fastica_centered
Matthew Stephens
2025-11-14
Last updated: 2025-11-16
Checks: 7 0
Knit directory: misc/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(1) was run prior to running the
code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version bd2737c. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.RData
Ignored: analysis/.Rhistory
Ignored: analysis/ALStruct_cache/
Ignored: data/.Rhistory
Ignored: data/methylation-data-for-matthew.rds
Ignored: data/pbmc/
Ignored: data/pbmc_purified.RData
Untracked files:
Untracked: .dropbox
Untracked: Icon
Untracked: analysis/GHstan.Rmd
Untracked: analysis/GTEX-cogaps.Rmd
Untracked: analysis/PACS.Rmd
Untracked: analysis/Rplot.png
Untracked: analysis/SPCAvRP.rmd
Untracked: analysis/abf_comparisons.Rmd
Untracked: analysis/admm_02.Rmd
Untracked: analysis/admm_03.Rmd
Untracked: analysis/bispca.Rmd
Untracked: analysis/cache/
Untracked: analysis/cholesky.Rmd
Untracked: analysis/compare-transformed-models.Rmd
Untracked: analysis/cormotif.Rmd
Untracked: analysis/cp_ash.Rmd
Untracked: analysis/eQTL.perm.rand.pdf
Untracked: analysis/eb_power2.Rmd
Untracked: analysis/eb_prepilot.Rmd
Untracked: analysis/eb_var.Rmd
Untracked: analysis/ebpmf1.Rmd
Untracked: analysis/ebpmf_sla_text.Rmd
Untracked: analysis/ebspca_sims.Rmd
Untracked: analysis/explore_psvd.Rmd
Untracked: analysis/fa_check_identify.Rmd
Untracked: analysis/fa_iterative.Rmd
Untracked: analysis/flash_cov_overlapping_groups_init.Rmd
Untracked: analysis/flash_test_tree.Rmd
Untracked: analysis/flashier_newgroups.Rmd
Untracked: analysis/flashier_nmf_triples.Rmd
Untracked: analysis/flashier_pbmc.Rmd
Untracked: analysis/flashier_snn_shifted_prior.Rmd
Untracked: analysis/greedy_ebpmf_exploration_00.Rmd
Untracked: analysis/ieQTL.perm.rand.pdf
Untracked: analysis/lasso_em_03.Rmd
Untracked: analysis/m6amash.Rmd
Untracked: analysis/mash_bhat_z.Rmd
Untracked: analysis/mash_ieqtl_permutations.Rmd
Untracked: analysis/methylation_example.Rmd
Untracked: analysis/mixsqp.Rmd
Untracked: analysis/mr.ash_lasso_init.Rmd
Untracked: analysis/mr.mash.test.Rmd
Untracked: analysis/mr_ash_modular.Rmd
Untracked: analysis/mr_ash_parameterization.Rmd
Untracked: analysis/mr_ash_ridge.Rmd
Untracked: analysis/mv_gaussian_message_passing.Rmd
Untracked: analysis/nejm.Rmd
Untracked: analysis/nmf_bg.Rmd
Untracked: analysis/nonneg_underapprox.Rmd
Untracked: analysis/normal_conditional_on_r2.Rmd
Untracked: analysis/normalize.Rmd
Untracked: analysis/pbmc.Rmd
Untracked: analysis/pca_binary_weighted.Rmd
Untracked: analysis/pca_l1.Rmd
Untracked: analysis/poisson_nmf_approx.Rmd
Untracked: analysis/poisson_shrink.Rmd
Untracked: analysis/poisson_transform.Rmd
Untracked: analysis/qrnotes.txt
Untracked: analysis/ridge_iterative_02.Rmd
Untracked: analysis/ridge_iterative_splitting.Rmd
Untracked: analysis/samps/
Untracked: analysis/sc_bimodal.Rmd
Untracked: analysis/shrinkage_comparisons_changepoints.Rmd
Untracked: analysis/susie_cov.Rmd
Untracked: analysis/susie_en.Rmd
Untracked: analysis/susie_z_investigate.Rmd
Untracked: analysis/svd-timing.Rmd
Untracked: analysis/temp.RDS
Untracked: analysis/temp.Rmd
Untracked: analysis/test-figure/
Untracked: analysis/test.Rmd
Untracked: analysis/test.Rpres
Untracked: analysis/test.md
Untracked: analysis/test_qr.R
Untracked: analysis/test_sparse.Rmd
Untracked: analysis/tree_dist_top_eigenvector.Rmd
Untracked: analysis/z.txt
Untracked: code/coordinate_descent_symNMF.R
Untracked: code/multivariate_testfuncs.R
Untracked: code/rqb.hacked.R
Untracked: data/4matthew/
Untracked: data/4matthew2/
Untracked: data/E-MTAB-2805.processed.1/
Untracked: data/ENSG00000156738.Sim_Y2.RDS
Untracked: data/GDS5363_full.soft.gz
Untracked: data/GSE41265_allGenesTPM.txt
Untracked: data/Muscle_Skeletal.ACTN3.pm1Mb.RDS
Untracked: data/P.rds
Untracked: data/Thyroid.FMO2.pm1Mb.RDS
Untracked: data/bmass.HaemgenRBC2016.MAF01.Vs2.MergedDataSources.200kRanSubset.ChrBPMAFMarkerZScores.vs1.txt.gz
Untracked: data/bmass.HaemgenRBC2016.Vs2.NewSNPs.ZScores.hclust.vs1.txt
Untracked: data/bmass.HaemgenRBC2016.Vs2.PreviousSNPs.ZScores.hclust.vs1.txt
Untracked: data/eb_prepilot/
Untracked: data/finemap_data/fmo2.sim/b.txt
Untracked: data/finemap_data/fmo2.sim/dap_out.txt
Untracked: data/finemap_data/fmo2.sim/dap_out2.txt
Untracked: data/finemap_data/fmo2.sim/dap_out2_snp.txt
Untracked: data/finemap_data/fmo2.sim/dap_out_snp.txt
Untracked: data/finemap_data/fmo2.sim/data
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.config
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.k
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.k4.config
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.k4.snp
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.ld
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.snp
Untracked: data/finemap_data/fmo2.sim/fmo2.sim.z
Untracked: data/finemap_data/fmo2.sim/pos.txt
Untracked: data/logm.csv
Untracked: data/m.cd.RDS
Untracked: data/m.cdu.old.RDS
Untracked: data/m.new.cd.RDS
Untracked: data/m.old.cd.RDS
Untracked: data/mainbib.bib.old
Untracked: data/mat.csv
Untracked: data/mat.txt
Untracked: data/mat_new.csv
Untracked: data/matrix_lik.rds
Untracked: data/paintor_data/
Untracked: data/running_data_chris.csv
Untracked: data/running_data_matthew.csv
Untracked: data/temp.txt
Untracked: data/y.txt
Untracked: data/y_f.txt
Untracked: data/zscore_jointLCLs_m6AQTLs_susie_eQTLpruned.rds
Untracked: data/zscore_jointLCLs_random.rds
Untracked: explore_udi.R
Untracked: output/fit.k10.rds
Untracked: output/fit.nn.pbmc.purified.rds
Untracked: output/fit.nn.rds
Untracked: output/fit.nn.s.001.rds
Untracked: output/fit.nn.s.01.rds
Untracked: output/fit.nn.s.1.rds
Untracked: output/fit.nn.s.10.rds
Untracked: output/fit.snn.s.001.rds
Untracked: output/fit.snn.s.01.nninit.rds
Untracked: output/fit.snn.s.01.rds
Untracked: output/fit.varbvs.RDS
Untracked: output/fit2.nn.pbmc.purified.rds
Untracked: output/glmnet.fit.RDS
Untracked: output/snn07.txt
Untracked: output/snn34.txt
Untracked: output/test.bv.txt
Untracked: output/test.gamma.txt
Untracked: output/test.hyp.txt
Untracked: output/test.log.txt
Untracked: output/test.param.txt
Untracked: output/test2.bv.txt
Untracked: output/test2.gamma.txt
Untracked: output/test2.hyp.txt
Untracked: output/test2.log.txt
Untracked: output/test2.param.txt
Untracked: output/test3.bv.txt
Untracked: output/test3.gamma.txt
Untracked: output/test3.hyp.txt
Untracked: output/test3.log.txt
Untracked: output/test3.param.txt
Untracked: output/test4.bv.txt
Untracked: output/test4.gamma.txt
Untracked: output/test4.hyp.txt
Untracked: output/test4.log.txt
Untracked: output/test4.param.txt
Untracked: output/test5.bv.txt
Untracked: output/test5.gamma.txt
Untracked: output/test5.hyp.txt
Untracked: output/test5.log.txt
Untracked: output/test5.param.txt
Unstaged changes:
Modified: .gitignore
Modified: analysis/eb_snmu.Rmd
Modified: analysis/ebnm_binormal.Rmd
Modified: analysis/ebpower.Rmd
Modified: analysis/flashier_log1p.Rmd
Modified: analysis/flashier_sla_text.Rmd
Modified: analysis/logistic_z_scores.Rmd
Modified: analysis/mr_ash_pen.Rmd
Modified: analysis/nmu_em.Rmd
Modified: analysis/susie_flash.Rmd
Modified: analysis/tap_free_energy.Rmd
Modified: misc.Rproj
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/fastica_centered.Rmd) and
HTML (docs/fastica_centered.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | bd2737c | Matthew Stephens | 2025-11-16 | workflowr::wflow_publish("fastica_centered.Rmd") |
| html | 9584c9b | Matthew Stephens | 2025-11-14 | Build site. |
| Rmd | 58374e8 | Matthew Stephens | 2025-11-14 | workflowr::wflow_publish("fastica_centered.Rmd") |
Introduction
I found previously that the original fastICA algorithm seems to have trouble finding groups that are unbalanced. I believe that this is because the log-cosh objective function favours symmetry. ie maximizing or minimizing \(\log \cosh (X'w)\) (subject to \(w'w=1\), which implies ||X’w||= 1 if \(XX'=I\)) tends to find symmetric \(X'w\). I want to try to fix this using an intercept. That is, maximize \(\log \cosh (X'w + c)\) over both \(w\) and \(c\).
I gave Gemini this problem and it gave me the following response and code.
“This adds a new parameter, \(c\), that we also need to optimize. We can derive the update rules by taking the partial derivatives of the objective function (negentropy approximation) with respect to both \(w\) and \(c\) and setting them to zero (this is the Karush-Kuhn-Tucker or KKT-based approach).
This gives us two conditions to satisfy:
For \(w\): \(E[X g(w^TX + c)] - \lambda w = 0\)
For \(c\): \(E[g(w^TX + c)] = 0\)
Where \(g\) is your contrast function (\(\tanh\)) and \(\lambda\) is a scalar (Lagrange multiplier).
Condition (2) is new and very intuitive: it means the algorithm must find an intercept \(c\) that “centers” the transformed projection \(g(P)\) to have a mean of zero. This leads to a simple two-step update within each iteration:
Update c: First, for a fixed w, find the c that solves \(E[g(w^TX + c)] = 0\).
Update w: Then, using that new c, perform the standard FastICA update for w.”
#' Single-unit FastICA update with an intercept term 'c'
#'
#' @param X The pre-processed (centered and whitened) data matrix (p x n).
#' @param w The current weight vector (p x 1).
#' @param c The current intercept scalar.
#' @param c_update_steps How many Newton steps to take for 'c' per iteration.
#' 1 is usually sufficient.
#' @return A list containing the updated 'w' (unnormalized) and 'c'.
fastica_r1update_wc = function(X, w, c, c_update_steps = 1) {
# 1. Normalize w (as before)
w = w / sqrt(sum(w^2))
# --- Step 1: Update the intercept 'c' ---
# Get the current projection *without* the intercept
# P_current is an (n x 1) vector
P_current = as.vector(t(X) %*% w)
# We want to find 'c' that solves E[g(P_current + c)] = 0
# We use one (or more) Newton steps: c_new = c - f(c) / f'(c)
for (i in 1:c_update_steps) {
P_shifted = P_current + c
# g(P_current + c)
G_c = tanh(P_shifted)
# g'(P_current + c)
G2_c = 1 - tanh(P_shifted)^2
# E[g(P_current + c)]
mean_G = mean(G_c)
# E[g'(P_current + c)]
mean_G2 = mean(G2_c)
# The Newton step (add epsilon for stability)
c = c - mean_G / (mean_G2 + 1e-6)
}
# --- Step 2: Update the weight vector 'w' ---
# Now we use the *updated* 'c' to update 'w'
# P = w'X + c (using the new 'c' from Step 1)
P = P_current + c
# g(P)
G = tanh(P)
# g'(P)
G2 = 1 - tanh(P)^2
# The FastICA update rule for 'w':
# w_new = E[X * g(P)] - E[g'(P)] * w
# (Note: We use sum(G2) as an estimate for E[g'(P)] * n)
# (and X %*% G as an estimate for E[X*g(P)] * n)
# The 'n' scaling factor cancels, as 'w' is normalized next iteration.
w_new = X %*% G - sum(G2) * w
# 4. Return the updated vector and intercept
return(list(w = w_new, c = c))
}
#'
#' This is the function that the "profiled" and "skew" update rules
#' are trying to find an extremum (max/min) of.
#'
#' @param X The pre-processed (centered and whitened) data matrix (p x n).
#' @param w The weight vector (p x 1).
#' @param c The intercept scalar.
#' @return The scalar value of the objective function.
compute_objective = function(X, w, c) {
# 1. Normalize w to stay on the ||w||=1 constraint
w = w / sqrt(sum(w^2))
# 2. Calculate the projection
P = as.vector(t(X) %*% w) # (n x 1) vector
# 3. Create the shifted projection
S = P + c
# 5. Apply G(u) = log(cosh(u))
G_S = log(cosh(S))
# 6. Compute the expectation (as a sample mean)
objective_value = mean(G_S)
return(objective_value)
}Preprocessing code
preprocess = function(X, n.comp=10){
n <- nrow(X)
p <- ncol(X)
X <- scale(X, scale = FALSE)
X <- t(X)
## This appears to be equivalant to X1 = t(svd(X)$v[,1:n.comp])
V <- X %*% t(X)/n
s <- La.svd(V)
D <- diag(c(1/sqrt(s$d)))
K <- D %*% t(s$u)
K <- matrix(K[1:n.comp, ], n.comp, p)
X1 <- K %*% X
return(X1)
}Simulate data: 3 small groups
Here I simulate 3 groups, with only 20 members each, which I previously found that fastICA had trouble with.
