SCZ - Brain Cortex
sheng Qian
2021-2-6
Last updated: 2022-03-14
Checks: 5 2
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/ | data |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/ctwas_config.R | code/ctwas_config.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 4c71b11. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .ipynb_checkpoints/
Ignored: data/AF/
Untracked files:
Untracked: Rplot.png
Untracked: analysis/.ipynb_checkpoints/
Untracked: analysis/SCZ_2014_EUR_Brain_Amygdala.Rmd
Untracked: analysis/SCZ_2014_EUR_Brain_Anterior_cingulate_cortex_BA24.Rmd
Untracked: analysis/SCZ_2014_EUR_Brain_Caudate_basal_ganglia.Rmd
Untracked: analysis/SCZ_2014_EUR_Brain_Cerebellar_Hemisphere.Rmd
Untracked: analysis/SCZ_2014_EUR_Brain_Cerebellum.Rmd
Untracked: analysis/SCZ_2014_EUR_Brain_Cortex.Rmd
Untracked: analysis/SCZ_2014_EUR_Brain_Frontal_Cortex_BA9.Rmd
Untracked: analysis/SCZ_2014_EUR_Brain_Hippocampus.Rmd
Untracked: analysis/SCZ_2014_EUR_Brain_Hypothalamus.Rmd
Untracked: analysis/SCZ_2014_EUR_Brain_Nucleus_accumbens_basal_ganglia.Rmd
Untracked: analysis/SCZ_2014_EUR_Brain_Putamen_basal_ganglia.Rmd
Untracked: analysis/SCZ_2014_EUR_Brain_Spinal_cord_cervical_c-1.Rmd
Untracked: analysis/SCZ_2014_EUR_Brain_Substantia_nigra.Rmd
Untracked: analysis/SCZ_2020_Brain_Cortex.Rmd
Untracked: analysis/SCZ_2020_Brain_Frontal_Cortex_BA9.Rmd
Untracked: analysis/SCZ_2020_Brain_Hypothalamus.Rmd
Untracked: analysis/SCZ_2020_Brain_Putamen_basal_ganglia.Rmd
Untracked: analysis/SCZ_Cross_Tissue_Analysis.Rmd
Untracked: code/.ipynb_checkpoints/
Untracked: code/AF_out/
Untracked: code/Autism_out/
Untracked: code/BMI_S_out/
Untracked: code/BMI_out/
Untracked: code/Glucose_out/
Untracked: code/LDL_S_out/
Untracked: code/SCZ_2014_EUR_out/
Untracked: code/SCZ_2020_out/
Untracked: code/SCZ_S_out/
Untracked: code/SCZ_out/
Untracked: code/T2D_out/
Untracked: code/ctwas_config.R
Untracked: code/mapping.R
Untracked: code/out/
Untracked: code/run_AF_analysis.sbatch
Untracked: code/run_AF_analysis.sh
Untracked: code/run_AF_ctwas_rss_LDR.R
Untracked: code/run_Autism_analysis.sbatch
Untracked: code/run_Autism_analysis.sh
Untracked: code/run_Autism_ctwas_rss_LDR.R
Untracked: code/run_BMI_analysis.sbatch
Untracked: code/run_BMI_analysis.sh
Untracked: code/run_BMI_analysis_S.sbatch
Untracked: code/run_BMI_analysis_S.sh
Untracked: code/run_BMI_ctwas_rss_LDR.R
Untracked: code/run_BMI_ctwas_rss_LDR_S.R
Untracked: code/run_Glucose_analysis.sbatch
Untracked: code/run_Glucose_analysis.sh
Untracked: code/run_Glucose_ctwas_rss_LDR.R
Untracked: code/run_LDL_analysis_S.sbatch
Untracked: code/run_LDL_analysis_S.sh
Untracked: code/run_LDL_ctwas_rss_LDR_S.R
Untracked: code/run_SCZ_2014_EUR_analysis.sbatch
Untracked: code/run_SCZ_2014_EUR_analysis.sh
Untracked: code/run_SCZ_2014_EUR_ctwas_rss_LDR.R
Untracked: code/run_SCZ_2020_analysis.sbatch
Untracked: code/run_SCZ_2020_analysis.sh
Untracked: code/run_SCZ_2020_ctwas_rss_LDR.R
Untracked: code/run_SCZ_analysis.sbatch
Untracked: code/run_SCZ_analysis.sh
Untracked: code/run_SCZ_analysis_S.sbatch
Untracked: code/run_SCZ_analysis_S.sh
Untracked: code/run_SCZ_ctwas_rss_LDR.R
Untracked: code/run_SCZ_ctwas_rss_LDR_S.R
Untracked: code/run_T2D_analysis.sbatch
Untracked: code/run_T2D_analysis.sh
Untracked: code/run_T2D_ctwas_rss_LDR.R
Untracked: code/wflow_build.R
Untracked: code/wflow_build.sbatch
Untracked: data/.ipynb_checkpoints/
Untracked: data/BMI/
Untracked: data/PGC3_SCZ_wave3_public.v2.tsv
Untracked: data/SCZ/
Untracked: data/SCZ_2014_EUR/
Untracked: data/SCZ_2020/
Untracked: data/SCZ_S/
Untracked: data/T2D/
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/mashr_sqtl/
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: data/untitled.txt
Unstaged changes:
Modified: analysis/SCZ_Brain_Amygdala.Rmd
Modified: analysis/SCZ_Brain_Anterior_cingulate_cortex_BA24.Rmd
Modified: analysis/SCZ_Brain_Caudate_basal_ganglia.Rmd
Modified: analysis/SCZ_Brain_Cerebellar_Hemisphere.Rmd
Modified: analysis/SCZ_Brain_Cerebellum.Rmd
Modified: analysis/SCZ_Brain_Cortex.Rmd
Modified: analysis/SCZ_Brain_Frontal_Cortex_BA9.Rmd
Modified: analysis/SCZ_Brain_Hippocampus.Rmd
Modified: analysis/SCZ_Brain_Hypothalamus.Rmd
Modified: analysis/SCZ_Brain_Nucleus_accumbens_basal_ganglia.Rmd
Modified: analysis/SCZ_Brain_Putamen_basal_ganglia.Rmd
Modified: analysis/SCZ_Brain_Spinal_cord_cervical_c-1.Rmd
Modified: analysis/SCZ_Brain_Substantia_nigra.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with wflow_publish() to start tracking its development.
