WhiteBlood - WholeBlood gene expression
sheng Qian
2023-2-1
Last updated: 2023-02-03
Checks: 5 2
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/ctwas_config_b38.R | code/ctwas_config_b38.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 66590cb. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .ipynb_checkpoints/
Untracked files:
Untracked: Proposal plots.R
Untracked: RGS14.pdf
Untracked: RNF186.pdf
Untracked: SCZ_annotation.xlsx
Untracked: SLC8B1.pdf
Untracked: analysis/.ipynb_checkpoints/
Untracked: cache/
Untracked: code/.ipynb_checkpoints/
Untracked: data/.ipynb_checkpoints/
Untracked: data/FUMA_output/
Untracked: data/GO_Terms/
Untracked: data/GTEx_Analysis_v8_eQTL.tar
Untracked: data/G_list.RData
Untracked: data/IBD_ME/
Untracked: data/LDL/
Untracked: data/LDL_E_S/
Untracked: data/LDL_M/
Untracked: data/LDL_S/
Untracked: data/LDL_multi/
Untracked: data/PGC3_SCZ_wave3_public.v2.tsv
Untracked: data/RedBlood_M/
Untracked: data/SCZ/
Untracked: data/SCZ_2014_EUR/
Untracked: data/SCZ_2014_EUR_ME/
Untracked: data/SCZ_2018/
Untracked: data/SCZ_2018_ME/
Untracked: data/SCZ_2018_S/
Untracked: data/SCZ_2020/
Untracked: data/SCZ_S/
Untracked: data/Supplementary Table 15 - MAGMA.xlsx
Untracked: data/Supplementary Table 20 - Prioritised Genes.xlsx
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/WhiteBlood_E_S_M/
Untracked: data/White_Blood_M/
Untracked: data/eqtl/
Untracked: data/gencode.v26.GRCh38.genes.gtf
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/gwas_sumstats/
Untracked: data/magma.genes.out
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/mashr_sqtl/
Untracked: data/mqtl/
Untracked: data/multigroup/
Untracked: data/notes.txt
