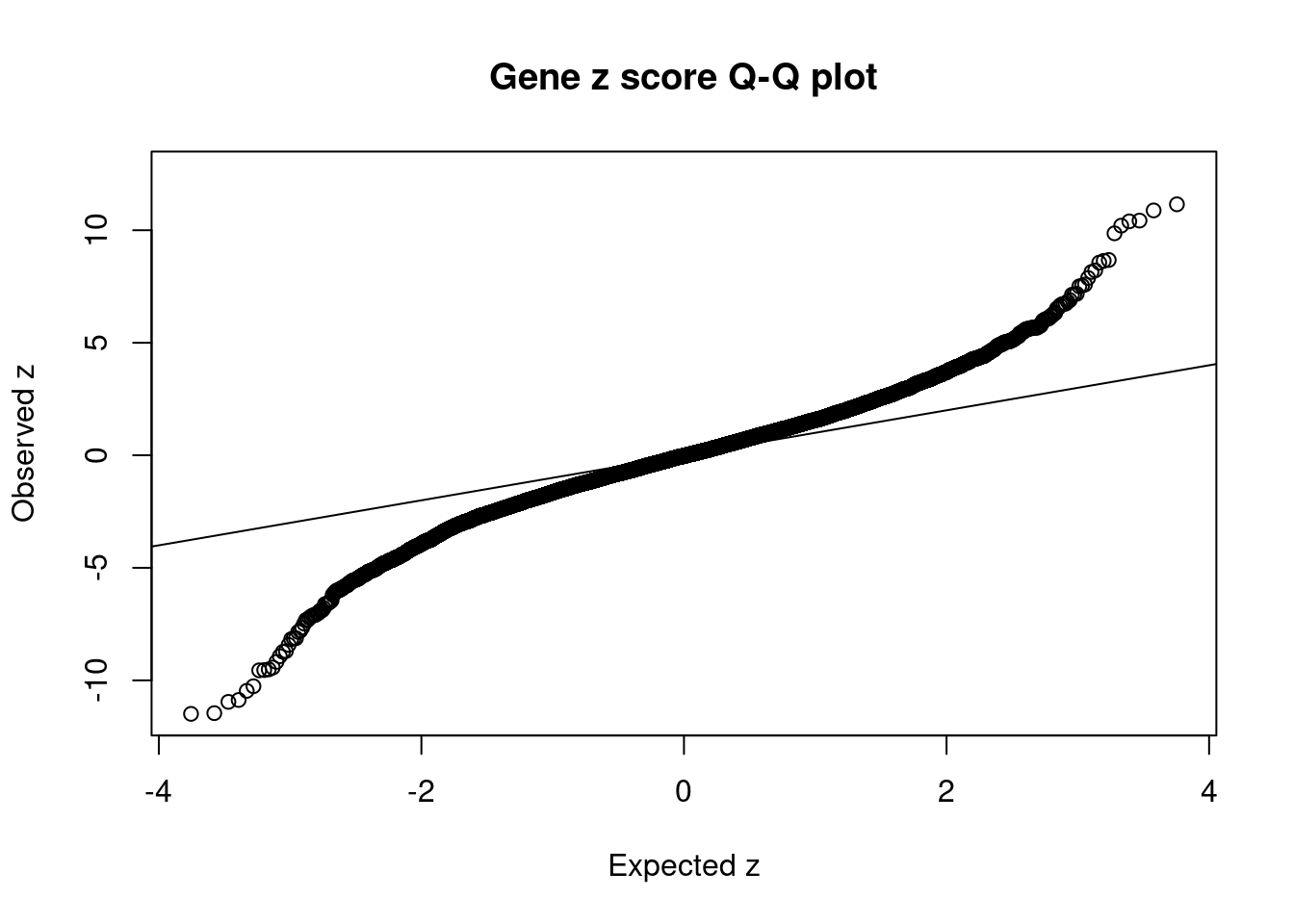
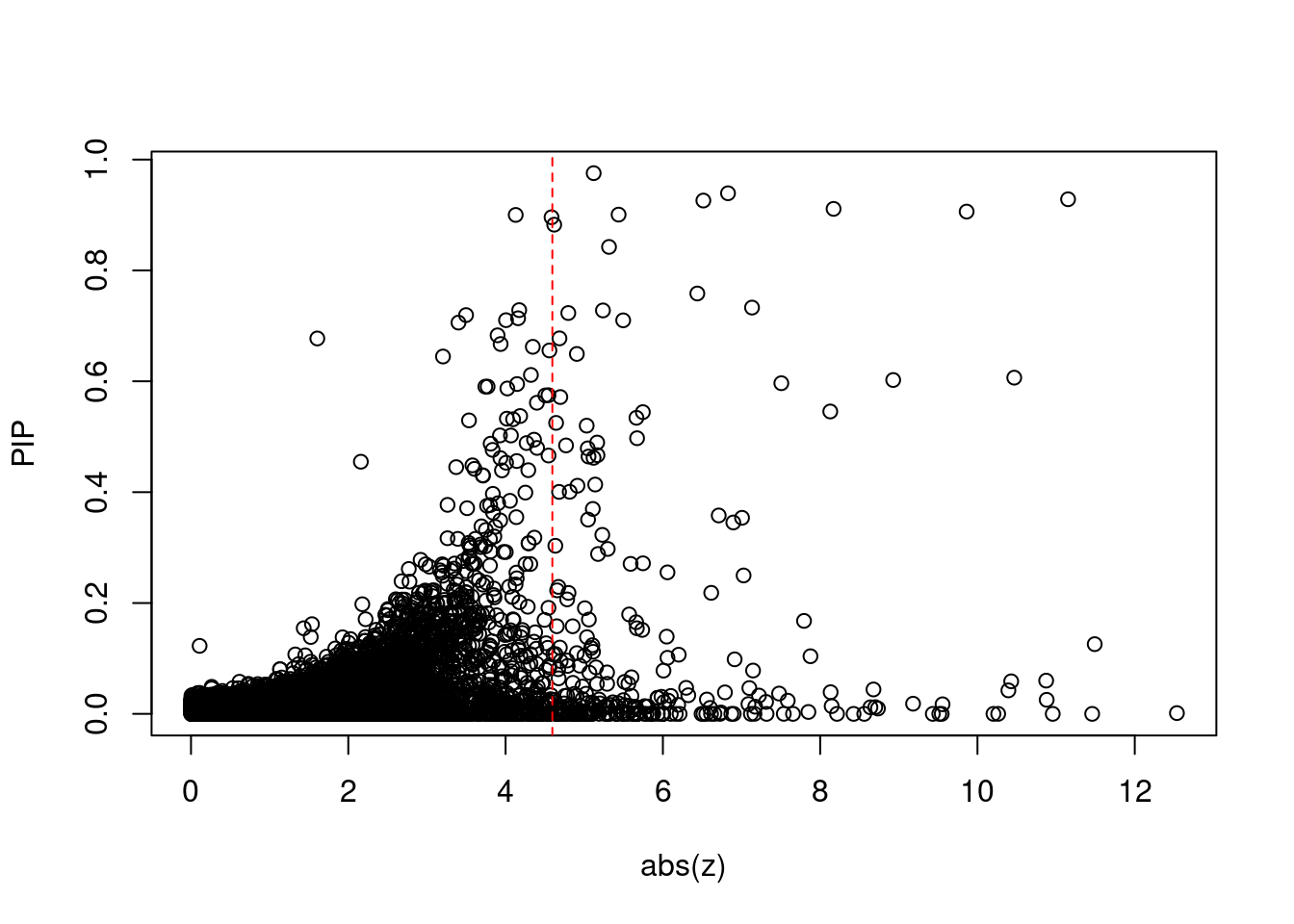
[1] 0.0235 genename region_tag susie_pip mu2 PVE z num_eqtl
34 RBM6 3_35 1.402e-03 914.63 3.816e-06 12.536 1
9289 KCTD13 16_24 1.258e-01 109.37 4.093e-05 -11.491 1
7735 MST1R 3_35 1.838e-10 233.55 1.277e-13 -11.458 2
8986 C1QTNF4 11_29 9.284e-01 1287.45 3.556e-03 11.152 2
7729 RNF123 3_35 1.686e-11 829.60 4.161e-14 -10.957 1
1860 MAPK3 16_24 2.536e-02 97.55 7.360e-06 10.880 1
11712 NDUFS3 11_29 5.998e-02 1353.72 2.416e-04 -10.874 1
9411 NUPR1 16_23 6.065e-01 63.68 1.149e-04 -10.468 2
12230 NPIPB7 16_23 5.871e-02 62.12 1.085e-05 10.429 1
8623 INO80E 16_24 4.239e-02 86.81 1.095e-05 10.393 2
