SCZ - Brain Cerebellar Hemisphere
sheng Qian
2021-2-6
Last updated: 2022-03-16
Checks: 5 2
Knit directory: cTWAS_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211220) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/data/ | data |
| /project2/xinhe/shengqian/cTWAS/cTWAS_analysis/code/ctwas_config.R | code/ctwas_config.R |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version d57314b. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .ipynb_checkpoints/
Ignored: data/AF/
Untracked files:
Untracked: Rplot.png
Untracked: analysis/.ipynb_checkpoints/
Untracked: analysis/SCZ_2020_Brain_Amygdala.Rmd
Untracked: analysis/SCZ_2020_Brain_Anterior_cingulate_cortex_BA24.Rmd
Untracked: analysis/SCZ_2020_Brain_Caudate_basal_ganglia.Rmd
Untracked: analysis/SCZ_2020_Brain_Cerebellar_Hemisphere.Rmd
Untracked: analysis/SCZ_2020_Brain_Cerebellum.Rmd
Untracked: analysis/SCZ_2020_Brain_Hippocampus.Rmd
Untracked: analysis/SCZ_2020_Brain_Nucleus_accumbens_basal_ganglia.Rmd
Untracked: analysis/SCZ_2020_Brain_Spinal_cord_cervical_c-1.Rmd
Untracked: analysis/SCZ_2020_Brain_Substantia_nigra.Rmd
Untracked: code/.ipynb_checkpoints/
Untracked: code/AF_out/
Untracked: code/Autism_out/
Untracked: code/BMI_S_out/
Untracked: code/BMI_out/
Untracked: code/Glucose_out/
Untracked: code/LDL_S_out/
Untracked: code/SCZ_2014_EUR_out/
Untracked: code/SCZ_2020_out/
Untracked: code/SCZ_S_out/
Untracked: code/SCZ_out/
Untracked: code/T2D_out/
Untracked: code/ctwas_config.R
Untracked: code/mapping.R
Untracked: code/out/
Untracked: code/run_AF_analysis.sbatch
Untracked: code/run_AF_analysis.sh
Untracked: code/run_AF_ctwas_rss_LDR.R
Untracked: code/run_Autism_analysis.sbatch
Untracked: code/run_Autism_analysis.sh
Untracked: code/run_Autism_ctwas_rss_LDR.R
Untracked: code/run_BMI_analysis.sbatch
Untracked: code/run_BMI_analysis.sh
Untracked: code/run_BMI_analysis_S.sbatch
Untracked: code/run_BMI_analysis_S.sh
Untracked: code/run_BMI_ctwas_rss_LDR.R
Untracked: code/run_BMI_ctwas_rss_LDR_S.R
Untracked: code/run_Glucose_analysis.sbatch
Untracked: code/run_Glucose_analysis.sh
Untracked: code/run_Glucose_ctwas_rss_LDR.R
Untracked: code/run_LDL_analysis_S.sbatch
Untracked: code/run_LDL_analysis_S.sh
Untracked: code/run_LDL_ctwas_rss_LDR_S.R
Untracked: code/run_SCZ_2014_EUR_analysis.sbatch
Untracked: code/run_SCZ_2014_EUR_analysis.sh
Untracked: code/run_SCZ_2014_EUR_ctwas_rss_LDR.R
