SCZ 2018 - Brain_Hypothalamus
sheng Qian
2021-2-6
Last updated: 2022-05-19
Checks: 5 2
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/ | data |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/ctwas_config.R | code/ctwas_config.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 7d08c9b. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .ipynb_checkpoints/
Untracked files:
Untracked: G_list.RData
Untracked: Rplot.png
Untracked: SCZ_annotation.xlsx
Untracked: analysis/.ipynb_checkpoints/
Untracked: analysis/ttt.Rmd
Untracked: code/.ipynb_checkpoints/
Untracked: code/AF_out/
Untracked: code/Autism_out/
Untracked: code/BMI_S_out/
Untracked: code/BMI_out/
Untracked: code/Glucose_out/
Untracked: code/LDL_S_out/
Untracked: code/SCZ_2014_EUR_out/
Untracked: code/SCZ_2018_S_out/
Untracked: code/SCZ_2018_out/
Untracked: code/SCZ_2020_Single_out/
Untracked: code/SCZ_2020_out/
Untracked: code/SCZ_S_out/
Untracked: code/SCZ_out/
Untracked: code/T2D_out/
Untracked: code/ctwas_config.R
Untracked: code/mapping.R
Untracked: code/out/
Untracked: code/process_scz_2018_snps.R
Untracked: code/run_AF_analysis.sbatch
Untracked: code/run_AF_analysis.sh
Untracked: code/run_AF_ctwas_rss_LDR.R
Untracked: code/run_Autism_analysis.sbatch
Untracked: code/run_Autism_analysis.sh
Untracked: code/run_Autism_ctwas_rss_LDR.R
Untracked: code/run_BMI_analysis.sbatch
Untracked: code/run_BMI_analysis.sh
Untracked: code/run_BMI_analysis_S.sbatch
Untracked: code/run_BMI_analysis_S.sh
Untracked: code/run_BMI_ctwas_rss_LDR.R
Untracked: code/run_BMI_ctwas_rss_LDR_S.R
Untracked: code/run_Glucose_analysis.sbatch
Untracked: code/run_Glucose_analysis.sh
Untracked: code/run_Glucose_ctwas_rss_LDR.R
Untracked: code/run_LDL_analysis_S.sbatch
Untracked: code/run_LDL_analysis_S.sh
Untracked: code/run_LDL_ctwas_rss_LDR_S.R