Weight QC
#number of imputed weights
nrow(qclist_all)[1] 11379#number of imputed weights by chromosome
table(qclist_all$chr)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
1142 801 644 426 537 655 549 427 432 458 680 647 221 367 377 522
17 18 19 20 21 22
695 179 851 353 123 293 #number of imputed weights without missing variants
sum(qclist_all$nmiss==0)[1] 8519#proportion of imputed weights without missing variants
mean(qclist_all$nmiss==0)[1] 0.7487Check convergence of parameters
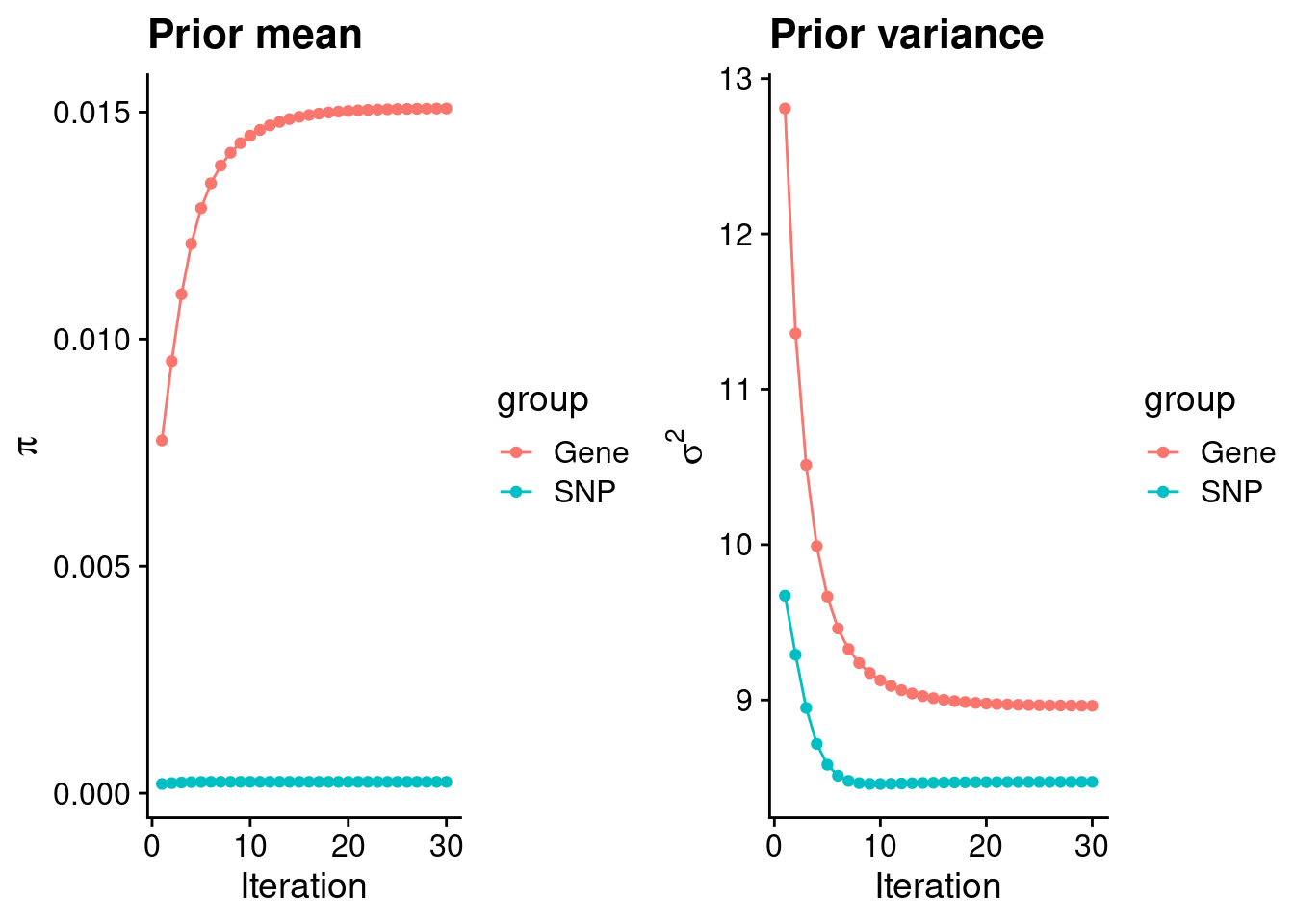
#estimated group prior
estimated_group_prior <- group_prior_rec[,ncol(group_prior_rec)]
names(estimated_group_prior) <- c("gene", "snp")
estimated_group_prior["snp"] <- estimated_group_prior["snp"]*thin #adjust parameter to account for thin argument
print(estimated_group_prior) gene snp
0.0150804 0.0002497 #estimated group prior variance
estimated_group_prior_var <- group_prior_var_rec[,ncol(group_prior_var_rec)]
names(estimated_group_prior_var) <- c("gene", "snp")
print(estimated_group_prior_var) gene snp
8.963 8.473 #report sample size
print(sample_size)[1] 77096#report group size
group_size <- c(nrow(ctwas_gene_res), n_snps)
print(group_size)[1] 11379 7352670#estimated group PVE
estimated_group_pve <- estimated_group_prior_var*estimated_group_prior*group_size/sample_size #check PVE calculation
names(estimated_group_pve) <- c("gene", "snp")
print(estimated_group_pve) gene snp
0.01995 0.20174 #compare sum(PIP*mu2/sample_size) with above PVE calculation
c(sum(ctwas_gene_res$PVE),sum(ctwas_snp_res$PVE))[1] 0.1379 1.7253Genes with highest PIPs
genename region_tag susie_pip mu2 PVE z num_eqtl
958 NT5C2 10_66 0.9990 2876.64 0.0372745 -8.190 1
1518 DDTL 22_6 0.9987 32.10 0.0004159 -3.206 2
11314 ZNF823 19_10 0.9889 29.71 0.0003811 5.560 2
13518 LINC01415 18_30 0.9807 42.34 0.0005386 -7.512 2
13679 RP11-230C9.4 6_102 0.9414 21.83 0.0002666 -4.558 2
3165 SF3B1 2_117 0.9029 43.80 0.0005130 6.784 1
3085 SPCS1 3_36 0.9013 33.68 0.0003938 -6.382 1
11176 PCBP2 12_33 0.8743 21.44 0.0002432 4.496 1
