Visualize and interpret topics in PBMC data
Peter Carbonetto
Last updated: 2020-08-22
Checks: 7 0
Knit directory: single-cell-topics/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2.9000). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(1) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 8daa131. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: data/droplet.RData
Ignored: data/pbmc_68k.RData
Ignored: data/pbmc_purified.RData
Ignored: data/pulseseq.RData
Ignored: output/droplet/fits-droplet.RData
Ignored: output/droplet/rds/
Ignored: output/pbmc-68k/fits-pbmc-68k.RData
Ignored: output/pbmc-68k/rds/
Ignored: output/pbmc-purified/fits-pbmc-purified.RData
Ignored: output/pbmc-purified/rds/
Ignored: output/pulseseq/fits-pulseseq.RData
Ignored: output/pulseseq/rds/
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/plots_pbmc.Rmd) and HTML (docs/plots_pbmc.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 8daa131 | Peter Carbonetto | 2020-08-22 | workflowr::wflow_publish(“plots_pbmc.Rmd”) |
| html | 7900d17 | Peter Carbonetto | 2020-08-22 | Revamping the process of defining clusters in plots_pbmc using PCA. |
| Rmd | 14dd774 | Peter Carbonetto | 2020-08-22 | workflowr::wflow_publish(“plots_pbmc.Rmd”) |
| Rmd | 944ce0c | Peter Carbonetto | 2020-08-22 | Working on new PCA plots for purified PBMC c3 subclusters. |
| html | b05232d | Peter Carbonetto | 2020-08-22 | Build site. |
| Rmd | d0e9f6e | Peter Carbonetto | 2020-08-22 | workflowr::wflow_publish(“plots_pbmc.Rmd”) |
| html | fbb0697 | Peter Carbonetto | 2020-08-21 | Build site. |
| Rmd | cdca13e | Peter Carbonetto | 2020-08-21 | workflowr::wflow_publish(“plots_pbmc.Rmd”) |
| html | 216027a | Peter Carbonetto | 2020-08-21 | Re-built plots_pbmc webpage with structure plots. |
| Rmd | 98888b2 | Peter Carbonetto | 2020-08-21 | Added structure plots to plots_pbmc.R. |
| html | 9310993 | Peter Carbonetto | 2020-08-21 | Revised the PCA plots in pbmc_plots. |
| Rmd | 3a746ff | Peter Carbonetto | 2020-08-21 | workflowr::wflow_publish(“plots_pbmc.Rmd”) |
| Rmd | 1091dd0 | Peter Carbonetto | 2020-08-21 | Added code for Ternary plots to plots_pbmc. |
| html | 6d3d7e4 | Peter Carbonetto | 2020-08-20 | Added 68k PCA plots to plots_pbmc. |
| Rmd | 6ec82ca | Peter Carbonetto | 2020-08-20 | workflowr::wflow_publish(“plots_pbmc.Rmd”) |
| html | 38f07a2 | Peter Carbonetto | 2020-08-20 | A few small revisions to the plots_pbmc analysis. |
| Rmd | fd0316f | Peter Carbonetto | 2020-08-20 | workflowr::wflow_publish(“plots_pbmc.Rmd”) |
| html | 606cd97 | Peter Carbonetto | 2020-08-20 | Added basic PCA plots to plots_pbmc. |
| Rmd | 1b41a60 | Peter Carbonetto | 2020-08-20 | workflowr::wflow_publish(“plots_pbmc.Rmd”) |
| html | d6e5d39 | Peter Carbonetto | 2020-08-20 | Added PCA plot with purified PBMC clustering to plots_pbmc analysis. |
| Rmd | 99301a7 | Peter Carbonetto | 2020-08-20 | workflowr::wflow_publish(“plots_pbmc.Rmd”) |
| Rmd | bf23ca0 | Peter Carbonetto | 2020-08-20 | Added manual labeling of purified PBMC data to plots_pbmc analysis. |
| html | 0c1b570 | Peter Carbonetto | 2020-08-20 | First build on plots_pbmc page. |
| Rmd | eb7f776 | Peter Carbonetto | 2020-08-20 | workflowr::wflow_publish(“plots_pbmc.Rmd”) |
TO DO: Add introductory text here.
Load the packages used in the analysis below.
library(dplyr)
library(fastTopics)
library(ggplot2)
library(cowplot)
library(Ternary)
source("../code/plots.R")Load the sample annotations. (The count data are no longer needed at this stage of the analysis, so I have removed them.)
load("../data/pbmc_purified.RData")
samples_purified <- samples
load("../data/pbmc_68k.RData")
samples_68k <- samples
rm(genes,counts)Load the \(k = 6\) Poisson NMF model fits for both PBMC data sets. In the descriptions below, I refer to these Poisson NMF model fits as the “purified” and “68k” fits.