K=3
p = 1000
n = 100
set.seed(1)
L = matrix(0,nrow=n,ncol=K)
for(i in 1:K){L[sample(1:n,20),i]=1}
FF = matrix(rnorm(p*K), nrow = p, ncol=K)
X = L %*% t(FF) + rnorm(n*p,0,0.01)
plot(X %*% FF[,1])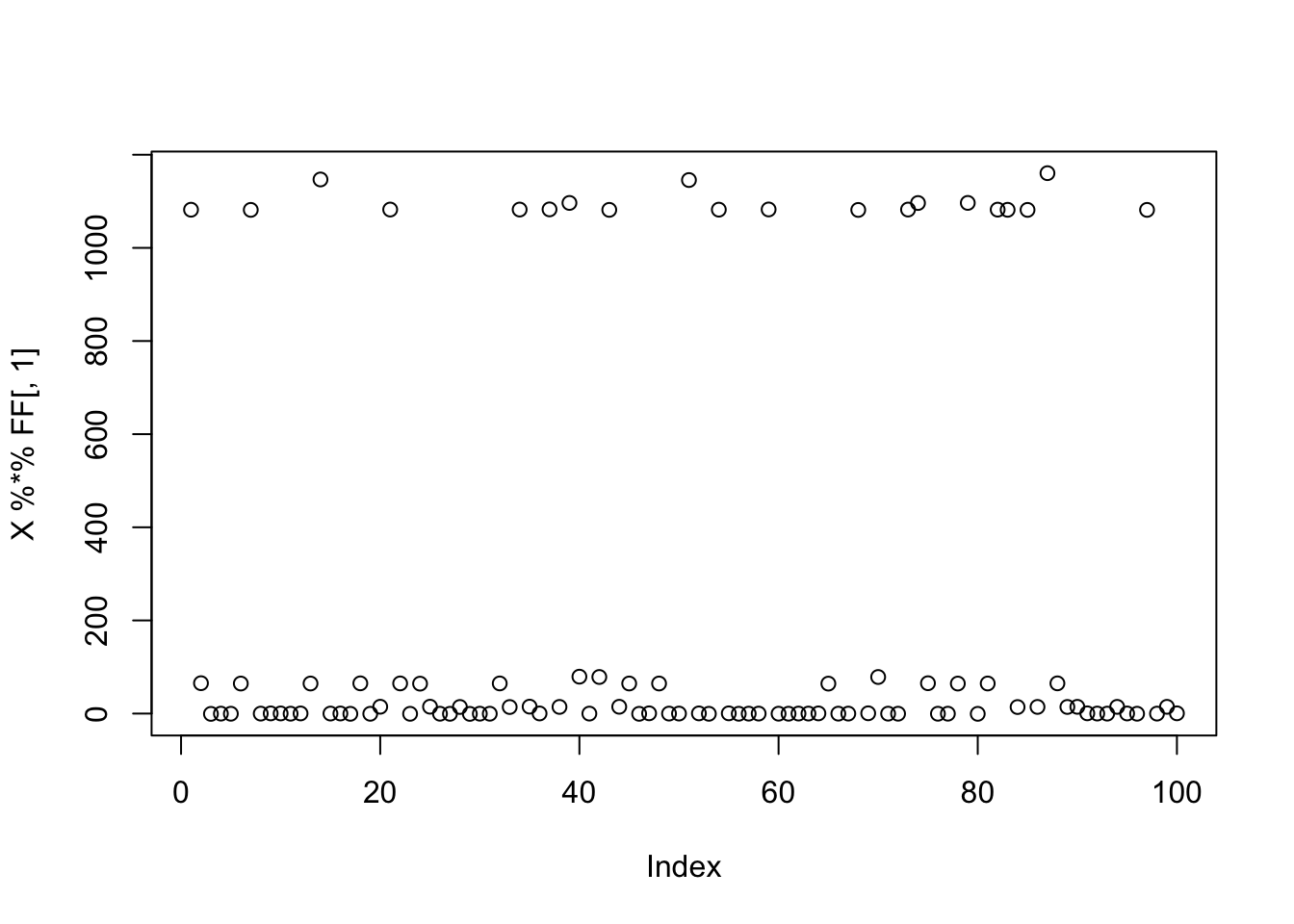
| Version | Author | Date |
|---|---|---|
| 9584c9b | Matthew Stephens | 2025-11-14 |
centered fastICA (random start)
When I initialize these new updates with a random w, it does not pick out a single source:
X1 = preprocess(X)
w = rnorm(nrow(X1))
c = 0 # this does not matter as c is updated first
res = list(w=w,c=c)
for(i in 1:100)
res = fastica_r1update_wc(X1,res$w,res$c)
cor(L,t(X1) %*% res$w) [,1]
[1,] -0.2146838
[2,] -0.1209332
[3,] 0.3947973plot(as.vector(t(X1) %*% res$w))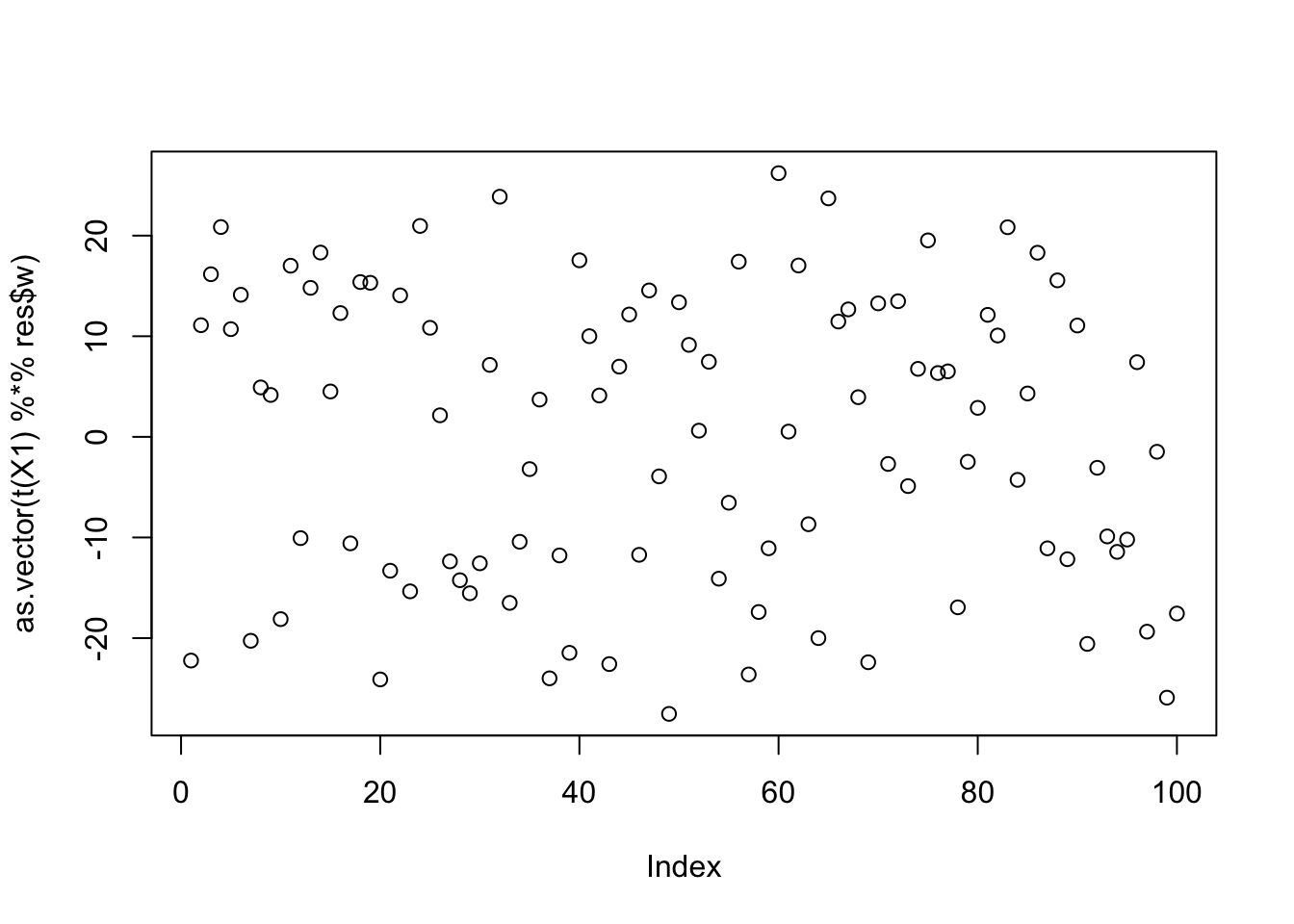
| Version | Author | Date |
|---|---|---|
| 9584c9b | Matthew Stephens | 2025-11-14 |
res$c[1] -0.03965451compute_objective(X1,res$w,res$c)[1] 0.4035189centered fastICA (true factor start)
Now I try initializing at (or close to) a true factor: it converges to the solution I wanted, and more importantly it has a much higher objective than then random start. This confirms that the changed objective function is having the desired effect.
w = X1 %*% L[,1]
plot(t(X1) %*% w)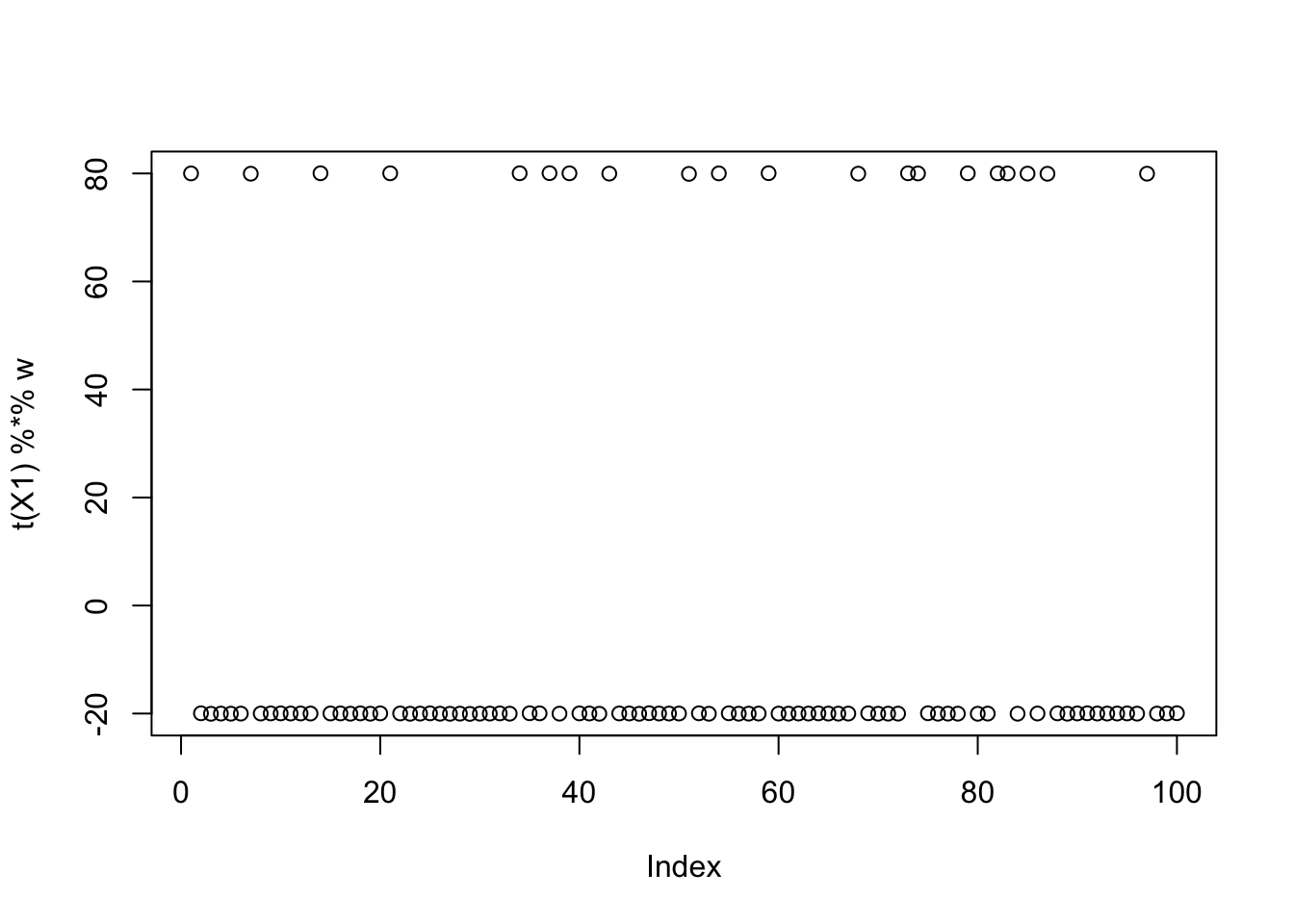
| Version | Author | Date |
|---|---|---|
| 9584c9b | Matthew Stephens | 2025-11-14 |
c = 0 # note this does not matter much because c gets updated first in the algorithm
res = list(w=w,c=c)
for(i in 1:100)
res = fastica_r1update_wc(X1,res$w,res$c)
cor(L,t(X1) %*% res$w) [,1]
[1,] 9.999997e-01
[2,] -7.339242e-06
[3,] -6.260554e-02plot(as.vector(t(X1) %*% res$w))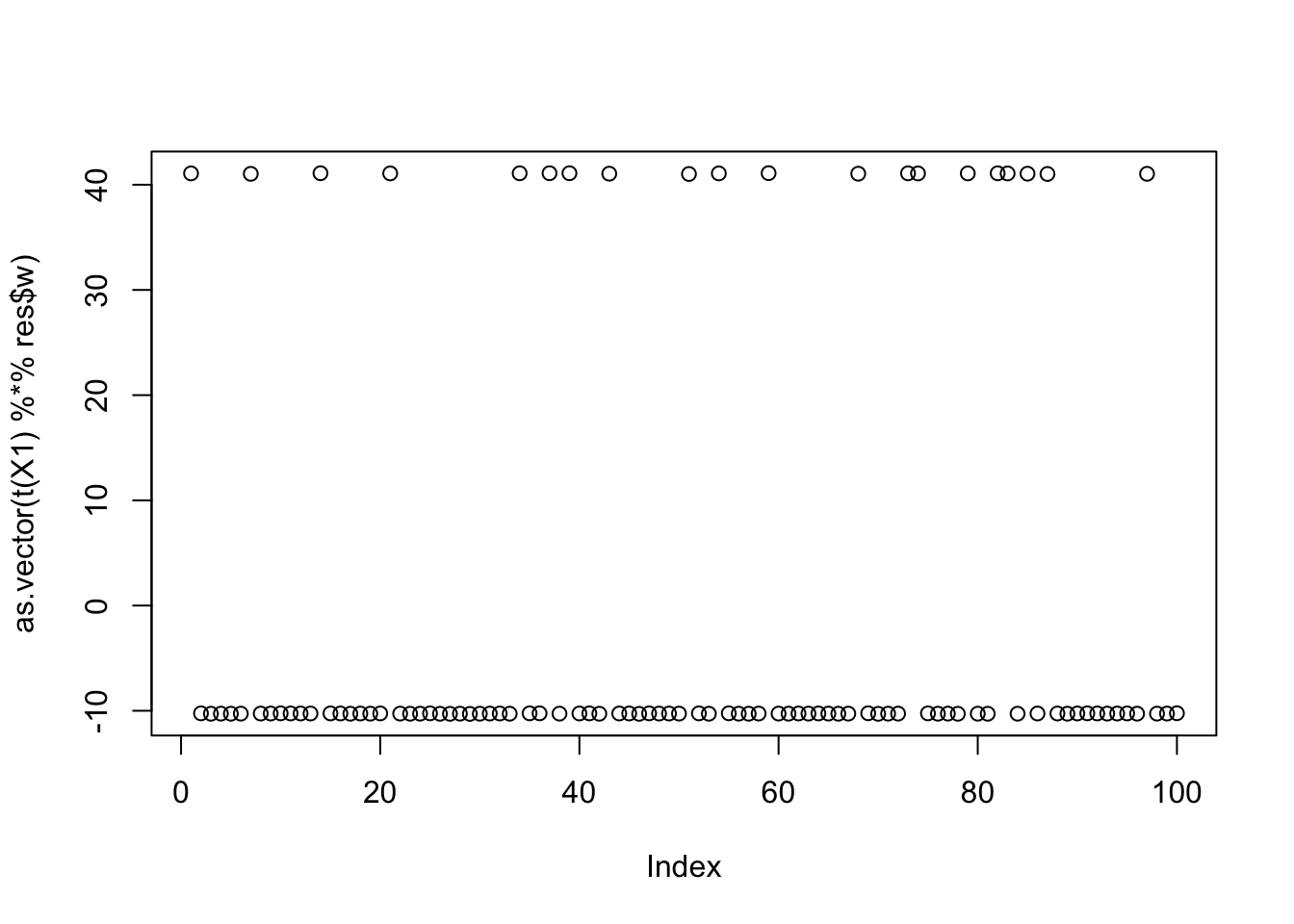
| Version | Author | Date |
|---|---|---|
| 9584c9b | Matthew Stephens | 2025-11-14 |
print(compute_objective(X1,res$w,res$c), digits=20)[1] 1.1605289119177262247Try initializing from a different true factor - we see the objective function is almost identical as for the first factor in this case (but slightly lower).