Untracked: code/run_SCZ_2014_EUR_analysis.sbatch
Untracked: code/run_SCZ_2014_EUR_analysis.sh
Untracked: code/run_SCZ_2014_EUR_ctwas_rss_LDR.R
Untracked: code/run_SCZ_2018_analysis.sbatch
Untracked: code/run_SCZ_2018_analysis.sh
Untracked: code/run_SCZ_2018_analysis_S.sbatch
Untracked: code/run_SCZ_2018_analysis_S.sh
Untracked: code/run_SCZ_2018_ctwas_rss_LDR.R
Untracked: code/run_SCZ_2018_ctwas_rss_LDR_S.R
Untracked: code/run_SCZ_2020_Single_analysis.sbatch
Untracked: code/run_SCZ_2020_Single_analysis.sh
Untracked: code/run_SCZ_2020_Single_ctwas_rss_LDR.R
Untracked: code/run_SCZ_2020_analysis.sbatch
Untracked: code/run_SCZ_2020_analysis.sh
Untracked: code/run_SCZ_2020_ctwas_rss_LDR.R
Untracked: code/run_SCZ_analysis.sbatch
Untracked: code/run_SCZ_analysis.sh
Untracked: code/run_SCZ_analysis_S.sbatch
Untracked: code/run_SCZ_analysis_S.sh
Untracked: code/run_SCZ_ctwas_rss_LDR.R
Untracked: code/run_SCZ_ctwas_rss_LDR_S.R
Untracked: code/run_T2D_analysis.sbatch
Untracked: code/run_T2D_analysis.sh
Untracked: code/run_T2D_ctwas_rss_LDR.R
Untracked: code/wflow_build.R
Untracked: code/wflow_build.sbatch
Untracked: data/.ipynb_checkpoints/
Untracked: data/GO_Terms/
Untracked: data/PGC3_SCZ_wave3_public.v2.tsv
Untracked: data/SCZ/
Untracked: data/SCZ_2014_EUR/
Untracked: data/SCZ_2018/
Untracked: data/SCZ_2018_S/
Untracked: data/SCZ_2020/
Untracked: data/SCZ_S/
Untracked: data/Supplementary Table 15 - MAGMA.xlsx
Untracked: data/Supplementary Table 20 - Prioritised Genes.xlsx
Untracked: data/T2D/
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/mashr_sqtl/
Untracked: data/scz_2018.RDS
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: data/untitled.txt
Untracked: top_genes_32.txt
Untracked: top_genes_37.txt
Untracked: top_genes_43.txt
Untracked: top_genes_81.txt
Untracked: z_snp_pos_SCZ.RData
