BMI - Brain Cortex
sheng Qian
2021-2-6
Last updated: 2022-02-22
Checks: 6 1
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/ | data |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/ctwas_config.R | code/ctwas_config.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 1058990. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .ipynb_checkpoints/
Untracked files:
Untracked: Rplot.png
Untracked: analysis/.ipynb_checkpoints/
Untracked: analysis/Glucose_Adipose_Subcutaneous.Rmd
Untracked: analysis/Glucose_Adipose_Visceral_Omentum.Rmd
Untracked: analysis/Splicing_Test.Rmd
Untracked: code/.ipynb_checkpoints/
Untracked: code/AF_out/
Untracked: code/BMI_S_out/
Untracked: code/BMI_out/
Untracked: code/Glucose_out/
Untracked: code/LDL_S_out/
Untracked: code/T2D_out/
Untracked: code/ctwas_config.R
Untracked: code/mapping.R
Untracked: code/out/
Untracked: code/run_AF_analysis.sbatch
Untracked: code/run_AF_analysis.sh
Untracked: code/run_AF_ctwas_rss_LDR.R
Untracked: code/run_BMI_analysis.sbatch
Untracked: code/run_BMI_analysis.sh
Untracked: code/run_BMI_analysis_S.sbatch
Untracked: code/run_BMI_analysis_S.sh
Untracked: code/run_BMI_ctwas_rss_LDR.R
Untracked: code/run_BMI_ctwas_rss_LDR_S.R
Untracked: code/run_Glucose_analysis.sbatch
Untracked: code/run_Glucose_analysis.sh
Untracked: code/run_Glucose_ctwas_rss_LDR.R
Untracked: code/run_LDL_analysis_S.sbatch
Untracked: code/run_LDL_analysis_S.sh
Untracked: code/run_LDL_ctwas_rss_LDR_S.R
Untracked: code/run_T2D_analysis.sbatch
Untracked: code/run_T2D_analysis.sh
Untracked: code/run_T2D_ctwas_rss_LDR.R
Untracked: data/.ipynb_checkpoints/
Untracked: data/AF/
Untracked: data/BMI/
Untracked: data/BMI_S/
Untracked: data/Glucose/
Untracked: data/LDL_S/
Untracked: data/T2D/
Untracked: data/TEST/
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/mashr_sqtl/
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: data/untitled.txt
Unstaged changes:
Modified: analysis/BMI_Brain_Cortex.Rmd
Modified: analysis/LDL_Liver_S.Rmd
Modified: analysis/index.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/BMI_Brain_Cortex_S.Rmd) and HTML (docs/BMI_Brain_Cortex_S.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 1058990 | sq-96 | 2022-02-22 | update |
| html | 9ef0786 | sq-96 | 2022-02-22 | Build site. |
| Rmd | e6ae626 | sq-96 | 2022-02-22 | update |
Weight QC
#number of imputed weights
nrow(qclist_all)[1] 27311#number of imputed weights by chromosome
table(qclist_all$chr)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
2515 1948 1633 1028 1120 1411 1528 995 1079 1185 1612 1380 526 907 911 1244
17 18 19 20 21 22
1938 360 1923 874 342 852 #number of imputed weights without missing variants
sum(qclist_all$nmiss==0)[1] 24860#proportion of imputed weights without missing variants
mean(qclist_all$nmiss==0)[1] 0.9103#add z scores to results
