Comparing Munro st & Susie st models in multi-group analysis
XSun
2025-04-07
Last updated: 2025-04-07
Checks: 6 1
Knit directory: multigroup_ctwas_analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown is untracked by Git. To know which version of the R
Markdown file created these results, you’ll want to first commit it to
the Git repo. If you’re still working on the analysis, you can ignore
this warning. When you’re finished, you can run
wflow_publish to commit the R Markdown file and build the
HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20231112) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version f119060. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: cv/
Untracked files:
Untracked: analysis/comparing_munro_susie_st.Rmd
Unstaged changes:
Modified: analysis/index.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
There are no past versions. Publish this analysis with
wflow_publish() to start tracking its development.
Introduction
The Susie st models are trained here https://sq-96.github.io/multigroup_ctwas_analysis/data_weight_training_fusionscale_rsqupdated_allhaveqtl.html
The Munro’s model are from this paper https://www.nature.com/articles/s41467-024-54840-8
library(ctwas)
library(ggplot2)
library(tidyverse)
source("/project/xinhe/xsun/multi_group_ctwas/data/samplesize.R")
folder_results_munroST <- "/project/xinhe/shengqian/ctwas_GWAS_analysis/snakemake_outputs/"
folder_results_susieST <- "/project/xinhe/xsun/multi_group_ctwas/15.susie_weights/snakemake_outputs/"
folder_results_woST <- "/project/xinhe/shengqian/ctwas_GWAS_analysis/snakemake_outputs/"
# mapping_predictdb <- readRDS("/project2/xinhe/shared_data/multigroup_ctwas/weights/mapping_files/PredictDB_mapping.RDS")
# mapping_munro <- readRDS("/project2/xinhe/shared_data/multigroup_ctwas/weights/mapping_files/Munro_mapping.RDS")
# mapping_two <- rbind(mapping_predictdb,mapping_munro)
colors <- c( "#1f77b4", "#ff7f0e", "#2ca02c", "#d62728", "#9467bd", "#8c564b", "#e377c2", "#7f7f7f", "#bcbd22", "#17becf", "#f7b6d2", "#c5b0d5", "#9edae5", "#ffbb78", "#98df8a", "#ff9896" )
plot_piechart <- function(ctwas_parameters, colors, by, title) {
# Create the initial data frame
data <- data.frame(
category = names(ctwas_parameters$prop_heritability),
percentage = ctwas_parameters$prop_heritability
)
# Split the category into context and type
data <- data %>%
mutate(
context = sub("\\|.*", "", category),
type = sub(".*\\|", "", category)
)
# Aggregate the data based on the 'by' parameter
if (by == "type") {
data <- data %>%
group_by(type) %>%
summarize(percentage = sum(percentage)) %>%
mutate(category = type) # Use type as the new category
} else if (by == "context") {
data <- data %>%
group_by(context) %>%
summarize(percentage = sum(percentage)) %>%
mutate(category = context) # Use context as the new category
} else {
stop("Invalid 'by' parameter. Use 'type' or 'context'.")
}
# Calculate percentage labels for the chart
data$percentage_label <- paste0(round(data$percentage * 100, 1), "%")
# Create the pie chart
pie <- ggplot(data, aes(x = "", y = percentage, fill = category)) +
geom_bar(stat = "identity", width = 1) +
coord_polar("y", start = 0) +
theme_void() + # Remove background and axes
geom_text(aes(label = percentage_label),
position = position_stack(vjust = 0.5), size = 3) + # Adjust size as needed
scale_fill_manual(values = colors) + # Custom colors
labs(fill = "Category") + # Legend title
ggtitle(title) # Title
return(pie)
}LDL-ukb-d-30780_irnt
Setting: shared_all, thin = 0.1
trait <- "LDL-ukb-d-30780_irnt"
thin <- 0.1
var_struc <- "shared_all"
gwas_n <- samplesize[trait]st <- "without_ST"
param_woST <- readRDS(paste0(folder_results_woST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_woST <- summarize_param(param_woST, gwas_n)
total_nonSNPpve_woST <- 1- ctwas_parameters_woST$prop_heritability["SNP"]
pve_pie_by_type_woST <- plot_piechart(ctwas_parameters = ctwas_parameters_woST, colors = colors, by = "type", title = paste0("withoutST - nonSNP %h2g:",round(total_nonSNPpve_woST, digits = 4)))st <- "with_susieST"
param_susieST <- readRDS(paste0(folder_results_susieST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_susieST <- summarize_param(param_susieST, gwas_n)
total_nonSNPpve_susieST <- 1- ctwas_parameters_susieST$prop_heritability["SNP"]
pve_pie_by_type_susieST <- plot_piechart(ctwas_parameters = ctwas_parameters_susieST, colors = colors, by = "type", title = paste0("susieST - nonSNP %h2g:",round(total_nonSNPpve_susieST, digits = 4)))st <- "with_ST"
param_munroST <- readRDS(paste0(folder_results_munroST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
total_nonSNPpve_munroST <- 1- ctwas_parameters_munroST$prop_heritability["SNP"]
pve_pie_by_type_munroST <- plot_piechart(ctwas_parameters = ctwas_parameters_munroST, colors = colors, by = "type", title = paste0("munroST - nonSNP %h2g:",round(total_nonSNPpve_munroST, digits = 4)))gridExtra::grid.arrange(pve_pie_by_type_woST, pve_pie_by_type_susieST,pve_pie_by_type_munroST,
ncol = 3, top = paste0(trait,"-thin",thin,"-",var_struc)) 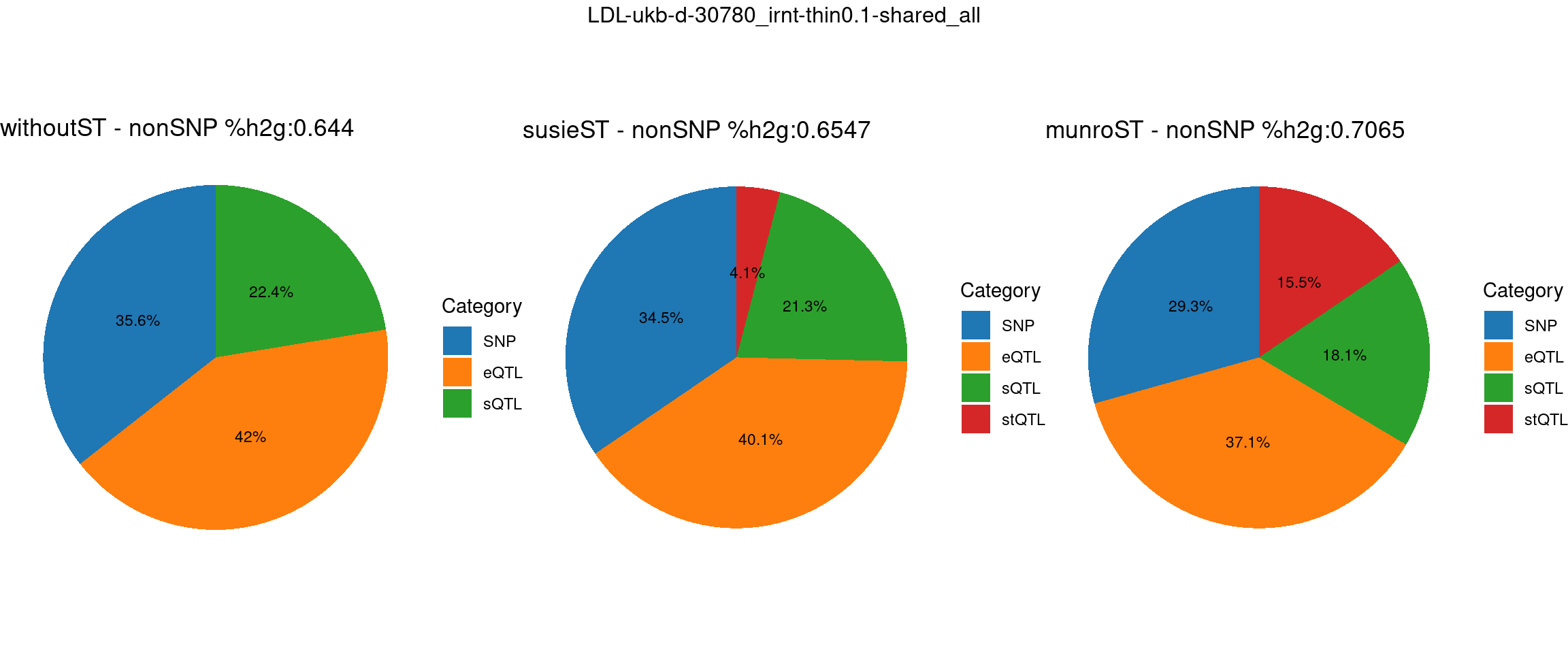
Setting: shared_all, thin = 0.5
trait <- "LDL-ukb-d-30780_irnt"
thin <- 0.5
var_struc <- "shared_all"
gwas_n <- samplesize[trait]st <- "without_ST"
param_woST <- readRDS(paste0(folder_results_woST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_woST <- summarize_param(param_woST, gwas_n)
total_nonSNPpve_woST <- 1- ctwas_parameters_woST$prop_heritability["SNP"]
pve_pie_by_type_woST <- plot_piechart(ctwas_parameters = ctwas_parameters_woST, colors = colors, by = "type", title = paste0("withoutST - nonSNP %h2g:",round(total_nonSNPpve_woST, digits = 4)))st <- "with_susieST"
