Gene analysis using topic modeling and DA results (v2) for Cusanovich et al (2018) scATAC-seq data
Kaixuan Luo
Last updated: 2022-02-24
Checks: 7 0
Knit directory: scATACseq-topics/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it's best to always run the code in an empty environment.
The command set.seed(20200729) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 0ffb5c6. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: output/plotly/Buenrostro_2018_Chen2019pipeline_v2/
Untracked files:
Untracked: analysis/analysis_Buenrostro2018_k10.Rmd
Untracked: analysis/clusters_Buenrostro2018_k10_Chen2019pipeline.Rmd
Untracked: analysis/process_data_Buenrostro2018_Chen2019.Rmd
Untracked: analysis/selected_figures_Buenrostro2018.Rmd
Untracked: analysis/test_structure_plots_Buenrostro2018_k11_Chen2019pipeline.Rmd
Untracked: gsea_b_cells.html
Untracked: output/clustering-Cusanovich2018.rds
Untracked: paper/
Untracked: scripts/fit_all_models_Buenrostro_2018_chromVar_scPeaks_filtered.sbatch
Untracked: scripts/fit_models_Cusanovich2018_tissues.sh
Untracked: scripts/postfit_Buenrostro2018_Chen2019pipeline_v2.sh
Untracked: scripts/postfit_Cusanovich2018_v2.sh
Untracked: scripts/postfit_DA_analysis.sbatch
Untracked: topic1.html
Unstaged changes:
Modified: .gitignore
Modified: analysis/clusters_Cusanovich2018_k13.Rmd
Modified: analysis/gene_analysis_Buenrostro2018_Chen2019pipeline.Rmd
Modified: analysis/gene_analysis_Cusanovich2018.Rmd
Modified: analysis/motif_analysis_Buenrostro2018_Chen2019pipeline.Rmd
Modified: analysis/motif_analysis_Cusanovich2018.Rmd
Modified: analysis/plots_Cusanovich2018.Rmd
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_10_tss-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_11_tss-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_12_tss-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_13_tss-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_1_tss-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_2_tss-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_3_tss-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_4_tss-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_5_tss-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_6_tss-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_7_tss-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_8_tss-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/gsea_topic_9_tss-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_10_tss-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_11_tss-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_12_tss-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_13_tss-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_1_tss-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_2_tss-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_3_tss-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_4_tss-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_5_tss-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_6_tss-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_7_tss-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_8_tss-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_genebody-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_genebody-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_genebody-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_genebody-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_genebody-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_genebody-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_genebody-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_genebody-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_genebody-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_genebody-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_genebody-sum_files/typedarray-0.1/typedarray.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_tss-sum_files/crosstalk-1.1.0.1/css/crosstalk.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_tss-sum_files/crosstalk-1.1.0.1/js/crosstalk.min.js.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_tss-sum_files/htmlwidgets-1.5.3/htmlwidgets.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_tss-sum_files/jquery-1.11.3/jquery-AUTHORS.txt
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_tss-sum_files/jquery-1.11.3/jquery.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_tss-sum_files/jquery-1.11.3/jquery.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_tss-sum_files/jquery-1.11.3/jquery.min.map
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_tss-sum_files/plotly-binding-4.9.2.1/plotly.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_tss-sum_files/plotly-htmlwidgets-css-1.52.2/plotly-htmlwidgets.css
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_tss-sum_files/plotly-main-1.52.2/plotly-latest.min.js
Deleted: output/plotly/Cusanovich2018/volcano_topic_9_tss-sum_files/typedarray-0.1/typedarray.min.js
Modified: scripts/fit_all_models_Cusanovich2018.sh
Modified: scripts/postfit_Buenrostro2018_Chen2019pipeline.sh
Modified: scripts/postfit_Cusanovich2018.sh
Modified: scripts/postfit_gene_analysis.sbatch
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/gene_analysis_Cusanovich2018_v2.Rmd) and HTML (docs/gene_analysis_Cusanovich2018_v2.html) files. If you've configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 0ffb5c6 | kevinlkx | 2022-02-24 | wflow_publish("analysis/gene_analysis_Cusanovich2018_v2.Rmd") |
| html | c010fd7 | kevinlkx | 2022-02-22 | Build site. |
| Rmd | a6b27b6 | kevinlkx | 2022-02-22 | added gene volcano plots |
| html | 4b5247c | kevinlkx | 2022-02-17 | Build site. |
| Rmd | 57ff8a0 | kevinlkx | 2022-02-17 | updated with v2 of DA analysis |
Here we perform gene analysis for the Cusanovich et al (2018) scATAC-seq result inferred from the multinomial topic model with \(k = 13\).
Load packages and some functions used in this analysis
library(Matrix)
library(fastTopics)
library(pathways)
library(dplyr)
library(tidyr)
library(ggplot2)
library(ggrepel)
library(cowplot)
library(plotly)
library(htmlwidgets)
library(DT)
library(reshape)
source("code/plots.R")Load samples and topic model (\(k = 13\)) results.
data.dir <- "/project2/mstephens/kevinluo/scATACseq-topics/output/Cusanovich_2018"
samples <- readRDS(paste0(data.dir, "/samples-clustering-Cusanovich2018.rds"))fit.dir <- "/project2/mstephens/kevinluo/scATACseq-topics/output/Cusanovich_2018"
fit <- readRDS(file.path(fit.dir, "/fit-Cusanovich2018-scd-ex-k=13.rds"))$fit
fit <- poisson2multinom(fit)Visualize by Structure plot grouped by tissues
set.seed(10)
colors_topics <- c("#a6cee3","#1f78b4","#b2df8a","#33a02c","#fb9a99","#e31a1c",
"#fdbf6f","#ff7f00","#cab2d6","#6a3d9a","#ffff99","#b15928",
"gray")
rows <- sample(nrow(fit$L),4000)
samples$tissue <- as.factor(samples$tissue)
p.structure <- structure_plot(select(fit,loadings = rows),
grouping = samples[rows, "tissue"],n = Inf,gap = 40,
perplexity = 50,colors = colors_topics,
num_threads = 4,verbose = FALSE)
print(p.structure)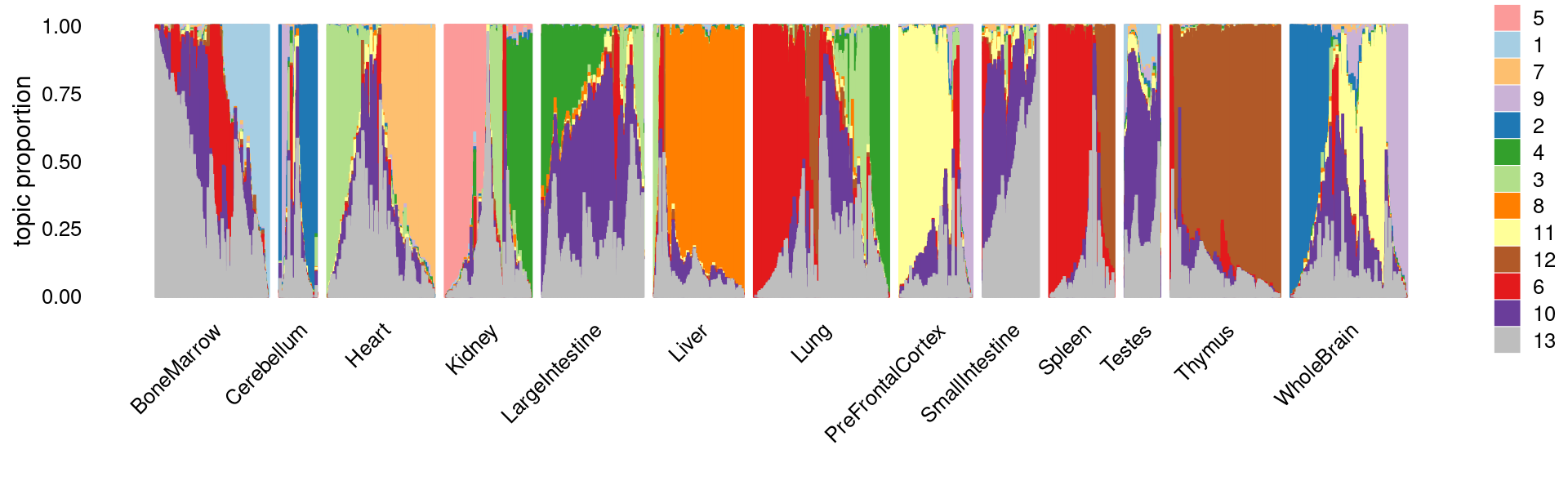
| Version | Author | Date |
|---|---|---|
| 4b5247c | kevinlkx | 2022-02-17 |
Load the clustering results
set.seed(10)
rows <- sample(nrow(fit$L),4000)
p.structure.kmeans <- structure_plot(select(fit,loadings = rows),
grouping = samples$cluster_kmeans[rows],n = Inf,gap = 40,
perplexity = 50,colors = colors_topics,
num_threads = 4,verbose = FALSE)
print(p.structure.kmeans)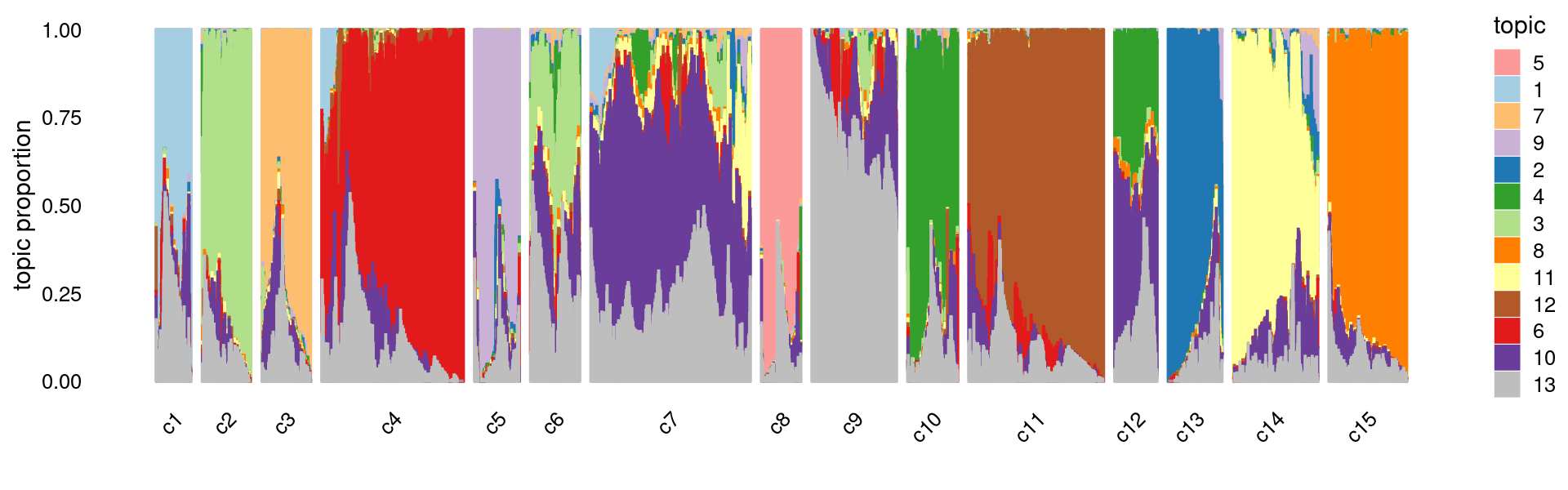
| Version | Author | Date |
|---|---|---|
| 4b5247c | kevinlkx | 2022-02-17 |
Distribution of tissue labels by cluster.
freq_table_cluster_tissue <- with(samples,table(tissue,cluster_kmeans))
freq_table_cluster_tissue <- as.data.frame.matrix(freq_table_cluster_tissue)
# DT::datatable(freq_table_cluster_tissue,
# options = list(pageLength = nrow(freq_table_cluster_tissue)),
# rownames = T, caption = "Number of cells")
create_celllabel_cluster_heatmap(samples$tissue, samples$cluster_kmeans, normalize_by = "column")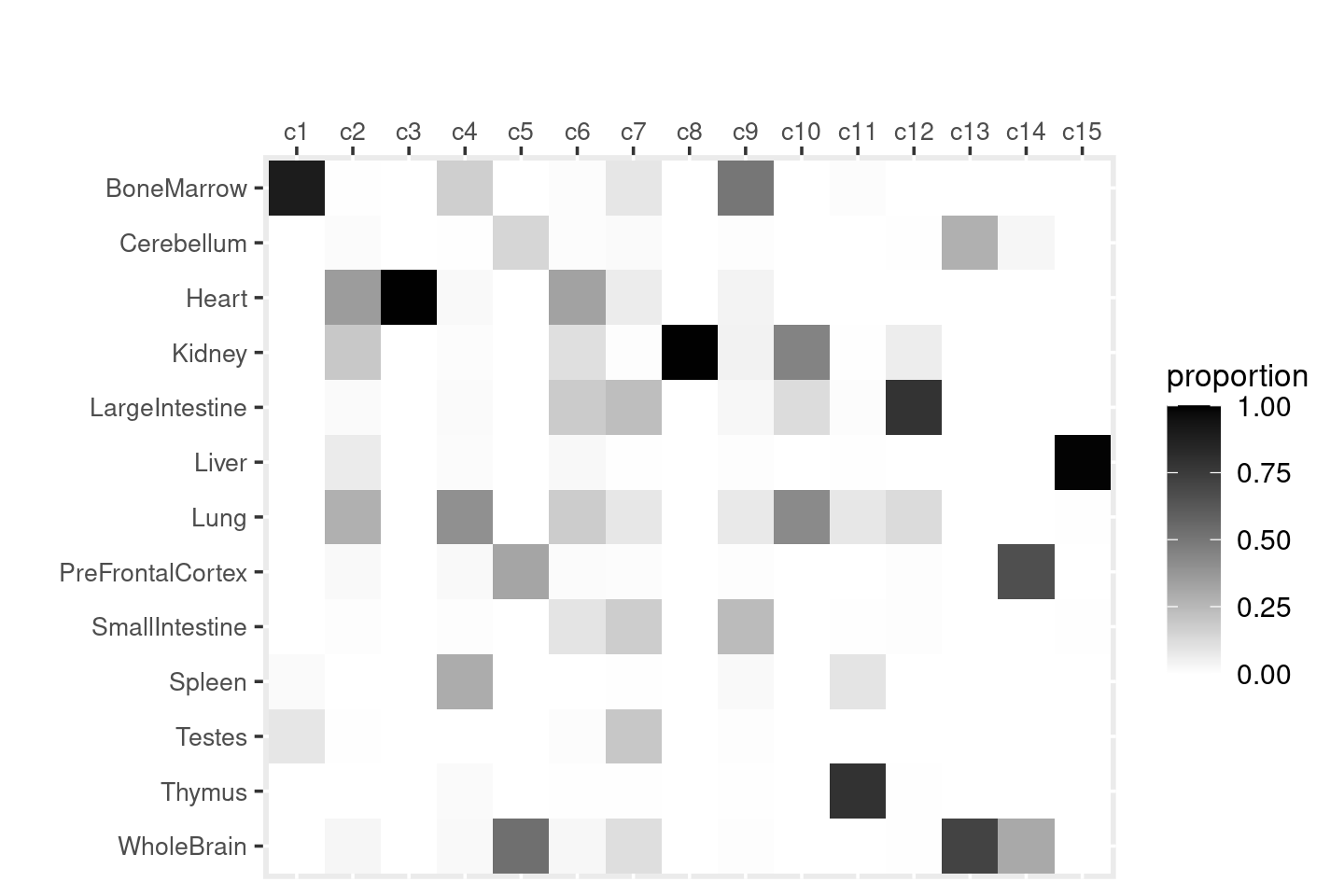
| Version | Author | Date |
|---|---|---|
| 4b5247c | kevinlkx | 2022-02-17 |
Distribution of cell labels by cluster.