w = X1 %*% L[,2]
plot(t(X1) %*% w)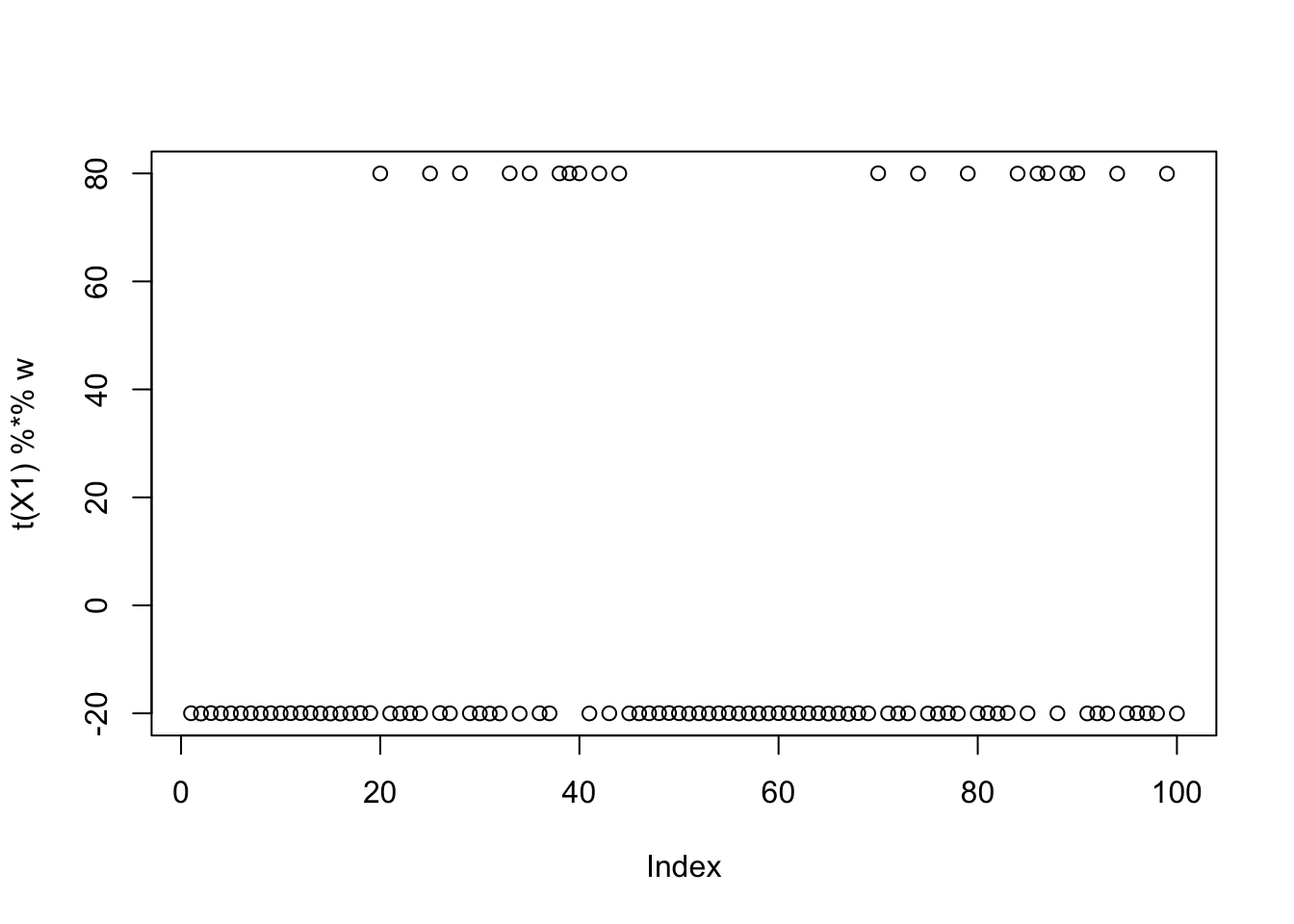
| Version | Author | Date |
|---|---|---|
| 9584c9b | Matthew Stephens | 2025-11-14 |
c = 0 # note this does not matter much because c gets updated first in the algorithm
res = list(w=w,c=c)
for(i in 1:100)
res = fastica_r1update_wc(X1,res$w,res$c)
cor(L,t(X1) %*% res$w) [,1]
[1,] 2.587794e-05
[2,] 9.999997e-01
[3,] 9.652197e-05plot(as.vector(t(X1) %*% res$w))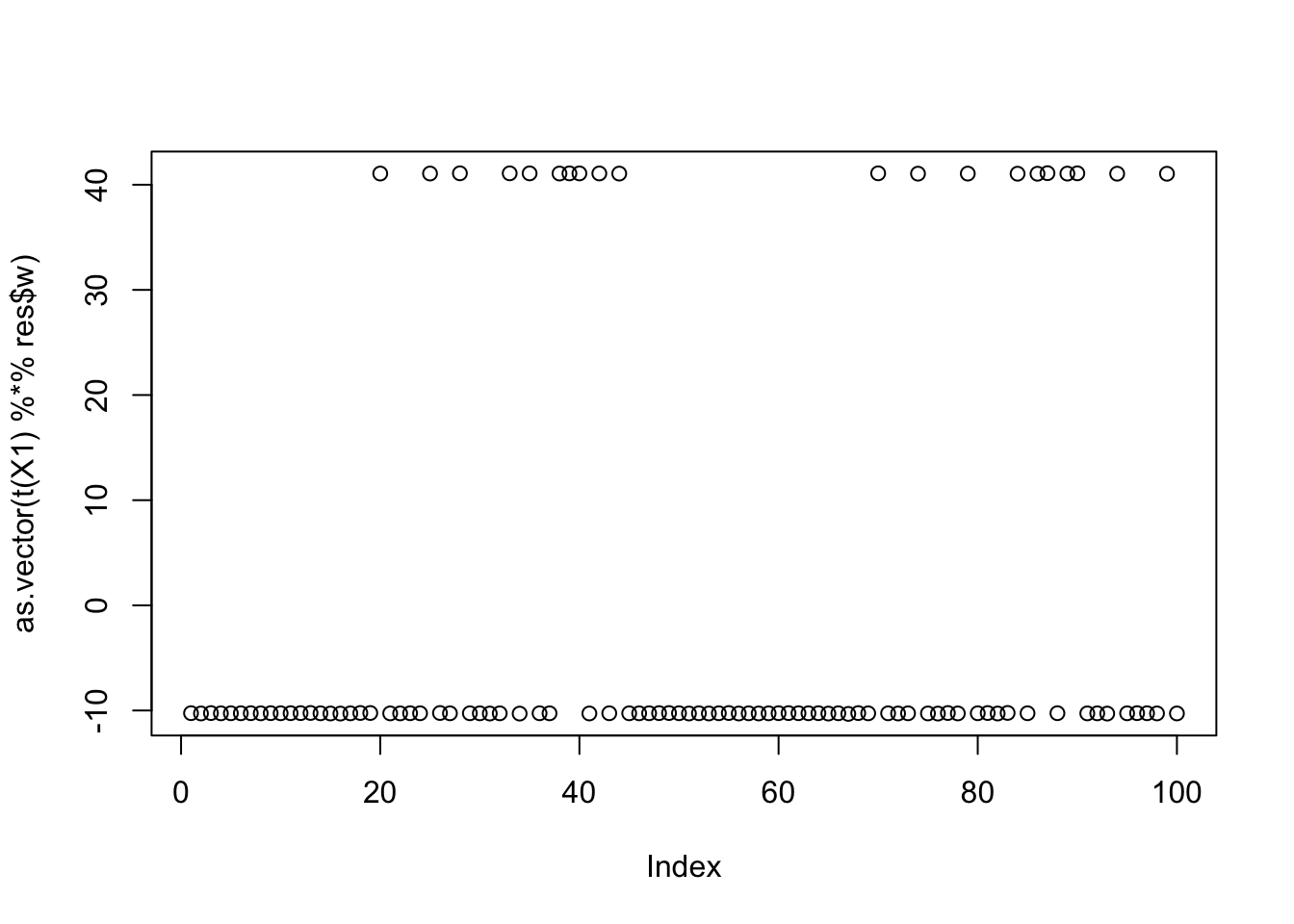
print(compute_objective(X1,res$w,res$c), digits=20)[1] 1.1605281825359521353Same for the third factor: so the objective function for all 3 factors is approximately equal. This is not a coincidence; in general the objective function for all binary factors of equal size should be approximately equal with this centering approach. (But the objective function will tend to favor unbalanced factors.)
w = X1 %*% L[,3]
plot(t(X1) %*% w)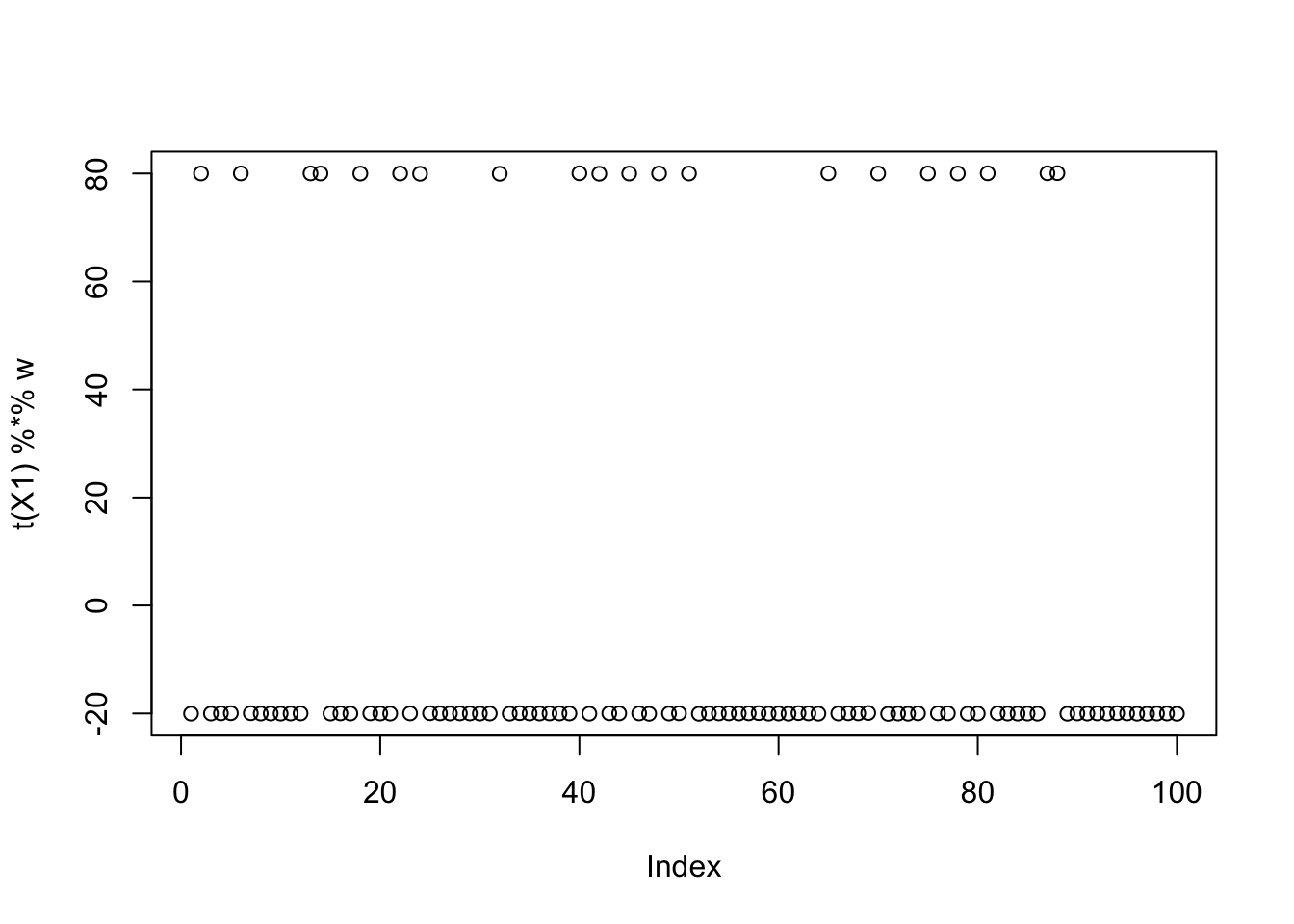
| Version | Author | Date |
|---|---|---|
| 9584c9b | Matthew Stephens | 2025-11-14 |
c = 0 # note this does not matter much because c gets updated first in the algorithm
res = list(w=w,c=c)
for(i in 1:100)
res = fastica_r1update_wc(X1,res$w,res$c)
cor(L,t(X1) %*% res$w) [,1]
[1,] -6.247381e-02
[2,] 9.005038e-06
[3,] 9.999997e-01plot(as.vector(t(X1) %*% res$w))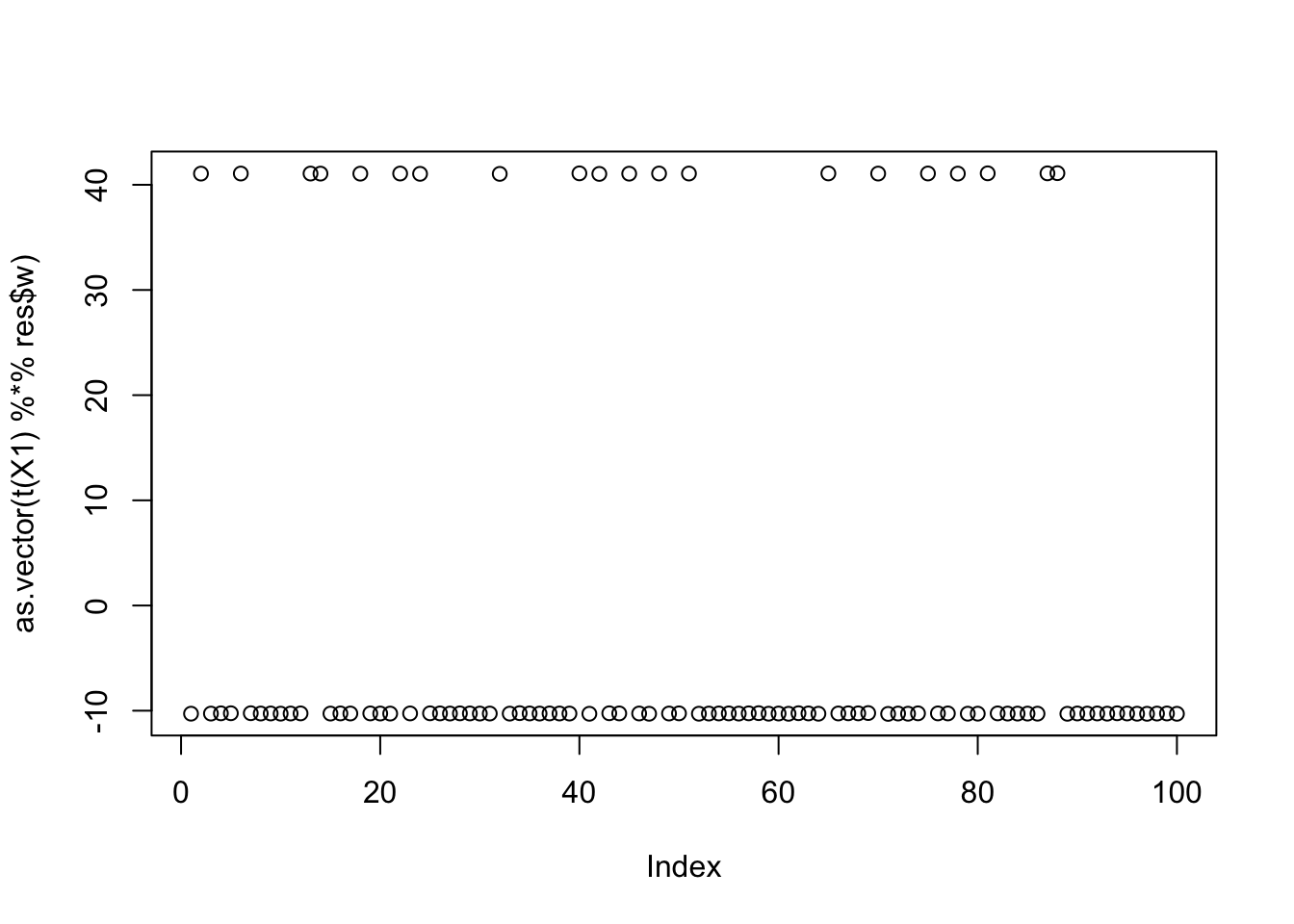
print(compute_objective(X1,res$w,res$c),digits=20)[1] 1.1605283389603271438centered fastICA (many random starts)
Here I try with 100 random starts - there are lots of different results, and some of them reach objectives similar to that from the true starts.