Untracked: code/run_SCZ_2020_analysis.sbatch
Untracked: code/run_SCZ_2020_analysis.sh
Untracked: code/run_SCZ_2020_ctwas_rss_LDR.R
Untracked: code/run_SCZ_analysis.sbatch
Untracked: code/run_SCZ_analysis.sh
Untracked: code/run_SCZ_analysis_S.sbatch
Untracked: code/run_SCZ_analysis_S.sh
Untracked: code/run_SCZ_ctwas_rss_LDR.R
Untracked: code/run_SCZ_ctwas_rss_LDR_S.R
Untracked: code/run_T2D_analysis.sbatch
Untracked: code/run_T2D_analysis.sh
Untracked: code/run_T2D_ctwas_rss_LDR.R
Untracked: code/wflow_build.R
Untracked: code/wflow_build.sbatch
Untracked: data/.ipynb_checkpoints/
Untracked: data/BMI/
Untracked: data/PGC3_SCZ_wave3_public.v2.tsv
Untracked: data/SCZ/
Untracked: data/SCZ_2014_EUR/
Untracked: data/SCZ_2020/
Untracked: data/SCZ_S/
Untracked: data/T2D/
Untracked: data/UKBB/
Untracked: data/UKBB_SNPs_Info.text
Untracked: data/gene_OMIM.txt
Untracked: data/gene_pip_0.8.txt
Untracked: data/mashr_Heart_Atrial_Appendage.db
Untracked: data/mashr_sqtl/
Untracked: data/summary_known_genes_annotations.xlsx
Untracked: data/untitled.txt
Unstaged changes:
Modified: analysis/SCZ_2020_Brain_Cortex.Rmd
Modified: analysis/SCZ_2020_Brain_Frontal_Cortex_BA9.Rmd
Modified: analysis/SCZ_2020_Brain_Hypothalamus.Rmd
Modified: analysis/SCZ_2020_Brain_Putamen_basal_ganglia.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with wflow_publish() to start tracking its development.
Weight QC
#number of imputed weights
nrow(qclist_all)[1] 11328#number of imputed weights by chromosome
table(qclist_all$chr)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
1087 766 651 425 543 628 553 423 439 442 695 635 209 381 371 537
17 18 19 20 21 22
707 170 905 332 134 295 #number of imputed weights without missing variants
sum(qclist_all$nmiss==0)[1] 8719#proportion of imputed weights without missing variants
mean(qclist_all$nmiss==0)[1] 0.7697Check convergence of parameters
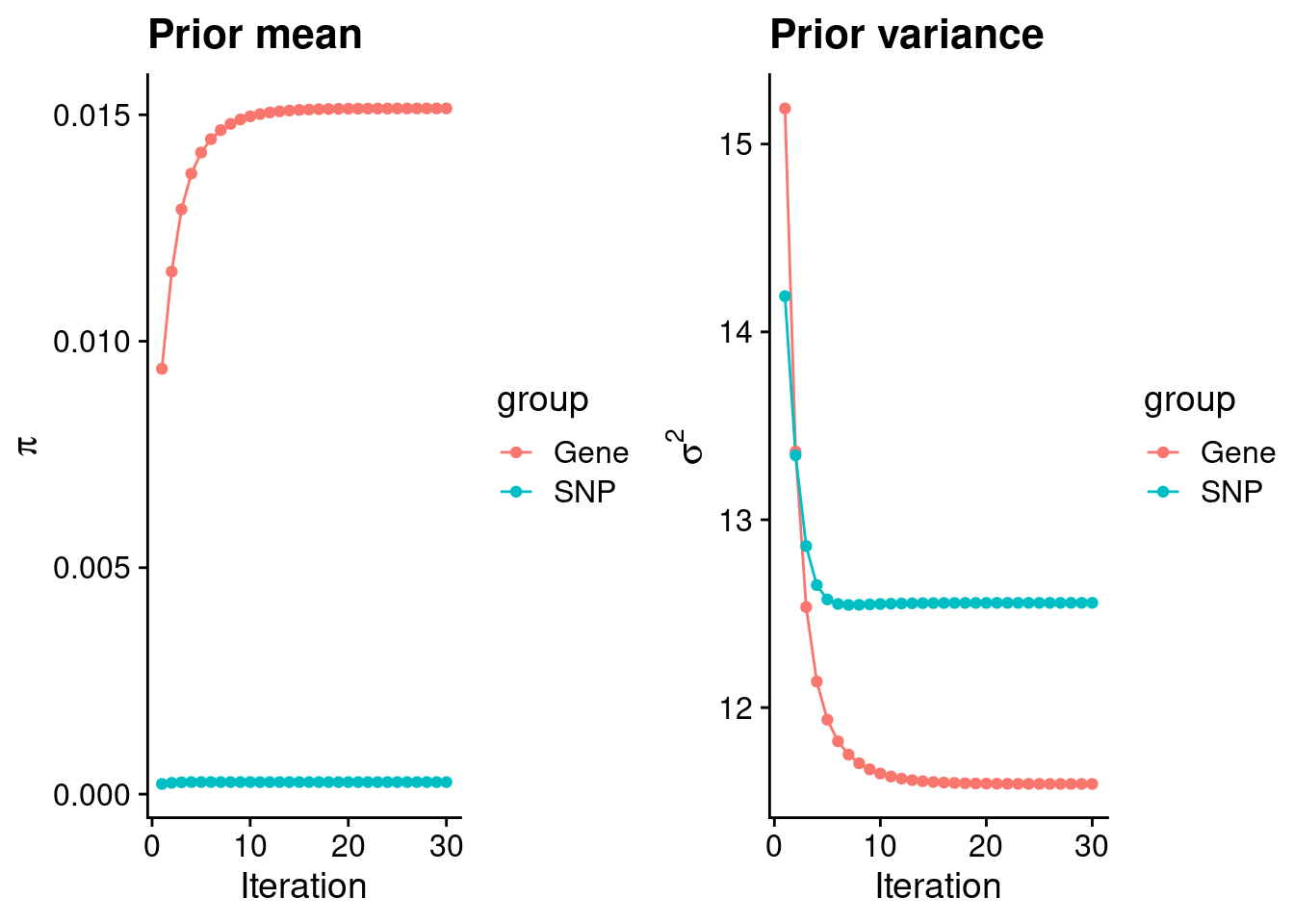
#estimated group prior
estimated_group_prior <- group_prior_rec[,ncol(group_prior_rec)]
names(estimated_group_prior) <- c("gene", "snp")
estimated_group_prior["snp"] <- estimated_group_prior["snp"]*thin #adjust parameter to account for thin argument
print(estimated_group_prior) gene snp
0.0151400 0.0002659 #estimated group prior variance
estimated_group_prior_var <- group_prior_var_rec[,ncol(group_prior_var_rec)]
names(estimated_group_prior_var) <- c("gene", "snp")