param_susieST <- readRDS(paste0(folder_results_susieST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_susieST <- summarize_param(param_susieST, gwas_n)
total_nonSNPpve_susieST <- 1- ctwas_parameters_susieST$prop_heritability["SNP"]
pve_pie_by_type_susieST <- plot_piechart(ctwas_parameters = ctwas_parameters_susieST, colors = colors, by = "type", title = paste0("susieST - nonSNP %h2g:",round(total_nonSNPpve_susieST, digits = 4)))st <- "with_ST"
param_munroST <- readRDS(paste0(folder_results_munroST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
total_nonSNPpve_munroST <- 1- ctwas_parameters_munroST$prop_heritability["SNP"]
pve_pie_by_type_munroST <- plot_piechart(ctwas_parameters = ctwas_parameters_munroST, colors = colors, by = "type", title = paste0("munroST - nonSNP %h2g:",round(total_nonSNPpve_munroST, digits = 4)))gridExtra::grid.arrange(pve_pie_by_type_woST, pve_pie_by_type_susieST,pve_pie_by_type_munroST,
ncol = 3, top = paste0(trait,"-thin",thin,"-",var_struc)) 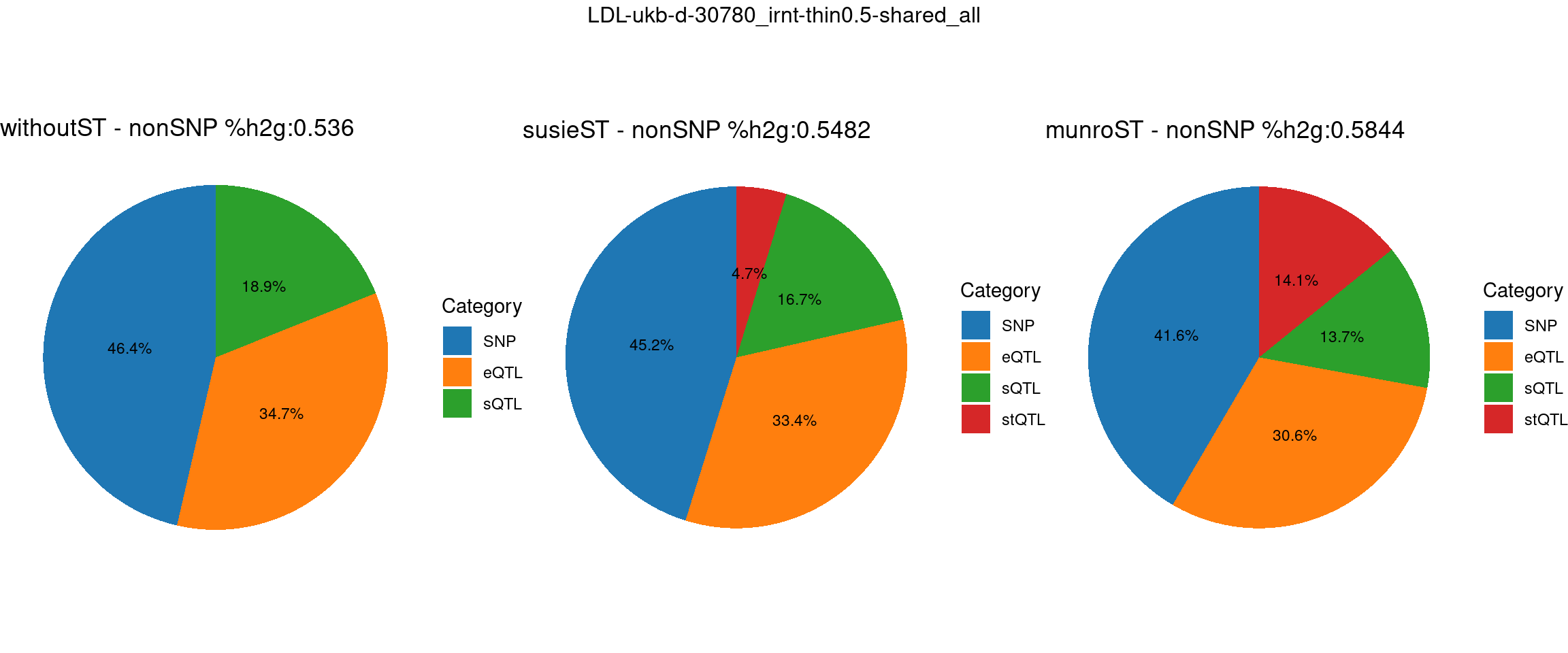
Setting: shared_all, thin = 1
trait <- "LDL-ukb-d-30780_irnt"
thin <- 1
var_struc <- "shared_all"
gwas_n <- samplesize[trait]st <- "without_ST"
param_woST <- readRDS(paste0(folder_results_woST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_woST <- summarize_param(param_woST, gwas_n)
total_nonSNPpve_woST <- 1- ctwas_parameters_woST$prop_heritability["SNP"]
pve_pie_by_type_woST <- plot_piechart(ctwas_parameters = ctwas_parameters_woST, colors = colors, by = "type", title = paste0("withoutST - nonSNP %h2g:",round(total_nonSNPpve_woST, digits = 4)))st <- "with_susieST"
param_susieST <- readRDS(paste0(folder_results_susieST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_susieST <- summarize_param(param_susieST, gwas_n)
total_nonSNPpve_susieST <- 1- ctwas_parameters_susieST$prop_heritability["SNP"]
pve_pie_by_type_susieST <- plot_piechart(ctwas_parameters = ctwas_parameters_susieST, colors = colors, by = "type", title = paste0("susieST - nonSNP %h2g:",round(total_nonSNPpve_susieST, digits = 4)))st <- "with_ST"
param_munroST <- readRDS(paste0(folder_results_munroST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
total_nonSNPpve_munroST <- 1- ctwas_parameters_munroST$prop_heritability["SNP"]
pve_pie_by_type_munroST <- plot_piechart(ctwas_parameters = ctwas_parameters_munroST, colors = colors, by = "type", title = paste0("munroST - nonSNP %h2g:",round(total_nonSNPpve_munroST, digits = 4)))gridExtra::grid.arrange(pve_pie_by_type_woST, pve_pie_by_type_susieST,pve_pie_by_type_munroST,
ncol = 3, top = paste0(trait,"-thin",thin,"-",var_struc)) 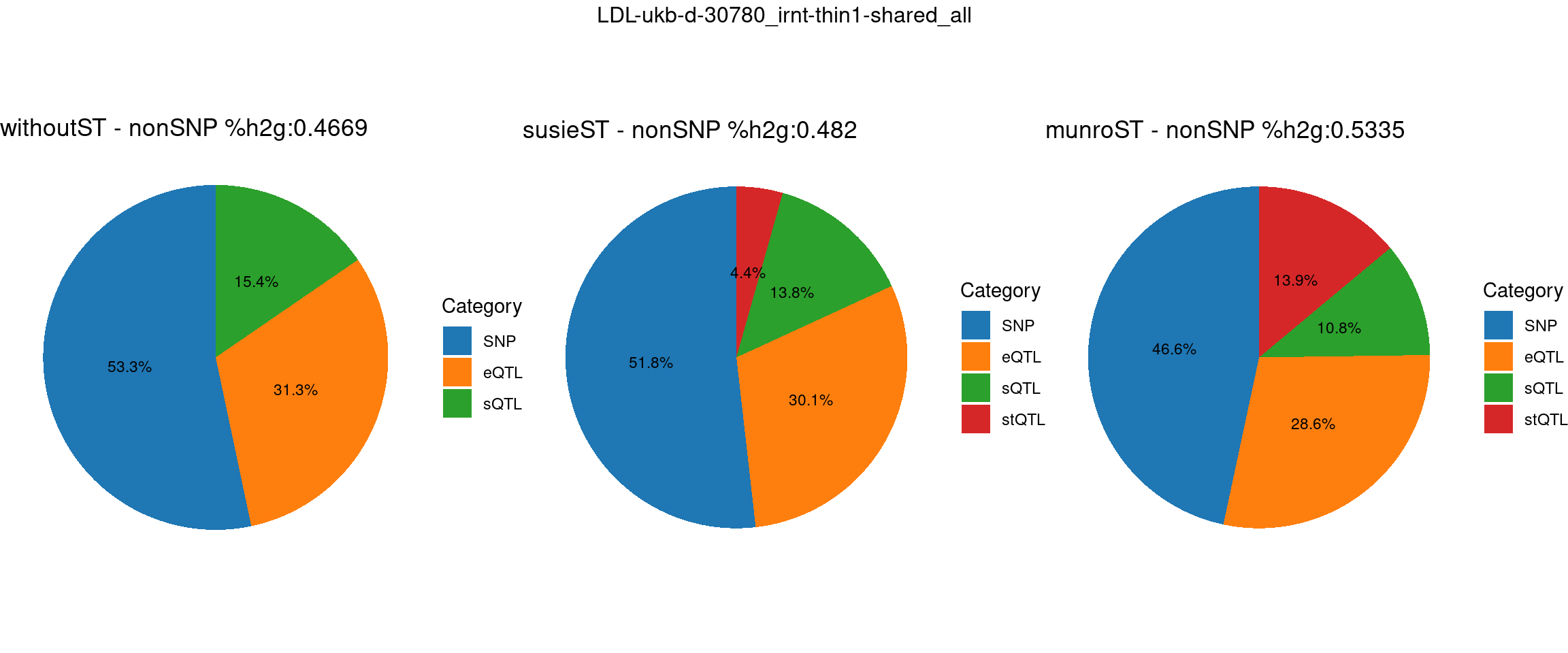
Setting: shared_type, thin = 0.1
trait <- "LDL-ukb-d-30780_irnt"
thin <- 0.1
var_struc <- "shared_type"
gwas_n <- samplesize[trait]st <- "without_ST"
param_woST <- readRDS(paste0(folder_results_woST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_woST <- summarize_param(param_woST, gwas_n)
total_nonSNPpve_woST <- 1- ctwas_parameters_woST$prop_heritability["SNP"]
pve_pie_by_type_woST <- plot_piechart(ctwas_parameters = ctwas_parameters_woST, colors = colors, by = "type", title = paste0("withoutST - nonSNP %h2g:",round(total_nonSNPpve_woST, digits = 4)))st <- "with_susieST"
param_susieST <- readRDS(paste0(folder_results_susieST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_susieST <- summarize_param(param_susieST, gwas_n)
total_nonSNPpve_susieST <- 1- ctwas_parameters_susieST$prop_heritability["SNP"]
pve_pie_by_type_susieST <- plot_piechart(ctwas_parameters = ctwas_parameters_susieST, colors = colors, by = "type", title = paste0("susieST - nonSNP %h2g:",round(total_nonSNPpve_susieST, digits = 4)))st <- "with_ST"
param_munroST <- readRDS(paste0(folder_results_munroST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
total_nonSNPpve_munroST <- 1- ctwas_parameters_munroST$prop_heritability["SNP"]
pve_pie_by_type_munroST <- plot_piechart(ctwas_parameters = ctwas_parameters_munroST, colors = colors, by = "type", title = paste0("munroST - nonSNP %h2g:",round(total_nonSNPpve_munroST, digits = 4)))gridExtra::grid.arrange(pve_pie_by_type_woST, pve_pie_by_type_susieST,pve_pie_by_type_munroST,
ncol = 3, top = paste0(trait,"-thin",thin,"-",var_struc)) 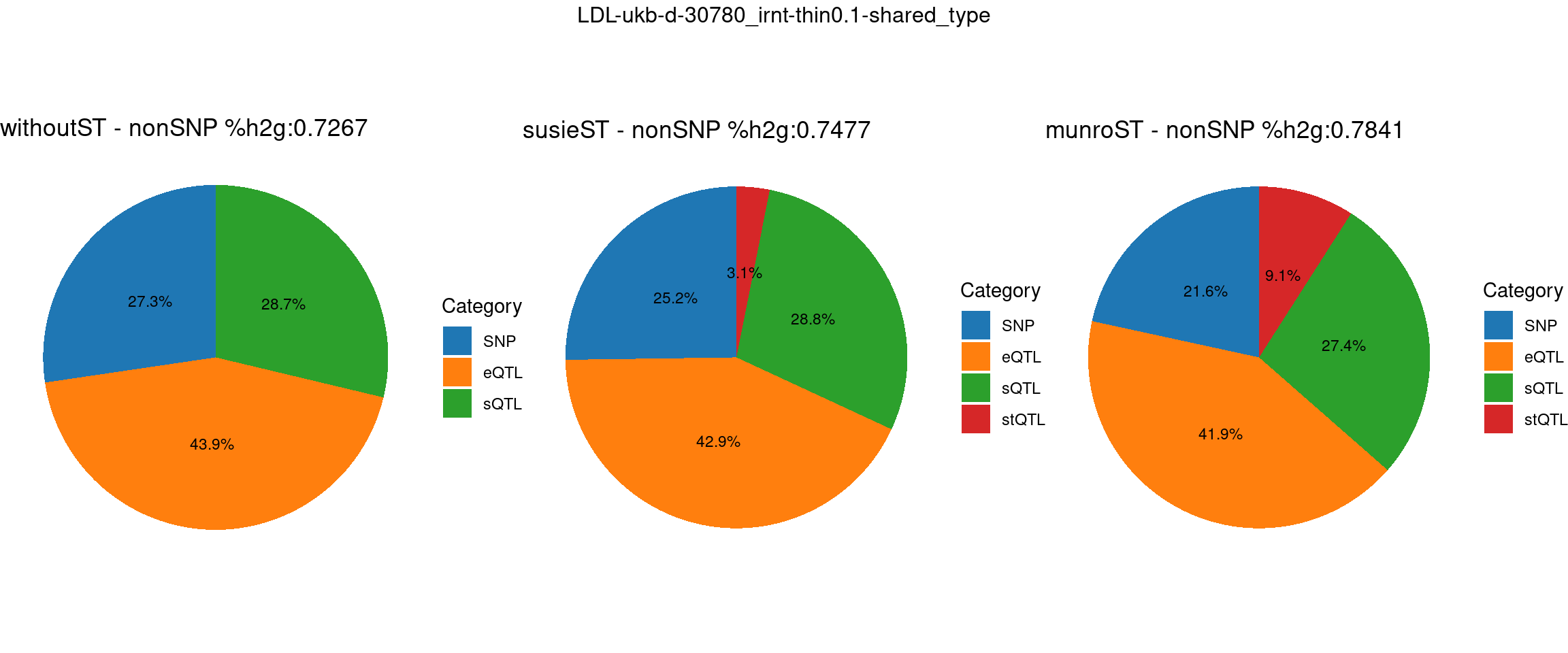
Setting: shared_type, thin = 0.5
trait <- "LDL-ukb-d-30780_irnt"
thin <- 0.5
var_struc <- "shared_type"
gwas_n <- samplesize[trait]st <- "without_ST"
param_woST <- readRDS(paste0(folder_results_woST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_woST <- summarize_param(param_woST, gwas_n)
total_nonSNPpve_woST <- 1- ctwas_parameters_woST$prop_heritability["SNP"]
pve_pie_by_type_woST <- plot_piechart(ctwas_parameters = ctwas_parameters_woST, colors = colors, by = "type", title = paste0("withoutST - nonSNP %h2g:",round(total_nonSNPpve_woST, digits = 4)))st <- "with_susieST"
param_susieST <- readRDS(paste0(folder_results_susieST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_susieST <- summarize_param(param_susieST, gwas_n)
total_nonSNPpve_susieST <- 1- ctwas_parameters_susieST$prop_heritability["SNP"]
pve_pie_by_type_susieST <- plot_piechart(ctwas_parameters = ctwas_parameters_susieST, colors = colors, by = "type", title = paste0("susieST - nonSNP %h2g:",round(total_nonSNPpve_susieST, digits = 4)))st <- "with_ST"
param_munroST <- readRDS(paste0(folder_results_munroST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
total_nonSNPpve_munroST <- 1- ctwas_parameters_munroST$prop_heritability["SNP"]