Untracked: z_snp_pos_SCZ_2014_EUR.RData
Untracked: z_snp_pos_SCZ_2018.RData
Untracked: z_snp_pos_SCZ_2020.RData
Unstaged changes:
Deleted: analysis/BMI_S_results.Rmd
Modified: analysis/SCZ_2018_Brain_Amygdala_S.Rmd
Modified: analysis/SCZ_2018_Brain_Anterior_cingulate_cortex_BA24_S.Rmd
Modified: analysis/SCZ_2018_Brain_Caudate_basal_ganglia_S.Rmd
Modified: analysis/SCZ_2018_Brain_Cerebellar_Hemisphere_S.Rmd
Modified: analysis/SCZ_2018_Brain_Cerebellum_S.Rmd
Modified: analysis/SCZ_2018_Brain_Cortex_S.Rmd
Modified: analysis/SCZ_2018_Brain_Frontal_Cortex_BA9_S.Rmd
Modified: analysis/SCZ_2018_Brain_Hippocampus_S.Rmd
Modified: analysis/SCZ_2018_Brain_Hypothalamus_S.Rmd
Modified: analysis/SCZ_2018_Brain_Nucleus_accumbens_basal_ganglia_S.Rmd
Modified: analysis/SCZ_2018_Brain_Putamen_basal_ganglia_S.Rmd
Modified: analysis/SCZ_2018_Brain_Spinal_cord_cervical_c-1_S.Rmd
Modified: analysis/SCZ_2018_Brain_Substantia_nigra_S.Rmd
Modified: analysis/SCZ_Annotation_Analysis.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/SCZ_2018_Brain_Hypothalamus_S.Rmd) and HTML (docs/SCZ_2018_Brain_Hypothalamus_S.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 7d08c9b | sq-96 | 2022-05-18 | update |
| html | 7d08c9b | sq-96 | 2022-05-18 | update |
| Rmd | 2749be9 | sq-96 | 2022-05-12 | update |
| html | 2749be9 | sq-96 | 2022-05-12 | update |
| html | 011327d | sq-96 | 2022-05-12 | update |
| Rmd | 6c6abbd | sq-96 | 2022-05-12 | update |
library(reticulate)
use_python("/scratch/midway2/shengqian/miniconda3/envs/PythonForR/bin/python",required=T)Weight QC
#number of imputed weights
nrow(qclist_all)[1] 19902#number of imputed weights by chromosome
table(qclist_all$chr)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
1843 1406 1208 781 846 1042 1147 677 814 921 1170 1073 396 705 657 781
17 18 19 20 21 22
