kompute algorithm test - OF data
Coby Warkentin and Donghyung Lee
2021-11-03
Last updated: 2021-11-03
Checks: 6 1
Knit directory: komputeExamples/
This reproducible R Markdown analysis was created with workflowr (version 1.6.2). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
The R Markdown file has unstaged changes. To know which version of the R Markdown file created these results, you’ll want to first commit it to the Git repo. If you’re still working on the analysis, you can ignore this warning. When you’re finished, you can run wflow_publish to commit the R Markdown file and build the HTML.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20211027) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version dacf723. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Unstaged changes:
Modified: analysis/index.Rmd
Modified: analysis/kompute_test_102121_CC.Rmd
Modified: analysis/kompute_test_102121_OF.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made to the R Markdown (analysis/kompute_test_102121_OF.Rmd) and HTML (docs/kompute_test_102121_OF.html) files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | b7ebb1f | GitHub | 2021-10-29 | Add files via upload |
Load packages
rm(list=ls())
library(data.table)
library(dplyr)
Attaching package: 'dplyr'The following objects are masked from 'package:data.table':
between, first, lastThe following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
intersect, setdiff, setequal, union#library(kableExtra)
library(reshape2)
Attaching package: 'reshape2'The following objects are masked from 'package:data.table':
dcast, meltlibrary(ggplot2)
library(tidyr) #spread
Attaching package: 'tidyr'The following object is masked from 'package:reshape2':
smiths#library(pheatmap)
library(RColorBrewer)
#library(GGally) #ggpairs
library(irlba) # partial PCALoading required package: Matrix
Attaching package: 'Matrix'The following objects are masked from 'package:tidyr':
expand, pack, unpack#library(gridExtra)
#library(cowplot)
library(circlize)========================================
circlize version 0.4.13
CRAN page: https://cran.r-project.org/package=circlize
Github page: https://github.com/jokergoo/circlize
Documentation: https://jokergoo.github.io/circlize_book/book/
If you use it in published research, please cite:
Gu, Z. circlize implements and enhances circular visualization
in R. Bioinformatics 2014.
This message can be suppressed by:
suppressPackageStartupMessages(library(circlize))
========================================library(ComplexHeatmap)Loading required package: grid========================================
ComplexHeatmap version 2.10.0
Bioconductor page: http://bioconductor.org/packages/ComplexHeatmap/
Github page: https://github.com/jokergoo/ComplexHeatmap
Documentation: http://jokergoo.github.io/ComplexHeatmap-reference
If you use it in published research, please cite:
Gu, Z. Complex heatmaps reveal patterns and correlations in multidimensional
genomic data. Bioinformatics 2016.
The new InteractiveComplexHeatmap package can directly export static
complex heatmaps into an interactive Shiny app with zero effort. Have a try!
This message can be suppressed by:
suppressPackageStartupMessages(library(ComplexHeatmap))
========================================#options(max.print = 3000)Raw Control Phenotype
Read all control phenotype data
# load(file="~/Google Drive Miami/Miami_IMPC/data/v10.1/AllControls_small.Rdata")
load(file="G:/.shortcut-targets-by-id/1SeBOMb4GZ2Gkldxp4QNEnFWHOiAqtRTz/Miami_IMPC/data/v10.1/AllControls_small.Rdata")
dim(allpheno)[1] 12025349 15head(allpheno) biological_sample_id procedure_stable_id
1 242736 IMPC_ACS_003
2 155527 IMPC_GRS_001
3 72957 IMPC_OFD_001
4 183476 IMPC_GRS_001
5 81327 IMPC_OFD_001
6 226658 IMPC_ACS_003
procedure_name parameter_stable_id
1 Acoustic Startle and Pre-pulse Inhibition (PPI) IMPC_ACS_036_001
2 Grip Strength IMPC_GRS_008_001
3 Open Field IMPC_OFD_020_001
4 Grip Strength IMPC_GRS_009_001
5 Open Field IMPC_OFD_007_001
6 Acoustic Startle and Pre-pulse Inhibition (PPI) IMPC_ACS_034_001
parameter_name data_point sex
1 % Pre-pulse inhibition - PPI4 88.0373 female
2 Forelimb grip strength measurement mean 75.3333 female
3 Distance travelled - total 6130.3000 female
4 Forelimb and hindlimb grip strength measurement mean 120.3970 male
5 Whole arena resting time 607.0000 female
6 % Pre-pulse inhibition - PPI2 68.0742 female
age_in_weeks weight phenotyping_center date_of_experiment strain_name
1 9 18.15 JAX 2014-08-18T00:00:00Z C57BL/6NJ
2 9 20.00 BCM 2018-02-20T00:00:00Z C57BL/6N
3 9 21.20 RBRC 2015-08-03T00:00:00Z C57BL/6NTac
4 9 28.40 WTSI 2015-03-02T00:00:00Z C57BL/6N
5 9 22.62 KMPC 2018-01-30T00:00:00Z C57BL/6NTac
6 10 18.54 JAX 2015-12-16T00:00:00Z C57BL/6NJ
developmental_stage_name observation_type data_type
1 Early adult unidimensional FLOAT
2 Early adult unidimensional FLOAT
3 Early adult unidimensional FLOAT
4 Early adult unidimensional FLOAT
5 Early adult unidimensional FLOAT
6 Early adult unidimensional FLOATCorrect procedure and phenotype names, filter out time series data
We use OF only.
allpheno = allpheno %>%
filter(procedure_name=="Open Field") %>%
mutate(proc_short_name=recode(procedure_name, "Open Field"="OF")) %>%
mutate(parameter_name=recode(parameter_name,
"center - average Speed"="Center average speed",
"center - distance"="Center distance travelled",
"center - permanence time"="Center permanence time",
"center - resting time"="Center resting time",
"Latency to enter in the center"="Latency to center entry",
"Number of entries in the center"="Number of center entries",
"Periphery - average Speed"="Periphery average speed",
"Periphery - distance"="Periphery distance travelled",
"Periphery - Permanence Time"="Periphery permanence time",
"Periphery - resting time"="Periphery resting time",
"Whole Arena - average Speed"="Whole arena average speed",
"Whole Arena - Permanence Time"="Whole arena permanence",
"Whole Arena - resting time"="Whole arena resting time")) %>%
mutate(proc_param_name=paste0(proc_short_name,"_",parameter_name)) %>%
mutate(proc_param_name_stable_id=paste0(proc_short_name,"_",parameter_name,"_",parameter_stable_id))
## Extract time series data and find out parameter names
ts <- allpheno %>% filter(observation_type=="time_series")
table(ts$proc_param_name)
OF_Center average speed series OF_Center distance travelled series
36167 36328
OF_Center permanence time series OF_Center resting time series
38864 30452
OF_Distance Traveled - 5min OF_Distance travelled
9410 87970
OF_Number of center entries series OF_Number of Rears
10692 51194
OF_Number of rears - 5min OF_Periphery average speed series
9310 36324
OF_Periphery distance travelled series OF_Periphery permanence time series
36328 38864
OF_Periphery resting time series OF_Whole arena average speed series
30452 38876
OF_Whole arena resting time series
36340 # Filter out time series data
allpheno <- allpheno %>% filter(observation_type!="time_series")
table(allpheno$proc_param_name)
OF_Center average speed OF_Center distance travelled
23007 23789
OF_Center permanence time OF_Center resting time
24750 17849
OF_Distance travelled - total OF_Latency to center entry
21956 18034
OF_Number of center entries OF_Number of rears - total
18043 12814
OF_Percentage center time OF_Periphery average speed
21733 23008
OF_Periphery distance travelled OF_Periphery permanence time
23790 24751
OF_Periphery resting time OF_Whole arena average speed
17850 24754
OF_Whole arena permanence OF_Whole arena resting time
23972 24744 Heatmap showing measured phenotypes
This heatmaps show phenotypes measured for each control mouse. Columns represent mice and rows represent phenotypes.
mtest <- table(allpheno$proc_param_name_stable_id, allpheno$biological_sample_id)
mtest <-as.data.frame.matrix(mtest)
dim(mtest)[1] 51 24604if(FALSE){
nmax <-max(mtest)
library(circlize)
col_fun = colorRamp2(c(0, nmax), c("white", "red"))
col_fun(seq(0, nmax))
pdf("~/Google Drive Miami/Miami_IMPC/output/measured_phenotypes_controls_OF.pdf", width = 12, height = 8)
ht = Heatmap(as.matrix(mtest), cluster_rows = FALSE, cluster_columns = FALSE, show_column_names = F, col = col_fun,
row_names_gp = gpar(fontsize = 8), name="Count")
draw(ht)
dev.off()
}Remove phenotypes with num of obs < 15000
mtest <- table(allpheno$proc_param_name, allpheno$biological_sample_id)
dim(mtest)[1] 16 24604head(mtest[,1:10])
21653 21713 21742 21745 21747 21751 21753 21756
OF_Center average speed 1 1 1 1 1 1 1 1
OF_Center distance travelled 1 1 1 1 1 1 1 1
OF_Center permanence time 1 1 1 1 1 1 1 1
OF_Center resting time 1 1 1 1 1 1 1 1
OF_Distance travelled - total 0 0 0 0 0 0 0 0
OF_Latency to center entry 1 1 1 1 1 1 1 1
21759 21800
OF_Center average speed 1 1
OF_Center distance travelled 1 1
OF_Center permanence time 1 1
OF_Center resting time 1 1
OF_Distance travelled - total 0 0
OF_Latency to center entry 1 1mtest0 <- mtest>0
head(mtest0[,1:10])
21653 21713 21742 21745 21747 21751 21753 21756
OF_Center average speed TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
OF_Center distance travelled TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
OF_Center permanence time TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
OF_Center resting time TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
OF_Distance travelled - total FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
OF_Latency to center entry TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
21759 21800
OF_Center average speed TRUE TRUE
OF_Center distance travelled TRUE TRUE
OF_Center permanence time TRUE TRUE
OF_Center resting time TRUE TRUE
OF_Distance travelled - total FALSE FALSE
OF_Latency to center entry TRUE TRUErowSums(mtest0) OF_Center average speed OF_Center distance travelled
22856 23638
OF_Center permanence time OF_Center resting time
24599 17698
OF_Distance travelled - total OF_Latency to center entry
21805 17883
OF_Number of center entries OF_Number of rears - total
17892 12814
OF_Percentage center time OF_Periphery average speed
21730 22857
OF_Periphery distance travelled OF_Periphery permanence time
23639 24600
OF_Periphery resting time OF_Whole arena average speed
17699 24603
OF_Whole arena permanence OF_Whole arena resting time
23821 24593 rmv.pheno.list <- rownames(mtest)[rowSums(mtest0)<15000]
rmv.pheno.list[1] "OF_Number of rears - total"dim(allpheno)[1] 344844 18allpheno <- allpheno %>% filter(!(proc_param_name %in% rmv.pheno.list))
dim(allpheno)[1] 332030 18# number of phenotypes left
length(unique(allpheno$proc_param_name))[1] 15Romove samples with num of measured phenotypes < 10
mtest <- table(allpheno$proc_param_name, allpheno$biological_sample_id)
dim(mtest)[1] 15 24604head(mtest[,1:10])
21653 21713 21742 21745 21747 21751 21753 21756
OF_Center average speed 1 1 1 1 1 1 1 1
OF_Center distance travelled 1 1 1 1 1 1 1 1
OF_Center permanence time 1 1 1 1 1 1 1 1
OF_Center resting time 1 1 1 1 1 1 1 1
OF_Distance travelled - total 0 0 0 0 0 0 0 0
OF_Latency to center entry 1 1 1 1 1 1 1 1
21759 21800
OF_Center average speed 1 1
OF_Center distance travelled 1 1
OF_Center permanence time 1 1
OF_Center resting time 1 1
OF_Distance travelled - total 0 0
OF_Latency to center entry 1 1mtest0 <- mtest>0
head(mtest0[,1:10])
21653 21713 21742 21745 21747 21751 21753 21756
OF_Center average speed TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
OF_Center distance travelled TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
OF_Center permanence time TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
OF_Center resting time TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
OF_Distance travelled - total FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
OF_Latency to center entry TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
21759 21800
OF_Center average speed TRUE TRUE
OF_Center distance travelled TRUE TRUE
OF_Center permanence time TRUE TRUE
OF_Center resting time TRUE TRUE
OF_Distance travelled - total FALSE FALSE
OF_Latency to center entry TRUE TRUEsummary(colSums(mtest0)) Min. 1st Qu. Median Mean 3rd Qu. Max.
1.00 11.00 15.00 13.41 15.00 15.00 rmv.sample.list <- colnames(mtest)[colSums(mtest0)<10]
length(rmv.sample.list)[1] 1747dim(allpheno)[1] 332030 18allpheno <- allpheno %>% filter(!(biological_sample_id %in% rmv.sample.list))
dim(allpheno)[1] 319816 18# number of observations to use
length(unique(allpheno$biological_sample_id))[1] 22857Heapmap of measured phenotypes after filtering
if(FALSE){
mtest <- table(allpheno$proc_param_name, allpheno$biological_sample_id)
dim(mtest)
mtest <-as.data.frame.matrix(mtest)
nmax <-max(mtest)
library(circlize)
col_fun = colorRamp2(c(0, nmax), c("white", "red"))
col_fun(seq(0, nmax))
pdf("~/Google Drive Miami/Miami_IMPC/output/measured_phenotypes_controls_after_filtering_OF.pdf", width = 10, height = 5)
ht = Heatmap(as.matrix(mtest), cluster_rows = FALSE, cluster_columns = FALSE, show_column_names = F, col = col_fun,
row_names_gp = gpar(fontsize = 5), name="Count")
draw(ht)
dev.off()
}Reshape the data (long to wide format)
ap.mat <- allpheno %>%
dplyr::select(biological_sample_id, proc_param_name, data_point, sex, phenotyping_center, strain_name) %>%
##consider weight or age in weeks
arrange(biological_sample_id) %>%
distinct(biological_sample_id, proc_param_name, .keep_all=TRUE) %>% ## remove duplicates, maybe mean() is better.
spread(proc_param_name, data_point) %>%
tibble::column_to_rownames(var="biological_sample_id")
head(ap.mat) sex phenotyping_center strain_name
21653 female WTSI C57BL/6Brd-Tyr<c-Brd> * C57BL/6N
21713 female WTSI C57BL/6Brd-Tyr<c-Brd> * C57BL/6N
21742 male WTSI C57BL/6Brd-Tyr<c-Brd> * C57BL/6N
21745 male WTSI C57BL/6Brd-Tyr<c-Brd> * C57BL/6N
21747 male WTSI C57BL/6Brd-Tyr<c-Brd> * C57BL/6N
21751 male WTSI C57BL/6Brd-Tyr<c-Brd> * C57BL/6N
OF_Center average speed OF_Center distance travelled
21653 51.5 4259
21713 40.1 3266
21742 51.0 710
21745 38.0 2580
21747 31.4 3022
21751 14.9 1723
OF_Center permanence time OF_Center resting time
21653 102 20
21713 87 6
21742 17 3
21745 100 33
21747 134 39
21751 240 130
OF_Distance travelled - total OF_Latency to center entry
21653 NA 5.0
21713 NA 6.1
21742 NA 24.0
21745 NA 8.3
21747 NA 6.6
21751 NA 18.7
OF_Number of center entries OF_Percentage center time
21653 193 NA
21713 221 NA
21742 73 NA
21745 165 NA
21747 210 NA
21751 75 NA
OF_Periphery average speed OF_Periphery distance travelled
21653 35.3 12923
21713 31.9 11625
21742 19.1 5769
21745 25.1 7910
21747 26.0 8208
21751 11.5 2150
OF_Periphery permanence time OF_Periphery resting time
21653 498 135
21713 513 152
21742 583 292
21745 500 191
21747 466 154
21751 360 184
OF_Whole arena average speed OF_Whole arena permanence
21653 38.3 600
21713 33.4 600
21742 20.5 600
21745 27.4 600
21747 27.2 600
21751 12.8 600
OF_Whole arena resting time
21653 155
21713 158
21742 295
21745 224
21747 193
21751 314dim(ap.mat)[1] 22857 18summary(colSums(is.na(ap.mat[,-1:-3]))) Min. 1st Qu. Median Mean 3rd Qu. Max.
