svd_single_cell_data
Matthew Stephens
2021-05-25
Last updated: 2021-05-27
Checks: 7 0
Knit directory: stat34800/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20180411) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 8130c58. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Untracked files:
Untracked: analysis/currency_analysis.Rmd
Untracked: analysis/haar.Rmd
Untracked: analysis/stocks_analysis.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/svd_single_cell_data.Rmd) and HTML (docs/svd_single_cell_data.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 8130c58 | Matthew Stephens | 2021-05-27 | workflowr::wflow_publish(“analysis/svd_single_cell_data.Rmd”) |
Introduction
I wanted to illutrate SVD on the single cell data we used in the 2021 midterm.
Read in the data:
df = read.csv("../data/cell_data.csv",sep=",")
X = as.matrix(df[,-1])
head(df) cell_type MALAT1 RPL13 RPS2 RPL10 RPL13A RPS6 RPS18 RPS27 RPL32 RPS14 B2M
1 cytotoxic 15 16 19 14 15 7 9 16 11 10 4
2 cytotoxic 69 38 35 31 31 24 30 29 27 28 8
3 cytotoxic 55 33 26 22 16 23 30 26 15 13 19
4 cytotoxic 52 33 25 31 29 24 21 16 16 20 13
5 cytotoxic 44 24 12 13 13 10 19 13 10 6 20
6 cytotoxic 60 20 21 15 17 6 15 10 8 13 29
RPL3 RPS19 RPS12 RPS4X RPS3 TMSB4X RPLP2 RPL11 RPL21 RPL18A RPL19 RPLP1 RPS3A
1 11 15 11 9 14 2 7 6 5 10 8 6 6
2 25 30 15 25 28 14 16 18 24 16 27 16 21
3 18 19 15 14 17 22 14 19 12 15 15 8 8
4 18 14 16 15 11 18 20 26 15 17 24 15 6
5 13 14 10 4 11 18 13 10 9 11 8 3 8
6 7 6 8 6 9 19 10 3 9 5 6 3 2
RPL34 RPS15 RPS15A RPS27A RPL31 RPL28 RPL23A TMSB10 RPL12 RPL15 RPL18 RPL27A
1 8 6 7 12 4 6 10 2 5 4 3 3
2 20 16 16 17 13 14 20 10 19 8 14 10
3 5 15 5 13 9 13 18 15 4 13 16 11
4 7 14 7 5 13 9 16 11 12 17 12 14
5 3 16 9 9 8 3 5 8 5 9 7 10
6 8 14 7 5 8 3 12 7 3 4 6 4
RPS8 RPL7 RPS23 RPS25 RPL26 RPL10A RPS9 RPS16 RPL6 RPL9 RPS5 RPL35A RPL29
