An ASH approach to finding structure in count data
Jason Willwerscheid
2/12/2019
Last updated: 2019-02-13
workflowr checks: (Click a bullet for more information)-
✔ R Markdown file: up-to-date
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
-
✔ Environment: empty
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
-
✔ Seed:
set.seed(20180714)The command
set.seed(20180714)was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible. -
✔ Session information: recorded
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
-
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.✔ Repository version: 5eaa875
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can usewflow_publishorwflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.Ignored files: Ignored: .DS_Store Ignored: .Rhistory Ignored: .Rproj.user/ Ignored: docs/.DS_Store Ignored: docs/figure/.DS_Store Untracked files: Untracked: analysis/count_preproc_r1.Rmd Untracked: analysis/gd_notes.Rmd Untracked: code/count_sim.R Untracked: code/pathways.R Untracked: data/lowrank/ Untracked: data/tmp14.rds Untracked: data/tmpdata.rds Untracked: data/tmplfsr.rds Untracked: docs/figure/count_notes.Rmd/ Untracked: temp_debug.RDS
Expand here to see past versions:
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 5eaa875 | Jason Willwerscheid | 2019-02-13 | wflow_publish(“analysis/count_shrinkage.Rmd”) |
Introduction
Let’s say that we’re interested in finding structure in a matrix of counts \(Y\). The usual approach is to set \(X = \log(Y + \alpha)\) for some pseudocount \(\alpha > 0\) and then look for low-rank structure in \(X\).
Here I propose a different method that uses ashr to shrink the counts \(Y_{ij}\).
Model for the data-generating process
One can consider the individual counts \(Y_{ij}\) as Poisson random variables with (unknown) rate parameters \(\lambda_{ij}\). And in fact, it’s structure in \(\Lambda\) that we’re primarily interested in, not structure in \(Y\).
The simplest model is that \[ \Lambda = \exp(LF'), \] but in most applications one wouldn’t expect the matrix of log-rates to be low-rank. A more useful model puts \[ \Lambda = \exp(LF' + E), \] where \(E_{ij} \sim N(0, \sigma_{ij}^2)\) (with some structure in the matrix of variances \(\Sigma\)).
Fitting \(LF'\) using ASH and FLASH
I propose a three-step approach to estimating \(LF'\):
Since we’re really interested in \(\Lambda\) (not \(Y\)), I propose that we first estimate \(\Lambda\) using
ashr. The ASH model is \[ Y_{ij} \sim \text{Poisson}(\lambda_{ij});\ \lambda_{ij} \sim g, \] where \(g\) is a unimodal prior to be estimated. (One can also runashrseparately on each row or column of \(Y\) to get row-wise or column-wise priors.) Conveniently,ashrdirectly gives estimates for posterior means \(\mathbb{E} (\lambda_{ij})\) and posterior variances \(\text{Var}(\lambda_{ij})\).Transform the ASH estimates using the approximations \[ X_{ij} := \mathbb{E} (\log \lambda_{ij}) \approx \log \mathbb{E}(\lambda_{ij}) - \frac{\text{Var}(\lambda_{ij})}{2(\mathbb{E}(\lambda_{ij}))^2}\] and \[ S_{ij}^2 := \text{Var} (\log \lambda_{ij}) \approx \frac{\text{Var}(\lambda_{ij})}{(\mathbb{E}(\lambda_{ij}))^2} \] (Importantly, the posterior means are all non-zero so that one can directly take logarithms. No pseudo-counts are needed.)
Run FLASH on the data \((X, S)\) with the additional variance in \(E\) specified as a “noisy” variance structure. In other words, the FLASH model is \[ X_{ij} = LF' + E^{(1)} + E^{(2)} \] where \(E_{ij}^{(1)} \sim N(0, S_{ij}^2)\) (with the \(S_{ij}\)s fixed) and \(E_{ij}^{(2)} \sim N(0, 1 / \tau_{ij})\) (with the \(\tau_{ij}\)s to be estimated). (And, as usual, there are priors on each column of \(L\) and \(F\).) The variance structure in \(E^{(2)}\) matches the assumed noise structure in \(\log (\Lambda)\).