0 0 1 1677 3882 5159 Distribution of each phenotype
ggplot(melt(ap.mat), aes(x=value)) +
geom_histogram() +
facet_wrap(~variable, scales="free", ncol=5)+
theme(strip.text.x = element_text(size = 6))Using sex, phenotyping_center, strain_name as id variables`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.Warning: Removed 25156 rows containing non-finite values (stat_bin).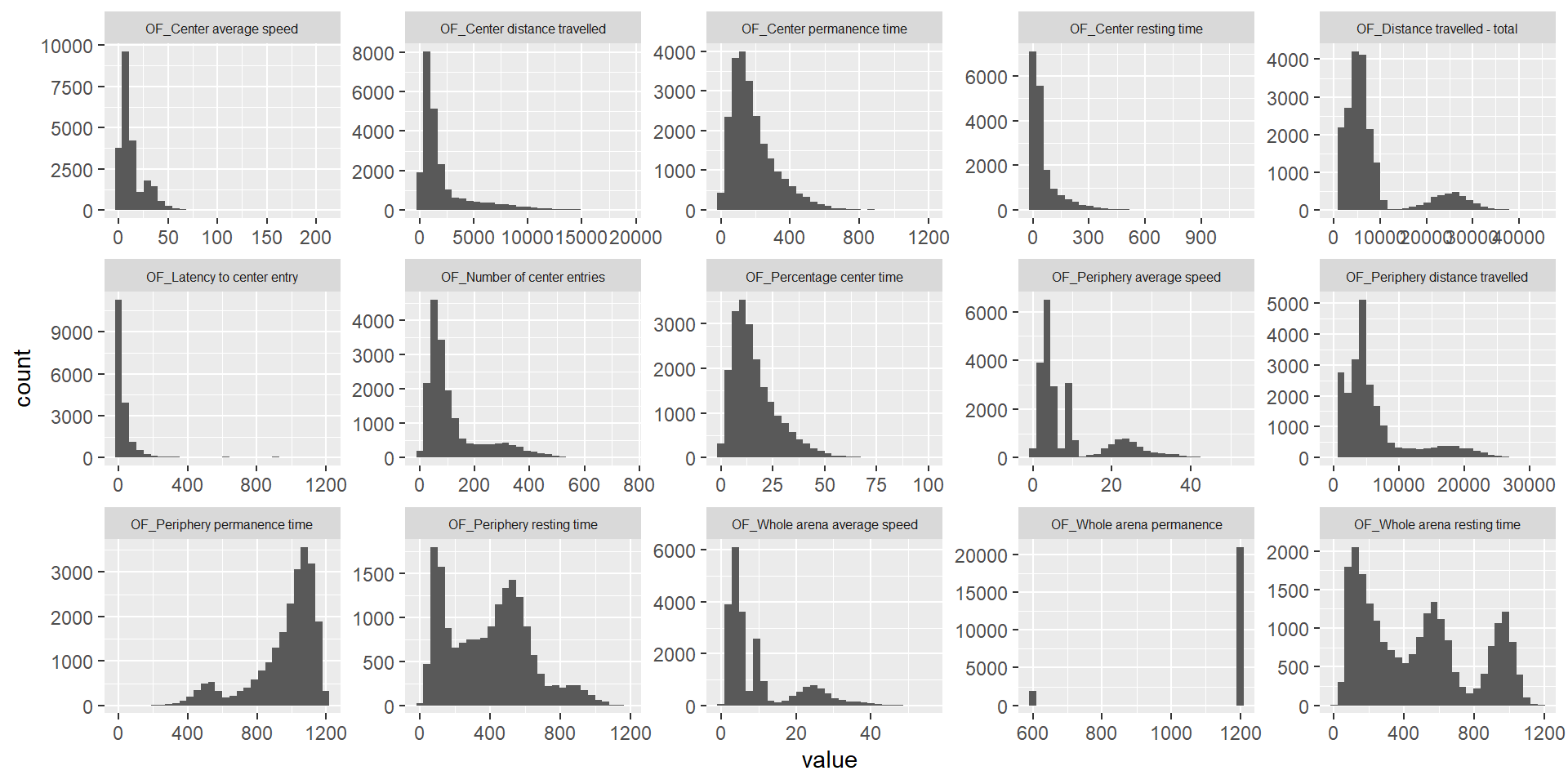
Rank Z transformation
library(RNOmni)
ap.mat.rank <- ap.mat
dim(ap.mat.rank)[1] 22857 18ap.mat.rank <- ap.mat.rank[complete.cases(ap.mat.rank),]
dim(ap.mat.rank)[1] 14863 18#rankZ <- function(x){ qnorm((rank(x,na.last="keep")-0.5)/sum(!is.na(x))) }
#ap.mat.rank <- ap.mat
#dim(ap.mat.rank)
#ap.mat.rank <- ap.mat.rank[complete.cases(ap.mat.rank),]
#dim(ap.mat.rank)
#library(RNOmni)
#ap.mat.rank <- cbind(ap.mat.rank[,1:3], apply(ap.mat.rank[,-1:-3], 2, RankNorm))
ap.mat.rank <- cbind(ap.mat.rank[,1:3], apply(ap.mat.rank[,-1:-3], 2, RankNorm))
ggplot(melt(ap.mat.rank), aes(x=value)) +
geom_histogram() +
facet_wrap(~variable, scales="free", ncol=5)+
theme(strip.text.x = element_text(size = 6))Using sex, phenotyping_center, strain_name as id variables`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.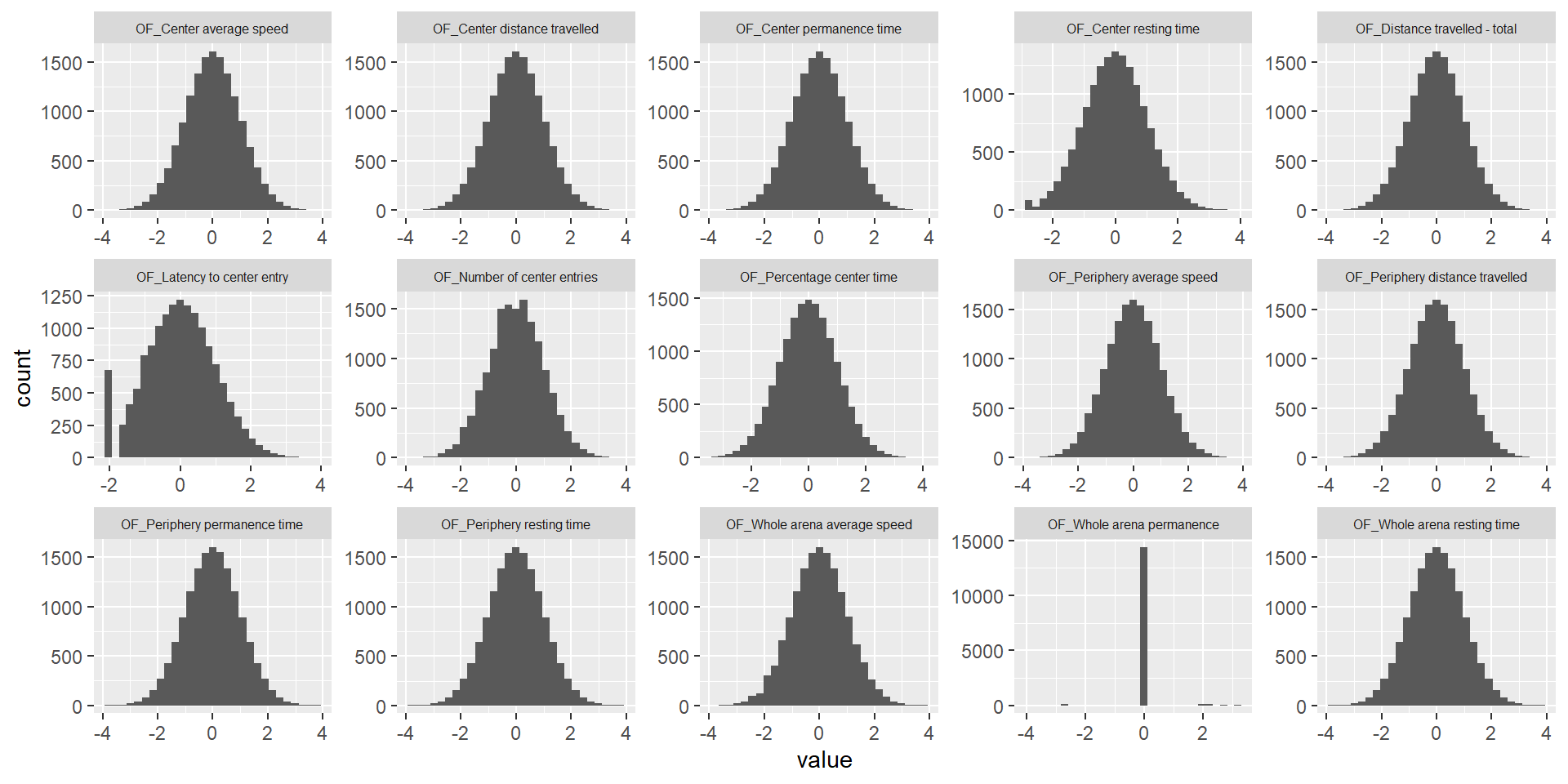
[TASK 1] Principal Variance Component Analysis
Please conduct a PVCA analysis on the phenotype matrix data (op.mat[,-1:-3]). I think you can measure the proportion of variance explained by each important covariate (sex, phenotyping_center, strain_name)
# source("~/Google Drive Miami/Miami_IMPC/reference/PVCA/examples/PVCA.R")
source("G:/.shortcut-targets-by-id/1SeBOMb4GZ2Gkldxp4QNEnFWHOiAqtRTz/Miami_IMPC/reference/PVCA/examples/PVCA.R")
meta <- ap.mat.rank[,1:3] ## looking at covariates sex, phenotyping_center, and strain_name
head(meta) sex phenotyping_center strain_name
39638 female MRC Harwell C57BL/6NTac
39639 female HMGU C57BL/6NCrl
39640 female HMGU C57BL/6NTac
39641 male HMGU C57BL/6NCrl
39642 female MRC Harwell C57BL/6NTac
39643 female HMGU C57BL/6NCrldim(meta)[1] 14863 3summary(meta) # variables are still characters sex phenotyping_center strain_name
Length:14863 Length:14863 Length:14863
Class :character Class :character Class :character
Mode :character Mode :character Mode :character meta[sapply(meta, is.character)] <- lapply(meta[sapply(meta, is.character)], as.factor)
summary(meta) # now all variables are converted to factors sex phenotyping_center strain_name
female:7428 MRC Harwell:4532 C57BL/6N :3655
male :7435 HMGU :3119 C57BL/6NCrl:3510
ICS :2417 C57BL/6NJcl: 459
RBRC :1323 C57BL/6NTac:7239
CCP-IMG :1141
TCP :1093
(Other) :1238 chisq.test(meta[,1],meta[,2])
Pearson's Chi-squared test
data: meta[, 1] and meta[, 2]
X-squared = 3.7637, df = 7, p-value = 0.8066chisq.test(meta[,2],meta[,3])
Pearson's Chi-squared test
data: meta[, 2] and meta[, 3]
X-squared = 29526, df = 21, p-value < 2.2e-16meta<-meta[,-3] # phenotyping_center and strain_name strongly associated and this caused confouding in PVCA analysis so strain_name dropped.
G <- t(ap.mat.rank[,-1:-3]) ## phenotype matrix data
set.seed(09302021)
# Perform PVCA for 10 random samples of size 1000 (more computationally efficient)
pvca.res <- matrix(nrow=10, ncol=3)
for (i in 1:10){
sample <- sample(1:ncol(G), 1000, replace=FALSE)
pvca.res[i,] <- PVCA(G[,sample], meta[sample,], threshold=0.6, inter=FALSE)
}boundary (singular) fit: see ?isSingular
boundary (singular) fit: see ?isSingular
boundary (singular) fit: see ?isSingular
boundary (singular) fit: see ?isSingular
boundary (singular) fit: see ?isSingular
boundary (singular) fit: see ?isSingular
boundary (singular) fit: see ?isSingular# Average effect size across samples
pvca.means <- colMeans(pvca.res)
names(pvca.means) <- c(colnames(meta), "resid")
# Plot PVCA
pvca.plot <- PlotPVCA(pvca.means, "PVCA of Phenotype Matrix Data")
pvca.plot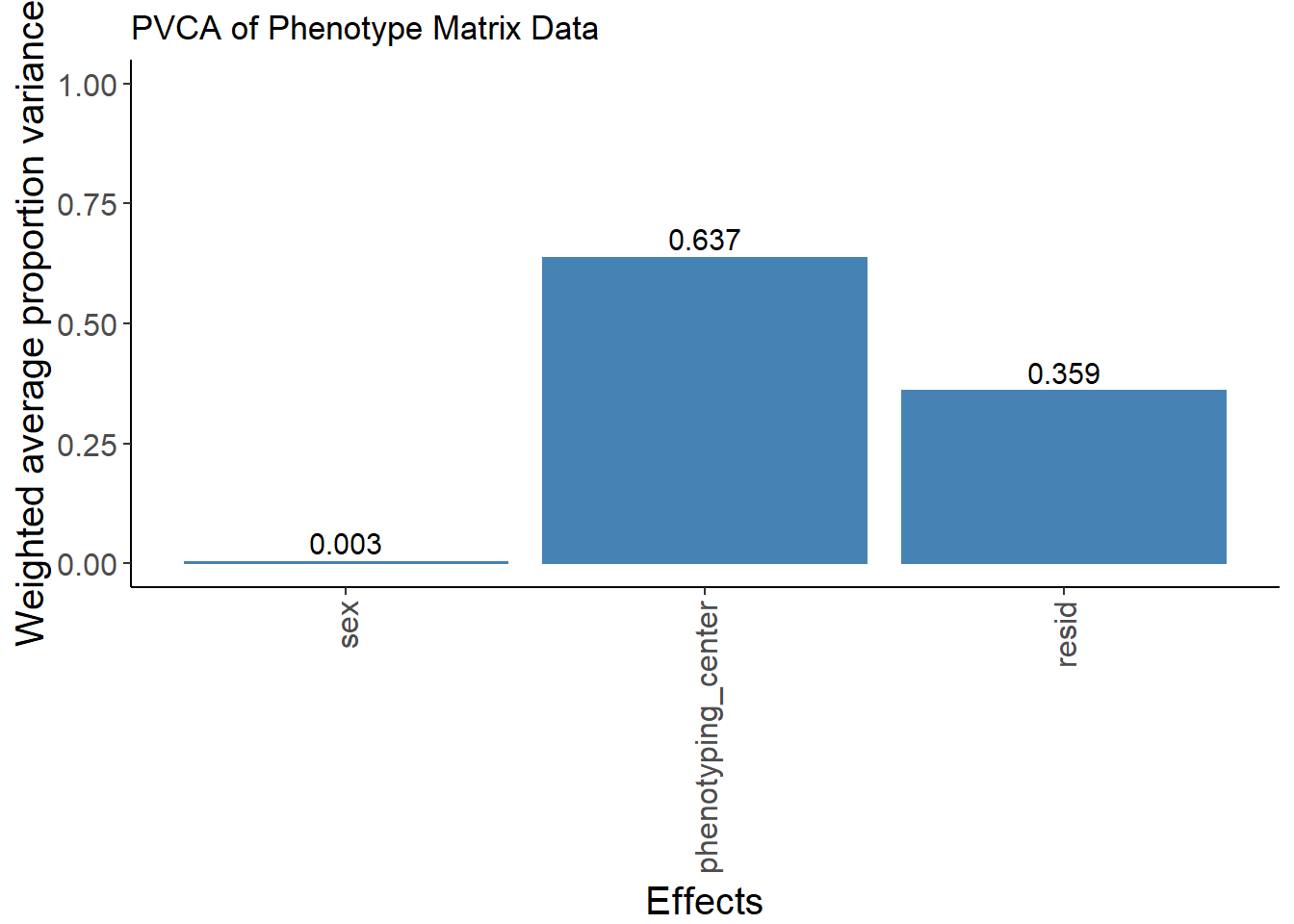
#ggsave(filename = "pvca_plot.png", pvca.plot, width=8, height=6)[TASK 2] ComBat analysis - Removing batch effects
If a large proportion of variance is explained by these covariats, we need to remove their effects from the data.