pve_pie_by_type_munroST <- plot_piechart(ctwas_parameters = ctwas_parameters_munroST, colors = colors, by = "type", title = paste0("munroST - nonSNP %h2g:",round(total_nonSNPpve_munroST, digits = 4)))gridExtra::grid.arrange(pve_pie_by_type_woST, pve_pie_by_type_susieST,pve_pie_by_type_munroST,
ncol = 3, top = paste0(trait,"-thin",thin,"-",var_struc)) 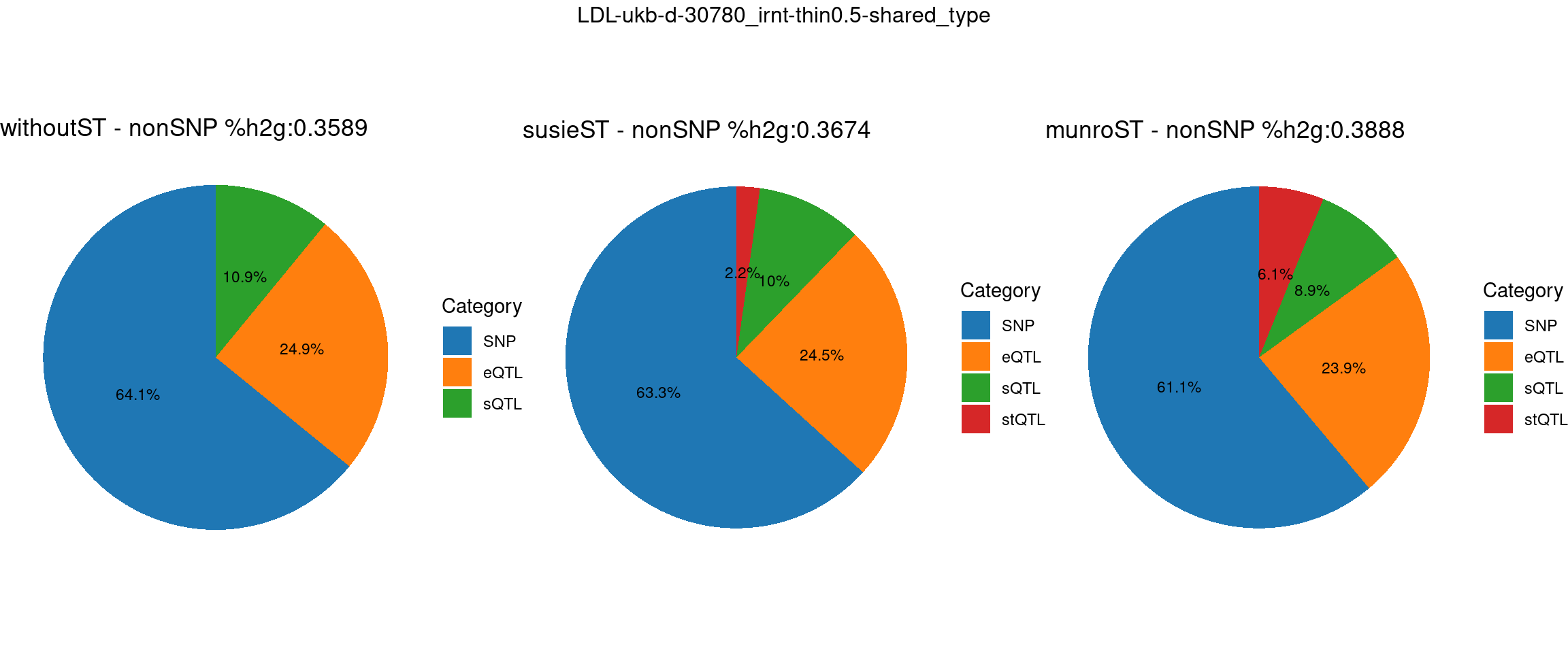
Setting: shared_type, thin = 1
trait <- "LDL-ukb-d-30780_irnt"
thin <- 1
var_struc <- "shared_type"
gwas_n <- samplesize[trait]st <- "without_ST"
param_woST <- readRDS(paste0(folder_results_woST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_woST <- summarize_param(param_woST, gwas_n)
total_nonSNPpve_woST <- 1- ctwas_parameters_woST$prop_heritability["SNP"]
pve_pie_by_type_woST <- plot_piechart(ctwas_parameters = ctwas_parameters_woST, colors = colors, by = "type", title = paste0("withoutST - nonSNP %h2g:",round(total_nonSNPpve_woST, digits = 4)))st <- "with_susieST"
param_susieST <- readRDS(paste0(folder_results_susieST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_susieST <- summarize_param(param_susieST, gwas_n)
total_nonSNPpve_susieST <- 1- ctwas_parameters_susieST$prop_heritability["SNP"]
pve_pie_by_type_susieST <- plot_piechart(ctwas_parameters = ctwas_parameters_susieST, colors = colors, by = "type", title = paste0("susieST - nonSNP %h2g:",round(total_nonSNPpve_susieST, digits = 4)))st <- "with_ST"
param_munroST <- readRDS(paste0(folder_results_munroST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
total_nonSNPpve_munroST <- 1- ctwas_parameters_munroST$prop_heritability["SNP"]
pve_pie_by_type_munroST <- plot_piechart(ctwas_parameters = ctwas_parameters_munroST, colors = colors, by = "type", title = paste0("munroST - nonSNP %h2g:",round(total_nonSNPpve_munroST, digits = 4)))gridExtra::grid.arrange(pve_pie_by_type_woST, pve_pie_by_type_susieST,pve_pie_by_type_munroST,
ncol = 3, top = paste0(trait,"-thin",thin,"-",var_struc)) 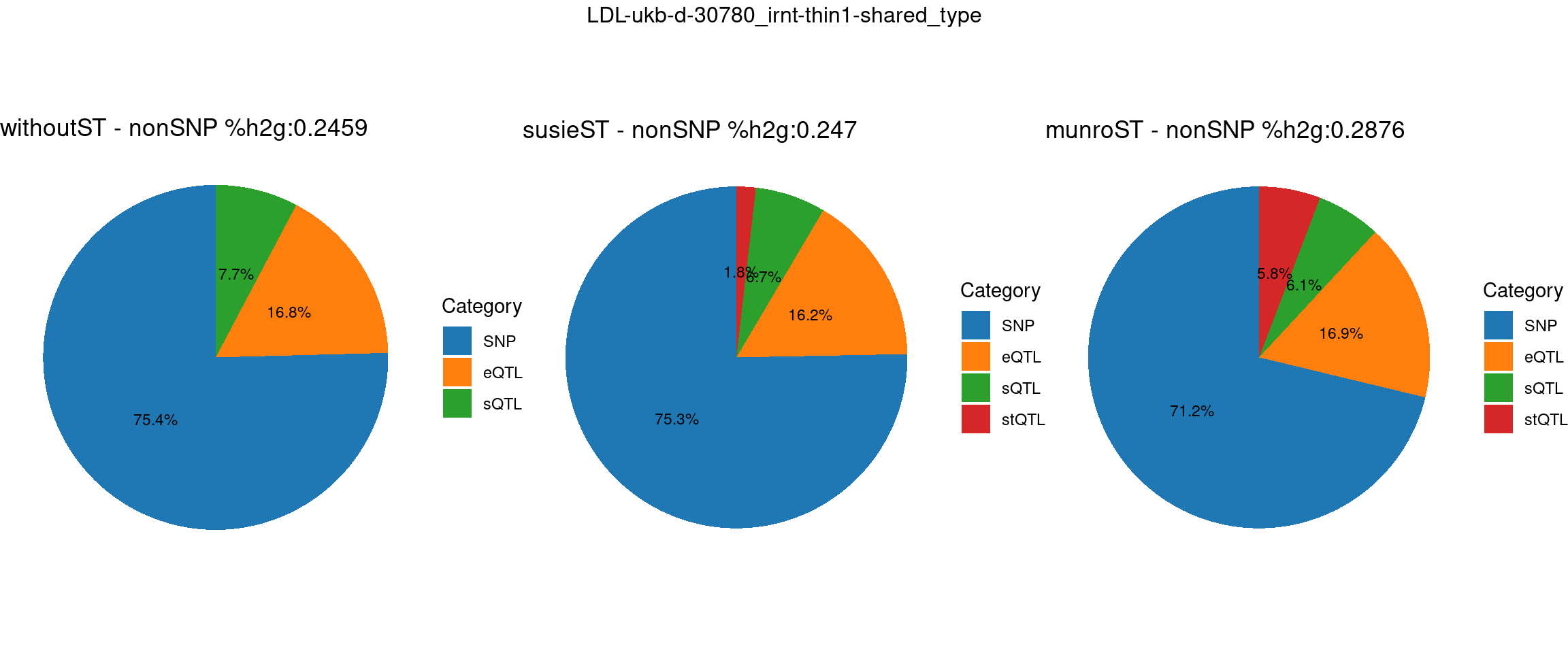
IBD-ebi-a-GCST004131
Setting: shared_all, thin = 0.1
trait <- "IBD-ebi-a-GCST004131"
thin <- 0.1
var_struc <- "shared_all"
gwas_n <- samplesize[trait]st <- "without_ST"
param_woST <- readRDS(paste0(folder_results_woST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_woST <- summarize_param(param_woST, gwas_n)
total_nonSNPpve_woST <- 1- ctwas_parameters_woST$prop_heritability["SNP"]
pve_pie_by_type_woST <- plot_piechart(ctwas_parameters = ctwas_parameters_woST, colors = colors, by = "type", title = paste0("withoutST - nonSNP %h2g:",round(total_nonSNPpve_woST, digits = 4)))st <- "with_susieST"
param_susieST <- readRDS(paste0(folder_results_susieST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_susieST <- summarize_param(param_susieST, gwas_n)
total_nonSNPpve_susieST <- 1- ctwas_parameters_susieST$prop_heritability["SNP"]
pve_pie_by_type_susieST <- plot_piechart(ctwas_parameters = ctwas_parameters_susieST, colors = colors, by = "type", title = paste0("susieST - nonSNP %h2g:",round(total_nonSNPpve_susieST, digits = 4)))st <- "with_ST"
param_munroST <- readRDS(paste0(folder_results_munroST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
total_nonSNPpve_munroST <- 1- ctwas_parameters_munroST$prop_heritability["SNP"]
pve_pie_by_type_munroST <- plot_piechart(ctwas_parameters = ctwas_parameters_munroST, colors = colors, by = "type", title = paste0("munroST - nonSNP %h2g:",round(total_nonSNPpve_munroST, digits = 4)))gridExtra::grid.arrange(pve_pie_by_type_woST, pve_pie_by_type_susieST,pve_pie_by_type_munroST,
ncol = 3, top = paste0(trait,"-thin",thin,"-",var_struc)) 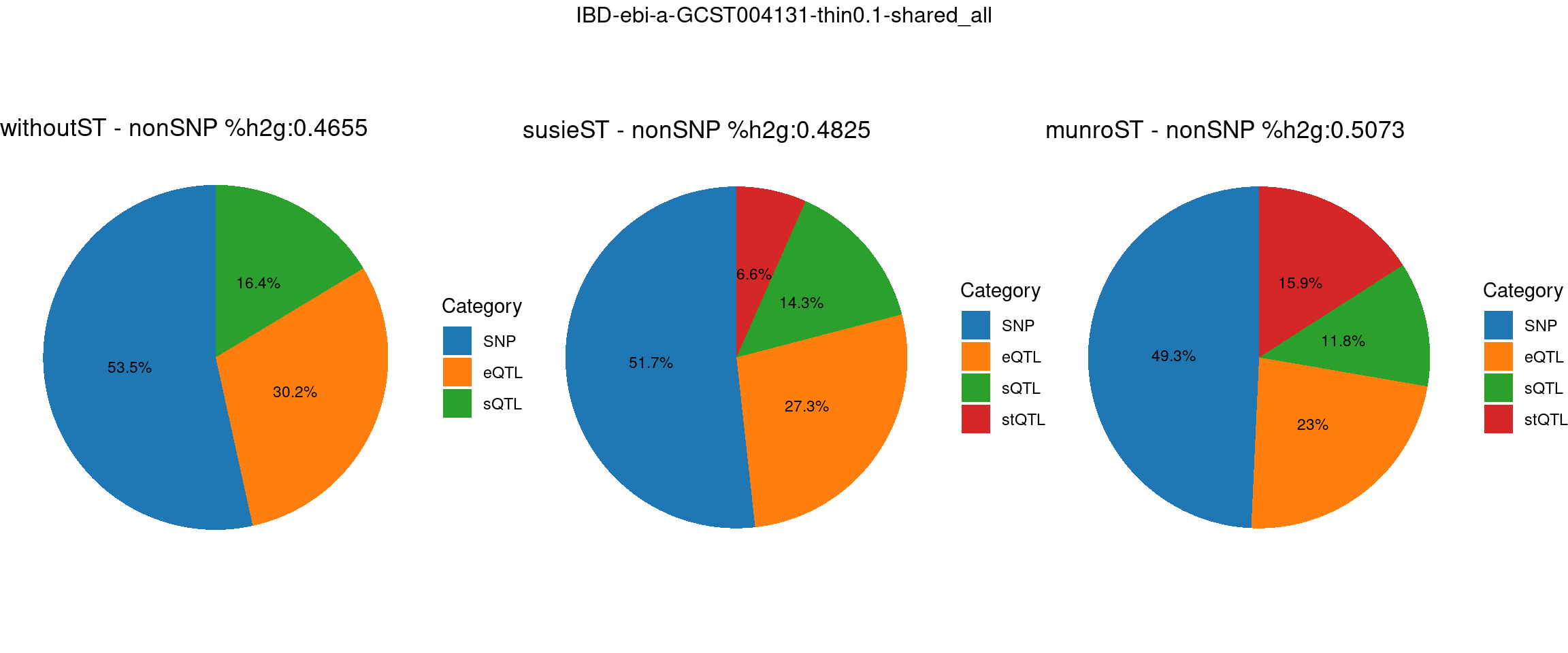
Setting: shared_all, thin = 0.5
trait <- "LDL-ukb-d-30780_irnt"
thin <- 0.5
var_struc <- "shared_all"
gwas_n <- samplesize[trait]st <- "without_ST"
param_woST <- readRDS(paste0(folder_results_woST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_woST <- summarize_param(param_woST, gwas_n)
total_nonSNPpve_woST <- 1- ctwas_parameters_woST$prop_heritability["SNP"]
pve_pie_by_type_woST <- plot_piechart(ctwas_parameters = ctwas_parameters_woST, colors = colors, by = "type", title = paste0("withoutST - nonSNP %h2g:",round(total_nonSNPpve_woST, digits = 4)))st <- "with_susieST"
param_susieST <- readRDS(paste0(folder_results_susieST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_susieST <- summarize_param(param_susieST, gwas_n)
total_nonSNPpve_susieST <- 1- ctwas_parameters_susieST$prop_heritability["SNP"]
pve_pie_by_type_susieST <- plot_piechart(ctwas_parameters = ctwas_parameters_susieST, colors = colors, by = "type", title = paste0("susieST - nonSNP %h2g:",round(total_nonSNPpve_susieST, digits = 4)))st <- "with_ST"
param_munroST <- readRDS(paste0(folder_results_munroST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
total_nonSNPpve_munroST <- 1- ctwas_parameters_munroST$prop_heritability["SNP"]
pve_pie_by_type_munroST <- plot_piechart(ctwas_parameters = ctwas_parameters_munroST, colors = colors, by = "type", title = paste0("munroST - nonSNP %h2g:",round(total_nonSNPpve_munroST, digits = 4)))gridExtra::grid.arrange(pve_pie_by_type_woST, pve_pie_by_type_susieST,pve_pie_by_type_munroST,
ncol = 3, top = paste0(trait,"-thin",thin,"-",var_struc)) 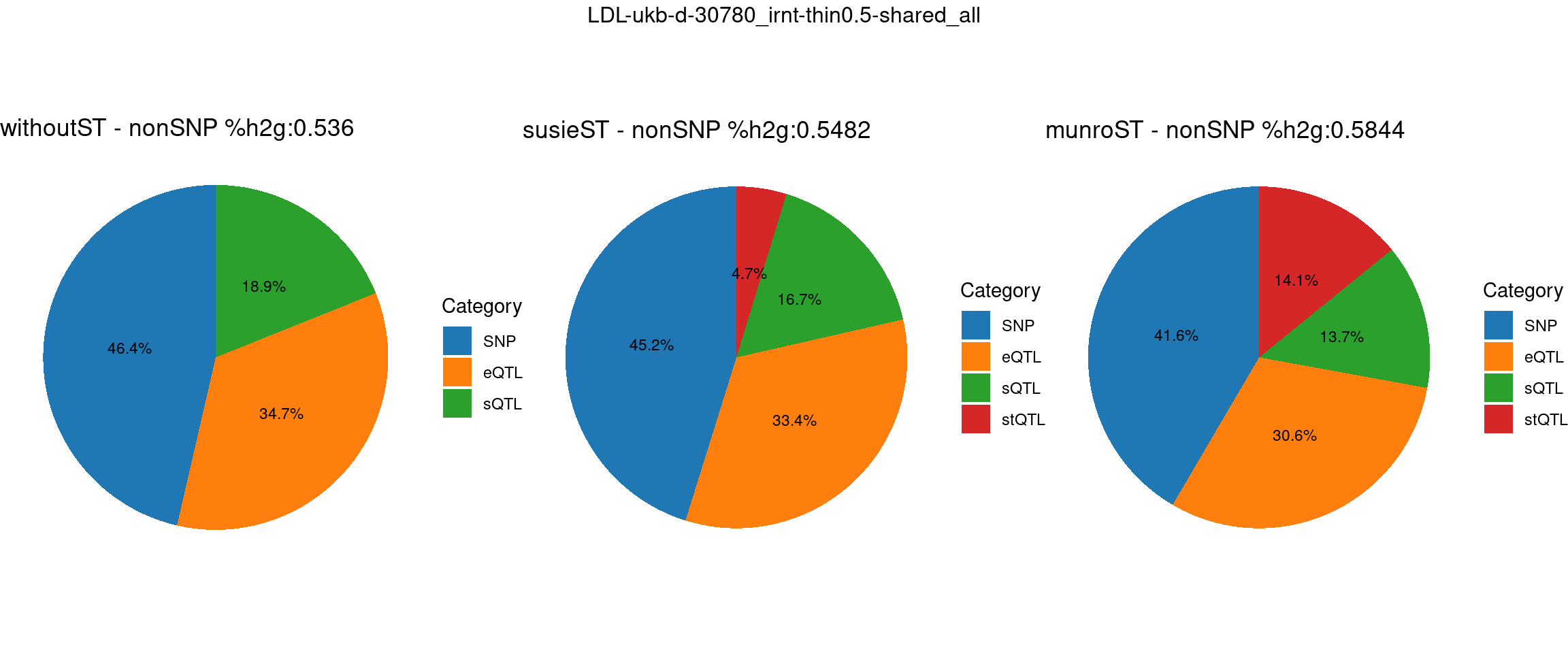
Setting: shared_all, thin = 1
trait <- "LDL-ukb-d-30780_irnt"
thin <- 1
var_struc <- "shared_all"
gwas_n <- samplesize[trait]st <- "without_ST"
param_woST <- readRDS(paste0(folder_results_woST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_woST <- summarize_param(param_woST, gwas_n)
total_nonSNPpve_woST <- 1- ctwas_parameters_woST$prop_heritability["SNP"]
