Look at pi0 under null for n=13500
Matthew Stephens
2017-04-18
Last updated: 2017-04-18
Code version: a34b949
Background
We saw on some simulated null data sets from RNA-seq data that the ash estimate of pi0 was often <1 - more often than in the New Deal paper. I wanted to check whether this could be due to the larger sample size in these simulations (13,500) compared with the paper (1,000). (An alternative is that ash is reacting to some subtle “non-nullness” creeping into the simulated null data).
Simulation
We just simulate 10 datasets with n=13500.
set.seed(1)
nsim=10
a = list()
pi0 = rep(0,nsim)
for(i in 1:nsim){
z = rnorm(13500)
a[[i]] = ashr::ash(z,1)
pi0[i] = ashr::get_pi0(a[[i]])
}
plot(pi0)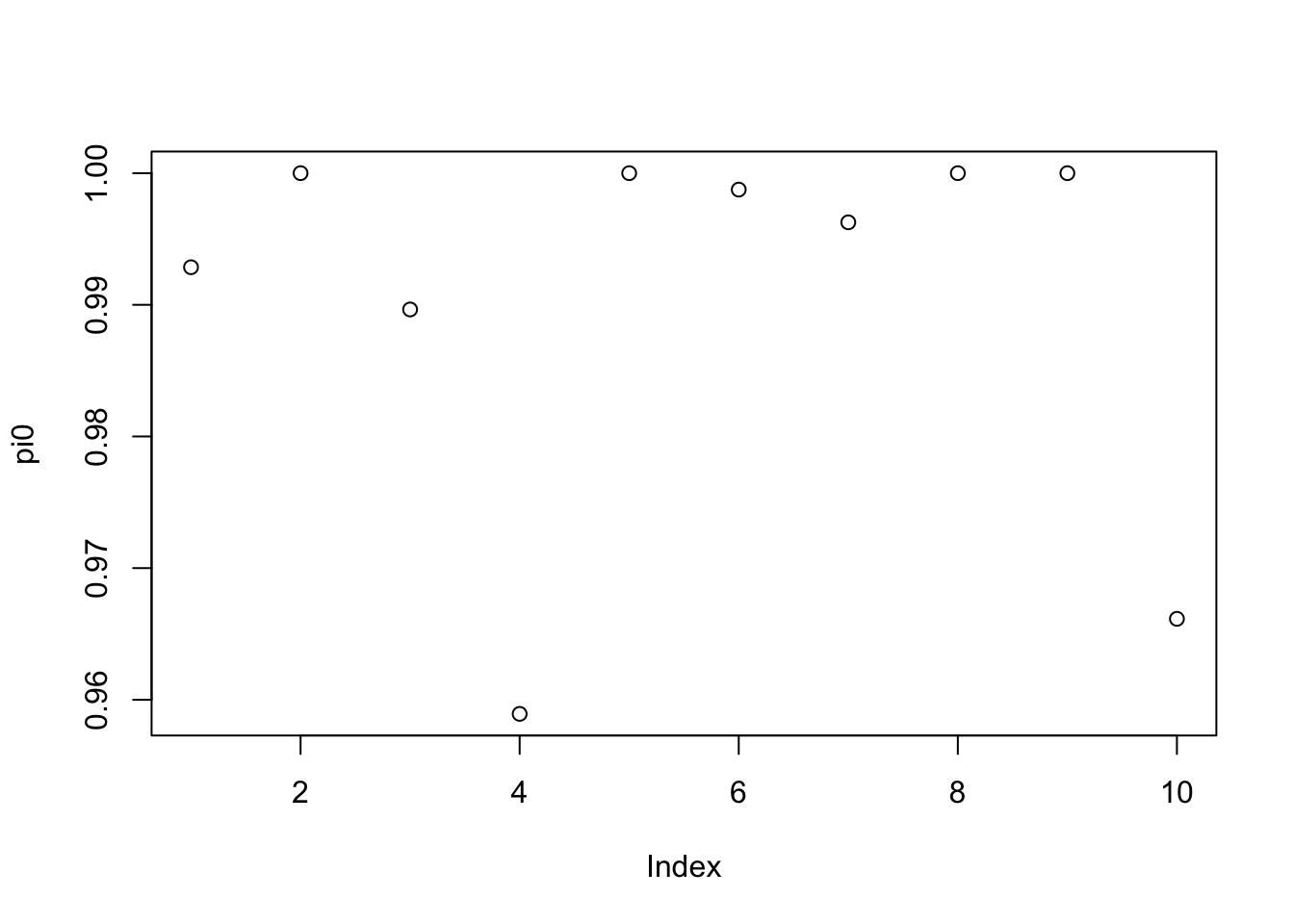
This looks not as bad to me as in the simulated RNA-seq data, so I’m guessing it is not a sample size issue. I also just wanted to check the lfsrs for the two datasets where pi0 is around 0.96; I reassuringly found nothing very significant:
min(ashr::get_lfsr(a[[4]]))[1] 0.5764098min(ashr::get_lfsr(a[[10]]))[1] 0.8275591And as a check I checked the log-likelihood
ashr::calc_loglik(g=ashr::normalmix(1,0,0),a[[4]]$data)[1] -19380.83ashr::calc_loglik(g=ashr::get_fitted_g(a[[4]]),a[[4]]$data)[1] -19377.65Session information
sessionInfo()R version 3.3.2 (2016-10-31)
Platform: x86_64-apple-darwin13.4.0 (64-bit)
Running under: OS X El Capitan 10.11.6
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] workflowr_0.4.0 rmarkdown_1.4
loaded via a namespace (and not attached):
[1] Rcpp_0.12.10 rstudioapi_0.6 knitr_1.15.1
[4] magrittr_1.5 REBayes_0.73 MASS_7.3-45
[7] doParallel_1.0.10 pscl_1.4.9 SQUAREM_2016.8-2
[10] lattice_0.20-34 foreach_1.4.3 ashr_2.1-8
[13] stringr_1.2.0 tools_3.3.2 parallel_3.3.2
[16] grid_3.3.2 git2r_0.18.0 htmltools_0.3.5
[19] iterators_1.0.8 assertthat_0.2.0 yaml_2.1.14
[22] rprojroot_1.2 digest_0.6.12 Matrix_1.2-8
[25] codetools_0.2-15 rsconnect_0.7 evaluate_0.10
[28] stringi_1.1.2 Rmosek_7.1.2 backports_1.0.5
[31] truncnorm_1.0-7 This R Markdown site was created with workflowr