library(sva)Loading required package: mgcvLoading required package: nlme
Attaching package: 'nlme'The following object is masked from 'package:lme4':
lmListThe following object is masked from 'package:dplyr':
collapseThis is mgcv 1.8-36. For overview type 'help("mgcv-package")'.Loading required package: genefilter
Attaching package: 'genefilter'The following object is masked from 'package:ComplexHeatmap':
dist2Loading required package: BiocParallelcombat_komp = ComBat(dat=G, batch=meta$phenotyping_center, par.prior=TRUE, prior.plots=TRUE, mod=NULL)Found 1 genes with uniform expression within a single batch (all zeros); these will not be adjusted for batch.Found8batchesAdjusting for0covariate(s) or covariate level(s)Standardizing Data across genesFitting L/S model and finding priorsFinding parametric adjustmentsAdjusting the Data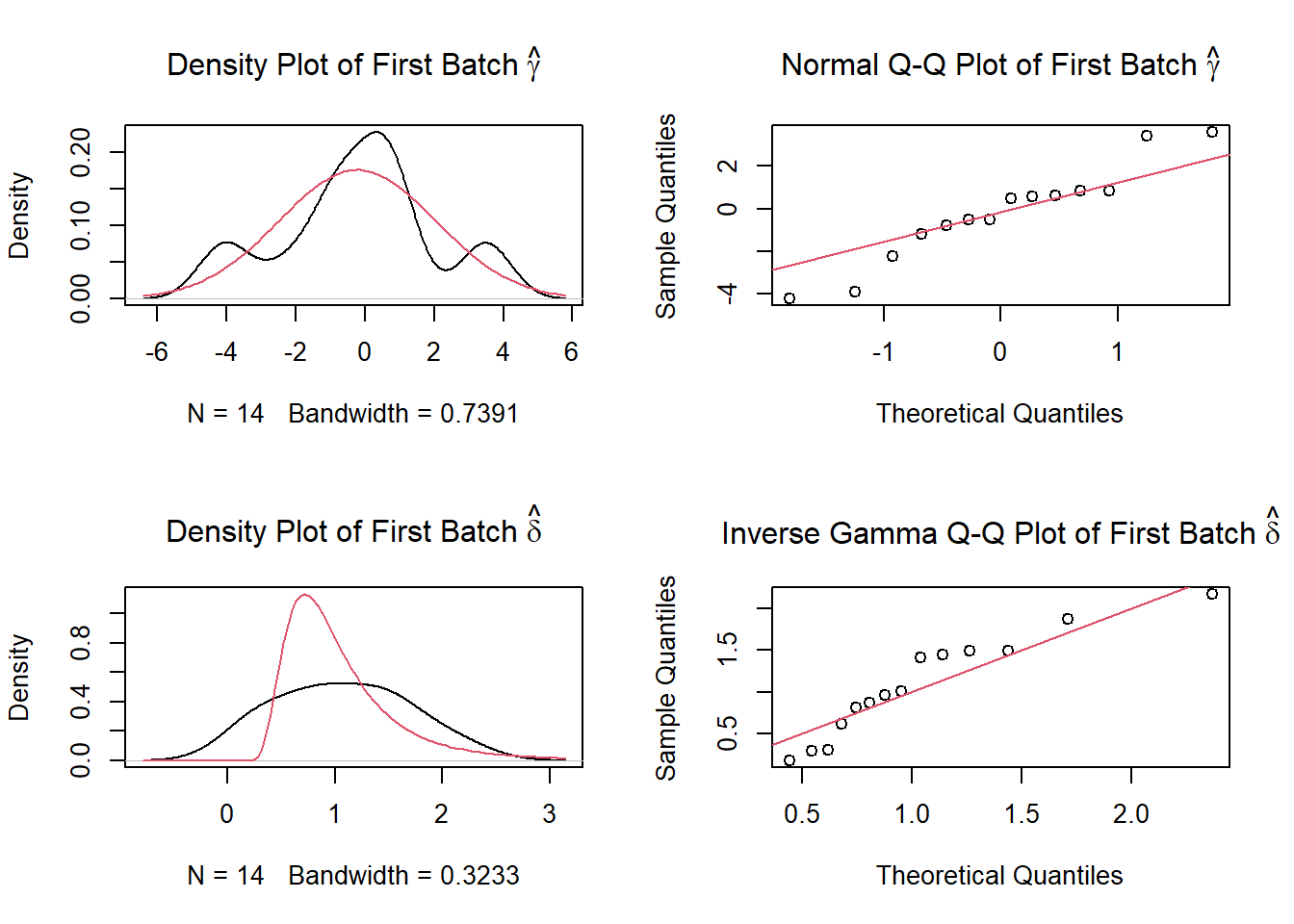
combat_komp[,1:3] 39638 39639 39640
OF_Center average speed 0.59446136 -0.10467275 0.12751142
OF_Center distance travelled 0.49851881 -0.39612726 0.26323301
OF_Center permanence time 0.05536160 -0.57404681 0.28108074
OF_Center resting time 0.09818595 -0.94195175 0.58940330
OF_Distance travelled - total 0.03620577 -0.07057520 0.03808406
OF_Latency to center entry -1.14540129 -0.37957832 1.26354212
OF_Number of center entries 0.47965785 -0.53107162 0.46539687
OF_Percentage center time 0.06084609 -0.57376582 0.28083754
OF_Periphery average speed -0.25956343 -0.13452539 -0.14510006
OF_Periphery distance travelled -0.22719214 0.24178808 -0.14416337
OF_Periphery permanence time 0.06091739 0.58467616 -0.29226506
OF_Periphery resting time 0.40380327 -0.05837431 -0.19729518
OF_Whole arena average speed 0.06158083 -0.15087855 -0.01278584
OF_Whole arena permanence -0.01863647 -0.01863647 -0.01863647
OF_Whole arena resting time 0.31830086 -0.20745469 0.09044585G[,1:3] # for comparison, combat_komp is same form and same dimensions as G 39638 39639 39640
OF_Center average speed 0.425513501 1.234197021 1.51675303
OF_Center distance travelled 0.129374222 0.997641495 1.58080810
OF_Center permanence time -0.515972599 0.009697299 0.85747405
OF_Center resting time 0.001686461 -1.202492878 0.34834230
OF_Distance travelled - total -0.470073474 1.298879853 1.41209135
OF_Latency to center entry -1.302416269 -0.496228459 0.76207343
OF_Number of center entries -0.024203070 0.906493664 1.75413567
OF_Percentage center time -0.514624462 0.010540573 0.85820501
OF_Periphery average speed -0.937318619 1.215628110 1.20336211
OF_Periphery distance travelled -0.581360130 1.624666701 1.21668801
OF_Periphery permanence time 0.568527427 0.023612640 -0.84970544
OF_Periphery resting time 0.836591193 -1.419211432 -1.56339922
OF_Whole arena average speed -0.590470467 1.199717422 1.35690828
OF_Whole arena permanence -0.018636466 -0.018636466 -0.01863647
OF_Whole arena resting time 0.811487506 -1.297508958 -0.92018584PVCA on residuals from ComBat and plot it (center effect should be much lower)
set.seed(09302021)
# Perform PVCA for 10 samples (more computationally efficient)
pvca.res.nobatch <- matrix(nrow=10, ncol=3)
for (i in 1:10){
sample <- sample(1:ncol(combat_komp), 1000, replace=FALSE)
pvca.res.nobatch[i,] <- PVCA(combat_komp[,sample], meta[sample,], threshold=0.6, inter=FALSE)
}boundary (singular) fit: see ?isSingular
boundary (singular) fit: see ?isSingular
boundary (singular) fit: see ?isSingular
boundary (singular) fit: see ?isSingular
boundary (singular) fit: see ?isSingular
boundary (singular) fit: see ?isSingular
boundary (singular) fit: see ?isSingular
boundary (singular) fit: see ?isSingular
boundary (singular) fit: see ?isSingular
boundary (singular) fit: see ?isSingular# Average effect size across samples
pvca.means.nobatch <- colMeans(pvca.res.nobatch)
names(pvca.means.nobatch) <- c(colnames(meta), "resid")
# Plot PVCA
pvca.plot.nobatch <- PlotPVCA(pvca.means.nobatch, "PVCA of Phenotype Matrix Data with Reduced Batch Effect")
pvca.plot.nobatchWarning: Removed 1 rows containing missing values (geom_text).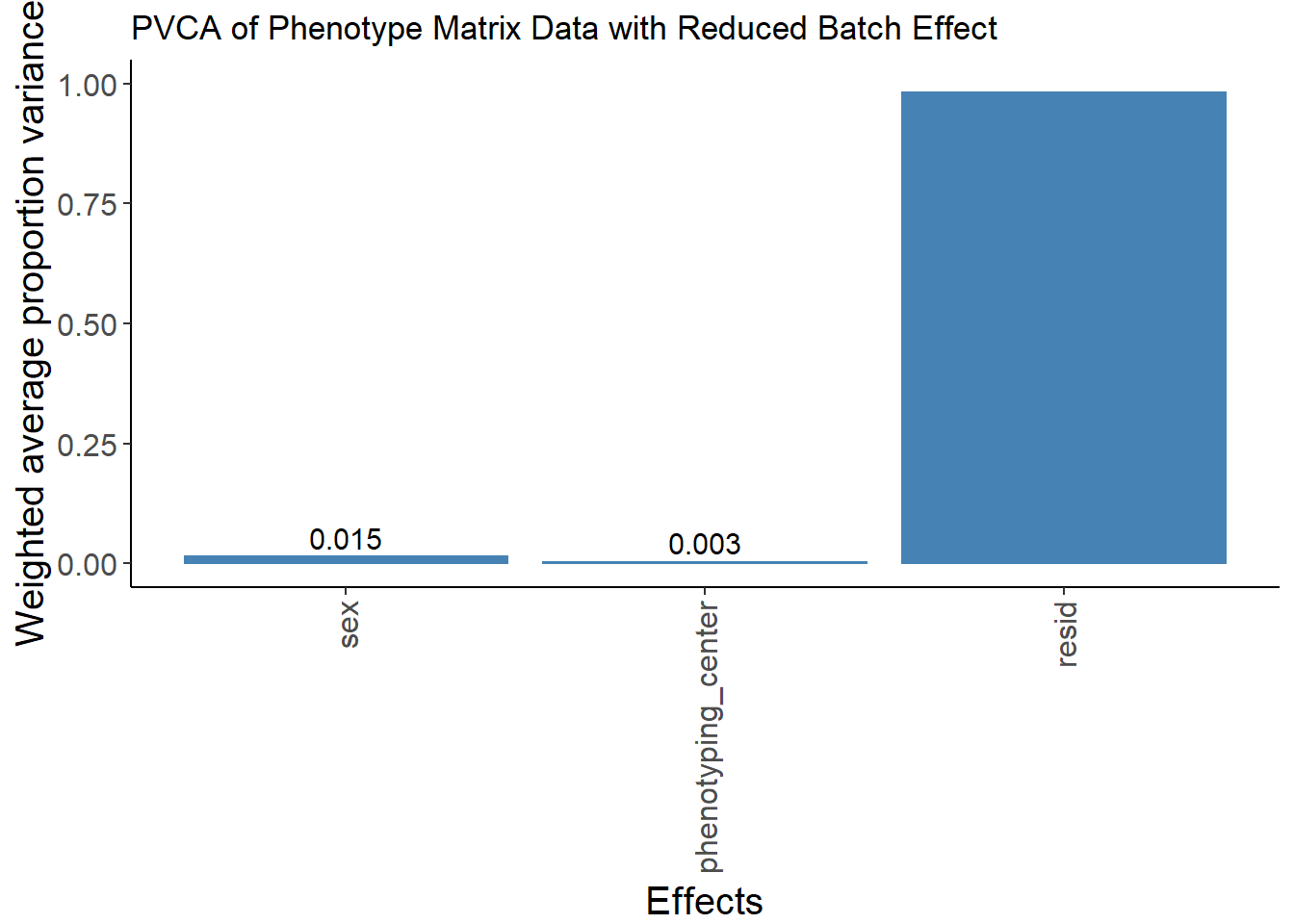
Compute correlations between CSD, GS, OF, PPI phenotypes
ap.cor.rank <- cor(ap.mat.rank[,-1:-3], use="pairwise.complete.obs") # pearson correlation coefficient
#ap.cor <- cor(ap.mat[,-1:-3], use="pairwise.complete.obs") # pearson correlation coefficient
ap.cor <- cor(ap.mat[,-1:-3], use="pairwise.complete.obs", method="spearman")
ap.cor.combat <- cor(t(combat_komp), use="pairwise.complete.obs")
#ap.cor <- cor(ap.mat[,-1:-3], use="pairwise.complete.obs", method="spearman") # use original phenotype data
#ap.cor <- cor(ap.mat.rank[,-1:-3], use="pairwise.complete.obs", method="spearman") # use rankZ transformed phenotype data
#col <- colorRampPalette(c("steelblue", "white", "darkorange"))(100)
#ap.cor.out <- pheatmap(ap.cor, cluster_rows = T, cluster_cols=T, show_colnames=F, col=col, fontsize = 7)
#col <- colorRampPalette(c("white","darkorange"))(100)
#pheatmap(abs(op.cor), cluster_rows = T, cluster_cols=T, show_colnames=F, col=col)
if(FALSE){
#pdf("~/Google Drive Miami/Miami_IMPC/output/genetic_corr_btw_phenotypes.pdf", width = 11, height = 8)
pdf("~/Google Drive Miami/Miami_IMPC/output/genetic_corr_btw_phenotypes_Pearson_OF.pdf", width = 5, height = 2.7)
ht = Heatmap(ap.cor, show_column_names = F, row_names_gp = gpar(fontsize = 9), name="Corr")
draw(ht)
dev.off()
pdf("~/Google Drive Miami/Miami_IMPC/output/genetic_corr_btw_phenotypes_rankZ_OF.pdf", width = 5, height = 2.7)
ht = Heatmap(ap.cor.rank, show_column_names = F, row_names_gp = gpar(fontsize = 9), name="Corr")
draw(ht)
dev.off()
pdf("~/Google Drive Miami/Miami_IMPC/output/genetic_corr_btw_phenotypes_ComBat_Adjusted_OF.pdf", width = 5, height = 2.7)
ht = Heatmap(ap.cor.combat, show_column_names = F, row_names_gp = gpar(fontsize = 9), name="Corr")
draw(ht)
dev.off()
}
pheno.list <- rownames(ap.cor)KOMPV10.1 association summary stat
Read KOMPv10.1
# KOMPv10.1.file = "~/Google Drive Miami/Miami_IMPC/data/v10.1/IMPC_ALL_statistical_results.csv.gz"
KOMPv10.1.file = "G:/.shortcut-targets-by-id/1SeBOMb4GZ2Gkldxp4QNEnFWHOiAqtRTz/Miami_IMPC/data/v10.1/IMPC_ALL_statistical_results.csv.gz"
KOMPv10.1 = fread(KOMPv10.1.file, header=TRUE, sep=",")
KOMPv10.1$parameter_name <- trimws(KOMPv10.1$parameter_name) #remove white spaces
KOMPv10.1$proc_param_name <- paste0(KOMPv10.1$procedure_name,"_",KOMPv10.1$parameter_name)
#head(KOMPv10.1, 10)
#sort(table(KOMPv10.1$procedure_name))
#sort(table(KOMPv10.1$proc_param_name), decreasing = TRUE)[1:100]
#sort(table(KOMPv10.1$procedure_name))
#table(KOMPv10.1$procedure_name, KOMPv10.1$parameter_name)
#table(KOMPv10.1$procedure_name, KOMPv10.1$statistical_method)
table(KOMPv10.1$procedure_name, KOMPv10.1$data_type)
adult-gross-path
Acoustic Startle and Pre-pulse Inhibition (PPI) 0
Acoustic Startle&PPI 0
Allergy (GMC) 0
Anti-nuclear antibody assay 0
Antigen Specific Immunoglobulin Assay 0
Auditory Brain Stem Response 0
Body Composition (DEXA lean/fat) 0
Body Weight 0
Bodyweight (GMC) 0
Bone marrow immunophenotyping 0
Buffy coat peripheral blood leukocyte immunophenotyping 0
Calorimetry 0
Challenge Whole Body Plethysmography 0
Clinical Chemistry 0
Clinical chemistry (GMC) 0
Combined SHIRPA and Dysmorphology 0
Cortical Bone MicroCT 0
DEXA 0
Dexa-scan analysis 0
DSS Histology 0
Dysmorphology 0
Ear epidermis immunophenotyping 0
ECG (Electrocardiogram) (GMC) 0
Echo 0
Electrocardiogram (ECG) 0
Electroconvulsive Threshold Testing 0
Electroretinography 0
ELISA (GMC) 0
ERG (Electroretinogram) (GMC) 0
Eye Morphology 0
Eye size (GMC) 0
FACS (GMC) 0
FACs Analysis 0
Fasted Clinical Chemistry 0
Fear Conditioning 0
Femoral Microradiography 0
Fertility of Homozygous Knock-out Mice 0
Food efficiency (GMC) 0
Grip-Strength 0
Grip Strength 0
Grip Strength (GMC) 0
Gross Morphology Embryo E12.5 0
Gross Morphology Embryo E14.5-E15.5 0
Gross Morphology Embryo E18.5 0
Gross Morphology Embryo E9.5 0
Gross Morphology Placenta E12.5 0
Gross Morphology Placenta E14.5-E15.5 0
Gross Morphology Placenta E18.5 0
Gross Morphology Placenta E9.5 0
Gross Pathology and Tissue Collection 633844
Haematology 0
Haematology (GMC) 0
Haematology test 0
Heart Dissection 0
Heart Weight 0
Heart weight/tibia length 0
Hematology 0
Hole-board Exploration 0
Holeboard (GMC) 0
Hot Plate 0
Immunoglobulin 0
Indirect Calorimetry 0
Indirect ophthalmoscopy 0
Insulin Blood Level 0
Intraperitoneal glucose tolerance test (IPGTT) 0
IPGTT 0
Light-Dark Test 0
Mesenteric Lymph Node Immunophenotyping 0
Modified SHIRPA 0
Nociception Hotplate (GMC) 0
Open-field 0
Open Field 0
Ophthalmoscope 0
Organ Weight 0
pDexa (GMC) 0
Plasma Chemistry 0
Rotarod 0
Rotarod A (GMC) 0
Shirpa (GMC) 0
Simplified IPGTT 0
Sleep Wake 0
Slit Lamp 0
Spleen Immunophenotyping 0
Spontaneous breathing (GMC) 0
Tail Suspension 0
Three-point Bend 0
Trabecular Bone MicroCT 0
Trichuris 0
Urinalysis 0
Vertebra Compression 0
Vertebral Microradiography 0
Viability E12.5 Secondary Screen 0
Viability E14.5-E15.5 Secondary Screen 0
Viability E18.5 Secondary Screen 0
Viability E9.5 Secondary Screen 0
Viability Primary Screen 0
Whole blood peripheral blood leukocyte immunophenotyping 0
X-ray 0
X-Ray 0
X-Ray (GMC) 0
categorical embryo
Acoustic Startle and Pre-pulse Inhibition (PPI) 0 0
Acoustic Startle&PPI 0 0
Allergy (GMC) 0 0
Anti-nuclear antibody assay 984 0
Antigen Specific Immunoglobulin Assay 0 0
Auditory Brain Stem Response 0 0
Body Composition (DEXA lean/fat) 0 0
Body Weight 0 0
Bodyweight (GMC) 0 0
Bone marrow immunophenotyping 0 0