freq_table_cluster_celllabel <- with(samples,table(cell_label,cluster_kmeans))
freq_table_cluster_celllabel <- as.data.frame.matrix(freq_table_cluster_celllabel)
# DT::datatable(freq_table_cluster_celllabel,
# options = list(pageLength = nrow(freq_table_cluster_celllabel)),
# rownames = T, caption = "Number of cells")
create_celllabel_cluster_heatmap(samples$cell_label, samples$cluster_kmeans, normalize_by = "column")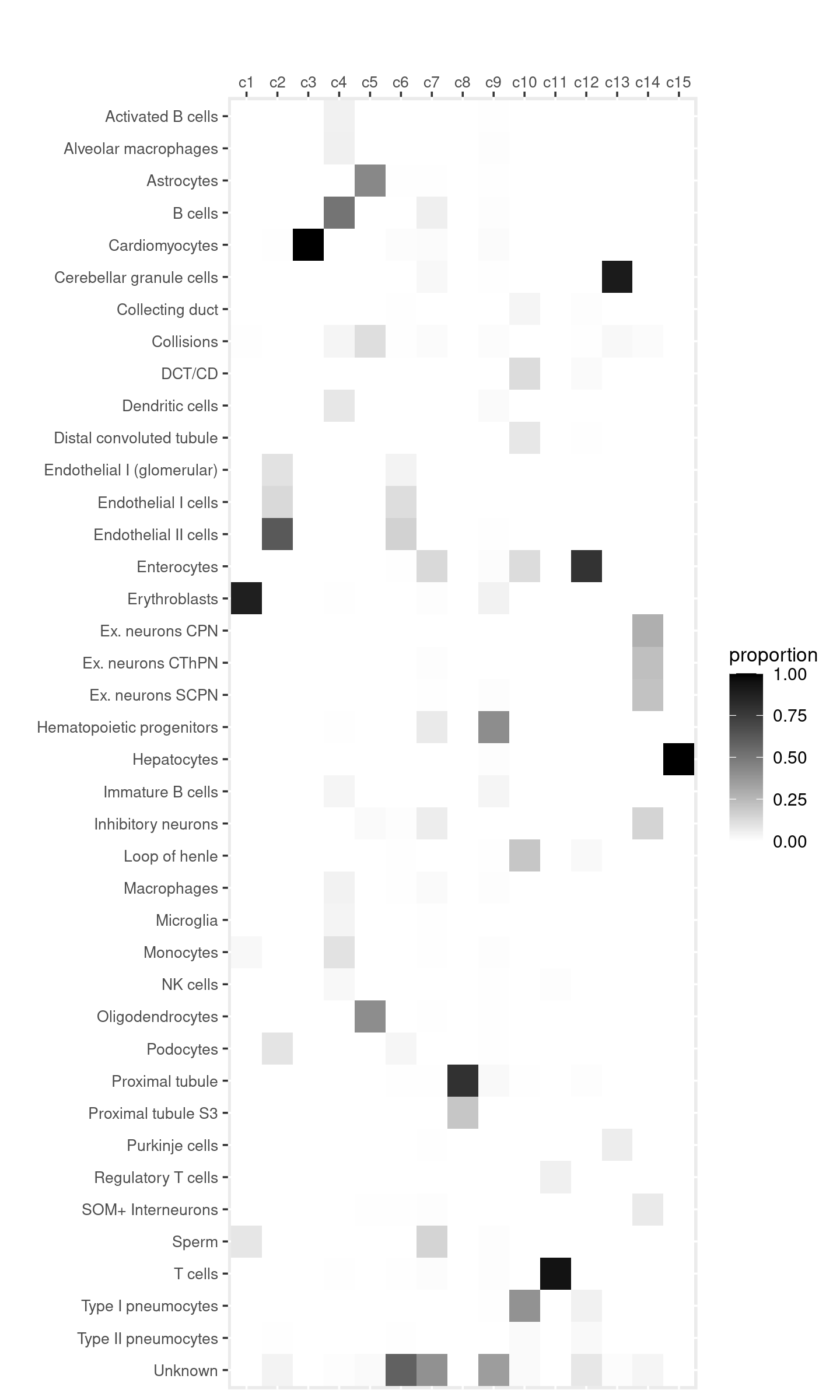
| Version | Author | Date |
|---|---|---|
| 4b5247c | kevinlkx | 2022-02-17 |
Top 5 cell types
top_celltypes_table <- data.frame(matrix(nrow=5, ncol = ncol(freq_table_cluster_celllabel)))
colnames(top_celltypes_table) <- colnames(freq_table_cluster_celllabel)
for (k in 1:ncol(freq_table_cluster_celllabel)){
top_celltypes <- rownames(freq_table_cluster_celllabel)[head(order(freq_table_cluster_celllabel[,k], decreasing=TRUE), 5)]
freq_top_celltypes <- freq_table_cluster_celllabel[top_celltypes, k]
percent_top_celltypes <- freq_table_cluster_celllabel[top_celltypes, k]/sum(freq_table_cluster_celllabel[,k])
top_celltypes_table[,k] <- sprintf("%s (%.1f%%)", top_celltypes, percent_top_celltypes*100)
}
DT::datatable(top_celltypes_table, rownames = T, caption = "Top 5 cell types in each cluster")We can see the major cell types in the clusters (topics):
- cluster 1 (topic 1): Erythroblasts
- cluster 2 (topic 3): Endothelial cells
- cluster 3 (topic 7): Cardiomyocytes cells
- cluster 4 (topic 6): B cells, Monocytes, Dendritic cells
- cluster 5 (topic 9): Astrocytes, Oligodendrocytes
- cluster 8 (topic 5): Proximal tubule
- cluster 10 (topic 4): a mixture of pneumocytes, Loop of henle, Enterocytes, DCT/CD
- cluster 11 (topic 12): T cells
- cluster 13 (topic 2): Cerebellar granule cells
- cluster 14 (topic 11): Ex. neurons and Inhibitory neurons
- cluster 15 (topic 8): Hepatocytes
Differential accessbility analysis of the ATAC-seq regions for the topics
Load results from differential accessbility analysis for the topics
out.dir <- "/project2/mstephens/kevinluo/scATACseq-topics/output/Cusanovich_2018/postfit_v2"
cat(sprintf("Load results from %s \n", out.dir))
DA_res <- readRDS(file.path(out.dir, paste0("DAanalysis-Cusanovich2018-k=13/DA_regions_topics_10000iters.rds")))# Load results from /project2/mstephens/kevinluo/scATACseq-topics/output/Cusanovich_2018/postfit_v2Gene score analysis
Set output directory
out.dir <- "/project2/mstephens/kevinluo/scATACseq-topics/output/Cusanovich_2018/postfit_v2"
fig.dir <- "output/plotly/Cusanovich2018_v2"
dir.create(fig.dir, showWarnings = F, recursive = T)TSS model ver1
Gene scores were computed using TSS based method as in Lareau et al Nature Biotech, 2019 as well as the model 21 in archR paper. This model weights chromatin accessibility around gene promoters by using bi-directional exponential decays from the TSS.
- TSS model
- use abs(z) scores
normalized by the sum of weights
Top genes
gene.dir <- paste0(out.dir, "/geneanalysis-Cusanovich2018-k=13-TSS-absZ-sum")
cat(sprintf("Directory of gene analysis result: %s \n", gene.dir))
genescore_tss_res <- readRDS(file.path(gene.dir, "genescore_result.rds"))
genescore_res <- genescore_tss_res
genes <- genescore_res$genes
gene_scores <- genescore_res$Z
gene_logFC <- genescore_res$logFC
topics <- colnames(gene_scores)
top_genes <- data.frame(matrix(nrow=10, ncol = ncol(gene_scores)))