1 2 5 7 6 7 8 5 1 8 3 2 7 5
2 14 13 15 16 11 13 12 12 9 11 15 18 4
3 9 2 7 10 11 9 9 10 11 3 8 8 8
4 10 1 11 7 8 6 10 12 9 7 5 8 12
5 4 3 5 6 5 9 10 5 4 6 9 5 5
6 6 5 6 4 5 6 7 4 2 2 6 7 3
RPL8 RPS13 RPS7 RPL14 RPL36 RPL30 RPS28 HLA.B EEF1A1 RPL35 TPT1 RPL5 ACTB
1 4 5 7 9 7 7 7 4 5 5 2 8 0
2 15 12 9 6 11 11 12 7 12 6 15 8 5
3 6 3 13 6 5 5 12 10 6 9 8 5 5
4 8 14 11 9 7 9 10 9 7 4 6 6 4
5 1 2 7 3 7 5 5 7 1 8 3 0 8
6 2 3 7 5 6 4 5 7 4 5 2 2 7
MT.CO1 FAU GNB2L1 RPL7A RPSA RPS20 RPL4 UBA52 HLA.A FTL MT.CO3 RPL37A RPL27
1 2 3 2 4 1 5 3 2 3 2 1 0 2
2 5 11 8 10 7 15 10 3 6 2 2 3 6
3 4 7 4 10 8 3 6 4 9 9 5 2 2
4 9 5 9 5 4 7 5 5 5 3 6 6 7
5 4 6 1 4 2 3 3 3 2 8 0 4 1
6 8 1 3 2 6 2 4 2 7 3 3 0 0
RPLP0 PTMA HLA.C EEF1D RPS26 RPL37 MT.CO2 NACA RPS24 PFN1 RPL24 EIF1 JUNB
1 2 0 2 2 1 1 3 3 3 1 4 1 1
2 9 8 3 2 10 5 1 3 9 2 2 4 0
3 2 3 7 5 3 8 2 2 6 3 4 2 2
4 2 5 3 3 1 7 1 1 3 3 4 6 4
5 2 4 13 3 4 0 3 5 1 5 1 5 0
6 0 16 4 3 2 2 6 1 0 5 3 7 2
RPS11 RPS10 MT.ND4 MT.CYB RPS29 FTH1 EEF1B2 RPS21 RPL36A COX4I1 RPL38 LTB
1 1 1 1 3 2 1 2 3 2 0 2 1
2 5 3 3 3 8 2 2 2 7 3 8 2
3 4 3 3 2 4 5 0 3 3 5 6 4
4 5 5 0 1 6 4 3 5 4 7 2 4
5 2 1 1 1 4 2 0 7 4 4 3 0
6 1 5 6 7 3 3 1 1 3 1 2 0
CD52 ARHGDIB MT.ND2 PFDN5 RPL17 MT.ND1 GLTSCR2 CFL1 BTG1 RPL22 BTF3 SERF2
1 0 2 4 3 0 4 2 1 1 2 2 2
2 2 4 6 4 4 7 3 3 1 6 4 1
3 7 2 1 3 0 7 6 3 1 0 2 2
4 3 2 2 2 3 5 2 4 0 2 1 3
5 1 1 0 2 2 3 1 1 8 0 0 3
6 2 3 7 4 0 6 2 1 1 1 2 1
NPM1 SH3BGRL3 RPL23 HNRNPA1 PABPC1 LDHB RPL41 RPL36AL ACTG1 CD3D SLC25A6
1 0 0 0 1 1 1 3 0 1 0 2
2 3 0 3 1 3 3 4 3 0 2 3
3 2 4 2 2 0 2 2 1 2 1 0
4 4 0 1 1 3 3 1 5 0 2 0
5 0 2 1 1 1 0 3 2 2 1 1
6 1 1 0 2 1 0 0 1 1 1 1
H3F3B IL32 UBC CCL5 MYL12A HLA.E UBB CORO1A EEF2 TOMM7 CD3E CYBA DDX5 PTPRCAP
1 0 0 1 1 0 1 1 0 1 2 1 0 0 0
2 0 3 3 0 1 1 4 5 5 2 1 1 5 0
3 2 0 2 2 1 0 0 1 1 0 0 3 2 0
