Eight schools data
Matthew Stephens
April 11, 2018
Last updated: 2018-05-21
workflowr checks: (Click a bullet for more information)-
✔ R Markdown file: up-to-date
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
-
✔ Environment: empty
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
-
✔ Seed:
set.seed(20180411)The command
set.seed(20180411)was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible. -
✔ Session information: recorded
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
-
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.✔ Repository version: 9548ba6
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can usewflow_publishorwflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.Ignored files: Ignored: .DS_Store Ignored: .Rhistory Ignored: .Rproj.user/ Ignored: .sos/ Ignored: exams/ Ignored: temp/ Untracked files: Untracked: analysis/hmm.Rmd Untracked: analysis/neanderthal.Rmd Untracked: analysis/pca_cell_cycle.Rmd Untracked: analysis/ridge_mle.Rmd Untracked: data/reduced.chr12.90-100.data.txt Untracked: data/reduced.chr12.90-100.snp.txt Untracked: docs/figure/hmm.Rmd/ Untracked: docs/figure/pca_cell_cycle.Rmd/ Untracked: homework/fdr.aux Untracked: homework/fdr.log Untracked: tempETA_1_parBayesC.dat Untracked: temp_ETA_1_parBayesC.dat Untracked: temp_mu.dat Untracked: temp_varE.dat Untracked: tempmu.dat Untracked: tempvarE.dat Unstaged changes: Modified: analysis/cell_cycle.Rmd Modified: analysis/density_est_cell_cycle.Rmd Modified: analysis/eb_vs_soft.Rmd Modified: analysis/svd_zip.Rmd
Expand here to see past versions:
Introduction
Here I apply the adaptive shrinkage EB method to the eight schools data at (http://andrewgelman.com/2014/01/21/everything-need-know-bayesian-statistics-learned-eight-schools/)
Enter the data
x = c(28,8,-3,7,-1,1,18,12)
s = c(15,10,16,11,9,11,10,18)Plot the data
Notice how the intervals very much overlap… suggesting the data may be consistent with no differences
library(rmeta)
metaplot(x, s)
Apply adaptive shrinkage
Here we apply the adaptive shrinkage package, which solves the normal means problem with unimodal prior. By default the mode is 0, which is not appropriate for these data. So we estimate the mode:
library(ashr)
a = ash(x,s,mode="estimate")