To aid presentation of the results, topics in the 68k fit are reordered to better align with the topics in purified Poisson NMF fit.
fit_purified <-
readRDS("../output/pbmc-purified/rds/fit-pbmc-purified-scd-ex-k=6.rds")$fit
fit_68k <- readRDS("../output/pbmc-68k/rds/fit-pbmc-68k-scd-ex-k=6.rds")$fit
cols <- c(4,1,5,3,6,2)
fit_68k$F <- fit_68k$F[,cols]
fit_68k$L <- fit_68k$L[,cols]
colnames(fit_68k$F) <- paste0("k",1:6)
colnames(fit_68k$L) <- paste0("k",1:6)We begin by exploring structure in the data as inferred by the topic model. We will visualize this structure by plotting principal components (PCs) of the topic proportions. Although PCA is simple, we will see that it quite effective, and avoids the complications of nonlinear dimensionality reduction techniques such as t-SNE and UMAP.
We begin with the mixture of FACS-purified PBMC data.
fit <- poisson2multinom(fit_purified)
pca <- prcomp(fit$L)$xThree large clusters are clearly evident from first two PCs (there is also finer-scale structure which we will examine shortly). We label these clusters as “A”, “B” and “C”.
n <- nrow(pca)
x <- rep("C",n)
pc1 <- pca[,"PC1"]