10945 C6orf106 6_28 4.877e-05 118.65 1.722e-08 -10.264 1
640 UHRF1BP1 6_28 1.556e-05 97.69 4.523e-09 10.203 2
12533 ETV5 3_114 9.061e-01 94.53 2.549e-04 9.862 1
1952 BCKDK 16_24 1.729e-02 67.73 3.484e-06 -9.556 2
7733 CAMKV 3_35 0.000e+00 1461.86 0.000e+00 -9.545 2
2608 MTCH2 11_29 3.575e-14 508.58 5.409e-17 -9.514 1
10920 FAM180B 11_29 1.743e-14 504.82 2.618e-17 -9.432 1
1953 KAT8 16_24 1.836e-02 63.60 3.473e-06 -9.181 2
8987 NEGR1 1_46 6.023e-01 44.67 8.005e-05 -8.928 1
10248 APOBR 16_23 9.618e-03 41.38 1.184e-06 -8.735 1GO enrichment analysis for genes with PIP>0.5
#number of genes for gene set enrichment
length(genes)[1] 52Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"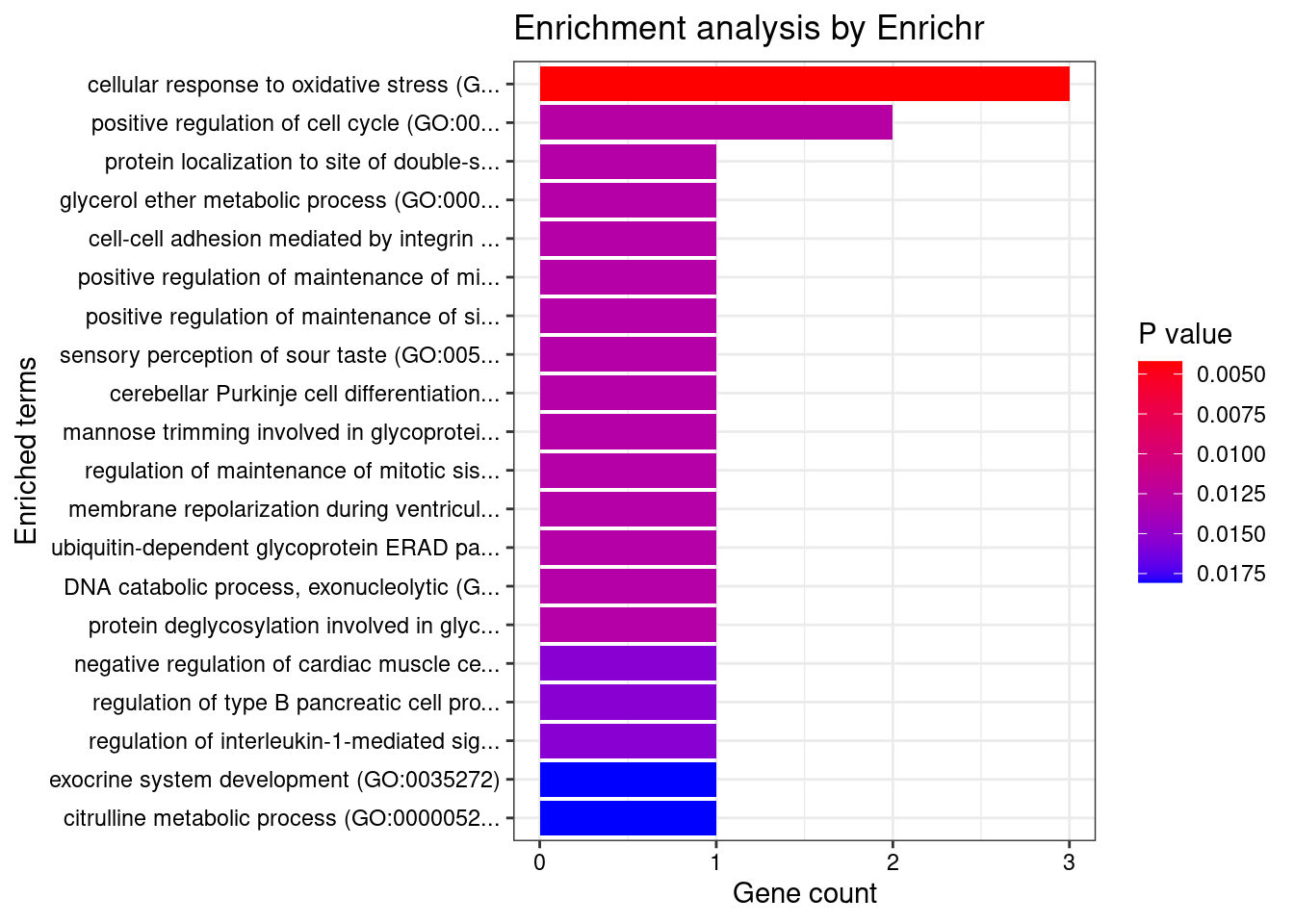
