RNA-stability
unawaz1996
2023-05-05
Last updated: 2023-05-08
Checks: 6 1
Knit directory: NMD-analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230314) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| /home/neuro/Documents/NMD_analysis/Analysis/NMD-analysis/data/data2.txt | data/data2.txt |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version f4e0d39. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/Differential-transcript-usage.nb.html
Ignored: analysis/Enichment-analysis-fgsea.nb.html
Ignored: analysis/Enichment-analysis-goseq.nb.html
Untracked files:
Untracked: PCA.png
Untracked: PCA_plot.pdf
Untracked: PCA_transcript.png
Untracked: analysis/Differential-transcript-expression.Rmd
Untracked: analysis/Differential-transcript-usage.Rmd
Untracked: analysis/UPF3B_KD.Rmd
Untracked: analysis/figures.Rmd
Untracked: analysis/transcript-preprocessing.Rmd
Untracked: code/eisaR.R
Untracked: code/external_code/
Untracked: data/LTK_Sample Metafile_V3.txt
Untracked: data/Mus_musculus.GRCm39.105__nifs.tsv
Untracked: data/data.txt
Untracked: data/data2.txt
Untracked: data/fastqc/
Untracked: data/nif_output/
Untracked: data/samples.txt
Untracked: output/DEG-limma-results.Rda
Untracked: output/DEG-list.Rda
Untracked: output/DEG/
Untracked: output/EISA/
Untracked: output/ISAR/
Untracked: output/QC/
Untracked: output/Transcript/
Untracked: output/isoformSwitchAnalyzeR_isoform_AA_complete.fasta
Untracked: output/isoformSwitchAnalyzeR_isoform_AA_subset_1_of_3.fasta
Untracked: output/isoformSwitchAnalyzeR_isoform_AA_subset_2_of_3.fasta
Untracked: output/isoformSwitchAnalyzeR_isoform_AA_subset_3_of_3.fasta
Untracked: output/isoformSwitchAnalyzeR_isoform_nt.fasta
Untracked: output/limma-matrices.Rda
Untracked: tmp/
Unstaged changes:
Modified: analysis/Enichment-analysis-fgsea.Rmd
Modified: analysis/_site.yml
Modified: code/functions.R
Modified: code/libraries.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/RNA-stability.Rmd) and
HTML (docs/RNA-stability.html) files. If you’ve configured
a remote Git repository (see ?wflow_git_remote), click on
the hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | f4e0d39 | unawaz1996 | 2023-05-08 | wflow_publish("analysis/RNA-stability.Rmd") |
Introduction
RNA-stability analysis
It has been suggested that, while exonic read counts in RNA-seq data correspond to steady-state mRNA abundance, changes in the abundance of intronic reads can be used to estimate the change in transcription rate The REMBRANDTS algorithm is a refinement of earlier methods based on the idea that a change in exonic reads without a corresponding change in intronic reads is diagnostic of differential RNA stability, while concurrent changes in both exonic and intronic reads suggest altered transcription. Therefore, RNA-seq reflects a snapshot of both steady-state mRNA level and transcriptional activity, providing the possibility for deconvolving the mRNA decay rate from RNA-seq data.
[1] "Analyzing exonic reads ..."
[1] "Analyzing batch 1"[1] "Analyzing intronic reads ..."
[1] "Analyzing batch 1"[1] "Optimizing read count cutoff at stringency 0.99 ..."[1] "Total correlation is 0.372551660128126"[1] "Total number of genes is 13455"[1] "Maximum correlation is 0.465630414816467"[1] "Selected threshold is 10.3628798754118"[1] "Number of remaining genes is 2101"png
2 png
2 png
2 png
2 png
2 'data.frame': 2101 obs. of 18 variables:
$ 212: num -0.0219 -0.0657 0.1234 -0.1129 -0.0349 ...
$ 213: num -0.09234 -0.05181 -0.06052 0.00876 -0.00139 ...
$ 214: num 0.00123 -0.01991 0.12214 0.02702 0.05739 ...
$ 215: num -0.024 -0.1024 0.0851 -0.1704 0.0242 ...
$ 216: num -0.07 -0.1353 -0.059 -0.0524 -0.0843 ...
$ 217: num 0.00202 -0.01629 -0.01338 -0.01292 -0.12356 ...
$ 218: num 0.0548 0.1069 0.0084 0.0547 0.0776 ...
$ 219: num 0.0434 -0.0381 -0.1663 0.0935 0.0428 ...
$ 220: num 0.0984 0.1741 0.0756 0.152 0.1472 ...
$ 221: num 0.05883 0.01973 -0.06929 -0.01619 -0.00779 ...
$ 222: num 0.0254 -0.0877 -0.0837 0.0103 -0.0219 ...
$ 223: num 0.072 -0.0697 -0.0474 -0.1061 0.1929 ...
$ 224: num 0.0988 0.1446 0.0295 0.1087 -0.0155 ...
$ 225: num -0.0982 -0.023 -0.0967 0.0906 -0.032 ...
$ 226: num -0.00716 0.14733 0.11268 0.05907 0.01315 ...
$ 227: num -0.0142 -0.0436 0.0786 -0.0398 -0.0806 ...
$ 228: num -0.0518 -0.0275 -0.0332 -0.0895 -0.075 ...
$ 229: num -0.07525 0.08832 -0.00597 -0.00438 -0.07835 ...[1] TRUE'data.frame': 2101 obs. of 8 variables:
$ GeneID : chr "ENSMUSG00000000001" "ENSMUSG00000000028" "ENSMUSG0000000"..
$ Estimate : num -0.037 -0.104 -0.0223 -0.0923 -0.0305 ...
$ Std. Error: num 0.0439 0.0737 0.0616 0.0822 0.0465 ...
$ t value : num -0.843 -1.411 -0.362 -1.123 -0.655 ...
$ pvalue : num 0.447 0.231 0.736 0.324 0.548 ...
$ FDR : num 0.627 0.432 0.836 0.524 0.707 ...
$ gene : Named chr "Gnai3" "Cdc45" "Xpo6" "Rtca" ...
..- attr(*, "names")= chr [1:2101] "ENSMUSG00000000001" "ENSMUSG00000000028"..
$ entrez : Named chr "14679" "12544" "74204" "66368" ...