Untracked: data/scz_2018.RDS
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: temp_LDR/
Untracked: top_genes_32.txt
Untracked: top_genes_37.txt
Untracked: top_genes_43.txt
Untracked: top_genes_54.txt
Untracked: top_genes_81.txt
Untracked: z_snp_pos_SCZ.RData
Untracked: z_snp_pos_SCZ_2014_EUR.RData
Untracked: z_snp_pos_SCZ_2018.RData
Untracked: z_snp_pos_SCZ_2020.RData
Unstaged changes:
Deleted: analysis/Atrial_Fibrillation_Heart_Atrial_Appendage.Rmd
Deleted: analysis/Atrial_Fibrillation_Heart_Left_Ventricle.Rmd
Deleted: analysis/Autism_Brain_Amygdala.Rmd
Deleted: analysis/Autism_Brain_Anterior_cingulate_cortex_BA24.Rmd
Deleted: analysis/Autism_Brain_Caudate_basal_ganglia.Rmd
Deleted: analysis/Autism_Brain_Cerebellar_Hemisphere.Rmd
Deleted: analysis/Autism_Brain_Cerebellum.Rmd
Deleted: analysis/Autism_Brain_Cortex.Rmd
Deleted: analysis/Autism_Brain_Frontal_Cortex_BA9.Rmd
Deleted: analysis/Autism_Brain_Hippocampus.Rmd
Deleted: analysis/Autism_Brain_Hypothalamus.Rmd
Deleted: analysis/Autism_Brain_Nucleus_accumbens_basal_ganglia.Rmd
Deleted: analysis/Autism_Brain_Putamen_basal_ganglia.Rmd
Deleted: analysis/Autism_Brain_Spinal_cord_cervical_c-1.Rmd
Deleted: analysis/Autism_Brain_Substantia_nigra.Rmd
Deleted: analysis/BMI_Brain_Amygdala.Rmd
Deleted: analysis/BMI_Brain_Amygdala_S.Rmd
Deleted: analysis/BMI_Brain_Anterior_cingulate_cortex_BA24.Rmd
Deleted: analysis/BMI_Brain_Anterior_cingulate_cortex_BA24_S.Rmd
Deleted: analysis/BMI_Brain_Caudate_basal_ganglia.Rmd
Deleted: analysis/BMI_Brain_Caudate_basal_ganglia_S.Rmd
Deleted: analysis/BMI_Brain_Cerebellar_Hemisphere.Rmd
Deleted: analysis/BMI_Brain_Cerebellar_Hemisphere_S.Rmd
Deleted: analysis/BMI_Brain_Cerebellum.Rmd
