M&M RSS
Yuxin Zou
07/21/2020
Last updated: 2020-09-21
Checks: 2 0
Knit directory: mmbr-rss-dsc/
This reproducible R Markdown analysis was created with workflowr (version 1.6.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Untracked files:
Untracked: data/tiny_data_211.config
Untracked: data/tiny_data_211.ld
Untracked: data/tiny_data_211.log
Untracked: data/tiny_data_211.master
Untracked: data/tiny_data_211.snp
Untracked: data/tiny_data_211_post
Untracked: data/tiny_data_211_set
Untracked: output/mnm_rss_lite_output.20200227.rds
Untracked: output/mnm_rss_lite_output.20200310.rds
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 493c703 | zouyuxin | 2020-09-21 | wflow_publish(“analysis/mmbr_rss_simulation_3.Rmd”) |
| Rmd | a5d9bee | zouyuxin | 2020-09-21 | wflow_publish(“analysis/mmbr_rss_simulation_3.Rmd”) |
This is result from our M&M RSS simulation. There are 300 datasets, each with 1000 SNPs. Per signal PVE is 0.05.
For each dataset, we simulate signals using 2 type of priors:
- Artificial mixture: 50 conditions.
The detail about prior is here.
The oracle residual variance is a diagonal matrix.
- GTEx mixture: 50 conditions.
The detail about prior is here.
The oracle residual variance is a dense matrix.
We estimate prior weights using ‘EM’ method.
Overall: Ignoring correlation between conditions in residual matrix results in poor fit.
PIP calibration
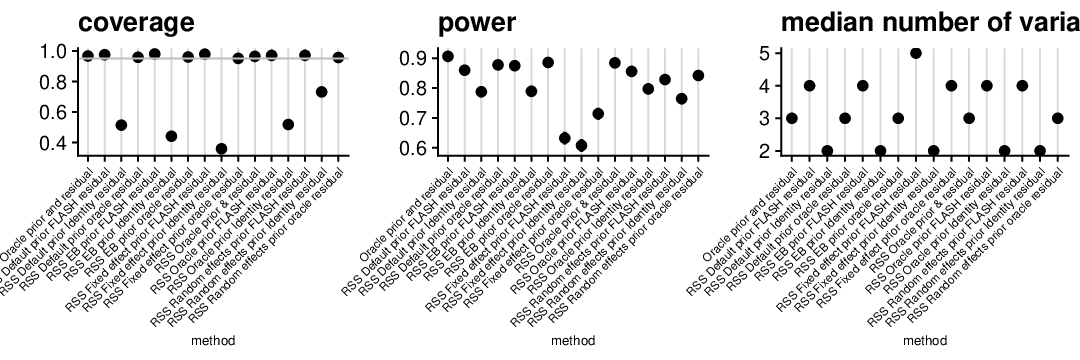
Artificial Mixture

GTEx Mixture

Power
Artificial Mixture
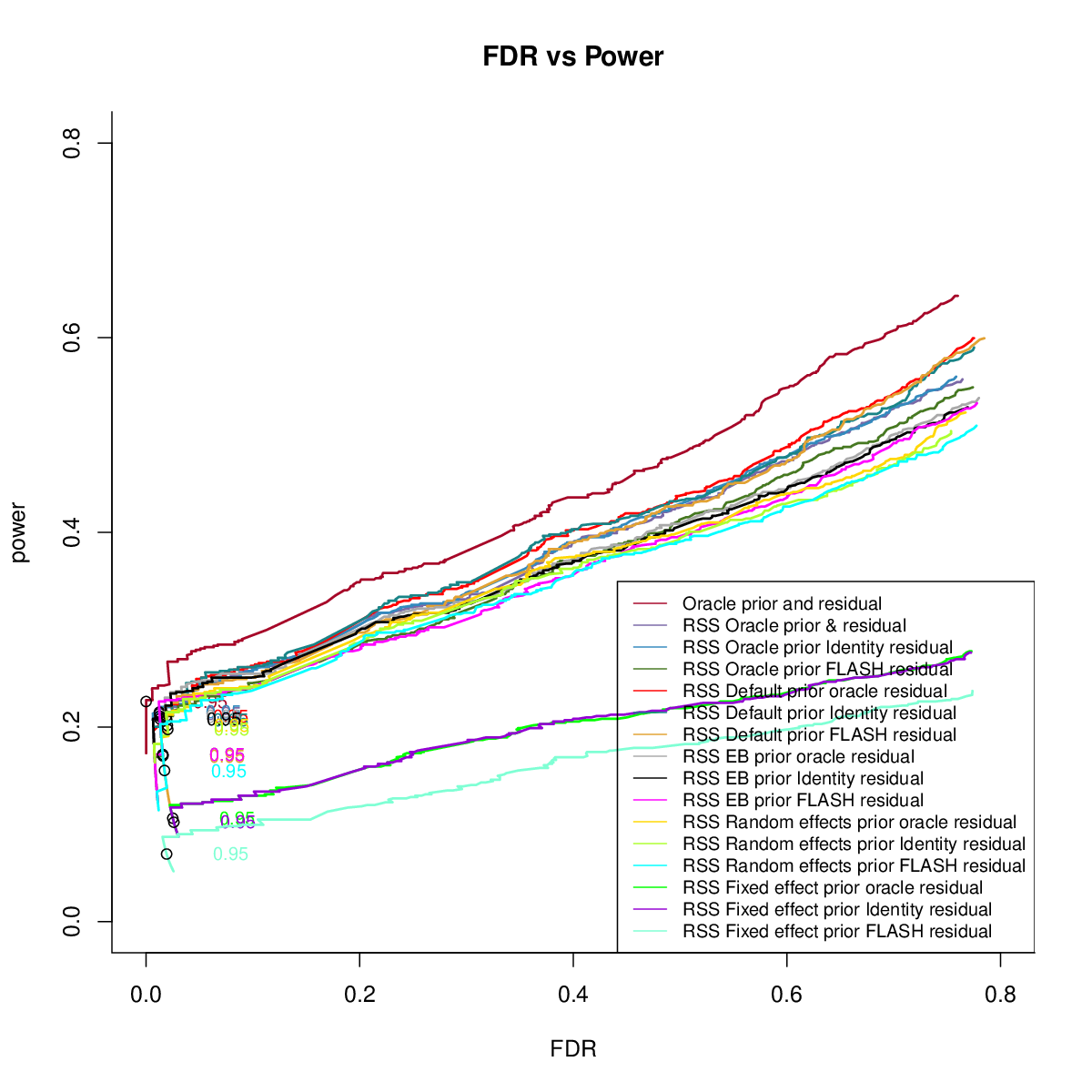
GTEx Mixture
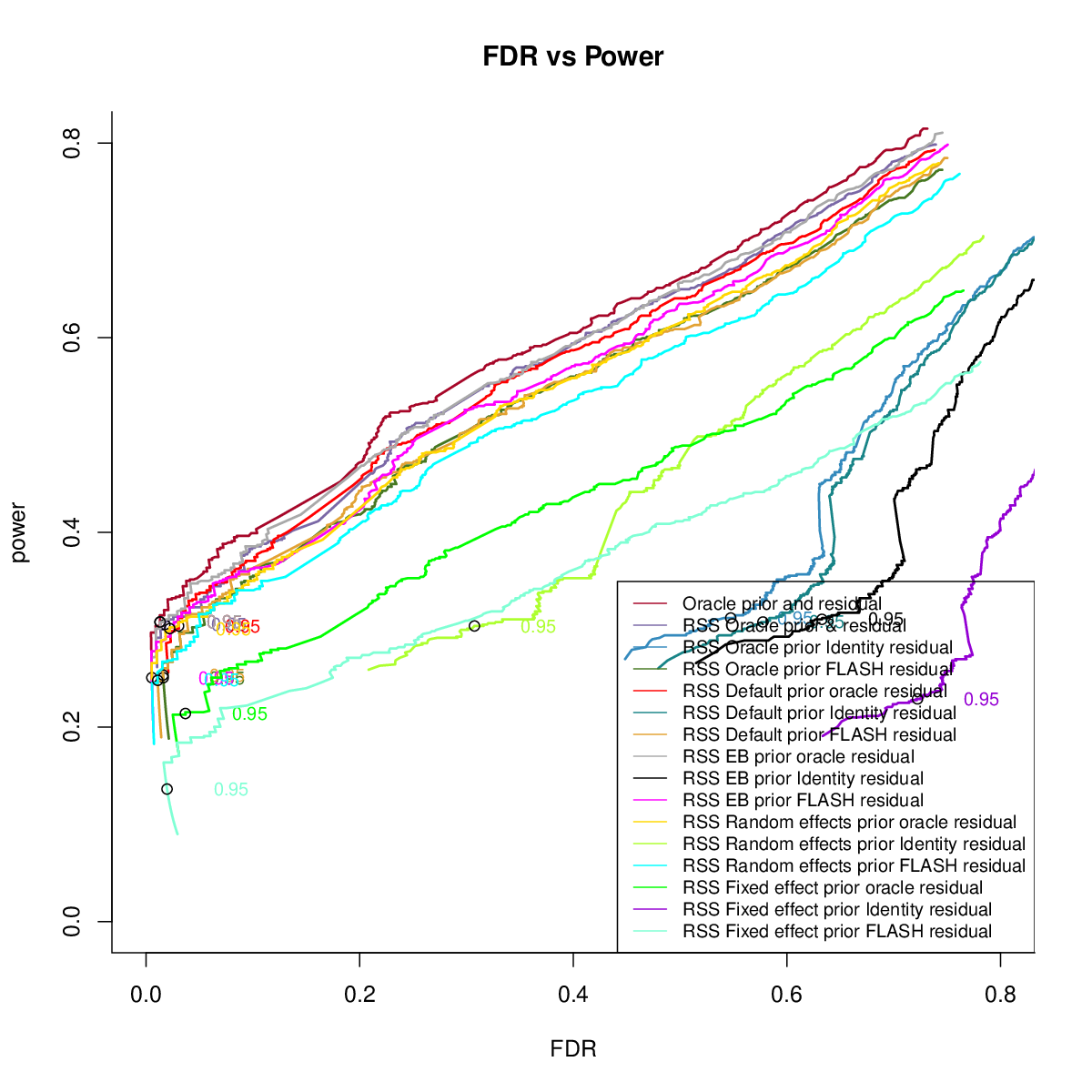
CS
Artificial Mixture
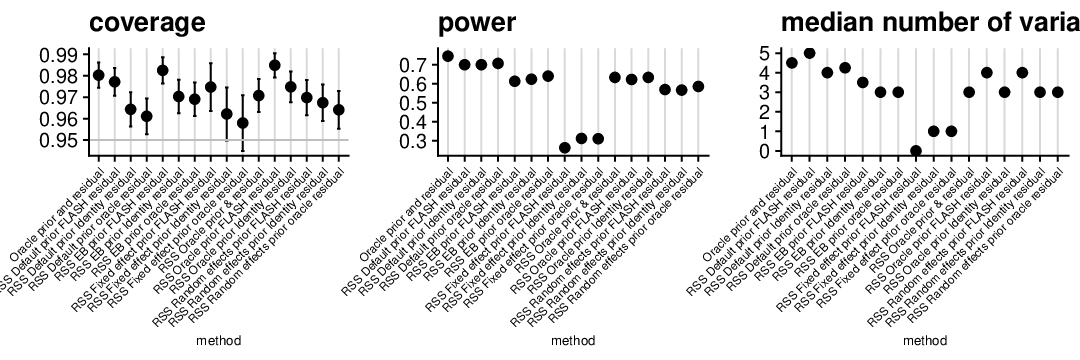
GTEx Mixture