BMI - Brain Substantia nigra
sheng Qian
2021-2-6
Last updated: 2022-02-21
Checks: 6 1
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/ | data |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/ctwas_config.R | code/ctwas_config.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version bbf6737. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .ipynb_checkpoints/
Untracked files:
Untracked: Rplot.png
Untracked: analysis/Glucose_Adipose_Subcutaneous.Rmd
Untracked: analysis/Glucose_Adipose_Visceral_Omentum.Rmd
Untracked: analysis/Splicing_Test.Rmd
Untracked: code/.ipynb_checkpoints/
Untracked: code/AF_out/
Untracked: code/BMI_S_out/
Untracked: code/BMI_out/
Untracked: code/Glucose_out/
Untracked: code/LDL_S_out/
Untracked: code/T2D_out/
Untracked: code/ctwas_config.R
Untracked: code/mapping.R
Untracked: code/out/
Untracked: code/run_AF_analysis.sbatch
Untracked: code/run_AF_analysis.sh
Untracked: code/run_AF_ctwas_rss_LDR.R
Untracked: code/run_BMI_analysis.sbatch
Untracked: code/run_BMI_analysis.sh
Untracked: code/run_BMI_analysis_S.sbatch
Untracked: code/run_BMI_analysis_S.sh
Untracked: code/run_BMI_ctwas_rss_LDR.R
Untracked: code/run_BMI_ctwas_rss_LDR_S.R
Untracked: code/run_Glucose_analysis.sbatch
Untracked: code/run_Glucose_analysis.sh
Untracked: code/run_Glucose_ctwas_rss_LDR.R
Untracked: code/run_LDL_analysis_S.sbatch
Untracked: code/run_LDL_analysis_S.sh
Untracked: code/run_LDL_ctwas_rss_LDR_S.R
Untracked: code/run_T2D_analysis.sbatch
Untracked: code/run_T2D_analysis.sh
Untracked: code/run_T2D_ctwas_rss_LDR.R
Untracked: data/.ipynb_checkpoints/
Untracked: data/AF/
Untracked: data/BMI/
Untracked: data/BMI_S/
Untracked: data/Glucose/