Deleted: analysis/BMI_Brain_Cerebellum_S.Rmd
Deleted: analysis/BMI_Brain_Cortex.Rmd
Deleted: analysis/BMI_Brain_Cortex_S.Rmd
Deleted: analysis/BMI_Brain_Frontal_Cortex_BA9.Rmd
Deleted: analysis/BMI_Brain_Frontal_Cortex_BA9_S.Rmd
Deleted: analysis/BMI_Brain_Hippocampus.Rmd
Deleted: analysis/BMI_Brain_Hippocampus_S.Rmd
Deleted: analysis/BMI_Brain_Hypothalamus.Rmd
Deleted: analysis/BMI_Brain_Hypothalamus_S.Rmd
Deleted: analysis/BMI_Brain_Nucleus_accumbens_basal_ganglia.Rmd
Deleted: analysis/BMI_Brain_Nucleus_accumbens_basal_ganglia_S.Rmd
Deleted: analysis/BMI_Brain_Putamen_basal_ganglia.Rmd
Deleted: analysis/BMI_Brain_Putamen_basal_ganglia_S.Rmd
Deleted: analysis/BMI_Brain_Spinal_cord_cervical_c-1.Rmd
Deleted: analysis/BMI_Brain_Spinal_cord_cervical_c-1_S.Rmd
Deleted: analysis/BMI_Brain_Substantia_nigra.Rmd
Deleted: analysis/BMI_Brain_Substantia_nigra_S.Rmd
Deleted: analysis/BMI_S_results.Rmd
Deleted: analysis/Glucose_Adipose_Subcutaneous.Rmd
Deleted: analysis/Glucose_Adipose_Visceral_Omentum.Rmd
Modified: analysis/WhiteBlood_WholeBlood_E.Rmd
Modified: analysis/WhiteBlood_WholeBlood_E_S_M.Rmd
Deleted: code/run_IBD_ctwas_rss_LDR_ME.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/WhiteBlood_WholeBlood_E.Rmd) and HTML (docs/WhiteBlood_WholeBlood_E.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 66590cb | sq-96 | 2023-02-03 | update |
| html | 66590cb | sq-96 | 2023-02-03 | update |
| Rmd | 9b01dad | sq-96 | 2023-02-01 | update |
| html | 9b01dad | sq-96 | 2023-02-01 | update |
Weight QC
#number of imputed weights
nrow(qclist_all)[1] 11095#number of imputed weights by chromosome
table(qclist_all$chr)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
