SCZ - Brain Hypothalamus
sheng Qian
2021-2-6
Last updated: 2022-03-16
Checks: 5 2
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/ | data |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/ctwas_config.R | code/ctwas_config.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version d57314b. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .ipynb_checkpoints/
Ignored: data/AF/
Untracked files:
Untracked: Rplot.png
Untracked: analysis/.ipynb_checkpoints/
Untracked: analysis/SCZ_2020_Brain_Amygdala.Rmd
Untracked: analysis/SCZ_2020_Brain_Anterior_cingulate_cortex_BA24.Rmd
Untracked: analysis/SCZ_2020_Brain_Caudate_basal_ganglia.Rmd
Untracked: analysis/SCZ_2020_Brain_Cerebellar_Hemisphere.Rmd
Untracked: analysis/SCZ_2020_Brain_Cerebellum.Rmd
Untracked: analysis/SCZ_2020_Brain_Hippocampus.Rmd
Untracked: analysis/SCZ_2020_Brain_Nucleus_accumbens_basal_ganglia.Rmd
Untracked: analysis/SCZ_2020_Brain_Spinal_cord_cervical_c-1.Rmd
Untracked: analysis/SCZ_2020_Brain_Substantia_nigra.Rmd
Untracked: code/.ipynb_checkpoints/
Untracked: code/AF_out/
Untracked: code/Autism_out/
Untracked: code/BMI_S_out/
Untracked: code/BMI_out/
Untracked: code/Glucose_out/
Untracked: code/LDL_S_out/
Untracked: code/SCZ_2014_EUR_out/
Untracked: code/SCZ_2020_out/
Untracked: code/SCZ_S_out/
Untracked: code/SCZ_out/
Untracked: code/T2D_out/
Untracked: code/ctwas_config.R
Untracked: code/mapping.R
Untracked: code/out/
Untracked: code/run_AF_analysis.sbatch
Untracked: code/run_AF_analysis.sh
Untracked: code/run_AF_ctwas_rss_LDR.R
Untracked: code/run_Autism_analysis.sbatch
Untracked: code/run_Autism_analysis.sh
Untracked: code/run_Autism_ctwas_rss_LDR.R
Untracked: code/run_BMI_analysis.sbatch
Untracked: code/run_BMI_analysis.sh
Untracked: code/run_BMI_analysis_S.sbatch
Untracked: code/run_BMI_analysis_S.sh
Untracked: code/run_BMI_ctwas_rss_LDR.R
Untracked: code/run_BMI_ctwas_rss_LDR_S.R
Untracked: code/run_Glucose_analysis.sbatch
Untracked: code/run_Glucose_analysis.sh
Untracked: code/run_Glucose_ctwas_rss_LDR.R
Untracked: code/run_LDL_analysis_S.sbatch
Untracked: code/run_LDL_analysis_S.sh
Untracked: code/run_LDL_ctwas_rss_LDR_S.R