pve_pie_by_type_woST <- plot_piechart(ctwas_parameters = ctwas_parameters_woST, colors = colors, by = "type", title = paste0("withoutST - nonSNP %h2g:",round(total_nonSNPpve_woST, digits = 4)))st <- "with_susieST"
param_susieST <- readRDS(paste0(folder_results_susieST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_susieST <- summarize_param(param_susieST, gwas_n)
total_nonSNPpve_susieST <- 1- ctwas_parameters_susieST$prop_heritability["SNP"]
pve_pie_by_type_susieST <- plot_piechart(ctwas_parameters = ctwas_parameters_susieST, colors = colors, by = "type", title = paste0("susieST - nonSNP %h2g:",round(total_nonSNPpve_susieST, digits = 4)))st <- "with_ST"
param_munroST <- readRDS(paste0(folder_results_munroST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
total_nonSNPpve_munroST <- 1- ctwas_parameters_munroST$prop_heritability["SNP"]
pve_pie_by_type_munroST <- plot_piechart(ctwas_parameters = ctwas_parameters_munroST, colors = colors, by = "type", title = paste0("munroST - nonSNP %h2g:",round(total_nonSNPpve_munroST, digits = 4)))gridExtra::grid.arrange(pve_pie_by_type_woST, pve_pie_by_type_susieST,pve_pie_by_type_munroST,
ncol = 3, top = paste0(trait,"-thin",thin,"-",var_struc)) 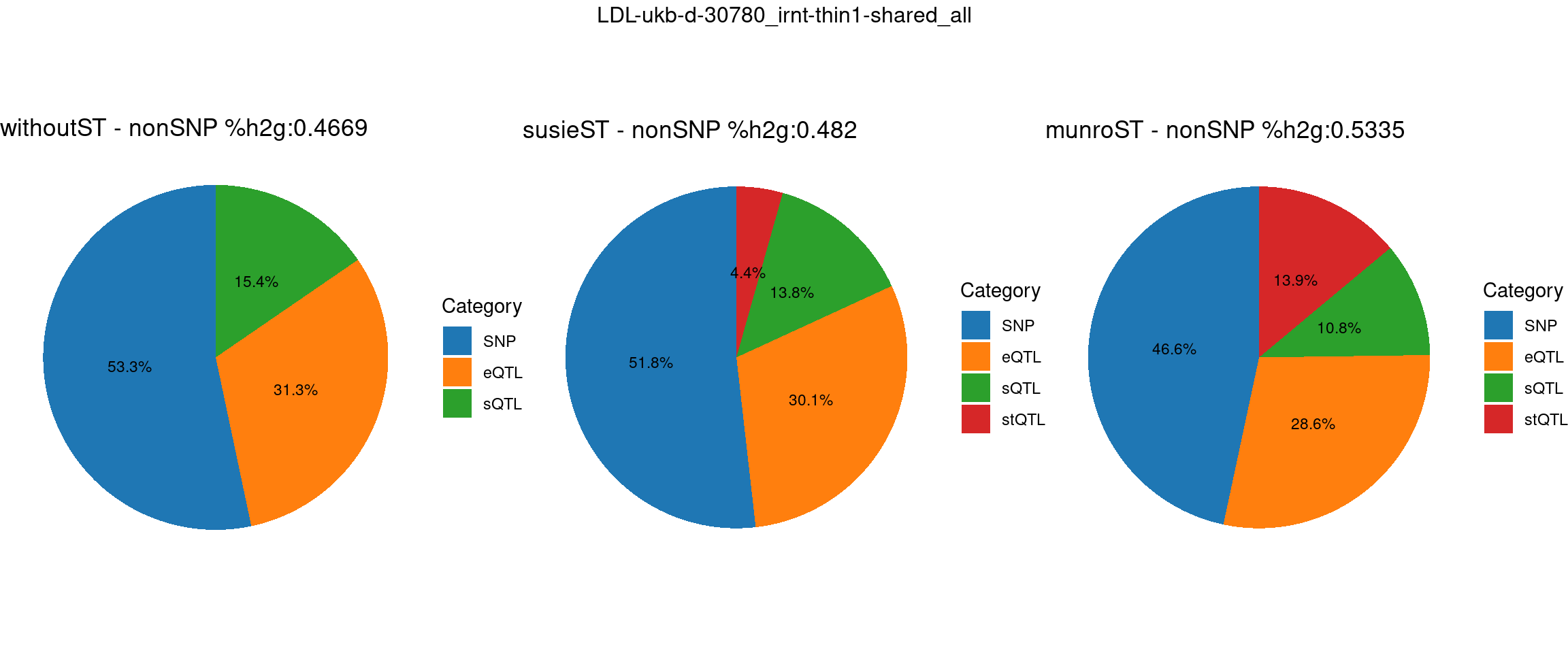
Setting: shared_type, thin = 0.1
trait <- "LDL-ukb-d-30780_irnt"
thin <- 0.1
var_struc <- "shared_type"
gwas_n <- samplesize[trait]st <- "without_ST"
param_woST <- readRDS(paste0(folder_results_woST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_woST <- summarize_param(param_woST, gwas_n)
total_nonSNPpve_woST <- 1- ctwas_parameters_woST$prop_heritability["SNP"]
pve_pie_by_type_woST <- plot_piechart(ctwas_parameters = ctwas_parameters_woST, colors = colors, by = "type", title = paste0("withoutST - nonSNP %h2g:",round(total_nonSNPpve_woST, digits = 4)))st <- "with_susieST"
param_susieST <- readRDS(paste0(folder_results_susieST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_susieST <- summarize_param(param_susieST, gwas_n)
total_nonSNPpve_susieST <- 1- ctwas_parameters_susieST$prop_heritability["SNP"]
pve_pie_by_type_susieST <- plot_piechart(ctwas_parameters = ctwas_parameters_susieST, colors = colors, by = "type", title = paste0("susieST - nonSNP %h2g:",round(total_nonSNPpve_susieST, digits = 4)))st <- "with_ST"
param_munroST <- readRDS(paste0(folder_results_munroST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
total_nonSNPpve_munroST <- 1- ctwas_parameters_munroST$prop_heritability["SNP"]
pve_pie_by_type_munroST <- plot_piechart(ctwas_parameters = ctwas_parameters_munroST, colors = colors, by = "type", title = paste0("munroST - nonSNP %h2g:",round(total_nonSNPpve_munroST, digits = 4)))gridExtra::grid.arrange(pve_pie_by_type_woST, pve_pie_by_type_susieST,pve_pie_by_type_munroST,
ncol = 3, top = paste0(trait,"-thin",thin,"-",var_struc)) 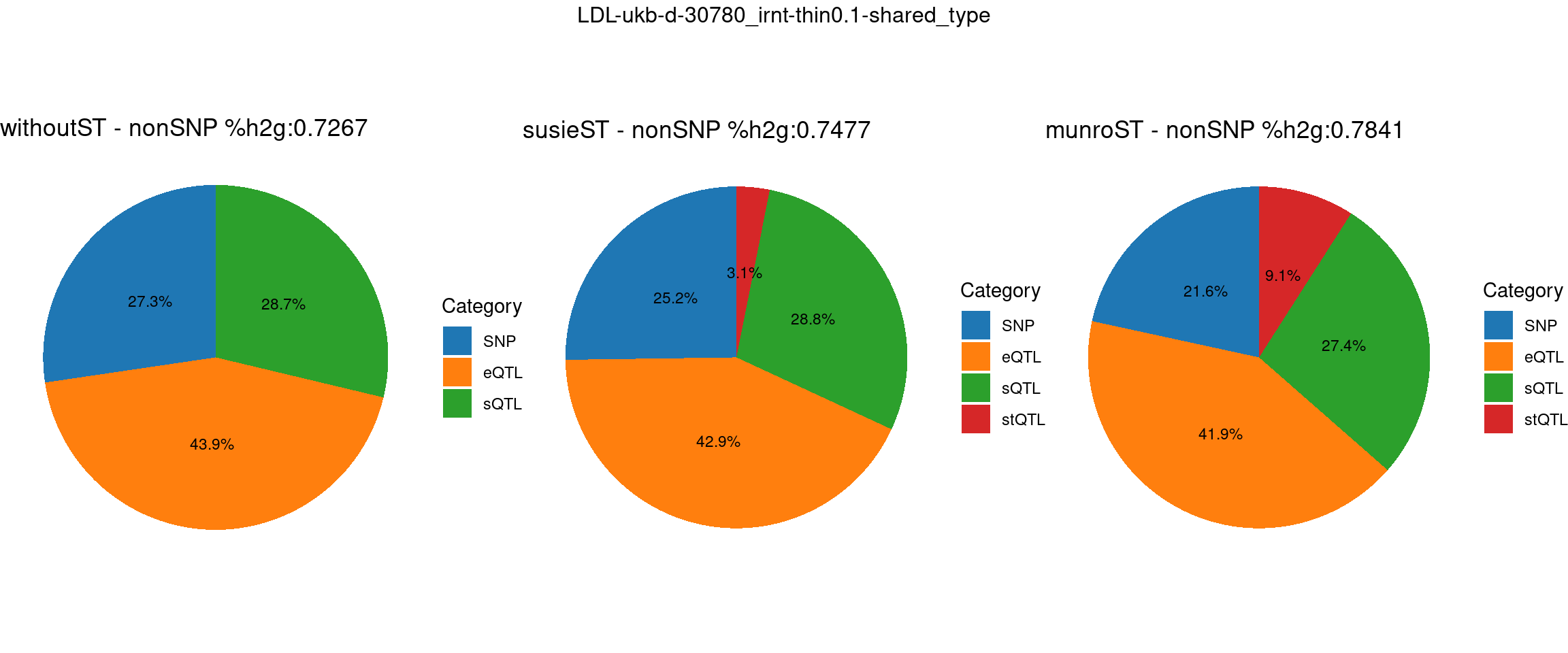
Setting: shared_type, thin = 0.5
trait <- "LDL-ukb-d-30780_irnt"
thin <- 0.5
var_struc <- "shared_type"
gwas_n <- samplesize[trait]st <- "without_ST"
param_woST <- readRDS(paste0(folder_results_woST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_woST <- summarize_param(param_woST, gwas_n)
total_nonSNPpve_woST <- 1- ctwas_parameters_woST$prop_heritability["SNP"]
pve_pie_by_type_woST <- plot_piechart(ctwas_parameters = ctwas_parameters_woST, colors = colors, by = "type", title = paste0("withoutST - nonSNP %h2g:",round(total_nonSNPpve_woST, digits = 4)))st <- "with_susieST"
param_susieST <- readRDS(paste0(folder_results_susieST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_susieST <- summarize_param(param_susieST, gwas_n)
total_nonSNPpve_susieST <- 1- ctwas_parameters_susieST$prop_heritability["SNP"]
pve_pie_by_type_susieST <- plot_piechart(ctwas_parameters = ctwas_parameters_susieST, colors = colors, by = "type", title = paste0("susieST - nonSNP %h2g:",round(total_nonSNPpve_susieST, digits = 4)))st <- "with_ST"
param_munroST <- readRDS(paste0(folder_results_munroST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
total_nonSNPpve_munroST <- 1- ctwas_parameters_munroST$prop_heritability["SNP"]
pve_pie_by_type_munroST <- plot_piechart(ctwas_parameters = ctwas_parameters_munroST, colors = colors, by = "type", title = paste0("munroST - nonSNP %h2g:",round(total_nonSNPpve_munroST, digits = 4)))gridExtra::grid.arrange(pve_pie_by_type_woST, pve_pie_by_type_susieST,pve_pie_by_type_munroST,
ncol = 3, top = paste0(trait,"-thin",thin,"-",var_struc)) 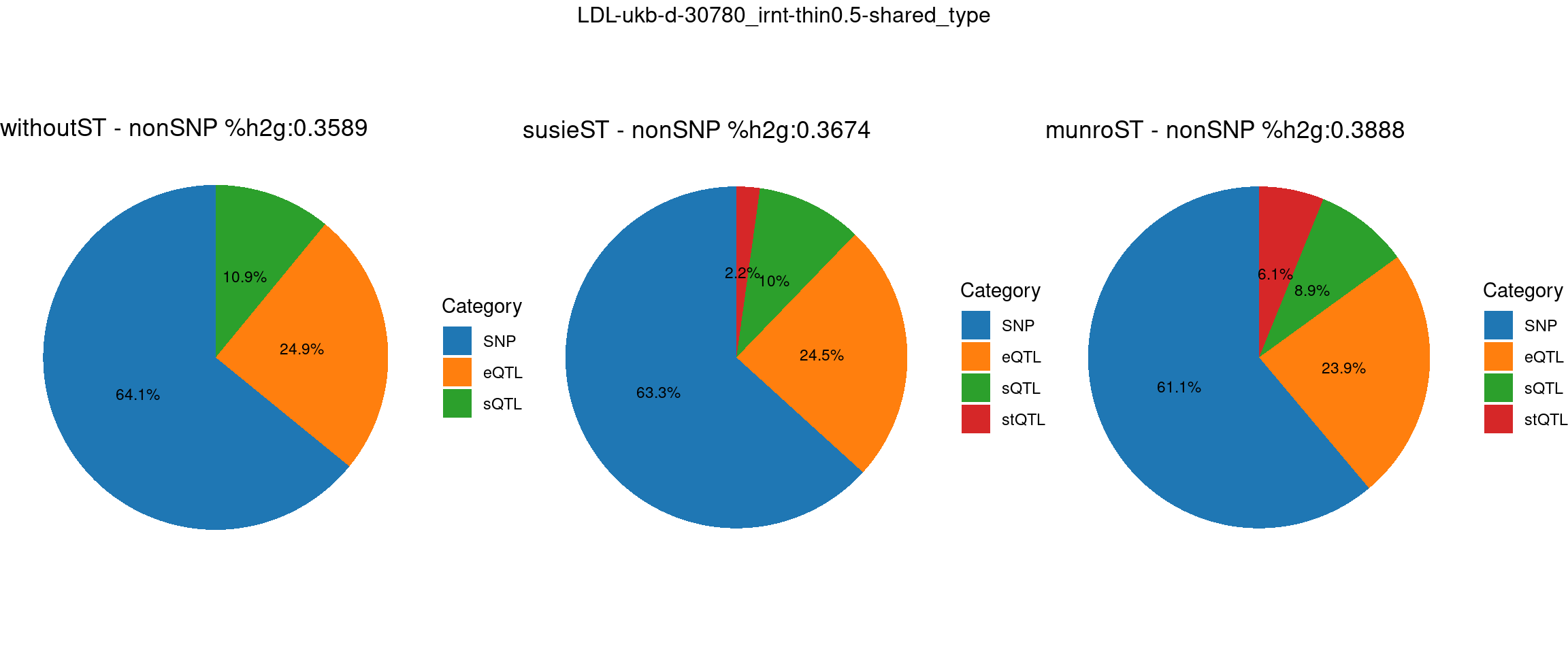
Setting: shared_type, thin = 1
trait <- "LDL-ukb-d-30780_irnt"
thin <- 1
var_struc <- "shared_type"
gwas_n <- samplesize[trait]st <- "without_ST"
param_woST <- readRDS(paste0(folder_results_woST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_woST <- summarize_param(param_woST, gwas_n)
total_nonSNPpve_woST <- 1- ctwas_parameters_woST$prop_heritability["SNP"]
pve_pie_by_type_woST <- plot_piechart(ctwas_parameters = ctwas_parameters_woST, colors = colors, by = "type", title = paste0("withoutST - nonSNP %h2g:",round(total_nonSNPpve_woST, digits = 4)))st <- "with_susieST"
param_susieST <- readRDS(paste0(folder_results_susieST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_susieST <- summarize_param(param_susieST, gwas_n)
total_nonSNPpve_susieST <- 1- ctwas_parameters_susieST$prop_heritability["SNP"]
pve_pie_by_type_susieST <- plot_piechart(ctwas_parameters = ctwas_parameters_susieST, colors = colors, by = "type", title = paste0("susieST - nonSNP %h2g:",round(total_nonSNPpve_susieST, digits = 4)))st <- "with_ST"
param_munroST <- readRDS(paste0(folder_results_munroST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
total_nonSNPpve_munroST <- 1- ctwas_parameters_munroST$prop_heritability["SNP"]