4 3 4 1 0 2 3 2 1 1 3 0 0 1 3
5 3 3 3 13 2 1 4 2 0 0 0 1 1 0
6 1 5 0 7 1 1 5 2 1 2 0 1 2 1
FXYD5 TMEM66 IER2 S100A4 ATP5L OAZ1 HCST JUN EIF3K ATP5G2 S100A6 CD8B COX7C
1 1 0 2 0 2 0 1 1 1 0 1 1 0
2 1 0 0 1 0 0 2 0 1 0 1 0 1
3 2 2 1 0 0 0 5 0 2 2 1 1 2
4 1 3 2 0 2 3 1 0 1 0 0 1 2
5 1 2 0 1 1 1 2 4 0 1 0 0 3
6 2 0 0 2 0 2 3 1 2 3 4 0 1
ATP5E GAPDH TMA7 NOSIP MT.ATP6 COMMD6 ITM2B MYL6 YBX1 HINT1 SNRPD2 VIM CD7
1 1 2 1 0 0 0 2 2 0 1 0 1 2
2 1 1 0 0 0 1 2 2 0 1 1 1 1
3 2 1 2 0 2 1 3 2 1 1 4 2 1
4 3 3 2 2 0 1 1 0 0 1 1 0 1
5 2 0 2 0 1 0 3 0 1 0 0 2 0
6 2 3 1 1 2 2 2 1 2 1 0 0 0
LIMD2 EDF1 DUSP1 GMFG PPDPF GPSM3 SRP14 GYPC CIRBP C6orf48 IFITM2 PSME1 CALM1
1 0 0 0 0 0 0 2 0 0 0 1 0 0
2 3 2 1 2 1 0 1 3 1 1 0 0 1
3 3 0 0 0 5 1 1 2 0 0 0 4 1
4 0 1 1 4 1 1 1 1 0 1 1 3 1
5 1 0 3 1 1 0 1 2 0 0 0 0 2
6 0 0 1 1 0 0 1 1 0 0 2 2 1
EIF3H SRSF5 CD37 HSPA8 RAC2 AES SSR2 EIF3F CHCHD2 OST4 GIMAP7 ARPC3 ARPC1B
1 1 1 0 1 0 0 1 1 0 0 0 0 3
2 0 1 0 1 0 3 2 1 1 2 0 1 0
3 1 1 0 2 0 0 1 1 2 0 3 0 0
4 1 1 1 1 0 2 1 1 1 1 1 0 1
5 0 1 1 0 1 0 2 1 2 1 0 0 0
6 0 1 0 1 0 0 1 0 1 1 1 1 0
NKG7 PPIA EIF4A2 ARPC2 C19orf43 UQCR11 S100A10 UXT EVL EIF3L TXNIP MZT2B
1 0 0 0 0 1 0 0 0 0 0 0 1
2 1 0 0 0 1 0 1 0 0 2 4 1
3 0 0 4 1 0 2 2 0 0 1 0 1
4 0 0 4 1 2 4 0 1 1 1 3 1
5 4 2 0 2 1 2 1 0 0 0 0 0
6 7 0 0 1 0 1 1 0 0 0 0 1
ATP5D ZFAS1 EMP3 SSR4 HNRNPA2B1 EIF3E HSP90AA1 APRT FOS CD48 TSC22D3 CD27
1 0 0 0 0 0 0 0 0 0 0 0 0
2 0 0 1 0 0 1 2 1 0 0 2 0
3 0 0 0 2 0 0 1 0 1 1 0 0
4 0 0 1 0 1 0 0 1 0 1 1 1
5 0 0 2 0 1 0 0 0 0 0 0 3
6 0 0 2 0 0 0 1 0 2 1 5 0
CTSW VAMP2 NDUFA4 PNRC1 HNRNPDL UQCRH GNLY COX6B1 ZFP36 MT.ND5 MYL12B EIF3G
1 0 0 0 0 0 0 0 0 1 2 0 3
2 0 0 0 4 0 0 0 0 0 1 0 1
3 1 2 0 0 0 0 0 0 0 1 1 2
4 1 2 0 3 0 2 0 0 0 0 0 1
5 0 1 1 3 1 1 0 1 1 0 0 0
6 1 1 0 0 1 0 4 2 0 2 0 0
UBXN1 HIGD2A RGS10 ATP5O ERP29 CNBP ISG20 ATP6V1G1 ALDOA LAPTM5 NAP1L1 COX6C