..- attr(*, "names")= chr [1:2101] "ENSMUSG00000000001" "ENSMUSG00000000028"..Check distribution of significant changes
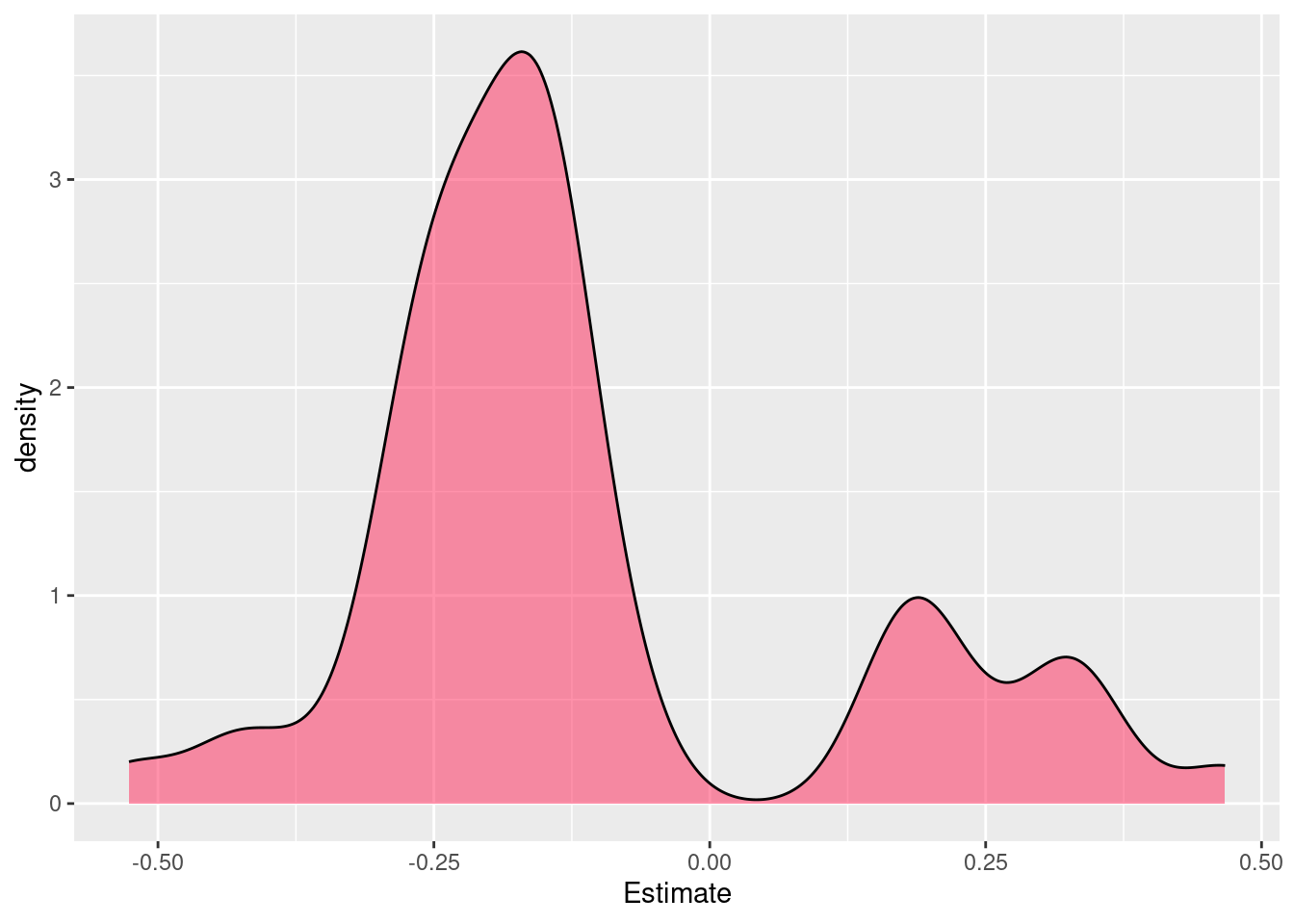
GeneID Estimate Std. Error t value pvalue
1 ENSMUSG00000000838 -0.44214121 0.037977667 -11.642137 3.111397e-04
2 ENSMUSG00000003814 -0.28437380 0.033807346 -8.411598 1.093418e-03
3 ENSMUSG00000004460 -0.20889555 0.025400609 -8.224037 1.191712e-03
4 ENSMUSG00000005481 -0.15616153 0.018183678 -8.588006 1.009976e-03
5 ENSMUSG00000005981 -0.21863277 0.024293287 -8.999719 8.439336e-04
6 ENSMUSG00000011179 -0.27175580 0.029024510 -9.362977 7.247196e-04
7 ENSMUSG00000014959 -0.11718934 0.011520766 -10.172010 5.260751e-04
8 ENSMUSG00000015002 -0.26406954 0.016418955 -16.083212 8.740760e-05
9 ENSMUSG00000018377 -0.22612401 0.026543394 -8.519031 1.041629e-03
10 ENSMUSG00000020015 -0.25354481 0.011015350 -23.017409 2.110962e-05
11 ENSMUSG00000020368 -0.27558954 0.013587272 -20.282920 3.488383e-05
12 ENSMUSG00000020869 -0.11843468 0.014121271 -8.386970 1.105737e-03
13 ENSMUSG00000021096 -0.13170771 0.008637619 -15.248150 1.078777e-04
14 ENSMUSG00000021701 -0.23346878 0.020719151 -11.268260 3.533936e-04
15 ENSMUSG00000021831 -0.28935344 0.030514847 -9.482382 6.901577e-04
16 ENSMUSG00000021868 -0.17895385 0.022264889 -8.037491 1.300557e-03
17 ENSMUSG00000025133 -0.15863591 0.017533947 -9.047359 8.269808e-04
18 ENSMUSG00000025144 -0.26253251 0.030686584 -8.555286 1.024841e-03
19 ENSMUSG00000026547 -0.17539664 0.021854728 -8.025570 1.307923e-03
20 ENSMUSG00000029290 -0.18132195 0.021782661 -8.324141 1.137950e-03
21 ENSMUSG00000029407 -0.14852216 0.018473729 -8.039641 1.299234e-03
22 ENSMUSG00000030082 -0.17583482 0.019909796 -8.831573 9.073210e-04
23 ENSMUSG00000032458 -0.40615507 0.040248301 -10.091235 5.425802e-04
24 ENSMUSG00000032518 -0.25890467 0.027892550 -9.282216 7.493182e-04
25 ENSMUSG00000033161 -0.23255782 0.009616724 -24.182645 1.734608e-05
26 ENSMUSG00000033184 -0.27578724 0.027060233 -10.191606 5.221652e-04
27 ENSMUSG00000034593 -0.15590310 0.014912768 -10.454337 4.730728e-04
28 ENSMUSG00000035365 -0.08024076 0.008367290 -9.589815 6.607934e-04
29 ENSMUSG00000035851 -0.11398884 0.012212464 -9.333812 7.334856e-04
30 ENSMUSG00000036737 -0.13064992 0.009451063 -13.823833 1.587217e-04
31 ENSMUSG00000041769 -0.16871494 0.017534358 -9.621963 6.523092e-04
32 ENSMUSG00000042305 -0.52626506 0.039777215 -13.230314 1.885855e-04
33 ENSMUSG00000043962 -0.13155424 0.016046953 -8.198082 1.206162e-03
34 ENSMUSG00000047714 -0.15147312 0.017503917 -8.653670 9.809424e-04
35 ENSMUSG00000050017 -0.15680392 0.016141444 -9.714367 6.286646e-04
36 ENSMUSG00000053510 -0.06223988 0.005221488 -11.919953 2.837565e-04
37 ENSMUSG00000058600 -0.20317381 0.022718734 -8.943008 8.646804e-04
38 ENSMUSG00000071650 -0.21175025 0.022351389 -9.473696 6.926020e-04
39 ENSMUSG00000072889 -0.13700644 0.008020255 -17.082555 6.887835e-05
40 ENSMUSG00000073563 -0.14600464 0.015254490 -9.571256 6.657535e-04
41 ENSMUSG00000079111 -0.21277051 0.013553709 -15.698324 9.617892e-05
42 ENSMUSG00000079614 -0.22064058 0.022385364 -9.856466 5.943423e-04
43 ENSMUSG00000114797 -0.34371644 0.032462591 -10.588078 4.502857e-04
FDR gene entrez
1 0.04085654 Fmr1 14265
2 0.04879249 Calr 12317
3 0.04968915 Dnajb11 67838
4 0.04863250 Ddx39a 68278
5 0.04780772 Trap1 68015
6 0.04498050 Odc1 18263
7 0.04498050 Gorasp2 70231
8 0.02543159 Efr3a 76740
9 0.04863250 Vezf1 22344
10 0.02217565 Cdk17 237459
11 0.02443031 Canx 12330
12 0.04879249 Lrrc59 98238
13 0.02543159 Ppm1a 19042
14 0.04367529 Plk2 20620
15 0.04498050 Ero1a 50527
16 0.04996267 Ppif 105675
17 0.04780772 Ints4 101861
18 0.04863250 Cenpx 20892
19 0.04996267 Tagln2 21346
20 0.04879249 Zfp326 54367
21 0.04996267 Uso1 56041
22 0.04810163 Sec61a1 53421
23 0.04498050 Copb2 50797
24 0.04498050 Rpsa 16785
25 0.02217565 Atp1a1 11928
26 0.04498050 Tmed7 66676
27 0.04498050 Myo5a 17918
28 0.04498050 Parpbp 75317
29 0.04498050 Ythdc1 231386
30 0.03031585 Oxsr1 108737
31 0.04498050 Ppp2r2d 52432
32 0.03301819 Tmem183a 57439
33 0.04968915 Thrap3 230753
34 0.04863250 Ppp1r2 66849
35 0.04498050 Pitpnb 56305
36 0.04085654 Nrd1 230598
37 0.04780772 Rpl30 19946
38 0.04498050 Ganab 14376
39 0.02543159 Nfxl1 100978
40 0.04498050 Csnk1g3 70425
41 0.02543159 Kdelr2 66913
42 0.04498050 Seh1l 72124
43 0.04498050 Nupl1 71844Using the RNA-decay paper - DiffRAC
Initializing DiffRAC framework...
Estimating size factors and dispersions...
Optimizing the bias constant...
0.381966011250105 : 443084.805542457
0.618033988749895 : 451660.927947527
0.76393202250021 : 447436.811077347
0.606281593377457 : 451755.93420737
0.581622715887764 : 451809.600721294
0.587245246809309 : 451815.294125421
0.587578588893278 : 451815.288818795
0.586911904725339 : 451815.259613898
0.587245246809309 : 451815.294125421
The bias constant is 0.587245246809309
Re-estimating dispersion...