Untracked: code/run_SCZ_2014_EUR_analysis.sbatch
Untracked: code/run_SCZ_2014_EUR_analysis.sh
Untracked: code/run_SCZ_2014_EUR_ctwas_rss_LDR.R
Untracked: code/run_SCZ_2020_analysis.sbatch
Untracked: code/run_SCZ_2020_analysis.sh
Untracked: code/run_SCZ_2020_ctwas_rss_LDR.R
Untracked: code/run_SCZ_analysis.sbatch
Untracked: code/run_SCZ_analysis.sh
Untracked: code/run_SCZ_analysis_S.sbatch
Untracked: code/run_SCZ_analysis_S.sh
Untracked: code/run_SCZ_ctwas_rss_LDR.R
Untracked: code/run_SCZ_ctwas_rss_LDR_S.R
Untracked: code/run_T2D_analysis.sbatch
Untracked: code/run_T2D_analysis.sh
Untracked: code/run_T2D_ctwas_rss_LDR.R
Untracked: code/wflow_build.R
Untracked: code/wflow_build.sbatch
Untracked: data/.ipynb_checkpoints/
Untracked: data/BMI/
Untracked: data/PGC3_SCZ_wave3_public.v2.tsv
Untracked: data/SCZ/
Untracked: data/SCZ_2014_EUR/
Untracked: data/SCZ_2020/
Untracked: data/SCZ_S/
Untracked: data/T2D/
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/mashr_sqtl/
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: data/untitled.txt
Unstaged changes:
Modified: analysis/SCZ_2020_Brain_Cortex.Rmd
Modified: analysis/SCZ_2020_Brain_Frontal_Cortex_BA9.Rmd
Modified: analysis/SCZ_2020_Brain_Hypothalamus.Rmd
Modified: analysis/SCZ_2020_Brain_Putamen_basal_ganglia.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/SCZ_2020_Brain_Hypothalamus.Rmd) and HTML (docs/SCZ_2020_Brain_Hypothalamus.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | d57314b | sq-96 | 2022-03-15 | update |
Weight QC
#number of imputed weights
nrow(qclist_all)[1] 11075#number of imputed weights by chromosome
table(qclist_all$chr)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
1102 753 663 439 556 620 531 428 412 428 658 631 212 356 369 488
17 18 19 20 21 22
679 167 846 327 134 276 #number of imputed weights without missing variants
sum(qclist_all$nmiss==0)[1] 8842#proportion of imputed weights without missing variants
mean(qclist_all$nmiss==0)[1] 0.7984Check convergence of parameters

#estimated group prior
estimated_group_prior <- group_prior_rec[,ncol(group_prior_rec)]
names(estimated_group_prior) <- c("gene", "snp")