obj = rep(0,100)
for(seed in 1:100){
set.seed(seed)
res = list(w = rnorm(nrow(X1)), c=0)
for(i in 1:100)
res = fastica_r1update_wc(X1,res$w,res$c)
obj[seed] = compute_objective(X1,res$w,res$c)
}
plot(obj)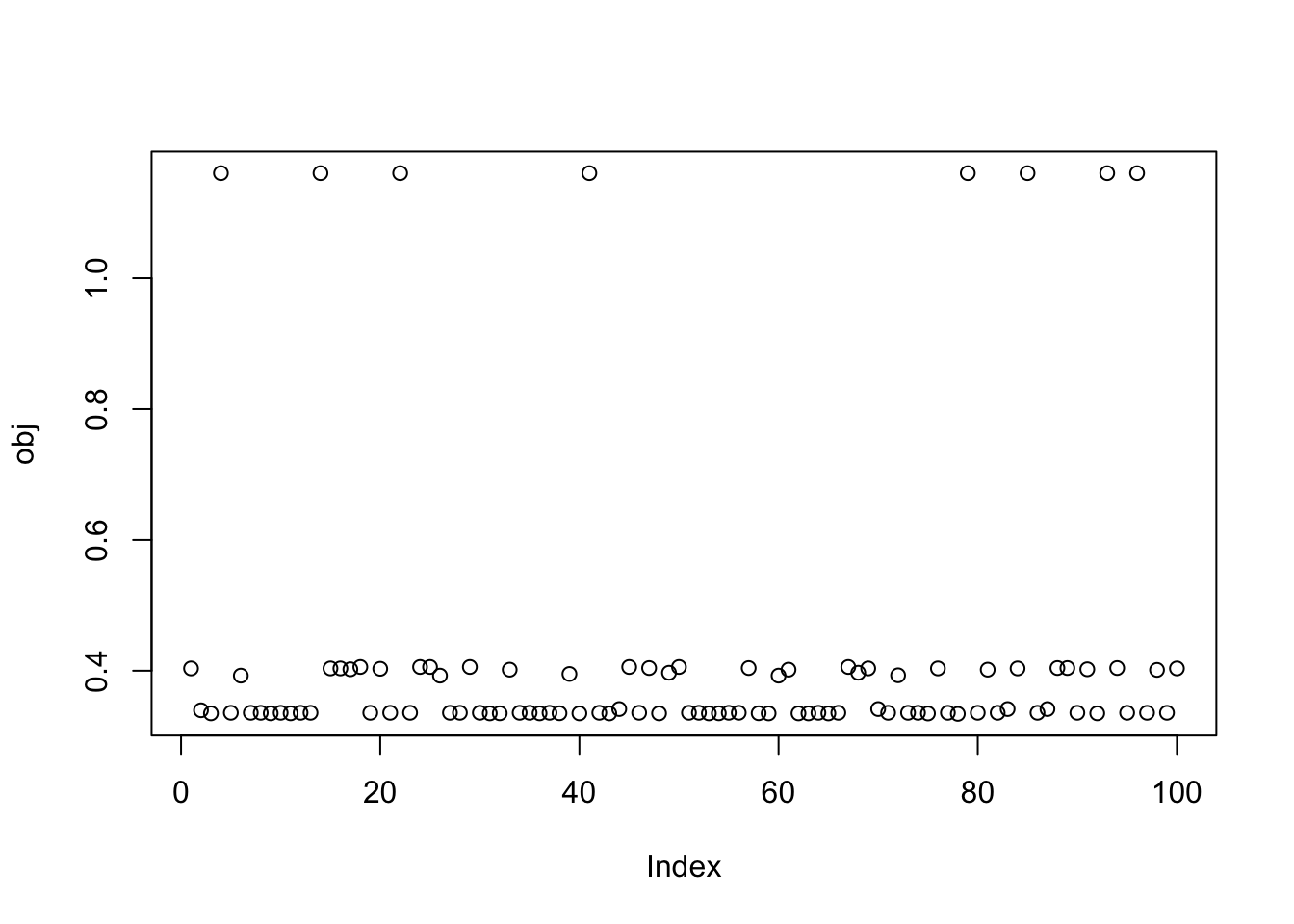
print(sort(obj,decreasing=TRUE)[1:15],digits=20) [1] 1.16052891191772644675 1.16052891191772622470 1.16052891191772555857
[4] 1.16052833896032758787 1.16052833896032714378 1.16052833894469764608
[7] 1.16052818253595213527 1.16052818253595169118 0.40573150817501507648
[10] 0.40573150817501502097 0.40573150817501502097 0.40573150817501502097
[13] 0.40573150817501496546 0.40573150817501496546 0.40573150817501424381Close inspection of the objective values suggest that the top 7 seeds find the first factor, seeds 8-10 find the third factor, and seeds 11-14 find the second factor. This turns out to be correct! For example, here are the results for the seed achieving the max objective.
seed= order(obj,decreasing=TRUE)[1]
set.seed(seed)
res = list(w = rnorm(nrow(X1)), c=0)
for(i in 1:100)
res = fastica_r1update_wc(X1,res$w,res$c)
cor(L,t(X1) %*% res$w) [,1]
[1,] -9.999997e-01
[2,] 7.339242e-06
[3,] 6.260554e-02plot(as.vector(t(X1) %*% res$w))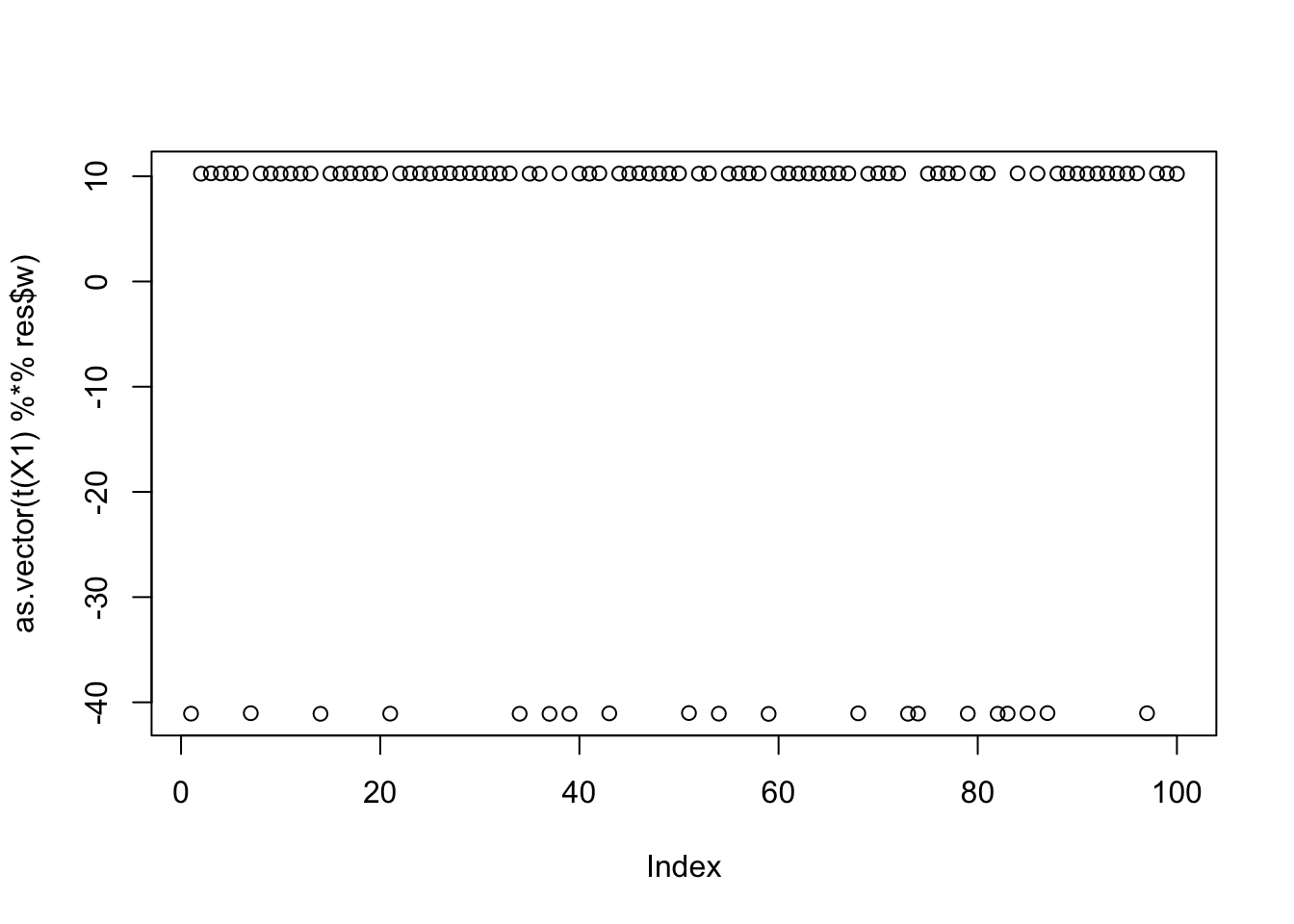
| Version | Author | Date |
|---|---|---|
| 9584c9b | Matthew Stephens | 2025-11-14 |
max(obj)[1] 1.160529Here is the seed for the 8th biggest objective (finds the third factor)
seed= order(obj,decreasing=TRUE)[8]
set.seed(seed)
res = list(w = rnorm(nrow(X1)), c=0)
for(i in 1:100)
res = fastica_r1update_wc(X1,res$w,res$c)
cor(L,t(X1) %*% res$w) [,1]
[1,] -2.587794e-05
[2,] -9.999997e-01
[3,] -9.652197e-05plot(as.vector(t(X1) %*% res$w))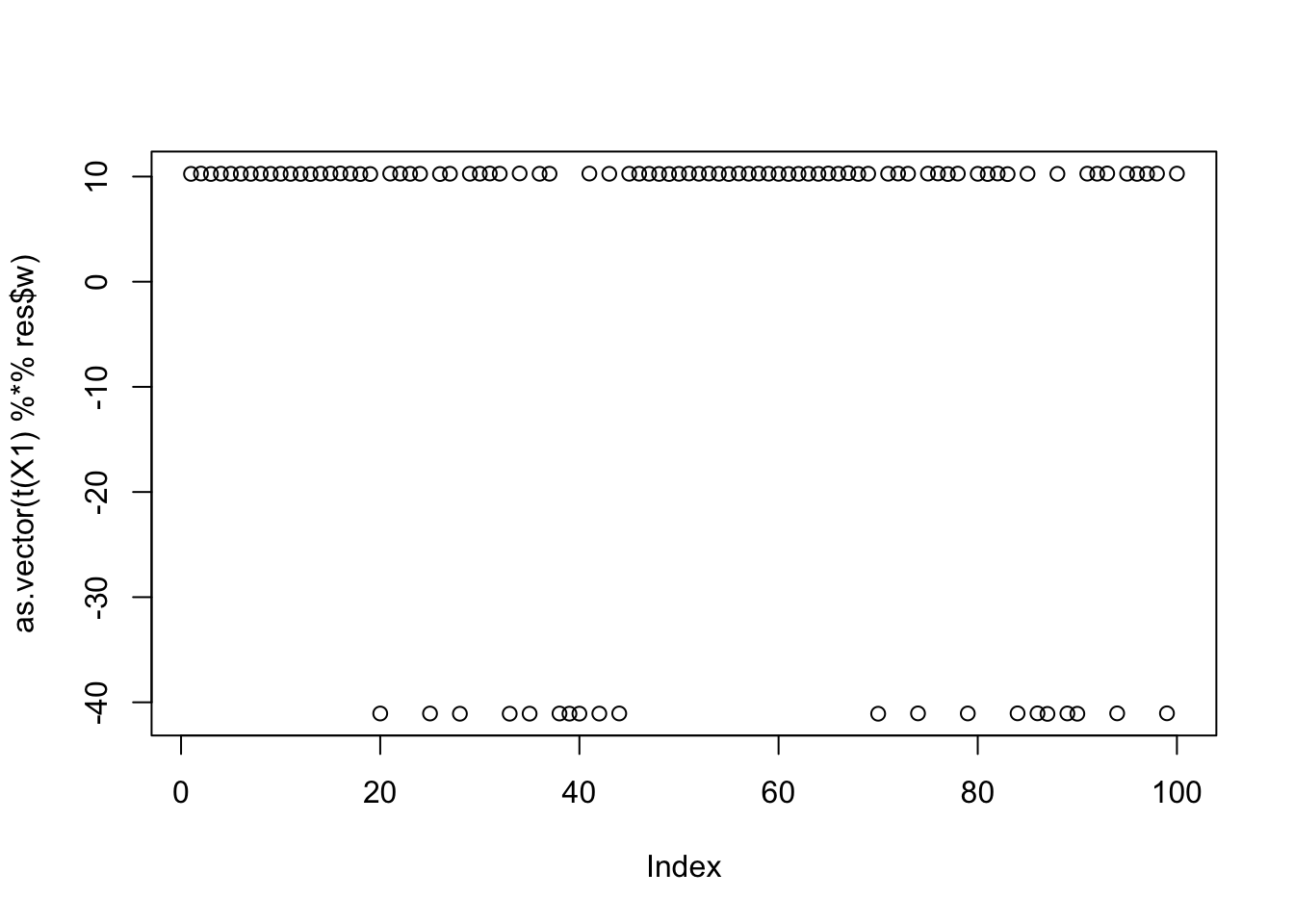
| Version | Author | Date |
|---|---|---|
| 9584c9b | Matthew Stephens | 2025-11-14 |
Here is the seed for the 11th biggest objective (finds the second factor)
seed= order(obj,decreasing=TRUE)[11]
set.seed(seed)
res = list(w = rnorm(nrow(X1)), c=0)
for(i in 1:100)
res = fastica_r1update_wc(X1,res$w,res$c)
cor(L,t(X1) %*% res$w) [,1]
[1,] 0.04258654
[2,] -0.11148268
[3,] -0.10750148plot(as.vector(t(X1) %*% res$w))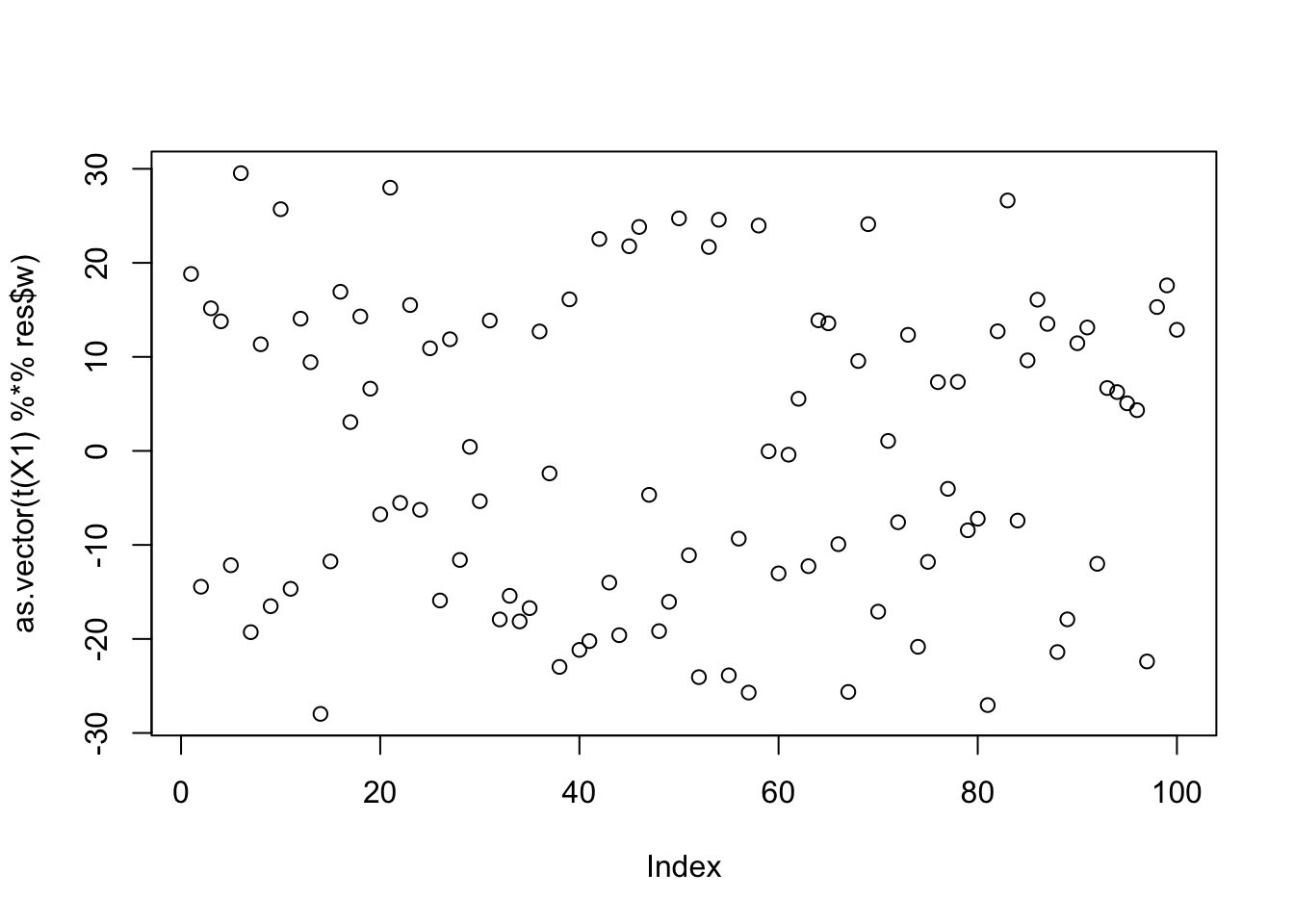
| Version | Author | Date |
|---|---|---|
| 9584c9b | Matthew Stephens | 2025-11-14 |
centered fastICA (PC starts)
We would probably like to find better ways to initialize so that we can find all the three solutions with fewer starts. I thought maybe it could be helpful to initialize from the PCs (ie w = (1,0,0..0) etc). There are 10 of these because I’m whitening to 10 dimensions. The first two PCs do indeed both find a group, and a different group for each one. This is promising - it suggests maybe we can get good starting points using the top PCs.
obj = rep(0,10)
for(pc in 1:10){
w = rep(0,10)
w[pc] = 1
res = list(w = w, c=0)
for(i in 1:100)
res = fastica_r1update_wc(X1,res$w,res$c)
obj[pc] = compute_objective(X1,res$w,res$c)
}
plot(obj)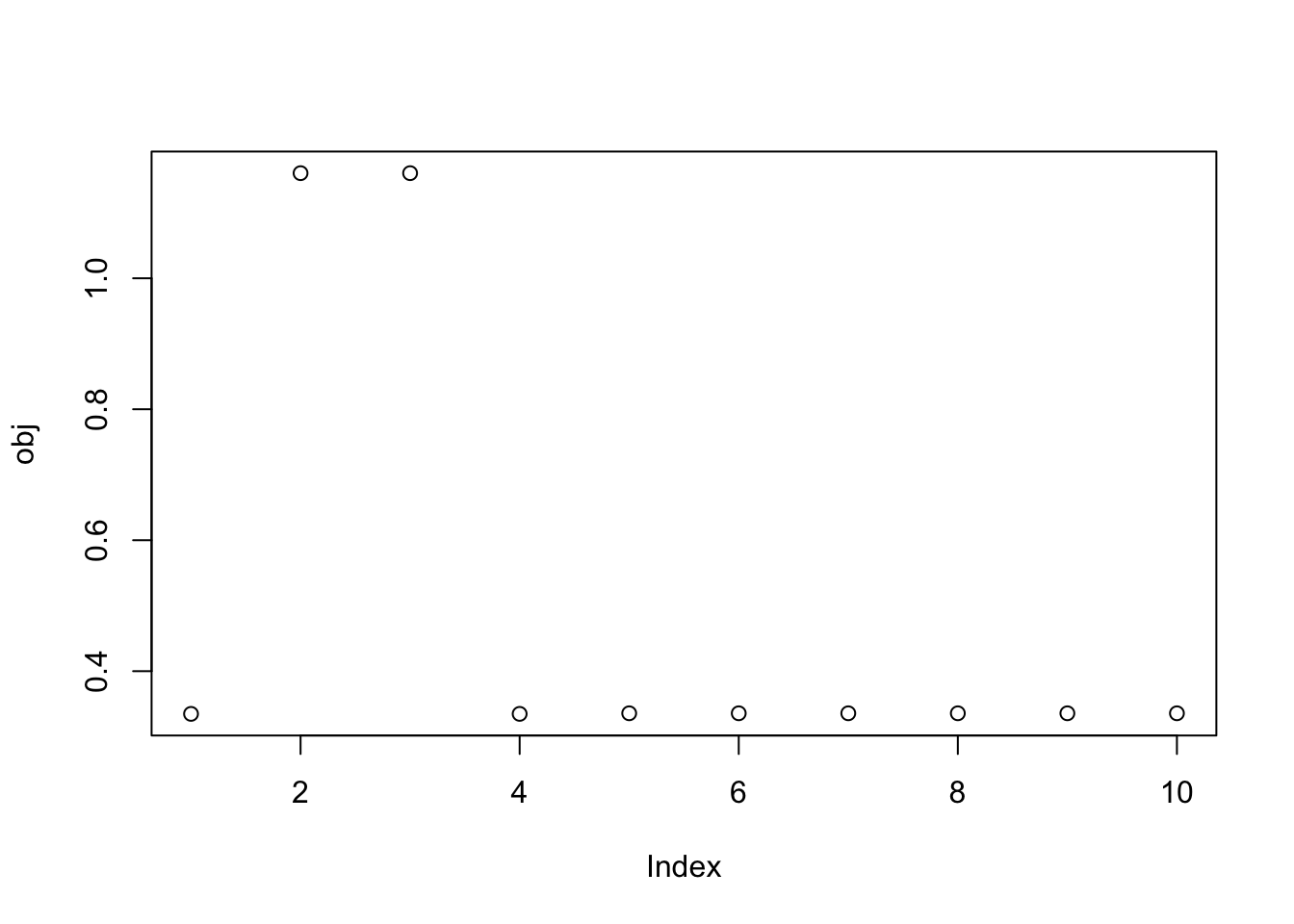
print(sort(obj,decreasing=TRUE)[1:10],digits=20) [1] 1.16052891191772578061 1.16052833896032780991 0.33569309204304309535
[4] 0.33569236181006040232 0.33569236180855116514 0.33569236180844630457
[7] 0.33569236180806799608 0.33569236180661360391 0.33479965888634471982
[10] 0.33478918679229641153Here I try random starts, but only weighting the top 3 PCs (which should be the ones with the signals). This successfully enriches for finding a good solution. It is a bit hard to see but these random starts do find all 3 of the different factors.
