T2D - Pancreas
sheng Qian
2021-2-6
Last updated: 2022-02-13
Checks: 6 1
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/ | data |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/ctwas_config.R | code/ctwas_config.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 87fee8b. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .ipynb_checkpoints/
Untracked files:
Untracked: code/.ipynb_checkpoints/
Untracked: code/AF_out/
Untracked: code/BMI_out/
Untracked: code/T2D_out/
Untracked: code/ctwas_config.R
Untracked: code/mapping.R
Untracked: code/out/
Untracked: code/run_AF_analysis.sbatch
Untracked: code/run_AF_analysis.sh
Untracked: code/run_AF_ctwas_rss_LDR.R
Untracked: code/run_BMI_analysis.sbatch
Untracked: code/run_BMI_analysis.sh
Untracked: code/run_BMI_ctwas_rss_LDR.R
Untracked: code/run_T2D_analysis.sbatch
Untracked: code/run_T2D_analysis.sh
Untracked: code/run_T2D_ctwas_rss_LDR.R
Untracked: data/.ipynb_checkpoints/
Untracked: data/AF/
Untracked: data/BMI/
Untracked: data/T2D/
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: data/untitled.txt
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/T2D_Pancreas.Rmd) and HTML (docs/T2D_Pancreas.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 87fee8b | sq-96 | 2022-02-13 | update |
Introduction
Weight QC
qclist_all <- list()
qc_files <- paste0(results_dir, "/", list.files(results_dir, pattern="exprqc.Rd"))
for (i in 1:length(qc_files)){
load(qc_files[i])
chr <- unlist(strsplit(rev(unlist(strsplit(qc_files[i], "_")))[1], "[.]"))[1]
qclist_all[[chr]] <- cbind(do.call(rbind, lapply(qclist,unlist)), as.numeric(substring(chr,4)))
}
qclist_all <- data.frame(do.call(rbind, qclist_all))
colnames(qclist_all)[ncol(qclist_all)] <- "chr"
rm(qclist, wgtlist, z_gene_chr)
#number of imputed weights
nrow(qclist_all)[1] 11277#number of imputed weights by chromosome
table(qclist_all$chr)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
1099 800 650 449 509 634 558 399 424 442 714 630 218 369 389 519
17 18 19 20 21 22
698 159 865 346 120 286 #number of imputed weights without missing variants
sum(qclist_all$nmiss==0)[1] 8525#proportion of imputed weights without missing variants
mean(qclist_all$nmiss==0)[1] 0.7559635Load ctwas results
Check convergence of parameters
library(ggplot2)
library(cowplot)