12719 HLA-DMB 6_27 0.8111 393.39 0.0041389 -8.273 1
421 TRIT1 1_25 0.8089 20.37 0.0002137 -4.060 3
9752 ZNF354C 5_108 0.8015 20.29 0.0002110 -4.154 1
5055 RCBTB1 13_21 0.7780 20.45 0.0002064 -4.141 2
6035 METTL21A 2_122 0.7368 22.25 0.0002126 -4.391 1
451 FAM120A 9_47 0.7113 22.53 0.0002079 -4.571 1
4616 DLG4 17_6 0.6780 22.05 0.0001939 3.863 2
3159 KCNJ13 2_137 0.6742 36.58 0.0003199 6.658 1
6435 ARFGAP2 11_29 0.6699 25.00 0.0002172 4.839 1
9648 LY6H 8_94 0.6544 20.80 0.0001766 4.118 1
6579 FAM177A1 14_9 0.6489 20.82 0.0001753 -4.548 1
947 FMO4 1_84 0.6487 22.82 0.0001920 3.839 1Genes with largest effect sizes
genename region_tag susie_pip mu2 PVE z num_eqtl
958 NT5C2 10_66 9.990e-01 2876.6 3.727e-02 -8.1897 1
6413 CNNM2 10_66 6.582e-05 2786.0 2.378e-06 -7.8764 1
6404 INA 10_66 5.789e-06 2049.1 1.539e-07 -7.1401 1
12375 HLA-DPA1 6_27 2.730e-08 1004.3 3.557e-10 5.3356 2
9334 USMG5 10_66 7.687e-11 409.2 4.079e-13 2.4174 1
12719 HLA-DMB 6_27 8.111e-01 393.4 4.139e-03 -8.2728 1
11611 HLA-DMA 6_27 3.220e-15 327.3 1.367e-17 0.4948 1
8214 WBP1L 10_66 5.766e-11 289.3 2.164e-13 2.1272 2
6414 PDCD11 10_66 3.156e-10 261.5 1.070e-12 3.0363 1
9405 MSL2 3_84 1.040e-06 196.8 2.655e-09 5.8137 2
11907 CLIC1 6_26 3.079e-04 176.8 7.062e-07 9.5362 2
5342 CALHM2 10_66 5.007e-11 162.6 1.056e-13 -1.7655 1
11645 LY6G6C 6_26 1.657e-05 151.0 3.247e-08 8.8896 1
11634 ZBTB12 6_26 3.831e-06 146.1 7.259e-09 8.7124 1
11640 HSPA1L 6_26 8.730e-12 140.4 1.590e-14 -7.6575 1
12122 C4B 6_26 3.225e-07 138.7 5.804e-10 -8.4450 1
11638 C6orf48 6_26 3.936e-13 137.8 7.036e-16 7.2997 1
12783 C4A 6_26 2.555e-07 137.2 4.546e-10 8.4728 3
12471 TRIM26 6_24 4.330e-15 135.7 7.619e-18 -5.4551 1
13651 HCG17 6_24 4.552e-15 134.5 7.943e-18 5.5087 1Genes with highest PVE
genename region_tag susie_pip mu2 PVE z num_eqtl
958 NT5C2 10_66 0.9990 2876.64 0.0372745 -8.190 1
12719 HLA-DMB 6_27 0.8111 393.39 0.0041389 -8.273 1
13518 LINC01415 18_30 0.9807 42.34 0.0005386 -7.512 2
3165 SF3B1 2_117 0.9029 43.80 0.0005130 6.784 1
1518 DDTL 22_6 0.9987 32.10 0.0004159 -3.206 2
