OxHb
sheng Qian
2023-10-14
Last updated: 2023-10-14
Checks: 5 2
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/OxHb/ | data/OxHb |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version d04dec9. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .ipynb_checkpoints/
Untracked files:
Untracked: LDL_LDLR_locus1.pdf
Untracked: LDL_TEME199_genetrack.pdf
Untracked: LDL_TEME199_locus.pdf
Untracked: Proposal plots.R
Untracked: RGS14.pdf
Untracked: RNF186.pdf
Untracked: Rplots.pdf
Untracked: SCZ_annotation.xlsx
Untracked: SLC8B1.pdf
Untracked: analysis/.ipynb_checkpoints/
Untracked: cache/
Untracked: code/.ipynb_checkpoints/
Untracked: data/.ipynb_checkpoints/
Untracked: data/FUMA_output/
Untracked: data/GO_Terms/
Untracked: data/GTEx_Analysis_v8_eQTL.tar
Untracked: data/G_list.RData
Untracked: data/IBD_ME/
Untracked: data/LB/
Untracked: data/LDL/
Untracked: data/LDL_E_S/
Untracked: data/LDL_E_S_M/
Untracked: data/LDL_M/
Untracked: data/LDL_S/
Untracked: data/OxHb/
Untracked: data/PGC3_SCZ_wave3_public.v2.tsv
Untracked: data/Predictive_Models/
Untracked: data/Supplementary Table 15 - MAGMA.xlsx
Untracked: data/Supplementary Table 20 - Prioritised Genes.xlsx
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/WhiteBlood_E/
Untracked: data/WhiteBlood_E_M/
Untracked: data/WhiteBlood_E_S_M/
Untracked: data/WhiteBlood_E_S_M_PC/
Untracked: data/WhiteBlood_M/
Untracked: data/WhiteBlood_M_compare/
Untracked: data/WhiteBlood_M_enet/
Untracked: data/cpg_annot.RData
Untracked: data/eqtl/
Untracked: data/gencode.v26.GRCh38.genes.gtf
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/gwas_sumstats/
Untracked: data/magma.genes.out
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/mashr_sqtl/
Untracked: data/mqtl/
Untracked: data/notes.txt
Untracked: data/scz_2018.RDS
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: data/test/
Untracked: hist.pdf
Untracked: ld_pip.pdf
Untracked: submit.sh
Untracked: temp_LDR/
Untracked: test-B1.snpgwas.txt
Unstaged changes:
Deleted: analysis/Atrial_Fibrillation_Heart_Atrial_Appendage.Rmd
Deleted: analysis/Atrial_Fibrillation_Heart_Left_Ventricle.Rmd
Deleted: analysis/Autism_Brain_Amygdala.Rmd
Deleted: analysis/Autism_Brain_Anterior_cingulate_cortex_BA24.Rmd
Deleted: analysis/Autism_Brain_Caudate_basal_ganglia.Rmd
Deleted: analysis/Autism_Brain_Cerebellar_Hemisphere.Rmd
Deleted: analysis/Autism_Brain_Cerebellum.Rmd
Deleted: analysis/Autism_Brain_Cortex.Rmd