Untracked: data/LDL_S/
Untracked: data/T2D/
Untracked: data/TEST/
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/mashr_sqtl/
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: data/untitled.txt
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/BMI_Brain_Substantia_nigra.Rmd) and HTML (docs/BMI_Brain_Substantia_nigra.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | bbf6737 | sq-96 | 2022-02-21 | update |
| html | 91f38fa | sq-96 | 2022-02-13 | Build site. |
| Rmd | eb13ecf | sq-96 | 2022-02-13 | update |
| html | e6bc169 | sq-96 | 2022-02-13 | Build site. |
| Rmd | 87fee8b | sq-96 | 2022-02-13 | update |
Weight QC
#number of imputed weights
nrow(qclist_all)[1] 10051#number of imputed weights by chromosome
table(qclist_all$chr)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
966 710 602 390 478 579 472 391 399 396 602 539 215 330 338 461 604 154 780 312
21 22
112 221 #number of imputed weights without missing variants
sum(qclist_all$nmiss==0)[1] 8301#proportion of imputed weights without missing variants
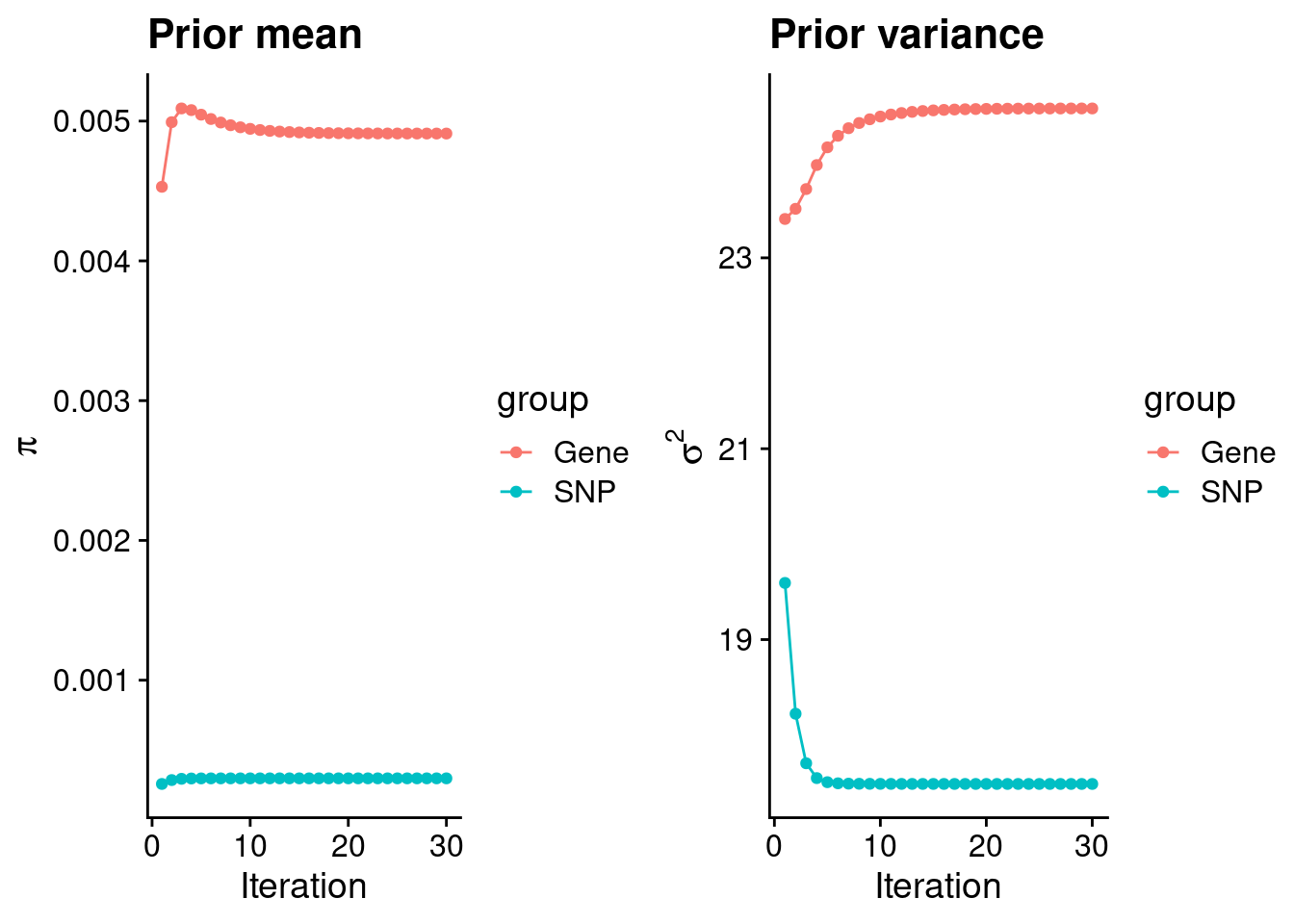
mean(qclist_all$nmiss==0)[1] 0.8259Check convergence of parameters
| Version | Author | Date |
|---|---|---|
| e6bc169 | sq-96 | 2022-02-13 |
#estimated group prior
estimated_group_prior <- group_prior_rec[,ncol(group_prior_rec)]
names(estimated_group_prior) <- c("gene", "snp")
estimated_group_prior["snp"] <- estimated_group_prior["snp"]*thin #adjust parameter to account for thin argument
print(estimated_group_prior) gene snp
0.0049107 0.0002978 #estimated group prior variance
estimated_group_prior_var <- group_prior_var_rec[,ncol(group_prior_var_rec)]
names(estimated_group_prior_var) <- c("gene", "snp")