********************************************************Note: As of version 1.0.0, cowplot does not change the default ggplot2 theme anymore. To recover the previous behavior, execute:
theme_set(theme_cowplot())********************************************************load(paste0(results_dir, "/", analysis_id, "_ctwas.s2.susieIrssres.Rd"))
df <- data.frame(niter = rep(1:ncol(group_prior_rec), 2),
value = c(group_prior_rec[1,], group_prior_rec[2,]),
group = rep(c("Gene", "SNP"), each = ncol(group_prior_rec)))
df$group <- as.factor(df$group)
df$value[df$group=="SNP"] <- df$value[df$group=="SNP"]*thin #adjust parameter to account for thin argument
p_pi <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(pi)) +
ggtitle("Prior mean") +
theme_cowplot()
df <- data.frame(niter = rep(1:ncol(group_prior_var_rec), 2),
value = c(group_prior_var_rec[1,], group_prior_var_rec[2,]),
group = rep(c("Gene", "SNP"), each = ncol(group_prior_var_rec)))
df$group <- as.factor(df$group)
p_sigma2 <- ggplot(df, aes(x=niter, y=value, group=group)) +
geom_line(aes(color=group)) +
geom_point(aes(color=group)) +
xlab("Iteration") + ylab(bquote(sigma^2)) +
ggtitle("Prior variance") +
theme_cowplot()
plot_grid(p_pi, p_sigma2)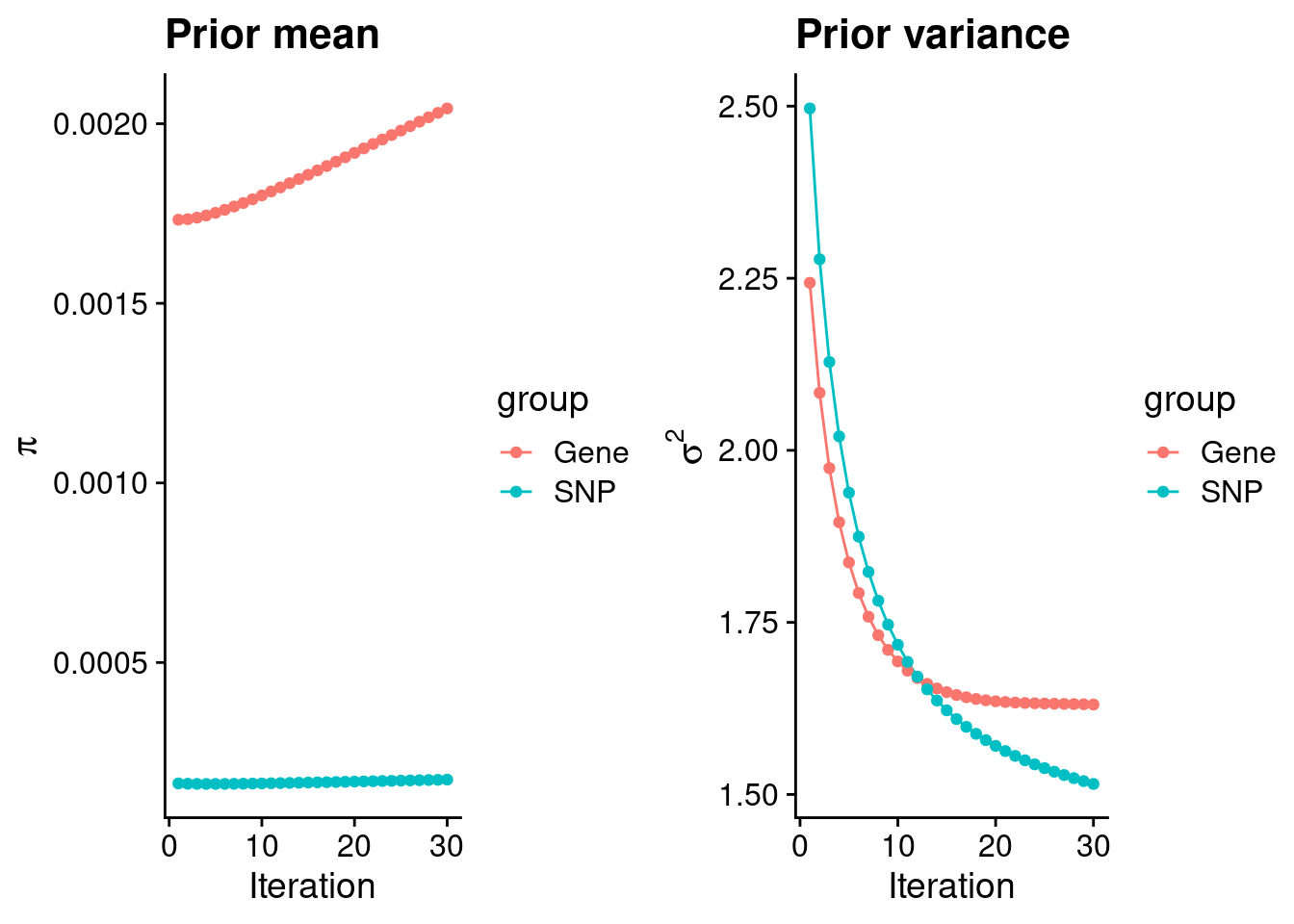
#estimated group prior
estimated_group_prior <- group_prior_rec[,ncol(group_prior_rec)]
names(estimated_group_prior) <- c("gene", "snp")
estimated_group_prior["snp"] <- estimated_group_prior["snp"]*thin #adjust parameter to account for thin argument
print(estimated_group_prior) gene snp
0.0020424708 0.0001739611 #estimated group prior variance
estimated_group_prior_var <- group_prior_var_rec[,ncol(group_prior_var_rec)]
names(estimated_group_prior_var) <- c("gene", "snp")
print(estimated_group_prior_var) gene snp
1.630669 1.515279 #report sample size
print(sample_size)[1] 337159#report group size
group_size <- c(nrow(ctwas_gene_res), n_snps)
print(group_size)[1] 11277 7535010#estimated group PVE
estimated_group_pve <- estimated_group_prior_var*estimated_group_prior*group_size/sample_size #check PVE calculation
names(estimated_group_pve) <- c("gene", "snp")
print(estimated_group_pve) gene snp
0.0001113988 0.0058910670 #compare sum(PIP*mu2/sample_size) with above PVE calculation
c(sum(ctwas_gene_res$PVE),sum(ctwas_snp_res$PVE))[1] 0.001961437 0.108209800Genes with highest PIPs
#distribution of PIPs
hist(ctwas_gene_res$susie_pip, xlim=c(0,1), main="Distribution of Gene PIPs")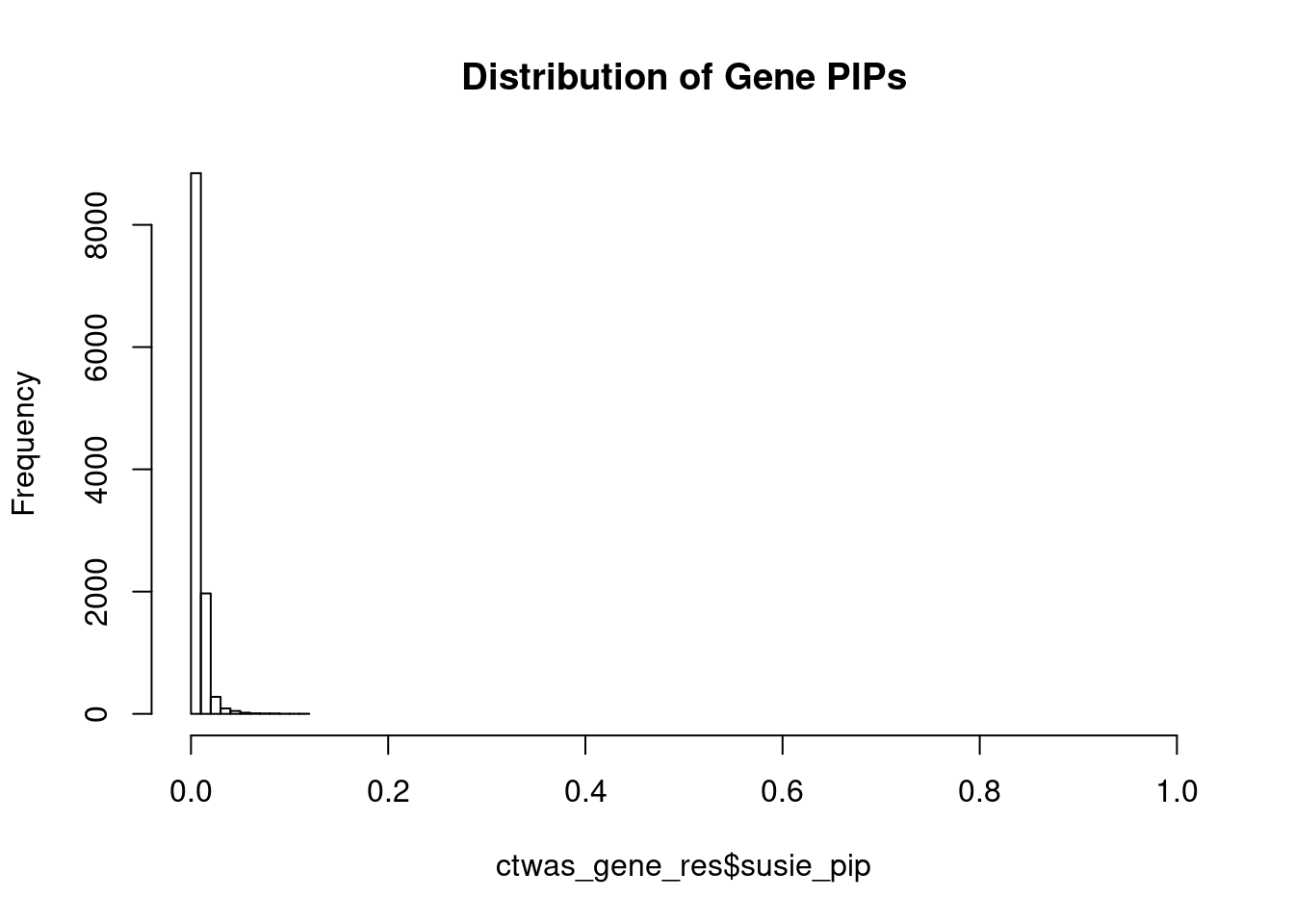
#genes with PIP>0.8 or 20 highest PIPs
