Last updated: 2019-06-04
Checks: 7 0
Knit directory: daarem/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.3.0.9000). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(1) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: docs/figure/
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | a1e8367 | Peter Carbonetto | 2019-06-04 | ridge workflowr analysis builds successfully. |
| Rmd | 844461a | Peter Carbonetto | 2019-06-04 | s0 is now the prior s.d. rather than the prior variance. |
In this small demo, we show how the DAAREM method can be used to accelerate a simple co-ordinate ascent algorithm for computing maximum a posteriori estimates of the coefficients in a linear regression with a simple normal prior on the coefficients (“ridge regression”).
Analysis settings
These variables specify how the data are generated (n is the number of simulated samples, p is the number of simulated predictors, na is the number of simulated predictors that have a nonzero effect, se is the variance oft he residual), and the prior on the regression coefficients (the prior is normal with zero mean and standard deviation s0).
n <- 200
p <- 500
na <- 10
se <- 2
s0 <- 1/seSet up environment
Load some packages and function definitions used in the example below.
library(MASS)
library(daarem)
# Warning: package 'daarem' was built under R version 3.4.4
library(ggplot2)
# Warning: package 'ggplot2' was built under R version 3.4.4
library(cowplot)
#
# Attaching package: 'cowplot'
# The following object is masked from 'package:ggplot2':
#
# ggsave
source("../code/misc.R")
source("../code/datasim.R")
source("../code/ridge.R")Initialize the sequence of pseudorandom numbers.
set.seed(1)Simulate data
Simulate the predictors.
X <- simulate_predictors_decaying_corr(n,p,s = 0.5)
X <- scale(X,center = TRUE,scale = FALSE)Generate additive effects for the markers so that exactly na of them have a nonzero effect on the trait.
i <- sample(p,na)
b <- rep(0,p)
b[i] <- rnorm(na)Simulate the continuous outcomes.
y <- drop(X %*% b + se*rnorm(n))
y <- y - mean(y)Set initial estimate of mixture proportions.
b0 <- rep(0,p)Run basic co-ordinate ascent updates
Fit ridge regression with basic co-ordinate ascent updates.
out <- system.time(fit1 <- ridge(X,y,b0,s0,numiter = 100))
f1 <- ridge.objective(X,y,fit1$b,s0)
cat(sprintf("Computation took %0.2f seconds.\n",out["elapsed"]))
cat(sprintf("Objective value at solution is %0.12f.\n",f1))