1 0 1 0 0 1 0 0 0 0 0 0 0
2 2 1 0 0 0 1 2 0 0 0 0 0
3 0 2 0 0 0 1 0 0 1 0 0 0
4 0 0 0 2 1 1 1 0 0 0 1 0
5 0 0 0 0 1 2 0 1 1 1 0 0
6 0 0 0 0 0 0 1 0 0 1 1 1
ANAPC16 LCK SOD1 RPSAP58 LSP1 GZMM CAPZB C19orf53 EIF4A1 NDUFB11 CUTA HMGN1
1 0 0 0 0 0 0 0 1 0 0 0 1
2 0 0 1 0 0 0 0 0 0 1 0 0
3 0 1 0 0 2 0 1 1 2 1 1 0
4 2 0 0 0 1 1 0 0 0 0 1 0
5 0 1 2 0 2 2 0 1 1 0 1 0
6 0 2 0 0 0 0 1 1 1 1 1 0
RARRES3 ZFP36L2 HMGB1 LY6E SMDT1 PSMA7 S100B TRAF3IP3 C11orf31 ARL6IP4 MIF
1 0 0 1 0 0 0 0 0 0 2 0
2 0 0 0 0 2 0 3 0 0 0 0
3 0 1 1 1 0 0 0 2 1 0 0
4 0 3 1 1 0 1 0 0 0 1 0
5 1 0 1 2 0 0 0 0 1 1 0
6 1 1 2 0 1 0 0 0 1 0 1
NDUFS5 C12orf57 CXCR4 TCEB2 ST13 PCBP2 NFKBIA CCNI GUK1 NBEAL1 GPX4 ENO1
1 0 0 0 0 0 0 0 1 0 0 0 1
2 0 0 0 0 0 1 1 1 0 0 0 1
3 0 0 1 1 0 0 2 0 0 0 0 1
4 1 0 1 0 0 0 0 1 2 2 0 0
5 3 1 0 3 0 0 0 1 1 0 0 1
6 0 0 0 0 0 0 0 0 0 0 1 0
CCDC85B HSP90AB1 ACAP1 NEDD8 RBM3 IFITM1 SERP1 UBE2D2 TAGLN2 IL2RG C9orf16
1 0 1 0 0 0 1 0 0 0 1 0
2 0 1 0 1 0 0 1 0 0 0 1
3 1 0 0 0 3 0 0 0 0 0 0
4 0 0 0 1 4 0 0 0 0 1 0
5 1 0 0 0 1 0 2 0 1 0 0
6 0 0 1 0 1 0 0 0 0 1 0
COX5B SAP18 LAMTOR4 UBL5 PSME2 SKP1 RP11.291B21.2 SNHG8 ISCU UBE2D3 NDUFA11
1 1 1 1 0 1 0 0 0 0 0 0
2 0 1 0 0 0 0 0 3 1 3 0
3 0 0 0 0 0 0 0 1 1 1 0
4 0 0 0 3 2 0 0 1 1 0 0
5 0 2 0 0 2 0 0 0 2 1 1
6 1 0 0 0 0 0 0 0 1 0 0
HNRNPA0 FBL DRAP1 GTF3A ID2 SF3B5 PRR13 ATP6V0E1 CD8A KLF2 TSTD1 TPI1 NDUFB9
1 1 1 1 0 1 0 0 0 0 0 0 0 1
2 1 1 1 0 0 1 0 0 0 1 0 0 0
3 1 0 0 0 1 0 0 0 2 0 1 0 0
4 2 2 0 0 0 0 0 0 0 0 1 0 1
5 0 1 0 0 0 0 1 0 0 0 0 1 0
6 0 0 1 0 0 0 0 1 0 1 0 0 1
CCND3 VAMP8 CD99 SUMO2 NDUFB8 ICAM3 ANP32B SELL SLC25A3 CD74 COX6A1 TRMT112
1 0 0 0 0 0 0 1 0 0 0 0 1
2 0 0 1 0 0 1 0 0 1 0 0 0
3 0 0 0 1 1 0 1 1 0 0 0 1
4 0 0 0 1 2 0 1 0 0 0 1 1
5 0 1 1 0 0 0 0 0 1 0 0 0
6 0 0 1 1 0 1 0 0 0 2 1 0
IL7R COX7A2 HSPB1 TBC1D10C NDUFA13 PARK7 JTB UCP2 SEPT1 PCBP1 COTL1 CALM3
