Clustering Buenrostro et al (2018) scATAC-seq data using topic proportions with K = 11
Kaixuan Luo
Last updated: 2021-02-05
Checks: 7 0
Knit directory: scATACseq-topics/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it's best to always run the code in an empty environment.
The command set.seed(20200729) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version a797b10. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: output/plotly/Buenrostro_2018_Chen2019pipeline/
Untracked files:
Untracked: analysis/process_data_Buenrostro2018_Chen2019.Rmd
Untracked: output/clustering-Cusanovich2018.rds
Untracked: output/plotly/Cusanovich2018/gsea_topic_10_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_10_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_10_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_10_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_10_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_10_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_10_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_10_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_10_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_10_tss-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_11_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_11_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_11_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_11_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_11_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_11_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_11_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_11_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_11_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_11_tss-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_12_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_12_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_12_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_12_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_12_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_12_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_12_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_12_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_12_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_12_tss-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_13_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_13_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_13_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_13_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_13_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_13_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_13_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_13_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_13_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_13_tss-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_1_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_1_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_1_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_1_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_1_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_1_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_1_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_1_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_1_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_1_tss-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_2_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_2_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_2_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_2_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_2_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_2_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_2_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_2_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_2_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_2_tss-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_3_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_3_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_3_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_3_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_3_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_3_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_3_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_3_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_3_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_3_tss-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_4_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_4_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_4_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_4_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_4_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_4_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_4_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_4_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_4_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_4_tss-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_5_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_5_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_5_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_5_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_5_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_5_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_5_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_5_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_5_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_5_tss-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_6_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_6_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_6_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_6_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_6_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_6_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_6_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_6_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_6_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_6_tss-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_7_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_7_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_7_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_7_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_7_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_7_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_7_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_7_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_7_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_7_tss-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_8_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_8_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_8_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_8_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_8_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_8_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_8_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_8_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_8_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_8_tss-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_9_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_9_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_9_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_9_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_9_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_9_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_9_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_9_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/gsea_topic_9_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/gsea_topic_9_tss-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_10_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_10_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_10_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_10_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_10_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_10_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_10_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_10_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_10_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_10_tss-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_11_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_11_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_11_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_11_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_11_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_11_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_11_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_11_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_11_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_11_tss-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_12_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_12_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_12_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_12_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_12_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_12_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_12_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_12_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_12_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_12_tss-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_13_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_13_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_13_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_13_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_13_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_13_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_13_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_13_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_13_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_13_tss-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_1_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_1_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_1_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_1_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_1_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_1_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_1_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_1_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_1_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_1_tss-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_2_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_2_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_2_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_2