load(paste0(results_dir, "/", analysis_id, "_expr_z_gene.Rd"))
ctwas_gene_res$z <- z_gene[ctwas_gene_res$intron_id,]$z
z_snp <- z_snp[z_snp$id %in% ctwas_snp_res$id,]
ctwas_snp_res$z <- z_snp$z[match(ctwas_snp_res$id, z_snp$id)]
#merge gene and snp results with added information
ctwas_snp_res$genename=NA
ctwas_snp_res$gene_type=NA
ctwas_snp_res$intron_id=NA
ctwas_res <- rbind(ctwas_gene_res,
ctwas_snp_res[,colnames(ctwas_gene_res)])
#get number of eQTL for geens
num_eqtl <- c()
for (i in 1:22){
load(paste0(results_dir, "/", analysis_id, "_expr_chr", i, ".exprqc.Rd"))
num_eqtl <- c(num_eqtl, unlist(lapply(wgtlist, nrow)))
}
ctwas_gene_res$num_eqtl <- num_eqtl[ctwas_gene_res$intron_id]
#store columns to report
report_cols <- colnames(ctwas_gene_res)[!(colnames(ctwas_gene_res) %in% c("type", "region_tag1", "region_tag2", "cs_index", "gene_type", "z_flag", "id", "chrom", "pos"))]
first_cols <- c("genename", "region_tag")
report_cols <- c(first_cols, report_cols[!(report_cols %in% first_cols)])
report_cols_snps <- c("id", report_cols[-1])
report_cols_snps <- report_cols_snps[!(report_cols_snps %in% "num_eqtl")]
#get number of SNPs from s1 results; adjust for thin argument
ctwas_res_s1 <- data.table::fread(paste0(results_dir, "/", analysis_id, "_ctwas.s1.susieIrss.txt"))
n_snps <- sum(ctwas_res_s1$type=="SNP")/thin
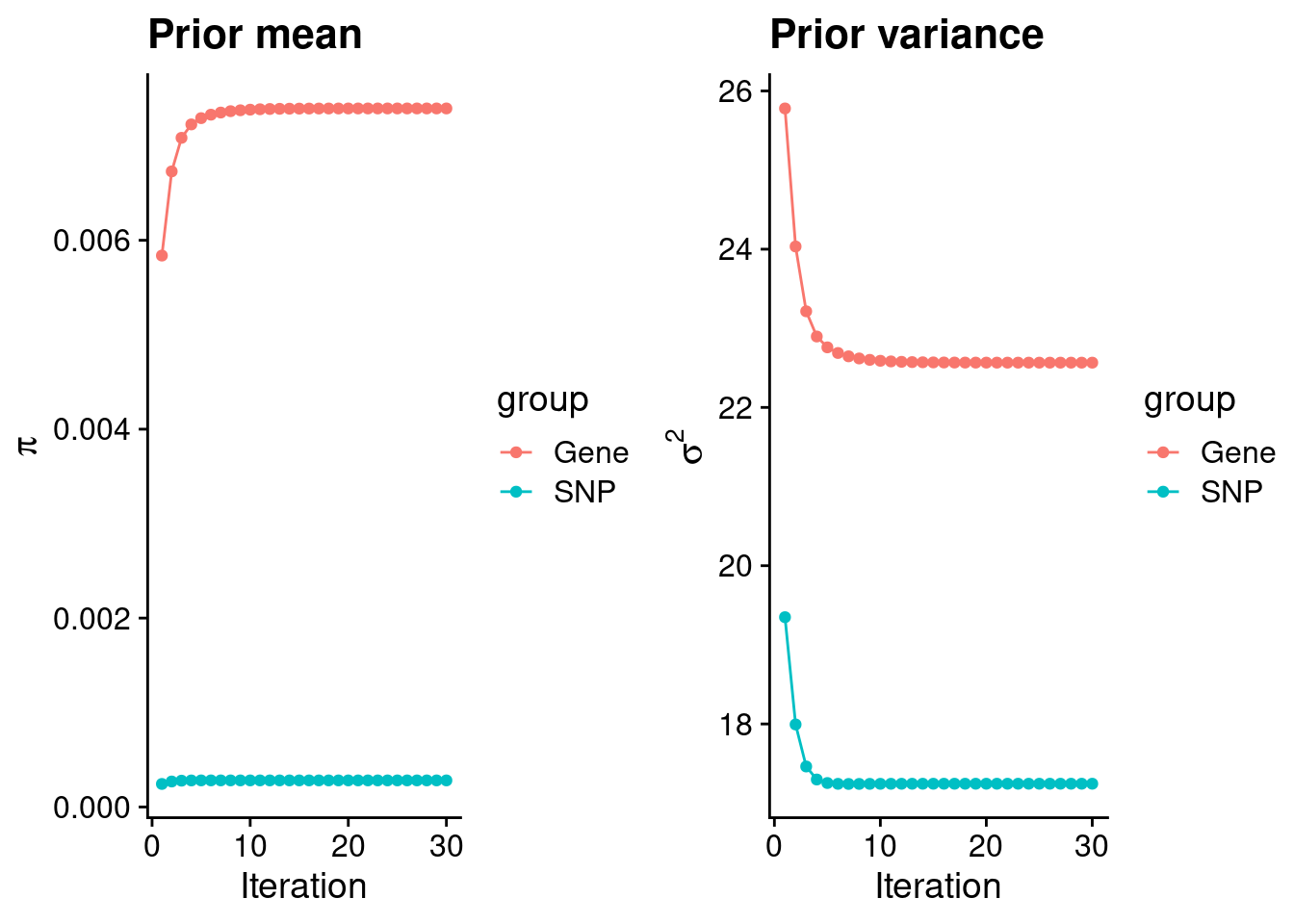
rm(ctwas_res_s1)Check convergence of parameters
| Version | Author | Date |
|---|---|---|
| 9ef0786 | sq-96 | 2022-02-22 |
#estimated group prior
estimated_group_prior <- group_prior_rec[,ncol(group_prior_rec)]
names(estimated_group_prior) <- c("gene", "snp")
estimated_group_prior["snp"] <- estimated_group_prior["snp"]*thin #adjust parameter to account for thin argument
print(estimated_group_prior) gene snp
0.0073957 0.0002813 #estimated group prior variance
estimated_group_prior_var <- group_prior_var_rec[,ncol(group_prior_var_rec)]
names(estimated_group_prior_var) <- c("gene", "snp")
print(estimated_group_prior_var) gene snp
22.57 17.24 #report sample size
print(sample_size)[1] 336107#report group size
group_size <- c(nrow(ctwas_gene_res), n_snps)
print(group_size)[1] 27311 7535010#estimated group PVE
estimated_group_pve <- estimated_group_prior_var*estimated_group_prior*group_size/sample_size #check PVE calculation
names(estimated_group_pve) <- c("gene", "snp")
print(estimated_group_pve) gene snp
0.01356 0.10874 #compare sum(PIP*mu2/sample_size) with above PVE calculation
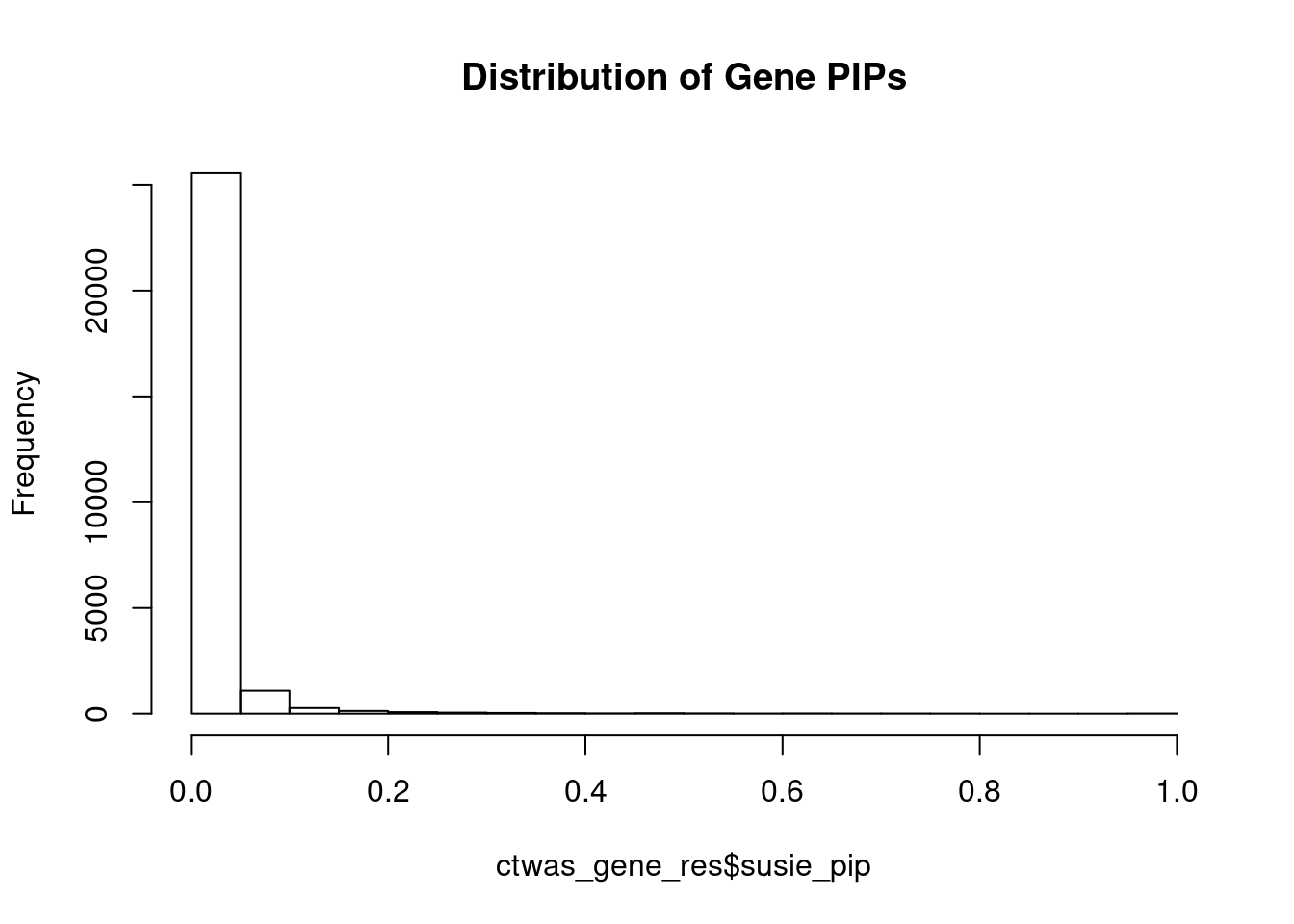
c(sum(ctwas_gene_res$PVE),sum(ctwas_snp_res$PVE))[1] 0.5009 15.8815Genes with highest PIPs
| Version | Author | Date |
|---|---|---|
| 9ef0786 | sq-96 | 2022-02-22 |
genename region_tag susie_pip mu2 PVE
25970 PIK3R3 1_28 1.0000 34091.07 1.014e-01
26275 CCDC127 5_1 1.0000 14934.79 4.443e-02
26744 SLC38A2 12_29 1.0000 14459.24 4.302e-02
