Identify clusters in pulse-seq data using topic model
Peter Carbonetto
Last updated: 2020-09-18
Checks: 7 0
Knit directory: single-cell-topics/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2.9000). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(1) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version ded741c. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: data/droplet.RData
Ignored: data/pbmc_68k.RData
Ignored: data/pbmc_purified.RData
Ignored: data/pulseseq.RData
Ignored: output/droplet/fits-droplet.RData
Ignored: output/droplet/rds/
Ignored: output/pbmc-68k/fits-pbmc-68k.RData
Ignored: output/pbmc-68k/rds/
Ignored: output/pbmc-purified/fits-pbmc-purified.RData
Ignored: output/pbmc-purified/rds/
Ignored: output/pulseseq/fits-pulseseq.RData
Ignored: output/pulseseq/rds/
Unstaged changes:
Modified: analysis/plots_tracheal_epithelium.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/clusters_pulseseq.Rmd) and HTML (docs/clusters_pulseseq.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | ded741c | Peter Carbonetto | 2020-09-18 | workflowr::wflow_publish(“clusters_pulseseq.Rmd”) |
| html | 4b4b233 | Peter Carbonetto | 2020-09-18 | Make some improvements to clusters_pulseseq analysis. |
| Rmd | d2cb602 | Peter Carbonetto | 2020-09-16 | workflowr::wflow_publish(“clusters_pulseseq.Rmd”) |
| html | 13a5956 | Peter Carbonetto | 2020-09-16 | Built clusters_pulseseq page. |
| Rmd | 5e0ee23 | Peter Carbonetto | 2020-09-16 | Completed first rough draft of clustering in clusters_pulseseq.Rmd. |
| Rmd | 337d6fc | Peter Carbonetto | 2020-09-16 | Added clusters identified in PCs 5 and 6 of k=11 pulse-seq fit. |
| Rmd | c0a27bd | Peter Carbonetto | 2020-09-16 | Added clustering of pulseseq data along PCs 3 and 4. |
| html | e7383b2 | Peter Carbonetto | 2020-09-16 | Produced first rendering of clusters_pulseseq analysis. |
| Rmd | 1dd20d4 | Peter Carbonetto | 2020-09-16 | workflowr::wflow_publish(“clusters_pulseseq.Rmd”) |
| Rmd | da9ac09 | Peter Carbonetto | 2020-09-16 | Added hexbin plots to clusters_pulseseq analysis. |
| Rmd | 485639a | Peter Carbonetto | 2020-09-16 | Working on clusters_pulseseq analysis. |
| Rmd | c8dd3af | Peter Carbonetto | 2020-09-16 | Implemented basic_pca_plot; improved labeled_pca_plot function. |
Here we perform PCA on the topic proportions to identify clusters in the pulse-seq data.
Load the packages used in the analysis below, as well as additional functions that we will use to generate some of the plots.
library(dplyr)
library(fastTopics)
library(ggplot2)
library(cowplot)
source("../code/plots.R")Load data and results
Load the pulse-seq data. The UMI counts are not needed for this analysis.
load("../data/pulseseq.RData")
x <- as.character(samples$tissue)
x[x == "club (hillock-associated)"] <- "hillock"
x[x == "goblet.1" | x == "goblet.2" | x == "goblet.progenitor"] <- "goblet"
x[x == "tuft.1" | x == "tuft.2" | x == "tuft.progenitor"] <- "tuft"
samples$tissue <- factor(x)
rm(counts)Load the \(k = 11\) Poisson NMF fit.
fit <- readRDS("../output/pulseseq/rds/fit-pulseseq-scd-ex-k=11.rds")$fitIdentify clusters from principal components
To identify clusters, we begin by plotting PCs computed from the rows of the topic proportions matrix. (Note that only 10 PCs are needed for the multinomial topic model with \(k = 11\) topics.)