_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_2_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_2_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_2_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_2_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_2_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_2_tss-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_3_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_3_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_3_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_3_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_3_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_3_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_3_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_3_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_3_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_3_tss-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_4_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_4_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_4_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_4_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_4_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_4_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_4_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_4_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_4_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_4_tss-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_5_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_5_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_5_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_5_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_5_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_5_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_5_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_5_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_5_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_5_tss-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_6_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_6_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_6_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_6_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_6_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_6_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_6_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_6_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_6_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_6_tss-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_7_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_7_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_7_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_7_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_7_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_7_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_7_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_7_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_7_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_7_tss-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_8_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_8_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_8_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_8_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_8_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_8_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_8_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_8_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_8_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_8_tss-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_9_genebody-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_9_genebody-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_9_genebody-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_9_genebody-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_9_genebody-sum_files/plotly-main-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_9_tss-sum_files/crosstalk-1.1.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_9_tss-sum_files/jquery-3.5.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_9_tss-sum_files/plotly-binding-4.9.3/
Untracked: output/plotly/Cusanovich2018/volcano_topic_9_tss-sum_files/plotly-htmlwidgets-css-1.57.1/
Untracked: output/plotly/Cusanovich2018/volcano_topic_9_tss-sum_files/plotly-main-1.57.1/
Untracked: scripts/fit_all_models_Buenrostro_2018_chromVar_scPeaks_filtered.sbatch
Untracked: scripts/fit_models_Cusanovich2018_tissues.sh
Unstaged changes:
Modified: analysis/assess_fits_Cusanovich2018.Rmd
Modified: analysis/gene_analysis_Cusanovich2018.Rmd
Modified: analysis/index.Rmd
Modified: analysis/motif_analysis_Buenrostro2018_Chen2019pipeline.Rmd
Modified: analysis/plots_Cusanovich2018.Rmd
Modified: analysis/process_data_Cusanovich2018.Rmd
Modified: output/plotly/Cusanovich2018/gsea_topic_10_genebody-sum.html
Modified: output/plotly/Cusanovich2018/gsea_topic_10_tss-sum.html
Modified: output/plotly/Cusanovich2018/gsea_topic_11_genebody-sum.html
Modified: output/plotly/Cusanovich2018/gsea_topic_11_tss-sum.html
Modified: output/plotly/Cusanovich2018/gsea_topic_12_genebody-sum.html
Modified: output/plotly/Cusanovich2018/gsea_topic_12_tss-sum.html
Modified: output/plotly/Cusanovich2018/gsea_topic_13_genebody-sum.html
Modified: output/plotly/Cusanovich2018/gsea_topic_13_tss-sum.html
Modified: output/plotly/Cusanovich2018/gsea_topic_1_genebody-sum.html
Modified: output/plotly/Cusanovich2018/gsea_topic_1_tss-sum.html
Modified: output/plotly/Cusanovich2018/gsea_topic_2_genebody-sum.html
Modified: output/plotly/Cusanovich2018/gsea_topic_2_tss-sum.html
Modified: output/plotly/Cusanovich2018/gsea_topic_3_genebody-sum.html
Modified: output/plotly/Cusanovich2018/gsea_topic_3_tss-sum.html
Modified: output/plotly/Cusanovich2018/gsea_topic_4_genebody-sum.html
Modified: output/plotly/Cusanovich2018/gsea_topic_4_tss-sum.html
Modified: output/plotly/Cusanovich2018/gsea_topic_5_genebody-sum.html
Modified: output/plotly/Cusanovich2018/gsea_topic_5_tss-sum.html
Modified: output/plotly/Cusanovich2018/gsea_topic_6_genebody-sum.html
Modified: output/plotly/Cusanovich2018/gsea_topic_6_tss-sum.html
Modified: output/plotly/Cusanovich2018/gsea_topic_7_genebody-sum.html
Modified: output/plotly/Cusanovich2018/gsea_topic_7_tss-sum.html
Modified: output/plotly/Cusanovich2018/gsea_topic_8_genebody-sum.html
Modified: output/plotly/Cusanovich2018/gsea_topic_8_tss-sum.html
Modified: output/plotly/Cusanovich2018/gsea_topic_9_genebody-sum.html
Modified: output/plotly/Cusanovich2018/gsea_topic_9_tss-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_10_genebody-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_10_tss-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_11_genebody-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_11_tss-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_12_genebody-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_12_tss-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_13_genebody-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_13_tss-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_1_genebody-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_1_tss-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_2_genebody-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_2_tss-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_3_genebody-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_3_tss-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_4_genebody-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_4_tss-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_5_genebody-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_5_tss-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_6_genebody-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_6_tss-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_7_genebody-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_7_tss-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_8_genebody-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_8_tss-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_9_genebody-sum.html
Modified: output/plotly/Cusanovich2018/volcano_topic_9_tss-sum.html
Modified: scripts/fit_all_models_Cusanovich2018.sh
Modified: scripts/fit_poisson_nmf.sbatch
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/clusters_Buenrostro2018_k11_Chen2019pipeline.Rmd) and HTML (docs/clusters_Buenrostro2018_k11_Chen2019pipeline.html) files. If you've configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | a797b10 | kevinlkx | 2021-02-05 | cleaned the clustering results, and tried k-means of pca results |
| html | bc5fc36 | kevinlkx | 2020-12-02 | Build site. |
| Rmd | 6f28b44 | kevinlkx | 2020-12-02 | update data.dir and out.dir |
| Rmd | 6a9dead | kevinlkx | 2020-11-19 | wflow_rename("analysis/clusters_Buenrostro2018_k11.Rmd", "analysis/clusters_Buenrostro2018_k11_Chen2019pipeline.Rmd") |
| html | 6a9dead | kevinlkx | 2020-11-19 | wflow_rename("analysis/clusters_Buenrostro2018_k11.Rmd", "analysis/clusters_Buenrostro2018_k11_Chen2019pipeline.Rmd") |
Here we explore the structure in the Buenrostro et al (2018) scATAC-seq data inferred from the multinomial topic model with \(k = 11\).
Load packages and some functions used in this analysis.
library(Matrix)
library(dplyr)
library(ggplot2)
library(cowplot)
library(plyr)
library(dplyr)
library(RColorBrewer)
library(fastTopics)
library(DT)
library(reshape)
source("code/plots.R")Load the data. The counts are no longer needed at this stage of the analysis.
data.dir <- "/project2/mstephens/kevinluo/scATACseq-topics/data/Buenrostro_2018/processed_data_Chen2019pipeline/"
load(file.path(data.dir, "Buenrostro_2018_binarized_counts.RData"))
cat(sprintf("%d x %d counts matrix.\n",nrow(counts),ncol(counts)))
rm(counts)
samples$cell <- rownames(samples)
samples$label <- as.factor(samples$label)
# 2034 x 101172 counts matrix.Load the model fit
fit.dir <- "/project2/mstephens/kevinluo/scATACseq-topics/output/Buenrostro_2018_Chen2019pipeline/binarized/"
fit <- readRDS(file.path(fit.dir, "/fit-Buenrostro2018-binarized-scd-ex-k=11.rds"))$fit
fit_multinom <- poisson2multinom(fit)Structure plots
The structure plots below summarize the topic proportions in the samples grouped by different tissues.
Visualize by Structure plot grouped by tissues
set.seed(10)
colors_topics <- c("#a6cee3","#1f78b4","#b2df8a","#33a02c","#fb9a99","#e31a1c",
"#fdbf6f","#ff7f00","#cab2d6","#6a3d9a","#ffff99")
samples$label <- as.factor(samples$label)
p.structure.1 <- structure_plot(fit_multinom,
grouping = samples[, "label"],
n = Inf,gap = 20,
perplexity = 50,topics = 1:11,colors = colors_topics,
num_threads = 4,verbose = FALSE)
print(p.structure.1)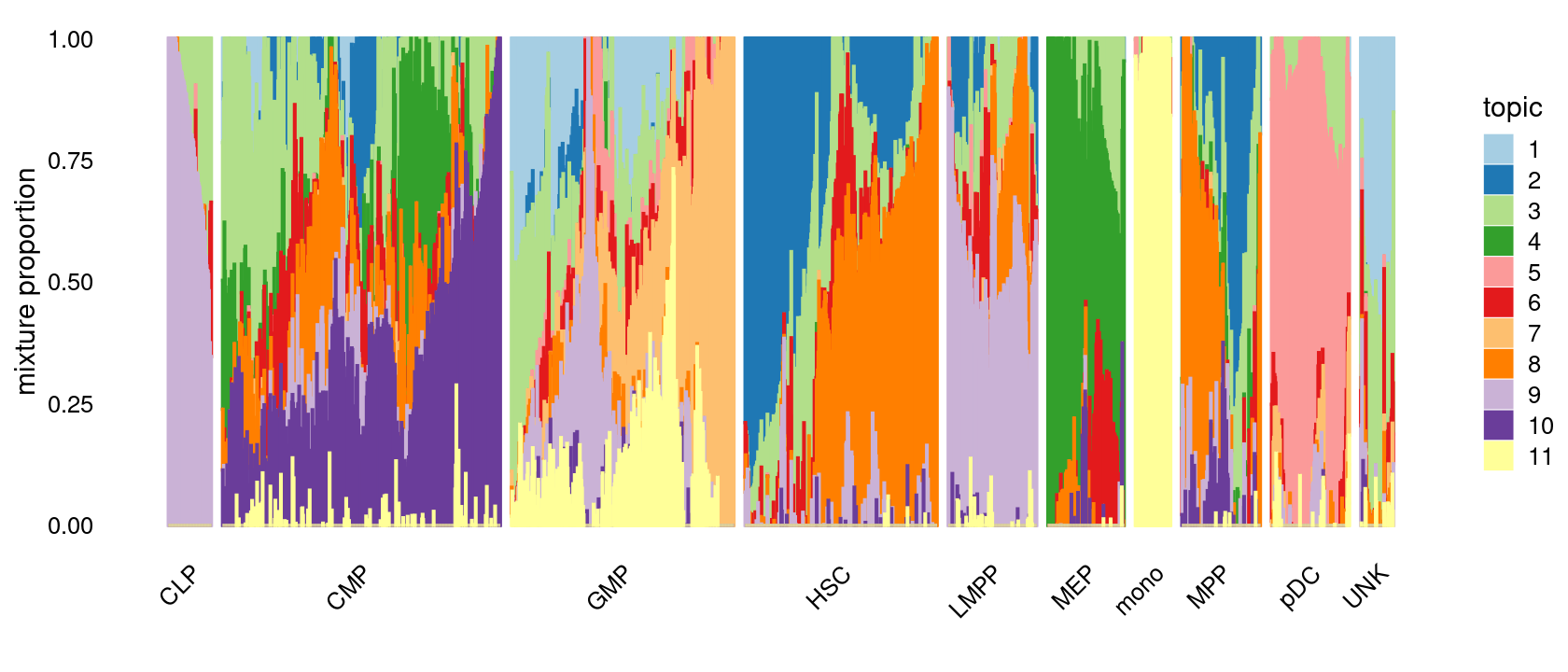
k-means clustering on topic proportions
Define clusters using k-means, and then create structure plot based on the clusters from k-means.
