SCZ - Brain Cerebellar Hemisphere
sheng Qian
2021-2-6
Last updated: 2022-04-19
Checks: 5 2
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/ | data |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/ctwas_config.R | code/ctwas_config.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version ba919ab. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .ipynb_checkpoints/
Ignored: data/AF/
Untracked files:
Untracked: Rplot.png
Untracked: analysis/.ipynb_checkpoints/
Untracked: code/.ipynb_checkpoints/
Untracked: code/AF_out/
Untracked: code/Autism_out/
Untracked: code/BMI_S_out/
Untracked: code/BMI_out/
Untracked: code/Glucose_out/
Untracked: code/LDL_S_out/
Untracked: code/SCZ_2014_EUR_out/
Untracked: code/SCZ_2018_out/
Untracked: code/SCZ_2020_Single_out/
Untracked: code/SCZ_2020_out/
Untracked: code/SCZ_S_out/
Untracked: code/SCZ_out/
Untracked: code/T2D_out/
Untracked: code/ctwas_config.R
Untracked: code/mapping.R
Untracked: code/out/
Untracked: code/process_scz_2018_snps.R
Untracked: code/run_AF_analysis.sbatch
Untracked: code/run_AF_analysis.sh
Untracked: code/run_AF_ctwas_rss_LDR.R
Untracked: code/run_Autism_analysis.sbatch
Untracked: code/run_Autism_analysis.sh
Untracked: code/run_Autism_ctwas_rss_LDR.R
Untracked: code/run_BMI_analysis.sbatch
Untracked: code/run_BMI_analysis.sh
Untracked: code/run_BMI_analysis_S.sbatch
Untracked: code/run_BMI_analysis_S.sh
Untracked: code/run_BMI_ctwas_rss_LDR.R
Untracked: code/run_BMI_ctwas_rss_LDR_S.R
Untracked: code/run_Glucose_analysis.sbatch
Untracked: code/run_Glucose_analysis.sh
Untracked: code/run_Glucose_ctwas_rss_LDR.R
Untracked: code/run_LDL_analysis_S.sbatch
Untracked: code/run_LDL_analysis_S.sh
Untracked: code/run_LDL_ctwas_rss_LDR_S.R
Untracked: code/run_SCZ_2014_EUR_analysis.sbatch
Untracked: code/run_SCZ_2014_EUR_analysis.sh
Untracked: code/run_SCZ_2014_EUR_ctwas_rss_LDR.R
Untracked: code/run_SCZ_2018_analysis.sbatch
Untracked: code/run_SCZ_2018_analysis.sh
Untracked: code/run_SCZ_2018_ctwas_rss_LDR.R
Untracked: code/run_SCZ_2020_Single_analysis.sbatch
Untracked: code/run_SCZ_2020_Single_analysis.sh
Untracked: code/run_SCZ_2020_Single_ctwas_rss_LDR.R
Untracked: code/run_SCZ_2020_analysis.sbatch
Untracked: code/run_SCZ_2020_analysis.sh
Untracked: code/run_SCZ_2020_ctwas_rss_LDR.R
Untracked: code/run_SCZ_analysis.sbatch