p1 <- basic_pca_plot(fit,1:2)
p2 <- basic_pca_plot(fit,3:4)
p3 <- basic_pca_plot(fit,5:6)
p4 <- basic_pca_plot(fit,7:8)
p5 <- basic_pca_plot(fit,9:10)
plot_grid(p1,p2,p3,p4,p5,nrow = 2,ncol = 3)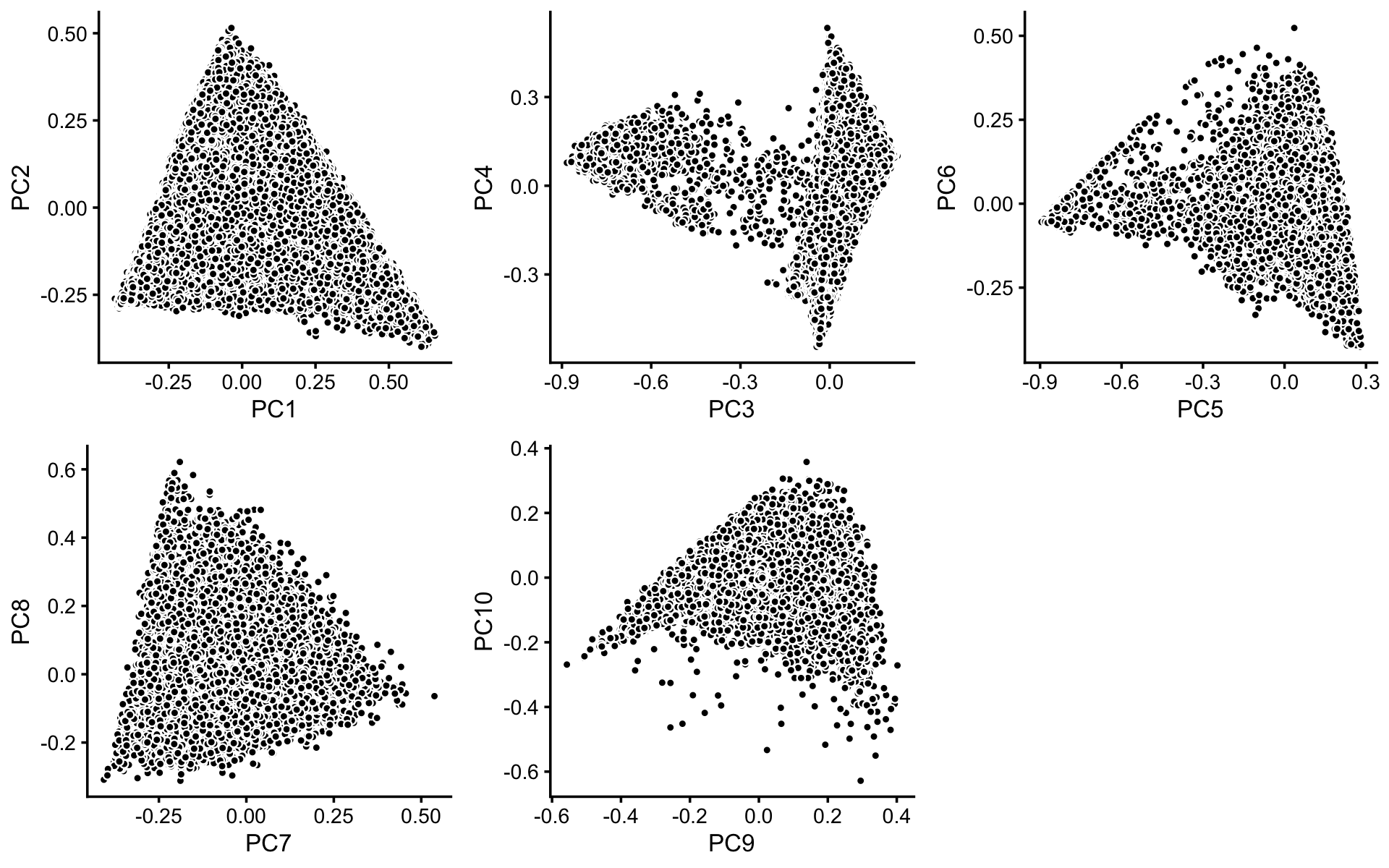
| Version | Author | Date |
|---|---|---|
| e7383b2 | Peter Carbonetto | 2020-09-16 |
Structure is more evident from the following “hexbin” plots showing the density of the points in the PCA projection. For example, the two clusters in PCs 1 and 2 is not evident from the scatterplot, but emerges in the hexbin plots:
p6 <- pca_hexbin_plot(fit,1:2) + guides(fill = "none")