pc2 <- pca[,"PC2"]
x[pc1 + 0.2 > pc2] <- "A"
x[pc2 > 0.25] <- "B"
x[(pc1 + 0.4)^2 + (pc2 + 0.1)^2 < 0.07] <- "C"
samples_purified$cluster <- x
p1 <- pca_plot_with_labels(fit_purified,c("PC1","PC2"),
samples_purified$cluster) +
labs(fill = "cluster")
print(p1)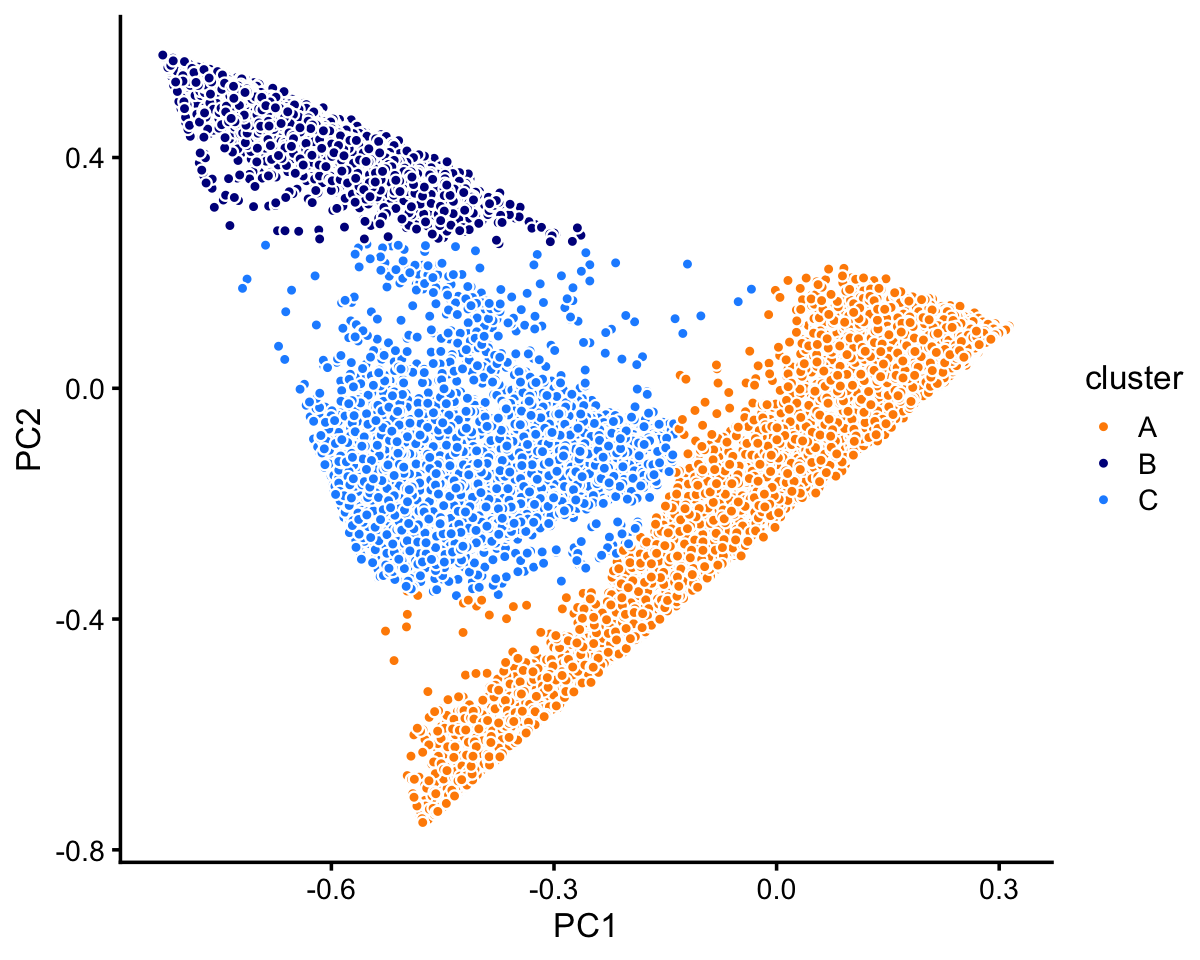
Most of the samples are in cluster A:
table(x)
# x
# A B C
# 72614 10439 11602In cluster C, there are two well-defined subclusters, labeled “C1” and “C2”. There are other possible subclusters that are less defined, but here we focus on the most obvious clustering:
rows <- which(samples_purified$cluster == "C")
fit <- select(poisson2multinom(fit_purified),loadings = rows)
pca <- prcomp(fit$L)$x
n <- nrow(pca)
x <- rep("C3",n)
pc1 <- pca[,1]
pc2 <- pca[,2]
x[pc1 < 0 & pc2 < 0.4] <- "C1"
x[pc1 > 0.5 & pc2 < 0.3] <- "C2"
samples_purified$cluster[rows] <- x
p5 <- pca_plot_with_labels(fit,c("PC1","PC2"),x) +
labs(fill = "cluster")
print(p5)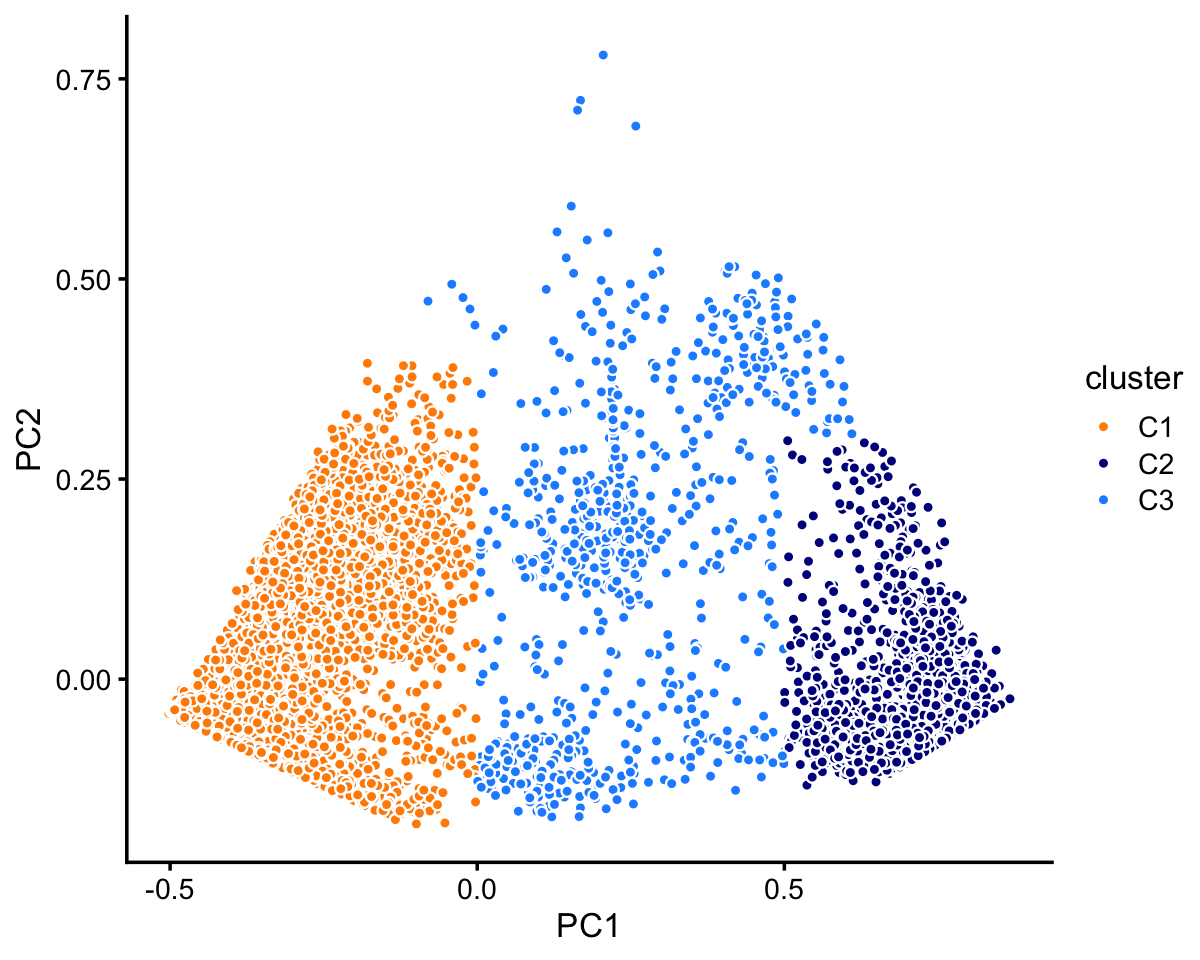
| Version | Author | Date |
|---|---|---|
| 7900d17 | Peter Carbonetto | 2020-08-22 |
The two subclusters C1 and C2 account for most of the samples:
table(x)