Example
To illustrate why this approach is a good idea, I consider a simple example with a low-intensity baseline and a block of high intensity:
set.seed(666)
n <- 120
p <- 160
log.lambda <- (-1 + outer(c(rep(2, n / 4), rep(0, 3 * n / 4)),
c(rep(2, p / 4), rep(0, 3 * p / 4)))
+ 0.5 * rnorm(n * p))
Y <- matrix(rpois(n * p, exp(log.lambda)), n, p)
# Define some variables to make analysis easier.
hi.rows <- rep(FALSE, n)
hi.rows[1:(n / 4)] <- TRUE
hi.cols <- rep(FALSE, p)
hi.cols[1:(p / 4)] <- TRUE
# Show heatmap.
image(x = 1:n, y = 1:p, z = log.lambda, xlab = "x index", ylab = "y index")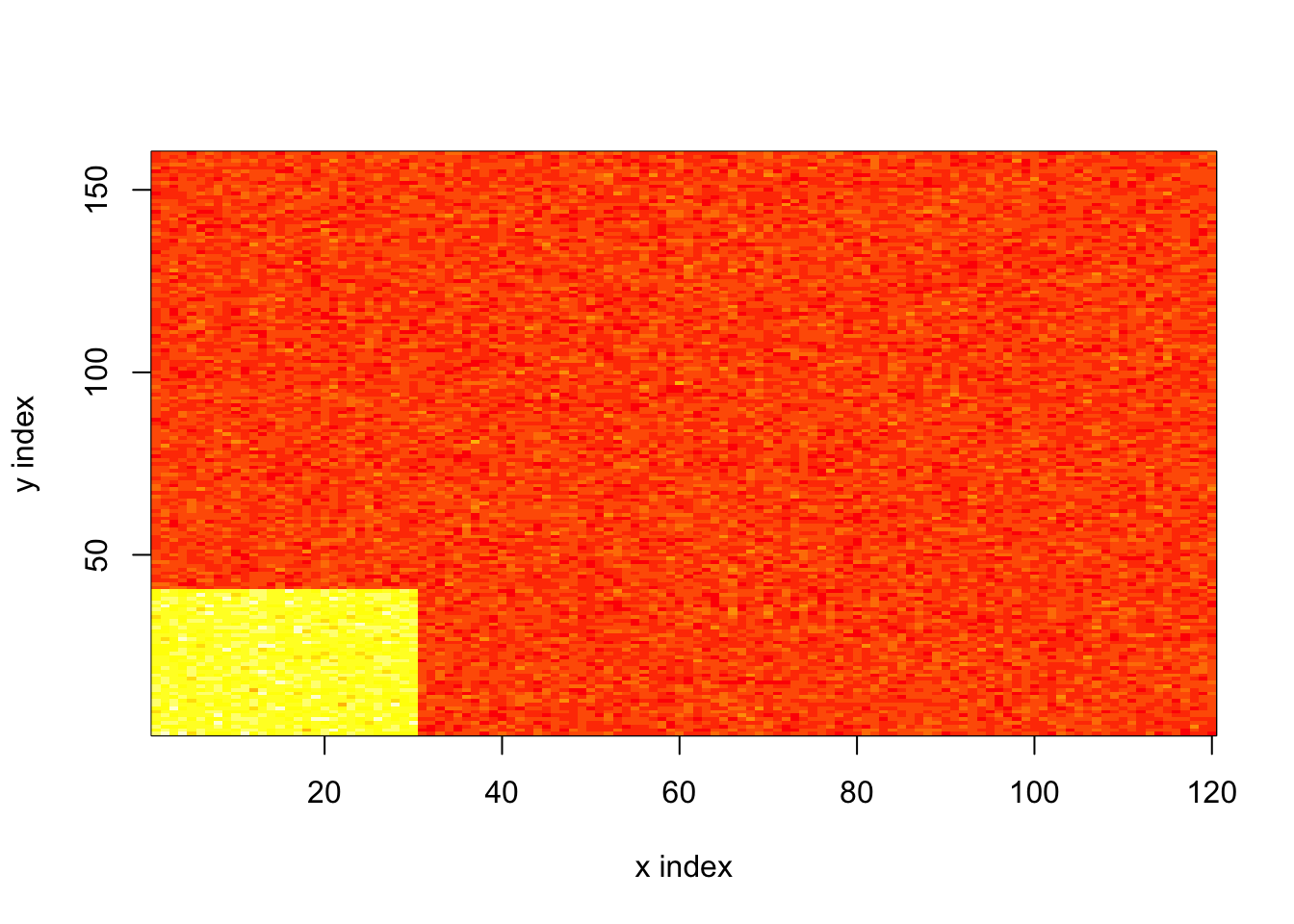
The usual approach would run FLASH as follows.