Untracked: code/run_SCZ_analysis.sh
Untracked: code/run_SCZ_analysis_S.sbatch
Untracked: code/run_SCZ_analysis_S.sh
Untracked: code/run_SCZ_ctwas_rss_LDR.R
Untracked: code/run_SCZ_ctwas_rss_LDR_S.R
Untracked: code/run_T2D_analysis.sbatch
Untracked: code/run_T2D_analysis.sh
Untracked: code/run_T2D_ctwas_rss_LDR.R
Untracked: code/wflow_build.R
Untracked: code/wflow_build.sbatch
Untracked: data/.ipynb_checkpoints/
Untracked: data/BMI/
Untracked: data/GO_Terms/
Untracked: data/PGC3_SCZ_wave3_public.v2.tsv
Untracked: data/SCZ/
Untracked: data/SCZ_2014_EUR/
Untracked: data/SCZ_2018/
Untracked: data/SCZ_2020/
Untracked: data/SCZ_2020_Single/
Untracked: data/SCZ_S/
Untracked: data/Supplementary Table 15 - MAGMA.xlsx
Untracked: data/Supplementary Table 20 - Prioritised Genes.xlsx
Untracked: data/T2D/
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/mashr_sqtl/
Untracked: data/scz_2018.RDS
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: data/untitled.txt
Untracked: top_genes_32.txt
Untracked: top_genes_37.txt
Untracked: top_genes_43.txt
Untracked: top_genes_81.txt
Unstaged changes:
Modified: analysis/SCZ_2018_Brain_Amygdala.Rmd
Modified: analysis/SCZ_2018_Brain_Anterior_cingulate_cortex_BA24.Rmd
Modified: analysis/SCZ_2018_Brain_Caudate_basal_ganglia.Rmd
Modified: analysis/SCZ_2018_Brain_Cerebellar_Hemisphere.Rmd
Modified: analysis/SCZ_2018_Brain_Cerebellum.Rmd
Modified: analysis/SCZ_2018_Brain_Cortex.Rmd
Modified: analysis/SCZ_2018_Brain_Frontal_Cortex_BA9.Rmd
Modified: analysis/SCZ_2018_Brain_Hippocampus.Rmd
Modified: analysis/SCZ_2018_Brain_Hypothalamus.Rmd
Modified: analysis/SCZ_2018_Brain_Nucleus_accumbens_basal_ganglia.Rmd
Modified: analysis/SCZ_2018_Brain_Putamen_basal_ganglia.Rmd
Modified: analysis/SCZ_2018_Brain_Spinal_cord_cervical_c-1.Rmd
Modified: analysis/SCZ_2018_Brain_Substantia_nigra.Rmd
Modified: analysis/SCZ_Annotation_Analysis.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/SCZ_2018_Brain_Cerebellar_Hemisphere.Rmd) and HTML (docs/SCZ_2018_Brain_Cerebellar_Hemisphere.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 9ddc9c4 | sq-96 | 2022-04-18 | update |
| Rmd | f6e7062 | sq-96 | 2022-04-17 | update |
| html | f6e7062 | sq-96 | 2022-04-17 | update |
Weight QC
#number of imputed weights
nrow(qclist_all)[1] 9945#number of imputed weights by chromosome