# Computation took 3.74 seconds.
# Objective value at solution is -20.573760535831.Run accelerated co-ordinate ascent updates
Fitting ridge regression with accelerated co-ordinate ascent updates
out <- system.time(fit2 <- daarridge(X,y,b0,s0,numiter = 100))
f2 <- ridge.objective(X,y,fit2$b,s0)
cat(sprintf("Computation took %0.2f seconds.\n",out["elapsed"]))
cat(sprintf("Objective value at solution is %0.12f.\n",f2))
# Computation took 3.73 seconds.
# Objective value at solution is -20.332771749786.Plot improvement in solution over time
bhat <- drop(solve(t(X) %*% X + diag(p)/s0,t(X) %*% y))
f <- ridge.objective(X,y,bhat,s0)
pdat <-
rbind(data.frame(iter = 1:100,dist = f - fit1$value,method = "basic"),
data.frame(iter = 1:100,dist = f - fit2$value,method = "accelerated"))
p <- ggplot(pdat,aes(x = iter,y = dist,col = method)) +
geom_line(size = 1) +
scale_y_continuous(trans = "log10",breaks = 10^seq(-8,4)) +
scale_color_manual(values = c("darkorange","dodgerblue")) +
labs(x = "iteration",y = "distance from solution")
print(p)
# Warning in self$trans$transform(x): NaNs produced
# Warning: Transformation introduced infinite values in continuous y-axis
# Warning: Removed 74 rows containing missing values (geom_path).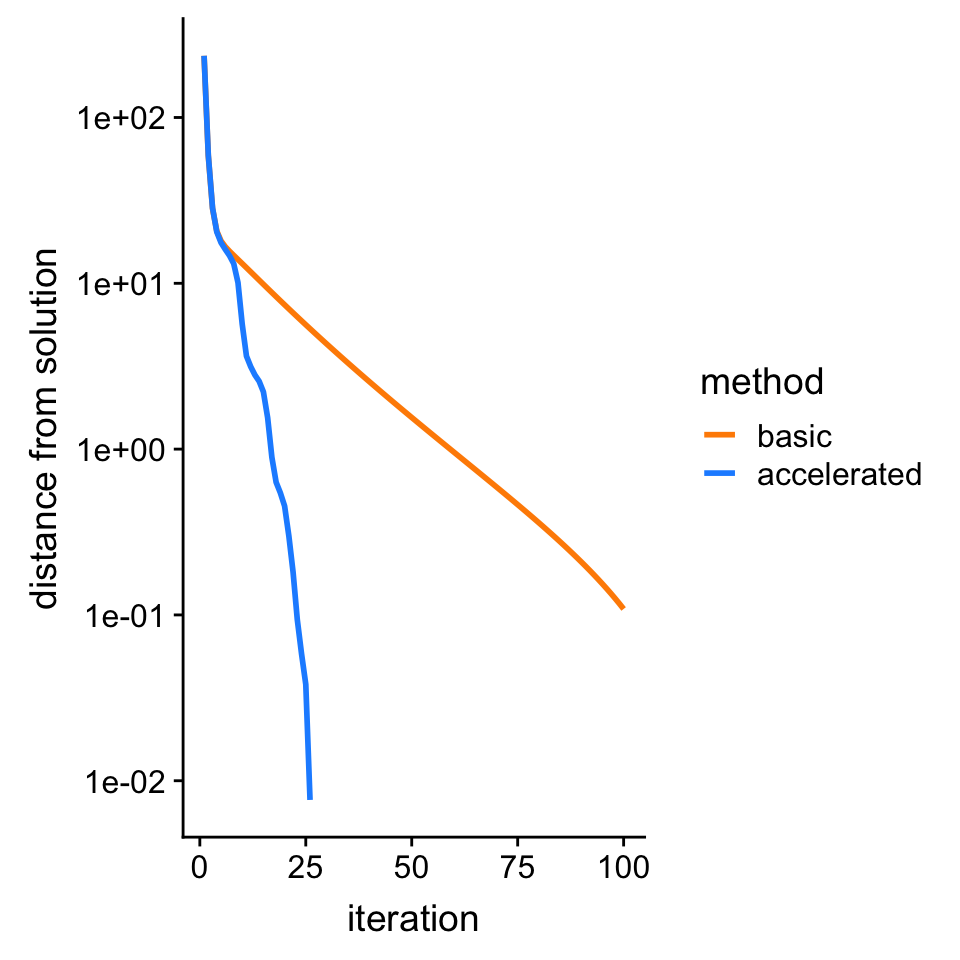
sessionInfo()
# R version 3.4.3 (2017-11-30)
# Platform: x86_64-apple-darwin15.6.0 (64-bit)
# Running under: macOS High Sierra 10.13.6
#
# Matrix products: default
# BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
# LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
#
# locale:
# [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
#
# attached base packages:
# [1] stats graphics grDevices utils datasets methods base
#
# other attached packages:
# [1] cowplot_0.9.4 ggplot2_3.1.0 daarem_0.3 MASS_7.3-48
#
# loaded via a namespace (and not attached):
# [1] Rcpp_1.0.1 knitr_1.20 whisker_0.3-2
# [4] magrittr_1.5 workflowr_1.3.0.9000 tidyselect_0.2.5
# [7] munsell_0.4.3 colorspace_1.4-0 R6_2.2.2
# [10] rlang_0.3.1 dplyr_0.8.0.1 stringr_1.3.1
# [13] plyr_1.8.4 tools_3.4.3 grid_3.4.3
# [16] gtable_0.2.0 withr_2.1.2 git2r_0.25.2.9007
# [19] htmltools_0.3.6 assertthat_0.2.0 lazyeval_0.2.1
# [22] yaml_2.2.0 rprojroot_1.3-2 digest_0.6.17
# [25] tibble_2.1.1 crayon_1.3.4 purrr_0.2.5
# [28] fs_1.2.6 glue_1.3.0 evaluate_0.11
# [31] rmarkdown_1.10 labeling_0.3 stringi_1.2.4
# [34] pillar_1.3.1 compiler_3.4.3 scales_0.5.0
# [37] backports_1.1.2 pkgconfig_2.0.2