head(ctwas_gene_res[order(-ctwas_gene_res$susie_pip),report_cols], max(sum(ctwas_gene_res$susie_pip>0.8), 20)) genename region_tag susie_pip mu2 PVE z
6813 NUS1 6_78 0.11190974 21.57983 7.162773e-06 3.692788
13639 LINC01126 2_27 0.11047242 22.33018 7.316634e-06 4.620415
3328 HPCAL4 1_24 0.10705683 20.92925 6.645587e-06 3.385568
619 HIPK2 7_85 0.10560943 20.59309 6.450441e-06 -3.356192
9179 HPSE 4_56 0.09090366 20.16349 5.436411e-06 -3.109502
7711 PPM1K 4_59 0.09063283 19.36279 5.204977e-06 3.048188
1140 FAT1 4_120 0.08898826 19.79091 5.223526e-06 3.124110
11775 KCTD11 17_6 0.08889942 19.53064 5.149683e-06 3.073744
7483 MAGOH 1_33 0.08556249 18.78052 4.766025e-06 3.091534
12626 RP11-675F6.3 8_34 0.08551601 19.54047 4.956185e-06 3.136535
11621 NT5M 17_15 0.08489430 18.42428 4.639107e-06 3.066208
12120 KLHL7-AS1 7_20 0.08230316 19.19736 4.686226e-06 3.456104
2430 SEC23IP 10_74 0.08213382 27.68458 6.744118e-06 -3.522402
1045 MOXD1 6_87 0.07975681 18.88876 4.468240e-06 2.971004
13004 IKBKE 1_105 0.07737702 18.94081 4.346861e-06 3.066183
922 MARK3 14_54 0.07500805 18.47044 4.109134e-06 3.322034
10756 ZNF559 19_9 0.07470959 18.40947 4.079274e-06 -3.038294
2836 SERINC1 6_82 0.07341538 18.38638 4.003580e-06 -3.103585
7860 AGGF1 5_45 0.07202300 18.10416 3.867363e-06 -3.189044
7087 AP3S2 15_41 0.07086689 18.12677 3.810036e-06 -3.581700
num_eqtl
6813 2
13639 1
3328 1
619 1
9179 1
7711 1
1140 1
11775 1
7483 1
12626 2
11621 2
12120 1
2430 1
1045 3
13004 2
922 2
10756 1
2836 2
7860 2
7087 1Genes with largest effect sizes
#plot PIP vs effect size
plot(ctwas_gene_res$susie_pip, ctwas_gene_res$mu2, xlab="PIP", ylab="mu^2", main="Gene PIPs vs Effect Size")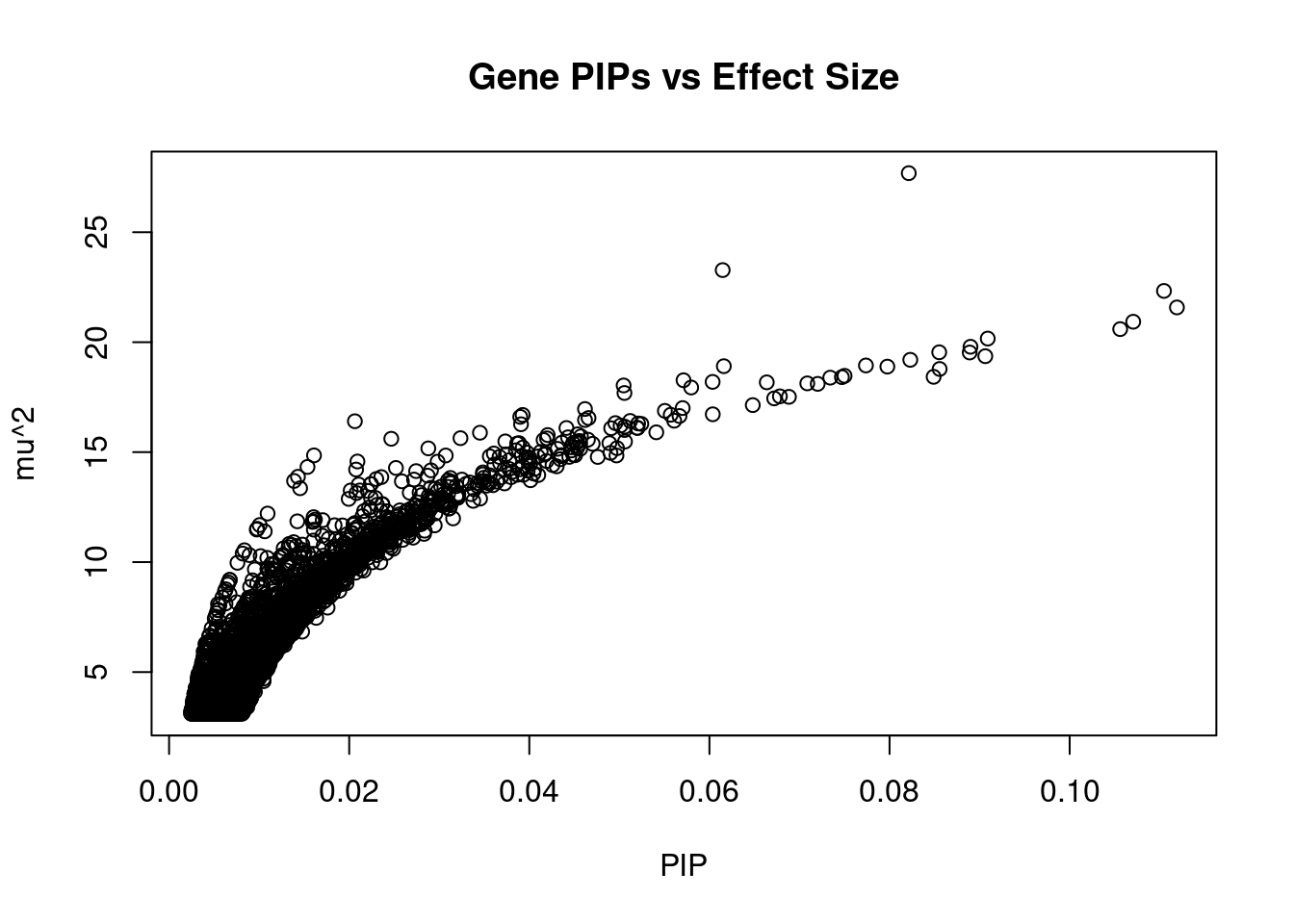
#genes with 20 largest effect sizes
head(ctwas_gene_res[order(-ctwas_gene_res$mu2),report_cols],20) genename region_tag susie_pip mu2 PVE z
2430 SEC23IP 10_74 0.08213382 27.68458 6.744118e-06 -3.522402
11263 SDAD1 4_52 0.06147185 23.27689 4.243914e-06 -3.799166
13639 LINC01126 2_27 0.11047242 22.33018 7.316634e-06 4.620415
6813 NUS1 6_78 0.11190974 21.57983 7.162773e-06 3.692788
3328 HPCAL4 1_24 0.10705683 20.92925 6.645587e-06 3.385568
619 HIPK2 7_85 0.10560943 20.59309 6.450441e-06 -3.356192
9179 HPSE 4_56 0.09090366 20.16349 5.436411e-06 -3.109502
1140 FAT1 4_120 0.08898826 19.79091 5.223526e-06 3.124110
12626 RP11-675F6.3 8_34 0.08551601 19.54047 4.956185e-06 3.136535