colnames(top_genes) <- topics
for (k in topics){
top_genes[,k] <- genes$SYMBOL[head(order(abs(gene_scores[,k]), decreasing=TRUE), 10)]
}
DT::datatable(data.frame(rank = 1:10, top_genes), rownames = F, caption = "Top 10 genes by abs(gene z-scores)")# Directory of gene analysis result: /project2/mstephens/kevinluo/scATACseq-topics/output/Cusanovich_2018/postfit_v2/geneanalysis-Cusanovich2018-k=13-TSS-absZ-sum- Volcano plots
genescore_volcano_plot(genescore_res, label_above_quantile = 0.99,
labels = genescore_res$genes$SYMBOL, max.overlaps = 20,
subsample_below_quantile = 0.5, subsample_rate = 0.1)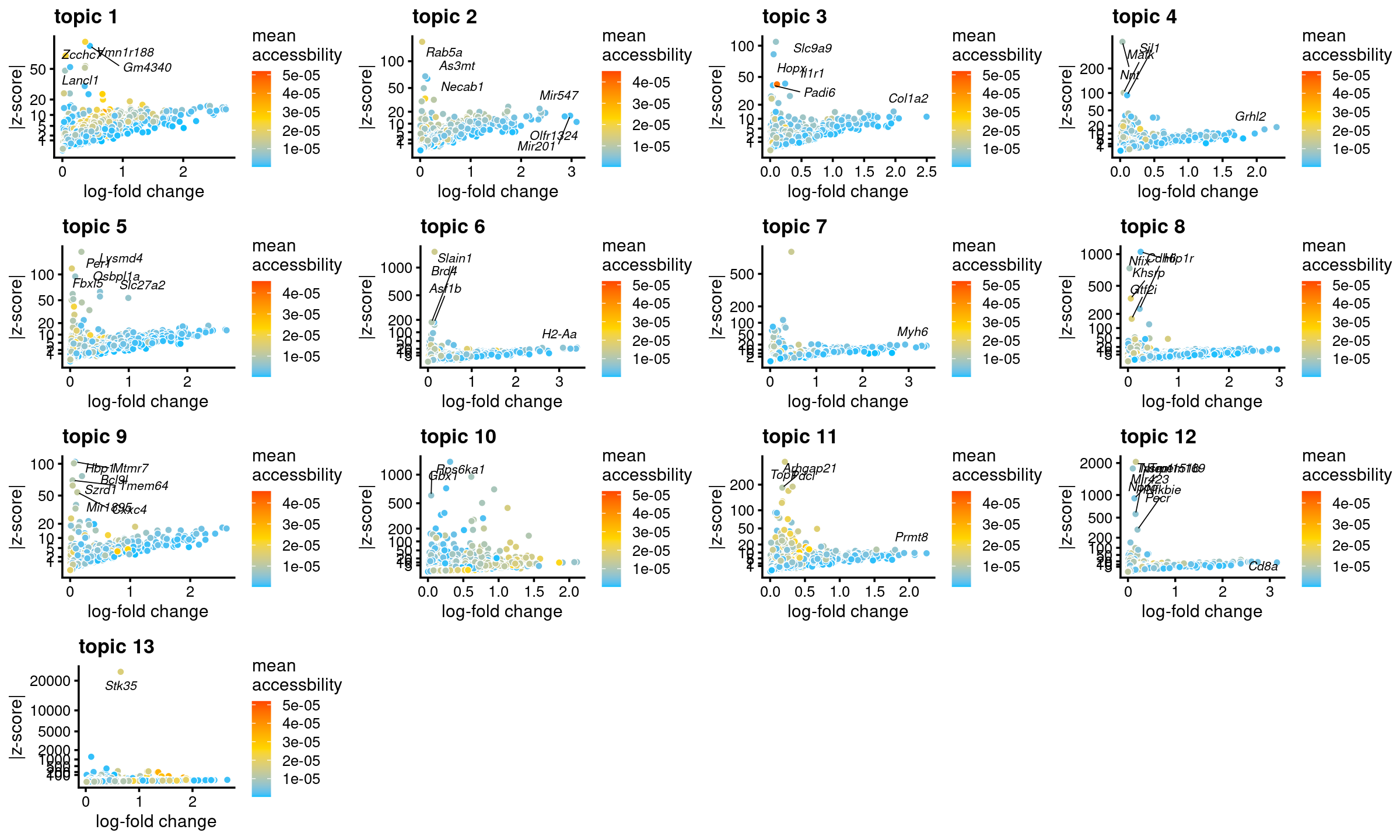
| Version | Author | Date |
|---|---|---|
| c010fd7 | kevinlkx | 2022-02-22 |
Topic 1 (Erythroblasts)
genescore_volcano_plot(genescore_res, k = 1, label_above_quantile = 0.99,
labels = genescore_res$genes$SYMBOL, max.overlaps = 20,
subsample_below_quantile = 0.5, subsample_rate = 0.1)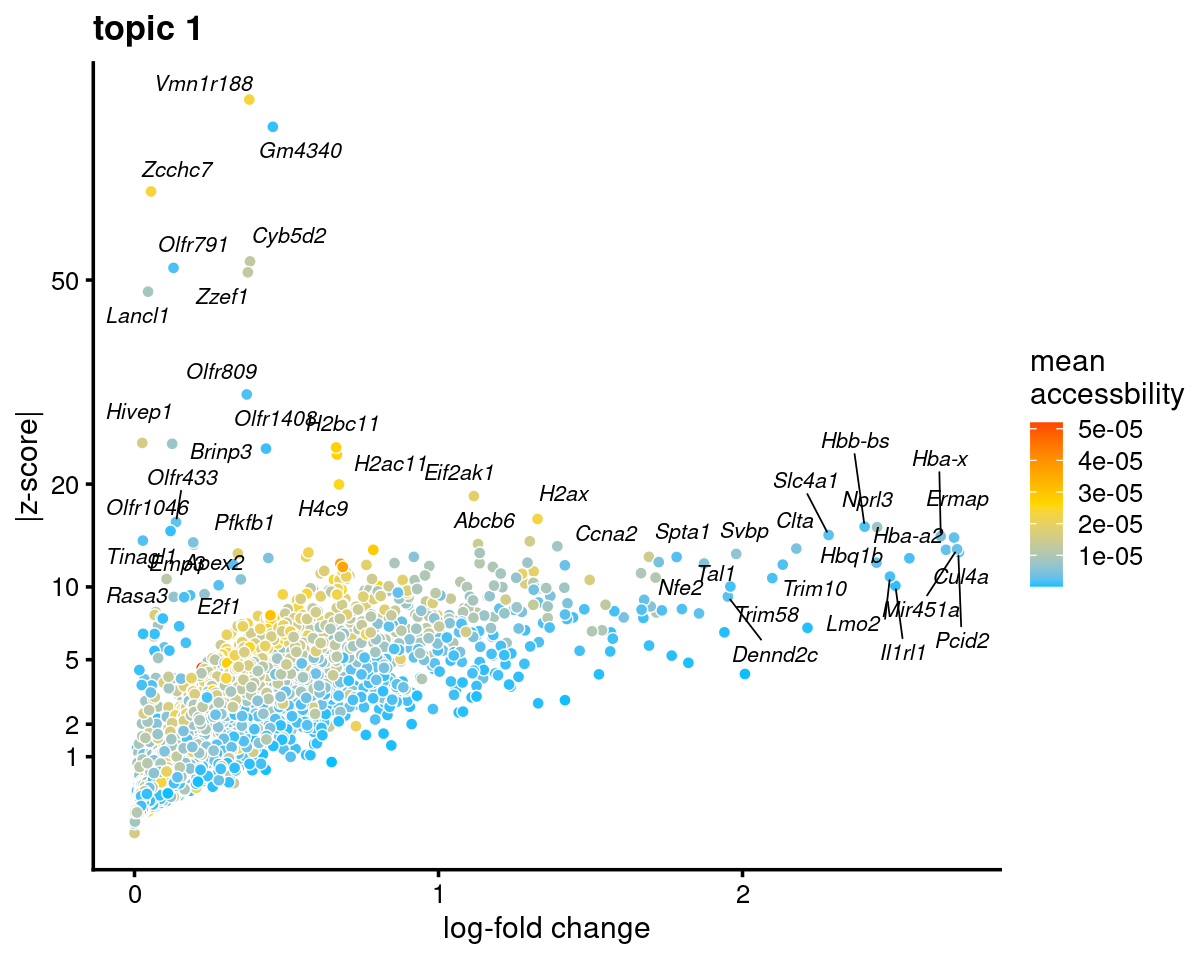
| Version | Author | Date |
|---|---|---|
| c010fd7 | kevinlkx | 2022-02-22 |
Check some known marker genes
marker_genes <- c("Hbb-b1", "Hbb-b2", "Gypa")
gene_scores <- genescore_res$Z
rownames(gene_scores) <- genescore_res$genes$SYMBOL
marker_gene_scores <- gene_scores[grep(paste(sprintf("^%s$", marker_genes), collapse = "|"), rownames(gene_scores), ignore.case = T),]
par(mfrow = c(ceiling(nrow(marker_gene_scores)/2),2))
for(i in 1:nrow(marker_gene_scores)){
barplot(marker_gene_scores[i,], xlab = "topics", ylab = "gene score", main = rownames(marker_gene_scores)[i], col = colors_topics)
}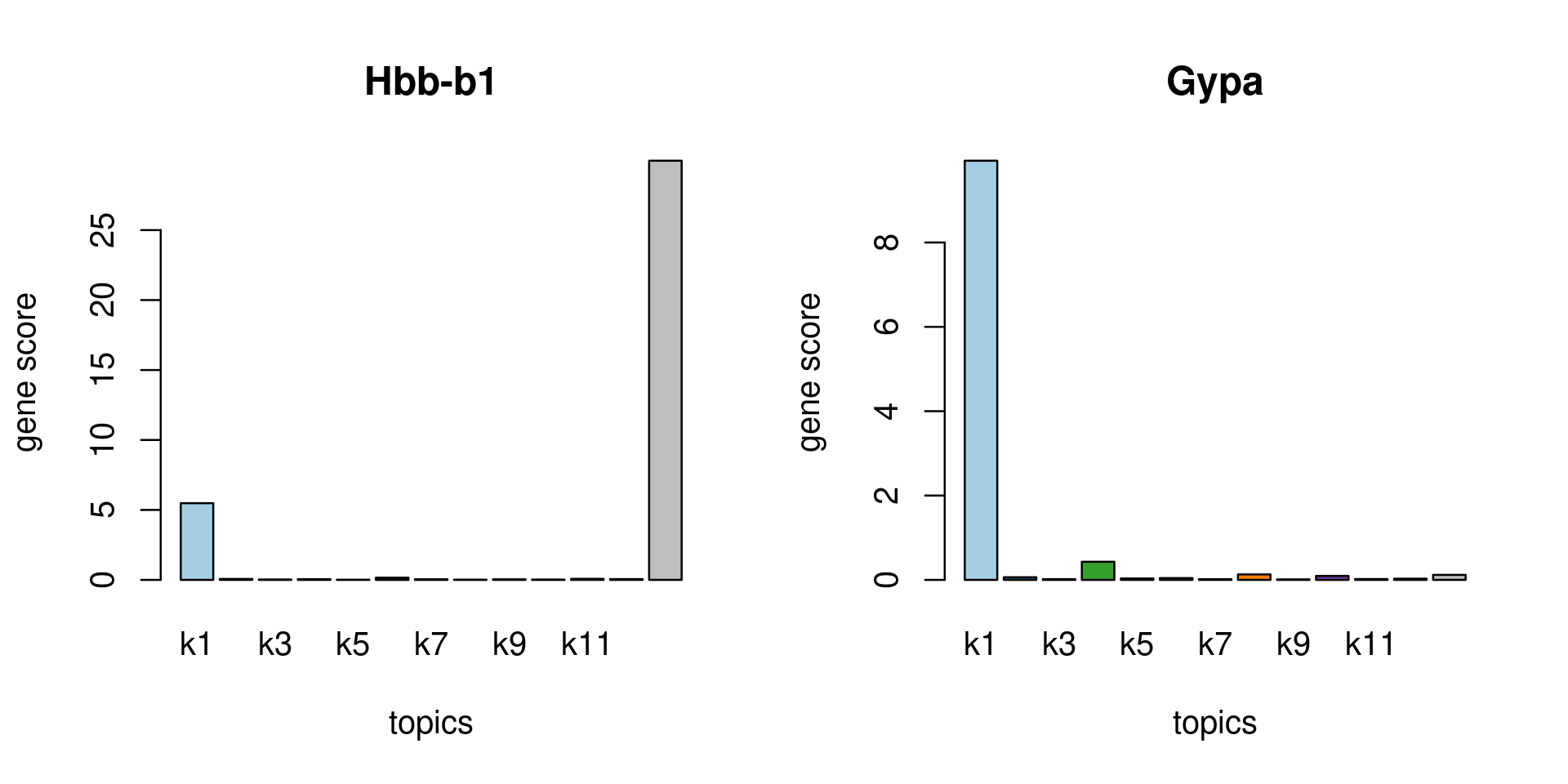
Topic 3 (Endothelial cells)
genescore_volcano_plot(genescore_res, k = 3, label_above_quantile = 0.99,
labels = genescore_res$genes$SYMBOL, max.overlaps = 20,
subsample_below_quantile = 0.5, subsample_rate = 0.1)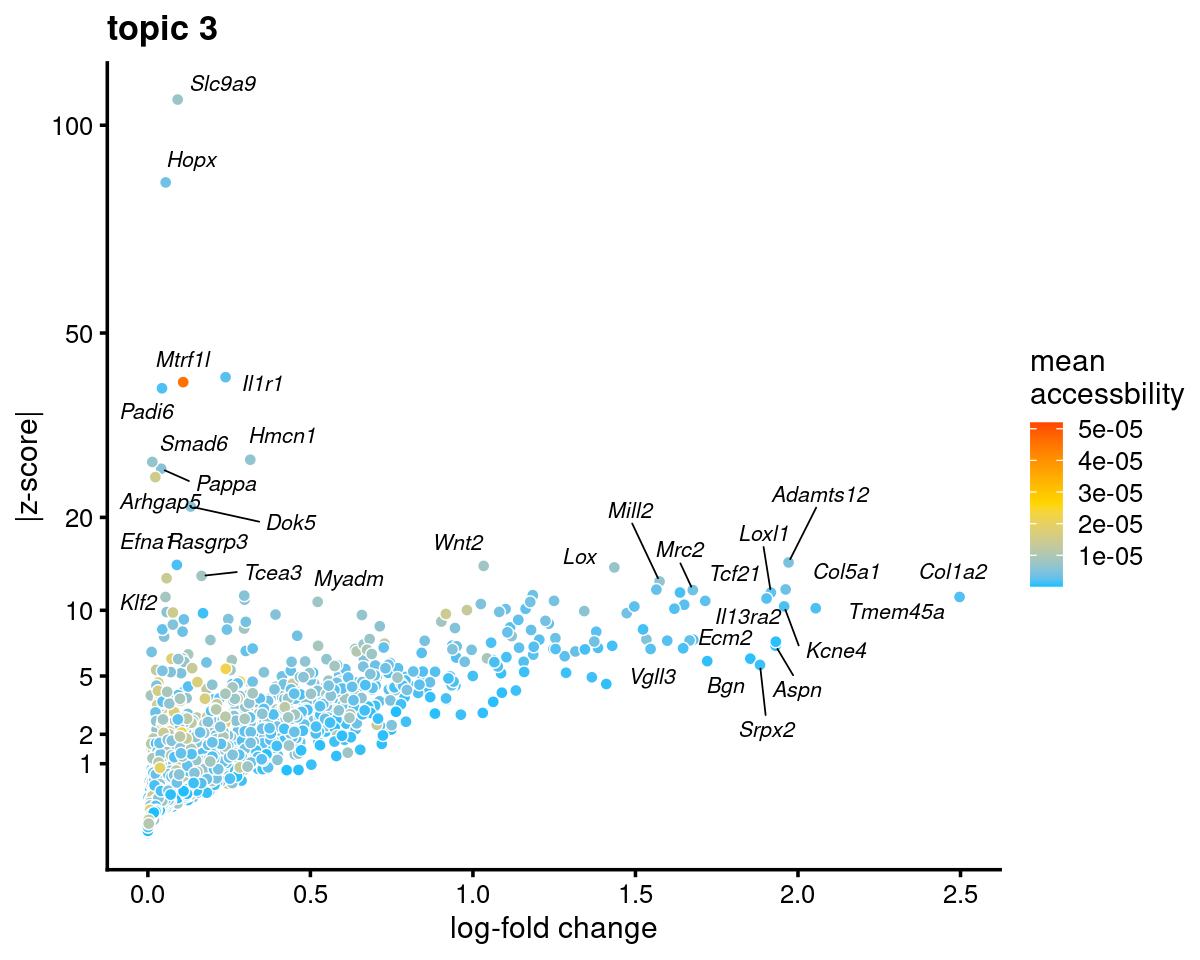
| Version | Author | Date |
|---|---|---|
| c010fd7 | kevinlkx | 2022-02-22 |
Check some known marker genes
marker_genes <- c("PECAM1", "CD106", "CD62E", "Sele", "Kdr", "ENG")
gene_scores <- genescore_res$Z
rownames(gene_scores) <- genescore_res$genes$SYMBOL
marker_gene_scores <- gene_scores[grep(paste(sprintf("^%s$", marker_genes), collapse = "|"), rownames(gene_scores), ignore.case = T),]
par(mfrow = c(ceiling(nrow(marker_gene_scores)/2),2))
for(i in 1:nrow(marker_gene_scores)){
barplot(marker_gene_scores[i,], xlab = "topics", ylab = "gene score", main = rownames(marker_gene_scores)[i], col = colors_topics)
}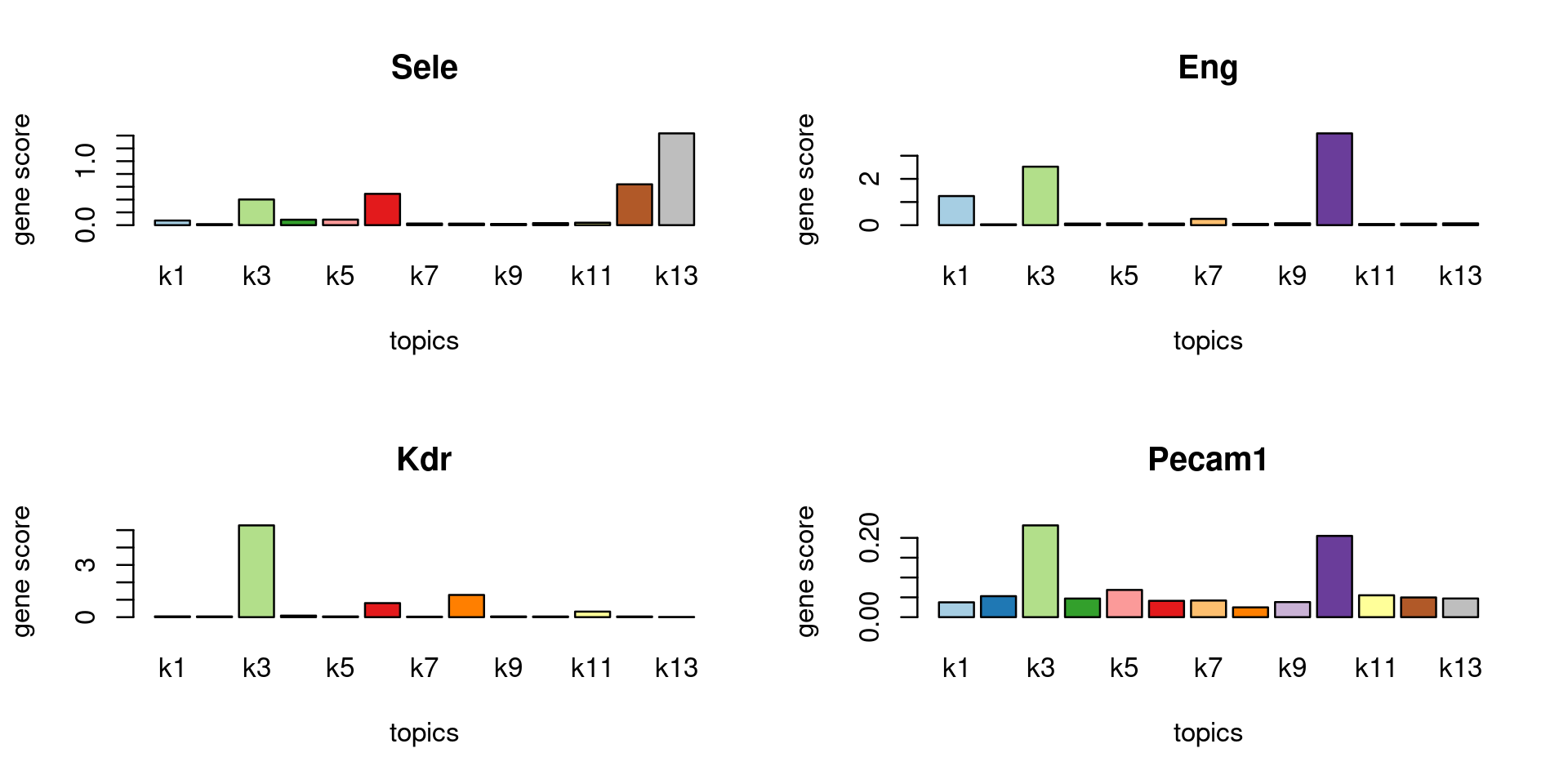
Topic 5 (Proximal tubule)
genescore_volcano_plot(genescore_res, k = 5, label_above_quantile = 0.99,
labels = genescore_res$genes$SYMBOL, max.overlaps = 20,
subsample_below_quantile = 0.5, subsample_rate = 0.1)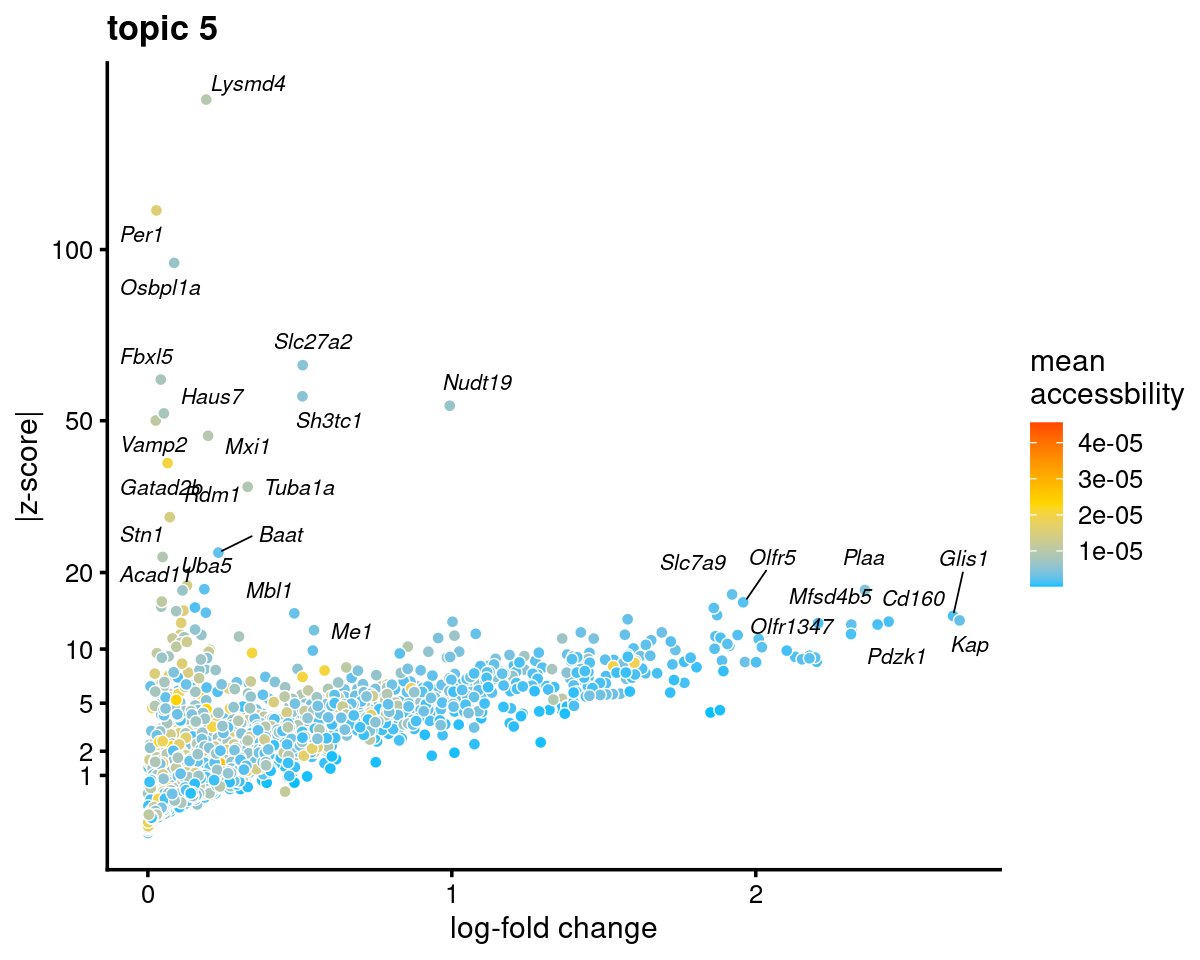
| Version | Author | Date |
|---|---|---|
| c010fd7 | kevinlkx | 2022-02-22 |
Check some known marker genes
marker_genes <- c("PALDOB", "CUBN", "LRP2", "SLC34A1")
gene_scores <- genescore_res$Z
rownames(gene_scores) <- genescore_res$genes$SYMBOL
marker_gene_scores <- gene_scores[grep(paste(sprintf("^%s$", marker_genes), collapse = "|"), rownames(gene_scores), ignore.case = T),]
par(mfrow = c(ceiling(nrow(marker_gene_scores)/2),2))
for(i in 1:nrow(marker_gene_scores)){
barplot(marker_gene_scores[i,], xlab = "topics", ylab = "gene score", main = rownames(marker_gene_scores)[i], col = colors_topics)