Deleted: analysis/Autism_Brain_Frontal_Cortex_BA9.Rmd
Deleted: analysis/Autism_Brain_Hippocampus.Rmd
Deleted: analysis/Autism_Brain_Hypothalamus.Rmd
Deleted: analysis/Autism_Brain_Nucleus_accumbens_basal_ganglia.Rmd
Deleted: analysis/Autism_Brain_Putamen_basal_ganglia.Rmd
Deleted: analysis/Autism_Brain_Spinal_cord_cervical_c-1.Rmd
Deleted: analysis/Autism_Brain_Substantia_nigra.Rmd
Deleted: analysis/BMI_Brain_Amygdala.Rmd
Deleted: analysis/BMI_Brain_Amygdala_S.Rmd
Deleted: analysis/BMI_Brain_Anterior_cingulate_cortex_BA24.Rmd
Deleted: analysis/BMI_Brain_Anterior_cingulate_cortex_BA24_S.Rmd
Deleted: analysis/BMI_Brain_Caudate_basal_ganglia.Rmd
Deleted: analysis/BMI_Brain_Caudate_basal_ganglia_S.Rmd
Deleted: analysis/BMI_Brain_Cerebellar_Hemisphere.Rmd
Deleted: analysis/BMI_Brain_Cerebellar_Hemisphere_S.Rmd
Deleted: analysis/BMI_Brain_Cerebellum.Rmd
Deleted: analysis/BMI_Brain_Cerebellum_S.Rmd
Deleted: analysis/BMI_Brain_Cortex.Rmd
Deleted: analysis/BMI_Brain_Cortex_S.Rmd
Deleted: analysis/BMI_Brain_Frontal_Cortex_BA9.Rmd
Deleted: analysis/BMI_Brain_Frontal_Cortex_BA9_S.Rmd
Deleted: analysis/BMI_Brain_Hippocampus.Rmd
Deleted: analysis/BMI_Brain_Hippocampus_S.Rmd
Deleted: analysis/BMI_Brain_Hypothalamus.Rmd
Deleted: analysis/BMI_Brain_Hypothalamus_S.Rmd
Deleted: analysis/BMI_Brain_Nucleus_accumbens_basal_ganglia.Rmd
Deleted: analysis/BMI_Brain_Nucleus_accumbens_basal_ganglia_S.Rmd
Deleted: analysis/BMI_Brain_Putamen_basal_ganglia.Rmd
Deleted: analysis/BMI_Brain_Putamen_basal_ganglia_S.Rmd
Deleted: analysis/BMI_Brain_Spinal_cord_cervical_c-1.Rmd
Deleted: analysis/BMI_Brain_Spinal_cord_cervical_c-1_S.Rmd
Deleted: analysis/BMI_Brain_Substantia_nigra.Rmd
Deleted: analysis/BMI_Brain_Substantia_nigra_S.Rmd
Deleted: analysis/BMI_S_results.Rmd
Deleted: analysis/Glucose_Adipose_Subcutaneous.Rmd
Deleted: analysis/Glucose_Adipose_Visceral_Omentum.Rmd
Modified: analysis/OxHB.Rmd
Deleted: code/WhiteBlood_M_ener_out/WhiteBlood_WholeBlood.out
Deleted: code/White_Blood_M_out/White_Blood_BreastMammary.err
Deleted: code/White_Blood_M_out/White_Blood_BreastMammary.out
Deleted: code/White_Blood_M_out/White_Blood_ColonTransverse.err
Deleted: code/White_Blood_M_out/White_Blood_ColonTransverse.out
Deleted: code/White_Blood_M_out/White_Blood_KidneyCortex.err
Deleted: code/White_Blood_M_out/White_Blood_KidneyCortex.out
Deleted: code/White_Blood_M_out/White_Blood_Lung.err
Deleted: code/White_Blood_M_out/White_Blood_Lung.out
Deleted: code/White_Blood_M_out/White_Blood_MuscleSkeletal.err