11775 KCTD11 17_6 0.08889942 19.53064 5.149683e-06 3.073744
7711 PPM1K 4_59 0.09063283 19.36279 5.204977e-06 3.048188
12120 KLHL7-AS1 7_20 0.08230316 19.19736 4.686226e-06 3.456104
13004 IKBKE 1_105 0.07737702 18.94081 4.346861e-06 3.066183
10530 PIP5K1C 19_4 0.06159915 18.90786 3.454478e-06 3.055707
1045 MOXD1 6_87 0.07975681 18.88876 4.468240e-06 2.971004
7483 MAGOH 1_33 0.08556249 18.78052 4.766025e-06 3.091534
922 MARK3 14_54 0.07500805 18.47044 4.109134e-06 3.322034
11621 NT5M 17_15 0.08489430 18.42428 4.639107e-06 3.066208
10756 ZNF559 19_9 0.07470959 18.40947 4.079274e-06 -3.038294
2836 SERINC1 6_82 0.07341538 18.38638 4.003580e-06 -3.103585
num_eqtl
2430 1
11263 2
13639 1
6813 2
3328 1
619 1
9179 1
1140 1
12626 2
11775 1
7711 1
12120 1
13004 2
10530 1
1045 3
7483 1
922 2
11621 2
10756 1
2836 2Genes with highest PVE
#genes with 20 highest pve
head(ctwas_gene_res[order(-ctwas_gene_res$PVE),report_cols],20) genename region_tag susie_pip mu2 PVE z
13639 LINC01126 2_27 0.11047242 22.33018 7.316634e-06 4.620415
6813 NUS1 6_78 0.11190974 21.57983 7.162773e-06 3.692788
2430 SEC23IP 10_74 0.08213382 27.68458 6.744118e-06 -3.522402
3328 HPCAL4 1_24 0.10705683 20.92925 6.645587e-06 3.385568
619 HIPK2 7_85 0.10560943 20.59309 6.450441e-06 -3.356192
9179 HPSE 4_56 0.09090366 20.16349 5.436411e-06 -3.109502
1140 FAT1 4_120 0.08898826 19.79091 5.223526e-06 3.124110
7711 PPM1K 4_59 0.09063283 19.36279 5.204977e-06 3.048188
11775 KCTD11 17_6 0.08889942 19.53064 5.149683e-06 3.073744
12626 RP11-675F6.3 8_34 0.08551601 19.54047 4.956185e-06 3.136535
7483 MAGOH 1_33 0.08556249 18.78052 4.766025e-06 3.091534
12120 KLHL7-AS1 7_20 0.08230316 19.19736 4.686226e-06 3.456104
11621 NT5M 17_15 0.08489430 18.42428 4.639107e-06 3.066208
1045 MOXD1 6_87 0.07975681 18.88876 4.468240e-06 2.971004
13004 IKBKE 1_105 0.07737702 18.94081 4.346861e-06 3.066183
11263 SDAD1 4_52 0.06147185 23.27689 4.243914e-06 -3.799166
922 MARK3 14_54 0.07500805 18.47044 4.109134e-06 3.322034
10756 ZNF559 19_9 0.07470959 18.40947 4.079274e-06 -3.038294
2836 SERINC1 6_82 0.07341538 18.38638 4.003580e-06 -3.103585
7860 AGGF1 5_45 0.07202300 18.10416 3.867363e-06 -3.189044
num_eqtl
13639 1
6813 2
2430 1
3328 1
619 1
9179 1
1140 1
7711 1
11775 1
12626 2
7483 1
12120 1
11621 2
1045 3
13004 2
11263 2
922 2
10756 1
2836 2
7860 2Genes with largest z scores
#genes with 20 largest z scores
head(ctwas_gene_res[order(-abs(ctwas_gene_res$z)),report_cols],20) genename region_tag susie_pip mu2 PVE z