p7 <- pca_hexbin_plot(fit,3:4) + guides(fill = "none")
p8 <- pca_hexbin_plot(fit,5:6) + guides(fill = "none")
p9 <- pca_hexbin_plot(fit,7:8) + guides(fill = "none")
p10 <- pca_hexbin_plot(fit,9:10) + guides(fill = "none")
plot_grid(p6,p7,p8,p9,p10,nrow = 2,ncol = 3)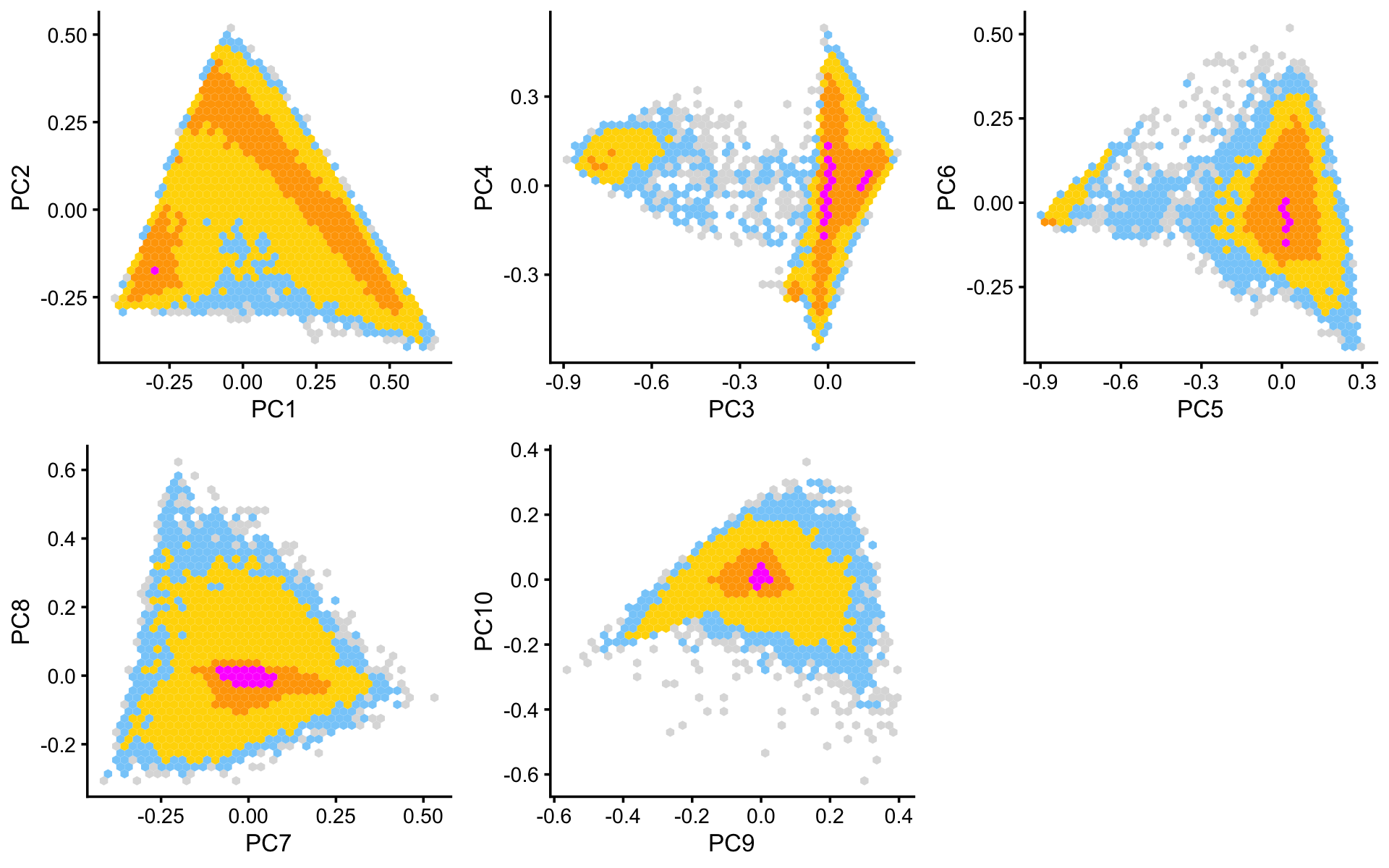
| Version | Author | Date |
|---|---|---|
| e7383b2 | Peter Carbonetto | 2020-09-16 |
From the PC plots above, we define 4 clusters, labeled A, C, Cil and T+N. (The reason for these labels will become more clear later on.) Points that do not fit in any of these clusters are assigned to a “background cluster” (this subset is labeled U for “Unknown”).