Buffy coat peripheral blood leukocyte immunophenotyping 0 0
Calorimetry 0 0
Challenge Whole Body Plethysmography 0 0
Clinical Chemistry 0 0
Clinical chemistry (GMC) 0 0
Combined SHIRPA and Dysmorphology 156103 0
Cortical Bone MicroCT 0 0
DEXA 0 0
Dexa-scan analysis 0 0
DSS Histology 21 0
Dysmorphology 17289 0
Ear epidermis immunophenotyping 0 0
ECG (Electrocardiogram) (GMC) 0 0
Echo 0 0
Electrocardiogram (ECG) 0 0
Electroconvulsive Threshold Testing 0 0
Electroretinography 0 0
ELISA (GMC) 0 0
ERG (Electroretinogram) (GMC) 0 0
Eye Morphology 85613 0
Eye size (GMC) 0 0
FACS (GMC) 0 0
FACs Analysis 0 0
Fasted Clinical Chemistry 0 0
Fear Conditioning 0 0
Femoral Microradiography 0 0
Fertility of Homozygous Knock-out Mice 0 0
Food efficiency (GMC) 0 0
Grip-Strength 0 0
Grip Strength 0 0
Grip Strength (GMC) 0 0
Gross Morphology Embryo E12.5 0 40122
Gross Morphology Embryo E14.5-E15.5 0 13670
Gross Morphology Embryo E18.5 0 21665
Gross Morphology Embryo E9.5 0 23135
Gross Morphology Placenta E12.5 0 4623
Gross Morphology Placenta E14.5-E15.5 0 2685
Gross Morphology Placenta E18.5 0 2976
Gross Morphology Placenta E9.5 0 3827
Gross Pathology and Tissue Collection 0 0
Haematology 0 0
Haematology (GMC) 0 0
Haematology test 0 0
Heart Dissection 570 0
Heart Weight 0 0
Heart weight/tibia length 103 0
Hematology 0 0
Hole-board Exploration 0 0
Holeboard (GMC) 0 0
Hot Plate 0 0
Immunoglobulin 0 0
Indirect Calorimetry 0 0
Indirect ophthalmoscopy 3040 0
Insulin Blood Level 0 0
Intraperitoneal glucose tolerance test (IPGTT) 0 0
IPGTT 0 0
Light-Dark Test 0 0
Mesenteric Lymph Node Immunophenotyping 0 0
Modified SHIRPA 20876 0
Nociception Hotplate (GMC) 0 0
Open-field 0 0
Open Field 0 0
Ophthalmoscope 8246 0
Organ Weight 0 0
pDexa (GMC) 0 0
Plasma Chemistry 0 0
Rotarod 0 0
Rotarod A (GMC) 0 0
Shirpa (GMC) 0 0
Simplified IPGTT 0 0
Sleep Wake 0 0
Slit Lamp 14988 0
Spleen Immunophenotyping 0 0
Spontaneous breathing (GMC) 0 0
Tail Suspension 1 0
Three-point Bend 0 0
Trabecular Bone MicroCT 0 0
Trichuris 10 0
Urinalysis 0 0
Vertebra Compression 0 0
Vertebral Microradiography 0 0
Viability E12.5 Secondary Screen 0 422
Viability E14.5-E15.5 Secondary Screen 0 210
Viability E18.5 Secondary Screen 0 176
Viability E9.5 Secondary Screen 0 316
Viability Primary Screen 0 0
Whole blood peripheral blood leukocyte immunophenotyping 0 0
X-ray 60705 0
X-Ray 1791 0
X-Ray (GMC) 47 0
line
Acoustic Startle and Pre-pulse Inhibition (PPI) 0
Acoustic Startle&PPI 0
Allergy (GMC) 0
Anti-nuclear antibody assay 0
Antigen Specific Immunoglobulin Assay 0
Auditory Brain Stem Response 0
Body Composition (DEXA lean/fat) 0
Body Weight 0
Bodyweight (GMC) 0
Bone marrow immunophenotyping 0
Buffy coat peripheral blood leukocyte immunophenotyping 0
Calorimetry 0
Challenge Whole Body Plethysmography 0
Clinical Chemistry 0
Clinical chemistry (GMC) 0
Combined SHIRPA and Dysmorphology 0
Cortical Bone MicroCT 0
DEXA 0
Dexa-scan analysis 0
DSS Histology 0
Dysmorphology 0
Ear epidermis immunophenotyping 0
ECG (Electrocardiogram) (GMC) 0
Echo 0
Electrocardiogram (ECG) 0
Electroconvulsive Threshold Testing 0
Electroretinography 0
ELISA (GMC) 0
ERG (Electroretinogram) (GMC) 0
Eye Morphology 0
Eye size (GMC) 0
FACS (GMC) 0
FACs Analysis 0
Fasted Clinical Chemistry 0
Fear Conditioning 0
Femoral Microradiography 0
Fertility of Homozygous Knock-out Mice 7512
Food efficiency (GMC) 0
Grip-Strength 0
Grip Strength 0
Grip Strength (GMC) 0
Gross Morphology Embryo E12.5 0
Gross Morphology Embryo E14.5-E15.5 0
Gross Morphology Embryo E18.5 0
Gross Morphology Embryo E9.5 0
Gross Morphology Placenta E12.5 0
Gross Morphology Placenta E14.5-E15.5 0
Gross Morphology Placenta E18.5 0
Gross Morphology Placenta E9.5 0
Gross Pathology and Tissue Collection 0
Haematology 0
Haematology (GMC) 0
Haematology test 0
Heart Dissection 0
Heart Weight 0
Heart weight/tibia length 0
Hematology 0
Hole-board Exploration 0
Holeboard (GMC) 0
Hot Plate 0
Immunoglobulin 0
Indirect Calorimetry 0
Indirect ophthalmoscopy 0
Insulin Blood Level 0
Intraperitoneal glucose tolerance test (IPGTT) 0
IPGTT 0
Light-Dark Test 0
Mesenteric Lymph Node Immunophenotyping 0
Modified SHIRPA 0
Nociception Hotplate (GMC) 0
Open-field 0
Open Field 0
Ophthalmoscope 0
Organ Weight 0
pDexa (GMC) 0
Plasma Chemistry 0
Rotarod 0
Rotarod A (GMC) 0
Shirpa (GMC) 0
Simplified IPGTT 0
Sleep Wake 0
Slit Lamp 0
Spleen Immunophenotyping 0
Spontaneous breathing (GMC) 0
Tail Suspension 0
Three-point Bend 0
Trabecular Bone MicroCT 0
Trichuris 0
Urinalysis 0
Vertebra Compression 0
Vertebral Microradiography 0
Viability E12.5 Secondary Screen 0
Viability E14.5-E15.5 Secondary Screen 0
Viability E18.5 Secondary Screen 0
Viability E9.5 Secondary Screen 0
Viability Primary Screen 7362
Whole blood peripheral blood leukocyte immunophenotyping 0
X-ray 0
X-Ray 0
X-Ray (GMC) 0
unidimensional
Acoustic Startle and Pre-pulse Inhibition (PPI) 20676
Acoustic Startle&PPI 8356
Allergy (GMC) 27
Anti-nuclear antibody assay 982
Antigen Specific Immunoglobulin Assay 482
Auditory Brain Stem Response 2149
Body Composition (DEXA lean/fat) 42582
Body Weight 7263
Bodyweight (GMC) 27
Bone marrow immunophenotyping 8043
Buffy coat peripheral blood leukocyte immunophenotyping 7560
Calorimetry 731
Challenge Whole Body Plethysmography 46
Clinical Chemistry 99693
Clinical chemistry (GMC) 548
Combined SHIRPA and Dysmorphology 3645
Cortical Bone MicroCT 18
DEXA 5529
Dexa-scan analysis 4516
DSS Histology 133
Dysmorphology 0
Ear epidermis immunophenotyping 3112
ECG (Electrocardiogram) (GMC) 130
Echo 16196
Electrocardiogram (ECG) 32701
Electroconvulsive Threshold Testing 409
Electroretinography 5
ELISA (GMC) 96
ERG (Electroretinogram) (GMC) 148
Eye Morphology 3263
Eye size (GMC) 34
FACS (GMC) 185
FACs Analysis 3644
Fasted Clinical Chemistry 3744
Fear Conditioning 420
Femoral Microradiography 12
Fertility of Homozygous Knock-out Mice 0
Food efficiency (GMC) 135
Grip-Strength 2472
Grip Strength 19766
Grip Strength (GMC) 19
Gross Morphology Embryo E12.5 0
Gross Morphology Embryo E14.5-E15.5 0
Gross Morphology Embryo E18.5 0
Gross Morphology Embryo E9.5 0
Gross Morphology Placenta E12.5 0
Gross Morphology Placenta E14.5-E15.5 0
Gross Morphology Placenta E18.5 0
Gross Morphology Placenta E9.5 0
Gross Pathology and Tissue Collection 0
Haematology 4731
Haematology (GMC) 428
Haematology test 6038
Heart Dissection 1510
Heart Weight 4312
Heart weight/tibia length 776
Hematology 49606
Hole-board Exploration 991
Holeboard (GMC) 513
Hot Plate 2044
Immunoglobulin 929
Indirect Calorimetry 4919
Indirect ophthalmoscopy 0
Insulin Blood Level 2081
Intraperitoneal glucose tolerance test (IPGTT) 12990
IPGTT 408
Light-Dark Test 11077
Mesenteric Lymph Node Immunophenotyping 44829
Modified SHIRPA 1245
Nociception Hotplate (GMC) 91
Open-field 7458
Open Field 50546
Ophthalmoscope 757
Organ Weight 1671
pDexa (GMC) 321
Plasma Chemistry 894
Rotarod 1798
Rotarod A (GMC) 6
Shirpa (GMC) 197
Simplified IPGTT 622
Sleep Wake 5333
Slit Lamp 0
Spleen Immunophenotyping 56372
Spontaneous breathing (GMC) 576
Tail Suspension 534
Three-point Bend 30
Trabecular Bone MicroCT 28
Trichuris 0
Urinalysis 1034
Vertebra Compression 21
Vertebral Microradiography 14
Viability E12.5 Secondary Screen 0
Viability E14.5-E15.5 Secondary Screen 0
Viability E18.5 Secondary Screen 0
Viability E9.5 Secondary Screen 0
Viability Primary Screen 0
Whole blood peripheral blood leukocyte immunophenotyping 0
X-ray 2543
X-Ray 231
X-Ray (GMC) 0
unidimensional-ReferenceRange
Acoustic Startle and Pre-pulse Inhibition (PPI) 0
Acoustic Startle&PPI 0
Allergy (GMC) 0
Anti-nuclear antibody assay 0
Antigen Specific Immunoglobulin Assay 0
Auditory Brain Stem Response 26586
Body Composition (DEXA lean/fat) 0
Body Weight 0
Bodyweight (GMC) 0
Bone marrow immunophenotyping 0
Buffy coat peripheral blood leukocyte immunophenotyping 0
Calorimetry 0
Challenge Whole Body Plethysmography 0
Clinical Chemistry 0
Clinical chemistry (GMC) 0
Combined SHIRPA and Dysmorphology 0
Cortical Bone MicroCT 0
DEXA 0
Dexa-scan analysis 0
DSS Histology 0
Dysmorphology 0
Ear epidermis immunophenotyping 0
ECG (Electrocardiogram) (GMC) 0
Echo 0
Electrocardiogram (ECG) 0
Electroconvulsive Threshold Testing 0
Electroretinography 0
ELISA (GMC) 0
ERG (Electroretinogram) (GMC) 0
Eye Morphology 0
Eye size (GMC) 0
FACS (GMC) 0
FACs Analysis 0
Fasted Clinical Chemistry 0
Fear Conditioning 0
Femoral Microradiography 0
Fertility of Homozygous Knock-out Mice 0
Food efficiency (GMC) 0
Grip-Strength 0
Grip Strength 0
Grip Strength (GMC) 0
Gross Morphology Embryo E12.5 0
Gross Morphology Embryo E14.5-E15.5 0
Gross Morphology Embryo E18.5 0
Gross Morphology Embryo E9.5 0
Gross Morphology Placenta E12.5 0
Gross Morphology Placenta E14.5-E15.5 0
Gross Morphology Placenta E18.5 0
Gross Morphology Placenta E9.5 0
Gross Pathology and Tissue Collection 0
Haematology 0
Haematology (GMC) 0
Haematology test 0
Heart Dissection 0
Heart Weight 0
Heart weight/tibia length 0
Hematology 0
Hole-board Exploration 0
Holeboard (GMC) 0
Hot Plate 0
Immunoglobulin 0
Indirect Calorimetry 0
Indirect ophthalmoscopy 0
Insulin Blood Level 0
Intraperitoneal glucose tolerance test (IPGTT) 0
IPGTT 0
Light-Dark Test 0
Mesenteric Lymph Node Immunophenotyping 0
Modified SHIRPA 0
Nociception Hotplate (GMC) 0
Open-field 0
Open Field 0
Ophthalmoscope 0
Organ Weight 0
pDexa (GMC) 0
Plasma Chemistry 0
Rotarod 0
Rotarod A (GMC) 0
Shirpa (GMC) 0
Simplified IPGTT 0
Sleep Wake 0
Slit Lamp 0
Spleen Immunophenotyping 0
Spontaneous breathing (GMC) 0
Tail Suspension 0