table(qclist_all$chr)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
941 692 574 371 468 558 511 377 411 395 633 567 188 341 340 419 633 153 782 293
21 22
31 267 #number of imputed weights without missing variants
sum(qclist_all$nmiss==0)[1] 6702#proportion of imputed weights without missing variants
mean(qclist_all$nmiss==0)[1] 0.6739Check convergence of parameters
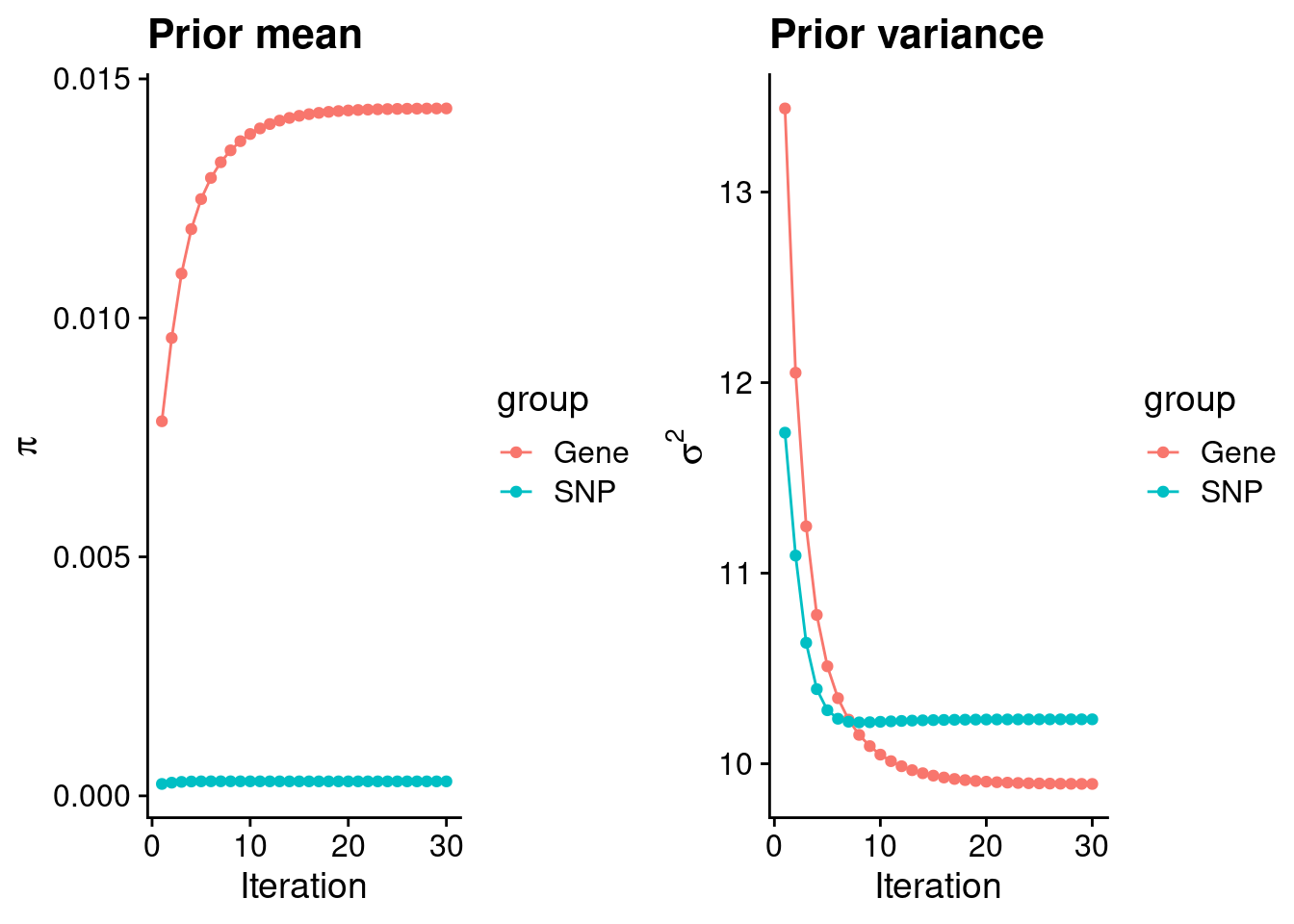
#estimated group prior
estimated_group_prior <- group_prior_rec[,ncol(group_prior_rec)]
names(estimated_group_prior) <- c("gene", "snp")
estimated_group_prior["snp"] <- estimated_group_prior["snp"]*thin #adjust parameter to account for thin argument
print(estimated_group_prior) gene snp
0.0143819 0.0003031 #estimated group prior variance
estimated_group_prior_var <- group_prior_var_rec[,ncol(group_prior_var_rec)]
names(estimated_group_prior_var) <- c("gene", "snp")
print(estimated_group_prior_var) gene snp
9.894 10.233 #report sample size
print(sample_size)[1] 105318#report group size
group_size <- c(nrow(ctwas_gene_res), n_snps)
print(group_size)[1] 9945 6309950#estimated group PVE
estimated_group_pve <- estimated_group_prior_var*estimated_group_prior*group_size/sample_size #check PVE calculation
names(estimated_group_pve) <- c("gene", "snp")
print(estimated_group_pve) gene snp
0.01344 0.18584 #compare sum(PIP*mu2/sample_size) with above PVE calculation
c(sum(ctwas_gene_res$PVE),sum(ctwas_snp_res$PVE))[1] 0.06308 1.04445Genes with highest PIPs
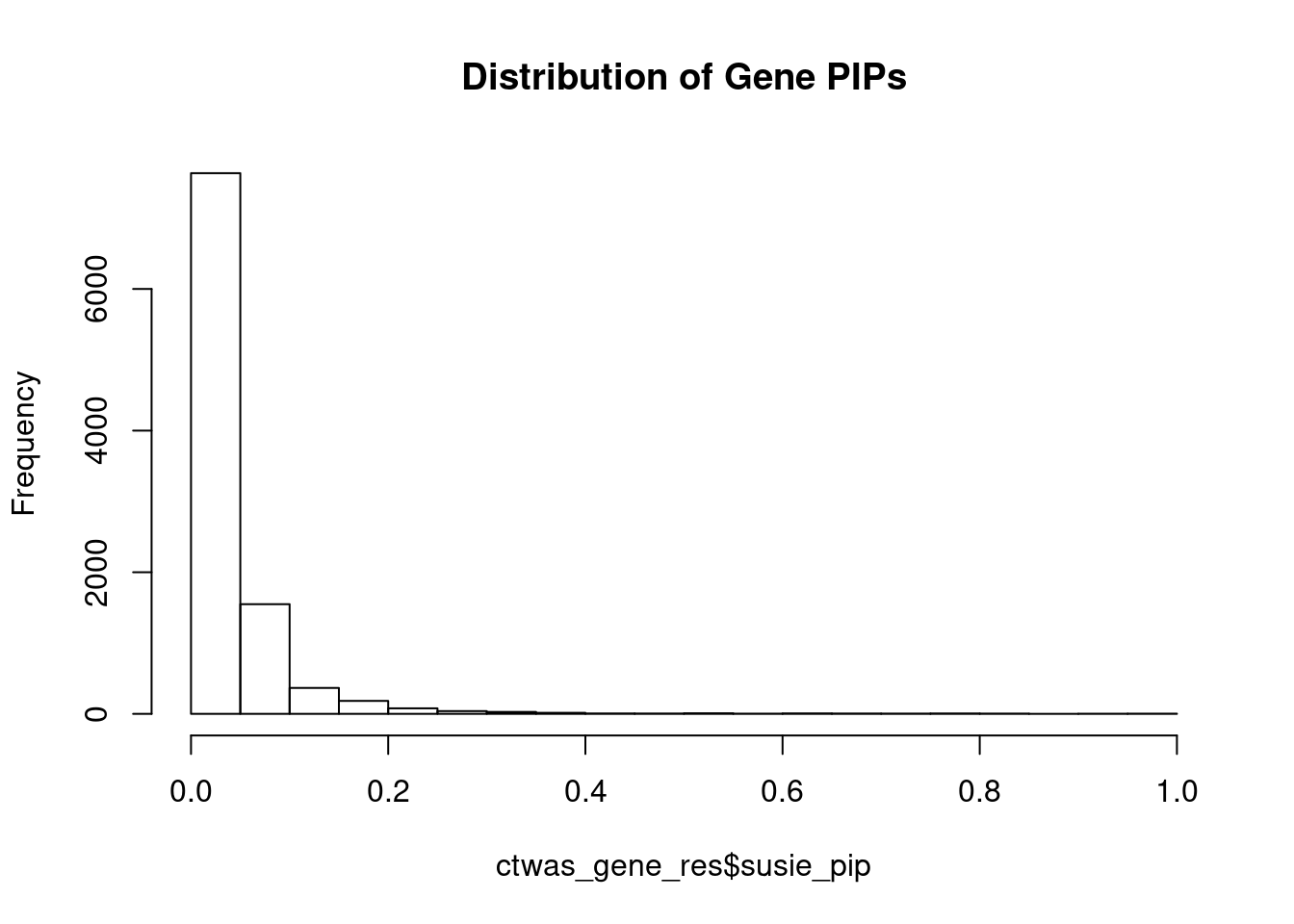
genename region_tag susie_pip mu2 PVE z num_eqtl
10988 ZNF823 19_10 0.9851 36.62 0.0003425 6.211 2
5783 GALNT2 1_117 0.9649 25.23 0.0002311 5.083 1
4143 FEZF1 7_74 0.9547 23.85 0.0002162 -4.812 1
12095 AC012074.2 2_15 0.9494 21.97 0.0001981 4.655 1
13452 RP11-408A13.3 9_12 0.9238 22.50 0.0001974 4.536 1
111 ELAC2 17_11 0.8205 21.18 0.0001650 4.752 1
3099 SF3B1 2_117 0.8183 47.43 0.0003685 7.265 1
5667 SYTL1 1_19 0.8065 22.96 0.0001758 4.272 2
10337 TMEM222 1_19 0.8055 22.21 0.0001699 4.303 1
174 ZNF207 17_19 0.7940 23.35 0.0001760 4.599 1
6255 DRD2 11_68 0.7935 26.30 0.0001981 -5.632 1
5591 ZCCHC2 18_34 0.7821 19.60 0.0001456 -3.877 1
1221 EDEM2 20_21 0.7595 20.25 0.0001460 4.057 2
11339 DISP3 1_8 0.7578 20.02 0.0001440 3.696 2
9372 LY6H 8_94 0.7552 21.68 0.0001555 4.186 1