1 0 1 0 0 0 0 1 0 0 0 0 0
2 0 0 0 0 0 0 0 0 0 0 0 0
3 2 0 1 0 1 0 0 0 2 0 0 0
4 0 0 0 1 0 0 0 1 0 0 2 0
5 0 0 0 0 0 0 0 0 1 0 1 0
6 0 1 0 0 1 0 1 0 0 0 0 0
ATPIF1 LAT SLC25A5 POLR2L ATP5A1 CLIC1 CALM2 RPL39 PRDX2 PEBP1 SEPT7 DUSP2
1 0 0 1 0 1 0 0 0 1 0 0 1
2 0 0 0 0 0 0 0 1 0 1 0 0
3 0 0 0 0 1 0 1 0 0 1 0 1
4 0 0 0 0 1 1 0 0 1 0 0 1
5 0 0 0 0 0 0 0 0 0 0 0 0
6 0 1 1 0 1 2 1 0 0 2 1 1
CD53 EIF3D RSL1D1 GSTK1 SRSF3 C9orf142 PPP1CA TPM3 NDFIP1 NDUFA1 LSM7 UFC1
1 0 0 0 0 0 0 0 1 0 0 1 0
2 0 0 0 1 0 0 0 0 1 0 0 0
3 0 0 0 0 2 1 0 0 1 0 1 0
4 0 0 0 1 0 0 1 0 3 1 0 0
5 1 0 0 0 0 1 0 0 1 0 1 1
6 0 0 0 0 0 0 1 1 0 1 0 0
COX7A2L SPCS1 NDUFB2 CIB1 ALKBH7 HNRNPK LEPROTL1 SNHG7 NUCB2 TMEM258 MZT2A
1 0 0 0 0 0 0 0 0 0 0 0
2 0 0 0 0 0 0 1 2 1 1 0
3 0 0 1 0 0 2 0 0 0 0 0
4 0 1 0 0 1 0 0 2 0 0 1
5 0 0 0 0 0 0 0 0 2 0 0
6 0 0 0 0 0 0 0 0 0 1 0
PIK3IP1 MORF4L1 ARPC5 FLT3LG RAN PNISR COPE C4orf3 TRAPPC1 C19orf60 RABAC1
1 0 0 0 0 1 0 0 0 0 0 0
2 1 0 0 0 0 1 0 0 0 0 0
3 0 0 0 0 3 1 0 0 0 0 1
4 1 0 0 0 1 1 0 0 2 0 0
5 0 0 0 0 0 0 3 0 0 0 0
6 0 0 1 0 0 0 1 0 0 0 1
TBCA OCIAD2 CD3G CNN2 FKBP8 TSPO ATP5H SCAND1 SRSF7 BRK1 WDR83OS TMBIM6 PRMT2
1 1 0 1 1 0 0 0 0 0 0 0 0 0
2 0 0 0 0 1 0 0 0 1 0 0 0 0
3 2 0 0 0 1 0 1 0 0 0 0 1 1
4 0 0 0 0 1 0 0 1 0 1 0 1 1
5 1 0 0 0 0 1 0 0 0 0 0 0 0
6 0 0 0 1 0 1 0 0 0 0 0 0 0
PSMB1 YWHAB UQCRQ PRKCQ.AS1 GPX1 ATP6V1F PGK1 KRT10 STK17A PRDX6 POLD4 GSTP1
1 0 2 0 1 0 0 1 0 0 0 0 1
2 1 0 0 0 0 0 0 0 0 0 0 0
3 0 0 0 1 0 0 0 0 0 0 0 0
4 0 1 2 0 1 0 0 0 0 1 0 0
5 0 0 1 0 1 0 0 0 1 0 0 1
6 0 1 0 0 0 0 0 0 1 0 0 0
EIF3M LITAF UQCR10 PPA1 HNRNPC PTPRC ATP5J SON PSMB9 C11orf58 CSTB MRPS21
1 0 0 0 0 0 1 0 0 0 0 0 0
2 1 0 1 0 0 0 1 0 0 0 0 0
3 0 0 1 0 0 0 0 1 1 2 0 0
4 1 1 1 0 0 0 1 0 0 0 0 0
5 0 1 0 0 0 0 1 0 0 0 0 2
6 0 0 1 3 0 0 0 0 1 1 0 0