estimated_group_prior["snp"] <- estimated_group_prior["snp"]*thin #adjust parameter to account for thin argument
print(estimated_group_prior) gene snp
0.0092792 0.0002797 #estimated group prior variance
estimated_group_prior_var <- group_prior_var_rec[,ncol(group_prior_var_rec)]
names(estimated_group_prior_var) <- c("gene", "snp")
print(estimated_group_prior_var) gene snp
16.99 12.46 #report sample size
print(sample_size)[1] 161405#report group size
group_size <- c(nrow(ctwas_gene_res), n_snps)
print(group_size)[1] 11075 7394310#estimated group PVE
estimated_group_pve <- estimated_group_prior_var*estimated_group_prior*group_size/sample_size #check PVE calculation
names(estimated_group_pve) <- c("gene", "snp")
print(estimated_group_pve) gene snp
0.01082 0.15970 #compare sum(PIP*mu2/sample_size) with above PVE calculation
c(sum(ctwas_gene_res$PVE),sum(ctwas_snp_res$PVE))[1] 0.04506 0.80755Genes with highest PIPs
genename region_tag susie_pip mu2 PVE z num_eqtl
679 RASSF1 3_35 0.9998 1010.97 6.262e-03 4.532 1
5324 FURIN 15_42 0.9865 93.98 5.744e-04 -9.913 1
10843 ZNF823 19_10 0.9768 40.83 2.471e-04 6.310 2
11997 AC012074.2 2_15 0.9739 31.12 1.878e-04 5.469 2
3872 IRF3 19_35 0.9153 56.18 3.186e-04 -7.641 1
2551 TRPV4 12_66 0.8672 24.86 1.336e-04 4.416 1
6907 ACE 17_38 0.8341 35.37 1.828e-04 -5.876 1
4594 DAGLA 11_34 0.8069 22.96 1.148e-04 -4.263 1
11948 HIST1H2BN 6_21 0.7920 180.20 8.842e-04 13.182 1
10850 RPL12 9_66 0.7825 24.72 1.198e-04 4.671 2
10823 HIST4H4 12_12 0.7792 23.09 1.115e-04 -4.038 2
13339 CTA-246H3.12 22_7 0.7627 23.19 1.096e-04 4.019 2
8828 KAT5 11_36 0.7462 32.09 1.483e-04 5.224 1
5639 RIT1 1_76 0.7327 24.28 1.102e-04 -4.023 1
2933 APC2 19_2 0.7277 24.44 1.102e-04 4.108 1
8406 CALML6 1_1 0.7157 23.43 1.039e-04 4.505 2
7356 SERPINI1 3_103 0.7084 23.39 1.027e-04 -4.520 1
3844 MAX 14_30 0.7044 22.80 9.951e-05 4.329 1
783 ACADVL 17_6 0.7037 22.96 1.001e-04 -4.325 1
3951 ZNF835 19_38 0.6839 28.50 1.208e-04 5.136 1Genes with largest effect sizes
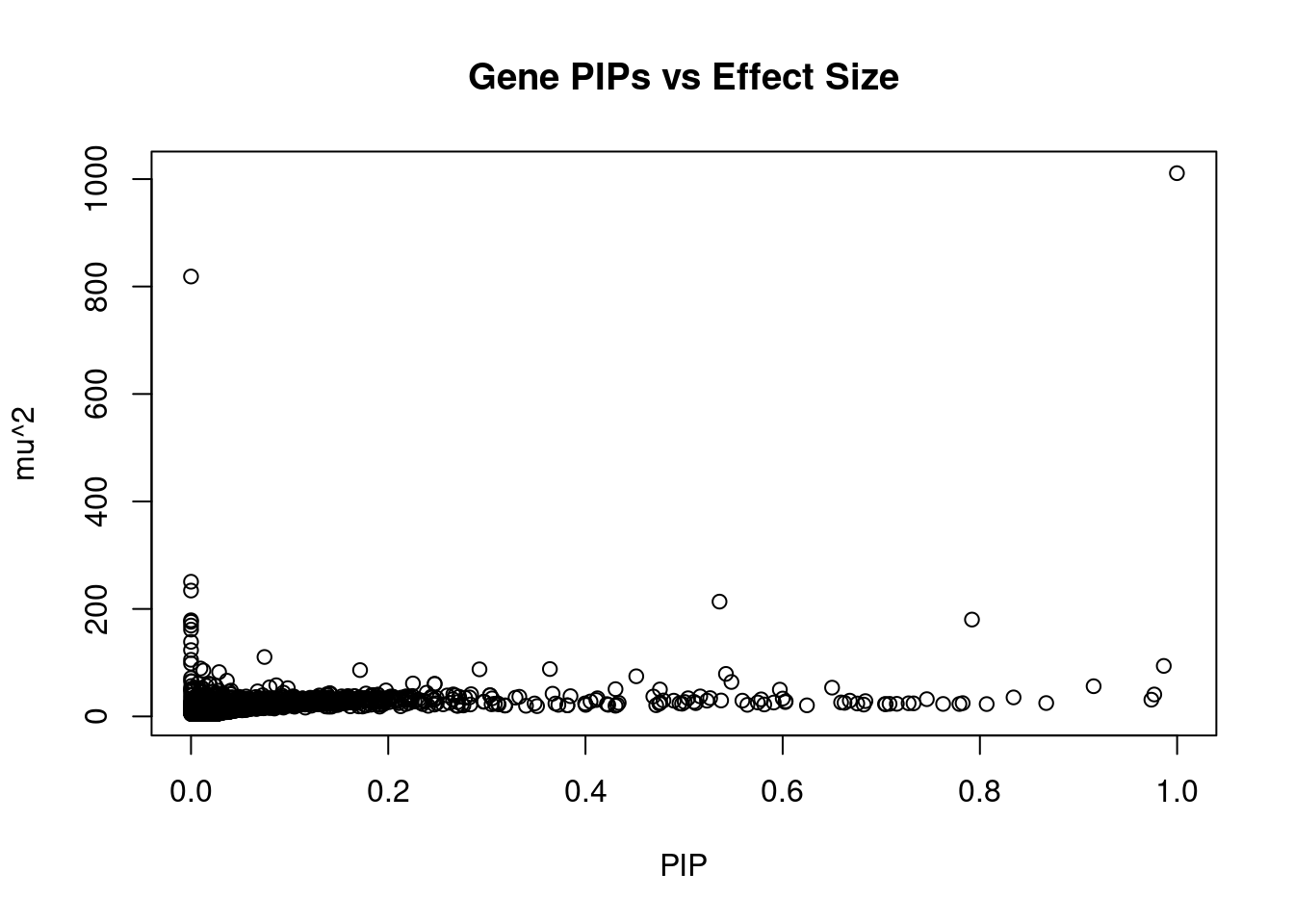
genename region_tag susie_pip mu2 PVE z num_eqtl
679 RASSF1 3_35 9.998e-01 1010.97 6.262e-03 4.5324 1