13639 LINC01126 2_27 0.11047242 22.33018 7.316634e-06 4.620415
11263 SDAD1 4_52 0.06147185 23.27689 4.243914e-06 -3.799166
6813 NUS1 6_78 0.11190974 21.57983 7.162773e-06 3.692788
7087 AP3S2 15_41 0.07086689 18.12677 3.810036e-06 -3.581700
2430 SEC23IP 10_74 0.08213382 27.68458 6.744118e-06 -3.522402
1650 RBX1 22_17 0.05667685 16.64306 2.797720e-06 -3.521311
9101 DNAJB7 22_17 0.05202223 16.09983 2.484137e-06 3.462008
12120 KLHL7-AS1 7_20 0.08230316 19.19736 4.686226e-06 3.456104
12694 RP11-108O10.2 11_66 0.04429810 15.67381 2.059326e-06 3.442454
3328 HPCAL4 1_24 0.10705683 20.92925 6.645587e-06 3.385568
619 HIPK2 7_85 0.10560943 20.59309 6.450441e-06 -3.356192
10196 SH2D7 15_36 0.05013328 16.21181 2.410586e-06 3.348970
922 MARK3 14_54 0.07500805 18.47044 4.109134e-06 3.322034
4899 ISCA1 9_44 0.06636759 18.17383 3.577403e-06 3.269765
7860 AGGF1 5_45 0.07202300 18.10416 3.867363e-06 -3.189044
10276 CRELD2 22_24 0.04657461 16.55146 2.286393e-06 3.185929
3756 KLHL7 7_20 0.04651000 15.56592 2.147268e-06 3.138791
12626 RP11-675F6.3 8_34 0.08551601 19.54047 4.956185e-06 3.136535
474 BCAR1 16_40 0.03966336 14.74818 1.734975e-06 3.130231
1617 CHKB 22_24 0.06035504 18.19411 3.256939e-06 -3.125165
num_eqtl
13639 1
11263 2
6813 2
7087 1
2430 1
1650 1
9101 1
12120 1
12694 2
3328 1
619 1
10196 1
922 2
4899 1
7860 2
10276 1
3756 3
12626 2
474 1
1617 2Comparing z scores and PIPs
#set nominal signifiance threshold for z scores
alpha <- 0.05
#bonferroni adjusted threshold for z scores
sig_thresh <- qnorm(1-(alpha/nrow(ctwas_gene_res)/2), lower=T)
#Q-Q plot for z scores
obs_z <- ctwas_gene_res$z[order(ctwas_gene_res$z)]
exp_z <- qnorm((1:nrow(ctwas_gene_res))/nrow(ctwas_gene_res))
plot(exp_z, obs_z, xlab="Expected z", ylab="Observed z", main="Gene z score Q-Q plot")
abline(a=0,b=1)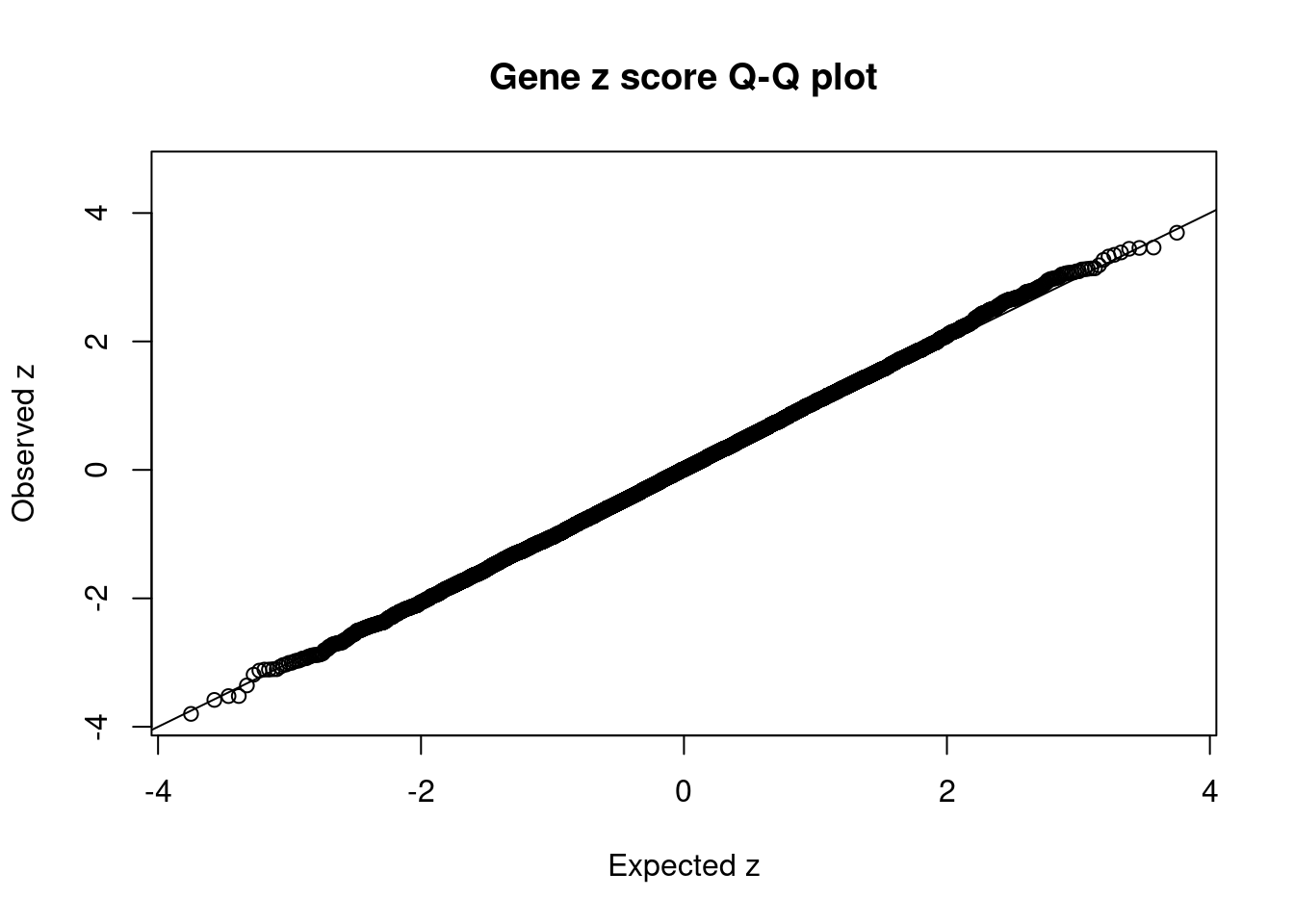
#plot z score vs PIP
plot(abs(ctwas_gene_res$z), ctwas_gene_res$susie_pip, xlab="abs(z)", ylab="PIP")
abline(v=sig_thresh, col="red", lty=2)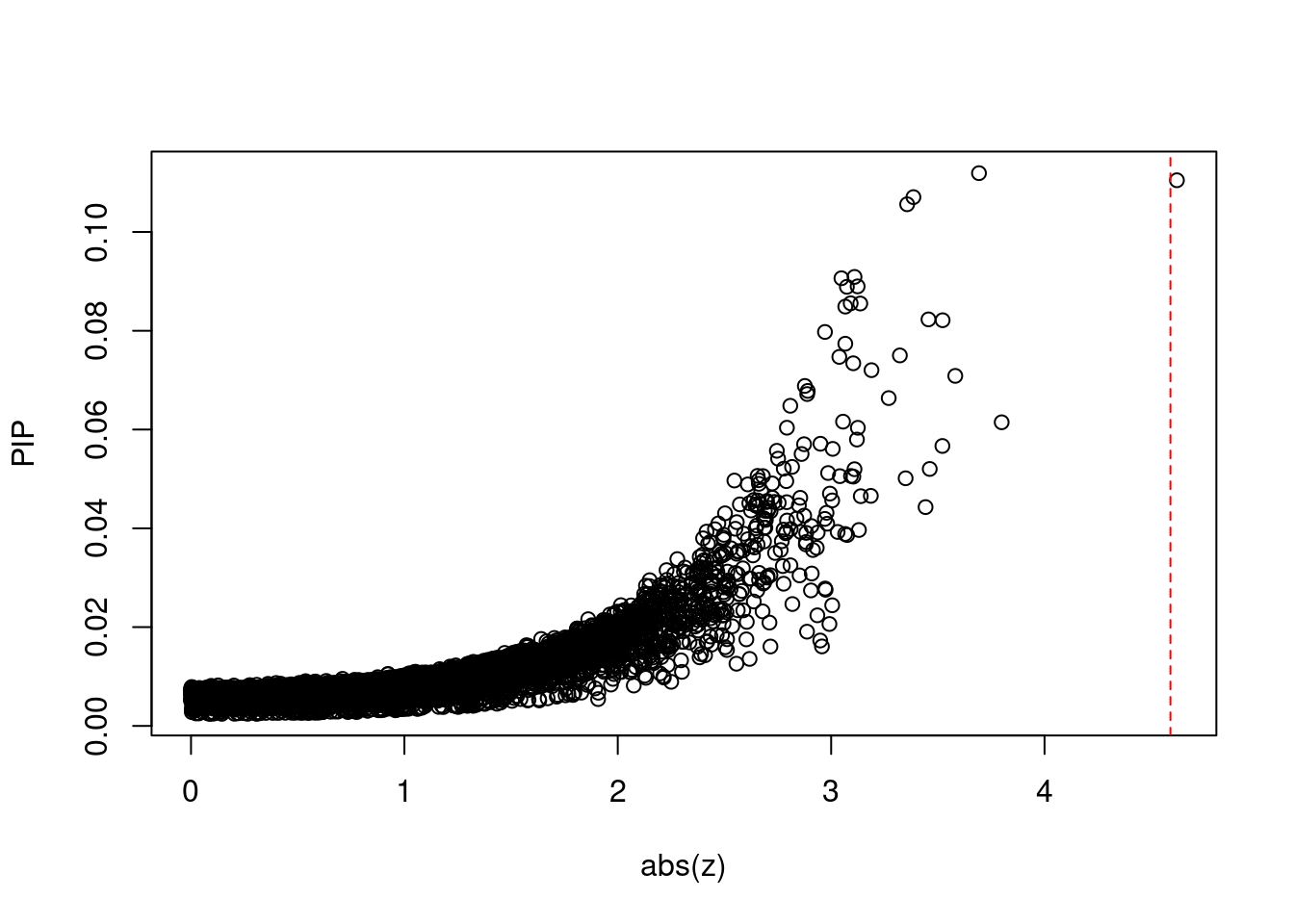
#proportion of significant z scores
mean(abs(ctwas_gene_res$z) > sig_thresh)[1] 8.867607e-05#genes with most significant z scores
head(ctwas_gene_res[order(-abs(ctwas_gene_res$z)),report_cols],20) genename region_tag susie_pip mu2 PVE z
13639 LINC01126 2_27 0.11047242 22.33018 7.316634e-06 4.620415
11263 SDAD1 4_52 0.06147185 23.27689 4.243914e-06 -3.799166