Fitting model parameters... (Intercept) UPF3AKD_Rep1_213 UPF3A_OE_Rep1_214
Control_Rep1_212:Denominator 1 0.0000000 0.0000000
UPF3AKD_Rep1_213:Denominator 1 1.0000000 0.0000000
UPF3A_OE_Rep1_214:Denominator 1 0.0000000 1.0000000
UPF3BKD_Rep1_215:Denominator 1 0.0000000 0.0000000
UPF3dKD_Rep1_216:Denominator 1 0.0000000 0.0000000
UPF3AOE_BKD_Rep1_217:Denominator 1 0.0000000 0.0000000
Control_Rep2_218:Denominator 1 0.0000000 0.0000000
UPF3AKD_Rep2_219:Denominator 1 0.0000000 0.0000000
UPF3A_OE_Rep2_220:Denominator 1 0.0000000 0.0000000
UPF3BKD_Rep2_221:Denominator 1 0.0000000 0.0000000
UPF3dKD_Rep2_222:Denominator 1 0.0000000 0.0000000
UPF3AOE_BKD_Rep2_223:Denominator 1 0.0000000 0.0000000
Control_Rep3_224:Denominator 1 0.0000000 0.0000000
UPF3AKD_Rep3_225:Denominator 1 0.0000000 0.0000000
UPF3A_OE_Rep3_226:Denominator 1 0.0000000 0.0000000
UPF3BKD_Rep3_227:Denominator 1 0.0000000 0.0000000
UPF3dKD_Rep3_228:Denominator 1 0.0000000 0.0000000
UPF3AOE_BKD_Rep3_229:Denominator 1 0.0000000 0.0000000
Control_Rep1_212:Numerator 1 0.0000000 0.0000000
UPF3AKD_Rep1_213:Numerator 1 0.5872452 0.0000000
UPF3A_OE_Rep1_214:Numerator 1 0.0000000 0.5872452
UPF3BKD_Rep1_215:Numerator 1 0.0000000 0.0000000
UPF3dKD_Rep1_216:Numerator 1 0.0000000 0.0000000
UPF3AOE_BKD_Rep1_217:Numerator 1 0.0000000 0.0000000
Control_Rep2_218:Numerator 1 0.0000000 0.0000000
UPF3AKD_Rep2_219:Numerator 1 0.0000000 0.0000000
UPF3A_OE_Rep2_220:Numerator 1 0.0000000 0.0000000
UPF3BKD_Rep2_221:Numerator 1 0.0000000 0.0000000
UPF3dKD_Rep2_222:Numerator 1 0.0000000 0.0000000
UPF3AOE_BKD_Rep2_223:Numerator 1 0.0000000 0.0000000
Control_Rep3_224:Numerator 1 0.0000000 0.0000000
UPF3AKD_Rep3_225:Numerator 1 0.0000000 0.0000000
UPF3A_OE_Rep3_226:Numerator 1 0.0000000 0.0000000
UPF3BKD_Rep3_227:Numerator 1 0.0000000 0.0000000
UPF3dKD_Rep3_228:Numerator 1 0.0000000 0.0000000
UPF3AOE_BKD_Rep3_229:Numerator 1 0.0000000 0.0000000
UPF3BKD_Rep1_215 UPF3dKD_Rep1_216
Control_Rep1_212:Denominator 0.0000000 0.0000000
UPF3AKD_Rep1_213:Denominator 0.0000000 0.0000000
UPF3A_OE_Rep1_214:Denominator 0.0000000 0.0000000
UPF3BKD_Rep1_215:Denominator 1.0000000 0.0000000
UPF3dKD_Rep1_216:Denominator 0.0000000 1.0000000
UPF3AOE_BKD_Rep1_217:Denominator 0.0000000 0.0000000
Control_Rep2_218:Denominator 0.0000000 0.0000000
UPF3AKD_Rep2_219:Denominator 0.0000000 0.0000000
UPF3A_OE_Rep2_220:Denominator 0.0000000 0.0000000
UPF3BKD_Rep2_221:Denominator 0.0000000 0.0000000
UPF3dKD_Rep2_222:Denominator 0.0000000 0.0000000
UPF3AOE_BKD_Rep2_223:Denominator 0.0000000 0.0000000
Control_Rep3_224:Denominator 0.0000000 0.0000000
UPF3AKD_Rep3_225:Denominator 0.0000000 0.0000000
UPF3A_OE_Rep3_226:Denominator 0.0000000 0.0000000
UPF3BKD_Rep3_227:Denominator 0.0000000 0.0000000
UPF3dKD_Rep3_228:Denominator 0.0000000 0.0000000
UPF3AOE_BKD_Rep3_229:Denominator 0.0000000 0.0000000
Control_Rep1_212:Numerator 0.0000000 0.0000000
UPF3AKD_Rep1_213:Numerator 0.0000000 0.0000000
UPF3A_OE_Rep1_214:Numerator 0.0000000 0.0000000
UPF3BKD_Rep1_215:Numerator 0.5872452 0.0000000
UPF3dKD_Rep1_216:Numerator 0.0000000 0.5872452
UPF3AOE_BKD_Rep1_217:Numerator 0.0000000 0.0000000
Control_Rep2_218:Numerator 0.0000000 0.0000000
UPF3AKD_Rep2_219:Numerator 0.0000000 0.0000000
UPF3A_OE_Rep2_220:Numerator 0.0000000 0.0000000
UPF3BKD_Rep2_221:Numerator 0.0000000 0.0000000
UPF3dKD_Rep2_222:Numerator 0.0000000 0.0000000
UPF3AOE_BKD_Rep2_223:Numerator 0.0000000 0.0000000
Control_Rep3_224:Numerator 0.0000000 0.0000000
UPF3AKD_Rep3_225:Numerator 0.0000000 0.0000000
UPF3A_OE_Rep3_226:Numerator 0.0000000 0.0000000
UPF3BKD_Rep3_227:Numerator 0.0000000 0.0000000
UPF3dKD_Rep3_228:Numerator 0.0000000 0.0000000
UPF3AOE_BKD_Rep3_229:Numerator 0.0000000 0.0000000
UPF3AOE_BKD_Rep1_217 Control_Rep2_218
Control_Rep1_212:Denominator 0.0000000 0.0000000
UPF3AKD_Rep1_213:Denominator 0.0000000 0.0000000
UPF3A_OE_Rep1_214:Denominator 0.0000000 0.0000000
UPF3BKD_Rep1_215:Denominator 0.0000000 0.0000000
UPF3dKD_Rep1_216:Denominator 0.0000000 0.0000000
UPF3AOE_BKD_Rep1_217:Denominator 1.0000000 0.0000000
Control_Rep2_218:Denominator 0.0000000 1.0000000
UPF3AKD_Rep2_219:Denominator 0.0000000 0.0000000
UPF3A_OE_Rep2_220:Denominator 0.0000000 0.0000000
UPF3BKD_Rep2_221:Denominator 0.0000000 0.0000000