print(estimated_group_prior_var) gene snp
24.57 17.49 #report sample size
print(sample_size)[1] 336107#report group size
group_size <- c(nrow(ctwas_gene_res), n_snps)
print(group_size)[1] 10051 7535010#estimated group PVE
estimated_group_pve <- estimated_group_prior_var*estimated_group_prior*group_size/sample_size #check PVE calculation
names(estimated_group_pve) <- c("gene", "snp")
print(estimated_group_pve) gene snp
0.003607 0.116746 #compare sum(PIP*mu2/sample_size) with above PVE calculation
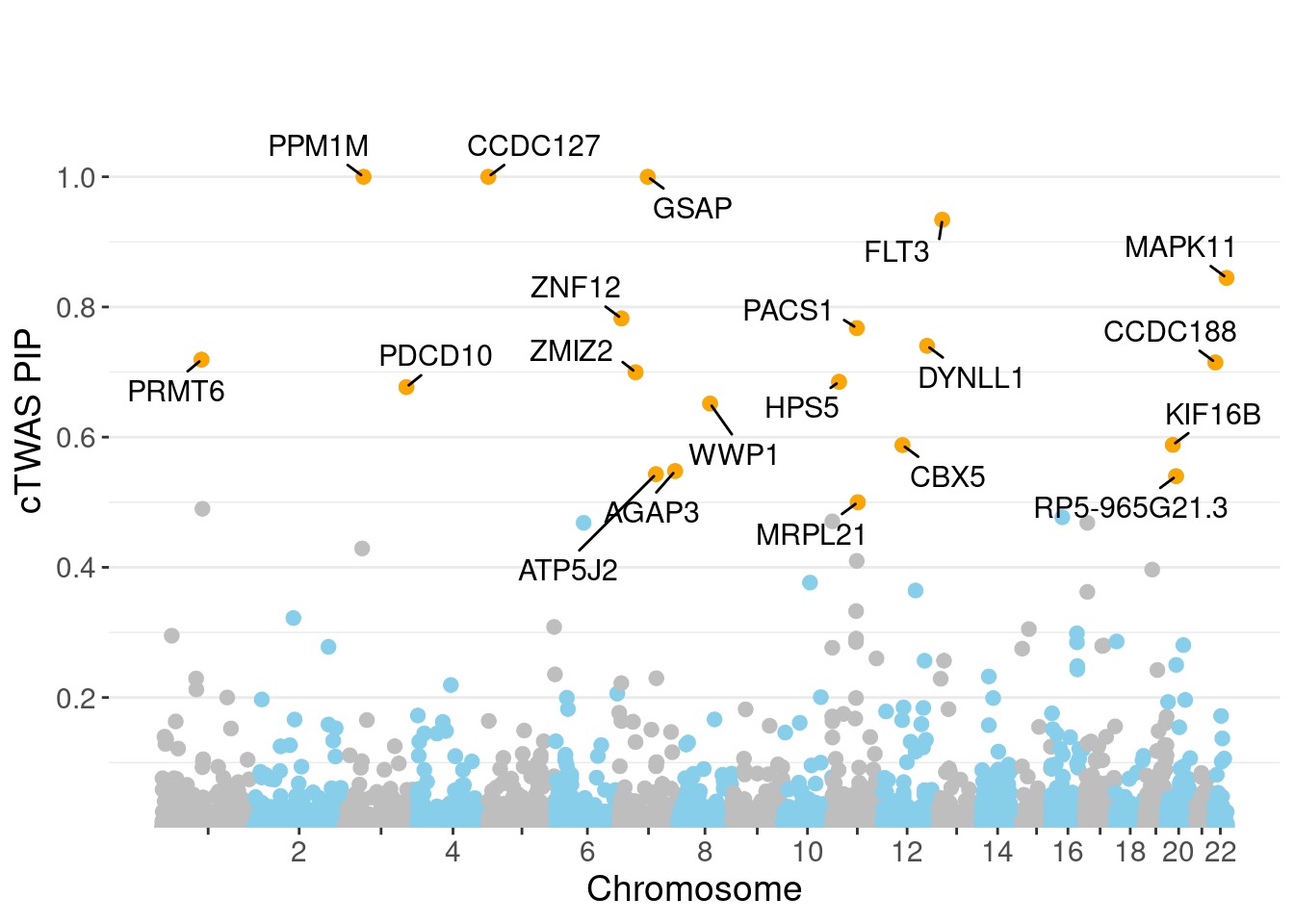
c(sum(ctwas_gene_res$PVE),sum(ctwas_snp_res$PVE))[1] 0.2472 16.0925Genes with highest PIPs
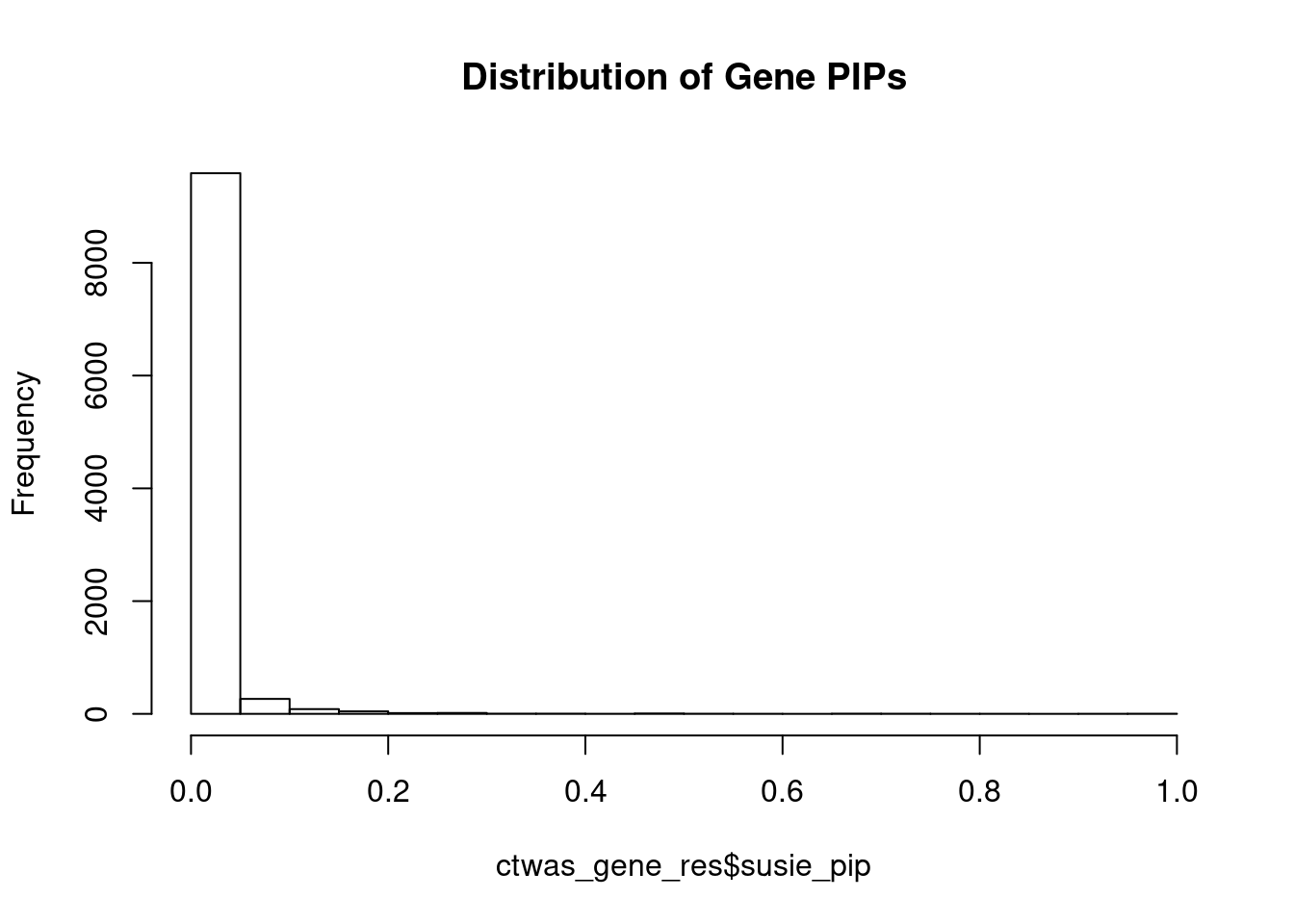
genename region_tag susie_pip mu2 PVE z num_eqtl
6994 CCDC127 5_1 1.0000 17290.64 5.144e-02 3.012 1
9381 GSAP 7_49 1.0000 33042.01 9.831e-02 5.260 1
6940 PPM1M 3_36 1.0000 883.39 2.628e-03 5.130 3
3273 FLT3 13_7 0.9340 33.54 9.319e-05 -5.360 1
9286 MAPK11 22_24 0.8447 26.72 6.716e-05 4.904 1
7028 ZNF12 7_9 0.7824 27.51 6.403e-05 5.114 2
8362 PACS1 11_36 0.7676 30.12 6.880e-05 5.121 2
1149 DYNLL1 12_74 0.7403 37.63 8.288e-05 -5.806 1
10169 PRMT6 1_66 0.7190 33.46 7.157e-05 5.528 1
11016 CCDC188 22_4 0.7147 25.33 5.386e-05 4.590 1
3302 ZMIZ2 7_33 0.6997 66.53 1.385e-04 -8.105 1
2386 HPS5 11_13 0.6850 25.31 5.159e-05 -4.584 2
2668 PDCD10 3_103 0.6769 24.04 4.841e-05 -4.059 2
3354 WWP1 8_61 0.6516 1124.54 2.180e-03 5.312 2
1166 KIF16B 20_12 0.5881 25.03 4.379e-05 -4.620 1
1275 CBX5 12_33 0.5878 25.63 4.483e-05 -4.691 1
4143 AGAP3 7_94 0.5479 26.84 4.375e-05 -5.031 2
11192 ATP5J2 7_61 0.5431 53.47 8.640e-05 -7.117 1
12531 RP5-965G21.3 20_19 0.5401 36.60 5.881e-05 -5.901 2