6813 NUS1 6_78 0.11190974 21.57983 7.162773e-06 3.692788
7087 AP3S2 15_41 0.07086689 18.12677 3.810036e-06 -3.581700
2430 SEC23IP 10_74 0.08213382 27.68458 6.744118e-06 -3.522402
1650 RBX1 22_17 0.05667685 16.64306 2.797720e-06 -3.521311
9101 DNAJB7 22_17 0.05202223 16.09983 2.484137e-06 3.462008
12120 KLHL7-AS1 7_20 0.08230316 19.19736 4.686226e-06 3.456104
12694 RP11-108O10.2 11_66 0.04429810 15.67381 2.059326e-06 3.442454
3328 HPCAL4 1_24 0.10705683 20.92925 6.645587e-06 3.385568
619 HIPK2 7_85 0.10560943 20.59309 6.450441e-06 -3.356192
10196 SH2D7 15_36 0.05013328 16.21181 2.410586e-06 3.348970
922 MARK3 14_54 0.07500805 18.47044 4.109134e-06 3.322034
4899 ISCA1 9_44 0.06636759 18.17383 3.577403e-06 3.269765
7860 AGGF1 5_45 0.07202300 18.10416 3.867363e-06 -3.189044
10276 CRELD2 22_24 0.04657461 16.55146 2.286393e-06 3.185929
3756 KLHL7 7_20 0.04651000 15.56592 2.147268e-06 3.138791
12626 RP11-675F6.3 8_34 0.08551601 19.54047 4.956185e-06 3.136535
474 BCAR1 16_40 0.03966336 14.74818 1.734975e-06 3.130231
1617 CHKB 22_24 0.06035504 18.19411 3.256939e-06 -3.125165
num_eqtl
13639 1
11263 2
6813 2
7087 1
2430 1
1650 1
9101 1
12120 1
12694 2
3328 1
619 1
10196 1
922 2
4899 1
7860 2
10276 1
3756 3
12626 2
474 1
1617 2Sensitivity, specificity and precision for silver standard genes
library("readxl")
known_annotations <- read_xlsx("data/summary_known_genes_annotations.xlsx", sheet="T2D")
known_annotations <- unique(known_annotations$`Gene Symbol`)
unrelated_genes <- ctwas_gene_res$genename[!(ctwas_gene_res$genename %in% known_annotations)]
#number of genes in known annotations
print(length(known_annotations))[1] 72#number of genes in known annotations with imputed expression
print(sum(known_annotations %in% ctwas_gene_res$genename))[1] 40#assign ctwas, TWAS, and bystander genes
ctwas_genes <- ctwas_gene_res$genename[ctwas_gene_res$susie_pip>0.8]
twas_genes <- ctwas_gene_res$genename[abs(ctwas_gene_res$z)>sig_thresh]
novel_genes <- ctwas_genes[!(ctwas_genes %in% twas_genes)]
#significance threshold for TWAS
print(sig_thresh)[1] 4.589937#number of ctwas genes
length(ctwas_genes)[1] 0#number of TWAS genes
length(twas_genes)[1] 1#show novel genes (ctwas genes with not in TWAS genes)
ctwas_gene_res[ctwas_gene_res$genename %in% novel_genes,report_cols][1] genename region_tag susie_pip mu2 PVE z num_eqtl
<0 rows> (or 0-length row.names)#sensitivity / recall