# x
# C1 C2 C3
# 7822 2990 790Let’s now look more closely at cluster A. There is a large, much less distinct subcluster, which we label as “A1”. Otherwise, there are no other clearly well-defined clusters.
rows <- which(samples_purified$cluster == "A")
fit <- select(poisson2multinom(fit_purified),loadings = rows)
pca <- prcomp(fit$L)$x
n <- nrow(fit$L)
x <- rep("A2",n)
pc1 <- pca[,1]
pc2 <- pca[,2]
x[pc1 > 0.6 - pc2] <- "A1"
samples_purified$cluster[rows] <- x
p6 <- pca_plot_with_labels(fit,c("PC1","PC2"),x) +
labs(fill = "cluster")
print(p6)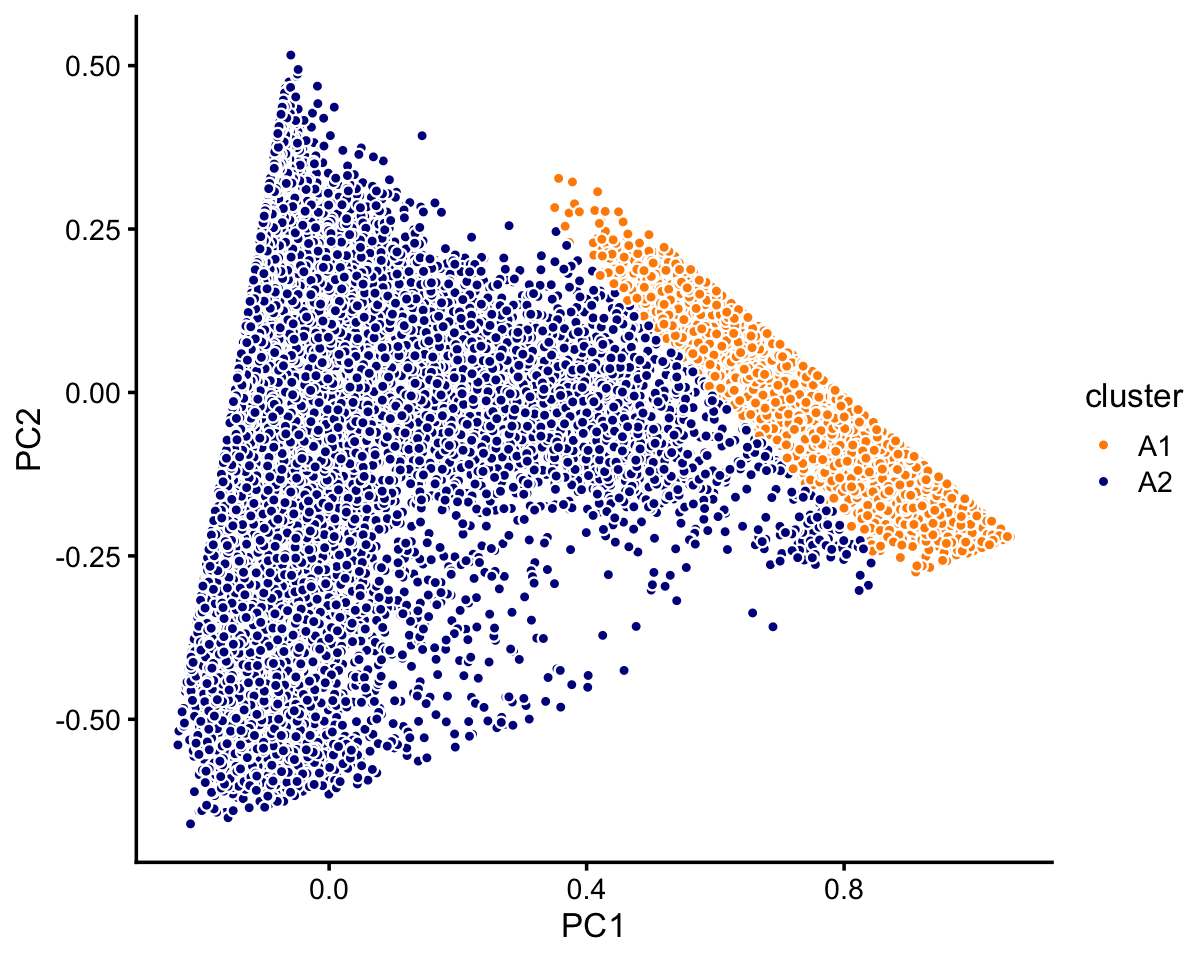
| Version | Author | Date |
|---|---|---|
| 7900d17 | Peter Carbonetto | 2020-08-22 |
In summary, we have subdivided these data into 6 clusters:
samples_purified$cluster <- factor(samples_purified$cluster)
table(samples_purified$cluster)
#
# A1 A2 B C1 C2 C3
# 8146 64468 10439 7822 2990 790Next, we turn to the 68k fit.
fit <- poisson2multinom(fit_68k)
pca <- prcomp(fit$L)$xIn this case, we find least three distinct clusters in the projection onto PCs 3 and 4:
n <- nrow(pca)
x <- rep("A",n)
pc3 <- pca[,"PC3"]
pc4 <- pca[,"PC4"]