9907 MRPL21 11_38 0.5000 27942.14 4.157e-02 4.379 1Genes with largest effect sizes
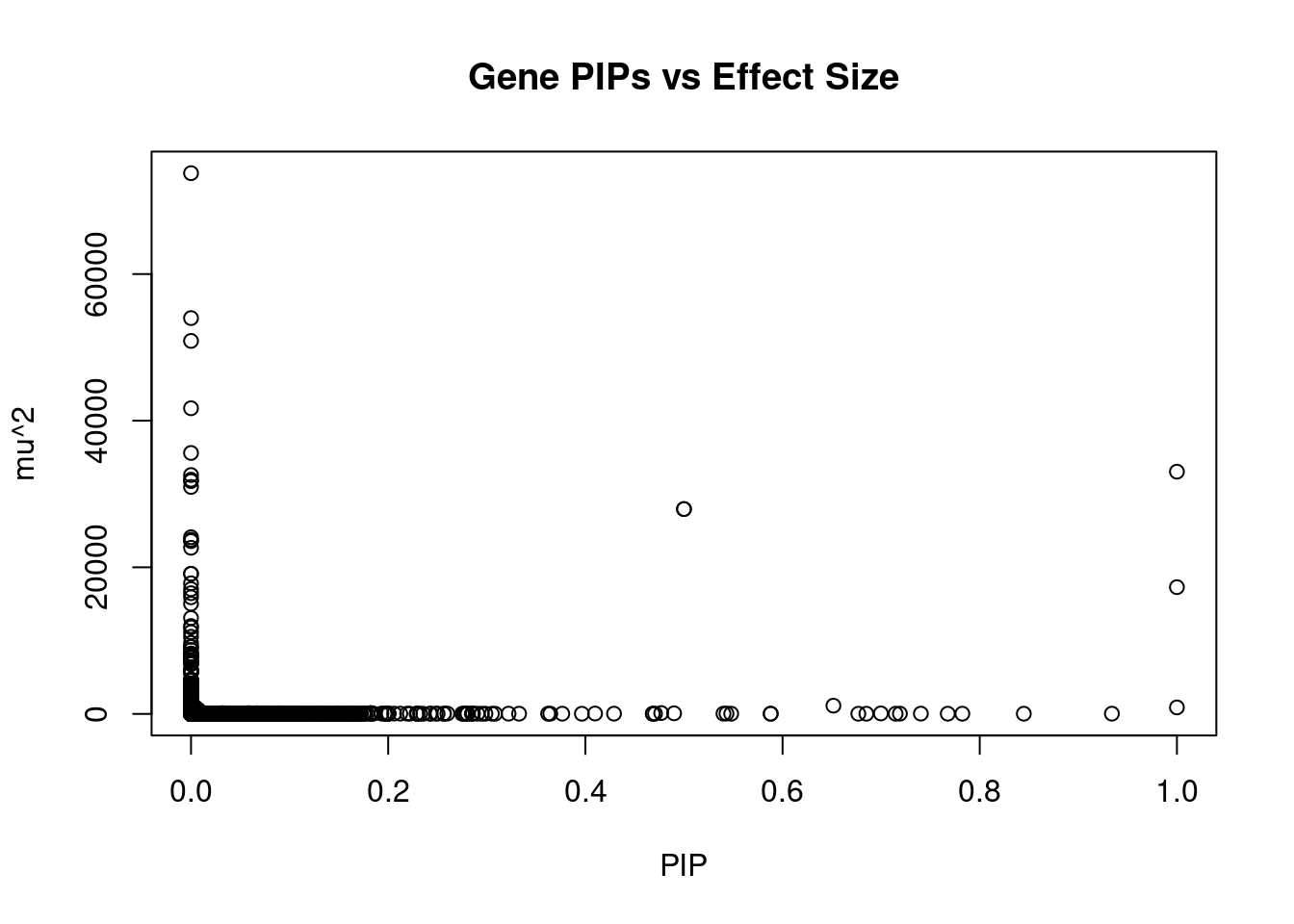
genename region_tag susie_pip mu2 PVE z num_eqtl
9 SEMA3F 3_35 0.000e+00 73763 0.000e+00 7.681 1
6937 CAMKV 3_35 0.000e+00 53990 0.000e+00 -9.848 1
7091 CCDC171 9_13 0.000e+00 50879 0.000e+00 8.043 2
33 RBM6 3_35 0.000e+00 41693 0.000e+00 12.536 1
6938 MST1R 3_35 0.000e+00 35597 0.000e+00 -12.626 1
9381 GSAP 7_49 1.000e+00 33042 9.831e-02 5.260 1
8677 DHFR2 3_59 0.000e+00 32585 0.000e+00 5.146 1
2783 CHST10 2_58 3.786e-10 31946 3.598e-11 4.807 1
8680 STX19 3_59 0.000e+00 31753 0.000e+00 -5.106 1
10629 SLC35E2 1_1 0.000e+00 30962 0.000e+00 5.161 1
4077 IGHMBP2 11_38 5.000e-01 27942 4.157e-02 -4.379 1
9907 MRPL21 11_38 5.000e-01 27942 4.157e-02 4.379 1
4918 MFAP1 15_16 2.759e-12 24102 1.979e-13 4.303 1
4364 HEY2 6_84 0.000e+00 23771 0.000e+00 3.066 1
6934 RNF123 3_35 0.000e+00 23572 0.000e+00 -10.959 1
11239 NAT6 3_35 0.000e+00 22664 0.000e+00 -7.156 2
4757 TMOD3 15_21 0.000e+00 19109 0.000e+00 5.412 1
2819 PLCL1 2_117 0.000e+00 19108 0.000e+00 -5.642 1
6966 RNF180 5_39 0.000e+00 17815 0.000e+00 -3.717 2
6994 CCDC127 5_1 1.000e+00 17291 5.144e-02 3.012 1Genes with highest PVE
genename region_tag susie_pip mu2 PVE z num_eqtl
9381 GSAP 7_49 1.0000 33042.01 9.831e-02 5.260 1
6994 CCDC127 5_1 1.0000 17290.64 5.144e-02 3.012 1
9907 MRPL21 11_38 0.5000 27942.14 4.157e-02 4.379 1
4077 IGHMBP2 11_38 0.5000 27942.14 4.157e-02 -4.379 1
6940 PPM1M 3_36 1.0000 883.39 2.628e-03 5.130 3
3354 WWP1 8_61 0.6516 1124.54 2.180e-03 5.312 2
8340 ASPHD1 16_24 0.4769 118.33 1.679e-04 -11.849 1
3302 ZMIZ2 7_33 0.6997 66.53 1.385e-04 -8.105 1
6156 GPR61 1_67 0.4900 80.05 1.167e-04 8.755 1