UPF3dKD_Rep2_222:Denominator 0.0000000 0.0000000
UPF3AOE_BKD_Rep2_223:Denominator 0.0000000 0.0000000
Control_Rep3_224:Denominator 0.0000000 0.0000000
UPF3AKD_Rep3_225:Denominator 0.0000000 0.0000000
UPF3A_OE_Rep3_226:Denominator 0.0000000 0.0000000
UPF3BKD_Rep3_227:Denominator 0.0000000 0.0000000
UPF3dKD_Rep3_228:Denominator 0.0000000 0.0000000
UPF3AOE_BKD_Rep3_229:Denominator 0.0000000 0.0000000
Control_Rep1_212:Numerator 0.0000000 0.0000000
UPF3AKD_Rep1_213:Numerator 0.0000000 0.0000000
UPF3A_OE_Rep1_214:Numerator 0.0000000 0.0000000
UPF3BKD_Rep1_215:Numerator 0.0000000 0.0000000
UPF3dKD_Rep1_216:Numerator 0.0000000 0.0000000
UPF3AOE_BKD_Rep1_217:Numerator 0.5872452 0.0000000
Control_Rep2_218:Numerator 0.0000000 0.5872452
UPF3AKD_Rep2_219:Numerator 0.0000000 0.0000000
UPF3A_OE_Rep2_220:Numerator 0.0000000 0.0000000
UPF3BKD_Rep2_221:Numerator 0.0000000 0.0000000
UPF3dKD_Rep2_222:Numerator 0.0000000 0.0000000
UPF3AOE_BKD_Rep2_223:Numerator 0.0000000 0.0000000
Control_Rep3_224:Numerator 0.0000000 0.0000000
UPF3AKD_Rep3_225:Numerator 0.0000000 0.0000000
UPF3A_OE_Rep3_226:Numerator 0.0000000 0.0000000
UPF3BKD_Rep3_227:Numerator 0.0000000 0.0000000
UPF3dKD_Rep3_228:Numerator 0.0000000 0.0000000
UPF3AOE_BKD_Rep3_229:Numerator 0.0000000 0.0000000
UPF3AKD_Rep2_219 UPF3A_OE_Rep2_220
Control_Rep1_212:Denominator 0.0000000 0.0000000
UPF3AKD_Rep1_213:Denominator 0.0000000 0.0000000
UPF3A_OE_Rep1_214:Denominator 0.0000000 0.0000000
UPF3BKD_Rep1_215:Denominator 0.0000000 0.0000000
UPF3dKD_Rep1_216:Denominator 0.0000000 0.0000000
UPF3AOE_BKD_Rep1_217:Denominator 0.0000000 0.0000000
Control_Rep2_218:Denominator 0.0000000 0.0000000
UPF3AKD_Rep2_219:Denominator 1.0000000 0.0000000
UPF3A_OE_Rep2_220:Denominator 0.0000000 1.0000000
UPF3BKD_Rep2_221:Denominator 0.0000000 0.0000000
UPF3dKD_Rep2_222:Denominator 0.0000000 0.0000000
UPF3AOE_BKD_Rep2_223:Denominator 0.0000000 0.0000000
Control_Rep3_224:Denominator 0.0000000 0.0000000
UPF3AKD_Rep3_225:Denominator 0.0000000 0.0000000
UPF3A_OE_Rep3_226:Denominator 0.0000000 0.0000000
UPF3BKD_Rep3_227:Denominator 0.0000000 0.0000000
UPF3dKD_Rep3_228:Denominator 0.0000000 0.0000000
UPF3AOE_BKD_Rep3_229:Denominator 0.0000000 0.0000000
Control_Rep1_212:Numerator 0.0000000 0.0000000
UPF3AKD_Rep1_213:Numerator 0.0000000 0.0000000
UPF3A_OE_Rep1_214:Numerator 0.0000000 0.0000000
UPF3BKD_Rep1_215:Numerator 0.0000000 0.0000000
UPF3dKD_Rep1_216:Numerator 0.0000000 0.0000000
UPF3AOE_BKD_Rep1_217:Numerator 0.0000000 0.0000000
Control_Rep2_218:Numerator 0.0000000 0.0000000
UPF3AKD_Rep2_219:Numerator 0.5872452 0.0000000
UPF3A_OE_Rep2_220:Numerator 0.0000000 0.5872452
UPF3BKD_Rep2_221:Numerator 0.0000000 0.0000000
UPF3dKD_Rep2_222:Numerator 0.0000000 0.0000000
UPF3AOE_BKD_Rep2_223:Numerator 0.0000000 0.0000000
Control_Rep3_224:Numerator 0.0000000 0.0000000
UPF3AKD_Rep3_225:Numerator 0.0000000 0.0000000
UPF3A_OE_Rep3_226:Numerator 0.0000000 0.0000000
UPF3BKD_Rep3_227:Numerator 0.0000000 0.0000000
UPF3dKD_Rep3_228:Numerator 0.0000000 0.0000000
UPF3AOE_BKD_Rep3_229:Numerator 0.0000000 0.0000000
UPF3BKD_Rep2_221 UPF3dKD_Rep2_222
Control_Rep1_212:Denominator 0.0000000 0.0000000
UPF3AKD_Rep1_213:Denominator 0.0000000 0.0000000
UPF3A_OE_Rep1_214:Denominator 0.0000000 0.0000000
UPF3BKD_Rep1_215:Denominator 0.0000000 0.0000000
UPF3dKD_Rep1_216:Denominator 0.0000000 0.0000000
UPF3AOE_BKD_Rep1_217:Denominator 0.0000000 0.0000000
Control_Rep2_218:Denominator 0.0000000 0.0000000
UPF3AKD_Rep2_219:Denominator 0.0000000 0.0000000
UPF3A_OE_Rep2_220:Denominator 0.0000000 0.0000000
UPF3BKD_Rep2_221:Denominator 1.0000000 0.0000000
UPF3dKD_Rep2_222:Denominator 0.0000000 1.0000000
UPF3AOE_BKD_Rep2_223:Denominator 0.0000000 0.0000000
Control_Rep3_224:Denominator 0.0000000 0.0000000
UPF3AKD_Rep3_225:Denominator 0.0000000 0.0000000
UPF3A_OE_Rep3_226:Denominator 0.0000000 0.0000000
UPF3BKD_Rep3_227:Denominator 0.0000000 0.0000000
UPF3dKD_Rep3_228:Denominator 0.0000000 0.0000000
UPF3AOE_BKD_Rep3_229:Denominator 0.0000000 0.0000000
Control_Rep1_212:Numerator 0.0000000 0.0000000
UPF3AKD_Rep1_213:Numerator 0.0000000 0.0000000
UPF3A_OE_Rep1_214:Numerator 0.0000000 0.0000000
UPF3BKD_Rep1_215:Numerator 0.0000000 0.0000000
UPF3dKD_Rep1_216:Numerator 0.0000000 0.0000000