1129 747 624 400 479 621 560 383 404 430 682 652 192 362 331 551
17 18 19 20 21 22
725 159 911 313 130 310 #number of imputed weights without missing variants
sum(qclist_all$nmiss==0)[1] 8463#proportion of imputed weights without missing variants
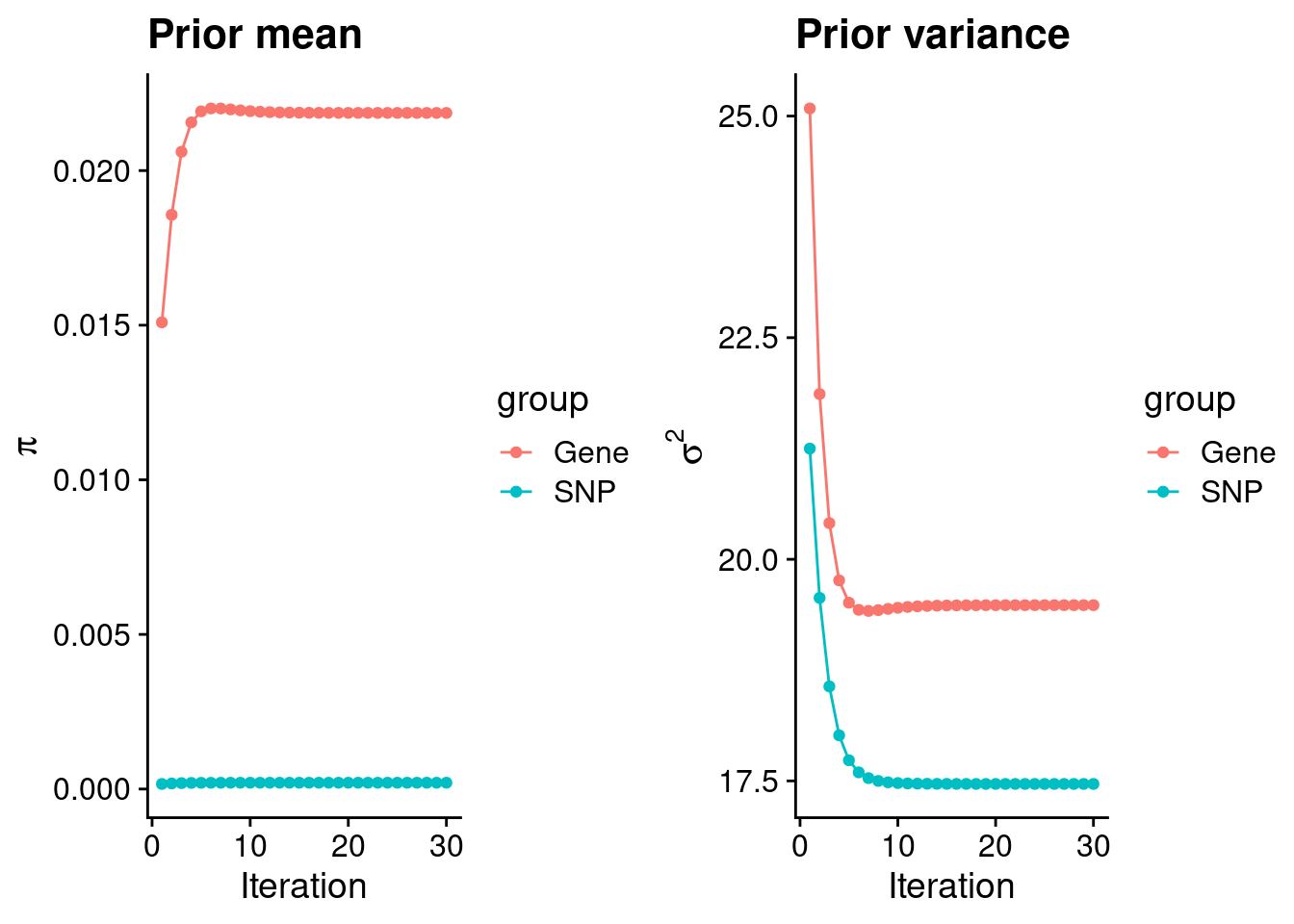
mean(qclist_all$nmiss==0)[1] 0.7628Check convergence of parameters
| Version | Author | Date |
|---|---|---|
| 9b01dad | sq-96 | 2023-02-01 |
#estimated group prior
estimated_group_prior <- group_prior_rec[,ncol(group_prior_rec)]
names(estimated_group_prior) <- c("gene", "snp")
estimated_group_prior["snp"] <- estimated_group_prior["snp"]*thin #adjust parameter to account for thin argument
print(estimated_group_prior) gene snp
0.021865 0.000204 #estimated group prior variance
estimated_group_prior_var <- group_prior_var_rec[,ncol(group_prior_var_rec)]
names(estimated_group_prior_var) <- c("gene", "snp")
print(estimated_group_prior_var) gene snp
19.48 17.47 #report sample size
print(sample_size)[1] 350470#report group size
group_size <- c(nrow(ctwas_gene_res), n_snps)
print(group_size)[1] 11095 8696600#estimated group PVE
estimated_group_pve <- estimated_group_prior_var*estimated_group_prior*group_size/sample_size #check PVE calculation
names(estimated_group_pve) <- c("gene", "snp")
print(estimated_group_pve) gene snp
0.01349 0.08840 #compare sum(PIP*mu2/sample_size) with above PVE calculation
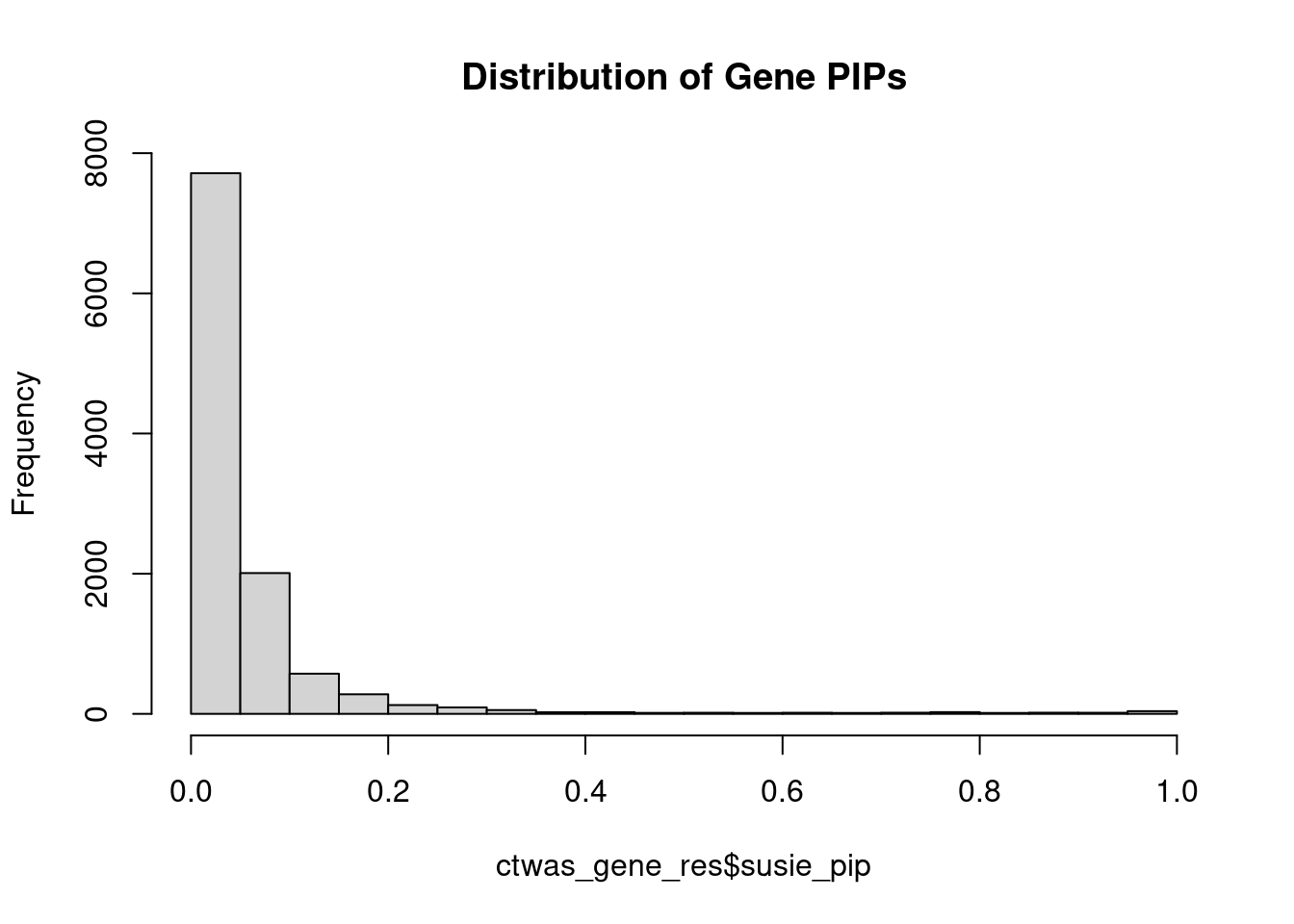
c(sum(ctwas_gene_res$PVE),sum(ctwas_snp_res$PVE))[1] 0.04734 1.38376Genes with highest PIPs
| Version | Author | Date |
|---|---|---|
| 9b01dad | sq-96 | 2023-02-01 |
genename region_tag susie_pip mu2 PVE z num_eqtl
9507 FES 15_43 1.0000 69.96 1.996e-04 -8.565 3
