Elastic Net cross-validation demo
Peter Carbonetto
Last updated: 2022-12-06
Checks: 7 0
Knit directory: cv/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0.3). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(1) was run prior to running the
code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version f30d795. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
working directory clean
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/elastic_net_demo.Rmd) and
HTML (docs/elastic_net_demo.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | f30d795 | Peter Carbonetto | 2022-12-06 | workflowr::wflow_publish("elastic_net_demo.Rmd") |
| html | 0900606 | Peter Carbonetto | 2022-12-06 | Filled out text in elastic_net_demo. |
| Rmd | 75bbb83 | Peter Carbonetto | 2022-12-06 | workflowr::wflow_publish("elastic_net_demo.Rmd") |
| Rmd | a07774d | Peter Carbonetto | 2022-12-06 | Filled out main code for elastic_net_demo. |
| html | d182ffd | Peter Carbonetto | 2022-12-06 | First build of elastic_net_demo example. |
| Rmd | 51a904e | Peter Carbonetto | 2022-12-06 | workflowr::wflow_publish("elastic_net_demo.Rmd") |
Here we illustrate how to use the perform_cv
cross-validation interface to estimate the penalty strength paramter in
the Elastic Net model in glmnet. Of course, glmnet already has a
cross-validation interface, so we can use the existing cross-validation
function in glmnet to verify our implementation. Indeed,
perform_cv almost exactly reproduces the
cv.glmnet output in this example.
Load a couple packages and the perform_cv code.
library(glmnet)
library(parallel)
source("../code/cv.R")Initialize the sequence of pseudorandom numbers.
set.seed(1)Simulate a regression data set.
n <- 200
p <- 1000
X <- matrix(rnorm(n*p),n,p)
b <- rep(0,p)
b[sample(p,4)] <- c(1,-1,1,-1)
y <- X %*% b + rnorm(n)To perform cross-validation, we need to define three functions: (1) a function to fit an Elastic Net model; (2) a function to predict Y using the fitted Elastic Net model; and (3) a function to evaluate the accuracy of the predicted Y (here we use the mean-squared error, which is also what is used in the glmnet package to evaluate the predictions).
This function fits an Elastic Net model:
fit_glmnet <- function (x, y, cvpar, noncvpar, init)
glmnet(x,y,lambda = cvpar,alpha = 0.5)This function predicts Y using the fitted Elastic Net model:
predict_glmnet <- function (x, model)
predict(model,x)This function computes the mean squared error (MSE) between the predicted and true Y:
compute_mse <- function (pred, true)
mean((pred - true)^2)Having defined these three functions, we are ready to use
perform_cv:
lambda <- round(rev(exp(seq(-3.75,0.85,length.out = 100))),digits = 4)
t0 <- proc.time()
cv <- perform_cv(fit_glmnet,predict_glmnet,compute_mse,X,y,lambda,nc = 2)
t1 <- proc.time()
print(t1 - t0)
# user system elapsed
# 5.291 0.549 3.475Compare the result with the result obtained from running
cv.glmnet on the same data (the dark blue line is the
cv.glmnet result, and the dashed orange line is our
result):
par(mar = c(4,4,0,0))
res <- cv.glmnet(X,y,alpha = 0.5)
plot(res$lambda,res$cvm,type = "l",col = "darkblue",lwd = 2,log = "x",
xlab = "lambda",ylab = "mse")
lines(lambda,rowMeans(cv),col = "darkorange",lwd = 2,lty = "dashed")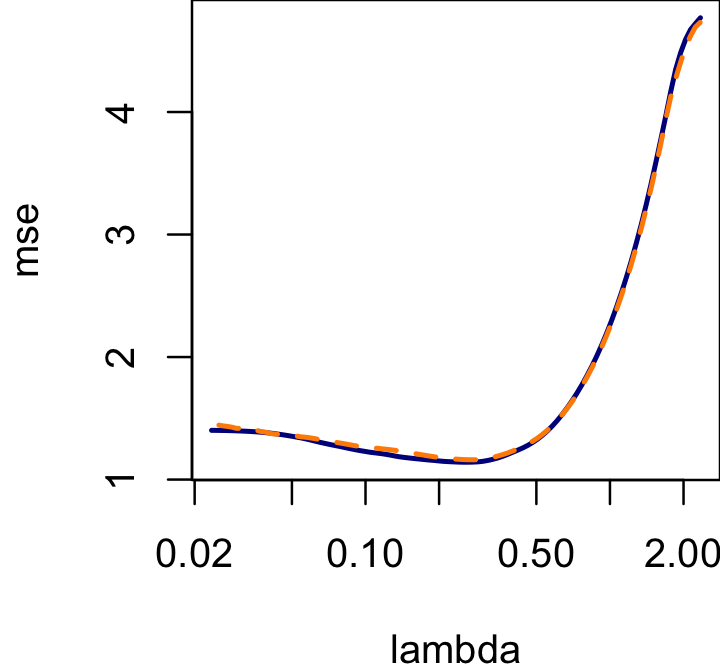
| Version | Author | Date |
|---|---|---|
| 0900606 | Peter Carbonetto | 2022-12-06 |
sessionInfo()
# R version 3.6.2 (2019-12-12)
# Platform: x86_64-apple-darwin15.6.0 (64-bit)
# Running under: macOS Catalina 10.15.7
#
# Matrix products: default
# BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
# LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
#
# locale:
# [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
#
# attached base packages:
# [1] parallel stats graphics grDevices utils datasets methods
# [8] base
#
# other attached packages:
# [1] glmnet_4.0-2 Matrix_1.4-2
#
# loaded via a namespace (and not attached):
# [1] Rcpp_1.0.8 highr_0.8 pillar_1.6.2 compiler_3.6.2
# [5] bslib_0.3.1 later_1.0.0 jquerylib_0.1.4 git2r_0.29.0
# [9] workflowr_1.7.0.3 iterators_1.0.12 tools_3.6.2 digest_0.6.23
# [13] lattice_0.20-38 jsonlite_1.7.2 evaluate_0.14 lifecycle_1.0.0
# [17] tibble_3.1.3 pkgconfig_2.0.3 rlang_0.4.11 foreach_1.4.7
# [21] yaml_2.2.0 xfun_0.29 fastmap_1.1.0 stringr_1.4.0
# [25] knitr_1.37 fs_1.5.2 vctrs_0.3.8 sass_0.4.0
# [29] rprojroot_1.3-2 grid_3.6.2 glue_1.4.2 R6_2.4.1
# [33] fansi_0.4.0 survival_3.1-8 rmarkdown_2.11 magrittr_2.0.1
# [37] whisker_0.4 splines_3.6.2 codetools_0.2-16 backports_1.1.5
# [41] promises_1.1.0 ellipsis_0.3.2 htmltools_0.5.2 shape_1.4.4
# [45] httpuv_1.5.2 utf8_1.1.4 stringi_1.4.3 crayon_1.4.1