pve_pie_by_type_munroST <- plot_piechart(ctwas_parameters = ctwas_parameters_munroST, colors = colors, by = "type", title = paste0("munroST - nonSNP %h2g:",round(total_nonSNPpve_munroST, digits = 4)))gridExtra::grid.arrange(pve_pie_by_type_woST, pve_pie_by_type_susieST,pve_pie_by_type_munroST,
ncol = 3, top = paste0(trait,"-thin",thin,"-",var_struc)) 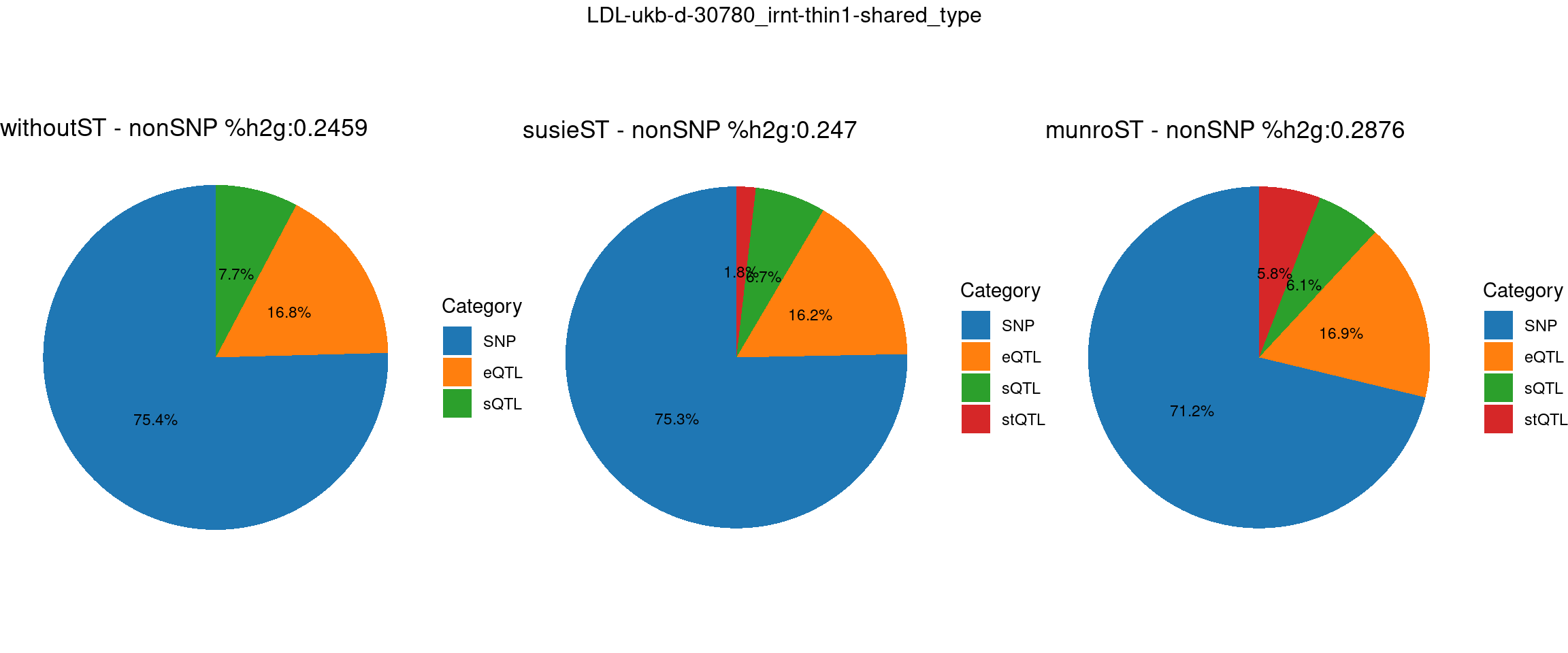
T2D-panukb
Setting: shared_all, thin = 0.1
trait <- "T2D-panukb"
thin <- 0.1
var_struc <- "shared_all"
gwas_n <- samplesize[trait]st <- "without_ST"
param_woST <- readRDS(paste0(folder_results_woST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_woST <- summarize_param(param_woST, gwas_n)
total_nonSNPpve_woST <- 1- ctwas_parameters_woST$prop_heritability["SNP"]
pve_pie_by_type_woST <- plot_piechart(ctwas_parameters = ctwas_parameters_woST, colors = colors, by = "type", title = paste0("withoutST - nonSNP %h2g:",round(total_nonSNPpve_woST, digits = 4)))st <- "with_susieST"
param_susieST <- readRDS(paste0(folder_results_susieST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_susieST <- summarize_param(param_susieST, gwas_n)
total_nonSNPpve_susieST <- 1- ctwas_parameters_susieST$prop_heritability["SNP"]
pve_pie_by_type_susieST <- plot_piechart(ctwas_parameters = ctwas_parameters_susieST, colors = colors, by = "type", title = paste0("susieST - nonSNP %h2g:",round(total_nonSNPpve_susieST, digits = 4)))st <- "with_ST"
param_munroST <- readRDS(paste0(folder_results_munroST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
total_nonSNPpve_munroST <- 1- ctwas_parameters_munroST$prop_heritability["SNP"]
pve_pie_by_type_munroST <- plot_piechart(ctwas_parameters = ctwas_parameters_munroST, colors = colors, by = "type", title = paste0("munroST - nonSNP %h2g:",round(total_nonSNPpve_munroST, digits = 4)))gridExtra::grid.arrange(pve_pie_by_type_woST, pve_pie_by_type_susieST,pve_pie_by_type_munroST,
ncol = 3, top = paste0(trait,"-thin",thin,"-",var_struc)) 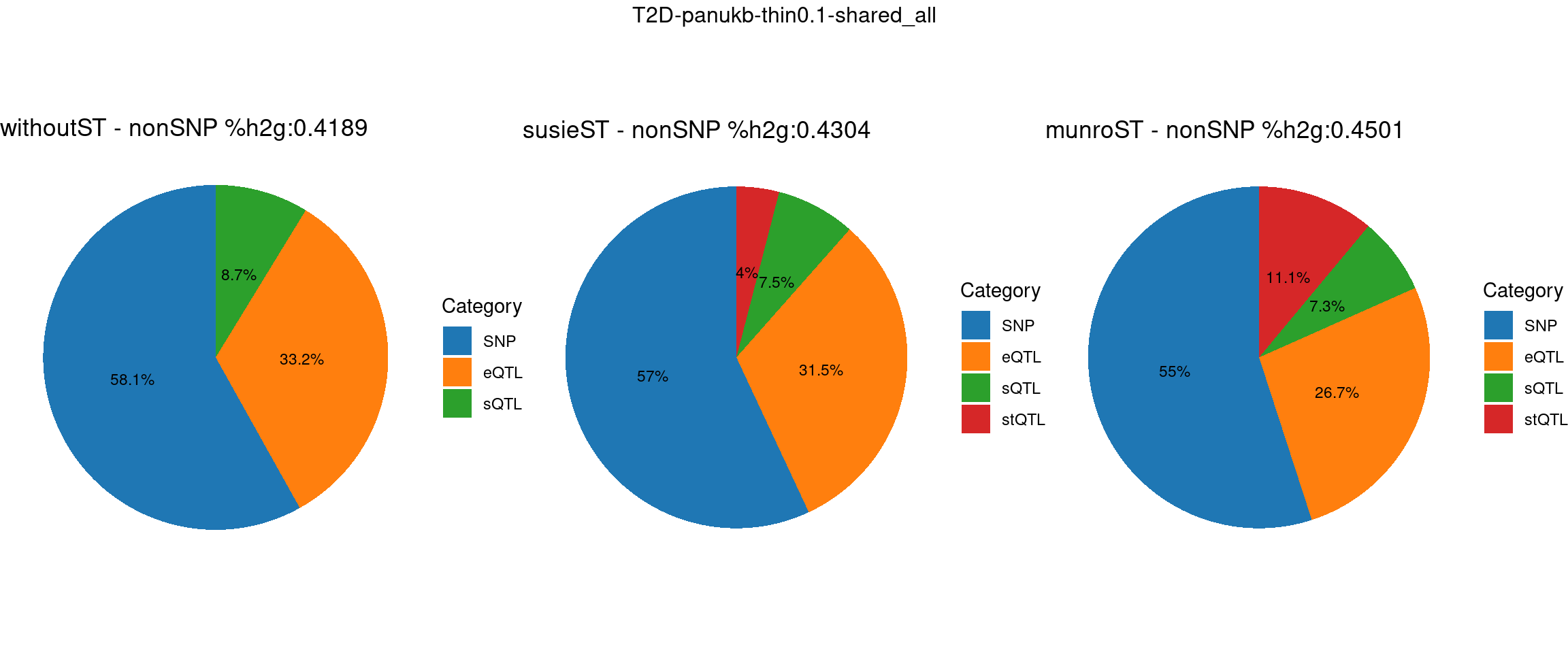
Setting: shared_all, thin = 0.5
trait <- "LDL-ukb-d-30780_irnt"
thin <- 0.5
var_struc <- "shared_all"
gwas_n <- samplesize[trait]st <- "without_ST"
param_woST <- readRDS(paste0(folder_results_woST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_woST <- summarize_param(param_woST, gwas_n)
total_nonSNPpve_woST <- 1- ctwas_parameters_woST$prop_heritability["SNP"]
pve_pie_by_type_woST <- plot_piechart(ctwas_parameters = ctwas_parameters_woST, colors = colors, by = "type", title = paste0("withoutST - nonSNP %h2g:",round(total_nonSNPpve_woST, digits = 4)))st <- "with_susieST"
param_susieST <- readRDS(paste0(folder_results_susieST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_susieST <- summarize_param(param_susieST, gwas_n)
total_nonSNPpve_susieST <- 1- ctwas_parameters_susieST$prop_heritability["SNP"]
pve_pie_by_type_susieST <- plot_piechart(ctwas_parameters = ctwas_parameters_susieST, colors = colors, by = "type", title = paste0("susieST - nonSNP %h2g:",round(total_nonSNPpve_susieST, digits = 4)))st <- "with_ST"
param_munroST <- readRDS(paste0(folder_results_munroST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
total_nonSNPpve_munroST <- 1- ctwas_parameters_munroST$prop_heritability["SNP"]
pve_pie_by_type_munroST <- plot_piechart(ctwas_parameters = ctwas_parameters_munroST, colors = colors, by = "type", title = paste0("munroST - nonSNP %h2g:",round(total_nonSNPpve_munroST, digits = 4)))gridExtra::grid.arrange(pve_pie_by_type_woST, pve_pie_by_type_susieST,pve_pie_by_type_munroST,
ncol = 3, top = paste0(trait,"-thin",thin,"-",var_struc)) 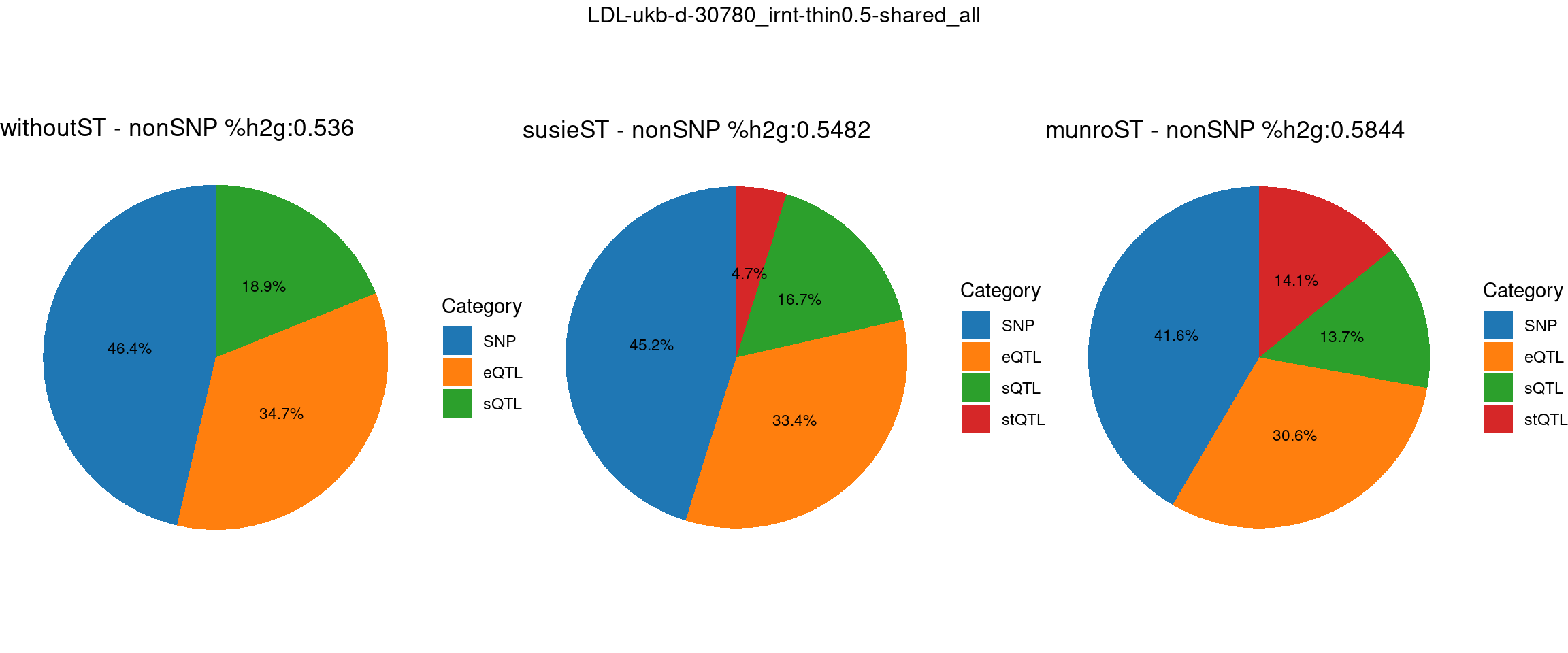
Setting: shared_all, thin = 1
trait <- "LDL-ukb-d-30780_irnt"
thin <- 1
var_struc <- "shared_all"
gwas_n <- samplesize[trait]st <- "without_ST"
param_woST <- readRDS(paste0(folder_results_woST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_woST <- summarize_param(param_woST, gwas_n)
total_nonSNPpve_woST <- 1- ctwas_parameters_woST$prop_heritability["SNP"]
pve_pie_by_type_woST <- plot_piechart(ctwas_parameters = ctwas_parameters_woST, colors = colors, by = "type", title = paste0("withoutST - nonSNP %h2g:",round(total_nonSNPpve_woST, digits = 4)))st <- "with_susieST"
param_susieST <- readRDS(paste0(folder_results_susieST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_susieST <- summarize_param(param_susieST, gwas_n)
total_nonSNPpve_susieST <- 1- ctwas_parameters_susieST$prop_heritability["SNP"]
pve_pie_by_type_susieST <- plot_piechart(ctwas_parameters = ctwas_parameters_susieST, colors = colors, by = "type", title = paste0("susieST - nonSNP %h2g:",round(total_nonSNPpve_susieST, digits = 4)))st <- "with_ST"
param_munroST <- readRDS(paste0(folder_results_munroST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
total_nonSNPpve_munroST <- 1- ctwas_parameters_munroST$prop_heritability["SNP"]
pve_pie_by_type_munroST <- plot_piechart(ctwas_parameters = ctwas_parameters_munroST, colors = colors, by = "type", title = paste0("munroST - nonSNP %h2g:",round(total_nonSNPpve_munroST, digits = 4)))gridExtra::grid.arrange(pve_pie_by_type_woST, pve_pie_by_type_susieST,pve_pie_by_type_munroST,
ncol = 3, top = paste0(trait,"-thin",thin,"-",var_struc)) 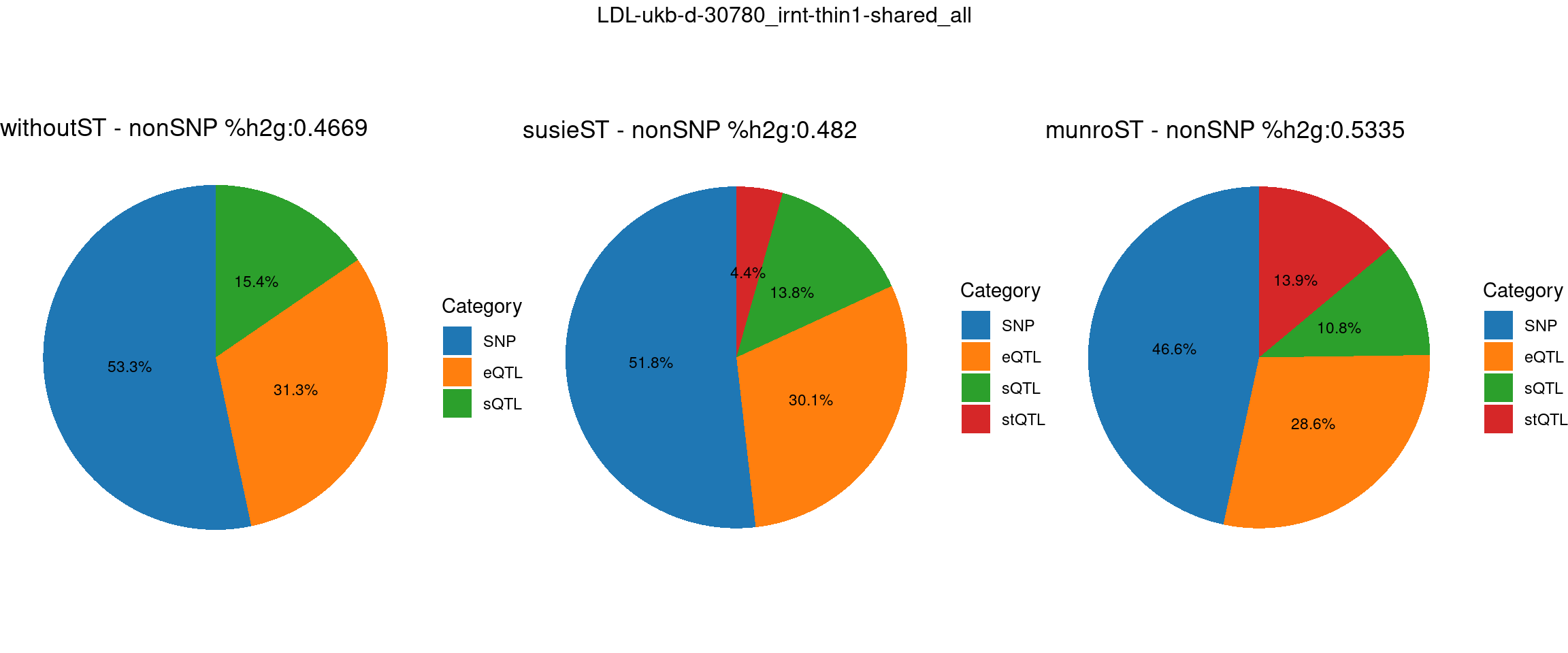
Setting: shared_type, thin = 0.1
trait <- "LDL-ukb-d-30780_irnt"
thin <- 0.1
var_struc <- "shared_type"
gwas_n <- samplesize[trait]st <- "without_ST"
param_woST <- readRDS(paste0(folder_results_woST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_woST <- summarize_param(param_woST, gwas_n)
total_nonSNPpve_woST <- 1- ctwas_parameters_woST$prop_heritability["SNP"]