Three-point Bend 0
Trabecular Bone MicroCT 0
Trichuris 0
Urinalysis 0
Vertebra Compression 0
Vertebral Microradiography 0
Viability E12.5 Secondary Screen 0
Viability E14.5-E15.5 Secondary Screen 0
Viability E18.5 Secondary Screen 0
Viability E9.5 Secondary Screen 0
Viability Primary Screen 0
Whole blood peripheral blood leukocyte immunophenotyping 29520
X-ray 10450
X-Ray 414
X-Ray (GMC) 0#dat <- KOMPv10.1 %>% select(procedure_name=="Gross Pathology and Tissue Collection")
# extract unidimensional data only.
dim(KOMPv10.1)[1] 1779903 88KOMPv10.1.ud <- KOMPv10.1 %>% filter(data_type=="unidimensional")
dim(KOMPv10.1.ud)[1] 580001 88Heatmap Gene - Pheno
Subset OF data and generate Z-score
table(allpheno$procedure_name)
Open Field
319816 #"Auditory Brain Stem Response"
#"Clinical Chemistry"
#"Body Composition (DEXA lean/fat)"
#"Intraperitoneal glucose tolerance test (IPGTT)"
#"Hematology"
# count the number of tests in each phenotype
proc.list <- table(KOMPv10.1.ud$procedure_name)
#proc.list <- proc.list[proc.list>1000]
proc.list
Acoustic Startle and Pre-pulse Inhibition (PPI)
20676
Acoustic Startle&PPI
8356
Allergy (GMC)
27
Anti-nuclear antibody assay
982
Antigen Specific Immunoglobulin Assay
482
Auditory Brain Stem Response
2149
Body Composition (DEXA lean/fat)
42582
Body Weight
7263
Bodyweight (GMC)
27
Bone marrow immunophenotyping
8043
Buffy coat peripheral blood leukocyte immunophenotyping
7560
Calorimetry
731
Challenge Whole Body Plethysmography
46
Clinical Chemistry
99693
Clinical chemistry (GMC)
548
Combined SHIRPA and Dysmorphology
3645
Cortical Bone MicroCT
18
DEXA
5529
Dexa-scan analysis
4516
DSS Histology
133
Ear epidermis immunophenotyping
3112
ECG (Electrocardiogram) (GMC)
130
Echo
16196
Electrocardiogram (ECG)
32701
Electroconvulsive Threshold Testing
409
Electroretinography
5
ELISA (GMC)
96
ERG (Electroretinogram) (GMC)
148
Eye Morphology
3263
Eye size (GMC)
34
FACS (GMC)
185
FACs Analysis
3644
Fasted Clinical Chemistry
3744
Fear Conditioning
420
Femoral Microradiography
12
Food efficiency (GMC)
135
Grip-Strength
2472
Grip Strength
19766
Grip Strength (GMC)
19
Haematology
4731
Haematology (GMC)
428
Haematology test
6038
Heart Dissection
1510
Heart Weight
4312
Heart weight/tibia length
776
Hematology
49606
Hole-board Exploration
991
Holeboard (GMC)
513
Hot Plate
2044
Immunoglobulin
929
Indirect Calorimetry
4919
Insulin Blood Level
2081
Intraperitoneal glucose tolerance test (IPGTT)
12990
IPGTT
408
Light-Dark Test
11077
Mesenteric Lymph Node Immunophenotyping
44829
Modified SHIRPA
1245
Nociception Hotplate (GMC)
91
Open-field
7458
Open Field
50546
Ophthalmoscope
757
Organ Weight
1671
pDexa (GMC)
321
Plasma Chemistry
894
Rotarod
1798
Rotarod A (GMC)
6
Shirpa (GMC)
197
Simplified IPGTT
622
Sleep Wake
5333
Spleen Immunophenotyping
56372
Spontaneous breathing (GMC)
576
Tail Suspension
534
Three-point Bend
30
Trabecular Bone MicroCT
28
Urinalysis
1034
Vertebra Compression
21
Vertebral Microradiography
14
X-ray
2543
X-Ray
231 length(proc.list)[1] 79pheno.list <- table(KOMPv10.1.ud$proc_param_name)
pheno.list <- pheno.list[pheno.list>1000] # find list of phenotypes with more than 1000 tests (i.e. 1000 mutants tested)
pheno.list <- names(pheno.list)
pheno.list [1] "Acoustic Startle and Pre-pulse Inhibition (PPI)_% Pre-pulse inhibition - Global"
[2] "Acoustic Startle and Pre-pulse Inhibition (PPI)_% Pre-pulse inhibition - PPI1"
[3] "Acoustic Startle and Pre-pulse Inhibition (PPI)_% Pre-pulse inhibition - PPI2"
[4] "Acoustic Startle and Pre-pulse Inhibition (PPI)_% Pre-pulse inhibition - PPI3"
[5] "Acoustic Startle and Pre-pulse Inhibition (PPI)_% Pre-pulse inhibition - PPI4"
[6] "Acoustic Startle and Pre-pulse Inhibition (PPI)_Response amplitude - S"
[7] "Body Composition (DEXA lean/fat)_BMC/Body weight"
[8] "Body Composition (DEXA lean/fat)_Body length"
[9] "Body Composition (DEXA lean/fat)_Bone Area"
[10] "Body Composition (DEXA lean/fat)_Bone Mineral Content (excluding skull)"
[11] "Body Composition (DEXA lean/fat)_Bone Mineral Density (excluding skull)"
[12] "Body Composition (DEXA lean/fat)_Fat mass"
[13] "Body Composition (DEXA lean/fat)_Fat/Body weight"
[14] "Body Composition (DEXA lean/fat)_Lean mass"
[15] "Body Composition (DEXA lean/fat)_Lean/Body weight"
[16] "Body Weight_Body Weight"
[17] "Clinical Chemistry_Alanine aminotransferase"
[18] "Clinical Chemistry_Albumin"
[19] "Clinical Chemistry_Alkaline phosphatase"
[20] "Clinical Chemistry_Alpha-amylase"
[21] "Clinical Chemistry_Aspartate aminotransferase"
[22] "Clinical Chemistry_Calcium"
[23] "Clinical Chemistry_Chloride"
[24] "Clinical Chemistry_Creatine kinase"
[25] "Clinical Chemistry_Creatinine"
[26] "Clinical Chemistry_Free fatty acids"
[27] "Clinical Chemistry_Fructosamine"
[28] "Clinical Chemistry_Glucose"
[29] "Clinical Chemistry_Glycerol"
[30] "Clinical Chemistry_HDL-cholesterol"
[31] "Clinical Chemistry_Iron"
[32] "Clinical Chemistry_LDL-cholesterol"
[33] "Clinical Chemistry_Magnesium"
[34] "Clinical Chemistry_Phosphorus"
[35] "Clinical Chemistry_Potassium"
[36] "Clinical Chemistry_Sodium"
[37] "Clinical Chemistry_Total bilirubin"
[38] "Clinical Chemistry_Total cholesterol"
[39] "Clinical Chemistry_Total protein"
[40] "Clinical Chemistry_Triglyceride"
[41] "Clinical Chemistry_Triglycerides"
[42] "Clinical Chemistry_Urea"
[43] "Clinical Chemistry_Urea (Blood Urea Nitrogen - BUN)"
[44] "Combined SHIRPA and Dysmorphology_Locomotor activity"
[45] "Echo_Cardiac Output"
[46] "Echo_Ejection Fraction"
[47] "Echo_Fractional Shortening"
[48] "Echo_HR"
[49] "Echo_LVAWd"
[50] "Echo_LVIDd"
[51] "Echo_LVIDs"
[52] "Echo_LVPWd"
[53] "Echo_LVPWs"
[54] "Echo_Stroke Volume"
[55] "Electrocardiogram (ECG)_CV"
[56] "Electrocardiogram (ECG)_HR"
[57] "Electrocardiogram (ECG)_HRV"
[58] "Electrocardiogram (ECG)_PQ"
[59] "Electrocardiogram (ECG)_PR"
[60] "Electrocardiogram (ECG)_QRS"
[61] "Electrocardiogram (ECG)_QTc"
[62] "Electrocardiogram (ECG)_QTc Dispersion"
[63] "Electrocardiogram (ECG)_rMSSD"
[64] "Electrocardiogram (ECG)_RR"
[65] "Electrocardiogram (ECG)_ST"
[66] "Grip Strength_Forelimb and hindlimb grip strength measurement mean"
[67] "Grip Strength_Forelimb and hindlimb grip strength normalised against body weight"
[68] "Grip Strength_Forelimb grip strength measurement mean"
[69] "Grip Strength_Forelimb grip strength normalised against body weight"
[70] "Heart Weight_Heart weight"
[71] "Hematology_Basophil cell count"
[72] "Hematology_Basophil differential count"
[73] "Hematology_Eosinophil cell count"
[74] "Hematology_Eosinophil differential count"
[75] "Hematology_Hematocrit"
[76] "Hematology_Hemoglobin"
[77] "Hematology_Lymphocyte cell count"
[78] "Hematology_Lymphocyte differential count"
[79] "Hematology_Mean cell hemoglobin concentration"
[80] "Hematology_Mean cell volume"
[81] "Hematology_Mean corpuscular hemoglobin"
[82] "Hematology_Mean platelet volume"
[83] "Hematology_Monocyte cell count"
[84] "Hematology_Monocyte differential count"
[85] "Hematology_Neutrophil cell count"
[86] "Hematology_Neutrophil differential count"
[87] "Hematology_Platelet count"
[88] "Hematology_Red blood cell count"
[89] "Hematology_Red blood cell distribution width"
[90] "Hematology_White blood cell count"
[91] "Hot Plate_Time of first response"
[92] "Indirect Calorimetry_Respiratory Exchange Ratio"
[93] "Insulin Blood Level_Insulin"
[94] "Intraperitoneal glucose tolerance test (IPGTT)_Area under glucose response curve"
[95] "Intraperitoneal glucose tolerance test (IPGTT)_Fasted blood glucose concentration"
[96] "Intraperitoneal glucose tolerance test (IPGTT)_Initial response to glucose challenge"
[97] "Light-Dark Test_Dark side time spent"
[98] "Light-Dark Test_Fecal boli"
[99] "Light-Dark Test_Latency to first transition into dark"
[100] "Light-Dark Test_Light side time spent"
[101] "Light-Dark Test_Percent time in dark"
[102] "Light-Dark Test_Percent time in light"
[103] "Light-Dark Test_Side changes"
[104] "Light-Dark Test_Time mobile dark side"
[105] "Light-Dark Test_Time mobile light side"
[106] "Modified SHIRPA_Locomotor activity"
[107] "Open Field_Center average speed"
[108] "Open Field_Center distance travelled"
[109] "Open Field_Center permanence time"
[110] "Open Field_Center resting time"
[111] "Open Field_Distance travelled - total"
[112] "Open Field_Latency to center entry"
[113] "Open Field_Number of center entries"
[114] "Open Field_Number of rears - total"
[115] "Open Field_Percentage center time"
[116] "Open Field_Periphery average speed"
[117] "Open Field_Periphery distance travelled"
[118] "Open Field_Periphery permanence time"
[119] "Open Field_Periphery resting time"
[120] "Open Field_Whole arena average speed"
[121] "Open Field_Whole arena resting time"
[122] "X-ray_Tibia length" length(pheno.list) #122[1] 122# Use phenotypes with more than 1000 tests (i.e. 1000 mutants tested)
dim(KOMPv10.1.ud)[1] 580001 88ap.stat <- KOMPv10.1.ud %>% filter(proc_param_name %in% pheno.list)
dim(ap.stat)[1] 358088 88mtest <- table(ap.stat$proc_param_name, ap.stat$marker_symbol)
mtest <-as.data.frame.matrix(mtest)
dim(mtest)[1] 122 5954if(FALSE){
nmax <-max(mtest)
library(circlize)
col_fun = colorRamp2(c(0, nmax), c("white", "red"))
col_fun(seq(0, nmax))
pdf("~/Google Drive Miami/Miami_IMPC/output/KMOPv10.1_heatmap_gene_vs_pheno_after_filtering.pdf", width = 10, height = 10)
ht = Heatmap(as.matrix(mtest), cluster_rows = FALSE, cluster_columns = FALSE, show_column_names = F, col = col_fun,
row_names_gp = gpar(fontsize = 5), name="Count")
draw(ht)
dev.off()
}