3646 BAZ2B 2_96 1.0000 86.85 2.478e-04 11.470 2
5673 PSEN2 1_116 1.0000 43.18 1.232e-04 -6.932 3
7481 TAGAP 6_103 0.9999 63.68 1.817e-04 -8.331 2
1640 KIAA0391 14_9 0.9997 47.58 1.357e-04 7.386 2
7070 LAPTM5 1_20 0.9995 70.88 2.021e-04 9.228 3
8131 RNF181 2_54 0.9995 2388.15 6.810e-03 -5.029 1
2611 ALDH2 12_67 0.9989 100.35 2.860e-04 -13.934 3
5966 VLDLR 9_3 0.9988 44.52 1.269e-04 6.949 4
893 ARHGAP15 2_85 0.9986 35.02 9.979e-05 7.184 3
1603 SPTLC2 14_36 0.9985 29.11 8.293e-05 -5.000 2
7222 CXCR1 2_129 0.9978 124.38 3.541e-04 11.321 3
2131 ATP13A1 19_15 0.9976 43.80 1.247e-04 6.372 2
8253 TPST1 7_43 0.9966 40.69 1.157e-04 -6.915 2
5908 CREB5 7_24 0.9966 367.89 1.046e-03 -20.722 1
5665 CNIH4 1_114 0.9954 93.65 2.660e-04 -9.203 2
4571 CD101 1_72 0.9950 39.62 1.125e-04 6.256 3
9672 UBE2F 2_141 0.9915 34.17 9.668e-05 -5.523 3
9863 LAMP1 13_62 0.9912 39.43 1.115e-04 -6.303 1
10100 SELL 1_83 0.9901 25.85 7.302e-05 3.837 3
412 ARAP2 4_30 0.9891 68.79 1.942e-04 -8.413 2
8044 TTC39C 18_12 0.9837 40.20 1.128e-04 5.211 1
1102 SLC25A24 1_67 0.9831 34.05 9.552e-05 5.832 3
1459 SPECC1L 22_6 0.9815 23.81 6.667e-05 5.337 2
9899 KIF18B 17_26 0.9810 26.88 7.525e-05 5.374 1
2312 LIPA 10_57 0.9755 40.48 1.127e-04 6.306 4
5360 NLRC5 16_31 0.9737 43.54 1.210e-04 6.504 2
2818 SLC12A7 5_2 0.9724 39.25 1.089e-04 5.708 4
5767 MED12L 3_93 0.9721 25.67 7.121e-05 -4.689 2
6064 PTPRJ 11_29 0.9710 68.03 1.885e-04 -9.800 2
6686 HIST1H2BD 6_20 0.9654 64.52 1.777e-04 9.575 1
9272 ZFPM1 16_54 0.9623 36.63 1.006e-04 -4.645 1
10454 ELANE 19_2 0.9618 25.17 6.906e-05 -4.762 2
3293 KLF12 13_36 0.9603 39.60 1.085e-04 -6.340 1
9410 DDX60L 4_109 0.9602 21.62 5.923e-05 4.427 5
811 ACAP1 17_6 0.9592 62.97 1.723e-04 7.734 2