x[pc4 > 0.13 | pc3/1.9 - 0.17 > -pc4] <- "B"
x[pc4 > 0.75] <- "C"
samples_68k$cluster <- x
p2 <- pca_plot_with_labels(fit_68k,c("PC3","PC4"),x) +
labs(fill = "cluster")
print(p2)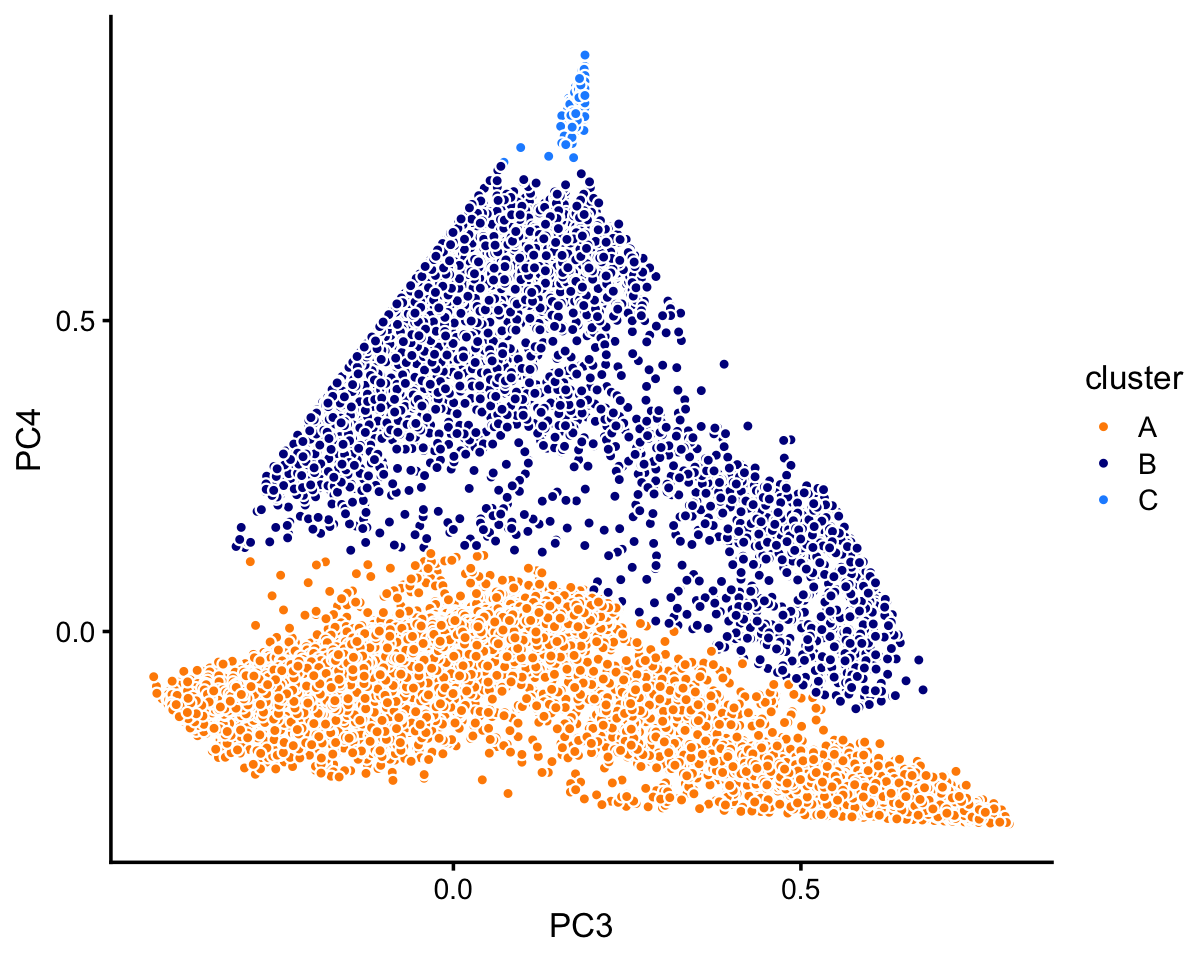
The vast majority of the cells are in cluster A:
table(samples_68k$cluster)
#
# A B C
# 63408 5006 165TO DO: Add text here.
set.seed(1)
pbmc_purified_topics <- c(2,5,1,3,4,6)
pbmc_purified_topic_colors <- c("gold","forestgreen","dodgerblue",
"gray","greenyellow","magenta")
rows <- sort(c(sample(which(samples_purified$cluster == "c1"),1200),
sample(which(samples_purified$cluster == "c2"),600),
sample(which(samples_purified$cluster == "c3"),600)))
p3 <- structure_plot(select(poisson2multinom(fit_purified),loadings = rows),
grouping = samples_purified[rows,"cluster"],
topics = pbmc_purified_topics,
colors = pbmc_purified_topic_colors[pbmc_purified_topics],
gap = 40,num_threads = 4,verbose = FALSE)
print(p3)TO DO: Add text here.
set.seed(1)
pbmc_68k_topics <- c(2,5,1,3,4,6)
pbmc_68k_topic_colors <- c("darkorange","olivedrab","dodgerblue",
"gray","royalblue","magenta")
rows <- sort(c(sample(which(samples_68k$cluster == "c1"),1400),