pca <- prcomp(poisson2multinom(fit)$L)$x
x <- rep("U",nrow(pca))
pc3 <- pca[,3]
pc4 <- pca[,4]
pc5 <- pca[,5]
pc6 <- pca[,6]
x[pc4 < 5.5*pc3 + 0.5] <- "A"
x[(pc3 + 0.7)^2 + (pc4 - 0.1)^2 < 0.18^2] <- "Cil"
x[x == "A" & pc6 > 1.3*pc5 + 0.87] <- "T+N"
x[x == "A" & pc5 > -0.4 & pc6 < 1.3*pc5 + 0.49] <- "C"We can refine cluster A somewhat by plotting PCs computed from the samples in cluster A only:
rows <- which(x == "A")
fit2 <- select(poisson2multinom(fit),loadings = rows)
p11 <- basic_pca_plot(fit2,1:2)
print(p11)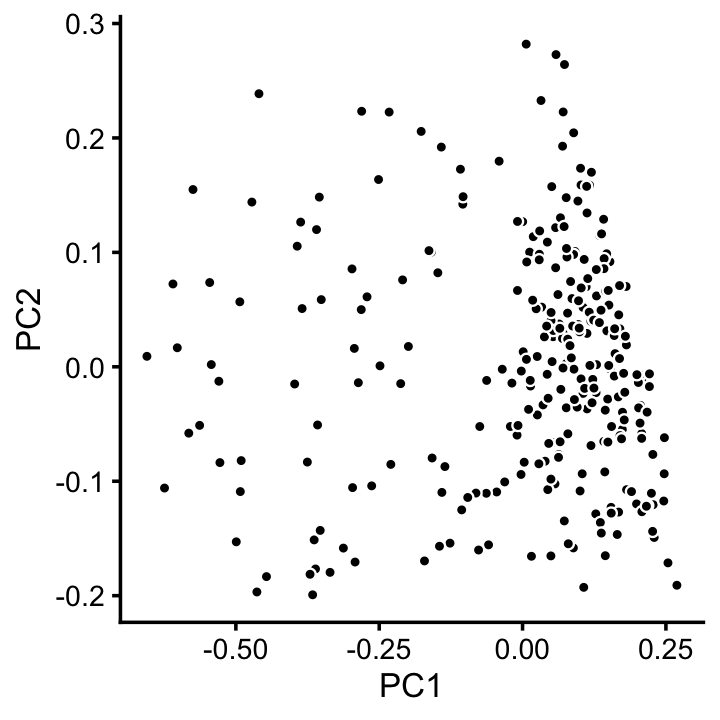
| Version | Author | Date |
|---|---|---|
| 13a5956 | Peter Carbonetto | 2020-09-16 |
We label this refined cluster as “I”, and the rest are added to the background cluster (U).