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Cellular_Component_2021"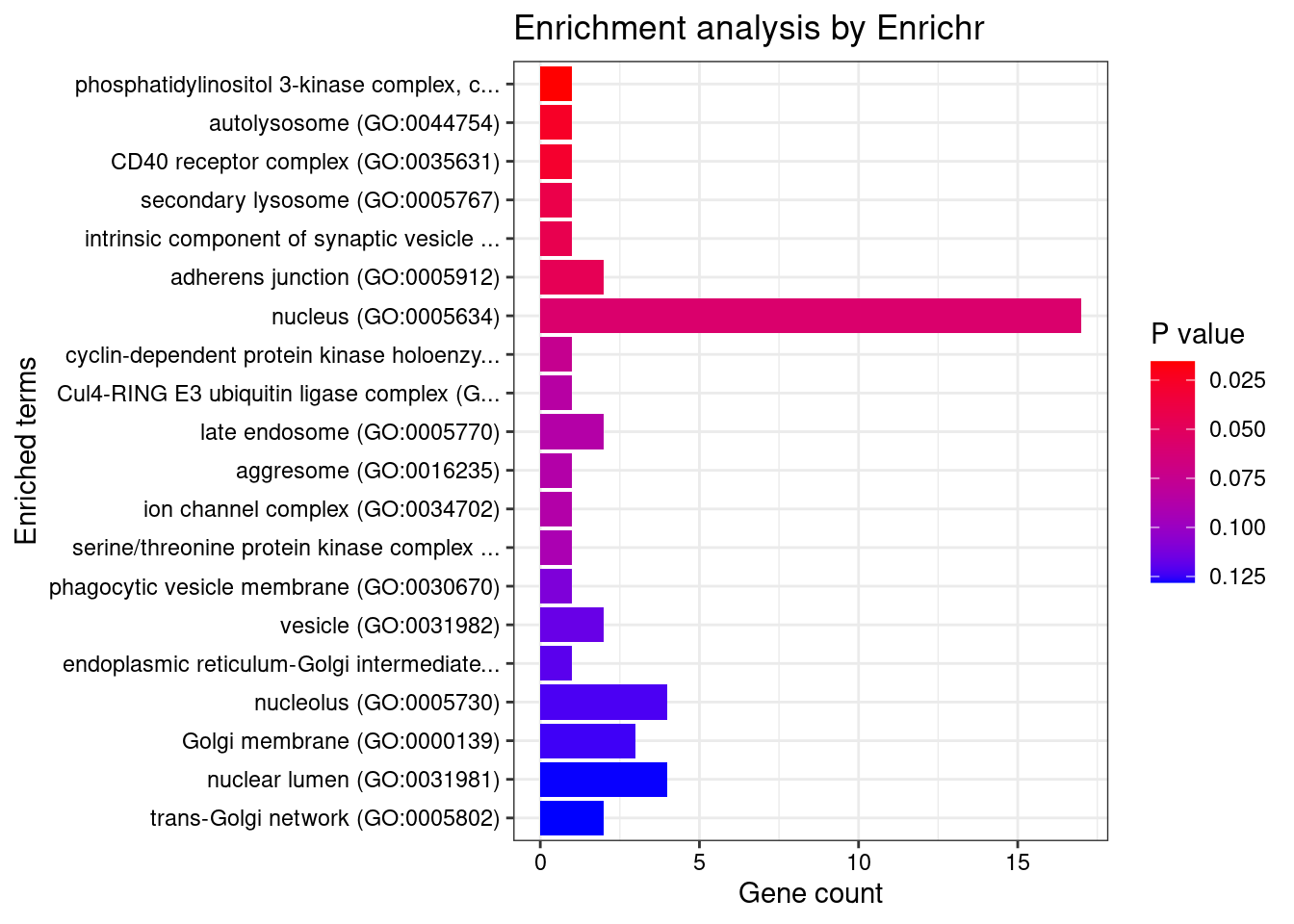
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Molecular_Function_2021"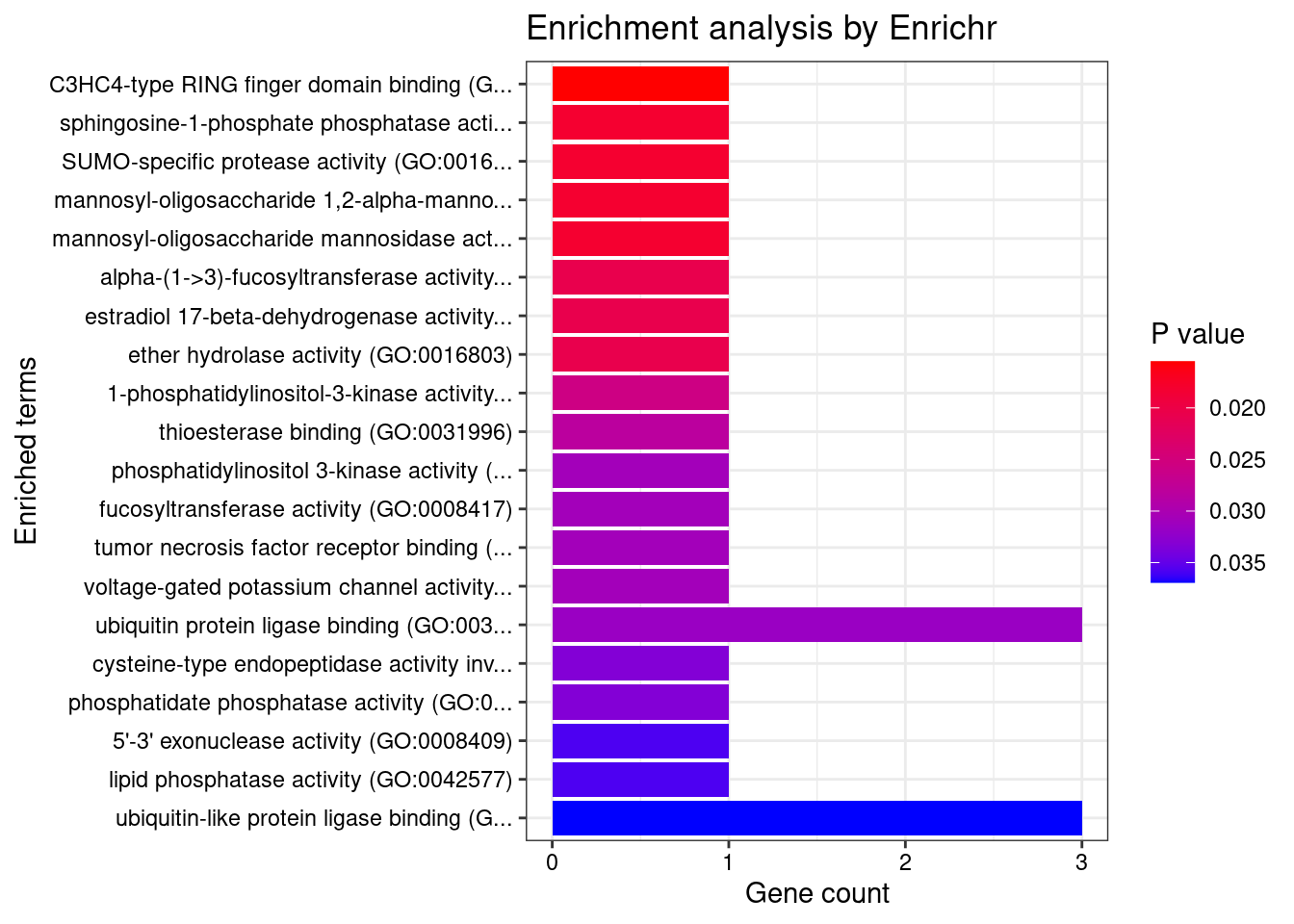
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)DisGeNET enrichment analysis for genes with PIP>0.5
Description
83 Carbamoyl-Phosphate Synthase I Deficiency Disease
107 Familial encephalopathy with neuroserpin inclusion bodies
119 MENTAL RETARDATION, AUTOSOMAL DOMINANT 7
121 ENCEPHALOPATHY, ACUTE, INFECTION-INDUCED (HERPES-SPECIFIC), SUSCEPTIBILITY TO, 5
122 MENTAL RETARDATION, AUTOSOMAL DOMINANT 17