}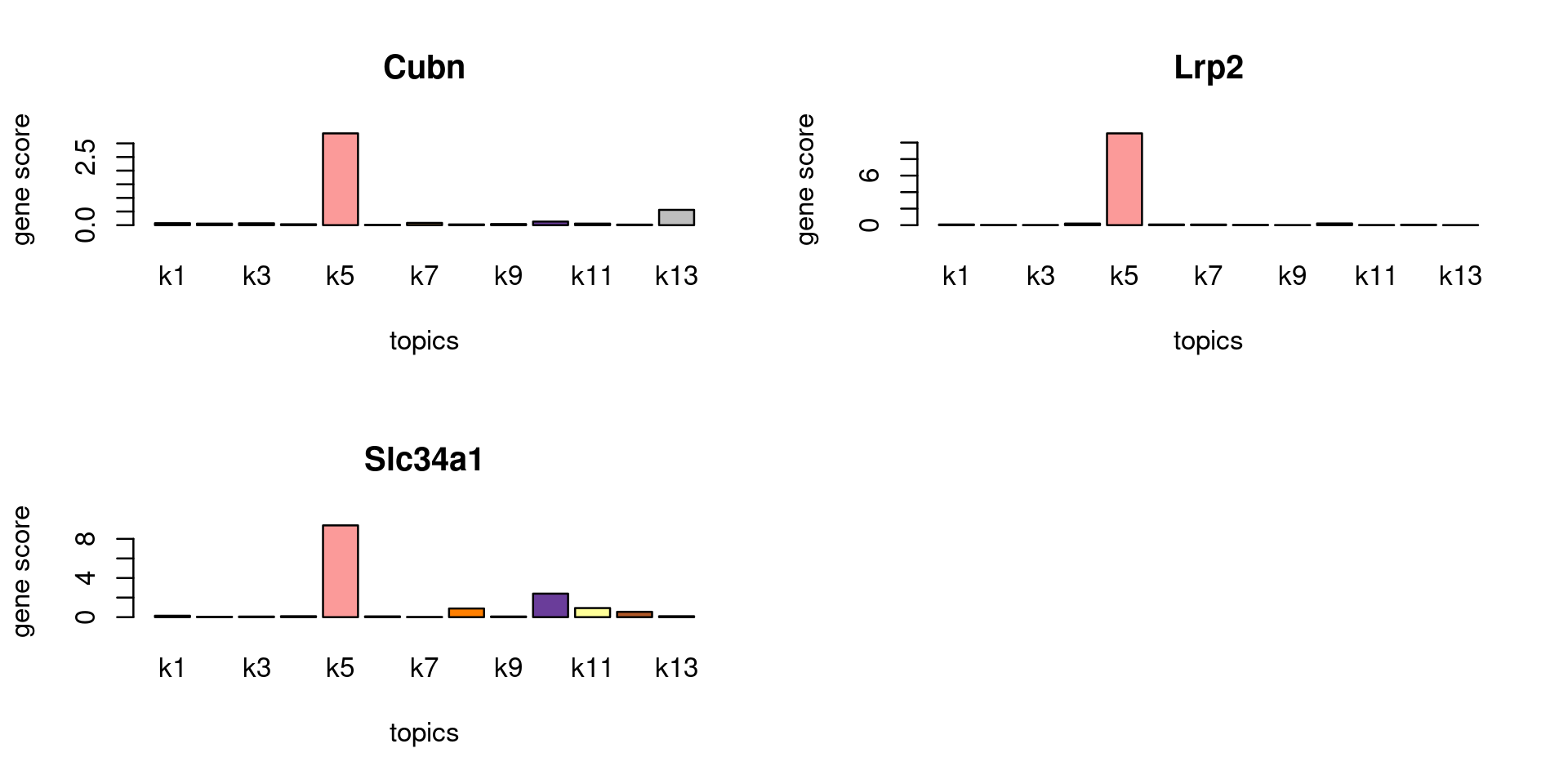
Topic 7 (Cardiomyocytes)
genescore_volcano_plot(genescore_res, k = 7, label_above_quantile = 0.99,
labels = genescore_res$genes$SYMBOL, max.overlaps = 20,
subsample_below_quantile = 0.5, subsample_rate = 0.1)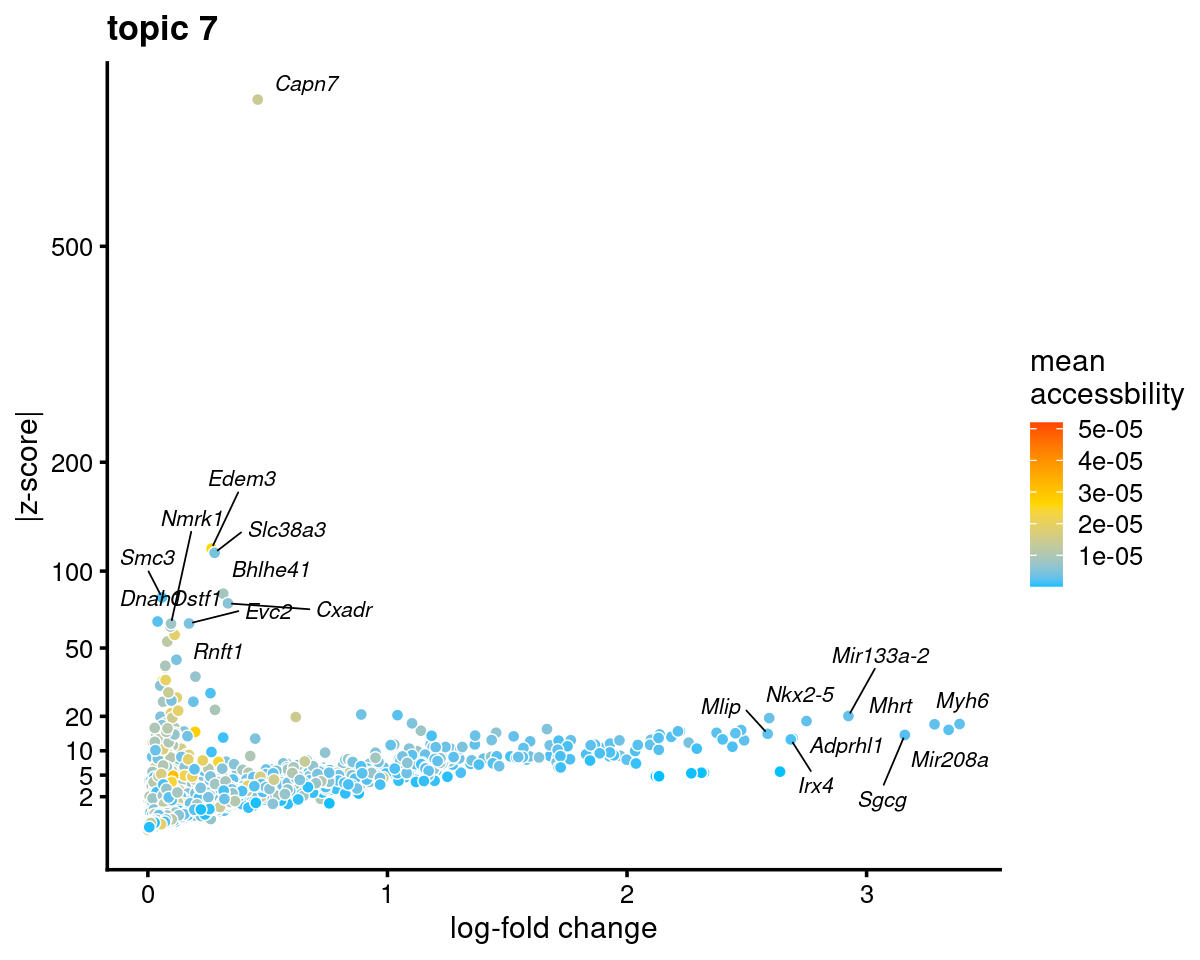
| Version | Author | Date |
|---|---|---|
| c010fd7 | kevinlkx | 2022-02-22 |
Check some known marker genes
marker_genes <- c("Nppa", "Myl4", "SLN", "PITX2", "Myl7", "Gja5", "Myl2", "Myl3", "IRX4", "HAND1", "HEY2")
gene_scores <- genescore_res$Z
rownames(gene_scores) <- genescore_res$genes$SYMBOL
marker_gene_scores <- gene_scores[grep(paste(sprintf("^%s$", marker_genes), collapse = "|"), rownames(gene_scores), ignore.case = T),]
par(mfrow = c(ceiling(nrow(marker_gene_scores)/3),3))
for(i in 1:nrow(marker_gene_scores)){
barplot(marker_gene_scores[i,], xlab = "topics", ylab = "gene score", main = rownames(marker_gene_scores)[i], col = colors_topics)
}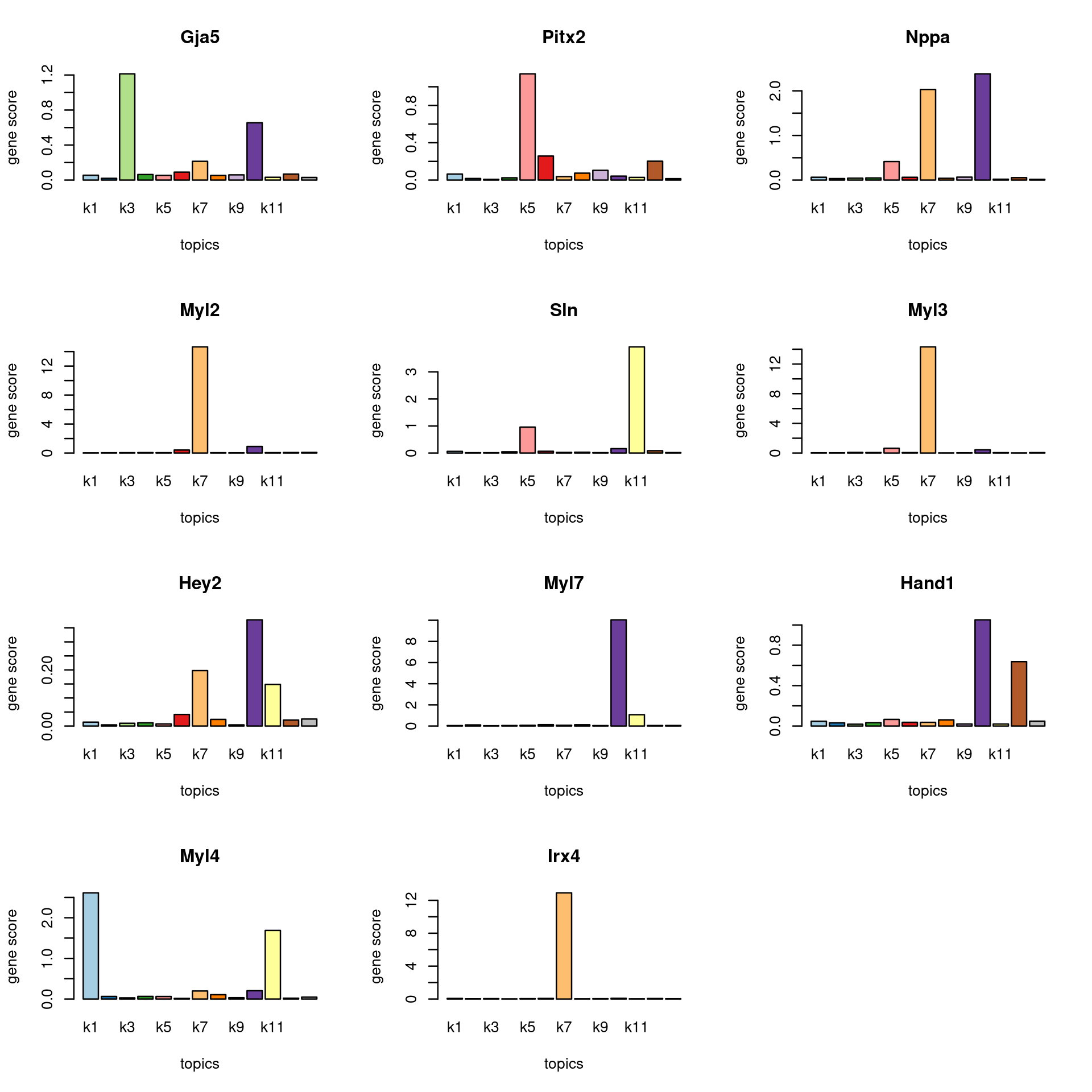
Topic 8 (Hepatocytes)
genescore_volcano_plot(genescore_res, k = 8, label_above_quantile = 0.99,
labels = genescore_res$genes$SYMBOL, max.overlaps = 20,
subsample_below_quantile = 0.5, subsample_rate = 0.1)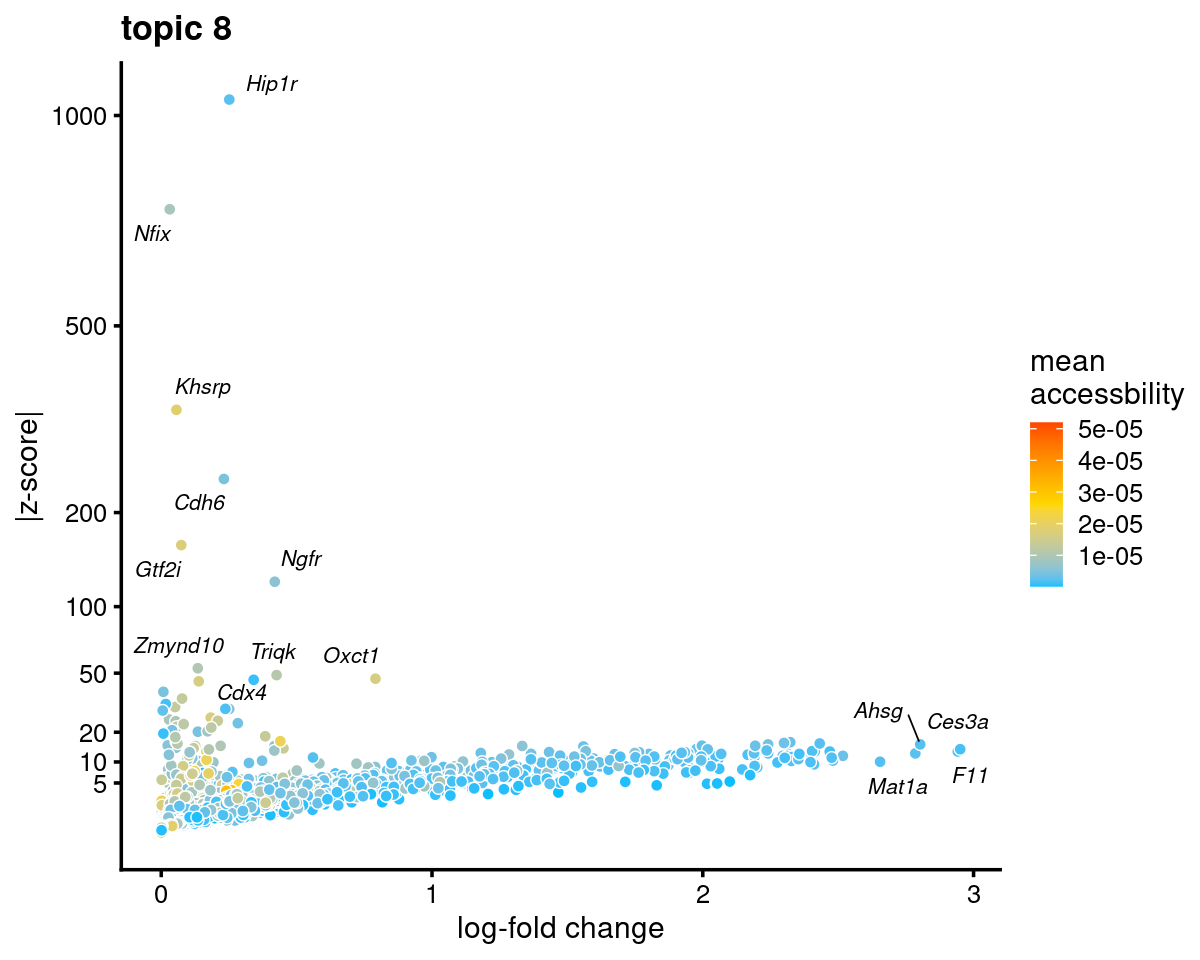
| Version | Author | Date |
|---|---|---|
| c010fd7 | kevinlkx | 2022-02-22 |
Check some known marker genes
marker_genes <- c("SERPINA1", "TTR", "ALB","AFP","CYP3A4","CYP7A1","FABP1","ALR","Glut1","MET","FoxA1","FoxA2","CD29","PTP4A2","Prox1", "HNF1B")
gene_scores <- genescore_res$Z
rownames(gene_scores) <- genescore_res$genes$SYMBOL
marker_gene_scores <- gene_scores[grep(paste(sprintf("^%s$", marker_genes), collapse = "|"), rownames(gene_scores), ignore.case = T),]
par(mfrow = c(ceiling(nrow(marker_gene_scores)/3),3))
for(i in 1:nrow(marker_gene_scores)){
barplot(marker_gene_scores[i,], xlab = "topics", ylab = "gene score", main = rownames(marker_gene_scores)[i], col = colors_topics)
}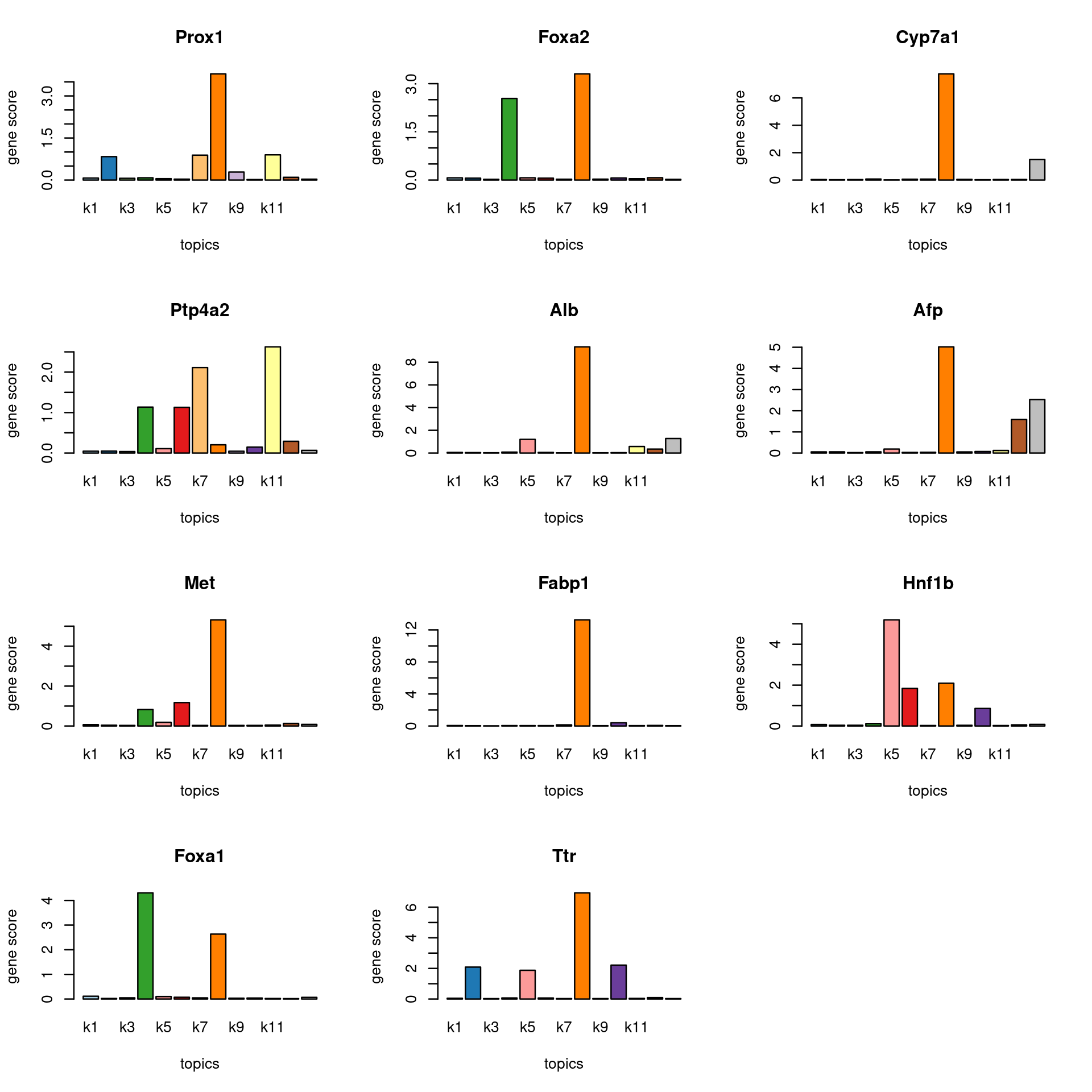
TSS model ver2
- use Z scores
normalized by the sum of weights
Top genes
gene.dir <- paste0(out.dir, "/geneanalysis-Cusanovich2018-k=13-TSS-Z-sum")
cat(sprintf("Directory of gene analysis result: %s \n", gene.dir))
genescore_tss_res <- readRDS(file.path(gene.dir, "genescore_result.rds"))
genescore_res <- genescore_tss_res
genes <- genescore_res$genes
gene_scores <- genescore_res$Z
gene_logFC <- genescore_res$logFC
topics <- colnames(gene_scores)
top_genes <- data.frame(matrix(nrow=10, ncol = ncol(gene_scores)))
colnames(top_genes) <- topics
for (k in topics){
top_genes[,k] <- genes$SYMBOL[head(order(abs(gene_scores[,k]), decreasing=TRUE), 10)]
}
DT::datatable(data.frame(rank = 1:10, top_genes), rownames = F, caption = "Top 10 genes by abs(gene z-scores)")# Directory of gene analysis result: /project2/mstephens/kevinluo/scATACseq-topics/output/Cusanovich_2018/postfit_v2/geneanalysis-Cusanovich2018-k=13-TSS-Z-sum- Volcano plots
genescore_volcano_plot(genescore_res, label_above_quantile = 0.99,
labels = genescore_res$genes$SYMBOL, max.overlaps = 20,
subsample_below_quantile = 0.5, subsample_rate = 0.1)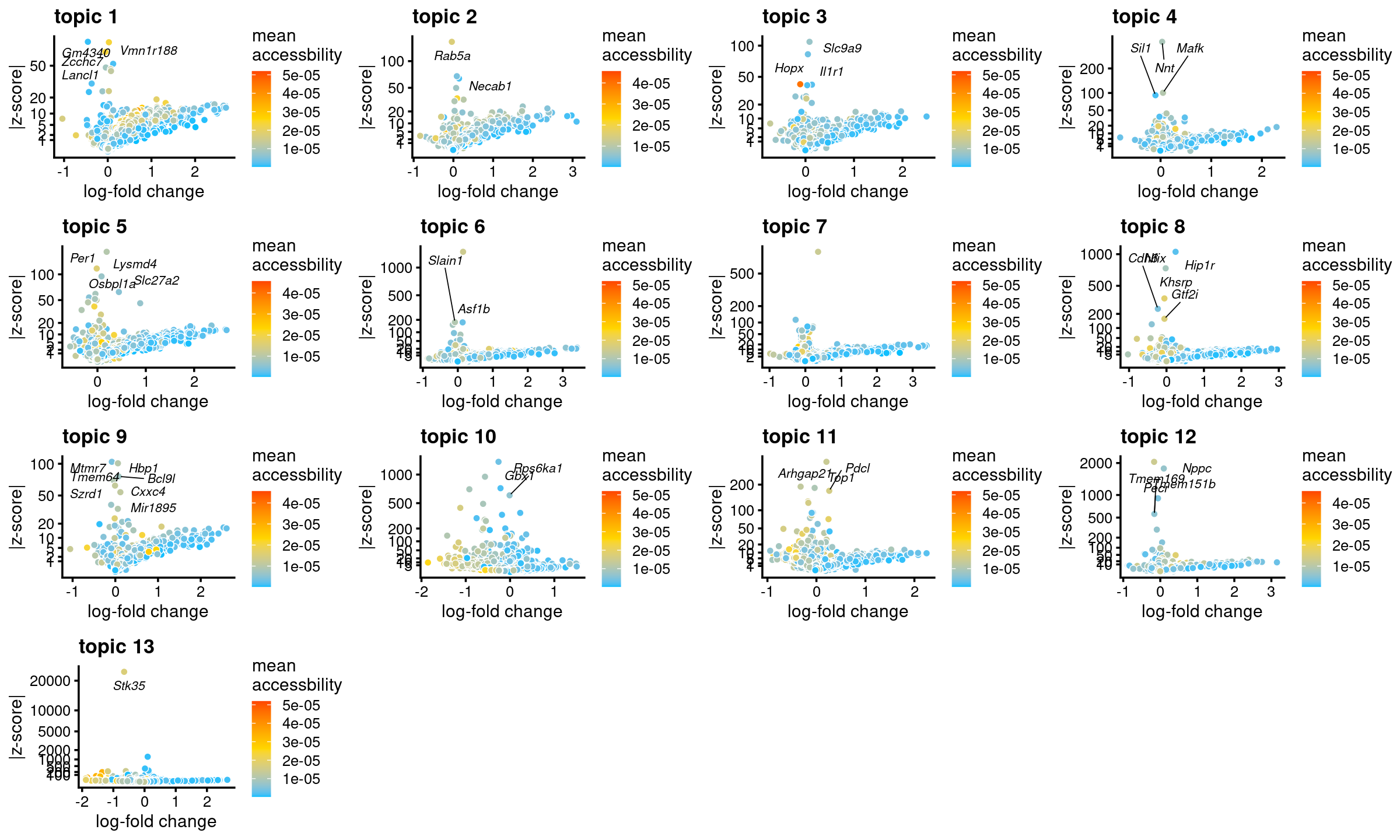
Topic 1 (Erythroblasts)
genescore_volcano_plot(genescore_res, k = 1, label_above_quantile = 0.99,
labels = genescore_res$genes$SYMBOL, max.overlaps = 20,