UPF3AOE_BKD_Rep1_217:Numerator 0.0000000 0.0000000
Control_Rep2_218:Numerator 0.0000000 0.0000000
UPF3AKD_Rep2_219:Numerator 0.0000000 0.0000000
UPF3A_OE_Rep2_220:Numerator 0.0000000 0.0000000
UPF3BKD_Rep2_221:Numerator 0.5872452 0.0000000
UPF3dKD_Rep2_222:Numerator 0.0000000 0.5872452
UPF3AOE_BKD_Rep2_223:Numerator 0.0000000 0.0000000
Control_Rep3_224:Numerator 0.0000000 0.0000000
UPF3AKD_Rep3_225:Numerator 0.0000000 0.0000000
UPF3A_OE_Rep3_226:Numerator 0.0000000 0.0000000
UPF3BKD_Rep3_227:Numerator 0.0000000 0.0000000
UPF3dKD_Rep3_228:Numerator 0.0000000 0.0000000
UPF3AOE_BKD_Rep3_229:Numerator 0.0000000 0.0000000
UPF3AOE_BKD_Rep2_223 Control_Rep3_224
Control_Rep1_212:Denominator 0.0000000 0.0000000
UPF3AKD_Rep1_213:Denominator 0.0000000 0.0000000
UPF3A_OE_Rep1_214:Denominator 0.0000000 0.0000000
UPF3BKD_Rep1_215:Denominator 0.0000000 0.0000000
UPF3dKD_Rep1_216:Denominator 0.0000000 0.0000000
UPF3AOE_BKD_Rep1_217:Denominator 0.0000000 0.0000000
Control_Rep2_218:Denominator 0.0000000 0.0000000
UPF3AKD_Rep2_219:Denominator 0.0000000 0.0000000
UPF3A_OE_Rep2_220:Denominator 0.0000000 0.0000000
UPF3BKD_Rep2_221:Denominator 0.0000000 0.0000000
UPF3dKD_Rep2_222:Denominator 0.0000000 0.0000000
UPF3AOE_BKD_Rep2_223:Denominator 1.0000000 0.0000000
Control_Rep3_224:Denominator 0.0000000 1.0000000
UPF3AKD_Rep3_225:Denominator 0.0000000 0.0000000
UPF3A_OE_Rep3_226:Denominator 0.0000000 0.0000000
UPF3BKD_Rep3_227:Denominator 0.0000000 0.0000000
UPF3dKD_Rep3_228:Denominator 0.0000000 0.0000000
UPF3AOE_BKD_Rep3_229:Denominator 0.0000000 0.0000000
Control_Rep1_212:Numerator 0.0000000 0.0000000
UPF3AKD_Rep1_213:Numerator 0.0000000 0.0000000
UPF3A_OE_Rep1_214:Numerator 0.0000000 0.0000000
UPF3BKD_Rep1_215:Numerator 0.0000000 0.0000000
UPF3dKD_Rep1_216:Numerator 0.0000000 0.0000000
UPF3AOE_BKD_Rep1_217:Numerator 0.0000000 0.0000000
Control_Rep2_218:Numerator 0.0000000 0.0000000
UPF3AKD_Rep2_219:Numerator 0.0000000 0.0000000
UPF3A_OE_Rep2_220:Numerator 0.0000000 0.0000000
UPF3BKD_Rep2_221:Numerator 0.0000000 0.0000000
UPF3dKD_Rep2_222:Numerator 0.0000000 0.0000000
UPF3AOE_BKD_Rep2_223:Numerator 0.5872452 0.0000000
Control_Rep3_224:Numerator 0.0000000 0.5872452
UPF3AKD_Rep3_225:Numerator 0.0000000 0.0000000
UPF3A_OE_Rep3_226:Numerator 0.0000000 0.0000000
UPF3BKD_Rep3_227:Numerator 0.0000000 0.0000000
UPF3dKD_Rep3_228:Numerator 0.0000000 0.0000000
UPF3AOE_BKD_Rep3_229:Numerator 0.0000000 0.0000000
UPF3AKD_Rep3_225 UPF3A_OE_Rep3_226
Control_Rep1_212:Denominator 0.0000000 0.0000000
UPF3AKD_Rep1_213:Denominator 0.0000000 0.0000000
UPF3A_OE_Rep1_214:Denominator 0.0000000 0.0000000
UPF3BKD_Rep1_215:Denominator 0.0000000 0.0000000
UPF3dKD_Rep1_216:Denominator 0.0000000 0.0000000
UPF3AOE_BKD_Rep1_217:Denominator 0.0000000 0.0000000
Control_Rep2_218:Denominator 0.0000000 0.0000000
UPF3AKD_Rep2_219:Denominator 0.0000000 0.0000000
UPF3A_OE_Rep2_220:Denominator 0.0000000 0.0000000
UPF3BKD_Rep2_221:Denominator 0.0000000 0.0000000
UPF3dKD_Rep2_222:Denominator 0.0000000 0.0000000
UPF3AOE_BKD_Rep2_223:Denominator 0.0000000 0.0000000
Control_Rep3_224:Denominator 0.0000000 0.0000000
UPF3AKD_Rep3_225:Denominator 1.0000000 0.0000000
UPF3A_OE_Rep3_226:Denominator 0.0000000 1.0000000
UPF3BKD_Rep3_227:Denominator 0.0000000 0.0000000
UPF3dKD_Rep3_228:Denominator 0.0000000 0.0000000
UPF3AOE_BKD_Rep3_229:Denominator 0.0000000 0.0000000
Control_Rep1_212:Numerator 0.0000000 0.0000000
UPF3AKD_Rep1_213:Numerator 0.0000000 0.0000000
UPF3A_OE_Rep1_214:Numerator 0.0000000 0.0000000
UPF3BKD_Rep1_215:Numerator 0.0000000 0.0000000
UPF3dKD_Rep1_216:Numerator 0.0000000 0.0000000
UPF3AOE_BKD_Rep1_217:Numerator 0.0000000 0.0000000
Control_Rep2_218:Numerator 0.0000000 0.0000000
UPF3AKD_Rep2_219:Numerator 0.0000000 0.0000000
UPF3A_OE_Rep2_220:Numerator 0.0000000 0.0000000
UPF3BKD_Rep2_221:Numerator 0.0000000 0.0000000
UPF3dKD_Rep2_222:Numerator 0.0000000 0.0000000
UPF3AOE_BKD_Rep2_223:Numerator 0.0000000 0.0000000
Control_Rep3_224:Numerator 0.0000000 0.0000000
UPF3AKD_Rep3_225:Numerator 0.5872452 0.0000000
UPF3A_OE_Rep3_226:Numerator 0.0000000 0.5872452
UPF3BKD_Rep3_227:Numerator 0.0000000 0.0000000
UPF3dKD_Rep3_228:Numerator 0.0000000 0.0000000
UPF3AOE_BKD_Rep3_229:Numerator 0.0000000 0.0000000