# Use my own branch due to bug in stephens999/master.
devtools::load_all("~/Github/ashr")Loading ashrdevtools::load_all("~/Github/flashier")Loading flashierfl.log1p <- flashier(log1p(Y), var.type = 0,
greedy.Kmax = 10, verbose = 1)Initializing flash object...
Adding factor 1 to flash object...
Adding factor 2 to flash object...
Adding factor 3 to flash object...
Factor doesn't increase objective and won't be added.
Nullchecking 2 factors...
Wrapping up...
Done.My proposed approach is the following.
# 1. Get ASH estimates for lambda (posterior means and SDs).
Y.ash <- ashr::ash(betahat = rep(0, n * p), sebetahat = 1,
lik = ashr::lik_pois(as.vector(Y)), mode = 0,
method = "shrink")
pm <- Y.ash$result$PosteriorMean
psd <- Y.ash$result$PosteriorSD
# 2. Transform to log scale.
X <- matrix(log(pm) - psd^2 / pm^2, n, p)
S <- matrix(psd / pm, n, p)
# 3. Run FLASH.
fl.ash <- flashier(X, S = S, var.type = 0,
greedy.Kmax = 10, verbose = 1)Initializing flash object...
Adding factor 1 to flash object...
Adding factor 2 to flash object...
Adding factor 3 to flash object...
An iteration decreased the objective by 1.75e+00. Try backfitting with warmstarts.
Factor doesn't increase objective and won't be added.
Nullchecking 2 factors...
Wrapping up...
Done.For comparison, I also run ashr separately on each column of \(Y\).
colwise.pm <- array(0, dim = dim(Y))
colwise.psd <- array(0, dim = dim(Y))
for (i in 1:p) {
# For a fair comparison, I use the same grid that was selected by Y.ash.
col.ash <- ashr::ash(betahat = rep(0, n), sebetahat = 1,
lik = ashr::lik_pois(Y[, i]), mode = 0,
method = "shrink", mixsd = Y.ash$fitted_g$b)
colwise.pm[, i] <- col.ash$result$PosteriorMean
colwise.psd[, i] <- col.ash$result$PosteriorSD
}
colw.X <- log(colwise.pm) - colwise.psd^2 / colwise.pm^2
colw.S <- colwise.psd / colwise.pm
fl.colw <- flashier(colw.X, S = colw.S, var.type = 0,
greedy.Kmax = 10, verbose = 1)Initializing flash object...
Adding factor 1 to flash object...
Adding factor 2 to flash object...
Adding factor 3 to flash object...
Factor doesn't increase objective and won't be added.
Nullchecking 2 factors...
Wrapping up...