208 SEMA3B 3_35 0.000e+00 818.63 0.000e+00 1.4290 2
10406 SLC38A3 3_35 5.910e-13 250.62 9.176e-16 -2.7756 1
125 CACNA2D2 3_35 0.000e+00 234.19 0.000e+00 -0.1392 1
38 RBM6 3_35 5.359e-01 213.63 7.093e-04 4.4688 1
11948 HIST1H2BN 6_21 7.920e-01 180.20 8.842e-04 13.1822 1
7487 CAMKV 3_35 1.002e-07 178.89 1.110e-10 1.7107 1
10243 HYAL3 3_35 3.250e-13 176.06 3.545e-16 -2.5066 1
2875 HEMK1 3_35 0.000e+00 169.08 0.000e+00 -0.8999 1
7489 MST1R 3_35 2.336e-05 161.53 2.337e-08 -4.0250 1
12211 NAT6 3_35 3.077e-12 138.78 2.646e-15 1.5617 2
13397 LINC02019 3_35 0.000e+00 123.39 0.000e+00 0.3148 2
2736 PRSS16 6_21 7.452e-02 110.59 5.106e-05 -10.0002 1
7484 RNF123 3_35 2.220e-16 105.34 1.449e-19 -2.3622 1
2876 CISH 3_35 0.000e+00 98.55 0.000e+00 -0.8833 1
5324 FURIN 15_42 9.865e-01 93.98 5.744e-04 -9.9133 1
9592 HIST1H2BC 6_20 9.537e-03 89.18 5.270e-06 -7.9928 1
11166 MSH5 6_26 3.639e-01 88.14 1.987e-04 10.7348 2
11169 ABHD16A 6_26 2.925e-01 87.60 1.588e-04 10.7104 1
11174 APOM 6_26 1.715e-01 86.17 9.154e-05 10.6484 1Genes with highest PVE
genename region_tag susie_pip mu2 PVE z num_eqtl
679 RASSF1 3_35 0.9998 1010.97 0.0062622 4.532 1
11948 HIST1H2BN 6_21 0.7920 180.20 0.0008842 13.182 1
38 RBM6 3_35 0.5359 213.63 0.0007093 4.469 1
5324 FURIN 15_42 0.9865 93.98 0.0005744 -9.913 1
3872 IRF3 19_35 0.9153 56.18 0.0003186 -7.641 1
11131 HLA-DMA 6_27 0.5425 79.13 0.0002660 -9.408 1
10843 ZNF823 19_10 0.9768 40.83 0.0002471 6.310 2
7453 GNL3 3_36 0.5480 64.08 0.0002176 9.162 2
2970 SF3B1 2_117 0.6501 53.51 0.0002155 7.605 1
2829 PCCB 3_84 0.4515 74.67 0.0002089 -7.445 1
11166 MSH5 6_26 0.3639 88.14 0.0001987 10.735 2
11997 AC012074.2 2_15 0.9739 31.12 0.0001878 5.469 2
10797 NMB 15_39 0.5971 49.81 0.0001843 7.121 1
6907 ACE 17_38 0.8341 35.37 0.0001828 -5.876 1
11169 ABHD16A 6_26 0.2925 87.60 0.0001588 10.710 1
8828 KAT5 11_36 0.7462 32.09 0.0001483 5.224 1
1685 PPP1R16B 20_23 0.4755 50.16 0.0001478 7.550 1
6178 TAOK2 16_24 0.4305 50.75 0.0001353 7.474 1
2551 TRPV4 12_66 0.8672 24.86 0.0001336 4.416 1
8764 FUT9 6_65 0.6000 33.25 0.0001236 5.446 1Genes with largest z scores
genename region_tag susie_pip mu2 PVE z num_eqtl