6920 CNNM4 2_57 0.7331 23.15 0.0001612 -4.456 1
7051 ZNF235 19_31 0.7271 20.54 0.0001418 -4.002 2
13014 TBC1D29 17_18 0.7266 22.91 0.0001581 -4.592 1
5459 RLBP1 15_41 0.7242 22.94 0.0001577 -4.280 1
2207 RUNDC3B 7_54 0.6982 23.87 0.0001582 5.102 1Genes with largest effect sizes

genename region_tag susie_pip mu2 PVE z num_eqtl
11296 C6orf48 6_26 1.273e-04 210.63 2.546e-07 11.5418 1
11553 CLIC1 6_26 1.217e-04 209.14 2.416e-07 11.5063 1
12355 C4A 6_26 9.091e-05 207.34 1.790e-07 11.4403 2
11281 RNF5 6_26 8.237e-06 160.76 1.257e-08 9.7754 2
11986 CYP21A2 6_26 3.463e-07 153.54 5.049e-10 -9.0790 2
11284 PRRT1 6_26 5.771e-05 146.17 8.010e-08 10.0611 1
11280 AGER 6_26 5.527e-06 104.88 5.504e-09 -9.0708 1
5086 PGBD1 6_22 1.907e-02 95.36 1.726e-05 -10.2310 1
11884 HCG11 6_20 2.727e-02 95.33 2.469e-05 11.0152 1
12879 CTA-14H9.5 6_20 2.727e-02 95.33 2.469e-05 11.0152 1
9601 HLA-DQB1 6_26 3.543e-07 92.42 3.109e-10 1.4260 2
11279 NOTCH4 6_26 6.558e-06 89.95 5.601e-09 7.8425 2
12178 HLA-DQA2 6_26 3.764e-07 87.21 3.117e-10 0.9484 1
10334 BTN3A2 6_20 2.289e-02 85.48 1.858e-05 10.5362 1
5083 FLOT1 6_24 4.328e-02 80.14 3.293e-05 -10.9813 1
11282 AGPAT1 6_26 3.493e-07 77.56 2.572e-10 -4.4655 1
6038 ABT1 6_20 5.969e-02 75.30 4.268e-05 9.6693 1
11554 DDAH2 6_26 2.056e-05 73.58 1.436e-08 8.1494 1
11298 HSPA1A 6_26 2.419e-05 71.50 1.643e-08 8.0745 1
2826 TRIM38 6_20 2.508e-02 70.09 1.669e-05 -9.5422 2Genes with highest PVE
genename region_tag susie_pip mu2 PVE z num_eqtl
3099 SF3B1 2_117 0.8183 47.43 0.0003685 7.265 1
10988 ZNF823 19_10 0.9851 36.62 0.0003425 6.211 2
9199 ATG13 11_28 0.6200 44.07 0.0002594 -6.977 1
5783 GALNT2 1_117 0.9649 25.23 0.0002311 5.083 1
4143 FEZF1 7_74 0.9547 23.85 0.0002162 -4.812 1
10942 NMB 15_39 0.6196 34.32 0.0002019 5.881 1
6255 DRD2 11_68 0.7935 26.30 0.0001981 -5.632 1
12095 AC012074.2 2_15 0.9494 21.97 0.0001981 4.655 1
13452 RP11-408A13.3 9_12 0.9238 22.50 0.0001974 4.536 1
174 ZNF207 17_19 0.7940 23.35 0.0001760 4.599 1
5667 SYTL1 1_19 0.8065 22.96 0.0001758 4.272 2
10337 TMEM222 1_19 0.8055 22.21 0.0001699 4.303 1
111 ELAC2 17_11 0.8205 21.18 0.0001650 4.752 1
8510 INO80E 16_24 0.3995 43.47 0.0001649 6.852 1
6920 CNNM4 2_57 0.7331 23.15 0.0001612 -4.456 1
9728 FAM83H 8_94 0.6009 27.99 0.0001597 5.057 2
2207 RUNDC3B 7_54 0.6982 23.87 0.0001582 5.102 1