pca <- prcomp(fit2$L)$x
y <- rep("U",nrow(pca))
pc1 <- pca[,1]
y[pc1 > -0.1] <- "I"
x[rows] <- yLikewise, we inspect cluster C for further structure:
rows <- which(x == "C")
fit2 <- select(poisson2multinom(fit),loadings = rows)
p12 <- basic_pca_plot(fit2,1:2)
p13 <- pca_hexbin_plot(fit2,1:2) + guides(fill = "none")
plot_grid(p12,p13)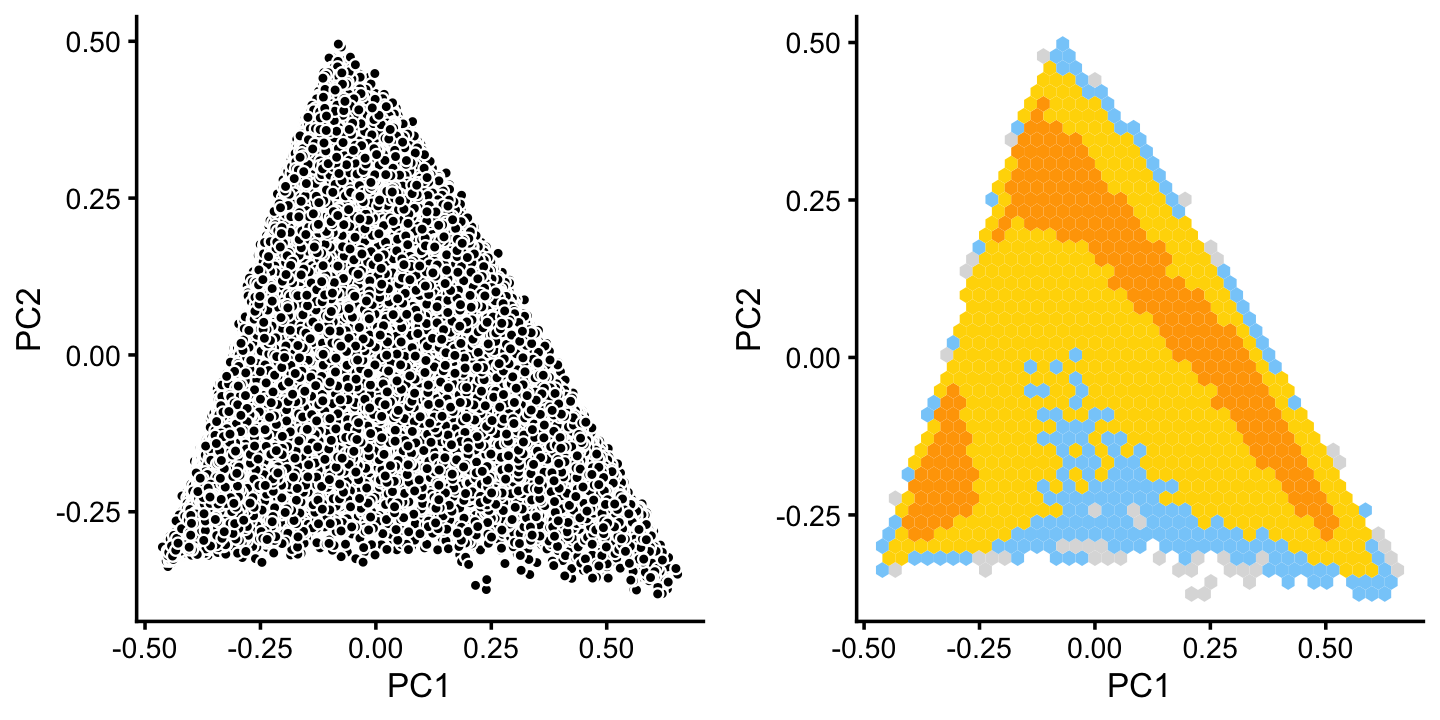
| Version | Author | Date |
|---|---|---|
| 13a5956 | Peter Carbonetto | 2020-09-16 |
The hexbin plot suggests two clusters. Although these clusters are not distinct, it may nonetheless be useful to subdivide these points (somewhat arbitrarily) into two subsets, labeled B and C.