27018 ZNF559-ZNF177 19_9 1.0000 15583.86 4.637e-02
26367 NSD1 5_106 1.0000 13207.48 3.930e-02
26836 TMOD3 15_21 1.0000 28879.15 8.592e-02
26865 TNFSF12-TNFSF13 17_7 0.9416 29.03 8.132e-05
27140 GIPR 19_32 0.9414 5582.93 1.564e-02
26489 PMS2P3 7_48 0.8710 93.55 2.425e-04
26114 EIF2B5 3_113 0.8332 37.82 9.377e-05
5664 SERPINI1 3_103 0.7891 23.68 5.560e-05
15679 POU6F1 12_31 0.7852 30.85 7.209e-05
13315 C10orf32-ASMT 10_66 0.7770 34.48 7.970e-05
10486 KCNH2 7_93 0.7703 40.82 9.356e-05
14010 BDNF-AS 11_19 0.7547 119.88 2.692e-04
7086 POC5 5_44 0.7469 93.32 2.074e-04
401 RP1-37C10.3 1_12 0.7444 28.18 6.240e-05
10814 SLC25A37 8_24 0.7400 26.40 5.812e-05
15751 CBX5 12_33 0.7372 25.84 5.669e-05
4376 SETD5 3_7 0.7324 33.39 7.275e-05
intron_id z num_eqtl
25970 intron_1_46055971_46061929 4.441 1
26275 intron_5_205958_216729 3.000 1
26744 intron_12_46366945_46367076 2.953 1
27018 intron_19_9364948_9376316 -3.338 1
26367 intron_5_177235945_177239756 2.957 1
26836 intron_15_51924578_51931031 -4.528 1
26865 intron_17_7559702_7560049 -4.211 2
27140 intron_19_45677779_45677906 10.801 1
26489 intron_7_75514779_75515718 10.211 2
26114 intron_3_184278313_184283785 6.065 2
5664 intron_3_167735823_167789111 -4.409 2
15679 intron_12_51206883_51217642 -4.747 1
13315 intron_10_102874661_102876954 -5.664 1
10486 intron_7_150947878_150948444 -6.380 1
14010 intron_11_27658462_27659171 -13.138 1
7086 intron_5_75705787_75712854 -11.305 1
401 intron_1_16986358_16986805 4.886 1
10814 intron_8_23529212_23566108 5.008 1
15751 intron_12_54257692_54280008 -4.732 1
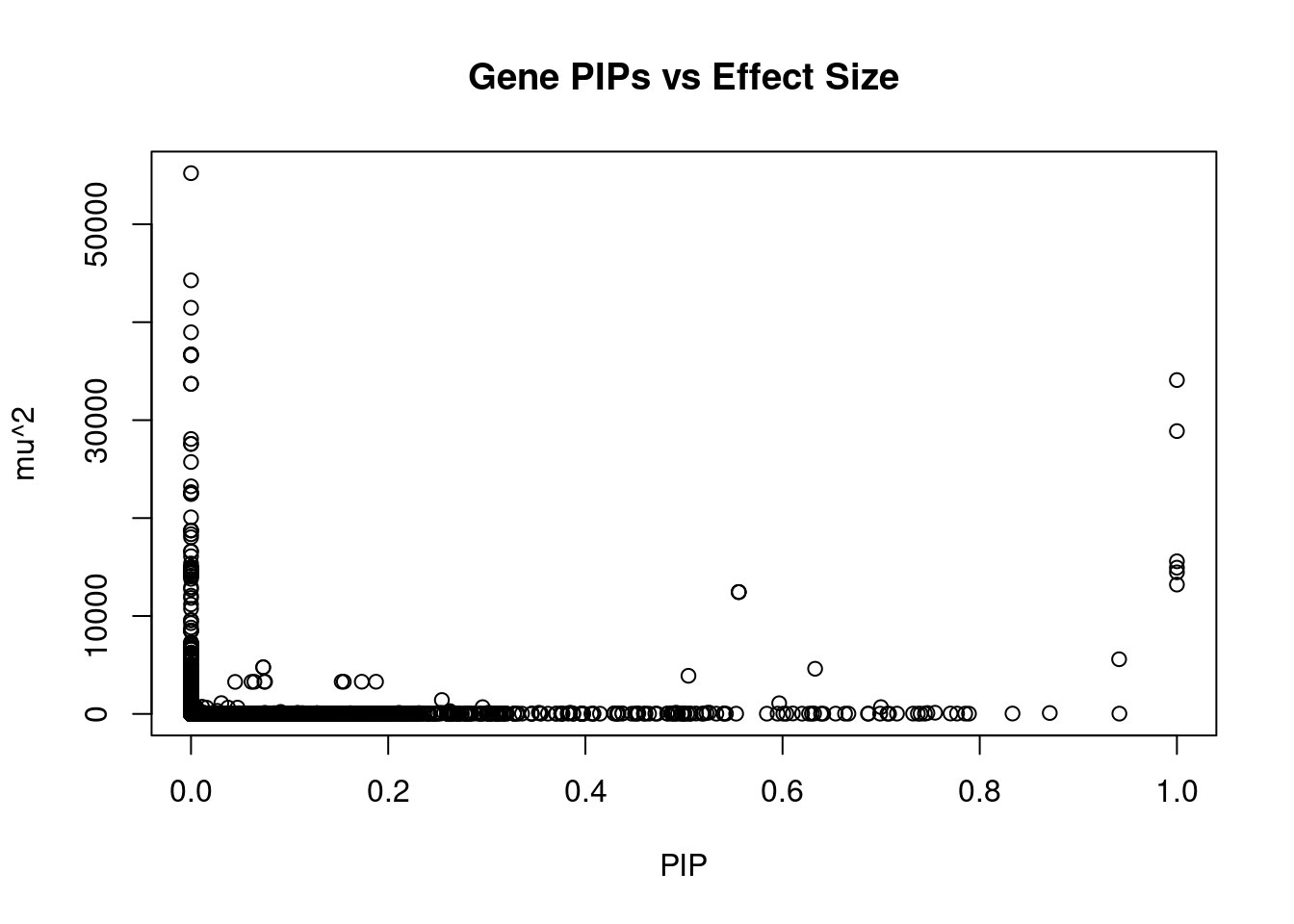
4376 intron_3_9464672_9468519 -5.700 1Genes with largest effect sizes
| Version | Author | Date |
|---|---|---|
| 9ef0786 | sq-96 | 2022-02-22 |
genename region_tag susie_pip mu2 PVE
22969 MAST3 19_14 0.000e+00 55219 0.000e+00
22971 MAST3 19_14 0.000e+00 44272 0.000e+00
11486 CCDC171 9_13 0.000e+00 41479 0.000e+00
11485 CCDC171 9_13 0.000e+00 38969 0.000e+00
22975 MAST3 19_14 0.000e+00 36721 0.000e+00
22976 MAST3 19_14 0.000e+00 36694 0.000e+00
26089 ANKRD28 3_11 0.000e+00 36605 0.000e+00
25970 PIK3R3 1_28 1.000e+00 34091 1.014e-01
25964 MAST2 1_28 0.000e+00 33718 0.000e+00
25963 MAST2 1_28 0.000e+00 33696 0.000e+00
26836 TMOD3 15_21 1.000e+00 28879 8.592e-02
25958 NASP 1_28 0.000e+00 28073 0.000e+00
25955 NASP 1_28 0.000e+00 27573 0.000e+00
25956 NASP 1_28 0.000e+00 27573 0.000e+00
22970 MAST3 19_14 0.000e+00 25724 0.000e+00
26837 TMOD3 15_21 0.000e+00 23242 0.000e+00
14607 MRPL21 11_38 2.220e-16 22652 1.496e-17
14611 IGHMBP2 11_38 2.331e-15 22511 1.562e-16
14612 IGHMBP2 11_38 2.331e-15 22511 1.562e-16
18242 CATSPER2P1 15_16 0.000e+00 22452 0.000e+00
intron_id z num_eqtl
22969 intron_19_18107618_18121685 6.803 1
22971 intron_19_18118254_18121685 -6.233 1
11486 intron_9_15889101_15920270 -7.233 1
11485 intron_9_15888088_15888983 -7.315 1