print(estimated_group_prior_var) gene snp
11.59 12.56 #report sample size
print(sample_size)[1] 161405#report group size
group_size <- c(nrow(ctwas_gene_res), n_snps)
print(group_size)[1] 11328 7394310#estimated group PVE
estimated_group_pve <- estimated_group_prior_var*estimated_group_prior*group_size/sample_size #check PVE calculation
names(estimated_group_pve) <- c("gene", "snp")
print(estimated_group_pve) gene snp
0.01232 0.15296 #compare sum(PIP*mu2/sample_size) with above PVE calculation
c(sum(ctwas_gene_res$PVE),sum(ctwas_snp_res$PVE))[1] 0.0538 0.7832Genes with highest PIPs
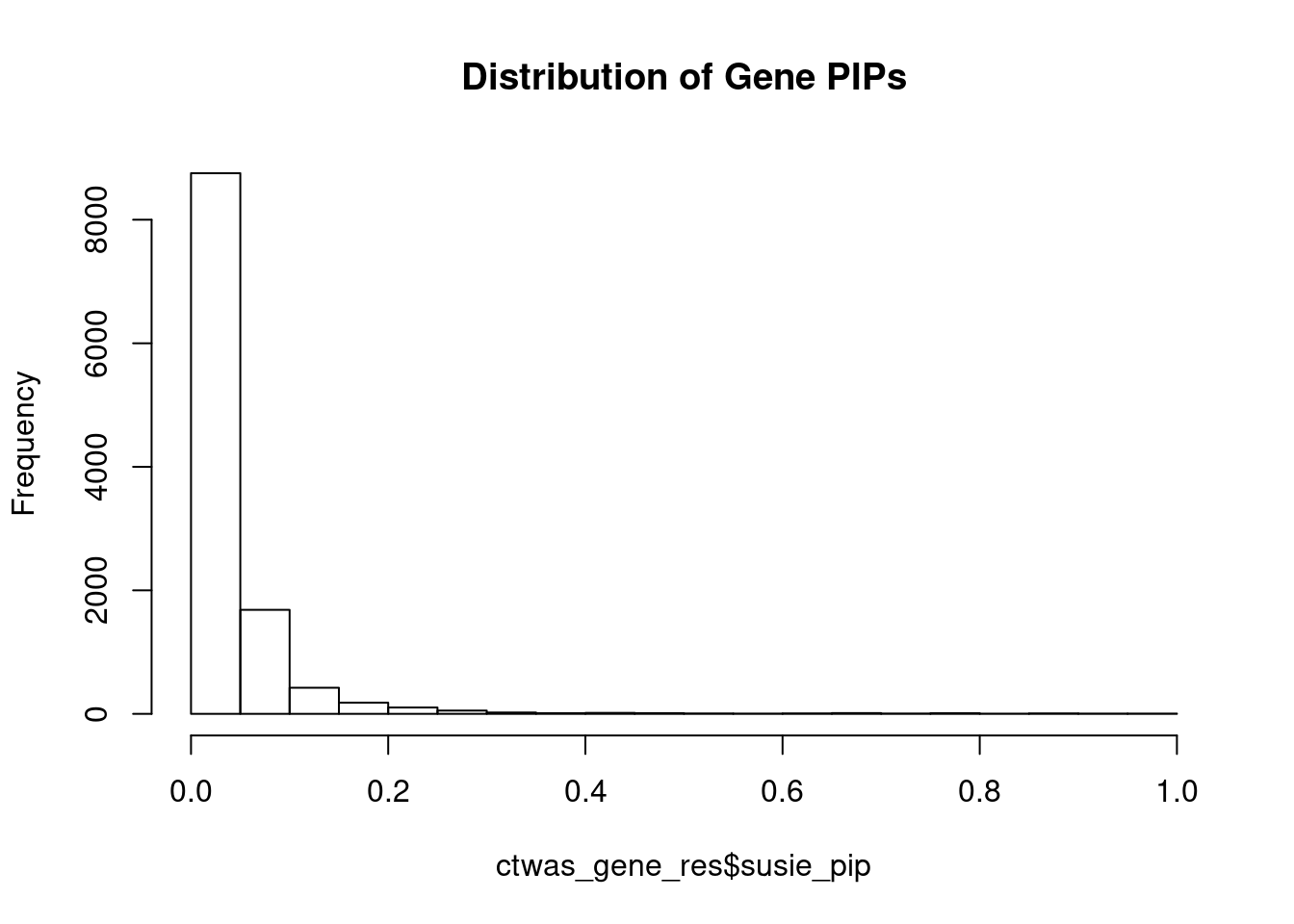
genename region_tag susie_pip mu2 PVE z num_eqtl
2523 YWHAE 17_2 0.9821 51.13 0.0003111 0.1008 2
12095 AC012074.2 2_15 0.9818 29.78 0.0001812 5.4694 1
5783 GALNT2 1_117 0.9806 31.50 0.0001914 5.6989 1
10988 ZNF823 19_10 0.9672 38.21 0.0002290 6.1841 2
4143 FEZF1 7_74 0.9631 23.24 0.0001387 -4.6555 1
1283 MLF2 12_7 0.9499 26.45 0.0001557 -4.9033 1
380 NSUN2 5_6 0.9400 22.60 0.0001316 -4.2792 3
2207 RUNDC3B 7_54 0.9365 27.25 0.0001581 5.4540 1
3040 ACTR1B 2_57 0.9188 30.79 0.0001753 -5.5513 1
929 KLHL20 1_85 0.8931 36.68 0.0002029 -5.7996 1
8411 SNTB2 16_37 0.8895 26.82 0.0001478 -4.8247 2
6611 SEMA3D 7_53 0.8861 21.87 0.0001201 -4.0278 3
9042 C11orf80 11_37 0.8847 19.92 0.0001092 -4.0883 2
7758 SLC4A2 7_94 0.8764 20.09 0.0001091 -4.0287 2
12301 HLA-DMB 6_27 0.8737 75.80 0.0004103 -9.6790 1
417 RETSAT 2_54 0.8699 20.37 0.0001098 3.9626 1
8715 KCNMB3 3_110 0.8539 19.36 0.0001024 -3.8661 1
3479 SLF2 10_64 0.8402 24.28 0.0001264 -4.5939 2
7031 ACE 17_37 0.8299 32.91 0.0001692 -5.8021 1
3381 ABCG2 4_59 0.8289 20.15 0.0001035 -3.9541 1Genes with largest effect sizes
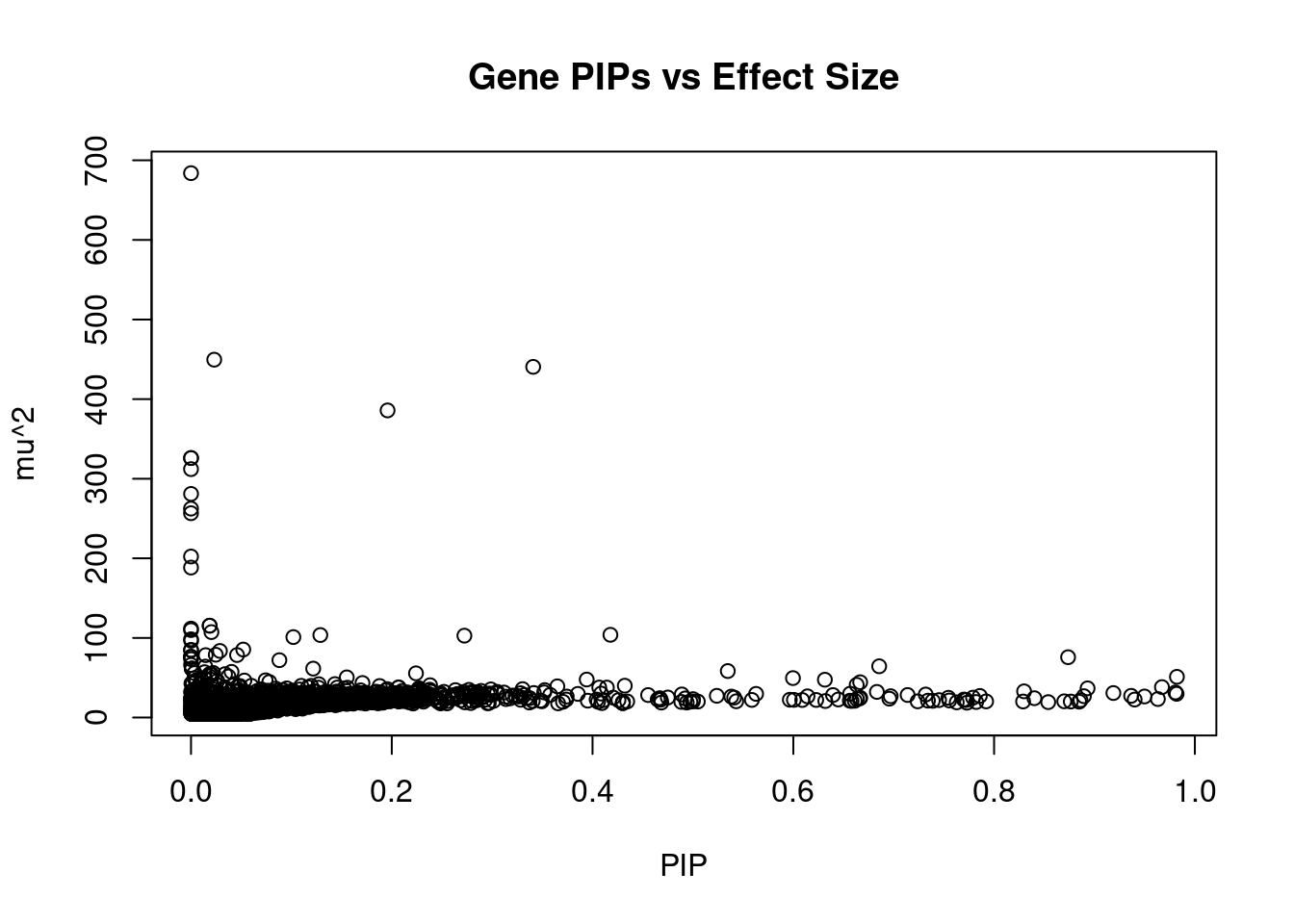
genename region_tag susie_pip mu2 PVE z num_eqtl
10533 SLC38A3 3_35 1.465e-05 683.9 6.206e-08 -2.77559 1
33 RBM5 3_35 2.313e-02 449.6 6.443e-05 3.98715 1
41 RBM6 3_35 3.409e-01 440.5 9.304e-04 4.46875 1
9614 LSMEM2 3_35 1.958e-01 385.8 4.681e-04 -4.27088 1
10363 HYAL3 3_35 4.474e-05 326.0 9.037e-08 -2.50662 1
11633 IFRD2 3_35 4.474e-05 326.0 9.037e-08 -2.50662 1
724 RASSF1 3_35 1.644e-05 312.1 3.178e-08 4.32685 1
11946 U73166.2 3_35 5.847e-05 280.9 1.018e-07 -4.59660 1
7604 RNF123 3_35 1.491e-05 262.4 2.424e-08 -2.32524 1
2981 CYB561D2 3_35 1.486e-05 256.7 2.362e-08 -2.19612 2
9825 UBA7 3_35 1.710e-05 202.3 2.143e-08 -1.08001 1
12318 NAT6 3_35 1.488e-05 188.4 1.737e-08 0.79523 2