11948 HIST1H2BN 6_21 0.7919507 180.20 8.842e-04 13.182 1
11166 MSH5 6_26 0.3639443 88.14 1.987e-04 10.735 2
11169 ABHD16A 6_26 0.2925358 87.60 1.588e-04 10.710 1
11174 APOM 6_26 0.1714513 86.17 9.154e-05 10.648 1
12252 C4A 6_26 0.0284680 82.47 1.455e-05 10.418 1
2736 PRSS16 6_21 0.0745211 110.59 5.106e-05 -10.000 1
5324 FURIN 15_42 0.9864665 93.98 5.744e-04 -9.913 1
11131 HLA-DMA 6_27 0.5424704 79.13 2.660e-04 -9.408 1
11142 RNF5 6_26 0.0069454 60.98 2.624e-06 9.267 1
7453 GNL3 3_36 0.5480209 64.08 2.176e-04 9.162 2
11172 GPANK1 6_26 0.0026378 52.42 8.567e-07 8.879 1
7454 PBRM1 3_36 0.0190804 59.72 7.060e-06 -8.722 1
9715 ARL6IP4 12_75 0.0005083 65.04 2.048e-07 8.615 1
8184 SMIM4 3_36 0.0156714 56.91 5.525e-06 -8.494 1
6103 DGKZ 11_28 0.2250490 61.32 8.550e-05 8.064 1
9577 HARBI1 11_28 0.2470633 60.56 9.269e-05 8.046 1
9085 ATG13 11_28 0.2470633 60.56 9.269e-05 -8.046 1
11139 NOTCH4 6_26 0.0364096 66.48 1.500e-05 8.033 3
9592 HIST1H2BC 6_20 0.0095369 89.18 5.270e-06 -7.993 1
2505 MDK 11_28 0.0864733 57.98 3.107e-05 -7.898 1Comparing z scores and PIPs


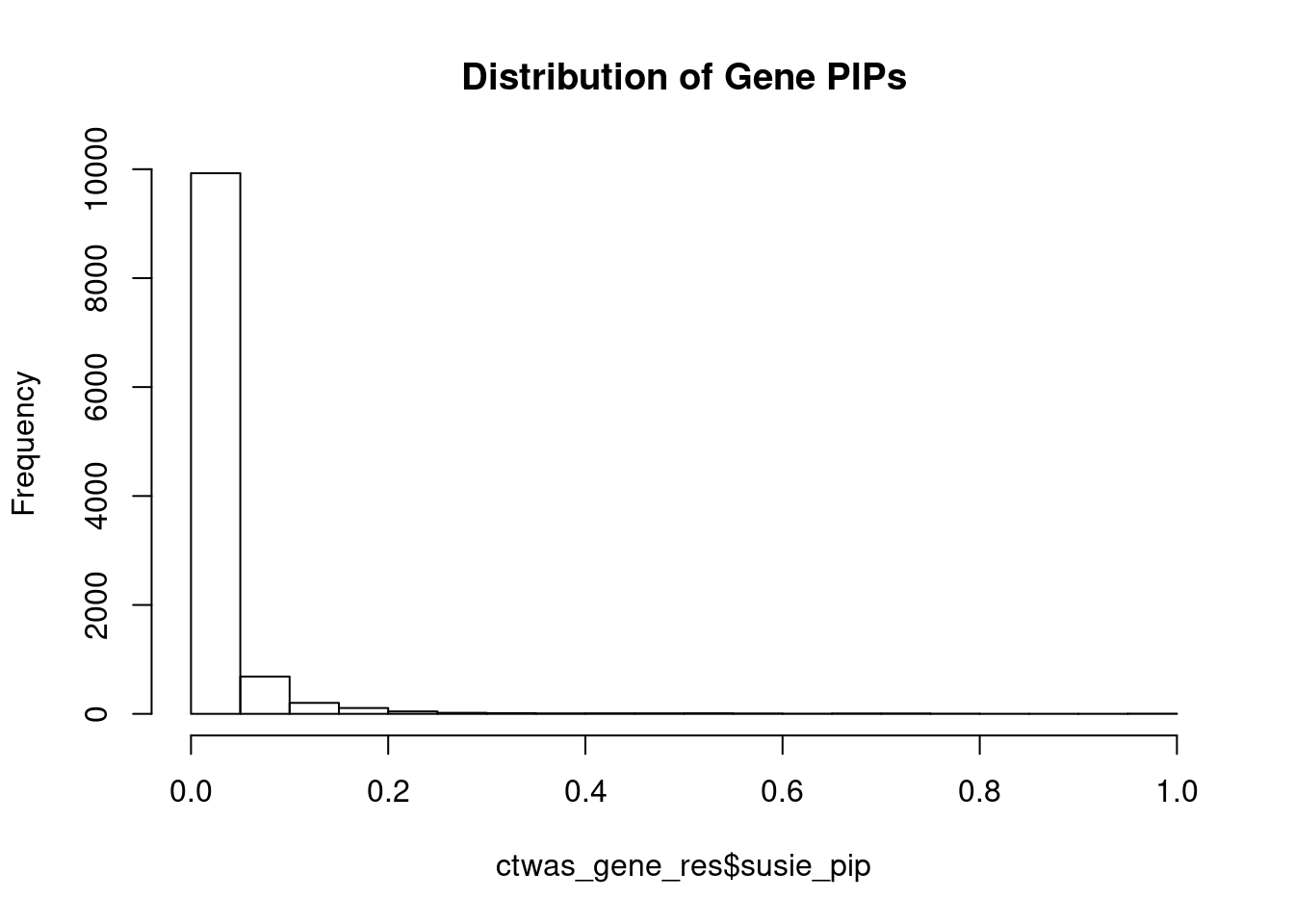
[1] 0.01652GO enrichment analysis for genes with PIP>0.5
#number of genes for gene set enrichment
length(genes)[1] 48Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Cellular_Component_2021"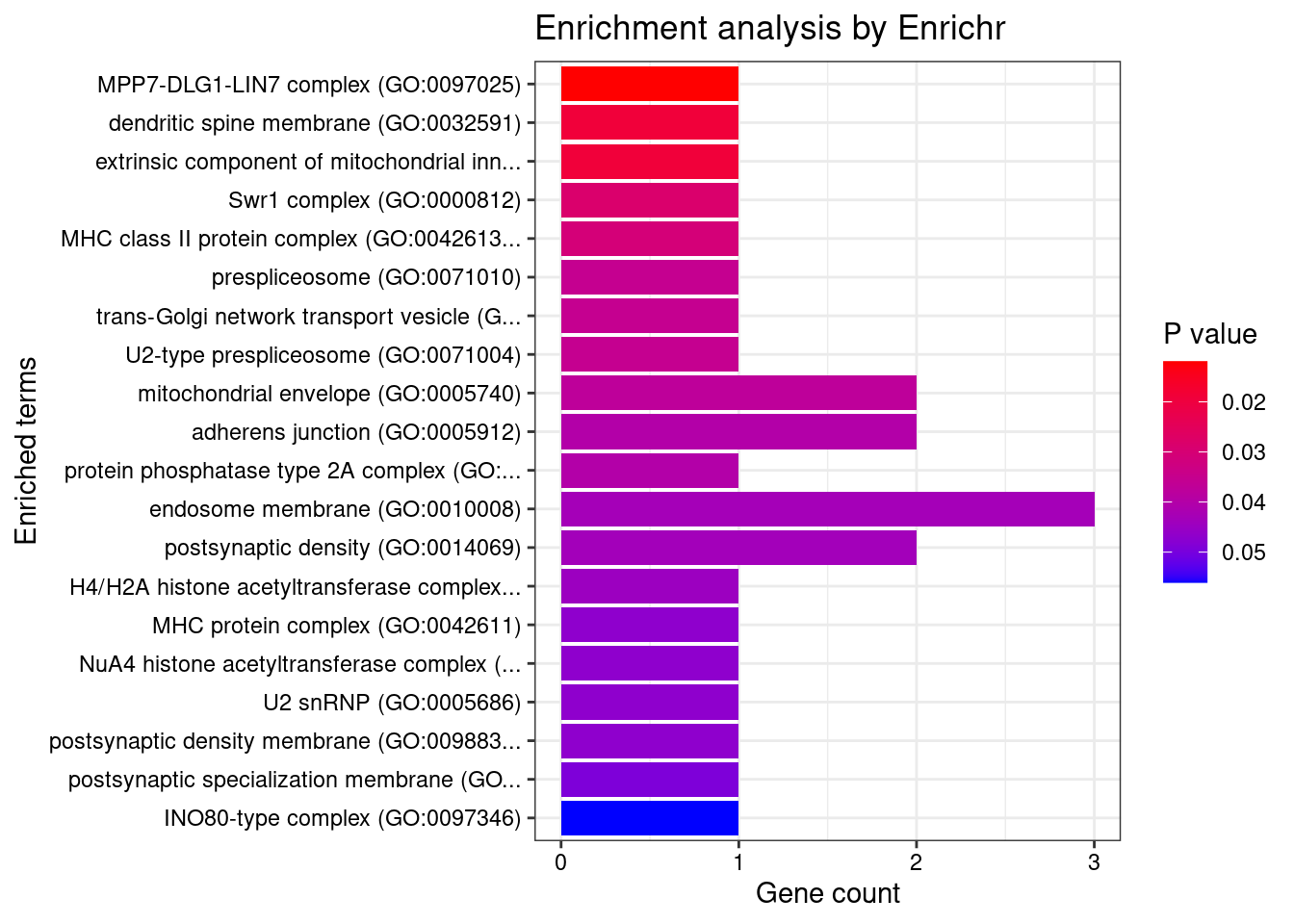
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Molecular_Function_2021"
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)DisGeNET enrichment analysis for genes with PIP>0.5
Description FDR Ratio BgRatio
32 Confusion 0.01977 1/21 1/9703
50 Gingival Hypertrophy 0.01977 1/21 1/9703
63 Infant, Premature, Diseases 0.01977 1/21 1/9703
75 Chronic Obstructive Airway Disease 0.01977 2/21 33/9703
99 Pneumonia, Viral 0.01977 1/21 1/9703
145 Speech impairment 0.01977 1/21 1/9703
146 Derealization 0.01977 1/21 1/9703
154 Spondylometaphyseal dysplasia, Kozlowski type 0.01977 1/21 1/9703
155 Metatropic dwarfism 0.01977 1/21 1/9703
182 Brachyolmia Type 3 0.01977 1/21 1/9703WebGestalt enrichment analysis for genes with PIP>0.5
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum =
minNum, : No significant gene set is identified based on FDR 0.05!NULLPIP Manhattan Plot
Warning: ggrepel: 3 unlabeled data points (too many overlaps). Consider
increasing max.overlaps
Sensitivity, specificity and precision for silver standard genes
#number of genes in known annotations
print(length(known_annotations))[1] 130#number of genes in known annotations with imputed expression
print(sum(known_annotations %in% ctwas_gene_res$genename))[1] 58#significance threshold for TWAS
print(sig_thresh)[1] 4.586#number of ctwas genes
length(ctwas_genes)[1] 8#number of TWAS genes
length(twas_genes)[1] 183#show novel genes (ctwas genes with not in TWAS genes)
ctwas_gene_res[ctwas_gene_res$genename %in% novel_genes,report_cols] genename region_tag susie_pip mu2 PVE z num_eqtl
679 RASSF1 3_35 0.9998 1010.97 0.0062622 4.532 1
4594 DAGLA 11_34 0.8069 22.96 0.0001148 -4.263 1
2551 TRPV4 12_66 0.8672 24.86 0.0001336 4.416 1#sensitivity / recall
print(sensitivity) ctwas TWAS
0.03077 0.15385 #specificity
print(specificity) ctwas TWAS
0.9996 0.9852 #precision / PPV
print(precision) ctwas TWAS
0.5000 0.1093 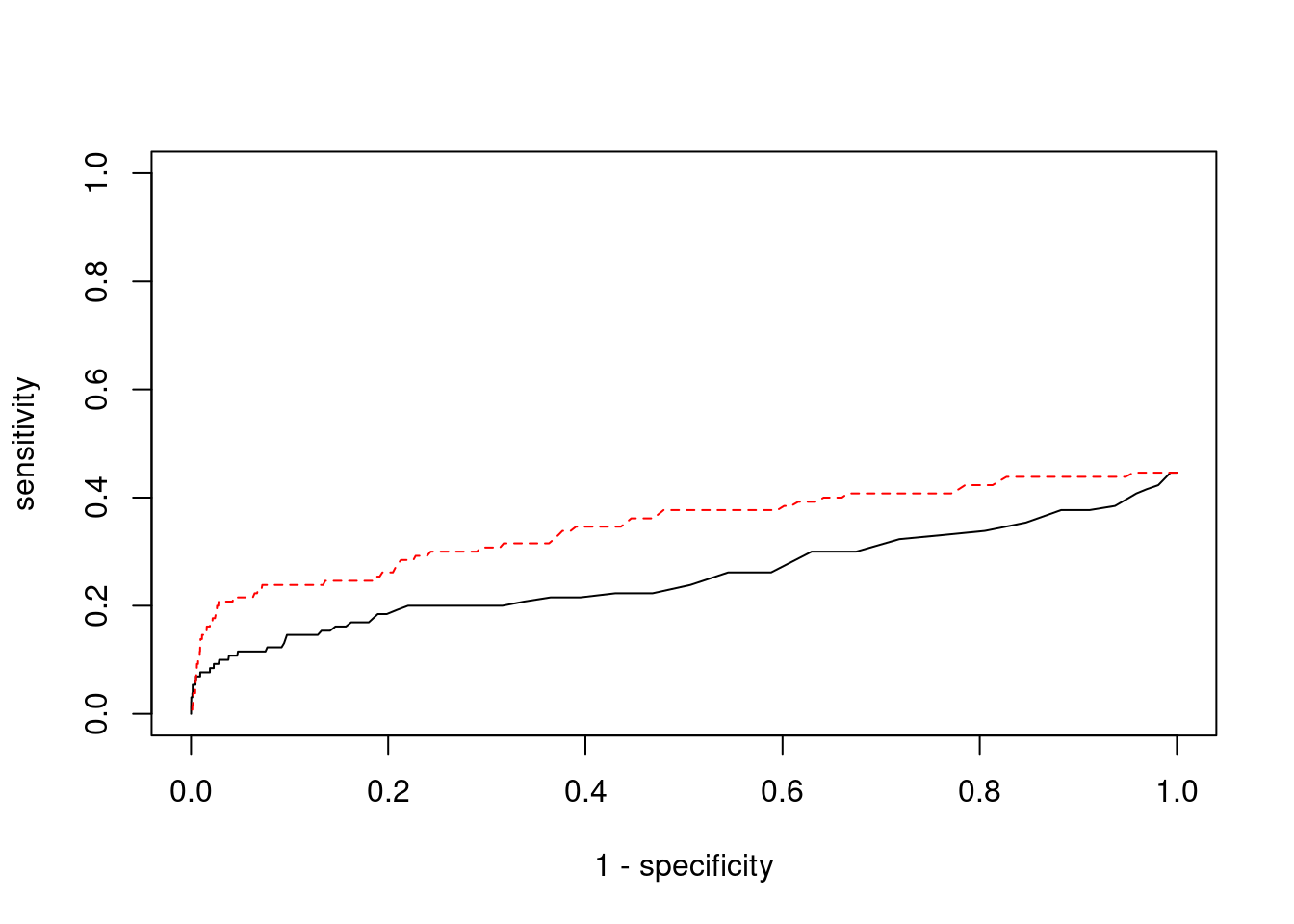
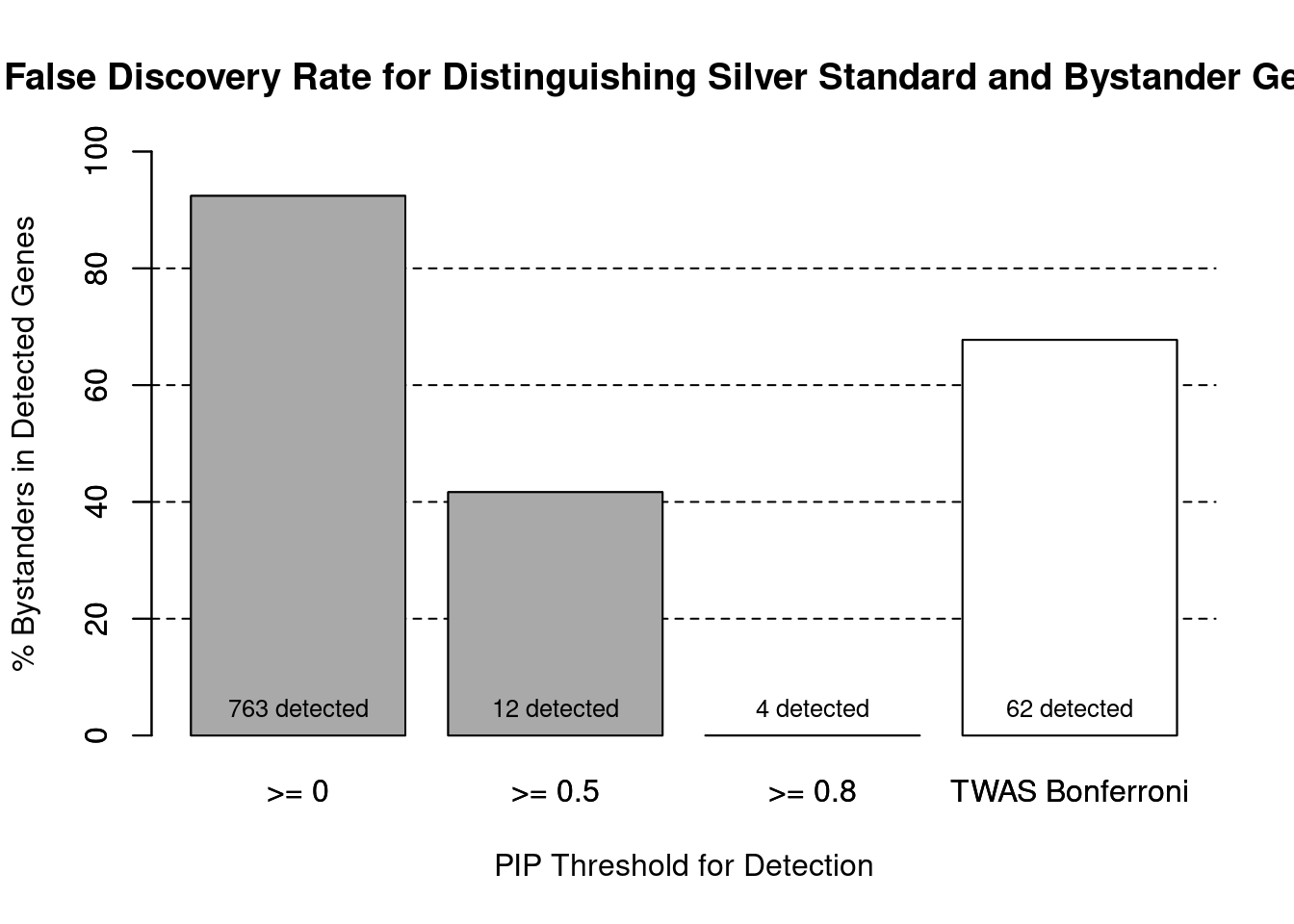
cTWAS is more precise than TWAS in distinguishing silver standard and bystander genes
#number of genes in known annotations (with imputed expression)
print(length(known_annotations))[1] 58#number of bystander genes (with imputed expression)
print(length(unrelated_genes))[1] 704#subset results to genes in known annotations or bystanders
ctwas_gene_res_subset <- ctwas_gene_res[ctwas_gene_res$genename %in% c(known_annotations, unrelated_genes),]
#assign ctwas and TWAS genes
ctwas_genes <- ctwas_gene_res_subset$genename[ctwas_gene_res_subset$susie_pip>0.8]
twas_genes <- ctwas_gene_res_subset$genename[abs(ctwas_gene_res_subset$z)>sig_thresh]
#significance threshold for TWAS
print(sig_thresh)[1] 4.586#number of ctwas genes (in known annotations or bystanders)
length(ctwas_genes)[1] 4#number of TWAS genes (in known annotations or bystanders)
length(twas_genes)[1] 62#sensitivity / recall
sensitivity ctwas TWAS
0.06897 0.34483 #specificity / (1 - False Positive Rate)
specificity ctwas TWAS