125 PULMONARY HYPERTENSION, NEONATAL, SUSCEPTIBILITY TO
128 MEGALENCEPHALY-POLYMICROGYRIA-POLYDACTYLY-HYDROCEPHALUS SYNDROME 3
129 Hyperammonemia Due to Carbamoyl Phosphate Synthetase 1 Deficiency
130 Carbamoyl Phosphate Synthase 1 Deficiency
43 Persistent Fetal Circulation Syndrome
FDR Ratio BgRatio
83 0.03247 1/21 1/9703
107 0.03247 1/21 1/9703
119 0.03247 1/21 1/9703
121 0.03247 1/21 1/9703
122 0.03247 1/21 1/9703
125 0.03247 1/21 1/9703
128 0.03247 1/21 1/9703
129 0.03247 1/21 1/9703
130 0.03247 1/21 1/9703
43 0.05307 1/21 2/9703WebGestalt enrichment analysis for genes with PIP>0.5
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum =
minNum, : No significant gene set is identified based on FDR 0.05!NULLPIP Manhattan Plot
Warning: ggrepel: 13 unlabeled data points (too many overlaps). Consider
increasing max.overlaps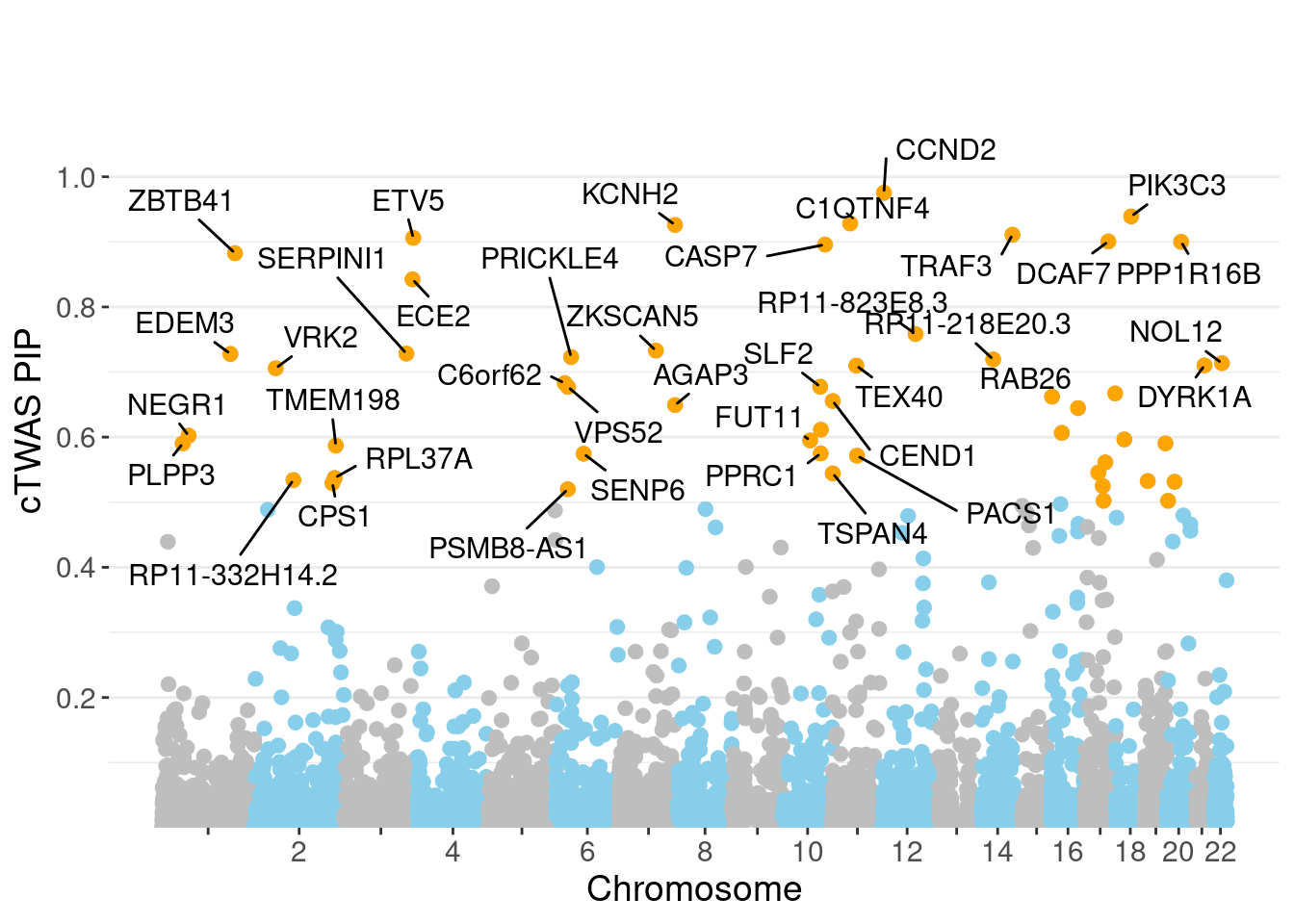
Sensitivity, specificity and precision for silver standard genes
#number of genes in known annotations
print(length(known_annotations))[1] 41#number of genes in known annotations with imputed expression
print(sum(known_annotations %in% ctwas_gene_res$genename))[1] 25#significance threshold for TWAS
print(sig_thresh)[1] 4.595#number of ctwas genes
length(ctwas_genes)[1] 11#number of TWAS genes
length(twas_genes)[1] 271#show novel genes (ctwas genes with not in TWAS genes)