Deleted: code/White_Blood_M_out/White_Blood_MuscleSkeletal.out
Deleted: code/White_Blood_M_out/White_Blood_Ovary.err
Deleted: code/White_Blood_M_out/White_Blood_Ovary.out
Deleted: code/White_Blood_M_out/White_Blood_Prostate.err
Deleted: code/White_Blood_M_out/White_Blood_Prostate.out
Deleted: code/White_Blood_M_out/White_Blood_Testis.err
Deleted: code/White_Blood_M_out/White_Blood_Testis.out
Deleted: code/White_Blood_M_out/White_Blood_WholeBlood.err
Deleted: code/White_Blood_M_out/White_Blood_WholeBlood.out
Deleted: code/run_IBD_ctwas_rss_LDR_ME.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/OxHB.Rmd) and HTML (docs/OxHB.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | d04dec9 | sq-96 | 2023-10-14 | update |
| html | d04dec9 | sq-96 | 2023-10-14 | update |
| html | f442f0e | sq-96 | 2023-10-14 | update |
| Rmd | 6a15ad1 | sq-96 | 2023-10-14 | update |
| html | 6a15ad1 | sq-96 | 2023-10-14 | update |
| Rmd | e0d796f | sq-96 | 2023-10-13 | update |
Artery Coronary
#estimated prior inclusion probability
ctwas_parameters$group_prior SNP gene
0.0003789661 0.0048196252 #estimated prior effect size
ctwas_parameters$group_prior_var SNP gene
0.6727008 0.5860888 #estimated enrichment of genes over variants
ctwas_parameters$enrichment gene
12.71783 #PVE explained by genes and variants
ctwas_parameters$group_pve SNP gene
0.71103798 0.01352419 #total heritability (sum of PVE)
ctwas_parameters$total_pve[1] 0.7245622#attributable heritability
ctwas_parameters$attributable_pve SNP gene
0.98133467 0.01866533 #plot convergence
ctwas_parameters$convergence_plot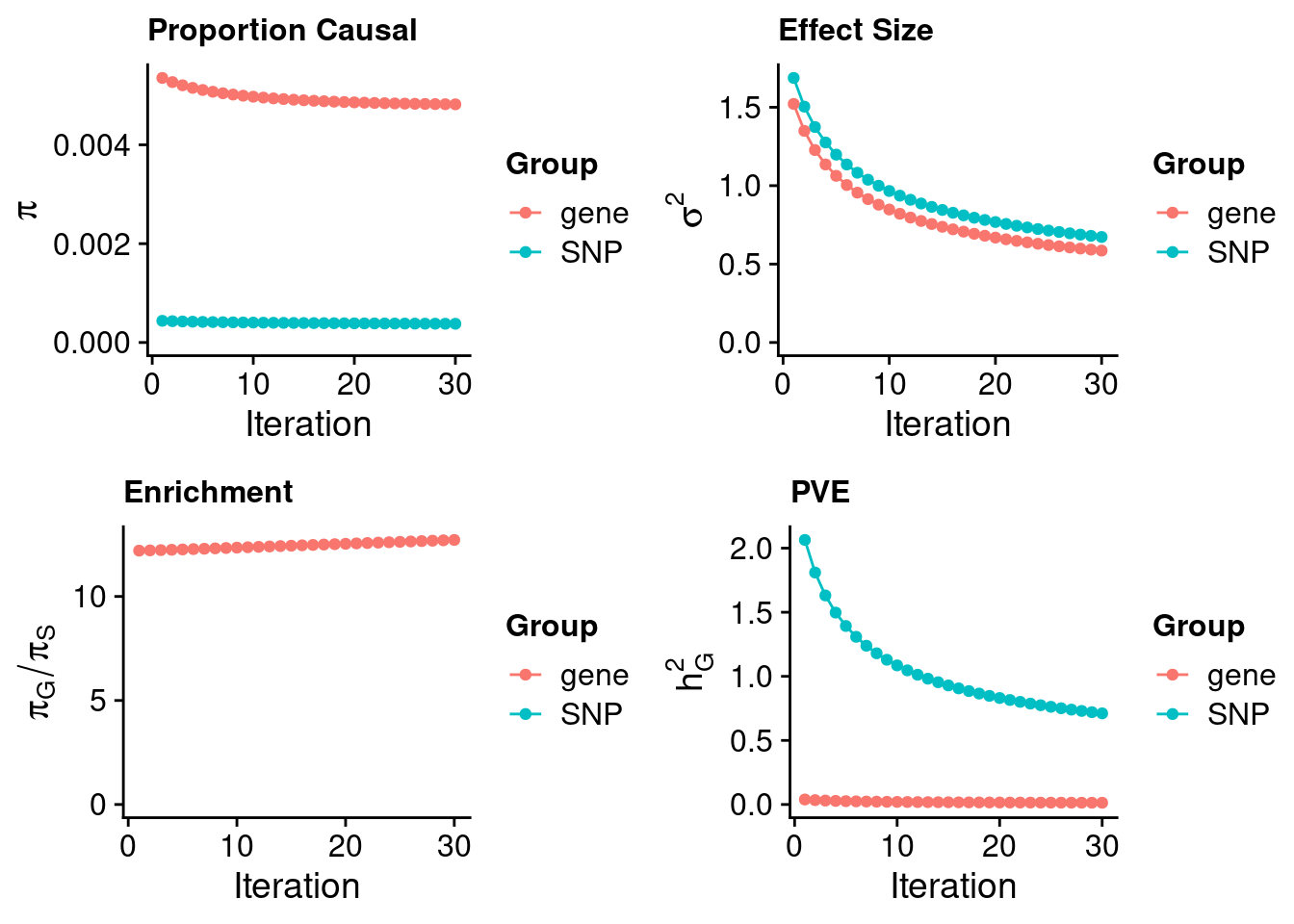
| Version | Author | Date |
|---|---|---|
| 6a15ad1 | sq-96 | 2023-10-14 |
#print top genes
head(ctwas_res[ctwas_res$type=="gene",]) chrom id pos type region_tag1 region_tag2 cs_index
164440 9 ENSG00000136868.10 113209149 gene 9 58 0
263152 20 ENSG00000089177.18 16581461 gene 20 12 0
272677 22 ENSG00000184381.18 38145259 gene 22 15 0
261678 20 ENSG00000275491.1 3807832 gene 20 3 0
90495 5 ENSG00000132356.11 40798520 gene 5 27 0
117969 6 ENSG00000178425.13 116099964 gene 6 77 0
susie_pip mu2 genename z
164440 0.11803156 9.216227 SLC31A1 -3.933203
263152 0.09730827 8.050336 KIF16B -3.085439
272677 0.08515657 7.548930 PLA2G6 3.053686
261678 0.08398874 7.466874 LINC01730 -2.945771
90495 0.08384722 7.723628 PRKAA1 -3.439407
117969 0.08334959 7.383582 NT5DC1 3.118395Artery Tibial
#estimated prior inclusion probability