obj = rep(0,100)
for(seed in 1:100){
set.seed(seed)
res = list(w = c(rnorm(3),rep(0,7)), c=0)
for(i in 1:100)
res = fastica_r1update_wc(X1,res$w,res$c)
obj[seed] = compute_objective(X1,res$w,res$c)
}
plot(log(max(obj)-obj))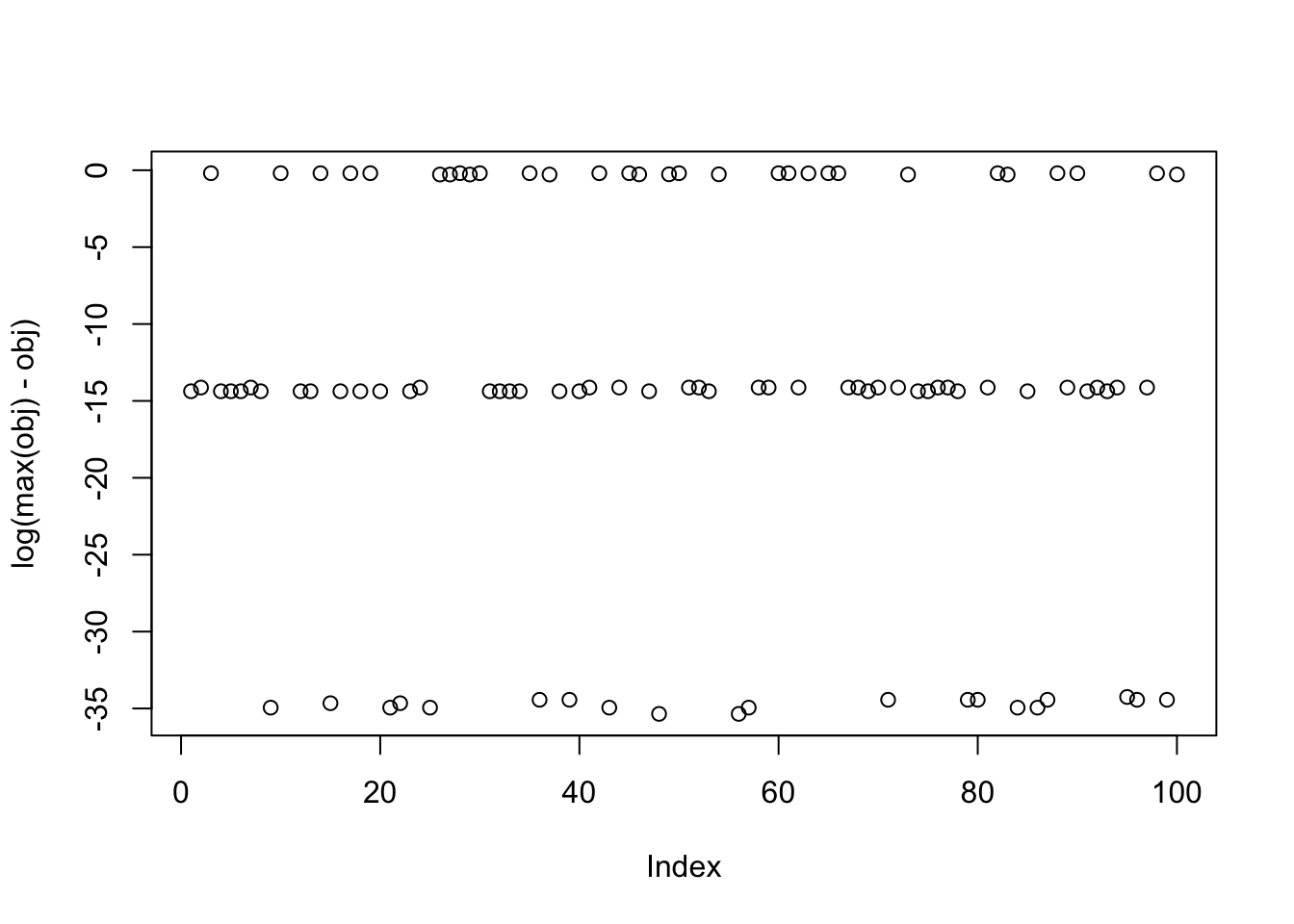
print(sort(obj-max(obj),decreasing=TRUE),digits=20) [1] 0.0000000000000000000e+00 0.0000000000000000000e+00
[3] 0.0000000000000000000e+00 -4.4408920985006261617e-16
[5] -4.4408920985006261617e-16 -6.6613381477509392425e-16
[7] -6.6613381477509392425e-16 -6.6613381477509392425e-16
[9] -6.6613381477509392425e-16 -6.6613381477509392425e-16
[11] -6.6613381477509392425e-16 -6.6613381477509392425e-16
[13] -8.8817841970012523234e-16 -8.8817841970012523234e-16
[15] -1.1102230246251565404e-15 -1.1102230246251565404e-15
[17] -1.1102230246251565404e-15 -1.1102230246251565404e-15
[19] -1.1102230246251565404e-15 -1.1102230246251565404e-15
[21] -1.1102230246251565404e-15 -1.1102230246251565404e-15
[23] -1.3322676295501878485e-15 -5.7295739863683081694e-07
[25] -5.7295739863683081694e-07 -5.7295739863683081694e-07
[27] -5.7295739863683081694e-07 -5.7295739908092002679e-07
[29] -5.7295739930296463172e-07 -5.7295739930296463172e-07
[31] -5.7295739930296463172e-07 -5.7295739930296463172e-07
[33] -5.7295739930296463172e-07 -5.7295739930296463172e-07
[35] -5.7295739930296463172e-07 -5.7295739952500923664e-07
[37] -5.7295739952500923664e-07 -5.7295739952500923664e-07
[39] -5.7295739952500923664e-07 -5.7295739952500923664e-07
[41] -5.7295739952500923664e-07 -5.7295739952500923664e-07
[43] -5.7295739952500923664e-07 -5.7295739952500923664e-07
[45] -5.7295739974705384157e-07 -5.7295739974705384157e-07
[47] -5.7295739974705384157e-07 -5.7295739974705384157e-07
[49] -5.7295739996909844649e-07 -7.2938177386738800578e-07
[51] -7.2938177408943261071e-07 -7.2938177408943261071e-07
[53] -7.2938177408943261071e-07 -7.2938177431147721563e-07
[55] -7.2938177431147721563e-07 -7.2938177431147721563e-07
[57] -7.2938177431147721563e-07 -7.2938177431147721563e-07
[59] -7.2938177431147721563e-07 -7.2938177431147721563e-07
[61] -7.2938177453352182056e-07 -7.2938177453352182056e-07
[63] -7.2938177453352182056e-07 -7.2938177453352182056e-07
[65] -7.2938177453352182056e-07 -7.2938177475556642548e-07
[67] -7.2938177475556642548e-07 -7.2938177475556642548e-07
[69] -7.2938177475556642548e-07 -7.2938177497761103041e-07
[71] -7.5641382430351467026e-01 -7.5641930268669210768e-01
[73] -7.5701004594153054050e-01 -7.5701004600101584607e-01
[75] -7.5701004984525943620e-01 -7.5765238076761654007e-01
[77] -7.5888592804964782879e-01 -7.6483359217003621389e-01
[79] -7.6754568945918366651e-01 -7.6802334733171306880e-01
[81] -8.1911547324708500195e-01 -8.1911547327230171955e-01
[83] -8.2480729140233477459e-01 -8.2483523861430496638e-01
[85] -8.2483586062517444404e-01 -8.2483652447530753093e-01
[87] -8.2483654468173117635e-01 -8.2483654787839433276e-01
[89] -8.2483654806860839948e-01 -8.2483654950253981752e-01
[91] -8.2483655010470391389e-01 -8.2483655011049950012e-01
[93] -8.2483655011380496713e-01 -8.2567807766327172558e-01
[95] -8.2571756401397278236e-01 -8.2572693829891297135e-01
[97] -8.2574645378866295964e-01 -8.2586640225611351873e-01
[99] -8.2598187574810000289e-01 -8.2653432775865576243e-01Tree simulations
Here is a simple tree-based simulations; it’s a 4-leaf tree with two bifurcating branches.