3273 FLT3 13_7 0.9340 33.54 9.319e-05 -5.360 1
11192 ATP5J2 7_61 0.5431 53.47 8.640e-05 -7.117 1
1149 DYNLL1 12_74 0.7403 37.63 8.288e-05 -5.806 1
10169 PRMT6 1_66 0.7190 33.46 7.157e-05 5.528 1
8362 PACS1 11_36 0.7676 30.12 6.880e-05 5.121 2
10689 VPS52 6_28 0.1824 125.80 6.827e-05 1.654 2
9286 MAPK11 22_24 0.8447 26.72 6.716e-05 4.904 1
8106 EFEMP2 11_36 0.4098 53.04 6.467e-05 -7.485 2
7028 ZNF12 7_9 0.7824 27.51 6.403e-05 5.114 2
12531 RP5-965G21.3 20_19 0.5401 36.60 5.881e-05 -5.901 2
11016 CCDC188 22_4 0.7147 25.33 5.386e-05 4.590 1Genes with largest z scores
genename region_tag susie_pip mu2 PVE z num_eqtl
6938 MST1R 3_35 0.000e+00 35597.35 0.000e+00 -12.626 1
33 RBM6 3_35 0.000e+00 41693.00 0.000e+00 12.536 1
8340 ASPHD1 16_24 4.769e-01 118.33 1.679e-04 -11.849 1
8341 KCTD13 16_24 5.853e-02 113.47 1.976e-05 -11.491 1
8339 SEZ6L2 16_24 3.177e-02 111.72 1.056e-05 -11.407 1
6934 RNF123 3_35 0.000e+00 23571.65 0.000e+00 -10.959 1
5905 POC5 5_44 1.482e-02 92.05 4.059e-06 -10.428 1
9879 SULT1A2 16_23 6.661e-02 96.31 1.909e-05 -10.415 1
9829 C6orf106 6_28 3.808e-05 122.40 1.387e-08 -10.264 1
7444 ZNF668 16_24 7.817e-02 79.05 1.838e-05 10.000 1
7445 ZNF646 16_24 7.817e-02 79.05 1.838e-05 -10.000 1
1759 KAT8 16_24 1.420e-02 75.60 3.193e-06 -9.874 2
1758 BCKDK 16_24 1.464e-02 75.72 3.299e-06 9.873 1
5100 SAE1 19_33 3.006e-03 100.74 9.010e-07 9.849 1
6937 CAMKV 3_35 0.000e+00 53990.32 0.000e+00 -9.848 1
8065 C1QTNF4 11_29 6.441e-03 88.85 1.703e-06 9.564 1
10961 RP11-196G11.6 16_24 7.677e-03 69.97 1.598e-06 9.354 2
7210 PSMC3 11_29 6.912e-03 77.52 1.594e-06 -8.866 1
7209 SLC39A13 11_29 6.350e-03 76.24 1.440e-06 -8.831 1
8448 NUPR1 16_23 8.723e-03 68.70 1.783e-06 -8.775 1Comparing z scores and PIPs