ctwas_parameters$group_prior SNP gene
0.0003778214 0.0040115252 #estimated prior effect size
ctwas_parameters$group_prior_var SNP gene
0.6684344 0.5565171 #estimated enrichment of genes over variants
ctwas_parameters$enrichment gene
10.61752 #PVE explained by genes and variants
ctwas_parameters$group_pve SNP gene
0.70439414 0.01309908 #total heritability (sum of PVE)
ctwas_parameters$total_pve[1] 0.7174932#attributable heritability
ctwas_parameters$attributable_pve SNP gene
0.98174327 0.01825673 #plot convergence
ctwas_parameters$convergence_plot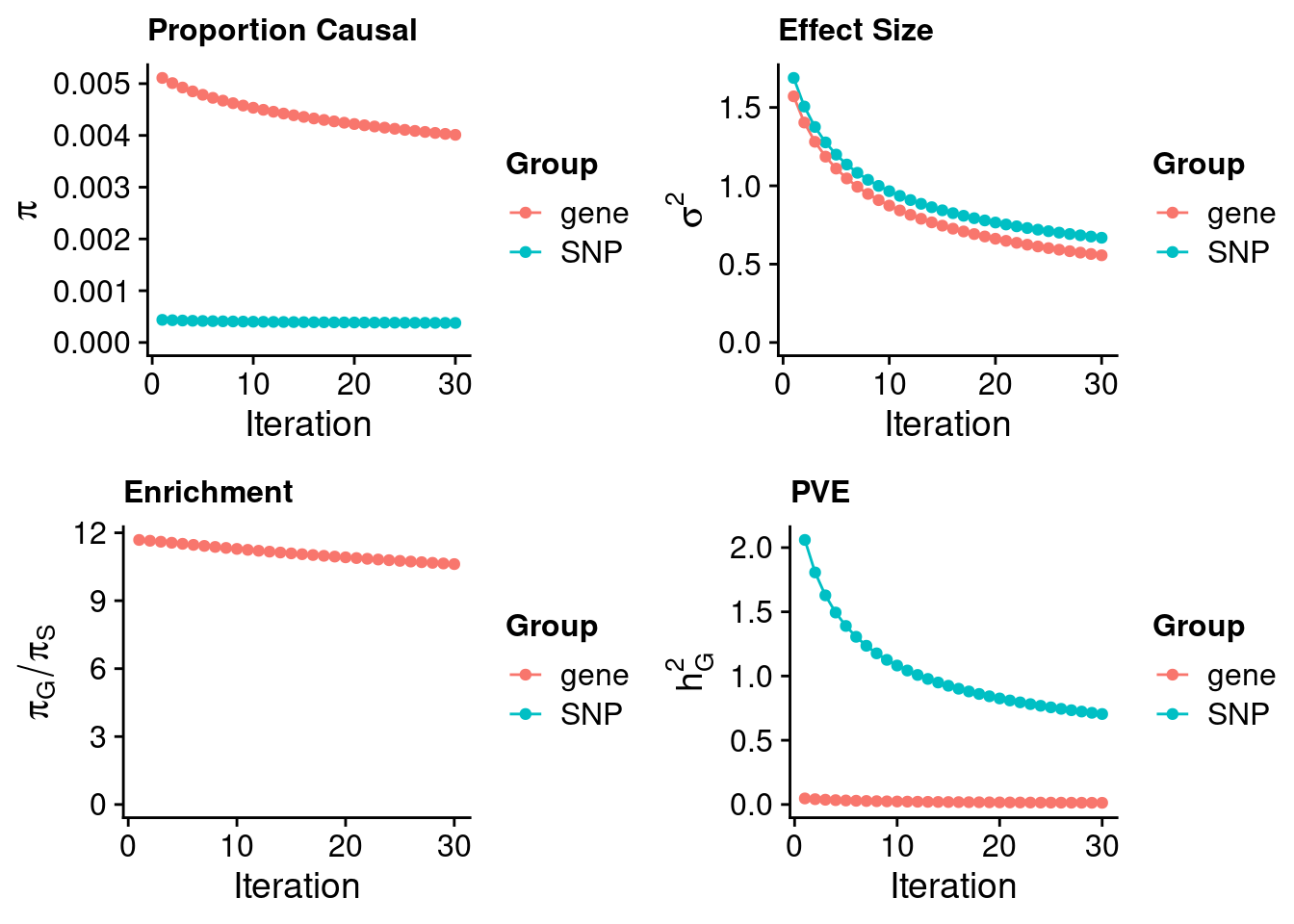
| Version | Author | Date |
|---|---|---|
| 6a15ad1 | sq-96 | 2023-10-14 |
head(ctwas_res[ctwas_res$type=="gene",]) chrom id pos type region_tag1 region_tag2 cs_index
114696 6 ENSG00000005700.14 82247064 gene 6 56 0
272419 22 ENSG00000159496.14 23679302 gene 22 6 0
179945 10 ENSG00000151923.17 119601669 gene 10 74 0
151888 8 ENSG00000164841.4 108787887 gene 8 74 0
241123 16 ENSG00000040199.18 71804965 gene 16 38 0
243700 17 ENSG00000174292.12 7380407 gene 17 6 0
susie_pip mu2 genename z
114696 0.11468988 9.126035 IBTK -3.787795
272419 0.08483873 7.971765 RGL4 3.286735
179945 0.08104538 7.839875 TIAL1 3.480318
151888 0.07962949 7.989882 TMEM74 -3.623924
241123 0.07790796 7.513540 PHLPP2 -3.126064
243700 0.07591772 7.535001 TNK1 3.186510Heart Atrial Appendage
#estimated prior inclusion probability
ctwas_parameters$group_prior SNP gene
0.0003778214 0.0040115252 #estimated prior effect size
ctwas_parameters$group_prior_var SNP gene
0.6684344 0.5565171 #estimated enrichment of genes over variants
ctwas_parameters$enrichment gene
10.61752 #PVE explained by genes and variants
ctwas_parameters$group_pve SNP gene
0.70439414 0.01309908 #total heritability (sum of PVE)
ctwas_parameters$total_pve[1] 0.7174932#attributable heritability
ctwas_parameters$attributable_pve SNP gene
0.98174327 0.01825673 #plot convergence
ctwas_parameters$convergence_plot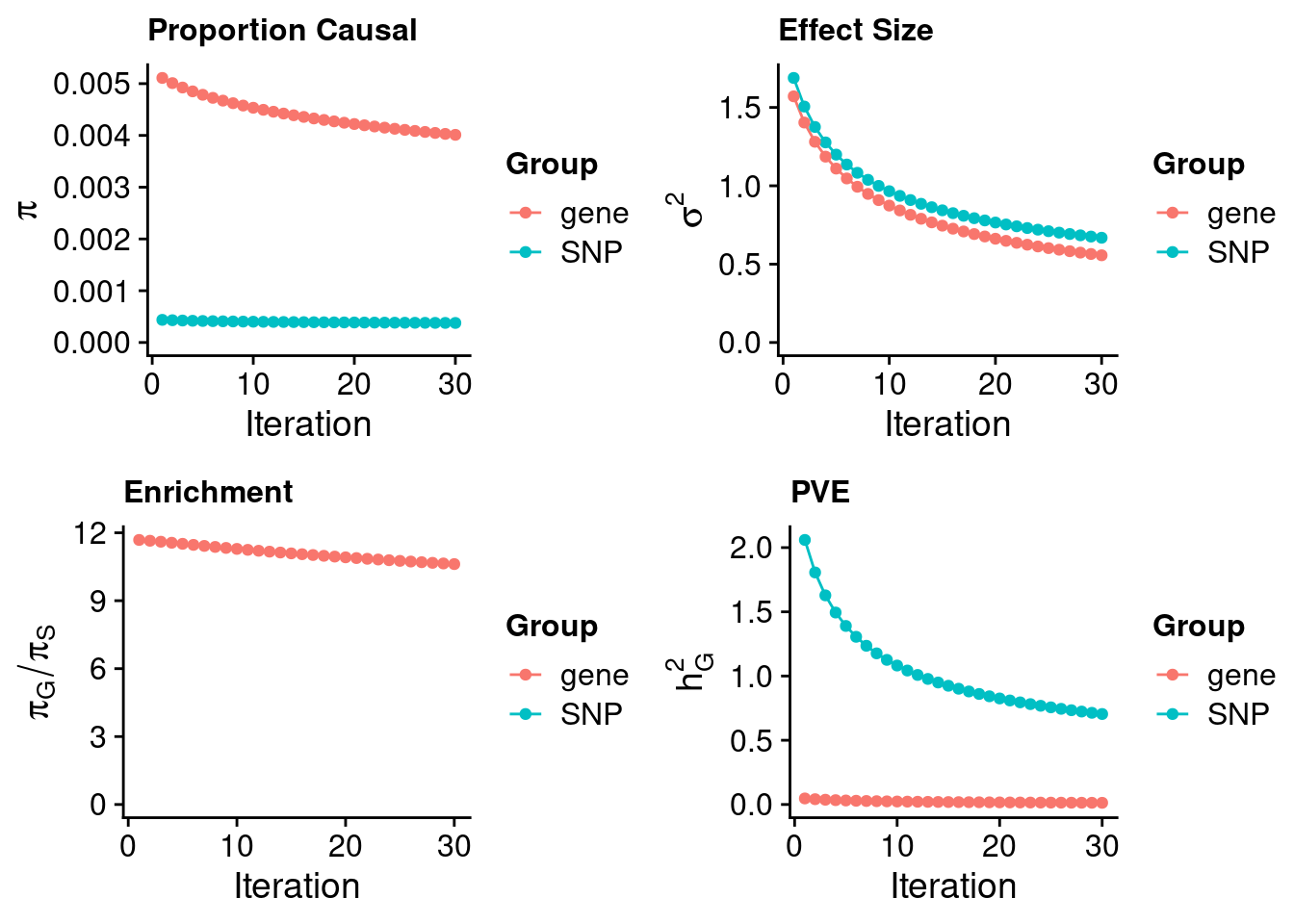
| Version | Author | Date |
|---|---|---|
| 6a15ad1 | sq-96 | 2023-10-14 |
head(ctwas_res[ctwas_res$type=="gene",]) chrom id pos type region_tag1 region_tag2 cs_index
114696 6 ENSG00000005700.14 82247064 gene 6 56 0
272419 22 ENSG00000159496.14 23679302 gene 22 6 0
179945 10 ENSG00000151923.17 119601669 gene 10 74 0
151888 8 ENSG00000164841.4 108787887 gene 8 74 0
241123 16 ENSG00000040199.18 71804965 gene 16 38 0
243700 17 ENSG00000174292.12 7380407 gene 17 6 0