k-means clustering (using 15 clusters) on topic proportions
set.seed(10)
clusters <- factor(kmeans(fit_multinom$L,centers = 15,iter.max = 100)$cluster)
summary(clusters)
p.structure.kmeans <- structure_plot(fit_multinom,
grouping = clusters,n = Inf,gap = 20,
perplexity = 50,topics = 1:11,colors = colors_topics,
num_threads = 4,verbose = FALSE)
print(p.structure.kmeans)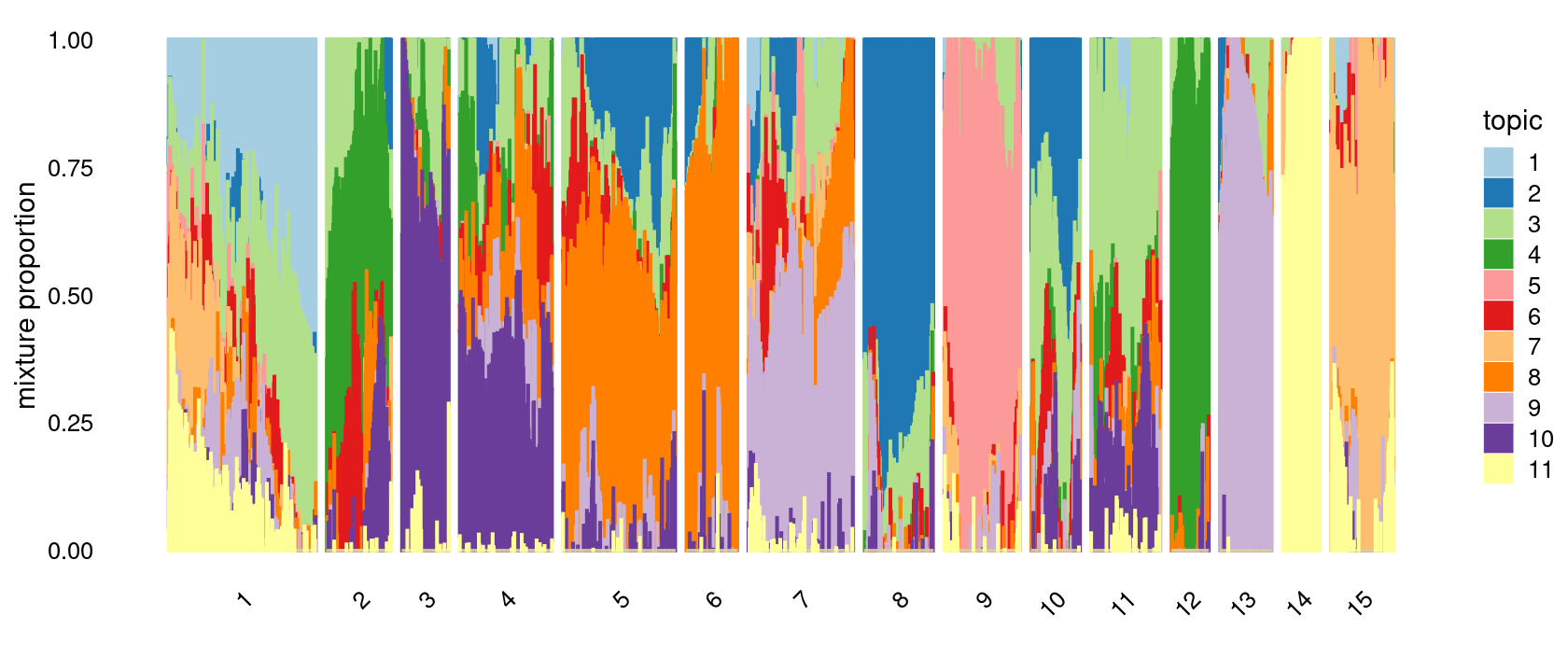
# 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15
# 279 122 89 175 213 97 199 131 144 93 132 71 99 71 119First do PCA on topic proportions and then do k-means clustering. Results are the same as the results from running k-means directly on the topic proportions.