UPF3BKD_Rep3_227 UPF3dKD_Rep3_228
Control_Rep1_212:Denominator 0.0000000 0.0000000
UPF3AKD_Rep1_213:Denominator 0.0000000 0.0000000
UPF3A_OE_Rep1_214:Denominator 0.0000000 0.0000000
UPF3BKD_Rep1_215:Denominator 0.0000000 0.0000000
UPF3dKD_Rep1_216:Denominator 0.0000000 0.0000000
UPF3AOE_BKD_Rep1_217:Denominator 0.0000000 0.0000000
Control_Rep2_218:Denominator 0.0000000 0.0000000
UPF3AKD_Rep2_219:Denominator 0.0000000 0.0000000
UPF3A_OE_Rep2_220:Denominator 0.0000000 0.0000000
UPF3BKD_Rep2_221:Denominator 0.0000000 0.0000000
UPF3dKD_Rep2_222:Denominator 0.0000000 0.0000000
UPF3AOE_BKD_Rep2_223:Denominator 0.0000000 0.0000000
Control_Rep3_224:Denominator 0.0000000 0.0000000
UPF3AKD_Rep3_225:Denominator 0.0000000 0.0000000
UPF3A_OE_Rep3_226:Denominator 0.0000000 0.0000000
UPF3BKD_Rep3_227:Denominator 1.0000000 0.0000000
UPF3dKD_Rep3_228:Denominator 0.0000000 1.0000000
UPF3AOE_BKD_Rep3_229:Denominator 0.0000000 0.0000000
Control_Rep1_212:Numerator 0.0000000 0.0000000
UPF3AKD_Rep1_213:Numerator 0.0000000 0.0000000
UPF3A_OE_Rep1_214:Numerator 0.0000000 0.0000000
UPF3BKD_Rep1_215:Numerator 0.0000000 0.0000000
UPF3dKD_Rep1_216:Numerator 0.0000000 0.0000000
UPF3AOE_BKD_Rep1_217:Numerator 0.0000000 0.0000000
Control_Rep2_218:Numerator 0.0000000 0.0000000
UPF3AKD_Rep2_219:Numerator 0.0000000 0.0000000
UPF3A_OE_Rep2_220:Numerator 0.0000000 0.0000000
UPF3BKD_Rep2_221:Numerator 0.0000000 0.0000000
UPF3dKD_Rep2_222:Numerator 0.0000000 0.0000000
UPF3AOE_BKD_Rep2_223:Numerator 0.0000000 0.0000000
Control_Rep3_224:Numerator 0.0000000 0.0000000
UPF3AKD_Rep3_225:Numerator 0.0000000 0.0000000
UPF3A_OE_Rep3_226:Numerator 0.0000000 0.0000000
UPF3BKD_Rep3_227:Numerator 0.5872452 0.0000000
UPF3dKD_Rep3_228:Numerator 0.0000000 0.5872452
UPF3AOE_BKD_Rep3_229:Numerator 0.0000000 0.0000000
UPF3AOE_BKD_Rep3_229 Numerator
Control_Rep1_212:Denominator 0.0000000 0
UPF3AKD_Rep1_213:Denominator 0.0000000 0
UPF3A_OE_Rep1_214:Denominator 0.0000000 0
UPF3BKD_Rep1_215:Denominator 0.0000000 0
UPF3dKD_Rep1_216:Denominator 0.0000000 0
UPF3AOE_BKD_Rep1_217:Denominator 0.0000000 0
Control_Rep2_218:Denominator 0.0000000 0
UPF3AKD_Rep2_219:Denominator 0.0000000 0
UPF3A_OE_Rep2_220:Denominator 0.0000000 0
UPF3BKD_Rep2_221:Denominator 0.0000000 0
UPF3dKD_Rep2_222:Denominator 0.0000000 0
UPF3AOE_BKD_Rep2_223:Denominator 0.0000000 0
Control_Rep3_224:Denominator 0.0000000 0
UPF3AKD_Rep3_225:Denominator 0.0000000 0
UPF3A_OE_Rep3_226:Denominator 0.0000000 0
UPF3BKD_Rep3_227:Denominator 0.0000000 0
UPF3dKD_Rep3_228:Denominator 0.0000000 0
UPF3AOE_BKD_Rep3_229:Denominator 1.0000000 0
Control_Rep1_212:Numerator 0.0000000 1
UPF3AKD_Rep1_213:Numerator 0.0000000 1
UPF3A_OE_Rep1_214:Numerator 0.0000000 1
UPF3BKD_Rep1_215:Numerator 0.0000000 1
UPF3dKD_Rep1_216:Numerator 0.0000000 1
UPF3AOE_BKD_Rep1_217:Numerator 0.0000000 1
Control_Rep2_218:Numerator 0.0000000 1
UPF3AKD_Rep2_219:Numerator 0.0000000 1
UPF3A_OE_Rep2_220:Numerator 0.0000000 1
UPF3BKD_Rep2_221:Numerator 0.0000000 1
UPF3dKD_Rep2_222:Numerator 0.0000000 1
UPF3AOE_BKD_Rep2_223:Numerator 0.0000000 1
Control_Rep3_224:Numerator 0.0000000 1
UPF3AKD_Rep3_225:Numerator 0.0000000 1
UPF3A_OE_Rep3_226:Numerator 0.0000000 1
UPF3BKD_Rep3_227:Numerator 0.0000000 1
UPF3dKD_Rep3_228:Numerator 0.0000000 1
UPF3AOE_BKD_Rep3_229:Numerator 0.5872452 1
GroupUPF3A_KD:Ratio GroupUPF3A_OE:Ratio
Control_Rep1_212:Denominator 0 0
UPF3AKD_Rep1_213:Denominator 0 0
UPF3A_OE_Rep1_214:Denominator 0 0
UPF3BKD_Rep1_215:Denominator 0 0
UPF3dKD_Rep1_216:Denominator 0 0
UPF3AOE_BKD_Rep1_217:Denominator 0 0
Control_Rep2_218:Denominator 0 0
UPF3AKD_Rep2_219:Denominator 0 0
UPF3A_OE_Rep2_220:Denominator 0 0
UPF3BKD_Rep2_221:Denominator 0 0
UPF3dKD_Rep2_222:Denominator 0 0
UPF3AOE_BKD_Rep2_223:Denominator 0 0
Control_Rep3_224:Denominator 0 0
UPF3AKD_Rep3_225:Denominator 0 0
UPF3A_OE_Rep3_226:Denominator 0 0
UPF3BKD_Rep3_227:Denominator 0 0
UPF3dKD_Rep3_228:Denominator 0 0
UPF3AOE_BKD_Rep3_229:Denominator 0 0
Control_Rep1_212:Numerator 0 0
UPF3AKD_Rep1_213:Numerator 1 0
UPF3A_OE_Rep1_214:Numerator 0 1
UPF3BKD_Rep1_215:Numerator 0 0
UPF3dKD_Rep1_216:Numerator 0 0
UPF3AOE_BKD_Rep1_217:Numerator 0 0
Control_Rep2_218:Numerator 0 0
UPF3AKD_Rep2_219:Numerator 1 0
UPF3A_OE_Rep2_220:Numerator 0 1