subsample_below_quantile = 0.5, subsample_rate = 0.1)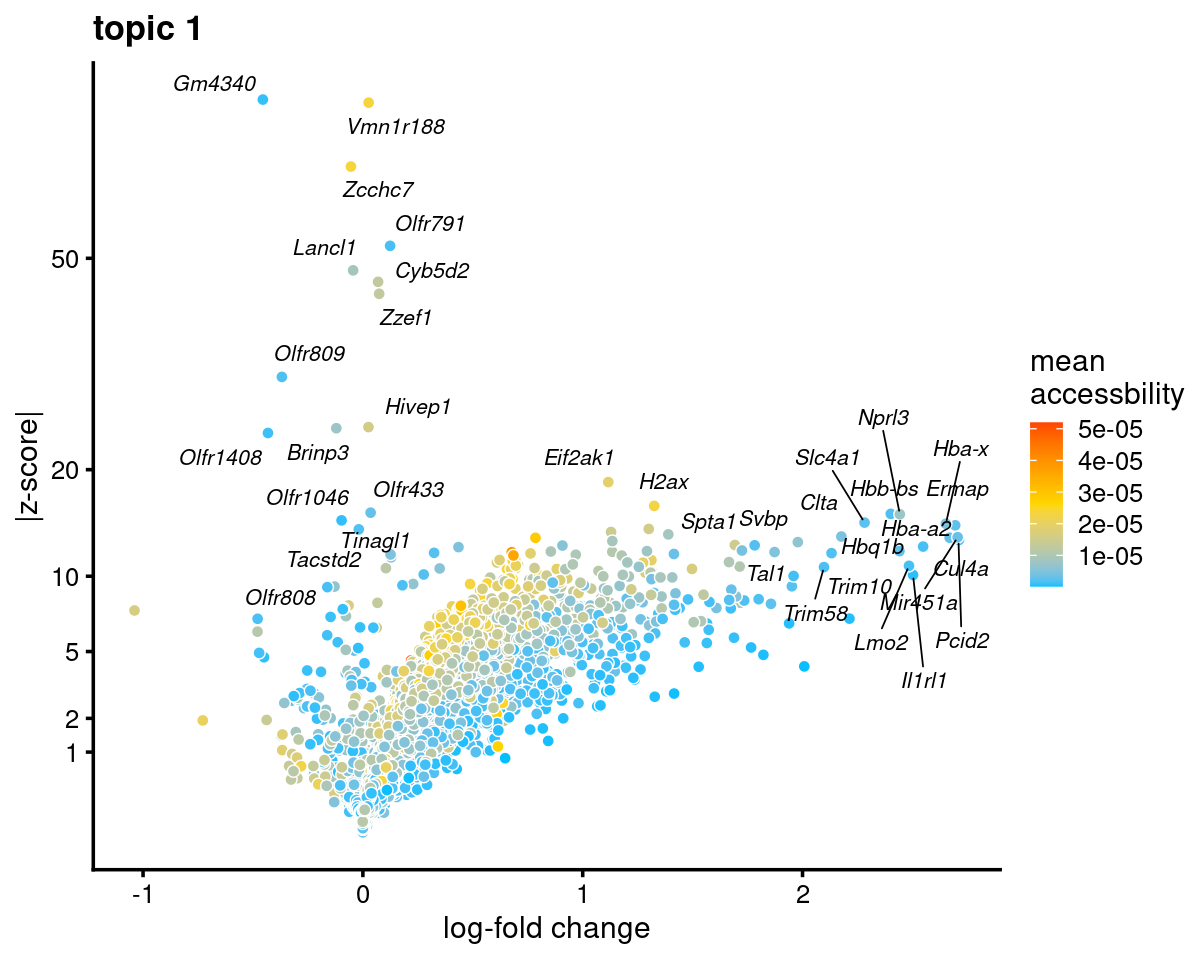
Check some known marker genes
marker_genes <- c("Hbb-b1", "Hbb-b2", "Gypa")
gene_scores <- genescore_res$Z
rownames(gene_scores) <- genescore_res$genes$SYMBOL
marker_gene_scores <- gene_scores[grep(paste(sprintf("^%s$", marker_genes), collapse = "|"), rownames(gene_scores), ignore.case = T),]
par(mfrow = c(ceiling(nrow(marker_gene_scores)/2),2))
for(i in 1:nrow(marker_gene_scores)){
barplot(marker_gene_scores[i,], xlab = "topics", ylab = "gene score", main = rownames(marker_gene_scores)[i], col = colors_topics)
}
Topic 3 (Endothelial cells)
genescore_volcano_plot(genescore_res, k = 3, label_above_quantile = 0.99,
labels = genescore_res$genes$SYMBOL, max.overlaps = 20,
subsample_below_quantile = 0.5, subsample_rate = 0.1)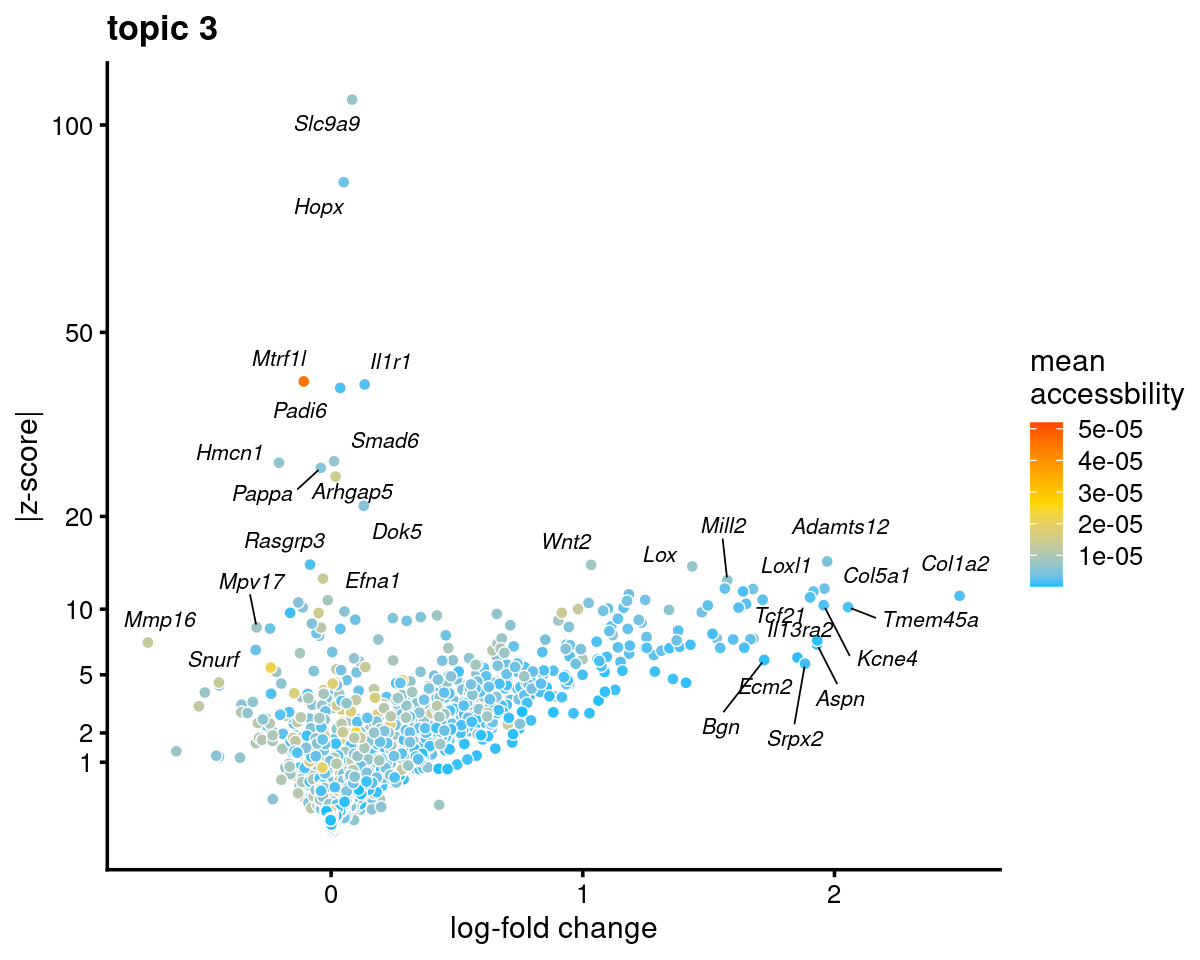
Check some known marker genes
marker_genes <- c("PECAM1", "CD106", "CD62E", "Sele", "Kdr", "ENG")
gene_scores <- genescore_res$Z
rownames(gene_scores) <- genescore_res$genes$SYMBOL
marker_gene_scores <- gene_scores[grep(paste(sprintf("^%s$", marker_genes), collapse = "|"), rownames(gene_scores), ignore.case = T),]
par(mfrow = c(ceiling(nrow(marker_gene_scores)/2),2))
for(i in 1:nrow(marker_gene_scores)){
barplot(marker_gene_scores[i,], xlab = "topics", ylab = "gene score", main = rownames(marker_gene_scores)[i], col = colors_topics)
}
Topic 5 (Proximal tubule)
genescore_volcano_plot(genescore_res, k = 5, label_above_quantile = 0.99,
labels = genescore_res$genes$SYMBOL, max.overlaps = 20,
subsample_below_quantile = 0.5, subsample_rate = 0.1)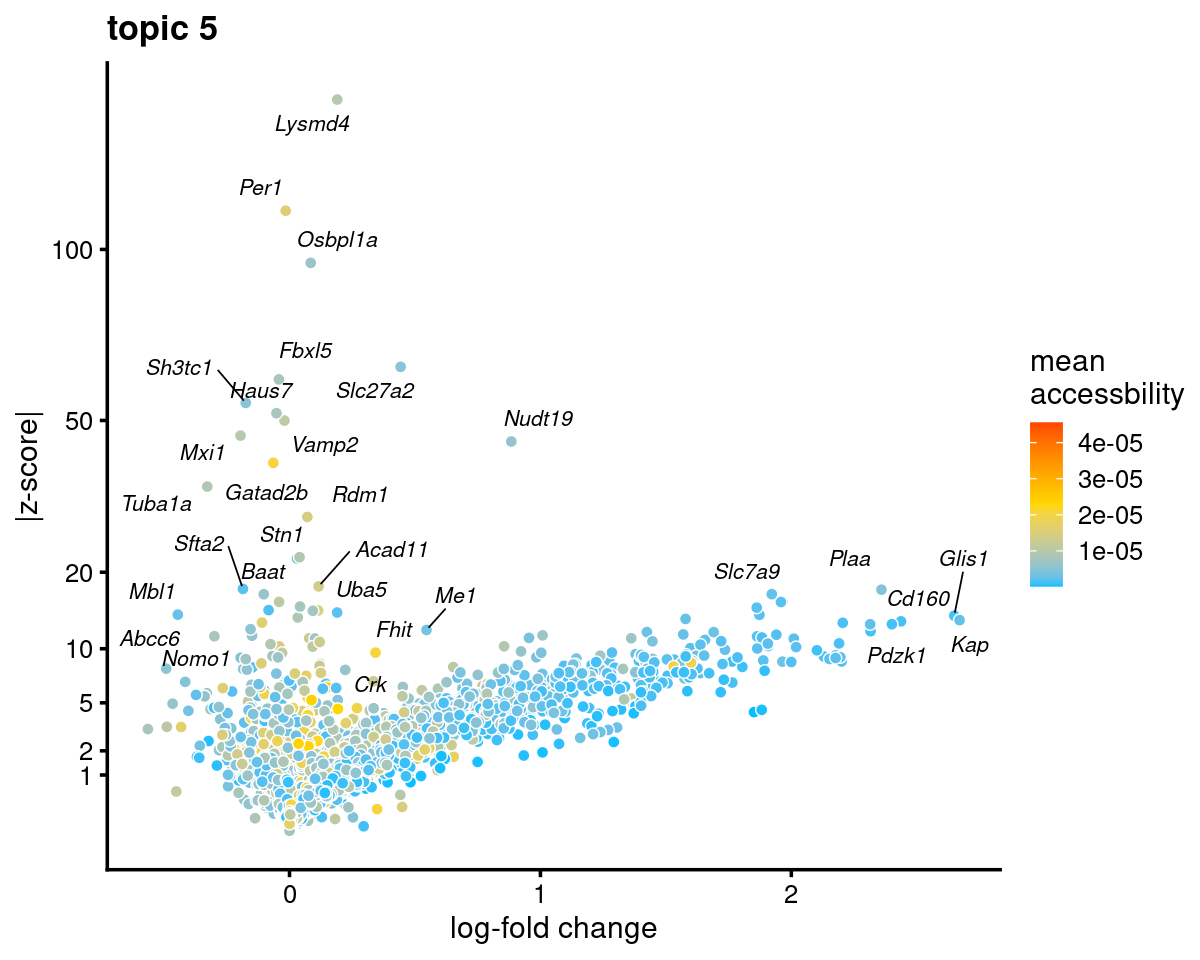
Check some known marker genes
marker_genes <- c("PALDOB", "CUBN", "LRP2", "SLC34A1")
gene_scores <- genescore_res$Z
rownames(gene_scores) <- genescore_res$genes$SYMBOL
marker_gene_scores <- gene_scores[grep(paste(sprintf("^%s$", marker_genes), collapse = "|"), rownames(gene_scores), ignore.case = T),]
par(mfrow = c(ceiling(nrow(marker_gene_scores)/2),2))
for(i in 1:nrow(marker_gene_scores)){
barplot(marker_gene_scores[i,], xlab = "topics", ylab = "gene score", main = rownames(marker_gene_scores)[i], col = colors_topics)
}
Topic 7 (Cardiomyocytes)
genescore_volcano_plot(genescore_res, k = 7, label_above_quantile = 0.99,
labels = genescore_res$genes$SYMBOL, max.overlaps = 20,
subsample_below_quantile = 0.5, subsample_rate = 0.1)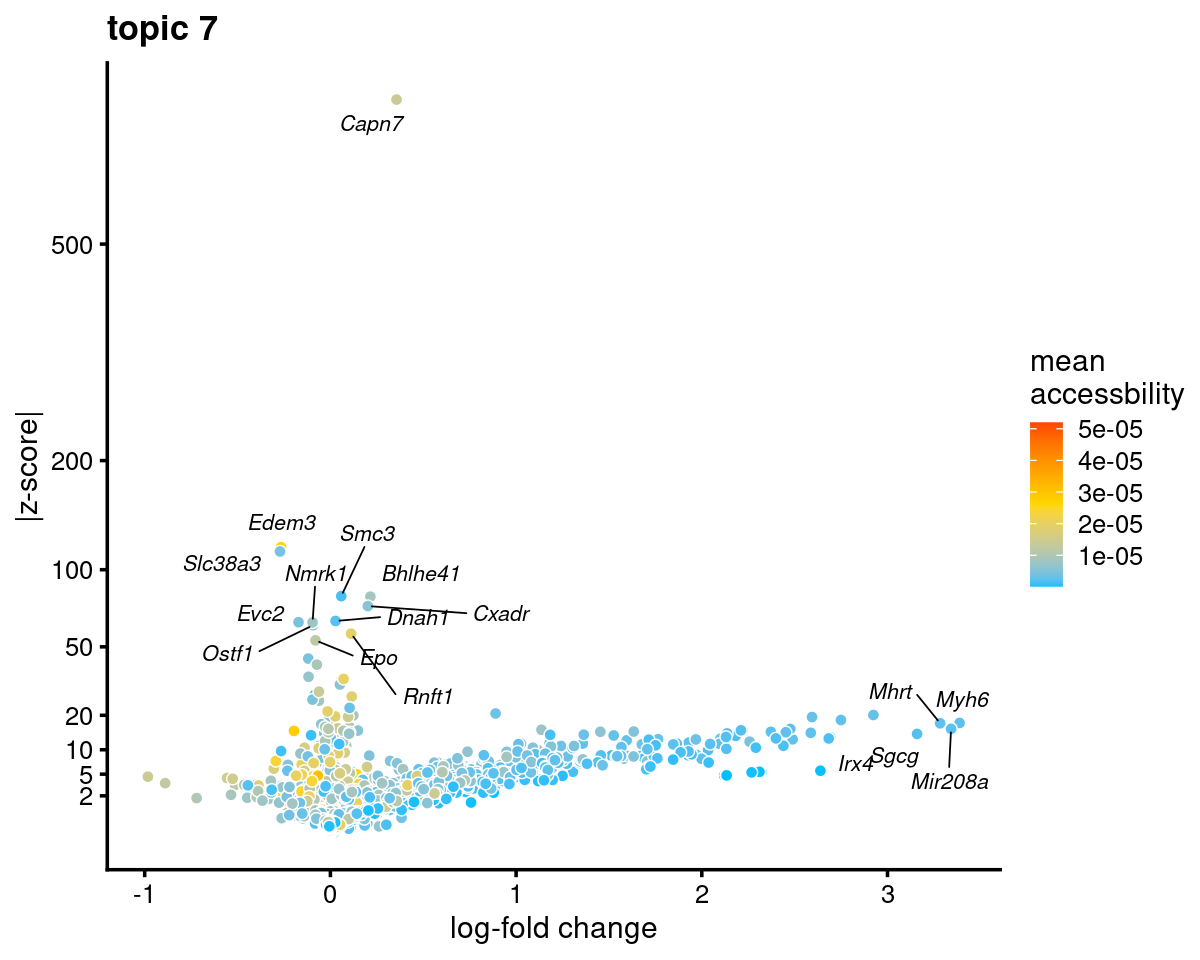
Check some known marker genes
marker_genes <- c("Nppa", "Myl4", "SLN", "PITX2", "Myl7", "Gja5", "Myl2", "Myl3", "IRX4", "HAND1", "HEY2")
gene_scores <- genescore_res$Z
rownames(gene_scores) <- genescore_res$genes$SYMBOL
marker_gene_scores <- gene_scores[grep(paste(sprintf("^%s$", marker_genes), collapse = "|"), rownames(gene_scores), ignore.case = T),]
par(mfrow = c(ceiling(nrow(marker_gene_scores)/3),3))
for(i in 1:nrow(marker_gene_scores)){
barplot(marker_gene_scores[i,], xlab = "topics", ylab = "gene score", main = rownames(marker_gene_scores)[i], col = colors_topics)
}
Topic 8 (Hepatocytes)
genescore_volcano_plot(genescore_res, k = 8, label_above_quantile = 0.99,
labels = genescore_res$genes$SYMBOL, max.overlaps = 20,
subsample_below_quantile = 0.5, subsample_rate = 0.1)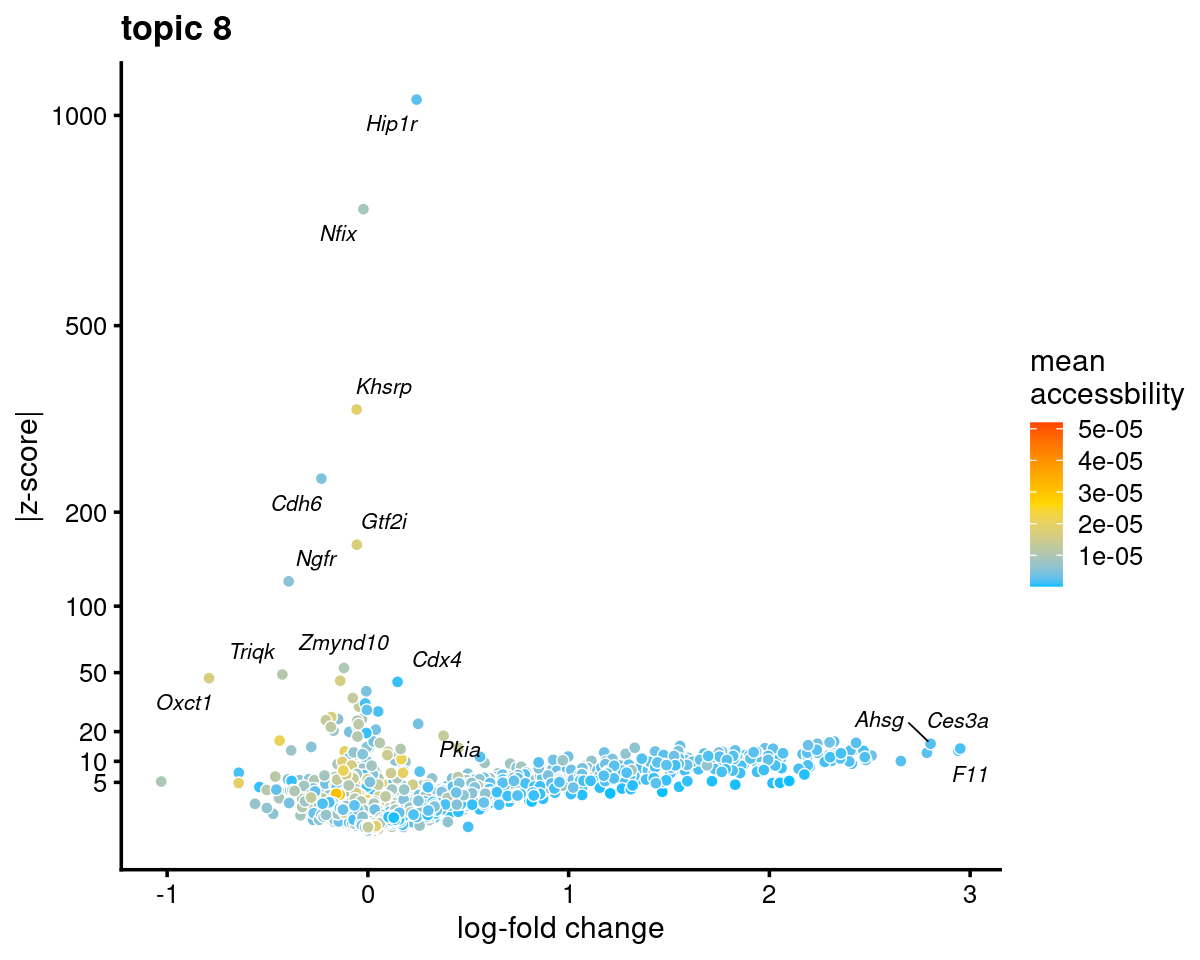
Check some known marker genes