Done.I calculate the root mean-squared error and the mean shrinkage obtained using each method. I calculate separately for large \(\lambda_{ij}\), small \(\lambda_{ij}\) in columns where all values are small, and small \(\lambda_{ij}\) in columns where some values are large.
get.res <- function(fl) {
preds <- flashier:::lowrank.expand(get.EF(fl$fit))
hi.resid <- preds[hi.rows, hi.cols] - log.lambda[hi.rows, hi.cols]
lo.resid <- preds[, !hi.cols] - log.lambda[, !hi.cols]
mix.resid <- preds[!hi.rows, hi.cols] - log.lambda[!hi.rows, hi.cols]
res <- list(rmse.hi = sqrt(mean((hi.resid)^2)),
rmse.lo = sqrt(mean((lo.resid)^2)),
rmse.mix = sqrt(mean((mix.resid)^2)),
shrnk.hi = -mean(hi.resid),
shrnk.lo = -mean(lo.resid),
shrnk.mix = -mean(mix.resid))
res <- lapply(res, round, 2)
return(res)
}
res <- data.frame(cbind(get.res(fl.log1p), get.res(fl.ash), get.res(fl.colw)))
var.names <- c("RMSE (lg vals)",
"RMSE (sm vals)",
"RMSE (sm vals in lg cols)",
"Mean shrinkage (lg vals)",
"Mean shrinkage (sm vals)",
"Mean shrinkage (sm vals in lg cols)")
meth.names <- c("log1p", "ASH", "col-wise ASH")
row.names(res) <- var.names
colnames(res) <- meth.names
knitr::kable(res, digits = 2)| log1p | ASH | col-wise ASH | |
|---|---|---|---|
| RMSE (lg vals) | 0.48 | 0.49 | 0.49 |
| RMSE (sm vals) | 1.34 | 0.6 | 0.57 |
| RMSE (sm vals in lg cols) | 1.36 | 0.59 | 1.11 |
| Mean shrinkage (lg vals) | 0 | -0.08 | -0.05 |
| Mean shrinkage (sm vals) | -1.25 | 0.33 | 0.2 |
| Mean shrinkage (sm vals in lg cols) | -1.27 | 0.31 | 0.89 |
If we care about controlling FDR, then the RMSE is unimportant on its own. Indeed, while the log1p and column-wise ASH methods perform similarly in terms of RMSE, the log1p approach tends to yield anti-conservative estimates for small \(\lambda_{ij}\), while the column-wise ASH approach “errs” by shrinking the estimates towards zero. The latter is much more desirable from the perspective of FDR control.
I’m surprised that ashr shrinks small \(\lambda_{ij}\) more in columns where there are some large \(\lambda_{ij}\): I expected the opposite. For a reason that at present eludes me, ashr is more likely to put mass on the null component when some values are large (when all are small, then it rarely puts any mass on the null component).
Session information
sessionInfo()R version 3.4.3 (2017-11-30)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS High Sierra 10.13.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] flashier_0.1.0 ashr_2.2-29
loaded via a namespace (and not attached):
[1] Rcpp_1.0.0 highr_0.7 compiler_3.4.3
[4] git2r_0.21.0 workflowr_1.0.1 R.methodsS3_1.7.1
[7] R.utils_2.6.0 iterators_1.0.10 tools_3.4.3
[10] testthat_2.0.1 digest_0.6.18 etrunct_0.1
[13] evaluate_0.12 memoise_1.1.0 lattice_0.20-35
[16] rlang_0.3.0.1 Matrix_1.2-14 foreach_1.4.4
[19] commonmark_1.4 yaml_2.2.0 parallel_3.4.3
[22] ebnm_0.1-17 xfun_0.4 withr_2.1.2.9000
[25] stringr_1.3.1 roxygen2_6.0.1.9000 xml2_1.2.0
[28] knitr_1.21.6 devtools_1.13.4 rprojroot_1.3-2
[31] grid_3.4.3 R6_2.3.0 rmarkdown_1.11
[34] mixsqp_0.1-97 magrittr_1.5 whisker_0.3-2
[37] backports_1.1.2 codetools_0.2-15 htmltools_0.3.6
[40] MASS_7.3-48 assertthat_0.2.0 stringi_1.2.4
[43] doParallel_1.0.14 pscl_1.5.2 truncnorm_1.0-8
[46] SQUAREM_2017.10-1 R.oo_1.21.0 This reproducible R Markdown analysis was created with workflowr 1.0.1