LDHA BIN2 PPIB ATP5J2 NHP2L1 ITGB2 SNRPB BIN1 LSM4 PPP1R15A COX8A ATP5F1
1 0 0 0 0 0 0 0 0 0 0 0 0
2 0 1 1 0 0 0 0 1 0 0 0 1
3 1 0 0 0 1 1 0 0 0 0 0 1
4 2 0 0 1 0 1 0 0 0 0 0 1
5 0 0 1 0 1 1 0 0 0 1 1 1
6 0 0 0 0 0 1 1 0 0 0 0 0
PTGES3 BTG2 RHOA SEPW1 KRTCAP2 PRELID1 C1QBP SEC62 RWDD1 CD2 CCR7 PSMB8 COX5A
1 0 2 0 0 0 0 0 0 1 0 0 0 0
2 0 0 0 1 1 0 0 1 0 0 0 0 0
3 0 0 0 0 0 0 0 0 2 0 0 1 0
4 0 0 1 0 1 0 1 1 0 1 0 0 1
5 2 0 0 1 0 0 0 0 0 1 0 1 0
6 1 0 0 0 0 0 0 0 2 2 0 0 0
MYEOV2 GADD45GIP1 AIF1 C9orf78 DNAJB1 LEF1 NHP2 ATP5G3 CD44 MPHOSPH8 NDUFB10
1 0 0 0 0 0 0 1 0 0 0 0
2 0 0 0 1 2 0 1 1 0 0 0
3 1 0 0 0 0 0 1 0 0 0 0
4 1 1 0 0 1 1 0 0 0 0 0
5 0 0 0 0 0 0 0 0 0 1 0
6 0 0 0 0 0 0 0 0 0 0 0
FYB FIS1 TUBA1B LBH TUBA1A SYF2 GZMA CRIP1 DDT SUN2 GLRX HNRNPM HLA.F PKM
1 1 0 0 0 0 0 0 0 0 1 0 0 0 0
2 0 0 0 0 1 1 0 0 1 0 1 0 0 0
3 0 0 0 0 0 0 0 0 1 0 0 0 0 0
4 0 1 0 1 1 0 0 0 0 0 1 1 0 0
5 0 0 0 1 0 0 3 1 0 0 0 0 0 2
6 0 1 0 1 4 1 1 1 1 0 0 0 0 0
SEPT9 TMEM59 TMEM230 CHCHD10 CAP1 ECH1 PRDX5 CD69 ERGIC3 SSU72 EID1 SERBP1
1 0 1 0 0 0 0 1 0 0 0 0 0
2 0 0 0 1 0 0 0 0 1 0 0 1
3 0 2 1 0 0 0 0 1 0 0 0 0
4 0 0 0 0 0 1 0 0 1 0 0 0
5 0 0 1 1 0 1 0 0 0 0 2 0
6 0 0 0 0 0 0 0 1 1 0 0 0
GZMK DAD1 CMPK1 RNASET2 PAIP2 ATP5B CST7 H2AFZ PSMC5 CCDC109B PA2G4 RASGRP2
1 0 0 1 0 0 0 0 0 0 0 0 0
2 0 0 0 1 2 1 0 0 0 0 1 0
3 0 0 0 0 0 1 0 0 0 0 0 0
4 0 0 0 0 0 0 0 0 0 0 0 0
5 4 0 0 1 0 0 2 1 0 0 1 0
6 2 0 0 0 0 1 0 0 0 1 0 0
MINOS1 RPS19BP1 C19orf70 TMEM123 TUFM MT.ND3 AURKAIP1 U2AF1 VPS28 DDIT4
1 0 0 1 0 1 0 0 0 1 0
2 0 0 0 0 0 0 0 0 0 0
3 1 0 1 0 1 0 1 0 1 0
4 0 0 0 1 0 0 0 0 0 0
5 0 1 1 0 0 0 0 1 0 0
6 0 0 0 0 0 1 0 0 0 0
MRPL23 RBM39
1 0 0
2 0 0
3 0 0
4 0 2
5 0 0
6 0 0Here I simply run svd and plot the first two eigenvectors against one another. (One could imagine wanting to standardize the columsn first, but I did not do that for now).