3085 SPCS1 3_36 0.9013 33.68 0.0003938 -6.382 1
11314 ZNF823 19_10 0.9889 29.71 0.0003811 5.560 2
3159 KCNJ13 2_137 0.6742 36.58 0.0003199 6.658 1
2682 MDK 11_28 0.5855 38.23 0.0002904 -6.344 1
7729 THOC7 3_43 0.6093 34.55 0.0002730 -5.844 4
13679 RP11-230C9.4 6_102 0.9414 21.83 0.0002666 -4.558 2
11176 PCBP2 12_33 0.8743 21.44 0.0002432 4.496 1
6507 TMEM219 16_24 0.5089 34.44 0.0002273 6.164 1
376 CUL3 2_132 0.6023 28.00 0.0002187 -5.422 1
6435 ARFGAP2 11_29 0.6699 25.00 0.0002172 4.839 1
421 TRIT1 1_25 0.8089 20.37 0.0002137 -4.060 3
6035 METTL21A 2_122 0.7368 22.25 0.0002126 -4.391 1
9752 ZNF354C 5_108 0.8015 20.29 0.0002110 -4.154 1
451 FAM120A 9_47 0.7113 22.53 0.0002079 -4.571 1
5055 RCBTB1 13_21 0.7780 20.45 0.0002064 -4.141 2Genes with largest z scores
genename region_tag susie_pip mu2 PVE z num_eqtl
11907 CLIC1 6_26 3.079e-04 176.84 7.062e-07 9.536 2
11645 LY6G6C 6_26 1.657e-05 151.03 3.247e-08 8.890 1
11129 ZSCAN16 6_22 1.319e-02 79.39 1.358e-05 8.813 1
11634 ZBTB12 6_26 3.831e-06 146.08 7.259e-09 8.712 1
2927 PRSS16 6_21 4.903e-02 42.22 2.685e-05 -8.631 2
12783 C4A 6_26 2.555e-07 137.19 4.546e-10 8.473 3
12122 C4B 6_26 3.225e-07 138.73 5.804e-10 -8.445 1
12719 HLA-DMB 6_27 8.111e-01 393.39 4.139e-03 -8.273 1
958 NT5C2 10_66 9.990e-01 2876.64 3.727e-02 -8.190 1
11649 GPANK1 6_26 3.819e-14 103.62 5.133e-17 7.973 1
12395 CYP21A2 6_26 7.677e-10 129.01 1.285e-12 -7.953 2
6413 CNNM2 10_66 6.582e-05 2786.03 2.378e-06 -7.876 1
11908 DDAH2 6_26 1.042e-13 127.56 1.725e-16 7.661 1
11640 HSPA1L 6_26 8.730e-12 140.43 1.590e-14 -7.658 1
11620 AGER 6_26 2.220e-16 70.05 2.017e-19 -7.547 1
13518 LINC01415 18_30 9.807e-01 42.34 5.386e-04 -7.512 2
11638 C6orf48 6_26 3.936e-13 137.83 7.036e-16 7.300 1
6404 INA 10_66 5.789e-06 2049.08 1.539e-07 -7.140 1
11621 RNF5 6_26 2.220e-16 45.12 1.300e-19 7.104 2
12511 ZSCAN31 6_22 2.921e-02 41.08 1.556e-05 -7.066 3Comparing z scores and PIPs
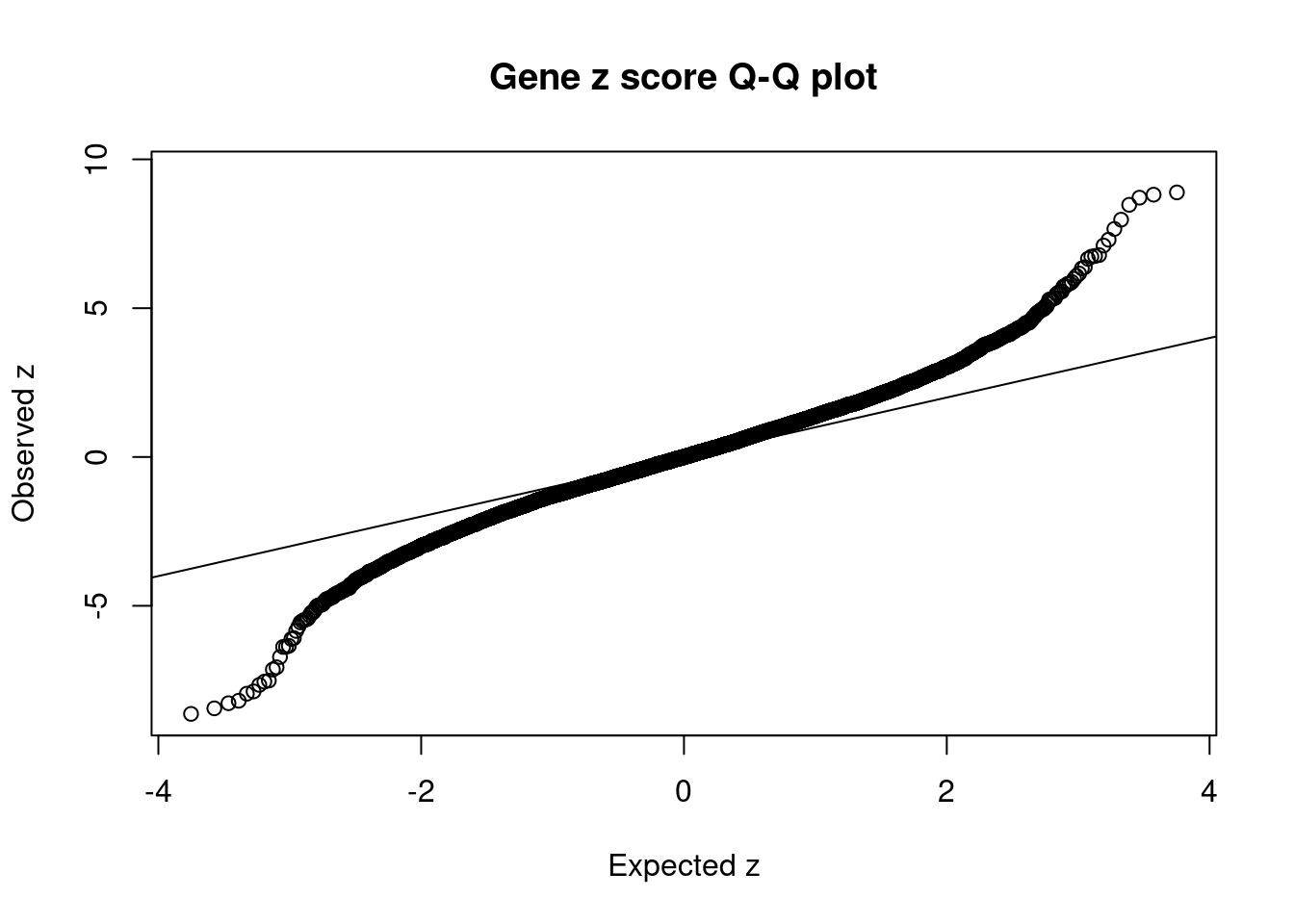
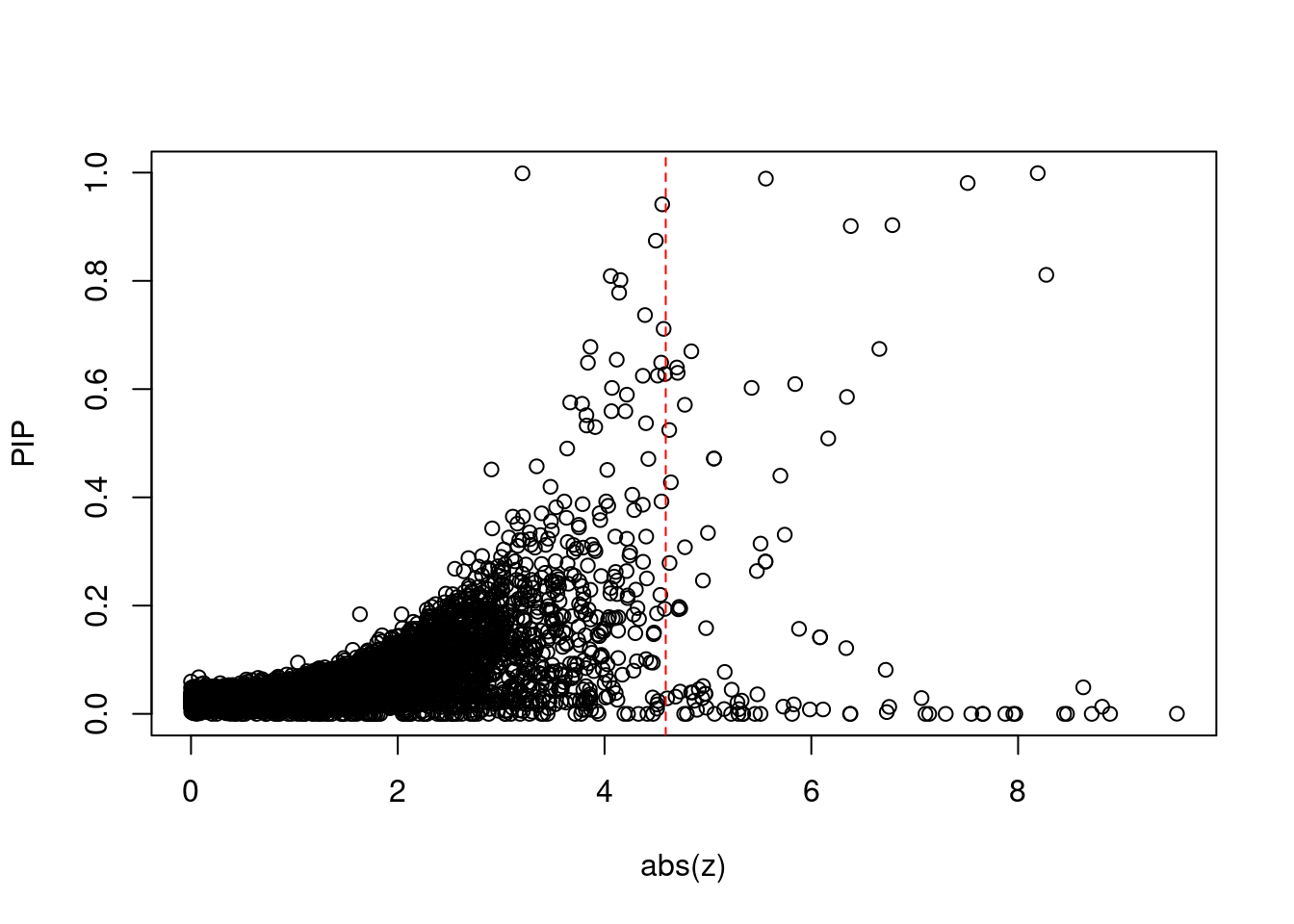
[1] 0.008173GO enrichment analysis for genes with PIP>0.5