UPF3BKD_Rep2_221:Numerator 0 0
UPF3dKD_Rep2_222:Numerator 0 0
UPF3AOE_BKD_Rep2_223:Numerator 0 0
Control_Rep3_224:Numerator 0 0
UPF3AKD_Rep3_225:Numerator 1 0
UPF3A_OE_Rep3_226:Numerator 0 1
UPF3BKD_Rep3_227:Numerator 0 0
UPF3dKD_Rep3_228:Numerator 0 0
UPF3AOE_BKD_Rep3_229:Numerator 0 0
GroupUPF3B_KD:Ratio
Control_Rep1_212:Denominator 0
UPF3AKD_Rep1_213:Denominator 0
UPF3A_OE_Rep1_214:Denominator 0
UPF3BKD_Rep1_215:Denominator 0
UPF3dKD_Rep1_216:Denominator 0
UPF3AOE_BKD_Rep1_217:Denominator 0
Control_Rep2_218:Denominator 0
UPF3AKD_Rep2_219:Denominator 0
UPF3A_OE_Rep2_220:Denominator 0
UPF3BKD_Rep2_221:Denominator 0
UPF3dKD_Rep2_222:Denominator 0
UPF3AOE_BKD_Rep2_223:Denominator 0
Control_Rep3_224:Denominator 0
UPF3AKD_Rep3_225:Denominator 0
UPF3A_OE_Rep3_226:Denominator 0
UPF3BKD_Rep3_227:Denominator 0
UPF3dKD_Rep3_228:Denominator 0
UPF3AOE_BKD_Rep3_229:Denominator 0
Control_Rep1_212:Numerator 0
UPF3AKD_Rep1_213:Numerator 0
UPF3A_OE_Rep1_214:Numerator 0
UPF3BKD_Rep1_215:Numerator 1
UPF3dKD_Rep1_216:Numerator 0
UPF3AOE_BKD_Rep1_217:Numerator 0
Control_Rep2_218:Numerator 0
UPF3AKD_Rep2_219:Numerator 0
UPF3A_OE_Rep2_220:Numerator 0
UPF3BKD_Rep2_221:Numerator 1
UPF3dKD_Rep2_222:Numerator 0
UPF3AOE_BKD_Rep2_223:Numerator 0
Control_Rep3_224:Numerator 0
UPF3AKD_Rep3_225:Numerator 0
UPF3A_OE_Rep3_226:Numerator 0
UPF3BKD_Rep3_227:Numerator 1
UPF3dKD_Rep3_228:Numerator 0
UPF3AOE_BKD_Rep3_229:Numerator 0
GroupUPF3A_KD_UPF3B_KD:Ratio
Control_Rep1_212:Denominator 0
UPF3AKD_Rep1_213:Denominator 0
UPF3A_OE_Rep1_214:Denominator 0
UPF3BKD_Rep1_215:Denominator 0
UPF3dKD_Rep1_216:Denominator 0
UPF3AOE_BKD_Rep1_217:Denominator 0
Control_Rep2_218:Denominator 0
UPF3AKD_Rep2_219:Denominator 0
UPF3A_OE_Rep2_220:Denominator 0
UPF3BKD_Rep2_221:Denominator 0
UPF3dKD_Rep2_222:Denominator 0
UPF3AOE_BKD_Rep2_223:Denominator 0
Control_Rep3_224:Denominator 0
UPF3AKD_Rep3_225:Denominator 0
UPF3A_OE_Rep3_226:Denominator 0
UPF3BKD_Rep3_227:Denominator 0
UPF3dKD_Rep3_228:Denominator 0
UPF3AOE_BKD_Rep3_229:Denominator 0
Control_Rep1_212:Numerator 0
UPF3AKD_Rep1_213:Numerator 0
UPF3A_OE_Rep1_214:Numerator 0
UPF3BKD_Rep1_215:Numerator 0
UPF3dKD_Rep1_216:Numerator 1
UPF3AOE_BKD_Rep1_217:Numerator 0
Control_Rep2_218:Numerator 0
UPF3AKD_Rep2_219:Numerator 0
UPF3A_OE_Rep2_220:Numerator 0
UPF3BKD_Rep2_221:Numerator 0
UPF3dKD_Rep2_222:Numerator 1
UPF3AOE_BKD_Rep2_223:Numerator 0
Control_Rep3_224:Numerator 0
UPF3AKD_Rep3_225:Numerator 0
UPF3A_OE_Rep3_226:Numerator 0
UPF3BKD_Rep3_227:Numerator 0
UPF3dKD_Rep3_228:Numerator 1
UPF3AOE_BKD_Rep3_229:Numerator 0
GroupUPF3A_OE_UPF3B_KD:Ratio
Control_Rep1_212:Denominator 0
UPF3AKD_Rep1_213:Denominator 0
UPF3A_OE_Rep1_214:Denominator 0
UPF3BKD_Rep1_215:Denominator 0
UPF3dKD_Rep1_216:Denominator 0
UPF3AOE_BKD_Rep1_217:Denominator 0
Control_Rep2_218:Denominator 0
UPF3AKD_Rep2_219:Denominator 0
UPF3A_OE_Rep2_220:Denominator 0
UPF3BKD_Rep2_221:Denominator 0
UPF3dKD_Rep2_222:Denominator 0
UPF3AOE_BKD_Rep2_223:Denominator 0
Control_Rep3_224:Denominator 0
UPF3AKD_Rep3_225:Denominator 0
UPF3A_OE_Rep3_226:Denominator 0
UPF3BKD_Rep3_227:Denominator 0
UPF3dKD_Rep3_228:Denominator 0
UPF3AOE_BKD_Rep3_229:Denominator 0
Control_Rep1_212:Numerator 0
UPF3AKD_Rep1_213:Numerator 0
UPF3A_OE_Rep1_214:Numerator 0
UPF3BKD_Rep1_215:Numerator 0
UPF3dKD_Rep1_216:Numerator 0
UPF3AOE_BKD_Rep1_217:Numerator 1
Control_Rep2_218:Numerator 0
UPF3AKD_Rep2_219:Numerator 0
UPF3A_OE_Rep2_220:Numerator 0
UPF3BKD_Rep2_221:Numerator 0
UPF3dKD_Rep2_222:Numerator 0
UPF3AOE_BKD_Rep2_223:Numerator 1
Control_Rep3_224:Numerator 0
UPF3AKD_Rep3_225:Numerator 0
UPF3A_OE_Rep3_226:Numerator 0
UPF3BKD_Rep3_227:Numerator 0
UPF3dKD_Rep3_228:Numerator 0
UPF3AOE_BKD_Rep3_229:Numerator 1Comparison with DEG results
[1] 136 9[1] 418 31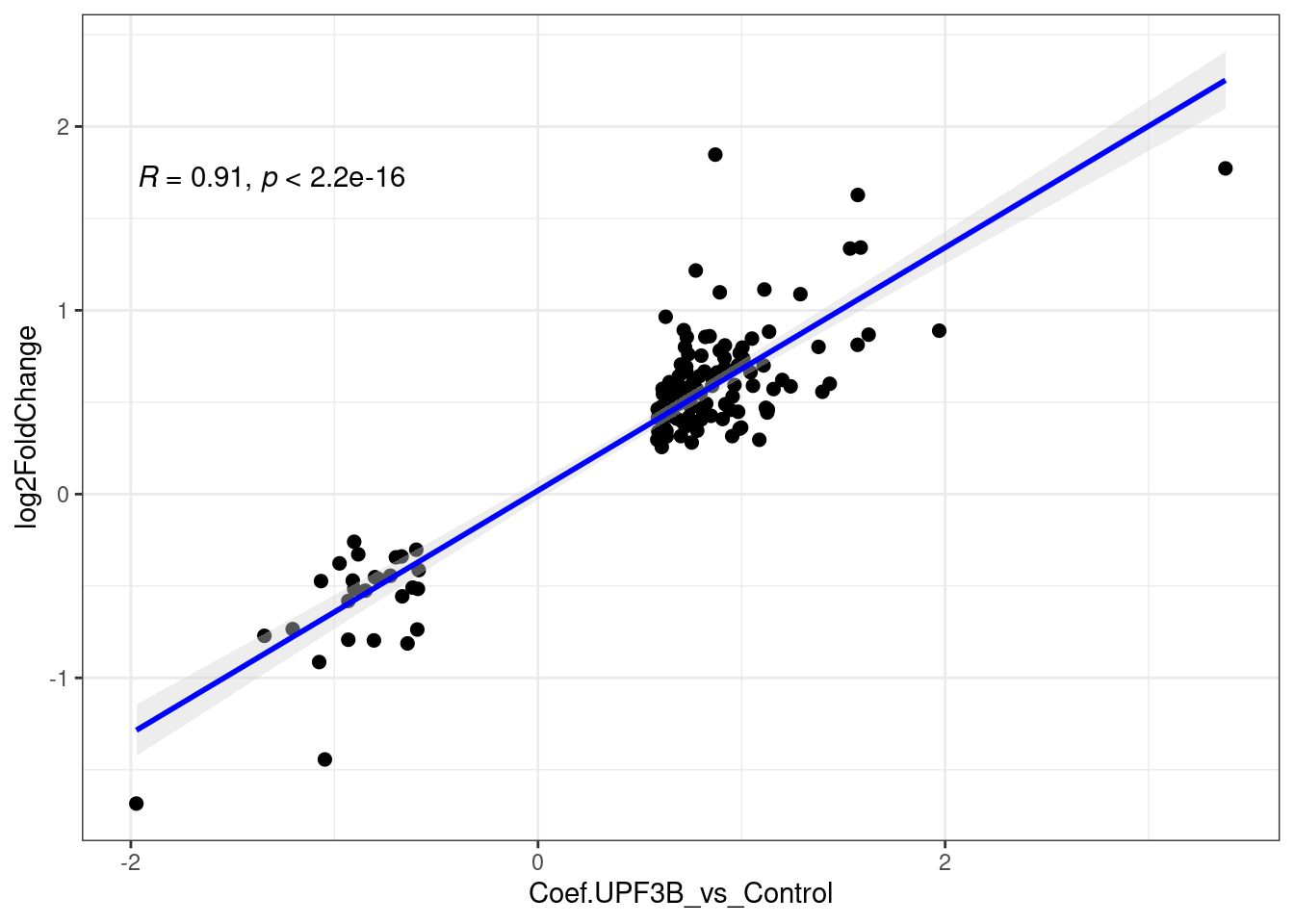
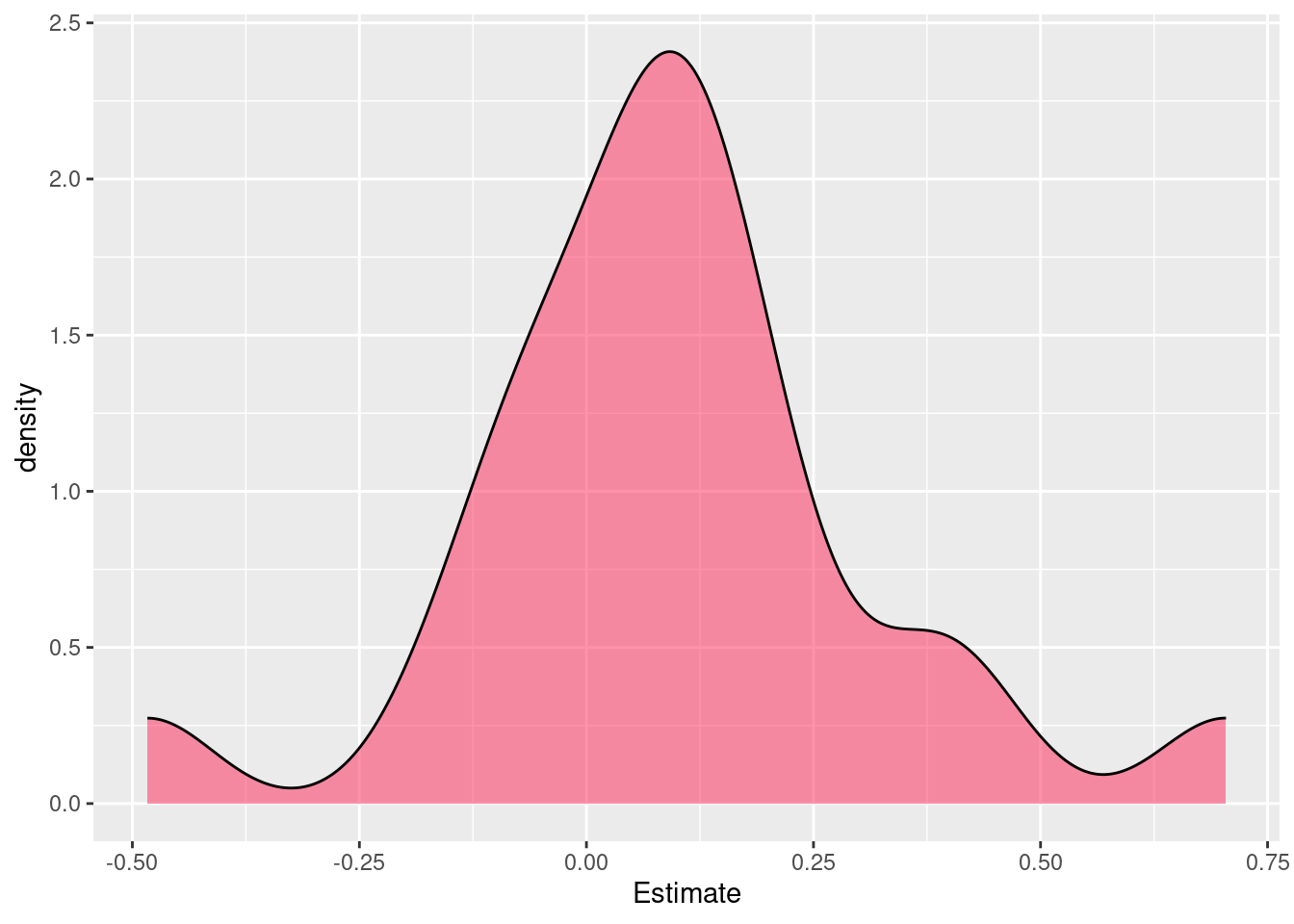
[1] 2101 8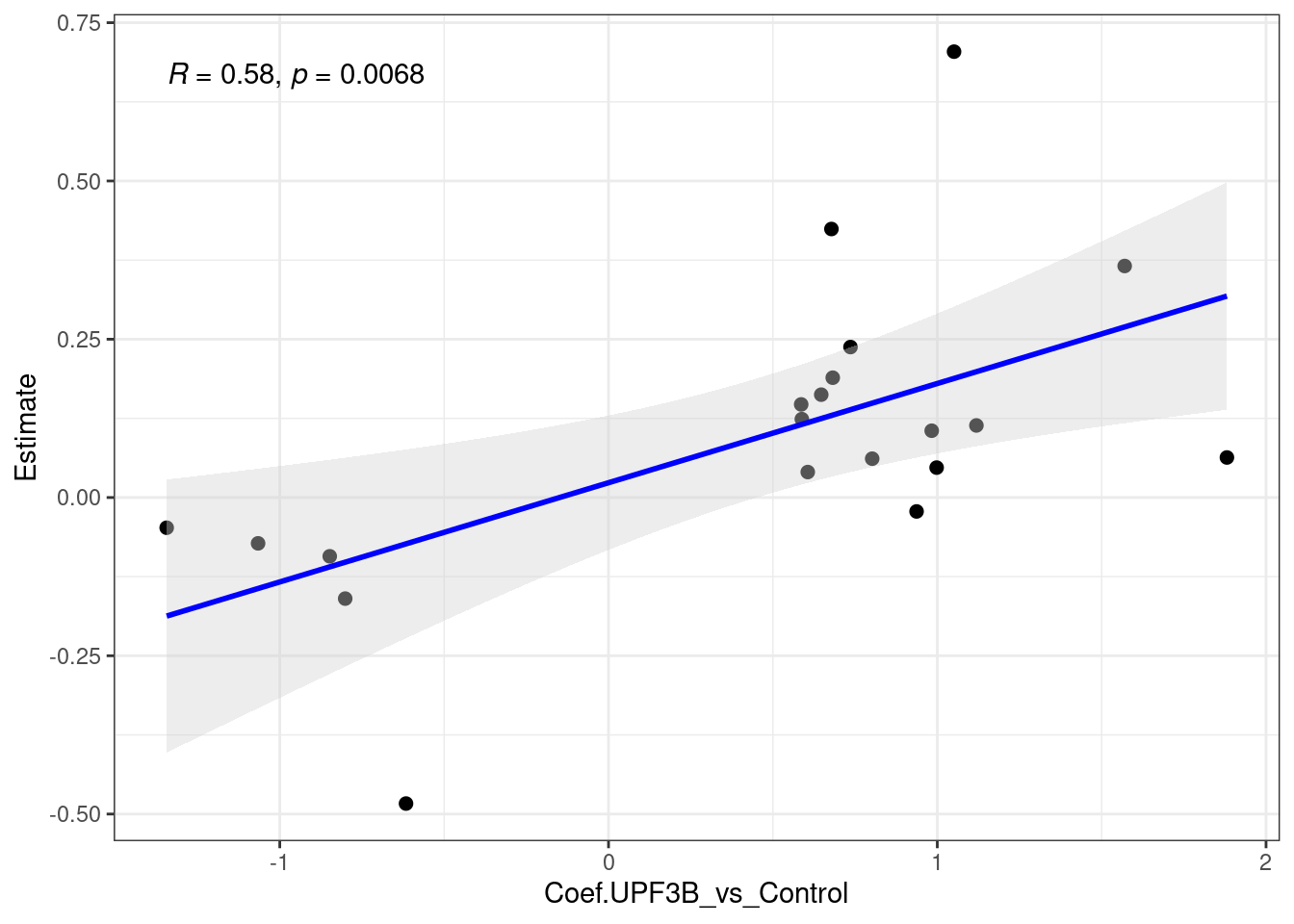
R version 4.2.2 Patched (2022-11-10 r83330)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 22.04.2 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.10.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.10.0
locale:
[1] LC_CTYPE=en_AU.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_AU.UTF-8 LC_COLLATE=en_AU.UTF-8
[5] LC_MONETARY=en_AU.UTF-8 LC_MESSAGES=en_AU.UTF-8