13014 TBC1D29 17_18 0.7266 22.91 0.0001581 -4.592 1
5459 RLBP1 15_41 0.7242 22.94 0.0001577 -4.280 1
4124 RNF112 17_16 0.5946 27.75 0.0001567 5.122 2Genes with largest z scores
genename region_tag susie_pip mu2 PVE z num_eqtl
11296 C6orf48 6_26 1.273e-04 210.63 2.546e-07 11.542 1
11553 CLIC1 6_26 1.217e-04 209.14 2.416e-07 11.506 1
12355 C4A 6_26 9.091e-05 207.34 1.790e-07 11.440 2
11884 HCG11 6_20 2.727e-02 95.33 2.469e-05 11.015 1
12879 CTA-14H9.5 6_20 2.727e-02 95.33 2.469e-05 11.015 1
5083 FLOT1 6_24 4.328e-02 80.14 3.293e-05 -10.981 1
10334 BTN3A2 6_20 2.289e-02 85.48 1.858e-05 10.536 1
5086 PGBD1 6_22 1.907e-02 95.36 1.726e-05 -10.231 1
11284 PRRT1 6_26 5.771e-05 146.17 8.010e-08 10.061 1
11281 RNF5 6_26 8.237e-06 160.76 1.257e-08 9.775 2
6038 ABT1 6_20 5.969e-02 75.30 4.268e-05 9.669 1
2826 TRIM38 6_20 2.508e-02 70.09 1.669e-05 -9.542 2
11986 CYP21A2 6_26 3.463e-07 153.54 5.049e-10 -9.079 2
11280 AGER 6_26 5.527e-06 104.88 5.504e-09 -9.071 1
12301 HLA-DMB 6_27 1.197e-01 69.61 7.910e-05 -8.812 1
11273 HLA-DMA 6_27 5.302e-02 63.51 3.197e-05 -8.778 2
6221 CNNM2 10_66 9.532e-02 41.33 3.740e-05 -8.161 1
11554 DDAH2 6_26 2.056e-05 73.58 1.436e-08 8.149 1
11298 HSPA1A 6_26 2.419e-05 71.50 1.643e-08 8.075 1
10488 ZSCAN23 6_22 7.885e-02 47.46 3.553e-05 -7.854 1Comparing z scores and PIPs
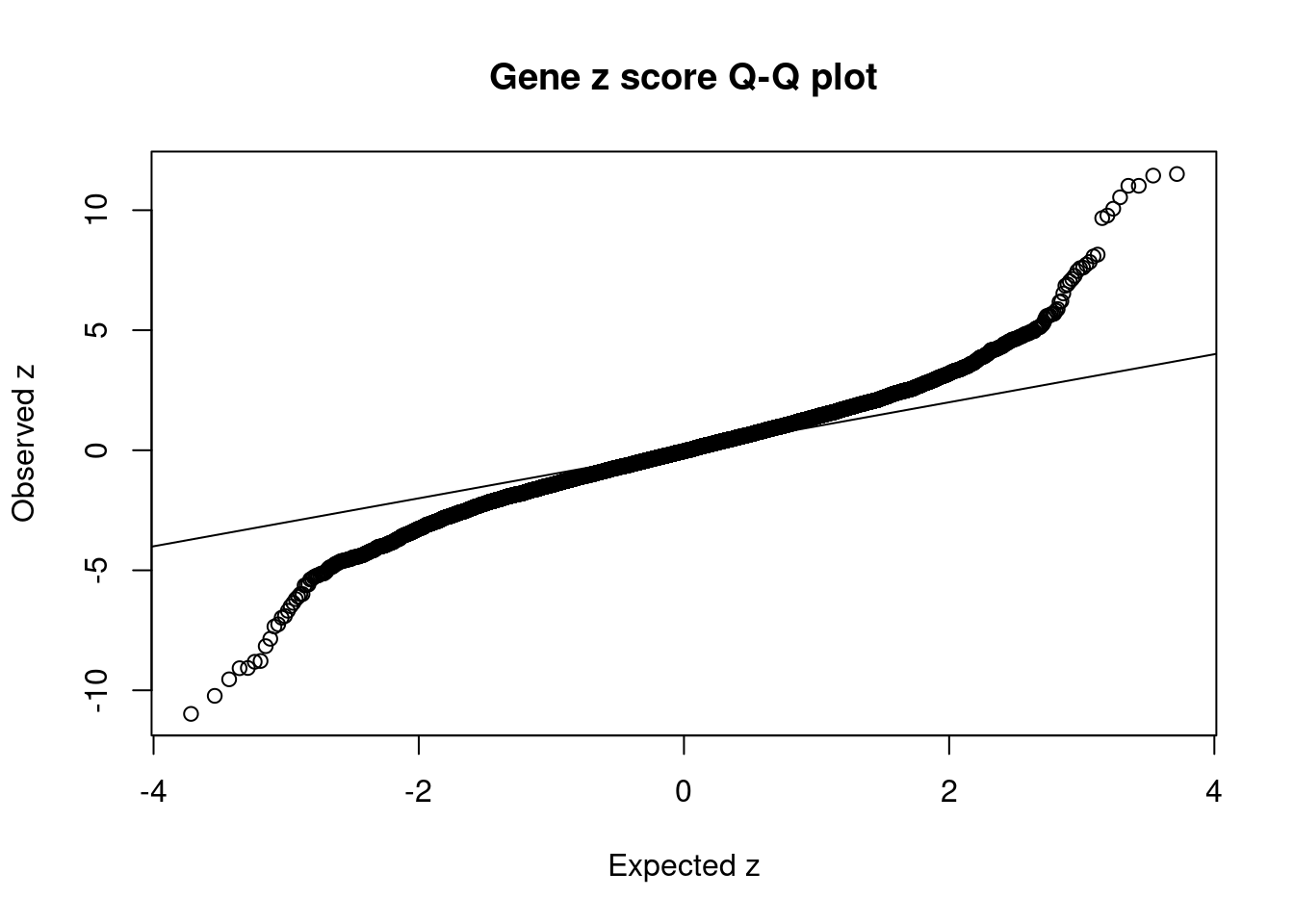
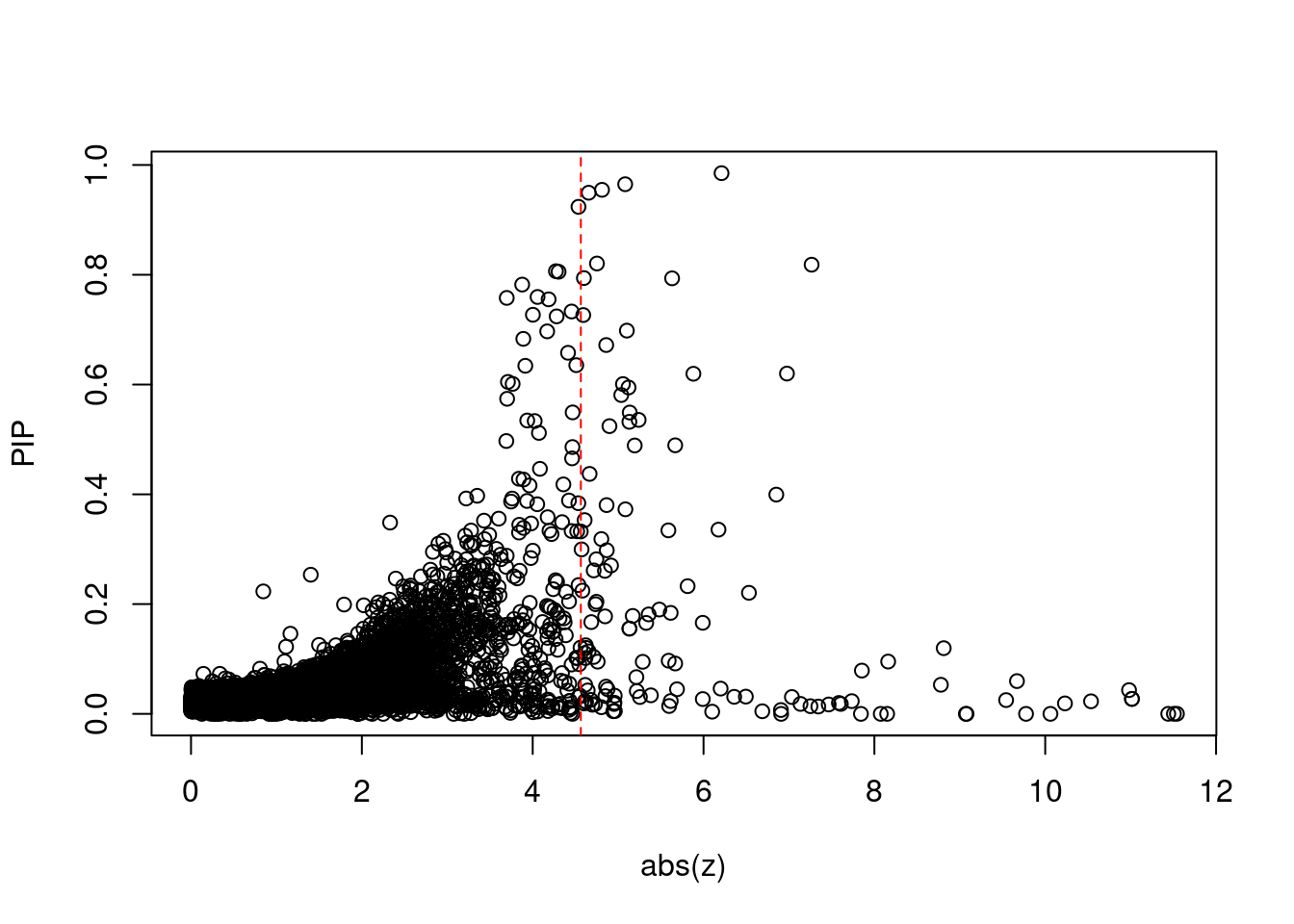
[1] 0.01227GO enrichment analysis for genes with PIP>0.5
#number of genes for gene set enrichment
length(genes)[1] 42Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"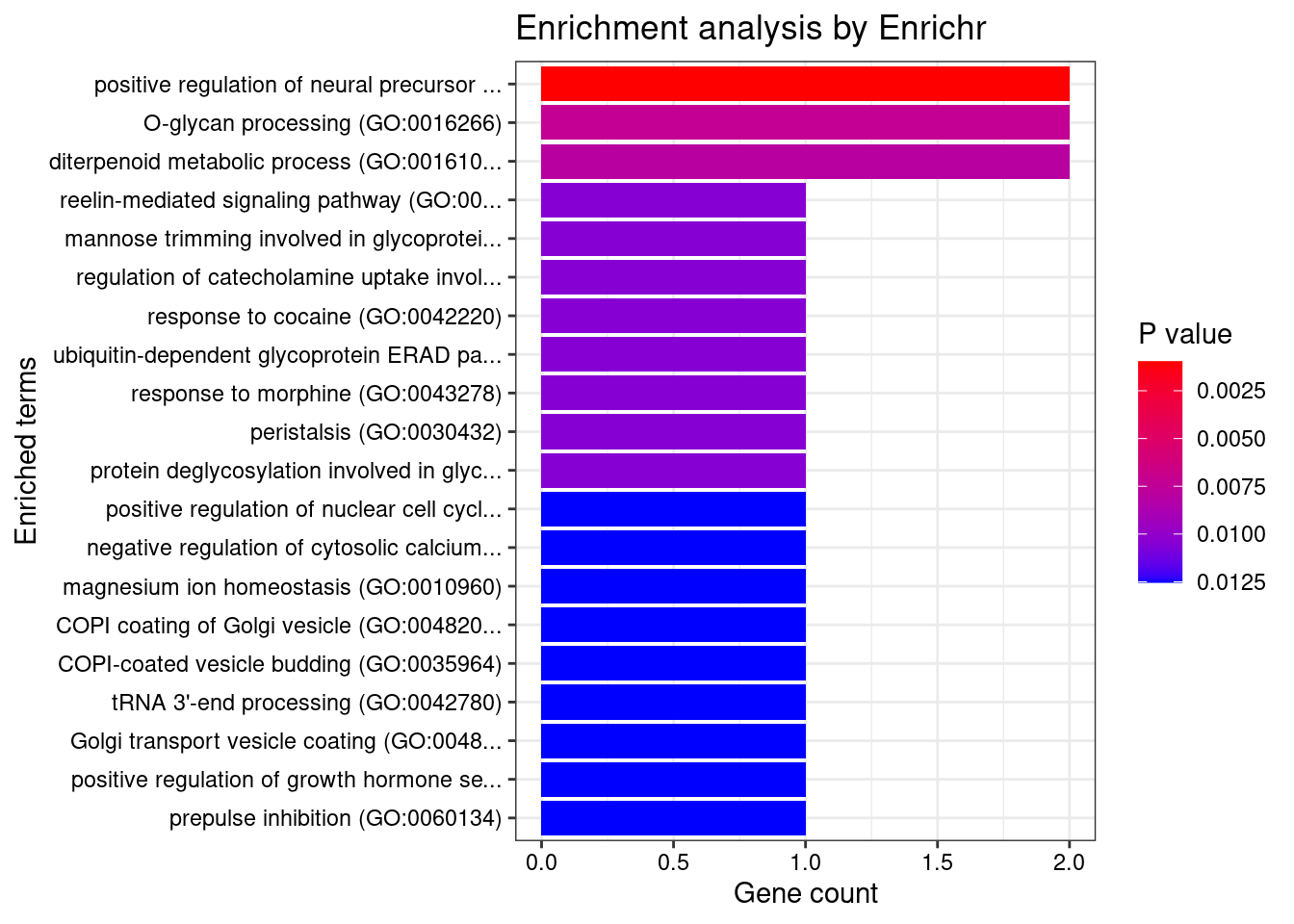
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Cellular_Component_2021"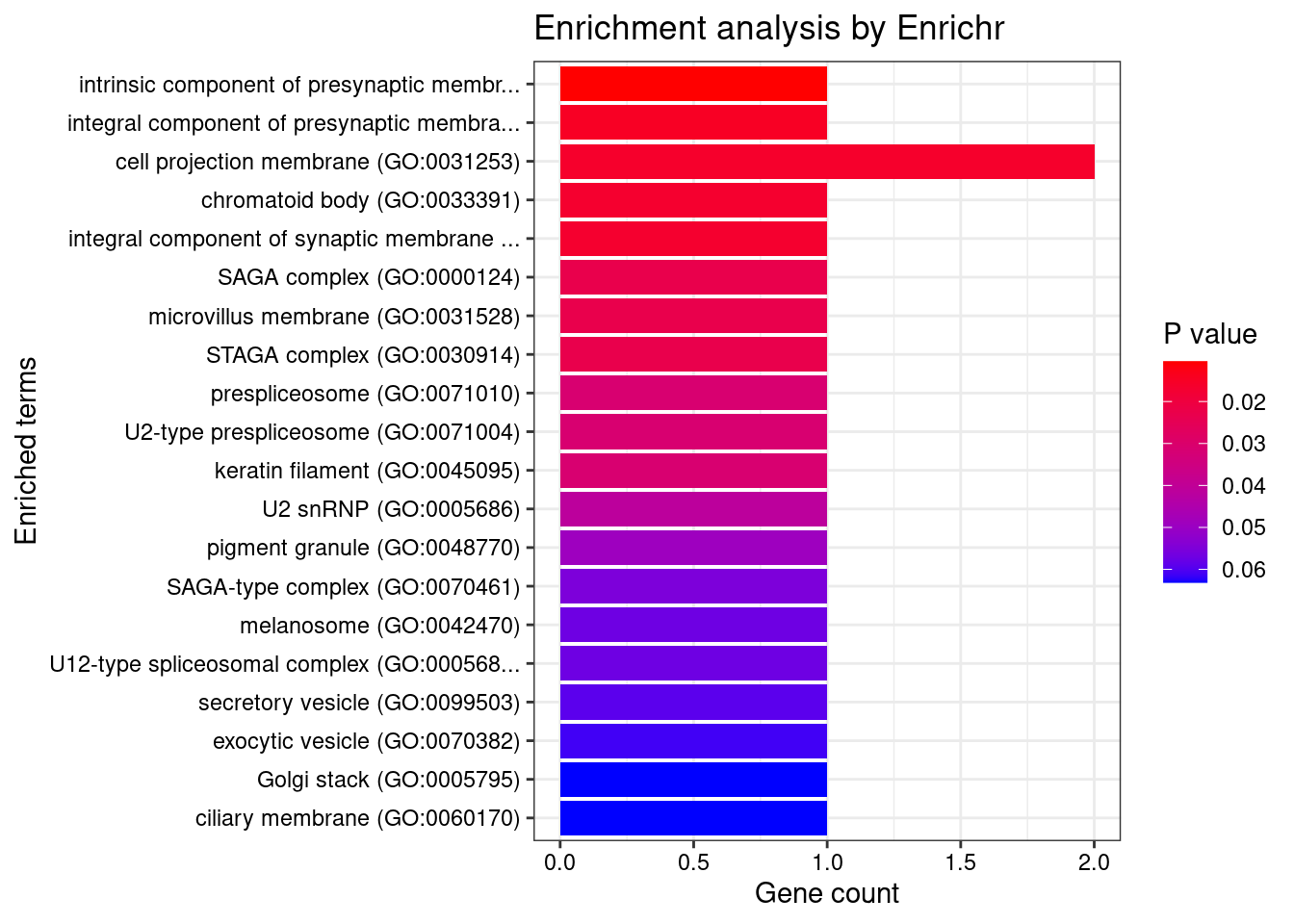
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Molecular_Function_2021"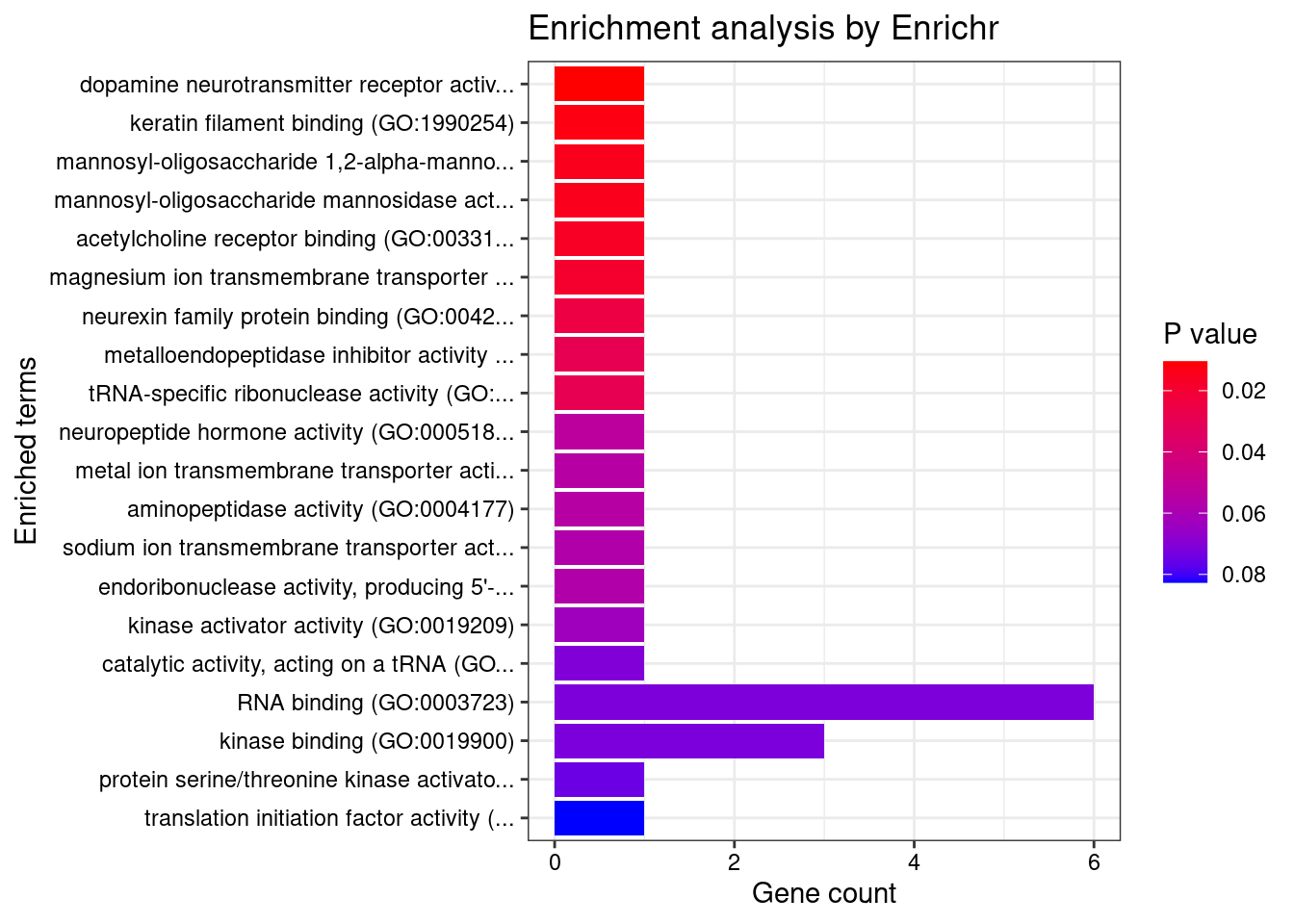
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)DisGeNET enrichment analysis for genes with PIP>0.5
Description FDR Ratio
161 Newfoundland Rod-Cone Dystrophy 0.03575 1/15
162 Bothnia Retinal Dystrophy 0.03575 1/15
163 Amaurosis hypertrichosis 0.03575 1/15
166 Cone rod dystrophy amelogenesis imperfecta 0.03575 1/15
169 Jalili syndrome 0.03575 1/15
171 PROSTATE CANCER, HEREDITARY, 2 0.03575 1/15
173 COMBINED OXIDATIVE PHOSPHORYLATION DEFICIENCY 17 0.03575 1/15