set.seed(10)
pca <- prcomp(fit_multinom$L)$x
clusters2 <- factor(kmeans(pca,centers = 15,iter.max = 100)$cluster)
summary(clusters2)
p.structure.kmeans <- structure_plot(fit_multinom,
grouping = clusters2,n = Inf,gap = 20,
perplexity = 50,topics = 1:11,colors = colors_topics,
num_threads = 4,verbose = FALSE)
print(p.structure.kmeans)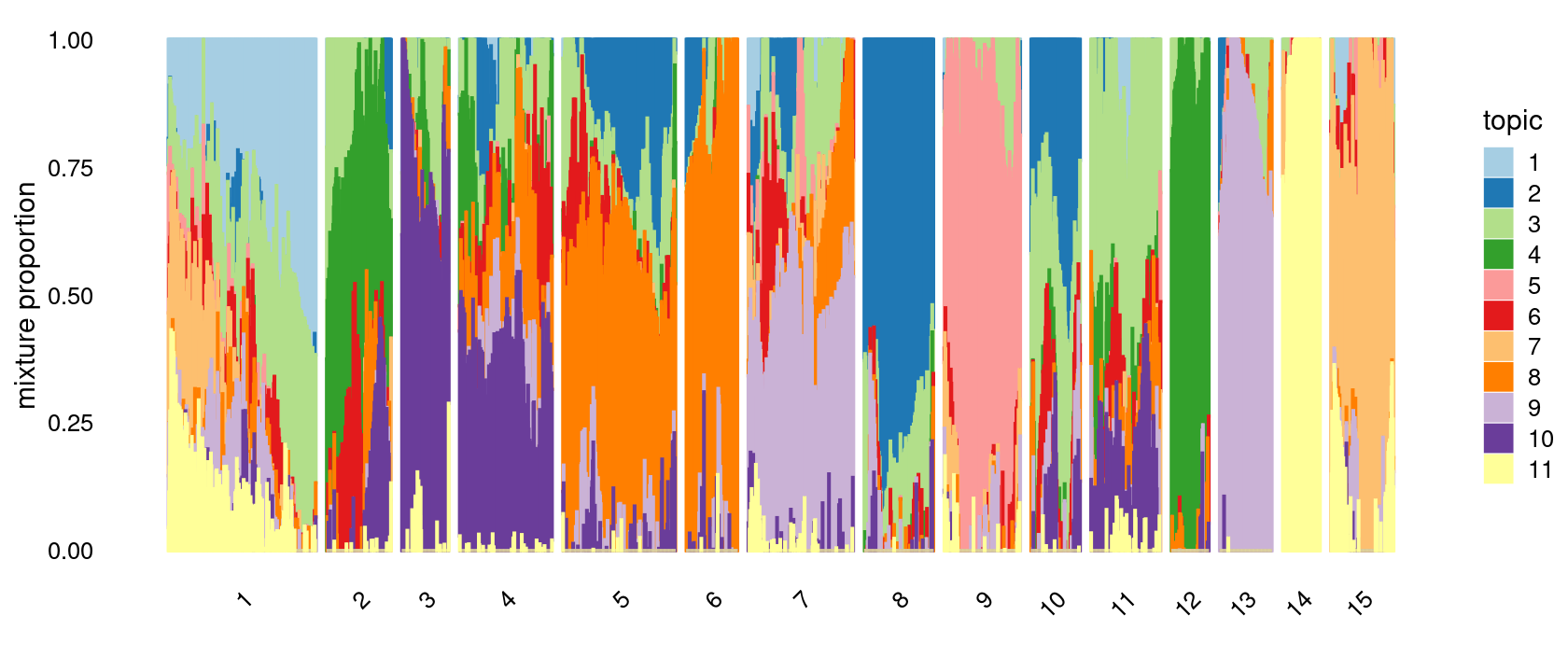
length(which(clusters != clusters2))
# 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15
# 279 122 89 175 213 97 199 131 144 93 132 71 99 71 119
# [1] 0Define clusters using k-means with \(k = 20\): k-means clustering (using 15 clusters) on topic proportions
set.seed(10)
clusters.20 <- factor(kmeans(fit_multinom$L,centers = 20,iter.max = 100)$cluster)
summary(clusters.20)
p.structure.kmeans <- structure_plot(fit_multinom,
grouping = clusters.20,n = Inf,gap = 20,
perplexity = 50,topics = 1:11,colors = colors_topics,
num_threads = 4,verbose = FALSE)
print(p.structure.kmeans)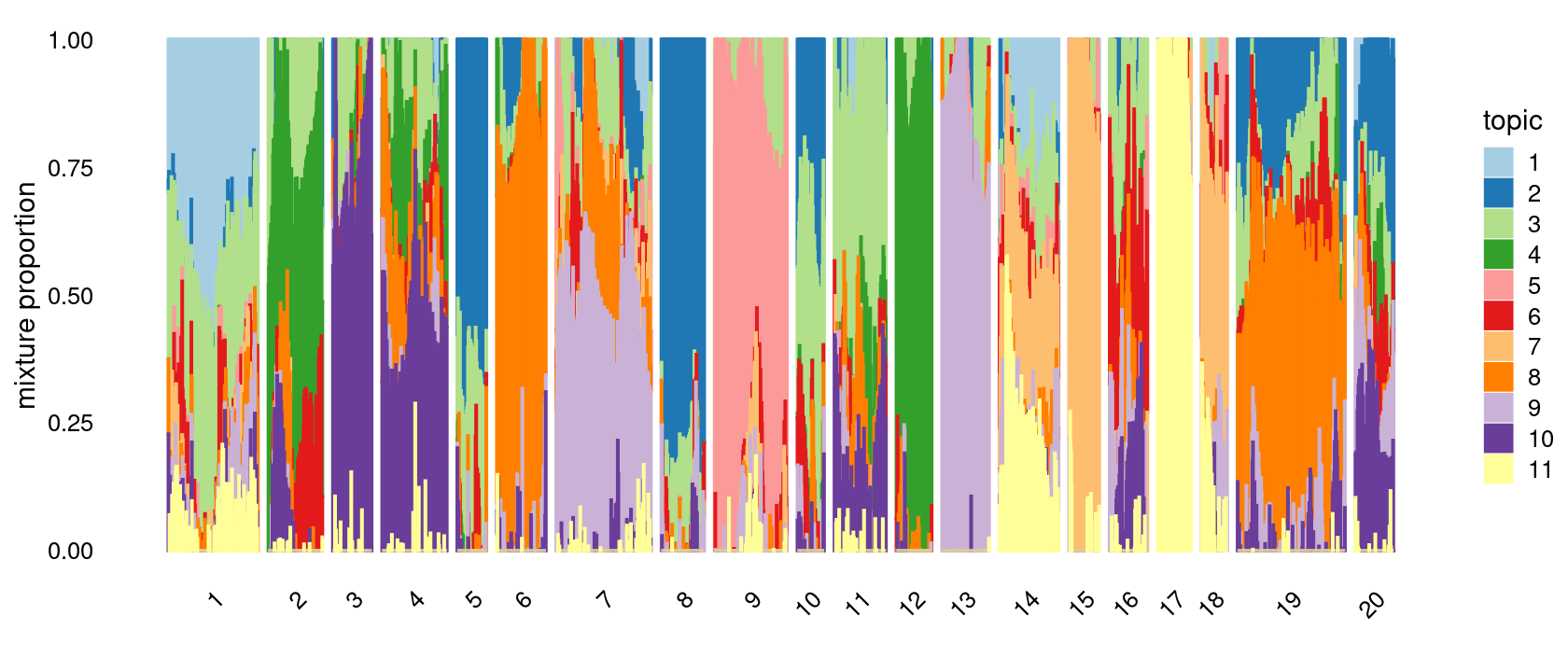
# 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
# 177 107 77 128 58 97 187 85 143 53 102 70 93 117 60 75 66 51 212 76First do PCA on topic proportions and then do k-means clustering. Results are again the same as the results from running k-means directly on the topic proportions.