3323 NEK6 9_64 0.9571 25.90 7.072e-05 5.717 2
9755 UBOX5 20_5 0.9532 27.74 7.546e-05 -4.863 1
1160 ADD1 4_4 0.9513 33.21 9.014e-05 -7.073 1
2969 SPTBN1 2_36 0.9492 47.30 1.281e-04 6.814 3
3758 ATXN1 6_13 0.9474 65.33 1.766e-04 8.173 1
1273 GLG1 16_40 0.9445 24.70 6.657e-05 4.680 2
8108 TET2 4_69 0.9438 25.10 6.759e-05 -5.355 2
2410 MLX 17_25 0.9435 56.41 1.519e-04 7.856 2
9287 CITED4 1_25 0.9421 27.10 7.285e-05 -4.751 2
736 HDHD5 22_1 0.9368 21.46 5.735e-05 3.481 3
4883 HS6ST1 2_75 0.9349 20.24 5.398e-05 -4.140 1
7272 ATXN7 3_43 0.9331 24.57 6.541e-05 -3.706 3
4385 TBC1D14 4_8 0.9297 29.01 7.697e-05 6.255 1
171 UQCRC1 3_34 0.9295 56.67 1.503e-04 -5.030 1
982 CDC14A 1_61 0.9259 19.56 5.168e-05 3.829 2
4658 OSTF1 9_35 0.9251 20.45 5.399e-05 4.056 3
10114 PAQR9 3_87 0.9249 21.18 5.590e-05 -4.049 2
1408 MYO9B 19_14 0.9038 28.58 7.369e-05 5.238 1
1145 ACHE 7_62 0.9002 37.30 9.582e-05 -3.852 1
323 RABEP1 17_5 0.8978 60.04 1.538e-04 8.715 2
4670 ADAM19 5_93 0.8975 23.14 5.926e-05 4.198 2
2033 TIMM50 19_26 0.8936 38.72 9.873e-05 -6.048 2
6935 CPSF4 7_61 0.8884 52.14 1.322e-04 -7.268 2
380 RAI14 5_23 0.8876 19.25 4.876e-05 3.788 1
9299 CCR8 3_28 0.8855 21.89 5.531e-05 -2.931 1
162 TRAF3IP3 1_106 0.8853 24.59 6.211e-05 4.778 2
1386 ITPR3 6_28 0.8796 37.90 9.511e-05 6.171 5
10088 C19orf35 19_4 0.8770 26.89 6.728e-05 -4.583 3
5598 RORC 1_74 0.8766 20.28 5.073e-05 4.101 1
11526 TNFSF12 17_7 0.8729 40.18 1.001e-04 -3.244 3
208 PPP5C 19_32 0.8726 25.17 6.266e-05 -4.940 2
2053 CCDC9 19_33 0.8697 46.13 1.145e-04 6.833 3
755 JMJD6 17_43 0.8670 24.25 5.999e-05 4.742 1
5834 TNFAIP8 5_72 0.8645 54.30 1.340e-04 7.624 1
1473 SLC25A1 22_3 0.8611 20.59 5.058e-05 -4.055 2
2437 SLC9A3R1 17_42 0.8539 46.97 1.144e-04 -7.630 1