1382 282 1439 658 36 638 #number of imputed weights without missing variants
sum(qclist_all$nmiss==0)[1] 17601#proportion of imputed weights without missing variants
mean(qclist_all$nmiss==0)[1] 0.8844INFO:numexpr.utils:Note: NumExpr detected 56 cores but "NUMEXPR_MAX_THREADS" not set, so enforcing safe limit of 8.finish
Attaching package: 'dplyr'The following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
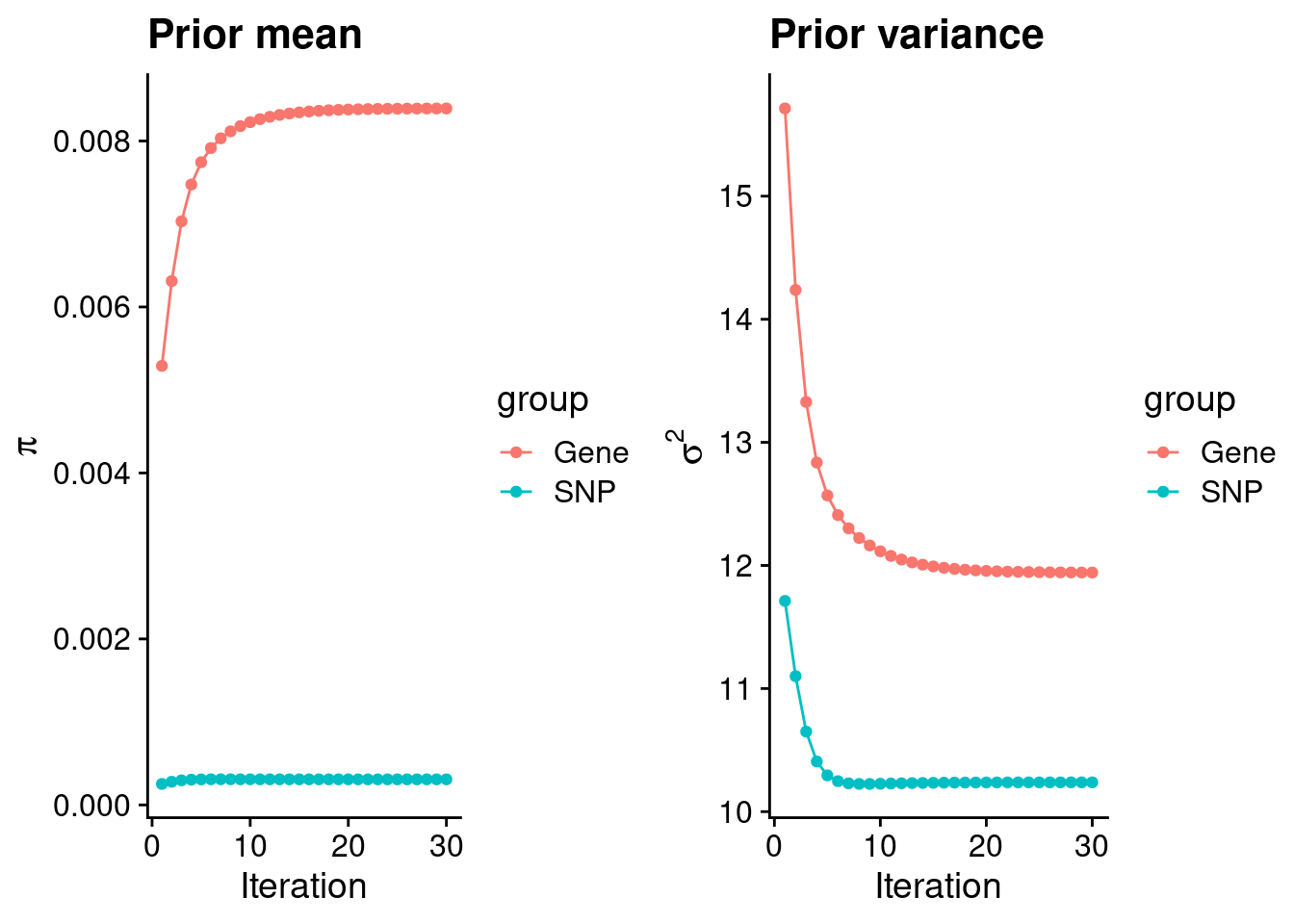
intersect, setdiff, setequal, unionCheck convergence of parameters
| Version | Author | Date |
|---|---|---|
| 2749be9 | sq-96 | 2022-05-12 |
gene snp
0.0083920 0.0003085 gene snp
11.94 10.24 [1] 105318[1] 7334 6309950 gene snp
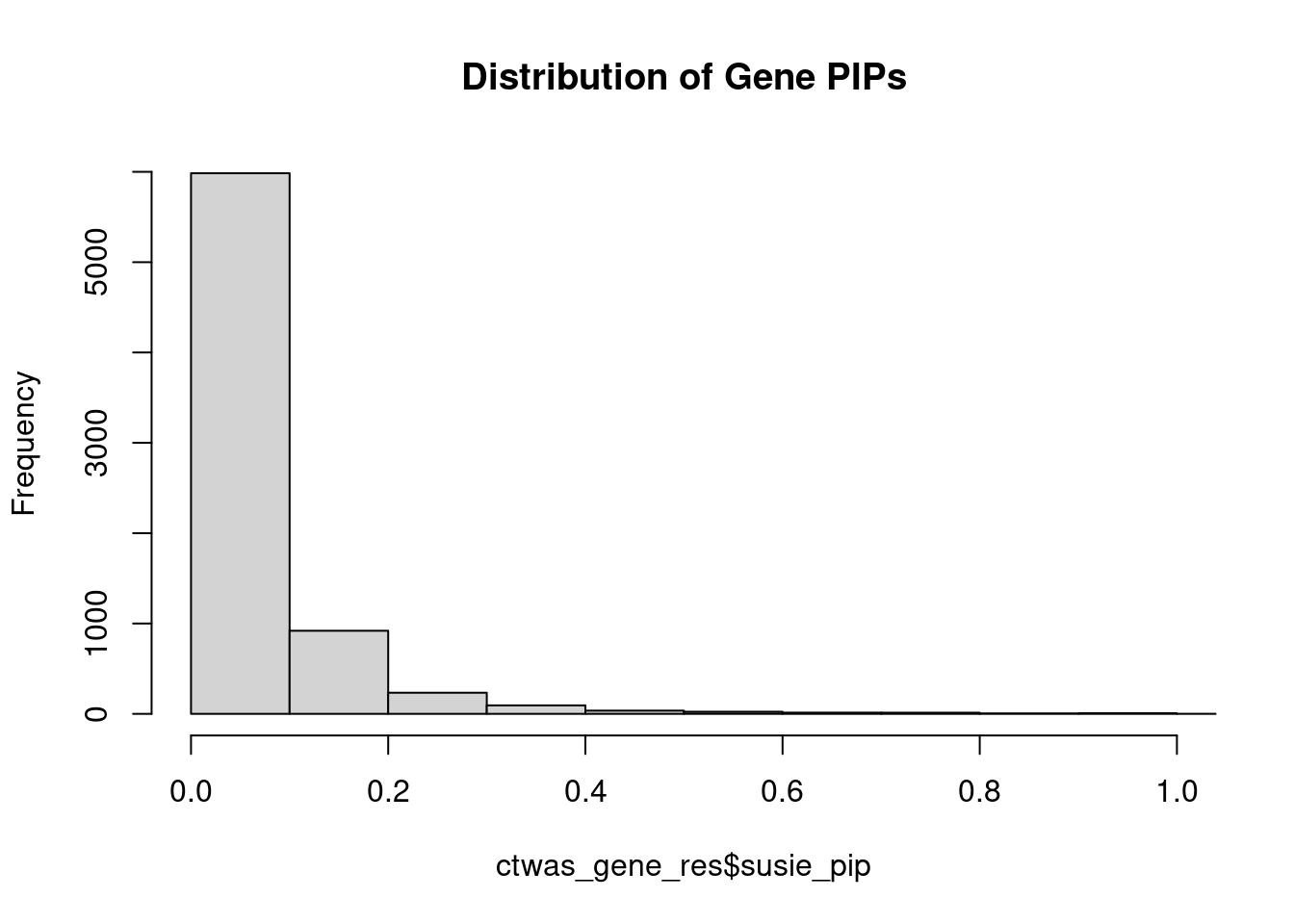
0.006979 0.189243 [1] 0.01838 1.05816Genes with highest PIPs
| Version | Author | Date |
|---|---|---|
| 2749be9 | sq-96 | 2022-05-12 |
genename region_tag susie_pip mu2 PVE z num_intron num_sqtl
4991 R3HDM2 12_36 1.1229 43.52 0.0004828 -6.634 4 4
2390 FEZF1 7_74 1.0238 24.62 0.0002438 -4.812 3 3
3351 LINC00320 21_6 0.9653 29.24 0.0002429 -5.336 5 5
3449 LRP8 1_33 0.9575 23.82 0.0002046 4.654 3 4
286 AKT3 1_128 0.9508 34.79 0.0002780 -6.291 8 8
2566 GIGYF2 2_137 0.9417 56.62 0.0004418 8.128 3 3
712 BDNF 11_19 0.9141 23.31 0.0001849 -4.348 2 2
4239 NRXN2 11_36 0.9073 24.81 0.0001918 4.723 3 3
7140 ZIC4 3_91 0.9031 23.46 0.0001755 -4.221 3 4
6335 THAP8 19_25 0.8792 19.47 0.0001426 3.847 2 2
4951 PTPRF 1_27 0.8792 37.18 0.0002644 6.680 4 4
1545 CRTAP 3_24 0.8764 19.92 0.0001445 3.929 2 2
148 ACTR1B 2_57 0.8316 19.24 0.0001263 -3.978 4 4
2565 GIGYF1 7_63 0.8088 28.55 0.0001764 -5.266 3 3
3873 MRPS33 7_87 0.7742 23.44 0.0001275 -4.304 4 5
6663 TSNARE1 8_93 0.7719 28.75 0.0001581 5.782 7 10
1922 DPYSL3 5_86 0.7459 22.30 0.0001178 4.157 1 1
2181 ETF1 5_82 0.7438 33.82 0.0001776 6.112 1 1