175 HYPOGONADOTROPIC HYPOGONADISM 22 WITH OR WITHOUT ANOSMIA 0.03575 1/15
118 Acquired Language Disorders 0.05716 1/15
152 Refractory anemia with ringed sideroblasts 0.05716 1/15
BgRatio
161 1/9703
162 1/9703
163 1/9703
166 1/9703
169 1/9703
171 1/9703
173 1/9703
175 1/9703
118 2/9703
152 2/9703WebGestalt enrichment analysis for genes with PIP>0.5
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum =
minNum, : No significant gene set is identified based on FDR 0.05!NULLPIP Manhattan Plot
Warning: ggrepel: 2 unlabeled data points (too many overlaps). Consider
increasing max.overlaps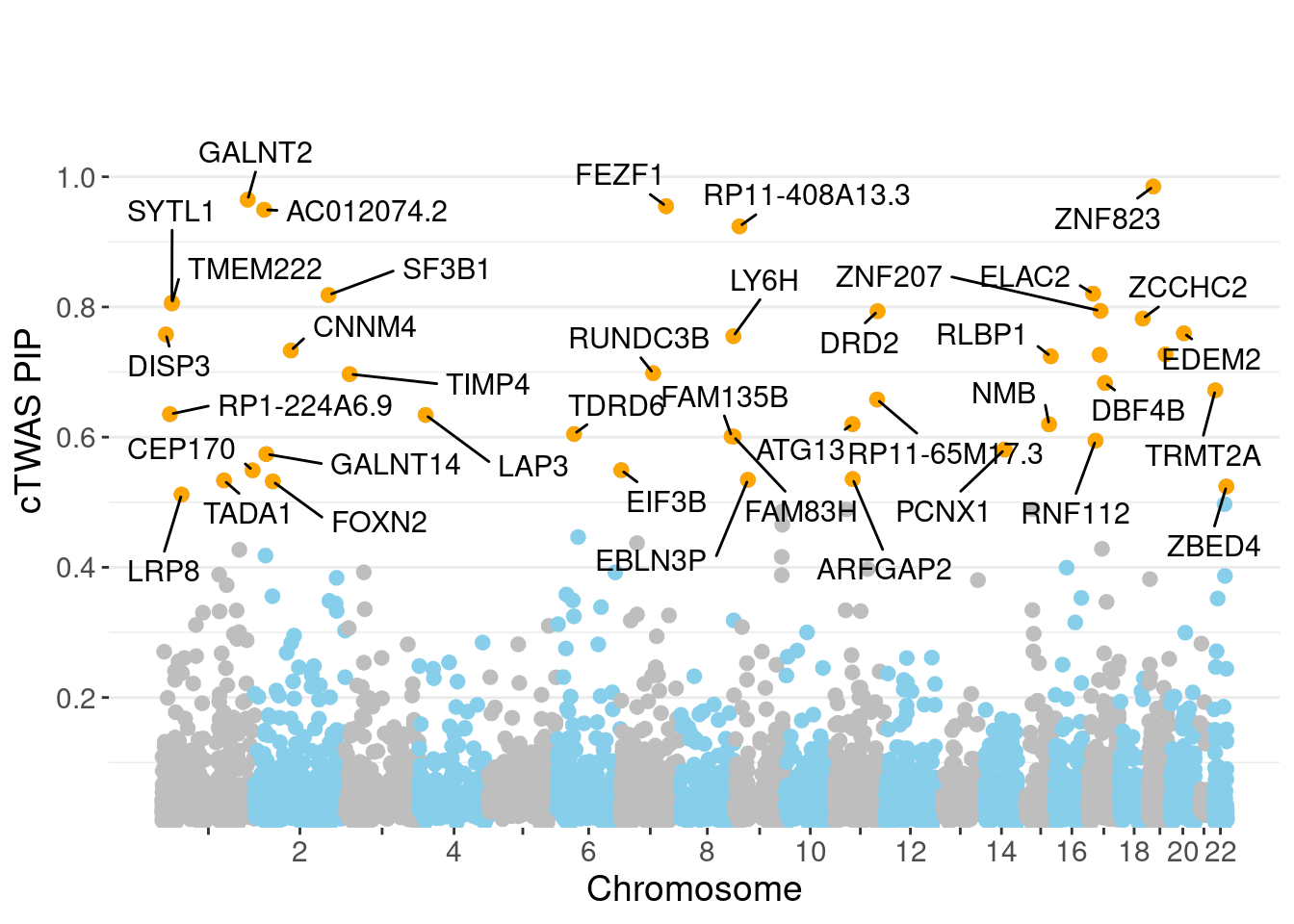
Sensitivity, specificity and precision for silver standard genes
#number of genes in known annotations
print(length(known_annotations))[1] 130#number of genes in known annotations with imputed expression
print(sum(known_annotations %in% ctwas_gene_res$genename))[1] 55#significance threshold for TWAS
print(sig_thresh)[1] 4.564#number of ctwas genes
length(ctwas_genes)[1] 9#number of TWAS genes
length(twas_genes)[1] 122#show novel genes (ctwas genes with not in TWAS genes)
ctwas_gene_res[ctwas_gene_res$genename %in% novel_genes,report_cols] genename region_tag susie_pip mu2 PVE z num_eqtl
5667 SYTL1 1_19 0.8065 22.96 0.0001758 4.272 2
10337 TMEM222 1_19 0.8055 22.21 0.0001699 4.303 1
13452 RP11-408A13.3 9_12 0.9238 22.50 0.0001974 4.536 1#sensitivity / recall
print(sensitivity) ctwas TWAS
0.02308 0.12308 #specificity
print(specificity) ctwas TWAS
0.9994 0.9893 #precision / PPV
print(precision) ctwas TWAS
0.3333 0.1311 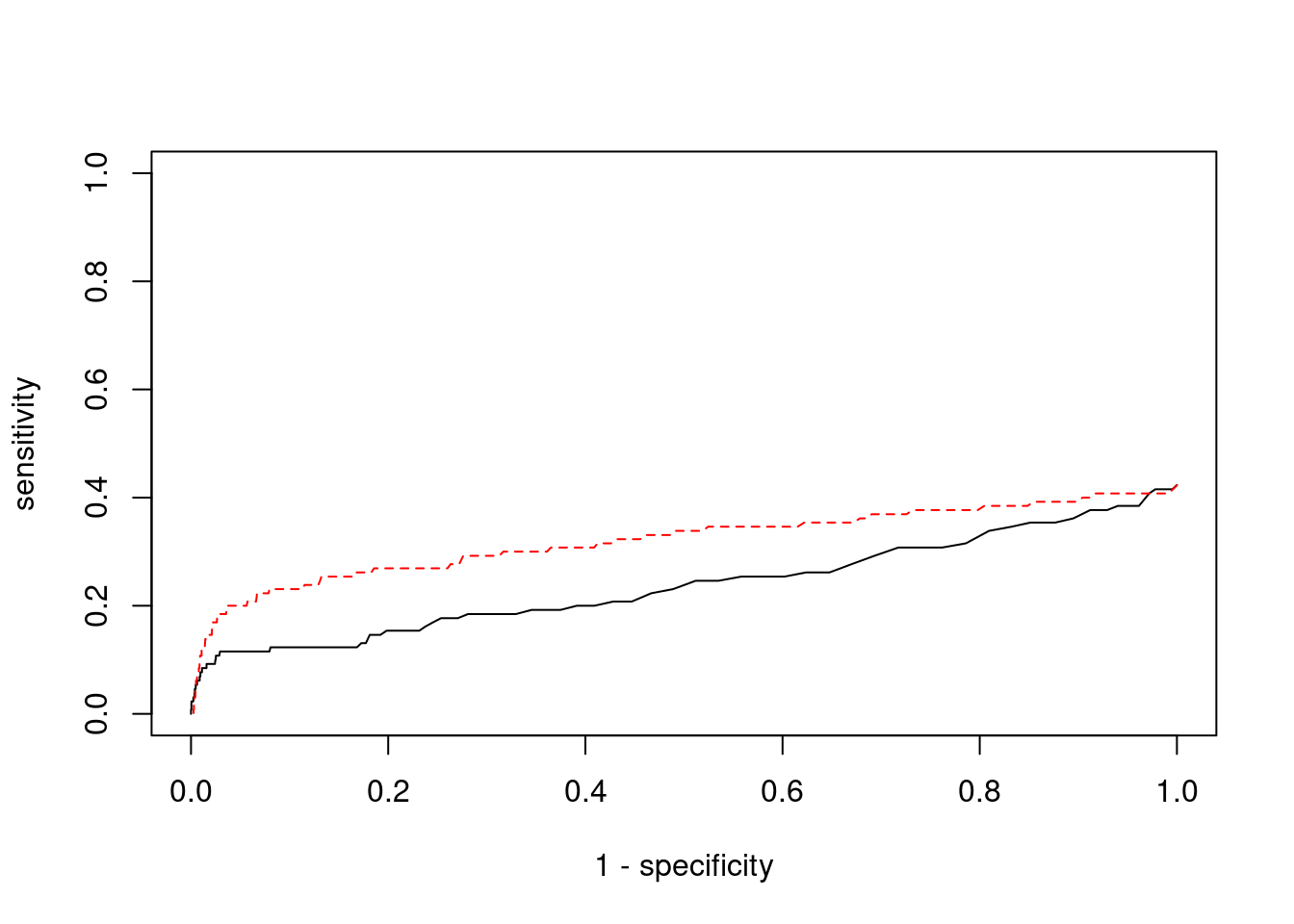
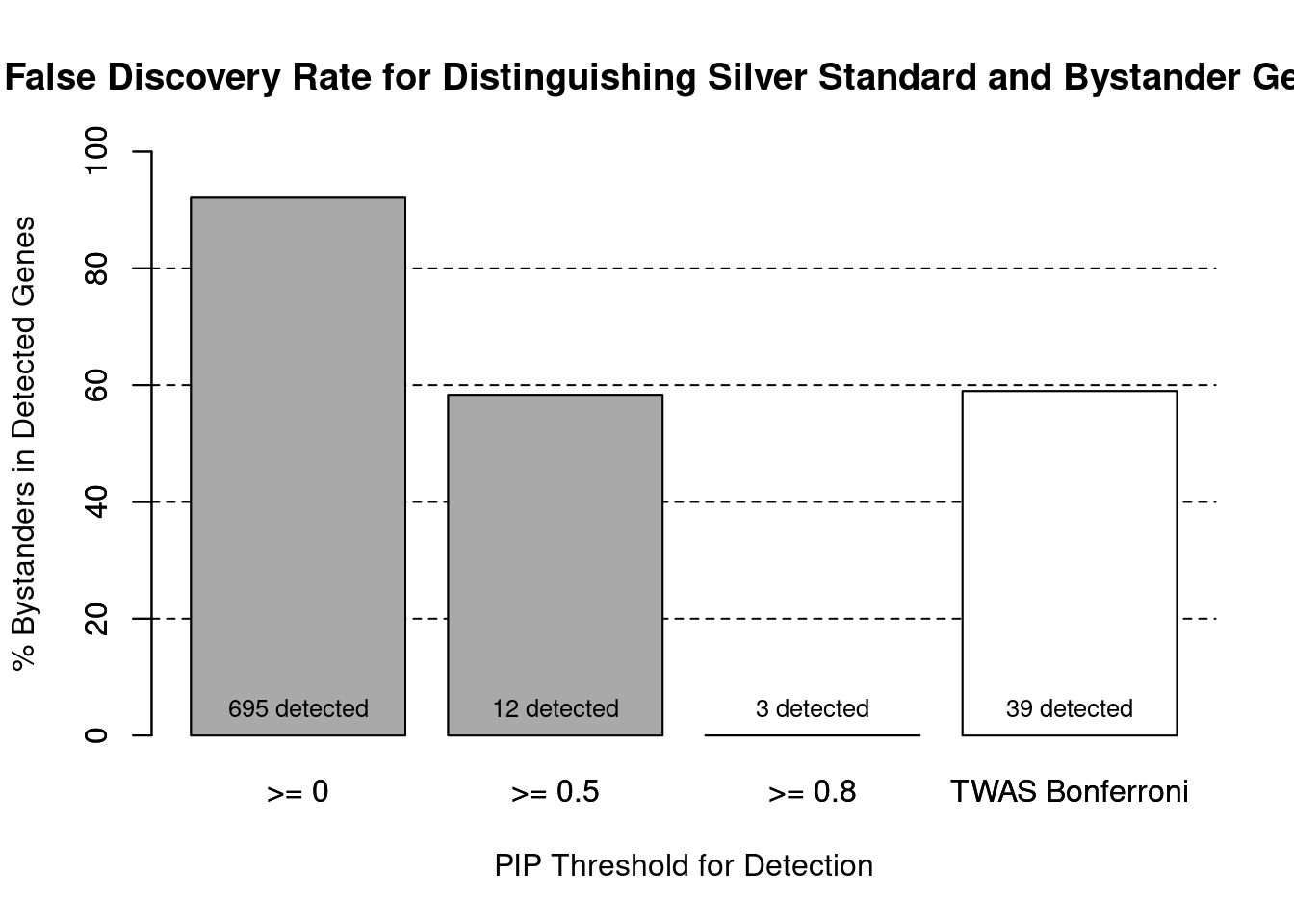
cTWAS is more precise than TWAS in distinguishing silver standard and bystander genes
#number of genes in known annotations (with imputed expression)
print(length(known_annotations))[1] 55#number of bystander genes (with imputed expression)
print(length(unrelated_genes))[1] 639#subset results to genes in known annotations or bystanders
ctwas_gene_res_subset <- ctwas_gene_res[ctwas_gene_res$genename %in% c(known_annotations, unrelated_genes),]
#assign ctwas and TWAS genes
ctwas_genes <- ctwas_gene_res_subset$genename[ctwas_gene_res_subset$susie_pip>0.8]
twas_genes <- ctwas_gene_res_subset$genename[abs(ctwas_gene_res_subset$z)>sig_thresh]
#significance threshold for TWAS