7233 EOMES 3_20 0.8459 55.80 1.347e-04 7.596 1
10656 RCSD1 1_82 0.8384 22.55 5.395e-05 4.395 3
9832 ZFP36L1 14_33 0.8319 57.06 1.354e-04 8.072 2
6143 MTMR12 5_22 0.8301 20.79 4.925e-05 -4.003 1
5668 CDC42BPA 1_116 0.8301 23.45 5.555e-05 5.108 2
253 RALBP1 18_7 0.8261 21.01 4.953e-05 -3.959 4
2813 NPR3 5_22 0.8197 21.28 4.977e-05 4.146 1
8907 LRRC25 19_15 0.8190 27.67 6.465e-05 -4.768 1
11105 MEG3 14_52 0.8119 33.89 7.851e-05 5.342 1
1074 REST 4_41 0.8090 96.14 2.219e-04 9.019 1
574 CA11 19_33 0.8058 31.37 7.212e-05 -5.480 2
9085 GPR4 19_32 0.8057 21.02 4.832e-05 4.252 1
7003 MED11 17_4 0.8049 22.21 5.102e-05 4.984 3GO enrichment analysis for genes with PIP>0.5
#number of genes for gene set enrichment
length(genes)[1] 85Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"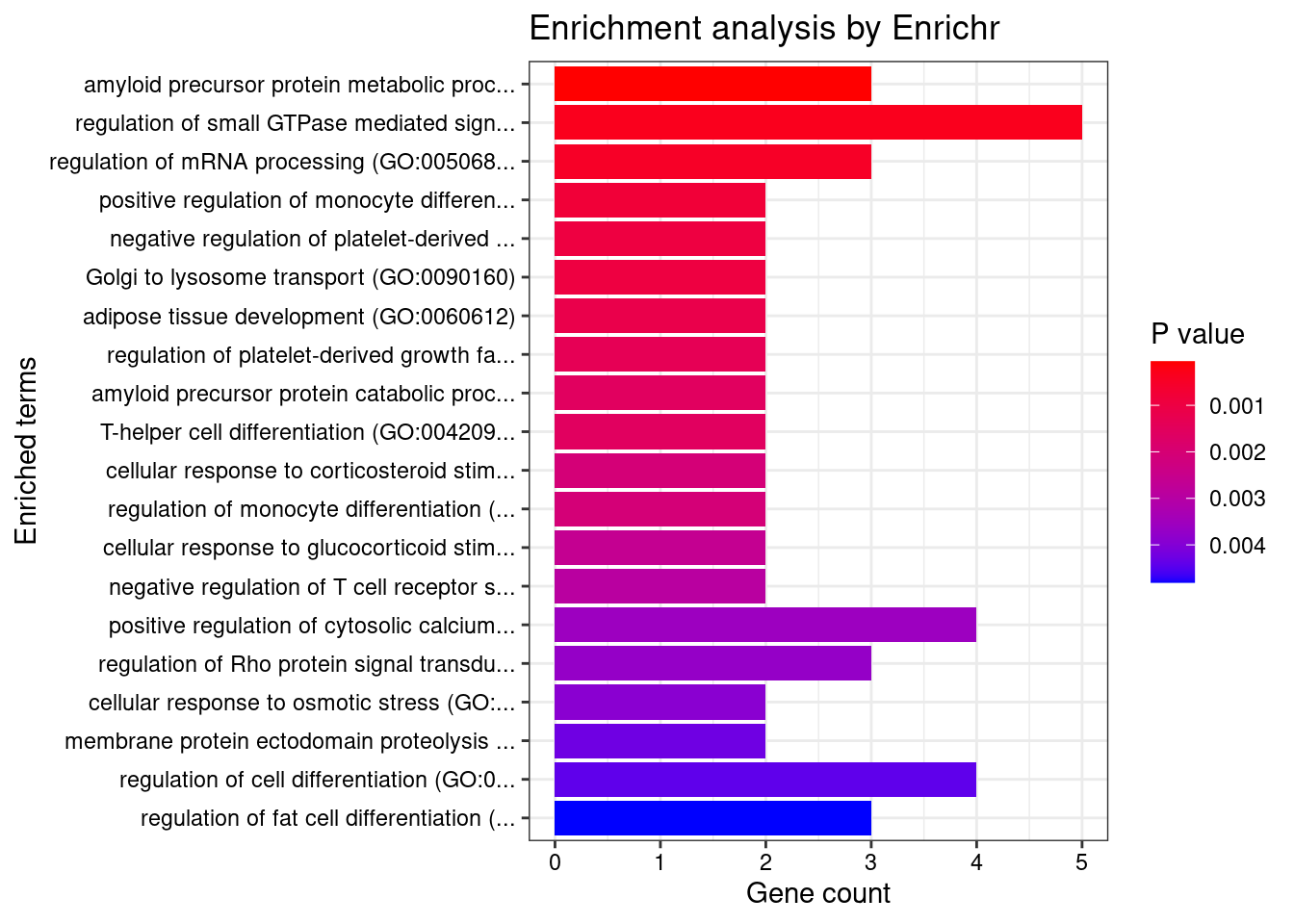
Term Overlap
1 amyloid precursor protein metabolic process (GO:0042982) 3/18
Adjusted.P.value Genes
1 0.04849 ADAM19;ACHE;PSEN2
[1] "GO_Cellular_Component_2021"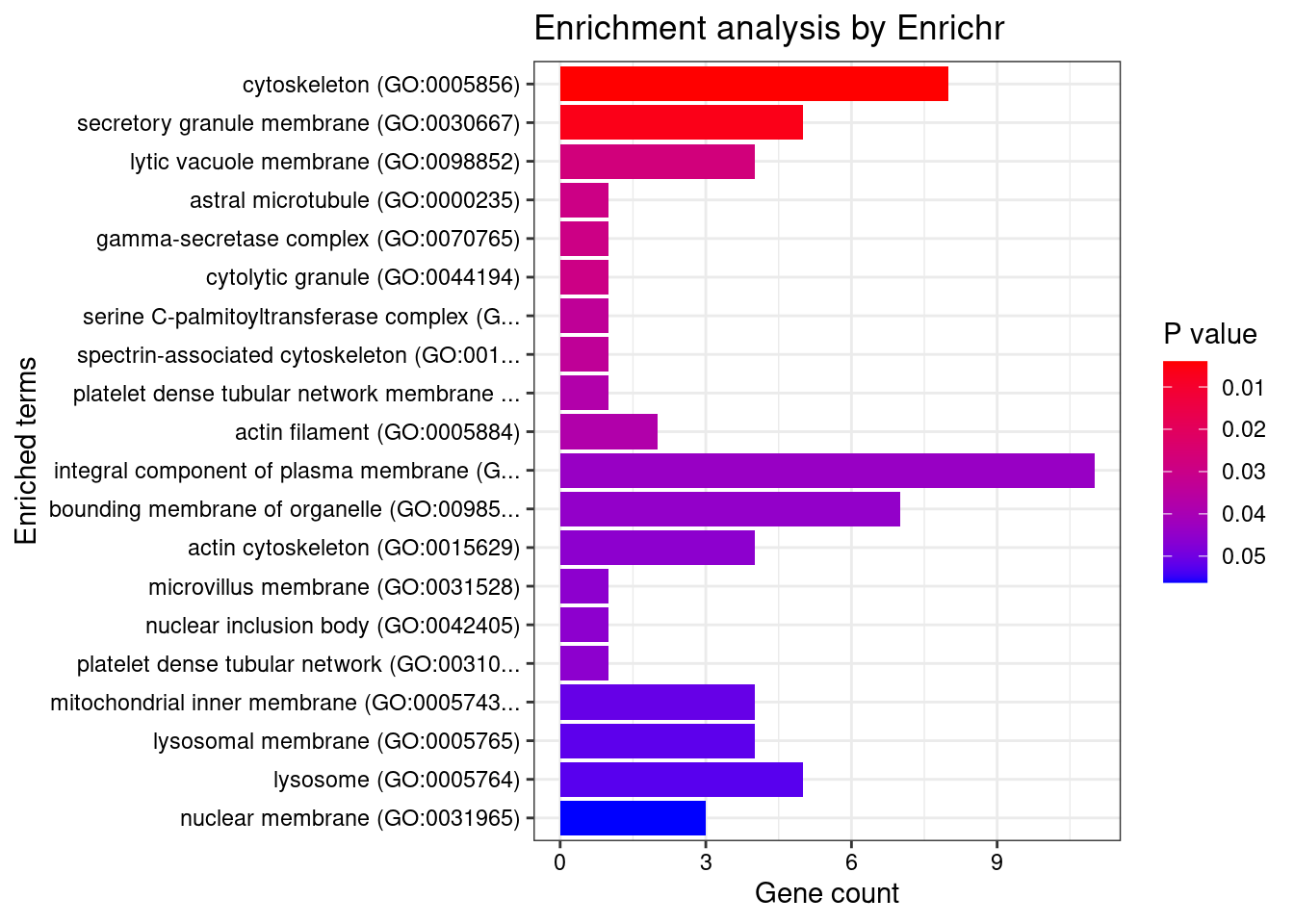
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Molecular_Function_2021"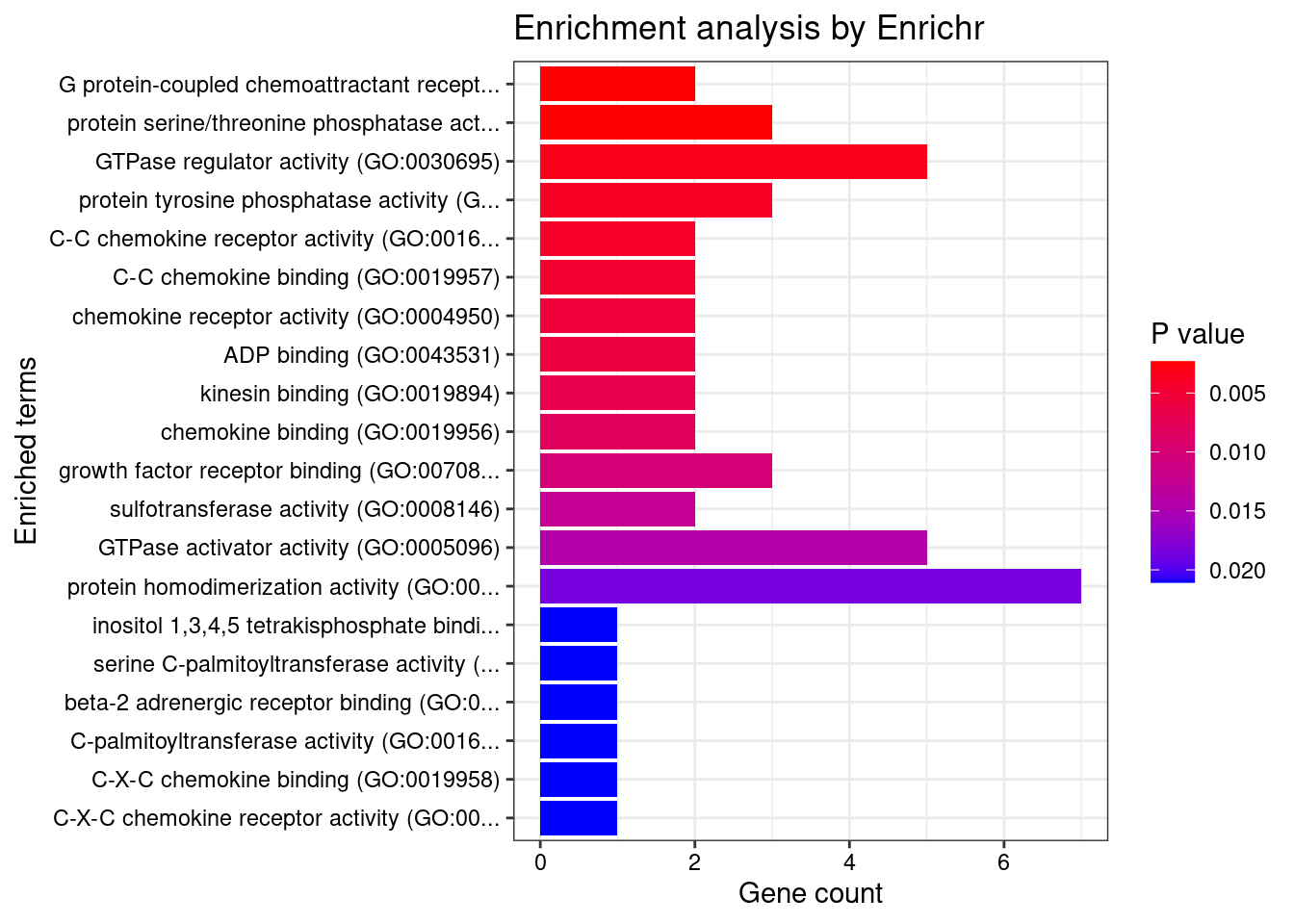
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)DisGeNET enrichment analysis for genes with PIP>0.5
Description FDR Ratio BgRatio
11 Refractory anaemia with excess blasts 0.05699 1/50 1/9703
29 Cholesterol Ester Storage Disease 0.05699 1/50 1/9703
50 Freckles 0.05699 1/50 1/9703
71 Melanosis 0.05699 1/50 1/9703
72 Chloasma 0.05699 1/50 1/9703
111 Wolman Disease 0.05699 1/50 1/9703
131 Cyclic neutropenia 0.05699 1/50 1/9703
132 Cerebellar Gait Ataxia 0.05699 1/50 1/9703
163 Alcohol-Induced Disorders 0.05699 1/50 1/9703