get_pm(a) # get the posterior mean[1] 7.685617 7.685617 7.685617 7.685617 7.685617 7.685617 7.685617 7.685617We can see that ash shrinks all the estimates to the same value, meaning the data are “most” consistent with no variation in effect.
Check adaptive shrinkage
We might be worried maybe that we mis-used the software, or that it is not working right. Does it always overshrink like that?
When you get an unexpected result it is important to go back and check your understanding. A good way to do this is to make a prediction and then test it.
For example, here I predict that if the standard errors for these data had been much smaller (with same x) then ash should not shrink as much, because the data would be more convincing of true variation in effect.
I can test this prediction by dividing s by 10 say:
metaplot(x,s/10)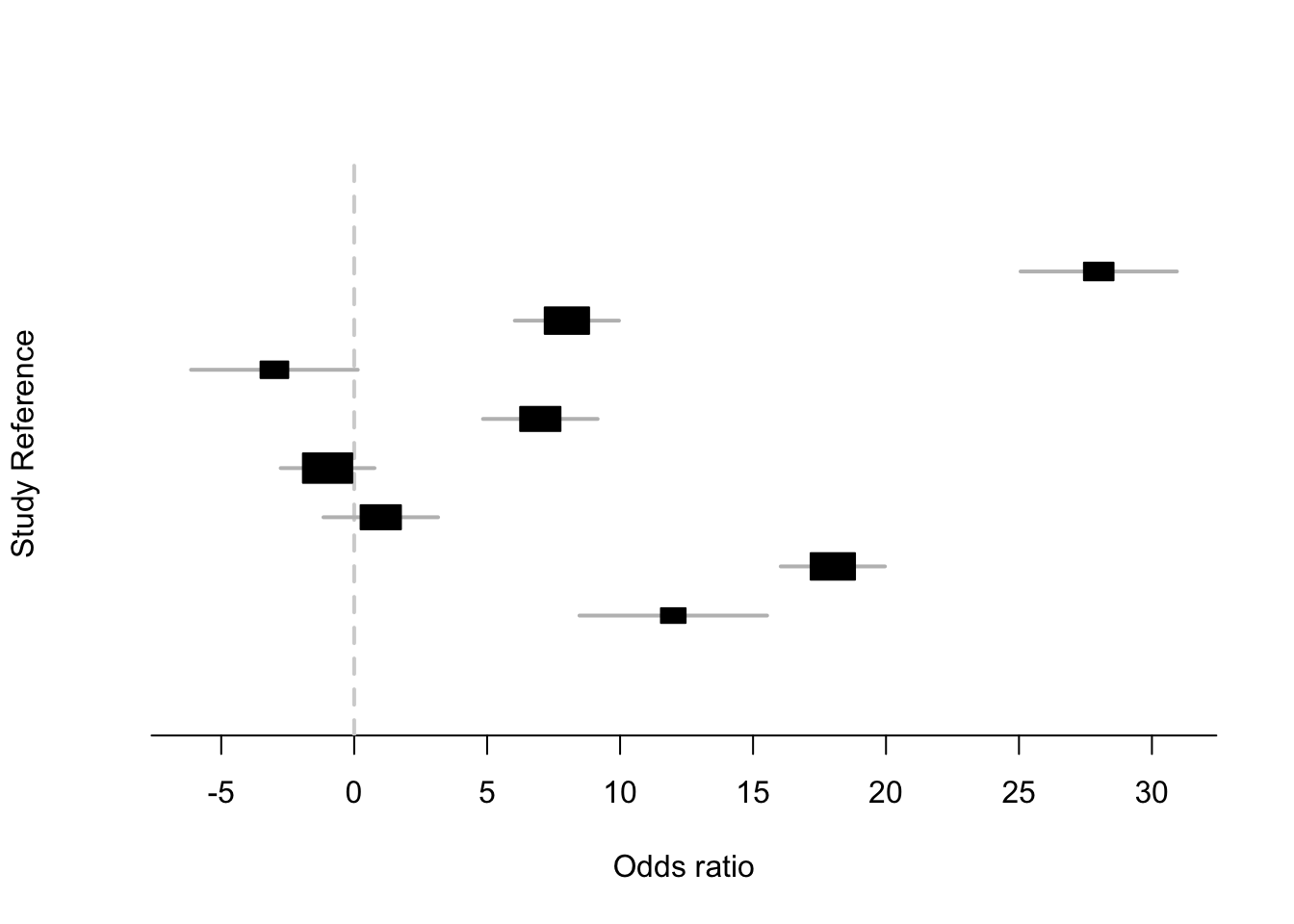
a = ash(x,s/10,mode="estimate")
get_pm(a)[1] 26.766778 7.971568 -2.957616 7.918593 -1.000000 1.000000 17.999958
[8] 9.721773We can see my prediction was correct. So this gives me more confidence that I used the software correctly and that it is behaving sensibly.
Remember: The most valuable opportunties to learn come from when you see something that you did not expect! Do not ignore unexpected results! They will sometimes just be silly errors, but other times they will reflect a gap in your understanding. And - sometimes - a gap in the understanding of the whole research community.
Session information
sessionInfo()R version 3.3.2 (2016-10-31)
Platform: x86_64-apple-darwin13.4.0 (64-bit)
Running under: OS X El Capitan 10.11.6
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] ashr_2.2-7 rmeta_3.0
loaded via a namespace (and not attached):
[1] Rcpp_0.12.16 knitr_1.20 whisker_0.3-2
[4] magrittr_1.5 workflowr_1.0.1 REBayes_1.3
[7] MASS_7.3-49 pscl_1.5.2 doParallel_1.0.11
[10] SQUAREM_2017.10-1 lattice_0.20-35 foreach_1.4.4
[13] stringr_1.3.0 tools_3.3.2 parallel_3.3.2
[16] grid_3.3.2 R.oo_1.22.0 git2r_0.21.0
[19] htmltools_0.3.6 iterators_1.0.9 assertthat_0.2.0
[22] yaml_2.1.18 rprojroot_1.3-2 digest_0.6.15
[25] Matrix_1.2-14 codetools_0.2-15 R.utils_2.6.0
[28] evaluate_0.10.1 rmarkdown_1.9 stringi_1.1.7
[31] Rmosek_7.1.2 backports_1.1.2 R.methodsS3_1.7.1
[34] truncnorm_1.0-7 This reproducible R Markdown analysis was created with workflowr 1.0.1