ctwas_gene_res[ctwas_gene_res$genename %in% novel_genes,report_cols] genename region_tag susie_pip mu2 PVE z num_eqtl
8020 CASP7 10_71 0.8958 24.20 6.450e-05 4.584 1
1797 PPP1R16B 20_23 0.9001 21.05 5.638e-05 -4.129 1#sensitivity / recall
print(sensitivity)ctwas TWAS
0.000 0.122 #specificity
print(specificity) ctwas TWAS
0.9990 0.9769 #precision / PPV
print(precision) ctwas TWAS
0.00000 0.01845 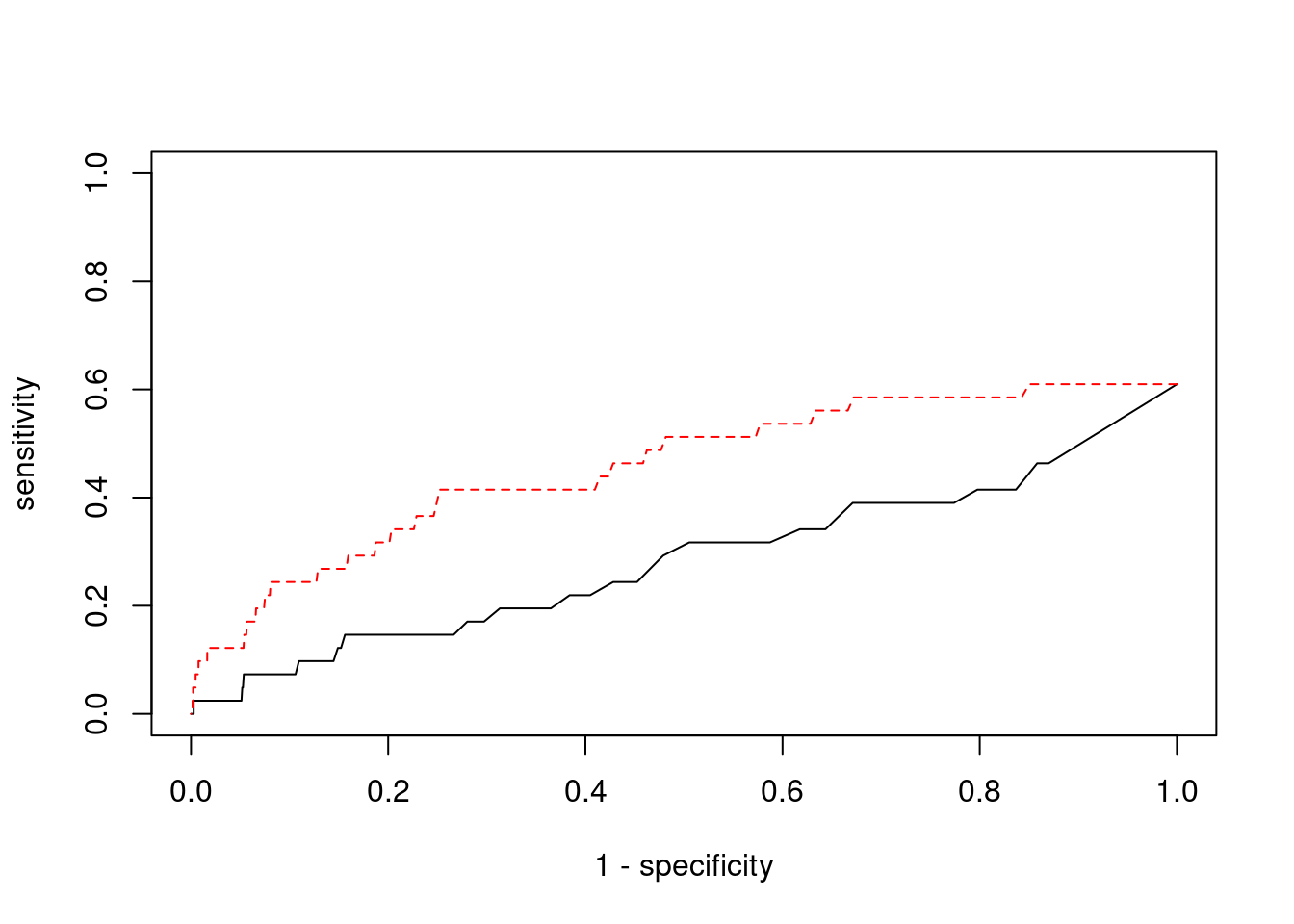
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] readxl_1.3.1 forcats_0.5.1 stringr_1.4.0 dplyr_1.0.7
[5] purrr_0.3.4 readr_2.1.1 tidyr_1.1.4 tidyverse_1.3.1
[9] tibble_3.1.6 WebGestaltR_0.4.4 disgenet2r_0.99.2 enrichR_3.0