#number of genes for gene set enrichment
length(genes)[1] 41Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Cellular_Component_2021"
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Molecular_Function_2021"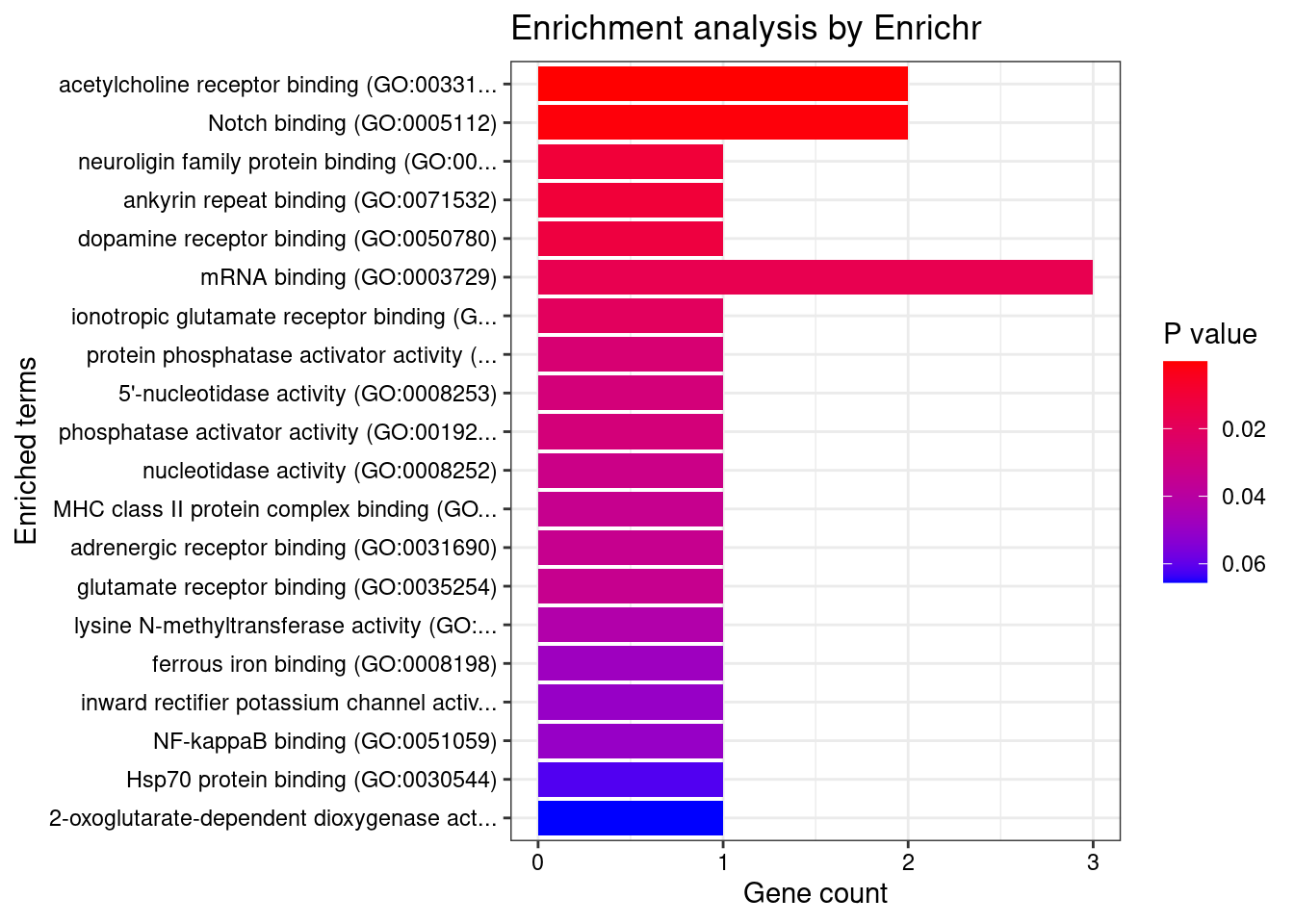
Term Overlap Adjusted.P.value
1 acetylcholine receptor binding (GO:0033130) 2/8 0.007632
2 Notch binding (GO:0005112) 2/24 0.036841
Genes
1 DLG4;LY6H
2 CUL3;HIF1ANDisGeNET enrichment analysis for genes with PIP>0.5
Description FDR Ratio
20 Measles 0.01082 1/15
64 Disproportionate tall stature 0.01082 1/15
65 Snowflake vitreoretinal degeneration 0.01082 1/15
66 Reticular Dystrophy Of Retinal Pigment Epithelium 0.01082 1/15
69 HEMOLYTIC UREMIC SYNDROME, ATYPICAL, SUSCEPTIBILITY TO, 2 0.01082 1/15
71 LEBER CONGENITAL AMAUROSIS 16 0.01082 1/15
73 PSEUDOHYPOALDOSTERONISM, TYPE IIE 0.01082 1/15
78 SPASTIC PARAPLEGIA 45, AUTOSOMAL RECESSIVE 0.01082 1/15
79 CONE-ROD DYSTROPHY 20 0.01082 1/15
82 SPASTIC PARAPLEGIA 62, AUTOSOMAL RECESSIVE 0.01082 1/15
BgRatio
20 1/9703
64 1/9703
65 1/9703
66 1/9703
69 1/9703
71 1/9703
73 1/9703
78 1/9703
79 1/9703
82 1/9703WebGestalt enrichment analysis for genes with PIP>0.5
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum =
minNum, : No significant gene set is identified based on FDR 0.05!NULLPIP Manhattan Plot
Warning: 'timedatectl' indicates the non-existent timezone name 'n/a'Warning: Your system is mis-configured: '/etc/localtime' is not a symlinkWarning: It is strongly recommended to set envionment variable TZ to 'America/
Chicago' (or equivalent)Warning: ggrepel: 6 unlabeled data points (too many overlaps). Consider
increasing max.overlaps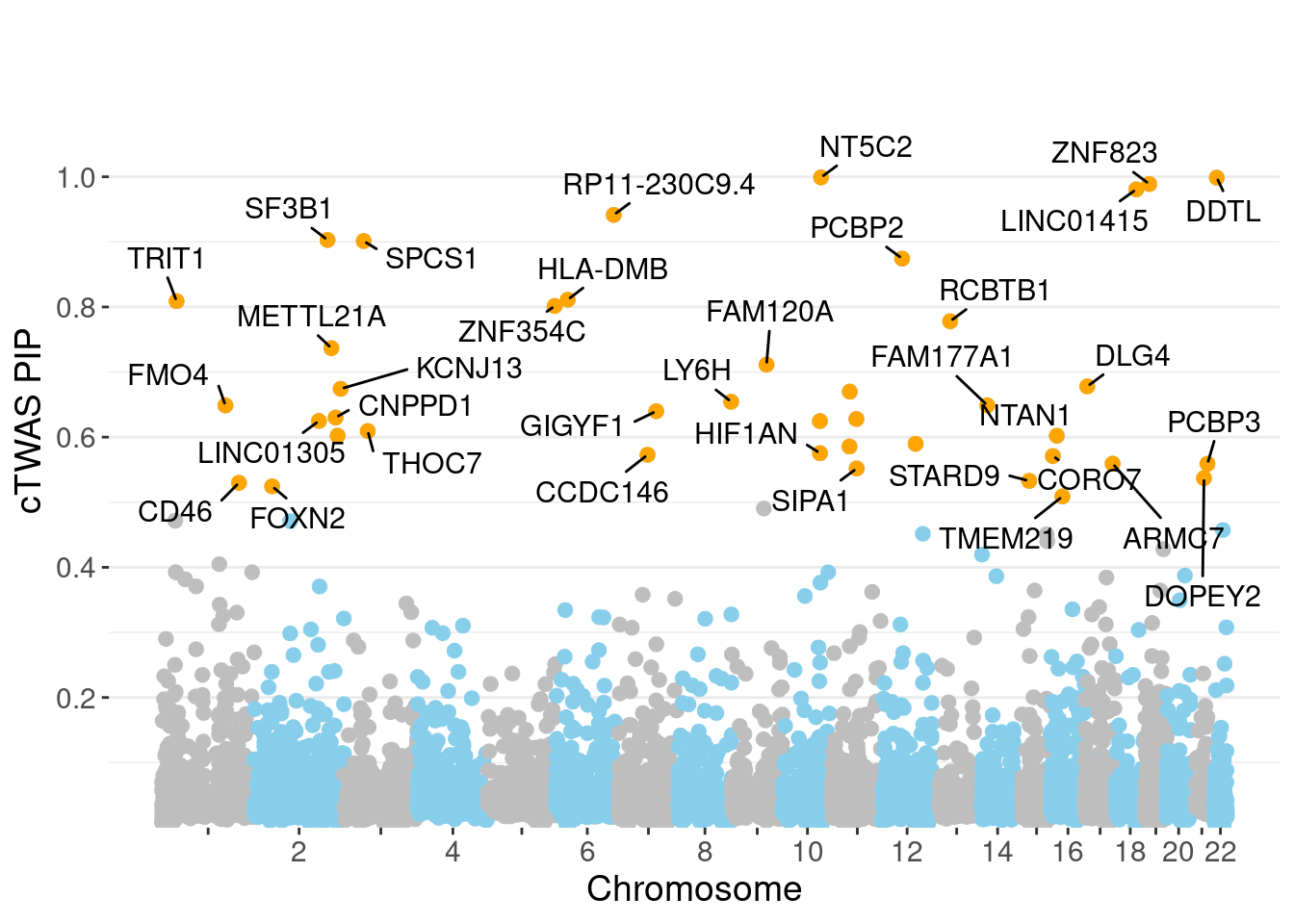
Sensitivity, specificity and precision for silver standard genes
#number of genes in known annotations
print(length(known_annotations))[1] 130#number of genes in known annotations with imputed expression
print(sum(known_annotations %in% ctwas_gene_res$genename))[1] 65#significance threshold for TWAS