11884 HCG11 6_20 1.858e-02 115.4 1.328e-05 9.84429 1
12879 CTA-14H9.5 6_20 1.858e-02 115.4 1.328e-05 9.84429 1
12038 GPX1 3_35 1.524e-05 111.7 1.055e-08 -0.09215 2
130 CACNA2D2 3_35 6.978e-05 109.8 4.748e-08 -0.10441 1
10334 BTN3A2 6_20 2.035e-02 107.3 1.353e-05 9.14834 2
11553 CLIC1 6_26 4.178e-01 103.9 2.691e-04 10.73111 1
2870 PRSS16 6_21 1.288e-01 103.6 8.272e-05 -10.00016 1
11296 C6orf48 6_26 2.724e-01 102.9 1.736e-04 10.68269 1Genes with highest PVE
genename region_tag susie_pip mu2 PVE z num_eqtl
41 RBM6 3_35 0.3409 440.55 0.0009304 4.4688 1
9614 LSMEM2 3_35 0.1958 385.82 0.0004681 -4.2709 1
12301 HLA-DMB 6_27 0.8737 75.80 0.0004103 -9.6790 1
2523 YWHAE 17_2 0.9821 51.13 0.0003111 0.1008 2
7572 GNL3 3_36 0.6855 64.38 0.0002735 9.4161 2
11553 CLIC1 6_26 0.4178 103.94 0.0002691 10.7311 1
10988 ZNF823 19_10 0.9672 38.21 0.0002290 6.1841 2
929 KLHL20 1_85 0.8931 36.68 0.0002029 -5.7996 1
9199 ATG13 11_28 0.5347 58.39 0.0001934 -8.0462 1
5783 GALNT2 1_117 0.9806 31.50 0.0001914 5.6989 1
10942 NMB 15_39 0.6314 47.51 0.0001859 7.1213 1
3099 SF3B1 2_117 0.5996 49.54 0.0001841 7.6053 1
5201 ARL3 10_66 0.6666 44.35 0.0001832 -9.6347 1
12095 AC012074.2 2_15 0.9818 29.78 0.0001812 5.4694 1
3040 ACTR1B 2_57 0.9188 30.79 0.0001753 -5.5513 1
11296 C6orf48 6_26 0.2724 102.85 0.0001736 10.6827 1
7031 ACE 17_37 0.8299 32.91 0.0001692 -5.8021 1
11777 PLEKHM1 17_27 0.6631 41.12 0.0001689 6.4041 2
2207 RUNDC3B 7_54 0.9365 27.25 0.0001581 5.4540 1
1283 MLF2 12_7 0.9499 26.45 0.0001557 -4.9033 1Genes with largest z scores
genename region_tag susie_pip mu2 PVE z num_eqtl
11553 CLIC1 6_26 0.4177893 103.94 2.691e-04 10.731 1
11296 C6orf48 6_26 0.2723513 102.85 1.736e-04 10.683 1
12355 C4A 6_26 0.1019645 101.06 6.384e-05 10.542 2
2870 PRSS16 6_21 0.1288225 103.64 8.272e-05 -10.000 1
11884 HCG11 6_20 0.0185817 115.38 1.328e-05 9.844 1
12879 CTA-14H9.5 6_20 0.0185817 115.38 1.328e-05 9.844 1
6221 CNNM2 10_66 0.2381535 40.61 5.992e-05 -9.686 1
12301 HLA-DMB 6_27 0.8737245 75.80 4.103e-04 -9.679 1
5201 ARL3 10_66 0.6666110 44.35 1.832e-04 -9.635 1
7572 GNL3 3_36 0.6855456 64.38 2.735e-04 9.416 2
11273 HLA-DMA 6_27 0.0880475 72.06 3.931e-05 -9.408 1
11281 RNF5 6_26 0.0142254 63.98 5.639e-06 9.278 2
11284 PRRT1 6_26 0.0135224 56.97 4.773e-06 9.276 1
11986 CYP21A2 6_26 0.0146039 78.38 7.091e-06 -9.197 1
10334 BTN3A2 6_20 0.0203464 107.34 1.353e-05 9.148 2