164 Tall stature 0.05699 1/50 1/9703WebGestalt enrichment analysis for genes with PIP>0.5
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum =
minNum, : No significant gene set is identified based on FDR 0.05!NULL
sessionInfo()R version 4.1.0 (2021-05-18)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] WebGestaltR_0.4.4 disgenet2r_0.99.2 enrichR_3.1 cowplot_1.1.1
[5] ggplot2_3.4.0 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] httr_1.4.4 sass_0.4.4 vroom_1.6.0 bit64_4.0.5
[5] jsonlite_1.8.4 foreach_1.5.2 bslib_0.4.1 assertthat_0.2.1
[9] getPass_0.2-2 highr_0.9 doRNG_1.8.2 blob_1.2.3
[13] yaml_2.3.6 pillar_1.8.1 RSQLite_2.2.19 lattice_0.20-44
[17] glue_1.6.2 digest_0.6.31 promises_1.2.0.1 colorspace_2.0-3
[21] htmltools_0.5.4 httpuv_1.6.7 Matrix_1.3-3 plyr_1.8.8
[25] pkgconfig_2.0.3 scales_1.2.1 svglite_2.1.0 processx_3.8.0
[29] whisker_0.4.1 later_1.3.0 tzdb_0.3.0 git2r_0.30.1
[33] tibble_3.1.8 generics_0.1.3 farver_2.1.0 ellipsis_0.3.2
[37] cachem_1.0.6 withr_2.5.0 cli_3.4.1 crayon_1.5.2
[41] magrittr_2.0.3 memoise_2.0.1 evaluate_0.19 ps_1.7.2
[45] apcluster_1.4.10 fs_1.5.2 fansi_1.0.3 doParallel_1.0.17
[49] tools_4.1.0 data.table_1.14.6 hms_1.1.2 lifecycle_1.0.3
[53] stringr_1.5.0 munsell_0.5.0 rngtools_1.5.2 callr_3.7.3
[57] compiler_4.1.0 jquerylib_0.1.4 systemfonts_1.0.4 rlang_1.0.6
[61] grid_4.1.0 iterators_1.0.14 rstudioapi_0.14 rjson_0.2.21
[65] igraph_1.3.5 labeling_0.4.2 rmarkdown_2.19 gtable_0.3.1
[69] codetools_0.2-18 DBI_1.1.3 curl_4.3.2 reshape2_1.4.4
[73] R6_2.5.1 knitr_1.41 dplyr_1.0.10 fastmap_1.1.0
[77] bit_4.0.5 utf8_1.2.2 rprojroot_2.0.3 readr_2.1.3
[81] stringi_1.7.8 parallel_4.1.0 Rcpp_1.0.9 vctrs_0.5.1
[85] tidyselect_1.2.0 xfun_0.35