sample(which(samples_68k$cluster == "c2"),500),
which(samples_68k$cluster == "c3")))
p4 <- structure_plot(select(poisson2multinom(fit_68k),loadings = rows),
grouping = samples_68k[rows,"cluster"],
topics = pbmc_68k_topics,
colors = pbmc_68k_topic_colors[pbmc_68k_topics],
gap = 40,num_threads = 4,verbose = FALSE)
print(p4)TO DO: Before comparing to Zheng et al (2017) labeling, do more manual identification of clusters from the PCA plots.
with(samples_purified,table(celltype,cluster))Comparison to Zheng et al (2017) cell-type labeling of the FACS-purified PBMC data. In fit_purified, cluster 2 captures B-cells; cluster 3 is almost exclusively CD34+ and CD14+ monocytes:
Now let’s look more closely at cluster 1 in fit_purified:
par(mar = c(0,0,0,0))
rows <- which(samples_purified$cluster == "c1")
TernaryPlot(alab = "k1",blab = "k4",clab = "k6",
grid.col = "gainsboro",grid.minor.lines = 0)
TernaryPoints(pdat[rows,c(1,4,6)],pch = 21,cex = 0.65,col = "white",
bg = as.character(pdat[rows,"celltype"]))In fit_68k, cluster 2 mostly consists of CD14+ monocyte and dendritic cells, whereas cluster 3 is a small population of CD34+ cells.