7573 PBRM1 3_36 0.0221295 56.08 7.688e-06 -8.722 1
11754 C4B 6_26 0.0520478 85.41 2.754e-05 -8.656 2
6038 ABT1 6_20 0.0288050 83.66 1.493e-05 8.650 1
1323 PITPNM2 12_75 0.0008402 61.35 3.194e-07 -8.615 1
2710 OGFOD2 12_75 0.0007551 61.45 2.875e-07 8.615 1Comparing z scores and PIPs
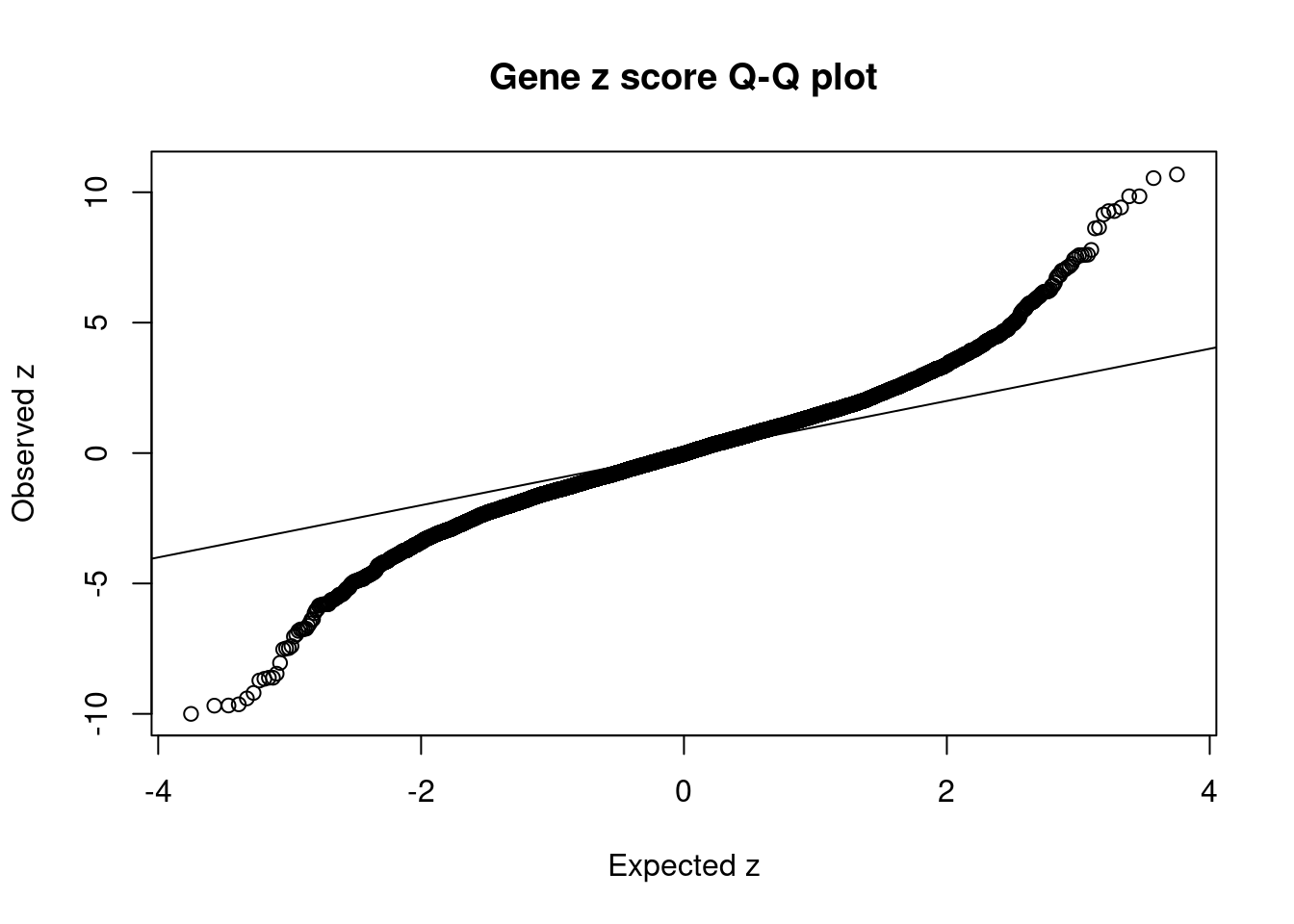
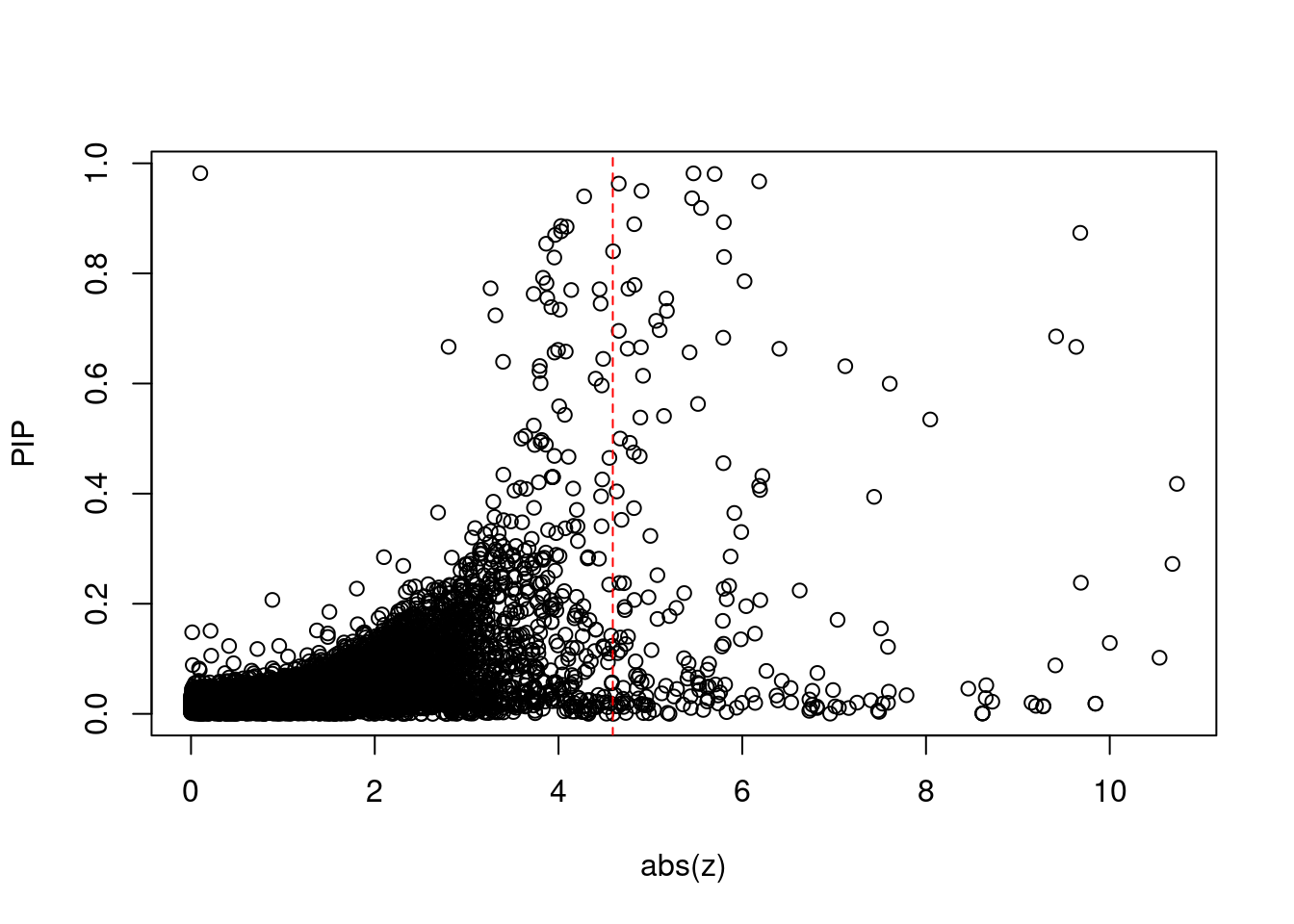
[1] 0.01677GO enrichment analysis for genes with PIP>0.5
#number of genes for gene set enrichment
length(genes)[1] 69Uploading data to Enrichr... Done.
Querying GO_Biological_Process_2021... Done.
Querying GO_Cellular_Component_2021... Done.
Querying GO_Molecular_Function_2021... Done.
Parsing results... Done.
[1] "GO_Biological_Process_2021"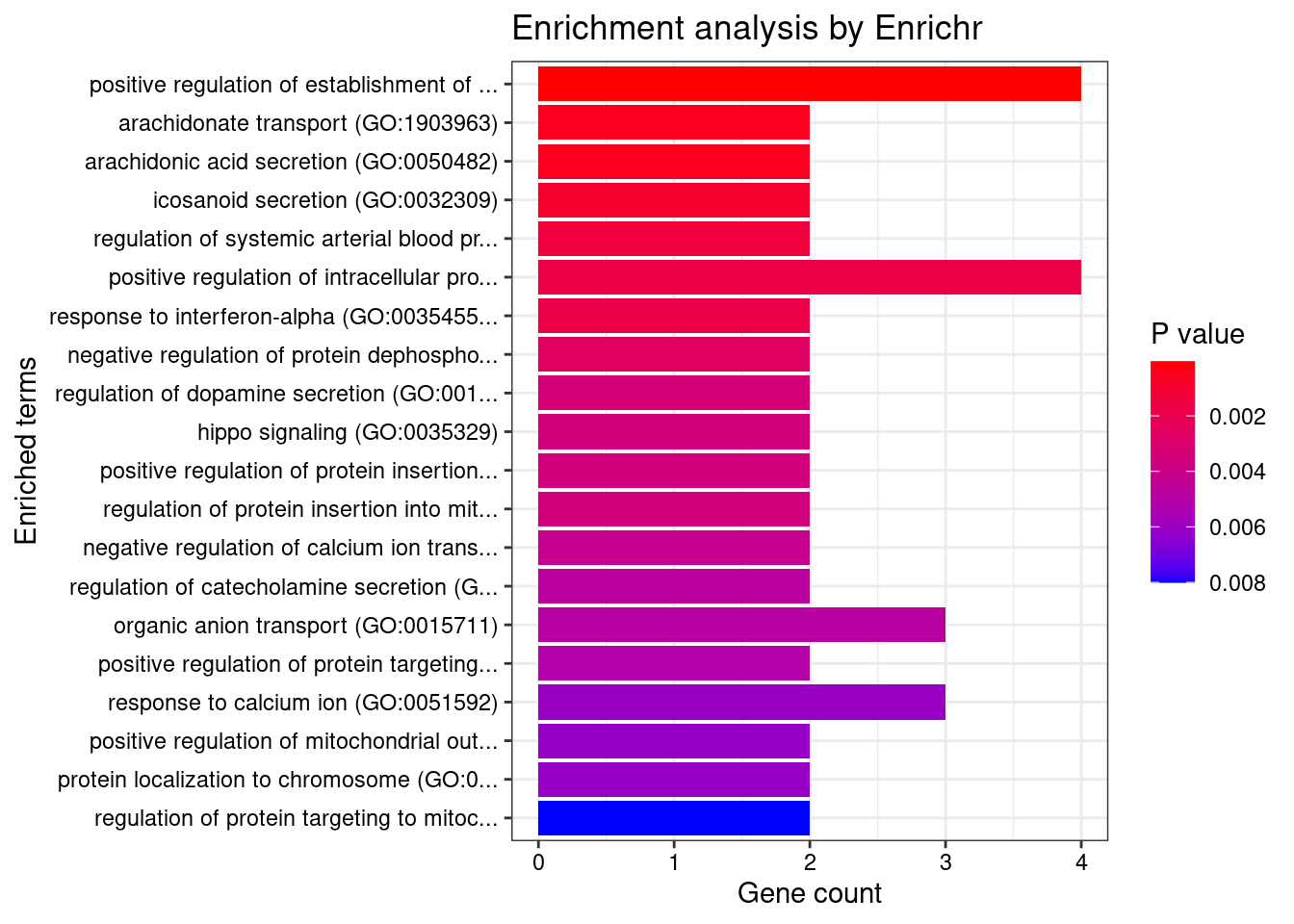
Term
1 positive regulation of establishment of protein localization to mitochondrion (GO:1903749)
Overlap Adjusted.P.value Genes
1 4/56 0.02547 YWHAE;USP36;YWHAB;ATG13
[1] "GO_Cellular_Component_2021"
[1] Term Overlap Adjusted.P.value Genes
<0 rows> (or 0-length row.names)
[1] "GO_Molecular_Function_2021"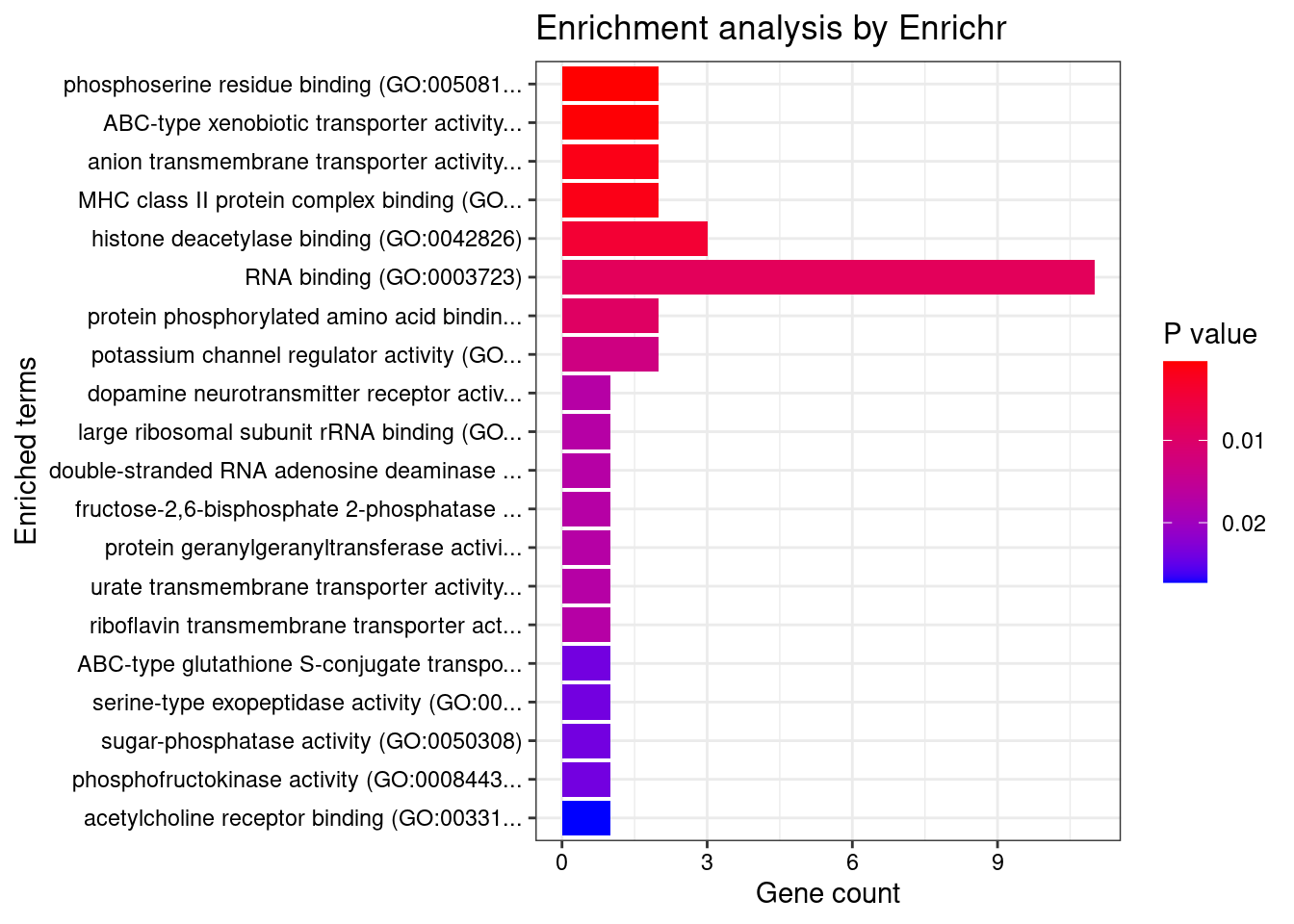
Term Overlap
1 phosphoserine residue binding (GO:0050815) 2/9
2 ABC-type xenobiotic transporter activity (GO:0008559) 2/11
3 anion transmembrane transporter activity (GO:0008509) 2/17
4 MHC class II protein complex binding (GO:0023026) 2/17
Adjusted.P.value Genes
1 0.03351 YWHAE;YWHAB
2 0.03351 ABCC10;ABCG2
3 0.04088 SLC4A2;ABCG2
4 0.04088 YWHAE;HLA-DMBDisGeNET enrichment analysis for genes with PIP>0.5
Description FDR Ratio
56 Gingival Hypertrophy 0.04643 1/32
71 Infant, Premature, Diseases 0.04643 1/32
110 Pneumonia, Viral 0.04643 1/32
118 Schizophrenia 0.04643 9/32
206 Gorlin Chaudhry Moss syndrome 0.04643 1/32
216 Symmetrical dyschromatosis of extremities 0.04643 1/32
284 Severe Acute Respiratory Syndrome 0.04643 1/32
303 URIC ACID CONCENTRATION, SERUM, QUANTITATIVE TRAIT LOCUS 1 0.04643 1/32
304 Ehlers-Danlos syndrome caused by tenascin-X deficiency 0.04643 1/32
305 Familial encephalopathy with neuroserpin inclusion bodies 0.04643 1/32
BgRatio
56 1/9703
71 1/9703
110 1/9703
118 883/9703
206 1/9703
216 1/9703
284 1/9703
303 1/9703
304 1/9703
305 1/9703WebGestalt enrichment analysis for genes with PIP>0.5
Loading the functional categories...
Loading the ID list...
Loading the reference list...
Performing the enrichment analysis...Warning in oraEnrichment(interestGeneList, referenceGeneList, geneSet, minNum =