set.seed(10)
pca <- prcomp(fit_multinom$L)$x
clusters.20.2 <- factor(kmeans(pca,centers = 20,iter.max = 100)$cluster)
summary(clusters.20.2)
p.structure.kmeans <- structure_plot(fit_multinom,
grouping = clusters.20.2,n = Inf,gap = 20,
perplexity = 50,topics = 1:11,colors = colors_topics,
num_threads = 4,verbose = FALSE)
print(p.structure.kmeans)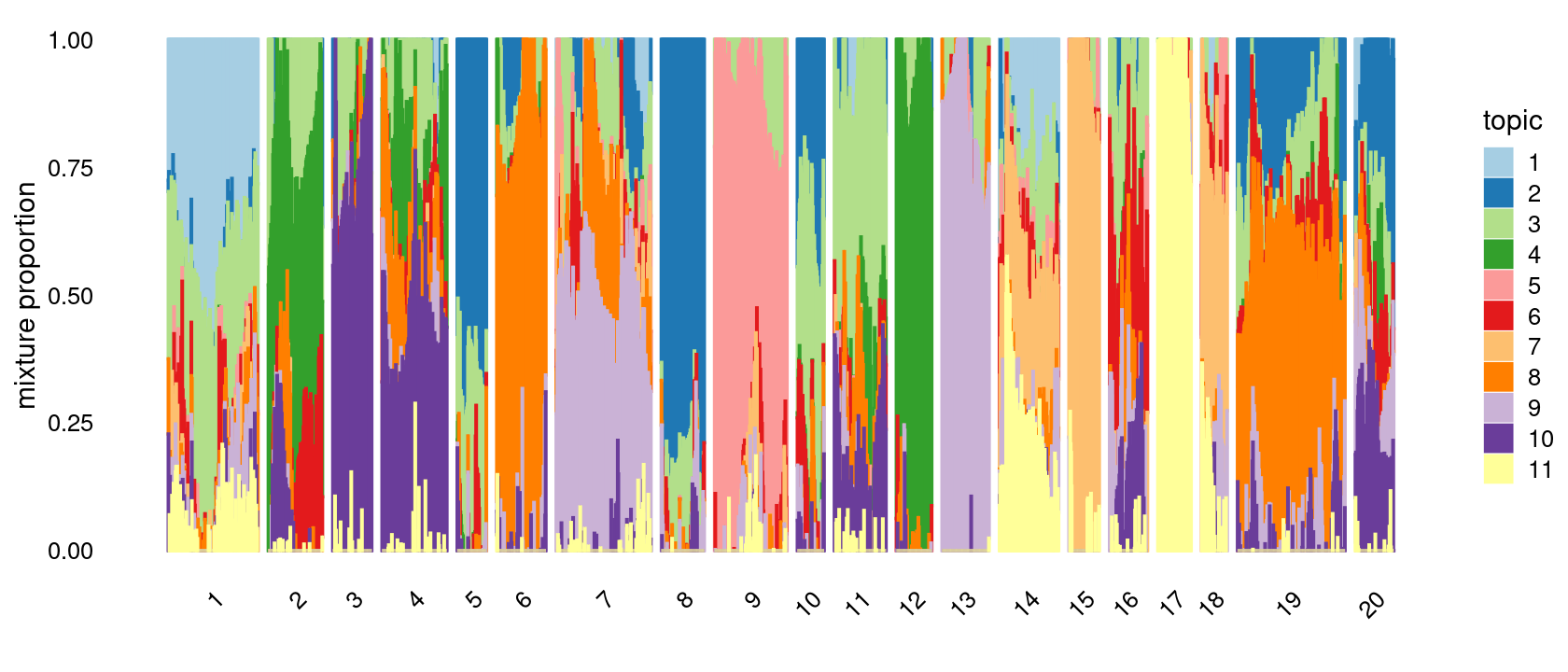
length(which(clusters.20 != clusters.20.2))
# 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
# 177 107 77 128 58 97 187 85 143 53 102 70 93 117 60 75 66 51 212 76
# [1] 0Refine clusters
We then further refine the clusters based on k-means result with \(k = 20\): merge clusters 3, 4; merge clusters 5 and 8; merge clusters 6 and 19; merge clusters 14 and 18; merge clusters 16 and 20
clusters.merged <- clusters.20
clusters.merged[clusters.20 %in% c(3,4)] <- 3
clusters.merged[clusters.20 %in% c(5,8)] <- 5
clusters.merged[clusters.20 %in% c(6,19)] <- 6
clusters.merged[clusters.20 %in% c(14,18)] <- 14
clusters.merged[clusters.20 %in% c(16,20)] <- 16
clusters.merged <- factor(clusters.merged, labels = paste0("c", c(1:length(unique(clusters.merged)))))
samples$cluster_kmeans <- clusters.merged
table(clusters.merged)
# clusters.merged
# c1 c2 c3 c4 c5 c6 c7 c8 c9 c10 c11 c12 c13 c14 c15
# 177 107 205 143 309 187 143 53 102 70 93 168 60 151 66p.structure.refined <- structure_plot(fit_multinom,
grouping = clusters.merged,
n = Inf,gap = 20,
perplexity = 50,topics = 1:11,colors = colors_topics,
num_threads = 4,verbose = FALSE)
# Perplexity automatically changed to 34 because original setting of 50 was too large for the number of samples (107)
# Perplexity automatically changed to 46 because original setting of 50 was too large for the number of samples (143)
# Perplexity automatically changed to 46 because original setting of 50 was too large for the number of