table(ap.stat$procedure_name)
Acoustic Startle and Pre-pulse Inhibition (PPI)
20676
Body Composition (DEXA lean/fat)
42582
Body Weight
1242
Clinical Chemistry
95963
Combined SHIRPA and Dysmorphology
3645
Echo
12435
Electrocardiogram (ECG)
32701
Grip Strength
18995
Heart Weight
3657
Hematology
48130
Hot Plate
1329
Indirect Calorimetry
2215
Insulin Blood Level
2081
Intraperitoneal glucose tolerance test (IPGTT)
12990
Light-Dark Test
11077
Modified SHIRPA
1245
Open Field
45326
X-ray
1799 table(allpheno$procedure_name)
Open Field
319816 ap.stat = ap.stat %>%
dplyr::select(phenotyping_center, procedure_name, parameter_name, zygosity, allele_symbol,
genotype_effect_parameter_estimate, genotype_effect_stderr_estimate,
genotype_effect_p_value, phenotyping_center, allele_name, marker_symbol) %>%
filter(procedure_name == "Open Field") %>%
mutate(procedure_name=recode(procedure_name, "Open Field"="OF")) %>%
mutate(z_score = genotype_effect_parameter_estimate/genotype_effect_stderr_estimate,
proc_param_name=paste0(procedure_name,"_",parameter_name),
gene_pheno = paste0(parameter_name, "_", allele_symbol))
table(ap.stat$parameter_name, ap.stat$procedure_name)
OF
Center average speed 3335
Center distance travelled 3591
Center permanence time 3632
Center resting time 1802
Distance travelled - total 3550
Latency to center entry 1818
Number of center entries 1816
Number of rears - total 2772
Percentage center time 3375
Periphery average speed 3335
Periphery distance travelled 3591
Periphery permanence time 3633
Periphery resting time 1803
Whole arena average speed 3637
Whole arena resting time 3636length(unique(ap.stat$marker_symbol)) #3362[1] 3362length(unique(ap.stat$allele_symbol)) #3412[1] 3412length(unique(ap.stat$proc_param_name)) #15 # number of phenotypes in association statistics data set[1] 15length(unique(allpheno$proc_param_name)) #15 # number of phenotypes in final control data[1] 15pheno.list.stat <- unique(ap.stat$proc_param_name)
pheno.list.ctrl <- unique(allpheno$proc_param_name)
sum(pheno.list.stat %in% pheno.list.ctrl)[1] 14sum(pheno.list.ctrl %in% pheno.list.stat)[1] 14## extract common phenotype list
common.pheno.list <- sort(intersect(pheno.list.ctrl, pheno.list.stat))
common.pheno.list [1] "OF_Center average speed" "OF_Center distance travelled"
[3] "OF_Center permanence time" "OF_Center resting time"
[5] "OF_Distance travelled - total" "OF_Latency to center entry"
[7] "OF_Number of center entries" "OF_Percentage center time"
[9] "OF_Periphery average speed" "OF_Periphery distance travelled"
[11] "OF_Periphery permanence time" "OF_Periphery resting time"
[13] "OF_Whole arena average speed" "OF_Whole arena resting time" length(common.pheno.list)[1] 14# Use summary statistics of common phenotypes
dim(ap.stat)[1] 45326 13ap.stat <- ap.stat %>% filter(proc_param_name %in% common.pheno.list)
dim(ap.stat)[1] 42554 13length(unique(ap.stat$proc_param_name))[1] 14Find duplicates in gene-phenotype pair
mtest <- table(ap.stat$proc_param_name, ap.stat$marker_symbol)
mtest <-as.data.frame.matrix(mtest)
nmax <-max(mtest)
col_fun = colorRamp2(c(0, nmax), c("white", "red"))
col_fun(seq(0, nmax)) [1] "#FFFFFFFF" "#FFEEE7FF" "#FFDCD0FF" "#FFCBB9FF" "#FFB9A2FF" "#FFA78CFF"
[7] "#FF9576FF" "#FF8161FF" "#FF6D4CFF" "#FF5636FF" "#FF3A1FFF" "#FF0000FF"ht = Heatmap(as.matrix(mtest), cluster_rows = FALSE, cluster_columns = FALSE, show_column_names = F, col = col_fun,
row_names_gp = gpar(fontsize = 8), name="Count")`use_raster` is automatically set to TRUE for a matrix with more than
2000 columns You can control `use_raster` argument by explicitly
setting TRUE/FALSE to it.
Set `ht_opt$message = FALSE` to turn off this message.'magick' package is suggested to install to give better rasterization.
Set `ht_opt$message = FALSE` to turn off this message.draw(ht)
Using Stouffer’s method, merge multiple z-scores of a gene-phenotype pair into a z-score
## sum(z-score)/sqrt(# of zscore)
sumz <- function(z){ sum(z)/sqrt(length(z)) }
ap.z = ap.stat %>%
dplyr::select(marker_symbol, proc_param_name, z_score) %>%
na.omit() %>%
group_by(marker_symbol, proc_param_name) %>%
summarize(zscore = sumz(z_score)) ## combine z-scores`summarise()` has grouped output by 'marker_symbol'. You can override using the `.groups` argument.dim(ap.z)[1] 35836 3Make z-score matrix (long to wide)
nan2na <- function(df){
out <- data.frame(sapply(df, function(x) ifelse(is.nan(x), NA, x)))
colnames(out) <- colnames(df)
out
}
ap.zmat = dcast(ap.z, marker_symbol ~ proc_param_name, value.var = "zscore",
fun.aggregate = mean) %>% tibble::column_to_rownames(var="marker_symbol")
ap.zmat = nan2na(ap.zmat) #convert nan to na
dim(ap.zmat)[1] 3360 14id.mat <- 1*(!is.na(ap.zmat)) # multiply 1 to make this matrix numeric
nrow(as.data.frame(colSums(id.mat)))[1] 14dim(id.mat)[1] 3360 14# heatmap of gene - phenotype (red: tested, white: untested)
if(FALSE){
pdf("~/Google Drive Miami/Miami_IMPC/output/missing_tests_after_filtering_OF.pdf", width = 6, height = 2.5)
ht = Heatmap(t(id.mat),
cluster_rows = T, clustering_distance_rows ="binary",
cluster_columns = T, clustering_distance_columns = "binary",
show_row_dend = F, show_column_dend = F, # do not show dendrogram
show_column_names = F, col = c("white","red"),
row_names_gp = gpar(fontsize = 10), name="Missing")
draw(ht)
dev.off()
}Association Z-score Distribution
We plot a association Z-score distribution for each phenotype.
ggplot(melt(ap.zmat), aes(x=value)) +
geom_histogram() +
facet_wrap(~variable, scales="free", ncol=5)+
theme(strip.text.x = element_text(size = 6))No id variables; using all as measure variables`stat_bin()` using `bins = 30`. Pick better value with `binwidth`.Warning: Removed 11204 rows containing non-finite values (stat_bin).
Estimate genetic correlation matrix between phenotypes using Zscores
Here, we estimate the genetic correlations between phenotypes using association Z-score matrix (num of genes:5479, num of phenotypes 14).
ap.zmat <- ap.zmat[,common.pheno.list]
ap.zcor = cor(ap.zmat, use="pairwise.complete.obs")
#col <- colorRampPalette(c("steelblue", "white", "darkorange"))(100)
#pheatmap(op.zcor, cluster_rows = T, cluster_cols=T, show_colnames=F, col=col)
#op.cor.order <- op.cor.out$tree_row[["order"]]
#op.zcor.org <- op.zcor # this will be used in correlation matrix test
#op.zcor <- op.zcor[op.cor.order,]
#op.zcor <- op.zcor[,op.cor.order]
#pheatmap(ap.zcor, cluster_rows = F, cluster_cols=F, show_colnames=F, col=col)
ht = Heatmap(ap.zcor, cluster_rows = T, cluster_columns = T, show_column_names = F, #col = col_fun,
row_names_gp = gpar(fontsize = 10),
#name="Genetic corr (Z-score)"
name="Genetic Corr (Zscore)"
)
draw(ht)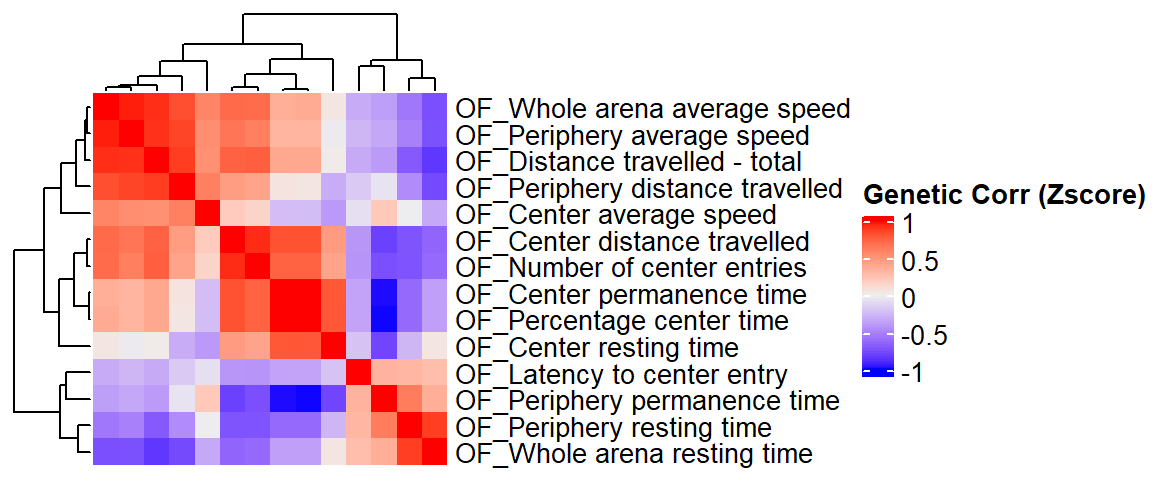
#pheno.order <- row_order(ht)
#ap.zcor <- ap.zcor[pheno.order,pheno.order]phenotype corr VS genetic corr btw phenotypes
We compare a correlation matrix obtained using control mice phenotype data v.s. a genetic correlation matrix estimated using association Z-scores. As you can see, both correlation heatmaps have similar correlation pattern.
ap.cor.rank.fig <- ap.cor.rank[common.pheno.list,common.pheno.list]
ap.cor.fig <- ap.cor[common.pheno.list,common.pheno.list]
ap.cor.combat.fig <- ap.cor.combat[common.pheno.list, common.pheno.list]
ap.zcor.fig <- ap.zcor
ht = Heatmap(ap.cor.rank.fig, cluster_rows = TRUE, cluster_columns = TRUE, show_column_names = F, #col = col_fun,
show_row_dend = F, show_column_dend = F, # do not show dendrogram
row_names_gp = gpar(fontsize = 8), column_title="Phenotype Corr (RankZ, Pearson)", column_title_gp = gpar(fontsize = 8),
name="Corr")
pheno.order <- row_order(ht)Warning: The heatmap has not been initialized. You might have different results
if you repeatedly execute this function, e.g. when row_km/column_km was
set. It is more suggested to do as `ht = draw(ht); row_order(ht)`.draw(ht)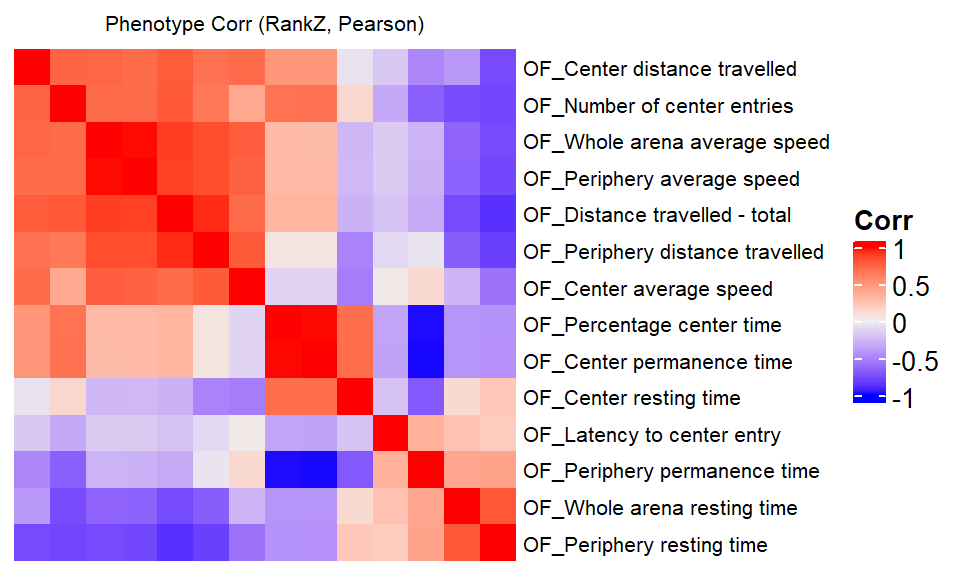
if(FALSE){
pdf("~/Google Drive Miami/Miami_IMPC/output/comp_pheno_corr_gene_corr_OF.pdf", width = 10, height = 2.5)
ap.cor.rank.fig <- ap.cor.rank.fig[pheno.order,pheno.order]
ht1 = Heatmap(ap.cor.rank.fig, cluster_rows = FALSE, cluster_columns = FALSE, show_column_names = F, #col = col_fun,
show_row_dend = F, show_column_dend = F, # do not show dendrogram
row_names_gp = gpar(fontsize = 8), column_title="Phenotype Corr (RankZ, Pearson)", column_title_gp = gpar(fontsize = 8),
name="Corr")
ap.cor.fig <- ap.cor.fig[pheno.order,pheno.order]
ht2 = Heatmap(ap.cor.fig, cluster_rows = FALSE, cluster_columns = FALSE, show_column_names = F, #col = col_fun,
row_names_gp = gpar(fontsize = 8), column_title="Phenotype Corr (Spearman)", column_title_gp = gpar(fontsize = 8),
name="Corr")
ap.cor.combat.fig <- ap.cor.combat.fig[pheno.order,pheno.order]
ht3 = Heatmap(ap.cor.combat.fig, cluster_rows = FALSE, cluster_columns = FALSE, show_column_names = F, #col = col_fun,
row_names_gp = gpar(fontsize = 8), column_title="Phenotype Corr (Combat, Pearson)", column_title_gp = gpar(fontsize = 8),
name="Corr")
ap.zcor.fig <- ap.zcor.fig[pheno.order,pheno.order]
ht4 = Heatmap(ap.zcor.fig, cluster_rows = FALSE, cluster_columns = FALSE, show_column_names = F, #col = col_fun,
row_names_gp = gpar(fontsize = 8), column_title="Genetic Corr (Pearson)", column_title_gp = gpar(fontsize = 8),
name="Corr"
)
draw(ht1+ht2+ht3+ht4)
dev.off()
}Test of the correlation between genetic correlation matrices
It looks like Jenrich (1970) test is too conservative here. Instead, we use Mantel test testing the correlation between two distance matrices.