22975 intron_19_18147017_18147443 -5.836 2
22976 intron_19_18147044_18147443 5.832 2
26089 intron_3_15737233_15751750 5.255 1
25970 intron_1_46055971_46061929 4.441 1
25964 intron_1_45997799_46000959 -4.325 1
25963 intron_1_45882395_45959386 -4.311 1
26836 intron_15_51924578_51931031 -4.528 1
25958 intron_1_45617591_45618061 4.297 1
25955 intron_1_45591270_45600385 3.951 1
25956 intron_1_45591270_45602255 -3.951 1
22970 intron_19_18110366_18110652 5.647 1
26837 intron_15_51938260_51947292 -2.837 2
14607 intron_11_68898012_68900535 4.245 2
14611 intron_11_68929357_68933299 4.486 1
14612 intron_11_68930434_68933299 -4.486 1
18242 intron_15_43744219_43744395 -4.508 1Genes with highest PVE
genename region_tag susie_pip mu2 PVE
25970 PIK3R3 1_28 1.00000 34091.1 0.101429
26836 TMOD3 15_21 1.00000 28879.1 0.085922
27018 ZNF559-ZNF177 19_9 1.00000 15583.9 0.046366
26275 CCDC127 5_1 1.00000 14934.8 0.044435
26744 SLC38A2 12_29 1.00000 14459.2 0.043020
26367 NSD1 5_106 1.00000 13207.5 0.039295
16747 SPG20 13_13 0.55556 12445.3 0.020571
16746 SPG20 13_13 0.55556 12445.3 0.020571
16748 SPG20 13_13 0.55556 12445.3 0.020571
27140 GIPR 19_32 0.94136 5582.9 0.015637
3948 LANCL1 2_124 0.63302 4612.3 0.008687
1268 LRRC8B 1_54 0.50455 3893.3 0.005845
7641 SFXN1 5_105 0.59658 1091.4 0.001937
2152 TATDN3 1_108 0.18764 3289.4 0.001836
2149 TATDN3 1_108 0.17321 3288.3 0.001695
2147 TATDN3 1_108 0.15497 3287.2 0.001516
2148 TATDN3 1_108 0.15284 3287.2 0.001495
7639 SFXN1 5_105 0.69978 693.6 0.001444
26525 LY6H 8_94 0.25445 1420.8 0.001076
26323 TMEM161B-AS1 5_52 0.07322 4761.5 0.001037
intron_id z num_eqtl
25970 intron_1_46055971_46061929 4.4412 1
26836 intron_15_51924578_51931031 -4.5284 1
27018 intron_19_9364948_9376316 -3.3380 1
26275 intron_5_205958_216729 2.9999 1
26744 intron_12_46366945_46367076 2.9534 1
26367 intron_5_177235945_177239756 2.9570 1
16747 intron_13_36335832_36346225 -2.8188 1
16746 intron_13_36335832_36336287 2.8188 1
16748 intron_13_36335832_36346582 2.8188 1
27140 intron_19_45677779_45677906 10.8005 1
3948 intron_2_210476412_210477450 -3.4970 1
1268 intron_1_89584789_89592771 3.3161 1
7641 intron_5_175516663_175521919 3.5182 1
2152 intron_1_212812328_212815013 3.2955 1
2149 intron_1_212807848_212812227 -3.3006 1
2147 intron_1_212804429_212804596 3.2995 1
2148 intron_1_212804651_212807736 3.2998 1
7639 intron_5_175512196_175513463 0.2221 2
26525 intron_8_143159709_143160198 3.8794 1
26323 intron_5_88270585_88282042 -8.0478 1Genes with largest z scores
genename region_tag susie_pip mu2 PVE
14009 BDNF-AS 11_19 2.532e-01 118.86 8.955e-05
14010 BDNF-AS 11_19 7.547e-01 119.88 2.692e-04
4898 RBM6 3_35 2.556e-03 897.70 6.826e-06
4899 RBM6 3_35 2.556e-03 897.70 6.826e-06
7086 POC5 5_44 7.469e-01 93.32 2.074e-04
2630 DNAJC27-AS1 2_15 6.927e-05 190.37 3.924e-08
2631 DNAJC27-AS1 2_15 6.927e-05 190.37 3.924e-08