samples (143)
# Perplexity automatically changed to 16 because original setting of 50 was too large for the number of samples (53)
# Perplexity automatically changed to 32 because original setting of 50 was too large for the number of samples (102)
# Perplexity automatically changed to 22 because original setting of 50 was too large for the number of samples (70)
# Perplexity automatically changed to 29 because original setting of 50 was too large for the number of samples (93)
# Perplexity automatically changed to 18 because original setting of 50 was too large for the number of samples (60)
# Perplexity automatically changed to 49 because original setting of 50 was too large for the number of samples (151)
# Perplexity automatically changed to 20 because original setting of 50 was too large for the number of samples (66)
print(p.structure.refined)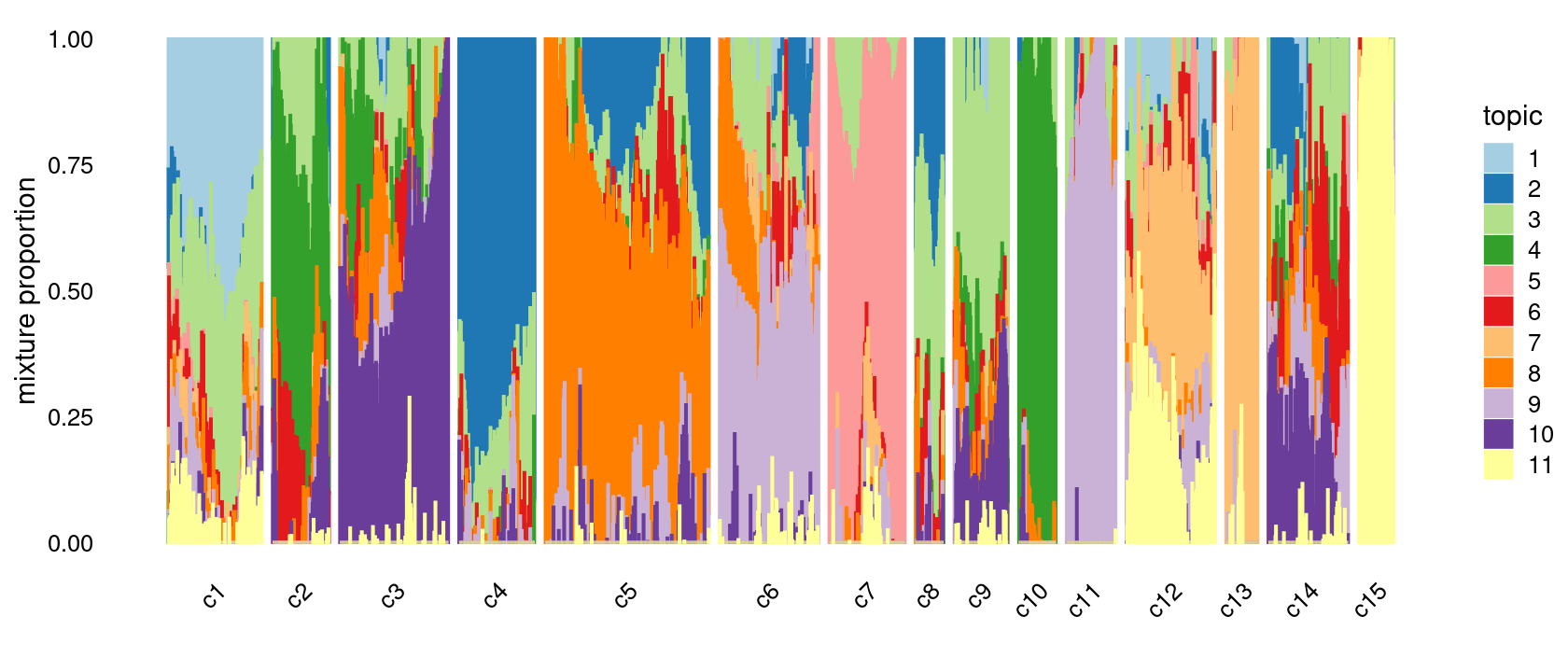
Save the clustering results to an RDS file
out.dir <- "/project2/mstephens/kevinluo/scATACseq-topics/output/Buenrostro_2018_Chen2019pipeline/binarized/"
saveRDS(samples, paste0(out.dir, "/samples-clustering-Buenrostro2018.rds"))
cat("Result saved to:", paste0(out.dir, "/samples-clustering-Buenrostro2018.rds"), "\n")
samples <- readRDS(paste0(out.dir, "/samples-clustering-Buenrostro2018.rds"))
print(table(samples$cluster_kmeans))
# Result saved to: /project2/mstephens/kevinluo/scATACseq-topics/output/Buenrostro_2018_Chen2019pipeline/binarized//samples-clustering-Buenrostro2018.rds
#
# c1 c2 c3 c4 c5 c6 c7 c8 c9 c10 c11 c12 c13 c14 c15
# 177 107 205 143 309 187 143 53 102 70 93 168 60 151 66Plot the distribution of cell type labels in each cluster
samples$label <- as.factor(samples$label)
cat(length(levels(samples$label)), "cell labels. \n")
table(samples$label)
freq_cluster_cells <- with(samples,table(label,cluster_kmeans))
print(freq_cluster_cells)