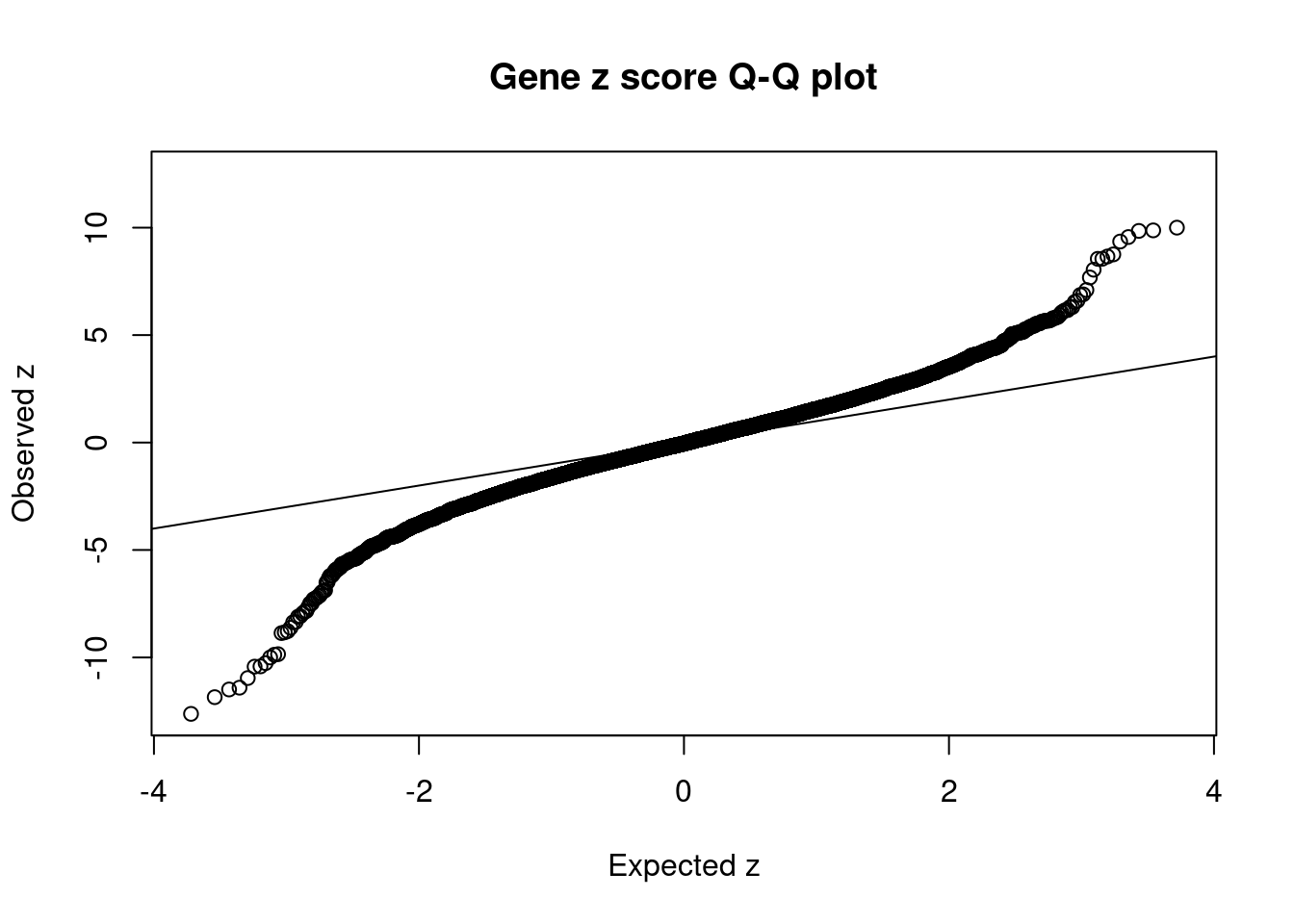
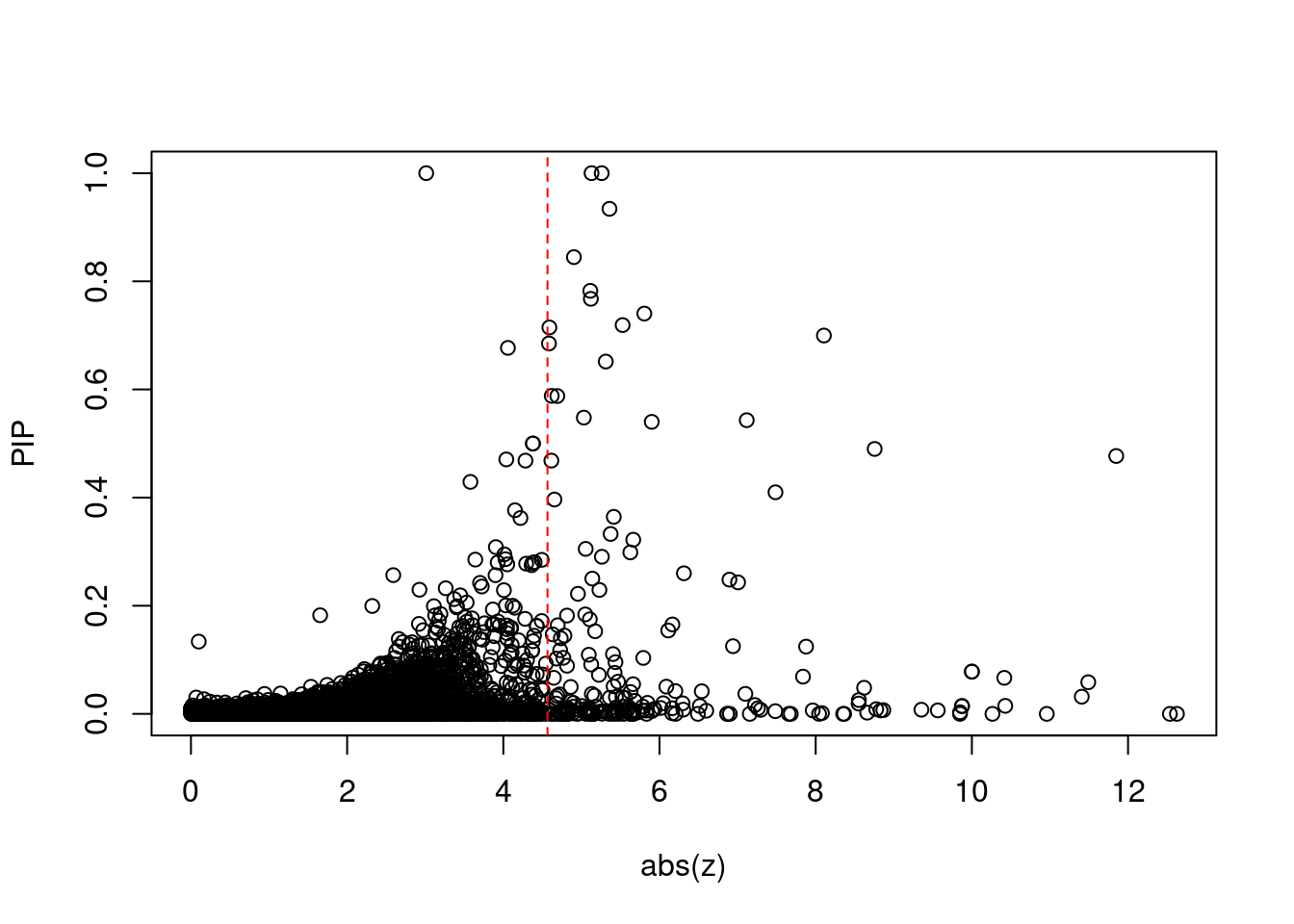
[1] 0.0201 genename region_tag susie_pip mu2 PVE z num_eqtl
6938 MST1R 3_35 0.000e+00 35597.35 0.000e+00 -12.626 1
33 RBM6 3_35 0.000e+00 41693.00 0.000e+00 12.536 1
8340 ASPHD1 16_24 4.769e-01 118.33 1.679e-04 -11.849 1
8341 KCTD13 16_24 5.853e-02 113.47 1.976e-05 -11.491 1
8339 SEZ6L2 16_24 3.177e-02 111.72 1.056e-05 -11.407 1
6934 RNF123 3_35 0.000e+00 23571.65 0.000e+00 -10.959 1
5905 POC5 5_44 1.482e-02 92.05 4.059e-06 -10.428 1
9879 SULT1A2 16_23 6.661e-02 96.31 1.909e-05 -10.415 1
9829 C6orf106 6_28 3.808e-05 122.40 1.387e-08 -10.264 1
7444 ZNF668 16_24 7.817e-02 79.05 1.838e-05 10.000 1
7445 ZNF646 16_24 7.817e-02 79.05 1.838e-05 -10.000 1
1759 KAT8 16_24 1.420e-02 75.60 3.193e-06 -9.874 2
1758 BCKDK 16_24 1.464e-02 75.72 3.299e-06 9.873 1
5100 SAE1 19_33 3.006e-03 100.74 9.010e-07 9.849 1
6937 CAMKV 3_35 0.000e+00 53990.32 0.000e+00 -9.848 1
8065 C1QTNF4 11_29 6.441e-03 88.85 1.703e-06 9.564 1
10961 RP11-196G11.6 16_24 7.677e-03 69.97 1.598e-06 9.354 2
7210 PSMC3 11_29 6.912e-03 77.52 1.594e-06 -8.866 1
7209 SLC39A13 11_29 6.350e-03 76.24 1.440e-06 -8.831 1
8448 NUPR1 16_23 8.723e-03 68.70 1.783e-06 -8.775 1GO enrichment analysis for genes with PIP>0.5
#number of genes for gene set enrichment
length(genes)[1] 20Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"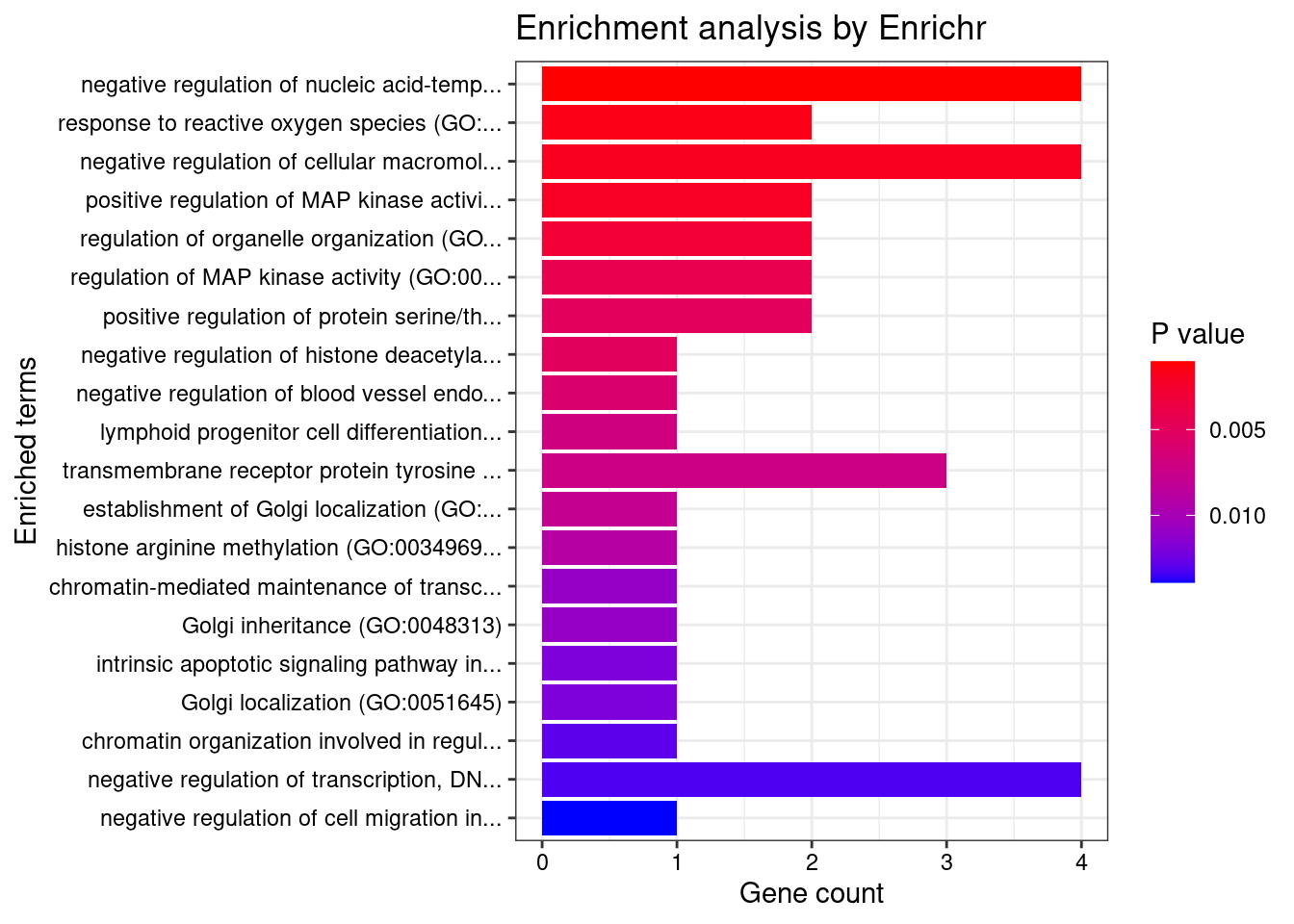
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Cellular_Component_2021"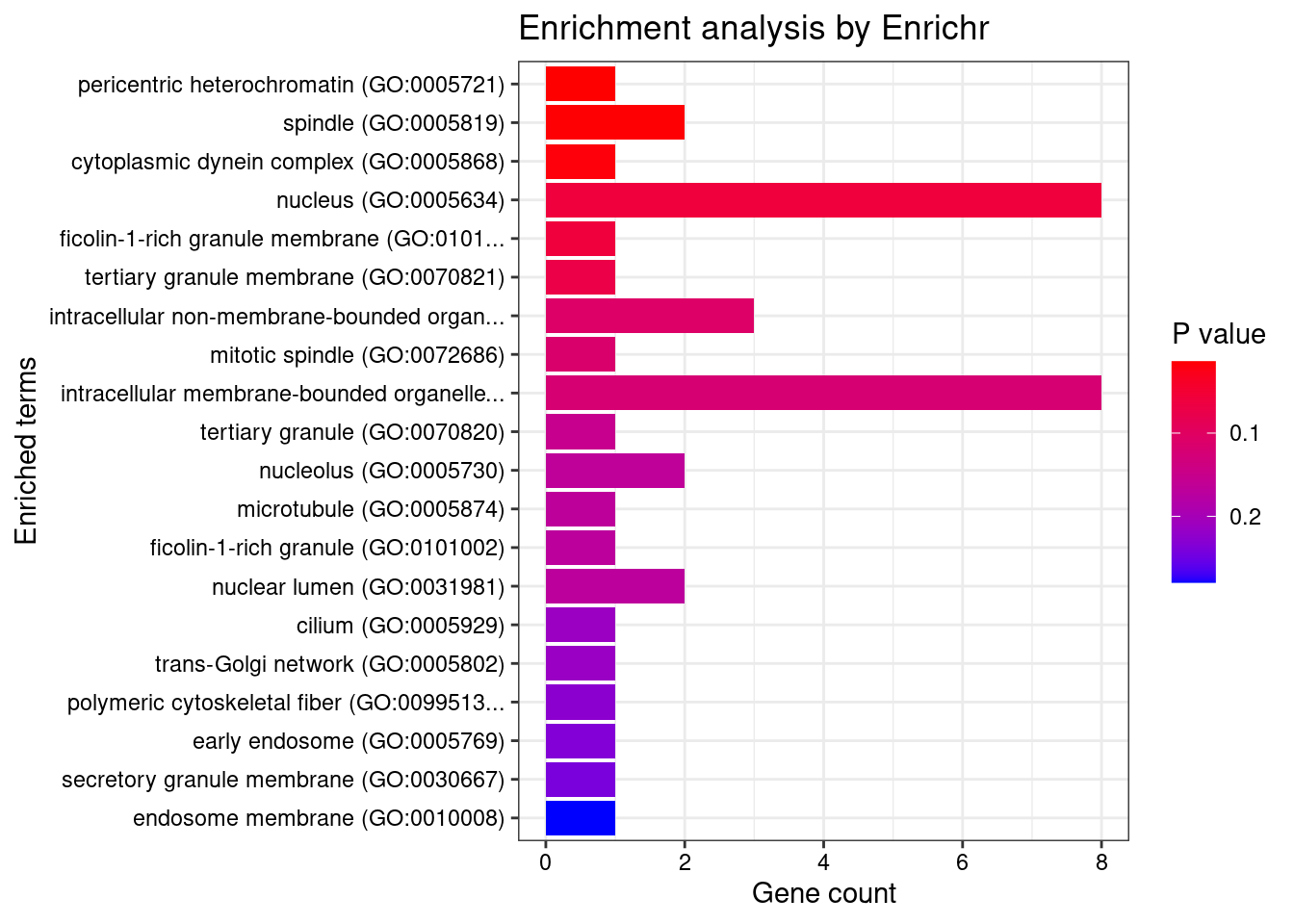
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Molecular_Function_2021"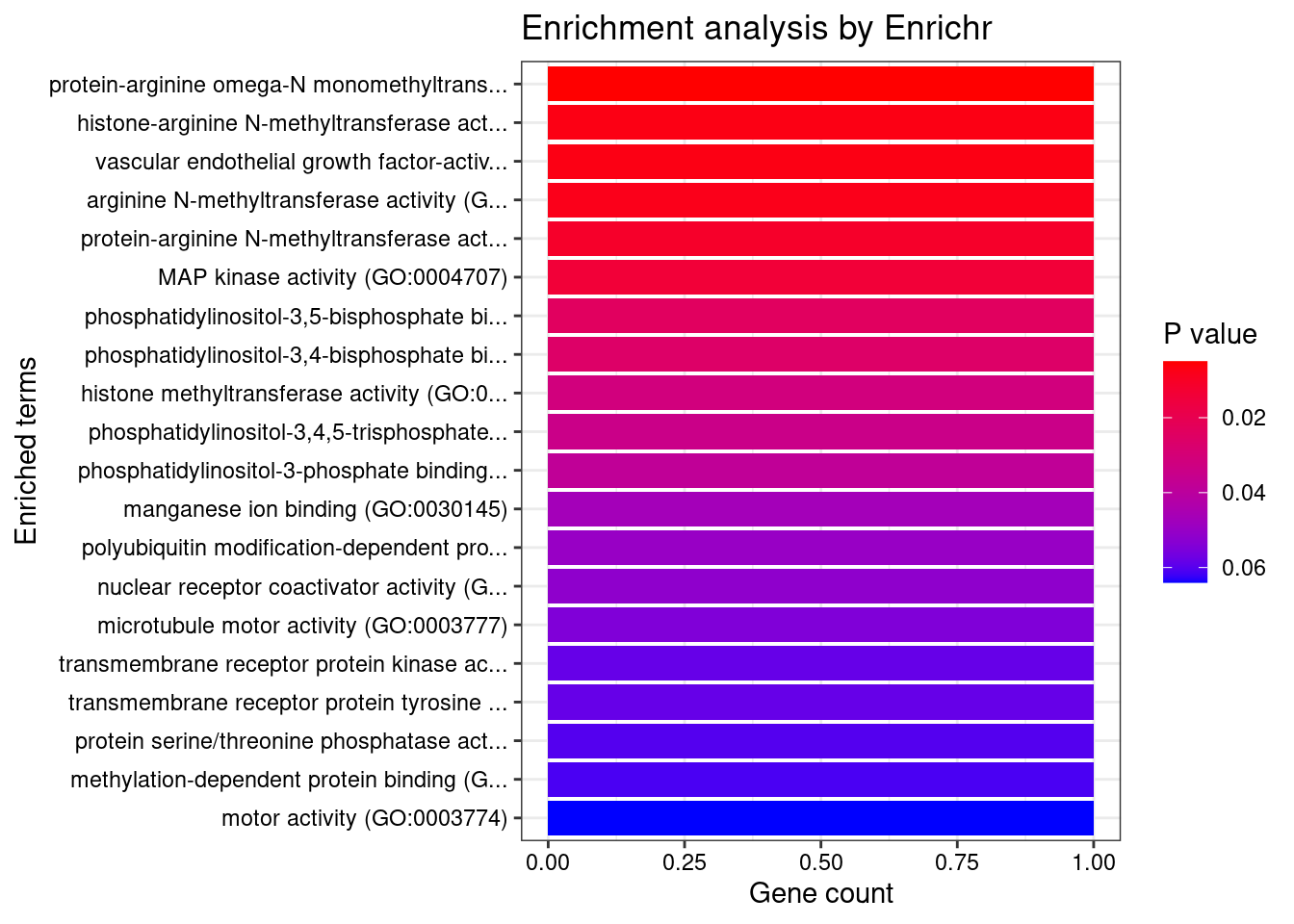