pca <- prcomp(fit2$L)$x
y <- rep("C",nrow(pca))
pc1 <- pca[,1]
pc2 <- pca[,2]
y[pc2 > -pc1 - 0.15] <- "B"
x[rows] <- yIn cluster B, there is some interesting structure along PCs 5 and 6:
rows <- which(x == "B")
fit2 <- select(poisson2multinom(fit),loadings = rows)
p14 <- basic_pca_plot(fit2,5:6)
p15 <- pca_hexbin_plot(fit2,5:6) + guides(fill = "none")
plot_grid(p14,p15)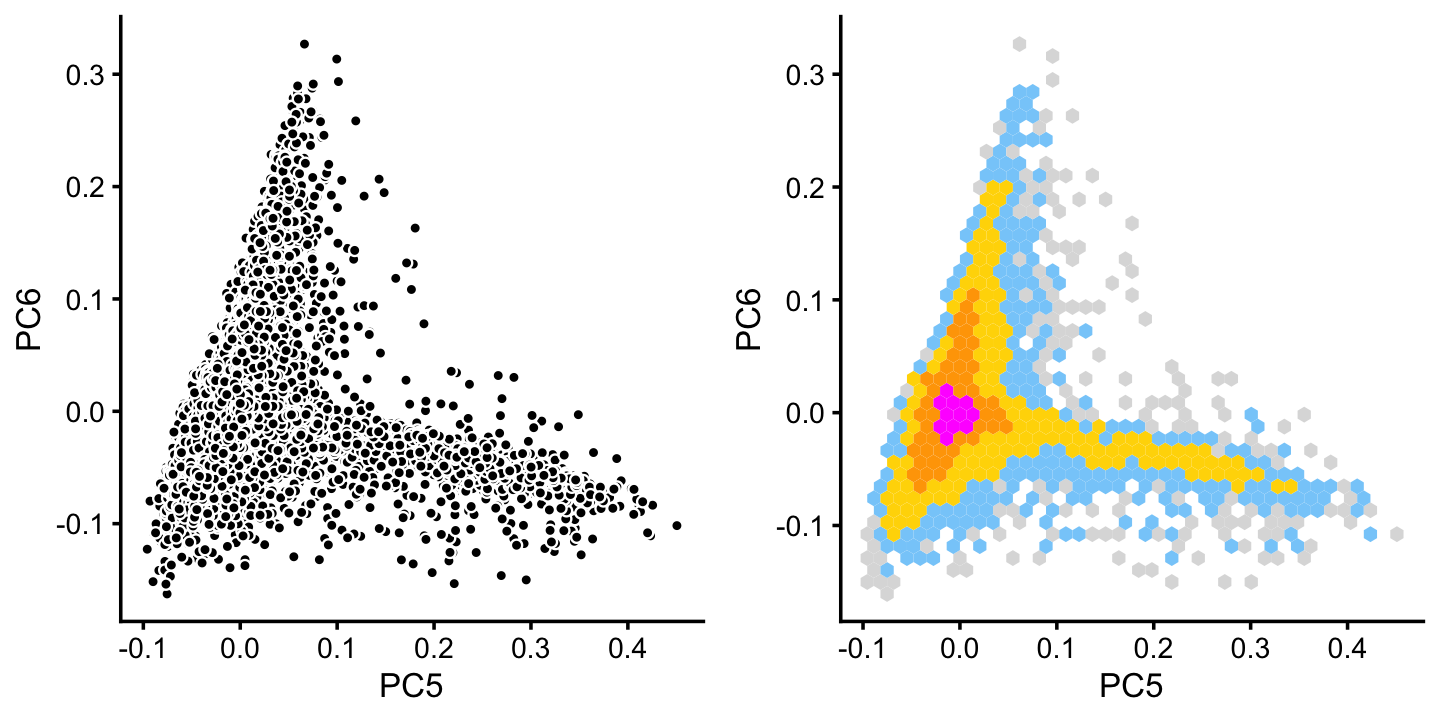
| Version | Author | Date |
|---|---|---|
| 13a5956 | Peter Carbonetto | 2020-09-16 |
We define a new cluster, “P”, recognizing that this cluster is again not very distinct.