# 10 cell labels.
#
# CLP CMP GMP HSC LMPP MEP mono MPP pDC UNK
# 78 502 402 347 160 138 64 142 141 60
# cluster_kmeans
# label c1 c2 c3 c4 c5 c6 c7 c8 c9 c10 c11 c12 c13 c14 c15
# CLP 0 0 0 0 0 12 0 0 1 0 63 0 0 2 0
# CMP 16 33 202 3 25 7 0 12 89 5 0 1 0 109 0
# GMP 102 1 0 0 0 41 3 0 3 0 10 165 60 15 2
# HSC 0 0 0 109 210 0 0 22 1 0 0 0 0 5 0
# LMPP 1 0 0 0 1 125 0 0 1 0 20 2 0 10 0
# MEP 0 70 2 0 0 0 0 0 1 65 0 0 0 0 0
# mono 0 0 0 0 0 0 0 0 0 0 0 0 0 0 64
# MPP 0 3 1 31 73 1 0 19 5 0 0 0 0 9 0
# pDC 0 0 0 0 0 0 140 0 0 0 0 0 0 1 0
# UNK 58 0 0 0 0 1 0 0 1 0 0 0 0 0 0Cell-type composition in each cluster:
freq_cluster_celltype <- count(samples, vars=c("cluster_kmeans","label"))
n_colors <- length(levels(samples$label))
colors_labels <- brewer.pal(10, "Set3")
p.barplot <- ggplot(freq_cluster_celltype, aes(fill=label, y=freq, x=cluster_kmeans)) +
geom_bar(position="fill", stat="identity") +
theme_classic() + xlab("Cluster") + ylab("Proportion of cells") +
scale_fill_manual(values = colors_labels) +
guides(fill=guide_legend(ncol=2)) +
theme(
legend.title = element_text(size = 10),
legend.text = element_text(size = 8)
)
print(p.barplot)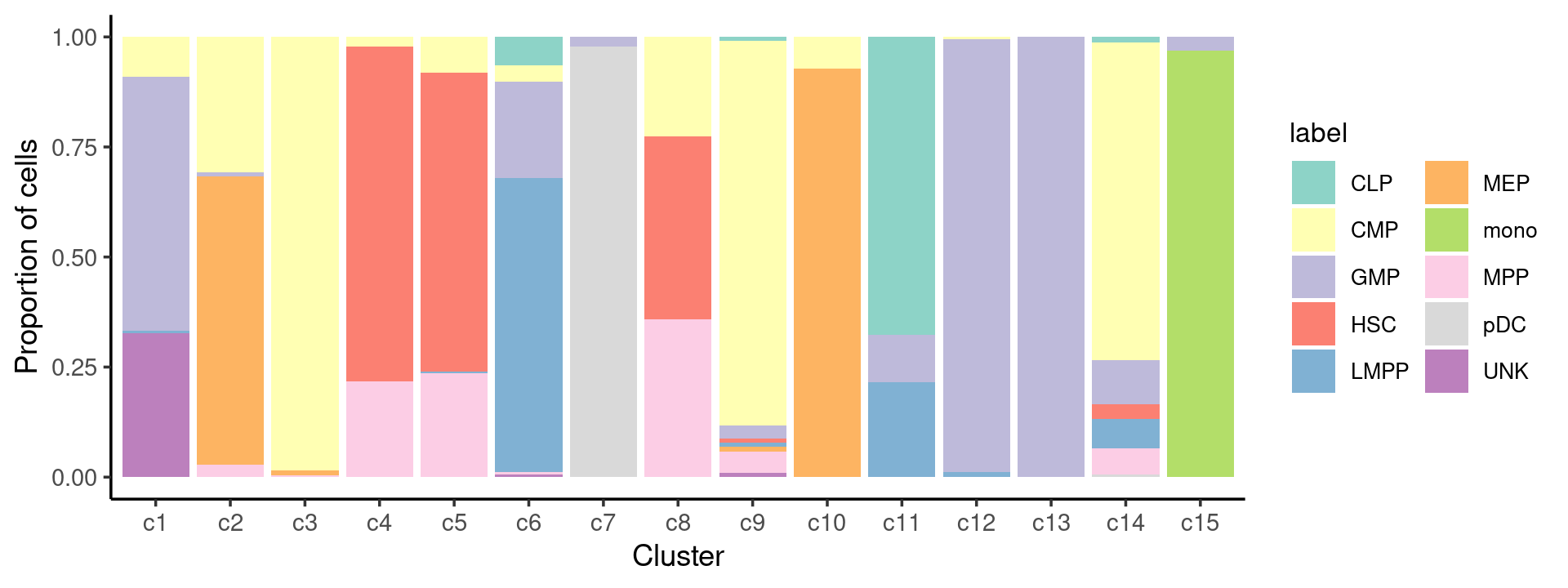
PCA of the topic proportions
Plot PCs of the topic proportions.
breaks <- c(0,1,5,10,100,Inf)
p.pca2.1 <- pca_hexbin_plot(fit_multinom, pcs = 1:2, breaks = breaks) + guides(fill = "none")
p.pca2.2 <- pca_hexbin_plot(fit_multinom, pcs = 3:4, breaks = breaks) + guides(fill = "none")
p.pca2.3 <- pca_hexbin_plot(fit_multinom, pcs = 5:6, breaks = breaks) + guides(fill = "none")
p.pca2.4 <- pca_hexbin_plot(fit_multinom, pcs = 7:8, breaks = breaks) + guides(fill = "none")
p.pca2.5 <- pca_hexbin_plot(fit_multinom, pcs = 9:10, breaks = breaks) + guides(fill = "none")
plot_grid(p.pca2.1,p.pca2.2,p.pca2.3,p.pca2.4,p.pca2.5)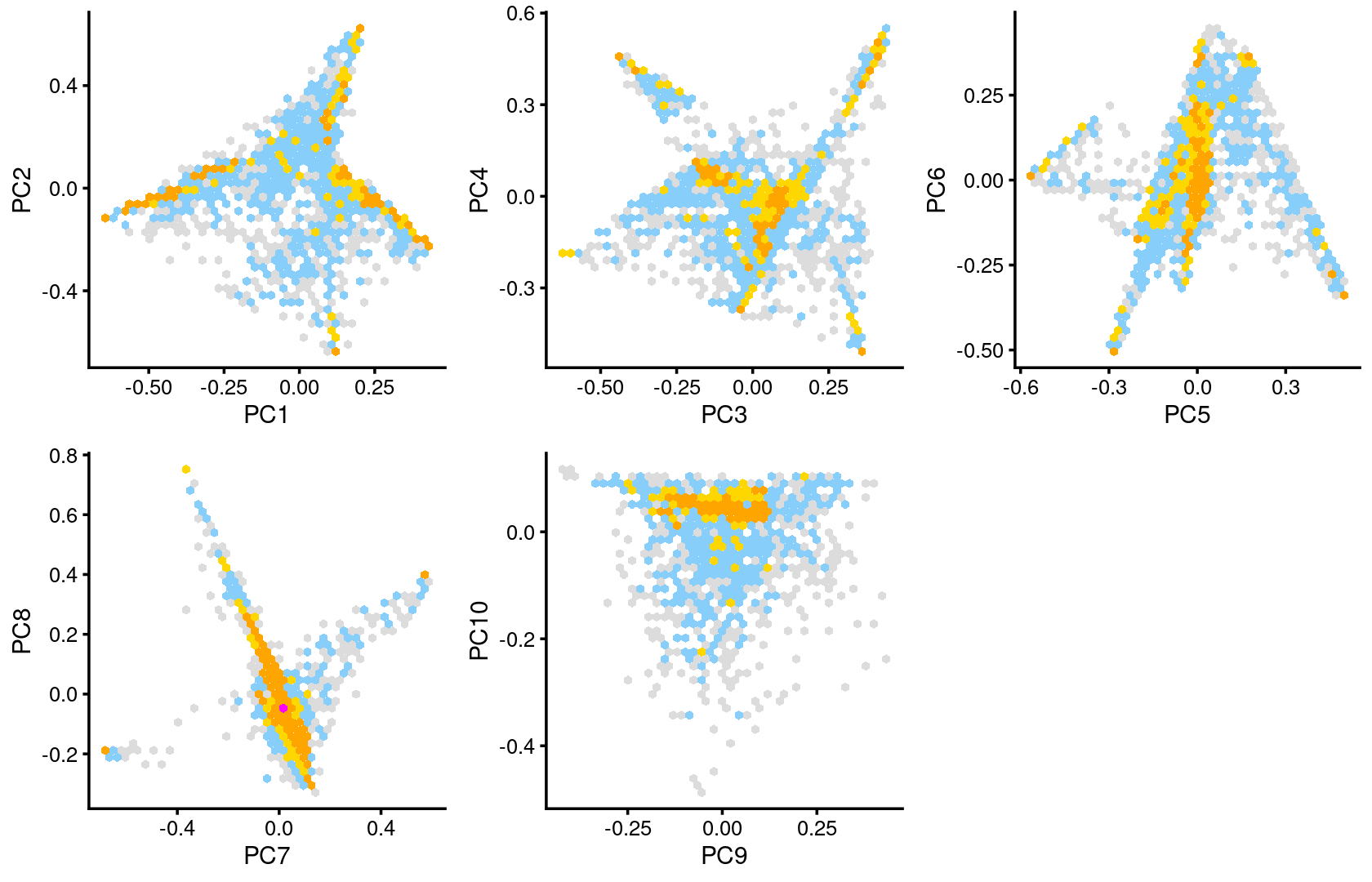
Layer the cell labels onto the PC plots.
p.pca3.1 <- labeled_pca_plot(fit,1:2,samples$label,font_size = 7,
legend_label = "Cell labels")
p.pca3.2 <- labeled_pca_plot(fit,3:4,samples$label,font_size = 7,
legend_label = "Cell labels")
p.pca3.3 <- labeled_pca_plot(fit,5:6,samples$label,font_size = 7,
legend_label = "Cell labels")