# set up L to be a tree with 4 tips and 7 branches (including top shared branch)
set.seed(1)
n = 100
p = 1000
L = matrix(0,nrow=n,ncol=6)
L[1:50,1] = 1 #top split L
L[51:100,2] = 1 # top split R
L[1:25,3] = 1
L[26:50,4] = 1
L[51:75,5] = 1
L[76:100,6] = 1
FF = matrix(rnorm(p*6),nrow=p)
X = L %*% t(FF) + rnorm(n*p,0,0.01)
image(X%*% t(X))
X1 = preprocess(X)Run 100 seeds, weighting only the top 3 PCs in the initialization (note: when I did the same weighting the top 4 PCs I got more mixed branch solutions). We see that the top seeds pick out a single group. Next seeds are the top branch, but then after that we have a bunch of seeds that combine two groups that are not adjacent on the tree - these solutions are less attractive to us. We need to find a way to find more of the single-branch solutions, and, ideally, avoid these mixed-branch solutions (although it might be possible to filter these somehow at the end). One thing I don’t quite understand is that the objective for the mixed branch solutions seems to be consistently, but very slightly, lower than for the single-branch solution. That would be good to understand better.
obj = rep(0,100)
Lhat = matrix(nrow=100,ncol=n)
for(seed in 1:100){
set.seed(seed)
res = list(w = c(rnorm(3),rep(0,7)), c=0)
for(i in 1:100)
res = fastica_r1update_wc(X1,res$w,res$c)
obj[seed] = compute_objective(X1,res$w,res$c)
Lhat[seed,] = t(X1) %*% res$w
}
plot(obj)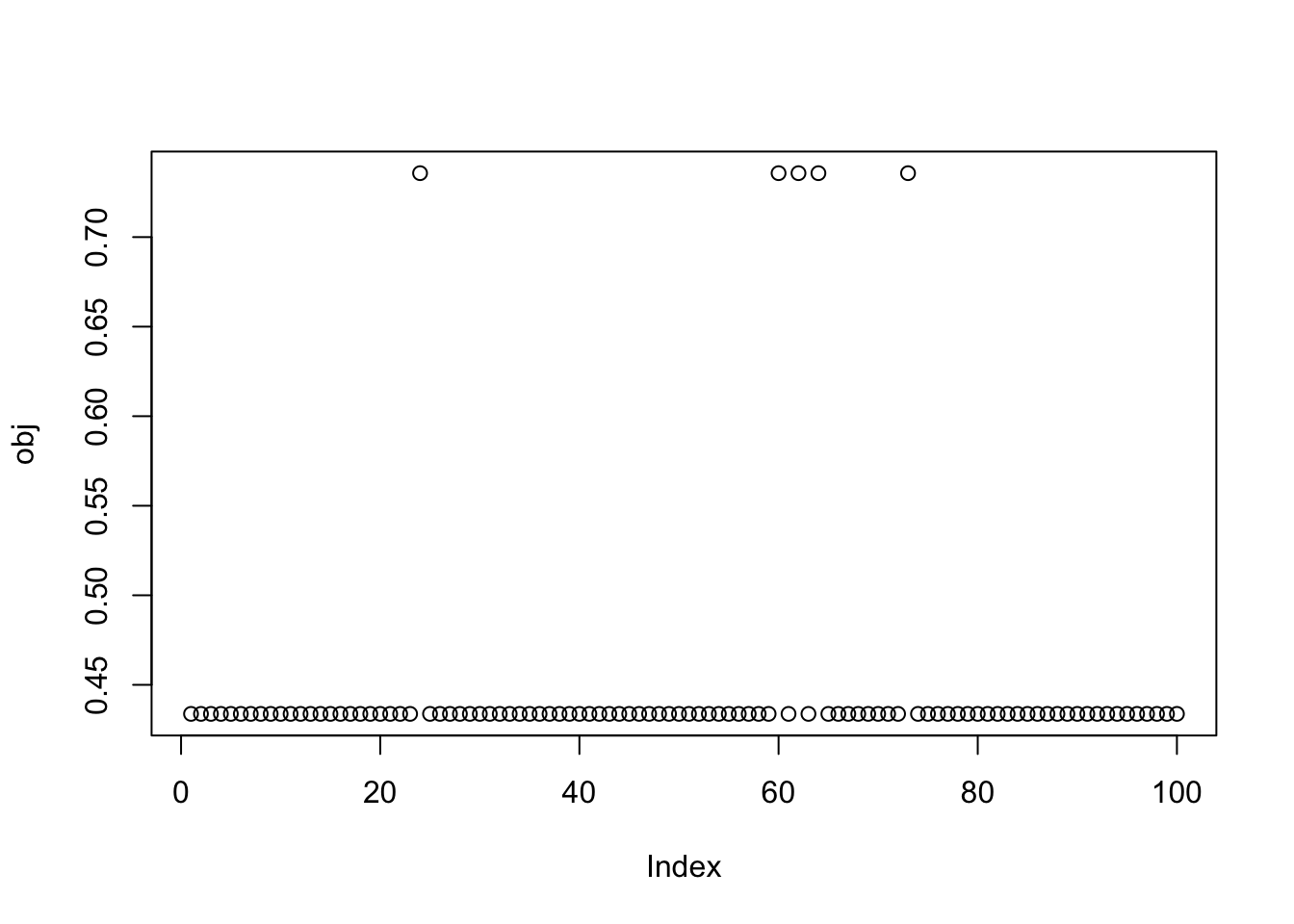
image(Lhat[order(obj,decreasing=TRUE),])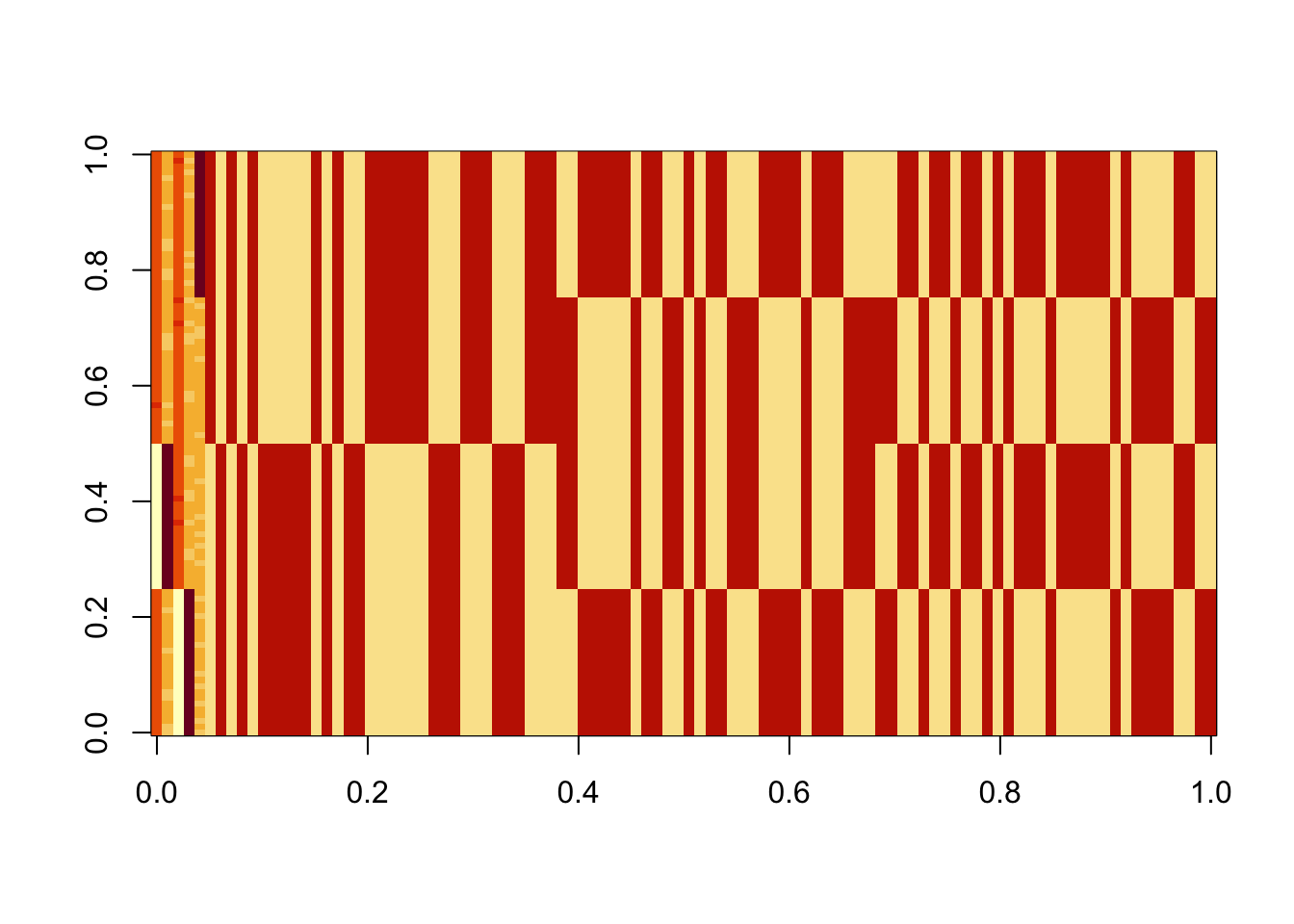
The top seed is a single group:
seed = order(obj,decreasing=TRUE)[1]
set.seed(seed)
res = list(w = c(rnorm(3),rep(0,7)), c=0)
for(i in 1:100)
res = fastica_r1update_wc(X1,res$w,res$c)
cor(L,t(X1) %*% res$w) [,1]
[1,] -0.5773503
[2,] 0.5773503
[3,] 0.3333332
[4,] -0.9999999
[5,] 0.3333333
[6,] 0.3333333plot(as.vector(t(X1) %*% res$w))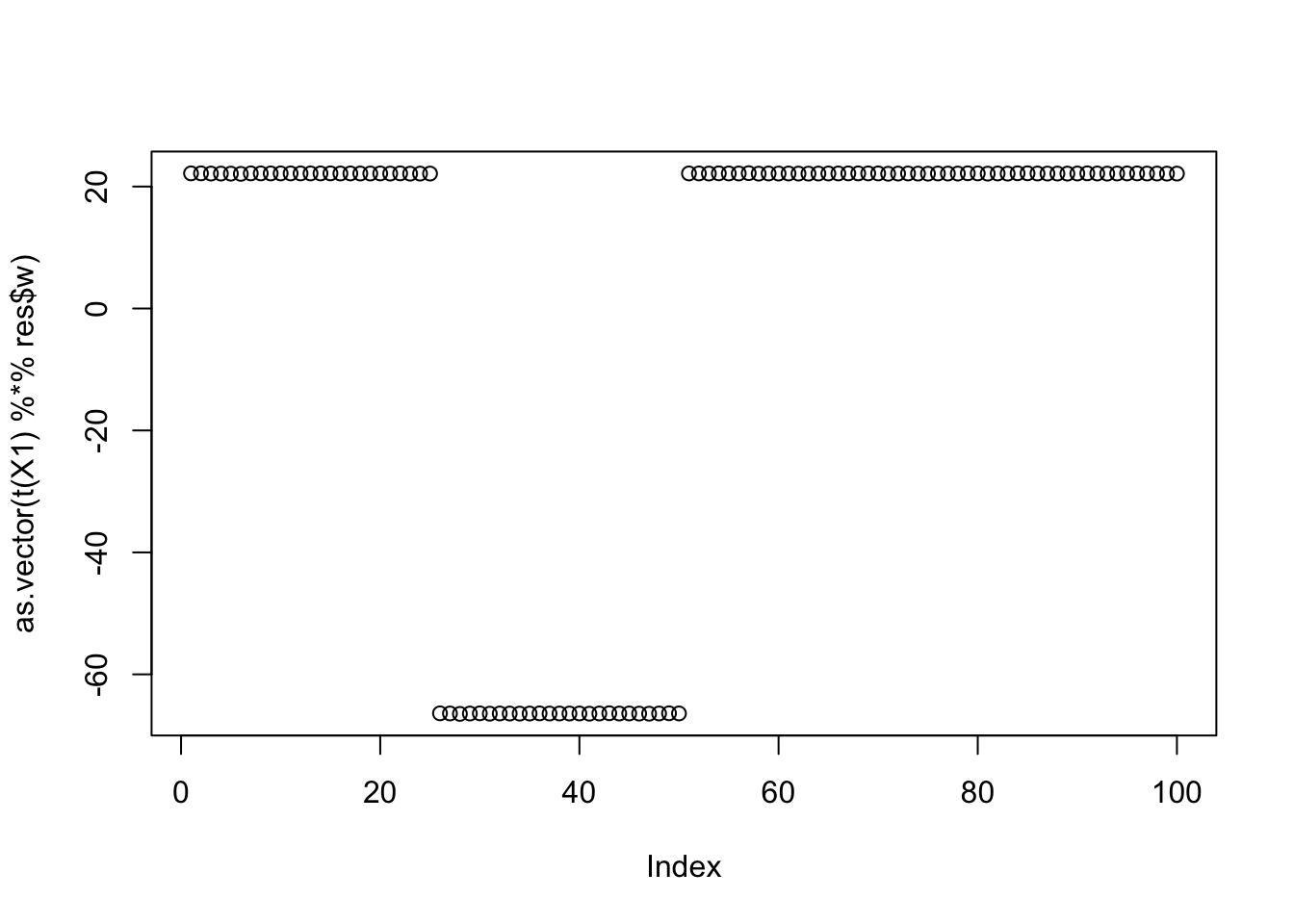
Results for the third biggest seed:
seed = order(obj,decreasing=TRUE)[3]
set.seed(seed)
res = list(w = c(rnorm(3),rep(0,7)), c=0)
for(i in 1:100)
res = fastica_r1update_wc(X1,res$w,res$c)
cor(L,t(X1) %*% res$w) [,1]
[1,] -0.5773502
[2,] 0.5773502
[3,] -0.9999999
[4,] 0.3333332
[5,] 0.3333334
[6,] 0.3333333plot(as.vector(t(X1) %*% res$w))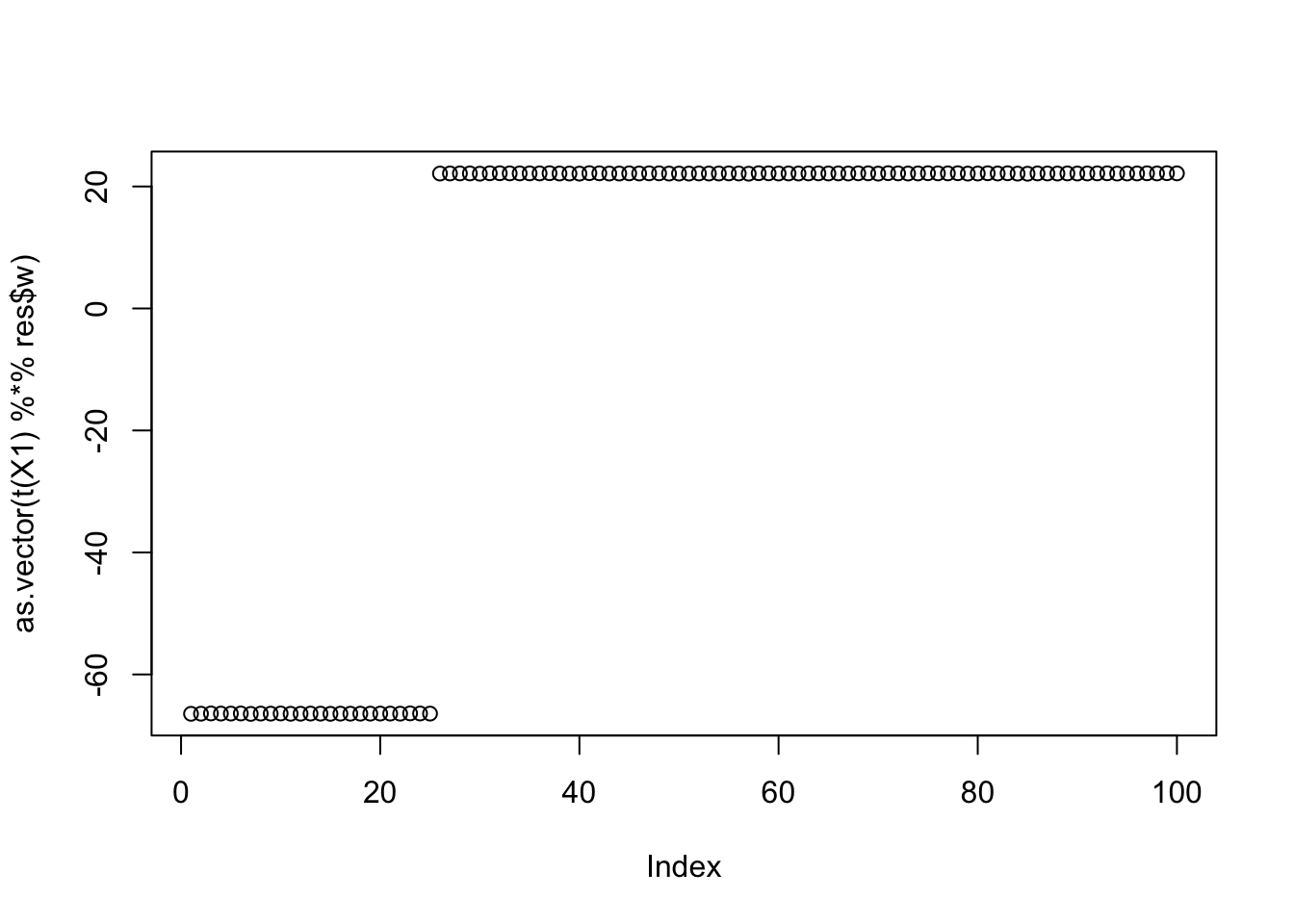
Noting here an idea to investigate: maybe initialize w “orthogonal” to previous ones (but don’t impose orthogonality during iterations) to try to find different solutions? Here’s what I mean. The problem is that there are 5 solutions we want to find here - not sure how to try to find all of them.
set.seed(1)
w.init = c(rnorm(3),rep(0,7))
w.init = w.init - sum(w.init * res$w)/(sum(res$w^2)) * res$w