sensitivity <- rep(NA,2)
names(sensitivity) <- c("ctwas", "TWAS")
sensitivity["ctwas"] <- sum(ctwas_genes %in% known_annotations)/length(known_annotations)
sensitivity["TWAS"] <- sum(twas_genes %in% known_annotations)/length(known_annotations)
sensitivityctwas TWAS
0 0 #specificity
specificity <- rep(NA,2)
names(specificity) <- c("ctwas", "TWAS")
specificity["ctwas"] <- sum(!(unrelated_genes %in% ctwas_genes))/length(unrelated_genes)
specificity["TWAS"] <- sum(!(unrelated_genes %in% twas_genes))/length(unrelated_genes)
specificity ctwas TWAS
1.000000 0.999911 #precision / PPV
precision <- rep(NA,2)
names(precision) <- c("ctwas", "TWAS")
precision["ctwas"] <- sum(ctwas_genes %in% known_annotations)/length(ctwas_genes)
precision["TWAS"] <- sum(twas_genes %in% known_annotations)/length(twas_genes)
precisionctwas TWAS
NaN 0 #ROC curves
pip_range <- (0:1000)/1000
sensitivity <- rep(NA, length(pip_range))
specificity <- rep(NA, length(pip_range))
for (index in 1:length(pip_range)){
pip <- pip_range[index]
ctwas_genes <- ctwas_gene_res$genename[ctwas_gene_res$susie_pip>=pip]
sensitivity[index] <- sum(ctwas_genes %in% known_annotations)/length(known_annotations)
specificity[index] <- sum(!(unrelated_genes %in% ctwas_genes))/length(unrelated_genes)
}
plot(1-specificity, sensitivity, type="l", xlim=c(0,1), ylim=c(0,1))
sig_thresh_range <- seq(from=0, to=max(abs(ctwas_gene_res$z)), length.out=length(pip_range))
for (index in 1:length(sig_thresh_range)){
sig_thresh_plot <- sig_thresh_range[index]
twas_genes <- ctwas_gene_res$genename[abs(ctwas_gene_res$z)>=sig_thresh_plot]
sensitivity[index] <- sum(twas_genes %in% known_annotations)/length(known_annotations)
specificity[index] <- sum(!(unrelated_genes %in% twas_genes))/length(unrelated_genes)
}
lines(1-specificity, sensitivity, xlim=c(0,1), ylim=c(0,1), col="red", lty=2)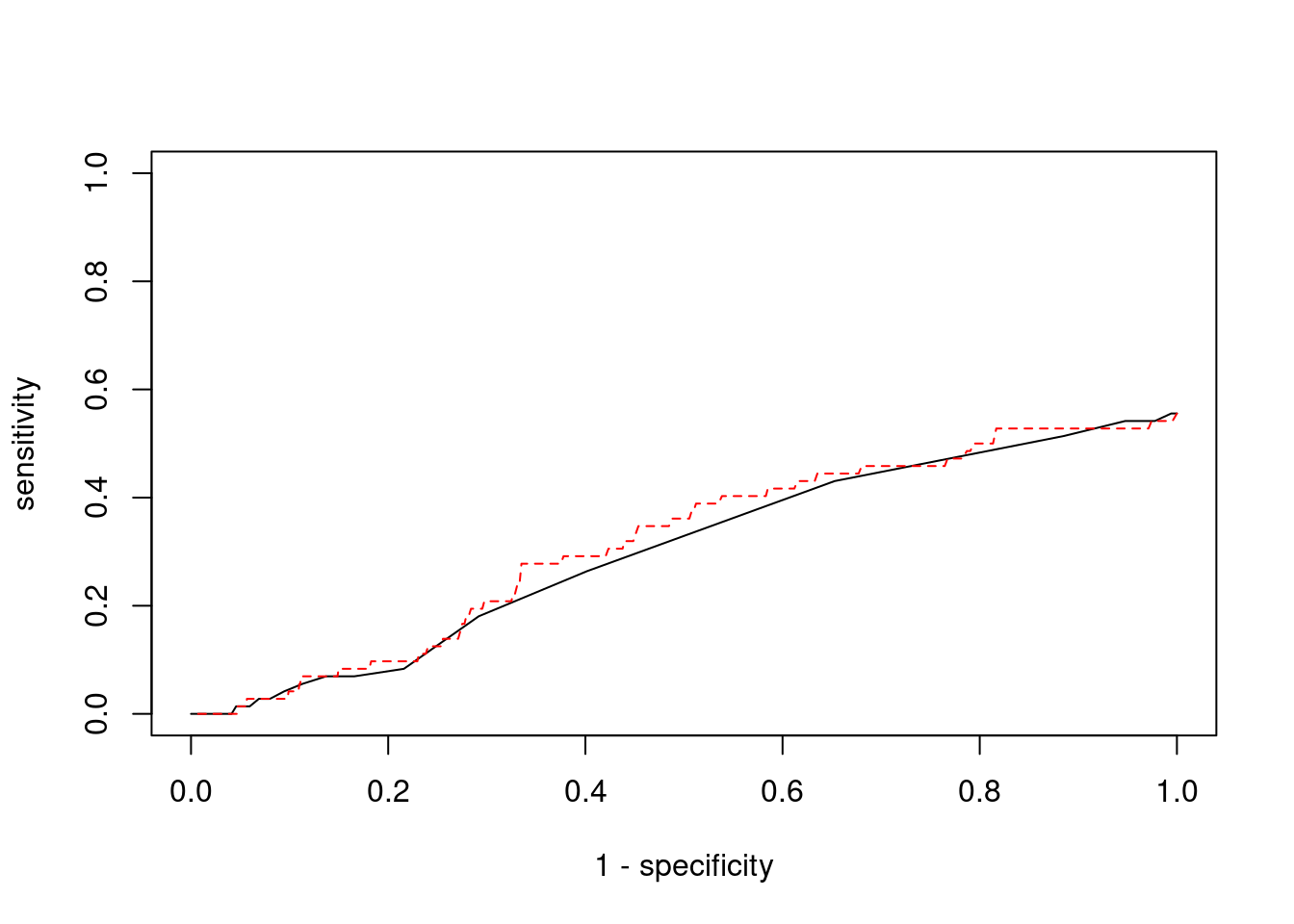
Sensitivity, specificity and precision for silver standard genes - bystanders only
This section first uses all silver standard genes to identify bystander genes within 1Mb. The silver standard and bystander gene lists are then subset to only genes with imputed expression in this analysis. Then, the ctwas and TWAS gene lists from this analysis are subset to only genes that are in the (subset) silver standard and bystander genes. These gene lists are then used to compute sensitivity, specificity and precision for ctwas and TWAS.