5639 SF3B1 2_117 0.7398 45.62 0.0002320 7.053 2 2
6848 UQCRC2 16_19 0.7381 22.09 0.0001143 4.716 2 2Genes with highest PVE
genename region_tag susie_pip mu2 PVE z num_intron num_sqtl
4991 R3HDM2 12_36 1.1229 43.52 0.0004828 -6.634 4 4
2566 GIGYF2 2_137 0.9417 56.62 0.0004418 8.128 3 3
286 AKT3 1_128 0.9508 34.79 0.0002780 -6.291 8 8
4951 PTPRF 1_27 0.8792 37.18 0.0002644 6.680 4 4
2390 FEZF1 7_74 1.0238 24.62 0.0002438 -4.812 3 3
3351 LINC00320 21_6 0.9653 29.24 0.0002429 -5.336 5 5
5639 SF3B1 2_117 0.7398 45.62 0.0002320 7.053 2 2
442 APOPT1 14_54 0.6940 46.11 0.0002066 7.429 4 7
3449 LRP8 1_33 0.9575 23.82 0.0002046 4.654 3 4
4239 NRXN2 11_36 0.9073 24.81 0.0001918 4.723 3 3
712 BDNF 11_19 0.9141 23.31 0.0001849 -4.348 2 2
2181 ETF1 5_82 0.7438 33.82 0.0001776 6.112 1 1
2565 GIGYF1 7_63 0.8088 28.55 0.0001764 -5.266 3 3
7140 ZIC4 3_91 0.9031 23.46 0.0001755 -4.221 3 4
6663 TSNARE1 8_93 0.7719 28.75 0.0001581 5.782 7 10
6221 TAOK2 16_24 0.6069 47.40 0.0001572 -7.024 5 5
1545 CRTAP 3_24 0.8764 19.92 0.0001445 3.929 2 2
6335 THAP8 19_25 0.8792 19.47 0.0001426 3.847 2 2
3873 MRPS33 7_87 0.7742 23.44 0.0001275 -4.304 4 5
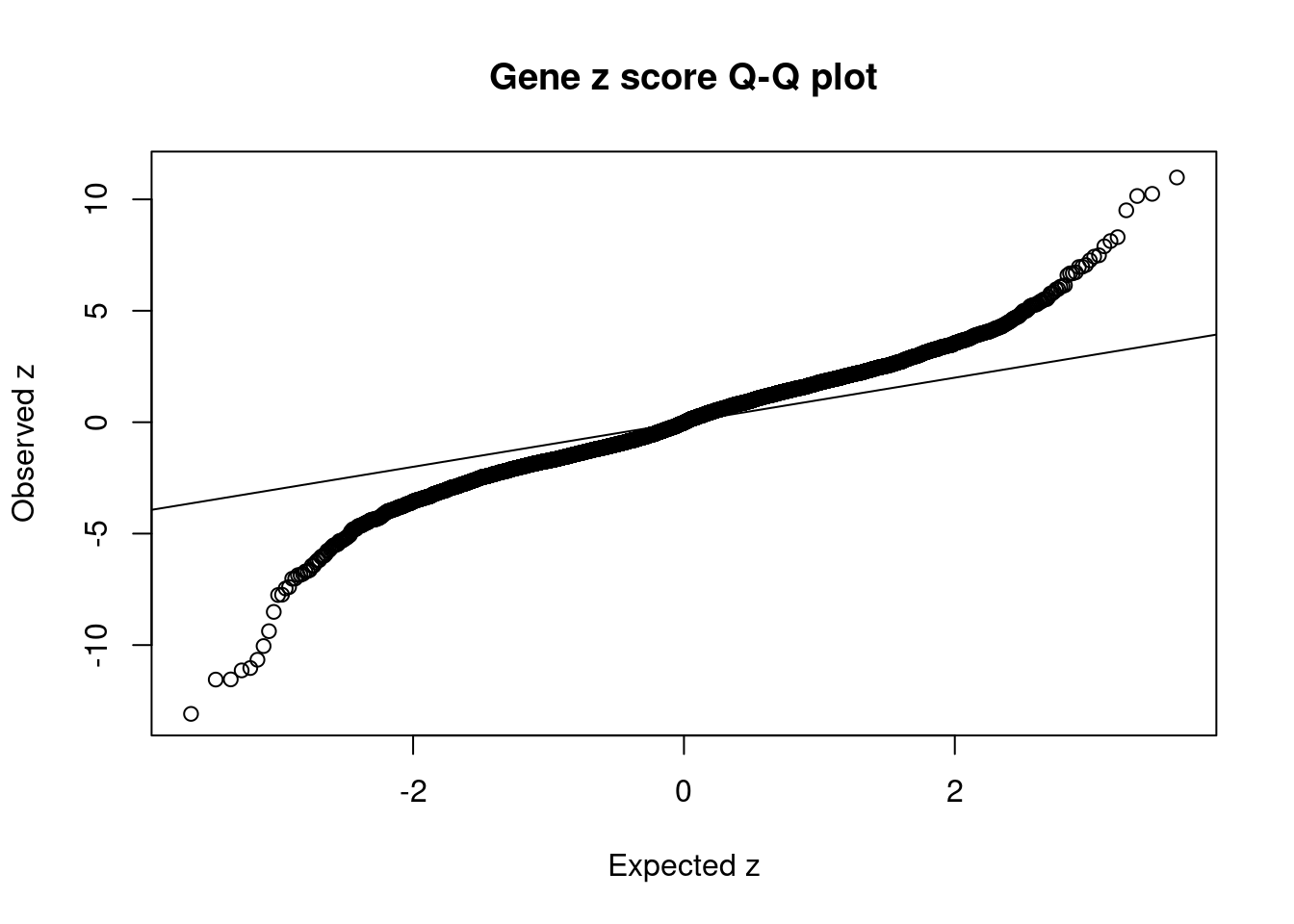
148 ACTR1B 2_57 0.8316 19.24 0.0001263 -3.978 4 4Comparing z scores and PIPs
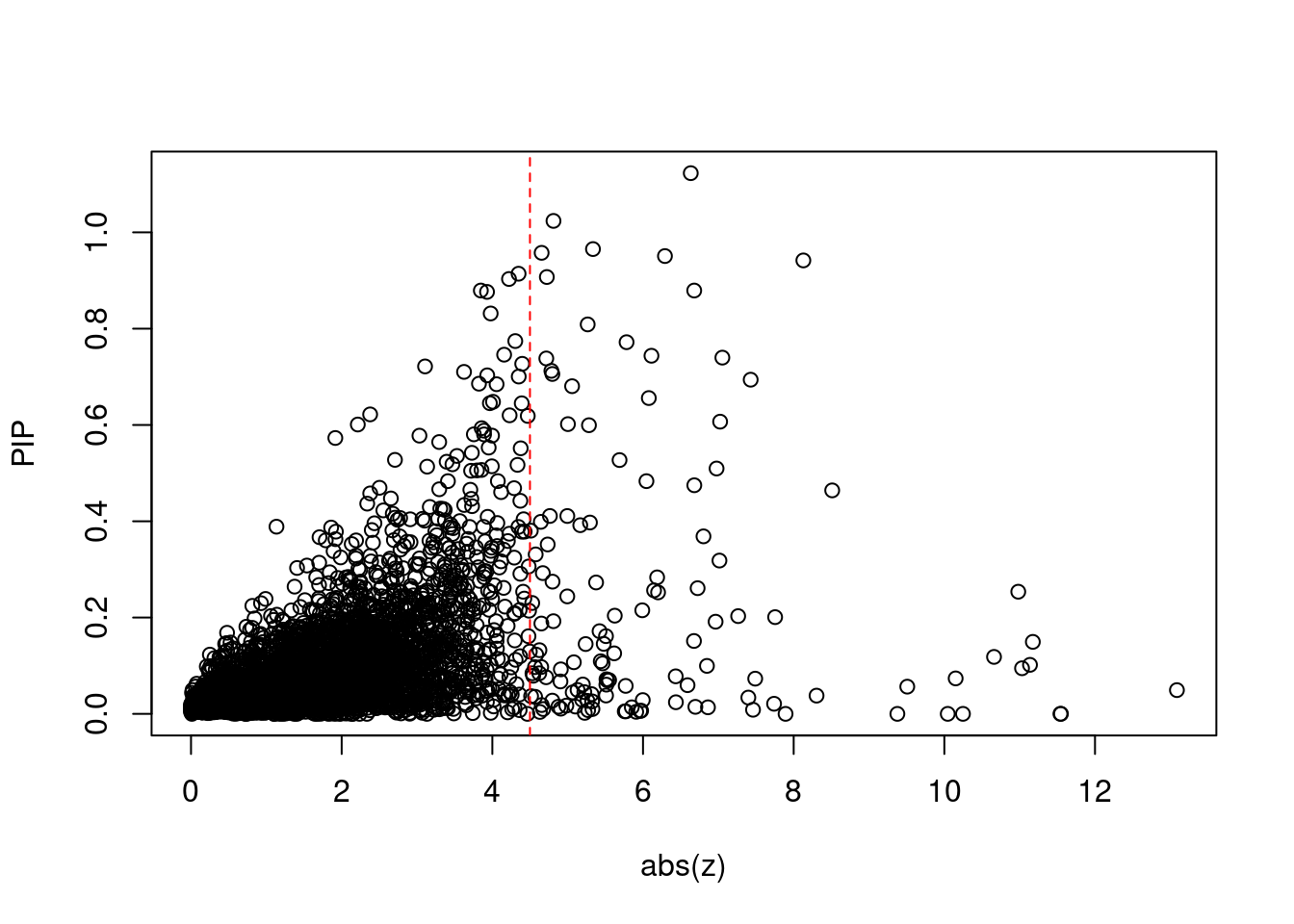
| Version | Author | Date |
|---|---|---|
| 2749be9 | sq-96 | 2022-05-12 |
| Version | Author | Date |
|---|---|---|
| 2749be9 | sq-96 | 2022-05-12 |
[1] 0.01773 genename region_tag susie_pip mu2 PVE z num_intron
4531 PGBD1 6_22 4.933e-02 161.09 1.444e-06 -13.087 2
6901 VARS 6_26 8.163e-05 217.29 1.375e-11 -11.548 1
441 APOM 6_26 8.357e-05 217.08 1.434e-11 -11.541 2
1747 DDR1 6_25 1.495e-01 101.78 2.106e-05 11.175 2
6902 VARS2 6_25 1.018e-01 100.66 9.907e-06 -11.137 1
914 C6orf136 6_24 9.472e-02 80.92 6.894e-06 -11.031 2