2632 DNAJC27-AS1 2_15 6.927e-05 190.37 3.924e-08
8197 C6orf106 6_28 3.529e-01 120.59 1.266e-04
8200 SNRPC 6_28 1.081e-01 117.97 3.795e-05
4887 RNF123 3_35 2.511e-11 813.45 6.077e-14
4885 RNF123 3_35 2.337e-11 813.03 5.653e-14
4888 RNF123 3_35 2.080e-11 812.45 5.028e-14
8198 C6orf106 6_28 1.271e-02 116.28 4.397e-06
8199 C6orf106 6_28 1.271e-02 116.28 4.397e-06
2005 LMOD1 1_103 3.020e-01 113.57 1.020e-04
19470 SH2B1 16_23 2.258e-01 67.15 4.512e-05
27140 GIPR 19_32 9.414e-01 5582.93 1.564e-02
19500 DOC2A 16_24 2.221e-02 95.07 6.282e-06
19454 SULT1A2 16_23 2.054e-01 64.91 3.967e-05
intron_id z num_eqtl
14009 intron_11_27640005_27659171 13.37 2
14010 intron_11_27658462_27659171 -13.14 1
4898 intron_3_49999513_50046716 12.54 1
4899 intron_3_49999513_50048245 -12.54 1
7086 intron_5_75705787_75712854 -11.31 1
2630 intron_2_24972232_24972626 -11.29 1
2631 intron_2_24972232_24979060 11.29 1
2632 intron_2_24972232_25001415 11.29 1
8197 intron_6_34654779_34688841 11.24 1
8200 intron_6_34757954_34762595 -11.13 1
4887 intron_3_49712656_49713513 10.96 1
4885 intron_3_49698822_49698980 -10.96 1
4888 intron_3_49714174_49715575 -10.95 1
8198 intron_6_34654779_34696446 -10.90 1
8199 intron_6_34688933_34696446 10.90 1
2005 intron_1_201900751_201946080 -10.89 1
19470 intron_16_28869383_28870709 -10.81 2
27140 intron_19_45677779_45677906 10.80 1
19500 intron_16_30007090_30007173 -10.76 1
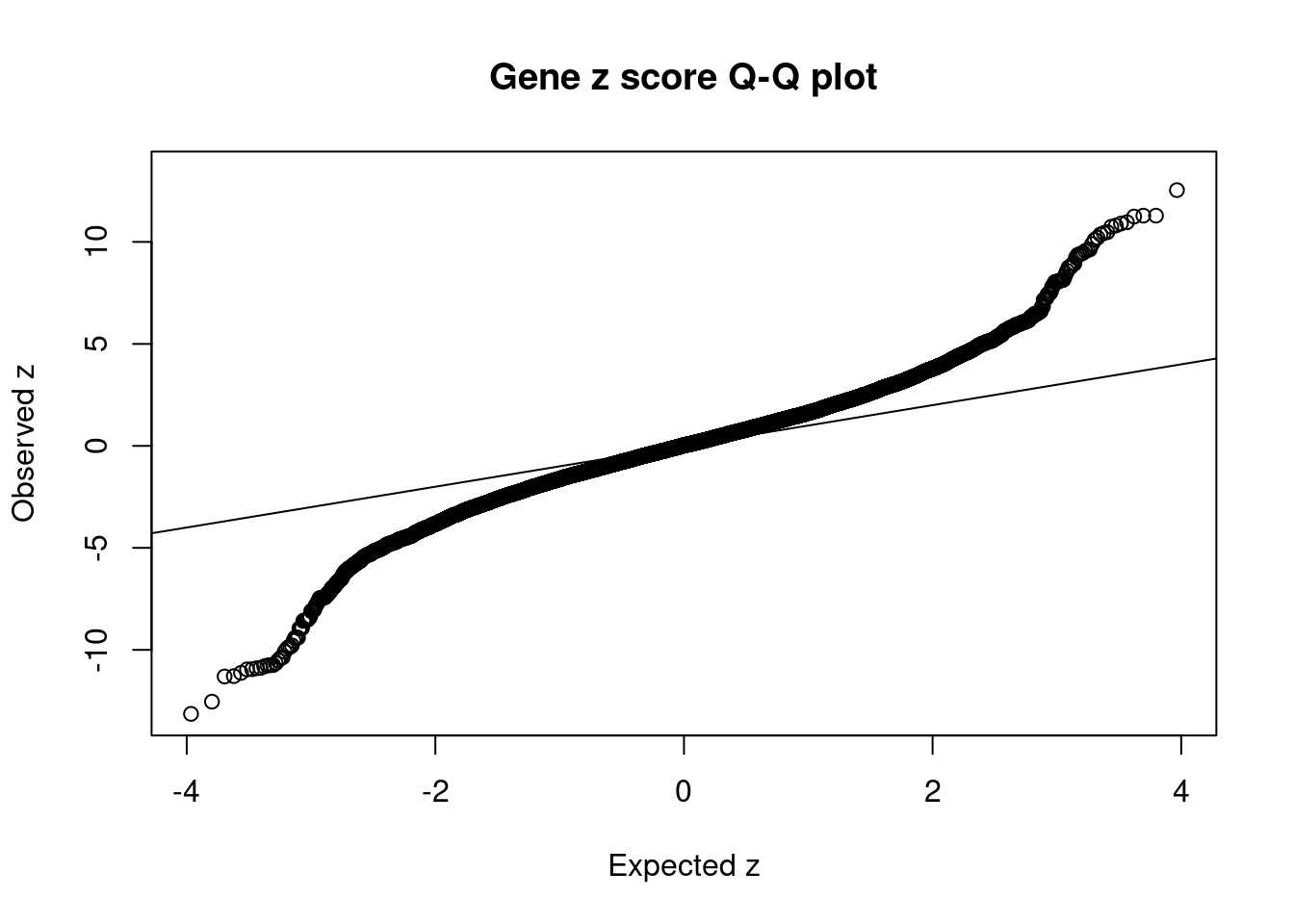
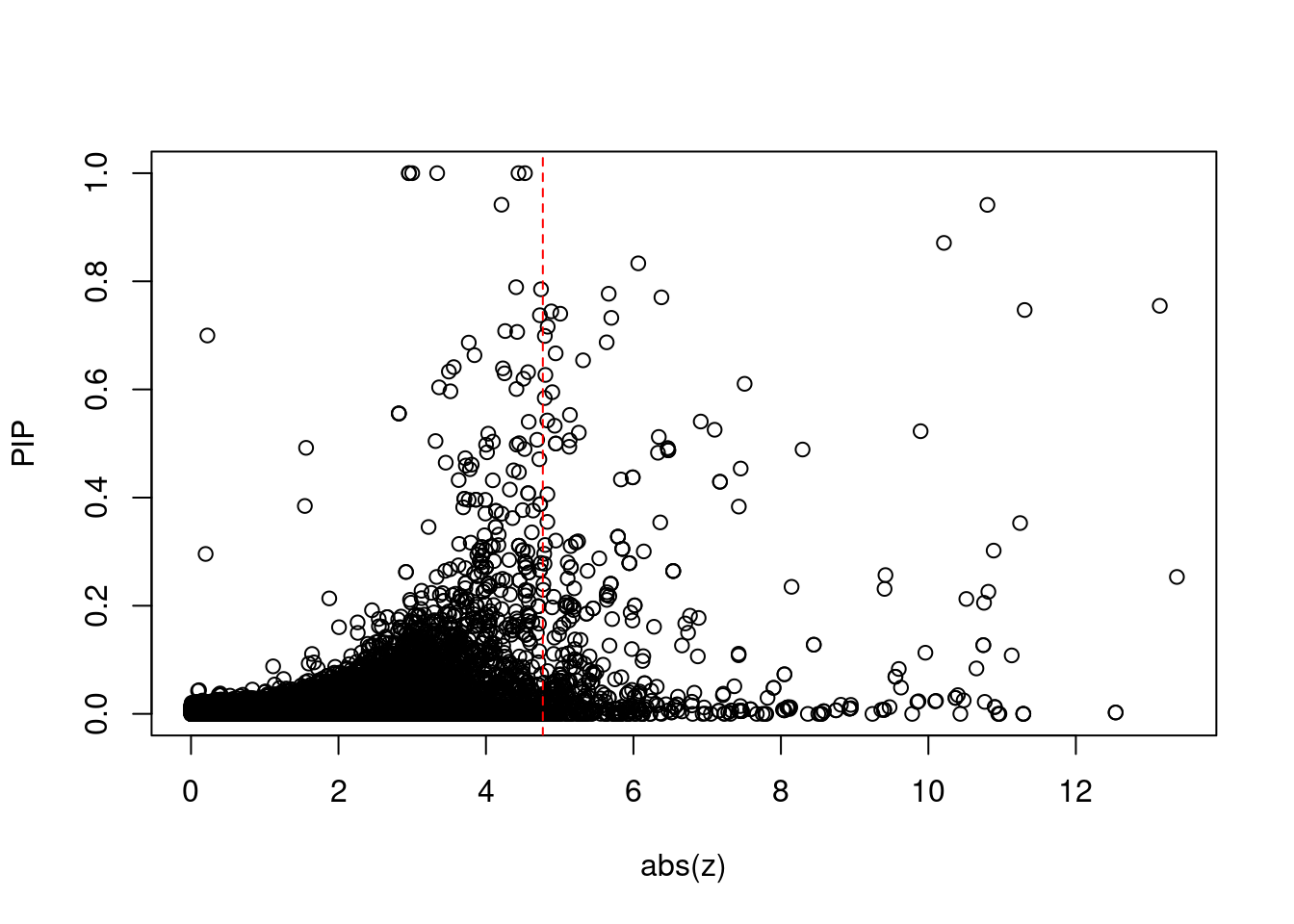
19454 intron_16_28592443_28593252 -10.75 2Comparing z scores and PIPs
| Version | Author | Date |
|---|---|---|
| 9ef0786 | sq-96 | 2022-02-22 |
| Version | Author | Date |
|---|---|---|
| 9ef0786 | sq-96 | 2022-02-22 |
[1] 0.01915 genename region_tag susie_pip mu2 PVE
14009 BDNF-AS 11_19 2.532e-01 118.86 8.955e-05
14010 BDNF-AS 11_19 7.547e-01 119.88 2.692e-04
4898 RBM6 3_35 2.556e-03 897.70 6.826e-06
4899 RBM6 3_35 2.556e-03 897.70 6.826e-06
7086 POC5 5_44 7.469e-01 93.32 2.074e-04
2630 DNAJC27-AS1 2_15 6.927e-05 190.37 3.924e-08
2631 DNAJC27-AS1 2_15 6.927e-05 190.37 3.924e-08
2632 DNAJC27-AS1 2_15 6.927e-05 190.37 3.924e-08
8197 C6orf106 6_28 3.529e-01 120.59 1.266e-04
8200 SNRPC 6_28 1.081e-01 117.97 3.795e-05
4887 RNF123 3_35 2.511e-11 813.45 6.077e-14
4885 RNF123 3_35 2.337e-11 813.03 5.653e-14
4888 RNF123 3_35 2.080e-11 812.45 5.028e-14
8198 C6orf106 6_28 1.271e-02 116.28 4.397e-06
8199 C6orf106 6_28 1.271e-02 116.28 4.397e-06
2005 LMOD1 1_103 3.020e-01 113.57 1.020e-04
19470 SH2B1 16_23 2.258e-01 67.15 4.512e-05
27140 GIPR 19_32 9.414e-01 5582.93 1.564e-02
19500 DOC2A 16_24 2.221e-02 95.07 6.282e-06
19454 SULT1A2 16_23 2.054e-01 64.91 3.967e-05
intron_id z num_eqtl
14009 intron_11_27640005_27659171 13.37 2
14010 intron_11_27658462_27659171 -13.14 1
4898 intron_3_49999513_50046716 12.54 1
4899 intron_3_49999513_50048245 -12.54 1