library(biomaRt)
library(GenomicRanges)Loading required package: stats4Loading required package: BiocGenericsLoading required package: parallel
Attaching package: 'BiocGenerics'The following objects are masked from 'package:parallel':
clusterApply, clusterApplyLB, clusterCall, clusterEvalQ,
clusterExport, clusterMap, parApply, parCapply, parLapply,
parLapplyLB, parRapply, parSapply, parSapplyLBThe following objects are masked from 'package:stats':
IQR, mad, sd, var, xtabsThe following objects are masked from 'package:base':
anyDuplicated, append, as.data.frame, basename, cbind, colnames,
dirname, do.call, duplicated, eval, evalq, Filter, Find, get, grep,
grepl, intersect, is.unsorted, lapply, Map, mapply, match, mget,
order, paste, pmax, pmax.int, pmin, pmin.int, Position, rank,
rbind, Reduce, rownames, sapply, setdiff, sort, table, tapply,
union, unique, unsplit, which, which.max, which.minLoading required package: S4Vectors
Attaching package: 'S4Vectors'The following object is masked from 'package:base':
expand.gridLoading required package: IRangesLoading required package: GenomeInfoDbensembl <- useEnsembl(biomart="ENSEMBL_MART_ENSEMBL", dataset="hsapiens_gene_ensembl")
G_list <- getBM(filters= "chromosome_name", attributes= c("hgnc_symbol","chromosome_name","start_position","end_position","gene_biotype"), values=1:22, mart=ensembl)
G_list <- G_list[G_list$hgnc_symbol!="",]
G_list <- G_list[G_list$gene_biotype %in% c("protein_coding","lncRNA"),]
G_list$start <- G_list$start_position
G_list$end <- G_list$end_position
G_list_granges <- makeGRangesFromDataFrame(G_list, keep.extra.columns=T)
known_annotations_positions <- G_list[G_list$hgnc_symbol %in% known_annotations,]
half_window <- 1000000
known_annotations_positions$start <- known_annotations_positions$start_position - half_window
known_annotations_positions$end <- known_annotations_positions$end_position + half_window
known_annotations_positions$start[known_annotations_positions$start<1] <- 1
known_annotations_granges <- makeGRangesFromDataFrame(known_annotations_positions, keep.extra.columns=T)
bystanders <- findOverlaps(known_annotations_granges,G_list_granges)
bystanders <- unique(subjectHits(bystanders))
bystanders <- G_list$hgnc_symbol[bystanders]
bystanders <- bystanders[!(bystanders %in% known_annotations)]
unrelated_genes <- bystanders
#number of genes in known annotations
print(length(known_annotations))[1] 72#number of genes in known annotations with imputed expression
print(sum(known_annotations %in% ctwas_gene_res$genename))[1] 40#number of bystander genes
print(length(unrelated_genes))[1] 1847#number of bystander genes with imputed expression
print(sum(unrelated_genes %in% ctwas_gene_res$genename))[1] 893#remove genes without imputed expression from gene lists
known_annotations <- known_annotations[known_annotations %in% ctwas_gene_res$genename]
unrelated_genes <- unrelated_genes[unrelated_genes %in% ctwas_gene_res$genename]
#assign ctwas and TWAS genes
ctwas_genes <- ctwas_gene_res$genename[ctwas_gene_res$susie_pip>0.8]
twas_genes <- ctwas_gene_res$genename[abs(ctwas_gene_res$z)>sig_thresh]
#significance threshold for TWAS
print(sig_thresh)[1] 4.589937#number of ctwas genes
length(ctwas_genes)[1] 0#number of ctwas genes in known annotations or bystanders
sum(ctwas_genes %in% c(known_annotations, unrelated_genes))[1] 0#number of ctwas genes
length(twas_genes)[1] 1#number of TWAS genes
sum(twas_genes %in% c(known_annotations, unrelated_genes))[1] 0#remove genes not in known or bystander lists from results
ctwas_genes <- ctwas_genes[ctwas_genes %in% c(known_annotations, unrelated_genes)]
twas_genes <- twas_genes[twas_genes %in% c(known_annotations, unrelated_genes)]
#sensitivity / recall
sensitivity <- rep(NA,2)
names(sensitivity) <- c("ctwas", "TWAS")
sensitivity["ctwas"] <- sum(ctwas_genes %in% known_annotations)/length(known_annotations)
sensitivity["TWAS"] <- sum(twas_genes %in% known_annotations)/length(known_annotations)
sensitivityctwas TWAS
0 0 #specificity
specificity <- rep(NA,2)
names(specificity) <- c("ctwas", "TWAS")
specificity["ctwas"] <- sum(!(unrelated_genes %in% ctwas_genes))/length(unrelated_genes)
specificity["TWAS"] <- sum(!(unrelated_genes %in% twas_genes))/length(unrelated_genes)
specificityctwas TWAS
1 1 #precision / PPV
precision <- rep(NA,2)
names(precision) <- c("ctwas", "TWAS")
precision["ctwas"] <- sum(ctwas_genes %in% known_annotations)/length(ctwas_genes)
precision["TWAS"] <- sum(twas_genes %in% known_annotations)/length(twas_genes)
precisionctwas TWAS
NaN NaN
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] GenomicRanges_1.36.1 GenomeInfoDb_1.20.0 IRanges_2.18.1
[4] S4Vectors_0.22.1 BiocGenerics_0.30.0 biomaRt_2.40.1
[7] readxl_1.3.1 cowplot_1.0.0 ggplot2_3.3.5
[10] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rcpp_1.0.7 prettyunits_1.1.1 assertthat_0.2.1
[4] rprojroot_2.0.2 digest_0.6.29 utf8_1.2.2
[7] R6_2.5.1 cellranger_1.1.0 RSQLite_2.2.8
[10] evaluate_0.14 httr_1.4.2 highr_0.9
[13] pillar_1.6.4 zlibbioc_1.30.0 progress_1.2.2
[16] rlang_0.4.12 curl_4.3.2 data.table_1.14.2
[19] whisker_0.3-2 jquerylib_0.1.4 blob_1.2.2
[22] rmarkdown_2.11 labeling_0.4.2 stringr_1.4.0
[25] RCurl_1.98-1.5 bit_4.0.4 munsell_0.5.0
[28] compiler_3.6.1 httpuv_1.5.1 xfun_0.29
[31] pkgconfig_2.0.3 htmltools_0.5.2 tidyselect_1.1.1
[34] GenomeInfoDbData_1.2.1 tibble_3.1.6 XML_3.99-0.3
[37] fansi_0.5.0 crayon_1.4.2 dplyr_1.0.7
[40] withr_2.4.3 later_0.8.0 bitops_1.0-7
[43] grid_3.6.1 gtable_0.3.0 lifecycle_1.0.1
[46] DBI_1.1.1 git2r_0.26.1 magrittr_2.0.1
[49] scales_1.1.1 stringi_1.7.6 cachem_1.0.6
[52] XVector_0.24.0 farver_2.1.0 fs_1.5.2
[55] promises_1.0.1 ellipsis_0.3.2 generics_0.1.1
[58] vctrs_0.3.8 tools_3.6.1 bit64_4.0.5
[61] Biobase_2.44.0 glue_1.5.1 purrr_0.3.4
[64] hms_1.1.1 fastmap_1.1.0 yaml_2.2.1
[67] AnnotationDbi_1.46.0 colorspace_2.0-2 memoise_2.0.1
[70] knitr_1.36