print(sig_thresh)[1] 4.592#number of ctwas genes
length(ctwas_genes)[1] 11#number of TWAS genes
length(twas_genes)[1] 93#show novel genes (ctwas genes with not in TWAS genes)
ctwas_gene_res[ctwas_gene_res$genename %in% novel_genes,report_cols] genename region_tag susie_pip mu2 PVE z num_eqtl
421 TRIT1 1_25 0.8089 20.37 0.0002137 -4.060 3
9752 ZNF354C 5_108 0.8015 20.29 0.0002110 -4.154 1
13679 RP11-230C9.4 6_102 0.9414 21.83 0.0002666 -4.558 2
11176 PCBP2 12_33 0.8743 21.44 0.0002432 4.496 1
1518 DDTL 22_6 0.9987 32.10 0.0004159 -3.206 2#sensitivity / recall
print(sensitivity) ctwas TWAS
0.01538 0.06923 #specificity
print(specificity) ctwas TWAS
0.9992 0.9926 #precision / PPV
print(precision) ctwas TWAS
0.18182 0.09677 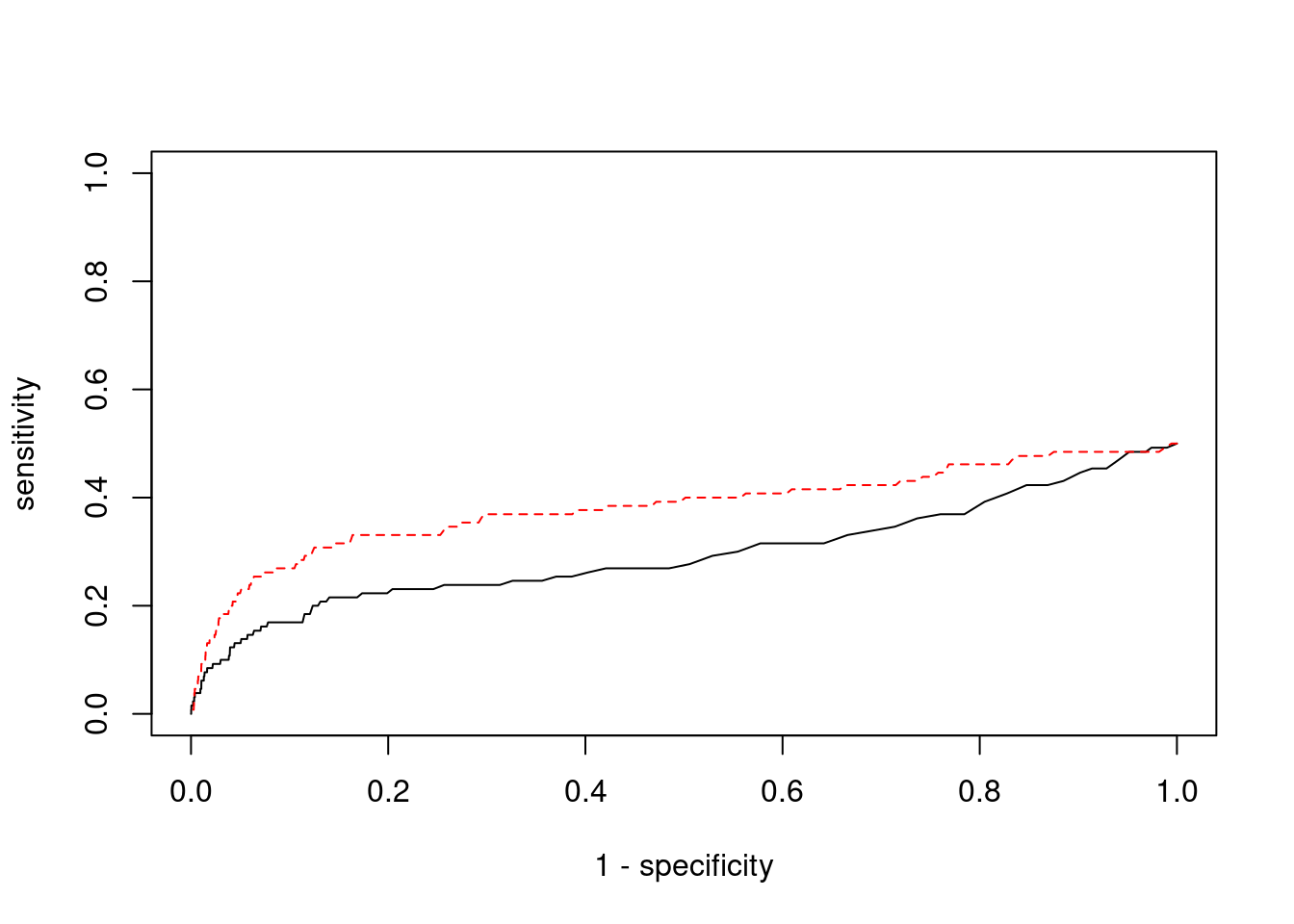
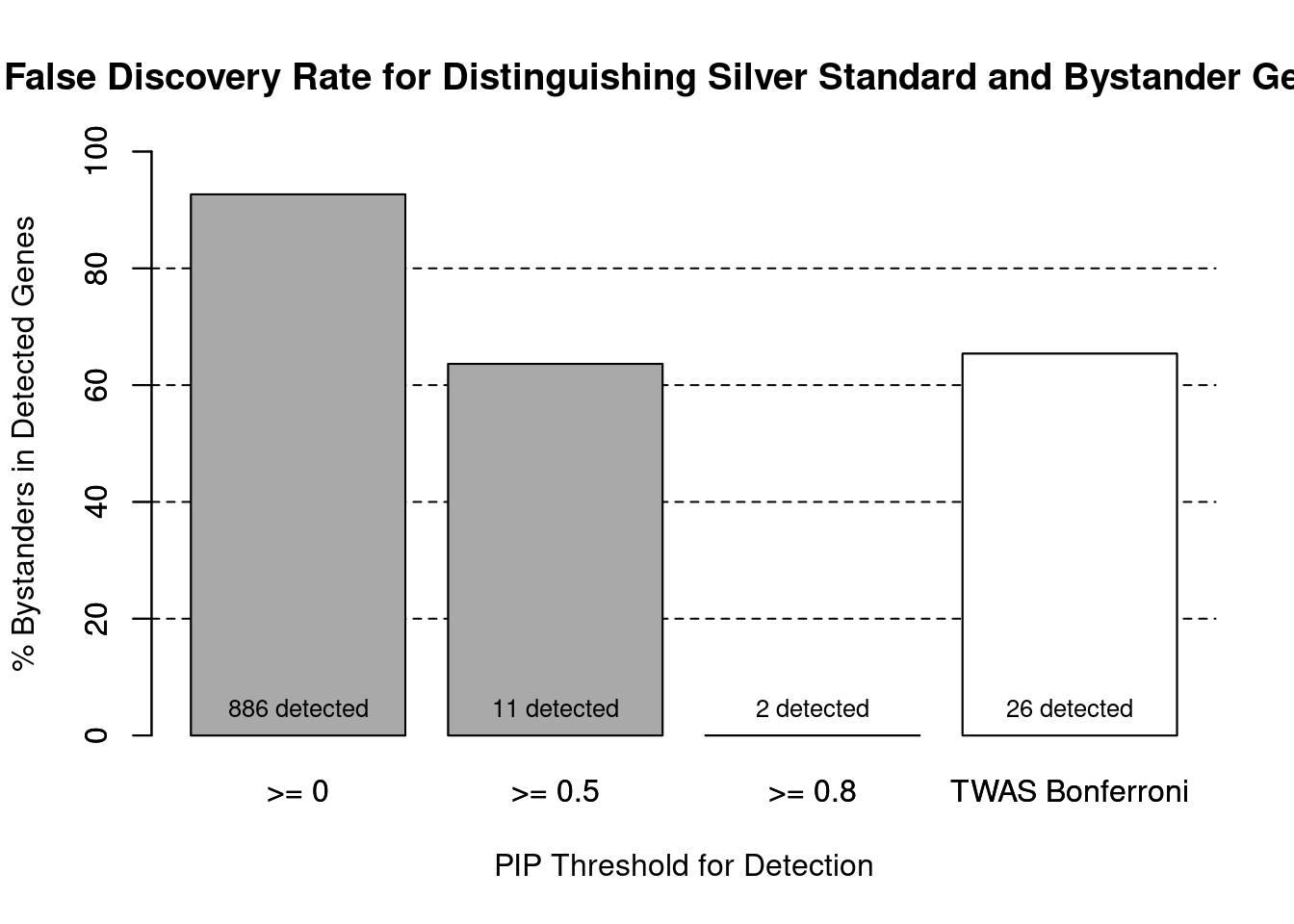
cTWAS is more precise than TWAS in distinguishing silver standard and bystander genes
#number of genes in known annotations (with imputed expression)
print(length(known_annotations))[1] 65#number of bystander genes (with imputed expression)
print(length(unrelated_genes))[1] 820#subset results to genes in known annotations or bystanders
ctwas_gene_res_subset <- ctwas_gene_res[ctwas_gene_res$genename %in% c(known_annotations, unrelated_genes),]
#assign ctwas and TWAS genes
ctwas_genes <- ctwas_gene_res_subset$genename[ctwas_gene_res_subset$susie_pip>0.8]
twas_genes <- ctwas_gene_res_subset$genename[abs(ctwas_gene_res_subset$z)>sig_thresh]
#significance threshold for TWAS
print(sig_thresh)[1] 4.592#number of ctwas genes (in known annotations or bystanders)
length(ctwas_genes)[1] 2#number of TWAS genes (in known annotations or bystanders)
length(twas_genes)[1] 26#sensitivity / recall
sensitivity ctwas TWAS
0.03077 0.13846 #specificity / (1 - False Positive Rate)
specificity ctwas TWAS
1.0000 0.9793 #precision / PPV / (1 - False Discovery Rate)
precision ctwas TWAS
1.0000 0.3462 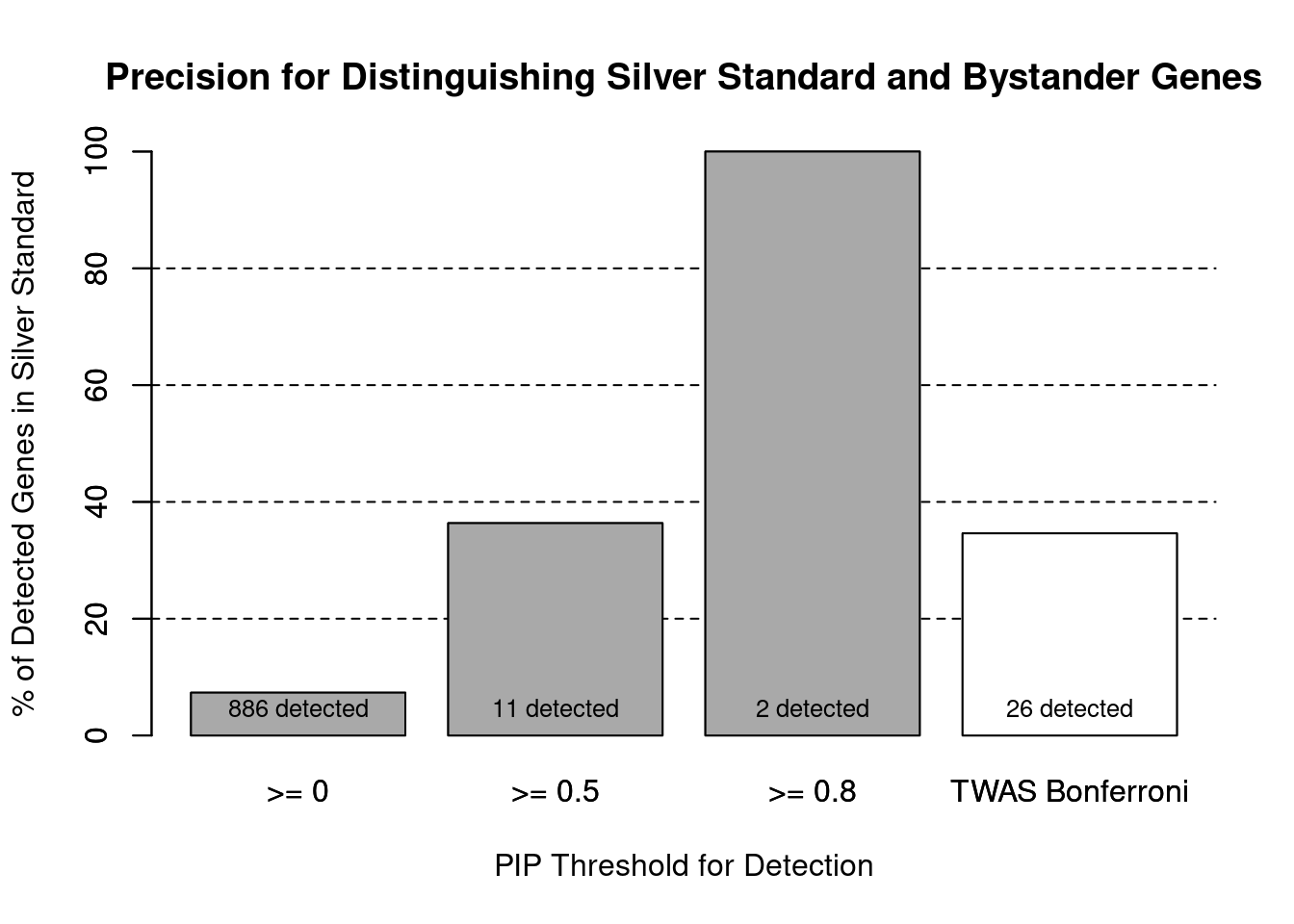
pip_range <- (0:1000)/1000
sensitivity <- rep(NA, length(pip_range))
specificity <- rep(NA, length(pip_range))