minNum, : No significant gene set is identified based on FDR 0.05!NULLPIP Manhattan Plot
Warning: 'timedatectl' indicates the non-existent timezone name 'n/a'Warning: Your system is mis-configured: '/etc/localtime' is not a symlinkWarning: It is strongly recommended to set envionment variable TZ to 'America/
Chicago' (or equivalent)Warning: ggrepel: 12 unlabeled data points (too many overlaps). Consider
increasing max.overlaps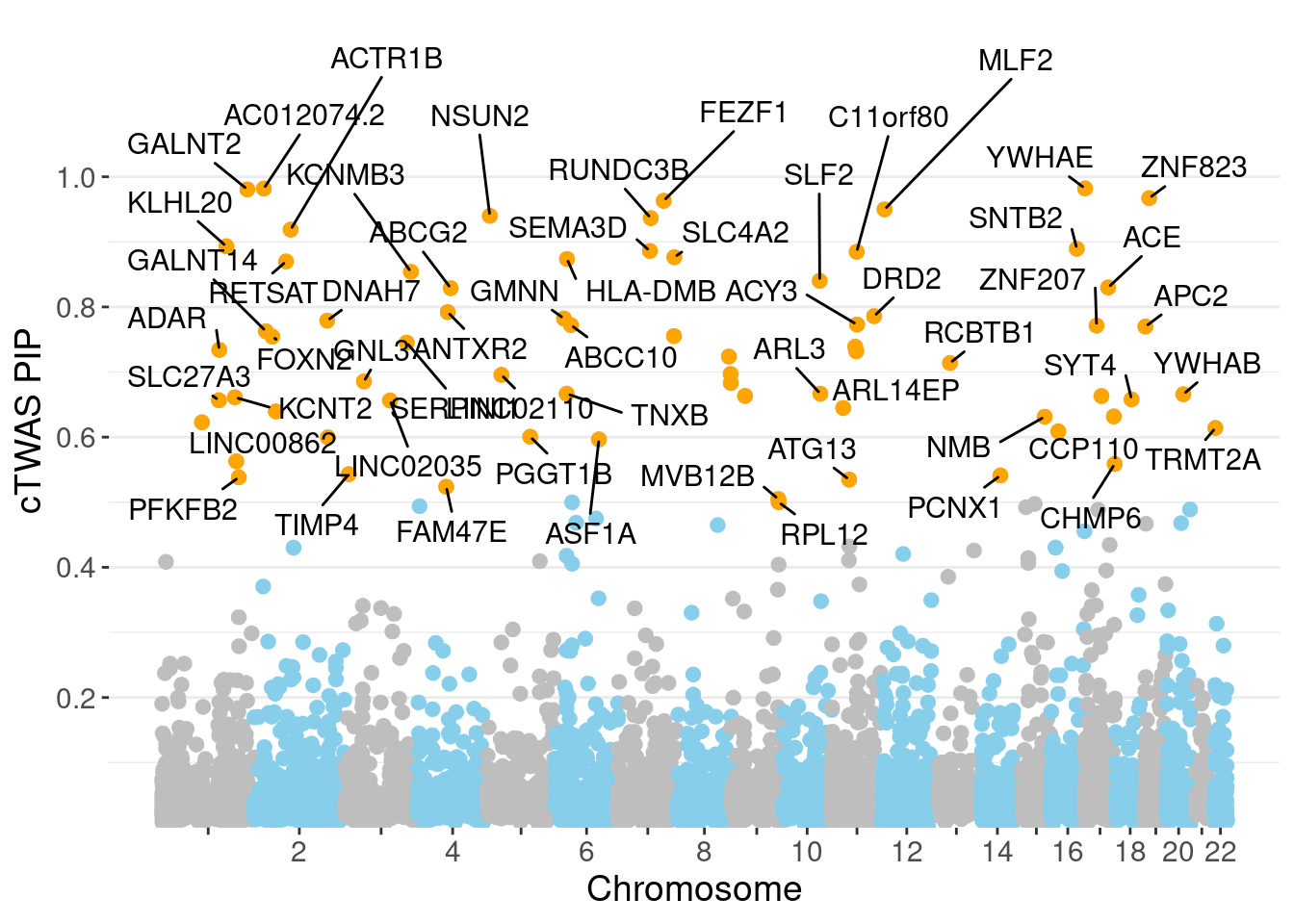
Sensitivity, specificity and precision for silver standard genes
#number of genes in known annotations
print(length(known_annotations))[1] 130#number of genes in known annotations with imputed expression
print(sum(known_annotations %in% ctwas_gene_res$genename))[1] 64#significance threshold for TWAS
print(sig_thresh)[1] 4.591#number of ctwas genes
length(ctwas_genes)[1] 20#number of TWAS genes
length(twas_genes)[1] 190#show novel genes (ctwas genes with not in TWAS genes)
ctwas_gene_res[ctwas_gene_res$genename %in% novel_genes,report_cols] genename region_tag susie_pip mu2 PVE z num_eqtl
417 RETSAT 2_54 0.8699 20.37 0.0001098 3.9626 1
8715 KCNMB3 3_110 0.8539 19.36 0.0001024 -3.8661 1
3381 ABCG2 4_59 0.8289 20.15 0.0001035 -3.9541 1
380 NSUN2 5_6 0.9400 22.60 0.0001316 -4.2792 3
6611 SEMA3D 7_53 0.8861 21.87 0.0001201 -4.0278 3
7758 SLC4A2 7_94 0.8764 20.09 0.0001091 -4.0287 2
9042 C11orf80 11_37 0.8847 19.92 0.0001092 -4.0883 2
2523 YWHAE 17_2 0.9821 51.13 0.0003111 0.1008 2#sensitivity / recall
print(sensitivity) ctwas TWAS
0.02308 0.18462 #specificity
print(specificity) ctwas TWAS
0.9985 0.9853 #precision / PPV
print(precision) ctwas TWAS
0.1500 0.1263 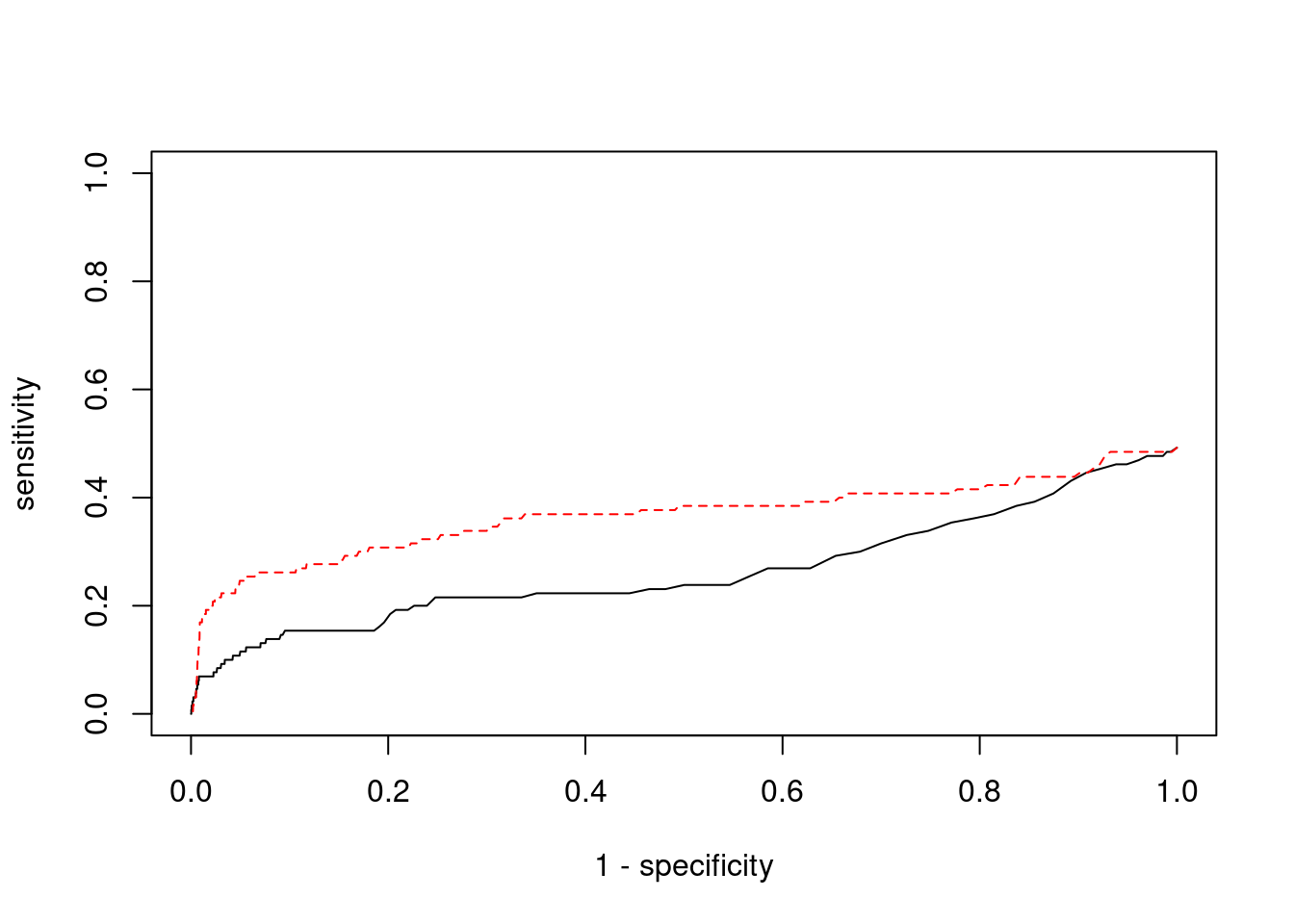
cTWAS is more precise than TWAS in distinguishing silver standard and bystander genes
#number of genes in known annotations (with imputed expression)
print(length(known_annotations))[1] 64#number of bystander genes (with imputed expression)
print(length(unrelated_genes))[1] 785#subset results to genes in known annotations or bystanders
ctwas_gene_res_subset <- ctwas_gene_res[ctwas_gene_res$genename %in% c(known_annotations, unrelated_genes),]
#assign ctwas and TWAS genes
ctwas_genes <- ctwas_gene_res_subset$genename[ctwas_gene_res_subset$susie_pip>0.8]
twas_genes <- ctwas_gene_res_subset$genename[abs(ctwas_gene_res_subset$z)>sig_thresh]
#significance threshold for TWAS