2421 FLOT1 6_24 2.537e-01 79.57 4.851e-05 10.981 7
781 BTN3A2 6_20 1.183e-01 91.37 4.454e-06 -10.659 6
668 BAG6 6_26 3.175e-05 166.08 1.360e-12 10.247 7
2849 HLA-B 6_25 7.365e-02 79.14 1.418e-06 10.150 10
5245 RNF5 6_26 2.893e-05 150.34 1.195e-12 -10.045 1
1123 CCHCR1 6_25 5.652e-02 66.51 1.192e-06 9.508 9
2682 GPSM3 6_26 1.971e-06 122.29 4.509e-15 -9.377 1
4257 NT5C2 10_66 4.641e-01 48.79 9.244e-05 -8.511 8
5383 RP5-874C20.8 6_22 3.788e-02 46.81 4.314e-07 8.304 4
2566 GIGYF2 2_137 9.417e-01 56.62 4.418e-04 8.128 3
3887 MSH5 6_26 1.908e-05 72.41 1.864e-13 7.892 3
814 C12orf65 12_75 2.009e-01 55.60 2.131e-05 -7.754 1
7141 ZKSCAN3 6_22 2.099e-02 36.54 8.960e-08 -7.740 2
780 BTN3A1 6_20 7.314e-02 47.48 7.925e-07 7.490 5
num_sqtl
4531 3
6901 1
441 2
1747 2
6902 1
914 2
2421 8
781 7
668 10
2849 19
5245 1
1123 13
2682 1
4257 12
5383 5
2566 3
3887 3
814 1
7141 2
780 5GO enrichment analysis for genes with PIP>0.5
#number of genes for gene set enrichment
length(genes)[1] 65Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"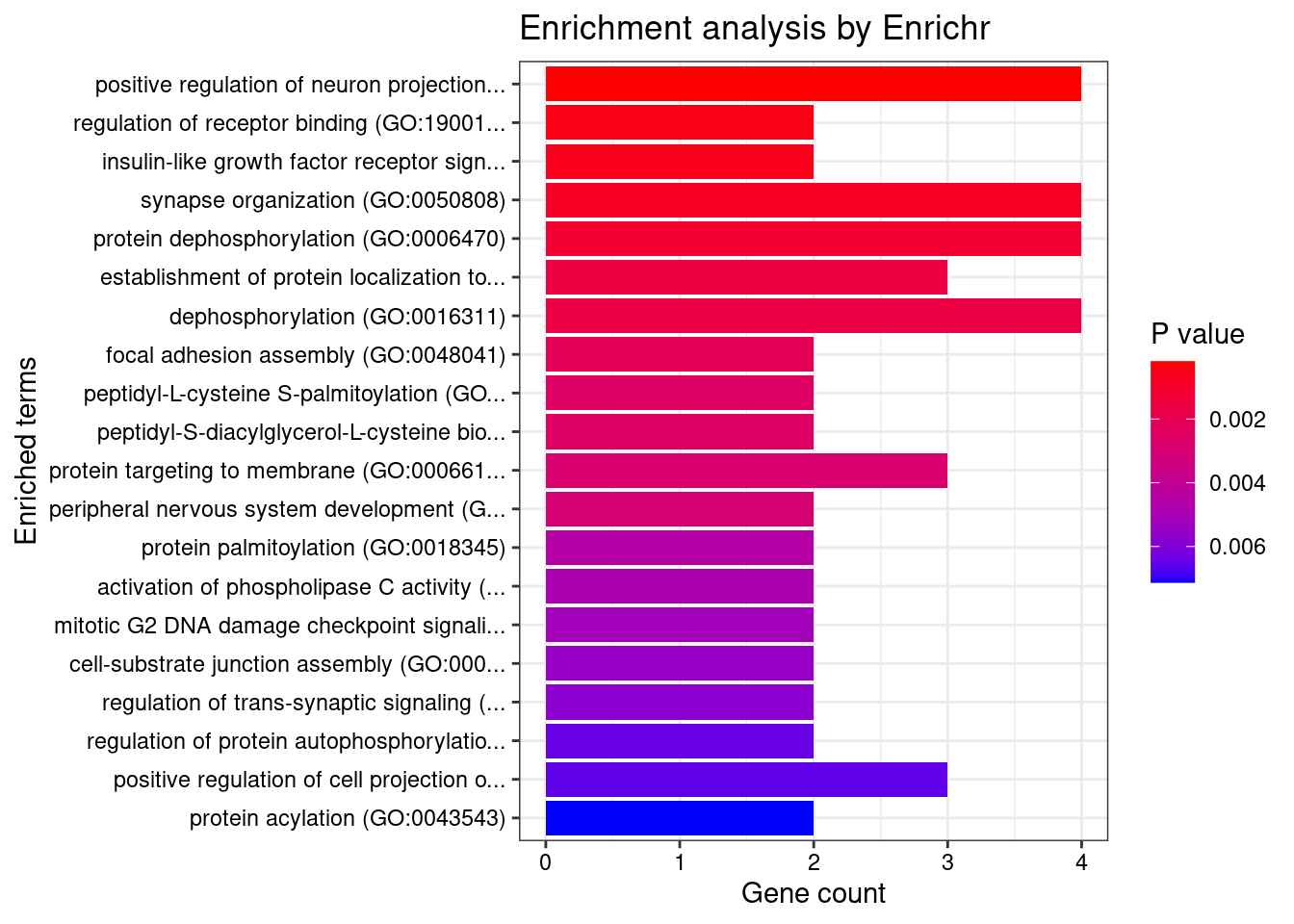
| Version | Author | Date |
|---|---|---|
| 2749be9 | sq-96 | 2022-05-12 |
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Cellular_Component_2021"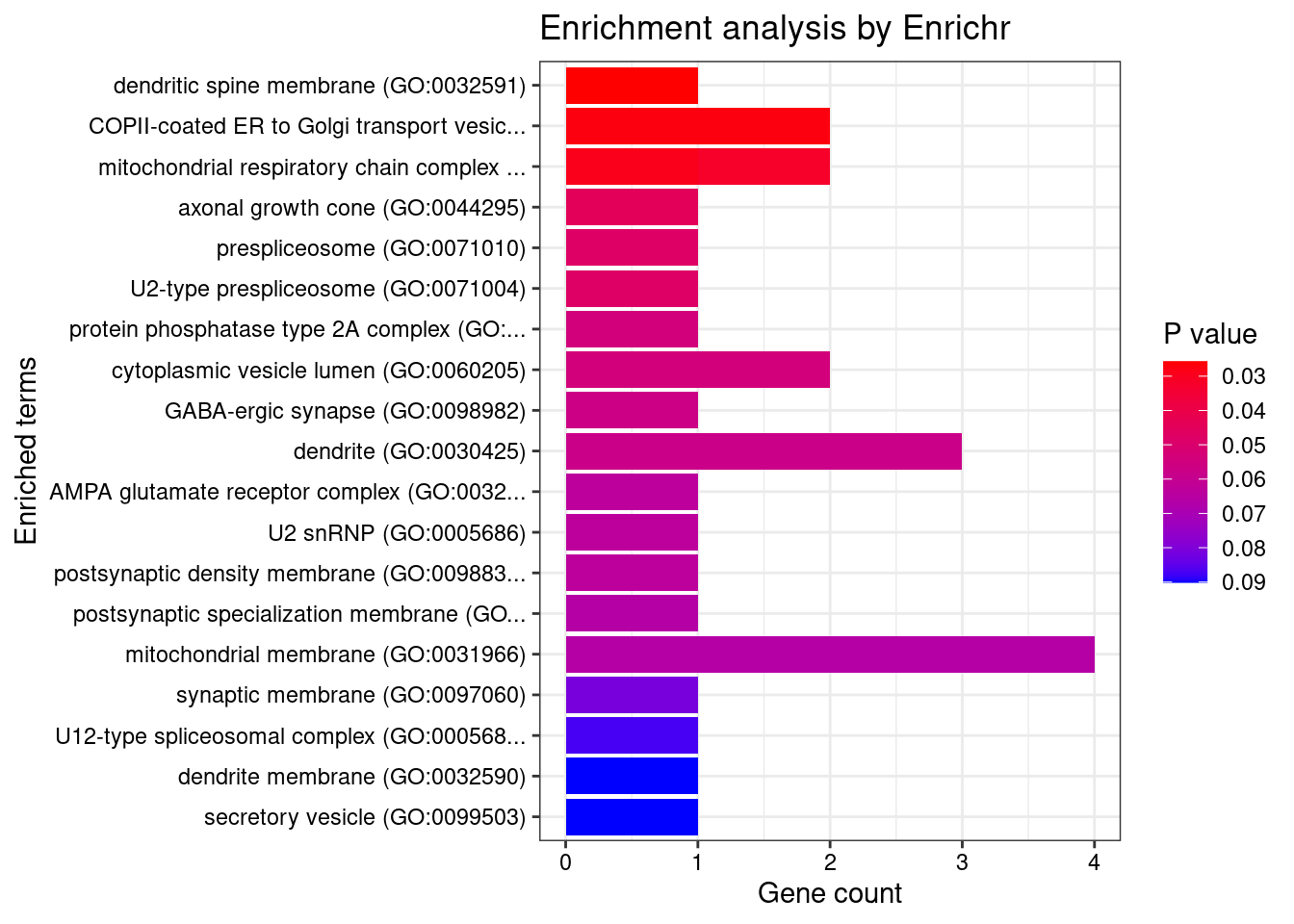
| Version | Author | Date |
|---|---|---|
| 2749be9 | sq-96 | 2022-05-12 |
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Molecular_Function_2021"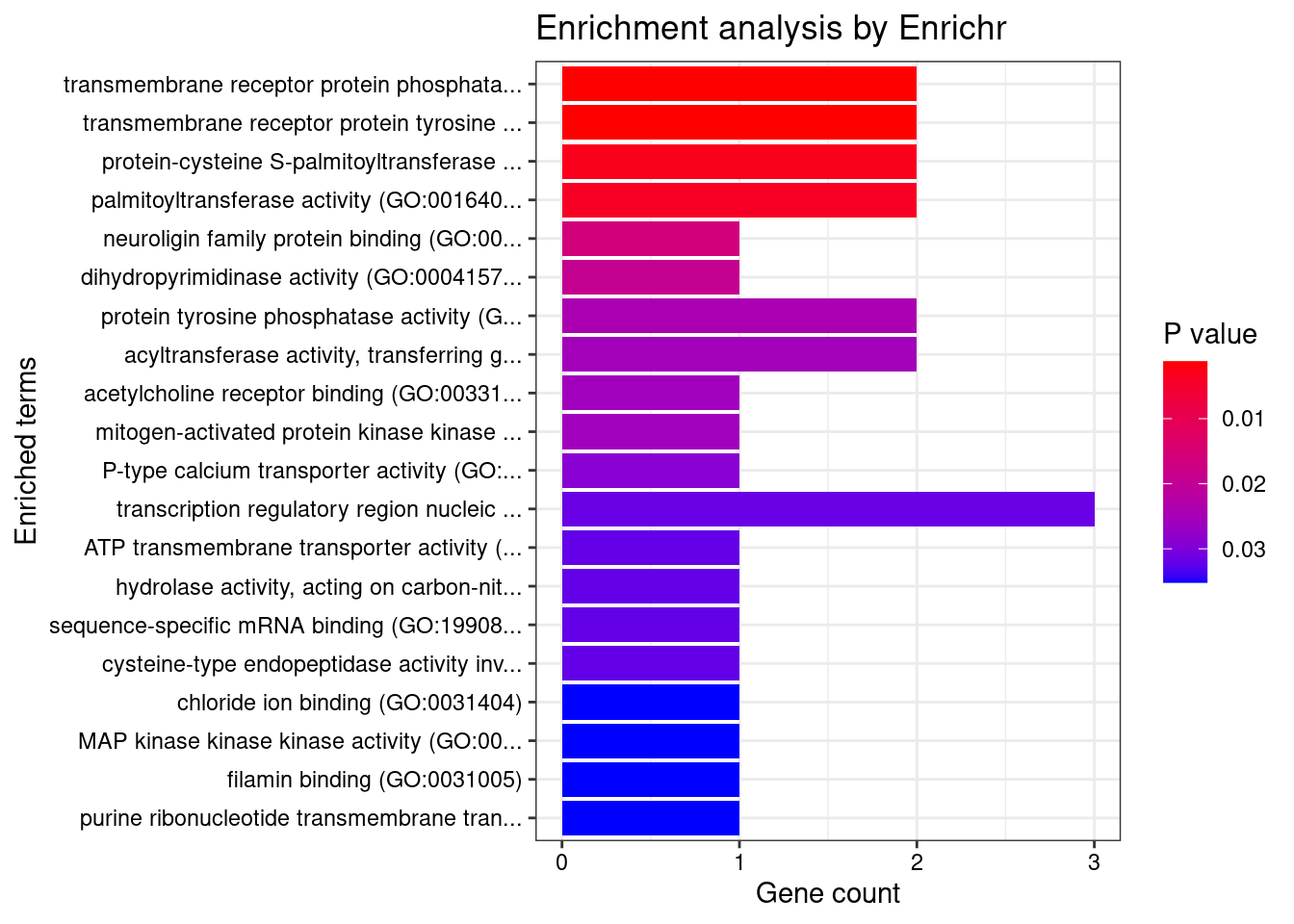
| Version | Author | Date |
|---|---|---|
| 2749be9 | sq-96 | 2022-05-12 |
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)DisGeNET enrichment analysis for genes with PIP>0.5
Description FDR Ratio
73 Schizophrenia 0.02013 11/32
49 Measles 0.03821 1/32
90 Electroencephalogram abnormal 0.03821 1/32
96 Congenital absent nipple 0.03821 1/32