pve_pie_by_type_woST <- plot_piechart(ctwas_parameters = ctwas_parameters_woST, colors = colors, by = "type", title = paste0("withoutST - nonSNP %h2g:",round(total_nonSNPpve_woST, digits = 4)))st <- "with_susieST"
param_susieST <- readRDS(paste0(folder_results_susieST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_susieST <- summarize_param(param_susieST, gwas_n)
total_nonSNPpve_susieST <- 1- ctwas_parameters_susieST$prop_heritability["SNP"]
pve_pie_by_type_susieST <- plot_piechart(ctwas_parameters = ctwas_parameters_susieST, colors = colors, by = "type", title = paste0("susieST - nonSNP %h2g:",round(total_nonSNPpve_susieST, digits = 4)))st <- "with_ST"
param_munroST <- readRDS(paste0(folder_results_munroST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
total_nonSNPpve_munroST <- 1- ctwas_parameters_munroST$prop_heritability["SNP"]
pve_pie_by_type_munroST <- plot_piechart(ctwas_parameters = ctwas_parameters_munroST, colors = colors, by = "type", title = paste0("munroST - nonSNP %h2g:",round(total_nonSNPpve_munroST, digits = 4)))gridExtra::grid.arrange(pve_pie_by_type_woST, pve_pie_by_type_susieST,pve_pie_by_type_munroST,
ncol = 3, top = paste0(trait,"-thin",thin,"-",var_struc)) 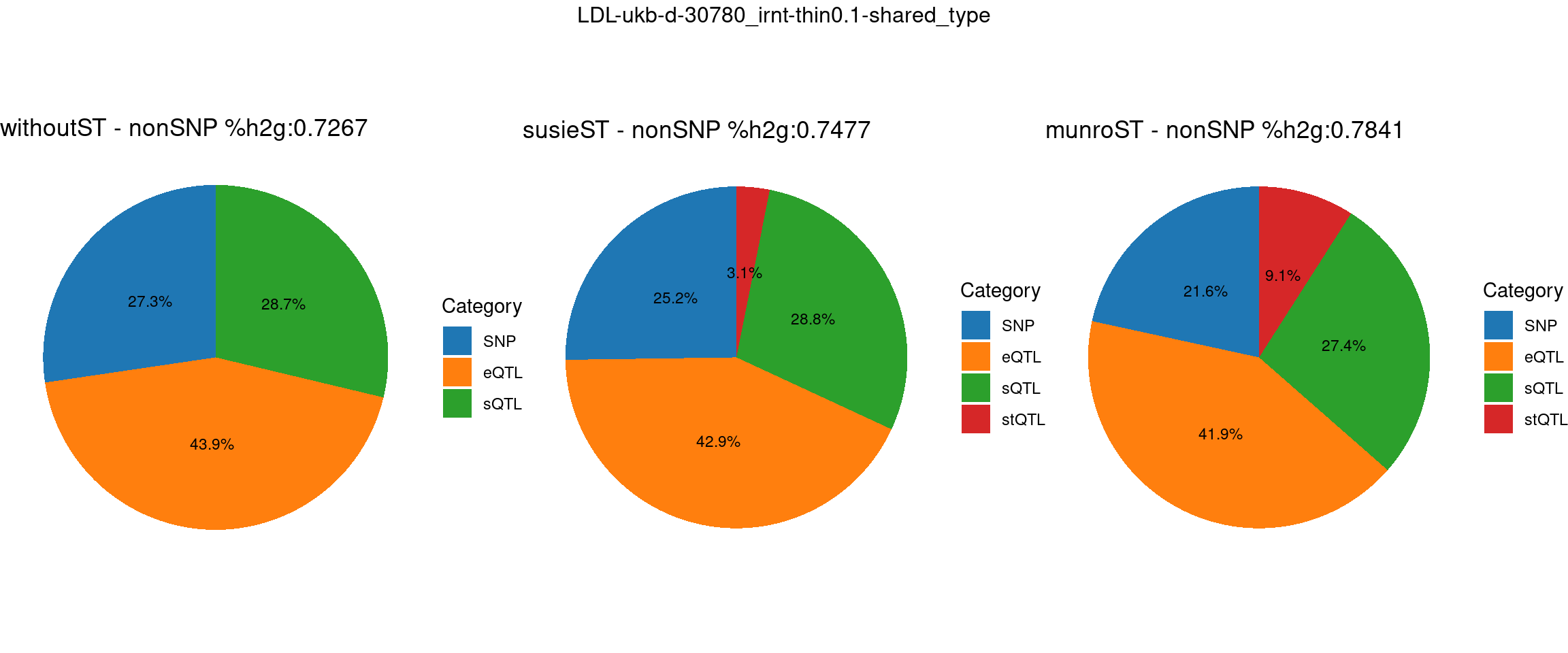
Setting: shared_type, thin = 0.5
trait <- "LDL-ukb-d-30780_irnt"
thin <- 0.5
var_struc <- "shared_type"
gwas_n <- samplesize[trait]st <- "without_ST"
param_woST <- readRDS(paste0(folder_results_woST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_woST <- summarize_param(param_woST, gwas_n)
total_nonSNPpve_woST <- 1- ctwas_parameters_woST$prop_heritability["SNP"]
pve_pie_by_type_woST <- plot_piechart(ctwas_parameters = ctwas_parameters_woST, colors = colors, by = "type", title = paste0("withoutST - nonSNP %h2g:",round(total_nonSNPpve_woST, digits = 4)))st <- "with_susieST"
param_susieST <- readRDS(paste0(folder_results_susieST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_susieST <- summarize_param(param_susieST, gwas_n)
total_nonSNPpve_susieST <- 1- ctwas_parameters_susieST$prop_heritability["SNP"]
pve_pie_by_type_susieST <- plot_piechart(ctwas_parameters = ctwas_parameters_susieST, colors = colors, by = "type", title = paste0("susieST - nonSNP %h2g:",round(total_nonSNPpve_susieST, digits = 4)))st <- "with_ST"
param_munroST <- readRDS(paste0(folder_results_munroST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
total_nonSNPpve_munroST <- 1- ctwas_parameters_munroST$prop_heritability["SNP"]
pve_pie_by_type_munroST <- plot_piechart(ctwas_parameters = ctwas_parameters_munroST, colors = colors, by = "type", title = paste0("munroST - nonSNP %h2g:",round(total_nonSNPpve_munroST, digits = 4)))gridExtra::grid.arrange(pve_pie_by_type_woST, pve_pie_by_type_susieST,pve_pie_by_type_munroST,
ncol = 3, top = paste0(trait,"-thin",thin,"-",var_struc)) 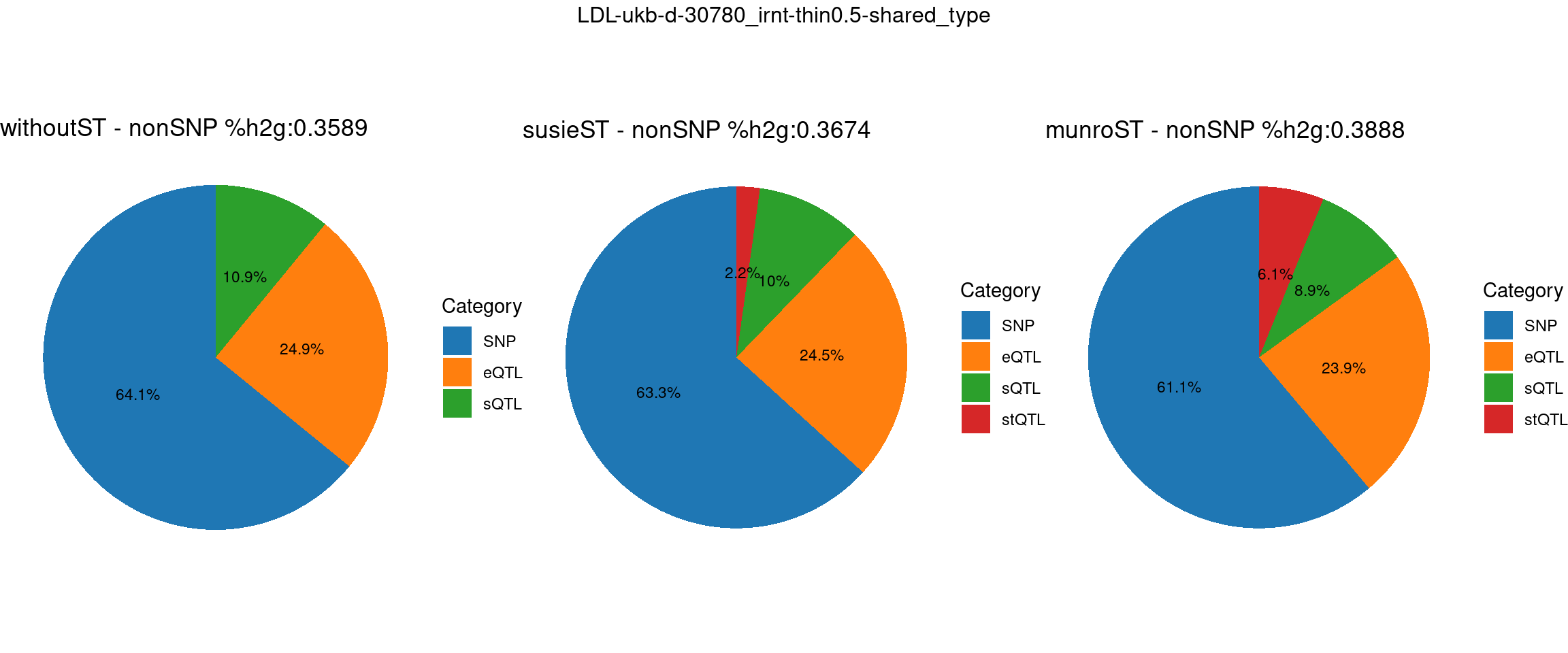
Setting: shared_type, thin = 1
trait <- "LDL-ukb-d-30780_irnt"
thin <- 1
var_struc <- "shared_type"
gwas_n <- samplesize[trait]st <- "without_ST"
param_woST <- readRDS(paste0(folder_results_woST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_woST <- summarize_param(param_woST, gwas_n)
total_nonSNPpve_woST <- 1- ctwas_parameters_woST$prop_heritability["SNP"]
pve_pie_by_type_woST <- plot_piechart(ctwas_parameters = ctwas_parameters_woST, colors = colors, by = "type", title = paste0("withoutST - nonSNP %h2g:",round(total_nonSNPpve_woST, digits = 4)))st <- "with_susieST"
param_susieST <- readRDS(paste0(folder_results_susieST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_susieST <- summarize_param(param_susieST, gwas_n)
total_nonSNPpve_susieST <- 1- ctwas_parameters_susieST$prop_heritability["SNP"]
pve_pie_by_type_susieST <- plot_piechart(ctwas_parameters = ctwas_parameters_susieST, colors = colors, by = "type", title = paste0("susieST - nonSNP %h2g:",round(total_nonSNPpve_susieST, digits = 4)))st <- "with_ST"
param_munroST <- readRDS(paste0(folder_results_munroST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
total_nonSNPpve_munroST <- 1- ctwas_parameters_munroST$prop_heritability["SNP"]
pve_pie_by_type_munroST <- plot_piechart(ctwas_parameters = ctwas_parameters_munroST, colors = colors, by = "type", title = paste0("munroST - nonSNP %h2g:",round(total_nonSNPpve_munroST, digits = 4)))gridExtra::grid.arrange(pve_pie_by_type_woST, pve_pie_by_type_susieST,pve_pie_by_type_munroST,
ncol = 3, top = paste0(trait,"-thin",thin,"-",var_struc)) 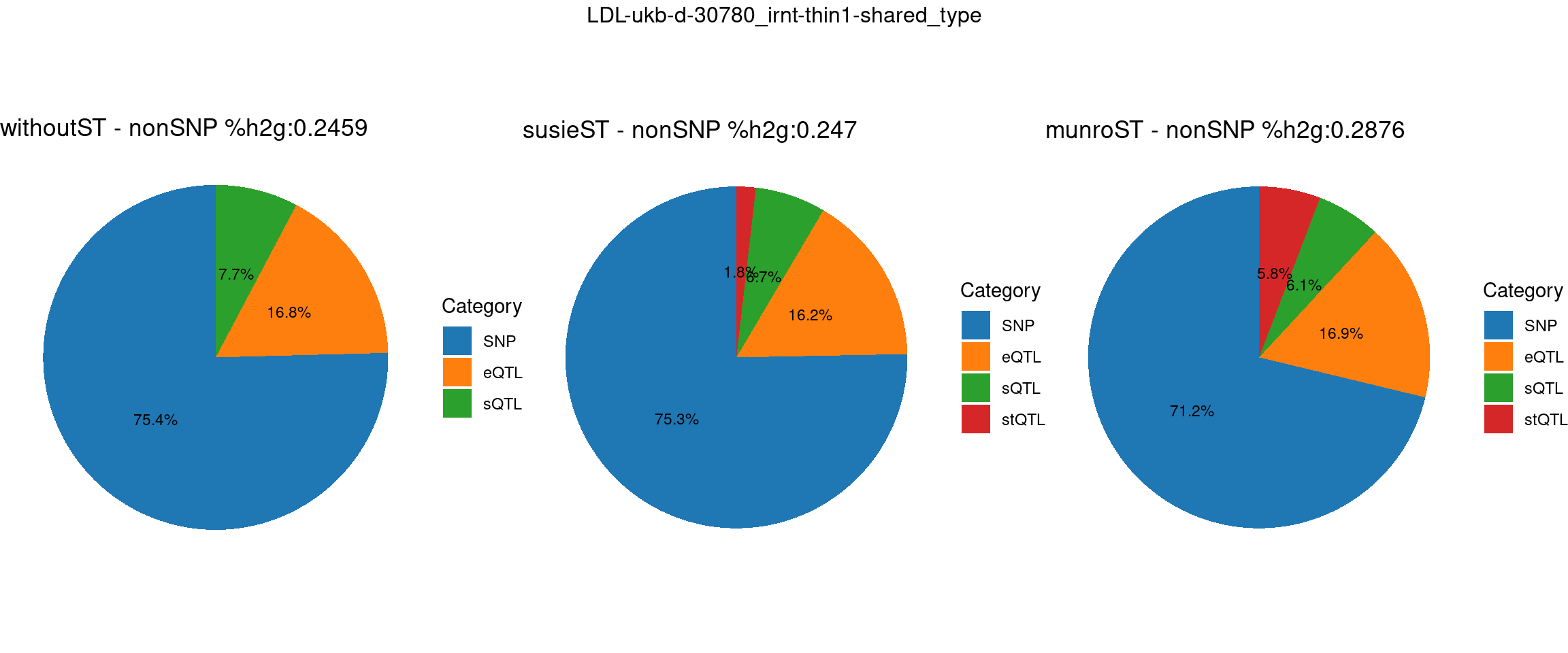
BMI-panukb
Setting: shared_all, thin = 0.1
trait <- "BMI-panukb"
thin <- 0.1
var_struc <- "shared_all"
gwas_n <- samplesize[trait]st <- "without_ST"
param_woST <- readRDS(paste0(folder_results_woST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_woST <- summarize_param(param_woST, gwas_n)
total_nonSNPpve_woST <- 1- ctwas_parameters_woST$prop_heritability["SNP"]
pve_pie_by_type_woST <- plot_piechart(ctwas_parameters = ctwas_parameters_woST, colors = colors, by = "type", title = paste0("withoutST - nonSNP %h2g:",round(total_nonSNPpve_woST, digits = 4)))st <- "with_susieST"
param_susieST <- readRDS(paste0(folder_results_susieST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_susieST <- summarize_param(param_susieST, gwas_n)
total_nonSNPpve_susieST <- 1- ctwas_parameters_susieST$prop_heritability["SNP"]
pve_pie_by_type_susieST <- plot_piechart(ctwas_parameters = ctwas_parameters_susieST, colors = colors, by = "type", title = paste0("susieST - nonSNP %h2g:",round(total_nonSNPpve_susieST, digits = 4)))st <- "with_ST"
param_munroST <- readRDS(paste0(folder_results_munroST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
total_nonSNPpve_munroST <- 1- ctwas_parameters_munroST$prop_heritability["SNP"]