7086 intron_5_75705787_75712854 -11.31 1
2630 intron_2_24972232_24972626 -11.29 1
2631 intron_2_24972232_24979060 11.29 1
2632 intron_2_24972232_25001415 11.29 1
8197 intron_6_34654779_34688841 11.24 1
8200 intron_6_34757954_34762595 -11.13 1
4887 intron_3_49712656_49713513 10.96 1
4885 intron_3_49698822_49698980 -10.96 1
4888 intron_3_49714174_49715575 -10.95 1
8198 intron_6_34654779_34696446 -10.90 1
8199 intron_6_34688933_34696446 10.90 1
2005 intron_1_201900751_201946080 -10.89 1
19470 intron_16_28869383_28870709 -10.81 2
27140 intron_19_45677779_45677906 10.80 1
19500 intron_16_30007090_30007173 -10.76 1
19454 intron_16_28592443_28593252 -10.75 2GO enrichment analysis for genes with PIP>0.5
#number of genes for gene set enrichment
length(genes)[1] 61Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"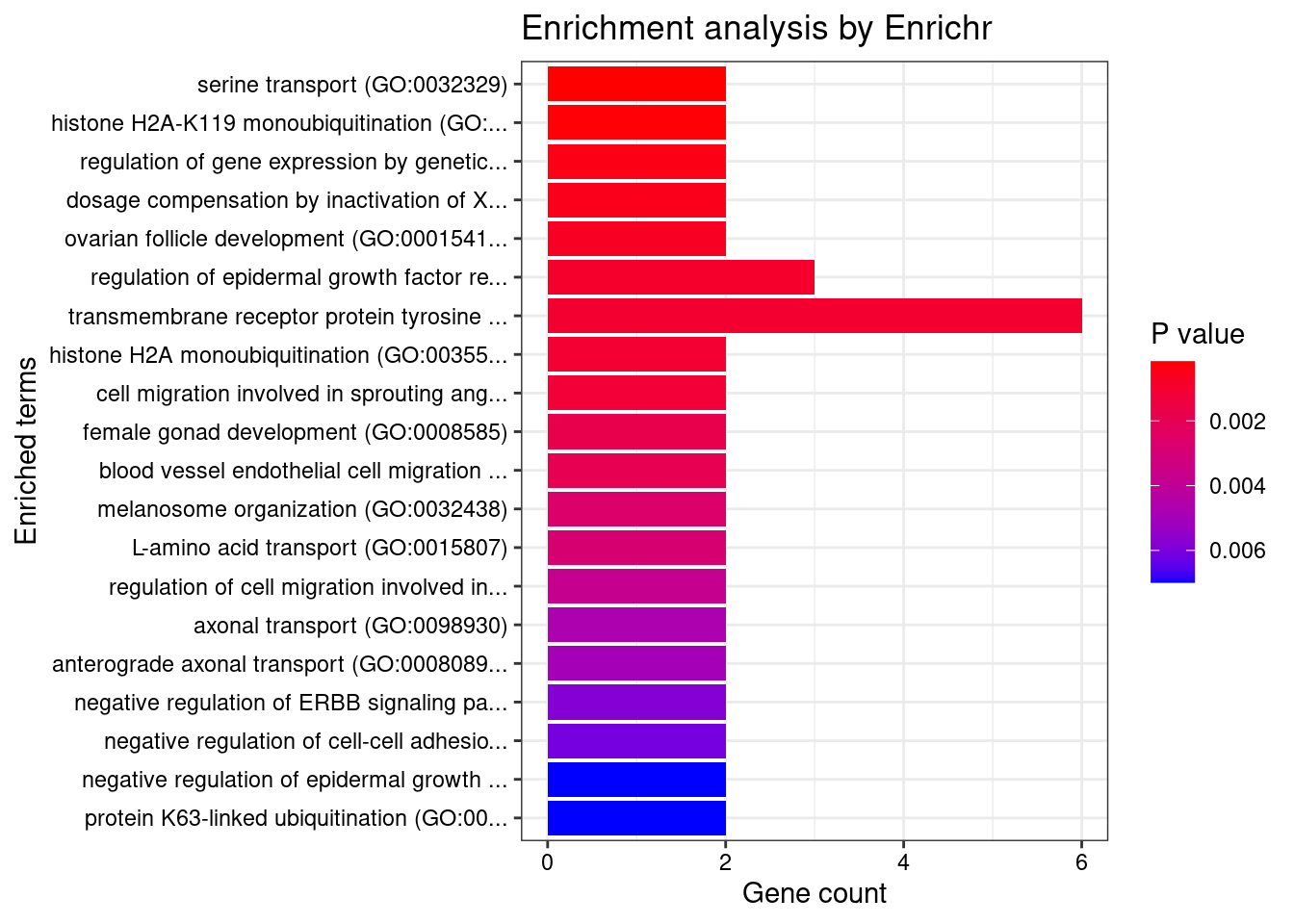
| Version | Author | Date |
|---|---|---|
| 9ef0786 | sq-96 | 2022-02-22 |
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Cellular_Component_2021"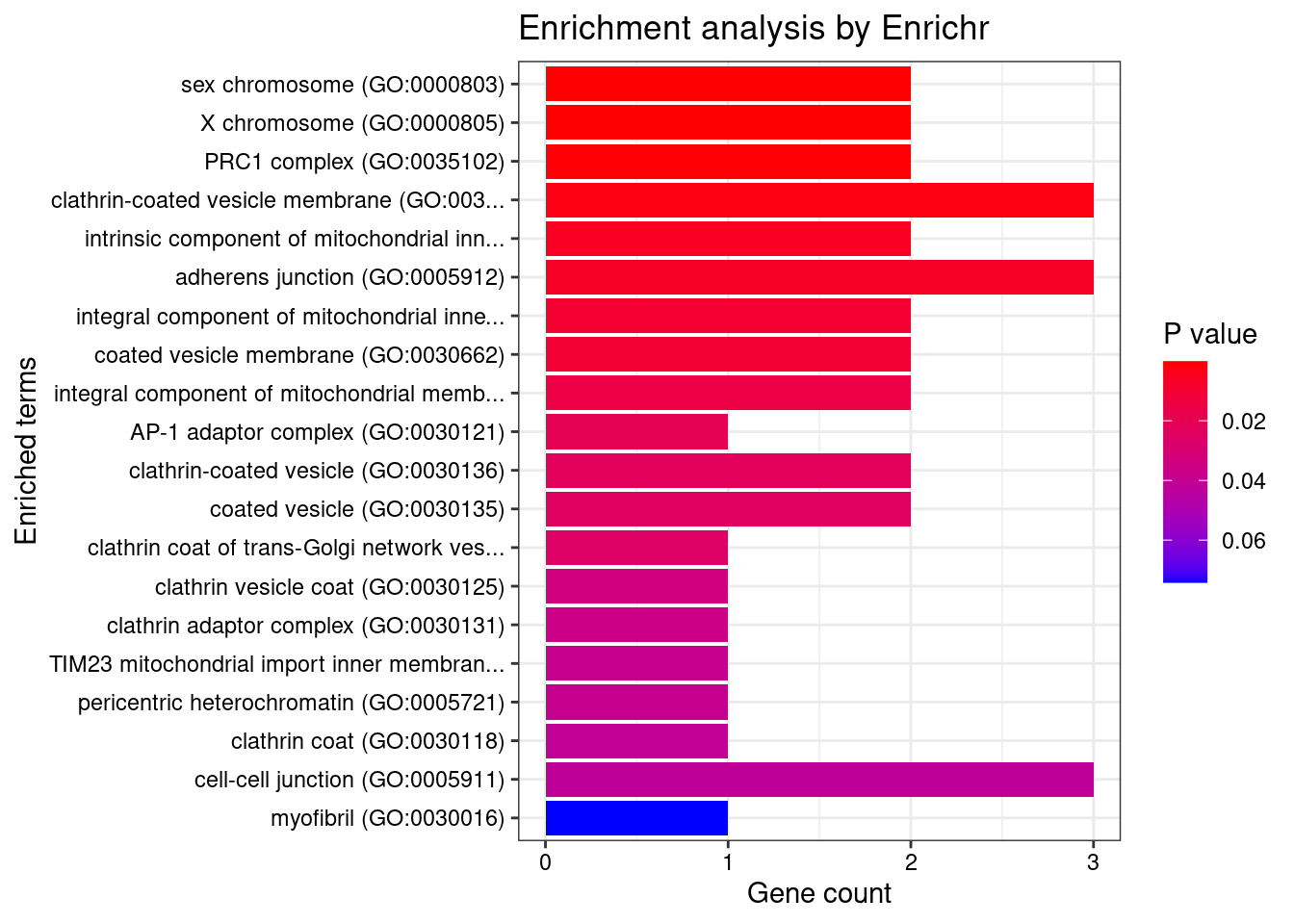
| Version | Author | Date |
|---|---|---|
| 9ef0786 | sq-96 | 2022-02-22 |
Term Overlap Adjusted.P.value
1 sex chromosome (GO:0000803) 2/6 0.004816
2 X chromosome (GO:0000805) 2/7 0.004816
3 PRC1 complex (GO:0035102) 2/15 0.015818
4 clathrin-coated vesicle membrane (GO:0030665) 3/90 0.031365
Genes
1 PCGF5;PCGF3
2 PCGF5;PCGF3
3 PCGF5;PCGF3
4 NECAP2;AP1G1;DENND1A
[1] "GO_Molecular_Function_2021"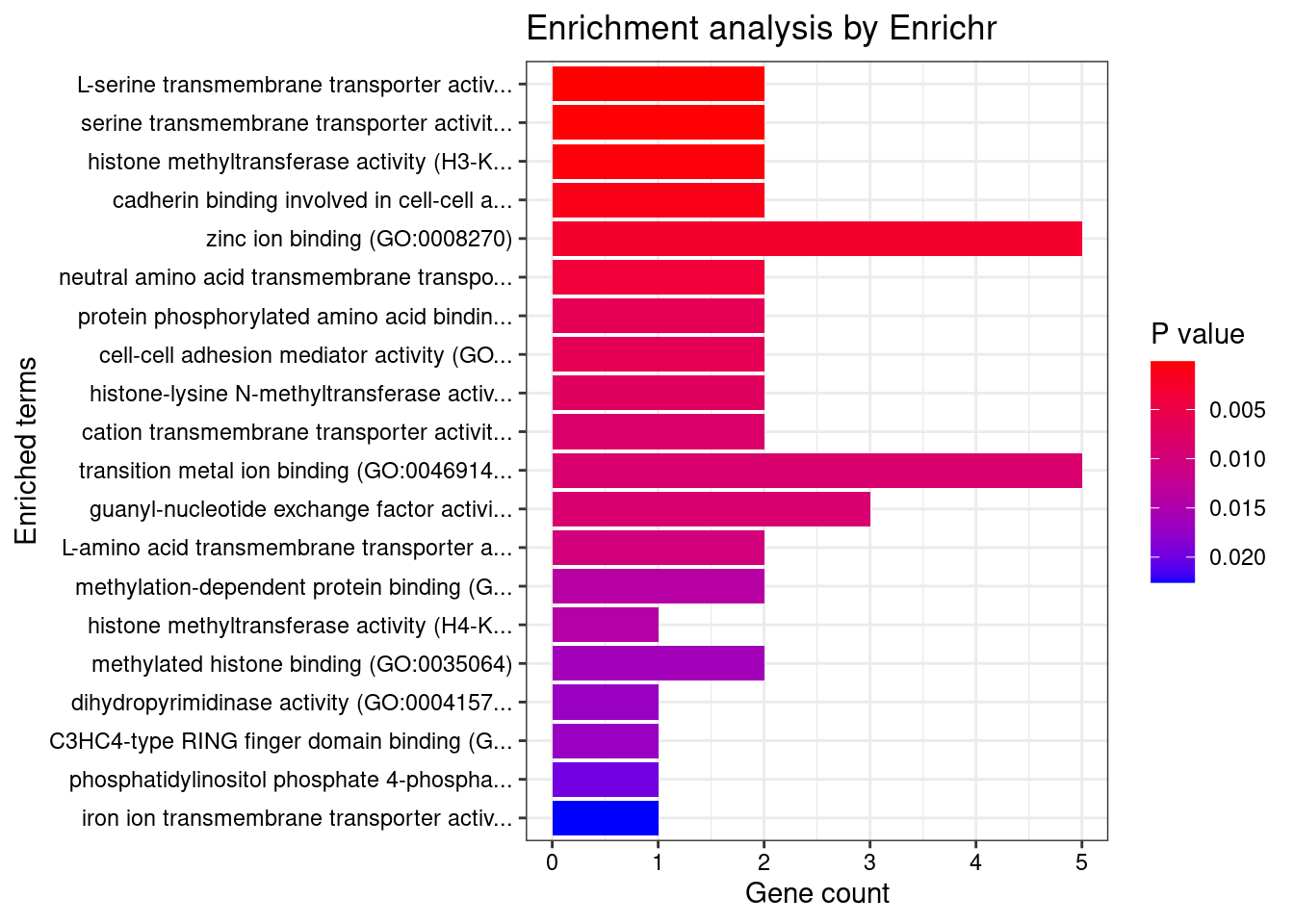
| Version | Author | Date |
|---|---|---|
| 9ef0786 | sq-96 | 2022-02-22 |
Term Overlap
1 L-serine transmembrane transporter activity (GO:0015194) 2/6
2 serine transmembrane transporter activity (GO:0022889) 2/8
3 histone methyltransferase activity (H3-K36 specific) (GO:0046975) 2/11
4 cadherin binding involved in cell-cell adhesion (GO:0098641) 2/18
Adjusted.P.value Genes
1 0.01425 SFXN1;SLC38A2
2 0.01425 SFXN1;SLC38A2
3 0.01856 SETD5;NSD1
4 0.03824 TMOD3;BAIAP2DisGeNET enrichment analysis for genes with PIP>0.5
Warning in disease_enrichment(entities = genes, vocabulary = "HGNC", database =
"CURATED"): Removing duplicates from input list. Description
16 Body Weight
136 Diffuse mesangial sclerosis (disorder)
152 Progressive cerebellar ataxia
163 Choroidal Neovascularization
220 Pierson syndrome
221 HERMANSKY-PUDLAK SYNDROME 2
223 Familial encephalopathy with neuroserpin inclusion bodies
234 MICROVASCULAR COMPLICATIONS OF DIABETES, SUSCEPTIBILITY TO, 1(finding)
240 NEPHROTIC SYNDROME, TYPE 5, WITH OR WITHOUT OCULAR ABNORMALITIES
242 Familial mesangial sclerosis
FDR Ratio BgRatio
16 0.03393 2/18 15/9703
136 0.03393 1/18 1/9703
152 0.03393 1/18 1/9703
163 0.03393 1/18 1/9703
220 0.03393 1/18 1/9703
221 0.03393 1/18 1/9703
223 0.03393 1/18 1/9703
234 0.03393 1/18 1/9703
240 0.03393 1/18 1/9703