####################
# Use Mantel test
# https://stats.idre.ucla.edu/r/faq/how-can-i-perform-a-mantel-test-in-r/
# install.packages("ade4")
library(ade4)
to.upper<-function(X) X[upper.tri(X,diag=FALSE)]
a1 <- to.upper(ap.cor.fig)
a2 <- to.upper(ap.cor.rank.fig)
a3 <- to.upper(ap.cor.combat.fig)
a4 <- to.upper(ap.zcor.fig)
plot(a4, a1)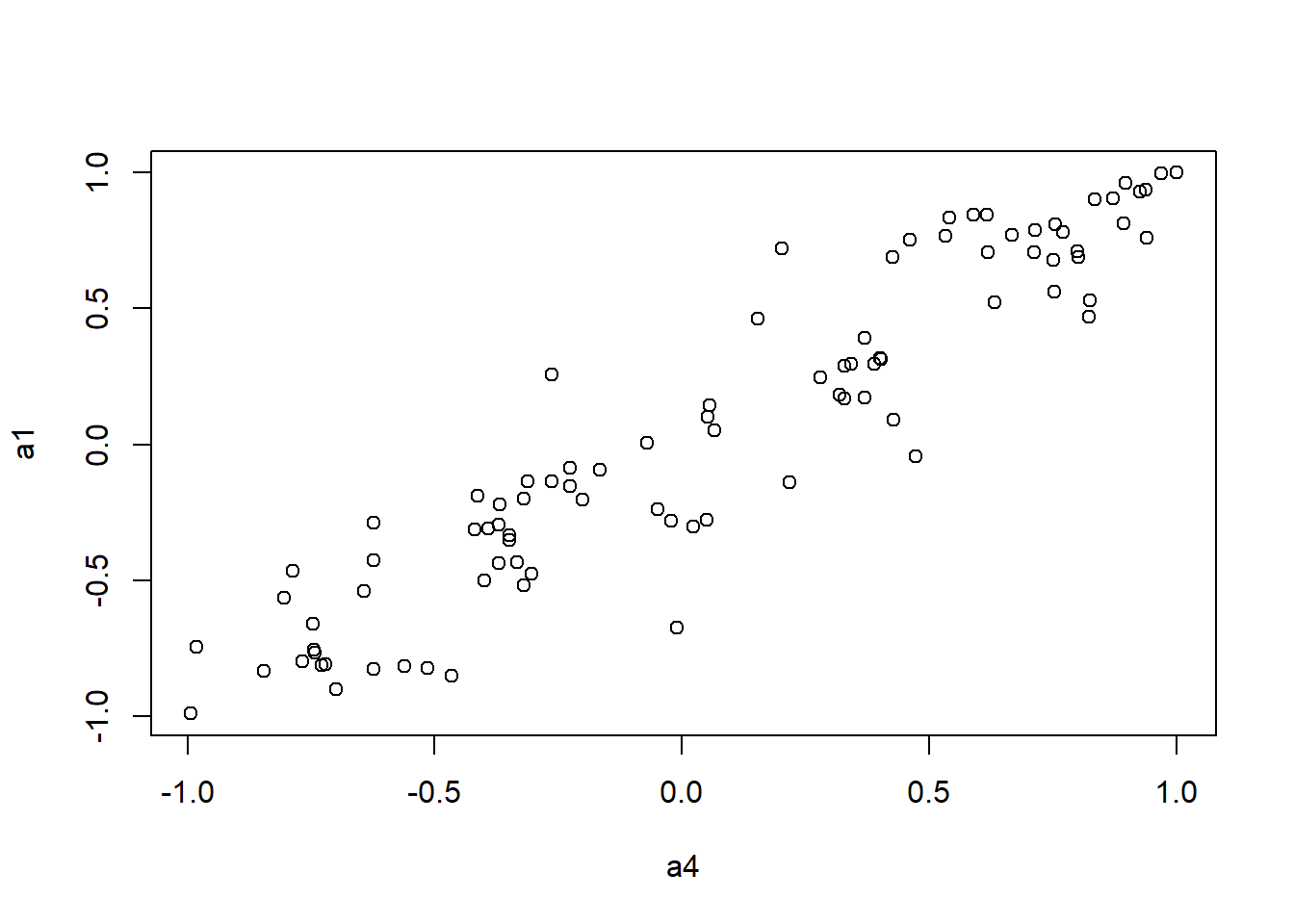
plot(a4, a2)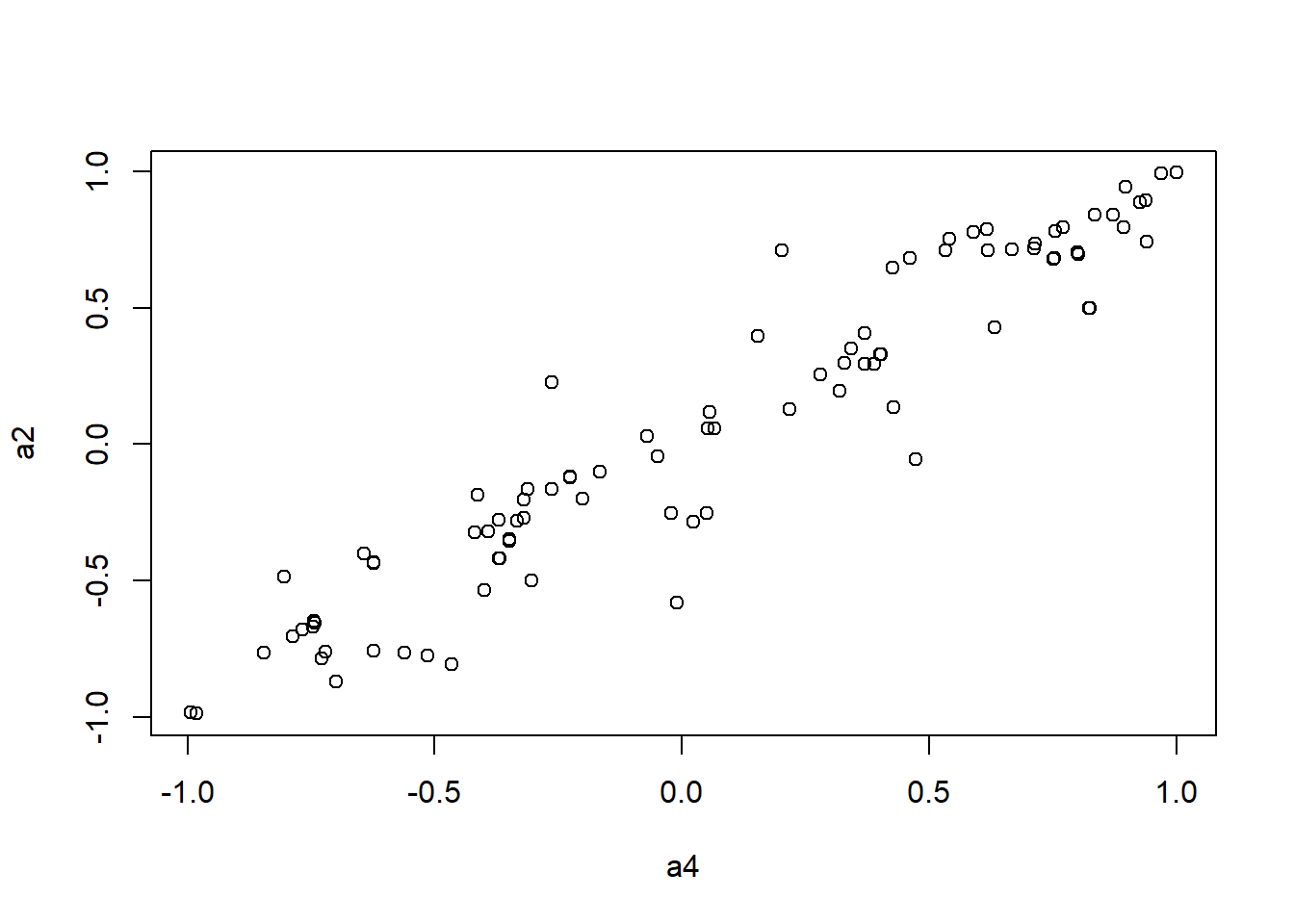
plot(a4, a3)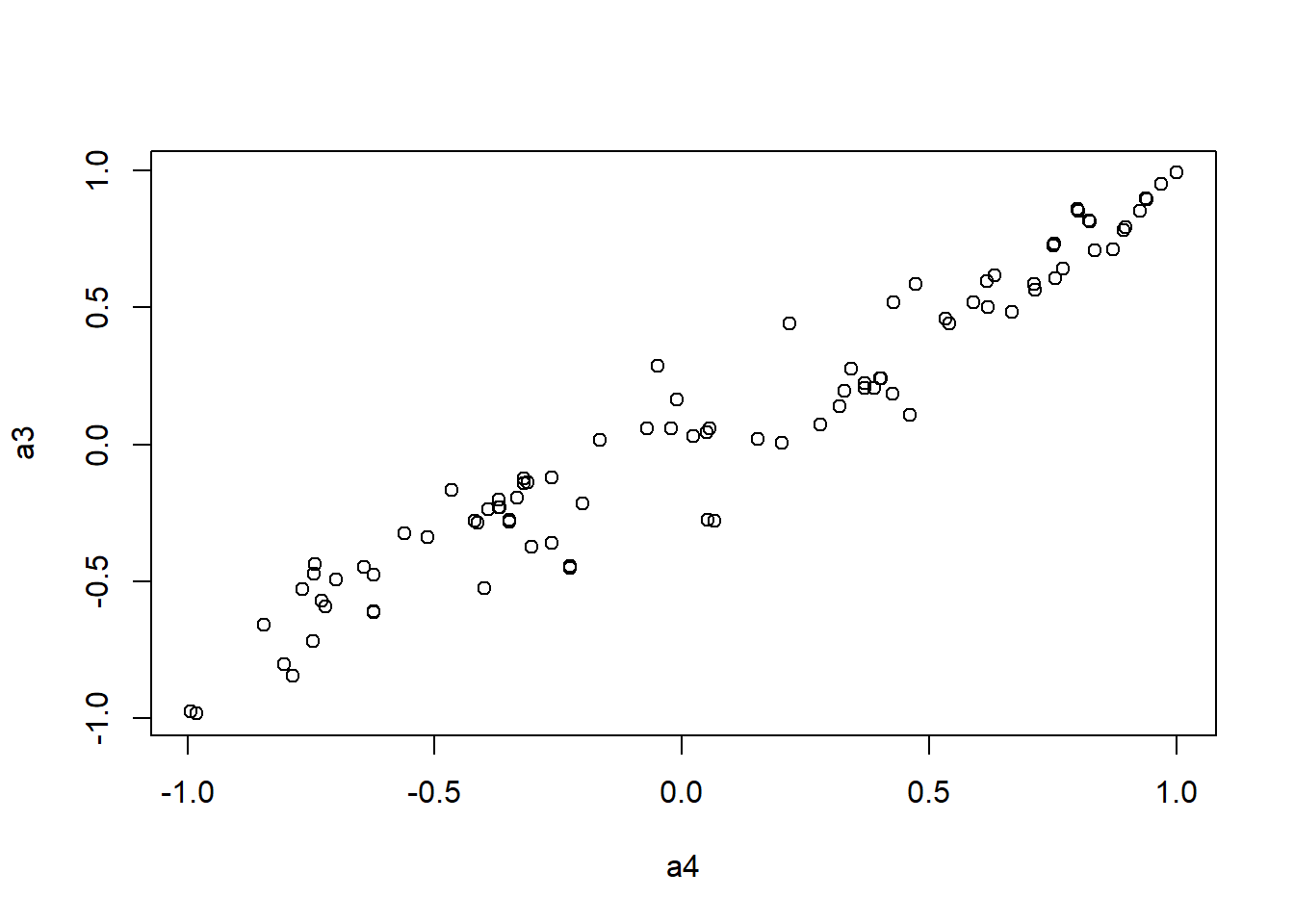
mantel.rtest(as.dist(1-ap.cor.fig), as.dist(1-ap.zcor.fig), nrepet = 9999) #nrepet = number of permutationsMonte-Carlo test
Call: mantelnoneuclid(m1 = m1, m2 = m2, nrepet = nrepet)
Observation: 0.9377157
Based on 9999 replicates
Simulated p-value: 1e-04
Alternative hypothesis: greater
Std.Obs Expectation Variance
8.2371110249 0.0007368824 0.0129392679 mantel.rtest(as.dist(1-ap.cor.rank.fig), as.dist(1-ap.zcor.fig), nrepet = 9999)Monte-Carlo test
Call: mantelnoneuclid(m1 = m1, m2 = m2, nrepet = nrepet)
Observation: 0.9513592
Based on 9999 replicates
Simulated p-value: 1e-04
Alternative hypothesis: greater
Std.Obs Expectation Variance
8.200659732 0.001238584 0.013423319 mantel.rtest(as.dist(1-ap.cor.combat.fig), as.dist(1-ap.zcor.fig), nrepet = 9999)Monte-Carlo test
Call: mantelnoneuclid(m1 = m1, m2 = m2, nrepet = nrepet)
Observation: 0.9660052
Based on 9999 replicates
Simulated p-value: 1e-04
Alternative hypothesis: greater
Std.Obs Expectation Variance
8.7304435478 -0.0008632403 0.0122648468 Test imputation algorithm
KOMPute algorithm
Impute z-scores of untested gene-pheno pair using phenotype correlation matrix
if(FALSE){
library(devtools)
devtools::install_github("dleelab/kompute")
}
library(kompute)Simulation study - imputed vs measured
We randomly select measured gene-phenotype association z-scores, mask those, impute them using kompute algorithm. Then we compare the imputed z-scores to the measured ones.
zmat <-t(ap.zmat)
dim(zmat)[1] 14 3360#filter genes with na < 20
zmat0 <- is.na(zmat)
num.na<-colSums(zmat0)
summary(num.na) Min. 1st Qu. Median Mean 3rd Qu. Max.