with(samples_68k,table(celltype,cluster))Let’s look more closely at cluster 2 in fit_68k:
par(mar = c(0,0,0,0))
pbmc_68k_celltype_colors <-
c("forestgreen", # CD14+ Monocyte
"dodgerblue", # CD19+ B
"palegreen", # CD34+"
"yellowgreen", # CD4+ T Helper2
"gold", # CD4+/CD25 T Reg
"darkorange", # CD4+/CD45RA+/CD25- Naive T
"gold", # CD4+/CD45RO+ Memory"
"gray", # CD56+ NK
"tomato", # CD8+ Cytotoxic T
"magenta", # CD8+/CD45RA+ Naive Cytotoxic"
"darkmagenta") # Dendritic"
L <- poisson2multinom(fit_68k)$L
pdat <- cbind(L,samples_68k["celltype"])
rows <- which(samples_68k$cluster == "c2")
levels(pdat$celltype) <- pbmc_68k_celltype_colors
TernaryPlot(alab = "k3",blab = "k5",clab = "k6",
grid.col = "gainsboro",grid.minor.lines = 0)
TernaryPoints(pdat[rows,c(3,5,6)],pch = 21,cex = 0.65,col = "white",
bg = as.character(pdat[rows,"celltype"]))Finally, let’s look more closely at cluster 1 in fit_68k:
par(mar = c(0,0,0,0))
rows <- which(samples_68k$cluster == "c1")
TernaryPlot(alab = "k2",blab = "k4",clab = "k6",
grid.col = "gainsboro",grid.minor.lines = 0)
TernaryPoints(pdat[rows,c(2,4,6)],pch = 21,cex = 0.65,col = "white",
bg = as.character(pdat[rows,"celltype"]))p4 <- pca_plot_with_labels(fit_purified,c("PC1","PC2"),
samples_purified$celltype,
purified_celltype_colors) + labs(fill = "celltype")
p5 <- pca_plot_with_labels(fit_purified,c("PC4","PC5"),
samples_purified$celltype,
purified_celltype_colors) + labs(fill = "celltype")pbmc_purified_celltype_colors <-
c("dodgerblue", # CD19+ B
"forestgreen", # CD14+ Monocyte
"palegreen", # CD34+
"plum", # CD4+ T Helper2
"gray", # CD56+ NK
"tomato", # CD8+ Cytotoxic T
"gold", # CD4+/CD45RO+ Memory
"magenta", # CD8+/CD45RA+ Naive Cytotoxic
"darkorange", # CD4+/CD45RA+/CD25- Naive T
"yellowgreen") # CD4+/CD25 T RegLoadings plot:
loadings_plot(poisson2multinom(fit_purified),samples_purified$celltype)
loadings_plot(poisson2multinom(fit_68k),samples_68k$celltype)PCA plot:
clrs <- c("forestgreen", # CD14+ Monocyte
"dodgerblue", # CD19+ B
"darkmagenta", # CD34+"
"yellowgreen", # CD4+ T Helper2
"gold", # CD4+/CD25 T Reg
"limegreen", # CD4+/CD45RA+/CD25- Naive T
"orange", # CD4+/CD45RO+ Memory"
"gray", # CD56+ NK
"tomato", # CD8+ Cytotoxic T
"magenta", # CD8+/CD45RA+ Naive Cytotoxic"
"darkblue") # Dendritic"
fit2 <- poisson2multinom(fit)
pca <- prcomp(fit2$L)
pdat <- cbind(samples,pca$x)
ggplot(pdat,aes(x = PC3,y = PC4,fill = celltype)) +
geom_point(shape = 21,color = "white",size = 1.5) +
scale_fill_manual(values = clrs) +
theme_cowplot(font_size = 10)t-SNE plot:
set.seed(1)
p2 <- tsne_plot(fit,n = 8000,num_threads = 4)Differential count analysis:
diff_count_res <- diff_count_analysis(fit,counts)Volcano plots:
p3 <- volcano_plot(diff_count_res,labels = genes$symbol,
label_above_quantile = 0.995)Structure plots:
set.seed(1)
fit2 <- select(poisson2multinom(fit),
loadings = which(samples$celltype == "CD19+ B"))
p4 <- structure_plot(fit2,n = 2000,num_threads = 4) # B-cells.
set.seed(1)
fit2 <- select(poisson2multinom(fit),
loadings = which(samples$celltype == "CD56+ NK"))
p5 <- structure_plot(fit2,n = 2000,num_threads = 4) # NK cells.