242 0.03393 1/18 1/9703WebGestalt enrichment analysis for genes with PIP>0.5
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum =
minNum, : No significant gene set is identified based on FDR 0.05!NULLPIP Manhattan Plot
Warning: ggrepel: 18 unlabeled data points (too many overlaps). Consider
increasing max.overlaps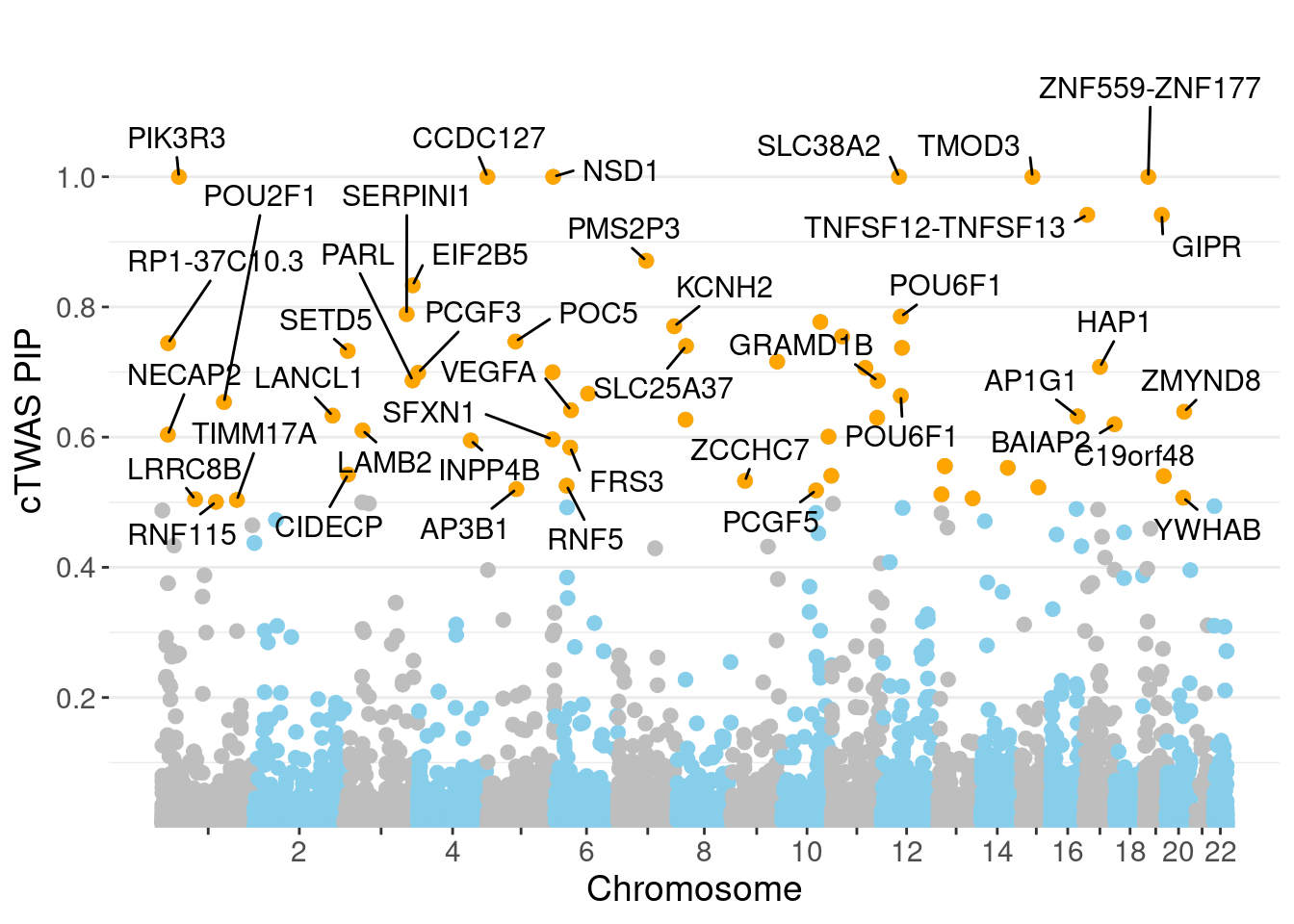
| Version | Author | Date |
|---|---|---|
| 9ef0786 | sq-96 | 2022-02-22 |
Sensitivity, specificity and precision for silver standard genes
#number of genes in known annotations
print(length(known_annotations))[1] 41#number of genes in known annotations with imputed expression
print(sum(known_annotations %in% ctwas_gene_res$genename))[1] 22#significance threshold for TWAS
print(sig_thresh)[1] 4.771#number of ctwas genes
length(ctwas_genes)[1] 10#number of TWAS genes
length(twas_genes)[1] 523#show novel genes (ctwas genes with not in TWAS genes)
ctwas_gene_res[ctwas_gene_res$genename %in% novel_genes,report_cols] genename region_tag susie_pip mu2 PVE
25970 PIK3R3 1_28 1.000e+00 34091.066 1.014e-01
26275 CCDC127 5_1 1.000e+00 14934.792 4.443e-02
26276 CCDC127 5_1 1.302e-12 14733.940 5.705e-14
26365 NSD1 5_106 0.000e+00 3481.694 0.000e+00
26366 NSD1 5_106 0.000e+00 5085.407 0.000e+00
26367 NSD1 5_106 1.000e+00 13207.484 3.930e-02
26744 SLC38A2 12_29 1.000e+00 14459.237 4.302e-02
26745 SLC38A2 12_29 0.000e+00 10735.175 0.000e+00
26836 TMOD3 15_21 1.000e+00 28879.147 8.592e-02
26837 TMOD3 15_21 0.000e+00 23241.921 0.000e+00
26862 TNFSF12-TNFSF13 17_7 5.896e-03 7.031 1.233e-07
26863 TNFSF12-TNFSF13 17_7 7.015e-03 9.520 1.987e-07
26864 TNFSF12-TNFSF13 17_7 7.015e-03 9.520 1.987e-07
26865 TNFSF12-TNFSF13 17_7 9.416e-01 29.027 8.132e-05
26866 TNFSF12-TNFSF13 17_7 7.015e-03 9.520 1.987e-07
26867 TNFSF12-TNFSF13 17_7 1.087e-02 10.232 3.308e-07
26868 TNFSF12-TNFSF13 17_7 1.087e-02 10.232 3.308e-07
26869 TNFSF12-TNFSF13 17_7 1.087e-02 10.232 3.308e-07
27018 ZNF559-ZNF177 19_9 1.000e+00 15583.862 4.637e-02
27019 ZNF559-ZNF177 19_9 0.000e+00 939.194 0.000e+00
intron_id z num_eqtl
25970 intron_1_46055971_46061929 4.44123 1
26275 intron_5_205958_216729 2.99985 1
26276 intron_5_216859_218093 2.90019 1
26365 intron_5_177136030_177136873 -0.32376 2
26366 intron_5_177136030_177191884 0.07789 2
26367 intron_5_177235945_177239756 2.95697 1
26744 intron_12_46366945_46367076 2.95342 1
26745 intron_12_46367340_46370512 -1.61310 1
26836 intron_15_51924578_51931031 -4.52839 1
26837 intron_15_51938260_51947292 -2.83650 2
26862 intron_17_7559297_7559846 1.61382 2
26863 intron_17_7559297_7559851 1.33178 1
26864 intron_17_7559702_7559846 -1.33178 1
26865 intron_17_7559702_7560049 -4.21078 2
26866 intron_17_7559893_7560049 1.33178 1
26867 intron_17_7560488_7560724 1.23420 1
26868 intron_17_7560817_7560999 -1.23420 1
26869 intron_17_7560872_7560999 -1.23420 1
27018 intron_19_9364948_9376316 -3.33796 1