susie_pip mu2 genename z
114696 0.11468988 9.126035 IBTK -3.787795
272419 0.08483873 7.971765 RGL4 3.286735
179945 0.08104538 7.839875 TIAL1 3.480318
151888 0.07962949 7.989882 TMEM74 -3.623924
241123 0.07790796 7.513540 PHLPP2 -3.126064
243700 0.07591772 7.535001 TNK1 3.186510Heart Left Ventricle
#estimated prior inclusion probability
ctwas_parameters$group_prior SNP gene
0.0003778214 0.0040115252 #estimated prior effect size
ctwas_parameters$group_prior_var SNP gene
0.6684344 0.5565171 #estimated enrichment of genes over variants
ctwas_parameters$enrichment gene
10.61752 #PVE explained by genes and variants
ctwas_parameters$group_pve SNP gene
0.70439414 0.01309908 #total heritability (sum of PVE)
ctwas_parameters$total_pve[1] 0.7174932#attributable heritability
ctwas_parameters$attributable_pve SNP gene
0.98174327 0.01825673 #plot convergence
ctwas_parameters$convergence_plot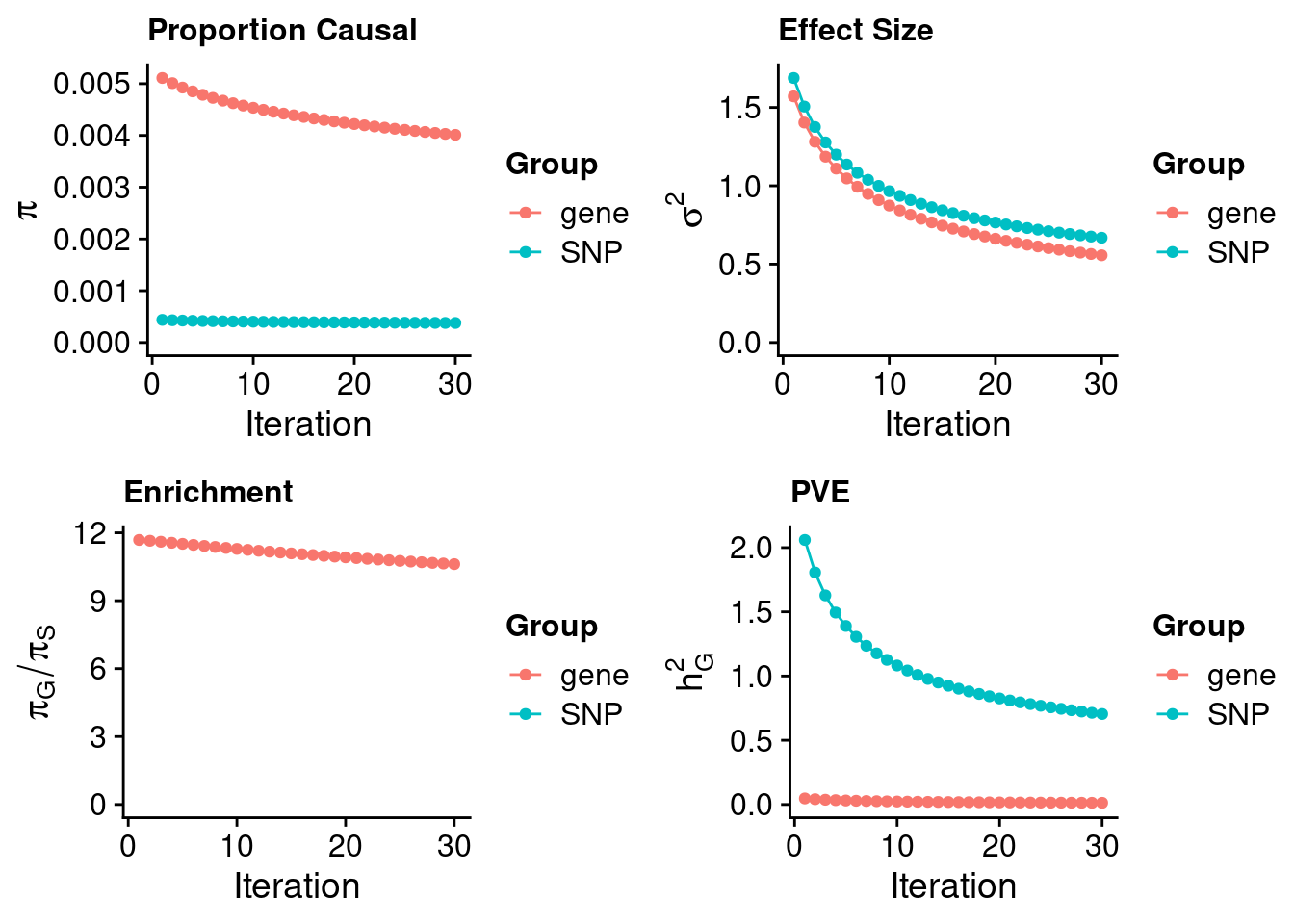
| Version | Author | Date |
|---|---|---|
| 6a15ad1 | sq-96 | 2023-10-14 |
head(ctwas_res[ctwas_res$type=="gene",]) chrom id pos type region_tag1 region_tag2 cs_index
114696 6 ENSG00000005700.14 82247064 gene 6 56 0
272419 22 ENSG00000159496.14 23679302 gene 22 6 0
179945 10 ENSG00000151923.17 119601669 gene 10 74 0
151888 8 ENSG00000164841.4 108787887 gene 8 74 0
241123 16 ENSG00000040199.18 71804965 gene 16 38 0
243700 17 ENSG00000174292.12 7380407 gene 17 6 0
susie_pip mu2 genename z
114696 0.11468988 9.126035 IBTK -3.787795
272419 0.08483873 7.971765 RGL4 3.286735
179945 0.08104538 7.839875 TIAL1 3.480318
151888 0.07962949 7.989882 TMEM74 -3.623924
241123 0.07790796 7.513540 PHLPP2 -3.126064
243700 0.07591772 7.535001 TNK1 3.186510Kidney Cortex
#estimated prior inclusion probability
ctwas_parameters$group_prior SNP gene
0.0003778214 0.0040115252 #estimated prior effect size
ctwas_parameters$group_prior_var SNP gene
0.6684344 0.5565171 #estimated enrichment of genes over variants
ctwas_parameters$enrichment gene
10.61752 #PVE explained by genes and variants
ctwas_parameters$group_pve SNP gene
0.70439414 0.01309908 #total heritability (sum of PVE)
ctwas_parameters$total_pve[1] 0.7174932#attributable heritability
ctwas_parameters$attributable_pve SNP gene
0.98174327 0.01825673 #plot convergence
ctwas_parameters$convergence_plot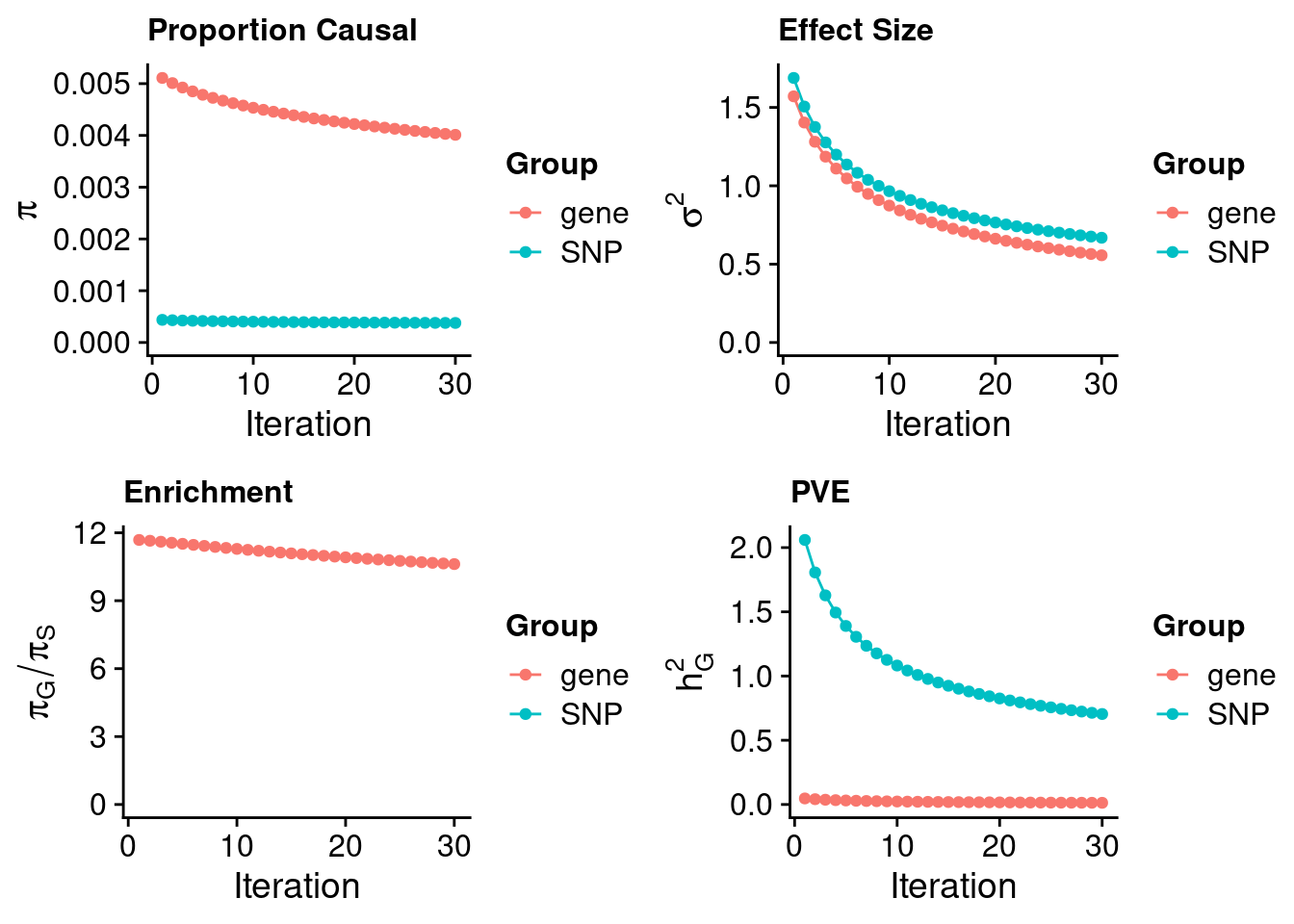
| Version | Author | Date |
|---|---|---|
| 6a15ad1 | sq-96 | 2023-10-14 |
head(ctwas_res[ctwas_res$type=="gene",]) chrom id pos type region_tag1 region_tag2 cs_index