print(sig_thresh)[1] 4.564#number of ctwas genes (in known annotations or bystanders)
length(ctwas_genes)[1] 3#number of TWAS genes (in known annotations or bystanders)
length(twas_genes)[1] 39#sensitivity / recall
sensitivity ctwas TWAS
0.05455 0.29091 #specificity / (1 - False Positive Rate)
specificityctwas TWAS
1.000 0.964 #precision / PPV / (1 - False Discovery Rate)
precision ctwas TWAS
1.0000 0.4103 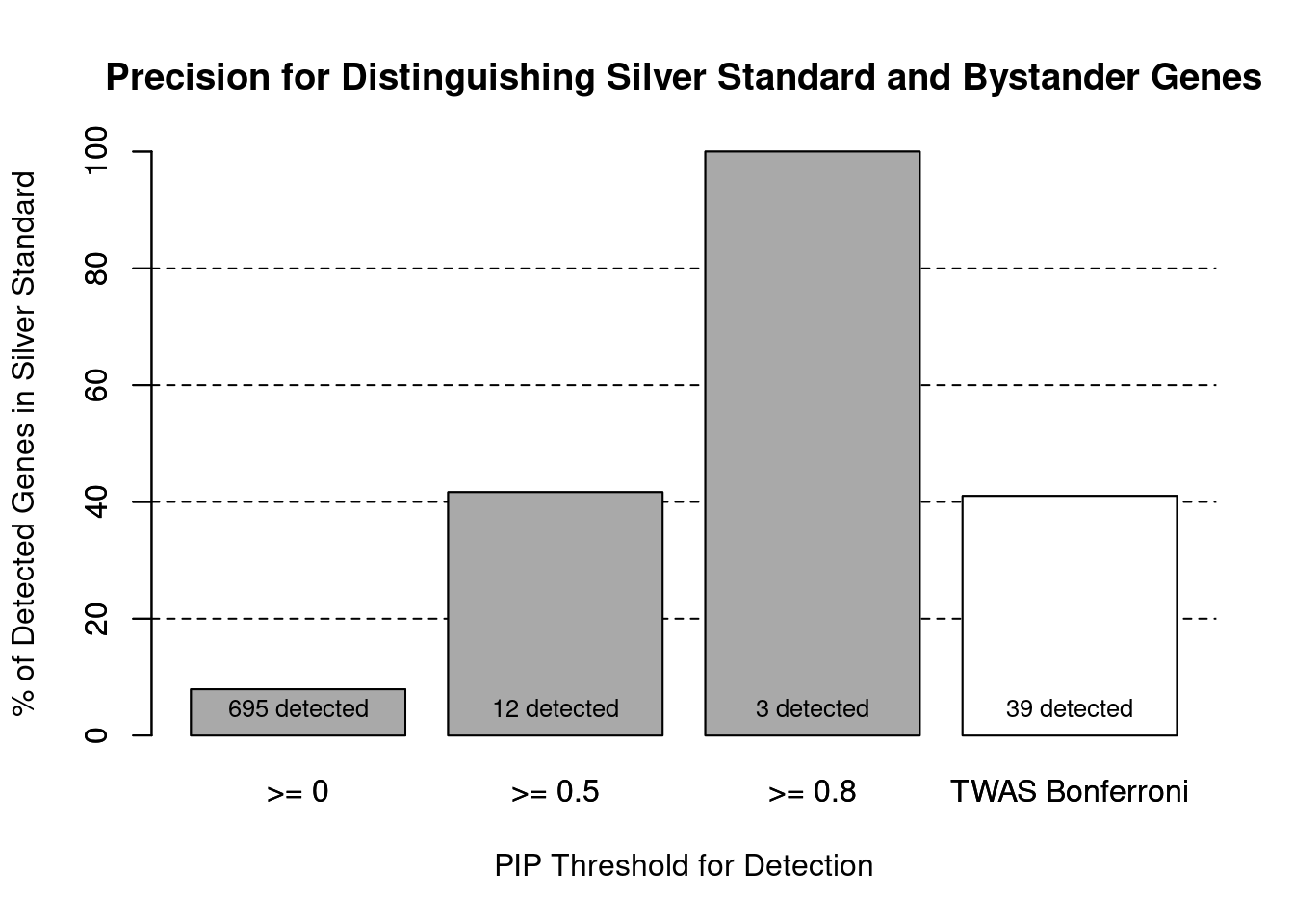
pip_range <- (0:1000)/1000
sensitivity <- rep(NA, length(pip_range))
specificity <- rep(NA, length(pip_range))
for (index in 1:length(pip_range)){
pip <- pip_range[index]
ctwas_genes <- ctwas_gene_res_subset$genename[ctwas_gene_res_subset$susie_pip>=pip]
sensitivity[index] <- sum(ctwas_genes %in% known_annotations)/length(known_annotations)
specificity[index] <- sum(!(unrelated_genes %in% ctwas_genes))/length(unrelated_genes)
}
plot(1-specificity, sensitivity, type="l", xlim=c(0,1), ylim=c(0,1), main="", xlab="1 - Specificity", ylab="Sensitivity")
title(expression("ROC Curve for cTWAS (black) and TWAS (" * phantom("red") * ")"))
title(expression(phantom("ROC Curve for cTWAS (black) and TWAS (") * "red" * phantom(")")), col.main="red")
sig_thresh_range <- seq(from=0, to=max(abs(ctwas_gene_res_subset$z)), length.out=length(pip_range))
for (index in 1:length(sig_thresh_range)){
sig_thresh_plot <- sig_thresh_range[index]
twas_genes <- ctwas_gene_res_subset$genename[abs(ctwas_gene_res_subset$z)>=sig_thresh_plot]
sensitivity[index] <- sum(twas_genes %in% known_annotations)/length(known_annotations)
specificity[index] <- sum(!(unrelated_genes %in% twas_genes))/length(unrelated_genes)
}
lines(1-specificity, sensitivity, xlim=c(0,1), ylim=c(0,1), col="red", lty=1)
abline(a=0,b=1,lty=3)
#add previously computed points from the analysis
ctwas_genes <- ctwas_gene_res_subset$genename[ctwas_gene_res_subset$susie_pip>0.8]
twas_genes <- ctwas_gene_res_subset$genename[abs(ctwas_gene_res_subset$z)>sig_thresh]
points(1-specificity_plot["ctwas"], sensitivity_plot["ctwas"], pch=21, bg="black")
points(1-specificity_plot["TWAS"], sensitivity_plot["TWAS"], pch=21, bg="red")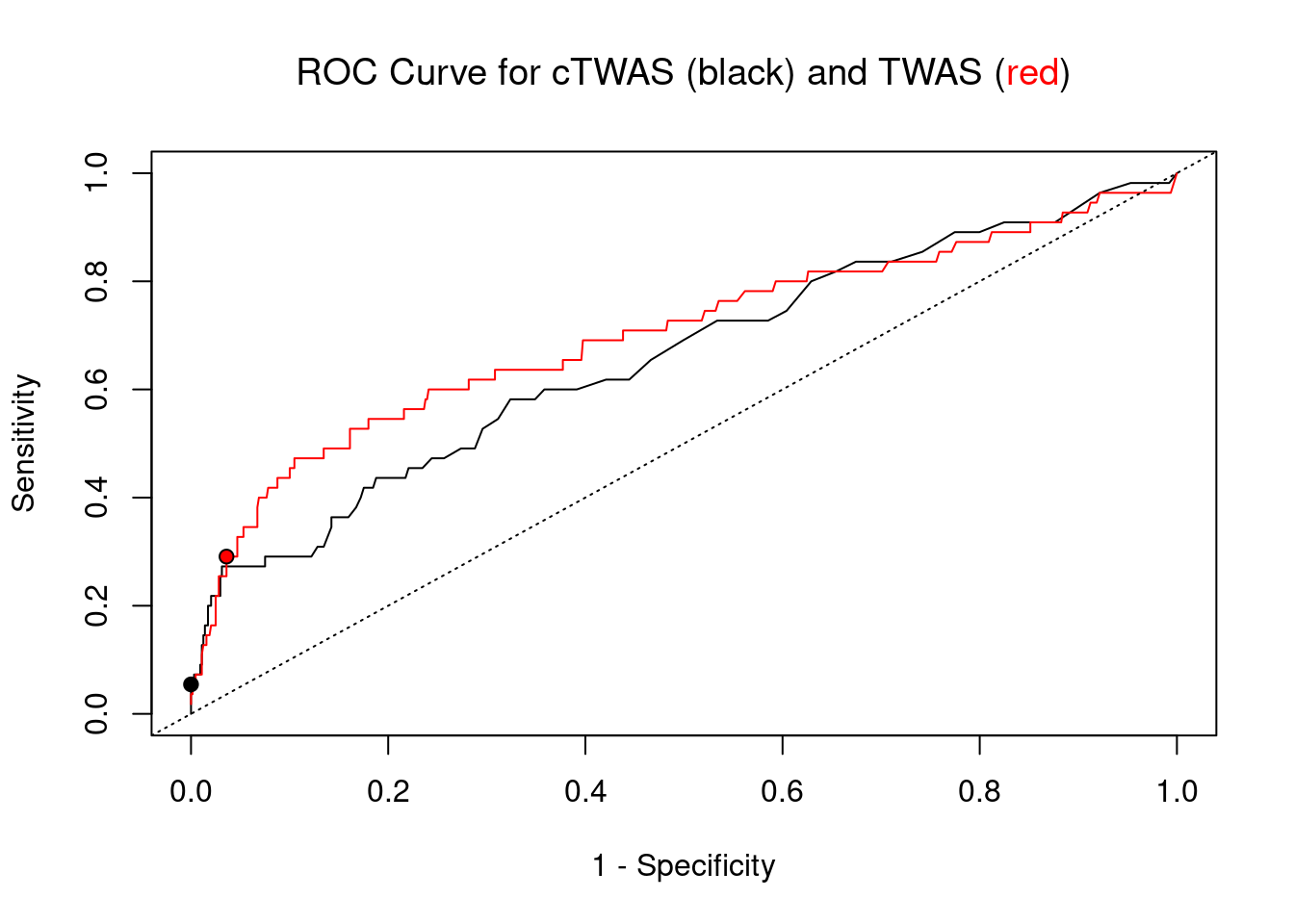
Undetected silver standard genes have low TWAS z-scores or stronger signal from nearby variants
#table of outcomes for silver standard genes