155 Congenital absence of breast with absent nipple 0.03821 1/32
212 Sporadic Breast Carcinoma 0.03821 1/32
215 Primary peritoneal carcinoma 0.03821 1/32
221 Osteogenesis Imperfecta Type VII 0.03821 1/32
222 Familial encephalopathy with neuroserpin inclusion bodies 0.03821 1/32
228 BREAST-OVARIAN CANCER, FAMILIAL, SUSCEPTIBILITY TO, 1 0.03821 1/32
BgRatio
73 883/9703
49 1/9703
90 1/9703
96 1/9703
155 1/9703
212 1/9703
215 1/9703
221 1/9703
222 1/9703
228 1/9703WebGestalt enrichment analysis for genes with PIP>0.5
Warning: replacing previous import 'lifecycle::last_warnings' by
'rlang::last_warnings' when loading 'hms'Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum =
minNum, : No significant gene set is identified based on FDR 0.05!NULLPIP Manhattan Plot
Warning: ggrepel: 21 unlabeled data points (too many overlaps). Consider
increasing max.overlaps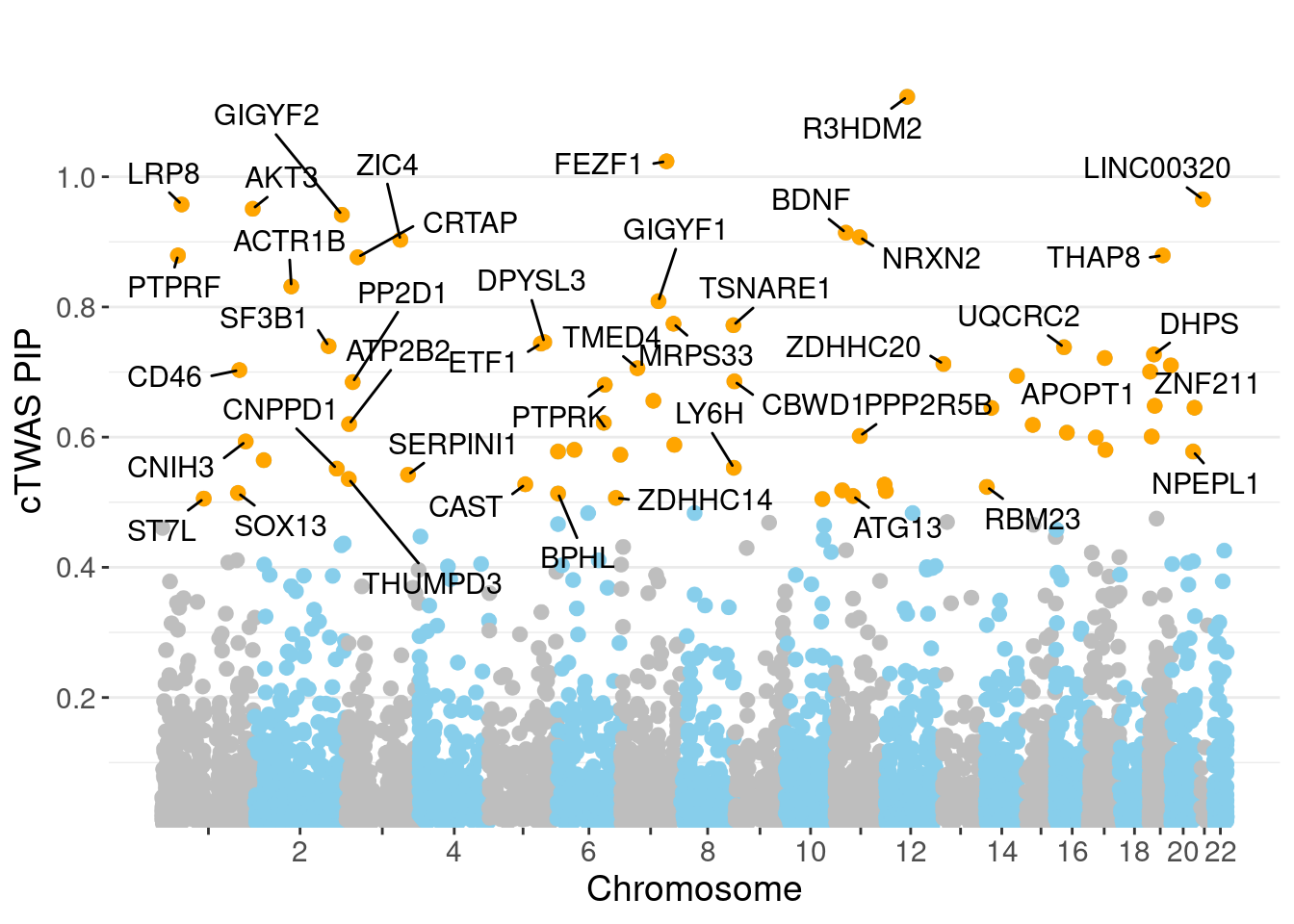
Sensitivity, specificity and precision for silver standard genes
#number of genes in known annotations
print(length(known_annotations))[1] 130#number of genes in known annotations with imputed expression
print(sum(known_annotations %in% ctwas_gene_res$genename))[1] 50#significance threshold for TWAS
print(sig_thresh)[1] 4.499#number of ctwas genes
length(ctwas_genes)[1] 14#number of TWAS genes
length(twas_genes)[1] 130#show novel genes (ctwas genes with not in TWAS genes)
ctwas_gene_res[ctwas_gene_res$genename %in% novel_genes,report_cols] genename region_tag susie_pip mu2 PVE z num_intron num_sqtl
148 ACTR1B 2_57 0.8316 19.24 0.0001263 -3.978 4 4
712 BDNF 11_19 0.9141 23.31 0.0001849 -4.348 2 2
1545 CRTAP 3_24 0.8764 19.92 0.0001445 3.929 2 2
6335 THAP8 19_25 0.8792 19.47 0.0001426 3.847 2 2
7140 ZIC4 3_91 0.9031 23.46 0.0001755 -4.221 3 4#sensitivity / recall
print(sensitivity) ctwas TWAS
0.03846 0.13077 #specificity