114696 6 ENSG00000005700.14 82247064 gene 6 56 0
272419 22 ENSG00000159496.14 23679302 gene 22 6 0
179945 10 ENSG00000151923.17 119601669 gene 10 74 0
151888 8 ENSG00000164841.4 108787887 gene 8 74 0
241123 16 ENSG00000040199.18 71804965 gene 16 38 0
243700 17 ENSG00000174292.12 7380407 gene 17 6 0
susie_pip mu2 genename z
114696 0.11468988 9.126035 IBTK -3.787795
272419 0.08483873 7.971765 RGL4 3.286735
179945 0.08104538 7.839875 TIAL1 3.480318
151888 0.07962949 7.989882 TMEM74 -3.623924
241123 0.07790796 7.513540 PHLPP2 -3.126064
243700 0.07591772 7.535001 TNK1 3.186510Liver
#estimated prior inclusion probability
ctwas_parameters$group_prior SNP gene
0.0003778214 0.0040115252 #estimated prior effect size
ctwas_parameters$group_prior_var SNP gene
0.6684344 0.5565171 #estimated enrichment of genes over variants
ctwas_parameters$enrichment gene
10.61752 #PVE explained by genes and variants
ctwas_parameters$group_pve SNP gene
0.70439414 0.01309908 #total heritability (sum of PVE)
ctwas_parameters$total_pve[1] 0.7174932#attributable heritability
ctwas_parameters$attributable_pve SNP gene
0.98174327 0.01825673 #plot convergence
ctwas_parameters$convergence_plot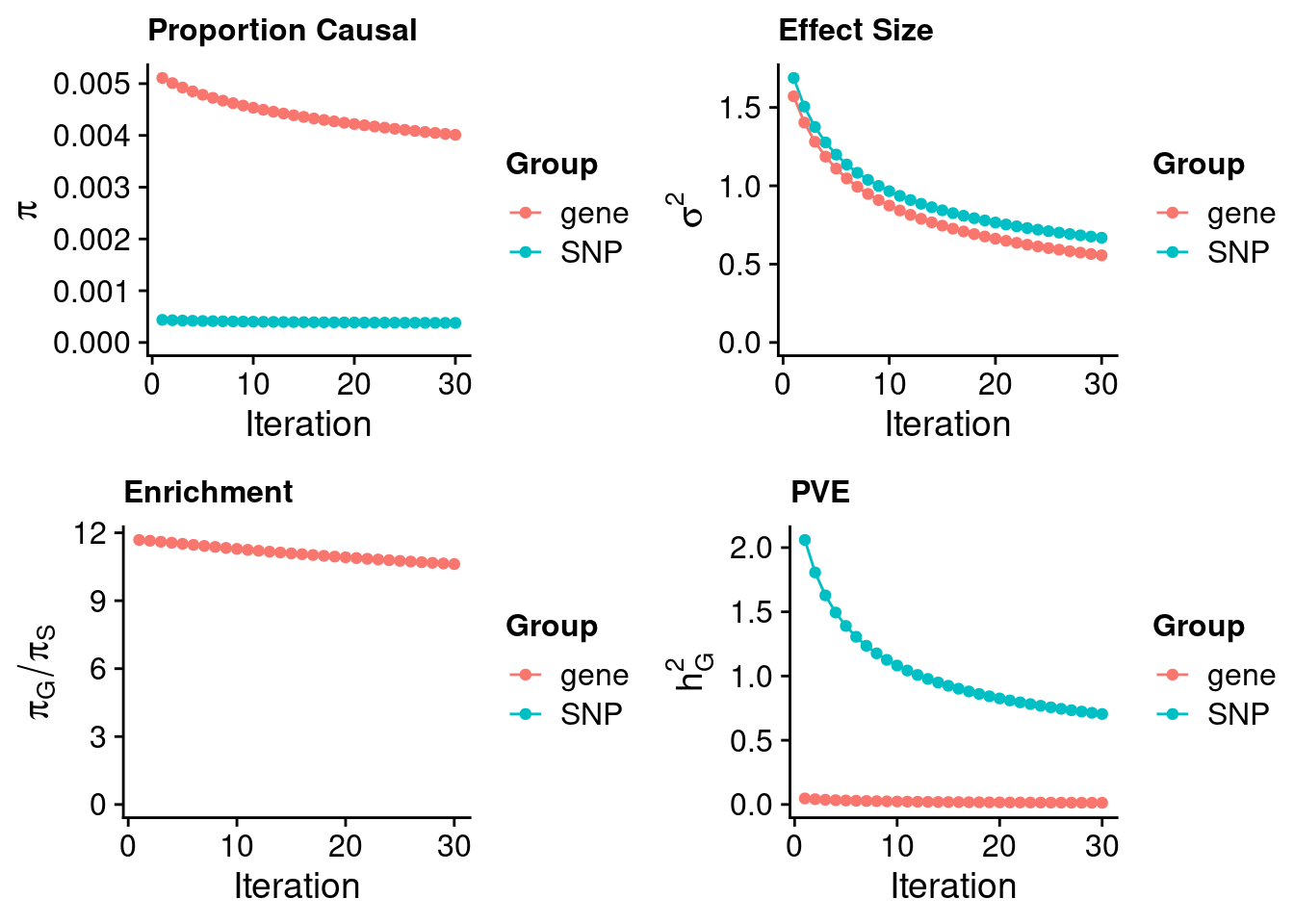
| Version | Author | Date |
|---|---|---|
| 6a15ad1 | sq-96 | 2023-10-14 |
head(ctwas_res[ctwas_res$type=="gene",]) chrom id pos type region_tag1 region_tag2 cs_index
114696 6 ENSG00000005700.14 82247064 gene 6 56 0
272419 22 ENSG00000159496.14 23679302 gene 22 6 0
179945 10 ENSG00000151923.17 119601669 gene 10 74 0
151888 8 ENSG00000164841.4 108787887 gene 8 74 0
241123 16 ENSG00000040199.18 71804965 gene 16 38 0
243700 17 ENSG00000174292.12 7380407 gene 17 6 0
susie_pip mu2 genename z
114696 0.11468988 9.126035 IBTK -3.787795
272419 0.08483873 7.971765 RGL4 3.286735
179945 0.08104538 7.839875 TIAL1 3.480318
151888 0.07962949 7.989882 TMEM74 -3.623924
241123 0.07790796 7.513540 PHLPP2 -3.126064
243700 0.07591772 7.535001 TNK1 3.186510Lung
#estimated prior inclusion probability
ctwas_parameters$group_prior SNP gene
0.0003778214 0.0040115252 #estimated prior effect size
ctwas_parameters$group_prior_var SNP gene
0.6684344 0.5565171 #estimated enrichment of genes over variants
ctwas_parameters$enrichment gene
10.61752 #PVE explained by genes and variants
ctwas_parameters$group_pve SNP gene
0.70439414 0.01309908 #total heritability (sum of PVE)
ctwas_parameters$total_pve[1] 0.7174932#attributable heritability