marker_genes <- c("SERPINA1", "TTR", "ALB","AFP","CYP3A4","CYP7A1","FABP1","ALR","Glut1","MET","FoxA1","FoxA2","CD29","PTP4A2","Prox1", "HNF1B")
gene_scores <- genescore_res$Z
rownames(gene_scores) <- genescore_res$genes$SYMBOL
marker_gene_scores <- gene_scores[grep(paste(sprintf("^%s$", marker_genes), collapse = "|"), rownames(gene_scores), ignore.case = T),]
par(mfrow = c(ceiling(nrow(marker_gene_scores)/3),3))
for(i in 1:nrow(marker_gene_scores)){
barplot(marker_gene_scores[i,], xlab = "topics", ylab = "gene score", main = rownames(marker_gene_scores)[i], col = colors_topics)
}
Gene body model
Gene scores were computed using the gene score model (model 42) in the archR paper with some modifications. This model uses bi-directional exponential decays from the gene TSS (extended upstream by 5 kb by default) and the gene transcription termination site (TTS). Note: the current version of the function does not account for neighboring gene boundaries.
- Top genes
gene.dir <- paste0(out.dir, "/geneanalysis-Cusanovich2018-k=13-genebody-absZ-sum")
cat(sprintf("Directory of gene analysis result: %s \n", gene.dir))
genescore_gb_res <- readRDS(file.path(gene.dir, "genescore_result.rds"))
genescore_res <- genescore_gb_res
genes <- genescore_res$genes
gene_scores <- genescore_res$Z
gene_logFC <- genescore_res$logFC
topics <- colnames(gene_scores)
top_genes <- data.frame(matrix(nrow=10, ncol = ncol(gene_scores)))
colnames(top_genes) <- topics
for (k in topics){
top_genes[,k] <- genes$SYMBOL[head(order(abs(gene_scores[,k]), decreasing=TRUE), 10)]
}
DT::datatable(data.frame(rank = 1:10, top_genes), rownames = F, caption = "Top 10 genes by abs(gene z-scores)")# Directory of gene analysis result: /project2/mstephens/kevinluo/scATACseq-topics/output/Cusanovich_2018/postfit_v2/geneanalysis-Cusanovich2018-k=13-genebody-absZ-sum- Volcano plots
genescore_volcano_plot(genescore_res, label_above_quantile = 0.99,
labels = genescore_res$genes$SYMBOL, max.overlaps = 20,
subsample_below_quantile = 0.5, subsample_rate = 0.1)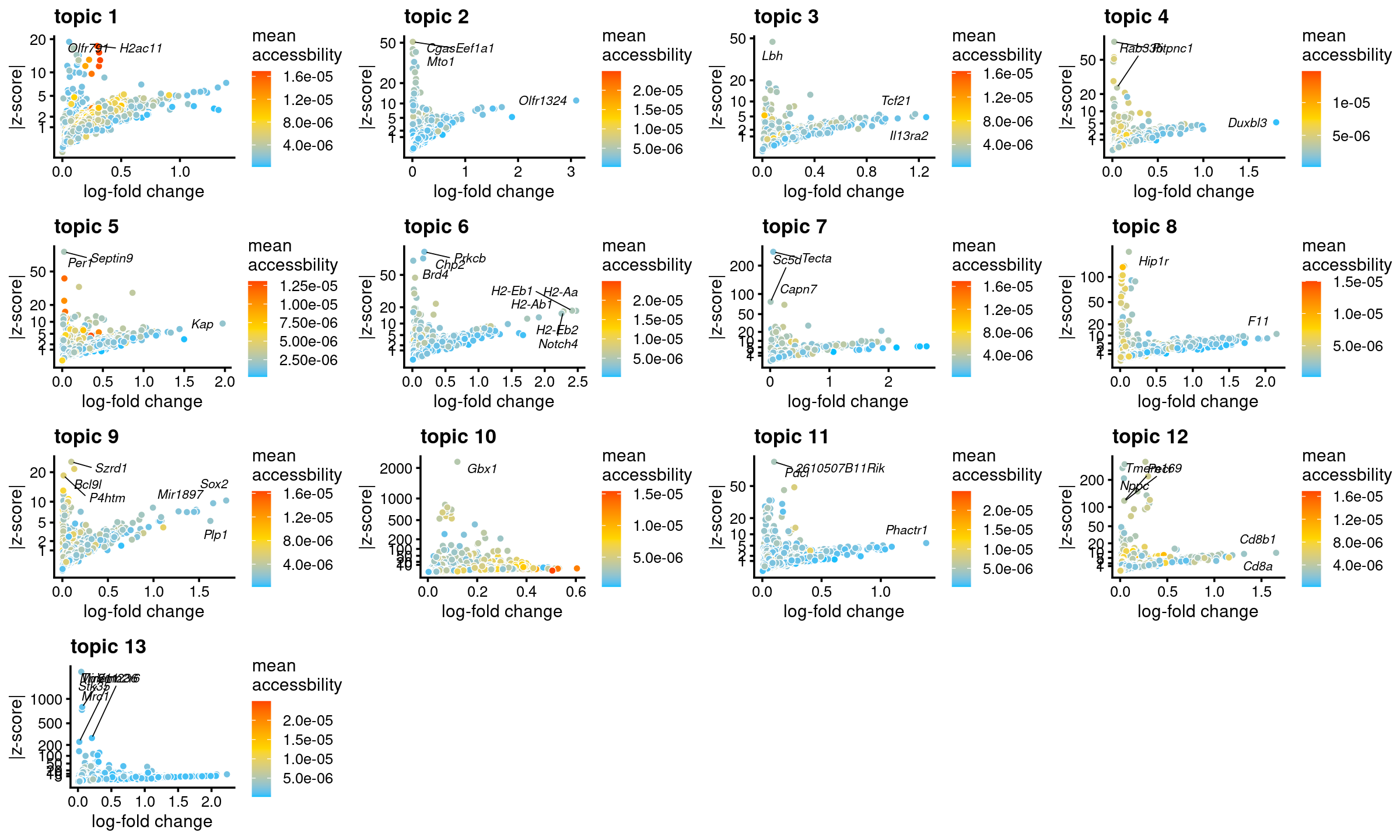
| Version | Author | Date |
|---|---|---|
| c010fd7 | kevinlkx | 2022-02-22 |
Topic 1 (Erythroblasts)
genescore_volcano_plot(genescore_res, k = 1, label_above_quantile = 0.99,
labels = genescore_res$genes$SYMBOL, max.overlaps = 20,
subsample_below_quantile = 0.5, subsample_rate = 0.1)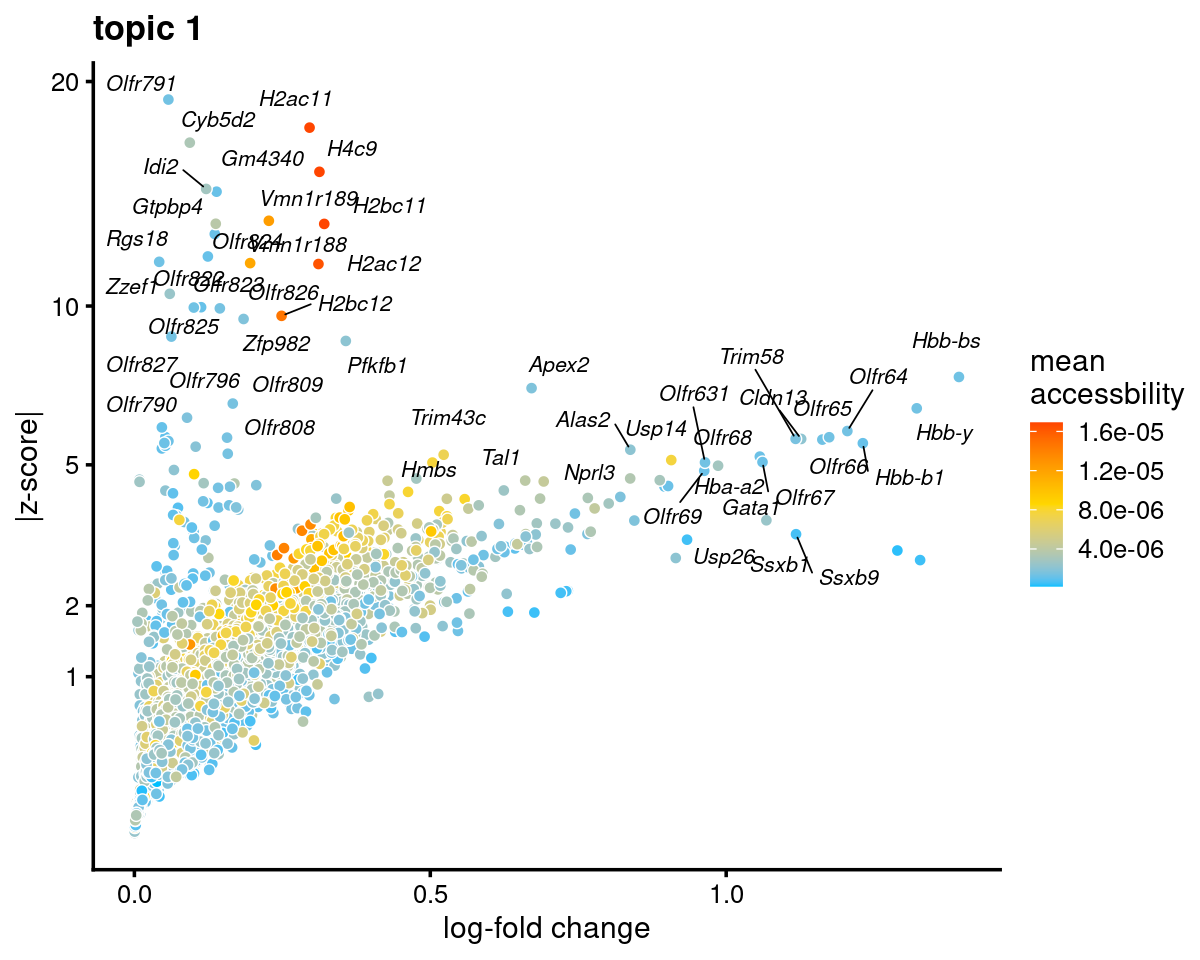
| Version | Author | Date |
|---|---|---|
| c010fd7 | kevinlkx | 2022-02-22 |
Check some known marker genes
marker_genes <- c("Hbb-b1", "Hbb-b2", "Gypa")
gene_scores <- genescore_res$Z
rownames(gene_scores) <- genescore_res$genes$SYMBOL
marker_gene_scores <- gene_scores[grep(paste(sprintf("^%s$", marker_genes), collapse = "|"), rownames(gene_scores), ignore.case = T),]
par(mfrow = c(ceiling(nrow(marker_gene_scores)/2),2))
for(i in 1:nrow(marker_gene_scores)){
barplot(marker_gene_scores[i,], xlab = "topics", ylab = "gene score", main = rownames(marker_gene_scores)[i], col = colors_topics)
}
Topic 3 (Endothelial cells)
genescore_volcano_plot(genescore_res, k = 3, label_above_quantile = 0.99,
labels = genescore_res$genes$SYMBOL, max.overlaps = 20,
subsample_below_quantile = 0.5, subsample_rate = 0.1)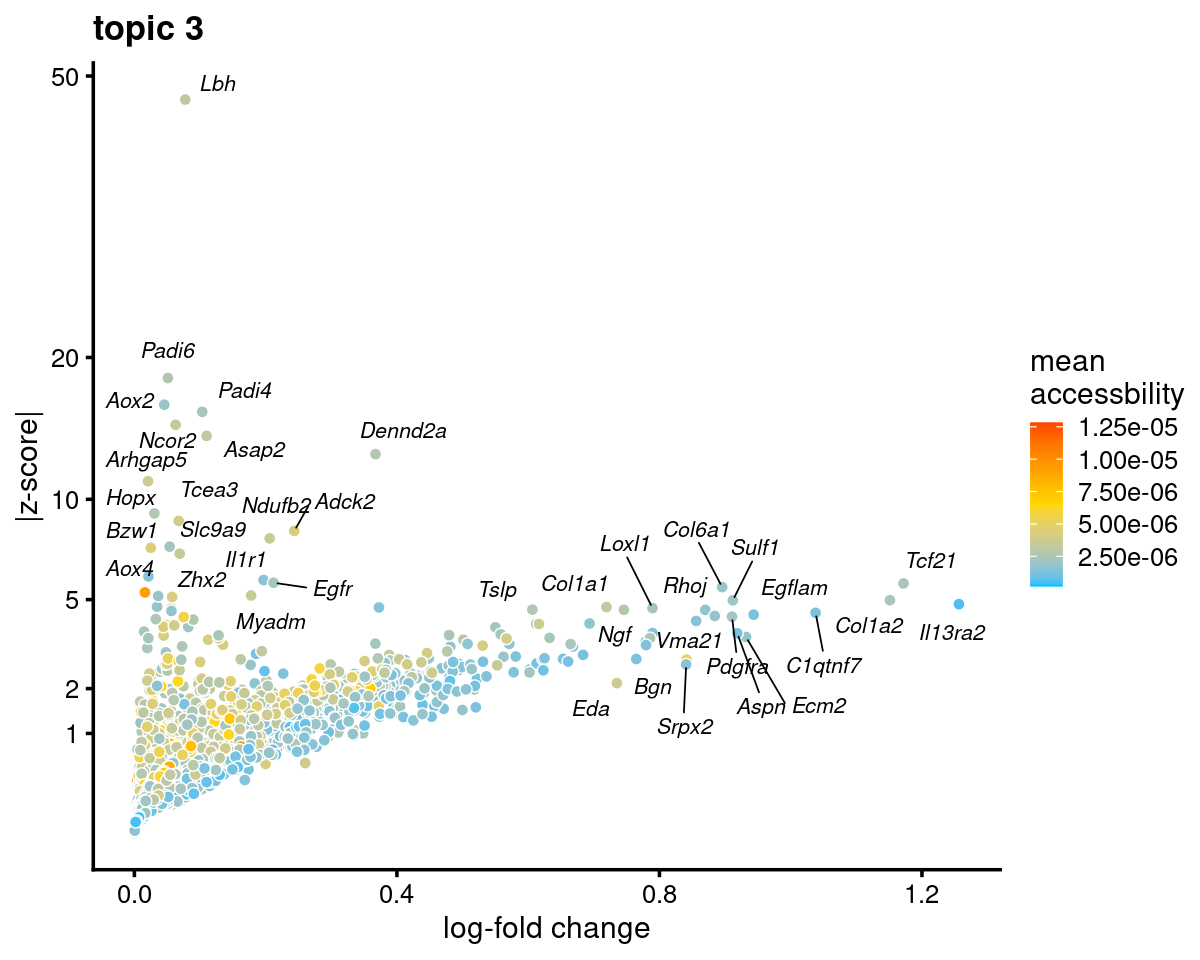
| Version | Author | Date |
|---|---|---|
| c010fd7 | kevinlkx | 2022-02-22 |
Check some known marker genes
marker_genes <- c("PECAM1", "CD106", "CD62E", "Sele", "Kdr", "ENG")