pca <- prcomp(fit2$L)$x
y <- rep("B",nrow(pca))
pc5 <- pca[,5]
pc6 <- pca[,6]
y[pc5 > 0.1 & pc6 < 0] <- "P"
x[rows] <- yIn summary, we have subdivided the pulse-seq samples into 7 subsets, including a background cluster (“U”):
colors <- c("royalblue","forestgreen","firebrick","darkmagenta",
"darkorange","peru","gainsboro")
p16 <- labeled_pca_plot(fit,1:2,x,colors) + guides(fill = "none")
p17 <- labeled_pca_plot(fit,3:4,x,colors) + guides(fill = "none")
p18 <- labeled_pca_plot(fit,5:6,x,colors) + guides(fill = "none")
p19 <- labeled_pca_plot(fit,1:2,x,colors,"tissue") +
xlim(0,0) + ylim(0,0)
plot_grid(p16,p17,p18,p19)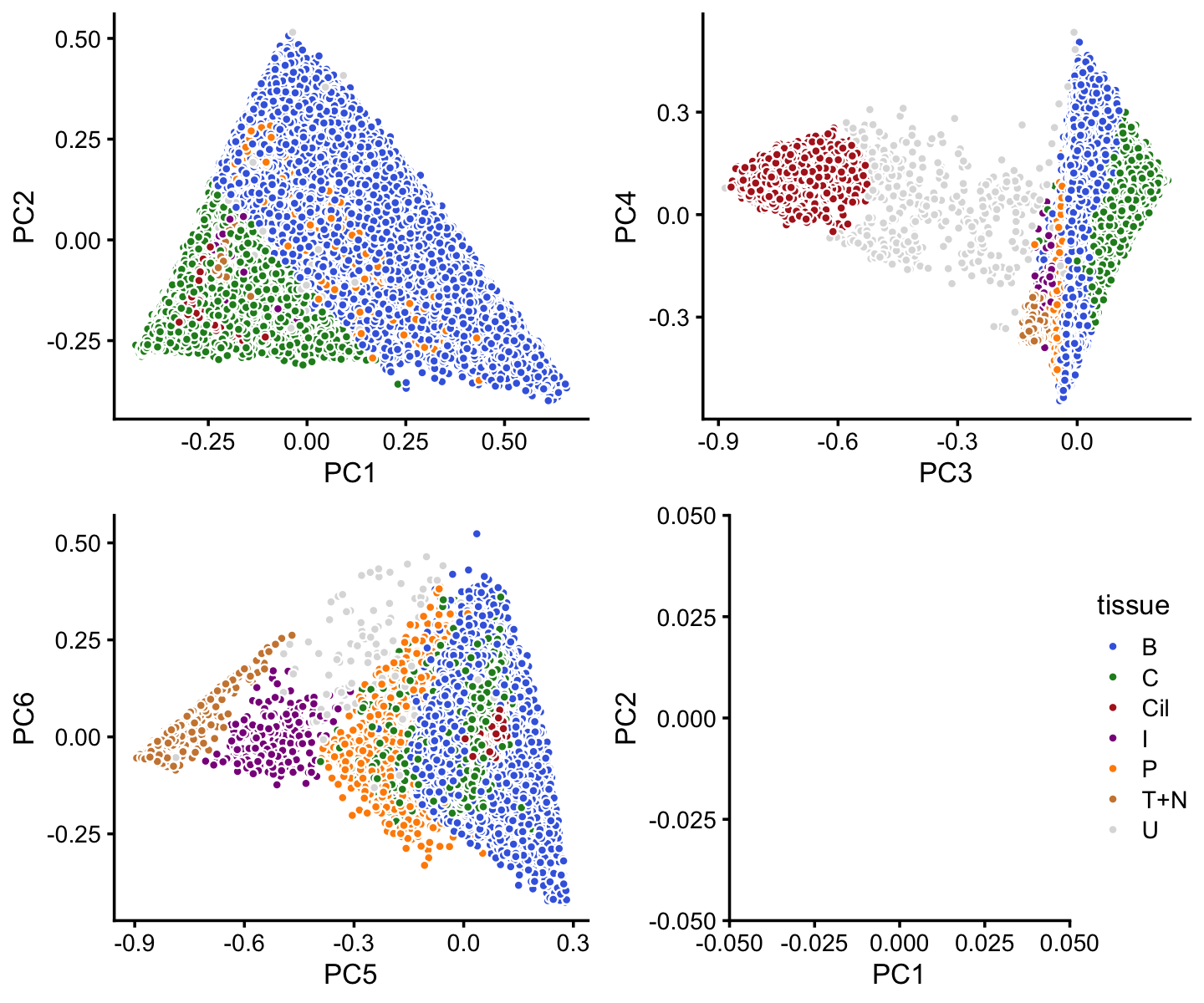
| Version | Author | Date |
|---|---|---|
| 4b4b233 | Peter Carbonetto | 2020-09-18 |
Comparing this to the Montoro et al (2018) clustering, we see that some close correspondence (e.g., B and basal cells, P and proliferating cells). The B and C clusters appear to contain additional structure that cannot be captured by distinct clusters. We will explore this structure in a subsequent analysis.