1.0000 0.9403 #precision / PPV / (1 - False Discovery Rate)
precision ctwas TWAS
1.0000 0.3226 
pip_range <- (0:1000)/1000
sensitivity <- rep(NA, length(pip_range))
specificity <- rep(NA, length(pip_range))
for (index in 1:length(pip_range)){
pip <- pip_range[index]
ctwas_genes <- ctwas_gene_res_subset$genename[ctwas_gene_res_subset$susie_pip>=pip]
sensitivity[index] <- sum(ctwas_genes %in% known_annotations)/length(known_annotations)
specificity[index] <- sum(!(unrelated_genes %in% ctwas_genes))/length(unrelated_genes)
}
plot(1-specificity, sensitivity, type="l", xlim=c(0,1), ylim=c(0,1), main="", xlab="1 - Specificity", ylab="Sensitivity")
title(expression("ROC Curve for cTWAS (black) and TWAS (" * phantom("red") * ")"))
title(expression(phantom("ROC Curve for cTWAS (black) and TWAS (") * "red" * phantom(")")), col.main="red")
sig_thresh_range <- seq(from=0, to=max(abs(ctwas_gene_res_subset$z)), length.out=length(pip_range))
for (index in 1:length(sig_thresh_range)){
sig_thresh_plot <- sig_thresh_range[index]
twas_genes <- ctwas_gene_res_subset$genename[abs(ctwas_gene_res_subset$z)>=sig_thresh_plot]
sensitivity[index] <- sum(twas_genes %in% known_annotations)/length(known_annotations)
specificity[index] <- sum(!(unrelated_genes %in% twas_genes))/length(unrelated_genes)
}
lines(1-specificity, sensitivity, xlim=c(0,1), ylim=c(0,1), col="red", lty=1)
abline(a=0,b=1,lty=3)
#add previously computed points from the analysis
ctwas_genes <- ctwas_gene_res_subset$genename[ctwas_gene_res_subset$susie_pip>0.8]
twas_genes <- ctwas_gene_res_subset$genename[abs(ctwas_gene_res_subset$z)>sig_thresh]
points(1-specificity_plot["ctwas"], sensitivity_plot["ctwas"], pch=21, bg="black")
points(1-specificity_plot["TWAS"], sensitivity_plot["TWAS"], pch=21, bg="red")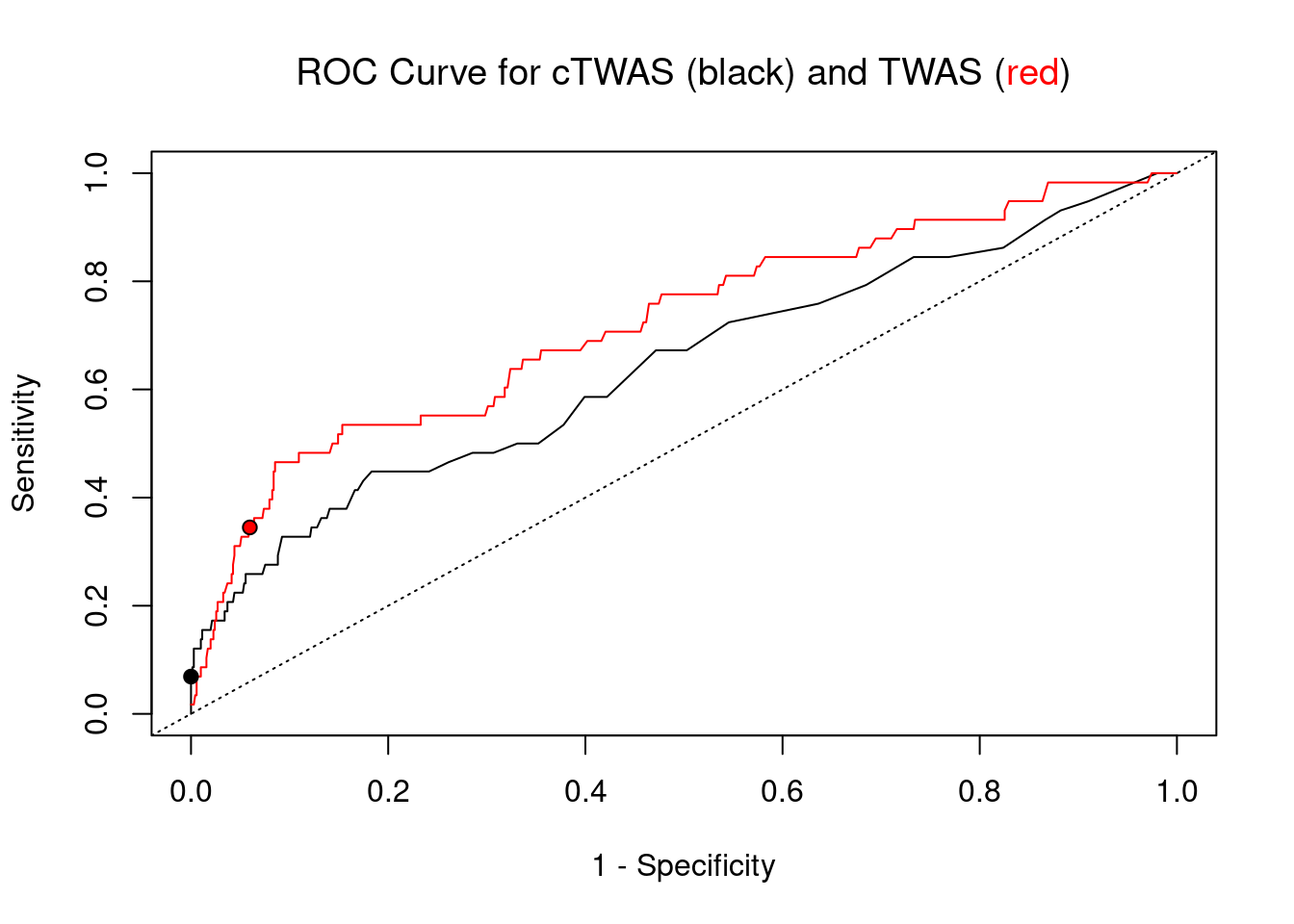
Undetected silver standard genes have low TWAS z-scores or stronger signal from nearby variants
#table of outcomes for silver standard genes
-sort(-table(silver_standard_case))silver_standard_case
Not Imputed Insignificant z-score Nearby SNP(s)
72 38 16
Detected (PIP > 0.8)
4 #show inconclusive genes