print(sig_thresh)[1] 4.591#number of ctwas genes (in known annotations or bystanders)
length(ctwas_genes)[1] 3#number of TWAS genes (in known annotations or bystanders)
length(twas_genes)[1] 62#sensitivity / recall
sensitivity ctwas TWAS
0.04688 0.37500 #specificity / (1 - False Positive Rate)
specificity ctwas TWAS
1.0000 0.9516 #precision / PPV / (1 - False Discovery Rate)
precision ctwas TWAS
1.0000 0.3871 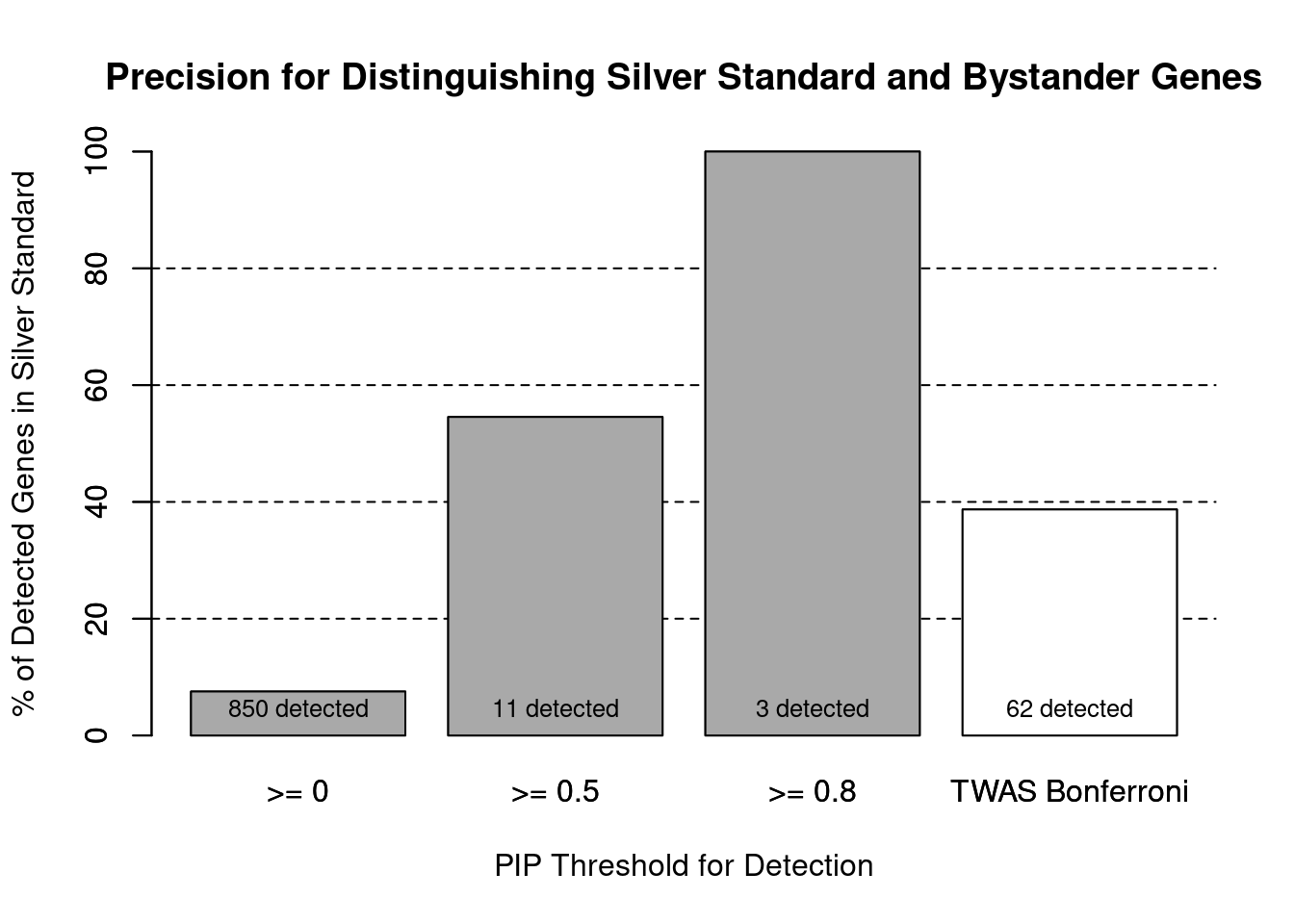
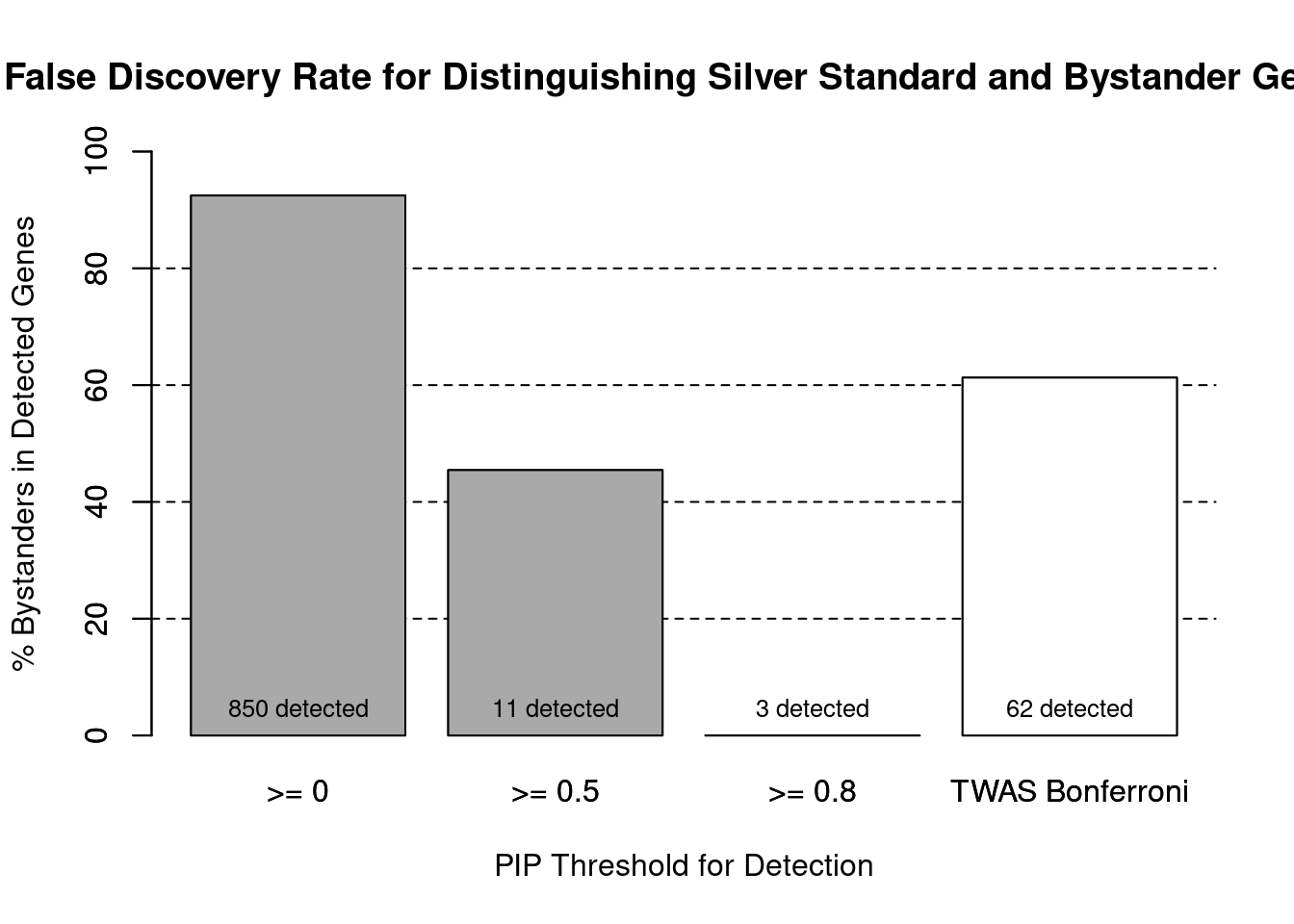
pip_range <- (0:1000)/1000
sensitivity <- rep(NA, length(pip_range))
specificity <- rep(NA, length(pip_range))
for (index in 1:length(pip_range)){
pip <- pip_range[index]
ctwas_genes <- ctwas_gene_res_subset$genename[ctwas_gene_res_subset$susie_pip>=pip]
sensitivity[index] <- sum(ctwas_genes %in% known_annotations)/length(known_annotations)
specificity[index] <- sum(!(unrelated_genes %in% ctwas_genes))/length(unrelated_genes)
}
plot(1-specificity, sensitivity, type="l", xlim=c(0,1), ylim=c(0,1), main="", xlab="1 - Specificity", ylab="Sensitivity")
title(expression("ROC Curve for cTWAS (black) and TWAS (" * phantom("red") * ")"))
title(expression(phantom("ROC Curve for cTWAS (black) and TWAS (") * "red" * phantom(")")), col.main="red")
sig_thresh_range <- seq(from=0, to=max(abs(ctwas_gene_res_subset$z)), length.out=length(pip_range))
for (index in 1:length(sig_thresh_range)){
sig_thresh_plot <- sig_thresh_range[index]
twas_genes <- ctwas_gene_res_subset$genename[abs(ctwas_gene_res_subset$z)>=sig_thresh_plot]
sensitivity[index] <- sum(twas_genes %in% known_annotations)/length(known_annotations)
specificity[index] <- sum(!(unrelated_genes %in% twas_genes))/length(unrelated_genes)
}
lines(1-specificity, sensitivity, xlim=c(0,1), ylim=c(0,1), col="red", lty=1)
abline(a=0,b=1,lty=3)
#add previously computed points from the analysis
ctwas_genes <- ctwas_gene_res_subset$genename[ctwas_gene_res_subset$susie_pip>0.8]
twas_genes <- ctwas_gene_res_subset$genename[abs(ctwas_gene_res_subset$z)>sig_thresh]
points(1-specificity_plot["ctwas"], sensitivity_plot["ctwas"], pch=21, bg="black")
points(1-specificity_plot["TWAS"], sensitivity_plot["TWAS"], pch=21, bg="red")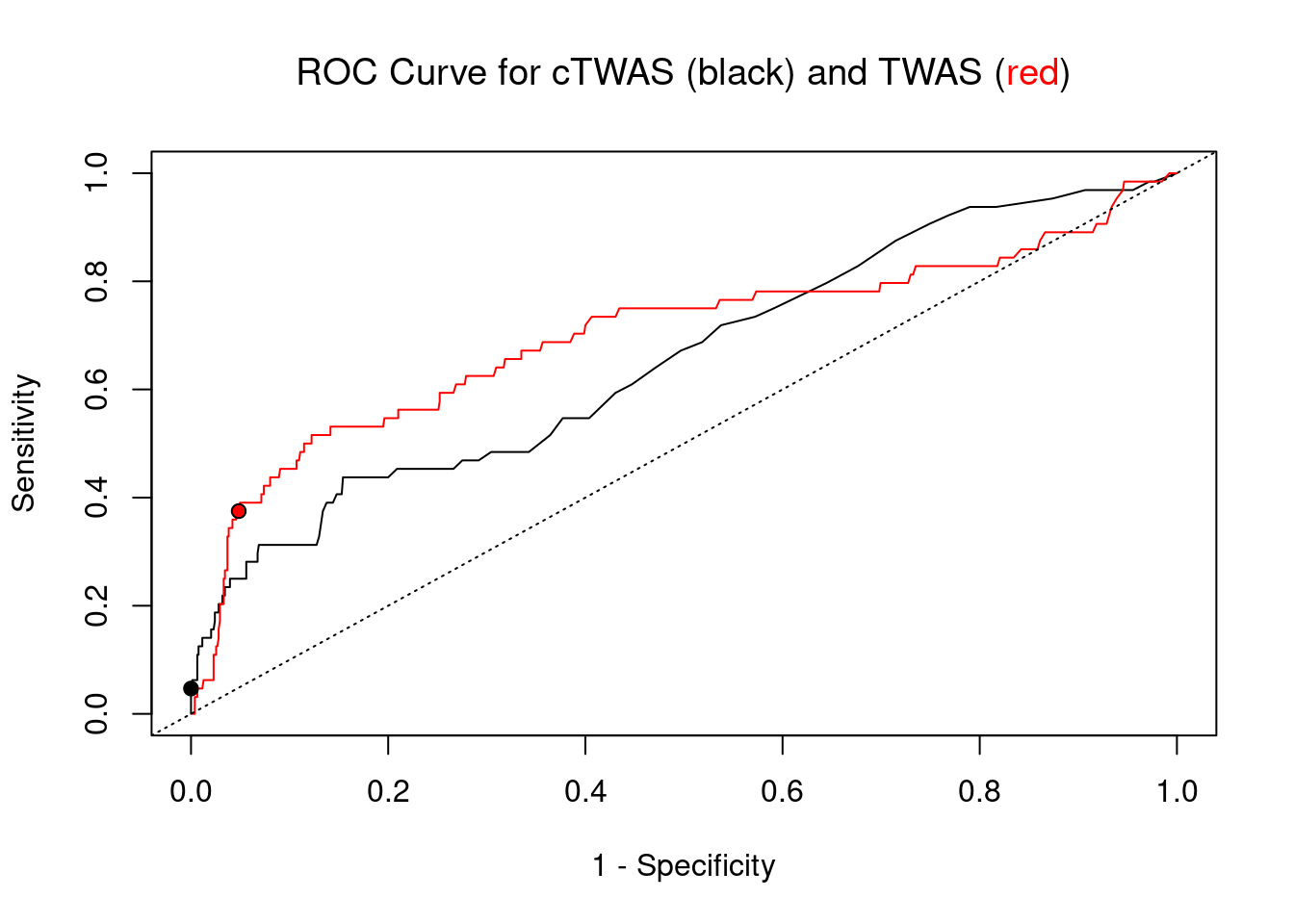
Undetected silver standard genes have low TWAS z-scores or stronger signal from nearby variants
#table of outcomes for silver standard genes
-sort(-table(silver_standard_case))silver_standard_case