set.seed(1)
fit2 <- select(poisson2multinom(fit),
loadings = which(samples$celltype == "CD34+"))
p6 <- structure_plot(fit2,num_threads = 4,perplexity = 50) # CD34+ cells
set.seed(1)
fit2 <- select(poisson2multinom(fit),
loadings = which(samples$celltype == "CD14+ Monocyte"))
p7 <- structure_plot(fit2,num_threads = 4) # CD14+ monocytes
set.seed(1)
fit2 <- select(poisson2multinom(fit),
loadings = which(samples$celltype == "Dendritic"))
p8 <- structure_plot(fit2,num_threads = 4) # dendritic cells
plot_grid(p7,p8,nrow = 2)
set.seed(1)
fit2 <- select(poisson2multinom(fit),
loadings = which(samples$celltype == "CD4+ T Helper2"))
p9 <- structure_plot(fit2,num_threads = 4,perplexity = 30) +
ggtitle("CD4+ T Helper2") +
set.seed(1)
fit2 <- select(poisson2multinom(fit),
loadings = which(samples$celltype == "CD4+/CD45RA+/CD25- Naive T"))
p10 <- structure_plot(fit2,num_threads = 4) +
ggtitle("CD4+/CD45RA+/CD25- Naive T")
set.seed(1)
fit2 <- select(poisson2multinom(fit),
loadings = which(samples$celltype == "CD4+/CD45RO+ Memory"))
p11 <- structure_plot(fit2,num_threads = 4) +
ggtitle("CD4+/CD45RO+ Memory")
set.seed(1)
fit2 <- select(poisson2multinom(fit),
loadings = which(samples$celltype == "CD4+/CD25 T Reg"))
p12 <- structure_plot(fit2,num_threads = 4) +
ggtitle("CD4+/CD25 T Reg")
fit2 <- select(poisson2multinom(fit),
loadings = which(samples$celltype == "CD8+/CD45RA+ Naive Cytotoxic"))
p13 <- structure_plot(fit2,num_threads = 4) +
ggtitle("CD8+/CD45RA+ Naive Cytotoxic")
fit2 <- select(poisson2multinom(fit),
loadings = which(samples$celltype == "CD8+ Cytotoxic T"))
p14 <- structure_plot(fit2,num_threads = 4) +
ggtitle("CD8+ Cytotoxic T")
plot_grid(p9,p10,p11,p12,p13,p14,nrow = 6)Another structure plot:
p15 <- structure_plot(fit,num_threads = 4)
sessionInfo()
# R version 3.6.2 (2019-12-12)
# Platform: x86_64-apple-darwin15.6.0 (64-bit)
# Running under: macOS Catalina 10.15.5
#
# Matrix products: default
# BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
# LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
#
# locale:
# [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
#
# attached base packages:
# [1] stats graphics grDevices utils datasets methods base
#
# other attached packages:
# [1] Ternary_1.1.4 cowplot_1.0.0 ggplot2_3.3.0 fastTopics_0.3-163
# [5] dplyr_0.8.3
#
# loaded via a namespace (and not attached):
# [1] ggrepel_0.9.0 Rcpp_1.0.3 lattice_0.20-38
# [4] tidyr_1.0.0 prettyunits_1.1.1 assertthat_0.2.1
# [7] zeallot_0.1.0 rprojroot_1.3-2 digest_0.6.23
# [10] R6_2.4.1 backports_1.1.5 MatrixModels_0.4-1
# [13] evaluate_0.14 coda_0.19-3 httr_1.4.1
# [16] pillar_1.4.3 rlang_0.4.5 progress_1.2.2
# [19] lazyeval_0.2.2 data.table_1.12.8 irlba_2.3.3
# [22] SparseM_1.78 whisker_0.4 Matrix_1.2-18
# [25] rmarkdown_2.3 labeling_0.3 Rtsne_0.15
# [28] stringr_1.4.0 htmlwidgets_1.5.1 munsell_0.5.0
# [31] compiler_3.6.2 httpuv_1.5.2 xfun_0.11
# [34] pkgconfig_2.0.3 mcmc_0.9-6 htmltools_0.4.0
# [37] tidyselect_0.2.5 tibble_2.1.3 workflowr_1.6.2.9000
# [40] quadprog_1.5-8 viridisLite_0.3.0 crayon_1.3.4
# [43] withr_2.1.2 later_1.0.0 MASS_7.3-51.4
# [46] grid_3.6.2 jsonlite_1.6 gtable_0.3.0
# [49] lifecycle_0.1.0 git2r_0.26.1 magrittr_1.5
# [52] scales_1.1.0 RcppParallel_4.4.4 stringi_1.4.3
# [55] farver_2.0.1 fs_1.3.1 promises_1.1.0
# [58] vctrs_0.2.1 tools_3.6.2 glue_1.3.1
# [61] purrr_0.3.3 hms_0.5.2 yaml_2.2.0
# [64] colorspace_1.4-1 plotly_4.9.2 knitr_1.26
# [67] quantreg_5.54 MCMCpack_1.4-5