silver_standard_case[silver_standard_case=="Inconclusive"]named character(0)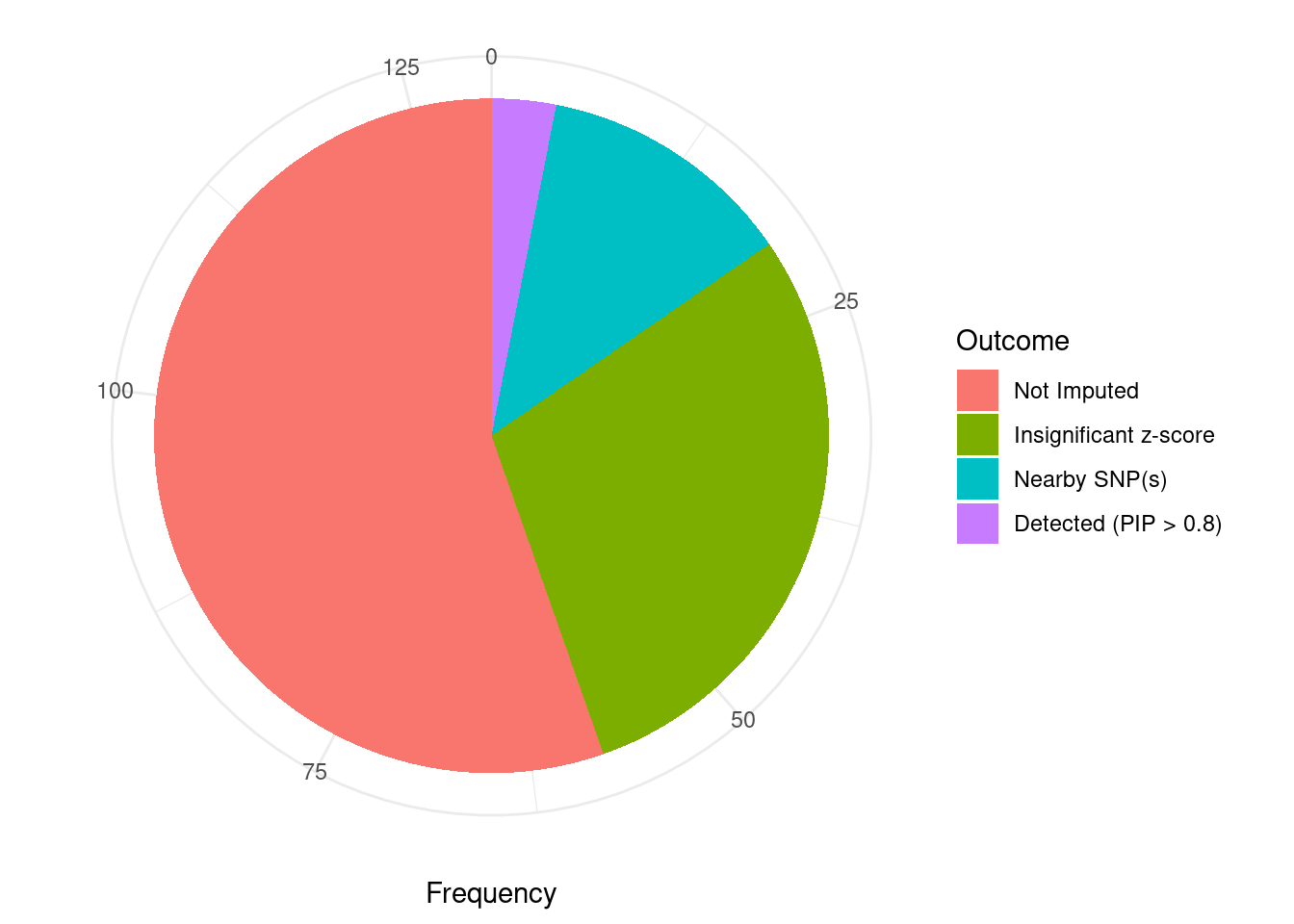
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] GenomicRanges_1.36.1 GenomeInfoDb_1.20.0 IRanges_2.18.1
[4] S4Vectors_0.22.1 BiocGenerics_0.30.0 biomaRt_2.40.1
[7] readxl_1.3.1 forcats_0.5.1 stringr_1.4.0
[10] dplyr_1.0.7 purrr_0.3.4 readr_2.1.1
[13] tidyr_1.1.4 tidyverse_1.3.1 tibble_3.1.6
[16] WebGestaltR_0.4.4 disgenet2r_0.99.2 enrichR_3.0
[19] cowplot_1.1.1 ggplot2_3.3.5 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] ggbeeswarm_0.6.0 colorspace_2.0-2 rjson_0.2.20
[4] ellipsis_0.3.2 rprojroot_2.0.2 XVector_0.24.0
[7] fs_1.5.2 rstudioapi_0.13 farver_2.1.0
[10] ggrepel_0.9.1 bit64_4.0.5 AnnotationDbi_1.46.0
[13] fansi_1.0.2 lubridate_1.8.0 xml2_1.3.3
[16] codetools_0.2-16 doParallel_1.0.17 cachem_1.0.6
[19] knitr_1.36 jsonlite_1.7.2 apcluster_1.4.8
[22] Cairo_1.5-12.2 broom_0.7.10 dbplyr_2.1.1
[25] compiler_3.6.1 httr_1.4.2 backports_1.4.1
[28] assertthat_0.2.1 Matrix_1.2-18 fastmap_1.1.0
[31] cli_3.1.0 later_0.8.0 prettyunits_1.1.1
[34] htmltools_0.5.2 tools_3.6.1 igraph_1.2.10
[37] GenomeInfoDbData_1.2.1 gtable_0.3.0 glue_1.6.2
[40] reshape2_1.4.4 doRNG_1.8.2 Rcpp_1.0.8
[43] Biobase_2.44.0 cellranger_1.1.0 jquerylib_0.1.4
[46] vctrs_0.3.8 svglite_1.2.2 iterators_1.0.14
[49] xfun_0.29 ps_1.6.0 rvest_1.0.2
[52] lifecycle_1.0.1 rngtools_1.5.2 XML_3.99-0.3
[55] zlibbioc_1.30.0 getPass_0.2-2 scales_1.1.1
[58] vroom_1.5.7 hms_1.1.1 promises_1.0.1
[61] yaml_2.2.1 curl_4.3.2 memoise_2.0.1
[64] ggrastr_1.0.1 gdtools_0.1.9 stringi_1.7.6
[67] RSQLite_2.2.8 highr_0.9 foreach_1.5.2
[70] rlang_1.0.1 pkgconfig_2.0.3 bitops_1.0-7
[73] evaluate_0.14 lattice_0.20-38 labeling_0.4.2
[76] bit_4.0.4 processx_3.5.2 tidyselect_1.1.1
[79] plyr_1.8.6 magrittr_2.0.2 R6_2.5.1
[82] generics_0.1.1 DBI_1.1.2 pillar_1.6.4
[85] haven_2.4.3 whisker_0.3-2 withr_2.4.3
[88] RCurl_1.98-1.5 modelr_0.1.8 crayon_1.5.0
[91] utf8_1.2.2 tzdb_0.2.0 rmarkdown_2.11
[94] progress_1.2.2 grid_3.6.1 data.table_1.14.2
[97] blob_1.2.2 callr_3.7.0 git2r_0.26.1
[100] reprex_2.0.1 digest_0.6.29 httpuv_1.5.1
[103] munsell_0.5.0 beeswarm_0.2.3 vipor_0.4.5