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)DisGeNET enrichment analysis for genes with PIP>0.5
Description FDR Ratio BgRatio
34 Acute myeloid leukemia, minimal differentiation 0.01020 1/9 1/9703
41 Cerebral Cavernous Malformations 3 0.01020 1/9 1/9703
51 Familial cerebral cavernous malformation 0.01020 1/9 1/9703
52 MENTAL RETARDATION, AUTOSOMAL DOMINANT 17 0.01020 1/9 1/9703
54 HERMANSKY-PUDLAK SYNDROME 5 0.01020 1/9 1/9703
45 Mixed phenotype acute leukemia 0.01700 1/9 2/9703
46 Cavernous Hemangioma of Brain 0.02185 1/9 3/9703
15 Acute myelomonocytic leukemia 0.02907 1/9 5/9703
17 Leukocytosis 0.02907 1/9 6/9703
30 Pleocytosis 0.02907 1/9 6/9703WebGestalt enrichment analysis for genes with PIP>0.5
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum =
minNum, : No significant gene set is identified based on FDR 0.05!NULLPIP Manhattan Plot
Sensitivity, specificity and precision for silver standard genes
#number of genes in known annotations
print(length(known_annotations))[1] 41#number of genes in known annotations with imputed expression
print(sum(known_annotations %in% ctwas_gene_res$genename))[1] 19#significance threshold for TWAS
print(sig_thresh)[1] 4.566#number of ctwas genes
length(ctwas_genes)[1] 5#number of TWAS genes
length(twas_genes)[1] 202#show novel genes (ctwas genes with not in TWAS genes)
ctwas_gene_res[ctwas_gene_res$genename %in% novel_genes,report_cols] genename region_tag susie_pip mu2 PVE z num_eqtl
6994 CCDC127 5_1 1 17291 0.05144 3.012 1#sensitivity / recall
print(sensitivity) ctwas TWAS
0.00000 0.04878 #specificity
print(specificity) ctwas TWAS
0.9995 0.9801 #precision / PPV
print(precision) ctwas TWAS
0.000000 0.009901 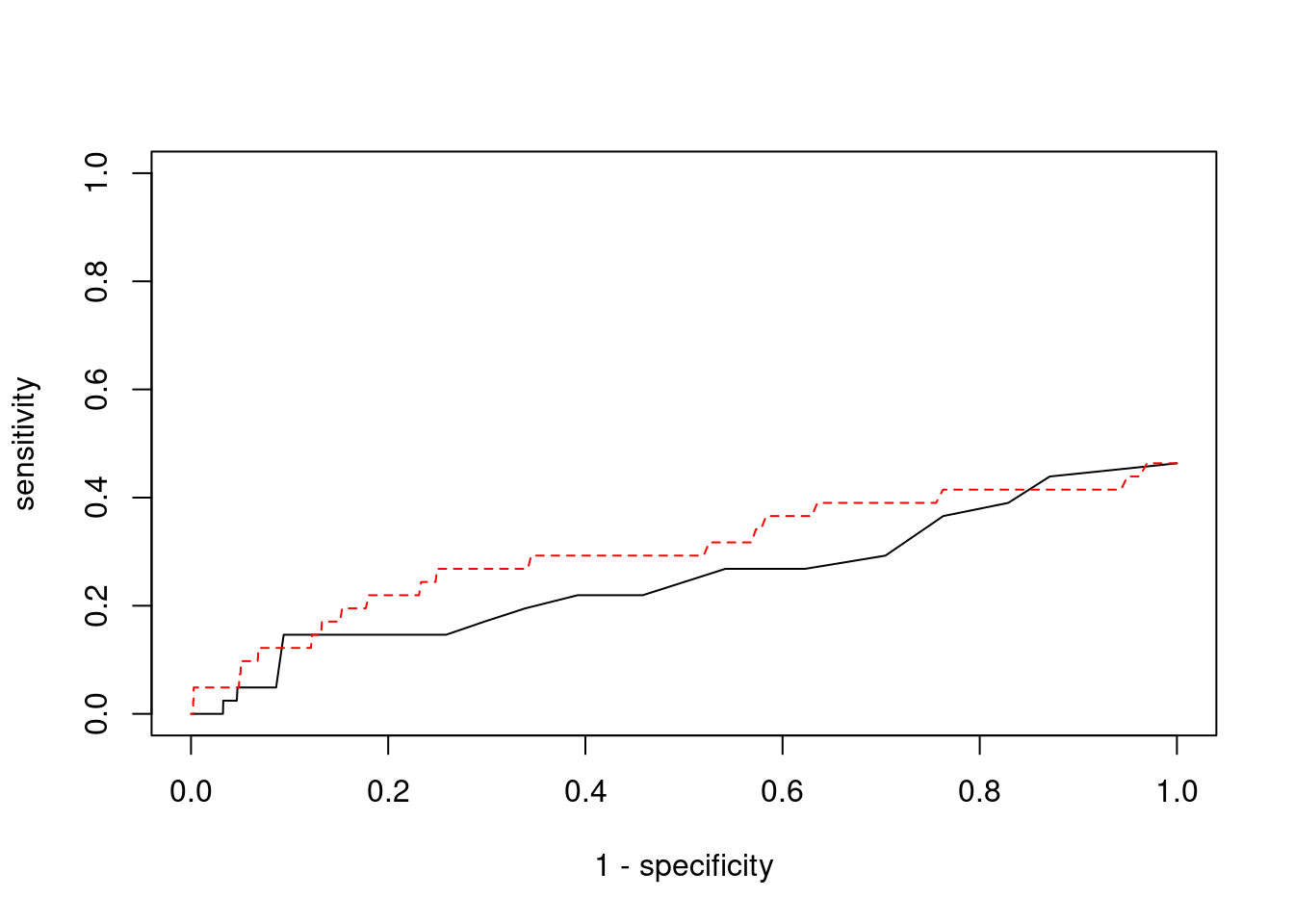