pve_pie_by_type_munroST <- plot_piechart(ctwas_parameters = ctwas_parameters_munroST, colors = colors, by = "type", title = paste0("munroST - nonSNP %h2g:",round(total_nonSNPpve_munroST, digits = 4)))gridExtra::grid.arrange(pve_pie_by_type_woST, pve_pie_by_type_susieST,pve_pie_by_type_munroST,
ncol = 3, top = paste0(trait,"-thin",thin,"-",var_struc)) 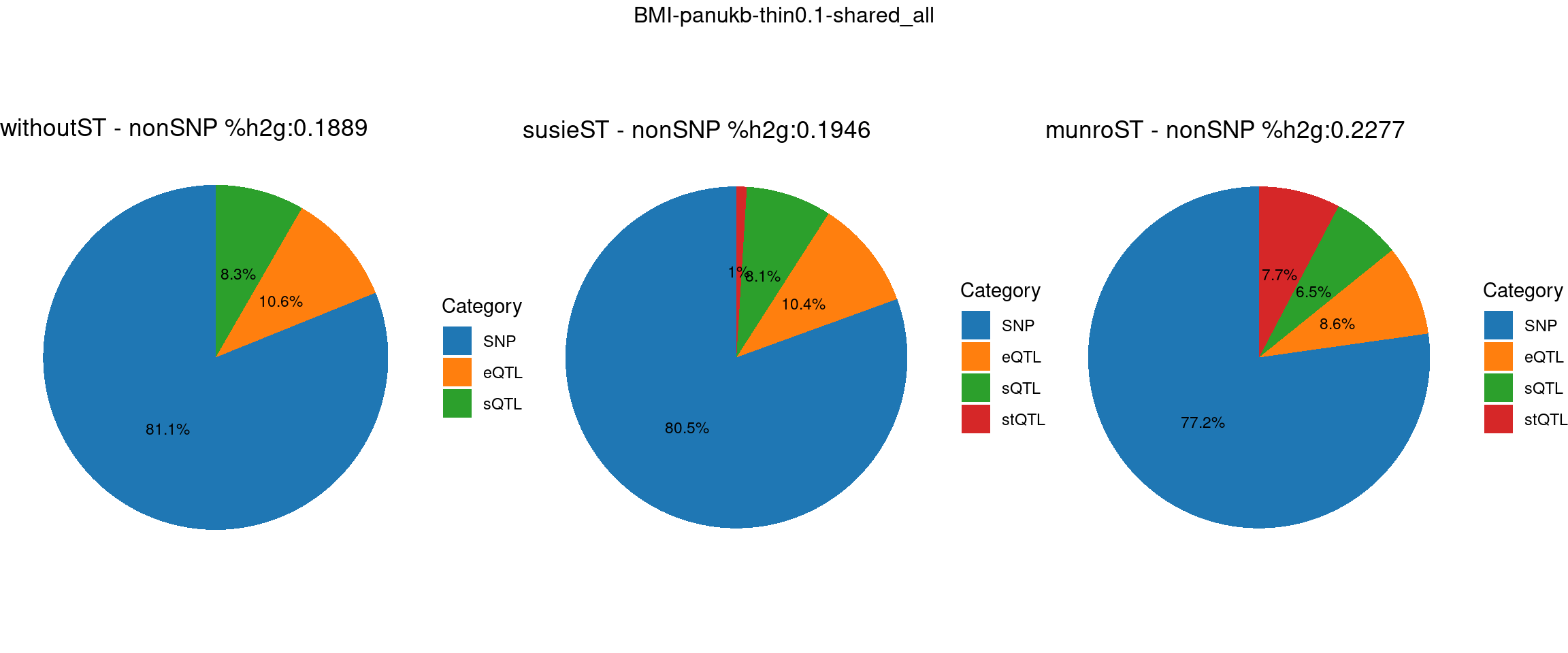
Setting: shared_all, thin = 0.5
trait <- "LDL-ukb-d-30780_irnt"
thin <- 0.5
var_struc <- "shared_all"
gwas_n <- samplesize[trait]st <- "without_ST"
param_woST <- readRDS(paste0(folder_results_woST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_woST <- summarize_param(param_woST, gwas_n)
total_nonSNPpve_woST <- 1- ctwas_parameters_woST$prop_heritability["SNP"]
pve_pie_by_type_woST <- plot_piechart(ctwas_parameters = ctwas_parameters_woST, colors = colors, by = "type", title = paste0("withoutST - nonSNP %h2g:",round(total_nonSNPpve_woST, digits = 4)))st <- "with_susieST"
param_susieST <- readRDS(paste0(folder_results_susieST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_susieST <- summarize_param(param_susieST, gwas_n)
total_nonSNPpve_susieST <- 1- ctwas_parameters_susieST$prop_heritability["SNP"]
pve_pie_by_type_susieST <- plot_piechart(ctwas_parameters = ctwas_parameters_susieST, colors = colors, by = "type", title = paste0("susieST - nonSNP %h2g:",round(total_nonSNPpve_susieST, digits = 4)))st <- "with_ST"
param_munroST <- readRDS(paste0(folder_results_munroST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
total_nonSNPpve_munroST <- 1- ctwas_parameters_munroST$prop_heritability["SNP"]
pve_pie_by_type_munroST <- plot_piechart(ctwas_parameters = ctwas_parameters_munroST, colors = colors, by = "type", title = paste0("munroST - nonSNP %h2g:",round(total_nonSNPpve_munroST, digits = 4)))gridExtra::grid.arrange(pve_pie_by_type_woST, pve_pie_by_type_susieST,pve_pie_by_type_munroST,
ncol = 3, top = paste0(trait,"-thin",thin,"-",var_struc)) 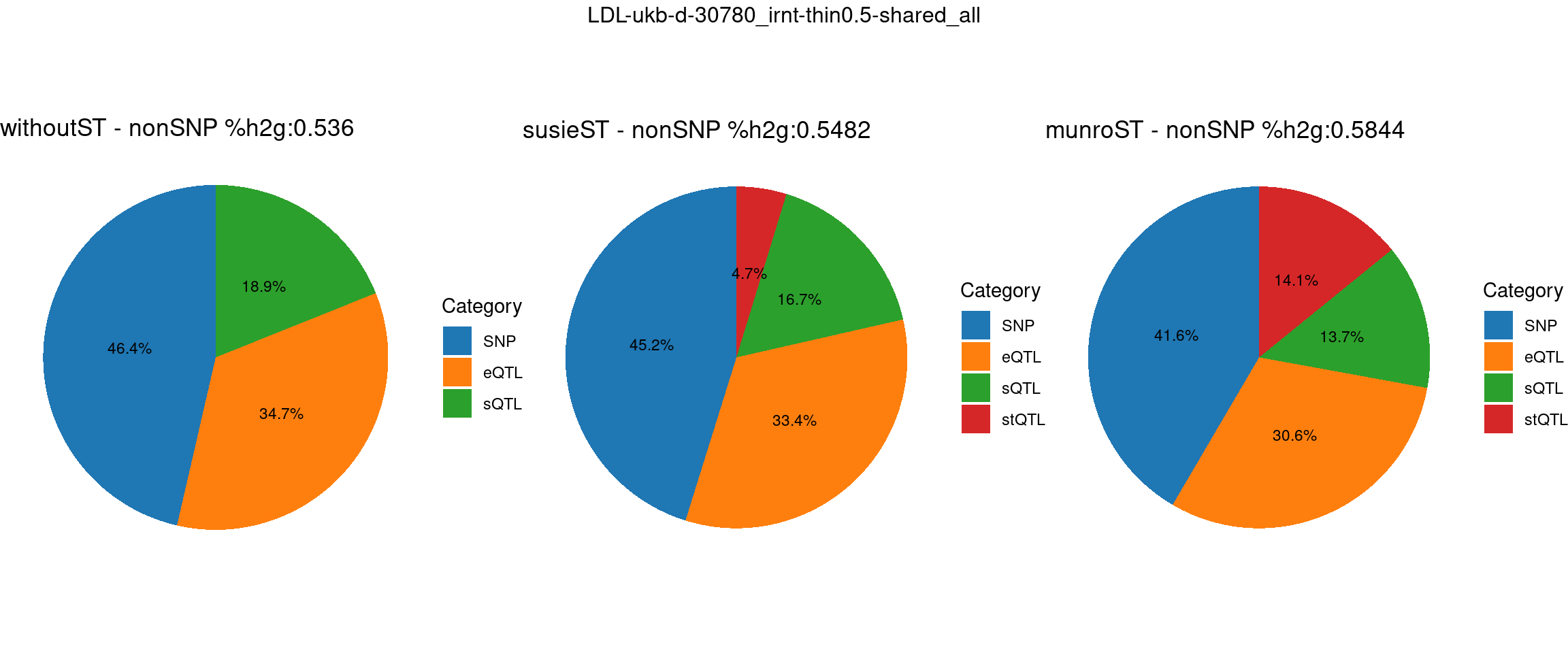
Setting: shared_all, thin = 1
trait <- "LDL-ukb-d-30780_irnt"
thin <- 1
var_struc <- "shared_all"
gwas_n <- samplesize[trait]st <- "without_ST"
param_woST <- readRDS(paste0(folder_results_woST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_woST <- summarize_param(param_woST, gwas_n)
total_nonSNPpve_woST <- 1- ctwas_parameters_woST$prop_heritability["SNP"]
pve_pie_by_type_woST <- plot_piechart(ctwas_parameters = ctwas_parameters_woST, colors = colors, by = "type", title = paste0("withoutST - nonSNP %h2g:",round(total_nonSNPpve_woST, digits = 4)))st <- "with_susieST"
param_susieST <- readRDS(paste0(folder_results_susieST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_susieST <- summarize_param(param_susieST, gwas_n)
total_nonSNPpve_susieST <- 1- ctwas_parameters_susieST$prop_heritability["SNP"]
pve_pie_by_type_susieST <- plot_piechart(ctwas_parameters = ctwas_parameters_susieST, colors = colors, by = "type", title = paste0("susieST - nonSNP %h2g:",round(total_nonSNPpve_susieST, digits = 4)))st <- "with_ST"
param_munroST <- readRDS(paste0(folder_results_munroST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
total_nonSNPpve_munroST <- 1- ctwas_parameters_munroST$prop_heritability["SNP"]
pve_pie_by_type_munroST <- plot_piechart(ctwas_parameters = ctwas_parameters_munroST, colors = colors, by = "type", title = paste0("munroST - nonSNP %h2g:",round(total_nonSNPpve_munroST, digits = 4)))gridExtra::grid.arrange(pve_pie_by_type_woST, pve_pie_by_type_susieST,pve_pie_by_type_munroST,
ncol = 3, top = paste0(trait,"-thin",thin,"-",var_struc)) 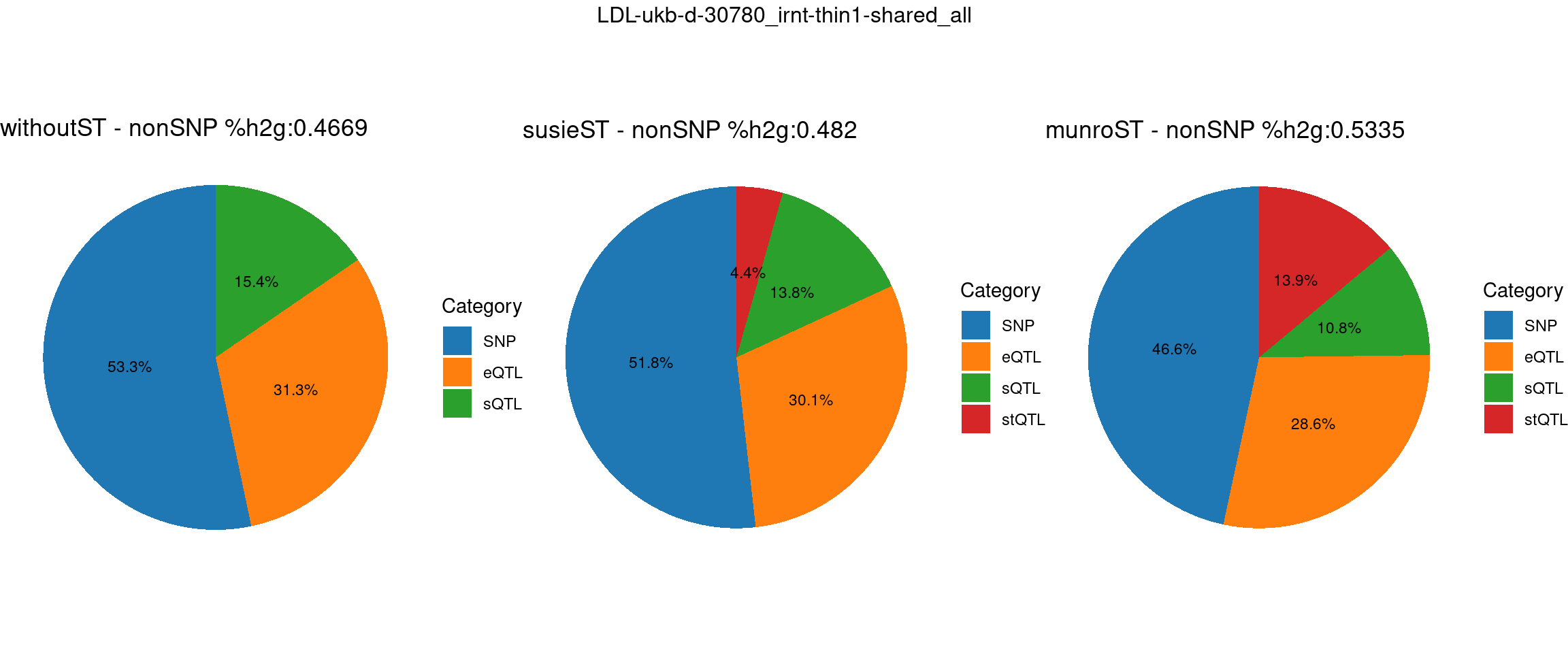
Setting: shared_type, thin = 0.1
trait <- "LDL-ukb-d-30780_irnt"
thin <- 0.1
var_struc <- "shared_type"
gwas_n <- samplesize[trait]st <- "without_ST"
param_woST <- readRDS(paste0(folder_results_woST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_woST <- summarize_param(param_woST, gwas_n)
total_nonSNPpve_woST <- 1- ctwas_parameters_woST$prop_heritability["SNP"]
pve_pie_by_type_woST <- plot_piechart(ctwas_parameters = ctwas_parameters_woST, colors = colors, by = "type", title = paste0("withoutST - nonSNP %h2g:",round(total_nonSNPpve_woST, digits = 4)))st <- "with_susieST"
param_susieST <- readRDS(paste0(folder_results_susieST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_susieST <- summarize_param(param_susieST, gwas_n)
total_nonSNPpve_susieST <- 1- ctwas_parameters_susieST$prop_heritability["SNP"]
pve_pie_by_type_susieST <- plot_piechart(ctwas_parameters = ctwas_parameters_susieST, colors = colors, by = "type", title = paste0("susieST - nonSNP %h2g:",round(total_nonSNPpve_susieST, digits = 4)))st <- "with_ST"
param_munroST <- readRDS(paste0(folder_results_munroST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
total_nonSNPpve_munroST <- 1- ctwas_parameters_munroST$prop_heritability["SNP"]
pve_pie_by_type_munroST <- plot_piechart(ctwas_parameters = ctwas_parameters_munroST, colors = colors, by = "type", title = paste0("munroST - nonSNP %h2g:",round(total_nonSNPpve_munroST, digits = 4)))gridExtra::grid.arrange(pve_pie_by_type_woST, pve_pie_by_type_susieST,pve_pie_by_type_munroST,
ncol = 3, top = paste0(trait,"-thin",thin,"-",var_struc)) 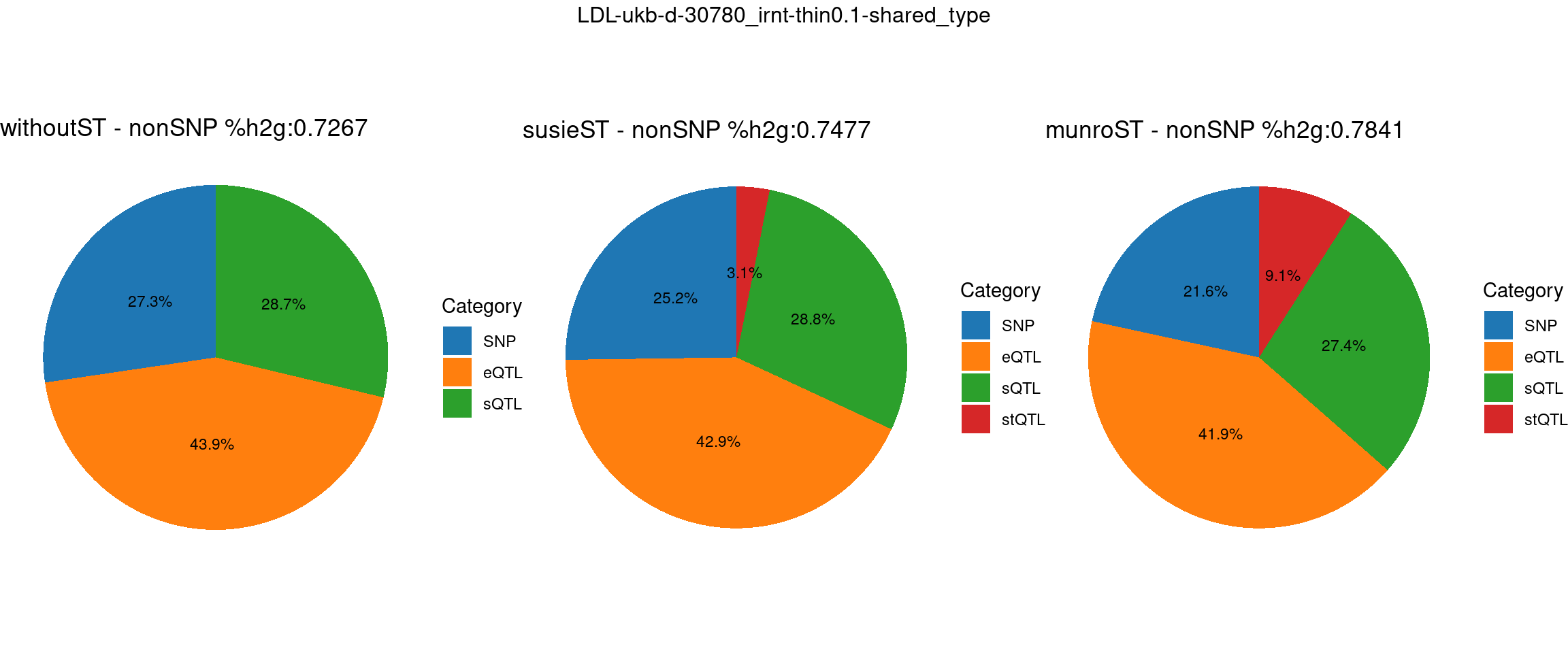
Setting: shared_type, thin = 0.5
trait <- "LDL-ukb-d-30780_irnt"
thin <- 0.5
var_struc <- "shared_type"
gwas_n <- samplesize[trait]st <- "without_ST"
param_woST <- readRDS(paste0(folder_results_woST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_woST <- summarize_param(param_woST, gwas_n)
total_nonSNPpve_woST <- 1- ctwas_parameters_woST$prop_heritability["SNP"]