gene_scores <- genescore_res$Z
rownames(gene_scores) <- genescore_res$genes$SYMBOL
marker_gene_scores <- gene_scores[grep(paste(sprintf("^%s$", marker_genes), collapse = "|"), rownames(gene_scores), ignore.case = T),]
par(mfrow = c(ceiling(nrow(marker_gene_scores)/2),2))
for(i in 1:nrow(marker_gene_scores)){
barplot(marker_gene_scores[i,], xlab = "topics", ylab = "gene score", main = rownames(marker_gene_scores)[i], col = colors_topics)
}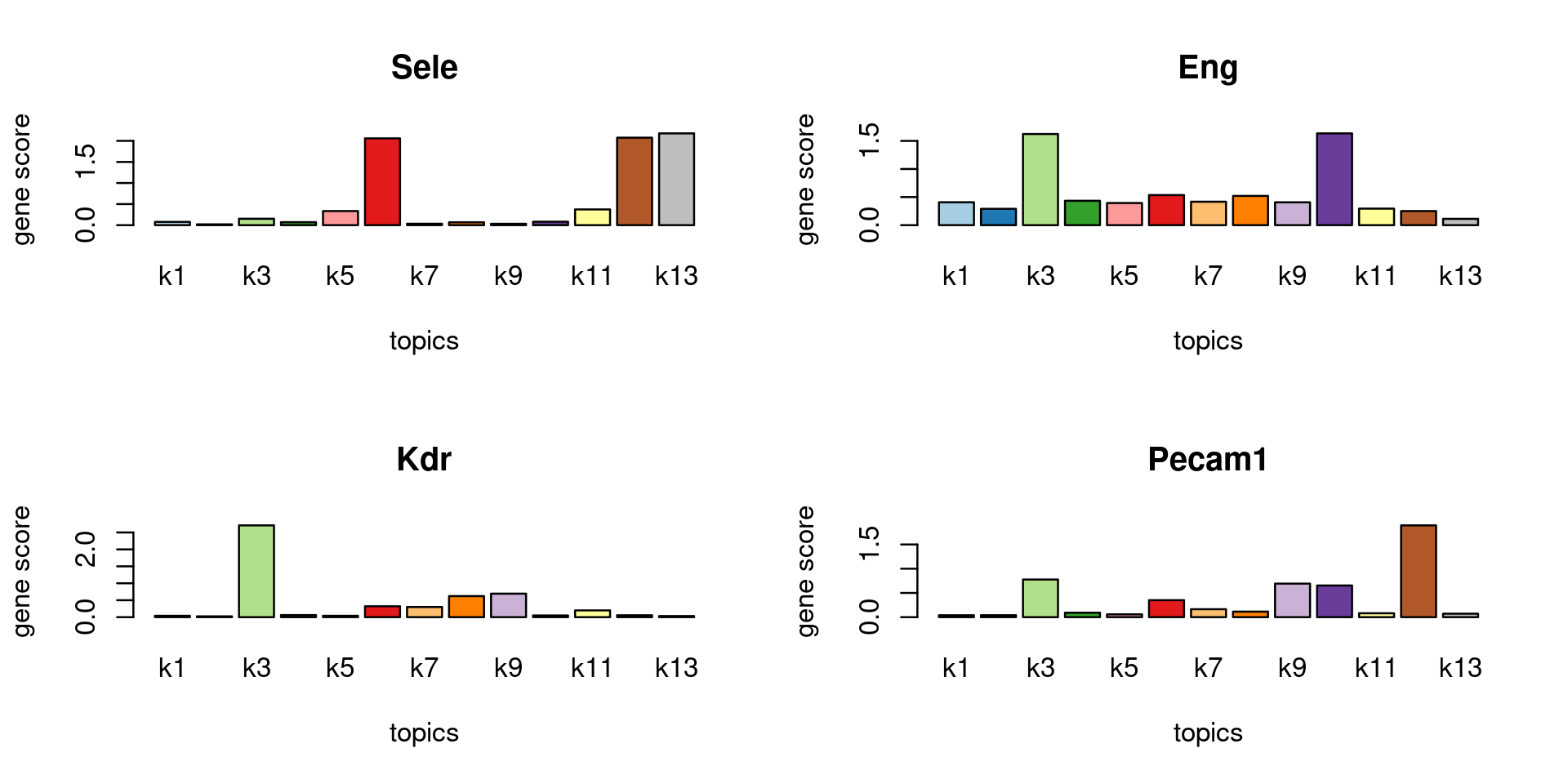
Topic 5 (Proximal tubule)
genescore_volcano_plot(genescore_res, k = 5, label_above_quantile = 0.99,
labels = genescore_res$genes$SYMBOL, max.overlaps = 20,
subsample_below_quantile = 0.5, subsample_rate = 0.1)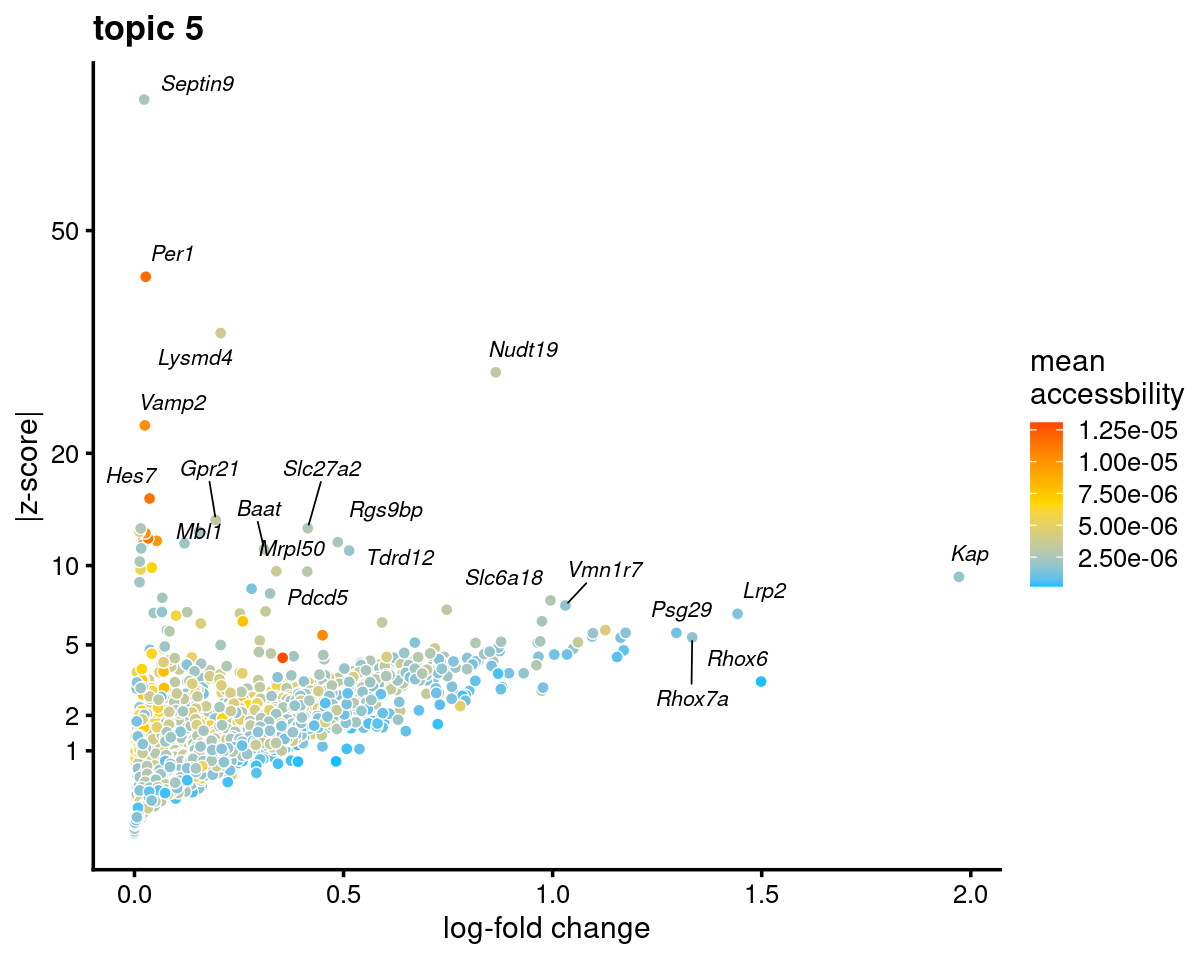
| Version | Author | Date |
|---|---|---|
| c010fd7 | kevinlkx | 2022-02-22 |
Check some known marker genes
marker_genes <- c("PALDOB", "CUBN", "LRP2", "SLC34A1")
gene_scores <- genescore_res$Z
rownames(gene_scores) <- genescore_res$genes$SYMBOL
marker_gene_scores <- gene_scores[grep(paste(sprintf("^%s$", marker_genes), collapse = "|"), rownames(gene_scores), ignore.case = T),]
par(mfrow = c(ceiling(nrow(marker_gene_scores)/2),2))
for(i in 1:nrow(marker_gene_scores)){
barplot(marker_gene_scores[i,], xlab = "topics", ylab = "gene score", main = rownames(marker_gene_scores)[i], col = colors_topics)
}
Topic 7 (Cardiomyocytes)
genescore_volcano_plot(genescore_res, k = 7, label_above_quantile = 0.99,
labels = genescore_res$genes$SYMBOL, max.overlaps = 20,
subsample_below_quantile = 0.5, subsample_rate = 0.1)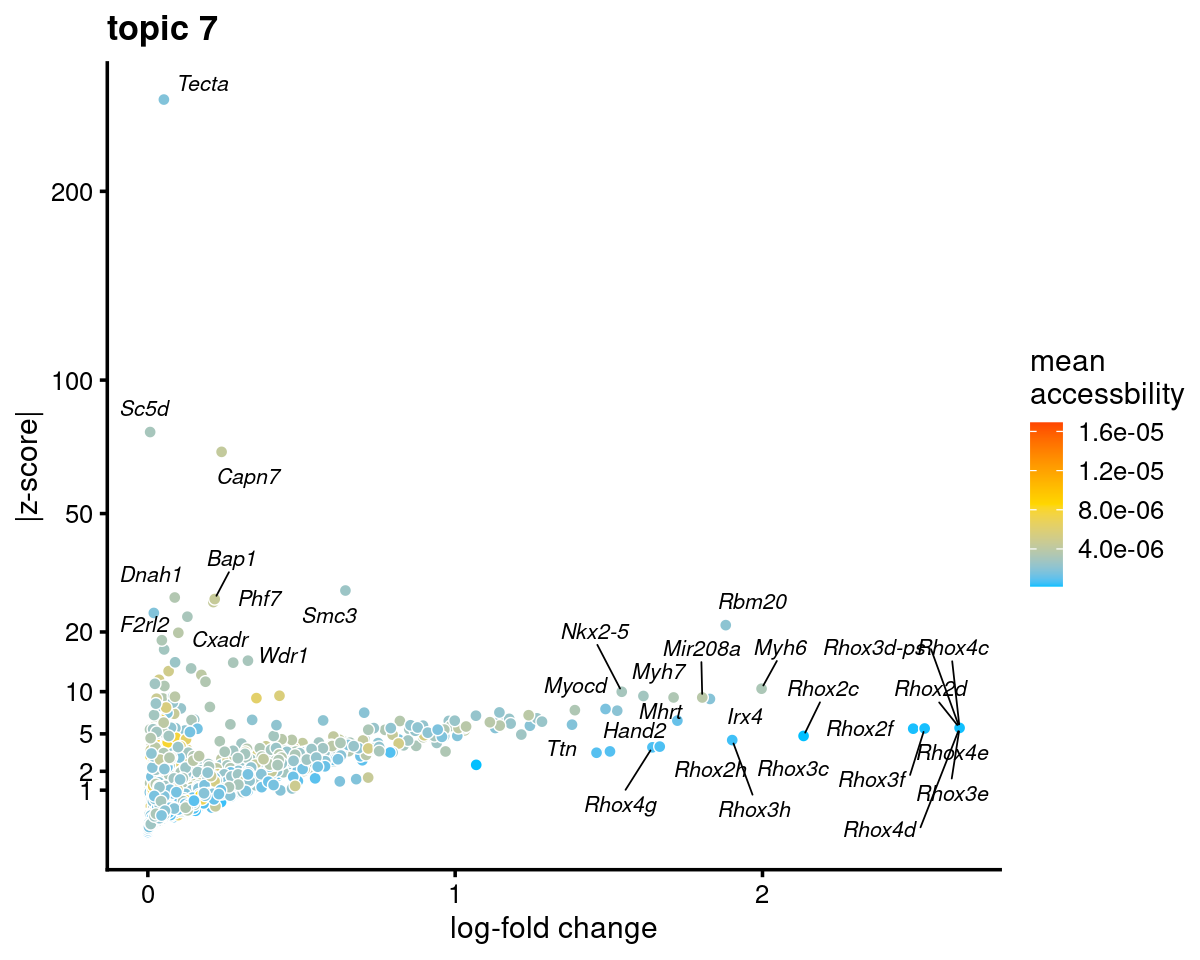
| Version | Author | Date |
|---|---|---|
| c010fd7 | kevinlkx | 2022-02-22 |
Check some known marker genes
marker_genes <- c("Nppa", "Myl4", "SLN", "PITX2", "Myl7", "Gja5", "Myl2", "Myl3", "IRX4", "HAND1", "HEY2")
gene_scores <- genescore_res$Z
rownames(gene_scores) <- genescore_res$genes$SYMBOL
marker_gene_scores <- gene_scores[grep(paste(sprintf("^%s$", marker_genes), collapse = "|"), rownames(gene_scores), ignore.case = T),]
par(mfrow = c(ceiling(nrow(marker_gene_scores)/3),3))
for(i in 1:nrow(marker_gene_scores)){
barplot(marker_gene_scores[i,], xlab = "topics", ylab = "gene score", main = rownames(marker_gene_scores)[i], col = colors_topics)
}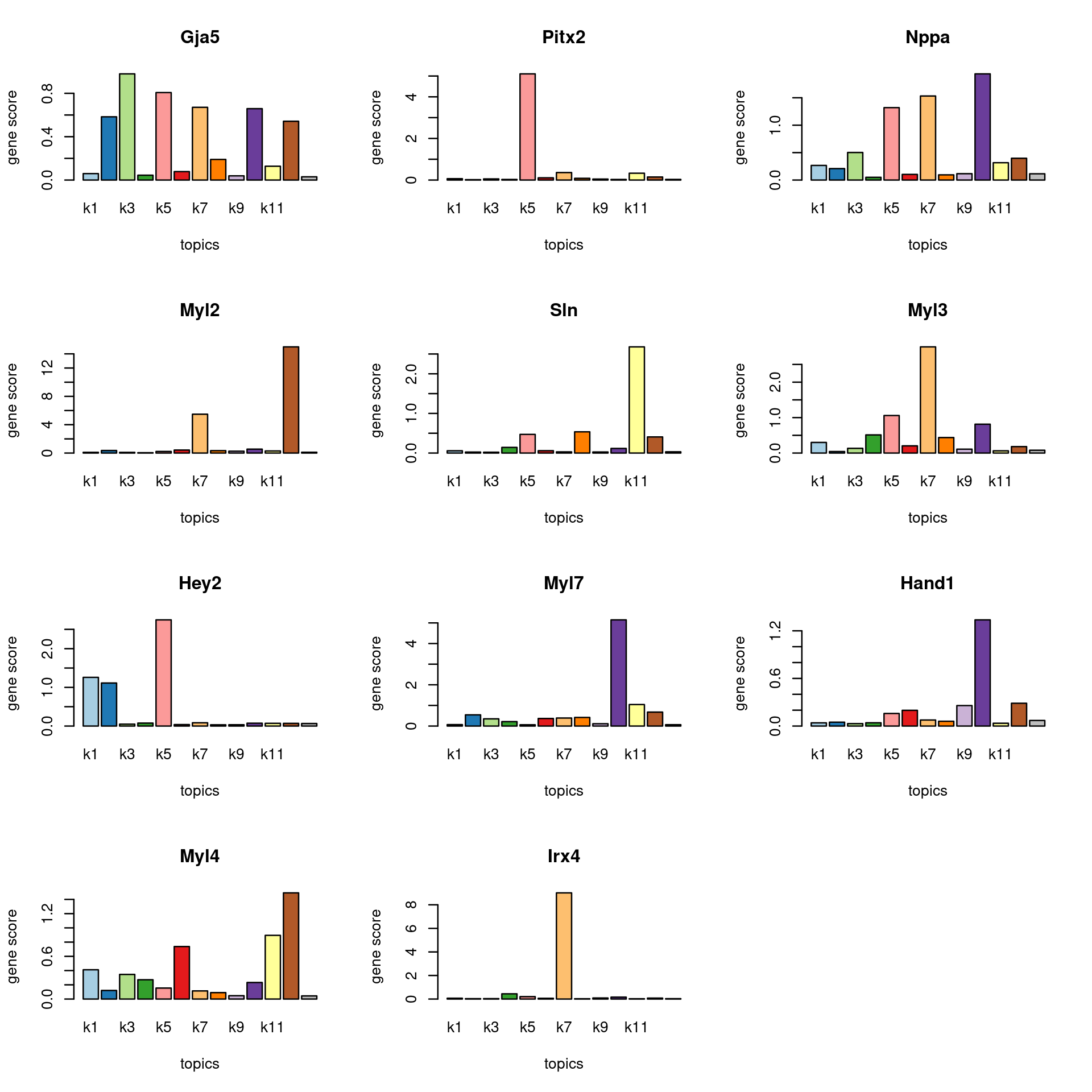
Topic 8 (Hepatocytes)
genescore_volcano_plot(genescore_res, k = 8, label_above_quantile = 0.99,
labels = genescore_res$genes$SYMBOL, max.overlaps = 20,
subsample_below_quantile = 0.5, subsample_rate = 0.1)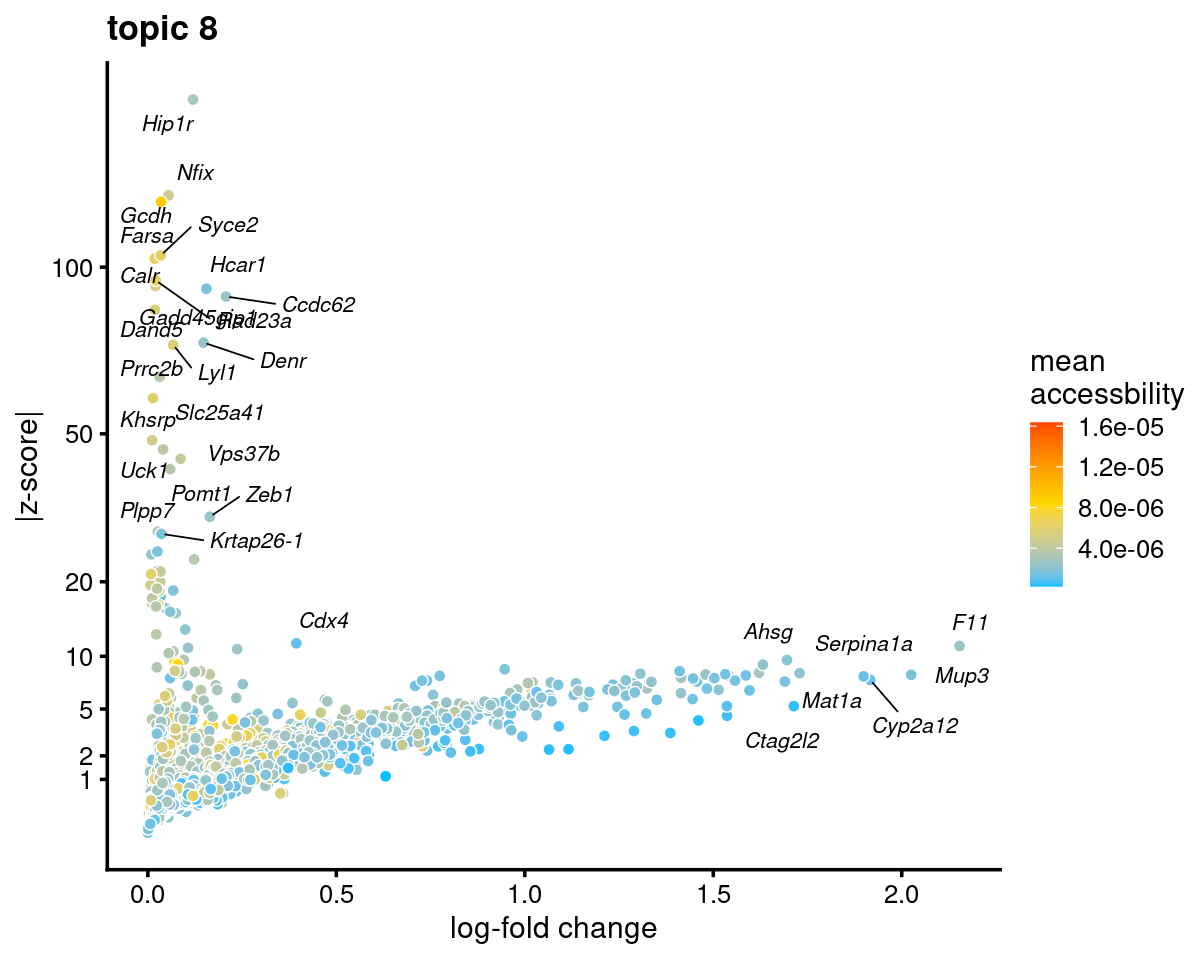
| Version | Author | Date |
|---|---|---|
| c010fd7 | kevinlkx | 2022-02-22 |
Check some known marker genes for Hepatocytes
marker_genes <- c("SERPINA1", "TTR", "ALB","AFP","CYP3A4","CYP7A1","FABP1","ALR","Glut1","MET","FoxA1","FoxA2","CD29","PTP4A2","Prox1", "HNF1B")
gene_scores <- genescore_res$Z
rownames(gene_scores) <- genescore_res$genes$SYMBOL
marker_gene_scores <- gene_scores[grep(paste(sprintf("^%s$", marker_genes), collapse = "|"), rownames(gene_scores), ignore.case = T),]
par(mfrow = c(ceiling(nrow(marker_gene_scores)/3),3))
for(i in 1:nrow(marker_gene_scores)){
barplot(marker_gene_scores[i,], xlab = "topics", ylab = "gene score", main = rownames(marker_gene_scores)[i], col = colors_topics)
}
Compare gene scores from the gene-body model vs. the TSS model.