for (index in 1:length(pip_range)){
pip <- pip_range[index]
ctwas_genes <- ctwas_gene_res_subset$genename[ctwas_gene_res_subset$susie_pip>=pip]
sensitivity[index] <- sum(ctwas_genes %in% known_annotations)/length(known_annotations)
specificity[index] <- sum(!(unrelated_genes %in% ctwas_genes))/length(unrelated_genes)
}
plot(1-specificity, sensitivity, type="l", xlim=c(0,1), ylim=c(0,1), main="", xlab="1 - Specificity", ylab="Sensitivity")
title(expression("ROC Curve for cTWAS (black) and TWAS (" * phantom("red") * ")"))
title(expression(phantom("ROC Curve for cTWAS (black) and TWAS (") * "red" * phantom(")")), col.main="red")
sig_thresh_range <- seq(from=0, to=max(abs(ctwas_gene_res_subset$z)), length.out=length(pip_range))
for (index in 1:length(sig_thresh_range)){
sig_thresh_plot <- sig_thresh_range[index]
twas_genes <- ctwas_gene_res_subset$genename[abs(ctwas_gene_res_subset$z)>=sig_thresh_plot]
sensitivity[index] <- sum(twas_genes %in% known_annotations)/length(known_annotations)
specificity[index] <- sum(!(unrelated_genes %in% twas_genes))/length(unrelated_genes)
}
lines(1-specificity, sensitivity, xlim=c(0,1), ylim=c(0,1), col="red", lty=1)
abline(a=0,b=1,lty=3)
#add previously computed points from the analysis
ctwas_genes <- ctwas_gene_res_subset$genename[ctwas_gene_res_subset$susie_pip>0.8]
twas_genes <- ctwas_gene_res_subset$genename[abs(ctwas_gene_res_subset$z)>sig_thresh]
points(1-specificity_plot["ctwas"], sensitivity_plot["ctwas"], pch=21, bg="black")
points(1-specificity_plot["TWAS"], sensitivity_plot["TWAS"], pch=21, bg="red")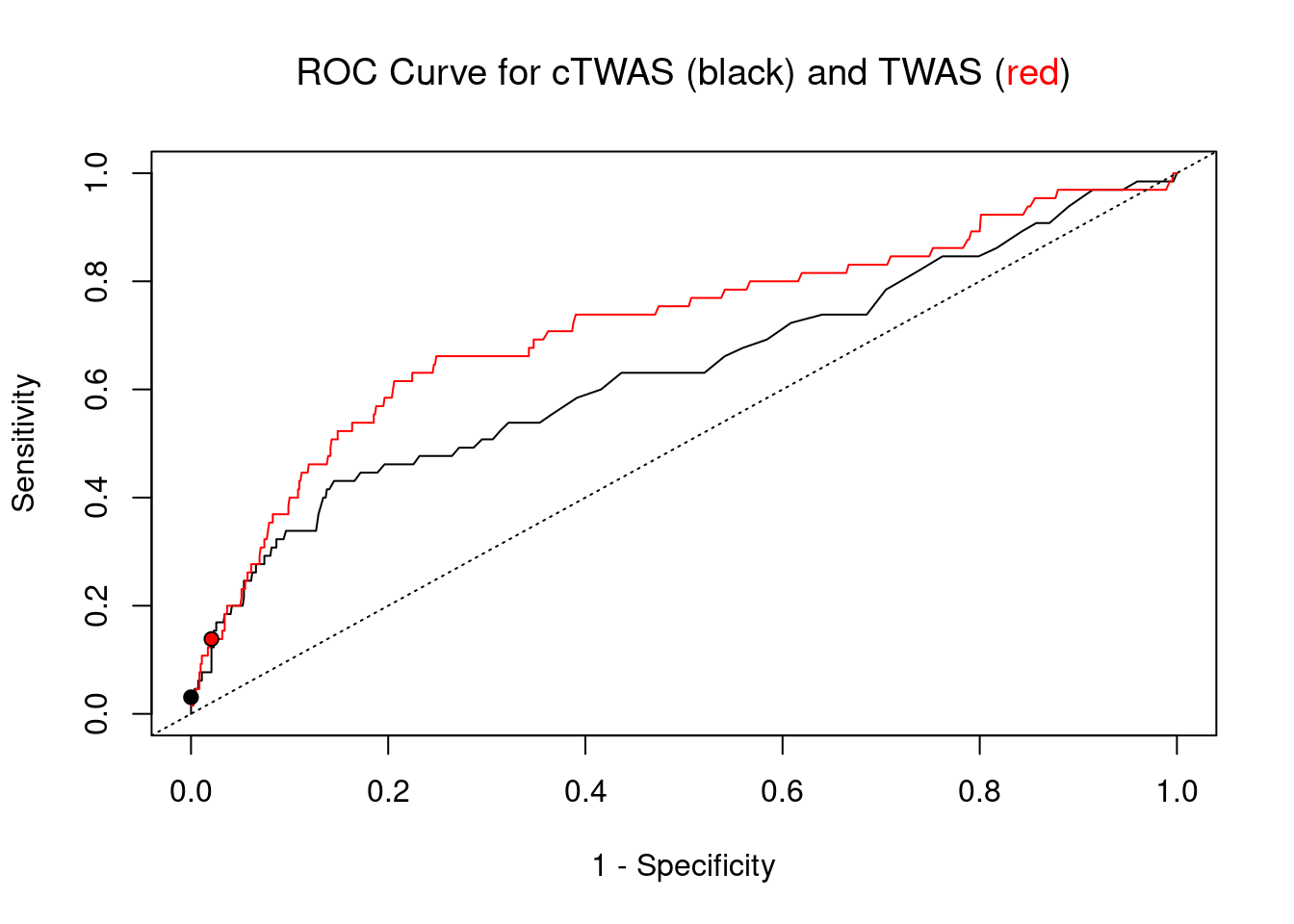
Undetected silver standard genes have low TWAS z-scores or stronger signal from nearby variants
#table of outcomes for silver standard genes
-sort(-table(silver_standard_case))silver_standard_case
Not Imputed Insignificant z-score Nearby SNP(s)
65 56 7
Detected (PIP > 0.8)
2 #show inconclusive genes
silver_standard_case[silver_standard_case=="Inconclusive"]named character(0)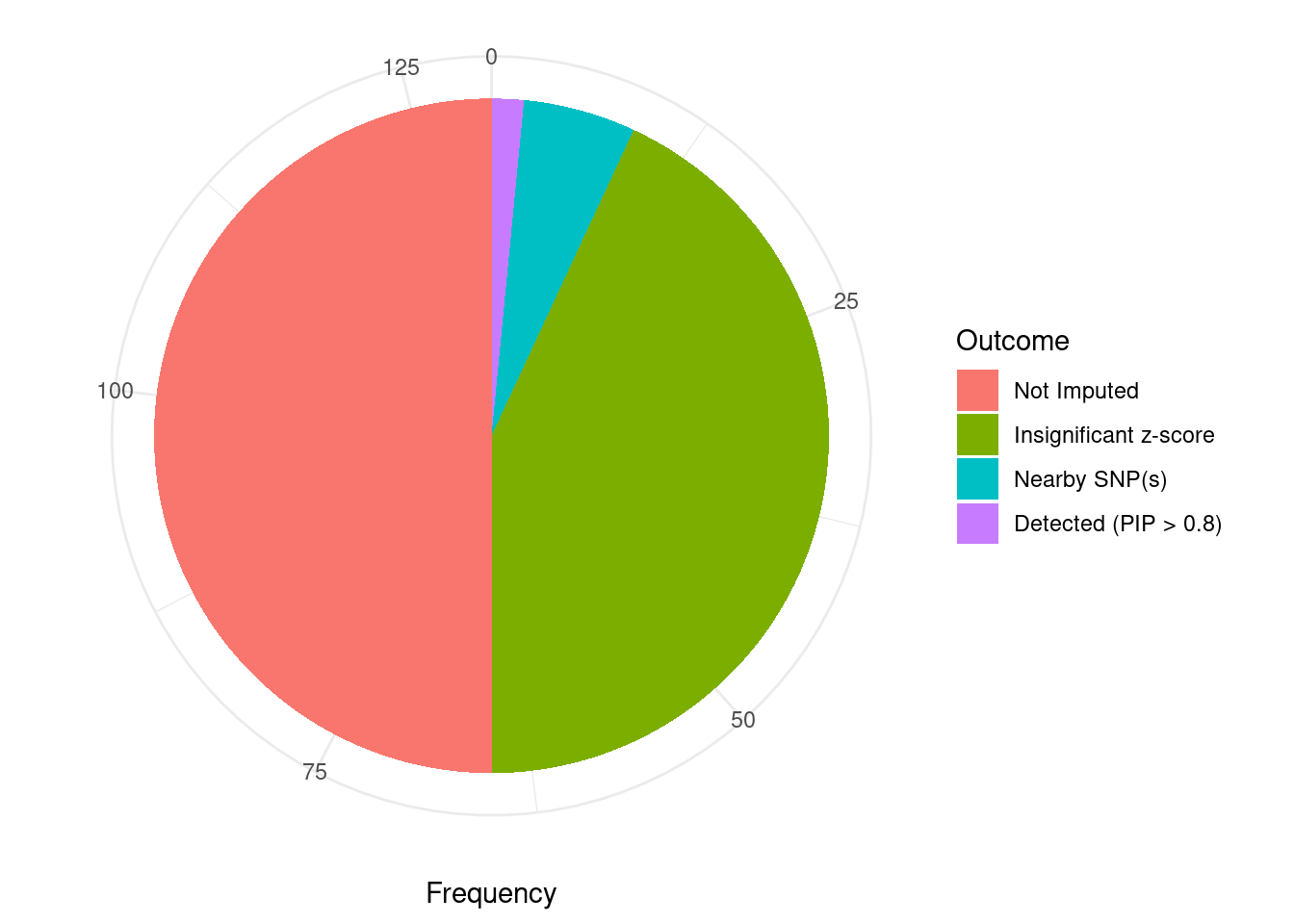