res = list(w = w.init, c=0)
for(i in 1:100)
res = fastica_r1update_wc(X1,res$w,res$c)
cor(L,t(X1) %*% res$w) [,1]
[1,] 0.9999999
[2,] -0.9999999
[3,] 0.5773503
[4,] 0.5773502
[5,] -0.5773503
[6,] -0.5773502plot(as.vector(t(X1) %*% res$w))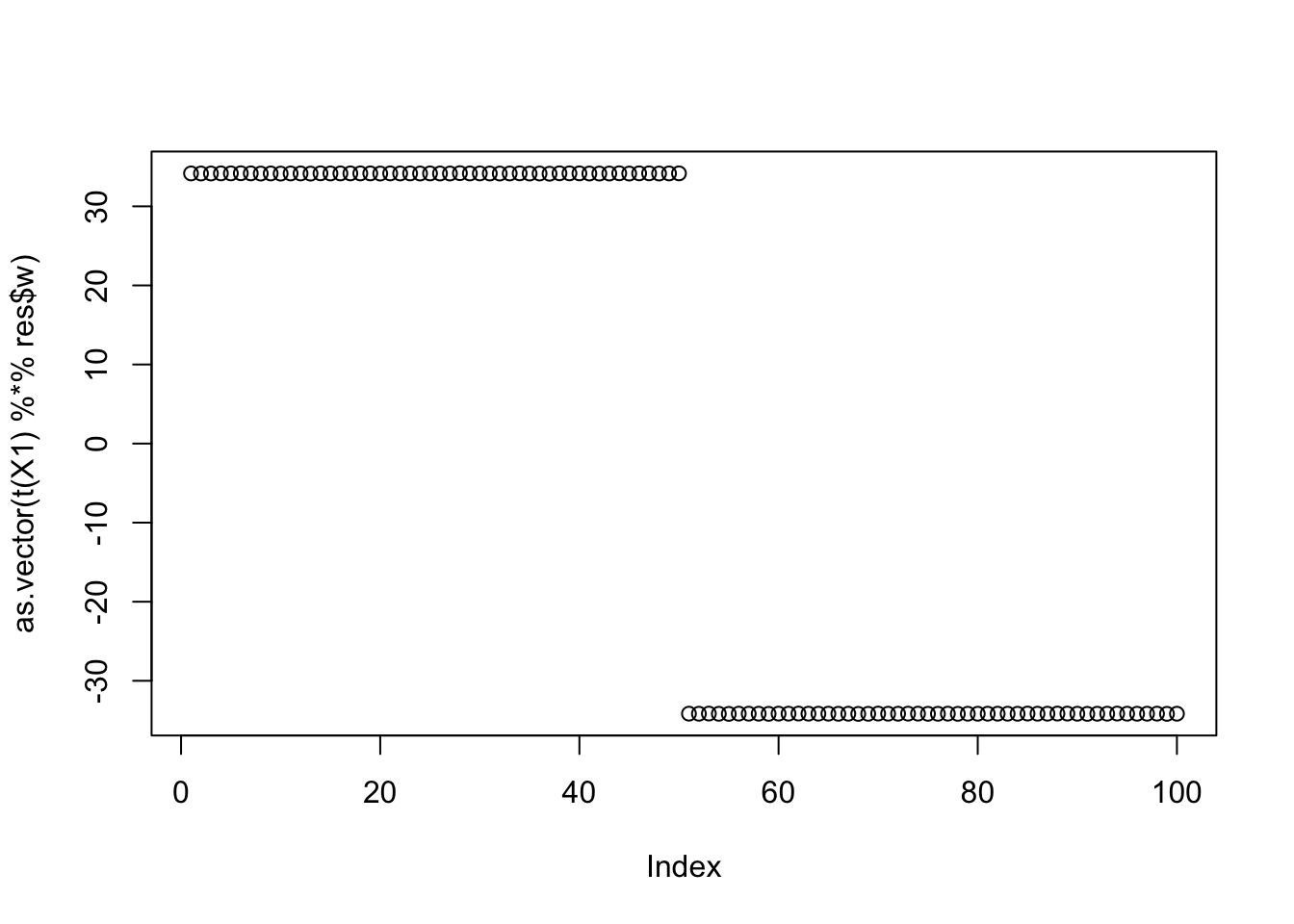
Joint updates (Ignore for now?)
After some more conversations with Gemini, we came up with another approach that uses the total derivative of the profiled objective \(J(w, c(w))\) to update \(w\). We also came up with an approximation for \(\hat{c}(w)\) as the negative cubed root of \(E(w^3)\). I am not sure that the update is correct, but Gemini gave me a new update rule for \(w\) that includes a correction term accounting for the dependence of \(c\) on \(w\), and also using the approximation for \(c\). I try this empirically here and include for completeness, but I think there are a number of issues and don’t think this is right. (It could be useful to investigate the approximation for c and the new update separately rather than doing both at once as here.) Just a placeholder for the idea that maybe we should jointly update w,c rather than block updates?
#'
#' This is the function that the "profiled" and "skew" update rules
#' are trying to find an extremum (max/min) of.
#'
#' @param X The pre-processed (centered and whitened) data matrix (p x n).
#' @param w The weight vector (p x 1).
#' @return The scalar value of the objective function.
compute_profiled_objective = function(X, w) {
# 1. Normalize w to stay on the ||w||=1 constraint
w = w / sqrt(sum(w^2))
# 2. Calculate the projection
P = as.vector(t(X) %*% w) # (n x 1) vector
# 3. Approximate c(w) from skewness (same as in your update rule)
skewness_estimate = mean(P^3)
c_approx = - sign(skewness_estimate) * (abs(skewness_estimate))^(1/3)
# 4. Create the shifted projection
S = P + c_approx
# 5. Apply G(u) = log(cosh(u))
G_S = log(cosh(S))
# 6. Compute the expectation (as a sample mean)
objective_value = mean(G_S)
return(objective_value)
}
#' Single-unit FastICA update using the total derivative of the
#' profiled objective J(w, c(w))
#'
#' @param X The pre-processed (centered and whitened) data matrix (p x n).
#' @param w The current weight vector (p x 1).
#' @return The updated, unnormalized weight vector 'w'.
fastica_r1update_profiled = function(X, w){
n_samples = ncol(X)
# 1. Normalize w
w = w / sqrt(sum(w^2))
# 2. Get projection and approximate c(w)
P = as.vector(t(X) %*% w)
skewness_estimate = mean(P^3)
c_approx = - sign(skewness_estimate) * (abs(skewness_estimate))^(1/3)
# 3. Create the shifted projection
P_shifted = P + c_approx
# 4. Calculate g(P) and g'(P)
G = tanh(P_shifted)