m <- ncol(genescore_gb_res$Z)
plots <- vector("list",m)
names(plots) <- colnames(genescore_gb_res$Z)
for (i in 1:m) {
dat <- data.frame(genebody = genescore_gb_res$Z[,i], tss = genescore_tss_res$Z[,i])
plots[[i]] <-
ggplot(dat,aes_string(x = "genebody",y = "tss")) +
geom_point(shape = 21, na.rm = TRUE, size = 1, alpha = 1/10) +
geom_abline(intercept = 0, slope = 1, color="blue") +
labs(x = "gene body model",y = "TSS model",
title = paste("topic",i)) +
theme_cowplot(9)
}
do.call(plot_grid,plots)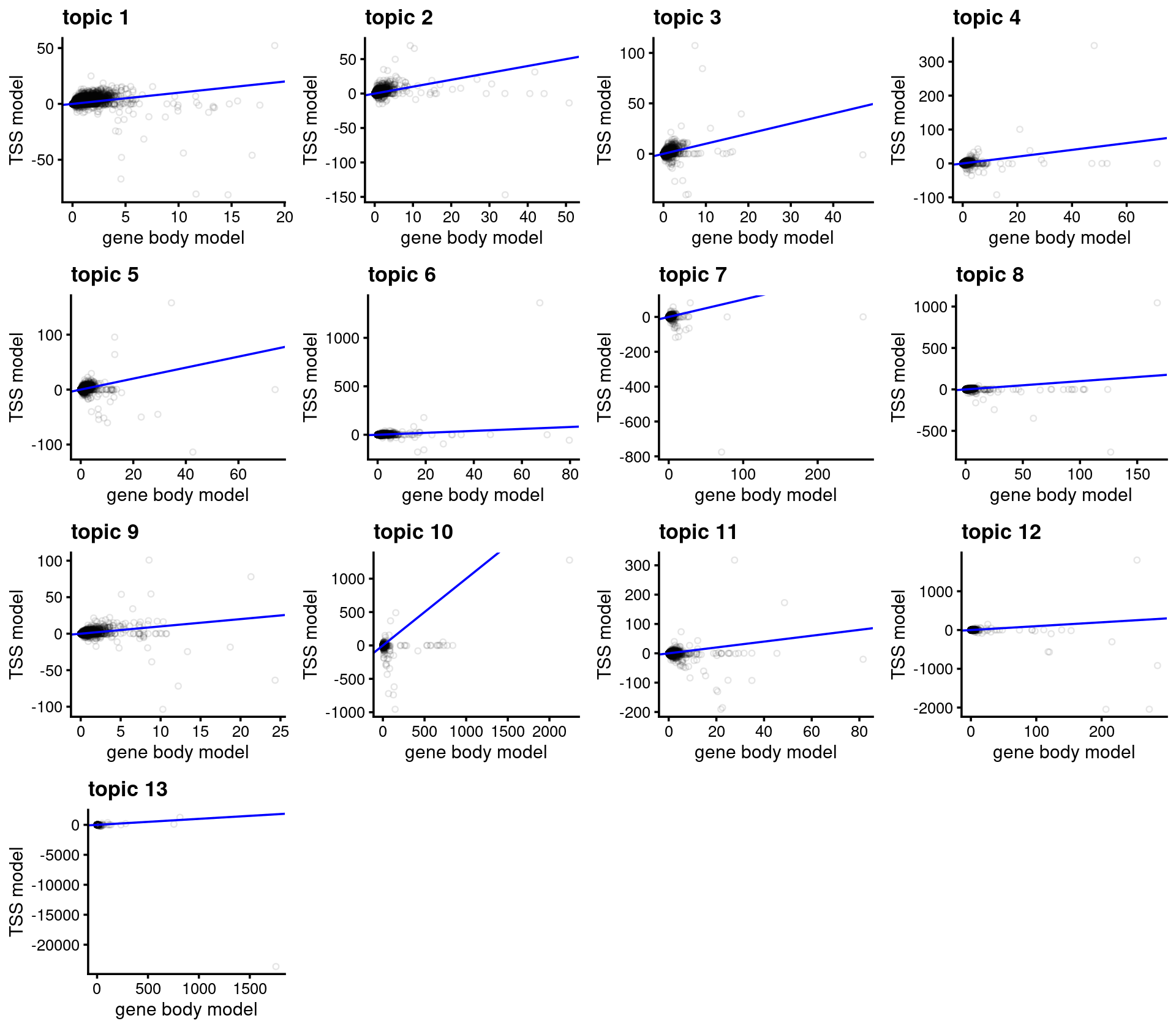
Gene-set enrichment analysis (GSEA)
Loading gene set data
cat("Loading mouse gene set data.\n")
data(gene_sets_mouse)
gene_sets <- gene_sets_mouse$gene_sets
gene_set_info <- gene_sets_mouse$gene_set_info# Loading mouse gene set data.TSS model
Top gene sets/pathways
gene.dir <- paste0(out.dir, "/geneanalysis-Cusanovich2018-k=13-TSS-absZ-sum")
cat(sprintf("Directory of gene analysis result: %s \n", gene.dir))
gsea_res <- readRDS(file.path(gene.dir, "gsea_result.rds"))
top_pathways_up <- top_pathways_down <- data.frame(matrix(nrow=10, ncol = ncol(gsea_res$pval)))
colnames(top_pathways_up) <- colnames(top_pathways_down) <- colnames(gsea_res$pval)
for (k in 1:ncol(gsea_res$pval)){
gsea_topic <- data.frame(pathway = rownames(gsea_res$pval),
pval = gsea_res$pval[,k],
log2err = gsea_res$log2err[,k],
ES = gsea_res$ES[,k],
NES = gsea_res$NES[,k])
gsea_up <- gsea_topic[gsea_topic$ES > 0,]
top_IDs_up <- as.character(gsea_up[head(order(gsea_up$pval), 10), "pathway"])
top_IDs_up <- gene_set_info[match(top_IDs_up, gene_set_info$id),c("name", "id")]
top_pathways_up[,k] <- paste0(top_IDs_up$name, "(", top_IDs_up$id, ")")
gsea_down <- gsea_topic[gsea_topic$ES < 0,]
top_IDs_down <- as.character(gsea_down[head(order(gsea_down$pval), 10), "pathway"])
top_IDs_down <- gene_set_info[match(top_IDs_down, gene_set_info$id),c("name", "id")]
top_pathways_down[,k] <- paste0(top_IDs_down$name, "(", top_IDs_down$id, ")")
}
DT::datatable(data.frame(rank = 1:10, top_pathways_up), rownames = F,
caption = "Top 10 pathways enriched at the top of the gene rank list.")# Directory of gene analysis result: /project2/mstephens/kevinluo/scATACseq-topics/output/Cusanovich_2018/postfit_v2/geneanalysis-Cusanovich2018-k=13-TSS-absZ-sumGene body model
- Top gene sets
gene.dir <- paste0(out.dir, "/geneanalysis-Cusanovich2018-k=13-genebody-absZ-sum")
cat(sprintf("Directory of gene analysis result: %s \n", gene.dir))
gsea_res <- readRDS(file.path(gene.dir, "gsea_result.rds"))
top_pathways_up <- top_pathways_down <- data.frame(matrix(nrow=10, ncol = ncol(gsea_res$pval)))
colnames(top_pathways_up) <- colnames(top_pathways_down) <- colnames(gsea_res$pval)
for (k in 1:ncol(gsea_res$pval)){
gsea_topic <- data.frame(pathway = rownames(gsea_res$pval),
pval = gsea_res$pval[,k],
log2err = gsea_res$log2err[,k],
ES = gsea_res$ES[,k],
NES = gsea_res$NES[,k])
gsea_up <- gsea_topic[gsea_topic$ES > 0,]
top_IDs_up <- as.character(gsea_up[head(order(gsea_up$pval), 10), "pathway"])
top_IDs_up <- gene_set_info[match(top_IDs_up, gene_set_info$id),c("name", "id")]
top_pathways_up[,k] <- paste0(top_IDs_up$name, "(", top_IDs_up$id, ")")
gsea_down <- gsea_topic[gsea_topic$ES < 0,]
top_IDs_down <- as.character(gsea_down[head(order(gsea_down$pval), 10), "pathway"])
top_IDs_down <- gene_set_info[match(top_IDs_down, gene_set_info$id),c("name", "id")]
top_pathways_down[,k] <- paste0(top_IDs_down$name, "(", top_IDs_down$id, ")")
}
DT::datatable(data.frame(rank = 1:10, top_pathways_up), rownames = F,
caption = "Top 10 pathways enriched at the top of the gene rank list.")# Directory of gene analysis result: /project2/mstephens/kevinluo/scATACseq-topics/output/Cusanovich_2018/postfit_v2/geneanalysis-Cusanovich2018-k=13-genebody-absZ-sum
sessionInfo()# R version 4.0.4 (2021-02-15)
# Platform: x86_64-pc-linux-gnu (64-bit)
# Running under: Scientific Linux 7.4 (Nitrogen)
#
# Matrix products: default
# BLAS/LAPACK: /software/openblas-0.3.13-el7-x86_64/lib/libopenblas_haswellp-r0.3.13.so
#
# locale:
# [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
# [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
# [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
# [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
# [9] LC_ADDRESS=C LC_TELEPHONE=C
# [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
#
# attached base packages:
# [1] stats graphics grDevices utils datasets methods base
#
# other attached packages:
# [1] reshape_0.8.8 DT_0.20 htmlwidgets_1.5.4 plotly_4.10.0
# [5] cowplot_1.1.1 ggrepel_0.9.1 ggplot2_3.3.5 tidyr_1.1.4
# [9] dplyr_1.0.8 pathways_0.1-20 fastTopics_0.6-97 Matrix_1.4-0
# [13] workflowr_1.7.0
#
# loaded via a namespace (and not attached):
# [1] fgsea_1.21.0 Rtsne_0.15 colorspace_2.0-3
# [4] ellipsis_0.3.2 class_7.3-20 rprojroot_2.0.2
# [7] fs_1.5.2 rstudioapi_0.13 farver_2.1.0
# [10] listenv_0.8.0 MatrixModels_0.5-0 prodlim_2019.11.13
# [13] fansi_1.0.2 lubridate_1.8.0 codetools_0.2-18
# [16] splines_4.0.4 knitr_1.37 jsonlite_1.7.3
# [19] pROC_1.18.0 mcmc_0.9-7 caret_6.0-90
# [22] ashr_2.2-47 uwot_0.1.11 compiler_4.0.4
# [25] httr_1.4.2 assertthat_0.2.1 fastmap_1.1.0
# [28] lazyeval_0.2.2 cli_3.2.0 later_1.3.0
# [31] prettyunits_1.1.1 htmltools_0.5.2 quantreg_5.86
# [34] tools_4.0.4 coda_0.19-4 gtable_0.3.0
# [37] glue_1.6.2 reshape2_1.4.4 fastmatch_1.1-3
# [40] Rcpp_1.0.8 jquerylib_0.1.4 vctrs_0.3.8
# [43] nlme_3.1-155 conquer_1.2.1 crosstalk_1.2.0
# [46] iterators_1.0.13 timeDate_3043.102 gower_0.2.2
# [49] xfun_0.29 stringr_1.4.0 globals_0.14.0
# [52] ps_1.6.0 lifecycle_1.0.1 irlba_2.3.5
# [55] future_1.23.0 getPass_0.2-2 MASS_7.3-55
# [58] scales_1.1.1 ipred_0.9-12 hms_1.1.1
# [61] promises_1.2.0.1 parallel_4.0.4 SparseM_1.81
# [64] yaml_2.2.2 gridExtra_2.3 pbapply_1.5-0
# [67] sass_0.4.0 rpart_4.1-15 stringi_1.7.6
# [70] SQUAREM_2021.1 highr_0.9 foreach_1.5.1
# [73] BiocParallel_1.24.1 lava_1.6.10 truncnorm_1.0-8
# [76] rlang_1.0.1 pkgconfig_2.0.3 matrixStats_0.61.0
# [79] evaluate_0.14 lattice_0.20-45 invgamma_1.1
# [82] purrr_0.3.4 labeling_0.4.2 recipes_0.1.17
# [85] processx_3.5.2 tidyselect_1.1.2 parallelly_1.30.0
# [88] plyr_1.8.6 magrittr_2.0.2 R6_2.5.1
# [91] generics_0.1.2 DBI_1.1.2 pillar_1.7.0
# [94] whisker_0.4 withr_2.4.3 survival_3.2-13
# [97] mixsqp_0.3-43 nnet_7.3-17 tibble_3.1.6
# [100] future.apply_1.8.1 crayon_1.5.0 utf8_1.2.2
# [103] rmarkdown_2.11 progress_1.2.2 grid_4.0.4
# [106] data.table_1.14.2 callr_3.7.0 git2r_0.29.0
# [109] ModelMetrics_1.2.2.2 digest_0.6.29 httpuv_1.6.5
# [112] MCMCpack_1.6-0 RcppParallel_5.1.5 stats4_4.0.4
# [115] munsell_0.5.0 viridisLite_0.4.0 bslib_0.3.1
# [118] quadprog_1.5-8