27019 intron_19_9371752_9376316 -2.29657 1#sensitivity / recall
print(sensitivity) ctwas TWAS
0.02439 0.19512 #specificity
print(specificity) ctwas TWAS
0.9978 0.9582 #precision / PPV
print(precision) ctwas TWAS
0.1000 0.0153 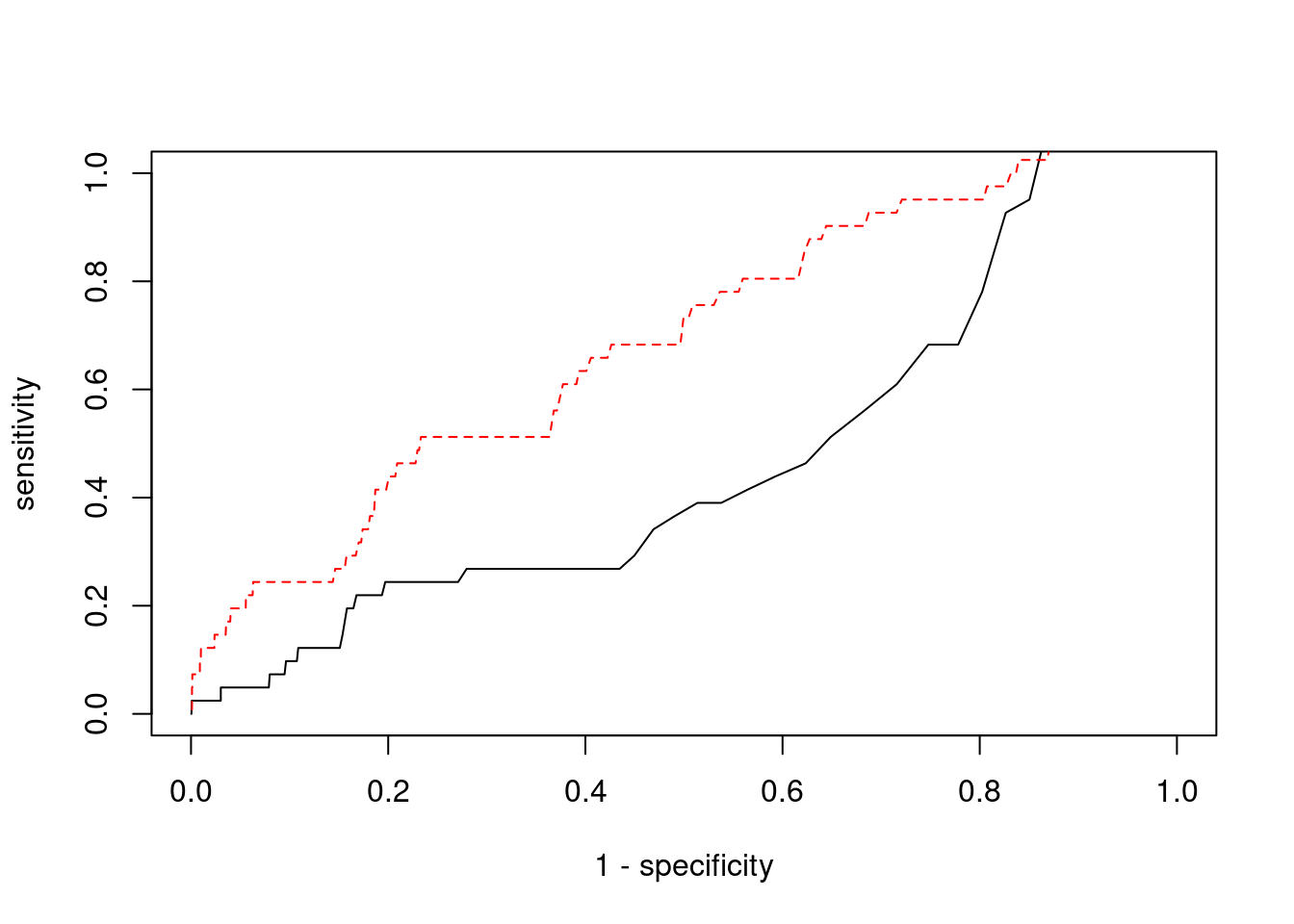
| Version | Author | Date |
|---|---|---|
| 9ef0786 | sq-96 | 2022-02-22 |
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats4 parallel stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] readxl_1.3.1 forcats_0.5.1
[3] stringr_1.4.0 dplyr_1.0.7
[5] purrr_0.3.4 readr_2.1.1
[7] tidyr_1.1.4 tidyverse_1.3.1
[9] tibble_3.1.6 WebGestaltR_0.4.4
[11] disgenet2r_0.99.2 enrichR_3.0
[13] cowplot_1.0.0 ggplot2_3.3.5
[15] EnsDb.Hsapiens.v79_2.99.0 ensembldb_2.8.0
[17] AnnotationFilter_1.8.0 GenomicFeatures_1.36.3
[19] AnnotationDbi_1.46.0 Biobase_2.44.0
[21] GenomicRanges_1.36.1 GenomeInfoDb_1.20.0
[23] IRanges_2.18.1 S4Vectors_0.22.1
[25] BiocGenerics_0.30.0 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] ggbeeswarm_0.6.0 colorspace_2.0-2
[3] rjson_0.2.20 ellipsis_0.3.2
[5] rprojroot_2.0.2 XVector_0.24.0
[7] fs_1.5.2 rstudioapi_0.13
[9] farver_2.1.0 ggrepel_0.9.1
[11] bit64_4.0.5 lubridate_1.8.0
[13] fansi_0.5.0 xml2_1.3.3
[15] codetools_0.2-16 doParallel_1.0.16
[17] cachem_1.0.6 knitr_1.36
[19] jsonlite_1.7.2 apcluster_1.4.8
[21] Cairo_1.5-12.2 Rsamtools_2.0.0
[23] broom_0.7.10 dbplyr_2.1.1
[25] compiler_3.6.1 httr_1.4.2
[27] backports_1.4.1 assertthat_0.2.1
[29] Matrix_1.2-18 fastmap_1.1.0
[31] lazyeval_0.2.2 cli_3.1.0
[33] later_0.8.0 htmltools_0.5.2
[35] prettyunits_1.1.1 tools_3.6.1
[37] igraph_1.2.10 gtable_0.3.0
[39] glue_1.5.1 GenomeInfoDbData_1.2.1
[41] reshape2_1.4.4 doRNG_1.8.2
[43] Rcpp_1.0.7 cellranger_1.1.0
[45] jquerylib_0.1.4 vctrs_0.3.8
[47] Biostrings_2.52.0 svglite_1.2.2
[49] rtracklayer_1.44.4 iterators_1.0.13
[51] xfun_0.29 rvest_1.0.2
[53] lifecycle_1.0.1 rngtools_1.5.2
[55] XML_3.99-0.3 zlibbioc_1.30.0
[57] scales_1.1.1 vroom_1.5.7
[59] hms_1.1.1 promises_1.0.1
[61] ProtGenerics_1.16.0 SummarizedExperiment_1.14.1
[63] yaml_2.2.1 curl_4.3.2
[65] memoise_2.0.1 ggrastr_1.0.1
[67] gdtools_0.1.9 biomaRt_2.40.1
[69] stringi_1.7.6 RSQLite_2.2.8
[71] highr_0.9 foreach_1.5.1
[73] BiocParallel_1.18.0 rlang_0.4.12
[75] pkgconfig_2.0.3 matrixStats_0.57.0
[77] bitops_1.0-7 evaluate_0.14
[79] lattice_0.20-38 GenomicAlignments_1.20.1
[81] labeling_0.4.2 bit_4.0.4
[83] tidyselect_1.1.1 plyr_1.8.6
[85] magrittr_2.0.1 R6_2.5.1
[87] generics_0.1.1 DelayedArray_0.10.0
[89] DBI_1.1.1 haven_2.4.3
[91] pillar_1.6.4 whisker_0.3-2
[93] withr_2.4.3 RCurl_1.98-1.5
[95] modelr_0.1.8 crayon_1.4.2
[97] utf8_1.2.2 tzdb_0.2.0
[99] rmarkdown_2.11 progress_1.2.2
[101] grid_3.6.1 data.table_1.14.2
[103] blob_1.2.2 git2r_0.26.1
[105] reprex_2.0.1 digest_0.6.29
[107] httpuv_1.5.1 munsell_0.5.0
[109] beeswarm_0.2.3 vipor_0.4.5