G2 = 1 - tanh(P_shifted)^2
# 5. Calculate all the expectation terms
# (Note: We use sums/matrix products for (n * E[...]))
E_Xg = X %*% G # n * E[X g(P)]
E_g2 = sum(G2) # n * E[g'(P)]
# --- New terms required for the correction ---
E_g = sum(G) # n * E[g(P)]
E_Xg2 = X %*% G2 # n * E[X g'(P)]
# Calculate the scalar correction factor
# (E_g / n) / (E_g2 / n) = E_g / E_g2
# Add epsilon for stability
correction_scalar = E_g / (E_g2 + 1e-6)
# 6. Calculate the new correction term vector
correction_term = correction_scalar * E_Xg2
# 7. Apply the full, new update rule
# w_new = (E_Xg - correction_term) - E_g2 * w
w_new = (E_Xg - correction_term) - E_g2 * w
return(w_new)
}set.seed(1)
w = X1 %*% L[,1] + rnorm(nrow(X1),0,5)
plot(t(X1) %*% w)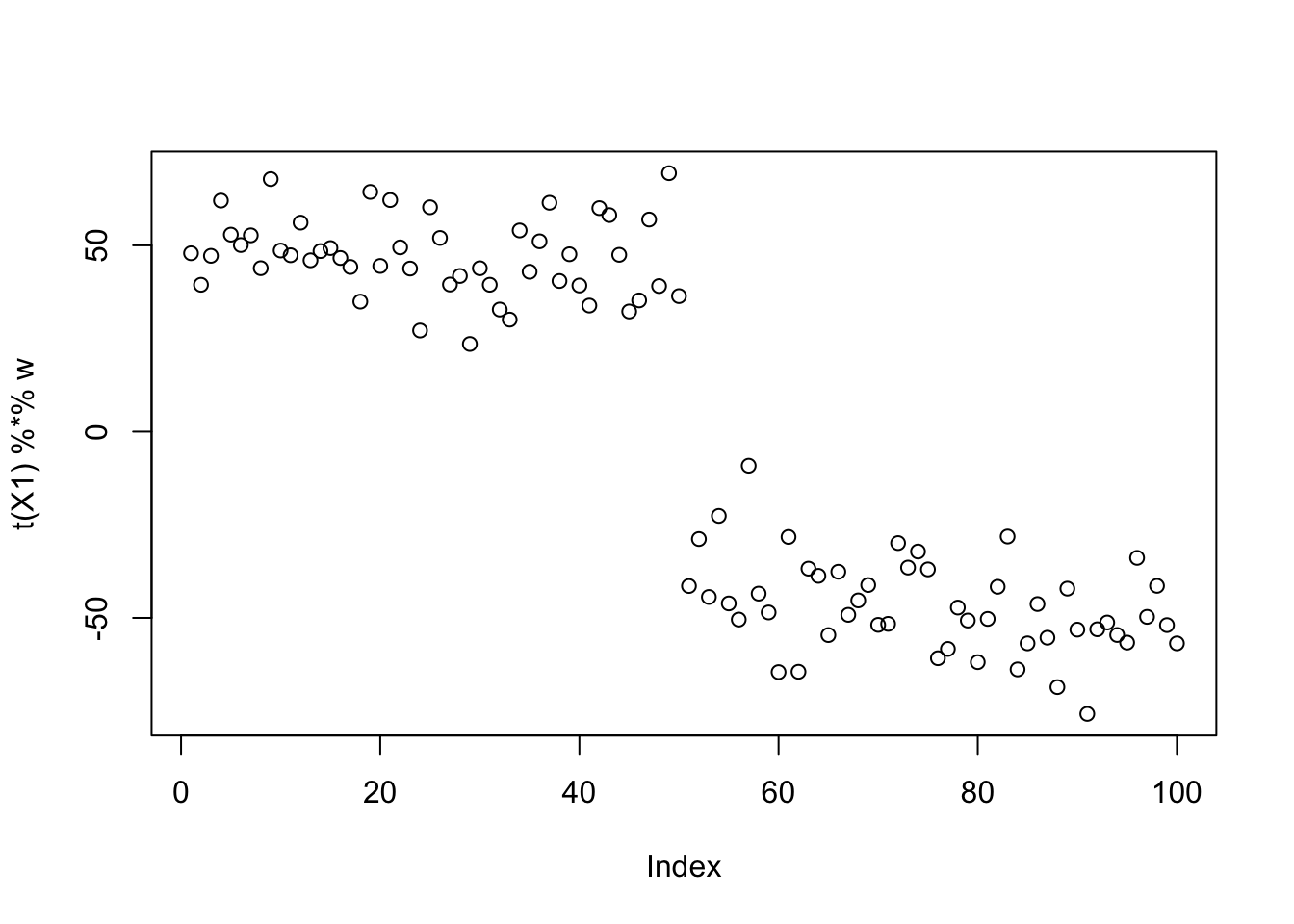
init_val = t(X1) %*% w
init_val = init_val/sqrt(mean(init_val^2))
for(i in 1:100)
w = fastica_r1update_profiled(X1,w)
cor(L,t(X1) %*% w) [,1]
[1,] 0.9999999
[2,] -0.9999999
[3,] 0.5773503
[4,] 0.5773502
[5,] -0.5773503
[6,] -0.5773502plot(as.vector(t(X1) %*% w))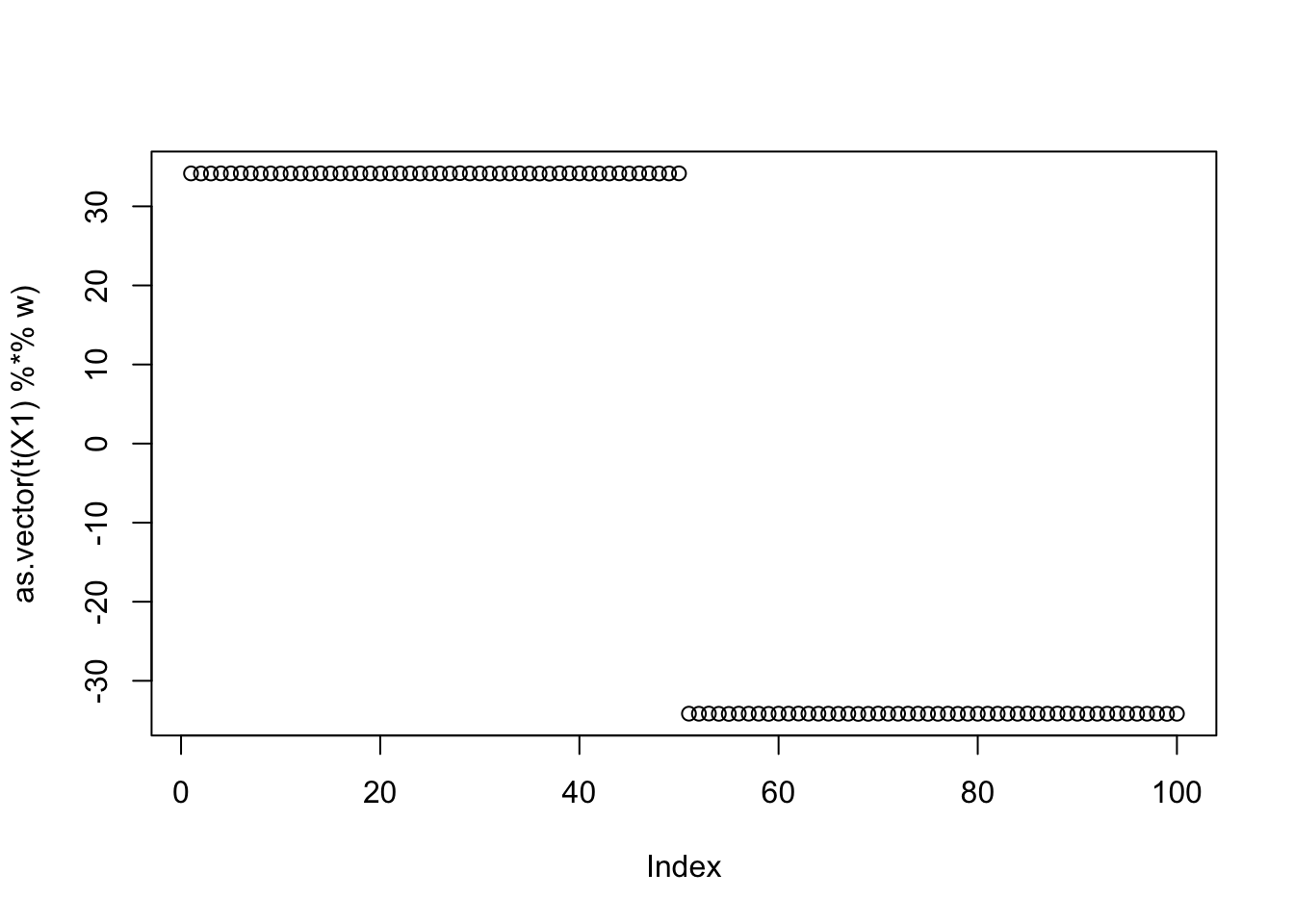
compute_profiled_objective(X1,w)[1] 0.4337842Try with more random start and several random seeds - there are lots of different results, and none of them reach as big a value of the objective.
obj = rep(0,100)
for(seed in 1:100){
set.seed(seed)
w = rnorm(nrow(X1))
for(i in 1:100)
w = fastica_r1update_profiled(X1,w)
obj[seed] = compute_profiled_objective(X1,w)
}
plot(obj)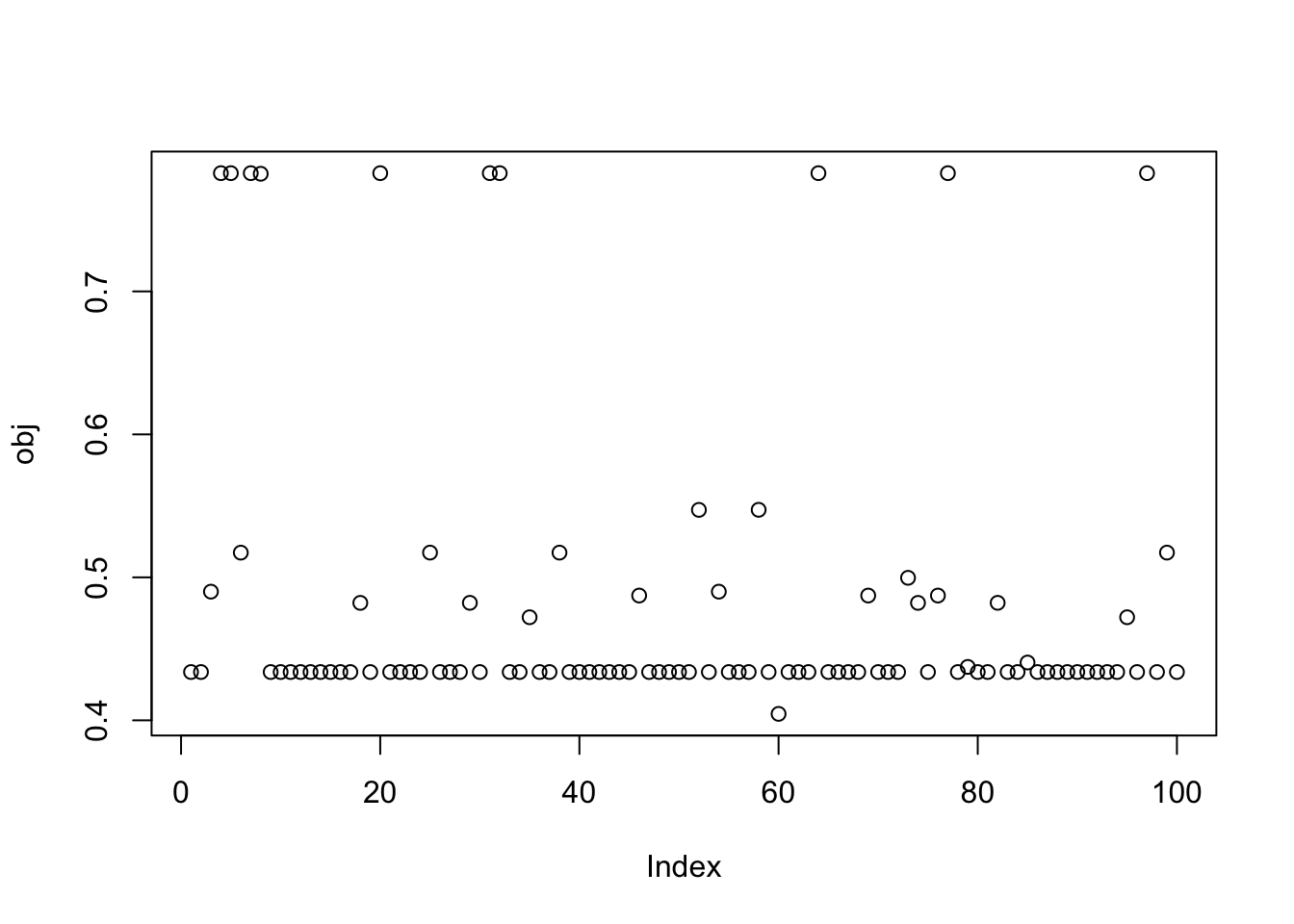
Here are the results for the seed achieving the max. It roughly combines two of the groups.
seed= which.max(obj)
set.seed(seed)
w = rnorm(nrow(X1))
for(i in 1:100)
w = fastica_r1update_profiled(X1,w)
cor(L,t(X1) %*% w) [,1]
[1,] -0.5773504
[2,] 0.5773504
[3,] 0.3333331
[4,] -0.9999999
[5,] 0.3333334
[6,] 0.3333334plot(as.vector(t(X1) %*% w))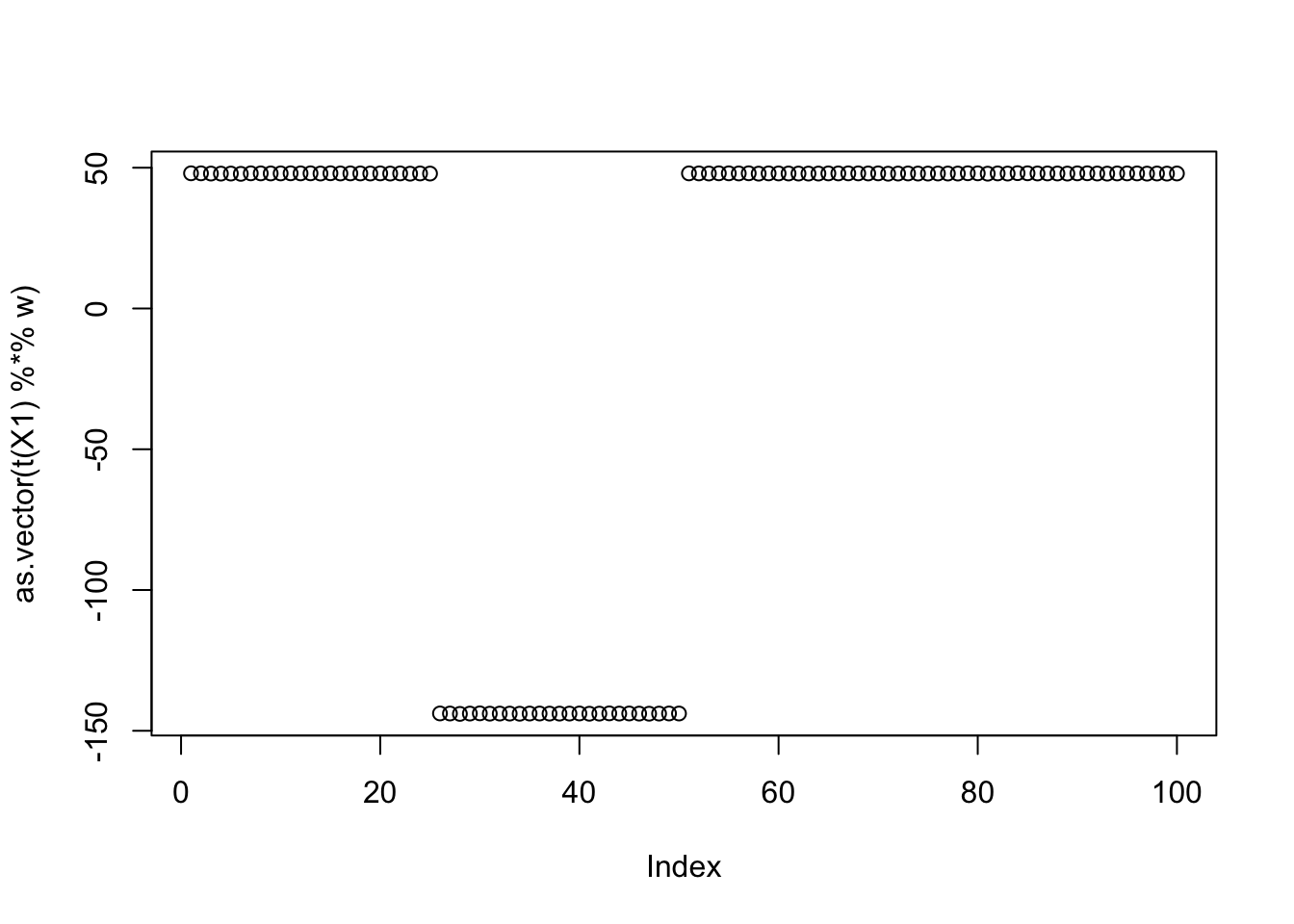
sessionInfo()R version 4.4.2 (2024-10-31)
Platform: aarch64-apple-darwin20
Running under: macOS Sequoia 15.6.1
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: America/Chicago
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
loaded via a namespace (and not attached):
[1] vctrs_0.6.5 cli_3.6.5 knitr_1.50 rlang_1.1.6
[5] xfun_0.52 stringi_1.8.7 promises_1.3.3 jsonlite_2.0.0
[9] workflowr_1.7.1 glue_1.8.0 rprojroot_2.0.4 git2r_0.35.0
[13] htmltools_0.5.8.1 httpuv_1.6.15 sass_0.4.10 rmarkdown_2.29
[17] evaluate_1.0.4 jquerylib_0.1.4 tibble_3.3.0 fastmap_1.2.0
[21] yaml_2.3.10 lifecycle_1.0.4 whisker_0.4.1 stringr_1.5.1
[25] compiler_4.4.2 fs_1.6.6 Rcpp_1.0.14 pkgconfig_2.0.3
[29] rstudioapi_0.17.1 later_1.4.2 digest_0.6.37 R6_2.6.1
[33] pillar_1.10.2 magrittr_2.0.3 bslib_0.9.0 tools_4.4.2
[37] cachem_1.1.0