Not Imputed Insignificant z-score Nearby SNP(s)
66 40 21
Detected (PIP > 0.8)
3 #show inconclusive genes
silver_standard_case[silver_standard_case=="Inconclusive"]named character(0)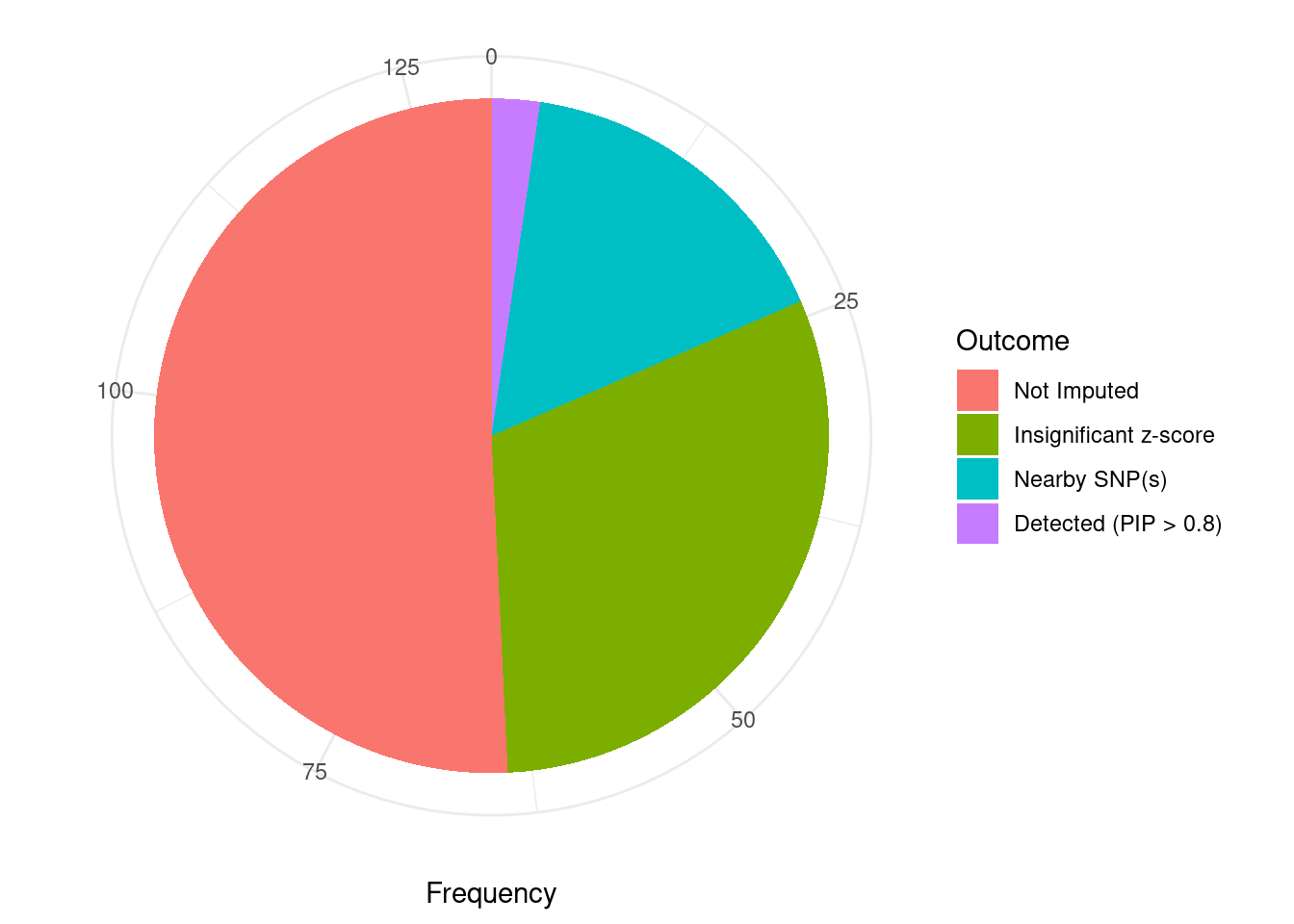
sessionInfo()R version 3.6.1 (2019-07-05)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Scientific Linux 7.4 (Nitrogen)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] GenomicRanges_1.36.1 GenomeInfoDb_1.20.0 IRanges_2.18.1
[4] S4Vectors_0.22.1 BiocGenerics_0.30.0 biomaRt_2.40.1
[7] readxl_1.3.1 forcats_0.5.1 stringr_1.4.0
[10] dplyr_1.0.7 purrr_0.3.4 readr_2.1.1
[13] tidyr_1.1.4 tidyverse_1.3.1 tibble_3.1.6
[16] WebGestaltR_0.4.4 disgenet2r_0.99.2 enrichR_3.0
[19] cowplot_1.1.1 ggplot2_3.3.5 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] ggbeeswarm_0.6.0 colorspace_2.0-2 rjson_0.2.20
[4] ellipsis_0.3.2 rprojroot_2.0.2 XVector_0.24.0
[7] fs_1.5.2 rstudioapi_0.13 farver_2.1.0
[10] ggrepel_0.9.1 bit64_4.0.5 AnnotationDbi_1.46.0
[13] fansi_1.0.2 lubridate_1.8.0 xml2_1.3.3
[16] codetools_0.2-16 doParallel_1.0.17 cachem_1.0.6
[19] knitr_1.36 jsonlite_1.7.2 apcluster_1.4.8
[22] Cairo_1.5-12.2 broom_0.7.10 dbplyr_2.1.1
[25] compiler_3.6.1 httr_1.4.2 backports_1.4.1
[28] assertthat_0.2.1 Matrix_1.2-18 fastmap_1.1.0
[31] cli_3.1.0 later_0.8.0 prettyunits_1.1.1
[34] htmltools_0.5.2 tools_3.6.1 igraph_1.2.10
[37] GenomeInfoDbData_1.2.1 gtable_0.3.0 glue_1.6.2
[40] reshape2_1.4.4 doRNG_1.8.2 Rcpp_1.0.8
[43] Biobase_2.44.0 cellranger_1.1.0 jquerylib_0.1.4
[46] vctrs_0.3.8 svglite_1.2.2 iterators_1.0.14
[49] xfun_0.29 ps_1.6.0 rvest_1.0.2
[52] lifecycle_1.0.1 rngtools_1.5.2 XML_3.99-0.3
[55] zlibbioc_1.30.0 getPass_0.2-2 scales_1.1.1
[58] vroom_1.5.7 hms_1.1.1 promises_1.0.1
[61] yaml_2.2.1 curl_4.3.2 memoise_2.0.1
[64] ggrastr_1.0.1 gdtools_0.1.9 stringi_1.7.6
[67] RSQLite_2.2.8 highr_0.9 foreach_1.5.2
[70] rlang_1.0.1 pkgconfig_2.0.3 bitops_1.0-7
[73] evaluate_0.14 lattice_0.20-38 labeling_0.4.2
[76] bit_4.0.4 processx_3.5.2 tidyselect_1.1.1
[79] plyr_1.8.6 magrittr_2.0.2 R6_2.5.1
[82] generics_0.1.1 DBI_1.1.2 pillar_1.6.4
[85] haven_2.4.3 whisker_0.3-2 withr_2.4.3
[88] RCurl_1.98-1.5 modelr_0.1.8 crayon_1.5.0
[91] utf8_1.2.2 tzdb_0.2.0 rmarkdown_2.11
[94] progress_1.2.2 grid_3.6.1 data.table_1.14.2
[97] blob_1.2.2 callr_3.7.0 git2r_0.26.1
[100] reprex_2.0.1 digest_0.6.29 httpuv_1.5.1
[103] munsell_0.5.0 beeswarm_0.2.3 vipor_0.4.5