X.svd = svd(X)
plot(X.svd$u[,1],X.svd$u[,2],col=c(rep(1,1000),rep(2,1000)), main = "first PC vs second PC, colored by cell type")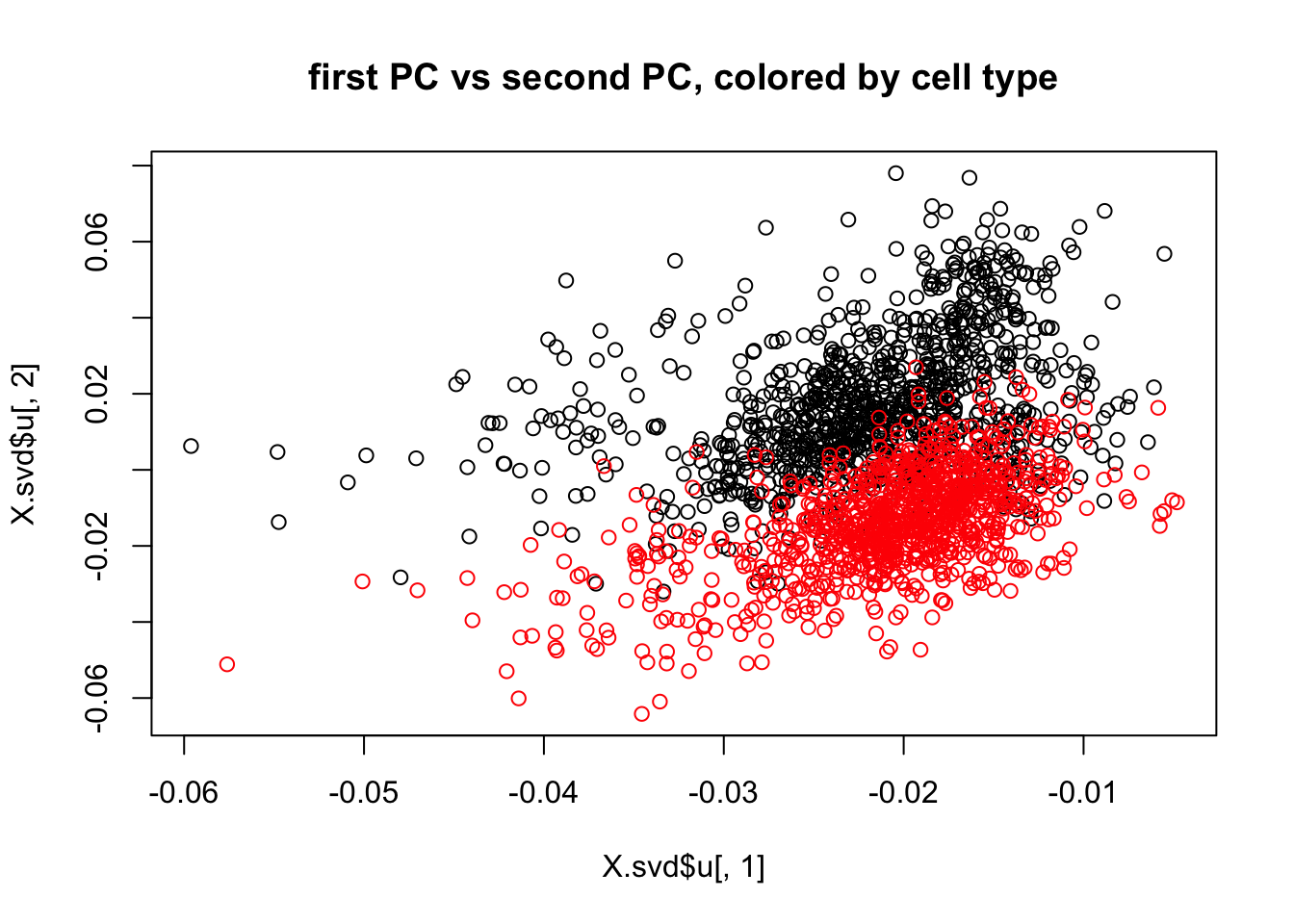
So it is seems that the cell types separate somewhat in PC space (even though there is considerable overlap). And yet the EM algorithm for Poisson clustering did not result in these two clusters. Why not?