p.pca3.4 <- labeled_pca_plot(fit,7:8,samples$label,font_size = 7,
legend_label = "Cell labels")
p.pca3.5 <- labeled_pca_plot(fit,9:10,samples$label,font_size = 7,
legend_label = "Cell labels")
plot_grid(p.pca3.1,p.pca3.2,p.pca3.3,p.pca3.4,p.pca3.5)
sessionInfo()
# R version 3.6.1 (2019-07-05)
# Platform: x86_64-pc-linux-gnu (64-bit)
# Running under: Scientific Linux 7.4 (Nitrogen)
#
# Matrix products: default
# BLAS/LAPACK: /software/openblas-0.2.19-el7-x86_64/lib/libopenblas_haswellp-r0.2.19.so
#
# locale:
# [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
# [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
# [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
# [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
# [9] LC_ADDRESS=C LC_TELEPHONE=C
# [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
#
# attached base packages:
# [1] stats graphics grDevices utils datasets methods base
#
# other attached packages:
# [1] reshape_0.8.8 DT_0.16 fastTopics_0.4-29 RColorBrewer_1.1-2
# [5] plyr_1.8.6 cowplot_1.1.1 ggplot2_3.3.3 dplyr_1.0.3
# [9] Matrix_1.2-18 workflowr_1.6.2
#
# loaded via a namespace (and not attached):
# [1] ggrepel_0.9.1 Rcpp_1.0.6 lattice_0.20-41 tidyr_1.1.2
# [5] prettyunits_1.1.1 rprojroot_2.0.2 digest_0.6.27 R6_2.5.0
# [9] MatrixModels_0.4-1 evaluate_0.14 coda_0.19-4 httr_1.4.2
# [13] pillar_1.4.7 rlang_0.4.10 progress_1.2.2 lazyeval_0.2.2
# [17] data.table_1.13.6 irlba_2.3.3 SparseM_1.78 hexbin_1.28.1
# [21] whisker_0.4 rmarkdown_2.6 labeling_0.4.2 Rtsne_0.15
# [25] stringr_1.4.0 htmlwidgets_1.5.3 munsell_0.5.0 compiler_3.6.1
# [29] httpuv_1.5.4 xfun_0.19 pkgconfig_2.0.3 mcmc_0.9-7
# [33] htmltools_0.5.1.1 tidyselect_1.1.0 tibble_3.0.6 quadprog_1.5-8
# [37] matrixStats_0.58.0 viridisLite_0.3.0 crayon_1.4.0 conquer_1.0.2
# [41] withr_2.4.1 later_1.1.0.1 MASS_7.3-53 grid_3.6.1
# [45] jsonlite_1.7.2 gtable_0.3.0 lifecycle_0.2.0 DBI_1.1.0
# [49] git2r_0.27.1 magrittr_2.0.1 scales_1.1.1 RcppParallel_5.0.2
# [53] stringi_1.5.3 farver_2.0.3 fs_1.3.1 promises_1.1.1
# [57] ellipsis_0.3.1 generics_0.1.0 vctrs_0.3.6 tools_3.6.1
# [61] glue_1.4.2 purrr_0.3.4 hms_1.0.0 yaml_2.2.1
# [65] colorspace_2.0-0 plotly_4.9.3 knitr_1.30 quantreg_5.83
# [69] MCMCpack_1.5-0