ctwas_parameters$attributable_pve SNP gene
0.98174327 0.01825673 #plot convergence
ctwas_parameters$convergence_plot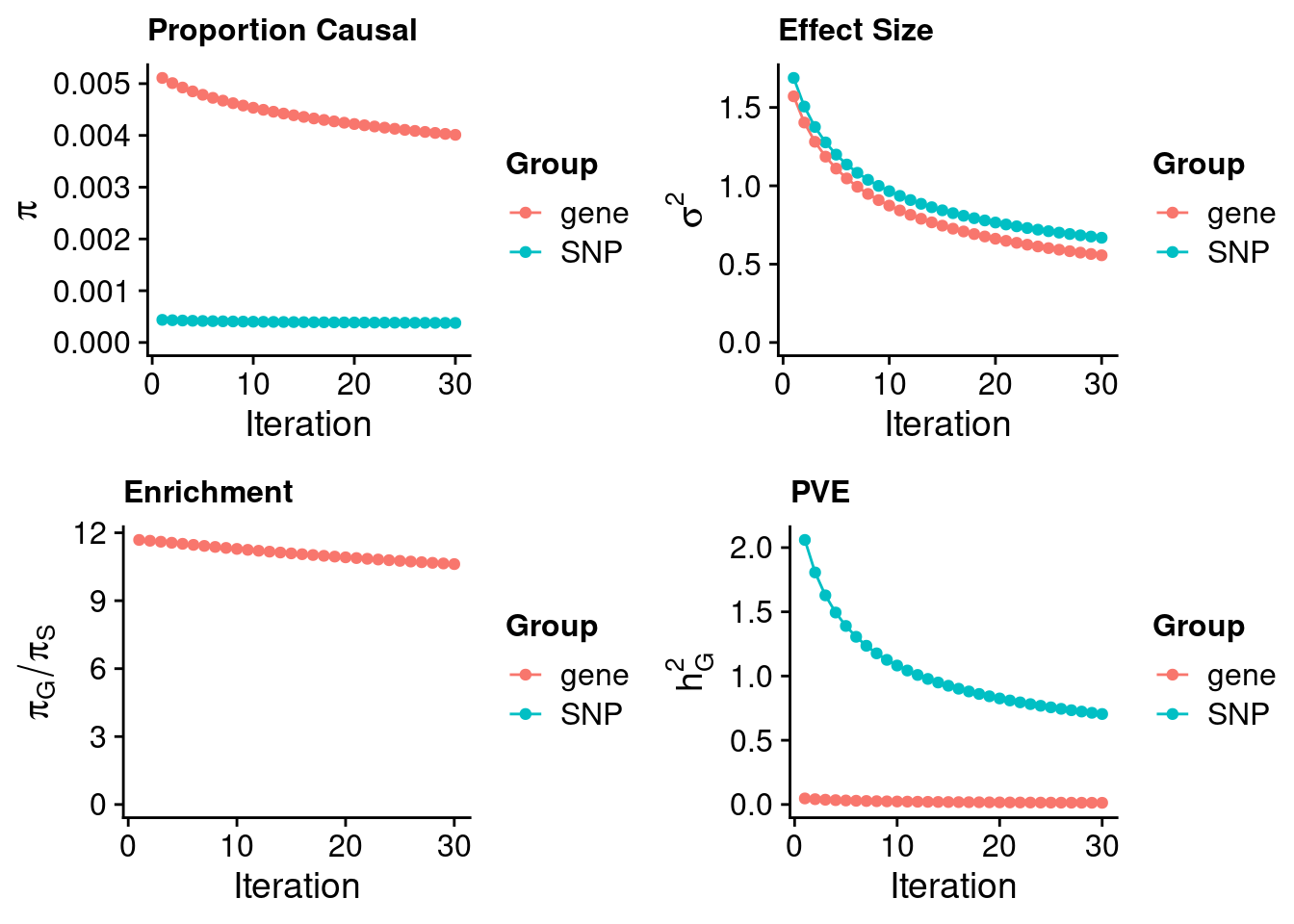
| Version | Author | Date |
|---|---|---|
| 6a15ad1 | sq-96 | 2023-10-14 |
head(ctwas_res[ctwas_res$type=="gene",]) chrom id pos type region_tag1 region_tag2 cs_index
114696 6 ENSG00000005700.14 82247064 gene 6 56 0
272419 22 ENSG00000159496.14 23679302 gene 22 6 0
179945 10 ENSG00000151923.17 119601669 gene 10 74 0
151888 8 ENSG00000164841.4 108787887 gene 8 74 0
241123 16 ENSG00000040199.18 71804965 gene 16 38 0
243700 17 ENSG00000174292.12 7380407 gene 17 6 0
susie_pip mu2 genename z
114696 0.11468988 9.126035 IBTK -3.787795
272419 0.08483873 7.971765 RGL4 3.286735
179945 0.08104538 7.839875 TIAL1 3.480318
151888 0.07962949 7.989882 TMEM74 -3.623924
241123 0.07790796 7.513540 PHLPP2 -3.126064
243700 0.07591772 7.535001 TNK1 3.186510Pancreas
#estimated prior inclusion probability
ctwas_parameters$group_prior SNP gene
0.0003778214 0.0040115252 #estimated prior effect size
ctwas_parameters$group_prior_var SNP gene
0.6684344 0.5565171 #estimated enrichment of genes over variants
ctwas_parameters$enrichment gene
10.61752 #PVE explained by genes and variants
ctwas_parameters$group_pve SNP gene
0.70439414 0.01309908 #total heritability (sum of PVE)
ctwas_parameters$total_pve[1] 0.7174932#attributable heritability
ctwas_parameters$attributable_pve SNP gene
0.98174327 0.01825673 #plot convergence
ctwas_parameters$convergence_plot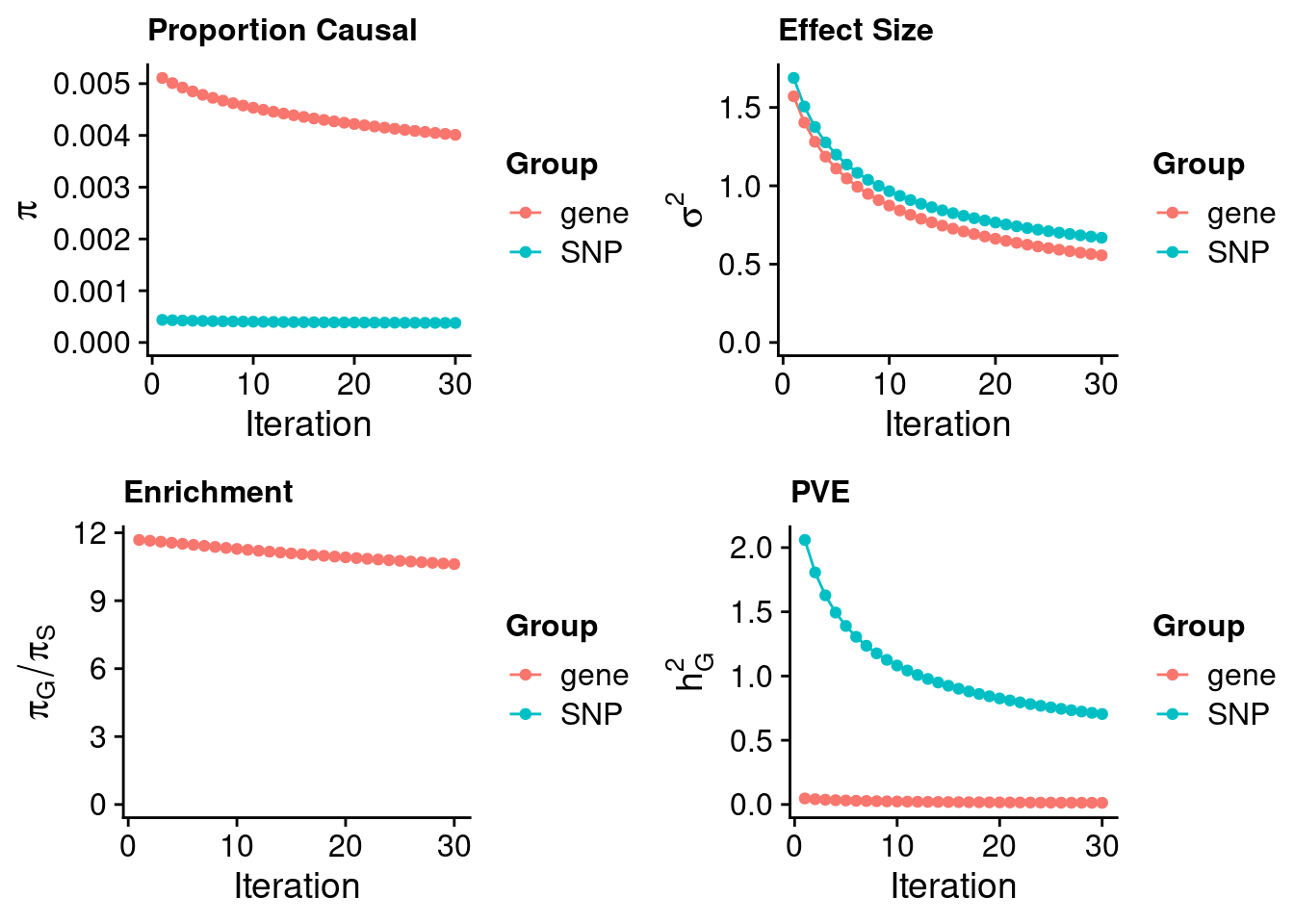
| Version | Author | Date |
|---|---|---|
| 6a15ad1 | sq-96 | 2023-10-14 |
head(ctwas_res[ctwas_res$type=="gene",]) chrom id pos type region_tag1 region_tag2 cs_index
114696 6 ENSG00000005700.14 82247064 gene 6 56 0
272419 22 ENSG00000159496.14 23679302 gene 22 6 0
179945 10 ENSG00000151923.17 119601669 gene 10 74 0
151888 8 ENSG00000164841.4 108787887 gene 8 74 0
241123 16 ENSG00000040199.18 71804965 gene 16 38 0
243700 17 ENSG00000174292.12 7380407 gene 17 6 0
susie_pip mu2 genename z
114696 0.11468988 9.126035 IBTK -3.787795
272419 0.08483873 7.971765 RGL4 3.286735
179945 0.08104538 7.839875 TIAL1 3.480318
151888 0.07962949 7.989882 TMEM74 -3.623924
241123 0.07790796 7.513540 PHLPP2 -3.126064
243700 0.07591772 7.535001 TNK1 3.186510Whole_Blood
#estimated prior inclusion probability
ctwas_parameters$group_prior SNP gene
0.0003778214 0.0040115252 #estimated prior effect size