0.000 0.000 4.000 3.335 5.000 13.000 zmat <- zmat[,num.na<10]
dim(zmat)[1] 14 3244#pheno.cor <- ap.cor.fig
#pheno.cor <- ap.cor.rank.fig
pheno.cor <- ap.cor.combat.fig
#pheno.cor <- ap.zcor.fig
zmat <- zmat[rownames(pheno.cor),,drop=FALSE]
rownames(zmat) [1] "OF_Center average speed" "OF_Center distance travelled"
[3] "OF_Center permanence time" "OF_Center resting time"
[5] "OF_Distance travelled - total" "OF_Latency to center entry"
[7] "OF_Number of center entries" "OF_Percentage center time"
[9] "OF_Periphery average speed" "OF_Periphery distance travelled"
[11] "OF_Periphery permanence time" "OF_Periphery resting time"
[13] "OF_Whole arena average speed" "OF_Whole arena resting time" rownames(pheno.cor) [1] "OF_Center average speed" "OF_Center distance travelled"
[3] "OF_Center permanence time" "OF_Center resting time"
[5] "OF_Distance travelled - total" "OF_Latency to center entry"
[7] "OF_Number of center entries" "OF_Percentage center time"
[9] "OF_Periphery average speed" "OF_Periphery distance travelled"
[11] "OF_Periphery permanence time" "OF_Periphery resting time"
[13] "OF_Whole arena average speed" "OF_Whole arena resting time" colnames(pheno.cor) [1] "OF_Center average speed" "OF_Center distance travelled"
[3] "OF_Center permanence time" "OF_Center resting time"
[5] "OF_Distance travelled - total" "OF_Latency to center entry"
[7] "OF_Number of center entries" "OF_Percentage center time"
[9] "OF_Periphery average speed" "OF_Periphery distance travelled"
[11] "OF_Periphery permanence time" "OF_Periphery resting time"
[13] "OF_Whole arena average speed" "OF_Whole arena resting time" npheno <- nrow(zmat)
# percentage of missing Z-scores in the original data
100*sum(is.na(zmat))/(nrow(zmat)*ncol(zmat)) # 43%[1] 21.92179nimp <- 2000 # # of missing/imputed Z-scores
set.seed(1111)
# find index of all measured zscores
all.i <- 1:(nrow(zmat)*ncol(zmat))
measured <- as.vector(!is.na(as.matrix(zmat)))
measured.i <- all.i[measured]
# mask 2000 measured z-scores
mask.i <- sort(sample(measured.i, nimp))
org.z = as.matrix(zmat)[mask.i]
zvec <- as.vector(as.matrix(zmat))
zvec[mask.i] <- NA
zmat.imp <- matrix(zvec, nrow=npheno)
rownames(zmat.imp) <- rownames(zmat)Run KOMPute
kompute.res <- kompute(zmat.imp, pheno.cor, 0.01)
KOMPute running...# of genes: 3244# of phenotypes: 14# of imputed Z-scores: 11956# measured vs imputed
length(org.z)[1] 2000imp.z <- as.matrix(kompute.res$zmat)[mask.i]
imp.info <- as.matrix(kompute.res$infomat)[mask.i]
plot(imp.z, org.z)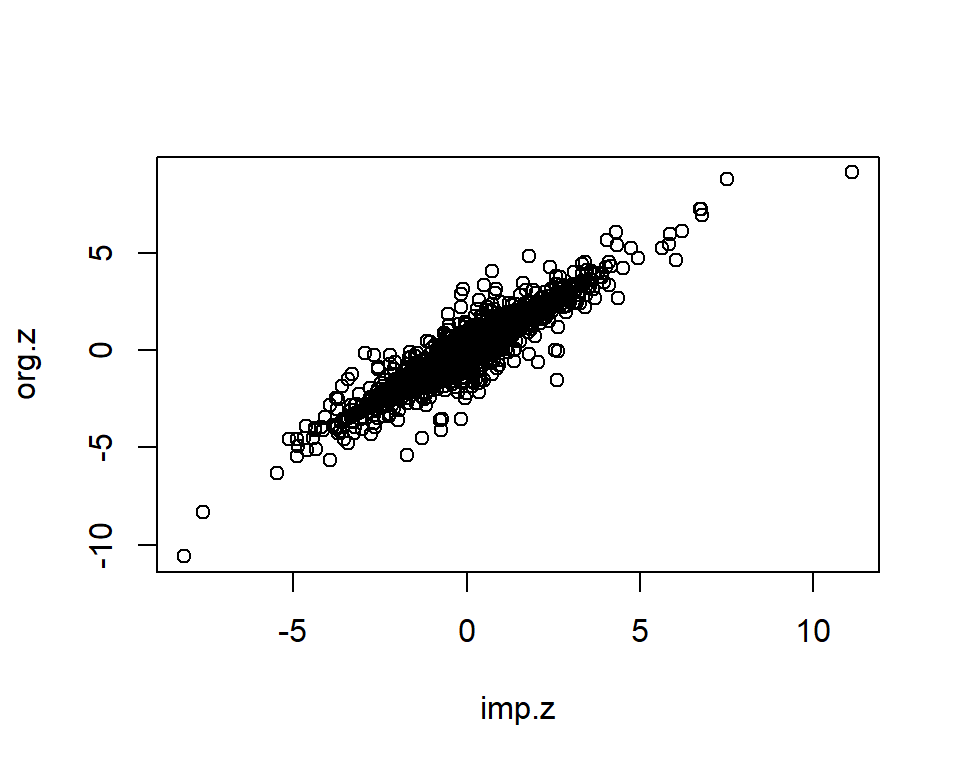
imp <- data.frame(org.z=org.z, imp.z=imp.z, info=imp.info)
dim(imp)[1] 2000 3imp <- imp[complete.cases(imp),]
imp <- subset(imp, info>=0 & info <= 1)
dim(imp)[1] 2000 3cor.val <- round(cor(imp$imp.z, imp$org.z), digits=3)
cor.val[1] 0.935plot(imp$imp.z, imp$org.z)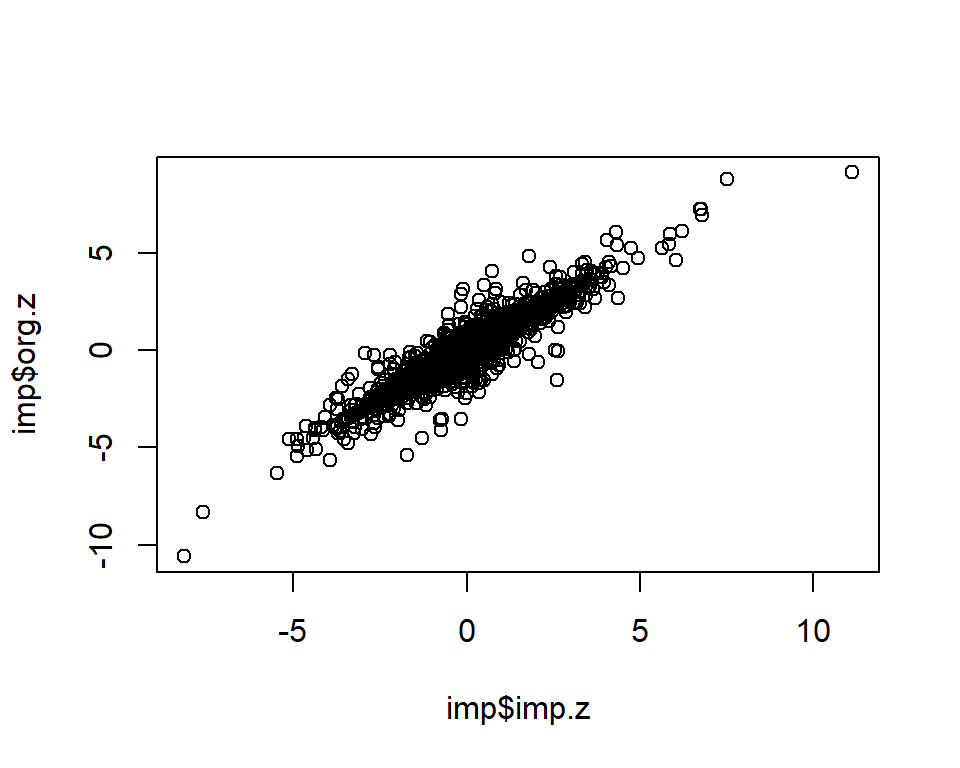
info.cutoff <- 0.9
imp.sub <- subset(imp, info>info.cutoff)
dim(imp.sub)[1] 1013 3summary(imp.sub$imp.z) Min. 1st Qu. Median Mean 3rd Qu. Max.
-5.45353 -1.00154 -0.02325 -0.02372 0.92737 6.79400 summary(imp.sub$info) Min. 1st Qu. Median Mean 3rd Qu. Max.
0.9005 0.9360 0.9536 0.9540 0.9785 0.9878 cor.val <- round(cor(imp.sub$imp.z, imp.sub$org.z), digits=3)
cor.val[1] 0.986g <- ggplot(imp.sub, aes(x=imp.z, y=org.z, col=info)) +
geom_point() +
labs(title=paste0("IMPC Behavior Data, Info>", info.cutoff, ", Cor=",cor.val),
x="Imputed Z-scores", y = "Measured Z-scores", col="Info") +
theme_minimal()
g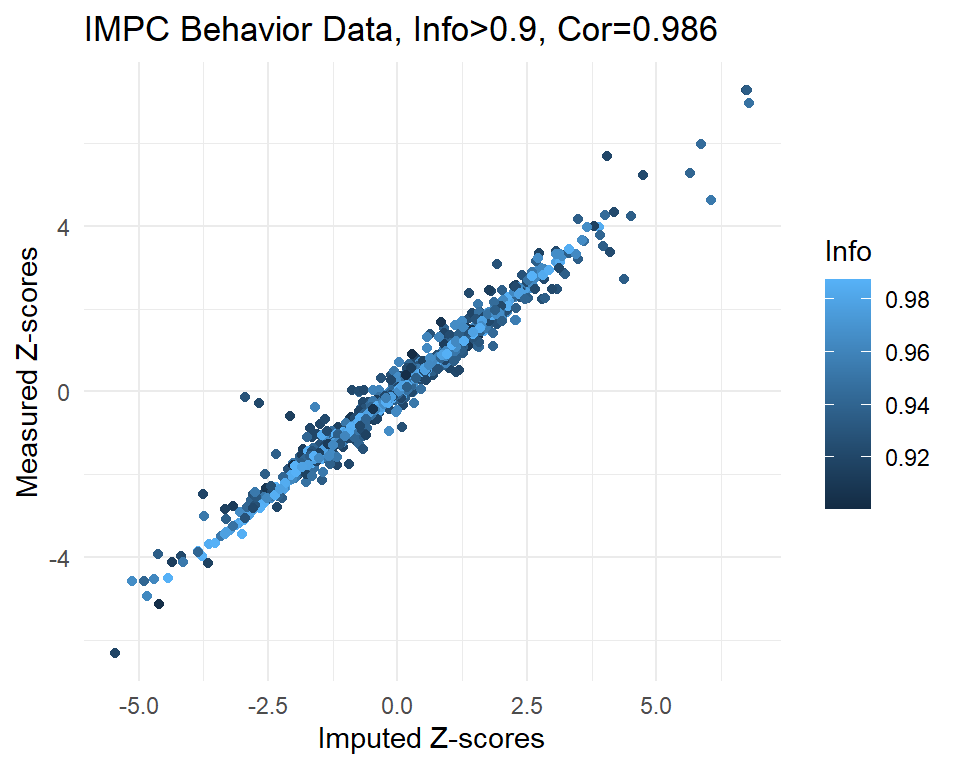
#filename <- "~/Google Drive Miami/Miami_IMPC/output/realdata_measured_vs_imputed_info_OF.pdf"
#ggsave(filename, plot=g, height=4, width=5)
sessionInfo()R version 4.1.1 (2021-08-10)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19042)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.1252
[2] LC_CTYPE=English_United States.1252
[3] LC_MONETARY=English_United States.1252
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.1252
attached base packages:
[1] grid stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] kompute_0.1.0 ade4_1.7-18 sva_3.42.0
[4] BiocParallel_1.28.0 genefilter_1.76.0 mgcv_1.8-36
[7] nlme_3.1-152 lme4_1.1-27.1 RNOmni_1.0.0
[10] ComplexHeatmap_2.10.0 circlize_0.4.13 irlba_2.3.3
[13] Matrix_1.3-4 RColorBrewer_1.1-2 tidyr_1.1.4
[16] ggplot2_3.3.5 reshape2_1.4.4 dplyr_1.0.7
[19] data.table_1.14.2
loaded via a namespace (and not attached):
[1] bitops_1.0-7 matrixStats_0.61.0 fs_1.5.0
[4] bit64_4.0.5 doParallel_1.0.16 httr_1.4.2
[7] GenomeInfoDb_1.30.0 rprojroot_2.0.2 tools_4.1.1
[10] utf8_1.2.2 R6_2.5.1 DBI_1.1.1
[13] BiocGenerics_0.40.0 colorspace_2.0-2 GetoptLong_1.0.5
[16] withr_2.4.2 tidyselect_1.1.1 bit_4.0.4
[19] compiler_4.1.1 git2r_0.28.0 Biobase_2.54.0
[22] labeling_0.4.2 scales_1.1.1 stringr_1.4.0
[25] digest_0.6.28 minqa_1.2.4 R.utils_2.11.0
[28] rmarkdown_2.11 XVector_0.34.0 pkgconfig_2.0.3
[31] htmltools_0.5.2 limma_3.50.0 fastmap_1.1.0
[34] highr_0.9 rlang_0.4.12 GlobalOptions_0.1.2
[37] RSQLite_2.2.8 shape_1.4.6 jquerylib_0.1.4
[40] farver_2.1.0 generics_0.1.1 R.oo_1.24.0
[43] RCurl_1.98-1.5 magrittr_2.0.1 GenomeInfoDbData_1.2.7
[46] Rcpp_1.0.7 munsell_0.5.0 S4Vectors_0.32.0
[49] fansi_0.5.0 R.methodsS3_1.8.1 lifecycle_1.0.1
[52] edgeR_3.36.0 stringi_1.7.5 whisker_0.4
[55] yaml_2.2.1 zlibbioc_1.40.0 MASS_7.3-54
[58] plyr_1.8.6 blob_1.2.2 parallel_4.1.1
[61] promises_1.2.0.1 crayon_1.4.1 lattice_0.20-44
[64] Biostrings_2.62.0 splines_4.1.1 annotate_1.72.0
[67] KEGGREST_1.34.0 locfit_1.5-9.4 knitr_1.36
[70] pillar_1.6.4 boot_1.3-28 rjson_0.2.20
[73] codetools_0.2-18 stats4_4.1.1 XML_3.99-0.8
[76] glue_1.4.2 evaluate_0.14 png_0.1-7
[79] vctrs_0.3.8 nloptr_1.2.2.2 httpuv_1.6.3
[82] foreach_1.5.1 gtable_0.3.0 purrr_0.3.4
[85] clue_0.3-60 assertthat_0.2.1 cachem_1.0.6
[88] xfun_0.27 xtable_1.8-4 later_1.3.0
[91] survival_3.2-11 tibble_3.1.5 iterators_1.0.13
[94] memoise_2.0.0 AnnotationDbi_1.56.0 IRanges_2.28.0
[97] workflowr_1.6.2 cluster_2.1.2 ellipsis_0.3.2