[7] LC_PAPER=en_AU.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_AU.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] grid stats4 tools stats graphics grDevices utils
[8] datasets methods base
other attached packages:
[1] plyr_1.8.8 gplots_3.1.3
[3] coin_1.4-2 survival_3.5-5
[5] IsoformSwitchAnalyzeR_2.01.04 pfamAnalyzeR_0.99.0
[7] sva_3.46.0 genefilter_1.80.3
[9] mgcv_1.8-42 nlme_3.1-162
[11] satuRn_1.6.0 DEXSeq_1.44.0
[13] BiocParallel_1.32.6 ggrepel_0.9.3
[15] pander_0.6.5 msigdbr_7.5.1
[17] cowplot_1.1.1 ngsReports_2.0.3
[19] patchwork_1.1.2 VennDiagram_1.7.3
[21] futile.logger_1.4.3 UpSetR_1.4.0
[23] fgsea_1.24.0 GOplot_1.0.2
[25] RColorBrewer_1.1-3 gridExtra_2.3
[27] ggdendro_0.1.23 AnnotationHub_3.6.0
[29] BiocFileCache_2.6.1 dbplyr_2.3.2
[31] openxlsx_4.2.5.2 ggiraph_0.8.7
[33] wasabi_1.0.1 sleuth_0.30.1
[35] DT_0.27 VennDetail_1.14.0
[37] msigdb_1.6.0 GSEABase_1.60.0
[39] graph_1.76.0 annotate_1.76.0
[41] XML_3.99-0.14 pheatmap_1.0.12
[43] ggvenn_0.1.10 MetBrewer_0.2.0
[45] ggpubr_0.6.0 venn_1.11
[47] viridis_0.6.2 viridisLite_0.4.1
[49] tximeta_1.16.1 tximport_1.26.1
[51] goseq_1.50.0 geneLenDataBase_1.34.0
[53] BiasedUrn_2.0.9 org.Mm.eg.db_3.16.0
[55] EnsDb.Mmusculus.v79_2.99.0 ensembldb_2.22.0
[57] AnnotationFilter_1.22.0 GenomicFeatures_1.50.4
[59] AnnotationDbi_1.60.2 biomaRt_2.54.1
[61] edgeR_3.40.2 limma_3.54.2
[63] DESeq2_1.38.3 SummarizedExperiment_1.28.0
[65] Biobase_2.58.0 MatrixGenerics_1.10.0
[67] matrixStats_0.63.0 GenomicRanges_1.50.2
[69] GenomeInfoDb_1.34.9 IRanges_2.32.0
[71] S4Vectors_0.36.2 BiocGenerics_0.44.0
[73] corrplot_0.92 lubridate_1.9.2
[75] forcats_1.0.0 purrr_1.0.1
[77] readr_2.1.4 tidyverse_2.0.0
[79] stringr_1.5.0 tidyr_1.3.0
[81] scales_1.2.1 data.table_1.14.8
[83] readxl_1.4.2 tibble_3.2.1
[85] magrittr_2.0.3 reshape2_1.4.4
[87] ggplot2_3.4.2 dplyr_1.1.1
[89] eisaR_1.10.0 workflowr_1.7.0
loaded via a namespace (and not attached):
[1] rappdirs_0.3.3 rtracklayer_1.58.0
[3] bit64_4.0.5 knitr_1.42
[5] multcomp_1.4-23 DelayedArray_0.24.0
[7] hwriter_1.3.2.1 KEGGREST_1.38.0
[9] RCurl_1.98-1.12 generics_0.1.3
[11] TH.data_1.1-1 callr_3.7.3
[13] lambda.r_1.2.4 RSQLite_2.3.1
[15] bit_4.0.5 tzdb_0.3.0
[17] xml2_1.3.3 httpuv_1.6.9
[19] xfun_0.38 hms_1.1.3
[21] jquerylib_0.1.4 babelgene_22.9
[23] evaluate_0.20 promises_1.2.0.1
[25] fansi_1.0.4 restfulr_0.0.15
[27] progress_1.2.2 caTools_1.18.2
[29] DBI_1.1.3 geneplotter_1.76.0
[31] htmlwidgets_1.6.2 ellipsis_0.3.2
[33] backports_1.4.1 libcoin_1.0-9
[35] locfdr_1.1-8 vctrs_0.6.1
[37] abind_1.4-5 cachem_1.0.7
[39] withr_2.5.0 BSgenome_1.66.3
[41] GenomicAlignments_1.34.1 prettyunits_1.1.1
[43] lazyeval_0.2.2 crayon_1.5.2
[45] labeling_0.4.2 pkgconfig_2.0.3
[47] ProtGenerics_1.30.0 rlang_1.1.0
[49] lifecycle_1.0.3 sandwich_3.0-2
[51] filelock_1.0.2 cellranger_1.1.0
[53] rprojroot_2.0.3 Matrix_1.5-3
[55] carData_3.0-5 boot_1.3-28.1
[57] Rhdf5lib_1.20.0 zoo_1.8-11
[59] whisker_0.4.1 processx_3.8.0
[61] png_0.1-8 rjson_0.2.21
[63] bitops_1.0-7 getPass_0.2-2
[65] KernSmooth_2.23-20 rhdf5filters_1.10.1
[67] Biostrings_2.66.0 blob_1.2.4
[69] rstatix_0.7.2 ggsignif_0.6.4
[71] memoise_2.0.1 zlibbioc_1.44.0
[73] compiler_4.2.2 BiocIO_1.8.0
[75] Rsamtools_2.14.0 cli_3.6.1
[77] XVector_0.38.0 pbapply_1.7-0
[79] ps_1.7.4 formatR_1.14
[81] MASS_7.3-58.3 tidyselect_1.2.0
[83] stringi_1.7.12 highr_0.10
[85] yaml_2.3.7 locfit_1.5-9.7
[87] sass_0.4.5 fastmatch_1.1-3
[89] timechange_0.2.0 parallel_4.2.2
[91] rstudioapi_0.14 uuid_1.1-0
[93] git2r_0.31.0 farver_2.1.1
[95] digest_0.6.31 BiocManager_1.30.20
[97] shiny_1.7.4 Rcpp_1.0.10
[99] car_3.1-2 broom_1.0.4
[101] BiocVersion_3.16.0 later_1.3.0
[103] httr_1.4.5 colorspace_2.1-0
[105] fs_1.6.1 splines_4.2.2
[107] statmod_1.5.0 plotly_4.10.1
[109] systemfonts_1.0.4 xtable_1.8-4
[111] jsonlite_1.8.4 futile.options_1.0.1
[113] modeltools_0.2-23 R6_2.5.1
[115] pillar_1.9.0 htmltools_0.5.5
[117] mime_0.12 glue_1.6.2
[119] fastmap_1.1.1 interactiveDisplayBase_1.36.0
[121] codetools_0.2-19 mvtnorm_1.1-3
[123] utf8_1.2.3 lattice_0.20-45
[125] bslib_0.4.2 curl_5.0.0
[127] gtools_3.9.4 zip_2.2.2
[129] GO.db_3.16.0 admisc_0.31
[131] rmarkdown_2.21 munsell_0.5.0
[133] rhdf5_2.42.0 GenomeInfoDbData_1.2.9
[135] gtable_0.3.3