ctwas_parameters$group_prior_var SNP gene
0.6684344 0.5565171 #estimated enrichment of genes over variants
ctwas_parameters$enrichment gene
10.61752 #PVE explained by genes and variants
ctwas_parameters$group_pve SNP gene
0.70439414 0.01309908 #total heritability (sum of PVE)
ctwas_parameters$total_pve[1] 0.7174932#attributable heritability
ctwas_parameters$attributable_pve SNP gene
0.98174327 0.01825673 #plot convergence
ctwas_parameters$convergence_plot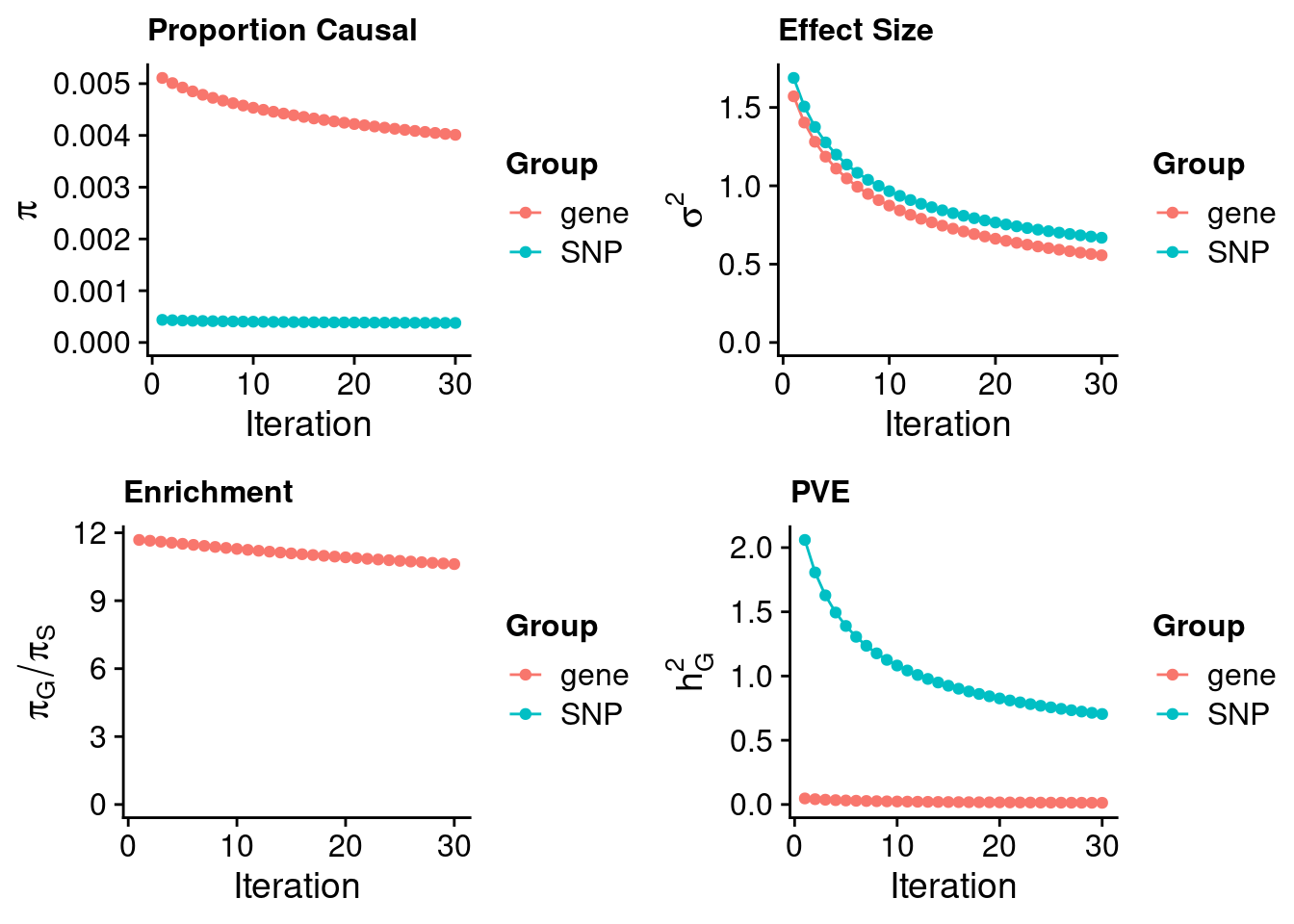
| Version | Author | Date |
|---|---|---|
| 6a15ad1 | sq-96 | 2023-10-14 |
head(ctwas_res[ctwas_res$type=="gene",]) chrom id pos type region_tag1 region_tag2 cs_index
114696 6 ENSG00000005700.14 82247064 gene 6 56 0
272419 22 ENSG00000159496.14 23679302 gene 22 6 0
179945 10 ENSG00000151923.17 119601669 gene 10 74 0
151888 8 ENSG00000164841.4 108787887 gene 8 74 0
241123 16 ENSG00000040199.18 71804965 gene 16 38 0
243700 17 ENSG00000174292.12 7380407 gene 17 6 0
susie_pip mu2 genename z
114696 0.11468988 9.126035 IBTK -3.787795
272419 0.08483873 7.971765 RGL4 3.286735
179945 0.08104538 7.839875 TIAL1 3.480318
151888 0.07962949 7.989882 TMEM74 -3.623924
241123 0.07790796 7.513540 PHLPP2 -3.126064
243700 0.07591772 7.535001 TNK1 3.186510
sessionInfo()R version 4.1.0 (2021-05-18)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] cowplot_1.1.1 ggplot2_3.4.0 ctwas_0.1.40 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] Rcpp_1.0.9 lattice_0.20-44 getPass_0.2-2 ps_1.7.2
[5] assertthat_0.2.1 rprojroot_2.0.3 digest_0.6.31 foreach_1.5.2
[9] utf8_1.2.2 R6_2.5.1 RSQLite_2.2.19 evaluate_0.19
[13] httr_1.4.4 highr_0.9 pillar_1.8.1 rlang_1.1.1
[17] rstudioapi_0.14 data.table_1.14.6 blob_1.2.3 whisker_0.4.1
[21] callr_3.7.3 jquerylib_0.1.4 Matrix_1.3-3 rmarkdown_2.19
[25] labeling_0.4.2 stringr_1.5.0 bit_4.0.5 munsell_0.5.0
[29] compiler_4.1.0 httpuv_1.6.7 xfun_0.35 pkgconfig_2.0.3
[33] htmltools_0.5.4 tidyselect_1.2.0 tibble_3.1.8 logging_0.10-108
[37] codetools_0.2-18 fansi_1.0.3 dplyr_1.0.10 withr_2.5.0
[41] later_1.3.0 grid_4.1.0 jsonlite_1.8.4 gtable_0.3.1
[45] lifecycle_1.0.3 DBI_1.1.3 git2r_0.30.1 magrittr_2.0.3
[49] scales_1.2.1 cli_3.6.1 stringi_1.7.8 cachem_1.0.6
[53] farver_2.1.0 fs_1.5.2 promises_1.2.0.1 pgenlibr_0.3.2
[57] bslib_0.4.1 vctrs_0.6.3 generics_0.1.3 iterators_1.0.14
[61] tools_4.1.0 bit64_4.0.5 glue_1.6.2 processx_3.8.0
[65] fastmap_1.1.0 yaml_2.3.6 colorspace_2.0-3 memoise_2.0.1
[69] knitr_1.41 sass_0.4.4