table(samples$tissue,x)
# x
# B C Cil I P T+N U
# basal 40389 1468 0 0 199 0 37
# ciliated 0 0 2896 0 6 0 114
# club 1677 11834 0 6 32 1 18
# goblet 3 396 0 0 2 0 2
# hillock 89 4036 0 0 4 0 3
# ionocyte 0 45 0 193 15 8 15
# neuroendocrine 0 1 0 7 0 619 3
# proliferating 61 194 9 4 914 0 231
# tuft 0 18 0 12 4 691 9Save results
Add text here.
sessionInfo()
# R version 3.6.2 (2019-12-12)
# Platform: x86_64-apple-darwin15.6.0 (64-bit)
# Running under: macOS Catalina 10.15.6
#
# Matrix products: default
# BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
# LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
#
# locale:
# [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
#
# attached base packages:
# [1] stats graphics grDevices utils datasets methods base
#
# other attached packages:
# [1] cowplot_1.0.0 ggplot2_3.3.0 fastTopics_0.3-175 dplyr_0.8.3
#
# loaded via a namespace (and not attached):
# [1] ggrepel_0.9.0 Rcpp_1.0.5 lattice_0.20-38
# [4] tidyr_1.0.0 prettyunits_1.1.1 assertthat_0.2.1
# [7] zeallot_0.1.0 rprojroot_1.3-2 digest_0.6.23
# [10] R6_2.4.1 backports_1.1.5 MatrixModels_0.4-1
# [13] evaluate_0.14 coda_0.19-3 httr_1.4.1
# [16] pillar_1.4.3 rlang_0.4.5 progress_1.2.2
# [19] lazyeval_0.2.2 data.table_1.12.8 irlba_2.3.3
# [22] SparseM_1.78 hexbin_1.28.0 whisker_0.4
# [25] Matrix_1.2-18 rmarkdown_2.3 labeling_0.3
# [28] Rtsne_0.15 stringr_1.4.0 htmlwidgets_1.5.1
# [31] munsell_0.5.0 compiler_3.6.2 httpuv_1.5.2
# [34] xfun_0.11 pkgconfig_2.0.3 mcmc_0.9-6
# [37] htmltools_0.4.0 tidyselect_0.2.5 tibble_2.1.3
# [40] workflowr_1.6.2.9000 quadprog_1.5-8 viridisLite_0.3.0
# [43] crayon_1.3.4 withr_2.1.2 later_1.0.0
# [46] MASS_7.3-51.4 grid_3.6.2 jsonlite_1.6
# [49] gtable_0.3.0 lifecycle_0.1.0 git2r_0.26.1
# [52] magrittr_1.5 scales_1.1.0 RcppParallel_4.4.2
# [55] stringi_1.4.3 farver_2.0.1 fs_1.3.1
# [58] promises_1.1.0 vctrs_0.2.1 tools_3.6.2
# [61] glue_1.3.1 purrr_0.3.3 hms_0.5.2
# [64] yaml_2.2.0 colorspace_1.4-1 plotly_4.9.2
# [67] knitr_1.26 quantreg_5.54 MCMCpack_1.4-5