print(specificity) ctwas TWAS
0.9988 0.9845 #precision / PPV
print(precision) ctwas TWAS
0.3571 0.1308
sessionInfo()R version 4.1.0 (2021-05-18)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] readxl_1.4.0 forcats_0.5.1 stringr_1.4.0 purrr_0.3.4
[5] readr_1.4.0 tidyr_1.1.3 tidyverse_1.3.1 tibble_3.1.7
[9] WebGestaltR_0.4.4 disgenet2r_0.99.2 enrichR_3.0 cowplot_1.1.1
[13] ggplot2_3.3.5 dplyr_1.0.7 reticulate_1.25 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] fs_1.5.0 lubridate_1.7.10 doParallel_1.0.16 httr_1.4.2
[5] rprojroot_2.0.2 tools_4.1.0 backports_1.2.1 doRNG_1.8.2
[9] bslib_0.2.5.1 utf8_1.2.1 R6_2.5.0 vipor_0.4.5
[13] DBI_1.1.1 colorspace_2.0-2 withr_2.4.2 ggrastr_1.0.1
[17] tidyselect_1.1.1 processx_3.5.2 curl_4.3.2 compiler_4.1.0
[21] git2r_0.28.0 rvest_1.0.0 cli_3.0.0 Cairo_1.5-15
[25] xml2_1.3.2 labeling_0.4.2 sass_0.4.0 scales_1.1.1
[29] callr_3.7.0 systemfonts_1.0.4 apcluster_1.4.9 digest_0.6.27
[33] rmarkdown_2.9 svglite_2.0.0 pkgconfig_2.0.3 htmltools_0.5.1.1
[37] dbplyr_2.1.1 highr_0.9 rlang_1.0.2 rstudioapi_0.13
[41] jquerylib_0.1.4 farver_2.1.0 generics_0.1.0 jsonlite_1.7.2
[45] magrittr_2.0.1 Matrix_1.3-3 ggbeeswarm_0.6.0 Rcpp_1.0.7
[49] munsell_0.5.0 fansi_0.5.0 lifecycle_1.0.0 stringi_1.6.2
[53] whisker_0.4 yaml_2.2.1 plyr_1.8.6 grid_4.1.0
[57] ggrepel_0.9.1 parallel_4.1.0 promises_1.2.0.1 crayon_1.4.1
[61] lattice_0.20-44 haven_2.4.1 hms_1.1.0 knitr_1.33
[65] ps_1.6.0 pillar_1.7.0 igraph_1.2.6 rjson_0.2.20
[69] rngtools_1.5 reshape2_1.4.4 codetools_0.2-18 reprex_2.0.0
[73] glue_1.4.2 evaluate_0.14 getPass_0.2-2 modelr_0.1.8
[77] data.table_1.14.0 png_0.1-7 vctrs_0.3.8 httpuv_1.6.1
[81] foreach_1.5.1 cellranger_1.1.0 gtable_0.3.0 assertthat_0.2.1
[85] xfun_0.24 broom_0.7.8 later_1.2.0 iterators_1.0.13
[89] beeswarm_0.4.0 ellipsis_0.3.2 here_1.0.1