pve_pie_by_type_woST <- plot_piechart(ctwas_parameters = ctwas_parameters_woST, colors = colors, by = "type", title = paste0("withoutST - nonSNP %h2g:",round(total_nonSNPpve_woST, digits = 4)))st <- "with_susieST"
param_susieST <- readRDS(paste0(folder_results_susieST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_susieST <- summarize_param(param_susieST, gwas_n)
total_nonSNPpve_susieST <- 1- ctwas_parameters_susieST$prop_heritability["SNP"]
pve_pie_by_type_susieST <- plot_piechart(ctwas_parameters = ctwas_parameters_susieST, colors = colors, by = "type", title = paste0("susieST - nonSNP %h2g:",round(total_nonSNPpve_susieST, digits = 4)))st <- "with_ST"
param_munroST <- readRDS(paste0(folder_results_munroST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
total_nonSNPpve_munroST <- 1- ctwas_parameters_munroST$prop_heritability["SNP"]
pve_pie_by_type_munroST <- plot_piechart(ctwas_parameters = ctwas_parameters_munroST, colors = colors, by = "type", title = paste0("munroST - nonSNP %h2g:",round(total_nonSNPpve_munroST, digits = 4)))gridExtra::grid.arrange(pve_pie_by_type_woST, pve_pie_by_type_susieST,pve_pie_by_type_munroST,
ncol = 3, top = paste0(trait,"-thin",thin,"-",var_struc)) 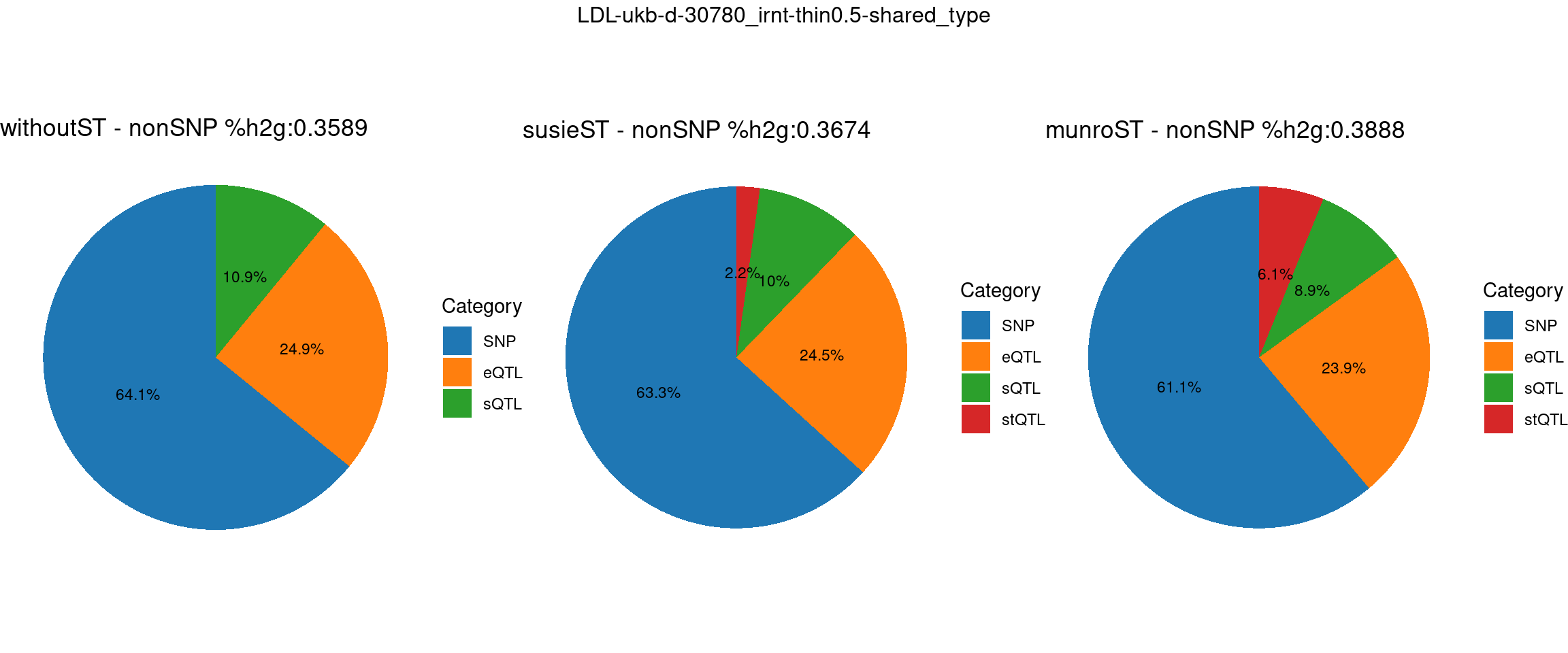
Setting: shared_type, thin = 1
trait <- "LDL-ukb-d-30780_irnt"
thin <- 1
var_struc <- "shared_type"
gwas_n <- samplesize[trait]st <- "without_ST"
param_woST <- readRDS(paste0(folder_results_woST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_woST <- summarize_param(param_woST, gwas_n)
total_nonSNPpve_woST <- 1- ctwas_parameters_woST$prop_heritability["SNP"]
pve_pie_by_type_woST <- plot_piechart(ctwas_parameters = ctwas_parameters_woST, colors = colors, by = "type", title = paste0("withoutST - nonSNP %h2g:",round(total_nonSNPpve_woST, digits = 4)))st <- "with_susieST"
param_susieST <- readRDS(paste0(folder_results_susieST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_susieST <- summarize_param(param_susieST, gwas_n)
total_nonSNPpve_susieST <- 1- ctwas_parameters_susieST$prop_heritability["SNP"]
pve_pie_by_type_susieST <- plot_piechart(ctwas_parameters = ctwas_parameters_susieST, colors = colors, by = "type", title = paste0("susieST - nonSNP %h2g:",round(total_nonSNPpve_susieST, digits = 4)))st <- "with_ST"
param_munroST <- readRDS(paste0(folder_results_munroST,"/",trait,"/",trait,".",st,".thin",thin,".",var_struc,".param.RDS"))
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
ctwas_parameters_munroST <- summarize_param(param_munroST, gwas_n)
total_nonSNPpve_munroST <- 1- ctwas_parameters_munroST$prop_heritability["SNP"]
pve_pie_by_type_munroST <- plot_piechart(ctwas_parameters = ctwas_parameters_munroST, colors = colors, by = "type", title = paste0("munroST - nonSNP %h2g:",round(total_nonSNPpve_munroST, digits = 4)))gridExtra::grid.arrange(pve_pie_by_type_woST, pve_pie_by_type_susieST,pve_pie_by_type_munroST,
ncol = 3, top = paste0(trait,"-thin",thin,"-",var_struc)) 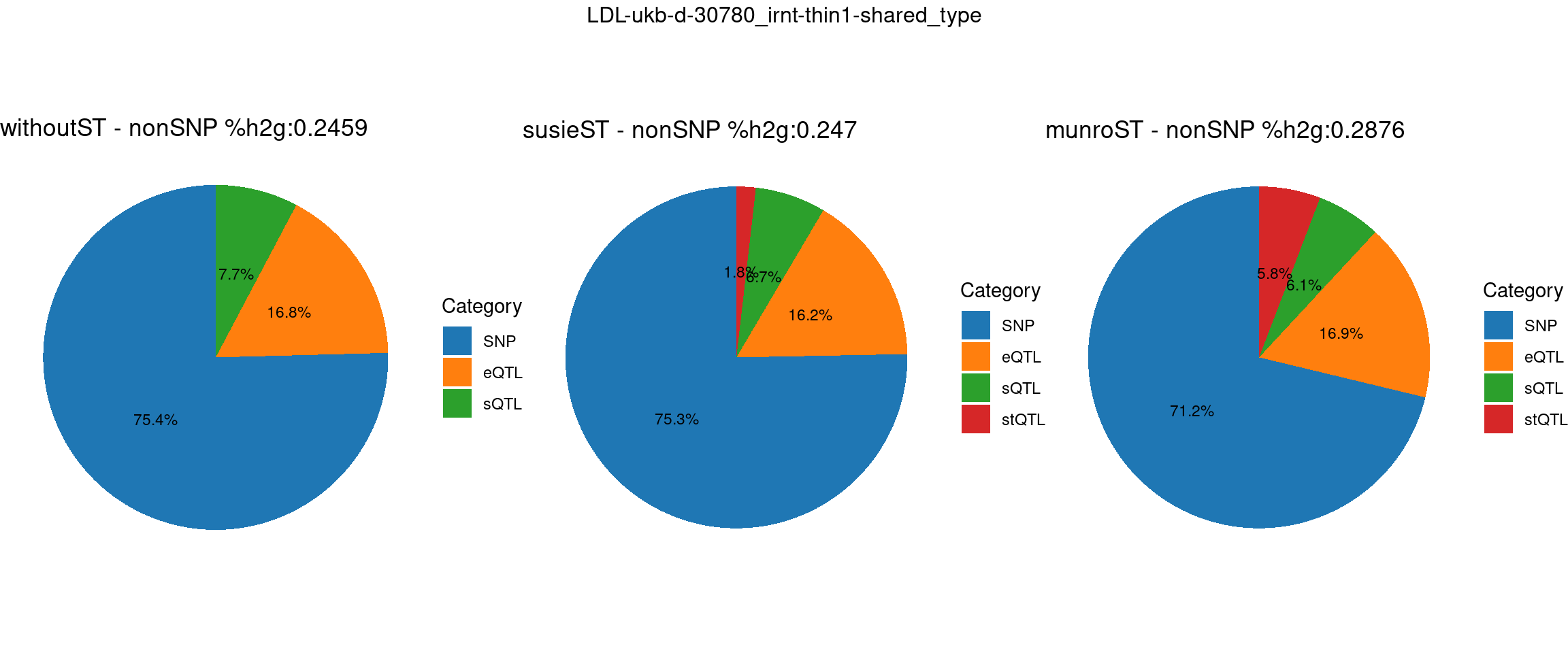
sessionInfo()R version 4.2.0 (2022-04-22)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
locale:
[1] C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] forcats_0.5.1 stringr_1.5.1 dplyr_1.1.4 purrr_1.0.2
[5] readr_2.1.2 tidyr_1.3.0 tibble_3.2.1 tidyverse_1.3.1
[9] ggplot2_3.5.1 ctwas_0.5.4.9000
loaded via a namespace (and not attached):
[1] colorspace_2.0-3 rjson_0.2.21
[3] ellipsis_0.3.2 rprojroot_2.0.3
[5] XVector_0.36.0 locuszoomr_0.2.1
[7] GenomicRanges_1.48.0 base64enc_0.1-3
[9] fs_1.5.2 rstudioapi_0.13
[11] farver_2.1.0 ggrepel_0.9.1
[13] bit64_4.0.5 lubridate_1.8.0
[15] AnnotationDbi_1.58.0 fansi_1.0.3
[17] xml2_1.3.3 codetools_0.2-18
[19] logging_0.10-108 cachem_1.0.6
[21] knitr_1.39 jsonlite_1.8.0
[23] workflowr_1.7.0 Rsamtools_2.12.0
[25] broom_0.8.0 dbplyr_2.1.1
[27] png_0.1-7 compiler_4.2.0
[29] httr_1.4.3 backports_1.4.1
[31] assertthat_0.2.1 Matrix_1.5-3
[33] fastmap_1.1.0 lazyeval_0.2.2
[35] cli_3.6.1 later_1.3.0
[37] htmltools_0.5.2 prettyunits_1.1.1
[39] tools_4.2.0 gtable_0.3.0
[41] glue_1.6.2 GenomeInfoDbData_1.2.8
[43] rappdirs_0.3.3 Rcpp_1.0.12
[45] Biobase_2.56.0 cellranger_1.1.0
[47] jquerylib_0.1.4 vctrs_0.6.5
[49] Biostrings_2.64.0 rtracklayer_1.56.0
[51] xfun_0.41 rvest_1.0.2
[53] irlba_2.3.5 lifecycle_1.0.4
[55] restfulr_0.0.14 ensembldb_2.20.2
[57] XML_3.99-0.14 zlibbioc_1.42.0
[59] zoo_1.8-10 scales_1.3.0
[61] gggrid_0.2-0 hms_1.1.1
[63] promises_1.2.0.1 MatrixGenerics_1.8.0
[65] ProtGenerics_1.28.0 parallel_4.2.0
[67] SummarizedExperiment_1.26.1 AnnotationFilter_1.20.0
[69] LDlinkR_1.2.3 yaml_2.3.5
[71] curl_4.3.2 gridExtra_2.3
[73] memoise_2.0.1 sass_0.4.1
[75] biomaRt_2.54.1 stringi_1.7.6
[77] RSQLite_2.3.1 highr_0.9
[79] S4Vectors_0.34.0 BiocIO_1.6.0
[81] GenomicFeatures_1.48.3 BiocGenerics_0.42.0
[83] filelock_1.0.2 BiocParallel_1.30.3
[85] repr_1.1.4 GenomeInfoDb_1.39.9
[87] rlang_1.1.2 pkgconfig_2.0.3
[89] matrixStats_0.62.0 bitops_1.0-7
[91] evaluate_0.15 lattice_0.20-45
[93] labeling_0.4.2 GenomicAlignments_1.32.0
[95] htmlwidgets_1.5.4 cowplot_1.1.1
[97] bit_4.0.4 tidyselect_1.2.0
[99] magrittr_2.0.3 AMR_2.1.1
[101] R6_2.5.1 IRanges_2.30.0
[103] generics_0.1.2 DelayedArray_0.22.0
[105] DBI_1.2.2 haven_2.5.0
[107] withr_2.5.0 pgenlibr_0.3.3
[109] pillar_1.9.0 KEGGREST_1.36.3
[111] RCurl_1.98-1.7 mixsqp_0.3-43
[113] modelr_0.1.8 crayon_1.5.1
[115] utf8_1.2.2 BiocFileCache_2.4.0
[117] plotly_4.10.0 tzdb_0.4.0
[119] rmarkdown_2.25 progress_1.2.2
[121] readxl_1.4.0 grid_4.2.0
[123] data.table_1.14.2 blob_1.2.3
[125] git2r_0.30.1 reprex_2.0.1
[127] digest_0.6.29 httpuv_1.6.5
[129] stats4_4.2.0 munsell_0.5.0
[131] viridisLite_0.4.0 skimr_2.1.4
[133] bslib_0.3.1