Have a look at PC1. It turns out that this is basically the total counts for each cell:
plot(X.svd$u[,1],rowSums(X))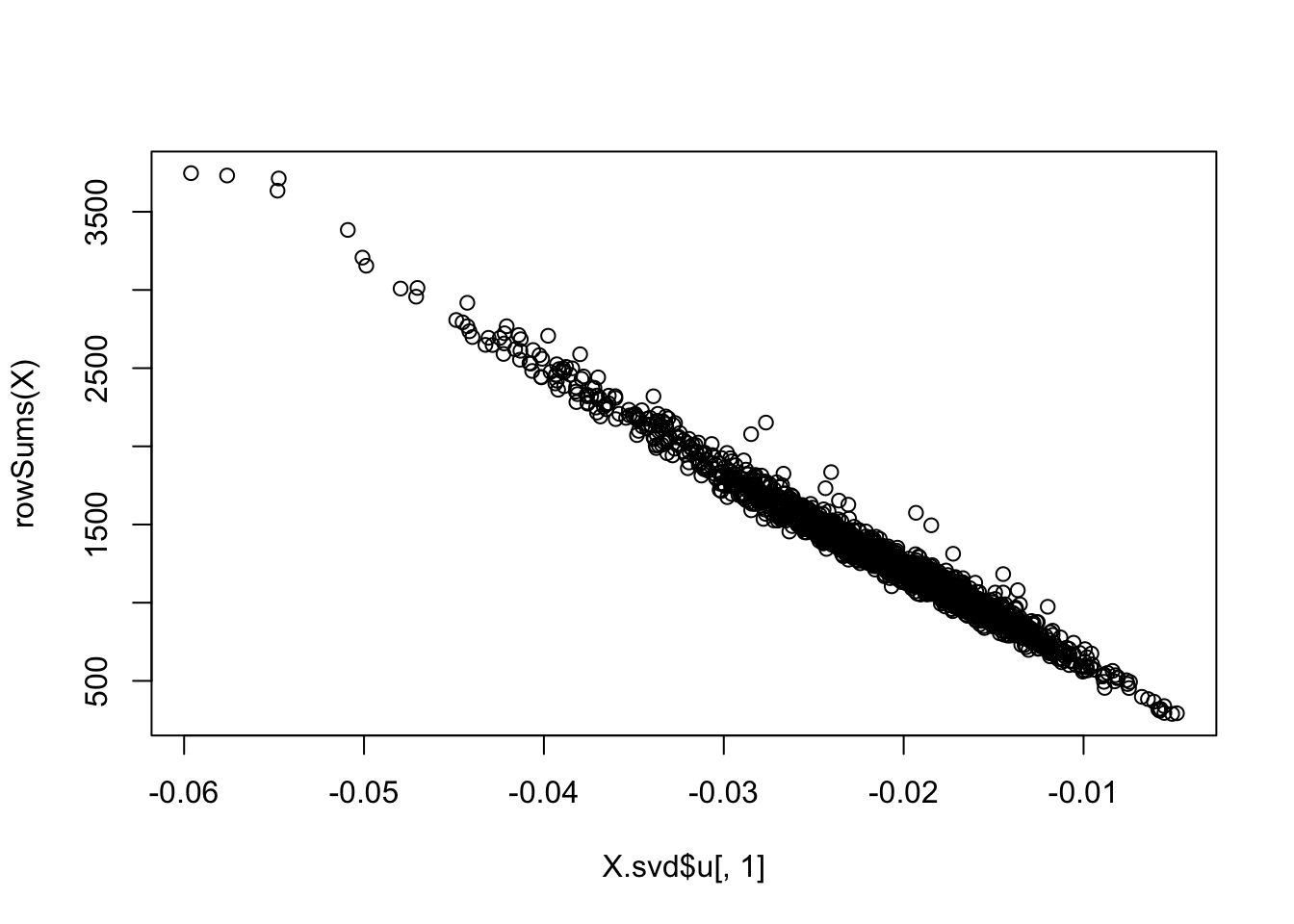
So what is going on is this. the Poisson cluster model ends up splitting the cells on PC1, basically splitting the cells by the total counts. This is like splitting documents into groups based on how long they are (how many words) rather than what the words say… PC2 is more about what the words say.
Basically, if we want to model based approach to have a chance to split into two groups we need to control for the fact that different cells have different total counts in the model.
sessionInfo()R version 3.6.0 (2019-04-26)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS 10.16
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] workflowr_1.6.2
loaded via a namespace (and not attached):
[1] Rcpp_1.0.6 rstudioapi_0.13 whisker_0.4 knitr_1.29
[5] magrittr_2.0.1 R6_2.4.1 rlang_0.4.10 stringr_1.4.0
[9] tools_3.6.0 xfun_0.16 git2r_0.27.1 htmltools_0.5.0
[13] ellipsis_0.3.1 rprojroot_1.3-2 yaml_2.2.1 digest_0.6.27
[17] tibble_3.0.4 lifecycle_1.0.0 crayon_1.3.4 later_1.1.0.1
[21] vctrs_0.3.8 promises_1.1.1 fs_1.5.0 glue_1.4.2
[25] evaluate_0.14 rmarkdown_2.3 stringi_1.4.6 compiler_3.6.0
[29] pillar_1.4.6 backports_1.1.10 httpuv_1.5.4 pkgconfig_2.0.3