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] readxl_1.3.1 forcats_0.5.1 stringr_1.4.0 dplyr_1.0.7
[5] purrr_0.3.4 readr_2.1.1 tidyr_1.1.4 tidyverse_1.3.1
[9] tibble_3.1.6 WebGestaltR_0.4.4 disgenet2r_0.99.2 enrichR_3.0
[13] cowplot_1.0.0 ggplot2_3.3.5 workflowr_1.6.2
loaded via a namespace (and not attached):
[1] fs_1.5.2 lubridate_1.8.0 bit64_4.0.5 doParallel_1.0.16
[5] httr_1.4.2 rprojroot_2.0.2 tools_3.6.1 backports_1.4.1
[9] doRNG_1.8.2 utf8_1.2.2 R6_2.5.1 vipor_0.4.5
[13] DBI_1.1.1 colorspace_2.0-2 withr_2.4.3 ggrastr_1.0.1
[17] tidyselect_1.1.1 bit_4.0.4 curl_4.3.2 compiler_3.6.1
[21] git2r_0.26.1 cli_3.1.0 rvest_1.0.2 Cairo_1.5-12.2
[25] xml2_1.3.3 labeling_0.4.2 scales_1.1.1 apcluster_1.4.8
[29] digest_0.6.29 rmarkdown_2.11 svglite_1.2.2 pkgconfig_2.0.3
[33] htmltools_0.5.2 dbplyr_2.1.1 fastmap_1.1.0 highr_0.9
[37] rlang_0.4.12 rstudioapi_0.13 RSQLite_2.2.8 jquerylib_0.1.4
[41] farver_2.1.0 generics_0.1.1 jsonlite_1.7.2 vroom_1.5.7
[45] magrittr_2.0.1 Matrix_1.2-18 ggbeeswarm_0.6.0 Rcpp_1.0.7
[49] munsell_0.5.0 fansi_0.5.0 gdtools_0.1.9 lifecycle_1.0.1
[53] stringi_1.7.6 whisker_0.3-2 yaml_2.2.1 plyr_1.8.6
[57] grid_3.6.1 blob_1.2.2 ggrepel_0.9.1 parallel_3.6.1
[61] promises_1.0.1 crayon_1.4.2 lattice_0.20-38 haven_2.4.3
[65] hms_1.1.1 knitr_1.36 pillar_1.6.4 igraph_1.2.10
[69] rjson_0.2.20 rngtools_1.5.2 reshape2_1.4.4 codetools_0.2-16
[73] reprex_2.0.1 glue_1.5.1 evaluate_0.14 data.table_1.14.2
[77] modelr_0.1.8 vctrs_0.3.8 tzdb_0.2.0 httpuv_1.5.1
[81] foreach_1.5.1 cellranger_1.1.0 gtable_0.3.0 assertthat_0.2.1
[85] cachem_1.0.6 xfun_0.29 broom_0.7.10 later_0.8.0
[89] iterators_1.0.13 beeswarm_0.2.3 memoise_2.0.1 ellipsis_0.3.2