-sort(-table(silver_standard_case))silver_standard_case
Not Imputed Insignificant z-score Nearby SNP(s)
75 39 13
Detected (PIP > 0.8)
3 #show inconclusive genes
silver_standard_case[silver_standard_case=="Inconclusive"]named character(0)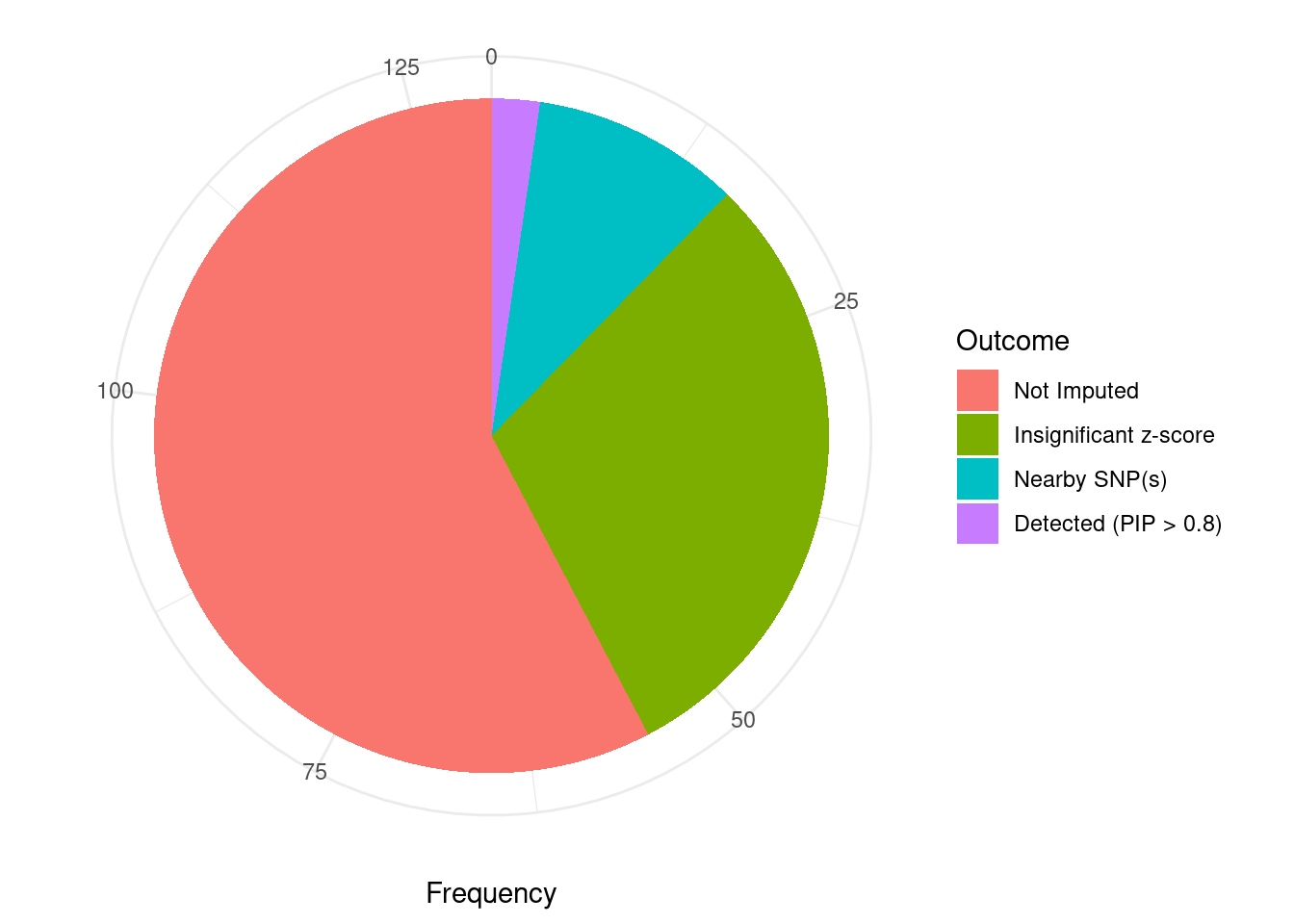
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] GenomicRanges_1.36.1 GenomeInfoDb_1.20.0 IRanges_2.18.1
[4] S4Vectors_0.22.1 BiocGenerics_0.30.0 biomaRt_2.40.1
[7] readxl_1.3.1 forcats_0.5.1 stringr_1.4.0
[10] dplyr_1.0.7 purrr_0.3.4 readr_2.1.1
[13] tidyr_1.1.4 tidyverse_1.3.1 tibble_3.1.6
[16] WebGestaltR_0.4.4 disgenet2r_0.99.2 enrichR_3.0
[19] cowplot_1.1.1 ggplot2_3.3.5 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] ggbeeswarm_0.6.0 colorspace_2.0-2 rjson_0.2.20
[4] ellipsis_0.3.2 rprojroot_2.0.2 XVector_0.24.0
[7] fs_1.5.2 rstudioapi_0.13 farver_2.1.0
[10] ggrepel_0.9.1 bit64_4.0.5 AnnotationDbi_1.46.0
[13] fansi_1.0.2 lubridate_1.8.0 xml2_1.3.3
[16] codetools_0.2-16 doParallel_1.0.17 cachem_1.0.6
[19] knitr_1.36 jsonlite_1.7.2 apcluster_1.4.8
[22] Cairo_1.5-12.2 broom_0.7.10 dbplyr_2.1.1
[25] compiler_3.6.1 httr_1.4.2 backports_1.4.1
[28] assertthat_0.2.1 Matrix_1.2-18 fastmap_1.1.0
[31] cli_3.1.0 later_0.8.0 prettyunits_1.1.1
[34] htmltools_0.5.2 tools_3.6.1 igraph_1.2.10
[37] GenomeInfoDbData_1.2.1 gtable_0.3.0 glue_1.6.2
[40] reshape2_1.4.4 doRNG_1.8.2 Rcpp_1.0.8
[43] Biobase_2.44.0 cellranger_1.1.0 jquerylib_0.1.4
[46] vctrs_0.3.8 svglite_1.2.2 iterators_1.0.14
[49] xfun_0.29 ps_1.6.0 rvest_1.0.2
[52] lifecycle_1.0.1 rngtools_1.5.2 XML_3.99-0.3
[55] zlibbioc_1.30.0 getPass_0.2-2 scales_1.1.1
[58] vroom_1.5.7 hms_1.1.1 promises_1.0.1
[61] yaml_2.2.1 curl_4.3.2 memoise_2.0.1
[64] ggrastr_1.0.1 gdtools_0.1.9 stringi_1.7.6
[67] RSQLite_2.2.8 highr_0.9 foreach_1.5.2
[70] rlang_1.0.1 pkgconfig_2.0.3 bitops_1.0-7
[73] evaluate_0.14 lattice_0.20-38 labeling_0.4.2
[76] bit_4.0.4 processx_3.5.2 tidyselect_1.1.1
[79] plyr_1.8.6 magrittr_2.0.2 R6_2.5.1
[82] generics_0.1.1 DBI_1.1.2 pillar_1.6.4
[85] haven_2.4.3 whisker_0.3-2 withr_2.4.3
[88] RCurl_1.98-1.5 modelr_0.1.8 crayon_1.5.0
[91] utf8_1.2.2 tzdb_0.2.0 rmarkdown_2.11
[94] progress_1.2.2 grid_3.6.1 data.table_1.14.2
[97] blob_1.2.2 callr_3.7.0 git2r_0.26.1
[100] reprex_2.0.1 digest_0.6.29 httpuv_1.5.1
[103] munsell_0.5.0 beeswarm_0.2.3 vipor_0.4.5