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] GenomicRanges_1.36.1 GenomeInfoDb_1.20.0 IRanges_2.18.1
[4] S4Vectors_0.22.1 BiocGenerics_0.30.0 biomaRt_2.40.1
[7] readxl_1.3.1 forcats_0.5.1 stringr_1.4.0
[10] dplyr_1.0.7 purrr_0.3.4 readr_2.1.1
[13] tidyr_1.1.4 tidyverse_1.3.1 tibble_3.1.6
[16] WebGestaltR_0.4.4 disgenet2r_0.99.2 enrichR_3.0
[19] cowplot_1.0.0 ggplot2_3.3.5 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] ggbeeswarm_0.6.0 colorspace_2.0-2 rjson_0.2.20
[4] ellipsis_0.3.2 rprojroot_2.0.2 XVector_0.24.0
[7] fs_1.5.2 rstudioapi_0.13 farver_2.1.0
[10] ggrepel_0.9.1 bit64_4.0.5 AnnotationDbi_1.46.0
[13] fansi_1.0.2 lubridate_1.8.0 xml2_1.3.3
[16] codetools_0.2-16 doParallel_1.0.17 cachem_1.0.6
[19] knitr_1.36 jsonlite_1.7.2 apcluster_1.4.8
[22] Cairo_1.5-12.2 broom_0.7.10 dbplyr_2.1.1
[25] compiler_3.6.1 httr_1.4.2 backports_1.4.1
[28] assertthat_0.2.1 Matrix_1.2-18 fastmap_1.1.0
[31] cli_3.1.0 later_0.8.0 prettyunits_1.1.1
[34] htmltools_0.5.2 tools_3.6.1 igraph_1.2.10
[37] GenomeInfoDbData_1.2.1 gtable_0.3.0 glue_1.6.2
[40] reshape2_1.4.4 doRNG_1.8.2 Rcpp_1.0.8
[43] Biobase_2.44.0 cellranger_1.1.0 jquerylib_0.1.4
[46] vctrs_0.3.8 svglite_1.2.2 iterators_1.0.14
[49] xfun_0.29 ps_1.6.0 rvest_1.0.2
[52] lifecycle_1.0.1 rngtools_1.5.2 XML_3.99-0.3
[55] zlibbioc_1.30.0 getPass_0.2-2 scales_1.1.1
[58] vroom_1.5.7 hms_1.1.1 promises_1.0.1
[61] yaml_2.2.1 curl_4.3.2 memoise_2.0.1
[64] ggrastr_1.0.1 gdtools_0.1.9 stringi_1.7.6
[67] RSQLite_2.2.8 highr_0.9 foreach_1.5.2
[70] rlang_1.0.1 pkgconfig_2.0.3 bitops_1.0-7
[73] evaluate_0.14 lattice_0.20-38 labeling_0.4.2
[76] bit_4.0.4 processx_3.5.2 tidyselect_1.1.1
[79] plyr_1.8.6 magrittr_2.0.2 R6_2.5.1
[82] generics_0.1.1 DBI_1.1.2 pillar_1.6.4
[85] haven_2.4.3 whisker_0.3-2 withr_2.4.3
[88] RCurl_1.98-1.5 modelr_0.1.8 crayon_1.5.0
[91] utf8_1.2.2 tzdb_0.2.0 rmarkdown_2.11
[94] progress_1.2.2 grid_3.6.1 data.table_1.14.2
[97] blob_1.2.2 callr_3.7.0 git2r_0.26.1
[100] reprex_2.0.1 digest_0.6.29 httpuv_1.5.1
[103] munsell_0.5.0 beeswarm_0.2.3 vipor_0.4.5