[13] cowplot_1.0.0 ggplot2_3.3.5 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] fs_1.5.2 lubridate_1.8.0 bit64_4.0.5 doParallel_1.0.16
[5] httr_1.4.2 rprojroot_2.0.2 tools_3.6.1 backports_1.4.1
[9] doRNG_1.8.2 utf8_1.2.2 R6_2.5.1 vipor_0.4.5
[13] DBI_1.1.1 colorspace_2.0-2 withr_2.4.3 ggrastr_1.0.1
[17] tidyselect_1.1.1 bit_4.0.4 curl_4.3.2 compiler_3.6.1
[21] git2r_0.26.1 cli_3.1.0 rvest_1.0.2 Cairo_1.5-12.2
[25] xml2_1.3.3 labeling_0.4.2 scales_1.1.1 apcluster_1.4.8
[29] digest_0.6.29 rmarkdown_2.11 svglite_1.2.2 pkgconfig_2.0.3
[33] htmltools_0.5.2 dbplyr_2.1.1 fastmap_1.1.0 highr_0.9
[37] rlang_0.4.12 rstudioapi_0.13 RSQLite_2.2.8 jquerylib_0.1.4
[41] farver_2.1.0 generics_0.1.1 jsonlite_1.7.2 vroom_1.5.7
[45] magrittr_2.0.1 Matrix_1.2-18 ggbeeswarm_0.6.0 Rcpp_1.0.7
[49] munsell_0.5.0 fansi_0.5.0 gdtools_0.1.9 lifecycle_1.0.1
[53] stringi_1.7.6 whisker_0.3-2 yaml_2.2.1 plyr_1.8.6
[57] grid_3.6.1 blob_1.2.2 ggrepel_0.9.1 parallel_3.6.1
[61] promises_1.0.1 crayon_1.4.2 lattice_0.20-38 haven_2.4.3
[65] hms_1.1.1 knitr_1.36 pillar_1.6.4 igraph_1.2.10
[69] rjson_0.2.20 rngtools_1.5.2 reshape2_1.4.4 codetools_0.2-16
[73] reprex_2.0.1 glue_1.5.1 evaluate_0.14 data.table_1.14.2
[77] modelr_0.1.8 vctrs_0.3.8 tzdb_0.2.0 httpuv_1.5.1
[81] foreach_1.5.1 cellranger_1.1.0 gtable_0.3.0 assertthat_0.2.1
[85] cachem_1.0.6 xfun_0.29 broom_0.7.10 later_0.8.0
[89] iterators_1.0.13 beeswarm_0.2.3 memoise_2.0.1 ellipsis_0.3.2